- 1Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Beiing Key Laboratory of Forestry Food Processing and Safety, Department of Food Science, Beijing Forestry University, Beijing, China
- 3Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (North China), Ministry of Agriculture and Rural Affairs, Beijing, China
Basic/helix-loop-helix (bHLH) transcription factors play critical roles in the regulation of plant secondary metabolism, but the molecular mechanisms involved in monoterpene synthesis by the grape bHLH family have not been systematically resolved. In this research, 12 VvbHLHs were detected in the grape genome, encoding 468–696 amino acids with molecular masses ranging from 52.5 to 77.5 kDa. Phylogenetic analyses showed that grapevine bHLH family members are closely related to dicotyledonous plants such as Pyrus bretschneideri and Malus domestica. These VvbHLHs were mainly localized in the nucleus, and their promoter regions were enriched for cis-acting elements linked to developmental stages and stress resistance. Spatio-temporal expression profiling indicated that VvbHLHs were significantly upregulated during fruit ripening, with the expression of VvbHLH9 (VvbHLH041) significantly and positively correlated with monoterpene accumulation. Subcellular localization analysis showed that VvbHLH9 was localized in the nucleus; and transient overexpression assays revealed that VvbHLH9 significantly activated monoterpene skeleton genes (VvDXS1 and VvDXS4) and synthase genes (VvTPS31, VvTPS32, and VvTPS35) to promote the accumulation of monoterpene metabolites, such as terpinolene, allo-ocimene, nerol, geraniol, cis-furan linalool oxide and linalool. Notably, VvbHLH9 lacked the typical bHLH-MYC_N structural domain, suggesting that it may affect the monoterpene metabolic network through a non-classical regulatory mechanism. The present study uncovers the modulatory function of grapevine bHLHs on the synthesis of monoterpene metabolites, providing new targets for grapevine fruit quality improvement and molecular breeding.
1 Introduction
As an important fruit crop widely cultivated around the world, grapevine (Vitis vinifera L.) is not only used for fresh consumption, winemaking and food processing, but also for pharmaceutical and industrial applications due to its richness in polyphenols, terpenoids and other active components (Chedea et al., 2022; Sabo et al., 2022). Recent studies have shown that the market value of grapes is strongly tied to the presence of metabolites (Wang et al., 2022a; Fan et al., 2023). Terpenoids, among the most abundant plant secondary metabolites, not only impart unique sensory characteristics to grapes but also have gained recognition for their remarkable therapeutic properties, include antimicrobial and anticancer properties (Mele et al., 2021). In addition, terpenoids are key regulators of plant developmental stages and stress resistance (Boncan et al., 2020; Wang et al., 2022b). However, terpenoids generally accumulate at low levels in plants, and how to optimize their synthetic pathways through genetic engineering has become an vital research direction to enhance the economic traits of grapes and develop terpenoid resources.
Transcription factors can achieve precise regulation of biosynthetic pathways through specific interactions with cis-acting elements in the promoters of target genes (Liu et al., 2022a). bHLH is the second largest gene family after MYB, and its members are crucial for light signal transduction, response to adversity, and secondary metabolism regulation through the formation of homo- or heterodimers (Ke et al., 2020b). Typical bHLH proteins contain two functional domains: the N-terminal DNA-binding domain consisting of 10–20 basic amino acids, which recognizes the E-box (CANNTG) sequences in the promoters of target genes; and the C-terminal HLH domain, which mediates protein dimerization through hydrophobic interactions, thereby regulating downstream gene transcription (Zhang et al., 2020). Up to now, the phylogenetic history of the bHLHs in the plant kingdom has been extensively investigated, including the model plant Arabidopsis (Toledo-Ortiz et al., 2003) and tobacco (Bano et al., 2021), as well as the common crops maize (Zhang et al., 2018b), barley (Ke et al., 2020a), foxtail millet (Fan et al., 2021), populus trichocarpa (Zhang et al., 2024a), and grape (Gao et al., 2019). For example, at least 162 bHLHs have been previously identified in Arabidopsis, which can be categorized into 12 different subfamilies, bHLH I to XII (Heim et al., 2003). In addition, another study identified 159 bHLH protein-coding genes in the tomato genome, and the SlbHLH genes were classified into 21 subfamilies by phylogenetic tree analysis (Sun et al., 2015). These plant bHLHs orchestrate development and adaptation to environmental stresses.
Numerous investigations have elucidated the functions of bHLHs across diverse plant species, demonstrating that certain bHLHs are key regulators of secondary metabolic pathways, such as anthocyanin, terpenoid, and alkaloid biosynthesis (Hichri et al., 2011). As a major class of transcriptional regulators, bHLHs participate in modulating terpene biosynthesis in plant systems, but most studies have been limited by the fact that they have only been carried out in model plants such as Arabidopsis (Hao et al., 2021). For many years, bHLHs have been found in Arabidopsis. For instance, the Arabidopsis bHLH transcription factor AtMYC2 binds directly to the promoters of AtTPS21 and AtTPS11, activating their expression and promoting the synthesis of sesquiterpenes (Hong et al., 2012). In addition, a regulatory role of bHLHs has been found in the terpene MVA pathway in plants. Mertens et al. (2016) identified two JA-induced bHLHs TSAR1 and TSAR2 in Medicago truncatula, which can activate the relevant genes of the MVA pathway through different activation modes, resulting in enhanced accumulation of triterpene saponins. In grapevine, scarce studies have predominantly emphasized the participation of bHLHs in metabolic control and growth processes. A recent investigation identified VdbHLH037 as a member of subfamily III (f), which interacts with anthocyanin biosynthesis-related genes and contributes to anthocyanin production in spine grapes (Li et al., 2021). This result indicates that some grape bHLHs are associated with the production of anthocyanins. However, studies investigating the role of bHLHs in grape monoterpene metabolism are still scarce.
This study conducted whole-genome analysis of the grape bHLHs, mapped the distribution of bHLHs on chromosomes and performed covariance analysis; made statistical predictions of the physicochemical properties of the proteins and constructed their three-dimensional structural models. Phylogenetic evolutionary trees were constructed by sequence comparison of bHLH protein family members from seven species; and separate developmental evolutionary trees, gene structures and motif analyses were performed for VvbHLHs from grapevine. In addition, the high monoterpene variety ‘Ruidu Xiangyu’ at different developmental periods was selected as the test material for the study, and the expression pattern of grape VvbHLHs and its relationship with monoterpene synthesis were further explored by RT-qPCR analysis and GC-MS analysis. Especially importantly, we discovered for the first time that the expression level of VvbHLH9 displayed a pronounced positive linkage with monoterpene production, and preliminarily explored its function through subcellular localization analysis and transient overexpression assays. The current results offer fresh perspectives into the regulatory mechanisms of the grape bHLHs, and offer theoretical support for enhancing grape quality through transcription factor engineering.
2 Materials and methods
2.1 Plant materials
The material was ‘Ruidu Xiangyu’ grape variety, taken from Beijing Forestry and Fruit Tree Research Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences. Row spacing 2.5 m, plant spacing 0.75 m open field cultivation, inclined horizontal dragon vine, single-arm hedge shaping. Conventional cultivation and management measures were used. 25 May 2024, ‘Ruidu Xiangyu’ grapes bloomed, and fruit samples were collected 30 d after flowering (25 June). Samples were then taken at 15 d intervals until 10 August 2024 (ripening), a total of four times, labelled S1 (young fruit), S2 (expansion), S3 (veraison) and S4 (ripening), respectively. Sampling should be done on both the shaded and sunny sides, and on the shoulders, center and top of each cluster.
2.2 Identification and screening of grape bHLH
Firstly, we downloaded the protein data of grape in Emsembl plants (ftp://ftp.ensemblgenomes.org/) and obtained the protein sequence of Arabidopsis bHLH transcription factor in PlantTFDB5.0. The analysis was conducted using a combination of the HMMER 3.0 software and the native Blastp method, with a limited E-value of 1 × e-10 for HMMER 3.0 and an E-value set at 1 × e-5 for Blastp. Subsequently, BatchCD-search in the NCBI online website (https://www.ncbi.nlm.nih.gov/) was used to retrieve the conserved structural domains of the bHLH family and collected all the results of the query with E-value less than 0.01, which were the predicted grape VvbHLH genes. The conserved structural domains of VvbHLHs were analyzed using the sequence analysis software DNAMAN 6.0 and visualized using Tbtools (v1.098774) (Chen et al., 2020).
2.3 Chromosomal localization and covariance analysis of grape bHLH
With the help of (gff3) file of grape genome annotation information, the bHLHs were located in the chromosome, and then the Gene location visualize from gff3 function of Tbtools (v1.098774) was used to visualize and analyze the structural results of the bHLH gene. Gene replication events were analyzed by the MCScanX program and KaKs_Calculator2.0 (Wang et al., 2024), and grape bHLH family members as well as covariance between VvbHLHs and Arabidopsis AtbHLHs were visualized using Tbtools (v1.098774).
2.4 Physicochemical property analysis and spatial structure prediction of grape bHLH
Using ProtParam software (https://web.expasy.org/protparam) analyze the physicochemical properties of bHLHs in grapes, including the number of amino acids, theoretical isoelectric point, and molecular weight (Eslami Bojnourdi et al., 2023). The subcellular localization of grape bHLHs was predicted using the WoLFPSORT (https://wolfpsort.hgc.jp/) (Salih et al., 2021). Prediction of protein secondary structure of grape bHLHs using SOPMA tool (http://npsa-pbil.ibcp.fr/); the tertiary structure of grape VvbHLH protein was predicted and analyzed using the SWISS-MODEL (https://swissmodel.expasy.org/) website (Liang et al., 2023).
2.5 Phylogenetic analysis, gene structure and conserved motif analysis
The bHLH sequences of grape and the bHLH sequences of Arabidopsis, Zea mays, Pyrus bretschneideri, Malus domestica, Populus trichocarpa and Oryza sativa species were subjected to multiple sequence comparisons using the MUSCLE tool in the MEGA11 software, and the phylogenetic tree was drawn by the neighbor-joining method (Zhang et al., 2021), and the constructed phylogenetic tree was trimmed and beautified using the Evolview (http://www.evolgenius.info/evolview/#/) online tool to modify and beautify the constructed phylogenetic tree. Gene sequence intron and UTR analyses were performed on the GSDS website (http://gsds.gao-lab.org/). Protein motif analysis was performed on the MEME website (http://meme-suite.org/tools/meme) (Zhang et al., 2021). The phylogenetic tree, conserved structural domains and gene structure results of VvbHLH were visualized in evolutionary tree order using Tbtools (v1.098774).
2.6 Cis-element analysis of grape bHLH promoter
The 2000 bp upstream of the start codon of the bHLH family gene was extracted from the grape genome file as the promoter region, and promoter cis-element analysis was performed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Zhang et al., 2024c). Tbtools (v1.098774) was used for promoter visualization of the results.
2.7 Determination of monoterpene content
Free-state monoterpenes were extracted and determined with reference to the method of Liu et al. (2022b). Retention indices (RIs) and mass spectral information were obtained by calculation using the Automated Mass Spectrometry Deconvolution and Identification Software (AMDIS), and the results of mass spectral analyses were searched by matching the results with the NIST Substance Retrieval Spectral Library, as well as the retention indices of the substance standards to identify the substances. The concentration determination of grape terpenoids is carried out through a standard curve. The standard curves were prepared and plotted with reference to the method of Zhou et al. (2022). For monoterpenes for which no standard curve is available, quantitative analyses should be carried out by means of standard curves for substances with a similar number of carbon atoms and similar chemical structure (Supplementary Table 1).
2.8 RNA extraction and RT-qPCR analysis
Primers specific for the grape bHLH family gene were designed using Premier 6 software (Supplementary Table 2), and total RNA was extracted using the TIANGEN RNAplant kit (TIANGEN, China). The yield and quality of RNA were determined using a NanoDrop spectrophotometer (ND-7000, NanoDrop Technologies, Inc., USA). The cDNA was synthesized using All-in-one 1st Strand cDNA Synthesis Super Mix (gDNA Purge) Reverse Transcription Kit (Novoprotein, China). cDNA was diluted 5-fold at a concentration of 200 ng/uL and used for RT-qPCR reaction. RT-qPCR Reactions were performed using SYBR qPCR mixture (GenStar, China) and CFX-connected real-time PCR detection system (Bio-Rad, USA). The PCR cycles were: 95°C for 2 min, 40 cycle numbers, 95°C for 15 s, and 60°C for 34 s. Each reaction was performed with 3 biological replicates. VvUbiquitin was used as an internal reference gene. Relative expression levels were counted using the 2-ΔΔCT method (Harshitha and Arunraj, 2021).
2.9 Correlation analysis between grape bHLH gene expression level and monoterpene content
Based on the changes of monoterpene content in grape berries and the relative expression of grape bHLHs at different periods, Pearson correlation analysis was performed using SPSS 27.0 (SPSS, USA). The significance of correlations was assessed at P < 0.05 and P < 0.01 levels, with adjustments for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) method. Heatmaps were drawn using ChiPlot (https://www.chiplot.online/) to visualize the correlation between the changes of monoterpene content and the gene expression results were visualized.
2.10 VvbHLH9 gene cloning, plasmid construction and subcellular localization
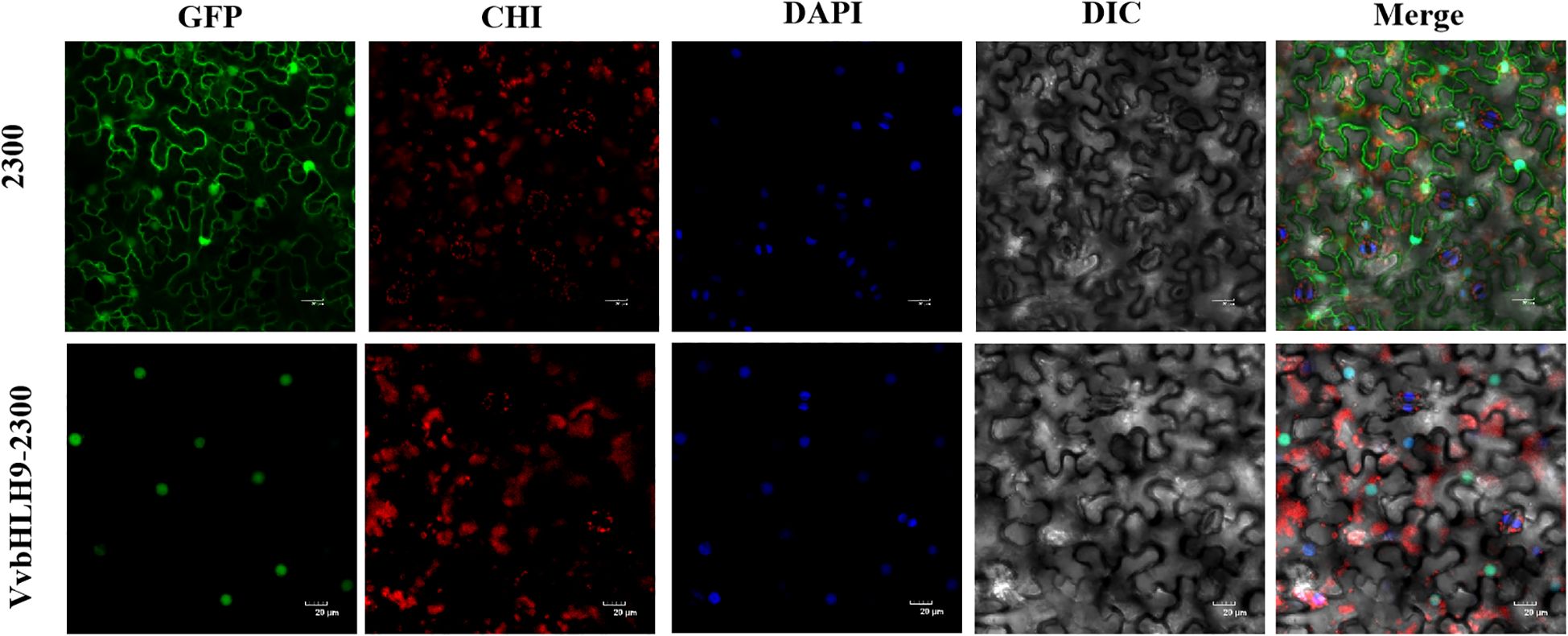
We generated cDNA by reverse transcription of total RNA from grape berries and cloned the VvbHLH9 gene. Subcellular localization of the VvbHLH9 gene was performed in order to determine the true positional information of the gene. We constructed a recombinant plasmid incorporating the GFP tag and the VvbHLH9 protein, and injected the recombinant vector into N. benthamiana leaves using Agrobacterium-mediated transformation, and used the empty PCAMBIA2300-GFP vector with a GFP tag as a control. After overnight dark incubation, the culture was then cultivation under light for 12 hours. About 3 days after leaf infiltration, the leaves were placed on slides and GFP fluorescence in transgenic N. benthamiana leaf cells was observed using a FV10-ASW laser confocal microscope. GFP stands for green fluorescence field, CHI stands for chloroplast autofluorescence field, DAPI stands for DAPI field (cytosolic staining), DIC stands for bright field, and Merge stands for superimposed field.
2.11 Transient overexpression in leaves
The fragment of VvbHLH9 was cloned into pCAMBIA2300-GFP vector, and the recombinant vector was used for transient overexpression analysis. Fresh ‘Alden’ variety grapevine leaves of similar size and without obvious mechanical damage were selected for infestation, and six leaves were infested with each sample as six technical parallels, and the same number of leaves were infested with Agrobacterium transfected into the pCAMBIA2300-GFP empty vector as the control group. The bacteriophage solution was incubated for 12 h until the OD600 was 0.6-1.0. The bacteriophage OD600 was adjusted to 0.4 using infestation buffer [MES 2.132 g/L, MgCl2·6H2O 2.033 g/L, sucrose 5 g/L, and acetosyringone at a final concentration of 200 mg/L, pH 5.9], and left to stand for 4 h. After infiltration, the leaves were incubated under low temperature and light protection for 3 days. The expression levels of VvbHLH9 and monoterpene synthesis-related genes were obtained by RT-qPCR, and the monoterpene substances were detected by GC-MS. The primers are shown in Supplementary Table 3.
3 Results
3.1 Identification of grape bHLH gene
Using HMMER software and TBtools local BLAST analysis, we identified 12 grape bHLH protein sequences that met the screening criteria (Figure 1A). According to the chromosomal location, these bHLH sequences were named VvbHLH1~VvbHLH12. Based on the conserved structural domains, we classified the grape bHLH family into bHLH_SF superfamily, bHLH-MYC_N, ACT superfamily, bHLH_AtAIB_like, bHLH_ AtAMS_like, ACT_UUR-ACR-like, and bHLH_AtbHLH_like, which are seven types. Except for VvbHLH9, the remaining 11 VvbHLHs contain the bHLH-MYC_N structural domain at the N-terminus (Figure 1B). Analysis of the HLH conserved structural domains of the 12 grape bHLHs using DNAMAN software showed that the C-terminal HLH structural domains were enriched in the hydrophobic amino acids leucine (L) and proline (P), of which, L was highly conserved and was important for maintaining the stability of the secondary structure of the bHLH protein (Figure 1C).
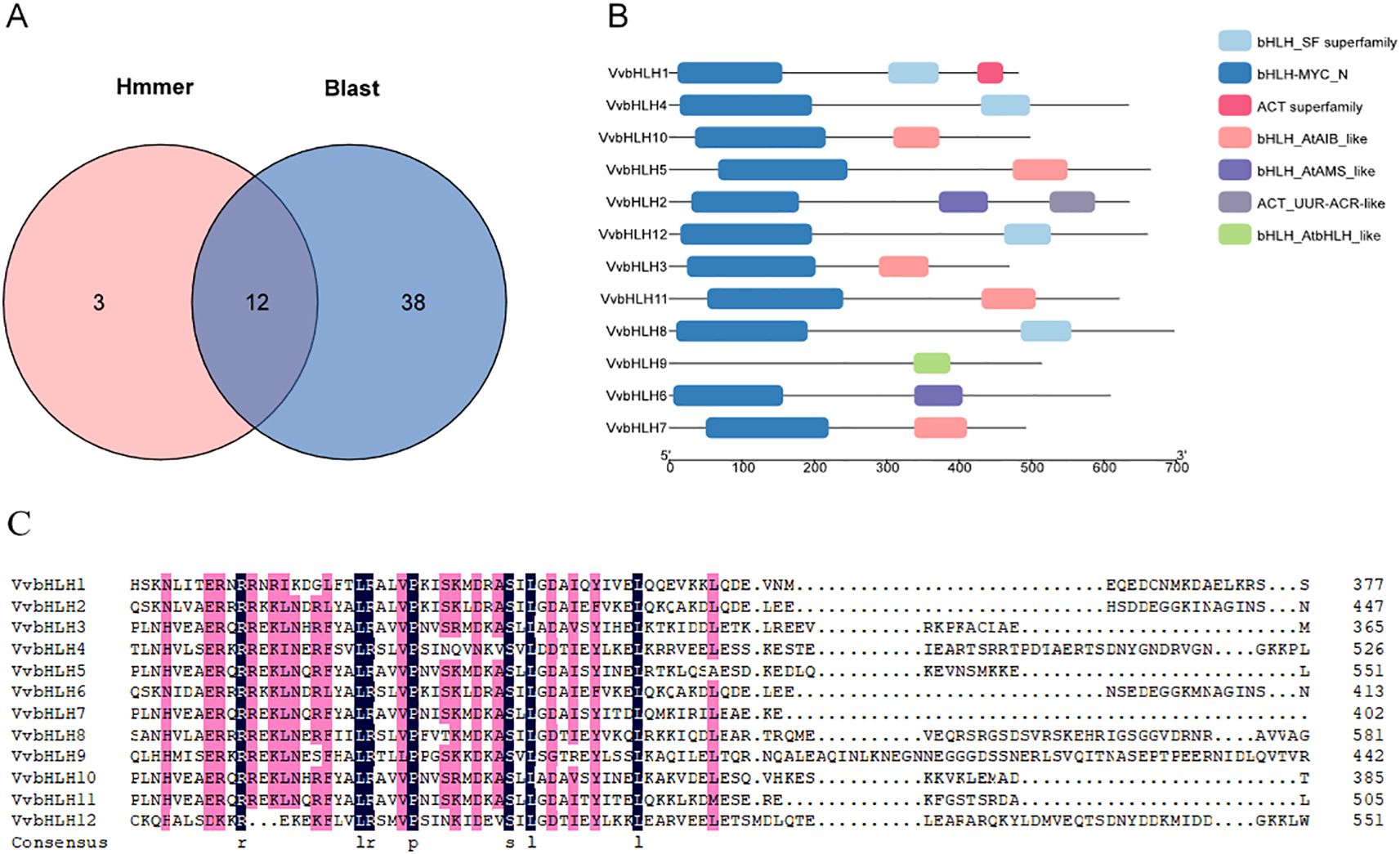
Figure 1. Identification of grape VvbHLHs and analysis of protein conserved structural domains. (A) Identification results of grape VvbHLH gene family. Hmmer: genes identified by HMMER software; Blast: genes compared by Tbtools software. (B) Analysis of grape VvbHLH protein conserved structural domains. (C) Comparative analysis of HLH conserved structural domains of grape VvbHLH gene family members. Black and pink colours indicate 100% and 75% amino acid conservation, respectively; hyphens indicate intervals.
3.2 Chromosomal localization and covariance analysis of grapevine bHLH gene
To further confirm the evolutionary duplication events of VvbHLH, we analyzed their chromosomal distribution and syntenic relationships. According to the grape genome annotation gff3 file, the 12 grape VvbHLHs identified were localized to six grape chromosomes (Figure 2). The 12 VvbHLHs were irregularly distributed, and the genes were distributed in different densities on different chromosome skeletons. Among them, chromosomes 2 and 15 in grapes contained the highest number of bHLHs with three; chromosomes 1 and 7 both contained two bHLH genes; and chromosomes 3 and 11 contained the lowest number of bHLH genes with one.
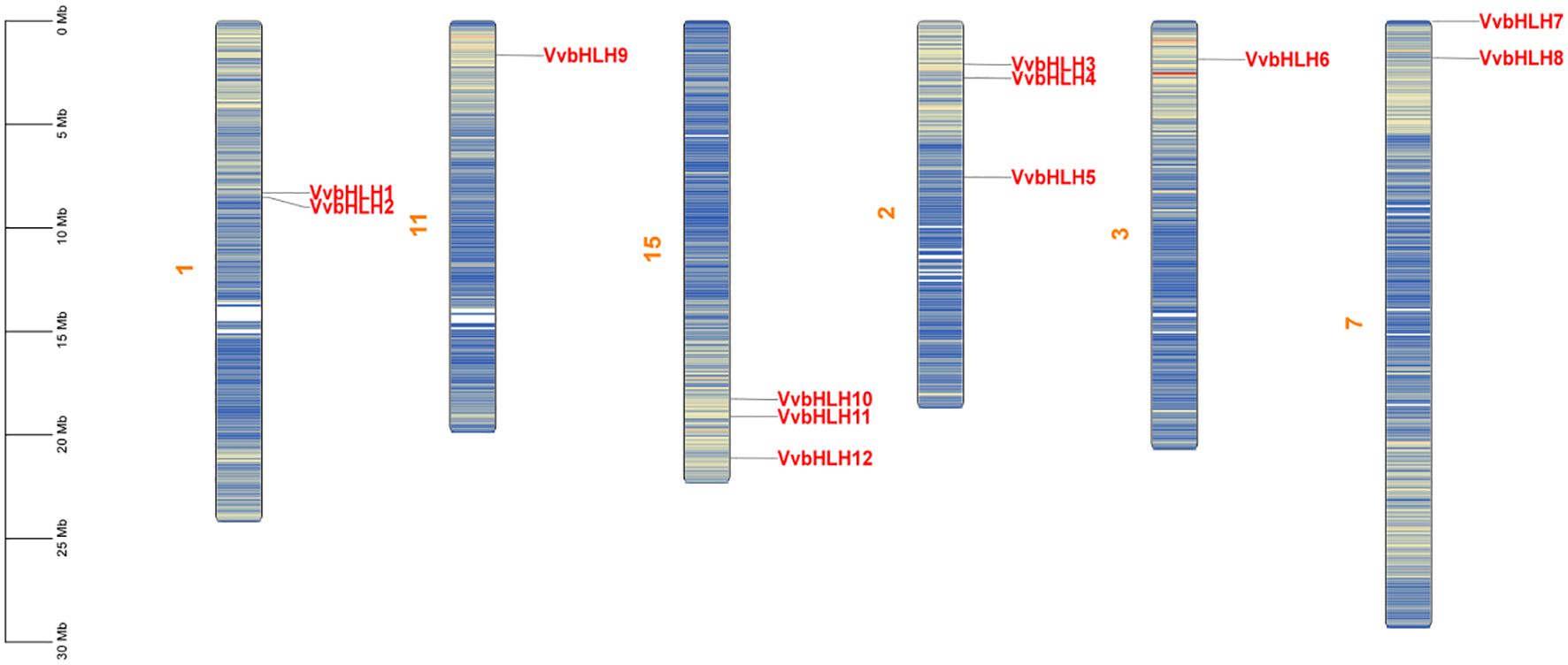
Figure 2. Chromosome distribution pattern of grape bHLH gene family members. Yellow characters are chromosome names, red characters are gene names, the chromosome colour gradient from blue to red represents gene density from low to high, and blank regions indicate the lack of gene distribution on the chromosome.
The collinearity analysis revealed that the two pairs of grape bHLHs exhibit a syntenic relationship (Figure 3A; Supplementary Table 4), VvbHLH3 and VvbHLH10, and VvbHLH4 and VvbHLH12, which can be inferred to have been formed through the replication of large segments of the genes. To further understand the evolutionary links among the bHLHs, we conducted collinearity analysis of bHLHs between grapes and Arabidopsis (Figure 3B; Supplementary Table 4). It was found that a total of eight VvbHLH genes in grape shared covariance with Arabidopsis bHLH genes. Among them, three VvbHLH genes showed homology with one Arabidopsis bHLH gene, including VvbHLH6, VvbHLH9, and VvbHLH10; four VvbHLH genes showed homology with two Arabidopsis bHLH genes, including VvbHLH3, VvbHLH5, VvbHLH11, and VvbHLH12; one VvbHLH gene showed homology with three Arabidopsis bHLH genes, VvbHLH4 (Supplementary Table 4).
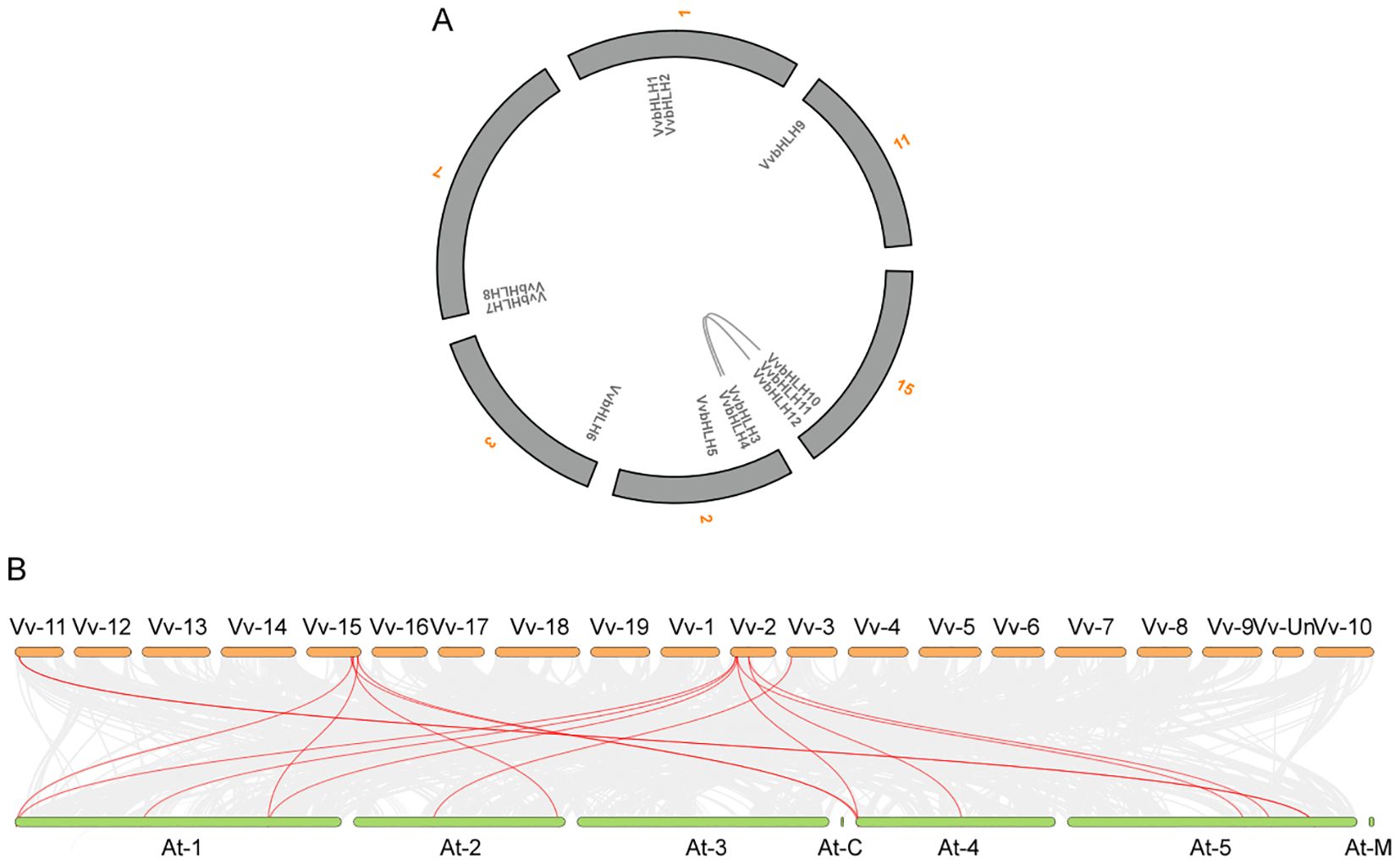
Figure 3. Analysis of covariance of VvbHLH within grape and between grape and Arabidopsis. (A) Covariance analysis of grape VvbHLH. The gray text in the figure represents 12 bHLH genes, and the orange numbers represent different chromosomes. (B) Covariance analysis of VvbHLH between grape and Arabidopsis. Vv represents Vitis vinifera, At represents Arabidopsis, and the numbers represent different chromosomes.
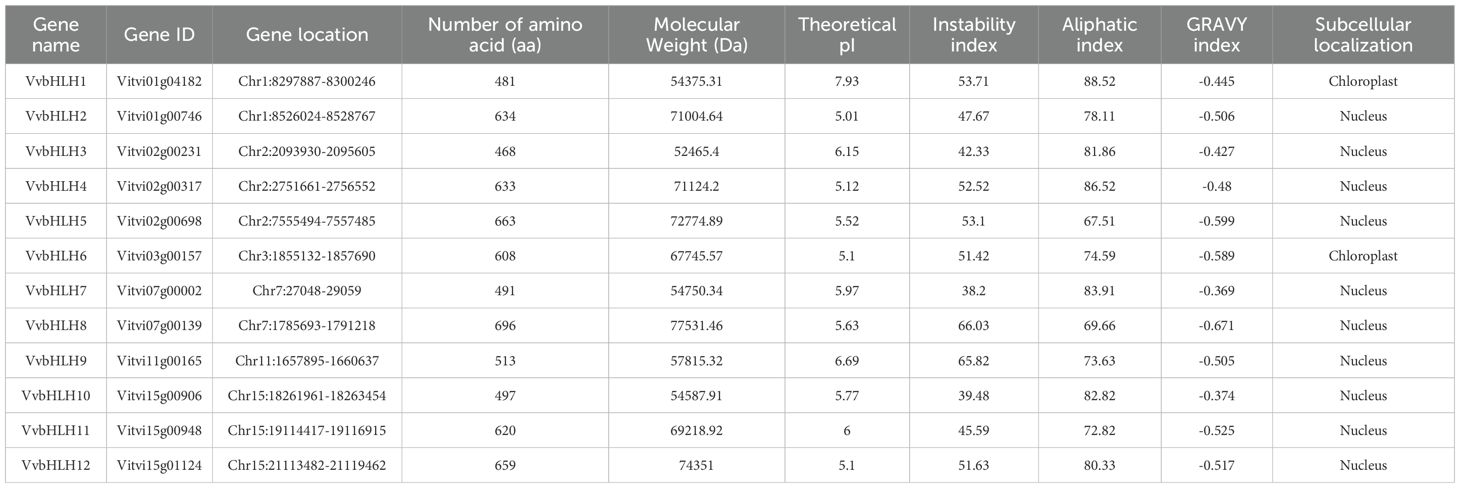
3.3 Physicochemical property analysis of grape bHLH family proteins
Analysis of the physicochemical properties of grape bHLH proteins showed (Table 1) that the length of grape bHLHs ranged from 468 aa (VvbHLH3) to 696 aa (VvbHLH8). The molecular mass of the 12 bHLHs ranged from 52,465.4 Da to 77,531.46 Da, with the highest molecular mass of the VvbHLH8 protein, and the lowest molecular mass of the VvbHLH3 protein. VvbHLH8 protein had the highest molecular mass and VvbHLH3 protein had the lowest. The pI of the grape bHLHs ranged from 5.01 (VvbHLH2) to 7.93 (VvbHLH1), and 11 bHLH proteins had pIs between 5 and 7, indicating that most of the grape bHLH family proteins had theoretically acidic pIs. The instability coefficients ranged from 38.2 (VvbHLH7) to 66.03 (VvbHLH8), indicating that most of the members were unstable proteins (II > 40); only two belonged to stable proteins (II < 40), with instability coefficients of 38.2 (VvbHLH7) and 39.48 (VvbHLH10), respectively. The fat coefficients were 67.51 (VvbHLH5) to 88.52 (VvbHLH1). In addition, the GRAVY index of all bHLH proteins was less than 0, revealing that these proteins exhibit predominantly hydrophilic properties, but the hydrophilicity varied among different proteins. Subcellular localization prediction revealed that most of the bHLHs were subcellularly localized in the nucleus and a few in the chloroplasts.
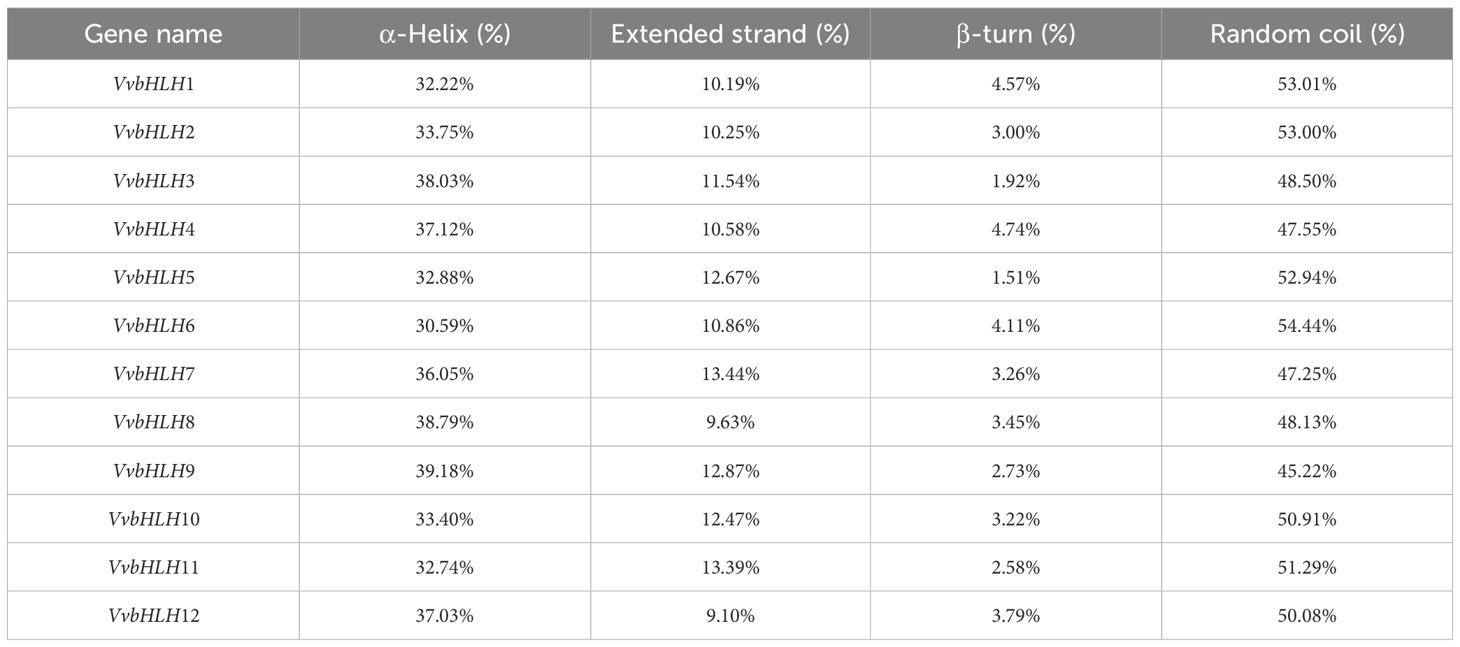
3.4 Spatial structure analysis of grape bHLH protein
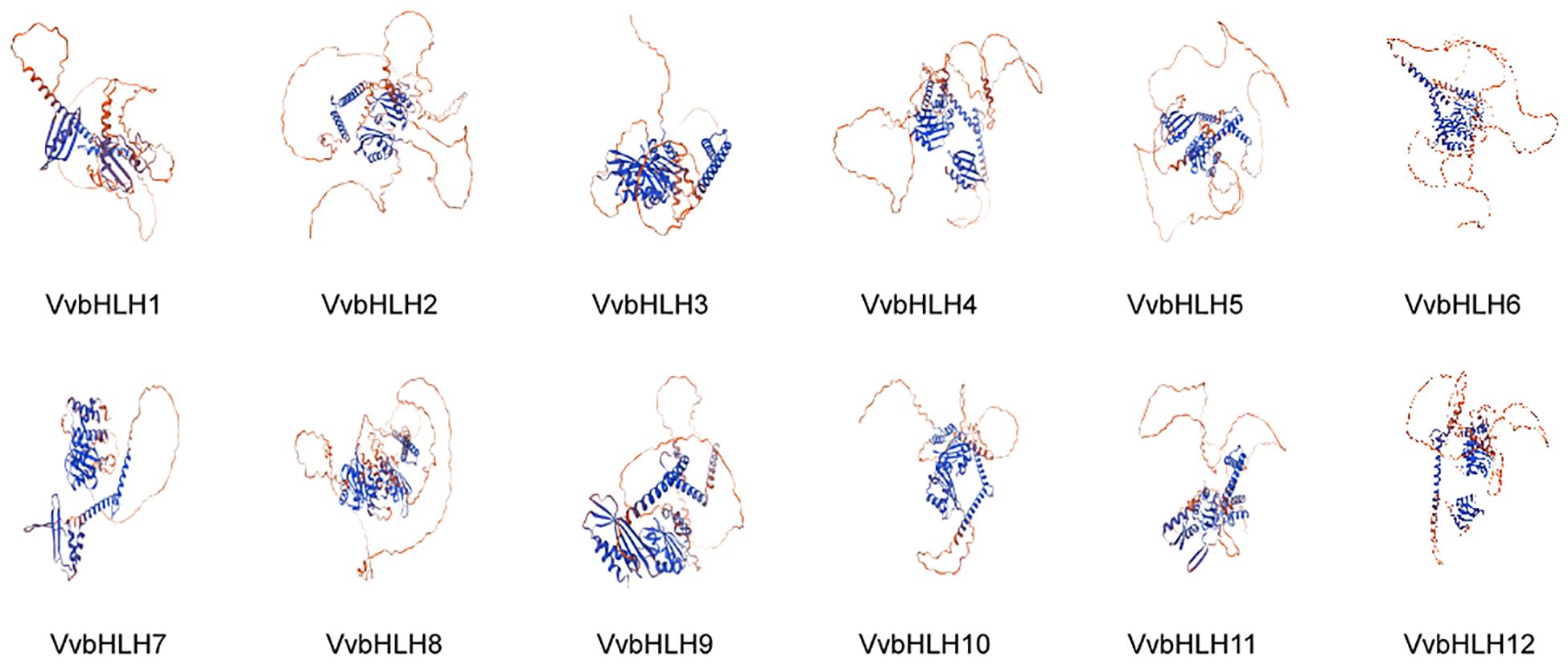
The amino acid sequences of the bHLHs were predicted and analyzed for secondary structure by SOPMA. Research has found that the secondary structure of grape bHLHs mainly consisted of α-helix (Hh), random coil (Cc), β-turn (Tt) and extended strand (Ee) (Table 2). Among them, random coils and α-helices accounted for the largest proportion, and β-turns accounted for the smallest proportion. α-helices structures had a number proportion of 30.59% (VvbHLH6) to 39.18% (VvbHLH9), extended strand structures had a number proportion of 9.10% (VvbHLH12) to 13.44% (VvbHLH7), and β-turns structures had a number proportion of 1.51% (VvbHLH5) to 4.74% (VvbHLH4), and the proportion of the number of random coil structures is 45.22% (VvbHLH9) to 54.44% (VvbHLH6). The sequences of the grape bHLHs were analyzed for the prediction of tertiary structures through the SWISS-MODEL website. The analysis revealed that the model match similarity exceeded 30% for all members, and the protein structures of the same subfamily had high similarity, but there were also differences between individuals (Figure 4).
3.5 Phylogenetic analysis, gene structure and conserved motif analysis
To elucidate the phylogenetic relationship between grape and other plant bHLHs, we constructed an evolutionary tree by comparing the protein sequences of the grape bHLHs with those of Arabidopsis, Zea mays, Pyrus bretschneideri, Malus domestica, Populus trichocarpa and Oryza sativa (Figure 5). Based on evolutionary relationships, a total of 132 bHLHs from different species were classified into 12 subfamilies (Group 1 ~ Group 12), with the smallest subfamily, Group 11, containing only one member, and the largest subfamilies, Group 3 and Group 7, with 22 members. Phylogenetic analysis of grapevine bHLHs showed that the 12 VvbHLH protein sequences could be divided into 3 groups: Group 3 contained 8 VvbHLH proteins, Group 4 contained 3 VvbHLH proteins, and Group 6 contained 1 VvbHLH protein. As can be seen from the figure, all grape VvbHLH genes are always clustered together with those of Pyrus bretschneideri, Malus domestica and Arabidopsis with close affinity, while they are relatively distantly related to the monocotyledonous plants Zea mays and Oryza sativa.
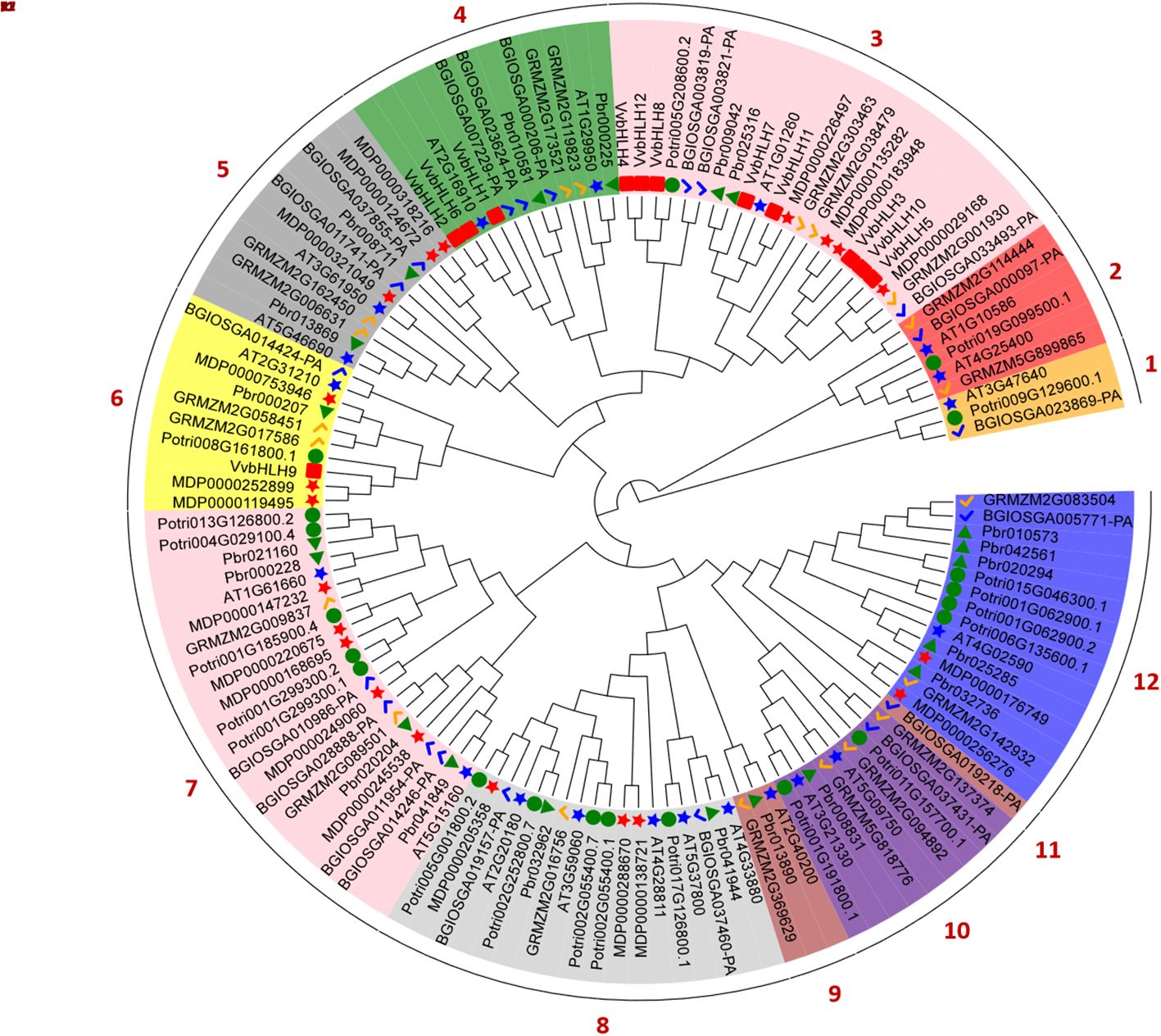
Figure 5. Phylogenetic evolutionary tree of bHLHs from Vitis vinifera, Arabidopsis, Zea mays, Pyrus bretschneideri, Malus domestica, Populus trichocarpa and Oryza sativa. Vv. Vitis vinifera; At. Arabidopsis thaliana; Zm. Zea mays; Pbr. Pyrus bretschneideri; Mdp. Malus domestica; Pot. Populus trichocarpa; Bgi. Oryza sativa.
In order to better analyze the bHLHs of grapevine, the study was carried out with separate developmental evolutionary trees, gene structure and motif analyses (Figure 6). It can be seen that VvbHLHs include 3 classes, which aligning with the protein phylogenetic tree classification. It can be found that all grape bHLH family members have coding sequences (CDS), and all have untranslated regions (UTR) except VvbHLH1, VvbHLH2, VvbHLH5, VvbHLH6, VvbHLH8, VvbHLH10 and VvbHLH11, and those that don’t may not be annotated. Meanwhile, grape bHLH family members all contain Motif 1, indicating the relative conservatism of the bHLH family. In addition, bHLH members containing the same kinds of conserved motifs are evolutionarily more closely related. Based on Motif differences, it was found that Group 3 subfamily members all contained Motif 1, Motif 2, Motif 3, Motif 5, and Motif 7; Group 4 subfamily members all contained Motif 1, Motif 2, Motif 3, Motif 5, Motif 7, and Motif 8; and Group 6 subfamily members all contained Motif1, Motif2 and Motif 7. It is hypothesized that the differences in motifs contained in the different branches may be one of the reasons for the functional divergence of VvbHLHs during the evolutionary process.
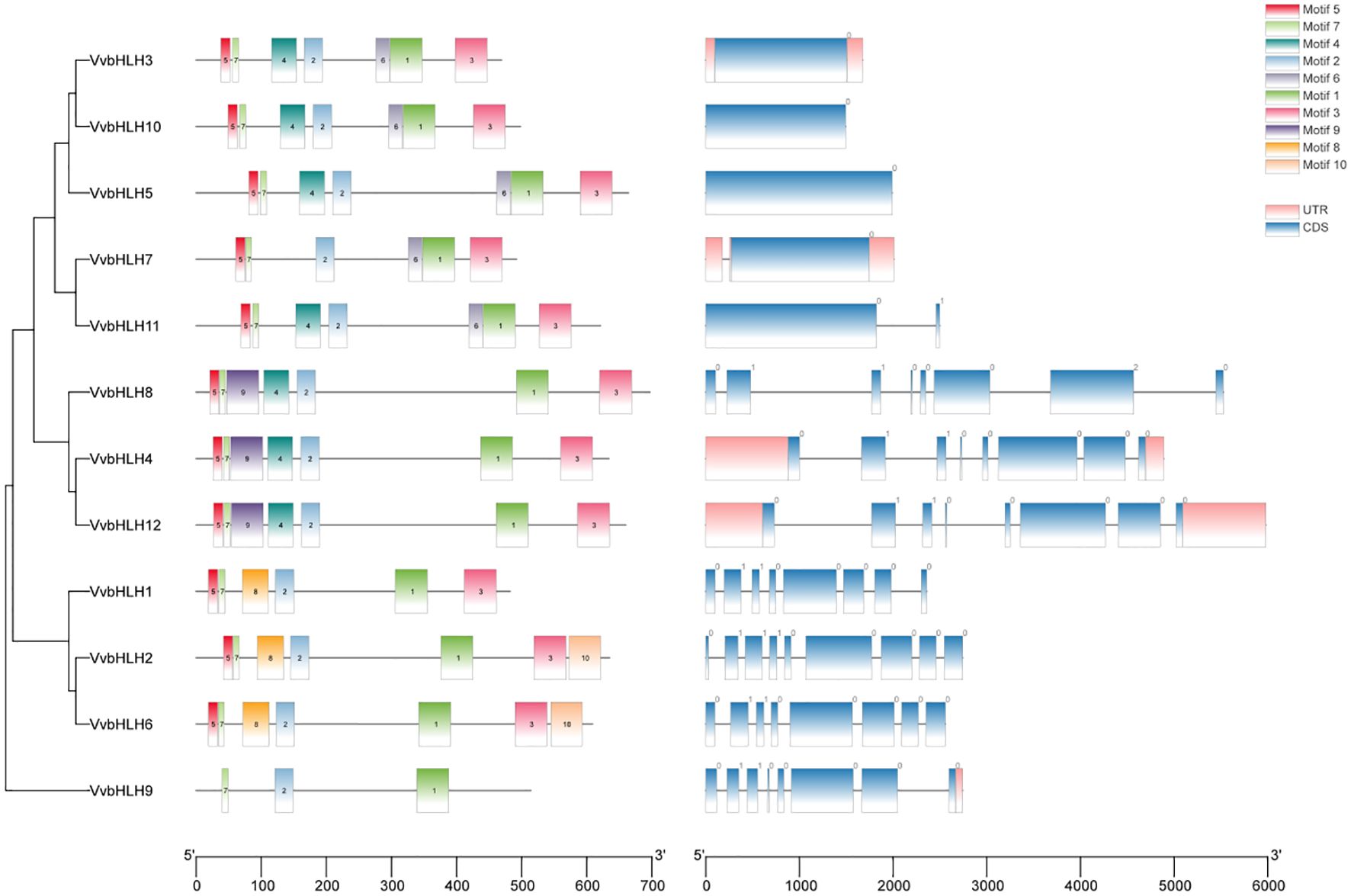
Figure 6. Phylogenetic analysis, gene structure and motif analysis of the grape bHLH protein family.
3.6 Cis promoter action elements of grape bHLHs
To understand the transcriptional regulation of grape bHLHs, in this study, we extracted the grape family gene sequences 2000 bp upstream of the start codon by using TBtools, and analyzed and counted the cis-acting elements in conjunction with the PlantCARE database, and then visualized and analyzed the results by using the TBtools after removing the redundant and irrelevant sequences manually. The results showed that the promoters of VvbHLHs differed in the type and number of elements contained in their promoters (Figure 7). The promoter region of the VvbHLHs contains many functional elements related to abiotic stresses, such as: circadian response elements, cis-regulatory elements involved in light response, cis-elements involved in low-temperature response, drought stress cis-responsive elements, and hypoxia-specific inducible elements (Figure 7). It was found that all bHLH family members contained light-responsive elements, and it was hypothesized that their functions were closely related to the light-responsive pathway. Hormone-related response elements accounted for more in grapes, with abscisic acid and methyl jasmonate response elements being the most numerous, with 23 and 24, respectively (Figure 7A). Therefore, it can be speculated that bHLHs are involved in the transmission of hormone signals such as auxin, abscisic acid, and methyl jasmonate. In addition, we found that six VvbHLH genes (bHLH2, bHLH4, bHLH6, bHLH7, bHLH8, and bHLH9) were involved in the regulation of zeinolysin metabolism in plants, which suggests that VvbHLHs are involved in metabolically regulated processes in plants (Figure 7A).
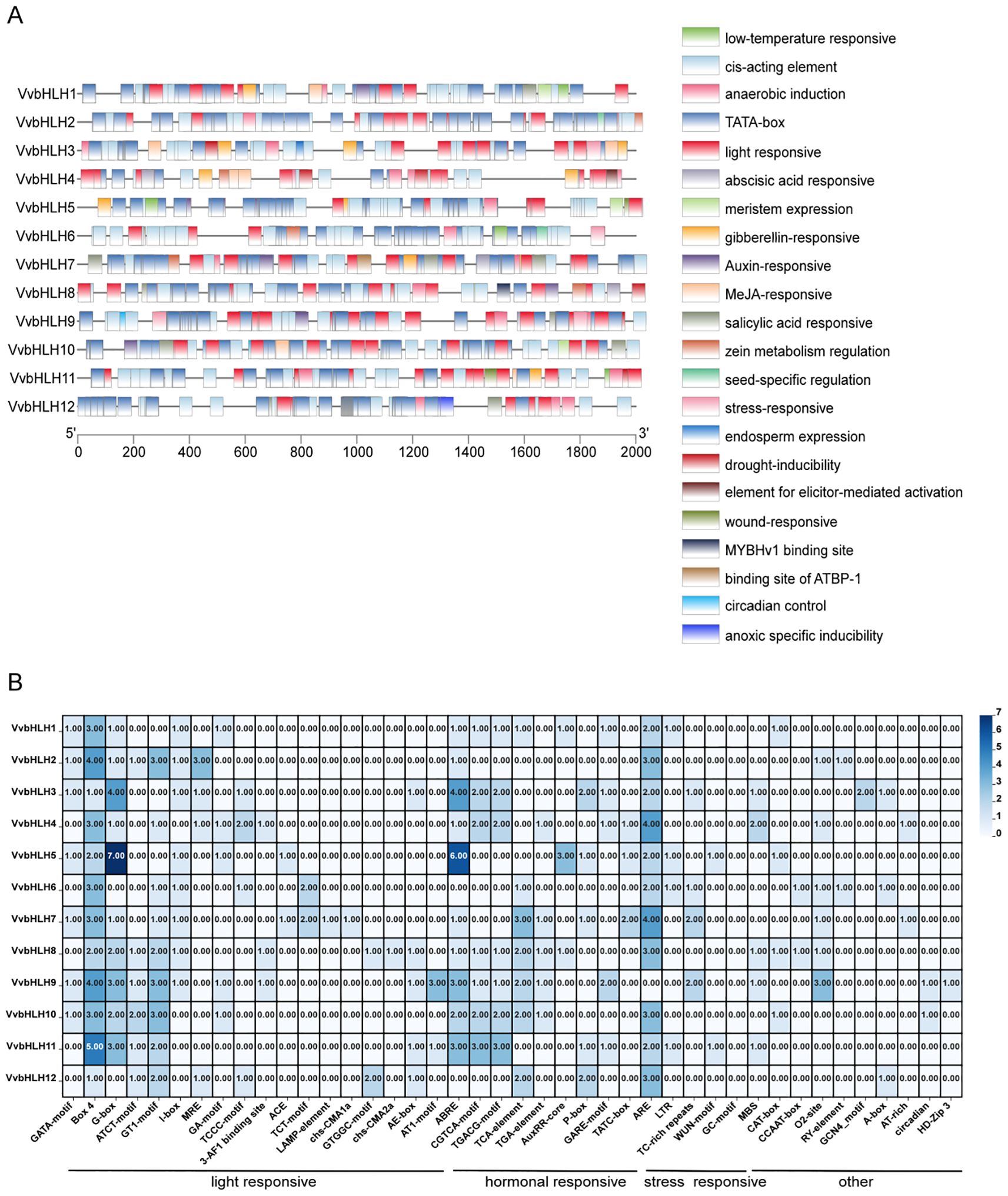
Figure 7. Analysis of the type and number of cis-acting elements in the promoter of grape bHLH gene. (A) Schematic diagram of cis-acting elements in the promoter of VvbHLH. (B) Heat map analysis of the number of cis-acting elements in the promoter of VvbHLH.
The study further analyzed the number of cis-acting elements in the VvbHLH promoter (Figure 7B). The results suggested that the major response elements included Box 4 and antioxidant response element (ARE), both of which were 1–5 and 2–4 in the VvbHLH promoter, respectively. Light-responsive elements mainly consisted of Box 4 and G-box elements, with VvbHLH2, VvbHLH9, and VvbHLH11 containing more than three Box 4 elements; VvbHLH5 contained up to seven G-box elements. The hormone response elements mainly consisted of abscisic acid response elements (ABRE), which were high in VvbHLH3, VvbHLH5, VvbHLH9, and VvbHLH11, with 4, 6, 3, and 3 elements, respectively. The stress response elements were mainly antioxidant response element (ARE) followed by defense and stress response element (TC-rich repeats) elements. Other acting elements mainly included maize alcohol soluble protein metabolic regulatory element (O2-site) and drought response element (MBS), etc.
3.7 Analysis of grapevine bHLHs expression patterns
We analyzed the spatio-temporal expression pattern of VvbHLHs during grapevine fruit development using RT-qPCR. Research has demonstrated that the expression of VvbHLH2 had significant spatio-temporal dynamics compared to other VvbHLH genes during different developmental periods in the plant, and the gene showed a higher expression level during the S4 (ripening) period of the fruit (Figure 8). Among the 12 VvbHLH genes, five VvbHLH genes (VvbHLH4, VvbHLH5, VvbHLH6, VvbHLH7, and VvbHLH8) had a significantly high expression in the fruit during the S3 (veraison) period, when grapes were in the veraison stage, which was hypothesized to be possibly associated with the accumulation of monoterpenes in grapes (Figure 8). In addition, five VvbHLH genes (VvbHLH1, VvbHLH2, VvbHLH3, VvbHLH9, and VvbHLH10) were up-regulated in gene expression during the S4 period, the VvbHLH11 gene was up-regulated during the S2 (expansion) period, and the VvbHLH12 gene was up-regulated during the S1 (young fruit) period. These findings reveal that these VvbHLHs exhibit distinct functions in development stages of grape berries.
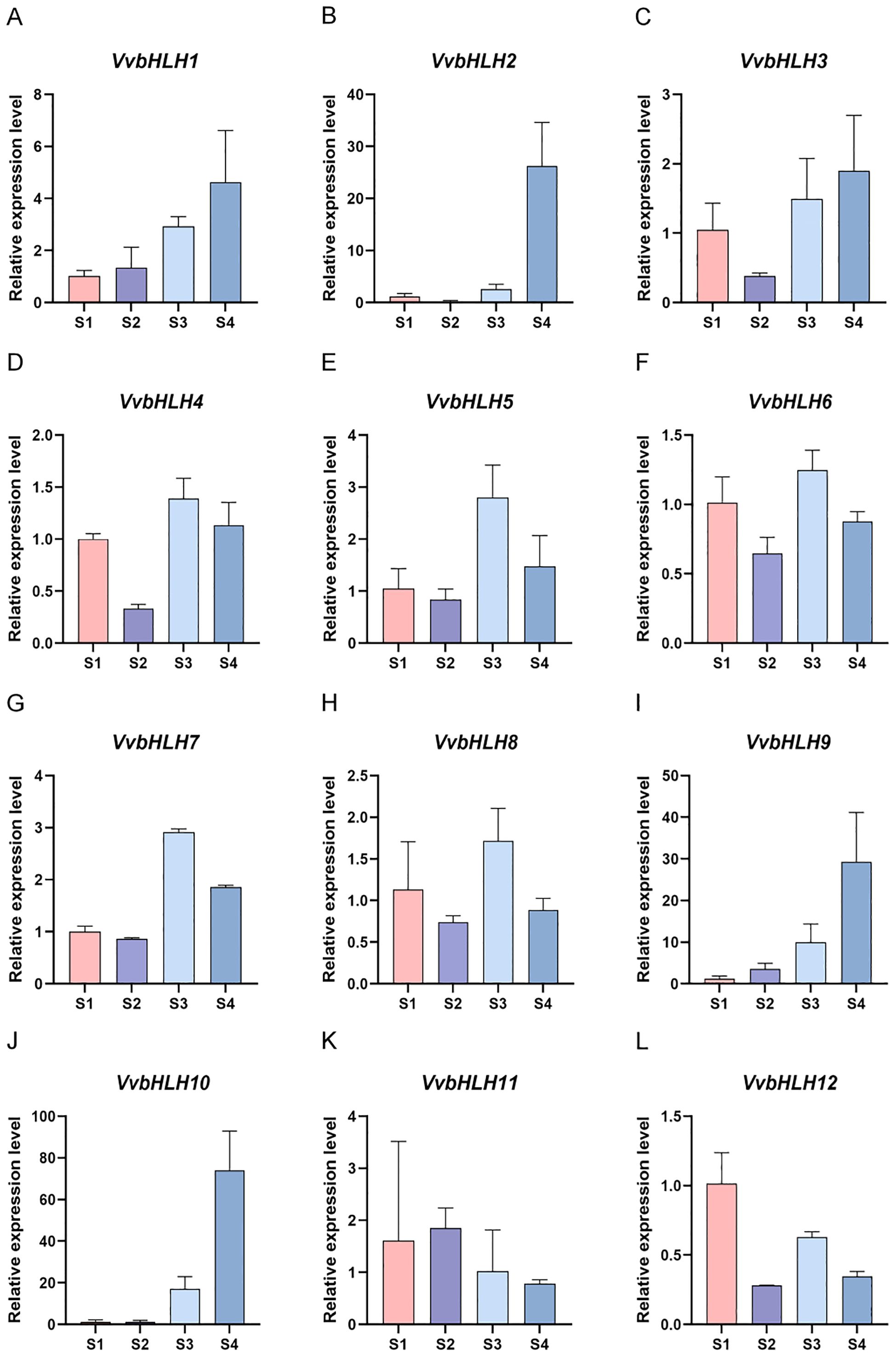
Figure 8. Expression pattern of bHLH gene family in ‘Ruidu Xiangyu’ grapes at different developmental stages. (A–L) Expression trends of 12 bHLH genes during four developmental stages (S1-S4).
3.8 Monoterpene content
Since monoterpene metabolites are mainly accumulated in grape berries, we investigated the accumulation of monoterpenes at four stages of grape berry development. The study identified 8 key monoterpene metabolites contained in all 4 developmental stages of ‘Ruidu Xiangyu’ grape berries, including terpinolene, cis-Rose oxide, cis-furan linalool oxide, linalool, hotrienol, β-Citronellol, nerol and geraniol. The results in Figure 9 show that the contents of the eight monoterpenes showed an overall increasing trend during the S1-S4 period, especially during the S3-S4 period when the rate of increase was more drastic. Specifically, except for nerolidol, the contents of the other seven monoterpenes showed a decreasing trend in the S1-S2 period.
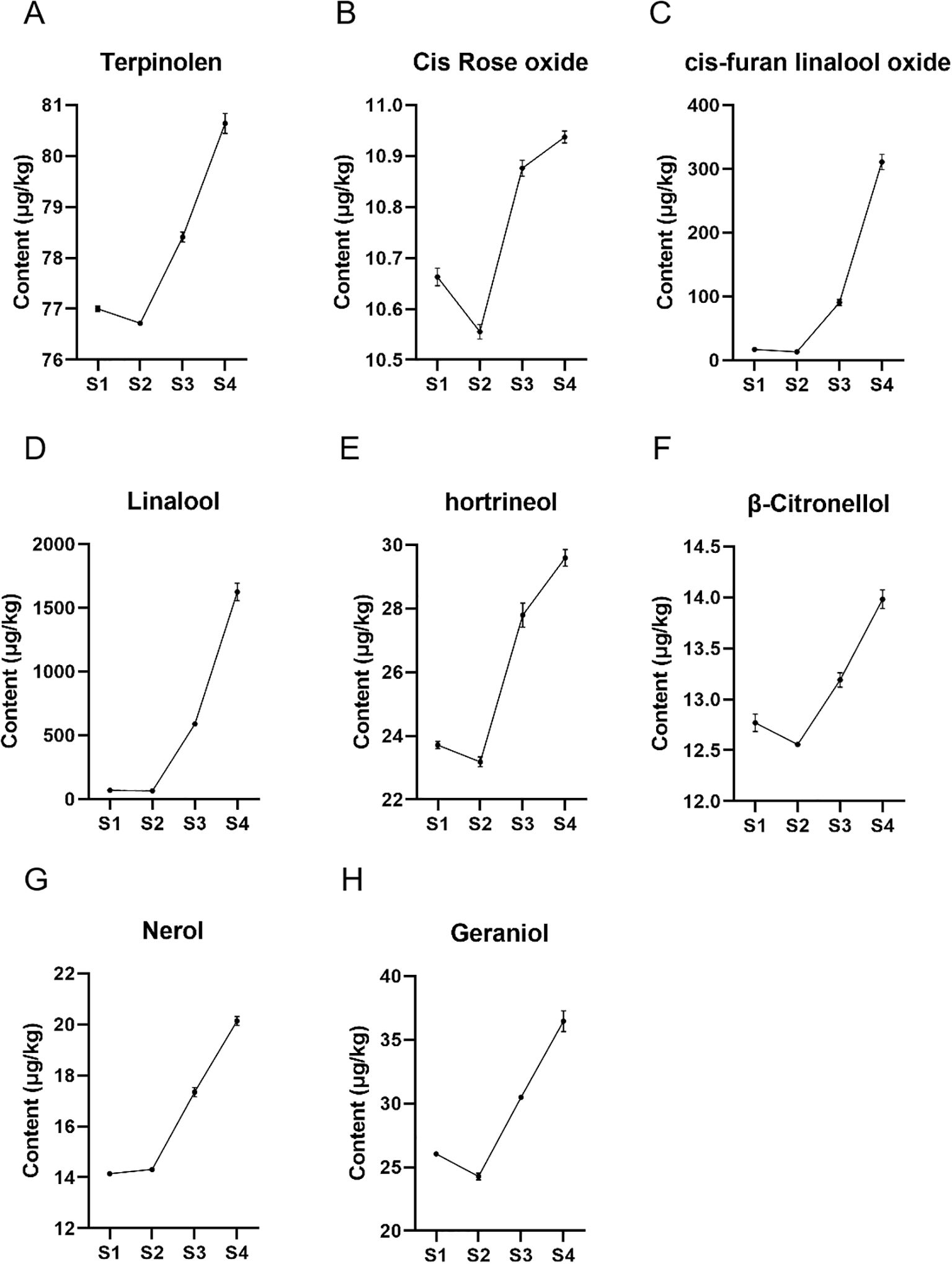
Figure 9. Monoterpene contents of ‘Ruidu Xiangyu’ grape berries in different periods. (A–H) 8 monoterpenes (terpinolene, cis-Rose oxide, cis-furan linalool oxide, linalool, hotrienol, β-Citronellol, nerol and geraniol) in developmental stages (S1–S4) of grape fruit. The horizontal axis represents developmental stages (S1–S4), and the vertical axis represents monoterpene concentrations (μg/kg). Data are presented as mean ± SD (n=3 biological replicates).
3.9 Correlation analysis
Since the monoterpene content in ‘Ruidu Xiangyu’ grape berries increased sharply in the S3 period (color change period), Pearson correlation analysis was conducted to assess the relationship between monoterpene content and the expression levels of the 12 VvbHLH genes across the S1–S4 developmental stages. It can be seen that there is also a correlation between the gene expression levels of the bHLH family members in grapes (Figure 10). It was found that VvbHLH1, VvbHLH2, VvbHLH3, VvbHLH9, and VvbHLH10 all displayed a pronounced positive linkage with monoterpene concentration in grape berries, while VvbHLH11 displayed a pronounced negative linkage with monoterpene content in grape berries (Figure 10; Supplementary Table 5). Specifically, VvbHLH1 displayed a pronounced positive linkage (P<0.05) with the increase in terpinolene, cis-furan linalool oxide, linalool, hotrienol, β-Citronellol and geraniol content in grapevine fruit, whereas it displayed a pronounced positive linkage (P<0.01) with the increase in nerol content, and VvbHLH2 displayed a pronounced positive linkage (P<0.01) with the increase in cis-furan linalool oxide and linalool concentration in grapevine fruit. VvbHLH3 displayed a pronounced positive linkage (P<0.05) with the growth of cis-Rose oxide content. VvbHLH9 and VvbHLH10 displayed a pronounced positive linkage (P<0.05) with terpinolene, β-Citronellol, nerol and geraniol content, and strongly positively correlated (P<0.01) with the growth of cis-furan linalool oxide and linalool content. VvbHLH11 displayed a pronounced negative linkage (P<0.05) for hotrienol content and for cis-Rose oxide content (P<0.01).
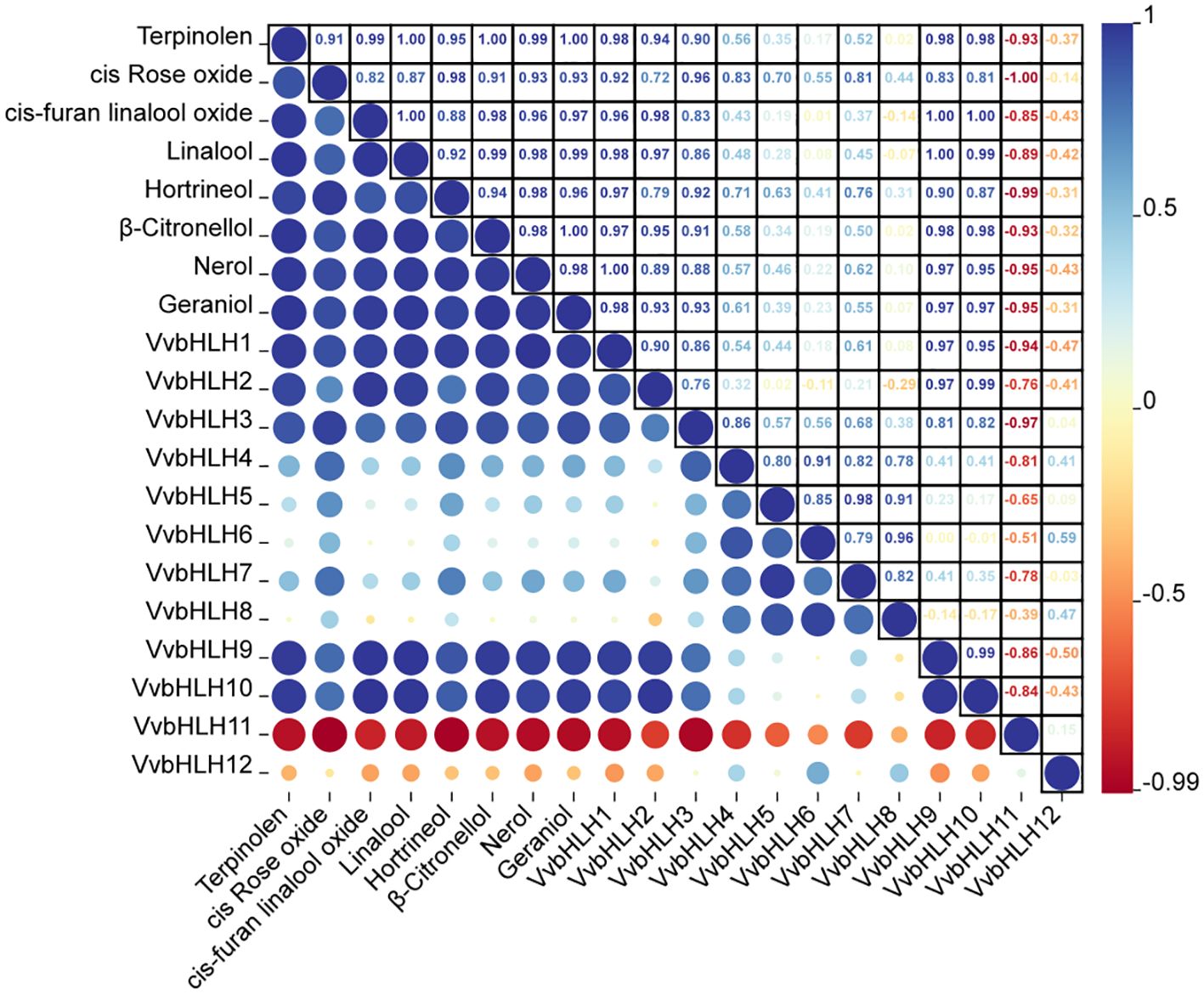
Figure 10. Correlation analysis of monoterpene content and gene expression in fruit of grapes at different periods of time.
3.10 Subcellular localization of VvbHLH9
To validate the predicted results of position of VvbHLH9 protein, we successfully cloned the full-length CDSs of VvbHLH9 gene and ligated them into PCAMBIA2300-GFP vector and transformed them into GV3101 receptor cells. Leaves of N. benthamiana were infested using the successfully tested Agrobacterium liquid, and the results were observed. Through subcellular localization, it was found that the green fluorescent protein without insertion of the VvbHLH9 gene was expressed in various organelles of N. benthamiana, but the green fluorescent protein fused with the VvbHLH9 protein was expressed only in the nucleus, which proved that the VvbHLH9 gene was expressed and functional in the nucleus (Figure 11).
3.11 Overexpression of VvbHLH9 promotes the synthesis of grape monoterpene metabolites
To further explore the contribution of VvbHLH9 in grape monoterpene production, the study was carried out to explore the overexpression of VvbHLH9 gene in ‘Alden’ grape leaves. It could be seen that the color of the infested leaves was significantly darker compared to the color of the non-infested leaves (Figure 12A). RT-qPCR data established that the gene markedly elevated the expression of critical enzyme-encoding genes involved in monoterpene formation, VvDXS1, VvDXS4, VvTPS31, VvTPS32 and VvTPS35 (Figure 12B). Quantification of monoterpene metabolites in grape leaves showed that overexpression of VvbHLH9 promoted an increase in the content of several monoterpenes including terpinolene, allo-ocimene, cis-furan linalool oxide, linalool, nerol and geraniol, with the increase in cis-furan linalool oxide and linalool being particularly significant (Figure 12C). This observation is in line with prior findings that VvbHLH9 displayed a pronounced positive linkage with the concentration of terpinolene, β-Citronellol, nerol, geraniol, cis-furan linalool oxide and linalool suggesting that VvbHLH9 positively regulates the synthesis of monoterpenes in grapes.
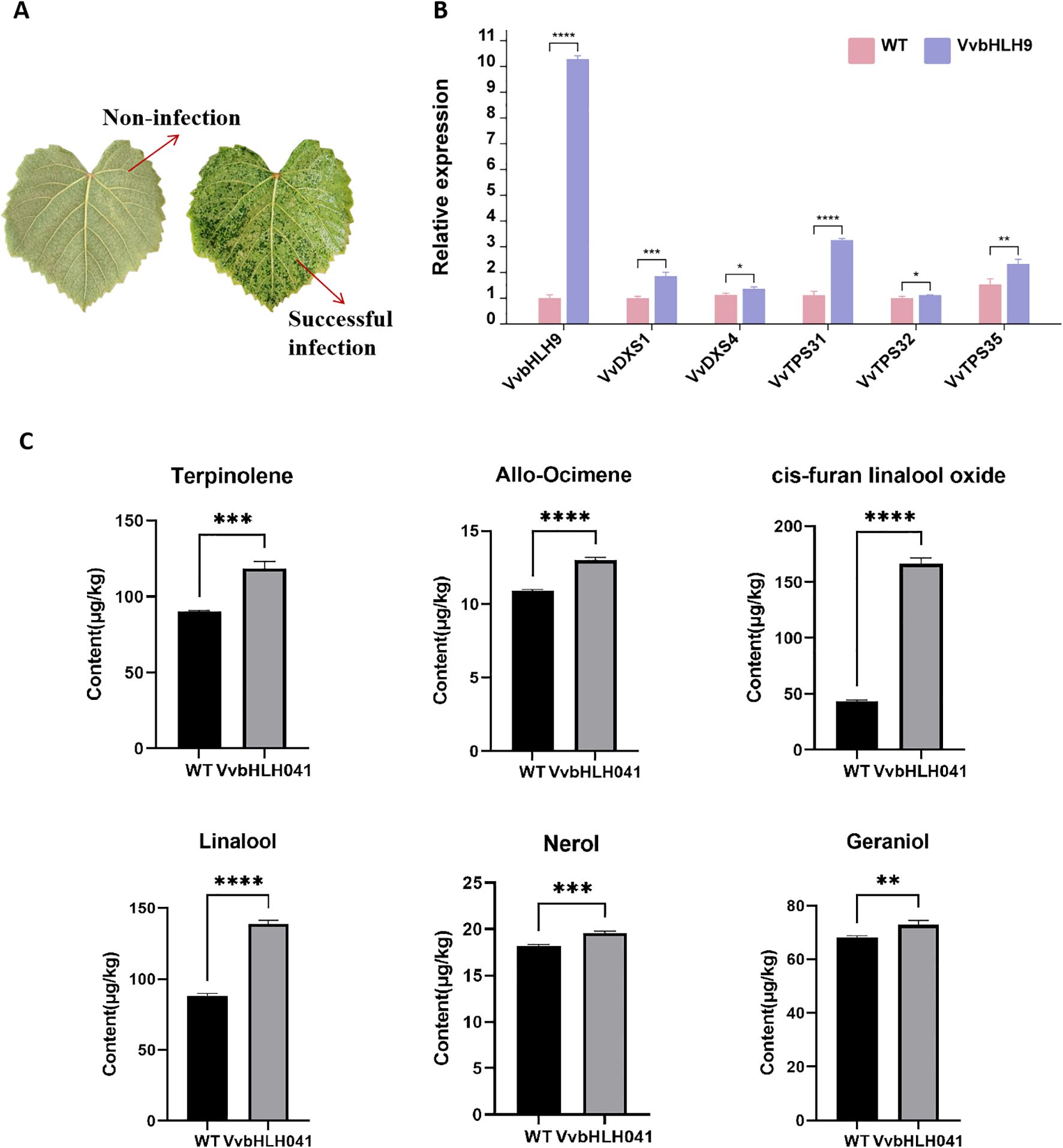
Figure 12. Functional analysis of VvbHLH9 in ‘Alden’ grape leaves. (A) Transient overexpression in grape leaves. Images show uninfected and infected leaves. (B) qRT-PCR results of monoterpene synthesis related genes in grape leaves before and after VvbHLH9 overexpression. Asterisk * represents significant level of p<0.1, asterisk ** represents significant level of p<0.01, asterisk *** represents significant level of p<0.001, and asterisk **** represents significant level of p<0.0001. (C) Concentration of monoterpenes (μg/kg) in ‘Alden’ grape leaves. WT represents the wild-type control; VvbHLH9 represents overexpressed VvbHLH9 group from infected leaves.
4 Discussion
As widespread transcriptional regulators in plants, the bHLH is strongly conserved in the regulation of metabolism (Wei and Chen, 2018). Carretero-Paulet et al. (2010) conducted a comprehensive phylogenetic analysis of bHLHs from Arabidopsis, poplar, rice, moss, and five algae species, classified the 638 identified bHLHs into 32 subfamilies, and defined the conserved regions of plant bHLHs, this opens up a feasible research pathway for identifying new bHLHs. In this research, the grape bHLHs was identified and analyzed based on the Arabidopsis bHLHs, and 12 VvbHLHs were identified. Chromosomal localization revealed that the chromosomal distribution of grape bHLH transcription factors was characterized by non-uniformity, which was also present in Arabidopsis and apple (Li et al., 2006; Mao et al., 2017). Further covariance analyses showed that VvbHLH family amplification was mainly driven by segmental replication (VvbHLH3/10, VvbHLH4/12), while tandem replication events were not seen, which provided clues for resolving its evolutionary mechanism. Notably, the VvbHLHs are mostly located in the nucleus, and their secondary structures are dominated by α-helix and random coil, which are highly similar to the structures of bHLHs from Arabidopsis and Ginkgo biloba, suggesting a conserved functional mechanism (Zhou et al., 2020; Liang et al., 2022). Phylogenetic analyses confirms that members with identical intron numbers and conserved motifs tend to cluster preferentially, this result aligns with earlier studies (Ke et al., 2020a). Moreover, grape bHLHs are more closely related to dicotyledonous plants (e.g., apple, pear), further supporting the species-specific association of bHLH family function with evolutionary relationships.
Promoter is a DNA sequence fragment recognized, bound, and transcribed by RNA polymerase, analyzing the cis acting elements of its promoter can help infer the potential function of genes (Hernandez-Garcia and Finer, 2014). Previous studies have shown that the bHLHs are essential for plant adaptation to abiotic stresses (Wu et al., 2022). In wheat, overexpression of the TabHLH1 can alter stomatal movement, leaf water loss rate, and other growth traits under drought stress in plants, and regulate the abscisic acid pathway to improve drought adaptation (Yang et al., 2016). Our investigation revealed that all 12 bHLHs contained hypoxia-specific inducible elements, indicating their potential involvement in stress response mechanisms. In addition, six VvbHLH genes (VvbHLH2, VvbHLH4, VvbHLH6, VvbHLH7, VvbHLH8, and VvbHLH9) were able to participate in the regulation of zeinolysin metabolism in plants, which suggests that VvbHLHs are involved in metabolic regulatory processes in plants. Previous studies have shown that MdbHLH3 in apple directly binds to the promoter of MdcyMDH and activates its transcriptional expression, which promotes malic acid accumulation in fruits (Yu et al., 2021). Increased expression of the bHLH AtMYC2 in Arabidopsis substantially increased the number of glandular hairs, which in turn promoted the synthesis of terpenes in glandular hairs (Zhai et al., 2013). The results verify that bHLH family members serve as key mediators of metabolic network regulation in planta.
Plant bHLH transcription factors often collaborate with other regulatory proteins to control the synthesis of diverse specialized metabolites, including terpenoids, phenylpropanoids, and flavonoids, and are critical for mediating plant-environment interactions (Feller et al., 2011). Terpenoids represent essential metabolic products that are ubiquitously present in plants. Existing research has demonstrated that bHLHs are significant regulatory factors in plant terpenoid biosynthesis (Yi et al., 2022). As a economically significant fruit species, grape quality attributes are strongly associated with terpenoid biosynthesis pathways (Zhang et al., 2024b). For investigating the impact of VvbHLHs on monoterpenoid synthesis patterns in grapevine, the study choose the juicy ‘Ruidu Xiangyu’ berries as the test material, and correlation analyses of gene expression and monoterpene content during the developmental period of the berries were carried out. The results showed that VvbHLH11 and VvbHLH12 were mainly expressed at the early stage of fruit development, a result consistent with the tendency of CmbHLH32 in melon to be expressed at the early stage of fruit development (Tan et al., 2021). In addition, in this study, five VvbHLHs (VvbHLH1, VvbHLH2, VvbHLH3, VvbHLH9, and VvbHLH10) had similar expression patterns, all of which demonstrating a marked upregulation in their expression during fruit ripening, these results are consistent with the functions of some bHLHs in grape, apple, and peach, indicating their potential importance in fruit ripening. It is hypothesized that such bHLHs may be the key genes driving the marked increase in monoterpene levels in fruits (Yang et al., 2017; Wang et al., 2018; Zhang et al., 2018a). Notably, VvbHLH11 expression showed a significant negative correlation with monoterpene accumulation, suggesting its potential role as a repressor in monoterpene biosynthesis. This inhibition might occur through competitive interference with activator-type bHLHs or direct downregulation of VvTPS genes. A comparable regulatory mechanism has been observed in Salvia miltiorrhiza, where SmbZIP1 suppresses tanshinone biosynthesis by inhibiting SmGGPPS expression (Deng et al., 2020). Further validation through VvbHLH11 knockout or ChIP assays would clarify its repressive mechanism in grape monoterpene synthesis.
Significantly, our investigation revealed a gene situated on chromosome 2, designated as VvbHLH3 (Vitvi02g00231), which encodes the transcription factor MYC1. Extensive evidence has revealed that MYC1 serves as an essential regulator in the metabolic pathways of flavonoids and terpenoids (Quattrocchio et al., 2006; Hichri et al., 2010; Muhammad et al., 2023; Li et al., 2024; Yu et al., 2024). In plants such as maize, petunia and Arabidopsis, MYC1 can promote flavonoid production by binding with MYB (Quattrocchio et al., 2006; Muhammad et al., 2023). In grape, VvMYC1 by itself does not activate the promoters of the VvCHI and VvUFGT of the flavonoid biosynthesis pathway, but its synergistic interaction with MYB significantly enhances promoter activity (Hichri et al., 2010). The current study identified VvMYC1 as a novel regulator of grape monoterpene metabolism, suggesting that a similar synergistic regulatory mechanism may also operate in this pathway. In addition, Yu et al. (2024) found that tomato SlMYC1 mediated trichome development and terpene metabolite synthesis by directly binding to the promoter of SlTOR and activating its expression, Our spatiotemporal expression studies demonstrated that the gene was significantly induced during the stage of highest monoterpene synthesis in grapes (from veraison to ripening), and displayed a pronounced positive linkage with the concentration of cis-Rose oxide monoterpenes. The above functional diversity indicates that bHLHs likely participate in modulating diverse secondary metabolic processes via a complex interaction network. However, the mechanism regarding the regulation of grape monoterpene production by MYC-like bHLH transcription factors is not yet fully understood and needs to be further clarified by means of molecular biology studies such as yeast single-hybrid and dual-luciferase reporter experiments.
Unlike the conserved structural domains of the remaining 11 VvbHLH transcription factors in this study, VvbHLH9 (annotated as bHLH041 in the NCBI RefSeq database, accession XP_002279486.2) does not contain the bHLH-MYC_N structural domain but retains the intact bHLH_AtbHLH_like structural domain, which drew our attention. Previous studies on terpene biosynthesis-associated bHLHs in other plants (e.g., Arabidopsis and tomato) have been relatively scarce, focusing mainly on MYC2 (Kazan and Manners, 2013; Spyropoulou et al., 2014). Since then, researchers have identified MYC2-independent bHLHs (BIS1 and BIS2), in the medicinal plant periwinkle, which both activate the monoterpene formation of the MIA (Liu et al., 2021). This investigation uncovered high expression of VvbHLH9 during grape ripening and its strong correlation with the accumulation of key monoterpenes (e.g., linalool, terpinolene) suggests that it orchestrates monoterpene synthesis fluxes through developmental stage-specific transcription. To further validate the function of VvbHLH9 in monoterpene synthesis, subcellular localization of the gene was investigated and transgenic lines overexpressing the gene were established in ‘Alden’ grape leaves. The results confirmed that VvbHLH9 exerted its transcriptional regulatory function mainly in the nucleus and significantly activated the transcription of genes such as VvDXS1 and VvTPS31, and positively regulated the biosynthesis of a variety of monoterpene metabolites including linalool. This suggests that VvbHLH9 may exert regulatory effects on monoterpene metabolites by activating upstream synthetic modules. However, given that this gene lacks a typical MYC structural domain, whether its regulation is achieved through the formation of heterodimers or modification of chromatin state still needs to be further resolved by Co-IP combined mass spectrometry analysis.
5 Conclusion
This research carried out a systematic genomic characterization of the grapevine bHLHs through bioinformatics, with the goal of screening for VvbHLH genes linked to monoterpene production. The study identified 12 key bHLHs genes in the grapevine genome. The results of covariance analysis indicated intraspecific covariance between two pairs of VvbHLHs and interspecific covariance between eight pairs of VvbHLHs and Arabidopsis bHLHs. Using phylogenetic reconstruction, 12 VvbHLHs were divided into four main subfamilies. RT-qPCR results demonstrated that the VvbHLHs were expressed in different developmental periods in grape and showed spatio-temporal specificity. By correlating grape VvbHLHs with the results of monoterpene content measurements, five VvbHLHs (VvbHLH1, VvbHLH2, VvbHLH3, VvbHLH9, and VvbHLH10) were found to be closely related to a dramatic growth in monoterpene concentration. Among them, VvbHLH9 (VvbHLH9) was shown to be associated with the accumulation of monoterpene metabolites such as terpinolene, allo-ocimene, nerol, geraniol, cis-furan linalool oxide and linalool in grapevines. The investigation sheds light on the whole-genome identification of grape bHLHs and their evolutionary history, and has important implications for quality improvement and molecular breeding in grape.
Data availability statement
Existing datasets are available in a publicly accessible repository:Publicly available datasets were analyzed in this study. The original proteome file of Vitis vinifera can be found in the Ensembl Plants repository here: [https://ftp.ebi.ac.uk/ensemblgenomes/pub/release-59/plants/fasta/vitis_vinifera/pep/]. The protein sequences of Arabidopsis thaliana bHLH transcription factors were obtained from the PlantTFDB database: [https://planttfdb.gao-lab.org/tf.php?sp=Ath&did=AT1G01260.1].
Author contributions
QS: Data curation, Methodology, Visualization, Writing – original draft, Writing – review & editing, Formal Analysis. ZL: Formal Analysis, Investigation, Software, Validation, Writing – review & editing. XW: Formal Analysis, Validation, Visualization, Writing – review & editing. AY: Formal Analysis, Validation, Visualization, Writing – review & editing. JR: Formal Analysis, Validation, Visualization, Writing – review & editing. HW: Conceptualization, Data curation, Resources, Validation, Writing – review & editing. LS: Conceptualization, Funding acquisition, Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was supported by the BAAFS Funding for the Development of Distinguished Scientist JKZX202402; the National Natural Science Foundation of China (No. 32472677), the Earmarked Fund for Modern Agro-industry Technology Research System CARS-29; Beijing Academy of Agriculture and Forestry Sciences Innovation Capability Construction Special Project (KJCX20230118).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1653650/full#supplementary-material
References
Bano, N., Patel, P., Chakrabarty, D., and Bag, S. K. (2021). Genome-wide identification, phylogeny, and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum). Physiol. Mol. Biol. Plants 27, 1747–1764. doi: 10.1007/s12298-021-01042-x
Boncan, D. A. T., Tsang, S. S., Li, C., Lee, I. H., Lam, H.-M., Chan, T.-F., et al. (2020). Terpenes and terpenoids in plants: Interactions with environment and insects. Int. J. Mol. Sci. 21, 7382. doi: 10.3390/ijms21197382
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chedea, V. S., Macovei, O., Bocsan, I. C., Măgureanu, D. C., Levai, A. M., Buzoianu, A. D., et al. (2022). Grape pomace polyphenols as a source of compounds for management of oxidative stress and inflammation—A possible alternative for non-steroidal anti-inflammatory drugs? Molecules. 27, 6826. doi: 10.3390/molecules27206826
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Deng, C., Shi, M., Fu, R., Zhang, Y., Wang, Q., Zhou, Y., et al. (2020). ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 71, 5948–5962. doi: 10.1093/jxb/eraa295
Eslami Bojnourdi, N., Haddad, R., and Heidari-Japelaghi, R. (2023). Molecular cloning and in silico analysis of a GTP cyclohydrolase I gene from grape. J. Plant Mol. Breed. 11, 1–16. doi: 10.22058/jpmb.2023.1990428.1271
Fan, Y., Lai, D., Yang, H., Xue, G., He, A., Chen, L., et al. (2021). Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genomics 22, 1–18. doi: 10.1186/s12864-021-08095-y
Fan, M., Yuan, S., Li, L., Zheng, J., Zhao, D., Wang, C., et al. (2023). Application of terpenoid compounds in food and pharmaceutical products. Fermentation. 9, 119. doi: 10.3390/fermentation9020119
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Gao, M., Zhu, Y., Yang, J., Zhang, H., Cheng, C., Zhang, Y., et al. (2019). Identification of the grape basic helix–loop–helix transcription factor family and characterization of expression patterns in response to different stresses. Plant Growth Regulation. 88, 19–39. doi: 10.1007/s10725-019-00485-3
Hao, Y., Zong, X., Ren, P., Qian, Y., and Fu, A. (2021). Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int. J. Mol. Sci. 22, 7152. doi: 10.3390/ijms22137152
Harshitha, R. and Arunraj, D. R. (2021). Real-time quantitative PCR: A tool for absolute and relative quantification. Biochem. Mol. Biol. Educ. 49, 800–812. doi: 10.1002/bmb.21552
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Hernandez-Garcia, C. M. and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Hong, G.-J., Xue, X.-Y., Mao, Y.-B., Wang, L.-J., and Chen, X.-Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648. doi: 10.1105/tpc.112.098749
Kazan, K. and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Ke, Q., Tao, W., Li, T., Pan, W., Chen, X., Wu, X., et al. (2020a). Genome-wide identification, evolution and expression analysis of basic helix-loop-helix (bHLH) gene family in barley (Hordeum vulgare L.). Curr. Genomics 21, 624–644. doi: 10.2174/1389202921999201102165537
Ke, Y.-Z., Wu, Y.-W., Zhou, H.-J., Chen, P., Wang, M.-M., Liu, M.-M., et al. (2020b). Genome-wide survey of the bHLH super gene family in Brassica napus. BMC Plant Biol. 20, 1–16. doi: 10.1186/s12870-020-2315-8
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, M., Sun, L., Gu, H., Cheng, D., Guo, X., Chen, R., et al. (2021). Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 11, 6863. doi: 10.1038/s41598-021-85754-w
Li, H., Yang, Y., Zhang, W., Zheng, H., Xu, X., Li, H., et al. (2024). Promoter replication of grape MYB transcription factor is associated with a new red flesh phenotype. Plant Cell Rep. 43, 136. doi: 10.1007/s00299-024-03225-8
Liang, J., Fang, Y., An, C., Yao, Y., Wang, X., Zhang, W., et al. (2023). Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 225, 389–403. doi: 10.1016/j.ijbiomac.2022.11.076
Liang, Y., Ma, F., Li, B., Guo, C., Hu, T., Zhang, M., et al. (2022). A bHLH transcription factor, SlbHLH96, promotes drought tolerance in tomato. Hortic. Res. 9, uhac198. doi: 10.1093/hr/uhac198
Liu, G.-S., Li, H.-L., Grierson, D., and Fu, D.-Q. (2022a). NAC transcription factor family regulation of fruit ripening and quality: a review. Cells 11, 525. doi: 10.3390/cells11030525
Liu, Y., Patra, B., Singh, S. K., Paul, P., Zhou, Y., Li, Y., et al. (2021). Terpenoid indole alkaloid biosynthesis in Catharanthus roseus: effects and prospects of environmental factors in metabolic engineering. Biotechnol. Lett. 43 (11), 1–19. doi: 10.1007/s10529-021-03179-x
Liu, S., Shan, B., Zhou, X., Gao, W., Liu, Y., Zhu, B., et al. (2022b). Transcriptome and metabolomics integrated analysis reveals terpene synthesis genes controlling linalool synthesis in grape berries. J. Agric. Food Chem. 70, 9084–9094. doi: 10.1021/acs.jafc.2c00368
Mao, K., Dong, Q., Li, C., Liu, C., and Ma, F. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00480
Mele, M. A., Kang, H.-M., Lee, Y.-T., and Islam, M. Z. (2021). Grape terpenoids: Flavor importance, genetic regulation, and future potential. Crit. Rev. Food Sci. Nutr. 61, 1429–1447. doi: 10.1080/10408398.2020.1760203
Mertens, J., Pollier, J., Vanden Bossche, R., Lopez-Vidriero, I., Franco-Zorrilla, J. M., and Goossens, A. (2016). The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 170, 194–210. doi: 10.1104/pp.15.01645
Muhammad, N., Uddin, N., Khan, M. K. U., Ali, N., Ali, K., and Jones, D. A. (2023). Diverse role of basic Helix-Loop-Helix (bHLH) transcription factor superfamily genes in the fleshy fruit-bearing plant species. Czech J. Genet. Plant Breed. 59, 1–13. doi: 10.17221/2/2022-CJGPB
Quattrocchio, F., Verweij, W., Kroon, A., Spelt, C., Mol, J., and Koes, R. (2006). PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18, 1274–1291. doi: 10.1105/tpc.105.034041
Sabo, J., Farkasová, S., Droppa, M., Žiarovská, J., and Kačániová, M. (2022). Molecular fingerprinting and microbiological characterisation of selected Vitis vinifera L. Varieties. Plants 11, 3375. doi: 10.3390/plants11233375
Salih, H., Tan, L., and Htet, N. N. W. (2021). Genome-wide identification, characterization of bHLH transcription factors in mango. Trop. Plant Biol. 14, 72–81. doi: 10.1007/s12042-020-09277-w
Spyropoulou, E. A., Haring, M. A., and Schuurink, R. C. (2014). RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics 15, 1–16. doi: 10.1186/1471-2164-15-402
Sun, H., Fan, H.-J., and Ling, H.-Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16, 1–12. doi: 10.1186/s12864-014-1209-2
Tan, C., Qiao, H., Ma, M., Wang, X., Tian, Y., Bai, S., et al. (2021). Genome-wide identification and characterization of melon bHLH transcription factors in regulation of fruit development. Plants 10, 2721. doi: 10.3390/plants10122721
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Wang, P., Su, L., Gao, H., Jiang, X., Wu, X., Li, Y., et al. (2018). Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00064
Wang, Y., Tang, H., Wang, X., Sun, Y., Joseph, P. V., and Paterson, A. H. (2024). Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 19, 2206–2229. doi: 10.1038/s41596-024-00968-2
Wang, J., VanderWeide, J., Yan, Y., Tindjau, R., Pico, J., Deluc, L., et al. (2022b). Impact of hormone applications on ripening-related metabolites in Gewürztraminer grapes (Vitis vinifera L.): The key role of jasmonates in terpene modulation. Food Chem. 388, 132948. doi: 10.1016/j.foodchem.2022.132948
Wang, C., Wang, L., Ye, J., and Xu, F. (2022a). Fruit quality of Vitis vinifera: How plant metabolites are affected by genetic, environmental, and agronomic factors. Sci. Hortic. 305, 111404. doi: 10.1016/j.scienta.2022.111404
Wei, K. and Chen, H. (2018). Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 18, 1–21. doi: 10.1186/s12870-018-1529-5
Wu, Y., Wu, S., Wang, X., Mao, T., Bao, M., Zhang, J., et al. (2022). Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mume. Hortic. Plant J. 8, 531–544. doi: 10.1016/j.hpj.2022.01.004
Yang, J., Gao, M., Huang, L., Wang, Y., van Nocker, S., Wan, R., et al. (2017). Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 7, 28. doi: 10.1038/s41598-017-00040-y
Yang, T., Yao, S., Hao, L., Zhao, Y., Lu, W., and Xiao, K. (2016). Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 35, 2309–2323. doi: 10.1007/s00299-016-2036-5
Yi, X., Wang, X., Wu, L., Wang, M., Yang, L., Liu, X., et al. (2022). Integrated analysis of basic helix loop helix transcription factor family and targeted terpenoids reveals candidate AarbHLH genes involved in terpenoid biosynthesis in Artemisia argyi. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.811166
Yu, J. Q., Gu, K. D., Sun, C. H., Zhang, Q. Y., Wang, J. H., Ma, F. F., et al. (2021). The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 19, 285–299. doi: 10.1111/pbi.13461
Yu, L., Zhang, Y., Ding, Q., Wang, H., Meng, X., Fan, H., et al. (2024). The slMYC1-TOR module regulates trichome formation and terpene biosynthesis in tomatoes (Solanum lycopersicum L.). J. Plant Growth Regul. 43, 3282–3294. doi: 10.1007/s00344-024-11292-0
Zhai, Q., Yan, L., Tan, D., Chen, R., Sun, J., Gao, L., et al. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PloS Genet. 9, e1003422. doi: 10.1371/journal.pgen.1003422
Zhang, Z., Chen, C., Jiang, C., Lin, H., Zhao, Y., and Guo, Y. (2024c). VvWRKY5 positively regulates wounding-induced anthocyanin accumulation in grape by interplaying with VvMYBA1 and promoting jasmonic acid biosynthesis. Hortic. Res. 11, uhae083. doi: 10.1093/hr/uhae083
Zhang, C., Feng, R., Ma, R., Shen, Z., Cai, Z., Song, Z., et al. (2018a). Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PloS One 13, e0195974. doi: 10.1371/journal.pone.0195974
Zhang, Y., Liu, F., Wang, B., Wu, H., Wu, J., Liu, J., et al. (2021). Identification, characterization and expression analysis of anthocyanin biosynthesis-related bHLH genes in blueberry (Vaccinium corymbosum L.). Int. J. Mol. Sci. 22, 13274. doi: 10.3390/ijms222413274
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018b). Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18, 1–14. doi: 10.1186/s12870-018-1441-z
Zhang, X.-Y., Qiu, J.-Y., Hui, Q.-L., Xu, Y.-Y., He, Y.-Z., Peng, L.-Z., et al. (2020). Systematic analysis of the basic/helix-loop-helix (bHLH) transcription factor family in pummelo (Citrus grandis) and identification of the key members involved in the response to iron deficiency. BMC Genomics 21, 1–16. doi: 10.1186/s12864-020-6644-7
Zhang, H., Ye, S., Wang, N., Xu, Z., and Gong, S. (2024a). Analyses of the bHLH gene family in Populus trichocarpa reveal roles of four PtbHLHs in regulating the drought stress response. Environ. Exp. Bot. 228, 106046. doi: 10.1016/j.envexpbot.2024.106046
Zhang, K., Zhang, J., Zheng, T., Gu, W., Zhang, Y., Li, W., et al. (2024b). Preharvest application of MeJA enhancing the quality of postharvest grape berries via regulating terpenes biosynthesis and phenylpropanoid metabolisms. Food Chem. 438, 137958. doi: 10.1016/j.foodchem.2023.137958
Zhou, X., Liao, Y., Kim, S.-U., Chen, Z., Nie, G., Cheng, S., et al. (2020). Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 10, 13723. doi: 10.1038/s41598-020-69305-3
Keywords: grape, bHLH gene family, bioinformatics, gene expression, monoterpene accumulation, subcellular localization, transient overexpression
Citation: Sun Q, Liu Z, Wang X, Yan A, Ren J, Wang H and Sun L (2025) Whole-genome identification and functional analysis of grape bHLH transcription factors: new insights into the regulation of monoterpene biosynthesis. Front. Plant Sci. 16:1653650. doi: 10.3389/fpls.2025.1653650
Received: 25 June 2025; Accepted: 30 July 2025;
Published: 16 September 2025.
Edited by:
Pan Liao, Hong Kong Baptist University, Hong Kong SAR, ChinaReviewed by:
Dahe Qiao, Guizhou Academy of Agricultural Sciences (CAAS), ChinaSongyu Liu, The University of Auckland, New Zealand
Copyright © 2025 Sun, Liu, Wang, Yan, Ren, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiling Wang, d2FuZ2h1aWxpbmdAYmFhZnMubmV0LmNu; Lei Sun, c3VubGVpQGJhYWZzLm5ldC5jbg==
 Qian Sun
Qian Sun Zhenhua Liu1,3
Zhenhua Liu1,3 Lei Sun
Lei Sun