Abstract
Nicotiana tabacum BY-2 cell suspension cultures are a powerful platform for producing recombinant glycoproteins such as immunoglobulins. Extensive efforts have been devoted to engineering the N-glycosylation pathway of BY-2 cells to overcome differences between mammalian and plant N-glycans. However, the mucin-type O-glycosylation pathway is absent in plant cells. This modification, which consists of glycan attachment to serine or threonine residues, is important in many human proteins, but is also highly complex. In this regard, plants offer a unique opportunity to engineer this pathway de novo without the interplay of many competing enzymes. In this study, transgenic BY-2 cell lines expressing the enzymes responsible for the formation of Core 1 and Tn antigen glycans were generated. First, GalNAc-O-glycosylation was initiated by the expression of human GalNAcT2. This O-glycan was then elongated by co-expression of Drosophila C1GalT1 to form the core 1 structure. Human IgA1 was produced in these engineered BY-2 cell lines and the presence of mucin-type O-glycans was confirmed by lectin blotting. The precise O-glycosylation profile of the hinge region was determined by mass spectrometry and showed the almost complete disappearance of pentoses and the presence of core 1 O-glycans.
1 Introduction
Suspension cultures of plant cells, particularly Nicotiana tabacum BY-2 cells, are an interesting platform for the production of biopharmaceuticals (Santos et al., 2016; Karki et al., 2021; Xu et al., 2025). This system combines the cost effectiveness and safety of plant cells with the industry GMP compliance, and some human enzymes produced in plant suspension cultures have been approved by regulatory agencies (Elelyso® and Elfabrio®). Therapeutic proteins are often glycosylated, and this modification has a critical impact on their properties. N-glycoengineering in BY-2 cells has already been well documented. In particular, the removal of non-human core alpha(1,3)-fucose and beta(1,2)-xylose structures has been successfully achieved in BY-2 cells by edition of alpha(1,3)-fucosyltranferase and beta(1,2)-xylosyltransferase genes by CRISPR/Cas9 (Hanania et al., 2017; Mercx et al., 2017; Sheva et al., 2020), allowing the production of glycoproteins with human-like complex N-glycans in XylT/FucT-KO (ΔXT/FT) BY-2 cells.
In humans, GalNAc-O-glycosylation on the oxygen atom of serine and threonine, also known as mucin-type, is the most abundant type of O-glycosylation, but this O-glycosylation pathway is absent in plants. The initiation of mucin-type GalNAc-O-glycosylation has been successfully demonstrated in plants. The expression of human GalNAcT2 (or GalNAcT4) in Nicotiana benthamiana leaves or transgenic BY-2 cells is functional on relevant glycoproteins such as MUC1, MUC16, interferon alpha 2b, EPO or G-CSF (Yang et al., 2012b, Yang et al., 2012a; Castilho et al., 2012; Ramírez-Alanis et al., 2018; Daskalova et al., 2010). The elongation of the GalNAc-O glycan in core 1 O-glycan structure has also been established for EPO produced in N. benthamiana leaves by co-expression of Drosophila melanogaster C1GalT1 (Castilho et al., 2012). Another important O-glycosylated protein is human IgA1, which displays a long hinge region with three to six O-glycan chains attached to some of the nine potential threonine/serine O-glycosylation sites (Ohyama et al., 2020). A few studies have described the successful engineering of IgA1 hinge region O-glycosylation upon co-expression of enzymes from the mucin O-glycosylation machinery in wild type or ΔXT/FT N. benthamiana leaves (Uetz et al., 2024; Dicker et al., 2016).
In this study, we generated a transgenic N. tabacum ΔXT/FT BY-2 cell line capable of producing Tn antigen or core 1 mucin-type O-glycans. Coexpression of human GalNAcT2 with Drosophila C1GalT in BY-2 cells resulted in the production of a human IgA1 variant of Trastuzumab with a hinge region displaying up to five core 1 O-glycan structures.
2 Material and methods
2.1 Cloning and expression vectors
The genetic constructs were obtained using the Golden Gate plant toolkit (Engler et al., 2014) following the protocol described by (Marillonnet and Grützner, 2020). The cloning of the level 0 plasmids pICH41258-SPPDI, pICH41295-pPMA4, pGEMT-tPMP1 and pICH41331-SAR and the level 1 pICH47732-nptII and pICH47732-hptI is described in (Navarre et al., 2023).The ORFs encoding human GalNAcT2 (NP_004472.1) and D. melanogaster C1GalT1 (NP_609258.1) were designed to be introduced into the level 0 CDS1 Golden Gate plasmid (pICH41308): AATG and GCTT fusion sites were added with BsaI restriction sites in 5’ and 3’, respectively. The sequences were synthesized by Genewiz (GalNAcT2) and Genscript (C1GalT1) and cloned into pICH41308. Next, expression cassettes were constructed into level 1 plasmids under the control of p35S promoter (pICH51288) and tNOS terminator (pICH41421). Different level 1 plasmids were used to define the correct position of the expression cassettes in the final constructs: pICH47742 (position 2, GalNAcT2 or C1GalT1) and pICH47751 (position 3, C1GalT1). In addition, the neomycin phosphotransferase II expression cassette from the kit (pICSL70004) was transferred in the plasmid pICH47732 at position 1. Finally, the genetic insulator of Rb7 scaffold attachment region (SAR) of N. tabacum (1.2 kb), which should reduce positional and silencing effects (Diamos and Mason, 2018), was transferred into level 1 at position 3 (pICH47751), 4 (pICH47761) or 5 (pICH47772). The expression cassettes were then assembled into the level M pAGM8031 plasmid to generate pAGM8031-nptII-GalNAcT2-SAR and pAGM8031-nptII-GalNAcT2-C1GalT1-SAR.
Sequences coding for IgA1 HC and LC were provided by Strasser’s laboratory in pEAQ (Göritzer et al., 2017) and PCR-amplified to add 5’-AATG and GCTT-3’ and BpiI restriction sites for cloning into the level 0 CDS1 plasmid pICH41308. The nucleotide sequence corresponding to the intronless N. tabacum Extensin terminator (tExt) was PCR amplified from pBYR2eKMd-GFP (Diamos and Mason, 2018) to add 5’-GCTT and CGCT-3’ and BsaI restriction sites and cloned into pICH41276. The ORFs were then assembled with pPMA4 and tExt in different level 1 plasmids: pICH47742 and pICH47761 for HC; pICH47751 and pICH47772 for LC. The HC and LC expression cassettes were then assembled with the nptII, GalNAcT2 and C1GalT1 expression cassettes into the level M plasmid pAGM8031 to generate: pAGM8031-nptII-HC-LC, pAGM8031-nptII-HC-LC-GalNAcT2 and pAGM8031-nptII-GalNAcT2-C1GalT1-HC-LC.
2.2 Transformation of BY-2 cells
Agrobacterium tumefaciens LBA4404 VirG-mediated stable transformation of ΔXT/FT BY-2 cells was carried out as described by (Navarre and Chaumont, 2022) with nptII resistance marker gene. Selection of transformed cell lines was carried out on MS-agar plates containing 500 µg/mL cefotaxime, 400 µg/mL carbenicillin and 100 µg/mL kanamycin.
Samples used for western blotting were harvested from liquid MS cultures obtained after at least two passages on solid selective medium (1–2 months) and at least two passages in liquid selective medium. D11b cultures (Vasilev et al., 2013) were grown for ten days in 50 mL of medium without cyclodextrin, in 250 mL Erlenmeyer flasks.
2.3 IgA1 protein purification
IgA1 purification was adapted from the protocol described by (Mercx et al., 2017). Briefly, 50–250 mL of 10-day-old BY-2 suspension culture grown in D11b medium (Vasilev et al., 2013) without cyclodextrin was filtered on three layers of Miracloth and the filtrate was centrifuged (13,900 g, 30 min). The supernatant was recovered, supplemented with 10% 1 M Tris-Cl pH 8.0 and incubated for 16 h at 4 °C with 400 µL of Pierce R Protein A Plus Agarose (Thermoscientific # 22812) previously washed three times with 4 mL of 0.1 M Tris-Cl pH 8.0. The sample was centrifuged at ~50 g for 5 min and the supernatant was discarded. The beads were washed twice with 10 mL of 0.1 M Tris-Cl pH 8.0 and transferred into a 10 mL Poly-Prep chromatography column (Bio-Rad #7311550). IgA1 was eluted with 5 × 500 µL 0.1 M glycine-HCl pH 3.0 and immediately buffered with 10% 1 M Tris-HCl pH 8.0.
2.4 Protein extraction
Two mL of the indicated BY-2 cultures were filtered through three layers of Miracloth by centrifugation (2,900 g for 5 min). The filtered BY-2 cells were used to harvest cellular total soluble proteins (TSP) and microsomal fractions (MF). The cell packs were transferred into a 2-mL screw cap microtube containing 0.5 glass beads (0.85-1.23 mm) and frozen in liquid nitrogen. Next, 700 µL of homogenization buffer (250 mM sorbitol, 2 mM Na2EDTA, 60 mM Tris-HCl pH 8.0) supplemented with 1 mM PMSF and protease inhibitor cocktail (leupeptin, aprotinin, antipain, pepstatin and chymostatin, each at 2 µg/mL) were added. The cells were ground for 3 x 40 s at 5,000 rpm (Precellys 24 tissue homogenizer, Bertin Technologies) with 2 min pauses on ice. The samples were first centrifuged for 5 min at 2,800 g at 4 °C, then for 7 min at 10,000 g at 4 °C. To separate TSP from the MF, the samples were centrifuged for 15 min at 130,000 g at 4 °C. The supernatant (TSP) was collected and the pellet (MF), was resuspended by sonication in 50 µL resuspension buffer (3 mM KH2PO4, 330 mM sucrose, 3 mM KCl, pH 7.8 (KOH)). Protein concentrations were determined according to Bradford (Bradford, 1976) using BSA as standard.
2.5 SDS-PAGE, western blotting and lectin blotting
The indicated amount of TSP, MF or purified IgA1 was analyzed by reducing SDS-PAGE after solubilization (5 min at 100 °C for TSP or purified IgA1; 15 min at 56 °C for MF). For non-reducing SDS-PAGE, purified IgA1 samples were solubilized for 30 min at room temperature. Gels were stained with Coomassie Blue G-250 (SERVA, 17524) or transferred onto PVDF membrane (Bio-Rad laboratories).
For western blotting, the membranes were saturated into Tris Buffer Saline containing 0.5% Tween®20 (TBST) and 3% (w/v) milk powder, before incubation with rabbit polyclonal antibodies against hGalNAcT2 (Sino Biologicals, 13764-T62, 1:2,000) and anti-rabbit HRP polyclonal antibodies (SynAbs, LO-RG-1-HRP, 1:10,000), or goat polyclonal antibodies against human IgA (alpha heavy chain) couple to HRP (Invitrogen, 31417, 1:25,000).
For lectin blotting, the PVDF membranes were saturated in TBST containing 2% (w/v) BSA before incubation with biotinylated Helix pomatia agglutinin (Sigma-Aldrich L6512, 1:2,500) or biotinylated peanut agglutinin (Vector Laboratories B1075, 1:2,500), followed by incubation with Streptavidin HRP (Roche 11089153001, 1:5,000).
Chemiluminescence was detected after revelation with BM chemilumiscence blotting substrate (Roche 11500694001) using Amersham Imager 600 (GE Healthcare).
2.6 In vitro deglycosylation
Purified IgA1 protein (125 ng) was denatured by heating for 10 min at 100 °C in the manufacturer’s glycoprotein denaturing buffer to ensure optimal glycosite accessibility. Samples were then incubated for 1 h at 37 °C with 50 U of EndoH (New England Biolabs P0703) or PNGase F (New England Biolabs P0704). Control samples were run in parallel, but without the glycosidase. The samples were then visualized by SDS-PAGE followed by western blotting using goat polyclonal antibodies raised against human IgA (alpha heavy chain) couple to HRP (Invitrogen 31417, 1:25,000) and revealed with BM chemilumiscence blotting substrate (Roche 11500694001). Chemiluminescence was detected using the Amersham Imager 600 (GE Healthcare).
2.7 Thermal shift assay
The thermal stability of the three Trastuzumab IgA1 O-glycovariants was analyzed by a different scanning fluorimetry assay following the protocol described by (Herman et al., 2023). Briefly, purified IgA1 samples (1.2 µg) were diluted twice in phosphate buffered saline (100 mM phosphate, 150 mM NaCl, pH 7.0) containing 5x SYPRO Orange Protein Gel Stain (S6650; Invitrogen, Waltham, MA, USA). The samples were transferred into a MicroAmpTM Optical 96-Well Reaction Plate (N8010560; Applied Biosystems, Waltham, MA, USA). Each sample was run as a technical triplicate. DSF measurements were performed using a temperature increase from 15 °C to 85 °C with a ramp of 1 °C/min (StepOnePlusTM Real-Time PCR System; Applied Biosystems). The mean melting temperatures were calculated as the temperature corresponding to the maximum value of the mean derivative curves. Mean derivative curves were obtained as the first derivative of the mean melt curves.
2.8 Mass spectrometry analysis IgA1 glycopeptides
The IgA1 O-glycovariants were digested in-solution. The proteins were S-alkylated with iodoacetamide and digested with trypsin (Promega) overnight. The peptide samples were then digested with GluC (Promega) for 4 h. The digested samples were loaded on a nanoEase C18 column (nanoEase M/Z HSS T3 Column, 100Å, 1.8 μm, 300 μm X 150 mm, Waters) using 0.1% formic acid as the aqueous solvent. A gradient from 1% B (B: 80% acetonitrile, 0.1% formic acid) to 40% B in 50 min was applied, followed by a 5 min gradient from 40% B to 95% B that facilitates elution of large peptides, at a flow rate of 6 μL/min. Detection was performed with an Orbitap MS (Exploris 480, Thermo) equipped with the standard H-ESI source in positive ion, DDA mode (= switching to MSMS mode for eluting peaks). MS scans were recorded (range: 350–3200 m/z) and the 8 highest peaks were selected for fragmentation. Instrument calibration was performed using Pierce FlexMix Calibration Solution (Thermo Scientific). The possible glycopeptides were identified as sets of peaks consisting of the peptide moiety and the attached N-glycan varying in the number of N-acetylhexosamine (HexNAc) units, hexose (Hex), pentose (Pent) and deoxyhexose (dHex) residues, as well as additional hydroxyproline modifications for the O-glycosylation site. The theoretical masses of these glycopeptides were determined with a spread sheet using the monoisotopic masses for amino acids and monosaccharides. Manual glycopeptide searches were made using FreeStyle 1.8 (Thermo). For the relative quantification of the different glycoforms, the peak intensities of the deconvoluted spectra were compared (using an in-house software tool. (Freestyle_parser_v0.3.R)).
3 Results
3.1 Stable engineering of mucin-type O-glycosylation in humanized BY-2 cells
The ORFs encoding human GalNAcT2 and D. melanogaster C1GalT1 were cloned under the control of the 35S promotor. The two constructs were assembled into binary vectors to generate two different plasmids encoding either GalNAcT2 alone (GalNAc) or both GalNAcT2 and C1GalT1 (Core1) (Figure 1A). The humanized ΔXT/FT BY-2 cell line, which shows the absence of beta(1,2)-xylose and core alpha(1,3)-fucose in N-glycans (Mercx et al., 2017), was transformed with the two constructs and several transgenic BY-2 cell lines were selected.
Figure 1

GalNAc-O-glycosylation in BY-2 cells. (A). Schematic overview of the O-glycosylation machinery constructs. The coding sequences for human GalNAc-transferase GalNAcT2 and D. melanogaster core 1 beta(1,3)-galactosyltransferase C1GalT1 were cloned under the control of p35S and tnos in level 1 Golden Gate vectors. The expression cassettes were assembled with the nptII kanamycin resistance cassette and a SAR genetic insulator to generate two different constructs (GalNAcT2 alone or GalNAcT2 + C1GalT1). (B). Initiation of GalNAc-O-glycosylation in ΔXT/FT BY-2 cells. (B1) Twenty-five µg TSP were analyzed by biotinylated H. pomatia lectin blotting followed by detection HRP-labeled Streptavidin. (B2) Twenty-five µg of microsomal fractions were analyzed by western blotting using anti-GalNAcT2 antibodies. A sample derived from the parental ΔXT/FT BY-2 cell line was loaded as a negative control. A sample corresponding to N. benthamiana leaves agroinfiltrated with the construct GalNAcT2 alone was used as a positive control (C+). (C). Generation of core 1 structures in ΔXTFT BY-2 cells. (C1) Twenty-five µg TSP of transgenic cell lines were analyzed by biotinylated peanut agglutinin blotting followed by detection HRP-labeled Streptavidin. (C2) Twenty-five µg of microsomal fractions were analyzed by western blotting using anti-GalNAcT2 antibodies. A sample derived from the parental ΔXTFT BY-2 cell line (C-) or a GalNAcT2 transgenic cell line were used as controls.
First, the initiation of GaNAc-O-glycosylation pathway was introduced in ΔXT/FT BY-2 cells by expressing the human GalNAcT2. We tested GalNAc attachment to endogenous proteins. Total soluble proteins were analyzed by lectin-probed western blotting using H. pomatia Agglutinin (HPA), which binds to terminal alpha- and beta-GalNAc residues. A clear HPA signal in all the GalNAc lines was observed compared to the parental ΔXT/FT BY-2 line (first lane in each panel) (Figure 1B1). Western blotting of the microsomal fraction of the cell lines probed with anti-GalNAcT2 antibodies confirmed hGalNACT2 expression in all the GalNAc cell lines tested (Figure 1B2).
Elongation of the O-glycans in the core 1 structure was achieved by co-expression of hGalNAcT2 and DmC1GalT1 in ΔXT/FT BY-2 cells. Lectin blots of TSPs were probed with peanut (Arachis hypogaea) agglutinin (PNA), which has activity toward unsubstituted core 1 (Figure 1C1). Strong, albeit highly variable, signals were observed when DmC1GalT1 was expressed (the membrane was cut due to non-specific signals). The expression of hGalNAcT2 was verified by western blotting (Figure 1C2).
3.2 Expression of IgA1 in BY-2 cells expressing mucin-type O-glycosylation enzymes
The genes encoding the heavy and light chains of the chimeric human IgA1 with the variable region from Trastuzumab were cloned under the control of the engineered Nicotiana plumbaginifolia PMA4 promoter and combined with GalNAcT2 and C1GalT1 expression cassettes in binary plasmids. A total of three constructs were generated: IgA1 alone (IgA1), IgA1 + GalNAcT2 (IgA1G) and IgA1 + GalNAcT2 + C1GalT1 (IgA1C) (Figure 2A). The three IgA1 constructs were used to transform ΔXT/FT BY-2 cells and stable transgenic cell lines were screened by western blotting with human anti-Fab to identify the best IgA1 producing lines in the three formats (data not shown). Extracellular medium was collected from D11b cultures by filtration and used to purify the three IgA1 O-glycovariants by affinity on Protein A agarose beads. The purified IgA1 O-glycovariants were analyzed using SDS-PAGE (Figure 2B1). Under non-reducing conditions, IgA1, IgA1G and IgA1C showed a band at a molecular mass of approximately 160 kDa corresponding to the fully assembled monomeric molecule, as well as two additional bands at 120 kDa and 100 kDa, a pattern similar to that of the same IgA1 antibody expressed in N. benthamiana leaves (Göritzer et al., 2017). These three bands were detected by anti-human IgA (heavy chain). Reducing SDS-PAGE confirmed the presence of the alpha heavy chain and the kappa light chain. Two additional bands that could correspond to proteolytic degradation products of the alpha chain were observed (Figure 2B2).
Figure 2
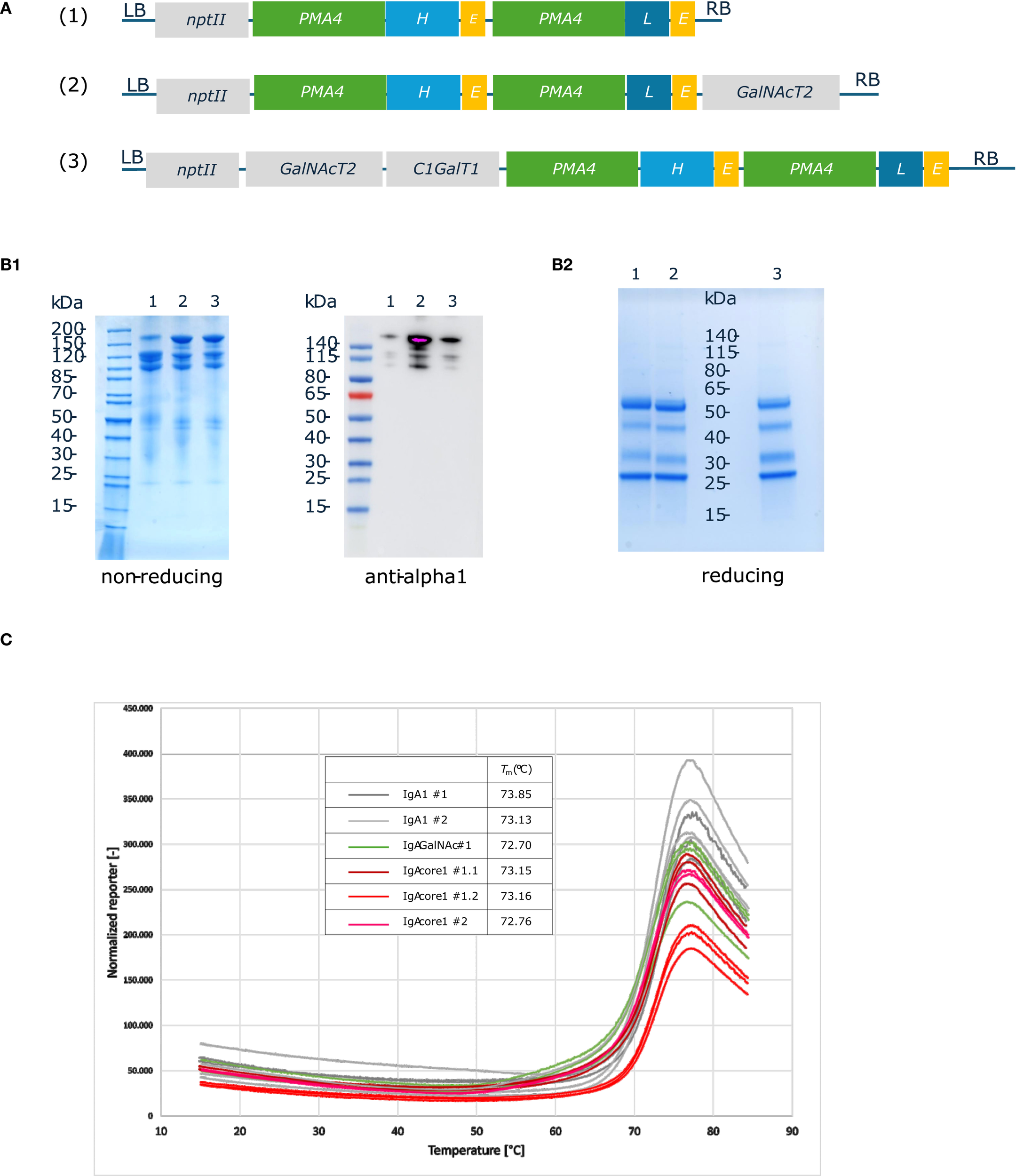
Production and purification of human IgA1 O-glycovariants in BY-2 cells. (A). IgA1 O-glycovariants constructs. The coding sequences from chimeric human Trastuzumab IgA1 HC and LC were cloned under the control of pPMA4 and tExtensin in Level 1 Golden Gate vectors. The expression cassettes were assembled with the O-glycosyltransferase expression cassettes (GalNAcT2 alone or GalNAcT2 + C1GalT1) and the nptII kanamycin resistance cassette to generate three different constructs (IgA1, IgA1 GalNAc alone or IgA1 core 1). (B). SDS-PAGE and immunoblotting of protein A-purified IgA1 O-glycovariants: 1 (IgA1); 2 (IgA1 + GalNAc); 3 (IgA1 core1). Purified samples were analyzed by Coomassie blue staining (2.5 µg) or by western blotting with anti-human IgA (alpha heavy chain) (1: 0.2 µg; 2: 0.4 µg; 3: 0.3 µg). (C). Thermal stability of IgA1 O-glycovariants measured using a thermal shift assay. Each O-glycovariants (two biological replicates #1, #2) was measured in triplicates at temperatures ranging from 15 °C to 85 °C. Melting temperatures were calculated based on the first derivative of the melting curves.
The thermal stability of all three IgA1 O-glycovariants was determined, but no significant differences in melting temperatures were observed (Figure 2C). The Tm value was approximately 73 °C for the three samples, a value similar to what was observed for other IgA1s produced in N. benthamiana leaves (Uetz et al., 2024).
3.3 Characterization of the N-glycosylation status of the alpha chain
IgA1 is highly glycosylated and characterized by its long 19 residues hinge region, which is GalNAc-O-glycosylated with three to six (Thr225, Thr228, Ser232 and Ser230, Thr233, Thr236) sialylated core 1 O-glycans in human serum (Ohyama et al., 2020). IgA1 also contains two N-glycosylation sites: N263 located in the CH2 and N459 in the tail piece (Göritzer et al., 2020).
We first assessed the N-glycosylation status of purified IgA1 produced in ΔXT/FT BY-2 cells by in vitro deglycosylation with PNGaseF and EndoH. EndoH cleaves high-mannose N-glycans while PNGaseF can cleave all glycan types except those containing core alpha(1,3)-fucose. A slight molecular mass shift of the alpha heavy chain was observed by western blotting in samples treated with PNGaseF, but not with EndoH (Figure 3A), indicating that the N-glycans are complex or hybrid types lacking core alpha(1,3)-fucose. This type of profile was previously observed for the alpha chain of sIgA1 purified from ΔXT/FT N. benthamiana leaves (Dicker et al., 2016).
Figure 3
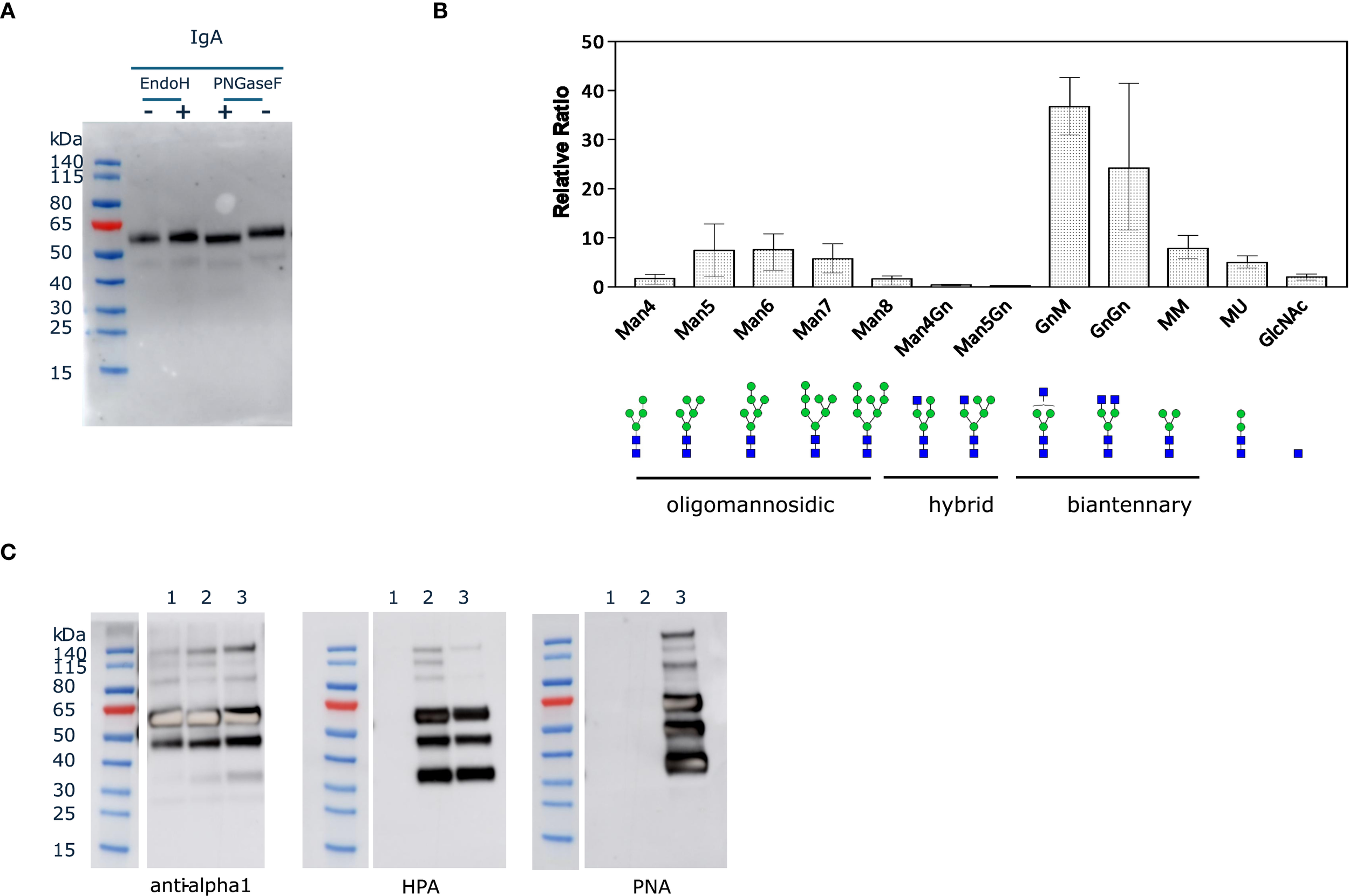
N- and O-glycosylation status of IgA1 in ΔXT/FT BY-2 cells. (A). Purified IgA1 (125 ng) was denatured for 10 min at 100 °C and treated for 1h at 37 °C with EndoH (50 U) or PNGaseF (50 U). Control samples (-) were run in parallel but without the glycosidase. (B). Quantification of the relative abundance of glycoforms corresponding to the glycopeptide ANLTCTLTGLR from the CH2 domain detected on the three IgA1 O-glycovariants. N-glycans are abbreviated according to ProGlycAn system (Altmann et al., 2024). (C). Purified IgA1 O-glycovariants (1.25 µg) were analyzed under reducing conditions by western blotting with anti-human IgA (alpha heavy chain) or lectin blotting with HPA and PNA (1: IgA1; 2: IgA1 + GalNAc; 3: IgA1 core1).
Then, the identity of the N-glycans was determined accurately by LC-MS/MS after trypsin and GluC digestion of the purified IgA1 O-glycovariants. The mass spectra of glycopeptides corresponding to the N263 site in CH2 (ANLTCTLTGLR) are shown in Figure 3B. The overall profile was broadly similar for the three O-glycovariants, with biantennary complex-type structures like GlcNAc2Man3GlcNAc2 (GnGn) and GlcNAc1Man3GlcNAc2 (GnM/MGn) being the most abundant N-glycans (50-75%), in agreement with the in vitro N-deglycosylation assay. The other N-glycans detected were different oligomannosidic (Man4 to Man8) and truncated N-glycans Man3GlcNAc2 (MM). The spectra also showed a small amount of Man2GlcNAc2 (MU) (± 5%) and single Gn (<3%) glycans. No ANLTCTLTGLR peptide without N-glycan was detected, suggesting that N263 was fully occupied.
Glycosylation analysis of the tail piece site (LAGKPTHVNVSVVMAE) yielded poor quality results. This glycosylation site is located at the C-terminus of the protein in a disordered region and was found to be cleaved and underglycosylated for IgA1 expressed in N. benthamiana (Dicker et al., 2016; Göritzer et al., 2020).
3.4 O-Glycan analysis of the IgA1 hinge region
To assess the identity of the glycans on each IgA1 O-glycovariant, blots of purified IgA1 were probed using anti-human IgA (alpha heavy chain) antibody, as well as HPA and PNA lectins (Figure 3C). The same bands were detected with the lectins and the anti-alpha chain antibody, suggesting that O-glycans were found on the HC, probably in the hinge region. The lectin blots clearly indicated that IgA1 was O-glycosylated with mucin-type O-glycans when the appropriate enzymes were co-expressed in ΔXT/FT BY-2 cells. The PNA-probe blot showed that co-expression of hGalNAcT2 and DmC1GalT1 was necessary for detection of core 1 O-glycans on alpha chain of IgA1C. In addition, the HPA-probe blot showed that expression of hGalNAcT2 was necessary to detect GalNAc residues on alpha chain of IgA1G and that both GalNAc and core 1 O-glycans could coexist on IgA1C (when both hGalNAcT2 and DmC1GalT1 were expressed), as both HPA and PNA signals were observed for IgA1C (Figure 3C). Alternatively, the signals could be due to a small reactivity of HPA toward core 1 glycans as previously reported (Sanchez et al., 2006).
Mass spectrometry analysis was then carried out on the O-glycan containing glycopeptides corresponding to the hinge region (HYTNPSQDVTVPCPVPSTPPTPSPSTPPTPSPSCCHPR). The PTM search included HexNAc and Hex for GalNAc-O-glycosylation as well as Pro hydroxylation and pentoses.
Figure 4 and Supplementary Table 1 show the mass spectra and relative intensities of two different IgA1 O-glycovariants hinge regions. When IgA1 was expressed alone in ΔXT/FT BY-2 cells, the hinge region was heavily modified with plant-specific hydroxyproline glycosylation (Figure 4A). Indeed, it was found that more than 90% of the peptide was modified with at least one Hyp and 80% were glycosylated with up to 18 pentoses, with the most abundant glycopeptides being 6 Hyp-9, 10, 12, and 13 Pentoses.
Figure 4
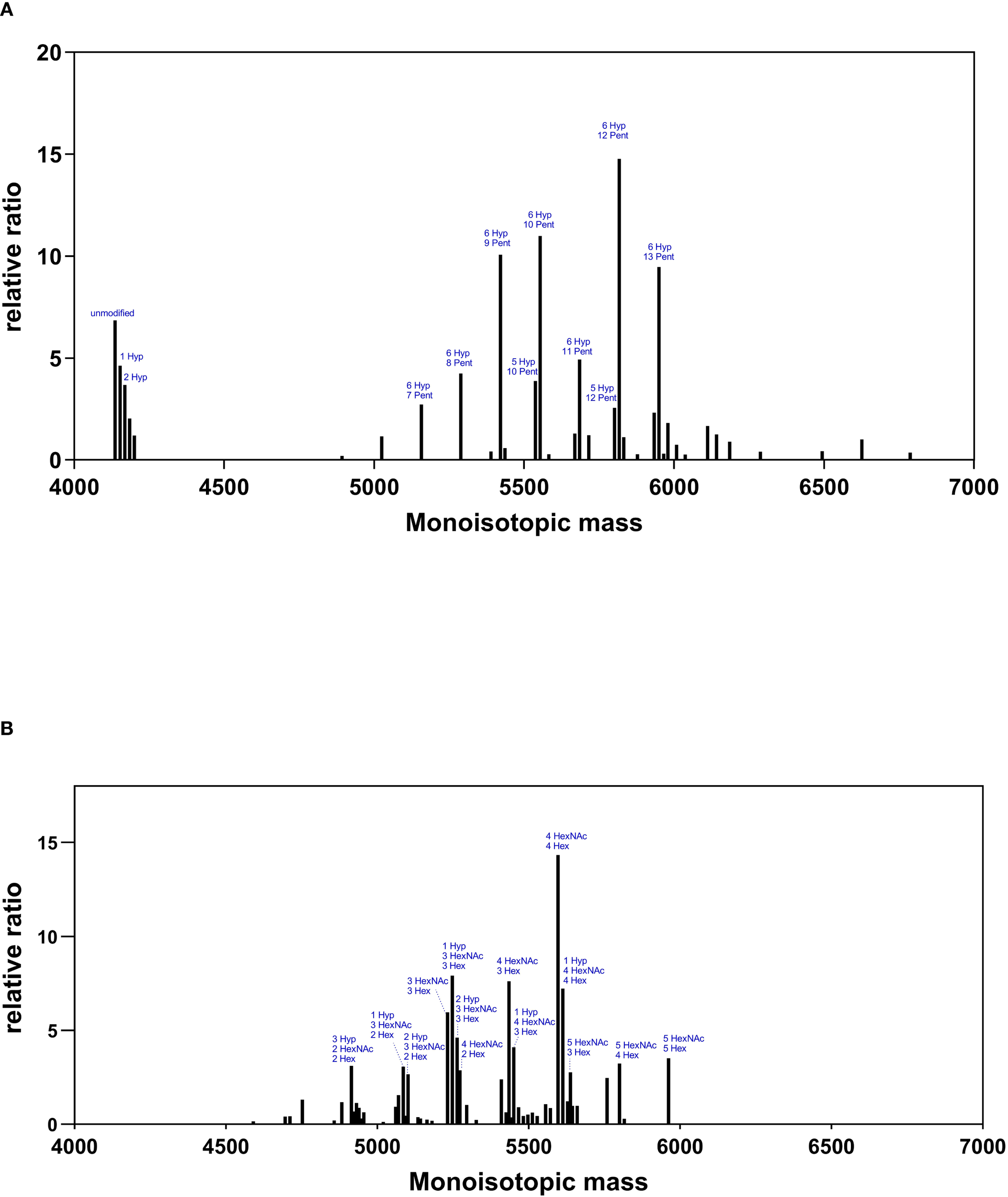
O-glycosylation profiles of IgA1 O-glycovariants in BY-2 cells. Mass spectra of the hinge region peptide (HYTNPSQDVTVPCPVPSTPPTPSPSTPPTPSPSCCHPR) for IgA1 (Panel (A)) and IgA1C (Panel (B)). Glycosylated peaks with a relative abundance >2.5 are indicated: pentoses (Pent), hydroxyproline (Hyp), N-acetylgalactosamine (HexNAc), and hexoses (Hex).
When hGalNAcT2 and DmC1GalT1were co-expressed in ΔXT/FT BY-2 cells, mass shifts corresponding to the addition of HexNAc and Hex were detected, supporting the initiation and elongation of GalNAc-O-glycosylation on the hinge region of IgA1 (Figure 4B). Three to five O-sites were occupied, in accordance with the glycosylation status of the same IgA1 observed in HEK 293 (Göritzer et al., 2017). In addition, all identified peptides were modified and showed a strong reduction of Pro hydroxylation and a nearly complete disappearance of pentoses.
4 Discussion
We have engineered mucin-type O-glycosylation in N. tabacum BY-2 cells. To our knowledge, there is only one example of introducing the first GalNAc step of mucin-type O-glycosylation in transgenic BY-2 cells (Yang et al., 2012a). In this report, a 2A-linked polycistronic construct composed of the human GalNAcT2 and the cytoplasmic Glc (NAc) C4-epimerase from Pseudomonas aeruginosa was used because previous data of transient expression in N. benthamiana leaves showed no detectable O-glycosylation of the MUC1-YFP reporter when only hGalNAcT2 was expressed (Yang et al., 2012b). It was indeed postulated that UDP-GalNAc concentration in the Golgi apparatus could not be sufficient to promote efficient GalNAc O-glycosylation of proteins expressed at high levels in N. benthamiana leaves (Daskalova et al., 2010). However, additional studies in N. benthamiana leaves showed that transient expression of hGalNAcT2 alone was sufficient to achieve efficient GalNAc O-glycosylation of EPO-Fc (Castilho et al., 2012; Kriechbaum et al., 2020) or human IgA1 (Dicker et al., 2016; Uetz et al., 2024). Our results demonstrated that hGalNAcT2 is sufficient when expressed alone to initiate GalNAc O-glycosylation of endogenous proteins as well as recombinant human IgA1 hinge region in transgenic ΔXT/FT BY-2 cells. However, it is unclear whether heavily glycosylated substrates such as MUC16 (30 sites) would benefit from the co-expression of a Glc (NAc) C4-epimerase.
The elongation of GalNAc-O-glycan to core 1 in yeast (Amano et al., 2008) and N. benthamiana leaves (Castilho et al., 2012) was achieved by co-expressing hGalNAcT2 with DmC1GalT1, a core 1 beta1–3 galactosyltransferase which does not require the presence of a specific chaperone COSMC. We used the same approach in transgenic ΔXT/FT BY-2 cells and confirmed the presence of core 1 O-glycan structure on BY-2 cells endogenous proteins and the human IgA1 hinge region. We detected glycopeptides containing 3, 4 and 5 core 1 O-glycan structures and glycopeptides containing both core 1 and Tn O-glycans. The formation of the core 1 O-glycans appeared to be highly efficient with most of the GalNAc being elongated with galactose (0.93 Hex/HexNAc residue ratio). Interestingly, unlike the O-glycosylation analysis of hinge IgA1 produced in N. benthamiana leaves co-expressing the core 1 O-glycosylation pathway (Uetz et al., 2024; Dicker et al., 2016), we detected no unmodified hinge glycopeptide, suggesting that GalNAc-O glycosylation was very efficient when hGalNAcT2 and DmC1GalT1 were constitutively expressed in transgenic BY-2 cells. The presence of unmodified IgA1 hinge observed in N. benthamiana leaves could be related to the high expression of IgA1 under transient expression or to the fact that IgA1C was purified from extracellular medium of BY-2 cells, i.e. at the very last step of the secretion process. It is likely that the IgA1C protein sample purified from BY-2 extracellular medium was more homogenous than the IgA1 protein sample from total leaf extracts, which consists of a mixture of IgA1 proteins from the entire secretory pathway (Dicker et al., 2016; Uetz et al., 2024). The use of transgenic BY-2 suspension cultures also facilitates downstream processing by starting from the extracellular medium and offers the possibility of tailoring the culture medium composition, which could complement genetic O-glycoengineering.
In addition, the modification of the hinge region with mucin-type O-glycans resulted in a strong reduction in the abundance of Hyps in the hinge region, suggesting that Pro hydroxylation was somehow prevented. Nevertheless, Pro hydroxylation and further Hyp glycosylation still occurred and may be deleterious for the properties of the antibody. Aberrant Hyp glycosylation was also detected on the hinge region of the same IgA1 produced in N. benthamiana leaves with glycopeptides containing 5 hydroxyprolines with 4 to 10 pentoses (Göritzer et al., 2017; Mócsai et al., 2021). Similarly, other IgA1 produced in N. benthamiana leaves revealed a similar O-glycosylation status with the presence of glycopeptides containing up to 6 hydroxyprolines with 8 and 10 pentoses (Karnoup et al., 2005; Dicker et al., 2016) or 5 hydroxyprolines and 4 to 10 pentoses (Uetz et al., 2024, Uetz et al., 2022; Göritzer et al., 2020). A promising strategy to reduce this plant-specific Hyp glycosylation is the silencing of P4Hs, the enzymes responsible for Pro hydroxylation, a strategy being explored in N. benthamiana leaves (Uetz et al., 2022, Uetz et al., 2024; Mócsai et al., 2021).
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
NB: Conceptualization, Investigation, Methodology, Writing – original draft, Data curation. CG-G: Data curation, Investigation, Methodology, Writing – original draft. MP: Investigation, Writing – original draft. FC: Funding acquisition, Supervision, Writing – review & editing. CN: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. NB was a recipient of a fellowship from the Fonds pour la Formation à la Recherche dans l’Industrie et l’Agriculture (1.E.026.21F, FRIA, Belgium). MP is a recipient of a fellowship from the Fonds National de la Recherche Scientifique (1.A.881.22F, FNRS, Belgium).
Acknowledgments
We are very grateful to Professor Richard Strasser from the University of Natural Resources and Life Sciences (BOKU, Vienna, Austria) for providing us with the pEAQ plasmid containing the IgA1 HC and LC. We acknowledge Adeline Courtoy (UCLouvain) for her excellent technical assistance. We are pleased to thank Professor Marc Boutry for his constant interest in our research and his careful reading of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1675517/full#supplementary-material
References
1
Altmann F. Helm J. Pabst M. Stadlmann J. (2024). Introduction of a human- and keyboard-friendly N-glycan nomenclature. Beilstein J. Org Chem.20, 607–620. doi: 10.3762/bjoc.20.53
2
Amano K. Chiba Y. Kasahara Y. Kato Y. Kaneko M. K. Kuno A. et al . (2008). Engineering of mucin-type human glycoproteins in yeast cells. Proc. Natl. Acad. Sci.105, 3232–3237. doi: 10.1073/pnas.0710412105
3
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem.72, 248–254. doi: 10.1016/0003-2697(76)90527-3
4
Castilho A. Neumann L. Daskalova S. Mason H. S. Steinkellner H. Altmann F. et al . (2012). Engineering of sialylated mucin-type O-glycosylation in plants. J. Biol. Chem.287, 36518–36526. doi: 10.1074/jbc.M112.402685
5
Daskalova S. M. Radder J. E. Cichacz Z. A. Olsen S. H. Tsaprailis G. Mason H. et al . (2010). Engineering of N. benthamiana L. plants for production of N-acetylgalactosamine-glycosylated proteins - towards development of a plant-based platform for production of protein therapeutics with mucin type O-glycosylation. BMC Biotechnol.10, 62. doi: 10.1186/1472-6750-10-62
6
Diamos A. G. Mason H. S. (2018). Chimeric 3’ flanking regions strongly enhance gene expression in plants. Plant Biotechnol. J.16, 1971–1982. doi: 10.1111/pbi.12931
7
Dicker M. Maresch D. Strasser R. (2016). Glyco-engineering for the production of recombinant IgA1 with distinct mucin-type O-glycans in plants. Bioengineered7, 484–489. doi: 10.1080/21655979.2016.1201251
8
Engler C. Youles M. Gruetzner R. Ehnert T. M. Werner S. Jones J. D. et al . (2014). A golden gate modular cloning toolbox for plants. ACS Synth Biol.3, 839–843. doi: 10.1021/sb4001504
9
Göritzer K. Goet I. Duric S. Maresch D. Altmann F. Obinger C. et al . (2020). Efficient N-glycosylation of the heavy chain tailpiece promotes the formation of plant-produced dimeric igA. Front. Chem.8, 346. doi: 10.3389/fchem.2020.00346
10
Göritzer K. Maresch D. Altmann F. Obinger C. Strasser R. (2017). Exploring site-specific N-glycosylation of HEK293 and plant-produced human IgA isotypes. J. Proteome Res.16, 2560–2570. doi: 10.1021/acs.jproteome.7b00121
11
Hanania U. Ariel T. Tekoah Y. Fux L. Sheva M. Gubbay Y. et al . (2017). Establishment of a tobacco BY2 cell line devoid of plant-specific xylose and fucose as a platform for the production of biotherapeutic proteins. Plant Biotechnol. J.15, 1120–1129. doi: 10.1111/pbi.12702
12
Herman X. Far J. Peeters M. Quinton L. Chaumont F. Navarre C. (2023). In vivo deglycosylation of recombinant glycoproteins in tobacco BY-2 cells. Plant Biotechnol. J. 21 (9), 1773–1784. doi: 10.1111/pbi.14074
13
Karki U. Fang H. Guo W. Unnold-Cofre C. Xu J. (2021). Cellular engineering of plant cells for improved therapeutic protein production. Plant Cell Rep.40, 1087–1099. doi: 10.1007/s00299-021-02693-6
14
Karnoup A. S. Turkelson V. Anderson W. H. K. (2005). O-Linked glycosylation in maize-expressed human IgA1. Glycobiology15, 965–981. doi: 10.1093/glycob/cwi077
15
Kriechbaum R. Ziaee E. Grünwald-Gruber C. Buscaill P. Van Der Hoorn R. A. L. Castilho A. (2020). BGAL1 depletion boosts the level of β-galactosylation of N- and O-glycans in N. benthamiana. Plant Biotechnol. J.18, 1537–1549. doi: 10.1111/pbi.13316
16
Marillonnet S. Grützner R. (2020). Synthetic DNA assembly using golden gate cloning and the hierarchical modular cloning pipeline. Curr. Protoc. Mol. Biol.130, e115. doi: 10.1002/cpmb.115
17
Mercx S. Smargiasso N. Chaumont F. De Pauw E. Boutry M. Navarre C. (2017). Inactivation of the β(1,2)-xylosyltransferase and the α(1,3)-fucosyltransferase genes in Nicotiana tabacum BY-2 Cells by a Multiplex CRISPR/Cas9 Strategy Results in Glycoproteins without Plant-Specific Glycans. Front. Plant Sci.8, 403. doi: 10.3389/fpls.2017.00403
18
Mócsai R. Göritzer K. Stenitzer D. Maresch D. Strasser R. Altmann F. (2021). Prolyl hydroxylase paralogs in Nicotiana benthamiana show high similarity with regard to substrate specificity. Front. Plant Sci.12. doi: 10.3389/fpls.2021.636597
19
Navarre C. Chaumont F. (2022). Production of recombinant glycoproteins in Nicotiana tabacum BY-2 suspension cells. Methods Mol. Biol.2480, 81–88. doi: 10.1007/978-1-0716-2241-4_5
20
Navarre C. Orval R. Peeters M. Bailly N. Chaumont F. (2023). Issue when expressing a recombinant protein under the control of p35S in Nicotiana tabacum BY-2 cells. Front. Plant Sci.14, 1266775. doi: 10.3389/fpls.2023.1266775
21
Ohyama Y. Yamaguchi H. Nakajima K. Mizuno T. Fukamachi Y. Yokoi Y. et al . (2020). Analysis of O-glycoforms of the IgA1 hinge region by sequential deglycosylation. Sci. Rep.10, 671. doi: 10.1038/s41598-020-57510-z
22
Ramírez-Alanis I. A. Renaud J. B. García-Lara S. Menassa R. Cardineau G. A. (2018). Transient co-expression with three O-glycosylation enzymes allows production of GalNAc-O-glycosylated Granulocyte-Colony Stimulating Factor in N. benthamiana. Plant Methods14, 98. doi: 10.1186/s13007-018-0363-y
23
Sanchez J. F. Lescar J. Chazalet V. Audfray A. Gagnon J. Alvarez R. et al . (2006). Biochemical and structural analysis of Helix pomatia agglutinin. A hexameric lectin with a novel fold. J. Biol. Chem.281 (29), 20171–20180. doi: 10.1074/jbc.M603452200
24
Santos R. B. Abranches R. Fischer R. Sack M. Holland T. (2016). Putting the spotlight back on plant suspension cultures. Front. Plant Sci.7, 297. doi: 10.3389/fpls.2016.00297
25
Sheva M. Hanania U. Ariel T. Turbovski A. Rathod V. K. R. Oz D. et al . (2020). Sequential genome editing and induced excision of the transgene in N. tabacum BY2 cells. Front. Plant Sci.11, 607174. doi: 10.3389/fpls.2020.607174
26
Uetz P. Göritzer K. Vergara E. Melnik S. Grünwald-Gruber C. Figl R. et al . (2024). Implications of O-glycan modifications in the hinge region of a plant-produced SARS-CoV-2-IgA antibody on functionality. Front. Bioeng Biotechnol.12, 1329018. doi: 10.3389/fbioe.2024.1329018
27
Uetz P. Melnik S. Grünwald-Gruber C. Strasser R. Stoger E. (2022). CRISPR/Cas9-mediated knockout of a prolyl-4-hydroxylase subfamily in Nicotiana benthamiana using DsRed2 for plant selection. Biotechnol. J.17, e2100698. doi: 10.1002/biot.202100698
28
Vasilev N. Grömping U. Lipperts A. Raven N. Fischer R. Schillberg S. (2013). Optimization of BY-2 cell suspension culture medium for the production of a human antibody using a combination of fractional factorial designs and the response surface method. Plant Biotechnol. J.11, 867–874. doi: 10.1111/pbi.12079
29
Xu J. Perezsanchez P. Sadravi S. (2025). Unlocking the full potential of plant cell-based production for valuable proteins: Challenges and innovative strategies. Biotechnol. Adv.79, 108526. doi: 10.1016/j.biotechadv.2025.108526
30
Yang Z. Bennett E. P. Jørgensen B. Drew D. P. Arigi E. Mandel U. et al . (2012a). Toward stable genetic engineering of human O-glycosylation in plants. Plant Physiol.160, 450–463. doi: 10.1104/pp.112.198200
31
Yang Z. Drew D. P. Jørgensen B. Mandel U. Bach S. S. Ulvskov P. et al . (2012b). Engineering mammalian mucin-type O-glycosylation in plants. J. Biol. Chem.287, 11911–11923. doi: 10.1074/jbc.M111.312918
Summary
Keywords
protein glycosylation, O-glycosylation, IgA1, Nicotiana tabacum BY-2 cells, recombinant glycoprotein
Citation
Bailly N, Grünwald-Gruber C, Peeters M, Chaumont F and Navarre C (2025) Production of recombinant IgA1 with defined mucin-type O-glycans in Nicotiana tabacum BY-2 cells. Front. Plant Sci. 16:1675517. doi: 10.3389/fpls.2025.1675517
Received
29 July 2025
Accepted
09 September 2025
Published
24 September 2025
Volume
16 - 2025
Edited by
Els Jm Van Damme, Ghent University, Belgium
Reviewed by
Kathrin Göritzer, University of Natural Resources and Life Sciences Vienna, Austria
Marcello Donini, Energy and Sustainable Economic Development (ENEA), Italy
Updates
Copyright
© 2025 Bailly, Grünwald-Gruber, Peeters, Chaumont and Navarre.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: François Chaumont, francois.chaumont@uclouvain.be; Catherine Navarre, catherine.navarre@uclouvain.be
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.