- 1School of Ocean, Yantai University, Yantai, China
- 2Laboratory of Marine Organism Taxonomy & Phylogeny, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 3Excellence Center for Biodiversity of Peninsular Thailand, Faculty of Science, Prince of Songkla University, Hat Yai, Songkhla, Thailand
Incipient (recently-evolved) species are morphologically distinct and have a relatively short evolutionary history from a common ancestor, yet comparative phylogeographic patterns of incipient seaweed species have seldom been explored. Here, we created 339 mitochondrial cox1, 339 cox3, 339 cox1+cox3 and 286 nuclear ITS2 sequences for Sargassum plagiophyllum (10 populations) and 326 cox1, 336 cox3, 310 cox1+cox3 and 341 ITS2 sequences for S. polycystum (14 populations) around the Thai-Malay Peninsula (TMP), with the aim to explore potential drivers in shaping population genetic structuring and diversity over space, including a phylogeographic signature of incipient speciation. Comparative analysis showed that the two Sargassum species around the TMP diverged from their most recent common ancestor at c. 0.17 Mya, followed by a demographic expansion at c. 0.015–0.060 Mya. Oceanic currents drove contemporary continuous south-to-north gene flow in the Malacca Strait, leading to most genetic variation partitioned within populations and among populations within groups. Interestingly, the incipient S. polycystum and S. plagiophyllum shared their most common haplotypes/ribotypes, and mitochondrial datasets revealed much higher phylogeographic diversity in S. polycystum than in S. plagiophyllum. These results imply that the late Quaternary sea-level fluctuations and contemporary oceanic currents co-contributed to population genetic structuring and demographic histories of S. polycystum and S. plagiophyllum around the TMP. Importantly, comparative phylogeographic analysis of S. polycystum and S. plagiophyllum shed lights on the existence of a few separate late Pleistocene glacial refugia, such as the Andaman Sea for S. plagiophyllum and the northern Malacca Strait for S. polycystum, particularly the revealing of important signatures of their incipient speciation.
Introduction
The Sunda shelf is located at the Indo-Pacific convergence region encompassing the Thai-Malay Peninsula (TMP) and the islands of Sumatra, Java, Borneo. Glacial and interglacial cycles during the Pleistocene epoch (1.8–0.012 million years ago (Mya)) caused the Sunda shelf to experience severe sea level fluctuations ranging from -120 m to 6 m compared to the present-day sea level, significantly altering the land bridge structure and waterbody connectivity in the Indo-Pacific convergence region (e.g. the disruption and reconnection between the east and west sides of the TMP, Hall, 1998; Voris, 2000). These geological configurations driven by ice cycles enabled the Sunda shelf to be an important marine biodiversity hotpot (Woodruff, 2010; Hall, 2012; Morley, 2012).
The TMP forms a natural biogeographical barrier between the Andaman Sea-Malacca Strait and the Gulf of Thailand that influences species distributions and genetic variability by hindering dispersal between its east and west coast (Gaither et al., 2010). Seasonal monsoons influence ocean surface currents significantly (Buranapratheprat, 2008), potentially driving species’ long-distance dispersal and population genetic structuring (Wee et al., 2014; 2020; Chan et al., 2022). During the northeast monsoon (December-February) a clockwise circulation forms in the Gulf of Thailand along with a clockwise flow in the Andaman Sea (Figure 1A). While during the southwest monsoon (June-August), some small-scale counterclockwise vortices develop along the west coast of the Gulf of Thailand (Figure 1B), reversing the direction of seawater flow through the Malacca Strait (Sojisuporn et al., 2010). The long-term interplay between ocean currents and paleoenvironmental changes considerably impacted speciation, biogeographic processes and population structuring of pelagic vertebrates and invertebrates around the TMP (Woodruff, 2010; Chan et al., 2022). Such a contribution is thus hypothesized to occur in marine sessile species such as macroalgae (Chan et al., 2013; Bulan et al., 2022).
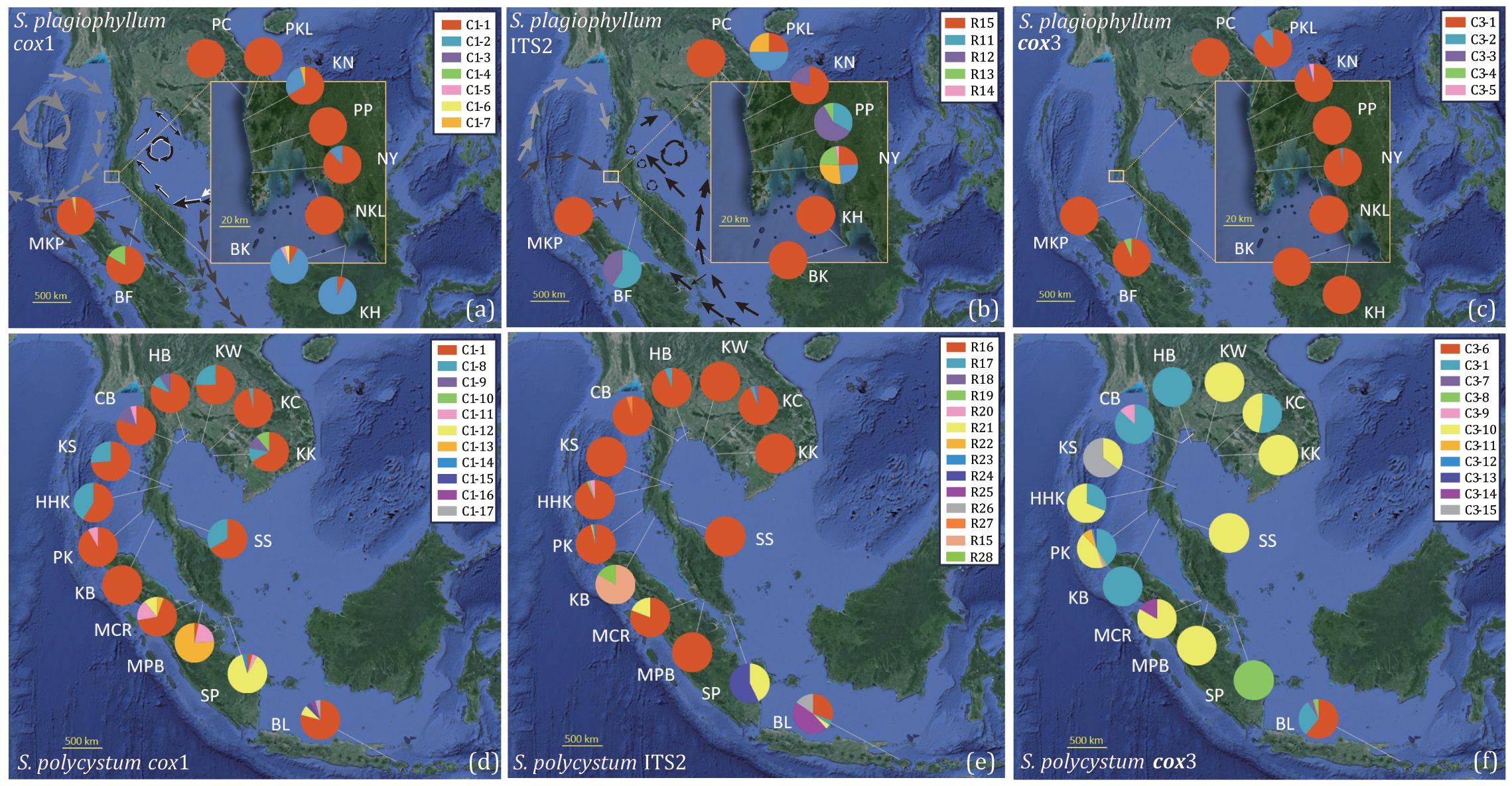
Figure 1. Geographical distribution and relative frequency of cox1 and cox3 haplotypes and ITS2 ribotypes for S. plagiophyllum (A–C) and S. polycystum (D–F). The ocean currents are plotted under the prevailing northeast (A) and southwest monsoons (B) (modified from Chan et al., 2022). a: Gray arrows indicate the clockwise circulation in the Andaman Sea during the northeast monsoon; dark gray arrows indicate the currents from the South China Sea flow through the Singapore Strait into the Malacca Strait and reach the northern part of Sumatra Island; white and black arrows indicate currents flowing from the Western Pacific and the Philippines into the Gulf of Thailand. b: light gray arrows indicate ocean currents from the Indian Ocean passing through the Andaman Islands and flowing towards the north coast of the Andaman Sea during the southwest monsoon; dark gray arrows indicate ocean currents from the Indian Ocean passing through the south of the Nicobar Islands and flowing towards the south of the Andaman Sea; black arrows indicate ocean currents from the Java Sea flowing northward towards Singapore and then along the east coast of the Malay Peninsula, and ultimately into the Gulf of Thailand.
The mapping of comparative phylogeographic structuring and diversity patterns served as a powerful tool for investigating evolutionary processes in a certain region (Bermingham and Moritz, 2010). Previous comparative studies exploring genetic diversity and phylogeographic patterns around the TMP, primarily focused on marine animals (Briggs, 2005; Reaka et al., 2008; Carpenter et al., 2011; Mirams et al., 2011). Recently, Chan et al. (2022) compared the distribution of intertidal Tetraclitas Schumacher barnacle species across the east and west sides of the TMP, concluding that geological vicariance during the Pleistocene glaciations and circulation-driven long-distance dispersal caused the predominant distributions of T. singaporensis Chan, Tsang & Chu in the Andaman Sea and T. squamosa Bruguière in the Gulf of Thailand, respectively. Seaweeds around the TMP also received increasing interest in the past decade, although in a non-comparative phylogeographic context. For example, the brown seaweed Padina boryana Thivy was found to be represented by two distinct lineages around the TMP of which one was restricted to the northern Gulf of Thailand (Wichachucherd et al., 2014). Muangmai et al. (2023) further detected a possible genetic break between populations of the red seaweed Gracilaria salicornia (C.Agardh) E.Y.Dawson in the Andaman Sea. More recently, Wang et al. (2025) found low phylogeographic diversity and shallow population structuring in the green alga Halimeda macroloba Decaisne around the TMP. These single-species based phylogeographic studies largely enhanced our understanding of how geological events and ocean currents interacted to affect geographic distribution and lineage structuring around the TMP. However, comparative phylogeographies of seaweeds, particularly for species originated in the mid-late Pleistocene (c. 0.8–0.1 Mya), are still poorly explored (e.g. Guillemin et al., 2018; Huanel et al., 2024).
Species of Sargassum C. Agardh, the most species-rich genus within the order Fucales (Phaeophyceae), serve as foundation species by providing essential ecological roles for other marine species (Faga and Gurgel, 2024). Sargassum has a diplontic life cycle, and the fertilized eggs remain attached to the receptacle for 2–3 days after fertilization (Draisma et al., 2010). The detached zygotes exhibit limited dispersal capabilities (<1 m) (Kendrick and Walker, 1995). However, Sargassum thalli exhibit gas-filled vesicles (aerocysts) which provide buoyancy for detached thalli (Komatsu et al., 2007; Mizuno et al., 2014), facilitating long-distance dispersal. This can partially explain why the genus Sargrassum currently has a cosmopolitan distribution spanning the Atlantic, Pacific, and Indian Oceans (Guiry and Guiry, 2025), whereas fucalean genera without aerocysts usually have a more limited distribution range (Draisma et al., 2010). The origination and diversification of the genus Sargassum was estimated to occur approximately at 4.3 Mya, and the longer history of diversification relative to other marine realms gave rise to higher species richness in the temperate northern Pacific (Yip et al., 2020).
Sargassum polycystum C. Agardh and S. plagiophyllum C. Agardh belong to the section Polycystae Mattio & Payri of the subgenus Sargassum and are prevalent throughout the Indo-Pacific convergence region (Soe-Htun et al., 2012). Geographically, S. polycystum occurs throughout the tropical Indo-West Pacific from Africa in the west to Tonga in the east (Guiry and Guiry, 2025). The distribution range of S. plagiophyllum is much smaller and stretches from India in the west to Guangxi (China) and Queensland (Australia) in the east (Guiry and Guiry, 2025). Sargassum polycystum and S. plagiophyllum may occur in sympatry (Soe-Htun et al., 2012; Stankovic et al., 2022). The two species can be distinguished by morphological differences such as the secondary holdfasts, with S. plagiophyllum forming cauline leaves, whereas S. polycystum developing primary branches (Kantachumpoo et al., 2015). In addition, the main branches of S. plagiophyllum are smooth, whereas they are distinctly muricate in S. polycystum (Soe-Htun et al., 2012). Sargassum polycystum and S. plagiophyllum have been calibrated to originate from a common ancestor at c. 0.4 Mya (95% HPD interval: 0.1–0.8 Mya, Yip et al., 2020), and their evolutionary history and population genetic structuring are likely associated with the late Pleistocene glacial fluctuations (e.g. the southwest of Hainan Island, China served as a glacial refugium for the survival of ancestral relics of S. polycystum, Hu et al., 2018) and contemporary marine conditions (Lin et al., 2024). Organellar-genome scale calibrations further revealed their divergence at c. 0.1–0.5 Mya (Zhang et al., 2022). These lines of evidence indicate that S. polycystum and S. plagiophyllum are recently-evolved (incipient) species, allowing us to hypothesize that kinds of organellar genes between them may share the same sequence composition. Therefore, comparing their phylogeographic patterns and processes of these two Sargassum species can bridge the knowledge gap between evolutionary processes and genetic signatures of the incipient speciation in seaweeds.
In this study, we sequenced the mitochondrial cytochrome c oxidase subunits I and III (cox1 and cox3) and the nuclear internal transcribed spacer 2 (ITS2) of S. polycystum and S. plagiophyllum populations around the TMP. Our aims are to test two hypotheses: i) the TMP acted as a geographical barrier contributing to population genetic connectivity of S. polycystum; ii) the late Pleistocene ice ages and contemporary ocean currents shaped the phylogeographic structuring and history of two Sargassum species around the TMP. By comparing population genetic variation and biogeographic patterns of two Sargassum species, we are also interested in exploring DNA-based phylogeographic signatures of recently-evolved species in the subgenus Sargassum. The results can offer valuable insights for managing and conserving Sargassum resources around the TMP, providing new knowledge to understand speciation, diversification and evolution in the Sunda Shelf.
Materials and methods
Sample collection, DNA extraction, amplification, and sequencing
Sargassum plagiophyllum has never been reported from the east coast of the TMP, and on the west coast Penang Island harbours the southmost known population. On the Thai west coast, S. polycystum is rare and only a single small population was found in the lower intertidal in Phuket (PK), where S. plagiophyllum also was collected (NY) (Table 1). This is the only site around the TMP where the two species grow in sympatry, but locally they were separated in water depth. Based on the predicted distribution range of two Sargassum species (Stankovic et al., 2022), S. plagiophyllum from 10 localities and S. polycystum from 14 localities were sampled respectively (Table 1, Figure 1). At each locality, 3–5 cm of the youngest branch (3–97 individuals per locality (Table 1), including leaves, aerocysts, and receptacles) that appeared without epiphytes were cut and preserved in silica gel. All populations sampled in the Gulf of Thailand grew in the shallow subtidal, whereas those collected on the west coast grew in the intertidal. The collection of Sargassum samples in the field complied with the national legislation of Thailand, Malaysia, Singapore and Indonesia and international guidelines. Dr. Draisma Stefano undertook the formal identification of S. polycystum and S. plagiophyllum, and all voucher specimens were deposited in Department of Marine Science, Ocean School, Yantai University.
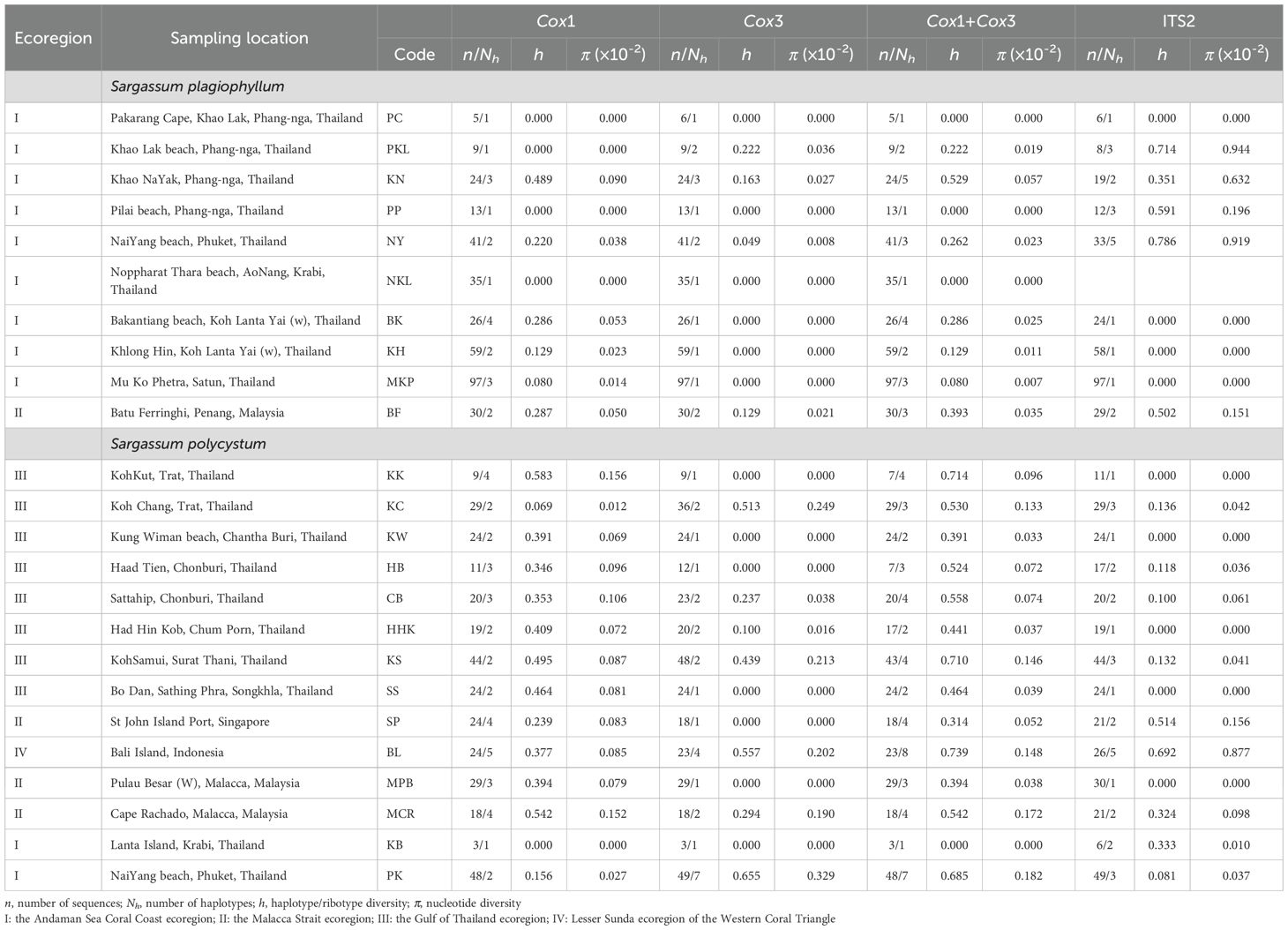
Table 1. Population genetic diversity indices of Sargassum polycystum and S. plagiophyllum in the Thai-Malay Peninsula based on mitochondrial cox1, cox3, cox1 + cox3 and nuclear ITS2.
Total genomic DNA was extracted from c. 0.3 g dried leaves using the FastPure Plant DNA Isolation Mini Kit (Vazyme Biotech Co., Ltd., Nanjing, China), following the manufacturer’s instructions. Mitochondrial cox1 and cox3, and nuclear ITS2 were selected as target markers according to previous phylogeographic studies of Sargassum (Hu et al., 2011; Li et al., 2017; Hu et al., 2018; Liang et al., 2022). The primer sets GazF2 and GazR2 (Lane et al., 2007), trnY-P1 and cox3-P2 (Kogishi et al., 2010) and 5.8S-BF and 25BR-2 (Yoshida et al., 2000) were utilized to amplify cox1, cox3 and ITS2, respectively (Supplementary Table S1). PCR protocols and purifications were conducted following Hu et al. (2018). PCR products were examined through 1% agarose gel electrophoresis and sequenced in both directions using the Applied Biosystems™ 3730XL at Bioengineering Co., Ltd. (Shanghai, China), using the same primer sets as PCR amplification.
Genetic diversity and population structuring
Cox1, cox3 and ITS2 sequences were aligned and trimmed using MEGA v10.1.8 (Kumar et al., 2016). Mitochondrial haplotypes (cox1, cox3) and nuclear ribotypes (ITS2) were identified for both datasets of S. plagiophyllum and S. polycystum using DnaSP v5.10 (Librado and Rozas et al., 2009). Genetic diversity indices, including haplotype number (Nh), haplotype diversity (h) and nucleotide diversity (π), were calculated using Arlequin v3.5 (Excoffier and Lischer, 2010).
Analysis of molecular variation (AMOVA) were conducted to partition genetic variation at different spatial scales. Sargassum plagiophyllum populations were categorized into two groups following the Marine Ecoregions of the World bioregionalization scheme proposed by Spalding et al. (2007): the Andaman Sea Coral Coast ecoregion (PC, PKL, KN, PP, NY, NKL, BK, KH, and MKP) and the Malacca Strait ecoregion (BF) (Figure 1). Sargassum polycystum populations were divided into three groups: the Gulf of Thailand ecoregion (KK, KC, KW, CB, HHK, KS, HB and SS which both were washed up), the Malacca Strait ecoregion (SP, MPB, and MCR), and the Andaman Sea Coral Coast ecoregion (PK, KB) (Figure 1). Population-level pairwise FST were estimated with Arlequin.
Subtle intraspecific genetic structuring between S. plagiophyllum and S. polycystum was performed using Structure v2.3.4 (Pritchard et al., 2000) based on an admixture model. For each dataset, five independent runs were performed for each value of K (number of clusters) from 1 to 5, based on the produced haplotype/ribotype networks and phylogenetic trees (see results below). The MCMC was set to 5×107 iterations with a burn-in of 5×105 iterations. The online tool Structure Selector (https://lmme.ac.cn/StructureSelector/) was used to calculate LnP(K) and ΔK (Evanno et al., 2005) to determine the optimal K value (Supplementary Figure S1) and draw clustering plots. Cox1 and cox3 datasets were also concatenated using MEGA to construct haplotype networks as well as to examine the consistence of population genetic structuring between S. plagiophyllum and S. polycystum.
Demographic history and spatial migration
Bayesian skyline plots (BSPs) were conducted with BEAST v1.10.4 (Suchard et al., 2018) to assess historical changes in effective population size for each of the two species. An uncorrelated lognormal relaxed molecular clock was applied, with tree priors set to coalescent Bayesian skyline. The MCMC chains were run for 2×108 iterations, with sampling conducted every 15,000 generations, and the first 15,000 iterations discarded as burn-in. Convergence of the treeset files was checked using Tracer v1.7.2 (Effective Sample Size (ESS) > 200) to generate a demographic trend chart.
Population-level migration driven by oceanic currents was estimated using Migrate-n v5.0.4 (Beerli and Felsenstein, 2001) for each dataset. The effective population size (θ = xNeμ, where Ne represents the effective population size, μ is the mutation rate per generation, and x is typically 4 for nuclear data and 1 for mitochondrial data) and the migration rate (M = m/μ, where m denotes the migration rate per generation and μ is the mutation rate) were substituted into the formula (Nm =θM/x) to calculate the effective number of migrants. The Monte Carlo Markov Chain (MCMC) was set to 100,000 iterations with a burn-in of 25,000 iterations, while heating parameters were configured as 1.0, 1.5, 3.0, and 1.0 under a static scheme.
Haplotype/ribotype network and molecular dating
To detect the relationships between the newly identified haplotypes/ribotypes around the TMP and the hypothesized late-Pleistocene glacial survived ancestral genotypes (Hu et al., 2018), 15 cox1 haplotypes, 12 cox3 haplotypes and 11 ITS2 ribotypes of S. polycystum from Hainan and Guangxi provinces, China (Hainan-Guangxi) were retrieved. The network diagrams of all combined haplotypes and ribotypes were created with a median-joining method using Network v10.2.0.0 (Bandelt et al., 1999). In addition, all cox1 and cox3 haplotypes and ITS2 genotypes were pooled together, respectively to examine if there are shared genotypes between S. polycystum and S. plagiophyllum as well as among the Andaman Sea, the Gulf of Thailand, the Malacca Strait and Hainan-Guangxi.
For each combined haplotype/ribotype dataset, Modelfinder (Kalyaanamoorthy et al., 2017) was used to determine the optimal nucleotide substitution model under Bayesian information criteria (BIC) (cox1: HKY+F+I, I=0.835; cox3: HKY+F+G4, G=0.107; ITS2: HKY+F). Beast v1.10.4 was used to calculate the intraspecific level of divergence times, selecting an uncorrelated lognormal relaxed molecular clock with tree priority set to the Yule model. Node time was set as 0.4 Mya (95% HPD: 0.1–0.8 Mya, Yip et al., 2020) for the Section Polycystae and 1.5 Mya (95% HPD: 0.6–2.6 Mya) for the Section Binderiana (Yip et al., 2020). Sargassum aquifolium (GenBank accession no. cox1/cox3: NC_033408, ITS2: HF572039) and S. patens (cox1/cox3: NC_052831, ITS2: KY935432) in the Section Binderiana were chosen as outgroups. The MCMC chains were run for 5×108 iterations, sampled every 5×104 generations, and the first 5×104 iterations discarded as burn-in. Tracer v1.7.2 was used to check convergence (ESS > 200). The topological structure and divergence times of the lineages were viewed using Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Population genetic diversity and variation
We obtained 339 cox1 (570 bp), 339 cox3 (618 bp), 339 cox1+cox3 (1188 bp), and 286 ITS2 (335 bp) sequences from 10 S. plagiophyllum populations (Table 1), which contained 6, 4, 10, and 8 variable sites, respectively. Cox1 and cox3 identified 7 and 5 haplotypes, respectively, with C1-1 (Figure 1A) and C3-1 (Figure 1C) as the most abundant and widely distributed. ITS2 identified 5 ribotypes, with R15 as the most abundant (Figure 1B). For S. polycystum, we obtained 326 cox1 (570 bp), 336 cox3 (618 bp), 310 cox1 + cox3 (1188 bp), and 341 ITS2 (335 bp) sequences from 14 populations (Table 1). The four datasets contained 9, 15, 22, and 14 variable sites, respectively. Cox1 and cox3 each identified 11 haplotypes, with C1-1 (Figure 1D) and C3-10 (Figure 1F) as the most abundant, and ITS2 identified 14 ribotypes, with R16 as the most common/frequent (Figure 1E). Concatenated cox1+cox3 yielded 11 and 24 haplotypes for S. plagiophyllum (Supplementary Figure S2A) and S. polycystum (Supplementary Figure S2b), respectively.
For S. plagiophyllum, cox1, cox3, and cox1+cox3 consistently identified a few site-specific haplotypes in population BF from Penang, Malaysia (Figures 1A, C, S2A). They also revealed relatively high haplotype and nucleotide diversity in the populations BF (Penang) and PKL and KN from Phang-Nga, Thailand (cox1+cox3: h=0.222–0.529; π=0.00019–0.00057, Table 1). ITS2 revealed much higher ribotype and nucleotide diversity in the populations PKL and NY (Phuket) from Thailand (h=0.714–0.786; π=0.00919–0.00944, Table 1). Other populations, particularly the PC (Phang-Nga), NKL (Krabi) and MKP (Satun) from in Thailand exhibited the lowest genetic diversity (h=0.000; π=0.00000, Table 1).
For S. polycystum, cox1, cox3, cox1+cox3, and ITS2 consistently identified site-specific rich haplotypes/ribotypes in population BL from Bali, Indonesia (Figures 1D–F, S2B). This enabled BL to harbour higher haplotype and nucleotide diversity (cox3: h=0.557; π=0.00202; cox1+cox3: h=0.739; π=0.00148) than other populations (cox3: the mean h=0.172; the mean π=0.00080; cox1+cox3: the h=0.482; the mean π=0.00083, Table 1). Except the KB, cox1 detected high level of genetic variation in Thai populations (h=0.069–0.583; π=0.00012–0.00156), whereas cox3 revealed rich genetic diversity only in the populations KC, KS and PK from Thailand (h=0.439–0.655; π=0.00213–0.00329, Table 1). ITS2 showed that all Thai populations exhibited much lower ribotype and nucleotide diversity (h=0.000–0.333; π=0.00000–0.00061) than the populations SP from Singapore (h=0.514; π=0.00156) and BL from Indonesia (h=0.692; π=0.00877) (Table 1).
Population genetic structuring
AMOVA revealed incompatible results among four datasets for S. plagiophyllum (Supplementary Table S2). Cox1 and cox1+cox3 identified approximate 80–90% of the total genetic variation occurred among populations within groups (p<0.001). Cox3 revealed 89.19% of the total genetic variation within populations, whereas ITS2 identified 57.99% of the genetic variation among groups, but both were statistically non-significant (Supplementary Table S2). For S. polycystum, AMOVA indicated that approximate 36–45% of the genetic variation occurred within populations (p<0.001), and the rest were distributed among populations within groups (p<0.001, Supplementary Table S2). Cox1+cox3 based FST estimates showed that BK and KH from Koh Lanta Yai, Thailand highly differentiated from other S. plaiophyllum populations (Supplementary Table S3). For S. polycystum, different datasets consistently revealed that SP from Singapore, BL from Indonesia and KB from Thailand significantly diverged from other populations (Supplementary Table S4).
Population structuring analysis of either combined (Figure 2) or separated dataset (Supplementary Figure S3), revealed the optimal Delta K-value (K=2), suggesting two genetically divided groups (S. plagiophyllum vs. S. polycystum). Cox3/ITS2-based clustering at K=2 indicated amounts of individual admixture between S. plagiophyllum and S. polycystum (Supplementary Figure S3B-S3C), despite cox3 showing that all S. plagiophyllum populations exclusively clustered as a separate group. Clustering analysis at K=3 revealed subtly structured populations in each Sargassum species, particularly for S. polycystum. This can mostly be ascribed to the unique genetic characteristics anchored in the populations SP from Singapore and BL from Indonesia (Figure 2).
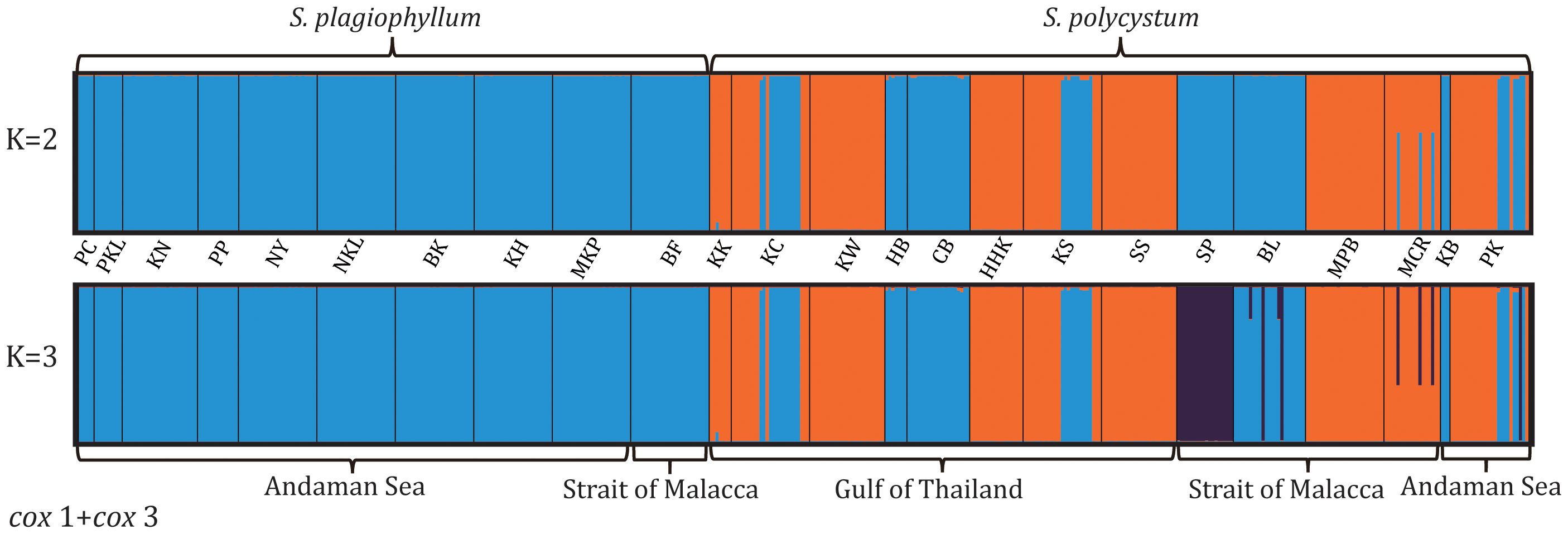
Figure 2. Structure analysis based on mitochondrial cox1+cox3 of S. plagiophyllum and S. polycystum at K = 2 (b) and K = 3 (d). Population codes are the same as in Table 1.
Historical demography and migration
BSPs results indicated that both Sargassum species experienced long-term stabilized population size since 0.35 Mya (Figure 3, Supplementary Figure S4). Cox1-inferred demographic expansion occurred at c. 0.015 Mya for S. plagiophyllum and at c. 0.025 Mya for S. polycystum (Figure 3). Cox3 and ITS2 analysis revealed similar demographic histories, with S. plagiophyllum expanded at c. 0.015–0.060 Mya (Supplementary Figure S4A-S4C) and S. polycystum expanded at c. 0.025–0.035 Mya (Supplementary Figure S4B-S4D).
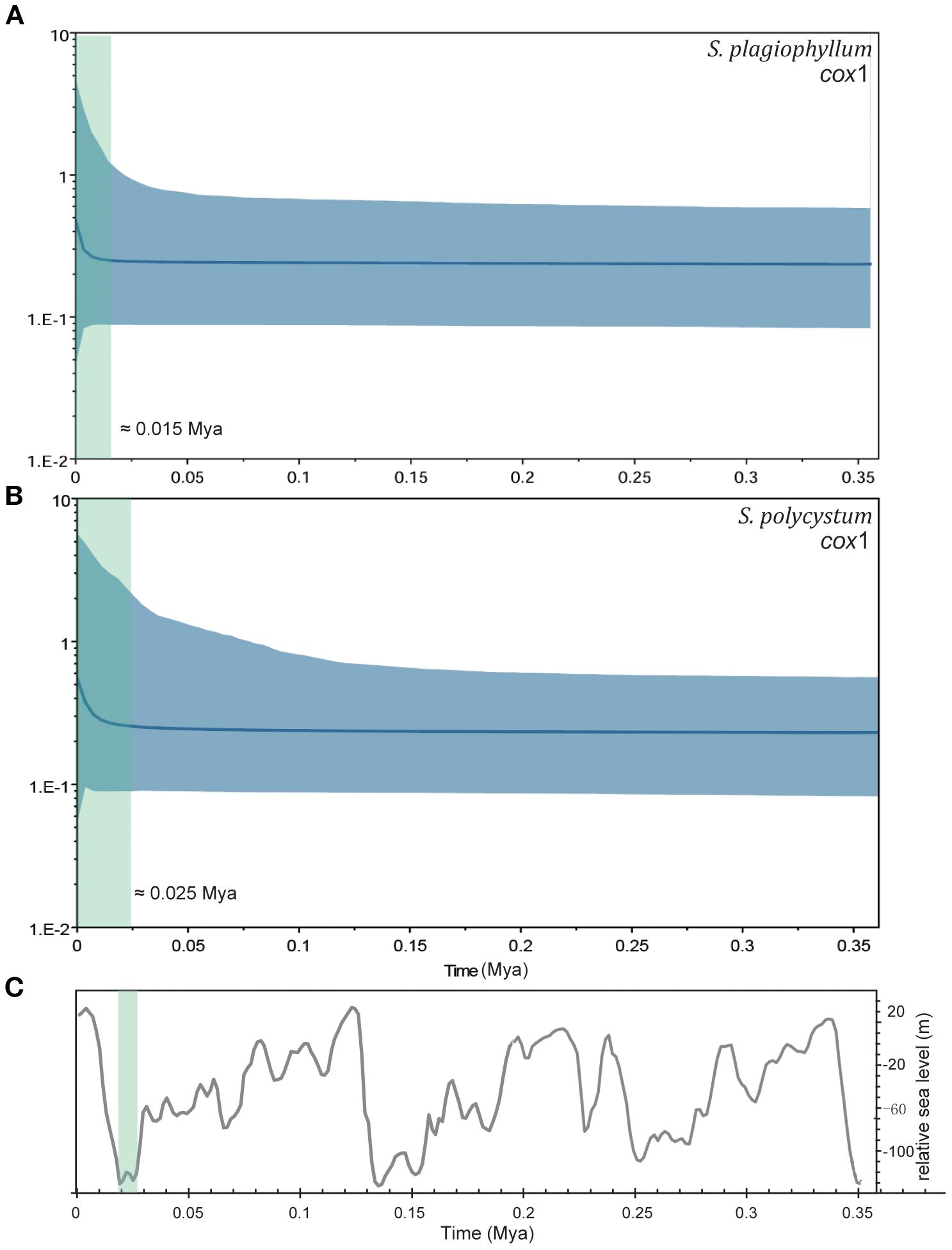
Figure 3. Bayesian skyline plots (BSPs) for S. plagiophyllum (A) and S. polycystum (B) based on cox1 dataset. Blue lines are the median posterior effective population size through time, blue shaded areas represent 95% confidence intervals. The green shadows represent the approximate demographic expansion time. (C) The global relative sea level curve during the late Pleistocene glacial cooling (modified from Waelbroeck et al., 2002).
For S. plagiophyllum, mitochondrial and nuclear datasets revealed a primarily south-to-north migration (Figures 4, S5A, S6A, S7A). Specifically, there were numerous migrations towards the west coast of Phuket Island (i.e. the PKL population) in the Andaman Sea from the northern Malacca Strait. Instead, the opposite migration from the Phuket Island (e.g. PC, PKL and PP) to other populations were generally weak (Figures 4A, S5A, S6A, S7A). For S. polycystum, molecular datasets revealed a large amount of clockwise migration between populations in the Gulf of Thailand, despite complementary anticlockwise migration occurred (Figures 4B, S5B, S6B, S7B). Intensive migration also occurred among the three populations HB, KK and HHK in the Gulf of Thailand. However, there was no clear migration spanning from Bali Island to the Andaman Sea.
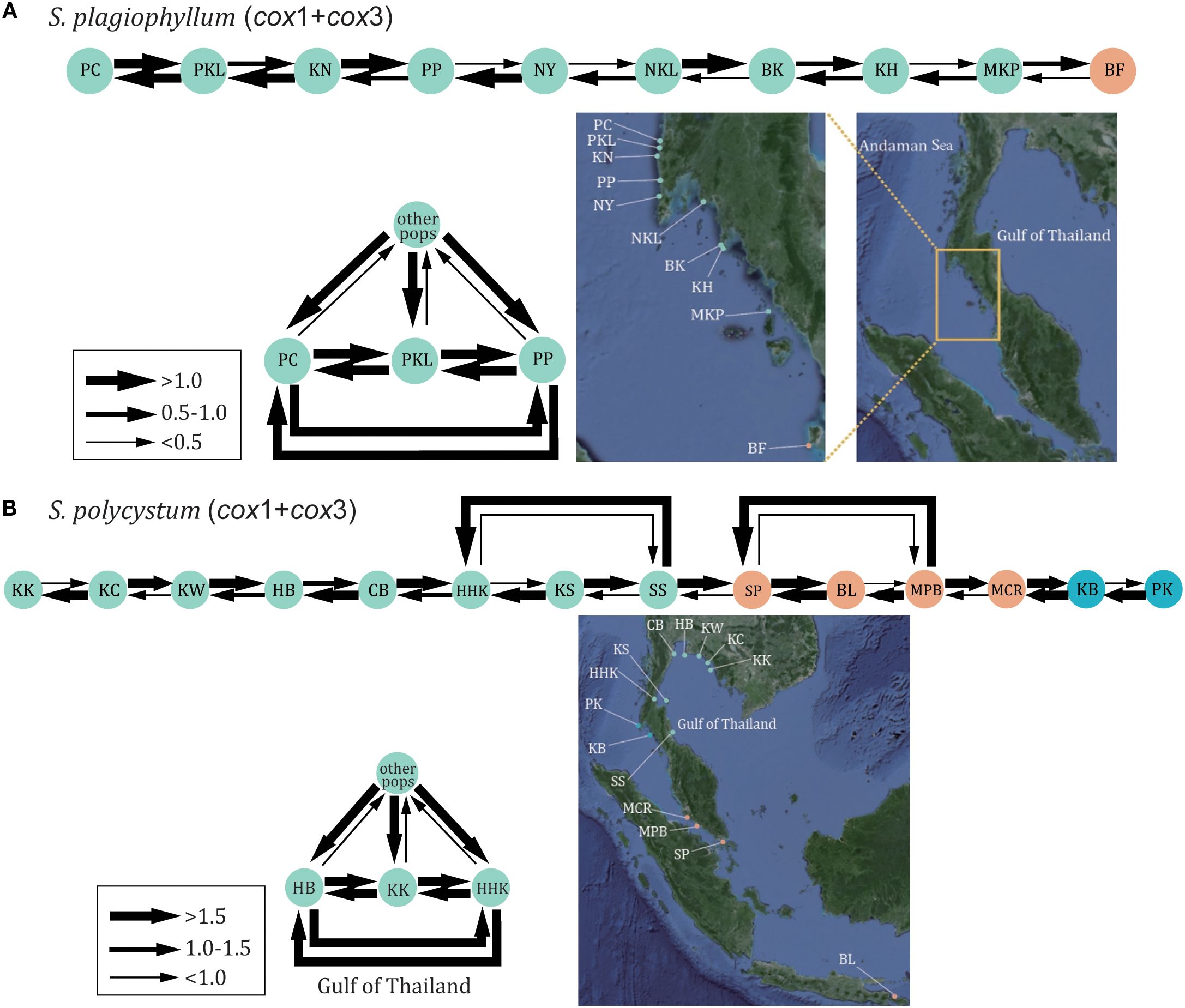
Figure 4. The estimated gene flow between S. plagiophyllum populations (A) and S. polycystum populations (B) in the Thai-Malay Peninsula based on cox1+cox3 dataset.
Network diagrams and molecular dating
When pooling each of the three datasets of S. plagiophyllum and S. polycystum with those from Hainan-Guangxi (China), we identified 30 cox1 (C1-1–C1-30, Figure 5A) and 25 cox3 haplotypes (C3-1–C3-25, Figure 5B), and 28 ITS2 ribotypes (R1-R28, Figure 5C). The network diagram showed that cox1-inferred C1-1, cox3-inferred C3-1 as well as ITS2-inferred R12 and R15, were each shared between S. plagiophyllum and S. polycystum in distribution (Figure 5), which accounted for 52.81% (442/837), 54.30% (467/860), and 26.32% (215/817) of the total individuals analyzed, respectively. Cox1, cox3 and ITS2 consistently showed that some haplotypes/ribotypes from Hainan-Guangxi (China) formed an independent structured lineage that was evidently diverged from other haplotypes/ribotypes (Supplementary Figure S8). The three datasets also indicated that the most common distributed cox1/cox3-inferred C1-1/C3-1 and ITS2-inferred R16 were co-occurred widely around the TMP populations (the Andaman Sea, the Gulf of Thailand and the Malacca Strait, Supplementary Figure S8).
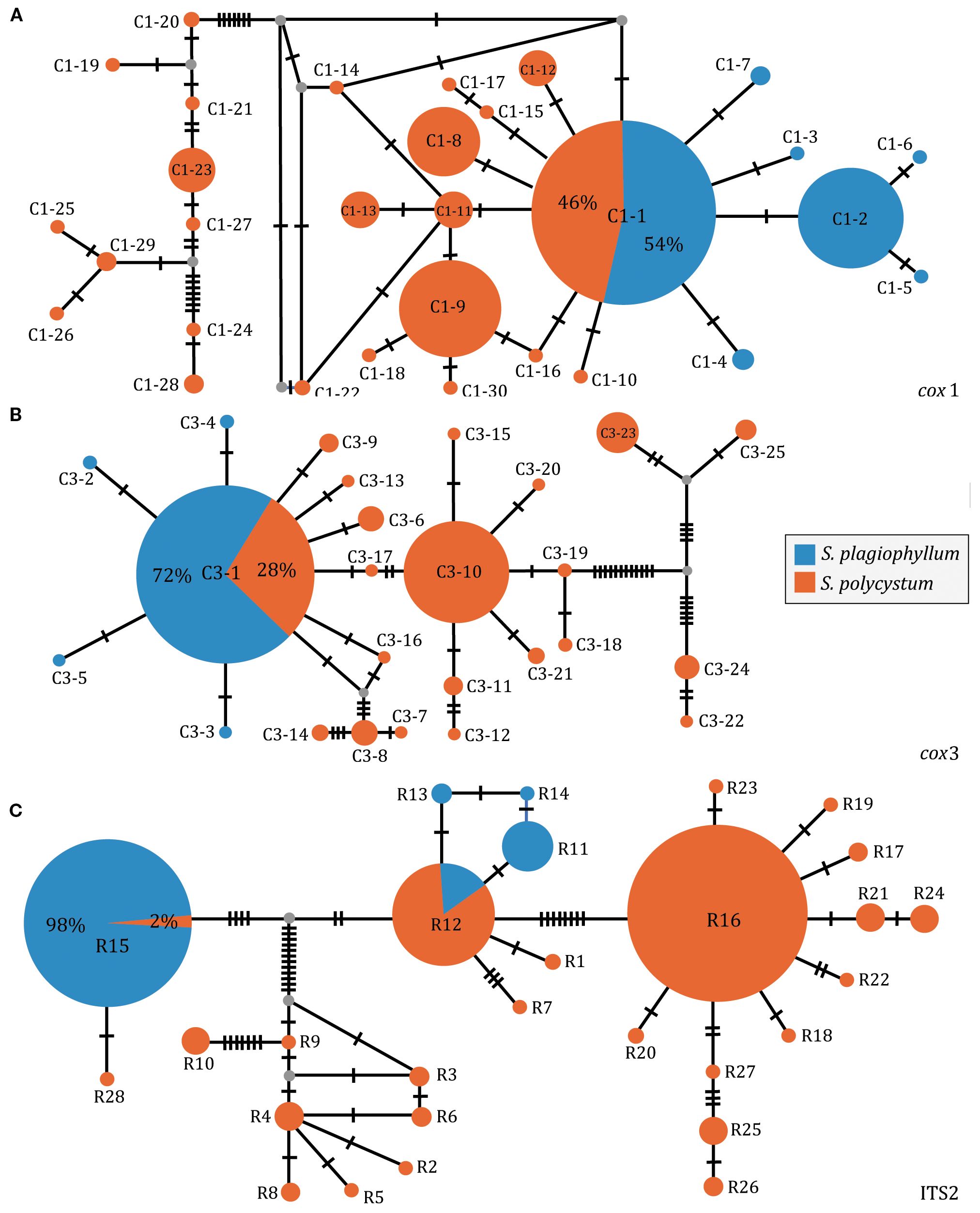
Figure 5. Parsimony median-joining network based on cox1 (A), cox3 (B) and ITS2 (C) dataset of S. plagiophyllum and S. polycystum. Circle size is proportional to population sample size. Each line between haplotypes/ribotypes represents one mutation step.
Molecular dating showed that the S. plagiophyllum-S. polycystum clade diverged c. 0.43–0.49 Mya (Supplementary Figures S9, S10, S11), which is compatible with the estimation by Yip et al. (2020). Building on this dating insight, the genetically isolated lineage of S. polycystum from Hainan-Guangxi (China) was estimated to occur and diversify c. 0.26–0.31 Mya. Over the TMP, S. polycystum haplotypes/ribotypes are nested within S. plagiophyllum stochastically, and started to diverge as a separate lineage c. 0.17–0.23 Mya (Supplementary Figures S9, S10, S11).
Discussion
The late Pleistocene glacial survival and post-glacial expansion
Phylogeographic evidence (Figures 1, 3, S2, S4, S9-S11) showed that the origination and demographic expansion of S. polycystum/S. plagiophyllum around the TMP were likely linked to sea level fluctuations driven by the late Pleistocene glacial-interglacial cycles. The highest numbers of endemic haplotypes/ribotypes and highest genetic diversity in PKL, KN and NY (Table 1, Figures 1, S2) suggest that the Andaman Sea (e.g. the west coast of the Phuket Island) probably harboured a glacial refugium for the ancestral population of S. plagiophyllum to survive (Provan et al., 2005; Maggs et al., 2008; Hu et al., 2018), when the maximum sea levels dropped to -120 meters in the Sunda shelf during the maximum glaciation (Voris, 2000; Woodruff, 2010).
For S. polycystum, two potential glacial refugia have been proposed for its ancestral survival during the late Pleistocene sea-level fluctuations (Hu et al., 2018), with one in the southwest of Hainan Island, China and the other distributed around the western Coral Triangle (e.g. the Bali Island, Indonesia). Here, the combined network diagrams (Supplementary Figure S8) and molecular dating of S. plagiophyllum/S. polycystum (Supplementary Figure S9; Supplementary Figure S11) revealed an evolutionary distinctiveness of Hainan-Guangxi haplotypes/ribotypes, providing additional evidence of a refugium existed near Hainan Island. Moreover, cox1-based S. polycystum haplotypes (C1-6–C1-11) were found only in the populations MCR, MPB and SP in the Malacca Strait (Figure 1D). Such a uniqueness of genetic diversity, together with the exclusive occurrence of haplotypes in the population BF for S. plagiophyllum (i.e. C1-4 in Figure 1A, C3-4 in Figure 1C, and H11 in Supplementary Figure S2A), suggest the likely existence of a third glacial refugium for S. polycystum in the northern Malacca Strait (Voris, 2000; Cannon et al., 2009; Wee et al., 2020). Alternatively, the rich genetic variation in the Malacca Strait probably arose from the secondary contact of other divergent lineages of S. polycystum as reported in the mangrove Sonneratia alba Sm (Yang et al., 2017). during inter-glacial periods. However, discerning these biogeographic scenarios depends on the integration of more evidence of paleogeographic coastal configuration during the ice ages, including more samplings of periphery populations like south Java, Sunda Strait, West Sumatra, Myanmar and Coral Triangle, together with phylogeographic illustrations from other species co-occurred around the TMP.
The S. polycystum and S. plagiophyllum clade was estimated to originate from the common ancestor at c. 0.43–0.49 Mya. Afterwards, the lineage of S. polycystum in Hainan-Guangxi (China) formed c. 0.26–0.31 Mya, and another lineage of combined S. polycystum and S. plagiophyllum around the TMP occurred c. 0.17–0.23 Mya (Supplementary Figures S9–S11). These timeframes correspond to the late Pleistocene epoch and are compatible to the latest organellar-genome based interspecific time-calibration (0.10–0.53 Mya) (Zhang et al., 2022), including the historical biogeographic dating of the most recent diverged section Polycystae at c. 0.40 Mya (Figure 1 in Yip et al., 2020). In addition, both Sargassum species around the TMP experienced slight demographic expansion c. 0.012–0.055 Mya (Figures 3, S4), which corresponds to the transition from the last glaciation (c. 0.115–0.012 Mya) in the late Pleistocene to early Holocene. It should be noted that the demographic expansion spectrums of S. polycystum around the TMP are much later than previous mismatch-distribution based estimation (0.41–0.62 Mya, Chan et al., 2013) and the cox1+cox3 based extend-BSPs results in Hainan-Guangxi (China) (0.60–0.80 Mya, Hu et al., 2018). It is also later than the expansion time in the congeneric S. aquifolium in Southeast Asia (0.25 Mya, Chan et al., 2014).
Ocean currents and population genetic connectivity
The most common haplotype/ribotype of S. polycystum (C1-1 in Figure 1D, R16 in Figure 1E, C3-10 in Figure 1F, H14 in Supplementary Figure S2B) distributes widely on both sides of the TMP. The lack of east-west differentiation indicates that the TMP has not served as a contemporary geographical barrier affecting genetic connectivity of S. polycystum. This contrasts with recent studies that illustrated a distinct genetic break across the TMP in mangroves with lower dispersal capabilities (e.g., Avicennia alba Blume and Sonneratia alba, Yang et al., 2017; Wee et al., 2020). However, our finding is consistent with the reported absence of an east-west genetic differentiation in the mangrove Rhizophora mucronata Lam. with high dispersal potential via progagules (Wee et al., 2014). The observed phylogeographic diversity and genetic structure do not present evidence of vicariance in S. polycystum/S. plagiophyllum (Wee et al., 2014; Tan et al., 2018). Instead, the widespread distribution of the ancestral haplotype/ribotype in the west of the TMP for S. plagiophyllum (C1-1 in Figure 1A, R15 in Figure 1B, C3-1 in Figure 1C, H4 in Supplementary Figure S2A), together with the structuring analysis (Supplementary Figure S3) and gene flow estimation (Figures 4, S5-S7), further allow us to propose that post-glacial genetic exchange driven by oceanic current may have re-established population connectivity in each Sargassum species across the TMP.
Regardless of migration pathway and recent migration rate, both Sargassum species exhibited a stepping-stone migration pattern along the Malacca Strait (Figures 4, S5-S7), which is basically in accordance with the correlation between geographical and genetic distance along a linear coastline observed in mangroves (Kennedy et al., 2017; Wee et al., 2020). The clear south-to-north historical migration route observed in S. plagiophyllum/S. polycystum along the Malacca Strait (Figures 4, S5-S7) can be explained by a strong contemporary gene flow via sea dispersal, as it concurs with the present-day ocean circulation patterns that predominantly flow from south to north in the Malacca Strait (Rizal et al., 2012). In the Gulf of Thailand, a high proportion of recent migration between distant S. polycystum populations indicates continuous contemporary gene flow driven by the clockwise circulation during the northeast monsoon season and a counterclockwise vortex during the southwest monsoon.
Sargassum species (e.g. S. plagiophyllum) around the TMP has a cycle of growth and reproduction throughout the year (Pongparadon et al., 2019) and reached its peak percentage of fertility at the end of the north-east monsoon season (Prathep et al., 2019). As the late Pleistocene ice age gradually ended, sea-level rose and connectivity between the east and west sides of the TMP was restored (Hall, 2009). The rise in temperature during the inter-glacial periods facilitated more favorable conditions enabling the detached vegetative branches of S. polycystum/S. plagiophyllum to float, survive and migrate (Komatsu et al., 2007; Woodruff, 2010). Particularly during the north-east and south-west monsoons, strong currents flow from the South China Sea via the Malacca Strait to the Indian Ocean (Figure 1). Thus, the long-distance dispersal can be considerably accelerated by seasonal monsoons that augment the ocean currents (Komatsu et al., 2007). These current systems not only prevented the mixing of waters at the boundary between the south of the Andaman Sea and the north of the Malacca Strait (Wee et al., 2014), but also likely contributed to S. polycystum and S. plagiophyllum to expand their distribution ranges north up to the Myanmar Andaman Sea and on the Gulf of Bengal coast (Soe-Htun et al., 2012).
Genetic signatures of incipient speciation
Repeated ecologically-driven transitions during the late Pleistocene glaciations are recognized as an important factor to generate genetic divergence among seaweed populations, ultimately leading to multiple speciation events via repeated isolation in allopatric refuges (Fraser et al., 2016; Ayres-Ostrock et al., 2019; Neiva et al., 2024). In the case of the sister-species S. polycystum and S. plagiophyllum, they occur largely allopatric around the TMP. Sargassum plagiophyllum occurs in abundance in the intertidal along the entire west coast of Thailand and is also recorded from the Andaman coast of Myanmar. Its southern limit is Penang Island (Malaysia) in the northern Malacca Strait. Sargassum polycystum occurs in the shallow subtidal of the Gulf of Thailand and in the intertidal in the southern Malacca Strait. It was not found in Lanta Island, where it was collected in the past (Kantachumpoo et al., 2015), which is likely linked to human-mediated change of the maximum sea surface salinity, dissolved oxygen and phosphate concentration (Sumandiarsa et al., 2021; Stankovic et al., 2022). The small intertidal S. polycystum population Phuket (PK) grew mixed with S. ilicifolium, close to the low water edge and would only be exposed during springtide. At the same site, but a little bit higher grew S. plagiophyllum (NY) in abundance. Sargassum polycystum is not found along the Andaman coast of Myanmar. Sargassum plagiophyllum grows all year round and reproduces during November-February (Pongparadon et al., 2019), whereas S. polycystum can only be found during the short reproductive season in July in Phuket (Thongroy et al., 2007). These ecological-niche and phenological transitions may be a directional selection process that was initiated by divergence in allopatric conditions during glacial phases (Fraser et al., 2016; Cocito et al., 2019), presenting important signatures of local adaptation and reproductive isolation in incipient species (Futuyma, 2005; Charron et al., 2014).
At present, the low mitochondrial resolution prevents confirmation of the degree of reproductive isolation of S. polycystum/S. plagiophyllum and, therefore, the underlying evolutionary scenarios behind their incipient speciation remain unclear. However, vicariance events, for example the repeated sea-level fluctuations in the Southeast Asia during the late Pleistocene (Benzie, 1998; Woodruff, 2010) followed by ‘founder takes all’ density-dependent processes during post-glacial range expansion (Waters et al., 2013) that can act as a plausible explanation to niche and reproductive season differentiation. In addition, parallel divergence underpinned by historical peripheral isolation and niche differentiation (e.g. temperature, Isa et al., 2020) seems a key force in driving allopatric speciation and radiation of hermaphrodite lineages across the TMP (Almeida et al., 2022), particularly for the dioecious S. plagiophyllum and S. polycystum with peripheral isolation of range edge and directional introgressive gene flow (BF in Figures 1C and S2A, Supplementary Table S3). The wide sharing of the common haplotypes/ribotypes between S. polycystum and S. plagiophyllum suggests that incipient reproductive isolation may have evolved as a by-product of divergence in allopatric speciation (Futuyma, 2005), representing an additional example of incipient reproductive speciation in the brown algae of the order Fucales (Neiva et al., 2024). In the future, genome-scale screening of independent replicating SNPs can be considered to identify clear evidence of recent or incipient speciation (Almeida et al., 2022).
Conclusion
Series of phylogeographic evidence illustrated that the biogeographic and genetic variation patterns of S. plagiophyllum/S. polycystum around the TMP likely resulted from ancestral survival during the late Pleistocene ice ages and following circulation-driven demographic expansion. Contemporary gene flow and population genetic structuring are maintained by ocean-current facilitating long-distance dispersal of detached rafting thalli. The prevailing ocean currents in the Gulf of Thailand and the Malacca Strait ostensibly explain the directions of population-level genetic continuity. Mitochondrial and nuclear datasets exhibited signature of incipient differentiation between S. plagiophyllum and S. polycystum. Future wider-sampling based phylogeographic studies of seaweeds are important and imperative, by incorporating oceanographic modelling approaches (Wee et al., 2014; Liang et al., 2022), to empirically confirm the influence of dispersal potential on the long-term patterns of population connectivity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
T-YZ: Writing – original draft, Investigation, Validation, Formal Analysis, Software, Methodology. Q-QW: Formal Analysis, Software, Validation, Methodology, Writing – review & editing, Resources. Z-MS: Validation, Supervision, Methodology, Funding acquisition, Writing – review & editing, Project administration. SD: Validation, Investigation, Data curation, Conceptualization, Resources, Writing – review & editing, Project administration, Funding acquisition, Methodology. Z-MH: Writing – review & editing, Supervision, Funding acquisition, Resources, Project administration, Validation, Methodology, Data curation, Conceptualization, Investigation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by National Natural Science Foundation of China (32371697 to Z-MH), the Thailand Research Fund (RDG6130002 to SGAD) and start-up funds from the Outstanding Talent Program at Yantai University (to Z-MH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1673650/full#supplementary-material
References
Almeida, S., Neiva, J., Sousa, F., Martins, N., Cox, C. J., Melo-Ferreira, J., et al. (2022). A low- latitude species pump: Peripheral isolation, parapatric speciation and mating- system evolution converge in a marine radiation. Mol. Ecol. 31, 4797–4817. doi: 10.1111/mec.16623
Ayres-Ostrock, L. M., Valero, M., Mauger, S., Oliveira, M. C., Plastino, E. M., Guilemin, M. L., et al. (2019). Dual influence of terrestrial and marine historical processes on the phylogeography of the Brazilian intertidal red alga Gracilaria caudata. J. Phycol. 55, 1096–1114. doi: 10.1111/jpy.12892
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Beerli, P. and Felsenstein, J. (2001). Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Nat. Acad. Sci. U.S.A. 98, 4563–4568. doi: 10.1073/pnas.081068098
Benzie, J. A. H. (1998). “Genetic structure of marine organisms and SE Asian biogeography,” in Biogeography and geological evolution of SE Asia. Eds. Hall, R. and Holloway, J. D. (Backhuys Publishers, Leiden, The Netherlands), 197–209.
Bermingham, E. and Moritz, C. (2010). Comparative phylogeography: concepts and applications. Mol. Ecol. 7, 367–369. doi: 10.1046/j.1365-294x.1998.00424.x
Briggs, J. C. (2005). The marine East Indies: diversity and speciation. J. Biogeogr.y 32, 1517–1522. doi: 10.1111/j.1365-2699.2005.01266.x
Bulan, J., Maneekat, S., Zuccarello, G. C., and Muangmai, N. (2022). Phylogeographic patterns in cryptic Bostrychia tenella species (Rhodomelaceae, Rhodophyta) across the Thai-Malay Peninsula. Algae 37, 123–133. doi: 10.4490/algae.2022.37.6.4
Buranapratheprat, A. (2008). Circulation in the upper Gulf of Thailand: a review. Burapha. Sci. J. 13, 75–83.
Cannon, C. H., Morley, R. J., and Bush, A. B. G. (2009). The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc. Nat. Acad. Sci. U.S.A. 106, 11188–11193. doi: 10.1073/pnas.0809865106
Carpenter, K. E., Barber, P. H., Crandall, E. D., Ablan-Lagman, M. C. A., Ambariyanto, Ngurah, M. G., et al. (2011). Comparative phylogeography of the coral triangle and implications for marine management. J. Mar. Biol. 2011, 1–14. doi: 10.1155/2011/396982
Chan, S. W., Cheang, C. C., Chirapart, A., Gerung, G., Tharith, C., and Ang, P. (2013). Homogeneous population of the brown alga Sargassum polycystum in Southeast Asia: possible role of recent expansion and asexual propagation. PloS One 8, e77662. doi: 10.1371/journal.pone.0077662
Chan, S. W., Cheang, C. C., Yeung, C. W., Chirapart, A., Gerung, G., and Ang, P. (2014). Recent expansion led to the lack of genetic structure of Sargassum aquifolium populations in Southeast Asia. Mar. Biol. 161, 785–795. doi: 10.1007/s00227-013-2377-3
Chan, B. K. K., Tsao, Y.-F., Wangkulangkul, K., Amjud, K., and Sukparangsi, W. (2022). Biogeography and biodiversity of the intertidal barnacle Tetraclita species in the Gulf of Thailand and Andaman Sea influences of oceanographic currents and Pleistocene glaciations. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.774041
Charron, G., Leducq, J. B., and Landry, C. R. (2014). Chromosomal variation segregates within incipient species and correlates with reproductive isolation. Mol. Ecol. 23, 4362–4372. doi: 10.1111/mec.12864
Cocito, L. L., Ceballos, S. G., and Fernández, D. A. (2019). Sharp phylogeographical differentiation near the southern range edge of the silverside Odontesthes nigricans: distinct peripheral populations and incipient speciation? Estua. Coast. Shelf Sci. 226, 106276. doi: 10.1016/j.ecss.2019.106276
Draisma, S. G. A., Ballesteros, E., Rousseau, F., and Thibaut, T. (2010). DNA sequence data demonstrate the polyphyly of the genus Cystoseira and other Sargassaceae genera (Phaeophyceae). J. Phycol. 46, 1329–1345. doi: 10.1111/j.1529-8817.2010.00891.x
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L. and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Faga, G. and Gurgel, C. F. D. (2024). Distributional range shifts of Western Atlantic benthic Sargassum species (Fucales, Phaeophyceae) under future climate change scenarios. Aquat. Bot. 190, 103705. doi: 10.1016/j.aquabot.2023.103705
Fraser, C. I., McGaughran, A., Chuah, A., and Waters, J. M. (2016). The importance of replicating genomic analyses to verify phylogenetic signal for recently evolved lineages. Mol. Ecol. 25, 3683–3695. doi: 10.1111/mec.13708
Gaither, M. R., Toonen, R. J., Robertson, D. R., Planes, S., and Bowen, B. W. (2010). Genetic evaluation of marine biogeographical barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus). J. Biogeogr. 37, 133–147. doi: 10.1111/j.1365-2699.2009.02188.x
Guillemin, M.-L., Dubrasquet, H., Reyes, J., and Valero, M. (2018). Comparative phylogeography of six red algae along the Antarctic Peninsula: extreme genetic depletion linked to historical bottlenecks and recent expansion. Polar Biol. 41, 827–837. doi: 10.1007/s00300-017-2244-7
Guiry, M. D. and Guiry, G. M. (2025). AlgaeBase (National University of Ireland, Galway: World-wide electronic publication). Available online at: http://www.algaebase.org.
Hall, R. (1998). “The plate tectonics of Cenozoic SE Asia and the distribution of land and sea,” in Biogeography and geological evolution of SE asia. Eds. Hall, R. and Holloway, J. D. (Leiden, the Northlands: Backhuys Publishers), 99–131.
Hall, R. (2009). Southeast Asia’s changing palaeogeography. Blumea–Biodivers. Evol. Biogeogr. Plants 54, 148–161. doi: 10.3767/000651909X475941
Hall, R. (2012). Late Jurassic–Cenozoic reconstructions of the Indonesian region and the Indian Ocean. Tectonophysics 570-571, 141. doi: 10.1016/j.tecto.2012.04.021
Hu, Z. M., Kantachumpoo, A., Liu, R. Y., Sun, Z. M., Yao, J. T., Komatsu, T., et al. (2018). A late Pleistocene marine glacial refugium in the Southwest of Hainan Island, China: Phylogeographical insights from the brown alga Sargassum polycystum. J. Biogeogr. 45, 355–366. doi: 10.1111/jbi.13130
Hu, Z. M., Uwai, S., Yu, S. H., Komatsu, T., Ajisaka, T., and Duan, D. L. (2011). Phylogeographic heterogeneity of the brown macroalga Sargassum horneri (Fucaceae) in the northwestern pacific in relation to late pleistocene glaciation and tectonic configurations. Mol. Ecol. 20, 3894–3909. doi: 10.1111/j.1365294X.2011.05220.x
Huanel, O. R., Montecinos, A. E., Sepúlveda-Espinoza, F., and Guillemin, M.-L. (2024). Impact of persistent barrier to gene flow and catastrophic events on red algae evolutionary history along the Chilean coast. Front. Genet. 15. doi: 10.3389/fgene.2024.1336427
Isa, N. S., Akhir, M. F., Kok, P. H., Daud, N. R., Khalil, I., and Roseli, N. H. (2020). Spatial and temporal variability of sea surface temperature during ElNiño Southern Oscillation and Indian Ocean dipole in the Strait of Malacca and Andaman Sea. Reg. Stud. Mar. Sci. 39, 101402. doi: 10.1016/j.rsma.2020.101402
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kantachumpoo, A., Uwai, S., Noiraksar, T., and Komatsu, T. (2015). Systematics of marine brown alga Sargassum from Thailand: A preliminary study based on morphological data and nuclear ribosomal internal transcribed spacer 2 (ITS2) sequences. Ocean. Sci. J. 50, 251–262. doi: 10.1007/s12601-015-0022-4
Kendrick, G. A. and Walker, D. I. (1995). Dispersal of propagules of Sargassum spp. (Sargassaceae: Phaeophyta): Observations of local patterns of dispersal and consequences for recruitment and population structure. J. Exp. Mar. Biol. Ecol. 192, 273–288. doi: 10.1016/0022-0981(95)00076-4
Kennedy, J. P., Garavelli, L., Truelove, N. K., Devlin, D. J., Box, S. J., Chérubin, L. M., et al. (2017). Contrasting genetic effects of red mangrove (Rhizophora mangle L.) range expansion along West and East Florida. J. Biogeogr. 44, 335–347. doi: 10.1111/jbi.12813
Kogishi, K., Kitayama, T., Miller, K. A., Hanyuda, T., and Kawai, H. (2010). Phylogeography of Cutleria cylindrica (Cutleriales, Phaeophyceae) in Northeastern Asia, and the identity of an introduced population in California. J. Phycol. 46, 553–558. doi: 10.1111/j.1529-8817.2010.00818.x
Komatsu, T., Tatsukawa, K., Filippi, J. B., Sagawa, T., Matsunaga, D., Mikami, A., et al. (2007). Distribution of drifting seaweeds in eastern East China Sea. J. Mar. Syst. 67, 245–252. doi: 10.1016/j.jmarsys.2006.05.018
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, C. E., Lindstrom, S. C., and Saunders, G. W. (2007). A molecular assessment of northeast Pacific Alaria species (Laminariales, Phaeophyceae) with reference to the utility of DNA barcoding. Mol. Phylogenet. Evol. 44, 634–648. doi: 10.1016/j.ympev.2007.03.016
Li, J. J., Hu, Z. M., Gao, X., Sun, Z. M., Choi, H. G., Duan, D. L., et al. (2017). Oceanic currents drove population genetic connectivity of the brown alga Sargassum thunbergii in the north-west Pacific. J. Biogeogr. 44, 230–242. doi: 10.1111/jbi.12856
Liang, Y., Zhang, S. S., Yan, C. X., Draisma, S. G. A., Kantachumpoo, A., Li, Z., et al. (2022). Influence of Indo-Pacific Ocean currents on the distribution and demographic patterns of the brown seaweed Sargassum polycystum in tropical East Asia. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.895554
Librado, P. and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, Y. D., Zhang, J., Du, Y. Q., Zhang, S. S., Liu, L., Draisma, S. G. A., et al. (2024). Marine conditions in Andaman Sea shape the unique genetic structure of Sargassum plagiophyllum C. Agardh. J. Appl. Phycol. 36, 501–511. doi: 10.1007/s10811-023-03144-3
Maggs, C. A., Castilho, R., Foltz, D., Henzler, C., Jolly, T., Kelly, J., et al. (2008). Evaluating signatures of glacial refugia for North Atlantic marine organisms. Ecology 89, S108–S122. doi: 10.1890/08-0257.1
Mirams, A. G. K., Treml, E. A., Shields, J. L., Liggins, L., and Riginos, C. (2011). Vicariance and dispersal across an intermittent barrier: population genetic structure of marine animals across the Torres Strait land bridge. Coral. Reefs. 30, 937–949. doi: 10.1007/s00338-011-0767-x
Mizuno, S., Ajisaka, T., Lahbib, S., Kokubu, Y., Alabsi, M. N., and Komatsu, T. (2014). Spatial distributions of floating seaweeds in the East China Sea from late winter to early spring. J. Appl. Phycol. 26, 1159–1167. doi: 10.1007/s10811-013-0139-8
Morley, R. J. (2012). “A review of the Cenozoic palaeoclimate history of Southeast Asia,” in Biotic evolution and environmental change in southeast asia. Eds. Gower, D., Johnson, K., Richardson, J., Rosen, B., Rüber, L., and Williams, S. (Cambridge, UK: Cambridge University Press), 79–114. doi: 10.1017/CBO9780511735882.006
Muangmai, N., Maneekat, S., Chirapart, A., and Zuccarello, G. C. (2023). Contrasting patterns of genetic diversity and population discontinuity in the common red seaweed Gracilaria salicornia (Gracilariaceae) along the coasts of Thailand. Phycologia 62, 452–461. doi: 10.1080/00318884.2023.2254621
Neiva, J., Assis, J., Fragkopoulou, E., Pearson, G. A., Raimondi, R. T., Anderson, L., et al. (2024). Trans-Arctic asymmetries, melting pots and weak species cohesion in the low-dispersal amphiboreal seaweed Fucus distichus. Front. Ecol. Evol. 12. doi: 10.3389/fevo.2024.1356987
Pongparadon, S., Tongwong, W., Panyawai, J., and Prathep, A. (2019). “Sargassum plagiophyllum C. Agardh (Fucales, Phaeophyta) population structure dynamic in Ko Lanta, Krabi Province,” in Abstract book of the 9th national conference on algae and plankton (Kasetsart University, Bangkok), 121.
Prathep, A. K., Pengnoo, S., and Pongparadon, S. (2019). The phenology of Sargassum plagiophyllum C. Agardh (Fucales, Pheaophyta) in Ko Lanta, Krabi province. Abstract book of The 9th National Conference on Algae and Plankton, Kasetsart University, Bangkok, pp 110
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Provan, J., Wattier, R. A., and Maggs, C. A. (2005). Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol. Ecol. 14793–803. doi: 10.1111/j.1365-294X.2005.02447.x
Reaka, M. L., Rodgers, P. J., and Kudla, A. U. (2008). “Speciation extinction dynamics and the topography of diversity on IndoWest Pacific coral reefs,” in Proceedings of the 11th international coral reef symposium, vol. 26. (Florida), 1377–1381.
Rizal, S., Damm, P., Wahid, M. A., Sundermann, J., Ilhamsyah, Y., Iskandar, T., et al. (2012). General circulation in the Malacca Strait and Andaman Sea: A numerical model study. Am. J. Environ. Sci. 8, 479–488. doi: 10.3844/ajessp.2012.479.488
Soe-Htun, U., Mya, K. W., Soe, P. P. K., Myo, M. T., and Hsu, M. O. (2012). The morphotaxonomy and phytogeographical distribution of the species of Sargassum section polycystae (fucales, phaeophyta) from Myanmar: Sargassum polycystum C. agardh and S. plagiophyllum C. agardh. Mawlamyine. Univers. Res. J. 4, 215–233.
Sojisuporn, P., Morimoto, A., and Yanagi, T. (2010). Seasonal variation of sea surface current in the Gulf of Thailand. Coast. Mar. Sci. 34, 91–102.
Spalding, M. D., Fox, H. E., llen, G. R., Davidson, N., Ferdaña, Z. A., Finlayson, M., et al. (2007). Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. BioScience 57, 573–583. doi: 10.1641/B570707
Stankovic, M., Draisma, S. G. A., Pongparadon, S., Wichachucherd, B., Noiraksar, T., and Hu, Z. M. (2022). Predicting macroalgal species distributions along the Thai-Malay Peninsula. Estua. Coast. Shelf Sci. 267, 107760. doi: 10.1016/j.ecss.2022.107760
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., and Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016. doi: 10.1093/ve/vey016
Sumandiarsa, I. K., Bengen, D. G., Santoso, J., and Januar, H. I. (2021). The impact of spatio-temporal variation on seawater quality and its effect on the domination of Sargassum polycystum on small islands in western Indonesian waters. Environ. Asia. 14, 80–92. doi: 10.14456/ea.2021.9
Tan, P. L., Lim, P. E., Lin, S. M., Phang, S. M., Draisma, S. G. A., and Liao, L. (2018). A genetic diversity assessment of Halymenia Malaysiana (Halymeniaceae, Rhodophyta) from Malaysia and the Philippines based on COI-5P and rbcL sequences. J. Appl. Phycol. 30, 3445–3454. doi: 10.1007/s10811-018-1484-4
Thongroy, P., Liao, L. M., and Prathep, A. (2007). Diversity, abundance and distribution of macroalgae at Sirinart Marine National Park, Phuket Province, Thailand. Bot. Mar. 50, 88–96. doi: 10.1515/BOT.2007.010
Voris, H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27, 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x
Waelbroeck, C., Labeyrie, L., Michel, E., Duplessy, J. C., and McManus, J. F. (2002). Sea-level and deep water temperature changes derived from benthic foraminifera isotopic records. Quaternary. Sci. Rev. 21, 295–305. doi: 10.1016/S0277-3791(01)00101-9
Wang, Q. Q., Zhang, T. Y., Liu, Y. J., Nguyen, X. V., Sun, Z. M., Draisma, S. G. A., et al (2025). Low phylogeographic diversity in the calcified green macroalga Halimeda macroloba (Bryopsidales) in Southeast Asia. Algae, 40 (2), 93–102. doi: 10.4490/algae.2025.40.5.15
Waters, J. M., Fraser, C. I., and Hewitt, G. M. (2013). Founder takes all: density-dependent processes structure biodiversity. Trend. Ecol. Evol. 28, 78–85. doi: 10.1016/j.tree.2012.08.024
Wee, A. K. S., Noreen, A. M. E., Ono, J., Takayama, K., Kumar, P. P., Tan, H. T. W., et al. (2020). Genetic structures across a biogeographical barrier reflect dispersal potential of four Southeast Asian mangrove plant species. J. Biogeogr. 47, 1258–1271. doi: 10.1111/jbi.13813
Wee, A. K. S., Takayama, K., Asakawa, T., Thompson, B., Onrizal, Sungkaew, S., et al. (2014). Oceanic currents, not land masses, maintain the genetic structure of the mangrove Rhizophora mucronate Lam. (Rhizophoraceae) in Southeast Asia. J. Biogeogr. 41, 954–964. doi: 10.1111/jbi.12263
Wichachucherd, B., Prathep, A., and Zuccarello, G. C. (2014). Phylogeography of Padina boryana (Dictyotales, Phaeophyceae) around the Thai-Malay Peninsula. Eur. J. Phycol. 49, 313–323. doi: 10.1080/09670262.2014.918658
Woodruff, D. S. (2010). Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodiver. Conserv. 19, 919–941. doi: 10.1007/s10531-010-9783-3
Yang, Y. C., Li, J. F., Yang, S. H., Li, X. N., Fang, L., Zhong, C. R., et al. (2017). Effects of Pleistocene sea-level fluctuations on mangrove populations dynamics: a lesson from Sonneratia alba. BMC Evol. Biol. 17, 22. doi: 10.1186/s12862-016-0849-z
Yip, Z. T., Quek, R. Z. B., and Huang, D. W. (2020). Historical biogeography of the widespread macroalga Sargassum (Fucales, Phaeophyceae). J. Phycol. 56, 300–309. doi: 10.1111/jpy.12945
Yoshida, T., Stiger, V., and Horiguchi, T. (2000). Sargassum boreale sp. nov. (Fucales, Phaeophyceae) from Hokkaido, Japan. Phycol. Res. 48, 125–131. doi: 10.1046/j.1440-1835.2000.00197.x
Keywords: genetic admixture, long-distance dispersal, ocean current, incipient speciation, phylogeographic diversity, sea-level fluctuation
Citation: Zhang T-Y, Wang Q-Q, Sun Z-M, Draisma SGA and Hu Z-M (2025) Comparative phylogeographic patterns and processes of the incipient brown seaweeds Sargassum polycystum and S. plagiophyllum around the Thai-Malay Peninsula. Front. Plant Sci. 16:1673650. doi: 10.3389/fpls.2025.1673650
Received: 26 July 2025; Accepted: 03 September 2025;
Published: 17 September 2025.
Edited by:
Daniel Pinero, National Autonomous University of Mexico, MexicoReviewed by:
Tang Yanbin, Ministry of Natural Resources, ChinaJane Edgeloe, University of Western Australia, Australia
Copyright © 2025 Zhang, Wang, Sun, Draisma and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano G. A. Draisma, c2dhZHJhaXNtYUB5YWhvby5jb20=; Zi-Min Hu, aHV6bUB5dHUuZWR1LmNu; aHV6aW1pbjk3MTJAMTYzLmNvbQ==
 Tong-Yun Zhang1
Tong-Yun Zhang1 Zhong-Min Sun
Zhong-Min Sun Stefano G. A. Draisma
Stefano G. A. Draisma Zi-Min Hu
Zi-Min Hu