- 1College of Agriculture, Qingdao Agricultural University, Qingdao, China
- 2Marine Agriculture Research Center, Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
- 3Qingdao Key Laboratory of Coastal Saline-alkali Land Resources Mining and Biological Breeding, Qingdao, China
- 4National Center of Technology Innovation for Comprehensive Utilization of Saline-Alkali Land, Dongying, China
Salt and alkali stress are major constraints on soybean productivity, but their distinct impacts during early development remain insufficiently understood. Wild soybean (Glycine soja), a valuable genetic resource for stress tolerance, was evaluated under salt (0.6% and 1.2% NaCl) and alkali (pH 9.16) stress by assessing germination, seedling traits, ion accumulation, and transcriptomic responses. Salt stress permitted partial germination, whereas alkali stress completely suppressed radicle emergence. Seedling growth and height showed tolerance under salinity, but high pH caused severe wilting and mortality. Ion profiling revealed root Na+ sequestration with stem K+ buffering in salinity, whereas alkali stress confined Na+ to roots, maintaining the highest stem K+/Na+ ratio. Bioaccumulation and translocation factors peaked at 0.6% NaCl in wild soybean. Transcriptome analysis identified 7,355 DEGs grouped into five clusters, enriched in phenylpropanoid/flavonoid biosynthesis and hormone signaling. Salt stress upregulated genes including FLS, F3H, and F3′5′H, whereas alkali stress induced CHS, peroxidase, and CYP75B1. Ion transport regulation differed, with HKT1 and KT11 activated under salinity and NIP5–1 under alkalinity. Among 385 TF-related DEGs, MYB, ERF, bHLH, and WRKY dominated (67% of total), with complex TF-gene networks observed under salt stress. Exogenous flavonoids (rutin, eriodictyol) treatment enhanced leaf area, root length, and plant height under salt and alkali stress treatments. These results suggest that G. soja mitigates ion toxicity via root Na+ sequestration, stress-responsive gene regulation, and flavonoid-mediated growth enhancement, providing insights into adaptive mechanisms under salt and alkali stress.
1 Introduction
Saline-alkaline stress resulting from soil salinization and/or alkalization has emerged as a major global environmental challenge, significantly compromising agricultural productivity and ecological stability (Zhao et al., 2022). According to GSAS map data (https://www.fao.org/global-soil-partnership/gsasmap/en/), over 6% of the world’s soils are already affected by salinity and alkalinity (Cai et al., 2022). Unsustainable irrigation practices and the excessive fertilizers application are further exacerbating land degradation through secondary salinization. It is estimated that approximately 50% of cultivated land may become salinized by 2050 (Wang et al., 2003; Zhou et al., 2023).
Saline-alkali soils are classified into neutral-pH soil, which are rich in NaCl and Na2SO4 (pH∼7), and high-pH sodic soils, characterized by high concentrations of Na2CO3 or NaHCO3 (pH>8.5) (Hou et al., 2023).While salt stress in soil typically induces osmotic stress and ion toxicity to plant tissues, alkaline stress imposes an additional detriment due to elevated pH, rendering it more severe than salt stress alone (Fang et al., 2021; Rao et al., 2023). The high concentrations of carbonates (CO32−) and bicarbonates (HCO3−) ions in alkaline soils lead to alkaline soil pH and reduced plant nutrients availability (Sagervanshi et al., 2022). Unlike neutral saline soils, high-pH saline-alkaline soils further inhibit the uptake of essential nutrients and sodium ion (Na+) exclusion (Zhang et al., 2023). In addition, high alkalinity stress induces excessive reactive oxygen species (ROS) accumulation, causing oxidative damage to cellular proteins, lipids, and DNA, and potentially leading to cell apoptosis (Javid et al., 2012; Sun et al., 2016; Zhang et al., 2017; An et al., 2020).
Cultivated soybean (Glycine max L.) was domesticated from its wild progenitor Glycine soja. During this domestication process, only approximately 50% of the genes present in wild soybean were retained, resulting in the loss of numerous stress-adaptive genes (Kofsky et al., 2018). Consequently, wild soybean represents a valuable genetic resource for enhancing salt-alkali tolerance in cultivated varieties. Studies have shown that wild soybean maintains higher genetic diversity and superior adaptability to abiotic stresses compared to cultivated varieties (Ge et al., 2010; Cai et al., 2022). For instance, under salt and alkali stress, wild soybean accumulates higher concentrations of osmo-protectants such as proline and glycine betaine, which contribute to maintaining osmotic balance and protecting cellular structures protection (Wu et al., 2014). Furthermore, wild soybean exhibits notably lower Na+ accumulation, higher K+ retention, and consequently a higher K+/Na+ ratio in both leaves and roots compared to salt-sensitive cultivated soybean germplasms under salt stress (Liu et al., 2017).
Nonetheless, prior research on wild soybean have focused exclusively on its response to either salt stress or alkali stress alone. Consequently, research directly comparing the physiological and molecular responses of wild soybean to both stresses remains limited (Sun et al., 2025). Identifying stress resistance genes in wild soybean and utilizing them into cultivated germplasm could enhance stress resistance and overall crop quality, while also addressing the issues of limited genetic diversity and inadequate environmental adaptability (Xie et al., 2019).The objective of this work is to (i) examine the responses of wild soybean to both saline and alkaline stresses at physiological, biochemical, and molecular levels, and (ii) investigate the mechanisms underlying its tolerance to these stresses. This work will not only contribute to the improvement of salt and alkali tolerance in soybean breeding but also offers significant insights into the broader molecular mechanisms underlying plant stress responses and environmental adaptability.
2 Materials and methods
2.1 Plant materials
Wild soybean seeds were collected from Dongying City (37°17053.4500 N, 118°37015.0400 E), Shandong Province, China. The seeds were the self-pollinated progeny of a single wild soybean plant and were stored under laboratory conditions for experimental use (Xu et al., 2020b).
2.2 Seeds pre-treatment, stress application, and plant growth conditions
To enhance the germination rate of wild soybean seeds, the seeds were soaked in concentrated sulfuric acid and repeatedly agitated to disrupt the mud coating. Following treatment, the seeds were thoroughly rinsed three times with deionized water and subsequently used for experimental purposes. Salt stress treatments was treated were administered using 0.6% and 1.2% NaCl solutions prepared in 1/20 strength Murashige and Skoog (MS) medium. Alkaline stress treatments were applied using Na2CO3-NaHCO3 buffer solution (also in 1/20 MS) at pH values of 9.16 (0.1M Na2CO3:0.1M NaHCO3 = 1:9), 9.90 (0.1M Na2CO3:0.1M NaHCO3 = 5:5) and 10.83 (0.1M Na2CO3:0.1M NaHCO3 = 8:2). A 1/20 MS solution without NaCl or alkali buffer served as the control.
For the growth experiment under salt and alkaline stress, seeds were sown in vermiculite and irrigated with 1/20 MS solution. After three weeks, uniform seedlings were exposed to the respective salt and alkali treatments. Each treatment comprised 32 seedlings per replicate, with three replicates per treatment. Photographs of the aerial portions were taken on the third and seventh days after treatment. After 7 days of treatments, root, stem, and leaf tissues were harvested, immediately immersed in liquid nitrogen, and stored in -80°C for subsequent physiological, biochemical, and transcriptomic analyses. Seed germination and plant growth conditions were consistent to previous protocols (Xu et al., 2020b), including a 16:8 h light:dark photoperiod, a temperature range of 20-22°C, and relative humidity of 65–75%.
2.3 Sodium and potassium content measurement
The concentrations of Na+ and K+ were determined following a modified protocol from a previous study (Xu et al., 2020a). In summary, 0.25 g of oven-dried and powdered plant tissues were digested in 5 mL of HNO3 at 110°C for approximately 6 hours until a clear, colorless solution was achieved. After cooling, the digested samples were diluted to a volume of 10 mL using deionized water. Na+ and K+ contents were measured using an optical emission spectrometer (ICP-OES, Optima 8000, PerkinElmer, USA).
2.4 Bioaccumulation factor and translocation factor calculation
The bioaccumulation factor (BF), representing the ability of plant to uptake and retain ions, was determined based on previously described methods with minor modifications. BF was calculated as the ratio of Na+ content in plant tissue (root, stem, and leaf; mg kg-1 dry matter) to) to its corresponding concentration in the soil (Yoon et al., 2006). The translocation factor (TF), indicating the efficiency of ion transport from roots to shoots, was calculated as the ratio of Na+ content in the shoots to that in the roots, both expressed on a dry matter basis (mg kg-1) (Padmavathiamma and Li, 2007; Zhou et al., 2013).
2.5 Transcriptome sequencing and KEGG enrichment analysis
Total RNA was extracted from root tissues of seedlings exposed to salt stress (0.6% NaCl and 1.2% NaCl), alkali stress (pH=9.16), and control conditions using an EasyPure®plant RNA kit (ER301-01, TransGen Biotech). RNA quality was assessed using 1% agarose gel, a 2100 Bioanalyzer (Agilent Technologies, USA), and a NanoDrop2000 spectrophotometer (Thermo FisherScientific, USA). High-quality RNA was used for library construction and sequencing, which was performed by Shanghai Majorbio Bio-Pharm Biotechnology Co., Ltd. (Shanghai, China). Raw reads were filtered using FASTP (https://github.com/OpenGene/fastp/), and clean reads were aligned to the reference genome using while HISAT2 (http://ccb.jhu.edu/software/hisat2/). Transcript assembly was carried out with StringTie (https://ccb.jhu.edu/software/stringtie/). Each treatment was represented by three biological replicates. Transcript expression levels were quantified using the transcripts per million reads (TPM) method. Differentially expressed genes (DEGs) were identified using DESeq2, DEGseq, edgeR, Limma, or NOIseq tools (threshold: P-adjust ≤ 0.05 and |log2 fold change| ≥ 1). Gene ontology(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted using the KOBAS database (http://bioinfo.org/kobas) to elucidate relevant metabolic pathways and biological functions associated with DEGs.
2.6 Flavonoids supplementation experiment
Wild soybean seedlings were cultured as described above. Half of the three-days-old seedlings received 100 μM of flavonoids rutin and eriodictyol for three days, then all the seedlings were treated with 0.6% NaCl, 1.2% NaCl, or a Na2CO3-NaHCO3 buffer solution (pH=9.16), while seedlings irrigated with 1/20 MS solution served as the control. Five days after stress exposure, plant height, root length, and leaf area were measured in all groups.
2.7 RNA extraction and reverse transcription quantitative PCR
Total RNA was isolated from the samples using RNA Kit reagent (Vazyme Biotech, China) following the manufacturer’s instructions. First-strand cDNA was synthesized using a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech, China) according to the manufacturer’s protocol. RT-qPCR was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech). GsGAPDH was used as a housekeeping gene. Each sample was analyzed in three biological replicates, and relative expression levels were obtained using the 2−△△Ct method.
2.8 Data analysis
All data were subjected to one-way analysis of variance (ANOVA), and means were compared using Tukey’s post hoc test at P < 0.05 level using SPSS software v 17.0 (SPSS Inc., Chicago, IL, USA).
3 Results
3.1 Salt and alkali stress inhibit germination and seedling development of wild soybean
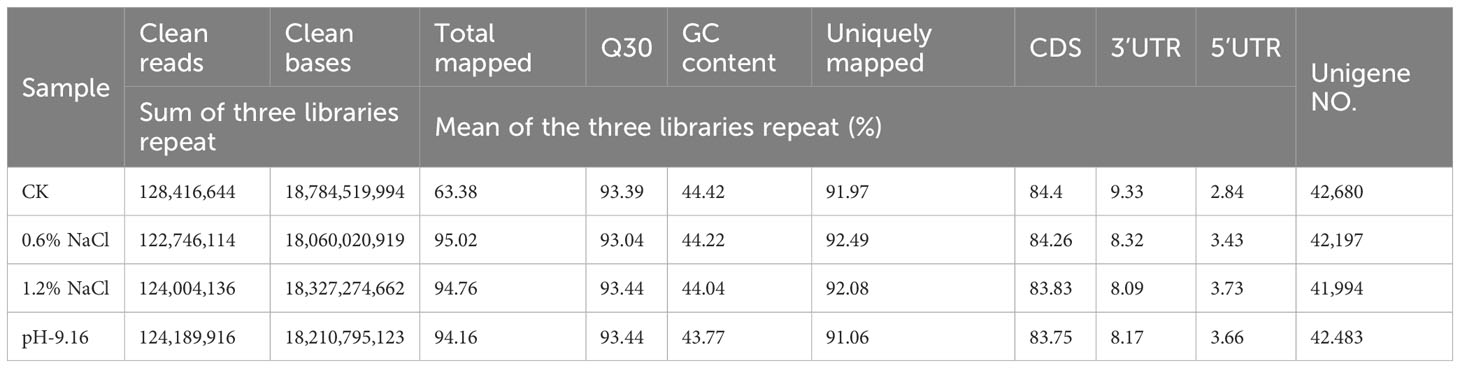
Salt and alkali stress significantly inhibited the germination and early development of wild soybean (G. soja). Notably, alkali stress exerted a greater inhibitory effect on seed germination than salt stress. At pH 10.83, no discernible indications of seed germination were observed. Under milder alkali treatments (pH 9.9 and pH 9.16), radicle emergence was limited to penetration of the seed coat, whereas control seeds developed fully extended radicles and elongated cotyledons. In contrast, seeds subjected to 0.6% and 1.2% NaCl showed radical emergence, indicating milder inhibition compared to alkali stress still reduced development relative to the control. Germination performance under 0.6% salt stress was significantly better than that under 1.2% salt stress (Supplementary Figure 1). At the seedling stage, wild soybean exhibited improved stress resistance compared to the germination stage. No apparent differences were observed among seedlings treated with salt (0.6% and 1.2% NaCl) and alkali (pH=9.16) stress when compared to the control. However, seedlings treated with pH 10.83 displayed severe wilting within 3 days approached complete mortality by day 7 (Supplementary Figure 2). After 7 days of stress exposure, measurements of plant height showed a slight reduction in treated plants compared to the control; however, the differences were not statistically significant (Figure 1A). These findings indicate that short-term salt and alkali stress had minimal impact on vertical growth during the early seedling stage.
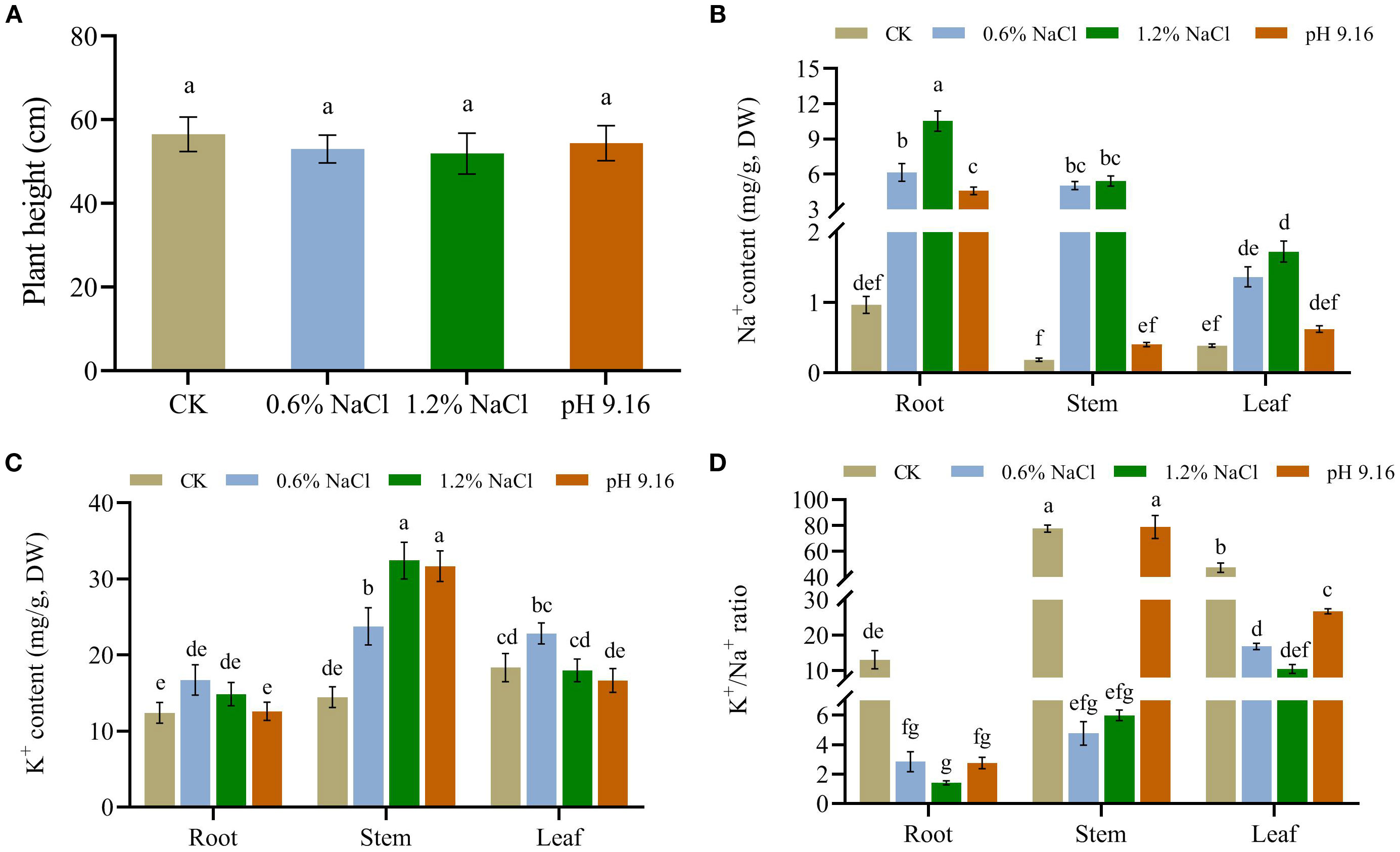
Figure 1. Plant height and ion distribution in wild soybean under salt and alkali stress. (A) Plant height; (B) Na+ content; (C) K+ content; and (D) K+/Na+ ratio in different tissues of wild soybean subjected to control (CK), salt (0.6% and 1.2% NaCl), and alkali (pH 9.16) treatments. Different letters above the bars represent significant differences among treatments (ANOVA and Tukey’s post hoc test, P < 0.05).
3.2 Root is the primary sodium-accumulating organ in wild soybean
Ion profiling under salt stress revealed a progressive increase in Na+ content from roots to leaves, with the highest accumulation observed in roots at 1.2% NaCl. In contrast, Na+ under alkali stress was predominantly confined to roots (Figure 1B). K+ was primarily concentrated in stems under all salt stress levels, with the highest levels detected in stems and leaves at 1.2% NaCl, followed by alkali stress (pH 9.16) in stems (Figure 1C). The K+/Na+ ratio was significantly reduced in all tissues under salt stress, particularly in roots, but showed a moderate increase toward stems and leaves. Under alkali stress, stems maintained the highest K+/Na+ ratios among all tissues, suggesting their critical role in ionic homeostasis(Figure 1D). These results suggest that wild soybean mitigates ion toxicity through root-localized Na+ retention and stem-based K+ buffering mechanisms.
Consistent with tissue ion content, the bioaccumulation factor (BF) was highest in roots across all treatments, exceeding a value of 1.44 under 0.6% NaCl, 1.23 under 1.2% NaCl, and 1.00 under alkaline conditions (pH 9.16). The maximum BF was observed in roots under 0.6% NaCl. Additionally, the stem BF under 0.6% NaCl also exceeded 1 (1.18). The highest translocation factor (TF) was recorded in plants subjected to 0.6% NaCl; however, TF values remained below 1 across all treatments, with the lowest observed under alkali stress. These results further indicate that roots function as the primary site of Na+ accumulation in wild soybean (Table 1).
Table 1. Bioaccumulation and translocation in root, stem, and leaf tissues of wild soybean under control, salt, and alkali stress conditions.
3.3 Genes enriched in wild soybean under salt and alkali stress
Since root was the primary Na+ storage organ, transcriptome sequencing was performed on root samples under salt, alkali, and control conditions. A total of 499,356,810 clean reads (73.38 Gb) were generated. The Q30 score exceeded 93.00% and GC content averaged ~44.00%. Over 92.00% of clean reads uniquely mapped to the G. soja reference genome (https://datahub.wildsoydb.org/). The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2025), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA028818) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Ultimately, about 42,000 unigenes were identified in each sample (Table 2), resulting in a total of 55,539 non-redundant unigenes (Supplementary Table 1). Among them, 55,534 were in at least one database, with the NR database showing the highest annotation rate (55,531) (Figure 2A; Supplementary Table 1). Principal component analysis (PCA) showed clear transcriptomic separation across all samples (Figure 2B). The samples subjected to alkali stress treatment clustered more closely to the control compared to those under salt stress, whereas the 1.2% NaCl treatment was distinctly separated from all other samples, indicating a greater divergence in response.
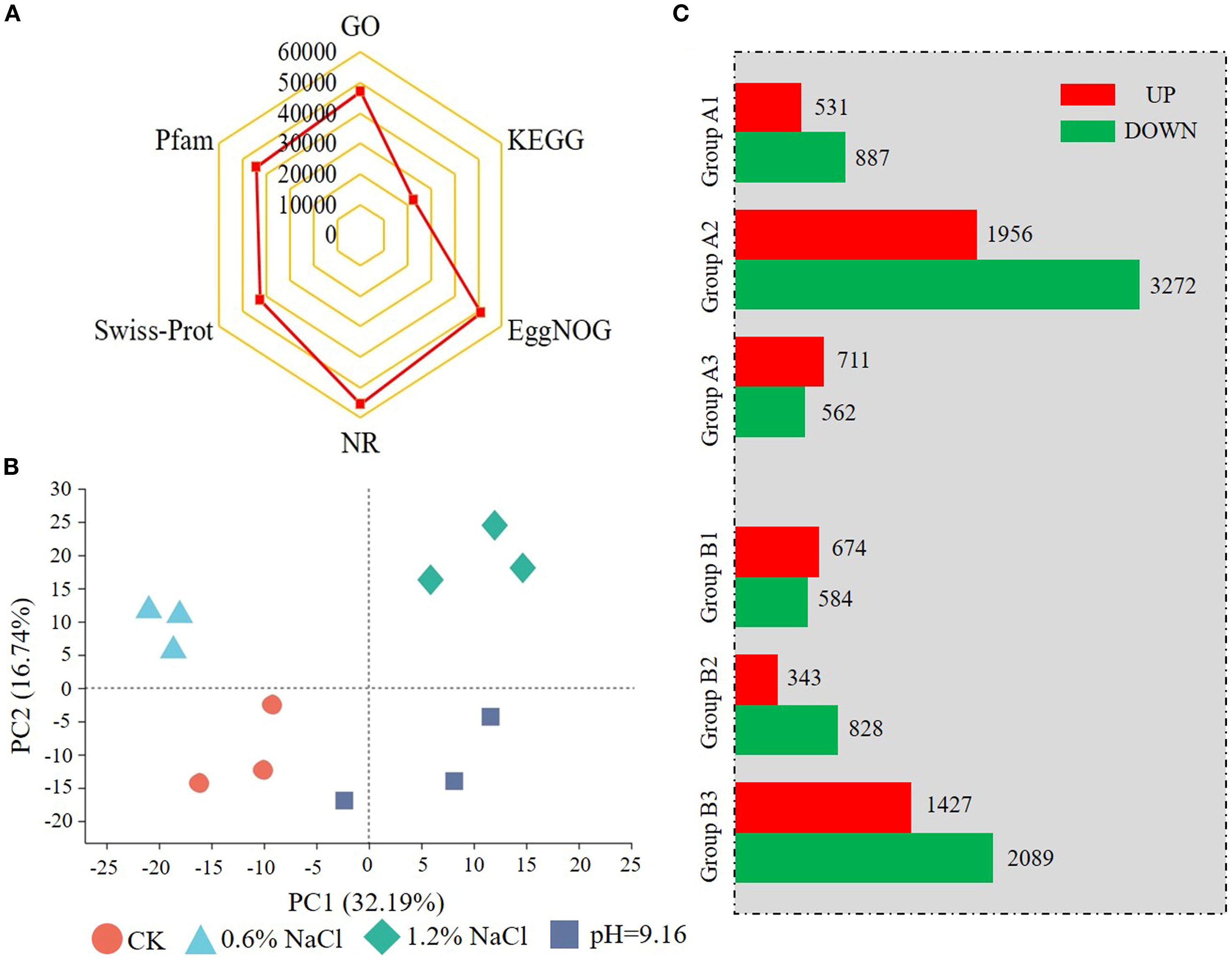
Figure 2. Principal component analysis (PCA), gene annotation statistics (A, B), and differentially expressed gene (DEG) statistics (C) of wild soybean under salt and alkali stress. In (C) DEGs were identified using thresholds of P < 0.05 and |fold change| ≥ 2. Comparisons include: A1 (0.6% NaCl vs CK), A2 (1.2% NaCl vs CK), A3 (1.2% NaCl vs 0.6% NaCl), B1 (pH 9.16 vs CK), B2 (pH 9.16 vs 0.6% NaCl), and B3 (pH 9.16 vs 1.2% NaCl).
DEGs were identified across six comparison groups. Group A consisted of comparisons among salt treatments and control, CK versus 0.6% NaCl (Group A1), 1.2% NaCl (Group A2), and 1.2% NaCl versus 0.6% NaCl (Group A3). While Group B compared alkali stress (pH 9.16) against CK (Group B1), 0.6% NaCl (Group B2), and 1.2% NaCl (Group B3). Group A2 had the highest number of DEGs (5,228), with 1,956 upregulated and 3,272 downregulated DEGs. Group B3 followed with 3,516 DEGs, including 1,427 up-regulated and 2,089 down-regulated (Figure 2C; Supplementary Table 2). These results indicate that high salt stress (1.2% NaCl) strongly alters gene expression in wild soybean.
3.4 DEGs clusters obtained from hierarchical clustering
A total of 7,355 non-redundant DEGs were identified across all comparison groups. Hierarchical clustering categorized these DEGs into five distinct expression clusters (Figure 3). Cluster I and IV were down-regulated under salt stress compared to both control and alkali stress. GO enrichment revealed that Cluster I was enriched in “lipid metabolic process” and Cluster IV in “plasma membrane”. Conversely, DEGs in Cluster III were up-regulated under salt stress relative to control. Enriched GO terms included “galactosyltransferase activity”, “transmembrane transporter activity”, and “transporter activity”, indicating involvement of transmembrane transporters and antioxidant-related enzymes in salt stress adaptation. Cluster II displayed down-regulation under both salt and alkali stress compared to the control, with “cell wall organization” as the most enriched term. Cluster V showed exclusive and markedly elevated expression under 1.2% NaCl treatment, with a secondary response observed under 0.6% NaCl and alkali stress treatments. In conclusion, DEGs in Clusters IV were exclusively upregulated under alkali stress but downregulated under salt stress, whereas DEGs in Cluster I and III were specifically different expressed under salt stress. Accordingly, Clusters I, III, and IV were selected for further analysis and downstream investigation.
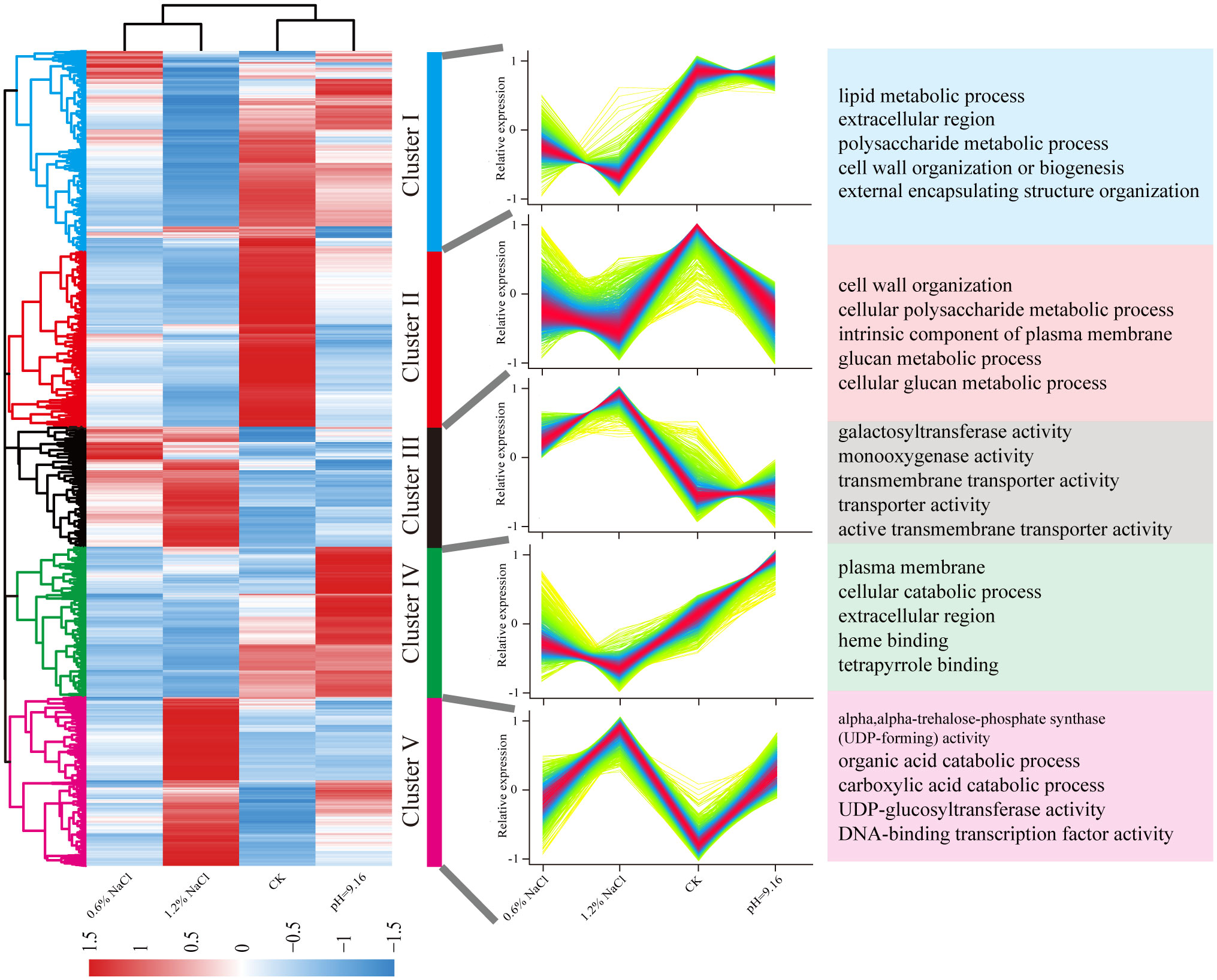
Figure 3. Hierarchical clustering of differentially expressed genes based on normalized TPM values. Heatmap color gradients indicate expression levels, and subclusters represent gene groups enriched in specific GO terms. DEGs expression is visualized as heatmap based on log2(fold change), with red and blue indicating high and low expression, respectively.
3.5 DEGs enriched in metabolic pathways under salt and alkali stress
GO enrichment analysis showed that DEGs in Clusters I and IV exhibited similar expression patterns, while DEGs in Cluster III displayed distinct expression profiles under salt stress treatments. The DEGs in Cluster IV were upregulated under alkali stress treatment. KEGG enrichment of the Clusters I (specifically downregulated under salt stress) and Cluster IV(downregulated under salt stress and upregulated under alkali stress) revealed pathways associated with flavonoid metabolism, including “phenylpropanoid biosynthesis” and “flavonoid biosynthesis”. The KEGG of Cluster III (upregulated under salt stress) were enriched in the pathways including “Galactose metabolism”, “Plant hormone signal transduction” and “ABC transporters” (Figure 4).
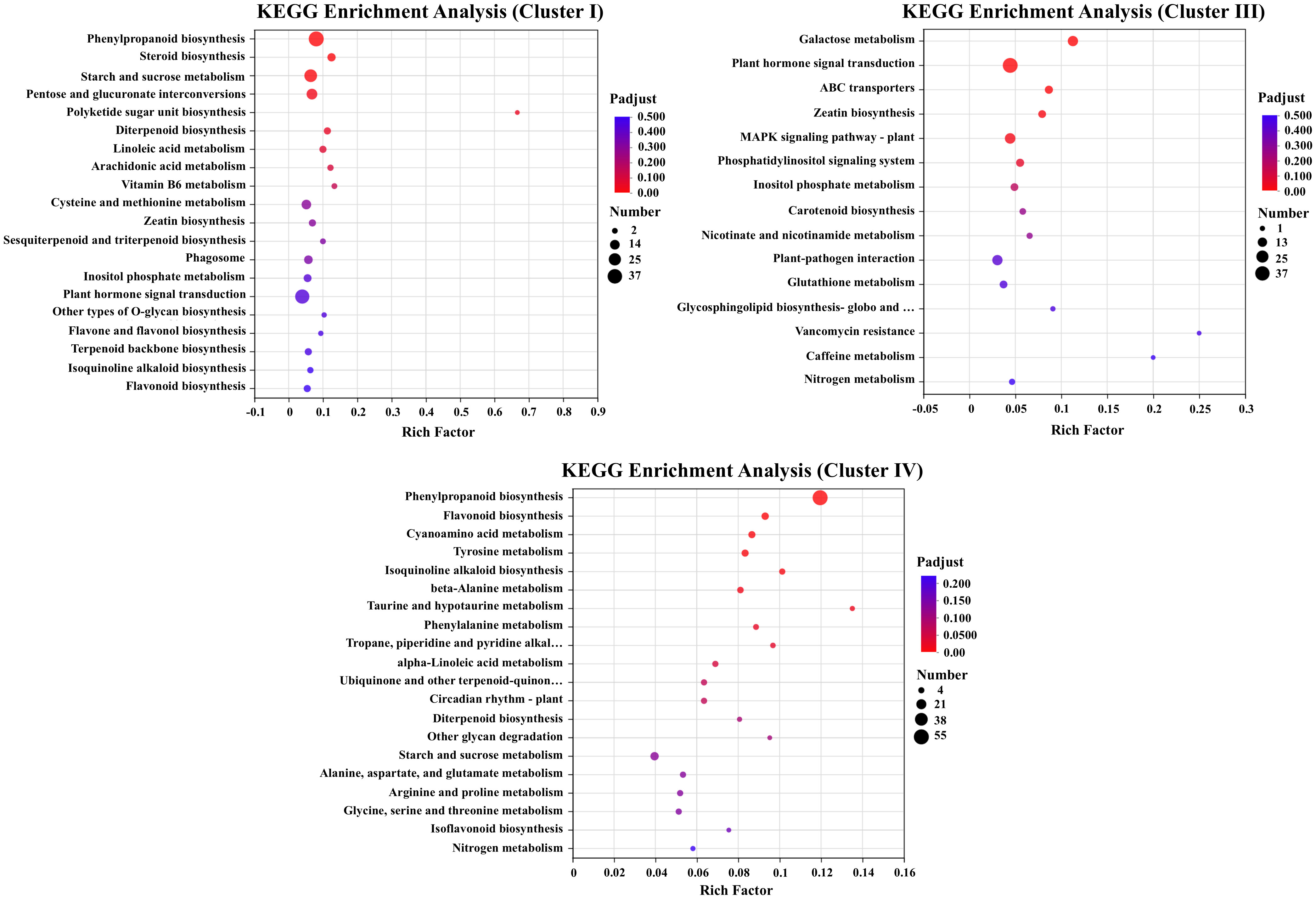
Figure 4. KEGG pathway enrichment analysis of DEGs in Cluster I, Cluster III, and Cluster IV, highlighting significantly enriched biological pathways under salt and alkali stress.
3.6 Differential expression of phenylpropanoid and flavonoid biosynthesis genes
To further elucidate the differential metabolic responses, we analyzed expression changes in structural genes within phenylpropanoid and flavonoid biosynthesis pathways. Under alkali stress, a phenylalanine ammonialyase (PAL) gene and 30 peroxidase genes were specifically upregulated. Moreover, under alkali stress, higher expression was observed for 13 chalcone synthase (CHS) genes and 5 chalcone isomerase (CHI) genes. In addition, one UDP-glycosyltransferase 79B1 (UGT79B1) and one flavonoid 3’-monooxygenase (CYP75B1) were exclusively regulated under this treatment. In contrary, two flavone synthase (FLS) genes and flavonoid 3’,5’-hydroxylase (F3’5’H) were specifically induced under salt stress. These results suggest that flavonoids may play a more critical role in alkali stress adaptation in wild soybean (Figure 5; Supplementary Table 3).

Figure 5. Gene expression profiles of the DEGs annotated to phenylpropanoid and flavonoid biosynthesis pathways in wild soybean under salt and alkali stress. Heatmap shows log2(fold change), with red and blue indicating high and low expression, respectively. Key enzymes include PAL, phenylalanine ammonia lyase; 4CL, 4-coumaroyl CoA ligase; HCT, shikimate O-hydroxycinnamoyltransferase; CAD, cinnamyl-alcohol dehydrogenase; COMT, caffeic acid 3-O-methyltransferase; UGT72E, coniferyl-alcohol glucosyltransferase; F6H, flavonoid 6-hydroxylase; F3H, flavanone 3 -hydroxylase; F3’5’H, flavonoid 3’,5’-hydroxylase; CHS, chalcone isomerase; CHI, chalcone isomerase; FLS, flavonol synthase; CYP81E, isoflavone 2’-and 3’-hydroxylases.
3.7 Sodium transport-related regulatory network under salt and alkali stress
In this study, we identified 29 DEGs associated with Na+ influx and storage, most of which were upregulated under salt stress, particularly in the 1.2% NaCl vs. CK comparison group (Table 3). This differential expression correlated with observed Na+ accumulation levels. Notably, Glysoja.03G007427 (encoding Sodium transporter HKT1) and Glysoja.15G041067 (encoding potassium transporter KT11) were specifically upregulated under salt stress. Conversely, Glysoja.20G054400 and Glysoja.10G027804 (encoding aquaporin NIP5-1) responded specifically to alkali stress (Table 3).
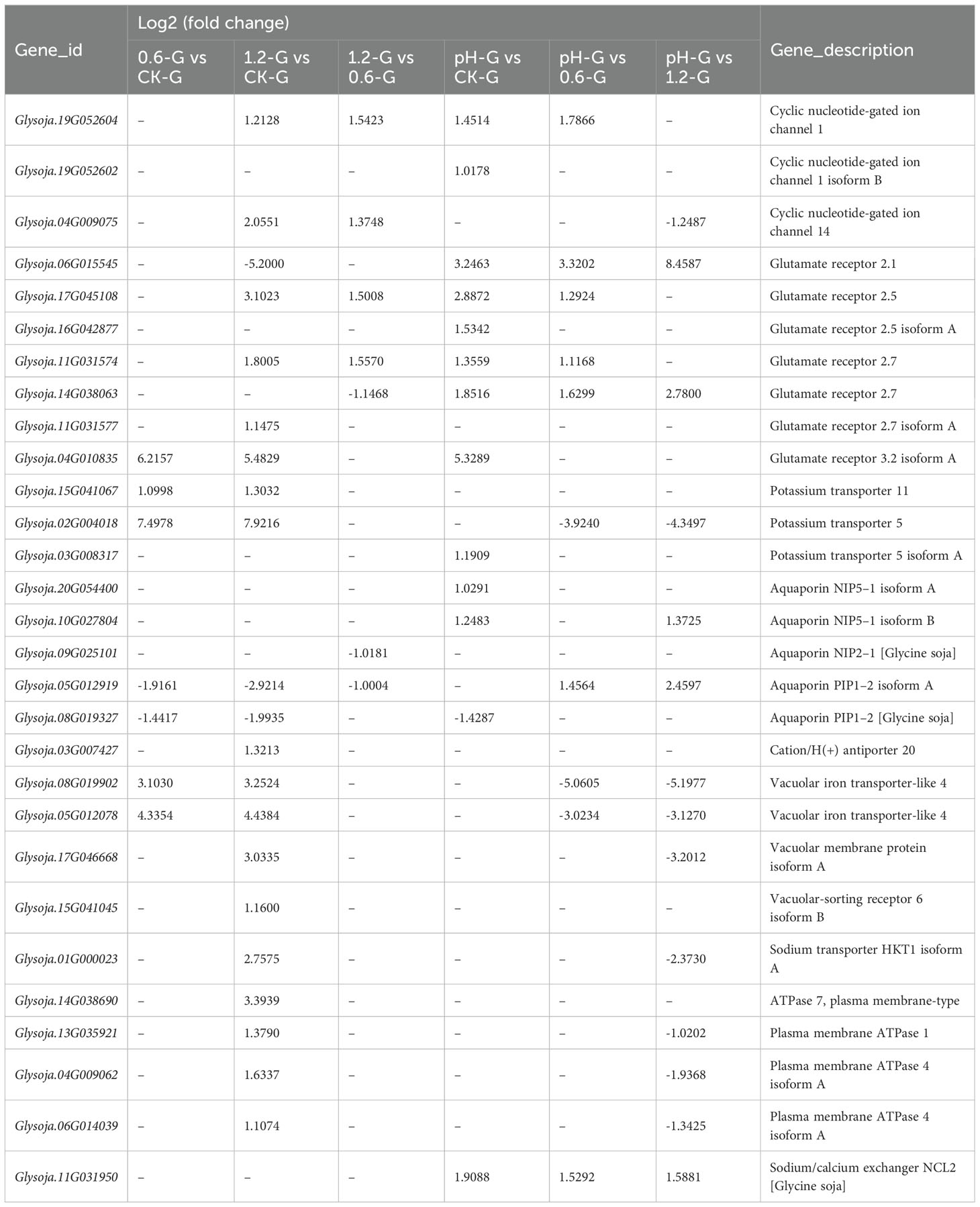
Table 3. Differentially expressed genes related to Na+ influx and storage across all sample comparisons, including log2 fold change, gene description, and gene ID.
Moreover, in total, 385 transcription factor (TF)-encoding DEGs were identified, predominantly from MYB (19%), ERF (17%), bHLH (17%), and WRKY (14%) families, collectively accounting for 67% of all TFs (Supplementary Figure 3). Co-expression network analysis revealed a more complex TF-gene regulatory interaction under salt stress than alkali stress (Figure 6). Salt-responsive TFs included Glysoja.11G029929, Glysoja.11G029815, and Glysoja.20G054770, whereas alkali-responsive TFs included Glysoja.06G015139, Glysoja.09G024775, and Glysoja.16G042505. Key Na+ transporter genes responsive under salt stress included Glysoja.08G019327, Glysoja.15G041067, and Glysoja.09G025101, while Glysoja.10G027804, Glysoja.15G041045, and Glysoja.15G041067 were associated with alkali stress.
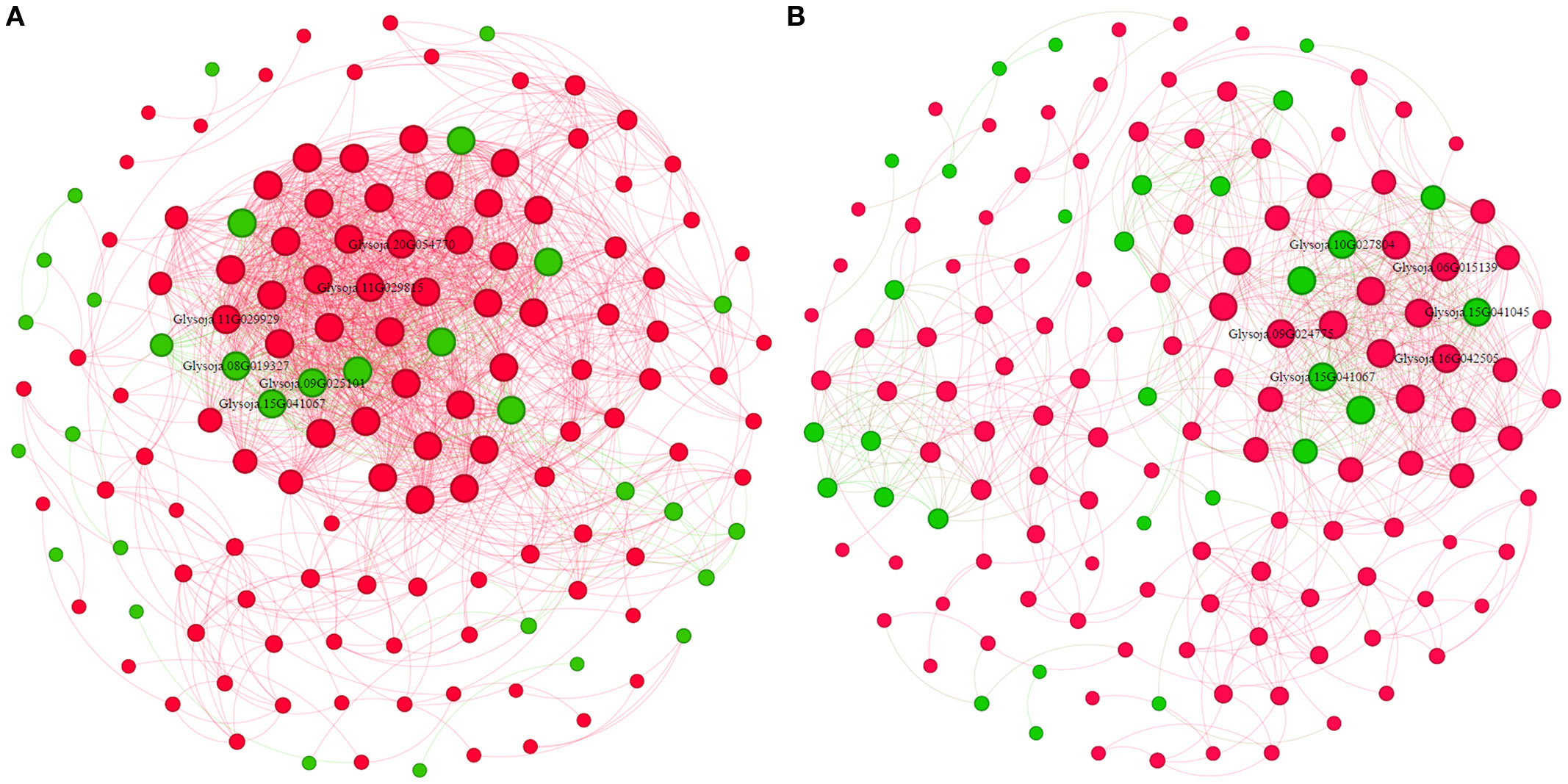
Figure 6. Co-expression network of transcription factors and Na+-related differentially expressed genes (DEGs) under (A) salt and (B) alkali stress. Green nodes indicate Na+ influx or storage genes; red nodes denote TFs.
3.8 qRT-PCR validation
Six DEGs related to flavonoid biosynthesis or Na+ influx and storage were randomly selected for qRT-PCR analysis, using specific primers designed using Primer Premier 5.0 (Supplementary Table 4), to determine the mRNA levels after exposure to salt (0.6% NaCl and 1.2% NaCl) and alkali (pH 9.16) stress treatments,. The results showed that the relative gene expression trends of qRT-PCR were consistent with RNA-seq, demonstrating the reliability of the RNA-seq data (Supplementary Figure 4).
3.9 Flavonoids supplementation improves salt and alkali stress tolerance
To further validate the functional relevance of flavonoids, exogenous applications of rutin and eriodictyol were conducted. Rutin (a flavonol glycoside) and eriodictyol (a central flavanone precursor) were selected due to their direct association with DEGs (CYP75B1, F3’5’H, F6H) in the flavonoid pathway. Under 1.2% NaCl and pH 9.16 treatments, flavonoid supplementation significantly increased plant height (Figures 7A, C). Root length was significantly enhanced under 0.6% NaCl stress, while leaf area showed a increase across all flavonoid-treated samples (Figure 7B, E). Collectively, these results demonstrate that flavonoids can enhance salt and alkali stress tolerance in wild soybean by promoting key growth parameters.
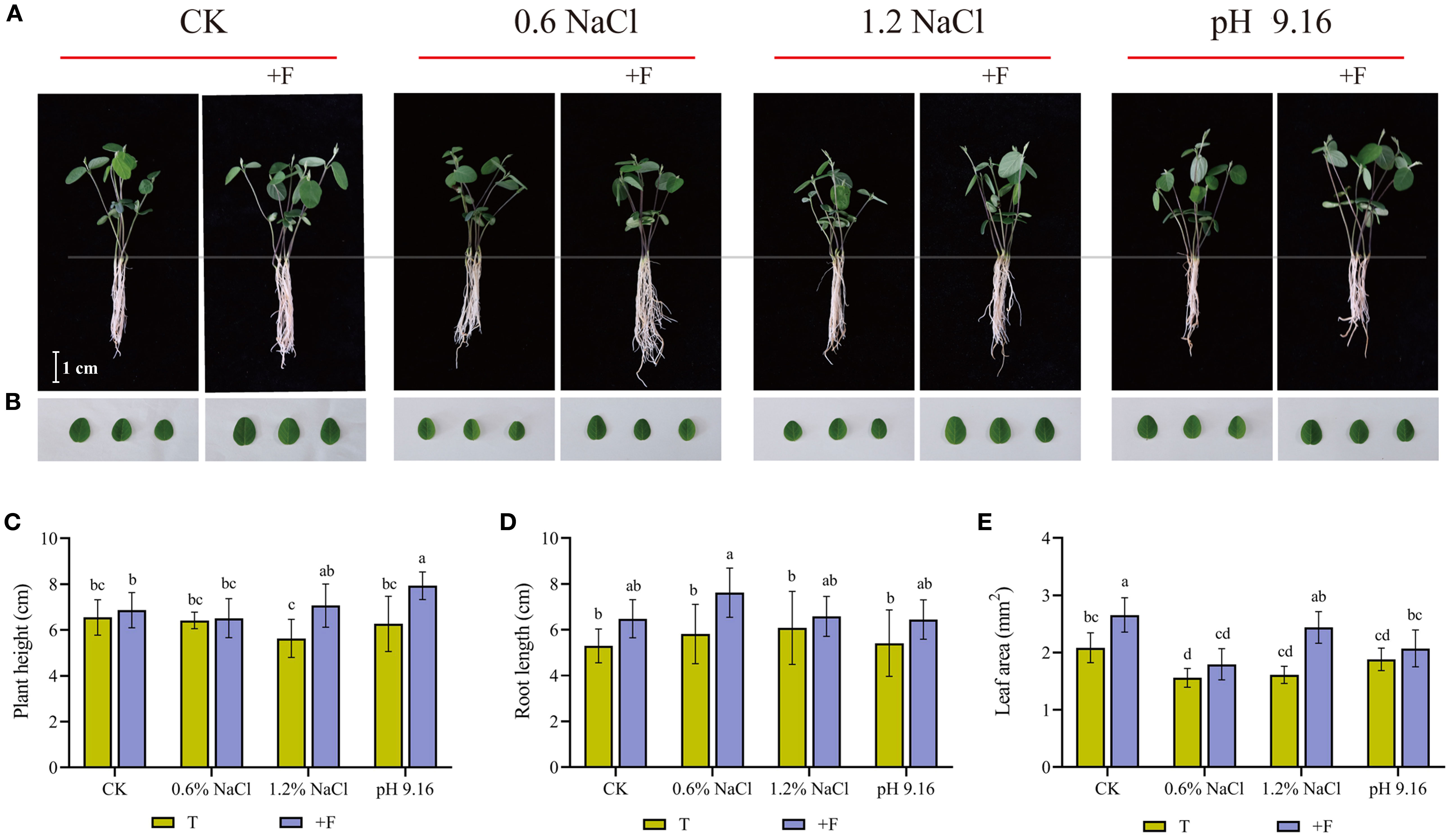
Figure 7. Assessment of morphological parameters in wild soybean under salt and alkali stress with and without flavonoid treatment. (A) Seedling morphology, (B) leaf appearance, (C) plant height, (D) root length, and (E) leaf area. Different lowercase letters denote statistically significant differences (ANOVA and Tukey’s post hoc test, P < 0.05).T, treatments without flavonoid; +F, treatments with flavonoid.
4 Discussion
Salt and alkali stress significantly hinder plant growth by disturbing ion equilibrium and inducing oxidative stress (Fang et al., 2021). A critical adaptive strategy observed in stress-tolerant plants involves the sequestration of excess Na+ in root tissues, thereby protecting photosynthetically active leaves (Wu et al., 2018). In this study, wild soybean germplasms showed significantly higher Na+ accumulation in roots than shoots under both salt and alkali stress. Na+ translocation was limited under alkali stress but more extensive under salt stress, indicating root-based Na+ sequestration as a key detoxification mechanism, particularly in high-pH environments (Jin et al., 2021). Transcriptomic analysis revealed that salt stress primarily upregulated Na+ sequestration-related genes, while alkali stress preferentially activated flavonoid biosynthesis pathways, suggesting a shift toward antioxidant and pH-buffering responses. These results underscore distinct stress-specific strategies in wild soybean, with root-localized ion retention and differential gene expression playing central roles in salinity and alkalinity tolerance.
Morphological observations revealed that wild soybean subjected to salt and alkali treatments exhibited suppressed seed germination and impaired seedling growth, with more pronounced inhibition under alkali stress. This is consistent with osmotic stress-induced growth restriction as previously reported (Li et al., 2025a). Although a slight reduction in plant height was observed, the difference was not statistically significant. These results suggested that alkali stress exerts a stronger inhibitory effect on early developmental stages than salt stress, likely due to the combined effects of high pH and osmotic imbalance (Paz et al., 2012).
Ion profiling under salt stress revealed a progressive increase in Na+ from roots to leaves, with roots showing the highest accumulation. Under alkali stress, Na+ was largely confined to roots. K+ was predominantly enriched in stems across all salt treatments, followed by alkali stress. The K+/Na+ ratio declined significantly under salt stress, especially in roots. In contrast, alkali stress maintained K+/Na+ ratios close to control levels, with stems showing the highest buffering capacity. This tissue-specific ion partitioning reflects a key mechanism for maintaining ionic homeostasis under saline-alkaline stress, consistent with previous reports in soybean and cotton (Gossypium hirsutum L.) that highlight the importance of ion homeostasis in stress tolerance (Guo et al., 2020; Sun et al., 2021a; Cai et al., 2022). This pattern was further supported by bioaccumulation (BF) and translocation (TF) factors. BF values were highest in roots across all treatments, particularly under high salinity, indicating effective root-based Na+ sequestration, a trend consistent with previous studies in soybean and rice (Li et al., 2016; Wang et al., 2022a). TF values were elevated in stems under salt stress, reflecting active Na+ translocation, as similarly reported by Xue et al. (2022). In contrast, under alkali stress, TF remained low across all tissues, suggesting restricted ion mobility likely due to high pH-induced ion transport inhibition (Yang et al., 2024). These results confirm that wild soybean relies on root ion retention and limited translocation under alkali conditions, whereas salt stress permits greater Na+ mobility, reflecting distinct ion management strategies in response to different stress types.
To further elucidate the regulatory mechanisms underlying salt and alkali stress responses in wild soybean, transcriptomic profiling of root tissues was conducted. DEGs were grouped into five clusters via hierarchical clustering. Clusters I and IV showed elevated expression under control and alkali stress conditions and were significantly enriched in lipid metabolism, cellular catabolic processes, and flavonoid biosynthesis pathways. This suggests a shift toward antioxidant defense and metabolic adjustment to mitigate high-pH-induced oxidative and structural stress. Such responses are consistent with previous findings demonstrating the protective role of flavonoids and lipid remodeling under alkali stress conditions (Xu et al., 2021; Gao et al., 2022). Flavonoids, synthesized via the phenylpropanoid pathway, act as key antioxidants and modulators of osmotic stress responses (Liu et al., 2024; Wang et al., 2024). Under alkali stress, significant enrichment of phenylpropanoid and flavonoid biosynthesis pathways was observed, involving key genes such as PAL, 4CL, COMT, CAD, UGT72E, and POD, which are associated with lignin and flavonoid accumulation, contributing to enhanced structural integrity and reactive oxygen species (ROS) scavenging (Chen et al., 2019; Yan et al., 2025) (Preisner et al., 2014; Han et al., 2021; Liang et al., 2022; Ma et al., 2023). This response reflects a common protective strategy in plants under high-pH conditions, as previously reported in soybean and rice (Shao et al., 2016; Lu et al., 2022). Collectively, these findings highlight the central role of secondary metabolism in reinforcing alkali stress tolerance in wild soybean. In contrast, Cluster III was specifically upregulated under salt stress and enriched in plant hormone signal transduction and ABC transporter pathways, indicating an active role of transmembrane transport and hormone-mediated regulation in salinity adaptation. This is supported by earlier reports highlighting the involvement of ABC transporters and hormone crosstalk in salt tolerance mechanisms (Wang et al., 2023; Sahakyan and Sahakyan, 2025). The transcriptome analysis also revealed upregulation of genes involved in ion transport, particularly Na+/H+ antiporters and high-affinity K+ transporters, supporting their established roles in ionic detoxification and homeostasis in soybean (Sun et al., 2021b). While Clusters I and IV were enriched in genes related to secondary metabolism and oxidative defense under alkali stress, Cluster III showed predominant expression of ion transporters under salt treatment, highlighting a transcriptional divergence between stress types. These findings suggest that wild soybean roots coordinate distinct molecular responses, limiting Na+ mobility and enhancing ROS defense under alkali stress and activating ion transport mechanisms under salinity. This stress-specific partitioning of transcriptional programs underscores the adaptive flexibility of wild soybean to contrasting soil conditions.
Co-expression network analysis between DEGs and TFs revealed significant upregulation of key Na+ transport and homeostasis-related genes under salt stress. These included HKT1 (Glysoja.01G000023), Na+/H+ antiporters, and H+-ATPases, which facilitate cytosolic Na+ exclusion and vacuolar compartmentalization, an essential mechanism for mitigating ion toxicity (Horie et al., 2009; Yang and Guo, 2018; Wang et al., 2022b). Salt stress enhances transmembrane electrochemical gradients, facilitating Na+ influx into root cells via non-selective cation channels (NSCCs), cyclic nucleotide-gated channels (CNGCs), glutamate receptors (GLRs), and high-affinity K+ transporters (HKTs) (Demidchik and Maathuis, 2007; Keisham et al., 2018). Excess Na+ disrupts ion homeostasis, triggering responses to maintain Na+/K+ ratios (Munns and Tester, 2008; Jia et al., 2017). HKT1 transporters play a central role in regulating long-distance Na+ transport and limiting root-to-shoot translocation. Decreased VcHKT1;1 transcript levels in blueberry plants led to increased Na+ concentrations in xylem sap and higher leaf Na+ contents compared with wild-type plants, indicating that VcHKT1;1 promotes leaf Na+ exclusion by retrieving Na+ from xylem sap (Song et al., 2023). In addition to xylem Na+ unloading, ScHKT1;2 was reported to be involved in Na+ uploading into the phloem, promoting Na+ recirculation from aerial parts to the roots in tomato (Romero-Aranda et al., 2021). The salt-specific upregulation of HKT1 in this study suggests its involvement in root-localized Na+ retention in wild soybean. TFs enriched in the MYB, ERF, bHLH, and WRKY families were co-expressed with Na+ transporters under salt stress and with flavonoid biosynthesis genes under alkali stress, consistent with their established roles in ion homeostasis and oxidative stress responses (Wang et al., 2023). Notably, the regulatory networks under salt stress were more complex than those under alkali stress, suggesting that salt stress elicits a broader transcriptional reprogramming involving both ion transport and regulatory pathways. This highlights the distinct and dynamic coordination of TFs and transporters in modulating soybean responses to different types of ionic stress.
Exogenous application of flavonoids (rutin and eriodictyol) enhanced tolerance to both salt and alkali stress, validating their functional relevance. The selection of rutin and eriodictyol for exogenous flavonoid supplementation in soybean was based on their central roles in the flavonoid biosynthetic pathway and their involvement in stress responses (Gautam et al., 2023). Rutin, a flavonol glycoside, and eriodictyol, a core flavanone, are directly or indirectly synthesized and co-expressed through the activity of key genes such as flavonoid 3’-hydroxylase (CYP75B1, F3’H), which converts naringenin to eriodictyol, and flavonoid 3’,5’-hydroxylase (CYP75A, F3’5’H), which hydroxylates the B-ring of flavonoids (Forouzanfar et al., 2025; Li et al., 2025b; Liu et al., 2025). Under salt stress, upregulation of these genes enhances flavonoid accumulation, promoting reactive oxygen species (ROS) scavenging and improving plant tolerance (Li et al., 2025b; Wu et al., 2025). Treated wild soybean exhibited increased plant height under alkali stress, longer roots under both stresses, and larger leaf area under salt stress, indicating stress-specific improvements in growth. These findings align with previous reports linking flavonoid accumulation to enhanced salt tolerance in legumes (Praxedes et al., 2010; Duan et al., 2025; Wu et al., 2025).
In summary, wild soybean exhibited distinct transcriptional responses to salt and alkali stress. Salt stress primarily upregulated genes related to Na+ transport and homeostasis, along with co-expressed stress-responsive TFs, promoting root-based Na+ retention and ionic balance. Conversely, alkali stress induced flavonoid biosynthetic genes, likely enhancing antioxidant defenses against high-pH and bicarbonate-induced oxidative stress. This was supported by exogenous flavonoid supplementation, which mitigated growth inhibition, particularly under alkali conditions. These findings reveal stress-specific adaptive mechanisms and identify molecular targets for enhancing legume tolerance to saline-alkaline environments.
5 Conclusion
In conclusion, our findings demonstrate that wild soybean employs distinct molecular strategies to cope with salt and alkali stress. Salt stress primarily activates ion transport and homeostasis pathways to maintain cytosolic Na+/K+ balance, whereas alkali stress predominantly induces flavonoid biosynthesis to counteract oxidative damage under elevated pH conditions. The enhanced tolerance observed with exogenous flavonoid application further confirms their functional role in alkali stress mitigation. These insights not only deepen our understanding of stress-specific plant responses but also provide valuable targets for breeding salt-alkali resilient soybean cultivars.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MW: Conceptualization, Formal Analysis, Methodology, Writing – original draft. YQ: Investigation, Validation, Writing – original draft. XL: Data curation, Investigation, Validation, Writing – original draft. SG: Investigation, Validation, Writing – review & editing. YS: Data curation, Writing – original draft. YB: Data curation, Investigation, Writing – original draft. YL: Funding acquisition, Project administration, Writing – review & editing. CZ: Validation, Writing – review & editing. ZX: Data curation, Funding acquisition, Project administration, Resources, Writing – review & editing. CM: Funding acquisition, Project administration, Software, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program Young Scientist Project (2023YFD1901900), Dongying Key Special Project for Scientific and Technological Innovation (2023ZDJH94), International Foundation of Tobacco Research Institute of Chinese Academy of Agricultural Sciences (IFT202301), National Natural Science Foundation of China (32171948), Agricultural Science and Technology Innovation Program (ASTIP-TRIC06, ASTIP-TRIC-ZD05, CAAS-ZDRW202407).
Conflict of interest
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1675559/full#supplementary-material
Supplementary Figure 1 | Phenotype of wild soybean at seed germination stage under normal, salt stress (0.6% NaCl and 1.2% NaCl) and alkali stress (pH 9.16 and pH 10.83) treatments. Thirty seeds were used for each treatment, and three replicates were performed for each treatment.
Supplementary Figure 2 | Phenotype of seedling growth of wild soybean under normal, salt stress (0.6% NaCl and 1.2% NaCl) and alkali stress (pH 9.16 and pH 10.83) treatments for three and seven days.
Supplementary Figure 3 | Types and proportion of differentially expressed transcription factors.
Supplementary Figure 4 | Validation of RNA-Seq results using qRT-PCR. Comparing the log2FC values of RNA-seq and qRT-PCR (log2FC ≥ 1 indicates upregulation, log2FC ≤ 1 indicates downregulation). The gray bar charts show the results of qRT-PCR, and the line graphs show the results of RNA-seq. Error bars represent the SD of the mean(n = 3).
Supplementary Table 1 | Non-redundant unigenes annotated in the transcriptome analysis.
Supplementary Table 2 | Different expressed genes between different treatments.
Supplementary Table 3 | Different expressed genes related to phenylpropanoid and flavonoid biosynthesis under salt and alkali stress treatments.
Supplementary Table 4 | Primers for qRT-PCR.
References
An, M. J., Wang, X. L., Chang, D. D., Wang, S., Hong, D. S., Fan, H., et al. (2020). Application of compound material alleviates saline and alkaline stress in cotton leaves through regulation of the transcriptome. BMC Plant Biol. 20, 462. doi: 10.1186/s12870-020-02649-0
Cai, X. X., Jia, B. W., Sun, M. Z., and Sun, X. L. (2022). Insights into the regulation of wild soybean tolerance to salt-alkaline stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1002302
Chen, X. H., Wang, H. T., Li, X. Y., Ma, K., Zhan, Y. G., and Zeng, F. S. (2019). Molecular cloning and functional analysis of 4-Coumarate : CoA ligase 4(4CL-like 1)from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biol. 19, 231. doi: 10.1186/s12870-019-1812-0
Demidchik, V. and Maathuis, F. J. M. (2007). Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 175, 387–404. doi: 10.1111/j.1469-8137.2007.02128.x
Duan, G., Liang, C., Su, J., Liang, Y., Li, W., Zhang, J., et al. (2025). Integrative transcriptomic and metabolomic analysis elucidates the vital pathways underlying the differences in salt stress responses between two chickpea (Cicer arietinum L.) varieties. BMC Plant Biol. 25, 903. doi: 10.1186/s12870-025-06910-2
Fang, S., Hou, X., and Liang, X. (2021). Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.667458
Forouzanfar, F., Sahranavard, T., Tsatsakis, A., Iranshahi, M., and Rezaee, R. (2025). Rutin: a pain-relieving flavonoid. Inflammopharmacology. 33, 1289–1301. doi: 10.1007/s10787-025-01671-8
Gao, Y., Jin, Y., Guo, W., Xue, Y., and Yu, L. (2022). Metabolic and physiological changes in the roots of two oat cultivars in response to complex saline-alkali stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.835414
Gautam, H., Sharma, A., and Trivedi, P. K. (2023). The role of flavonols in insect resistance and stress response. Curr. Opin. Plant Biol. 73, 102353. doi: 10.1016/j.pbi.2023.102353
Ge, Y., Li, Y., Zhu, Y. M., Bai, X., Lv, D. K., Guo, D., et al. (2010). Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 10, 153. doi: 10.1186/1471-2229-10-153
Guo, H., Huang, Z., Li, M., and Hou, Z. (2020). Growth, ionic homeostasis, and physiological responses of cotton under different salt and alkali stresses. Sci. Rep. 10, 21844. doi: 10.1038/s41598-020-79045-z
Han, M. H., Yang, N., Wan, Q. W., Teng, R. M., Duan, A. Q., Wang, Y. H., et al. (2021). Exogenous melatonin positively regulates lignin biosynthesis in Camellia sinensis. Int. J. Biol. Macromol 179, 485–499. doi: 10.1016/j.ijbiomac.2021.03.025
Horie, T., Hauser, F., and Schroeder, J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660–668. doi: 10.1016/j.tplants.2009.08.009
Hou, R., Yang, L., Tana, W., Chen, S., and Zhang, L. (2023). Genes related to osmoregulation and antioxidation play important roles in the response of Trollius chinensis seedlings to saline-alkali stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1080504
Javid, M., Ford, R., and Nicolas, M. E. (2012). Tolerance responses of Brassica juncea to salinity, alkalinity and alkaline salinity. Funct. Plant Biol. 39, 699–707. doi: 10.1071/FP12109
Jia, B., Sun, M., DuanMu, H., Ding, X., Liu, B., Zhu, Y., et al. (2017). GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci. Rep. 7, 9423. doi: 10.1038/s41598-017-09772-3
Jin, J., Wang, J., Li, K., Wang, S., Qin, J., Zhang, G., et al. (2021). Integrated physiological, transcriptomic, and metabolomic analyses revealed molecular mechanism for salt resistance in soybean roots. Int. J. Mol. Sci. 22, 12848. doi: 10.3390/ijms222312848
Keisham, M., Mukherjee, S., and Bhatla, S. C. (2018). Mechanisms of sodium transport in plants-progresses and challenges. Int. J. Mol. Sci. 19, 647. doi: 10.3390/ijms19030647
Kofsky, J., Zhang, H., and Song, B. H. (2018). The untapped genetic reservoir: the past, current, and future applications of the wild soybean (Glycine soja). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00949
Li, Q., Yang, A., and Zhang, W.-H. (2016). Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativaL.). J. Exp. Bot. 67, 6431–6444. doi: 10.1093/jxb/erw407
Li, S., Xu, L., Li, Y., Waqar, A., Hu, Z., Yang, M., et al. (2025a). Advances in salinity tolerance of soybean: molecular mechanism and breeding strategy. Food Energy Secur 14, e70073. doi: 10.1002/fes3.70073
Li, Y., Wu, Y., Pu, R., Li, X., Bai, T., Li, N., et al. (2025b). Metabolomic and transcriptomic analyses of flavonoid biosynthesis in Dendrobium devonianum flowers. Genes 16, 264. doi: 10.3390/genes16030264
Liang, S., Xu, S., Qu, D., Yang, L., Wang, J., Liu, H., et al. (2022). Identification and functional analysis of the caffeic acid O-methyltransferase (COMT) gene family in rice (Oryza sativa L.). Int. J. Mol. Sci. 23, 8491. doi: 10.3390/ijms23158491
Liu, X., Ma, H., Liu, X., Wang, X., Chen, Z., Yang, J., et al. (2025). Transcriptomic and metabolomic analyses provide insights into the flavonoid biosynthesis in Dangshen. Phytochemical Anal. 36, 1063–1078. doi: 10.1002/pca.3492
Liu, L., Si, L., Zhang, L., Guo, R., Wang, R., Dong, H., et al. (2024). Metabolomics and transcriptomics analysis revealed the response mechanism of alfalfa to combined cold and saline-alkali stress. Plant J. 119, 1900–1919. doi: 10.1111/tpj.16896
Liu, H., Song, J., Dong, L., Wang, D., Zhang, S., and Liu, J. (2017). Physiological responses of three soybean species (Glycine soja, G. gracilis and G. max cv. Melrose) to salinity stress. J. Plant Res. 130, 723–733. doi: 10.1007/s10265-017-0929-1
Lu, X., Min, W., Shi, Y., Tian, L., Li, P., Ma, T., et al. (2022). Exogenous melatonin alleviates alkaline stress by removing reactive oxygen species and promoting antioxidant defence in rice seedlings. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.849553
Ma, J., Li, X., He, M., Li, Y., Lu, W., Li, M., et al. (2023). A joint transcriptomic and metabolomic analysis reveals the regulation of shading on lignin biosynthesis in asparagus. Int. J. Mol. Sci. 24, 1539. doi: 10.3390/ijms24021539
Munns, R. and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Padmavathiamma, P. K. and Li, L. Y. (2007). Phytoremediation technology: hyper-accumulation metals in plants. Water Air Soil Poll 184, 105–126. doi: 10.1007/s11270-007-9401-5
Paz, R. C., Rocco, R. A., Reinoso, H., Menéndez, A. B., Pieckenstain, F. L., and Ruiz, O. A. (2012). Comparative study of alkaline, saline, and mixed saline-alkaline stresses with regard to their effects on growth, nutrient accumulation, and root morphology of lotus tenuis. J. Plant Growth Regul. 31, 448–459. doi: 10.1007/s00344-011-9254-4
Praxedes, S. C., De Lacerda, C. F., Damatta, F. M., Prisco, J. T., and Gomes-Filho, E. (2010). Salt tolerance is associated with differences in ion accumulation, biomass allocation and photosynthesis in cowpea cultivars. J. Agron. Crop Scie 196, 193–204. doi: 10.1111/j.1439-037x.2009.00412.x
Preisner, M., Kulma, A., Zebrowski, J., Dyminska, L., Hanuza, J., Arendt, M., et al. (2014). Manipulating cinnamyl alcohol dehydrogenase (CAD) expression in flax affects fibre composition and properties. BMC Plant Biol. 14, 50. doi: 10.1186/1471-2229-14-50
Rao, Y., Peng, T., and Xue, S. (2023). Mechanisms of plant saline-alkaline tolerance. J. Plant Physiol. 281, 153916. doi: 10.1016/j.jplph.2023.153916
Romero-Aranda, M. R., Espinosa, J., Gonzalez-Fernandez, P., Jaime-Fernandez, E., Traverso, J. A., Asins, M. J., et al. (2021). Role of Na(+) transporters HKT1;1 and HKT1;2 in tomato salt tolerance. I. Function loss of cheesmaniae alleles in roots and aerial parts. Plant Physiol. Biochem. 168, 282–293. doi: 10.1016/j.plaphy.2021.10.018
Sagervanshi, A., Geilfus, C. M., Kaiser, H., and Muehling, K. H. (2022). Alkali salt stress causes fast leaf apoplastic alkalinization together with shifts in ion and metabolite composition and transcription of key genes during the early adaptive response of Vicia faba L. Plant Sci. 319, 111253. doi: 10.1016/j.plantsci.2022.111253
Sahakyan, G. and Sahakyan, N. (2025). ABC Proteins as Regulators of Plant Tolerance to Biotic and Abiotic Stresses (Boca Raton, FL: CRC Press), 203–222.
Shao, S., Li, M., Yang, D., Zhang, J., and Shi, L. (2016). The physiological variations of adaptation mechaniam in Glycine soja seedlings under saline and alkaline stresses. Pak. J. Bot. 48, 2183–2193.
Song, H., Cao, Y., Zhao, X., and Zhang, L. (2023). Na+-preferential ion transporter HKT1;1 mediates salt tolerance in blueberry. Plant Physiol. 194, 511–529. doi: 10.1093/plphys/kiad510
Sun, J., Li, S., Guo, H., and Hou, Z. (2021a). Ion homeostasis and Na+ transport-related gene expression in two cotton (Gossypium hirsutum L.) varieties under saline, alkaline and saline-alkaline stresses. PloS One 16, e0256000. doi: 10.1371/journal.pone.0256000
Sun, T., Ma, N., Wang, C., Fan, H., Wang, M., Zhang, J., et al. (2021b). A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/na+ Ratio in soybean. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.638340
Sun, Y., Shu, H., Lu, D., Zhang, T., Li, M. X., Guo, J. X., et al. (2025). Wild soybean cotyledons at the emergence stage tolerate alkali stress by maintaining carbon and nitrogen metabolism, and accumulating organic acids. Physiol. Plant 177, e70117. doi: 10.1111/ppl.70117
Sun, X., Sun, M., Jia, B., Qin, Z., Yang, K., Chen, C., et al. (2016). A Glycine soja methionine sulfoxide reductase B5a interacts with the Ca2+ /CAM-binding kinase GsCBRLK and activates ROS signaling under carbonate alkaline stress. Plant J. 86, 514–529. doi: 10.1111/tpj.13187
Wang, Y., Cao, Y., Liang, X., Zhuang, J., Wang, X., Qin, F., et al. (2022b). A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 13, 2222. doi: 10.1038/s41467-022-29809-0
Wang, Y., Jiang, W., Li, C., Wang, Z., Lu, C., Cheng, J., et al. (2024). Integrated transcriptomic and metabolomic analyses elucidate the mechanism of flavonoid biosynthesis in the regulation of mulberry seed germination under salt stress. BMC Plant Biol. 24, 132. doi: 10.1186/s12870-024-04804-3
Wang, G., Shen, W., Zhang, Z., Guo, S., Hu, J., Feng, R., et al. (2022a). The effect of neutral salt and alkaline stress with the same na+ Concentration on root growth of soybean (Glycine max (L.) merr.) seedlings. Agronomy 12, 2708. doi: 10.3390/agronomy12112708
Wang, W. X., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, B., Wang, J., Yang, T., Wang, J., Dai, Q., Zhang, F., et al. (2023). The transcriptional regulatory network of hormones and genes under salt stress in tomato plants (Solanum lycopersicum L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1115593
Wu, H., Shabala, L., Azzarello, E., Huang, Y., Pandolfi, C., Su, N., et al. (2018). Na+ extrusion from the cytosol and tissue-specific Na+ sequestration in roots confer differential salt stress tolerance between durum and bread wheat. J. Exp. Bot. 69, 3987–4001. doi: 10.1093/jxb/ery194
Wu, J., Wang, X., Xu, J., Li, T., Shan, G., Zhang, L., et al. (2025). Overexpression of soybean flavonoid 3’-hydroxylase enhances plant salt tolerance by promoting ascorbic acid biosynthesis. J. Adv. Res S2090-1232(25)00302-9. doi: 10.1016/j.jare.2025.05.009
Wu, G., Zhou, Z. D., Chen, P., Tang, X. L., Shao, H. B., and Wang, H. Y. (2014). Comparative ecophysiological study of salt stress for wild and cultivated soybean species from the yellow river delta, China. Sci. World J. 2014, 651745. doi: 10.1155/2014/651745
Xie, M., Chung, C. Y., Li, M. W., Wong, F. L., Wang, X., Liu, A., et al. (2019). A reference-grade wild soybean genome. Nat. Commun. 10, 1216. doi: 10.1038/s41467-019-09142-9
Xu, Z. C., Ren, T. T., Marowa, P., You, X. W., Lu, X. L., Li, Y. Q., et al. (2020b). Establishment of a cultivation mode of glycine soja, the bridge of phytoremediation and industrial utilization. Agronomy 10, 595. doi: 10.3390/agronomy10040595
Xu, X., Zhang, J., Yan, B., Wei, Y., Ge, S., Li, J., et al. (2021). The adjustment of membrane lipid metabolism pathways in maize roots under saline-alkaline stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.635327
Xu, Z., Zhou, J., Ren, T., Du, H., Liu, H., Li, Y., et al. (2020a). Salt stress decreases seedling growth and development but increases quercetin and kaempferol content in Apocynum venetum. Plant Biol. (Stuttg) 22, 813–821. doi: 10.1111/plb.13128
Xue, Z. C., Wang, Y., and Liu, J. (2022). Systematic salt tolerance-related physiological mechanisms of wild soybean and their role in the photosynthetic activity and Na+ distribution of grafted soybean plants. Photosynthetica 60, 400–407. doi: 10.32615/ps.2022.030
Yan, S., Zhao, L. L., Zhao, D. Y., Xu, G. X., Wang, Y. F., Zhou, Z. Q., et al. (2025). Transcriptomic and metabolomic analyses reveal phenolic metabolism regulated by melatonin in pear peel. Chem. Biol. Technol. Agric. 12, 45. doi: 10.1186/S40538-025-00763-5
Yang, Y. and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, S., Xu, Y., Tang, Z., Jin, S., and Yang, S. (2024). The impact of alkaline stress on plant growth and its alkaline resistance mechanisms. Int. J. Mol. Sci. 25, 13719. doi: 10.3390/ijms252413719
Yoon, J., Cao, X., Zhou, Q., and Ma, L. Q. (2006). Accumulation of Pb, Cu, and Zn in native plants growing on a contaminated Florida site. Sci. Total Environ. 368, 456–464. doi: 10.1016/j.scitotenv.2006.01.016
Zhang, H., Liu, X. L., Zhang, R. X., Yuan, H. Y., Wang, M. M., Yang, H. Y., et al. (2017). Root Damage under Alkaline Stress Is Associated with Reactive Oxygen Species Accumulation in Rice (Oryza sativa L.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01580
Zhang, H., Yu, F., Xie, P., Sun, S., Qiao, X., Tang, S., et al. (2023). A Ggamma protein regulates alkaline sensitivity in crops. Science 379, eade8416. doi: 10.1126/science.ade8416
Zhao, Q., Chen, S., Wang, G., Du, Y., Zhang, Z., Yu, G., et al. (2022). Exogenous melatonin enhances soybean (Glycine max (L.) Merr.) seedling tolerance to saline-alkali stress by regulating antioxidant response and DNA damage repair. Physiol. Plant 174, e13731. doi: 10.1111/ppl.13731
Zhou, Y., Xu, K., Gao, H., Yao, W., Zhang, Y., Zhang, Y., et al. (2023). Comparative proteomic analysis of two wild soybean (Glycine soja) genotypes reveals positive regulation of saline-alkaline stress tolerance by tonoplast transporters. J. Agric. Food Chem. 71, 14109–14124. doi: 10.1021/acs.jafc.3c02111
Keywords: ion homeostasis, flavonoids, abiotic stress, ion compartmentalization, wild soybean
Citation: Wang M, Qu Y, Lu X, Gillani SW, Song Y, Bai Y, Li Y, Zhang C, Xu Z and Meng C (2025) Root-centered sodium sequestration and transcriptomic regulation under salt and alkali stress in wild soybean (Glycine soja). Front. Plant Sci. 16:1675559. doi: 10.3389/fpls.2025.1675559
Received: 31 July 2025; Accepted: 03 September 2025;
Published: 18 September 2025.
Edited by:
Jihong Zhang, Shihezi University, ChinaReviewed by:
Min Chen, Shandong Normal University, ChinaYing Zhao, Northeast Agricultural University, China
Wang Chaofan, Hebei Agricultural University, China
Copyright © 2025 Wang, Qu, Lu, Gillani, Song, Bai, Li, Zhang, Xu and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongchang Xu, eHV6b25nY2hhbmdAY2Fhcy5jbg==; Chen Meng, bWVuZ2NoZW4wMUBjYWFzLmNu
†These authors have contributed equally to this work
 Meng Wang1†
Meng Wang1† Syeda Wajeeha Gillani
Syeda Wajeeha Gillani Yiqiang Li
Yiqiang Li Chengsheng Zhang
Chengsheng Zhang Chen Meng
Chen Meng