- 1Julius Kühn Institute, Institute for Plant Protection in Field Crops and Grassland, Braunschweig, Germany
- 2Faculty of Organic Agricultural Sciences, section Ecological Plant Protection, Universität Kassel, Witzenhausen, Germany
- 3INRAE, Université Côte d’Azur, CNRS, Institut Sophia-Agrobiotech, Sophia Antipolis, France
The root-knot nematode Meloidogyne enterolobii poses a significant challenge in agricultural production systems due to its damage potential and the ability to overcome plant resistance genes, which are effective against other root-knot nematode species. With little plant resistance available, few nematicides still allowed, crop rotation with non- or poor host plants is the only option for managing M. enterolobii. As virulence and pathogenicity can vary between Meloidogyne populations, determination of the intrinsic host range and pathogenicity of M. enterolobii populations is crucial for the implementation of effective management strategies in the future. In greenhouse experiments, the host range and pathogenicity of seven M. enterolobii populations were tested on 19 plant species. In addition, two populations of M. incognita, virulent against tomato Mi-1resistance gene, were included in this study, as they had demonstrated a similar range of reproductive potential and damage compared to M. enterolobii. The study revealed that tomato, eggplant, pepper, tobacco, cucumber, potato, bean, melon, sugar beet, yellow mustard, and soybean were good hosts for all tested Meloidogyne populations. However, variations in reproduction among populations were observed in carrot, cotton, phacelia, fodder radish, maize, sunflower, and peanut. In rose, none of the M. enterolobii populations reproduced (reproduction factor: RF< 0.1). However, virulent M. incognita populations allowed some multiplication with RF > 0.1, but below 1.0. Curiously, three M. enterolobii populations (M.ent3, 4 and5) showed a lower RF compared to the remaining populations, but were more damaging, resulting in reduced root and shoot fresh weight of the majority of the host plants tested. This is the first study comparing multiple populations of M. enterolobii, including the two type populations, from different geographic regions with a large panel of plant species. This study provides crucial information to develop new and sustainable control strategies against the quarantine nematode M. enterolobii.
Introduction
Meloidogyne enterolobii was initially described by Yang and Eisenback in 1983 from samples obtained from the roots of Enterolobium contortisiliquum (pacara earpod tree) in Hainan province, China. In 1988, a morphologically very similar species, Meloidogyne mayaguensis, was described by Rammah and Hirschmann who found this species on roots of Solanum melongena (eggplant) in Puerto Rico. Due to the similarities in morphological features, it was long suspected that they were both M. enterolobii and based on molecular data Karssen et al. (2012) confirmed that M. mayaguensis was as a junior synonym for M. enterolobii. Reports in recent years have shown the global distribution and broad host range of M. enterolobii, affecting a variety of vegetables, field crops, tree crops, ornamental plants, and weeds (Khanal and Harshman, 2021). Advances in molecular diagnostics have facilitated the accurate detection, identification, and confirmation of M. enterolobii, resulting in a significant increase in records of new host plants. In 2022 and 2023 alone, eight new reports were published (e.g. European and Mediterranean Plant Protection Organization, 2022; Anes et al., 2023; Ibrahim et al., 2023; Naveenkumar et al., 2023). Meloidogyne enterolobii causes significant yield losses of up to 65% (Philbrick et al., 2020). Its ability to reproduce on many commercial crop cultivars regardless of the source of resistance to other RKN makes crop production difficult (Sikandar et al., 2023). Meloidogyne enterolobii is not controlled by resistance in Capsicum annuum (N gene, Tabasco gene and rootstock ‘Snooker’ carrying the Mi1 and Mi3/Mi7 genes), Vigna unguiculata (Rk gene), Glycine max (Mir1 gene), Gossypium hirsutum, Ipomoea batatas, Solanum lycopersicum (Mi1 gene) as well as Solanum tuberosum (Mh gene; European and Mediterranean Plant Protection Organization, 2014; Sikandar et al., 2023).
In the European Union it has been concluded that M. enterolobii fulfilled the conditions provided in Article 3 and Section 1 of Annex I to Regulation (EU) 2016/2031 with respect to the Union territory and is listed now in Part A of Annex II to Implementing Regulation (EU) 2019/2072 as Union quarantine pest (Anonymous, 2021). This decision emphasizes that M. enterolobii transitioned from an “emerging” species (Elling, 2013) to one of the most detrimental root-knot nematode species worldwide, leaving few options for control (Sikandar et al., 2023). To manage plant parasitic nematode populations, resistant cultivars or crop rotation with non-hosts can be very effective (European and Mediterranean Plant Protection Organization, 2014; Sikandar et al., 2023). Given the declining availability of chemical nematicides and the lack of resistance in major crops with the exception of certain Psidium guajava root-stocks (Freitas et al., 2014; Prem et al., 2023), crop rotation with non- or poor host plants will be a primary strategy to manage M. enterolobii.
Despite numerous reports of M. enterolobii damaging various crops in different geographical regions around the world, the intrinsic host range of M. enterolobii populations from different geographical regions and host origins has not been investigated. The intrinsic host range is hereby referred to as the natural or inherent range of plant species that M. enterolobii can successfully reproduce on. Therefore, the objective of this greenhouse study was to determine if the extremely wide host range of M. enterolobii is intrinsic, therefore determined by its genetic composition or due to other factors such as selection or adaptation. To achieve this, seven M. enterolobii populations were used to infect 19 plant species previously reported as major, poor, or non-hosts. In order to maximize the reproduction potential, single egg mass lines of M. enterolobii populations, pre-selected for the highest level of virulence were used in these experiments. In this way, declaration of a plant species as non- or poor host due to an intrinsically low reproduction of M. enterolobii was avoided. In addition, near isogenic lines from two M. incognita populations highly virulent against the tomato Mi1 gene were included in these experiments as they showed a comparable level of virulence on resistant tomato cultivars when compared to M. enterolobii.
Materials and methods
Greenhouse experiments were conducted under quarantine conditions at the Julius Kühn Institute (JKI) in Braunschweig, Germany. The seven M. enterolobii populations originated from Africa, Asia and North America, while the two virulent M. incognita populations originated from Germany (Table 1). To determine the host range of these populations, 19 plant species were tested for their potential to serve as a host plant (Table 2). All populations were maintained in a greenhouse on tomato cvs. Moneymaker (susceptible) or Phantasia (carrying Mi1 resistance gene; Volmary GmbH, Münster, DE) throughout this study.
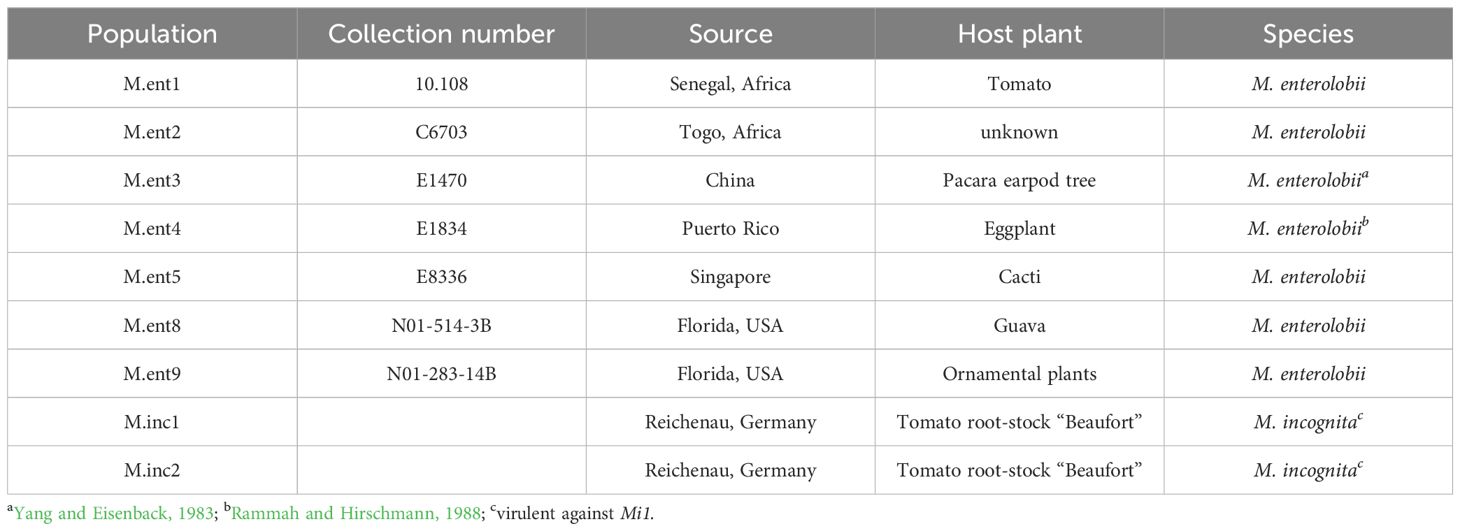
Table 1. Populations of Meloidogyne enterolobii and virulent Meloidogyne incognita from different geographical regions and maintained on tomato under greenhouse conditions, used in this study.
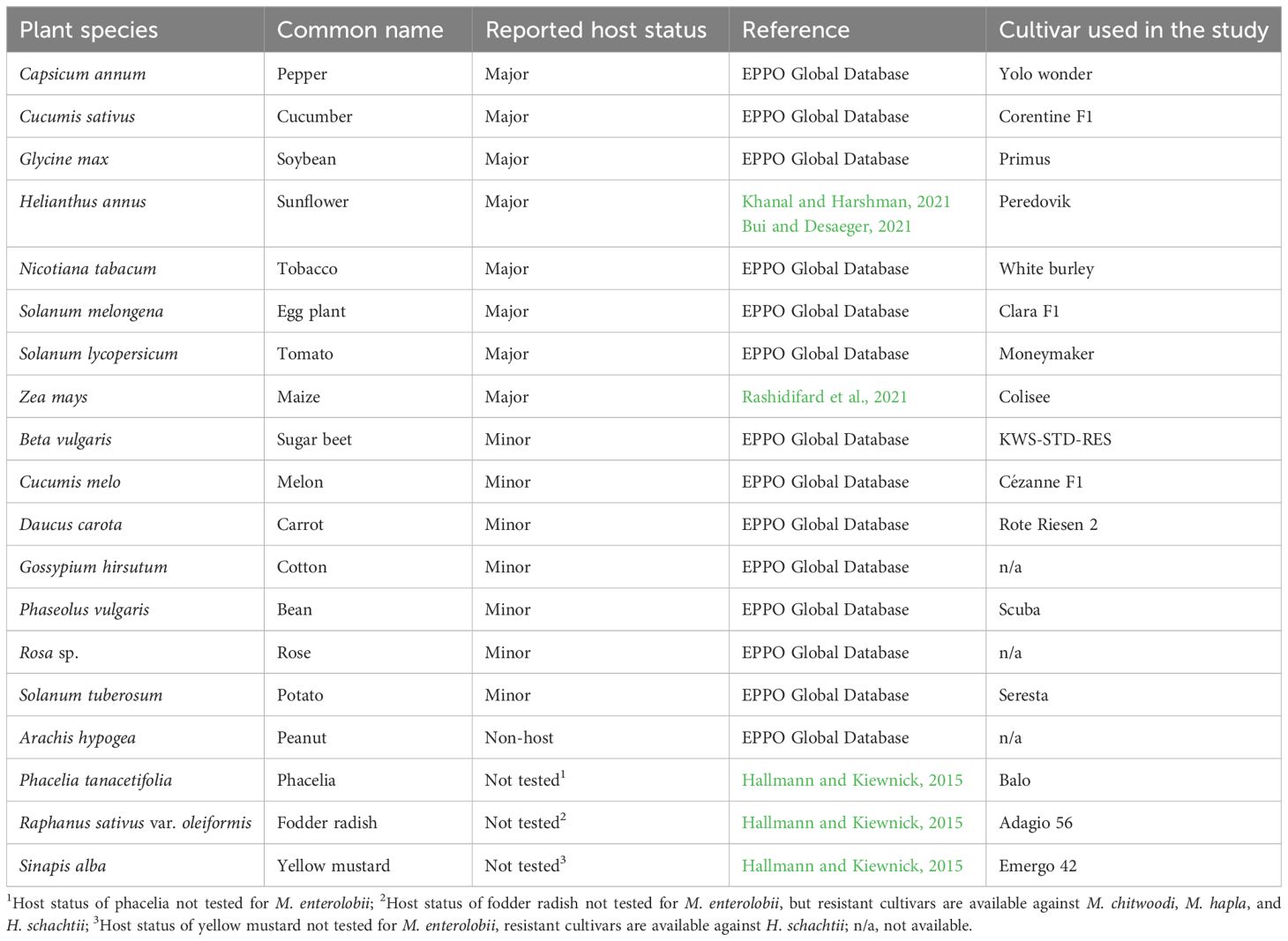
Table 2. Plant species used in greenhouse experiments to evaluate the reproductive potential of Meloidogyne enterolobii and virulent Meloidogyne incognita populations.
For the experiments, single egg-mass lines were generated from each population. Ten individual females with egg mass were hand-picked and multiplied individually on the tomato cultivar ‘Phantasia’, carrying the Mi-1 resistance gene. After eight weeks, the reproduction was determined and the line with the highest reproduction factor was selected for further experiments. In addition, species identification for each line was confirmed using Real-time (Kiewnick et al., 2015) and species-specific PCR (Tigano et al., 2010).
To produce inoculum for the experiments, the single egg-mass lines were multiplied twice on tomato cv. Moneymaker. Greenhouse experiments were conducted using plastic pots (11x11x11.5 cm Pöppelmann®, Lohne, DE) containing 750ml quartz sand (0.3-1mm) supplemented with slow release fertilizer, Osmocote (1.5g/L). Seeds were germinated in seedling trays and then transplanted into pots. One week after transplanting, plants were inoculated with suspensions containing on average 3800 eggs and second stage juveniles (E+J2) with at least 70% eggs (containing J2s) and 20% J2s. Plants were maintained in a greenhouse during summer at 25 ± 2°C and the winter month at 20 ± 2°C with 16h of light and 8h of darkness. Plants were watered daily and received additional Wuxal® super solution (8:8:6; N: P: K, Hauert MANNA, Nürnberg, DE) once per week. Each host plant species was tested with five replicates and each experiment was conducted twice (summer and winter). Eight weeks after inoculation, the root and shoot fresh weight was recorded and eggs and juveniles (E+J2) extracted from the roots using 0.7% chlorine solution (Stetina et al., 1997) to determine the reproduction factor (RF). Based on the RF values, the host status of tested plant species was defined. Categorization of RF classes was as follows: RF class 0 (non-host) when RF between 0 and ≤0.1; RF class 1 (poor host) when RF >0.1and ≤1; RF class 2 (host) when RF >1 and ≤ 2 and RF class 3 (good host) when RF >2. Although plants with RF >1 are already considered a host (Sasser et al., 1984), the additional class (RF >1 and <2) allows for a more nuanced differentiation of the host plant status.
Statistical analysis
Statistical analysis was carried out in R (version 4.3.1) software (R Core Team, 2021). Normality of the data was tested using the Shapiro-Wilk test, and homogeneity of variances was checked with the Breusch-Pagan test (P<0.05). Due to the observed heteroscedasticity, data was considered to be non-parametric and analyzed using ANOVA on ranks. Since no interaction between experiments for parameter RF class was observed, data from two experiments were pooled for this variable. The heat map depicting the reproduction factor (RF) of M. enterolobii and virulent M. incognita populations was created based on four RF class values. Hierarchical clustering of populations was done by ‘hclust’ function and ‘Ward.D2’ method using RF class values. However, as the interactions between experiments were significant for parameters RF, root fresh weight (RFW) and shoot fresh weight (SFW), data from each experiment was analyzed separately (Kruskal-Wallis test and Dunn’s test). The data for RFW and SFW from two experiments were normalized using Z-scores to account for the differences in mean weights between plant species. These normalized scores were then used to illustrate the effects of various nematode populations on different plant species. The phylogenetic data for 19 plant species were obtained from the TimeTree database (Kumar et al., 2022). The classification tree for these plant species, based on their host status with respect to Meloidogyne populations, was generated using Ward clustering in R followed by comparison of the topologies for species phylogeny and the classification tree.
Results
Based on similarities in reproduction factors on 19 host plants, the M. enterolobii populations grouped into one cluster containing M.ent3, 4 and 5 and a second cluster with M.ent1, 2, 8 and 9, whereas M.inc1 and 2 formed an outgroup relative to all M. enterolobii populations. Across all plant species tested, 13 were hosts for both Meloidogyne species, whereas cotton, maize, sunflower and roses significantly differed in their response to the Meloidogyne species tested. Phacelia and fodder radish revealed a potential as host or poor host for M. incognita and some of the M. enterolobii populations (Figure 1).
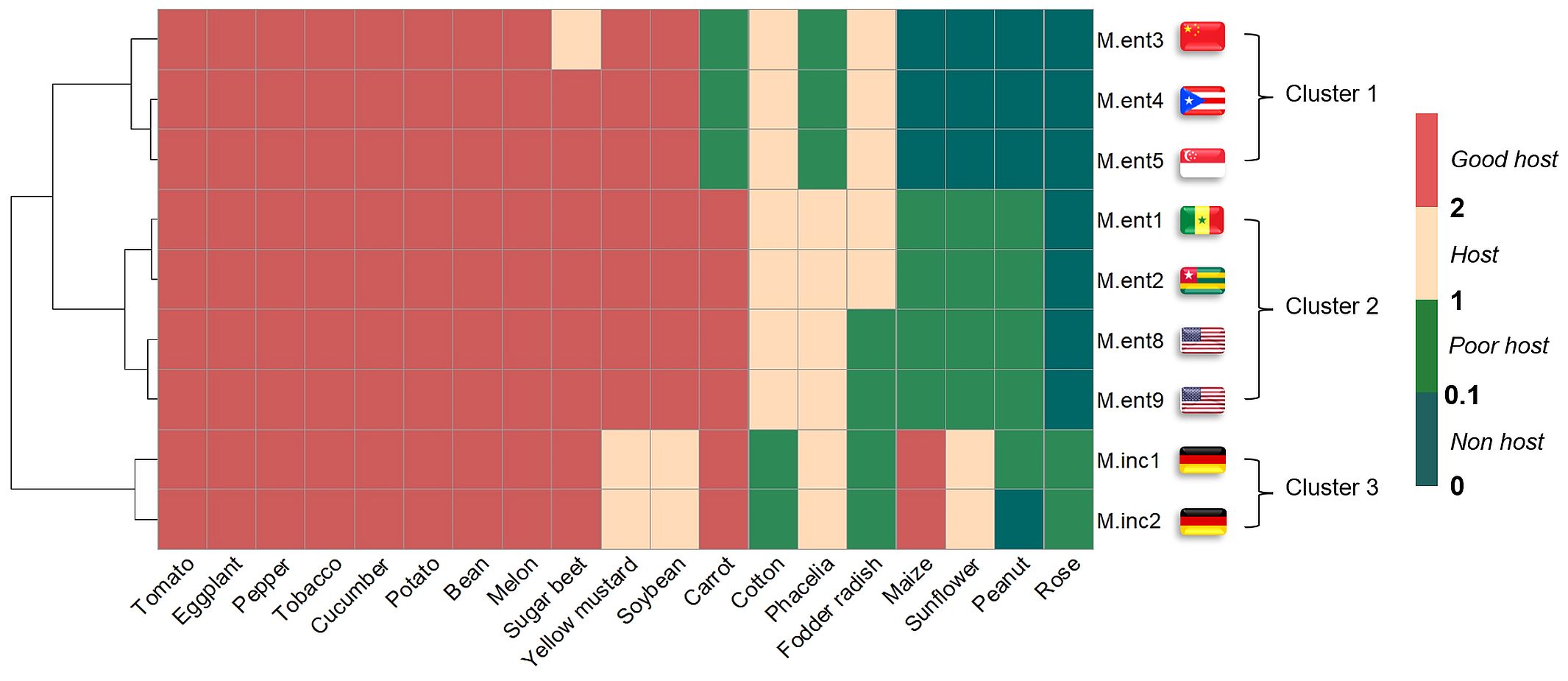
Figure 1. Heat map representing the reproduction factor (RF) of seven Meloidogyne enterolobii (M.ent) and two virulent Meloidogyne incognita (M.inc) populations across 19 different plant species. The RF class was derived from the RF means and standard deviation of individual treatments, used to depict clustering and the host status for different populations. The RF class was categorised into four groups: RF class 0 (non-host) when RF = 0 and ≤0.1; RF class 1 (poor host) when RF >0.1and ≤1; RF class 2 (host) when RF >1 and ≤ 2 and RF class 3 (good host) when RF >2. Data from two experiments were pooled (n=10) and subjected to non-parametric Kruskal-Wallis test and pairwise Wilcoxon test. The clustering of populations was performed by the ‘hclust’ function and the ‘Ward.D2’ method. Flags represent the geographical origin of populations.
Of the 19 plant species challenged with M. enterolobii, 15 were hosts (RF>1) for at least two of the M. enterolobii populations, whereas 12 were hosts for all populations (Figure 1). The remaining plant species revealed differences in their host status towards the M. enterolobii populations. In particular, carrots and phacelia were hosts for populations M.ent1, 2, 8 and 9, but poor hosts for populations M.ent3, 4 and 5. In contrast, fodder radish was a host plant for populations M.ent3, 4, 5, 1 and 2, but a poor host for M.ent8 and 9. Maize, sunflower and peanut were poor hosts for populations M.ent1, 2, 8 and 9 and allowed no reproduction when challenged with populations M.ent3, 4 or 5. None of the M. enterolobii populations tested was able to reproduce substantially on roses.
For the two virulent M. incognita populations, 15 out of 19 plant species were suitable hosts. Cotton, fodder radish and roses were poor hosts for both populations and peanut proved to be a poor host for M.inc1, but a non-host for M.inc2 (Figure 1).
On average, the ensemble of M. enterolobii and M. incognita populations showed a high reproductive potential across the 19 plant species tested, resulting in RF values ranging from 11 to 37 when greenhouse experiments were performed during summer (Figure 2a) and 15 to 25 during winter (Figure 2b). RF values for populations M.ent3, 4 and 5 were significantly different to populations M.ent1 and 2 when greenhouse temperatures were higher (Figure 2a). Furthermore, data pooled per cluster revealed significant differences between the three clusters (Figure 2). Greenhouse experiments conducted during winter resulted in similar reproduction factors across all populations tested (Figure 2b).
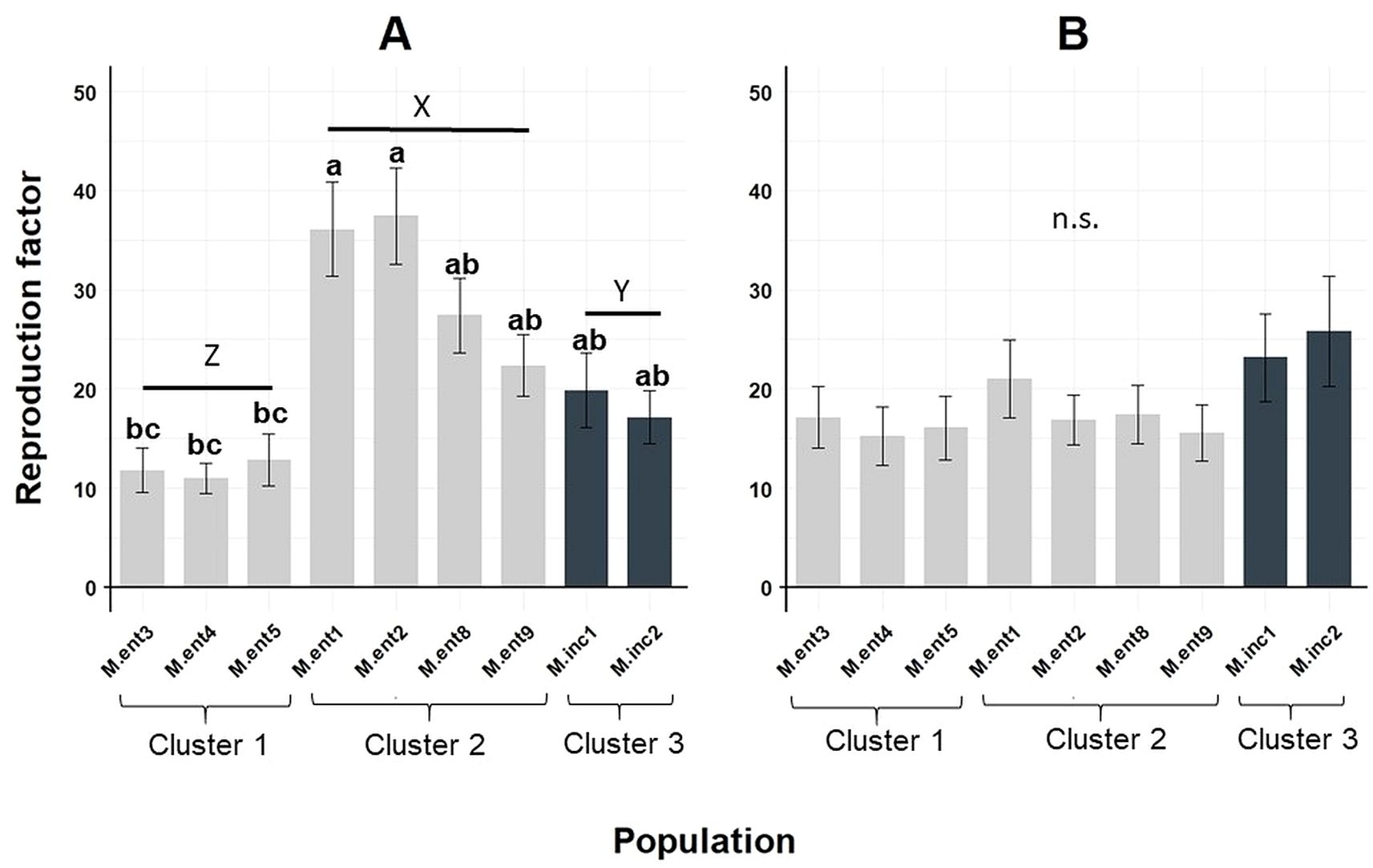
Figure 2. Means of reproduction factor (RF) of seven Meloidogyne enterolobii (M.ent) and two virulent M. incognita (M.inc) populations averaged across 19 host plant species. Bars indicate the mean RF values ± standard error; (a) greenhouse experiment conducted during summer (n=95); (b) greenhouse experiment conducted during winter (n=95). Means showing different letters are significantly based on non-parametric Kruskal-Wallis test and Wilcoxon Rank-Sum Test (P < 0.05); Data were LN(X + 1) transformed before analysis, but original non-transformed data are shown. Data pooled per Cluster showing different capital letters are significantly different according to non-parametric Kruskal-Wallis test and Wilcoxon Rank-Sum Test (P < 0.05).
When comparing the reproductive potential across all M. enterolobii populations on tomato to the remaining host plant species, all but eggplant, pepper, cucumber and potato showed significantly lower RF values during summer experiments (Figure 3a). However, RF values obtained in winter experiments were significantly greater than on tomato for eggplant and pepper, whereas with the exception of tobacco, cucumber and potato, the remaining host plants revealed significantly lower RF values compared to tomato (Figure 3b).
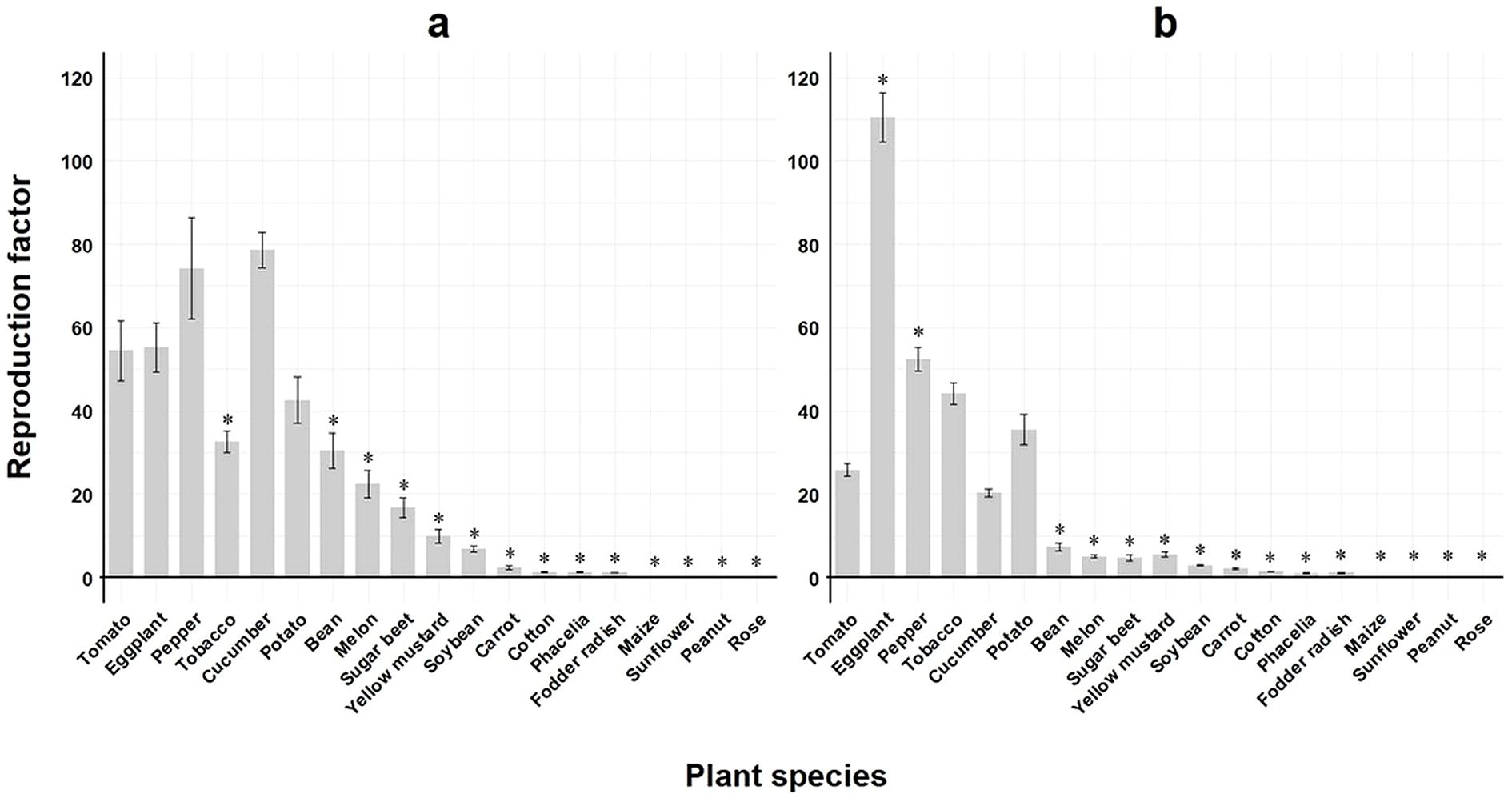
Figure 3. Means of reproduction factor (RF) of seven Meloidogyne enterolobii populations across 19 different plant species. Bars indicate the mean RF values ± standard error; (a) greenhouse experiment conducted during summer; (b) greenhouse experiment conducted during winter (n=35); *significantly different compared to tomato as control according to on non-parametric Kruskal-Wallis test and Dunnet’s t-sided t-test (p< 0.05).
In comparison to M. enterolobii, M. incognita populations showed significantly lower RF values on 15 out of 18 plant species compared to tomato during summer experiments (Figure 4a). During winter experiments, eggplant and tobacco demonstrated a significant, more than 3-fold increase in reproduction compared to tomato, whereas 15 of the plant species showed significantly lower RF values (Figure 4b).
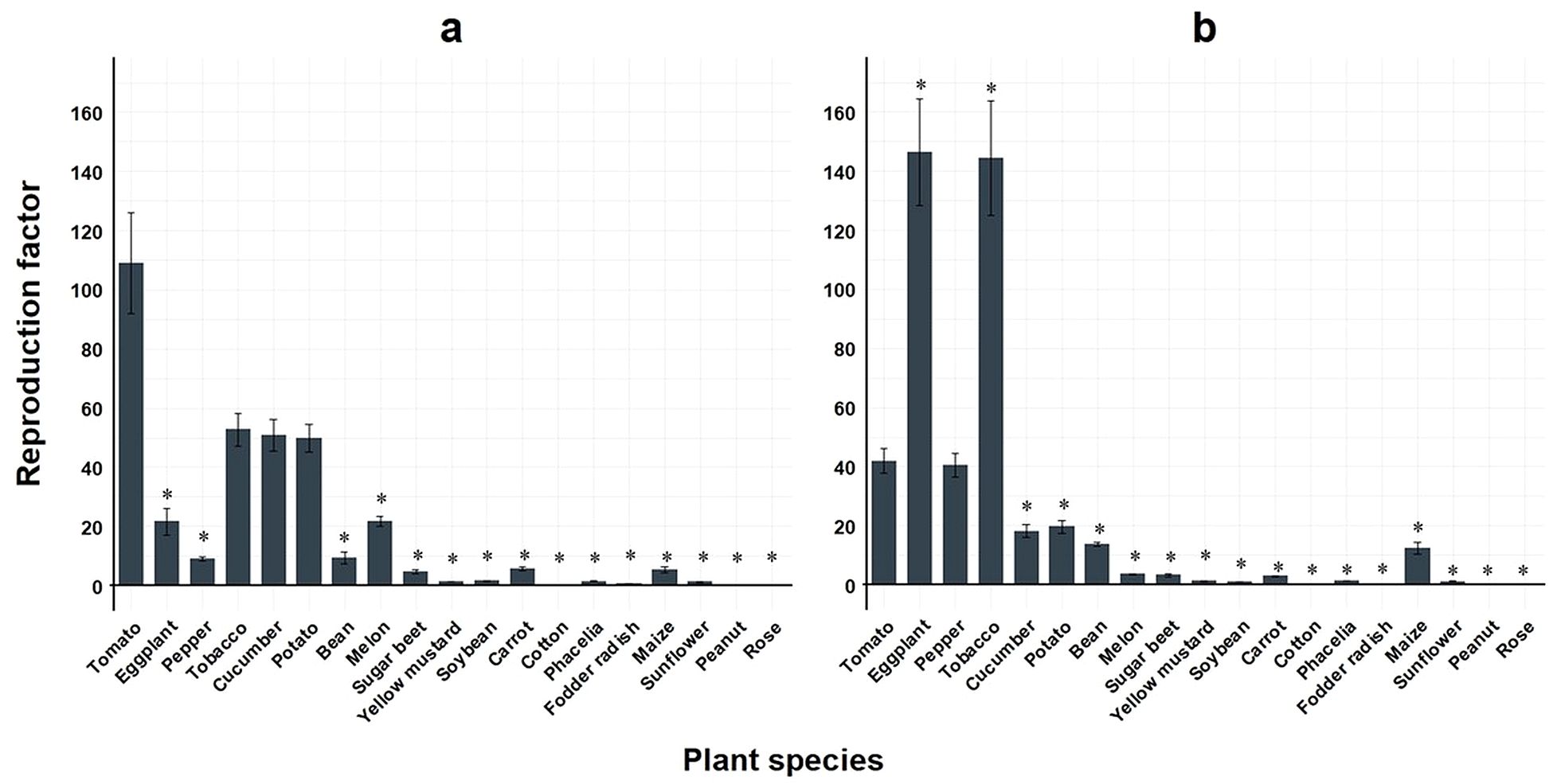
Figure 4. Means of reproduction factor (RF) of two virulent Meloidogyne incognita populations across 19 different plant species. Bars indicate the mean RF values ± standard error; (a) greenhouse experiment conducted during summer (n=35); (b) greenhouse experiment conducted during winter (n=35); *significantly different compared to tomato as control according to non-parametric Kruskal-Wallis test and Dunnet’s t-sided t-test (p< 0.05).
To estimate the damage potential caused by different M. enterolobii and M. incognita populations, Z-Score values were calculated (Figures 5, 6). Based on the average Z-Score for root fresh weight (Figure 5), a distinct effect by the M. enterolobii populations M.ent3, 4 and 5 was observed across all tested plant species except for soybean, maize, sunflower and rose (Figure 5). With respect to shoot fresh weight, 11 out of 19 plant species revealed a negative response due to populations M.ent3, 4 and 5. In contrast, cucumber, bean, yellow mustard, carrot, fodder radish, maize and sunflower revealed no negative effects by populations M.ent 3, 4 and 5. As an exception, rose responded with reduced shoot weight when being challenged by these three M. enterolobii populations, although no reproduction was found (Figures 1, 6).
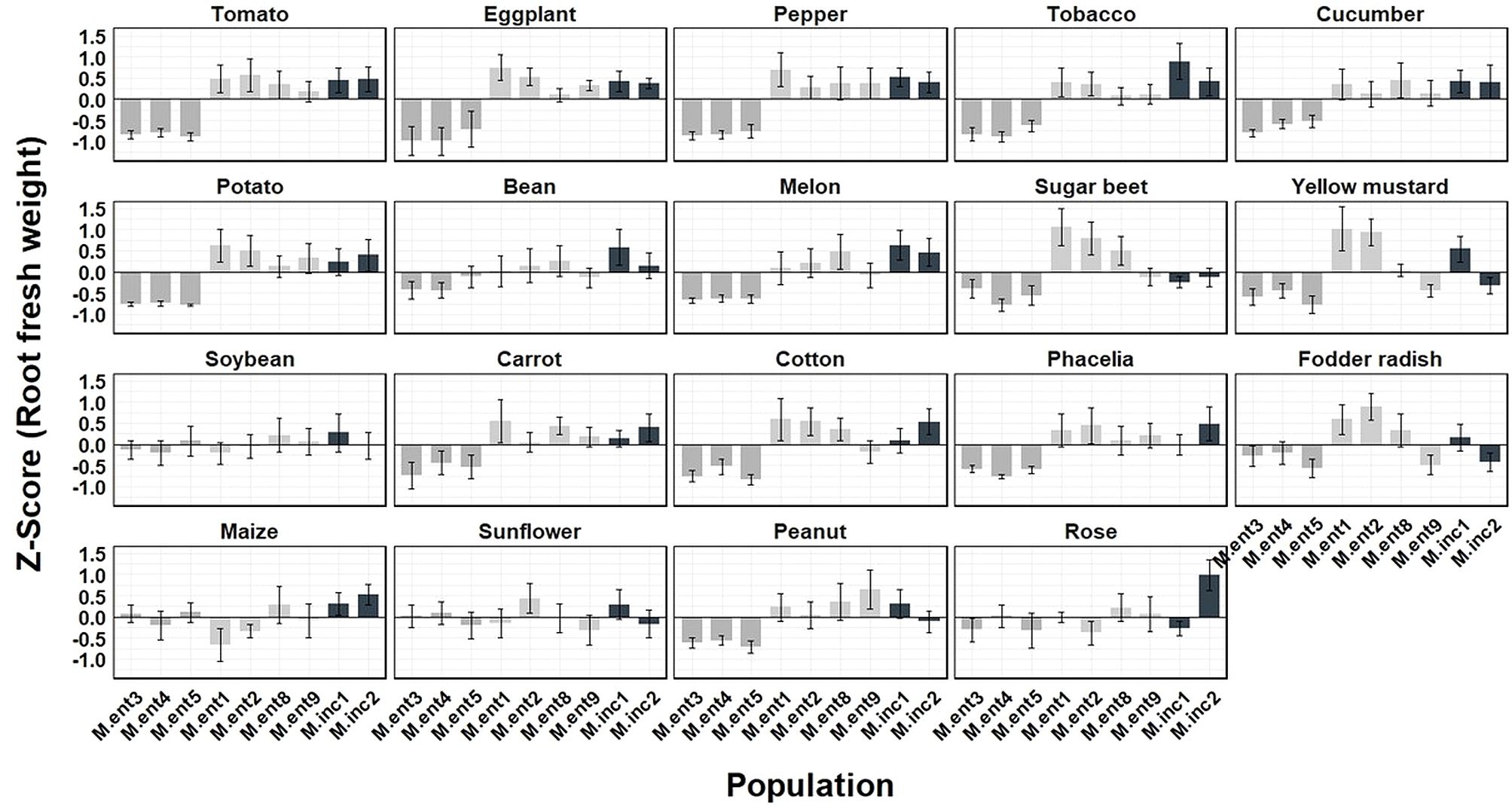
Figure 5. Mean Z score of root fresh weight (g) of 19 plant species challenged with seven Meloidogyne enterolobii (M.ent) and two virulent M. incognita (M.inc) populations under greenhouse conditions. Data from two experiments were used (n=10). Scoring was achieved using the formula: Z score = (the individual observation - mean of plant species)/standard deviation of plant species. Z = 0: The value is exactly at the mean. Z > 0: The value is above the mean. Z < 0: The value is below the mean. Z > 2 or Z < -2: The value is an outlier.  = cluster 1 (population M.ent3, M.ent4 and M.ent5);
= cluster 1 (population M.ent3, M.ent4 and M.ent5);  = cluster 2 (population M.ent1, M.ent2, M.ent8 and M.ent9);
= cluster 2 (population M.ent1, M.ent2, M.ent8 and M.ent9);  = cluster 3 (population M.inc1 and M.inc2).
= cluster 3 (population M.inc1 and M.inc2).
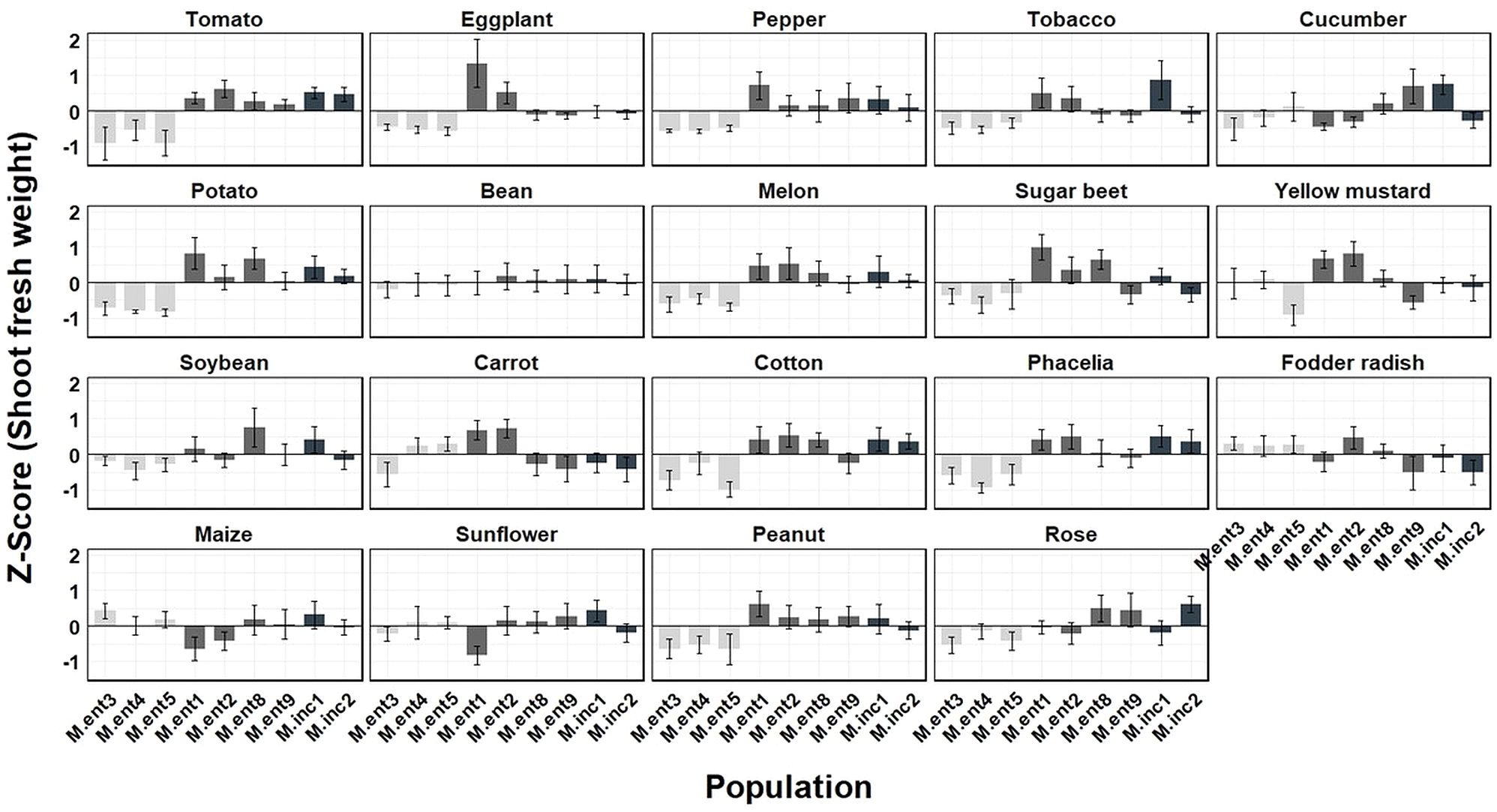
Figure 6. Mean Z score of shoot fresh weight (g) of 19 plant species challenged with seven Meloidogyne enterolobii (M.ent) and two virulent M. incognita (M.inc) populations under greenhouse conditions. Data from two experiments were used (n=10). Scoring was achieved using the formula Z score = (the individual observation - mean of plant species)/standard deviation of plant species. Z = 0: The value is exactly at the mean. Z > 0: The value is above the mean. Z < 0: The value is below the mean. Z > 2 or Z < -2: The value is an outlier.  = cluster 1 (population M.ent3, M.ent4 and M.ent5);
= cluster 1 (population M.ent3, M.ent4 and M.ent5);  = cluster 2 (population M.ent1, M.ent2, M.ent8 and M.ent9);
= cluster 2 (population M.ent1, M.ent2, M.ent8 and M.ent9);  = cluster 3 (population M.inc1 and M.inc2).
= cluster 3 (population M.inc1 and M.inc2).
When the phylogenetic distance of tested plant species was compared to their host plant status towards M. enterolobii or M. incognita, no clear correlation became evident (Figure 7). As could be expected, plants of the Solanaceae family showed a very similar pattern of host status and grouped together. Cucumber showed a pattern very similar to that of Solanaceae although being separated by 125 Million years (Ma) of evolution between the Solanaceae and Cucurbitaceae. The distantly related monocot maize and the dicot sunflower (160 Ma of divergence) had the same poor and non-host pattern with respect to the M. enterolobii populations while they were hosts of M. incognita. Similar patterns were observed for peanut and rose (113 Ma), fodder radish and cotton (101 Ma) as well as carrot and phacelia (111 Ma; Figure 7).
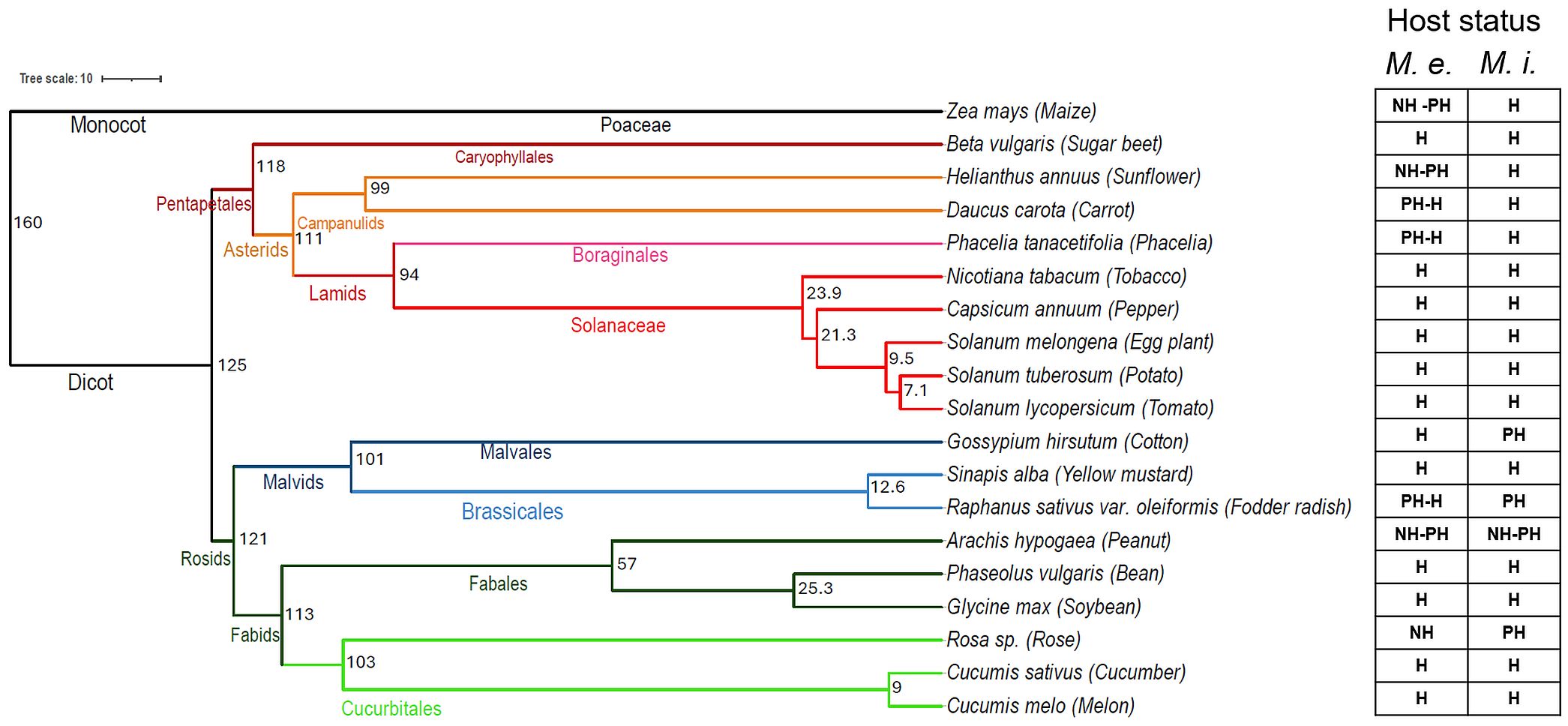
Figure 7. Phylogenetic tree showing the categorization of plants according to the species combined with their range of host status to Meloidogyne enterolobii (M. e.) and Meloidogyne incognita (M. i.) The numbers within the classification denote the evolutionary period in million years and the colours indicate the families of the plant species. The dated species phylogeny of 19 crops was obtained from the TimeTree database (Kumar et al., 2022) and the host status classification of plant species was generated using Ward clustering in R; NH, non host; PH, poor host; H, host.
Discussion
Meloidogyne enterolobii, although regulated as quarantine species in many countries, has become a major challenge for all agricultural production systems worldwide. The increase in the number of reports of new host plants across the world demonstrates the rise of this species from the status “emerging” to a real threat for many main crops on a global scale (Sikandar et al., 2023). Recent reviews documented the increase in reports describing new hosts of M. enterolobii where significant damage was observed, in particular crops with resistance to other RKN species (Sikandar et al., 2023).
This study investigated, for the first time, the intrinsic host range of M. enterolobii populations from different geographic and host origins. In particular, single egg-mass lines were used to infect a panel of 19 plants species to evaluate the host range of different M. enterolobii populations, including the two type populations from China and Puerto Rico (Yang and Eisenback, 1983; Rammah and Hirschmann, 1988). For comparison, we included two highly virulent M. incognita populations, with a similar level of damage and reproduction potential (Hallmann and Kiewnick, 2018).
The majority of the plant species tested in this study reacted as expected when challenged with diverse M. enterolobii populations. However, a subset of plant species differed in their response and resulted in clustering of the populations M.ent 3, 4 and 5 versus M.ent 1, 2, 8 and 9. The two M. incognita populations M.inc1 and 2, obviously belonged to race 2 (Hartman and Sasser, 1985) with tobacco allowing reproduction, but cotton and peanut were poor to non-hosts. Consequently, these two populations formed an additional distinct cluster.
In previous studies, only M. enterolobii populations from the same geographical region were compared for virulence and pathogenicity on different host plants or host plant genotypes (Schwarz et al., 2020; Gaudin et al., 2023; Salazar-Mesta et al., 2023), but no or only minor differences were observed.
In our study, we found deviations from published reports for carrot, maize, sunflower and rose. Brito et al. (2007) identified carrot as a poor host when they tested 14 host plants against M. enterolobii (syn. M. mayaguensis) populations from Florida, USA. Conversely, our study revealed high reproduction rates for populations M.ent1, 2, 8 and 9, which included the two populations originating from Florida (Table 1, Figure 1). As these populations were most likely obtained from different original host plants, it could explain this different response. The distinct cluster containing both reference populations M.ent3 and 4 as well as population M.ent5 (Figure 1) showed only poor reproduction rates on carrot. Consequently, this is the first report of differences in the host range of M. enterolobii populations, based on their geographic and host origin. Further discrepancies concerning potential host plants of M. enterolobii were found for sunflower and maize. In contrast to Bui and Desaeger (2021); Khanal and Harshman (2021) or Rashidifard et al. (2021), sunflower and maize were non- or poor hosts for all M. enterolobii populations tested, despite using similar cultivars. However, in contrast to carrots, these differences were obviously caused by the intrinsic host range of the M. enterolobii populations used in this study.
In the year 2008, a consignment of rose rootstocks from China intended for commercial markets was intercepted in the Netherlands due to the presence of M. enterolobii. As one consequence rose was listed as a minor host for M. enterolobii (European and Mediterranean Plant Protection Organization, 2008). However, in our study, we identified rose as a non-host for all seven M. enterolobii populations (Figures 1, 3). This confirms the study by Mendes and Dickson (2016) who demonstrated that Fortuniana rose rootstock, which is also used for the European market, was a non-host for all tropical Meloidogyne spp., including M. enterolobii. No further studies are available confirming the host status of rose toward M. enterolobii. Therefore, further research is needed to investigate the host status of roses. However, interceptions of ornamental plants in the Netherlands and Italy confirmed possible routes of introduction for M. enterolobii by Ficus microcarpa (European and Mediterranean Plant Protection Organization, 2023). These findings suggest that imported horticultural plants, such as rose or Ficus spp., are potential sources for introduction into Europe. Often, these imported plants are briefly maintained or multiplied in commercial greenhouses, which are commonly used for growing major crops such as tomato, pepper, or cucumber, before distribution (European and Mediterranean Plant Protection Organization, 2023).
Phacelia, a cover crop commonly grown in temperate regions for weed suppression and green manure as well as yellow mustard and fodder radish were previously identified as maintenance hosts (RF values of 1.0 ± 0.5) for M. hapla (Viaene and Abawi, 1998). Resistant cultivars of fodder radish can be used to effectively control M. chitwoodi, M. fallax, M. hapla, and Heterodera schachtii (Hallmann and Kiewnick, 2015). Yellow mustard, grown as either an oilseed or green manure crop, is widely cultivated in Europe (Mitrović et al., 2020) with resistant cultivars available to control H. schachtii (Hallmann and Kiewnick, 2015). Despite their demonstrated neutrality or resistance against other Meloidogyne species and H. schachtii, phacelia, fodder radish, and yellow mustard supported the reproduction of M. enterolobii to varying extents.
When reproduction factors were compared across all Meloidogyne populations tested, the three M. enterolobii populations M.ent3, 4 and 5 stood out as they showed the lowest reproduction during the summer experiments when temperatures averaged 25 ± 2 °C. Greenhouse experiments conducted during winter showed no differences in reproduction. However, M. incognita reproduction was not different from M. enterolobii and even greater compared to the above mentioned three populations in the summer experiments. This contradicts previous findings where M. enterolobii always showed higher reproduction rates compared to other tropical Meloidogyne species (Kiewnick et al., 2009). The effect of temperature on the reproduction of Meloidogyne spp. is well documented and therefore expected (Wong and Mai, 1973; Greco and Vito, 2009; Dinardo-Miranda and Fracasso, 2010; Fernandez et al., 2014; Daramola et al., 2021). In contrast, the observed host plant status did not differ between experiments, but in some cases reduced root damage resulted in increased RF values. Overall, the results confirm the findings by Velloso et al. (2022) who showed high reproduction rates for both M. enterolobii and M. incognita under varying temperature regimes. For few hosts, such as eggplant, the lower greenhouse temperatures (20 ± 2°C) allowed for high reproduction rates of M. enterolobii and M. incognita, respectively (Figures 3b, 4b).
For some host plant/Meloidogyne population combinations, positive Z-scores for root fresh weight were observed (Figure 5) as nematode infection stimulates plant growth and the development of secondary roots and galls despite supporting large nematode populations (Tandingan et al., 1996; Parveen, 2006; Guarneri et al., 2022). However, the populations M.ent3, 4 and 5 stood out as they clearly affected the root and shoot fresh weight of several of the tested plant species negatively (Figures 5, 6). As these three populations formed a cluster based on their host range (Figure 1), it might indicate a correlation between the intrinsic host range of a given population and its damage potential.
The phylogenetic analysis of the host plant species tested in this study demonstrated the wide reproductive potential of M. enterolobii populations on crops with evolutionary distances of up to 160 million years. No correlation was found between the evolutionary distance of plant species and their status as host plant to M. enterolobii (Figure 7). These findings support the wide host range, varying levels of aggressiveness, and diverse host compatibility of M. enterolobii. Few plant species covering the full range of phylogenetic distances such as maize, sunflower, and cotton were able to separate M. enterolobii from M. incognita. Overall, these findings confirm the wide range of host plant species that M. enterolobii is able to reproduce on and cause significant damage. As its intrinsic host range is obviously not restricted to a certain group of closely related plant species it explains the constantly increasing number of reports of new host plant species around the globe (Sikandar et al., 2023).
In conclusion, this is the first study investigating a collection of M. enterolobii populations from different geographical and host origins for their intrinsic host range with 19 different host plant species. We confirmed the wide host range of M. enterolobii which is comparable to M. incognita, currently described as the root-knot nematode species causing the highest economic damage worldwide (Jones et al., 2013). Based on the presented results, current prediction models (e.g. Pan et al., 2023) considering only the impact caused by abiotic, but not biological factors, should be adjusted accordingly. This will lead to predicting wider suitable areas for the spread and establishment of M. enterolobii in context of global warming, which has resulted in a northward migration of previously undetected ‘tropical’ RKN species (Gerič Stare et al., 2018). Based on the obtained results, farmers will have few options to use poor- or non-host plants to manage nematode populations once M. enterolobii is introduced into Europe. Therefore, further studies on suitable intercrops are needed in support of farmers, as plant resistance is not yet available to control M. enterolobii. Discrepancies between published reports on host plants and this study indicate the intrinsic potential of M. enterolobii to multiply on all mayor crops worldwide although the use of single eggmass lines potentially have restricted the range. Recently, a high-quality genome assembly of M. enterolobii for the type population from Puerto Rico has been published (Poullet et al., 2025). This constitutes a reliable resource for within- and between-species comparative genomics and for the identification of genomic variations in relation with the host range of this quarantine nematode species in support of plant health and to develop new measures for control.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
HK: Investigation, Writing – original draft, Writing – review & editing. MF: Supervision, Writing – original draft, Writing – review & editing. MB-B: Writing – original draft, Writing – review & editing, Formal analysis. ED: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision. SK: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing, Investigation, Validation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The project was financially supported by the German Research Foundation (DFG) and the French National Research Agency (ANR) “AEGONE”, reference: 431627824 and ANR-19-CE35-0017.
Acknowledgments
We are grateful for the colleagues from the Netherlands Institute for Vectors, Invasive plants and Plant health (NIVIP), Florida Department of Agriculture and Consumer Services and French Agency for Food (FDACS), Environmental and Occupational Health & Safety (ANSES) for providing the M. enterolobii populations for this study. We would like to thank Johannes Hallmann for his valuable comments and improvement of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anes, K. M., Babu, M., and Rajkumar, J. A. (2023). Root-knot nematode, Meloidogyne enterolobii Yang and Eisenback 1983 infecting guava intercropped with coconut plantation in Kerala, India. Indian J. Nematol. 53, 15–20. doi: 10.5958/0974-4444.2023.00003.3
Anonymous (2021). The European Commission Implementing Regulation (EU) 2021/2285 of 14 December 2021 amending Implementing Regulation (EU) 2019/2072 as regards the listing of pests, prohibitions and requirements for the introduction into, and movement within, the Union of plants, plant products and other objects, and repealing Decisions 98/109/EC and 2002/757/EC and Implementing Regulations (EU) 2020/885 and (EU) 2020/1292. Off. J. Eur. Union. L 458, 173–283.
Brito, J., Stanley, J., Mendes, M., Cetintas, R., and Dickson, D. (2007). Host status of selected cultivated plants to Meloidogyne mayaguensis in Florida. Nematropica 37, 65–71. Available online at: http://ojs-test.fcla.edu/index.php/nematropica/article/view/64416 (Accessed January 21, 2024).
Bui, H. X. and Desaeger, J. A. (2021). Host suitability of summer cover crops to Meloidogyne arenaria, M. enterolobii, M. incognita and M. javanica. Nematology 24, 171–179. doi: 10.1163/15685411-bja10122
Daramola, F. Y., Malgas, R., and Malan, A. P. (2021). Occurrence and seasonal changes in the population of root-knot nematodes on honeybush (Cyclopia Sp.). Helminthologia 58, 202–212. doi: 10.2478/helm-2021-0018
Dinardo-Miranda, L. L. and Fracasso, J. V. (2010). Spatial distribution of plant-parasitic nematodes in sugarcane fields. Bragantia 69, 39–52. doi: 10.1590/S0103-90162009000200007
Elling, A. (2013). Major emerging problems with minor Meloidogyne species. Phytopathology 103, 1092–1102. doi: 10.1094/PHYTO-01-13-0019-RVW
European and Mediterranean Plant Protection Organization (2008). Meloidogyne enterolobii: Reporting services - pest & disease, Paris. No.5. Available online at: https://gd.eppo.int/media/data/reporting/rs-2008-05-en.pdf (Accessed January 21, 2024).
European and Mediterranean Plant Protection Organization (2014). Meloidogyne enterolobii. Datasheets on pests recommended for regulation. EPPO. Bull. 44, 159–163. doi: 10.1111/epp.12120
European and Mediterranean Plant Protection Organization (2022). EPPO global database (Paris, France: EPPO). Available online at: https://gd.eppo.int/ (Accessed January 21, 2024).
European and Mediterranean Plant Protection Organization (2023). Meloidogyne enterolobii: Reporting services. Available online at: https://gd.eppo.int/taxon/MELGMY/reporting (Accessed January 21, 2024).
Fernandez, L., Cabasan, M. T. N., and De Waele, D. (2014). Life cycle of the rice root-knot nematode Meloidogyne graminicola at different temperatures under non-flooded and flooded conditions. Arch. Phytopathol. Plant Prot. 47, 1042–1049. doi: 10.1080/03235408.2013.829627
Freitas, V. M., Correa, V. R., Motta, F. C., Sousa, M. G., Gomes, A. C. M. M., Carneiro, M. D. G., et al. (2014). Resistant accessions of wild Psidium spp. to Meloidogyne enterolobii and histological characterization of resistance. Plant Pathol. 63, 738–746. doi: 10.1111/ppa.12149
Gaudin, A. G., Wubben, M. J., McCarty, J. C., and Jenkins, J. N. (2023). Virulence of two isolates of Meloidogyne enterolobii (guava root-knot nematode) from North Carolina on cotton lines resistant to southern root-knot nematode (M. incognita) and reniform nematode (Rotylenchulus reniformis). J. Nematolo. 55, 1–11. doi: 10.2478/jofnem-2023-0021
Gerič Stare, B., Strajnar, P., Širca, S., Susič, N., and Urek, G. (2018). Record of a new location for tropical root knot nematode Meloidogyne luci in Slovenia. EPPO. Bull. 48, 135–137. doi: 10.1111/epp.12443
Greco, N. and Vito, M. D. (2009). “Population dynamics and damage levels,” in Root-knot nematodes (CABI, Wallingford UK), 246–274.
Guarneri, N., Willig, J.-J., Sterken, M. G., Zhou, W., Hasan, M. S., Sharon, L., et al. (2022). Root architecture plasticity in response to endoparasitic cyst nematodes is mediated by damage signalling. New Phytol. 237, 807–822. doi: 10.1111/nph.18570
Hallmann, J. and Kiewnick, S. (2015). “Diseases caused by nematodes in organic agriculture,” in Plant disease and their management in organic agriculture. Eds. Finckh, M. R., van Bruggen, A. H. C., and Tamm, L. (St. Paul, Minnesota, USA: APS press), 91–105. doi: 10.1094/9780890544785.008
Hallmann, J. and Kiewnick, S. (2018). Virulence of Meloidogyne incognita populations and Meloidogyne enterolobii on resistant cucurbitaceous and solanaceous plant genotypes. J. Plant Dis. Prot. 125, 415–424. doi: 10.1007/s41348-018-0165-5
Hartman, K. M. and Sasser, J. N. (1985). “). Identification of Meloidogyne species on the basis of differential host test and perineal pattern morphology,” in An Advanced Treatise on Meloidogyne. Eds. Sasser, J. N. and Carter, C. C. (North Carolina State University, Raleigh, NC), 69–77.
Ibrahim, D. S. S., Zawam, H. S., El Deriny, M., Riad, S. N., Castillo, P., and Palomares-Rius, J. E. (2023). First report of Meloidogyne enterolobii Yang & Eisenback 1983 (Guava root-knot nematode) infecting guava (Psidium guajava) in Egypt. Plant Dis. 107, 1637. doi: 10.1094/PDIS-09-22-2171-PDN
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Karssen, G., Liao, J., Kan, Z., van Heese, E. Y., and Nijs, L.J.d. (2012). On the species status of the root-knot nematode Meloidogyne mayaguensis Rammah & Hirschmann 1988. Zookeys 181, 67–77. doi: 10.3897/zookeys.181.2787
Khanal, C. and Harshman, D. (2021). Evaluation of summer cover crops for host suitability of Meloidogyne enterolobii. Crop Prot. 151, 105821. doi: 10.1016/j.cropro.2021.105821
Kiewnick, S., Dessimoz, M., and Franck, L. (2009). Effects of the Mi-1 and the N root-knot nematode-resistance gene on infection and reproduction of Meloidogyne enterolobii on tomato and pepper cultivars. J. Nematol. 41, 134–139.
Kiewnick, S., Frey, J. E., and Braun-Kiewnick, A. (2015). Development and validation of LNA-based quantitative real-time PCR assays for detection and identification of the root-knot nematode Meloidogyne enterolobii in complex DNA backgrounds. Phytopathol 105, 1245–1249. doi: 10.1094/PHYTO-12-14-0364-R
Kumar, S., Suleski, M., Craig, J. M., Kasprowicz, A. E., Sanderford, M., Li, M., et al. (2022). TimeTree 5: An expanded resource for species divergence times. Mol. Biol. Evol. 39, msac174. doi: 10.1093/molbev/msac174
Mendes, M. L. and Dickson, D. (2016). An overview on Fortuniana rose resistance to root-knot nematodes. J. Nematolo. 48, 351.
Mitrović, P. M., Stamenković, O. S., Banković-Ilić, I., Djalović, I. G., Nježić, Z. B., Farooq, M., et al. (2020). White mustard (Sinapis alba L.) oil in biodiesel production: A review. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00299
Naveenkumar, K. R., Dash, M., Khan, M. R., Satya, K., Pasupuleti, S., Kundu, A., et al. (2023). A survey of guava orchards of Uttarakhand, Uttar Pradesh and Haryana states in northern India to assess the spread of root-knot nematodes. Indian Phytopathol. 76, 541–549. doi: 10.1007/s42360-023-00608-x
Pan, S., Peng, D., Li, Y., Chen, Z., Zhai, Y., Liu, C., et al. (2023). Potential global distribution of the guava root-knot nematode Meloidogyne enterolobii under different climate change scenarios using MaxEnt ecological niche modelling. J. @ Int. Agric. 22, 2138–2150. doi: 10.1016/j.jia.2023.06.022
Parveen, R. (2006). Studies on Ocimum sanctum (L) infected with root-knot nematode, Meloidogyne incognita (Kofoid and White) Chitwood (Aligarh: Aligarh Muslim University).
Philbrick, A. N., Adhikari, T. B., Louws, F. J., and Gorny, A. M. (2020). Meloidogyne enterolobii, a major threat to tomato production: Current status and future prospects for its management. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.606395
Poullet, M., Konigopal, H., Rancurel, C., Sallaberry, M., Lopez-Roques, C., Mota, A. P. Z., et al. (2025). High-fidelity annotated triploid genome of the quarantine root-knot nematode, Meloidogyne enterolobii. Sci. Data. 12, 184. doi: 10.1038/s41597-025-04434-w
Prem, R., Prabhu, S., Muthuvel, I., Shanmuga Sundaram, K. A., and Seenivasan, N. (2023). Meloidogyne enterolobii resistance in cultivated and wild guava: A review. Pharma. Innov. 12, 1323–1329.
Rammah, A. and Hirschmann, H. (1988). Meloidogyne mayaguensis n. sp. (Meloidogynidae), a root- knot nematode from Puerto Rico. J. Nematolo. 20, 58–69.
Rashidifard, M., Ashrafi, S., Claassens, S., Thünen, T., and Fourie, H. (2021). A pilot approach investigating the potential of crop rotation with sainfoin to reduce Meloidogyne enterolobii infection of maize under greenhouse conditions. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.659322
R Core Team (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available online at: https://www.R-project.org/ (Accessed January 21, 2024).
Salazar-Mesta, R. J., Facio, J., Retes-Manjarrez, J., García-Estrada, R., León-Félix, J., Mora-Romero, A., et al. (2023). Characterization, pathogenicity, and reproduction of Meloidogyne enterolobii populations parasitizing vegetables in Sinaloa, Mexico. Trop. Plant Pathol. 48, 394–407. doi: 10.1007/s40858-023-00576-7
Sasser, J. N., Carter, C. C., and Hartman, K. M. (1984). Standardisation of host suitability studies and reporting of resistance to root-knot nematodes (Raleigh, N.C., USA: International Nematology Network Newsletter, (15) Raleigh, N.C. Dept. of Plant Pathology, North Carolina State University).
Schwarz, T., Li, C., Ye, W., and Davis, E. (2020). Distribution of Meloidogyne enterolobii in eastern North Carolina and comparison of four isolates. Plant Health Prog. 21, 91–96. doi: 10.1094/PHP-12-19-0093-RS
Sikandar, A., Jia, L., Wu, H., and Yang, S. (2023). Meloidogyne enterolobii risk to agriculture, its present status and future prospective for management. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1093657
Stetina, S. R., McGawley, E. C., and Russin, J. S. (1997). Extraction of root-associated Meloidogyne incognita and Rotylenchulus reniformis. J. Nematol. 29, 209–215.
Tandingan, I. C., Prot, J.-C., and Davide, R. G. (1996). Influence of water management on tolerance of rice cultivars for Meloidogyne graminicola. Fundam. Appl. Nematol. 19, 189–192.
Tigano, M., De Siqueira, K., Castagnone-Sereno, P., Mulet, K., Queiroz, P., Dos Santos, M., et al. (2010). Genetic diversity of the root-knot nematode Meloidogyne enterolobii and development of a SCAR marker for this guava-damaging species. Plant Pathol. 59, 1054–1061. doi: 10.1111/j.1365-3059.2010.02350.x
Velloso, J. A., Maquilan, M. A. D., Campos, V. P., Brito, J. A., and Dickson, D. W. (2022). Temperature effects on development of Meloidogyne enterolobii and M. floridensis. J. Nematolo. 54, 1–11. doi: 10.2478/jofnem-2022-0013
Viaene, N. M. and Abawi, G. S. (1998). Management of Meloidogyne hapla on lettuce in organic soil with Sudangrass as a cover crop. Plant Dis. 82, 945–952. doi: 10.1094/PDIS.1998.82.8.945
Wong, T. K. and Mai, W. F. (1973). Effect of temperature on growth, development and reproduction of Meloidogyne hapla in lettuce. J. Nematol. 5, 139–142.
Keywords: damage, greenhouse, reproduction factor, quarantine nematode, host plant
Citation: Konigopal H, Finckh MR, Bailly-Bechet M, Danchin EGJ and Kiewnick S (2025) Intrinsic host range of the root-knot nematode Meloidogyne enterolobii and virulent M. incognita populations. Front. Plant Sci. 16:1668191. doi: 10.3389/fpls.2025.1668191
Received: 17 July 2025; Accepted: 16 October 2025;
Published: 31 October 2025.
Edited by:
Charith Raj Adkar-Purushothama, Université de Sherbrooke, CanadaReviewed by:
Tariq Mukhtar, Pir Mehr Ali Shah Arid Agriculture University, PakistanJuvenil Cares, University of Brasilia, Brazil
Copyright © 2025 Konigopal, Finckh, Bailly-Bechet, Danchin and Kiewnick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Kiewnick, c2ViYXN0aWFuLmtpZXduaWNrQGp1bGl1cy1rdWVobi5kZQ==
†These authors have contributed equally to this work and share last authorship
 Hemanth Konigopal
Hemanth Konigopal Maria R. Finckh
Maria R. Finckh Marc Bailly-Bechet3
Marc Bailly-Bechet3 Etienne G.J. Danchin
Etienne G.J. Danchin Sebastian Kiewnick
Sebastian Kiewnick