- 1Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China
- 2Nanfan Research Institute, CAAS, Sanya, Hainan, China
- 3The National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 4Northwest Agricultural and Forestry University, Yangling, China
The soil-borne fungi Rhizoctonia cerealis is one of the major pathogens for the economically-important diseases sharp eyespot of common wheat (Triticum aestivum). Certain phytosulfokine receptor 1 (PSKR1) genes mediate resistance to diseases caused by biotrophic/hemibiotrophic pathogens in several plant species. Yet, none of wheat PSKR1 genes with positive effect on the innate immune responses to R. cerealis has been reported. Here, we characterize the phytosulfokine receptor 1-like gene TaPSKR1L-6A on wheat chromosome 6A, and determined functional role of it in wheat defense against R. cerealis. We used RNA-seq, BSMV-induced gene silencing and transgenic wheat to investigate the role of TaPSKR1L-6A in resistance to R. cerealis. The results showed that TaPSKR1L-6A has a resistance effect on R. cerealis. Transcription analysis showed that its expression level was upregulated when infected with sheath blight. Overexpression of TaPSKR1L-6A significantly increased sheath blight resistance, upregulated the expression of lignin synthesis and metabolism pathway genes, and increased lignin accumulation, which was opposite to the results of silencing TaPSKR1L-6A. Collectively, these results clearly suggest that TaPSKR1L-6A mediates resistance to sharp eyespot by activating the expression of several lignin synthesis-related genes and increasing lignin accumulation. Thus, TaPSKR1L-6A is a promising gene for improving wheat resistance against sharp eyespot. This study sheds light on wheat defense mechanisms against R. cerealis.
Introduction
Bread wheat (Triticum aestivum L.) is a globally vital staple crop, supplying approximately one-fifth of human dietary calories (International Wheat Genome Sequencing Consortium, 2018). Sharp eyespot, a serious disease primarily caused by the soil-borne fungus Rhizoctonia cerealis, affects grain cereals including bread wheat in major wheat-growing regions such as Australia, the USA, and China (Hamada et al., 2011). Therefore, the control of sharp eyespot is extremely urgent. Utilizing resistant wheat varieties represents a long-established, environmentally friendly, and economical control strategy. However, the majority of popular cultivars susceptible to this disease in China (Wu et al., 2017; Li et al., 2022). Therefore, identifying resistance genes and elucidating the underlying molecular mechanisms are vital for efficiently breeding wheat varieties with resistance to sharp eyespot.
Past studies indicate that resistance to sharp eyespot in wheat is controlled by multiple resistant genes (Wu et al., 2022). Several genes involved in the resistance response to sharp eyespot have been identified in wheat through approaches such as transcript expression analysis, gene silencing, and overexpression. For example, studies by Zhang et al. (2007) and Zhu et al. (2014) demonstrated that the pathogen-induced wheat ERF transcription factors TaERF3 and TaPIE1 transmit ethylene signals, upregulate the expression of defense genes, and consequently enhance wheat resistance to sharp eyespot. Shan et al (2016) and Liu et al. (2020) discovered, through experiments including Virus-Induced Gene Silencing (VIGS) and overexpression, that the R. cerealis-induced MYB transcription factor gene TaRIM1 and the GATA transcription factor gene TaGATA1 positively regulate wheat resistance to sharp eyespot. This regulation is associated with their ability to significantly upregulate the transcription levels of defense genes. Zhu et al. (2017) functionally analyzed a wheat R. cerealis-induced Nucleotide-Binding Site Leucine-Rich Repeat (NBS-LRR) class Resistance (R) gene, TaRCR1, using VIGS and transgenic wheat. They found that this gene enhances wheat resistance to the pathogen by promoting reactive oxygen species (ROS) scavenging and the expression of defense genes. Wang et al. (2020a), employing gene silencing and heterologous overexpression in Arabidopsis thaliana, revealed that TaELP4 increases the expression of defense genes and wheat plant resistance to sharp eyespot by regulating the acetylation of defense gene proteins. Wang et al. (2021) characterized the function of TaMKK5 in wheat resistance to sharp eyespot using overexpression and knockdown methods. They proposed that the TaMKK5-TaMPK3-TaERF3 signaling cascade regulates this resistance. Research by Guo et al (2021) indicated that the membrane protein TaCRK3 can directly inhibit the growth of R. cerealis through its extracellular DUF26 domain and activate the expression of disease resistance-related genes via its intracellular kinase domain. VIGS analysis by Qi et al. (2021a, 2021b) showed that silencing TaWAK7D and TaWAK-6D suppresses the expression of downstream defense response genes and attenuates wheat resistance to sharp eyespot. Zhu et al. (2022) found that the transcript abundance of the glycosyltransferase-encoding gene TaSTT3b-2B is positively correlated with wheat resistance to sharp eyespot. In summary, several candidate genes positively regulating wheat resistance to sharp eyespot have currently been identified. However, the resistance network of wheat against sharp eyespot remains to be fully elucidated.
To defend against pathogens, plants evolved an array of receptor-like kinases (RLKs) or receptor-like proteins to perceive pattern-associated molecular patterns (PAMPs) or damage-associated molecular patterns (Boutrot and Zipfel, 2017; Rhodes et al., 2021; Wang et al., 2020b; Zipfel, 2014). The disulfated pentapeptide Phytosulfokine (PSK) is an important signaling molecule, and can act as a PAMP in plant defenses to pathogen infection (Sauter, 2015; Yang et al., 2019). It is perceived by the LRR RLKs PSKR1 (phytosulfokine receptor 1) and PSKR2 (phytosulfokine receptor 2) (Matsubayashi et al., 2002; Zhang et al., 2018). PSK has been shown to bind to the island domain of PSKR1 that intersects between LRR17 and LRR18 of the extracellular receptor domain, from where the signal is transmitted to the intracelluar receptor kinase domain (Matsubayashi et al., 2002; Wang et al., 2015; Kaufmann et al., 2017). PSK binding enhances PSKR heterodimerization with somatic embryogenesis receptor-like kinases (SERKs) (Wang et al., 2015; Hohmann et al., 2018). Recently, PSKR1s in Arabidopsis, tomato and rice have been implicated in plant innate immune responses to pathogens. In Arabidopsis, AtPSKR1 could enhance defense against the necrotrophic fungal pathogen Alternaria brassicicola (Mosher et al., 2013). In tomato, SlPSKR1 participated in the auxin-mediated pathways and positively contributed to innate immunity to infection of the necrotrophic fungus Botrytis cinerea (Zhang et al., 2018). In rice, OsPSKR1 was positively involved in resistance to Xanthomonas oryzae pv. oryzicola by regulating the expression of pathogenesis-related genes in salicylic acid pathway (Yang et al., 2019). Our laboratory showed that TaRLK‐6A, a PSKR1-like gene locating on the chromosome 6A, could interact with TaSERK1 positively modulate the resistance to Fusarium crown rot in wheat (Qi et al., 2024). However, the functions of PSKR1 involved in resistance to R. cerealis, and lignin accumulation in wheat were scarcely reported.
In this study, by means of RNA-sequencing (RNA-Seq) of wheat against R. cerealis, barley stripe mosaic virus-induced VIGS, as well as transgenic assays, we found that the expression of TaPSKRL1-6A was significantly induced after R. cerealis infection and the induction was higher in sharp eyespot-resistant wheat cultivars than in susceptible cultivars. Overexpression of TaPSKRL1-6A significantly enhanced resistance to sharp eyespot, increasing the expression lignin synthesis genes, and lignin content in transgenic wheat, while silencing of TaPSKRL1-6A displayed the opposite effects. These findings not only demonstrate positive contribution of TaPSKRL1-6A to sharp eyespot resistance, but also provide insights on functional roles of PSKR1-like in innate immunity of plant species, especially wheat.
Results
TaPSKR1L-6A was involved in the wheat defense response to R. cerealis
By mining the RNA-seq data from resistant and susceptible lines of recombinant inbred lines (RILs) derived from the cross Shanhongmai × Wenmai 6, the gene with ID: TraesCS6A02G222000.1, named as TaPSKR1L-6A, was identified to be up-regulated in the resistant RILs relative to the susceptible RILs after R. cerealis strain Rc207 inoculation. The gene transcript level was higher by 2.18- and 1.14-fold in the resistant RILs relative to the susceptible lines at 4- and 10-days post inoculation (dpi) with the fungus, respectively (Figure 1A). RT-qPCR analysis showed that after inoculation with R. cerealis, TaPSKR1L-6A transcript level was significantly greater in resistant wheat cv.CI12633 than the susceptible wheat cultivar Wenmai 6 (Figure 1B), implying that the gene may be involved in the early defense response to the fungal infection. The analyses were in agreement with the gene transcript trend in the RNA-seq data. Importantly, among the four different wheat cultivars, the TaPSKR1L-6A transcript level was the highest in R. cerealis resistant wheat cultivar CI12633, followed by the resistant wheat Shanhongmai, but was the lowest in two highly susceptible cultivars Yangmai 9 and Wenmai 6 (Figure 1C). This result suggested that the transcript abundance of TaPSKR1L-6A was correlated to the resistant degree of the four wheat cultivars.
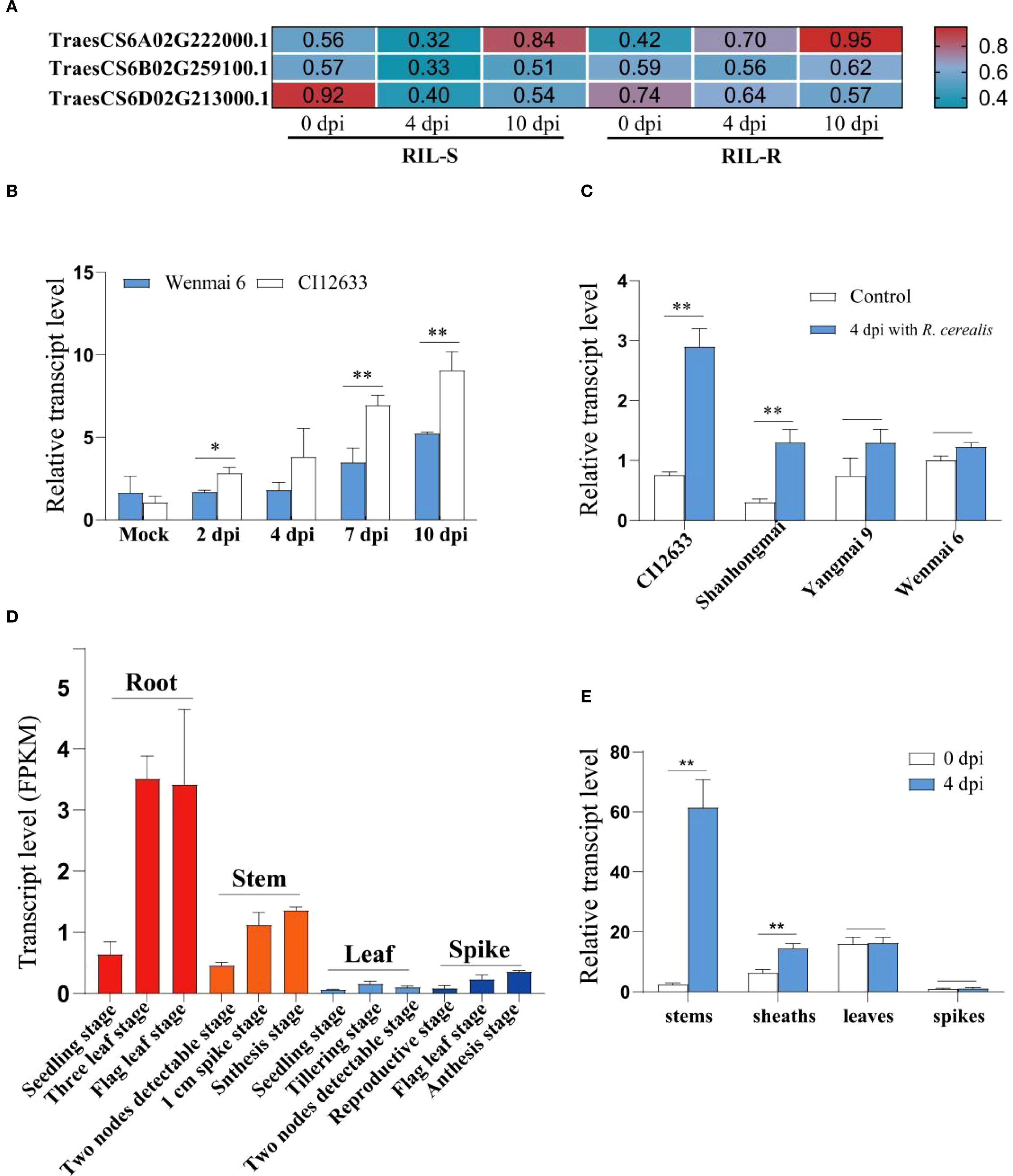
Figure 1. The expression pattern of TaPSKR1L-6A. (A) The transcript change of TaPSKR1L-6A in the recombinant inbred lines (RILs) upon R. cerealis infection. (B) Transcript patterns of TaPSKR1L-6A with R. cerealis infection. The TaPSKR1L-6A transcript level of wheat cultivar Wenmai 6 at non-treatment was set to 1. (C) Expression patterns of TaPSKR1L-6A in 4 wheat cultivars after R. cerealis infection. The expression level of TaPSKR1L-6A in Wenmai 6 at 0 dpi was set to 1. TaActin was used as an internal control gene (t-test: *P< 0.05; **P< 0.01). Bars indicate SEs of the mean. (D) Tissue-specific expression patterns of TaPSKR1L-6A, TaPSKR1L-6B, and TaPSKR1L-6D in the wheat cultivar Chinese Spring as determined from RNA-seq online data (http://www.wheat-expression.com, International Wheat Genome Sequencing Consortium (IWGSC), 2014). (E) Transcript pattern of TaPSKR1L-6A in stems, sheathes, leaves and spikes of CI12633 at 4 dpi with R. cerealis of non-treatment. TaActin was used as an internal control gene (t-test: *P< 0.05; **P< 0.01).
By querying the tissue-specific expression database of Chinese Spring wheat, it was found that the expression level of this gene is relatively high in root and stem tissues, but lower in leaves and spikes (Figure 1D). This expression pattern is consistent with the infection and occurrence sites of sharp eyespot. And the expression patterns of different tissues showed that at 4 dpi with R. cerealis, the higher-level change appeared at the stems and sheaths of CI12633 seedlings (Figure 1E), where sharp eyespot disease usually occurs. These data suggested that TaPSKR1L-6A might participate in wheat resistance responses to R. cerealis infection.
Sequencing of TaPSKR1L-6A promoter in 61 wheat cultivars indicated that a SNP (T to C) at -1191 sit formed two TaPSKR1L-6A haplotypes in this population (Supplementary Figure S1A). To compare the contributions of TaPSKR1L-6A Hap I and Hap II to wheat sharp eyespot resistance, we examined the resistance of the population to R. cerealis. As shown in Supplementary Figure S1B, the disease index of wheat cultivars containing Hap I was significantly lower than that of cultivars containing Hap II. These results suggest that the TaPSKR1L-6A Hap I allelic variant is associated with higher sharp eyespot resistance compared with TaPSKR1L-6A Hap II. Curiously, only 9.84% of wheat cultivars had Hap I (Supplementary Figure S1C), suggested that this haplotype was not fully selected during the modern wheat breeding process.
TaPSKR1L-6A positively regulates resistance to sharp eyespot in wheat
To investigate TaPSKR1L-6A-mediated resistance to sharp eyespot in wheat, we generated TaPSKR1L-6A-silenced plants by Barley stripe mosaic virus (BSMV) mediated silencing assay (VIGS). The TaPSKR1L-6A-silenced and BSMV: GFP-infected(control)wheat plants were individually inoculated with R. cerealis strain RC207. At 30 dpi with R. cerealis, compared with BSMV: GFP-infected CI12633 plants, the stems of TaPSKR1L-6A-silenced CI12633 plants exhibited more serious disease severity of sharp eyespot with larger necrotic areas (Figure 2A). In the disease tests of two VIGS batches, the disease indices of TaPSKR1L-6A-silenced CI12633 plants were 59.2 and 62, respectively, while that of BSMV: GFP-infected CI12633 plants were 43.8 and 41.6, respectively (Figure 2B). These data suggested that the expression of TaPSKR1L-6A is required for resistance of wheat to R. cerealis.
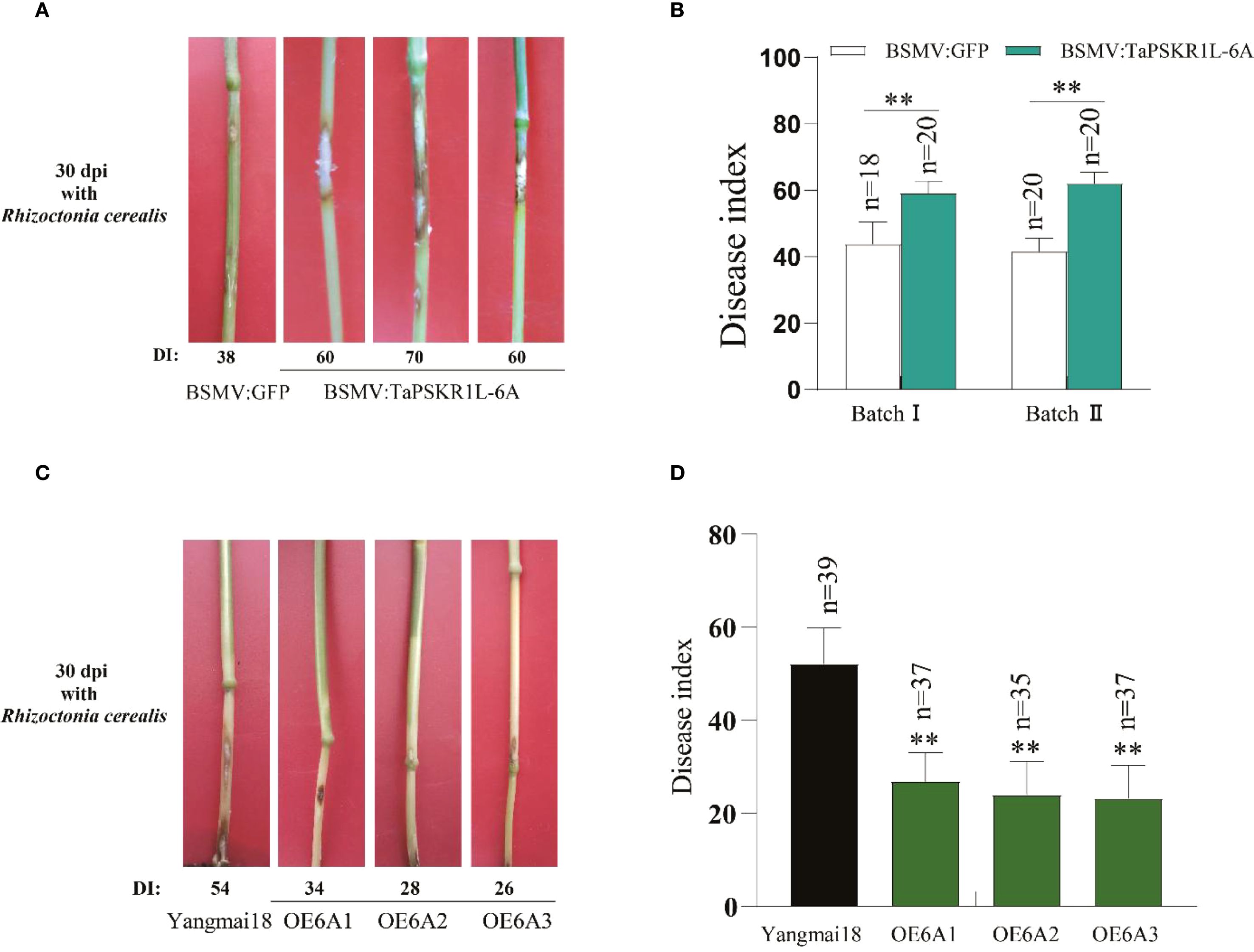
Figure 2. TaPSKR1L-6A positively regulates resistance to sharp eyespot in wheat. (A) sharp eyespot symptoms on TaPSKR1L-6A-silenced and BSMV: GFP-infected (control) CI12633 plants at 30 dpi with R. cerealis. (B) Disease index of TaPSKR1L-6A-silenced or control CI12633 plants at 30 dpi with R. cerealis in two independent batches (t-test: **P< 0.01). Bars indicate SEs of the mean. (C) Typical symptoms of sharp eyespot in the three TaPSKR1L-6A overexpressing and WT ‘Yangmai18’ lines at 30 dpi with R. cerealis. (D) Disease indexes of TaPSKR1L-6A overexpressing and WT ‘Yangmai18’ lines. Bars indicate SEs of the means (n varies for each column and is shown in each case directly on the graphs), and asterisks indicate significant differences between WT and transgenic lines using Student’s t-tests (**P< 0.01).
To further examine the function of TaPSKR1L-6A in wheat resistant response to sharp eyespot, we generated TaPSKR1L-6A transgenic plants. These plants in T2 generation were further inoculated with R. cerealis strain Rc207 to assess the resistance role of TaPSKR1L-6A. At 30 dpi, the sharp eyespot lesion sizes were smaller on the stems of TaPSKR1L-6A overexpressing wheat plants than the WT plants (Figure 2C). In disease assessments, the disease indexes of three TaPSKR1L-6A-6 overexpression wheat lines were 26.85, 23.93, and 23.25, respectively; whereas the disease index of the WT recipient ‘Yangmai18’ plants was 52.16 (Figure 2D; Supplementary Table S1). These results indicated that compared to WT plants, overexpression of TaPSKR1L-6A significantly increased resistance to sharp eyespot in the transgenic wheat lines.
TaPSKR1L-6A positively regulate lignin synthesis in wheat
During the evaluation on resistance of transgenic wheat to sharp eyespot, we found the stem strength in TaPSKR1L-6A overexpression wheat plants seemed to be higher than that in WT ‘Yangmai18’ plants. Therefore, we analyzed the lignin content of TaPSKR1L-6A overexpression and WT wheat plants. The results showed that the lignin content in the TaPSKR1L-6A overexpression transgenic plants was significantly higher than that in the WT plants (Figure 3A). Furthermore, hand-cut sections from leaf sheath of the TaPSKR1L-6A overexpression and WT plants were subjected to Wiesner and Mäule staining. As a result, compared to the WT samples, the samples of the TaPSKR1L-6A overexpression plants displayed more strengthened staining (red-brown) (Figure 3B), which was indicative of a higher lignin level. Additionally, compared with the WT plants, the TaPSKR1L-6A over-expression wheat plants had more sclerenchyma cells, which also can be detected by Mäule and Wiesner (Figures 3B, C) staining. These results indicated that overexpression of TaPSKR1L-6A enhanced lignin accumulation. Eventually, we measured the stem mechanical (breaking) strength of the second basal internode in TaPSKR1L-6A overexpressing and ‘Yangmai18’ wheat plants at harvest stage. The results showed that, TaPSKR1L-6A overexpressing lines exhibited significantly increased mechanical strength than those in ‘Yangmai18’ wheat plants (Supplementary Figure S2).
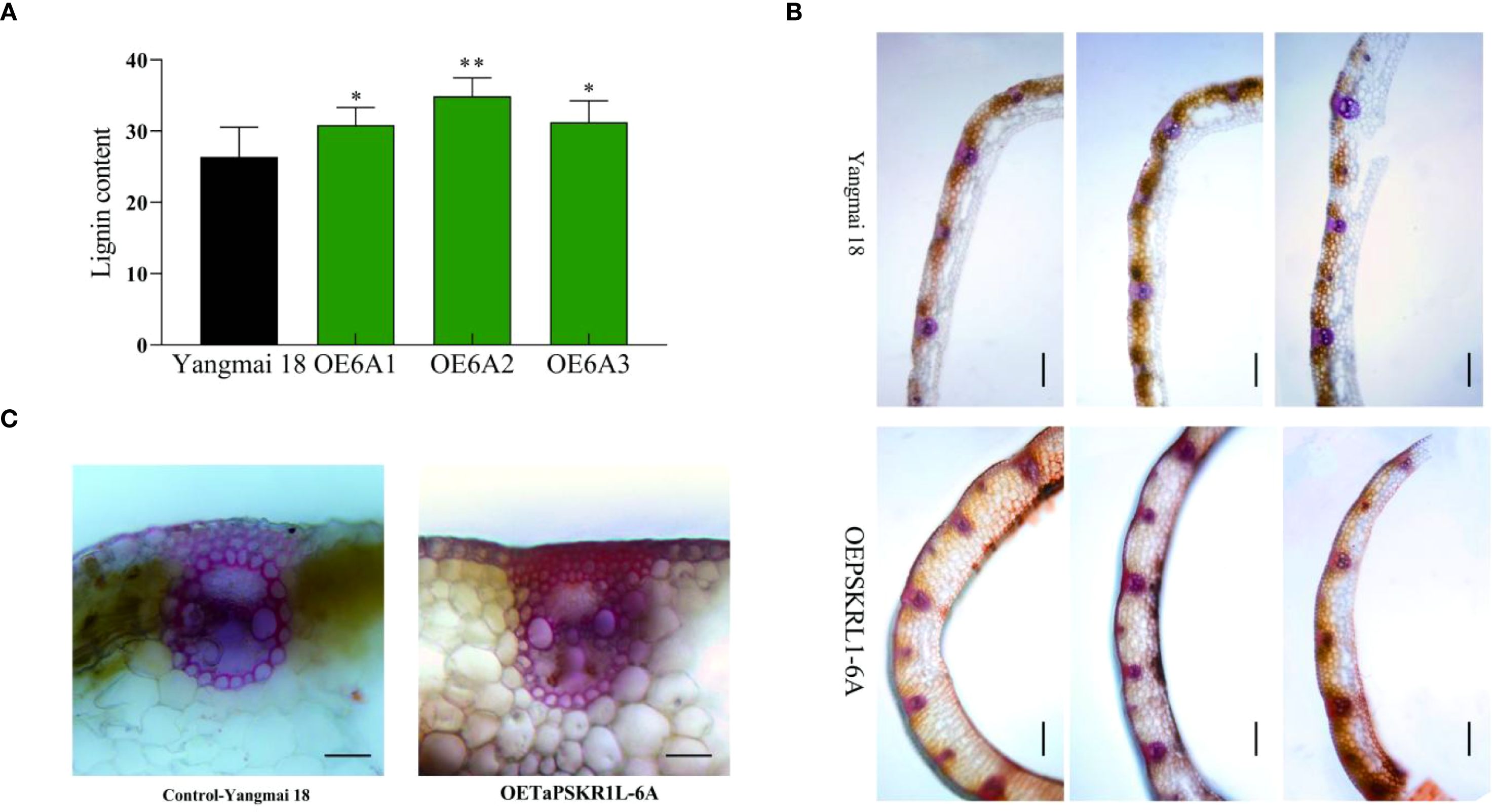
Figure 3. TaPSKR1L-6A overexpression increases lignin content in transgenic wheat plants. (A) Lignin content was higher in TaPSKR1L-6A overexpression plants than WT ‘Yangmai 18’ by quantitative measurement. Lignin content was quantitatively measured by using the lignin ELISA Kit (ChemFaces, China) according to the manufacturer’s protocol. Three biological replicates per line were averaged (t-test; **P< 0.01,*p<0.05). Bars indicate SEs of the means. (B) Lignin staining of the TaPSKR1L-6A overexpression and WT ‘Yangmai 18’ plants. Hand-cut sections from leaf sheaths were subjected to Mäule staining. Bar=100 μm. (C) The results of Wiesner staining showed that TaPSKR1L-6A overexpressing wheat plants had more sclerenchyma cells than WT ‘Yangmai 18’ plants. Bar=50 μm.
To investigate why the lignin content is higher in TaPSKR1L-6A overexpression plants, we measured the expression of several lignin synthesis-related genes in TaPSKR1L-6A- overexpression and silenced wheat plants as well as their controls. The results showed that transcript levels of lignin synthesis-related genes, TaPAL5, TaCOMT-3D, and TaCCR1, were higher in TaPSKR1L-6A overexpression plants than in the WT (control) plants (Figure 4A). While transcript levels of TaPAL5 and TaCOMT-3D were significantly lower in TaPSKR1L-6A-silenced plants than in control BSMV: GFP-infected plants, but transcript levels of TaCCR1 had no significant difference between TaPSKR1L-6A-silenced plants and control plants (Figure 4B). The results indicated TaPSKR1L-6A positively regulated lignin synthesis through modulating the expression of at least TaPAL5 and TaCOMT-3D.
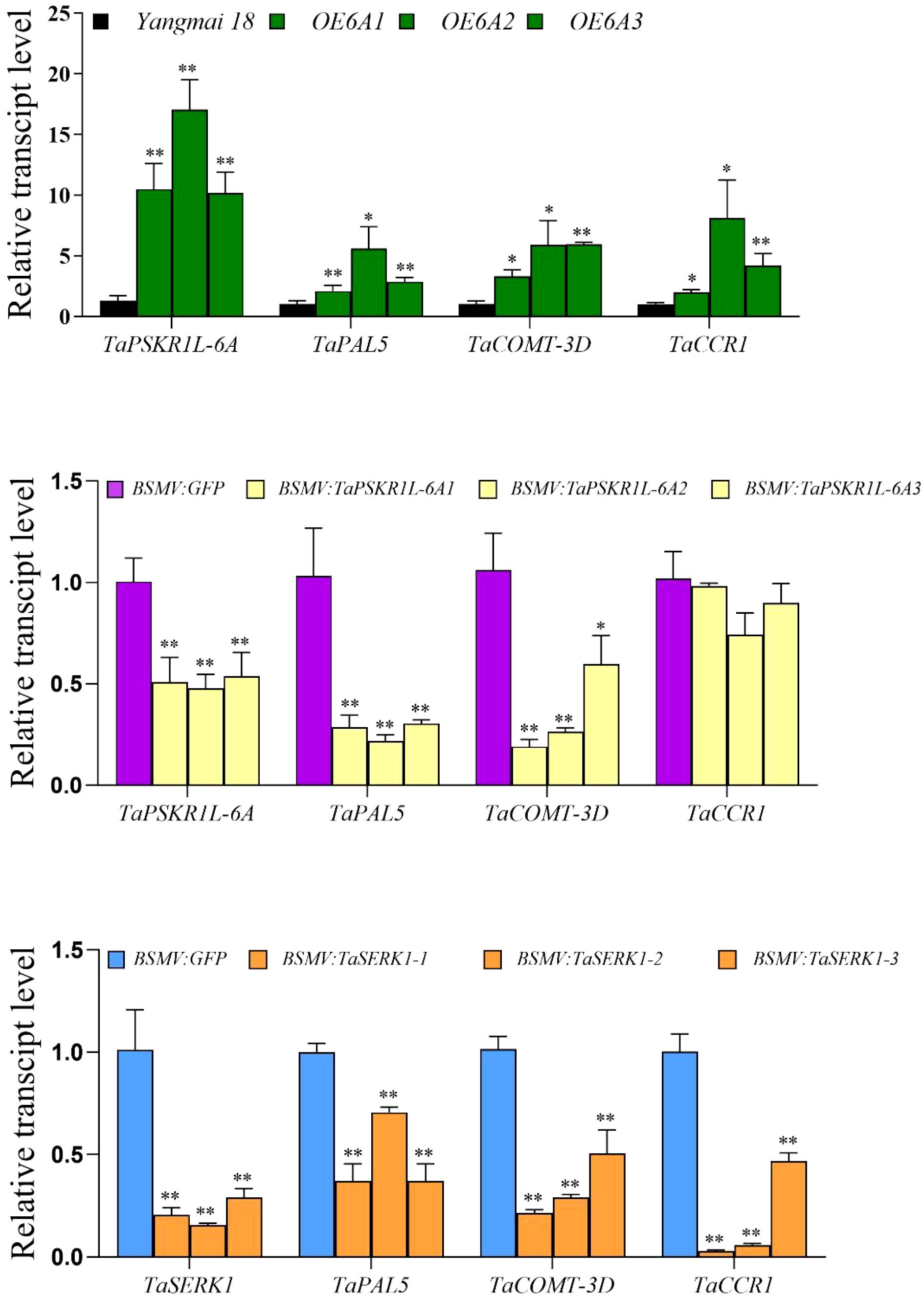
Figure 4. Lignin synthesis genes were regulated by TaPSKR1L-6A and TaSERK1. (A) Relative transcript abundances of three lignin synthesis genes in TaPSKR1L-6A over-expressing wheat plants and WT wheat ‘Yangmai 18’ plants. (B) Relative transcript abundances of three lignin synthesis genes in BSMV: GFP -infected and BSMV: TaPSKR1L-6A-silenced wheat CI12633 plants. (C) Relative transcript abundances of three lignin synthesis genes in BSMV: GFP-infected and BSMV: TaSERK1-silenced wheat plants at 4 dpi with R. cerealis. All samples were taken from sheaths of wheat plants. Statistically significant differences were determined based on three replications using a t-test (*P< 0.05; **P< 0.01). Bars indicate the SEs of the mean. TaActin was used as the internal reference gene to normalize the relative expression.
Our previous article demonstrated that TaPSKR1L-6A could interact with TaSERK1 to co-regulate wheat resistance to Fusarium crown rot (Qi et al., 2024). To analysis whether the TaSERK1 was related with lignin synthesis in wheat. We generated TaSERK1-silenced wheat plants and tested transcripts of the three lignin synthesis-related genes (TaPAL5, TaCOMT-3D, and TaCCR1) that are up-regulated by TaPSKR1L-6A. The results showed that transcript levels of TaPAL5, TaCOMT-3D, and TaCCR1 were significantly lower in TaSERK1-silenced plants compared with the control plants (Figure 4C). These data suggest that TaPSKR1L-6A and TaSERK1 co-regulated the lignin synthesis to enhance stem strength.
Discussion
Recently, sharp eyespot, mainly caused by soil-borne fungus R. cerealis, has become an important devastating disease of wheat in many regions of the world. Given the polygenic nature of sharp eyespot resistance, research should focus on expanding the wheat disease resistance network, identifying major QTL, and pyramiding multiple resistance genes using molecular markers to gradually enhance wheat resistance to this disease. Although the PSKR1 genes have been reported to involve in innate immunity against pathogens in Arabidopsis, tomato, and rice (Mosher et al., 2013; Zhang et al., 2018; Yang et al., 2019b), the function role of PSKR1 in wheat resistance responses has not been reported yet. In this study, we identified TaPSKR1L-6A as a candidate resistance gene of wheat against sharp eyespot, its transcript induction by infection of R. cerealis was higher in the tested sharp eyespot-resistant wheat cultivars than those in the susceptible wheat cultivars. Further functional dissection results showed that overexpression of TaPSKR1L-6A significantly enhanced wheat resistance to sharp eyespot, while silencing of TaPSKR1L-6A compromised resistance of wheat to sharp eyespot. Taken together, these results indicated that TaPSKR1L-6A confers wheat resistance to sharp eyespot and thus TaPSKR1L-6A is a promising gene for improving wheat resistance to sharp eyespot.
Downstream pathway of the PSKR-triggered immunity is not well understood (Yang et al., 2019). In Arabidopsis thaliana, AtCCR1 is important for lignin synthesis (Meester et al., 2018). In rice, OsPALs were reported to mediate resistance to brown plant hopper by regulating the biosynthesis and accumulation of salicylic acid and lignin (He et al., 2020). In wheat, overexpression of TaCOMT-3D was demonstrated to enhance wheat resistance against sharp eyespot and increase stem mechanical strength through promoting lignin accumulation, while silencing of TaCOMT-3D had opposite effects (Wang et al., 2018). In this study, the experiments indicated that overexpression of TaPSKR1L-6A elevated the expression of lignin synthesis genes (TaPAL5, TaCOMT-3D, and TaCCR1), increased lignin content/accumulation and the stem strength in the resistant transgenic wheat lines, while silencing of TaPSKR1L-6A had opposite effects. Moreover, the TaPSKR1L-6A overexpression plants had more sclerenchyma cells and deeper lignin staining compared to the WT plants, suggesting that overexpression of TaPSKR1L-6A enhanced lignin accumulation and promoted the development of vascular bundles.
Our previous article demonstrated that TaPSKR1L-6A could interact with TaSERK1 to co-regulate wheat resistance to Fusarium crown rot (Qi et al., 2024). In this study, we found the expression levels of TaPAL5, TaCOMT-3D, and TaCCR1 in TaSERK1-silenced plants were reduced compared to the control wheat plants. As the SERK proteins were reported to be related to the development of vascular bundles (Zhang et al., 2016; Li et al., 2019). Taken together, these results indicated that TaPSKR1L-6A, which interacted with TaSERK1, co-regulated lignin synthesis and stem strength, leading to enhanced resistance of wheat to sharp eyespot (Figure 5).
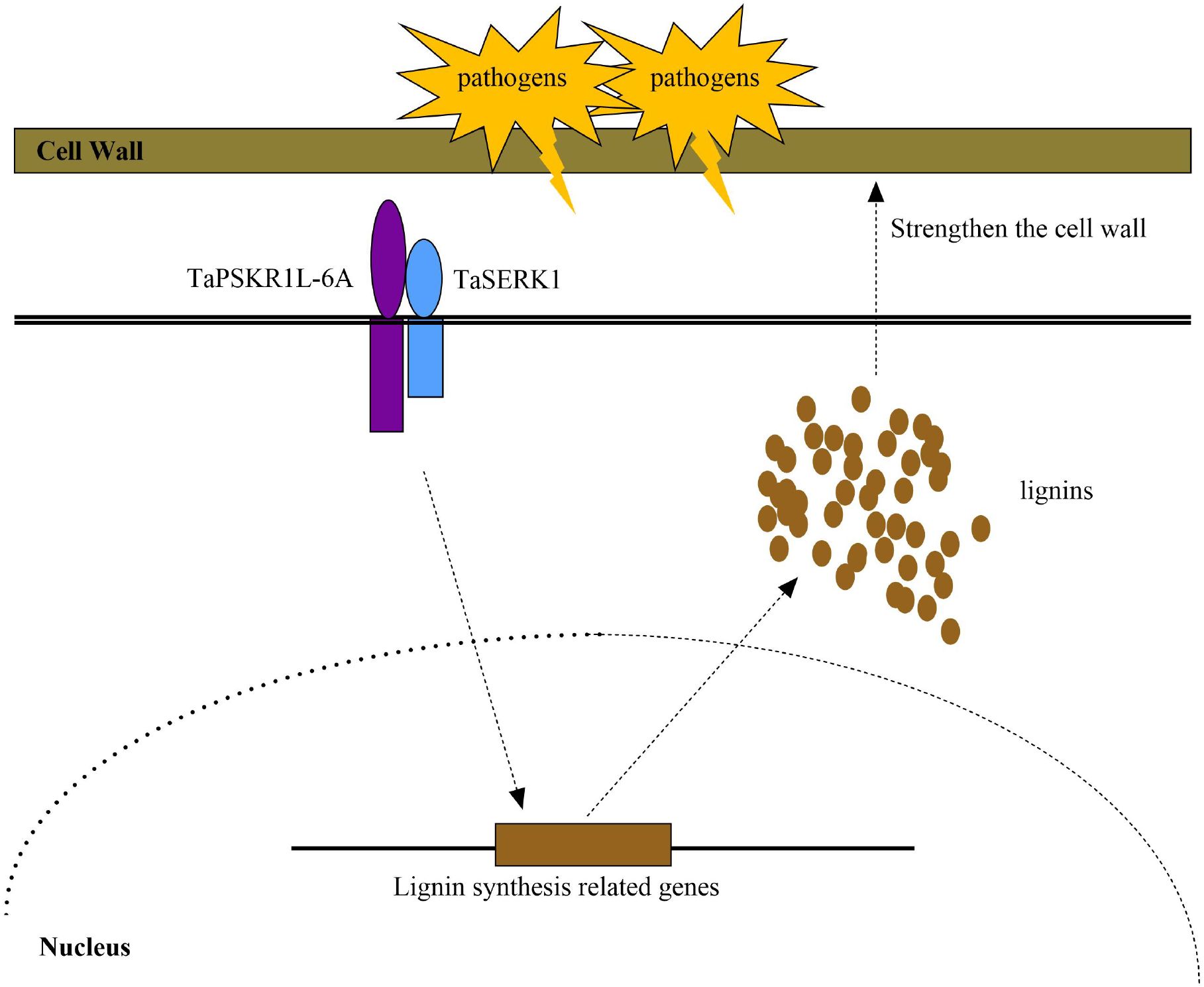
Figure 5. A working model of TaPSKR1L-6A improves resistance to sharp eyespot and increases lignin accumulation in wheat.
To our knowledge, this is the first investigation of PSKR1 or PSKR1L involvement in lignin synthesis and accumulation. The findings enrich the PSKR1/L signal pathway. Taken together, the current results enrich insights onto the PSKR-transducing signal pathways.
Conclusions
We demonstrated that TaPSKR1L-6A could confer sharp eyespot resistance through activating the expression of lignin synthesis-related genes and increasing lignin accumulation in wheat. Overexpression of TaPSKR1L-6A significantly enhanced wheat resistance against sharp eyespot and promoting lignin synthesis/accumulation. By contrast, silencing of TaPSKR1L-6A and TaSERK1 resulted in the opposite effects. To our knowledge, this is the first report about regulatory role of PSKR1/PSKR1L in lignin synthesis, and overexpression of a resistance gene improving wheat resistance to sharp eyespot. Thus, TaPSKR1L-6A was a promising gene for breeding wheat varieties with resistance to sharp eyespot. This study sheds light on wheat defense mechanisms against R. cerealis, and provides insights into functional roles of the PSKR1/PSKR1L in plant innate immunity, especially in the important crop wheat.
Materials and methods
Plant and fungal materials and treatments
Four wheat cultivars, including CI12633, Shanhongmai, Yangmai 9, and Wenmai 6, which exhibit different levels of resistance and susceptibility to sharp eyespot, were used to investigate TaPSKR1L-6A transcript profiles against R. cerealis. A middle resistant wheat cv. CI12633 was used for VIGS experiments. A middle susceptible wheat Yangmai 30 was used as the recipient material for genetic transformation experiments. All wheat seedlings were grown in a greenhouse at 23°C for 14 h in light, and 15°C for 10 h in darkness.
Rhizoctonia cerealis strain RC207, which is highly virulent in north China, was isolated by Prof. Jinfen Yu and Dr. Li Zhang (Shandong Agricultural University, China). The fungal strain was cultured on a PDA medium at 25°C in dark conditions. The sequences of all primers in this study are listed in Supplementary Table S2.
RNA-Seq and RT-qPCR analysis
In this study, the RILs derived from the cross Shanhongmai × Wenmai 6, were used for RNA-Seq (RNA-sequencing) analysis as described by Guo et al. (2021). Primers of target genes for RT-qPCR were designed by Primer Premier 5 software and are listed in Supplementary Table S2. The RT-qPCR was performed using SYBR Green SuperReal PreMix (TIANGEN, China) in an ABI Prism 7500 Real-Time PCR System (Life tech, USA). The TaActin gene was used as an internal control for RT-qPCR. The relative expression levels of the target genes were calculated using the comparative 2−ΔΔCT method (Livak and Schmittgen, 2001).
BSMV-induced gene silencing in wheat CI12633
Barley yellow dwarf virus-mediated VIGS has been successfully utilized to study gene function in barley and wheat. In this study, a 207 bp fragment of TaPSKR1L-6A was subcloned into the Nhe I site of the BSMV RNA γ cDNA, forming the BSMV: TaPSKR1L-6A recombinant constructs. BSMV carrying empty GFP (BSMV: GFP) was used as control treatment. Then, these viruses were individually inoculated according to previous described methods (Zhu et al., 2017). After 15 days inoculated with BSMV, gene silencing efficiency was tested by RT-qPCR, and the silencing plants were inoculated with R. cerealis for disease resistance evaluation.
Generation and identification of transgenic wheat
The full ORF sequence of TaPSKR1L-6A was amplified from cDNA of CI12633 plants. The full ORF sequence was added with a 6-His epitope tag and then was sub-cloned into monocot transformation vector pWMB110. The resulting pTaPSKR1L-6A-6*His vector, the transcript of the TaPSKR1L-6A-6*His was driven by the maize ubiquitin (Ubi) promoter and terminated by the 3’-non-transcribed region of the Agrobacterium tumefaciens nopalinesynthase (Tnos) gene (Wang et al., 2017). Subsequently, the pTaPSKR1L-6A-6*His vector was introduced into immature embryos of the wheat cv. ‘Yangmai 18’ via Agrobacterium mediated transformation. The transgenic wheat plants were detected via PCR with primers TaPSKR1L-6A-TF and Poly-R (Qi et al., 2024).
R. cerealis inoculation and wheat sharp eyespot evaluation
The sharp eyespot inoculated and sharp eyespot evaluation methods were referred to the existing work of research (Chen et al., 2008; Qi et al., 2021).
The disease index calculation formula is as follows:
The numbers 1–5 in the above formula represent the sharp eyespot infection types of different wheat plants, and X0-X9 represent the number of wheat plants in different disease infection types.
Lignin staining, lignin content and stem mechanical (breaking) strength measurement
At 4 dpi with R. cerealis, basal leaf sheaths of T1 transgenic or WT ‘Yangmai 18’ plants were collected and stained with lignin staining. Lignin content was quantitatively measured by using the lignin ELISA Kit (ChemFaces, China) according to the manufacturer’s protocols. For lignin staining, fresh hand-cut sections of leaf sheaths were prepared from basal stems of wheat plants. Wiesner and Mäule staining methods were performed as previously described (Wang et al., 2018) and observed under an optical microscope. At harvest stage, the stem mechanical (breaking) strength of the second basal internode in TaPSKR1L-6A overexpressing and ‘Yangmai18’ wheat plants were measured using a Texture Analyser (TA) with a three-point bend test setup according a previous study (Miller et al., 2016).
Data availability statement
Due to agreements with the project funding agency, we are currently not permitted to provide the transcriptome data. Should you require access to transcript data of relevant genes from this study, please feel free to contact the corresponding author.
Author contributions
HQ: Writing – original draft, Data curation, Investigation, Formal Analysis, Writing – review & editing. YH: Writing – review & editing, Investigation. XZ: Writing – review & editing, Funding acquisition, Visualization. ZZ: Supervision, Funding acquisition, Writing – review & editing, Methodology, Formal Analysis.
Funding
The author(s) declare financial support was received for the research, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (321 72004), the National Key Project for Research on Transgenic Biology (2016ZX08002-001-004), and the Innovation Program of Chinese Academy of Agricultural Sciences.
Acknowledgments
Authors are very grateful to Prof. Jinfen Yu and Dr. Li Zhang (Shandong Agricultural University, China) for providing Fusarium pseudograminearum strain WHF220, and to Prof. Derong Gao, and Dr. Xujiang Wu (Institute of Agricultural Science of Lixiahe District in Jiangsu Province), and Dr. Jizhong Wu (Jiangsu Academy of Agricultural Sciences) for providing the wheat cultivars.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1653282/full#supplementary-material
Supplementary Figure 1 | TaPSKR1L-6A haplotypes are associated with sharp eyespot resistance. (A) Haplotypes of TaPSKR1L-6A. The SNP polymorphism at -1191 sit is shown in red. (B) Disease indexes of TaPSKR1L-6A haplotypes among 61 wheat cultivars. (C) Distribution of TaPSKR1L-6A Hap I and Hap II haplotypes in wheat cultivars.
Supplementary Figure 2 | Stem mechanical (breaking) strength of the second basal internode in TaPSKR1L-6A overexpressing and ‘Yangmai18’ wheat plants at harvest stage.
Supplementary Table 1 | Infection types and disease indexes of sharp eyespot in TaPSKR1L-6A -overexpressing and WT (‘Yangmai18’) wheat plants in T2 generation. Student’s t-test (**P < 0.01).
Supplementary Table 2 | Primers used in this study.
References
Boutrot, F. and Zipfel, C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Guo, F., Wu, T., Shen, F., Xu, G., Qi, H., and Zhang, Z. (2021). The cysteine-rich receptor-like kinase TaCRK3 contributes to defense against Rhizoctonia cerealis in wheat. J. Exp. Bot. 72, 6904–6919. doi: 10.1093/jxb/erab328
Hamada, M. S., Yin, Y., Chen, H., and Ma, Z. (2011). The escalating threat of Rhizoctonia cerealis, the causal agent of sharp eyespot in wheat. Pest Manag. Sci. 67, 1411–1419. doi: 10.1002/ps.2236
He, J., Liu, Y., Yuan, D., Duan, M., Liu, Y., Shen, Z., et al. (2020). An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. U.S.A. 117, 271–277. doi: 10.1073/pnas.1902771116
Hohmann, U., Santiago, J., Nicolet, J., Olsson, V., Spiga, F., Hothorn, L., et al. (2018). Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. U.S.A. 115, 3488–3493. doi: 10.1073/pnas.1714972115
International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar 7191. doi: 10.1126/science.aar7191
Kaufmann, C., Motzkus, M., and Sauter, M. (2017). Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. J. Exp. Bot. 68, 1411–1423. doi: 10.1093/jxb/erx030
Li, H., Cai, Z., Wang, X., Li, M., Cui, Y., Cui, N., et al. (2019). SERK receptor-like kinases control division patterns of vascular precursors and ground tissue stem cells during embryo development in Arabidopsis. Mol. Plant 12, 984–1002. doi: 10.1016/j.molp.2019.04.011
Li, M. Y., Zhao, S. Q., Yang, J. Y., Ren, Y. J., Su, J. J., Zhao, X. P., et al. (2022). Exogenous expression of barley HvWRKY6 in wheat improves broad-spectrum resistance to leaf rust, Fusarium crown rot, and sharp eyespot. Int. J. Biol. Macromol. 218, 1002–1012. doi: 10.1016/j.ijbiomac.2022.07.138
Liu, X., Zhu, X. L., Wei, X. N., Lu, C. G., Shen, F. D., Zhang, X. W., et al. (2020). The wheat LLM-domain-containing transcription factor TaGATA1 positively modulates host immune response to Rhizoctonia cerealis. J. Exp. Bot. 71, 344–355. doi: 10.1093/jxb/erz409
Livak, K. and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsubayashi, Y., Ogawa, M., Morita, A., and Sakagami, Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472. doi: 10.1126/science.1069607
Meester, B., Vries, L., Özparpucu, M., Gierlinger, N., Corneillie, S., Pallidis, A., et al. (2018). Vessel-specific reintroduction of cinnamoyl-CoA reductase1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 176, 611–633. doi: 10.1104/pp.17.01462
Mosher, S., Seybold, H., Rodriguez, P., Stahl, M., Davies, K., Dayaratne, S., et al. (2013). The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 73, 469–482. doi: 10.1111/tpj.12050
Qi, H., Guo, F., Lv, L., Zhu, X., Zhang, L., Yu, J., et al. (2021a). The Wheat Wall-Associated Receptor-Like Kinase TaWAK-6D Mediates Broad Resistance to Two Fungal Pathogens Fusarium pseudograminearum and Rhizoctonia cerealis. Front. Plant Sci. 12, 758196. doi: 10.3389/fpls.2021.758196
Qi, H. J., Zhu, X. L., Guo, F. L., LV, L. J., and Zhang, Z. Y. (2021b). The wall-associated receptor-like kinase TaWAK7D is required for defense responses to Rhizoctonia cerealis in wheat. Int. J. Mol. Sci. 22, 5629. doi: 10.3390/ijms22115629
Qi, H. J., Zhu, X. L., Shen, W. B., Yang, X., Zhang, C. Z., Li, G. Y., et al. (2024). TaRLK-6A promotes Fusarium crown rot resistance in wheat. J. Integr. Plant Biol. 66, 12–16. doi: 10.1111/jipb.13596
Rhodes, J., Yang, H., Moussu, S., Boutrot, F., Santiago, J., and Zipfel, C. (2021). Perception of a divergent family of phytocytokines by the Arabidopsis receptor kinase MIK2. Nat. Commun. 12, 705. doi: 10.1038/s41467-021-20932-y
Sauter, M. (2015). Phytosulfokine peptide signaling. J. Exp. Bot. 66, 5161–5169. doi: 10.1093/jxb/erv071
Shan, T., Rong, W., Xu, H., Du, L., Liu, X., and Zhang, Z. (2016). Wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci Rep. 6, 28777. doi: 10.1038/srep28777
Wang, J., Li, H., Han, Z., Zhang, H., Wang, T., Lin, G., et al. (2015). Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525, 265–268. doi: 10.1038/nature14858
Wang, K., Liu, H. Y., Du, L. P., and Ye, X. G. (2017). Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 15, 614–623. doi: 10.1111/pbi.12660
Wang, K., Rong, W., Liu, Y. P., et al. (2020a). Wheat Elongator subunit 4 is required for epigenetic regulation of host immune response to Rhizoctonia cerealis. Crop J. 8, 565–576. doi: 10.1016/j.cj.2019.11.005
Wang, K., Shao, Z., Guo, F., Wang, K., and Zhang, Z. (2021). The mitogen-activated protein kinase kinase TaMKK5 mediates immunity via the TaMKK5-TaMPK3-TaERF3 module. Plant Physiol. 187, 2323–2337. doi: 10.1093/plphys/kiab227
Wang, M., Zhu, X., Wang, K., Lu, C., Luo, M., Shan, T., et al. (2018). A wheat caffeic acid 3-O-methyltransferase TaCOMT-3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 25 8, 6543. doi: 10.1038/s41598-018-24884-0
Wang, P., Zhou, L., Jamieson, P., Zhang, L., Zhao, Z., Babilonia, K., et al. (2020b). The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell 32, 3978–4001. doi: 10.1105/tpc.19.00950
Wu, X., Cheng, K., Zhao, R., Zang, S., Bie, T., Jiang, Z., et al. (2017). Quantitative trait loci responsible for sharp eyespot resistance in common wheat CI12633. Sci. Rep. 7, 11799. doi: 10.1038/s41598-017-12197-7
Wu, X., Wang, J., Wu, D., Jiang, W., Gao, Z., Li, D., et al. (2022). Identification of new resistance loci against wheat sharp eyespot through genome-wide association study. Front. Plant Sci. 13, 1056935. doi: 10.3389/fpls.2022.1056935
Yang, W., Zhang, B., Qi, G., Shang, L., Liu, H., Ding, X., et al. (2019). Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta 250, 1603–1612. doi: 10.1007/s00425-019-03238-8
Zhang, H., Hu, Z., Lei, C., Zheng, C., Wang, J., Shao, S., et al. (2018). A Plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 30, 652–667. doi: 10.1105/tpc.17.00537
Zhang, H., Lin, X., Han, Z., Wang, J., Qu, L., and Chai, J. (2016). SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol. Plant 9, 1406–1414. doi: 10.1016/j.molp.2016.07.004
Zhang, Z., Yao, W., Dong, N., Liang, H., Liu, H., and Huang, R. (2007) A novel ERF transcription activator in wheat and its induction kinetics after pathogen and hormone treatments. J Exp Bot. 58 (11), 2993–3003. doi: 10.1093/jxb/erm151
Zhu, X., Lu, C., Du, L., Ye, X., Liu, X., Coules, A., et al. (2017). The wheat NB-LRR gene TaRCR1 is required for host defense response to the necrotrophic fungal pathogen Rhizoctonia cerealis. Plant Biotechnol. J. 15, 674–687. doi: 10.1111/pbi.12665
Zhu, X., Qi, L., Liu, X., Cai, S., Xu, H., Huang, R., et al. (2014) The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164 (3), 1499–1514. doi: 10.1104/pp.113.229575
Zhu, X., Rong, W., Wang, K., Guo, W., Zhou, M., Wu, J., et al. (2022) Overexpression of TaSTT3b-2B improves resistance to sharp eyespot and increases grain weight in wheat. Plant Biotechnol J. 20 (4), 777–793. doi: 10.1111/pbi.13760
Keywords: common wheat (Triticum aestivum), lignin synthesis, phytosulfokine receptor 1-like, resistance, sharp eyespot
Citation: Qi H, Hao Y, Zhu X and Zhang Z (2025) Overexpression of TaPSKR1L-6A improves resistance to sharp eyespot and increases lignin accumulation in wheat. Front. Plant Sci. 16:1653282. doi: 10.3389/fpls.2025.1653282
Received: 24 June 2025; Accepted: 19 September 2025;
Published: 17 October 2025.
Edited by:
Udai B. Singh, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaReviewed by:
Veerendra Sharma, Oregon State University, United StatesMingyue Gou, Henan Agricultural University, China
Copyright © 2025 Qi, Hao, Zhu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengyan Zhang, emhhbmd6ZW5neWFuQGNhYXMuY24=; Xiuliang Zhu, emh1eGl1bGlhbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Haijun Qi
Haijun Qi Yuran Hao4†
Yuran Hao4†