- 1Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China
- 2Institute of Industrial Crops, Shandong Academy of Agricultural Sciences, Jinan, China
- 3Jilin Provincial Key Laboratory of Traditional Chinese Medicinal Materials Cultivation and Propagation, Changchun, China
Introduction: Continuous cropping (CC) poses significant challenges to the yield and quality of Salvia miltiorrhiza, a medicinally important plant with annual market demand of around 20 million kg. Previous studies have explored chemical mitigation methods, but concerns persist regarding environmental pollution and safety issues.
Methods: In this study, we evaluated three strains of the fungus Trichoderma (T. brevicompactum, T. viridescens, and T. velutinum) as ecofriendly alternatives for CC obstacle mitigation in S. miltiorrhiza cultivation.
Results: The Trichoderma treatments significantly enhanced the relative abundance of beneficial soil bacteria (Actinobacteria 9.25%–16.88%; Chloroflexi 6.41%–16.73%; Gemmatimonadetes 1.21%–3.16%), while decreasing the abundance of pathogenic Fusarium by 12.67%–31.75%. Soil analysis revealed substantial improvements in total organic carbon (47.35%–65.88%), nitrate nitrogen (91.38%–318.89%), available potassium (4.29%–17.16%), and available phosphorus (1.85%–11.86%) following Trichoderma treatment. T. brevicompactum demonstrated superior performance among the three tested strains, increasing the individual plant fresh weight by 17.79% and the survival rate by 13.33%. This treatment also significantly elevated (p < 0.05) the content of key bioactive compounds in S. miltiorrhiza root: tanshinone IIA (51.16% increase), cryptotanshinone (56.76%), tanshinone I (50.00%), and salvianolic acid B (18.43%).
Discussion: Trichoderma can effectively alleviate S. miltiorrhiza CC obstacles by improving the soil nutrient status and modulating the soil microbial community, thereby enhancing plant growth and stress resistance. This study provides a promising ecofriendly strategy for sustainable cultivation of S. miltiorrhiza and other medicinal plants facing similar challenges.
1 Introduction
Salvia miltiorrhiza is a perennial plant native to China and Japan, also known as red sage or Chinese sage. Its roots are a commonly used bulk medicinal material, which has demonstrated therapeutic efficacy in the treatment of pathologies including cardiovascular and cerebrovascular diseases, liver diseases, and diabetes (Feng et al., 2022; Zhang et al., 2024c). The annual market demand for S. miltiorrhiza is around 20 million kg, and this is increasing (Liu et al., 2020). With the increase in the planting area of S. miltiorrhiza, the problem of continuous cropping (CC) obstacles has gradually become prominent.
CC obstacles are a prevalent issue in cultivation of medicinal plants (Zeeshan Ul. Haq. et al., 2023), particularly for root-derived materials (Yang et al., 2024b). CC compromises plant growth because of deteriorating soil physicochemical properties, disrupted soil microbial communities, and increased soil-borne diseases, leading to decreased crop productivity and quality (Chen et al., 2022; Su et al., 2023). Previous studies demonstrated that the incidence of root diseases in S. miltiorrhiza reached approximately 10% after one year of cultivation, escalating to 60%–70% following three consecutive cropping cycles (Pu et al., 2022; Ju et al., 2023). Furthermore, excessive dependence on chemical fertilizers and pesticides has been documented to exacerbate soil degradation and lead to accumulation of pesticide residues (Yan et al., 2022). Consequently, CC has emerged as a critical constraint on sustainable S. miltiorrhiza production. Although preliminary insights into the CC obstacle mechanisms have been obtained, effective management strategies remain exploratory due to system complexity.
Previous studies have demonstrated that dazomet and chloropicrin fumigation can enhance the yield of crops and reduce the abundance of harmful microorganisms (Wang et al., 2023; Chen et al., 2023). However, the application of chemical methods has potential risks, including environmental pollution, ecosystem disruption, and consumer safety concerns. In contrast, research has confirmed that biocontrol microorganisms (e.g., Trichoderma spp.) promote plant biomass by modifying soil conditions (Meng et al., 2019; Wang et al., 2021), offering a nontoxic, ecofriendly alternative without chemical residues.
Trichoderma spp. are beneficial fungi in soil and plant root ecosystems (Kashyap et al., 2017). Numerous studies have shown that Trichoderma spp. have significant control effects on various plant diseases (Rees et al., 2022; Rodrigues et al., 2023), and promote plant growth and improve soil health (Wu et al., 2022; Zhang et al., 2024b). Trichoderma has been demonstrated to alleviate CC obstacles. Chen et al (2011, 2012) found that application of T. harzianum effectively degraded allelopathic substances in the rhizosphere of continuously cropped cucumber, diversified the soil microbial community, and controlled cucumber wilt disease. Similarly, Mao and Jiang (2021) reported that application of Trichoderma MHT1134 significantly decreased the incidence of Capsicum wilt and improved the soil microbiome composition in CC conditions. In five-year continuously cropped watermelon soil, T. asperellum M45a granules improved Fusarium wilt control efficacy by up to 67.44% during flowering and significantly enhanced soil nutrient conversion rates [total nitrogen (TN), nitrate-nitrogen (NO3—N), and available phosphorus (AP)] (Zhang et al., 2020). However, the effects of Trichoderma on S. miltiorrhiza remain unexplored.
In this study, three soil-isolated Trichoderma strains were selected to investigate their impacts on biomass, soil chemical properties, and microbial community structure for continuously cropped S. miltiorrhiza. Our findings may provide valuable insights for sustainable, pollution-free S. miltiorrhiza production.
2 Materials and methods
2.1 Fungal strains
Three Trichoderma strains were obtained from the Institute of Special Wild Economic Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China. They were isolated from ginseng roots in samples from Zuojia Town (Jilin City, Jilin Province, China). Following morphological and molecular analyses, these strains were identified as T. brevicompactum (CGMCC No. 23213), T. viridescens (CGMCC No. 23212), and T. velutinum (CGMCC No. 23211), and preserved at the China General Microbiological Culture Collection Center (CGMCC).
2.2 Experimental design
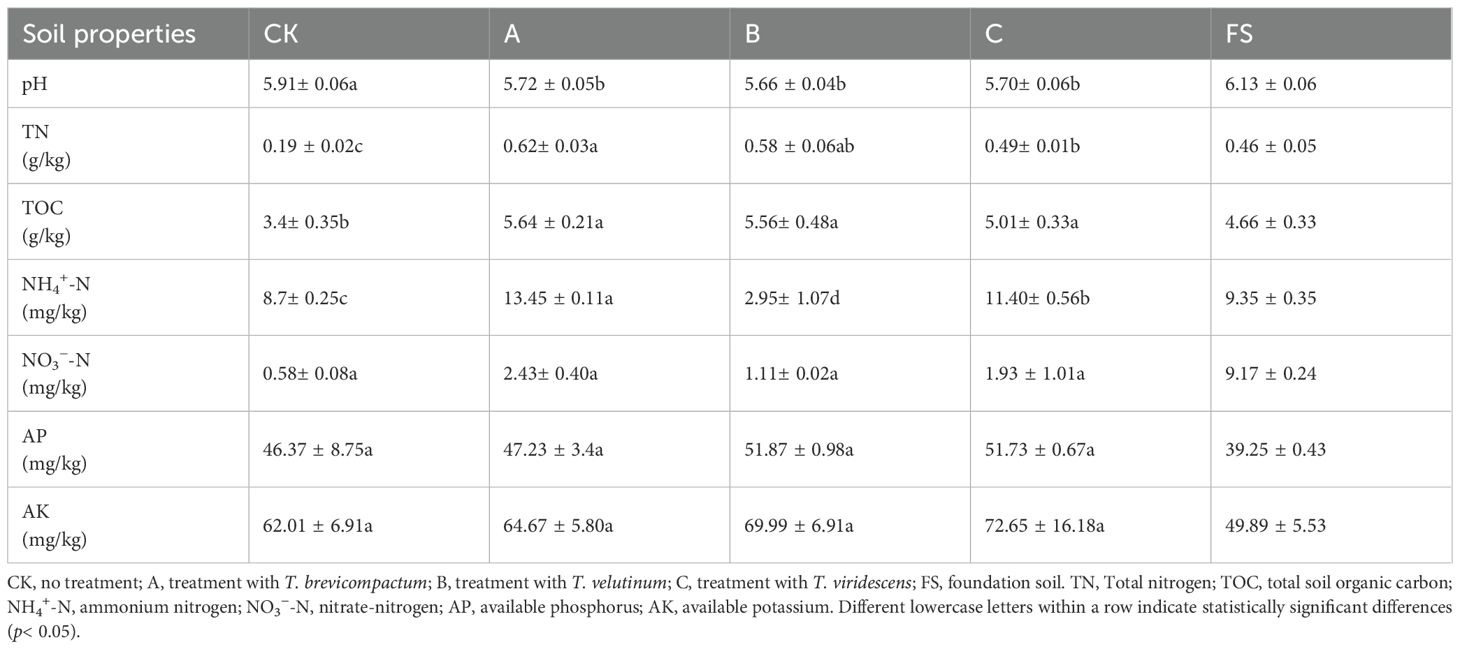
This study was carried out between May and November 2022 in Pingyi County, Linyi City, Shandong Province, China. This plot has been continuously planted with S. miltiorrhiza for 6 years. The physicochemical properties of the foundation soil are presented in Table 1. The experiments consisted of four treatments: T. brevicompactum (A); T. viridescens (B); T. velutinum (C); and untreated plants as control CK (D), with a plot area of 6 m2 for each treatment. Each treatment had three replicates, with 150 seedlings per plot. S. miltiorrhiza seedlings with consistent growth and plant height were selected for transplantation.
Trichoderma strain was inoculated on PDA medium and cultured at 25°C for 7 days, add 10 ml of sterile water to the petri dish 7 days after inoculation. The surface of the medium was gently scraped with an inoculation needle, and then the spore suspension was prepared after gauze filtration, precipitation and sterile water suspension. After transplantation, the treatment group was irrigated with a Trichoderma spore suspension (106 colony-forming units/ml) at a dosage of 30 ml per plant, once every 20 days, for a total of three times. The roots of the CK group were irrigated with the same amount of water per plant. Soil samples were obtained at the termination of the growth period of S. miltiorrhiza. Rhizosphere soil and bulk soil samples were collected in sterile plastic containers. Portions of each sample were preserved at −80°C for microbial DNA analysis, with the remainder air-dried for assessment of chemical properties. The fresh weight of whole S. miltiorrhiza plants was recorded after complete cleaning. Plant dry weight was measured after desiccation at 105°C to constant mass.
The T. brevicompactum strain that had the best comprehensive effect was made into a microbial preparation. It was applied in Pingyi County, Linyi City, Shandong Province, from May 2024 to November 2024. These experiments included two treatments: PA (T. brevicompactum) and PCK (untreated plants, control). After one growing season, S. miltiorrhiza samples were collected and determined for fresh weight, survival rate, and effective ingredient content.
2.3 Determination of soil chemical properties
Soil pH was measured in soil–water suspensions (1:2.5) using a pH meter. Total nitrogen (TN) and total soil organic carbon (TOC) contents were determined using an elemental analyzer (Vario EL, Germany). Ammonium nitrogen (NH4+-N) and nitrate-nitrogen (NO3−-N) contents in the soil were analyzed with a continuous flow analyzer (SEAL AA3, Germany) (Jin et al., 2022). The available phosphorus (AP) content was measured by performing lixiviating-molybdenum blue colorimetry after extraction with 0.5 M NaHCO3 for 30 min. The available potassium (AK) content was measured using the NH4OAc extraction-flame spectrophotometer method (Bao, 2000).
2.4 Soil DNA extraction, polymerase chain reaction, and sequencing
DNA extraction was conducted using a PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA). The bacterial 16S rRNA gene was amplified using primers 806R (GGACTACHVGGGTWTCTAAT) and 338F (ACTCCTACGGGAGGCAGCA), as described by Fu et al. (2023). PCR conditions: initial denaturation at 95°C (10 min), 40 amplification cycles (95°C for 15 s, 55°C for 60 s, and 72°C for 90 s), and a final extension at 72°C (7 min). The fungal internal transcribed spacer (ITS) region was amplified with primers ITS1F/ITS2 (CTTGGTCATTTAGAGGAAGTAA/GCTGCGTTCTTCATCGATGC) in conditions: initial denaturation at 95°C (5 min), and 35 amplification cycles (95°C for 1 min, 53°C for 45 s, and 72°C for 1 min). The amplicons were purified and sequenced on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). The raw sequencing data generated in this study have been deposited in the China National Center for Bioinformation (https://bigd.big.ac.cn/gsa/browse/CRA025401).
Sequence data were processed using Trimmomatic v0.33 for quality filtering and Cutadapt v1.9.1 for primer removal (Martin, 2011; Bolger et al., 2014). Merged reads were generated using USEARCH v10 (Edgar, 2013), followed by chimera removal with UCHIME v4.2 (Edgar et al., 2011) to obtain quality-filtered sequences. Sequences were clustered into operational taxonomic units (OTUs) by 97% similarity using USEARCH. OTUs with total counts below two were excluded from further analysis. Taxonomy annotation was performed using the Naive Bayes classifier (70% confidence threshold) in QIIME2 with the SILVA database (release 138.1) as the reference (Quast et al., 2013; Bolyen et al., 2019).
2.5 Analysis of effective ingredient content
Quantitative determination of bioactive constituents in S. miltiorrhiza root, including of tanshinones (cryptotanshinone, tanshinone I, and tanshinone IIA) and salvianolic acid B, was performed using high-performance liquid chromatography (HPLC). For the tanshinones, the chromatographic conditions involved separation in octadecylsilane-bonded silica gel using acetonitrile–0.02% phosphoric acid gradient elution (20°C) with detection at 270 nm. The theoretical plate number should not be less than 60,000, calculated based on the tanshinone IIA peak. Reference and test solutions were prepared, with the latter obtained by precisely weighing about 0.3 g of powdered sample, extracting with 50 mL of methanol via ultrasonication, and adjusting for the weight loss with more methanol. Both solutions were subjected to HPLC analysis. The relative retention times of cryptotanshinone and tanshinone I, referenced to tanshinone IIA, must be within ±5% of the specified values; their contents were calculated using the peak area of tanshinone IIA and respective correction factors.
Salvianolic acid B was analyzed in modified conditions: acetonitrile–0.1% phosphoric acid (22:78 v:v) at a flow-rate of 1.2 mL/min with detection at 286 nm. The theoretical plate number should not be less than 6,000, calculated based on the salvianolic acid B peak. Salvianolic acid B reference solution (0.10 mg/mL) was prepared in methanol–water (8:2 v:v). The test solution was prepared similarly but with 0.15 g of powdered sample and 50 mL of the methanol–water mixture. Both solutions were analyzed by HPLC in the established chromatographic conditions.
2.6 Statistical analysis
Statistical comparisons were performed using one-way analysis of variance followed by the least significance difference post-hoc test (p < 0.05) to compare the mean values of samples, with data variability expressed as standard error (n = 3). Redundancy analysis (RDA) was conducted using the vegan package (v2.6-8) in R software (v4.4.1). Pearson correlation analyses were implemented with the psych package (v2.4.6.26).
3 Results
3.1 Comparison of the fresh and dry weights of S. miltiorrhiza in different treatments
Compared with the fresh and dry weights of S. miltiorrhiza in the control (CK) group, those of S. miltiorrhiza in the A treatment group (T. brevicompactum) were increased by 20.73% and 49.02% (p< 0.05), respectively; those of S. miltiorrhiza in the B treatment group (T. viridescens) were increased by 12.60% and 45.10% (p < 0.05), respectively; and those of S. miltiorrhiza in the C treatment group (T. velutinum) were increased by 10.16% and 35.29% (p < 0.05), respectively (Figure 1). All the treatments markedly increased the dry weight, and treatment A substantially increased the fresh weight.
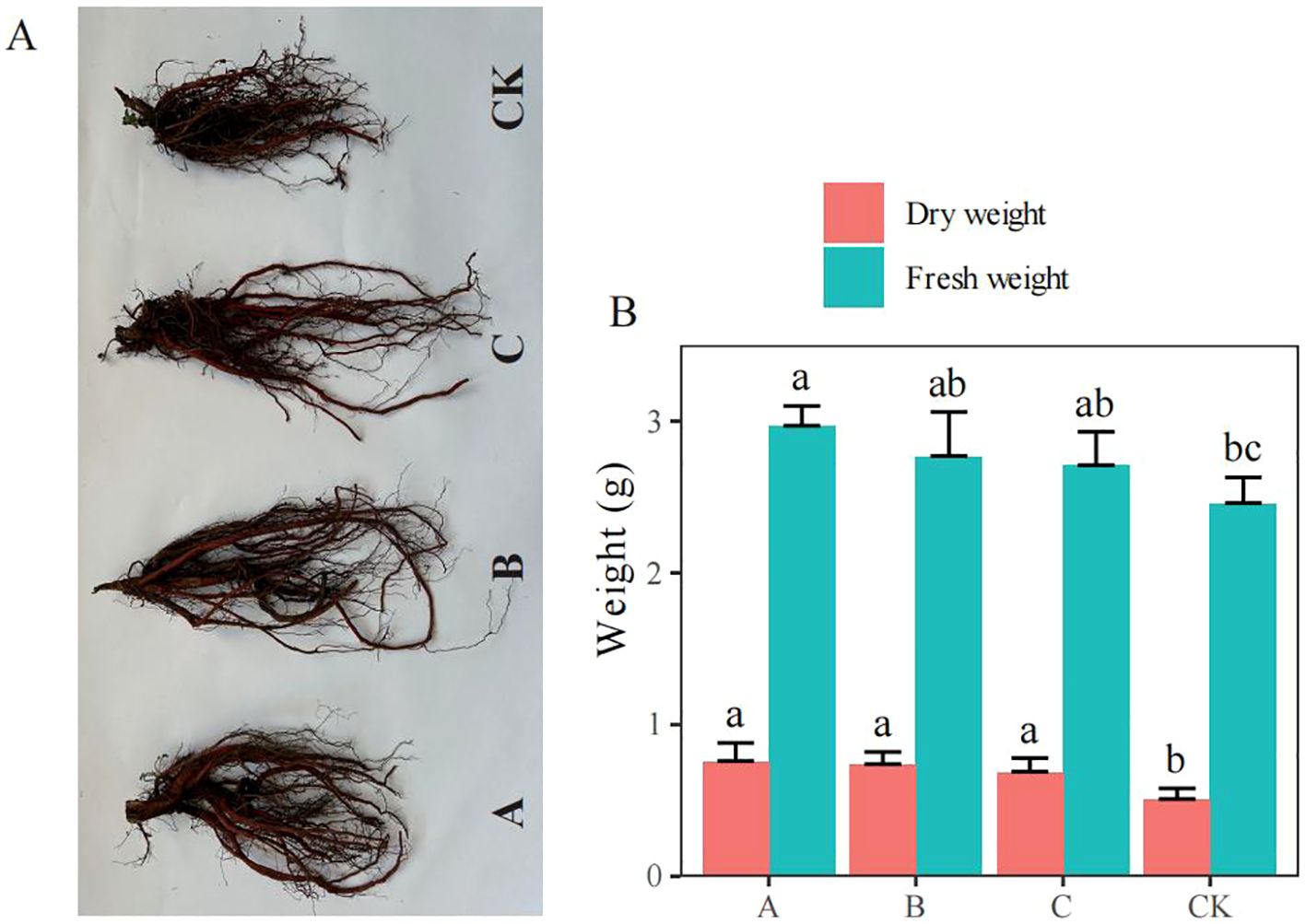
Figure 1. Effects of Trichoderma treatment on Salvia miltiorrhiza biomass production (mean ± SE). (A) Photographs of S. miltiorrhiza; (B) Fresh and dry weights of S. miltiorrhiza. CK, no treatment; A, T. brevicompactum treatment; B, T. velutinum treatment; C, T. viridescens treatment. Different lowercase letters above columns indicate significant differences (p < 0.05).
3.2 Effects of different treatments on soil chemical properties
The soil chemical properties in the Trichoderma treatment groups are shown in Table 1. Compared with the control (CK) group, the Trichoderma treatment groups notably decreased the soil pH value, and remarkably increased the TN and TOC contents, by 157.89%–226.32% and 47.35%–65.88%, respectively (p < 0.05). Treatments A and C significantly enhanced the NH4+-N content relative to the CK group (p < 0.05). Trichoderma treatment increased NO3--N levels by 91.38%–318.89%, AP by 1.85%–11.86%, and AK by 4.29%–17.16% relative to the CK group.
3.3 Variations in soil microbial community composition across treatment groups
3.3.1 Bacterial and fungal community structures at the phylum level
No significant differences were observed between treatments in the relative abundances of the top 10 bacterial phyla, including Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Gemmatimonadetes, Nitrospirae, Planctomycetes, Verrucomicrobia, Bacteroidetes, and Firmicutes. However, quantitative analysis revealed subtle differences in relative abundances between treatments. The three dominant phyla (Proteobacteria, Acidobacteria, and Actinobacteria) represented 74.32%–74.55% of the total bacterial composition (Figure 2A). Compared with the CK group, the relative abundances of Actinobacteria, Chloroflexi, and Gemmatimonadetes were increased in the Trichoderma treatment groups. The maximum relative abundances of Actinobacteria and Chloroflexi occurred in treatment B (T. viridescens), while treatment A (T. brevicompactum) contained the highest proportion of Gemmatimonadetes.
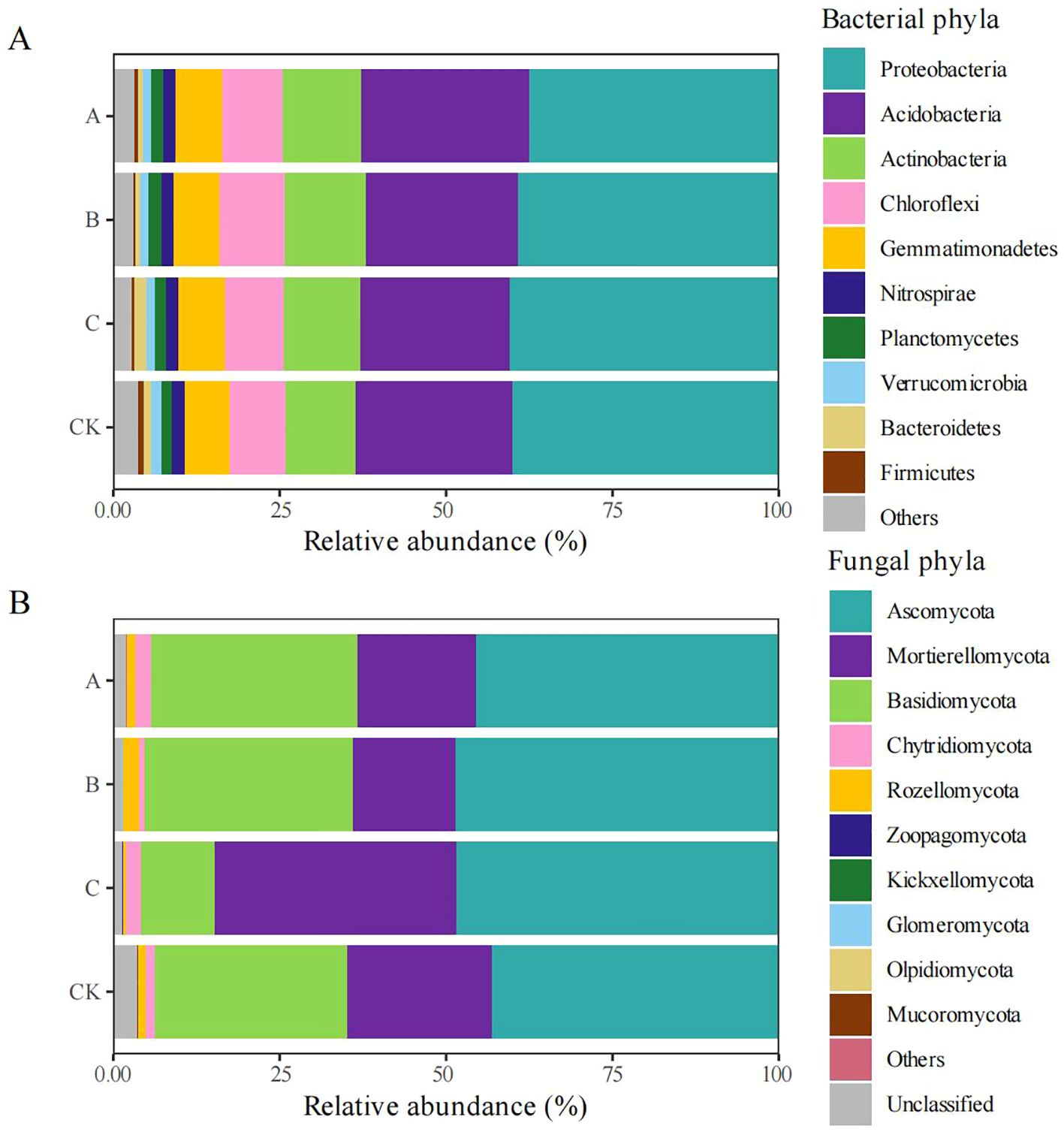
Figure 2. Relative abundance (%) of major bacterial and fungal phyla among treatment groups. The phylum abundance of bacteria (A) and fungi (B) in different groups: CK, no treatment; A, T. brevicompactum treatment; B, T. velutinum treatment; C, T. viridescens treatment.
The fungal community composition showed no significant variations in the top 10 phyla between treatments; these phyla were Ascomycota, Mortierellomycota, Basidiomycota, Chytridiomycota, Rozellomycota, Zoopagomycota, Kickxellomycota, Glomeromycota, Olpidiomycota, and Mucoromycota. However, the relative abundances of phyla exhibited some differences between treatments. Ascomycota, Mortierellomycota, and Basidiomycota collectively represented 94.41%–95.93% of fungal sequences (Figure 2B). The relative abundance of Ascomycota was significantly higher in the Trichoderma-treated groups than in the control (CK), reaching its maximum in treatment B (T. viridescens). Trichoderma inoculation suppressed Olpidiomycota and Mucoromycota populations, showing maximal suppression in treatment C (T. velutinum).
3.3.2 Classification of bacteria and fungi at the genus level
The top 10 bacterial genera showed stable relative abundances across treatments (Figure 3A); however, minor variations in relative abundance were observed. Trichoderma application enhanced the abundance of Gemmatimonas, uncultured_bacterium_o_Acidobacteriales, uncultured_bacterium_f_SC-I-84, uncultured_bacterium_o_Gaiellales, and uncultured_bacterium_c_KD4-96, compared with the control (CK) treatment.
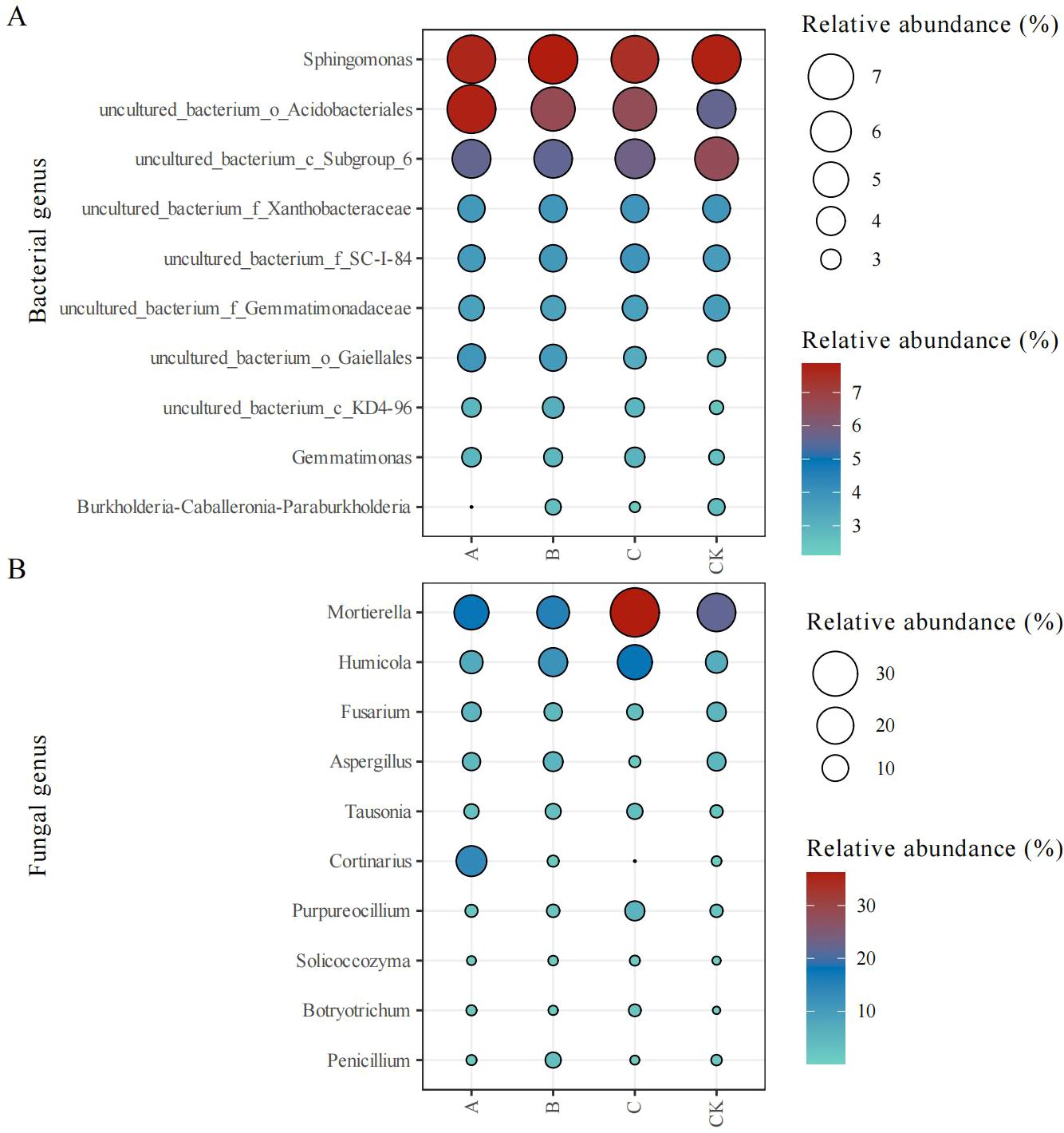
Figure 3. Relative abundance (%) of bacterial (A) and fungal (B) genera in different treatment groups. CK, no treatment; A, T. brevicompactum treatment; B, T. velutinum treatment; C, T. viridescens treatment.
The composition of the top 10 fungal genera was slightly different between treatment groups (Figure 3B). The Trichoderma treatment groups showed an increased relative abundance of Humicola, Tausonia, Solicoccozyma, and Botryotrichum and a decreased relative abundance of Fusarium relative to the CK group.
3.4 Redundancy analysis of microbial communities and nutrients in the soil under different treatments
Redundancy analysis illustrated associations between the soil bacterial community composition and physicochemical parameters; RDA axes 1 and 2 explained 52.12% and 29.83% of the total variance respectively, cumulatively accounting for 81.95% of the variation in the dataset (Figure 4A). AP, NO3−-N, and NH4+-N were the main physicochemical factors associated with soil bacterial communities. As indicated in Figure 4C, AP correlated positively with Proteobacteria and Planctomycetes, but negatively with Acidobacteria, Chloroflexi, Gemmatimonadetes, and Rokubacteria (p < 0.05); the soil pH was positively related to Firmicutes and Bacteroidetes, and negatively related to Actinobacteria (p < 0.05); TN was positively associated with Actinobacteria and negatively associated with Bacteroidetes (p < 0.05); TOC content showed a strong positive association with the abundance of Actinobacteria (p < 0.05).
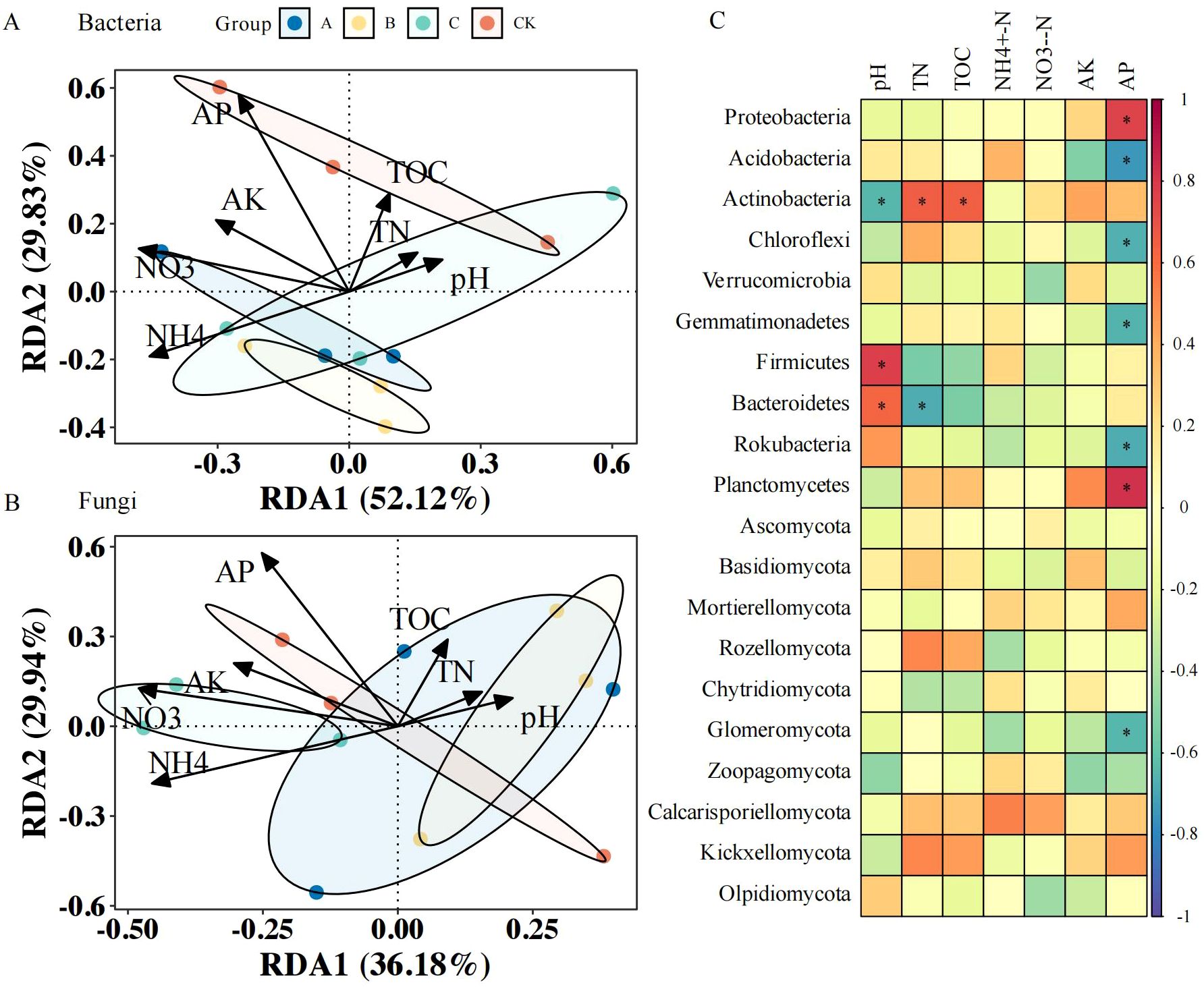
Figure 4. Soil nutrient–microbiome interactions in different treatments. (A) Redundancy analysis (RDA) of soil nutrients and bacterial community; (B) RDA of soil nutrients and fungal community; (C) Pearson’s correlation analysis of soil nutrients and bacterial and fungal phyla. CK, no treatment; A, T. brevicompactum treatment; B, T. velutinum treatment; C, T. viridescens treatment; TN, Total nitrogen; TOC, total soil organic carbon; NH4+-N, ammonium nitrogen; NO3−-N, nitrate-nitrogen; AP, available phosphorus; AK, available potassium. * provided p<0.05.
The soil fungal communities also showed significant covariation with physicochemical properties in RDA (Figure 4B). The RDA ordination explained 66.12% of total variance through axes 1 and 2 (36.18% and 29.94% respectively). AP, NO3—N, and NH4+-N were the main physicochemical factors associated with the soil fungal community. As shown in Figure 4B, the AP content was significantly negatively associated with Glomeromycota (p < 0.05).
3.5 Effect of T. brevicompactum application on secondary metabolite production in S. miltiorrhiza
Based on the experiments above, T. brevicompactum was identified as the most effective agent for improving S. miltiorrhiza cultivation. A further set of experiments was then conducted in which T. brevicompactum was applied to S. miltiorrhiza cultivation.
As shown in Figure 5, the fresh weight of individual plants in the PA group (108.90 g) was significantly increased, by 17.79%, compared with that in the PCK group (92.45 g; p< 0.05). The survival rate in the PA group (80.95%) also increased compared with that in the PCK group (71.43%; no significant difference).
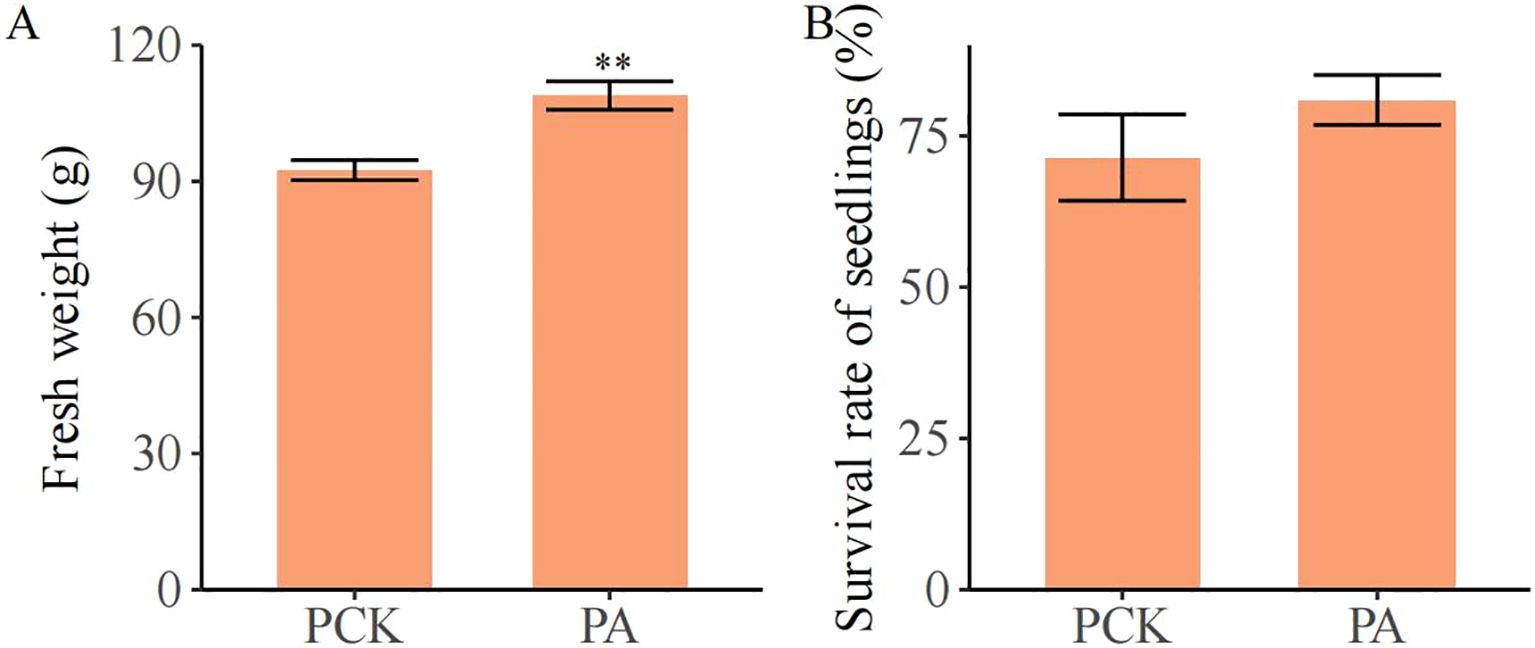
Figure 5. The effect of T. brevicompactum on (A) the fresh weight and (B) the survival rate of S. miltiorrhiza. PCK, control group; PA, T. brevicompactum treatment group. **p < 0.01.
There were significant variations in the contents of bioactive ingredients in S. miltiorrhiza roots among the treatment groups (Figure 6; p< 0.05). Compared with the PCK group, the PA group showed a higher content of tanshinone IIA (0.65% vs. 0.43%, 51.16% higher than in the PCK group), cryptotanshinone (0.58% vs. 0.37%, 56.76% higher than in the PCK group), tanshinone I (0.06% vs. 0.04%, 50.00% higher than in the PCK group), and salvianolic acid B (5.59% vs. 4.72%, 18.43% higher than in the PCK group).
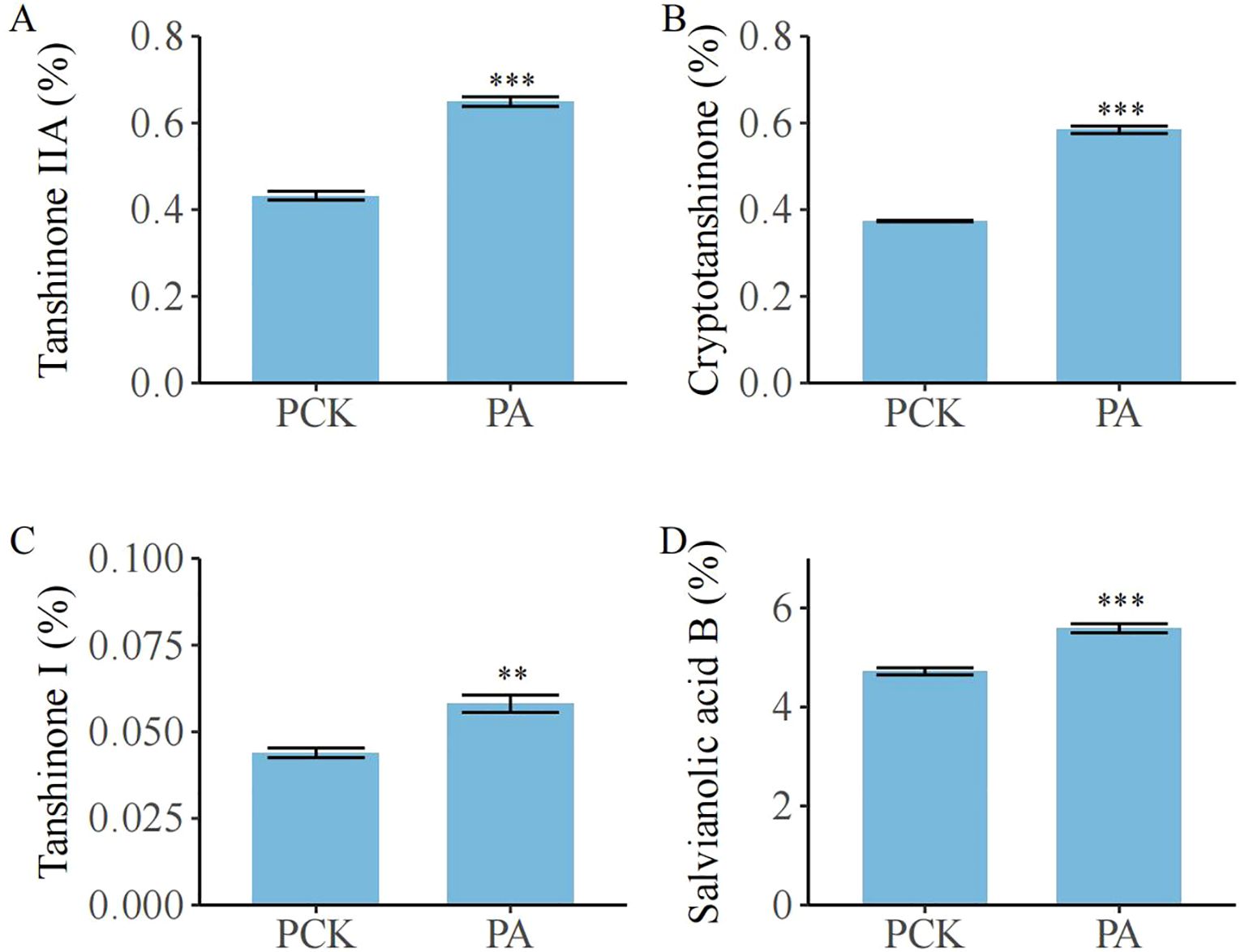
Figure 6. The effect of T. brevicompactum on the content of active components in S. miltiorrhiza root. (A) Tanshinone IIA, (B) cryptotanshinone, (C) tanshinone I, (D) salvianolic acid (B) PCK, control group; PA, T. brevicompactum treatment group. **p < 0.01, ***p < 0.001.
4 Discussion
A previous study has demonstrated that Trichoderma has a positive impact on soil physical and chemical properties (Mao and Jiang, 2021). In the present study, the application of Trichoderma resulted in a decrease in soil pH and an increase in available nutrient content in the soil. This may be attributable to Trichoderma increasing soil nutrient solubilization (Yedidia et al., 2001; Harman et al., 2004). Trichoderma can secrete a large amount of organic acids, which can lower soil pH, promote the transformation of plant nutrients (N, P, and K) from noneffective to effective forms, and promote plant growth (Zhang et al., 2019). Redundancy analysis revealed that in this work, AP, NO3--N, and NH4+-N were the dominant factors that shaped the soil microbial community. Furthermore, AP content correlated positively with both Proteobacteria and Planctomycetes, taxa that are key contributors to soil nutrient cycles (Arunrat et al., 2022; Hu et al., 2022). We speculate that the application of Trichoderma increased the relative abundance of Proteobacteria and Planctomycetes in the soil, increasing the amount of dissolved phosphorus and promoting the resistance and growth of S. miltiorrhiza.
Accumulating studies have shown that Trichoderma can improve the soil microbial community structure (Zhang et al., 2020; Mao and Jiang, 2021). This is consistent with our findings (Figure 1). Actinobacteria are widely distributed in soil; they decompose organic matter, produce antibiotics that inhibit various plant diseases (Lee et al., 2012; Fuentes et al., 2014; Zheng et al., 2020), and exhibit symbiotic nitrogen fixation and phosphorus solubilization effects (Juhnke et al., 1987; Xu et al., 2023). Chloroflexi are involved in the biogeochemical cycling of C, N, and S, and play a positive role in crop growth (Yang et al., 2024a). Gemmatimonas are involved in nitrogen metabolism and transformation (Zhang et al., 2024a); can reduce N2O in the soil to nitrate (Zou et al., 2023); and improve soil nutrients, including converting insoluble phosphorus into soluble phosphorus and enhancing nutrient absorption. Consistent with this, Trichoderma addition increased AP content in the present study (Table 1). Additionally, Gemmatimonas can induce plant stress resistance and produce antifungal compounds (Zhu et al., 2021), combat plant pathogens, inhibit soil-borne diseases, and promote plant growth.
Fusarium has been shown to be the main pathogen that causes root rot in S. miltiorrhiza, leading to a decrease in crop yield and quality (Sa et al., 2022). Fusarium in fact causes important diseases in many crops, including tomatoes, corn, cotton, and others (Ma et al., 2010; Srinivas et al., 2019; Liu et al., 2021). We suggest that Trichoderma can decrease the incidence of S. miltiorrhiza diseases by inhibiting the abundance of pathogens such as Fusarium, thereby increasing the yield of S. miltiorrhiza.
T. brevicompactum treatment significantly improved the content of bioactive components in the roots of S. miltiorrhiza. The potential mechanisms may include suppression of pathogenic microorganisms by Trichoderma, leading to decreased disease incidence and attenuated damage to the S. miltiorrhiza root system, thereby indirectly preserving the biosynthesis of its bioactive constituents (Contreras-Cornejo et al., 2014). Moreover, previous studies have shown that Trichoderma can activate the jasmonic acid (JA) and salicylic acid (SA) signaling pathways in plant roots (Islam et al., 2023; Peng et al., 2019), while external application of JA and SA stimulates an increase in secondary compounds in various plant species (Tafreshi et al., 2025). Furthermore, Trichoderma secretes organic acids that solubilize insoluble phosphorus and potassium in soil (Bononi et al., 2020), improving nutrient availability to facilitate root development (Chen et al., 2006) and secondary metabolism. Trichoderma can also synthesize indole-3-acetic acid, which modulates the root development and distribution of secondary metabolites in S. miltiorrhiza (Zhang et al., 2023; Contreras-Cornejo et al., 2024).
5 Conclusions
Overall, all three strains of Trichoderma tested here (T. brevicompactum, T. viridescens, and T. velutinum) can increase the fresh and dry weights of S. miltiorrhiza, improve soil nutrients, increase beneficial microbial abundance, and decrease the abundance of harmful microbial species, thereby alleviating the CC obstacles of S. miltiorrhiza. In particular, T. brevicompactum was superior to the other tested species. It can improve the survival rate and effective component content of continuous cropping S. miltiorrhiza, and has the potential for commercial application. Trichoderma can reduce the use of chemical pesticides and fertilizers by regulating the soil microecological balance, inhibiting pathogen reproduction, and enhancing soil fertility. These properties align with the demands of green agriculture and is of great significance for advancing sustainable agricultural development. This research offers a new biological control approach to address continuous cropping obstacles in medicinal plants, ensuring both the quality and safety of medicinal materials and maintaining soil health. Looking ahead, this technology is expected to be extended for use in more crops, supporting the development of resource-efficient and eco-friendly agricultural models. It will drive agriculture toward ecological and sustainable transformation, contributing to high-quality agricultural development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
LZ: Data curation, Validation, Methodology, Writing – original draft. XP: Investigation, Validation, Writing – original draft, Data curation, Methodology. YZ: Formal Analysis, Project administration, Methodology, Writing – review & editing, Investigation, Resources. YG: Supervision, Data curation, Writing – review & editing. YL: Writing – review & editing, Data curation, Investigation. QJ: Writing – review & editing, Formal Analysis, Investigation. KC: Formal Analysis, Writing – review & editing, Investigation. CS: Writing – review & editing, Funding acquisition, Resources, Supervision, Project administration. QW: Funding acquisition, Writing – review & editing, Conceptualization, Validation, Supervision, Project administration, Methodology, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by grants from the Agricultural Science and Technology Innovation Program (grant number CAAS-ASTIP-2021-ISAPS); the Shandong Province Modern Agricultural Technology System Chinese Herbal Medicine Industrial Innovation Team (grant number SDAIT-20); the Seed-Industrialized Development Program in Shandong Province (grant number 2021LZGC008); and the Jilin Provincial International Cooperation key laboratory for Science and Technology Innovation of Special Animal and Plants (grant number YDZJ202502CXJD080).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arunrat, N., Sansupa, C., Kongsurakan, P., Sereenonchai, S., and Hatano, R. (2022). Soil microbial diversity and community composition in rice–fish co-culture and rice monoculture farming system. Biology 11, 1242. doi: 10.3390/biology11081242
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., and Al-Ghalith, G. A. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bononi, L., Chiaramonte, J. B., Pansa, C. C., Moitinho, M. A., and Melo, I. S. (2020). Phosphorus-solubilizing Trichoderma spp. from Amazon soils improve soybean plant growth. Sci. Rep. 10, 2858. doi: 10.1038/s41598-020-59793-8
Chen, Y. D., Du, J. F., Li, Y., Tang, H., Yin, Z. Y., Yang, L., et al. (2022). Evolutions and managements of soil microbial community structure drove by continuous cropping. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.839494
Chen, L. H., Huang, X. Q., Zhang, F. G., Zhao, D. K., Yang, X. M., and Shen, Q. R. (2012). Application of Trichoderma harzianum SQR-T037 bio-organic fertiliser significantly controls Fusarium wilt and affects the microbial communities of continuously cropped soil of cucumber. J. Sci. Food Agric. 92, 2465–2470. doi: 10.1002/jsfa.5653
Chen, Y. P., Rekha, P. D., Arun, A. B., Shen, F. T., Lai, W.-A., and Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 34, 33–41. doi: 10.1016/j.apsoil.2005.12.002
Chen, B., Shao, G. G., Zhou, T., Fan, Q. H., Yang, N. L., Cui, M., et al. (2023). Dazomet changes microbial communities and improves morel mushroom yield under continuous cropping. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1200226
Chen, L. H., Yang, X. M., Raza, W., Li, J. H., Liu, Y. X., Qiu, M. H., et al. (2011). Trichoderma harzianum SQR-T037 rapidly degrades allelochemicals in rhizospheres of continuously cropped cucumbers. Appl. Microbiol. Biotechnol. 89, 1653–1663. doi: 10.1007/s00253-010-2948-x
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Alfaro-Cuevas, R., and López-Bucio, J. (2014). Trichoderma spp. improve growth of Arabidopsis seedlings under salt stress through enhanced root development, osmolite production, and Na+ elimination through root exudates. Mol. Plant Microbe Interact. 27, 503–514. doi: 10.1094/MPMI-09-13-0265-R
Contreras-Cornejo, H. A., Schmoll, M., Esquivel-Ayala, B. A., González-Esquivel, C. E., Rocha-Ramírez, V., and Larsen, J. (2024). Mechanisms for plant growth promotion activated by Trichoderma in natural and managed terrestrial ecosystems. Microbiol. Res. 281, 127621. doi: 10.1016/j.micres.2024.127621
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Feng, J., Liao, F., Kong, D. Y., Ren, R. H., Sun, T., Liu, W., et al. (2022). Genetic diversity of the cultivated Salvia miltiorrhiza populations revealed by four intergenic spacers. PloS One 17, e0266536. doi: 10.1371/journal.pone.0266536
Fu, J. C., Wu, J., Xu, M., Ma, J., Long, L. L., Chen, C., et al. (2023). Effects of simulated warming on soil properties and bacterial community composition in the Bashania faberi ecosystem. Appl. Soil Ecol. doi: 10.1016/j.apsoil.2023.105091
Fuentes, S., Méndez, V., Aguila, P., and Seeger, M. (2014). Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 98, 4781–4794. doi: 10.1007/s00253-014-5684-9
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Hu, X. J., Gu, H. D., Liu, J. J., Wei, D., Zhu, P., and Cui, X. A. (2022). Metagenomics reveals divergent functional profiles of soil carbon and nitrogen cycling under long-term addition of chemical and organic fertilizers in the black soil region. Geoderma 418, 115846. doi: 10.1016/j.geoderma.2022.115846
Islam, M. R., Chowdhury, R., Roy, A. S., Islam, M. N., Mita, M. M., and Bashar, S. (2023). Native Trichoderma Induced the Defense-Related Enzymes and Genes in Rice against Xanthomonas oryzae pv. oryzae (Xoo). Plants (Basel) 12, 1864. doi: 10.3390/plants12091864
Jin, Q., Zhang, Y. Y., Ma, Y. Y., Sun, H., Guan, Y. M., Liu, Z. B., et al. (2022). The composition and function of the soil microbial community and its driving factors before and after cultivation of Panax ginseng in farmland of different ages. Ecol. Indic. 145, 109748. doi: 10.1016/j.ecolind.2022.109748
Ju, J. D., Zhou, B. Q., Yang, G. H., Fu, X. Y., Wang, X., Guo, L. P., et al. (2023). Study on the metabolic process of phthalic acid driven proliferation of Rhizoctonia solani. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1266916
Juhnke, M. E., Mathre, D. E., and Sands, D. C. (1987). Identification and characterization of rhizosphere-competent bacteria of wheat. Appl. Environ. Microbiol. 53, 2793–2799. doi: 10.1128/aem.53.12.2793-2799.1987
Kashyap, P. L., Rai, P., Srivastava, A. K., and Kumar, S. (2017). Trichoderma for climate resilient agriculture. World J. Microbiol. Biotechnol. 33, 155. doi: 10.1007/s11274-017-2319-1
Lee, S. Y., Tindwa, H., Lee, Y. S., Naing, K. W., Hong, S. H., Nam, Y., et al. (2012). Biocontrol of anthracnose in pepper using chitinase, β-1,3 glucanase, and 2-furancarboxaldehyde produced by Streptomyces cavourensis SY224. J. Microbiol. Biotechnol. 22, 1359–1366. doi: 10.4014/jmb.1203.02056
Liu, H. Y., Niu, M., Zhu, S., Zhang, F., Liu, Q., Liu, Y., et al. (2020). Effect study of continuous monoculture on the quality of salvia miltiorrhiza bge roots. BioMed. Res. Int. 2020, 4284385. doi: 10.1155/2020/4284385
Liu, Z., Tang, J., Ren, X., and Schaeffer, S. M. (2021). Effects of phosphorus modified nzvi-biochar composite on emission of greenhouse gases and changes of microbial community in soil. Environ. pollut. 274, 116483. doi: 10.1016/j.envpol.2021.116483
Ma, L., Does, H., Borkovich, K., Coleman, J., Daboussi, M., Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. doi: 10.1038/nature08850
Mao, T. T. and Jiang, X. L. (2021). Changes in microbial community and enzyme activity in soil under continuous pepper cropping in response to Trichoderma hamatum MHT1134 application. Sci. Rep. 11, 21585. doi: 10.1038/S41598-021-00951-X
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Meng, X. H., Miao, Y. Z., Liu, Q. M., Ma, L., Guo, K., Liu, D. Y., et al. (2019). TgSWO from Trichoderma guizhouense NJAU4742 promotes growth in cucumber plants by modifying the root morphology and the cell wall architecture. Microb. Cell Fact. 18, 148. doi: 10.1186/s12934-019-1196-8
Peng, W., Ming, Q. L., Zhai, X., Zhang, Q., Rahman, K., Wu, S. J., et al. (2019). Polysaccharide Fraction Extracted from Endophytic Fungus Trichoderma atroviride D16 Has an Influence on the Proteomics Profile of the Salvia miltiorrhiza Hairy Roots. Biomolecules 9, 415. doi: 10.3390/biom9090415
Pu, C. J., Liu, S., Lu, Z. Y., Luo, Y. Z., Wang, Z. H., and Chen, M. L. (2022). Pathogenicity and induced systemic resistance of Fusarium oxysporum and Verticillium dahlia to Salvia miltiorrhiza. Zhongguo Zhong Yao Za Zhi. 47, 5832–5837. doi: 10.19540/j.cnki.cjcmm.20220712.103
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rees, H. J., Drakulic, J., Cromey, M. G., Bailey, A. M., and Foster, G. D. (2022). Endophytic Trichoderma spp. can protect strawberry and privet plants from infection by the fungus Armillaria mellea. PloS One 17, e0271622. doi: 10.1371/journal.pone.0271622
Rodrigues, A. O., May De Mio, L. L., and Soccol, C. R. (2023). Trichoderma as a powerful fungal disease control agent for a more sustainable and healthy agriculture: recent studies and molecular insights. Planta 257, 31. doi: 10.1007/s00425-022-04053-4
Sa, R. B., He, S., Han, D. D., Liu, M. J., Yu, Y. X., Shang, R. E., et al. (2022). Isolation and identification of a new biocontrol bacteria against Salvia miltiorrhiza root rot and optimization of culture conditions for antifungal substance production using response surface methodology. BMC Microbiol. 22, 231. doi: 10.1186/s12866-022-02628-5
Srinivas, C., Devi, D. N., Murthy, K. N., Mohan, C. D., and Nayaka, S. C. (2019). Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity–a review. Saudi J. Biol. Sci. 26, 1315–1324. doi: 10.1016/j.sjbs.2019.06.002
Su, Y. T., Zeeshan Ul. Haq., M., Liu, X. F., Li, Y., Yu, J., Yang, D. M., et al. (2023). A Genome-Wide Identification and Expression Analysis of the Casparian Strip Membrane Domain Protein-like Gene Family in Pogostemon cablin in Response to p-HBA-Induced Continuous Cropping Obstacles. Plants (Basel) 12, 3901. doi: 10.3390/plants12223901
Tafreshi, Y. M., Eghlima, G., Hatami, M., and Vafadar, M. (2025). Exploring the potential impact of salicylic acid and jasmonic acid in promoting seed oil content, vitamin C and antioxidant activity in rosehip (Rosa canina L.). BMC Plant Biol. 25, 249. doi: 10.1186/s12870-025-06251-0
Wang, R., Chen, D., Khan, R. A. A., Cui, J., Hou, J., and Liu, T. (2021). A novel Trichoderma asperellum strain DQ-1 promotes tomato growth and induces resistance to gray mold caused by Botrytis cinerea. FEMS Microbiol. Lett. 368, 125–128. doi: 10.1093/femsle/fnab140
Wang, X., Wang, Q., Li, W. J., Zhang, D. Q., Fang, W. S., Li, Y., et al. (2023). Long-term effects of chloropicrin fumigation on soil microbe recovery and growth promotion of Panax notoginseng. Front. Microbiol. 14. doi: 10.3389/fmicb.1225944
Wu, J. J., Zhu, J. H., Zhang, D. Q., Cheng, H. Y., Hao, B. Q., Cao, A. C., et al. (2022). Beneficial effect on the soil microenvironment of Trichoderma applied after fumigation for cucumber production. PloS One 17, e0266347. doi: 10.1371/journal.pone.0266347
Xu, Y., Ding, H., Zhang, G. C., Li, Z. L., Guo, Q., Feng, H., et al. (2023). Green manure increases peanut production by shaping the rhizosphere bacterial community and regulating soil metabolites under continuous peanut production systems. BMC Plant Biol. 23, 69. doi: 10.1186/s12870-023-04079-0
Yan, W. P., Cao, S. J., Wu, Y. G., Ye, Z. C., Zhang, C., Yao, G. L., et al. (2022). Integrated Analysis of Physiological, mRNA Sequencing, and miRNA Sequencing Data Reveals a Specific Mechanism for the Response to Continuous Cropping Obstacles in Pogostemon cablin Roots. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.853110
Yang, X. H., Li, Q., Lu, Y., Zhang, L. X., and Bian, X. B. (2024a). Insight into the short-term effects of TiO2 nanoparticles on the cultivation of medicinal plants: Comprehensive analysis of Panax ginseng physiological indicators, soil physicochemical properties and microbiome. Sci. Total Environ. 951, 175581. doi: 10.1016/j.scitotenv.2024.175581
Yang, Z. D., Qiu, D. Y., Jiang, K., Du, M. X., and Li, H. Y. (2024b). Abatement effects of different soil amendments on continuous cropping of Codonopsis pilosula. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1376362
Yedidia, I., Srivastva, A. K., Kapulnik, Y., and Chet, I. (2001). Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant Soil. 235, 235–242. doi: 10.1023/a:1011990013955
Zeeshan Ul. Haq., M., Yu, J., Yao, G. L., Yang, H. G., Iqbal, H. A., Tahir, H., et al. (2023). A systematic review on the continuous cropping obstacles and control strategies in medicinal plants. Int. J. Mol. Sci. 24, 12470. doi: 10.3390/ijms241512470
Zhang, L. Q., Feng, Y. D., Zhao, Z. H., Baoyin, B., Cui, Z. G., Wang, H. Y., et al. (2024a). Macrogenomics-based analysis of the effects of intercropped soybean photosynthetic characteristics and nitrogen-assimilating enzyme activities on yield at different nitrogen levels. Microorganisms 12, 1220. doi: 10.3390/microorganisms12061220
Zhang, L. L., Jin, Q., Guan, Y. M., Liu, Z. B., Pan, X. X., Zhang, Y., et al. (2024b). Trichoderma spp. promotes ginseng biomass by influencing the soil microbial community. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1283492
Zhang, S. C., Qiu, L., Zheng, Y. W., Wang, W., Zhao, H. G., and Yang, D. F. (2023). Comparative transcriptome analysis reveals the regulatory effects of exogenous auxin on lateral root development and tanshinone accumulation in Salvia miltiorrhiza. Planta 258, 33. doi: 10.1007/s00425-023-04193-1
Zhang, L. Y., Tao, S., Zhang, Y. F., Yang, Y. M., Peng, F., Liao, H. L., et al. (2024c). Study on the effect of compound cultivation on the growth feature and active ingredients content of Salvia miltiorrhiza. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1238896
Zhang, Y., Tian, C., Xiao, J. L., Wei, L., Tian, Y., and Liang, Z. H. (2020). Soil inoculation of Trichoderma asperellum M45a regulates rhizosphere microbes and triggers watermelon resistance to Fusarium wilt. AMB Express. 10, 189. doi: 10.1186/s13568-020-01126-z
Zhang, F. G., Xu, X. X., Huo, Y. Q., and Xiao, Y. (2019). Trichoderma-inoculation and mowing synergistically altered soil available nutrients, rhizosphere chemical compounds and soil microbial community, potentially driving alfalfa growth. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03241
Zheng, X. F., Wang, Z. R., Zhu, Y. J., Wang, J. P., and Liu, B. (2020). Effects of a microbial restoration substrate on plant growth and rhizosphere bacterial community in a continuous tomato cropping greenhouse. Sci. Rep. 10, 13729. doi: 10.1038/s41598-020-70737-0
Zhu, Y., Shao, T. Y., Zhou, Y. J., Zhang, X., Gao, X. M., Long, X. H., et al. (2021). Periphyton improves soil conditions and offers a suitable environment for rice growth in coastal saline alkali soil. Land. Degrad. Dev. 32, 2775–2788. doi: 10.1002/ldr.3944
Zou, X. X., Liu, Y., Huang, M. M., Li, F., Si, T., Wang, Y. F., et al. (2023). Rotational strip intercropping of maize and peanut enhances productivity by improving crop photosynthetic production and optimizing soil nutrients and bacterial communities. Field Crops Res. 291, 13. doi: 10.1016/j.fcr.2022.108770
Keywords: Trichoderma spp., Salvia miltiorrhiza, soil nutrient, microorganism, effective ingredient
Citation: Zhang L, Pan X, Zhang Y, Guan Y, Li Y, Jin Q, Cao K, Shan C and Wang Q (2025) Effects of addition of Trichoderma spp. on growth and soil microorganisms of continuously cropped Salvia miltiorrhiza. Front. Plant Sci. 16:1654385. doi: 10.3389/fpls.2025.1654385
Received: 27 June 2025; Accepted: 07 August 2025;
Published: 03 September 2025.
Edited by:
Maurizio Ruzzi, University of Tuscia, ItalyReviewed by:
Reda Abdelhameed, Zagazig University, EgyptChutima Kaewkrajay, Phranakhon Si Ayutthaya Rajabhat University, Thailand
Copyright © 2025 Zhang, Pan, Zhang, Guan, Li, Jin, Cao, Shan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenggang Shan, c2hhbmNoZW5nZ2FuZ0AxMjYuY29t; Qiuxia Wang, d2FuZ3FpdXhpYTEwMThAMTI2LmNvbQ==
 Linlin Zhang
Linlin Zhang Xiaoxi Pan1,2
Xiaoxi Pan1,2 Qiao Jin
Qiao Jin Chenggang Shan
Chenggang Shan Qiuxia Wang
Qiuxia Wang