- 1School of Breeding and Multiplication (Sanya Institute of Breeding and Multiplication), College of Tropical Agriculture and Forestry, Hainan University, Sanya, Hainan, China
- 2Department of Biological Sciences, Faculty of Science and Technology, Virtual University, Lahore, Pakistan
- 3Department of Plant Pathology, College of Agriculture, University of Sargodha, Sargodha, Pakistan
- 4College of Agriculture, Guangxi University, Nanning, China
- 5Department of Biology, College of Science, King Khalid University, Abha, Saudi Arabia
- 6Prince Sultan Bin Abdelaziz for Environmental Research and Natural Resources Sustainability Center, King Khalid University, Abha, Saudi Arabia
Introduction: Botryosphaeria dothidea (B. dothidea) is a catastrophic fungal pathogen that threatens fruit production worldwide. Secreted peptidases like serine carboxypeptidases (SCPs) are well known to be involved in fungal virulence, but their role in B. dothidea is unknown.
Methodology: Here, we identified and functionally characterized Bd-SCP10, a homolog of SCPs found in B. dothidea, which is a member of the S10 family, using a split marker strategy for gene knockout and complementation.
Results: Mutants exhibited substantial phenotypic changes, including reduced radial growth and compromised biomass production, as well as altered pathogenicity and stress tolerance in response to multiple stress conditions. In contrast, complementation restored these traits, suggesting a functional role of Bd-SCP10. Particularly, Bd-SCP10 contributes to maintaining growth, cell wall integrity and adaptation to host-induced stresses, highlighting its involvement in fungal survival and pathogenicity.
Discussion: This study provides the first functional evidence that secreted peptidases in B. dothidea are a key factor in vegetative growth, pathogenicity, and stress tolerance. The identification and functional characterization of Bd-SCP10 led us to believe that it is a promising molecular target for eco-friendly strategies to manage diseases caused by B. dothidea and related pathogens.
1 Introduction
B. dothidea infects many woody plants worldwide and damages most parts of plants, including leaves, fruits, branches, and stems. It can cause fruit rot, leaf spot, twig dieback, and stem and branch canker diseases, leading to the death of infected trees (Jash et al., 2025). It has caught the attention of fruit tree pathologists, as it can infect most economically significant fruit trees, such as apples, pears, grapes, peaches, and blueberries, and cause severe diseases resulting in substantial annual losses. In China, apple white rot caused by B. dothidea is considered as one of the most destructive diseases (Dong and Guo, 2020). Raindrops facilitated the spread of fungal conidia to the host tissue surface, and conidia primarily form a germ tube and appressorium. After the development of the appressoria appressorium, it begins to invade cells and expand inside the cell, then infects the cells and absorbs nutrients from the infected tissues (Kim et al., 2016). Eco-friendly strategies for disease control measures are still unknown for this devastating fungal pathogen. That is why it is necessary to explore sustainable approaches. The host defense system is considered the first line of defense against fungal invasion, and the plant cell wall can provide an adequate defense against fungal attacks (Looi et al., 2017; Munzert and Engelsdorf, 2025). Alternatively, fungal pathogens employ a range of strategies, including physical, enzymatic (such as hemicellulase, cutinase, pectinase, cellulase, lipases, secreted peptidases, and degradative enzymes), and chemical effectors to overcome barriers during invasion and infection (Zhao et al., 2013; Rauwane et al., 2020). Comparative genomic analysis of B. dothidea has suggested that virulence- and pathogenicity-related genes are involved in regulating metabolic pathways associated with the production of various enzymes, including those involved in plant cell wall degradation, biosynthesis, cytochrome P450, carbohydrate-active, and secreted peptidases (Wang et al., 2018). Plant cell wall degrading and secreted peptidases are considered the most important factors related to pathogenicity, and the former have been well characterized as responsible for virulence in many phytopathogenic fungi. While the latter remains enigmatic (Wang et al., 2018; Munzert and Engelsdorf, 2025). SCPs are proteolytic enzymes belonging to the peptidase S10 family, characterized by a conserved Ser–Asp–His catalytic triad (Oda et al., 2022) and hydrolyze peptide bonds from the C-terminus of peptides and proteins (Semenova et al., 2020), thereby contributing to protein processing, nutrient acquisition, and regulation of cellular functions (Chen et al., 2022). In fungi, SCPs are often secreted into the extracellular environment, where they interact with host proteins and cell wall components, facilitating the release of nutrients and promoting host colonization. Beyond their housekeeping functions, SCPs are increasingly recognized as essential virulence factors (Muszewska et al., 2017; Da Silva, 2018; Madhu et al., 2020). They vary in their characteristics, including substrate selectivity, active site, and catalytic mechanism, and are also involved in a wide range of complex physiological activities (Da Silva et al., 2006; Dong et al., 2021). Generally, fungal pathogens secrete peptidases to modify spore production and germination and these enzymes act as virulence regulators (Krishnan et al., 2018; Yang et al., 2023). In addition, fungal peptidases can inhibit the host defense system by modifying and inactivating protein components of the protection machinery of the host (Xia, 2004; Shi et al., 2025). Fusarium graminearum, FgSCP was shown to be essential for fungal growth, toxin biosynthesis, stress tolerance, pathogenicity, and suppression of host immunity (Liu et al., 2024). Similarly, in Penicillium expansum, secretome profiling identified a PeBgl1 and PeSCP as enzymes required for virulence on apple fruit, emphasizing their direct role in host invasion (Wang et al., 2025). Several old studies have also shown the importance of secreted peptidases in the regulation of virulence in fungal pathogens, e.g., SsNEP2 in Sclerotinia sclerotiorum (Yang et al., 2022), BcCGF1 in Botrytis cinerea (Zhang et al., 2020a), CpSge1 in Cryphonectria parasitica (Lin et al., 2025b), Fospc2 in Fusarium odoratissimum (Yang et al., 2023), and GcStuA in Glomerella cingulate (Chethana et al., 2021). These findings highlight the functional role of SCPs as key modulators of fungal physiology, managing growth, stress adaptation, and host immune suppression. Although the recognized roles of SCPs in other phytopathogenic fungi are well understood, their functional role in B. dothidea remains largely unknown. Therefore, identifying the function of Bd-SCP10 offers a key step toward understanding the mechanisms of pathogenicity and will help in developing innovative, eco-friendly strategies for disease control in fruit crops.
2 Materials and methods
2.1 Strains and cultural conditions
The B. dothidea wildtype strain (Bd-wt) was generously provided by Hafiz Husnain Nawaz (Hainan University, Haikou, China). For colony morphology analysis, the control [CK, representing uninoculated potato dextrose agar medium (PDA)] and Bd-wt were cultured and replicated 6 times on PDA for 5 days (d) at 25°C (Wei et al., 2019).
2.2 Nucleic acid manipulations and polymerase chain reaction
Fungal gDNA was isolated from 5 d old mycelium through the 2X Cetyltrimethylammonium bromide (CTAB) method (Jasim et al., 2024). The verification and identification of mutants and complementary strains were performed as previously described through polymerase chain reaction (PCR) (Huang et al., 2017a).
2.3 Bioinformatics analysis
The full-length nucleotide sequence of the Bd-SCP10 gene, along with upstream and downstream flanking sequences, was downloaded from B. dothidea (ASM1106463v1, GenBank: GCA_011064635.1, https://www.ncbi.nlm.nih.gov/assembly/GCA_011064635.1/). Oligo primer analysis v. 7.0 (Molecular Biology Insights, Inc., Colorado, USA) was used to design the primers. For phylogenetic analysis, the protein sequence of Bd-SCP10 (GenBank: KAF4310043.1) was obtained from NCBI, and the homologs of SCPs in 12 other phytopathogenic fungi were identified via MEROPS (https://www.ebi.ac.uk/Tools/services/web/toolresult.ebi?jobId=ncbiblast-I20210421-144041-0569-47522791-p2m) (Rawlings, 2020), followed by the multiple sequence alignment with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Finally, the phylogenetic tree was generated using MEGA software v. 7.0 (Kumar et al., 2016; Ahmad et al., 2021) with the neighbor-joining (NJ) method. The dendrograms were based on clustered homologs, determined through bootstrap analysis (with 1000 replicates), and displayed replication percentages on the respective branches. ORFFINDER (https://www.ncbi.nlm.nih.gov/orffinder/) was used for the analysis of the open reading frame (ORF) (Varabyou et al., 2023), and the protein domain was predicted using Pfam (https://pfam.xfam.org/) (Savojardo et al., 2021).
2.4 Targeted knockout of the Bd-SCP10 gene from a wildtype strain
The deletion of the Bd-SCP10 gene was conducted using a split marker strategy, in which the selectable marker HYG replaced the targeted gene (Yuan et al., 2023). Briefly, in the first round of PCR, the fragment of the upstream flanking sequence (UPS) of Bd-SCP10 (887 base pairs (bp) was amplified with Up-F and Up-R primers, while the downstream flanking sequence (DNS) of 789 bp was amplified with Dn-F and Dn-R primers. On the other hand, the pCX62 plasmid (Lin et al., 2025a) was used to amplify the HYG resistance cassette for a 767 bp HY fragment with HYG-F/HY-R primers and a 931 bp YG fragment with HYG-R/YG-F primers. In the second round or fusion PCR, the Up-F/HY-R primers were used to fuse the UPS fragment with the HY fragment, while the Dn-R/YG-F primers were used to fuse the DNS fragment with the YG fragment through splicing of overlapped sequence extension, and templates were used in equal proportion in fusion PCR. Fusion PCR cycling conditions were conducted initially at 95 °C for 5 minutes (min), followed by 35 cycles of 95 °C for 30 seconds (s), 60 °C for 45 s, and 72 °C for 90 s, and finally at 72 °C for 10 min. The products of fusion PCR were transformed through the PEG-mediated transformation method (Zhang et al., 2022) into the protoplasts (2×107 cells/mL) isolated from 5 d old mycelium of Bd-wt using the previously described method (Ning et al., 2022). The transformants were cultured on PDA supplemented with 50 µg/mL hygromycin B (HYG; Roche, USA), and the developed colonies were randomly selected and verified by standard PCR using the two sets of primers Bd-SCP10-F/Bd-SCP10-R and HYG-F/HYG-R.
2.5 Complementation of the Bd-SCP10 gene
The complementation of the mutant ΔBd-SCP10a was achieved by inserting the Bd-SCP10 gene, along with a NEO resistance cassette, into the knockout mutant through a split marker strategy (Gravelat et al., 2012). The first-round PCR was briefly conducted to amplify the 47 bp upstream flanking region (UPS) together with the full length of the Bd-SCP10 gene, resulting in a 2800 bp amplified fragment and the 832 bp amplified fragment of the downstream flanking region (DNS). Then, the pSELECT-neo plasmid (InvivoGen, USA) was used to amplify the NEO drug cassette for the NE fragment (975 bp) with NEO-F/NE-R primers and the EO fragment (543 bp) with NEO-R/EO-F primers. In the second round or fusion PCR, the cycling conditions were kept the same as those used for deleting the targeted gene. The corresponding primers UP-F/NE-R were used for the fusion of the UPS fragment with the NE fragment. In contrast, the Dn-R/EO-F primers fused the DNS fragment with the EO fragment by splicing an overlapped extension. The products of fusion PCR were transformed through the PEG-mediated transformation method (Zhang et al., 2022) into the protoplasts (1×105 cells/mL) isolated from 5 d old mycelium of mutant ΔBd-SCP10a using the previously described method (Ning et al., 2022). Finally, the complementary transformants were screened on PDA supplemented with 150 μg/mL of the antibiotic G418, and the generated colonies were randomly selected and verified by standard PCR using the two sets of primers Bd-SCP10-F/Bd-SCP10-R and NEO-F/NEO-R.
2.6 Phenotype analysis
For the growth rate analysis, mycelial plugs (5 mm diameter) excised from colony margins were placed in the center of PDA (at least 3 replicates) and then incubated for 5 d at 25 °C in the dark (Wei et al., 2019). The per day radial growth rate (Oledibe et al., 2023) was calculated with formula [(Colony diameter – Plug diameter ÷ 2) × Number of incubation days] and per day biomass production rate (Novotna et al., 2023) was calculated with formula (Colony weight ÷ Number of incubation days) and final measurements were taken at 5 days post-inoculation (dpi). The hyphal tips of fungal strains were observed under a Ni90 microscope (Nikon, Japan) after 5 dpi incubation, following the previous method (Aboelfotoh Hendy et al., 2019; Zhang et al., 2020b).
2.7 Virulence assay
Virulence assay was performed on pear fruits (Pyrus bretschneideri var. huangguan) with the mycelial plugs (5 mm diameter) derived from actively growing colonies (at least 6 replicates), and uncolonized PDA plugs were involved in parallel as a control (CK). Mechanically produced wounds with a sterile needle on the fruits were used for inoculation by reversely placing mycelial plugs. After inoculation, the fruits were incubated at 25 °C with 100% relative humidity. The resulting lesions were measured at 5 days post-infection (dpi), and photos were taken to document lesion development.
2.8 Stress response assays
For a stress response assays, mycelia of Bd-wt, complementary strains, and mutants were quantitatively analyzed after cultured on unamended PDA or amended PDA [supplemented with different stress agents (at least 6 replicates), including 0.04% sodium dodecyl sulfate (SDS), 0.5 M calcium chloride (CaCl2), 1 M glucose (C6H12O6), 1.5 M sodium chloride (NaCl), 1 M potassium chloride (KCl), and 0.05% hydrogen peroxide (H2O2)] for 5 d, respectively. The percentage growth inhibition rates were calculated with the formula [(Do – Dt ÷ Do) × 100], where Do represents the mean colony diameter on PDA, and Dt represents the mean colony diameter on amended PDA (Khanh et al., 2024).
2.9 Statistical analysis
The quantitative data were statistically analyzed using Statistix v. 8.1 (Analytical Software, Florida, USA) with ANOVA and the least significant difference (LSD) test at P ≤ 0.05. The bar graphs were drawn on GraphPad Prism v. 8.0 (GraphPad Software, California, USA).
3 Results
3.1 Phenotypic and virulence assay of the B. dothidea wildtype strain
For phenotypic analysis, the CK and Bd-wt were cultured on PDA for 5 d at 25 °C. CK culture showed a plain dish of PDA without fungal growth, while Bd-wt culture showed the whitish color of the colony with a fluffy cotton-like white margin and a relatively thick mycelium mat, as shown in Supplementary Figure S1A. For virulence assays, CK does not induce disease on pear fruits (var. huangguan, 6 replicates). However, lesions induced by Bd- wt can be seen in Supplementary Figure S1B after 5 dpi. Therefore, these results suggested that Bd-wt is a virulent strain.
3.2 Identification and characterization of the Bd-SCP10 gene
The MEROPS database analysis of amino acid sequences similar to SCPs belonging to the peptidase S10 family revealed its homolog in B. dothidea, which was tentatively named as Bd-SCP10. Bd-SCP10 shared the highest identity (40.9%, E-value = 8.9E-133) with the homolog in Microsporum canis (EEQ30006.1), a member of the family Arthrodermataceae. A phylogenetic tree was constructed based on the amino acid sequences of Bd-SCP10 together with other homologs of SCPs chosen from 12 fungal species based on percentage amino acid sequence identity and E-value (32% to 40.9%, and 8.9E-133 to 7.9E-57, respectively). The neighbor-joining phylogenetic tree showed that all the fungal SCPs were grouped into 3 clusters (a to c), and Bd-SCP10 was located in a separate clade far from other members in cluster A and that has been shown in Figure 1A. The multiple sequence alignment results depicted that Bd-SCP10 belongs to the peptidase S10 family (PF00450) and this family have 43 conserved amino acids among in all of the selected phytopathogenic fungi shown in Figure 1B.
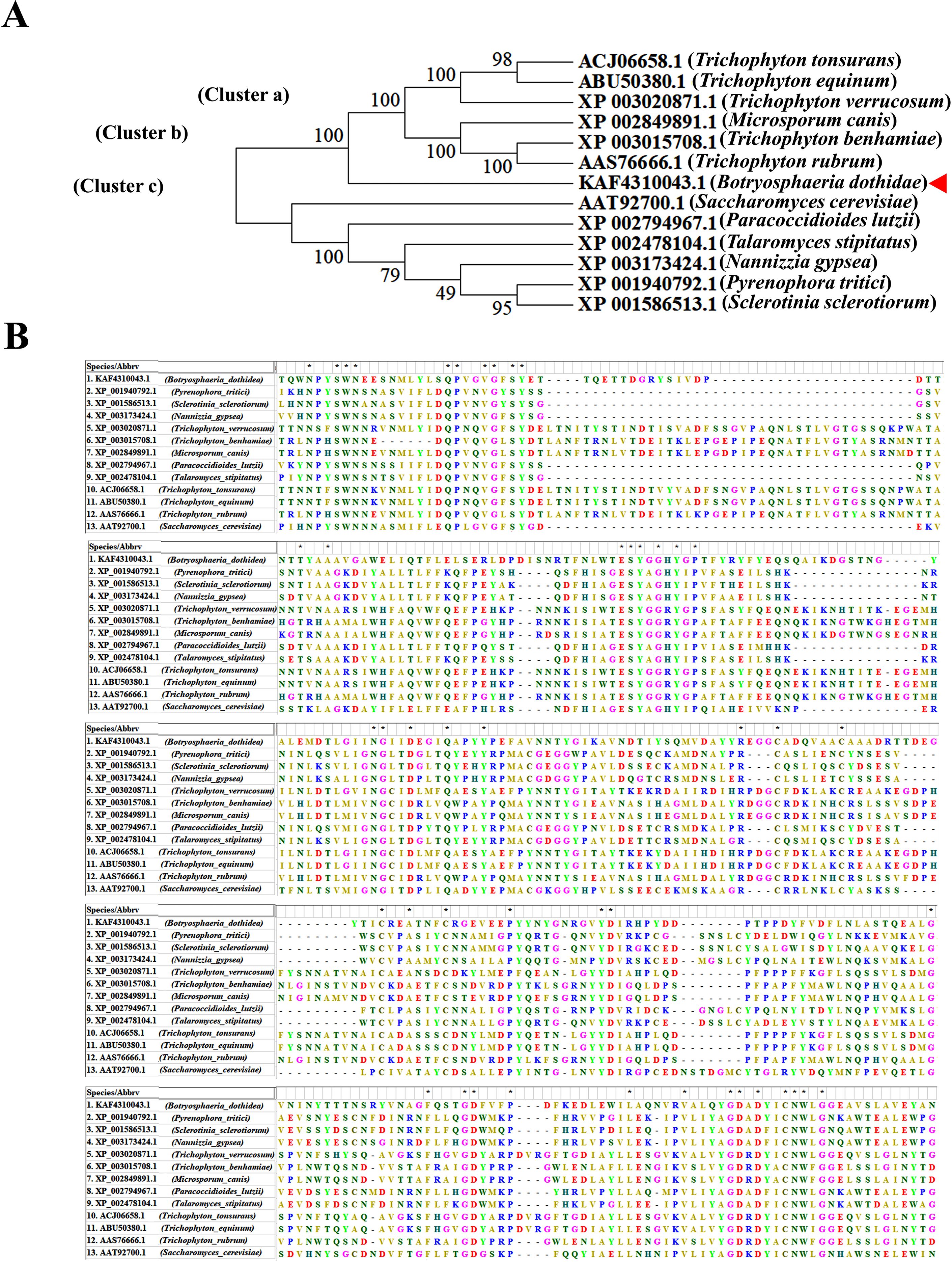
Figure 1. Phylogenetic analysis and multiple sequence alignment of Bd-SCP10 together with other fungal homologs of SCPs. (A) The phylogenetic tree was generated using MEGA v. 7.0 with the neighbor-joining method, and fungal SCPs were grouped into 3 clusters (a to c) as indicated by the arrow. The number beside the branch nodes refers to the bootstrap value, and the host’s fungal name is presented in brackets, followed by the homolog accession number. B. dothidea is indicated with a triangle ahead of its accession number. (B) The sequences of amino acids are aligned using Clustal Omega, and conserved amino acids among the selected pathogenic fungi are denoted by an asterisk (*).
3.3 Knockout of the Bd-SCP10 gene from a wildtype strain
To verify the gene function of Bd-SCP10, Bd-wt induced long lesions (approximately 35.8 mm) as inoculated on pear fruits (Pyrus bretschneideri var. huangguan) as shown in Supplementary Figures S1A, B, was subjected to knockout for the target gene through split marker strategy, i.e., Bd-SCP10 gene was replaced with a HYG resistance cassette through homologous recombination (Dong and Guo, 2020) of the flanking parts around the targeted gene and schematic representation has been shown in Figure 2A. The first-round PCR was conducted to amplify 887 bp UPS and 789 bp DNS fragments, as well as the HY (767 bp) and YG (931 bp) fragments of the HYG resistance cassette as shown in Figure 2B and Supplementary Figure S2A. The UPS fragment was further fused with the HY fragment using fusion PCR, generating the fragment termed as 5′HY (1654 bp). Similarly, fragment DNS was fused with YG to generate a fragment termed as 3′YG (1720 bp), which is shown in Figure 2C and Supplementary Figure S2B. The resulting DNA fragments 5′HY and 3′YG were purified and used to transfect the protoplasts of Bd-wt through PEG-mediated transformation. Upon transfection, a UPS-HYG-DNS combination replaced the Bd-SCP10 gene through homologous recombination. After transfection, 12 colonies were grown in media containing the antibiotic HYG, and 3 colonies named ΔBd-SCP10a, ΔBd-SCP10b, and ΔBd-SCP10c were selected for identification by a strict PCR strategy with Bd-SCP10-F/Bd-SCP10-R and HYG-F/HYG-R primers mentioned in Supplementary Table S1, targeting Bd-SCP10 and HYG fragments, respectively. The results showed that the Bd-SCP10 amplified fragment of 1200 bp was detected in Bd-wt except in mutants, while the HYG amplified fragment of 1383 bp was present in mutants except for Bd-wt, as depicted in Figure 2D, suggesting that Bd-SCP10 was successfully knocked out.
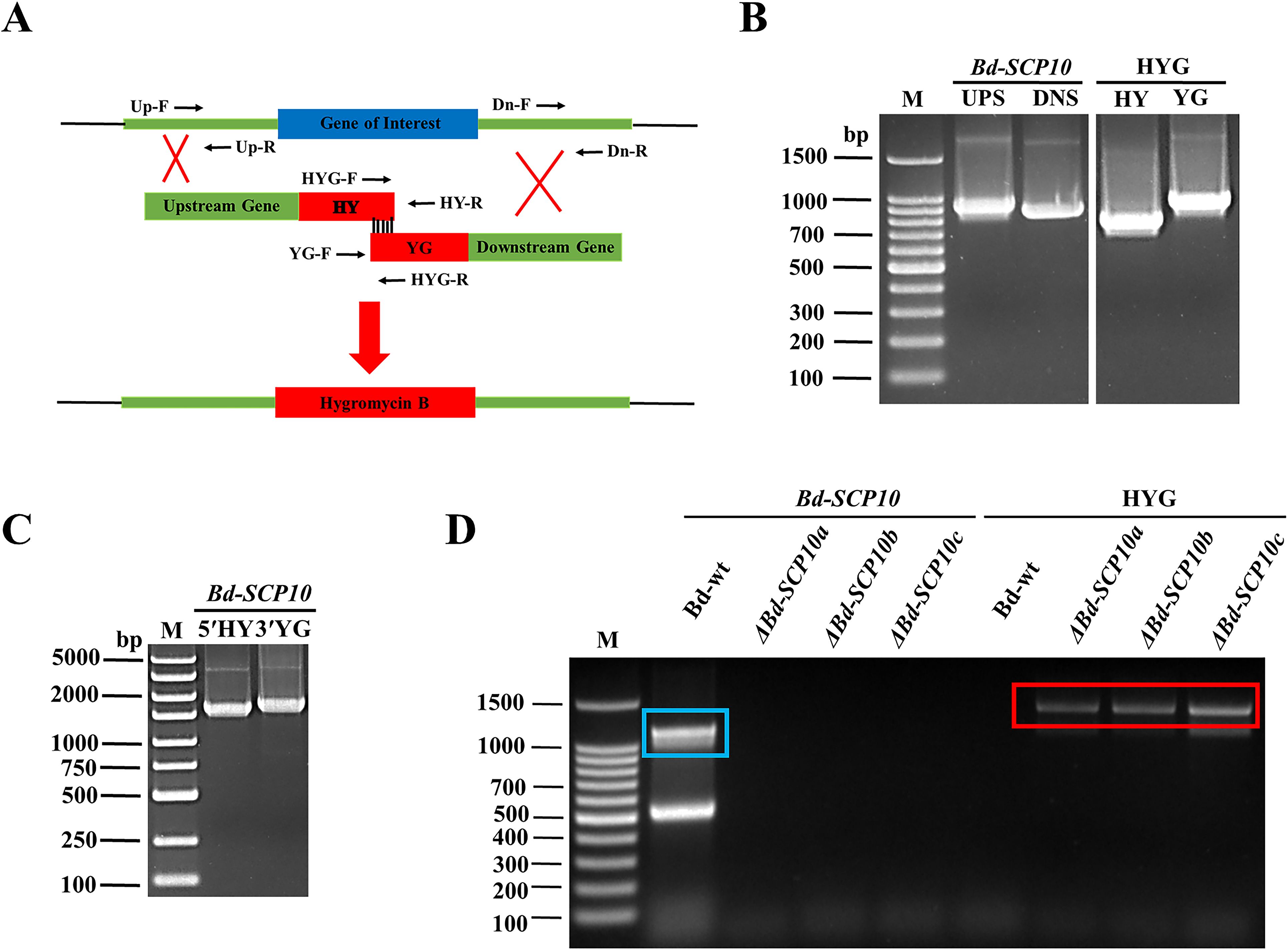
Figure 2. Targeted knockout of Bd-SCP10 from B. dothidea through split marker strategy and PCR identification of the mutants. (A) Schematic representation of the split marker strategy for deleting Bd-SCP10 from Bd-wt. (B) First-round PCR amplified fragments (UPS: 887 bp, DNS: 789 bp, HY: 767 bp, and YG: 931 bp). (C) Fusion PCR products showing the synthesis of the two recombinant fragments 5′HY (1654 bp) and 3′YG (1720 bp) used for transformation. (D) PCR identification for the Bd-SCP10 gene and HYG resistance cassette in Bd-wt and mutants. Amplification of Bd-SCP10 (blue box, amplified band of 1200 bp) was detected only in Bd-wt, while amplification of the HYG cassette (red box, amplified band of 1383 bp) was detected only in mutants (ΔBd-SCP10a, ΔBd-SCP10b, and ΔBd-SCP10c), confirming successful gene knockout. Here, M is denoted for DNA markers (Note: The original gel documentation figures for (B, C) can be seen in Supplementary Figure S2).
3.4 Complementation of the Bd-SCP10 gene
The mutant ΔBd-SCP10a was subjected for complementation of the targeted gene through split marker strategy, i.e., the HYG resistance cassette was replaced with gene Bd-SCP10 and NEO resistance cassette through homologous recombination (Dong and Guo, 2020) of the flanking parts around the HYG resistance cassette and schematic representation can been seen in Figure 3A. The first-round PCR was conducted to amplify the 2800 bp UPS and 832 bp DNS fragments, along with the NE (975 bp) and EO (543 bp) fragments of the NEO resistance cassette, as shown in Figure 3B and Supplementary Figure S3A. The UPS fragment was further fused with the NE fragment using fusion PCR, resulting in the 5′NE (3775 bp) fragment. At the same time, the DNS with EO generated the 3′EO (1375 bp) fragment, as depicted in Figure 3C and Supplementary Figure S3B. The resulting DNA fragments 5′NE and 3′EO were purified and used to transfect the protoplasts of the mutant ΔBd-SCP10a through PEG-mediated transformation. Upon transfection, a UPS-NEO-DNS combination replaced the HYG resistance cassette through homologous recombination. After transfection, 8 colonies were grown in the media containing G418 antibiotic, and colonies CΔBd-SCP10a, CΔBd-SCP10b, and CΔBd-SCP10c were picked for identification by PCR strategy with Bd-SCP10-F/Bd-SCP10-R and NEO-F/NEO-R primers described in Supplementary Table S1, targeting Bd-SCP10 and NEO fragments, respectively. The results showed that the amplified fragment of 1200 bp Bd-SCP10 was detected in Bd-wt together with complementary strains, while the NEO amplified fragment of 1200 bp was present in complementary strains except for Bd-wt, as shown in Figure 3D, suggesting that complementation of Bd-SCP10 was successful and complementary strains were generated from mutant ΔBd-SCP10a.
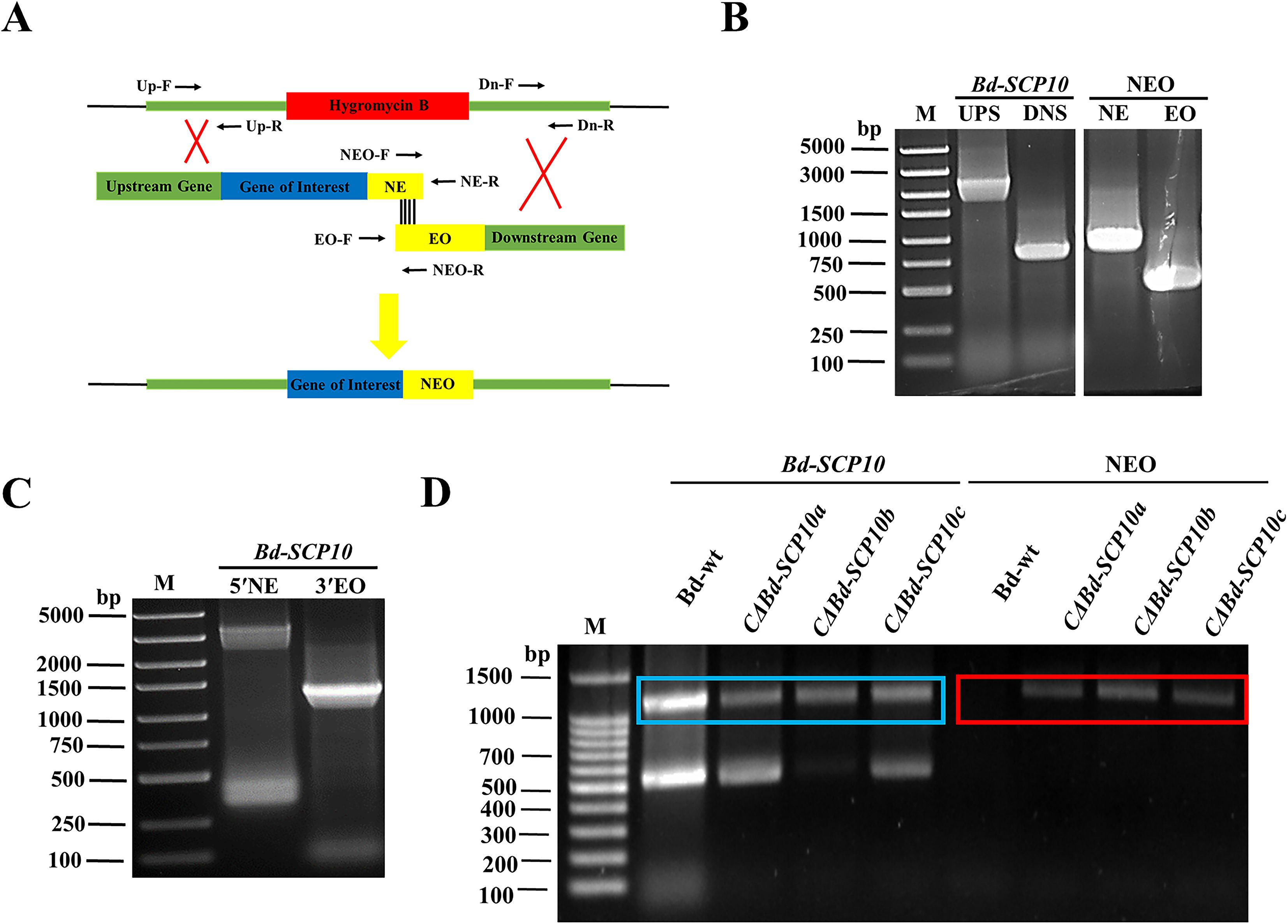
Figure 3. Complementation of the Bd-SCP10 mutant through the split marker strategy and PCR identification of the complementary strains. (A) Schematic representation of the complementation through the split marker strategy, in which Bd-SCP10 was reintroduced into mutant ΔBd-SCP10a with the NEO resistance cassette. (B) First-round PCR fragments (UPS: 2800 bp, DNS: 832 bp, NE: 975 bp, and EO: 543 bp). (C) Fusion PCR products, which showed recombinant fragments (5′NE, 3775 bp; 3′EO, 1375 bp), were used for transformation. (D) PCR identification for the Bd-SCP10 gene and NEO resistance cassette in Bd-wt and complementary strains. Bd-SCP10 (blue box, amplified band of 1200 bp) was detected in Bd-wt and complementary strains (CΔBd-SCP10a, CΔBd-SCP10b, and CΔBd-SCP10c), while the NEO cassette (red box, amplified band of 1200 bp) was detected only in complementary strains, confirming restoration of Bd-SCP10 expression in ΔBd-SCP10a. Here, M is denoted for DNA markers (Note: The original gel documentation figures for (B, C) are shown in Supplementary Figure S3).
3.5 Bd-SCP10 gene is required for vegetative growth and virulence
To test whether Bd-SCP10 is related to the fungal vegetative growth, the mutants and complementary strains, along with Bd-wt, were cultured on PDA in triplicate for 5 d and subjected to assessment of their phenotype, growth, and biomass. These mutants exhibited snowy white and compact aerial mycelium throughout the entire colonies, characterized by random radial grooves. In contrast, both complementary strains and Bd-wt showed a collapsed and cleared mycelial growth in the colony center. At the same time, upward fluffy and cottony mycelia in the margins can be seen in Figure 4A. Additionally, the mutants exhibited growth rates ranging from 2.1 mm/d (ΔBd-SCP10b) to 2.4 mm/d (ΔBd-SCP10c), which were substantially reduced compared to Bd-wt (15.9 mm/d). However, the growth rates for Bd-SCP10 complementary strains ranged from 15.4 mm/d (CΔBd-SCP10a) to 15.5 mm/d (CΔBd-SCP10c), which are nearly identical to Bd-wt, and are shown in Figure 4B and Supplementary Table S2. The biomass production for the mutants was observed, ranging from 0.018 g/d (ΔBd-SCP10b) to 0.019 g/d (ΔBd-SCP10c), which were significantly lower than Bd-wt (0.2 g/d), while complementary strains produced biomass approximately 0.2 g/d (CΔBd-SCP10b to CΔBd-SCP10c), similar to Bd-wt, as shown in Figure 4C and Supplementary Table S2. Moreover, the hyphal tips of Bd-wt and complementary strains have fewer septa at hyphal tips and more dense mycelial growth than mutants, as observed in Supplementary Figures S4A, B. The results showed that Bd-SCP10 is involved in the phenotypic development and vegetative growth of B. dothidea. To test whether Bd-SCP10 is associated with fungal pathogenicity, the mutants and complementary strains together with Bd-wt were assessed on the pear fruits (var. huangguan, 6 replicates). At 5 dpi, the Bd-SCP10 complementary strains induced lesions ranging from 28.4 mm (CΔBd-SCP10c) to 29.1 mm (CΔBd-SCP10b). It is almost similar to the lesion lengths (31.1 mm) induced by Bd-wt. In contrast, the mutants did not induce lesions, as shown in Figures 4A, D, and Supplementary Table S3. These results suggested that Bd-SCP10 is responsible for the pathogenicity of B. dothidea.
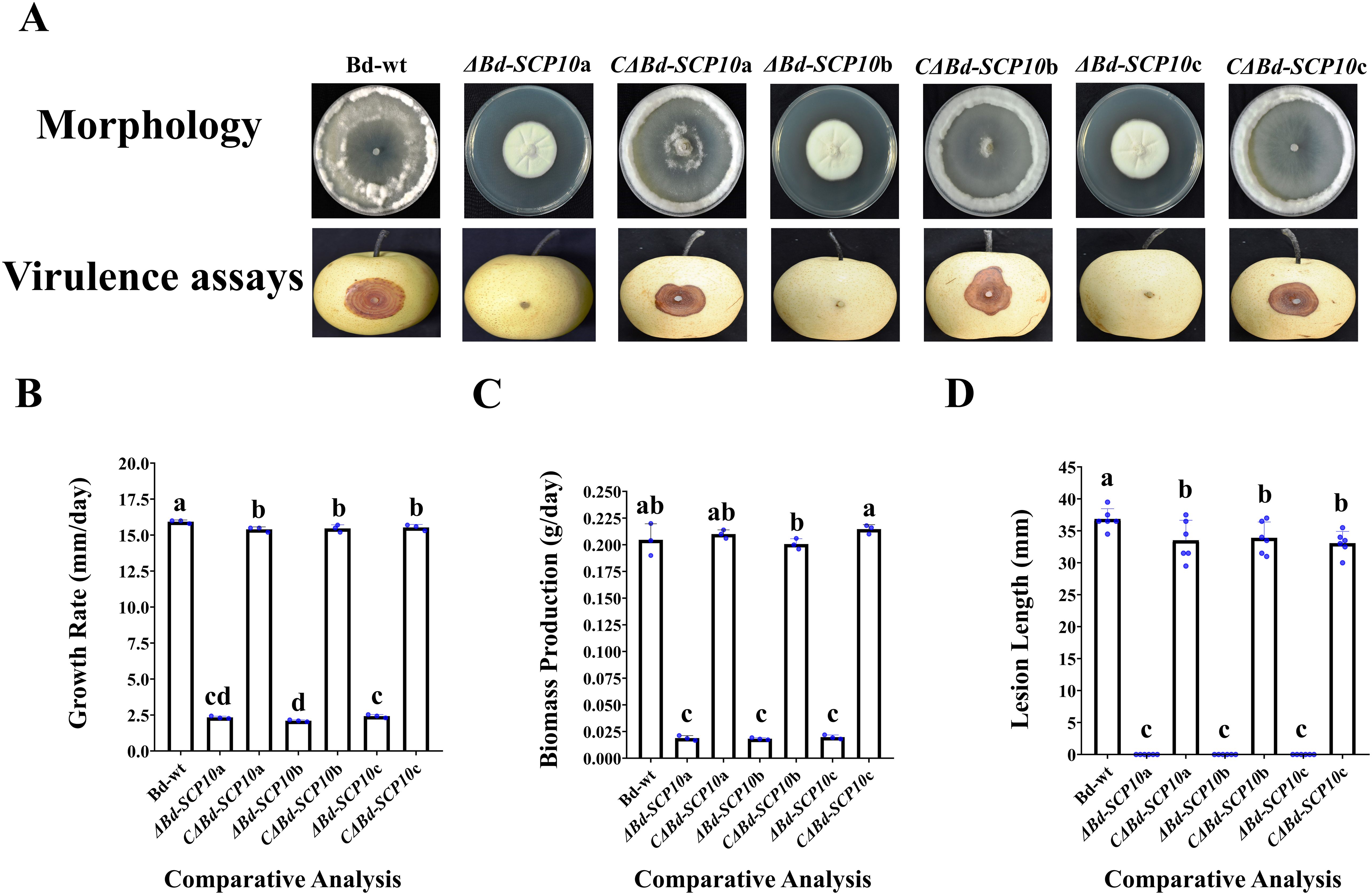
Figure 4. Comparative analysis of Bd-wt, mutants, and complementary strains for vegetative growth, biomass production, and virulence. (A) Phenotype and induced symptoms on the inoculated pear fruits (Pyrus bretschneideri var. huangguan) for Bd-wt, mutants, and complementary strains. (B-D) Bar graphs for the growth rates, biomass, and lesion lengths on the inoculated pear fruits for Bd-wt, mutants, and complementary strains, respectively. The alphabet on a column represented analysis of variance (ANOVA) with the least significant difference (LSD) at P ≤ 0.05. During this experimental process, 3 replicates were used for vegetative growth and biomass production, and 6 replicates were used for the virulence assay.
3.6 Bd-SCP10 gene plays a role in stress tolerance and cell wall integrity
Mutants and complementary strains, along with Bd-wt, were cultured on PDA amended with 0.04% SDS to analyze the level of tolerance to cell wall disruption. The results showed significantly higher growth inhibition rates ranging from 45.0% to 49.9% in mutants compared to Bd-wt (27.6%) and complementary strains (29.3% to 33.9%). Similarly, on PDA amended with chemicals used to response against hyperosmotic stresses (0.5 M CaCl2, 1 M C6H12O6, 1.5 M NaCl, and 1 M KCl) all mutants showed significantly reduced growth rates on PDA amended with 0.5 M CaCl2, 1 M C6H12O6, and 1 M KCl (42.8 to 49.3%, 15.9 to 25.2%, and 42.2 to 48%, respectively) as compared to Bd-wt (7.5%, 5%, and 15.2%) and the complementary strains (8.6 to 14.2%, 15.9 to 25.2%, and 22.8 to 30%). In contrast, the mutants did not show a significant difference in the growth inhibition rates on PDA amended with 1.5 M NaCl (72.4 to 74.7%) as compared with Bd-wt (75.31%) or the complementary strains (76 to 77.2%). Moreover, on PDA amended with 0.05% H2O2, the response of strains against oxidative stress was observed. The results highlighted that mutant had a significantly higher growth inhibition rate from 46% to 59% than Bd-wt (1.6%) and complementary strains (0.8 to 2.5%), as shown in Figures 5 and 6, along with Supplementary Tables S4 and S5. These results suggested that the Bd-SCP10 gene is involved in cell wall integrity and contributes to the resistance against hyperosmotic and oxidative stresses in B. dothidea.
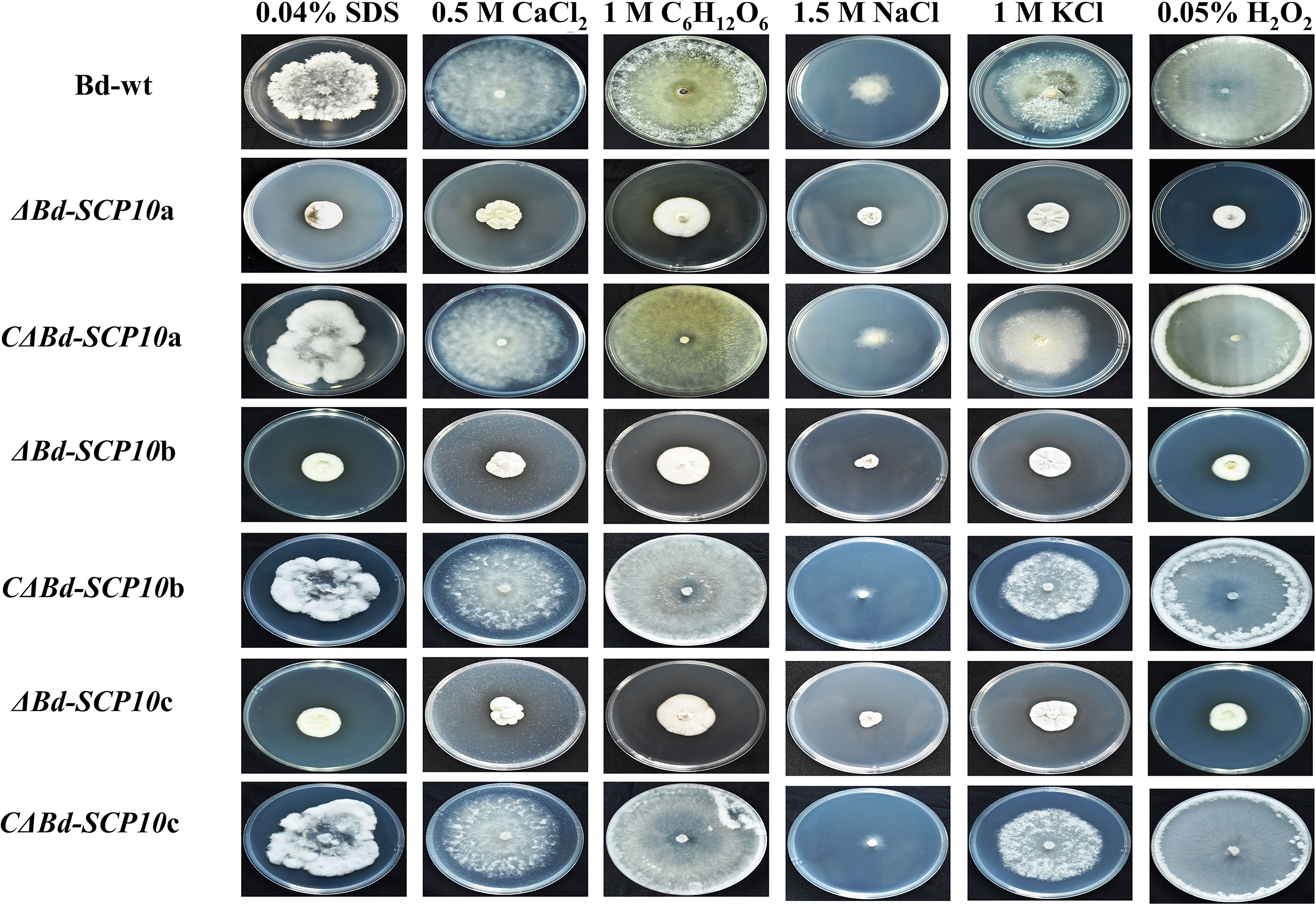
Figure 5. Phenotypic comparison of Bd-wt, mutant, and complementary strains on PDA amended with 0.04% SDS, 0.5 M CaCl2, 1 M C6H12O6, 1.5 M NaCl, 1 M KCl, and 0.05% H2O2 after 5 d at 25 °C. Mutants exhibited reduced growth relative to Bd-wt and complementary strains, particularly under SDS, CaCl2, C6H12O6, KCl, and H2O2 stress.
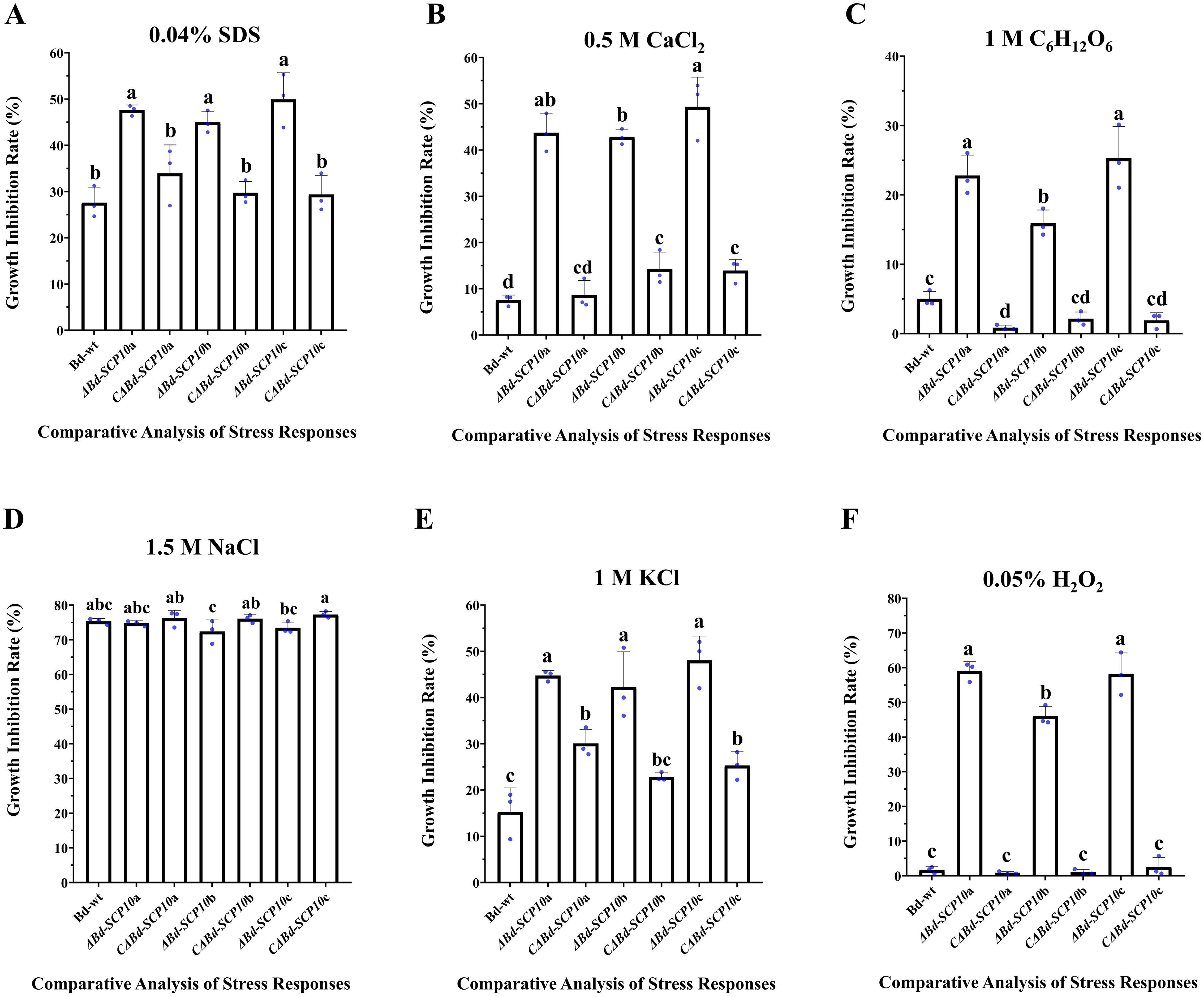
Figure 6. Comparative analysis of stress responses of Bd-wt, mutant, and complementary strains. Growth inhibition rates were measured on PDA amended with 0.04% SDS (A), 0.5 M CaCl2 (B), 1 M C6H12O6 (C), 1.5 M NaCl (D), 1 M KCl (E), and 0.05% H2O2 (F). Alphabet on a columns depicted ANOVA with the LSD at P ≤ 0.05, mutants exhibiting pronounced sensitivity to cell wall disrupting, osmotic, and oxidative stresses, underscoring the importance of Bd-SCP10 in fungal stress tolerance.
4 Discussion
SCPs were functionally reported to be involved in protein processing and degradation, as well as the production of secondary metabolites (Chen et al., 2022). This study aligns with emerging evidence that secreted peptidases, particularly SCPs belonging to the S10 family, are key virulence factors (Iqbal et al., 2018; Zhang et al., 2019) in phytopathogenic fungi (Semenova et al., 2020). SCPs were also identified as potential regulators of phenotypes, habitat adaptation, and growth (Iqbal et al., 2018). However, the functions of SCPs in the phytopathogenic fungus B. dothidea are still unknown. In this study, we performed functional characterization of the Bd-SCP10 gene, a homolog of the SCPs in B. dothidea. Our experimental data confirmed that Bd-SCP10 plays a key role in vegetative growth, pathogenicity, and stress response, as demonstrated by generating mutants and complementary strains using a split marker strategy. FgSCP was previously reported for its role in the virulence of F. graminearum, and affects fungal growth, stress tolerance, and pathogenicity. It also suppresses cell death triggered by the INF1 elicitor, indicating its role in modulating plant immune responses (Liu et al., 2024). The substantial reduction in radial growth of Bd-wt (2.1–2.4 mm/d vs. 15.9 mm/d) and biomass production (0.018–0.019 g/d vs. 0.2 g/d) in mutants underscores the importance of the Bd-SCP10 in fungal physiology. Similar observations were found in Botrytis cinerea and Sclerotinia sclerotiorum, highlighting that peptidases often facilitate nutrient acquisition by hydrolyzing host proteins into assimilable amino acids (Urbanek and Kaczmarek, 1985; Clark et al., 1997). Moreover, it was reported that Magnaporthe oryzae wildtype and complementary strains have denser mycelial development than mutants, as the hyphal tips of the wildtype strain have longer cell lengths and less septation as compared to the mutant, which has more septation and shorter cell lengths (Aboelfotoh Hendy et al., 2019; Zhang et al., 2020b). Our microscopic studies revealed that the thin and compact mycelia of mutants suggested that Bd-SCP10 regulates hyphal expansion or septation, processes that require precise proteolytic activity to remodel cell walls or recycle proteins (Zhang et al., 2020b). Furthermore, we did not find any differences in colony pigmentation among the mutants, suggesting that the production of secondary metabolites (e.g., melanin or polyketide-derived pigments) was not noticeably affected under PDA culture conditions. Even so, SCPs have been reported for the regulation of fungal secondary metabolism in other fungal species (Madhu et al., 2020). It suggested that the Bd-SCP10 gene is involved in the fungal growth and the phenotypic development of B. dothidea. SCPs regulate virulence in Phellinus sulphurascens (Williams et al., 2014) and Clonostachys rosea (Iqbal et al., 2018). A secreted peptidase, BEC1019, regulating a virulence factor, was identified in Blumeria graminis, which is required for haustorium development. Silencing of BEC1019 induced hypovirulence in the fungus (Zhang et al., 2019). An ortholog Rs-SCP1 was identified in the plant-parasitic nematode Radopholus similis and was involved in reducing pathogenicity (Huang et al., 2017b). Mutations in the FgSCP gene compromise nutritional growth and stress tolerance, and deletion leads to reduced pathogenicity in F. graminearum. Expression is upregulated during infection, suggesting involvement in invasion and potentially by suppressing host defense genes (Liu et al., 2024). The full loss of pathogenicity in mutants on pear fruits and the restoration of pathogenicity in complementary strains represented the essential role of Bd-SCP10 in infection. Mirror findings in Fusarium culmorum revealed that secreted peptidases degrade host defense proteins, enabling tissue colonization by Fusarium culmorum (Urbanek and Yirdaw, 1984). Notably, B. dothidea inoculation relied on wounded fruit surfaces, implying that Bd-SCP10 acts post penetration by degrading plant cell wall components or suppressing host immune responses. For instance, fungal proteases in Glomerella cingulata cleave pathogenesis-related (PR) proteins, neutralizing plant defenses (Clark et al., 1997). The hypersensitivity of ΔBd-SCP10 mutants to cell wall disrupting agents (0.04% SDS) and hyperosmotic stressors (0.5 M CaCl2, 1 M C6H12O6, and 1 M KCl) emphasized the role of Bd-SCP10 in maintaining cell wall integrity and osmotic homeostasis. These results align with studies related to Aspergillus fumigatus, in which SCPs were reported to stabilize the cell wall under stress by processing structural proteins (Gravelat et al., 2012). Mannoproteins present in the cell wall of Candida albicans play important roles in cell wall remodeling under stress. These proteins interact with cell wall integrity and stress-activated signaling pathways, such as the high-osmolarity glycerol (HOG) and mitogen-activated protein kinase (MAPK) pathways (e.g., Cek1 and Mkc1), which explain how cell wall protein processing ties into stress tolerance (Ibe and Munro, 2021). Although direct evidence connecting SCPs to the processing of fungal wall proteins is lacking, the moonlighting capacity of extracellular fungal proteases (Satala et al., 2023) supports the hypothesis that Bd-SCP10 may serve a functional role in vegetative growth, pathogenicity, and stress tolerance. However, the unaltered sensitivity to 1.5 M NaCl in mutants suggested that Bd-SCP10 selectively regulates osmotic stress pathways, possibly via interactions with calcium or potassium signaling cascades rather than sodium-specific transporters. In plants, the rapid production and accumulation of reactive oxygen species (ROS) are considered the initial response to invading pathogens (Shetty et al., 2007; Imam et al., 2016). The H2O2 is a critical ROS that host plants can produce in response to fungal infection and initiate lipid peroxidation, DNA damage, formation of hydroxyl radicals, and protein oxidation (Rolke et al., 2004). Therefore, proper cell wall integrity and stress tolerance are essential for successful fungal infection and thus required for pathogenicity in numerous phytopathogenic fungi (Kim et al., 2009). Similarly, oxidative stress hypersensitivity (0.05% H2O2) suggests that Bd-SCP10 may mitigate ROS generated during host invasion, a strategy employed by Magnaporthe oryzae to counteract plant oxidative bursts (Zhang et al., 2020b). The partial restoration of stress tolerance in complementary strains indicated the potential epistatic interactions or incomplete genetic rescue, warranting further transcriptomic analyses to map regulatory networks involving Bd-SCP10. In addition, the HOG pathway is associated with hyperosmotic stresses and the osmoregulation ability of fungal pathogens, primarily due to its involvement in an environment characterized by hyperosmotic stresses (Yaakoub et al., 2022). However, our studies found that mutants showed hypersensitivity in response to hyperosmotic stresses (0.5 M CaCl2, 1 M C6H12O6, and 1 M KCl, except for 1.5 M NaCl). Deletion of Bd-SCP10 from Bd-wt has no link with the osmoregulation of 1.5 M NaCl, and we found the same results as mutant Bdo-05381 from B. dothidea strains HTLW03 and ZY7 (Dong and Guo, 2020). These results indicate that Bd-SCP10 is a positive modulator of the HOG pathway for the hyperosmotic stress response against 0.5 M CaCl2, 1 M C6H12O6, and 1 M KCl but a negative modulator for the hyperosmotic stress response against 1.5 M NaCl. Phylogenetically, SCPs are conserved in phytopathogenic fungi (Lv et al., 2023). However, Bd-SCP10 clusters separately from SCPs homologs found in other fungi. These results suggest evolutionary divergence personalized to the host range or environmental niche of B. dothidea. This divergence may explain its unique regulatory roles, such as its apparent lack of relevance to NaCl tolerance. Comparative genomics could elucidate whether homologs of Bd-SCP10 in related pathogens share similar functional specializations. While this study provides compelling evidence for multifunctionality, limitations exist for Bd-SCP10. The split marker knockout strategy although is reliable method however it does not rule out off-target effects. CRISPR-Cas9 validation could strengthen conclusions. Additionally, in vitro, fruit assays may not fully replicate field conditions, where host-pathogen interactions involve complex environmental and immunological variables. Future studies should aim to identify the direct substrates of Bd-SCP10 through proteomic approaches to clarify its mechanistic role in host invasion. Host response profiling using transcriptomics or metabolomics could reveal how Bd-SCP10 modulates plant defense pathways, particularly ROS detoxification and PR protein activity. In planta, validation under natural conditions will be essential to assess its role in systemic colonization and disease spread. Moreover, exploring Bd-SCP10 associated stress regulatory networks and evolutionary comparisons with homologs of SCPs in related fungi may uncover unique adaptations. Finally, screening for specific inhibitors of Bd-SCP10 could provide novel, eco-friendly strategies for controlling B. dothidea infections.
5 Conclusion
This study provides the first functional characterization of Bd-SCP10, a homolog of serine carboxypeptidase belonging to the S10 family in B. dothidea. Our results demonstrate that Bd-SCP10 is essential for pathogenicity and stress tolerance, also revealing its role in maintaining growth and cell wall integrity. The complete loss of pathogenicity in mutants suggested that Bd-SCP10 is an essential determinant of pathogenicity. However, identifying Bd-SCP10 as a functional regulator of vegetative growth, pathogenicity, and stress tolerance, this work provides a foundation for developing eco-friendly control strategies against B. dothidea and related pathogens.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
MU: Data curation, Methodology, Writing – review & editing. NA: Validation, Writing – review & editing. MM: Conceptualization, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. YL: Project administration, Writing – review & editing. KA: Formal analysis, Writing – review & editing. AA: Formal analysis, Writing – review & editing. PL: Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by grants from the China NSFC Research Fund for International Young Scientists (grant number: 32250410291) and a Special Project for the Academician Team Innovation Centre of Hainan Province (grant number: YSPTZX202206).
Acknowledgments
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding work through the Large Research Project under grant number RGP2/316/45.
Conflict of interest
Th authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1678786/full#supplementary-material
References
Aboelfotoh Hendy, A., Xing, J., Chen, X., and Chen, X. L. (2019). The farnesyltransferase B-subunit ram1 regulates localization of RAS proteins and appressorium-mediated infection in Magnaporthe oryzae. Mol. Plant Pathol. 20, 1264–1278. doi: 10.1111/mpp.12838
Ahmad, S. U., Khan, M. S., Jan, Z., Khan, N., Ali, A., Rehman, N., et al. (2021). Genome wide association study and phylogenetic analysis of novel SARS-COV-2 virus among different countries. Pakistan J. Pharm. Sci. 34, 1305–1313.
Chen, Y., Liao, X., Zhang, C., Kong, X., and Hua, Y. (2022). Hydrolyzing behaviors of endogenous proteases on proteins in sesame milk and application for producing low-phytate sesame protein hydrolysate. Food Chem. 385, 132617. doi: 10.1016/j.foodchem.2022.132617
Chethana, K. W. T., Jayawardena, R. S., Chen, Y. J., Konta, S., Tibpromma, S., Abeywickrama, P. D., et al. (2021). Diversity and function of appressoria. Pathogens 10, 746. doi: 10.3390/pathogens10060746
Clark, S. J., Templeton, M. D., and Sullivan, P. A. (1997). A Secreted Aspartic Proteinase from Glomerella cingulata: Purification of the Enzyme and Molecular Cloning of the cDNA. Microbiol-SGM 143, 1395–1403. doi: 10.1099/00221287-143-4-1395
Da Silva, B. A., Dos Santos, A. L., Barreto-Bergter, E., and Pinto, M. R. (2006). Extracellular peptidase in the fungal pathogen Pseudallescheria boydii. Curr. Microbiol. 53, 18–22. doi: 10.1007/s00284-005-0156-1
Da Silva, R. R. (2018). Commentary: Fungal lifestyle reflected in serine protease repertoire. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00467
Dong, B.-Z. and Guo, L.-Y. (2020). An efficient gene disruption method for the woody plant pathogen Botryosphaeria dothidea. BMC Biotechnol. 20, 14. doi: 10.1186/s12896-020-00608-z
Dong, Z., Yang, S., and Lee, B. H. (2021). Bioinformatic mapping of a more precise Aspergillus Niger degradome. Sci. Rep. 11, 693. doi: 10.1038/s41598-020-80028-3
Gravelat, F. N., Askew, D. S., and Sheppard, D. C. (2012). Targeted Gene Deletion in Aspergillus fumigatus using the Hygromycin-Resistance Split-Marker Approach. Methods Mol. Biol. 845, 119–130. doi: 10.1007/978-1-61779-539-8_8
Huang, C., Cun, Y., Yu, H., Tong, Z., Xiao, B., Song, Z., et al. (2017a). Transcriptomic profile of tobacco in response to tomato zonate spot orthotospovirus infection. Virol. J. 14, 153. doi: 10.1186/s12985-017-0821-6
Huang, X., Xu, C. L., Chen, W. Z., Chen, C., and Xie, H. (2017b). Cloning and characterization of the first serine carboxypeptidase from a plant parasitic nematode, Radopholus similis. Sci. Rep. 7, 4815. doi: 10.1038/s41598-017-05093-7
Ibe, C. and Munro, C. A. (2021). Fungal cell wall proteins and signaling pathways form a cytoprotective network to combat stresses. J. Fungi (Basel) 7, 739. doi: 10.3390/jof7090739
Imam, J., Mandal, N. P., Variar, M., and Shukla, P. (2016). “Advances in molecular mechanism toward understanding plant-microbe interaction: A study of M. oryzae versus rice,” in Frontier discoveries and innovations in interdisciplinary microbiology (India: Springer), 79–96. doi: 10.1007/978-81-322-2610-9_6
Iqbal, M., Dubey, M., Gudmundsson, M., Viketoft, M., Jensen, D. F., and Karlsson, M. (2018). Comparative evolutionary histories of fungal proteases reveal gene gains in the mycoparasitic and nematode-parasitic fungus Clonostachys rosea. BMC Evol. Biol. 18, 171. doi: 10.1186/s12862-018-1291-1
Jash, S., Sarkar, A., Moinuddin, G., and Kundu, R. (2025). “Botryosphaeria,” in Compendium of Phytopathogenic Microbes in Agro-Ecology. Eds. Amaresan, N. and Kumar, K. (Springer Nature Switzerland, Cham), 43–74. doi: 10.1007/978-3-031-81770-0_3
Jasim, J. H. M., Othman, A. S., Nordin, F. A., and Talkah, N. S. M. (2024). High-quality genomic DNA extraction methods of Yellow Spathoglottis Blume complex for next-generation sequencing. Biodiversitas J. Biol. Diversity 25, 654–663. doi: 10.13057/biodiv/d250224
Khanh, N. V., Dutta, S., Kim, C. S., and Lee, Y. H. (2024). Features of bacterial and fungal communities in the rhizosphere of Gastrodia elata cultivated in greenhouse for early harvest. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1389907
Kim, K. W., Kim, K. R., and Park, E. W. (2016). Effects of interrupted wetness periods on conidial germination, germ tube elongation and infection periods of Botryosphaeria dothidea causing apple white rot. Plant Pathol. 32, 1–7. doi: 10.5423/ppj.Oa.07.2015.0131
Kim, J. E., Lee, H. J., Lee, J., Kim, K. W., Yun, S. H., Shim, W. B., et al. (2009). Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 55, 449–459. doi: 10.1007/s00294-009-0258-6
Krishnan, P., Ma, X., Mcdonald, B. A., and Brunner, P. C. (2018). Widespread signatures of selection for secreted peptidases in a fungal plant pathogen. BMC Evol. Biol. 18, 7. doi: 10.1186/s12862-018-1123-3
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lin, L., Wu, Q., Wang, S., Gong, Q., Huang, X., Abubakar, Y. S., et al. (2025a). Recycling of trans-golgi SNAREs promotes apoplastic effector secretion for effective host invasion in magnaporthe oryzae. Plant Cell Environ. 48, 6047–6065. doi: 10.1111/pce.15582
Lin, X., Yang, Y., Huang, C., Xiong, D., Qiu, X., and Tian, C. (2025b). Transcriptome analysis reveals that a Gti1/Pac2 family gene, CpSge1, regulates fungal growth, stress response, and virulence in Cryphonectria parasitica. Phytopathology 115, 521–534. doi: 10.1094/PHYTO-11-24-0354-R
Liu, K., Wang, X., Qi, Y., Li, Y., Shi, Y., Ren, Y., et al. (2024). Effector protein serine carboxypeptidase fgSCP is essential for full virulence in Fusarium graminearum and is involved in modulating plant immune responses. Phytopathology 114, 2131–2142. doi: 10.1094/PHYTO-02-24-0068-R
Looi, H. K., Toh, Y. F., Yew, S. M., Na, S. L., Tan, Y. C., Chong, P. S., et al. (2017). Genomic insight into pathogenicity of dematiaceous fungus Corynespora cassiicola. PeerJ 5, e2841. doi: 10.7717/peerj.2841
Lv, B., Zhao, X., Guo, Y., Li, S., and Sun, M. (2023). Serine protease CrKP43 interacts with MAPK and regulates fungal development and mycoparasitism in Clonostachys chloroleuca. Microbiol. Spectr. 11, e0244823. doi: 10.1128/spectrum.02448-23
Madhu, S. N., Sharma, S., and Gajjar, D. U. (2020). Identification of proteases: carboxypeptidase and aminopeptidase as putative virulence factors of Fusarium solani species complex. Open Microbiol. J. 14, 266–277. doi: 10.2174/1874434602014010266
Munzert, K. S. and Engelsdorf, T. (2025). Plant cell wall structure and dynamics in plant-pathogen interactions and pathogen defence. J. Exp. Bot. 76, 228–242. doi: 10.1093/jxb/erae442
Muszewska, A., Stepniewska-Dziubinska, M. M., Steczkiewicz, K., Pawlowska, J., Dziedzic, A., and Ginalski, K. (2017). Fungal lifestyle reflected in serine protease repertoire. Sci. Rep. 7, 9147. doi: 10.1038/s41598-017-09644-w
Ning, Y., Hu, B., Yu, H., Liu, X., Jiao, B., and Lu, X. (2022). Optimization of protoplast preparation and establishment of genetic transformation system of an arctic-derived fungus Eutypella sp. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.769008
Novotna, A., Mennicken, S., De Paula, C. C. P., Vogt-Schilb, H., Kotilinek, M., Tesitelova, T., et al. (2023). Variability in nutrient use by orchid mycorrhizal fungi in two medium types. J. Fungi (Basel) 9, 88. doi: 10.3390/jof9010088
Oda, K., Dunn, B. M., and Wlodawer, A. (2022). Serine-carboxyl peptidases, sedolisins: from discovery to evolution. Biochemistry 61, 1643–1664. doi: 10.1021/acs.biochem.2c00239
Oledibe, O. J., Enweani-Nwokelo, I. B., Okigbo, R. N., and Achugbu, A. N. (2023). Formulation of fungal media from local plant materials. Advanced Gut Microbiome Res. 2023, 1711026. doi: 10.1155/2023/1711026
Rauwane, M. E., Ogugua, U. V., Kalu, C. M., Ledwaba, L. K., Woldesemayat, A. A., and Ntushelo, K. (2020). Pathogenicity and virulence factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms 8, 305. doi: 10.3390/microorganisms8020305
Rawlings, N. D. (2020). Twenty-five years of nomenclature and classification of proteolytic enzymes. Biochim. Biophys. Acta Proteins Proteom. 1868, 140345. doi: 10.1016/j.bbapap.2019.140345
Rolke, Y., Liu, S., Quidde, T., Williamson, B., Schouten, A., Weltring, K. M., et al. (2004). Functional Analysis of H2O2-Generating Systems in Botrytis cinerea: the Major Cu-Zn-superoxide dismutase (BCSOD1) Contributes to Virulence on French bean, Whereas a Glucose Oxidase (BCGOD1) is Dispensable. Mol. Plant Pathol. 5, 17–27. doi: 10.1111/j.1364-3703.2004.00201.x
Satala, D., Bras, G., Kozik, A., Rapala-Kozik, M., and Karkowska-Kuleta, J. (2023). More than just protein degradation: the regulatory roles and moonlighting functions of extracellular proteases produced by fungi pathogenic for humans. J. Fungi (Basel) 9, 121. doi: 10.3390/jof9010121
Savojardo, C., Babbi, G., Martelli, P. L., and Casadio, R. (2021). Mapping OMIM disease–related variations on protein domains reveals an association among variation type, Pfam models, and disease classes. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.617016
Semenova, T. A., Dunaevsky, Y. E., Beljakova, G. A., and Belozersky, M. A. (2020). Extracellular peptidases of insect-associated fungi and their possible use in biological control programs and as pathogenicity markers. Fungal Biol. 124, 65–72. doi: 10.1016/j.funbio.2019.11.005
Shetty, N. P., Mehrabi, R., Lütken, H., Haldrup, A., Kema, G. H. J., Collinge, D. B., et al. (2007). Role of Hydrogen Peroxide during the Interaction Between the Hemibiotrophic Fungal Pathogen Septoria tritici and Wheat. New Phytol. 174, 637–647. doi: 10.1111/j.1469-8137.2007.02026.x
Shi, Y., Yang, B., Linghu, Y., Liu, Y., Li, H., Xing, Y., et al. (2025). Piezo1 senses hyphae and initiates host defense against pathogenic fungi. Cell Rep. 44, 115839. doi: 10.1016/j.celrep.2025.115839
Urbanek, H. and Kaczmarek, A. (1985). Extracellular proteinases of the isolate of Botrytis cinerea virulent to apple tissues. Acta Biochim. Pol. 32, 101–109.
Urbanek, H. and Yirdaw, G. (1984). Hydrolytic Ability of Acid Protease of Fusarium culmorum and its Possible Role in Phytopathogenesis. Acta Microbiol. Pol. 33, 131–136.
Varabyou, A., Erdogdu, B., Salzberg, S. L., and Pertea, M. (2023). Investigating open reading frames in known and novel transcripts using ORFanage. Nat. Comput. Sci. 3, 700–708. doi: 10.1038/s43588-023-00496-1
Wang, B., Liang, X., Gleason, M. L., Zhang, R., and Sun, G. (2018). Comparative genomics of Botryosphaeria dothidea and B. kuwatsukai, causal agents of apple ring rot, reveals both species expansion of pathogenicity-related genes and variations in virulence gene content during speciation. IMA Fungus 9, 243–257. doi: 10.5598/imafungus.2018.09.02.02
Wang, F., Shen, T., Cao, C., Shan, S., Sun, J., and Zhu, R. (2025). Virtual screening of inhibitor against Penicillium expansum and its application for controlling blue mold decay in apples. Food Biosci. 68, 106741. doi: 10.1016/j.fbio.2025.106741
Wei, S., Bian, R., Andika, I. B., Niu, E., Liu, Q., Kondo, H., et al. (2019). Symptomatic plant viroid infections in phytopathogenic fungi. Proc. Natl. Acad. Sci. 116, 13042–13050. doi: 10.1073/pnas.1900762116
Williams, H. L., Sturrock, R. N., Islam, M. A., Hammett, C., Ekramoddoullah, A. K., and Leal, I. (2014). Gene expression profiling of candidate virulence factors in the laminated root rot pathogen Phellinus sulphurascens. BMC Genom. 15, 603. doi: 10.1186/1471-2164-15-603
Xia, Y. (2004). Proteases in pathogenesis and plant defence. Cell Microbiol. 6, 905–913. doi: 10.1111/j.1462-5822.2004.00438.x
Yaakoub, H., Sanchez, N. S., Ongay-Larios, L., Courdavault, V., Calenda, A., Bouchara, J. P., et al. (2022). The high osmolarity glycerol (HOG) pathway in fungi(dagger). Crit. Rev. Microbiol. 48, 657–695. doi: 10.1080/1040841X.2021.2011834
Yang, C., Li, W., Huang, X., Tang, X., Qin, L., Liu, Y., et al. (2022). SsNEP2 contributes to the virulence of sclerotinia sclerotiorum. Pathogens 11, 446. doi: 10.3390/pathogens11040446
Yang, S., Zhuo, Y., Lin, Y., Huang, M., Tang, W., Zheng, W., et al. (2023). The signal peptidase FoSpc2 is required for normal growth, conidiation, virulence, stress response, and regulation of light sensitivity in Fusarium odoratissimum. Microbiol. Spectr. 11, e0440322. doi: 10.1128/spectrum.04403-22
Yuan, G., Lu, H., De, K., Hassan, M. M., Liu, Y., Islam, M. T., et al. (2023). Split selectable marker systems utilizing inteins facilitate gene stacking in plants. Commun. Biol. 6, 567. doi: 10.1038/s42003-023-04950-8
Zhang, S., Lin, C., Zhou, T., Zhang, L. H., and Deng, Y. Z. (2020b). Karyopherin MoKap119-mediated Nuclear Import of Cyclin-dependent kinase Regulator MoCks1 is Essential for Magnaporthe oryzae Pathogenicity. Cell Microbiol. 22, e13114. doi: 10.1111/cmi.13114
Zhang, M. Z., Sun, C. H., Liu, Y., Feng, H. Q., Chang, H. W., Cao, S. N., et al. (2020a). Transcriptome analysis and functional validation reveal a novel gene, BcCGF1, that enhances fungal virulence by promoting infection-related development and host penetration. Mol. Plant Pathol. 21, 834–853. doi: 10.1111/mpp.12934
Zhang, Y., Xu, K., Yu, D., Liu, Z., Peng, C., Li, X., et al. (2019). The highly conserved barley powdery mildew effector BEC1019 confers susceptibility to biotrophic and necrotrophic pathogens in wheat. Int. J. Mol. Sci. 20, 4376. doi: 10.3390/ijms20184376
Zhang, Q., Zhao, L., Shen, M., Liu, J., Li, Y., Xu, S., et al. (2022). Establishment of an Efficient Polyethylene Glycol (PEG)-Mediated Transformation System in Pleurotus eryngii var. ferulae Using Comprehensive Optimization and Multiple Endogenous Promoters. J. Fungi (Basel) 8, 186. doi: 10.3390/jof8020186
Keywords: Botryosphaeria dothidea, eco-friendly disease control strategies, functional genomics, pathogenicity, serine carboxypeptidase S10 family
Citation: Umer M, Anwar N, Mubeen M, Li Y, Alsyaad KM, Ahmed AE and Liu P (2025) Functional genome analysis reveals that serine carboxypeptidase Bd-SCP10 mediates vegetative growth, pathogenicity, and stress tolerance in Botryosphaeria dothidea. Front. Plant Sci. 16:1678786. doi: 10.3389/fpls.2025.1678786
Received: 03 August 2025; Accepted: 21 October 2025;
Published: 14 November 2025.
Edited by:
Zhen Wang, Yunnan Agricultural University, ChinaReviewed by:
Mubashar Raza, Xinjiang Academy of Agricultural Sciences, ChinaShayan Syed, Lithuanian Research Centre for Agriculture and Forestry, Lithuania
Copyright © 2025 Umer, Anwar, Mubeen, Li, Alsyaad, Ahmed and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Li, Z3h1bGl5dW5AZ3h1LmVkdS5jbg==; Pingwu Liu, aG51bHB3QGhhaW5hbnUuZWR1LmNu
†These authors have contributed equally to this work
 Muhammad Umer1†
Muhammad Umer1† Mustansar Mubeen
Mustansar Mubeen Ahmed Ezzat Ahmed
Ahmed Ezzat Ahmed Pingwu Liu
Pingwu Liu