- School of Life Sciences, Qufu Normal University, Qufu, Shandong, China
Heat stress severely limits the productivity of alfalfa (Medicago sativa L.). In this study, the defender against apoptotic death 1 (DAD1) gene, MsDAD1, was identified and functionally characterized as a key positive regulator of heat tolerance. The expression of MsDAD1 was specifically and strongly induced by heat stress, and phylogenetic analysis confirmed its high conservation across plant species. Ectopic overexpression of MsDAD1 in transgenic alfalfa significantly enhanced tolerance to heat stress. Compared to wild-type plants, MsDAD1-overexpressing lines (MsDAD1-OE) exhibited reduced leaf chlorosis and abscission, higher relative water content, lower electrolyte leakage, greater chlorophyll retention, and diminished accumulation of reactive oxygen species (H2O2, O2-) and malondialdehyde (MDA), suggesting improved membrane integrity and reduced oxidative damage. Transcriptome (RNA-seq) analysis and subsequent physiological validation indicated that MsDAD1 suppresses heat-induced accumulation of jasmonic acid (JA) and abscisic acid (ABA) by down-regulating key biosynthetic genes, LOX1 and NCED1. As a result, MsDAD1-OE plants displayed attenuated JA- and ABA-mediated leaf senescence under heat stress. Furthermore, MsDAD1 overexpression delayed heat-induced flowering, correlating with the repression of flowering-promoting genes such as FT and ELF4. Collectively, these findings demonstrate that MsDAD1 enhances alfalfa heat tolerance by mitigating oxidative stress, modulating JA and ABA biosynthesis to delay senescence, and altering flowering time under high-temperature conditions. MsDAD1 represents a promising genetic target for improving heat resilience in alfalfa.
1 Introduction
Temperature is a critical environmental factor influencing plant growth, development, geographical distribution, quality, and productivity (Zhou et al., 2024). Global temperatures are projected to rise by approximately 0.2°C per decade, potentially reaching 1.8–4.0°C above current levels by the year 2100. Recent studies have shown that for each 1°C increase in average temperature, yields of major crops such as wheat, rice, maize, and soybeans decline by 6.0%, 3.2%, 7.4%, and 3.1%, respectively (Arshad and Hannoufa, 2022; Battisti and Naylor, 2009; Zhao et al., 2017). Heat stress induces a wide range of often detrimental changes in plant growth, morphology, physiological processes, and ultimately yield (Arshad and Hannoufa, 2022). At the cellular and biochemical levels, heat stress disrupts metabolite homeostasis and inhibits numerous physiological and biochemical processes. These include alterations in water status, membrane stability, photosynthesis, secondary metabolite production, and hormone balance (Guo et al., 2023). Heat stress rapidly impairs photosynthesis by damaging chloroplast ultrastructure, reducing photosynthetic pigment content, and impairing photosystem II function (Li et al., 2018). Additionally, it affects protein synthesis and stability, compromises membrane integrity, and induces the accumulation of reactive oxygen species (ROS) (Correia et al., 2022). Heat stress also disrupts organelle function, alters hormone signaling pathways (Ding and Yang, 2022), and perturbs calcium and lipid signaling as well as kinase activity (e.g., MAPKs, CBKs, CDPKs). These responses are accompanied by transcriptomic reprogramming and widespread metabolomic shifts (Haider et al., 2021).
The plant hormone jasmonic acid (JA) plays a critical role in the response to heat stress. Studies have shown that heat stress induces the accumulation of JA and its derivatives, including jasmonoyl-isoleucine (JA-Ile) and 12-oxo-phytodienoic acid (OPDA), thereby enhancing cell viability and heat tolerance in Arabidopsis (Wang et al., 2023). Exogenous application of JA to wild-type plants prior to heat exposure alleviates heat-induced damage, indicating that JA directly contributes to heat stress protection (Hu et al., 2017). However, excessive JA accumulation can also promote premature senescence in plants. For instance, the expression levels of LOX1 (LIPOXYGENASE 1), LOX3, and LOX4 increase markedly during leaf senescence, resulting in significantly higher JA concentrations in senescent leaves compared to non-senescent ones (Hu et al., 2017). Furthermore, treatment with exogenous JA accelerates leaf senescence and induces the expression of senescence-associated genes (Lim et al., 2007).
In addition to its role in stress responses, jasmonic acid (JA) also plays a critical role in inflorescence and flower development (Yuan and Zhang, 2015). JA has been identified as a key phytohormone regulating diurnal flower-opening time (DFOT) in rice (Zhu et al., 2024). In Arabidopsis, peroxisomal β-oxidation enzymes—including ACYL-COA OXIDASE (ACX), MULTIFUNCTIONAL PROTEIN (MFP; possessing 2-trans-enoyl-CoA hydratase and L-3-hydroxyacyl-CoA dehydrogenase activities), and 3-KETOACYL-COA THIOLASE (KAT)—are essential for proper inflorescence patterning (Yuan and Zhang, 2015; Ghasemi Pirbalouti et al., 2014; Schaller and Stintzi, 2009; Wiszniewski et al., 2014). Exogenous application of methyl jasmonate (MeJA) has been shown to promote flowering time and influence floral organ development in oilseed rape (Brassica napus L.) (Pak et al., 2009). More recently, MeJA treatment was found to accelerate DFOT in rice, with the proportion of opened florets increasing in a concentration-dependent manner (Wang M et al., 2024). While the roles of JA in plant development and stress adaptation are well established, the molecular mechanisms underlying its regulation of temperature-dependent flowering time and heat-induced leaf senescence remain largely unknown.
The DAD1 protein was initially identified in the temperature-sensitive tsBN7 mutant cell line (Zhou et al., 1997). DAD1 functions as a subunit of the oligosaccharyltransferase (OST) complex, a key catalytic component of the endoplasmic reticulum (ER) (Wang et al., 2024). The OST complex catalyzes N-glycosylation in the ER, facilitating the attachment of oligosaccharides to specific asparagine residues on nascent polypeptides. This modification is essential for proper protein folding and subsequent export from the ER (Yan et al., 2005; Roboti and High, 2012; Zhang et al., 2016). Although DAD1 family proteins have been implicated in salinity tolerance, high-light responses, and disease resistance in plants, their roles in heat stress regulation remain poorly understood (Wang et al., 2022; Yan et al., 2019; Wang X et al., 2024; Beaugelin et al., 2019).
In this study, we identified MsDAD1 as a heat-inducible gene in alfalfa. Overexpression of MsDAD1 suppressed the heat-induced hyperaccumulation of jasmonic acid (JA) and abscisic acid (ABA), enhanced reactive oxygen species (ROS) scavenging, and delayed both leaf senescence and flowering under heat stress. We propose that MsDAD1 functions as a heat-responsive “senescence brake” in alfalfa, offering a promising genetic target for future breeding efforts aimed at improving thermotolerance.
2 Materials and methods
2.1 Plant material, growth conditions and stress treatment
The alfalfa (Medicago sativa L.) genotypes used in this study included the wild-type cultivar SY4D and MsDAD1 transgenic lines (OE#1 and OE#3), which were generated in the SY4D background. Rooted stem cuttings of both wild-type and transgenic plants were prepared and transplanted into 10×10 cm pots. Plants were grown under controlled environmental conditions: a 16/8 h light/dark photoperiod, a temperature of 23°C, relative humidity of 50–70%, and a light intensity of 300 μmol m-² s-¹.
To analyze the expression pattern of MsDAD1 under heat stress, four-week-old alfalfa seedlings were transferred to a climate chamber and exposed to high-temperature treatment (40°C) for 0, 1, 3, 6, 12, 24, and 48 hours. To evaluate the heat tolerance function of MsDAD1, transgenic and wild-type plants were subjected to a controlled heat stress regime in a growth chamber set at 32°C (night)/40°C (day), allowing for plant survival and the assessment of physiological responses. Phenotypic evaluations were conducted after six days of treatment.
For flowering time analysis under both normal and heat stress conditions, above ground part of wild-type and MsDAD1-overexpressing (OE) plants were cut off at the same time. Plants were divided into two groups: one maintained under normal conditions (20°C night/23°C day), and the other exposed to a heat stress regime of 30°C (night)/35°C (day), with identical photoperiod and light intensity settings as the control group.
For hormone treatment assays, healthy mature leaves were divided into two groups. One group was placed on half-strength Murashige and Skoog (½ MS) medium (control), and the other was treated with 50 μM jasmonic acid (JA) or abscisic acid (ABA), respectively. Unless otherwise stated, all experiments were performed with at least three biological replicates.
2.2 Gene isolation and sequence analysis
The full-length coding sequence (CDS) of MsDAD1 was amplified using gene-specific primers listed in Supplementary Table S1. PCR amplification was performed with an initial denaturation at 98°C for 3 minutes, followed by 35 cycles of 98°C for 10 seconds, 55°C for 30seconds, and 72°C for 1 minute, with a final extension at 72°C for 10 minutes. The amplified product was ligated into the pMD19-T vector (Takara) and verified by Sanger sequencing. Homologous polypeptide sequences of DAD1 were retrieved from the GenBank database. Phylogenetic analysis was conducted using the neighbor-joining method. Sequence alignment was performed with ClustalX (www.clustal.org), and the phylogenetic tree was constructed using MEGA version 6.0 (www.megasoftware.net) with 1000 bootstrap replicates to assess branch support and reliability.
2.3 RNA extraction and qRT‐PCR
Total RNA was extracted using the MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China), following the manufacturer’s protocol. First-strand cDNA synthesis was performed using the TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). Quantitative real-time PCR (qRT-PCR) was conducted using 2× SYBR Green Mix (Vazyme, Cat. No. Q711-03) according to the manufacturer’s instructions. The ACT2 gene was used as an internal reference for normalization. Relative transcript levels were calculated using the 2^–ΔΔCt method (Livak and Schmittgen, 2001). Primer sequences are listed in Supplementary Table S1. Each biological sample was analyzed in triplicate.
2.4 Plasmid construction and genetic transformation
To generate transgenic alfalfa seedlings, the coding sequence of MsDAD1 was cloned and inserted into the binary vector 35S::NOS::1300, under the control of the CaMV:: 35S promoter. The resulting construct was introduced into alfalfa via Agrobacterium tumefaciens-mediated transformation, following the protocol described in Transgenic Plants: Methods and Protocols (Jiang et al., 2019). Transgenic lines were selected on hygromycin-containing medium, and successful integration of MsDAD1 was confirmed by PCR analysis.
2.5 Physiological measurement
Relative water content (RWC) was determined using the leaf saturation method. Electrolyte leakage (EL) was measured with a conductivity meter (BELL, BEC-6600, Dalian, China). Briefly, six fully expanded, healthy leaves were collected from the middle canopy of each plant at the same developmental stage. The fresh weight of each sample was recorded prior to analysis and then incubated in 25 mL of double-distilled water. After 2 hours of gentle shaking, the initial conductivity of the solution was measured using a DIST-5 conductometer (Hanna Instruments). Samples were then boiled to release all electrolytes, and the final conductivity was recorded. EL was expressed as a percentage of the total conductivity and normalized to fresh weight. Chlorophyll content was measured using a SPAD chlorophyll meter (SPAD-502; Konica Minolta Sensing, Japan). Hydrogen peroxide (H2O2), malondialdehyde (MDA), and superoxide anion (O2-) levels were quantified using commercial assay kits (Jiancheng Bioengineering Institute, Nanjing, China). Endogenous levels of jasmonic acid (JA) and abscisic acid (ABA) were quantified using 20 mg of fresh plant tissue. Phytohormones were extracted with 10% (v/v) methanol in water (MeOH/H2O). A cocktail of stable isotope-labeled internal standards was added to validate the liquid chromatography–mass spectrometry (LC-MS) quantification. The extracts were purified using Oasis hydrophilic-lipophilic balanced (HLB) columns (30 mg/1 mL; Waters), and targeted analytes were eluted with 80% (v/v) methanol. The eluent, containing both neutral and acidic compounds, was gently evaporated to dryness under a stream of nitrogen. Chromatographic separation was carried out using an Acquity Ultra Performance Liquid Chromatography (UPLC) system (Waters) equipped with an Acquity UPLC BEH C18 column (100 × 2.1 mm, 1.7 µm; Waters). The effluent was introduced into the electrospray ionization (ESI) source of a Xevo TQ-S triple quadrupole mass spectrometer (Waters) for targeted quantification of JA and ABA.
2.6 Transcriptomic analysis
Two-week-old wild-type and MsDAD1-overexpressing (OE) seedlings were cultivated as previously described. Leaf tissues were harvested and immediately frozen in liquid nitrogen for total RNA extraction. For each sample, 1.5 μg of mRNA was used as input for library preparation. RNA sequencing libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs), following the manufacturer’s instructions. RNA concentration was measured using a NanoDrop 2000C spectrophotometer (Thermo Scientific, Mississauga, Canada), and RNA integrity was assessed with an Agilent 2100 Bioanalyzer using an RNA Nano chip (Agilent Technologies, Santa Clara, CA, USA). RNA libraries were constructed and sequenced using the Illumina HiSeq 2500 platform at the Centre for Applied Genomics, SickKids Hospital (Toronto, Canada), under a fee-for-service agreement. Differential gene expression analysis between MsDAD1-OE lines and wild-type plants was performed using the DESeq2 R package (version 1.20.0). P-values were adjusted for multiple testing, and genes with an adjusted p-value< 0.001 were defined as differentially expressed genes (DEGs). Gene Ontology (GO) enrichment analysis was performed, and GO terms with a corrected p-value < 0.05 were considered significantly enriched. Functional annotation of DEGs was carried out using the NR, GO, and KEGG databases.
2.7 Statistical analysis
All experiments and gene expression analyses were conducted with at least three independent biological replicates. Results are presented as mean values ± standard error (SE). Statistical analyses were performed using one-way analysis of variance (ANOVA). Asterisks above columns indicate statistically significant differences compared to the control: p < 0.05 (*) and p < 0.01 (**). Different letters above histogram bars denote significant differences among treatments at p < 0.05, as determined by post hoc multiple comparison tests.
3 Results
3.1 The expression of MsDAD1 was significantly induced under heat stress in alfalfa
Based on transcriptomic analysis of alfalfa (Medicago sativa L.) from our previous study (Dong et al., 2018), a gene encoding defender against apoptotic death 1 (DAD1), designated MsDAD1 was identified. Phylogenetic analysis revealed that this gene shares the highest sequence homology with MtDAD1 from Medicago truncatula, a model legume species, and was thus named MsDAD1 (Figure 1A). Expression analysis showed that MsDAD1 is significantly upregulated in response to heat stress (Figure 1B), suggesting its potential role in high-temperature adaptation. The coding sequence (CDS) of MsDAD1 was subsequently cloned using primers listed in Supplementary Table S1. Consistent with other DAD1 orthologs, MsDAD1 encodes a protein with three predicted transmembrane (TM) domains (TM I/II/III) and contains a conserved oligosaccharyltransferase (OST) subunit domain (Figure 1C). Phylogenetic analysis further demonstrated that MsDAD1 shares 98%, 92%, and 87% sequence identity with DAD1 orthologs from M. truncatula, soybean (Glycine max), rice (Oryza sativa), and Arabidopsis thaliana, respectively, indicating that DAD1 is highly conserved across plant species (Figure 1C; Supplementary Table S1).
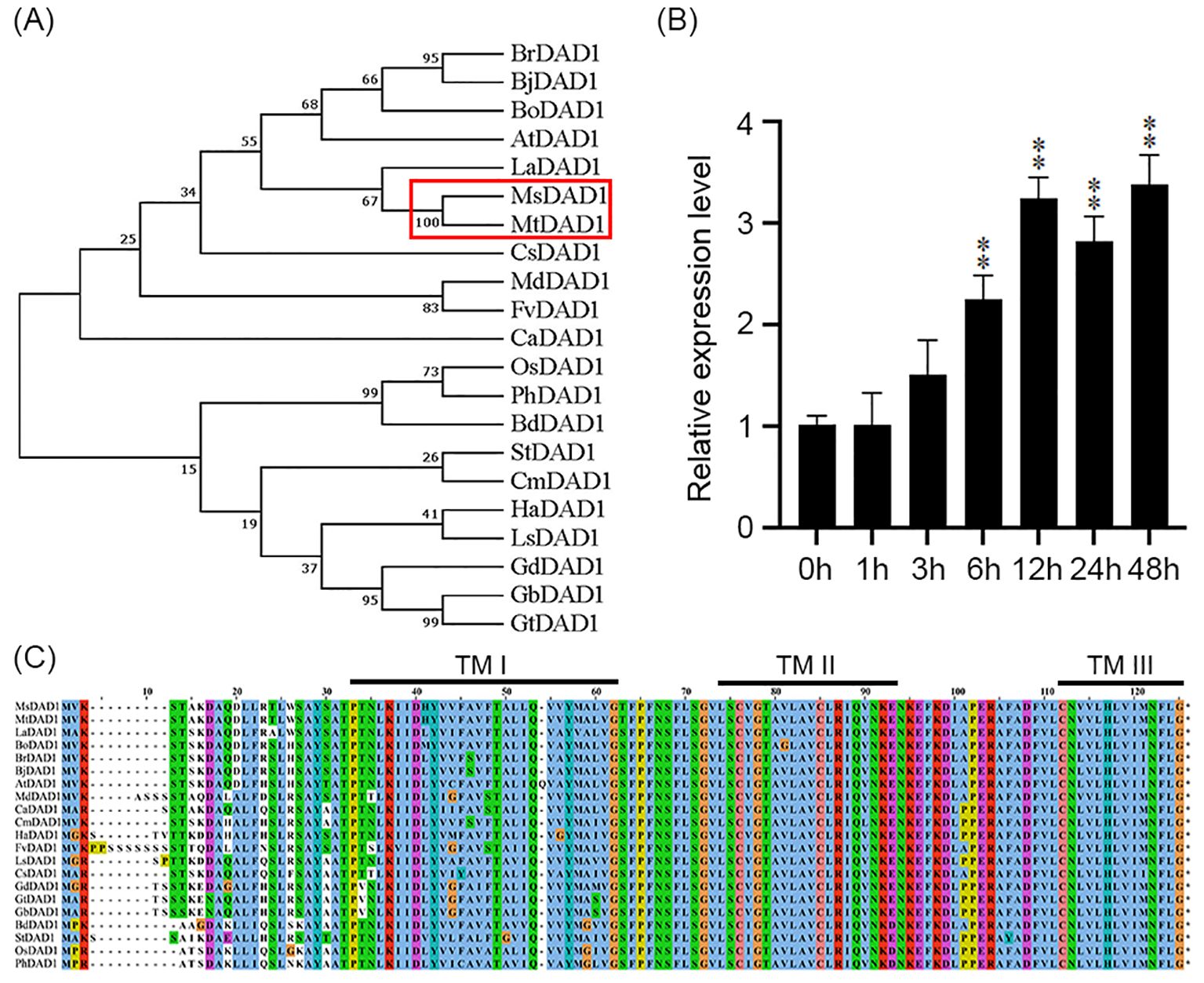
Figure 1. Sequence characteristics and heat stress response of MsDAD1. (A) Phylogenetic tree of DAD1 homologs from various plant species constructed using the neighbor-joining (NJ) method based on amino acid sequences. Bootstrap values were calculated from 1000 replicates. (B) Expression profile of MsDAD1 in alfalfa under heat stress (40°C) at different time points, as determined by qRT-PCR. Data represent mean ± SE of three biological replicates. (C) Conserved domain and structural features of MsDAD1 and its homologs, highlighting predicted transmembrane (TM) domains and oligosaccharyltransferase (OST) subunit regions. **Asterisks indicate that the expression level of MsDAD1 in samples treated with high-temperature stress shows a significant difference (p < 0.01) compared with the 0 h control.
3.2 Overexpression of MsDAD1 enhanced heat stress tolerance in alfalfa
To investigate the role of MsDAD1 in heat stress tolerance, transgenic alfalfa plants constitutively expressing MsDAD1 were developed through stable genetic transformation. qRT-PCR analysis of six independent transgenic lines revealed that MsDAD1-OE1 and MsDAD1-OE3 exhibited the highest transcript levels and were selected for subsequent experiments (Supplementary Figure S1). Phenotypic evaluation under heat stress showed that MsDAD1-OE plants were more resilient than wild-type (WT) plants. While WT plants displayed pronounced leaf chlorosis and abscission, MsDAD1-OE lines maintained greener, healthier foliage with reduced visible damage (Figures 2A, B).
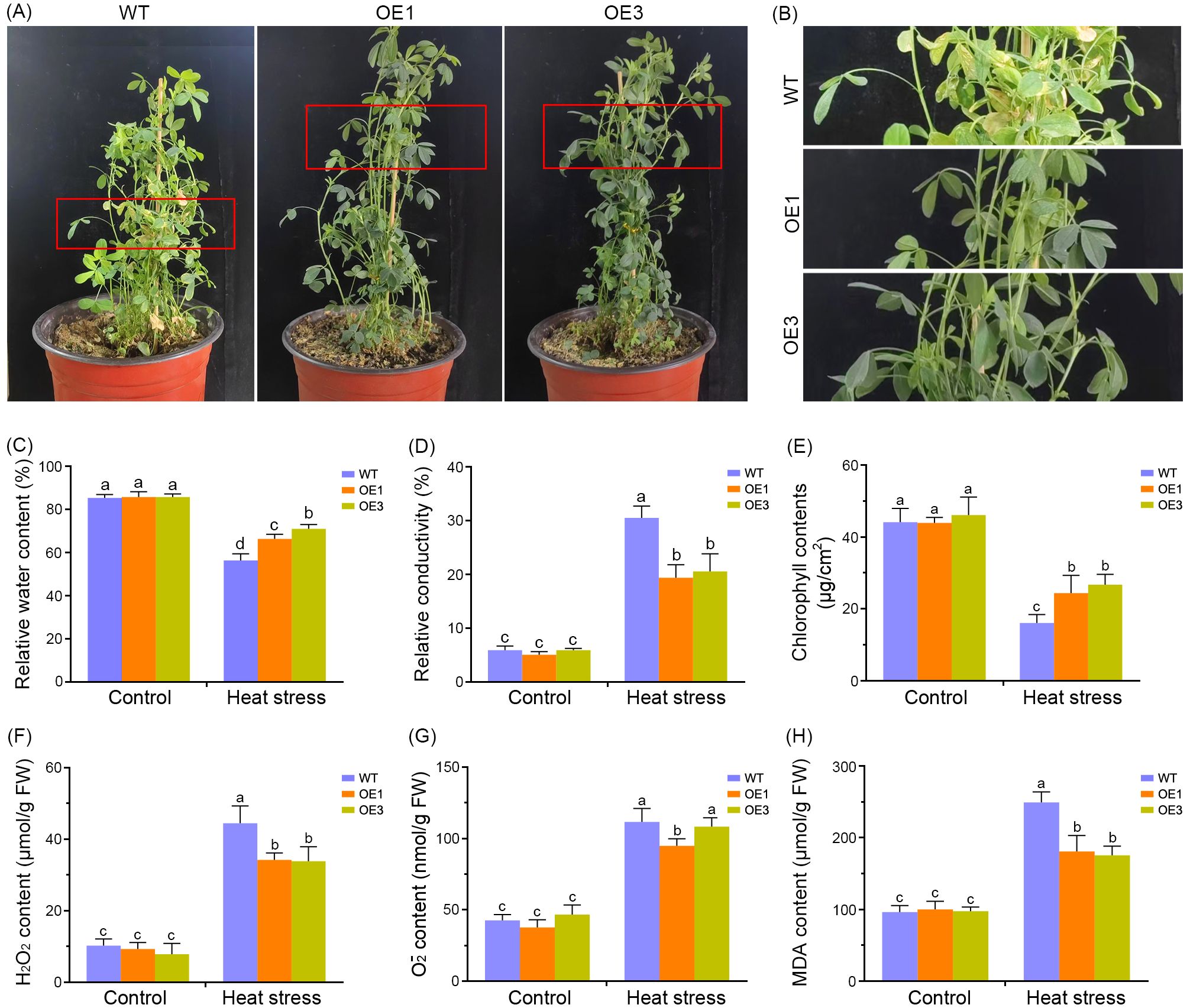
Figure 2. Overexpression of MsDAD1 enhances heat tolerance in alfalfa. (A) Phenotypic comparison of wild-type (WT) and MsDAD1-overexpressing (OE) lines after exposure to high-temperature stress (35°C night/40°C day) for six days. (B) Enlarged view of the red-boxed region from panel (A), highlighting leaf morphology differences. (C-H) Physiological responses of WT and MsDAD1-OE plants under control and heat stress conditions: (C) Relative water content (RWC), (D) Electrolyte leakage, (E) Chlorophyll content, (F) Hydrogen peroxide (H2O2) content, (G) Superoxide anion (O2-) content, (H) Malondialdehyde (MDA) content.
Physiological responses of WT and MsDAD1-OE lines were further evaluated under both normal and heat stress conditions. Under non-stress conditions, no significant differences in relative water content (RWC) were observed between the genotypes (Figure 2C). However, upon exposure to heat stress, RWC declined in all plants, with MsDAD1-OE lines maintaining significantly higher RWC compared to WT (Figure 2C). Electrolyte leakage, assessed via ion conductivity, was significantly elevated in both genotypes under heat stress, but the increase was more pronounced in WT plants, indicating greater membrane damage (Figure 2D). Correspondingly, MsDAD1-OE lines retained higher chlorophyll content than WT under heat stress, consistent with delayed senescence phenotypes (Figure 2E).
As heat stress disrupts reactive oxygen species (ROS) homeostasis, the accumulation of hydrogen peroxide (H2O2), superoxide anion (O2-), and malondialdehyde (MDA) were quantified. Under heat stress, MsDAD1-OE lines exhibited significantly lower levels of H2O2 and O2- compared to WT (Figures 2F, G). MDA content, a marker of lipid peroxidation and oxidative damage, was also markedly reduced in MsDAD1-OE plants relative to WT (Figure 2H). These results suggest that MsDAD1 overexpression mitigates heat-induced oxidative damage by enhancing ROS scavenging capacity.
Collectively, these findings demonstrate that MsDAD1-OE lines outperform WT under heat stress, exhibiting enhanced physiological stability, reduced oxidative damage, and improved stress tolerance.
3.3 MsDAD1 regulates the biosynthesis of jasmonic acid and abscisic acid in alfalfa
To elucidate the transcriptional changes regulated by MsDAD1, RNA-seq analysis on wild-type and MsDAD1-overexpressing (OE) alfalfa plants was performed. After quality control filtering, high-quality reads were retained for downstream analysis. A total of 1,088 differentially expressed genes (DEGs) were identified between WT and MsDAD1-OE lines, using thresholds of |Log2FoldChange| ≥ 1 and adjusted p-value < 0.05 (Figure 3A). Among these, 611 genes were upregulated and 477 genes were downregulated in MsDAD1-OE plants. qRT-PCR validation of six randomly selected genes confirmed the RNA-seq results, showing a high degree of consistency (Figures 3B–G).

Figure 3. RNA-seq analysis of wild-type and MsDAD1-OE alfalfa seedlings. (A) Volcano plot showing differentially expressed genes (DEGs) between wild-type (WT) and MsDAD1-overexpressing (OE) seedlings. DEGs were defined by |Log2FoldChange| ≥ 1 and adjusted p < 0.05. (B-G) Validation of selected DEGs by qRT-PCR. Expression levels are shown relative to WT. Data represent mean ± SE from three biological replicates. (H) Gene Ontology (GO) enrichment analysis of DEGs. Significantly enriched GO terms are shown based on biological processes. (I) KEGG pathway enrichment analysis of DEGs. Dot size indicates the number of genes associated with each pathway, while color intensity reflects the adjusted p-value (Padj).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses revealed that DEGs were primarily associated with cellular and metabolic processes, biosynthesis of secondary metabolites, plant–pathogen interactions, MAPK signaling, plant hormone signal transduction, glutathione metabolism, and flavonoid and phenylpropanoid biosynthesis (Figures 3H, I; Supplementary Tables S2, S3).
Given the critical role of plant hormones in heat stress responses, we focused on hormone-related genes. Transcriptome data revealed that MsDAD1-OE plants exhibited reduced expression of key genes involved in jasmonic acid (JA) and abscisic acid (ABA) biosynthesis specifically, LIPOXYGENASE1 (LOX1) and 9-cis-EPOXYCAROTENOID DIOXYGENASE1 (NCED1), respectively. This suggests that MsDAD1 may modulate JA and ABA metabolism during heat stress. To validate this, the expression of LOX1 and NCED1 in WT and MsDAD1-OE plants were examined under control and heat stress conditions. Under normal conditions, both genes showed slightly lower expression in MsDAD1-OE plants compared to WT, but the differences were not statistically significant (p >0.05). Upon exposure to high temperatures, expression of both LOX1 and NCED1 was significantly upregulated in both genotypes; however, the induction was notably stronger in WT plants (Figures 4B–E, G). LOX1 encodes a key enzyme in the JA biosynthetic pathway, catalyzing the conversion of α-linolenic acid to 13-hydroperoxyoctadecatrienoic acid (13-HPOT), a critical initial step in JA production (Figure 4A). NCED1 is the rate-limiting enzyme in ABA biosynthesis, catalyzing the oxidative cleavage of carotenoids to produce xanthoxin, a precursor of ABA (Figure 4F). To determine whether altered gene expression translated into changes in hormone levels, endogenous JA and ABA concentrations under both control and heat stress conditions were quantified. Heat stress significantly increased the accumulation of both hormones in WT and MsDAD1-OE plants; however, the increase was significantly greater in WT plants (Figures 4I, J).
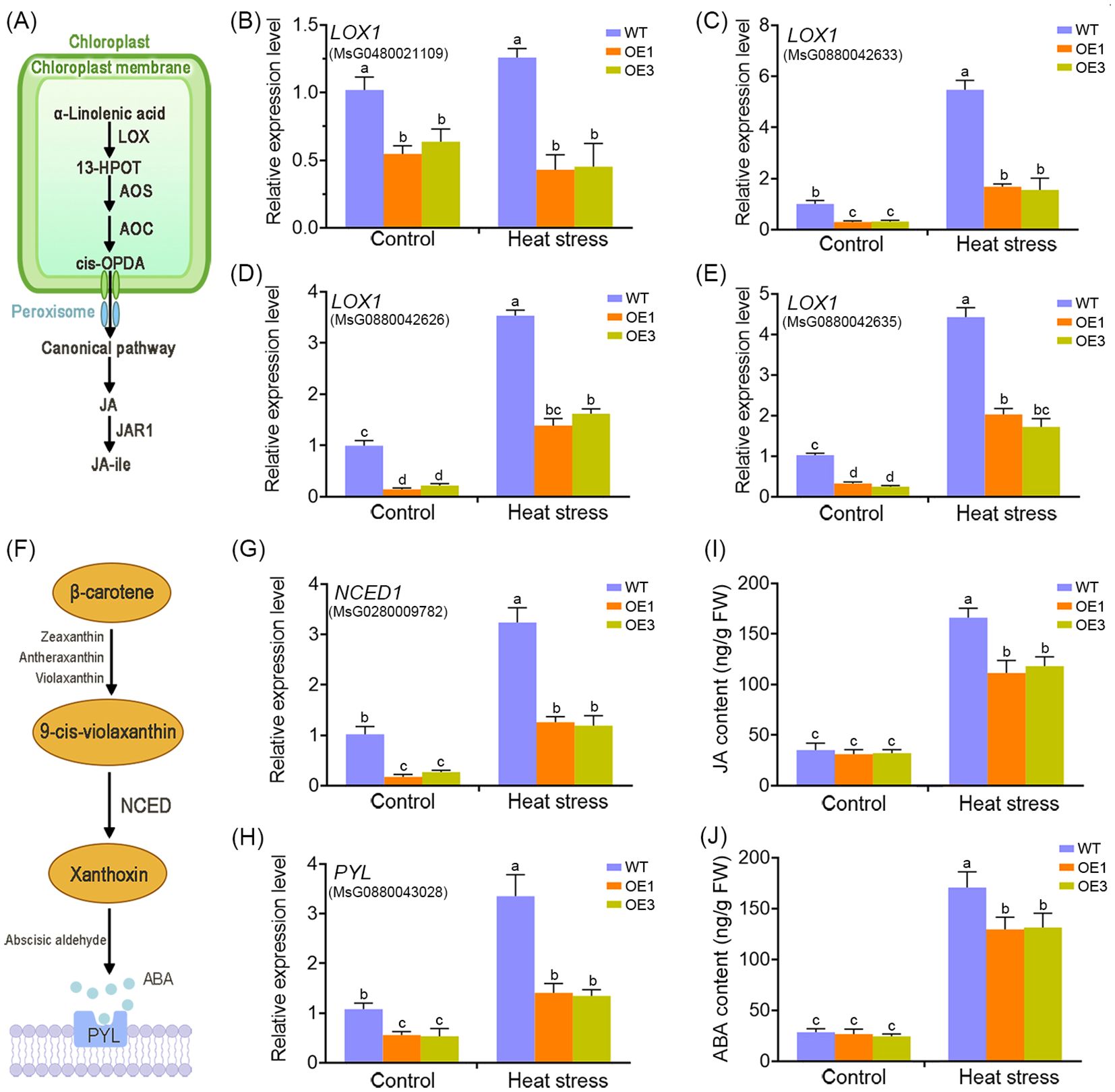
Figure 4. MsDAD1 regulates the biosynthesis of jasmonic acid (JA) and abscisic acid (ABA) in alfalfa. (A) Simplified schematic representation of the JA biosynthetic pathway in plants. (B-E) Relative expression levels of LOX1 in WT and MsDAD1-OE seedlings under normal and heat-stress conditions, as determined by qRT-PCR. (F) Simplified schematic representation of the ABA biosynthetic pathway. (G, H) Relative expression levels of NCED1 (a key ABA biosynthesis gene) and PYL (an ABA receptor gene) in WT and MsDAD1-OE seedlings under normal and heat-stress conditions. (I, J) Endogenous levels of JA (I) and ABA (J) in WT and MsDAD1-OE seedlings under normal and heat-stress conditions.
Together, these results indicate that MsDAD1 negatively regulates the heat-induced accumulation of JA and ABA in alfalfa, likely through suppression of LOX1 and NCED1, contributing to enhanced stress tolerance.
3.4 Ectopic expression of MsDAD1 suppressed JA/ABA-induced leaf senescence
To assess the involvement of JA and ABA in regulating leaf senescence in alfalfa, senescence phenotypes were evaluated following exogenous hormone treatments. Compared to the control, application of either JA or ABA significantly accelerated chlorophyll degradation, leaf yellowing, electrolyte leakage, and malondialdehyde (MDA) accumulation in both wild-type and MsDAD1-OE plants (Figures 5A-D). However, the severity of these senescence-associated responses was moderately attenuated in the MsDAD1-OE lines relative to wild-type plants. Under JA and ABA treatments, MsDAD1-OE plants retained higher chlorophyll content and exhibited lower electrolyte leakage and MDA levels compared to the wild type (Figures 5B-D). These findings suggest that MsDAD1 delays the progression of leaf senescence by limiting excessive JA and ABA accumulation, particularly under heat stress conditions.

Figure 5. Overexpression of MsDAD1 significantly suppresses JA- and ABA-induced leaf senescence. (A) Leaf senescence phenotypes of wild-type (WT) and MsDAD1-OE seedlings following treatment with control (mock), exogenous jasmonic acid (JA), or abscisic acid (ABA). (B-D) Quantification of chlorophyll content (B), electrolyte leakage (C), and malondialdehyde (MDA) content (D) in leaves corresponding to panel (A).
3.5 MsDAD1 is a key regulator involved in heat-mediated flowering in alfalfa
Extensive evidence indicates that heat stress can significantly disrupt the vegetative-to-reproductive transition in plants. In this study, longitudinal monitoring of developmental progression revealed a marked delay in flowering time in MsDAD1-OE lines compared to wild-type (WT) plants (Figure 6A). Under normal growth conditions, MsDAD1-OE plants exhibited delayed flowering, as reflected by a longer time to floral initiation, increased node number, and greater plant height at flowering onset (Figures 6B–D). Although high-temperature stress accelerated flowering in alfalfa, this effect was notably attenuated in MsDAD1-OE lines, suggesting a role for MsDAD1 in modulating temperature-dependent flowering responses (Figures 6B–D).
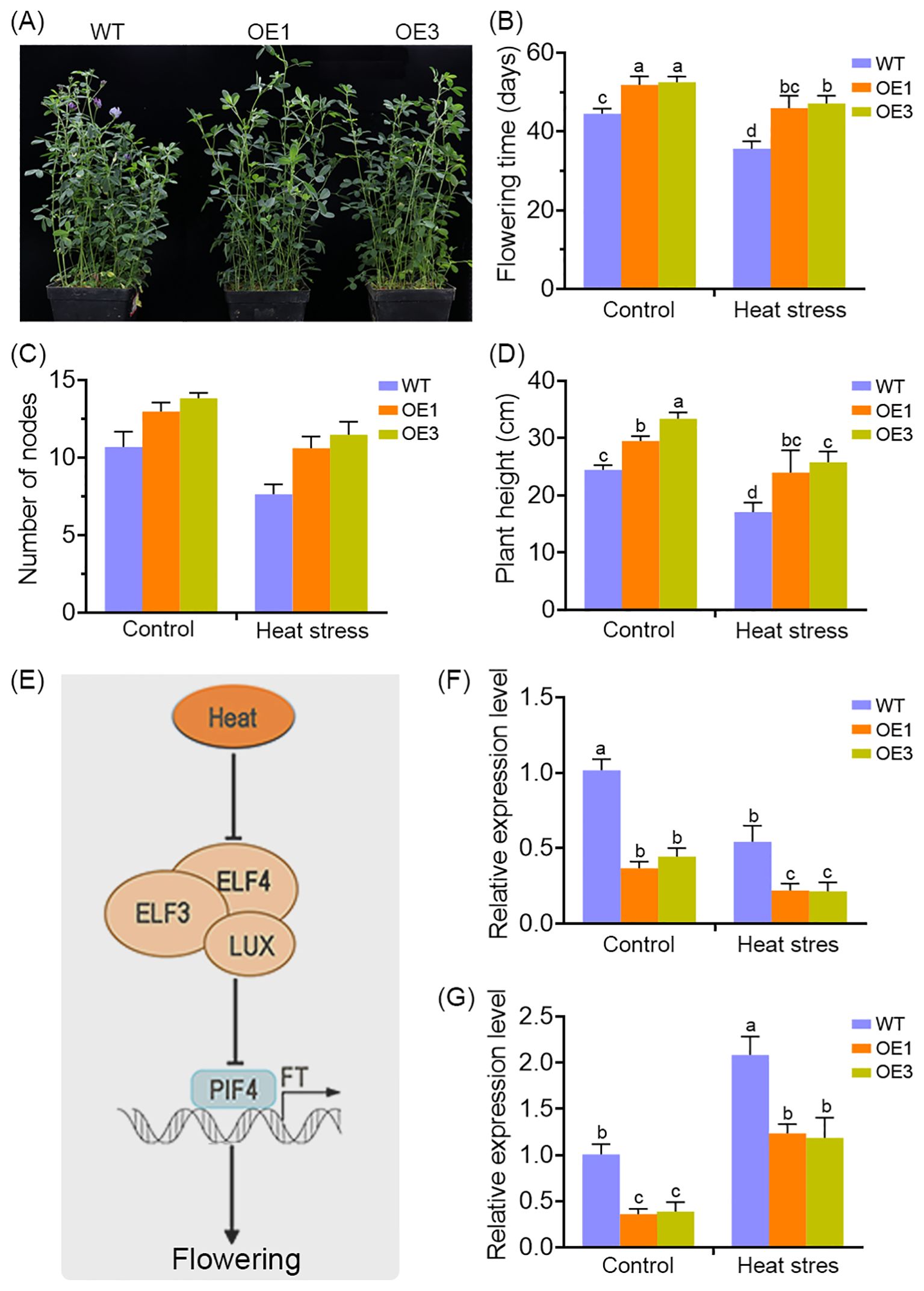
Figure 6. MsDAD1 modulates heat stress-associated flowering in alfalfa. (A) Overexpression of MsDAD1 delays flowering time in alfalfa under both normal and high-temperature conditions. (B-D) Quantification of flowering-related physiological traits in WT and MsDAD1-OE seedlings under control and heat stress conditions: (B) days to flowering, (C) node number at flowering, and (D) plant height at flowering. (E) Simplified schematic of the molecular module involved in heat stress-mediated flowering regulation, highlighting the roles of the evening complex (EC), PIF4, and FT. (F, G) Expression analysis of ELF4 (F) and FT (G) in WT and MsDAD1-OE seedlings under control and heat stress conditions, as determined by qRT-PCR.
Given the late-flowering phenotype of the MsDAD1-OE plants, the expression of flowering-related differentially expressed genes (DEGs) identified in our RNA-seq dataset were examined. Several key flowering regulators—such as FLOWERING LOCUS T (FT), EARLY FLOWERING 4 (ELF4), and FLOWERING TIME CONTROL GENE (FY)—were downregulated in MsDAD1-OE plants (Figure 6E). These genes are known to play critical roles in regulating flowering under heat stress conditions.
To validate these observations, we analyzed the expression of FT and ELF4 via qRT-PCR in both WT and MsDAD1-OE plants. Under normal conditions, expression levels of both genes were significantly lower in MsDAD1-OE lines compared to WT. Heat stress induced the expression of FT and ELF4 in both genotypes, but the induction was significantly stronger in WT plants (Figures 6F, G). These results suggest that MsDAD1 overexpression suppresses the heat-induced upregulation of flowering-promoting genes, thereby contributing to delayed flowering under elevated temperatures.
4 Discussion
Global warming has led to an increased frequency of extreme high-temperature events. Rising ambient temperatures driven by climate change are altering the geographical distribution of plant species and affecting a range of morphological and developmental traits, ultimately posing a serious threat to crop productivity (Matthews et al., 2019; Pausas, 2025). Alfalfa (Medicago sativa L.), though well-adapted to warm, semi-humid, and semi-arid environments, is particularly sensitive to high-temperature stress (Buzzanca et al., 2025). Exposure to heat combined with high humidity accelerates leaf senescence and triggers premature defoliation, significantly reducing both yield and forage quality (Arshad and Hannoufa, 2022).
In our previous transcriptomic analysis comparing salt-tolerant and salt-sensitive alfalfa genotypes, we identified MsDAD1 and MsDAD2 as salinity-induced genes (Dong et al., 2018). Functional studies revealed that overexpression of MsDAD2 enhanced salt tolerance in transgenic alfalfa (Wang X al., 2024). Despite sharing 95% sequence similarity, MsDAD1 and MsDAD2 display distinct stress-response profiles: MsDAD1 is strongly induced by high-temperature stress, while MsDAD2 is not. This divergence suggests that MsDAD1 and MsDAD2 may participate in separate abiotic stress signaling pathways.
The DAD1 gene was originally identified in a temperature-sensitive mutant cell line (tsBN7) in animals, where it was shown to play a role in programmed cell death suppression (Zhou et al., 1997). DAD1 encodes a subunit of the oligosaccharyltransferase (OST) complex, a central component of the endoplasmic reticulum (ER) machinery responsible for N-glycosylation of nascent proteins (Yan et al., 2005). This process involves the attachment of oligosaccharides to specific asparagine residues and is essential for correct protein folding and ER export (Kukuruzinska and Lennon, 1998). In plants, the functional roles of DAD1 homologs under abiotic stress remain incompletely understood. For instance, AtDAD1 has been shown to protect Arabidopsis protoplasts from UV-C-induced programmed cell death (PCD) (Danon et al., 2004), and in Gladiolus L., DAD1 expression sharply declines during petal senescence (Yamada et al., 2004). However, the molecular mechanisms by which DAD1 proteins mediate stress responses and developmental processes remain largely uncharacterized.
MsDAD1 suppresses the expression of key rate-limiting enzymes LOX1 and NCED1, involved in jasmonic acid (JA) and abscisic acid (ABA) biosynthesis, respectively (Figure 4). This suppression was particularly pronounced under high-temperature stress conditions. JA and ABA are widely recognized as stress hormones involved in plant responses to both biotic and abiotic stressors (Wang et al., 2025). In addition to their roles in stress signaling, these hormones regulate several physiological processes, including root elongation, reproductive organ development, and senescence (Wan et al., 2025; Varshney and Majee, 2021; Kim et al., 2018). Previous studies have shown that heat shock activates the JA signaling pathway and promotes JA accumulation, as observed in agarwood and Arabidopsis through increased expression of biosynthetic genes such as OPR3 (Wang et al., 2023; Xu et al., 2016; Tian et al., 2020). However, contrasting findings have been reported, suggesting a more nuanced role of JA in heat responses. For instance, Du et al. (2013) reported that genes involved in JA biosynthesis were downregulated under heat stress but upregulated during drought and cold stress. Similarly, Zhu et al. (2021) showed that elevated temperatures in Arabidopsis lead to reduced JA levels due to the upregulation of JOXs and ST2A, which degrade active JA. In cotton, high temperatures suppressed the expression of GhAOC2 in anthers, leading to reduced JA biosynthesis (Khan et al., 2023). These seemingly contradictory findings may stem from differences in plant species, experimental designs, stress intensity, or exposure duration. Thus, JA levels are not static during heat stress but are influenced by multiple factors. Short-term or moderate heat stress may elevate JA levels to promote stress tolerance, whereas prolonged or extreme heat stress can lead to excessive JA and ABA accumulation, which may trigger premature senescence and cell death. In Arabidopsis, OXI1 and DAD1 were shown to antagonistically regulate light-induced cell death through modulation of JA and salicylic acid (SA) levels (Beaugelin et al., 2019). Furthermore, many studies have demonstrated that ABA and JA can act synergistically under environmental stress conditions (Wang et al., 2025). In Arabidopsis and tobacco, ABA receptor proteins such as PYRABACTIN RESISTANCE1-Like (PYLs) regulate metabolic reprogramming via the JA signaling pathway (Aleman et al., 2016). These findings point to a complex JA–ABA crosstalk network that fine-tunes plant metabolism and growth.
Whether MsDAD1 participates directly in metabolic homeostasis or signaling crosstalk between JA and ABA remains to be determined. Future research is needed to elucidate how MsDAD1 specifically responds to high-temperature stress and modulates JA and ABA biosynthesis or signaling. In addition to its role in hormone regulation, MsDAD1 also appears to influence flowering time in alfalfa. Plants overexpressing MsDAD1 exhibited delayed flowering under both normal and heat stress conditions (Figures 6A-D). Transcriptome profiling revealed significant downregulation of the flowering-time regulators FLOWERING LOCUS T (FT) and EARLY FLOWERING 4 (ELF4), with a more pronounced effect under heat stress (Figures 6E-G). The role of FT in regulating flowering time is well established in various plant species, including alfalfa (Kang et al., 2019). In Arabidopsis thaliana, ELF3 functions as a central component in temperature sensing and thermomorphogenesis by participating in the evening complex (EC), together with ELF4 and LUX ARRYTHMO (LUX) (Zhu et al., 2023; Liu et al., 2024). Recent studies suggest that warm temperatures inhibit the EC complex’s DNA-binding activity by reducing the subnuclear localization of ELF3, thereby permitting PIF4 to interact with FT and promote flowering (Preston and Fjellheim, 2022). This EC–PIF4–FT module represents a critical mechanism in temperature-regulated flowering. Based on our current findings, we propose that MsDAD1 may modulate flowering time in alfalfa through regulation of the EC–PIF4–FT signaling axis. However, the precise molecular mechanism by which MsDAD1 interfaces with this pathway remains largely unexplored and warrants further investigation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YS: Funding acquisition, Writing – review & editing. XG:Investigation, Writing – original draft. LW: Investigation, Writing – original draft. YZ: Investigation, Writing – original draft. TL: Data curation, Writing – original draft. WD: Funding acquisition, Investigation, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant nos. 32371759, 32070304).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1664465/full#supplementary-material
Supplementary Figure 1 | qRT-PCR analysis of MsDAD1 in wild type and transgenic seedlings.
Supplementary Table 1 | List of primers used in this study.
Supplementary Table 2 | GO analysis of the DEGs between the wild-type and MsDAD1-OE line.
Supplementary Table 3 | KEGG analysis of DEGs between the wild-type and MsDAD1-OE line.
References
Aleman, F., Yazaki, J., Lee, M., Takahashi, Y., Kim, A. Y., Li, Z., et al. (2016). An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci. Rep. 6, 28941. doi: 10.1038/srep28941, PMID: 27357749
Arshad, I. M. and Hannoufa, A. (2022). Alfalfa transcriptome profiling provides insight into miR156-mediated molecular mechanisms of heat stress tolerance. Genome. 65, 315–330. doi: 10.1139/gen-2021-0099, PMID: 35298891
Battisti, D. S. and Naylor, R. L. (2009). Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 323, 240–244. doi: 10.1126/science.1164363, PMID: 19131626
Beaugelin, I., Chevalier, A., D'Alessandro, S., Ksas, B., Novák, O., Strnad, M., et al. (2019). OXI1 and DAD regulate light-induced cell death antagonistically through jasmonate and salicylate levels. Plant Physiol. 180, 1691–1708. doi: 10.1104/pp.19.00353, PMID: 31123095
Buzzanca, C., D'Amico, A., Gabrielli, P., Durazzo, A., Lucarini, M., and Di Stefano, V. (2025). Alfalfa (Medicago sativa L.): literature quantitative research analysis. Nat. Prod Res. 9, 1–10. doi: 10.1080/14786419.2025.2486335, PMID: 40340622
Correia, P. M. P., Cairo Westergaard, J., Bernardes da Silva, A., Roitsch, T., CarmoSilva, E., Marques da Silva, J., et al. (2022). High-throughput phenotyping of physiological traits for wheat resilience to high temperature and drought stress. J. Exp. Bot. 73, 5235–5251. doi: 10.1093/jxb/erac160, PMID: 35446418
Danon, A., Rotari, V. I., Gordon, A., Mailhac, N., and Gallois, P. (2004). Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J. Biol. Chem. 279, 779–787. doi: 10.1074/jbc.M304468200, PMID: 14573611
Ding, Y. and Yang, S. (2022). Surviving and thriving: how plants perceive and respond to temperature stress. Dev. Cell. 57, 947–958. doi: 10.1016/j.devcel.2022.03.010, PMID: 35417676
Dong, W., Liu, X., Li, D., Gao, T., and Song, Y. (2018). Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa. PloS One 13, e0204033. doi: 10.1371/journal.pone.0204033, PMID: 30252877
Du, H., Liu, H., and Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00397, PMID: 24130566
Ghasemi Pirbalouti, A., Sajjadi, S. E., and Parang, K. (2014). A review (research and patents) on jasmonic acid and its derivatives. Arch. Pharm. (Weinheim). 347, 229–239. doi: 10.1002/ardp.201300287, PMID: 24470216
Guo, L., Tan, J., Deng, X., Mo, R., Pan, Y., Cao, Y., et al. (2023). Integrated analysis of metabolome and transcriptome reveals key candidate genes involved in ffavonoid biosynthesis in Pinellia ternata under heat stress. J. Plant Res. 136, 359–369. doi: 10.1007/s10265-023-01446-8, PMID: 36881276
Haider, S., Iqbal, J., Naseer, S., Yaseen, T., Shaukat, M., Bibi, H., et al. (2021). Molecular mechanisms of plant tolerance to heat stress: current landscape and future perspectives. Plant Cell Rep. 40, 2247–2271. doi: 10.1007/s00299-021-02696-3, PMID: 33890138
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004, PMID: 28201612
Jiang, Q., Fu, C., and Wang, Z. (2019). A Unified Agrobacterium-Mediated Transformation Protocol for Alfalfa (Medicago sativa L.) and Medicago truncatula. Methods Mol. Biol. 1864, 153–163. doi: 10.1007/978-1-4939-8778-811, PMID: 30415335
Kang, J., Zhang, T., Guo, T., Ding, W., Long, R., Yang, Q., et al. (2019). Isolation and functional characterization of msFTa, a FLOWERING LOCUS T homolog from alfalfa (Medicago sativa). Int. J. Mol. Sci. 20, 1968. doi: 10.3390/ijms20081968, PMID: 31013631
Khan, A. H., Ma, Y., Wu, Y., Akbar, A., Shaban, M., Ullah, A., et al. (2023). High-temperature stress suppresses allene oxide cyclase 2 and causes male sterility in cotton by disrupting jasmonic acid signaling. Crop J. 11, 33–45. doi: 10.1016/j.cj.2022.05.009
Kim, J. A., Bhatnagar, N., Kwon, S. J., Min, M. K., Moon, S. J., Yoon, I. S., et al. (2018). Transcriptome analysis of ABA/JA-dual responsive genes in rice shoot and root. Curr. Genomics 19, 4–11. doi: 10.2174/1389202918666170228134205, PMID: 29491728
Kukuruzinska, M. A. and Lennon, K. (1998). Protein N-glycosylation: molecular genetics and functional significance. Crit. Rev. Oral. Biol. Med. 9, 415–448. doi: 10.1177/10454411980090040301, PMID: 9825220
Li, B., Gao, K., Ren, H., and Tang, W. (2018). Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 60, 757–779. doi: 10.1111/jipb.12701, PMID: 30030890
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316, PMID: 17177638
Liu, Z., Liu, W., Wu, Q., Xie, Z., Qi, K., Zhang, S., et al. (2024). Dual roles of pear EARLY FLOWERING 4 -like genes in regulating flowering and leaf senescence. BMC Plant Biol. 24, 1117. doi: 10.1186/s12870-024-05850-7, PMID: 39581970
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262, PMID: 11846609
Matthews, C., Arshad, M., and Hannoufa, A. (2019). Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant 165, 830–842. doi: 10.1111/ppl.12787, PMID: 29923601
Pak, H., Guo, Y., Chen, M., Chen, K., Li, Y., Hua, S., et al. (2009). The effect of exogenous methyl jasmonate on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in the development of oilseed rape flowers (Brassica napus L.). Planta 231, 79–91. doi: 10.1007/s00425-009-1029-9, PMID: 19826836
Pausas, J. G. (2025). The onset of phenological plant response to climate warming. New Phytol. doi: 10.1111/nph.70268, PMID: 40495351
Preston, J. C. and Fjellheim, S. (2022). Flowering time runs hot and cold. Plant Physiol. 190, 5–18. doi: 10.1093/plphys/kiac111, PMID: 35274728
Roboti, P. and High, S. (2012). The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J. Cell Sci. 125, 3474–3484. doi: 10.1242/jcs.103952, PMID: 22467853
Schaller, A. and Stintzi, A. (2009). Enzymes in jasmonate biosynthesis structure, function, regulation. Phytochemistry 7, 1532–1538. doi: 10.1016/j.phytochem.2009.07.032, PMID: 19703696
Tian, X., Wang, F., Zhao, Y., Lan, T., Yu, K., Zhang, L., et al. (2020). Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnol. J. 18, 1109–1111. doi: 10.1111/pbi.13268, PMID: 31559685
Varshney, V. and Majee, M. (2021). JA shakes hands with ABA to delay seed germination. Trends Plant Sci. 26, 764–766. doi: 10.1016/j.tplants, PMID: 34053891
Wan, Q., Yao, R., Zhao, Y., and Xu, L. (2025). JA and ABA signaling pathways converge to protect plant regeneration in stress conditions. Cell Rep. 44, 115423. doi: 10.1016/j.celrep.2025.115423, PMID: 40088448
Wang, M., Fan, X., and Ding, F. (2023). Jasmonate: A hormone of primary importance for temperature stress response in plants. Plants (Basel). 12, 4080. doi: 10.3390/plants12244080, PMID: 38140409
Wang, X., Tang, H., Lu, T., Shen, P., Chen, J., Dong, W., et al. (2024). Novel underlying regulatory mechanism of the MsDAD2-mediated salt stress response in alfalfa. Biochem. Biophys. Res. Commun. 690, 149252. doi: 10.1016/j.bbrc.2023.149252, PMID: 37995452
Wang, X., Tang, C., Zhang, H., Xu, J., Liu, B., Lv, J., et al. (2022). TaDAD2, a negative regulator of programmed cell death, is important for the interaction between wheat and the stripe rust fungus, Mol. Plant Microbe Interact. 24, 79–90. doi: 10.1094/MPMI-06-10-0131, PMID: 20795855
Wang, M., Zhu, X., Huang, Z., Chen, M., Xu, P., Liao, S., et al. (2024). Controlling diurnal flower-opening time by manipulating the jasmonate pathway accelerates development of indica-japonica hybrid rice breeding. Plant Biotechnol. J. 22, 2267–2281. doi: 10.1111/pbi.14343, PMID: 38526838
Wang, Y. B., Zou, Y. L., Wei, Y. T., and Meng, L. S. (2025). Crosstalk between ethylene and JA/ABA/sugar signalling in plants under physiological and stress conditions. Mol. Plant Pathol. 26, e70048. doi: 10.1111/mpp.70048, PMID: 40059084
Wiszniewski, A. A., Bussell, J. D., Long, R. L., and Smith, S. M. (2014). Knockout of the two evolutionarily conserved peroxisomal 3-ketoacyl-CoA thiolases in Arabidopsis recapitulates the abnormal inflorescence meristem 1 phenotype. J. Exp. Bot. 65, 6723–6733. doi: 10.1093/jxb/eru397, PMID: 25297549
Xu, Y. H., Liao, Y. C., Zhang, Z., Liu, J., Sun, P. W., Gao, Z. H., et al. (2016). Jasmonic acid is a crucial signal transducer in heat shock induced sesquiterpene formation in Aquilaria sinensis. Sci. Rep. 6, 21843. doi: 10.1038/srep21843, PMID: 26902148
Yamada, T., Takatsu, Y., Kasumi, M., MArubashi, W., and Ichimura, K. (2004). A homolog of the defender against apoptotic death gene (DAD1) in senescing gladiolus petals is down-regulated prior to the onset of programmed cell death. J. Plant Physiol. 161, 1281–1283. doi: 10.1016/j.jplph.2004.06.005, PMID: 15602820
Yan, Q., Si, J., Cui, X., Peng, H., Jing, M., Chen, X., et al. (2019). GmDAD1, a conserved defender against cell death 1 (DAD1) from soybean, positively regulates plant resistance against phytophthora pathogens, Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00107, PMID: 30800138
Yan, A., Wu, E., and Lennarz, W. J. (2005). Studies of yeast oligosaccharyl transferase subunits using the split-ubiquitin system: topological features and in vivo interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 7121–6. doi: 10.1073/pnas.0502669102, PMID: 15886282
Yuan, Z. and Zhang, D. (2015). Roles of jasmonate signalling in plant inflorescence and flower development. CurrOpinPlantBiol. 27, 44–51. doi: 10.1016/j.pbi.2015.05.024, PMID: 26125498
Zhang, Y., Cui, C., and Lai, Z. C. (2016). The defender against apoptotic cell death 1 gene is required for tissue growth and efficient N-glycosylation in Drosophila melanogaster. Dev. Biol. 420, 186–195. doi: 10.1016/j.ydbio.2016.09.021, PMID: 27693235
Zhao, C., Liu, B., Piao, S. L., Wang, X. H., Lobell, D. B., Huang, Y., et al. (2017). Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U.S.A. 114, 9326–9331. doi: 10.1073/pnas.1701762114, PMID: 28811375
Zhou, P., Qian, L., Kozopas, K. M., and Craig, R. W. (1997). Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood. 89, 630–643. doi: 10.1182/blood.V89.2.630, PMID: 9002967
Zhou, J., Tang, X., Li, J., Dang, S., Ma, H., and Zhang, Y. (2024). Comparative transcriptomic and metabolomic analyses provide insights into the responses to high temperature stress in Alfalfa (Medicago sativa L.). BMC Plant Biol. 24, 776. doi: 10.1186/s12870-024-05494-7, PMID: 39143536
Zhu, Z., Esche, F., Babben, S., Trenner, J., Serfling, A., Pillen, K., et al. (2023). An exotic allele of barley EARLY FLOWERING 3 contributes to developmental plasticity at elevated temperatures. J. Exp. Bot. 74, 2912–2931. doi: 10.1093/jxb/erac470, PMID: 36449391
Zhu, T., Herrfurth, C., Xin, M., Savchenko, T., Feussner, I., Goossens, A., et al. (2021). Warm temperature triggers JOX and ST2A-mediated jasmonate catabolism to promote plant growth. Nat. Commun. 12, 4804. doi: 10.1038/s41467-021-24883-2, PMID: 34376671
Keywords: MsDAD1, heat stress, senescence, flowering, Medicago sativa
Citation: Song Y, Guo X, Wang L, Zheng Y, Li T and Dong W (2025) MsDAD1 acts as a heat-induced “senescence brake” in alfalfa. Front. Plant Sci. 16:1664465. doi: 10.3389/fpls.2025.1664465
Received: 12 July 2025; Accepted: 11 August 2025;
Published: 05 September 2025.
Edited by:
Guangqiang Zhang, Heze University, ChinaCopyright © 2025 Song, Guo, Wang, Zheng, Li and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Dong, ZHdlaUBxZm51LmVkdS5jbg==
 Yuguang Song
Yuguang Song Xinying Guo
Xinying Guo Wei Dong
Wei Dong