- 1Key Laboratory of Plant Genetics and Molecular Breeding, Henan Key Laboratory of Crop Molecular Breeding and Bioreactor, Henan International Joint Laboratory of Translational Biology, Zhoukou Normal University, Zhoukou, Henan, China
- 2College of Agronomy, Henan Agricultural University, Zhengzhou, Henan, China
Nitrogen (N) plays a crucial role in various aspects of crop growth, development, yield, and quality. It is essential for processes ranging from protein synthesis and photosynthesis to crop adaptation and stress tolerance, thereby having a profound impact on crop production. Crops primarily absorb N in the forms of ammonium (NH4+) and nitrate (NO3-), with NH4+ being the predominant form absorbed by flooded crops such as rice. This review focuses on rice and highlights recent significant advances in the mechanisms of N uptake and utilization, including the roles of NO3- and NH4+ transporters. Key transporters such as OsAMT1.1 and OsNRT1.1B play central roles in enhancing N uptake and improving N use efficiency (NUE). Furthermore, natural allelic variations in genes such as DNR1 and OsWRKY23 underlie the differences in NUE between indica and japonica subspecies. We also discuss the potential of multi-gene pyramiding strategies, such as OsAMT1.2×OsGS1.2×OsAS1, to synergistically improve NUE through coordinated regulation of N uptake, assimilation, and remobilization. Collectively, this review systematically summarizes the functions and regulatory mechanisms of key NUE-related genes in rice, providing valuable gene resources and a theoretical foundation for the molecular breeding of N-efficient rice varieties.
1 Introduction
Nitrogen (N) is a major limiting factor for crop growth and high grain yield, as it is a key component of numerous essential biomolecules, including nucleic acids, enzymes, amino acids, and proteins. Hence, it is often referred to as the ‘element of life’ (Crawford, 1995; Stitt, 1999). N deficiency is a key limiting factor in crop yield formation. However, excessive N fertilizer application not only increases economic costs but also causes serious environmental damage. Therefore, elucidating the genetic basis of N use efficiency (NUE) in crops and breeding improved varieties with both high yield and enhanced NUE is essential for reducing N fertilizer demand and promoting sustainable agricultural development. The mechanisms of efficient N uptake and utilization in plants involve multiple processes. Firstly, plant roots absorb various forms of N from the soil; secondly, N is transported and transformed within the plant, and finally, it is assimilated through the action of various enzymes. This constitutes a complex process regulated by multiple factors at different stages of crop growth and development. Plant roots absorb and assimilate different forms of N, including inorganic N (such as NH4+ and NO3-) and organic N (such as amino acids and peptides), through transmembrane transporters or ion channels. In aerobic soils, nitrate (NO3-) is the predominant form of inorganic N, whereas in flooded wetlands or acidic soils, ammonium (NH4+) is the main inorganic N form (Sasakawa and Yamamoto, 1978). NO3- and NH4+ are absorbed through NO3- transporters (NPF/NRTs) and NH4+ transporters (AMTs), respectively. The absorption of NO3- or NH4+ by plant roots typically induces rhizosphere acidification or alkalization, thereby further affecting the bioavailability of soil N to plants. To cope with the heterogeneity and dynamic changes in NO3- or NH4+ ion concentrations in soil solutions, plants have evolved both high-affinity transport systems (HATS) and low-affinity transport systems (LATS) for NH4+ and NO3-. These systems are distributed in different plant tissues and cooperatively regulate N uptake and distribution (Crawford, 1995; Glass et al., 1992; Crawford and Glass, 1998; Forde, 2000). In rice, the HATS for NH4+ belong to the OsAMT1 family, while the HATS for NO3- belong to the OsNRT2 family and its partner proteins, the OsNAR2 family.
In most plants, a small portion of the absorbed NO3- is assimilated in the roots, while the majority is transported to the shoots, where it is reduced to nitrite by nitrate reductase (NR) in the cytosol. It is then transported into plastids and chloroplasts and further reduced to NH4+ by nitrite reductase (NiR) (Xu et al., 2012). NH4+ derived from NO3- reduction or directly absorbed by AMTs is toxic and must be assimilated in the roots via the glutamine synthetase (GS)/glutamate synthase (GOGAT) cycle into glutamine (Gln) and glutamate (Glu), which are the core molecules in plant N metabolism. Subsequently, Glu can be converted into aspartate (Asp) via aspartate aminotransferase (AAT), and Gln can be converted into asparagine (Asn) by asparagine synthetase (AS). These four amino acids (Glu, Gln, Asp, and Asn) play crucial roles in N transport within plants, transferring N from absorption sites to tissues where it is required (Xu et al., 2012).
In recent years, NO3- and NH4+ transporters have been identified and their functions characterized in the model crop rice. Meanwhile, the regulatory mechanisms of N uptake, transport, and assimilation have also been extensively studied. This review focuses on rice and highlights recent significant advances in the mechanisms of N uptake and utilization, including the roles of NO3- and NH4+ transporters. In addition, the functions and regulatory mechanisms of key genes related to NUE in rice are systematically summarized, providing gene resources and theoretical foundations for the molecular improvement of N-efficient rice varieties. Finally, this article emphasizes the challenges of improving NUE and advocates an integrated research approach combining molecular mechanisms, advanced technologies, and agronomic practices. By precisely coordinating N uptake, transport, assimilation, and remobilization with rice developmental responses to N availability, it is possible to ensure efficient N use, thereby contributing to global food security and the sustainable development of agriculture.
2 Functions of NH4+ transporters in rice
2.1 Classification and transport characteristics
Although NH4+ has long been recognized as the primary form of N absorbed by rice, research on NH4+ transporters have remained relatively limited. With the advancement of genomics, at least 12 potential NH4+ transporters (AMTs) have been identified in the rice genome. These transporters are classified into five subfamilies: OsAMT1 (OsAMT1.1, OsAMT1.2, and OsAMT1.3), OsAMT2 (OsAMT2.1, OsAMT2.2, and OsAMT2.3), OsAMT3 (OsAMT3.1, OsAMT3.2, and OsAMT3.3), OsAMT4 (OsAMT4.1), and OsAMT5 (OsAMT5.1 and OsAMT5.2) (Suenaga et al., 2003; Li et al., 2009a). The OsAMT1 subfamily functions as a high-affinity NH4+ transporter, operating under low NH4+ concentrations and exhibiting saturation kinetics. In contrast, the OsAMT2, OsAMT3, and OsAMT4 families are classified as low-affinity transporters, predominantly active under high NH4+ concentrations (1–40 mM), and do not display saturation kinetics (Gaur et al., 2012). Studies have demonstrated that OsAMT1.1, OsAMT1.2, OsAMT1.3, OsAMT2.1, and OsAMT5.1 all possess NH4+ transport capacity (Bu et al., 2011) (Figure 1).
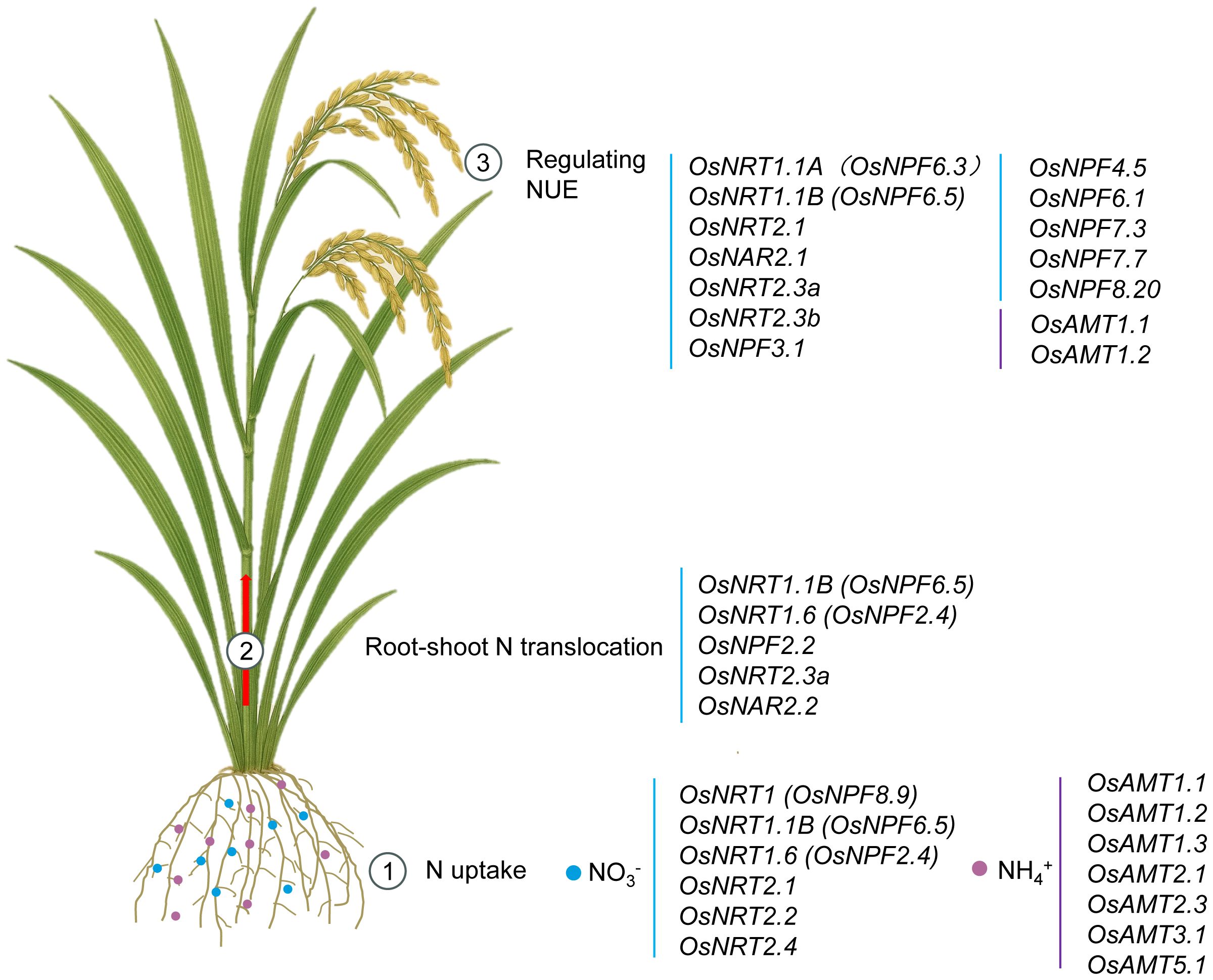
Figure 1. Integrative model to illustrate physiological functions of NO3- and NH4+ transporters in rice. Detailed illustration of NO3- and NH4+ uptake, translocation, and utilization in rice. Rice NO3- uptake is orchestrated by transporters OsNRT1, OsNRT1.1B, OsNRT1.6, OsNRT2.1, OsNRT2.2 and OsNRT2.4. For NH4+, OsAMT1.1, OsAMT1.2, OsAMT1.3, OsAMT2.1, OsAMT2.3, OsAMT3.1 and OsAMT5.1 are principal. OsNRT1.1B, OsNRT1.6, OsNPF2.2, OsNRT2.3a and OsNAR2.2 are crucial for NO3- translocation to shoots. Among the currently identified members of the rice NRT/NPF family, OsNRT1.1A, OsNRT1.1B, OsNRT2.1, OsNAR2.1, OsNRT2.3a, OsNRT2.3b, OsNPF3.1, OsNPF4.5, OsNPF6.1, OsNPF7.7, and OsNPF8.20 have all been shown to enhance NUE, whereas OsNPF7.3 decreased NUE at high NH4+ supply. In the AMT family, only OsAMT1.1 and OsAMT1.2 have been identified as capable of improving NUE.
2.2 Spatial and N-responsive expression of OsAMTs
The uptake of low concentrations of NH4+ in rice roots requires the coordinated activity of three OsAMT1 members, which share high amino acid sequence homology. Among these, OsAMT1.1 makes the largest contribution to N accumulation (Sonoda et al., 2003a; Zhao et al., 2014). Spatial expression analyses revealed that NH4+ exposure induces the upregulation of OsAMT1.1 and OsAMT1.2, and the downregulation of OsAMT1.3 (Konishi and Ma, 2021). OsAMT1.1, OsAMT3.2, and OsAMT3.3 are constitutively expressed in both roots and stems, while OsAMT1.1 expression is promoted by NH4+. OsAMT1.2 is root-specific and induced by NH4+, whereas OsAMT1.3 is root-specific but suppressed by N. OsAMT2.1, OsAMT2.2, OsAMT2.3, and OsAMT3.1 are mainly expressed in aerial tissues, with relatively higher expression in stems than other genes. The expression of OsAMT3 family genes is generally higher in shoots than in roots, suggesting that AMT3 members may participate in the translocation and distribution of NH4+ within leaves (Suenaga et al., 2003; Gaur et al., 2012; Sonoda et al., 2003a; Li et al., 2012; Li and Shi, 2006). Additionally, the expression patterns of OsAMT1 genes show strong correlations with Gln levels in root tissues (OsAMT1.1 and OsAMT1.2 positively correlated, OsAMT1.3 negatively correlated), but not with NH4+ content (Sonoda et al., 2003b). OsAMT5.1 is specifically expressed in leaves, with its expression enhanced by increasing NH4+ concentrations (Deng et al., 2007).
2.3 Functional divergence and regulatory mechanisms of key OsAMT1 genes
At present, research on the regulation of AMT genes in rice has primarily focused on three OsAMT1 family genes. Among them, OsAMT1.1 plays a central role in N absorption and utilization in rice. It significantly promotes NH4+ uptake under both low and high NH4+ conditions, maintains N-potassium (K) homeostasis, and enhances NUE, plant growth, and grain yield under suboptimal to optimal N supply. Moreover, this gene underwent strong selection from wild rice to cultivated rice in response to soil conditions (Figure 1; Table 1) (Lee et al., 2020; Ranathunge et al., 2014; Ding et al., 2011). Simultaneous activation of OsAMT1.2 and the glutamate synthase gene (OsGOGAT1) improve tolerance to N limitation and enhances NH4+ uptake and N remobilization at the whole-plant level (Figure 1; Table 1) (Lee et al., 2020). In contrast, overexpression of OsAMT1.3 causes imbalances in carbon (C)-N metabolism, leading to poor plant growth and reduced yield (Bao et al., 2015).
2.4 Perspectives on the regulatory and metabolic roles of AMT genes in rice
The AMT gene family plays a critical role in NH4+ uptake in rice. To date, studies on the phylogeny, expression patterns, and functions of rice AMT genes have provided preliminary insights into their roles in N absorption. However, the regulation of AMT genes is not limited to N uptake but may also be involved in N assimilation, translocation, and other N-related metabolic processes. Therefore, future studies should expand our understanding of AMT gene regulation, environmental adaptability, and especially their integrative roles in N transport and metabolism. Such research will help uncover the potential applications of these genes in improving NUE and crop productivity in rice, ultimately offering new strategies for sustainable agricultural production and environmental protection.
3 Functions of NO3- transporters in rice
The absorption of NO3- is an active process driven by H+/NO3- co-transporters (Miller et al., 2007). Although rice is a plant that prefers NH4+, under the action of soil microorganisms, NH4+ can be converted into NO3- through nitrification. In addition, N fertilizers applied to the soil are also partially converted into NO3-, which can then be absorbed and utilized by rice. As a result, about 25-40% of the total N absorbed by rice exists in the form of NO3- (Xu et al., 2012; Li et al., 2008; Kirk and Kronzucker, 2005). Compared with NH4+, the mechanisms of NO3- absorption have been more extensively studied in rice, and the corresponding transporters have been thoroughly identified. In rice, NO3- transporters are generally classified into two families: the low-affinity NRT1 family and the high-affinity NRT2 family, enabling rice to adapt to changes in N availability in the environment.
3.1 NRT1/PTR family: low-affinity NO3- transporters
The number of NO3- transporter 1/peptide transporter (NRT1/PTR, also known as NPF) family members in rice has been confirmed by several genomic analyses, with approximately 93 NPF genes identified. However, the functions of only a few NPF family members have been characterized to date (Léran et al., 2014). Different members of the NPF family perform distinct functions in rice. NRT1.1 in rice, an important NO3- transporter belonging to the NPF6 subfamily, mainly includes three homologs: OsNRT1.1A (OsNPF6.3), OsNRT1.1B (OsNPF6.5), and OsNRT1.1C (OsNPF6.4), which play critical roles in N uptake, transport, signaling, and NUE (Wang et al., 2020b).
OsNRT1.1A exhibits NH4+-induced expression and can significantly upregulate the expression of various genes related to NO3- and NH4+ utilization. Overexpression of OsNRT1.1A significantly improves NUE and grain yield and also shortens the rice maturity period, providing a feasible approach for breeding high-yield, early-maturing rice varieties (Wang et al., 2018c). Among the NRT1 family members in rice, only OsNRT1.1B possesses dual-affinity transport properties and functions across a wide range of NO3- concentrations. Under low N (LN) conditions, OsNRT1.1B enables plants to accumulate more N and promotes rice growth, whereas OsNRT1.1A lacks such functionality in rice (Fan et al., 2016a). The NO3- uptake activity of indica rice is higher than that of japonica, and genetic variation in OsNRT1.1B significantly influences differences in NUE between indica and japonica by regulating NO3- uptake and rhizosphere microbiota. Moreover, introducing the NRT1.1Bindica allele into japonica could potentially enhance the NUE of japonica rice (Hu et al., 2015; Zhang et al., 2019) (Figure 1; Figure 2; Table 1). The functional differentiation between OsNRT1.1A and OsNRT1.1B helps rice coordinate internal and external N signals and improve its adaptability to complex N environments (Wang et al., 2020b). OsNRT1.3 promoter responds to drought stress, potentially participating in basic NO3- uptake and stress responses (Hu et al., 2006).
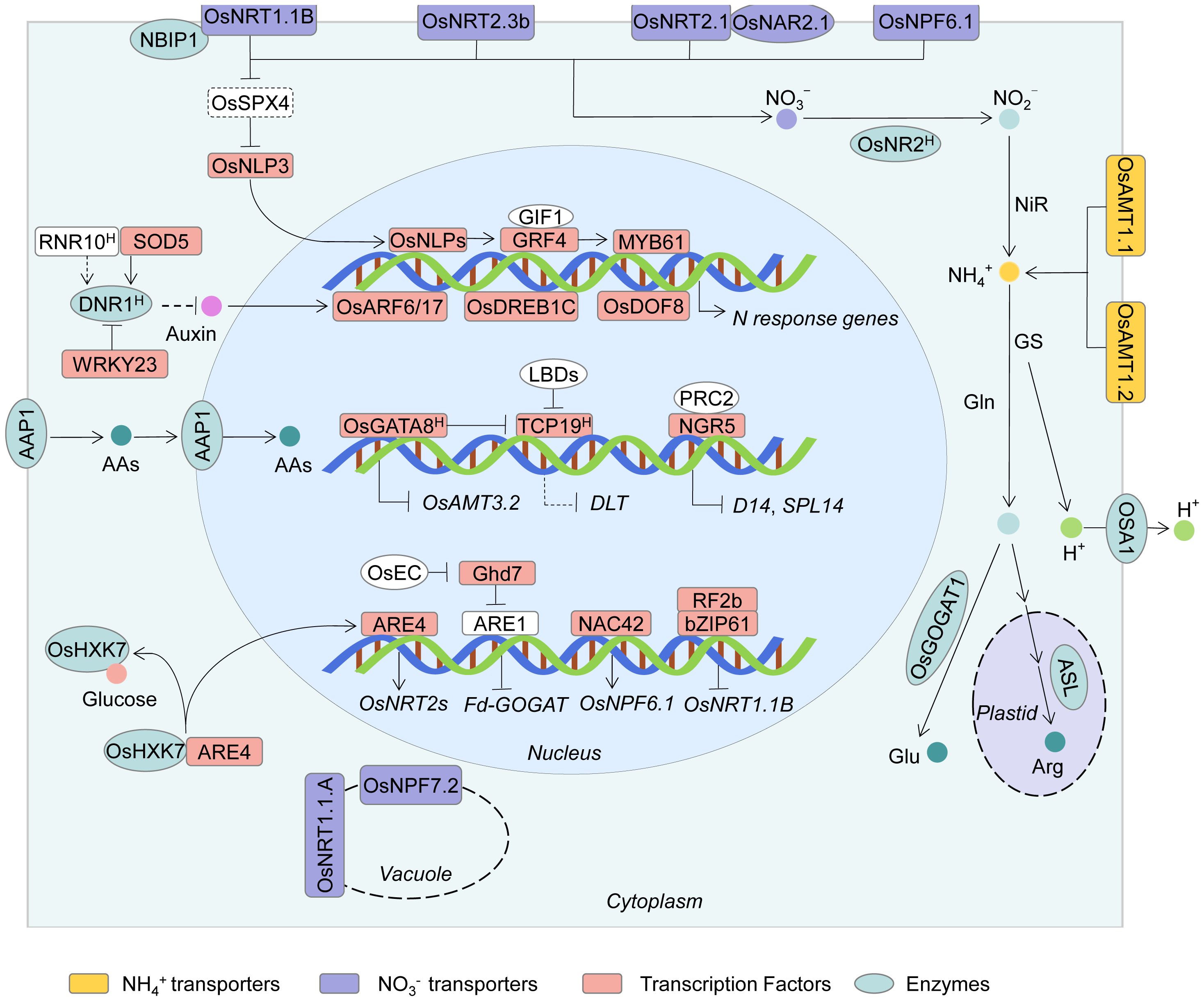
Figure 2. Schematic representation of key regulatory pathways and major genes involved in NUE in rice. This model summarizes major molecular players and signaling pathways involved in NO3- and NH4+ uptake, assimilation, and systemic regulation of NUE in rice. NO3⁻ and NH4+ are absorbed via NRT/NPF and AMT transporter families, respectively. A central NO3- signaling module comprising OsNRT1.1B-OsSPX4-OsNLP3/NLP4 integrates NO3- sensing and transcriptional responses, regulating genes involved in N uptake and assimilation. Key transcription factors, including OsDREB1C, GRF4, NGR5, WRKY23, GATA8, and ARE4, modulate NUE through direct or indirect regulation of transporter genes and metabolic enzymes. Auxin-mediated pathways also contribute to NUE control: DNR1 antagonizes auxin accumulation and NO3⁻ responsiveness, while RNR10, SOD5, and WRKY23 regulate DNR1 expression or stability. The GRF4-MYB61, Ghd7-ARE1, and NGR5-PRC2 modules further coordinate NUE with plant development and chromatin dynamics. Additional regulators include OsDOF18, which activates AMT genes, OsGATA8 and OsTCP19, which link N status to tillering, and bZIP transcription factors OsRF2b/OsbZIP61, which negatively regulate OsNRT1.1B. The amino acid transporter OsAAP1, located on both the plasma membrane and nuclear membrane, is capable of absorbing and transporting amino acids, thereby enhancing NUE. Together, these components form a complex regulatory network integrating nutrient signaling, transcriptional control, hormone crosstalk, and epigenetic regulation to optimize NUE and sustain rice yield under variable N inputs.
Within the rice NPF family, OsNPF2.2 can unload NO3- from the xylem, thereby affecting NO3- transport in the root-stem and plant development (Li et al., 2015). OsNPF2.4 (OsNRT1.6) is a pH-dependent low-affinity transporter functioning in NO3- uptake, long-distance transport, and redistribution, while its altered expression indirectly affects K reutilization in roots and stems (Xia et al., 2015). A coding region mutation in OsNPF3.1 affects NUE differences between wild and cultivated rice and can improve NUE and biomass yield (Yang et al., 2023). OsNPF4.1 (SP1) encodes a putative peptide transporter highly expressed in the phloem of young panicle branches, controlling panicle size (Li et al., 2009b). Mycorrhizal rice could receive more than 40% of its N via the mycorrhizal pathway, and the arbuscular mycorrhizal-specific NO3- transporter OsNPF4.5 accounted for approximately 45% of the mycorrhizal NO3- uptake. Enhanced expression of NPF4.5 can significantly improve NUE and promote rice growth (Wang et al., 2020a). The NO3- transporter OsNPF5.16 positively regulates rice tillering and yield by regulating cytokinin levels (Wang et al., 2022). OsNPF6.1 is NO3–inducible and has two haplotypes: OsNPF6.1HapA and OsNPF6.1HapB. OsNPF6.1HapB enhances NO3- absorption and improves NUE. Furthermore, OsNPF6.1 enhances viral resistance by upregulating the expression of NO3- reductase (OsNR2) and subsequently promoting nitric oxide (NO) biosynthesis (Tang et al., 2019; Xu et al., 2025) (Figure 1; Table 1).
Among the OsNPF7 and OsNPF8 subfamilies, OsNPF7.1 (OsPTR4) and OsNPF7.4 show opposite expression patterns in tiller buds under different N concentrations. Overexpression of either OsNPF7.1 or OsNPF7.4 promotes NO3- absorption, although biomass is reduced in OsNPF7.4-overexpressing plants (Huang et al., 2019b). OsNPF7.2 acts as a positive regulator of NO3- influx and concentration, with overexpression lines showing significant increases in tiller number and yield (Hu et al., 2016; Wang et al., 2018a). OsNPF7.3 (OsPTR6) mainly transports di- and tripeptides (e.g., Gly-His, Gly-His-Gly). While its overexpression promotes rice growth, its effect on NUE is limited (Fan et al., 2014); however, subsequent studies revealed that overexpression of both OsNPF7.3 and OsNPF7.7 increases rice tiller number, NUE, and yield (Fang et al., 2017; Huang et al., 2018). OsNPF8.1 (OsPTR7) mediates stress-induced organic N transport, contributing to balanced plant growth and enhanced tolerance to salt/drought stress and N deficiency (Qiu et al., 2023). OsNPF8.9 (OsNRT1) is the first low-affinity NO3- transporter identified in rice, functioning under high NO3- conditions (Lin et al., 2000). The di-/tripeptide and low-affinity NO3- transporter OsNPF8.20 (OsPTR9) enhances NH4+ uptake, promotes lateral root formation, and increases grain yield when its expression is upregulated (Fang et al., 2013) (Figure 1; Table 1).
3.2 NRT2/NAR2 family: high-affinity NO3- transporters
The HATS play a crucial role in rice N uptake. To date, five NRT2 (OsNRT2.1, OsNRT2.2, OsNRT2.3a, OsNRT2.3b, OsNRT2.4) and two NAR2 (OsNAR2.1, OsNAR2.2) high-affinity NO3- transporters have been identified in rice (Araki and Hasegawa, 2006; Feng et al., 2011; Cai et al., 2008). The rice NRT2 and NAR2 family members exhibit distinct functions. Some NRT2 members require the partner protein NAR2 for NO3- transport within relatively low concentration ranges (Cai et al., 2008).
OsNRT2.3 generates two transcripts, OsNRT2.3a and OsNRT2.3b, through mRNA splicing, with 94.2% amino acid sequence identity and identical coding regions but different 5′ and 3′ untranslated regions. OsNRT2.3a is primarily expressed in roots and induced by NO3-, whereas OsNRT2.3b is mainly expressed in shoots. Further research revealed that under LN supply, OsNRT2.3a plays a key role in long-distance NO3- transport from roots to shoots, and OsMADS57 regulates NO3- transport through OsNRT2.3a (Huang et al., 2019a; Tang et al., 2012). Allelic variation in the 5’ untranslated region of OsNRT2.3 leads to elevated OsNRT2.3b protein levels under high-temperature stress, increasing yield (Zhang et al., 2022). Additionally, high OsNRT2.3b expression enhances pH buffering capacity and improves the uptake of N, iron, and phosphorus (Fan et al., 2016b; Feng et al., 2017).
OsNAR2.1 interacts with OsNRT2.1/2.2 and OsNRT2.3a to mediate NO3- uptake (Feng et al., 2011; Yan et al., 2011). OsNAR2.2, localized to the endoplasmic reticulum (ER), was recently shown to regulate NO3- transport from roots to stems and control spikelet number, yield, and NUE in rice (Hou et al., 2025). OsNRT2.1, OsNRT2.2, and OsNAR2.1 are promising candidate genes for breeding high NUE rice cultivars (Araki and Hasegawa, 2006). Enhancing OsNAR2.1 expression via its native promoter, or increasing OsNRT2.1 expression under the control of the OsNAR2.1 promoter, or co-overexpressing OsNAR2.1 and OsNRT2.3a can all improve NO3- uptake, yield, and NUE in rice (Figure 1; Table 1) (Chen et al., 2016, Chen et al., 2017, Chen et al., 2020). OsNRT2.4, a dual-affinity NO3- transporter, participates in regulating NO3- uptake and allocation between roots and shoots and promotes plant growth and development under NO3- regulation (Wei et al., 2018).
3.3 NO3- sensing and signal transduction
In rice, NO3- acts not only as a nutrient but also as a signaling molecule. OsNRT1.1B has been confirmed to sense external NO3- signal (Hu et al., 2015). Additionally, studies have shown that the NO3- sensor OsNRT1.1B physically interacts with the phosphate signaling repressor OsSPX4; the presence of NO3- enhances this interaction and promotes the recruitment of NRT1.1B-Interacting Protein 1 (NBIP1, an E3 ubiquitin ligase), leading to the ubiquitination and degradation of OsSPX4. The core NO3- signaling transcription factor NLP3 is also regulated by SPX4. This OsNRT1.1B-OsSPX4-OsNLP3 regulatory module fills the gap between plasma membrane NO3- sensing and downstream NO3- responses in the nucleus (Figure 2) (Wang et al., 2020b; Hu et al., 2019).
4 Key genes regulating nitrogen use efficiency in rice
4.1 NUE-associated genes identified by QTL and map-based cloning
During crop domestication, many advantageous variant loci are retained by natural or artificial selection. Identifying these natural variant loci can provide theoretical support for crop genetic improvement (Hu et al., 2015; Li et al., 2018). In modern rice cultivars, NUE-related quantitative trait loci (QTL) or genes have been identified through map-based cloning methods. According to the varying N absorption capacity among different varieties, key genes controlling NUE, such as OsNRT1.1B, OsNR2, DNR1, and OsWRKY23, have been cloned (Hu et al., 2015; Li et al., 2018; Zhang et al., 2021b; Gao et al., 2019; Zhang et al., 2025b).
OsNRT1.1B and OsNR2 in indica have significant improvement in NUE and grain yield than those in japonica (Hu et al., 2015; Gao et al., 2019). Auxin Response Factor OsARFs mediate the promotion of N metabolism by auxin, DNR1 participates in the regulation of auxin homeostasis, and reflects differences in NO3- uptake, N assimilation, and yield enhancement between indica and japonica. The variation in the promoter of DNR1 in indica decreased expression levels and a higher auxin content, which triggers ARF-activated the transcription of NO3- uptake and assimilation-related genes, leading to improving the grain yield and NUE (Zhang et al., 2021b). RNR10 encodes an F-box protein that interacts with DNR1. RNR10 monoubiquitinates DNR1 and inhibits its degradation, thus antagonizing auxin accumulation, which results in reduced root responsivity to N and NO3- uptake (Huang et al., 2023). SOD5 directly binds to the DNR1 promoter, activates its expression, and further inhibits auxin accumulation. Notably, knockout of SOD5 significantly improves NUE and grain yield, especially under LN conditions (Zhang et al., 2025a). OsWRKY23 is a key regulator of the differences in NO3- absorption rate and NUE between indica and japonica rice. OsWRKY23indica exhibits reduced transcriptional activation of DNR1, leading to higher auxin levels, improved NO3- absorption and assimilation, and ultimately enhanced NUE and yield (Zhang et al., 2025b).
Different varieties exhibit developmental differences due to their varying sensitivity to N supply, which can be observed in factors such as root length, biomass, and yield. Map-based cloning has been used to identify the genetic loci responsible for N modulation of plant growth, such as MYB61, through analysis of phenotypic values of these traits or ratios under different N levels supplied (Gao et al., 2020; Obara et al., 2010; Lian et al., 2005). The transcription factor MYB61 is regulated by GROWTH-REGULATING FACTOR4 (GRF4) and coordinates the production of cellulosic biomass and N utilization. The indica allele of MYB61 shows strong transcriptional activity, leading to improved NUE and higher grain yield under reduced N supply compared to the japonica allele (Gao et al., 2020) (Figure 2; Table 1).
4.2 NUE-associated genes identified by GWAS
In recent years, genome-wide association study (GWAS) has also been used to locate some N-efficient genes. For example, using NUE-related agronomic traits, the GWAS identified the excellent variation of OsNPF6.1HapB, which originated from the variation of wild rice. This excellent allele was transcriptionally activated by NAC42, and it enhances the ability of N absorption capacity and improves NUE under LN conditions (Tang et al., 2019). Through GWAS analysis of NUE-related traits (effective panicle number and yield per plant) in natural populations of rice, combined with transcript data under high N (HN) and LN conditions, OsNLP4 was identified as a transcription factor that regulates NUE. Simultaneously, OsNLP4 can promote the transcription of nitrite reductase gene OsNiR and N transport-related genes (NRTs and AMT1.1) (Yu et al., 2021; Wu et al., 2021), achieving coordinated regulation of N uptake and assimilation in rice. Accordingly, the localization of OsNLP3 and OsNLP4 in rice cells is affected by NO3- supply levels. With the application of NO3-, the localization of OsNLP3 and OsNLP4 in cells is shifted from cytoplasm to nucleus (Hu et al., 2019; Wu et al., 2021), indicating that OsNLP3 and OsNLP4 are central regulatory factors in the N signaling pathway of rice.
Using a multiparent advanced generation intercross (MAGIC) population, GWAS of NUE-related traits (tillering) under HN and LN conditions identified the OsSTP28 as a key regulator of N-responsive tillering and yield formation in rice (Zhang et al., 2024). A GWAS for N-responsive tillering in rice identified OsTCP19 as a regulatory factor; a 29-bp indel in the promoter of OsTCP19 represents a natural variation that determines tiller number under LN conditions in different rice varieties. OsTCP19 as a modulator of tillering response to N through its transcriptional response to N and its targeting to the tiller-promoting gene DWARF AND LOW-TILLERING (DLT), the OsLBD37/39-OsTCP19-DLT pathway is a key regulatory cascade governing N response and tillering in rice (Liu et al., 2021). Furthermore, the transcription factor OsGATA8 was identified as a critical regulator of N uptake and tiller formation in rice. OsGATA8 negatively regulates N absorption by repressing OsAMT3.2 transcription, while promoting tiller formation by inhibiting the transcription of the negative tillering regulator OsTCP19. The OsGATA8H haplotype displays high NUE, with enhanced N uptake and a higher proportion of productive tillers (Wu et al., 2024) (Figure 2; Table 1).
4.3 NUE-associated genes identified by mutant identification
In recent years, several NUE-related genes have been cloned from mutant identification, such as ARE1, ARE4, OsELF3-1, and OsDOF18. ARE1 is a negative regulator of N assimilation, encoding a chloroplast-localized protein, and is transcriptionally inhibited by Ghd7. Loss-of-function mutations in ARE1 cause delayed senescence and grain yield increases, hence enhance NUE under LN conditions (Wang et al., 2018b). OsELF3–1 forms a ternary complex (OsEC) with OsELF4s and OsLUX, repressing the expression of Ghd7, which in turn directly inhibits ARE1 expression and promotes N absorption (Wang et al., 2021; Tsednee, 2024; Sun et al., 2024). ARE4, a MYB-related transcription factor, coordinates glucose signaling with NUE in rice. It is kept in the cytosol by interacting with the glucose sensor OsHXK7. Upon sensing a glucose signal, ARE4 is released, translocated into the nucleus, and activates the expression of a group of high-affinity NO3- transporter genes, resulting in increased NO3- uptake and accumulation (Ma et al., 2023). In the osdof18 mutant, the expression of OsAMT1.1, OsAMT1.3, OsAMT2.1, and OsAMT4.1 is reduced, indicating that these NH4+ transporter genes function downstream of the transcription factor OsDOF18. The findings demonstrate that OsDOF18 mediates NH4+ transport and N allocation, thereby influencing NUE (Wu et al., 2017) (Figure 2; Table 1).
4.4 Regulation of cellular pH homeostasis enhances NUE in rice
Excessive absorption of NH4+ by plants leads to cellular acidification, while excessive NO3- uptake leads to cellular alkalization. Therefore, excessive uptake of a single N source affects the pH balance in plant cells, causing enzyme dysfunction and ultimately impacting crop growth and yield. In a recent study, overexpression of the N transport gene OsNRT2.3b helps counteract pH changes in rice plants, thus improving NUE and rice yield (Fan et al., 2016b). Plasma membrane (PM) H+-ATPase facilitates the transport of various nutrients, such as NO3-, phosphate (Pi), and K, and maintains cytosolic H+ homeostasis by pumping H+ outside the cells. In previous studies, overexpression of Oryza sativa PM H+-ATPase 1 (OSA1) in rice enhances NH4+ uptake and assimilation, leading to increased grain yield and NUE (Zhang et al., 2021a). The assimilation of NH4+ in root cells requires a C skeleton as the substrate for the synthesis of amino acids through the GS/GOGAT cycle. The assimilation of one molecule of NH4+ generates two molecules of H+ in the cytoplasm. This inhibits the growth and development of plant roots and reduces NUE (Jia et al., 2020; Hachiya et al., 2021). A recent study identified a mutant that exhibited root hypersensitivity to NH4+ due to a missense mutation in the gene encoding argininosuccinate lyase (ASL), which localizes to plastids and mitigates NH4+-induced inhibition of root elongation by converting excess glutamine into arginine. Natural variations in ASL alleles between the japonica and indica subspecies of rice demonstrate ASL expression is positively correlated with NUE and yield (Xie et al., 2023). These results suggest that the H+ produced during the mitigation of NH4+ assimilation can improve NUE in rice (Figure 2; Table 1).
4.5 Other key regulatory genes involved in NUE in rice
The transcription factor OsDREB1C is identified through RNA-seq as co-induced by light and LN supply, directly targets OsNR2, OsNRT2.4, and OsNRT1.1B, and simultaneously enhances the efficiency of photosynthesis and NUE, significantly improving rice yield (Wei et al., 2022). The bZIP transcription factor OsRF2b, identified through biochemical screening, interacts with OsbZIP61 to form heterodimers. This complex directly binds to the OsNRT1.1B promoter region and represses its expression, acting as a negative regulator of NUE and grain yield (Li et al., 2025). OsNR1.2 encodes an NADH-dependent NO3- reductase, essential for achieving high NUE in rice (Han et al., 2022). Furthermore, a batch of key genes involved in NUE has also been identified, such as rice OsBT1, OsBT2, and AAP genes (Araus et al., 2016; Lu et al., 2018; Pereira et al., 2022), crop yield and NUE can be improved by changing the expression levels of these genes (Figure 2; Table 1).
4.6 Dissecting NUE pathways in green revolution varieties
Apart from using the methods mentioned above to identify N-efficient genes, analyzing the mechanism of low NUE in ‘Green Revolution’ varieties (GRVs) that limit efficient N use, and mining N-efficient genes from such varieties have also proven effective. The ‘Green Revolution’ gene sd1, which encodes the GA20 oxidase 2 (GA20ox2) enzyme, an important synthetic enzyme in the gibberellin (GA) synthesis pathway, is widely used in indica breeding. The mutated type of sd1 causes a decrease in endogenous GA activity and GA signal suppressor DELLAs protein (SLR1) accumulation in rice, which leads to a reduction in rice plant height (Ashikari et al., 2002).
A rice transcription factor GRF4 interacts with the transcriptional activator GIF1 to promote the expression of N transport and assimilation-related genes (such as OsAMT1.1, OsGS1.2, OsNRT1.1B) (Li et al., 2018). SLR1 competitively represses the GRF4-GIF1 interaction, inhibiting the formation of the GRF4-GIF1 protein complex, which leads to a reduction in NUE of rice. Introduction of the excellent allele gene GRF4ngr2 into semi-dwarf and high-yielding rice varieties can achieve a coordinated increase in the yield and NUE of rice without changing their plant height (Li et al., 2018). The key repressor DWARF 53 (D53) of the SL signalling interacts with GRF4 and prevents GRF4 from binding to its target gene promoters, and negatively regulates NUE (Sun et al., 2023).
Subsequently, NGR5 is a new target of GA-GIBBERELLIN-INSENSITIVE DWARF1 (GID1)-mediated proteasomal destruction, and SLR1 competes with NGR5 for interaction with GID1, in the case of NGR5, with stabilized SLR1 of rice GRVs promoting stabilization of NGR5, thus explaining why GRVs exhibit increased tillering. N status affects chromatin function through modification of histones, a process in which the transcription factor NGR5 recruits polycomb repressive complex 2 (PRC2) to inhibit tiller genes, including OsD14 and OsSPL14, through repressive H3K27me3 modifications (Wu et al., 2020). Additionally, SLR1 competes with NGR5 for interaction with GID1. In the case of NGR5 with stabilized SLR1 of rice (GRVs), promoting the stabilization of NGR5 leads to increased tillering, explaining why GRVs exhibit enhanced tillering. Furthermore, pyramiding of sd1 elite NGR5 alleles can enhance NUE, leading to reduced N fertilizer usage and increased grain yield, without affecting the beneficial semi-dwarfism (Wu et al., 2020) (Figure 2; Table 1). This suggests that manipulation of plant development and NUE co-modulation would drive modern breeding for sustainable food security.
5 Conclusion and future perspectives
5.1 Identification of key NO3- and NH4+ transporters enhancing NUE in rice
Over the past two decades, one of the most significant advances in understanding N utilization regulation in rice has been the identification of NO3- and NH4+ transporters, as well as transcription factors involved in NUE. In rice, OsNRT1, OsNRT1.1B, OsNRT1.6, OsNRT2.1, OsNRT2.2 and OsNRT2.4 are responsible for NO3- uptake. OsAMT1.1, OsAMT1.2, OsAMT1.3, OsAMT2.1, OsAMT2.3, OsAMT3.1 and OsAMT5.1 are principal for NH4+ uptake. OsNRT1.1B, OsNRT1.6, OsNPF2.2, OsNRT2.3a and OsNAR2.2 are crucial for NO3- translocation to shoots. In addition, among the currently identified members of the rice NRT/NPF family, OsNRT1.1A, OsNRT1.1B, OsNRT2.1, OsNAR2.1, OsNRT2.3a, OsNRT2.3b, OsNPF3.1, OsNPF4.5, OsNPF6.1, OsNPF7.7, and OsNPF8.20 have all been shown to enhance NUE, whereas OsNPF7.3 decreased NUE at high NH4+ supply. Compared to the substantial advances made in NO3- transporters, the progress achieved with NH4+ transporter proteins in improving NUE has remained relatively limited. In the AMT family, only OsAMT1.1 and OsAMT1.2 have been identified as capable of improving NUE (Figure 1; Table 1).
5.2 Genetic and molecular strategies for enhancing NUE in rice
Significant genetic variation in NUE exists within rice germplasm resources, providing a valuable foundation for the precise breeding of cultivars with enhanced NUE. During rice domestication, natural variations in several key loci genes have been identified (including OsNRT1.1B, OsNR2, DNR1, OsWRKY23, and MYB61), playing important roles in regulating NUE and contributing to yield differences between indica and japonica cultivars. In the future, it remains essential to further dissect the candidate genes responsible for NUE variation among rice germplasm resources. GWAS has already cloned multiple key NUE-regulating genes, such as OsNPF6.1, OsNLP4, OsSTP28, OsTCP19, and OsGATA8, which enhance N uptake, assimilation, and tillering ability in rice, thereby improving both NUE and grain yield under varying N conditions. These findings have provided important genetic resources for the molecular breeding of rice cultivars with improved NUE. Moreover, maintaining cellular pH homeostasis is crucial for achieving high NUE in rice. Genes such as OsNRT2.3b, OSA1, and ASL play pivotal roles in regulating proton flux, nutrient transport, and N assimilation, ultimately enhancing NUE and grain yield under N stress conditions. Moreover, introducing advantageous alleles such as GRF4ngr2 or NGR5 into the genetic background of the ‘Green Revolution’ sd1 can simultaneously enhance NUE and grain yield while maintaining a desirable dwarf plant architecture. The synergistic effect of sd1-GRF4-NGR5 enables the coordinated improvement of both NUE and yield, representing an ideal strategy for future molecular breeding (Figure 2; Table 1).
5.3 Multi-gene co-regulation strategies for enhancing NUE in rice
Despite the successive identification and characterization of key genes involved in N uptake, transport, and utilization in rice, the effect of a single gene on improving N absorption or NUE remains limited. By adopting a multi−gene co−regulation strategy, rice NUE and yield can be further enhanced. Studies have shown that co−overexpression of genes for N transport, uptake, and assimilation increases both rice yield and NUE. For instance, the OsNPF8.9a×OsNR2, OsAMT1.2×OsGS1.2×OsAS1, and OsGS2×OsAS2×OsANT3 combinations respectively optimize NO3- uptake, NH4+ conversion, and N recycling. Notably, combining OsAMT1.2, OsGS1.2, and OsAS1 overexpression represents a promising breeding strategy (Luo et al., 2023).
Looking forward, the success of such multi-gene strategies will benefit greatly from the integration of advanced biotechnological tools. CRISPR/Cas-based multiplex genome editing allows for precise and simultaneous modification of multiple target genes, while transgenic stacking enables coordinated expression of gene cassettes. When combined with high-throughput phenotyping and omics-assisted selection, these approaches provide a robust framework for the rational design of rice cultivars with enhanced NUE and yield potential. This systems-level breeding strategy offers an effective path toward reducing N fertilizer input while maintaining high productivity, contributing to more sustainable and environmentally friendly rice production.
5.4 Integration of NUE regulation with environmental stress responses
In the context of global climate change and increasingly variable field conditions, the regulation of NUE in rice must be understood not only under optimal environments but also under abiotic stress conditions. Recent studies have demonstrated that N uptake and assimilation are not only genetically regulated but also highly responsive to environmental cues. Under abiotic stresses such as drought, salinity, and extreme temperatures, N transporter expression and NR activity or other enzymes involved in N metabolism are frequently suppressed, leading to reduced NUE (Henckel, 1964; Plaut, 1974; Guo et al., 2003; Robredo et al., 2011; Goel and Singh, 2015; Han et al., 2015; Meng et al., 2016).
However, the application of key genes has been shown to be significant potential for improving NUE under stress conditions. For instance, DST (Drought and Salt Tolerance)-OsNR1.2 regulatory module has been shown to be involved in the suppression of NO3- assimilation under drought tolerance. Given that DST negatively regulates stomatal closure while positively regulating N assimilation, it likely mediates a coupling between N metabolism and stomatal movement. This mechanism offers a promising target for developing drought-tolerant crops with improved NUE (Han et al., 2022). OsDREB1C, a member of the AP2/EREBP transcription factor family, was initially identified for its role in cold stress responses in rice, recent studies have demonstrated that overexpression of OsDREB1C shortens the growth duration, enhances NUE, and promotes more effective resource allocation, suggesting a potential regulatory link between stress response pathways and nutrient efficiency (Mao and Chen, 2012; Wei et al., 2022).
Moreover, agronomic practices such as irrigation regimes, fertilization strategies, and soil amendments can influence the expression and function of key N-related genes, thereby affecting NUE in crops (Shoji et al., 2001; Yang et al., 2015; Sajjad et al., 2024). Furthermore, in paddy fields, NO3- and NH4+ availability fluctuates significantly in time and space, necessitating root responses to diverse and changing environmental cues. It has been found that NO3- supply enhances NH4+ uptake in rice (Zhao et al., 2008). Therefore, a deeper understanding of the interaction between NO3- and NH4+, and their roles in physiological and biochemical regulation of N uptake, is crucial for improving NUE. In summary, a systematic understanding of the dynamic interactions between genetic regulatory networks and management practices will provide a theoretical foundation and practical guidance for developing N management strategies that are both high-yielding and environmentally sustainable under variable environmental conditions.
5.5 Integrative strategies for future NUE improvement
In summary, we have outlined the genetic regulatory factors involved in the transport of NO3- and NH4+, which contribute to efficient N absorption and translocation. We further discussed the key genes regulating NUE in rice, highlighting their potential to significantly improve both crop yield and NUE. Although substantial progress has been made in understanding the genetic architecture and molecular mechanisms underlying NUE in rice, there remain significant gaps in our knowledge of the complex genetic networks governing NUE regulation. This calls for an in-depth exploration of the genes and regulatory elements affecting NUE through advanced genomic technologies and bioinformatics tools. Future studies should focus on elucidating the functions of these genes, their interactions, and their responses to nitrogen availability under varying environmental conditions. Ultimately, by leveraging strategies such as multi-gene pyramiding, in-depth analysis of signaling regulatory networks, and the mining of elite genetic resources, it is expected that rice NUE can be further improved, facilitating the development of high-yield, environmentally sustainable rice varieties.
Author contributions
XG: Data curation, Project administration, Validation, Resources, Visualization, Formal analysis, Software, Writing – review & editing, Investigation, Supervision, Writing – original draft, Conceptualization. JZ: Writing – review & editing, Funding acquisition, Investigation. FM: Writing – review & editing, Investigation. PL: Writing – review & editing, Investigation. YM: Writing – review & editing. KX: Supervision, Writing – review & editing. TL: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was funded by the Department of Science and Technology Planning Project of Henan Province (252102111147 and 252102110328).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Araki, R. and Hasegawa, H. (2006). Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breed. Sci. 56, 295–302. doi: 10.1270/jsbbs.56.295
Araus, V., Vidal, E. A., Puelma, T., Alamos, S., Mieulet, D., Guiderdoni, E., et al. (2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171, 1523–1532. doi: 10.1104/pp.15.01731
Ashikari, M., Sasaki, A., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Datta, S., et al. (2002). Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘green revolution’. Breed. Sci. 52, 143–150. doi: 10.1270/jsbbs.52.143
Bao, A., Liang, Z., Zhao, Z., and Cai, H. (2015). Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Int. J. Mol. Sci. 16, 9037–9063. doi: 10.3390/ijms16059037
Bu, Y., Takano, T., Nemoto, K., and Liu, S. (2011). Research progress of ammonium transporter in rice plants. G. A. B. 2, 373–379. doi: 10.5376/gab.2011.02.0003
Cai, C., Wang, J. Y., Zhu, Y. G., Shen, Q. R., Li, B., Tong, Y. P., et al. (2008). Gene structure and expression of the high-affinity nitrate transport system in rice roots. J. Integr. Plant Biol. 50, 443–451. doi: 10.1111/j.1744-7909.2008.00642.x
Chen, J., Fan, X., Qian, K., Zhang, Y., Song, M., Liu, Y., et al. (2017). pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 15, 1273–1283. doi: 10.1111/pbi.12714
Chen, J., Liu, X., Liu, S., Fan, X., Zhao, L., Song, M., et al. (2020). Co-overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01245
Chen, J., Zhang, Y., Tan, Y., Zhang, M., Zhu, L., Xu, G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14, 1705–1715. doi: 10.1111/pbi.12531
Crawford, N. M. (1995). Nitrate: nutrient and signal for plant growth. Plant Cell. 7, 859–868. doi: 10.1105/tpc.7.7.859
Crawford, N. M. and Glass, A. D. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
Deng, R. L., Gu, J. T., Lu, W. J., Xu, H. R., and Xiao, K. (2007). Characterization, function and expression analysis of ammonium transporter gene OsAMT1;4 and OsAMT5 in rice (Oryza sativa). Sci. Agric. Sin. 40, 2395–2402. Available online at: https://api.semanticscholar.org/CorpusID:87104589 (Accessed July 11, 2025).
Ding, Z., Wang, C., Chen, S., and Yu, S. (2011). Diversity and selective sweep in the OsAMT1;1 genomic region of rice. BMC Evol. Biol. 11, 61. doi: 10.1186/1471-2148-11-61
Fan, X., Feng, H., Tan, Y., Xu, Y., Miao, Q., and Xu, G. (2016a). A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 58, 590–599. doi: 10.1111/jipb.12382
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016b). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. 113, 7118–7123. doi: 10.1073/pnas.1525184113
Fan, X., Xie, D., Chen, J., Lu, H., Xu, Y., Ma, C., et al. (2014). Over-expression of OsPTR6 in rice increased plant growth at different nitrogen supplies but decreased nitrogen use efficiency at high ammonium supply. Plant Sci. 227, 1–11. doi: 10.1016/j.plantsci.2014.05.013
Fang, Z., Bai, G., Huang, W., Wang, Z., Wang, X., and Zhang, M. (2017). The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01338
Fang, Z., Xia, K., Yang, X., Grotemeyer, M. S., Meier, S., Rentsch, D., et al. (2013). Altered expression of the PTR/NRT1 homologue OsPTR9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol. J. 11, 446–458. doi: 10.1111/pbi.12031
Feng, H., Li, B., Zhi, Y., Chen, J., Li, R., Xia, X., et al. (2017). Overexpression of the nitrate transporter, OsNRT2.3b, improves rice phosphorus uptake and translocation. Plant Cell Rep. 36, 1287–1296. doi: 10.1007/s00299-017-2153-9
Feng, H., Yan, M., Fan, X., Li, B., Shen, Q., Miller, A. J., et al. (2011). Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 62, 2319–2332. doi: 10.1007/s00299-017-2153-9
Forde, B. G. (2000). Nitrate transporters in plants: structure, function and regulation. Biochim. Biophys. Acta 1465, 219–235. doi: 10.1016/s0005-2736(00)00140-1
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10, 5207. doi: 10.1038/s41467-019-13110-8
Gao, Y., Xu, Z., Zhang, L., Li, S., Wang, S., Yang, H., et al. (2020). MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat. Commun. 11, 5219. doi: 10.1038/s41467-020-19019-x
Gaur, V. S., Singh, U. S., Gupta, A. K., and Kumar, A. (2012). Understanding the differential nitrogen sensing mechanism in rice genotypes through expression analysis of high and low affinity ammonium transporter genes. Mol. Biol. Rep. 39, 2233–2241. doi: 10.1007/s11033-011-0972-2
Glass, A. D., Shaff, J. E., and Kochian, L. V. (1992). Studies of the uptake of nitrate in barley: IV. Electrophysiology. Plant Physiol. 99, 456–463. doi: 10.1104/pp.99.2.456
Goel, P. and Singh, A. K. (2015). Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea L. PLoS One 10, e0143645. doi: 10.1371/journal.pone.0143645
Guo, F. Q., Young, J., and Crawford, N. M. (2003). The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell. 15, 107–117. doi: 10.1105/tpc.006312
Hachiya, T., Inaba, J., Wakazaki, M., Sato, M., Toyooka, K., Miyagi, A., et al. (2021). Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in. Arabidopsis thaliana. Nat. Commun. 12, 4944. doi: 10.1038/s41467-021-25238-7
Han, M., Lv, Q., Zhang, J., Wang, T., Zhang, C., Tan, R., et al. (2022). Decreasing nitrogen assimilation under drought stress by suppressing DST-mediated activation of Nitrate Reductase 1.2 in rice. Mol. Plant 15, 167–178. doi: 10.1016/j.molp.2021.09.005
Han, M., Okamoto, M., Beatty, P. H., Rothstein, S. J., and Good, A. G. (2015). The genetics of nitrogen use efficiency in crop plants. Annu. Rev. Genet. 49, 269–289. doi: 10.1146/annurev-genet-112414-055037
Henckel, P. A. (1964). Physiology of plants under drought. Annu. Rev. Plant Physiol. 15, 363–386. doi: 10.1146/annurev.pp.15.060164.002051
Hou, L., Chen, D., Pan, X., Jiang, S., Liu, J., Li, Q., et al. (2025). 9311 allele of OsNAR2.2 enhances nitrate transport to improve rice yield and nitrogen use efficiency. Plant Biotechnol. J. 23, 1–11. doi: 10.1111/pbi.70073
Hu, T., Cao, K., Xia, M., and Wang, X. (2006). Functional characterization of a putative nitrate transporter gene promoter from rice. Acta. Biochim. Biophys. Sin. 38, 795–802. doi: 10.1111/j.1745-7270.2006.00225.x
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat.Plants. 5, 637. doi: 10.1038/s41477-019-0384-1
Hu, R., Qiu, D., Chen, Y., Miller, A. J., Fan, X., Pan, X., et al. (2016). Knock-down of a tonoplast localized low-affinity nitrate transporter OsNPF7.2 affects rice growth under high nitrate supply. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01529
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Huang, W., Bai, G., Wang, J., Zhu, W., Zeng, Q., Lu, K., et al. (2018). Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00300
Huang, Y., Ji, Z., Tao, Y., Wei, S., Jiao, W., Fang, Y., et al. (2023). Improving rice nitrogen-use efficiency by modulating a novel monouniquitination machinery for optimal root plasticity response to nitrogen. Nat. Plants. 9, 1902–1914. doi: 10.1038/s41477-023-01533-7
Huang, S., Liang, Z., Chen, S., Sun, H., Fan, X., Wang, C., et al. (2019a). A transcription factor, OsMADS57, regulates long-distance nitrate transport and root elongation. Plant Physiol. 180, 882–895. doi: 10.1104/pp.19.00142
Huang, W., Nie, H., Feng, F., Wang, J., Lu, K., and Fang, Z. (2019b). Altered expression of OsNPF7.1 and OsNPF7.4 differentially regulates tillering and grain yield in rice. Plant Sci. 283, 23–31. doi: 10.1016/j.plantsci.2019.01.019
Jia, L., Xie, Y., Wang, Z., Luo, L., Zhang, C., Pélissier, P. M., et al. (2020). Rice plants respond to ammonium stress by adopting a helical root growth pattern. Plant J. 104, 1023–1037. doi: 10.1111/tpj.14978
Kirk, G. J. and Kronzucker, H. J. (2005). The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: a modelling study. Ann. Bot. 96, 639–646. doi: 10.1093/aob/mci216
Konishi, N. and Ma, J. F. (2021). Three polarly localized ammonium transporter 1 members are cooperatively responsible for ammonium uptake in rice under low ammonium condition. New Phytol. 232, 1778–1792. doi: 10.1111/nph.17679
Lee, S., Marmagne, A., Park, J., Fabien, C., Yim, Y., Kim, S. J., et al. (2020). Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 103, 7–20. doi: 10.1111/tpj.14794
Léran, S., Varala, K., Boyer, J., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Li, Y. L., Fan, X. R., and Shen, Q. R. (2008). The relationship between rhizosphere nitrification and nitrogen-use efficiency in rice plants. Plant Cell Environ. 31, 73–85. doi: 10.1111/j.1365-3040.2007.01737.x
Li, S. M., Li, B. Z., and Shi, W. M. (2012). Expression patterns of nine ammonium transporters in rice in response to N status. Pedosphere. 22, 860–869. doi: 10.1016/S1002-0160(12)60072-1
Li, B., Merrick, M., Li, S., Li, H., Zhu, S., Shi, W., et al. (2009a). Molecular basis and regulation of ammonium transporter in rice. Rice Sci. 16, 314–322. doi: 10.1016/S1672-6308(08)60096-7
Li, Y., Ouyang, J., Wang, Y., Hu, R., Xia, K., Duan, J., et al. (2015). Disruption of the rice nitrate transporter OsNPF2.2 hinders root-to-shoot nitrate transport and vascular development. Sci. Rep. 5, 9635. doi: 10.1038/srep09635
Li, S., Qian, Q., Fu, Z., Zeng, D., Meng, X., Kyozuka, J., et al. (2009b). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58, 592–605. doi: 10.1111/j.1365-313X.2009.03799.x
Li, S. M. and Shi, W. M. (2006). Quantitative characterization of nitrogen regulation of OsAMT1;1, OsAMT1;2, and OsAMT2;2 expression in rice seedlings. Russian J. Plant Physiol. 53, 837–843. doi: 10.1134/S102144370606015X
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Li, C., Wen, H., Wu, Y., Li, Y., Feng, X., Li, X., et al. (2025). OsRF2b interacting with OsbZIP61 modulates nitrogen use efficiency and grain yield via heterodimers in rice. Plant Biotechnol. J. 23, 3300–3312. doi: 10.1111/pbi.70136
Lian, X., Xing, Y., Yan, H., Xu, C., Li, X., and Zhang, Q. (2005). QTLs for low nitrogen tolerance at seedling stage identified using a recombinant inbred line population derived from an elite rice hybrid. Theor. Appl. Genet. 112, 85–96. doi: 10.1007/s00122-005-0108-y
Lin, C. M., Koh, S., Stacey, G., Yu, S. M., Lin, T. Y., and Tsay, Y. F. (2000). Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol. 122, 379–388. doi: 10.1104/pp.122.2.379
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Lu, K., Wu, B., Wang, J., Zhu, W., Nie, H., Qian, J., et al. (2018). Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechno. J. 16, 1710–1722. doi: 10.1111/pbi.12907
Luo, J., Hang, J., Wu, B., Wei, X., Zhao, Q., and Fang, Z. (2023). Co-overexpression of genes for nitrogen transport, assimilation, and utilization boosts rice grain yield and nitrogen use efficiency. Crop J. 11, 785–799. doi: 10.1016/j.cj.2023.01.005
Ma, X., Nian, J., Yu, H., Zhang, F., Feng, T., Kou, L., et al. (2023). Linking glucose signaling to nitrogen utilization by the OsHXK7-ARE4 complex in rice. Dev. Cell 58, 1489–1501. e5. doi: 10.1016/j.devcel.2023.06.003
Mao, D. and Chen, C. (2012). Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS One 7, e47275. doi: 10.1371/journal.pone.0047275
Meng, S., Zhang, C., Su, L., Li, Y., and Zhao, Z. (2016). Nitrogen uptake and metabolism of Populus simonii in response to PEG-induced drought stress. Environ. Exp. Bot. 123, 78–87. doi: 10.1016/j.envexpbot.2015.11.005
Miller, A. J., Fan, X., Orsel, M., Smith, S. J., and Wells, D. M. (2007). Nitrate transport and signalling. J. Exp. Bot. 58, 2297–2306. doi: 10.1093/jxb/erm066
Obara, M., Tamura, W., Ebitani, T., Yano, M., Sato, T., and Yamaya, T. (2010). Fine-mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4+ concentrations in hydroponic conditions. Theor. Appl. Genet. 121, 535–547. doi: 10.1007/s00122-010-1328-3
Pereira, E. G., Bucher, C. P. C., Bucher, C. A., Santos, L. A., Lerin, J., Catarina, C. S., et al. (2022). The amino acid transporter OsAAP1 regulates the fertility of spikelets and the efficient use of N in rice. Plant Soil 480, 507–521. doi: 10.1007/s11104-022-05598-9
Plaut, Z. (1974). Nitrate reductase-activity of wheat seedlings during exposure to and recovery from water stress and salinity. Physiol. Plant. 30, 212–217. doi: 10.1111/j.1399-3054.1974.tb03646.x
Qiu, D., Hu, R., Li, J., Li, Y., Ding, J., Xia, K., et al. (2023). Peptide transporter OsNPF8.1 contributes to sustainable growth under salt and drought stresses, and grain yield under nitrogen deficiency in rice. Rice Sci. 30, 113–126. doi: 10.1016/j.rsci.2023.01.004
Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Robredo, A., Pérez-López, U., Miranda-Apodaca, J., Lacuesta, M., Mena-Petite, A., and Muñoz-Rueda, A. (2011). Elevated CO2 reduces the drought effect on nitrogen metabolism in barley plants during drought and subsequent recovery. Environ. Exp. Bot. 71, 399–408. doi: 10.1016/j.envexpbot.2011.02.011
Sajjad, M., Hussain, K., Wajid, S. A., and Saqib, Z. A. (2024). The impact of split nitrogen fertilizer applications on the productivity and nitrogen use efficiency of rice. Nitrogen 6, 1. doi: 10.3390/nitrogen6010001
Sasakawa, H. and Yamamoto, Y. (1978). Comparison of the uptake of nitrate and ammonium by rice seedlings: influences of light, temperature, oxygen concentration, exogenous sucrose, and metabolic inhibitors. Plant Physiol. 62, 665–669. doi: 10.1104/pp.62.4.665
Shoji, S., Delgado, J., Mosier, A., and Miura, Y. (2001). Use of controlled release fertilizers and nitrification inhibitors to increase nitrogen use efficiency and to conserve air andwater quality. Commun. Soil Sci. Plan 32, 1051–1070. doi: 10.1081/CSS-100104103
Sonoda, Y., Ikeda, A., Saiki, S., Wirén, N. V., Yamaya, T., and Yamaguchi, J. (2003a). Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in Rice. Plant Cell Physiol. 44, 726–734. doi: 10.1093/pcp/pcg083
Sonoda, Y., Ikeda, A., Saiki, S., Yamaya, T., and Yamaguchi, J. (2003b). Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant Cell Physiol. 44, 1396–1402. doi: 10.1093/pcp/pcg169
Stitt, M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2, 178–186. doi: 10.1016/S1369-5266(99)80033-8
Suenaga, A., Moriya, K., Sonoda, Y., Ikeda, A., Von Wirén, N., Hayakawa, T., et al. (2003). Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 44, 206–211. doi: 10.1093/pcp/pcg017
Sun, H., Guo, X., Zhu, X., Gu, P., Zhang, W., Tao, W., et al. (2023). Strigolactone and gibberellin signaling coordinately regulate metabolic adaptations to changes in nitrogen availability in rice. Mol. Plant 16, 588–598. doi: 10.1016/j.molp.2023.01.009
Sun, Q., Yu, Z., Wang, X., Chen, H., Lu, J., Zhao, C., et al. (2024). EARLY FLOWERING3–1 represses Grain number, plant height, and heading date7 to promote ABC1 REPRESSOR1 and regulate nitrogen uptake in rice. Plant Physiol. 196, 1857–1868. doi: 10.1093/plphys/kiae416
Tang, Z., Fan, X., Li, Q., Feng, H., Miller, A. J., Shen, Q., et al. (2012). Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 160, 2052–2063. doi: 10.1104/pp.112.204461
Tang, W., Ye, J., Yao, X., Zhao, P., Xuan, W., Tian, Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10, 5279. doi: 10.1038/s41467-019-13187-1
Tsednee, M. (2024). Linking timing to nitrogen use efficiency: Rice OsEC-Ghd7-ARE1 module works on it. Plant Physiol. 196, 1720–1721. doi: 10.1093/plphys/kiae488
Wang, S., Chen, A., Xie, K., Yang, X., Luo, Z., Chen, J., et al. (2020a). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. 117, 16649–16659. doi: 10.1073/pnas.2000926117
Wang, W., Hu, B., Li, A., and Chu, C. (2020b). NRT1.1s in plants: functions beyond nitrate transport. J. Exp. Bot. 71, 4373–4379. doi: 10.1093/jxb/erz554
Wang, W., Hu, B., Yuan, D., Liu, Y., Che, R., Hu, Y., et al. (2018c). Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30, 638–651. doi: 10.1105/tpc.17.00809
Wang, J., Lu, K., Nie, H., Zeng, Q., Wu, B., Qian, J., et al. (2018a). Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice 11, 12. doi: 10.1186/s12284-018-0205-6
Wang, Q., Nian, J., Xie, X., Yu, H., Zhang, J., Bai, J., et al. (2018b). Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat. Commun. 9, 735. doi: 10.1038/s41467-017-02781-w
Wang, Q., Su, Q., Nian, J., Zhang, J., Guo, M., Dong, G., et al. (2021). The Ghd7 transcription factor represses ARE1 expression to enhance nitrogen utilization and grain yield in rice. Mol. Plant 14, 1012–1023. doi: 10.1016/j.molp.2021.04.012
Wang, J., Wan, R., Nie, H., Xue, S., and Fang, Z. (2022). OsNPF5.16, a nitrate transporter gene with natural variation, is essential for rice growth and yield. Crop J. 10, 397–406. doi: 10.1016/j.cj.2021.08.005
Wei, S., Li, X., Lu, Z., Zhang, H., Ye, X., Zhou, Y., et al. (2022). A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 377, eabi8455. doi: 10.1126/science.abi8455
Wei, J., Zheng, Y., Feng, H., Qu, H., Fan, X., Yamaji, N., et al. (2018). OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. J. Exp. Bot. 69, 1095–1107. doi: 10.1093/jxb/erx486
Wu, W., Dong, X., Chen, G., Lin, Z., Chi, W., Tang, W., et al. (2024). The elite haplotype OsGATA8-H coordinates nitrogen uptake and productive tiller formation in rice. Nat. Genet. 56, 1516–1526. doi: 10.1038/s41588-024-01795-7
Wu, K., Wang, S., Song, W., Zhang, J., Wang, Y., Liu, Q., et al. (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367, eaaz2046. doi: 10.1126/science.aaz2046
Wu, Y., Yang, W., Wei, J., Yoon, H., and An, G. (2017). Transcription factor OsDOF18 controls ammonium uptake by inducing ammonium transporters in rice roots. Mol. Cells 40, 178–185. doi: 10.14348/molcells.2017.2261
Wu, J., Zhang, Z. S., Xia, J. Q., Alfatih, A., Song, Y., Huang, Y. J., et al. (2021). Rice NIN-LIKE PROTEIN 4 plays a pivotal role in nitrogen use efficiency. Plant Biotechnol. J. 19, 448–461. doi: 10.1111/pbi.13475
Xia, X., Fan, X., Wei, J., Feng, H., Qu, H., Xie, D., et al. (2015). Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 66, 317–331. doi: 10.1093/jxb/eru425
Xie, Y., Lv, Y., Jia, L., Zheng, L., Li, Y., Zhu, M., et al. (2023). Plastid-localized amino acid metabolism coordinates rice ammonium tolerance and nitrogen use efficiency. Nat. Plants 9, 1514–1529. doi: 10.1038/s41477-023-01494-x
Xu, G., Fan, X., and Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Xu, S., Wei, Y., Zhao, P., Sun, Y., Gao, K., Yin, C., et al. (2025). A nitrate transporter OsNPF6.1 promotes nitric oxide signaling and virus resistance. Plant Cell Environ. 48, 6493-6508. doi: 10.1111/pce.15626
Yan, M., Fan, X., Feng, H., Miller, A. J., Shen, Q., and Xu, G. (2011). Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 34, 1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x
Yang, X., Nong, B., Chen, C., Wang, J., Xia, X., Zhang, Z., et al. (2023). OsNPF3.1, a member of the NRT1/PTR family, increases nitrogen use efficiency and biomass production in rice. Crop J. 11, 108–118. doi: 10.1016/j.cj.2022.07.001
Yang, S., Peng, S., Xu, J., He, Y., and Wang, Y. (2015). Effects of water saving irrigation and controlled release nitrogen fertilizer managements on nitrogen losses from paddy fields. Paddy Water Environ. 13, 71–80. doi: 10.1007/s10333-013-0408-9
Yu, J., Xuan, W., Tian, Y., Fan, L., Sun, J., Tang, W., et al. (2021). Enhanced OsNLP4-OsNiR cascade confers nitrogen use efficiency by promoting tiller number in rice. Plant Biotechnol. J. 19, 167–176. doi: 10.1111/pbi.13450
Zhang, S., Huang, Y., Ji, Z., Fang, Y., Tian, Y., Shen, C., et al. (2025a). Discovery of SOD5 as a novel regulator of nitrogen-use efficiency and grain yield via altering auxin level. New Phytol. 246, 1084–1095. doi: 10.1111/nph.70038
Zhang, S., Ji, Z., Jiao, W., Shen, C., Qin, Y., Huang, Y., et al. (2025b). Natural variation of OsWRKY23 drives difference in nitrate use efficiency between indica and japonica rice. Nat. Commun. 16, 1420. doi: 10.1038/s41467-025-56752-7
Zhang, J., Liu, Y., Zhang, N., Hu, B., Jin, T., Xu, H., et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684. doi: 10.1038/s41587-019-0104-4
Zhang, Y., Tateishi-Karimata, H., Endoh, T., Jin, Q., Li, K., Fan, X., et al. (2022). High-temperature adaptation of an OsNRT2.3 allele is thermoregulated by small RNAs. Sci. Adv. 8, eadc9785. doi: 10.1126/sciadv.adc9785
Zhang, M., Wang, Y., Chen, X., Xu, F., Ding, M., Ye, W., et al. (2021a). Plasma membrane H+-ATPase overexpression increases rice yield via simultaneous enhancement of nutrient uptake and photosynthesis. Nat. Commun. 12, 735. doi: 10.1038/s41467-021-20964-4
Zhang, J., Zhang, Y., Chen, J., Xu, M., Guan, X., Wu, C., et al. (2024). Sugar transporter modulates nitrogen-determined tillering and yield formation in rice. Nat. Commun. 15, 9233. doi: 10.1038/s41467-024-53651-1
Zhang, S., Zhu, L., Shen, C., Ji, Z., Zhang, H., Zhang, T., et al. (2021b). Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell 33, 566–580. doi: 10.1093/plcell/koaa037
Zhao, S. P., Ye, X. Z., and Shi, W. M. (2014). Expression of OsAMT1 (1.1-1.3) in rice varieties differing in nitrogen accumulation. Russ. J. Plant Physiol. 61, 707–713. doi: 10.1134/S1021443714040220
Keywords: rice, ammonium, nitrate, transport, nitrogen use efficiency
Citation: Guo X, Zhang J, Ma F, Li P, Ma Y, Xu K and Liu T (2025) Regulatory network of ammonium and nitrate uptake and utilization in rice. Front. Plant Sci. 16:1656041. doi: 10.3389/fpls.2025.1656041
Received: 29 June 2025; Accepted: 24 September 2025;
Published: 20 October 2025.
Edited by:
Dong-Wei Di, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Abdul Waheed, Chinese Academy of Agricultural Sciences, ChinaSheikh Shanawaz Bashir, Jamia Hamdard University, India
Copyright © 2025 Guo, Zhang, Ma, Li, Ma, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kedong Xu, eHVrZDExMDdAMTI2LmNvbQ==; Ti Liu, dGlsaXU0NTQyQDE2My5jb20=
 Xiaoli Guo
Xiaoli Guo Ju Zhang
Ju Zhang Feilong Ma1
Feilong Ma1 Kedong Xu
Kedong Xu