- 1Yuelushan Laboratory, College of Life Science and Technology, Central South University of Forestry and Technology, Changsha, China
- 2Key Laboratory of Plant Carbon Capture, Chinese Academy of Sciences (CAS) Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China
- 3State Key Laboratory for Quality and Safety of Agro-Products, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 4Key Laboratory of Biodiversity Conservation in Southwest, State Forestry Administration, Southwest Forestry University, Kunming, China
Introduction: Wood is primarily made up of secondary xylem cell walls, with lignin, cellulose, and hemicellulose as the main chemical components. The presence of lignin represents recalcitrance to wood pulping and biofuel conversion. Consequently, reducing lignin content is a key approach to improving wood properties and optimizing its processing.
Methods: In this study, we suppressed lignin biosynthesis by overexpressing a mutated transcription repressor PdLTF1AA and enhanced cellulose synthesis simultaneously by introducing cellulose synthase genes, PdCesA4, PdCesA7A, or PdCesA8A, specifically in xylem fiber cells.
Results and discussion: The transgenic plants exhibited decreased lignin content and a significant increase in cellulose content. Transcriptome analysis indicated that expression of PdLTF1AA along with PdCesA4, PdCesA7A, or PdCesA8A in fiber cells resulted in transcriptional alterations in the genes associated with cell wall remodeling and polysaccharide synthesis during xylem development. The results also indicated that the diameter of wood fiber cells within the xylem is increased, which leads to a larger stem diameter in the transgenic plants. This study suggests that the biosynthesis of lignin and cellulose can be simultaneously modified, which presents a new strategy for modifying wood fiber characteristics for more efficient fiber and biomass processing.
1 Introduction
Wood, as a renewable biomass resource, is widely used in pulping and papermaking, as well as the potential for biofuel production (Luo and Li, 2022; Zhu and Li, 2024). Its basic structural components are cellulose, lignin, and hemicellulose. Lignin, among these components, represents significant barriers in biomass processing and usage due to its high chemical stability. The presence of lignin complicates the processing of biomass and significantly increases associated economic expenses (Ye and Zhong, 2015; Li et al., 2024). As a result, reducing lignin content has been a major study emphasis in the field of wood property modification and enhancement.
Lignin biosynthesis is derived from phenylpropanoid pathway (Boerjan et al., 2003; Li et al., 2024). Starting from phenylalanine, the pathway involves a series of enzymatic reactions catalyzed by key enzymes, including phenylalanine ammonia lyase (PAL), 4-coumarate: CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD), which convert phenylalanine into various intermediates for monolignol synthesis. The activities of these enzymes directly influence the rate and efficiency of lignin synthesis, thus playing a crucial role in determining the quantity and quality of lignin synthesized (Hu et al., 1999; Li et al., 2003; Bryant et al., 2020; Cao et al., 2020; De Meester et al., 2022; Zhu and Li, 2024). Numerous studies have reported reducing lignin content in poplar trees through genetic manipulation of the monolignol biosynthesis pathway genes (Chanoca et al., 2019; Li et al., 2024; Zhu and Li, 2024). For instance, the expression of the 4CL gene was successfully suppressed in Populus, resulting in a significant reduction of lignin content in transgenic trees (Hu et al., 1999; Li et al., 2003; Wang et al., 2012; Cao et al., 2020). Similarly, the inhibition of 4CL and C4H gene expression via antisense RNA technology led to significant decreases in lignin content and the syringyl lignin (S-lignin) in transgenic Populus trees under both greenhouse and field conditions (Bjurhager et al., 2010; Voelker et al., 2010). Furthermore, the downregulation of C3H and CAD gene expression using RNA interference technology also achieved a substantial reduction in lignin content in transgenic trees (Ralph et al., 2012; Van Acker et al., 2017). Additionally, the knockout of PAL, CCoAOMT1/2, and CCR2 genes using CRISPR-Cas9 technology has been shown to effectively reduce lignin content (De Meester et al., 2020; Sulis et al., 2023).
Direct manipulation of genes in the monolignol biosynthesis pathway leads to alterations in lignin accumulation but also disrupts phenylpropanoid metabolic flow, affecting plant growth (Du and Groover, 2010; Voelker et al., 2011).
Lignin biosynthesis is intricately regulated by a multitude of transcription factors and microRNAs. Therefore, modification of the regulatory factors may help to bypass the disruption of metabolic flow caused by directly modifying the lignin biosynthesis genes.
Overexpression of miR408 has been shown to significantly reduce lignin content, decrease the ratio of syringyl lignin (S-lignin) to guaiacyl lignin (G-lignin) monomers, and consequently enhance the utilization efficiency of wood (Guo et al., 2023). Overexpression of MYB115 can suppress the expression of lignin biosynthesis genes, resulting in a reduction in both the content of S-lignin monomers and the total lignin content (Fan et al., 2022). In our studies, a lignin-related transcription factor, PdLTF1, that modulates the entire monolignol biosynthesis pathway, is identified in Populus for its repressive role in lignin biosynthesis (Gui et al., 2019). Because the PdLTF1 repression function can be released via phosphorylation, mutation of PdLTF1 at its phosphorylation sites (PdLTF1AA) transforms it into a stable repressor (Gui et al., 2019). Thus, the mutant PdLTF1AA can be utilized to engineer the monolignol biosynthesis pathway while not showing growth penalty (Gui et al., 2020).
Concurrently, cellulose is synthesized by cellulose synthase (CesA). It is believed that multiple CesAs aggregate into a cellulose synthase complex (CSC) localized on the plasma membrane to catalyze the synthesis of cellulose microfibrils (Paredez et al., 2006; Turner and Kumar, 2018; Tai et al., 2023). Each CesA subunit synthesizes a cellulose chain, and multiple subunits within the CSC collaborate to generate cellulose microfibrils. The CSC elongates cellulose chains by integrating glucose units from UDP-glucose through β-1,4-glycosidic linkages (Turner and Kumar, 2018; Pedersen et al., 2024).
The Populus genome contains 18 CesA genes (Suzuki et al., 2006). Compared with Arabidopsis, which has 10 CesA genes, Populus has almost twice as many CesA genes as Arabidopsis, with two homologous genes for each Arabidopsis CesA gene. Two types of CSCs have been identified in Populus. Type I CSCs include CesA4, CesA7A, CesA7B, CesA8A, and CesA8B, while Type II CSCs comprise CesA1A, CesA1B, CesA3C, CesA3D, CesA6E, and CesA6F. The two types of CSCs facilitate cellulose synthesis in wood xylem cells (Suzuki et al., 2006; Song et al., 2010). It is noteworthy that the two types of CSCs showed an effect on the formation of distinct structural properties of cellulose microfibrils in Populus wood (Xi et al., 2017; Abbas et al., 2020; Xu et al., 2021). In Populus, CesA4, CesA7A, CesA7B, CesA8A, and CesA8B may form a type of CSC that is associated with the deposition of highly crystalline cellulose (Song et al., 2010; Xi et al., 2017). Studies also showed that suppression of PtrCesA7A expression results in a significant reduction in cellulose crystallinity, whereas suppression of PtrCesA3D expression leads to an increase in cellulose crystallinity (Abbas et al., 2020). The temporal and spatial expression patterns of these CesA genes exhibit significant differences and may be related to their functional roles in cellulose deposition (Suzuki et al., 2006). These findings indicate the functional differentiation in the deposition of crystalline cellulose and the regulatory significance of CesAs in Populus, displaying their critical functions in wood cellulose deposition.
This study utilized a fiber cell-specific promoter to express PdLTF1AA for the repression of lignin biosynthesis while concurrently introducing cellulose synthase genes (PdCesA4, PdCesA7A, or PdCesA8A), which are highly expressed in xylem and may be related to the deposition of high crystalline cellulose to modify cellulose properties in xylem fiber cells. Results indicate that the biosynthesis of lignin and cellulose can be concurrently modified, resulting in changes to the wood fiber properties in Populus.
2 Materials and methods
2.1 Conditions for plant growth
This study utilized the hybrid Populus tree (Populus deltoides × P. euramericana cv. “Nanlin895”) (Pd), grown in a controlled-environment phytotron with the following parameters: a photoperiod of 14 hours of light and 10 hours of darkness, a relative humidity of 60%, and a constant temperature of 25°C.
2.2 Constructs and transformation
Based on a previous study (Gui et al., 2020), we constructed a vector named PdDUF579-9p-35Smini-PdLTF1AA. This vector utilizes the fiber cell-specific promoter PdDUF579-9p to drive the expression of PdLTF1AA, a mutant form of PdLTF1 that can stably inhibit lignin synthesis. Meanwhile, the genes encoding PdCesA4 (Potri.002G257900), PdCesA7A (Potri.006G181900), and PdCesA8A (Potri.011G069600) were cloned from Nanlin895. Using a fiber cell-specific promoter, PdWND1B (Potri.001G448400), the expression cassettes (PdWND1Bp-35Smini-PdCesA4, PdWND1Bp-35Smini-PdCesA7A, and PdWND1Bp-35Smini-PdCesA8A) were constructed for fiber cell-specific modification of cellulose in Populus. Following verification of the constructs’ accuracy through sequencing, they were introduced into Populus trees in accordance with the established protocol (Li et al., 2003). Supplementary Table S1 lists the primers used for vector construction.
2.3 Quantitative RT-PCR
According to the previously described protocols (Li et al., 2025), xylem tissues were collected from the 15th internodes. Total RNA was extracted, and reverse transcription PCR (RT-PCR) was performed using the kit in accordance with the manufacturer’s protocol. The specific primers employed for each gene are provided in Supplementary Table S1.
2.4 Morphological analysis
Forty independent transgenic lines were generated. Among them, thirty-two independent transgenic lines were integrated with a single construct, five lines with two constructs, two lines with three constructs, and one line with four constructs were identified. The transgenic lines that had both PdLTF1AA and CesA cassettes were cloned using micro-cutting to generate multiple copies, which were then used as biological replicates. The transgenics grown in a phytotron were phenotypically assessed.
2.5 Microscopic observation of stem structure
Stem samples from the 15th internode were prepared for paraffin sectioning. In brief, stem samples were subjected to dehydration in a graded ethanol series, followed by clearing in xylene, and then immersed in paraffin. The stem tissues were sectioned at a thickness of 12 μm, stained with 1% toluidine blue for 20 minutes, and mounted with gum. The sections were then observed under an optical microscope. The thickness of the secondary cell wall (SCW) was examined via transmission electron microscopy. In brief, stem samples were sectioned transversely at 1 mm thickness and fixed under vacuum in 2.5% glutaraldehyde. They were then washed three times with 0.1 M phosphate buffered saline (PBS) buffer, fixed with 0.5% osmium tetroxide on a shaker, and washed again three times with 0.1 M PBS buffer. Subsequently, the samples were dehydrated in a graded ethanol series, embedded in epoxy resin, and polymerized at 50°C. Ultrathin sections were stained with lead citrate and uranyl acetate and then examined using a transmission electron microscope. The thickness of SCW was measured with ImageJ software.
2.6 Fiber length determination
The stem (the 15th internode) had its bark removed, was cut into 2-cm pieces, and then soaked overnight in a mixture of glacial acetic acid and 30% hydrogen peroxide at 60°C. After that, it was dyed with 1% safranine for 10 minutes and then photographed with a light microscope (Olympus, BX53). Next, the lengths of the fibers were measured using the Lorentzen & Wettre (LW) Fiber Tester from AB Lorentzen & Wettre in Kista, Sweden (Li et al., 2025).
2.7 Wood composition determination
Cellulose detection was performed as previously detailed (Gui et al., 2011). Xylem tissue from 16th-20th internode cellulose content was measured as previously described (Foster et al., 2010b). Lignin detection was performed using phloroglucinol-HCl staining, following the described methods (Gui et al., 2011). Xylem tissue from 16th-20th internode lignin content was measured as previously described (Foster et al., 2010a). Analytical thioacidolysis on alcohol insoluble residue (AIR) samples was performed as described previously (Yamamura et al., 2012). In brief, 5 mg of the AIR sample was treated with a reaction mixture composed of dioxane, ethanethiol, and boron trifluoride (35:4:1, v/v) to release lignin monomers. The lignin monomers were subsequently detected and quantified via gas chromatography-mass spectrometry (GC-MS). Tetracosane was used as an internal standard for quantification.
2.8 RNA sequencing
According to the previously described protocols (Li et al., 2025), total RNA was extracted from the xylem tissue of the 15th internode. The quality of the extracted RNA was verified, and mRNA was enriched using Oligo dT, fragmented, and then reverse transcribed into cDNA. After ligation with adaptors, the cDNA was sequenced on the NovaSeq X Plus platforms. Genes with an expression fold change of more than 2 were considered as differentially expressed genes (DEGs). Functional enrichment analysis and functional annotation were performed on the Meiji Cloud platform (https://analysis.majorbio.com/).
2.9 Statistical analysis
In this study, data were subjected to significance analysis using GraphPad Prism software, with one-way analysis of variance (ANOVA) utilized as the statistical method. Heat maps were visualized using TBtools.
3 Result
3.1 Morphological analysis of transgenic plants
In earlier studies, we showed that the overexpression of PdLTF1AA, controlled by a fiber cell-specific promoter, effectively reduced lignin content without compromising normal plant growth (Gui et al., 2020). In this study, we investigate the specific expression of PdLTF1AA in fiber cells, in conjunction with PdCesA4, PdCesA7A, or PdCesA8A in Populus. We generated around 60 independent transgenic lines. After examination, the transgenic plants that had both PdLTF1AA and CesA genes, specifically the line called LTF/4/7 with PdLTF1AA, PdCesA4, and PdCesA7A, the line named LTF/7/8 with PdLTF1AA, PdCesA7A, and PdCesA8A, and the line labeled LTF/4/7/8 with PdLTF1AA, PdCesA4, PdCesA7A, and PdCesA8A, were selected and cloned using micro-cutting. The propagated transgenics were grown in a phytotron for detail evaluation. In comparison to the wild type (WT), the transgenic plants LTF/4/7, LTF/7/8, and LTF/4/7/8 exhibited comparable height growth but exhibited variations in stem diameter in a two-month period of growth (Figure 1A, B). The height and number of internodes showed no significant difference. However, the stem diameter increased by 13.7%, 12.2%, and 14.3% in LTF/4/7, LTF/7/8, and LTF/4/7/8, respectively (Figure 1C-E). In the transgenic plants, the integrated PdLTF1AA, PdCesA4, PdCesA7A, and PdCesA8A genes were expressed correctly in LTF/4/7, LTF/7/8, and LTF/4/7/8, respectively (Figure 1F-I).
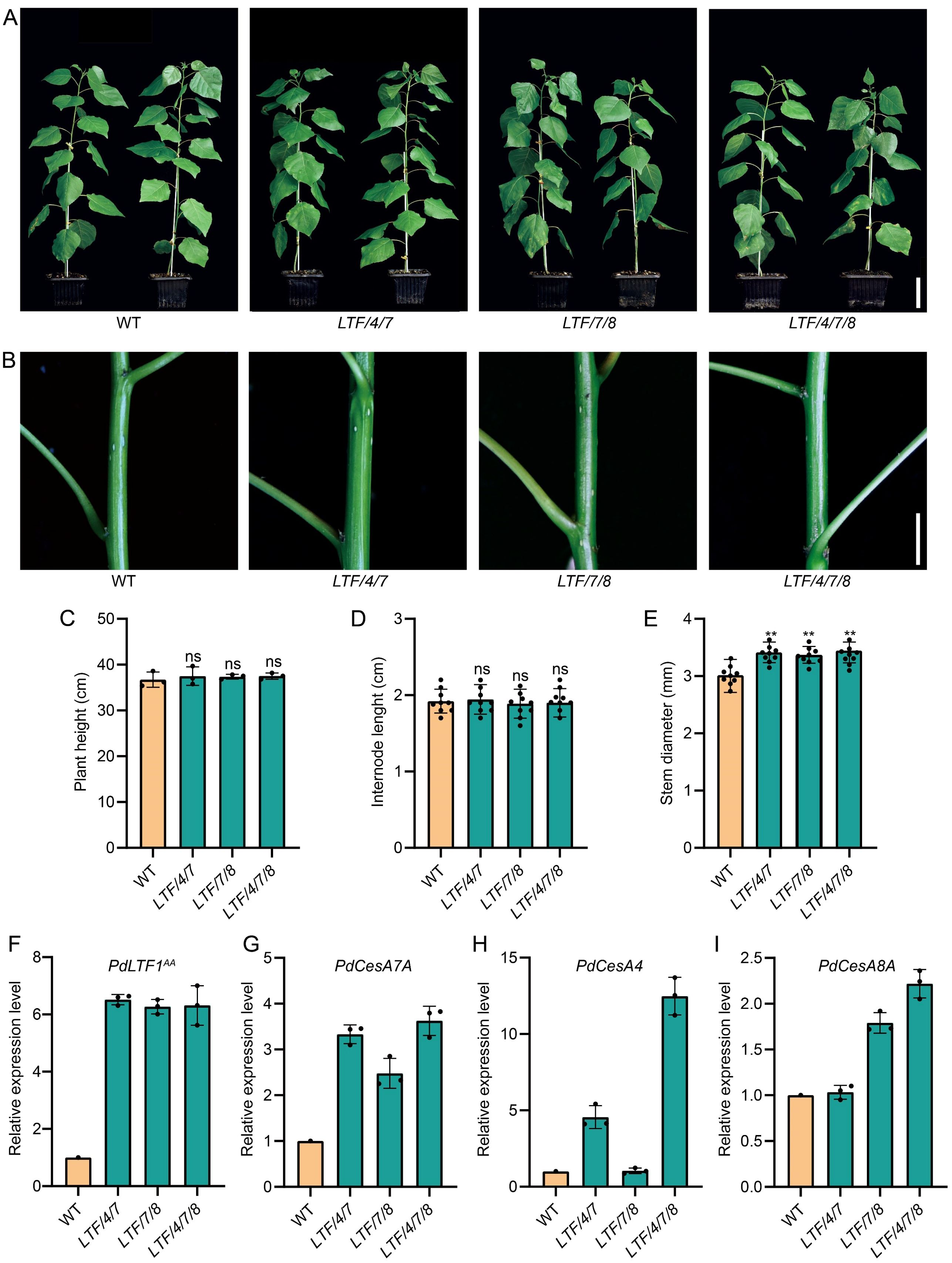
Figure 1. The growth morphologies of the fiber cell-specific regulation of PdLTF1AA, PdCesA4, PdCesA7A, and PdCesA8A in Populus. (A, B) Transgenics of LTF/4/7, LTF/7/8, and LTF/4/7/8 at 2 months old. Scale bar, 6 cm in (A) and 0.6 cm in (B). (C) Plant height, (D) Internode length (the 15th internode), (E) Stem diameter (the 15th internode). The PdLTF1AA (F), PdCesA7A (G), PdCesA4 (H), and PdCesA8A (I) expression in the transgenic xylem tissue compared to that in WT (the expression in WT is set to 1). The actin gene (Potri.001G309500) is used as a reference for expression normalization. The data are the mean values. The means ± standard deviation (SD) is calculated based on data obtained from three clonally propagated plants. Asterisks (**) indicate significant differences, P <0.01 (Student’s t-test). “ns” means no significant difference.
3.2 Anatomical features of the transgenic stems
The altered stem diameter morphology observed in the transgenic plants suggests potential modifications in xylem development. To investigate this further, we examined the development of the stem internode. Results indicated that the transgenics displayed an increase in xylem width (Figure 2A, B). In contrast, the number of cell layers within the xylem tissue was similar to the WT (Figure 2C). Further examination of the size of cells in the xylem tissue showed that the diameter of fiber cells increased, but the diameter of vessel cells did not change much (Figure 2D, E). To further elucidate the characteristics of fiber cells in the transgenic lines, we performed longitudinal dissections of the stems. This analysis indicated that fiber cell length did not show a significant difference from that of the WT (Figure 3A, B). However, the transgenics LTF/4/7, LTF/7/8, and LTF/4/7/8 exhibited a notable reduction in fiber cell wall thickness (Figure 3C-E). Overall, these results show that the transgenics LTF/4/7, LTF/7/8, and LTF/4/7/8 have fiber cells that are wider but have thinner walls.
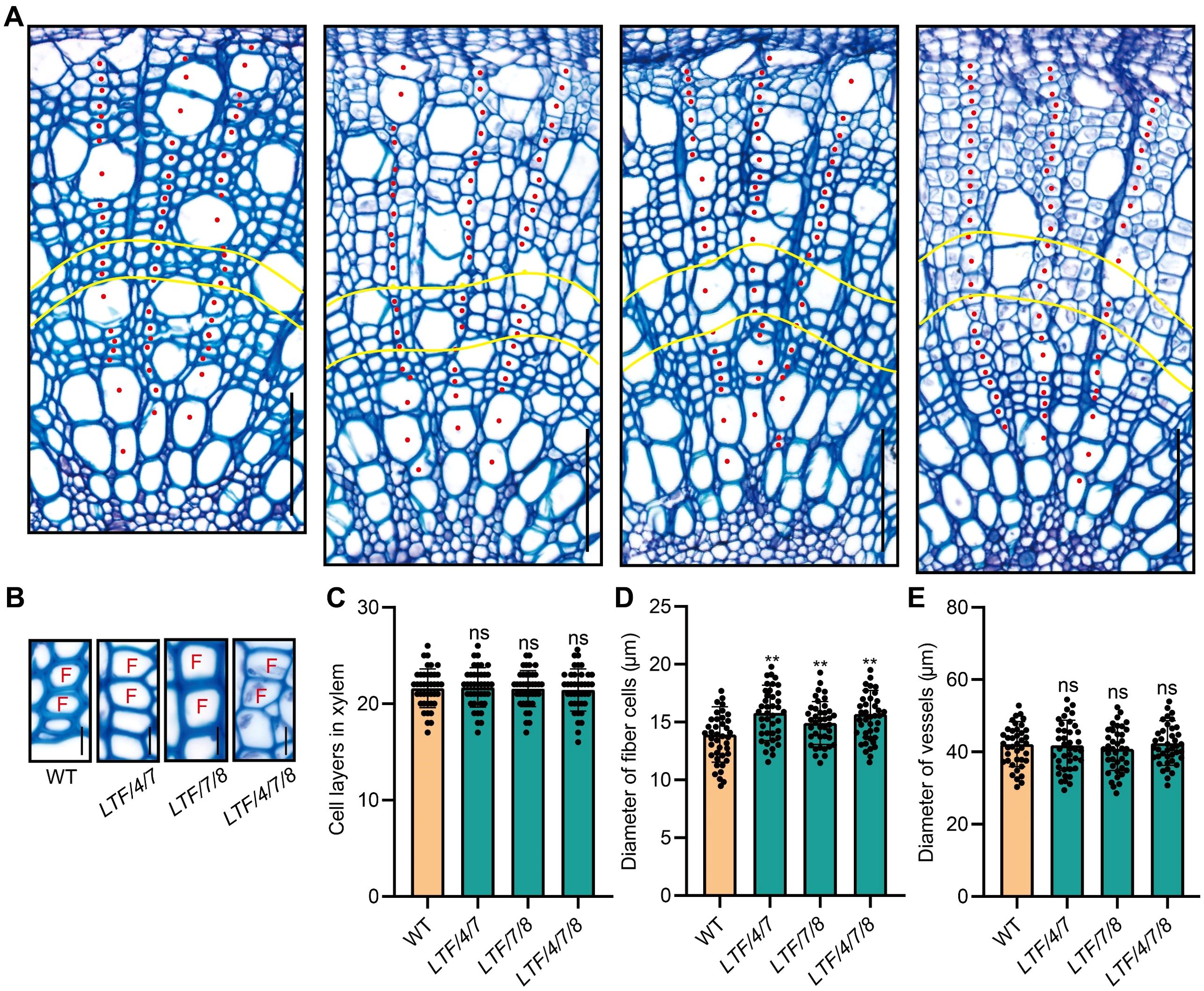
Figure 2. Stem cross anatomy of the LTF/4/7, LTF/7/8, and LTF/4/7/8. (A) Stem cross sections (the 15th internode) of LTF/4/7, LTF/7/8, and LTF/4/7/8 stained with toluidine blue. The red dots indicate xylem cells arranged in a file. The yellow curve marks an area of the xylem cells at the 12th ~ 15th layers in a file that was examined for cell morphology. Scale bar, 100 μm, (B) Cross view of xylem fibers. Scale bar, 13 μm. F, fiber cell. (C) Cell layers in xylem, (D) Diameter of fiber cells. (E) Diameter of vessels. The results are mean ± SD based on the determination of 40 cells from each of three clonally propagated plants. Asterisks (**) indicate significant differences, P <0.01 (Student’s t-test). “ns” means no significant difference.
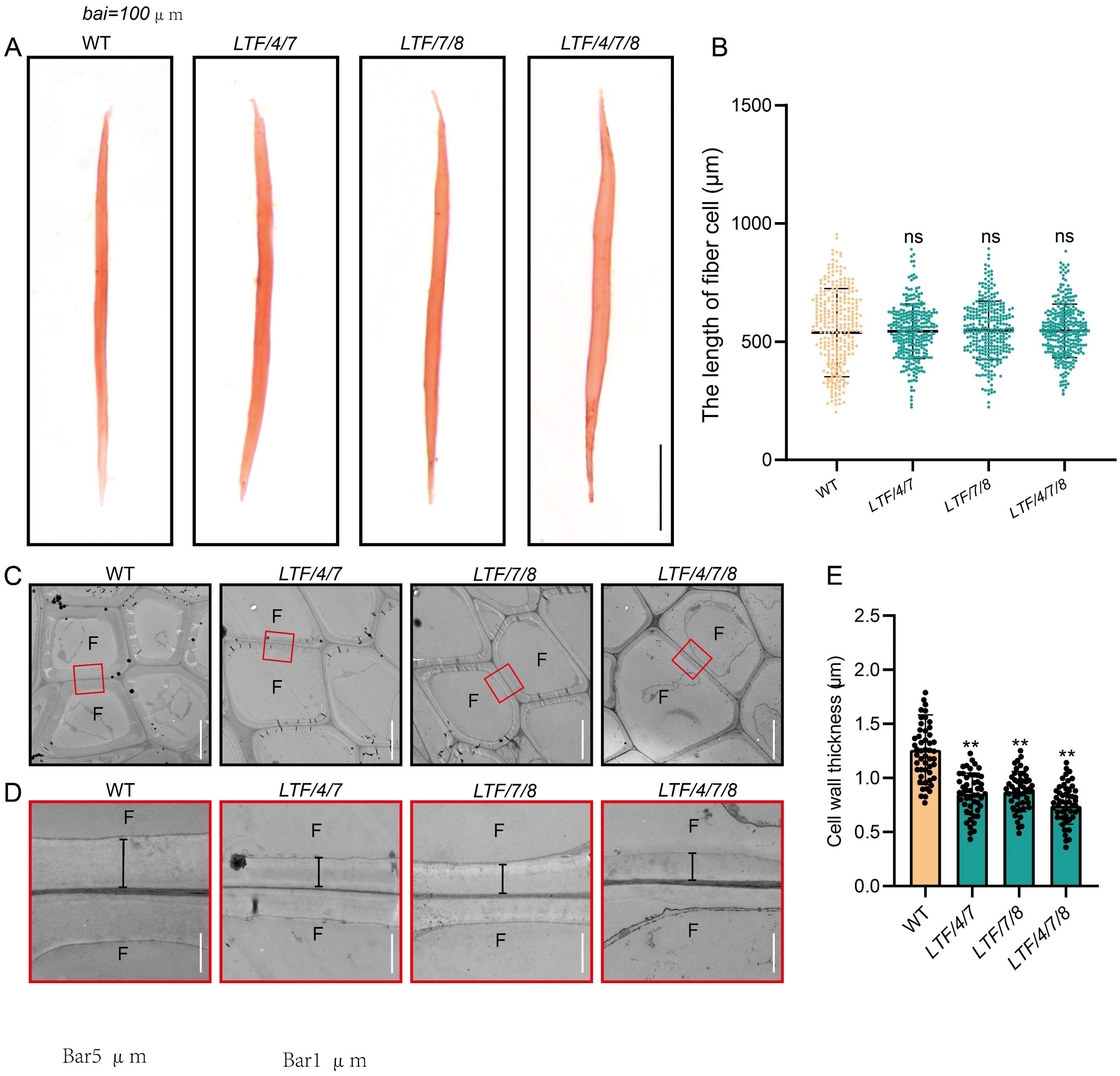
Figure 3. The 15th internode of fiber cell length and cell wall thickness in LTF/4/7, LTF/7/8, and LTF/4/7/8. (A) Dissociated fibers from the 15th internode were observed under an optical microscope. Scale bar, 100 μm. (B) The length of the dissociated fibers. (C, D) Transmission electron micrographs of the 15th internode fiber cell wall in LTF/4/7, LTF/7/8, and LTF/4/7/8. F, fiber cell. Magnification of the fiber cell wall in marked red rectangles. The black lines indicate the thickness of the cell wall. Scale bar, 5 μm in (C) and 1 μm in (D). (E) Cell wall thickness in LTF/4/7, LTF/7/8, and LTF/4/7/8. The length of fiber cells: mean ± SD, based on 200 fiber cells measured from the 15th internode of transgenics. Cell wall thickness: means ± SD of 50 fiber cells from the 15th internode of transgenics. Asterisks (**) indicate significant differences, P <0.01 (Student’s t-test). “ns” indicates no significant difference.
3.3 Cell wall composition in the transgenics
To investigate the composition of fiber cell walls in transgenic plants, cellulose staining demonstrated that the 14th internode of these transgenic lines exhibited significantly stronger staining intensity compared to that of the WT (Figure 4A). Quantitative analysis of crystalline cellulose content corroborated these results, revealing increases of 33.3%, 30.6%, and 38.1% in LTF/4/7, LTF/7/8, and LTF/4/7/8, respectively (Figure 4B). Phloroglucinol staining indicated a significant reduction in staining intensity in the transgenics compared to the WT, suggesting a decrease in lignin content (Figure 4C). Lignin content analysis was consistent with the phloroglucinol staining results, showing reductions of 19.1%, 11.7%, and 27.9% in LTF/4/7A, LTF/7/8, and LTF/4/7/8, respectively (Figure 4D). Additionally, quantitative analysis of lignin monomers in the transgenic plants revealed a decrease in both G-type and S-type lignin monomer contents. Notably, the decrease in S-type lignin monomers was significantly more pronounced than that of G-type monomers, leading to a substantial reduction in the S/G ratio (Figure 4E). Among the transgenic lines, LTF/4/7/8 exhibited the most significant reduction in S-type lignin monomer content, with a decrease of 44.2% (Figure 4E). These findings indicate that the overexpression of PdLTF1AA, PdCesA4, PdCesA7A, and PdCesA8A modified xylem fiber cell wall biosynthesis, thereby leading to an increase in fiber cell diameter.
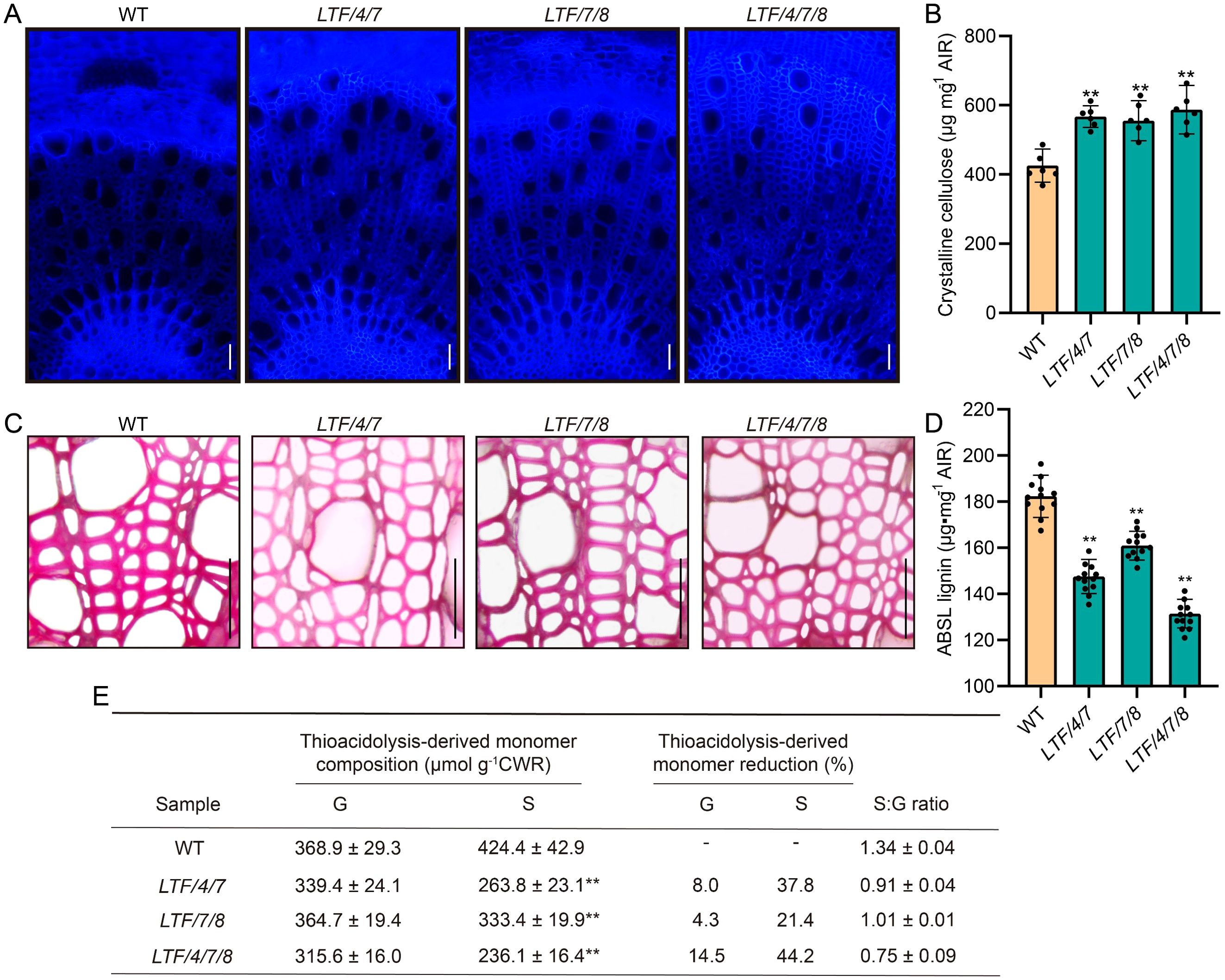
Figure 4. The cell wall composition was measured in the 16th to 20th internode of LTF/4/7, LTF/7/8, and LTF/4/7/8. (A) The cross-sectional stems of LTF/4/7, LTF/7/8, and LTF/4/7/8 were stained with cellulose dyes. Bar, 100μm. (B) Crystalline cellulose. (C) The cross-sectional stems of LTF/4/7, LTF/7/8, and LTF/4/7/8 were stained with phloroglucinol. Bar, 100μm. (D) Lignin content. (E) Lignin monomers. Cellulose content: means ± SD of 6 samples from three clonally propagated plants. Lignin content: means ± SD of 12 samples. Lignin monomers: means ± SD of 3 samples. Asterisks (**) indicate significant differences (P < 0.01, Student’s t-test).
3.4 The transcriptional landscape in the developing xylem of the transgenic plants was altered
Given the observed modifications in fiber cell dimension in the transgenics, we conducted transcriptomic analysis to elucidate how transcriptional activities were regulated by integration of PdLTF1AA, PdCesA4, PdCesA7A, and PdCesA8A genes. Developing xylem tissue was harvested for RNA sequencing analysis (Figure 5A). RNA sequencing analysis identified a substantial number of differential expressed genes (DEGs), which had a fold change > 2 in the transgenic plants compared with the WT. Specifically, the LTF/4/7 exhibited 1,922 DEGs, including 1,429 genes that were significantly upregulated and 493 genes that were significantly downregulated. The LTF/7/8 exhibited a total of 2,664 DEGs, consisting of 1,368 significantly upregulated genes and 1,296 significantly downregulated genes. The LTF/4/7/8 exhibited 2,011 DEGs, comprising 973 genes that were considerably upregulated and 1,038 genes that were significantly downregulated (Figure 5B, Supplementary Table S2). Through the examination of DEGs in LTF/4/7, LTF/7/8, and LTF/4/7/8 via a Venn diagram, we identified that 402 DEGs were shared among these transgenic plants (Figure 5C). The common DEGs predominantly participate in the metabolism of polysaccharides, as well as the biosynthesis of phenylpropanoids and associated secondary metabolites (Figure 5D). Conversely, it is noteworthy that the distinct DEGs found in the three types of transgenic plants were associated with various biological activities related to cell wall formation in an analogous distribution (Figure 5E). Among the common DEGs, those related to SCW deposition, such as PdNAC1, and lignin biosynthesis, including Pd4CL1, PdCAD6, and PdCOMT, were significantly downregulated (Figures 6A–E). This downregulation is consistent with the observed changes in the characteristics of the fiber cell wall. Moreover, the expression of genes associated with cell wall expansion, such as PdXTH4 and PdEXPA13, was significantly upregulated (Figures 6F–G). This upregulation correlates with the observed increase in fiber cell diameter in the transgenic plants. The data demonstrate that modification of lignin biosynthesis, in conjunction with regulation of various CesA genes in fiber cells, led to changes in the transcriptional activity associated with cellulose and lignin deposition. Because the transgenic plants did not fully produce all the CesA combinations, it is important to further investigate how the different CesAs control cellulose deposition.
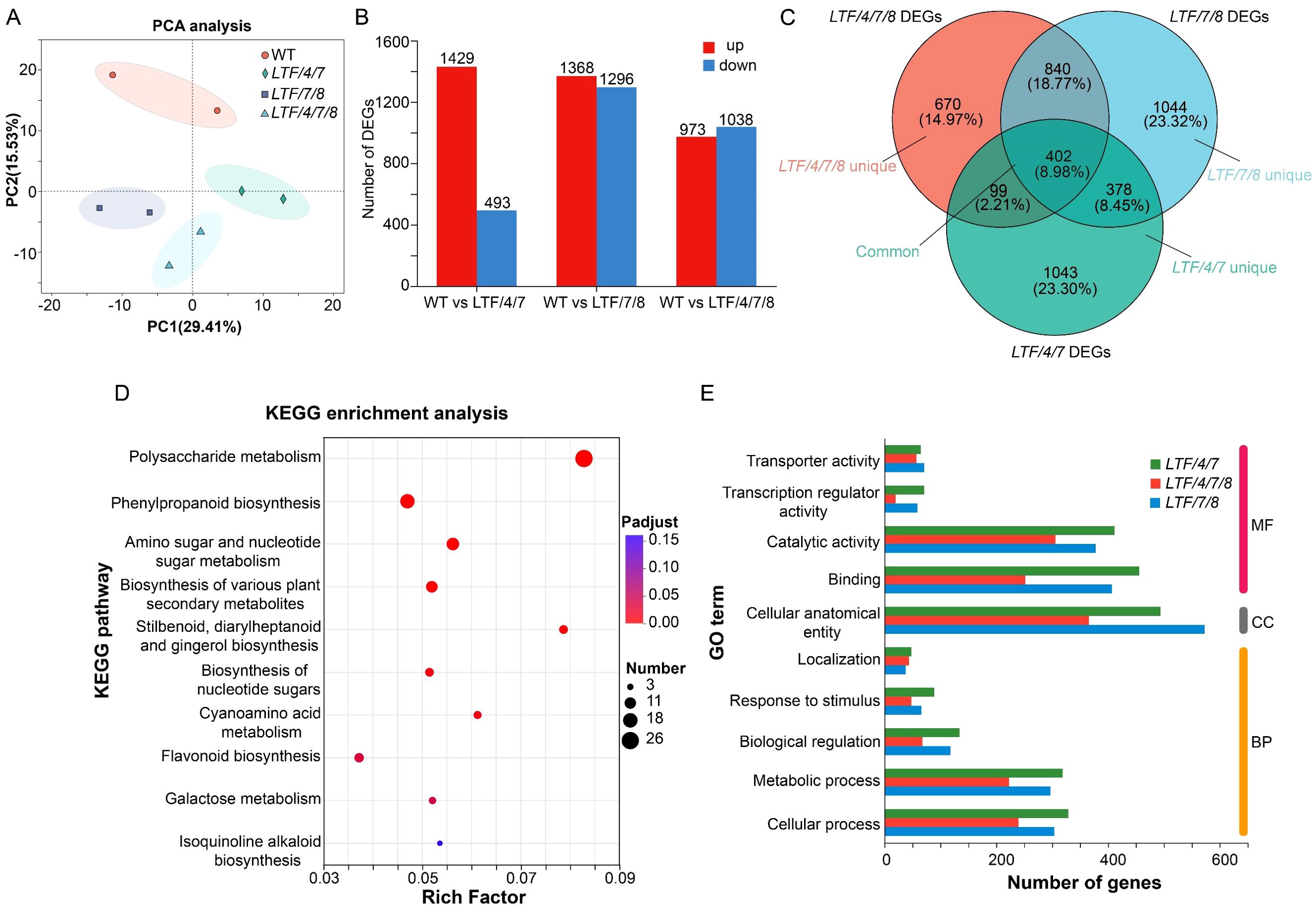
Figure 5. RNA sequencing of the stem xylem tissue in LTF/4/7, LTF/7/8, LTF/4/7/8, and WT. (A) PCA analysis of WT, LTF/4/7, LTF/7/8, and LTF/4/7/8 samples. The horizontal axis represents the contribution degree of principal component 1 (PC1) in the two-dimensional graph to the distinguished samples, and the vertical axis represents the contribution degree of principal component 2 (PC2) in the two-dimensional graph to the distinguished samples. (B) DEGs were identified between LTF/4/7 and WT, LTF/7/8 and WT, and LTF/4/7/8 and WT. (C) The Venn diagram analysis of DEGs in LTF/4/7, LTF/7/8, and LTF/4/7/8. (D) KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis was performed on the common DEGs in LTF/4/7, LTF/7/8, and LTF/4/7/8. p-adjust and rich factor are indicated. (E) GO annotation was performed on the unique DEGs in LTF/4/7, LTF/7/8, and LTF/4/7/8. MF, molecular function. CC, cellular component. BP, biological process.
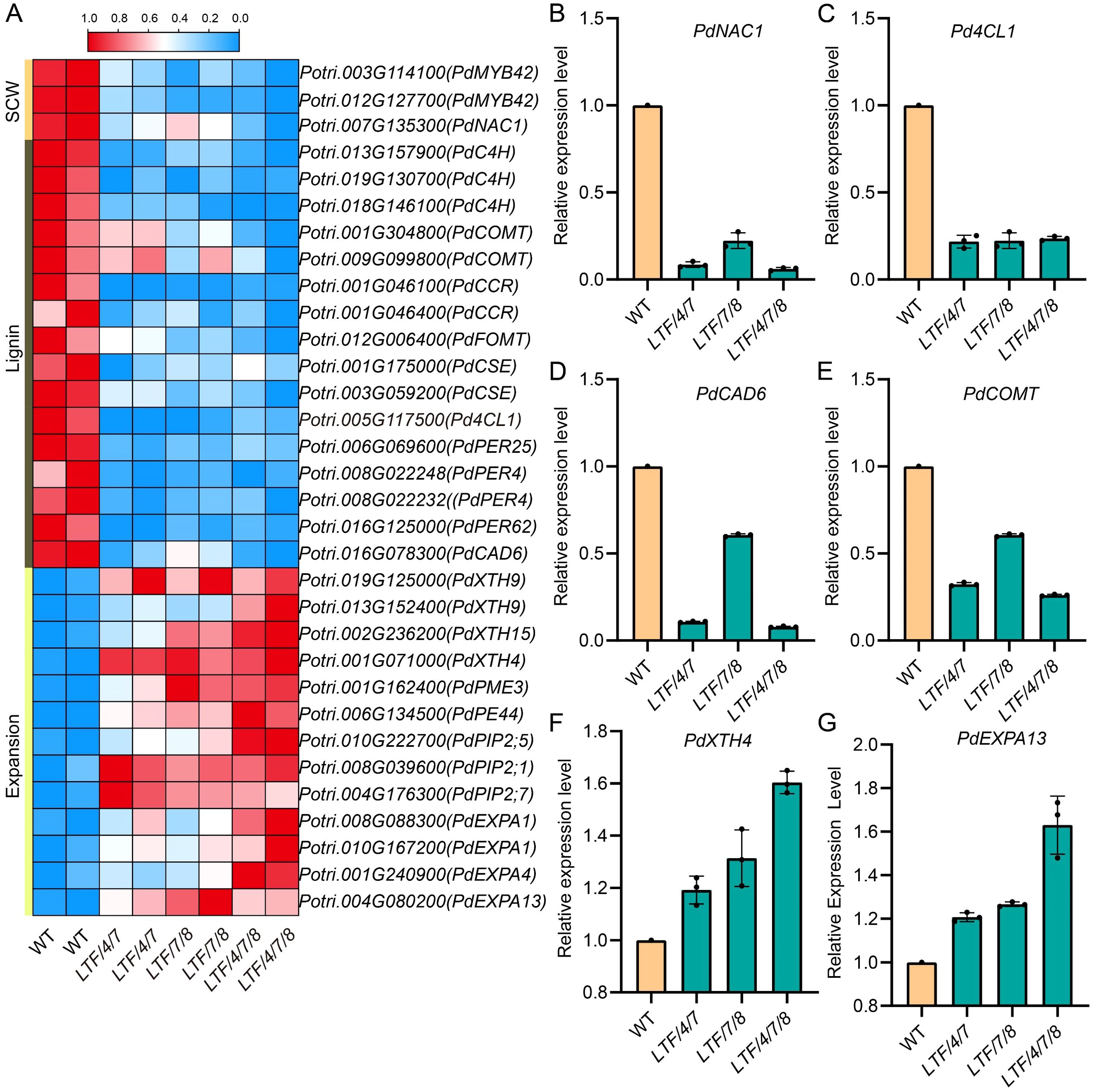
Figure 6. The common DEGs were enriched in cell wall organization and lignin biosynthesis. (A) Heat maps of the genes associated with cell wall organization and lignin biosynthesis. (B) RT-PCR validation of the differential expression of key genes involved in secondary cell wall biosynthesis (PdNAC1), lignin biosynthesis (Pd4CL1, PdCAD6, PdCOMT), expansion (PdXTH4, PdEXPA13). Data are means ± SD of 3 independent plants.
4 Discussion
Wood, a natural and renewable resource, is widely used in various fields, including pulping and papermaking, construction and furniture manufacturing, and energy and biomass uses. Wood comprises many cell types, with fiber cells constituting the primary structural component. The chemical composition of fiber cell walls directly affects the characteristics of wood. Thus, genetic engineering of fiber cell walls is regarded as an effective approach to improve wood characteristics (Chanoca et al., 2019; Zhu and Li, 2024).
The amounts of lignin and cellulose, as primary constituents of wood cell walls, significantly influence the qualities and applications of wood. Therefore, regulating lignin and cellulose synthesis to generate wood with appropriate cell wall components is essential for enhancing wood application value. Many studies have been undertaken in the past thirty years on the regulation of lignin biosynthesis and the genetic modification of lignin composition and structure (Boerjan et al., 2003; Chanoca et al., 2019; Li et al., 2024; Zhu and Li, 2024). The regulation of lignin deposition often influences field growth (Voelker et al., 2010; Zhu and Li, 2024). Such an effect may be due to the utilization of constitutive promoters to regulate lignin deposition, potentially altering the lignin composition in all cells, including vessel cell walls, which are crucial for the long-distance transport of water within the vessel. Recent studies indicate that controlling lignin deposition specifically in fiber cells has little effect on growth (Cao et al., 2020; De Meester et al., 2020; Gui et al., 2020). Altering lignin production in the fiber cell wall did not affect the lignin composition in the vessel cell wall, meaning it did not impact long-distance water transport and therefore did not affect growth in field conditions (Cao et al., 2020; Gui et al., 2020).
Meanwhile, studies have also been conducted on modifying cellulose biosynthesis in trees (Coleman et al., 2009; Maloney and Mansfield, 2010; Joshi et al., 2011; Yu et al., 2014; Nayeri et al., 2022). Our previous studies identified two types of CSCs participating in wood formation, with distinct gene influences on crystalline cellulose synthesis (Song et al., 2010; Xi et al., 2017). One type of CSA was linked to the deposition of crystalline cellulose, while the other may be linked to the biosynthesis of cellulose with a low degree of crystallinity. Evidence indicates that the regulation of cellulose synthase genes influences both cellulose content and cellulose crystalline properties (Yu et al., 2014; Xi et al., 2017; Abbas et al., 2020; Pedersen et al., 2024). In this study, we enhanced the expression of cellulose synthase genes that are related to a high degree of crystalline cellulose in fiber cells while inhibiting lignin biosynthesis. The transgenics showed that the lignin content decreased and the cellulose content increased, especially the increase of crystalline cellulose. The results of this study further suggest that cellulose with different crystalline structures may be related to different CesA genes; still, this conclusion warrants further corroboration through more comprehensive studies.
While lignin and cellulose can be individually modified, there have been no previous studies published on the concurrent regulation of lignin and cellulose biosynthesis. The results of the current study indicated modifications in both cell wall constituents, characterized by a reduction in lignin and an increase in cellulose. The study revealed the possibility of inhibiting lignin biosynthesis metabolism while simultaneously enhancing cellulose synthesis. The biosynthesis of cell wall components involves the coordinative function of multiple levels of gene networks, encompassing transcription factors, hormone signaling, cell wall biosynthesis enzyme genes, etc (Zhong et al., 2019; Zhang et al., 2020; Zhu and Li, 2024). It appears that both suppressing the lignin biosynthesis pathway and enhancing the cellulose synthesis process can happen at the same time; thus, manipulating several processes that produce cell walls at the same time could be crucial, enabling the development of various strategies to bioengineer trees with well-designed cell walls.
5 Conclusion
Lignin and cellulose are the primary components of wood cell walls, significantly influencing the characteristics and uses of wood. Many studies have indicated that the lignin and cellulose content of wood can be altered using genetic engineering; nevertheless, it remains to be established if lignin and cellulose can be concurrently modified to provide wood raw materials with desirable chemical composition. This study demonstrated how the deposition of cellulose and lignin in fiber cells, which are the main cells in wood, can be controlled, allowing both of these components in wood to be modified at the same time. Transgenic plants exhibited an enlarged diameter of wood fiber cells, accompanied by transcriptional changes in genes related to cell wall remodeling and polysaccharide synthesis during xylem development. These findings enable the development of an innovative technical strategy for engineering chemical composition in wood and for the bioengineering of trees with designated wood characteristics.
Data availability statement
The accession number for our deposited data is CRA028446. The data is publicly accessible at the following link: https://ngdc.cncb.ac.cn/gsa.
Author contributions
JL: Methodology, Formal Analysis, Visualization, Software, Writing – original draft, Writing – review & editing. YH: Writing – original draft, Validation, Resources, Methodology, Formal Analysis. XL: Writing – original draft, Data curation, Methodology, Validation. XT: Methodology, Resources, Writing – original draft. JS: Resources, Writing – original draft, Methodology. ML: Funding acquisition, Writing – original draft, Formal Analysis. LL: Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was financially supported by the National Key Research and Development Program (Grant No. 2021YFD2200204), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB0630203), and the National Natural Science Foundation of China (Grant No. 32130072) awarded to LL. Furthermore, it was also supported by the Hunan Natural Science Funding (Grant No. 2024JJ5634) granted to ML.
Acknowledgments
We are grateful to Junhui Shen for the management of experimental reagents. We also thank Xiaoyan Gao and Zhiping Zhang for their assistance in the preparation and observation of ultrathin sections for transmission electron microscopy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1646316/full#supplementary-material
References
Abbas, M., Peszlen, I., Shi, R., Kim, H., Katahira, R., Kafle, K., et al. (2020). Involvement of CesA4, CesA7-A/B and CesA8-A/B in secondary wall formation in Populus trichocarpa wood. Tree Physiol. 40, 73–89. doi: 10.1093/treephys/tpz020
Bjurhager, I., Olsson, A., Zhang, B., Gerber, L., Kumar, M., Berglund, L., et al. (2010). Ultrastructure and mechanical properties of populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase. Biomacromolecules 11, 2359–2365. doi: 10.1021/bm100487e
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bryant, N., Pu, Y., Tschaplinski, T., Tuskan, G., Muchero, W., Kalluri, U., et al. (2020). Transgenic poplar designed for biofuels. Trends Plant Sci. 25, 881–896. doi: 10.1016/j.tplants.2020.03.008
Cao, S., Huang, C., Luo, L., Zheng, S., Zhong, Y., Sun, J., et al. (2020). Cell-specific suppression of 4-coumarate-coa ligase gene reveals differential effect of lignin on cell physiological function in Populus. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.589729
Chanoca, A., De Vries, L., and Boerjan, W. (2019). Lignin engineering in forest trees. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00912
Coleman, H., Yan, J., and Mansfield, S. (2009). Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. U.S.A. 106, 13118–13123. doi: 10.1073/pnas.0900188106
De Meester, B., Madariaga, B., De Vries, L., Pollier, J., Goeminne, G., Van Doorsselaere, J., et al. (2020). Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient CINNAMOYL-CoA REDUCTASE2 allele. Nat. Commun. 11, 5020. doi: 10.1038/s41467-020-18822-w
De Meester, B., Vanholme, R., Mota, T., and Boerjan, W. (2022). Lignin engineering in forest trees: from gene discovery to field trials. Plant Commun. 3, 100465. doi: 10.1016/j.xplc.2022.100465
Du, J. and Groover, A. (2010). Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 52, 17–27. doi: 10.1111/j.1744-7909.2010.00901.x
Foster, C., Martin, T., and Pauly, M. (2010a). Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) part I: lignin. J. Vis. Exp. 37), 1745. doi: 10.3791/1745
Foster, C., Martin, T., and Pauly, M. (2010b). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J. Vis. Exp. 37), 1837. doi: 10.3791/1837
Gui, J., Lam, P., Tobimatsu, Y., Sun, J., Huang, C., Cao, S., et al. (2020). Fibre-specific regulation of lignin biosynthesis improves biomass quality in Populus. New Phytol. 226, 1074–1087. doi: 10.1111/nph.16411
Gui, J., Luo, L., Zhong, Y., Sun, J., Umezawa, T., and Li, L. (2019). Phosphorylation of LTF1, an myb transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol. Plant 12, 1325–1337. doi: 10.1016/j.molp.2019.05.008
Gui, J., Shen, J., and Li, L. (2011). Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 157, 574–586. doi: 10.1104/pp.111.178301
Hu, W., Harding, S., Lung, J., Popko, J., Ralph, J., Stokke, D., et al. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17, 808–812. doi: 10.1038/11758
Joshi, C., Thammannagowda, S., Fujino, T., Gou, J., Avci, U., Haigler, C., et al. (2011). Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen. Mol. Plant 4, 331–345. doi: 10.1093/mp/ssq081
Li, J., Guo, X., Lu, X., Sun, J., Jin, Y., Li, M., et al. (2025). Fiber-specific expression of PdCel9A6 modifies the characteristics of wood fibers in Populus. Tree Physiol., tpaf051. doi: 10.1093/treephys/tpaf051
Li, L., Zhou, Y., Cheng, X., Sun, J., Marita, J., Ralph, J., et al. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. U.S.A. 100, 4939–4944. doi: 10.1073/pnas.0831166100
Li, W., Lin, Y. J., Chen, Y., Zhou, C., Li, S., De Ridder, N., et al. (2024). Woody plant cell walls: fundamentals and utilization. Mol. Plant 17, 112–140. doi: 10.1016/j.molp.2023.12.008
Luo, L. and Li, L. (2022). Molecular understanding of wood formation in trees. For Res. 2, 5. doi: 10.48130/FR-2022-0005
Maloney, V. and Mansfield, S. (2010). Characterization and varied expression of a membrane-bound endo-β-1, 4-glucanase in hybrid poplar. Plant Biotechnol. J. 8, 294–307. doi: 10.1111/j.1467-7652.2009.00483.x
Nayeri, S., K., B., Ahmadikhah, A., and Mahna, N. (2022). CRISPR/Cas9-mediated P-CR domain-specific engineering of CESA4 heterodimerization capacity alters cell wall architecture and improves saccharification efficiency in Poplar. Plant Biotechnol. J. 20, 1197–1212. doi: 10.1111/pbi.13803
Paredez, A., Somerville, C., and Ehrhardt, D. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Pedersen, G., Blaschek, L., Frandsen, K., Noack, L., and Persson, S. (2024). Cellulose synthesis in land plants. Mol. Plant 17, 359–361. doi: 10.1016/j.molp.2023.12.013
Ralph, J., Akiyama, T., Coleman, H., and Mansfield, S. (2012). Effects on lignin structure of coumarate 3-hydroxylase downregulation in Poplar. Bioenergy Res. 5, 1009–1019. doi: 10.1007/s12155-012-9218-y
Song, D., Shen, J., and Li, L. (2010). Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol. 187, 777–790. doi: 10.1111/j.1469-8137.2010.03315.x
Sulis, D., Jiang, X., Yang, C., Marques, B., Matthews, M., Miller, Z., et al. (2023). Multiplex CRISPR editing of wood for sustainable fiber production. Science 381, 216–221. doi: 10.1126/science.add4514
Suzuki, S., Li, L., Sun, Y., and Chiang, V. (2006). The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142, 1233–1245. doi: 10.1104/pp.106.086678
Tai, H., Chang, C., Cai, W., Lin, J., Huang, S., Lin, Q., et al. (2023). Wood cellulose microfibrils have a 24-chain core-shell nanostructure in seed plants. Nat. Plants 9, 1154–1168. doi: 10.1038/s41477-023-01430-z
Turner, S. and Kumar, M. (2018). Cellulose synthase complex organization and cellulose microfibril structure. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 376, 20170048. doi: 10.1098/rsta.2017.0048
Van Acker, R., Dejardin, A., Desmet, S., Hoengenaert, L., Vanholme, R., Morreel, K., et al. (2017). Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol. 175, 1018–1039. doi: 10.1104/pp.17.00834
Voelker, S., Lachenbruch, B., Meinzer, F., Jourdes, M., Ki, C., Patten, A., et al. (2010). Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown Poplar. Plant Physiol. 154, 874–886. doi: 10.1104/pp.110.159269
Voelker, S., Lachenbruch, B., Meinzer, F., Kitin, P., and Strauss, S. (2011). Transgenic poplars with reduced lignin show impaired xylem conductivity, growth efficiency and survival. Plant Cell Environ. 34, 655–668. doi: 10.1111/j.1365-3040.2010.02270.x
Wang, H., Xue, Y., Chen, Y., Li, R., and Wei, J. (2012). Lignin modification improves the biofuel production potential in transgenic Populus tomentosa. Ind. Crops Prod. 37, 170–177. doi: 10.1016/j.indcrop.2011.12.014
Xi, W., Song, D., Sun, J., Shen, J., and Li, L. (2017). Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus. Plant Mol. Biol. 93, 419–429. doi: 10.1007/s11103-016-0570-8
Xu, W., Cheng, H., Zhu, S., Cheng, J., Ji, H., Zhang, B., et al. (2021). Functional understanding of secondary cell wall cellulose synthases in Populus trichocarpa via the Cas9/gRNA-induced gene knockouts. New Phytol. 231, 1478–1495. doi: 10.1111/nph.17338
Yamamura, M., Hattori, T., Suzuki, S., Shibata, D., and Umezawa, T. (2012). Microscale thioacidolysis method for the rapid analysis of β-O-4 substructures in lignin. Plant Biotechnol. 29, 419–423. doi: 10.5511/plantbiotechnology.12.0627a
Ye, Z. and Zhong, R. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66, 4119–4131. doi: 10.1093/jxb/erv081
Yu, L., Chen, H., Sun, J., and Li, L. (2014). PtrKOR1 is required for secondary cell wall cellulose biosynthesis in Populus. Tree Physiol. 34, 1289–1300. doi: 10.1093/treephys/tpu020
Zhang, J., Tuskan, G., Tschaplinski, T., Muchero, W., and Chen, J. (2020). Transcriptional and post-transcriptional regulation of lignin biosynthesis pathway genes in Populus. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00652
Zhong, R., Cui, D., and Ye, Z. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Keywords: lignin, Populus, cellulose, cell wall, fiber cell
Citation: Li J, Han Y, Lu X, Tang X, Sun J, Li M and Li L (2025) Simultaneous regulation of both lignin and cellulose biosynthesis modifies xylem fiber properties in Populus. Front. Plant Sci. 16:1646316. doi: 10.3389/fpls.2025.1646316
Received: 13 June 2025; Accepted: 14 July 2025;
Published: 04 August 2025.
Edited by:
Aimin Wu, South China Agricultural University, ChinaCopyright © 2025 Li, Han, Lu, Tang, Sun, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Li, bGltZW5nMDQyMkBjc3VmdC5lZHUuY24=; Laigeng Li, bGdsaUBjZW1wcy5hYy5jbg==
†These authors have contributed equally to this work
 Jian Li
Jian Li Yu Han
Yu Han Xianwen Lu4
Xianwen Lu4 Xinwei Tang
Xinwei Tang Jiayan Sun
Jiayan Sun Meng Li
Meng Li Laigeng Li
Laigeng Li