- 1Department of Plant Pathology, Institute of Food and Agricultural Sciences (IFAS), University of Florida, Gainesville, FL, United States
- 2Department of Biology, University of Mississippi, Oxford, MS, United States
- 3Integrative Bioinformatics, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, Durham, NC, United States
As part of an armory against pathogens, plants carry resistance (R) genes despite the fitness costs they can incur. While these detrimental effects have been associated with the presence and interactions of numerous R genes in various plant species, molecular models do not exist for the mechanisms underlying R gene-mediated fitness costs. The rice R gene Xa21, encoding a cell-surface immune receptor, specifies robust resistance to Xanthomonas oryzae pv. oryzae. Here, we demonstrate that Xa21 expression causes drastic fertility defects, including reduced pollen viability, impaired anther dehiscence, and severe grain loss, at a low temperature (24°C) and in a dose-dependent manner. Under such growth conditions, Xa21 plants displayed abundant accumulation of reactive oxygen species in their anthers and decreased expression of genes related to jasmonate biosynthesis, signaling, and response in their spikelets during anthesis. Consequently, jasmonate contents in XA21 spikelets were lower than those in the control. The exogenous application of methyl jasmonate largely rescued the anther dehiscence of Xa21 plants. Given the key roles of lipid-derived jasmonates in stamen development and maturation in plants, our findings link R gene expression, jasmonic acid (JA) signaling, and fertility defects; identify temperature as an environmental factor influencing the range of R gene functions; and explain the abundant accumulation of 17 transposable-like elements previously observed in the Xa21 locus.
Introduction
Extensive research over the past three decades has led to the molecular characterization of diverse plant species of more than 300 disease resistance (R) genes that confer resistance to a variety of pathogens (Kourelis and van der Hoorn, 2018). Major families of characterized R genes include those encoding intracellular receptors containing nucleotide-binding and leucine-rich repeat (NLR) domains (Jones et al., 2016, 2024), cell-surface receptor-like proteins (RLPs), and receptor-like kinases (RLKs) (Thomas et al., 1998; Sun et al., 2004; Chen et al., 2006; Hu et al., 2017; Ercoli et al., 2022; Jones et al., 2024). Many R loci were identified in wild relatives during the breeding of disease-resistant crops (Jones et al., 2024). Often, R loci contain small gene families with alleles present in disease-susceptible individuals (Song et al., 1997; Parniske et al., 1997; Deng et al., 2017). In natural populations, both resistant and susceptible alleles may have coexisted for millions of years.
R genes are beneficial to host survival only when pathogen invasion occurs, while accumulating evidence shows that the presence of R genes can impose negative impacts on plant growth and reproduction in the absence of obvious infection (Kruger et al., 2002; Karasov et al., 2017; Calvo-Baltanas et al., 2020; Gao et al., 2024). This phenomenon is known as the fitness cost of resistance, which forms an important component of a more prevalent observation called growth-defense trade-offs (Huot et al., 2014; He et al., 2022). Pioneered by Vanderplank’s studies in the 1960s, fitness costs were first used to explain the lack of more durable partial resistance in potato varieties to late blight (caused by Phytophthora infestans) (Vanderplank, 1963; Brown and Rant, 2013). The barley recessive gene mlo confers strong, durable resistance to powdery mildew (Büschges et al., 1997) and has been widely deployed through plant breeding in Europe and other areas, but mlo resistance incurs a 5%–15% grain loss, causes necrotic leaf spotting, and increases susceptibility to other diseases (Jørgensen, 1992; Brown and Rant, 2013). Likewise, transgenic Arabidopsis plants harboring RPM1 or RPS5 (two NLR-type R genes countering Pseudomonas bacteria) produce 5%–10% fewer seeds than the susceptible controls in field trials (Tian et al., 2003; Karasov et al., 2014). Such costs of resistance have likely prevented fixation of R alleles within populations. Similarly, yield reduction has been observed in rice cultivars carrying either the NLR gene Pi-ta (a yield penalty of 12%) or the pyramided NLR genes Pib, Pi25, and Pi54, which specify resistance against rice blast disease caused by the fungal pathogen Magnaporthe oryzae (Jia et al., 2004; Dean et al., 2012; Wang et al., 2015; Ning et al., 2017; Peng et al., 2021; Tan et al., 2023). A large-scale genome-wide association study of 1,495 hybrid and parental rice lines revealed a correlation between high yields and alleles responsible for susceptibility to blight and blast diseases, reflecting a trade-off between yield performance and disease resistance to both bacteria and fungi in this agronomically important crop (Huang et al., 2015). Despite the broad importance of disease resistance in both evolution and agriculture, little is known about the underlying physiological and molecular mechanisms behind R gene-mediated fitness costs, particularly those leading to defective seed development.
Anther dehiscence is the last stage of stamen maturation in flowering plants and enables the release of mature pollen grains from the opened anther. This key step in pollination influences subsequent seed set (Wilson et al., 2011). Jasmonic acid (JA) and its derivatives (collectively called jasmonates) are required for stamen maturation and reproductive development, although JA signaling generally plays a negative role in vegetative growth and acts antagonistically with growth-promoting hormones to modulate growth-defense conflicts in plants (Wilson et al., 2011; Yang et al., 2012; Huot et al., 2014; He et al., 2022; Dhakarey et al., 2016; Acosta and Przybyl, 2019). Mutation of genes involved in JA biosynthesis and perception in Arabidopsis and rice often leads to male sterility, with deficiencies in pollen viability and anther dehiscence (McConn and Browse, 1996; Xie et al., 1998; Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; Riemann et al., 2003; Schilmiller et al., 2007; Nguyen et al., 2023). Although the pathways for JA biosynthesis and signaling have largely been elucidated in model species, our understanding of the regulatory networks controlling JA content and JA response in stamen is far from complete.
The rice Xa21 gene confers resistance to the Gram-negative bacterium Xanthomonas oryzae pv. oryzae (Xoo) that causes bacterial leaf blight disease (Khush et al., 1990; Song et al., 1995). Xa21 was originally isolated from a Mali accession of the African wild rice species longistaminata, which differs from many rice varieties in that it is perennial, its flowers are partially self-incompatible, and it can propagate asexually through its rhizomes (Khush et al., 1990; Tong et al., 2023). Xa21 encodes an RLK protein (XA21) that is mainly localized on the plasma membrane and the endoplasmic reticulum of rice cells (Park et al., 2010a; Chen et al., 2010). XA21 binds to the sulfated Xoo peptide RaxX (RaxX-sY, required for activation of XA21-mediated immunity X, tyrosine-sulfated) that is homologous to phytohormones in the plant Peptide-containing Sulfated tYrosine (PSY) family, with eight members (OsPSY1–8) in rice (Pruitt et al., 2015, 2017; Luu et al., 2019). The activation of XA21 by RaxX-sY triggers various defense responses, including the production of reactive oxygen species (ROS) (Pruitt et al., 2015; Chen et al., 2021).
Rice is a tropical/subtropical crop that produces grains normally between 22 °C and 28 °C (Su et al., 2023). XA21-mediated resistance is dose-dependent (Zhang et al., 2024) and can be primed using a low-temperature treatment of 23 °C to 27°C (Chen et al., 2019). Under low-temperature conditions, the steady-state level of the XA21 protein is not significantly affected. We have recently reported that the XA21 protein can be cleaved by a spikelet-expressed rhomboid protease, OsRBL3b (Vergish et al., 2025). The elevated accumulation of XA21 caused by OsRBL3b mutations coincides with male sterility and yield reduction. In this article, we demonstrate that Xa21 in the wild-type OsRBL3b background impairs anther dehiscence, decreases pollen viability, and reduces grain set at a lower temperature. Moreover, our data reveal a function of the immune receptor in negatively regulating JA levels and JA response and in positively modulating ROS production in reproductive tissues of rice.
Materials and methods
Plant materials and growth conditions
Transgenic lines used in this study were in either the Oryza sativa L. ssp. japonica cv. Taipei 309 (TP309) or cv. Kitaake (Kitaake) background. Seed germination on half-strength Murashige and Skoog medium supplemented with 30 g/L sucrose and 50 μg/mL hygromycin and growth in a greenhouse in the subtropical climate of Gainesville, FL, USA (29°39′55″N, 82°20′10″W) were described previously (Shamsunnaher et al., 2020).
Plasmid construction and rice transformation
A 2,204-bp promoter fragment of the Xa21 gene was PCR-amplified with the Xa21Pro-1/Xa21Pro-2 primers and cloned into the binary vector pCmH-GUS using HindIII–BamHI. The resulting construct (Xa21pro:GUS) was transformed into rice cultivar TP309 using Agrobacterium-mediated transformation according to the procedure described previously (Vergish et al., 2025).
Plant fertility, pollen viability, and anther dehiscence assays
Plants were first grown to the early booting stage and then moved to a temperature-controlled growth chamber [under LED light (200 μmol m−2 s−1) with a 13-h light (24°C)/11-h dark (21°C) photoperiod and 70% relative humidity] without pre-acclimation treatment.
Plant fertility was measured based on the number of filled and empty grains within the two uppermost panicles from at least five plants per line. To determine pollen viability, approximately 10 anthers from five plants per line were randomly harvested and squashed in a centrifuge tube. Five replicates per line were prepared. After staining with 1% (w/v) iodine–potassium iodide (I2−KI), the released pollen grains were visualized using a BX43 LED Fluorescence microscope (Olympus, Breinigsville, PA, USA). To determine anther dehiscence/indehiscence, at least 100 spikelets per line were harvested 2 hours after anthesis (HAA) from five plants. The dissected spikelets were stained with I2−KI solution and visualized using the Olympus microscope to determine the number of pollen grains on the stigmas. Anther indehiscence of a spikelet was scored when I2−KI-stained pollen grains were found inside the anthers but not on the stigmas. Cross-sectioning of anthers was performed after paraffin embedding. Briefly, collected anthers were fixed in Dietrich’s Formalin Acetic Acid for 16 hours at room temperature. Samples were processed using a Leica tissue processor. After embedding in paraffin, samples were sectioned to 10 μm using a Rotary Microtome (HM 355 S) and visualized using the Olympus microscope as above.
RNA-seq analysis
Twenty rice spikelets were harvested at anthesis from five plants per line and pooled to minimize individual variations. Three biological replicates were prepared. Total RNA was extracted using TRIzol reagent (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The RNA integrity (RIN) of each sample was assessed to ensure RIN values greater than eight. To enrich mRNA, poly(A) selection was performed using the KAPA mRNA HyperPrep kit (Roche, Indianapolis, IN, USA). The purified RNA was then used for RNA-seq library construction and paired-end (2 × 151 base read length) sequencing using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) at the Comprehensive Cancer Center (Monrovia, CA, USA). Approximately 60 million paired reads were generated per sample. Following trimming for adapters using the TrimGalore program (version 0.6.10), the cleaned reads were aligned to the O. sativa ‘Nipponbare’ reference genome (Kawahara et al., 2013) using HISAT2 (version v2.2.1) (Kim et al., 2015). Ambiguous reads that mapped to more than one region in the genome or those with a MAPQ score below 10 were removed using the SAMtools software (Genome Research Limited). Transcript quantification was performed using the Partek Genomics Suite (version 7.18) to obtain raw read counts and normalized read counts [reads per kilobase per million mapped reads (RPKM)] (Mortazavi et al., 2008). Differential gene expression was determined using generalized linear model approaches implemented in the Bioconductor package edgeR (McCarthy et al., 2012). The differentially expressed genes (DEGs) were assessed for significance based on the following criteria: absolute fold change of over 2 and false discovery rate (FDR) q-value below 0.05. Genes with a fold change greater than or equal to 2 were considered upregulated, whereas those with a fold change less than or equal to 2 were considered downregulated.
RT-qPCR analysis
Total RNA was extracted using TRIzol reagent (Ambion) followed by purification using an RNAeasy MiniElute Cleanup Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Single-stranded cDNA was synthesized with 2 μg total RNA using a QuantiTect Reverse Transcription Kit (Qiagen). qPCR was carried out using a LightCycler 480 System (Roche) according to the manufacturer’s instructions under the following conditions: 95°C, 2 min; 45 cycles of 95°C, 5 sec and 60°C, 20 sec, and 72°C, 5 min. The gene expression levels were calculated by the ΔΔCt method using the geometric mean of rice UBIQUITIN 5 and ACTIN expression levels to normalize the data (Jain et al., 2018). Primers used for q-PCR are listed in Supplementary Table 1.
LC–tandem mass spectrometry quantification of jasmonates
For JA quantification, spikelets that just opened were collected (stage 14 of anther development). Twenty spikelets (~160 mg, fresh weight) from five plants per line were transferred to a 2-mL screw cap tube (Cole-Parmer, Vernon Hills, IL, USA) and immediately frozen in liquid nitrogen. Four biological replicates/samples per line were performed. For each sample tube, a 6-mm stainless steel grinding ball (Cole-Parmer) was added. The sample was homogenized in a Spex® HG-400 MiniG® Homogenizer (Cole-Parmer) at 1,000 rpm for 5 min, with 1-min intervals. During grinding, liquid nitrogen was added to the sample trays to maintain the low temperature. After homogenization, internal standards (10 μL of 100 μM lidocaine and 10 μL of 100 μM 10-camphorsulfonic acid) were added to each sample. A serial extraction was performed with three pre-cooled solvents: extraction solvent I (acetonitrile:isopropanol:water, 3:3:2), extraction solvent II (acetonitrile:water, 1:1), and extraction solvent III (80% methanol). For each solvent, 800 μL was added, followed by 10 min of mixing by vortex and sonication in ice water for 6 min in 2-min intervals. After each extraction, samples were centrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were collected. The three extracts were combined, lyophilized to dryness, and stored at −20 °C until further analysis. For liquid chromatography–tandem mass spectrometer (LC-MS/MS) analysis, 100 μL of 0.1% formic acid in water was added to each dried extract, followed by solubilization and centrifugation (15,000 × g, 4 °C, 10 min). The supernatants were lyophilized again and reconstituted in 25 μL of 0.1% formic acid in water. A final centrifugation step was performed under the same conditions, and the supernatants were transferred to LC–MS vials for analysis. The internal standards and authentic standards of JA and JA-isoleucine (JA-Ile) were purchased from Sigma (St. Louis, MO, USA).
Quantification analysis was performed using a microflow UPLC-ZenoTOF 7600 mass spectrometer (MS) (SCIEX, Toronto, ON, Canada), equipped with an OptiFlow™ Turbo V ion source. Chromatographic separation was achieved on a nanoEase™ M/Z Symmetry® C18 column (5 μm, 300 μm i.d. × 50 mm, 100 Å) at a flow rate of 7 μL/min. The column chamber was maintained at 40°C, and the autosampler was maintained at 6°C. The injection volume for all the samples was 1 μL. The total run time was 10 min. The LC gradient consisted of mobile phase A (water with 0.1% formic acid) and mobile phase B (acetonitrile with 0.1% formic acid) and started at 90% A (0.0–0.5 min), decreased to 10% A (by 5 min), and, following a hold for 2 min, returned to 90% A (by 10 min). The eluent was introduced into the MS using the OptiFlow Turbo V source with a microflow probe under electrospray ionization (ESI) in positive mode. ESI source parameters were as follows: curtain gas, 35 psi; Collisionally Activated Dissociation (CAD) gas, 7 psi; GS1, 30 psi; GS2, 35 psi; and source temperature, 200°C. The MS/MS spectra for JA and JA-Ile were acquired using the function of multiple reaction monitoring high resolution (MRMhr) by including the [M + H]+ precursor ion. The precursor of JA at m/z 211.13 [M + H]+ yields product ions m/z 133.10 (quantifier) and 69.07 (qualifier) at a collision energy of 20 eV. The precursor of JA-Ile at m/z 324.21 [M + H]+ yields product ions m/z 151.11 (quantifier) and 86.09 (qualifier) at a collision energy of 20 eV. The declustering potential was set at 40 V, and the accumulation time was 0.05 ms for all MRMhr transitions. A five-point calibration curve (2.56, 12.8, 64, 320, and 1,600 pg/μL) was used for quantification. Data acquisition and processing were performed using the SciexOS 3.1 software (SCIEX, 2015).
Dehiscence restoration by exogenous MeJA
To determine whether the dehiscence could be rescued, Myc-Xa21L and Myc-Xa21H plants were treated with exogenous methyl jasmonate (MeJA). Two panicles per plant (five plants of each line) were sprayed with either MeJA solution [200 μM MeJA (PhytoTech Labs, Lenexa, KS, USA) and 0.1% Tween] or mock solution (0.1% Tween) in the morning. At least 100 spikelets were harvested 2 HAA and dissected to determine anther dehiscence/indehiscence as described above.
Histochemical GUS staining
GUS staining was carried out as described (Anbu and Arul, 2013). Freshly collected spikelets were immersed in GUS staining solution [100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% (v/v) Triton X-100, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 20% (v/v) methanol, 2 mM X-Gluc], vacuum-infiltrated for 10 min, and incubated overnight at 37 °C. The stained samples were incubated in 100% (v/v) ethanol for 10 hours to remove chlorophyll.
ROS assays
Staining with 3,3′-diaminobenzidine (DAB) was performed as described (Li et al., 2023). Freshly collected spikelets were immersed in DAB solution (1 mg/mL, pH 3.8) overnight and then boiled in ethanol for 10 min, followed by several washes in ethanol. Seven DAB-stained anthers per line were used. Approximate borders of DAB-stained areas in each anther were traced. The corresponding areas were measured in pixels using the image processing software Fiji/ImageJ (ImageJ, RRID: SCR_003070). Relative DAB-stained areas were calculated by dividing the number of DAB-stained pixels by the total number of anther pixels.
Results
XA21 drastically reduces grain set in a dose-dependent manner at a lower ambient temperature (24°C)
Since lower temperatures prime XA21 resistance to Xoo (Chen et al., 2019), we reasoned that low temperature may be sufficient to induce the XA21-dependent fertility defects even in the presence of the functional OsRBL3b. To test this hypothesis, we used two previously characterized transgenic lines (Myc-Xa21L and Myc-Xa21H in the background of TP309) carrying the same construct encoding an N-terminal c-Myc-tagged XA21 (Myc-XA21) expressed from its native promoter (Xu et al., 2006; Wang et al., 2006; Chen et al., 2019; Vergish et al., 2025). The Myc-XA21 protein accumulated to a higher level in line Myc-Xa21H than in line Myc-Xa21L in spikelets at 28°C (Vergish et al., 2025). We normally grow these plants in our outdoor greenhouse in Florida during the summer, with daytime temperatures ranging from 30 °C to 43°C.
When grown at a lower temperature in a growth chamber [under LED light (200 μmol m−2 s−1) with a 13-h light (24°C)/11-h dark (21°C) photoperiod under 70% relative humidity, a growth condition labeled 24°C], these plants displayed drastically reduced grain set (Figure 1A) in the absence of the bacterial pathogen Xoo (Xoo is a quarantine pathogen in the USA). The average rates of grain set for the Myc-Xa21L and Myc-Xa21H plants were 30.52% and 2.71%, respectively, compared to 79.53% for the empty vector control line A36 (Figure 1B). These rates in Xa21 plants were inversely correlated to the transcript levels of the Xa21 gene in the spikelets at anthesis from plants grown at 24°C (Figure 1C). Apart from the fertility defect, Xa21 plants appeared normal (Figure 1D).
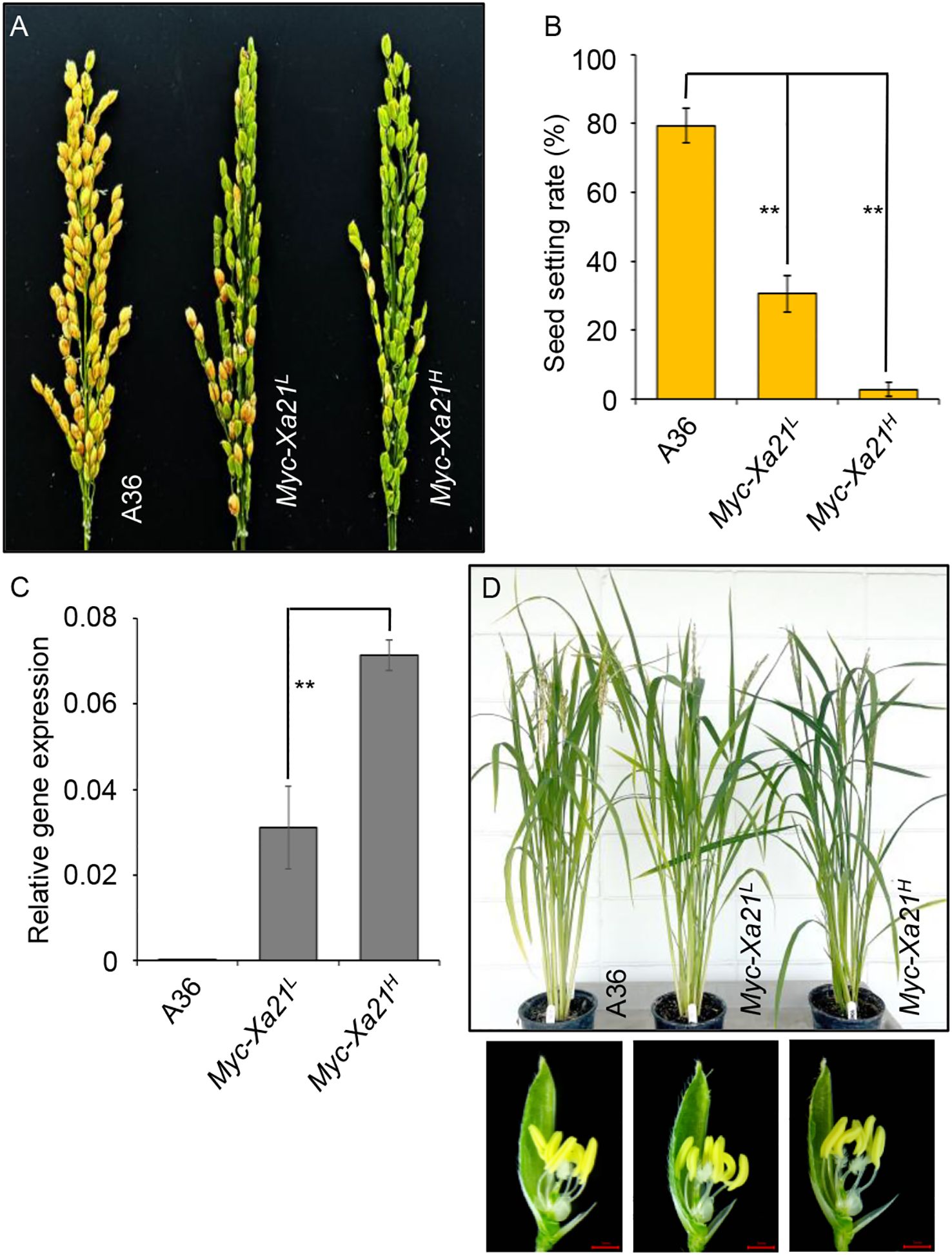
Figure 1. Xa21 decreases grain set in rice at 24°C. (A) Panicle phenotypes of the rice lines A36 (empty vector control), Myc-Xa21L, and Myc-Xa21H. (B) Grain-setting rates of plants in (A) Values are shown as the mean ± SD from five independent plants with two panicles each. Statistical analyses were performed using Student’s t-test. Asterisks denote statistically significant differences (**p < 0.01). (C) Phenotypes of mature plants (top) and floral structures (bottom) of the indicated lines in (A) Scale bars, 1 mm. The experiments were repeated twice with similar results. (D) Relative transcript levels of the Xa21 gene in the spikelets of the indicated lines grown at 24°C at anthesis. Results were normalized relative to levels of the mRNAs for UBIQUITIN 5 and ACTIN. Values are means ± SD of three biological replicates, each with three technical replicates. Statistical analyses were performed using Student’s t-test. Asterisks denote statistically significant differences (**p < 0.01).
When Myc-Xa21 plants were grown in the greenhouse during the summer, no significant differences in grain set were observed (Vergish et al., 2025). Similarly, a significant reduction in grain set was also observed from the previously characterized transgenic line 20-1 (average rate of grain set: 62.70%), which expresses Myc-Xa21 under its native promoter (Park et al., 2010b), relative to the wild-type control O. sativa ssp. japonica variety Kitaake (Kitaake, average rate of grain set: 86.01%) when grown at 24°C (Supplementary Figure S1). Compared to TP309, Kitaake is shorter in stature and has a shorter life cycle of approximately 9 weeks (Kunihiro et al., 1989; Kim et al., 2013). Together, these findings indicate that XA21 expression can induce a strong fertility defect in a dose- and temperature-dependent manner.
XA21 plants display compromised pollen viability and anther dehiscence at 24°C
Our recent study showed that osrbl3b Myc-Xa21 mutants accumulate increased levels of the Myc-XA21 protein and exhibit pollen and dehiscence defects (Vergish et al., 2025). We examined whether XA21 expression affects pollen viability and anther dehiscence at 24°C. The Myc-Xa21 plants produced lower levels of viable pollen grains than the control (A36), as evidenced by starch staining with iodine–potassium iodide (I2−KI) solution (Figures 2A, B).
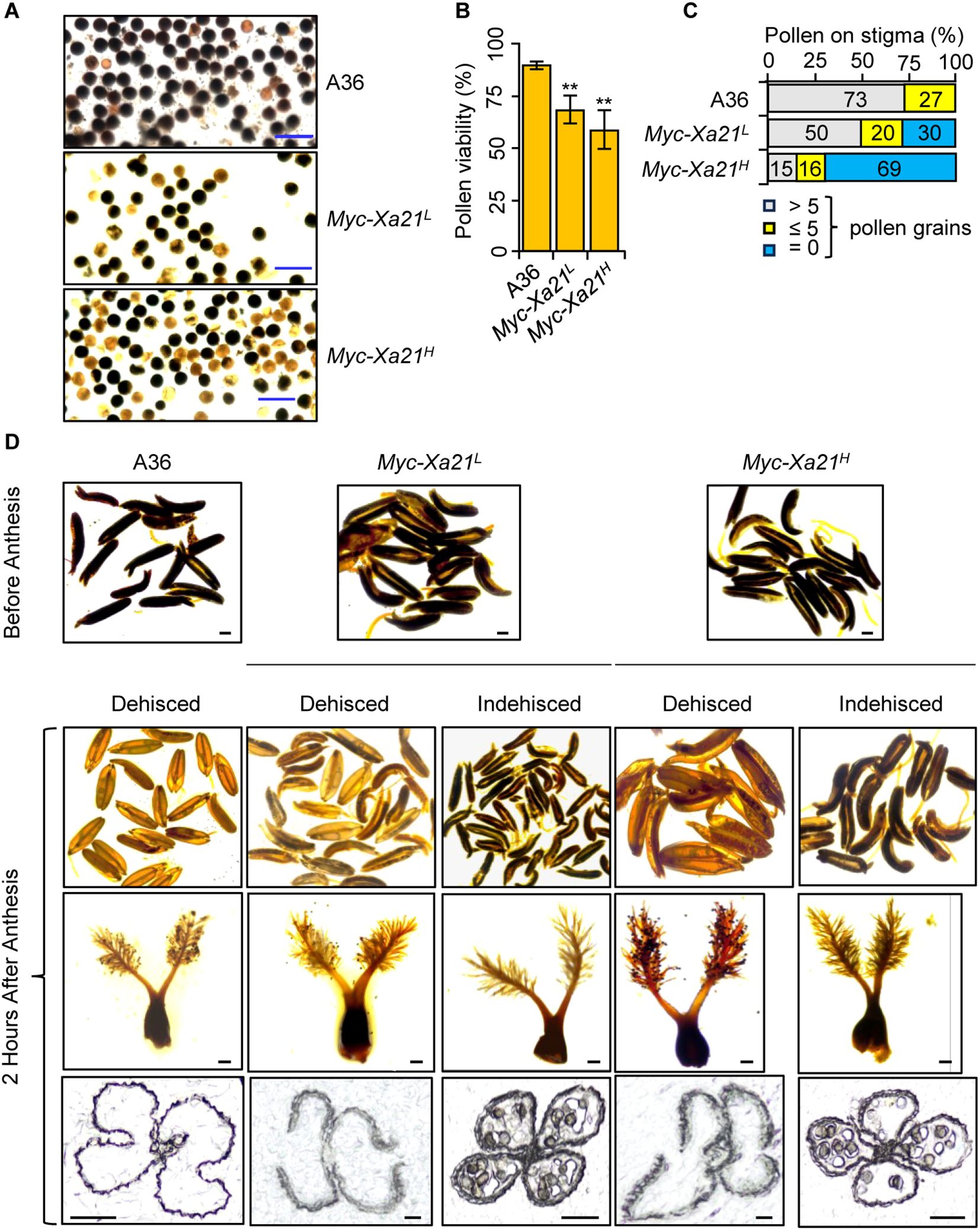
Figure 2. Xa21 impairs pollen viability and anther dehiscence at 24°C. (A, B) Myc-Xa21 plants exhibit reduced starch accumulation in their pollen grains. I2−KI-stained pollen grains from the indicated lines (A). Scale bars, 100 μM. Pollen viability was determined based on the number of stained pollen grains (dark color in A) relative to the total pollen counted (B). Values are means ± SD, n = 5 (five independent samplings). Statistical analysis was performed using Student’s t-test. Asterisks indicate statistically significant differences (**p < 0.01). This experiment was repeated two times with similar results. (C) Pollen grains on the stigmas of the indicated lines after anthesis. The graph shows the distribution of dehiscent and indehiscent spikelets among 100 random flower samples chosen from five individual plants of each indicated line. (D) Presence or absence of pollen grains on the stigmas of the indicated lines after anthesis. (Top 2 rows) I2−KI staining of pollen grains in anthers of the indicated lines before and after anthesis. Scale bars, 500 μM. (Third row) Pollen grains on the stigmas of the indicated lines after anthesis. Scale bars, 200 μM. (Bottom row) Cross-sections of anthers from the indicated lines after anthesis. Scale bars, 100 μM.
To assess anther dehiscence, we first examined the presence of pollen grains (stained with I2−KI) in the anthers of Myc-Xa21 and A36 plants before anthesis and 2 HAA. To confirm anther dehiscence, we further examined the presence of pollen grains on the stigmas and the breakage of anther walls 2 HAA. Among more than 100 randomly chosen spikelets, most anthers opened in A36 spikelets, and indehiscent ones were rare (Figures 2C, D). In contrast, 30%–69% of spikelets from the Myc-Xa21L and Myc-Xa21H lines were indehiscent 2 HAA. These data demonstrate that Myc-Xa21 plants have a defect in anther dehiscence, which can partially explain the reduced grain set.
XA21 downregulates the expression of a subset of JA-related genes at 24°C
The sterile osrbl3b-b Myc-Xa21H mutant at higher temperatures shows the downregulation of JA-responsive and JA-signaling genes relative to the fertile line Myc-Xa21H, although no obvious enrichment of transcripts related to JA biosynthesis or transport is evident in the mutant (Vergish et al., 2025). To understand the mechanisms underlying the XA21-mediated fertility defects, we sequenced the transcripts in spikelets at anthesis from the temperature-sensitive Myc-Xa21H and the fertile A36 lines grown at 24°C. We identified a total of 4,792 DEGs between these two lines using an FDR value of <0.05 and log2FC cut-off criteria of >1 and <−1. The downregulated genes included 11 JA-responsive and JA-signaling genes in Myc-Xa21H, which encode nine JAZ transcriptional repressors (Chini et al., 2007; Thines et al., 2007; Ye et al., 2009) and the transcription factors OsbHLH148 (Seo et al., 2011) and OsWRKY71 (Liu et al., 2007) (Figure 3A). Among the 15 JAZ genes identified in the rice genome (Ye et al., 2009), only OsJAZ15 was upregulated in Myc-Xa21H spikelets.
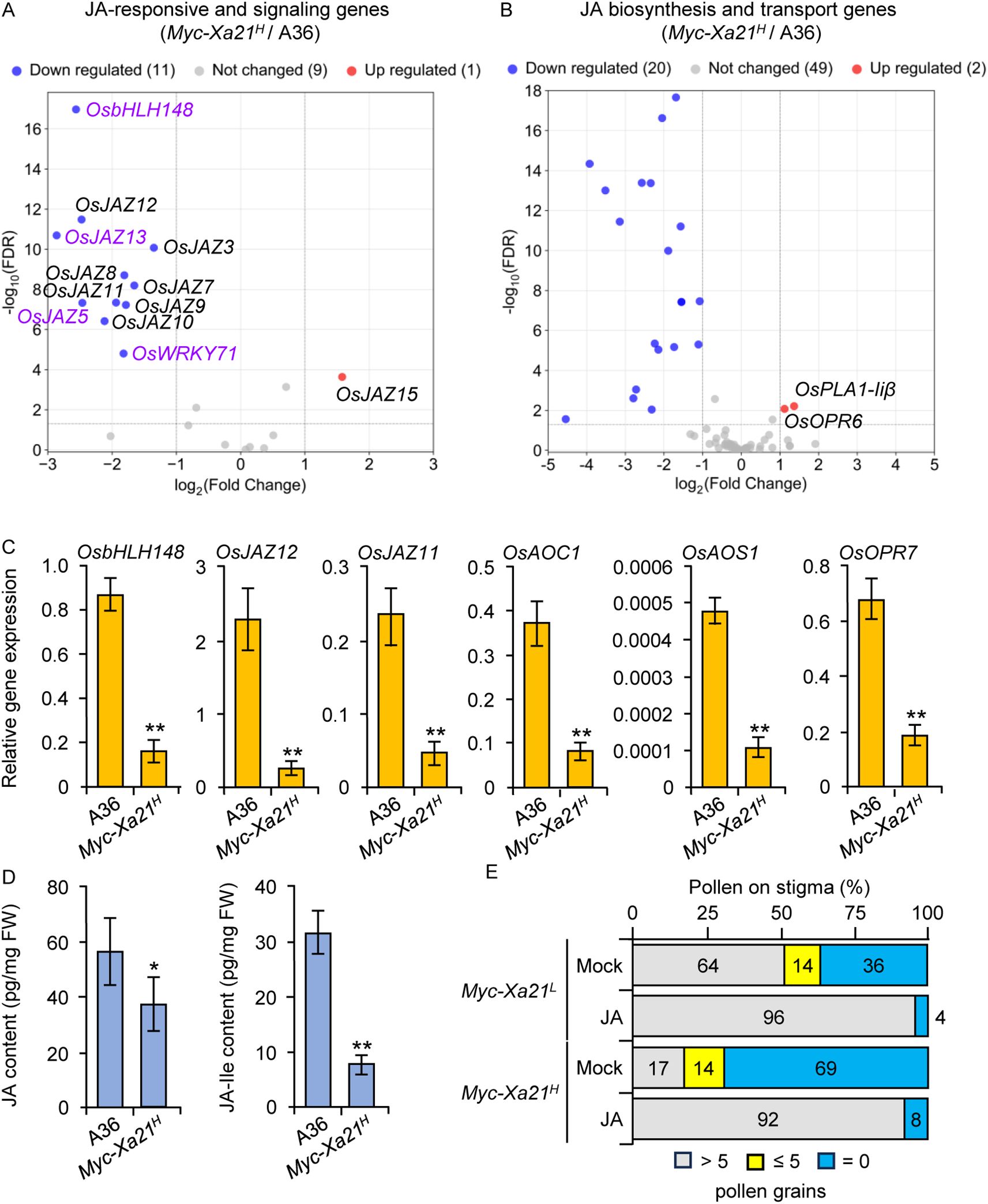
Figure 3. Downregulation of jasmonic acid (JA)-related genes in the indehiscent Myc-Xa21H spikelets relative to the dehiscent A36 spikelets at anthesis at 24°C. (A) RNA-seq volcano plot showing differentially expressed JA-responsive and JA-signaling genes. The genes highlighted in purple are also downregulated in the spikelets of osrblb-b Myc-Xa21H mutant during anthesis (Vergish et al., 2025). (B) RNA-seq volcano plot showing differentially expressed JA biosynthesis and transport genes. Downregulated genes in (B) are OsPLA1α1, OsPLA1α3, OsPLA1β1, OsPLA1β2, OsLOX5, OsLOX6, OsLOX9, OsLOX11, OsLOX12, OsLOXL-2, OsAOS1, OsAOS2, OsAOS3, OsAOC1, OsOPR1, OsOPR7, OsJMT1, OsABCG1, OsABCG22, and OsABCG23. (C) Relative transcript levels, quantified by RT-qPCR, of indicated JA-responsive and JA biosynthesis genes in (A, B) Results were normalized relative to transcript levels of UBIQUITIN 5 and ACTIN. Values are means ± SD of three biological replicates, each with three technical replicates. Statistical analyses were performed using Student’s t-test. Asterisks denote significant differences (**p < 0.01). (D) Relative abundances of JA and JA-Ile, quantified by MS/MS, in the spikelets of the fertile A36 and the semi-sterile Myc-Xa21H. Values are means ± SD of four biological replicates. Statistical analyses were performed using Student’s t-test. Asterisks denote significant differences (**p < 0.01; *p < 0.05). A36 is a transgenic empty vector control. (E) Restoration of anther dehiscence of XA21 plants by exogenous JA treatment at 24°C. Mature panicles of indicated plants were sprayed with either 200 μM MeJA or mock solution. The graph shows the distribution of dehiscent and indehiscent spikelets among 100 random flower samples chosen from five plants of each indicated line at 2 hours after anthesis (HAA).
Interestingly, among the 70 genes related to JA biosynthesis and transport in the rice genome (Marla and Singh, 2012; Matsuda et al., 2012; Singh et al., 2012; Fukumoto et al., 2013; Riemann et al., 2013; Huang et al., 2014; Qi et al., 2016; Liu et al., 2017; Deepika and Singh, 2021), 20 were significantly downregulated (only two were upregulated) in the spikelets of Myc-Xa21H compared to those of the control (A36) (Figure 3B). The downregulated genes included the key JA biosynthetic genes OsAOC1, OsAOS1, and OsOPR7 (Riemann et al., 2003, 2013; Hibara et al., 2016; Pak et al., 2021; Fan et al., 2023; Wang et al., 2024). The downregulation of six of the above genes was verified using qRT-PCR analysis (Figure 3C). Therefore, the XA21-mediated downregulation of JA-responsive genes may occur through the reduction of JA levels.
To test this, the abundance of JA and JA-Ile (the most bioactive form of jasmonates) was quantified in the spikelets of Myc-Xa21H and A36 plants grown at 24°C at anthesis using LC–MS/MS analysis. JA and JA-Ile were identified in all the samples using MS/MS spectral matching between authentic standards and metabolites from the samples (Supplementary Figure S2). This is level 1 structural identification, the most confident level based on the well-accepted Metabolomics Standards Initiative (Sumner et al., 2007). Based on the MS/MS fragmentation pattern of JA and JA-Ile, specific multiple reaction monitoring (MRM) transitions were selected for targeted metabolite quantification and validation. Relative peak areas were normalized using lidocaine as the internal standard, and then spikelet fresh weight was taken into account for quantitative analysis. As shown in Figure 3D, JA levels were significantly lower in Myc-Xa21H than in A36 spikelets, and JA-Ile was also significantly decreased in Myc-Xa21H relative to A36 spikelets.
To further test the above hypothesis, we sprayed the mature panicles of Myc-Xa21L and Myc-Xa21H plants with either 200 μM MeJA or a mock control. The dehiscence of male-sterile JA-related mutants can be restored by treatment with exogenous MeJA (Ishiguro et al., 2001; Song et al., 2018). Indeed, JA treatment largely restored the anther dehiscence of Myc-Xa21 plants at 24°C (Figure 3E). Together, these findings support that the compromised anther dehiscence in Myc-Xa21 plants may be due to a deficiency in JA levels within rice spikelets.
Xa21 is preferentially expressed in the anther filaments, veins, and rachillae of rice spikelets
To determine the expression sites of Xa21 in spikelets, transgenic rice lines expressing the uidA (GUS reporter) gene were generated under the control of the Xa21 promoter. This 2.0-kb promoter sequence is sufficient for directing Xa21 functions (Song et al., 1995; Vergish et al., 2025). GUS assays detected strong activities in the anther filaments, the veins of the palea/lemma, and the rachillae of the spikelets (Figure 4). Notably, little GUS activity was observed in anthers.
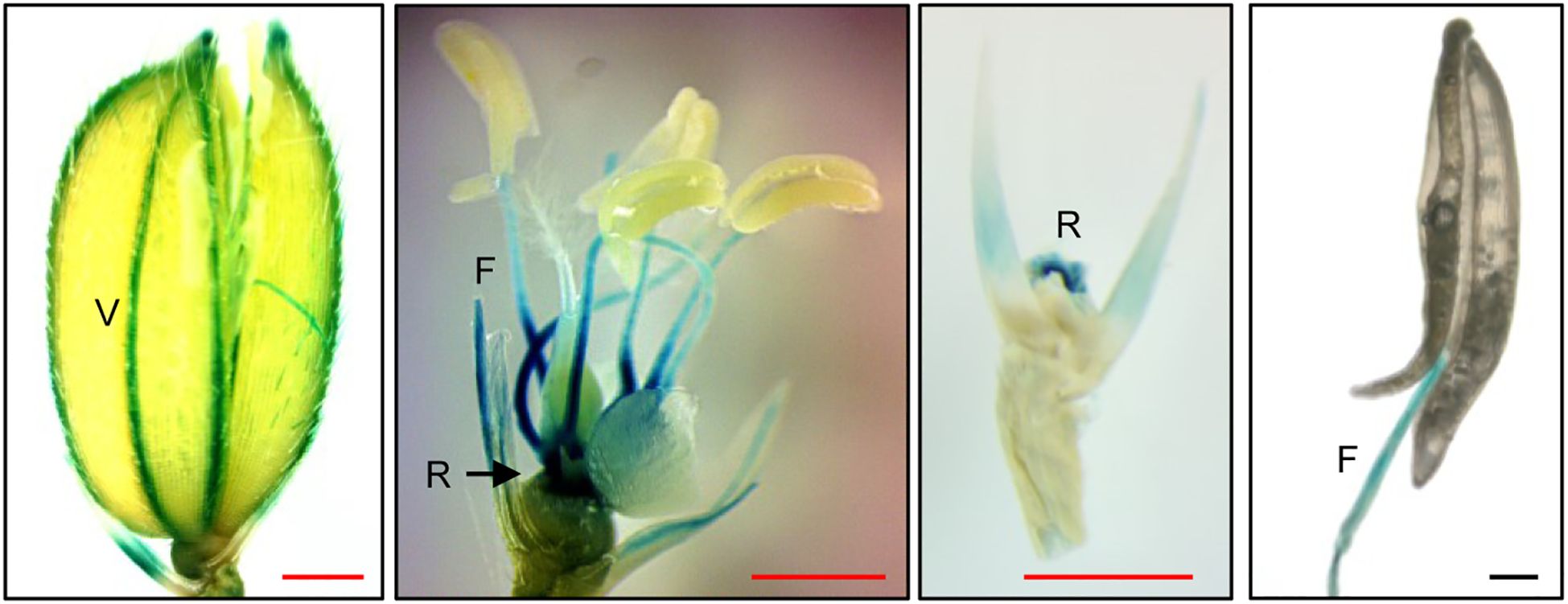
Figure 4. Histochemical analysis of GUS activity from Xa21pro:GUS reporter lines in spikelets. GUS activity was detectable in veins (V) of lemma and palea, in rachillae (R), and in anther filament (F) in the Xa21pro:GUS lines. Scale bars, 1 mm (red), 100 μm (black). Similar results were obtained from two independent (Xa21pro:GUS) lines.
ROS levels increase in the anther of Myc-Xa21 lines at anthesis at 24°C
The treatment of rice leaves expressing XA21 with RaxX-sY induces the robust production of ROS (Pruitt et al., 2015, 2017; Chen et al., 2021). We examined ROS accumulation in the anthers of XA21-expressing lines and their control (A36) using DAB staining, an assay that mainly detects the accumulation of hydrogen peroxide in tissues (Daudi and O'Brien, 2012). As shown in Figure 5, increasing levels of oxidized DAB were visible in the Myc-Xa21L and Myc-Xa21H anthers at anthesis in the absence of Xoo. This finding indicates that ROS production is shared by the RaxX-sY-triggered XA21 defense response in the leaf and the XA21-dependent alterations in the anther.
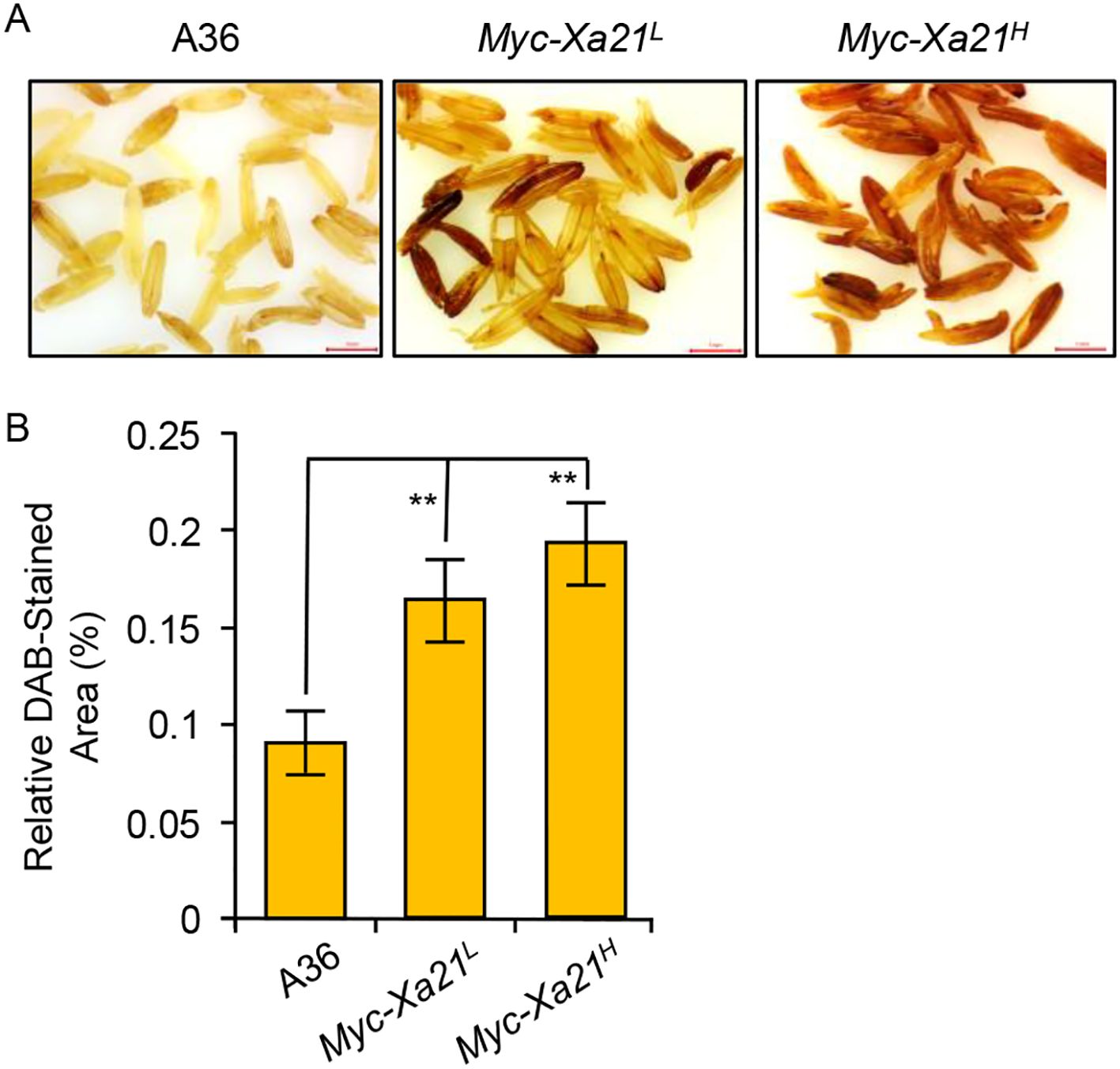
Figure 5. Reactive oxygen species (ROS) production in the anthers of Myc-Xa21 plants at 24°C. (A) 3,3′-Diaminobenzidine (DAB) staining for H2O2 in anthers harvested at anthesis from A36 (empty vector control), Myc-Xa21L, and Myc-Xa21H plants grown. Samples were incubated with DAB solution overnight at room temperature. Scale bars, 1 mm. The experiments were repeated twice with similar results. (B) Quantification of ROS in the DAB-stained anthers of the indicated lines. Values are shown as the mean ± SD from seven anthers. Statistical analyses were performed using Student’s t-test. Asterisks denote statistically significant differences (**p < 0.01).
Discussion
R genes with high fitness costs in host plants rarely propagate/survive through evolutionary selection. However, increasing evidence indicates that plants have developed sophisticated mechanisms by which significant R genes are retained to fight against pathogen invasion. For instance, the Cf-2 gene discovered in the wild tomato species Lycopersicon pimpinellifolium specifies resistance against the fungus Cladosporium fulvum (Dixon et al., 1996). In plants lacking a functional Rcr3 gene (e.g., in the cultivated tomato Lycopersicon esculentum), which encodes a papain-like cysteine endoprotease, Cf-2 activates autonecrosis (Kruger et al., 2002). The rice gene PigmR confers broad-spectrum resistance to M. oryzae, but its expression also causes a decrease in grain weight by approximately 2% (Deng et al., 2017). This detrimental effect is compensated for by PigmS, a gene located in the same locus (called Pigm), which is capable of increasing grain set by approximately 5%. Consequently, the Pigm locus with both PigmR and PigmS has been used for rice breeding for more than five decades without an overall yield penalty. We recently reported a rhomboid-controlled, post-translational regulatory mechanism through which rice prevents the over-accumulation of the R protein XA21, and potentially other transmembrane domain proteins, in spikelets to avoid fertility defects (Vergish et al., 2025).
We demonstrate here that XA21 decreases pollen viability and anther dehiscence. This study reveals the potent effect of temperature [24°C (daytime)] on the XA21 function. These findings are in line with our recent observation that an optimal daytime temperature (28 °C) facilitates the male sterility observed in the osrbl3b Myc-Xa21L mutant lines with a mutant rhomboid protease and an increased abundance of Myc-XA21 (Vergish et al., 2025) and with the previous discovery that a 4 °C decrease in temperature (from 31 °C to 27°C) fully activates the developmentally regulated Xa21 resistance to the incompatible Xoo strain PXO99A at the seedling stage (Chen et al., 2019). In contrast, high temperatures (31°C or above) can suppress Xa21 functions (Chen et al., 2019; Vergish et al., 2025). As an intermediate level of resistance was observed at 29°C, we speculate that 24°C may not represent a “low temperature” threshold for XA21-induced fertility defects. Since its cloning in 1995 (Song et al., 1995), Xa21 has been genetically engineered into various rice cultivars for the assessment of resistance and deleterious effects on plant growth, presumably under normal growth conditions. No developmental defects were reported in most of these studies (Li et al., 2001; Gao et al., 2013; Gayen et al., 2016; Zhang et al., 2024). Therefore, high-temperature tropical environments seem to mitigate the effects of XA21 on rice fertility. Since environmental conditions can greatly influence disease resistance in various plants (Cheng et al., 2019; Cohen and Leach, 2020), it is tempting to speculate that more examples of the costs associated with R genes may exist when plants are grown under distinct environments.
XA21-mediated fertility defects are dose-dependent and strongly modified by ambient temperature. Dependent on the low temperature, the Myc-Xa21H line (with a higher level of the XA21 protein) exhibits more severe fertility-related phenotypes than the Myc-Xa21L line. Independent of the temperature, the osrbl3b-b Myc-Xa21H mutant, which has even more abundant XA21 than Myc-Xa21H, is sterile (Vergish et al., 2025). These findings suggest a potential threshold of XA21 abundance that induces fertility defects. In an empirical field study, marked yield loss was observed in an Xa21 transgenic line with high resistance (Hao et al., 2009). Such yield reduction could be attributed to a potentially very high level of Xa21 expression (Zhang et al., 2024), although it is unclear whether temperature (e.g., growth seasons) may be another factor in this study. Notably, the highly resistant Pi-d2 and Pi-d3 lines also displayed yield penalties (Hao et al., 2009). Pi-d2 and Pi-d3 confer resistance to the fungus M. oryzae and encode an RLK with an extracellular domain of a bulb-type mannose-specific binding lectin and an NLR protein, respectively (Chen et al., 2006; Shang et al., 2009). Like Xa21, these genes, which encode distinct protein structures, may invoke the costs of fitness in a dose-dependent manner.
To our knowledge, this work is the first to reveal the physiological and molecular mechanisms underlying R gene-instigated detrimental effects on reproduction. While the observed impairments to anther dehiscence and pollen viability can partially explain the semi-sterility of Myc-Xa21 plants, the compromised JA signaling (Figure 3) could provide, at least in part, a molecular basis for Xa21-mediated male sterility (see Figure 6 for a model). Among the 11 downregulated JA-responsive and JA-signaling genes in the semi-sterile line Myc-Xa21H at 24°C, four genes were also differentially regulated in the sterile line osrbl3b-b Myc-Xa21H under greenhouse conditions during the summer (Figure 3; Vergish et al., 2025), suggesting that altered JA signaling may have a role in the fertility defects of both Myc-Xa21H and osrbl3b-b Myc-Xa21H. Furthermore, the sharp enrichment of differentially downregulated JA biosynthesis genes in Myc-Xa21H in this study suggests that XA21 signaling mediates the downregulation of JA responsiveness, which may lead to fertility defects (Figure 6). This hypothesis is supported by our observations that JA and JA-Ile contents were decreased in Myc-Xa21H spikelets during anthesis relative to the control (A36) and that exogenous treatment with MeJA was capable of restoring Myc-Xa21 anther dehiscence at 24°C. Further support of the hypothesis comes from the promoter GUS reporter assay, in which the Xa21 promoter was found to be active in the anther filaments, the veins of the palea/lemma, and the rachillae of the spikelets (Figure 4). Consistently, the Arabidopsis Defective in Anther Dehiscence (DAD1) gene, key to JA biosynthesis and anther dehiscence, is exclusively expressed in anther filaments before anthesis (Ishiguro et al., 2001). It has been hypothesized that JA accumulation in anther filaments promotes water movement from anther locules to the filaments. This dehydration of anther locules facilitates the maturation of pollen grains and induces anther dehiscence (Ishiguro et al., 2001; Acosta and Przybyl, 2019). In addition, transgenic rice lines ectopically expressing Xa21 from the constitutive maize ubiquitin promoter show very high levels of XA21 and resistance to Xoo in the leaf but normal grain set (Park et al., 2010b), indicating the need for the native promoter to condition Xa21 costs. A related finding is our discovery that the exogenous application of JA compromises XA21-mediated resistance to Xoo at the 2-week-old seedling stage (Chen et al., 2025), suggesting that XA21 function and JA signaling interact antagonistically in rice.
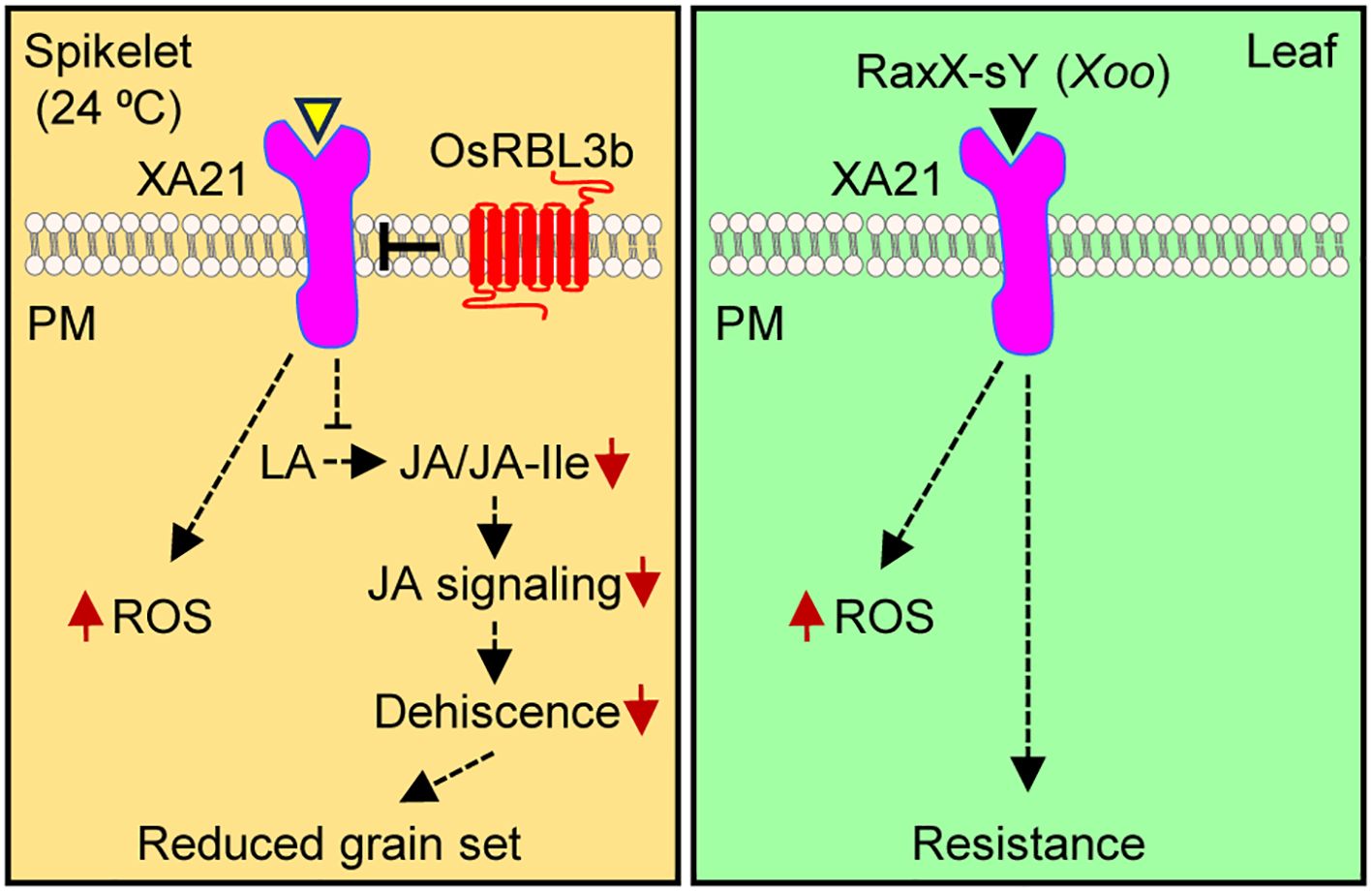
Figure 6. Simplified models of XA21-mediated signaling in spikelets and leaves. In spikelets during anthesis at 24°C, XA21 signaling is activated by an unidentified rice-derived signal (yellow triangle), leading to the production of reactive oxygen species (ROS) and to the suppression of jasmonic acid (JA) biosynthesis from its precursor linolenic acid (LA), which in turn represses JA signaling. Deceased JA signaling compromises anther dehiscence, ultimately resulting in reduced grain set. In leaves, XA21-mediated signaling is activated by the Xoo peptide RaxX-sY, which results in ROS production and resistance to Xoo. As described recently (Vergish et al., 2025), the rhomboid protease OsRBL3b prevents the over-accumulation of XA21 in rice spikelets.
It may be worth noting that the signaling mechanisms in the semi-sterile line Myc-Xa21H at 24°C and the sterile mutant osrbl3b-b Myc-Xa21H at high temperatures (greenhouse conditions) may differ from each other. Unlike Myc-Xa21H, no significant enrichment of the altered expression of JA biosynthesis genes was observed in the sterile osrbl3b-b Myc-Xa21H spikelets compared to the fertile Myc-Xa21H spikelets at anthesis at high temperatures (Vergish et al., 2025). In contrast to the osrbl3b-b Myc-Xa21H mutant, the enrichment of the upregulated expression of NLR genes is not significant in Myc-Xa21H (Supplementary Figure S3; Vergish et al., 2025). These discrepancies may be due to the potential cleavage of additional signaling regulators by OsRBL3b (Vergish et al., 2025) and the suppression effects of elevated temperatures on XA21 signaling (Chen et al., 2019), which collectively lead to the activation of distinct signaling pathways in the osrbl3b-b Myc-Xa21H mutants.
The increased accumulation of ROS in spikelets may also contribute to Xa21-mediated male sterility. During anther development, proper levels of ROS trigger tapetal degradation, which provides nutrients for pollen (Zhang et al., 2021; Xie et al., 2022). However, the abnormal accumulation of ROS can damage tapetal function and pollen development, leading to male sterility. XA21-mediated fertility defects may be caused by the activation of the receptor by an endogenous signal in rice spikelets (Figure 6). Previous studies have shown that neither low-temperature treatment nor abundant XA21 produced from ectopic expression by the strong maize ubiquitin promoter can activate the receptor, as XA21 plants are susceptible to the compatible Xoo strains under the above conditions (Park et al., 2010b; Chen et al., 2019; Zhang et al., 2024). Given that ROS production in XA21 leaves is triggered by the Xoo peptide RaxX-sY (Pruitt et al., 2015; Chen et al., 2021), our observation of XA21-dependent ROS accumulation in anthers at 24°C suggests that the receptor may be activated by an endogenous signal, although the impact of abundant ROS on grain development remains to the determined. Low temperature (24°C) treatment here likely primes the activation of XA21 signaling, as suggested previously (Chen et al., 2019). XA21 is among a set of immune receptors that are present in both plants and animals (Ronald and Beutler, 2010; Ercoli et al., 2022). Autoimmunity and autoimmune diseases (e.g., type I diabetes) have emerged as a worldwide threat to public health (e.g., estimated yearly increase in the prevalence of human autoimmune diseases is 12.5%) (Miller, 2023). Current views underscore the importance of environmental cues (including temperature) and genetic risk factors for the rise of autoimmune disorders; however, mechanistic insights remain elusive (Miller, 2023). Further studies may lead to a deeper understanding of autoimmunity and to the development of strategies to control such disorders.
In the 1990s, we identified a total of 17 transposable-like elements (TEs) in the Xa21 locus, which harbors at least six Xa21-related family members originating from O. longistaminata, with member D being 98% identical to Xa21 at the DNA level (Song et al., 1997, 1998). Interestingly, D provides partial resistance to the same spectrum of Xoo strains as Xa21 does, likely because it is compromised by insertion of a retrotransposon (called Retrofit) into its coding region (Wang et al., 1998). At the time, the selective advantage of the accumulation and movement of these TEs was unclear (Song et al., 1998). Our findings in this study suggest that the TEs may represent a genomic tool by which rice controls the deleterious function of Xa21 and possibly member D. This idea is in line with the prevalent hypothesis, proposed by Barbara McClintock and others, that TEs may facilitate genome responses to challenges (McClintock, 1984; Wessler, 1996; Slotkin and Martienssen, 2007).
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJEB102018.
Author contributions
BF: Methodology, Investigation, Visualization, Writing – original draft, Formal Analysis, Validation, Data curation. SV: Data curation, Investigation, Visualization, Validation, Writing – review & editing, Methodology. YS: Data curation, Methodology, Writing – review & editing, Investigation, Formal Analysis, Resources. J-LL: Investigation, Resources, Formal Analysis, Writing – review & editing, Methodology, Funding acquisition. S: Writing – review & editing, Investigation. G-LD: Investigation, Data curation, Writing – review & editing. SC: Resources, Supervision, Validation, Writing – review & editing, Conceptualization, Methodology, Formal Analysis. W-YS: Writing – review & editing, Investigation, Visualization, Conceptualization, Funding acquisition, Validation, Methodology, Writing – original draft, Resources, Supervision, Formal Analysis, Project administration, Data curation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The work was supported by the National Science Foundation (no. 2114833 to W.Y.S.) and the Intramural Research Program of the National Institute of Environmental Health Sciences (ZIC ES103371 to J.L.L.).
Acknowledgments
We thank Anita K. Snyder for invaluable discussion and critical reading of the manuscript; Dr. Frank White for helpful discussion; Drs. Qi Li, Liya Pi, and Apekshya Parajuli for technical assistance; and Dr. Pamela C. Ronald for providing the 4021-3, 20-1, and Kitaake lines.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1673821/full#supplementary-material
Supplementary Figure 1 | Xa21 decreases grain-set in Oryza sativa ssp. japonica variety Kitaake at 24 °C. (A) Panicle phenotypes of the rice lines Kitaake (wild-type control) and 20-1 (also called 20-1-18–1 or 18–1 in the lab) that expresses Myc-Xa21 under its native promoter. (B) Phenotypes of mature plants of the indicated lines in (A). (C) Grain-setting rates of plants in (A). Values are shown as the mean ± SD from 5 independent plants with top two panicles each. Statistical analyses were performed using Student’s t-test. Asterisks denote statistically significant differences (**P < 0.01). (D) Dehusked grains harvested from the indicated lines in (A) showing the difference in grain set between these two lines.
Supplementary Figure 2 | Mass spectrometry identification of jasmonic acid (JA) and JA-isoleucine (JA-Ile). In each panel, the top MS/MS spectrum in blue represents the authentic standard spectrum, and the bottom spectrum in purple is from the samples.
Supplementary Figure 3 | RNA-seq volcano plot showing differentially expressed rice NLR genes. A complete list of transcript IDs detected is shown previously (Vergish et al., 2025). Cut-off criteria for data: log2[Myc-Xa21H/A36] ≥ 1 and ≤ -1 and FDR-q value < 0.05.
References
Acosta, I. F. and Przybyl, M. (2019). Jasmonate signaling during Arabidopsis stamen maturation. Plant Cell Physiol. 60, 2648–2659. doi: 10.1093/pcp/pcz201
Anbu, P. and Arul, L. (2013). Beta glucuronidase activity in early stages of rice seedlings and callus: A comparison with Escherichia coli beta glucuronidase expressed in the transgenic rice. Int. J. Biotechnol. Mol. Biol. Res. 4, 061–068. doi: 10.5897/IJBMBR2013.0152
Brown, J. K. M. and Rant, J. C. (2013). Fitness costs and trade-offs of disease resistance and their consequences for breeding arable crops. Plant Pathol. 62, 83–95. doi: 10.1111/ppa.12163
Büschges, R., Hollricher, H., Panstruga, R., Simons, G., Wolter, M., Frijters, A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. doi: 10.1016/s0092-8674(00)81912-1
Calvo-Baltanas, V., Wang, J., and Chae, E. (2020). Hybrid incompatibility of the plant immune system: An opposite force to heterosis equilibrating hybrid performances. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.576796
Chen, F., Gao, M. J., Miao, Y. S., Yuan, Y. X., Wang, M. Y., Li, Q., et al. (2010). Plasma membrane localization and potential endocytosis of constitutively expressed XA21 proteins in transgenic rice. Mol. Plant 3, 917–926. doi: 10.1093/mp/ssq038
Chen, Q., Chen, X., and Song, W. Y. (2025). Jasmonic acid and abscisic acid compromise XA21-mediated immunity in rice to Xanthomonas oryzae pv. oryzae. Eur. J. Plant Pathol. 173, 429–438. doi: 10.1007/s10658-025-03064-7
Chen, Q., Huang, X., Chen, X., Shamsunnher, and Song, W. Y. (2019). Reversible activation of XA21-mediated resistance by temperature. Eur. J. Plant Pathol. 153, 1177–1184. doi: 10.1007/s10658-018-01634-6
Chen, T. C., Chern, M., Steinwand, M., Ruan, D., Wang, Y., Isharani, A., et al. (2021). Paladin, a tyrosine phosphatase-like protein, is required for XA21-mediated immunity in rice. Plant Commun. 2, 100215. doi: 10.1016/j.xplc.2021.100215
Chen, X., Shang, J., Chen, D., Lei, C., Zou, Y., Zhai, W., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. doi: 10.1111/j.1365-313X.2006.02739.x
Cheng, Y. T., Zhang, L., and He, S. Y. (2019). Plant-microbe interactions facing environmental challenge. Cell Host Microbe 26, 183–192. doi: 10.1016/j.chom.2019.07.009
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Cohen, S. P. and Leach, J. E. (2020). High temperature-induced plant disease susceptibility: More than the sum of its parts. Curr. Opin. Plant Biol. 56, 235–241. doi: 10.1016/j.pbi.2020.02.008
Daudi, A. and O'Brien, J. A. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc. 2, e263. doi: 10.21769/BioProtoc.263
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Deepika and Singh, A. (2021). Expression dynamics indicate the role of jasmonic acid biosynthesis pathway in regulating macronutrient (N, P and K(+)) deficiency tolerance in rice (Oryza sativa L.). Plant Cell Rep. 40, 1495–1512. doi: 10.1007/s00299-021-02721-5
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Dhakarey, R., Kodackattumannil Peethambaran, P., and Riemann, M. (2016). Functional analysis of jasmonates in rice through mutant approaches. Plants 5, 15. doi: 10.3390/plants5010015
Dixon, M. S., Jones, D. A., Keddie, J. S., Thomas, C. M., Harrison, K., and Jones, J. D. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. doi: 10.1016/s0092-8674(00)81290-8
Ercoli, M. F., Luu, D. D., Rim, E. Y., Shigenaga, A., Teixeira De Araujo, A., Chern, M., et al. (2022). Plant immunity: Rice XA21-mediated resistance to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 119, e2121568119. doi: 10.1073/pnas.2121568119
Fan, X., Tang, H., Chen, X., Zeng, F., Chen, G., Chen, Z. H., et al. (2023). Allene oxide synthase 1 contributes to limiting grain arsenic accumulation and seedling detoxification in rice. Stress Biol. 3, 52. doi: 10.1007/s44154-023-00136-8
Fukumoto, K., Alamgir, K., Yamashita, Y., Mori, I. C., Matsuura, H., and Galis, I. (2013). Response of rice to insect elicitors and the role of OsJAR1 in wound and herbivory-induced JA-Ile accumulation. J. Integr. Plant Biol. 55, 775–784. doi: 10.1111/jipb.12057
Gao, L., Cao, Y., Xia, Z., Jiang, G., Liu, G., Zhang, W., et al. (2013). Do transgenesis and marker-assisted backcross breeding produce substantially equivalent plants? A comparative study of transgenic and backcross rice carrying bacterial blight resistant gene Xa21. BMC Genomics 14, 738. doi: 10.1186/1471-2164-14-738
Gao, M., Hao, Z., Ning, Y., and He, Z. (2024). Revisiting growth-defence trade-offs and breeding strategies in crops. Plant Biotechnol. J. 22, 1198–1205. doi: 10.1111/pbi.14258
Gayen, D., Paul, S., Sarkar, S. N., Datta, S. K., and Datta, K. (2016). Comparative nutritional compositions and proteomics analysis of transgenic Xa21 rice seeds compared to conventional rice. Food Chem. 203, 301–307. doi: 10.1016/j.foodchem.2016.02.058
Hao, Z. N., Wang, J., Wang, L. P., and Tao, R. X. (2009). Influences of the disease resistance conferred by the individual transgenes, Pi-d2, Pi-d3 and Xa21, on the transgenic rice plants in yield and grain quality. Afr. J. Biotechnol. 8, 4845–4848.
He, Z., Webster, S., and He, S. Y. (2022). Growth-defense trade-offs in plants. Curr. Biol. 32, R634–R639. doi: 10.1016/j.cub.2022.04.070
Hibara, K., Isono, M., Mimura, M., Sentoku, N., Kojima, M., Sakakibara, H., et al. (2016). Jasmonate regulates juvenile-to-adult phase transition in rice. Development 143, 3407–3416. doi: 10.1242/dev.138602
Hu, K., Cao, J., Zhang, J., Xia, F., Ke, Y., Zhang, H., et al. (2017). Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009. doi: 10.1038/nplants.2017.9
Huang, J., Cai, M., Long, Q., Liu, L., Lin, Q., Jiang, L., et al. (2014). OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 23, 643–655. doi: 10.1007/s11248-014-9803-2
Huang, X., Yang, S., Gong, J., Zhao, Y., Feng, Q., Gong, H., et al. (2015). Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 6, 6258. doi: 10.1038/ncomms7258
Huot, B., Yao, J., Montgomery, B. L., and He, S. Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. doi: 10.1105/tpc.010192
Jain, N., Vergish, S., and Khurana, J. P. (2018). Validation of house-keeping genes for normalization of gene expression data during diurnal/circadian studies in rice by RT-qPCR. Sci. Rep. 8, 3203. doi: 10.1038/s41598-018-21374-1
Jia, Y., Wang, Z., Fjellstrom, R. G., Moldenhauer, K. A., Azam, M. A., Correll, J., et al. (2004). Rice Pi-ta gene confers resistance to the major pathotypes of the rice blast fungus in the United States. Phytopathology 94, 296–301. doi: 10.1094/PHYTO.2004.94.3.296
Jones, J. D. G., Staskawicz, B. J., and Dangl, J. L. (2024). The plant immune system: From discovery to deployment. Cell 187, 2095–2116. doi: 10.1016/j.cell.2024.03.045
Jones, J. D., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395. doi: 10.1126/science.aaf6395
Jørgensen, J. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152. doi: 10.1007/BF00023919
Karasov, T. L., Chae, E., Herman, J. J., and Bergelson, J. (2017). Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29, 666–680. doi: 10.1105/tpc.16.00931
Karasov, T. L., Kniskern, J. M., Gao, L., DeYoung, B. J., Ding, J., Dubiella, U., et al. (2014). The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512, 436–440. doi: 10.1038/nature13439
Kawahara, Y., De La Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6, 4. doi: 10.1186/1939-8433-6-4
Khush, G. S., Esperanza, B., and Ogawa, T. (1990). A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. Newsl. 7, 2.
Kim, S. L., Choi, M., Jung, K. H., and An, G. (2013). Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J. Exp. Bot. 64, 4169–4182. doi: 10.1093/jxb/ert226
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kourelis, J. and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 Years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Kruger, J., Thomas, C. M., Golstein, C., Dixon, M. S., Smoker, M., Tang, S., et al. (2002). A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296, 744–747. doi: 10.1126/science.1069288
Kunihiro, Y., Ebe, Y., Wada, S., Shinbashi, N., Honma, A., Sasaki, T., et al. (1989). The new rice variety Kita-ake Vol. 59 (Bulletin of Hokkaido prefectural agricultural experiment stations) 59, 4. Available online at: https://www.cabidigitallibrary.org/doi/full/10.5555/19891605850.
Li, X. B., Yi, C. D., Zhai, W. X., Yang, Z. Y., and Zhu, L. H. (2001). A genetically modified japonica restorer line, C418-Xa21, and its hybrid rice with bacterial blight resistance. Sheng Wu Gong Cheng Xue Bao 17, 380–384.
Li, Q., Zhou, M., Chhajed, S., Yu, F., Chen, S., Zhang, Y., et al. (2023). N-hydroxypipecolic acid triggers systemic acquired resistance through extracellular NAD(P). Nat. Commun. 14, 6848. doi: 10.1038/s41467-023-42629-0
Liu, X., Bai, X., Wang, X., and Chu, C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Liu, L., Zou, Z., Qian, K., Xia, C., He, Y., Zeng, H., et al. (2017). Jasmonic acid deficiency leads to scattered floret opening time in cytoplasmic male sterile rice Zhenshan 97A. J. Exp. Bot. 68, 4613–4625. doi: 10.1093/jxb/erx251
Luu, D. D., Joe, A., Chen, Y., Parys, K., Bahar, O., Pruitt, R., et al. (2019). Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc. Natl. Acad. Sci. U. S. A. 116, 8525–8534. doi: 10.1073/pnas.1818275116
Marla, S. S. and Singh, V. K. (2012). LOX genes in blast fungus (Magnaporthe grisea) resistance in rice. Funct. Integr. Genomics 12, 265–275. doi: 10.1007/s10142-012-0268-1
Matsuda, S., Funabiki, A., Furukawa, K., Komori, N., Koike, M., Tokuji, Y., et al. (2012). Genome-wide analysis and expression profiling of half-size ABC protein subgroup G in rice in response to abiotic stress and phytohormone treatments. Mol. Genet. Genomics 287, 819–835. doi: 10.1007/s00438-012-0719-3
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. doi: 10.1126/science.15739260
McConn, M. and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. doi: 10.1105/tpc.8.3.403
Miller, F. W. (2023). The increasing prevalence of autoimmunity and autoimmune diseases: an urgent call to action for improved understanding, diagnosis, treatment, and prevention. Curr. Opin. Immunol. 80, 102266. doi: 10.1016/j.coi.2022.102266
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nguyen, H. T., Cheaib, M., Fournel, M., Rios, M., Gantet, P., Laplaze, L., et al. (2023). Genetic analysis of the rice jasmonate receptors reveals specialized functions for OsCOI2. PloS One 18, e0291385. doi: 10.1371/journal.pone.0291385
Ning, Y., Liu, W., and Wang, G. L. (2017). Balancing immunity and yield in crop plants. Trends Plant Sci. 22, 1069–1079. doi: 10.1016/j.tplants.2017.09.010
Pak, H., Wang, H., Kim, Y., Song, U., Tu, M., Wu, D., et al. (2021). Creation of male-sterile lines that can be restored to fertility by exogenous methyl jasmonate for the establishment of a two-line system for the hybrid production of rice (Oryza sativa L.). Plant Biotechnol. J. 19, 365–374. doi: 10.1111/pbi.13471
Park, C. J., Bart, R., Chern, M., Canlas, P. E., Bai, W., and Ronald, P. C. (2010a). Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PloS One 5, e9262. doi: 10.1371/journal.pone.0009262
Park, C.-J., Lee, S.-W., Chern, M., Sharma, R., Canlas, P. E., Song, M.-Y., et al. (2010b). Ectopic expression of rice Xa21 overcomes developmentally controlled resistance to Xanthomonas oryzae pv. oryzae. Plant Sci. 179, 466–471. doi: 10.1046/j.1365-313x.2002.01328.x
Park, J. -H., Halitschke, R., Kim, H. B., Baldwin, I. T., Feldmann, K. A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. doi: 10.1046/j.1365-313x.2002.01328.x
Parniske, M., Hammond-Kosack, K. E., Golstein, C., Thomas, C. M., Jones, D. A., Harrison, K., et al. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. doi: 10.1016/s0092-8674(00)80470-5
Peng, M., Lin, X., Xiang, X., Ren, H., Fan, X., and Chen, K. (2021). Characterization and evaluation of transgenic rice pyramided with the Pi genes Pib, Pi25 and Pi54. Rice (N Y) 14, 78. doi: 10.1186/s12284-021-00512-w
Pruitt, R. N., Joe, A., Zhang, W., Feng, W., Stewart, V., Schwessinger, B., et al. (2017). A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytol. 215, 725–736. doi: 10.1111/nph.14609
Pruitt, R. N., Schwessinger, B., Joe, A., Thomas, N., Liu, F., Albert, M., et al. (2015). The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 1, e1500245. doi: 10.1126/sciadv.1500245
Qi, J., Li, J., Han, X., Li, R., Wu, J., Yu, H., et al. (2016). Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 58, 564–576. doi: 10.1111/jipb.12436
Riemann, M., Haga, K., Shimizu, T., Okada, K., Ando, S., Mochizuki, S., et al. (2013). Identification of rice allene oxide cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238. doi: 10.1111/tpj.12115
Riemann, M., Muller, A., Korte, A., Furuya, M., Weiler, E. W., and Nick, P. (2003). Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol. 133, 1820–1830. doi: 10.1104/pp.103.027490
Ronald, P. C. and Beutler, B. (2010). Plant and animal sensors of conserved microbial signatures. Science 330, 1061–1064. doi: 10.1126/science.1189468
Sanders, P. M., Lee, P. Y., Biesgen, C., Boone, J. D., Beals, T. P., Weiler, E. W., et al. (2000). The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1061. doi: 10.1105/tpc.12.7.1041
Schilmiller, A. L., Koo, A. J., and Howe, G. A. (2007). Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol. 143, 812–824. doi: 10.1104/pp.106.092916
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Shamsunnaher, Chen, X., Zhang, X., Wu, X. X., Huang, X., and Song., W. Y. (2020). Rice immune sensor XA21 differentially enhances plant growth and survival under distinct levels of drought. Sci. Rep. 10, 16938. doi: 10.1038/s41598-020-73128-7
Shang, J., Tao, Y., Chen, X., Zou, Y., Lei, C., Wang, J., et al. (2009). Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site–leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182, 1303–1311. doi: 10.1534/genetics.109.102871
Singh, A., Baranwal, V., Shankar, A., Kanwar, P., Ranjan, R., Yadav, S., et al. (2012). Rice phospholipase A superfamily: organization, phylogenetic and expression analysis during abiotic stresses and development. PloS One 7, e30947. doi: 10.1371/journal.pone.0030947
Slotkin, R. K. and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Song, S., Chen, Y., Liu, L., See, Y. H. B., Mao, C., Gan, Y., et al. (2018). OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat. Plants 4, 495–504. doi: 10.1038/s41477-018-0175-0
Song, W. Y., Pi, L. Y., Bureau, T. E., and Ronald, P. C. (1998). Identification and characterization of 14 transposon-like elements in the noncoding regions of members of the Xa21 family of disease resistance genes in rice. Mol. Gen. Genet. 258, 449–456. doi: 10.1007/s004380050755
Song, W. Y., Pi, L. Y., Wang, G. L., Gardner, J., Holsten, T., and Ronald, P. C. (1997). Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9, 1279–1287. doi: 10.1105/tpc.9.8.1279
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Stintzi, A. and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630. doi: 10.1073/pnas.190264497
Su, Q., Rohila, J. S., Ranganathan, S., and Karthikeyan, R. (2023). Rice yield and quality in response to daytime and nighttime temperature increase - A meta-analysis perspective. Sci. Total Environ. 898, 165256. doi: 10.1016/j.scitotenv.2023.165256
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221. doi: 10.1007/s11306-007-0082-2
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. doi: 10.1046/j.1365-313x.2003.01976.x
Tan, J., Zhao, H., Li, J., Gong, Y., and Li, X. (2023). The devastating rice blast airborne pathogen Magnaporthe oryzae-A review on genes studied with mutant analysis. Pathogens 12, 379. doi: 10.3390/pathogens12030379
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Thomas, C. M., Dixon, M. S., Parniske, M., Golstein, C., and Jones, J. D. (1998). Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Philos. Trans. R Soc. Lond. B Biol. Sci. 353, 1413–1424. doi: 10.1098/rstb.1998.0296
Tian, D., Traw, M. B., Chen, J. Q., Kreitman, M., and Bergelson, J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423, 74–77. doi: 10.1038/nature01588
Tong, S., Ashikari, M., Nagai, K., and Pedersen, O. (2023). Can the wild perennial, rhizomatous rice species Oryza longistaminata be a candidate for de novo domestication? Rice (N Y) 16, 13. doi: 10.1186/s12284-023-00630-7
Vergish, S., Huang, X., Zhang, G., De Toledo Franceschi, B., Li, J. L., Wu, X. X., et al. (2025). Rhomboid-mediated cleavage of the immune receptor XA21 protects grain set and male fertility in rice. Proc. Natl. Acad. Sci. U.S.A. 122, e2502025122. doi: 10.1073/pnas.2502025122
Wang, X., Jia, M. H., Ghai, P., Lee, F. N., and Jia, Y. (2015). Genome-wide association of rice blast disease resistance and yield-related components of rice. Mol. Plant Microbe Interact. 28, 1383–1392. doi: 10.1094/MPMI-06-15-0131-R
Wang, Y. S., Pi, L. Y., Chen, X., Chakrabarty, P. K., Jiang, J., De Leon, A. L., et al. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18, 3635–3646. doi: 10.1105/tpc.106.046730
Wang, G. L., Ruan, D. L., Song, W. Y., Sideris, S., Chen, L., Pi, L. Y., et al. (1998). Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10, 765–779. doi: 10.1105/tpc.10.5.765
Wang, M., Zhu, X., Huang, Z., Chen, M., Xu, P., Liao, S., et al. (2024). Controlling diurnal flower-opening time by manipulating the jasmonate pathway accelerates development of indica-japonica hybrid rice breeding. Plant Biotechnol. J. 22, 2267–2281. doi: 10.1111/pbi.14343
Wessler, S. R. (1996). Turned on by stress. Plant retrotransposons. Curr. Biol. 6, 959–961. doi: 10.1016/s0960-9822(02)00638-3
Wilson, Z. A., Song, J., Taylor, B., and Yang, C. (2011). The final split: the regulation of anther dehiscence. J. Exp. Bot. 62, 1633–1649. doi: 10.1093/jxb/err014
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Xie, D. L., Zheng, X. L., Zhou, C. Y., Kanwar, M. K., and Zhou, J. (2022). Functions of redox signaling in pollen development and stress response. Antioxidants 11, 287. doi: 10.3390/antiox11020287
Xu, W. H., Wang, Y. S., Liu, G. Z., Chen, X., Tinjuangjun, P., Pi, L. Y., et al. (2006). The autophosphorylated Ser686, Thr688, and Ser689 residues in the intracellular juxtamembrane domain of XA21 are implicated in stability control of rice receptor-like kinase. Plant J. 45, 740–751. doi: 10.1111/j.1365-313X.2005.02638.x
Yang, D. L., Yao, J., Mei, C. S., Tong, X. H., Zeng, L. J., Li, Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200. doi: 10.1073/pnas.1201616109
Ye, H., Du, H., Tang, N., Li, X., and Xiong, L. (2009). Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71, 291–305. doi: 10.1007/s11103-009-9524-8
Zhang, N., Dong, X., Jain, R., Ruan, D., De Araujo Junior, A. T., Li, Y., et al. (2024). XA21-mediated resistance to Xanthomonas oryzae pv. oryzae is dose dependent. Peer J. 12, e17323. doi: 10.7717/peerj.17323
Keywords: fertility defect, anther dehiscence, jasmonate signaling, disease resistance, innate immune receptor, Oryza sativa
Citation: de Toledo Franceschi B, Vergish S, Singh Y, Li J-L, Shamsunnaher, Ding G-L, Chen S and Song W-Y (2025) The immune receptor XA21 causes semi-male sterility and grain loss in rice. Front. Plant Sci. 16:1673821. doi: 10.3389/fpls.2025.1673821
Received: 26 July 2025; Accepted: 30 September 2025;
Published: 10 November 2025.
Edited by:
Katarzyna Otulak-Kozieł, Warsaw University of Life Sciences, PolandReviewed by:
Md Ahasanur Rahman, Howard University, United StatesHafiz Muhammad Usman Aslam, Colorado State University, United States
Copyright © 2025 de Toledo Franceschi, Vergish, Singh, Li, Shamsunnaher, Ding, Chen and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Yuan Song, d3NvbmdAaWZhcy51ZmwuZWR1
†Present address: Satyam Vergish, Department of Plant Pathology, Citrus Research and Education Center, IFAS, University of Florida, Lake Alfred, FL, United States
‡These authors have contributed equally to this work
 Beatriz de Toledo Franceschi
Beatriz de Toledo Franceschi Satyam Vergish
Satyam Vergish Yatendra Singh
Yatendra Singh Jian-Liang Li
Jian-Liang Li Shamsunnaher
Shamsunnaher Gao-Lu Ding
Gao-Lu Ding Sixue Chen
Sixue Chen Wen-Yuan Song
Wen-Yuan Song