- Department of Biology, Luoyang Normal University, Luoyang, Henan, China
This study systematically identifies and characterizes the GLK gene family in melon (Cucumis melo), identifying 49 GLK genes within the melon genome, which are subsequently named CmGLK1 to CmGLK49 based on their chromosomal locations. The CmGLK family members exhibit significant variation in physicochemical properties, including amino acid length, molecular weight, and isoelectric point. Phylogenetic analysis classified the CmGLK genes into six groups (I-VI), demonstrating a high degree of conservation of GLK genes across melon, cucumber, and watermelon. Motif and domain analyses indicate that all CmGLK family members possess the characteristic GLK transcription factor domain. Moreover, genes within the same phylogenetic group display similar protein motifs and gene structures, suggesting that these genes may have a common function in regulating plant growth and development. Chromosomal localization reveals that CmGLK genes are unevenly distributed across 12 chromosomes, with nine pairs of segmental duplications and one pair of tandem repeats, indicating that gene duplication likely plays a key role in the expansion of this gene family. Additionally, GLK homologs were identified between melon and Arabidopsis thaliana, Oryza sativa, Cucumis sativus, and Citrullus lanatus, with the highest homology found between melon and cucumber, as well as melon and watermelon. Cis-regulatory element analysis of CmGLK promoters reveals a high abundance of elements associated with light response, hormone regulation, and stress responses, suggesting that the CmGLK family may be involved in the regulation of melon growth, development, and environmental stress responses. Expression profiling shows that CmGLK genes are expressed in a tissue-specific manner, with the highest expression levels found in the root, stem, and leaf tissues. Further Quantitative fluorescence analysis reveals that several CmGLK family members exhibit significant expression changes under Fusarium wilt and drought stress, suggesting their potential involvement in melon’s stress response mechanisms. This study provides a comprehensive analysis of the structure, evolution, and stress-responsive expression of the CmGLK gene family. Our findings offer new insights into the potential molecular mechanisms of melon’s stress adaptation and highlight candidate CmGLK genes for future functional studies aimed at improving disease resistance.
1 Introduction
Chloroplasts are essential organelles in plants, serving as the primary site for photosynthesis and the production of energy and metabolites. Their proper formation is crucial for plant growth and development (Razi and Muneer, 2021). As semi-autonomous organelles, chloroplasts can independently replicate DNA, transcribe, and translate a portion of the proteins they require. However, they are also tightly regulated by nuclear-encoded gene products (Mayfield et al., 1995). Among these gene products, the GOLDEN2-LIKE (GLK or G2-like) transcription factors play a key role in chloroplast formation, development, and senescence. The coordination between nuclear and chloroplast genes ensures proper chloroplast development, thereby optimizing photosynthesis in plants (Shen et al., 2022).
The GLK gene family was first identified in genes responsible for yellowing in maize (Hall et al., 1998) and is classified as part of the GARP superfamily (Riechmann et al., 2000). Typical GLK transcription factors contain two conserved domains: a Myb-DNA binding domain (DBD) and a C-terminal domain with a conserved GCT-box (Rossini et al., 2001). The Myb-DNA binding domain includes a helix-loop-helix (HLH) structure, where the first helix contains the PELHRR motif, the second helix contains the NI/VASHLQ motif, and the loop connecting the helices consists of 22 amino acids (Liu et al., 2016).
Existing studies have demonstrated that GLK transcription factors are primarily involved in regulating processes such as chloroplast development, senescence, and chlorophyll accumulation (Fitter et al., 2010). For instance, GLK1 and GLK2 are critical for chloroplast formation and development in A. thaliana (Kobayashi et al., 2013; Waters et al., 2009). Moreover, GLK family members are associated with responses to both biotic and abiotic stresses. Through interactions with other factors, GLK transcription factors regulate the expression of resistance-related genes, thereby enhancing photosynthesis under stress conditions (Han et al., 2016). For example, under fungal or viral infections, A. thaliana shows a significant increase in GLKgene expression, and overexpression of GLK1 enhances resistance to Fusarium graminearum (Savitch et al., 2007). Transformation of maize GLK genes into rice reduces photoinhibition and improves CO2 fixation efficiency under high-light conditions (Xia et al., 2020). These findings suggest that GLK genes may play an important role in stress tolerance. Reports have also indicated that GLK1 and GLK2 in the GLK gene family are differentially involved in chloroplast development in C3 and C4 photosynthetic plants (Peng et al., 2017). In maize, ZmGLK1 is predominantly expressed in mesophyll cells, while ZmGLK2 exhibits stronger expression in bundle sheath cells, indicating spatial compartmentalization essential for chloroplast development in these cell types (Chang et al., 2012). In tomato, SlGLK1 and SlGLK2 enhance photosynthesis in fruits and increase the expression of chloroplast development genes, leading to higher carbohydrate and carotenoid content in mature fruits, which provides greater resistance to cold, drought, and heat stress (Nguyen et al., 2014).
Melon (C. melo), commonly known as “honeydew melon” or “muskmelon,” is a rich source of glucose, minerals, and vitamins, providing health benefits such as cooling, nutritional supplementation, and diuretic effects (Gao, 2015). However, the yield and quality of melons are influenced by various factors, including genetic traits, cultivation practices, and environmental stressors. F. wilt, caused by F. oxysporum, is a major fungal disease that severely impacts melon production in Henan, often resulting in plant wilting and death, which adversely affects both yield and fruit quality (Herman and Perl-Treves, 2007). Moreover, drought stress in the region exacerbates the challenges associated with melon cultivation. Some studies have been conducted on the response of various gene families in melon to F. wilt stress (Zheng et al., 2024; 2025). Current research on the GLK gene family in response to biotic stress is primarily focused on A. thaliana (Murmu et al., 2014; Schreiber et al., 2011), while a relevant investigation in watermelon has also reported the involvement of GLK genes in stress responses (Rahman et al., 2021).However, there is a lack of research on the role of the GLK gene family in F. wilt stress or drought stress in melon. Bioinformatics methods were used to analyze the chromosomal localization, phylogenetic relationships, synteny, gene structure, protein motifs, expression patterns, and cis-regulatory elements of the melon GLK gene family. Furthermore, the expression of these genes under F. wilt (biotic Stress) and drought (abiotic Stress) stresses was examined, providing a foundation for understanding the functional roles of the GLK gene family in melon and offering molecular targets for its genetic improvement.
2 Materials and methods
2.1 Identification of GLK gene family in melon
The melon DHL92 v4.0 genome was downloaded from the CuGenDB database (http://cucurbitgenomics.org/) for GLK gene identification (Garcia-Mas et al., 2012). Protein sequences of 36 A. thaliana GLK family members were obtained from the UniProt database (https://www.uniprot.org/) (UniProt Consortium et al., 2021). Using the BLAST tool, the melon genome protein data were aligned with the A. thaliana GLK protein sequences, and duplicates were removed, retaining genes with an e-value ≤ e−5 (Ye et al., 2006). Domain identification was performed using the NCBI Conserved Domain Database (CDD), and genes containing the GOLDEN2-LIKE domain were selected. In total, 49 GLK genes were identified in the melon genome. The physicochemical properties of the proteins were then analyzed using ExPASy (http://web.expasy.org/protparam/) (Wilkins et al., 1999), and subcellular localization predictions were conducted using WoLF PSORT II (https://www.genscript.com/wolf-psort.html?src=leftbar) (Horton et al., 2007). Additionally, 55 GLK family members were respectively identified in the cucumber and watermelon genomes using the same method.
2.2 Phylogenetic tree construction and classification
Multiple sequence alignment of GLK family proteins from melon, cucumber, watermelon, and A. thaliana was performed using MEGA 7 software and the MUSCLE algorithm. The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates, employing the Poisson correction model and pairwise deletion (Kumar et al., 2016). Based on the phylogenetic tree, the GLK family members were classified into six groups: Group I, Group II, Group III, Group IV, Group V, and Group VI.
2.3 Conserved motifs, domains, and gene structure
Conserved motifs in the 49 CmGLK protein sequences were identified using the MEME tool (http://meme-suite.org/tools/meme), with a maximum of 10 motifs and default parameters (Bailey et al., 2009). The positions and numbers of GLK domains in the CmGLK family proteins were determined using the NCBI CDD online tool (https://www.ncbi.nlm.nih.gov/cdd/) (Marchler-Bauer et al., 2015). The positions and numbers of exons and introns in the CmGLK genes were extracted from the melon genome data. Finally, the phylogenetic tree, motifs, domains, and gene structures of the CmGLK family members were clustered and visualized using TBtools (Chen et al., 2020).
2.4 Gene localization, duplication, and Ka/Ks ratio
The chromosomal distribution of CmGLK genes was mapped using the TBtools software. Fragment duplication and tandem repeat gene analyses were analyzed with the MCScanX software, and the chromosomal synteny map was generated with the Circos software (Wang et al., 2012; Krzywinski et al., 2009). The GLK orthologous gene pairs between melon and A. thaliana, as well as between melon and rice, were identified. The Ka (nonsynonymous substitution rate), Ks (synonymous substitution rate), and Ka/Ks ratio for all types of duplicated genes were determined with the KaKs_Calculator (Zhang et al., 2006).
2.5 Promoter annotation
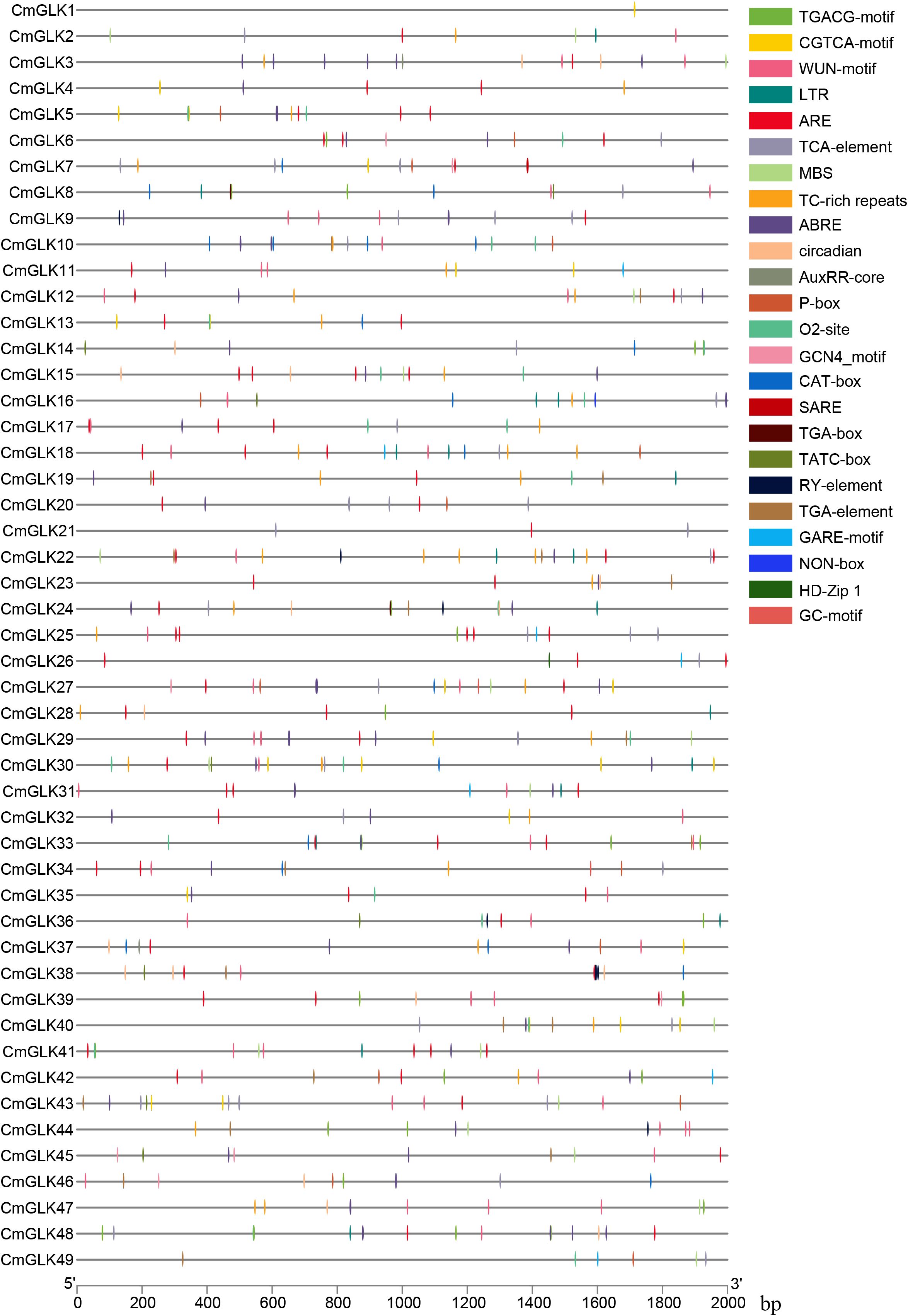
The 2000 bp upstream promoter sequences of the 49 CmGLK genes were extracted and their cis-regulatory elements annotated using the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The cis-regulatory elements associated with abiotic and biotic stresses, phytohormone responsiveness, and plant growth and development, as identified in previous studies, were analyzed for positional distribution (Zhang et al., 2024).
2.6 Expression profiling
The expression profiles of CmGLK family members in 13 different tissues—Root, Middle stem, Upside stem, Shoot apex, Young leaves, 12th leaves, Tendril, Anther male flower, Anther female flower, Petal female flower, Stigma female flower, Ovary DAF4, and Fruit flesh DAF50—were obtained by analyzing the FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) values from the melon transcriptome database (http://cucurbitgenomics.org/rnaseq/home). Genes with an FPKM value greater than or equal to 0.8 were defined as significantly expressed (Zhang et al., 2024; Yu et al., 2023).
2.7 RNA extraction, cDNA synthesis, and qRT-PCR analysis of the melon GLK gene family
Seeds of the melon cultivar “Super Sweet White Sugar Melon” were soaked, germinated, and planted. The plants were grown in a greenhouse until the three-leaf and one-heart stage, with a light cycle of 16/8 hours (day/night) and temperatures of 28/18 °C (day/night). F. wilt was isolated from infected melon plants. A spore suspension (1 × 10^8 spores/mL) was applied to the plants through root irrigation, with 10 mL of suspension per plant. For the control (CK) group, the same root irrigation method was used, and each plant was irrigated with 10 mL of sterile water (distilled water) instead of the F. wilt spore suspension. Real-time quantitative PCR (qRT-PCR) was used to investigate the expression levels of 15 CmGLK genes at four time points (0, 12, 24, and 48 hours) following drought stress treatment (leaves; three biological replicates) and at four time points (0, 24, 48, and 96 hours) following F. wilt infection (stems; three biological replicates).
RNA quality and concentration were assessed using the Nanodrop 2000 instrument. First-strand cDNA was synthesized using the SweScript All-in-One First-Strand cDNA Synthesis Kit (TRANS, G3337). Quantitative PCR (qPCR) analysis was performed using the 2×SYBR Green qPCR Master Mix (without ROX) (TRANS, G3320) on the Bio-Rad CFX96 Real-Time PCR Detection System. XM_008449635.3 was used as the internal reference gene for data normalization. Three biological and three technical replicates were performed for each sample. The relative expression levels of the GLK family genes were calculated using the 2−ΔΔCT method. The statistical significance of differences among treatment groups was determined by one-way ANOVA followed by the least significant difference (LSD) post-hoc test using SPSS 19.0, as shown in Figures 2, 3.
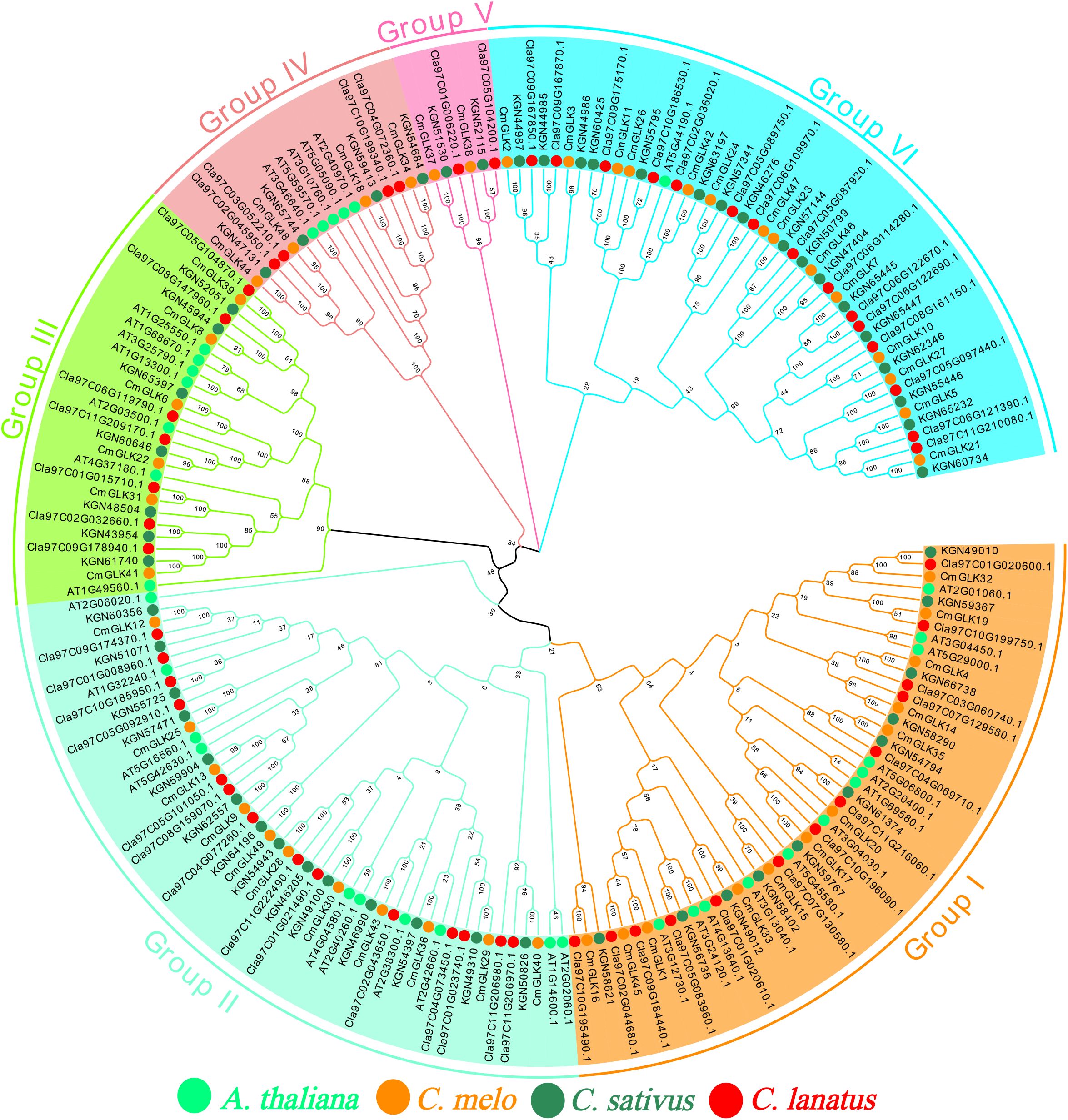
Figure 1. Chromosomal localization of the CmGLK gene family. The six different colors represent the six Groups, and the solid circles in four colors represent the GLK genes from four species.
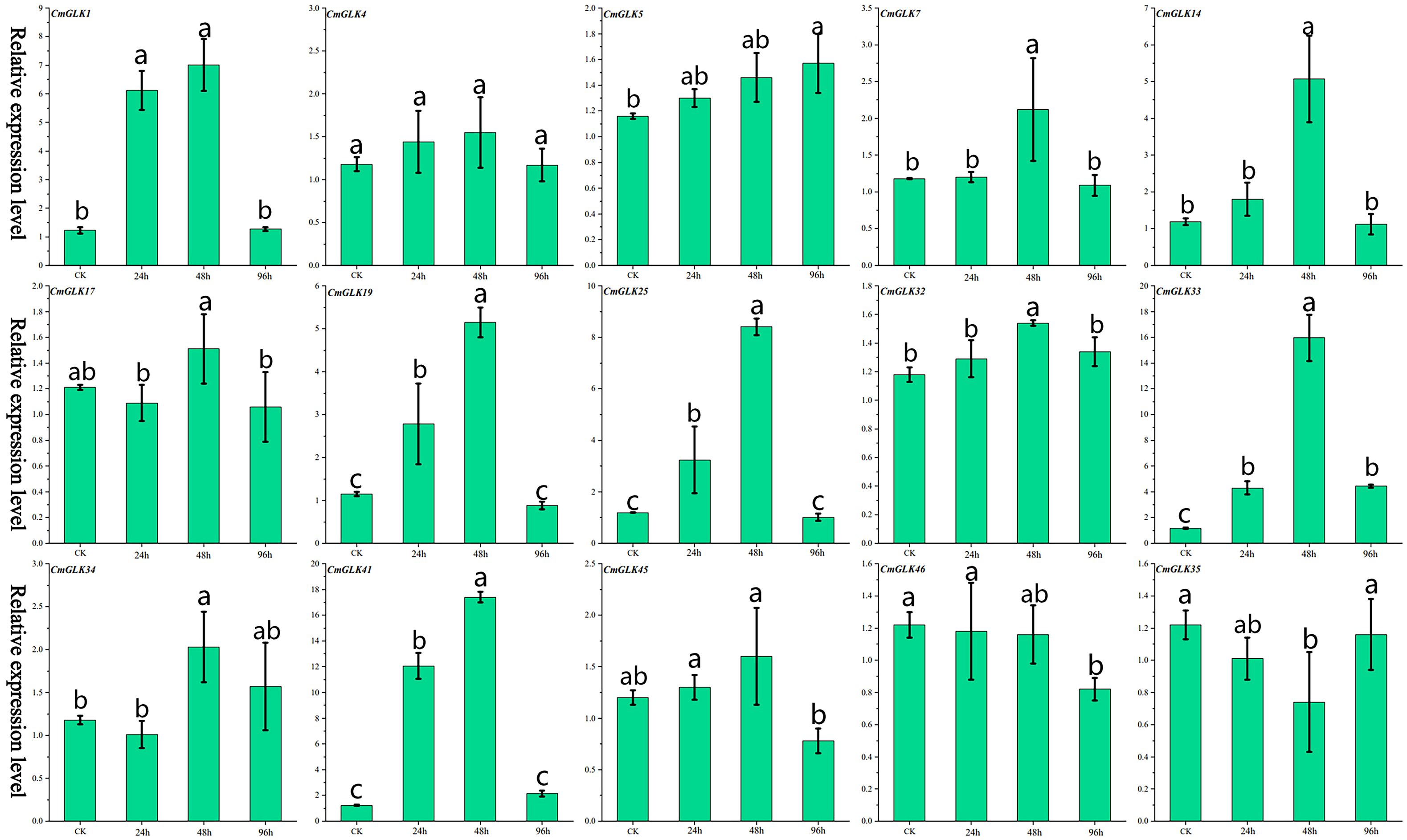
Figure 2. Relative expression levels of 15 CmGLK genes at four time points under F. wilt stress. Values are presented as mean ± SD (n = 3). Bars labeled with different lowercase letters indicate statistically significant differences among treatment groups (p < 0.05) as determined by one-way ANOVA followed by the least significant difference (LSD) post-hoc test.
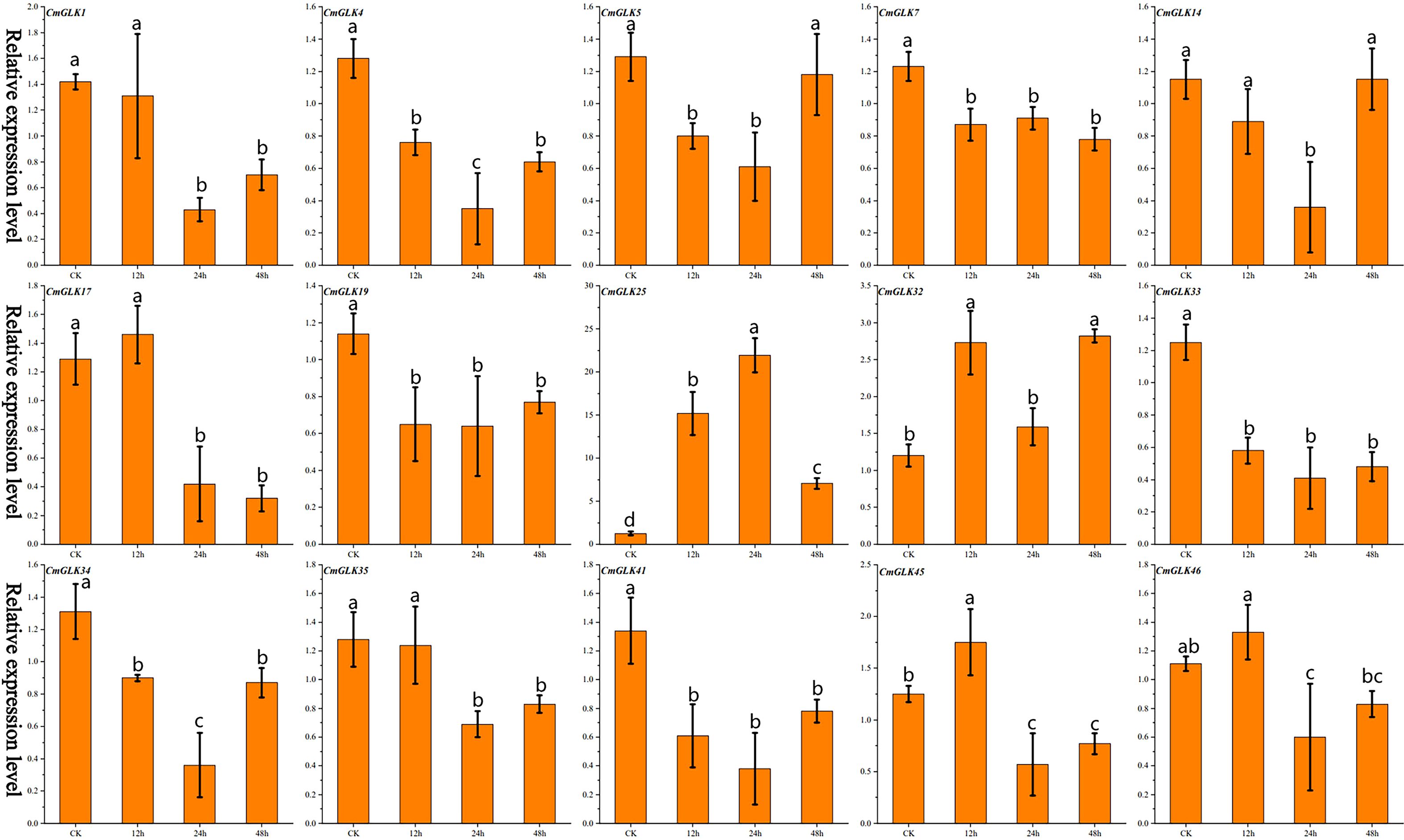
Figure 3. Relative expression levels of 15 CmGLK genes at four time points under drought stress, represented as a bar chart. Values are shown as mean ± SD (n=3).Bars labeled with different lowercase letters indicate statistically significant differences among treatment groups (p < 0.05) as determined by one-way ANOVA followed by the least significant difference (LSD) post-hoc test.
3 Results and analysis
3.1 Identification and analysis of the melon GLK gene family
A total of 49 GLK genes were identified in the melon genome, which were renamed according to their chromosomal positions as CmGLK1 to CmGLK49. The physicochemical properties of the family members revealed considerable differences among them (Supplementary Table S1). The number of amino acids in the CmGLK family members ranged from 92 aa (CmGLK20) to 699 aa (CmGLK21), with protein molecular weights ranging from 10,573.15 Da (CmGLK20) to 76,840.49 Da (CmGLK21). The theoretical isoelectric points of the CmGLK family members ranged from 5.13 to 10.07, with 27 members having an isoelectric point below 7 and 22 members having an isoelectric point above 7. All CmGLK transcription factors were hydrophobic proteins. Subcellular localization analysis indicated that 47 CmGLK members were localized in the nucleus, while 2 were located in the mitochondria.
3.2 Phylogenetic analysis of CmGLK
A Neighbor-Joining phylogenetic tree of GLK family members from melon, cucumber, watermelon, and A. thaliana was constructed using the MEGA 7 tool (Figure 1). The tree was divided intosix groups: Group I, Group II, Group III, Group IV, Group V, and Group VI. Group I to Group VI contained 12, 11, 6, 4, 2, and 14 CmGLK genes, respectively. The clustering results showed that GLK family members from melon, cucumber, and watermelon were highly conserved.
3.3 Analysis of conserved motifs, domains, and gene structure in CmGLK family members
We integrated the Neighbor-Joining phylogenetic tree, conserved motifs, domains, and gene structure of CmGLK family members for visualization. Figure 4A shows the Neighbor-Joining phylogenetic tree of the CmGLK family members. A total of 10 conserved motifs were identified in the CmGLK family members. The NCBI CDD database identified Motif 1 and Motif 2 as sequences characteristic of the GOLDEN2-LIKE transcription factor. All CmGLK family members contained Motif 1, while the only exception was CmGLK37, which lacked Motif 2. The other 48 members contained Motif 2. Members within the same group exhibited highly conserved motifs in both type and number (Figure 4B). Figure 4C indicates that all CmGLK family members contained the PLN03162 domain (GOLDEN2-LIKE like transcription factor), confirming that these genes belong to the GLK type.
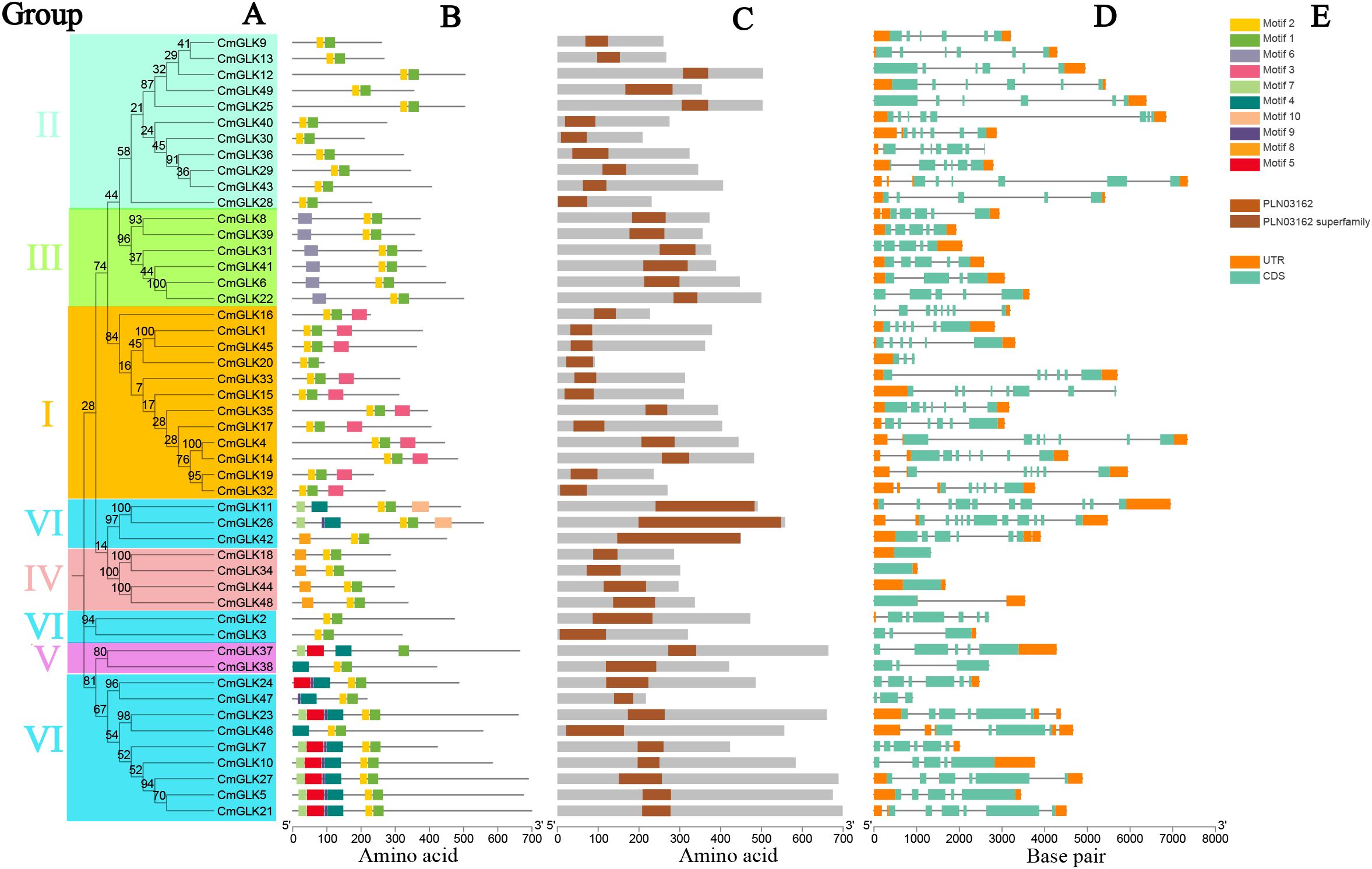
Figure 4. Phylogenetic tree, conserved motifs, domains, and gene structure analysis of CmGLK family members. (A) Neighbor-Joining phylogenetic tree. (B) Conserved protein motifs. (C) Distribution of GLK domains. (D) Exon and intron distribution. (E) Color-coded rectangles representing the types of motifs, domains, exons, and introns.
The gene structure of CmGLK family members was diverse, with variations in the number of exons and introns within the same group (Figure 4D). In Group I, the number of exons ranged from 3 to 8, and the number of introns ranged from 2 to 7. In Group II, exons ranged from 5 to 8, and introns ranged from 4 to 7. In Group III, the number of exons ranged from 4 to 5, and introns ranged from 3 to 4. All four CmGLK members in Group IV contained only 1 exon. In Group V, the number of exons ranged from 3 to 5, and introns ranged from 2 to 4. In Group VI, the number of exons ranged from 3 to 11, and introns ranged from 2 to 10. Figure 4E presents the color-coded rectangles corresponding to motifs, domains, and exon/intron structures. In conclusion, the diversity in protein motifs and gene structures suggests that CmGLK family members may have a broad range of functions in regulating the growth and development of melon.
3.4 Chromosomal localization, duplication, and Ka/Ks ratio of CmGLK genes
The CmGLK family members were unevenly distributed across 12 chromosomes (Figure 5). Specifically, chromosomes chr1 to chr12 contained 3, 4, 3, 9, 3, 4, 2, 8, 1, 3, 7, and 2 CmGLK genes, respectively. According to the chromosomal density map, CmGLK family members were primarily located in regions with a high density of genes on the chromosomes.
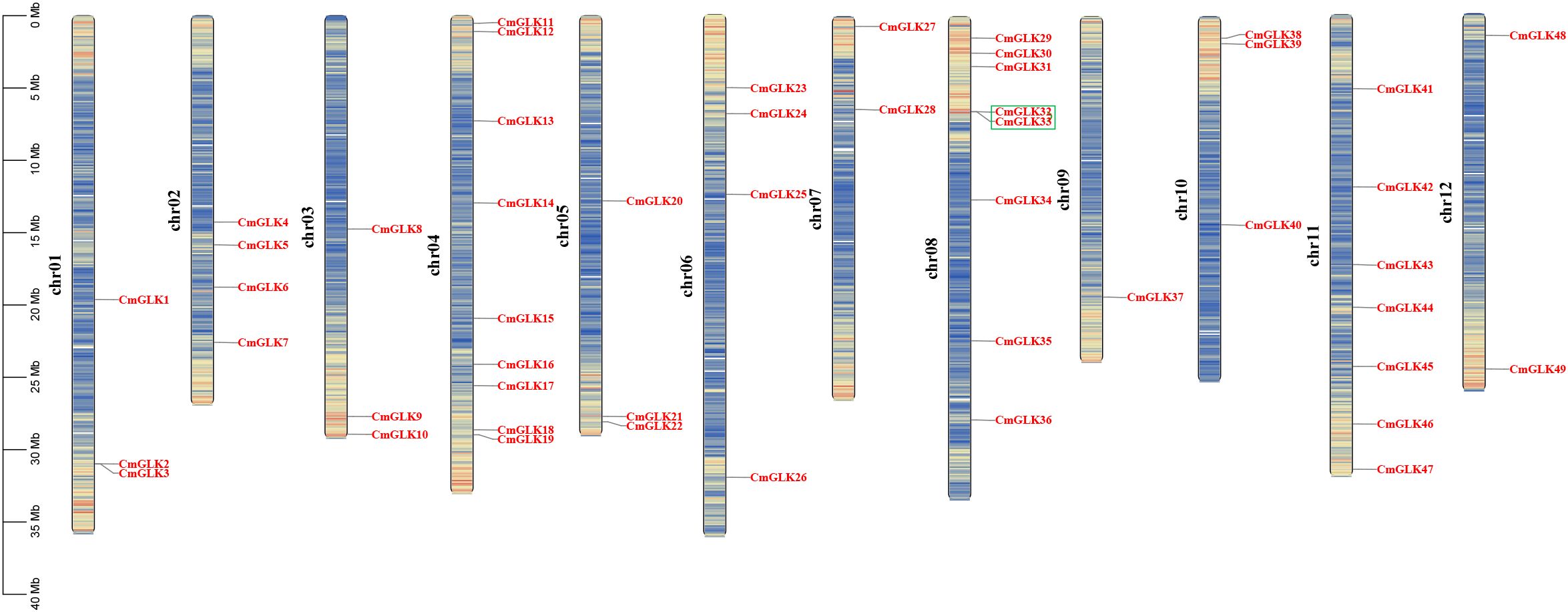
Figure 5. Chromosomal localization of CmGLK family members. The left scale represents chromosome length, with “chr” denoting chromosomes. Gene density for each chromosome was calculated at intervals of 200 kb. A color gradient from blue (low gene density) to red (high gene density) indicates gene density, while blank regions represent areas with no gene distribution information. Green boxes denote tandem duplication gene pairs.
Nine pairs of segmental duplication genes within the CmGLK family were identified: CmGLK1/CmGLK45, CmGLK2/CmGLK46, CmGLK5/CmGLK27, CmGLK6/CmGLK22, CmGLK8/CmGLK39, CmGLK11/CmGLK26, CmGLK23/CmGLK46, CmGLK37/CmGLK38, and CmGLK44/CmGLK48 (Figure 6; Supplementary Table S2). Additionally, one tandem duplication gene pair, CmGLK32/CmGLK33 (Figure 5), was identified. Homologous GLK gene pairs between melon and four other species (A. thaliana, rice, cucumber, and watermelon)were also identified, with 19, 19, 68, and 69 pairs of homologous genes, respectively (Figure 7; Supplementary Table S2). Notably, a high level of homology was observed between the GLK family members in melon, cucumber, and watermelon. Several CmGLK members, such as CmGLK31 and CmGLK39, exhibited homology with distinct cucumber or watermelon GLK members. Finally, the majority of segmental duplication gene pairs, tandem duplication gene pairs, and homologous gene pairs between melon and cucumber, as well as melon and watermelon, exhibited Ka/Ks values of less than 1 (Figure 7; Supplementary Table S2), suggesting purifying selection. However, for most homologous gene pairs between melon and A. thaliana, as well as melon and rice, Ka/Ks values could not be determined.
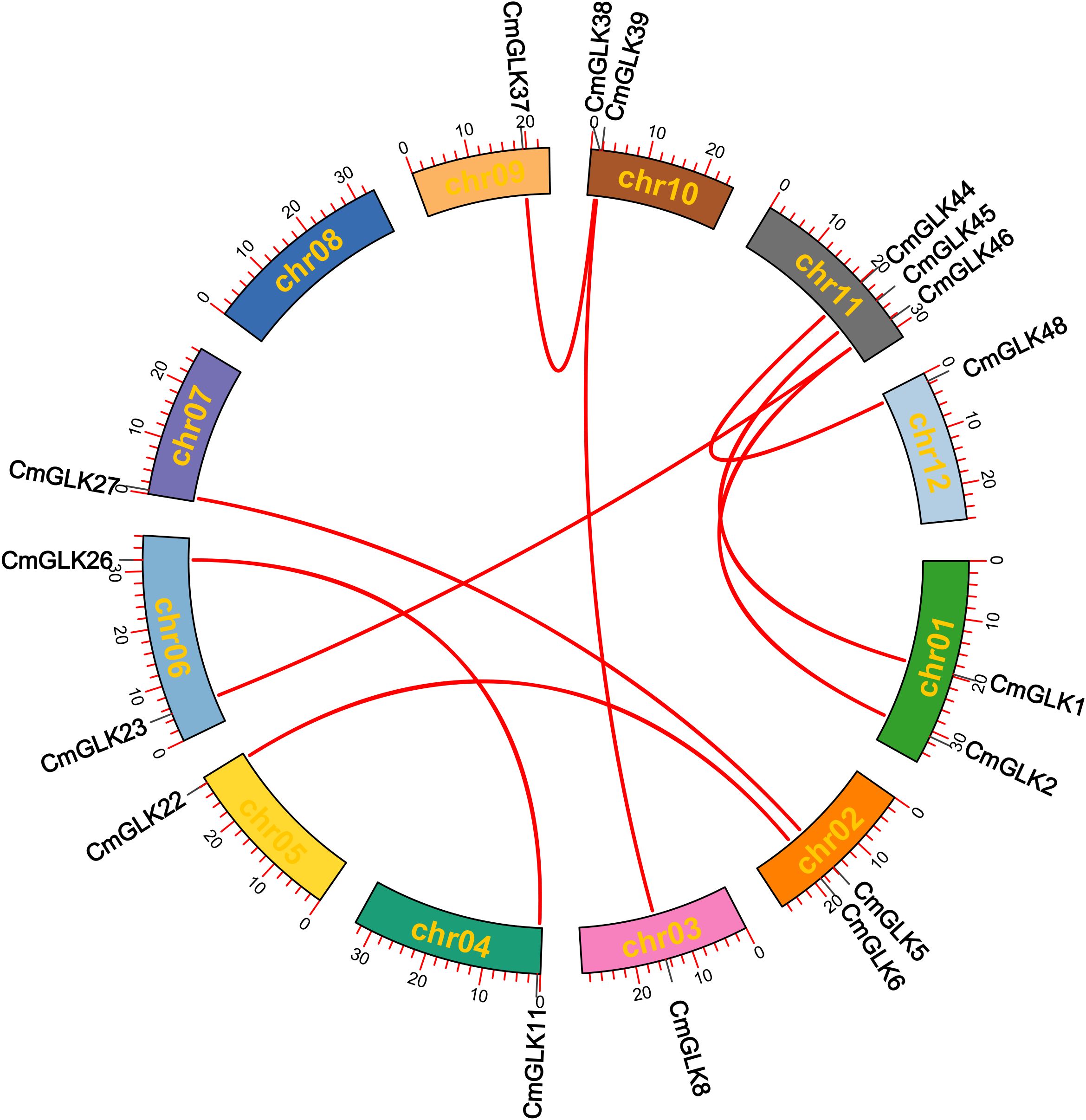
Figure 6. Collinearity analysis of CmGLK family members within chromosomes red lines represent segmental duplication gene pairs.
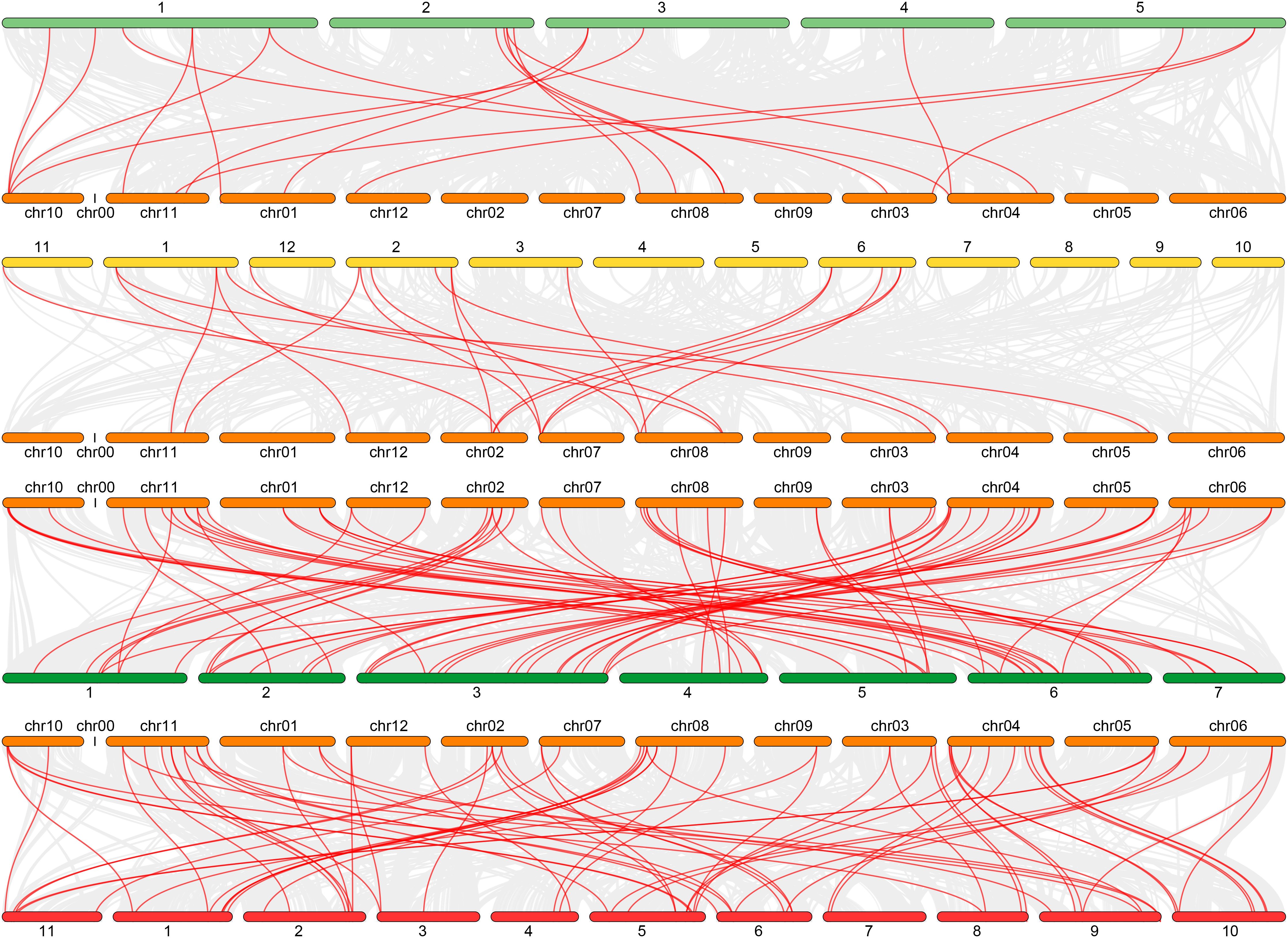
Figure 7. Distribution of GLK family homologous gene pairs. (A) A. thaliana and C. melo. (B) O. sativa and C. melo. (C) C. melo and C. sativus. (D) C. melo and C.lanatus. Red lines represent homologous gene pairs. Orange segments represent melon chromosomes, cyan segments represent A. thaliana chromosomes, yellow segments represent rice chromosomes, dark green segments represent cucumber chromosomes, and red segments represent watermelon chromosomes. Numbers represent the corresponding chromosome numbers.
3.5 CmGLK family member promoter analysis
A total of 107 types of cis-regulatory elements in the CmGLK family were identified, including numerous light-responsive elements, such as ACE, chs-CMA1a, and GT1-motif (Supplementary Table S3). In addition to the basic and light-related elements, these elements also include cis-regulatory elements associated with abiotic and biotic stresses, phytohormone responses, and plant growth and development (Figure 8). The abiotic and biotic stress-related cis-regulatory elements include six types: TC-rich repeats, LTR, ARE, GC-motif, MBS, and WUN-motif. The phytohormone response-related elements consist of eleven types: TGA-element, TATC-box, TCA-element, SARE, ABRE, AuxRR-core, TGACG-motif, CGTCA-motif, P-box, GARE-motif, and TGA-box. The plant growth and development-related elements consist of nine types:: MSA-like, circadian, RY-element, O2-site, CAT-box, NON-box, GCN4_motif, HD-Zip 1, and MBSI. In conclusion, CmGLK family members may stress responses.
3.6 Expression pattern analysis of CmGLK family members
FPKM values for CmGLK family members were obtained in 13 melon tissues: root, middle stem, upside stem, shoot apex, young leaves, 12th leaves, tendril, anther male flower, another female flower, petal female flower, stigma female flower, ovary DAF4, and fruit flesh DAF50. A heatmap was generated to visualize the expression patterns (Figure 9; Supplementary Table S4). The results showed that the number of CmGLK genes significantly expressed was higher in root and middle stem tissues, with 15 and 16 genes, respectively. In the remaining 11 tissues, 4 to 9 CmGLK genes were significantly expressed. Notably, 9 genes exhibited high expression levels in the 12th leaves. Several genes exhibited tissue-specific expression patterns. For instance, CmGLK29, CmGLK32, CmGLK36, and CmGLK42 were highly expressed in leaves. CmGLK8, CmGLK10, CmGLK27, CmGLK28, and CmGLK49 were highly expressed in roots. CmGLK8, CmGLK17, CmGLK20, and CmGLK26 were highly expressed in stems. CmGLK21, CmGLK23, CmGLK24, CmGLK46, and CmGLK48 were highly expressed in flowers, most of which belong to Group VI. Among the previously identified paralogous gene pairs, five pairs (CmGLK1/CmGLK45, CmGLK23/CmGLK46, CmGLK5/CmGLK27, CmGLK6/CmGLK22, CmGLK32/CmGLK33) exhibited similar or identical expression patterns across tissues, suggesting that duplicated genes have retained similar evolutionary fates. These findings indicate that CmGLK family members exhibit significant tissue-specific expression characteristics, hormone regulation, and stress responses.
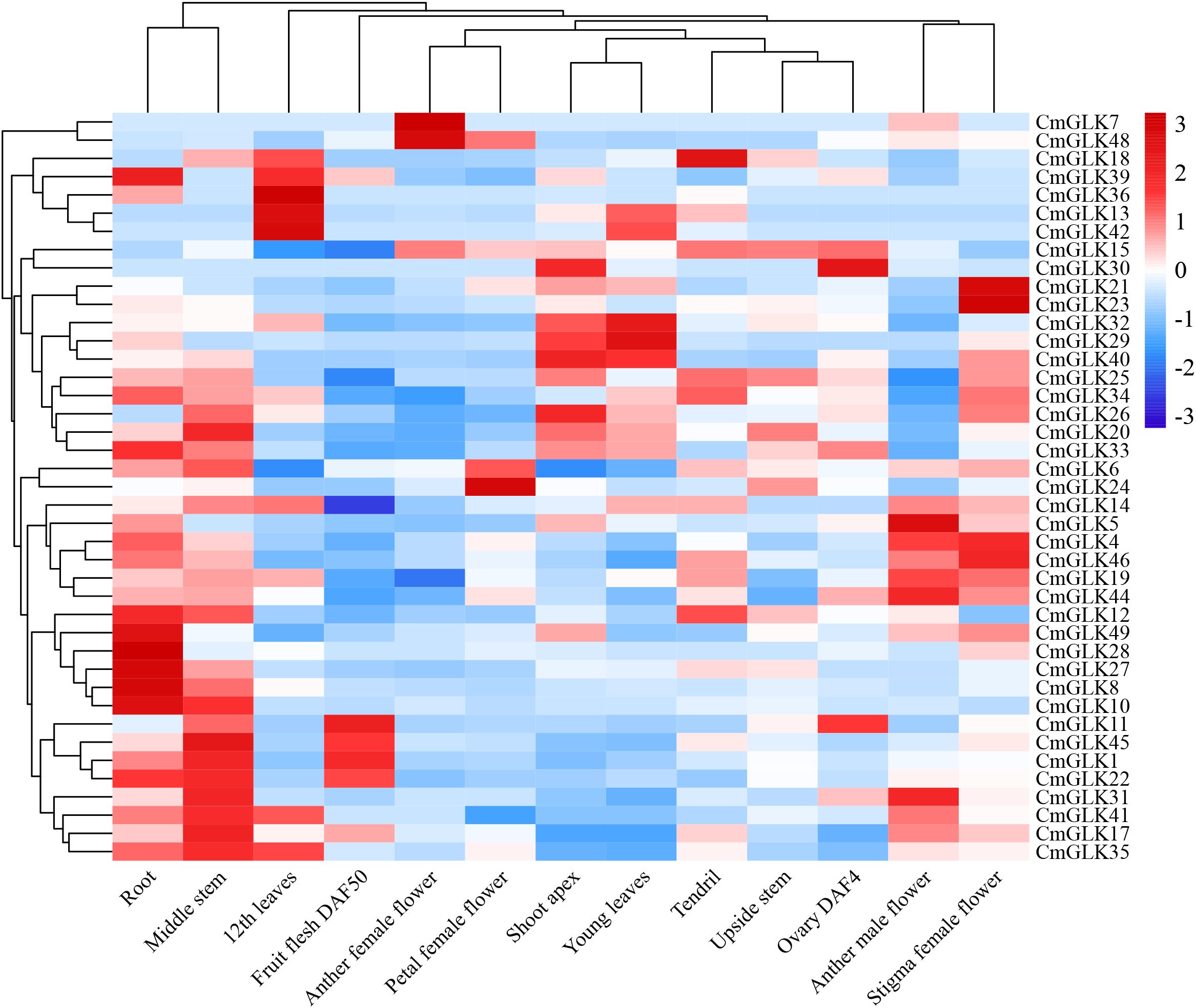
Figure 9. Expression heatmap of CmGLK family members in 13 melon tissues. The heatmap was generated using the average row normalization method. Red indicates high expression, while blue indicates low expression. Numbers on the map represent significance levels.
3.7 Quantitative fluorescence expression analysis of CmGLK family members under F. wilt stress
We used qRT-PCR to analyze the expression changes of 15 CmGLK genes in melon stems under F. wilt stress. The results (Figure 2; Supplementary Table S5) indicated that the 15 CmGLK genes exhibited varying degrees of expression changes at different time points after F. wilt inoculation. Most CmGLK genes exhibited a trend of increasing expression levels from 0 to 48 hours, followed by a decrease from 48h to 96h. CmGLK1, CmGLK19, CmGLK25, CmGLK33, and CmGLK41 exhibited significantly elevated expression levels at the 24h and 48h time points, surpassing twice the expression levels of the CK group. Notably, CmGLK33 and CmGLK41 reached expression levels approximately 16 and 17 times higher than the CK group at 48 hours, respectively. In contrast, the remaining 10 genes exhibited relatively smaller changes in expression at the 24h, 48h, and 96h time points compared to the CK group. These results suggest that CmGLK family members may play regulatory roles in melon’s response to F. wilt stress.
3.8 Quantitative fluorescent expression analysis of CmGLK family members under drought stress
We used qRT-PCR to analyze the expression changes of 15 CmGLK genes in melon leaves under drought stress. The results (Figure 3; Supplementary Table S5) revealed varying degrees of expression changes among the 15 CmGLK genes at different time points after drought stress. CmGLK25 exhibited significantly elevated expression levels at the 12h, 24h, and 48h time points, with the expression level at 24 hours reaching 21.93 times that of the CK group. CmGLK32 exhibited expression levels twice those of the CK group at the 12h and 48h time points. The remaining 13 genes exhibited either relatively lower increases or decreases in expression levels at different time points under drought stress. These results suggest that CmGLK family members may play a role in enhancing the response of melon to drought stress.
4 Discussion
GLK transcription factors belong to the recently classified GARP superfamily and play crucial roles in chloroplast development, senescence, and optimizing photosynthesis under biotic and abiotic stress (Kobayashi et al., 2013; Han et al., 2016; Savitch et al., 2007; Nguyen et al., 2014). Previous studies have identified 44, 46, 66, 78, 130, 59, and 66 GLK genes in forsythia (Zhao Z et al., 2022), grape (Xu et al., 2022), tomato (Wang et al., 2022), moso bamboo (Wu et al., 2022), soybean (Alam et al., 2022), foxtail millet (Chen et al., 2022), and tea (Wang et al., 2024), respectively. In this study, we identified 49 GLK genes in the melon genome, which were classified into six groups (I–VI) based on the classification of A. thaliana GLK genes. Members of the same CmGLK subfamily exhibited highly conserved protein motifs and gene structures, confirming the reliability of the grouping. Generally, proteins within the same subgroup tend to have more similar functions. In Group II, we found three A. thaliana KANADI (KAN) genes (AT1G32240.1, AT5G16560.1, AT5G42630.1), which are expressed in the shoot apical meristem (SAM) and inhibit auxin biosynthesis, transport, and signaling to suppress organ morphogenesis (Ram et al., 2020). It is speculated that CmGLK9, CmGLK12, and CmGLK25, which belong to the same group in the phylogenetic tree, may have similar functions. Group IV included AtLUX (AT3G46640.1), AtBOA (AT5G59570.1), and AtMYBC1 (AT2G40970.1), which are associated with circadian rhythm oscillations. CmGLK18, CmGLK48, CmGLK44, and CmGLK34 in the same group may also perform similar functions (Zhao X et al., 2022).
Gene duplication is a crucial mechanism for gene expansion during plant genome evolution and helps plants adapt to diverse environments. Using bioinformatics techniques, we identified one tandem duplication pair and nine segmental duplication pairs in the melon genome, indicating that gene duplication significantly contributed to the expansion of the GLK family in melon. For example, moso bamboo has 31 segmental and 1 tandem duplication pair (Wu et al., 2022), grape has 5 segmental and 1 tandem duplication pair (Xu et al., 2022), and tea has 17 segmental and 1 tandem duplication pair (Wang et al., 2024). Segmental duplication is a primary mechanism for the expansion of the melon GLK family, as it has been proven to be more common than tandem duplication and plays an essential role in long-term evolution. The Ka/Ks values of segmental duplication gene pairs, tandem duplication gene pairs, and homologous gene pairs between melon and cucumber, as well as between melon and watermelon, were less than 1, suggesting strong purifying selection. These findings indicate that gene duplication and evolutionary conservation among Cucurbitaceae species contributed to the expansion of the CmGLK gene family.
Transcriptome data revealed FPKM values of CmGLK genes across 13 melon tissues. Expression profiling showed that CmGLK genes displayed significant and diverse expression patterns among tissues, with noticeable tissue-specific expression, which is consistent with a potential role in melon growth and development. In many plants, such as A. thaliana, rice, and maize, GLK genes are highly expressed in leaves but less so in stems, flowers (inflorescences), and roots (Fitter et al., 2010; Yeh et al., 2022), which is consistent with their central role in chloroplast development. Interestingly, our study revealed a distinct expression pattern in melon, where a considerable number of CmGLK genes exhibited higher expression levels in root and stem tissues. This divergence from the canonical pattern observed in model plants suggests potential neofunctionalization or subfunctionalization of the CmGLK family in melon, possibly reflecting unique physiological adaptations of this horticultural crop.”
“The biological significance of this root/stem-biased expression may be closely linked to organ-specific stress adaptation. It is established that GLK transcription factors can function beyond leaf chloroplast development. For instance, they have been documented to participate in stress response signaling pathways, sometimes independently of their photosynthetic role (Murmu et al., 2014). Moreover, the presence of functional chloroplasts in non-foliar tissues like roots, whose development can be influenced by GLK proteins, has been implicated in local energy production and stress resilience (Kobayashi et al., 2013). Therefore, we hypothesize that the high expression of specific CmGLK genes in melon roots could represent an adaptive strategy to enhance local defenses against soil-borne pathogens such as F. oxysporum, potentially by modulating redox homeostasis or energy metabolism. Similarly, their elevated expression in stems might bolster vascular function and photosynthetic capacity critical for surviving drought stress. While this compelling hypothesis requires future functional validation, it provides a rational framework for interpreting our data and underscores the functional diversity of the GLK family across plant species.”
GLK transcription factors have been extensively studied for their role in enhancing plant resistance to biotic stresses. Overexpression of AtGLK1 in A. thaliana significantly improves resistance to F. graminearum (Schreiber et al., 2011) and Botrytis cinerea (Murmu et al., 2014). Similarly, ectopic expression of peanut AhGLK1b in A. thaliana enhances resistance to bacterial pathogen Pst DC3000 and Ralstonia solanacearum (Ali et al., 2020). The Turnip Yellow Mosaic Virus P69 interacts with AtGLKs, inhibiting their transcriptional activation activity, and suppressing GLK-targeted genes involved in light harvesting and chlorophyll biosynthesis, which affects normal plant growth (Ni et al., 2017). GLKs may mediate disease resistance through the SA signaling pathway. NPR1, an SA receptor, induces SIB1 expression to initiate defense responses. SIB1 mutants show reduced resistance to B. cinerea, whereas overexpression enhances resistance (Lai et al., 2011). SIB1 interacts with GLKs in the nucleus to enhance transcriptional activation of PhANGs genes, while in chloroplasts, it interacts with SIG1 to inhibit PhAPGs expression, leading to the accumulation of ROS and 1O2. This accumulation activates plastid-nucleus retrograde signaling, triggering programmed cell death (PCD) and immunity (Lv et al., 2019). LSD1 also interacts with GLKs to suppress their binding to target promoters, reducing transcriptional activity. Overexpression of LSD1 suppresses PhANGs expression and chloroplast development, while lsd1 mutants exhibit enhanced disease resistance (Li et al., 2021).
F. wilt, caused by F. oxysporum, is characterized by typical symptoms such as wilting and plant death. This pathogen is a soil-borne fungus that tends to occur and spread in warm, humid environments (Fravel et al., 2003).The multiple cis-regulatory elements in promoters play a critical role in resistance signaling pathways, and their interactions regulate the complex processes of disease resistance (Wood, 2002).The promoters of CmGLK genes contain various hormone response elements (Figure 8; Supplementary Table S3), including IAA (auxin) response elements (TGA-element, AuxRR-core), ABA (abscisic acid) response elements (ABRE), MeJA (methyl jasmonate) response elements (CGTCA-motif, TGACG-motif), SA (salicylic acid) response elements (TCA-element, TGA-element, SARE), and GA (gibberellin) response elements (TATC-box, GARE-motif, P-box), among others. Plant hormones not only regulate plant growth and development but also act as important signaling molecules in plant resistance responses (Bari and Jones, 2009). The interactions between these hormones can enhance the expression of resistance genes, helping plants better cope with adverse conditions. Cis-regulatory element analysis shows that CmGLK1, CmGLK19, CmGLK25, CmGLK33, and CmGLK41 in the melon genome contain both methyl jasmonate and salicylic acid response elements, and their expression is significantly elevated during F. wilt infection. This suggests that these genes may be involved in the methyl jasmonate and salicylic acid-mediated resistance pathways, thus enhancing the plant’s resistance. Specifically, we can leverage the integrated expression data to propose more precise functional hypotheses. For instance, CmGLK33 and CmGLK41 not only possess these defense-related promoter elements but also exhibit particularly strong induction upon pathogen attack and are highly expressed in root tissues—the primary site of F. oxysporum infection. This co-localization of expression and function suggests a compelling model: we hypothesize that CmGLK33 and CmGLK41 act as key regulators in the root, where they are activated by JA/SA signaling pathways to coordinate downstream defense responses, potentially through modulating the expression of photosynthesis-related nuclear genes as part of a holistic defense strategy.
Additionally, the combined action of methyl jasmonate and abscisic acid can improve plant stress tolerance, as seen in RT-qPCR validation where many resistance genes in banana showed upregulated expression following ABA and MeJA treatment (Luo et al., 2023). Notably, CmGLK19, CmGLK33, and CmGLK41 contain both methyl jasmonate and abscisic acid response elements, indicating their potential involvement in the resistance pathways mediated by these hormones during F. wilt infection. Furthermore, this combined analysis approach can be extended to other stresses. The promoter of CmGLK25—the most responsive gene under drought stress—is enriched in ABRE elements alongside light-responsive motifs. Given its concurrent high expression in stems, we hypothesize that CmGLK25 may function in vascular tissues to integrate ABA-dependent drought signaling with photosynthetic adjustments, thereby maintaining energy balance under water deficit. These specific, data-driven hypotheses provide clear targets for future functional validation of CmGLK genes in melon stress adaptation.
To date, extensive research has been conducted on GLK gene regulation in response to drought stress in various species. In maize, studies indicate that GLK genes may be associated with cold and drought stress responses (Liu et al., 2016). In cotton, the GhGLK1 gene appears to play a role in drought and low-temperature stress regulation (Liu et al., 2021), with three genes showing potential negative regulation under drought stress. Significant changes have also been observed in GhGLK46 and GhGLK55 genes (Zhao et al., 2021). In peanuts, AhGLK1 acts as a transcription factor that upregulates AhPORA expression during drought recovery, stimulating chlorophyll biosynthesis and photosynthesis (Liu et al., 2018). In millet, SiGLK30, SiGLK38, SiGLK48, and SiGLK52 play significant roles in regulating salt, cold, and drought stress responses (Chen et al., 2022). The 15 CmGLK genes in melon show varied expression changes at different time points under drought stress, with CmGLK25 showing expression levels over 20 times higher than the CK group at 24h. Based on these findings, we hypothesize that CmGLK genes may contribute to the regulation of melon’s drought stress resistance.
It is also important to consider the phylogenetic framework of our study. Our classification was anchored using A. thaliana sequences as a reference. Given the considerable evolutionary divergence between the Brassicaceae and Cucurbitaceae families, some recently evolved, lineage-specific GLK genes in melon might not have been identified if they lack clear homologs in A. thaliana. Moreover, while clustering with A. thaliana genes provides valuable initial functional clues, the precise biological roles of the CmGLK genes may have diverged during evolution. Therefore, functional predictions based on these distant homologs should be interpreted with caution and require direct experimental validation in melon. Nevertheless, the strong phylogenetic conservation and synteny observed among the cucurbit species (melon, cucumber, and watermelon) confirm that our analysis provides a robust and reliable framework for studying the GLK family within the Cucurbitaceae.
In conclusion, this study provides a comprehensive genomic identification and expression atlas of the CmGLK gene family. It is crucial to note that the observed expression changes under stress establish a correlative, not causative, relationship. Thus, the definitive functional roles of significantly induced genes like CmGLK25, CmGLK33, and CmGLK41 must be confirmed through future genetic experiments, such as knockout or overexpression in melon. Despite this, our work lays a solid foundation and offers a set of high-priority candidate genes. The resources and hypotheses generated here will undoubtedly guide and accelerate future functional characterization efforts aimed at enhancing stress tolerance in melon.”
5 Conclusion
This study identified and systematically analyzed 49 members of the CmGLK gene family in the melon genome (DHL92 v4.0). The analysis included their physicochemical properties, phylogenetic relationships, gene structures, protein motifs, gene duplication, and cis-regulatory elements in promoter regions. The CmGLK family was classified into six subgroups, with high conservation in gene structure and protein motifs within each subgroup. Gene duplication appears to play a critical role in the expansion of the CmGLK family. Expression profiling revealed tissue-specific expression patterns of CmGLK genes in melon and significant responses to drought and F. wilt stress. Collectively, our findings on tissue-specific expression and stress-induced expression changes suggest that the CmGLK family is likely involved in melon growth, development, and stress resistance. This study lays a foundation for future studies on the functional characterization of CmGLK genes and provides valuable candidates for molecular breeding efforts aimed at enhancing stress tolerance in melon.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
LZ: Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. YW: Data curation, Formal Analysis, Writing – review & editing. FL: Formal Analysis, Methodology, Writing – original draft. JH: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The work was funded by Science and Technology Key Project of Henan Province (Project No. 232102110046); Luoyang Core Technology Research and Development Public Welfare Special Project (Project No. 2302036A).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1679452/full#supplementary-material
References
Alam, I., Zhang, Y., Chen, L., Xie, Q., Liu, S., Li, Z., et al. (2022). Identification of GOLDEN2-like transcription factor genes in soybeans and their role in regulating plant development and metal ion stresses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1052659
Ali, N., Chen, H., Zhang, C., Wang, J., Zhang, Z., Xu, Q., et al. (2020). Ectopic expression of AhGLK1b (golden 2-like transcription factor) in Arabidopsis confers dual resistance to fungal and bacterial pathogens. Genes 11, 343. doi: 10.3390/genes11030343
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bari, R. and Jones, J. D. G. (2009). Role of plant hormones in plant defense responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Chang, Y. M., Liu, W. Y., Shih, A. C. C., Chiang, H. P., and Chen, T. H. H. (2012). Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160, 165–177. doi: 10.1104/pp.112.205360
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Zhang, Y., Zhao, S., Liu, J., He, Y., Yang, Z., et al. (2022). Identification and evolutionary analysis of the GOLDEN 2-LIKE gene family in foxtail millet. Trop. Plant Biol. 15, 301–318. doi: 10.1007/s12042-022-09324-8
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., and Langdale, J. A. (2010). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727. doi: 10.1111/j.1365-313X.2002.01316.x
Fravel, D., Olivain, C., and Alabouvette, C. (2003). Fusarium oxysporum and its biocontrol. New Phytol. 157, 493–502. doi: 10.1046/j.1469-8137.2003.00700.x
Gao, Q. X. (2015). High-quality and efficient cultivation technology of melon. Jilin Veg 6, 5–6. doi: CNKI:SUN:JLSS.0.2015-06-006
Garcia-Mas, J., Benjak, A., Sanseverino, W., Bourgeois, M., Mir, G., González, V. M., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. U.S.A. 109, 11872–11877. doi: 10.1073/pnas.1205415109
Hall, L. N., Rossini, L., Cribb, L., and Langdale, J. A. (1998). GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10, 925–936. doi: 10.1105/tpc.10.6.925
Han, X. Y., Li, P. X., Zou, L. J., Du, Z., Zhang, J., and Yang, J. (2016). GOLDEN2-LIKE transcription factors coordinate the tolerance to cucumber mosaic virus in arabidopsis. Biochem. Biophys. Res. Commun. 477, 626–632. doi: 10.1016/j.bbrc.2016.06.063
Herman, R. and Perl-Treves, R. (2007). Characterization and Inheritance of a New Source of Resistance to Fusarium oxysporum f. sp. Melonis Race 1.2 in Cucumis melo. Plant Dis. 91, 1180–1186. doi: 10.1094/PDIS-91-9-1180
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm316
Kobayashi, K., Sasaki, D., Noguchi, K., Fujinuma, D., Komatsu, H., Kobayashi, M., et al. (2013). Photosynthesis of root chloroplasts developed in arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors. Plant Cell Physiol. 54, 1365–1377. doi: 10.1093/pcp/pct094
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lai, Z. B., Li, Y., Wang, F., Li, H., Zhang, X., Zhang, J., et al. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23, 3824–3841. doi: 10.1105/tpc.111.090571
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, M., Lee, K. P., Liu, T., Wang, X., Xie, Y., Zhang, M., et al. (2021). Antagonistic modules regulate photosynthesis-associated nuclear genes via golden 2-like transcription factors. Plant Physiol. 187, 2140–2155. doi: 10.1093/plphys/kiab600
Liu, X., Li, L., Li, M., Su, L., Lian, S., Zhang, B., et al. (2018). AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci. Rep. 8, 2250. doi: 10.1038/s41598-018-20542-7
Liu, J., Mehari, T. G., Xu, Y., Umer, M. J., Hou, Y., Wang, Y., et al. (2021). GhGLK1 a key candidate gene from the GARP family enhances cold and drought stress tolerance in cotton. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.759312
Liu, F., Xu, Y., Han, G., Wang, W., Li, X., Cheng, Y., et al. (2016). Molecular evolution and genetic variation of G2-like transcription factor genes in maize. PloS One 11, e0161763. doi: 10.1371/journal.pone.0161763
Luo, B., Zhang, H., Li, D., He, W., Li, J., Zhao, T., et al. (2023). Genome-wide identification of the banana GLR gene family and its expression analysis in response to low temperature and abscisic acid/methyl jasmonate. Chin. J. Biotechnol. 39, 2874–2896. doi: 10.13345/j.cjb.220972
Lv, R. Q., Li, Z. H., Li, M. P., Yang, T., Chen, X., Xu, X., et al. (2019). Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell 31, 210–230. doi: 10.1105/tpc.18.00813
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Mayfield, S. P., Yohn, C. B., Cohen, A., and Taylor, W. C. (1995). Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 147–166. doi: 10.1146/annurev.pp.46.060195.001051
Murmu, J., Wilton, M., Allard, G., Sun, M., and Atkinson, H. (2014). Arabidopsis GOLDEN2-LIKE (GLK) Transcription Factors Activate Jasmonic Acid (JA)-Dependent Disease Susceptibility to the Biotrophic Pathogen Hyaloperonospora arabidopsidis, as well as JA-Independent Plant Immunity against the Necrotrophic Pathogen Botrytis cinerea. Mol. Plant Pathol. 15, 174–184. doi: 10.1111/mpp.12113
Nguyen, C. V., Vrebalov, J. T., Gapper, N. E., Lang, L., Colletti, P., DellaPenna, D., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26, 585–601. doi: 10.1105/tpc.113.121666
Ni, F. R., Wu, L., Wang, Q., Li, X., Li, S., Zhang, Y., et al. (2017). Turnip yellow mosaic virus P69 interacts with and suppresses GLK transcription factors to cause pale-green symptoms in arabidopsis. Mol. Plant 10, 764–766. doi: 10.1016/j.molp.2016.12.003
Peng, W., Roxana, K., Shanta, K., Jing, L., Zhang, Q., Xie, M., et al. (2017). Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr. Biol. 27, 3278–3287. doi: 10.1016/j.cub.2017.09.016
Rahman, M. Z., Ahmad, K., and Bashir Kutawa, A. (2021). Biology, Diversity, Detection and Management of Fusarium oxysporum f. sp. niveum Causing Vascular Wilt Disease of Watermelon (Citrullus lanatus): A Review. Agronomy 11, 1310. doi: 10.3390/agronomy11071310
Ram, H., Sahadevan, S., Gale, N., Caggiano, M. P., Yu, X., Ohno, C., et al. (2020). An integrated analysis of cell-type specific gene expression reveals genes regulated by REVOLUTA and KANADI1 in the Arabidopsis shoot apical meristem. PloS Genet. 16, e1008661. doi: 10.1371/journal.pgen.1008661
Razi, K. and Muneer, S. (2021). Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 41, 669–691. doi: 10.1080/07388551.2020.1740532
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rossini, L., Cribb, L., Martin, D. J., and Langdale, J. A. (2001). The maize GOLDEN2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13, 1231–1244. doi: 10.1105/tpc.13.5.1231
Savitch, L. V., Subramaniam, R., Allard, G. C., Rachlin, J., Ohlrogge, J. B., and Osmond, B. (2007). The GLK1 ‘Regulon’ Encodes disease defense-related proteins and confers resistance to fusarium graminearum in arabidopsis. Biochem. Biophys. Res. Commun. 359, 234–238. doi: 10.1016/j.bbrc.2007.05.187
Schreiber, K. J., Nasmith, C. G., Allard, G., McLaren, D. L., Houde, M., Hogue, R., et al. (2011). Found in translation: high-throughput chemical screening in arabidopsis thaliana identifies small molecules that reduce fusarium head blight disease in wheat. Mol. Plant-Microbe Interact. 24, 640–648. doi: 10.1094/MPMI-02-11-0042
Shen, S., Yuan, J., Xu, Y., Yang, S., and Wang, H. (2022). Biological function and molecular mechanism of the transcription factor GLKs in plants: A review. Chin. J. Biotechnol. 38, 2700–2712. doi: 10.13345/j.cjb.220119
UniProt Consortium (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi: 10.1093/nar/gkaa1100
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, H., Xu, F., Zhang, J., Zhao, Y., Li, X., Chen, H., et al. (2024). Identification and expression analysis of the GLK gene family in tea plant (Camellia sinensis) and a functional study of CsGLK54 under low-temperature stress. Sci. Rep. 14, 12465. doi: 10.1038/s41598-024-63323-1
Wang, Z., Zhao, S., Liu, J., Zhang, H., Li, X., Lin, Y., et al. (2022). Genome-wide identification of tomato golden 2-like transcription factors and screening of abiotic stress-related members. BMC Plant Biol. 22, 82. doi: 10.1186/s12870-022-03460-9
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065365
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the exPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/0-89603-321-6:531
Wood, N. T. (2002). Synthetic promoters illuminate roles of cis-acting elements in plant defense. Trends Plant Sci. 7, 279–284. doi: 10.1016/s1360-1385(02)02287-2
Wu, R., Guo, L., Wang, R., Zhang, W., Li, Z., Zhang, L., et al. (2022). Genome-wide identification and characterization of G2-like transcription factor genes in Moso Bamboo (Phyllostachys edulis). Molecules 27, 5491. doi: 10.3390/molecules27175491
Xia, L., Peng, W., Jing, L., Zhi, H., Zhang, Q., Qi, L., et al. (2020). Maize GOLDEN2-LIKE genes enhance biomass and grain yields in rice by improving photosynthesis and reducing photoinhibition. Commun. Biol. 3, 151. doi: 10.1038/s42003-020-0954-5
Xu, C., Li, Q. Y., Wan, J. N., Liang, Z., Zheng, X., Guo, Y., et al. (2022). Identification and expression analysis of the golden2-like transcription factor family of vitis vinifera L. Plant Sci. J. 40, 205–215. doi: CNKI:SUN:WZXY.0.2022-02-009
Ye, J., McGinnis, S., and Madden, T. L. (2006). BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34, W6–W9. doi: 10.1093/nar/gkl209
Yeh, S. Y., Lin, H. H., Chang, Y. M., Kuo, M., Wu, F., Lee, M. Y., et al. (2022). Maize golden 2-like transcription factors boost rice chloroplast development, photosynthesis, and grain yield. Plant Physiol. 188, 442–459. doi: 10.1093/plphys/kiab511
Yu, J., Wu, S., Sun, H., Wang, X., Tang, X., Guo, S., et al. (2023). CuGenDBv2: an updated database for cucurbit genomics. Nucleic Acids Res. 51, D1457–D1464. doi: 10.1093/nar/gkac921
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K., and Yu, J. (2006). KaKs_Calculator: calculating ka and ks through model selection and model averaging. Genomics Proteomics Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhang, D., Yu, Z., Zeng, B., and Liu, X. (2024). Genome-wide analysis of the ABC gene family in almond and functional predictions during flower development, freezing stress, and salt stress. BMC Plant Biol. 24, 12. doi: 10.1186/s12870-024-04296-2
Zhao, Z., Shuang, J., Li, Z., Xiao, H., Liu, Y., Wang, T., et al. (2021). Identification of the Golden-2-like transcription factors gene family in Gossypium hirsutum. PeerJ 9, e12484. doi: 10.7717/peerj.12484
Zhao, X., Yang, J., Li, X., Li, G., Sun, Z., Chen, Y., et al. (2022). Identification and expression analysis of GARP superfamily genes in response to nitrogen and phosphorus stress in Spirodela polyrhiza. BMC Plant Biol. 22, 308. doi: 10.1186/s12870-022-03696-5
Zhao, Z., Zhang, X., Cheng, T., Li, Z., Li, H., Liu, Y., et al. (2022). Identification of the GLK gene family in forsythia and expression analysis in response to light. Henan Agric. Univ. J. 56, 759–769. doi: 10.16445/j.cnki.1000-2340.20220915.001
Zheng, L., Dai, H., Mu, Y., Li, J., Cheng, Y., and Han, J. (2025). Genome-wide identification and expression analysis of C3H gene family in melon. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1500429
Keywords: melon, GLK gene family, Fusarium wilt stress, drought stress, expression analysis, bioinformatics
Citation: Zheng L, Wang Y, Lv F and Han J (2025) Genome-wide identification of the GLK transcription factor family in melon and its expression analysis under biotic and abiotic stresses. Front. Plant Sci. 16:1679452. doi: 10.3389/fpls.2025.1679452
Received: 04 August 2025; Accepted: 14 October 2025;
Published: 29 October 2025.
Edited by:
Bin Wu, Chengdu University of Technology, ChinaReviewed by:
Masood Jan, University of Florida, United StatesXingxu Zhang, Lanzhou University, China
Copyright © 2025 Zheng, Wang, Lv and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianming Han, aGFuamlhbm1pbmdAbHludS5lZHUuY24=
 Ling Zheng
Ling Zheng Yuna Wang
Yuna Wang