- Panxi Crop Improvement Key Laboratory of Sichuan Province, College of Agricultural Science, Xichang University, Xichang, Sichuan, China
Introduction: To elucidate the physiological and molecular responses of peanut (Arachis hypogaea L L. c.v. ‘Haihua No. 1’) to copper stress, this study aimed to investigate the changes in root morphology, ion content, oxidative stress, and gene expression under copper stress conditions.
Methods: Seedlings were exposed to 0 (control) or 50 mg/L CuSO₄ solution, with three biological replicates for each treatment. Root length and biomass were measured quantitatively, along with tissue contents of eight ions (K+, Na+, Mg2+, Ca2+, Fe3+, Mn2+, Cu2+, Zn2+), secondary oxidative stress indices, and activities of key antioxidant enzymes. RNA-seq and qPCR validation were performed to analyze transcriptional changes and identify specific gene-response modules in peanut seedling roots under copper stress.
Results: Copper stress significantly induced the expression of MPK4, a key component of the MPK4 pathway. Post-translationally, MPK4 likely phosphorylated two critical protein classes: NAC and LBD. NAC functioned as a core transcription factor, directly regulating the transcription of copper defense-related genes. LBD directly down-regulated genes associated with lateral root growth, indirectly promoting the expression of genes involved in GSH-dependent heavy metal detoxification and secondary oxidative stress (e.g., GST and POD), thereby enhancing the plant's detoxification and antioxidant capacity.
Discussion: This study provides insights into the regulatory mechanisms that peanut plants employ to cope with copper stress. The findings highlight the roles of MPK4, NAC, and LBD in the plant's response to copper stress and suggest that these genes could be targeted in breeding programs to develop copper-tolerant peanut cultivars. The results may provide theoretical support for the development of such cultivars.
Introduction
With the rapid acceleration of industrialization and continuing population growth, heavy-metal contamination of soils has escalated into a critical global environmental concern (Li et al., 2020). According to the Survey Bulletin on the National Soil Contamination Status (Ministry of Environmental Protection and Ministry of Land and Resources, April 2014), 16.1% of all monitored sites across China exceeded national environmental quality standards; among these non-compliant sites, 82.8% were attributed to inorganic pollutants, and copper (Cu2+) alone exhibited a site-exceedance rate of 2.1% (Wu et al., 2021). In China, the Southwest region constitutes a major non-ferrous metallogenic belt, and mining as well as smelting operations within this region are the dominant sources of soil contamination by Cu2+, Fe3+, Zn2+, and other heavy metals (Borkert and Cox, 1999). For example, soils surrounding the Dongchuan and Luchun copper mines in Yunnan Province exhibit pronounced heavy-metal enrichment, with concentrations markedly exceeding regional geochemical background values (Afzal et al., 2022). Such contamination not only suppresses local vegetation growth but also poses a potential threat to human health via trophic transfer through the food web (Demecsová et al., 2020).
Peanut (Arachis hypogaea L.) is cultivated worldwide and serves as a pivotal economic and oilseed crop in southwest China; its yield and quality directly determine agricultural productivity and food safety in peanut-producing regions (Kobayashi et al., 2019). Nevertheless, in Cu2+-contaminated soils both vegetative growth and kernel quality are severely compromised. Excess Cu2+ not only suppresses plant development, but also compromises the safety of peanuts destined for human consumption (Sharma et al., 2021). Therefore, elucidating the response mechanisms of peanut to Cu2+ stress is essential for ensuring regional agricultural sustainability and food security.
During long-term evolution, plants have developed a sophisticated network of heavy-metal stress responses, including antioxidant defense, signal transduction, and ion-homeostatic regulation to adapt to their surrounding geochemical environments (Cai et al., 2023). Under metal stress, excessive reactive oxygen species (ROS) are scavenged through the activation of antioxidant enzymes such as superoxide dismutase (SOD), glutathione peroxidase (GPX), and catalase (CAT) (Wang et al., 2021). Concomitantly, plants maintain intracellular metal homeostasis via finely tuned ion-balance mechanisms; for instance, under Fe3+ deficiency, the expression of FERRIC REDUCTION OXIDASE 2 (FRO2) and IRON-REGULATED TRANSPORTER 1 (IRT1) is up-regulated to enhance Fe uptake and translocation (Zhai et al., 2017).
Among the signaling networks that orchestrate these responses, the mitogen-activated protein kinase (MAPK) cascade functions as a pivotal pathway enabling plants to perceive and transduce external stress cues (Yadav et al., 2021). MAPK modules typically operate through sequential phosphorylation (MAPKKK → MAPKK → MAPK) to regulate downstream transcription factors, thereby initiating defense mechanisms against metal toxicity (Takatsuji, 1998). In Arabidopsis, the MEKK1–MKK4/MKK5–MPK3/MPK6 cascade confers resistance to both abiotic and biotic stresses (Xu et al., 2019). Similarly, in rice, infection by Magnaporthe oryzae triggers endoplasmic-reticulum-stress-mediated activation of MAPK cascades to enhance disease resistance (Li et al., 2021). In potato (Solanum tuberosum), the StMEK1-mediated MAPK cascade plays a pivotal role in immunity against pathogens (Khan et al., 2021) whereas in cotton (Gossypium hirsutum) the GhMAP3K65 gene modulates pathogen perception via salicylic acid, jasmonic acid, and ethylene signaling, as well as ROS homeostasis; silencing GhMAP3K65 significantly enhances resistance to Ralstonia solanacearum (Ammar et al., 2025).
Beyond biotic stresses, MAPK pathways are also integral to heavy-metal tolerance. Under Zn2+ stress, MYB72 protein orchestrates Zn2+ uptake and detoxification (Peralta et al., 2022); Cd2+ exposure activates MPK3 and MPK6 via ROS accumulation to enhance Cd2+ tolerance in Arabidopsis (Liu et al., 2010); Cr3+ stress induces the bZIP transcription factor TGA3 to promote H2S biosynthesis through transcriptional activation of LCD, thereby improving Cr3+ resistance (Fang et al., 2017); Pb2+ exposure up-regulates RsWRKY to modulate the antioxidant system (Khedia et al., 2019); Ni2+ stress is mitigated by SbMYB15 regulated antioxidant enzyme activities (Sapara et al., 2019); and As tolerance is conferred by OsARM1-mediated control of As uptake and root-to-shoot translocation (Wang et al., 2017). Collectively, these studies demonstrate that MAPK cascades initiate heavy-metal stress defense by phosphorylation-dependent activation of downstream transcription factors.
Consistent cross-species multi-omics evidence has established the MAPK cascade as the convergent node that integrates ROS, NO, and hormonal signals during heavy-metal stress, directly phosphorylating key transcription factors such as WRKY, MYB, and bZIP to activate antioxidant and ion-homeostatic networks (Li et al., 2022). Nevertheless, the expression patterns of the MAPK module and its Cu2+-specific defense logic in peanut under Cu2+ stress remain undocumented.
To address this knowledge gap, we exposed peanut seedlings to 0 and 50 mg/L Cu2+ for 48 hours using a hydroponic system. By combining root transcriptomics with physiological readouts, we aim to elucidate the response mechanisms of the MAPK signaling pathway, antioxidant enzymes, and transcription factor DEGs in peanut roots to copper stress, thereby providing a theoretical basis for breeding heavy-metal-resistant crops.
Materials and methods
Material cultivation
Seeds of peanut cultivar ‘Haihua No. 1’ were used. Intact, uniformly sized seeds were selected on the basis of 100-seed weight and kernel diameter using a caliper and analytical balance. Selected seeds were surface-sterilized with 70% (v/v) ethanol for 10 min, rinsed three times with sterile deionized water, and germinated on moist filter paper in Petri dishes at 28 ± 1 °C in darkness. Distilled water was replenished every 12 h to maintain constant moisture. After 72 h, the seedlings were cultured in distilled water for 4 days at 25 °C to be used as experimental material.
Experimental design
The experiment consisted of two treatment groups: a distilled water control (CK) and a copper stress treatment (Cu). The copper stress treatment was administered using 50 mg/L of CuSO4·H2O. Each treatment was applied for 48 hours with three biological replicates.
After the treatment, we photographed the samples to document their growth status. Subsequently, we harvested the roots. A portion of the root samples was immediately frozen in liquid nitrogen and stored at -80 °C for subsequent physiological, transcriptome (RNA-seq), and RT-qPCR analyses. The remaining samples were blanched at 90 °C for 30 minutes, then oven-dried to a constant weight at 60 °C to measure the content of various ions in the tissues.
Metrics and analysis methods
Growth phenotype of peanut seedling roots
The growth status of the peanut seedlings was documented by taking high-resolution photos with a smartphone. Subsequently, the roots were harvested, and root length was measured with a caliper, while fresh root weight was determined using an electronic balance.
Analysis of oxidative stress indicators and antioxidant enzyme activities in peanut seedling roots
Frozen peanut root samples stored at -80 °C were used to determine several stress-related physiological indices. These included the content of reactive oxygen species (ROS), specifically superoxide anion (O2-) and hydrogen peroxide (H2O2), as well as malondialdehyde (MDA). We also measured the activities of three antioxidant enzymes: superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) (Dong et al., 2023).
Absolute quantification of eight cations in peanut seedling roots
Dried samples were digested in 5 mL HNO3/HClO4 solution (5:1, v/v) at 180 °C until the solution became clear. The resulting digest was then brought to a final volume of 25 mL with ultrapure water. The absolute content of the eight cations (K+, Na+, Ca2+, Mg2+, Fe3+, Mn2+, Cu2+, and Zn2+) was determined using flame atomic absorption spectrophotometry (AAS, PinAAcle 900T, PerkinElmer, USA). The measurements were validated using matrix-matched standards and a certified reference material (NIST 1573a).
High-throughput transcriptome sequencing (RNA-seq)
Total RNA was extracted from peanut roots stored at −80 °C using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA integrity was assessed using an Agilent Bioanalyzer 2100 system with the RNA Nano 6000 LabChip kit (Agilent Technologies, Santa Clara, CA, USA), and only samples with an RNA integrity number (RIN) ≥ 8.0 were used for subsequent steps.
Strand-specific cDNA libraries were constructed from 2 µg of total RNA using the TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s guidelines. Briefly, poly(A)-enriched mRNA was purified with oligo(dT)-conjugated magnetic beads, fragmented, and reverse-transcribed into cDNA. The cDNA was then end-repaired, A-tailed, and ligated to indexed adapters. After 12 cycles of PCR amplification (98 °C for 30 s; 12 cycles of 98 °C for 10 s, 60 °C for 30 s, 72 °C for 30 s; 72 °C for 5 min), the libraries were quantified with a Qubit 3.0 fluorometer (Thermo Fisher Scientific) and validated on the Bioanalyzer 2100. Paired-end 150-bp sequencing was performed on the Illumina HiSeq 4000 platform by LC Sciences (Hangzhou, China), generating approximately 6 Gb of clean data per sample (Dong et al., 2023). Our raw sequencing data have been uploaded to the NCBI database (https://www.ncbi.nlm.nih.gov/). The accession ID for the Sequence Read Archive (SRA) dataset is SUB15570566.
Quantitative real-time polymerase chain reaction assay
Total RNA was extracted from root samples, which had been snap-frozen in liquid nitrogen and stored at −80 °C to prevent RNA degradation. The extraction was performed using the RNAiso Plus reagent (Takara Bio, Kusatsu, Japan) according to the manufacturer’s instructions. Subsequently, mRNA was enriched from the total RNA using oligo(dT)-conjugated magnetic beads (Invitrogen, Carlsbad, CA, USA) and further purified via probe-based hybridization. First-strand cDNA was synthesized with the GoScript™ Reverse Transcription System (Promega, Beijing, China) from 1 µg of purified mRNA.
Quantitative real-time PCR was performed on a QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) using GoTaq® qPCR Master Mix (Promega, Beijing, China) in a 20 µL reaction volume containing 2 µL of 1:10-diluted cDNA, 0.4 µM each primer, and 1 × master mix. Cycling conditions were 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Melting-curve analysis (65–95 °C, 0.5 °C increment) confirmed amplicon specificity.
Actin-7 (GenBank ID: LOC112715878) served as the internal reference gene. Relative mRNA abundance was calculated by the 2(−ΔΔCt) method (Sun et al., 2024). Primer sequences and amplicon sizes are listed in Supplementary Table S1.
Data processing and statistical analysis
Raw data for root growth parameters, physiological indices, and RT-qPCR were organized and normalized in Microsoft Excel 2016.
After quality filtering with Trimmomatic v0.39, clean reads were aligned to the Arachis hypogaea reference genome (NCBI Taxonomy ID 3818) using HISAT2 v2.2.1 (Kim et al., 2019) Gene-level counts were generated with featureCounts v2.0.3 (Liao et al., 2014) Differentially expressed genes (DEGs) were identified using edgeR v3.38.0 (Robinson et al., 2010) under thresholds of |log2 fold change| ≥ 1 and FDR < 0.05.
GO and KEGG enrichment of DEGs and was conducted using clusterProfiler v4.0 with a hypergeometric test (p < 0.05, Benjamini–Hochberg correction).
For physiological phenotypes, ion contents, and RT-qPCR data, one-way analysis of variance (ANOVA) was performed in R v4.3.1 using the aov() function, followed by Duncan’s multiple range test (α = 0.05).
Data visualization was performed as follows: bar plots of physiological data and KEGG enrichment results were generated in Origin 2021; heatmaps were constructed with TBtools-II (Chen et al., 2023).
Results
Root growth, oxidative stress, and antioxidant responses in peanut seeding roots under copper stress
Excess Cu2+ markedly inhibited root elongation and biomass accumulation in peanut seedlings. Compared with the control (CK), 50 mg/L Cu2+ treatment reduced mean primary root length by 32.9% (CK: 6.540 cm vs. Cu2+: 4.390 cm) and fresh root weight by 18.6% (CK: 0.590 g vs. Cu2+: 0.480 g). Concurrently, Cu2+ stress significantly elevated antioxidant enzyme activities (Figures 1A–C). Superoxide dismutase (SOD) activity increased from 587.077 U g-¹ h-¹ in CK to 654.867 U g-¹ h-¹ under Cu2+ stress; catalase (CAT) activity rose from 318.723 to 369.33 U g-¹ min-¹, while peroxidase (POD) activity exhibited the most pronounced induction, increasing 1.75-fold (from 769.807 to 1 345. 14 U g-¹ min-¹). The oxidative burst profile revealed that superoxide anion (O2-) content declined slightly (CK: 12.44 μg/g; Cu2+: 10.873 μg/g), whereas hydrogen peroxide (H2O2) and malondialdehyde (MDA) accumulated markedly. H2O2 content increased >16-fold (CK: 22.08 μg/g; Cu2+: 372.533 μg/g), and MDA concentration rose 1.59-fold (CK: 7.06 μmol/g; Cu2+: 11.2 μmol/g) (Figures 1D–F). These data demonstrate that excess Cu2+ elicits secondary oxidative stress in peanut roots, triggering a compensatory up-regulation of the enzymatic antioxidant system (SOD, CAT, and POD) to mitigate Cu2+-induced ROS accumulation and membrane lipid peroxidation.
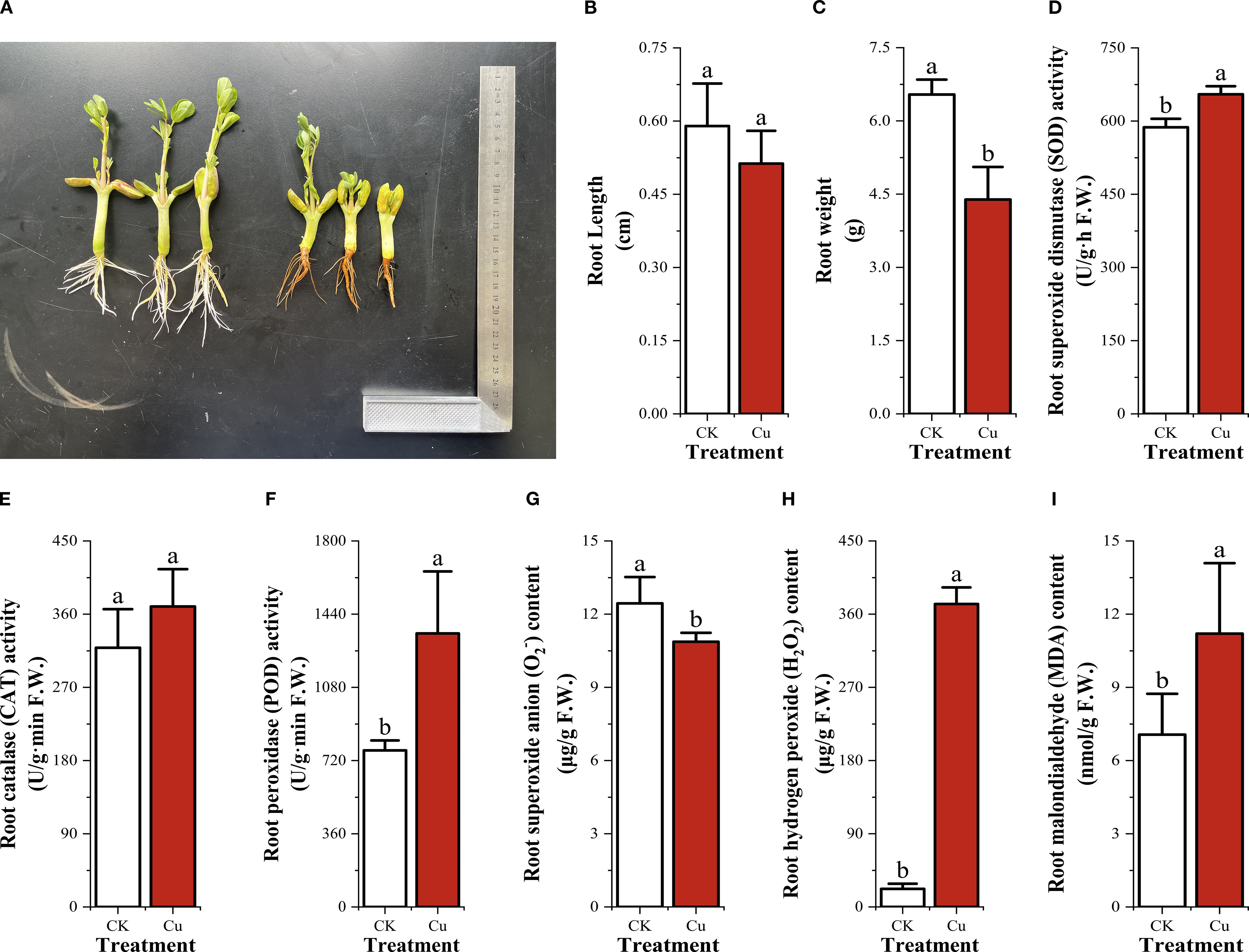
Figure 1. Root growth, secondary oxidative stress and antioxidant enzyme activities in peanut seedling roots under two treatments. (A) Root growth photographs; (B) Root length; (C) Root weight; (D) Superoxide dismutase (SOD) activity; (E) Catalase (CAT) activity; (F) Peroxidase (POD) activity; (G) Superoxide anion (O2-) content; (H) Hydrogen peroxide (H2O2) content; (I) Malondialdehyde (MDA) content.
Eight metal ion concentrations in peanut seeding roots under copper stress
After 48 h of exposure, ionomic profiling revealed marked shifts in root metal.
contents (Figure 2). Sodium (Na+) exhibited the sharpest decline, dropping by 61.6%from 130.517 mg/kg in the control (CK) to 50.033 mg/kg under Cu2+ stress. Potassium (K+) and calcium (Ca2+) showed modest variations: K+ decreased from 11.407 to 9.577mg/g (-16.0%), whereas Ca²+ slightly increased from 7.77 to 8.557 mg/g (+10.1%), although neither change reached statistical significance (p < 0.05).
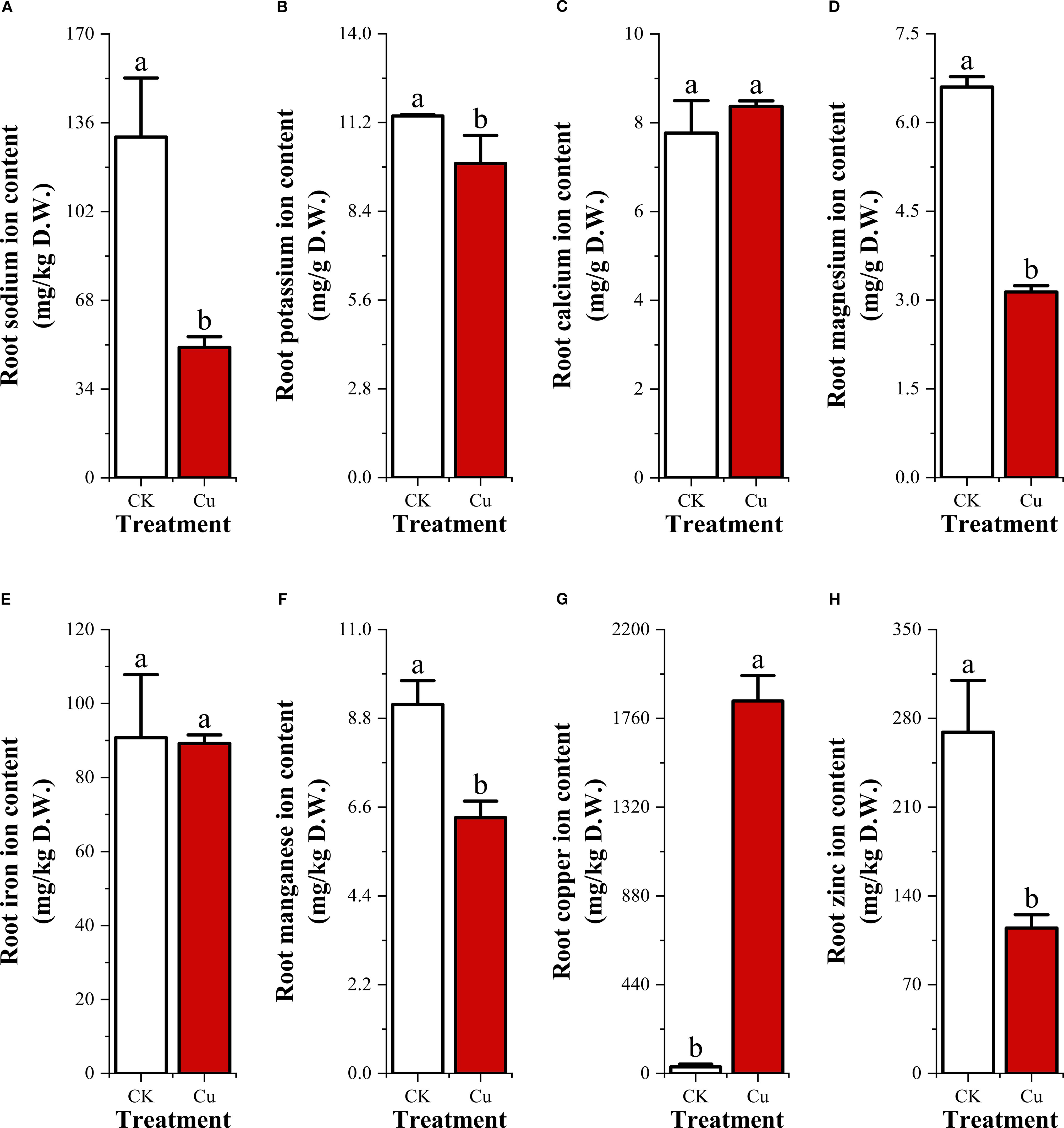
Figure 2. Contents of 8 metal cations in peanut seedling roots under two treatments. (A) Sodium ion (Na+); (B) Potassium ion (K+); (C) Calcium ion (Ca2+); (D) Magnesium ion (Mg2+); (E) Zinc ion (Zn2+); (F) Manganese ion (Mn2+); (G) Iron ion (Fe3+); (H) Copper ion (Cu2+).
Among divalent micronutrients, magnesium (Mg2+) fell by 52.5% (CK:6.6 mg/g; Cu2+:3.137 mg/g), zinc (Zn2+) by 57.4% (269.1 vs 114.547 mg/kg), and manganese (Mn2+) by 30.7% (9.143 vs 6.34 mg/kg). Iron (Fe3+) displayed only a marginal reduction of 1.8% (90.793 vs 89.173 mg/kg).
In contrast, copper (Cu2+) accumulation was dramatic. Root Cu2+ concentration surged 57.9-fold from 31.867mg/kg in CK to 1846.603 mg/kg under Cu2+ stress.
Collectively, the data indicate that excess exogenous Cu²+ not only caused extreme Cu2+ accumulation but also triggered substantial losses of Na+, K+, Mg2+, Zn2+, and Mn2+, while Ca2+ and Fe3+ remained relatively stable. This ionic imbalance underscores the severity of Cu2+-induced nutrient deficiency stress in peanut roots.
DEGs and enrichment analysis of peanut seedling roots under copper stress
Following 48 h of Cu2+ exposure, volcano plots revealed a striking transcriptional reprogramming (Figure 3). A total of 9901 DEGs were identified, with 2700 genes up-regulated and 7201 genes down-regulated, indicating that Cu2+ stress exerts a predominantly repressive effect on root gene expression.
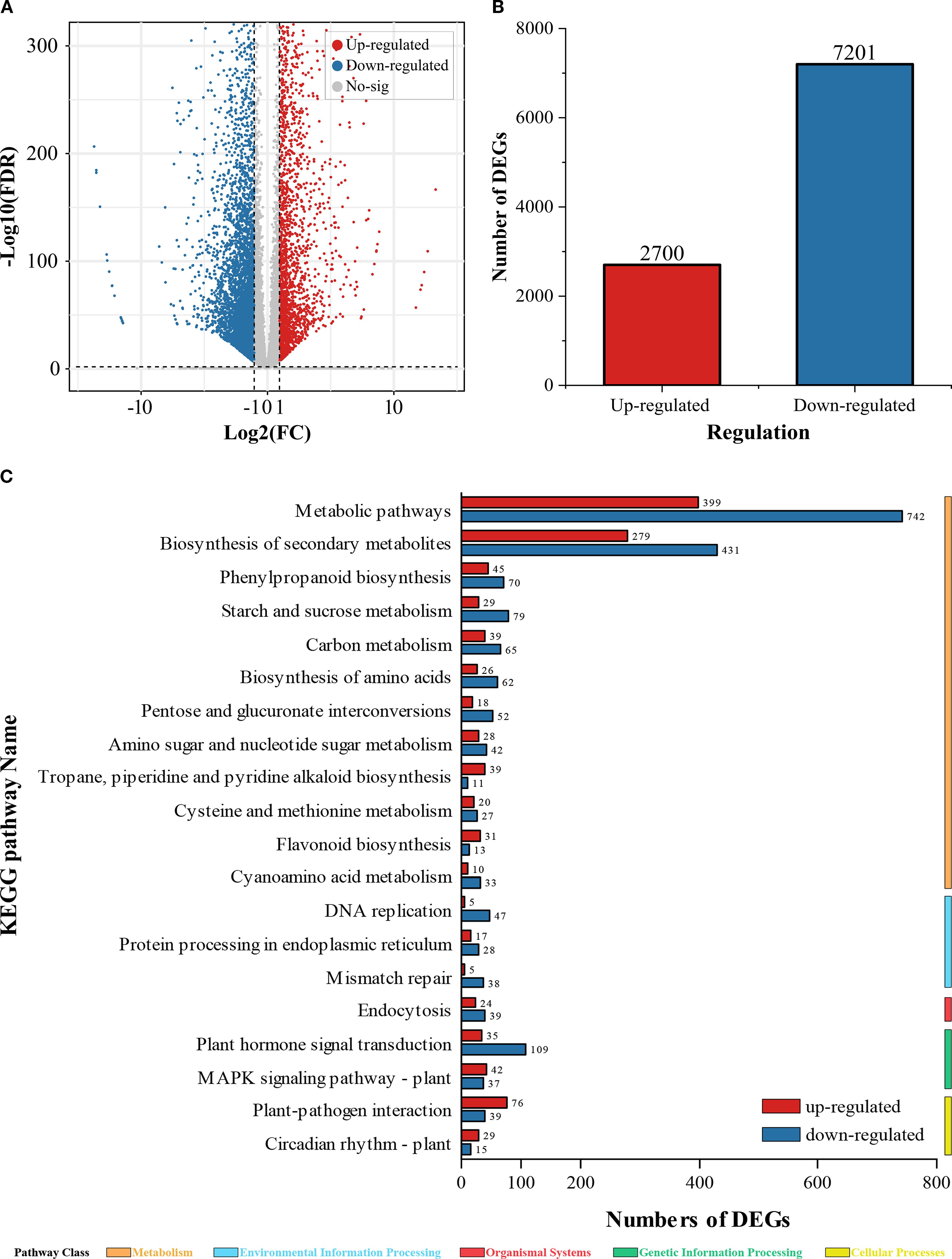
Figure 3. DEGs and KEGG enrichment analysis. (A) Volcano plot of DEGs; (B) Total count of up-regulated and down-regulated DEGs; (C) KEGG enrichment analysis of DEGs.
KEGG pathway enrichment analysis further uncovered significant perturbations in primary and secondary metabolism. Within the global “metabolic pathways” (ko01100), 743 genes were suppressed while only 399 were induced (down/up ratio = 1.86). This pronounced skew suggests a broad inhibition of carbohydrate, amino-acid, and energy metabolism, likely compromising ATP-dependent ion uptake and exacerbating nutrient deficiency.
Similarly, the “biosynthesis of secondary metabolites” (ko01110) displayed 431 down-regulated versus 279 up-regulated genes (down/up ratio =1.54). The disproportionate suppression of this pathway implies that Cu2+ stress hampers the production of flavonoids, phenylpropanoids, and other stress-protective compounds thereby weakening the root’s antioxidant defense network.
These transcriptomic signatures reveal that excess Cu2+ not only triggers massive transcriptional repression but also targets metabolic and secondary-metabolite pathways that are central to energy homeostasis and oxidative stress mitigation in peanut roots.
Figure 4 summarizes the Gene Ontology (GO) enrichment of differentially expressed genes (DEGs) in peanut roots exposed to 48 h of Cu2+ stress, focusing on the 20 terms with the smallest p-values. These categories illuminate the principal biological responses triggered by excess Cu2+.
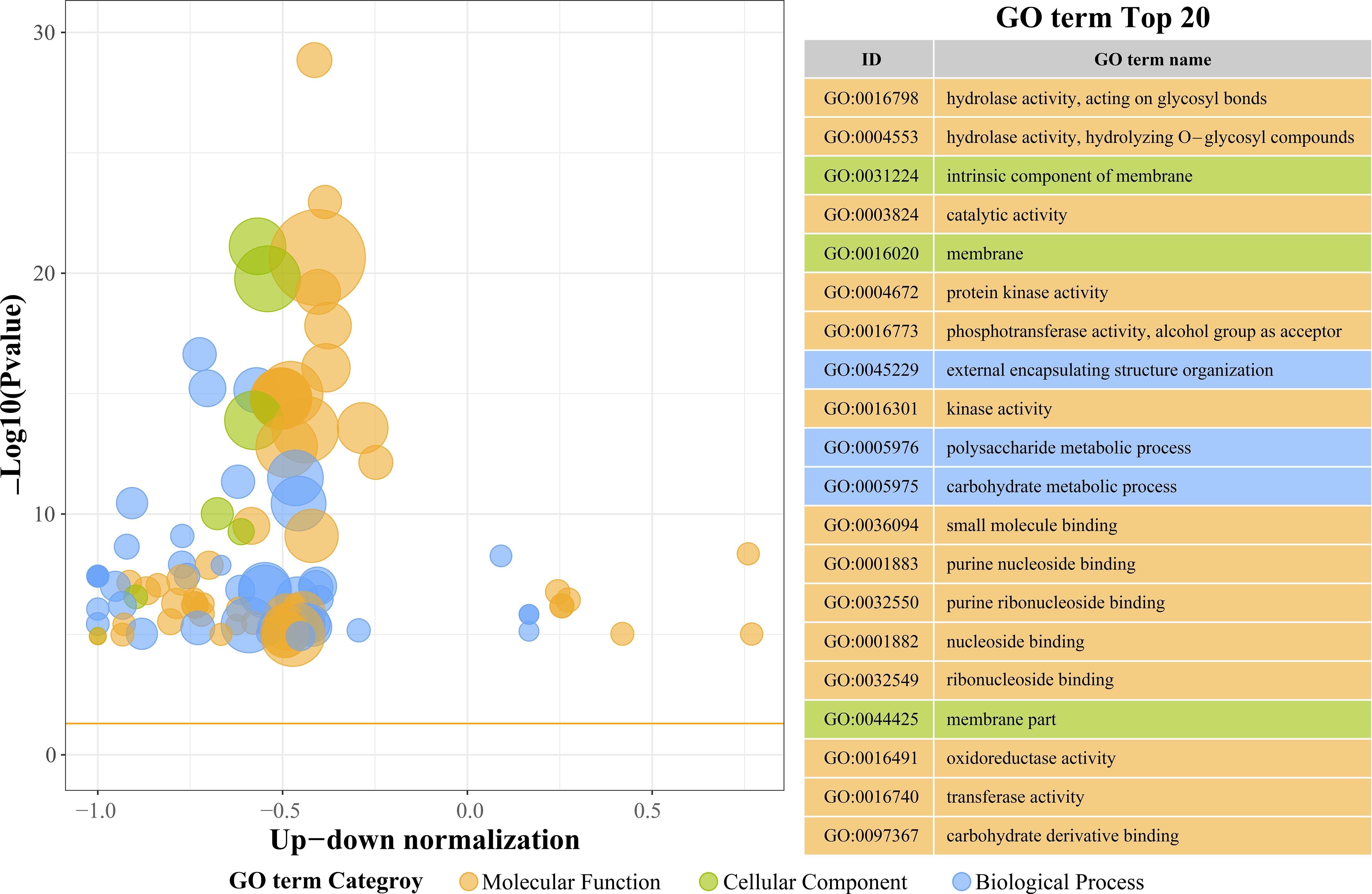
Figure 4. The top 20 GO terms with the smallest p-values in the Gene Ontology (GO) enrichment enrichment analysis.
From a Molecular Function (MF) perspective, we found the terms for hydrolase activity acting on glycosidic bonds (GO:0016798), hydrolase activity, hydrolyzing O-glycosyl compounds (GO:0004553), and general catalytic activity (GO:0003824). This means that hydrolase, oxidoreductase, and transferase pathways were significantly activated during Cu²+ stress. It is worth noting that oxidoreductase activity (GO:0016491) and transferase activity (GO:0016740) showed relatively weaker enrichment, which suggests that plants may primarily utilize glycosyltransferases and related enzymes to synthesize secondary metabolites to promote Cu²+ chelation and detoxification. Meanwhile, terms such as intrinsic component of membrane (GO:0031224) and plasma membrane (GO:0016020) were significantly enriched, which emphasizes the critical role of membrane-associated structures in Cu²+ perception and stress signaling. The extracellular region (GO:0044425) also showed a modest enrichment, indicating that membrane-resident peroxidative repair systems contribute to cell wall integrity under Cu²+ stress. Additionally, in terms of Biological Process (BP) terms, external encapsulating structure organization (GO:0045229) was the most prominent process, suggesting that plants rapidly strengthen their cell wall architecture to form a physical barrier, thereby limiting Cu²+ influx into root tissues. Polysaccharide metabolic process (GO:0005976) and carbohydrate metabolic process (GO:0005975) showed weaker enrichment, which indicates that short-term (48 h) Cu²+ stress prioritizes immediate defense responses, meaning that plant carbohydrate metabolism is affected.
Antioxidant-related DEGs in peanut seedling roots under copper stress
Some DEGs related to antioxidant enzyme coding and glutathione metabolism were listed in Figure 5. We found that under copper stress, only two genes encoding copper/zinc superoxide dismutase (Cu/Zn-SOD, CSD), LOC112706839 and LOC112772458, were transcriptionally upregulated. Interestingly, no DEGs encoding catalase (CAT) were detected. In stark contrast, the peroxidase (POD) superfamily exhibited a significant transcriptional response. A total of 53 DEGs encoding PER enzymes were identified, distributed across 20 subtypes, encoding PER3, PER4, PER52, and PER53. Notably, the PER52 subtype alone comprised 13 DEGs, suggesting a recent gene duplication event that may have conferred a selective advantage in reactive oxygen species (ROS) scavenging during Cu2+ stress. Additionally, two DEGs encoding L-ascorbate peroxidase (APX), LOC112747725 and LOC112800332, were also upregulated. This indicates that the transcriptional activation of POD enzyme-encoding genes plays a dominant role in the secondary oxidative stress response of peanut roots to copper stress.
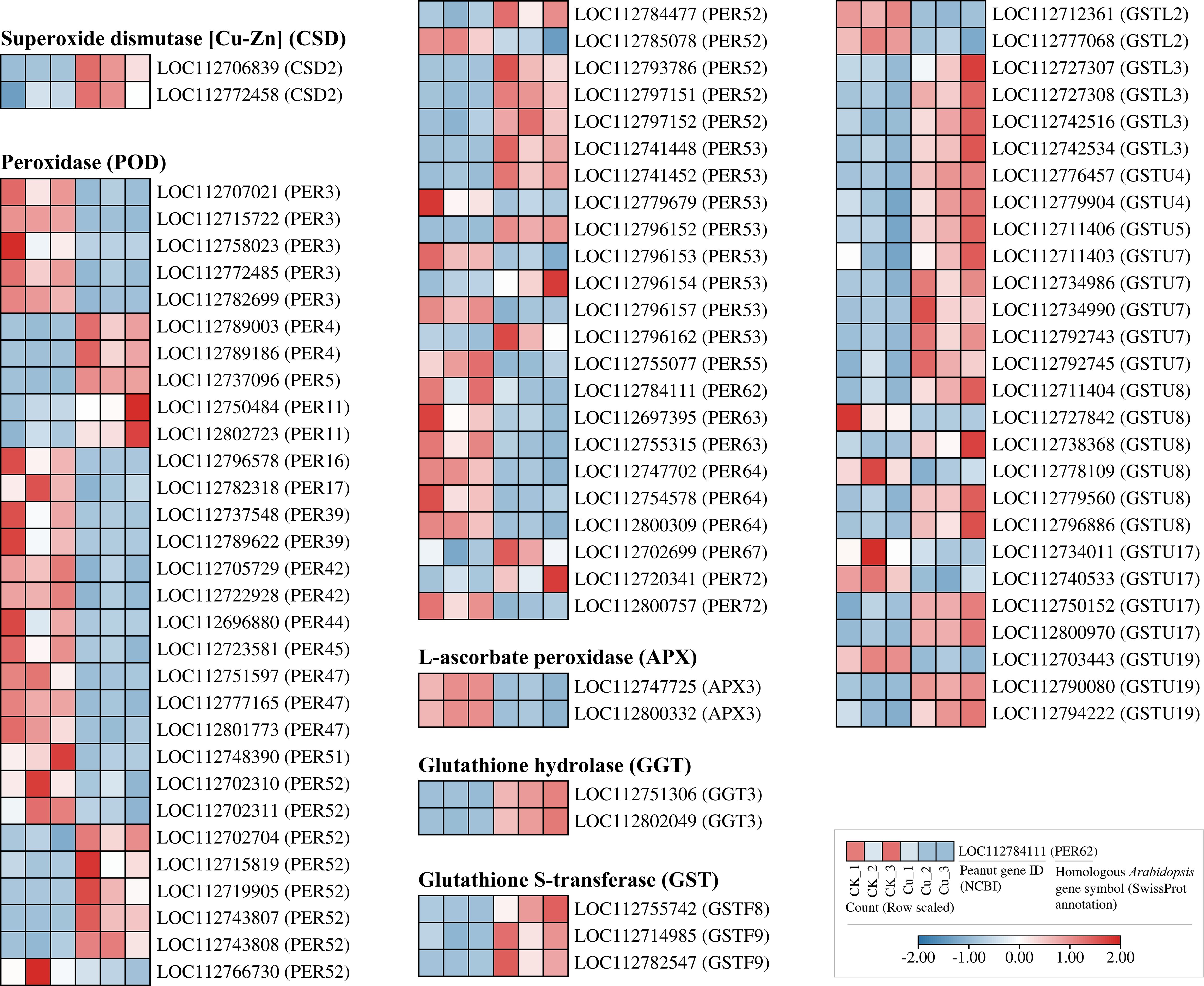
Figure 5. Heatmap of differentially expressed genes encoding antioxidant-related enzymes in peanut seedling roots under two treatments. CSD, Copper/Zinc Superoxide Dismutase; PER, Peroxidase; APX, Ascorbate Peroxidase; GGT, Gamma-Glutamyltransferase; GST, Glutathione S-Transferase; GSTU, Glutathione S-Transferase U.
The glutathione S-transferase (GST) superfamily contained 31 members, categorized into three subfamilies: GSTF (3 DEGs), GSTL (6 DEGs), and GSTU (21 DEGs). Almost all GST genes were upregulated, with the GSTU class showing the strongest response. Concurrently, two DEGs encoding glutathione hydrolase (GGT), LOC112751306 and LOC112802049, were also upregulated by copper stress. Collectively, these findings reveal that the conversion and hydrolysis of glutathione are crucial mechanisms for peanut roots to cope with copper stress, acting synergistically with the antioxidant enzyme system.
DEGs of mitogen-activated protein kinase cascade in peanut seedling roots under copper stress
Figure 6 listed some of the copper-responsive DEGs encoding MAPK signaling pathway proteins. We found that all five DEGs encoding MPK were upregulated by copper stress, and the upstream gene encoding MKK2, LOC112790474, was also upregulated. In contrast, most of the DEGs encoding MAPKKK isoforms were transcriptionally suppressed by Cu2+. Despite the transcriptional suppression of MAPKKK at the sampling time point, it is interesting to note that the transcription of its downstream MKK and MAPK modules was activated by copper stress. This finding may suggest that the core downstream modules of the MAPK signaling pathway are activated in response to copper stress, thereby playing a role in coping with the stress.
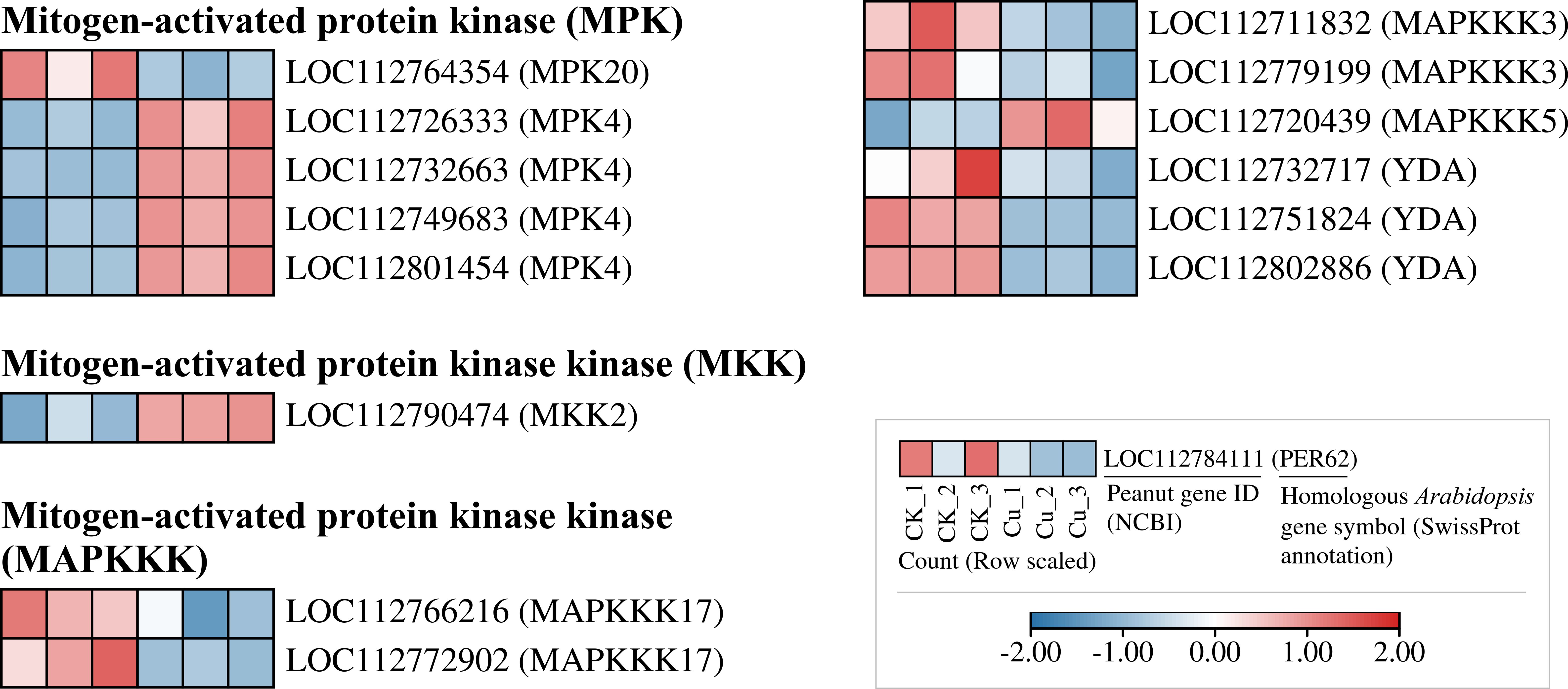
Figure 6. Heatmap of differentially expressed genes encoding MAPK signaling-related proteins in peanut seedling roots under two treatments. MPK, Mitogen-Activated Protein Kinase; MKK, Mitogen-Activated Protein Kinase Kinase; MAPKKK, Mitogen-Activated Protein Kinase Kinase Kinase; YDA, MAPKKK YODA.
Transcription Factor coding DEGs in peanut seedling roots under copper stress
We also investigated the response of transcription factor-encoding DEGs to copper stress, and in Figure 7, we found a large number of DEGs in eight common transcription factor (TF) families. Notably, some families showed a significant downregulation trend: 19 out of 21 DEGs in the bHLH family were consistently downregulated under copper stress. A similar trend was observed in the GRAS family (13 out of 15 DEGs) and the MYB family (13 out of 15 DEGs). In stark contrast, all DEGs from the NAC family (8 DEGs) and the LBD family (5 DEGs) were strongly upregulated by copper stress. This suggests that while most transcription factor genes are suppressed, members of the NAC and LBD families may be specifically responsible for regulating downstream stress and adaptive pathways in response to Cu2+ toxicity.
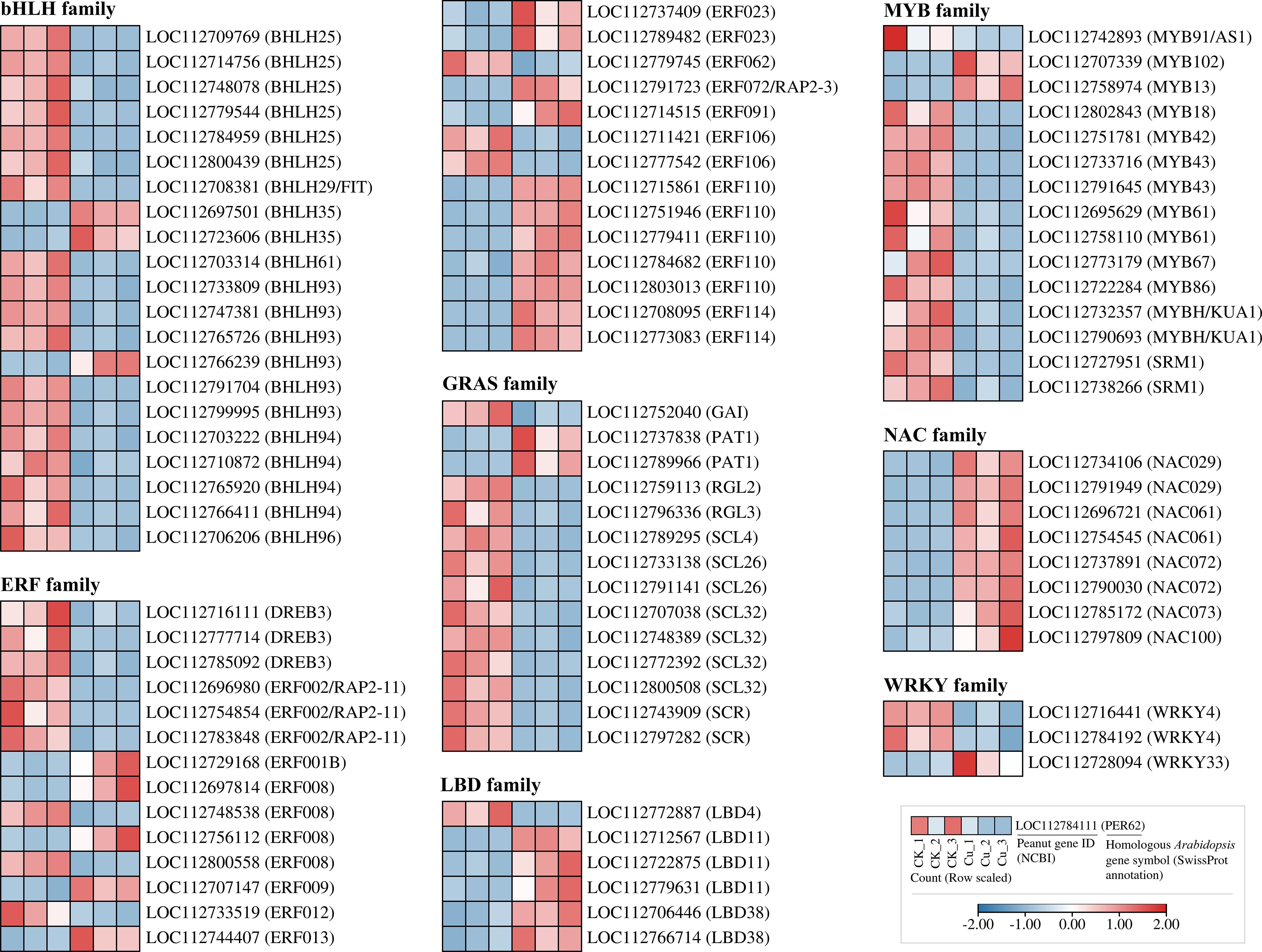
Figure 7. Heatmap of differentially expressed genes encoding transcription factors from seven families in peanut seedling roots under two treatments. bHLH, basic Helix-Loop-Helix; ERF, Ethylene Response Factor; DREB, Dehydration-Responsive Element-Binding Protein; RAP2, Related to AP2; GRAS, GRAS; GAI, GIBBERELLIN INSENSITIVE; PAT, PAT; RGL, RGA-LIKE; SCL, SCARECROW-LIKE; SCR, SCARECROW; LBD, Lateral Organ Boundaries Domain; MYB, Myeloblastosis; KUA, Kuan; SRM, Sorem; NAC, NAC domain protein; WRKY, WRKY domain protein.
Quantitative real-time polymerase chain reaction validation of high-throughput transcriptome sequencing (RNA-seq)
To validate the accuracy of the RNA sequencing (RNA-seq) data, we randomly selected 18 DEGs for quantitative real-time PCR (RT-qPCR) validation. These 18 genes included 4 DEGs related to the MAPK signaling pathway, 8 encoding transcription factor proteins, 2 encoding peroxidase (POD) enzymes, 2 encoding glutathione S-transferase, and 2 additional randomly selected genes.
As shown in Figure 8, among the 18 DEGs, the expression trends of 14 genes were consistent between the two testing methods, indicating a high reliability of the RNA-seq data. However, the RT-qPCR results for four DEGs (LOC112706158 (ERF071), LOC112726690 (ERF109), LOC112697019 (NAC037) and LOC112695735 (RRS1)) were inconsistent with the RNA-seq results, which might be attributed to technical differences between the methods or the dynamic nature of gene expression.
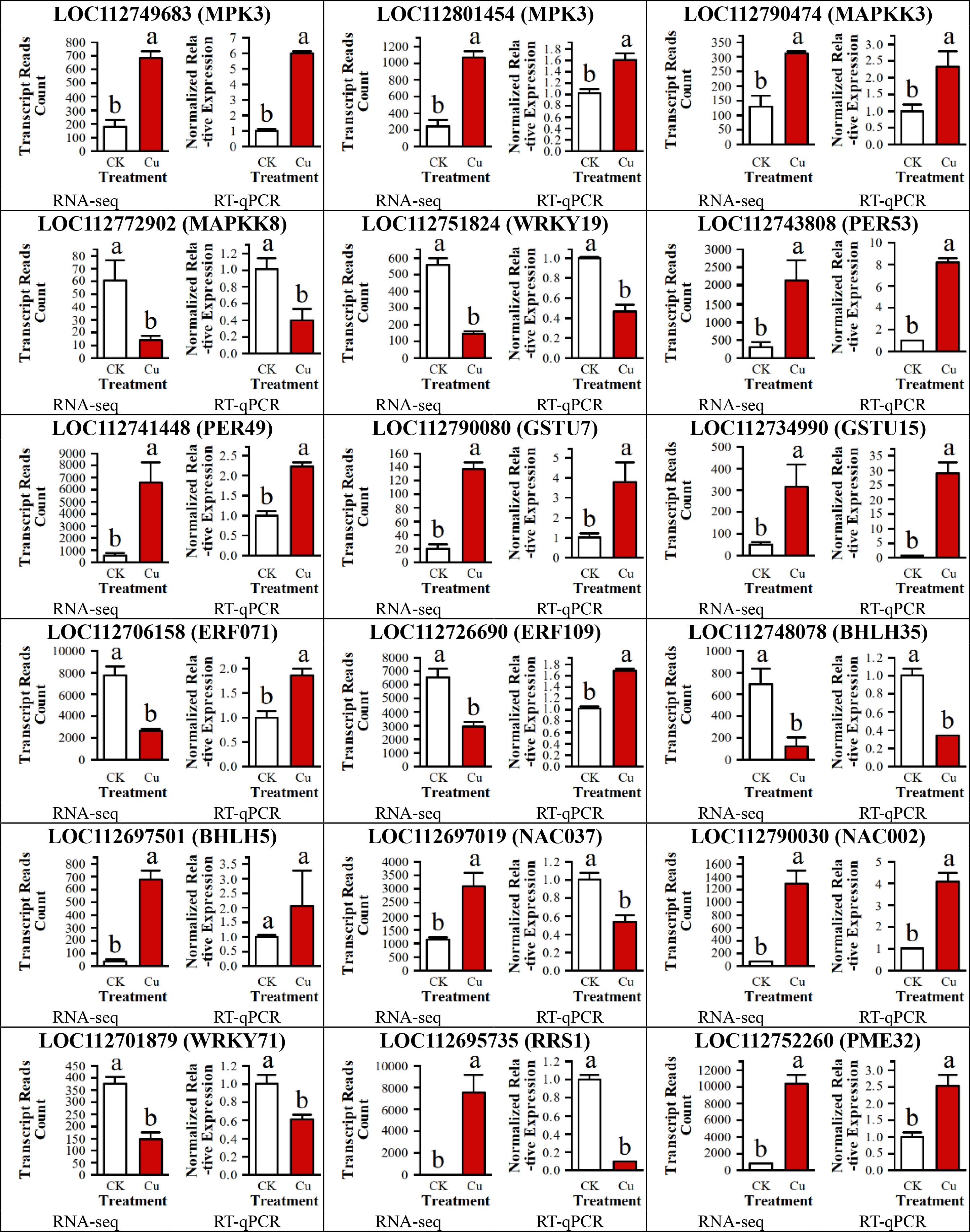
Figure 8. RT-qPCR validation of 18 DEGs in peanut seedling roots under two treatments. MAPKK, Mitogen-Activated Protein Kinase Kinase; WRKY, WRKY Transcription Factor; PER, Peroxidase; GSTU, Glutathione S-Transferase U; ERF, Ethylene Response Factor; bHLH, basic Helix-Loop-Helix; NAC, NAC Transcription Factor; RRS, Resistance to Root Rot Symptom; PME, Pectin Methylesterase.
Discussion
Secondary oxidative stress and antioxidation in peanut roots under copper stress
Elevated Cu2+ concentrations not only disrupt normal cellular physiology but also inhibit both cell division and elongation, ultimately retarding overall plant growth. In the present study, Cu2+ exposure induced pronounced oxidative stress in peanut roots, as evidenced by a 1.5-fold increase in malondialdehyde (MDA) content relative to the control. This elevation indicates extensive peroxidation of membrane lipids and, consequently, severe oxidative injury to root cell membranes (Saleem et al., 2020).
Interestingly, although superoxide anion (O2-) levels decreased, hydrogen peroxide (H2O2) accumulated 17-fold under Cu2+ stress. This apparent paradox aligns with the dual role of H2O2 as both a reactive oxygen species and a signaling molecule. At elevated concentrations, H2O2 operates in a dose-dependent manner to activate antioxidant defenses and to modulate the expression of stress-responsive genes, thereby exerting profound regulatory effects on plant growth and development (Khedia et al., 2019).
Under copper stress, the activities of three key antioxidant enzymes in peanut seedling roots increased markedly: superoxide dismutase (SOD) rose by 11%, catalase (CAT) by 16%, and peroxidase (POD) by 75% (Figure 1). This concerted up-regulation indicates that peanut initiates a robust enzymatic antioxidant defense to mitigate Cu2+-induced reactive oxygen species (ROS) accumulation and to maintain cellular redox homeostasis.
This pattern aligns with previous reports in which cadmium or other heavy-metal stresses similarly enhance SOD and CAT activities to quench ROS and limit oxidative damage (Liu et al., 2014). The disproportionately strong induction of POD observed here, however, suggests that peanut may deploy a distinct POD-centric detoxification strategy under Cu2+ stress. Whether this reflects an isoform-specific POD expansion, substrate preference, or interaction with Cu2+-chelating metabolites remains to be elucidated.
Oxygen ion content in peanut roots under copper stress
Copper stress elicited pronounced perturbations in the concentrations of eight metal ions in peanut roots (Figure 2). As we observed, Cu2+ exhibited a dramatic surge (≈ 58-fold), while the levels of Na+, K+, Mg2+, Zn2+, and Mn2+ declined significantly.
Integrated cation contents data and RNA-seq evidence suggests that this disruption of ion balance is mechanistically linked to the transcriptional regulation of cation-transport genes. The underlying mechanism involves competitive binding to shared transporters, such as cation channels, heavy-metal ATPases (HMAs), and natural resistance-associated macrophage proteins (NRAMPs). Specifically, the downregulation of shaker-type K+ channel transcripts (e.g., AKT/KAT homologs) and high-affinity K+ transporters (HKTs) directly reduces K+ uptake, while the concomitant suppression of ZIP and NRAMP family members restricts the acquisition of Zn2+ and Mn2+ (Gao et al., 2008; Kazemi-Dinan et al., 2014; Xu et al., 2015).
Concurrently, the plant initiates active defense mechanisms. We observed the upregulation of genes encoding Cu2+-chelating agents (e.g., metallothioneins, phytochelatin synthases) and Cu2+-transporting P-type ATPases (HMAs), which facilitates the intracellular sequestration of Cu2+ and its vacuolar compartmentalization. This process further exacerbates Cu2+ enrichment in the roots while simultaneously intensifying the deficiencies of competing cations due to impaired uptake. Furthermore, Cu2+-induced oxidative stress compromises membrane integrity and ion selectivity, and the inhibition of root elongation along with the reduction in root-hair density indirectly exacerbates these nutrient deficiencies.
Nevertheless, the marked decline in Zn2+ levels is likely to exert deleterious effects on plant growth and development, as zinc functions as an essential catalytic component of numerous enzymes and is indispensable for photosynthesis, respiration, and other vital physiological processes (Kazemi-Dinan et al., 2014). Collectively, these findings demonstrate that copper stress imposes severe negative impacts on peanut growth and, under extreme conditions, may accelerate the onset of senescence and death in peanut as well as in other plant species.
The effect of copper stress on gene transcription in peanut roots
RNA-seq analysis revealed extensive transcriptional reprogramming in peanut seedling roots under Cu2+ stress: 2700 genes were up-regulated and 7201 genes were down-regulated (Figure 3). Volcano plots and enrichment profiling indicated that these differentially expressed genes (DEGs) were predominantly enriched in “metabolic pathways” and “biosynthesis of secondary metabolites” (ko01100 and ko01110), corroborating that excess Cu2+ broadly perturbs primary and specialized metabolism (Bao et al., 2021), thereby impairing normal physiological functions.
Gene Ontology (GO) enrichment further highlighted significant over-representation of molecular functions related to oxidoreductase and transferase activities (Figure 4). The concordance between these GO terms and the observed increases in antioxidant enzyme activities substantiates that peanut enhances its ROS-scavenging capacity by transcriptional up-regulation of antioxidant-related genes, thereby improving survival under Cu2+ stress (Leng et al., 2015).
The MAPK cascade is a highly conserved signaling pathway that transmits extracellular stimuli through sequential phosphorylation events, thereby modulating gene expression, cell proliferation, differentiation, and programmed cell death (Seger and Krebs, 1995). In plants, this cascade has been shown to orchestrate responses to a wide array of abiotic stresses. For instance, early studies in Arabidopsis revealed that Fe deficiency triggers MAPK-mediated phosphorylation cascades that modulate downstream transcription factors (TFs), thereby initiating defense reactions (Jian et al., 2024). Similarly, in mammalian systems, JNK and p38-MAPKs govern inflammatory and apoptotic responses, whereas the ERK pathway primarily regulates cell growth and differentiation (Moustafa et al., 2014). In our study, the Cu²+-induced upregulation of MPK4 and MKK2 (Figure 6) and the strong upregulation of all 13 DEGs from the LBD and NAC transcription factor families (Figure 7) occurred concurrently. This co-expression pattern may suggest that under copper stress, MPK4 protein in peanut roots has the opportunity to phosphorylate members of the LBD and NAC families, thereby activating downstream stress-responsive genes for Cu²+ detoxification or adaptation (Guo et al., 2021).
NAC transcription factors are widely recognized for their significant role in improving plant tolerance to abiotic stresses such as drought and salinity (Shu et al., 2024). In recent years, an increasing number of studies have also begun to uncover their new functions in enhancing plant resistance to heavy metal stress. For instance, heterologous overexpression of the AemNAC2 gene from Aegilops markgrafii in wheat significantly enhanced the plant’s tolerance to cadmium stress while markedly reducing the cellular cadmium levels (Du et al., 2020). Similarly, in rice, overexpression of OsNAC300 increased the tolerance of transgenic rice to cadmium stress, whereas its knockdown resulted in increased sensitivity to the heavy metal (Liu et al., 2023). Furthermore, a study involving the heterologous expression of the EuNAC9 gene from Eucommia ulmoides in yeast demonstrated that it enhanced the yeast’s tolerance to both copper and manganese stress. This was accompanied by the upregulation of the yeast ScSMF1 and the ScSOD2 (Zhan et al., 2024). However, direct evidence is still scarce regarding the precise heavy metal-responsive genes whose transcription is regulated by these factors.
Although LBD proteins are widely recognized for their role in regulating lateral organ development (e.g., lateral roots and leaves) (Lee et al., 2009, Lee et al., 2019), some reports have also indicated their function in responding to salt and drought stresses (Guan et al., 2023; Chen et al., 2024). However, their role in enhancing plant heavy metal tolerance is seldom reported. Based on the findings of the present study (Figure 4 and Figure 5), the upregulation of LBD-encoding genes may have a deeper significance. Under copper stress, the plant might actively reduce lateral root growth by upregulating these genes. This strategy is likely aimed at reallocating limited biological resources from root morphological development to more critical defense mechanisms, such as GSH-dependent heavy metal detoxification, POD-mediated antioxidation and cell wall biosynthesis, thereby prioritizing cell survival and homeostasis.
Additionally, although authentic Cu2+ receptors remain unidentified, our RNA-seq revealed several DEGs encoding membrane proteins — such as receptor-like kinases (RLKs), wall-associated kinases (WAKLs), LRR-RLKs, and ZIP transporters (Supplementary Table S3) — as candidate upstream Cu2+-perception components for future validation.
In this study, the MAPK cascade is positioned as a central hub: upstream Cu2+ signals (extracellular Cu2+ → ROS/NO burst → MAPKKK activation) feed into the module, while downstream phosphorylated MPKs target WRKY, bHLH and ERF transcription factors (Figure 6, 7). RNA-seq identified eight differentially expressed MAPKKKs, one MAPKK and five MPKs, together with 75 TF genes (21 bHLH, 28 ERF, 8 NAC etc.), providing the first transcriptional evidence that this linear “Cu2+ → ROS/NO → MAPK → TF → defense gene” axis operates in peanut roots under graded Cu2+ stress. The precise molecular interactions among these MAPK components and their target TFs warrant further investigation.
Conclusions
Based on the integrated physiological and transcriptomic studies, this research definitively demonstrates that the gene response in peanut seedling roots exhibits remarkable specificity within the “Cu²+ → ROS/NO → MAPK → TF → defense gene” model. Specifically, we have shown that copper stress strongly induces the expression of MPK4, a key nodal point within the MAPK pathway. Post-translationally, MPK4 is highly likely to phosphorylate two critical protein classes: NAC and LBD. Within this unique regulatory network, NAC functions as a core transcription factor, directly regulating the transcription of copper defense-related genes. Concurrently, LBD directly down-regulates genes associated with lateral root growth. This action by LBD, through a reallocation of biological resources, indirectly promotes the increased expression of genes involved in GSH-dependent heavy metal detoxification and secondary oxidative stress (e.g., GST and POD), thereby cooperatively enhancing the plant’s detoxification and antioxidant capacity.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at the National Center for Biotechnology Information (NCBI) using accession number PRJNA1311253.
Author contributions
XB: Visualization, Resources, Formal Analysis, Funding acquisition, Validation, Writing – original draft, Project administration, Investigation, Methodology, Conceptualization. WN: Writing – review – editing, Software, Writing – original draft, Methodology. MH: Writing – original draft. ZG: Methodology, Software, Writing – original draft, Writing – review – editing. XD: Software, Writing – original draft, Resources, Project administration, Data curation, Methodology.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Open Project Program of Panxi Crop Improvement Key Laboratory of Sichuan Province (SZ22ZZ07), Liangshan Science and Technology Program (22ZDYF0186), Liangshan Science and Technology Program (25YYYJ0159).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1667121/full#supplementary-material
References
Afzal, A., Shafqat, A., Akhtar, S., Sultana, T., Kazmi, A., Ali, A., et al. (2022). Biosorbents removed copper heavy metal from agricultural land cultivated with vigna radiata (Mung bean). Int. J. Agron. 2022, 1–14. doi: 10.1155/2022/6067181
Ammar, W. B., Haouari, C. C., Rezgui, M., Nazim, M., and Soufan, W. (2025). Exogenous alpha-ketoglutarate (AKG) modulate physiological characteristics, photosynthesis, secondary metabolism and antioxidant defense system in peganum harmala L. under nickel stress. Phyton 94, 137–155. doi: 10.32604/phyton.2025.058851
Bao, X., Lin, G., Dong, X., Ma, X., Han, X., and Zhang, H. (2021). Synergistic effects of copper and ethylene on resveratrol synthesis in peanuts. Food Sci Nutr. 9, 1471–1479. doi: 10.1002/fsn3.2117
Borkert, C. M. and Cox, F. R. (1999). Effects of acidity at high soil zinc, copper, and manganese on peanut, rice, and soybean. Commun. Soil Sci Plant Anal. 30, 1371–1384. doi: 10.1080/00103629909370293
Cai, H., Wang, H., Zhou, L., Li, B., Zhang, S., He, Y., et al. (2023). Time-series transcriptomic analysis of contrasting rice materials under heat stress reveals a faster response in the tolerant cultivar. IJMS 24, 9408. doi: 10.3390/ijms24119408
Chen, L., Ji, X., Luo, C., Song, X., Leng, X., Ma, Y., et al. (2024). VvLBD39, a grape LBD transcription factor, regulates plant response to salt and drought stress. Environ. Exp. Bot. 226, 105918. doi: 10.1016/j.envexpbot.2024.105918
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Demecsová, L., Zelinová, V., Liptáková, Ľ., and Tamás, L. (2020). Mild cadmium stress induces auxin synthesis and accumulation, while severe cadmium stress causes its rapid depletion in barley root tip. Environ. Exp. Bot. 175, 104038. doi: 10.1016/j.envexpbot.2020.104038
Dong, X., Gao, Y., Bao, X., Wang, R., Ma, X., Zhang, H., et al. (2023). Multi-omics revealed peanut root metabolism regulated by exogenous calcium under salt stress. Plants 12, 3130. doi: 10.3390/plants12173130
Du, X., He, F., Zhu, B., Ren, M., and Tang, H. (2020). NAC transcription factors from Aegilops markgrafii reduce cadmium concentration in transgenic wheat. Plant Soil 449, 39–50. doi: 10.1007/s11104-019-04419-w
Fang, H., Liu, Z., Long, Y., Liang, Y., Jin, Z., Zhang, L., et al. (2017). The Ca2+/calmodulin2-binding transcription factor TGA 3 elevates LCD expression and H2 S production to bolster Cr6+ tolerance in Arabidopsis. Plant J. 91, 1038–1050. doi: 10.1111/tpj.13627
Gao, S., Yan, R., Cao, M., Yang, W., Wang, S., and Chen, F. (2008). Effects of copper on growth, antioxidant enzymes and phenylalanine ammonia-lyase activities in Jatropha curcas L. seedling. Plant Soil Environ. 54, 117–122. doi: 10.17221/2688-PSE
Guan, C., Wu, B., Ma, S., Zhang, J., Liu, X., Wang, H., et al. (2023). Genome-wide characterization of LBD transcription factors in switchgrass (Panicum virgatum L.) and the involvement of PvLBD12 in salt tolerance. Plant Cell Rep. 42, 735–748. doi: 10.1007/s00299-023-02989-9
Guo, J., Sun, B., He, H., Zhang, Y., Tian, H., and Wang, B. (2021). Current understanding of bHLH transcription factors in plant abiotic stress tolerance. IJMS 22, 4921. doi: 10.3390/ijms22094921
Jian, H., Sadau, S. B., Wei, F., Ahmad, A., Lu, Z., Ma, L., et al. (2024). GhMAPK3, a mitogen-activated protein kinase, enhance salt and drought tolerance in cotton (Gossypium hirsutum). Ind. Crops Products 214, 118492. doi: 10.1016/j.indcrop.2024.118492
Kazemi-Dinan, A., Thomaschky, S., Stein, R. J., Krämer, U., and Müller, C. (2014). Zinc and cadmium hyperaccumulation act as deterrents towards specialist herbivores and impede the performance of a generalist herbivore. New Phytol. 202, 628–639. doi: 10.1111/nph.12663
Khan, J., Malangisha, G. K., Ali, A., Mahmoud, A., Yang, J., Zhang, M., et al. (2021). Nitric oxide alleviates lead toxicity by inhibiting lead translocation and regulating root growth in watermelon seedlings. Hortic. Environ. Biotechnol. 62, 701–714. doi: 10.1007/s13580-021-00346-x
Khedia, J., Agarwal, P., and Agarwal, P. K. (2019). Deciphering hydrogen peroxide-induced signalling towards stress tolerance in plants. 3 Biotech. 9, 395. doi: 10.1007/s13205-019-1924-0
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kobayashi, T., Nozoye, T., and Nishizawa, N. K. (2019). Iron transport and its regulation in plants. Free Radical Biol. Med. 133, 11–20. doi: 10.1016/j.freeradbiomed.2018.10.439
Lee, H. W., Cho, C., Pandey, S. K., Park, Y., Kim, M.-J., and Kim, J. (2019). LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 19, 46. doi: 10.1186/s12870-019-1659-4
Lee, H. W., Kim, N. Y., Lee, D. J., and Kim, J. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in arabidopsis. Plant Physiol. 151, 1377–1389. doi: 10.1104/pp.109.143685
Leng, X., Jia, H., Sun, X., Shangguan, L., Mu, Q., Wang, B., et al. (2015). Comparative transcriptome analysis of grapevine in response to copper stress. Sci. Rep. 5, 17749. doi: 10.1038/srep17749
Li, S., Han, X., Lu, Z., Qiu, W., Yu, M., Li, H., et al. (2022). MAPK cascades and transcriptional factors: regulation of heavy metal tolerance in plants. IJMS 23, 4463. doi: 10.3390/ijms23084463
Li, X., Mao, X., Xu, Y., Li, Y., Zhao, N., Yao, J., et al. (2021). Comparative transcriptomic analysis reveals the coordinated mechanisms of Populus × canadensis ‘Neva’ leaves in response to cadmium stress. Ecotoxicology Environ. Saf. 216, 112179. doi: 10.1016/j.ecoenv.2021.112179
Li, X., Zhang, J., Gong, Y., Liu, Q., Yang, S., Ma, J., et al. (2020). Status of copper accumulation in agricultural soils across China–2016). Chemosphere 244, 125516. doi: 10.1016/j.chemosphere.2019.125516
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, X.-M., Kim, K. E., Kim, K.-C., Nguyen, X. C., Han, H. J., Jung, M. S., et al. (2010). Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species. Phytochemistry 71, 614–618. doi: 10.1016/j.phytochem.2010.01.005
Liu, J., Qiao, Y., Li, C., and Hou, B. (2023). The NAC transcription factors play core roles in flowering and ripening fundamental to fruit yield and quality. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1095967
Liu, J. J., Wei, Z., and Li, J. H. (2014). Effects of copper on leaf membrane structure and root activity of maize seedling. Bot. Stud. 55, 47. doi: 10.1186/s40529-014-0047-5
Moustafa, K., AbuQamar, S., Jarrar, M., Al-Rajab, A. J., and Trémouillaux-Guiller, J. (2014). MAPK cascades and major abiotic stresses. Plant Cell Rep. 33, 1217–1225. doi: 10.1007/s00299-014-1629-0
Peralta, J. M., Travaglia, C., Romero-Puertas, M. C., Molina-Moya, E., Furlan, A., Castro, S., et al. (2022). Decoding the antioxidant mechanisms underlying arsenic stress in roots of inoculated peanut plants. Plant Growth Regul. 97, 77–90. doi: 10.1007/s10725-022-00803-2
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saleem, M. H., Fahad, S., Rehman, M., Saud, S., Jamal, Y., Khan, S., et al. (2020). Morpho-physiological traits, biochemical response and phytoextraction potential of short-term copper stress on kenaf (Hibiscus cannabinus L.) seedlings. PeerJ 8, e8321. doi: 10.7717/peerj.8321
Sapara, K. K., Khedia, J., Agarwal, P., Gangapur, D. R., and Agarwal, P. K. (2019). SbMYB15 transcription factor mitigates cadmium and nickel stress in transgenic tobacco by limiting uptake and modulating antioxidative defence system. Funct. Plant Biol. 46, 702. doi: 10.1071/FP18234
Seger, R. and Krebs, E. G. (1995). The MAPK signaling cascade. FASEB J. 9, 726–735. doi: 10.1096/fasebj.9.9.7601337
Sharma, S. S., Kumar, V., and Dietz, K.-J. (2021). Emerging trends in metalloid-dependent signaling in plants. Trends Plant Sci 26, 452–471. doi: 10.1016/j.tplants.2020.11.003
Shu, L., Li, L., Jiang, Y.-Q., and Yan, J. (2024). Advances in membrane-tethered NAC transcription factors in plants. Plant Sci 342, 112034. doi: 10.1016/j.plantsci.2024.112034
Sun, X., Chen, Y.-L., Xin, F., and Zhang, S. (2024). Transcriptome-wide identification and analysis reveals m6A regulation of metabolic reprogramming in shrimp (Marsupenaeus japonicus) under virus infection. BMC Genomics 25, 1103. doi: 10.1186/s12864-024-11032-4
Takatsuji, H. (1998). Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. (CMLS) 54, 582–596. doi: 10.1007/s000180050186
Dong, X., Gao, Y., Bao, X., Wang, R., Ma, X., Zhang, H., et al. (2023). Multi-omics revealed peanut root metabolism regulated by exogenous calcium under salt stress. Plants 12, 3130. doi: 10.3390/plants12173130
Wang, F.-Z., Chen, M.-X., Yu, L.-J., Xie, L.-J., Yuan, L.-B., Qi, H., et al. (2017). OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01868
Wang, K., Shao, Z., Guo, F., Wang, K., and Zhang, Z. (2021). The mitogen-activated protein kinase kinase TaMKK5 mediates immunity via the TaMKK5–TaMPK3–TaERF3 module. Plant Physiol. 187, 2323–2337. doi: 10.1093/plphys/kiab227
Wu, B., Peng, H., Sheng, M., Luo, H., Wang, X., Zhang, R., et al. (2021). Evaluation of phytoremediation potential of native dominant plants and spatial distribution of heavy metals in abandoned mining area in Southwest China. Ecotoxicology Environ. Saf. 220, 112368. doi: 10.1016/j.ecoenv.2021.112368
Xu, Z., Dong, M., Peng, X., Ku, W., Zhao, Y., and Yang, G. (2019). New insight into the molecular basis of cadmium stress responses of wild paper mulberry plant by transcriptome analysis. Ecotoxicology Environ. Saf. 171, 301–312. doi: 10.1016/j.ecoenv.2018.12.084
Xu, X., Huang, Z., Wang, C., Zhong, L., Tian, Y., Li, D., et al. (2015). Toxicological effects, mechanisms, and implied toxicity thresholds in the roots of Vicia faba L. seedlings grown in copper-contaminated soil. Environ. Sci. pollut. Res. 22, 13858–13869. doi: 10.1007/s11356-015-5073-7
Yadav, B., Chhaya, D. R., Gnanasekaran, P., and Narayan, O. P. (2021). OMICS approaches towards understanding plant’s responses to counterattack heavy metal stress: An insight into molecular mechanisms of plant defense. Plant Gene 28, 100333. doi: 10.1016/j.plgene.2021.100333
Zhai, N., Jia, H., Liu, D., Liu, S., Ma, M., Guo, X., et al. (2017). GhMAP3K65, a cotton raf-like MAP3K gene, enhances susceptibility to pathogen infection and heat stress by negatively modulating growth and development in transgenic nicotiana benthamiana. IJMS 18, 2462. doi: 10.3390/ijms18112462
Keywords: Arachis hypogaea, copper stress, physiological response, MAPK pathway, gene expression
Citation: Bao X, Ning W, Gao Z, Hu M and Dong X (2025) MAPK-dependent copper tolerance mechanisms revealed by integrated physiology and transcriptomics in peanut. Front. Plant Sci. 16:1667121. doi: 10.3389/fpls.2025.1667121
Received: 16 July 2025; Accepted: 22 September 2025;
Published: 21 October 2025.
Edited by:
Mariela Torres, Instituto Nacional de Tecnología Agropecuaria (Argentina), ArgentinaReviewed by:
Rohit Joshi, Institute of Himalayan Bioresource Technology (CSIR), IndiaThounaojam Thorny Chanu, Assam Don Bosco University, India
Copyright © 2025 Bao, Ning, Gao, Hu and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuan Dong, eGNjMjAyNDAwMDlAeGNjLmVkdS5jbg==
 XueFeng Bao
XueFeng Bao WeiMin Ning
WeiMin Ning ZhiQiang Gao
ZhiQiang Gao Xuan Dong
Xuan Dong