- 1Department of Plant Sciences, Centro de Investigación y Tecnología Agroalimentaria de Aragón (CITA), Zaragoza, Spain
- 2Instituto Agroalimentario de Aragón-IA2 (CITA-Universidad de Zaragoza), Zaragoza, Spain
- 3Department of Pomology, Estación Experimental de Aula Dei (EEAD-CSIC), Zaragoza, Spain
- 4Fundación Agencia Aragonesa para la Investigación y el Desarrollo (ARAID), Zaragoza, Spain
Evergrowing phenotypes in deciduous trees have only been described in three unrelated species: peach, hazelnut and pomegranate. These genotypes are a useful tool for forward genetics approaches aimed at understanding the processes that regulate seasonal growth and dormancy. Research in evergrowing peach lead to the identification of the DORMANCY ASSOCIATED MADS BOX transcription factors (DAMs) as regulators of dormancy in stone and pome fruits. In this work we present the breeding and preliminary characterization of an evergrowing (evg) sweet cherry genotype. This individual was obtained from in vitro embryo rescue of self-fertilization seeds, of a local self-compatible landrace. Unlike wild type sweet cherry, evg sweet cherry does not set buds in autumn and continues to grow throughout the winter. In contrast to evergrowing peach, no major structural deletion was observed in the tandemly arranged PavDAMs genes. However, specific expression profiles of these genes were observed in the evg sweet cherry. The specific polymorphisms previously observed in the PavDAMs of the parental cultivar, and the high level of inbreeding depression resulting from self-fertilization, suggest that the expression of homozygous recessive alleles might be the cause of the evergrowing phenotype. Ongoing work to fully characterize the underlying molecular mechanism regulating evg sweet cherry phenotype is discussed, highlighting its importance and utility as a research and breeding tool.
1 Introduction
Sweet cherry (Prunus avium L.) is a temperate fruit tree species of high economic interest. It belongs to the Prunus genus together with other important temperate fruit species like peach (P. persica), plum (P. domestica), apricot (P. armeniaca) and almond (P. dulcis); and to the Rosaceae family, which also includes economically important pome fruit trees such as apple (Malus domestica) and pear (Pyrus communis). These species undergo a dormancy period during the winter, when growth stops and resumes in spring, once the cold and warmth needs are satisfied. Environmental cues, such as temperature and photoperiod, are sensed to adjust growth and phenophases (Singh et al., 2017). In temperate fruit trees, optimum production relies on the adaptation of the plant material to environmental conditions. In particular, winter chill requirements and bloom date are phenological traits on which the choice of cultivars is based to ensure adequate flowering and fruit development in a given region. Current and future climate scenarios, with mild winter temperatures, are about to alter this finely tuned plant-environment interaction (Kovaleski, 2024). Dormancy and blooming time are therefore becoming important breeding traits in temperate Rosaceous fruit trees, and great efforts are being made to understand their underlying molecular mechanisms.
Recent advances in the understanding of flowering time control using model plant species have led to the identification of several pathways that cross talk through a few major transcription factors called floral integrators. These include several MIKCc-type transcription factors such as FLOWERING LOCUS C (FLC), and SHORT VEGETATIVE PHASE (SVP), that repress the expression of the often referred to as florigen FLOWERING LOCUS T (FT), and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), resulting in the inhibition of the transition to flowering (reviewed in Teotia and Tang, 2015 and Xu and Chong, 2018; Wang et al., 2025). Interestingly, some of these flowering pathways also regulate seasonal growth in woody plant species. Homologous or orthologous genes to floral integrators have been shown to mediate autumn growth cessation, winter dormancy and spring sprouting in model forest tree species (e.g. Böhlenius et al., 2006; Singh et al., 2018, 2019, André et al., 2022; Ding et al., 2024). Environmental cue sensing that underly growth cessation and dormancy has been shown to be different in some Rosaceae temperate fruit tree species (Singh et al., 2017; Zhang et al., 2025), warranting similar strategies in these species.
The progress in understanding flowering time and seasonal growth regulation in temperate fruit trees is lagging, mainly because of the limited number of mutants and the difficulty of regenerating knock-out and overexpression transformants, especially in Prunus species (see Vergara et al., 2021 for recent progress). However, important progress has been made through the characterization of natural spontaneous evergrowing mutants. In peach, the identification, characterization and mapping of an evergrowing genotype (Rodriguez-A et al., 1994; Wang et al., 2002) revolutionized the understanding of dormancy in the Amygdaloideae subfamily by uncovering a prominent role of a tandem set of MIKCc-type transcription factors within the SVP/AGL24 clade, named DAM genes. Deletion of 4 of the 6 DAM genes in the evergrowing peach leads to the non-dormant evergrowing phenotype (Bielenberg et al., 2004; 2008). However, pathway(s) involving DAM genes, and function are not yet fully understood. In subtropical pomegranate, the recent characterization of an evergrowing phenotype revealed that the phenotype is also caused by a mutation in a MIKC-type MADS box gene, highly similar to the Arabidopsis FLC-clade member Agamous-like 27 (AGL27), also known as Flowering Locus M 1 (FLM1) or MADS Affecting Flowering 1 (MAF1), with a unique glutamine repeat motif (PolyQ) in its N-terminal region (Harel-Beja et al., 2022). While these results highlight the prominent role played by MIKCc-type transcription factors of the FLC and SVP/AGL24 clades in the perception of environmental cues that allow trees to adjust their growth and flowering, more mutants and mutagenesis approaches are needed in fruit tree species, to resolve the underlying molecular mechanisms.
In 2008-2009, the development of a self-fertilization population (F2) from the self-compatible local sweet cherry cultivar (‘Cristobalina’), for genetic mapping and genetic analyses (Calle et al., 2018), led to the unexpected obtention of evergrowing plants (A. Wünsch, unpublished results). These plants were planted in the field but did not survive in field growing conditions. In 2021, as part of a sweet cherry breeding project aimed at generating new phenotypic diversity with low chilling requirements, self-fertilization seeds of the same cultivar were newly germinated to obtain the same evergrowing phenotype. One individual exhibiting an evergrowing phenotype was obtained. In this work, we present a preliminary morphological and molecular characterization of this valuable plant material and discuss its possible causes and biotechnological usefulness. As a perspective, we propose that inbreeding in this highly heterozygous species may be an important source for generating unique genotypes for forward genetics approaches.
2 Materials and methods
2.1 Obtention and phenotypic characterization of sweet cherry evg
Sweet cherry cv ‘Cristobalina’ (wt-p; Table 1) trees located in CITA de Aragón, sweet cherry cultivar collection in Zaragoza (Spain) were used in this work. Fully mature fruits (n=173) from the open pollination of these trees were collected in May 2021 (66 days after full bloom, regular maturity period of this cultivar; Calle and Wünsch, 2020). Seeds were extracted and disinfected by washing with Sodium hypochlorite (10 g/l active chlorine) for 15 min and rinsed (3 times) with sterile distilled water. Integuments were removed aseptically, and well developed embryos (n=72; 42% of total fruits collected) were transferred to test tubes with C2D culture medium (Chée and Pool, 1987; Arbeloa et al., 2009). Test tubes were placed at 5.5 °C in the dark for 6 weeks, and then at 23 °C under cool-white, fluorescent light (50 μmol.m-2.s-1; 16h light/8h dark). Shoot tips from germinated embryos (n=24, 33% of cultured embryos, 16% of total fruits collected) were micropropagated ex vitro (Andreu and Marín, 2005) in DKW culture medium (Driver and Kuniyuki, 1984) to allow shoot multiplication. Shoots were rooted ex vitro (Castillo and Marín, 1994) after a quick dip (30 s) in K-IBA (1 mM) and grown with a peat–perlite substrate (1:1) under 100% RH in the greenhouse. Rooted shoots were acclimated (Marin, 2003) and transplanted into pots with the same substrate. Hence, several clonal plants were obtained of the different germinated embryos.
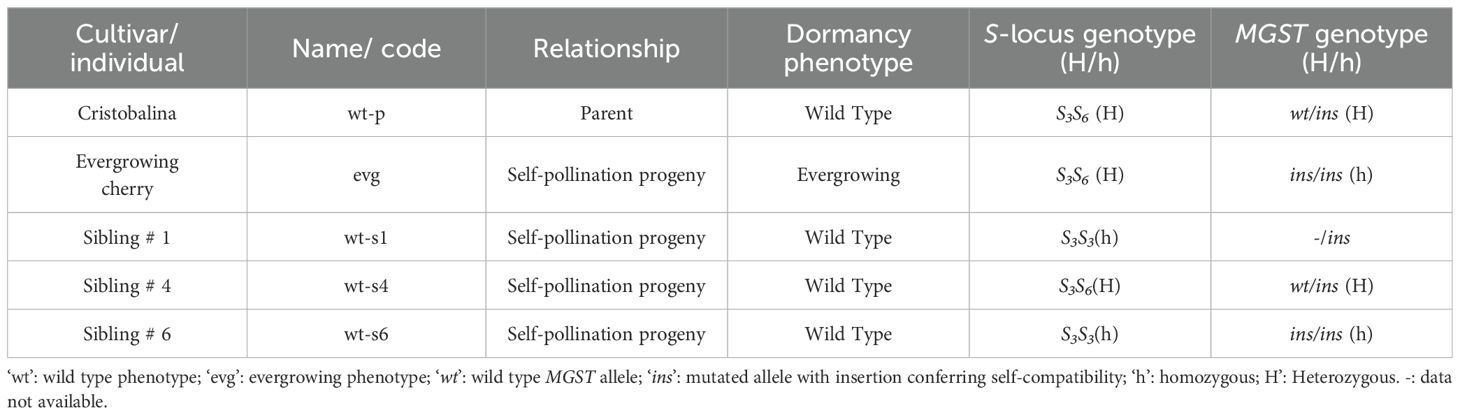
Table 1. Information on plant materials, dormancy phenotype and self-incompatibility genotype. S-locus and MGST genotypes analyzed to establish hybrid identity.
During the first year of growth, evg clonal plants and three randomly selected wild type full-siblings (wt-s1, -s4, -s6; Table 1) were monitored in a temperature-controlled greenhouse and in an outdoor shade-house. Preliminary phenotypic profiling consisted of a morphological characterization throughout the dormancy period monitoring leaf development, bud set, leaf fall, bud break and re-growth.
2.2 Preliminary genotypic characterization
Genomic DNA was extracted from leaf samples of the four genotypes (evg, wt-s1, -s4, -s6; individual potted plants as experimental unit), and S-locus analysis was carried out by PCR amplification of the S-locus genes S-RNase and SFB, using the primers PaConsI-F/PaConsIR2 and FBOX5´A/F-BOXintronR (Sonneveld et al., 2006; Vaughan et al., 2006) following the protocol described by Cachi and Wünsch (2014). Testing for the presence of insertion within the MGST gene conferring self-compatibility (SC) from ‘Cristobalina’, was carried out as described by Čmejlová et al. (2023) with modifications. Fragment analyses of all PCRs were carried out by capillary electrophoresis using SeqStudio (Thermo Fisher) genetic analyzer, size standard GeneScan 500-LIZ (Thermo Fisher), and analyzed and visualized using Genenious Prime® 2025.0.3.
2.3 PavDAMs genes conservations and expression in evg
Genomic DNA of the evg sweet cherry, the parental genotype (wt-p), and two siblings (wt-4 and -6) were used to check the conservation of PavDAM genes by PCR analysis. PCR was carried out for each PavDAM gene using specific primers (Supplementary Table 1), at an annealing temperature of 59°C. PCR fragments were analyzed by gel electrophoresis. Reverse Transcription PCR (RT-PCR) was used to analyze the expression of the DAM genes in the same genotypes. Shoot apices or buds (depending on the growth stage of each individual at each sampling date) were sampled and immediately frozen in liquid nitrogen. Sampling was carried out at regular intervals during seasonal growth and dormancy (September to May). RNA was extracted using the ‘RNeasy Plant Mini Kit’ (Qiagen) following the manufacturer’s instructions. Reverse transcription was carried with using 2 µg of RNA using the ‘High-Capacity cDNA Reverse Transcription Kit’ (Applied Biosystems) following the manufacturer’s instructions. cDNA PCR was carried out for each PavDAM and a reference gene Actin using specific primers (Supplementary Table 2). PCR products were amplified with the following conditions: 94 °C for 4 min, 34 cycles of 94 °C for 45 s, 59°C (for Actin, PavDAM-1,-4,-6) or 57°C (for PavDAM-2,-3,-5) for 45 s, and 72 °C for 2 min, followed by a final extension at 72 °C for 7 min. All PCR fragments were separated on 1.7% (w/v) agarose gels and 1x TBE buffer run a 7 V/cm and stained with GelRed. The experimental units consisted of individual orchard trees (wt-p) or individual potted plants (evg, wt-4, and wt-6).
3 Results and discussion
3.1 An evergrowing (evg) cherry
Among germinated plants from embryo rescue of open pollinated ‘Cristobalina’ seed progenies (Figures 1A–C) an individual with a strikingly different phenotype was observed (Figure 1D). After acclimation, potting and transferring to greenhouse, while all normal-looking seedlings showed a typical cherry seedling phenotype, this offspring showed unusually continuous growth throughout the year and did not shed its leaves in autumn (Figures 1E–J). Left unpruned under warm greenhouse conditions, while wild type seedling reached an average height of 48 cm (SD 14), evg clones reached 160 cm (SD 36) cm at the at the end of growing season (Figure 1E). This phenotype showed profuse leaf and lateral shoots development (Figure 1F), and few flowers, developed from lateral shoots (Figure 1G), appeared at the end of its first growth cycle. Aged leaves develop a browning all around the leaf limb (Figure 1G) before falling. Under shade house conditions the following years, although the growth rate appeared similar between the wt and evg replica plants, the evg phenotype was maintained (Figure 1J) but flowers did not occur. This phenotype is similar to evergrowing individuals described in peach (Rodriguez-A et al., 1994).

Figure 1. In vitro embryo culture and establishment of sweet cherry evg. (A) Germinated embryo. (B) Micropropagation of seedlings. (C) A rooted and acclimated seedling. (D) Acclimated seedlings in tray. (E) Seedlings in their first-year dormancy cycle. Notice the difference in height between wild type (right) and evg seedlings (left). (F–I) Close-ups of abundant leaf development (F), flowering (G), leaf margin desiccation before shedding (H), and lateral vegetative buds (I) in evg plants. (J) Comparison of 2 wild type individuals that shed their leaves (left) at dormancy onset (October 2024), with two evg replica plants retaining their leaves (right). Bars = 2 cm.
‘Cristobalina’ is an exceptionally early flowering cultivar for which there is no flowering overlap with other cultivars in the collection and growing conditions. Thus, open pollinated seedlings are expected to originate from self-pollination. To confirm this assumption and confirm the origin of evg from self-pollination of ‘Cristobalina’, we used two complementary genetic analysis approaches. First, evg and its wild type siblings (wt-s1, -s4, and -s6) were analyzed with molecular markers to identify the self-incompatibility genotype at the S-locus. Since the wild type parental cultivar ‘Cristobalina’ (wt-p) S-genotype is S3S6 (Wünsch and Hormaza, 2004), inbred seedlings are expected to be S3S3, S3S6, or S6S6(Table 1). The evg and all three wt seedlings had indeed the expected genotypes (Table 1, Supplementary Figure 1), with evg S-genotype being S3S6. Second, the self-compatibility of ‘Cristobalina’ is originated from a pollen part mutation unlinked to the S-locus (Wünsch and Hormaza, 2004, Cachi and Wünsch, 2011), namely an insertion in the MGST gene (Ono et al., 2018). ‘Cristobalina’ is heterozygote for the mutation, and only pollen carrying the mutation, either S3 or S6, is capable of siring seeds. As a result, self-fertilization offspring should also carry the self-compatibility mutation. All full-siblings (wt-s1, -s4, and -s6), as well as evg were found to carry the mutated MGST allele, either in homozygosis or heterozygosis (Table 1, Supplementary Figure 1), further confirming the inbred origin of evg and wt-analyzed siblings.
3.2 Sweet cherry evg does not set buds or go dormant
To characterize in more detail the behavior of evg during dormancy, three wild type full-siblings and four evg replicas plants were weekly photographed paying special attention to the terminal shoot meristem and the first axillary buds below (Figure 2). At the onset of the developmental time series, on early October, while the three wild type individuals already had terminal buds and presented the typical brown leaf senescence spots that preceded their fall, the four evg plants were still growing actively and developing young leaves (Figure 2). Leaf fall dynamics varied among wt individuals, ranging from complete leaf fall in early November (wt-s1) to progressive leaf fall throughout most of the winter (wt-s6) (Figure 2). In evg plants, as in wt individuals, only the old leaves -away from the shoot tip- are shed. Although evg plants did not fully set the typical closed, compact, brown terminal buds, they appeared to slow their growth during the December to January period. However, the terminal and axillary shoots that appeared to have initiated the transition to closed bud structure, never matured into closed buds before the resumption of growth with increasing temperature. In fact, with increasing temperatures in February, evg individuals resumed growth earlier and faster than wt individuals.

Figure 2. Sweet cherry evg does not set buds or go dormant. A developmental time series during the 2022–2023 dormancy cycle for 3 wild type individuals and 4 evg clonal plants. Whereas all wild-type individuals set terminal buds before the onset of the developmental time series and lost their leaves by the end of December, evg plants neither set terminal buds nor lost their leaves. In December, an apparent slowing of evg growth was observed (white arrows indicate buds not fully set), but it continued to grow throughout the dormancy period. .
3.3 PavDAM genes are distinctly and differentially expressed in evg
The evergrowing phenotype of peach is caused by a major deletion affecting four of the six tandemly arranged DAM genes (Bielenberg et al., 2008). Our previous structural analysis of the PavDAM gene array in the sweet cherry cv ‘Cristobalina’, the wild-type parental cultivar of evg, revealed unique mutations when compared with other cultivars (Calle et al., 2021). Namely, specific SNPs in the coding sequences, a large deletion upstream of the PavDAM1 gene, and various INDELS and SNPs in the UTR regions between PavDAM4 and PavDAM5, that may be altering PavDAMs expression. These structural variants were shown to segregate with early blooming and are putatively associated with low chilling requirements and early flowering from this genotype (Calle et al., 2021). However, the precise cause of the low chilling phenotype of ‘Cristobalina’ remains to be elucidated. To confirm the structural conservation of the ‘Cristobalina’ PavDAM gene array in the wt progenies and the evg genotype, genomic DNA from all six DAM genes was amplified in these plant materials (Supplementary Figure 2). All six genes yielded the same expected PCR fragments of the gene body, in the tree types of materials (wild-type parental, wild-type seedlings, and evg cherry), indicating the absence of large structural mutations in the tandem array (Supplementary Figure 2). These results confirm that the evg cherry phenotype is not caused by the deletion of DAM genes as discovered in peach, and therefore the cause of the evg cherry phenotype is different from that of peach.
However, we cannot rule out that differential expression in DAM genes may be the cause of sweet cherry evg phenotype. Alterations in DAM genes underlie the evergrowing phenotype in other Rosaceous fruit trees (Bielenberg et al., 2008; Moser et al., 2020), and DAMs are generally considered to be the major integrators of winter dormancy in stone and pome fruits, so their expression could be affected in evg. To test this hypothesis, we performed RT-PCR gene expression analyses comparing the wild type (parental) and evg phenotype (Figure 3). Time-series RNA sampling of shoot apical meristem was carried in spring (May) and in the period between bud set in autumn (September) and bud burst the following spring (February). Consistent expression was observed for the reference gene Actin at the different genotypes, samples and times. However, differential expressions were observed between the evg and the wild type phenotypes for the DAM genes. Specifically, three distinct differential expression patterns could be distinguished. In the first pattern, affecting DAM1, DAM5, and DAM6, variable levels of down-regulation and time-limited expression was observed in the sweet cherry evg phenotype during the dormancy period. In the second pattern, involving DAM2 and DAM3, a complete down-regulation is observed throughout the time series, including the spring sample, for the evg phenotype. In the third pattern, affecting DAM4, in addition to a slightly down-regulated expression observed in evg phenotype at some sampling times, differential alternative splicing appears to be at play with distinct differential patterns for both phenotypes. Thus, our results confirm the hypothesis of an putative impaired expression of DAM genes in the evg phenotype, but further research is needed to decipher which or to what extent these expression patterns are associated with or cause the evg phenotype.
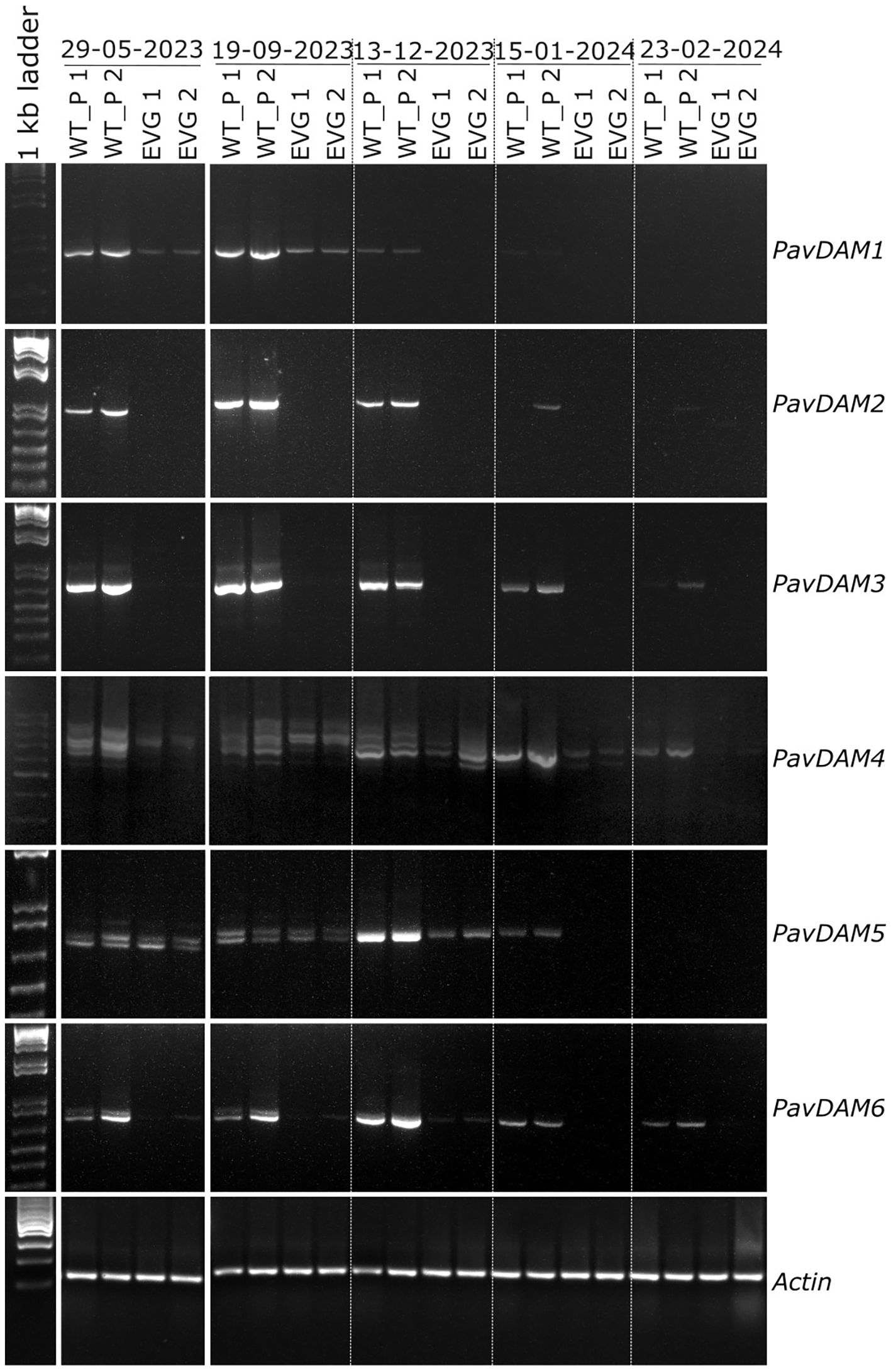
Figure 3. Time series RT-PCRs of PavDAM genes. RT-PCR was carried out for two wild type parental plants (wt-p1, wt-p2) and two evg clonal plants (evg-1, evg-2) in spring (May 2023) and throughout the 2023–2024 winter dormancy period (September to February). For better understanding, the individual images have been cropped and rearranged so that the time series is presented in chronological order (i.e., in the original images the May 2023 samples occupy the last lanes).
Evergrowing phenotypes in Rosaceous woody plant species have been reported to be caused by a major deletion affecting four (DAM1-4) of the six DAM genes in peach (Bielenberg et al., 2008) and by RNAi-mediated down-regulation of DAM1 in apple (Moser et al., 2020). It was shown that the DAM1–4 genes consistently reach their maximum expression during bud set (Falavigna et al., 2019), and, thus, emerged as good candidates to explain the observed phenotype of absent bud set in evergrowing. But more generally, DAM genes have been shown to exhibit distinct seasonal expression patterns in several Rosaceous species (Falavigna et al., 2019), suggesting that different genes play roles in successive environmentally mediated seasonal growth regulation events, starting with bud set and growth cessation, then establishment and maintenance of endodormancy, and finally release from dormancy and bud break. Based on this assumption, DAM5 and DAM6, which reach their maximum expression during winter dormancy (Falavigna et al., 2019), probably regulate dormancy maintenance and bud break (Falavigna et al., 2019; Lloret et al., 2021; Zhao et al., 2023). But the observation that overexpression of DAM6 from Japanese apricot induces earlier growth cessation and bud set in poplar (Sasaki et al., 2011) and apple (Yamane et al., 2019) potentially challenges those assumptions. Thus, in addition to the known effects of DAM1-4, whether the sweet cherry evg phenotype is also associated with any of the patterns observed in DAM5–6 warrants further research. Future studies should therefore focus on unravelling the function of each DAM gene, identifying their respective major interaction partners (e.g., if they interact with each other or with FLC or SVP), and ascertaining their regulatory networks.
3.4 Inbreeding depression as a cause of sweet cherry evg
Sweet cherry, like other Roseaceous species, is a highly heterozygous species. Heterozygosity is reinforced by a strict self-incompatibility system (gametopytic self-incompatibility) that prevents self- and cross-fertilization of genetically close individuals (reviewed in Herrero et al., 2017). Self-compatible mutants are scarce in this species, but the parental cultivar in this work is a local self-compatible landrace (Wünsch and Hormaza, 2004). The SNP genotyping of an F2 population (self-fertilization) from this cultivar, for the construction of a genetic map, revealed high levels of homozygosity, with large homozygous regions in the F2 population individuals, but also in the parental cultivar (Calle et al., 2018). Phenotypically, this population also exhibits symptoms of inbreeding depression like low vigor, early aging, and female and male sterility (Calle et al., 2022). In highly heterozygous species, high levels of homozygosity can lead to inbreeding depression due to overdominance, but mostly due to the expression of recessive deleterious alleles (Charlesworth and Charlesworth, 1999). The evg phenotype studied herein is a full-sib of this inbred population and its singular phenotype may be due to homozygosity in recessive dormancy-regulating alleles. Alternatively, inbreeding depression can also be driven by epigenetic changes (Vergeer et al., 2012). Furthermore, the low proportion of evergrowing phenotypes obtained after selfing (approx. 1/25) rules out the hypothesis of a single gene or locus (where 25% of the progeny should express the phenotype) and would rather point to a polygenic cause. Therefore, genes other than those in the DAMs cluster, both upstream and downstream of the dormancy regulating pathway, may be involved in the expression of the evg phenotype. To address this hypothesis, SNP and structural variant analyses involving evg versus a bulk of wild-type genotypes are warranted.
4 Conclusions
In summary, the sweet cherry evg phenotype reported here is the fourth natural mutant described in woody plant species (Thompson et al., 1985; Bielenberg et al., 2008; Harel-Beja et al., 2022). Although the precise molecular mechanism underlying this phenotype is still unknown, our preliminary analysis of DAM genes expression revealed expression patterns that -individually or in combination- may explain the observed phenotype. Importantly, this sweet cherry evergrowing phenotype is not due to a major deletion within the tandem set of DAM genes, as was observed in peach, making it valuable plant material to unravel DAM genes regulation of growth cessation and bud set, onsetting dormancy and bud break. Insight in this biological molecular mechanism(s) has great potential for understanding how woody plant species suspend their growth and development just at the right time to withstand the cold winter season before resuming growth with rising spring temperatures in temperate regions. This is especially important in the case of temperate Rosaceous fruit trees, where breeding for new varieties with low winter chill requirements became a major breeding goal for expanding cultivation areas and in the context of global warming.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
AH: Supervision, Writing – review & editing, Conceptualization, Methodology, Writing – original draft, Visualization. NM: Investigation, Validation, Writing – review & editing. AG: Writing – review & editing, Investigation. JM: Supervision, Writing – review & editing. AA: Writing – review & editing, Resources, Supervision. EG: Writing – review & editing, Validation, Investigation. AW: Funding acquisition, Project administration, Writing – review & editing, Resources, Conceptualization, Supervision.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Grant PID2019-103985RR-I00 funded by MICIU/AEI/10.13039/501100011033. Grant PID2022-137105OR-I00 funded by MICIU/AEI/10.13039/501100011033 and by the “European Union Next GenerationEU/PRTR”. Grant PREP2022-000496 funded by MICIU/AEI/10.13039/501100011033 and by “ESF+”. Gobierno de Aragon A12_23R Research Group funding.
Acknowledgments
Acknowledgments to the technical staff (lab and field work) of the Department of Plant Science of CITA de Aragón.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1677862/full#supplementary-material
References
André, D., Zambrano, J. A., Zhang, B., Lee, K. C., Rühl, M., Marcon, A., et al. (2022). Populus SVL acts in leaves to modulate the timing of growth cessation and bud set. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.823019
Andreu, P. and Marín, J. A. (2005). In vitro culture establishment and multiplication of the Prunus rootstock ‘Adesoto 101′ (P. insititia L.) as affected by the type of propagation of the donor plant and by the culture medium composition. Sci. Hortic. 106, 258–267. doi: 10.1016/j.scienta.2005.03.008
Arbeloa, A., Daorden, M. E., García, M. E., Andreu, P., and Marín, J. A. (2009). In vitro culture of ‘Myrobalan’ (Prunus cerasifera ehrh.) embryos. HortScience 44, 1672–1674. doi: 10.21273/hortsci.44.6.1672
Bielenberg, D. G., Wang, Y., Fan, S., Reighard, G. L., Scorza, R., and Abbott, A. G. (2004). A deletion affecting several gene candidates is present in the evergrowing peach mutant. J. Hered. 95, 436–444. doi: 10.1093/jhered/esh057
Bielenberg, D. G., Wang, Y., Li, Z., Zhebentyayeva, T., Fan, S., Reighard, G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4, 495–507. doi: 10.1007/s11295-007-0126-9
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Cachi, A. M. and Wünsch, A. (2011). Characterization and mapping of non-s gametophytic self-compatibility in sweet cherry (Prunus avium L.). J. Exp. Bot. 62, 1847–1856. doi: 10.1093/jxb/erq374
Cachi, A. M. and Wünsch, A. (2014). ‘S-genotyping of sweet cherry varieties from Spain and S-locus diversity in Europe’. Euphytica 197, 229–236. doi: 10.1007/s10681-014-1061-0
Calle, A., Cai, L., Iezzoni, A., and Wünsch, A. (2018). High density linkage maps constructed in sweet cherry (Prunus avium L.) using cross- and self-pollination populations reveal chromosomal homozygosity in inbred families and non-syntenic regions with the peach genome. Tree Genet. Genomes 14, 37. doi: 10.1007/s11295-018-1252-2
Calle, A., Grimplet, J., Le Dantec, L., and Wünsch, A. (2021). Identification and characterization of DAMs mutations associated with early blooming in sweet cherry and validation of DNA-based markers for selection. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.621491
Calle, A., Santolaria, N., Hedhly, A., and Wünsch, A. (2022). Characterization of female and male sterility in sweet cherry (Prunus avium L.). Acta Hortic. 1342, 63–69. doi: 10.17660/ActaHortic.2022.1342.9
Calle, A. and Wünsch, A. (2020). Multiple-population QTL mapping of maturity and fruit-quality traits reveals LG4 region as a breeding target in sweet cherry (Prunus avium L.). Hort Res. 7, 127. doi: 10.1038/s41438-020-00349-2
Castillo, M. and Marín, J. A. (1994). Enraizamiento in vivo de patrones frutales micropropagados. ITEA 15, 138–144.
Charlesworth, B. and Charlesworth, D. (1999). The genetic basis of inbreeding depression. Genet. Res. 74, 329–340. doi: 10.1017/s0016672399004152
Chée, R. and Pool, M. (1987). Improved inorganic media constituents for in vitro shoot multiplication of Vitis. Sci. Hortic. 32, 85–95. doi: 10.1016/0304-4238(87)90019-7
Čmejlová, J., Paprštein, F., Suran, P., Zelený, L., and Cmejla, R. (2023). A new one-tube reaction assay for the universal determination of sweet cherry (Prunus avium L.) self-(In)Compatible MGST- and S-alleles using capillary fragment analysis. Int. J. Mol. Sci. 24, 6931. doi: 10.3390/ijms24086931
Ding, J., Wang, K., Pandey, S., Perales, M., Allona, I., Khan, M. R. I., et al. (2024). Molecular advances in bud dormancy in trees. J. Exp. Bot. 75 (19), 6063–6075. doi: 10.1093/jxb/erae183
Driver, J. A. and Kuniyuki, A. H. (1984). In vitro propagation of Paradox walnut rootstock. HortScience 19, 507–509. doi: 10.21273/hortsci.19.4.507
Falavigna, V., da, S., Guitton, B., Costes, E., and Andrés, F. (2019). I want to (bud) break free: the potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01990
Harel-Beja, R., Ophir, R., Sherman, A., Eshed, R., Rozen, A., Trainin, T., et al. (2022). The pomegranate deciduous trait is genetically controlled by a pgPolyQ-MADS gene. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.870207
Herrero, M., Rodrigo, J., and Wünsch, A. (2017). “Flowering, Fruit Set and Development,” in Cherries: botany, production and uses. Eds. Quero-Garcia, J., Iezzoni, A., Pulawska, J., and Lang, G. (CABI International, Wallingford, UK), 14–35. doi: 10.1079/9781780648378.0014
Kovaleski, A. P. (2024). The potential for an increasing threat of unseasonal temperature cycles to dormant plants. New Phytol. 244, 377–383. doi: 10.1111/nph.20052
Lloret, A., Quesada-Traver, C., Conejero, A., Arbona, V., Gómez-Mena, C., Petri, C., et al. (2021). Regulatory circuits involving bud dormancy factor PpeDAM6. Hortic. Res. 8, 261. doi: 10.1038/s41438-021-00706-9
Marin, J. A. (2003). High survival rates during acclimatization of micropropagated fruit tree rootstocks by increasing exposures to low relative humidity. Acta Hortic. 616, 139–142. doi: 10.17660/ActaHortic.2003.616.13
Moser, M., Asquini, E., Miolli, G. V., Weigl, K., Hanke, M. V., Flachowsky, H., et al. (2020). The MADS-Box gene MdDAM1 controls growth cessation and bud dormancy in apple. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01003
Ono, K., Akagi, K., Morimoto, T., Wünsch, A., and Tao, R. (2018). Genome re-sequencing of diverse sweet cherry (Prunus avium) individuals reveals a modifier gene mutation conferring pollen-part self-compatibility. Plant Cell Physiol. 59, 1265–1275. doi: 10.1093/pcp/pcy068
Rodriguez-A, J., Sherman, W. B., Scorza, R., Wisniewski, M., and Okie, W. R. (1994). ‘Evergreen’ peach, its inheritance and dormant behavior. J. Am. Soc Hortic. Sci. 119, 789–792. doi: 10.21273/jashs.119.4.789
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157, 485–497. doi: 10.1104/pp.111.181982
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkovič, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9, 4173. doi: 10.1038/s41467-018-06696-y
Singh, R. K., Miskolczi, P., Maurya, J. P., and Bhalerao, R. P. (2019). A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 29, 128–133. doi: 10.1016/j.cub.2018.11.006
Singh, R. K., Svystun, T., AlDahmash, B., Jönsson, A. M., and Bhalerao, R. P. (2017). Photoperiod-and temperature-mediated control of phenology in trees–a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Sonneveld, T., Robbins, T. P., and Tobutt, K. R. (2006). Improved discrimination of self-incompatibility S-RNase alleles in cherry and high throughput genotyping by automated sizing intron PCR products. Plant Breed. 125, 305–307. doi: 10.1111/j.1439-0523.2006.01205.x
Teotia, S. and Tang, G. (2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol. Plant 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Thompson, M. M., Smith, D. C., and Burgess, J. E. (1985). Nondormant mutants in a temperate tree species, Corylus avellana L. Theor. Appl. Genet. 70, 687–692. doi: 10.1007/bf00252298
Vaughan, S. P., Russell, K., Sargent, D. J., and Tobutt, K. R. (2006). Isolation of S-locus F-box alleles in Prunus avium and their application in a novel method to determine self-incompatibility genotype. Theor. Appl. Genet. 112, 856–866. doi: 10.1007/s00122-005-0187-9
Vergara, R., Olivares, F., Olmedo, B., Toro, C., Muñoz, M., Zúñiga, C., et al. (2021). “Gene editing in Prunus Spp.: The challenge of adapting regular gene transfer procedures for precision breeding,” in Prunus - Recent Advances. Eds. Küden, A. B. and Küden, A. (IntechOpen, United Kingdom). doi: 10.5772/intechopen.98843
Vergeer, P., Wagemaker, N., and Ouborg, N. J. (2012). Evidence for an epigenetic role in inbreeding depression. Biol. Lett. 8, 798–801. doi: 10.1098/rsbl.2012.0494
Wang, Y., Georgi, L. L., Reighard, G. L., Scorza, R., and Abbott, A. G. (2002). Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch. J. Hered. 93, 352–358. doi: 10.1093/jhered/93.5.352
Wang, Y., Lv, T., Fan, T., Zhou, Y., and Tian, C. E. (2025). Research progress on delayed flowering under short-day condition in Arabidopsis thaliana. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1523788
Wünsch, A. and Hormaza, J. I. (2004). Genetic and molecular analysis in Cristobalina sweet cherry, a spontaneous self-compatible mutant. Sex Plant Reprod. 17, 203–210. doi: 10.1007/s00497-004-0234-8
Xu, S. and Chong, K. (2018). Remembering winter through vernalization. Nat. Plants 4, 997–1009. doi: 10.1038/s41477-018-0301-z
Yamane, H., Wada, M., Honda, C., Matsuura, T., Ikeda, Y., Hirayama, T., et al. (2019). Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PloS One 14. doi: 10.1371/journal.pone.0214788
Zhang, B., Lee, K. C., Romañach, L. G., Ding, J., Marcon, A., Nilsson, O., et al. (2025). Phytochrome B and phytochrome-interacting-factor4 modulate tree seasonal growth in cold environments. Nat. Commun 16, 8114. doi: 10.1038/s41467-025-63391-5
Keywords: Prunus avium, evergrowing, evergreen, seasonal growth, dormancy, dormancy associated MADS-box genes, DAMs, inbreeding
Citation: Hedhly A, Martínez-Romera N, Gracia AP, Marin J, Arbeloa A, García E and Wünsch A (2025) An evergrowing sweet cherry for research and breeding. Front. Plant Sci. 16:1677862. doi: 10.3389/fpls.2025.1677862
Received: 01 August 2025; Accepted: 03 November 2025;
Published: 26 November 2025.
Edited by:
Andrea Miyasaka Almeida, Universidad Mayor, ChileReviewed by:
Magdi A. A. Mousa, King Abdulaziz University, Saudi ArabiaHisayo Yamane, Kyoto University, Japan
Copyright © 2025 Hedhly, Martínez-Romera, Gracia, Marin, Arbeloa, García and Wünsch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Wünsch, YXd1bnNjaEBhcmFnb24uZXM=
 Afif Hedhly
Afif Hedhly Nerea Martínez-Romera
Nerea Martínez-Romera Ana Pilar Gracia
Ana Pilar Gracia Juan Marin3
Juan Marin3 Ana Wünsch
Ana Wünsch