- The Energy Plant Research and Development Center, Wuhan Rundo Biotechnology LLC., Wuhan, China
Introduction: Arundo species have long served as vital raw materials for human livelihoods, yet their phylogenetic relationships remained poorly resolved until recent decades.
Methods: This study identifies a novel species, Arundo smaragdina, through integrative analyses of morphology, multiple nucleotide polymorphism (MNP) markers, and chloroplast genome data, and elucidates its phylogenetic placement within the genus.
Results: A. smaragdina is characterized by 72 chromosomes (2n=72), solid and limited rhizomes, erect and branched culms, glabrous nodes, and inflorescences emerging in early September. Mature inflorescences contain 2–5 florets per spikelet, with lemma hairs perpendicularly inserted at the basal region, and the pollen germination rate averages 12.7%. Within the genus, A. formosana is confirmed as the basal species. A. donax (a species potentially of Asian origin) differs from A. smaragdina in having spreading rhizomes and lemma hairs obliquely distributed on the lower quarter, although both species share morphological convergence and similar yields. While A. smaragdina is distinct from the Mediterranean A. plinii complex (including A. plinii, A. donaciformis, and A. micrantha), which possesses hollow rhizomes and 1(2) florets per spikelet, it shares similar pollen germination rates and chromosome numbers with some clones of A. plinii, and exhibits parallels with A. micrantha in yield, chromosome count, and branched culm architecture. At the molecular level, MNP markers confirm the genomic distinctiveness of A. smaragdina from A. donax, while chloroplast phylogeny reveals its intermediate phylogenetic position between A. donax and the A. plinii complex. Molecular dating estimates divergence times of approximately 2.29 million years ago (Mya) from A. plinii and ~2.9 Mya from A. donax.
Discussion: The congruent morphological and molecular evidence suggests that A. smaragdina may have played a pivotal role in the evolution of Arundo species.
1 Introduction
Species identification plays a crucial role in biological research, biodiversity conservation, and species utilization (Wagensommer, 2023; Perrino et al., 2024). Traditional morphological identification methods, still largely used (Di Pietro et al., 2016; Masoumi et al., 2025), while foundational, present limitations as they require extensive taxonomic expertise, access to holotype specimens, and continuous updates to taxonomic keys (Balakrishnan, 2005). With the development of DNA sequencing techniques, DNA markers and chloroplast genome sequencing have been recognized as powerful tools for species discrimination and discovery (Hebert et al., 2003; Antil et al., 2023; Dobrogojski et al., 2020).
Chloroplast genomes, which are maternally inherited in most plants, serve as another valuable tool for species identification, phylogenetics, and evolution studies (Ahmad et al., 2023; Dobrogojski et al., 2020). Chloroplast DNA typically constitutes 5-10% of total cellular DNA extracts (Ahmed, 2015) and has been shown to be fully transcribed (Shi et al., 2016). Multiple tools are currently available for chloroplast genome assembly (Freudenthal et al., 2020), and some studies have successfully reconstructed near-complete chloroplast genomes directly from leaf transcriptome data (Osuna-Mascaró et al., 2018; Senthilkumar et al., 2021). These advancements could significantly improve the resolution of phylogenetic analyses.
Arundo belongs to the subfamily Arundinoideae (Teisher et al., 2017). Molecular dating analyses suggest that Arundo formosana Hack. is the basal species within the genus (Jike et al., 2020). A recent study by Luo et al. (2024) proposes that A. formosana may have diverged from the other Arundo species, and A. donax L. cv. Lvzhou No. 1, collected from Fujian Province, China, may represent an ecotype with a different genetic origin from the other A. donax varieties. Notably, genetic evidence demonstrates that A. donax likely originated in Asia and subsequently dispersed globally via human migration (Mariani et al., 2010; Hardion et al., 2014; Canavan et al., 2017). In the Chinese mainland, we inferred the existence of three groups (common, emerald and versicolor) of A. donax (Ren et al., 2023a, 2024). But cytogenetic analyses revealed 118 chromosomes in the common group and the versicolor group, whereas the emerald group possesses 72 chromosomes. Whole-genome sequencing estimated a divergence time of 25.03 million years (MYA) between clones 0004 (the common group) and 0408 (the emerald group), and multiple nucleotide polymorphism (MNP) markers detected over 90% polymorphic loci (the percentage of different loci) between the two groups (Ren et al., 2023a, 2023b). These findings suggest that the emerald group likely represents a distinct species within Arundo, rather than belonging to A. donax.
However, while these preliminary findings from our group suggested the emerald group was a distinct taxonomic entity, a formal taxonomic treatment integrating comprehensive data was lacking. This study was therefore designed to address this knowledge gap by aiming to: 1) Conduct comparative morphological analyses and MNP marker assessments to characterize the emerald and common groups. 2) Further validate the reliability of the near-complete chloroplast genome assembled from transcriptome data, and verify the currently known chloroplast genomes within the genus Arundo. 3) Confirm that the emerald group (e.g., clone 0408) represents a novel species within the genus Arundo based on chloroplast genome data, and distinguish its morphological differences from other Arundo species.
2 Materials and methods
2.1 Morphological and cytological trait survey and analysis
A total of 118 Arundo clones collected from the Chinese mainland, comprising 49 emerald group clones and 69 common group clones, were subjected to morphological analyses. Morphological analysis was divided into two parts: field observation and laboratory analysis.
Field studies were conducted at the Yueyang Experimental Station (113.01, 29.47; WGS84), Hunan Province, China. Clones were planted in April 2020 in 94 m² plots (1.4 × 1.4 m spacing), with 2 m buffer zones between plots. During autumn 2023, morphometric assessments were performed. For each clone, 11 traits were quantified in 10 randomly selected individuals. These traits were selected from a prior investigation (data not shown) of 31 traits across 9 clones due to their suitability for large-scale field measurement and their observed variation between the two groups. The selected traits including: culm height (CH), number of culm internodes per culm (NOCIPC), middle ten culm internodes length (excluding inflorescence) (MTCIL), basal culm stem internode diameter (BCSID), basal culm internode wall thickness (BCIWT), leaf length of the sixth leaf from top (LLOTSLFT), leaf width of the sixth leaf from top (LWOTSLFT), aspect ratio of leaf (AROL), inflorescence width (IW), inflorescence length (excluding peduncle) (ILEP), aspect ratio of inflorescence (AROI). PCA (using ade4) and box-and-whisker plots (generated via ggplot2) were applied to analyze morphological traits and detect potential clusters among clones.
Laboratory analyses were conducted for the inflorescence structure, rhizome growth patterns, and chromosome number of the common and emerald groups. Pollen viability in the common and emerald groups was assessed via I2-KI staining: normal, fertile pollen grains exhibited dark blue staining, whereas abnormal pollen grains remained unstained. Following the method of Hardion et al. (2015), we additionally performed pollen germination experiments.
2.2 MNP marker analysis
Leaf samples were collected from 118 clones, and genotyped using 1,100 MNP markers developed by Ren et al. (2023a). Following the method of Liu J. et al. (2024), markers with 100% amplification success across all clones were selected. Amplified sequences of clones by same maker were aligned using MAFFT v7.526 (Katoh and Standley, 2013) with default parameters. Concatenated sequences, generated by Concatenator v0.3.1 (Vences et al., 2022), served as input for phylogenetic reconstruction in IQ-TREE v2.4.0 (Minh et al., 2020). The best-fit model (GTR +F+R4) was determined by IQ-TREE according to the Bayesian Information Criterion (BIC), and 1,000 ultrafast bootstrap replicates were performed to assess node support. The final tree was visualized using iTOL (Letunic and Bork, 2024). Subsequently, genetic similarity (GS) among clones from the two groups was assessed using the formula proposed by Liu J. et al. (2024), and visualized as heatmaps via ggplot2.
2.3 Chloroplast genome assembly and comparative analysis
For chloroplast genome assembly, transcriptome data from BioProject PRJEB36611 (Jike et al., 2020) were used to assemble near-complete chloroplast genomes for five Arundo species (Arundo plinii Turra; Arundo donaciformis (Loisel.) Hardion, Verlaque & B. Vila; Arundo micrantha Lam.; Arundo formosana Hack.; Arundo donax L.) and three outgroup species (Molinia caerulea L.; Hakonechloa macra Makino; Phragmites australis (Cav.) Trin. ex Steud). Whole-genome sequencing and transcriptome data for clones 0004 and 0408, obtained from our previous studies (Ren et al., 2023b, 2024), were utilized to assemble complete and near-complete chloroplast genomes for these two clones. BioProject PRJNA974205 (Luo et al., 2024) provided data for the reassembly of the complete chloroplast genome of A. donax cv. Lvzhou No.1.
Additionally, complete chloroplast genomes of three Arundo species—A. formosana (NC_054211.1, MZ620725.1), A. plinii (NC_034652.1), and A. donax (NC_037077.1)—along with three outgroup species (M. caerulea NC_033980.1, H. macra NC_025235.1, P. australis NC_022958.1) from subfamily Arundinoideae (Poaceae), were retrieved from NCBI GenBank to infer interspecific evolutionary relationships. The boundary of A. formosana (NC_054211.1) follows the description by Feng et al. (2021).
Complete chloroplast genomes were assembled using GetOrganelle v1.7.7.0 (Jin et al., 2020) with default parameters. Near-complete chloroplast genomes were assembled following the methodology of Osuna-Mascaró et al. (2018), with A. donax (NC_037077.1) as reference. The starting positions of the assembled chloroplast genomes were standardized to align with the reference genome of A. donax (NC_037077.1). Chloroplast genome annotation was performed using CPGAVAS2 (Shi et al., 2019), while tRNA detection employed tRNAscan-SE v1.21 (Schattner et al., 2005). The circular chloroplast genome map was subsequently generated using CPGView (Liu et al., 2023). Chloroplast genome comparisons were performed using mVISTA (LAGAN algorithm mode) with A. donax (NC_037077.1) as the reference genome (Frazer et al., 2004). Sequence similarity between A. donax and other Arundo species with complete chloroplast genomes was further assessed via Circoletto (Darzentas, 2010) under an E-value threshold of 1×10⁻10. Genome junction sites were visualized using CPJSdraw (Li et al., 2023).
2.4 Phylogenetic analysis and divergence time estimation
All assembled chloroplast genomes were analyzed to resolve the evolutionary relationships within Arundo. Sequences were aligned with MAFFT v7.526 (Katoh and Standley, 2013), trimmed using TrimAL v1.4 (Capella-Gutiérrez et al., 2009), and used to reconstruct a maximum-likelihood tree in IQ-TREE v2.4.0 (Minh et al., 2020), with the best-fit model (GTR+F+I+G4) determined by IQ-TREE according to the BIC and 1,000 ultrafast bootstrap replicates performed to assess node support. The final tree was visualized using iTOL (Letunic and Bork, 2024).
Divergence time estimation included four complete Arundo chloroplast genomes (A. plinii, clone 0004, clone 0408 and A. donax) with P. australis as the outgroup. We employed the Phragmites-Arundo divergence time of 29.0 Mya (95% CI: 19.6-38.3) proposed by Christin et al. (2013), which was validated by Hardion et al. (2015) in A. formosana ecotypes. This molecular dating exhibited temporal concordance with the orogenesis of the center mountain range (~2 Ma). The divergence time estimates were performed using the MCMCTree v4.9e program (Yang, 2007).
3 Results
3.1 Morphological and cytological differentiation across clones
Field observations comparing the emerald and common groups identified significant divergence in morphological and phenological traits. The emerald group exhibited a conspicuously brighter green leaf coloration, which emerged as the most diagnostic feature enabling effortless visual distinction between the two groups in field conditions. In terms of flowering period, the common group initiated flowering approximately one and a half months earlier than the emerald group (Figure 1A). The inflorescence of the emerald group was shorter than that of the common group (Figure 1B; Supplementary Table S1), but both had branched culms (Figure 1C). A distinct contrast was observed in rhizome growth patterns (Figure 1F). Unlike the common group whose rhizomes grew upwards in a spreading pattern, the emerald group displayed limited rhizome growth without upward tendencies, though both groups had solid (non-hollow) rhizomes (Figure 1G).

Figure 1. Comparison morphological and cytological traits between the common and the emerald group. (A) Plants after 2 years (photographed in mid-September 2024): Common (left), emerald group (right). (B) Fully emerged inflorescence: Common (left two), emerald group (right). (C) Branched culms: Common (left), emerald group (right). (D) Glumes (top: lower glumes [exterior and interior], bottom: upper glumes [exterior and interior]): Common (left), emerald group (right). (E) Inflorescence structures (from left to right: secondary branch, spikelet, expanded spikelet, glumes, three florets): Common (left), emerald group (right). (F) Rhizomes after 1 year: Common (left), emerald group (right). (G) Rhizome cross-sections: Common (left), emerald group (right). (H) Chromosomes (100×): Common (left), emerald group (right). (I) Pollen germination of the emerald group. (J) Pollen staining of the emerald group. (K) Glabrous node (top three: common group, bottom three: emerald group).
Laboratory observations focused on reproductive structures provided additional insights into the divergence between the two groups. Both groups bore 2–5 florets per spikelet. However, significant differences were noted in the lemma hair distribution and orientation. The lemma hair of the emerald group was primarily distributed in the middle to lower regions of the lemma and exhibited a non-erect orientation, in contrast to the common group (Figure 1E). No discernible differences were observed in the glumes between the two groups (Figure 1D).
In terms of chromosome number, the emerald group had a lower chromosome count (2n = 72) compared to the common group (2n = 108; Figure 1H). Pollen viability assays further differentiated the two groups. Clones of the common group predominantly produced small, irregularly shaped pollen grains. In contrast, the emerald group clones (e.g., clone 0408) exhibited partial pollen staining and higher pollen germination (approximately 12.7%; Figures 1I, J).
To further investigate the differences between the two groups, PCA was performed on 11 morphological traits, revealing that the 118 clones were grouped into two phenotypically divergent groups (Supplementary Figure S1; Supplementary Table S2). Box-and-whisker plots analysis of traits further revealed significant distribution variations among the two groups, with substantial overlapping observed in most traits (Supplementary Figure S2). In terms of culm height, the common group was higher than the emerald group, which was consistent with the finding that the length of the middle ten internodes in the common group was greater than that in the emerald group. Interestingly, the emerald group had a larger number of internodes compared to the common group. Notably, no obvious differences were found in the basal stem thickness between the two groups.
Differences in flowering periods, lemma hair distributions, and pollen viability suggest potential reproductive isolation between the two groups. The PCA of 11 traits and significant rhizome morphological divergence indicate adaptation to distinct environmental conditions. These results clearly distinguish the emerald group from the common group.
3.2 Phylogenomic relationships via MNP markers
To further confirm that the emerald group does not belong to the common group at the genomic level, MNP markers were employed to genotype the two groups. The number of shared MNP markers among different clones ranged from 495 to 947 (Supplementary Table S3). A total of 304 shared MNP markers were used to classified 118 clones into two distinct groups: Clade I consisted exclusively of clones from the common group, whereas Clade II was composed solely of emerald group clones (Figure 2). These results indicate that the emerald group is genomically distinct from the common group at the genome level.
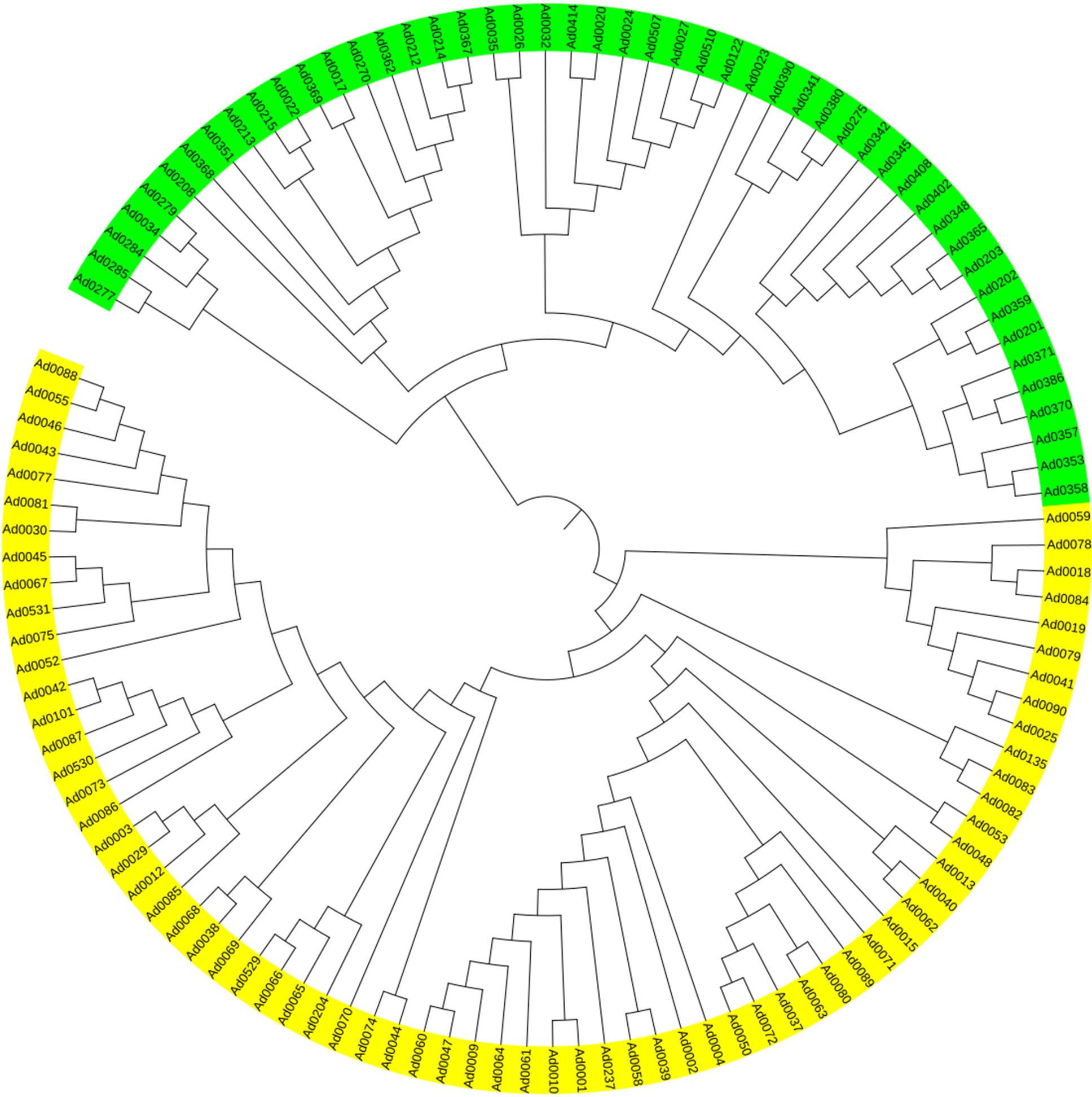
Figure 2. Clustering dendrogram based on MNP markers for 118 clones. Yellow and green represent the common group and the emerald group, respectively.
Based on genetic similarity, the emerald group was also be distinguished from the common group. The genetic similarity between the common group and the emerald group was only 6.05%-10.32%, demonstrating a profound genomic divergence consistent with species-level separation (Figure 3). Within-group genetic similarity was markedly higher in the common group (96.2%-100%; Supplementary Figure S3) than in the emerald group (58.9%-100%; Supplementary Figure S4), indicating that the emerald group possesses a more diverse gene pool. This pattern aligns with morphological observations, where the common group exhibited significantly lower pollen viability compared to the emerald group (Figure 1I). These findings collectively demonstrate that the emerald group represents a genomically distinct entity from the common group (A. donax). This genomic differentiation aligns with morphological and cytological observations, suggesting the emerald group represents a novel taxon in the Chinese mainland.
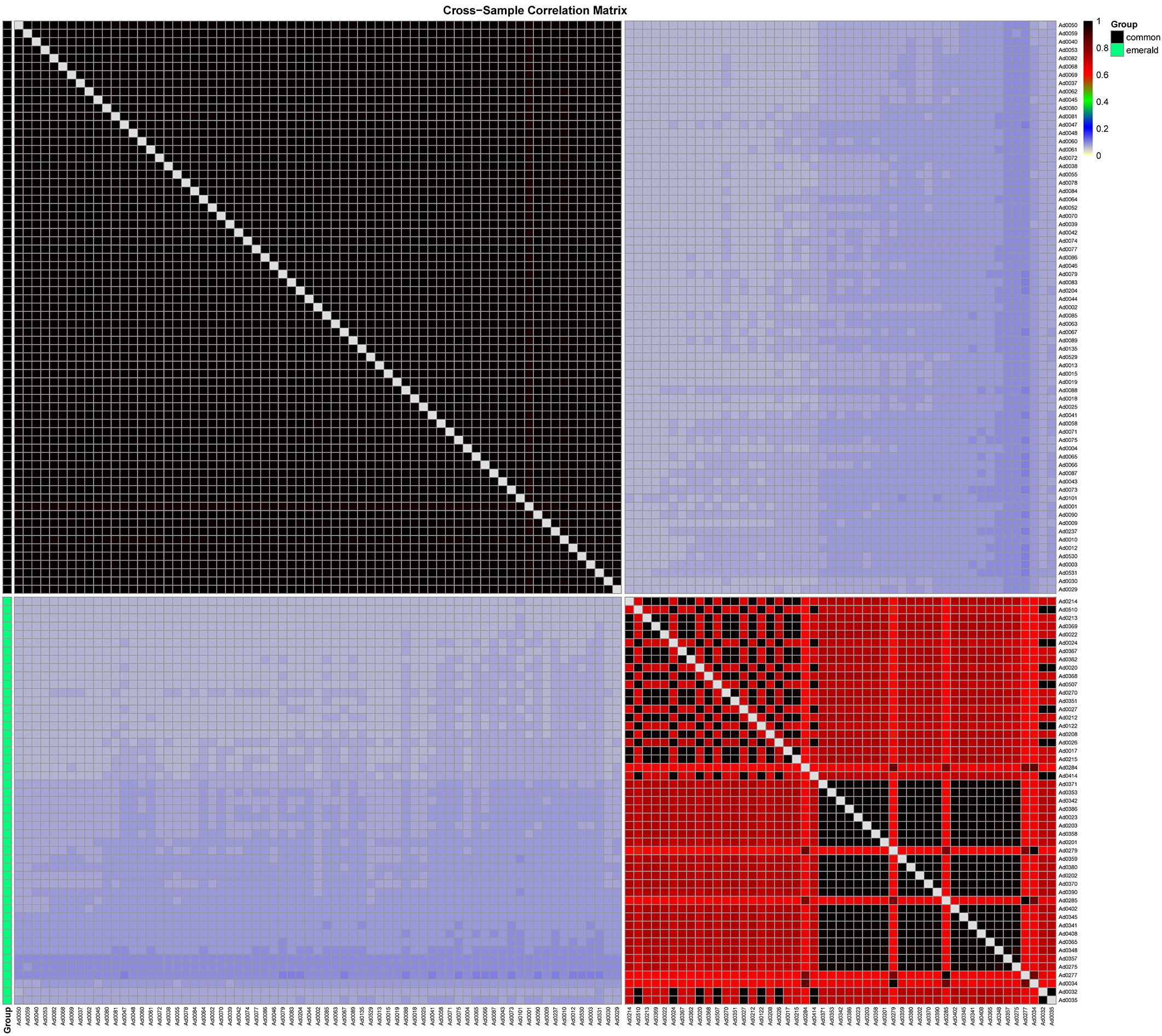
Figure 3. Heatmap of genetic similarity between two groups. The common and emerald groups are color-coded in black and green, respectively, on the left side. The genetic similarity values are provided in the top-right corner.
3.3 Chloroplast genome assembly
To further confirm that the emerald group does not belong to A. donax or the other four species within the genus Arundo, further research was conducted at the chloroplast genome level. Based on NGS sequencing data, the complete chloroplast genomes of clones 0408 and 0004 were successfully assembled. Additionally, the complete chloroplast genome of A. donax cv. Lvzhou No. 1 was also successfully reassembled from its raw data. Assembly of transcriptome data yielded 12 near-complete chloroplast genomes. All assembled chloroplast genomes are listed in Supplementary Table S4.
The complete chloroplast genome of clone 0408 (137,185 bp) exhibited a typical quadripartite angiosperm structure, comprising large single-copy (LSC: 82,092 bp), small single-copy (SSC: 12,613 bp), and inverted repeat (IR: 21,240 bp) regions (Supplementary Figure S5). The complete chloroplast genome of clone 0004 was identical to that of A. donax cv. Lvzhou No. 1 as reassembled (Supplementary Figure S6). Published A. formosana genomes (MZ620725.1, NC_054211.1) are completely identical (Supplementary Table S5).
3.4 Chloroplast genome comparison
Compared with the complete chloroplast genomes of clones 0004 and 0408, the near-complete chloroplast genomes exhibited significantly shorter lengths across four regions. Notably, all nine near-complete chloroplast genomes of Arundo species shared identical low-similarity and missing regions, including extensive sequence deletions near 84–87 k bp, 92–93 and 132–134 k bp (Supplementary File S1). Furthermore, sequencing strategy significantly influenced genome assembly quality. For example, the near-complete chloroplast genome assembled from BioProject PRJEB36611 exhibited deletions in the 46–48 and 64–65 k bp regions, whereas clones 0004 and 0408 displayed no such gaps, highlighting the impact of sequencing methodology on sequence integrity and confirming the feasibility of assembling a complete chloroplast genome from transcriptome data.
Using A. donax (NC_037077.1) as the reference, a comprehensive genomic variation analysis of the complete chloroplast genomes across Arundo species was performed (Figure 4). Results revealed that all analyzed genomes, except A. formosana, exhibited high similarity to A. donax. The inclusion of chloroplast genomes assembled from transcriptome data in the comparative analysis similarly yielded consistent results (Supplementary File S1), further confirming that the emerald group (e.g., clone 0408) belongs to the genus Arundo and demonstrating the reliability of chloroplast genomes assembled from transcriptome data.
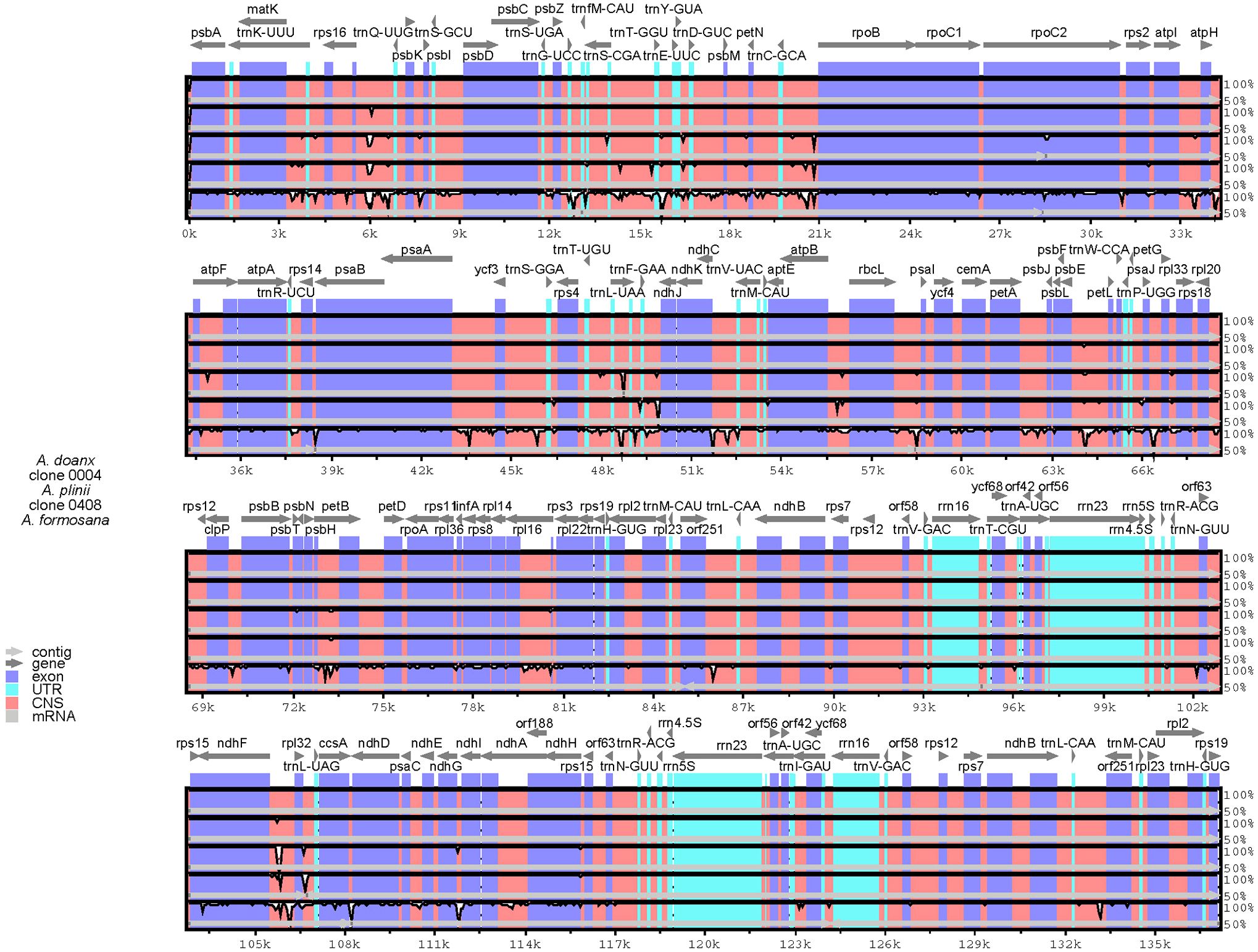
Figure 4. Sequence variation plots among the Arundo chloroplast genomes. Annotated genes are displayed on the top. The vertical scale represents the percentage within 50–100% homogeneity. The color legend is summarized in the lower left-hand corner.
Notably, in contrast to the reference genome A. formosana (NC_054211.1), the near-complete chloroplast genome of A. formosana assembled from transcriptome data showed higher overall similarity to A. donax (Supplementary File S1). Furthermore, collinearity analysis (Supplementary Figure S7) revealed that the reference genome A. formosana (NC_054211.1) exhibited lower similarity to A. donax than P. australis or M. caerulea, with most regions showing less than 25% collinearity. The reference genome A. formosana (NC_054211.1) may warrant further investigation. In contrast, A. plinii showed the best collinearity with A. donax, followed by clone 0408, indicating that clone 0408 differed from both A. plinii and A. donax at the chloroplast genome level.
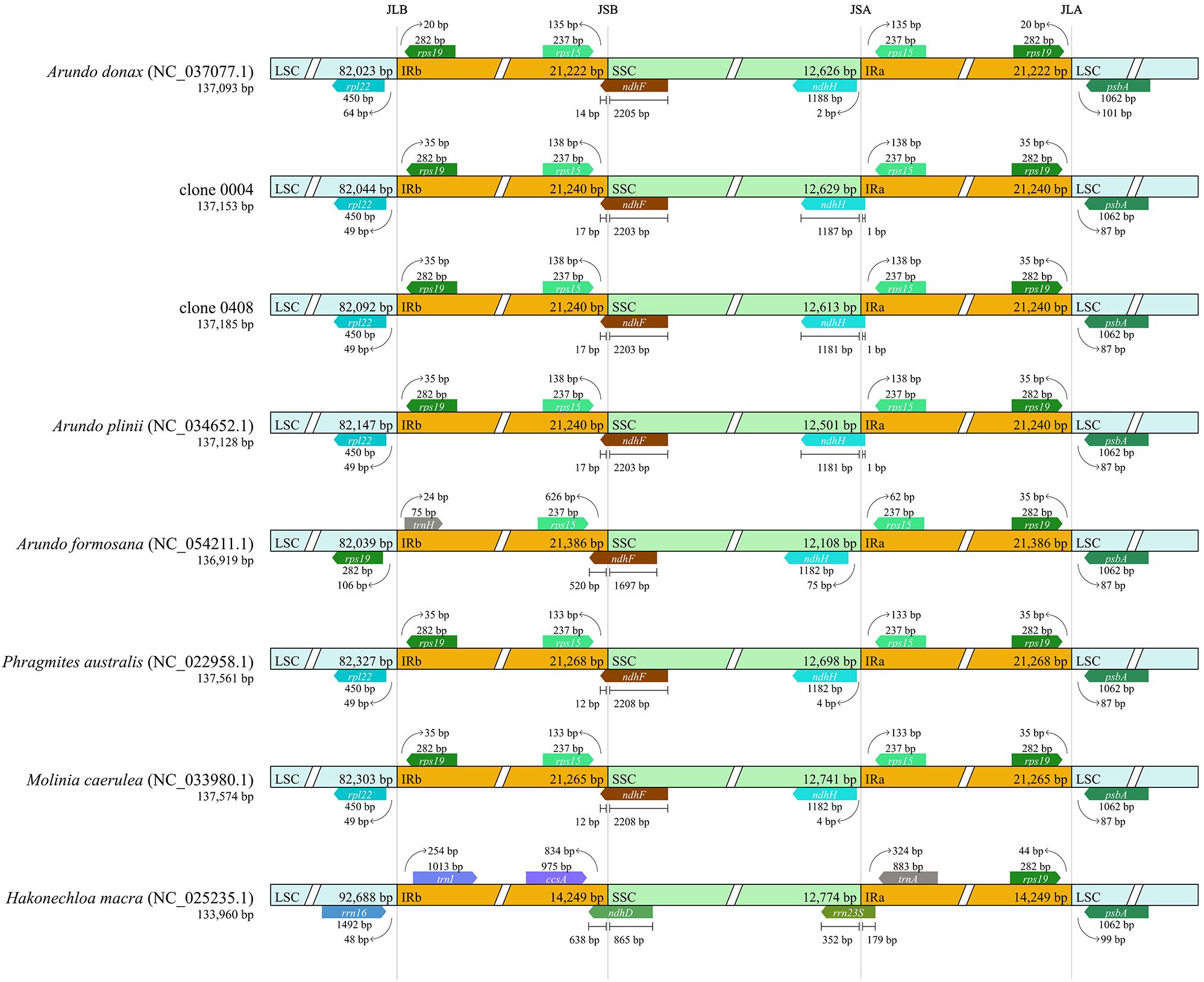
Inverted repeat (IR) boundary analysis revealed that A. plinii, clones 0004 and 0408 within the genus Arundo shared identical IR boundaries and lengths. In contrast, the Italian A. donax (NC_037077.1) exhibited an IR contraction compared to clone 0004, a Chinese A. donax clone (Figure 5; Supplementary Table S6). The observed IR contraction in the Italian A. donax could reflect an evolutionary change following geographic isolation, though its functional significance requires further study.
The high similarity at the chloroplast genome level further confirmed that the emerald group belongs to the genus Arundo. However, collinearity analysis revealed significant differences in the chloroplast genomes between the emerald group (e.g., clone 0408) and other Arundo species, providing further evidence that it is likely a new species within the genus.
3.5 Phylogenetic resolution of Arundo
To determine the phylogenetic relationships between the emerald group and other species within the genus Arundo, a chloroplast genome-based phylogenetic tree was constructed. In the maximum likelihood (ML) phylogenetic tree (Figure 6; Supplementary Figure S8), the near-complete chloroplast genomes, with the exception of A. formosana, clustered with their corresponding complete reference genomes, confirming the reliability of transcriptome-derived chloroplast data.
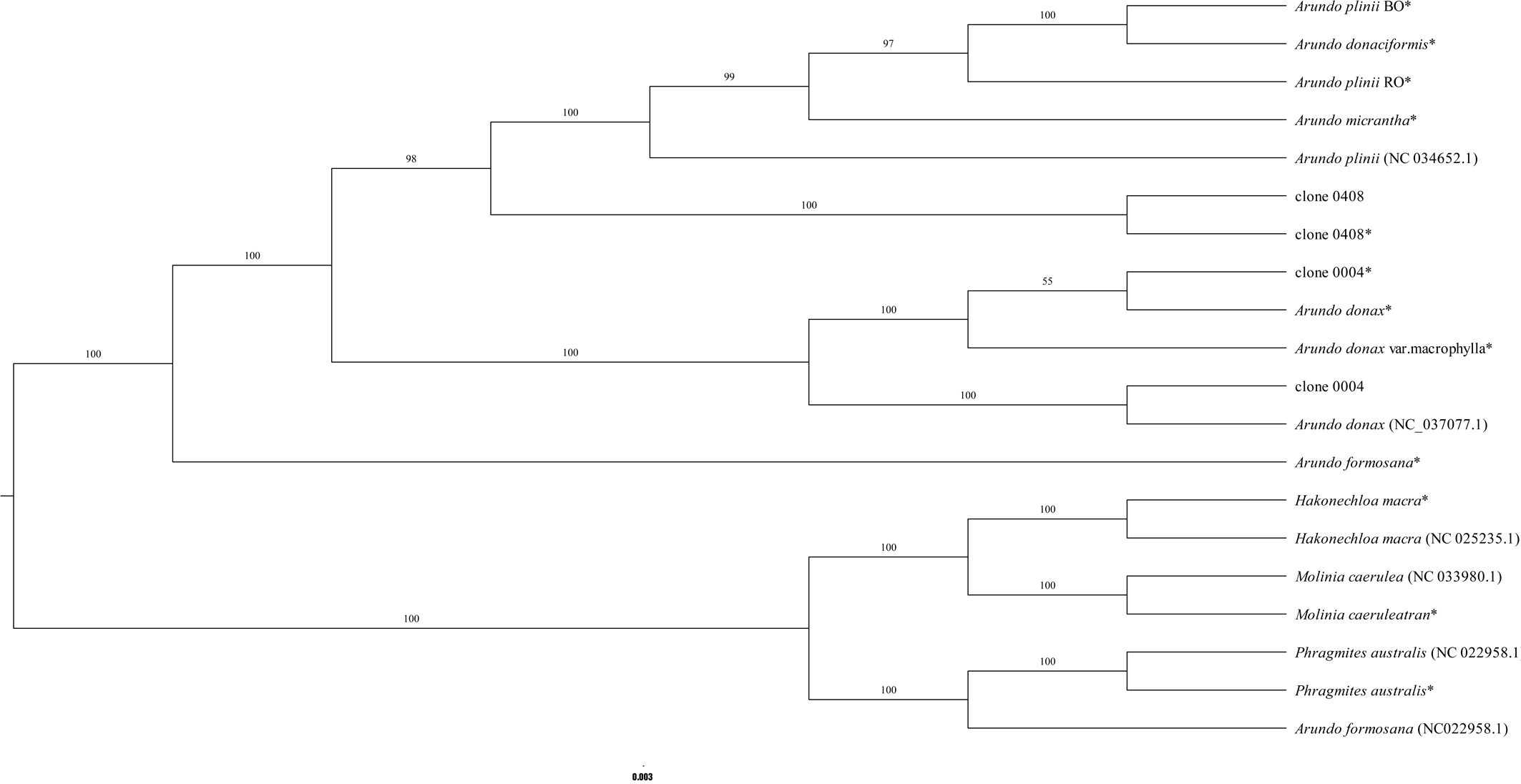
Figure 6. Phylogenetic relationships of Arundo species inferred from chloroplast genomes. “*” indicates a nearly complete chloroplast genome assembled from transcriptome data.
Clone 0408 did not cluster with any recognized Arundo species, forming an independent lineage that robustly supports its designation as a novel species. Clone 0004, whose chloroplast genome sequence is identical to that of A. donax cv. Lvzhou No.1, formed a clade with A. donax (NC_037077.1). A notable discrepancy was observed in the phylogenetic placement of A. formosana. While the near-complete chloroplast genome was clustered within the genus Arundo as expected, the published reference chloroplast genome of A. formosana (NC_054211.1) formed a distinct clade with P. australis (NC_022958.1) and other outgroup species (Figure 6; Supplementary Figure S8). This anomalous phylogenetic placement, inconsistent with the transcriptome-based assembly, reinforces the concern raised in section 3.4 that the reference genome of A. formosana (NC_054211.1) may warrant further investigation.
Chloroplast genome divergence provides further validation that the emerald group (e.g., clone 0408) represents a novel species. Divergence from other species in nuclear and chloroplast genomes, morphological traits, and cytological features indicates adaptive modifications during its historical evolutionary process. We propose the name Arundo smaragdina for this species, reflecting its distinctive leaf pigmentation.
3.6 Divergence time analysis
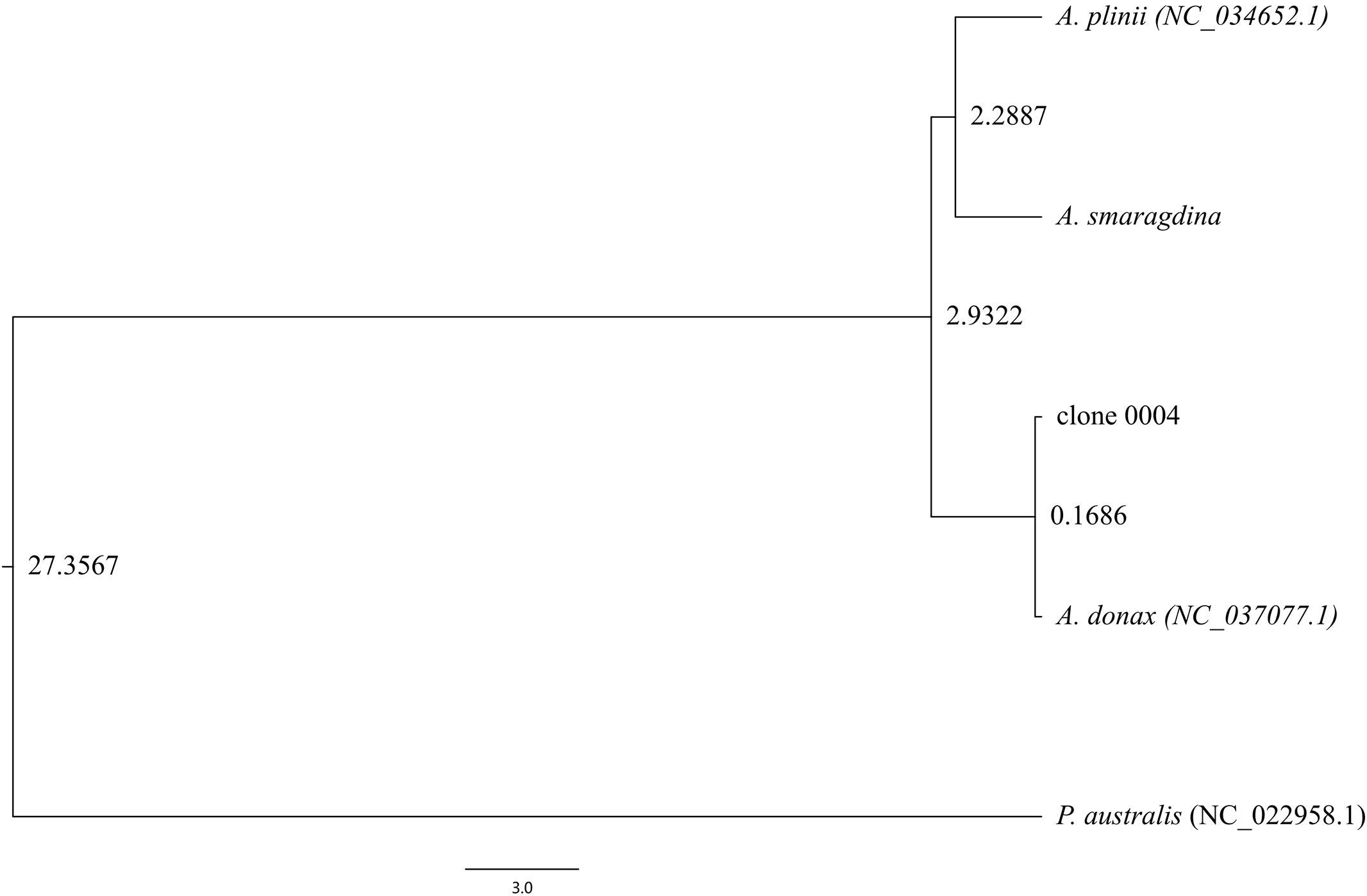
Integration of the A. smaragdina chloroplast genome with published Arundo genomes and the outgroup P. australis revealed key divergence events within the genus (Figure 7). Phylogenetic analysis indicated that A. smaragdina and A. plinii diverged 2.29 million years ago (Mya), with this lineage splitting from A. donax 2.9 Mya. Within A. donax, the Italian clone (NC_037077.1) and clone 0004 diverged more recently, approximately 0.17 Mya.
This temporal evidence, consistent with the morphological, cytological, and genomic divergences documented in previous sections, offers robust and multi-layered confirmation of A. smaragdina as a distinct species within the genus Arundo.
4 Discussion
4.1 MNP markers support genomic divergence between A. smaragdina and A. donax
Previous studies have employed various molecular markers and partial chloroplast genes to explore intraspecific diversity within A. donax, but these approaches demonstrated limited discriminatory power for distinguishing A. donax clones (Bucci et al., 2013; Pilu et al., 2013; Touchell et al., 2016; Liu et al., 2019; Tarin et al., 2013). To address this, we developed 1,100 MNP markers, which mapped uniformly across the A. donax reference genome (Ren et al., 2023a, 2024; Supplementary Figure S9). MNP marker technology has emerged as a powerful tool for variety identification, demonstrating success across diverse species including Oryza sativa L., Dendranthema morifolium (Ramat.) Tzvelev, Lentinula edodes (Berk.) Pegler, and Vitis vinifera L (Fang et al., 2021; Ling et al., 2023; Liu J. et al., 2024; Liu Y. et al., 2024). Phylogenetic clustering of A. smaragdina and A. donax into distinct groups underscores their divergence at the genomic level (Figure 2).
The significantly lower genetic similarity among different clones of A. smaragdina, compared to the high clonal uniformity within A. donax (Supplementary Figures S3, S4; Supplementary Table S3), suggests a history of sexual reproduction and the maintenance of a more diverse gene pool, which is consistent with the extensive phenotypic variation (e.g., in leaf morphology, tiller number, and flowering time) observed in A. smaragdina in the field. However, A. donax exhibits relatively limited phenotypic variation in the field. The most common phenomenon is that some individuals of the common group transform into the versicolor groups due to somatic mutations resulting foliar variegation (Antal et al., 2018; Guarino et al., 2019; Danelli et al., 2020; Ren et al., 2023b). SRAP and TE-based molecular markers failed to detect genetic differentiation between these morphotypes (Ahmad et al., 2008). Comprehensive differences in genetic (MNP markers), cytological (chromosome number), reproductive (pollen germination rate), and ecological (ecotype diversity) traits demonstrate potential early reproductive isolation mechanisms, indicating that A. smaragdina and A. donax split into distinct species early in their evolutionary history.
4.2 Morphological and cytological evidence for A. smaragdina as a new species
The genus Arundo exhibits substantial morphological plasticity (Ngernsaengsaruay et al., 2023; Cantaluppi et al., 2015). In this genus, A. smaragdina exhibits distinct morphological differences compared to the other five species.
Compared to other Arundo species, A. formosana exhibits a prostrate growth habit, ~1 m height, and is endemic as a pioneer grass in estuarine environments (Hardion et al., 2017a). Its restricted distribution further underscores its taxonomic uniqueness within the genus (Liu and Sylvia, 2006; Lee et al., 2022).
The A. plinii complex (A. plinii, A. donaciformis, and A. micrantha) represents a circum-Mediterranean taxon, distinguished from A. donax and A. smaragdina by its spikelet structure, which bears 1(2) florets compared to 3–5 florets in the latter two (Figure 1E; Danin, 2004; Hardion et al., 2012a). Additionally, the A. plinii complex exhibits thinner rhizomes characterized by a parenchymous cross-section with a central lumen (Danin and Naenny, 2008), contrasting sharply with the solid, lumenless rhizomes of A. donax and A. smaragdina (Figure 1G).
Among the complex members, A. micrantha is further differentiated by a larger culm diameter exceeding 5 mm under the panicle (vs. <4 mm in A. plinii and A. donaciformis; Danin, 2004; Hardion et al., 2012a); and greater similarity to A. smaragdina (4.7-12.9 mm) and A. donax (9.3-18.2 mm). Intriguingly, A. smaragdina shares rhizome growth patterns with A. micrantha (Figure 1F; Danin, 2004). However, the inflorescence architecture of A. micrantha more closely resembles that of A. donax, differ significantly from A. smaragdina (Figure 1B; Hardion et al., 2012b; Tomàs et al., 2019; Ferrer-Gallego and Hardion, 2025). The branched culms of A. micrantha distinguish it very well from A. plinii (Danin, 2004); however, A. smaragdina and A. donax also have branched culms (Figure 1C).
A. donaciformis exhibits pubescent nodes and hairy upper glumes, while A. micrantha and A. plinii s.str. are marked by glabrous nodes and glabrous upper glumes, respectively (Hardion et al., 2012a). Moreover, A. donax and A. smaragdina are glabrous at the nodes, lower glumes, and upper glumes (Figures 1D, K). Additionally, in A. smaragdina, hairs are primarily distributed in the middle to lower regions of lemma and exhibit a non-erect orientation similar to A. donaciformis, whereas in A. donax, hairs are notably longer and closely appressed to the lemma surface (Figure 1E; Hardion et al., 2012a).
Regarding Italy and Asian A. donax (e.g., clone 0004), the two exhibit no significant morphological differences in traits such as the lemma and root system (Figure 1; Danin, 2004; Hardion et al., 2012a), indicating the reliability of morphological identification for species differentiation. Furthermore, both this study and the research by Hardion et al. (2012a) demonstrate that PCA and box-and-whisker plots analysis are also important tools for species discrimination within the genus.
Pollen germination and chromosome number also provide valuable insights for species identification. Germination rates of A. smaragdina align closely with those of A. plinii and A. donaciformis within the genus (Figure 1I; Supplementary Table S7; Hardion et al., 2015). However, unlike A. donax populations in the Middle East that exhibit seed set (Danin, 2004; Hardion et al., 2014), no seeds were observed in A. smaragdina or A. donax in the Chinese mainland under ex situ conditions. This sterility in A. smaragdina may mirror the reproductive behavior of A. plinii, which produces seeds exclusively in its native habitat (Hardion et al., 2015). Cytogenetic analyses further differentiate A. smaragdina: its karyotype (2n=72) contrasts with A. donax (2n=108) and A. donaciformis (2n=108), while aligning with A. micrantha (2n=70-72) and some cytotypes of A. plinii (2n=72, 74, 76, 108, 114; Hardion et al., 2015).
Collectively, A. smaragdina is morphologically distinct from all known Arundo species. However, its morphological convergence with A. donax—including shared traits such as plant height and perennial habit—likely resulted in its historical misclassification as a non-distinct taxon within the genus.
4.3 Chloroplast genome support A. smaragdina as a new species
Species-specific clustering validated the utility of transcriptome-derived chloroplast genomes, as these genomes consistently grouped with their complete genome counterparts, consistent with previous studies (Figure 6; Supplementary Figure S8; Osuna-Mascaró et al., 2018; Senthilkumar et al., 2021). In contrast to studies using partial chloroplast genes for interspecific comparisons, nearly complete chloroplast genomes provide more comprehensive genetic information, particularly when species divergence times are short. For example, Jike et al. (2020) analyzed Arundo species using five intergenic regions, yielding results slightly divergent from nuclear genome-based studies (specifically the phylogenetic relationships among three species in the A. plinii complex). In contrast, our findings based on nearly complete chloroplast genomes showed strong consistency with nuclear genome analyses (Figure 6, Supplementary Figure S8; Jike et al., 2020). Furthermore, the near-complete chloroplast genome of A. smaragdina clustered with its complete genome, supporting its novel species status.
A comparative analysis of the chloroplast genome between A. smaragdina and other Arundo species revealed high sequence similarity between A. smaragdina and other species, whereas notable differences were observed in the IR regions: A. smaragdina exhibited greater similarity to the IR regions of A. plinii and the Chinese mainland A. donax, while the IR region of Italian A. donax was contracted. These results imply that A. smaragdina contributed to the evolutionary radiation of Arundo species.
4.4 Interspecific relationships within genus Arundo
Phylogenetic topology aligned with prior studies, positioning A. formosana as the basal taxon within the genus Arundo (Figure 6; Supplementary Figure S8; Jike et al., 2020). Interestingly, the published A. formosana (NC_054211.1) chloroplast genome did not cluster with the near-complete chloroplast genome, but instead grouped with P. australis and other outgroup species, a finding consistent with other phylogenetic studies that also reported its clustering with species such as Crinipes abyssinicus Hochst. and Crinipes longifolius C.E.Hubb., outside the core Arundo lineage (Supplementary Figure S8; Luo et al., 2024). The genomic comparisons (Supplementary File S1) also consistently demonstrate significant differences between the A. formosana reference genome (NC_054211.1) and A. donax (NC_037077.1). Furthermore, similarity analysis (Supplementary Figure S7) indicates that their degree of similarity is even lower than that between A. donax and either P. australis or M. caerulea. These data strongly suggest a fundamental issue with the taxonomic identity of the source material used for its assembly. Notably, this published genome was reportedly sampled from Yunnan Province (Feng et al., 2021). Meanwhile, two A. formosana specimens from Sichuan Province are archived at the Chengdu Institute of Biology, Chinese Academy of Sciences (https://www.cvh.ac.cn/spms/detail.php?id=d7e77a3a), underscoring the need to validate distribute localities for A. formosana (Liu and Sylvia, 2006; Lee et al., 2022). For A. donax, integrating the reassembled chloroplast genomes of A. donax cv. Lvzhou No.1 with the datasets used by Luo et al. (2024) revealed that it clustered with other A. donax chloroplast genomes, contradicting Luo et al.’s hypothesis that “A. donax cv. Lvzhou No.1 (OQ993163.1) may represent a variety with a different genetic origin from the other A. donax” (Supplementary File S2; Supplementary Figure S8; Luo et al., 2024). Crucially, clone 0004 exhibited distinct morphological traits (e.g., sprawling growth habit, taller culms) despite chloroplast genome identity with A. donax cv. Lvzhou No.1 (Ren et al., 2023b; Luo et al., 2024). Molecular clock analyses reveal synchronized divergence events between East Asian and Mediterranean lineages: P. australis populations in China’s Hexi Corridor diverged from European counterparts 0.186 Mya (Qiu and Cui, 2021), contemporaneous with the ~0.17 Mya split between A. donax lineages (Figure 7). This temporal synchronicity reinforces hypotheses of human-mediated dispersal facilitating Mediterranean A. donax expansion (Hardion et al., 2017b).
For the A. plinii complex, divergence time estimation indicates that A. smaragdina and A. plinii diverged approximately 2.29 MYA (Figure 7). This period, characterized by initial cooling and mild rainfall reduction followed by the consolidation of an arid Mediterranean climate, temporally coincides with both the divergence of A. plinii complex taxa and analogous speciation patterns observed in Avena species (Liu et al., 2017). Furthermore, the divergence among the three species within the A. plinii complex requires further clarification. Neither nuclear genome nor chloroplast genome analyses have clustered A. plinii BO and A. plinii RO into a single clade (Figure 6; Jike et al., 2020), likely attributable to both the slower evolutionary rate of the chloroplast genome and the limited nuclear gene dataset analyzed (comprising only 144 genes).
Regarding A. micrantha within this complex, Jike et al. (2020) proposed that it likely originated from hybridization between A. plinii and a low-ploidy fertile Asian A. donax. The identification of A. smaragdina provides evidence supporting an alternative hypothesis: A. micrantha more likely originated from hybridization between A. plinii and A. smaragdina. The supporting evidence is as follows (Supplementary Table S7): (1) Morphologically, A. smaragdina shares similarities with A. micrantha in rhizome growth patterns, exhibiting limited growth, which contrasts with the spreading growth habit observed in A. donax. Furthermore, the culm diameter under the panicle of A. micrantha (>5mm) is partially similar to that of A. smaragdina (4.7-12.9 mm), exceeding 5 mm. This characteristic also helps distinguish A. micrantha from A. plinii (< 4 mm) and A. donaciformis (< 4 mm). (2) The nearly identical pollen germination rates between A. smaragdina (12.7%) and A. plinii (7.8%-8%) suggest reproductive compatibility, which is a prerequisite for successful hybridization. In contrast, the pollen germination rate for A. donax is 0%. (3) Cytological analysis reveals that the chromosome numbers of both A. smaragdina and A. micrantha are approximately 72. This number falls within the range (72-114) reported for some A. plinii accessions, indicating a shared cytological background that could facilitate hybridization. (4) Phylogenetic reconstruction based on chloroplast genomes indicates a closer genetic relationship among A. micrantha, A. plinii, and A. smaragdina than with A. donax.
In conclusion, integrative analyses of morphological traits, chromosome numbers, MNP markers, and chloroplast genomes provide compelling evidence that A. smaragdina represents a distinct species within Arundo, diverging from A. donax and other species. At the nuclear genome level, A. smaragdina exhibits genetic divergence from A. donax based on MNP analysis, whereas at the chloroplast genome level, it shares structural similarities with the Chinese mainland A. donax and A. plinii. Morphologically, A. smaragdina combines high-yield characteristics of A. donax and A. micrantha with pollen germination traits similar to A. plinii. Collectively, these findings demonstrate that A. smaragdina occupies a pivotal evolutionary role within the genus Arundo. The discovery of this species provides a critical missing link for elucidating both the evolutionary history and biogeographic dispersal of the genus, thereby establishing a robust foundation for future initiatives in breeding programs, conservation strategies, and sustainable utilization of its members.
Data availability statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA027255) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Author contributions
FL: Writing – review & editing, Writing – original draft. BL: Validation, Writing – review & editing. QH: Investigation, Writing – review & editing. AZ: Investigation, Writing – review & editing. BX: Investigation, Writing – review & editing. XS: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Wuhan Rundo Biotechnology, LLC.
Conflict of interest
All authors were employed by Wuhan Rundo Biotechnology, LLC. during the research.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1660442/full#supplementary-material
References
Ahmad, W., Asaf, S., Al-Rawahi, A., Al-Harrasi, A., and Khan, A. L. (2023). Comparative plastome genomics, taxonomic delimitation and evolutionary divergences of Tetraena hamiensis var. Qatarensis and Tetraena simplex (Zygophyllaceae). Sci. Rep. 13, 1. doi: 10.1038/s41598-023-34477-1
Ahmad, R., Liow, P. S., Spencer, D. F., and Jasieniuk, M. (2008). Molecular evidence for a single genetic clone of invasive Arundo donax in the United States. Aquat. Bot. 88, 113–120. doi: 10.1016/j.aquabot.2007.08.015
Ahmed, I. (2015). Chloroplast genome sequencing: Some reflections. J. Next Generation Sequencing Appl. 2, 2. doi: 10.4172/2469-9853.1000119
Antal, G., Fári, M. G., and Domokos-Szabolcsy, É. (2018). Obtention of new ornamental leaf variants of giant reed (Arundo donax L.) originated from somatic embryogenesis and their photosynthetic parameters. Int. J. Hortic. Sci. 24, 1. doi: 10.31421/ijhs/24/1-2./1542
Antil, S., Abraham, J. S., Sripoorna, S., Maurya, S., Dagar, J., Makhija, S., et al. (2023). DNA barcoding, an effective tool for species identification: A review. Mol. Biol. Rep. 50, 761–775. doi: 10.1007/s11033-022-08015-7
Balakrishnan, R. (2005). Species concepts, species boundaries and species identification: A view from the tropics. Systematic Biol. 54, 689–693. doi: 10.1080/10635150590950308
Bucci, A., Cassani, E., Landoni, M., Cantaluppi, E., and Pilu, R. (2013). Analysis of chromosome number and speculations on the origin of Arundo donax L. (Giant Reed). Cytology Genet. 47, 237–241. doi: 10.3103/s0095452713040038
Canavan, K., Paterson, I. D., and Hill, M. P. (2017). Exploring the origin and genetic diversity of the giant reed, Arundo donax in south Africa. Invasive Plant Science and Management. 10, 53–60. doi: 10.1017/inp.2016.5
Cantaluppi, E., Cassani, E., Puglisi, D., Corno, L., Munaro, M., Landoni, M., et al. (2015). Study on the inflorescences of Arundo donax L. clones sampled in Italy. Braz. J. Bot. 39, 275–285. doi: 10.1007/s40415-015-0205-3
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Christin, P.-A., Spriggs, E., Osborne, C. P., Stromberg, C. A. E., Salamin, N., and Edwards, E. J. (2013). Molecular dating, evolutionary rates, and the age of the grasses. Systematic Biol. 63, 153–165. doi: 10.1093/sysbio/syt072
Danelli, T., Laura, M., Savona, M., Landoni, M., Adani, F., and Pilu, R. (2020). Genetic improvement of Arundo donax L.: opportunities and challenges. Plants 9, 1584. doi: 10.3390/plants9111584
Danin, A. (2004). Arundo (Gramineae) in the mediterranean reconsidered. Willdenowia 34, 361. doi: 10.3372/wi.34.34204
Danin, A. and Naenny, W. (2008). Contribution to the recognition of reeds by their rhizome anatomy. Fl. Medit. 18, 385–392.
Darzentas, N. (2010). Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 26, 2620–2621. doi: 10.1093/bioinformatics/btq484
Di Pietro, R., Di Marzio, P., Medagli, P., Misano, G., Silletti, G. N., Wagensommer, R. P., et al. (2016). Evidence from multivariate morphometric study of the Quercus pubescens complex in southeast Italy. Botanica Serbica 40, 83–100. doi: 10.5281/zenodo.48865
Dobrogojski, J., Adamiec, M., and Luciński, R. (2020). The chloroplast genome: A review. Acta Physiologiae Plantarum 42, 6. doi: 10.1007/s11738-020-03089-x
Fang, Z., Li, L., Zhou, J., You, A., Gao, L., Li, T., et al. (2021). Multiple nucleotide polymorphism DNA markers for the accurate evaluation of genetic variations (Cold Spring Harbor Laboratory). Available online at: https://www.biorxiv.org/content/10.1101/2021.03.09.434561v1.full.pdf (Accessed June 15, 2025). Preprint.
Feng, L.-Y., Shi, C., and Gao, L.-Z. (2021). The complete chloroplast genome sequence of Arundo formosana Hack. (Poaceae). Mitochondrial DNA Part B 6, 2819–2821. doi: 10.1080/23802359.2021.1972865
Ferrer-Gallego, P. and Hardion, L. (2025). Typifications in the genus Arundo L. (Poaceae, arundinoideae). (Harvard University). Available online at: https://www.authorea.com/users/877557/articles/1256983-typifications-in-the-genus-arundo-l-poaceae-arundinoideae (Accessed November 4, 2025). Preprint.
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, 273–279. doi: 10.1093/nar/gkh458
Freudenthal, J. A., Pfaff, S., Terhoeven, N., Korte, A., Ankenbrand, M. J., and Förster, F. (2019). A systematic comparison of chloroplast genome assembly tools. Genome Biol. 24 (1), 254. doi: 10.1186/s13059-020-02153-6
Guarino, F., Cicatelli, A., Brundu, G., Improta, G., Triassi, M., and Castiglione, S. (2019). The use of MSAP reveals epigenetic diversity of the invasive clonal populations of Arundo donax L. PLoS One 14, e0215096. doi: 10.1371/journal.pone.0215096
Hardion, L., Verlaque, R., Baumel, A., Juin, M., and Vila, B. (2012a). Revised systematics of Mediterranean Arundo (Poaceae) based on AFLP fingerprints and morphology. Taxon 61, 1217–1226. doi: 10.1002/tax.616004
Hardion, L., Verlaque, R., Callmander, M. W., and Vila, B. (2012b). Arundo Micrantha Lam. (Poaceae), the correct name for Arundo Mauritanica Desf. and Arundo Mediterranea Danin. Candollea. 67, 131. doi: 10.15553/c2012v671a11
Hardion, L., Verlaque, R., Haan-Archipoff, G., Cahen, D., Hoff, M., and Vila, B. (2017b). Cleaning up the grasses dustbin: Systematics of the Arundinoideae subfamily (Poaceae). Plant Systematics Evol. 303, 1331–1339. doi: 10.1007/s00606-017-1451-6
Hardion, L., Verlaque, R., Rosato, M., Rosselló, J. A., and Vila, B. (2015). Impact of polyploidy on fertility variation of Mediterranean Arundo L. (Poaceae). Comptes Rendus. Biologies 338, 298–306. doi: 10.1016/j.crvi.2015.03.013
Hardion, L., Verlaque, R., Saltonstall, K., Leriche, A., and Vila, B. (2014). Origin of the invasive Arundo donax (Poaceae): A trans-Asian expedition in herbaria. Ann. Bot. 114, 455–462. doi: 10.1093/aob/mcu143
Hardion, L., Verlaque, R., Vorontsova, M. S., Combroux, I., Chen, C.-W., Takamizo, T., et al. (2017a). Does infraspecific taxonomy match species evolutionary history? A phylogeographic study of Arundo formosana (Poaceae). Botanical J. Linn. Soc. 183, 236–249. doi: 10.1093/botlinnean/bow006
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Ser. B 270, 313–321. doi: 10.1098/rspb.2002.2218
Jike, W., Li, M., Zadra, N., Barbaro, E., Sablok, G., Bertorelle, G., et al. (2020). Phylogenomic proof of recurrent demipolyploidization and evolutionary stalling of the “Triploid bridge” in Arundo (Poaceae). Int. J. Mol. Sci. 21, 5247. doi: 10.3390/ijms21155247
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome biology. 21 (1), 241. doi: 10.1186/s13059-020-02154-5
Katoh, K. and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lee, J.-T., Shih, C.-Y., Wang, J.-T., Liang, Y.-H., Hsu, Y.-S., and Lee, M.-J. (2022). Root traits and erosion resistance of three endemic grasses for estuarine sand drift control. Sustainability 14, 4672. doi: 10.3390/su14084672
Letunic, I. and Bork, P. (2024). Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic acids research. 52 (1), 78–82. doi: 10.1093/nar/gkae268
Li, H., Guo, Q., Xu, L., Gao, H., Liu, L., and Zhou, X. (2023). CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 11, e15326. doi: 10.7717/peerj.15326
Ling, Y., Zhang, M., Ling, Z., Cao, B., Wu, X., Peng, H., et al. (2023). Evolutionary relationship and a novel method of efficient identification of Lentinula edodes cultivars in China. Mycosphere 13, 56–85. doi: 10.5943/mycosphere/si/1f/3
Liu, Q., Lin, L., Zhou, X., Peterson, P. M., and Wen, J. (2017). Unraveling the evolutionary dynamics of ancient and recent polyploidization events in Avena (Poaceae). Sci. Rep. 7, 1. doi: 10.1038/srep41944
Liu, S., Ni, Y., Li, J., Zhang, X., Yang, H., Chen, H., et al. (2023). CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 23, 694–704. doi: 10.1111/1755-0998.13729
Liu, L. and Sylvia, M. P. (2006). Poaceae. In flora of China Vol. 22 (Beijing: Science Press; St. Louis: Missouri Botanical Garden Press), 447–448.
Liu, J., Wang, H., Fan, X., Zhang, Y., Sun, L., Liu, C., et al. (2024a). Establishment and application of a Multiple nucleotide polymorphism molecular identification system for grape cultivars. Scientia Hortic. 325, 112642. doi: 10.1016/j.scienta.2023.112642
Liu, Y.-N., Xiao, X.-Y., and Guo, Z.-H. (2019). Identification of indicators of giant reed (Arundo donax L.) ecotypes for phytoremediation of metal-contaminated soil in a non-ferrous mining and smelting area in southern China. Ecol. Indic. 101, 249–260. doi: 10.1016/j.ecolind.2019.01.029
Liu, Y., Zhao, Q., Li, T., Teng, C., Peng, H., Yao, Z., et al. (2024b). Availability evaluation and application of MNP (multiple nucleotide polymorphism) markers in variety identification of chrysanthemum. Horticulturae 10, 845. doi: 10.3390/horticulturae10080845
Luo, L., Qu, Q., Lin, H., Chen, J., Lin, Z., Shao, E., et al. (2024). Exploring the Evolutionary History and Phylogenetic Relationships of Giant Reed (Arundo donax) through Comprehensive Analysis of Its Chloroplast Genome. Int. J. Mol. Sci. 25, 7936. doi: 10.3390/ijms25147936
Mariani, C., Cabrini, R., Danin, A., Piffanelli, P., Fricano, A., Gomarasca, S., et al. (2010). Origin, diffusion and reproduction of the giant reed (Arundo donax L.): A promising weedy energy crop. Ann. Appl. Biol. 157, 191–202. doi: 10.1111/j.1744-7348.2010.00419.x
Masoumi, A., Faghir, M. B., and Rikan, M. H. (2025). Fruit micromorphology of the genus Valerianella (Caprifoliaceae) in Iran. Phytotaxa 717, 1–30. doi: 10.11646/phytotaxa.717.1.1
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Ngernsaengsaruay, C., Puangsin, B., Leksungnoen, N., Khantayanuwong, S., Chanton, P., Thaepthup, T., et al. (2023). Morphology, taxonomy, culm internode and leaf anatomy, and palynology of the giant reed (Arundo donax L.), poaceae, growing in Thailand. Plants 12, 1850. doi: 10.3390/plants12091850
Osuna-Mascaró, C., Rubio de Casas, R., and Perfectti, F. (2018). Comparative assessment shows the reliability of chloroplast genome assembly using RNA-seq. Sci. Rep. 8, 1. doi: 10.1038/s41598-018-35654-3
Perrino, E. V., Wagensommer, R. P., Mezzapesa, G. N., and Trani, A. (2024). Stachys italica Mill.: Synecology, functional compounds and potential use of an Italian endemic taxon. Planta 260 (6), 138. doi: 10.1007/s00425-024-04571-3
Pilu, R., Cassani, E., Landoni, M., Badone, F. C., Passera, A., Cantaluppi, E., et al. (2013). Genetic characterization of an Italian Giant Reed (Arundo donax L.) clones collection: Exploiting clonal selection. Euphytica 196, 169–181. doi: 10.1007/s10681-013-1022-z
Qiu, T. and Cui, S. (2021). Evolutionary analysis for Phragmites ecotypes based on full-length plastomes. Aquat. Bot. 170, 103349. doi: 10.1016/j.aquabot.2020.103349
Ren, M., Han, X., and Liu, F. (2023a). MNP Marker Loci, primer combinations, and kits for identification of Arundo donax varieties, and their applications (Beijing, DC: C.N. National intellectual Property Administration). C. N. Patent No. CN117089644A.
Ren, M., Liu, F., Han, X., Wu, D., and Peng, H. (2023b). Chromosome-scale genome assembly of the allopolyploid Arundo donax. (Cold Spring Harbor Laboratory). Available online at: https://www.biorxiv.org/content/10.1101/2023.06.18.544523v4
Ren, M., Liu, F., Han, X., Wu, D., and Peng, H. (2024). Chromosome-scale genome assembly of the autoalloenneaploid Arundo donax. Grassland Res. 3, 230–242. doi: 10.1002/glr2.12091
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, 686–689. doi: 10.1093/nar/gki366
Senthilkumar, S., Ulaganathan, K., and Ghosh Dasgupta, M. (2021). Reference-based assembly of chloroplast genome from leaf transcriptome data of Pterocarpus santalinus. 3 Biotech. 11, 8. doi: 10.1038/srep30135
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, 65–73. doi: 10.1093/nar/gkz345
Shi, C., Wang, S., Xia, E. H., Jiang, J. J., Zeng, F. C., and Gao, L. Z. (2016). Full transcription of the chloroplast genome in photosynthetic eukaryotes. Scientific reports. 6, 30135. doi: 10.1038/srep30135
Tarin, D., Pepper, A. E., Goolsby, J. A., Moran, P. J., Arquieta, A. C., Kirk, A. E., et al. (2013). Microsatellites uncover multiple introductions of clonal giant reed (Arundo donax). Invasive Plant Sci. Manage. 6, 328–338. doi: 10.1614/ipsm-d-12-00085.1
Teisher, J. K., McKain, M. R., Schaal, B. A., and Kellogg, E. A. (2017). Polyphyly of Arundinoideae (Poaceae) and evolution of the twisted geniculate lemma awn. Ann. Bot. 120, 725–738. doi: 10.1093/aob/mcx058
Tomàs, J., Mateu, J., Gil, L., Boira, H., and Llorens, L. (2019). Arundo micrantha Lam. as an alternative to Arundo donax L. as energy crop in saline soils irrigated with treated urban wastewaters. Plant Biosyst. - Int. J. Dealing All Aspects Plant Biol. 154, 560–567. doi: 10.1080/11263504.2019.1651779
Touchell, D. H., Ranney, T. G., Panthee, D. R., Gehl, R. J., and Krings, A. (2016). Genetic diversity, cytogenetics, and biomass yields among taxa of giant reeds (Arundo species). J. Am. Soc. Hortic. Sci. 141, 256–263. doi: 10.21273/jashs.141.3.256
Vences, M., Patmanidis, S., Kharchev, V., and Renner, S. S. (2022). Concatenator, a user-friendly program to concatenate DNA sequences, implementing graphical user interfaces for MAFFT and FastTree. Bioinf. Adv. 2, 1. doi: 10.1093/bioadv/vbac050
Wagensommer, R. P. (2023). Floristic studies in the light of biodiversity knowledge and conservation. Plants 12, 2973. doi: 10.3390/plants12162973
Keywords: Arundo smaragdina, morphological differentiation, MNP markers, chloroplast genome, transcriptome data
Citation: Liu F, Li B, He Q, Zhao A, Xi B and Shen X (2025) Arundo smaragdina (Poaceae): a novel species revealed by integrative taxonomy and its implications for the phylogeny of the genus. Front. Plant Sci. 16:1660442. doi: 10.3389/fpls.2025.1660442
Received: 06 July 2025; Accepted: 27 October 2025;
Published: 17 November 2025.
Edited by:
Robert Philipp Wagensommer, Free University of Bozen, ItalyReviewed by:
Leonardo Alfredo Ornella, EVOGENIX LTD, United KingdomEnrico Vito Perrino, University of Foggia, Italy
Copyright © 2025 Liu, Li, He, Zhao, Xi and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaotong Shen, V1JCVF9EUkAxNjMuY29t
 Fupeng Liu
Fupeng Liu Boyu Li
Boyu Li