- 1College of Forestry, Central South University of Forestry and Technology, Changsha, China
- 2Key Laboratory of Cultivation and Protection for Non-wood Forest Trees, Ministry of Education, Central South University of Forestry and Technology, Changsha, China
- 3Key Laboratory of Non-wood Forest Products of State Forestry Administration, Central South University of Forestry and Technology, Changsha, China
- 4Engineering Technology Research Center of Southern Hilly and Mountainous Ecological Non-Wood Forest Industry of Hunan Province, Central South University of Forestry and Technology, Changsha, China
- 5Lutou National Station for Scientific Observation and Research of Forest Ecosystem in Hunan Province Changsha, Changsha, China
Plant abiotic stress refers to the unfavorable effects on plants caused by any abiotic factors in a specific environment, such as drought, high temperature, low temperature, etc., which cause disruption of plant physiology and metabolism, and seriously affect the growth and yield of plants. Mounting evidence demonstrates that WRKY transcription factors modulate plant abiotic stress responses by regulating sugar metabolic pathways. Sugar metabolism pathway plays an essential role in plant stress resistance, and WRKY transcription factors, as an important class of regulatory factors, have attracted wide attention for their mechanism of action in abiotic stress. Therefore, this review primarily aims to analyze the structure and classification of WRKY transcription factors, summarize the research progress on how WRKY transcription factors themselves respond to stress, and how they participate in regulating plant stress responses through sugar metabolism pathways. Through in-depth investigation of the relationship between WRKY transcription factors and sugar metabolic pathways we uncovered novel abiotic stress-related gene regulatory networks providing theoretical basis and practical guidance for genetic improvement of plants under abiotic stress.
1 Introduction
Plant abiotic stress has become one of the key constraints to agricultural production and food security. Among them, drought, cold, and salinity stress are primary abiotic factors that impair plant growth and development and constrain their geographic distribution. These stresses often cause similar effects on plants, such as disrupting cellular osmotic balance, damaging cell membrane structures, and impairing antioxidant defense systems (Qu et al., 2019). To survive adverse environmental conditions, plants have evolved intricate signaling and gene regulatory pathways. These sophisticated mechanisms enable adaptation and mitigation against the detrimental impacts of abiotic stresses. Sugar metabolism, as one of these pathways, plays a crucial role in the process by which plants resist abiotic stress (Qin et al., 2018; Ma et al., 2019; Yoon et al., 2020). Sugars not only provide energy and carbon sources but also participate in signal transduction and the regulation of physiological processes. Although a large number of studies have shown that sugar metabolism is extensively involved in plant stress, relatively few studies have been conducted on how sugar mediates stress mechanisms.
Transcription factors, as key components of signal transduction, play the role of “molecular switches” in the transcriptional regulatory networks of abiotic stress responses. WRKY transcription factors (TFs), a unique class of proteins in higher plants, are widely involved in regulating various physiological and metabolic pathways. They are capable of highly specific recognition and binding to cis-acting elements called W-box on DNA sequences. This binding directly regulates the transcription levels of target genes, including self and other stress-related genes, and thus plays a key role in plant response to abiotic stresses. In addition, WRKY TFs can also bind to cis-acting elements in the promoter regions of sugar metabolism genes and regulate the expression of sugar metabolism-related genes (Sun et al., 2003; Govardhana and Kumudini, 2020; Goyal et al., 2020). Through regulation of the sugar metabolism pathway, they mediate responses to stress, thus improving plant tolerance to abiotic stresses and injury, and mitigating the damage inflicted by stress on plants (Wu et al., 2020).
Therefore, this paper mainly analyzed the structure and classification of WRKY transcription factors, summarized the research results and reviewed the regulatory mechanisms of WRKY transcription factors themselves in response to abiotic stresses as well as their involvement in abiotic stresses in plants through sugar metabolism pathways, with a view to providing a theoretical basis for the genetic improvement of plant stress tolerance, and providing a technological safeguard for the enhancement of agricultural production and the assurance of food safety.
2 Structural characteristics and classification of WRKY TFs
WRKY TFs are among the largest families of transcription factors in higher plants and are designated as the “central regulators” of the abiotic stress response. The first WRKY TFs, SPF1, was originally identified and isolated from Ipomoea batatas (Ishiguro and Nakamura, 1994). Subsequent studies have characterized numerous WRKY TFs across diverse plant species, including. Arabidopsis (Glöckner et al., 2002), Oryza sativa (Kim et al., 2000), soybean (Schmutz et al., 2010), and Hordeum vulgare (Mangelsen et al., 2008). Comprehensive research has elucidated the extensive membership and multifaceted regulatory mechanisms characterizing the WRKY transcription factor family. By constructing complex signaling networks, WRKY TFs play a crucial role in plant growth, development, and stress responses (ülker and Somssich, 2004; Rushton et al., 2010).
WRKY TFs derive their nomenclature from the characteristic WRKY domain, defined by the highly conserved WRKYGQK motif (Wu et al., 2020). This family of proteins is characterized by the fact that all family members contain at least one WRKY structural domain consisting of about 60 highly conserved amino acids, the N-terminal end of which contains the highly conserved WRKYGQK heptapeptide sequence, and the C-terminal end of which has a zinc-finger motif of either the C2H2 or the C2HC type (Wang et al., 2014b). WRKY TFs specifically recognize W-box cis-elements (A/TAACCA; C/TAACG/TG) in target gene promoters, thereby modulating transcription. Depending on the number of conserved WRKY domains and the type of zinc finger structure, WRKY transcription factors are usually divided into three families: family I contains two WRKY domains and two C2H2 zinc finger structures, family II contains one WRKY domain and one C2H2 zinc finger structure, and family III contains one WRKY domain and one C2H2 zinc finger structure. Family II is subdivided into five subfamilies: a, b, c, d, and e. Family II WRKY proteins are involved in the regulation of plant growth and development, such as senescence, seed dormancy, and germination; they are also involved in plant responses to drought, salt stress, and cold damage (Rushton et al., 2012). WRKYs cannot form homologous or heterodimers if they do not have LZ (leucine zipper) motifs (Narusaka et al., 2016). In addition to the above structural domains, WRKY transcription factor families have many other structures, such as kinase domains, glutamine rich regions, proline rich regions, nuclear localization signals, and so on. The existence of these structural domains makes it possible for these WRKY proteins to regulate the expression of target genes through the formation of homodimers or heterodimers by protein-protein interactions (Zhang and Wang, 2005; Agarwal et al., 2011) (Figure 1).
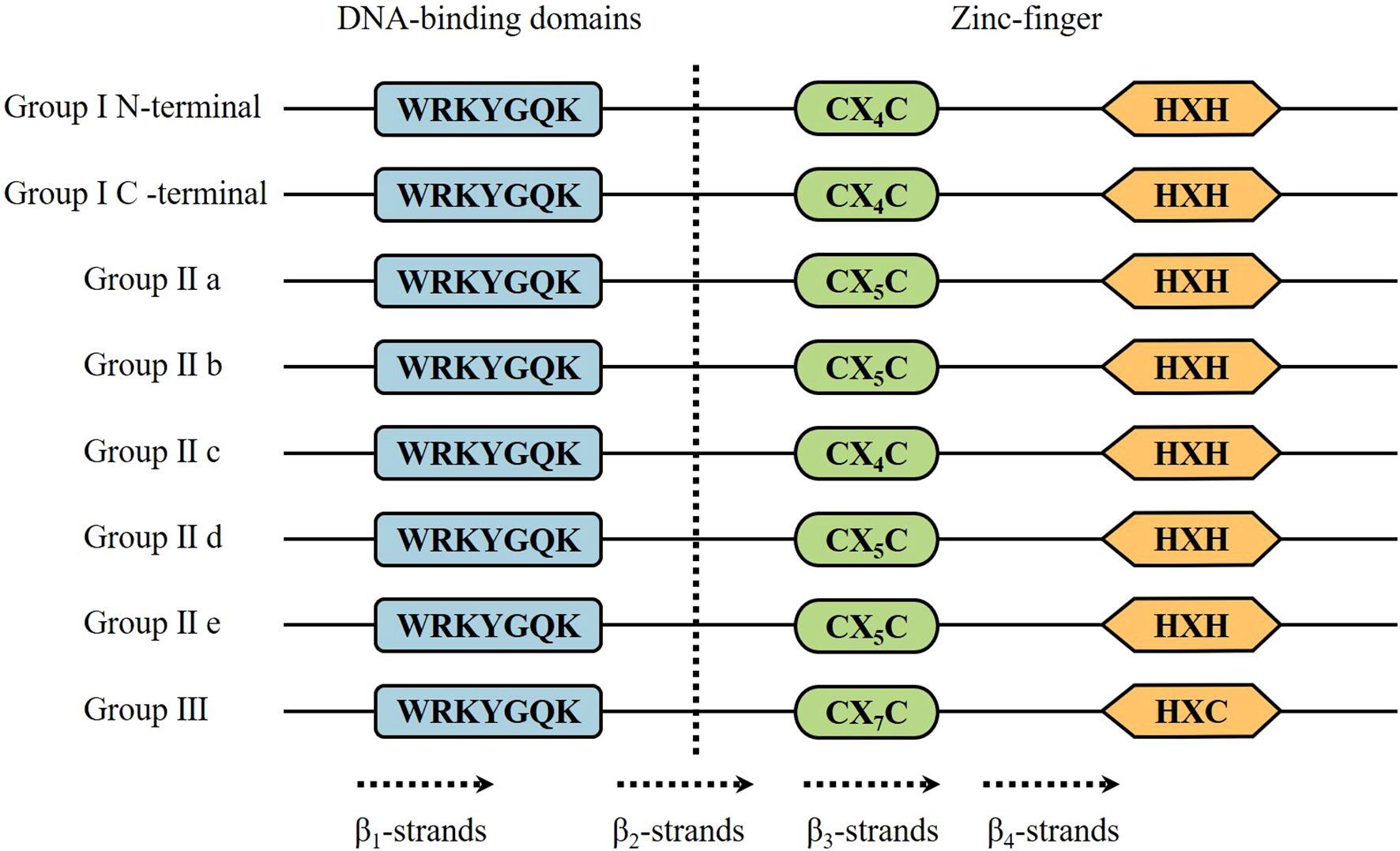
Figure 1. Domain structures of different WRKY subfamilies in higher plants. The WRKY motif, the cysteines, and the histidines that form the zinc finger are shown in boxes. I N and I C denote the N-terminal and C-terminal domains from Group I WRKY proteins, respectively. The 4 β-strands are shown with dashed arrows.
3 WRKY TFs involved in abiotic stress responses
In recent years, as climate change and extreme weather events have increased, the impact of abiotic stress on crop production has become more pronounced, resulting in slowed growth, deteriorating quality, and reduced yields (Hrmova and Hussain, 2021; Khoso et al., 2022). Consequently, over the course of long-term natural selection and evolutionary processes, plants have developed a complex and finely tuned regulatory network that enables them to detect and respond effectively to various environmental stresses (Su et al., 2023). Facing abiotic stress, WRKY TFs could dynamically modulate downstream gene expression, either enhancing transcriptional activation or imposing repression, directly regulating the expression of genes involved in stress response, or participate in other signaling pathways and regulatory networks to manage the stress response (Figure 2). This activation of defense mechanisms helps enhance crop resilience against abiotic stress (Ma and Hu, 2024). Given that extensive and in-depth studies and reviews have already been conducted on WRKY TFs’ roles in responding to abiotic stress (Table 1), this paper provides only a concise summary of the key findings.
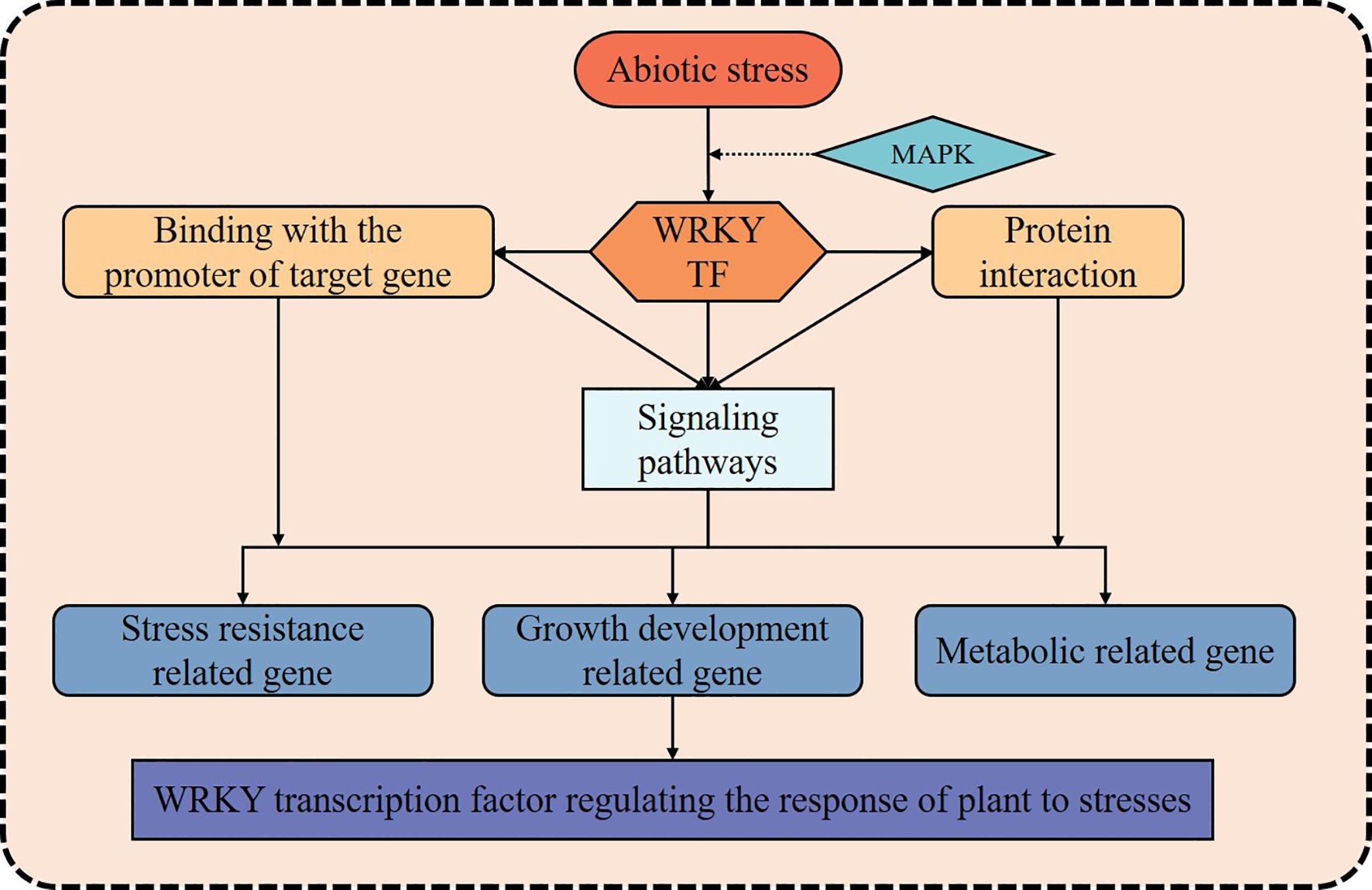
Figure 2. The diagram of WRKY transcription factor regulating stress responses in plants. The solid black arrows indicate that WRKYs regulating plant stress response pathway; The dotted black arrow indicates that WRKY transcription factors might be activated by the MAPK cascade and thus participates in the regulation of stress response.
Drought stress, as an essential abiotic stress, poses a serious threat to plant growth, development, and yield. Recent studies have identified multiple WRKY TFs as key regulators of drought tolerance. IgWRKY50 and IgWRKY32 in Iris germanica, which can enhance drought resistance in transgenic Arabidopsis by coordinated up-regulation of drought-responsive downstream genes (Zhang et al., 2022a). In Glycine max, GmWRKY17 directly binds to promoters of drought-inducible genes GmDREB1D and GmABA2, activating their transcription under water deficit (Yi and Yueping, 2023). SbWRKY30 in Sorghum bicolor directly activates the drought-response gene SbRD19, conferring improved growth and survival rates under drought stress (Yang et al., 2020).
Salt stress critically constrains plant growth and development, wherein WRKY transcription factors (TFs) execute pivotal regulatory roles. ZmWRKY104 in Zea mays overexpression enhances salt tolerance via positive regulation of ZmSOD4, reducing ROS accumulation, MDA content, and electrolyte leakage (Yan et al., 2022).In Gossypium hirsutum, GhWRKY34 confers salt tolerance by modulating selective Na+/K+ uptake and maintaining low Na+/K+ ratios in leaves/roots (Zhou et al., 2015).GmWRKY54 in Glycine max activates transcription in response to salt stress by binding to the W-box elements of the promoters of the DREB2A and STZ/ZAT10 genes, which are key transcription factors in the ABA-independent pathway that regulates osmoprotective substance synthesis, and STZ/ZAT10, which is involved in ROS scavenging and ion homeostasis maintenance (Zhou et al., 2008).
WRKY TFs also have a significant job in responding to heavy metal stresses. In Oryza sativa, OsWRKY74 regulates the expression of a set of downstream genes involved in phosphorus uptake, transport, and metabolism, collectively enhancing rice tolerance to phosphorus starvation (Dai et al., 2016). Similarly, OsWRKY72 negatively regulates lignin synthesis and accumulation by inhibiting the expression of OsGLP8-7 (germin-like protein), thereby reducing the ability of the cell wall to retain heavy metal ions and realizing the regulation of Cd/Cu toxicity (Shangguan et al., 2024). TaWRKY70 in Triticum aestivum reduces root Cd²+influx by re-pressing the expression of AtNRAMP5, AtHMA3, AtYSL3, and AtIRT1 heavy-metal transporter genes, which act as Cd transporters, while TaWRKY70 activates the expression of the TaCAT5 promoter by directly binding to its W-box motif to enhance catalase activity, reduce membrane lipid peroxidation, scavenge ROS, and confer resistance to Cd stress in transgenic Arabidopsis (Jia et al., 2021).
In addition, with global climate change in recent years, both high and low temperatures have become significant agricultural meteorological disasters, severely limiting normal plant and crop development. Heat stress triggers upregulation of WRKY25 and WRKY26 yet downregulates WRKY33 in Arabidopsis thaliana. Molecular evidence reveals that these three factors synergistically enhance thermotolerance by coordinating ethylene signaling activation with heat shock protein (HSP) pathways, leveraging functional crosstalk and complementary effects (Li et al., 2011). In Capsicum annuum, the transcription of CaWRKY40 is induced by Ralstonia solanacearum and high temperature. Heat stress induces an upregulation of CaWRKY40 expression, and its overexpression enhances heat stress tolerance, likely through the regulation of antioxidant systems and maintenance of cell membrane stability (Dang et al., 2013). Regarding low temperature, Zhang et al. (2022b) proposed a transcriptional regulatory cascade model involving OsWRKY63–OsWRKY76–OsDREB1B, where OsWRKY63 acts as a transcriptional repressor by inhibiting the expression of OsWRKY76, thereby suppressing the activation of OsDREB1B, which leads to reduced cold tolerance (Zhang et al., 2022b). The expression of VbWRKY32 was significantly increased in Verbena leaves under cold stress. Overexpression of the VbWRKY32 gene in Verbena and comparison of the expression profiles of cold-responsive genes between overexpressed and wild-type plants under cold stress revealed that VbWRKY32 acted as a positive regulator to enhance the cold resistance of plants by up-regulating the transcript levels of cold-responsive genes (Wang et al., 2020). In Camellia sinensis, CsWRKY6, CsWRKY31, CsWRKY48 were induced to be up-regulated under 4 °C cold treatment, indicating that they acted as positive regulators involved in the regulatory pathway of Camellia sinensis in response to cold (Wang et al., 2019). All these studies confirmed that WRKY TFs plays an essential role in defense against abiotic stresses.
4 Sugar metabolism involved in abiotic stress responses
Adversities such as low temperature, high temperature, drought, and salinity usually produce water stress in plants, and osmoregulation is one of the important physiological mechanisms for plants to resist such abiotic stresses. Plants through the regulation of various physiological metabolism in body accumulation of a wide range of organic or inorganic substances to increase the concentration of cell membranes, reduce the osmotic potential, and enhance the cellular water absorption or retention capacity. Sugars are an effective carbohydrate in plant response to abiotic stress, which not only keep the cellular osmotic balance, but also participate in the signaling molecules for the perception and conduction of adversity signals, and regulate the growth and development of plants and their ability to cope with adversity (Sun et al., 2015).
4.1 The mechanism by which sugar responds to abiotic stress
The role played by sugar compounds in response to abiotic stresses in plants is characterized by four main aspects: First, under abiotic stress, plants accumulate sugars within cytoplasmic and vacuolar compartments. This solute accumulation elevates cytosolic concentration, modulates tissue osmotic potential, depresses freezing point, and mitigates cellular dehydration—collectively establishing a physical defense barrier against environmental adversities (Sun et al., 2015; Hu et al., 2016). Second, sugar compounds provide protective effects on biomembranes and macromolecules. Specifically, fructans demonstrate membrane-stabilizing properties by reducing the phase transition temperature between lipid gel and liquid crystalline states (Hincha et al., 2002). This phenomenon facilitates enhanced molecular interactions between sugar moieties and membrane phospholipids, thereby preserving membrane structural integrity under stress conditions. Third, plant sugar catabolism integrates bio-oxidative pathways with oxidative phosphorylation systems. This integrative mechanism can provide not only sufficient reducing power and energy for other biosynthetic processes, it also produces other protective substances that protect plant organizations from stress-induced damage (Savitch et al., 2010). Fourth, sugar compounds form complex signaling networks with other signaling molecules, regulating the expression of stress-related genes involved in metabolic activities, thereby helping plants respond to adverse environmental conditions (Smeekens et al., 2010). For instance, low-temperature stress upregulates key sucrose metabolic enzymes such as sucrose synthase (SUS) and sucrose phosphate synthase (SPS), enhancing their catalytic activity and thereby driving sucrose accumulation in plants. On the one hand, sucrose acts as an osmotic regulator, maintaining cellular osmotic potential under low temperatures and preventing cell freezing. On the other hand, as an important signaling molecule and antioxidant, sucrose can induce the expression of cold tolerance genes and key enzymes in the antioxidant system (Turhan and Ergin, 2012; Cao et al., 2014). Therefore, sugar compounds play an indispensable and crucial role in plant stress resistance, and they help plants maintain their normal physiological functions and growth status in the face of various abiotic stresses at multiple levels and through the utilization of different regulatory mechanisms.
4.2 The response of glucose metabolism to abiotic stress
Regarding the study of the pathway of sugar metabolism involved in stress tolerance, a large number of studies have shown that the pathway of sugar metabolism involved in stress tolerance is mainly related to the accumulation of saccharides, and that the higher the content of soluble sugars in the body of a plant, the stronger its cold tolerance will be (Ouyang et al., 2019). By accumulating soluble sugar content, plants are able to increase cellular osmotic potential, which in turn enhances cellular water retention capacity (Ouyang et al., 2019). Current research predominantly focuses on abiotic stress-induced accumulation of soluble sugars. For instance, cold acclimation elevates levels of soluble sugars like sucrose, glucose, trehalose, and raffinose in Camellia sinensis leaves, concomitantly increasing cold tolerance (Yue, 2015). In Medicago sativa, low-temperature stress induces up-regulated expression of the Galactinol Synthase (GoLS) and improves cold tolerance in Medicago sativa (Zhuo et al., 2013). Low temperature induces enhanced SPS activity, leading to increased sucrose accumulation (Lundmark et al., 2006; Miao et al., 2007). Similarly, overexpression of ZmSUS1, a key enzyme in sugar metabolism, has been found to increase drought tolerance in Zea mays by regulating sucrose metabolism and soluble sugar content (Xiao et al., 2024). Furthermore, seaweed extract-based bio-stimulants mitigate drought stress in Saccharum officinarum by enhancing leaf metabolic activity and total sugar levels (Jacomassi et al., 2022). Therefore, soluble sugars and their metabolic pathways play a pivotal role in plant responses to abiotic stress. By regulating the synthesis and metabolism of soluble sugars, plants bolster their osmotic adjustment capacity and antioxidant defenses, thereby enhancing their overall adaptability to abiotic stress.
5 WRKY TFs are involved in plant abiotic stress responses mediated by sugar metabolism
5.1 The mechanism of WRKY TFs regulate sugar metabolism
WRKY TFs regulate plant sugar metabolism through multiple distinct pathways. First, WRKY TFs directly bind to W-box cis-elements within the promoters of sugar-metabolic genes, enabling their transcriptional regulation (Chen et al., 2019). Second, WRKY TFs indirectly modulate sugar metabolism by integrating into intricate plant signaling networks. These transcription factors are activated by diverse signals, including plant hormones, biotic stresses, and abiotic stresses, subsequently modulating sugar metabolic pathways via signaling cascades. Furthermore, WRKY TFs can form complexes with other proteins, achieving cooperative regulation of sugar metabolism through cross-family collaboration with other transcription factor families or interaction with epigenetic regulators, integrating signals and expanding the target range (Li et al., 2025). Here, this paper concludes the modes of action of WRKY TFs involved in the regulation of plant sugar metabolism and classifies them into three types: direct regulation, indirect regulation, and cooperative regulation (Figure 3) (Li et al., 2025). Elucidating the regulatory mechanisms of WRKY TFs in sugar metabolism will deepen our understanding of the intricate relationship between WRKY TFs and sugar metabolism.
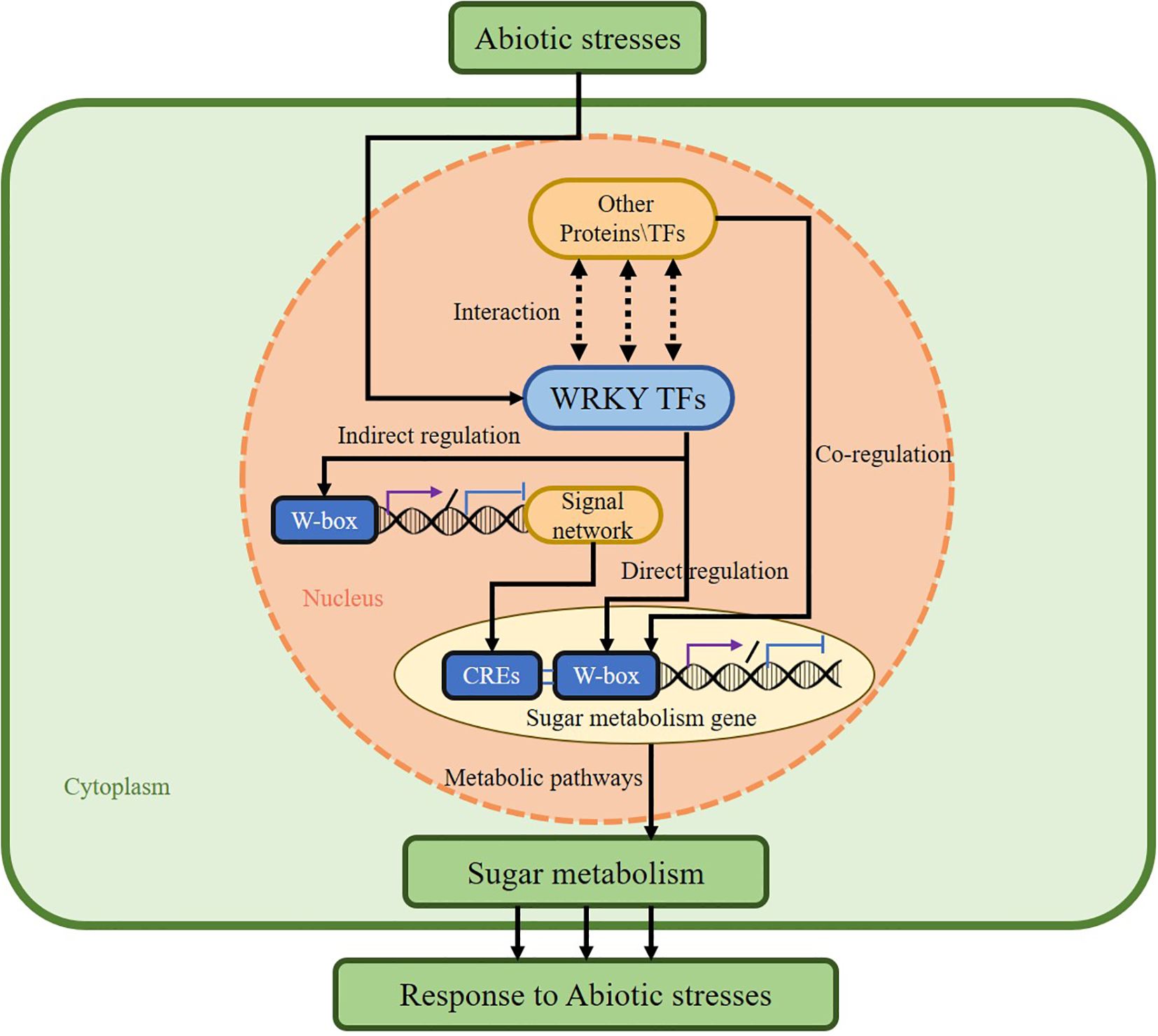
Figure 3. The mechanism by which WRKY TFs regulate glucose metabolism and mediate abiotic stress. We categorized the modes of regulation of WRKY transcription factors in plant sugar metabolism into three different types: Direct regulation, Indirect regulation, and Cooperative regulation.
5.1.1 Direct regulation
WRKY TFs directly regulate the expression of genes encoding sugar-metabolic enzymes by binding to cis-regulatory elements (e.g., W-box or CRT/DRE motifs) within their promoters. This modulates sugar biosynthesis, degradation, and transport, ultimately enhancing plant stress tolerance. For example, SUSIBA2, a WRKY-like transcription factor isolated from barley, binds not merely to the W-box on the ISO1 (isoamylase) promoter (Sun et al., 2003), and also to SURE (Sugar Metabolism Cis-Acting Element), thereby regulating starch synthesis. MdWRKY126 in Malus dasyphylla enhances the activity of the SPS enzyme by directly binding to the promoter region of the SPS gene and upregulating its expression level (Zhang et al., 2025). In Oryza sativa, OsWRKY71 binds to W-box sequences in the promoter of α-amylase genes, repressing gibberellin-induced expression of Amy32b (Zhang et al., 2004). Furthermore, AtWRKY18 and AtWRKY53 in Arabidopsis directly couple to the promoters of sugar-responsive genes and trigger their expression upon glucose treatment (Chen et al., 2019). In Pitaya (Hylocereus), WRKY TFs (e.g., HpWRKY3, HpWRKY18, and HpWRKY44) are associated with up-regulated expression of genes involved in betalain biosynthesis and sugar metabolism (HpCytP450-like1, HpSS2, and HpAI2) (Cheng, 2018).Similarly, HpWRKY3 activates the expression of HpINV2 and HpSuSy1, suggesting it may directly target their promoters to modulate transcription and regulate sucrose metabolism (Cheng, 2018; Wei et al., 2019).
5.1.2 Indirect regulation
Plant sugar metabolism exhibits intricate cross-talk with hormone signaling path-ways, including ABA, JA, and SA. WRKY TFs frequently serve as pivotal integrators within hormone signal transduction networks to indirectly modulate sugar metabolism. For instance, in Camellia sinensis, CsWRKY29 binds to the promoter of CsABI5, an ABA signaling component harboring a W-box element, and activates its expression. Subsequently, as a downstream regulator, CsABI5 binds to ABREs (ABA-responsive elements) within the promoters of CsHXK1 and CsSUS4 (Xue et al., 2024). This establishes a “CsWRKY29-CsABI5-HXK1/SUS4” regulatory cascade, mediating indirect control of sugar metabolism (Xue et al., 2024). In Oryza sativa, OsWRKY5 functions as a negative regulatory hub in the ABA pathway; it indirectly suppresses the activation of sugar metabolism-related genes by re-pressing OsMYB2 expression. Following OsWRKY5 knockout, the repression on the ABA signaling pathway is lifted, activating sugar metabolic pathways and leading to a significant increase in soluble sugar content (Lim et al., 2021). Beyond ABA signaling, Vitis vinifera VviWRKY10 and VviWRKY30 modulate the expression of sugar metabolism-related genes by engaging with SA and JA signaling pathways. VviWRKY10 primarily responds to SA signals, upregulating SUS and sucrose transporter (SUT) gene expression, thereby promoting sucrose accumulation to enhance osmotic adjustment capacity (Zhou et al., 2024). In contrast, VviWRKY30 acts through the JA signaling pathway to repress glycolysis, reducing glucose consumption and thereby prioritizing carbon allocation toward defense-related metabolism (Zhou et al., 2024). By integrating SA and JA signals, VviWRKY10 and VviWRKY30 cooperatively regulate powdery mildew resistance and sugar metabolism partitioning in grapevine (Zhou et al., 2024). This “bidirectional regulatory” mode exemplifies the finely tuned balance WRKY factors achieve between sugar metabolism and stress resilience (Zhou et al., 2024).
5.1.3 Cooperative regulation
WRKY transcription factors achieve coordinated regulation of sugar metabolism at multiple levels by forming complexes or interactions networks with other families of transcription factors or other proteins (Li et al., 2025). For instance, in Glycine max, GmWRKY27 assembles into a complex with MYB-family transcription factor GmMYB174, co-repressing the expression of NAC-family factor GmNAC29 to attenuate ABA biosynthesis while enhancing sucrose transporter GmSWEET15 expression (Li et al., 2025). In Vitis vinifera, VvWRKY22 interacts with sucrose non-fermenting-1-related kinases (VvSnRK1.1/VvSnRK1.2) to form a regulatory complex that phosphorylates downstream targets (VvTPP, VvHXK), thereby modulating glucose accumulation during cold stress (Huang et al., 2021). Furthermore, in Prunus persica, PpWRKY40 physically associates with NPR1 protein to activate PpPRs gene expression, while concurrently upregulating sucrose synthase (PpSS1) and sucrose phosphate synthase (PpSPS3) genes (Li et al., 2021). The WRKY structural domain of PoWRKY69 binds directly to the VQ motif of PoVQ11 to form a stable transcriptional regulatory complex; the PoWRKY69-PoVQ11 module shifts the carbon flow from glycolysis to fructose synthesis through the activation of PoFBA5 (fructose-1,6-bisphosphate aldolase gene), while inhibiting the sucrose synthase (SUS) activity and decreasing the sucrose consumption. thereby specifically elevating fructose levels (Luan et al., 2024).
5.2 The WRKY TFs respond to abiotic stress by regulating the glucose metabolism pathway
WRKY TFs, as an influential class of transcriptional regulators in plants, play a crucial role in plant responses to various environmental stresses. Recent studies have shown that the deep involvement of WRKY transcription factors in plant stress response is largely realized through the precise regulation of multiple pathways of sugar metabolism.
5.2.1 Drought stress
Drought stress seriously threatens plant growth, development and viability. Under water deficit conditions, water absorption by the plant root system is blocked and leaf stomata are closed to reduce water loss. At the same time, cell dehydration destroys membrane structural integrity and reactive oxygen species (ROS) accumulate in large quantities, accelerating cellular senescence and even cell death. WRKY transcription factors are able to target sugar metabolism-related genes through different pathways, coordinate sugar transport, synthesis and utilization, and enhance osmoprotection and ROS scavenging, thus improving plant drought resistance. For instance, in Paeonia ostii, the PoWRKY69-PoVQ11 transcription complex directly activates the key sugar metabolism gene PoFBA5 (fructose-1,6-bisphosphate aldolase), promoting efficient fructose accumulation via its reverse catalytic function (Luan et al., 2024). The accumulated fructose exerts dual core roles: as an osmolyte to reduce cellular osmotic potential and maintain water balance, and by activating the antioxidant enzyme system while directly quenching ROS to mitigate membrane lipid peroxidation damage (Luan et al., 2024). This physiologic defense network mediated by sugar metabolism ultimately confers significant enhancement of plant drought tolerance, confirming that PoFBA5 serves as an indispensable metabolic hub converting WRKY transcriptional regulation into drought-resistant phenotypes (Luan et al., 2024). In Oryza sativa, OsWRKY11 acts as a molecular switch, specifically activating the expression of the raffinose synthase gene to promote synthesis of raffinose precursors, leading to specific raffinose accumulation in leaves (Wu et al., 2009). The accumulated raffinose functions as an efficient osmolyte to significantly decrease cellular osmotic potential, maintain cell turgor and water balance, and mitigate oxidative damage by inhibiting ROS burst. This osmotic-antioxidant collaborative network mediated by sugar metabolism translates WRKY transcriptional regulation signals into cellular homeostasis protection, ultimately endowing plants with drought-resistant phenotypes.Under dehydration stress, the ABA signaling pathway rapidly activates BhWRKY1 transcription factor in Boea hygrometrica (Wang et al., 2009). This factor precisely recognizes and binds to the W-box cis-element in the BhGolS1 promoter, directly driving BhGolS1 transcription. With significantly enhanced BhGolS1 activity, massive galactinol accumulates, triggering synthesis of raffinose and stachyose, which together form key osmoprotectants (RFOs) (Wang et al., 2009). These RFOs exert drought resistance through multiple mechanisms: decreasing cellular osmotic potential and activating antioxidant enzyme systems, while promoting hydrogen bond interactions with phospholipid bilayers and membrane proteins to effectively resist dehydration-induced cell damage. Phenotypically, this results in improved cell viability, enhanced membrane structural integrity, and significantly optimized plant recovery capacity after rewatering. However, the regulatory mechanisms of WRKY transcription factors in response to drought stress show remarkable diversity. In addition to positively mediated mechanisms, some members negatively regulate sugar metabolism through distinct molecular pathways. Under drought stress, activation of AtWRKY53 specifically binds to the promoter W-box elements of the starch-degrading gene QQS and the antioxidant genes CAT2/CAT3, initiating reprogramming of sugar metabolism. Overexpression of QQS accelerates the hydrolysis of starch to soluble sugars, which promotes the accumulation of malic acid via the glycolytic pathway (Sun and Yu, 2015). Malic acid, as an osmotic substance, acted synergistically with potassium ions to regulate the osmotic potential and expansion pressure of defense cells, while the attenuation of ROS signaling blocked the stomatal closure cascade, which ultimately led to an abnormal increase in stomatal conductance, an increase in transpirational loss of water, and a significant reduction in drought tolerance. This coexistence of positive and negative regulatory mechanisms reveals that the sugar metabolism pathway, as a core effector, plays a key pivotal role in connecting the molecular transcriptional network with drought stress response under the differential regulation of WRKY transcription factors.
5.2.2 Salt stress
Salt stress on plants mainly manifests the triple effects of osmotic stress, ionic toxicity and oxidative damage. High-salt environments lead to elevated soil osmotic pressure, which prevents water uptake by the root system and causes drought-like physiological dehydration. Under salt stress-induced ion toxicity and osmotic stress, WRKY TFs alleviate osmotic imbalance by triggering starch-to-soluble sugar conversion. They orchestrate synergistic coordination between sugar signaling and ion transport systems (e.g., Na+/H+ antiporters), thereby reinforcing membrane integrity. Concurrently, sugar-derived metabolites modulate antioxidant defenses to mitigate oxidative injury. For instance, under salt stress, VvWRKY30 drives remodeling of sugar metabolism by specifically activating sugar transport genes (VvHT1/5) and metabolic genes (VvSS, VvHXK, VvTRE). On one hand, it promotes the accumulation of glucose, fructose, and trehalose as osmotic regulators to lower cellular osmotic potential and maintain turgor pressure. On the other hand, it enhances the pentose phosphate pathway to supply NADPH, which boosts the activities of antioxidant enzymes like superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) – effectively scavenging H2O2 and reducing membrane lipid peroxidation (Cao et al., 2025b). Additionally, HXK-mediated sugar signaling amplifies stress responses in a cascading manner, forming a positive feedback loop with ethylene signaling. This ultimately maintains photosynthetic function, safeguards reproductive development, and reduces biomass loss at the phenotypic level, achieving an integrated salt-tolerance mechanism from transcriptional regulation to physiological adaptation (Zhu et al., 2019). In Rosa rugosa, RrWRKY1 maintains cellular osmotic balance by regulating proline accumulation under salt stress (Zang et al., 2024). As an intermediate metabolite in sugar metabolism, proline is generated from glutamate via the glycolytic pathway. Experiments show that silencing RrWRKY1 leads to a significant decrease in proline content and an increase in malondialdehyde (MDA) content, indicating that this transcription factor enhances antioxidant capacity through sugar metabolism-related pathways to alleviate oxidative damage caused by salt stress (Zang et al., 2024). In conclusion, WRKY transcription factors regulate sugar metabolic pathways to not only cope with osmotic stress and ion toxicity from salt stress but also enhance plant antioxidant defense capabilities. This establishes multilayered salt-tolerance mechanisms spanning from gene expression regulation to physiological function adaptation, providing crucial molecular guarantees for plant survival in high-salt environments.
5.2.3 Cold stress
WRKY transcription factors play a central role in plant responses to low-temperature stress by dynamically regulating sugar metabolic pathways. Under low-temperature conditions, which disrupt membrane fluidity and inhibit photosynthesis, plants maintain osmotic balance and energy supply through the accumulation of soluble sugars. Members of the WRKY family activate the expression of key enzymes such as amylase and sucrose synthase, thereby promoting sugar accumulation and enhancing cold tolerance. Take the cold-inducible WRKY transcription factor CdWRKY2 as an example (Huang et al., 2022b). It directly binds to the W-box elements in the promoter regions of the sucrose phosphate synthase gene (CdSPS1) and the CBF1 gene, activating their transcriptional expression (Huang et al., 2022b). The product of CdSPS1, serving as both an osmolyte and a signaling molecule, exerts dual regulatory functions: it enhances cellular osmotic homeostasis by accumulating sucrose, which lowers the freezing point and maintains membrane integrity; meanwhile, it activates the pentose phosphate pathway to generate NADPH, thereby improving cellular antioxidant capacity. Additionally, CdWRKY2 collaborates with the core gene CdCBF1 of the CBF signaling pathway to coordinately regulate the expression of downstream cold-responsive genes. The synergistic action of these two regulatory pathways ultimately enhances the tolerance of transgenic Arabidopsis to low-temperature stress significantly. In Raphanus sativus, the RsWRKY40 transcription factor acts as a central regulatory hub to coordinate cold resistance mechanisms through a dual-function mode (Chen et al., 2025). It not only directly activates the sucrose phosphate synthase gene RsSPS1 to promote sucrose accumulation for osmotic protection and energy supply but also simultaneously induces the CBF signaling pathway to activate downstream antifreeze genes. Sucrose serves as a critical bridging molecule in this process: it functions as a protective metabolite to maintain membrane stability and reactive oxygen species (ROS) scavenging, while also reinforcing the CBF pathway and its own synthesis through feedback regulation. Research in Camellia sinensis has shown that CsWRKY29 activates sugar metabolic genes such as sucrose phosphate synthase (CsSPS1) and hexokinase (CsHXK1) under low temperature, promoting the synthesis and accumulation of sucrose and hexoses (Xue et al., 2024). This transcription factor orchestrates two parallel processes: one involves activating sucrose degradation and glycolysis to ensure adenosine triphosphate (ATP) supply, and the other promotes the synthesis of osmoprotective oligosaccharides (trehalose, raffinose) and flavonoid glycosides. The former maintains cellular osmotic balance and membrane stability, while the latter enhances antioxidant activity through glycosylation modification. Furthermore, CsWRKY29 strengthens the expression of sugar metabolic genes via the abscisic acid (ABA) signaling pathway, forming a “ABA-CsWRKY29-Sugar metabolism” positive feedback loop (Xue et al., 2024). It also synergistically activates the CBF-COR pathway, integrating sugar metabolism with antifreeze protein synthesis to achieve enhanced freezing tolerance through multi-pathway coordination. Beyond endogenous regulatory mechanisms, WRKY transcription factors may also mediate low-temperature stress responses through exogenous sugar application. For instance, exogenous sucrose supplementation compensates for the insufficient sucrose synthesis caused by RsWRKY40 silencing, indirectly demonstrating that exogenous sugars alleviate cold damage by regulating the RsWRKY40-mediated sugar metabolic network (Chen et al., 2025). In Cucumis sativus, treatment with exogenous trehalose significantly upregulates WRKY gene expression, induces soluble sugar synthesis, and thereby mitigates cold injury (Pan et al., 2022).
In summary, WRKY transcription factors can precisely regulate the metabolism of sugars by utilizing the “WRKY - Sugar Metabolism” module in order to participate in plant stress response, and this module plays a key role in drought, salt, low temperature, and a variety of abiotic stresses, which provides an important survival and reproduction of plants in harsh natural environments. However, significant gaps remain in understanding WRKY-mediated thermotolerance (Zhang et al., 2024b). Limited studies suggest WRKYs participate in heat stress by regulating anti-oxidant systems and membrane stability, but whether this involves sugar metabolism remains elusive, presenting a key avenue for future research (Dang et al., 2013).
6 Conclusions and perspectives
WRKY transcription factors are ubiquitously distributed across the plant kingdom and play pivotal roles in regulating plant growth, developmental programs, and stress-responsive mechanisms. In recent years, a growing body of research has focused on the mediation of abiotic stress by WRKY transcription factors through sugar metabolic pathways, with remarkable advancements achieved in research methodologies. Notwithstanding these progresses, this field still harbors significant prospects and unresolved research gaps that warrant systematic exploration.
First and foremost, future investigations should endeavor to elucidate in greater detail the specific molecular mechanisms underlying the interaction between WRKY transcription factors and sugar metabolic pathways. Although existing evidence has established that WRKY transcription factors modulate the expression of sugar metabolism-related genes, the intricate regulatory networks and precise target sites thereof remain incompletely characterized. To address this, future studies are expected to employ cutting-edge technical approaches, including gene-editing technologies (particularly multi-gene editing to circumvent functional redundancy) (Cao et al., 2025a), high-resolution protein-protein interaction analyses (such as in vivo co-immunoprecipitation, yeast two-hybrid library screening, and proximity labeling techniques) (Cao et al., 2024), and single-cell omics. These methodologies will facilitate the precise dissection of how WRKY proteins recognize and bind to the promoter regions of downstream sugar metabolism genes, as well as how their complexes with other transcription factors or coregulatory molecules exert fine-scale regulation over the activity of key enzymes, thereby influencing the dynamic homeostasis of critical sugar molecules.
Second, it is imperative to resolve the long-standing challenges of functional redundancy and specificity within the WRKY gene family. Comprising a large repertoire of members, the WRKY family often exhibits extensive functional redundancy or overlap, rendering traditional genetic approaches inadequate for accurately evaluating the contribution of individual members in sugar metabolism-stress response cascades. Moreover, distinct WRKY members may exhibit context-dependent functions under varying stress conditions, in different tissues/organs, or at specific developmental stages. Future research should integrate systems biology approaches with conditional gene-editing/inducible expression systems to meticulously dissect the specific roles of different WRKY members in regulating sugar metabolism under defined environmental and physiological contexts, along with the underlying molecular determinants.
Furthermore, more direct physiological and metabolic evidence is required to establish the causal relationship between WRKY-mediated sugar metabolism and enhanced stress tolerance. Current investigations predominantly focus on molecular-level analyses or terminal phenotypic observations, whereas the evidentiary chain for intermediate links remains fragmented. Specifically, following the regulation of specific sugar metabolism genes by WRKY, how do changes in sugar composition, concentration, spatiotemporal distribution, and cellular energy status directly impact key physiological processes, including stress signal perception, reactive oxygen species scavenging, osmotic adjustment, and maintenance of cell membrane integrity? Future studies should integrate metabolomics, enzyme activity assays, subcellular localization analyses, and live-cell imaging technologies to track, at high spatiotemporal resolution, how WRKY-mediated reprogramming of sugar metabolism translates into specific physiological and biochemical responses that underpin plant stress resilience.
In conclusion, the field of WRKY transcription factors mediating abiotic stress through sugar metabolic pathways presents substantial scope for advancement. Future research should prioritize the in-depth investigation of regulatory mechanisms, expansion of research frontiers, and comprehensive consideration of their roles across multiple metabolic networks, thereby fostering a holistic understanding of WRKY transcription factors in plant stress adaptation. Such endeavors are critical for the breeding of stress-tolerant crop varieties and the promotion of sustainable development in China’s forestry and agricultural sectors.
Author contributions
XZ: Visualization, Methodology, Formal analysis, Writing – original draft, Writing – review & editing, Software. WL: Visualization, Investigation, Writing – review & editing, Software. YY: Data curation, Investigation, Validation, Writing – review & editing. JZ: Validation, Data curation, Writing – review & editing, Investigation. JL: Methodology, Writing – review & editing, Supervision. XT: Writing – review & editing, Methodology, Supervision. LW: Conceptualization, Funding acquisition, Writing – review & editing, Project administration, Resources.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China for Young Scholars (grant number 32301640).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1646357/full#supplementary-material
References
Agarwal, P., Reddy, M. P., and Chikara, J. (2011). WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 38, 3883–3896. doi: 10.1007/s11033-010-0504-5
Bao, Y., Zou, Y., An, X., Liao, Y., Dai, L., Liu, L., et al. (2024). Overexpression of a Ramie (Boehmaeria nivea L. Gaud) Group I WRKY Gene, BnWRKY49, Increases Drought Resistance in Arabidopsis thaliana. Plants-Basel 13 (3), 379. doi: 10.3390/plants13030379
Cao, Y., Feng, X., Ding, B., Huo, H., Abdullah, M., Hong, J., et al. (2025a). Gap-free genome assemblies of two Pyrus bretschneideri cultivars and GWAS analyses identify a CCCH zinc finger protein as a key regulator of stone cell formation in pear fruit. Plant Commun. 6 (3), 101238. doi: 10.1016/j.xplc.2024.101238
Cao, Y., Hong, J., Wang, H., Lin, M., Cai, Y., Liao, L., et al. (2025b). Beyond glycolysis: multifunctional roles of glyceraldehyde-3-phosphate dehydrogenases in plants. Horticulture Res. 6, 6. doi: 10.1093/hr/uhaf070
Cao, Y., Hong, J., Yun, Z., Li, X., Feng, X., Wang, H., et al. (2024). De novo gene integration into regulatory networks via interaction with conserved genes in peach. Horticulture Res. 11 (12), 12. doi: 10.1093/hr/uhae252
Cao, Y., Yang, M., Li, X., Zhou, Z., Wang, X., and Bai, J. G. (2014). Exogenous sucrose increases chilling tolerance in cucumber seedlings by modulating antioxidant enzyme activity and regulating proline and soluble sugar contents. Scientia Hortic. 179, 67–77. doi: 10.1016/j.scienta.2014.09.016
Chen, Q., Xu, X., Xu, D., Zhang, H., and Li, G. (2019). WRKY18 and WRKY53 coordinate with HISTONE ACETYLTRANSFERASE1 to regulate rapid responses to sugar. Plant Physiol. 180, 2212–2226. doi: 10.1104/pp.19.00511
Chen, S., Xu, L., Wang, Y., Mao, B., Zhang, X., Song, Q., et al. (2025). RsWRKY40 coordinates the cold stress response by integrating RsSPS1-mediated sucrose accumulation and the CBF-dependent pathway in radish (Raphanus sativus L.). Mol. Horticulture 5 (1), 14. doi: 10.1186/s43897-024-00135-x
Cheng, M. N. (2018). Molecular mechanism of WRKY transcription factors regulating betalain synthesis and sugar metabolism in pitaya fruit. Guangzhou, China: South China Agricultural University.
Crispim, J. G., Souza, E. D. S., Antunes, M. F. K., Liu, H., Pandolfi, V., Morais, M. B. D., et al. (2023). Expression of cowpea vuWRKY21 and vuWRKY87 genes in arabidopsis thaliana confers plant tolerance to salt stress. DNA 3, 18. doi: 10.3390/dna3040014
Dai, X., Wang, Y., and Zhang, W. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Dang, F., Wang, Y., Yu, L., Eulgem, T., Lai, Y., Liu, Z., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Glöckner, G., Eichinger, L., Szafranski, K., Pachebat, J. A., Bankier, A. T., Dear, P. H., et al. (2002). Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 418, 79–85. doi: 10.1038/nature00847
Govardhana, M. and Kumudini, B. (2020). In-silico analysis of cucumber (Cucumis sativus L.) Genome for WRKY transcription factors and cis-acting elements. Comput. Biol. Chem. 85, 107212. doi: 10.1016/j.compbiolchem.2020.107212
Goyal, P., Manzoor, M., Vishwakarma, R., Sharma, D., Dhar, M., and Gupta, S. (2020). A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in glycyrrhiza glabra. Sci. Rep. 10 (1), 373. doi: 10.1038/s41598-019-57232-x
Guo, J. J., Yin, L. H., Xiao, Q., Xia, E. H., and Tong, W. (2024). Cloning and cold-resistant function of WRKY19 gene from tea plant. J. Tea Sci. 65, 20–27.
He, G., Saleem, M., Deng, T., Zhong, Z., He, T., and Wu, J. (2023). Unraveling the mechanism of stWRKY6 in potato (Solanum tuberosum)’s cadmium tolerance for ensuring food safety. Foods 12 (12), 2303. doi: 10.3390/foods12122303
Hincha, D. K., Ellen, Z., Hellwege, E. M., and Heyer, A. G. (2002). Specific effects of fructo- and gluco-oligosaccharides in the preservation of liposomes during drying. Glycobiology 2, 103–110. doi: 10.1093/glycob/12.2.103
Hrmova, M. and Hussain, S. S. (2021). Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 22 (11), 5662. doi: 10.3390/ijms22115662
Hu, J., Wu, W., Cao, Z., Wen, J., Shu, Q., and Fu, S. (2016). Morphological, physiological and biochemical responses of Camellia oleifera to low-temperature stress. Pakistan J. Bot. 48, 899–905.
Huang, X., Cao, L., Fan, J., Ma, G., and Chen, L. (2022b). CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in Bermudagrass. Plant Biotechnol. J. 23 (19), 660–675. doi: 10.1111/pbi.13745
Huang, J., Liu, F., Chao, D., Xin, B., Liu, K., Cao, S., et al. (2022a). The WRKY transcription factor osWRKY54 is involved in salt tolerance in rice. Int. J. Mol. Sci. 23 (19), 11999. doi: 10.3390/ijms231911999
Huang, T., Yu, D., and Wang, X. (2021). VvWRKY22 transcription factor interacts with VvSnRK1.1/VvSnRK1.2 and regulates sugar accumulation in grape. Biochem. Biophys. Res. Commun. 554, 193–198. doi: 10.1016/j.bbrc.2021.03.092
Ishiguro, S. and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. MGG 244, 563–571. doi: 10.1007/BF00282746
Jacomassi, L. M., Viveiros, J. O., Oliveira, M. P., et al. (2022). A seaweed extract-based biostimulant mitigates drought stress in sugarcane. Front. Plant Sci. 13, 865291. doi: 10.3389/fpls.2022.865291
Jia, Z., Li, M., Wang, H., Zhu, B., and Ren, M. (2021). TaWRKY70 positively regulates TaCAT5 enhanced Cd tolerance in transgenic Arabidopsis. Environ. Exp. Bot. 190, 104591. doi: 10.1016/j.envexpbot.2021.104591
Khoso, M., Hussain, A., Ritonga, F., Ali, Q., Channa, M., Alshegaihi, R., et al. (2022). WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1039329
Kim, C. Y., Lee, S. H., Park, H. C., Bae, C. G., Cheong, Y. H., Choi, Y. J., et al. (2000). Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant-Microbe Interact. 13, 470–474. doi: 10.1094/MPMI.2000.13.4.470
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, D., Gu, B., Huang, C., Shen, J., Wang, X., Guo, J., et al. (2023). Functional study of amorpha fruticosa WRKY20 gene in response to drought stress. Int. J. Mol. Sci. 24 (15), 12231. doi: 10.3390/ijms241512231
Li, M., Shao, Y., Pan, B., Liu, C., and Tan, H. (2025). Regulation of important natural products biosynthesis by WRKY transcription factors in plants. J. Advanced Res. doi: 10.1016/j.jare.2025.01.009
Li, C., Wang, K., Xu, F., Lei, C., Jiang, Y., and Zheng, Y. (2021). Sucrose metabolism and sensory evaluation in peach as influenced by β-aminobutyric acid (BABA)-induced disease resistance and the transcriptional mechanism involved. Postharvest Biol. Technol. 174, 111465. doi: 10.1016/j.postharvbio.2021.111465
Li, H., Xu, Y., Xiao, Y., Zhu, Z., Xie, X., Zhao, H., et al. (2010). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 232, 1325–1337. doi: 10.1007/s00425-010-1258-y
Lim, C., Kang, K., Shim, Y., Yoo, S. C., and Paek, N. C. (2021). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 188 (4), 1900–1916. doi: 10.1093/plphys/kiab492
Lin, H., Bai, L., Wei, W., Su, W., Wu, Y., Wu, R., et al. (2024). The role of maWRKY70 in regulating lipoxygenase gene transcription during chilling injury development in banana fruit. Foods 13, 14. doi: 10.3390/foods13060854
Liu, W., Wang, T., Wang, Y., Liang, X., Han, J., Hou, R., et al. (2023). The transcription factor mbWRKY46 in malus baccata (L.) borkh mediate cold and drought stress responses. Int. J. Mol. Sci. 24 (15), 12468. doi: 10.3390/ijms241512468
Lu, X. T., Yin, F. L., Liu, C. Y., Liang, Y. L., Liu, Y. F., and Shuai, L. (2025). Bioinformatics and expression analysis of the cucumber low-temperature stress-responsive gene WRKY51. Mol. Plant Breed. 1–19.
Luan, Y., Chen, Z., Fang, Z., Meng, J., Tao, J., and Zhao, D. (2024). PoWRKY69-PoVQ11 module positively regulates drought tolerance by accumulating fructose in Paeonia ostii. Plant J. 119, 18. doi: 10.1111/tpj.16884
Lundmark, M., Cavaco, A. M., Trevanion, S., and Hurry, V. (2006). Carbon partitioning and export in transgenic Arabidopsis thaliana with altered capacity for sucrose synthesis grown at low temperature: a role for metabolite transporters. Plant Cell Environ. 29 (9), 1703–1714. doi: 10.1111/j.1365-3040.2006.01543.x
Ma, Z. and Hu, L. (2024). WRKY transcription factor responses and tolerance to abiotic stresses in plants. Int. J. Mol. Sci. 25, 18. doi: 10.3390/ijms25136845
Ma, Q., Xia, Z., Cai, Z., Li, L., Cheng, Y., Liu, J., et al. (2019). GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in arabidopsis thaliana. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01979
Mangelsen, E., Kilian, J., Berendzen, K. W., Kolukisaoglu, Ü.H., Harter, K., Jansson, C., et al. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9, 194. doi: 10.1186/1471-2164-9-194
Miao, M., Xu, X., Chen, X., Xue, L., and Cao, B. (2007). Cucumber carbohydrate metabolism and translocation under chilling night temperature. J. Plant Physiol. 164, 621–628. doi: 10.1016/j.jplph.2006.02.005
Naoki, Y., Yuko, S., Shigeru, T., Tetsuya, C., Takafumi, S., Kazunori, O., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64 (16), 5085–5097. doi: 10.1093/jxb/ert298
Narusaka, M., Toyoda, K., Shiraishi, T., Iuchi, S., Takano, Y., Shirasu, K., et al. (2016). Leucine zipper motif in RRS1 is crucial for the regulation of Arabidopsis dual resistance protein complex RPS4/RRS1. Sci. Rep. 6, 18702. doi: 10.1038/srep18702
Ouyang, L., Leus, L., De Keyser, E., and Van Labeke, M. C. (2019). Seasonal changes in cold hardiness and carbohydrate metabolism in four garden rose cultivars. J. Plant Physiol. 232, 188–199. doi: 10.1016/j.jplph.2018.12.001
Pan, Y. J., Zhang, Y., Wu, Q. M., and L, Z. Q. (2022). Research progress of WRKY on sugar regulation of cold adaptation in horticultural crops. Biotechnol. Bull. 38 (03), 203–212. doi: 10.13560/j.cnki.biotech.bull.1985.2021-0691
Qin, X., Li, K., and Duan, Z. (2018). Research progress on plant cold stress signals. Mol. Plant Breed. 16 (21), 7187–7194. doi: 10.13271/j.mpb.016.007187
Qu, X., Wang, H., Chen, M., Liao, J., Yuan, J., and Niu, G. (2019). Drought stress–induced physiological and metabolic changes in leaves of two oil tea cultivars. J. Am. Soc. Hortic. Sci. 144, 439–447. doi: 10.21273/JASHS04775-19
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, D., Tripathi, P., Rabara, R., Lin, J., Ringler, P., Boken, A., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Savitch, L. V., Harney, T., and Huner, N. P. A. (2010). Sucrose metabolism in spring and winter wheat in response to high irradiance, cold stress and cold acclimation. Physiologia Plantarum 108, 270–278. doi: 10.1034/j.1399-3054.2000.108003270.x
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shangguan, X., Tian, Z., Wang, Y., Xiao, T., Yu, X., Jing, W., et al. (2024). Transcription factor OsWRKY72 is involved in Cu/Cd toxicity by regulating lignin synthesis in rice. Crop J. 12, 1471–1482. doi: 10.1016/j.cj.2024.09.002
Shi, G., Liu, G., Liu, H., Xu, N., Yang, Q., Song, Z., et al. (2023). WRKY Transcriptional Factor IlWRKY70 from Iris laevigata Enhances Drought and Salinity Tolerances in Nicotiana tabacum. Int. J. Mol. Sci. 24 (22), 16174. doi: 10.3390/ijms242216174
Smeekens, S., Ma, J., Hanson, J., and Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 273–278. doi: 10.1016/j.pbi.2009.12.002
Su, W. J., Cao, R. L., Zhou, Z. L., Zhao, N. H., Zhang, Y. Y., Hu, D. N., et al. (2023). Identification and expression analysis of the WRKY gene family in Camellia oleifera. J. Cent. South Univ. Forestry Technol. 43 (03), 155–166+174. doi: 10.14067/j.cnki.1673-923x.2023.03.017
Sun, Y. M., Liu, L. J., Feng, M. F., Wang, J. H., Cang, J., Li, S., et al. (2015). Research progress on sugar metabolism in plants under low temperature stress. J. Northeast Agric. Univ. 46 (07), 95–102+108.
Sun, C. X., Palmqvist, S., Olsson, H., Borén., M., and Jansson, A. C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092. doi: 10.1105/tpc.014597
Sun, Y. D. and Yu, D. Q. (2015). Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 34 (8), 1295–1306. doi: 10.1007/s00299-015-1787-8
Turhan, E. and Ergin, S. (2012). Soluble sugars and sucrose-metabolizing enzymes related to cold acclimation of sweet cherry cultivars grafted on different rootstocks. Sci. World J. 2012, 979682. doi: 10.1100/2012/979682
ülker, B. and Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
Wang, D., Chen, Q., Chen, W., Liu, X., Xia, Y., Guo, Q., et al. (2021). A WRKY transcription factor, ejWRKY17, from eriobotrya japonica enhances drought tolerance in transgenic arabidopsis. Int. J. Mol. Sci. 22 (11), 5593. doi: 10.3390/ijms22115593
Wang, M., Huang, Q., Lin, P., Zeng, Q., Li, Y., Liu, Q., et al. (2020). The overexpression of a transcription factor gene vbWRKY32 enhances the cold tolerance in verbena bonariensis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01746
Wang, Y., Li, M., Mu, Y., Guan, L., Wu, F., Liu, K., et al. (2024). PwuWRKY48 confers drought tolerance in populus wulianensis. Forests 15(2), 302. doi: 10.3390/f15020302
Wang, C., Ru, J., Liu, Y., Yang, J., Li, M., Xu, Z., et al. (2018). The maize WRKY transcription factor zmWRKY40 confers drought resistance in transgenic arabidopsis. Int. J. Mol. Sci. 19(9), 2580. doi: 10.3390/ijms19092580
Wang, M., Vannozzi, A., Wang, G., Liang, Y. H., Tornielli, G. B., Zenoni, S., et al. (2014a). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Horticulture Res. 1, 14016. doi: 10.1038/hortres.2014.16
Wang, P. J., Yue, C., Chen, D., Zheng, Y. C., Zheng, Z. L., Lin, S., et al. (2019). Isolation and expression analysis of CsWRKY6, CsWRKY31 and CsWRKY48 genes in tea plant (Camellia sinensis). J. Zhejiang Univ. (Agriculture Life Sciences) 45, 30–38. doi: CNKI:SUN:ZJNY.0.2019-01-007
Wang, N., Zhang, Z., Huang, F., Li, H., and Zhang, S. (2014b). Research progress on the involvement of WRKY transcription factors in plant responses to abiotic stresses. J. Nucl. Agric. Sci. (10), 9. doi: 10.11869/j.issn.100-8551.2014.10.1819
Wang, Z., Zhu, Y., Wang, L., Liu, X., Liu, Y., Phillips, J., et al. (2009). A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230, 1155–1166. doi: 10.1007/s00425-009-1014-3
Wei, W., Cheng, M., Ba, L., Zeng, R., Luo, D., Qin, Y., et al. (2019). Pitaya hpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes hpINV2 and hpSuSy1. Int. J. Mol. Sci. 20 (8), 1890. doi: 10.3390/ijms20081890
Wen, C., Zhang, Z., Shi, Q., Duan, X., Du, J., Wu, C., et al. (2023). Methyl jasmonate- and salicylic acid-induced transcription factor zjWRKY18 regulates triterpenoid accumulation and salt stress tolerance in jujube. Int. J. Mol. Sci. 24, 15. doi: 10.3390/ijms24043899
Wu, X. L., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Accumulation of raffinose in rice seedlings overexpressing OsWRKY11 in relation to desiccation tolerance. Plant Biotechnol. 26, 431–434. doi: 10.5511/plantbiotechnology.26.431
Wu, Y., Wu, J., Wang, Y., and Sun, Q. (2020). Research progress on the functions of WRKY transcription factors in plant stress responses. Mol. Plant Breed. 18 (22), 10.
Xiao, N., Ma, H., Wang, W., Sun, Z., Li, P., and Xia, T. (2024). Overexpression of ZmSUS1 increased drought resistance of maize (Zea mays L.) by regulating sucrose metabolism and soluble sugar content. Planta: Int. J. Plant Biol. 259 (2), 43.
Xu, H. Y. and Xu, Z. J. (2024). Cloning and bioinformatics analysis of the low - temperature responsive CwWRKY65 gene from Camellia brevistyla in Weining. Mol. Plant Breed., 1–16.
Xue, C., Huang, X., and Zhao, Y. (2024). CsWRKY29, a key transcription factor in tea plant for freezing tolerance, ABA sensitivity, and sugar metabolism. Sci. Rep. 14 (1), 28620.
Yan, J., Li, J., Zhang, H., Liu, Y., and Zhang, A. (2022). ZmWRKY104 positively regulates salt tolerance by modulating ZmSOD4 expression in maize. Crop J. 10, 555–564. doi: 10.1016/j.cj.2021.05.010
Yang, Z., Chi, X., Guo, F., Jin, X., and Sun, B. (2020). SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 246-247, 153142. doi: 10.1016/j.jplph.2020.153142
Yang, C., Jin, M., Zhang, L., Shen, J., and Guan, Q. (2023). Drought and salinity tolerance of the ptWRKY33 gene in populus. Forests 14 (10), 2039. doi: 10.3390/f14102039
Yi, L. and Yueping, C. (2023). GmWRKY17-mediated transcriptional regulation of GmDREB1D and GmABA2 controls drought tolerance in soybean. Plant Mol. Biol. 113, 157–170. doi: 10.1007/s11103-023-01380-2
Yoon, Y., Seo, D., Shin, H., Kim, H., Kim, C., and Jang, G. (2020). The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy-Basel 10 (6), 788. doi: 10.3390/agronomy10060788
Yue, C. (2015). Mining of carbohydrate-related genes in tea plants and their expression studies in tea plant cold acclimation. Beijing, China: Chinese Academy of Agricultural Sciences.
Zang, F., Wu, Q., Li, Z., Li, L., Xie, X., Tong, B., et al. (2024). RrWRKY1, a transcription factor, is involved in the regulation of the salt stress response in rosa rugosa. Plants 13 (21), 2973. doi: 10.3390/plants13212973
Zhang, P., Chao, R., Qiu, L., Ge, W., Liang, J., and Wen, P. (2024a). ChaWRKY40 enhances drought tolerance of ‘Dawei’ Hazelnuts by positively regulating proline synthesis. Forests 13 (21), 2973. doi: 10.3390/f15030407
Zhang, J., Huang, D., Zhao, X., Zhang, M., Wang, Q., Hou, X., et al. (2022a). Drought-responsive WRKY transcription factor genes IgWRKY50 and IgWRKY32 from Iris germanica enhance drought resistance in transgenic Arabidopsis. Front. Plant Sci. 13, 983600. doi: 10.3389/fpls.2022.983600
Zhang, Y. and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Zhang, Z. L., Xie, Z., Zou, X. L., Casaretto, J., Ho, T.-H. D., and Shen, Q. J. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. doi: 10.1104/pp.103.034967
Zhang, L., Xu, Y., Luo, Z., Lyu, L., Wang, C., Zhu, L., et al. (2025). Overexpression of a transcription factor MdWRKY126 altered soluble sugar accumulation in apple and tomato fruit. Hortic. Plant J. 11, 989–998. doi: 10.1016/j.hpj.2023.09.010
Zhang, Z., Yang, C., Xi, J., Wang, Y., Guo, J., Liu, Q., et al. (2024b). The MdHSC70-MdWRKY75 module mediates basal apple thermotolerance by regulating the expression of heat shock factor genes. Plant Cell 15 (3), 407, 3631–3653. doi: 10.1093/plcell/koae171
Zhang, M. X., Zhao, R., Huang, K., Huang, S., Wang, H., Wei, Z., et al. (2022b). The OsWRKY63-OsWRKY76-OsDREB1B module regulates chilling tolerance in rice. Plant J. 112, 383–398. doi: 10.1111/tpj.15950
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhou, L., Wang, N., Gong, S., Lu, R., Li, Y., and Li, X. (2015). Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 96, 311–320. doi: 10.1016/j.plaphy.2015.08.016
Zhou, M., Wang, H., Yu, X., Cui, K., Hu, Y., Xiao, S., et al. (2024). Transcription factors VviWRKY10 and VviWRKY30 co-regulate powdery mildew resistance in grapevine. Plant Physiol. 195, 446–461. doi: 10.1093/plphys/kiae080
Zhu, D., Hou, L., Xiao, P., Guo, Y., Deyholos, M. K., and Liu, X. (2019). VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant ence, 132–142. doi: 10.1016/j.plantsci.2018.03.018
Zhu, H., Jiang, Y., Guo, Y., Huang, J., and Qiao, L. (2021). A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 160, 175–183. doi: 10.1016/j.plaphy.2021.01.014
Zhuo, C., Wang, T., Lu, S., Zhao, Y., Li, X., and Guo, Z. (2013). A cold responsive galactinol synthase gene from Medicago falcata (MfGolS1) is induced by myo-inositol and confers multiple tolerances to abiotic stresses. Physiologia Plantarum 149, 67–78. doi: 10.1111/ppl.12019
Keywords: WRKY transcription factor, sugar metabolism, abiotic stress, regulation mechanism, plant growth and development
Citation: Zhang X, Liu W, Yin Y, Zheng J, Li J, Tan X and Wu L (2025) WRKY transcription factors participate in abiotic stress responses mediated by sugar metabolism. Front. Plant Sci. 16:1646357. doi: 10.3389/fpls.2025.1646357
Received: 13 June 2025; Accepted: 14 July 2025;
Published: 07 August 2025.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Peng Sun, Chinese Academy of Forestry, ChinaHeping Cao, United States Department of Agriculture (USDA), United States
Wenjie Lu, Henan Agricultural University, China
Copyright © 2025 Zhang, Liu, Yin, Zheng, Li, Tan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: LingLi Wu, d3VsaW5nbGkwMzA3QDE2My5jb20=
 XueYi Zhang
XueYi Zhang WanXia Liu1,2,3,4,5
WanXia Liu1,2,3,4,5 LingLi Wu
LingLi Wu