- 1The Germplasm Bank of Wild Species and Yunnan Key Laboratory of Crop Wild Relatives Omics, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2University of Chinese Academy of Sciences, Beijing, China
Structural variations in legume plastomes impact phylogenetic and evolutionary studies. In this study, we focus on the tribe Mimoseae by integrating a newly assembled plastome of Calliandra haematocephala (from PacBio sequencing data) with 15 previously published plastomes representing major lineages, to analyze structural rearrangements, repeat sequences, and selection pressures. The plastome of C. haematocephala revealed extensive structural rearrangements and a ca. 14-kb expansion of the inverted repeats (IRs) into the large single copy (LSC) region, resulting in IRs of 42,069 bp. It also contained a high abundance of clustered dispersed repeats (> 90 bp). These features potentially contribute to significant plastome rearrangements, making it the largest plastome (200,623 bp) recorded to date in Mimoseae and, more broadly, in Leguminosae. Nucleotide diversity (Pi) analysis identified several highly variable regions (Pi > 0.03), including the genes accD, rps18, clpP, and multiple non-coding loci, suggesting their potential as molecular markers. Selection pressure analyses detected positive selection (dN/dS > 1) in clpP, ycf2, and rps17, suggesting possible roles in adaptive evolution. Branch-specific positive selection was also found in genes such as rpoC1 and atpA within the Calliandra clade, indicating lineage-specific adaptive pressures. This study highlights the dynamic evolution of plastomes in Mimoseae and offers new insights into their structural diversity and adaptive evolution.
1 Introduction
The plastome is a circular molecule typically exhibiting a conserved quadripartite structure in most autotrophic plants: two inverted repeats (IRs) separate the large single copy (LSC) and the small single copy (SSC) regions (Jansen and Ruhlman, 2012; Wang et al., 2024). Plastome size generally ranges from 120 to 160 kb (Bock, 2007), though frequently influenced by IR expansion/contraction or loss (Jansen and Ruhlman, 2012). Extensive IR expansion occurs in plastomes of multiple taxa such as Pelargonium L’Hér. ex Aiton (Geraniaceae), where IRs exceed 87 kb, yielding plastomes > 242 kb (Chumley et al., 2006; Weng et al., 2017). IR contraction is also common, e.g., plastomes of Pentasachme caudata Wall. ex Wight (Apocynaceae) (Wang et al., 2023) and Dicorynia paraensis Benth. (Fabaceae) (Bai et al., 2020). Some plastomes of non-autotrophic plants lose one IR copy (Huang et al., 2011), concomitant with the loss of photosynthesis-related genes (Krause and Scharff, 2014), resulting in significant plastomic variations. IR loss also occurs in autotrophic seed plants, such as the Inverted Repeat-Lacking Clade (IRLC) and Camoensia Welw. ex Benth. of Fabaceae (Choi et al., 2019; Cauz-Santos et al., 2020), certain species in Geraniaceae (Guisinger et al., 2011; Blazier et al., 2016; Ruhlman et al., 2017), and some species of Cactaceae (Sanderson et al., 2015). These plastomes usually exhibit considerable plastomic changes. Some IR-retaining species also exhibit substantial plastome structural variations, such as those in certain species of Campanulaceae (Wu and Chaw, 2014), Oleaceae (Cosner et al., 2004), Plantaginaceae (Zhu et al., 2016), and Pelargonium (Lee et al., 2007). Plastome size and structural variation are influenced not only by IR dynamics but also by repetitive sequences, including tandem repeats (Jo et al., 2011; Dugas et al., 2015) and dispersed repeats (Cosner et al., 1997; Haberle et al., 2008; Jansen and Ruhlman, 2012).
Significant plastome structural variation occurs among legumes, particularly in subfamily Papilionoideae. A 50-kb inversion in the LSC region characterizes plastomes of most papilionoids, except a few early-diverged lineages (Doyle et al., 1996; Chen et al., 2025). Plastomes of Vigna radiata (L.) R.Wilczek and Phaseolus vulgaris L. exhibit an additional 78-kb inversion (Palmer et al., 1988), while an additional 36-kb inversion within the 50-kb segment is observed in Lupinus luteus L. (Martin et al., 2014). IRLC plastomes lack complete IRs (Palmer et al., 1987; Doyle et al., 1996; Wojciechowski et al., 2000), with many species exhibiting significant plastome structural rearrangements like gene/intron losses (e.g., Cicer arietinum L.) (Jansen et al., 2008). In addition to the above-mentioned two subfamilies, a 7.5-kb inversion was found in Tylosema esculentum (Burch.) A.Schreib. (Cercidoideae), and this structural variation was initially proposed to occur throughout Bauhinia s.l. (Kim and Cullis, 2017), but subsequent studies demonstrated that it is restricted to Tylosema (Schweinf.) Torre & Hillc. (Wang et al., 2017b).
Mimosoideae, redefined as tribe Mimoseae (subfamily Caesalpinioideae), comprises about 100 genera and 3500 species distributed in pantropical regions (Ringelberg et al., 2022; Bruneau et al., 2024). Many species are ecologically dominant in major tropical biomes (Lewis et al., 2005; LPWG, 2013). Some species are of high economic value as fodder crops [Leucaena leucocephala (Lam.) de Wit], ornamental plants (species of Albizia Durazz. and Calliandra Benth.), timber trees (species of Acacia Mill., Anadenanthera Speg., and Prosopis L.), and sources of food thickeners (species of Acacia) (Lewis et al., 2005). Plastomes of this tribe exhibit a relatively conserved structure. However, plastomes of a clade formed by tribe Ingeae and Acacia s.s. contain a ca. 13-kb IR expansion into the SSC (Dugas et al., 2015; Williams et al., 2015), representing the largest known legume plastomes (from 174,217 bp to 178,887 bp) (Wang et al., 2017a). This clade, designated as the Inverted Repeat-expanding clade (IREC) (Wang, 2017), exhibits additional structural variations such as IR-LSC junction alterations, intron losses, gene duplications, and inversions (Wang et al., 2017a). Our preliminary work revealed a large, highly variable plastome in the ornamental species Calliandra haematocephala Hassk. Given that Calliandra resides within the tribe Mimoseae (Bruneau et al., 2024) and its plastome variations remain uncharacterized, it presents an excellent opportunity for making an investigation.
In this study, we selected 15 species from representative genera across the major clades and grades within Mimoseae to capture the extent of the plastomic structural diversity. We investigated structural divergence by comparing the newly sequenced plastome of C. haematocephala with those of these 15 representative plastomes of this tribe. We conducted a comprehensive phylogenetic analysis using 16 plastomes to clarify the relationship within Mimoseae. The objectives of this study are: 1. To elucidate the unique plastome structural variations of the C. haematocephala plastome; 2. To identify hypervariable regions that may serve as informative molecular markers for future phylogenetic studies and species identifying in Mimoseae. We report the plastome of C. haematocephala, revealing extensive rearrangements despite IR retention. This resource facilitates further exploration of plastome structural dynamics in Mimoseae.
2 Materials and methods
2.1 Sampling, genomic DNA extraction, and sequencing
Our study incorporated 15 assembled plastomes from GenBank and one newly sequenced plastome, representing 16 clades or grades of the tribe Mimoseae (Supplementary Table S1). The 15 selected species, each representing a genus from the major clades or grades of Mimoseae, reflect the structural diversity of plastomes in this tribe. We selected Adenanthera microsperma Teijsm. & Binn., an early-divergent species of Mimoseae (Bruneau et al., 2024), as the outgroup for the taxa of interest; its plastome was retrieved from GenBank (Supplementary Table S1). Fresh leaf tissue of C. haematocephala was flash-frozen in Liquid nitrogen collected from Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (Menglun Town, Xishuangbanna Dai Autonomous Prefecture, Yunnan Province, China; voucher specimen deposited in the Kunming Institute of Botany, Chinese Academy of Sciences). Samples were subsequently sent to BIOMARKER Technologies Co., Ltd. (Beijing) for total DNA and RNA extraction. Target-enriched fragment selection was performed using BluePippin, followed by library preparation according to PacBio standard protocols. The sequencing libraries were ultimately sequenced for full-length sequencing on a PacBio Sequel II platform.
2.2 Assembly and annotation of plastomes
Oatk v1.0 (Zhou et al., 2024) and TIPPo v2.3 (Xian et al., 2025) were used to assemble the complete plastome of C. haematocephala from raw sequencing data, with assemblies cross-validated to ensure accuracy. All 17 plastomes (including the outgroup) were annotated using PGA (Qu et al., 2019; Zhang et al., 2025). Finally, manual adjustments of the annotation were performed using Geneious v.9.0.2 (Kearse et al., 2012). Physical maps were generated using the online tool OGDRAW (Greiner et al., 2019). Genome collinearity was analyzed with the progressiveMauve (Darling et al., 2004) plugin in Geneious (Kearse et al., 2012).
2.3 Phylogenetic analysis
Eighty protein-coding genes (PCGs) were extracted from 17 plastomes of Mimoseae, aligned using MAFFT v7.487 (Katoh et al., 2002), and concatenated into a data matrix. Concurrently, a whole plastome alignment (designated as Full-Con) was constructed using MAFFT and PhyloSuite v1.2.2 (Zhang et al., 2020). Maximum likelihood (ML) trees were inferred from both matrices using RAxML v8.2.12 (Stamatakis, 2014) under the GTRGAMMA model with 1000 rapid bootstrap replicates.
2.4 IR boundary analysis and nucleotide diversity
To characterize IR expansion/contraction across 16 plastomes from Mimoseae (excluding the outgroup), boundaries of IR/SC were visualized and compared using IRscope (Amiryousefi et al., 2018). Whole plastomes were aligned under the shuffle-LAGAN model and visualized using mVISTA (Frazer et al., 2004). Sixty-three PCGs and 86 non-coding regions including introns or intergenic spacers with a length exceeding 200 bp were aligned using MAFFT; nucleotide diversity (Pi) was subsequently calculated via sliding window analysis using DnaSP v. 6.10 (Rozas et al., 2017) with a window length of 600 bp and a step size of 200 bp.
2.5 Repeat sequences identification and codon usage analysis
Simple sequence repeats (SSRs) were identified using the online software MISA (Beier et al., 2017) (https://webblast.ipk-gatersleben.de/misa/) with minimum thresholds of 10, 5, 4, 3, 3, 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively. Long repeat sequences (forward, palindromic, reverse, and complement repeats) were detected with REPuter (Kurtz and Schleiermacher, 1999; Kurtz et al., 2001) (https://bibiserv.cebitec.uni-bielefeld.de/reputer/) using the following parameters: minimum repeat length = 30, Hamming distance = 3, minimum identity = 90%, and maximum computed repeats = 150.
2.6 Selection pressure analyses
The alignment of PCGs was generated using the MAFFT plugin with “Codon” mode, followed by format conversion to PML using the “Convert Sequence Format” tool in PhyloSuite v1.2.2 (Zhang et al., 2020). For phylogenetic reconstruction, sequences from all 16 species of Mimoseae were aligned using MAFFT v7.487 (Katoh et al., 2002) with default parameters. ML trees of these PCGs alignments were inferred respectively using RAxML v8.2.12 (Stamatakis, 2014) under the GTRGAMMA substitution model and 1000 rapid bootstrap replicates.
Non-synonymous/synonymous rate ratios (dN/dS) were calculated using CodeML in PAML v4.10.7 (Yang, 2007) with the ML tree and PML-formatted alignment files as inputs. Branch model analyses were employed to assess selective pressures. Comparative analyses between the one-ratio and the two-ratio models were performed to identify variation in selective pressure across branches in PCGs. Model comparisons were executed through likelihood ratio tests (LRT) (Yang et al., 2005), where designated clades were specified as foreground branches and remaining lineages as background. CodeML was run under branch models (runmode = 0, model = 0 or 2, NSsites = 0) as described in the PAML manual (Álvarez-Carretero et al., 2023). CodeML configuration files were established in accordance with official documentation (Álvarez-Carretero et al., 2023), applying this protocol to all tested clades (Lu et al., 2024).
3 Result
3.1 Plastome size and features
We assembled and annotated the newly sequenced plastome of C. haematocephala and re-annotated 15 additional plastomes of Mimoseae from GenBank. All plastomes exhibited a circular, quadripartite structure comprising the LSC region, SSC region, and IR regions. Total lengths ranged from 159,963 to 200,623 bp, with the LSC regions ranging from 87,462 to 110,424 bp, SSC regions ranging from 4,470 to 19,392 bp, and IR regions ranging from 25,341 to 42,069 bp. The total GC content of these plastomes ranged from 35.0% to 36.6%. The plastome size of C. haematocephala is 200,623 bp, with an LSC length of 110,424 bp, an SSC length of 6,061 bp, and a single IR length of 42,069 bp. The structure of the plastome of C. haematocephala is illustrated in Figure 1.
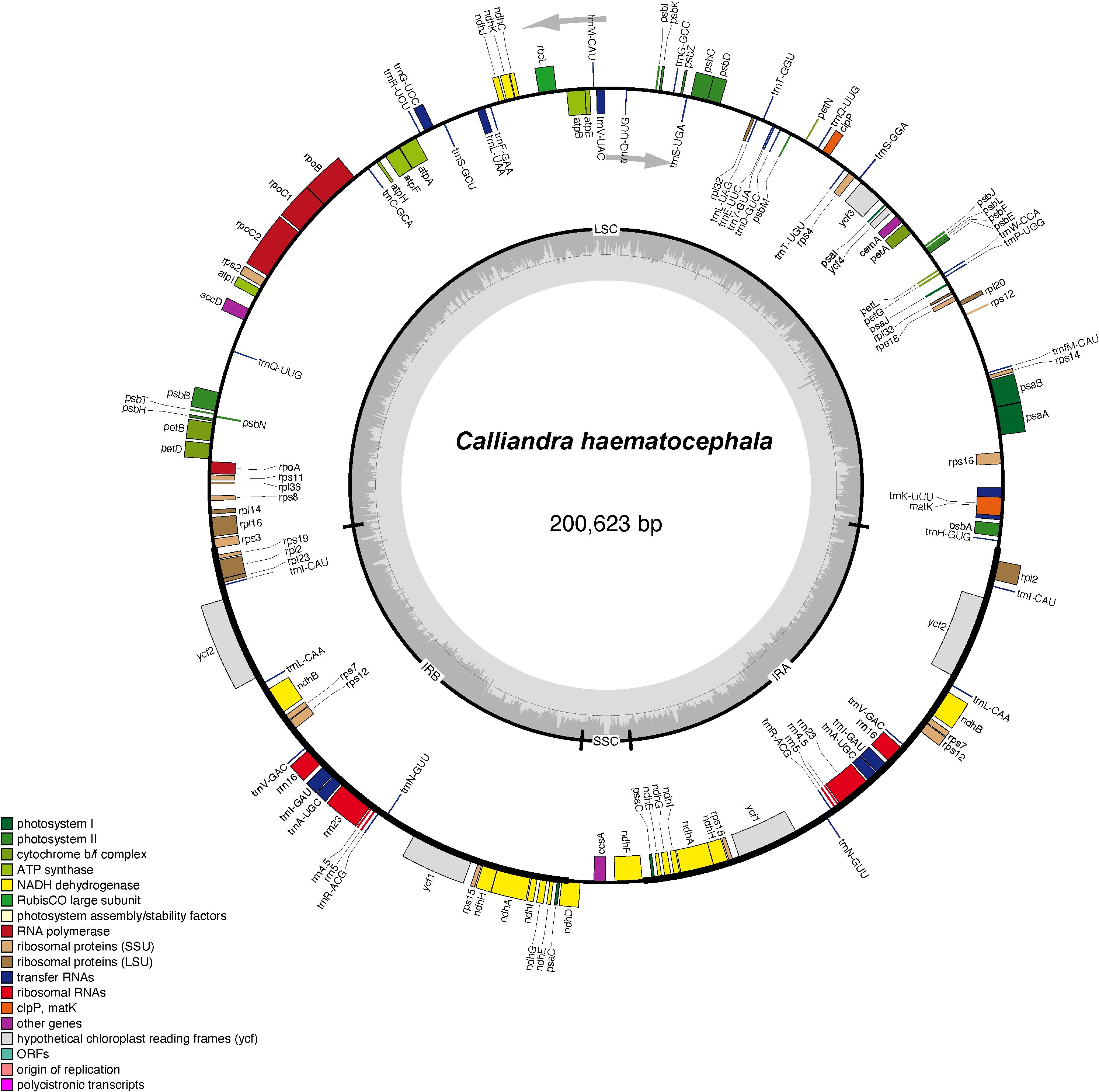
Figure 1. The circular plastome map of Calliandra haematocephala. Genes inside the circle are transcribed clockwise, while genes outside the circle are transcribed counterclockwise. Genes belonging to different functional groups are marked with different colors. The darker and lighter gray areas in the inner circle corresponds to the GC content and AT content, respectively.
The 16 plastomes of Mimoseae exhibited relatively high gene content and collinearity (Figure 2). Fifteen plastomes maintained nearly identical gene arrangements, while the plastome of C. haematocephala showed substantial structural variation. Annotation revealed that these 15 plastomes contained 128 to 142 genes, including 83 to 95 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. In contrast, C. haematocephala contained 137 genes, including 90 protein-coding genes (15 in IR regions), 39 tRNA genes (6 in IR regions), and 8 rRNA genes (4 in IR regions).
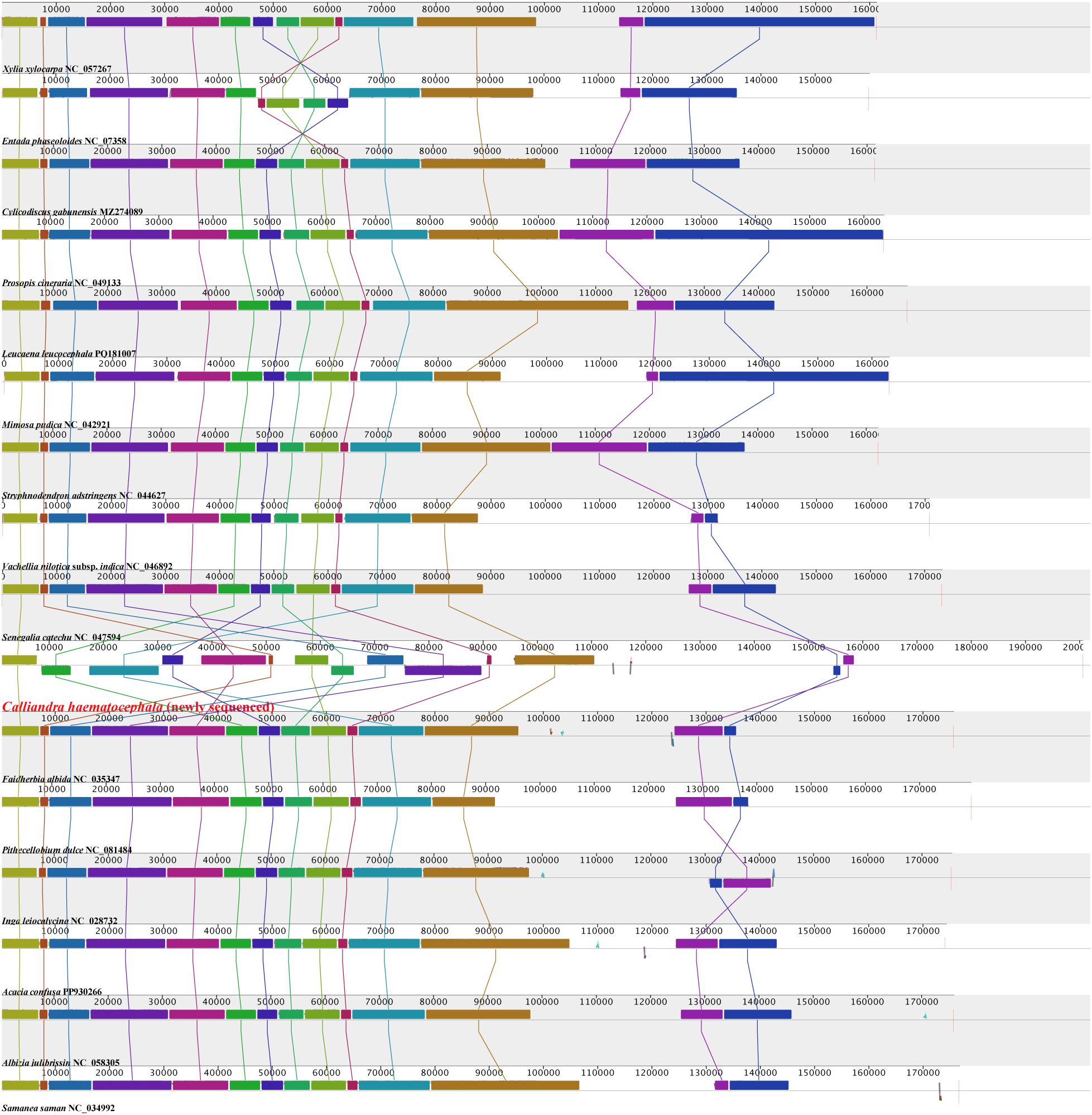
Figure 2. Collinearity comparison analysis of 16 plastomes from tribe Mimoseae. The horizontally arranged colored rectangles linked by sane colored line represent locally collinear blocks, indicating homologous regions with conserved gene order and sequence similarity.
Entada phaseoloides (L.) Merr. exhibited a rearrangement distance of 1, corresponding to a single inversion event involving four contiguous blocks. Inga leiocalycina Benth. showed a rearrangement distance of 1, associated with a single inversion of two contiguous blocks. Calliandra haematocephala displayed a rearrangement distance of 12, reflecting extensive genomic rearrangement involving multiple translocations and inversions (Figure 2; Supplementary Table S2).
3.2 IR expansion and contraction in 16 plastomes of Mimoseae
Among the 16 analyzed plastomes of Mimoseae, only plastomes of four species [Cylicodiscus gabunensis Harms, L. leucocephala, Prosopis cineraria (L.) Druce, and Stryphnodendron adstringens (Mart.) Coville] exhibited canonical IRs without significant expansion or contraction, with IR lengths ranging from 25,931 bp (P. cineraria) to 26,062 bp (S. adstringens). Nine species [Acacia confusa Merr., Albizia julibrissin Durazz., C. haematocephala, Faidherbia albida (Delile) A.Chev., I. leiocalycina, Pithecellobium dulce (Roxb.) Benth., Samanea saman (Jacq.) Merr., Senegalia catechu (L.f.) P.J.H.Hurter & Mabb., and Vachellia nilotica subsp. indica (Benth.) Kyal. & Boatwr.] exhibited substantial IR expansions of approximately 13–16 kb into the SSC region, resulting in IR lengths ranging from 39,347 bp (V. nilotica subsp. indica) to 42,069 bp (C. haematocephala).
Except for C. haematocephala, whose IRB/SSC junction (JSB) was located within ndhD, duplicating only eight complete protein-coding genes (from ycf1 to psaC), the IRs of the remaining eight species contained nine complete protein-coding genes (from ycf1 to ndhD). In A. confusa, A. julibrissin, F. albida, P. dulce, S. saman, and S. catechu, the JSB shifted into ndhF, duplicating its 3’ end (19–205 bp), while the SSC/IRA junction (JSA) relocated from ycf1 to between ccsA and ndhD (except F. albida having JSA within the stop codon of ccsA).
In C. haematocephala, the JSB shifted into ndhD, duplicating its 5’ end (538 bp), and the JSA relocated from ycf1 to between ndhF and psaC. In I. leiocalycina, JSB occurred between ndhD and ccsA, while JSA shifted into ndhF, duplicating its 3’ end (6 bp). In V. nilotica subsp. indica, JSB relocated between ndhD and ndhF, and JSA shifted to between ccsA and ndhD.
Four species (A. julibrissin, F. albida, I. leiocalycina, and S. saman) exhibited LSC/IRB junctions (JLB) within rps19, duplicating its 5’ end (100–105 bp). Calliandra haematocephala displayed an additional 0.7-kb IR expansion into LSC, shifting its JLB into rps3 and incorporating the entire rps19 and 31 bp of rps3. In P. dulce, a 1.7-kb IR expansion into the LSC positioned JLB between rps3 and rpl16, adding rps3 and rps19 to IR. S. catechu and V. nilotica subsp. indica showed a 1.2-kb IR expansion into LSC, relocating JLB between rps19 and rpl23 and incorporating rps19. Conversely, A. confusa exhibited a 0.3-kb IR contraction, shifting its JLB into rpl2 and transferring the entire rpl2 (4 bp) to LSC.
Three species [E. phaseoloides, Mimosa pudica L., and Xylia xylocarpa (Roxb.) W.Theob.] displayed IR contractions, with IR lengths ranging from 25,341 bp (E. phaseoloides) to 26,370 bp (X. xylocarpa), retaining entire ndhF in SSC. JSB of E. phaseoloides, M. pudica, and X. xylocarpa shifted between trnN and ndhF, while JSA remained within ycf1 (E. phaseoloides and X. xylocarpa) or relocated between ycf1 and trnN (M. pudica). The JLB of E. phaseoloides and M. pudica occurred within rps19, duplicating their 5’ ends (104 bp and 103 bp, respectively). Xylia xylocarpa exhibited a 0.2-kb IR expansion into LSC, positioning its JLB between rpl22 and rps19 and incorporating the entire rps19 into the IR (Figure 3).
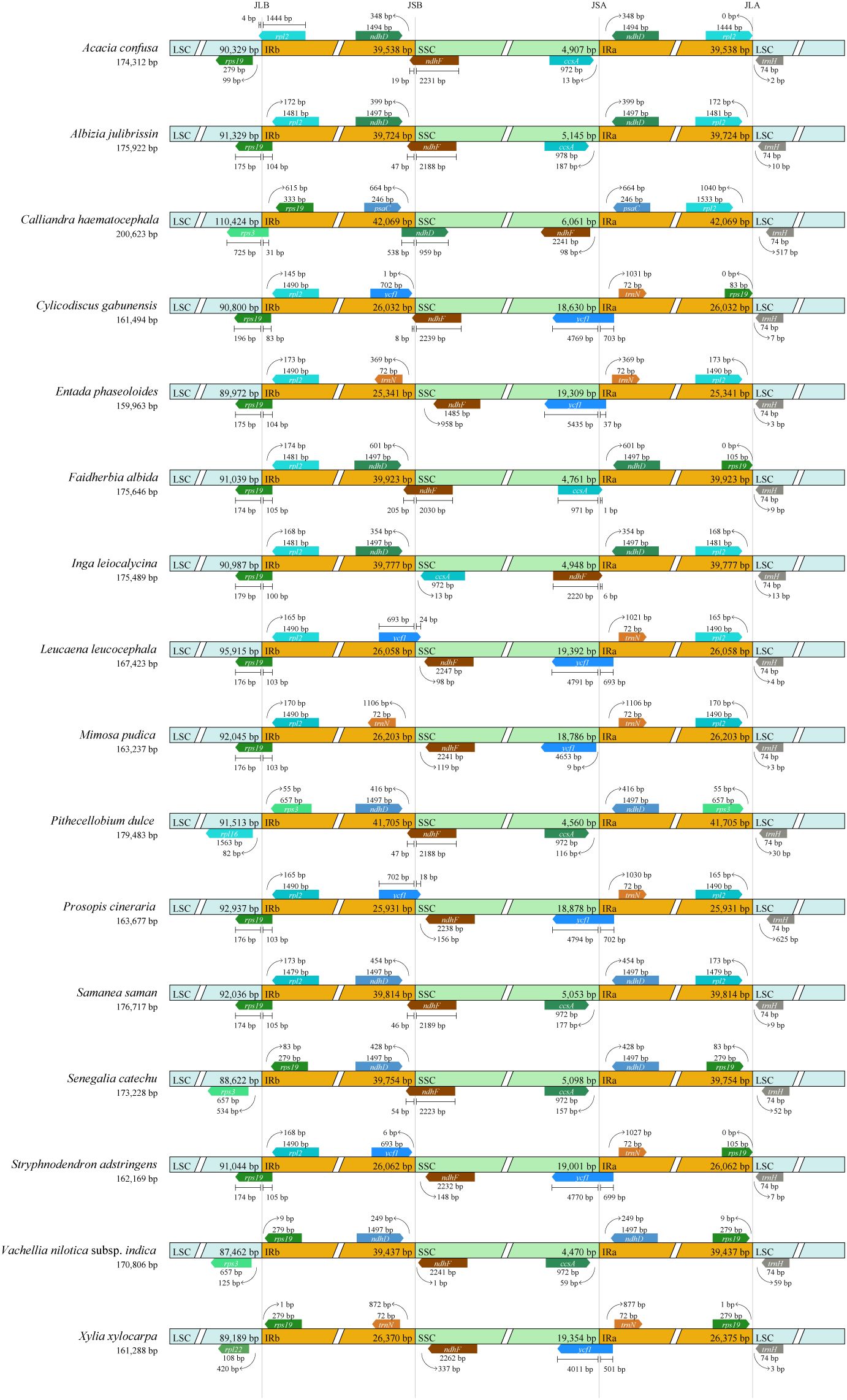
Figure 3. Comparison of the LSC, SSC, and IR boundaries of 16 plastomes from the tribe Mimoseae. JLB, JSB, JSA, and JLA refer to the junctions of LSC/IRB, SSC/IRB, SSC/IRA, and LSC/IRA, respectively. The boxes above and below the line refer to the forward and reverse genes.
3.3 Identification of divergence hotspot regions
Comparative analysis of the 16 plastomes of Mimoseae revealed higher sequence variability in single copy regions than in IR regions, and in non-coding regions than in coding regions. Nucleotide diversity (Pi) analysis identified several highly variable regions (Pi > 0.03): accD, rps18, rpl20, clpP, rps11, and rps3 (located in LSC), plus ccsA and ycf1 (located in SSC). Among non-coding regions, rps8-rpl14, trnS (GCU)-trnG (UCC), clpP_intron1, and rpl36-rps8 (located in LSC), as well as trnN (GUU)-ndhF and ycf1-trnN (GUU) (located in SSC) exhibited even higher nucleotide diversity (Pi > 0.09). Most other non-coding regions displayed Pi values between 0.03 and 0.09 (Figure 4).
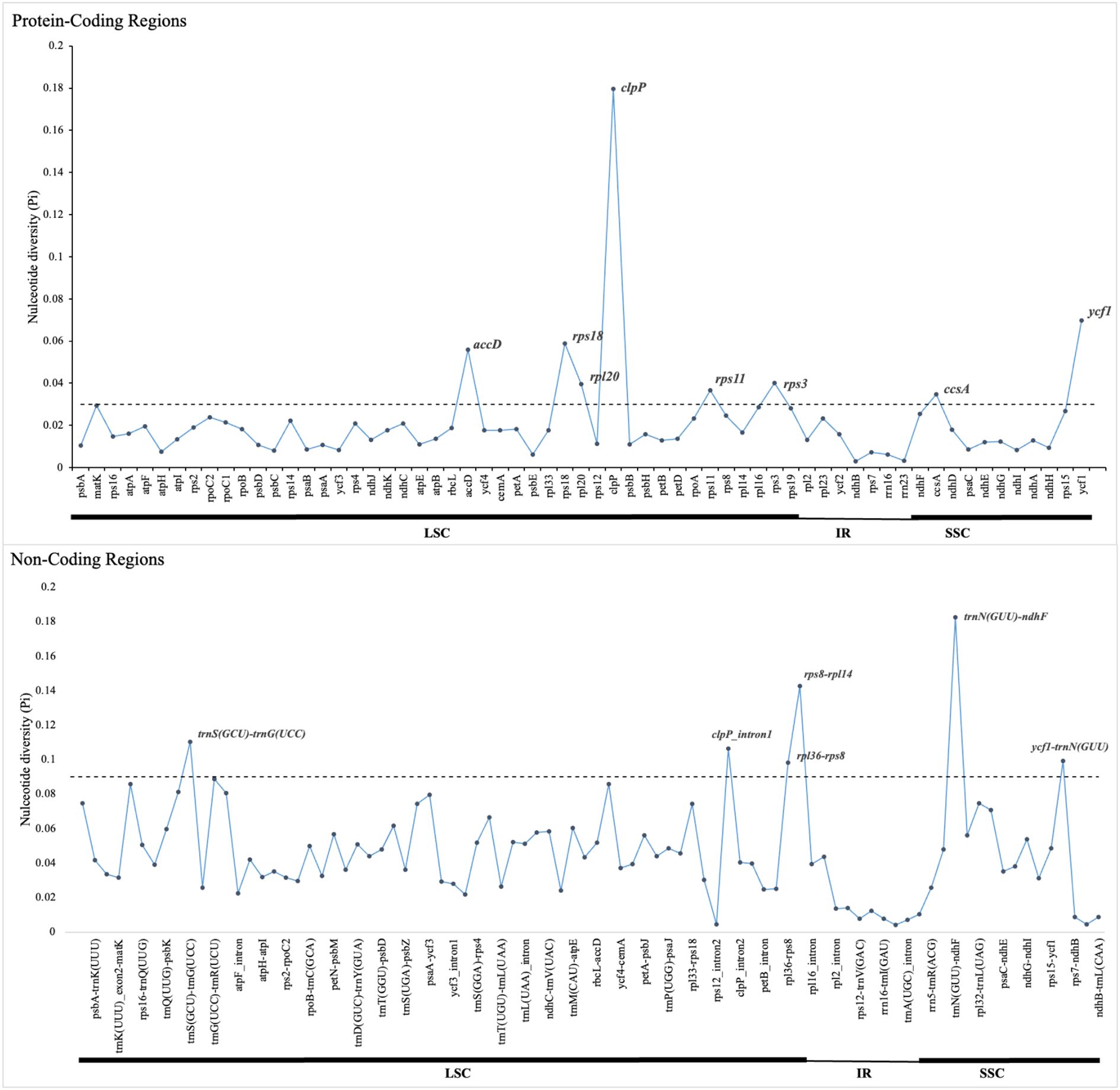
Figure 4. Nucleotide diversity (Pi) values of different regions of 16 plastomes from tribe Mimoseae. The dotted line indicates the threshold for high Pi values.
3.4 Repeat analysis
A total of 2,314 SSRs were identified across the 16 plastomes of Mimoseae, ranging from 85 in X. xylocarpa to 200 in A. julibrissin (Figure 5A). The majority of these SSRs (~70%) were located in non-coding regions (Figure 5B). Among six types of SSR, mononucleotides constituted the largest proportion, followed by dinucleotides and tetranucleotides. Trinucleotides were less frequent, while pentanucleotides and hexanucleotides appeared only in a subset of plastomes (Figure 5C).
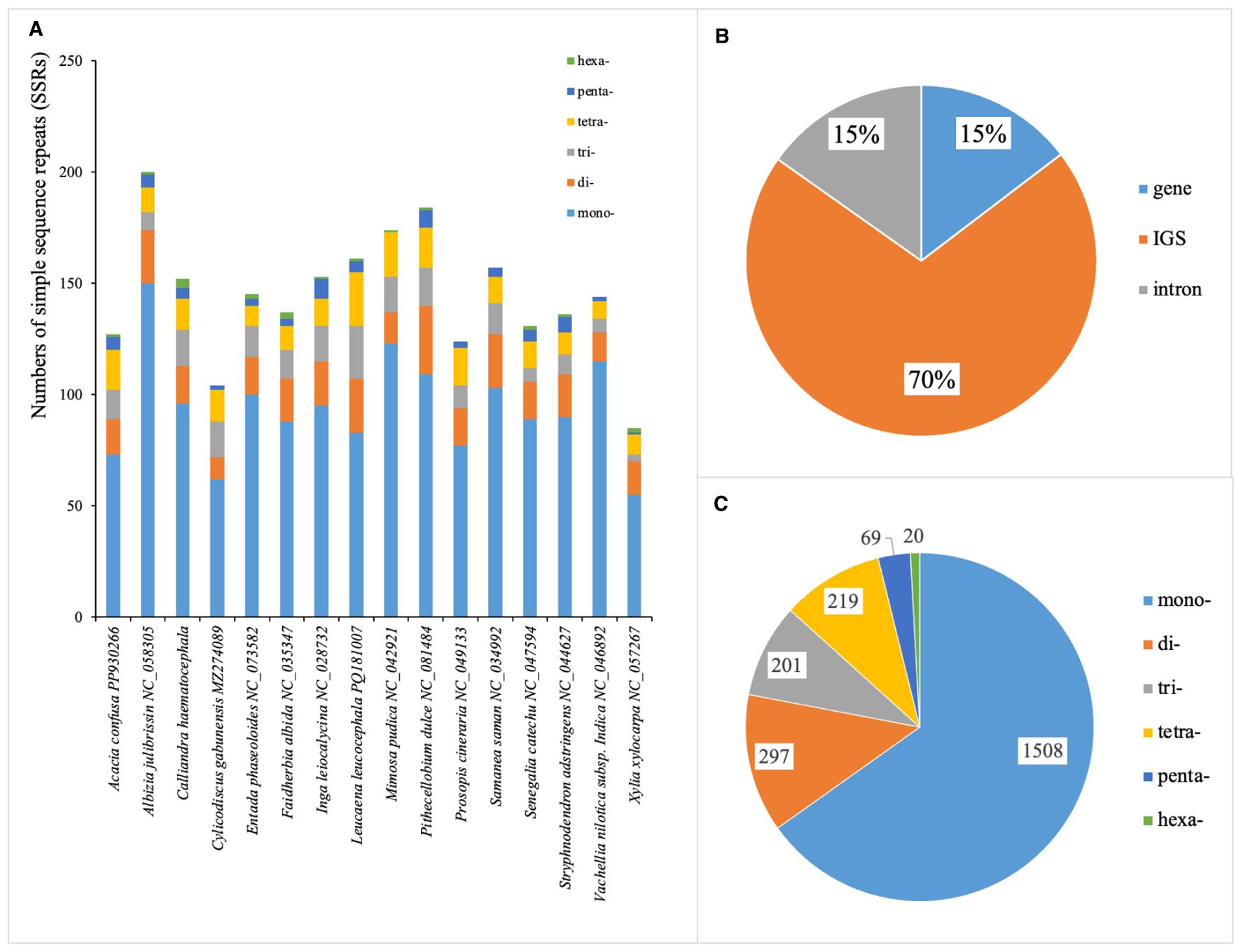
Figure 5. Distribution of simple sequence repeats (SSRs) in 16 plastomes of tribe Mimoseae. (A) Total numbers of SSRs detected in each species; (B) Distribution of SSRs in introns, coding sequences, and intergenic spacers (IGS); (C) Frequency of different SSR types.
Additionally, we identified 1,290 long repeat sequences, comprising 697 forward, 196 reverse, 340 palindromic, and 57 complement repeats. The number of long repeats per plastome varied substantially, from 30 in E. phaseoloides to 150 in P. dulce, L. leucocephala, and I. leiocalycina (Figure 6A). Despite this interspecific variation, the distribution patterns of repeat types and length categories were largely conserved across species, with most repeats occurring in non-coding regions. Repeats of 30–45 bp predominated, followed by 45–60 bp, > 90 bp, 60–75 bp, and 75–90 bp (Figure 6B).
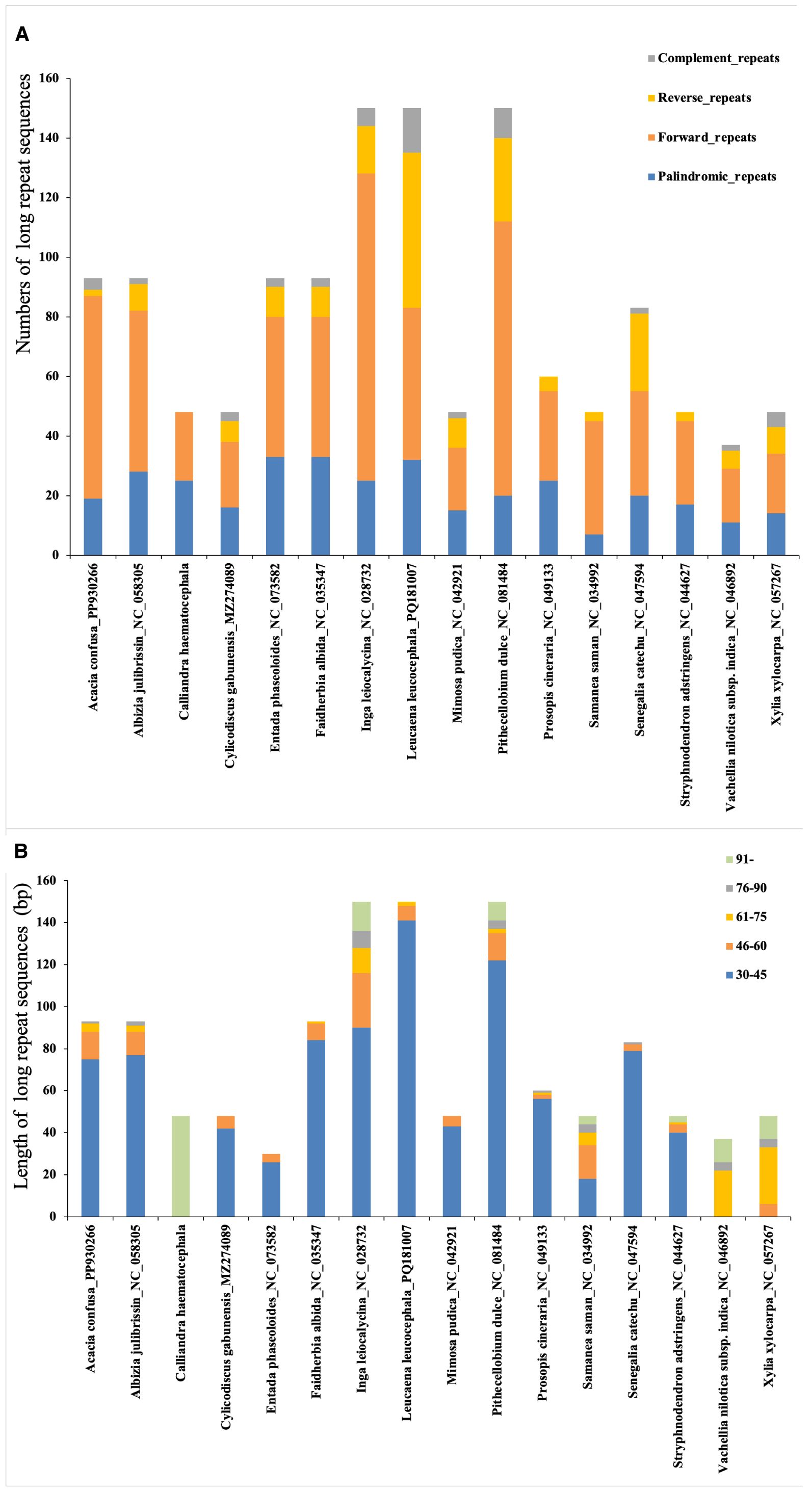
Figure 6. Analysis of long repeat sequences in 16 plastomes of tribe Mimoseae. (A) The types and abundance of long repeat sequences identified in each species; (B) Length distribution of these repeat sequences.
Notably, C. haematocephala exhibited an exceptional pattern, with 48 long repeat sequences identified. These included 25 palindromic and 23 forward repeats, while no reverse or complement repeats were detected. The lengths of these repeats ranged from 228 to 1,610 bp, all exceeding 90 bp.
3.5 Codon usage analyses
Protein-coding regions in the 16 plastomes of Mimoseae contained 22,226 to 22,697 codons. AUU (encoding Isoleucine) was the most frequent codon (980 – 1,026 occurrences). UUA showed the highest mean relative synonymous codon usage (RSCU) value (mean value = 1.93). Thirty codons exhibited RSCU > 1, with 29 ending with A/U. Two codons, AUG (methionine) and UGG (tryptophan), had RSCU values of 1 in all 16 species, indicating no codon bias (Figure 7).
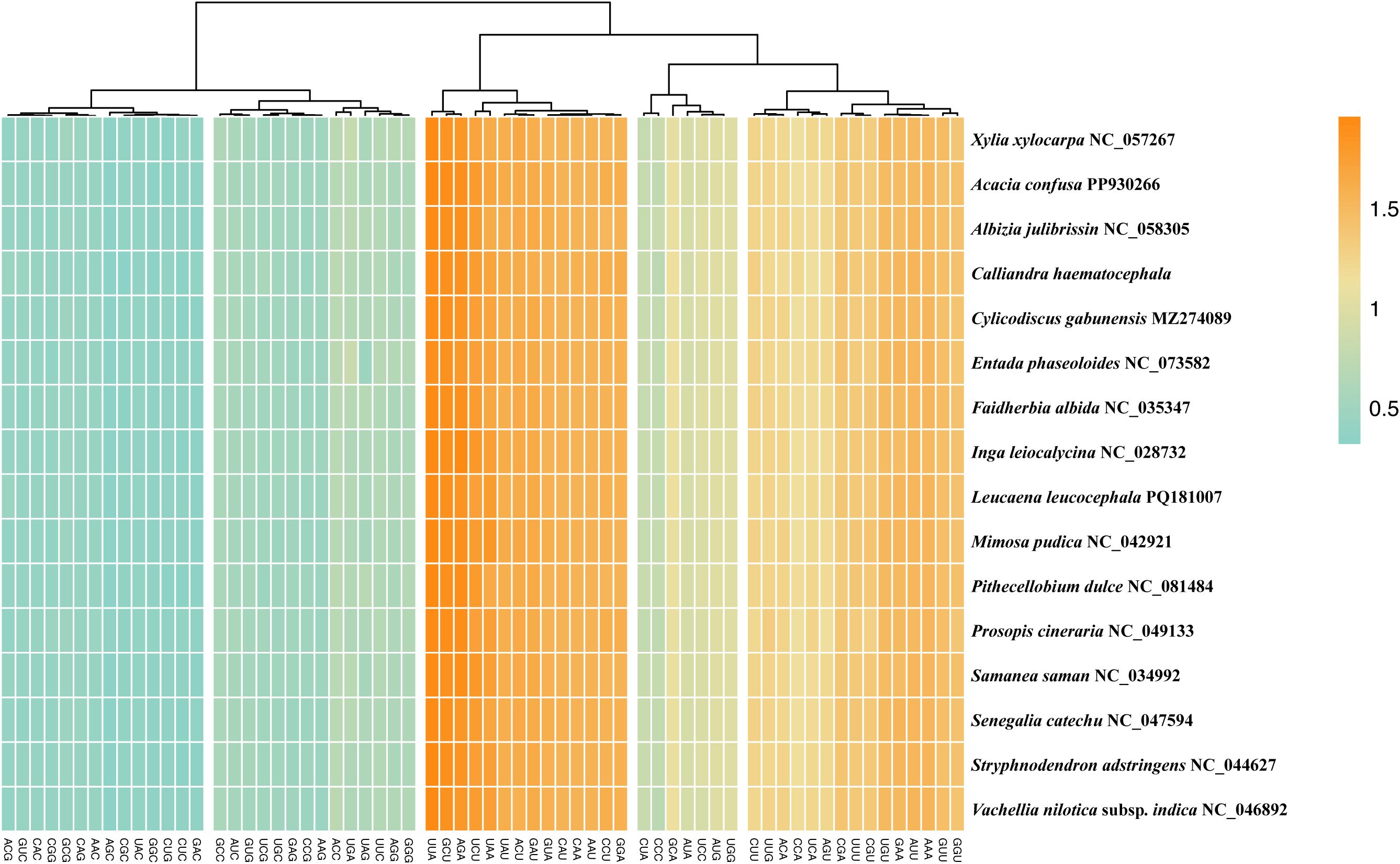
Figure 7. Relative synonymous codon usage (RSCU) analysis for protein-coding genes across 16 plastomes of the tribe Mimoseae.
3.6 Phylogenetic inferences
Phylogenetic trees reconstructed from the PCGs and CP matrices showed congruent topologies, with strong nodal support (bootstrap support, BS > 90%). The ML tree reconstructed from the whole plastome alignment (Figure 8) exhibited higher overall support, and displayed a topology largely congruent with previous studies (Bruneau et al., 2024; Queiroz et al., 2024), and served for subsequent selection pressure analyses.
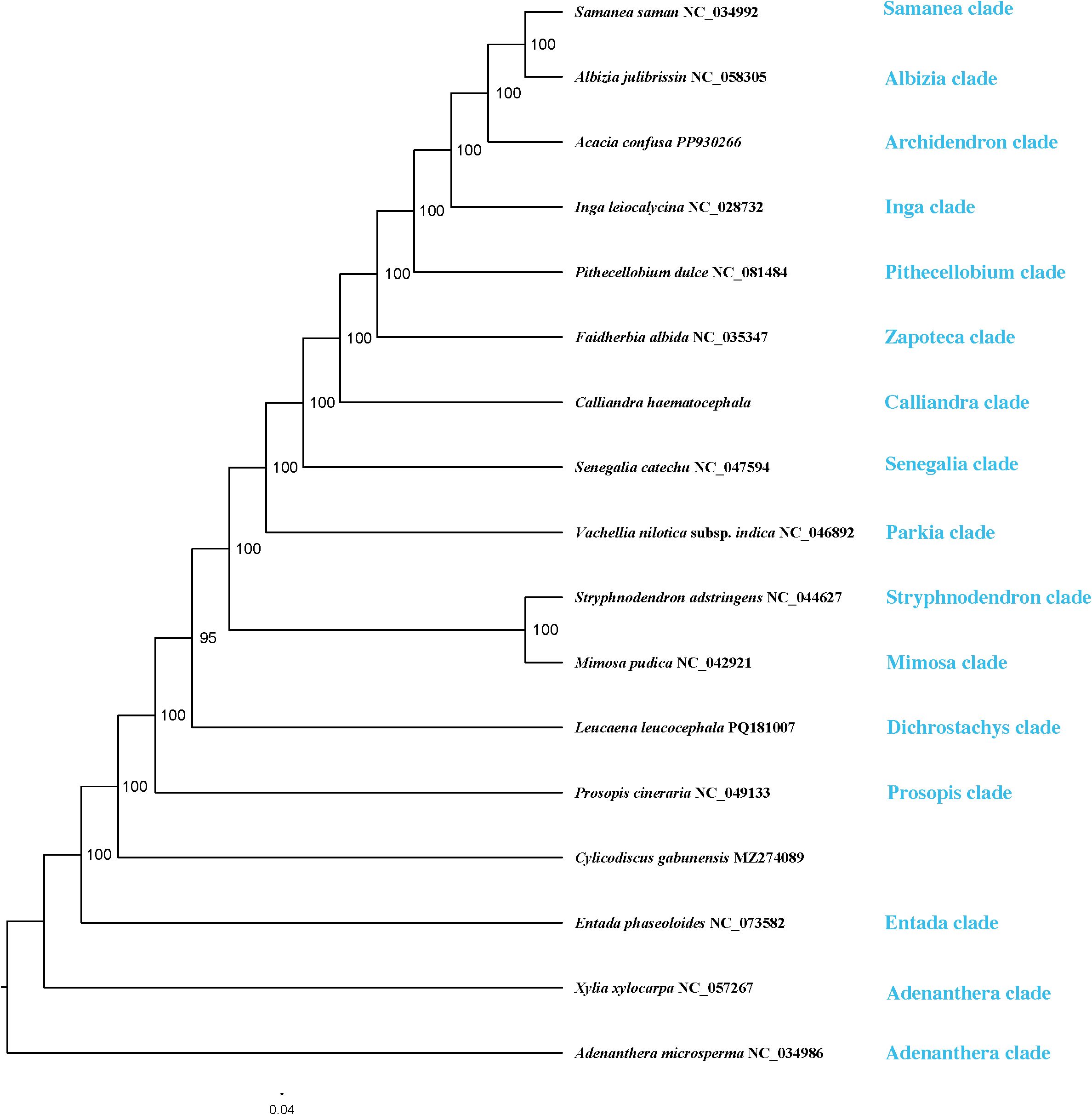
Figure 8. Maximum likelihood (ML) tree based on whole plastome sequences of 16 species of the tribe Mimoseae and one outgroup. Bootstrap values are shown at the branch nodes.
3.7 Selection pressure analyses
The dN/dS ratios for 76 PCGs across the 16 Mimoseae plastomes ranged from 0.0001 to 2.04416. Three genes, clpP, ycf2, and rps17, showed dN/dS > 1, indicating positive selection, while psaJ and psbI exhibited dN/dS ratios close to 0 (0.0001).
Branch model analyses assessed differential selection between foreground and background branches. Likelihood ratio tests comparing M0 (one-ratio) and M2 (two-ratio) models favored M0 for most PCGs. After excluding abnormal dN/dS ratios (ratio = 999), 12 genes (rpoC1, atpA, rpoB, rpoC2, rps11, rps12, atpB, clpP, rps18, rpoA, petB, and rpl14) better fit M2 in the Calliandra lineage. Specifically, rpoC1, atpA, rpoB, rps11, rps12, rps18, and rpl14 underwent positive selection; rpoC2, rpoA, and petB showed relaxed purifying selection; atpB experienced intensified purifying selection (Figure 9; Supplementary Table S3).
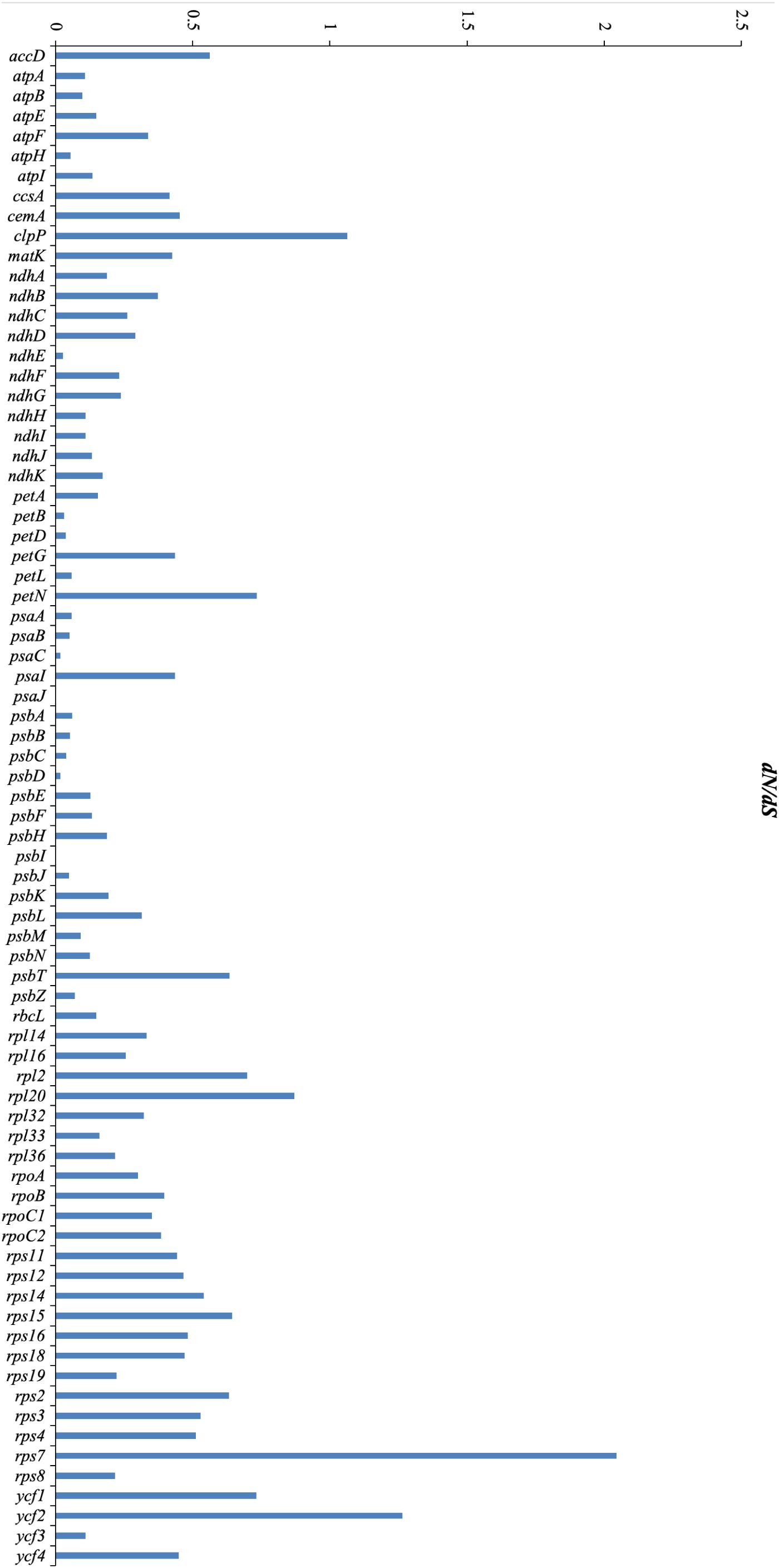
Figure 9. Distribution of non-synonymous/synonymous rate ratios (dN/dS ratios) for 76 protein-coding genes across 16 plastomes of the tribe Mimoseae.
4 Discussion
4.1 Selection analysis indicating potential adaptative evolution
Positive selection (dN/dS > 1) was detected in clpP, ycf2, and rps17. The gene clpP encodes the ClpP protease, which maintains organellar protein homeostasis and mediates environmental stress responses (Yu and Houry, 2007; Wicke et al., 2011; Hong et al., 2020), such as high temperature, drought, and salt stress (Deng et al., 2016). Given that most species of Mimoseae exhibit drought-tolerant traits, the positive selection observed in clpP may be related to their enhanced adaptability to arid conditions (Wicke et al., 2011; Deng et al., 2016). The protein encoded by ycf2 localizes to chloroplast membranes and participates in membrane assembly and homeostasis (Xing et al., 2025). The positive selection of the ycf2 gene indirectly indicates that the tribe Mimoseae has a more stable chloroplast structure, thus adapting to more complex environments (Xing et al., 2025). The species of Mimoseae occupy broad distribution ranges (Bruneau et al., 2024). These positively selected genes might be involved in unique environmental adaptations in Mimoseae.
When the Calliandra clade was designated as the foreground branch, positive selection was detected in rpoC1, atpA, rpoB, rps11, rps12, rps18, and rpl14. Functionally, rpoC1 encodes the β-subunit of chloroplast RNA polymerase in plants and is a core component of the chloroplast gene transcription mechanism, regulating the transcription of chloroplast genes (Wu et al., 2016; Peng et al., 2020), rps11, rps12, rps18, and rpl14 function in protein synthesis (Wicke et al., 2011), atpA functions in photosynthesis, and rpoB encodes the β-subunit of plastid-encoded RNA polymerase (PEP) (Wicke et al., 2011; Wu et al., 2024). Concurrently, atpB (involved in photosynthesis) exhibited intensified purifying selection, while rpoC2, rpoA (transcription), and petB (photosynthesis) displayed relaxed purifying selection. The primary role of atpB in photophosphorylation and energy homeostasis links it to abiotic stress responses (Li et al., 2010; Li et al., 2022). These selected genes are mainly related to chloroplast function including photosynthesis and may enhance the plant’s environmental adaptability by regulating photosynthetic efficiency (Wu et al., 2016; Peng et al., 2020; Wu et al., 2024). These genes likely experienced distinct evolutionary histories. In addition, our study found higher nucleotide diversity in non-coding regions than in coding regions, consistent with most previous studies (Wang, 2017; Contreras-Díaz et al., 2021). These identified hypervariable regions (both coding and non-coding) may complement previous studies and serve as valuable markers for phylogenetic, population genetic, and barcoding studies in Mimoseae or other plant taxa.
4.2 Significant structural variations indicating high plastome diversity in Mimoseae
The plastome of C. haematocephala exhibits significant structural features that highlight the plastomic diversity within Mimoseae. Most notably, it displays substantial plastome expansion and extensive structural rearrangements, closely associated with a ~14-kb expansion of the inverted repeat (IR) region into the large single-copy (LSC) region (Chumley et al., 2006). This IR expansion results in exceptionally large IRs (42,069 bp), contributing to a total plastome size of 200,623 bp—the largest reported to date within Mimoseae and across Leguminosae.
A prominent feature of this plastome is the high abundance of clustered dispersed repeats, ranging from 228 to 1,610 bp. These repeats likely serve as structural hotspots, promoting duplications, inversions, and the accumulation of additional repeat elements (Jin, 2019). Geneious-based (Kearse et al., 2012) visualization revealed that these repeats are non-randomly distributed and often clustered, potentially mediating large-scale rearrangements through intramolecular recombination.
Assembly of the C. haematocephala plastome posed challenges using short-read sequencing alone, resulting in a short 23-kb contig or fragmented contigs due to unresolved long repeats. In contrast, PacBio HiFi long reads enabled complete circular assembly using tools such as Oatk (Zhou et al., 2024) and TIPPo (Xian et al., 2025), confirming the accuracy and necessity of long-read sequencing for plastomes with complex repetitive structures. We recommend this approach for plastomes that cannot be circularized using short-read sequencing alone.
Collinearity analysis revealed extensive plastomic rearrangements in C. haematocephala. Although IR loss in the IRLC of Papilionoideae has been inferred to be associated with structural rearrangements (Palmer and Thompson, 1982; Wicke et al., 2011), some IR-lacking species (e.g., Medicago sativa L. and Wisteria floribunda (Willd.) DC.) exhibit limited structural variation (Wolfe et al., 1987; Lee et al., 2021). Conversely, substantial rearrangements have been observed in multiple IR-retaining lineages, including Campanulaceae (Wu and Chaw, 2014), Oleaceae (Cosner et al., 2004), Plantaginaceae (Zhu et al., 2016), and Pelargonium (Lee et al., 2007). Studies on Erodium further confirmed that plastome stability lacks a direct correlation with IR presence (Blazier et al., 2016; Wang et al., 2022). The extensive rearrangements observed in IR-retaining C. haematocephala are consistent with these findings.
In Mimosoideae, Wang (2017) revealed an approximately 13-kb IR expansion into the SSC region in the clade formed by tribes Ingeae and Acacia s.s (Dugas et al., 2015; Williams et al., 2015). Notably, C. haematocephala exhibited an additional 0.7-kb IR expansion into the LSC, shifting the JLB boundary into rps3. This IR expansion contributes to its exceptionally large plastome and further elucidates IR dynamics in Mimoseae. Phylogenetically, C. haematocephala belongs to the Calliandra clade, distinct from the Senegalia grade and the Zapoteca clade (Bruneau et al., 2024). Broader sampling of Calliandra plastomes is needed to assess whether IR expansion is characteristic of this lineage.
In comparison to previously reported plastome sizes in Leguminosae in GenBank such as Faidherbia albida (175,675 bp) (Mensous et al., 2017), and recently, Zhang et al. (2025) assembled 235 plastomes, with the largest circularized plastome being Pseudosamanea guachapele (Kunth) Harms (182,795 bp), our assembly of C. haematocephala (200,623 bp) now represents the largest plastome reported in Leguminosae. Elevated dispersed repeat abundance is known to drive plastomic rearrangements (Guisinger et al., 2008; Guisinger et al., 2011; Xu et al., 2023), and likely underlies the extensive structural variations in C. haematocephala. As in other Mimoseae species (Dugas et al., 2015; Wang, 2017), the predominant dispersed repeats in C. haematocephala are palindromic and forward repeats. Similarly, in Geraniaceae, plastome variations in Pelargonium × hortorum L.H.Bailey involved at least 12 inversions (Chumley et al., 2006). Weng et al. (2014) further showed that inversions were a major driver of plastome variability in 11 Geraniaceae species, with large insertions positively correlated with structural variation and distribution of repeat sequences strongly associated with breakpoints (Chumley et al., 2006). Previous researches demonstrated that plastome rearrangements in the Putranjivoids clade (Malpighialean) correlated with the abundance of repeat sequences (Wolfe et al., 1987; Jin et al., 2019).
Molecular mechanisms underlying plastome structural changes include repeat-mediated homologous recombination, slipped-strand mispairing, and occasional foreign-DNA integration (Cauz-Santos, 2025). Several studies have proposed that dispersed repeats (Ogihara et al., 1988; Wang et al., 2024; Cauz-Santos, 2025), particularly long forward repeats, are key mediators of inversions and rearrangements. For example, Wang et al. (2022) demonstrated that both the number and length of dispersed repeats are positively correlated with rearrangement frequency and magnitude. In C. haematocephala, all forward repeats exceed 90 bp, suggesting their active role in recombination-mediated plastome remodeling. IR dynamics are also recognized as major drivers of plastome structural evolution (Cauz-Santos, 2025). Shifts in IR boundaries through recombination or gene conversion can lead to expansions, contractions, or even gene duplication. A well-documented case in Nicotiana acuminata (Graham) Hook. involved a >12-kb IR expansion triggered by a double-strand break and recombination event (Goulding et al., 1996). In N. tabacum, experimental removal of the IR region led to altered gene dosage and increased plastome copy number (Krämer et al., 2024), emphasizing the IR’s role in plastome architecture and regulation.
In C. haematocephala, the 14-kb IR expansion may be associated with insertions, deletions, or duplications of genes near IR junctions. These changes, in combination with abundant dispersed repeats, likely contribute to the extensive plastomic rearrangements observed in this species. Notably, these findings support a growing consensus that plastome size and structure are determined by the interplay between repeat content and IR boundary dynamics, rather than IR presence or absence alone.
5 Conclusion
The plastome of C. haematocephala is remarkable in both structure and size. We found that it possesses the largest reported plastome in Mimoseae, and indeed within Leguminosae to date. The large increase in size and the high number of rearrangements are associated with a series of major IR expansions, as well as translocations and inversions. Furthermore, an abundance of clustered dispersed repeats has also been identified as a key factor contributing to the extensive plastomic rearrangements and the increase in plastome size. Selection pressure analyses identified positive selection in clpP, ycf2, and rps17, suggesting their potential roles in adaptive evolution. Branch-specific positive selection was also detected in genes such as rpoC1 and atpA within the Calliandra clade, indicating lineage-specific adaptive pressures.
Data availability statement
The original contributions presented in the study are publicly available. The newly sequenced plastome has been deposited in GenBank under accession number PX367310 (Supplementary Table S4). The supplementary tables of this study and data matrix used for phylogenetic analysis are available at Figshare (doi.org/10.6084/m9.figshare.30050251).
Author contributions
LZ: Conceptualization, Formal Analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. KA: Methodology, Visualization, Writing – review & editing. WG: Visualization, Writing – review & editing. QL: Visualization, Writing – review & editing. DW: Formal Analysis, Writing – review & editing. ZX: Formal Analysis, Writing – review & editing. RZ: Data curation, Funding acquisition, Writing – review & editing. TY: Conceptualization, Funding acquisition, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. We are grateful to the following institutes for providing materials: Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN), and The Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences (KIB, CAS). We are also grateful to the Molecular Biology Experiment Center, and the Germplasm Bank of Wild Species in Southwest China (KIB, CAS) for sequencing. This study was supported by the Science and Technology Basic Resources Investigation Program of China (2019FY100900), the Yunnan Revitalization Talent Support Program: Yunling Scholar Project (XDYC-YLXZ-2024-0021), National Natural Science Foundation of China (32270247), the Yunnan Applied & Basic Research Program (202301AT070310), Basic Research Project of Yunnan Province (202401BC070001), the National Natural Science Foundation of China, Key International (regional) Cooperative Research Project (31720103903).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1673127/full#supplementary-material
References
Álvarez-Carretero, S., Kapli, P., and Yang, Z. (2023). Beginner’s guide on the use of PAML to detect positive selection. Mol. Biol. Evol. 40, msad041. doi: 10.1093/molbev/msad041
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Bai, H. R., Oyebanji, O., Zhang, R., and Yi, T. S. (2020). Plastid phylogenomic insights into the evolution of subfamily Dialioideae (Leguminosae). Plant Diversity 43, 27–34. doi: 10.1016/j.pld.2020.06.008
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: A web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Blazier, J. C., Jansen, R. K., Mower, J. P., Govindu, M., Zhang, J., Weng, M. L., et al. (2016). Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 117, 1209–1220. doi: 10.1093/aob/mcw065
Bock, R. (2007). “Structure, function, and inheritance of plastid genomes,” in Cell and molecular biology of plastids. Ed. Bock, R. (Springer Berlin Heidelberg, Berlin, Heidelberg), 29–63.
Bruneau, A., de Queiroz, L. P., Ringelberg, J. J., Borges, L. M., Bortoluzzi, R. L. D. C., Brown, G. K., et al. (2024). Advances in legume systematics 14. Classification of Caesalpinioideae. Part 2: higher-level classification. PhytoKeys 240, 1–552. doi: 10.3897/phytokeys.240.101716
Queiroz, L. P., Koenen, E. J. M., Hughes, C. E., Luckow, M., Lewis, G. P., Ringelberg, J. J., et al. (2024). “Tribe Mimoseae,” in Advances in Legume Systematics 14. Classification of Caesalpinioideae, Part 2: Higher-level classification, eds. Bruneau A, Queiroz LP, Ringelberg JJ, PhytoKeys 240, 201–206. doi: 10.3897/phytokeys.240.101716
Cauz-Santos, L. A. (2025). Beyond conservation: the landscape of chloroplast genome rearrangements in angiosperms. New Phytol. 247, 2571–2580. doi: 10.1111/nph.70364
Cauz-Santos, L. A., da Costa, Z. P., Callot, C., Cauet, S., Zucchi, M. I., Bergès, H., et al. (2020). A repertory of rearrangements and the loss of an inverted repeat region in Passiflora chloroplast genomes. Genome Biol. Evol. 12, 1841–1857. doi: 10.1093/gbe/evaa155
Chen, X. F., Wu, Z. N., Yang, Y. T., Tao, Q. B., Na, N., Wan, W. Y., et al. (2025). The complete mitochondrial genome and phylogenetic analysis of Lotus corniculatus (Fabaceae, Papilionoideae). Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1555595
Choi, I. S., Jansen, R., and Ruhlman, T. (2019). Lost and found: Return of the inverted repeat in the legume clade defined by its absence. Genome Biol. Evol. 11, 1321–1333. doi: 10.1093/gbe/evz076
Chumley, T. W., Palmer, J. D., Mower, J. P., Fourcade, H. M., Calie, P. J., Boore, J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190. doi: 10.1093/molbev/msl089
Contreras-Dı́az, R., van den Brink, L., Navarrete-Fuentes, M. J., and Arias-Aburto, M. (2021). Characterization of the complete chloroplast genome of Prosopis tamarugo (Prosopis, Leguminosae), an endangered endemic tree species from the Atacama Desert. Bosque 42, 365–370. doi: 10.4067/S0717-92002021000300365
Cosner, M. E., Jansen, R. K., Palmer, J. D., and Downie, S. R. (1997). The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr. Genet. 31, 419–429. doi: 10.1007/s002940050225
Cosner, M. E., Raubeson, L. A., and Jansen, R. K. (2004). Chloroplast DNA rearrangements in Campanulaceae: Phylogenetic utility of highly rearranged genomes. BMC Evol. Biol. 4, 1–17. doi: 10.1186/1471-2148-4-27
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Deng, C. H., Kong, X. Y., Chen, G. X., Sui, J. M., Qiao, L. X., Wang, J. S., et al. (2016). Screening, clustering and response to salinity stress of Clp family genes in peanut (Arachis hypogaea L.). Shandong Agric. Sci. 48, 1–5. doi: 10.14083/j.issn.1001-4942.2016.12.001
Doyle, J. J., Doyle, J. L., Ballenger, J. A., and Palmer, J. D. (1996). The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogenet. Evol. 5, 429–438. doi: 10.1006/mpev.1996.0038
Dugas, D. V., Hernandez, D., Koenen, E. J. M., Schwarz, E., Straub, S., Hughes, C. E., et al. (2015). Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions and accelerated rate of evolution in clpP. Sci. Rep. 5, 16958. doi: 10.1038/srep16958
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Goulding, S. E., Wolfe, K. H., Olmstead, R. G., and Wolfe, K. H. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. MGG 252, 195–206. doi: 10.1007/BF02173220
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guisinger, M. M., Kuehl, J. V., Boore, J. L., and Jansen, R. K. (2008). Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. 105, 18424–18429. doi: 10.1073/pnas.0806759105
Guisinger, M. M., Kuehl, J. V., Boore, J. L., and Jansen, R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28, 583–600. doi: 10.1093/molbev/msq229
Haberle, R. C., Fourcade, H. M., Boore, J. L., and Jansen, R. K. (2008). Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. Mol. Evol. 66, 350–361. doi: 10.1007/s00239-008-9086-4
Hong, Z., Wu, Z. Q., Zhao, K. K., Yang, Z. J., Zhang, N. N., Guo, J. Y., et al. (2020). Comparative analyses of five complete chloroplast genomes from the genus Pterocarpus (Fabacaeae). Int. J. Mol. Sci. 21, 3758. doi: 10.3390/ijms21113758
Huang, X. Y., Guan, K. Y., and Li, A. (2011). Biological traits and their ecological significances of parasitic plants: A review. Chin. J. Ecol. 30, 1838–1844.
Jansen, R. K. and Ruhlman, T. A. (2012). “Plastid genomes of seed plants,” in Genomics of chloroplasts and mitochondria. Eds. Bock, R. and Knoop, V. (Springer Netherlands, Dordrecht), 103–126.
Jansen, R. K., Wojciechowski, M. F., Sanniyasi, E., Lee, S. B., and Daniell, H. (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48, 1204–1217. doi: 10.1016/j.ympev.2008.06.013
Jin, D. M. (2019). Plastome Structural Diversification of Malpighiales (Beijing: University of Chinese Academy of Sciences).
Jin, D. M., Gan, L., Jin, J. J., and Yi, T. S. (2019). The plastid genome of Klainedoxa Gabonensis Pierre ex Engl. (Malpighiales). Mitochondrial DNA Part B 4, 2541–2542. doi: 10.1080/23802359.2019.1639557
Jo, Y. D., Park, J., Kim, J., Song, W., Hur, C. G., Lee, Y. H., et al. (2011). Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 30, 217–229. doi: 10.1007/s00299-010-0929-2
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, Y. and Cullis, C. (2017). A novel inversion in the chloroplast genome of marama (Tylosema esculentum). J. Exp. Bot. 68, 2065–2072. doi: 10.1093/jxb/erw500
Krämer, C., Boehm, C. R., Liu, J. H., Ting, M. K. Y., Hertle, A. P., Forner, J., et al. (2024). Removal of the large inverted repeat from the plastid genome reveals gene dosage effects and leads to increased genome copy number. Nat. Plants 10, 923–935. doi: 10.1038/s41477-024-01709-9
Krause, K. and Scharff, L. B. (2014). Reduced genomes from parasitic plant plastids: Templates for minimal plastomes? Prog. Bot. 75, 97–115. doi: 10.1007/978-3-642-38797-5_3
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., Giegerich, R., et al. (2001). REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Kurtz, S. and Schleiermacher, C. (1999). REPuter: Fast computation of maximal repeats in complete genomes. Bioinf. (Oxford England) 15, 426–427. doi: 10.1093/bioinformatics/15.5.426
Lee, C., Choi, I. S., Cardoso, D., de Lima, H. C., de Queiroz, L. P., Wojciechowski, M. F., et al. (2021). The chicken or the egg? Plastome evolution and an independent loss of the inverted repeat in papilionoid legumes. Plant J. 107, 861–875. doi: 10.1111/tpj.15351
Lee, H. L., Jansen, R. K., Chumley, T. W., and Kim, K. J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. doi: 10.1093/molbev/msm036
Lewis, G., Schrire, B., Mackinder, B., and Lock, M. (2005). Legumes of the world (Kew, Richmond, U.K: Royal Botanic Gardens).
Li, J. L., Yuan, J. R., Li, Y. H., Sun, H. L., Ma, T. T., Huai, J. L., et al. (2022). The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 39, 110664. doi: 10.1016/j.celrep.2022.110664
Li, W. R., Zhang, S. Q., Ding, S. Y., and Shan, L. (2010). Root morphological variation and water use in alfalfa under drought stress. Acta Ecologica Sin. 30, 5140–5150.
LPWG (2013). Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248. doi: 10.12705/622.8
Lu, Q., Tian, Q., Gu, W., Yang, C. X., Wang, D. J., Yi, T. S., et al. (2024). Comparative genomics on chloroplasts of Rubus (Rosaceae). Genomics 116, 110845. doi: 10.1186/s12864-021-08225-6
Martin, G. E., Rousseau-Gueutin, M., Cordonnier, S., Lima, O., Michon-Coudouel, S., Naquin, D., et al. (2014). The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113, 1197–1210. doi: 10.1093/aob/mcu050
Mensous, M., Van de Paer, C., Manzi, S., Bouchez, O., Baali-Cherif, D., Besnard, G., et al. (2017). Diversity and evolution of plastomes in Saharan mimosoids: potential use for phylogenetic and population genetic studies. Tree Genet. Genomes 13, 48. doi: 10.1007/s11295-017-1131-2
Ogihara, Y., Terachi, T., and Sasakuma, T. (1988). Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. 85, 8573–8577. doi: 10.1073/pnas.85.22.8573
Palmer, J. D., Osorio, B., Aldrich, J., and Thompson, W. F. (1987). Chloroplast DNA evolution among legumes: Loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr. Genet. 11, 275–286. doi: 10.1007/BF00355401
Palmer, J. D., Osorio, B., and Thompson, W. F. (1988). Evolutionary significance of inversions in legume chloroplast DNAs. Curr. Genet. 14, 65–74. doi: 10.1007/BF00405856
Palmer, J. D. and Thompson, W. F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29, 537–550. doi: 10.1016/0092-8674(82)90170-2
Peng, Y., Su, Y. J., and Wang, T. (2020). Intron loss and molecular evolution rate of rpoC1 in ferns. Chin. Bull. Bot. 55, 287. doi: 10.11983/CBB19105
Qu, X. J., Moore, M. J., Li, D. Z., and Yi, T. S. (2019). PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 1–12. doi: 10.1186/s13007-019-0435-7
Ringelberg, J. J., Koenen, E. J. M., Iganci, J. R., de Queiroz, L. P., Murphy, D. J., Gaudeul, M., et al. (2022). “Phylogenomic analysis of 997 nuclear genes reveals the need for extensive generic re-delimitation in Caesalpinioideae (Leguminosae),” in Advances in Legume Systematics 14. Classification of Caesalpinioideae Part 1: New generic delimitations, vol. 205 . Eds. Hughes, C. E., Queiroz, L. P., and Lewis, G. P. (Leguminosae: PhytoKeys), 3–58. doi: 10.3897/phytokeys.205.85866
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruhlman, T. A., Zhang, J., Blazier, J. C., Sabir, J. S. M., and Jansen, R. K. (2017). Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am. J. Bot. 104, 559–572. doi: 10.3732/ajb.1600453
Sanderson, M. J., Copetti, D., Búrquez, A., Bustamante, E., Charboneau, J. L., Eguiarte, L. E., et al. (2015). Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 102, 1115–1127. doi: 10.3732/ajb.1500184
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Wang, Y. H. (2017). Plastid Phylogenomics of Fabaceae (Beijing: University of Chinese Academy of Sciences).
Wang, Z. X. (2022). Evolution of the Chloroplast Genome in IR-Lacking Lineages of Autotrophic Seed Plants (Beijing: University of Chinese Academy of Sciences).
Wang, J., Kan, S. L., Liao, X. Z., Zhou, J. W., Tembrock, L. R., Daniell, H., et al. (2024). Plant organellar genomes: much done, much more to do. Trends Plant Sci. 29, 754–769. doi: 10.1016/j.tplants.2023.12.014
Wang, Y. H., Qu, X. J., Chen, S. Y., Li, D. Z., and Yi, T. S. (2017a). Plastomes of Mimosoideae: Structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet. Genomes 13, 41. doi: 10.1007/s11295-017-1124-1
Wang, Z. X., Wang, D. J., and Yi, T. S. (2022). Does IR-loss promote plastome structural variation and sequence evolution? Front. Plant Sci. 13. doi: 10.3389/fpls.2022.888049
Wang, Y. H., Wang, H., Yi, T. S., and Wang, Y. H. (2017b). The complete chloroplast genomes of Adenolobus garipensis and Cercis glabra (Cercidoideae, Fabaceae). Conserv. Genet. Resour. 9, 635–638. doi: 10.1007/s12686-017-0744-y
Wang, Y., Zhang, C. F., Odago, W. O., Jiang, H., Yang, J. X., Hu, G. W., et al. (2023). Evolution of 101 Apocynaceae plastomes and phylogenetic implications. Mol. Phylogenet. Evol. 180, 107688. doi: 10.1016/j.ympev.2022.107688
Weng, M. L., Blazier, J. C., Govindu, M., and Jansen, R. K. (2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. doi: 10.1093/molbev/mst257
Weng, M. L., Ruhlman, T. A., and Jansen, R. K. (2017). Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214, 842–851. doi: 10.1111/nph.14375
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Williams, A. V., Boykin, L. M., Howell, K. A., Nevill, P. G., and Small, I. (2015). The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PLoS One 10, e0138367. doi: 10.1371/journal.pone.0125768
Wojciechowski, M. F., Sanderson, M. J., Steele, K. P., and Liston, A. (2000). “Molecular phylogeny of “the Btemperate herbaceous tribes” of papilionoid legumes: A supertree approach,” in Advances in legume systematics, part 9. Eds. Herendeen, P. S. and Bruneau, A. (Royal Botanic Gardens, Kew, Richmond, U.K.), 277–298.
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, C. S. and Chaw, S. M. (2014). Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol. J. 12, 344–353. doi: 10.1111/pbi.12141
Wu, X. X., Mu, W. H., Li, F., Sun, S. Y., Cui, C. J., Kim, C., et al. (2024). Cryo-EM structures of the plant plastid-encoded RNA polymerase. Cell 187, 1127–1144. doi: 10.1016/j.cell.2024.01.026
Wu, Y. M., Wang, S. Y., and Dong, S. R. (2016). Phylogenetic comparison between Spirulina and Arthrospira based on 16S rRNA and rpoC1 gene. Wei Sheng Wu Xue Bao= Acta Microbiologica Sin. 56, 232–240.
Xian, W. F., Bezrukov, I., Bao, Z. G., Vorbrugg, S., Gautam, A., Weigel, D., et al. (2025). TIPPo: A user-friendly tool for de novo assembly of organellar genomes with high-fidelity data. Mol. Biol. Evol. 42, msae247. doi: 10.1093/molbev/msae247
Xing, J. L., Pan, J. T., and Yang, W. Q. (2025). Chloroplast protein translocation complexes and their regulation. J. Integr. Plant Biol. 67, 912–925. doi: 10.1111/jipb.13875
Xu, S. J., Teng, K., Zhang, H., Gao, K., Wu, J. Y., Duan, L. H., et al. (2023). Chloroplast genomes of four Carex species: Long repetitive sequences trigger dramatic changes in chloroplast genome structure. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1100876
Yang, Z. H. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z. H., Wong, W. S. W., and Nielsen, R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yu, A. Y. H. and Houry, W. A. (2007). ClpP: A distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581, 3749–3757. doi: 10.1016/j.febslet.2007.04.076
Zhang, D., Gao, F. L., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, R., Stull, G. W., Jin, J. J., Wang, Y. H., Guo, Y., Yang, Z. Y., et al. (2025). Phylogenetic resolution and conflict in the species-rich flowering plant family leguminosae. Systematic Biol., syaf057. doi: 10.1093/sysbio/syaf057
Zhang, N. N., Stull, G. W., Zhang, X. J., Fan, S. J., Yi, T. S., Qu, X. J., et al. (2025). PlastidHub: An integrated analysis platform for plastid phylogenomics and comparative genomics. Plant Diversity. doi: 10.1016/j.pld.2025.05.005
Zhou, C. X., Brown, M., Blaxter, M, McCarthy, S. A., Durbin, R, and Darwin Tree of Life Project Consortium. (2024). Oatk: A de novo assembly tool for complex plant organelle genomes. bioRxiv. 26, 235. doi: 10.1101/2024.10.23.619857
Keywords: plastome evolution, inverted repeat expansion, rearrangement, nucleotide diversity, positive selection
Citation: Zhao L, An K, Gu W, Lu Q, Wang D-J, XiaHou Z, Zhang R and Yi T-S (2025) Dispersed repeats and inverted repeat expansion drive major plastomic rearrangements in Calliandra haematocephala (Leguminosae: Mimoseae). Front. Plant Sci. 16:1673127. doi: 10.3389/fpls.2025.1673127
Received: 25 July 2025; Accepted: 12 September 2025;
Published: 03 October 2025.
Edited by:
Zhiqiang Wu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Kai Zhao, Fujian Normal University, ChinaJia Minlong, Shanxi Agricultural University, China
Copyright © 2025 Zhao, An, Gu, Lu, Wang, XiaHou, Zhang and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting-Shuang Yi, dGluZ3NodWFuZ3lpQG1haWwua2liLmFjLmNu; Rong Zhang, emhhbmdyb25nYUBtYWlsLmtpYi5hYy5jbg==
 Libo Zhao1,2
Libo Zhao1,2 Ding-Jie Wang
Ding-Jie Wang Rong Zhang
Rong Zhang Ting-Shuang Yi
Ting-Shuang Yi