- Department of Cell Biology, Zunyi Medical University, Zunyi, Guizhou, China
Medicinal plants serve as a crucial source of traditional Chinese medicine and have garnered considerable attention due to their unique bioactive compounds and notable pharmacological properties. However, during natural growth, these plants are frequently susceptible to infection by various pathogenic microorganisms, pests and nematodes, leading to reduced yields and inconsistent accumulation of medicinal compounds, thereby significantly limiting their resource development and utilization. WRKY transcription factors (TFs) are central regulators of plant immunity that integrate pathogen-perception signals, coordinate signaling pathways, and transcriptionally control defense-gene expression. This review provides a systematic synthesis of current knowledge on the regulatory mechanisms of WRKY TFs in the immune responses of medicinal plants. Emphasis is placed on their roles in cellular metabolic regulation, activation of Mitogen-Activated Protein Kinase (MAPK) signaling pathways, integration of phytohormone signaling, and the biosynthesis of secondary metabolites. In addition, we highlight that WRKY TFs orchestrate immune responses at multiple levels through epigenetic mechanisms, including DNA methylation and histone modifications. Furthermore, it is proposed that transgenic approaches and Cut-Dip-Budding (CDB)-mediated transformation be integrated with gene editing technologies such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), in conjunction with artificial intelligence (AI)-assisted identification of key regulatory elements. This integrated strategy offers novel insights and theoretical support for establishing efficient immune regulatory networks and breeding disease-resistant medicinal plant varieties.
1 Introduction
Plants are naturally versatile and diverse, serving as essential sources of nutrients, pharmaceuticals, and chemical components (Owusu Adjei et al., 2021). Medicinal plants in particular contain natural compounds of important value in both traditional and modern medicine (Tanvir et al., 2024). They can be classified according to their medicinal parts, therapeutic effects, or main chemical constituents. For example, roots and rhizomes include Panax ginseng C.A.Mey. (Li et al., 2025a); flowers include Lonicera japonica Thunb. (Li et al., 2025b); leaves include Ginkgo biloba L. (Liu et al., 2022); fruits and seeds include Lycium barbarum L. (Shi et al., 2025); and whole herbs include Leonurus japonicus Houtt. (Wei et al., 2023). In terms of therapeutic effects, Artemisia annua L. is a typical antimalarial (Angupale et al., 2024), Curcuma longa L. is widely used for its notable anti-inflammatory effects (Tian et al., 2025), and Astragalus membranaceus Bunge is valued for its immunomodulatory potential (Wang et al., 2022a). These representative species are not only widely used in traditional medicine but also demonstrate significant immunological and therapeutic effects in modern pharmacological studies.
However, during their growth and development, medicinal plants are frequently attacked by viruses, pathogens, pests and nematodes (Han et al., 2025), which seriously affect their quality and medicinal value. Plants have developed sophisticated immune mechanisms in response to pathogen invasion (Yu et al., 2024). The plant immune system consists of two main layers of active defense. The first layer is triggered by the recognition of pathogen-associated molecular patterns (PAMPs) and host-derived damage-associated molecular patterns (DAMPs) by pattern recognition receptors (PRRs), which activate PAMP-triggered immunity (PTI). The second layer involves intracellular receptors called nucleotide-binding leucine-rich repeat receptors (NLRs), which trigger effector-triggered immunity (ETI) (Yuan et al., 2021). Although different receptors initiate PTI and ETI through separate signaling pathways, their downstream immune responses share significant overlap (Yuan et al., 2021). For example, PTI and ETI are closely linked through common signaling pathways like cell wall remodeling (Wan et al., 2021), activation of Mitogen-Activated Protein Kinase (MAPK) cascades, production of reactive oxygen species (ROS), and phytohormone signaling (Yang et al., 2025).
In plant immune response, transcription factors are central to the regulation of immune pathways (Xiang et al., 2025). The WRKY transcription factor (TF) family serves as a central regulator of plant immunity, modulating PTI and ETI responses either positively or negatively, while enhancing disease resistance through the regulation of secondary metabolite accumulation and epigenetic modifications (Chen et al., 2024a). Although the core components of plant immune mechanisms are largely conserved across species, medicinal plants display a unique characteristic: their immune signaling is closely integrated with secondary metabolite production, a feature that not only strengthens disease resistance but also directly affects their medicinal value (Li et al., 2025c; Zhao et al., 2023; Li et al., 2025d).
Currently, however, systematic understanding of WRKY transcription factors (TFs) in medicinal plant immunity remains limited. This study examines their regulatory role in innate immune responses and offers a foundation for enhancing disease resistance in medicinal plants.
2 From structure to function: the role of WRKY TFs in plant defense
WRKY TFs are a widely distributed family of plant-specific transcriptional regulators that recognize W-box sequences (TTGACT/C) in DNA and play crucial roles in diverse physiological processes, including seed germination, root development, stress adaptation, and immune defense (Wang et al., 2024a). In this paper, we specifically focus on their central role in mediating plant responses to pathogen invasion, emphasizing their key position as hubs within the defense regulatory network. WRKY proteins typically contain at least one WRKY domain, approximately 60 amino acids in length, featuring a highly conserved WRKYGQK motif at the N-terminus and a zinc finger motif at the C-terminus, both of which are essential for DNA binding (Zhang et al., 2023a). Based on structural characteristics, the WRKY family has been classified into three distinct groups. Group I contains two WRKY domains, each associated with a C2H2-type zinc finger motif at the C-terminus. Groups II and III possess a single WRKY domain, with C-terminal zinc finger motifs of the C2H2 and C2HC types, respectively (Rushton et al., 2010). These structural features enable WRKY TFs to recognize and bind specifically to W-box elements (TTGACT/C) in the promoters of downstream target genes, thereby precisely regulating gene expression and contributing to various biological processes (Li et al., 2024a), particularly those involved in plant immune responses (Liu et al., 2025).
During plant immune responses, WRKY TFs drive transcriptional reprogramming by recognizing and binding to W-box elements in the promoters of target genes, thereby activating key components of the salicylic acid (SA) signaling pathway, such as NPR1/3, TGA, and PR1, to enhance disease resistance (Li et al., 2024b). Studies have shown that RhWRKY30 directly binds to the W-box in the RhCAD1 promoter, promoting lignin biosynthesis and enhancing resistance to Botrytis cinerea Pers. in Rosa spp (Li et al., 2024c). Similarly, class IIc WRKYs bind to the W-box in the GhMKK2 promoter, thereby increasing Gossypium hirsutum L. resistance to Fusarium oxysporum Schltdl (Wang et al., 2022b). In addition, WRKY TFs often act synergistically with other TFs to regulate immune responses. For example, in Rheum palmatum L., WRKY and MYB factors synergistically activate genes involved in flavonoid biosynthesis, thereby promoting the accumulation of defensive secondary metabolites and enhancing both immune and chemical defenses (Zhou et al., 2022a).
Although WRKYs also participate in plant developmental processes, they establish relatively independent regulatory hubs during immune responses, with certain signaling pathways potentially shared with developmental networks (Liu, et al., 2024). This functional divergence enables the WRKY gene family to integrate multiple signals within complex transcriptional networks, thereby achieving precise reprogramming of immune-related gene expression and maintaining a central role in plant defense. To better illustrate these roles, we summarized the classification of WRKY TFs in medicinal plants and their immune mechanisms (Table 1).
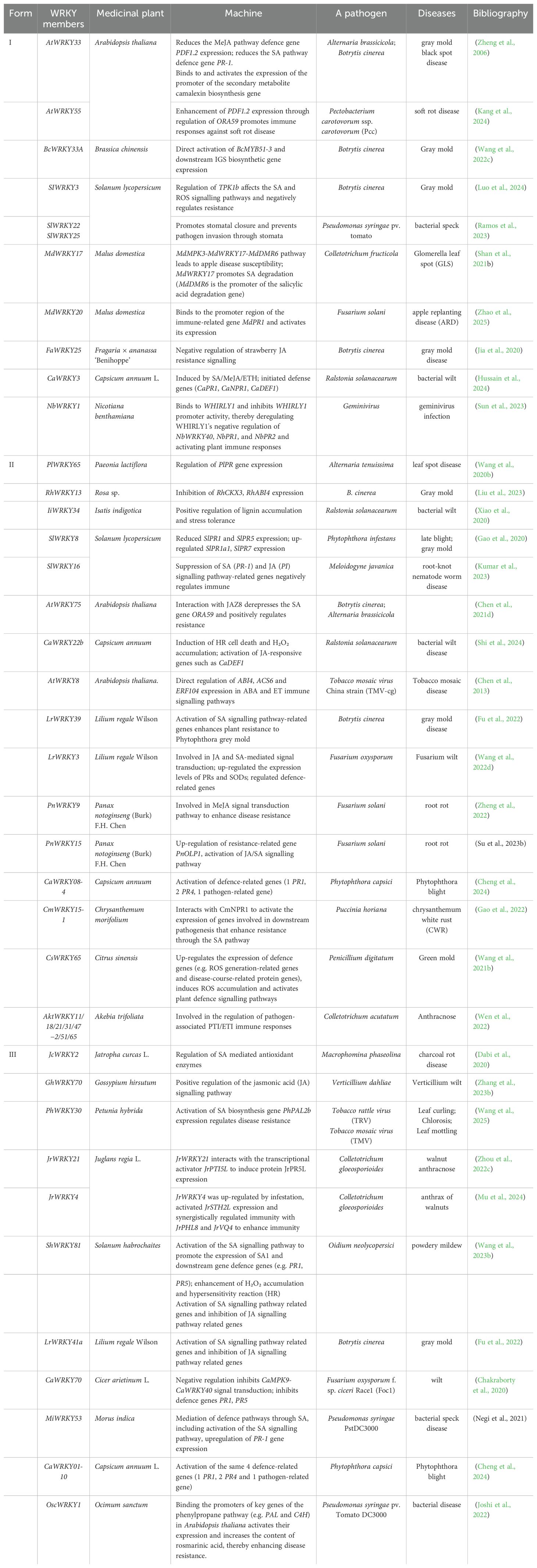
Table 1. Classification of medicinal plant WRKY transcription factors and their mechanism of action in plant immunity.
3 Mechanisms of WRKY-mediated immune responses in medicinal plants
WRKY TFs serve as central hubs in the immunoregulatory networks of medicinal plants, synergistically modulating multiple layers of defense, including structural barrier reinforcement, oxidative stress mitigation, signal transduction, and metabolic defenses in response to pathogen attack (Wang et al., 2024a; Chen et al., 2025a). In Arabidopsis thaliana (L.) Heynh., WRKY research has primarily elucidated their conserved roles in plant immunity (Wang et al., 2024a). By contrast, in medicinal plants such as Panax notoginseng (Burk.) F.H.Chen (Su et al., 2024), Gastrodia elata Bl. f. glauca S. Chow (Wang et al., 2020a), and Salvia miltiorrhiza Bunge (Yu et al., 2025), WRKY factors more prominently mediate the crosstalk between immune signaling networks and secondary metabolic pathways (Li et al., 2025c). Their downstream MAPK cascades and hormone signaling pathways exhibit species-specific responses, thereby tightly coupling defense reactions with the biosynthesis of medicinally active metabolites. This integration represents the defining feature that distinguishes immune research in medicinal plants from studies in other plant systems (Li et al., 2025c, e).
Mechanistically, WRKY TFs upregulate genes involved in lignin biosynthesis, thereby enhancing cell wall-mediated defense. They also modulate antioxidant enzyme systems to alleviate pathogen-induced ROS accumulation and reduce oxidative damage. At the level of signal transduction, WRKY TFs often act synergistically with the MAPK cascade to promote the activation of defense-related genes. In the hormonal signaling network, WRKYs finely regulate immune responses by interacting with key phytohormones, including jasmonic acid (JA), SA, and ethylene (Wang et al., 2024a; Javed and Gao, 2023). For example, PnWRKY9 in Panax notoginseng activates the JA signaling pathway, enhances the expression of the antimicrobial peptide gene PnDEFL1, and increases resistance to Fusarium solani (Zheng et al., 2022). Meanwhile, WRKY TFs have also been shown to directly or indirectly regulate genes involved in the biosynthesis of key secondary metabolites, such as flavonoids, terpenoids, and alkaloids, thereby enhancing metabolic defenses. For instance, EbWRKY30, EbWRKY31, and EbWRKY44 are co-expressed with structural genes involved in flavonoid biosynthesis in Erigeron breviscapus (Vaniot) Hand.-Mazz., leading to enhanced antioxidant capacity and disease resistance (Song et al., 2024d).
Collectively, these studies demonstrate that WRKY factors play a central, multidimensional, and synergistic role in the immune network of medicinal plants, providing novel insights for the molecular breeding of highly resistant medicinal plant varieties. These regulatory mechanisms are further illustrated in the immune signaling network of medicinal plants (Figure 1).
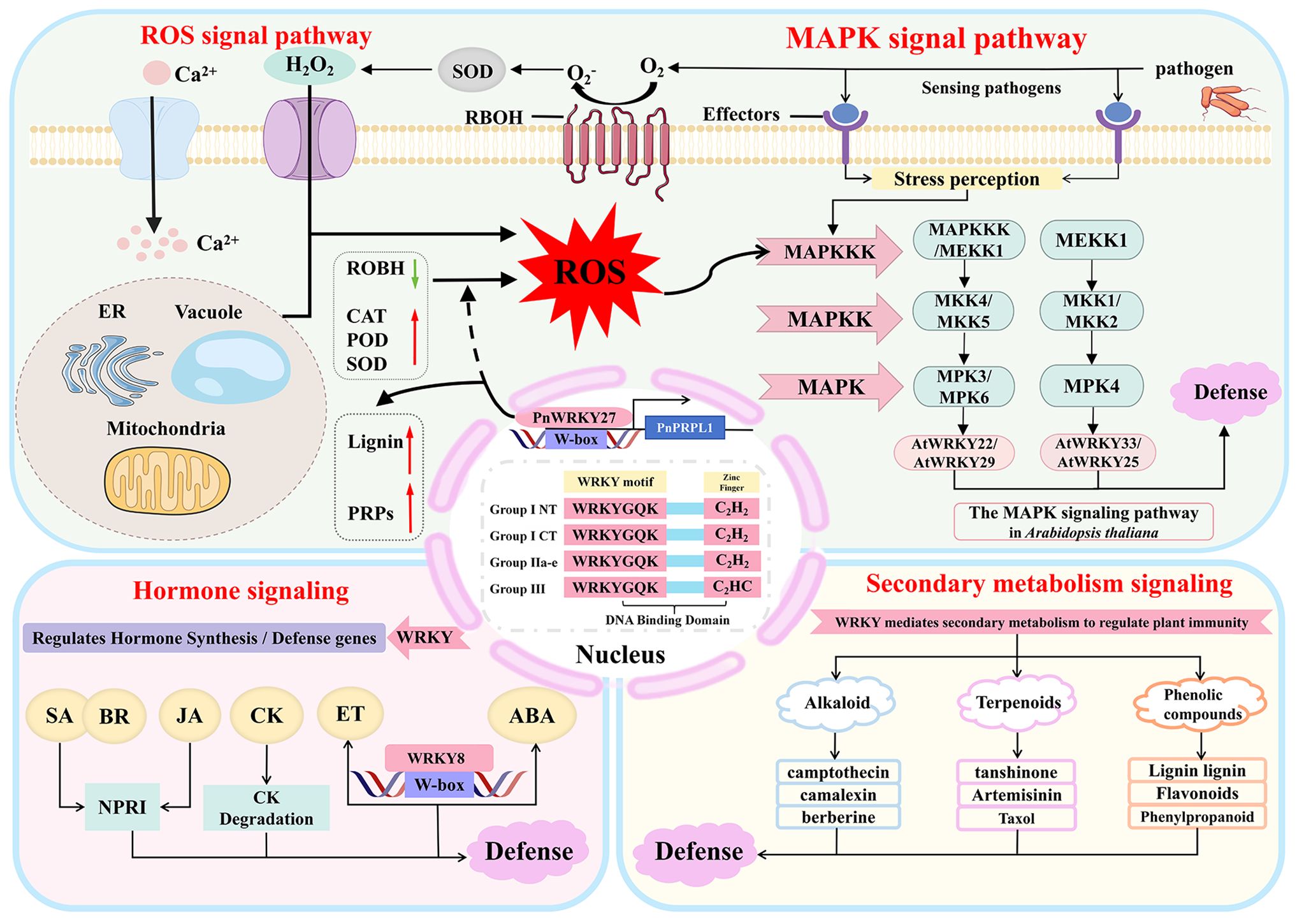
Figure 1. Regulatory mechanisms of WRKY transcription factors in the immune signaling network of medicinal plants. Dashed lines are speculative paths, solid lines are experimentally supported pathways; arrows indicate activation (→) or inhibition (⊣); ER, endoplasmic reticulum; ROS, Reactive Oxygen Species; CAT, Catalase; POD, Peroxidase; SOD, Superoxide Dismutase; RBOH, Respiratory Burst Oxidase Homolog; MAPK, Mitogen-Activated Protein Kinase; MAPKK/MKK, Mitogen-Activated Protein Kinase Kinase; MAPKKK, Mitogen-Activated Protein Kinase Kinase Kinase; MEKK1, Mitogen-Activated Protein Kinase Kinase Kinase 1; MKK2, Mitogen-Activated Protein Kinase Kinase 2; MPK3, Mitogen-Activated Protein Kinase 3; MPK4, Mitogen-Activated Protein Kinase 4; PRPs, Pathogenesis-Related Proteins; SA, Salicylic Acid; JA, Jasmonic Acid; ET-Ethylene; ABA, Abscisic Acid; CK, Cytokinin; BR, Brassinosteroids.
3.1 WRKY mediates the regulation of medicinal plant immune responses at the intracellular physiological and biochemical level
3.1.1 WRKY mediates lignin regulation of medicinal plant immune responses
In various medicinal plants, WRKY TFs have been shown to play a crucial role in lignin biosynthesis and pathogen defense. As a major component of plant secondary cell walls (Ma, 2024), lignin serves as a key marker of bio-induced immune responses (Xiao et al., 2022). It constitutes the first line of defense against pathogen invasion by interacting with cellulose and other cell wall components to enhance mechanical strength and reduce permeability (Ma, 2024). WRKY TFs contribute to plant immune responses by regulating the phenylpropanoid pathway, thereby promoting lignin accumulation (Xiao et al., 2023). For example, WRKY11 in Lilium regale Wilson enhances resistance to usarium oxysporum by suppressing the expression of the LrCel1 gene, thereby reducing cellulase activity and increasing lignin content (Chen et al., 2025b). SmWRKY40 in Salvia miltiorrhiza and NtWRKY28 in Nicotiana tabacum L. are both involved in regulating lignin biosynthesis. Studies have shown that SmWRKY40 is associated with phenylpropanoid metabolism and the stability of root cellular structures (Yu et al., 2025), while NtWRKY28 upregulates the expression of key lignin biosynthetic genes (such as CAD, CCR, and HCT) and promotes the accumulation of defense-related metabolites, including lignin and flavonoids, thereby significantly enhancing resistance to aphid infestation (Chu et al., 2025). Overall, WRKY TFs play a central role in immune response by promoting lignin biosynthesis, reinforcing mechanical barriers, and coordinating the regulation of secondary metabolic pathways, thereby enhancing environmental adaptability and stress tolerance.
3.1.2 WRKY-mediated regulation of antioxidant enzymes in medicinal plant immunity
Upon pathogen attack, plants not only establish a first line of defense by strengthening cell wall mechanical properties, but also rapidly activate an immune signaling network centered around ROS (Haghpanah et al., 2025). ROS function as key signaling molecules that initiate defense pathways during early immune responses, but their excessive accumulation induces oxidative stress and leads to cellular damage. To maintain ROS balance, plants regulate the expression of antioxidant enzymes (including superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD)) through WRKY TFs, thereby scavenging excess ROS and enhancing disease resistance.
PnWRKY27 in Panax notoginseng specifically binds to the PnPRPL1 promoter, promoting PnPRPL1 protein synthesis, which in turn regulates the expression and enzymatic activities of antioxidant enzymes (CAT, POD, and SOD), maintains intracellular ROS homeostasis, and enhances resistance to Fusarium root rot (Su et al., 2024). Overexpression of CsWRKY25 in Citrus spp. and heterologous expression of SpWRKY1 in Nicotiana tabacum upregulate the transcription and enzymatic activity of antioxidant enzymes such as SOD, CAT, and POD, promote ROS scavenging, and activate phosphorylation-related signaling pathways, thereby enhancing plant resistance to pathogens (Wang et al., 2021a; Li et al., 2015a). In Chrysanthemum morifolium, constitutive overexpression of CmWRKY48 markedly suppressed aphid population growth, indicating its pivotal role in aphid resistance (Li et al., 2015b).
It is noteworthy that not all WRKY TFs contribute to positive regulation of plant defense. For example, overexpression of CaWRKY20 suppressed the transcription of ROS scavenging-related enzyme genes (CaCAT, CaPOD, and CaSOD), thereby reducing the ROS scavenging capacity of cells, leading to excessive accumulation of H2O2, and weakening the resistance of plants to Colletotrichum spp (Li et al., 2025f). In addition, overexpression of CmWRKY53 suppressed POD gene expression, thereby increasing Chrysanthemum susceptibility to aphids and offering a molecular basis for its susceptibility mechanism (Zhang et al., 2020).
At the same time, ROS functions as an upstream signal in the MAPK cascade, triggering the phosphorylation and activation of MPK3/MPK6 (mitogen-activated protein kinase 3/6) and other kinases. WRKY TFs regulate ROS homeostasis and act as MAPK pathway targets, linking signal perception to gene expression and mediating plant immune responses.
3.2 WRKY-mediated protein kinase MAPK cascade pathway regulates immune responses in medicinal plants
The MAPK cascade response, which consists of three layers of kinases: Mitogen-Activated Protein Kinase Kinase Kinase (MAPKKK), Mitogen-Activated Protein Kinase Kinase (MAPKK), and MAPK, is one of the immune signaling pathways that is rapidly activated by plants upon sensing pathogens (Wu and Wang, 2024). Once activated, MAPKs regulate the expression of specific downstream immune-related genes by modulating the activity of various TFs, including WRKY, MYB, and ERF (Zhang and Zhang, 2022). Among these, WRKY TFs have been identified as primary targets of MAPKs and play a central role in immune signaling by bridging signal transduction with downstream gene expression (Laflamme, 2023).
In Arabidopsis thaliana, PAMP signaling activates two distinct MAPK-WRKY pathways. One is the MEKK1 (mitogen-activated protein kinase kinase kinase 1)-MKK4/5 (mitogen-activated protein kinase kinase 4/5)-MPK3/6 cascade, leading to the activation of WRKY22 and WRKY29, which enhances plant resistance to pathogens (Asai et al., 2002). The other is the MEKK1-MKK1/2-MPK4 pathway, in which MPK4 phosphorylates the transcriptional regulatory protein MAP kinase substrate 1 (MKS1). MKS1 subsequently regulates its interacting partner WRKY33, which negatively regulates the plant immune response to prevent excessive activation. However, under certain pathogen stresses, such as infection by Pseudomonas spp., WRKY33 remains active. In such cases, WRKY33 can mediate the expression of downstream defense-related genes (Kong et al., 2012). Additionally, WRKY33 is activated by MPK3/6-mediated phosphorylation, which promotes the expression of camalexin biosynthesis genes in coordination with ERF1, thereby enhancing Arabidopsis resistance to Botrytis cinerea (Zhou et al., 2022b).
This mechanism has also been observed in other plant species. In Nicotiana tabacum, NtWRKY4, NtWRKY6, and NtWRKY10 interact with the MAPK cascade and positively regulate immune responses against whitefly infestation (Yao et al., 2020a). Similarly, PnWRKY35 from Panax notoginseng has been shown to activate MAPK signaling and enhance disease resistance when ectopically expressed in Nicotiana tabacum (Li et al., 2025a). However, in Malus domestica, activation of the MKK4-MPK3-WRKY17 signaling pathway reduces SA levels, resulting in increased susceptibility to Glomerella leaf spot, indicating that this MAPK-WRKY module may function as a negative regulator in plant immunity (Shan et al., 2021).
In summary, MAPK-WRKY signaling modules play widespread roles in pathogen recognition and immune regulation across diverse plant species and can function in both positive and negative regulation, emphasizing the complexity and precise modulation of plant immune networks required for maintaining dynamic homeostasis.
3.3 WRKY mediates hormonal regulation of immune responses in medicinal plants
Upon pathogen attack, the MAPK cascade is rapidly activated, leading to the phosphorylation and activation of WRKY TFs, which serve as key hubs that link early pathogen recognition to downstream immune responses. WRKY TFs form a core regulatory network for disease resistance by modulating antagonistic and synergistic interactions among immune-related hormones such as SA, JA, and ET, and by coordinating signaling pathways involving gibberellin (GA), brassinosteroids (BR), auxin (IAA), and strigolactones (SL) to enhance precise pathogen recognition and improve environmental adaptability in plants (Wani et al., 2021; Wang et al., 2023a; Goyal et al., 2023). This complex regulatory framework is depicted in the map of WRKY-regulated hormonal immune defense mechanisms in medicinal plants (Figure 2).
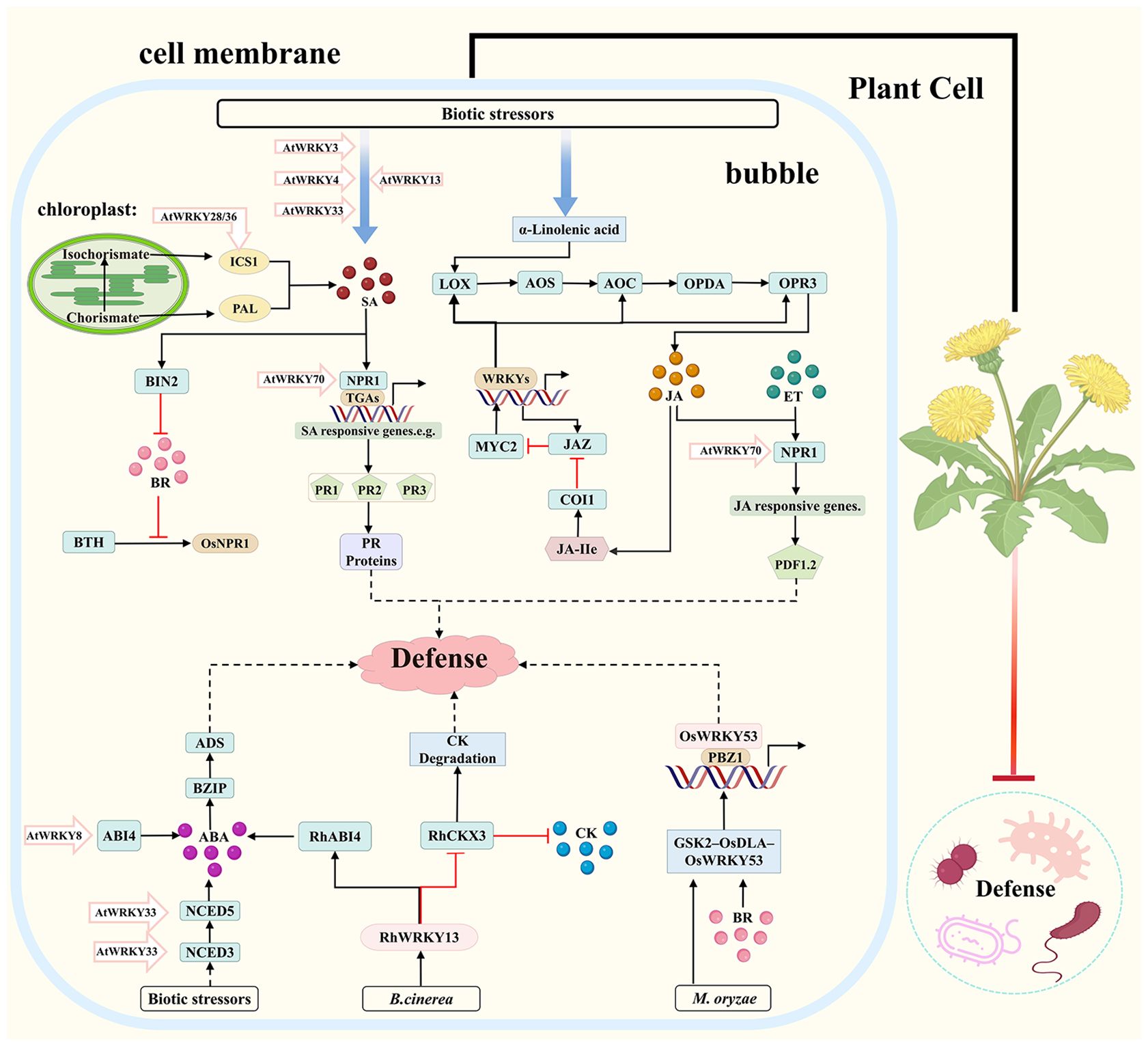
Figure 2. Map of WRKY-regulated hormonal immune defense mechanisms in medicinal plants. Dashed lines are speculative paths, solid lines are experimentally supported pathways; arrows indicate activation (→) or inhibition (⊣); JA, Jasmonic Acid; SA, Salicylic Acid; ET, Ethylene; ABA, Abscisic Acid; CK, Cytokinin; BR, Brassinosteroid; LOX, Lipoxygenase; AOS, Allene Oxide Synthase; AOC, Allene Oxide Cyclase; OPDA, 12-oxo-phytodienoic acid; OPR3, 12-oxo-phytodienoic acid reductase 3; MYC2, bHLH Transcription Factor MYC2; JAZ, Jasmonate ZIM, domain; COI1, Coronatine Insensitive 1; JAIIe, JA, Ile (Jasmonoyl, Isoleucine); ICSI, Isochorismate Synthase I; PAL, Phenylalanine Ammonia, Lyase; NPR1, Nonexpressor of Pathogenesis, Related genes 1; OsNPR1, Oryza sativa NPR1; TGAs, TGA transcription factors; BIN2, Brassinosteroid-Insensitive 2; BTH, Benzothiadiazole; PR Proteins, Pathogenesis-Related Proteins; ADS, Amorpha-4,11-diene Synthase; BZIP, Basic Leucine Zipper; ABI4, ABA-Insensitive 4; NCED5/3, 9-cis-Epoxycarotenoid Dioxygenase 3/5; RhCKX3, Rosa Cytokinin Oxidase/Dehydrogenase 3; RhABI4, Rosa ABA, Insensitive 4; GSK2-OsDLA-OsWRKY53, Glycogen Synthase Kinase2-DLA-WRKY53; PBZ1, Pathogenesis-related protein PBZ1; RhWRKY13, Rosa WRKY13; AtWRKY, Arabidopsis WRKY.
In the model plant Arabidopsis thaliana, WRKY70 serves as a pivotal node in the antagonistic regulation between SA and JA/ET, promoting the expression of SA-dependent resistance genes such as PR proteins while repressing genes in the JA/ET pathway (Jiang et al., 2015). In addition, WRKY25, WRKY33, WRKY11, and WRKY17 also participate in this crosstalk regulation (Li et al., 2004; Journot-Catalino et al., 2006; Zheng et al., 2007). In Nicotiana attenuata, NaWRKY3 functions as a key transcriptional regulator during Alternaria alternata infection, activating jasmonate biosynthetic genes (NaLOX3), ethylene biosynthetic genes (NaACS1, NaACO1), ROS-generating genes (NaRbohD), and defense-related secondary metabolite genes (NaF6’H1, NaBBL28), thereby enhancing antifungal immunity through the integration of hormonal, oxidative, and metabolic responses (Xu et al., 2023). These findings provide important insights into elucidating the immune mechanisms of medicinal plants.
In medicinal plants, WRKY TFs likewise serve as central hubs of hormone regulatory networks. For example, in Pinus massoniana, exogenous signaling molecules (MeJA, SA, etc.) rapidly induce the expression of PmWRKY31, which regulates PmLp8 to activate downstream hormone signaling and terpene biosynthesis genes, thereby elevating endogenous levels of MeJA, GA, SA, and abscisic acid (ABA), promoting the accumulation of terpenes and volatiles, and ultimately enhancing resistance to Dendrolimus punctatus (Chen et al., 2021a). In Panax ginseng, PnWRKY15 synergistically regulates the SA and JA pathways and activates the resistance gene PnOLP1, thereby strengthening resistance to root rot disease (Su et al., 2023);Similarly, in Paeonia lactiflora, PlWRKY65 induces the expression of defense genes such as PlPR1 and enhances systemic immune responses, possibly through the coordination of SA-JA signaling (Wang et al., 2020b).
In addition to the backbone hormones such as SA, JA, and ET, WRKY TFs are also widely involved in the defense regulation of hormones including ABA, cytokinin (CK), and BR. In Nicotiana attenuata, NaWRKY70 directly activates the biosynthetic genes of JA (NaAOS, NaJAR4) and ABA (NaNCED1, NaXD1-like), while simultaneously promoting the accumulation of NaF6’H1-mediated defense metabolites, scopoletin and scopolin, thereby rapidly initiating resistance against Alternaria alternata (Song and Wu, 2024b; Song and Wu, 2024c). Several studies have demonstrated that ABA and CK often act antagonistically in plant immunity. In Rosa hybrida, ABA enhances susceptibility, whereas exogenous CK increases resistance. Mechanistically, RhWRKY13 strengthens defense against grey mould by repressing the CK-degrading gene RhCKX3 and the ABA-responsive factor RhABI4, thereby enhancing CK signaling and suppressing ABA responses (Liu et al., 2023). Moreover, BR was also shown to improve rose petal resistance to Botrytis cinerea, possibly by regulating the expression of TFs such as WRKY, together with cell wall receptors and hormone signaling-related genes (Liu et al., 2018).
In summary, WRKY TFs, as key regulatory nodes of hormone signaling, not only coordinate synergism and antagonism among immune hormones, but also construct an efficient and dynamic immune network by regulating defense genes and metabolic pathways to help medicinal plants to cope with the complex pathogen environment.
3.4 WRKY mediates secondary metabolite synthesis to regulate immune responses in medicinal plants
In recent years, plant immunity research has gradually expanded from traditional focuses on pathogen recognition and signal transduction to defense strategies centering on secondary metabolite-mediated immunity. These metabolites not only have strong toxic inhibitory effects on pathogenic microorganisms, but also serve as important barriers for plants against multiple classes of stresses by modulating insect feeding behavior and nematode movement. Pathogen, pest or nematode infestation induces key TFs such as WRKY, MYB, bHLH, etc., which regulate multiple metabolic pathways and promote the accumulation of multiple classes of defensive metabolites such as alkaloids, terpenoids, phenolics (including flavonoids), and phytoalexins (Jahan et al., 2025; Ali et al., 2024; Monsalvo et al., 2024; Cai et al., 2023; Yang et al., 2024a). Among them, plant antitoxins (phytoalexins) are specific metabolites synthesized de novo during infection, originating from the phenylpropanoid pathway, terpenoid or indole pathways (Wu et al., 2023; Muñoz-Hoyos and Stam, 2023; Yadav et al., 2020), and are not only able to kill pathogens directly, but also act as signaling molecules to amplify host immune response (Zhao et al., 2023; Zhan et al., 2022; Adhikary and Dasgupta, 2023).
It has been shown that erucamide synthesized by Arabidopsis thaliana under stress blocks the assembly of the bacterial T3SS needle protein SctF, thereby reducing pathogenicity and establishing a metabolite-based defense system (Miao et al., 2025). In addition, the volatile secondary metabolite citral was found to down-regulate several effector genes (e.g., PcAvh137, PcAvh238, PcSCR5) in Phytophthora capsici, effectively reducing its infectivity (Song et al., 2023). These findings highlight the dual role of secondary metabolites in plant disease resistance and insect defense. Meanwhile, in Arabidopsis thaliana, AtWRKY33 promotes camalexin accumulation through the MAPK signaling pathway, and this metabolite not only enhances resistance to pathogens but also exerts an inhibitory effect on aphids (Zhou et al., 2022b; Kettles et al., 2013; Chen and Zhang, 2024b). In Nicotiana attenuata, NaWRKY70 activates the transcription of NaF6’H1, a key gene in coumarin biosynthesis, thereby promoting the accumulation of scopoletin and its glycoside scopolin, which enhances resistance to Alternaria alternata (Song and Wu, 2024b, c; Sun et al., 2014). Studies in these model plants provide an important foundation for elucidating WRKY-regulated, secondary metabolite-mediated immune mechanisms.
In medicinal plants, the defensive function of WRKY TFs is closely linked to the metabolic regulation of their unique active components, reflecting an integration of immune defense and pharmacological value. For example, in Withania somnifera, WsWRKY1 enhances resistance to insect feeding by regulating withanolide accumulation and phytosterol-mediated defense pathways (Singh et al., 2017). In Artemisia annua, AaWRKY1 and AaWRKY17 positively regulate the expression of artemisinin-synthesising genes (AaDBR2, AaCYP71AV1, AaADS), thereby strengthening immune responses against Pseudomonas syringae pv. tomato DC3000; meanwhile, artemisinin exerts anti-malarial effects by disrupting Plasmodium proteins (Han et al., 2014; Zhan et al., 2023; Chen et al., 2021b). In Taxus spp., TcWRKY1, TcWRKY33, and TcWRKY26 activate key genes such as DBAT to promote paclitaxel accumulation, which shows antimicrobial activity in vitro, though its direct role in enhancing resistance in planta remains unconfirmed (Li et al., 2013; Chen et al., 2021c, 2022).
Phenolic and flavonoid compounds exhibit antimicrobial activity, reinforce cell walls, and induce systemic acquired resistance (SAR), a crucial component of sustained defense (Saini et al., 2024; Li et al., 2025c). For example, in medicinal plants including Erigeron breviscapus (Song et al., 2024d), Passiflora edulis (Ma et al., 2024), Sophora flavescens (Li et al., 2024d), and Lycium barbarum (Tong et al., 2025), multiple WRKY TFs (e.g., EbWRKY44, PeWRKY30, SfWRKY29, LcWRKY3, and LcWRKY13) positively regulate flavonoid accumulation, while others, such as PeWRKY12, may act as negative regulators to maintain immune homeostasis. PpWRKY70 activates the promoters of 4CL and PAL, thereby increasing the synthesis of total phenolics, flavonoids, and lignin, and enhancing Prunus persica fruit resistance to Rhizopus stolonifer, highlighting the key regulatory role of WRKY TFs in the phenylalanine pathway and plant immunity (Ji et al., 2021).
In conclusion, WRKY TFs play a crucial role in enhancing the direct defense of medicinal plants against pathogens by regulating the synthesis of diverse classes of secondary metabolites, thus broadening the understanding of plant immune regulation. To further illustrate these regulatory relationships, we summarize the classification of WRKY-regulated secondary metabolites (Table 2).
4 WRKY mediates epigenetic regulation of immune responses in medicinal plants
Notably, WRKY TFs regulate their own expression as well as downstream defense genes through epigenetic mechanisms such as DNA methylation and histone modifications, enabling precise control of immune responses.
When plants are attacked by pathogens, epigenetic modifications, including DNA methylation, histone acetylation, and histone methylation, alter the chromatin state of WRKY genes and their targets, thereby precisely regulating immune responses. Under pathogen-infected conditions, WRKY TFs bind to regulatory elements introduced by domesticated transposable elements (TEs) and modulate these elements through H3K27me3 modifications and DNA methylation, enabling Arabidopsis to activate precise immune responses during pathogen attack (Barco et al., 2019; Halter et al., 2021; Hure et al., 2025; Li et al., 2023). In addition, acetylation of histones H3 and H4, as well as H3K4 methylation in the WRKY promoter region, may facilitate transcriptional initiation of WRKY genes during pathogen infection (Jaskiewicz et al., 2011). Following Pseudomonas syringae infection of wild-type Arabidopsis, Trithorax 1 (ATX1) activates WRKY70 by catalyzing trimethylation of histone H3 lysine 4 (H3K4me3), thereby enhancing SA signaling-mediated disease resistance (Alvarez-Venegas et al., 2007). Furthermore, Arabidopsis LDL1 and LDL2, homologous to human lysine demethylase 1-like 1, remodel chromatin accessibility by demethylating histone H3K4 at defense gene loci such as WRKY22, WRKY40, and WRKY70, thereby influencing the epigenetic regulation of plant immunity (Noh et al., 2021).
A growing body of evidence highlights the critical role of non-coding RNAs in plant immunity. For example, WRKY1 activates the expression of lncRNA33732, which in turn upregulates RBOH, leading to ROS, particularly H2O2 accumulation during the early immune response in tomato, thereby enhancing resistance to Phytophthora infestans (Cui et al., 2019). In rice, researchers identified a circular RNA named circ-WRKY9, which encodes a peptide of 88 amino acids (WRKY9-88aa). Overexpression of this peptide not only effectively inhibits rice stripe mosaic virus (RSMV) infection but also enhances immunity against rice blast and bacterial leaf blight (Pan et al., 2025).
5 Outlook
Medicinal plants harbor diverse bioactive compounds and exhibit strong responsiveness to environmental fluctuations and pathogen attacks. Diseases not only reduce plant growth and yield but also directly compromise the stability and quality of medicinal compounds. In recent years, the integration of CRISPR/Cas gene editing and synthetic biology with high-throughput transcriptomics, proteomics, and metabolomics has accelerated research on immune networks and key TFs in medicinal plants, offering novel theoretical frameworks and technical tools to improve disease resistance.
Existing studies have identified some immune regulatory modules through histological analyses. However, significant challenges remain, including unclear mechanisms and a disconnect between basic research and practical applications, making the transition to molecular breeding difficult. In the future, the integration of artificial intelligence and biotechnology is expected to overcome this bottleneck by enabling functional prediction of key immune genes, regulatory network modeling, and intelligent screening of the superior germplasm, thereby establishing a highly efficient and smart disease-resistant breeding system. By reconstructing transcription factor regulatory networks and optimizing signaling pathways, disease resistance in medicinal plants can be significantly enhanced, providing a solid foundation for the high-quality and sustainable development of the Chinese herbal medicine industry.
In conclusion, systematic analyses of key TFs’ immune functions in medicinal plants, integrated with multi-omics, gene editing, and artificial intelligence approaches, are anticipated to bridge the gap between basic research and breeding applications, thus facilitating the synergistic advancement of disease resistance research and the breeding of superior medicinal plant cultivars.
5.1 Molecular design breeding to accelerate transformation
To enhance the immunity of medicinal plants, immune-related factors can be heterologously expressed, overexpressed, or suppressed using transgenic breeding approaches utilizing advanced genetic transformation technologies, the Cut-Dip-Budding (CDB) technique. Such approaches not only confer desirable genetic traits to medicinal plants, facilitating gene function elucidation and targeted trait improvement, but also improve plant yield and enhance tolerance to pathogen infestation (Yan et al., 2022).
In transgenic research, commonly employed biological transformation methods include Agrobacterium-mediated and virus-mediated approaches. For instance, transferring WRKY disease resistance genes into medicinal plants through Agrobacterium-mediated transformation has been shown to effectively enhance their pathogen resistance. In papaya, overexpression of CpWRKY50 via Agrobacterium infiltration positively regulates anthracnose resistance by promoting JA signaling (Yang et al., 2024b). Similarly, Agrobacterium-mediated transformation of CsWRKY48 into tobacco enhanced its resistance to aphids (Wang et al., 2024b).
However, traditional genetic transformation methods are restricted to a limited number of medicinal plants and are often time-consuming. To overcome this limitation, the improved CDB technique was developed, allowing direct infection of medicinal plant organs, including the roots of Taraxacum mongolicum and Rehmannia, as well as the petiole of Salvia miltiorrhiza. This method not only enhances transformation efficiency but also prevents the formation of callus tissue and hairy roots (Cao et al., 2023, 2024). Through this approach, disease resistance-related genes can be efficiently delivered into medicinal plants, thereby improving their resistance to pathogens.
It is noteworthy that current genetic transformation systems are being continuously improved through RNA interference (RNAi) and gene editing technologies. The integration of these technologies with artificial intelligence applications can substantially improve the precision and efficiency of gene editing.
5.2 Artificial intelligence breakthroughs in medicinal plant immune networks
With the integration of gene editing and AI, research on medicinal plant breeding and immunity is entering a new phase of empirically driven innovation. AI has shown significant value across multiple key processes: from AlphaFold’s high-precision protein structure prediction, which enables the analysis of immune-related factors and the design of target sites (Ma et al., 2022), to novel tools such as CRISOT and CCLMoff that advance sgRNA optimization and off-target control. Collectively, these developments outline a promising technological pathway for achieving precise immunoediting in medicinal plants (Du et al., 2025; Chen et al., 2023; Lee, 2023).
In disease monitoring, AI-driven image recognition and environmental modeling are advancing rapidly. Experimental evidence shows that near-infrared and hyperspectral imaging provide high sensitivity and accuracy for early disease detection (Upadhyay et al., 2025). In addition, models based on transfer learning, such as You Only Look Once version 7 (YOLOv7) and version 8 (YOLOv8), can identify a wide range of diseases including powdery mildew, leaf spot and grey mold, and perform well on key metrics (mean accuracy mAP ≈ 91%, precision, recall, and F1 scores), underlining the potential of deep learning for fast and accurate identification. (YOLOv7) and version 8 (YOLOv8) can identify multiple diseases including powdery mildew, leaf spot, and grey mould, and perform well on key metrics (Mean Average Precision, mAP ≈ 91%; Precision; Recall; and F1-score), highlighting the potential of deep learning for fast and accurate identification (Sambana et al., 2025). These advances lay a foundation for dynamic monitoring and precise intervention in the immune networks of medicinal plants, and open possibilities for establishing a closed-loop system of monitoring, intervention, and verification to enhance disease resistance and ensure the stability of medicinal compounds.
Further, integrated prediction of genome and environment (iGEP), combining multi-omics data with machine learning, can optimize plant design at both macro and micro levels while capturing nonlinear features of high-dimensional data, thereby enabling accurate prediction of disease resistance mechanisms (Xu et al., 2022; Mohamedikbal et al., 2025). Although its application is still in the early stages, it has already provided important theoretical and technological support for AI-driven immune networks and “on-demand editing”.
Overall, integrating AI with multi-omics is shifting medicinal plant immunity research from passive resistance to proactive regulation, laying the foundation for intelligent and efficient medicinal plant breeding.
Author contributions
LL: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. XYZ: Conceptualization, Data curation, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. NC: Investigation, Software, Validation, Writing – original draft. HL: Conceptualization, Data curation, Formal Analysis, Software, Supervision, Writing – original draft, Writing – review & editing. YW: Conceptualization, Data curation, Investigation, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. ZG: Data curation, Software, Supervision, Writing – original draft. GS: Software, Supervision, Validation, Writing – original draft. XKZ: Project administration, Software, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was financially supported by the National Natural Science Foundation of China (31960074, 32260089), the Science and Technology Department Foundation of Guizhou Province (QKHJC-MS[2025]371, QKHJC-ZK[2023]496, QKHJC-MS[2025]359).
Acknowledgments
We used Microsoft Word to create the table. We thank you a lot for the assistance of figure drawing by BioRender.com and Scifig.bio. For Figures 1 and 2, we also used PowerPoint drawing in Microsoft Office; we are equally grateful to them for their assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adhikary, S. and Dasgupta, N. (2023). Role of secondary metabolites in plant homeostasis during biotic stress. Biocatal. Agric. Biotechnol. 50, 102712. doi: 10.1016/j.bcab.2023.102712
Ali, J., Tonğa, A., Islam, T., Mir, S., Mukarram, M., Konôpková, A. S., et al. (2024). Defense strategies and associated phytohormonal regulation in Brassica plants in response to chewing and sap-sucking insects. Front. Plant Sci 15–2024. doi: 10.3389/fpls.2024.1376917
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
An, J. P., Zhang, X. W., You, C. X., Bi, S. Q., Wang, X. F., and Hao, Y. J. (2019). MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 224, 380–395. doi: 10.1111/nph.16008
Angupale, J. R., Ajayi, C. O., Tusiimire, J., and Ngwuluka, N. C. (2024). Optimization of in vivo antimalarial efficacy of combinations of aqueous leaf extracts of Artemisia annua L., Vernonia amygdalina Del, and Microglossa pyrifolia (Lam.) Kuntze using factorial design. BMC Complement. Med. Ther. 24, 393. doi: 10.1186/s12906-024-04691-z
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Barco, B., Kim, Y., and Clay, N. K. (2019). Expansion of a core regulon by transposable elements promotes Arabidopsis chemical diversity and pathogen defense. Nat. Commun. 10, 3444. doi: 10.1038/s41467-019-11406-3
Cai, J., Jiang, Y., Ritchie, E. S., Macho, A. P., Yu, F., and Wu, D. (2023). Manipulation of plant metabolism by pathogen effectors: more than just food. FEMS Microbiol. Rev. 47, fuad007. doi: 10.1093/femsre/fuad007
Cao, X., Xie, H., Song, M., Lu, J., Ma, P., Huang, B., et al. (2023). Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture. Innovation (Cambridge (Mass.)). 4, 100345. doi: 10.1016/j.xinn.2022.100345
Cao, X., Xie, H., Song, M., Zhao, L., Liu, H., Li, G., et al. (2024). Simple method for transformation and gene editing in medicinal plants. J. Integr. Plant Biol. 66, 17–19. doi: 10.1111/jipb.13593
Chakraborty, J., Sen, S., Ghosh, P., Jain, A., and Das, S. (2020). Inhibition of multiple defense responsive pathways by CaWRKY70 transcription factor promotes susceptibility in chickpea under Fusarium oxysporum stress condition. BMC Plant Biol. 20, 319. doi: 10.1186/s12870-020-02527-9
Chen, Q., Chuai, G., Zhang, H., Tang, J., Duan, L., Guan, H., et al. (2023). Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints. Nat. Commun. 14, 7521. doi: 10.1038/s41467-023-42695-4
Chen, H., Hu, Y., Liang, X., Xie, J., Xu, H., Luo, Q., et al. (2021a). Roles of hormones, calcium and PmWRKY31 in the defense of Pinus massoniana Lamb. against Dendrolimus punctatus Walker. Forestry. Res. 1, 21. doi: 10.48130/fr-2021-0021
Chen, X.-h., Li, X.-m., Deng, J., Li, J.-m., and Liu, D.-q. (2025b). The inhibition of endo-β-1,4-glucanase is required for the resistance of Lilium regale Wilson to Fusarium wilt. Int. J. Biol. Macromol. 307, 142249. doi: 10.1016/j.ijbiomac.2025.142249
Chen, T., Li, Y., Xie, L., Hao, X., Liu, H., Qin, W., et al. (2021b). AaWRKY17, a positive regulator of artemisinin biosynthesis, is involved in resistance to Pseudomonas syringae in Artemisia annua. Horticult. Res. 8, 217. doi: 10.1038/s41438-021-00652-6
Chen, S., Tan, S., Jin, Z., Wu, J., Zhao, Y., Xu, W., et al. (2024a). The transcriptional landscape of Populus pattern/effector-triggered immunity and how PagWRKY18 involved in it. Plant. Cell Environ. 47, 2074–2092. doi: 10.1111/pce.14860
Chen, L., Wu, L., Yang, L., Yu, H., Huang, P., Wang, Y., et al. (2022). TcJAV3-tcWRKY26 cascade is a missing link in the jasmonate-activated expression of taxol biosynthesis gene DBAT in taxus chinensis. Int. J. Mol. Sci. 23, 13194. doi: 10.3390/ijms232113194
Chen, Y. and Zhang, J. (2024b). Multiple functions and regulatory networks of WRKY33 and its orthologs. Gene 931, 148899. doi: 10.1016/j.gene.2024.148899
Chen, L., Zhang, L., Li, D., Wang, F., and Yu, D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. United. States Am. 110, E1963–E1971. doi: 10.1073/pnas.1221347110
Chen, X., Zhang, T., Wang, H., Zhao, W., and Guo, Z. (2025a). Transcription factor WRKY complexes in plant signaling pathways. Phytopathol. Res. 7, 54. doi: 10.1186/s42483-025-00349-x
Chen, L., Zhang, L., Xiang, S., Chen, Y., Zhang, H., and Yu, D. (2021d). The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 72, 1473–1489. doi: 10.1093/jxb/eraa529
Chen, Y., Zhang, H., Zhang, M., Zhang, W., Ou, Z., Peng, Z., et al. (2021c). Salicylic acid-responsive factor tcWRKY33 positively regulates taxol biosynthesis in taxus chinensis in direct and indirect ways. Front. Plant Sci 12–2021. doi: 10.3389/fpls.2021.697476
Cheng, W., Wang, N., Li, Y., Zhou, X., Bai, X., Liu, L., et al. (2024). CaWRKY01–10 and caWRKY08–4 confer pepper’s resistance to phytophthora capsici infection by directly activating a cluster of defense-related genes. J. Agric. Food Chem. 72, 11682–11693. doi: 10.1021/acs.jafc.4c01024
Chu, L.-Y., Liu, T., Xia, P.-l., Li, J.-P., Tang, Z.-R., Zheng, Y.-L., et al. (2025). NtWRKY28 orchestrates flavonoid and lignin biosynthesis to defense aphid attack in tobacco plants. Plant Physiol. Biochem. 221, 109673. doi: 10.1016/j.plaphy.2025.109673
Cui, J., Jiang, N., Meng, J., Yang, G., Liu, W., Zhou, X., et al. (2019). LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J. 97, 933–946. doi: 10.1111/tpj.14173
Dabi, M., Agarwal, P., and Agarwal, P. K. (2020). Overexpression of JcWRKY2 confers increased resistance towards Macrophomina phaseolina in transgenic tobacco. 3 Biotech. 10, 490. doi: 10.1007/s13205-020-02490-0
Deng, C., Hao, X., Shi, M., Fu, R., Wang, Y., Zhang, Y., et al. (2019). Tanshinone production could be increased by the expression of SmWRKY2 in Salvia miltiorrhiza hairy roots. Plant Sci 284, 1–8. doi: 10.1016/j.plantsci.2019.03.007
Du, Y., Ma, H., Liu, Y., Gong, R., Lan, Y., Zhao, J., et al. (2024). Major quality regulation network of flavonoid synthesis governing the bioactivity of black wolfberry. New Phytol. 242, 558–575. doi: 10.1111/nph.19602
Du, W., Zhao, L., Diao, K., Zheng, Y., Yang, Q., Zhu, Z., et al. (2025). A versatile CRISPR/Cas9 system off-target prediction tool using language model. Commun. Biol. 8, 882. doi: 10.1038/s42003-025-08275-6
Fang, S., Zhang, C., Qiu, S., Xiao, Y., Chen, K., Lv, Z., et al. (2023). SbWRKY75- and SbWRKY41-mediated jasmonic acid signaling regulates baicalin biosynthesis. Front. Plant Sci 14–2023. doi: 10.3389/fpls.2023.1302112
Fu, Y., Li, J., Wu, H., Jiang, S., Zhu, Y., Liu, C., et al. (2022). Analyses of Botrytis cinerea-responsive LrWRKY genes from Lilium regale reveal distinct roles of two LrWRKY transcription factors in mediating responses to B. cinerea. Plant Cell Rep. 41, 995–1012. doi: 10.1007/s00299-022-02833-6
Gao, J., Chen, Y., Gao, M., Wu, L., Zhao, Y., and Wang, Y. (2023). LcWRKY17, a WRKY transcription factor from litsea cubeba, effectively promotes monoterpene synthesis. Int. J. Mol. Sci. 24, 7210. doi: 10.3390/ijms24087210
Gao, G., Jin, R., Liu, D., Zhang, X., Sun, X., Zhu, P., et al. (2022). CmWRKY15–1 promotes resistance to chrysanthemum white rust by regulating cmNPR1 expression. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.865607
Gao, Y. F., Liu, J. K., Yang, F. M., Zhang, G. Y., Wang, D., Zhang, L., et al. (2020). The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant. 168, 98–117. doi: 10.1111/ppl.12978
Goyal, P., Devi, R., Verma, B., Hussain, S., Arora, P., Tabassum, R., et al. (2023). WRKY transcription factors: evolution, regulation, and functional diversity in plants. Protoplasma 260, 331–348. doi: 10.1007/s00709-022-01794-7
Haghpanah, M., Namdari, A., Kaleji, M. K., Nikbakht-dehkordi, A., Arzani, A., and Araniti, F. (2025). Interplay between ROS and hormones in plant defense against pathogens. Plants (Basel). 14, 1297. doi: 10.3390/plants14091297
Halter, T., Wang, J., Amesefe, D., Lastrucci, E., Charvin, M., Singla Rastogi, M., et al. (2021). The Arabidopsis active demethylase ROS1 cis-regulates defence genes by erasing DNA methylation at promoter-regulatory regions. eLife 10, e62994. doi: 10.7554/eLife.62994
Han, Y., Sun, T., Tang, Y., Yang, M., Gao, W., Wang, L., et al. (2025). Root rot in medicinal plants: a review of extensive research progress. Front. Plant Sci. 15, 1504370. doi: 10.3389/fpls.2024.1504370
Han, J., Wang, H., Lundgren, A., and Brodelius, P. E. (2014). Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102, 89–96. doi: 10.1016/j.phytochem.2014.02.011
Hao, X., Xie, C., Ruan, Q., Zhang, X., Wu, C., Han, B., et al. (2021). The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila. Horticult. Res. 8, 7. doi: 10.1038/s41438-020-00437-3
Hu, Q., Xiao, S., Wang, X., Ao, C., Zhang, X., and Zhu, L. (2021). GhWRKY1-like enhances cotton resistance to Verticillium dahliae via an increase in defense-induced lignification and S monolignol content. Plant Sci 305, 110833. doi: 10.1016/j.plantsci.2021.110833
Huang, X., Jia, A., Huang, T., Wang, L., Yang, G., and Zhao, W. (2023). Genomic profiling of WRKY transcription factors and functional analysis of CcWRKY7, CcWRKY29, and CcWRKY32 related to protoberberine alkaloids biosynthesis in Coptis chinensis Franch. Front. Genet. 14. doi: 10.3389/fgene.2023.1151645
Hure, V., Piron-Prunier, F., Yehouessi, T., Vitte, C., Kornienko, A. E., Adam, G., et al. (2025). Alternative silencing states of transposable elements in Arabidopsis associated with H3K27me3. Genome Biol. 26, 11. doi: 10.1186/s13059-024-03466-6
Hussain, A., Qayyum, A., Farooq, S., Almutairi, S. M., Rasheed, R. A., Qadir, M., et al. (2024). Pepper immunity against Ralstonia solanacearum is positively regulated by CaWRKY3 through modulation of different WRKY transcription factors. BMC Plant Biol. 24, 522. doi: 10.1186/s12870-024-05143-z
Jahan, T., Huda, M. N., Zhang, K., He, Y., Lai, D., Dhami, N., et al. (2025). Plant secondary metabolites against biotic stresses for sustainable crop protection. Biotechnol. Adv. 79, 108520. doi: 10.1016/j.bioteChadv.2025.108520
Jaskiewicz, M., Conrath, U., and Peterhänsel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Javed, T. and Gao, S.-J. (2023). WRKY transcription factors in plant defense. Trends Genet. 39, 787–801. doi: 10.1016/j.tig.2023.07.001
Ji, N. N., Wang, J., Li, Y. F., Li, M. L., Jin, P., and Zheng, Y. H. (2021). Involvement of PpWRKY70 in the methyl jasmonate primed disease resistance against Rhizopus stolonifer of peaches via activating phenylpropanoid pathway. Postharvest. Biol. Technol. 174, 111466. doi: 10.1016/j.postharvbio.2021.111466
Jia, S., Wang, Y., Zhang, G., Yan, Z., and Cai, Q. (2020). Strawberry faWRKY25 transcription factor negatively regulated the resistance of strawberry fruits to botrytis cinerea. Genes 12, 56. doi: 10.3390/genes12010056
Jiang, C.-H., Huang, Z.-Y., Xie, P., Gu, C., Li, K., Wang, D.-C., et al. (2015). Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J. Exp. Bot. 67, 157–174. doi: 10.1093/jxb/erv445
Joshi, A., Jeena, G. S., Shikha, Kumar, R. S., Pandey, A., and Shukla, R. K. (2022). Ocimum sanctum, OscWRKY1, regulates phenylpropanoid pathway genes and promotes resistance to pathogen infection in Arabidopsis. Plant Mol. Biol. 110, 235–251. doi: 10.1007/s11103-022-01297-2
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kang, J. E., Kim, H., Song, K., Choi, C., Kim, Y. J., Hwang, D. J., et al. (2024). Arabidopsis WRKY55 transcription factor enhances soft rot disease resistance with ORA59. Plant Pathol J. 40, 537–550. doi: 10.5423/ppj.Oa.08.2024.0126
Kettles, G. J., Drurey, C., Schoonbeek, H. J., Maule, A. J., and Hogenhout, S. A. (2013). Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 198, 1178–1190. doi: 10.1111/nph.12218
Kong, Q., Qu, N., Gao, M., Zhang, Z., Ding, X., Yang, F., et al. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase in Arabidopsis. Plant Cell. 24, 2225–2236. doi: 10.1105/tpc.112.097253
Kumar, A., Sichov, N., Bucki, P., and Miyara, S. B. (2023). SlWRKY16 and SlWRKY31 of tomato, negative regulators of plant defense, involved in susceptibility activation following root-knot nematode Meloidogyne javanica infection. Sci. Rep. 13, 14592. doi: 10.1038/s41598-023-40557-z
Laflamme, B. (2023). WRKYng together: Coordination between kinase cascades and transcription factors contributes to immunity in rice. Plant Cell. 35, 1968–1969. doi: 10.1093/plcell/koad074
Lee, M. (2023). Deep learning in CRISPR-Cas systems: a review of recent studies. Front. Bioeng. Biotechnol. 11. doi: 10.3389/fbioe.2023.1226182
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 16, 319–331. doi: 10.1105/tpc.016980
Li, M., Che, X., Liang, Q., Li, K., Xiang, G., Liu, X., et al. (2025a). Genome-wide identification and characterization of WRKYs family involved in responses to Cylindrocarpon destructans in Panax notoginseng. BMC Genomics 26, 104. doi: 10.1186/s12864-025-11280-y
Li, H., Chen, N., Zhang, H., and Xu, D. (2025e). Multidimensional regulation of transcription factors: decoding the comprehensive signals of plant secondary metabolism. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1522278
Li, J., Huang, H. C., Zuo, Y. Q., Zhang, M. Y., He, M. L., and Xia, K. F. (2024e). PatWRKY71 transcription factor regulates patchoulol biosynthesis and plant defense response. BMC Plant Biol. 24, 8. doi: 10.1186/s12870-023-04660-7
Li, S., Ji, Q., An, X., Chen, C., Luo, X., Liu, T., et al. (2024a). Genome-wide analysis of WRKY gene family and the dynamic responses of key WRKY genes involved in cadmium stress in Brassica juncea. Front. Plant Sci 15–2024. doi: 10.3389/fpls.2024.1465905
Li, S. L., Khoso, M. A., Xu, H., Zhang, C., Liu, Z. Y., Wagan, S., et al. (2024b). WRKY transcription factors (TFs) as key regulators of plant resilience to environmental stresses: current perspective. Agronomy-Basel 14, 2421. doi: 10.3390/agronomy14102421
Li, J., Li, Y., Dang, M., Li, S., Chen, S., Liu, R., et al. (2022). Jasmonate-responsive transcription factors nnWRKY70a and nnWRKY70b positively regulate benzylisoquinoline alkaloid biosynthesis in lotus (Nelumbo nucifera). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.862915
Li, D., Li, X., Wang, Z., Wang, H., Gao, J., Liu, X., et al. (2024c). Transcription factors RhbZIP17 and RhWRKY30 enhance resistance to Botrytis cinerea by increasing lignin content in rose petals. J. Exp. Bot. 75, 1633–1646. doi: 10.1093/jxb/erad473
Li, Y., Li, Z., Xu, C., and Wang, Q. (2025d). WRKYs as regulatory hubs of secondary metabolic networks: Diverse inducers and distinct responses. Plant Commun. 6(9), 101438. doi: 10.1016/j.xplc.2025.101438
Li, J. B., Luan, Y. S., and Liu, Z. (2015a). Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 155, 248–266. doi: 10.1111/ppl.12315
Li, Y., Ma, X., Xiao, L. D., Yu, Y. N., and Gong, Z. H. (2025f). CaWRKY20 negatively regulates plant resistance to colletotrichum scovillei in pepper. Plant. Cell Environ. 48, 1514–1534. doi: 10.1111/pce.15205
Li, Z., Pi, B., Liu, S., Li, Y., Cai, N., Peng, J., et al. (2025b). Genome-wide analysis of WRKY transcription factors involved in abiotic stress in Lonicera japonica. Front. Plant Sci. 16, 1653750. doi: 10.3389/fpls.2025.1653750
Li, M., Shao, Y., Pan, B., Liu, C., and Tan, H. (2025c). Regulation of important natural products biosynthesis by WRKY transcription factors in plants. J. Adv. Res. doi: 10.1016/j.jare.2025.01.009
Li, P., Song, A., Gao, C., Jiang, J., Chen, S., Fang, W., et al. (2015b). The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem.: PPB. 95, 26–34. doi: 10.1016/j.plaphy.2015.07.002
Li, J., Wang, X., Lu, J., Song, H., Lei, H., Niu, T., et al. (2024d). Genome-wide identification and expression analysis of the WRKY gene family in Sophora flavescens during tissue development and salt stress. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1520786
Li, Z., Zhang, Y., Ding, C.-H., Chen, Y., Wang, H., Zhang, J., et al. (2023). LHP1-mediated epigenetic buffering of subgenome diversity and defense responses confers genome plasticity and adaptability in allopolyploid wheat. Nat. Commun. 14, 7538. doi: 10.1038/s41467-023-43178-2
Li, S., Zhang, P., Zhang, M., Fu, C., and Yu, L. (2013). Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. (Stuttgart Germany). 15, 19–26. doi: 10.1111/j.1438-8677.2012.00611.x
Liu, X., Cao, X., Shi, S., Zhao, N., Li, D., Fang, P., et al. (2018). Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet. 19, 62. doi: 10.1186/s12863-018-0668-x
Liu, D., Jelenska, J., Morgan, J.M., and Greenberg, J.T. (2024). Phytosulfokine downregulates defense-related WRKY transcription factors and attenuates pathogen-associated molecular pattern-triggered immunity. Plant Journal 120, 2367–2384. doi: 10.1111/tpj.17115
Liu, D., He, J., Li, Q., Zhang, X., Wang, Y., Sun, Q., et al. (2025). A WRKY transcription factor confers broad-spectrum resistance to biotic stresses and yield stability in rice. Proc. Natl. Acad. Sci. U.S.A. 122, e2411164122. doi: 10.1073/pnas.2411164122
Liu, Y., Xin, H., Zhang, Y., Che, F., Shen, N., and Cui, Y. (2022). Leaves, seeds and exocarp of Ginkgo biloba L. (Ginkgoaceae): A Comprehensive Review of Traditional Uses, phytochemistry, pharmacology, resource utilization and toxicity. J. Ethnopharmacol. 298, 115645. doi: 10.1016/j.jep.2022.115645
Liu, X., Zhou, X., Li, D., Hong, B., Gao, J., and Zhang, Z. (2023). Rose WRKY13 promotes disease protection to Botrytis by enhancing cytokinin content and reducing abscisic acid signaling. Plant Physiol. 191, 679–693. doi: 10.1093/plphys/kiac495
Lu, Z., Wang, X., Mostafa, S., Noor, I., Lin, X., Ren, S., et al. (2023). WRKY Transcription Factors in Jasminum sambac: An Insight into the Regulation of Aroma Synthesis. Biomolecules. 13, 1679. doi: 10.3390/biom13121679
Luo, D., Cai, J., Sun, W., Yang, Q., Hu, G., and Wang, T. (2024). Tomato SlWRKY3 Negatively Regulates Botrytis cinerea Resistance via TPK1b. Plants (Basel). 13, 1597. doi: 10.3390/plants13121597
Ma, Q. H (2024a). Lignin biosynthesis and its diversified roles in disease resistance 15, 295.doi: 10.3390/genes15030295
Ma, W., Zhang, S., Li, Z., Jiang, M., Wang, S., Lu, W., et al. (2022). Enhancing protein function prediction performance by utilizing alphaFold-predicted protein structures. J. Chem. Inf. Modeling. 62, 4008–4017. doi: 10.1021/acs.jcim.2c00885
Ma, F., Zhou, H., Yang, H., Huang, D., Xing, W., Wu, B., et al. (2024b). WRKY transcriptionfactors in passion fruit analysis reveals key PeWRKYs involved in abiotic stress and flavonoidbiosynthesis. Int. J. Biol. Macromol. 256, 128063. doi: 10.1016/j.ijbiomac.2023.128063
Miao, P., Wang, H., Wang, W., Wang, Z., Ke, H., Cheng, H., et al. (2025). A widespread plant defense compound disarms bacterial type III injectisome assembly. Sci (New York N.Y.) 387, eads0377. doi: 10.1126/science.ads0377
Mohamedikbal, S., Al-Mamun, H. A., Bestry, M. S., Batley, J., and Edwards, D. (2025). Integrating multi-omics and machine learning for disease resistance prediction in legumes. TAG. Theoretical and applied genetics. Theoretische. und Angewandte. Genetik. 138, 163. doi: 10.1007/s00122-025-04948-2
Monsalvo, I., Lin, J., and Kovinich, N. (2024). Phytoalexin gene regulation in Arabidopsis thaliana – On the verge of a paradigm shift? Curr. Plant Biol. 39, 100367. doi: 10.1016/j.cpb.2024.100367
Mu, Y., Dong, Y., Li, X., Gong, A., Yu, H., Wang, C., et al. (2024). JrPHL8-JrWRKY4-JrSTH2L module regulates resistance to Colletotrichum gloeosporioides in walnut. Horticult. Res. 11, uhae148. doi: 10.1093/hr/uhae148
Muñoz-Hoyos, L. and Stam, R. (2023). Metabolomics in plant pathogen defense: from single molecules to large-scale analysis. Phytopathology 113, 760–770. doi: 10.1094/phyto-11-22-0415-fi
Negi, N. and Khurana, P. (2021). A salicylic acid inducible mulberry WRKY transcription factor, MiWRKY53 is involved in plant defence response. Plant Cell Rep. 40, 2151–2171. doi: 10.1007/s00299-021-02710-8
Noh, S. W., Seo, R. R., Park, H. J., and Jung, H. W. (2021). Two arabidopsis homologs of human lysine-specific demethylase function in epigenetic regulation of plant defense responses. Front. Plant Sci 12. doi: 10.3389/fpls.2021.688003
Owusu Adjei, M., Zhou, X., Mao, M., Rafique, F., and Ma, J. (2021). MicroRNAs roles in plants secondary metabolism. Plant Signaling Behav. 16, 1915590. doi: 10.1080/15592324.2021.1915590
Pan, X., Xu, S., Cao, G., Chen, S., Zhang, T., Yang, B. B., et al. (2025). A novel peptide encoded by a rice circular RNA confers broad-spectrum disease resistance in rice plants. New Phytol. 246, 689–701. doi: 10.1111/nph.70018
Ramos, R. N., Zhang, N., Lauff, D. B., Valenzuela-Riffo, F., Figueroa, C. R., Martin, G. B., et al. (2023). Loss-of-function mutations in WRKY22 and WRKY25 impair stomatal-mediated immunity and PTI and ETI responses against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 112, 161–177. doi: 10.1007/s11103-023-01358-0
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Saini, N., Anmol, A., Kumar, S., Wani, A. W., Bakshi, M., and Dhiman, Z. (2024). Exploring phenolic compounds as natural stress alleviators in plants- a comprehensive review. Physiol. Mol. Plant Pathol. 133, 102383. doi: 10.1016/j.pmpp.2024.102383
Sambana, B., Nnadi, H. S., Wajid, M. A., Fidelia, N. O., Camacho-Zuñiga, C., Ajuzie, H. D., et al. (2025). An efficient plant disease detection using transfer learning approach. Sci. Rep. 15, 19082. doi: 10.1038/s41598-025-02271-w
Shan, D., Wang, C., Zheng, X., Hu, Z., Zhu, Y., Zhao, Y., et al. (2021). MKK4-MPK3-WRKY17-mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 186, 1202–1219. doi: 10.1093/plphys/kiab108
Shi, L., Fan, Y., Yang, Y., Yan, S., Qiu, Z., Liu, Z., et al. (2024). CaWRKY22b plays a positive role in the regulation of pepper resistance to ralstonia solanacearum in a manner associated with jasmonic acid signaling. Plants (Basel Switzerland) 13, 2081. doi: 10.3390/plants13152081
Shi, X., Wang, X., Zheng, Y., and Fu, L. (2025). Advances in the study of bioactive compounds and nutraceutical properties of goji berry (Lycium barbarum L.). Appl. Sci. 15, 262. doi: 10.3390/app15010262
Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S., Shasany, A. K., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215, 1115–1131. doi: 10.1111/nph.14663
Song, N. and Wu, J. (2024b). NaWRKY70 is a key regulator of Nicotiana attenuata resistance to Alternaria alternata through regulation of phytohormones and phytoalexins biosynthesis. New Phytol. 242, 1289–1306. doi: 10.1111/nph.19647
Song, N. and Wu, J. (2024c). Synergistic induction of phytoalexins in Nicotiana attenuata by jasmonate and ethylene signaling mediated by NaWRKY70. J. Exp. Bot. 75, 1063–1080. doi: 10.1093/jxb/erad415
Song, W., Yin, Z., Lu, X., Shen, D., and Dou, D. (2023). Plant secondary metabolite citral interferes with Phytophthora capsici virulence by manipulating the expression of effector genes. Mol. Plant Pathol. 24, 932–946. doi: 10.1111/mpp.13340
Song, W., Zhang, S., Li, Q., Xiang, G., Zhao, Y., Wei, F., et al. (2024d). Genome-wide profiling of WRKY genes involved in flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1412574
Su, L., Li, W., Chen, X., Wang, P., and Liu, D. (2024). Proline-rich protein PRPL1 enhances Panax notoginseng defence against Fusarium solani by regulating reactive oxygen species balance and strengthening the cell wall barrier. Plant. Cell Environ. 47, 2377–2395. doi: 10.1111/pce.14886
Su, L., Zheng, L., Wang, H., Qu, Y., Ge, F., and Liu, D. (2023). Panax notoginseng transcription factor WRKY15 modulates resistance to Fusarium solani by up-regulating osmotin-like protein expression and inducing JA/SA signaling pathways. BMC Plant Biol. 23, 362. doi: 10.1186/s12870-023-04373-x
Sun, S., Li, S., Zhou, X., and Yang, X. (2023). WRKY1 represses the WHIRLY1 transcription factor to positively regulate plant defense against geminivirus infection. PloS Pathogens. 19, e1011319. doi: 10.1371/journal.ppat.1011319
Sun, Y. Z., Niu, Y. Y., Xu, J., Li, Y., Luo, H. M., Zhu, Y. J., et al. (2013). Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tissue Organ Cult. 114, 269–277. doi: 10.1007/s11240-013-0323-1
Sun, H., Wang, L., Zhang, B., Ma, J., Hettenhausen, C., Cao, G., et al. (2014). Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 65, 4305–4315. doi: 10.1093/jxb/eru203
Sun, P. W., Xu, Y. H., Yu, C. C., Lv, F. F., Tang, X. L., Gao, Z. H., et al. (2020). WRKY44 represses expression of the wound-induced sesquiterpene biosynthetic gene ASS1 in Aquilaria sinensis. J. Exp. Bot. 71, 1128–1138. doi: 10.1093/jxb/erz469
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tanvir, R., Guo, L., Wu, H., and Li, L. (2024). Special issue: Manipulation/regulation of secondary metabolites in medicinal plants. Plant Physiol. Biochem.: PPB. 211, 108549. doi: 10.1016/j.plaphy.2024.108549
Tian, W.-W., Liu, L., Chen, P., Yu, D.-M., Li, Q.-M., Hua, H., et al. (2025). Curcuma Longa (turmeric): from traditional applications to modern plant medicine research hotspots. Chin. Med. 20, 76. doi: 10.1186/s13020-025-01115-z
Tong, L., Jiang, Y., Zhang, X., Zhang, X., Zhang, W., Ren, G., et al. (2025). Metabolic and molecular basis of flavonoid biosynthesis in Lycii fructus: An integration of metabolomic and transcriptomic analysis. J. Pharm. Biomed. Analysis. 255, 116653. doi: 10.1016/j.jpba.2024.116653
Upadhyay, A., Chandel, N. S., Singh, K. P., Chakraborty, S. K., Nandede, B. M., Kumar, M., et al. (2025). Deep learning and computer vision in plant disease detection: a comprehensive review of techniques, models, and trends in precision agriculture. Artif. Intell. Rev. 58, 92. doi: 10.1007/s10462-024-11100-x
Wan, J., He, M., Hou, Q., Zou, L., Yang, Y., Wei, Y., et al. (2021). Cell wall associated immunity in plants. Stress Biol. 1, 3. doi: 10.1007/s44154-021-00003-4
Wang, W., Cao, H., Wang, J., and Zhang, H. (2024a). Recent advances in functional assays of WRKY transcription factors in plant immunity against pathogens. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1517595
Wang, H., Cheng, X., Yin, D., Chen, D., Luo, C., Liu, H., et al. (2023a). Advances in the research on plant WRKY transcription factors responsive to external stresses. Curr. Issues Mol. Biol. 45, 2861–2880. doi: 10.3390/cimb45040187
Wang, Z., Deng, J., Liang, T., Su, L., Zheng, L., Chen, H., et al. (2022d). Lilium regale Wilson WRKY3 modulates an antimicrobial peptide gene, LrDef1, during response to Fusarium oxysporum. BMC Plant Biol. 22, 257. doi: 10.1186/s12870-022-03649-y
Wang, Y., Gao, Y., Zang, P., and Xu, Y. (2020a). Transcriptome analysis reveals underlying immune response mechanism of fungal (Penicillium oxalicum) disease in Gastrodia elata Bl. f. glauca S. chow (Orchidaceae). BMC Plant Biol. 20, 445. doi: 10.1186/s12870-020-02653-4
Wang, J., Gong, Y., Li, M., Bai, Y., and Wu, T. (2024b). A csWRKY48 gene from tea plants intercropped with chinese chestnut plays an important role in resistance to biotic and abiotic stresses. Int. J. Mol. Sci. 25, 13526. doi: 10.3390/ijms252413526
Wang, H., Gong, W., Wang, Y., and Ma, Q. (2023b). Contribution of a WRKY transcription factor, shWRKY81, to powdery mildew resistance in wild tomato. Int. J. Mol. Sci. 24, 2583. doi: 10.3390/ijms24032583
Wang, L., Guo, D., Zhao, G., Wang, J., Zhang, S., Wang, C., et al. (2022b). Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytol. 236, 249–265. doi: 10.1111/nph.18329
Wang, W., Li, T., Chen, Q., Deng, B., Deng, L., and Zeng, K. (2021b). Transcription factor csWRKY65 participates in the establishment of disease resistance of citrus fruits to penicillium digitatum. J. Agric. Food Chem. 69, 5671–5682. doi: 10.1021/acs.jafc.1c01411
Wang, W., Li, T., Chen, Q., Yao, S., Deng, L., and Zeng, K. (2021a). CsWRKY25 Improves Resistance of Citrus Fruit to Penicillium digitatum via Modulating Reactive Oxygen Species Production. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.818198
Wang, X., Li, J., Guo, J., Qiao, Q., Guo, X., and Ma, Y. (2020b). The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Horticult. Res. 7, 57. doi: 10.1038/s41438-020-0267-7
Wang, F., Li, X., Zuo, X., Li, M., Miao, C., Zhi, J., et al. (2021c). Transcriptome-wide identification of WRKY transcription factor and functional characterization of rgWRKY37 involved in acteoside biosynthesis in rehmannia glutinosa. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.739853
Wang, Y., Wang, X., Fang, J., Yin, W., Yan, X., Tu, M., et al. (2023c). VqWRKY56 interacts with VqbZIPC22 in grapevine to promote proanthocyanidin biosynthesis and increase resistance to powdery mildew. New Phytol. 237, 1856–1875. doi: 10.1111/nph.18688
Wang, X.-Y., Wang, R.-C., Qu, Z.-Y., Zhu, Y.-Z., and Li, Y.-L. (2022a). Advances on immunoregulation effect of astragalus polysaccharides. Front. Nat. Prod. 1, 971679. doi: 10.3389/fntpr.2022.971679
Wang, M., Yuan, Y., Zhao, Y., Hu, Z., Zhang, S., Luo, J., et al. (2025). PhWRKY30 activates salicylic acid biosynthesis to positively regulate antiviral defense response in petunia. Horticult. Res. 12, uhaf013. doi: 10.1093/hr/uhaf013
Wang, H., Zheng, Y., Xiao, D., Li, Y., Liu, T., and Hou, X. (2022c). BcWRKY33A Enhances Resistance to Botrytis cinerea via Activating BcMYB51–3 in Non-Heading Chinese Cabbage. Int. J. Mol. Sci. 23, 8222. doi: 10.3390/ijms23158222
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Wei, Q. H., Cao, X. X., Xu, D. F., Wang, S. T., Zhang, J. S., and Zhang, H. (2023). Anti-inflammatory labdane diterpenoids from the aerial parts of Leonurus japonicus. Phytochemistry 210, 113646. doi: 10.1016/j.phytochem.2023.113646
Wei, Y., Liu, G., Chang, Y., Lin, D., Reiter, R. J., He, C., et al. (2018). Melatonin biosynthesis enzymes recruit WRKY transcription factors to regulate melatonin accumulation and transcriptional activity on W-box in cassava. J. Pineal. Res. 65, e12487. doi: 10.1111/jpi.12487
Wen, F., Wu, X., Li, T., Jia, M., and Liao, L. (2022). Characterization of the WRKY gene family in Akebia trifoliata and their response to Colletotrichum acutatum. BMC Plant Biol. 22, 115. doi: 10.1186/s12870-022-03511-1
Wu, M., Northen, T. R., and Ding, Y. (2023). Stressing the importance of plant specialized metabolites: omics-based approaches for discovering specialized metabolism in plant stress responses. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1272363
Wu, G. and Wang, W. (2024). Recent advances in understanding the role of two mitogen-activated protein kinase cascades in plant immunity. J. Exp. Bot. 75, 2256–2265. doi: 10.1093/jxb/erae020
Xiang, H., Stojilkovic, B., and Gheysen, G. (2025). Decoding plant-pathogen interactions: A comprehensive exploration of effector-plant transcription factor dynamics. Mol. Plant Pathol. 26, e70057. doi: 10.1111/mpp.70057
Xiao, Y., Feng, J., Li, Q., Zhou, Y., Bu, Q., Zhou, J., et al. (2020). IiWRKY34 positively regulates yield, lignan biosynthesis and stress tolerance in Isatis indigotica. Acta Pharm. Sin. B. 10, 2417–2432. doi: 10.1016/j.apsb.2019.12.020
Xiao, S., Ming, Y., Hu, Q., Ye, Z., Si, H., Liu, S., et al. (2023). GhWRKY41 forms a positive feedback regulation loop and increases cotton defence response against Verticillium dahliae by regulating phenylpropanoid metabolism. Plant Biotechnol. J. 21, 961–978. doi: 10.1111/pbi.14008
Xiao, F., Xu, W., Hong, N., Wang, L., Zhang, Y., and Wang, G. (2022). A secreted lignin peroxidase required for fungal growth and virulence and related to plant immune response. Int J Mol Sci. 23, 6066. doi: 10.3390/ijms23116066
Xu, Y., Zhang, X., Li, H., Zheng, H., Zhang, J., Olsen, M. S., et al. (2022). Smart breeding driven by big data, artificial intelligence, and integrated genomic-enviromic prediction. Mol. Plant 15, 1664–1695. doi: 10.1016/j.molp.2022.09.001
Xu, Z., Zhang, S., and Wu, J. (2023). NaWRKY3 is a master transcriptional regulator of the defense network against brown spot disease in wild tobacco. J. Exp. Bot. 74, 4169–4188. doi: 10.1093/jxb/erad142
Yadav, V., Wang, Z., Wei, C., Amo, A., Ahmed, B., Yang, X., et al. (2020). Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathog. (Basel Switzerland) 9, 312. doi: 10.3390/pathogens9040312
Yan, Y., Zhu, X., Yu, Y., Li, C., Zhang, Z., and Wang, F. (2022). Nanotechnology strategies for plant genetic engineering. Adv. Mater. (Deerfield Beach Fla.). 34, e2106945. doi: 10.1002/adma.202106945
Yang, L., Fang, S., Liu, L., Zhao, L., Chen, W., Li, X., et al. (2025). WRKY transcription factors: Hubs for regulating plant growth and stress responses. J. Integr. Plant Biol. 67, 488–509. doi: 10.1111/jipb.13828
Yang, W., Zhang, L., Yang, Y., Xiang, H., and Yang, P. (2024a). Plant secondary metabolites-mediated plant defense against bacteria and fungi pathogens. Plant Physiol. Biochem. 217, 109224. doi: 10.1016/j.plaphy.2024.109224
Yang, M., Zhou, C., Kuang, R., Wu, X., Liu, C., He, H., et al. (2024b). Transcription factor CpWRKY50 enhances anthracnose resistance by promoting jasmonic acid signaling in papaya. Plant Physiol. 196, 2856–2870. doi: 10.1093/plphys/kiae479
Yao, L., Wang, J., Sun, J. C., He, J. P., Paek, K. Y., Park, S. Y., et al. (2020b). A WRKY transcription factor, PgWRKY4X, positively regulates ginsenoside biosynthesis by activating squalene epoxidase transcription in Panax ginseng. Ind. Crops Products. 154, 112671. doi: 10.1016/j.indcrop.2020.112671
Yao, D. M., Zou, C., Shu, Y. N., and Liu, S. S. (2020a). WRKY Transcription Factors in Nicotiana tabacum Modulate Plant Immunity against Whitefly via Interacting with MAPK Cascade Pathways. Insects 12, 16. doi: 10.3390/insects12010016
Yu, H., Liao, J., Jiang, Y., Zhong, M., Tao, S., Chai, S., et al. (2025). Ecotype-specific phenolic acid accumulation and root softness in Salvia miltiorrhiza are driven by environmental and genetic factors. Plant Biotechnol. J. 23, 2224–2241. doi: 10.1111/pbi.70048
Yu, X. Q., Niu, H. Q., Liu, C., Wang, H. L., Yin, W., and Xia, X. (2024). PTI-ETI synergistic signal mechanisms in plant immunity. Plant Biotechnol. J. 22, 2113–2128. doi: 10.1111/pbi.14332
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030. doi: 10.1016/j.pbi.2021.102030
Yuan, Y., Ren, S., Liu, X., Su, L., Wu, Y., Zhang, W., et al. (2022). SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 234, 164–178. doi: 10.1111/nph.17977
Zhan, W., Li, D., Subramanyaswamy, S. B., Liu, Y. J., Yang, C., Zhang, H., et al. (2023). Dual-pharmacophore artezomibs hijack the Plasmodium ubiquitin-proteasome system to kill malaria parasites while overcoming drug resistance. Cell Chem. Biol. 30, 457–469.e11. doi: 10.1016/j.chembiol.2023.04.006
Zhan, C., Shen, S., Yang, C., Liu, Z., Fernie, A. R., Graham, I. A., et al. (2022). Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 27, 981–1001. doi: 10.1016/j.tplants.2022.03.002
Zhang, S., Dong, L., Zhang, X., Fu, X., Zhao, L., Wu, L., et al. (2023b). The transcription factor GhWRKY70 from gossypium hirsutum enhances resistance to verticillium wilt via the jasmonic acid pathway. BMC Plant Biol. 23, 141. doi: 10.1186/s12870-023-04141-x
Zhang, W., Gao, T., Li, P., Tian, C., Song, A., Jiang, J., et al. (2020). Chrysanthemum CmWRKY53 negatively regulates the resistance of chrysanthemum to the aphid Macrosiphoniella sanborni. Horticult. Res. 7, 109. doi: 10.1038/s41438-020-0334-0
Zhang, M. and Zhang, S. (2022). Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 64, 301–341. doi: 10.1111/jipb.13215
Zhang, J., Zhao, H., Chen, L., Lin, J., Wang, Z., Pan, J., et al. (2023a). Multifaceted roles of WRKY transcription factors in abiotic stress and flavonoid biosynthesis. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1303667
Zhao, L., Liu, Y., Wang, M., Xiang, L., Wang, H., Chen, X., et al. (2025). MdWRKY20-MdPR1 module mediates resistance of apple to Fusarium solani. Fruit Res. 5, e001. doi: 10.48130/frures-0024-0033
Zhao, Y., Liu, G., Yang, F., Liang, Y., Gao, Q., Xiang, C., et al. (2023). Multilayered regulation of secondary metabolism in medicinal plants. Mol. Horticult. 3, 11. doi: 10.1186/s43897-023-00059-y
Zheng, Z., Mosher, S. L., Fan, B., Klessig, D. F., and Chen, Z. (2007). Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 7, 2. doi: 10.1186/1471-2229-7-2
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zheng, L., Qiu, B., Su, L., Wang, H., Cui, X., Ge, F., et al. (2022). Panax notoginseng WRKY Transcription Factor 9 Is a Positive Regulator in Responding to Root Rot Pathogen Fusarium solani. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.930644
Zhou, R., Dong, Y., Liu, X., Feng, S., Wang, C., Ma, X., et al. (2022c). JrWRKY21 interacts with JrPTI5L to activate the expression of JrPR5L for resistance to Colletotrichum gloeosporioides in walnut. Plant J. 111, 1152–1166. doi: 10.1111/tpj.15883
Zhou, J., Mu, Q., Wang, X., Zhang, J., Yu, H., Huang, T., et al. (2022b). Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell. 34, 3066–3087. doi: 10.1093/plcell/koac139
Zhou, T., Sun, J., Zhai, Y., Gao, C., Ruhsam, M., and Wang, X. (2022a). Transcriptome profiles of yellowish-white and fuchsia colored flowers in the Rheum palmatum complex reveal genes related to color polymorphism. Plant Mol. Biol. 110, 187–197. doi: 10.1007/s11103-022-01299-0
Zhou, W., Wang, C., Hao, X., Chen, F., Huang, Q., Liu, T., et al. (2024). A chromosome-level genome assembly of anesthetic drug-producing Anisodus acutangulus provides insights into its evolution and the biosynthesis of tropane alkaloids. Plant Commun. 5, 100680. doi: 10.1016/j.xplc.2023.100680
Keywords: medicinal plants, immune responses, WRKY transcription factors, molecular mechanisms, immune regulation
Citation: Zhong X, Chen N, Li H, Wang Y, Guo Z, Shi G, Zhan X and Li L (2025) Advances in WRKY regulation of immune responses in medicinal plants. Front. Plant Sci. 16:1659732. doi: 10.3389/fpls.2025.1659732
Received: 04 July 2025; Accepted: 18 September 2025;
Published: 01 October 2025.
Edited by:
João Paulo Rodrigues Marques, University of São Paulo, BrazilReviewed by:
Jinsong Wu, Chinese Academy of Sciences (CAS), ChinaPreeti Arya, Institute of Microbial Technology (CSIR), India
Lifang Sun, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2025 Zhong, Chen, Li, Wang, Guo, Shi, Zhan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Li, bGlsaW56bWMyMDE1QDE2My5jb20=
 Xinying Zhong
Xinying Zhong Nana Chen
Nana Chen Hongwei Li
Hongwei Li Ziyi Guo
Ziyi Guo Lin Li
Lin Li