- 1Research Center for Ecology and Ethnobiology, National Research and Innovation Agency, Cibinong, Bogor, Indonesia
- 2Research Center for Applied Botany, National Research and Innovation Agency, Cibinong, Bogor, Indonesia
- 3Department of Biology, Faculty of Mathematic and Natural Sciences, Universitas Indonesia, Depok, Indonesia
- 4Research Center for Biosystematics and Evolution, National Research and Innovation Agency, Cibinong, Bogor, Indonesia
- 5Herbarium Bandungense, School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia
- 6Directorate for Scientific Management Collection, National Research and Innovation Agency, Cibinong, Bogor, Indonesia
1 Introduction
Elaeodendron glaucum (Rottb.) Pers., a woody species in the family Celastraceae, is widely distributed across Sri Lanka to South East Asia Hou, 1960; (POWO, 2025). Despite its broad geographic range and presumed ecological adaptability, genomic data for this species remain scarce (Simmons et al., 2012). In recent years, chloroplast genome characterization has emerged as a powerful tool for resolving phylogenetic relationships and elucidating evolutionary patterns in angiosperms (Zhang and Ma, 2024). For example, a recent comparative analysis of complete chloroplast genomes from 13 species of the genus Celastrus successfully reconstructed a well-supported phylogenetic tree, clarified relationships within the genus, and confirmed that Celastrus forms a monophyletic group with Tripterygium as its closest sister lineage. The comparative genomic analysis pinpointed distinct variable regions suitable as molecular markers for species delimitation and demonstrated that C. tonkinensis Pit. and C. hindsii Benth. are conspecific (Liu et al., 2024).
Chloroplast genomes are particularly valuable due to their highly conserved structure, maternal inheritance, and moderate mutation rates, which make them effective molecular markers for species identification, comparative genomics, and systematic classification (Daniell et al., 2016; Nadeem et al., 2018). In this context, the complete sequencing and structural annotation of the E. glaucum chloroplast genome addresses a notable gap in genomic resources and offers a critical foundation for improving taxonomic resolution within the Celastraceae (Coughenour et al., 2010). This research is especially relevant given the persistent taxonomic ambiguities within the genus Elaeodendron Jacq., which have been confounded by morphological convergence and a lack of comprehensive molecular data (Simmons et al., 2012). By generating and analyzing the complete chloroplast genome of E. glaucum, the present study provides essential data to clarify phylogenetic relationships both within the genus and among closely related taxa. Comparative analyses with other chloroplast genomes may further reveal species-specific structural variations, genomic signatures, and adaptive traits, thereby contributing to a more refined and reliable classification framework for the family.
The chloroplast genome in most land plants is characterized by a conserved quadripartite circular structure comprising a large single-copy (LSC) region, a small single-copy (SSC) region, and two inverted repeat (IR) regions (Dobrogojski et al., 2020). However, structural exceptions such as the loss of IRs or SSCs have been reported (Wang et al., 2024). The overall size of chloroplast genomes typically ranges from approximately 19 to 217 kilobases, with IR regions spanning 20 to 26 kilobases (NCBI Organelle Genome Resources). The chloroplast proteome includes roughly 3,000 proteins involved in crucial metabolic pathways, including photosynthesis, as well as the biosynthesis of fatty acids, amino acids, nucleotides, vitamins, hormones, and secondary metabolites (Dobrogojski et al., 2020). Most of these proteins are encoded by nuclear genes, synthesized in the cytosol, and subsequently imported into the chloroplast, while a smaller fraction is encoded by the chloroplast genome itself (Fu et al., 2022).
Recent advancements in chloroplast genome engineering have facilitated detailed investigations into gene function, regulatory mechanisms, and targeted genome modification (An et al., 2022). These technologies are increasingly being applied to enhance photosynthetic performance, develop nutritionally improved crops, and produce high-value bioproducts (Daniell et al., 2021; Singhal et al., 2023). This study reports the de novo sequencing, annotation, and structural characterization of the complete chloroplast genome of E. glaucum, presenting a valuable genomic reference for future research in taxonomy, evolutionary biology, and biotechnological innovation.
2 Method
2.1 Plant material
Fresh leaf material of E. glaucum was collected from a cultivated individual maintained at the Kebun Raya Bogor (Bogor Botanic Gardens), West Java, Indonesia, under accession number III.G.189. The specimen’s original provenance is traced to Puger, East Java.
2.2 DNA extraction
Genomic DNA was extracted from young leaf tissue using a modified cetyltrimethylammonium bromide (CTAB) protocol following Doyle and Doyle (1987), with optimizations implemented to enhance yield and purity. DNA quality and quantity were initially assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific), and integrity was evaluated via 1% TBE agarose gel electrophoresis. For more accurate quantification, the Qubit dsDNA High Sensitivity Assay Kit (Thermo Scientific) was used. Fragment size distribution and integrity were further validated using the Agilent 4150 TapeStation system.
2.3 Whole genome sequencing
High-quality genomic DNA was subsequently subjected to library preparation. The DNA was enzymatically fragmented to produce insert-sized fragments appropriate for high-throughput sequencing. Following fragmentation, sequencing libraries were constructed and sequenced on the Illumina NextSeq 500 platform (Genetika Science Lab, Tangerang, Indonesia), generating paired-end reads of 150 base pairs. The sequencing run targeted a total yield of 10 gigabases, providing sufficient depth for comprehensive chloroplast genome assembly and downstream analyses.
2.3 Chloroplast genome assembly and annotation
Quality assessment of the raw sequencing reads was conducted using FastQC version 0.11.8 (Andrews, 2010), which provided diagnostic metrics including per-base quality scores, GC content, sequence length distribution, and indicators of potential contamination. To ensure high-fidelity reads, adapter sequences, low-quality bases (Phred score <30), and nucleotide biases at the 5′ and 3′ ends were removed using Trimmomatic version 0.39 (Bolger et al., 2014). The following trimming parameters were applied: ILLUMINACLIP: TruSeq3-PE.fa:2:30:10, SLIDINGWINDOW:4:28, LEADING:28, TRAILING:28, and MINLEN:20. These processes were executed through the Galaxy web platform (https://usegalaxy.org, The Galaxy Community, 2024).
High-quality trimmed reads were assembled de novo into a complete chloroplast genome using GetOrganelle version 1.7.7.1 (Jin et al., 2020), an organelle-specific assembler employing a k-mer-based graph approach optimized for high-coverage plastid genomes. Genome annotation was performed using CPGAVAS2 (Shi et al., 2019) through its online platform (http://47.96.249.172:16019/analyzer/annotate), with the chloroplast genome of Euonymus kiautschovicus Loes. (syn. Euonymus fortunei var. fortunei, GenBank accession: PQ397793) serving as the reference to guide gene prediction and structural feature identification.
To ensure annotation precision, subsequent manual curation and validation of coding sequences, intron-exon boundaries, and RNA genes were carried out using Unipro UGENE version 45.1 (Okonechnikov et al., 2012) and NCBI Genome Workbench version 3.8.2 (Kuznetsov and Bollin, 2021). Finally, the complete circular chloroplast genome map was visualized using OrganellarGenomeDRAW (OGDRAW) through the MPI-MP Chlorobox web server (Greiner et al., 2019), enabling clear graphical representation of gene content, orientation, and overall genome architecture.
3 Data
3.1 Characterization of Elaeodendron glaucum chloroplast genome
The complete chloroplast genome of E. glaucum was assembled as a circular molecule of 157,958 base pairs (bp) with an overall GC content of 37%. Its organization follows the typical quadripartite structure of angiosperm plastomes, consisting of a large single-copy (LSC) region of 86,485 bp with 35.21% GC content, a small single-copy (SSC) region of 18,363 bp with 31.80% GC content, and two inverted repeat (IR) regions of 26,555 bp each with 42.79% GC content (Figure 1). The total genome size of E. glaucum is slightly larger than those reported for closely related genera within the Celastraceae family, including Celastrus vaniotii (H.Lév.) Rehder (157,194 bp; GenBank accession: OR726632), E. kiautschovicus (157,611 bp; GenBank accession: PQ397793), Microtropis osmanthoides (Hand.-Mazz.) Hand.-Mazz. (156,659 bp; GenBank accession: NC 065714), and Parnassia faberi Oliv. (153,846 bp; GenBank accession: NC 061028), but shorter than Salacia menglaensis J.Y.Shen, L.C.Yan & Landrein (163,255 bp; GenBank accession: NC 047214).
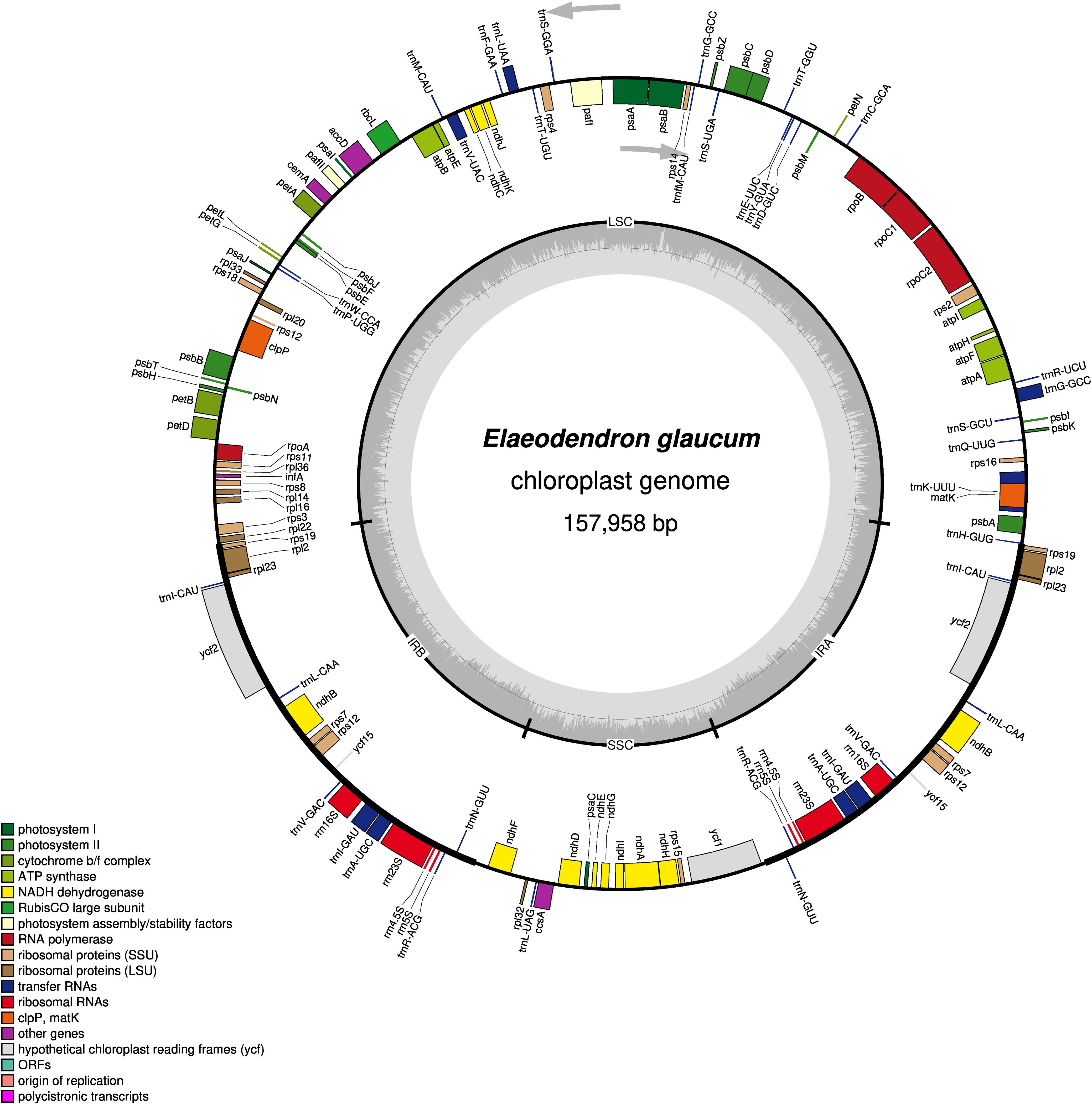
Figure 1. Circular gene map of the Elaeodendron glaucum chloroplast genome. Genes positioned on the inner track of the circular map are transcribed in the counterclockwise direction, while those on the outer track are transcribed clockwise. Distinct colors are used to indicate different functional categories of genes. The innermost circle illustrates the GC content in grey, with the lighter grey areas representing AT content.
3.2 Gene annotation of Elaeodendron glaucum chloroplast genome
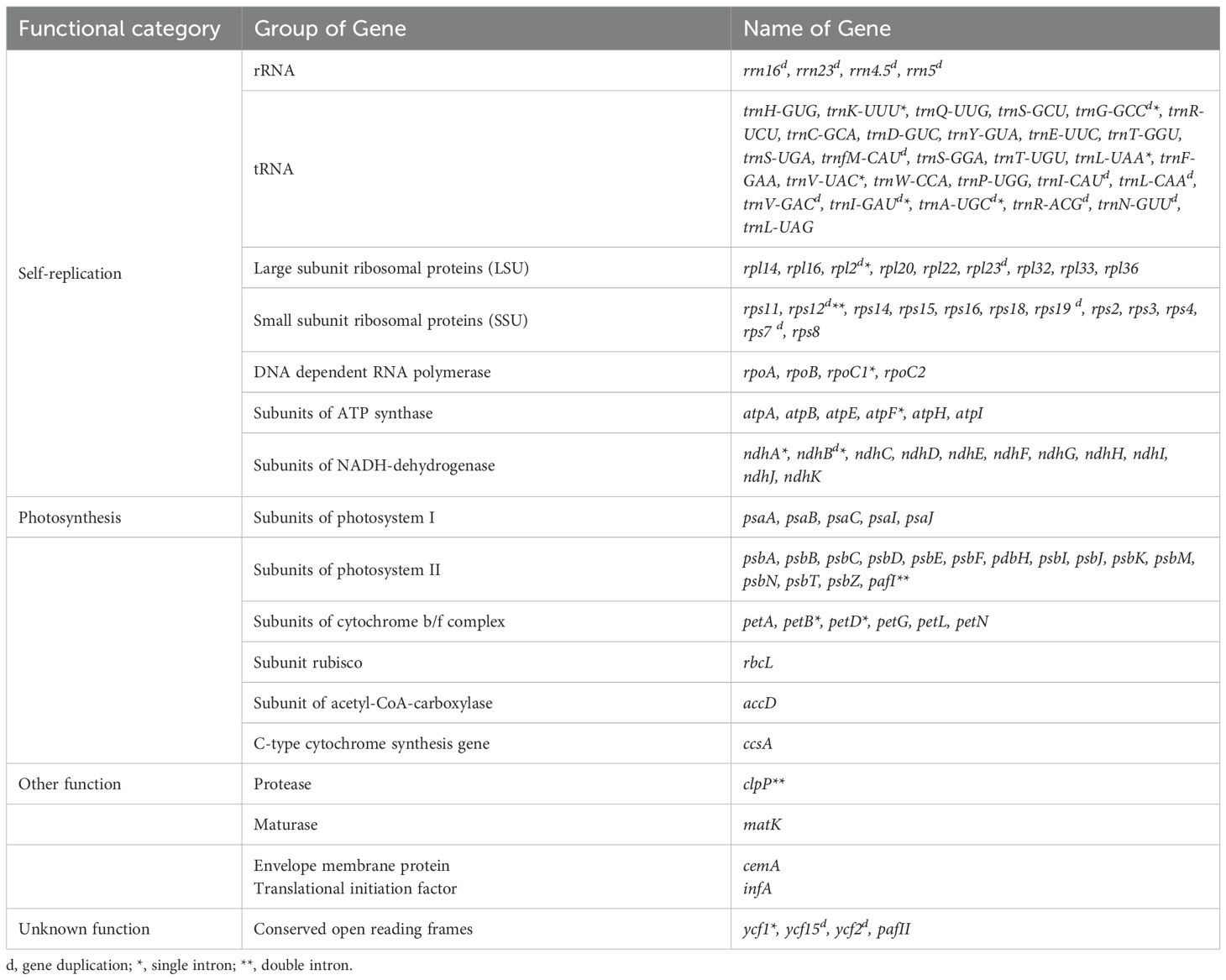
A total of 133 genes were annotated within the E. glaucum chloroplast genome, encompassing 112 unique genes. These include 88 protein-coding genes (79 unique), 37 transfer RNA (tRNA) genes (29 unique), and 8 ribosomal RNA (rRNA) genes (4 unique). Among these, 17 genes contain a single intron, while three genes—rps12, pafI, and clpP—each possess two introns (Table 1).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank, accession PV153503.
Author contributions
DSR: Writing – review & editing. IPAH: Writing – review & editing, Data curation. MM: Data curation, Writing – review & editing. IM: Formal analysis, Writing – review & editing, Methodology, Data curation. TYIW: Writing – review & editing. JTH: Writing – review & editing. DG: Writing – review & editing. NU: Writing – review & editing. R: Writing – review & editing. ASDI: Data curation, Writing – review & editing. RRI: Writing – review & editing. AS: Writing – review & editing. MRH: Data curation, Methodology, Conceptualization, Funding acquisition, Writing – review & editing, Formal analysis, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded through the Joint Collaboration Rumah Program of the Organization for Research in Life and Environmental Sciences (ORHL), National Research and Innovation Agency (BRIN) for the 2023 fiscal year under contract number B-651/III.5/PR.03.06/2/2023.
Acknowledgments
The authors express gratitude to the Directorate of Scientific Collection Management, as well as the management of Bogor Botanic Gardens, for their essential support and facilitation of this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, Y., Wang, Y., Wang, X., and Xiao, J. (2022). Development of chloroplast transformation and gene expression regulation technology in land plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1037038
Andrews, S. (2010). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed December 15, 2024).
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Coughenour, J. M., Simmons, M. P., Lombardi, J. A., and Cappa, J. J. (2010). Phylogeny of Celastraceae subfamily Salacioideae and tribe Lophopetaleae inferred from morphological characters and nuclear and plastid genes. Syst. Bot. 35, 358–367. doi: 10.1600/036364410791638289
Daniell, H., Jin, S., Zhu, X. G., Gitzendanner, M. A., Soltis, D. E., and Soltis, P. S. (2021). Green giant—a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol. J. 19, 430–447. doi: 10.1111/pbi.13556
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 1–29. doi: 10.1186/s13059-016-1004-2
Dobrogojski, J., Adamiec, M., and Lucinski, R. (2020). The chloroplast genome: a review. Act. Physiol. Plant 42, 98. doi: 10.1007/s11738-020-03089-x
Doyle, J. J. and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Fu, Y., Li, X., Fan, B., Zhu, C., and Chen, Z. (2022). Chloroplasts protein quality control and turnover: a multitude of mechanisms. Int. J. Mol. Sci. 23, 7760. doi: 10.3390/ijms23147760
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59eW64. doi: 10.1093/nar/gkz238
Hou, D. (1960). “Celastraceae—I,”, Flora Malesiana - Series 1, Spermatophyta vol. 6(1). (Djakarta: Noordhoff-Kolff) 6, 227–291.
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., DePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 1–31. doi: 10.1186/s13059-020-02154-5
Kuznetsov, A. and Bollin, C. J. (2021). “NCBI genome workbench: desktop software for comparative genomics, visualization, and GenBank data submission,” in Multiple Sequence Alignment: Methods and Protocols. Ed. Katoh, K. (Humana Press, New York), 261–295. doi: 10.1007/978-1-0716-1036-7_16
Liu, Q., Dai, J., Chen, J., Liu, Z., Lin, Y., Qiu, G., et al. (2024). Comparative analysis the chloroplast genomes of Celastrus (Celastraceae) species: Provide insights into molecular evolution, species identification and phylogenetic relationships. Phytomedicine 131, 155770.
Nadeem, M. A., Nawaz, M. A., Shahid, M. Q., Doğan, Y., Comertpay, G., Yıldız, M., et al. (2018). DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 32, 261–285. doi: 10.1080/13102818.2017.1400401
Okonechnikov, K., Golosova, O., Fursov, M., and Ugene Team (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28, 1166–1167. doi: 10.1093/bioinformatics/bts091
POWO (2025). Plants of the World Online (Richmond, United Kingdom: Facilitated by the Royal Botanic Gardens, Kew). Available online at: http://www.plantsoftheworldonline.org.
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–W73. doi: 10.1093/nar/gkz345
Simmons, M. P., Bacon, C. D., Cappa, J. J., and McKenna, M. J. (2012). Phylogeny of Celastraceae subfamilies Cassinoideae and Tripterygioideae inferred from morphological characters and nuclear and plastid loci. Syst. Bot. 37, 456–467. doi: 10.1600/036364412X635502
Singhal, R., Pal, R., and Dutta, S. (2023). Chloroplast engineering: fundamental insights and its application in amelioration of environmental stress. App. Biochem. Biotechnol. 195, 2463–2482. doi: 10.1007/s12010-022-03930-8
The Galaxy Community (2024). The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 52, W83–W94. doi: 10.1093/nar/gkae410
Wang, J., Kan, S., Liao, X., Zhou, J., Tembrock, L. R., Daniell, H., et al. (2024). Plant organellar genomes: much done, much more to do. Trends Plant Sci. 29, 754–769. doi: 10.1016/j.tplants.2023.12.014
Keywords: Celastrales, gene annotation, genome structure, illumina, plastome assembly
Citation: Rinandio DS, Husaini IPA, Magandhi M, Martiansyah I, Wulansari TYI, Hadiah JT, Girmansyah D, Utami N, Rugayah, Irsyam ASD, Irwanto RR, Suhatman A and Hariri MR (2025) Complete plastome of Elaeodendron glaucum (Rottb.) Pers.: genomic resources for Celastraceae systematics. Front. Plant Sci. 16:1649473. doi: 10.3389/fpls.2025.1649473
Received: 18 June 2025; Accepted: 11 August 2025;
Published: 01 September 2025.
Edited by:
Yuri Shavrukov, Flinders University, AustraliaReviewed by:
Leonardo Alfredo Ornella, Cubiqfoods SL, SpainHoang Dang Khoa Do, Nguyen Tat Thanh University, Vietnam
Qiang Wen, Jiangxi Academy of Forestry, China
Copyright © 2025 Rinandio, Husaini, Magandhi, Martiansyah, Wulansari, Hadiah, Girmansyah, Utami, Rugayah, Irsyam, Irwanto, Suhatman and Hariri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Rifqi Hariri, bXVoYW1tYWQucmlmcWkuaGFyaXJpQGJyaW4uZ28uaWQ=
 Dipta Sumeru Rinandio1
Dipta Sumeru Rinandio1 Mahat Magandhi
Mahat Magandhi Irfan Martiansyah
Irfan Martiansyah Muhammad Rifqi Hariri
Muhammad Rifqi Hariri