- Department of Plant Pathology, Washington State University, Tree Fruit Research and Extension Center, Wenatchee, WA, United States
Neonectria fruit rot (NFR) is primarily attributed to Neonectria ditissima, the causal agent of European canker, in many apple-growing regions globally. Between 2017 and 2019, NFR-like symptoms were observed in several surveyed apple and pear packinghouses in Washington State and Oregon. In this study, 52 Neonectria isolates were characterized using a multilocus sequence analysis (MLSA), pathogenicity assays, and morphological traits across various growth media and photoperiods. MLSA analysis of four DNA regions, i.e., β-TUB, TEF1, LSU, and ITS rDNA, identified the pathogen as Neonectria candida (syn. Neonectria ramulariae, anamorph Cylindrocarpon obtusiusculum). All the 52 N. candida isolates formed a distinct clade from N. ditissima and other Neonectria spp. with a Bayesian interference posterior probability of 0.98. Fruit pathogenicity assays showed that N. candida isolates caused light brown lesions on ‘Fuji’ apples and ‘Green d’Anjou’ pears both at room temperature (22°C) and cold storage (0.5 and 1.5°C), with NFR incidences ranging from 6 to 100% after 15 days to four months. N. candida isolates grew and sporulated profusely under a multitude of nutrient and photoperiod conditions in vitro. This study is a foundational step towards species identification and understanding the biology and epidemiology of NFR to support the development of effective management approaches.
1 Introduction
Over 100 fungal species have been reported to infect apples and pears at different phenological stages before harvest and during storage (Sutton et al., 2014). In the U.S. Pacific Northwest (PNW) and other pome fruit growing regions, the most economically import fungal pathogens include Penicillium spp. (Sanderson and Spotts, 1995), Botrytis spp. (Amiri and Ali, 2016), Neofabraea spp. (Henriquez et al., 2007), Mucor spp. (Bertrand and Saulie-Carter, 1980), Alternaria spp. (Filajdić and Sutton, 1991), Cladosporium spp. (Virginia et al., 2021), Phacidiopycnis spp. (Xiao and Boal, 2004), Sphaeropsis spp. (Kim and Xiao, 2008), Lambertella corni-marris (Wiseman et al., 2015), Colletotrichum spp. (Khodadadi et al., 2020), and Neonectria spp. (Holthusen and Weber, 2021). Beyond direct losses resulting from decay, some of these pathogens can taint processed pome fruit products with mycotoxins (Brian et al., 1956; Zhong et al., 2018) or trigger quarantine restrictions that block access to export markets (Hansen, 2014).
Among the above-mentioned pathogens, Neonectria spp. have a wide host range and can infect more than 60 tree and shrub species across the Myrtaceae, Pinaceae, Proteaceae, and Rosaceae families, producing a wide range of symptoms (Chaverri et al., 2011; Hirooka et al., 2013). In the Rosaceae, Neonectria and its anamorph Cylindrocarpon spp. cause cankers and dieback on trunks and young shoots, weakening them and reducing yields if unmanaged (Ghasemkhani et al., 2016). Neonectria belongs to the Nectriaceae, characterized by brightly colored, uniloculate perithecia (Tulasne and Tulasne, 1865). Species limits are complicated, prompting multilocus phylogenetics. Chaverri et al. (2011) analyzed 66 isolates with six loci (LSU, ITS, RPB1, RPB2, β-tubulin, TEF1, ACT), stabilizing Cylindrocarpon taxonomy and placing N. ditissima, N. candida, and N. fuckeliana in Neonectria sensu stricto. Among these species, N. ditissima, N. punicea, and N. candida have been identified as causal agents of Neonectria fruit rot (NFR) in pome fruits grown in England (Xu and Robinson, 2010), Netherlands (Wenneker et al., 2016), and China (Wu et al., 2022). NFR symptoms are seldom visible in the orchard, even in humid areas, and are virtually undetected in the arid central-Washington climate. Instead, circular, slightly sunken, light- to dark-brown lesions with firm to soft texture usually appear after several months of cold storage (Edwards, 2006; Xu and Robinson, 2010). Yellow- to white-pustulate sporulation follows as the rot advances (Munson, 1939). Early NFR caused by N. ditissima closely mimics bull’s-eye rot caused by Neofabraea spp., leading to frequent misdiagnosis (Xu and Robinson, 2010) and potential fruit rejection during transit or retail.
Recent reports have expanded the NFR complex, i.e., N. punicea on ‘Red Delicious’ and ‘Fuji’ apples in China (Wu et al., 2022) and N. candida (syn. N. ramulariae, anamorph Cylindrocarpon obtusiusculum) on ‘Conference’ pears in the Netherlands (Wenneker et al., 2016). In accordance with the “one fungus, one name” (Hawksworth, 2011), the name Neonectria candida is used instead of its synonyms or anamorphic names. Infections of the NFR originate through natural openings (stem-end, calyx-end, lenticels) or harvest wounds. Besides infecting pears, N. punicea and N. candida were reported to cause seed rot, stem dieback, or canker diseases on a variety of forest trees (Hirooka et al., 2012, 2013; Jankowiak et al., 2016; Karadžić et al., 2020). While the ability of N. ditissima, N. punicea, and N. candida to infect fruit is documented elsewhere, their capacity to produce cankers and fruit rot in PNW orchards, and their local distribution and epidemiology, remain largely unexplored. In large surveys conducted between 2016 and 2019 in several apple and pear packinghouses in Washington State and Oregon, a postharvest decay resembling Neonectria-like disease, was observed in fruit originating from several orchards (Amiri and Ali, 2016; Mellow and Amiri, 2023).
We conducted this study to address three primary objectives: i) identify the species causing Neonectria fruit rot (NFR) in pome fruit from the PNW through multi-locus phylogenetic analysis; ii) characterize the morphology of the isolates in vitro under varying media and photoperiod conditions; and iii) evaluate the pathogenicity and virulence of these species on detached apple and pear fruit.
2 Materials and methods
2.1 Collection, culturing, and characterization of Neonectria isolates
Decayed apples and pears were collected on the packing line after six to eight months of storage under standard commercial conditions between 2017 and 2019 from multiples apple and pear packinghouses in Washington State (WA) and Oregon (OR), United States (Figure 1). Fruits were transported in clean clamshells to the laboratory and decay types were identified based on key morphological characteristics (Amiri et al., 2017; Ali et al., 2018). Pure fungal cultures were made from each fruit on General Isolation (GI) medium consisting of half-strength potato dextrose agar (PDA) (Hardy Diagnostics, Santa Maria, CA), 7.5 g of bacto agar (Difco Laboratories, Sparks, MD), in 1L of distilled water supplemented with 200 mg/L streptomycin sulfate (Amresco, Solon, OH) and 100 mg/L ampicillin (MP Biomedicals, Solon, OH) added post-autoclave. The decayed fruits were cut open using an ethanol-sterilized knife, and a small piece of flesh at the margin between decayed and healthy fruit tissue was cut and transferred to GI plates (Amiri et al., 2017). Cultures were incubated at 22 °C for 10 days, and fungal isolates were characterized based on known colony morphology, and by spore shape and conidiophore structure according to Dugan (2006). Pure cultures were then preserved as mycelial plugs in 20% glycerol and stored at -80 °C in the Pathology Lab Washington State University (WSU) - Tree Fruit Research and Extension Center (TFREC), Wenatchee, WA. Conidial and conidiophore morphology was assessed microscopically for representative isolates. After 28 days on PDA, each culture was washed with 10 mL sterile water, scraped, and the resulting suspension filtered through cheesecloth; spore concentration was then determined with a hemacytometer on a Zeiss Axioskop microscope (Zeiss, Oberkochen, Germany). A 50 µl droplet of each suspension was placed on a PDA-coated slide, incubated at 23°C under a 12 hr photoperiod for three days (Hua’an et al., 1991), and examined with Olympus BX53 compound (Olympus, Central Valley, PA) and Zeiss Stemi 508 dissecting microscopes, with images captured on an AmScope camera (AmScope, Los Angeles, CA).
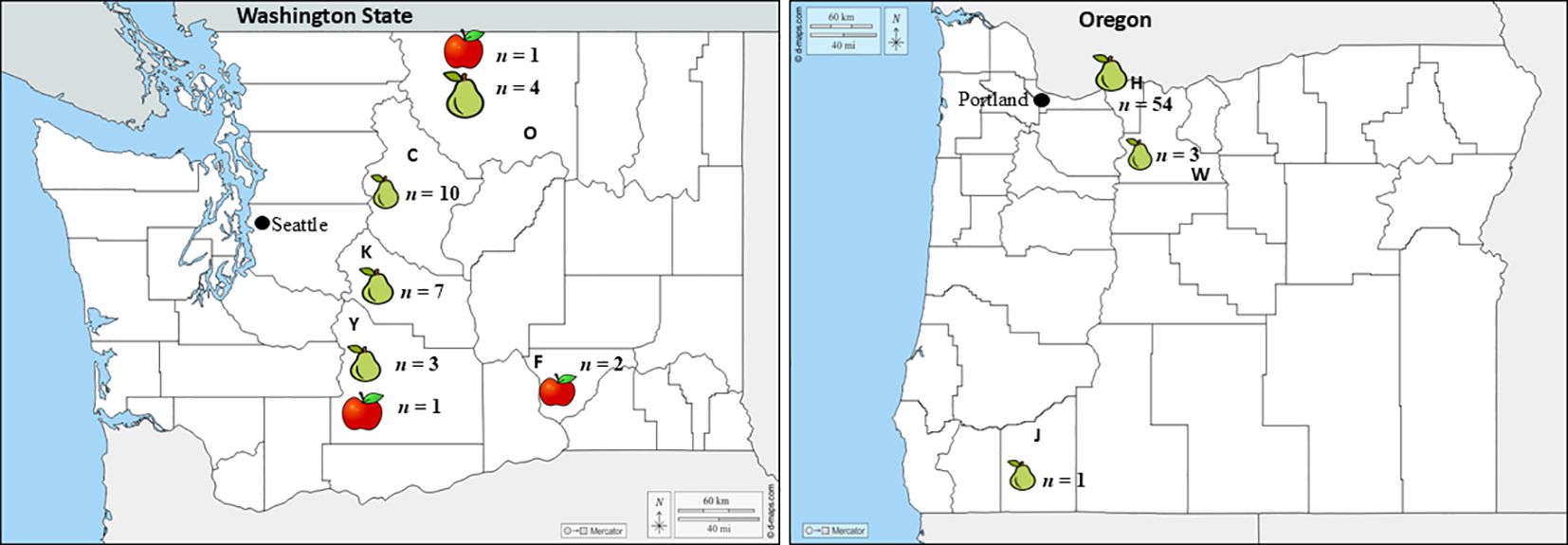
Figure 1. Geographic origin (state, county) of the isolates collected by host (apple or pear) in Washington and Oregon. Bold letters C, F, K, O, and Y represent Chelan, Franklin, Grant, Kittitas, Okanogan and Yakima counties in Washington, respectively, whereas H, J, and W indicates the Hood River and Jackson, Wasco counties in Oregon, respectively. The n next to each host in each county indicates the number of isolates collected.
2.2 Molecular characterization of Neonectria isolates
The Neonectria-like isolates (Table 1) were revived from -80°C storage on PDA and incubated for 14 days at 22°C. Peripheral mycelial plugs were transferred to 50 mL of sterile potato dextrose broth in flasks and shaken at 160 rpm for 5 days at 22°C. The resulting mycelia were harvested on 8 µm Whatman filter paper with a Büchner funnel, transferred to 1.5 mL tubes, and stored at -20°C. For DNA extraction, ~100 mg of thawed mycelium was combined with 200 mg of glass-silica beads in a 1.5 mL tube and homogenized in a FastPrep-24™ grinder (MP Biomedicals, Irvine, CA). Genomic DNA was then isolated using the alkaline-lysis method of Raeder and Broda (1985), resuspended in 100 µL sterile distilled water, quantified with a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA), and stored at -20°C.
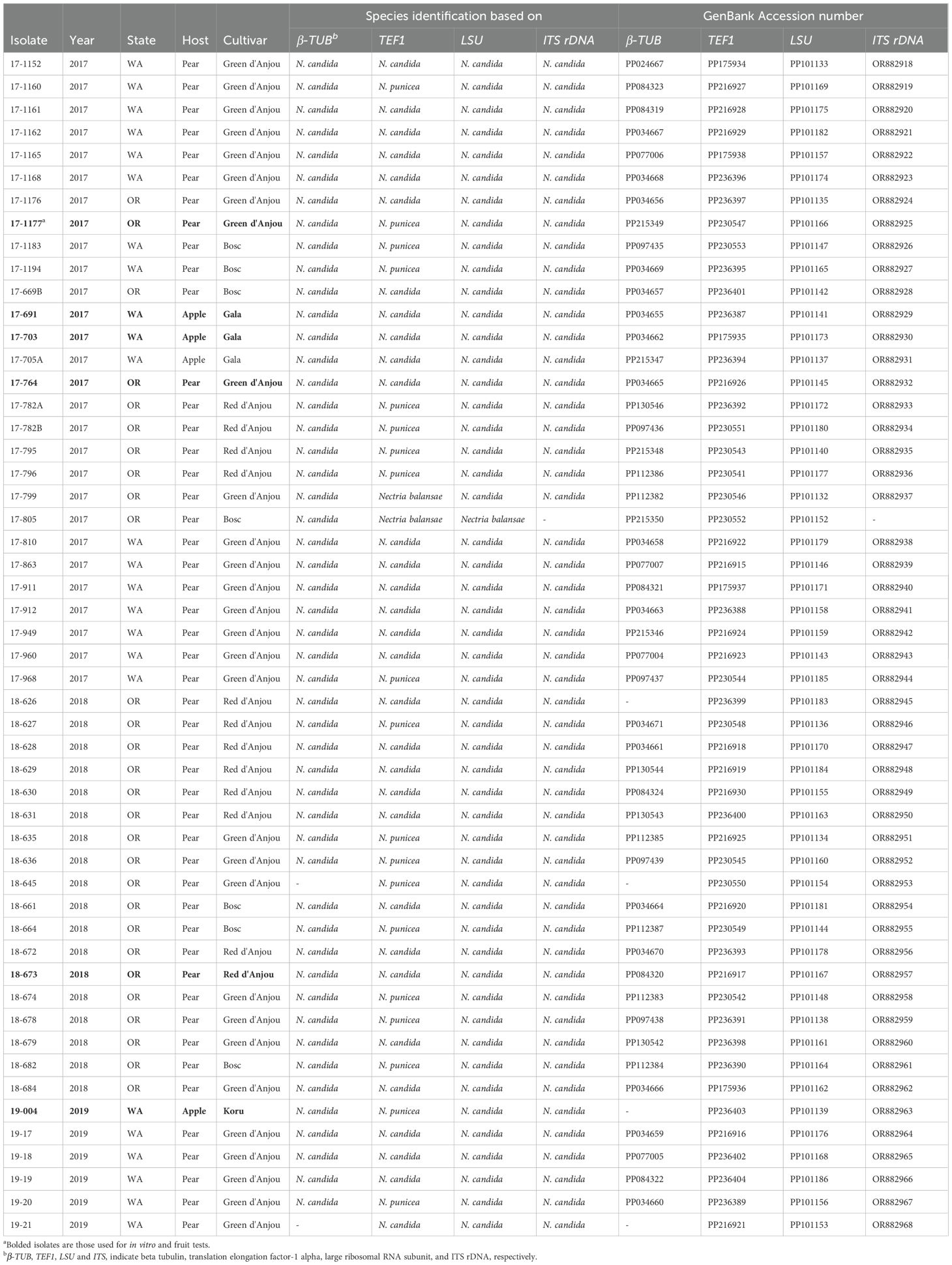
Table 1. Details about the isolates, species identifications and accession numbers used for the phylogenetic analysis.
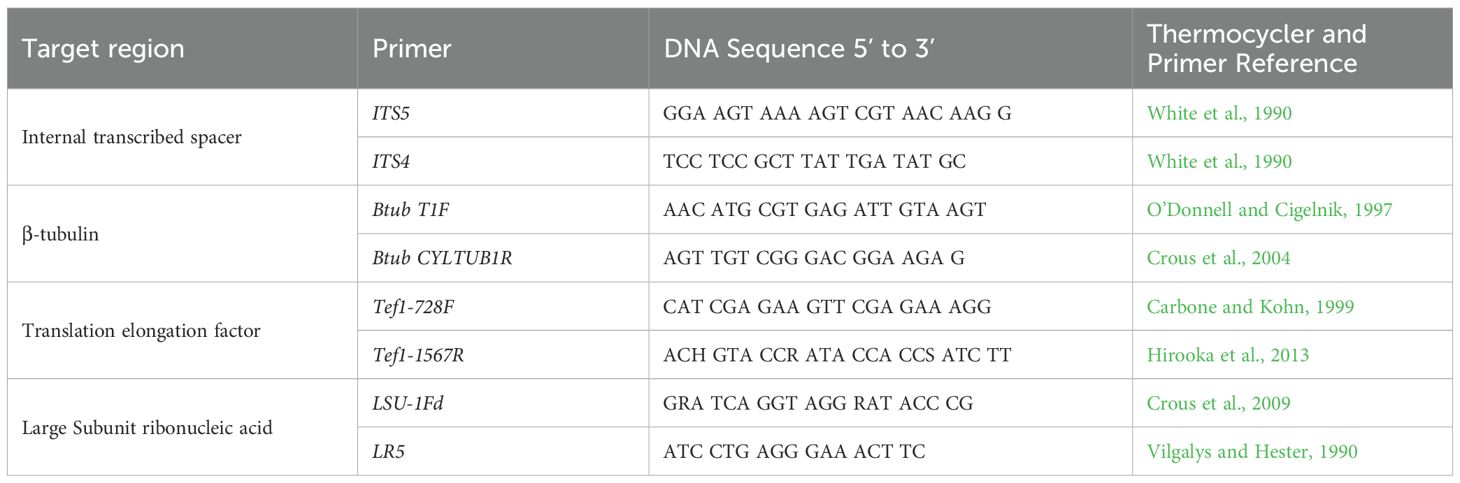
Molecular characterization of isolates was performed using multilocus sequence analysis (MLSA) of four DNA regions known to effectively distinguish Nectriaceae species (Crous et al., 2009; Hirooka et al., 2013; Lombard et al., 2015). The loci included the internal transcribed spacer ribosomal DNA (ITS rDNA [ITS1-5.8S-ITS2] (White et al., 1990), beta-tubulin (β-TUB) (Crous et al., 2004; O’Donnell and Cigelnik, 1997), translation elongation factor-1 alpha (TEF1) (Carbone and Kohn, 1999; Rehner and Buckley, 2005), and the large ribosomal RNA subunit (LSU) (Vilgalys and Hester, 1990; Crous et al., 2009).
Polymerase chain reaction (PCR) amplifications were performed in 25 µL reactions containing 12.5 µL of EconoTaq PLUS Green 2x Master Mix (Lucigen, Middleton, WI), 1 µL of each 10 μM primer (Table 2), 0.2 µL of dimethyl sulfoxide (DMSO), 1.2 µL of bovine serum albumin (BSA), 9.1 µL of nuclease-free water and 1 µL of 20 ng/µL of DNA template. Reactions were conducted using a BIO-RAD T100 thermal cycler (Bio-Rad Laboratories, Hercules, CA) under thermocycling conditions optimized for each primer pair, as previously described (Crous et al., 2004; O’Donnell and Cigelnik, 1997). PCR products were separated by electrophoresis on 1% agarose gels (Green BioResearch LLC, Baton Rouge, LA) in 1× Tris-acetate-EDTA (TAE) buffer, stained with GelRed (Biotium Inc., Fremont, CA), and visualized utilizing a UVP GelDoc-It 130 Imaging System (Analytik Jena U.S., Upland, CA). Successful amplicons were purified using the Wizard SV Gel and PCR Clean-Up System (Promega Corporation, Madison, WI), following the manufacturer’s protocol. Purified products were sequenced bidirectionally at Retrogen, Inc. (San Diego, CA). Raw sequences were trimmed, assembled, and aligned using Geneious Prime v2021.0.3 (Kearse et al., 2012). Final sequences were submitted to the National Center for Biotechnology Information (NCBI) GenBank, and corresponding accession numbers are listed in Table 1.
2.3 Phylogenetic analyses
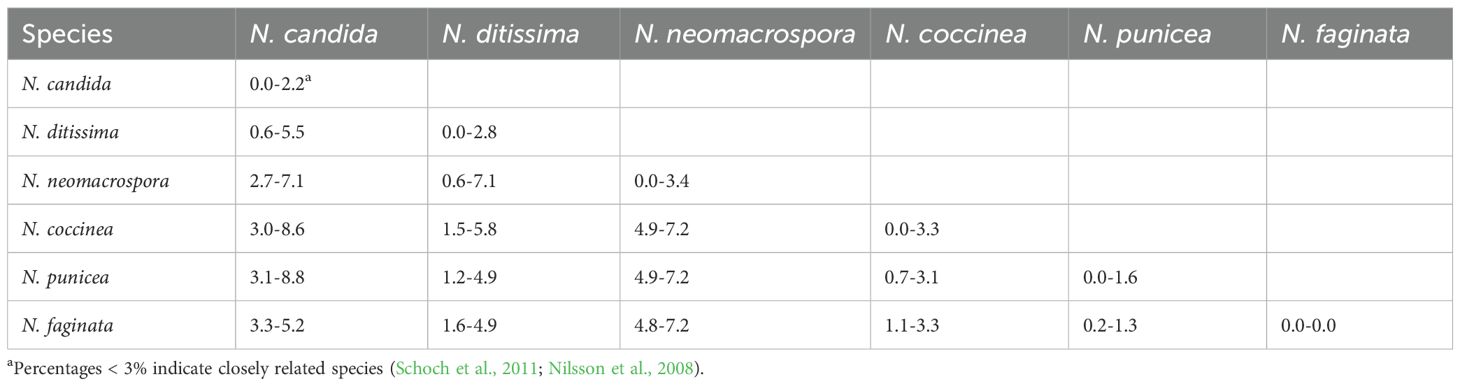
Single-locus (ITS rDNA, β-tubulin, TEF1-α, and LSU) and four-locus concatenated phylogenies were inferred for all 52 Neonectria isolates. Homologous reference sequences and outgroups (Supplementary Table S1) were downloaded from GenBank and aligned using Clustal Omega (Sievers and Higgins, 2014) through Geneious Prime v2021.0.3. Phylogenetic trees were inferred by Maximum-likelihood (ML) using RAxML v8.2.11 (Stamatakis, 2014) with 1,000 rapid bootstrap replicates. In addition, Bayesian inference (BI) trees were generated using MrBayes v3.2.6 (Ronquist and Huelsenbeck, 2012; Ronquist et al., 2012). BI trees were run for 2 million generations and trees were sampled every 1,000 generations using the GTR + G4 model through Geneious Prime v2021.0.3. Convergence was confirmed by an average split-frequency < 0.01 and ESS > 200 (Tracer v1.7). Trees were visualized in FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/). Nodes were considered robust when bootstrap percentages and posterior probabilities were >70% and >0.70, respectively. Topological congruence among the four loci was tested with the partition homogeneity test in PAUP* v4.0a169 (Swofford, 2003) using pairwise homogeneity test (PHT) with heuristic search (100 replicates, P < 0.01). To gauge species boundaries, pairwise ITS divergences were calculated in Geneious; isolates differing by < 3% were treated as conspecific (Nilsson et al., 2008; Schoch et al., 2011).
2.4 Mycelial growth and sporulation on different media under different photoperiod conditions
From the total of 52 Neonectria isolates characterized to the species level (Table 1), six representative isolates (each three isolates originated from apple and pear) were selected for detailed physiological assays. Radial growth, sporulation, and micro-/macroconidium production were assessed on PDA, apple-juice agar (AJA; 200 mL organic Fuji juice + 18 g agar + 800 mL H2O), V8 agar (V8A), and low-nutrient agar (LNA, Gerlach and Nirenberg, 1982) at 23 °C under three photoperiods: 12 h light/12 h dark, continuous darkness, and continuous light. A 5-mm plug was cut from 14-day PDA cultures of each isolate and placed mycelium-down on each medium (three replicates per isolate × medium × light regime). Colony diameters were measured along two perpendicular axes on days 7, 14, 21, and 28 days and growth rates (mm/day) were calculated from the 14–21-day interval. Plates were photographed (Canon EOS 30D) at each measurement point. After 28 days, conidial suspensions were prepared from the same plates as described above, and micro- and macroconidia were quantified and imaged microscopically following Chaverri et al. (2011). The trial was conducted twice.
2.5 Pathogenicity and virulence assays
Pathogenicity and virulence of six Neonectria isolates (Table 1) were evaluated on ‘Green d’Anjou’ pears and ‘Fuji’ apples harvested at commercial maturity in September and October 2022, respectively, from an experimental orchard in Rock Island, WA. These cultivars were selected because of their importance in commercial orchards in the PNW. Fruits were surface-disinfected in 1% sodium hypochlorite for 2 min, rinsed twice with tap water, and air-dried. Four equatorial wounds (3 mm diameter, 4 mm deep) were made per fruit, and each wound was inoculated with 30 µL of a 1 × 105 conidia/mL suspension of the appropriate isolate. For each isolate, 72 apples and 72 pears were arranged in a randomized complete-block design: two lots of 36 fruit (four replicates of nine). One lot was incubated at 22°C and assessed weekly for 28 days; the second lot was stored at 1.5°C (‘Fuji’) or 0.5°C (‘Green d’Anjou’) and assessed monthly for four months. Disease incidence was recorded as the proportion of wounds exhibiting decay, and disease severity as lesion diameter (average of two perpendicular measurements). To confirm the symptoms were caused by the inoculated pathogen, Koch’s postulates were completed by re-isolation on representative lesions from fruit held 28 days at 22°C or four months at cold temperatures as described above for the initial isolation.
2.6 Data analyses
Growth rate and conidial production were analyzed with a three-way ANOVA in which culture medium, light regime, and assessment interval served as fixed factors and isolate identity was treated as a random factor. Normal distribution of the data was assessed using the Shapiro-Wilk test. When the overall F-test was significant (α = 0.05), the means were separated with Student’s t-tests. For mycelial growth, post-hoc contrasts between every medium × light combination were evaluated. For the pathogenicity assay, lesion diameter (mm) and disease incidence were subjected to separate three-way ANOVAs for each host species, using storage temperature and inspection time as additional factors. Mean separation again employed Student’s t-tests at P < 0.05. All statistical analyses were performed in RStudio (R Core Team, 2023) with the “aov” function.
3 Results
3.1 Disease symptoms and isolation of Neonectria-like isolates
Symptoms first appeared as light- to dark-brown, soft, water-soaked and slightly sunken lesions (Figures 2A, D, E). The lesions expanded rapidly, especially on pears (Figures 2B, C). After four months at 0.5 °C, lesions on pear fruit were frequently blanketed by white to yellowish pustules and mycelial mats that covered the entire affected area (Figures 2A-C). Disease progression was slower on apple and no mycelia or sporulation was observed on ‘Gala’ and ‘Fuji’ apples stored for eight months at 1.5 °C (Figures 2D, E). From 2017 to 2019, 52 Neonectria-like isolates were obtained from multiple apple and pear packinghouses in Washington and Oregon (Table 1). Collections included 28, 19, and five isolates in 2017, 2018, and 2019, respectively, with 23 originating from Washington and 29 from Oregon. Pears accounted for most isolates (49; 94%), i.e., from different cultivars ‘Green d’Anjou’ (29; 59%), ‘Red d’Anjou’ (13; 27%), and ‘Bosc’ (7; 14%). Only three isolates were recovered from apples, all from the cultivar ‘Gala’ (Table 1). Isolates were collected from five counties in WA and three counties in OR (Figure 1).

Figure 2. Neonectria fruit rot (NFR) caused by Neonectria candida on "Green d'Anjou" pear after four (A) and eight (B) months on storage in regular atmosphere at 0.5°C and advanced NFR symptoms on "Green d'Anjou" pears after 8 months (C). Note the profuse sporulation on with-yellowish lesions, sunken NFR lesions on "Gala" (D) and "Fuji" apples (E) five months of storage in regular atmosphere at 1.5°C; Front (F) and reverse (G) colonies of N. candida isolate 17-703 grown on PDA, (F) on V8 (H) and on AJA (I) for 28 days at 23°C under 12 hour alternating photoperiod; sporodochia and conidiophores of N. candida (J), macroconidia (K) and microconidia (L). Scale bars in (J–L) are at 50 µm.
On PDA, the Neonectria-like isolates produced a white colony within seven days that turned light to dark brown after 14–28 days (Figure 2F, G). Concentric white mycelial rings developed, and sporodochia formed after about 21 days at 23°C (Figures 2F, G). Conidiophores, arising laterally from individual hyphae, were 35–40 µm long. Sporodochia contained abundant conidial masses (Figure 2J). Macroconidia were rare, hyaline, straight to slightly curved, cylindrical to ellipsoidal, aseptate to bisepate, with rounded ends, and measured 23–30 µm (Figures 2J, K). Microconidia (n = 50) were hyaline, aseptate, ovoid, and 3–5 ± 1.7 µm long (Figure 2L). Asci, ascospores, and perithecia were not observed on PDA for up to 60 days at 23°C.
3.2 Multilocus sequence analysis
To obtain a general taxonomic frame of the isolates, BLAST searches in GenBank returned ≥ 99% identity with N. candida for 50 of 52 isolates in the β-TUB and TEF1 datasets and for 51 of 52 isolates in the LSU and ITS datasets. All 52 isolates were identified as Neonectria candida (syn. N. ramulariae; anamorph Cylindrocarpon obtusiusculum) based on β-TUB, TEF1, LSU and ITS rDNA sequences (Table 1).
A pairwise homogeneity test detected no significant incongruence among the four loci (P = 0.01). Separate Bayesian (BI) analyses of the individual genes produced slightly different topologies (Supplementary Figures S1A–D), but the concatenated dataset resolved all 52 pome-fruit isolates in a well-supported clade with N. candida reference sequences with a BI posterior probability of 0.98 (Figure 3). This clade was sister to a group containing N. neomacrospora and N. ditissima. Within the N. candida clade, two subclades were evident: one comprising 26 isolates from this study and another comprising 23 isolates plus the reference sequences (posterior probability = 0.63). Three isolates from Oregon pears (18-626, 18-631, 18-679) formed a strongly supported minor sister group to the main N. candida cluster. Maximum-likelihood (ML) analysis of the concatenated alignment yielded an equivalent topology, distinguishing N. candida from non-candida reference sequences with 61% bootstrap support (Figure 3), in agreement with the individual-gene ML trees (63–81% bootstrap; Supplementary Figures S1A–D). Pairwise ITS sequence divergence averaged 0.6–5.5% between N. candida and N. ditissima and 2.7–7.7% between N. candida and N. neomacrospora (Table 3). No phylogenetic structure was associated with the state of origin (Washington vs. Oregon) or with host (apple vs. pear) with isolates from all sources being intermingled throughout the N. candida clade.
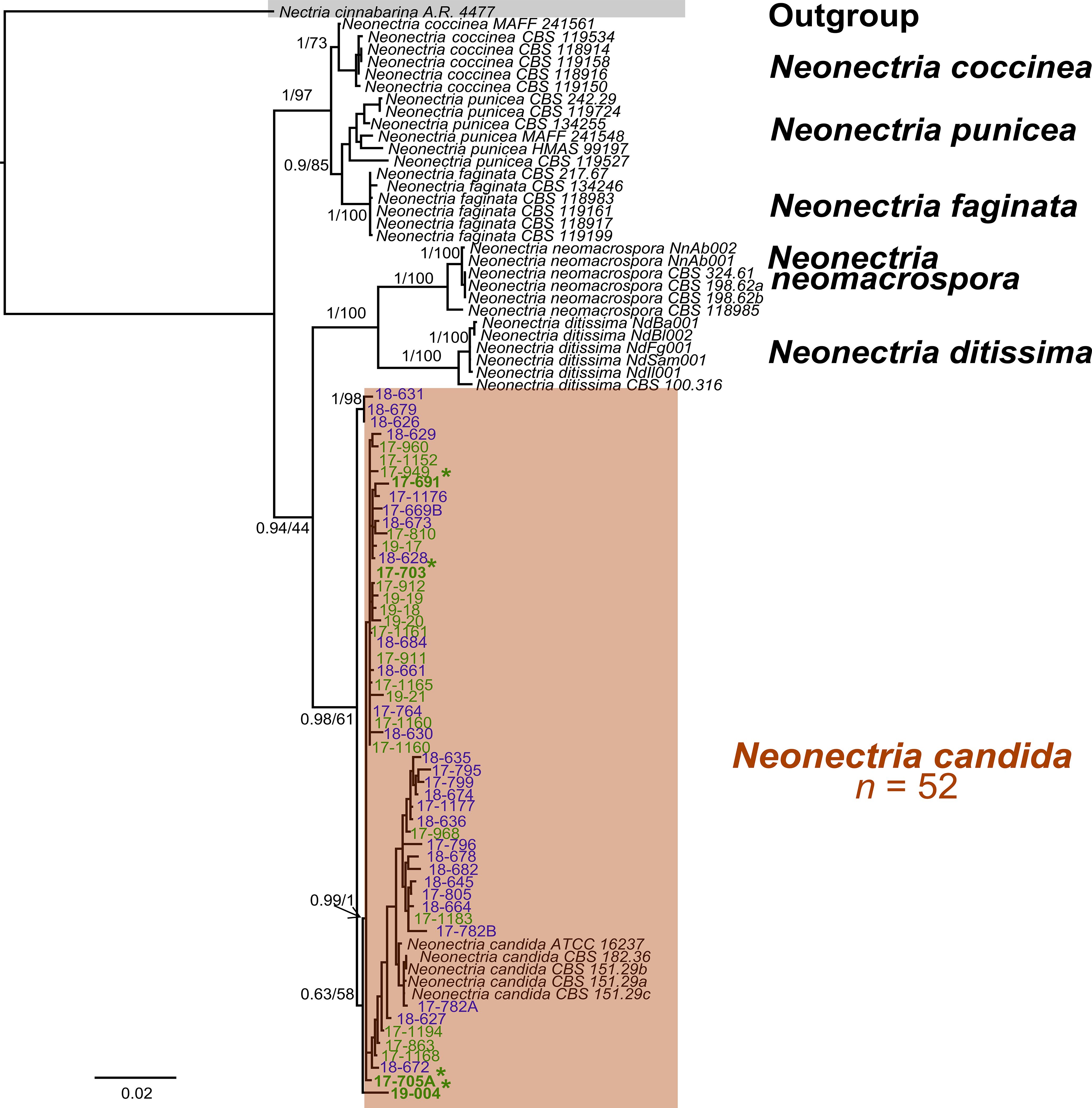
Figure 3. Phylogenetic tree from Bayesian inference (BI) analyses of the concatenated sequences of β-TUB, TEF1, LSU, and ITS rDNA from multiple Neonectria isolates collected from apple and pear fruit in Washington State and Oregon and other Neonectria reference sequences. BI posterior probabilities/Maximum likelihood bootstrap values are indicated at the branches. BI posterior probabilities >0.70 and ML bootstrap support >70% were deemed reliable. Asterisks indicate isolates from apple. Blue and green fonts indicate isolates from Oregon and Washington State, respectively.
3.3 Effect of photoperiod on growth and sporulation in vitro
Photoperiod and isolate each had a significant effect on radial growth of N. candida (P < 0.05), whereas growth medium alone did not (P = 0.143). Significant interactions were detected for medium × photoperiod, medium × isolate, and the three-way combination (P < 0.05); the photoperiod × isolate interaction was marginal (P = 0.05). After 21 days in continuous light, colonies grew fastest on AJA (27.8 mm/day) and V8A (27.7 mm/day), followed by LNA (21.3 mm/day) and PDA (20.5 mm/day) (Figure 4). The slowest growth of 9.6 mm/day occurred on AJA under a 12 h light/12 h dark cycle (Figure 4B). Overall isolates 17–703 from apples grew the least on LNA, AJA and V8A, especially under a 12 hr photoperiod (Figures 4A-C). Photoperiod did not noticeably alter colony pigmentation on any medium. Morphology varied with medium: isolate 17–703 formed hyaline, concentrically ringed colonies on LNA, but typical brown Neonectria colonies with abundant conidiophores and pycnidia on PDA and especially V8A. Colonies on AJA became light to dark brown only after 28 days (Figures 2F-I).
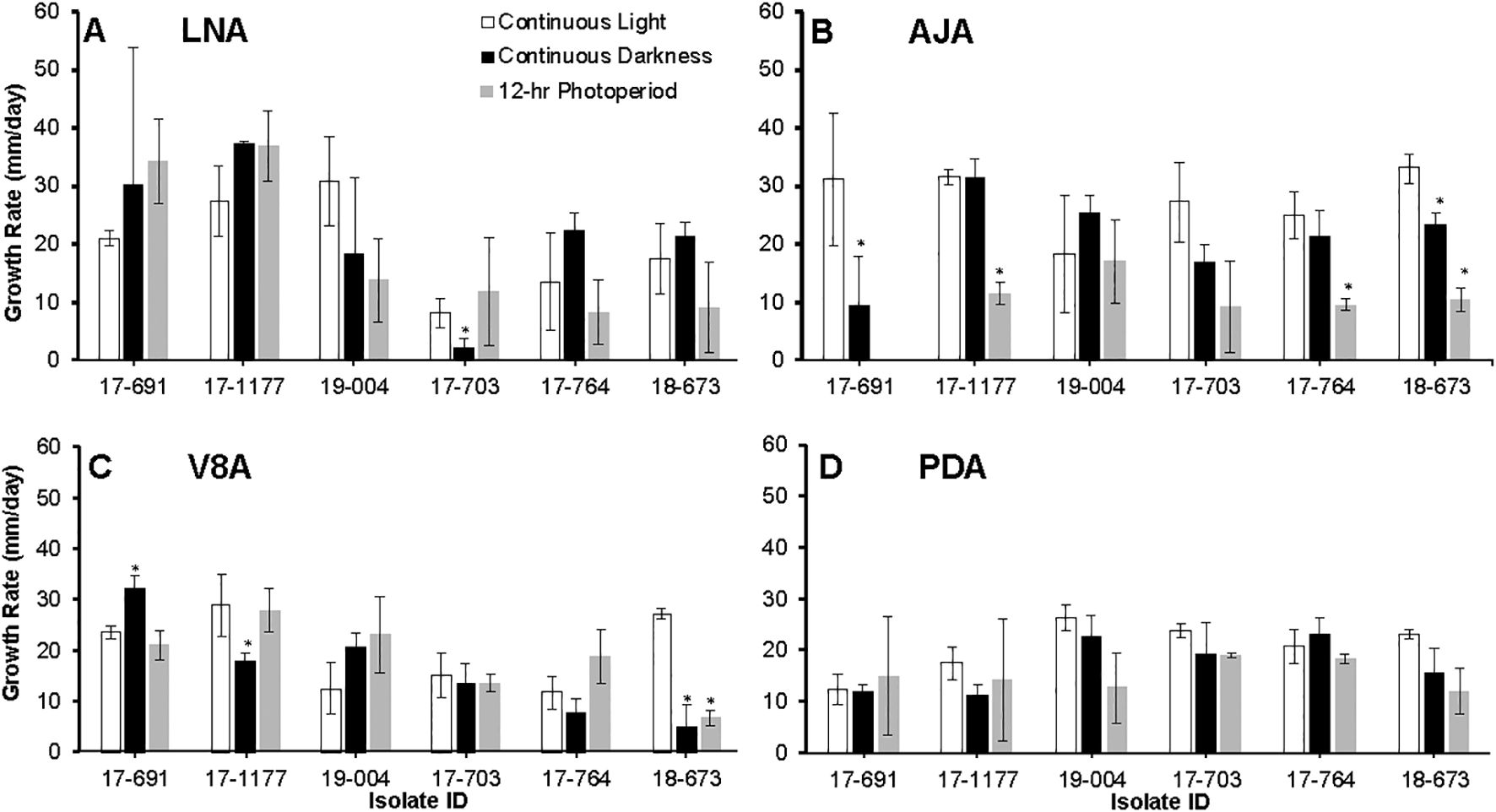
Figure 4. Growth rates of six Neonectria candida isolates on low nutrient agar (LNA, A), apple juice agar (AJA, B), V8 agar (V8A, C), and potato dextrose agar (PDA, D) for up to 21 days under continuous light, continuous darkness, and 12 hour alternating photoperiod. Data bars are the average of six values per treatment across two experimental runs. Vertical bars are standard deviations of the means. Asterisks indicate significant differences between media across isolates.
Sporulation differed significantly among culture media, photoperiods, isolates, and the medium × isolate interaction (P < 0.05). Interactions between medium × photoperiod, photoperiod × isolate, and the three-way combination were not significant (P > 0.05). Conidial production was significantly reduced under continuous darkness on most media but particularly on LNA and AJA (Figures 5A, B). Sporulation peaked on PDA and V8A under a 12 h light/12 h dark cycle (Figures 5C, D) with isolate 17–1177 sporulating the most.
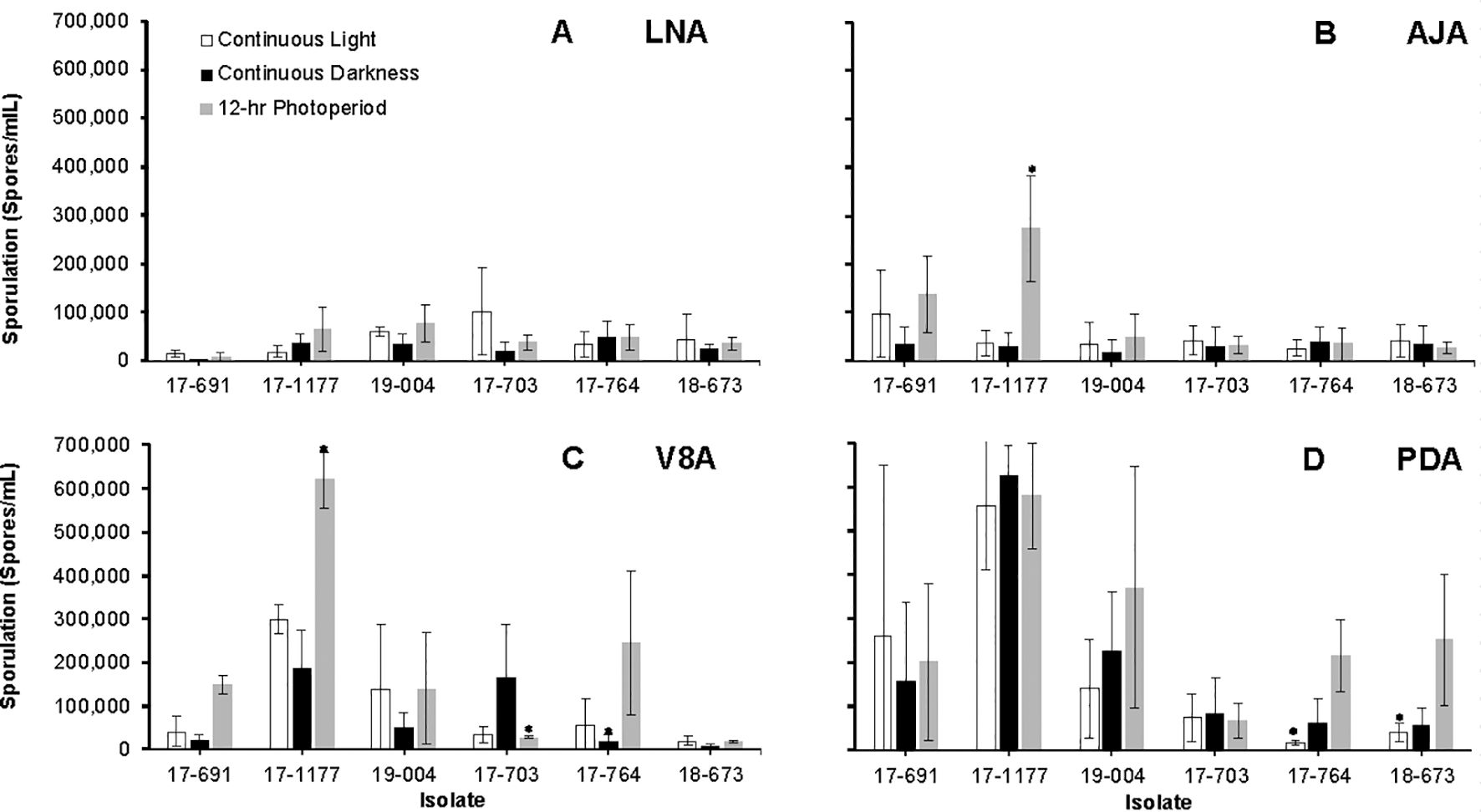
Figure 5. Average sporulation of six Neonectria candida isolates on low nutrient agar (LNA, A), apple juice agar (AJA, B), V8 agar (V8A, C), and potato dextrose agar (PDA, D) media under continuous light, continuous darkness, and a 12-hr alternating photoperiod, after 60 days of incubation at 23°C. Data bars are the average of six values per treatment across two experimental runs. Vertical bars indicate the standard deviations of the means. Asterisks indicate significant differences between media across isolates.
3.4 Pathogenicity and virulence on apple and pear fruit
All six N. candida isolates were pathogenic to apples and pears at both room and cold temperatures. There was no significant effect of the host (apple vs pear), isolate, temperature, and all associated interactions on Neonectria Fruit Rot (NFR) incidence. In contrast, virulence, expressed as the lesion size, was strongly influenced by all these factors and all associated interactions (P < 0.05). For most isolates, both NFR incidence and lesion diameter were greater on pear fruit at both temperatures except for isolate 17–691 which exhibited similar virulence on both hosts (Table 4). On ‘Fuji’ apples, NFR incidence ranged from 56% to 91.7% after 28 days at 22 °C and from 50% to 80.5% after 120 days at 1.5°C (Table 4). On ‘Green d’Anjou’, NFR incidence ranged from 5.6 to 94.4 after 28 days at 22 °C and from 80.6 to 100%, after four months at 0.5°C.
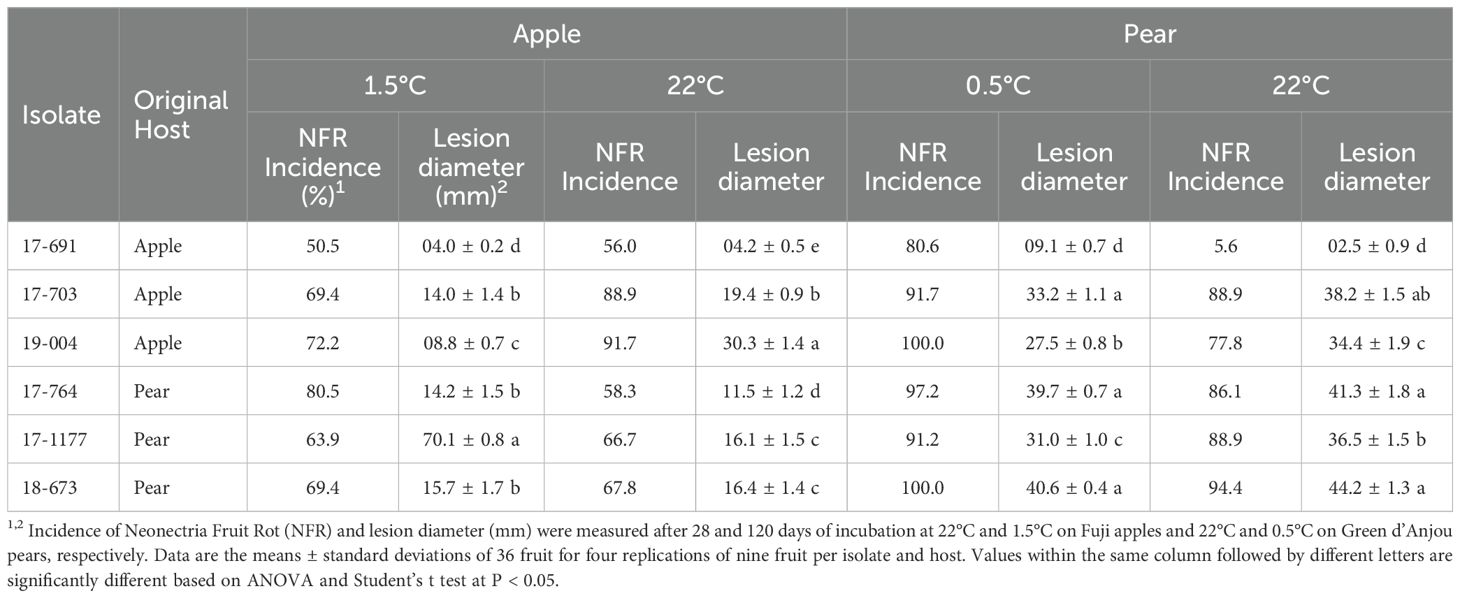
Table 4. Incidence of Neonectria fruit rot and lesion diameter caused by Neonectria candida on apple and pear fruit.
4 Discussion
Neonectria ditissima and N. punicea have previously been reported as causal agents of NFR on apples (Xu and Robinson, 2010; Wu et al., 2022; Wesche and Weber, 2023), while N. candida Wollenw (syn. N. ramulariae, anamorph Cylindrocarpon obtusiusculum) was associated with NFR on pears (Wenneker et al., 2016). In this study, Bayesian and Maximum Likelihood multilocus sequence analyses (MLSA) based on four concatenated and individual loci confirmed that all 52 Neonectria isolates collected from decayed apple and pear fruit clustered within a well-supported clade containing N. candida reference sequences, including isolates from Malus sylvestris from the UK and Portugal. This is the second report of N. candida causing NFR on pear following its initial report in the Netherlands (Wenneker et al., 2016) and represents the first report of N. candida as the causal agent of NFR on apple fruit globally. Bayesian phylogenetic analysis placed the N. candida clade as a sister group to clades containing N. ditissima and N. neomacrospora, while N. punicea, the third species previously linked to NFR on apples, was more distantly related. These relationships were further supported by pairwise ITS rDNA sequence divergence, which reached 5.5% between N. candida and N. ditissima, 7.1% with N. neomacrospora, and 8.1% with N. punicea.
The ecology and epidemiology of N. candida remain largely unresolved. First described as a root pathogen of alfalfa (Medicago sativa) (Cormack, 1937), the species has since been linked to cankers on almond seedlings (Prunus amygdalus) (Marek et al., 2013), bark cankers of beech in North America (Castlebury et al., 2006), beech seed rot in Japan (Hirooka et al., 2012), and postharvest decay of pear fruit in the Netherlands (Wenneker et al., 2016). Beyond its role as a plant pathogen, N. candida is considered a prominent colonizer of topsoil and leaf litter in European beech forests (Jankowiak et al., 2016) and a widespread soilborne fungus worldwide (Booth, 1966; Domsch et al., 2007). Whether potential reservoir hosts, such as American or Japanese beech, occur near the commercial pear and apple orchards surveyed in this study is unknown. Nevertheless, the fungus may persist in orchard soils or overwinter in surface litter, providing inoculum for the following season. Central Washington orchards commonly use sprinkler irrigation, which could splash N. candida conidia from the soil onto fruit, especially those low in the canopy. Wenneker et al. (2016) noted heavy soil contamination on pears with NFR in the Netherlands, underscoring the likely epidemiological role of soil and the value of orchard floor and bin sanitation in disease management. Moreover, although N. candida can induce cankers on almonds and beech (Castlebury et al., 2006; Marek et al., 2013), its capacity to produce cankers or dieback on apple and pear trees, and thereby generate primary inoculum for NFR, has not been studied in the PNW. In our study, in vitro cultures and inoculated fruit were observed for up to four months of cold storage, yet no perithecia with asci and ascospores developed. Determining whether the teleomorph occurs in regional orchards, and whether sexual reproduction contributes to the fungus’ life cycle, remains an important avenue for future research.
Both light regime and nutrient sources (agar media) significantly influenced the growth and sporulation of N. candida. Continuous light supported the most vigorous growth, followed by a 12-hour light/dark photoperiod, while continuous darkness was the least favorable. Among the media tested, V8A and LNA supported greater colony expansion compared to AJA or PDA. Across all lighting conditions, colonies on PDA developed yellowish to brown pigmentation after 14 days, consistent with previous observations (Lombard et al., 2015; Wenneker et al., 2016). Jankowiak et al. (2016), however, reported yellow to orange pigmentation under continuous darkness on PDA. Unlike N. ditissima, which requires specific light and media combinations to induce sporulation (Scheper et al., 2014), N. candida sporulated readily under all light regimes and on all media types tested, although it was more abundant under the 12-hour photoperiod on PDA and V8A. Conidia produced on PDA were predominantly two-septate, differing from the mostly one-septate conidia described by Jankowiak et al. (2016). While more isolates should be examined to confirm diagnostic traits, the conidia of N. candida, typically straight to ellipsoidal and approximately 30 µm long, may help distinguish this species morphologically from other NFR pathogens. In contrast, N. ditissima and N. punicea produce longer, crescent-shaped conidia ranging from 35 to 45 µm (Castlebury et al., 2006; Wu et al., 2022). Moreover, of the six isolates tested, only one (17–703 from apples) produced macro-conidia and did not cause significantly different NFR incidence, suggesting that micro and macroconidia of N. candida can equally infect pome fruit in contrast with N. ditissima (Wesche and Weber, 2023).
Pathogenicity tests conducted on detached, wounded, and artificially inoculated ‘Fuji’ apples and ‘Green d’Anjou’ pears showed that N. candida isolates from both apple and pear were pathogenic to fruit of both hosts. Infections produced similar symptoms across hosts: soft, brown lesions, while white to yellowish mycelia were mostly seen on pears. These findings on ‘Green d’Anjou’ pears are consistent with those previously reported for ‘Conference’ pears in the Netherlands (Wenneker et al., 2016) and confirms the pathogenicity of N. candida on apple fruit. Despite the difference in host origin, no evidence of host specificity was observed. However, broader screening across additional apple and pear cultivars is needed to fully evaluate host susceptibility. Notably, ‘Green d’Anjou’ and ‘Gala’ are the most widely planted pear and apple cultivars, respectively, in the PNW. Their demonstrated susceptibility to N. candida highlights a potential emerging pathogen to postharvest fruit quality in the region. In comparison, susceptibility of apple cultivars to N. ditissima, the causal agent of European canker, varies among apple cultivars with ‘Gala’, ‘Braeburn’, and ‘Red Delicious’ being the most susceptible, while ‘Golden Delicious’ and ‘Honeycrisp’ show reduced susceptibility (Dubin and English, 1974; Xu and Robinson, 2010; Gómez-Cortecero et al., 2016; Alves and Nunes, 2017). Whether such differences in susceptibility extend to fruit infections by N. candida remain unknown.
While pathogenicity studies of N. candida on other plant hosts are limited, previous research has documented 30% infection incidence in beech seeds (Hirooka et al., 2012) and a 0.2% incidence in beech litter (Jankowiak et al., 2016). In our study, isolates from pear were generally more virulent than those from apples, with disease incidence on pears ranging from 86.1% to 100%, compared to a broader range of 5.6% to 100% for apples. Notably, none of the apple isolates were obtained from ‘Fuji’, the cultivar used in pathogenicity assays, indicating a need for future virulence testing across additional apple cultivars. The ability of N. candida to cause cankers or shoot dieback on pome fruit trees remains untested. Given its association with cankers on beech trees (Castlebury et al., 2006), such potential should be investigated to clarify the pathogen’s epidemiology.
5 Conclusions
This study identifies N. candida as the causal agent of Neonectria fruit rot (NFR) in pear and documents the first confirmed case of NFR in apple globally. The pathogen was detected throughout the Pacific Northwest, from northern Washington to southern Oregon, demonstrating its capacity to thrive across diverse environmental conditions. Additional work is needed to clarify its etiology and epidemiology and to evaluate its sensitivity to current pre- and postharvest fungicides. Such research will be essential for designing effective management strategies that can curb future outbreaks and protect fruit quality.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JM: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. ARA: Data curation, Formal analysis, Investigation, Writing – review & editing. AA: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Funding for this research was provided by the U.S. Department of Agriculture, Foreign Agricultural Service, Technical Assistance for Specialty Crops (TASC) program (grant #2221-51300-34913) and by the Washington Tree Fruit Research Commission (grant # AP-16-105).
Acknowledgments
The authors gratefully acknowledge the Mike and Kathy Hambleton Fellowship for partial funding of this research, as well as David Rice, Washington State University, for assistance with statistical analysis and data visualization.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1661560/full#supplementary-material
References
Ali, E. M., Pandit, L. K., Mulvaney, K. A., and Amiri, A. (2018). Sensitivity of Phacidiopycnis spp. isolates from pome fruit to six pre- and postharvest fungicides. Plant Dis. 102, 533–539. doi: 10.1094/PDIS-07-17-1014-RE
Alves, S. A. M. and Nunes, C. C. (2017). Seasonal susceptibility of apple trees to Neonectria ditissima wound infections. New Zeal. Plant Prot. 70, 73–777. doi: 10.30843/nzpp.2017.70.30
Amiri, A. and Ali, M. E. (2016). “Prevalence of storage decays of apple: Lessons from the 2016 statewide survey,” in Fruit matters. Available online at: https://treefruit.wsu.edu/article/prevalence-of-storage-decays-of-apple-lessons-from-the-2016-statewide-survey/ (Accessed March 8, 2025).
Amiri, A., Hawkins, A. W., and Mulvaney, K. A. (2017). Study of fitness, virulence, and fungicide sensitivity of Lambertella corni-maris causing yellow rot on apple. Plant Dis. 101, 738–7743. doi: 10.1094/PDIS-08-16-1101-RE
Bertrand, P. F. and Saulie-Carter, J. (1980). Mucor rot of pears and apples (Corvallis, OR: Special Report 568, Agricultural Experiment Station, Oregon State University). Available online at: file:///C:/Users/a.amiri/Downloads/SR_no._568_ocr.pdf (Accessed February 22, 2025).
Brian, P. W., Elson, G. W., and Lowe, D. (1956). Production of patulin in apple fruits by Penicillium expansum. Nat. (London) 178, 263–2264. doi: 10.1128/am.28.4.589-593.1974
Carbone, I. and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Castlebury, L. A., Rossman, A. Y., and Hyten, S. A. (2006). Phylogenetic relationships of Neonectria/Cylindrocarpon on fagus in North America. Can. J. Bot. 84, 1417–11433. doi: 10.1139/b06-105
Chaverri, P., Salgado, C., Hirooka, Y., Rossman, A. Y., and Samuels, G. J. (2011). Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud. Mycol. 68, 57–578. doi: 10.3114/sim.2011.68.03
Cormack, M. W. (1937). Cylindrocarpon ehrenbergi w.r., and other species, as root parasites of alfalfa and sweet closer in Alberta. Can. J. Res. 15, 403–424. doi: 10.1139/cjr37c-031
Crous, P. W., Groenewald, J. Z., Risède, J.-M., Simoneau, P., and Hywel-Jones, N. L. (2004). Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Stud. Mycol. 50, 415–4430. doi: 10.3114/sim.55.1.213
Crous, P. W., Schoch, C. L., Hyde, K. D., Wood, A. R., Gueidan, C., de Hoog, G. S., et al. (2009). Phylogenetic lineages in the capnodiales. Stud. Mycol. 64, 17–47. doi: 10.3114/sim.2009.64.02
Domsch, K. H., Gams, W., and Anderson, T. H. (2007). Compendium of Soil Fungi, 2nd Taxonomically Revised Edition by W. Gams (Eching, Germany: IHW-Verlag), 672 pp.
Dubin, H. J. and English, H. (1974). Factors affecting apple leaf scar infection by Nectria galligena conidia. Phytopathology 64, 1201–11203. doi: 10.1094/Phyto-64-1201
Dugan, F. M. (2006). The identification of fungi: An illustrated introduction with keys, glossary, and guide to literature (St. Paul, MN, USA: APS Press), 176 pp.
Edwards, J. (2006). National diagnostic protocol for detection of Neonectria ditissima (European canker) (Canberra, Australia: Australian Government Department of Agriculture). Available online at: https://www.plantbiosecuritydiagnositcs.net.au/app/uploads/2018/11/NDP-21-European-canker-Neonectria-ditissima-V1.2.pdf (Accessed June 4, 2025).
Filajdić, N. and Sutton, T. B. (1991). Identification and distribution of Alternaria mali on apples in North Carolina and susceptibility of different varieties of apples to Alternaria blotch. Plant Dis. 75, 1045–11048. doi: 10.1094/PD-75-1045
Gerlach, W. and Nirenberg, H. (1982). The genus Fusarium: a pictorial atlas. Mitteil. Boil. Bundesanstalt fur Land- und Fortwirtschaft Berlin-Dahlem. 209, 1–406. Available online at: https://www.openagrar.de/servlets/MCRFileNodeServlet/openagrar_derivate_00040271/Mitt_BBA_209.pdf (Accessed March 8, 2025).
Ghasemkhani, M., Garkava-Gustavsson, L., Liljeroth, E., and Nyborn, H. (2016). Assessment of diversity and genetic relationships of Neonectria ditissima: the causal agent of fruit tree canker. Hereditas 153, 1–11. doi: 10.1186/s41065-016-0011-3
Gómez-Cortecero, A., Saville, R. J., Scheper, R. W. A., Bowen, J. K., De Medeiros, H. A., Kingsnorth, J., et al. (2016). Variation in host and pathogen in the Neonectria/Malus Interaction; toward an understanding of the genetic basis of resistance to European canker. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01365
Hansen, M. (2014). “China opens to Washington State apples,” in The Good Fruit Grower, vol. 10. Available online at: https://goodfruit.com/China-opens-to-washington-state-apples/ (Accessed June 4, 2025).
Hawksworth, D. L. (2011). A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. IMA Fungus 2, 155–162. doi: 10.5598/imafungus.2011.02.02.06
Henriquez, J. L., Sugar, D., and Spotts, R. A. (2007). Etiology of bull’s eye rot on pear caused by Neofabraea spp. in Oregon, Washington, and California. Plant Dis. 88, 1134–11138. doi: 10.1094/PDIS.2004.88.10.1134
Hirooka, Y., Ichihara, Y., Masuya, H., and Kubono, T. (2012). Seed rot, a new disease on beech tree caused by Neonectria ramulariae (anamorph: Cylindrocarpon obtusiusculum). J. Phytopathol. 160, 504–5506. doi: 10.1111/j.1439-0434.2012.01934.x
Hirooka, Y., Rossman, A. Y., Zhuang, W.-Y., Salgado-Salazar, C., and Chaverri, P. (2013). Species delimitation for Neonectria coccinea group including the causal agents of beech bark disease in Asia, Europe, and North America. Mycosystema 32, 485–517. Available online at: http://journals.im.ac.cn/jwxtcn/ch/reader/advance_query.aspx (Accessed March 8, 2025).
Holthusen, H. H. F. and Weber, R. W. S. (2021). Apple blossom-end rot due to Neonectria ditissima is initiated by infections at full flowering and incipient petal fall. New Zeal. Plant Prot. 74, S2–S8. doi: 10.30843/nzpp.2021.74.11727
Hua’an, Y., Sivasithamparam, K., and O’Brien, P. A. (1991). An improved technique for fluorescence staining of fungal nuclei and septa. Aust. Plant Pathol. 20, 192–1121. doi: 10.1071/app9910119
Jankowiak, R., Szwagrzyk, J., Bilański, P., and Stępniewska, H. (2016). Characterization of Cylindrocarpon-like species associated with litter in the old-growth beech forests of Central Europe. For. Pathol. 46, 582–594. doi: 10.1111/efp.12275
Karadžić, D., Stanivuković, Z., Milanovic, S., and Sikora, K. (2020). Development of Neonectria punicea pathogenic symptoms in juvenile Fraxinus excelsior. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.592260
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–11649. doi: 10.1093/bioinformatics/bts199
Khodadadi, F., González, J. B., Martin, P. L., Giroux, E., Bilodeau, G. J., Peter, K. A., et al. (2020). Identification and characterization of Colletotrichum species causing apple bitter rot in New York and description of C. noveboracense sp. nov. Sci. Rep. 10, 11043. doi: 10.1038/s41598-020-66761-9
Kim, Y. K. and Xiao, C. L. (2008). Distribution and incidence of Sphaeropsis rot in apple in Washington State. Plant Dis. 92, 940–9946. doi: 10.1094/PDIS-92-6-0940
Lombard, L., van der Merwe, N. A., Groenewald, J. Z., and Crous, P. W. (2015). Generic concepts in Nectriaceae. Stud. Mycol. 80, 189–245. doi: 10.1016/j.simyco.2014.12.002
Marek, S. M., Yaghmour, M. A., and Bostock, R. M. (2013). Fusarium spp., Cylindrocarpon spp., and environmental stress in the etiology of canker disease of cold-stored fruit and nut tree seedlings in California. Plant Dis. 97, 259–2270. doi: 10.1094/PDIS-04-12-0355-RE
Mellow, J. K. and Amiri, A. (2023). Characterization of the Neonectria spp. of pome fruit in Pacific Northwest. Phytopathology 113, 11–S3:63. doi: 10.1094/PHYTO-112-8-S2.22
Munson, R. G. (1939). Observations on apple canker: The discharge and germination of spores of Nectria galligena Bres. Ann. Appl. Biol. 26, 440–4456. doi: 10.1111/j.1744-7348.1939.tb06982.x
Nilsson, R. H., Kristiansson, E., Ryberg, M., Hallenberg, N., and Larsson, K. H. (2008). Intraspecific ITS variability in the Kingdom Fungi as expressed in the International Sequence Databases and its implications for molecular species identification. Evol. Bioinfor. 4, 193–1201. doi: 10.4137/EBO.S653
O’Donnell, K. and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are non orthologous. Mol. Phylog. Evol. 7, 103–1116. doi: 10.1006/mpev.1996.0376
Raeder, U. and Broda, P. (1985). Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–120. doi: 10.1111/j.1472-765X.1985.tb01479.x
R Core Team (2023). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available online at: https://www.R-project.org/ (Accessed February 22, 2025).
Rehner, S. A. and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–898. doi: 10.1080/15572536.2006.11832842
Ronquist, F. and Huelsenbeck, J. P. (2012). MrBayes 3: Bayesian phylogenetic inference under mixed mo. dels. Bioinf. 19, 1572–11574. doi: 10.1093/bioinformatics/btg180
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sanderson, P. G. and Spotts, R. A. (1995). Postharvest decay of winter pear and apple fruit caused by species of Penicillium. Phytopathology 85, 103–1110. doi: 10.1094/Phyto-85-103
Scheper, R. W. A., Fisher, B. M., Amponsah, N. T., and Walter, M. (2014). Effect of culture medium, light and air circulation on sporulation of Neonectria ditissima. New Zeal. Plant Prot. 67, 123–132. doi: 10.30843/nzpp.2014.67.5742
Schoch, C. L., Keith, A. S., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2011). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Nat. Acad. Sci. 109, 6241–66246. doi: 10.1073/pnas.1117018109
Sievers, F. and Higgins, D. G. (2014). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–1145. doi: 10.1002/pro.3290
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–11313. doi: 10.1093/bioinformatics/btu033
Sutton, T. B., Aldwinckle, H. S., Agnello, A. M., and Walgenbach, J. F. (2014). Compendium of apple and pear diseases and pest (St. Paul, MN. USA: APS Press), 218 pp.
Swofford, D. L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sunderland, MA, USA: Sinauer Associates). doi: 10.1002/0471650129.dob0522
Tulasne, L. R. and Tulasne, C. (1865). “Selecta fungorum carpologia: nectriei- phacidiei- pezizei (in Latin),” in Imperiali Typographeo Excudebatur, vol. 3. (Imperatoris Jussu, Paris, France), 3. doi: 10.5962/bhl.title.50436
Vilgalys, R. and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Crpytococcus species. J. Bacteriol. 172, 4238–44246. doi: 10.1128/jb.172.8.4238-4246.1990
Virginia, T. C., Néstor, A. J., Dario, C. A., and Noemi, P. G. (2021). Cladosporium species causing “Cladosporium rot” on “Bosc” pear fruit in Argentina. Rev. Argent. Microbiol. 53, 75–777. doi: 10.1016/j.ram.2019.11.006
Wenneker, M., Pham, K. T. K., Lemmers, M. E. C., de Boer, F. A., van der Lans, A. M., van Leeuwen, P. J., et al. (2016). First Report of Neonectria candida causing postharvest decay on “Conference” pears in the Netherlands. Plant Dis. 100, 1787. doi: 10.1094/PDIS-02-16-0247-PDN
Wesche, J. and Weber, R. W. S. (2023). Are microconidia infectious principles in Neonectria ditissima? J. Plant Dis. Prot. 130, 157–1162. doi: 10.1007/s41348-022-00669-6
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications (Academic Press, San Diego, CA), 315–322.
Wiseman, M. S., Dugan, F. M., Kim, Y. K., and Xiao, C. L. (2015). A postharvest fruit rot of apple caused by Lambertella corni-maris in Washington State. Plant Dis. 99, 201–2206. doi: 10.1094/PDIS-03-14-0327-RE
Wu, W., Luo, X., and Ren, M. (2022). First report of Neonectria punicea causing preharvest fruit rot of apple in Sichuan, China. Plant Dis. 106, 1759. doi: 10.1094/PDIS-10-21-2145-PDN
Xiao, C. L. and Boal, R. J. (2004). Prevalence and incidence of Phacidiopycnis rot in d’Anjou pears in Washington State. Plant Dis. 88, 413–4418. doi: 10.1094/PDIS.2004.88.4.413
Xu, X. M. and Robinson, J. D. (2010). Effects of fruit maturity and wetness on the infection of apple fruit by Neonectria galligena. Plant Pathol. 59, 542–5547. doi: 10.1111/j.1365-3059.2009.02232.x
Keywords: Neonectria spp., cylindrocarpon, pome fruit, virulence, fungal pathogen, postharvest, multilocus sequence analysis
Citation: Mellow JK, Arifin AR and Amiri A (2025) Morpho-phylogenic characterization of Neonectria candida as a causal agent of a postharvest rot of pome fruit in the U.S. Pacific Northwest. Front. Plant Sci. 16:1661560. doi: 10.3389/fpls.2025.1661560
Received: 08 July 2025; Accepted: 12 August 2025;
Published: 11 September 2025.
Edited by:
Carla M. R. Varanda, Research Centre for Natural Resources, Environment and Society (CERNAS), PortugalReviewed by:
Guillaume Legrand Ngolong Ngea, University of Molise, ItalyMaria Cristina Sosa, Universidad Nacional del Comahue, Argentina
Ilaria Martino, University of Turin, Italy
Copyright © 2025 Mellow, Arifin and Amiri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Achour Amiri, YS5hbWlyaUB3c3UuZWR1
 Joseph K. Mellow
Joseph K. Mellow Arild R. Arifin
Arild R. Arifin Achour Amiri
Achour Amiri