- 1Plant Protection and Biomolecular Diagnosis Department, Arid Lands Cultivation Research Institute, City of Scientific Research and Technological Applications, Alexandria, Egypt
- 2Plant Production Department, Arid Lands Cultivation Research Institute (ALCRI), City of Scientific Research and Technological Applications, Alexandria, Egypt
- 3Department of Integrated Pest Management of Fruit Trees and Vegetable Crops, Istituto Agronomico Mediterraneo di Bari, Bari, Italy
- 4Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 5Plant Protection Department, The National Institute of Horticultural Research, Skierniewice, Poland
The utilization of arbuscular mycorrhizal fungi (AMF) and Trichoderma spp. correlates with improved plant nutrition and the stimulation of systemic plant defenses in response to pathogen challenges. Nonetheless, studies examining the effects of AMF colonization and the foliar application of the Trichoderma viride isolate Tvd44 on viral infection are limited. By analyzing the phenotypic, biochemical, and transcriptional expression of eleven defense genes, we investigated the effects of AMF colonization, foliar application of Tvd44, and their combined (dual) application on tomato plants challenged with potato virus Y. Interestingly, the dual application significantly suppressed viral symptoms and decreased viral accumulation levels, disease incidence, and disease severity by 88.1%, 40%, and 53.4%, respectively. Furthermore, both single and dual treatments significantly enhanced the activity of antioxidant enzymes, chlorophyll concentration, and macronutrient levels in the tomato tissues. In the realm of transcriptional analyses, the CHS gene served as a master key in understanding the physiological and pathway relationships among various genes (F3’H, HQT, C3H, GST, JERF, CHI, WRKY-1, WRKY-19, FLS, and F3H) involved in plant defense. These results suggest a sophisticated network of interactions that governs multiple facets of plant defense responses, encompassing the biosynthesis of flavonoids and other secondary metabolites, as well as the activation of transcription factors related to defense mechanisms. The obtained data indicate that AMF colonization and T. viride foliar spraying enhance tomato resistance to PVY by activating defense systems, thereby affecting viral replication. This finding highlights the significance of AMF and T. viride within the ecosystem and their crucial role in managing plant viruses.
1 Introduction
Viruses affecting plants lead to significant reductions in crop yield and quality, posing serious threats to global food security and the sustainability of agricultural practices (Rabie et al., 2024). Many viral infections cause widespread damage, ultimately leading to the death of the plant as the virus spreads throughout its system (Abdelkhalek and Hafez, 2019). Viral infections lead to necrosis, chlorosis, and abnormal growth patterns (Mandal et al., 2025), while they significantly diminish host plant performance by inhibiting photosynthetic carbon absorption and altering various processes, including the cell cycle, transport mechanisms, secondary metabolism, protein alterations, hormone regulation, and isoprenoid production (Abdelkhalek et al., 2019a). These alterations diminish plants’ protective mechanisms and facilitate virus proliferation. Potato virus Y (PVY, Potyvirus yituberosi, genus Potyvirus, family Potyviridae) is one of the most damaging viruses to potatoes and other solanaceous crops, such as tomatoes and peppers (Morais et al., 2025). The viral genome is single-stranded, positive-sense RNA of about 10 kb in length. It encodes a single large polypeptide, which is cleaved by three virus-encoded proteases into nine distinct products (Tribodet et al., 2005). Unlike fungi or bacteria, which may be treated with antifungal or antibacterial treatments, plants cannot be cured or controlled after contracting a virus. Consequently, the main goals of disease management are to prevent virus infection in plants or to increase plant resistance to viral infection. Although agrochemicals are commonly employed to manage plant virus infections by controlling their insect vectors, their elevated costs and potential environmental damage raise considerable concerns (El-Bilawy et al., 2022; Al-Askar et al., 2023).
Developing a sustainable approach is essential for managing viral infections and reducing the reliance on chemical fertilizers in agricultural areas. Consequently, the potential for improving plant immunity to viruses through the application of beneficial microbes, such as arbuscular mycorrhizal fungi (AMF), warrants careful consideration and has been suggested as a viable and sustainable approach to mitigate plant viruses (Aseel et al., 2019; Chaudhary et al., 2025). AMF assists plants in agricultural soils in overcoming biotic and abiotic challenges. Although morphologically restricted to the roots, AM fungi significantly influence the overall physiology of the plant due to the metabolic and physiological alterations they induce in the root system (Li et al., 2025). Their reliance on carbohydrates and lipids present in plants enables fungi symbionts to function as effective carbon sinks in roots. Therefore, plant carbon balance is preserved by regulating leaf primary metabolism and photosynthesis (Metwally and Abdelhameed, 2024). Setting up a functional AMF symbiosis also causes significant changes in the secondary metabolism of plants. These changes include alterations in the amount of phenolic compounds and the accumulation of phytohormones, such as jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA) (Vasan et al., 2024). Despite evidence that AMF colonization mitigates the severity of diseases caused by several plant pathogens, it has received scant attention, and our comprehension of the influence of AMF on plant-virus interactions remains inadequate. The plant’s nutrition, the time of the encounter, and the lifestyle of the viral pathogen all influence the tripartite relationship between a plant, AMF, and a virus (Abid Mehmood et al., 2025).
Applying bio-inoculants containing Trichoderma as an antagonistic agent is among the most effective biological control methods in numerous countries (Ríos-Ruiz et al., 2025). The proportion of different Trichoderma species constitutes approximately 50-60% of the worldwide market for biological control agents (Yuan et al., 2025). Trichoderma induces local or systemic resistance in various plant pathogens, empowering plants to combat multiple diseases and enhancing overall plant productivity (Behiry et al., 2023). Trichoderma enhances the activation of defense responses in leaf tissue through the mediation of jasmonic acid (JA) (Aseel et al., 2023b). It produces enzymes and metabolites that can alter the ethylene levels in a plant’s root structure, enhancing nutrient uptake. Furthermore, it has the potential to bolster plant resistance, improve nutrient utilization efficiency, mitigate disease, stimulate plant growth, and remediate agrochemical contamination (Rodríguez-Martínez et al., 2025). The beneficial relationship between Trichoderma and plants can also induce the expression of genes related to plant defense mechanisms. Recently, it was reported that WRKYs regulate plant signaling by physically interacting with proteins involved in signaling, defense, transcription, and other cellular functions (Wang et al., 2025). Although the understanding of WRKY transcriptional activity during plant virus infection remains unclear, recent evidence suggests that WRKY transcription factors protect plants against virus infection (Nunna et al., 2025).
This investigation aimed to determine whether the combination of AMF and Trichoderma viride isolate Tvd44 enhances the resistance of tomato plants to biotic stress induced by PVY infection. Specifically, we aim to a) examine the impact of AMF on the colonization of tomato roots and/or the foliar application of Tvd44 to mitigate disease incidence and enhance tomato plant growth; b) investigate the transcriptomic expression of defense pathways, including WRKY transcription factors (WRKY1 and WRKY19), jasmonic acid/ethylene (JA/ET) signaling like Jasmonate and ethylene-response factor 3 (JERF3) and Glutathione S-transferase 1(GST1), flavonoids such as Flavanone 3-hydroxylase (F3H), Chalcone synthase (CHS), Flavonol synthase 1 (FLS), Chalcone isomerase 2 (CHI2), and Flavonoid 3′ hydroxylase (F3'H), and chlorogenic acid like Hydroxycinnamoyl-CoA quinate transferase (HQT) and p-coumarate 3-hydroxylase (C3H) genes in tomato plants in response to PVY infection and c) analyze the expression of secondary metabolites and the physiological traits network of plants inoculated with AMF, Tvd44, and/or dual treatments co-inoculated with both fungi, against PVY.
2 Materials and methods
2.1 Tomato cultivar and virus inoculum source
The Agriculture Research Center in Egypt supplied a virus-free tomato (Solanum lycopersicum) cultivar, GS-12, showing susceptibility to PVY infection. The PVY strain DA55 employed in this research has been previously documented (Aseel et al., 2023b). The purified PVY served as the viral inoculum for all treatments challenged with PVY. The concentration of PVY inoculum was 20 µg/mL, prepared in a 10 mM phosphate buffer at pH 7.2, with the addition of 0.1% sodium sulfite.
2.2 Fungal inoculum
AMF was kindly obtained from the Plant Pathology Research Institute, Agricultural Research Centre, Egypt. The inoculum used in this study was made up of equal amounts of spores from Rhizophagus irregularis (Blaszk., Wubet, Renker, and Buscot), Rhizoglomus clarum (Nicolson and Schenck), and Funneliformis mosseae (Nicolson and Gerd.). We cultivated the AMF inoculum in sterile soil beneath Sudan grass (Sorghum bicolor L.) as a potential host. The AMF inoculum, exhibiting 83.1% colonization, consists of root fragments, mycelia, spores, and rhizospheric soil. The T. viride isolate Tvd44 (Ac# OQ991378) utilized in this study was previously isolated and characterized (Aseel et al., 2023b). The Tvd44 culture filtrate (spraying solution) was prepared by inoculating 1 mL containing 1 × 10^9 conidia into 100 mL of potato dextrose broth and culturing it at 28°C for 6 days on a rotary shaker at 150 rpm. Next, we filtered the culture by using Whatman filter paper No. 1. The filtrate was subjected to a 0.2 μm pore biological membrane filter before application to the plant leaves.
2.3 Antiviral assay and experimental design
Before planting, tomato seeds were surface-sterilized with a 70% ethyl alcohol solution and a 0.5% sodium hypochlorite (NaOCl) solution. At 20 days after seeds were planted in sterilized peat moss soil, tomato seedlings were transferred to 25-cm-diameter pots filled with a mixture of equal parts sterilized soil, sand, and clay, with five seedlings in each pot. Each treatment had five replicates. Fertilization was not applied, and all pots received regular irrigation. The plants were grown in a controlled greenhouse environment maintained at 26-28°C during the day and night, with a humidity level of 65%. Seven treatments were administered. The first treatment (control) consisted of tomato plants that were foliarly treated with sterilized medium-free Tvd44 and mechanically inoculated with viral inoculation buffer. The second treatment (AMF) consisted of tomato plants colonized by AMF, supplemented with a foliar application of sterilized medium-free Tvd44, and mechanically inoculated using a viral inoculation buffer. The third treatment (Tvd44) consisted of tomato plants that were foliarly treated with T. viride culture filtrate and mechanically inoculated with viral inoculation buffer. The fourth treatment (PVY) included tomato plants that were foliarly treated with sterilized medium-free Tvd44 and mechanically rubbed with PVY. The mechanical inoculation involved dusting the upper two leaves of each tomato seedling with carborundum and gently rubbing with 1 mL of PVY using the forefinger technique (Omar et al., 2022). The fifth treatment (AMF+PVY) consisted of tomato plants colonized with AMF, foliarly treated with sterilized medium-free Tvd44, and mechanically inoculated with PVY. The sixth treatment (Tvd44+PVY) included tomato plants that were foliarly treated with Tvd44 culture filtrate and mechanically inoculated with PVY. The seventh treatment (dual+PVY) involved tomato plants colonized by AMF, foliarly treated with T. viride culture filtrate, and mechanically inoculated with PVY. In all AMF-treated plants, AMF was applied during the transfer of tomato seedlings into new pots (at 20 days post-germination) by incorporating 10 g of the AMF inoculum into each seedling bed. The viral inoculation was conducted on all PVY-inoculated plants fifteen days after seedling transplantation, corresponding to 35 days post-germination. In every instance of Tvd44-treated plants, the culture filtrate was administered to the tomato leaves 24 h before the inoculation with PVY, corresponding to 34 days post-germination (at two fully expanded true leaves). A handheld pressure sprayer was employed to apply culture filtrate or sterilized medium-free Tvd44 to the entire plant until runoff occurred. All plants were maintained in insect-proof greenhouses for more than three weeks, and the manifestation of viral symptoms was monitored daily (Abdelkhalek et al., 2022a).
2.4 Sample collection and disease estimation
Tomato plants were collected 25 days post-PVY inoculation (dpi) and underwent various physiological and biomolecular assessments. The plants collected from each group were subjected to multiple washes with running water. For further analysis, three upper leaves from each plant were collected from every pot (15 leaves per pot) and categorized as a biological sample within the same treatment group, resulting in 5 biological samples. Every biological sample underwent three technical replicates. The disease incidence and severity were evaluated by observing visual viral symptoms, utilizing a rating scale from 0 to 5, and assessing the extent of leaf damage (Imran et al., 2012; Aseel et al., 2023b).
2.5 Mycorrhizal colonization estimation
Five tomato roots from each applied treatment were estimated for mycorrhizal colonization with PVY at 25 dpi. Small segments (1 cm) of each root were prepared and stained with trypan blue (Sigma-Aldrich, St. Louis, USA), as previously described (Phillips and Hayman, 1970). Mycorrhizal colonization was assessed in root segments of each treatment using a light microscope, adhering to the previously described approach (Trouvelot, 1986).
2.6 Growth parameter estimation
At 25 dpi, all plants from each treatment were meticulously uprooted, washed under running water, and assessed for the number of leaves, root and shoot lengths (in cm), and shoot and root dry weights (in g). The dry weights were measured following a 72-hour drying period of the plant samples in an oven set to 40°C.
2.7 Estimation of macronutrient content
The nitrogen (N) content in the tomato leaves was determined through the Kjeldahl method (Horneck and Miller, 2019). As previously described, total sulfur (S) was determined by precipitating the sulfate with barium chloride and measuring turbidity using a spectrophotometer at 420 nm (Olsen and Sommers, 1982).
2.8 Biochemical estimation
The evaluation of photosynthetic chlorophyll a and b in tomato leaves was conducted as described previously (Harborne, 1998). The techniques for extracting and assessing the enzymes peroxidase (POX), polyphenol oxidase (PPO), and catalase (CAT) were established by Maxwell and Bateman (1967); Galeazzi et al. (1981), and Aeby (1984), respectively. The extraction and methodology for quantifying total protein content were conducted following Bradford (1976).
2.9 Transcriptomic profiles of the defense pathway genes
Total RNA was extracted from 100 mg of fresh tomato leaves collected at 25 days post-inoculation (dpi) using the RNeasy Plant Mini Kit according to the manufacturer’s instructions. Following the RNA concentration and purity assessment, a reverse transcription procedure was conducted to convert one µg of DNase-treated RNA into cDNA utilizing M-MuLV reverse transcriptase, as previously outlined (Aseel et al., 2023b). The cDNA was stored at -20°C until it was used. The evaluation of AMF colonization and T. viride treatments on the expression of defense-related genes and the accumulation level of the PVY-CP gene in tomato plants was conducted using real-time quantitative PCR (RT-qPCR). The expression ratio of the PVY-CP gene to the housekeeping gene in tomato control plants was utilized to determine the level of viral accumulation. The sequences of specific primers for tomato defense pathway genes WRKY1, WRKY19, JERF3, GST1, CHS, C3H, CHI2, FLS, F3H, F3'H, and HQT, as well as PVY-CP, are listed in Supplementary Table S1. The β-actin gene was selected as a housekeeping gene due to its stable expression in tomatoes, as reported in prior studies (Aseel et al., 2023a; Rabie et al., 2024). The RT-qPCR experiment utilized a CFX Connect TM Real-Time System (BIO-RAD, USA). Each reaction contained 10 µL of 2xSYBR Green RT Mix (Bioline, Germany), 7.4 µL of RNase-free water, 0.8 µL of each primer (10 pmol/µL), and 1 µL of cDNA. The RT-qPCR protocol consisted of an initial cycle at 95°C for 5 min, followed by 45 cycles of 95°C for 5 seconds, 60°C for 10 seconds, and 72°C for 15 seconds. The relative gene expression was determined using the comparative CT approach (2−ΔΔCT) as described previously (Schmittgen and Livak, 2008).
2.10 Statistical analysis
Volcano plots were created using the ggplot2 package, and a t-test was employed to determine the statistical significance of gene expression compared to the control treatment. The significance was expressed as -log10, while gene expression was expressed as log2 fold-change. Heatmap and cluster analysis figures of gene expression were generated using pheatmap package. The data were first transformed using log2; then, Euclidean distance was applied, followed by the Ward method of hard clustering. Thirty internal validation indexes were employed to determine the optimal number of clusters in the data (Charrad et al., 2014). Principal component analysis was conducted using function prcomp in stats package. The figures of contribution and biplot were built using the factoextra package. Gene expression and physiological trait networks were generated using the qgraph package, based on the Spearman rank correlation coefficient, to connect gene expression results with physiological data. All statistical analyses were performed using R software version 4.3.2 (2023).
3 Results
3.1 Viral symptoms development and disease assessment
Under controlled greenhouse conditions, the tomato plants treated with PVY exhibited characteristic viral symptoms at 19 dpi. The pronounced leaf mosaic, distinct veins, yellowing, and necrosis were distinctly observable at 22 dpi (Figure 1). Interestingly, the AMF+PVY, Tvd44+PVY, and dual+PVY treatments resulted in a delay of approximately 5 days in the appearance of symptoms, with mild symptoms observed at 25 dpi (Figure 1). No symptoms were observed in the control, AMF, and Tvd44 treatments. The findings demonstrated that the PVY treatment resulted in a 100% incidence of disease and a severity level reaching 96.8% (Table 1). The findings correspond to the accumulation level of the PVY-CP gene, indicating an approximately 27.8-fold increase in PVY-treated plants relative to control plants. Compared to PVV treatment, the AMF+PVY, Tvd44+PVY, and dual+PVY treatments exhibited significantly reduced PVY accumulation expressions, with relative expression levels of 4.82-, 5.84-, and 3.29-fold, respectively. The dual+PVY treatment resulted in a decrease in disease incidence and severity to 60% and 46.62%, respectively. The AMF+PVY and Tvd44+PVY exhibited disease severity levels of 51.96% and 54.64%, respectively, as shown in Table 1. Nonetheless, the data presented in Figure 2 indicated that the roots of infected plants with PVY were shorter than those treated with AMF+PVY, Tvd44+PVY, and/or the dual+PVY treatments, which exhibited a significantly greater length.
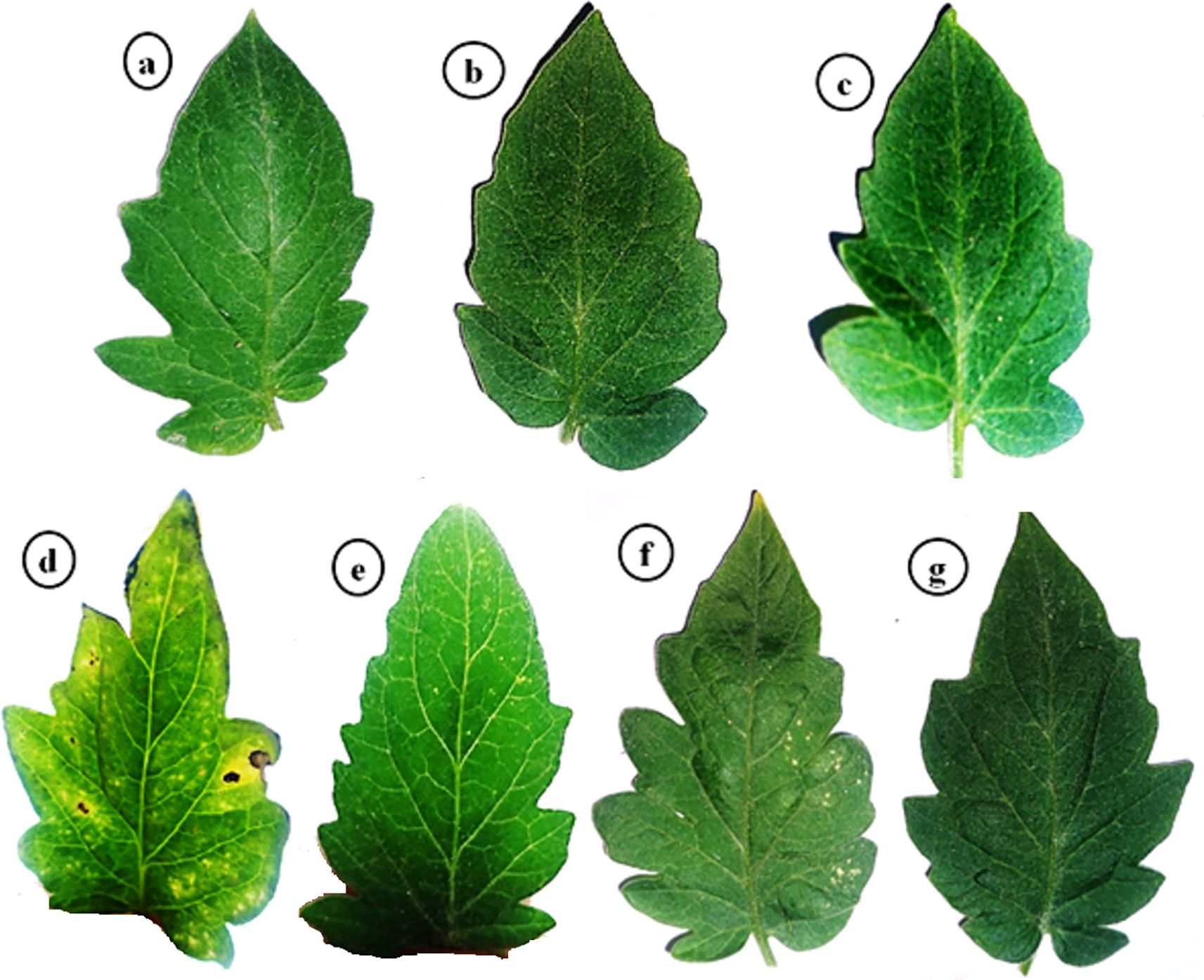
Figure 1. Impacts of arbuscular mycorrhizal and Trichoderma viride on PVY symptoms at 25 dpi. (a) control plants, (b) AMF plants colonized with AM fungi, (c) sprayed with Tvd44, (d) PVY-infected tomato plants, (e) colonized with AMF and infected with PVY, (f) sprayed with Tvd44 and infected with PVY, (g) colonized with AMF and infected with PVY and sprayed with Tvd44.
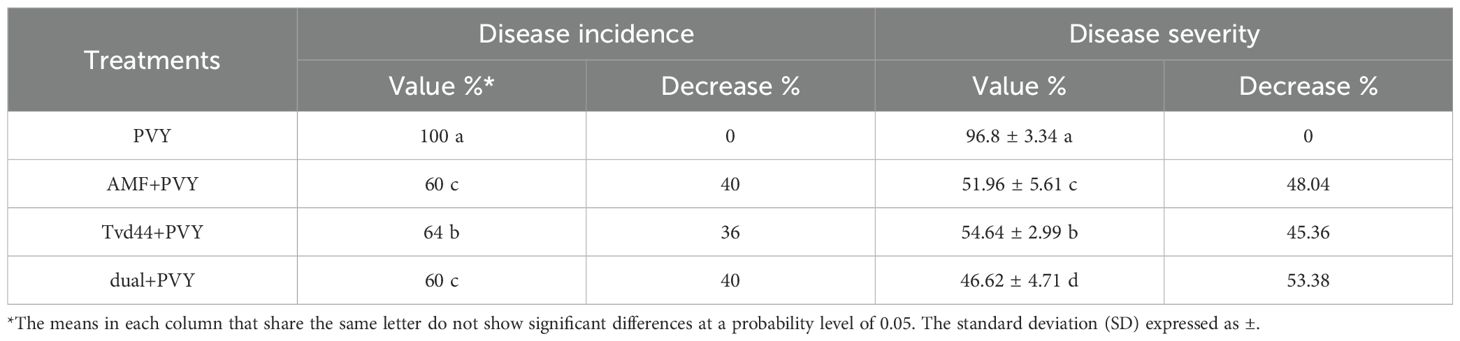
Table 1. Evaluation of disease incidence and disease severity of tomato plants upon PVY challenge and treatment with AMF and Tvd44 at 25 dpi.
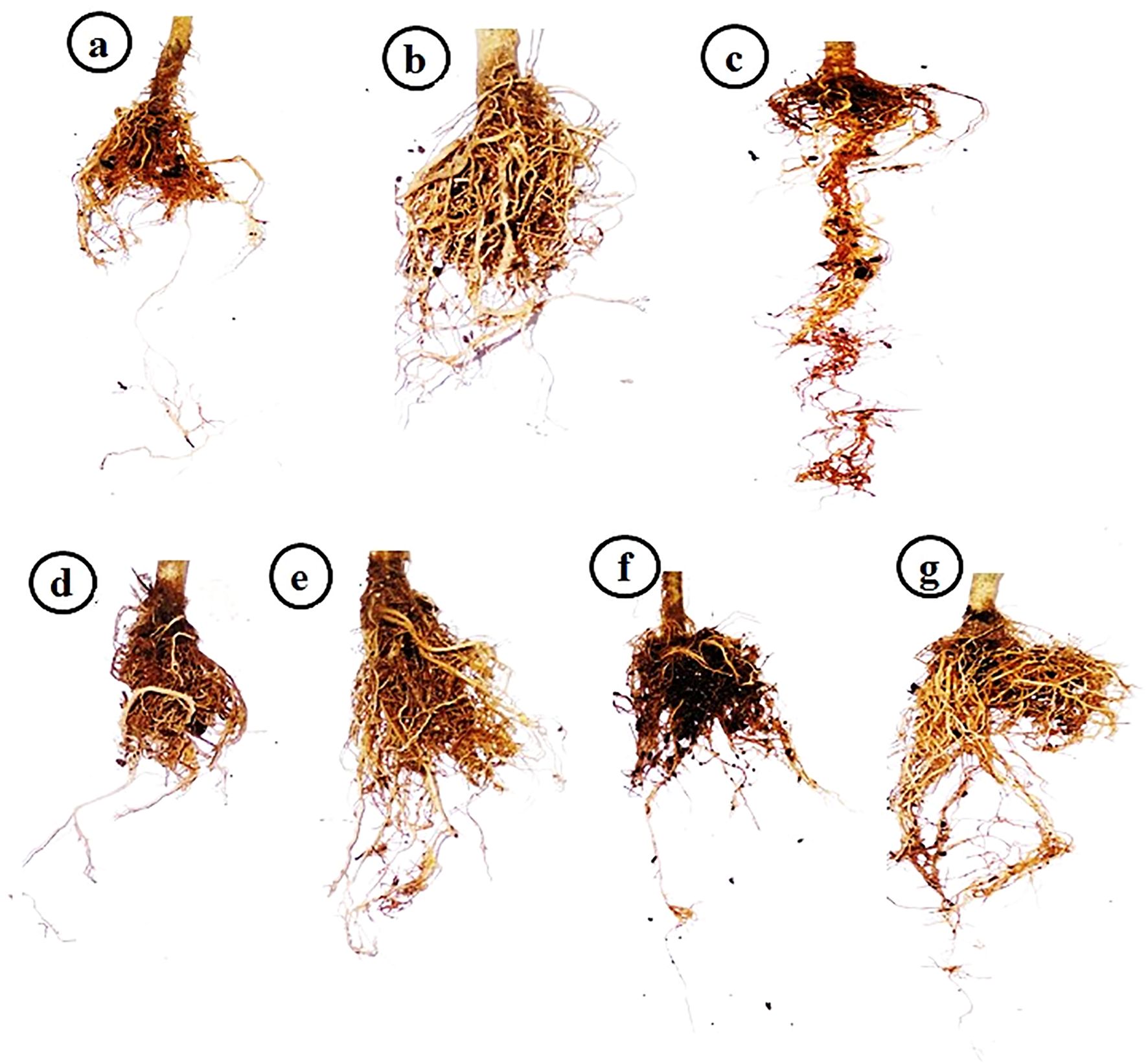
Figure 2. Impacts of arbuscular mycorrhizal and Trichoderma viride on tomato roots at 25 dpi. (a) control plants, (b) AMF plants colonized with AM fungi, (c) sprayed with Tvd44, (d) PVY-infected tomato plants, (e) colonized with AMF and infected with PVY, (f) sprayed with Tvd44 and infected with PVY, (g) colonized with AMF and infected with PVY and sprayed with Tvd44.
3.2 Mycorrhizal colonization estimation
Concerning mycorrhizal colonization in tomato roots infected with PVY and/or treated with Tvd44 at 25 dpi, the data revealed that no mycorrhizal colonization was detected in tomato plants that did not receive the AMF inoculum. On the contrary, the other treatments with AMF inoculum showed varying degrees of mycorrhizal colonization. Different typical mycorrhizal structures were observed in the tomato roots through microscopic examination (Figure 3). Successful colonization was confirmed by light microscopy investigation, which revealed the presence of both arbuscules and intraradical mycelium in the root cortex of the colonized plants (Figures 3b–d). Finally, as indicated by our results, PVY-uninfected tomato plants treated only with AMF inoculum exhibited the highest degree of colonization frequency (83.1%).
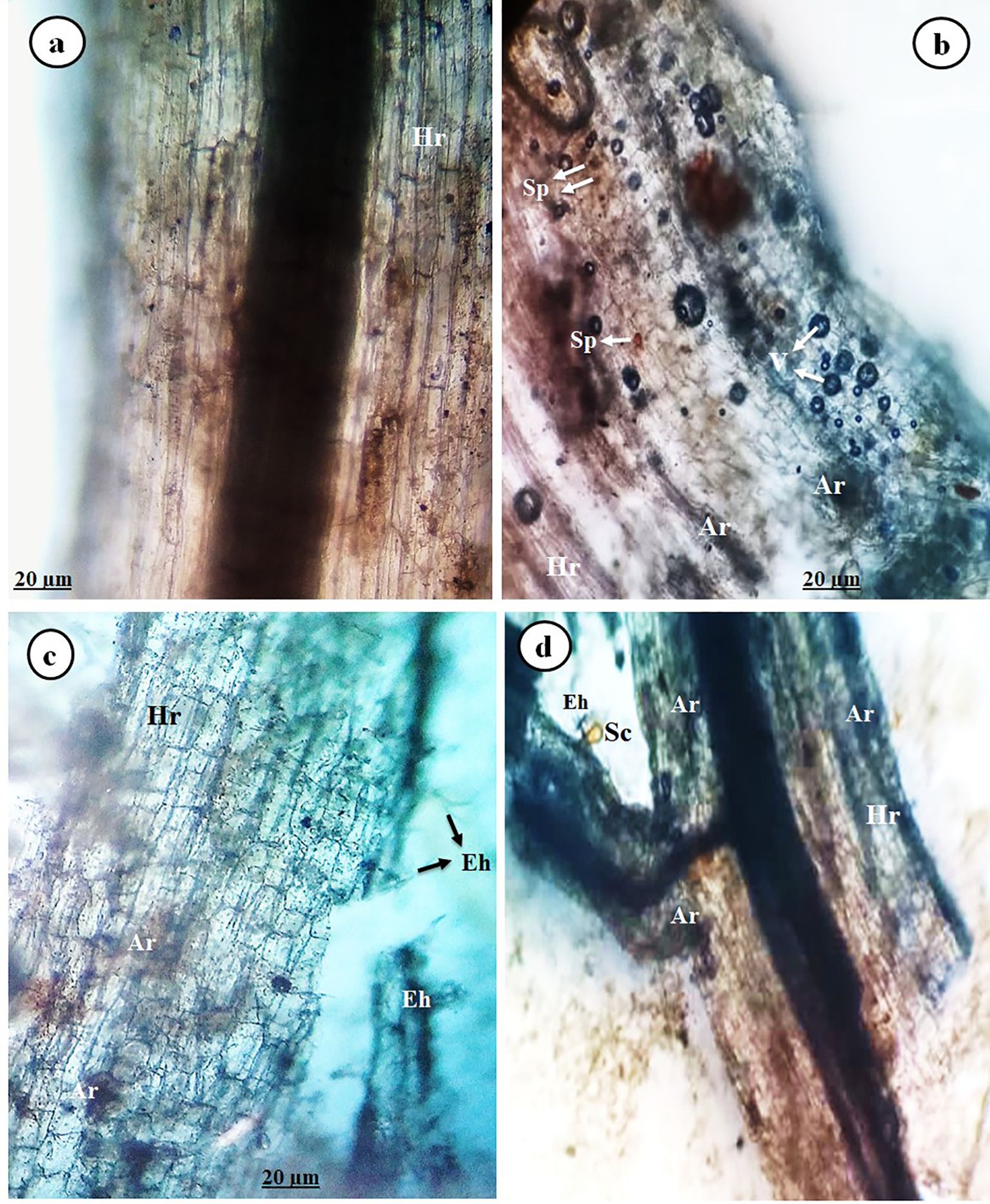
Figure 3. Image showing light micrographs of tomato roots colonized with AMF displaying typical mycorrhizal structures (at 25 dpi), control root (a), and AMF-colonized tomato roots (b–d), where Hr, host root; Ar, arbuscule; Eh, exteraradical hyphae; Sc, sporocarp; Sp, spore; and V, vesicle.
3.3 Growth parameters evaluation
Figure 4 displays the mean growth parameters of 25 dpi tomato plants in response to the various treatments. More precisely, treating tomato plants with AMF and foliar Tvd44 improved their shoot height, root length, fresh weight (both root and shoot), dry weight (both root and shoot), and leaf number compared to control tomato plants. Meanwhile, AMF root colonization promoted all assessed growth parameters. In contrast, PVY treatment significantly decreases all assessed growth parameters compared to control tomato plants. The colonization with AMF, foliar Tvd44, and dual treatments reduces the adverse impacts of PVY infection on the estimated parameters compared with untreated infected plants. Compared to PVY treatment, the dual+PVY treatment was more efficient than the AMF colonization and foliar Tvd44 alone. The dual+PVY treatment resulted in a significant increase of 180% and 254% in shoot and root fresh weight, respectively; additionally, leaf numbers increased by 185% (Figure 4).
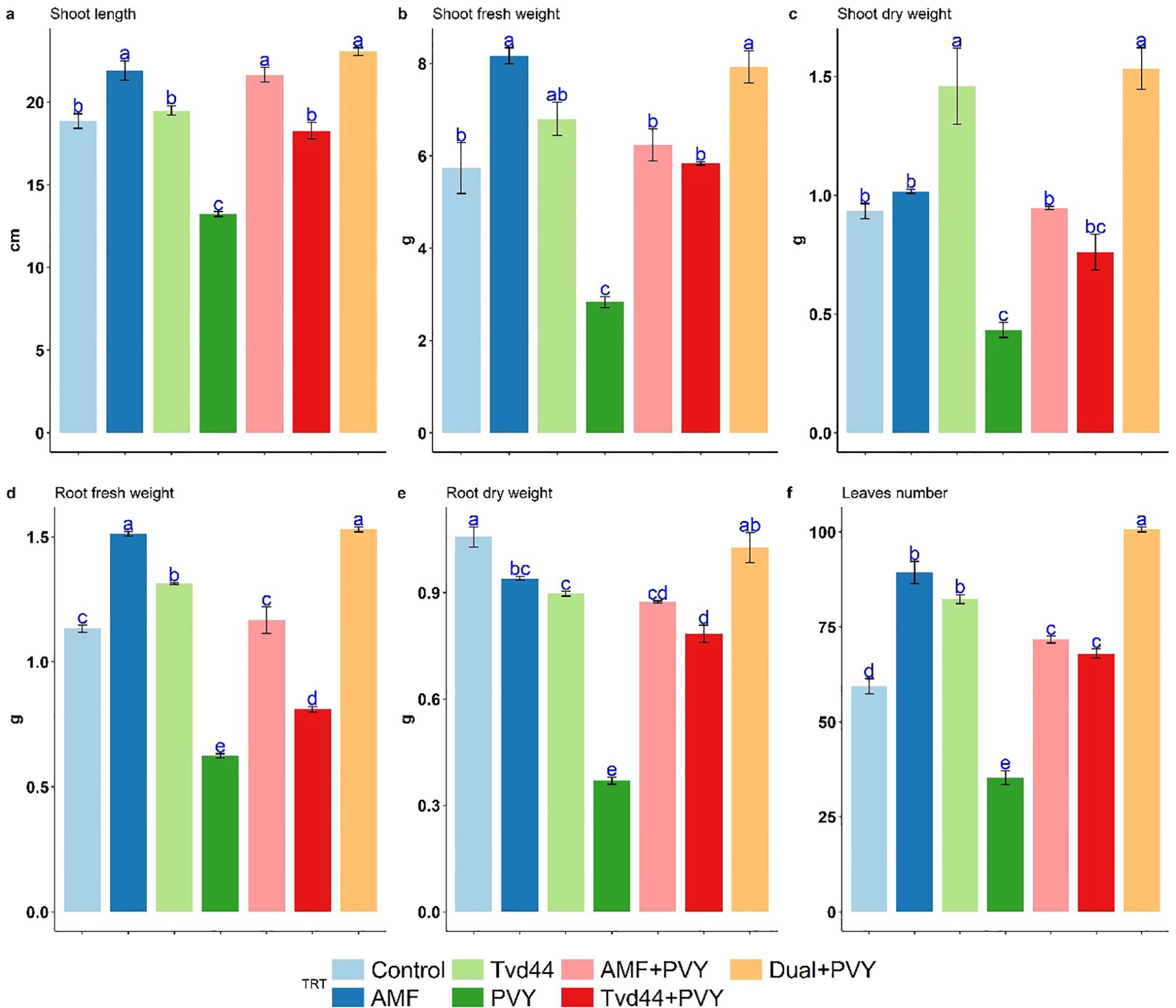
Figure 4. Impact of 7 treatments at 25 dpi on shoot length (cm) (a), shoot fresh weight (g) (b), root fresh weight (g) (c), shoot dry weight (g) (d), root dry weight (g) (e), and leaf number (f) traits using Tukey's HSD test at p ≤ 0.05. Statistical significance was indicated alphabetically above the histogram in ascending order, whereas a>b>c>d. The means in each column that share the same letter do not show significant differences.
3.4 Effect of treatments on macronutrient content and biochemical traits
The total nitrogen (TN) content in tomato leaves varied among the different treatments (Table 2). All treatments resulted in increased TN levels compared to the control treatment. The most significant increases in TN content were noted in the dual+PVY treatment at 3.84%, followed by AMF at 3.45% and Tvd44 at 3.39%. AMF colonization reached its peak in total carbon content at 34.95%, closely followed by Tvd44+PVY at 33.95%. Moreover, the majority of the treatments implemented resulted in elevated levels of total hydrogen (TH) and total sulfur (TS). The observed C/N ratios decreased across all treatments compared to the control (Table 2). Figure 5 presents the findings from the Tukey multiple comparison test concerning the effects of AMF and Tvd44 on biochemical traits. The dual+PVY treatment resulted in a significant increase in chlorophyll (a), (b), CAT, and TP compared to the PVY treatment, with enhancements of 191%, 91%, 147%, and 34%, respectively.
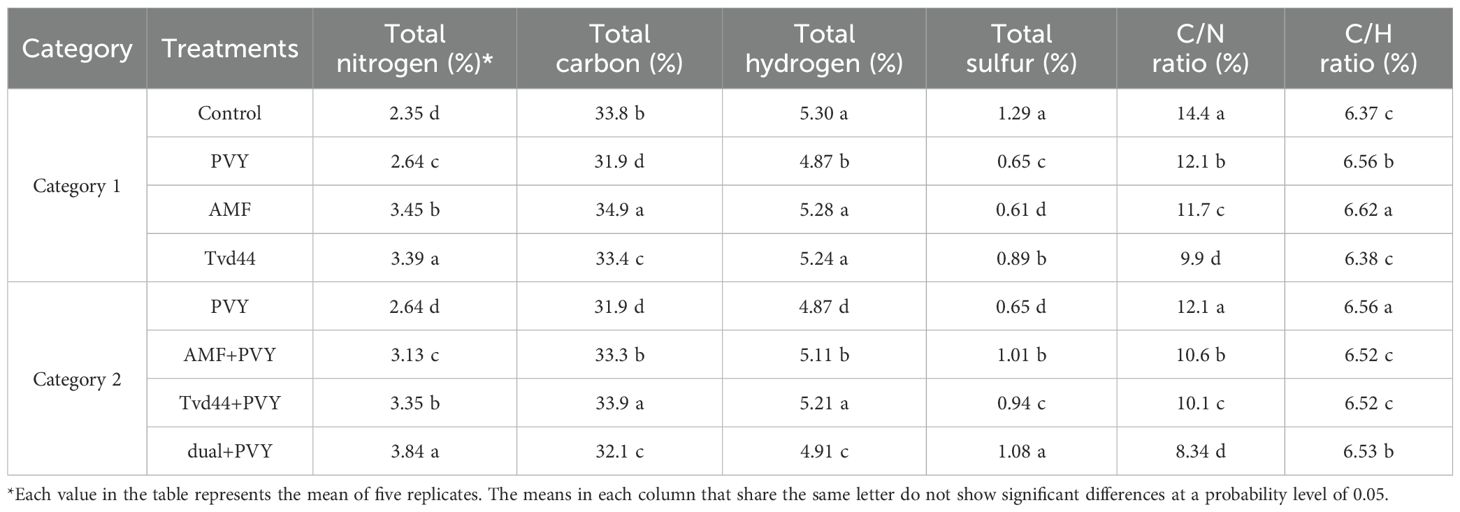
Table 2. Effect of AMF and T. viride isolate Tvd44 on the macronutrient contents in the tomato leaves.
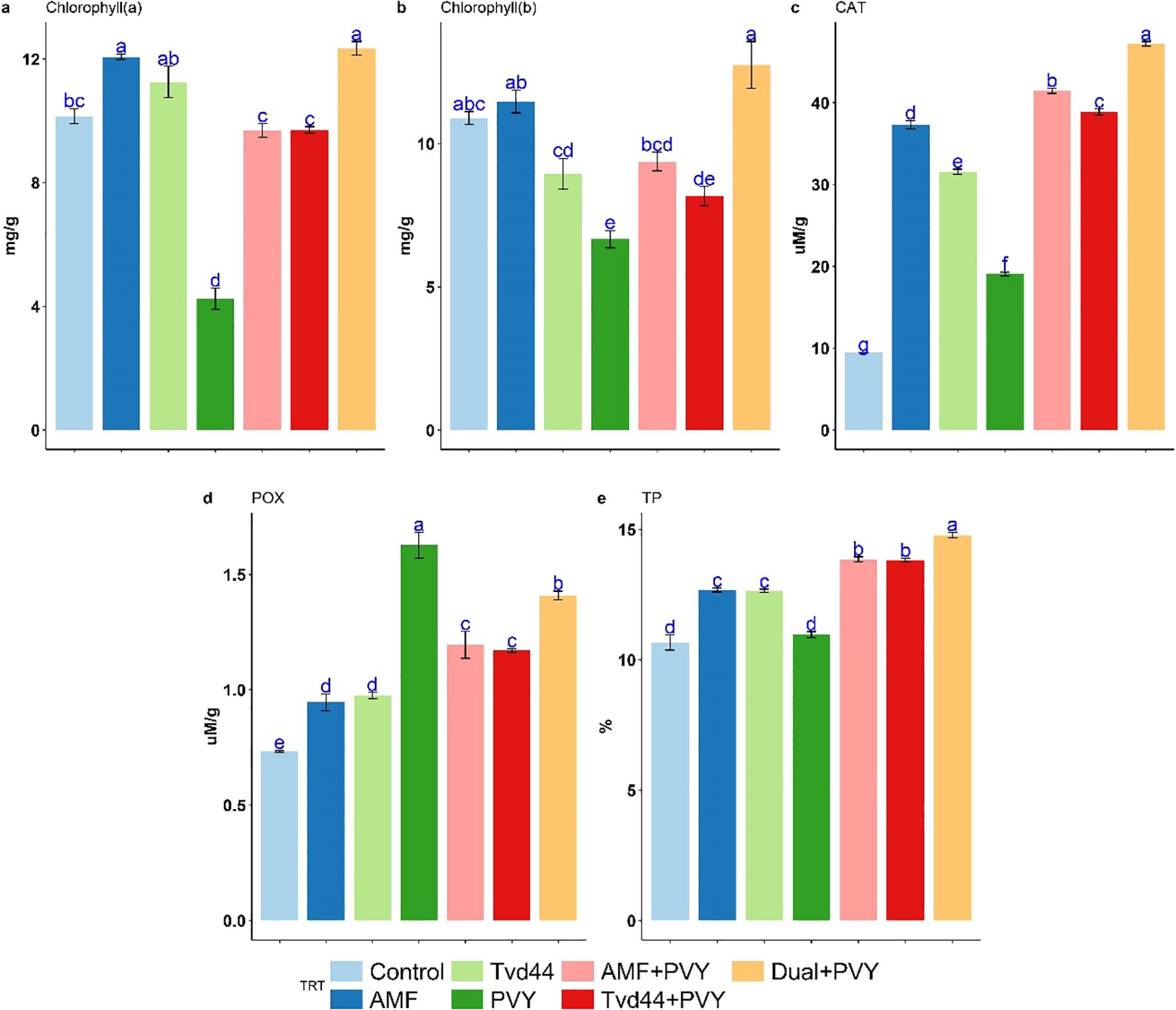
Figure 5. Impact of 7 treatments at 25 dpi on chlorophyll (a), chlorophyll (b), catalase (CAT) (c), peroxidase (POX) (d), and total protein (TP) (e) traits using Tukey's HSD test at p ≤ 0.05. Statistical significance was indicated alphabetically above the histogram in ascending order, whereas a>b>c>d. The means in each column that share the same letter do not show significant differences.
3.5 Transcriptional levels of defense-related genes
The present study assessed the relative expression levels of eleven genes involved in regulating the five key components of the defense pathways, using RT-qPCR at 25 dpi (Figure 6). The analysis revealed that the infection of tomato plants with PVY resulted in a 7.2-fold increase in the expression level of WRKY1 compared to the control group. Furthermore, the treatments AMF+PVY and Tvd44+PVY applications exhibited the highest expression levels of WRKY1, 66.3- and 71.9-fold increase, respectively (Figure 6). The expression levels of WRKY19 exhibited varying degrees of increase in response to PVY, AMF, and Tvd44 treatments compared to the control group. JERF3 exhibited stimulation following infection with PVY, AMF, and Tvd44 treatments compared to control plants. Concurrently, the expression levels of AMF+PVY, Tvd44+PVY, and the dual+PVY treatment increased by 2.9-, 2.9-, and 20.2-fold, respectively (Figure 6). Concerning GST1, the infected plants underwent treatment with AMV and Tvd44, resulting in a significant increase in the transcript level of GST1. Notably, the dual+PVY treatment elicited a more pronounced response than the treatments involving infection alone. The findings from Figure 6 indicate that all the treatments applied led to an increase in the expression levels of three genes: F3H and CHI2 at 25 dpi, with notable enhancement in the AMF+PVY, Tvd44+PVY, and dual+PVY treatment groups. Conversely, the PVY treatment showed no impact on the expression of FLS gene transcripts. In contrast, treatments involving AMF+PVY, Tvd44+PVY, and dual+PVY induction resulted in transcript gene expression levels of 59.9-, 45.7-, and 134.9-fold, respectively. The data presented in Figure 6 indicate no effect on the F3'H expression for the tomato plant treatments utilizing AMF or Tvd44 alone. However, an impact was noted for all applied treatments on the F3H gene expression, particularly in tomato plants infected with PVY, which exhibited a 3.54-fold increase. All applied treatments induce the expression of HQT gene transcripts in the chlorogenic pathway. In C3H, a downregulating effect was noted when treating AMV or Tvd44 individually, in contrast to all treatments infected with PVY, which increased gene transcript expression.
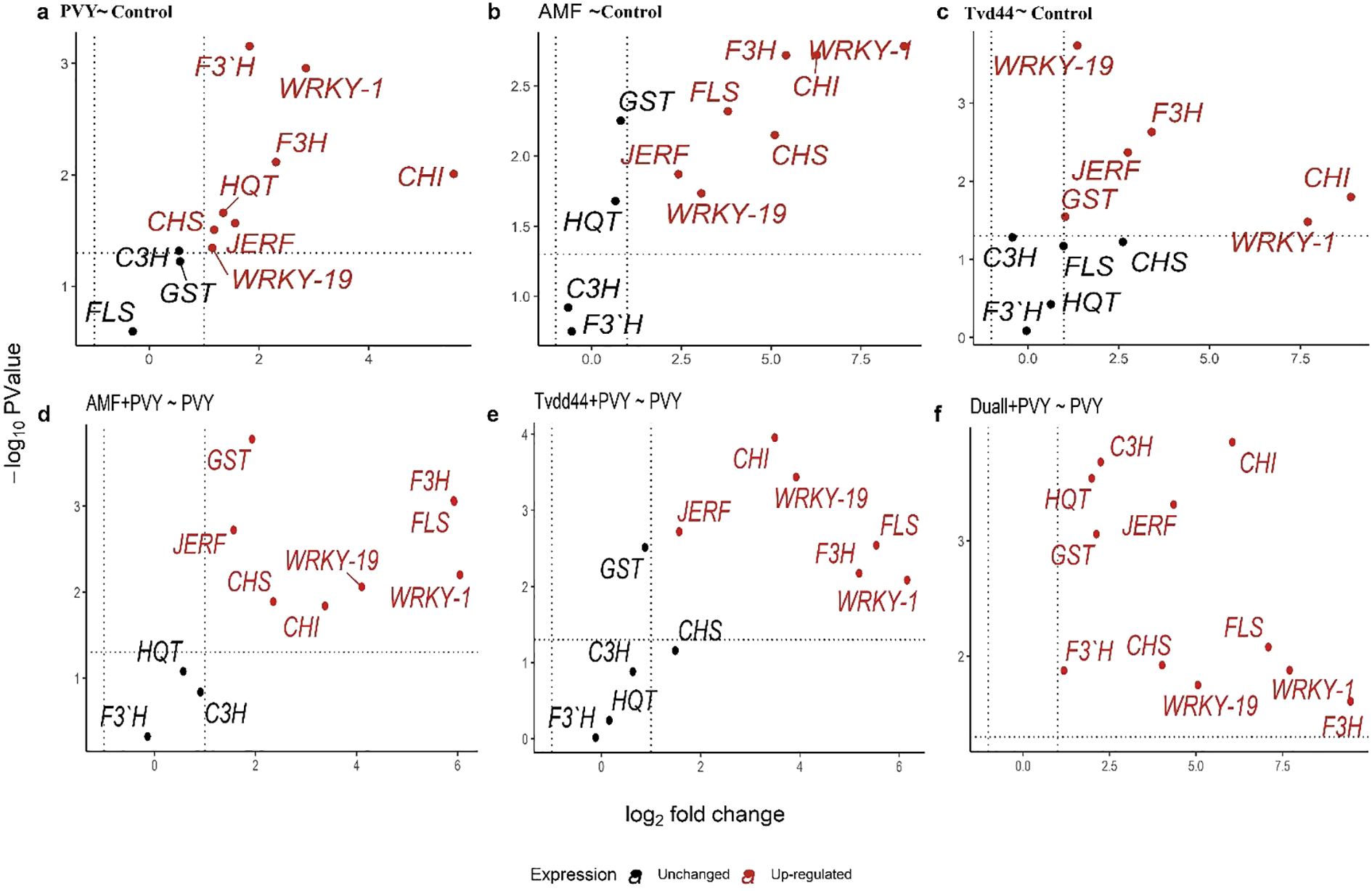
Figure 6. Volcano plots of change in gene expression for eleven genes in tomato leaves infected with PVY in response to AMF colonization and T. viride at 25 dpi. Where y-axis represents P-values (-log10), the x-axis represents fold change (log2), the black color represents unchanged genes, and the red color represents significantly upregulated genes. Because P-values on the y-axis in volcano plots were transformed and became negative, the higher the number on the y-axis, the smaller the P-value and the greater the significance. Threshold indicator dashed lines were drawn on volcano plots, where genes above the horizontal dashed line are significantly expressed at a p-value of 0.05. Genes beyond the right vertical dashed line are upregulated, genes behind the left vertical dashed line are downregulated, and genes between the two vertical dashed lines are unchanged. Relative gene expression changes among PVY and control (a), AMF and control (b), Tvd44 and control (c), AMF+PVY and PVY (d), Tvd44+PVY and PVY (e), and Dual+PVY and PVY (f) treatments.
3.6 Statistical significance of gene expression
Results from the volcano plots in Figure 6a showed that infected plants with the virus led to significant upregulation of all the studied genes except for GST1, C3H, and FLS. When plants were treated with AMF, as shown in Figure 6b, seven out of eleven genes were significantly upregulated (JERF3, WRKY1, WRKY19, CHS, CHI2, F3H, and FLS), while four genes (GST1, HQT, C3H, and F3’H) were unchanged. In Figure 6c, when treatment Tvd44 was applied, six genes were significantly upregulated (WRKY1, WRKY19, F3H, CHI2, JERF, and GST1), while five genes were unchanged (C3H, FLS, CHS, HQT, and F3’H). Figure 6d revealed that in plants treated with AMF+PVY, eight out of eleven genes were significantly upregulated (JERF3, WRKY1, WRKY19, CHS, CHI2, F3H, GST1, and FLS), while three genes (HQT, C3H, and F3’H) were unchanged. When the treatment Tvd44 was applied before virus infection (Tvd44+PVY), as shown in Figure 6e, six out of eleven genes were significantly upregulated (JERF3, WRKY1, WRKY19, CHI2, F3H, and FLS), while five genes (HQT, C3H, CHS, GST1, and F3’H) were unchanged. The application of dual+PVY treatments resulted in a significant upregulation of all eleven studied genes (Figure 6f).
3.7 Heatmap and cluster analysis of the differentially expressed genes
The heatmap in Figure 7 was constructed based on two treatment groups. The first group consists of treatments with PVY, AMF, and Tvd44 against the control, while the second group comprises treatments with AMF+PVY, Tvd44+PVY, and dual+PVY against PVY. The heatmap demonstrates that the eleven studied genes were grouped into four clusters based on their expression. Four genes in the second cluster (C3H, GST1, F3’H, and HQT) exhibited similar expression patterns across all treatments, meaning they responded similarly to each treatment. In contrast, two genes (WRKY1 and CHI2) in the third cluster and three genes (WRKY19, FLS, and F3H in the fourth cluster exhibited different expression patterns at each treatment. For example, in the dual+PVY~PVY treatment, the genes of the third and fourth clusters exhibited different expression levels (represented by different colors), with WRKY1 and F3H being highly expressed, followed by CHI2, FLS, and WRKY19. For the same treatment, the expression of the first cluster genes (JERF and CHS) was similar in their expression (same color). These results highlight the significant role of the third and fourth clusters of genes in response to PVY, compared to the first and second clusters.
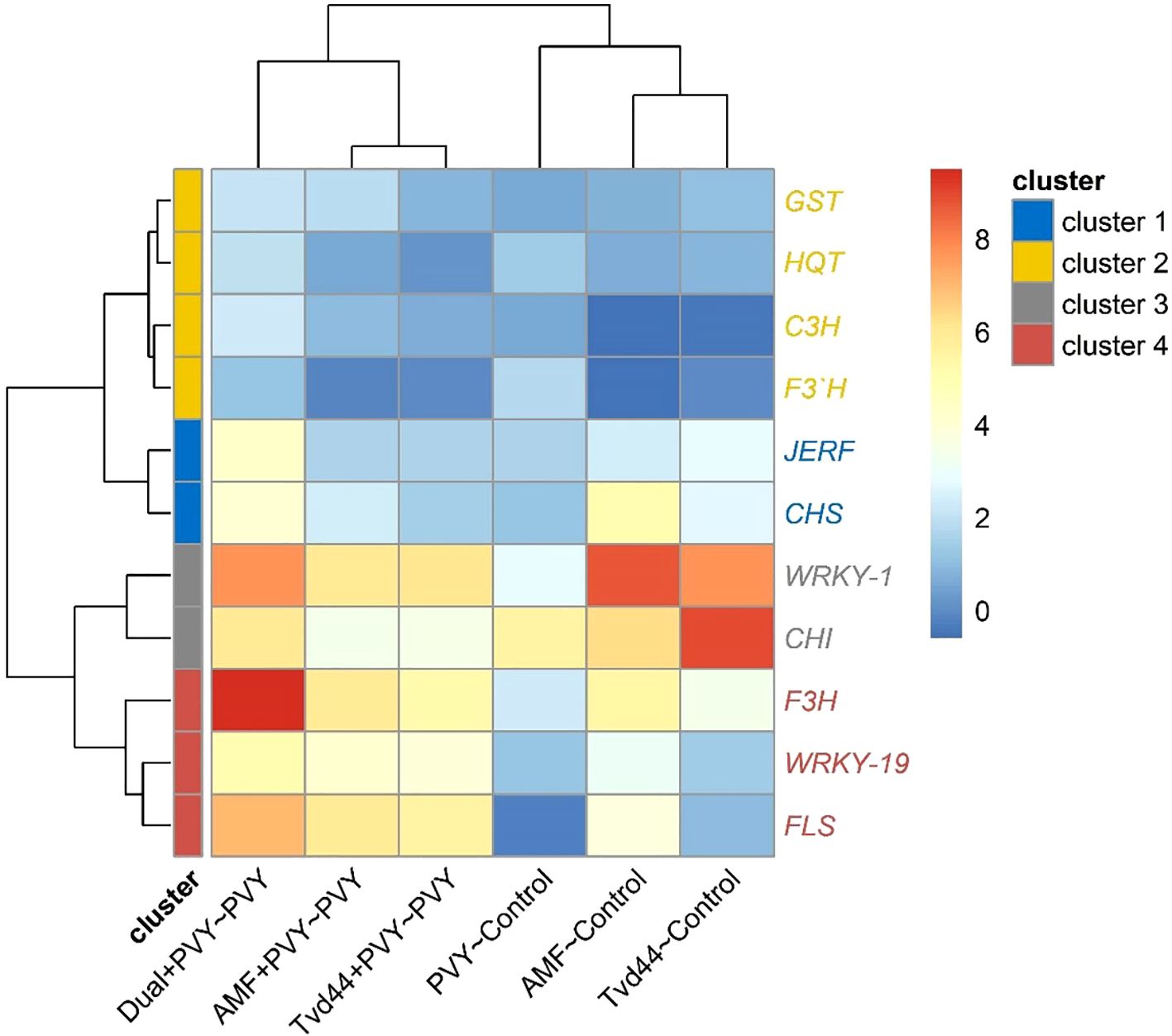
Figure 7. Heatmap and hierarchical clustering of change in gene expression for eleven genes in tomato leaves infected with PVY in response to AMF colonization and T. viride at 25 dpi. Cell color intensities were based on gene expression, represented as log2, where blue indicates downregulated genes and red indicates upregulated genes.
3.8 Principal component analysis
Figures 8a, b show that the first two principal components (PCs) account for 87.7% of the total variance, with 69.6% and 18.1%, respectively. The first principal component was represented by leaf number, chlorophyll (a), shoot length, shoot weight (both fresh and dry), and root dry weight, with higher loadings (longer arrows) located in the left area of the biplot. At the same time, the second principal component was represented by CAT, POX, and TP, which had higher loadings and were located in the upper center of the biplot (Figures 8c, d). These results illustrated that all traits were suitable for distinguishing between control and PVY treatments, as well as the rest.
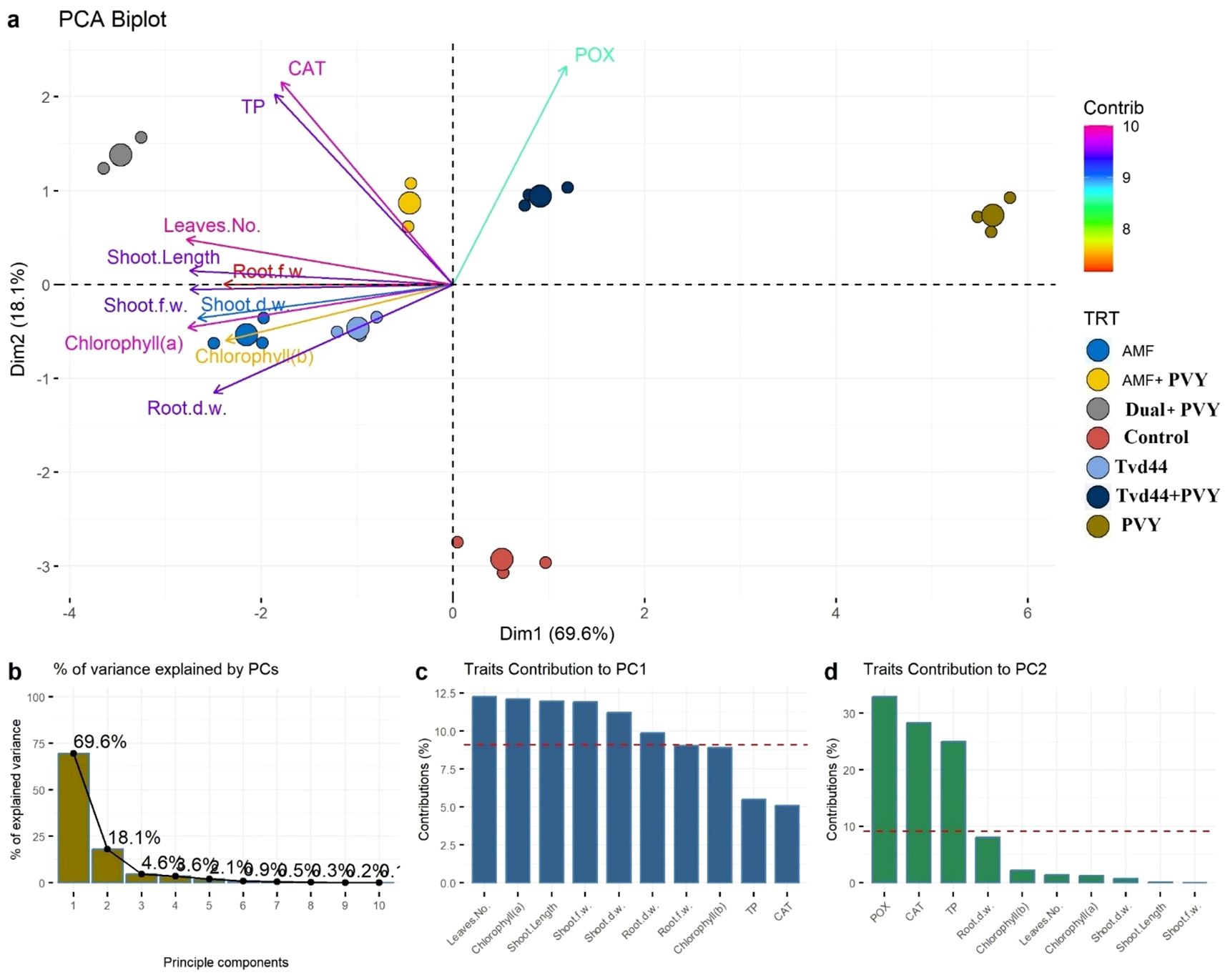
Figure 8. (a) Principal component analysis (PCA) biplot illustrates the contribution of each dimension to the total variance of 7 treatments and 11 physiological traits. Longer arrows specify higher contribution, while shorter arrows show lower contribution. (b) The bar plot indicates the percentage of contribution of PCs to the total variance. (c, d) bar plots of the contribution of the 11 studied traits to PCs where traits above the dashed red line specify significant contribution.
3.9 Gene expression and physiological traits network
Figures 9, 10 represent the network of eleven gene expressions and eleven physiological traits in response to the seven treatments. The network revealed that all eleven studied genes were significantly correlated, except for gene CH3 with JERF3, CHI2, HQT, C3H, F3’H, and CHI2 with F3’H. The least correlated physiological traits to both genes and other traits were root dry weight, POX, and chlorophyll (b). The most correlated were CAT, TP, and the number of leaves. On the other hand, the gene least correlated with both other genes and physiological traits was F3’H, while the most correlated ones were F3H, FLS, WRKY1, WRKY19, GST1, JERF3, and CHS. The most highly correlated gene with the physiological traits was CHS, which correlated with nine traits, followed by FLS, which correlated with eight traits, and CHI2, WRKY19, and F3H, which correlated with six traits each. GST1 and WRKY1 correlated with five traits, followed by JERF3, which correlated with four traits. Finally, HQT, F3’H, and C3H were the least correlated with the physiological traits, as they were correlated with only three traits. The network identified two local communities; the first community (nodes with blue borders) comprised 10 genes, excluding CHS, and three physiological traits (CAT, POX, and TP). The second community (nodes with green border) contained eight physiological traits, except for CAT, POX, TP, and one gene (CHS).
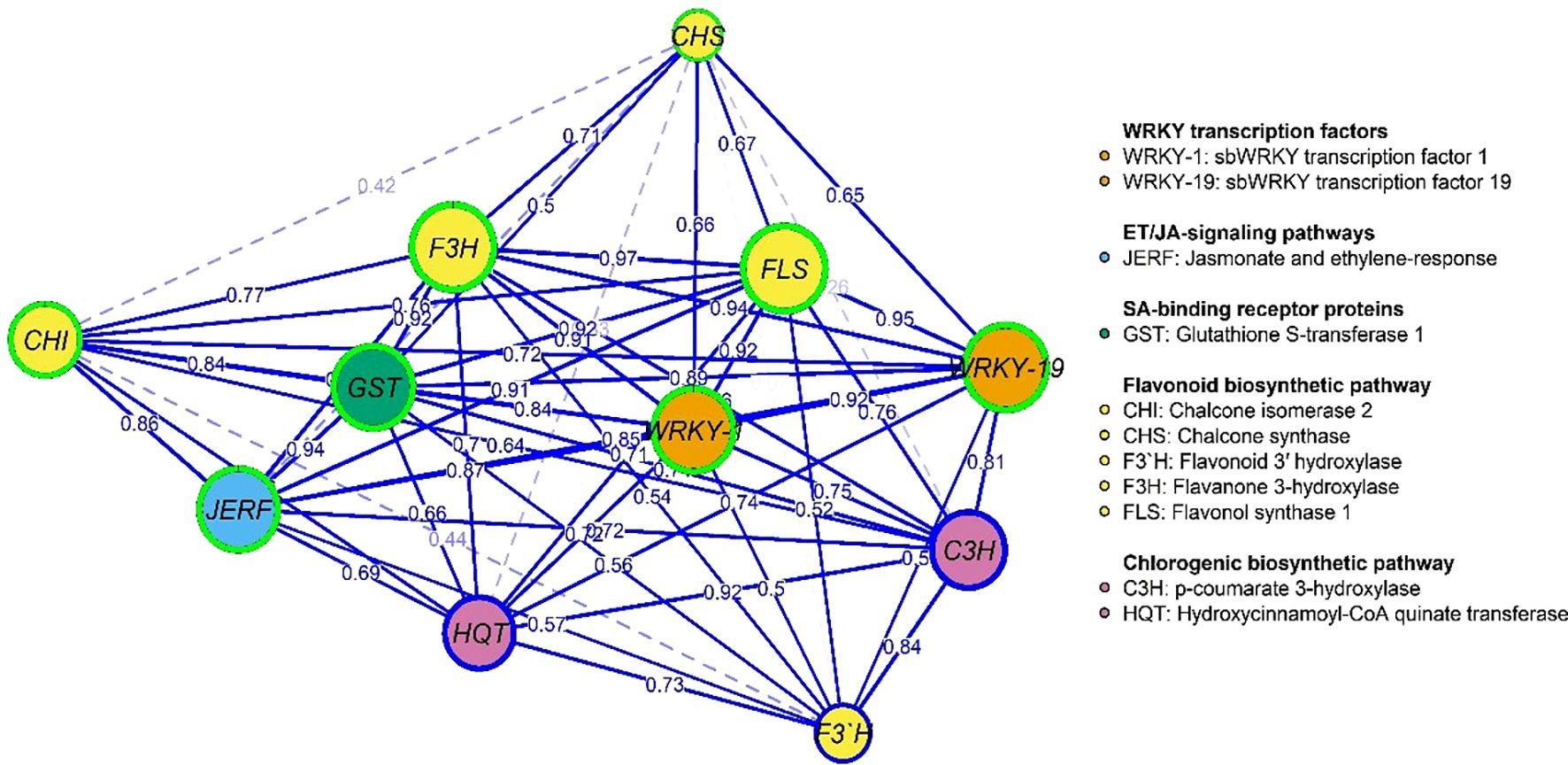
Figure 9. Gene co-expression network: nodes (circles) represent genes, while edges (lines) represent associations among genes based on the Spearman rank correlation coefficient. Faded lines represent non-significant correlations.
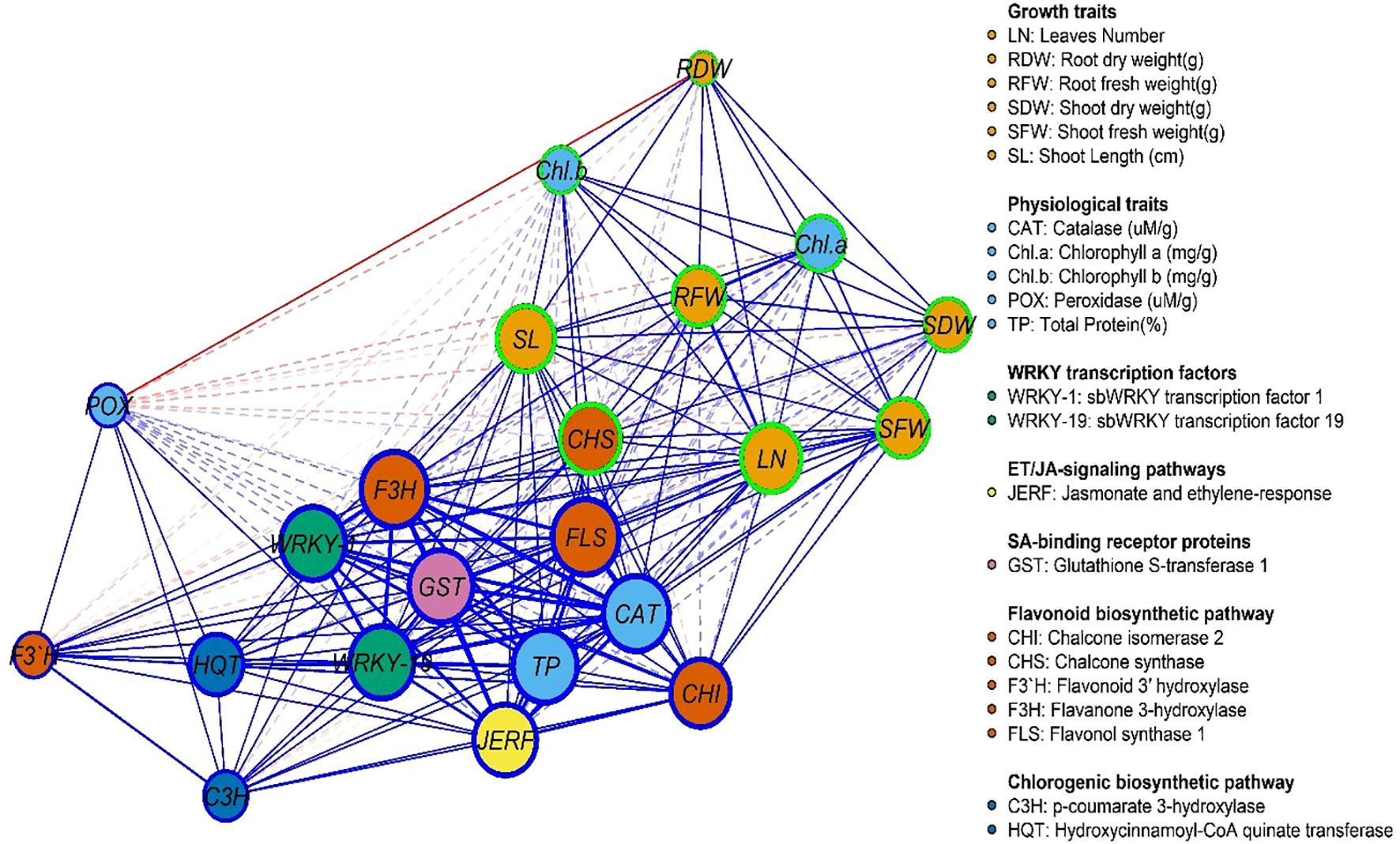
Figure 10. Gene and physiological traits network, where nodes (circles) represent genes and physiological traits, and edges (lines) represent associations among genes and physiological traits based on the Spearman rank correlation coefficient. Faded lines represent non-significant correlations.
4 Discussion
The effectiveness of defense priming in a biological system has been assessed based on several key criteria: enhanced defense responses, retention, better outcomes, and minimal suitability expenses (Riseh and Vazvani, 2024). Hormone-dependent signaling pathways are recognized for their potential to influence defense mechanisms during mycorrhiza formation (Pozo and Azcón-Aguilar, 2007). This mechanism, referred to as mycorrhiza-induced resistance, has the potential to “prime” plants, enabling them to respond to an impending stressor, such as a pathogen attack, with enhanced speed and strength. Arbuscular mycorrhizal fungi (AMF) and Trichoderma spp. represent two beneficial microorganisms frequently utilized as plant biostimulants to enhance crop yields, as they promote plant growth and mitigate rhizospheric infections (Chaithra et al., 2025). Alongside promoting plant growth, AMF enhances photosynthesis and boosts resilience against pests, diseases, and abiotic stress (Hashem et al., 2025). Trichoderma species enhance plant health by producing enzymes that break down fungal cell walls, thereby improving crop nutrition, growth, and stress resilience while triggering resistance mechanisms (Philip et al., 2024). Our analysis of the findings concerning these standards suggests that tomato plants can develop tolerance to PVY infection through mycorrhization and the use of Trichoderma spp. applications. Under greenhouse conditions, the foliar application of T. viride isolate Tvd44 on tomato leaves and the colonization of tomato roots with AMF before PVY inoculation reduced the mitigation of viral effects. Remarkably, the combined application (dual+PVY) significantly reduced disease incidence, disease severity, and viral accumulation levels by 40%, 53.4%, and 88.1%, respectively. The findings align with recent studies indicating that the use of AMF effectively manages zucchini yellow mosaic virus (ZYMV) in cucumber plants, leading to a reduction in both disease incidence and severity compared to untreated plants (Metwally and Abdelhameed, 2024). In the same context, Khoshkhatti et al. (2020) reported that at 20 dpi, tomato plants treated with Rhizoglomus irregularis significantly mitigated the symptoms of tomato bushy stunt virus (TBSV) compared to the untreated ones. Furthermore, Miozzi et al. (2020) noted that the colonization of Funneliformis mosseae influenced the tomato’s susceptibility to cucumber mosaic virus (CMV) infection at 14 dpi. Moreover, Aseel et al. (2023b) showed that T. viride activates the host’s innate immune response and/or initiates systemic acquired resistance, potentially diminishing PVY and/or inhibiting its accumulation. Maffei et al. (2014) noted that tomato plants colonized by AMF and infected with the tomato yellow leaf curl Sardinia virus (TYLCSV) exhibited reduced symptoms and a lower viral titer. Nonetheless, the colonization by AMF was insufficient to mitigate the reduction in root biomass caused by the virus.
The current study employed microscopic examination to reveal the various mycorrhizal structures present in the roots of tomatoes across all treatments colonized by AMF. Miozzi et al. (2020) observed that AMF colonization in virus-inoculated plants resulted in a slight increase in mycorrhization density and the ratio of arbuscules throughout the entire root system compared to healthy plants. Our finding aligns with a recent study demonstrating that AMF effectively colonizes plant roots, as observed through a light microscope (Metwally and Abdelhameed, 2024). Additionally, Khoshkhatti et al. (2020) found that the mean colonization ratio of mycorrhiza-inoculated plants before TBSV/ToMV infection was not significantly different from that of non-infected control plants with AMF. The findings from our treatments for AMF+PVY, Tvd44+PVY, and dual+PVY treatments demonstrated a significant enhancement in plant growth evaluation when compared to tomatoes that were solely inoculated with PVY. Furthermore, another study showed that under greenhouse conditions, AMF-root colonization significantly boosted all growth measurements compared to other treatments, with increases observed in shoot length, dry weight, and fresh weight even when compared to healthy plants that weren’t colonized (Metwally and Abdelhameed, 2024; Gaši et al., 2025; Subhash et al., 2025).
Plant viruses can infiltrate the intracellular spaces of plant cells, where they closely associate with cellular organelles and the cytoplasm, leading to oxidative stress (Abdelkhalek et al., 2025). Plants possess defense mechanisms that incorporate antioxidant enzymes within their cells, thereby shielding them from oxidative stress (Rabie et al., 2024). These enzymes play a crucial role in regulating reactive oxygen species (ROS) and mitigating oxidative stress to lipids, proteins, and nucleic acids. Moreover, PVY infection resulted in a reduction in photosynthetic pigments (chlorophyll a and b) and antioxidant enzymes, such as CAT, while simultaneously increasing POX levels and decreasing total phenolic content. Furthermore, these parameters were notably influenced by increased enhancements resulting from AMF root colonization and TVD44 across all treatments, including AMF+PVY, TVD44+PVY, and the dual+PVY treatments. Similarly, it was noted that tomato plants treated with T. viride and T. harzianum, when infected with ToMV, exhibited increased levels of chlorophyll (a and b) and enhanced CAT activity compared to healthy tomato plants inoculated with ToMV (Aseel et al., 2024). In a similar context, Venkatesan et al. (2010) noted that the presence of chlorotic and necrotic symptoms in all virus-infected plants correlates with a reduction in the net photosynthetic rate and chlorophyll levels. The viral infection leads to damage and aggregation of chloroplasts, ultimately causing the destruction or halt of chloroplast synthesis (Abdelkhalek et al., 2019b). The characteristics of the host plant, the specific virus isolate, environmental conditions, and the progression of disease development all contribute to the extent of photosynthetic suppression (Akbar et al., 2021). AMF induces a physiological state that enhances plants’ ability to react more swiftly and effectively to pathogen assaults. Aseel et al. (2019) demonstrated that tomato plants colonized by AM fungi exhibit reduced susceptibility to the tomato mosaic virus.
Concerning the effect of the treatments tested on PVY titer, our findings indicated that the application of AMF+PVY, Tvd44+PVY, and dual+PVY treatments resulted in a notable decrease in PVY-CP expression in the leaves of tomato plants. The findings are consistent with previously reported data, indicating that AMF-colonized plants can reduce PVY symptom severity and lower viral accumulation levels (Deja-Sikora et al., 2024). Similar results showed that using Trichoderma significantly lowered the viral accumulation levels in the treated plant tissues (Thiem et al., 2014; Aseel et al., 2023b). The application of AMF and Tvd44 resulted in a significant upregulation of all eleven genes studied across various pathways, including WRKYs, JA, SA, flavonoid, and chlorogenic acid. CHS serves as the first enzyme in the flavonoid pathway, and the primary metabolites it produces are essential for flavonoid synthesis across different plant tissues (André et al., 2009). A previous investigation revealed a notable buildup of isoflavonoid and flavonoid compounds exhibiting diverse antibacterial properties against various phytopathogens linked to the overexpression of CHS (Dao et al., 2011; Martínez et al., 2017), as well as the plant’s response to viral infections and vice versa (Niggeweg et al., 2004; Abdelkhalek et al., 2018). Chlorogenic acid helps plants defend against diseases and prevent infections, including those caused by viruses (Abdelkhalek et al., 2022b; Salmerón et al., 2025). We suggest that the increased transcription levels of CHS signify their antiviral characteristics, illustrating that tomato plants can utilize polyphenolic compounds as a unique defense mechanism against viral infection and spread.
A gene expression network was used to illustrate the pathway containing these genes, in which WRKY transcription factors (such as WRKY-1 and WRKY-19) control essential genes, including CHS, C3H, F3’H, and FLS, which in turn participate in the synthesis of flavonoids and other phenolic compounds crucial for defense. JERF factors also trigger defense responses through jasmonic acid signaling, which links to other plant immune responses (Roychowdhury et al., 2025). The detoxification of toxic chemicals and direct protection against infections are two functions of GSTs and CHI genes (Ellwanger et al., 2025). For instance, WRKY-TFs physically interact with proteins involved in plant defense, signaling, transcription, and other cellular processes to control and regulate plant signaling (Wani et al., 2021; Wang et al., 2025). The role of transcriptional factors from the WRKY family in plant virus infection remains inadequately explored. To fine-tune the defense response against various stresses, the defense signaling network, comprising numerous defense-associated genes, collaborates with WRKY members or other signaling proteins to regulate the overall expression of stress-responsive genes through autoregulation, cross-regulation, or protein-protein interactions (Freeborough et al., 2021). In this investigation, CHS gene is the master key in the physiological and pathway relationships of the genes (F3’H, HQT, C3H, GST, JERF, CHI, WRKY-1, WRKY-19, FLS, and F3H) in plant defense, which likely involves a complex network of interactions that regulate various aspects of plant defense responses, including the biosynthesis of flavonoids and other secondary metabolites and the activation of defense-related transcription factors. Overall, these genes create an interconnected system that governs the synthesis of secondary metabolites such as flavonoids and the management of defense response mechanisms, thereby enhancing the plant’s capacity to withstand pathogens and environmental stressors.
5 Conclusions
The current study investigated the impact of pre-inoculating tomato plants with arbuscular mycorrhizal fungi (AMF) and foliar application of T. viride against PVY. The colonization of AMF and the foliar application of T. viride significantly enhanced the tolerance of tomato plants to PVY by inducing systemic resistance and increasing the expression of defense-related genes and enzymes. The results indicated that the treatments improving the efficiency of defense responses in plants during pathogen attacks engage pathways governed by jasmonic acid (JA), salicylic acid (SA), and flavonoids. Consequently, treatments with AMF and T. viride alleviated the viral effects on the physiological and transcriptional disturbances induced by PVY infection. The symbiotic relationship between mycorrhizal fungi and most land plants suggests that these fungi, along with T. viride, could be viable, eco-friendly options for controlling viral diseases in crops.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
DA: Visualization, Data curation, Conceptualization, Writing – original draft, Methodology, Writing – review & editing. OI: Investigation, Validation, Writing – review & editing, Software, Formal Analysis, Writing – original draft. TE: Writing – review & editing. AA-A: Supervision, Writing – review & editing, Resources, Writing – original draft, Funding acquisition. AA: Writing – review & editing, Software, Conceptualization, Writing – original draft, Investigation, Validation, Methodology.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to extend their appreciation to the Ongoing Research Funding program (ORF-2025-505), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1650871/full#supplementary-material
References
Abdelkhalek, A., Al-Askar, A. A., Hamzah, K. A., Elbeaino, T., Moawad, H., El-Gendi, H., et al. (2025). Bacillus siamensis strain B30 as a biocontrol agent for enhancing systemic resistance and mitigating bean yellow mosaic virus infestation in faba bean plants. Eur. J. Plant Pathol. 171, 257–275. doi: 10.1007/s10658-024-02943-9
Abdelkhalek, A., Aseel, D. G., Király, L., Künstler, A., Moawad, H., and Al-Askar, A. A. (2022a). Induction of Systemic Resistance to Tobacco mosaic virus in Tomato through Foliar Application of Bacillus amyloliquefaciens Strain TBorg1 Culture Filtrate. Viruses 14, 1830. doi: 10.3390/v14081830
Abdelkhalek, A., Dessoky, E. S. E. S., and Hafez, E. (2018). Polyphenolic genes expression pattern and their role in viral resistance in tomato plant infected with Tobacco mosaic virus. Biosci. Res. 15, 3349–3356.
Abdelkhalek, A. and Hafez, E. (2019). “Plant viral diseases in Egypt and their control,” in Cottage Industry of Biocontrol Agents and Their Applications: Practical Aspects to Deal Biologically with Pests and Stresses Facing Strategic Crops (Cham, Switzerland: Springer), 403–421. doi: 10.1007/978-3-030-33161-0_13
Abdelkhalek, A., Ismail, I. A. I. A., Dessoky, E. S. E. S., El-Hallous, E. I. E. I., and Hafez, E. (2019a). A tomato kinesin-like protein is associated with Tobacco mosaic virus infection. Biotechnol. Biotechnol. Equip. 33, 1424–1433. doi: 10.1080/13102818.2019.1673207
Abdelkhalek, A., Király, L., Al-Mansori, A. N. A., Younes, H. A., Zeid, A., Elsharkawy, M. M., et al. (2022b). Defense Responses and Metabolic Changes Involving Phenylpropanoid Pathway and PR Genes in Squash (Cucurbita pepo L.) following Cucumber mosaic virus Infection. Plants 11, 1908. doi: 10.3390/plants11151908
Abdelkhalek, A., Qari, S. H. S. H., and Hafez, E. (2019b). Iris yellow spot virus–induced chloroplast malformation results in male sterility. J. Biosci. 44, 142. doi: 10.1007/s12038-019-9960-9
Abid Mehmood, M., Rauf, A., Abd-Elsalam, K. A., Ashfaq, M., Kayani, S. B., and Javeed, S. (2025). “The Multifaceted Biocontrol Mechanisms of Endophytic Fungi,” in Fungal Endophytes Volume II: Applications in Agroecosystems and Plant Protection (Singapore: Springer), 433–484.
Aeby, H. (1984). “Catalase in vitro,” in Methods in Enzymology (Academic Press, Elsevier), 105, 121–126. doi: 10.1016/s0076-6879(84)05016-3
Akbar, S., Yao, W., Qin, L., Yuan, Y., Powell, C. A., Chen, B., et al. (2021). Comparative analysis of sugar metabolites and their transporters in sugarcane following sugarcane mosaic virus (SCMV) infection. Int. J. Mol. Sci. 22, 13574. doi: 10.3390/ijms222413574
Al-Askar, A. A., Aseel, D. G., El-Gendi, H., Sobhy, S., Samy, M. A., Hamdy, E., et al. (2023). Antiviral Activity of Biosynthesized Silver Nanoparticles from Pomegranate (Punica granatum L.) Peel Extract against Tobacco Mosaic Virus. Plants 12, 2103. doi: 10.3390/plants12112103
André, C. M., Schafleitner, R., Legay, S., Lefèvre, I., Aliaga, C. A. A., Nomberto, G., et al. (2009). Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 70, 1107–1116. doi: 10.1016/j.phytochem.2009.07.008
Aseel, D. G., Alaa, A., Elsilk, S., and Gaafar, R. M. (2024). Protective and curative applications of some Trichoderma species to tomato plants infected by tomato mosaic virus. Egypt. J. Bot. 64, 109–129. doi: 10.21608/ejbo.2024.265650.2679
Aseel, D. G., Rashad, Y. M., and Hammad, S. M. (2019). Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against Tomato Mosaic Virus. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-46281-x
Aseel, D. G., Sobhy, S., Samy, M. A., Hamdy, E., Behiry, S. I., and Abdelkhalek, A. (2023a). Comparative analysis of the expression profiles of pathogenesis-related genes in tomato systemically infected with tobacco mosaic and cucumber mosaic viruses. Int. J. Plant Biol. 14, 458–473. doi: 10.3390/ijpb14020035
Aseel, D. G., Soliman, S. A., Al-Askar, A. A., Elkelish, A., Elbeaino, T., and Abdelkhalek, A. (2023b). Trichoderma viride Isolate Tvd44 Enhances Potato Growth and Stimulates the Defense System against Potato Virus Y. Horticulturae 9, 716. doi: 10.3390/horticulturae9060716
Behiry, S., Soliman, S. A., Massoud, M. A., Abdelbary, M., Kordy, A. M., Abdelkhalek, A., et al. (2023). Trichoderma pubescens Elicit Induced Systemic Resistance in Tomato Challenged by Rhizoctonia solani. J. Fungi 9, 167. doi: 10.3390/jof9020167
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chaithra, M., Nishmitha, K., Patel, P. S., Mitra, D., Gangaraj, R., and Sayyed, R. Z. (2025). “Fungal Bio-stimulants: Cutting-Edge Bioinoculants for Sustainable Agriculture,” in Plant Microbiome and Biological Control: Emerging trends and applications (Cham: Springer), 289–307. doi: 10.1007/978-3-031-75845-4_13
Charrad, M., Ghazzali, N., Boiteau, V., and Niknafs, A. (2014). NbClust: an R package for determining the relevant number of clusters in a data set. J. Stat. Software 61, 1–36. doi: 10.18637/jss.v061.i06
Chaudhary, A., Poudyal, S., and Kaundal, A. (2025). Role of arbuscular mycorrhizal fungi in maintaining sustainable agroecosystems. Appl. Microbiol. 5, 1. doi: 10.3390/applmicrobiol5010006
Dao, T. T. H., Linthorst, H. J. M., and Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi: 10.1007/s11101-011-9211-7
Deja-Sikora, E., Gołębiewski, M., and Hrynkiewicz, K. (2024). Transcriptomic responses of Solanum tuberosum cv. Pirol to arbuscular mycorrhiza and potato virus Y (PVY) infection. Plant Mol. Biol. 114, 123. doi: 10.1007/s11103-024-01519-9
El-Bilawy, E. H., Al-Mansori, A.-N. A., Alotibi, F. O., Al-Askar, A. A., Arishi, A. A., Teiba, I. I., et al. (2022). Antiviral and antifungal of Ulva fasciata extract: HPLC analysis of polyphenolic compounds. Sustainability 14, 12799. doi: 10.3390/su141912799
Ellwanger, J. H., Ziliotto, M., and Chies, J. A. B. (2025). Toxicogenomics of glutathione S-transferase (GST) gene family members: Chemical-gene interactions and potential implications of gene deletions. Comput. Biol. Med. 189, 110025. doi: 10.1016/j.compbiomed.2025.110025
Freeborough, W., Gentle, N., and Rey, M. E. C. (2021). WRKY transcription factors in cassava contribute to regulation of tolerance and susceptibility to cassava mosaic disease through stress responses. Viruses 13, 1820. doi: 10.3390/v13091820
Galeazzi, M. A. M., Sgarbieri, V. C., and CONSTANTINIDES, S. M. (1981). Isolation, purification and physicochemical characterization of polyphenoloxidases (PPO) from a dwarf variety of banana (Musa cavendishii, L). J. Food Sci. 46, 150–155. doi: 10.1111/j.1365-2621.1981.tb14551.x
Gaši, E., Radić, T., Gambino, G., Čarija, M., Matić, F., Balestrini, R., et al. (2025). Arbuscular mycorrhizal fungi modify temporal virus accumulation and distribution in different grapevine tissues. Phytobiomes J. 9, 139–148. doi: 10.1094/PBIOMES-06-24-0066-R
Harborne, A. J. (1998). Phytochemical methods a guide to modern techniques of plant analysis (Chapman & Hall: London, UK, Springer Science & Business Media), 1301. doi: 10.1007/978-94-009-5921-7
Hashem, A., Almutairi, K. F., Alshaikh, N. A., Kumar, A., Wu, Q.-S., Ibramimova, U., et al. (2025). “Role of arbuscular mycorrhizal fungi in plant growth promotion and biotic stress management,” in Management of Mycorrhizal Symbiosis for Mycoremediation and Phytostabilization (Elsevier), 145–155.
Horneck, D. A. and Miller, R. O. (2019). “Determination of total nitrogen in plant tissue,” in Handbook of reference methods for plant analysis (Boca Raton, FL, USA: CRC Press), 75–83.
Imran, M., Khan, M. A., Azeem, M., Ahmed, N., Binyamin, R., and Riaz, A. (2012). Screening of tomato germplasm for the source of resistance and its management against ToMV. Pak. J. Phytopathol. 24, 53–57.
Khoshkhatti, N., Eini, O., Koolivand, D., Pogiatzis, A., Klironomos, J. N., and Pakpour, S. (2020). Differential Response of Mycorrhizal Plants to Tomato bushy stunt virus and Tomato mosaic virus Infection. Microorganisms 8, 2038. doi: 10.3390/microorganisms8122038
Li, J., Zhou, L., Chen, G., Yao, M., Liu, Z., Li, X., et al. (2025). Arbuscular mycorrhizal fungi enhance drought resistance and alter microbial communities in maize rhizosphere soil. Environ. Technol. Innov. 37, 103947. doi: 10.1016/j.eti.2024.103947
Maffei, G., Miozzi, L., Fiorilli, V., Novero, M., Lanfranco, L., and Accotto, G. P. (2014). The arbuscular mycorrhizal symbiosis attenuates symptom severity and reduces virus concentration in tomato infected by Tomato yellow leaf curl Sardinia virus (TYLCSV). Mycorrhiza 24, 179–186. doi: 10.1007/s00572-013-0527-6
Mandal, A., Mukherjee, A., and Bandyopadhyay, R. (2025). “Plant Virus Disease Management Strategies: Conventional Versus Modern Techniques,” in Detection and Management of New and Emerging Mystery Plant Virus Sources (Palm Bay, FL, USA: Apple Academic Press), 207–238.
Martínez, G., Regente, M., Jacobi, S., Del Rio, M., Pinedo, M., and de la Canal, L. (2017). Chlorogenic acid is a fungicide active against phytopathogenic fungi. Pestic. Biochem. Physiol. 140, 30–35. doi: 10.1016/j.pestbp.2017.05.012
Maxwell, D. P. and Bateman, D. F. (1967). Changes in the activities of some oxidases in extracts of Rhizoctonia-infected bean hypocotyls in relation to lesion maturation. Phytopathology 57, 132–136.
Metwally, R. A. and Abdelhameed, R. E. (2024). Co-application of arbuscular mycorrhizal fungi and nano-ZnFe2O4 improves primary metabolites, enzymes and NPK status of pea (Pisum sativum L.) plants. J. Plant Nutr. 47, 468–486. doi: 10.1080/01904167.2023.2280121
Miozzi, L., Vaira, A. M., Brilli, F., Casarin, V., Berti, M., Ferrandino, A., et al. (2020). Arbuscular mycorrhizal symbiosis primes tolerance to cucumber mosaic virus in tomato. Viruses 12, 675. doi: 10.3390/v12060675
Morais, I. J., Silva, D. Y. M., Camargo, B. M., Lourenção, A. L., and Inoue-Nagata, A. K. (2025). Unraveling the dynamics of host specificity and resistance responses to potato virus Y, and implications for crop management. Trop. Plant Pathol. 50, 20. doi: 10.1007/s40858-025-00694-4
Niggeweg, R., Michael, A. J., and Martin, C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22, 746. doi: 10.1038/nbt966
Nunna, H., Palmer, N. A., Sarath, G., Wegulo, S. N., and Tatineni, S. (2025). Synergistic interaction between wheat streak mosaic virus and Triticum mosaic virus modulates wheat transcriptome to favor disease severity. Front. Plant Sci. 15, 1504482. doi: 10.3389/fpls.2024.1504482
Olsen, S. R. and Sommers, L. E. (1982). Phosphorus Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties. (Madison, WI, USA: American Society of Agronomy, Soil Science Society of America). doi: 10.2134/agronmonogr9.2.2ed.c24
Omar, A. Z., Hamdy, E., Hamed, E. A., Hafez, E., and Abdelkhalek, A. (2022). The curative activity of some arylidene dihydropyrimidine hydrazone against Tobacco mosaic virus infestation. J. Saudi Chem. Soc 26, 101504. doi: 10.1016/j.jscs.2022.101504
Philip, B., Behiry, S. I., Salem, M. Z. M., Amer, M. A., El-Samra, I. A., Abdelkhalek, A., et al. (2024). Trichoderma afroharzianum TRI07 metabolites inhibit Alternaria alternata growth and induce tomato defense-related enzymes. Sci. Rep. 14, 1874. doi: 10.1038/s41598-024-52301-2
Phillips, J. M. and Hayman, D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc 55, 158–IN18. doi: 10.1016/S0007-1536(70)80110-3
Pozo, M. J. and Azcón-Aguilar, C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398. doi: 10.1016/j.pbi.2007.05.004
Rabie, M., Aseel, D. G., Younes, H. A., Behiry, S. I., and Abdelkhalek, A. (2024). Transcriptional responses and secondary metabolites variation of tomato plant in response to tobacco mosaic virus infestation. Sci. Rep. 14, 19565. doi: 10.1038/s41598-024-69492-3
Ríos-Ruiz, W. F., Jave-Concepción, H. G., Torres-Chávez, E. E., Rios-Reategui, F., Padilla-Santa-Cruz, E., and Guevara-Pinedo, N. E. (2025). Plant-growth-promoting microorganisms: their impact on crop quality and yield, with a focus on rice. Int. J. Plant Biol. 16, 9. doi: 10.3390/ijpb16010009
Riseh, R. S. and Vazvani, M. G. (2024). Unveiling methods to stimulate plant resistance against pathogens. Front. Biosci. 29, 188. doi: 10.31083/j.fbl2905188
Rodríguez-Martínez, E. S., Torres-Torres, E., Guigón-López, C., and Alvarado-González, M. (2025). “Trichoderma Roles in Sustainable Agriculture,” in Sustainable Engineering and Agro-Food Processing (Palm Bay, FL, USA: Apple Academic Press), 147–187.
Roychowdhury, R., Hada, A., Biswas, S., Mishra, S., Prusty, M. R., Das, S. P., et al. (2025). Jasmonic acid (JA) in plant immune response: unravelling complex molecular mechanisms and networking of defence signalling against pathogens. J. Plant Growth Regul. 44, 89–114. doi: 10.1007/s00344-024-11264-4
Salmerón, A., del, M., Abreu, A. C., Tristán, A. I., Fernández, S., Gázquez-Expósito, J. E., et al. (2025). Metabolic profiling of tomato plants infected with tomato brown rugose fruit virus: insights into plant defense mechanisms and potential prebiotic interventions. ACS Agric. Sci. Technol. 5, 714–724. doi: 10.1021/acsagscitech.4c00557
Schmittgen, T. D. and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Subhash, A. P., Veena, S. S., Makeshkumar, T., and Anith, K. N. (2025). Piriformospora indica and Arbuscular Mycorrhizal Fungus suppress fungal root rot and mosaic diseases of cassava. Symbiosis 95, 241–254. doi: 10.1007/s13199-025-01044-3
Thiem, D., Szmidt-Jaworska, A., Baum, C., Muders, K., Niedojadlo, K., and Hrynkiewicz, K. (2014). Interactive physiological response of potato (Solanum tuberosum L.) plants to fungal colonization and Potato virus Y (PVY) infection. Acta Mycol. 49, 291–303. doi: 10.5586/am.2014.015
Tribodet, M., Glais, L., Kerlan, C., and Jacquot, E. (2005). Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. J. Gen. Virol. 86, 2101–2105. doi: 10.1099/vir.0.80926-0
Trouvelot, A., Kough, J. L., and Gianinazzi-Pearson, V. (1986). “Measure du taux de mycorrhization d’un systeme radiculaire. Recherche de methods d’estimation ayant une signification fonctionnelle.” in Proceedings of the Physiological and Genetical Aspects of Mycorrhizae. Proceedings of the 1st European Symposium on Mycorrhizae, eds Gianinazzi-Pearson, V. and Gianinazzi, S. (Paris: Institut National de la Recherche Agronomique), 217–221.
Vasan, S., Johny, L., Conlan, X. A., Singh, P. P., Cahill, D. M., and Adholeya, A. (2024). Differential phenolic patterns during arbuscular mycorrhizal symbiosis in tomato. Symbiosis 94, 139149. doi: 10.1007/s13199-024-01020-3
Venkatesan, S., Radjacommare, R., Nakkeeran, S., and Chandrasekaran, A. (2010). Effect of biocontrol agent, plant extracts and safe chemicals in suppression of mungbean yellow mosaic virus (MYMV) in black gram (Vigna mungo). Arch. Phytopathol. Plant Prot. 43, 59–72. doi: 10.1080/03235400701652508
Wang, W., Cao, H., Wang, J., and Zhang, H. (2025). Recent advances in functional assays of WRKY transcription factors in plant immunity against pathogens. Front. Plant Sci. 15, 1517595. doi: 10.3389/fpls.2024.1517595
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Keywords: arbuscular mycorrhizal, Trichoderma viride, PVY, defense genes, tomato
Citation: Aseel DG, Ibrahim OM, Elbeaino T, Al-Askar AA and Abdelkhalek A (2025) Impacts of arbuscular mycorrhizal and Trichoderma viride on enhancing physicochemical properties and triggering defense mechanisms of tomato plants challenged with potato virus Y. Front. Plant Sci. 16:1650871. doi: 10.3389/fpls.2025.1650871
Received: 20 June 2025; Accepted: 30 July 2025;
Published: 22 August 2025.
Edited by:
Katarzyna Otulak-Kozieł, Warsaw University of Life Sciences, PolandReviewed by:
Yan Liang, Northwest A&F University, ChinaDespoina Beris, Benaki Phytopathological Institute, Greece
Copyright © 2025 Aseel, Ibrahim, Elbeaino, Al-Askar and Abdelkhalek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dalia Gamil Aseel, ZGFsaWFnYW1pbDUyQGdtYWlsLmNvbQ==; Abdulaziz A. Al-Askar, YWFsYXNrYXJhQGtzdS5lZHUuc2E=; Ahmed Abdelkhalek, YWhtZWQuYWJkZWxraGFsZWtAaW5ob3J0LnBs
 Dalia Gamil Aseel
Dalia Gamil Aseel Omar M. Ibrahim
Omar M. Ibrahim Toufic Elbeaino
Toufic Elbeaino Abdulaziz A. Al-Askar
Abdulaziz A. Al-Askar Ahmed Abdelkhalek
Ahmed Abdelkhalek