- 1Key Laboratory of Biology and Genetic Improvement of Oil Crops, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Ministry of Agriculture and Rural Affairs, Wuhan, China
- 2Hubei Hongshan Laboratory, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
Cold stress is a major abiotic stress that seriously hinders plant growth and development, ultimately affecting crop yields. During the process of evolution, plants have evolved sophisticated adaptive strategies encompassing acclimation processes and tolerance mechanisms. Over the past two decades, substantial research breakthroughs have been made in elucidating the core components and complex regulatory networks underlying cold tolerance. This review systematically synthesizes the recent progress in three fundamental aspects: cold stress perception and signal transduction pathways, downstream physiological and molecular responses, and the pivotal regulatory roles of transcription factors (particularly CBF/DREB1 family) and cold-responsive miRNAs. In addition, we also investigated the intricate crosstalk between cold response and other biological processes including photoperiod sensing, flowering regulation, circadian rhythm, phytohormone signaling, and the dedicated discussion addresses how plants achieve metabolic and developmental trade-offs when allocating resources between cold defense and other vital traits. Looking forward, we propose four promising research directions: identifying novel cryo-sensors beyond currently known receptors, post-translational modification dynamics of CBF proteins, homeostatic control mechanisms among competing regulatory factors, and translational applications of cold stress pathways in precision breeding programs. Addressing these knowledge gaps will not only deepen our understanding of plant cold adaptation at molecular level, but also facilitate the development of climate-resilient crops through molecular design breeding.
1 Introduction
Cold stress is a significant factor limiting plant growth and development, which often leads to a slowdown or even arrest of plant growth, and ultimately results in a large-scale decline in grain crop yields (Shi et al., 2018). For example, cold stress can cause a decrease in tomato yield by 8–21%, rice yield by 15–35%, and chickpea, soybean, and mung bean yields by 45–61% respectively (El-Refaee et al., 2024; Oliveira et al., 2025). Based on the different physiological mechanisms that function at varying temperature levels, the stress can be divided into chilling stress (0-15 °C) and freezing stress (below 0 °C) (Chinnusamy et al., 2007). Chilling stress mainly results from osmotic dehydration caused by extracellular ice crystals, leading to the hardening of plant membranes, the destruction of organelles and the inhibition of plant growth (Wang et al., 2024c). However, plants suffer greater damage under freezing stress because water diffuses out of cells and forms ice crystals, decreasing extracellular water potential and causing severe dehydration and ultimately death (Villouta et al., 2021).
Plants have evolved adaptive strategies to combat cold stress through prolonged evolutionary processes. Tropical plants such as rice can enhance the cold tolerance after a period of cold treatment. Temperate plants such as rapeseed, wheat, and Arabidopsis usually exhibit a higher tolerance to cold stress than tropical plants. The phenomenon of improving cold tolerance through cold training is called cold acclimation (Kutsuno et al., 2023). During this period, many changes occur, including upregulation and downregulation of related genes, which will alter the levels of some proteins (such as AFPs), metabolites, and hormones in order to combat cold stress (Maruyama et al., 2012). In recent years, research on cold tolerance has attracted widespread attention, and many cold related genes have been discovered. However, there are few reports that comprehensively explain the molecular mechanisms of how plants respond to cold stress from multiple dimensions. Therefore, this article will focuses on the cutting-edge dynamics of plant cold tolerance mechanisms, including perception, signal transmission, and response. It emphasizes the analysis of key gene functions closely related to cold stress and provides a systematic classification and detailed explanation of related signaling pathways. It has irreplaceable value in deepening our understanding of how plants can cleverly avoid or reduce cold damage. Meanwhile, the article also sorted out problems that need to be urgently solved and, in combination with the existing cold stress related gene information, put forward practical and feasible technical suggestions for the practice of cold-resistant molecular breeding, aiming to provide useful references for scientific research and application in this field.
2 Cold signal perception
The cell membrane serves as the primary site for plants to perceive temperature, with cold signal perception commencing via multiple receptors located on the membrane. To date, several potential cold sensors/receptors have been identified. In Arabidopsis, the plasma membrane receptor-like kinase CRLK1 modulates Ca2+ channel activity, thereby triggering calcium signal transduction and activating the MAPK kinase cascade reaction during the period of cold stress. This ultimately leads to the upregulation of CBF1 expression, thereby enhancing plant cold tolerance (Zhao et al., 2017). Additionally, the cytoplasmic receptor-like kinase CRPK1, located on the plasma membrane (PM), facilitates its interaction with CBF proteins by specifically phosphorylating 14-3–3 proteins, and actively regulates cold tolerance (Liu et al., 2017). Recent studies have indicated that the formation of a complex between CRPK1 and the receptor protease KOIN influences the response of the 14-3-3-CBF module to cold stress (Zhang et al., 2025). These cold receptors connect cold perception with the subsequent cold transduction.
Cold sensors are equally vital for cold perception across different plant species. For instance, in rice, the G protein regulatory factor COLD1, which is located in the plasma membrane, forms a functional complex with RGA1. This complex not only mediates the initial perception of cold signals but also induces extracellular Ca2+ influx, thereby activating downstream signaling cascades (Ma et al., 2015b). Recent research indicates that OsSRO1c forms biomolecular aggregates through liquid-liquid phase separation (LLPS), recruiting the transcription factor OsDREB2B into nuclear aggregates. This dynamic phase transition significantly enhances cold tolerance in rice by promoting the expression of COLD1 (Hu et al., 2024b). In Vitis amurensis, VaCOLD1 also actively regulates cold stress. VaCOLD1 forms a complex with the G protein α subunit VaGPA1, activates Ca2+ channels, promotes an increase in intracellular Ca2+ concentration, and triggers the MAPK cascade reaction and the downstream CBF signaling pathway (Zheng et al., 2023). Collectively, COLD1 plays a conserved yet flexible role as a cold sensor in both monocotyledonous and dicotyledonous plants, which providing a crucial molecular target for cold tolerance crop breeding.
The Ca2+ channels can also function as cold receptors by sensing cold stress. These channels regulate the transmembrane flow of Ca2+ in cellular perception of cold signals. In Arabidopsis, the permeable transporter protein ANN1, located on the plasma membrane, mediates cold-induced Ca2+ influx into the cytoplasm, thereby establishing frost resistance (Liu et al., 2021a). The calcium-permeable channel AtMCA1/2 is involved in increasing cold-induced Ca2+ concentration and enhancing cold tolerance (Mori et al., 2018). The Ca2+-permeable channel CNGC20 actively regulates frost resistance in Arabidopsis by influencing cold-induced Ca2+ influx. Deletion of the CNGC20 gene results in reduced Ca2+ influx. Similarly, the rice homolog OsCNGC20 positively regulates plant cold tolerance by affecting Ca2+ flow. During initial cold exposure, receptor kinase PSY1R phosphorylates OsCNGC20, inducing cytosolic Ca2+ influx. In later stress phases, CRPK1 phosphorylates CNGC20 and triggers its degradation via the 26S proteasome. Thus, PSY1R and CRPK1 regulate OsCNGC20-mediated cold stress in an antagonistic manner (Peng et al., 2024a). Another channel, OsCNGC9, a homolog of the Arabidopsis OST1 gene, can be activated by phosphorylation, triggering cytoplasmic calcium influx and activating the downstream cold responsive genes (CORs) expression (Wang et al., 2021). The mechanism by which plants perceive cold stress through plasma membrane ion channels exhibits both significant conservation and diversity.
3 Cold signal transduction
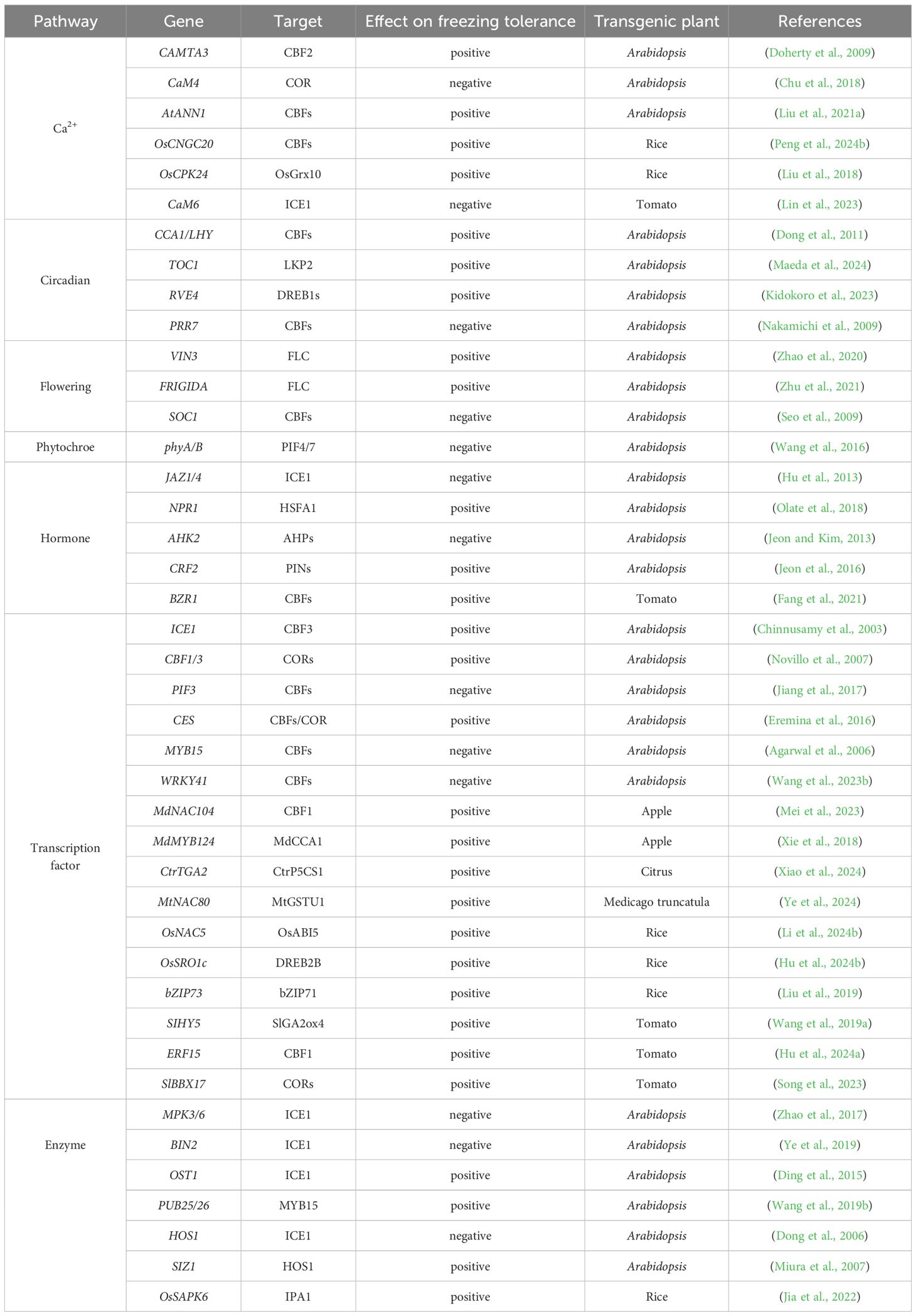
After being perceived, cold signals undergo a series of signal molecules and multiple transmission pathways to achieve cold signal transduction (Mortazavi et al., 2008). Secondary messengers like Ca2+, ROS, and NO are generated by cells to convey cold signals. These messengers control intracellular Ca2+ levels, resulting in Ca2+ influx and subsequent signal transduction (Figure 1).
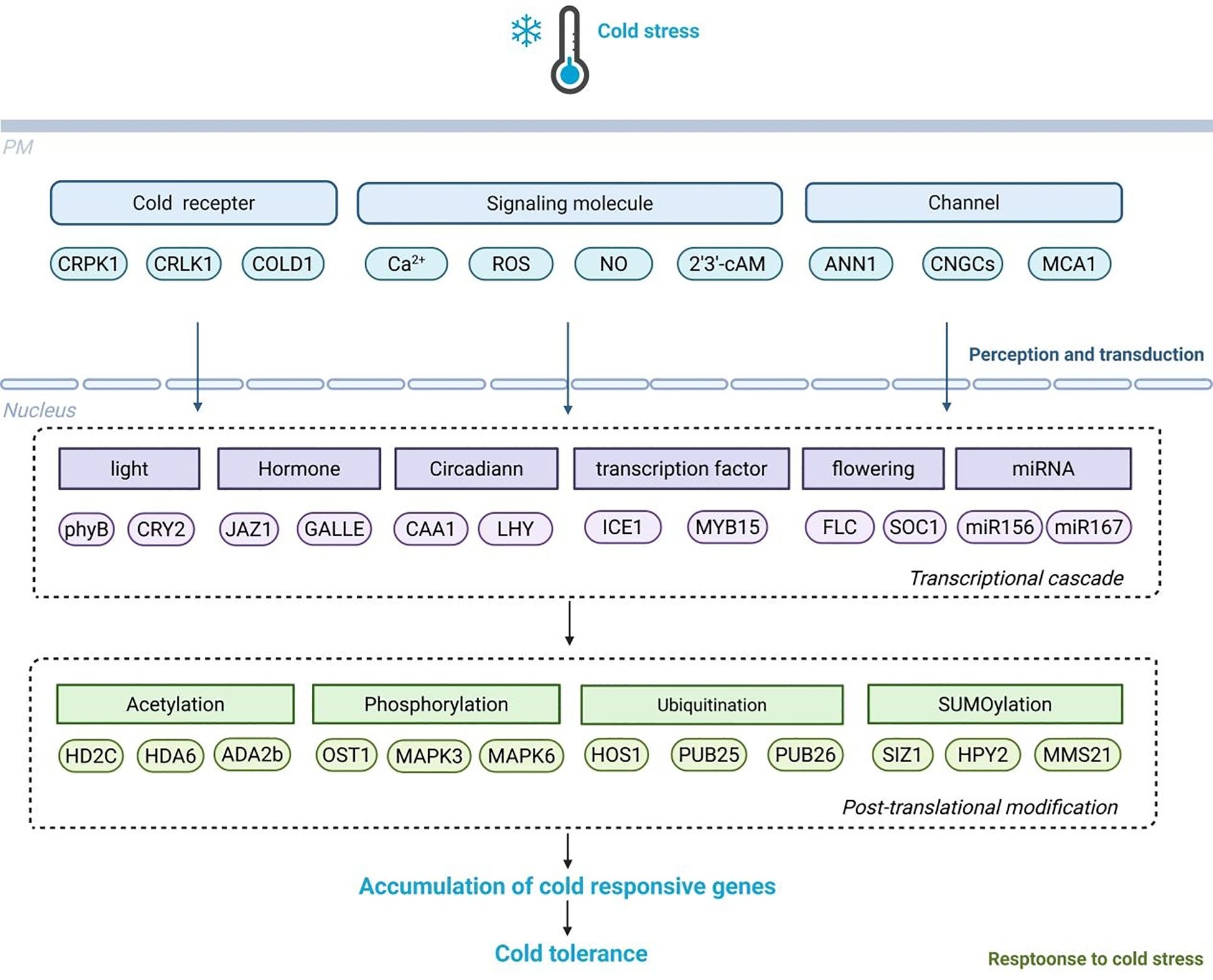
Figure 1. The signal perception, transduction, and response mechanisms of plant cold stress. Plants perceive cold signals through cold receptors such as COLD1, and conduct signal transduction through signaling molecules such as Ca2+, NO, ROS, as well as related channels such as ANN1 and MCA1. These signaling molecules activate a series of transcription factors, circadian rhythms, light, flowering, and enzyme related regulatory factors in the transcription cascade and post-translational modifications to induce the expression of COR genes under cold stress.
3.1 Ca2+ signal
Ca2+ serves as the second messenger in plant cells, mediating various developmental processes and responding to environmental stimuli by triggering primary signals (Yamazaki et al., 2008). Under cold stress, cell membrane channels are activated, causing Ca2+ influx. These Ca2+ signals are recognized by Ca2+-dependent protein kinases such as CIPKs, CaM, CAMTAs, and CDPKs, which interpret the signals and initiate subsequent events. In Arabidopsis, the Ca2+-dependent protein kinase CIPK3 is activated after cold stimulation, directly phosphorylating CNGC5/6 channels, enhancing their permeability, and promoting initial Ca2+ influx. CaM2 binds to CNGC5/6 after Ca2+ accumulation, inhibiting channel activity and preventing cell toxicity caused by Ca2+ overload (Ming et al., 2025). CPK28, another Ca2+-dependent protein kinase, can be rapidly activated and regulates the expression level of CBF genes via phosphorylating nin-like protein 7 (NLP7) (Ding et al., 2022). Additionally, Ca2+-dependent protein kinase CAMTA3/5 can induce the expression of the CBF1 gene, helping plants combat rapid temperature drops (Kidokoro et al., 2017). Based on the above research, it can be clearly seen that Ca2+ acts as a key mediator of cold signaling, and Ca2+-dependent protein kinases serve as a vital link between Ca2+ signal transduction and CBF gene expression.
The relationship between Ca2+ and cold stress in other plants has also garnered attention. In tomato, low temperatures can trigger Ca2+ influx. The Ca2+ signal is captured and decoded by SICaM6 before entering the nucleus, binds to ICE1 and inhibits its transcriptional activity, ultimately reducing tomato cold tolerance (Lin et al., 2023). In rice, OsCPK24 negatively regulates the cold tolerance via phosphorylating OsGrx10, maintaining high levels of glutathione and phosphorylation, and thereby enhancing cold tolerance (Liu et al., 2018). Despite significant progress in studying Ca2+ signaling mechanisms under cold stress, the principles and dynamic changes of Ca2+ production during cold stress process remain unclear. Exploring the spatiotemporal dynamics of Ca2+ is vital for elucidating the specific mechanisms of Ca2+ signal transduction in plant adaptation to cold stress.
3.2 Reactive oxygen species signal
Cold stress can cause damage to the cell membrane system, increase membrane permeability, lead to electron leakage, and induce the production of ROS, including hydrogen peroxide (H2O2), superoxide anion (O2-), hydroxyl radicals (OH-) (Mittler, 2002). The role of ROS depends on its concentration level. When plants are under optimal growth conditions, intracellular ROS levels are low. However, under cold stress, excessive accumulation of ROS can cause oxidative damage by attacking polyunsaturated fatty acids, leading to lipid peroxidation and altering membrane fluidity. ROS can also oxidase and modify amino acid residues in proteins, altering protein activity, thereby affecting the function of transcription factors, regulating downstream gene expression and ultimately causing serious impact on plant growth (Gill and Tuteja, 2010).
Maintaining a low ROS concentrations in plants is widely recognized as beneficial for growth. In rice, the mitochondrial protein SOP10 can regulate superoxide generation and positively regulate cold tolerance. Research has shown that the sop10 mutant exhibits significantly enhanced survival ability at low temperatures due to reduced ROS accumulation (Zu et al., 2023). In apple, MdNAC104 improves cold tolerance by regulating ROS levels and enhancing antioxidant capacity. Transgenic plants overexpressing MdNAC104 show decreased ion leakage and ROS accumulation under cold stress, while osmotic regulatory substances and antioxidant enzyme activity increase (Mei et al., 2023). These findings highlight the important role of ROS in plant cold stress responses, providing potential gene targets for developing cold-resistant plant varieties and promoting research in plant cold tolerance breeding.
3.3 Nitric oxide signal
NO also acts as a messenger molecule and is widely involved in cold response (Wilson et al., 2008). At low levels, NO acts as a signaling molecule, while at high levels, it induces cell damage and triggers NO stress. In Arabidopsis, cold acclimation promotes the production of endogenous NO and the accumulation of osmotic substance. In the nia1nia2 double mutant, the expression levels of CBF1/2/3, KIN1, and COR15a were significantly decreased, indicating that NO is a necessary signal for rapid upregulation of cold-induced genes (Cantrel et al., 2011). Further research reveals that NO actively regulates 14-3–3 protein expression and mediates the CBF/DREB1 transcription network (Yang et al., 2013b).
Previous studies have shown that NO-induced protein S-nitrosylation prevent cold damage by regulating plant antioxidant mechanisms. NO mediates the S-nitrosylation of superoxide dismutase (SOD), enabling it to regulate ROS detoxification through the antioxidant system (Zhao et al., 2009). When cold stimulation triggers the opening of the plasma membrane Ca2+ channel MCA1/2, NO activates calcium dependent protein kinase (CPK) through S-nitrosylation modification to enhance calcium transients and promote transcriptional bursts in the CBF regulatory network In wheat (Babuta et al., 2025). The precise interaction between NO and ROS also constitutes the defense line against cold in plants. Cold stress can trigger explosive accumulation of ROS, and NO acts not only as a scavenger for ROS but also maintain redox balance by activating antioxidant enzymes such as SOD. In dichondra, cold stress leads to a rapid increase of NO in cold resistant varieties, causing soluble sugar accumulation. The antioxidant system works together to achieve low ROS accumulation, while cold sensitive genotype varieties lack this linkage mechanism (Ling et al., 2025).
The relationship between NO and Ca2+ under cold conditions has attracted attention. At low temperatures, the concentration of Ca2+ rapidly increases in the cytoplasm, leading to an enhanced expression of nitrate reductase ClNR1 and a sharp increase in NO. In turn, NO induces the expression of the cyclic nucleotide-gated channel ClCNGC20, which further causes Ca2+ influx. This ultimately inhibits the negative regulatory module of ClCaM2/5/7-ClVDAC1, thereby maintaining high CBF pathway activity and significantly enhancing cold resistance in watermelon, indicating that Ca2+ and NO have a synergistic effect in plant cold response (Guo et al., 2025).
3.4 2’,3’-cAMP signal
2’,3’-cAMP is emerging as a novel second messenger. Earlier studies have revealed that 2’,3’-cAMP synthase activity resides in TIR domain proteins, which are pivotal in plant immune responses (Yu et al., 2022). Recent advances have highlighted the critical role of 2’,3’-cAMP in cold reaction. Under cold stress conditions, COLD6 forms a protein complex with cold-induced OSM1. This complex senses extracellular cold signals, triggers the accumulation of 2’,3’-cAMP and subsequently activates the COR genes expression (Luo et al., 2024). Notably, studies have observed a high degree of synchronization between the accumulation of 2’,3’-cAMP and the expression of COR genes in Arabidopsis over time. This finding further reinforces the importance of 2’,3’-cAMP in cold signal transduction. Moreover, the application of exogenous 2’,3’-cAMP analogs can mimic the effect of increased intracellular 2’,3’-cAMP levels and enhance cold tolerance. This suggests that 2’,3’-cAMP can function as a signaling molecule to trigger cellular cold defense responses. In addition, 2’,3’-cAMP may amplify cold signals and activate downstream COR gene expression by interacting with CaMs and CDPKs in the calcium signaling pathway. Future exploration of the relationship between 2’,3’-cAMP and other messenger molecules promises to be highly insightful.
4 Cold signal response
cold signals are transmitted from the PM to the nucleus, triggering transcription and post transcriptional regulation, inducing the expression of cold related genes, and ultimately affecting plant cold tolerance. In the past two decades, various regulatory pathways have been discovered in plants at different stages to respond to cold stress, and key genes in these pathways have been continuously revealed (Table 1). These regulatory pathways are crucial for plants to combat cold stress. Next, we will describe the regulation of transcription factor, flowering, circadian rhythm, hormone regulation, and miRNA in the transcriptional cascade, as well as the regulation of phosphorylation, ubiquitination, and acetylation in post-translational modifications. These regulatory factors collectively respond to cold stress.
4.1 Transcription factor responds to cold stress
Transcription factors are key regulatory proteins situated at the forefront of plant growth regulation networks. Numerous transcription factor families in plants, such as AP2/ERF, WRKY, NAC, MYB, and bZIP, are known to be activated in response to cold stress.
The CBF/DREB transcription factor is a member of the AP2/ERF superfamily and serves as a core regulatory hub for cold response. In Arabidopsis, CBF proteins bind to CRT/DRE cis-elements within the promoters of COR genes (including COR, LTI, and RD) to activate transcriptional programs that enhance freezing tolerance. Genetic evidence indicates that cbf1/cbf2/cbf3 triple mutants exhibit hypersensitivity to freezing stress, accompanied by compromised seed germination and impaired stress-responsive capacity. Notably, AtCBF4 has been identified primarily as a mediator of drought stress adaptation (Shi et al., 2018). Recent advancements have revealed that CBF protein accumulates under cold conditions and directly interacts with SKIP, a key component of spliceosomes. SKIP serves as an essential factor regulating alternative splicing. CBF facilitates the formation of nuclear liquid-liquid phase separation aggregates involving SKIP proteins through its intrinsic disordered domain (IDR). This process enriches transcripts from the COR gene family and significantly enhances their alternative splicing efficiency. However, disrupting the interactions between CBF and SKIP or inhibiting phase separation markedly diminishes plant frost resistance. These findings underscore that regulation of alternative splicing induced by cold stress is critical for effective cold response (Fu et al., 2025).
In other plants, CBFs also showed a positive effect in response to cold stress. In rice, OsCBF3 positively regulates cold tolerance (Wang et al., 2021). The expression of OsCBF1/2/3 is directly regulated by OsERF52. Further research has found that OsSAPK9 can directly phosphorylate the Ser261 site of OsERF52 to promote protein accumulation, thereby increasing the transcription level of OsCBF1/2/3 (Xu et al., 2024a). In sweet potato, Overexpression of CBF3 gene enhances the cold tolerance. This indicates that CBF3 positively regulates the cold tolerance (Jin et al., 2017). In summary, CBF transcription factors exhibit conservation and species diversity.
The CBF-dependent pathways, especially the ICE1-CBF-COR pathway, is a key regulatory pathway conserved across different plant species (Tang et al., 2020). Under cold stress, ICE1 is activated and induces CBF expression by binding to the CBF3 promoter. The CBF protein further activates downstream COR genes, thereby enhancing cold tolerance. ICE1 stability is regulated by ubiquitination via HOS1 and SUMOylation by SIZ1 (Dong et al., 2006; Miura et al., 2007). Homologs of ICE1, such as OsbHLH002 in rice, also enhance plant cold tolerance by regulating the OsCBFs expression (Zhang et al., 2017; Xu et al., 2024a). Recent studies have revealed that the transcription factor AIF2 acts as a key mediator of the CBF-dependent pathway. Specifically, AIF2 enhances the stability of ICE1, thereby promoting the transcription of CBF genes, which collectively increase the cold tolerance. Additionally, AIF2 physically interacts with and induces phosphorylation-dependent degradation of MPK3/6 (Kim et al., 2024), which were previously shown to negatively regulate cold tolerance by phosphorylating ICE1 and targeting it for proteasomal degradation (Li et al., 2017a).
Despite the CBF-dependent pathway is important, it only regulates approximately 12% of COR genes. Many COR genes exhibit CBF-independent expression under cold stress, suggesting that plants may employ a multi-level transcriptional regulatory network involving other transcription factor families, such as bZIP, MYB, NAC, and WRKY.
The bZIP transcription factor is highly conserved across multiple species and is widely involved in plant stress response and adverse conditions. The conserved sequence of bZIP proteins (PYCGTGG) can specifically bind to the ABRE element (ACGT) in the promoters of target genes. In Arabidopsis, the bZIP transcription factors elongated hypocotyl 5 (HY5) and its homolog HYH activate gene expression by binding to the G-box element (ACGTGTC) in the SIG5 promoter. This process regulates the transcription of the chloroplast gene psbD BLRP, maintaining the abundance of the PSII D2 protein and thereby sustaining photosynthetic efficiency under long-term low-temperature and short-term freezing conditions (Cano-Ramirez et al., 2023). Similarly, in maize, HY5 binds to the natural variation site of the COOL1 gene promoter to regulate COOL1 expression, which affects adaptability to high-latitude and cold stress. The CPK17 kinase, activated by cold stress, stabilizes the COOL1 protein through phosphorylation, thereby enhancing cold tolerance (Zeng et al., 2025). In asparagus beans, VunHY5 and VunMED2 synergistically bind to the G-box/BRE element in the VunERD14 promoter, inducing its expression and increasing the expression of antioxidant genes to actively regulate cold tolerance (Liang et al., 2025). Likewise, overexpression of SlHY5 in tomatoes can elevate the expression levels of antioxidant enzyme genes, reduce ROS content, and enhance plant cold tolerance. Notably, SlHY5 can also influence tomato cold tolerance by directly regulating the expression of COR genes. It is important to highlight that bZIP family members exhibit bidirectional regulatory characteristics in cold response. Recent studies have shown that SlHY5 can directly affect CBF transcription levels and indirectly regulate CBF expression by inhibiting MYB15 (Zhang et al., 2020). Additionally, the cold-activated MAPK family member SlMPK1/2 phosphorylates the SlBBX17 protein, enhancing its interaction with SlHY5 and thereby synergistically improving cold tolerance (Song et al., 2023). In contrast, in maize, bZIP68 acts as a negative regulatory factor. After phosphorylation modification of ZmMPK8, cold tolerance is reduced by inhibiting the expression of ZmDREB1 gene (Li et al., 2022). However, the natural variation of the HSF21 gene promoter can alleviate the transcriptional repression of bZIP68 and compensate for cold tolerance by increasing the expression level of HSF21 induced by cold stress. The research on rice further expands the regulatory dimension of bZIP. The heterodimer formed by bZIP73Jap and bZIP71 enhances cold tolerance during seedling stage by activating peroxidase gene expression, while during reproductive growth stage, it specifically enhances the expression of sugar transport related genes such as OsMST7, OsMST8, and OsINV4, significantly improving the seed setting rate under cold stress by promoting soluble sugar transport from anthers to pollen (Liu et al., 2019).
MYB transcription factors constitute one of plant biology’s most expansive TF families, functioning as master regulators coordinating developmental programs and abiotic stress adaptation pathways. In Arabidopsis, AtMYB15 acts as a crucial regulator in response to cold stress. Mutant plants lacking MYB15 exhibit enhanced tolerance to cold stress, whereas plants overexpressing MYB15 show reduced freezing tolerance. This is attributed to the ability of MYB15 to directly bind to the CBF promoter, thereby inhibiting the expression of CBF genes. Additionally, MYB15 can interact with the CBF regulatory factor ICE1 to modulate CBF genes expression (Wang et al., 2023c). However, studies on homologous mechanisms in tomatoes have revealed contrasting results. SlMYB15 in tomatoes directly binds to the CBF promoter and positively regulates the cold tolerance (Zhang et al., 2020). In rice, OsMYBS3 negatively regulates cold tolerance by inhibiting the expression of OsDREB1B (Su et al., 2010). In apple, MdMYB23 displays unique functions. It can directly bind to the promoters of MdCBF1/2, activating their expression. Moreover, it interacts with the promoter of MdANR, a key regulator of anthocyanin biosynthesis, to enhance MdANR expression, which in turn promotes anthocyanin accumulation and scavenging of ROS (An et al., 2018). Furthermore, MdMYB88/124 in apple activates CBF3 expression by directly binding to the promoter of MdCCA1. This ultimately induces the expression of COR genes, thereby enhancing cell membrane stability and osmotic regulation capabilities. Additionally, MdMYB88/124 can directly bind to the promoters of key anthocyanin synthesis genes, such as MdUFGT, promoting anthocyanin accumulation under cold conditions through a CBF-independent pathway (Liu et al., 2021b).
NAC transcription factors comprise the dominant plant-specific regulator family. NAC proteins regulate the expression of target genes by binding to the core sequence (CGTG/A) (Shao et al., 2015). In Arabidopsis, NAC056 positively modulates the expression of CBF genes, thereby enhancing plant cold tolerance (Xu et al., 2024b). In apple, MdNAC104 directly binds to the promoters of MdCBF1/3 to enhance the cold tolerance. Additionally, it stimulates anthocyanin accumulation under cold conditions by upregulating the expression of genes involved in anthocyanin synthesis (Mei et al., 2023). In rice, OsNAC5 actively regulates germination and seedling cold tolerance by directly activating the expression of OsABI5. The expression of COR genes is significantly upregulated in OsNAC5-overexpressing lines, while it is downregulated in abi5 knockout mutants (Li et al., 2024b). In short, NAC transcription factors in plants exhibit distinct but effective strategies to enhance plant cold tolerance.
The WRKY transcription factor family is a highly conserved group in plants. These factors specifically recognize W-box (TTGACC/T) elements in the promoter regions of target genes. In Arabidopsis, AtWRKY41 directly binds to the CBF promoter, negatively regulating the transcription and thereby reducing freezing tolerance (Wang et al., 2023b). In contrast, the homologous protein ScWRKY41 in potato functions as a positive regulator of cold stress. It recruits the histone acetyltransferase ScHAC1 to the promoter of the key flavonoid metabolism gene ScF3’H, significantly increasing the histone H3K9ac modification level and activating ScF3’H expression to enhance cold tolerance (Bao et al., 2025). Similarly, rice OsWRKY76 positively regulates cold tolerance by directly activating OsDREB1B. Meanwhile, its upstream inhibitory factor OsWRKY63 forms a negative feedback loop to finely control the activity of this pathway, ensuring a dynamic balance in the cold response (Zhang et al., 2022b). These findings highlight the significant functional differentiation of WRKY family transcription factors across different species.
This functional differentiation may represent an evolutionary strategy developed by plants to adapt to diverse growth environments and cope with various cold stress. Further research into the molecular mechanisms underlying the functional differentiation of WRKY and other transcription factors is of great significance. It can help to unravel the complex network of plant responses to cold stress and provide a theoretical basis for developing crop varieties with enhanced cold tolerance.
4.2 MiRNA responds to cold stress
MiRNAs represent a class of evolutionarily conserved, small non-coding RNA molecules that function as key post-transcriptional regulators in plants. A significant mode of action for miRNAs in cold response is through the CBF-dependent pathway (Figure 2). For instance, in Arabidopsis, miR319 targets the TCP transcription factor family, thereby modulating the expression of CBF genes and subsequently influencing the levels of COR genes (Yang et al., 2013a). Similarly, in rice, overexpression of miR319 has been shown to upregulate the expression of CBF genes. MiR319 can also target genes such as OsPCF6 and OsTCP21, contributing to enhanced cold tolerance following cold acclimation, which suggests that miR319 may partially regulate cold tolerance in rice via the CBF-dependent pathway (Yang et al., 2013b). Other miRNAs, such as miR394, positively regulate plant cold tolerance by activating the CBF-dependent pathway. Overexpression of CBF genes in miR394 transgenic plants significantly enhances their expression, further supporting the role of miR394 in cold response (Huo et al., 2022). Additionally, miR397 has been demonstrated to actively regulate cold tolerance through the CBF-dependent pathway. Overexpression of miR397a in Arabidopsis significantly reduces electrolyte leakage in leaves, thereby enhancing cold resistance. Further research revealed an enhanced expression of CBF in these transgenic plants (Huo et al., 2022). MiR156 is another important miRNA involved in cold stress response. Cold stress induces miR156 expression in Arabidopsis, which differentially regulates its SPL targets: suppressing SPL3/13 transcripts but activating SPL9. Notably, SPL9 positively modulates CBF2 expression to enhance the cold tolerance (Zhao et al., 2022). Similar observations have been made in bananas, where miR156e is induced under cold stress, and in sugarcane, where miR156 levels increase during cold treatment. In tomatoes, miR156 targets the cold-induced MYB15 gene, reducing cold tolerance. Overexpression of sly-MIR156e-3p increases sensitivity to cold stress, while silencing this miRNA through artificial microRNA interference enhances cold tolerance (Zhang et al., 2022a).
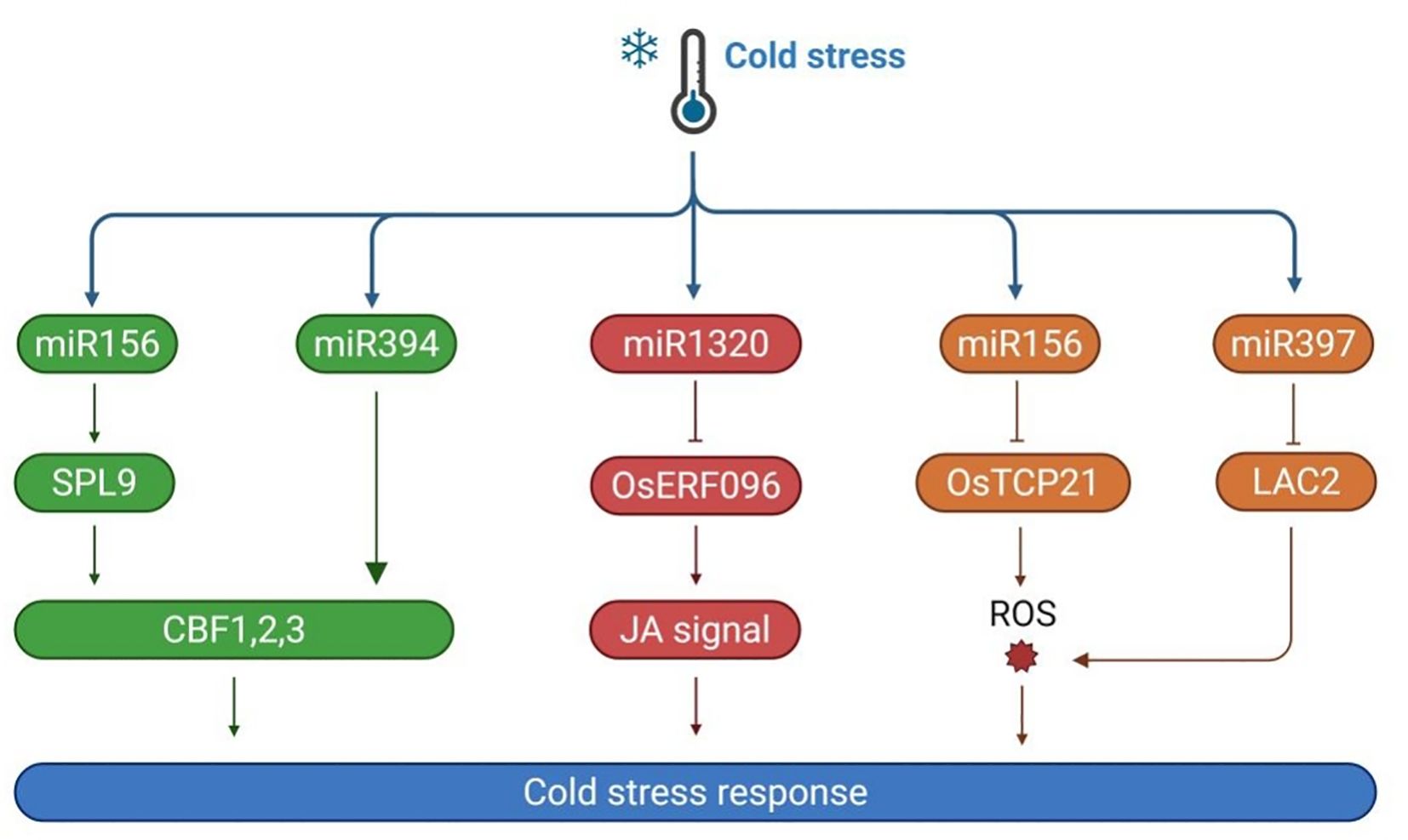
Figure 2. The mechanism of MiRNA responds to cold stress. When plants are subjected to cold stress, multiple miRNAs such as miR156, miR394, miR1320, and miR156 are activated. MiR156 regulates the expression of CBFs by targeting SPL9. Meanwhile, miR394 targets OsTCP21 and affects the production of ROS; MiR1320 targets OsERF096 and regulates the jasmonic acid (JA) signaling pathway. MiRNAs activate plant cold stress response mechanisms through multiple regulatory pathways, affecting plant cold tolerance.
In addition to the CBF-dependent pathway, miRNAs also regulate plant cold tolerance by modulating ROS levels. For instance, Bna-miR397a post-transcriptionally regulates the expression of the laccase gene BnaLAC2 in Brassica napus. This regulation enhances the adaptation to cold stress by reducing total lignin remodeling and maintaining ROS homeostasis (Hussain et al., 2025). The miR-397-LAC2 module has also been shown to enhance frost tolerance in Arabidopsis, highlighting its conserved role in cruciferous plants. MiR408 regulates cold tolerance by controlling ROS levels, with overexpression of miR408 enhancing the expression of antioxidant-related genes (Ma et al., 2015a). Furthermore, miRNAs respond to cold stress by regulating hormone signaling. In rice, miR1320 targets the ethylene-responsive factor OsERF096, normally degrading its mRNA and inhibiting its expression. Under cold stress, miR1320 expression is downregulated, leading to upregulation of OsERF096. This change inhibits jasmonic acid (JA) synthesis and the JA-mediated cold signaling pathway, making rice more sensitive to cold stress (Sun et al., 2022). Overall, miRNAs mediate plant cold response by integrating multiple pathways, including the CBF-dependent pathway, ROS regulation, and hormone signaling. These findings highlight the multifaceted roles of miRNAs in plant cold response and provide valuable insights for developing cold-resistant crops.
5 The relationship between cold stress and other factors
Beyond transcription factors and miRNAs, which are pivotal in plant cold stress responses, numerous factors including light, flowering, circadian rhythm, and hormones are also extensively implicated in plants’ intricate cold response mechanisms (Song et al., 2021).
5.1 Light signaling modulates cold stress response
Light is a vital factor influencing plant growth and stress responses. Cold stress is a prevalent abiotic stress that substantially impacts plant survival and productivity. Plants have evolved mechanisms to integrate light and cold signals (Figure 3). Key regulatory factors in plant light signaling include red light photoreceptors (PHYTOCHROMES), blue light photoreceptors (CRYPTOCHROMES), and phytochrome interacting factors (PIFs).
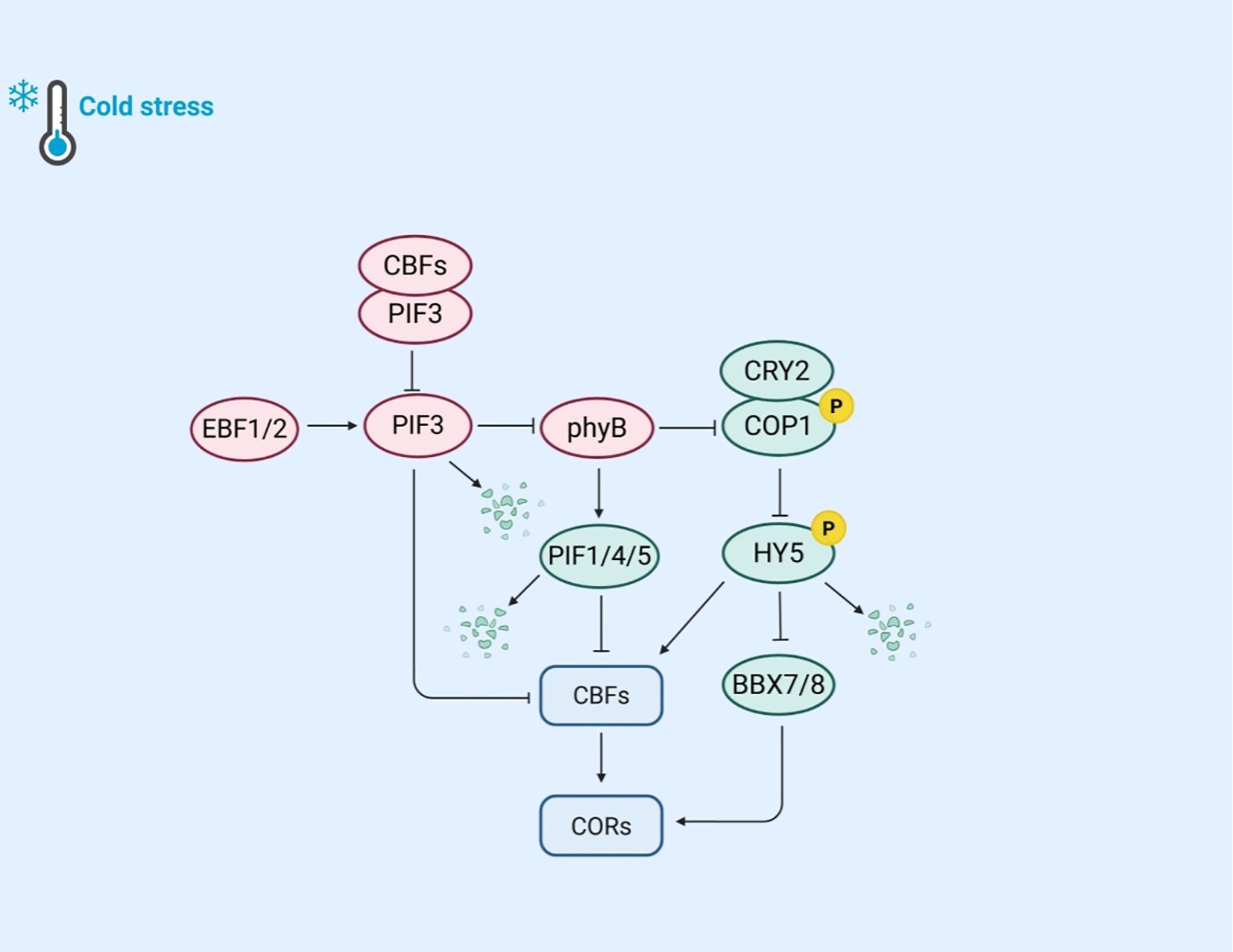
Figure 3. The mechanism of plant regulation of light and cold response gene expression under cold stress. Under cold stress, EBF1/2 ubiquitination degrades PIF3, reducing its inhibition of CBFs. When CBFs bind to PIF3, they can inhibit the co degradation pathway of PIF3 and phyB, thereby stabilizing the accumulation of phyB protein. Cold stable phyB further promotes the degradation of PIF1, PIF4, and PIF5, reducing the inhibition of CBF genes by PIFs. Meanwhile, phyB can inhibit the expression of COP1; The blue light receptor CRY2 phosphorylates COP1, effectively weakening the interaction between HY5 and COP1, thereby promoting the accumulation of HY5. HY5 responds to cold stress by directly binding to CBF promoters or by regulating the expression of COR genes through BBX7/8.
Phytochromes, including phyA and phyB, are essential red light photoreceptors that mediate plant responses to cold stress under specific light conditions. PhyB is particularly important in regulating photoperiodism and CBF expression. Under long-day conditions with red or white light, phyB mutants exhibit significantly reduced cold tolerance, whereas plants overexpressing phyB show enhanced cold tolerance. This highlights phyB’s critical role in light and cold signal transduction. The CBFs-PIFs-phyB module is vital for integrating light and cold signals in plants. Under warm light conditions, CBFs directly interact with PIF3, preventing the synergistic degradation of PIF3 and phyB proteins. When the temperature drops, PIF3 binds to the CBF gene promoter, downregulating CBF expression and acting as a negative regulator of cold tolerance. However, CBFs can inhibit the co-degradation pathway of PIF3 and phyB, stabilizing phyB protein levels. The stabilized phyB further promotes the degradation of PIF1, PIF4, and PIF5, regulates COR gene expression, enhances cold tolerance, and forms a positive feedback loop to strengthen frost resistance (Jiang et al., 2020). Similar regulatory mechanisms have also been found in other plants, such as in rice, where phyB inhibits the expression of OsPIL16 (PIF3 homologous gene), thereby suppressing the binding of OsPIL16 to the N-box region of the OsDREB1B promoter (He et al., 2016). Additionally, F-box proteins EBF1/2 are involved in this regulatory process. They target PIF3 for degradation via the 26S proteasome pathway. Previous research found that phyB interacts with EBF1/2, enhancing substrate E3 ligase interactions in a light-dependent manner to control EIN3 stability (Jiang et al., 2017).
CRY2 is a blue light photoreceptor whose degradation via the 26S proteasome pathway is activated by blue light under cold conditions. Studies have shown that the E3 ubiquitin ligase LRB interacts with CRY2, regulating its ubiquitination and stability, allowing plants to accurately respond to temperature changes (Ma et al., 2021). Other research indicates that CRY2 regulates plant cold tolerance through the COP1-HY5-BBX7/8 signaling module. Under cold stress, the blue light-induced phosphorylated form of CRY2 remains stable. The stable CRY2 competes with HY5 during light signal transduction, weakening the interaction between HY5 and COP1 and promoting HY5 accumulation (Li et al., 2021). BBX7/8, a B-BOX domain protein and a direct target of HY5, positively regulates frost resistance by controlling COR gene expression. In contrast, BBX29, another member of the BBX family, acts as a CBF-independent negative regulator of cold tolerance in Arabidopsis (Wang et al., 2023a). These findings suggest that CRY2 is jointly regulated by blue light and environmental temperature.
The PIF family serves as a pivotal regulator in light responses and is widely involved in cold adaptation, exhibiting diverse functions across different plant species. Under warm conditions, PIFs typically interact with phytochromes to modulate light-mediated signal transduction. However, under cold stress, PIFs adopt distinct roles. For instance, in rice, PIF family members such as OsPIL16 regulate the cold tolerance and grain shape (He et al., 2016). In maize, ZmPIF6 improves cold tolerance by reducing ROS content and enhancing cell membrane stability while increasing grain size (length and width) and thousand-grain weight (Li et al., 2024a). In tomatoes, the SlPIF4-silenced strain not only reduces fruit weight and yield, but also increases carotenoid content and accelerates ripening (Rosado et al., 2019).
5.2 Cold adaptation strategies in flowering regulation network
More and more evidence suggests that the significance of the flowering process in plants’ response to cold stress. Winter frost, a type of cold stress, can inflict fatal damage on plant flowers and developing seeds. To mitigate this risk, most plants have evolved mechanisms to avoid flowering during winter. These mechanisms involve sensing and responding to cold signals, regulating the expression of flowering-related genes, and delaying flowering until after an extended cold period (Figure 4).
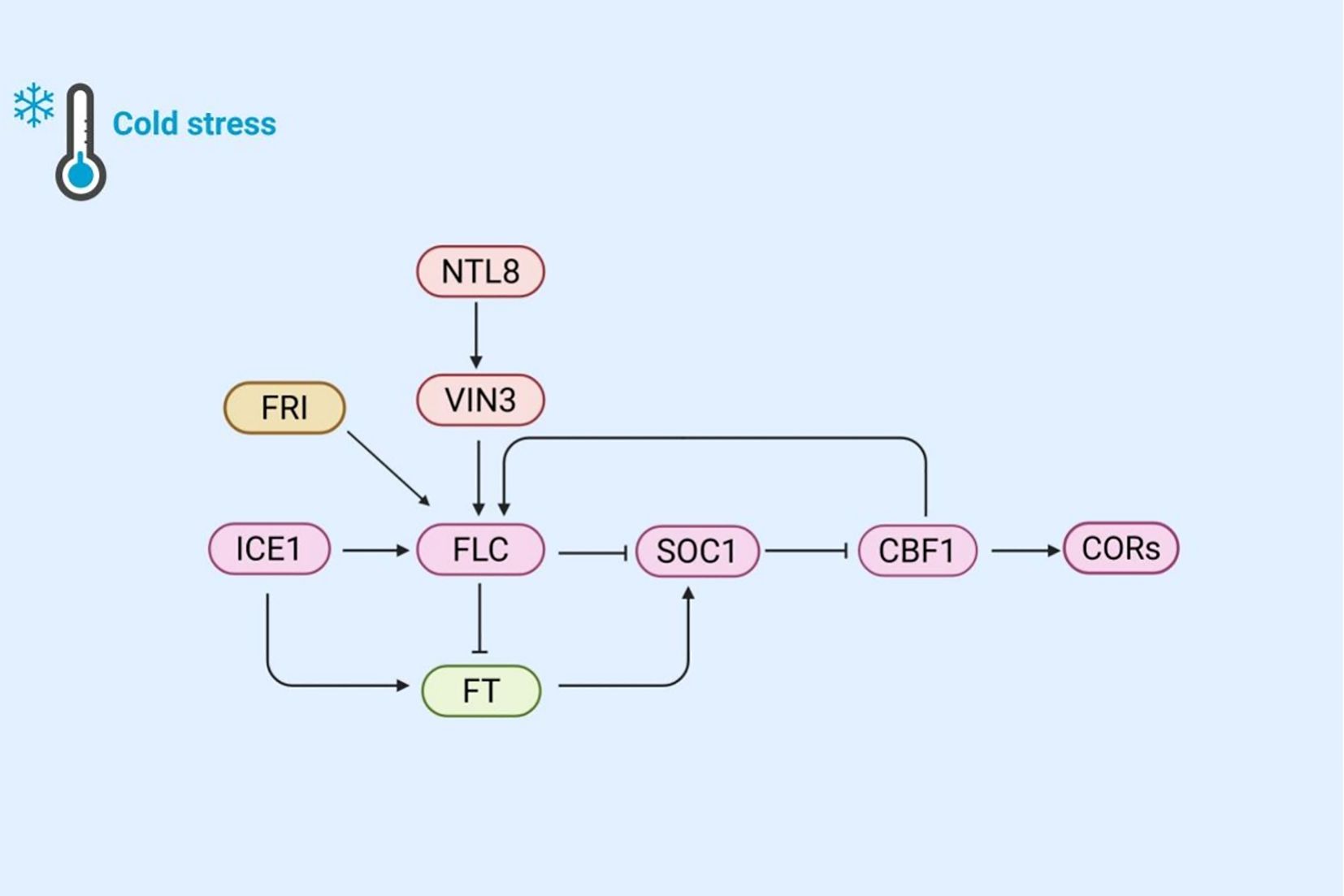
Figure 4. The mechanism of plant regulation of flowering time and cold response gene expression under cold stress. Cold stress activates NTL8, which in turn activates VIN3, participates in vernalization, and affects FLC expression. FRI inhibits FLC expression through VIN3, while ICE1 directly activates FLC. FLC inhibits SOC1 and delays flowering; SOC1 activates CBF1, promotes CORs expression to enhance cold tolerance. Meanwhile, ICE1 and SOC1 promote flowering through FT. This network coordinates the cold response and flowering time of plants.
Under warm conditions, rapid plant growth and frequent cell division lead to the dilution of NTL8. However, under cold stress, growth retardation reduces this dilution effect, resulting in increased accumulation of NTL8 during cold exposure and the activation of VIN3 expression (Zhao et al., 2020). The VIN3 gene encodes a protein that inhibits the flowering inhibitory factor (FLC), thereby indirectly promoting flowering. When FLC expression is downregulated, plants can bloom under warm conditions. FLC acts as an upstream negative regulator of the flowering time component SOC1, which is a key flower activation factor that integrates multiple flowering induction pathways and is required for downregulating CBF genes. Studies have shown that in soc1–2 knockout mutant, the expression of CBF genes is upregulated, whereas in SOC1 overexpression line, their expression is downregulated (Seo et al., 2009). This indicates that SOC1 negatively regulates the expression of cold responsive genes. Cold-induced overexpression of CBFs can lead to late flowering by increasing FLC expression. Thus, a feedback loop exists between cold adaptation and flowering time regulation. When cold exposure is brief (such as in autumn or early spring), flowering is delayed due to increased FLC expression. However, when cold induction genes are activated by SOC1 inhibition, the cold response is suppressed. Additionally, cold-activated ICE1 directly induces FLC expression, which in turn inhibits SOC1 expression and delays flowering (Lee et al., 2015). The upregulation of the flowering locus t (FT) gene in the ice1 mutant has garnered attention. FT is a key gene in the flowering pathway, and its product, the FT protein, can induce SOC1 expression (Yoo et al., 2005). Under normal conditions, FLC inhibits FT gene expression, thereby indirectly suppressing SOC1 expression and maintaining the balance of flowering time regulation. The upregulation of the FT gene may be related to the weakened SOC1-mediated cold response. Increased FT expression enhances SOC1 expression, which in turn negatively regulates COR genes. Consequently, the upregulation of the FT gene may further reduce SOC1’s inhibitory effect on cold-responsive genes, thereby affecting cold tolerance under cold stress. It is intriguing to explore whether FT is associated with the weakened cold response mediated by SOC1 and how ICE1 regulates FT gene expression.
Recent studies have shown that the frigida (FRI) gene regulates FLC expression at low temperatures. Cold stress can rapidly promote the formation of FRI nuclear condensates, which is associated with decreased FRI occupancy in the FLC promoter region and subsequent FLC inhibition (Zhu et al., 2021). This discovery reveals a potential new pathway under cold stress: the FRI-FLC-SOC1 pathway, which affects plant cold resistance. Through this pathway, plants can better balance flowering time and cold response under cold stress to adapt to environmental changes.
5.3 Circadian rhythm orchestrates cold stress response
The plant circadian rhythm system is composed of key genes, including circadian clock associated 1 (CCA1), late elongated hypocotyl (LHY), lux arrhythmo (LUX), and pseudo response regulator factors (PRRs). These genes act as a positive regulator of cold response (Figure 5). The promoter regions of genes induced by cold stress often contain EE (evening element) and EEL elements. EE/EEL serves as the binding site for CCA1/LHY, a core component of the circadian rhythm, and its highly similar RVE protein in the target gene promoter region. CCA1 and LHY are inhibited by PRRs in the morning and bind to CBF promoters, actively regulating cold tolerance. For example, the tea tree homolog of LHY, CsLHY, also positively affects cold stress. Research has shown that under cold stress, the expression level of CBFs increases in the prr9/7/5 mutant, and PRR5 inhibits the expression of CCA1 and LHY on cold nights (Nakamichi et al., 2009).
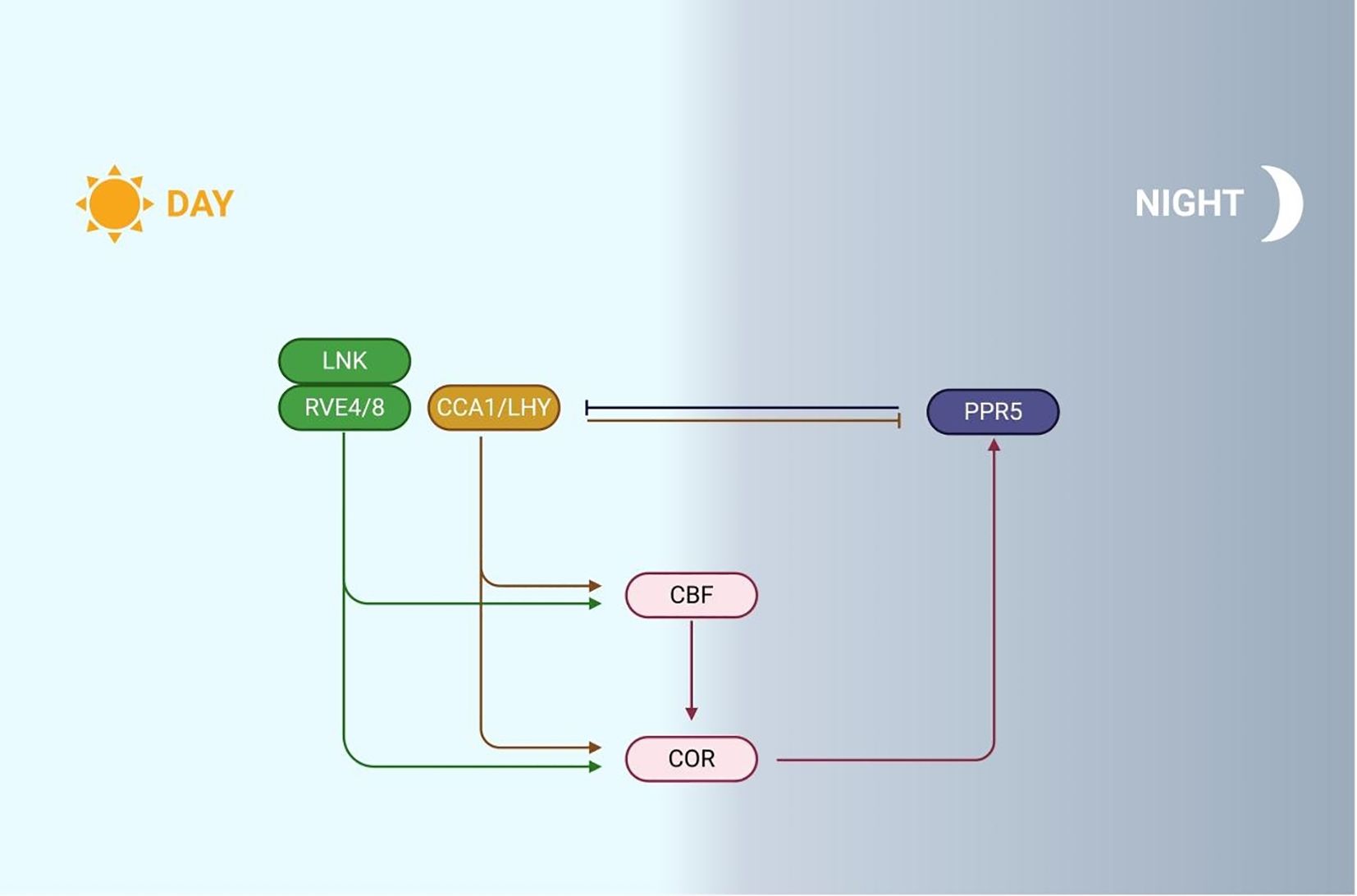
Figure 5. Mechanism of plant responds to cold stress through circadian rhythm. During the day, LNK works synergistically with RVE4/8 to activate CBF expression. In addition, high expression of CCA1/LHY inhibited the expression of PRRs, and as the day progressed, PRRs gradually accumulated and began to inhibit the expression of CCA1/LHY at night. At the moment, CCA1/LHY directly binds to CBF promoters, promoting COR genes expression and enhancing plant cold tolerance.
Under cold conditions, nuclear accumulation of CCA1 homologs RVE4/8 activates CBF3 expression through EE element binding in its promoter (Kidokoro et al., 2021). RVE8-LNK1/2 interaction coordinately regulates circadian oscillators and COR genes (Sorkin et al., 2023). Independent of CBF signaling, CCA1 directly targets COR27/28 promoters, suppressing cold-induced COR27 expression while these genes maintain circadian rhythms by repressing LUX/PRR5/TOC1, consequently reducing Arabidopsis freezing tolerance (Wang et al., 2017; Mikkelsen and Thomashow, 2009). CBF1 inhibits its transcription by directly binding to the LUX promoter, while in the tea tree, CsCBF1-mediated CsLUX activation creates a regulatory node connecting CBF and JA pathways. CsLUX both responds to CsCBF1 and directly modulates JA biosynthesis/signaling by binding CsLOX2 (JA synthesis enzyme) and CsJAZ1 promoters, forming a dynamic cold-response network (Wang et al., 2024b).
In addition, cold regulation of alternative splicing of CCA1 and LHY precursor mRNA. Under cold conditions, the splicing form of CCA1 α increases, while the splicing form of CCA1 β decreases. Overexpression of CBFs in CCA1 α materials increases the expression level and enhances plant frost tolerance. CCA1 β may inhibit its regulation of CBFs by interacting with CCA1 α, negatively regulating plant frost tolerance (Park et al., 2012). A recent study has shown that CCA1 and LHY respond to temperature drops and participate in activating the transcription of flowering regulatory factor VIN3 to cope with long-term cold stress. A vernalization reaction element composed of G-box and EE motifs was discovered in the VIN3 promoter, which can be recognized by transcription factors CCA1 and LHY, leading to the accumulation of VIN3 during cold stress conditions (Kyung et al., 2022). The results indicate that plants may have integrated their circadian rhythms and flowering components to effectively cope with cold stress. These studies indicate that circadian rhythms are closely related to cold stress. However, further research is warranted on how plants precisely regulate their circadian rhythms under cold stress, and whether there are specific time periods that make plants more sensitive to the circadian rhythm’s response to cold stress.
5.4 Hormonal networks coordinate cold stress response
Under cold stress, plant hormones interact to form a complex signaling pathway network that jointly regulates cold response signals. Among these hormones, the CBF gene acts as a vital regulator in hormone crosstalk during the cold stress response. Its expression is regulated by several major plant hormones, including abscisic acid (ABA), brassinosteroid (BR), ethylene (ETH), gibberellin (GA), jasmonic acid (JA). Under cold conditions, the hormonal balance in plants undergoes significant changes (Figure 6). The levels of inhibitory hormones, such as ABA, increase, while the levels of growth-promoting hormones, such as IAA and GA, decrease. Plants adapt to cold stress by regulating the proportions of these hormones.
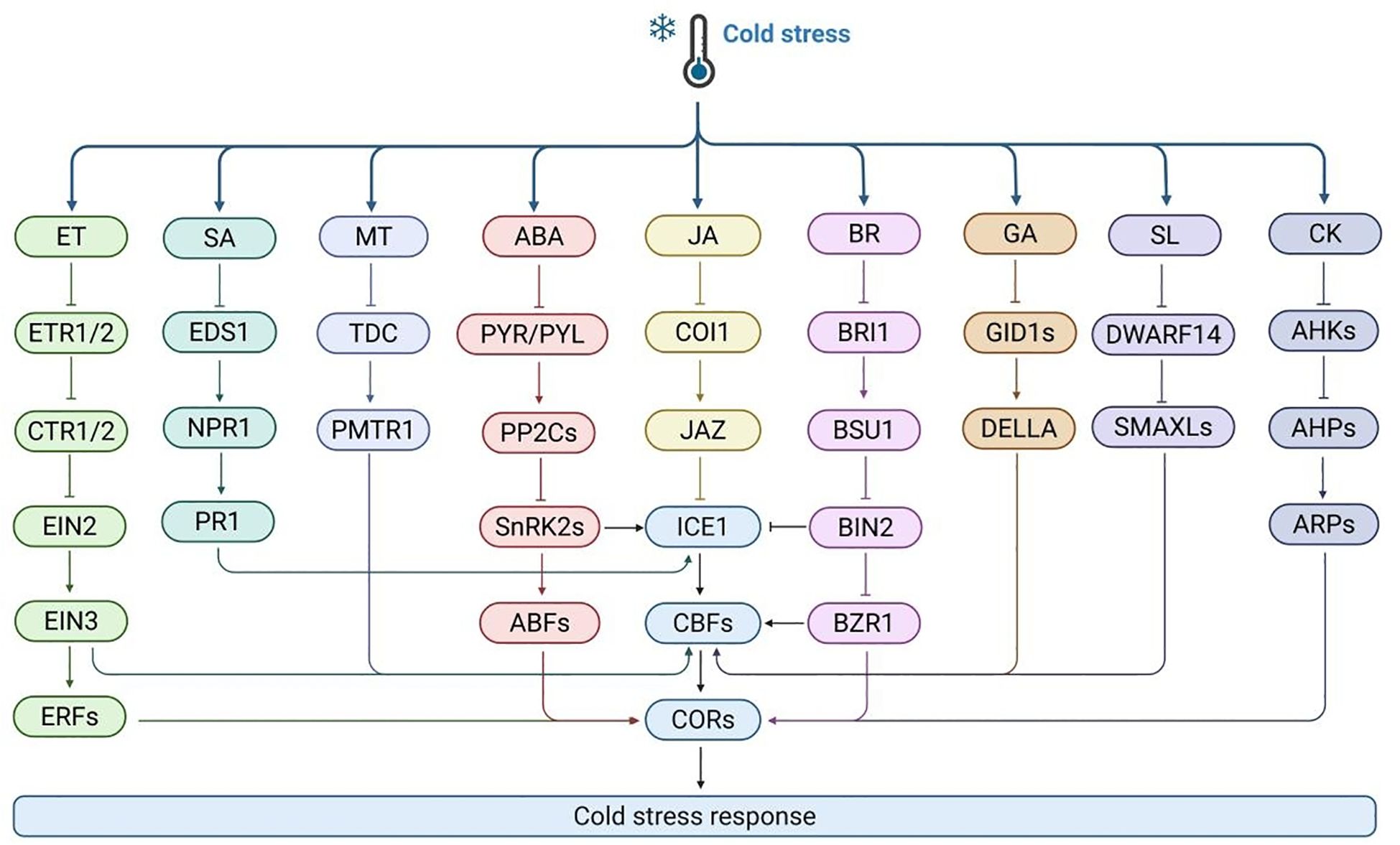
Figure 6. The mechanism of plant respond to cold stress through various hormone signaling pathways. Cold stress activates multiple hormone pathways, including ethylene (ET), salicylic acid (SA), methionine (MT), abscisic acid (ABA), jasmonic acid (JA), brassinosteroid (BR), gibberellin (GA), lactone (SL), and cytokinin (CK). Each pathway perceives signals through its specific receptors (such as ETR1/2, EDS1, PMTR1, PYR/PYL, COI1, BRI1, GID1s, DWARF14, AHKs) and transmits them through intermediate components (such as CTR1/2, NPR1, PP2Cs, SnRK2s, JAZ, BSU1, DELLA, SMAXLs, AHPs), ultimately converging to transcription factors (such as ICE1, CBFs) to activate CORs expression and affect plant cold tolerance.
ABA is an endogenous hormone in plants that is widely involved in plant growth, development, and stress response. After cold treatment, the application of exogenous ABA can usually enhance the cold tolerance. the ABA synthesis-deficient mutant aba3 in Arabidopsis exhibited reduced cold tolerance under cold stress, but this deficiency could be compensated for by exogenous ABA treatment (Xiong et al., 2001). The ABA signaling pathway comprises four core components: PYR/PYL/RCAR receptor proteins, PP2C phosphatases, SnRK2 kinases, and ABF/AREB transcription factors. Upon binding to ABA, PYR/PYL/RCAR proteins inhibit the activity of PP2C phosphatases, thereby activating the SnRK2 kinase cascade. SnRK2 kinases, such as OST1, can directly promote the expression of ICE1, facilitate the transcription of CBF genes, and ultimately enhance plant cold tolerance (Ding et al., 2015). In addition, ABA regulates cold stress response genes through the TOC1-MYB44 module in the CBF-independent pathway, balancing energy allocation and stress adaptation (Du et al., 2024). ABA is also widely involved in other plant responses to cold stress. For example, in rice, ABA induces OsDREB1B to enhance cold tolerance by regulating proline synthesis. Overexpression of ABA receptor PYL10 increases the cold tolerance (Verma et al., 2019). These studies indicate that ABA signaling acts as a crucial regulator in response to cold stress.
BR is a plant specific steroid hormone. After cold treatment, exogenous BR application enhances the expression of CBF genes in Arabidopsis, indicating that BRs improve frost resistance. BRs bind to the receptor kinase BRI1 and initiate intracellular phosphorylation cascades. In this process, the negative regulator BIN2 undergoes dephosphorylation, activating the transcription factors BES1 and BZR1. Activated BES1/BZR1 regulate many BR response genes. BRI1, after activation, detaches from the BR signal suppressor BKI1 and transmits signals to BSK1 via phosphorylation. BSK1 activates the phosphatase BSU1, which phosphorylates BIN2, inactivating it. This enhances the activity and stability of BZR1 and BES1, enabling them to directly regulate downstream BR gene transcription. Studies show that BR signal-defective mutants are highly freezing-sensitive, with or without cold acclimation. BIN2, a key negative BR signaling factor, undergoes autophosphorylation at low temperatures, inhibiting its binding to the CBF promoter and negatively regulating cold stress (Ye et al., 2019). BIN2 overexpression reduces frost resistance, while the bin2-3bil1-bil2 triple mutant increases it. Unlike BIN2, BES1 and BZR1 positively regulate cold responses (Kim et al., 2024). BZR1, a key BR signaling transcription factor, regulates CBF1/2 expression by binding to the E-box site in the promoter, actively regulating cold tolerance (Li et al., 2017b). BIN2 can phosphorylate ICE1, enhancing its interaction with HOS1 and destabilizing ICE1. Similar mechanisms exist in other plants. For example, in rice, OsGSK2, the BIN2 homolog, inhibits OsbHLH002 (an ICE1 homolog) transcriptional activity via phosphorylation, reducing OsCBF3 expression. However, it remains unclear how cold stress directly activate BIN2. CESTA and BZR1, regulated by BR, are BIN2 targets that bind to the CBF promoter to regulate constitutive CBF expression. OsBRI1, a key BR signaling receptor, causes cold sensitivity when dysfunctional.
Cytokinin is an important hormone in plants, whose transduction operates through a two-component system involving receptor histidine kinase (HK), histidine phosphotransferase (HP), and response regulator (RR). Cold stress can promote an increase in cytokinin levels in Arabidopsis roots, thereby activating response RR, which can induce the expression of SHY2 gene, thereby inhibiting the biosynthesis of auxin and the transport of auxin through PIN1, PIN3, and PIN7. This series of changes will lead to a decrease in auxin levels, causing abnormal root growth. In Arabidopsis, ARR1/12 is a key response regulator in the cytokinin signaling pathway. Research has found that ARR1/12 is involved in cold stress mediated root growth inhibition. Specifically, the root length and number of meristematic cells of arr1–3 and arr12–1 mutant seedlings are sensitive to cold stress (Zhu et al., 2015). In addition, the decrease in PIN1/3 transcript levels and auxin levels in the roots of arr1–3 arr12–1 mutant was significantly less. In addition, phosphorylated ARR1 can interact with HPY2, enhance acetylation modification of histone H3, and promote transcriptional activation of ARR1. In maize, under cold stress, the accumulation of ZmRR1 protein significantly increases and induces the expression of ZmDREB1s and ZmCesA genes, thereby enhancing the cold tolerance of maize (Zeng et al., 2021). Similarly, in rice, OsRR6 can actively respond to cold stress. In plants overexpressing OsRR6, the expression of DREB1A/CBF3, COR15A, KIN1, and RD29A was significantly upregulated, indicating that OsRR6 plays as a vital regulator in cold response (Bhaskar et al., 2021).
Ethylene signaling receptors include ETR1, ETR2, ERS1, ERS2, and EIN4. In Arabidopsis, ethylene typically exhibits negative regulatory effects under cold stress. When ethylene is present, receptors bind to ethylene to inhibit CTR1 kinase activity, activate downstream signal transduction, lead to the accumulation of EIN3 transcription factors, activate ERF expression, regulate downstream gene transcription, and enable plants to respond to ethylene signals. In Arabidopsis, overexpressed plants of ethylene signaling core factor EIN3 exhibit a phenotype of reduced frost resistance. Further research has shown that EIN3 can bind to the CBF promoter and inhibit its expression. In addition, the E3 ligase EBF1/EBF2 in the ethylene signaling pathway negatively regulates plant frost resistance by degrading its target protein EIN3 transcription factor. In rice, EIN family member OsEIN2 is a negative regulator of cold stress. Overexpressed plants of OsEIN2 exhibit severe stress symptoms at low temperatures, with excessive accumulation of ROS, while ein2 mutant plants show enhanced cold tolerance. Further research has shown that OsEIL1 and OsEIL2 can form heterodimers and synergistically inhibit the expression of OsICE1 by binding to their promoters. OsEIN2 and OsEIL1/2 activated by OsICE1 downregulated the expression of OsICE1 target genes, ROS related, and photosynthesis related genes (Zhai et al., 2024). In addition, overexpression of ethylene related transcription factor VaERF057 can also enhance the cold tolerance. This indicates that the role of ethylene in cold tolerance varies among different plants.
GA is a plant hormone whose signal transduction is centrally regulated by the GRAS protein DELLAs, and participates in response through the GA-GID1-DELLA module during cold stress. In Arabidopsis, overexpression of CBF1 enhances the accumulation of DELLAs by regulating the transcription of RGL3 (Lantzouni et al., 2020). The absence of GA is consistent with the improvement of frost resistance. DELLAs knockout lines gai-t6 and rga-24 are sensitive to freezing. Further research has shown that the growth retardation under cold stress is achieved by CBF regulating DELLAs proteins located in the nucleus. The promoting effect of CBF on growth depends on the degradation of DELLAs stimulated by GA. DELLAs protein is a repressor of the GA signaling pathway. GA induces the degradation of DELLAs repressor protein, thereby controlling many key developmental processes and responses to stress such as cold. DELLAs proteins modulate plant cold stress responses through interactions with multiple transcription factors. The interaction between DELLAs and PIF4 can regulate CBF transcription. In tomato, SlPIF4 not only actively regulates cold tolerance by directly activating the SlDELLA gene S1GAI4, but also activates the gene expression of SlCBF1 by directly binding to the SlCBF1 promoter. The crosstalk between PIF4 and DELLAs can regulate CBFs transcription and hormone homeostasis in tomato cold response. SlPIF4 not only directly binds to the promoter of SlCBF1 gene and activates its expression, but also regulates the biosynthesis and signal transduction of plant hormones including abscisic acid, jasmonic acid, and gibberellin to cope with cold stress. In addition, S1PIF4 directly activates the SlDELLA gene S1GAI4 under cold stress, while S1GAI4 positively regulates cold tolerance (Wang et al., 2020).
Melatonin is a widely present active molecule in plants, which helps maintain redox homeostasis by inducing the expression of antioxidant enzyme genes. Exogenous application of melatonin has been shown to enhance the activity of various key antioxidant enzymes. In Arabidopsis, MT can upregulate the expression of CBFs, COR15a and ROS related antioxidant genes, thereby actively regulating plant cold tolerance (Tang et al., 2022). After the melatonin receptor gene CAND2/PMTR1 binds to melanin on the plant cell membrane, it triggers a series of signaling pathways, such as the cAMP pathway and Ca2+ signal, to regulate the cold tolerance (Wang et al., 2022; Aghdam and Arnao, 2024). These findings indicate that melatonin can affect plant physiological processes through receptor-mediated signaling pathways. However, research on the molecular mechanisms of melatonin related genes in response to cold stress is limited. Future research on the molecular mechanisms of melatonin in response to cold stress will help us better understand the regulatory mechanisms of melatonin in response to cold stress.
JA is a lipid-derived plant hormone whose levels rise under cold stress. Applying exogenous JA can boost cold tolerance in Arabidopsis by increasing CBF expression, while impaired JA biosynthesis can make plants more susceptible to freezing. These changes in cold tolerance are mediated by JASMONATE ZIM-DOMAIN (JAZ) proteins, specifically JAZ1 and JAZ4, which act as inhibitors of JA signaling, interacting with ICE1/2 to suppress the transcriptional activity of ICE1 and the expression of the CBF1–3 genes (Hu et al., 2013). Additionally, JA-mediated cold-induced growth inhibition is achieved by stabilizing DELLAs proteins, which interact with transcription factors of growth regulatory factors (GRFs) to inhibit their activity. The apple orthologs MdJAZ1/2 exhibit analogous functions by disrupting the MdABI4-MdICE1 interaction. Notably, MdABI4 acts as a positive regulator of MdICE1’s transactivation capacity, with MdJAZ1/2-mediated inhibition consequently attenuating cold tolerance (An et al., 2022).
The SA signaling pathway is essential for cold stress adaptation in plants, where receptor complexes NPR-TGA constitute the central regulatory node. In Arabidopsis, SA signaling core receptor NPR1 can sensitively sense cold signals, leading to conformational changes, transitioning from an oligomeric state to a monomeric form and transferring to the nucleus. NPR1 precisely interacts with the transcription factor HSFA1, which can activate COR genes expression, significantly enhancing the cold adaptation ability and enabling them to survive and grow under cold conditions (Olate et al., 2018). At the same time, NPR1 can bind to the core transcription factor ICE1 and key transcription factor TGA3 involved in the cold signaling cascade to form a functional protein complex. This complex can directly bind to a specific region of the PR1 promoter downstream of SA, thereby efficiently activating the transcription of the PR1 gene. Research has shown that the binding of NPR1 and TGA3 can significantly enhance the transcriptional activation of PR1 by ICE1. This means that under cold conditions, plants can strengthen their immune response in this way, effectively resisting pathogen invasion, which is of great significance for the survival of plants in harsh environments. Similar functional mechanisms have also been found in citrus. CtrNPR3 can interact with CtrTGA2, inhibiting its function and weakening its activation of target genes, thereby exerting a negative regulatory effect on the cold resistance. Additionally, CtrTGA2 can specifically bind to the CtrP5CS1 promoter and activate the expression. The increased expression of the CtrP5CS1 gene promotes the synthesis and accumulation of proline, which can help to improve their cold tolerance. CtrTGA2 directly regulates the expression of the gene CtrICS1 involved in SA biosynthesis, thereby constructing a positive feedback loop (Xiao et al., 2024). In this circuit, the accumulation of SA can further enhance the transcriptional activation of CtrP5CS1 mediated by CtrTGA2, forming a self-enhancing regulatory mechanism and further improving the cold resistance of plants. Interestingly, external application of SA can partially alleviate this inhibitory effect, reactivate the function of CtrTGA2, and enhance the cold resistance of plants. This discovery provides important theoretical basis for regulating plant cold tolerance through exogenous SA.
SL is a sesquiterpene plant hormone that is recognized by D14 (α/β hydrolase) receptors (Umehara et al., 2008). Upon recognition, the D14-SCF-MAX2 complex is formed, targeting the degradation of D53/SMXLs (Waters et al., 2017). The degradation of SMXLs releases the inhibition of downstream CBFs, thereby enhancing plant frost resistance. In Arabidopsis, cold stress induce the expression of MAXs genes and increase endogenous SL levels. SL inhibits the expression of the WRKY41 gene and promotes the interaction between MAX2 and WRKY41. This interaction mediates the degradation of WRKY41 through the 26S proteasome pathway, alleviating the inhibition of WRKY41 on its target gene CBFs. As a result, the expression of CBFs and their downstream genes enhances the cold tolerance (Wang et al., 2023b).
Of course, there are numerous interactions between hormones and other regulatory factors that collectively respond to cold stress. BIN2 can form a complex with JA signal inhibitor JAZ1 to jointly inhibit ICE1 activity. Exogenous JA treatment can alleviate the cold sensitive phenotype of BIN2 overexpressing strains. Tomato SlWRKY50 promotes cold tolerance by positively regulating the jasmonic acid biosynthesis pathway, and its expression is directly activated by the JA signaling core transcription factor SlMYC2 (Wang et al., 2024a); And OsWRKY53 specifically inhibits the cold tolerance of anthers during the booting stage by antagonizing gibberellin (GA) metabolism. Multiple hormone signaling mediators can specifically attach to the CBF promoter region or interact with important regulatory factors of CBF expression, thereby promoting plant development during cold acclimation (Tang et al., 2022). However, the complex molecular mechanisms of hormone mediated signaling and its regulation of CBF in cold stress response still need to be elucidated.
6 Post-translational regulation in response to cold stress
In addition to transcriptional and post-transcriptional regulation, an increasing body of evidence highlights the significance of post-translational modifications in the cold response, which primarily encompass protein phosphorylation, ubiquitination, and acetylation (Kidokoro et al., 2022).
Phosphorylation represents a ubiquitous post-translational modification mechanism essential for signal transduction amplification. OST1, a member of the SNF1-related protein kinase family, is activated under cold stress and regulated by the protein phosphatase CLADE E GROWTH-REGULATING 2 (EGR2). OST1 interacts with the protein phosphatase PP2CG1, thereby inhibiting its activity and negatively regulating cold tolerance (Lv et al., 2021). Moreover, OST1 phosphorylates ICE1 to enhance its stability, thus positively responding to cold stress. OST1 also mediates the interaction between HOS1 and ICE1, which effectively prevents the degradation of ICE1 by HOS1 (Ding et al., 2015). Additionally, OST1 phosphorylates the U-box E3 ligases PUB25 and PUB26, thereby enhancing their E3 activity. This leads to the polyubiquitination and degradation of MYB15, which in turn boosts CBF expression under cold stress (Wang et al., 2019b). OST1 can also interact with a newly formed peptide-associated complex (NAC) protein BTF3s and phosphorylate it, promoting its interaction with CBF proteins to enhance frost resistance (Ding et al., 2018). Interestingly, OST1 can interact with AtANN1 and phosphorylate it, thereby augmenting its Ca2+ transport activity and enhancing Ca2+ signaling (Liu et al., 2021a). These findings demonstrate that OST1 responds to cold stress via multiple pathways. Recent research indicates that OST1 mediates acetylation in response to cold stress (Figure 7). Specifically, OST1 phosphorylates the histone acetyltransferase HAT1, promoting its interaction with HOS1. This interaction induces the ubiquitination and degradation of HAT1, thereby alleviating the inhibition of CBF genes by HAT1 and activating the cold stress response (Kang et al., 2025).
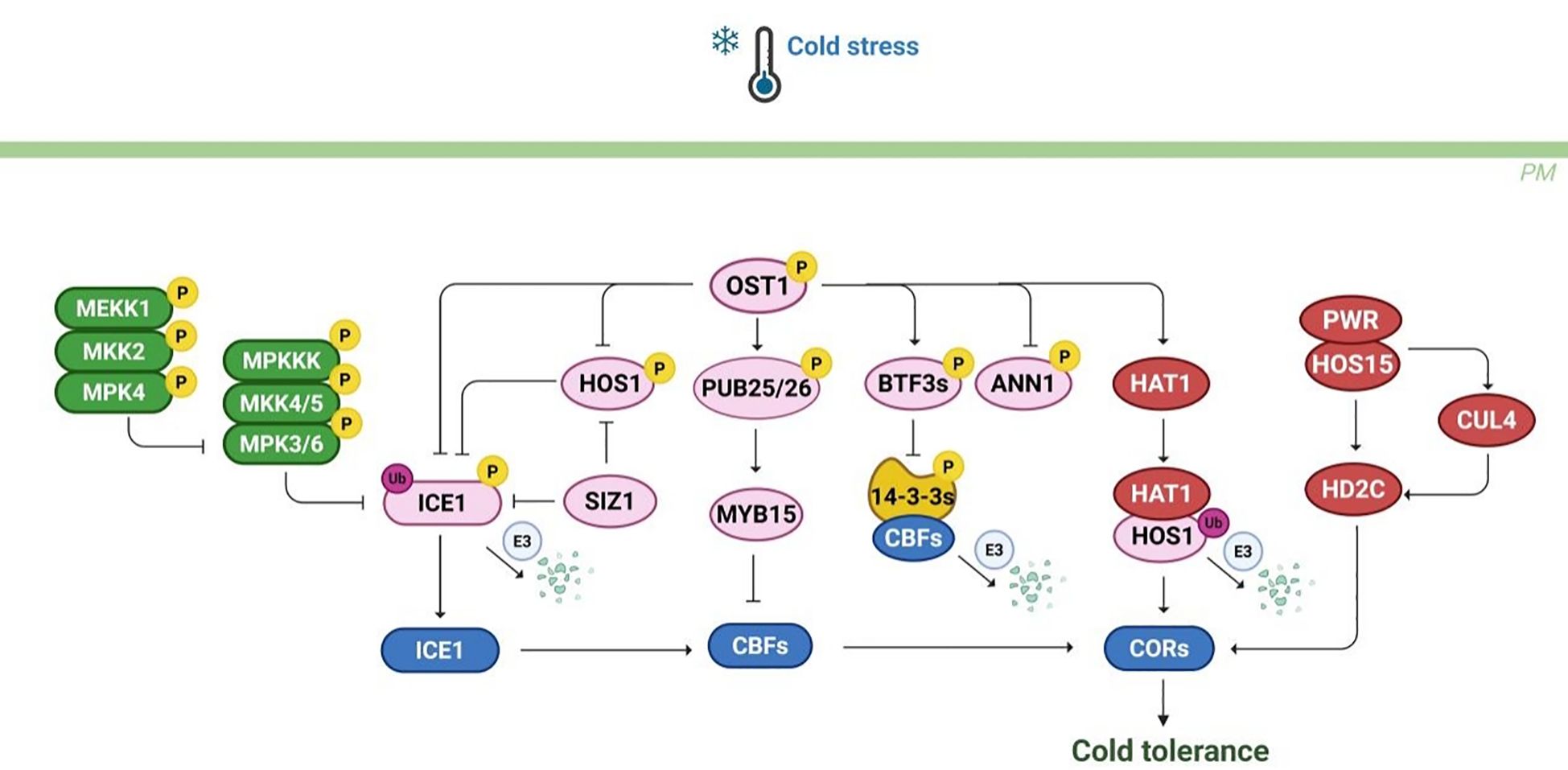
Figure 7. The post-translational modification regulatory network under cold stress. Cold stress activates the MEKK1-MKK2-MPKK4/5-MPK3/6 signaling cascade, phosphorylates ICE1 and promotes its ubiquitination degradation, negatively regulating plant cold tolerance. OST1 responds to cold stress by phosphorylating various regulatory factors such as PUB25/26, BTF3s, ANN1, HAT1. in addition, Regulatory factors such as PWR, HOS15, CUL4, and HD2C regulate CBF and COR genes through ubiquitination or acetylation modification. In summary, the integration of multiple post-translational modification pathways such as phosphorylation, ubiquitination, and acetylation in plants affects the cold tolerance.
The MAPK cascade also transduces cold stress signals in plants via sequential phosphorylation events. This cascade comprises three core kinase families: MAP3K, MAP2K, and MAPK. In Arabidopsis, two well-characterized MAPK cascades are the MEKK1-MKK1/2-MPK4 and MKK4/5-MPK3/6 pathways. Under cold stress conditions, MEKK1 kinase activity is upregulated, which leads to the phosphorylation of MKK2 and subsequently activates MPK4 and MPK6. Notably, the MEKK1-MKK2-MPK4 pathway positively regulates cold tolerance by inhibiting MPK3/MPK6 activity. In contrast, MPK3/6 negatively regulate cold responses by phosphorylating ICE1, thereby reducing its stability (Li et al., 2017a). Additionally, MPK6 phosphorylates MYB15, which negatively regulates CBF3 gene expression (Kim et al., 2017). MAPK signaling is also implicated in cold responses across various plant species. In tomato, SlMPK1 and SlMPK2 enhance cold tolerance by phosphorylating the transcription factor SlBBX17. In rice, OsMAPK3 improves cold resistance by phosphorylating OsICE1, which inhibits its ubiquitination and activates the expression of OsTPP1 (Zhang et al., 2017). Furthermore, the kinase OsCTK1 phosphorylates OsMPK1, which in turn dephosphorylates MAPK3/6, thereby positively regulating cold tolerance (Wu et al., 2024). Recent studies have identified the phosphatase OsPP2C27 negatively regulates cold tolerance by directly dephosphorylating OsMAPK3 and OsbHLH002 (Xia et al., 2021). In summary, the MAPK signaling cascade is critically involved in mediating plant cold response. However, further investigation is required to fully understand species-specific variations in MAPK-mediated cold signaling. Gaining deeper insights into these regulatory pathways will be essential for developing crop varieties with improved cold tolerance.
Ubiquitination is another important post-translational modification involved in cold stress. In Arabidopsis, E3 ubiquitin ligase HOS1 degrades ICE1 through ubiquitination, thereby reducing its stability at low temperatures (Dong et al., 2006). SIZ1 inhibits the 26S proteasome degradation of ICE1 protein by SUMOylation, reducing HOS1 mediated polyubiquitination to improve stability (Miura et al., 2007). Similarly, in apple, the interaction between MdSIZ1 and MdMYB2 significantly enhances plant cold tolerance. Further research suggests that inducing MdMYB2 at low temperatures can activate MdSIZ1 expression, thereby promoting the synthesis of anthocyanin biosynthesis (Jiang et al., 2022). However, the substrates regulated by ubiquitinases under cold stress are currently limited, and further exploration is needed for the identification of targets through ubiquitination modification. In addition, E3 ubiquitinase is a key factor in ubiquitination modification, but the regulatory mechanism of E3 ubiquitinase under cold stress is not fully understood.
Substantial evidence indicates that histone acetylation modifications directly correlate with cold tolerance capacity across plant species. This reversible modification system operates via the counterbalancing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs), which precisely tune acetylation homeostasis to regulate transcriptional reprogramming during cold stress. These enzymes modulate gene expression by interacting with transcription factors involved in cold signaling. Generally, histone acetylation promotes gene transcription, whereas deacetylation leads to transcriptional repression. In Arabidopsis, the histone deacetylase HD2C is closely linked to the CBF-dependent pathway. Under normal conditions, HD2C collaborates with HOS15 to suppress the expression of COR genes by deacetylating their promoters. However, under cold stress, the PWR-HOS15 complex facilitates the degradation of HD2C, enabling the recruitment of CBF to the COR gene promoters and promoting their expression (Lim et al., 2020). Additionally, HOS15 recruits the CUL4 ubiquitin ligase to degrade HD2C, thereby increasing H3 acetylation at COR promoters and enhancing cold tolerance. Similarly, in rice, OsHDA716 interacts with OsbZIP46, which leads to the deacetylation of the OsbZIP46 DNA-binding domain, reducing its binding affinity and transcriptional activity. Consequently, the stability of OsbZIP46 is compromised, resulting in decreased cold tolerance (Sun et al., 2024).
Beyond the CBF-dependent pathway, histone acetylation is also involved in CBF-independent cold stress responses. For instance, in Arabidopsis, the interaction between RHOMBOID-like protease 11 (RBL11) and fatty acid export protein 1 (FAX1) promotes the ubiquitination and degradation of FAX1, which is vital for cold adaptation (John et al., 2024). Moreover, the transcription factor ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) is regulated through phosphorylation and SUMOylation, which influence its activity and promote H3 acetylation, thereby modulating cold-responsive gene expression. In apple, the histone deacetylase MdHDA6 negatively regulates cold tolerance by deacetylating and repressing the expression of MdTCP15, a negative regulator of cold stress. MdTCP15 directly binds to the promoter of MdCOR47 and reduce cold tolerance (Guo et al., 2023). Collectively, these findings highlight the significant role of histone acetylation in plant responses to cold stress. However, the number of identified genes regulated by histone acetylation under cold conditions remains limited, and the underlying regulatory mechanisms require further investigation.
7 Balancing cold stress and multiple traits
When plants cope with cold stress, they need to finely balance and regulate cold tolerance with other traits. This trade-off mainly relies on two types of genes: “traditional” trade-off genes and “ideal” trade-off genes. The former is prone to negative effects when balancing traits, while the latter can achieve trait balance while maintaining the original state of the plant (Yang et al., 2025). Exploring “ideal” genes is significant for cultivating new varieties with cold tolerance.
CBF is a typical “traditional” trade-off gene. In Arabidopsis, overexpression of CBF constitutively activates cold stress response, which increases plant resistance to cold stress but also leads to growth inhibition, accumulation of DELLA protein, and inhibition of stem elongation (Achard et al., 2008). Similarly, high expression levels of CBF1/3 genes can lead to similar problems. Overexpression of CBF3 gene in cassava improves the cold resistance of transgenic cassava, but also introduces adverse effects such as growth retardation, leaf curling, shortened root length, and reduced yield (An et al., 2016).
In crops, mining and utilizing “ideal” trade-off genes could enhance cold tolerance in plants without compromising growth or yield, providing new strategies for cultivating crop varieties with traits such as cold tolerance and high yield. For example, the maize heat shock transcription factor HSF21 is considered as an “ideal” trade-off genes. HSF21 enhances the chilling tolerance by maintaining lipid metabolism homeostasis and modulating natural genetic variation under cold stress. Overexpression of HSF21 not only enhances maize cold tolerance, but also significantly increases maize field yield. Further analysis of high-throughput corn lipidomics revealed that HSF21 is a key gene regulating lipid metabolism homeostasis under cold stress, controlling the metabolic balance of unsaturated lipids under cold stress (Gao et al., 2024).
8 Discussion
Over recent decades, substantial advancements have been achieved in elucidating plant responses to cold stress. Early investigations primarily examined low-temperature-induced alterations in physiological and biochemical characteristics, such as membrane lipid phase changes. Subsequent progress in molecular techniques redirected attention to the isolation and functional characterization of cold-responsive genes. Critical regulators like CBF/DREB transcription factors were identified, offering mechanistic insights into cold adaptation pathways. Nevertheless, plant cold stress adaptation constitutes a multifaceted biological phenomenon, with existing knowledge representing merely an initial exploration of this intricate system.
From the perspective of research history, early studies often focused on a single gene, but plant cold response clearly involves the interaction of multiple genes and signaling pathways. Future research requires the use of systems biology methods to integrate multiple omics data and construct a complete cold responsive molecular network. Therefore, future research should focus on the following aspects: firstly, the initial perception mechanism of cold signals. The cell membrane is the main sensing site, but there is still no answer on how cold stress activates cold signal receptors and whether receptors depend on ROS or NO signaling. The second is the modification mechanism of CBF protein. The newly discovered modification pathway, such as histone acetylation, is involved in CBF regulation, but its specific mechanism is still unclear. The third is how plants balance key regulatory factors under cold stress. Light, flowering, circadian rhythm, hormones, and other factors are widely involved in the cold response, but their equilibrium mechanisms are still unclear.
At present, most existing research is based on the model plant Arabidopsis. With the intensification of global climate change, the frequency and intensity of extreme cold weather are fluctuating, posing new threats to crop production. Studying how crops perceive and respond to cold stress, and how to regulate the balance between growth and resistance, is crucial for cultivating cold resistant varieties and ensuring food security. At the same time, in actual agricultural production, there are many types of crops, and the cold tolerance of different crops varies greatly. It is necessary to strengthen the exploration of cold responsive genes specific to different crops and their application in breeding. This is expected to cultivate crop varieties with strong cold tolerance and excellent performance in key indicators such as yield, thereby promoting higher levels of agricultural production, meeting the growing demand for food in the future, and addressing complex and changing environmental challenges.
In addition, the long-term adaptation mechanism of plants to cold stress also deserves further research. Most studies have focused on the short-term response of plants to cold stress, lacking systematic research on how plants maintain growth and development, complete their life cycle and other issues in long-term cold condition. Meanwhile, the interaction between cold stress and other environmental stresses is also a direction for future research. In natural environments, plants often face a combination of various stresses, such as cold-drought, cold-salt stress, etc. Studying the response mechanisms of plants under these complex stresses is of great significance for a comprehensive understanding of plant environmental adaptability.
In summary, although the research on plants’ responses to cold stress has achieved fruitful results, it still faces many challenges. Through interdisciplinary collaboration and advanced research techniques and methods, we are expected to gradually uncover the mysteries of plant response to cold stress, providing a solid theoretical foundation and technical support for cultivating cold resistant crops and addressing climate change.
Author contributions
CP: Writing – original draft. WH: Writing – review & editing. JL: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This review was supported by grants from the Agricultural Science and Technology Innovation Project (CAAS-ZDRW202405), the Wuhan Science and Technology Major Project on Key techniques of biological breeding, Breeding of new varieties (2022021302024851) and the National Natural Science Foundation of China (Project No. 32372184).
Acknowledgments
The authors would like to express their sincere gratitude to Professor JL and Professor WH for their invaluable guidance and assistance throughout the research process. Their expertise and insights have been instrumental in shaping the direction of this work. We are also deeply appreciative of the support provided by the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences and the Hongshan Laboratory in Wuhan. Their resources and collaborative efforts have significantly contributed to the successful completion of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Aghdam, M. S. and Arnao, M. B. (2024). Phytomelatonin: from intracellular signaling to global horticulture market. J. Pineal Res. 76, e12990. doi: 10.1111/jpi.12990
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., and Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
An, D., Ma, Q., Yan, W., Zhou, W., Liu, G., and Zhang, P. (2016). Divergent regulation of CBF regulon on cold tolerance and plant phenotype in cassava overexpressing arabidopsis CBF3 gene. Front. Plant Sci. 7, 1866. doi: 10.3389/fpls.2016.01866
An, J. P., Xu, R. R., Liu, X., Su, L., Yang, K., Wang, X. F., et al. (2022). Abscisic acid insensitive 4 interacts with ICE1 and JAZ proteins to regulate ABA signaling-mediated cold tolerance in apple. J. Exp. Bot. 73, 980–997. doi: 10.1093/jxb/erab433
Babuta, P., Sougrakpam, Y., and Deswal, R. (2025). Nitric oxide cross-talks during low-temperature stress in plants. Plant Sci. 360, 112708. doi: 10.1016/j.plantsci.2025.112708
Bao, H., Yuan, L., Luo, Y., Zhang, J., Liu, X., Wu, Q., et al. (2025). The transcription factor WRKY41-FLAVONOID 3’-HYDROXYLASE module fine-tunes flavonoid metabolism and cold tolerance in potato. Plant Physiol. 197. doi: 10.1093/plphys/kiaf070
Bhaskar, A., Paul, L. K., Sharma, E., Jha, S., Jain, M., and Khurana, J. P. (2021). OsRR6, a type-A response regulator in rice, mediates cytokinin, light and stress responses when over-expressed in Arabidopsis. Plant Physiol. Biochem. 161, 98–112. doi: 10.1016/j.plaphy.2021.01.047
Cano-Ramirez, D. L., Panter, P. E., Takemura, T., De Fraine, T. S., De Barros Dantas, L. L., Dekeya, R., et al. (2023). Low-temperature and circadian signals are integrated by the sigma factor SIG5. Nat. Plants 9, 661–672. doi: 10.1038/s41477-023-01377-1
Cantrel, C., Vazquez, T., Puyaubert, J., REZé, N., Lesch, M., Kaiser, W. M., et al. (2011). Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. N. Phytol. 189, 415–427. doi: 10.1111/j.1469-8137.2010.03500.x
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Chu, M., Li, J., Zhang, J., Shen, S., Li, C., Gao, Y., et al. (2018). AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 69, 5241–5253. doi: 10.1093/jxb/ery278
Ding, Y., Jia, Y., Shi, Y., Zhang, X., Song, C., Gong, Z., et al. (2018). OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 37. doi: 10.15252/embj.201798228
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., and Yang, S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Ding, Y., Yang, H., Wu, S., Fu, D., Li, M., Gong, Z., et al. (2022). CPK28-NLP7 module integrates cold-induced Ca(2+) signal and transcriptional reprogramming in Arabidopsis. Sci. Adv. 8, eabn7901. doi: 10.1126/sciadv.abn7901
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad Sci. U. S. A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Dong, M. A., FARRé, E. M., and Thomashow, M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad Sci. U.S.A. 108, 7241–7246. doi: 10.1073/pnas.1103741108
Du, S. X., Wang, L. L., Yu, W. P., Xu, S. X., Chen, L., and Huang, W. (2024). Appropriate induction of TOC1 ensures optimal MYB44 expression in ABA signaling and stress response in Arabidopsis. Plant Cell Environ. 47, 3046–3062. doi: 10.1111/pce.14922
El-Refaee, Y. Z., Gharib, H. S., Badawy, S. A., Elrefaey, E. M., El-Okkiah, S. A. F., Okla, M. K., et al. (2024). Mitigating cold stress in rice: a study of genotype performance and sowing time. BMC Plant Biol. 24, 713. doi: 10.1186/s12870-024-05423-8
Eremina, M., Unterholzner, S. J., Rathnayake, A. I., Castellanos, M., Khan, M., Kugler, K. G., et al. (2016). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad Sci. U.S.A. 113, E5982–e5991. doi: 10.1073/pnas.1611477113
Fang, P., Wang, Y., Wang, M., Wang, F., Chi, C., Zhou, Y., et al. (2021). Crosstalk between brassinosteroid and redox signaling contributes to the activation of CBF expression during cold responses in tomato. Antioxid (Basel) 10, 509. doi: 10.3390/antiox10040509
Fu, D., Song, Y., Wu, S., Peng, Y., Ming, Y., Li, Z., et al. (2025). Regulation of alternative splicing by CBF-mediated protein condensation in plant response to cold stress. Nat. Plants 11, 505–517. doi: 10.1038/s41477-025-01933-x
Gao, L., Pan, L., Shi, Y., Zeng, R., Li, M., Li, Z., et al. (2024). Genetic variation in a heat shock transcription factor modulates cold tolerance in maize. Mol. Plant 17, 1423–1438. doi: 10.1016/j.molp.2024.07.015
Gill, S. S. and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Guo, Y., Li, J., Liu, L., Yang, W., Zhou, Y., Wei, C., et al. (2025). Nitric oxide cross-links calcium signals to enhance cold tolerance via inhibiting calmodulin expression in watermelon. Plant Physiol. 198. doi: 10.1093/plphys/kiaf243
Guo, M., Wang, S., Liu, H., Yao, S., Yan, J., Wang, C., et al. (2023). Histone deacetylase MdHDA6 is an antagonist in regulation of transcription factor MdTCP15 to promote cold tolerance in apple. Plant Biotechnol. J. 21, 2254–2272. doi: 10.1111/pbi.14128
He, Y., Li, Y., Cui, L., Xie, L., Zheng, C., Zhou, G., et al. (2016). Phytochrome B negatively affects cold tolerance by regulating osDREB1 gene expression through phytochrome interacting factor-like protein osPIL16 in rice. Front. Plant Sci. 7, 1963. doi: 10.3389/fpls.2016.01963
Hu, Y., Jiang, L., Wang, F., and Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Hu, C., Wang, M., Zhu, C., Wu, S., Li, J., Yu, J., et al. (2024a). A transcriptional regulation of ERF15 contributes to ABA-mediated cold tolerance in tomato. Plant Cell Environ. 47, 1334–1347. doi: 10.1111/pce.14816
Hu, D., Yao, Y., Lv, Y., You, J., Zhang, Y., Lv, Q., et al. (2024b). The OsSRO1c-OsDREB2B complex undergoes protein phase transition to enhance cold tolerance in rice. Mol. Plant 17, 1520–1538. doi: 10.1016/j.molp.2024.08.006
Huo, C., Zhang, B., and Wang, R. (2022). Research progress on plant noncoding RNAs in response to low-temperature stress. Plant Signal Behav. 17, 2004035. doi: 10.1080/15592324.2021.2004035
Hussain, M. A., Huang, Y., Luo, D., Mehmood, S. S., Raza, A., Duan, L., et al. (2025). Integrative analyses reveal Bna-miR397a-BnaLAC2 as a potential modulator of low-temperature adaptability in Brassica napus L. Plant Biotechnol. J. 23, 1968–1987. doi: 10.1111/pbi.70017
Jeon, J., Cho, C., Lee, M. R., Van Binh, N., and Kim, J. (2016). CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in arabidopsis. Plant Cell 28, 1828–1843. doi: 10.1105/tpc.15.00909
Jeon, J. and Kim, J. (2013). Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 161, 408–424. doi: 10.1104/pp.112.207621
Jia, M., Meng, X., Song, X., Zhang, D., Kou, L., Zhang, J., et al. (2022). Chilling-induced phosphorylation of IPA1 by OsSAPK6 activates chilling tolerance responses in rice. Cell Discov. 8, 71. doi: 10.1038/s41421-022-00413-2
Jiang, B., Shi, Y., Peng, Y., Jia, Y., Yan, Y., Dong, X., et al. (2020). Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in arabidopsis. Mol. Plant 13, 894–906. doi: 10.1016/j.molp.2020.04.006
Jiang, B., Shi, Y., Zhang, X., Xin, X., Qi, L., Guo, H., et al. (2017). PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad Sci. U.S.A. 114, E6695–e6702. doi: 10.1073/pnas.1706226114
Jiang, H., Zhou, L. J., Gao, H. N., Wang, X. F., Li, Z. W., and Li, Y. Y. (2022). The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 189, 2044–2060. doi: 10.1093/plphys/kiac211
Jin, R., Kim, B. H., Ji, C. Y., Kim, H. S., Li, H. M., Ma, D. F., et al. (2017). Overexpressing IbCBF3 increases low temperature and drought stress tolerance in transgenic sweetpotato. Plant Physiol. Biochem. 118, 45–54. doi: 10.1016/j.plaphy.2017.06.002
John, A., KRäMER, M., Lehmann, M., Kunz, H. H., Aarabi, F., Alseekh, S., et al. (2024). Degradation of FATTY ACID EXPORT PROTEIN1 by RHOMBOID-LIKE PROTEASE11 contributes to cold tolerance in Arabidopsis. Plant Cell 36, 1937–1962. doi: 10.1093/plcell/koae011
Kang, X., Wei, F., Chai, S., Peng, S., Huang, B., Han, Q., et al. (2025). The OST1-HOS1-HAT1 module regulates cold response in Arabidopsis thaliana. New Phytol. 247, 209–223. doi: 10.1111/nph.70189
Kidokoro, S., Hayashi, K., Haraguchi, H., Ishikawa, T., Soma, F., Konoura, I., et al. (2021). Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis. Proc. Natl. Acad Sci. U.S.A. 118, e2021048118. doi: 10.1073/pnas.2021048118
Kidokoro, S., Konoura, I., Soma, F., Suzuki, T., Miyakawa, T., Tanokura, M., et al. (2023). Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in Arabidopsis. Proc. Natl. Acad Sci. U.S.A. 120, e2216183120. doi: 10.1073/pnas.2216183120
Kidokoro, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 27, 922–935. doi: 10.1016/j.tplants.2022.01.008
Kidokoro, S., Yoneda, K., Takasaki, H., Takahashi, F., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29, 760–774. doi: 10.1105/tpc.16.00669
Kim, S. H., Kim, H. S., Bahk, S., An, J., Yoo, Y., Kim, J. Y., et al. (2017). Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 45, 6613–6627. doi: 10.1093/nar/gkx417
Kim, Y., Kim, S. H., Lim, J., and Kim, S. H. (2024). ATBS1-INTERACTING FACTOR 2 positively regulates freezing tolerance via INDUCER OF CBF EXPRESSION 1/C-REPEAT BINDING FACTOR-induced cold acclimation pathway. Plant Cell Physiol. 65, 1363–1376. doi: 10.1093/pcp/pcae072
Kutsuno, T., Chowhan, S., Kotake, T., and Takahashi, D. (2023). Temporal cell wall changes during cold acclimation and deacclimation and their potential involvement in freezing tolerance and growth. Physiol. Plant 175, e13837. doi: 10.1111/ppl.13837
Kyung, J., Jeon, M., Jeong, G., Shin, Y., Seo, E., Yu, J., et al. (2022). The two clock proteins CCA1 and LHY activate VIN3 transcription during vernalization through the vernalization-responsive cis-element. Plant Cell 34, 1020–1037. doi: 10.1093/plcell/koab304
Lantzouni, O., Alkofer, A., Falter-Braun, P., and Schwechheimer, C. (2020). GROWTH-REGULATING FACTORS interact with DELLAs and regulate growth in cold stress. Plant Cell 32, 1018–1034. doi: 10.1105/tpc.19.00784
Lee, J. H., Jung, J. H., and Park, C. M. (2015). INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS C-mediated flowering pathways in Arabidopsis. Plant J. 84, 29–40. doi: 10.1111/tpj.12956
Li, H., Ding, Y., Shi, Y., Zhang, X., Zhang, S., Gong, Z., et al. (2017a). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in arabidopsis. Dev. Cell 43, 630–642.e4. doi: 10.1016/j.devcel.2017.09.025
Li, Z., Fu, D., Wang, X., Zeng, R., Zhang, X., Tian, J., et al. (2022). The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 34, 2833–2851. doi: 10.1093/plcell/koac137
Li, Q., Hou, Y., Wang, Q., Pan, X., Sun, Y., Zhu, X., et al. (2024a). Phytochrome interacting factor ZmPIF6 simultaneously enhances chilling tolerance and grain size in rice. Plant Physiol. Biochem. 214, 108954. doi: 10.1016/j.plaphy.2024.108954
Li, Y., Shi, Y., Li, M., Fu, D., Wu, S., Li, J., et al. (2021). The CRY2-COP1-HY5-BBX7/8 module regulates blue light-dependent cold acclimation in Arabidopsis. Plant Cell 33, 3555–3573. doi: 10.1093/plcell/koab215
Li, R., Song, Y., Wang, X., Zheng, C., Liu, B., Zhang, H., et al. (2024b). OsNAC5 orchestrates OsABI5 to fine-tune cold tolerance in rice. J. Integr. Plant Biol. 66, 660–682. doi: 10.1111/jipb.13585
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., and Yang, S. (2017b). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in arabidopsis. Mol. Plant 10, 545–559. doi: 10.1016/j.molp.2017.01.004
Liang, L., Sui, X., Xiao, J., Tang, W., Song, X., Xu, Z., et al. (2025). ERD14 regulation by the HY5- or HY5-MED2 module mediates the cold signal transduction of asparagus bean. Plant J. 121, e17172. doi: 10.1111/tpj.17172
Lim, C. J., Park, J., Shen, M., Park, H. J., Cheong, M. S., Park, K. S., et al. (2020). The histone-modifying complex PWR/HOS15/HD2C epigenetically regulates cold tolerance. Plant Physiol. 184, 1097–1111. doi: 10.1104/pp.20.00439
Lin, R., Song, J., Tang, M., Wang, L., Yu, J., and Zhou, Y. (2023). CALMODULIN6 negatively regulates cold tolerance by attenuating ICE1-dependent stress responses in tomato. Plant Physiol. 193, 2105–2121. doi: 10.1093/plphys/kiad452
Ling, Y., Lin, J., Peng, D., and Li, Z. (2025). Nitric oxide production, sugar metabolism, and antioxidant capacity related to differential cold tolerance and recovery between two Dichondra repens genotypes. Ind. Crops Prod 231. doi: 10.1016/j.indcrop.2025.121132
Liu, Q., Ding, Y., Shi, Y., Ma, L., Wang, Y., Song, C., et al. (2021a). The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 40, e104559. doi: 10.15252/embj.2020104559
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3–3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128.e5. doi: 10.1016/j.molcel.2017.02.016
Liu, C., SCHLäPPI, M. R., Mao, B., Wang, W., Wang, A., and Chu, C. (2019). The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 17, 1834–1849. doi: 10.1111/pbi.13104
Liu, Y., Xu, C., Zhu, Y., Zhang, L., Chen, T., Zhou, F., et al. (2018). The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 60, 173–188. doi: 10.1111/jipb.12614
Liu, X., Zhao, C., Gao, Y., Xu, Y., Wang, S., Li, C., et al. (2021b). A multifaceted module of BRI1 ETHYLMETHANE SULFONATE SUPRESSOR1 (BES1)-MYB88 in growth and stress tolerance of apple. Plant Physiol. 185, 1903–1923. doi: 10.1093/plphys/kiaa116
Luo, W., Xu, Y., Cao, J., Guo, X., Han, J., Zhang, Y., et al. (2024). COLD6-OSM1 module senses chilling for cold tolerance via 2’,3’-cAMP signaling in rice. Mol. Cell 84, 4224–4238.e9. doi: 10.1016/j.molcel.2024.09.031
Lv, J., Liu, J., Ming, Y., Shi, Y., Song, C., Gong, Z., et al. (2021). Reciprocal regulation between the negative regulator PP2CG1 phosphatase and the positive regulator OST1 kinase confers cold response in Arabidopsis. J. Integr. Plant Biol. 63, 1568–1587. doi: 10.1111/jipb.13100
Ma, C., Burd, S., and Lers, A. (2015a). miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 84, 169–187. doi: 10.1111/tpj.12999
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015b). COLD1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi: 10.1016/j.cell.2015.01.046
Ma, L., Li, X., Zhao, Z., Hao, Y., Shang, R., Zeng, D., et al. (2021). Light-Response Bric-A-Brack/Tramtrack/Broad proteins mediate cryptochrome 2 degradation in response to low ambient temperature. Plant Cell 33, 3610–3620. doi: 10.1093/plcell/koab219
Maeda, A. E., Matsuo, H., Muranaka, T., and Nakamichi, N. (2024). Cold-induced degradation of core clock proteins implements temperature compensation in the Arabidopsis circadian clock. Sci. Adv. 10, eadq0187. doi: 10.1126/sciadv.adq0187
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Mei, C., Yang, J., Mei, Q., Jia, D., Yan, P., Feng, B., et al. (2023). MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 21, 2057–2073. doi: 10.1111/pbi.14112
Mikkelsen, M. D. and Thomashow, M. F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60, 328–339. doi: 10.1111/j.1365-313X.2009.03957.x
Ming, Y., Peng, Y., Liu, Q., Fu, D., Lin, Q., You, P., et al. (2025). Coordinated control of calcium signaling by CPK3 and CaM2 via CNGCs in response to cold stress in Arabidopsis. Dev. Cell. S1534-5807(25)00375-2. doi: 10.1016/j.devcel.2025.06.020
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Mori, K., Renhu, N., Naito, M., Nakamura, A., Shiba, H., Yamamoto, T., et al. (2018). Ca(2+)-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca(2+) increase and cold tolerance in Arabidopsis. Sci. Rep. 8, 550. doi: 10.1038/s41598-017-17483-y
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nakamichi, N., Kusano, M., Fukushima, A., Kita, M., Ito, S., Yamashino, T., et al. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. doi: 10.1093/pcp/pcp004
Novillo, F., Medina, J., and Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad Sci. U.S.A. 104, 21002–21007. doi: 10.1073/pnas.0705639105
Olate, E., JIMéNEZ-GóMEZ, J. M., Holuigue, L., and Salinas, J. (2018). NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat. Plants 4, 811–823. doi: 10.1038/s41477-018-0254-2
Oliveira, E. R., Nunes, A., Dutra, F. S., Azevedo, G. Z., Schneider, A. R., Dos Santos, B. R., et al. (2025). Marine and terrestrial biostimulant elicitors of tolerance to cold stress. Front. Plant Sci. 16, 1569516. doi: 10.3389/fpls.2025.1569516
Park, M. J., Seo, P. J., and Park, C. M. (2012). CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signal Behav. 7, 1194–1196. doi: 10.4161/psb.21300
Peng, Y., Ming, Y., Jiang, B., Zhang, X., Fu, D., Lin, Q., et al. (2024a). Differential phosphorylation of Ca2+-permeable channel CNGC20 modulates calcium-mediated freezing tolerance in Arabidopsis. Plant Cell. 36, 4356–4371. doi: 10.1093/plcell/koae177
Peng, Y., Ming, Y., Jiang, B., Zhang, X., Fu, D., Lin, Q., et al. (2024b). Differential phosphorylation of Ca2+-permeable channel CYCLIC NUCLEOTIDE-GATED CHANNEL20 modulates calcium-mediated freezing tolerance in Arabidopsis. Plant Cell 36, 4356–4371. doi: 10.1093/plcell/koae177
Rosado, D., Trench, B., Bianchetti, R., Zuccarelli, R., Rodrigues Alves, F. R., Purgatto, E., et al. (2019). Downregulation of PHYTOCHROME-INTERACTING FACTOR 4 influences plant development and fruit production. Plant Physiol. 181, 1360–1370. doi: 10.1104/pp.19.00833
Seo, E., Lee, H., Jeon, J., Park, H., Kim, J., Noh, Y. S., et al. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21, 3185–3197. doi: 10.1105/tpc.108.063883
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6, 902. doi: 10.3389/fpls.2015.00902
Shi, Y., Ding, Y., and Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Song, J., Lin, R., Tang, M., Wang, L., Fan, P., Xia, X., et al. (2023). SlMPK1- and SlMPK2-mediated SlBBX17 phosphorylation positively regulates CBF-dependent cold tolerance in tomato. New Phytol. 239, 1887–1902. doi: 10.1111/nph.19072
Song, Y., Zhang, X., Li, M., Yang, H., Fu, D., Lv, J., et al. (2021). The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 63, 1874–1887. doi: 10.1111/jipb.13161
Sorkin, M. L., Tzeng, S. C., King, S., Romanowski, A., Kahle, N., Bindbeutel, R., et al. (2023). COLD REGULATED GENE 27 and 28 antagonize the transcriptional activity of the RVE8/LNK1/LNK2 circadian complex. Plant Physiol. 192, 2436–2456. doi: 10.1093/plphys/kiad210
Su, C. F., Wang, Y. C., Hsieh, T. H., Lu, C. A., Tseng, T. H., and Yu, S. M. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153, 145–158. doi: 10.1104/pp.110.153015
Sun, M., Shen, Y., Chen, Y., Wang, Y., Cai, X., Yang, J., et al. (2022). Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 189, 2500–2516. doi: 10.1093/plphys/kiac208
Sun, Y., Xie, Z., Jin, L., Qin, T., Zhan, C., and Huang, J. (2024). Histone deacetylase OsHDA716 represses rice chilling tolerance by deacetylating OsbZIP46 to reduce its transactivation function and protein stability. Plant Cell 36, 1913–1936. doi: 10.1093/plcell/koae010
Tang, J., Tian, X., Mei, E., He, M., Gao, J., Yu, J., et al. (2022). WRKY53 negatively regulates rice cold tolerance at the booting stage by fine-tuning anther gibberellin levels. Plant Cell 34, 4495–4515. doi: 10.1093/plcell/koac253
Tang, K., Zhao, L., Ren, Y., Yang, S., Zhu, J. K., and Zhao, C. (2020). The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 62, 258–263. doi: 10.1111/jipb.12918
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
Verma, R. K., Santosh Kumar, V. V., Yadav, S. K., Pushkar, S., Rao, M. V., and Chinnusamy, V. (2019). Overexpression of ABA receptor PYL10 gene confers drought and cold tolerance to indica rice. Front. Plant Sci. 10, 1488. doi: 10.3389/fpls.2019.01488
Villouta, C., Workmaster, B. A., and Atucha, A. (2021). Freezing stress damage and growth viability in Vaccinium macrocarpon Ait. bud structures. Physiol. Plant 172, 2238–2250. doi: 10.1111/ppl.13457
Wang, L., Chen, H., Chen, G., Luo, G., Shen, X., Ouyang, B., et al. (2024a). Transcription factor SlWRKY50 enhances cold tolerance in tomato by activating the jasmonic acid signaling. Plant Physiol. 194, 1075–1090. doi: 10.1093/plphys/kiad578
Wang, F., Chen, X., Dong, S., Jiang, X., Wang, L., Yu, J., et al. (2020). Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 18, 1041–1055. doi: 10.1111/pbi.13272
Wang, P., Cui, X., Zhao, C., Shi, L., Zhang, G., Sun, F., et al. (2017). COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Integr. Plant Biol. 59, 78–85. doi: 10.1111/jipb.12512
Wang, X., Ding, Y., Li, Z., Shi, Y., Wang, J., Hua, J., et al. (2019b). PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev. Cell 51, 222–235.e5. doi: 10.1016/j.devcel.2019.08.008
Wang, F., Guo, Z., Li, H., Wang, M., Onac, E., Zhou, J., et al. (2016). Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 170, 459–471. doi: 10.1104/pp.15.01171
Wang, X., Li, Z., Shi, Y., Liu, Z., Zhang, X., Gong, Z., et al. (2023b). Strigolactones promote plant freezing tolerance by releasing the WRKY41-mediated inhibition of CBF/DREB1 expression. EMBO J. 42, e112999. doi: 10.15252/embj.2022112999
Wang, L. F., Lu, K. K., Li, T. T., Zhang, Y., Guo, J. X., Song, R. F., et al. (2022). Maize PHYTOMELATONIN RECEPTOR1 functions in plant tolerance to osmotic and drought stress. J. Exp. Bot. 73, 5961–5973. doi: 10.1093/jxb/erab553
Wang, J., Ren, Y., Liu, X., Luo, S., Zhang, X., Liu, X., et al. (2021). Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 14, 315–329. doi: 10.1016/j.molp.2020.11.022
Wang, S., Shen, Y., Deng, D., Guo, L., Zhang, Y., Nie, Y., et al. (2023a). Orthogroup and phylotranscriptomic analyses identify transcription factors involved in the plant cold response: A case study of Arabidopsis BBX29. Plant Commun. 4, 100684. doi: 10.1016/j.xplc.2023.100684
Wang, Y., Tong, W., Li, F., Samarina, L., Li, P., Yang, T., et al. (2024b). LUX ARRHYTHMO links CBF pathway and jasmonic acid metabolism to regulate cold tolerance of tea plants. Plant Physiol. 196, 961–978. doi: 10.1093/plphys/kiae337
Wang, F., Zhang, L., Chen, X., Wu, X., Xiang, X., Zhou, J., et al. (2019a). SlHY5 integrates temperature, light, and hormone signaling to balance plant growth and cold tolerance. Plant Physiol. 179, 749–760. doi: 10.1104/pp.18.01140
Wang, X., Zhang, X., Song, C. P., Gong, Z., Yang, S., and Ding, Y. (2023c). PUB25 and PUB26 dynamically modulate ICE1 stability via differential ubiquitination during cold stress in Arabidopsis. Plant Cell 35, 3585–3603. doi: 10.1093/plcell/koad159
Wang, Y., Zhang, M., Wu, C., Chen, C., Meng, L., Zhang, G., et al. (2024c). SlWRKY51 regulates proline content to enhance chilling tolerance in tomato. Plant Cell Environ. 47, 5104–5114. doi: 10.1111/pce.15081
Waters, M. T., Gutjahr, C., Bennett, T., and Nelson, D. C. (2017). Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. doi: 10.1146/annurev-arplant-042916-040925
Wilson, I. D., Neill, S. J., and Hancock, J. T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 31, 622–631. doi: 10.1111/j.1365-3040.2007.01761.x
Wu, J., Liu, H., Zhang, Y., Zhang, Y., Li, D., Liu, S., et al. (2024). A major gene for chilling tolerance variation in Indica rice codes for a kinase OsCTK1 that phosphorylates multiple substrates under cold. New Phytol. 242, 2077–2092. doi: 10.1111/nph.19696
Xia, C., Gong, Y., Chong, K., and Xu, Y. (2021). Phosphatase OsPP2C27 directly dephosphorylates OsMAPK3 and OsbHLH002 to negatively regulate cold tolerance in rice. Plant Cell Environ. 44, 491–505. doi: 10.1111/pce.13938
Xiao, W., Zhang, Y., Wang, Y., Zeng, Y., Shang, X., Meng, L., et al. (2024). The transcription factor TGA2 orchestrates salicylic acid signal to regulate cold-induced proline accumulation in Citrus. Plant Cell 37. doi: 10.1093/plcell/koae290
Xie, Y., Chen, P., Yan, Y., Bao, C., Li, X., Wang, L., et al. (2018). An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 218, 201–218. doi: 10.1111/nph.14952
Xiong, L., Ishitani, M., Lee, H., and Zhu, J. K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13, 2063–2083.
Xu, P., Ma, W., Feng, H., and Cai, W. (2024b). The NAC056 transcription factor confers freezing tolerance by positively regulating expression of CBFs and NIA1 in Arabidopsis. Plant Commun. 5, 100923. doi: 10.1016/j.xplc.2024.100923
Xu, L., Yang, L., Li, A., Guo, J., Wang, H., Qi, H., et al. (2024a). An AP2/ERF transcription factor confers chilling tolerance in rice. Sci. Adv. 10, eado4788. doi: 10.1126/sciadv.ado4788
Yamazaki, T., Kawamura, Y., Minami, A., and Uemura, M. (2008). Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20, 3389–3404. doi: 10.1105/tpc.108.062679
Yang, J. L., Chen, W. W., Chen, L. Q., Qin, C., Jin, C. W., Shi, Y. Z., et al. (2013b). The 14-3–3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. New Phytol. 197, 815–824. doi: 10.1111/nph.12057
Yang, L., Fang, S., Liu, L., Zhao, L., Chen, W., Li, X., et al. (2025). WRKY transcription factors: Hubs for regulating plant growth and stress responses. J. Integr. Plant Biol. 67, 488–509. doi: 10.1111/jipb.13828
Yang, C., Li, D., Mao, D., Liu, X., Ji, C., Li, X., et al. (2013a). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218. doi: 10.1111/pce.12130
Ye, K., Li, H., Ding, Y., Shi, Y., Song, C., Gong, Z., et al. (2019). Brassinosteroid-insensitive2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in arabidopsis. Plant Cell 31, 2682–2696. doi: 10.1105/tpc.19.00058
Ye, Q., Zheng, L., Liu, P., Liu, Q., Ji, T., Liu, J., et al. (2024). The S-acylation cycle of transcription factor MtNAC80 influences cold stress responses in Medicago truncatula. Plant Cell 36, 2629–2651. doi: 10.1093/plcell/koae103
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). Constans activates suppressor of overexpression of constans 1 through flowering locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Yu, D., Song, W., Tan, E. Y. J., Liu, L., Cao, Y., Jirschitzka, J., et al. (2022). TIR domains of plant immune receptors are 2’,3’-cAMP/cGMP synthetases mediating cell death. Cell 185, 2370–2386.e18. doi: 10.1016/j.cell.2022.04.032
Zeng, R., Li, Z., Shi, Y., Fu, D., Yin, P., Cheng, J., et al. (2021). Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 12, 4713. doi: 10.1038/s41467-021-25001-y
Zeng, R., Shi, Y., Guo, L., Fu, D., Li, M., Zhang, X., et al. (2025). A natural variant of COOL1 gene enhances cold tolerance for high-latitude adaptation in maize. Cell 188, 1315–1329.e13. doi: 10.1016/j.cell.2024.12.018
Zhai, M., Chen, Y., Pan, X., Chen, Y., Zhou, J., Jiang, X., et al. (2024). OsEIN2-OsEIL1/2 pathway negatively regulates chilling tolerance by attenuating OsICE1 function in rice. Plant Cell Environ. 47, 2561–2577. doi: 10.1111/pce.14900
Zhang, L., Jiang, X., Liu, Q., Ahammed, G. J., Lin, R., Wang, L., et al. (2020). The HY5 and MYB15 transcription factors positively regulate cold tolerance in tomato via the CBF pathway. Plant Cell Environ. 43, 2712–2726. doi: 10.1111/pce.13868
Zhang, Z., Li, J., Li, F., Liu, H., Yang, W., Chong, K., et al. (2017). OsMAPK3 phosphorylates osbHLH002/osICE1 and inhibits its ubiquitination to activate osTPP1 and enhances rice chilling tolerance. Dev. Cell 43, 731–743.e5. doi: 10.1016/j.devcel.2017.11.016
Zhang, X., Li, M., Zhang, X., Zeng, R., Peng, Y., Shi, Y., et al. (2025). A receptor-kinase cascade confers cold-induced root growth inhibition in Arabidopsis. Nat. Plants. doi: 10.1038/s41477-025-02034-5
Zhang, L., Song, J., Lin, R., Tang, M., Shao, S., Yu, J., et al. (2022a). Tomato SlMYB15 transcription factor targeted by sly-miR156e-3p positively regulates ABA-mediated cold tolerance. J. Exp. Bot. 73, 7538–7551. doi: 10.1093/jxb/erac370
Zhang, M., Zhao, R., Huang, K., Huang, S., Wang, H., Wei, Z., et al. (2022b). The OsWRKY63-OsWRKY76-OsDREB1B module regulates chilling tolerance in rice. Plant J. 112, 383–398. doi: 10.1111/tpj.15950
Zhao, Y., Antoniou-Kourounioti, R. L., Calder, G., Dean, C., and Howard, M. (2020). Temperature-dependent growth contributes to long-term cold sensing. Nature 583, 825–829. doi: 10.1038/s41586-020-2485-4
Zhao, M. G., Chen, L., Zhang, L. L., and Zhang, W. H. (2009). Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol. 151, 755–767. doi: 10.1104/pp.109.140996
Zhao, J., Shi, M., Yu, J., and Guo, C. (2022). SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in Arabidopsis thaliana. BMC Plant Biol. 22, 59. doi: 10.1186/s12870-022-03445-8
Zhao, C., Wang, P., Si, T., Hsu, C. C., Wang, L., Zayed, O., et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43, 618–629.e5. doi: 10.1016/j.devcel.2017.09.024
Zheng, Q., Yu, Q., Yao, W., Lv, K., Zhang, N., and Xu, W. (2023). Decoding vaCOLD1 function in grapevines: A membrane protein enhancing cold stress tolerance. J. Agric. Food Chem. 71, 19357–19371. doi: 10.1021/acs.jafc.3c05101
Zhu, P., Lister, C., and Dean, C. (2021). Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657–661. doi: 10.1038/s41586-021-04062-5
Zhu, J., Zhang, K. X., Wang, W. S., Gong, W., Liu, W. C., Chen, H. G., et al. (2015). Low temperature inhibits root growth by reducing auxin accumulation via ARR1/12. Plant Cell Physiol. 56, 727–736. doi: 10.1093/pcp/pcu217
Keywords: cold stress, signaling pathway, regulatory mechanism, intelligent breeding, cold responsive gene
Citation: Peng C, Hua W and Liu J (2025) Unraveling the signaling pathways of plant cold stress: current insights and future directions. Front. Plant Sci. 16:1666852. doi: 10.3389/fpls.2025.1666852
Received: 16 July 2025; Accepted: 31 August 2025;
Published: 24 September 2025.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Sarah Muniz Nardeli, Umeå University, SwedenBasharat Ahmad Bhat, University of Kashmir, India
Copyright © 2025 Peng, Hua and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liu, bGl1amluZzAxQGNhYXMuY24=
 Chen Peng
Chen Peng Wei Hua
Wei Hua Jing Liu
Jing Liu