- 1College of Resources and Environment, Shanxi Agricultural University, Taiyuan, China
- 2Soil Health Laboratory in Shanxi Province, Taiyuan, China
- 3College of Forestry, Beijing Forestry University, Beijing, China
Introduction: Fertilization is vital for improving grape (Vitis vinifera L.) yield and quality. Unlike traditional nitrogen fertilizers, the mechanisms by which sludge alkaline hydrolysate (SAH), a novel fertilizer, influences grape quality and yield are still poorly understood.
Methods: In this study, six treatments were established: 20% SAH + 80% urea (M1), 40% SAH + 60% urea (M2), 60% SAH + 40% urea (M3), 80% SAH + 20% urea (M4), pure SAH (M5), and pure urea (M6). The effects of applying SAH and urea mixtures to grapes were evaluated, with focus on performance parameters, soil nutrients, and microbial communities.
Results and discussion: The results show that 60–80% SAH application significantly enhanced grape stem thickness, chlorophyll content, photosynthetic efficiency, fruit quality, and increased yield. Concurrently, it elevated soil nutrient contents, improved microbial community structure, and altered nitrogen cycle gene copy numbers. Molecular ecological network analyses indicated that Firmicutes, Acidobacteriota, Gemmatimonadota, and Ascomycota were key taxa. Bacterial–fungal cooperation was the dominant interaction, accounting for 65.98–94.61% of all observed microbial interactions, compared to antagonistic interactions. Mantel analysis showed that bacterial community and nitrogen cycle genes (ammonia-oxidizing bacteria (AOB), nitrogen fixation hydrogenase (nifH)) were important for grape yield and quality. These findings offer guidance for the effective use of SAH in grape production. Future studies should elucidate how SAH regulates fruit quality-related gene expression to uncover its mechanisms and enable its full-scale use in viticulture.
1 Introduction
Grapes (Vitis vinifera L.) are a key component of Chinese agriculture. Grape quality and yield are critical for strengthening China’s position in international agriculture. With the rapid development of the grape industry, soil fertility has become a pivotal factor in constraining its sustainable development (Fan et al., 2022). Nitrogen is a key element for the growth and development of grapes, and has significant impacts on plant growth, fruit quality, and soil fertility (Fu et al., 2024). The nitrogen cycle refers to the conversion of nitrogen among various forms. This cycle is regulated by soil microorganisms through the secretion of enzymes encoded by functional genes (Das et al., 2025). These microbial activities influence both soil nitrogen content and plant growth (Han et al., 2024; Zhang et al., 2024a).
In grape cultivation, the application of nitrogen fertilizer plays a crucial role in regulating soil nitrogen content and maintaining the nitrogen cycle (Li and Li, 2025). Nevertheless, the current use of nitrogen fertilizer is accompanied by several challenges, such as the decline in fruit quality caused by over-fertilization and the yield limitations that arise from under-fertilization (Anas et al., 2020; Luo et al., 2024). Traditional grape cultivation often relies on quick-release chemical fertilizers (Kaya et al., 2024). However, the long-term use of these fertilizers can lead to soil acidification and reduced microbial activity (Pahalvi et al., 2021; Gao et al., 2023). Studies have shown that long-term chemical fertilizer use can lower soil pH, inhibit microbial activity, reduce the expression of genes related to the nitrogen cycle (AOB, nirK), and affect the sugar-acid ratio and uniformity of fruit coloring (Bai et al., 2023; Kiya et al., 2024). Thus, finding a sustainable and efficient nitrogen fertilizer alternative has become an important research direction in grape cultivation.
Sludge alkaline hydrolysate (SAH), a novel organic fertilizer, is increasingly recognized as a potential alternative to nitrogen fertilizers. It is rich in nitrogen, peptides, proteins, and small-molecule nutrients (Tang et al., 2022). These components help improve soil conditions and enhance fruit quality and yield (Wang et al., 2023). Studies have shown that applying SAH can increase vegetable yields (Wu et al., 2023a). When combined with nitrogen fertilizers, it can significantly boost levels of soluble sugars, organic acids, and vitamin C (VC) in tomato fruits, thereby improving their nutritional value and safety (Xue et al., 2023). Unlike annual vegetables, perennial fruit crops like grapes have more complex soil-microbe-root interactions, making it essential to evaluate long-term SAH impacts in such systems.
Given these gaps, we designed a study to assess the impact of varying SAH and urea ratios on grape performance, soil chemistry, and microbial dynamics. Specifically, the objectives are to (1) evaluate the effects of different treatments on grape performance (stem thickness, leaf area, fruit shape index, yield, chlorophyll, fruit quality and photosynthetic characteristics), (2) elucidate response mechanisms of soil nutrients, microbial communities, and nitrogen-cycling functional genes to different treatments; and (3) identify the key factors influencing grape performance across different treatments. This study demonstrates that blending the novel sludge alkaline hydrolysate fertilizer and urea enhances grape performance through multiple mechanisms, offering a sustainable approach for grape productivity improvement.
2 Materials and methods
2.1 Overview of the experimental area
This long-term experiment was performed in the TaiGu Comprehensive Experiment Station of the National Grape Industry Technology System in Jinzhong City, Shanxi Province, China (37°25′38.2″N, 112°32′40.5″E). This region features a temperate continental monsoon climate, with an average annual temperature of 10.4°C, annual precipitation of 397.1 mm, a sunshine duration of 2527.5 h, and an altitude of 1098 m. The experiment used a 14-year-old table grape (Zao Hei Bao). The grape plants were planted with a row spacing of 1.30 × 2.12 m and supported on a T-shaped frame.
The SAH was manufactured by Shanxi Jinlian Environmental Science and Technology Co., Ltd. through an alkaline hydrolysis process. By mixing quicklime with domestic sludge, organic nitrogen compounds were extracted to obtain a hot alkaline solution rich in nutrients, including proteins, nitrogen, phosphorus, and potassium. The primary components of this solution are shown in Supplementary Table S1. The heavy metal content is significantly lower than the limits stipulated in the Limits for Toxic and Harmful Substances in Fertilizers of the People’s Republic of China (GB 38400-2019).
2.2 Experiment design
The present study adopted a randomized experimental design with six treatments based on the principle of equal nitrogen application (176.85 kg/ha), with the application ratios of SAH and urea as follows: 20% SAH + 80% urea (M1), 40% SAH + 60% urea (M2), 60% SAH + 40% urea (M3), 80% SAH + 20% urea (M4), pure SAH (M5), and pure urea (M6). There were six replicates per treatment, resulting in a total of 36 plots, each with an area of 24.80 m2 and containing ten grapevines (Supplementary Figure S1). The nitrogen contents in urea and SAH were 46.40% and 7.33%, respectively. Following the application rates specified in Supplementary Table S2, fertilizer was applied during the stable fruiting (late May) and fruit-swelling (mid-July) periods.
2.3 Sample collection and measurement
Five grapevines with similar growth vigor were randomly selected as observation plants within each plot. In June 2024, during the flowering stage of grapevines, three healthy and fully expanded functional leaves were sampled from the upper, middle, and lower strata of every observed plant. Measurements were conducted on the sampled leaves for chlorophyll content, photosynthetic parameters (Ci: intercellular CO2 concentration, Tr: transpiration ratio, gs: stomatal conductance, Pn: net photosynthetic rate), as well as leaf length, leaf width and leaf area (LA). Chlorophyll was measured using a handheld SPAD-502 Plus chlorophyll meter (SPAD-502, Konica Minolta, Japan). The photosynthetic indicators were calculated using a LI-6800 photosynthesis instrument (LI-6800, Li-COR, USA). The stem thickness was measured with vernier calipers, and leaf length and width were measured using a tape measure (Li et al., 2022a). The LA was defined as described by Montgomery and Montgomery (1911) Equation 1:
where Length is the leaf length, Width is the leaf maximum width, and 0.75 is the LA coefficient.
In September 2024, during grape harvesting, one cluster of fruit was randomly collected from the upper, middle, and lower sections of each observed plant. From each cluster, ten berries were randomly selected, and their fruit length (FL) and fruit diameter (FD) were measured using a vernier caliper. The fruit shape index (FSI) was calculated as the ratio of FL to FD. Subsequently, all berries from each cluster were combined and juiced for fruit quality analysis (Ts: total sugar, Rs: reducing sugars, proteins, VC: Vitamin C) (Bao, 2000). Then, fruits from all six treatments were collected to determine the yield (Y). The following Equation 2 was used:
where m is the total weight of grapes harvested from each treatment, s is the area of each treatment, and 1000 is the conversion factor for m2 per hectare.
After the grapes were harvested, we collected soil samples from the 0–20 cm depth in each plot using a five-point sampling method, and then we composited the five samples into one. Soil samples were divided into three sections for real-time fluorescence quantitative PCR (qPCR), high-throughput sequencing after storage at -80 °C, and soil nutrient determination after being air-dried. The method of Bao (2000) was used to determine soil nutrient content (SOC: soil organic carbon, TN: total nitrogen, TP: total phosphorus, TK: total potassium, AP: available phosphorus, AK: available potassium, NH4+-N: ammonium nitrogen, NO3–N: nitrate nitrogen).
For soil nutrient indicators, we calculated the membership values using the simple linear scoring method. Adhering to the principle of higher is better, the highest measured value was assigned a membership value of 1. The membership values for other measurements were calculated as their ratios to the highest value (Li et al., 2020; Liebig et al., 2001), using Equation 3:
where f (x) is the membership value, x is the measured value of the indicator, and xmax is the highest measured value of the indicator.
2.4 DNA extraction, quantitative polymerase chain reaction, and sequence analysis
Soil genomic DNA was isolated with the OMEGA Soil DNA Kit (Omega BioTek, Norcross, GA, USA). Real-time fluorescent qPCR was performed to determine copy numbers of nifH, nirS, and AOB genes using the primers listed in Supplementary Table S3. The thermal cycling protocol consisted of initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s.
The concentration of DNA was quantified using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Bacterial communities were targeted by amplifying the V3–V4 hypervariable regions with primers 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’), while fungal communities were assessed through ITS1 region amplification using primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3’). Amplicons were pooled, size-selected via 2% agarose gel electrophoresis, and subsequently purified before quality assessment and quantification. Library preparation was conducted using the TruSeq Nano DNA LT Library Prep Kit (Illumina Inc., San Diego, CA, USA), followed by sequencing on an Illumina NovaSeq 6000 platform (Illumina) with SP Reagent Kit (500 cycles; Illumina). Raw sequences underwent quality control (QIIME2, 2022.11), including filtering, denoising, chimera removal, and assembly. High-quality sequences were clustered into amplicon sequence variants (ASVs) at 100% similarity. Taxonomic assignment of bacterial and fungal ASVs was conducted using the Greengenes2 (https://greengenes2.ucsd.edu/) and UNITE (https://unite.ut.ee/) databases, respectively.
2.5 Statistical analyses
One-way Analysis of Variance (ANOVA), followed by Tukey’s HSD test, was performed using IBM SPSS Statistics 21 (IBM Corp., Armonk, NY, USA) software to examine significant differences in grape and soil indicators under different treatments. The ‘linkET’ package (devtoolsinstall_github (“Hy4m/linkET”, force=TRUE) in R (4.4.2; R Foundation for Statistical Computing, Vienna, Austria) was used for Mantel test analysis to explore the relationships between bacterial and fungal communities, nitrogen cycling genes, soil nutrient contents, grape yield, and fruit quality.
To ensure comparability between samples and to reduce the effects of variability in sequencing depth, the microbial data were normalized to a uniform minimum sequencing depth using rarefaction before conducting ecological and network analyses Based on the random matrix theory, network construction was performed for the ASVs of bacteria and ASVs of fungi, and 0.94 was selected as the threshold for each group of bacterial and fungal networks to ensure comparability between the six processed networks (Zhou et al., 2023). We constructed the microbial networks and determined the network parameters using the Molecular Ecological Network Analysis Pipeline (MENAP) (Zheng et al., 2021). The network visualization and generation of node and edge files were performed using the Gephi (v.0.9.2) interactive platform (http://gephi.github.io/). The ZiPi analysis on nodes and edge files was performed using the “ggClusterNet” package in R 4.4.2 (Ding et al., 2016). Based on topological roles defined by within-module connectivity (Zi) and among-module connectivity (Pi) thresholds, nodes were classified into four categories: (i) peripherals (Zi < 2.5, Pi < 0.62); (ii) connectors (Zi < 2.5, Pi ≥ 0.62); (iii) module hubs (Zi ≥ 2.5, Pi < 0.62); and (iv) network hubs (Zi ≥ 2.5, Pi ≥ 0.62). Notably, module hubs, network hubs, and connectors were identified as keystone species in the microbial network (Wen et al., 2022).
3 Results
3.1 Grape performance under different treatments
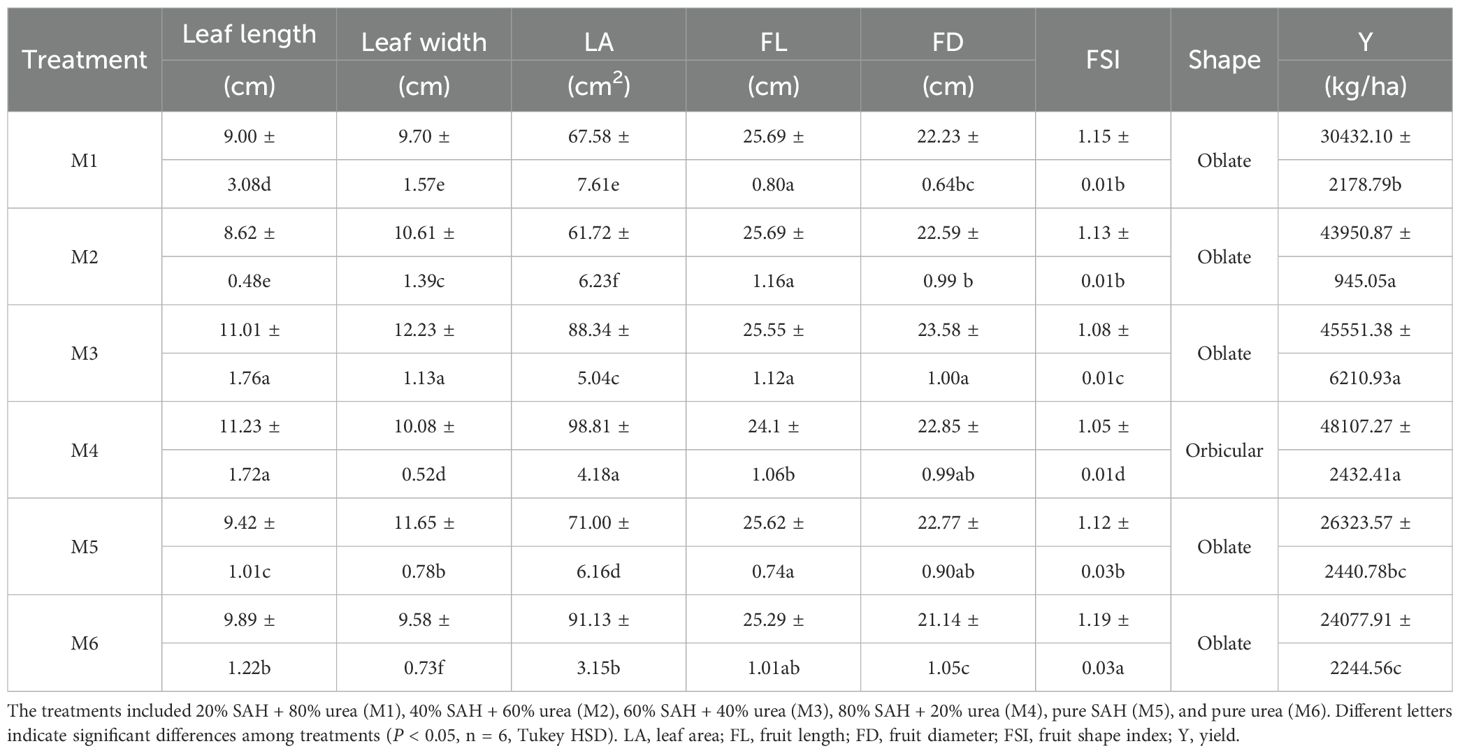
The M4 treatment achieved maximal values for stem thickness (50.78 cm), leaf area (LA, 98.81 cm2), fruit shape index (FSI, 1.05), and yield (48,107.27 kg/ha), with the yield being 99.80% greater than that in M6 (P < 0.05, n=6; Figure 1A; Table 1). In addition, there was no significant difference in chlorophyll among treatments (P > 0.05, n=6; Supplementary Figure S2).
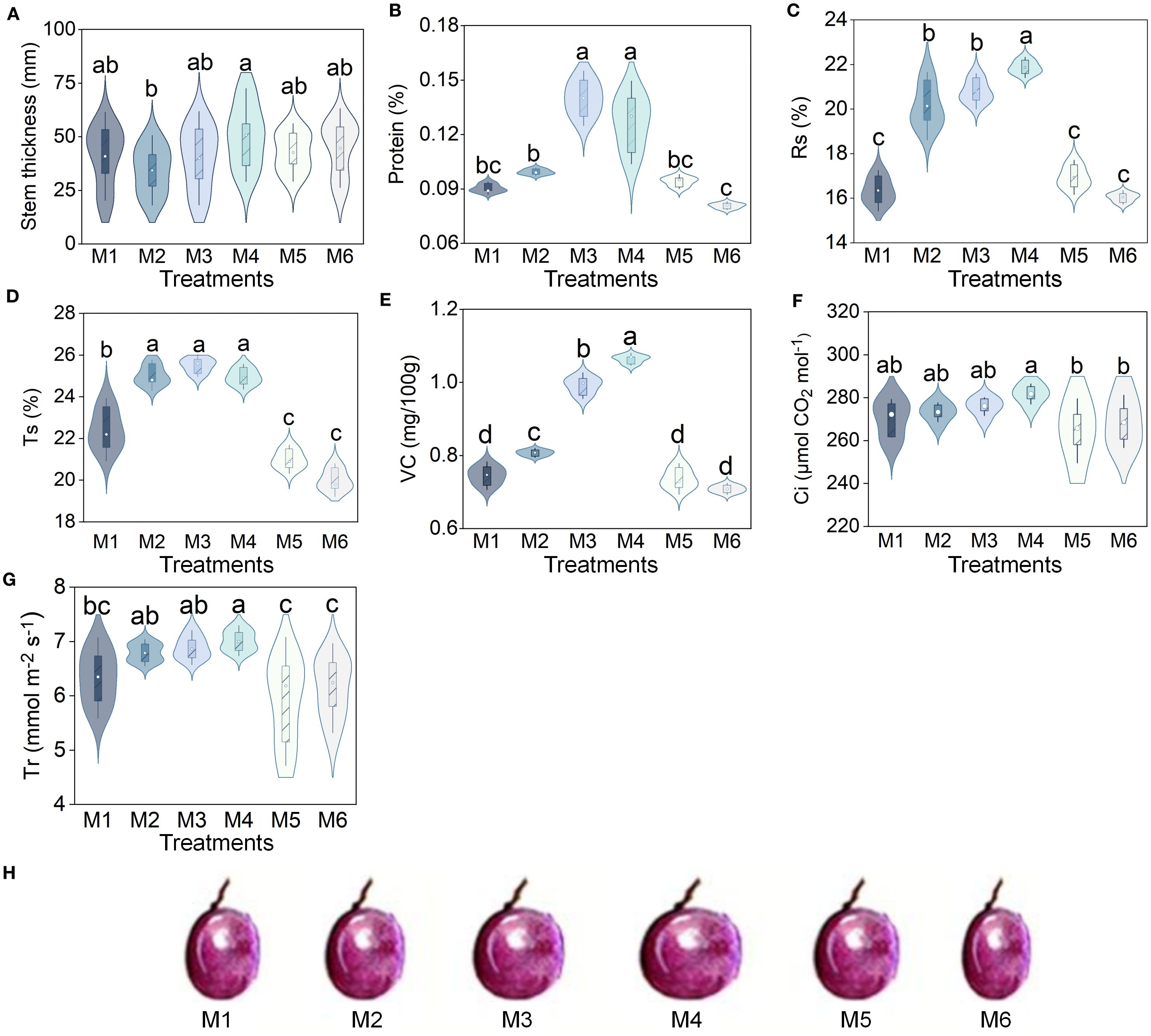
Figure 1. Grape performance under different treatments. (A) Stem thickness (B) Protein. (C) Rs, reducing sugars. (D) Ts, total sugars. (E) VC, vitamin C. (F) Ci, intercellular CO2 concentration. (G) Tr, transpiration ratio. (H) Schematic diagram of fruit type. The treatments included 20% SAH + 80% urea (M1), 40% SAH + 60% urea (M2), 60% SAH + 40% urea (M3), 80% SAH + 20% urea (M4), pure SAH (M5), and pure urea (M6). Different letters indicate significant differences among treatments (P < 0.05, n = 6, Tukey HSD). SAH, sludge alkaline hydrolysate.
Fruit quality (protein, Rs: reducing sugars, Ts: total sugar, VC: Vitamin C) showed a unimodal trend across treatments. The M4 treatment yielded the highest levels of Rs and VC (21.87% and 1.06%), which were 36.43% and 49.30% greater than those in the M6 treatment. Meanwhile, the M3 treatment produced the highest protein and Ts contents (0.14% and 25.50%), exceeding the M6 values by 48.94% and 27.50%. (P < 0.05, n=6; Figures 1B–E).
Photosynthetic characteristics (Tr, Ci) increased with rising SAH ratios up to M4, after which they declined. The Tr (6.97 mmol m−2 s−1) and Ci (281.75 µmol CO2 mol−1) concentrations were both highest under the M4 treatment, which was significantly increased by 12.60% and 17.94% over the values recorded in M5 and M6, respectively (P < 0.05, n=6; Figure 1). The gs and Pn content did not differ significantly among treatments (P > 0.05; Supplementary Figure S3).
3.2 Characterization of soil nutrients under different treatments
The contents of SOC (10.03–12.42 g/kg), TN (1.03–1.29 g/kg), TK (19.89–22.62 g/kg), NH4+-N (3.69–5.92 mg/kg), and NO3–N (29.10–74.67 mg/kg) were the highest under the M4 treatment and the lowest under the M6 treatment, which increased significantly by 23.83%, 25.24%, 13.73%, 60.43%, and 156.60%, respectively, compared to M6. In contrast, M3 exhibited superior AP (52.78–67.43 mg/kg) and AK (427.71–723.55 mg/kg) levels, exceeding M6 by 27.76% and 69.17% (P < 0.05, n=6; Supplementary Table S4). Subsequently, the membership degree values of each indicator were calculated, and the results consistently indicated that the soil nutrient conditions were superior under the M3 and M4 treatments (Figure 2).
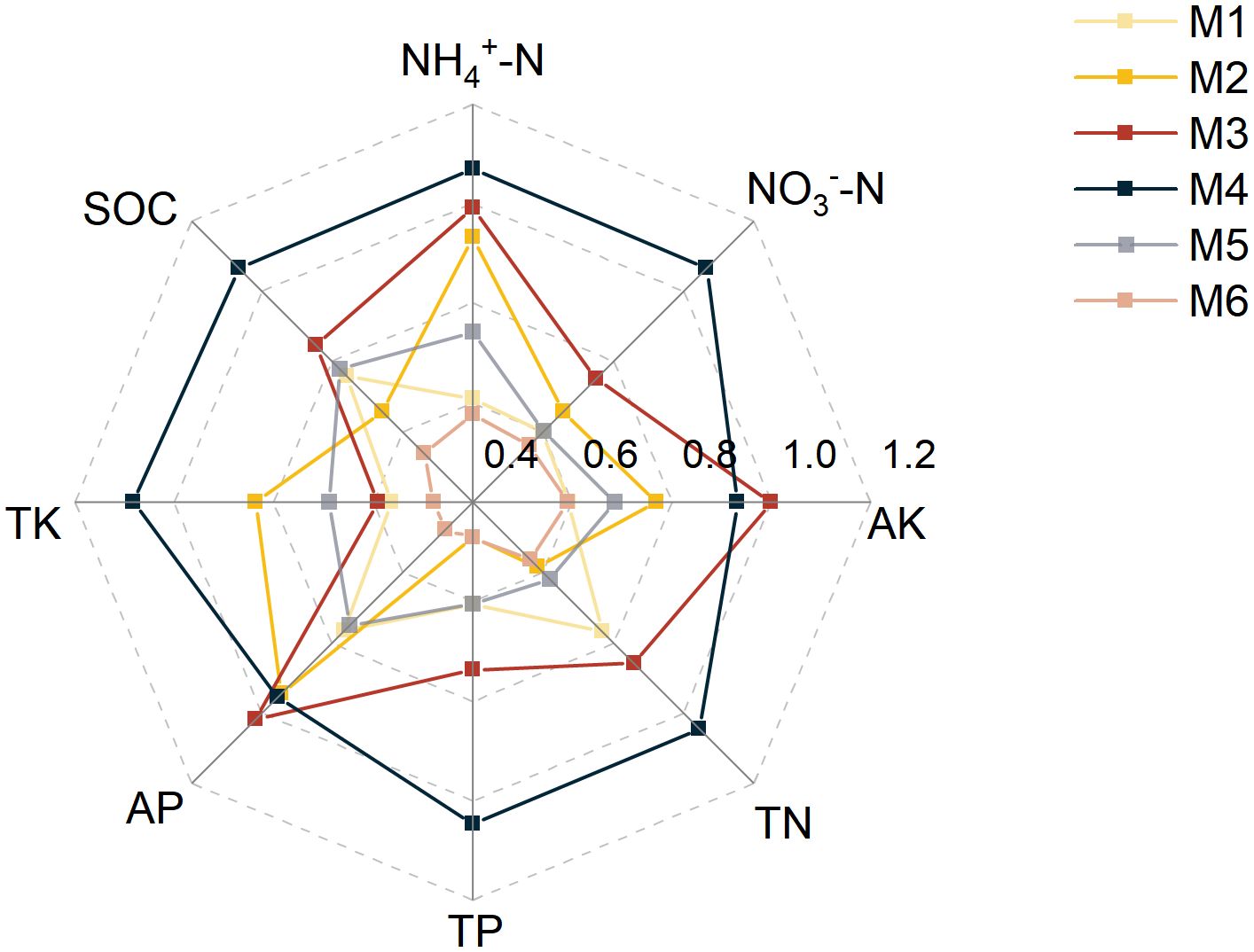
Figure 2. Radar plot of membership value of indicators. SOC, soil organic carbon, TN, total nitrogen; TP, total phosphorus; AP, available phosphorus; TK, total potassium; AK, available potassium; NH4+-N, ammonium nitrogen; NO3–N, nitrate nitrogen. The treatment codes, M1-M6, are consistent with those in Figure 1.
3.3 Changes in microbiological characteristics under different treatments
At the phylum level, the dominant bacterial taxa were Proteobacteria, Gemmatimonadota, and Actinobacteria. Proteobacteria had the highest relative abundance (28–32%) and were significantly higher in M3 than in M5 (+14.29%) (P < 0.05, n=6; Figure 3A and Supplementary Table S5). The QUBU01 was the dominant genus under the M4 treatment and belonged to the Proteobacteria phylum (P < 0.05, n=6; Figure 3B; Supplementary Table S7). The results suggested that treatments M3 and M4 provided more favorable conditions for the growth of Proteobacteria. For fungi, Ascomycota was the predominant phylum and showed higher abundance in M5 (Figure 3A; Supplementary Table S6).
Figure 3. Effects of different treatments on microbial communities. (A) Relative abundance of major bacterial (left) and fungal (right) phyla between different treatments. The large circle indicates high abundance. (B) Relative abundance of major bacterial (left) and fungal (right) genera between different treatments. (C) Comparison of bacterial and fungal community diversity between different treatment areas. (D) PCoA analysis of bacterial (left) and fungal (right) communities based on Bray-Curtis distance. (E) bacterial and fungal biomarker taxa composition under different treatments. Circles from inside to outside represent taxonomic levels from phylum to genus. Each small circle at a different taxonomic level represents a taxonomic unit at that level, and the size of the circle diameter is proportional to the size of the relative abundance. Species without significant differences are uniformly white. Species names indicated by letters in the figure are shown in the legend on the right, and only species with LDA scores of >3 for differences are shown. Different letters indicate significant differences among treatments (P < 0.05, n = 6, Tukey HSD). The treatment codes, M1-M6, are consistent with those in Figure 1.
The Chao1 index of both bacteria and fungi peaked in M4 (26.45% and 51.49% higher than in M6, respectively, P < 0.05). The Shannon index of bacteria and fungi was the highest under the M3 and M4 treatments, respectively (P < 0.05, n=6; Figure 3C). There were significant differences in soil microbial community structure among treatments (Figure 3D). Additionally, the alpha diversity of bacteria and fungi (Chao1, Shannon index) exhibited positive correlations with transpiration ratio (Tr), protein, Vitamin C (VC), reducing sugars (Rs), total sugar (Ts), and yield, but a negative correlation with the fruit shape index (FSI) (Supplementary Figure S4).
Biomarkers often represent taxa that perform critical ecological functions. The biomarker taxa showed significant differences among the six treatments. For bacteria, the number of biomarker taxa was the highest in M2 (16) and lowest in M3 (2). For fungi, it was the highest in M1 (12) and lowest in M4 (4) (Figure 3E). Furthermore, we found that most of these biomarkers belong to the Acidobacteria phylum (bacteria) and the Ascomycota phylum (fungi), indicating that they play a crucial role in the functioning of soil ecosystems.
3.4 Molecular ecological network analysis of bacterial and fungal communities
Microbial interactions across the six treatments were analyzed using a molecular ecological network. A total of 2, 10, 13, 9, 7, and 10 major modules were identified within each respective treatment, collectively accounting for over 90% of the network structure (Figure 4). Under the M1 treatment, average degree (avgK, 32.55) and average clustering coefficient (avgCC, 0.76) were higher, whereas modularity was significantly lower than in the other treatments. The highest connectedness (Con) index (1.00) occurred under the M4. The positive correlation between fungi and bacteria was generally higher than the negative correlation under all treatments. Its range was distributed between 65.98% (M4) and 94.61% (M5) (Supplementary Table S9). Besides, four microbial key taxa were identified at the phylum level, including three bacteria (Firmicutes, Acidobacteriota, and Gemmatimonadota) and one fungus (Ascomycota) (Supplementary Figure S5; Supplementary Table S10).
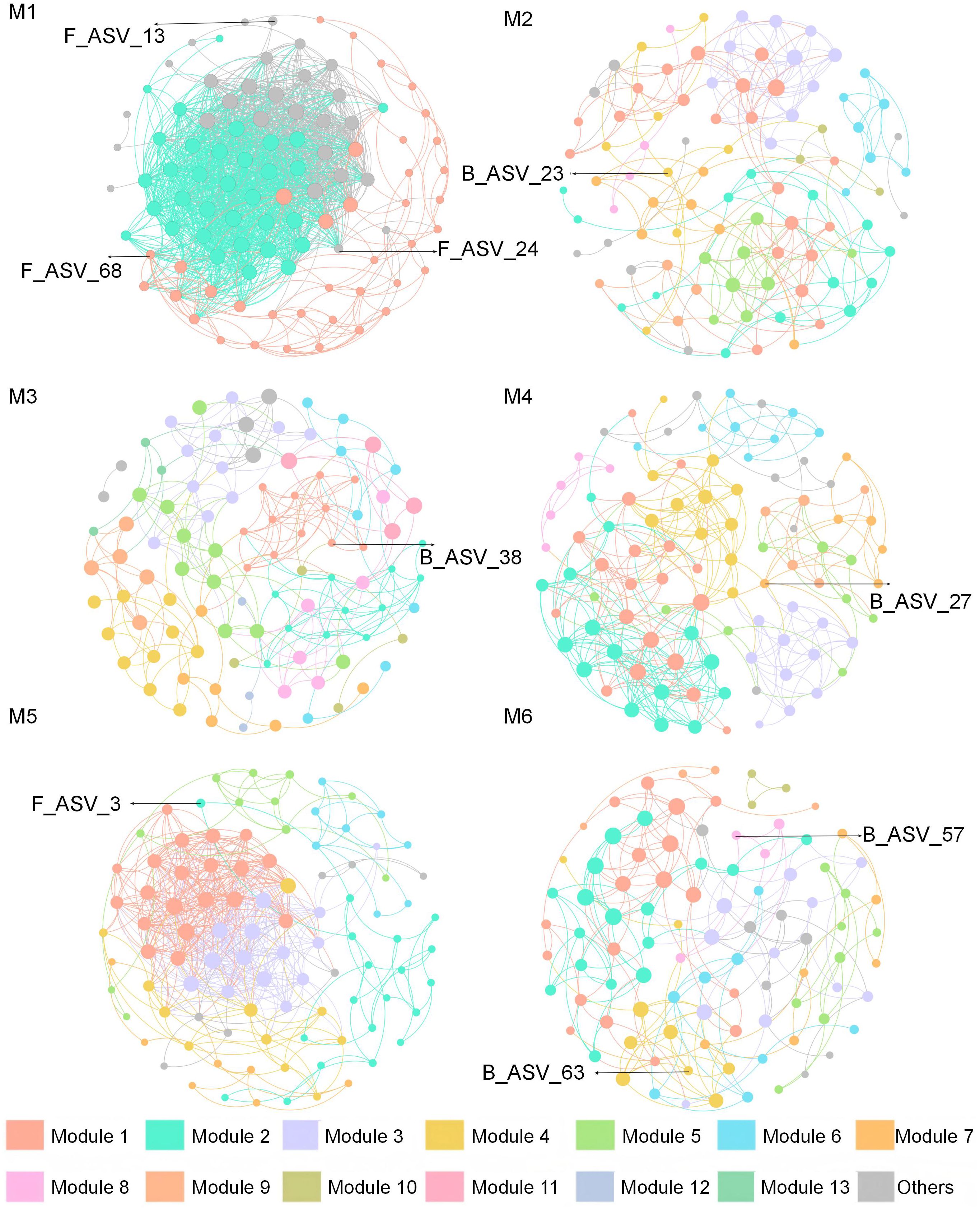
Figure 4. Bacterial and fungal co-occurring networks under each treatment. The treatment codes, M1-M6, are consistent with those in Figure 1.
3.5 Functional gene changes under different treatments
The functional genes involved in nitrogen cycling exhibited three distinct trends. The copies of nifH showed a bimodal pattern, peaking under M2 treatment with significant increases of 59.46% and 58.10% compared to the lowest levels in M3 and M4 (P < 0.05; Figure 5A). The copies of nirS demonstrated a progressive increase across treatments (P < 0.05, n=6; Figure 5B). The copy numbers of AOB followed a unimodal distribution, reaching their maximum in M4 (825.83 × 106 copies), representing a 155.46% elevation relative to the level in the sludge-free control (M6) (P < 0.05, n=6; Figure 5C). Furthermore, the average copies of AOB genes were orders of magnitude higher than those of nifH genes (64.90 times, Figure 5).
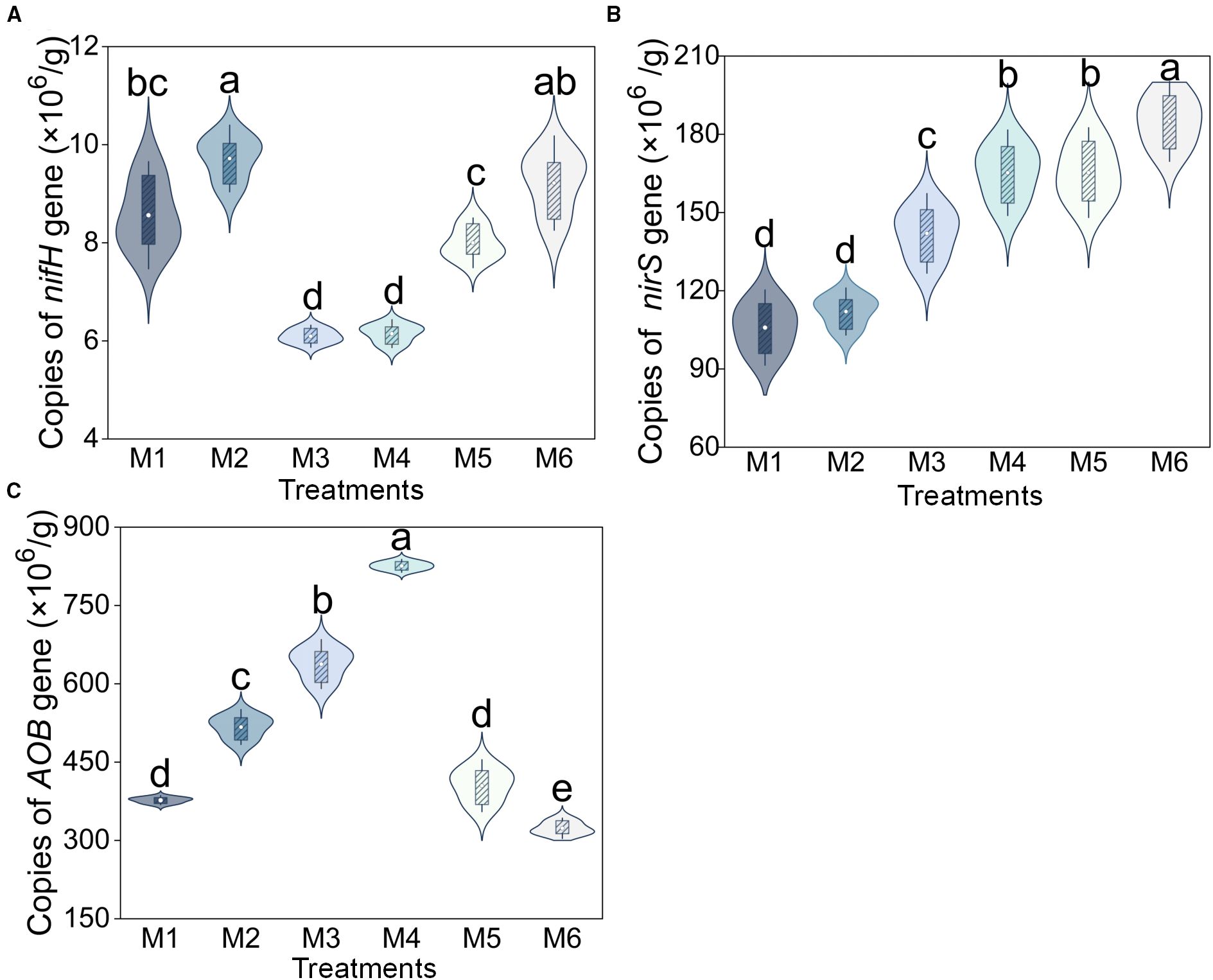
Figure 5. Absolute abundance of (A) nifH, (B) nirS, and (C) AOB gene under different treatments. Different letters indicate significant differences between treatments (P < 0.05, n = 6, Tukey HSD). The treatment codes, M1-M6, are consistent with those in Figure 1.
3.6 Driving factors of yield and quality
Bacterial and fungal species abundances were strongly correlated with grape performance and soil nutrients. Notably, bacterial species abundances exhibited significant correlations with over half of the indicators (SOC, TN, TK, AK, NH4+-N, NO3–N, yield, VC: Vitamin C, Rs: reducing sugars, and Ts: total sugar), demonstrating a particularly strong association with yield (r ≥ 0.4, P < 0.01, Figure 6A). However, fungi showed significant correlations with only a few indicators (AK, yield, VC, Rs, and Ts). This indicated that bacteria had a greater impact on grape performance and soil nutrients than fungi.
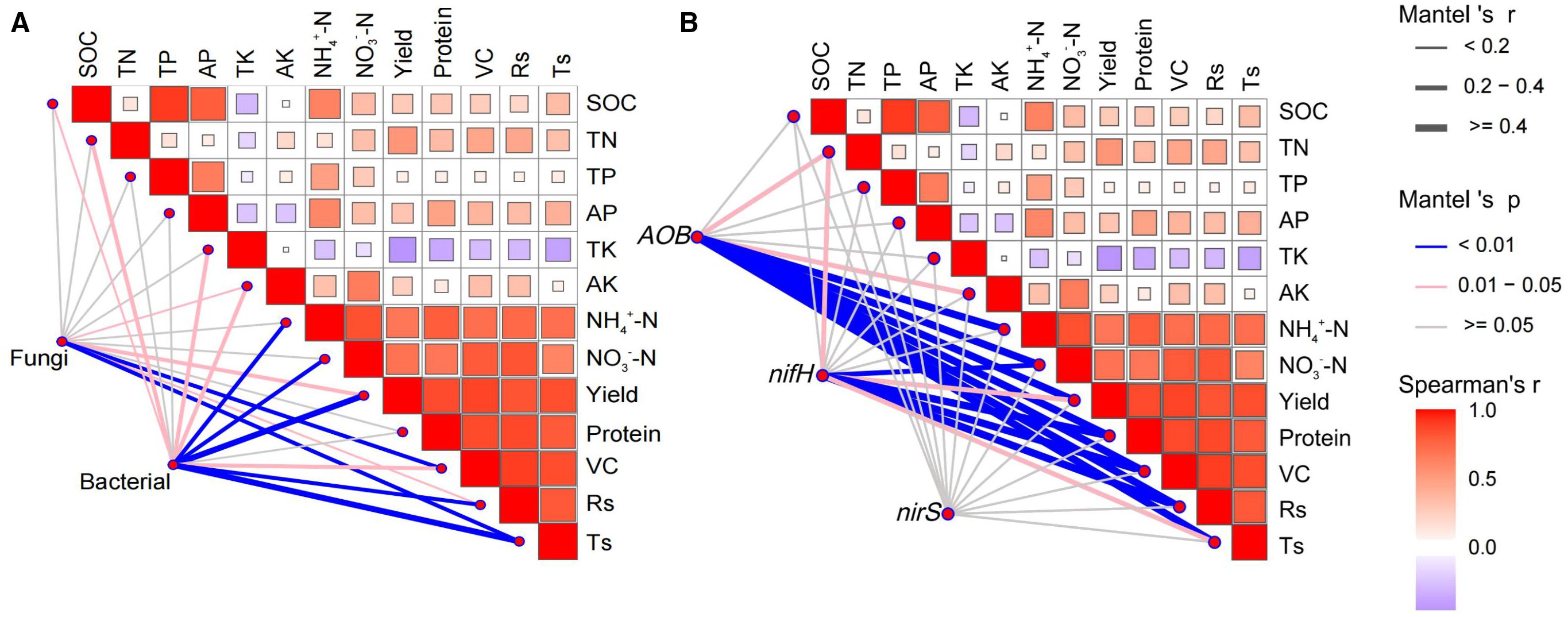
Figure 6. Relationships between (A) microbial communities, (B) nitrogen cycling genes, and fruit quality, yield, and soil nutrients based on Spearman correlation and Mantel test. SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus; AP, available phosphorus; TK, total potassium; AK, available potassium; NH4+-N, ammonium nitrogen; NO3–N, nitrate nitrogen; VC, Vitamin C, Rs, reducing sugar; Ts, total sugar.
The nitrogen cycle gene AOB copies strongly correlated with NH4+-N, NO3–N, yield, protein, VC, Rs, and Ts (r ≥ 0.4, P < 0.01) and was closely related to TN and AK (0.2 < r < 0.4, P < 0.05). Similarly, the copies of nifH showed strong correlations with NO3–N, yield, protein, VC, and Rs (r ≥ 0.4, P < 0.01) and were closely related to TN and Ts (0.2 < r < 0.4, P < 0.05, Figure 6B). In contrast, nirS showed no significant correlation with any indicator. In summary, these strong correlations demonstrate the key role of AOB gene copies in regulating grape yield and quality (protein, VC, Rs, Ts).
Additionally, soil nutrients (except TK) were positively correlated with grape performance, especially TN, NH4+-N, and NO3–N. In conclusion, the results indicated that applying SAH and urea mainly affected grape fruit quality and yield by regulating the bacterial community, AOB, and nifH gene copies (Figure 6).
4 Discussion
4.1 Effect of SAH-urea co-application on grape performance
Crop stem thickness and leaf area (LA) are associated with high crop yield and quality, and they are key indicators for assessing crop growth and predicting yield (Pei et al., 2024). Plants with thicker stems can store more water and nutrients, and they have high stress resistance and load-bearing capacity (Li et al., 2022b). The LA can regulate photosynthetic efficiency, further affecting yield (Ren et al., 2025). The fruit shape index (FSI), defined as the ratio of longitudinal to transverse diameter, serves as a key morphological indicator in fruit characterization (Croce et al., 2024). The FSI closer to 1 indicates a more orbicular fruit morphology. The contents of sugar, vitamins, and other nutrients in the fruit have a considerable influence on the taste and flavor of the fruit, and protein, Vitamin C (VC), reducing sugar (Rs), and total sugar (Ts), which are generally the main evaluation indices (Zhang et al., 2024b; Balık et al., 2023).
Studies have shown that fertilizer application significantly increased the LA of plants and improved the effective area for photosynthesis, thus affecting yield (Cheng et al., 2025). Homsa et al. (2023) examined garlic growth and found that strategic fertilization produced garlic with a larger LA, more photosynthesis, and the highest tuber growth. Wu et al. (2023b) demonstrated that the use of plant growth regulators altered the FSI of grapes (0.92). The content of VC and soluble sugars in radishes increased by 10.62% and 2.15% after the application of organic and inorganic fertilizers, respectively (Jin et al., 2024). The above results demonstrate a strong link between fertilizer application and crop nutrient content (Hu et al., 2024a).
In our study, applying 60–80% of SAH (M3, M4) achieved the best results for grapevine stem thickness, LA, photosynthesis, FSI, quality, and yield (Figure 1; Supplementary Figure S1, Table 1). This suggests that this dosage range can significantly enhance fruit development and canopy photosynthesis, leading to higher yields and overall plant performance. Moreover, the humic acid and amino acids in SAH can enhance the activity of enzymes involved in sugar metabolism, promote the accumulation and transformation of sugars, and increase the Rs and Ts content in fruits (Yuan et al., 2024). While the efficacy of chemical fertilizers in promoting plant growth is well-established, their overuse has led to a range of environmental issues (Pahalvi et al., 2021). Therefore, developing novel, environmentally friendly fertilizers from waste materials, which can replace or reduce chemical fertilizer application, is crucial for advancing sustainable viticulture. These findings provide strong empirical evidence for significantly reducing reliance on chemical fertilizers while enabling the valuable reuse of waste. To thoroughly evaluate the treatment’s effectiveness, we will next investigate how SAH affects the expression of genes related to fruit shape and quality using molecular approaches.
4.2 Effect of SAH-urea co-application on soil nutrient content
Soil nutrients have a strong effect on land productivity, and fertilization can replenish the nutrient stocks in the soil and affect soil fertility and plant growth (Tian et al., 2022). In our study, the contents of TN, NH4+-N, and NO3–N were the highest under the M4 treatment (Supplementary Table S4). Du et al. (2024) observed that the use of either nitrogen-only fertilizer or nitrogen–phosphorus–potassium compound fertilizer led to significant alterations in soil physicochemical characteristics. The N content was enhanced by 25.41% after N fertilizer application compared with the no fertilizer application. This is in line with our findings (Supplementary Table S4; Figure 2).
The AP and AK can be directly absorbed by the crop, affect the nutritional status of the crop, and are indicators of the intensity of the soil’s ability to supply P and K (Ye et al., 2024). Extended organic fertilization reportedly promotes P enrichment in soils (Wang et al., 2024a). Jiang et al. (2024) reported that fertilization significantly elevated soil nutrient levels, with AP increasing by 17.12% (nitrogen fertilizers) to 474.74% (nitrogen-phosphorus-organic fertilizers) and AK by 2.90% (nitrogen-phosphorus fertilizers) to 64.40% (nitrogen-phosphorus-organic fertilizers). Notably, the significant increases in AP and AK contents under the M3 and M4 treatments observed in this study did not originate from conventional chemical fertilizers, but rather from the mineralization of organic matter in SAH and its stimulatory effect on soil microbial activity (Supplementary Table S4). This organic waste-driven approach to enhancing soil fertility not only offers more sustained benefits but also effectively improves soil structure and mitigates compaction issues associated with sole chemical fertilizer application, representing a more sustainable strategy for nutrient management (Hu et al., 2024b; N’Dri et al., 2023).
4.3 Effect of SAH-urea co-application on microbial community
Fertilization can increase soil nutrients, regulate microbial community structure, and improve farm productivity (Fan et al., 2021; Hartmann and Six, 2023). Changes in fertilizer ratios influenced community structure and the distribution of microbial taxa. For example, the dominant bacterial phylum under the M3 treatment was Proteobacteria. Meanwhile, the dominant fungal phylum under M5 treatment was Ascomycota (Figure 3A). In addition, the highest microbial diversity indices were observed under the M3 and M4 treatments (Figure 3C), which was attributed to soil structure and nutrient conditions regulating microenvironmental heterogeneity, thus directly or indirectly controlling microbial composition (Hu et al., 2025; Laurent et al., 2023).
The biomarker and key taxa of microbiological under different treatments were characterized further by LEfSe analysis and molecular ecological network. There were differences in the biomarker and key taxa of microbiological under different treatments (Figures 3E, 4). Such variation likely arises from differential microbial colonization strategies across distinct soil environments (Zhang et al., 2022). Interestingly, we found that the Acidobacteriota and Ascomycetes belong to both biomarker and key taxa (Supplementary Figure S5; Supplementary Table S10). The phenomenon is primarily attributed to their unique metabolic complementarity; metabolites (e.g., organic acids) secreted by Ascomycetes facilitate Acidobacteriota proliferation, thereby enhancing soil nitrogen fixation (Kost et al., 2023). Such cross-kingdom interaction markedly improves community stability and nutrient-use efficiency, rendering such taxa ecologically indispensable within microbial networks (Jia et al., 2025). However, considering the dynamic nature of microbial communities, future studies should incorporate microbial data from different application durations to better assess the long-term impacts of SAH on soil microbiota.
Positive correlations in microbial networks can generally be interpreted as interactive symbiotic and mutualistic relationships between species (Liu et al., 2024). Notably, under M5 treatment, microbial cooperation was highest (Supplementary Table S9); however, the yield was lower (Table 1). Previous studies indicate that microbial cooperation is influenced by environmental nutrient conditions: under low-nutrient conditions, microorganisms enhance cooperation to resist stress and maintain survival (Dai et al., 2022). Thus, the high microbial cooperation observed in the M5 treatment represents a “defensive” strategy prioritizing survival under adverse conditions, rather than directly serving grape growth. This explains the decoupling between cooperation and yield. The M1 treatment exhibited maximal values for both average degree (avgK) and average clustering coefficient (avgCC). This indicates that the microbial network under this treatment has better adaptability and flexibility and is more closely connected in function (Du et al., 2025). It can cope with environmental changes through internal connections, reconnect, and adjust to maintain its functionality and structure with higher robustness and a more stable network (Zhang et al., 2024c; Al Musawi et al., 2023; Puente-Sánchez et al., 2024).
Under M4 treatment, AOB exhibited the highest gene copy number (Figure 5C), which may be related to SOC and TN content (Zhou et al., 2014; Li et al., 2023). Previous studies indicate that AOB abundance correlates positively with soil C, N, and NH4+-N contents. The elevated SOC and TN levels provide additional growth substrates for AOB, thereby enhancing their proliferation (Thion et al., 2016; Li et al., 2017; Song and Niu, 2022). Additionally, it is noteworthy that although the nifH gene copies peaked in M2, this trend contrasts sharply with the superior agronomic performance observed in M4. This is because the nifH gene copy number is relatively low (Figure 5), limiting its potential impact in soil (Li et al., 2022). Moreover, the association between AOB gene abundance and agronomic performance was stronger than that of nifH (Figure 6). The above results collectively indicate that the nifH gene is not a key factor influencing grape agronomic performance, while AOB-driven nitrification may be a more direct and efficient nitrogen supply pathway in soil, thereby more critically determining the final expression of agronomic performance (Li et al., 2018; Ma et al., 2021a).
4.4 Bacterial communities, AOB, and nifH genes mainly regulate the yield and nutritional quality
Grape yields and their quality are often closely related to microbial species and gene copy number (Guo et al., 2021; Asad et al., 2022). Close associations between bacterial communities, AOB, nifH genes, and yield and fruit nutritional quality were found following Mantel test analysis (Figure 6). Soil bacteria play an important role in breaking down crop residues, mineralizing, and fixing SOC (Shamshitov et al., 2024). Some bacterial populations can inhibit soil pathogen growth, protecting the health of the crop and improving yields (Liang et al., 2025). Wang et al. (2024b) showed that stover returns increased maize yield in arid and semi-arid regions mainly by regulating soil bacterial community abundance. Bacteria are 1.4–5 times more efficient than fungi in using simple organic compounds (Dini et al., 2025). Structural features of bacterial cell membranes enable rapid binding and utilization of small organic molecules in SAH, facilitating metabolic adaptation to maximize nutrient use and thereby influence crop yield (Akhtar et al., 2024). Due to the roles of AOB and nifH genes in nitrogen cycling, an increase in their gene abundance may imply more efficient nitrogen transformation, positively affecting crop nitrogen use and yield (Ma et al., 2021b). This suggests that changes in bacterial abundance and the number of AOB and nifH genes stimulated grape development and increased nutrient acquisition, leading to significant increases in key fruit quality indicators (e.g., Ts and Rs) and yield.
The results demonstrate that the combined application of SAH and urea establishes a more efficient and synergistically integrated soil nutrient delivery system. This improvement can be attributed to the optimized resource input strategy achieved through partial substitution of urea with SAH derived from organic waste. Therefore, this study not only presents an effective fertilization strategy but also highlights the significant potential of organic wastes in reducing chemical fertilizer application and promoting nutrient recycling, thereby providing important theoretical and practical support for sustainable viticulture.
5 Conclusions
Our study systeatically evaluated the influence of varying proportions of SAH and urea on multiple aspects of grape production systems (Figure 7). At SAH-to-urea fertilizer ratios of 60:40 and 80:20, grape performance, soil nutrients, microbial community structure, and nitrogen cycle functional genes reached optimal levels. The molecular ecological network showed that Firmicutes, Acidobacteriota, Gemmatimonadota, and Ascomycota were key taxa. Notably, bacteria and fungi showed strong cooperation (over 50%), which significantly enhanced the microbial network’s adaptability, modularity, and species coexistence. Mantel test showed that bacterial community and AOB and nifH gene copies are key factors regulating grape quality and yield. These findings demonstrate that SAH functions as an ecological regulator in grape cultivation, providing new insights for sustainable agricultural development.
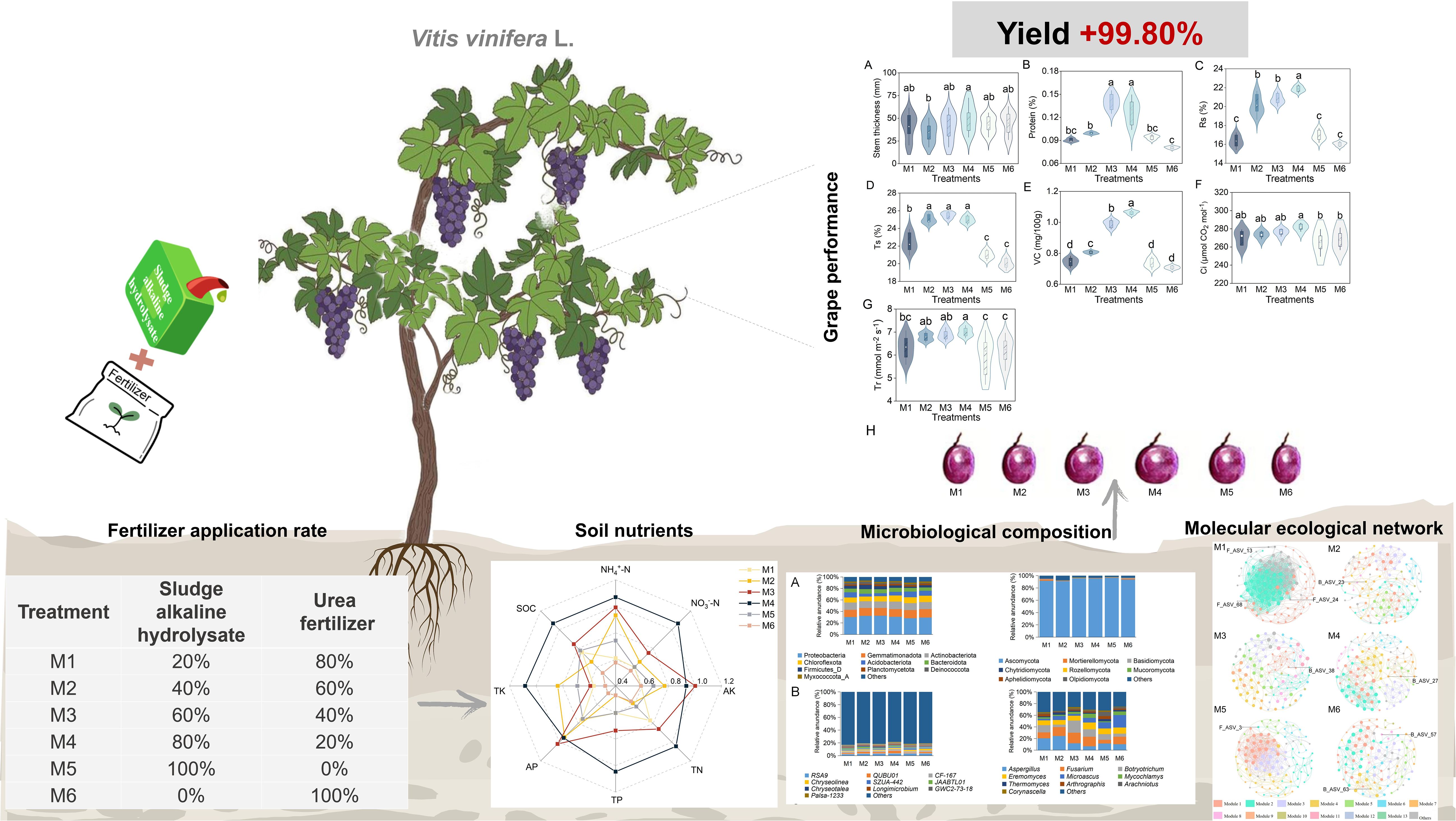
Figure 7. A concept figure illuminated the effects of varying proportions of SHA on grape performance, soil nutrients, and microbial communities. The application of SHA enhances soil nutrient availability, modulates microbial community composition, and accelerates plant growth and nutrient accumulation, thereby increasing yield. The treatment codes, M1-M6, are consistent with those in Figure 1.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
DX: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Conceptualization. YY: Software, Visualization, Writing – original draft, Methodology, Data curation. HZ: Writing – original draft, Data curation. YQ: Writing – original draft, Software. ZeL: Writing – review & editing, Investigation, Methodology. ZiL: Writing – review & editing, Data curation, Methodology. WW: Writing – review & editing. HB: Writing – review & editing, Visualization, Conceptualization, Methodology, Investigation, Data curation, Project administration, Funding acquisition. DJ: Writing – review & editing, Conceptualization, Investigation, Methodology, Project administration, Funding acquisition. QZ: Writing – review & editing, Conceptualization. MX: Writing – review & editing, Resources. ZY: Writing – review & editing, Conceptualization, Project administration, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was financially supported by the Major Science and Technology Special Projects in Shanxi Province Unveiled Projects (No. 202201140601028), the Shanxi Basic Research Project (No. 202303021212104), Shanxi Agricultural University School-Enterprise Cooperation Program (No. 2023HX006), Open Research Projects of the Engineering Technology Innovation Center for Ecological Protection and Restoration in the Middle Yellow River, Ministry of Natural (2025078), Resources the Shanxi Basic Research Project (No. 202303021222077).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1665661/full#supplementary-material
References
Akhtar, N., Shahzad, A., Ilyas, N., Bostan, N., Jameel, M. A., Mukhtar, S., et al. (2024). Nanobiotechnology and microbial influence on cold adaptation in plants. Nanotechnol Rev. 13, 160–176. doi: 10.1515/ntrev-2024-0059
Al Musawi, A. F., Satyaki, R., and Preetam, G. (2023). Examining indicators of complex network vulnerability across diverse attack scenarios. Sci. Rep. 13, 18208–18208. doi: 10.1038/s41598-023-45218-9
Anas, M., Liao, F., Verma, K. K., Sarwar, M. A., Mahmood, A., Chen, Z. L., et al. (2020). Fate of nitrogen in agriculture and environment: agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol. Res. 53, 47. doi: 10.1186/s40659-020-00312-4
Asad, N. I., Wang, X. B., Dozois, J., Azarbad, H., Constant, P., and Yergeau, E. (2022). Early season soil microbiome best predicts wheat grain quality. FEMS Microbiol. Ecol. 99. doi: 10.1093/femsec/fiac144
Bai, X., Li, Y., Jing, X., Zhao, X., and Zhao, P. (2023). Response mechanisms of bacterial communities and nitrogen cycle functional genes in millet rhizosphere soil to chromium stress. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1116535
Balık, S., Kaya, T., and Aslantaş, R. (2023). Fruit quality parameters, sugars, vitamin C, antioxidant activity, organic acids, and phenolic compounds for a new endemic apple variety, “Long apple. Horticulturae. 9. doi: 10.3390/horticulturae9111171
Cheng, Y., Chen, X. Y., Ren, H., Zhang, J. W., Zhao, B., Ren, B. Z., et al. (2025). Deep nitrogen fertilizer placement improves the yield of summer maize (Zea mays L.) by enhancing its photosynthetic performance after silking. BMC Plant Biol. 25, 1–16. doi: 10.1186/s12870-025-06145-1
Croce, R., Carmo-Silva, E., Cho, Y. B., Ermakova, M., Harbinson, J., Lawson, T., et al. (2024). Perspectives on improving photosynthesis to increase crop yield. J. Plant Cell Dev. 36. doi: 10.1093/plcell/koae132
Dai, T. J., Wen, D. H., Bates, C. T., Wu, L. W., Guo, X., Liu, S., et al. (2022). Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 13, 175–175. doi: 10.1038/s41467-021-27857-6
Das, P., Barker, C., Park, Y., Perreault, F., Westerhoff, P., and Penton, C. R. (2025). Impact of graphite nano amendments on soil enzyme activities, functional genes, and microbiome composition in a soil-plant system. Soil Biol. Biochem. 206, 109714. doi: 10.1016/j.soilbio.2025.109714
Ding, Y., Zhang, P., Qin, Y. J., Tu, Q. C., Yang, Y. F., He, Z. L., et al. (2016). Network succession reveals the importance of competition in response to emulsified vegetable oil amendment for uranium bioremediation. Environ. Microbiol. 18, 205–18. doi: 10.1111/1462-2920.12981
Dini, I., Troiano, E., Gigliotti, C., Vassetti, A., Lotito, D., Staropoli, A., et al. (2025). Combined metagenomic and metabolomic analysis to evaluate the comprehensive effects of trichoderma and 6PP on vineyard. Ecosystems. Agriculture. 13, e01565–25. doi: 10.1128/spectrum.01565-23
Du, Y., Yang, Y., Wu, S., Gao, X., He, X., and Dong, S. (2025). Core microbes regulate plant-soil resilience by maintaining network resilience during long-term restoration of alpine grasslands. Nat. Commun. 16 1, 3116. doi: 10.1111/gcb.17235
Du, L., Zhong, H. H., Guo, X. N., Li, H. N., Xia, J. X., and Chen, Q. (2024). Nitrogen fertilization and soil nitrogen cycling: Unraveling the links among multiple environmental factors, functional genes, and transformation rates. Sci. Total Environ. 951, 175561. doi: 10.1016/j.scitotenv.2024.175561
Fan, X. H., Chen, M. S., Liu, W. T., Lin, X., Weng, K. L., Wu, S. H., et al. (2022). Effect of combined chemical fertilizer reduction with organic fertilizer on yield and quality of grape and soil quality. Soil Fertil. Sci. China 03, 46–51. doi: 10.11838/sfsc.1673-6257.20741
Fan, K. K., Delgado-Baquerizo, M., Guo, X. S., Wang, D. Z., Zhu, Y. G., and Chu, H. Y. (2021). Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 15, 550–561. doi: 10.1038/s41396-020-00796-8
Fu, X., Chen, X., Chen, Y., Hui, Y., Wang, R., and Wang, Y. (2024). Foliar co-applications of nitrogen and iron on vines at different developmental stages impact wine grape (Vitis vinifera L.) composition. Plants. 13(16), 2203. doi: 10.3390/plants13162203
Gao, R. P., Duan, Y., Zhang, J., Ren, Y. F., Liang, J. M., Jing, Y. P., et al. (2023). Effects of long-term fertilization on soil microbial diversity and community structure in the agro-pastoral ecotone. Environ. Sci. 44, 1063–1073. doi: 10.13227/j.hjkx.202203143
Guo, S., Xiong, W., Hang, X. N., Gao, Z. L., Jiao, Z. X., Liu, H. J., et al. (2021). Protists as main indicators and determinants of plant performance. Microbiome. 9, 64–64. doi: 10.1186/s40168-021-01025-w
Han, P., Tang, X. F., Koch, H. N., Dong, X. Y., Hou, L. J., Wang, D. H., et al. (2024). Unveiling unique microbial nitrogen cycling and nitrification driver in coastal Antarctica. Nat. Commun. 15, 3143. doi: 10.1038/s41467-024-47392-4
Hartmann, M. and Six, J. (2023). Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 4–18. doi: 10.1038/s43017-022-00366-w
Homsa, A. R., Sidiq, R. M., Yanuar, S. E., Swardana, A., and Nafi’ah, H. H. (2023). Interaksi Tiga Faktor Pupuk Terhadap Pertumbuhan dan Hasil Bawang Putih (Allium sativum L.). Agric. Food Sci. 23, 51–64. doi: 10.31293/agrifor.v23i1.6973
Hu, Y., Li, Y., Liu, K. M., Shi, C. Q., Wang, W., Yang, Z. G., et al. (2025). Improving the stability of black soil microbial communities through long–term application of biochar to optimize the characteristics of DOM components. Biochar 7. doi: 10.1007/s42773–025–00473–z
Hu, C. C., Liu, X. Y., Driscoll, A. W., Kuang, Y. W., Brookshire, E. N. J., Lü, X. T., et al. (2024b). Global distribution and drivers of relative contributions among soil nitrogen sources to terrestrial plants. Nat. Commun. 15, 6407. doi: 10.1038/s41467-024-50674-6
Hu, W., Zhang, Y., Rong, X., Zhou, X., Fei, J., Peng, J., et al. (2024a). Biochar and organic fertilizer applications enhance soil functional microbial abundance and agroecosystem multifunctionality. Biochar. 6, 3. doi: 10.1007/s42773-023-00296-w
Jia, Y. L., Huang, D. Y., Lan, X. L., Sun, X. X., Lin, W. J., Sun, W. M., et al. (2025). Community structure and metabolic potentials of keystone taxa and their associated bacteriophages within rice root endophytic microbiome in response to metal(loid)s contamination. Environ. pollut. 372, 126028. doi: 10.1016/j.envpol.2025.126028
Jiang, M. H., Dong, C., Bian, W. P., Zhang, W. B., and Wang, Y. (2024). Effects of different fertilization practices on maize yield, soil nutrients, soil moisture, and water use efficiency in northern China based on a meta-analysis. Sci. Rep. 14, 6480–6480. doi: 10.1038/s41598-024-57031-z
Jin, D., Lu, Z. W., Song, X. C., Ahammed, G. J., Yan, Y., and Chen, S. C. (2024). Improvement of yield and quality properties of radish by the organic fertilizer application combined with the reduction of chemical fertilizer. Agronomy 14, 1847. doi: 10.3390/agronomy14081847
Kaya, O., Yilmaz, T., Ates, F., Çelik, S., Demir, M., Bakırcı, G., et al. (2024). Improving organic grape production: the effects of soil management and organic fertilizers on biogenic amine levels in Vitis vinifera cv. ‘Royal’ grapes. Chem. Biol. Technol. Agric. 11, 38. doi: 10.1186/s40538-024-00564-2
Kiya, A. T., Zhang, L., Zhe, S., Nano, A. D., Jiwen, L., Md, A. A., et al. (2024). Impacts of long-term chemical nitrogen fertilization on soil quality, crop yield, and greenhouse gas emissions: With insights into post-lime application responses. Sci. Total Environ. 944, 173827. doi: 10.1016/j.scitotenv.2024.173827
Kost, C., Pati, L. K. R., Friedman, J., Garcia, S. L., and Ralser, M. (2023). Metabolic exchanges are ubiquitous in natural microbial communities. Nat. Microbiol. 8, 2244–2252. doi: 10.1038/s41564-023-01511-x
Laurent, P., Claire, C., Andreas, K., Matthias, C. R., and Noah, F. (2023). The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 22, 226–239. doi: 10.1038/s41579-023-00980-5
Li, Y. Y., Chapman, S. J., Nicol, G. W., and Yao, H. Y. (2018). Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 116, 290–301. doi: 10.1016/j.soilbio.2017.03.025
Li, Y. and Li, Y. (2025). Nitrogen addition enhances soil carbon and nutrient dynamics in Chinese croplands: a machine learning and nationwide synthesis. Carbon Balance Manage. 20, 15. doi: 10.1186/s13021-025-00305-4
Li, Y. Y., Ming, B., Fan, P. P., Liu, Y., Wang, K. R., Hou, P., et al. (2022a). Quantifying contributions of leaf area and longevity to leaf area duration under increased planting density and nitrogen input regimens during maize yield improvement. Field Crops Res. 283. doi: 10.1016/j.fieldcropsres.2022.128526
Li, R. C., Ren, C. Y., Wu, L. K., Zhang, X. X., Mao, X. Y., Fan, Z., et al. (2023). Fertilizing-induced alterations of microbial functional profiles in soil nitrogen cycling closely associate with crop yield. Environ. Res. 231, 116194–116194. doi: 10.1038/s43247-025-02032-7
Li, Y., Shang, Y., and Yang, Y. (2017). Clustering coefficients of large networks. Inform. Sci. 382, 383350–383358. doi: 10.1016/j.ins.2016.12.027
Li, P., Wu, M. C., Kang, G. D., Zhu, B. J., Li, H. X., and Jiao, J. G. (2020). Soil quality response to organic amendments on dryland red soil in subtropical China. Geoderma. 373. doi: 10.1016/j.geoderma.2020.114416
Li, L. C., Yang, M. Y., Li, J. C., Roland, B., Du, Z. L., and Wu, D. (2022b). Potential denitrification activity response to long-term nitrogen fertilization - A global meta-analysis. J. Cleaner Prod. 336. doi: 10.1016/j.jclepro.2022.130451
Liang, X., Yu, S., Ju, Y., Wang, Y., and Yin, D. (2025). Integrated management practices foster soil health, productivity, and agroecosystem resilience. Agronomy. 15(8), 1816. doi: 10.3390/agronomy15081816
Liebig, M. A., Varvel, G., and Doran, J. W. (2001). A simple performance-based index for assessing multiple agro-ecosystem functions. Agron. J. 93, 313–318. Available online at: https://digitalcommons.unl.edu/agronomyfacpub/336/. (Accessed April 19, 2025).
Liu, X., Chu, H., Godoy, O., and Delgado-Baquerizo, M. (2024). Positive associations fuel soil biodiversity and ecological networks worldwide. Proc. Natl. Acad. Sci. U. S. A. 121, e2308769121–e2308769121. doi: 10.1073/pnas.2308769121
Luo, Y., Yin, H., Ma, Y., Wang, J. H., Che, Q. X., Zhang, M., et al. (2024). Optimizing nitrogen fertilizer for improved root growth, nitrogen utilization, and yield of cotton under mulched drip irrigation in southern Xinjiang, China. Sci. Rep. 14, 2045–2322. doi: 10.1038/s41598-024-73350-7
Ma, L., Gao, W., Luan, H. A., Tang, J. W., Li, M. Y., and Huang, S. W. (2021a). Effects of partial substitution of chemical fertilizer with manure and/or straw on the abundance of functional genes related to soil N-cycling. J. Plant Nutr. Fert. 27, 1767–1778. doi: 10.1016/j.eti.2022.102900
Ma, X., Song, Y., Song, C., Wang, X., Wang, N., Gao, S., et al. (2021b). Effect of nitrogen addition on soil microbial functional gene abundance and community diversity in permafrost peatland. Microorganisms. 9, 2498. doi: 10.3390/microorganisms9122498
Montgomery, E. B. and Montgomery, M. B. (1911). Correlation studies in corn. 24th Annu. Rep. 24, 108–159.
N’Dri, Y. B., Han, S. W., Li, H. R., Kouadio, Y. D., Ejaz, I., Virk, A., et al. (2023). Effects of fertilizer application strategies on soil organic carbon and total nitrogen storage under different agronomic practices: A meta-analysis. Land Degrad Dev. 34, 5889–5904. doi: 10.1002/saj2.70000
Pahalvi, H. N., Rafiya, L., Rashid, S., Nisar, B., and Kamili, A. N. (2021). “Chemical fertilizers and their impact on soil health,” in Microbiota and Biofertilizers Vol 2. Eds. Dar, G. H., Bhat, R. A., Mehmood, M. A., and Hakeem, K. R. (Springer, Cham). doi: 10.1007/978-3-030-61010-4_1
Pei, B., Zhang, Y., Liu, T., Cao, J., Ji, H., Hu, Z., et al. (2024). Effects of seaweed fertilizer application on crops’ yield and quality in field conditions in China-A meta-analysis. PLoS One 19, 15. doi: 10.1371/journal.pone.0307517
Puente-Sánchez, F., Pascual-García, A., Bastolla, U., Pedrós-Alió, C., and Tamames, J. (2024). Cross-biome microbial networks reveal functional redundancy and suggest genome reduction through functional complementarity. Commun. Biol. 7, 1046. doi: 10.1038/s42003-024-06616-5
Ren, W., Li, X., Liu, T., Chen, N., Xin, M., Qi, Q., et al. (2025). Controlled-release fertilizer affects leaf nitrogen allocation and photosynthesis to improve nitrogen use efficiency and yield in the sunflower field. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1622766
Shamshitov, A., Kadžienė, G., and Supronienė, S. (2024). The role of soil microbial consortia in sustainable cereal crop residue management. Plants. 13(6), 766–784. doi: 10.3390/plants13060766
Song, L. and Niu, S. L. (2022). Increased soil microbial AOB, amoA and narG abundances sustain long-term positive responses of nitrification and denitrification to N deposition. Soil Biol. Biochem. 170, 108539. doi: 10.1016/j.soilbio.2021.108539
Tang, Y. F., Xie, H., Sun, J., Li, X. O., Zhang, Y., and Dai, X. H. (2022). Alkaline thermal hydrolysis of sewage sludge to produce high-quality liquid fertilizer rich in nitrogen-containing plant-growth-promoting nutrients and biostimulants. Water Res. 211, 118036. doi: 10.1016/j.watres.2021.118036
Thion, C. E., Poirel, J. D., Cornulier, T., De, V. F. T., Bardgett, R. D., and Prosser, J. I. (2016). Plant nitrogen-use strategy as a driver of rhizosphere archaeal and bacterial ammonia oxidiser abundance. FEMS Microbiol. Ecol. 92, fiw091–fiw091. doi: 10.1093/femsec/fiw091
Tian, S. Y., Zhu, B. J., Yin, R., Wang, M. W., Jiang, Y., Zhang, C. Z., et al. (2022). Organic fertilization promotes crop productivity through changes in soil aggregation. Soil Biol. Biochem. 165. doi: 10.1016/j.soilbio.2021.108533
Wang, X. L., Qian, R., Han, Y. F., Ji, Z., Yang, Q. X., Wang, L. L., et al. (2024b). Straw return can increase maize yield by regulating soil bacteria and improving soil properties in arid and semi-arid areas. Eur. J. Agron. 161, 127389. doi: 10.1038/s41598-024-70404-8
Wang, L., Wang, J., Tang, Z. H., Wang, J. D., and Zhang, Y. C. (2024a). Long-term organic fertilization reshapes the communities of bacteria and fungi and enhances the activities of C-and P-cycling enzymes in calcareous alluvial soil. Appl. Soil Ecol. 194. doi: 10.1007/s11104-024-07169-6
Wang, Y. L., Wu, C. R., Xue, X. R., Sun, J. X., Liu, X. L., Yang, Z. P., et al. (2023). Effects of flushing alkaline thermal hydrolysis liquid to promote Brassica chinensis yield and nitrogen invertase activity mechanistic research. Chin. J. Eco-Agric. 31, 1768–1779. doi: 10.12357/cjea.20230028
Wen, T., Xie, P. H., Yang, S. D., Niu, G. Q., Liu, X. Y., Ding, Z. X., et al. (2022). ggClusterNet: An R package for microbiome network analysis and modularity-based multiple network layouts. IMeta. 1, e32–e32. doi: 10.1002/imt2.32
Wu, Y. S., Li, X. J., Zhang, W. W., Wang, L., Li, B., and Wang, S. P. (2023b). Aroma profiling of Shine Muscat grape provides detailed insights into the regulatory effect of gibberellic acid and N-(2-chloro-4-pyridinyl)-N-phenylurea applications on aroma quality. Food Res. Int. 170, 112950. doi: 10.1016/j.foodres.2023.112950
Wu, C. R., Xue, X. R., Wang, Y. L., Bai, J., Guo, C. X., Yang, Z. P., et al. (2023a). Effects of spraying alkaline thermal hydrolysis liquid on leafy vegetable yield and nitrogen invertase. J. Agro-Environ. Sci. 42, 1156–1165. doi: 10.11654/jaes.2022-1013
Xue, X. R., Liu, Z. B., Wu, C. R., Liu, X. L., Wang, Y. L., Bai, J., et al. (2023). Effects of combined application of sludge alkaline thermal hydrolyzate and nitrogen fertilizer on tomato yield, qualityand nitrogen conversion. Soil Fertil. Sci. China 10), 201–208. doi: 10.11838/sfsc.1673-6257.22584
Ye, C., Zheng, G., Tao, Y., Xu, Y., Chu, G., Xu, C., et al. (2024). Effect of soil texture on soil nutrient status and rice nutrient absorption in paddy soils. Agronomy. 14(6), 1339. doi: 10.3390/agronomy14061339
Yuan, Q., Jiang, Y., Yang, Q., Li, W., Gan, G., Cai, L., et al. (2024). Mechanisms and control measures of low temperature storage-induced chilling injury to solanaceous vegetables and fruits. Front. Plant Sci. 15, 1488666. doi: 10.3389/fpls.2024.1488666
Zhang, H. K., Fang, Y. Y., Chen, Y. C., Li, Y., Lin, Y. X., Wu, J. S., et al. (2022). Enhanced soil potential N2O emissions by land-use change are linked to AOB-amoA and nirK gene abundances and denitrifying enzyme activity in subtropics. Sci. Total Environ. 850, 158032. doi: 10.1016/j.scitotenv.2022.158032
Zhang, C., Lei, S., Wu, H., Liao, L., Wang, X., Zhang, L., et al. (2024c). Simplified microbial network reduced microbial structure stability and soil functionality in alpine grassland along a natural aridity gradient. Soil Biol. Biochem. doi: 10.1016/j.soilbio.2024.109366
Zhang, Y., Wang, H., Chen, H., Liu, R., Chen, H., Fang, X., et al. (2024b). The crucial evaluation indexes and relative measurement methods of edible value for fresh fruits and vegetables: A review. FPF 1, 222. doi: 10.1002/fpf2.12026
Zhang, H., Zhu, W., Zhang, J. B., Christoph, M., Wang, L. F., and Jiang, R. (2024a). Enhancing soil gross nitrogen transformation through regulation of microbial nitrogen-cycling genes by biodegradable microplastics. J. Hazard. Mater. 478, 0304–3894. doi: 10.1016/j.jhazmat.2025.02.001
Zheng, H. P., Yang, T. J., Bao, Y. Z., He, P. P., Yang, K. M., Mei, X. L., et al. (2021). Network analysis and subsequent culturing reveal keystone taxa involved in microbial litter decomposition dynamics. Soil Biol. Biochem. 157, 108230. doi: 10.1016/j.soilbio.2021.108230
Zhou, Z. F., Shi, X. J., Zheng, Y., Qin, Z. X., Xie, D. T., Li, Z. L., et al. (2014). Abundance and community structure of ammonia-oxidizing bacteria and archaea in purple soil under long-term fertilization. Eur. J. Soil Biol. 60, 24–33. doi: 10.1016/j.ejsobi.2014.07.002
Keywords: bacterial–fungal interactions, sludge alkaline hydrolysate, grape nutrition, sustainable fertilization, yield
Citation: Xue D, Yang Y, Zhang H, Quan Y, Li Z, Li Z, Wang W, Bo H, Jin D, Xu M, Zhang Q and Yang Z (2025) Blending sludge alkaline hydrolysate and urea affects grape yield and quality by regulating soil bacterial communities. Front. Plant Sci. 16:1665661. doi: 10.3389/fpls.2025.1665661
Received: 14 July 2025; Accepted: 08 September 2025;
Published: 30 September 2025.
Edited by:
Hanuman Singh Jatav, Sri Karan Narendra Agriculture University, IndiaReviewed by:
Javed Iqbal, Quaid-i-Azam University, PakistanMuhammad Salman Zahid, Shanghai Jiao Tong University, China
Copyright © 2025 Xue, Yang, Zhang, Quan, Li, Li, Wang, Bo, Jin, Xu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huijuan Bo, Ym9odWlqdWFuQHN4YXUuZWR1LmNu; Dongsheng Jin, c3hkeGpkc0AxMjYuY29t
 Donghe Xue1,2
Donghe Xue1,2 Zixu Li
Zixu Li Huijuan Bo
Huijuan Bo