- 1State Key Laboratory of Plant Environmental Resilience, College of Life Sciences, Zhejiang University, Hangzhou, China
- 2Hainan Institute of Zhejiang University, Yazhou Bay Science and Technology City, Sanya, Hainan, China
Phosphorus (P), an essential macronutrient critical for plant growth and development, faces significant availability constraints in agricultural soils, substantially limiting crop yield potential. Transcription factors (TFs) play pivotal roles in phosphate (Pi) starvation responses in plants. In this study, we identified OSH45 (Oryza sativa homeobox 45), a homeobox domain TF in rice (Oryza sativa L.), which was strongly induced in roots under Pi starvation. Subcellular localization assays indicated that OSH45 is a nuclear localized protein. OSH45 overexpression transgenic plants exhibited enhanced low-Pi tolerance, characterized by significantly higher Pi concentrations and increased shoot and root biomass compared to wild type (WT) under Pi-limited conditions. Whereas osh45 loss-of-function mutants displayed no significant difference in shoot and root biomass compared to WT under both Pi-sufficient and Pi-limited conditions, but showed lower Pi concentration under Pi-sufficient conditions. Through transcriptomic profiling, 2,406 differential expressed genes (DEGs) were identified in OSH45 overexpression plants versus WT under Pi-sufficient conditions. About 38% of Pi starvation-induced (PSI) genes were upregulated and 25% of Pi starvation-suppressed (PSS) genes were downregulated in OSH45 overexpression plants. The expression of phosphate transporters, such as OsPT1, OsPT2, OsPT4, OsPT8, and acid phosphatases was upregulated, while the expression of Pi signaling repressors OsSPX1-3 was suppressed in OSH45 overexpression plants. Conversely, osh45 displayed decreased expression of OsPT1 and OsPT8 compared to WT. Altogether, our findings demonstrated that OSH45 is a novel TF involved in Pi deficiency response, regulating a set of Pi starvation responsive (PSR) genes to optimize plant adaptation to Pi-limited environments. This mechanism provides a strategic target for engineering Pi-efficient crops.
1 Introduction
Phosphorus (P) is an essential macronutrient for plant growth and development, which is primarily absorbed as inorganic phosphate (Pi) in the forms H2PO₄−, HPO₄²−, or PO₄³− depending on soil pH (Brinch-Pedersen et al., 2002). Pi scarcity severely limits agricultural productivity, affecting approximately 50% of global arable lands (Lynch, 2011; Liu et al., 2024). Phosphorus fertilization is a common strategy for improving crop yields in Pi-limited soils. However, due to its low diffusion coefficient and strong chelation with metal ions, less than 25% of the phosphate fertilizer is directly available to crops (Johnston et al., 2014; Puga et al., 2024). Elucidating the molecular mechanisms governing Pi signaling pathways is crucial for engineering crops with improved phosphorus-use efficiency (PUE), which could significantly reduce fertilizer dependency while ensuring sustainable agricultural production (Paz-Ares et al., 2022; Madison et al., 2023).
To cope with Pi limitation, plants have evolved multifaceted strategies to enhance Pi acquisition and utilization efficiency (Ham et al., 2018; Wang et al., 2021; Poirier et al., 2022). Under Pi-deficient conditions, plants remodel root architecture to increase Pi foraging capacity and fine-tune the activity of phosphate transporters (PTs) through transcriptional and post-transcriptional regulation, thereby optimizing Pi uptake and redistribution (Wang et al., 2021). Although significant advances have been made to elucidate the molecular components of Pi deficiency responses, the detailed mechanism underlying plant adaptation to Pi limitation remains unclear. Notably, transcription factors (TFs) act as central players in orchestrating plant adaptation to Pi scarcity. In Arabidopsis, the Myeloblastosis (MYB)-type regulators PHR1 (Phosphate Starvation Response 1) and its homologs PHL1-4 serve as master regulators of Pi starvation signaling, while their orthologs OsPHR1-4 function as master regulators of Pi starvation response (PSR) in rice. These MYB-CC type TFs activate subsets of Pi starvation induced (PSI) genes by binding to the P1BS motif (GNATATNC) in target gene promoters (Rubio et al., 2001; Bustos et al., 2010; Chiou and Lin, 2011; Guo et al., 2015; Sun et al., 2016). The PHR-regulated genes coordinate Pi uptake, assimilation, and reallocation, while PHR activities are post-translationally modulated by SPX (SYG1/Pho81/XPR1) proteins that act as Pi sensors (Wang et al., 2014; Lv et al., 2014; Zhou et al., 2021; Guan et al., 2022). Alongside core PHR regulators, additional MYB family members participate in the regulation of Pi homeostasis. In rice, OsRLI1 (Regulator of leaf inclination 1) is involved in regulating leaf angle based on Pi status and activating PSR genes under Pi deficiency (Ruan et al., 2018; Zhang et al., 2021). OsMYB2P-1, OsMYB4P, and OsMYB5P regulate Pi homeostasis, partly through transcriptional control of phosphate transporter genes (Dai et al., 2012; Yang et al., 2014; Wu et al., 2018). Additional TF families including WRKY, GRAP, and bHLH are also involved in Pi signaling regulation. In Arabidopsis, AtWRKY75 and AtWRKY45 activate AtPHT1 genes through W-box binding, while in rice, OsWRKY21, OsWRKY74 and OsWRKY108 activate OsPT genes via the same mechanism (Devaiah et al., 2007; Zhang et al., 2020; Dai et al., 2016). Conversely, OsWRKY10 represses OsPT2 under Pi-sufficient conditions (Wang et al., 2022). GARP family TF NIGT1 (Nitrate-inducible GARP-type transcriptional repressor 1) coordinates nitrogen (N)-P crosstalk, enhancing Pi absorption while suppressing nitrate uptake (Wang et al., 2023a). bHLH TF OsPTF1 confers low-Pi tolerance (Yi et al., 2005), while OsbHLH6 interacts with OsSPX4 to release OsPHR2, thereby activating PSI genes (He et al., 2021). This multilayered transcriptional network underscores the complexity of Pi signaling regulation. Characterization of novel TFs will advance our understanding of the plant Pi signaling network and provide strategies for improving PUE.
Homeodomain (HD) TFs are pivotal regulators governing plant development, which are phylogenetically categorized into homeodomain-leucine zipper (HD-Zip) and three-amino-acid loop extension (TALE) families. The TALE superfamily includes knotted1-like homeobox (KNOX) transcription factors critical for meristem maintenance, organogenesis, and phytohormone regulation (Hake et al., 2004). The KNOX subfamily were divided into class I (Shoot Apical Meristem (SAM)-specific, essential for meristem maintenance) and class II (broader expression, functional diversification) based on phylogeny and expression (Furumizu et al., 2015). Class I KNOX genes, such as STM (Shoot meristemless) in Arabidopsis, KN1 (Knotted 1) in maize, OSH1 (Oryza sativa homeobox 1) in rice, have been well characterized for their roles in SAM development (Tsuda et al., 2011; Chen et al., 2023; Tsuda et al., 2014), whereas the functions of class II KNOX proteins remain less explored. In Arabidopsis, class II members (AtKNAT3, AtKNAT4, AtKNAT5 and AtKNAT7) participate in cytokinin response and secondary cell wall biosynthesis (Wang et al., 2020b). In rice, the class II subfamily comprises HOS58 (Homeobox oryza sativa 58), HOS59, OSH45, and OsKNAT7/HOS66. Current evidence shows OsKNAT7 regulates cell wall biosynthesis and grain size through interactions with OsNAC31 and OsGRF4 (Wang et al., 2020b; Yu, 2019). Recently, ChIP-seq analyses identified HOS59 as a transcriptional repressor of grain size regulators OsSPL13 and OsSPL18, yet comprehensive mechanistic insights into KNOX II TFs remain scarce (Sheng et al., 2022). Although several HD domain proteins were known as developmental regulators, whether HD domain proteins are involved in plant adaptation to P nutrient limitation remains unexplored.
Here, we identify OSH45, a class II KNOX TF transcriptionally upregulated under P deficiency. We create CRISPR/Cas9 mutants and overexpression transgenic lines in rice, and demonstrate that OSH45 promotes Pi acquisition by modulating the expression of genes encoding phosphate transporters and phosphatases, while suppressing the expression of Pi signaling repressors OsSPXs. These findings establish OSH45 as a new key regulator of PSR to optimize plant adaptation to Pi-limited environments, which will benefit engineering Pi-efficient crops.
2 Materials and methods
2.1 Plant materials and growth conditions
The wild-type (WT) japonica rice (Oryza sativa L.) cultivar ‘Nipponbare’ (NIP) was used in this study. All transgenic lines were generated in the NIP background. Rice plants were hydroponically grown in a glasshouse under controlled conditions: 30/22°C (day/night), 60-70% relative humidity, and a 14 h light/10 h dark photoperiod with a light intensity of ~300 μmol m-2 s-1 provided by spectrum-tunable LED plant-growth lamps. Hydroponic cultivation was performed using full-strength Kimura nutrient solution as previously described (Wang et al., 2020a). Pi treatments were set as: Pi-sufficient (HP, 200 μM Pi), Pi-limited (LP, 10 μM Pi), and Pi-deficient (–P, 0 μM Pi). Solutions were prepared by replacing the appropriate amount of KH2PO4 with an equimolar concentration of KCl. The pH of the nutrient solution was adjusted to 5.5 using 1 M HCl or 1 M NaOH.
2.2 Phylogenetic analysis of the KNOX subfamily
To identify close homologs of OSH45 in the rice genome, the full-length OSH45 protein sequence was queried against the NCBI database using BLASTP (https://www.ncbi.nlm.nih.gov/). Candidate homologs with ≥ 80% query coverage and ≥ 50% identity were selected for further analysis. Multiple sequence alignment of the retrieved full-length protein sequences was performed with Clustal X.
2.3 Vector construction and generation of transgenic plants
To generate the OSH45 overexpression vector, the 834-bp coding sequence (excluding the stop codon) of OSH45 (Os08g0292900) was amplified from NIP cDNA using KOD-FX DNA polymerase (Toyobo, Japan). The amplified fragment was subsequently cloned into a modified pCAMBIA1300-sGFP vector under the control of the cauliflower mosaic virus 35S promoter (Lv et al., 2014), resulting in a C-terminal GFP fusion protein. The two overexpression lines obtained after backcrossing with NIP were designated OSH45-OE1 and OSH45-OE2. For osh45 knockout mutants, a CRISPR-Cas9 system was employed following established protocols (Ma et al., 2015). Two target specific single-guide RNAs (sgRNAs; sequences listed in Supplementary Data Sheet 1) were designed and ligated into the pYLCRISPR/gRNA vector. The sgRNA cassettes were then ligated into the pYLCRISPR/Cas9-MN binary vectors as previously described (Ma et al., 2015). All constructs were validated by Sanger sequencing prior to transformation. Transgenic rice plants were generated via Agrobacterium tumefaciens (strain EHA105)-mediated transformation of embryogenic calli derived from mature NIP seeds, following standard procedures (Chen et al., 2015). The primers used for vector construction are provided in Supplementary Data Sheet 1.
2.4 Subcellular localization
For subcellular localization analysis, the 35S::OSH45-GFP construct was transiently expressed in Nicotiana benthamiana leaves via Agrobacterium tumefaciens -mediated infiltration (strain EHA105). Fluorescence signals from root tips of 10-day-old transgenic rice seedlings harboring the 35S::OSH45-GFP transgene were observed using a confocal laser-scanning microscope (LSM710, Zeiss). GFP signals were excited with a 488-nm argon-ion laser, and emission was collected between 493 to 542 nm. For DAPI, signals were excited with a 405-nm laser, and emission was collected between 420 to 480 nm. A 25 × water-immersion objective was used for confocal imaging.
2.5 RNA-sequencing and gene expression analysis
Seven-day-old NIP, osh45 and OSH45 overexpression seedlings cultured in full-strength Kimura nutrient solution were transferred to –P (0 μM Pi) and HP (200 μM Pi) solutions and cultured for a further 7 d. Roots from six independent plants were harvested, pooled together as one biological replicate, and used for RNA-sequencing (RNA-seq). Three biological repeats were performed per line and treatment. RNA-seq experiments were performed by LC-Bio Technologies Co., Ltd (Hangzhou, China). Total RNA was extracted using TRIzol reagent (Thermofisher, 15596018). mRNA libraries were constructed following the Illumina TruSeq Standed mRNA protocol and sequenced on Novaseq™ 6000 instrument. All paired-end reads were mapped to the Oryza sativa cv. Nipponbare reference genome using HISAT2 (v2-2.2.1). The rice reference genome and gene model annotations files (GFF files) were obtained from The Rice Annotation Project database (https://rapdb.dna.affrc.go.jp/). The raw sequencing data were deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (Accession No. CRA026415) (https://ngdc.cncb.ac.cn/gsa).
The mapped reads of each sample were assembled using StringTie (http://ccb.jhu.edu/software/stringtie/) with default parameters. After the final transcriptome was generated, StringTie and ballgown were used to estimate the expression levels of all transcripts and perform expression abundance for mRNAs by calculating FPKM (fragment per kilobase of transcript per million mapped reads) value. The R package DESeq2 was used for analyzing differentially expressed genes (DEGs). The genes with the parameter of q-value below 0.05 and absolute fold change ≥ 2 were considered as DEGs. DEGs were then subjected to enrichment analysis of Gene Ontology (GO) functions and KEGG pathways. Clustered genes were assigned to biological process categories based on GO analysis using the GO database (http://geneontology.org/). GO terms with q-value < 0.05 were considered significantly enriched for DEGs. Heatmaps and Venn diagrams were visualized using TBtools (Chen et al., 2020).
2.6 RT-qPCR analysis
Total RNA was isolated from roots of related plants using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. First-strand cDNA synthesis was initiated from 2 μg of total RNA with a PrimeScript™ RT reagent kit with gDNA Eraser (RR047A; Takara, Japan). qPCR was performed using SYBR Green I Master (Roche) on a LightCycler 480 Real-Time PCR system according to the manufacturer’s instructions. OsACTIN1 (Os03g0718100) was used to normalize the relative expression levels. Primers used for qPCR assays are listed in Supplementary Data Sheet 1.
2.7 Measurement of the concentration of Pi, total P, and other nutrient elements
Cellular Pi concentrations were measured as described previously (Wang et al., 2023b). Briefly, leaves, shoots and roots of 28-day-old seedlings hydroponically cultivated in Kimura nutrient solution containing 200 μM Pi were harvested. Pi concentrations were determined using a continuous flow analyzer (SAN++, SKALAR, Breda, the Netherlands) with molybdenum blue detection at 880 nm.
All plants, including WT and OSH45 mutants and overexpression lines (BC1F2 transgenic lines), were harvested after 4 weeks’ growth in Kimura nutrient solution with 200 μM Pi, separated into roots and shoots, rinsed thoroughly with deionized water, and oven dried at 70°C for 72 h to constant mass. Total elemental analysis was conducted on dried tissues. Samples underwent microwave-assisted digestion (Mars 6, CEM Corporation) with concentrated 68% HNO3 (v/v) and 30% H2O2 (v/v) in a 5:1 (v/v) ratio. Phosphorus (P), potassium (K), iron (Fe), calcium (Ca), and copper (Cu) concentrations were determined by inductively coupled plasma-optical emission spectrometry (Optima 7300DV; Perkin-Elmer).
2.8 Statistical analyses
Statistical analyses were conducted using the program SPSS Statistics v22.0 (IBM). Data visualization was generated using GraphPad Prism 8.0 (GraphPad Software).
3 Results
3.1 OSH45 is a Pi starvation-responsive TF
To discover novel regulators in plant adaptation to Pi limitation, we identified OSH45, which is significantly upregulated under Pi deficiency, from the Plant Public RNA-seq Database (Supplementary Figure S1). OSH45, which contains a homeodomain (HD) together with characteristic KNOX1 and KNOX2 subdomains, is classified within the KNOX subfamily of HD transcription factors (Supplementary Figure S2). Sequence alignment of OSH45 homologs in rice (retrieved via BLASTP with ≥ 50% sequence identity and ≥ 80% query coverage) demonstrated that the OSH45 homologs contain conserved C-terminal HD domains but divergence in N-terminal sequences (Supplementary Figure S2). To validate whether OSH45 responds to Pi deficiency, we performed RT-qPCR analysis using RNA extracted from roots of NIP under Pi-sufficient or Pi-deficient conditions. The result showed that the transcript level of OSH45 was significantly upregulated under Pi deficiency (Figure 1A), consistent with the result from RNA-seq database. RT-qPCR analysis of OSH45 expression across various tissues revealed that OSH45 is ubiquitously expressed in different tissues, with a significantly higher expression level in the leaf blade than in the root, shoot base, and leaf sheath (Figure 1B). To determine the subcellular localization of OSH45, the OSH45-GFP fusion construct was transiently expressed in N. benthamiana leaves via Agrobacterium-mediated transformation. The GFP fluorescence was predominantly accumulated in the nucleus (Figure 1C), indicating that OSH45 is a nuclear-localized protein. Furthermore, two independent OSH45 overexpression transgenic lines (OSH45-OE1/OE2), which exhibited significantly elevated OSH45 transcript levels (Figure 2A), displayed predominantly nuclear localized GFP fluorescence (Figure 1C), consistent with OSH45’s predicted function as a transcription regulator (Tamaoki et al., 1995).
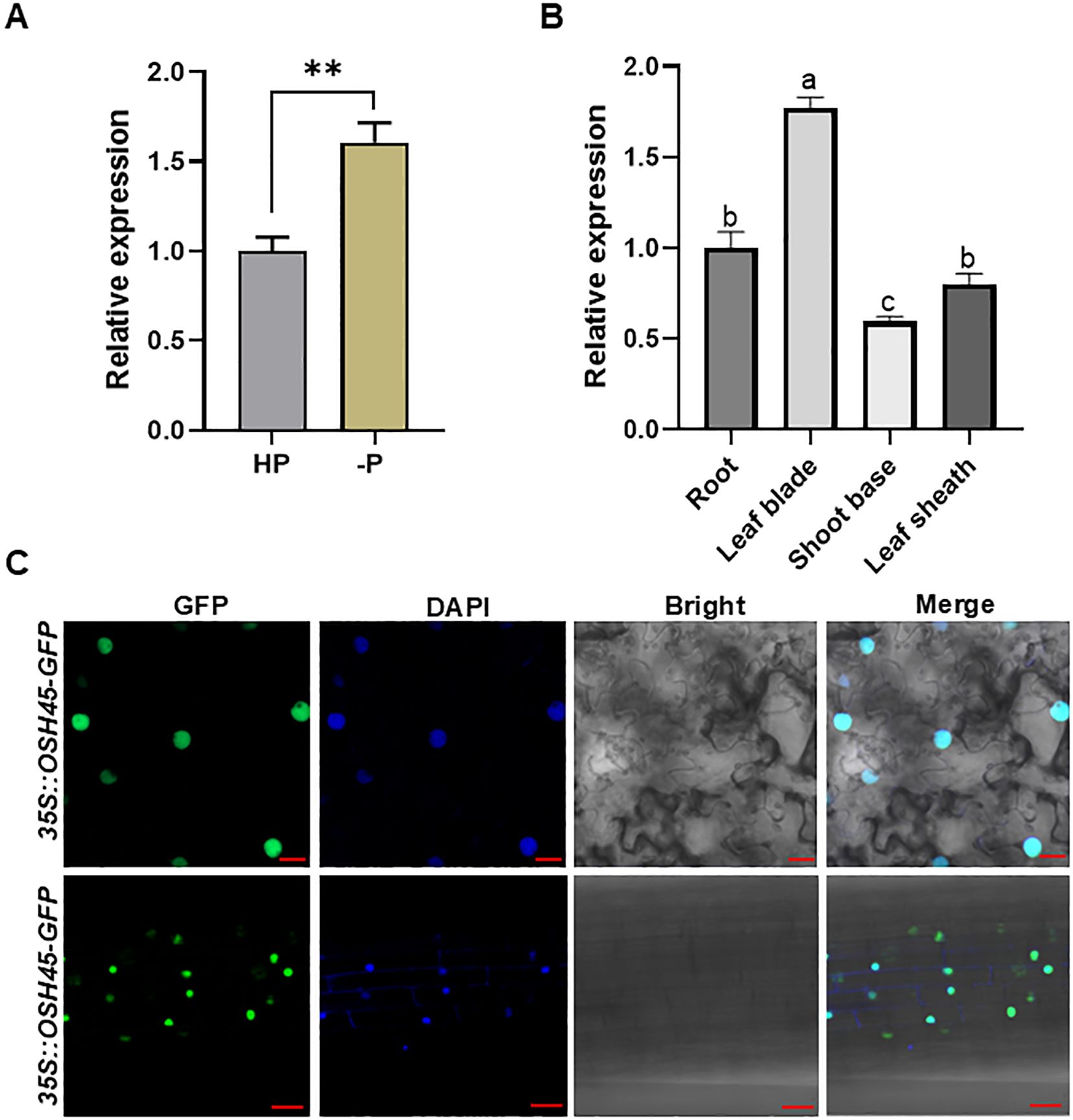
Figure 1. Expression pattern of OSH45 and subcellular localization of OSH45. (A) Expression of OSH45 in wild type (NIP) under HP (200 μM) or -P (0 μM Pi) treatment for 7 d. Seedlings were grown under normal hydroponic conditions for 7 d before -P treatment. (B) The relative expression of OSH45 in different tissues of 7-day-old rice seedlings under Pi-sufficient conditions revealed by RT-qPCR. ACTIN1 was used as an endogenous control in (A, B). Data were normalized to the expression of OSH45 in the NIP under Pi-sufficient conditions (which was set to 1). Data are means ± SD (n=3); asterisks in (A) indicate significant difference (**P < 0.01; Student’s t-test). Different lowercase letters in (B) indicate significant differences (P < 0.05; one-way ANOVA). (C) GFP fluorescence in N.benthamiana leaf cells transformed with 35S::OSH45-GFP construct (upper panel), and in root epidermal cells of 10-d-old transgenic rice seedlings harboring 35S::OSH45-GFP (lower panel). The blue signals indicate cell nucleus that were specifically stained with DAPI. Scale bars, 20 μm.
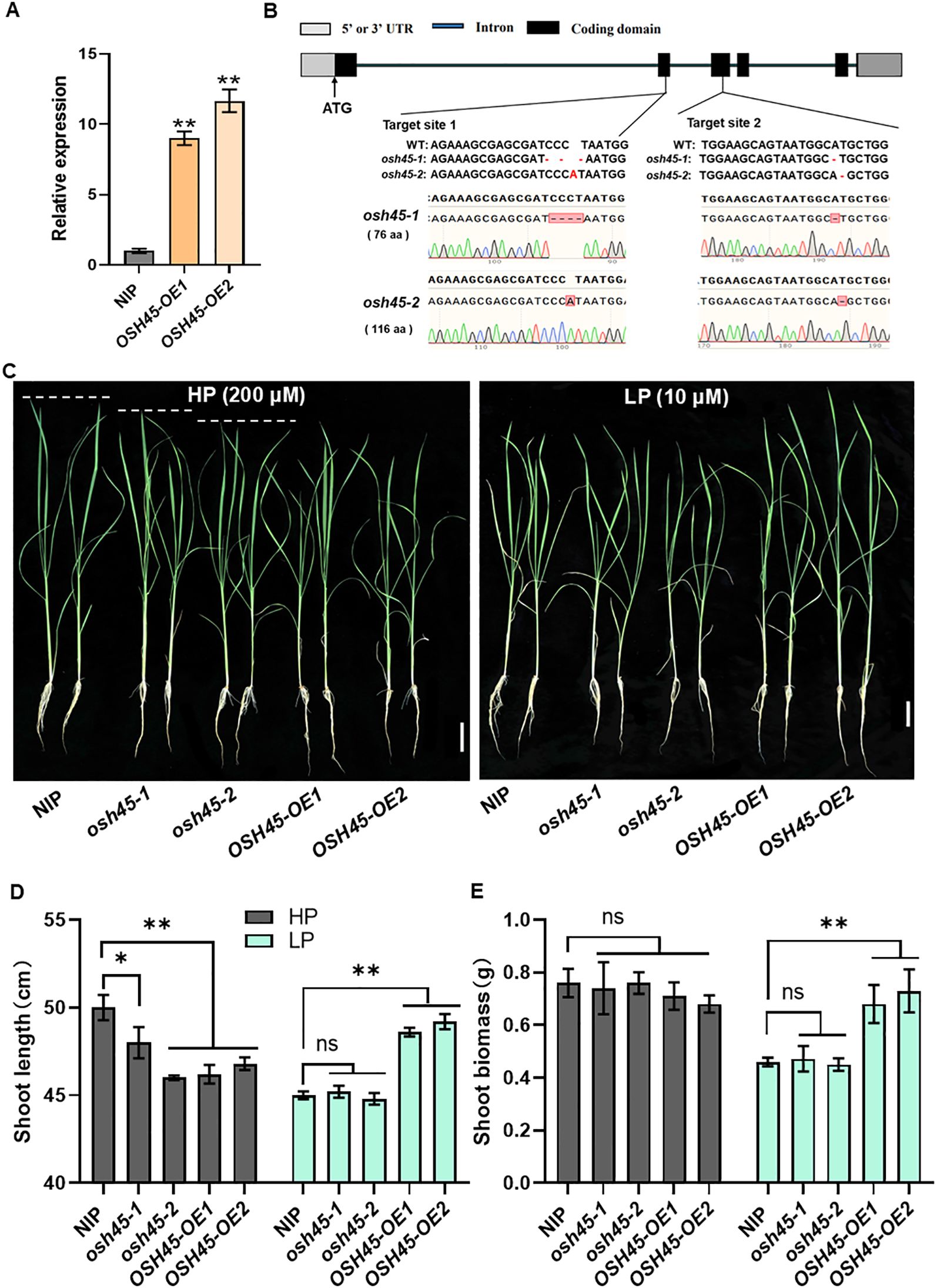
Figure 2. Phenotypes of OSH45 mutants and overexpression transgenic lines under different Pi conditions. (A) Expression of OSH45 in two independent OSH45 overexpression transgenic lines. Seedlings were grown under nutrient-replete conditions for 10 d. ACTIN1 was used as an endogenous control, OSH45 expression in the NIP was set to 1. Date are means ± SD (n=3). (B) Target sites and mutated sequences in osh45-1 and osh45-2 mutants generated using CRISPR/Cas9. (C) Phenotypes of 21-day-old wild-type (NIP), osh45 mutants, and OSH45 overexpression lines. Scale bars, 5 cm. (D, E) Shoot length (D) and shoot biomass (E) of related plants. Seven-day-old plants cultured in Pi-sufficient solution were transferred to HP (200 μM) or LP (10 μM) conditions and grown for another 14 d. Date are means ± SD (n=10); asterisks indicate significant differences compared to NIP (ns, no significant difference, *P < 0.05, **P < 0.01; Student’s t-test).
3.2 OSH45 promotes plant growth under Pi starvation
To investigate the role of OSH45 in rice growth under Pi-limited conditions, we generated loss-of-function mutants in the NIP background using clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-mediated genome editing with two single-guide RNAs (sgRNAs) targeting the exon regions of OSH45. Two independent homozygous mutant lines, designated osh45-1 and osh45-2, were identified and used for further analyses. osh45-1 exhibited a 4-bp deletion in the first sgRNA target site and a 1-bp deletion in the second target site, while osh45-2 exhibited a 1-bp insertion and a 1-bp deletion in the first and second target sites, respectively (Figure 2B). The mutants were backcrossed with NIP, and the homozygous osh45-1 or osh45-2 lines were identified from the BC1F2 to rule out the potential off-target mutations. To investigate the role of OSH45 in rice adaptation to Pi availability, 7-day-old NIP, osh45, and OSH45 overexpression lines were treated hydroponically under Pi-sufficient (HP; 200 μM) or Pi-limited (LP; 10 μM) conditions for 14 d. Under HP conditions, no significant differences in root length or shoot biomass were observed among WT, mutant, and overexpression lines (Figures 2C–E; Supplementary Figure S3). However, both mutant and overexpression lines exhibited a significant reduction in plant height compared to WT (Figures 2C, D). Under LP conditions, the OSH45 overexpression lines displayed superior growth performance compared to the WT, with an 8-10% increase in plant height and 47-58% increase in shoot biomass (Figures 2C–E). Notably, the osh45 plants showed no significant difference in shoot length, shoot biomass, root length and root biomass compared to the WT under LP conditions (Figures 2C–E; Supplementary Figure S3). These results suggested that OSH45 overexpression lines are tolerant to LP conditions compared to WT. After 21 d LP treatments, the OSH45 overexpression lines showed significantly higher shoot and root fresh weight compared to WT. Conversely, the OSH45 overexpression lines exhibited lower shoot and root fresh weight compared to WT under HP conditions (Supplementary Figure S4). Collectively, these results demonstrate that OSH45 overexpression enhances rice growth under Pi-limited conditions.
3.3 OSH45 regulates Pi acquisition in rice
To investigate the potential role of OSH45 in Pi acquisition, Pi concentrations in the leaves of 28-day-old WT, osh45, and OSH45 overexpression lines grown under HP conditions were quantified. The osh45 showed a slight reduction in leaf Pi concentrations compared to the WT, whereas OSH45 overexpression lines displayed 80-90% increase in Pi concentrations compared to the WT (Figure 3A). Whole-plant Pi concentration analysis further revealed that OSH45 overexpression plants increased shoot Pi concentration by approximately 30% and significantly enhanced root Pi accumulation compared to WT. Although the root Pi concentration did not differ significantly between osh45 and the WT, the shoot Pi concentration was reduced in osh45 plants (Supplementary Figure S5). Similarly, OSH45 overexpression lines exhibited significantly higher shoot total P concentration than the WT, whereas osh45 showed no significant difference with WT (Figure 3B). In addition, no significant differences were observed in shoot concentrations of K, Fe, Ca, or Cu among WT, osh45 and OSH45 overexpression lines (Figures 3C–F). These results demonstrate that OSH45 specifically regulates Pi acquisition but does not alter the uptake of other essential mineral nutrients.
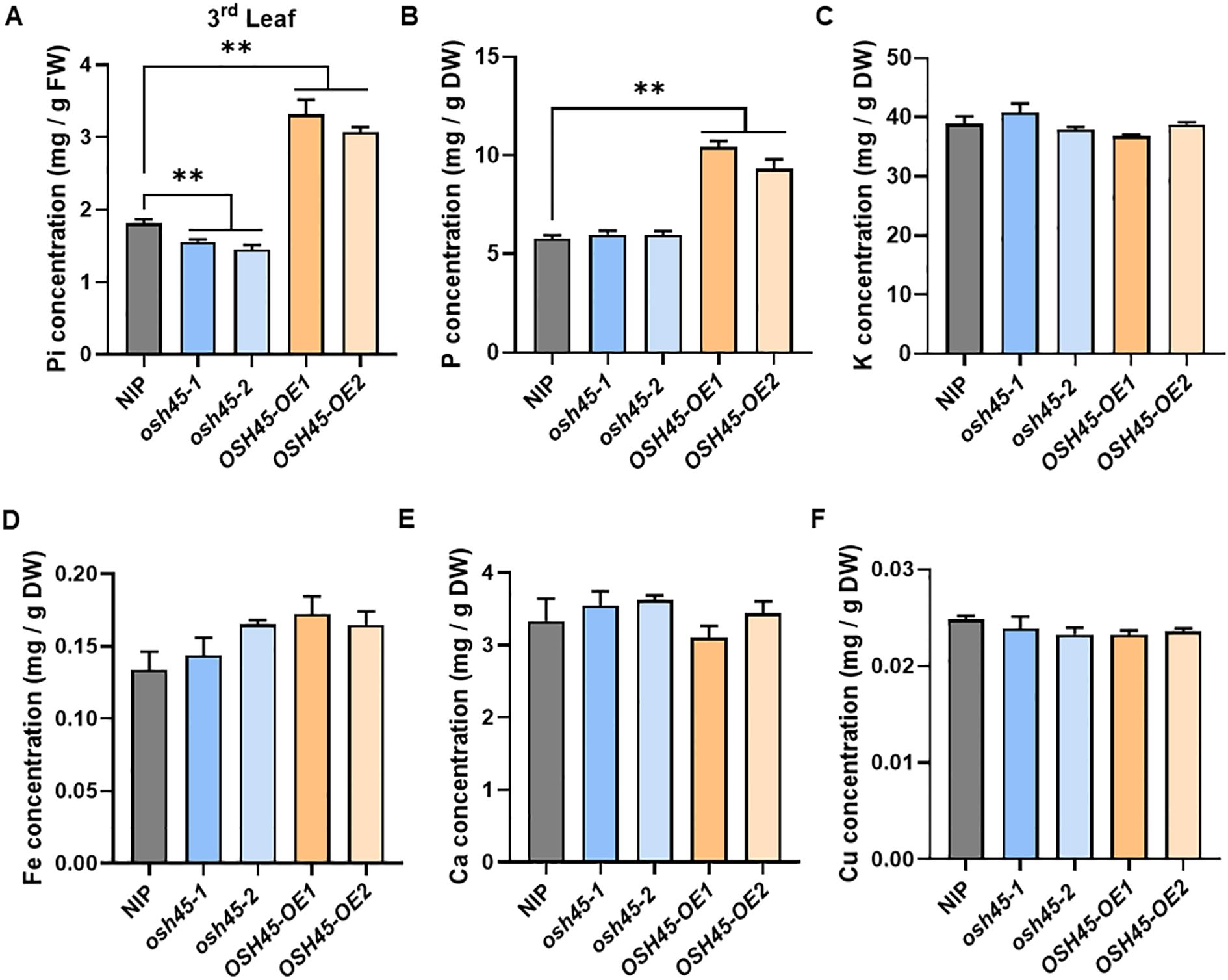
Figure 3. Pi concentration and element contents in osh45 mutants and OSH45 overexpression plants. (A) Leaf Pi concentration; (B–F) Total P concentration (B), K concentration (C), Fe concentration (D), Ca concentration (E), and Cu concentration (F) in the shoot of plants cultured in nutrient-sufficient conditions for 28 d. Date are means ± SD (n=5); asterisks indicate significant differences compared to wild-type NIP (**P < 0.01; Student’s t-test).
3.4 OSH45 regulates the expression of Pi starvation responsive genes
To understand whether OSH45 modulates the expression of PSR genes, a comparative transcriptome analysis was performed using the WT and OSH45 overexpression lines grown under HP or –P (0 μM Pi) conditions. There were 2,406 and 1,439 DEGs (fold-change [FC] ≥ 2, P-value < 0.05) in OSH45 overexpression plants compared to WT plants under HP and –P conditions, respectively (Figure 4A; Supplementary Table S1). Approximately 60% of DEGs were upregulated in OSH45 overexpression lines compared to the WT under both HP and –P conditions. Venn analysis indicated that 415 DEGs were uniformly changed in OSH45 overexpression line (OSH45-OE) versus WT under both HP and –P conditions (Supplementary Figure S6A). These uniformly changed DEGs were significantly enriched in ‘regulation of defense response’, ‘regulation of DNA-templated transcription’, ‘regulation of jasmonic acid mediated signaling pathway’, ‘nitrate assimilation’, etc. (Supplementary Figure S6C). However, most DEGs identified in OSH45 overexpression line versus WT under HP conditions (1,991) were not differentially expressed in OSH45 overexpression line compared to WT under –P conditions. The HP-specific DEGs were enriched in functional categories including ‘secondary cell wall biogenesis’, ‘carbohydrate metabolic process’, ‘protein phosphorylation’, ‘phosphate ion transport’, etc. (Supplementary Figure S6B). Collectively, these findings indicated that overexpression of OSH45 broadly affected gene expression, but the magnitude of this transcriptional reprogramming is attenuated under Pi-deficient conditions. To further dissect OSH45-mediated PSR, transcriptomic profiles of the WT and OSH45 overexpression lines under HP and –P conditions were compared. Strikingly, there were 3,370 DEGs between –P and HP treatments in WT, including 1,881 phosphate starvation induced (PSI) genes and 1,489 phosphate starvation suppressed (PSS) genes. In contrast, only 460 DEGs between –P and HP treatments were detected in the OSH45 overexpression line, including 283 PSI genes and 177 PSS genes (Figure 4A). While 97% (3,274 out of 3,370) of the PSR genes were not responsive to Pi starvation in the OSH45-OE line, only 96 common PSR genes were found between the OSH45-OE line and the WT (Supplementary Figure S7A). These 3,274 genes were significantly enriched in the following GO terms: ‘hydrogen peroxide catabolic process’, ‘cellular oxidant detoxification’, ‘response to oxidative stress’, ‘carbohydrate metabolic process’, and ‘phosphate ion transport’, etc. (Supplementary Figure S7B). It suggests that OSH45 overexpression decreased the sensitivity to Pi starvation. By comparing the PSR genes in WT (–P vs HP) and the DEGs in OSH45 overexpression lines vs WT under Pi sufficiency, we found that the expression of approximately 38% of the PSI genes (708/1881) were upregulated and about 25% of the PSS genes (366/1489) were down-regulated in OSH45 overexpression line under Pi-sufficient conditions (Figure 4B; Supplementary Table S2). Heatmap analysis revealed that the expression of the 708 PSI genes was upregulated in WT under Pi-deficient conditions and in the OSH45 overexpression lines compared to the WT under HP conditions (Figure 4C). Conversely, the 366 PSS genes were downregulated in WT under Pi starvation and in the OSH45 overexpression lines compared to the WT under HP conditions (Figure 4C). GO analysis showed that the 708 PSI genes upregulated in OSH45-OE plants and –P WT are significantly enriched in terms ‘cell surface receptor signaling pathway’, ‘defense response’, ‘carbohydrate metabolic process’, ‘phosphate ion transport’, ‘protein phosphorylation’, etc. (Figure 4D; Supplementary Table S3). In contrast, the 366 PSS genes downregulated in OSH45 overexpression lines and –P WT were enriched in GO term including ‘amino acid transport’, ‘negative regulation of mitotic cell cycle’, ‘asymmetric cell division’ and ‘maintenance of root meristem identify’, etc. (Figure 4E; Supplementary Table S3). All these results suggest that overexpression of OSH45 constitutively activates PSR.
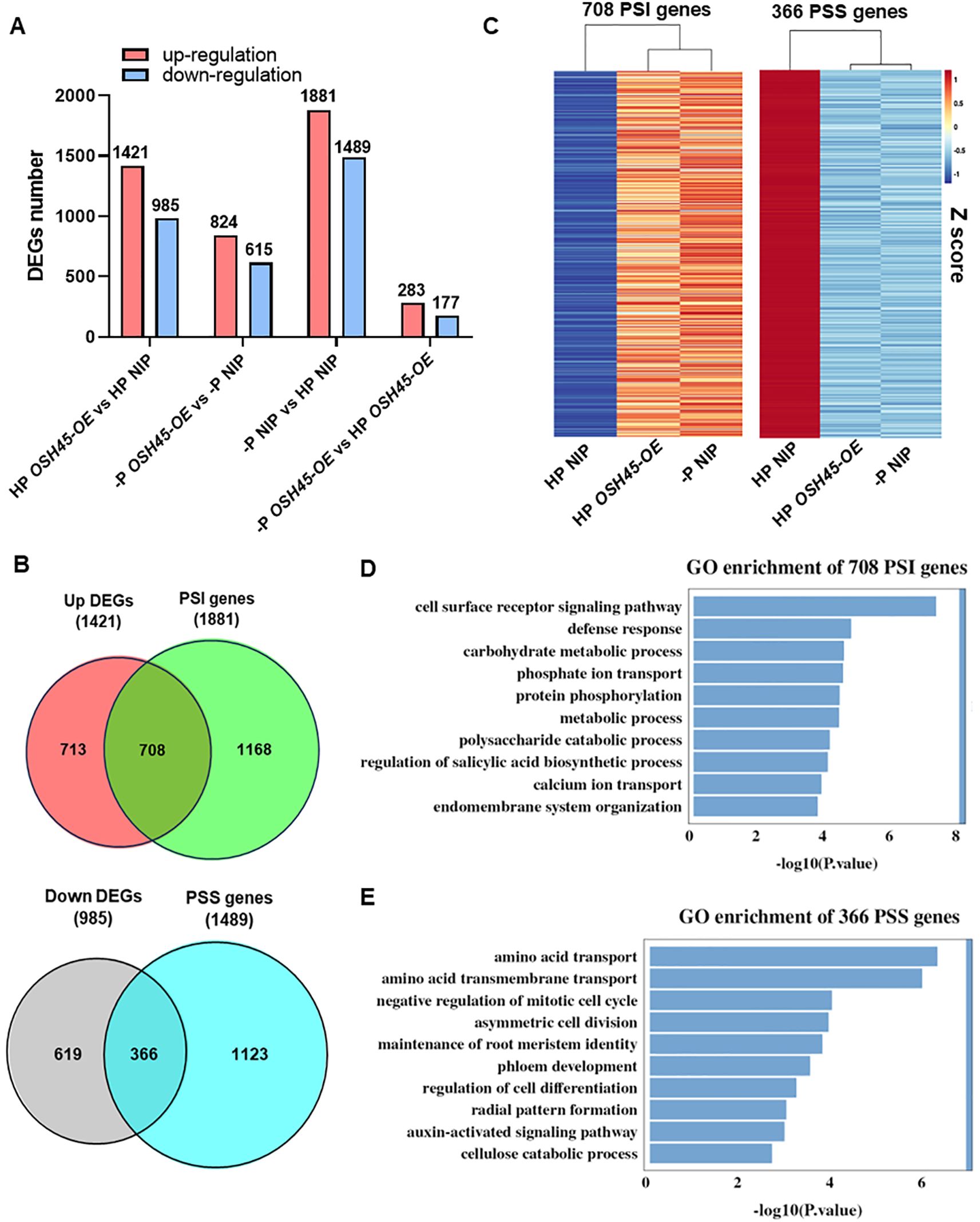
Figure 4. OSH45 regulates the expression of a series of phosphate starvation responsive (PSR) genes. (A) Numbers of differentially expressed genes (DEGs) in OSH45-OE lines relative to wild-type (WT) seedlings grown under high-Pi (HP, 200 μM Pi) or no-Pi (-P, 0 μM Pi) conditions. DEGs were defined as fold-change (FC) ≥ 2 and P-value < 0.05. Seedlings were cultured under HP conditions for 7 d and then transferred to HP or -P conditions for another 7 d before sampling and subsequent RNA-seq. (B) Venn diagrams showing the number of overlap DEGs in OSH45-OE compared to WT under HP conditions and the Pi starvation-induced (PSI) genes or the Pi starvation-suppressed (PSS) genes in WT. (C) Heatmaps showing the normalized expression levels of OSH45-affected 708 PSI genes and 366 PSS genes. Z-scores: red represents higher transcript levels, and blue represents lower transcript levels. (D, E) GO analysis of OSH45-affected 706 PSI genes (D) and 366 PSS genes (E). Bars represent P-values.
3.5 Overexpression of OSH45 affected the expression of a series of phosphate transporters
To delineate OSH45-regulated biological processes, we analyzed the DEGs of OSH45 overexpression line versus WT under Pi-sufficient conditions. The 1,421 upregulated DEGs in OSH45 overexpression line were enriched in GO terms ‘defense response’, ‘protein phosphorylation’, ‘regulation of DNA-templated transcription’, and ‘phosphate ion transport’, etc. (Figure 5A; Supplementary Table S4). The enriched term ‘phosphate ion transport’ in the upregulated DEGs suggests that OSH45 may be an important transcriptional regulator orchestrating Pi acquisition. In addition, the 985 downregulated DEGs in OSH45 overexpression plants were enriched in biological processes ‘plant-type secondary cell wall biogenesis’, ‘amino acid transport’, ‘hydrogen peroxide catabolic process’, ‘nitric oxide biosynthetic process’ and ‘nitrate assimilation’, etc. (Supplementary Figure S8; Supplementary Table S4). The enrichment of terms ‘amino acid transport’ and ‘nitrate assimilation’ suggests that OSH45 may exert a negative regulatory role in nitrogen assimilation pathways. We further analyzed the expression of known Pi signaling- or homeostasis-related genes, in the OSH45 overexpression lines. Among the known Pi signaling- or homeostasis-related genes, the transcript levels of phosphate transporters (OsPT1, PT2, PT4, PT8, PT10 and PT11), which are involved in Pi uptake from the soil, and the transcript levels of purple acid phosphatase OsPAP21b and OsPAP10c which are involved in Pi release from organic P, were increased in roots of OSH45 overexpression lines; whereas the transcript abundances of OsSPX1, OsSPX2 and OsSPX3, which are Pi starvation signaling suppressors, were decreased in OSH45 overexpression lines (Figure 5B). To verify the RNA-seq data and to investigate whether OSH45 directly regulates Pi uptake, we analyzed the expression of several phosphate transporter genes, SPX genes, and PHR genes in OSH45 overexpression lines and osh45 plants by RT-qPCR. The results showed that the transcript levels of phosphate transporters OsPT1, OsPT2, OsPT4, and OsPT8 were significantly upregulated, while OsSPX1, OsSPX2, and OsSPX3 were markedly downregulated in OSH45 overexpression plants compared to WT (Figures 6A–G). However, the expression of core Pi signaling regulators OsPHR1, OsPHR2, and OsPHR4 remained unchanged in both OSH45-OE and osh45 compared with WT (Supplementary Figures S9A–D). In osh45, the expression of OsPT1 and OsPT8 was decreased, while the transcript levels of OsPT2, OsPT4, and OsSPX1-3 remained unchanged compared to the WT (Figures 6A–G). We also analyzed the expression of these genes in WT, OSH45-OE, and osh45 plants under LP conditions. The expression of OsPT1, OsPT2, OsPT4, and OsPT8 remained significantly upregulated, whereas the expression of OsSPX1-3 were consistently decreased in OSH45-OE plants (Supplementary Figures S10A–G). In contrast, the expression of OsPT1, OsPT2, and OsPT8 were decreased in osh45 plants, while the expression levels of OsPT4 and OsSPX2-3 showed no significant change in osh45 compared to WT (Supplementary Figures S10A–G). These results confirm that OSH45 functions as a positive regulator, activating PTs while suppressing SPX genes. It is notable that the expression level of OsIPS1, a LP-induced long non-coding RNA targeting OsPHO2, was significantly lower in osh45 compared with the WT (Supplementary Figures S9E, F). These observations suggest that OSH45 participate in the regulation of Pi homeostasis. Additionally, the transcript level of nitrate transporter OsNRT2.1 was upregulated in osh45 but suppressed in OSH45 overexpression line (Figure 6H), suggesting an antagonistic regulation of nitrogen uptake genes.
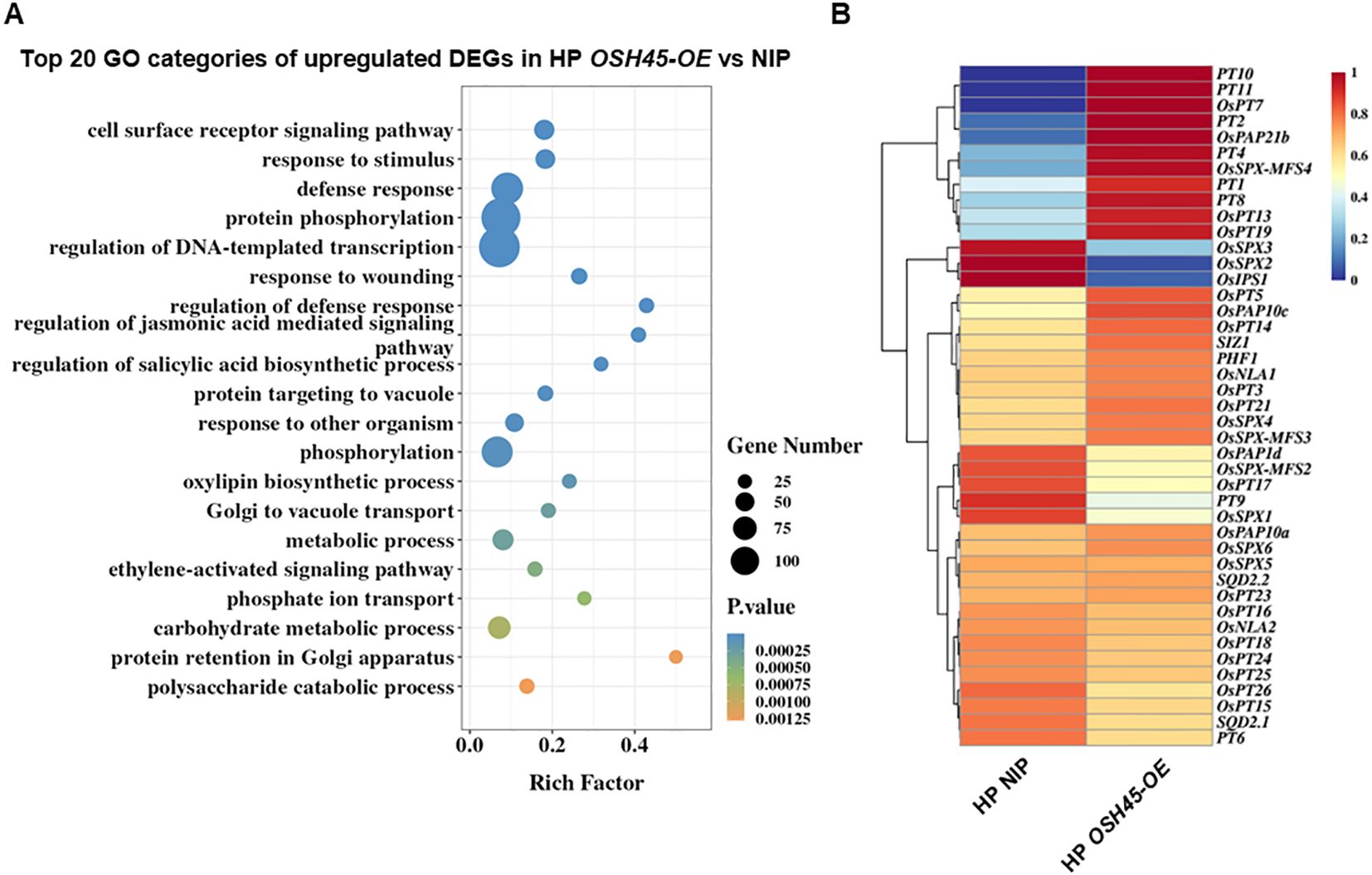
Figure 5. Biological processes of upregulated DEGs in OSH45-OE vs NIP and the expression of known Pi signaling related genes in OSH45-OE and NIP. (A) Gene ontology (GO) analysis of upregulated DEGs in OSH45 overexpression lines vs NIP under HP conditions. The top twenty significantly enriched GO terms (P-values for biological processes) are shown. (B) Heatmaps showing the normalized expression levels of OSH45-affected Pi signaling and transport related genes. Red represents higher transcript levels, and blue represents lower transcript levels.
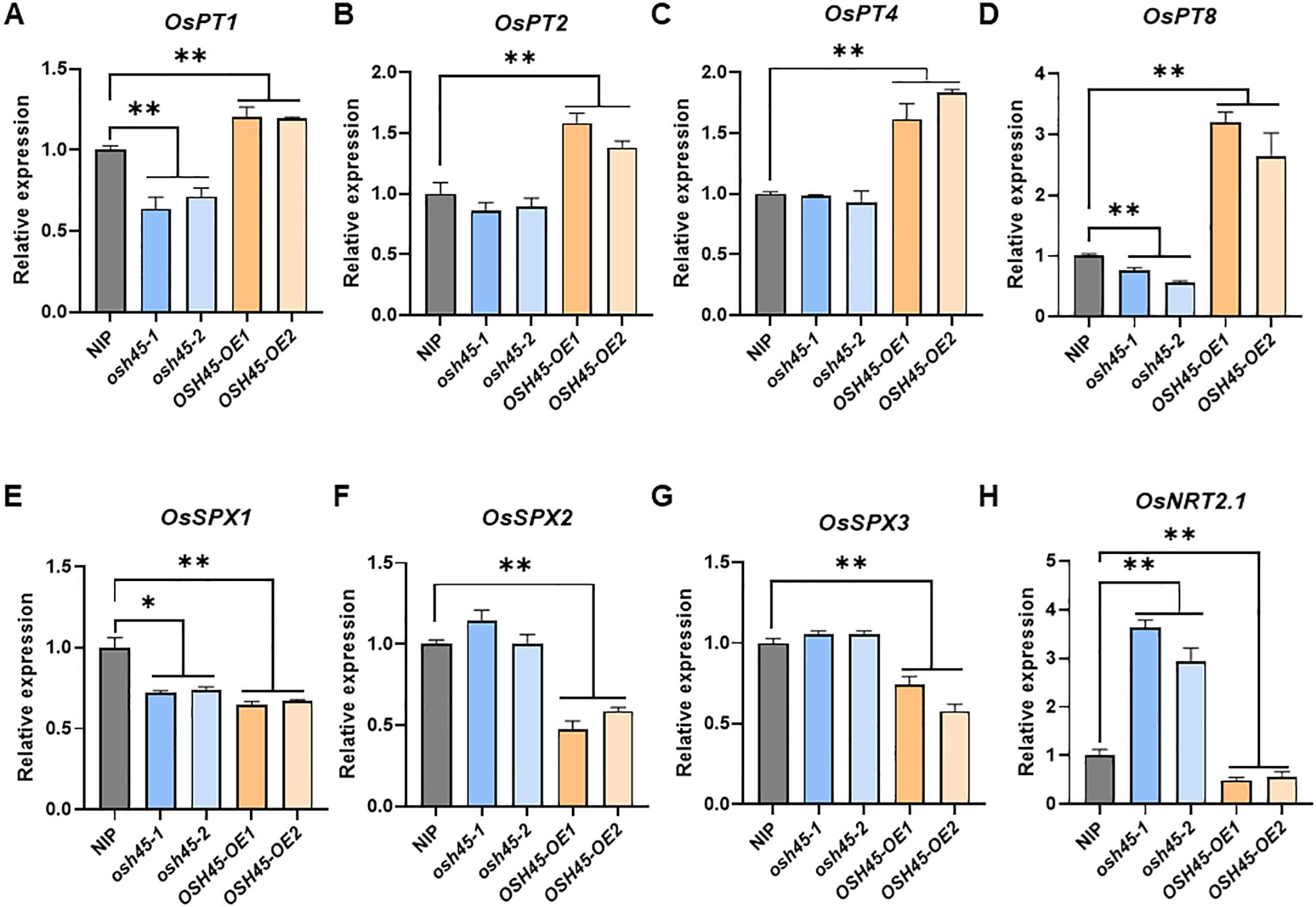
Figure 6. RT-qPCR analysis of the expression levels of OsPT1 (A), OsPT2 (B), OsPT4 (C), OsPT8 (D), OsSPX1 (E), OsSPX2 (F), OsSPX3 (G) and OsNRT2.1 (H) in NIP, osh45 mutants, and OSH45 overexpression plants cultured under HP conditions. ACTIN1 was used as an endogenous control. The expression level of each gene in the wild type NIP was set to 1. Data are means ± SD (n = 3); asterisks indicate significant differences compared to wild-type NIP (*P < 0.05; **P < 0.01; Student’s t-test).
4 Discussion
4.1 OSH45 modulates PSR through transcriptional regulation of PT and SPX genes
P is an essential macronutrient for plant growth and development. To adapt to Pi-limited environments, plants have evolved systemic adaptive strategies involving transcriptional reprogramming to improve Pi acquisition and utilization. Previous studies have demonstrated that several transcription factors, such as OsPHRs, OsPTF1, and OsWRKY74, are involved in plant adaptation to Pi deficiency (Yi et al., 2005; Ruan et al., 2016; Dai et al., 2016; Guo et al., 2015). In this study, we identified OSH45 as a novel transcription factor, orchestrating PSR in rice. The expression of OSH45 is upregulated under Pi-deficient conditions (Figure 1A; Supplementary Figure S1). Transgenic plants overexpressing OSH45 accumulated significantly higher inorganic Pi and total P in shoots compared to WT plants under Pi-sufficient conditions (Figures 3A, B; Supplementary Figure S5). Consistently, the transcript levels of PT genes OsPT1, OsPT2, OsPT4 and OsPT8 were elevated in OSH45-OE lines (Figures 6A–D), facilitating Pi uptake from the soil. Conversely, OSH45 loss-of-function mutants exhibited reduced leaf and shoot Pi concentrations compared to WT (Figures 3A; Supplementary Figure S5A), consistent with a concomitant decrease in OsPT1 and OsPT8 transcript levels (Figures 6A, D). Global transcriptomic profiling revealed that overexpression of OSH45 perturbed the expression of 32% of the PSR genes (Figure 4B; Supplementary Table S3), highlighting the important role of OSH45 in the regulation of PSR. Consistently, OSH45 overexpression plants showed markedly improved tolerance to Pi deprivation, as evidenced by increased shoot and root biomass and greater plant height under LP conditions compared to WT (Figure 2; Supplementary Figure S4). Collectively, these enhancements in growth under Pi starvation conditions suggest that OSH45 is a potential candidate for engineering crops with improved Pi uptake and use efficiency on Pi-limited soils.
4.2 OSH45 might regulate LP adaptation via SPX-PHR module
Under Pi-limited conditions, plants undergo transcriptional reprogramming to upregulate key functional genes, including Pi signaling components such as IPS1, Pi transporters (OsPT1-OsPT11), Pi remobilization enzymes (e.g., acid phosphatase-encoding genes), and intracellular Pi partitioning mediators (OsSPX-MFSs, OsVPE1/2), etc. These adaptive responses are mainly regulated by the master regulator OsPHR2 and its homologs (Lu et al., 2022; Wang et al., 2018), while the function of OsPHR2 is negatively regulated by SPX proteins (OsSPX1–OsSPX6) (Prathap et al., 2022). Transcriptome analysis revealed that about 38% of the PSI genes were upregulated and 25% of the PSS genes were downregulated in the OSH45 overexpression line (Figure 4B), suggesting that OSH45 is an important regulator of PSR genes. Considering that the central regulatory role of OsPHR2 in PSR genes expression and the phenotypic similarity in Pi hyperaccumulation between OSH45- and OsPHR2 overexpression lines, we hypothesize that OSH45 affects OsPHR2 function and thereby modulates the expression of PSR genes. Transcriptomic data and RT-qPCR analyses revealed that there were no significant changes in OsPHR2 transcript levels in either OSH45 knockout mutants or overexpression lines compared to WT plants (Supplementary Figure S9), indicating that OSH45 does not regulate the expression of OsPHR2. Intriguingly, the expression of key Pi signaling negative regulators OsSPX1, OsSPX2, and OsSPX3 were markedly downregulated in OSH45 overexpression lines (Figures 6E–G). This result suggests that OSH45 may enhance OsPHR2 function through downregulating the expression of OsSPXs, which inhibiting OsPHR2 function.
4.3 OSH45 may modulate P and nitrogen-associated transcriptional responses and plant development
The above results indicate that OSH45 positively regulates a series of PSR genes, including phosphate transporter genes. In parallel, GO analysis of the downregulated DEGs in OSH45 overexpression line (OSH45-OE vs NIP) revealed significant enrichment in categories related to “nitrate assimilation” and “amino acid transport” (Supplementary Figure S8). Specifically, the transcript levels of the high-affinity nitrate transporter OsNRT2.1 and nitrate reductase OsNIA1 were significantly reduced in OSH45-OE compared to WT (Figure 6H; Supplementary Table S2). These findings suggest that OSH45 may repress nitrogen uptake and assimilation. This repression is consistent with previous studies showing that Pi starvation alters nitrogen metabolism. In particular, SPX-PHR module and NIGT1 have been shown to integrate Pi and nitrate signaling, repressing NRT genes expression under LP conditions (Hu et al., 2019; Wang et al., 2020c; Ueda et al., 2020; Wang et al., 2023a). The simultaneous upregulation of Pi acquisition genes and downregulation of nitrate-associated genes in OSH45-OE lines may reflect a broader regulatory mechanism in maintaining nutrient balance under Pi starvation. In addition, OSH45 overexpression activated both the phosphate ion transport and plant defense response pathways (Figure 5A). This result implies potential crosstalk between nutrient signaling and immune pathways. Recently, OsSPX1 and OsSPX2 have been shown to fine-tune brassinosteroid signaling to balance growth and immunity depending on Pi availability (He et al., 2023). The dual regulation of Pi uptake and defense-related genes suggests that OSH45 may contribute to the coordination of nutrient status and stress responses to enhance plant fitness in nutrient-limited environments.
In summary, we characterized OSH45 as a positive regulator of PSR. Our data showed that OSH45 positively regulated Pi acquisition and plant growth under LP conditions, whereas OSH45 negatively regulated N acquisition and assimilation related genes. Our results expand the knowledge of molecular regulatory networks underlying plant adaptation to nutrient limitation and identify potential target for P-efficient crop breeding.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/gsa. Accession No. CRA026415.
Author contributions
HL: Formal Analysis, Writing – original draft, Data curation, Conceptualization, Writing – review & editing. KJ: Data curation, Formal Analysis, Writing – review & editing. YLL: Writing – review & editing. YW: Investigation, Writing – review & editing, Conceptualization. YL: Investigation, Conceptualization, Writing – review & editing. JX: Writing – review & editing, Conceptualization. CM: Investigation, Writing – review & editing, Writing – original draft, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Biological Breeding-National Science and Technology Major Project (2023ZD04072); the National Key Research and Development Program of China (2021YFF1000402); the Natural Science Foundation of Zhejiang Province, China (LD24C130002), the Fundamental Research Funds for the Central Universities (226-2024-00102), and the Ministry of Education and Bureau of Foreign Experts of China (B14027).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1654599/full#supplementary-material
References
Brinch-Pedersen, H., Sørensen, L. D., and Holm, P. B. (2002). Engineering crop plants: getting a handle on phosphate. Trends Plant Sci. 7, 118–125. doi: 10.1016/S1360-1385(01)02222-1
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Perez-Perez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PloS Genet. 6, e1001102. doi: 10.1371/journal.pgen.1001102
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Wang, Y., Wang, F., Yang, J., Gao, M., Li, C., et al. (2015). The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27, 711–723. doi: 10.1105/tpc.114.135335
Chen, Y., Qi, H., Yang, L., Xu, L., Wang, J., Guo, J., et al. (2023). The OsbHLH002/OsICE1-OSH1 module orchestrates secondary cell wall formation in rice. Cell Rep. 42, 112702. doi: 10.1016/j.celrep.2023.112702
Chiou, T. J. and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Dai, X., Wang, Y., Yang, A., and Zhang, W.-H. (2012). OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol. 159, 169–183. doi: 10.1104/pp.112.194217
Dai, X., Wang, Y., and Zhang, W. H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971
Furumizu, C., Alvarez, J. P., Sakakibara, K., and Bowman, J. L. (2015). Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PloS Genet. 11, e1004980. doi: 10.1371/journal.pgen.1004980
Guan, Z., Zhang, Q., Zhang, Z., Zuo, J., Chen, J., Liu, R., et al. (2022). Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex. Nat. Commun. 13, 1581. doi: 10.1038/s41467-022-29275-8
Guo, M., Ruan, W., Li, C., Huang, F., Zeng, M., Liu, Y., et al. (2015). Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol. 168, 1762–1776. doi: 10.1104/pp.15.00736
Hake, S., Smith, H. M. S., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of KNOX genes in plant development. Annu. Rev. Cell Dev. Biol. 20, 125–151. doi: 10.1146/annurev.cellbio.20.031803.093824
Ham, B. K., Chen, J., Yan, Y., and Lucas, W. J. (2018). Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49, 1–9. doi: 10.1016/j.copbio.2017.07.005
He, Q., Lu, H., Guo, H., Wang, Y., Zhao, P., Li, Y., et al. (2021). OsbHLH6 interacts with OsSPX4 and regulates the phosphate starvation response in rice. Plant J. 105, 649–667. doi: 10.1111/tpj.15061
He, Y., Zhao, Y., Hu, J., Wang, L., Li, L., Zhang, X., et al. (2023). The OsBZR1–OsSPX1/2 module fine-tunes the growth–immunity trade-off in adaptation to phosphate availability in rice. Mol. Plant 17, 258–276. doi: 10.1016/j.molp.2023.12.003
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Johnston, A. E., Poulton, P. R., Fixen, P. E., and Curtin, D. (2014). Phosphorus: its efficient use in agriculture. Adv. Agron. 123, 177–228. doi: 10.1016/B978-0-12-420225-2.00005-4
Liu, S., Xu, Z., Essemine, J., Liu, Y., Liu, C., Zhang, F., et al. (2024). GWAS unravels acid phosphatase ACP2 as a photosynthesis regulator under phosphate starvation conditions through modulating serine metabolism in rice. Plant Commun. 5, e100885. doi: 10.1016/j.xplc.2024.100885
Lu, H., Wang, F., Wang, Y., Lin, R., Wang, Z., and Mao, C. (2022). Molecular mechanisms and genetic improvement of low-phosphorus tolerance in rice. Plant Cell Environment. 46 (4), 1104–1119. doi: 10.1111/pce.14457
Lv, Q., Zhong, Y., Wang, Y., Wang, Z., Zhang, L., Shi, J., et al. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. doi: 10.1105/tpc.114.123208
Lynch, J. P. (2011). Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 156, 1041–1049. doi: 10.1104/pp.111.175414
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Madison, I., Gillan, L., Peace, J., Gabrieli, F., Van Den Broeck, L., Jones, J. L., et al. (2023). Phosphate starvation: response mechanisms and solutions. J. Exp. Bot. 74 (21), 6417–6430. doi: 10.1093/jxb/erad326
Paz-Ares, J., Puga, M. I., Rojas-Triana, M., Martinez-Hevia, I., Diaz, S., Poza-Carrion, C., et al. (2022). Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Mol. Plant 15, 104–124. doi: 10.1016/j.molp.2021.12.005
Poirier, Y., Jaskolowski, A., and Clúa, J. (2022). Phosphate acquisition and metabolism in plants. Curr. Biol. 32, R623–R629. doi: 10.1016/j.cub.2022.03.073
Prathap, V., Kumar, A., Maheshwari, C., and Tyagi, A. (2022). Phosphorus homeostasis: acquisition, sensing, and long-distance signaling in plants. Mol. Biol. Rep. 49, 8071–8086. doi: 10.1007/s11033-022-07354-9
Puga, M. I., Poza-Carrión, C., Martinez-Hevia, I., Perez-Liens, L., and Paz-Ares, J. (2024). Recent advances in research on phosphate starvation signaling in plants. J. Plant Res. 137, 315–330. doi: 10.1007/s10265-024-01545-0
Ruan, W., Guo, M., Wu, P., and Yi, K. (2016). Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol. Biol. 93, 327–340. doi: 10.1007/s11103-016-0564-6
Ruan, W., Guo, M., Xu, L., Wang, X., Zhao, H., Wang, J., et al. (2018). An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 30, 853–870. doi: 10.1105/tpc.17.00738
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Sheng, M., Ma, X., Wang, J., Xue, T., Li, Z., Cao, Y., et al. (2022). KNOX II transcription factor HOS59 functions in regulating rice grain size. Plant J. 110, 863–880. doi: 10.1111/tpj.15709
Sun, L., Song, L., Zhang, Y., Zheng, Z., and Liu, D. (2016). Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 170, 499–514. doi: 10.1104/pp.15.01336
Tamaoki, M., Tsugawa, H., Minami, E. I., Kayano, T., Yamamoto, N., Kano-Murakami, Y., et al. (1995). Alternative RNA products from a rice homeobox gene. Plant J. 7, 927–938. doi: 10.1046/j.1365-313X.1995.07060927.x
Tsuda, K., Ito, Y., Sato, Y., and Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381. doi: 10.1105/tpc.111.090050
Tsuda, K., Kurata, N., Ohyanagi, H., and Hake, S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26, 3488–3500. doi: 10.1105/tpc.114.129122
Ueda, Y., Kiba, T., and Yanagisawa, S. (2020). Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of SPX genes. Plant J. 102, 448–466. doi: 10.1111/tpj.14637
Wang, F., Deng, M., Chen, J., He, Q., Jia, X., Guo, H., et al. (2020a). CASEIN KINASE2-dependent phosphorylation of PHOSPHATE2 fine-tunes phosphate homeostasis in rice. Plant Physiol. 183, 250–262. doi: 10.1104/pp.20.00078
Wang, F., Deng, M., Xu, J., Zhu, X., and Mao, C. (2018). Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 74, 114–122. doi: 10.1016/j.semcdb.2017.06.013
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. United States America 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wang, X., Wang, H. F., Chen, Y., Sun, M. M., Wang, Y., and Chen, Y. F. (2020c). The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in Arabidopsis and Maize. Plant Cell 32, 3519–3534. doi: 10.1105/tpc.20.00361
Wang, Y., Wang, F., Lu, H., Lin, R., Liu, J., Liu, Y., et al. (2023b). Rice chromatin protein OsHMGB1 is involved in phosphate homeostasis and plant growth by affecting chromatin accessibility. New Phytologist. 240 (2), 727–743. doi: 10.1111/nph.19189
Wang, Y., Wang, F., Lu, H., Liu, Y., and Mao, C. (2021). Phosphate uptake and transport in plants: An elaborate regulatory system. Plant Cell Physiol. 62, 564–572. doi: 10.1093/pcp/pcab011
Wang, F., Wang, Y., Ying, L., Lu, H., Liu, Y., Liu, Y., et al. (2023a). Integrated transcriptomic analysis identifies coordinated responses to nitrogen and phosphate deficiency in rice. Front. Plant Sci. 14, 116441. doi: 10.3389/fpls.2023.1164441
Wang, S., Xu, T., Chen, M., Geng, L., Huang, Z., Dai, X., et al. (2022). The transcription factor OsWRKY10 inhibits phosphate uptake via suppressing OsPHT1;2 expression under phosphate-replete condition in rice. J. Exp. Bot. 74, 1074–1089. doi: 10.1093/jxb/erac456
Wang, S., Yamaguchi, M., Grienenberger, E., Martone, P. T., Samuels, A. L., and Mansfield, S. D. (2020b). The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 101, 293–309. doi: 10.1111/tpj.14541
Wu, K., Yang, W. T., Baek, D., Yun, D.-J., Lee, K. S., Hong, S. Y., et al. (2018). Rice OsMYB5P improves plant phosphate acquisition by regulation of phosphate transporter. PloS One 13, e0194628. doi: 10.1371/journal.pone.0194628
Yang, W. T., Baek, D., Yun, D.-J., Hwang, W. H., Park, D. S., Nam, M. H., et al. (2014). Overexpression of OsMYB4P, an R2R3-type MYB transcriptional activator, increases phosphate acquisition in rice. Plant Physiol. Biochem. 80, 259–267. doi: 10.1016/j.plaphy.2014.02.024
Yi, K., Wu, Z., Zhou, J., Du, L., Guo, L., Wu, Y., et al. (2005). OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 138, 2087–2096. doi: 10.1104/pp.105.063115
Yu, Y. (2019). OsKNAT7 bridges secondary cell wall formation and cell growth regulation. Plant Physiol. 181, 385–386. doi: 10.1104/pp.19.01018
Zhang, J., Gu, M., Liang, R., Shi, X., Chen, L., Hu, X., et al. (2020). OsWRKY21 and OsWRKY108 function redundantly to promote phosphate accumulation through maintaining the constitutive expression of OsPHT1;1 under phosphate-replete conditions. New Phytol. 229, 1598–1614. doi: 10.1111/nph.16931
Zhang, Z., Li, Z., Wang, W., Jiang, Z., Guo, L., Wang, X., et al. (2021). Modulation of nitrate-induced phosphate response by the MYB transcription factor RLI1/HINGE1 in the nucleus. Mol. Plant 14, 517–529. doi: 10.1016/j.molp.2020.12.005
Keywords: Oryza sativa L., phosphate starvation, knotted1-like homeobox protein, transcriptome, phosphate transporter
Citation: Lu H, Jin K, Luo Y, Wu Y, Liu Y, Xu J and Mao C (2025) OSH45, a homeobox transcription factor, coordinates low-phosphate adaptation in rice. Front. Plant Sci. 16:1654599. doi: 10.3389/fpls.2025.1654599
Received: 26 June 2025; Accepted: 01 August 2025;
Published: 28 August 2025.
Edited by:
Raul Huertas, The James Hutton Institute, United KingdomReviewed by:
Qing-Bin Chen, Shangqiu Normal University, ChinaPrathap V, Indian Agricultural Research Institute (ICAR), India
Enguang Nie, Yangzhou University, China
Copyright © 2025 Lu, Jin, Luo, Wu, Liu, Xu and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanzao Mao, bWN6QHpqdS5lZHUuY24=
 Hong Lu
Hong Lu Kangming Jin
Kangming Jin Yuelin Luo
Yuelin Luo Yunrong Wu1
Yunrong Wu1 Yu Liu
Yu Liu Jiming Xu
Jiming Xu Chuanzao Mao
Chuanzao Mao