- 1Jiangxi Provincial Key Laboratory of Conservation Biology, Jiangxi Agricultural University, Nanchang, China
- 2Research Center of Plant Functional Genes and Tissue Culture Technology, College of Bioscience and Bioengineering, Jiangxi Agricultural University, Nanchang, China
- 3Jiangxi Province Key Laboratory of Vegetable Cultivation and Utilization, Jiangxi Agricultural University, Nanchang, China
1 Introduction
Plants exhibit a remarkable capacity to regenerate tissue, organs, or entirely new individuals after wounding or under in vitro conditions (Eshed Williams, 2021; Doll and Ikeuchi, 2025). Plant regeneration in vitro involves a biphasic process encompassing the acquisition of cell pluripotency and subsequent de novo shoots regeneration (DNSR) or de novo root regeneration (DNRR) (Figure 1A; Kim and Seo, 2025; Youngstrom et al., 2025). Initially, explants are cultured on an auxin-enriched callus-inducing medium (CIM) to induce the formation of a pluripotent callus. The pluripotent callus then undergoes extensive cellular reprogramming and spatial cell identities reorganization to generate shoots or roots (Figure 1A). Accumulating evidence indicates the vital involvement of phytohormones such as auxin, jasmonic acid (JA), cytokinin (CK), ethylene in regulating plant regeneration (García-Gómez and Ten Tusscher, 2024; Asghar et al., 2025; Xu and Yang, 2025). In addition, a couple of key transcription factors (TFs) such as WUSCHEL-RELATED HOMEOBOX (WOX), PLETHORAs (PLTs) have been reported to play a key role in plant regeneration (Islam et al., 2023; Chen et al., 2024). Although this regenerative capability has been well-documented across numerous plant species, the underlying mechanisms remain largely elusive (Eshed Williams, 2021; Chen et al., 2024).
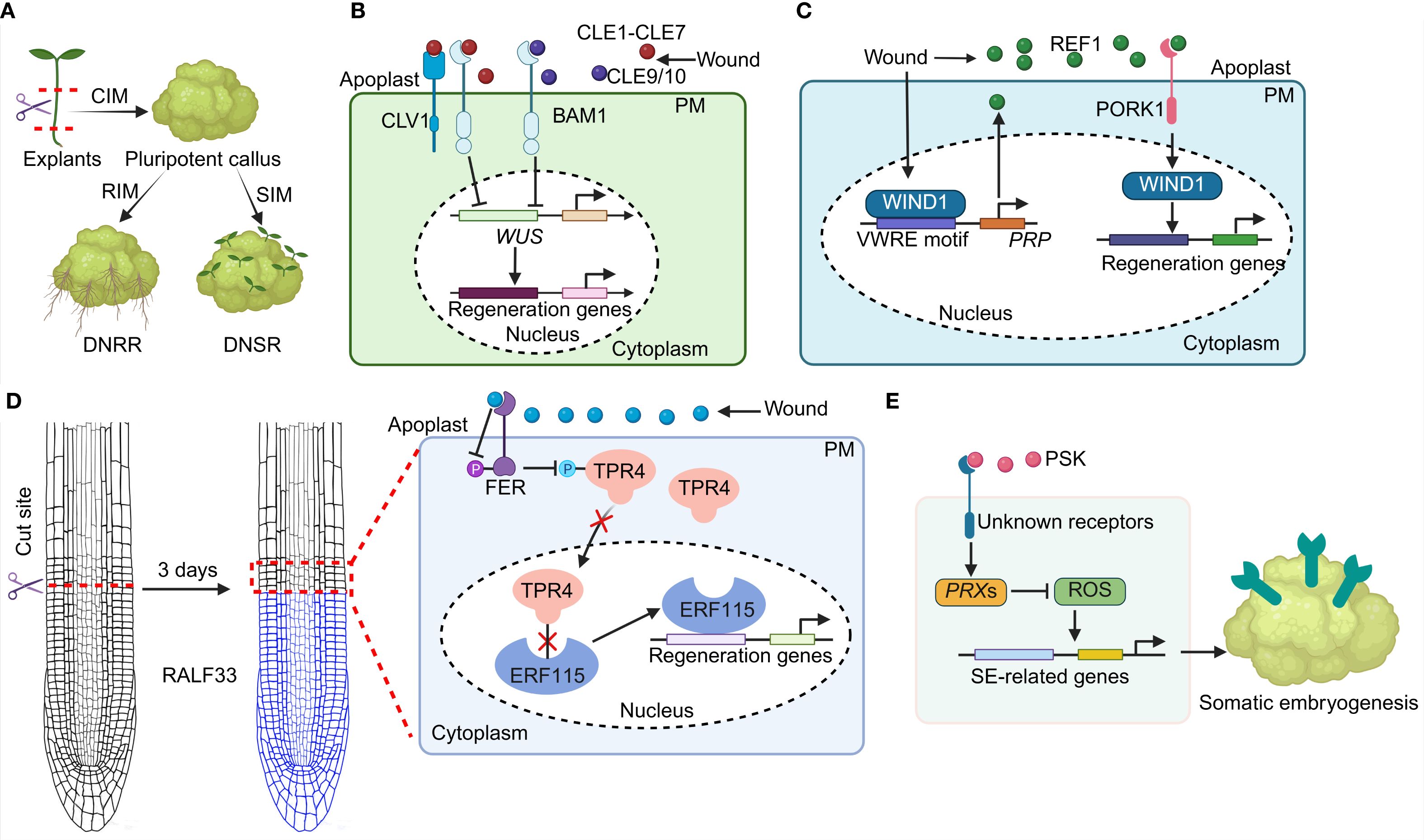
Figure 1. Small signaling peptides regulate plant regeneration potential. (A) A simplified plant DNSR and DNRR process. (B) CLE1-CLE7 and CLE9/10 peptides are perceived by the CLV1 and BAM1 receptors to regulate WUS transcription and its downstream targets of regeneration associated genes, thereby impeding shoot regeneration. (C) WIND1 binds to the PRP promoter via the VWRE motif to initiate PRP-dependent REF1 peptide synthesis. The REF1-PORK1 module in turn induces the transcription of WIND1, thereby facilitating plant regeneration. (D) The RALF33-FER modulates the phosphorylation status of TPR4. The altered TPR4 protein then releases the ERF115 activity and promotes root regeneration. P: phosphorylation. (E) PSK peptide modulates SE development via regulating ROS signaling.
Small signaling peptides represent a novel class of plant growth regulators, typically comprising fewer than 150 amino acids. These peptides are recognized by plasma membrane-localized receptors or co-receptors, which subsequently activate or deactivate specific regulatory pathways to modulate plant growth and stress adaptions (Ji et al., 2025; Xiao et al., 2025). Notably, several small signaling peptide-encoding genes, including CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE), C-TERMINALLY ENCODED PEP TIDE (CEP), phytosulfokine (PSK), and GOLVEN/ROOT MERISTEM GROWTH FACTOR/CLE-LIKE (GLV/RGF/CLEL) are differentially activated at specific stages of DNSR, as revealed by single-cell transcriptomic analysis, suggesting their potential roles in the plant shoot regeneration process (Zhai and Xu, 2021; Wang et al., 2022). Experimental data further demonstrate the regulatory functions of CLE1-CLE7 (Kang et al., 2022), CLE9/10 (Glazunova et al., 2025) and CLE46 (Ito et al., 2025), REGENERATION FACTOR1 (REF1) (Yang et al., 2024) and RAPID ALKALINIZATION FACTOR 33 (RALF33) peptides (Shen et al., 2025) in modulating plant regeneration capacity under in vitro conditions or in response to wounding signal as well as PSK peptides in the regulation of somatic embryogenesis (SE) (Hao et al., 2023; Luo et al., 2024) (Table 1; Figures 1B–E).
2 CLE-CLV1/BAM1 signaling module negatively regulates adventitious shoot regeneration
The CLE genes encode precursor proteins featuring an N-terminal signal sequence that targets them to the secretory pathway, a central variable region, and a highly conserved CLE domain at the C-terminus, which frequently undergoes posttranslational modifications to produce a functional polypeptide (Xie et al., 2022; Ji et al., 2025). Consistent with their pivotal functions in stem cell regulation (Selby and Jones, 2023; Shpak and Uzair, 2025), the expressions of CLE members are differentially activated at distinct phases of shoot regeneration (Zhai and Xu, 2021; Wang et al., 2022). Specifically, CLE1-CLE7 and CLE9/10 peptides have been demonstrated to negatively influence shoot regeneration (Figure 1B; Kang et al., 2022; Glazunova et al., 2025).
CLE1-CLE7 gene expression is significantly induced by CIM or shoot-inducing medium (SIM), and CRISPR-engineered cle1–7 septuple mutant exhibits an increased number of adventitious shoots (Kang et al., 2022). While most single cle mutants do not affect DNSR, the cle4 and cle7 mutants show enhanced DNSR (Kang et al., 2022). Conversely, application of synthetic CLE1–7 peptides exhibits a dose-dependent inhibition of DNSR, and overexpression of CLE4 or CLE7 also suppresses DNSR. In addition, CLE9/10 peptide also suppresses DNSR without affecting callus formation (Figure 1B; Glazunova et al., 2025).
Notably, the clv1 and bam1 mutants show increased shoot regeneration capabilities and are insensitive to CLE1-CLE7 peptide-mediated inhibition of shoot regeneration, confirming that CLAVATA1 (CLV1) and BARELY ANY MERISTEM1 (BAM1) receptors mediate the CLE1-CLE7 signal to negatively regulate DNSR. However, clv1 mutant respond normally to CLE9/10 peptide treatment, while bam1 mutant is insensitive to CLE9/10 peptide (Glazunova et al., 2025). Further studies reveal that the CLE-CLV1/BAM1 module restricts the transcriptional level of WUSCHEL (WUS), a critical regulator of plant regeneration (Ince and Sugimoto, 2023; Shpak and Uzair, 2025), thereby controlling shoot regeneration potential (Figure 1B).
3 REF1-PORK1 pathway promotes regeneration through activating WIND1 expression
Recent studies have identified REF1, a Pep peptide homolog in tomato, as a crucial regulator of plant regenerative capacity (Figure 1C; Yang et al., 2024). The absence of PRP, the precursor of REF1 peptide, results in impaired callus formation and shoot regeneration, while PRP overexpression markedly enhances regenerative capacity (Yang et al., 2024). The exogenous application of synthetic REF1 peptide also enhances callus formation and shoot regeneration in wild-type (WT) plants in a dose-responsive manner, and compensates for the regenerative deficits observed in prp mutants. These findings collectively underscore the key role of REF1 peptide in regulating plant regeneration (Yang et al., 2024). Additionally, mutation in PEPR1/2 ORTHOLOG RECEPTOR-LIKE KINASE 1 (PORK1) result in similar defects in callus formation and shoot regeneration that are observed in prp mutants. While PORK1 overexpression significantly boosts regenerative capacity. Additionally, pork1 mutant exhibits insensitivity to REF1 peptide treatments. Further investigations reveal that REF1 binds to PORK1, establishing PORK1 as the receptor for REF1 peptide (Yang et al., 2024).
WOUND-INDUCED DEDIFFERENTIATION 1 (WIND1), a prominent transcription factor implicated in plant regeneration (Iwase et al., 2017), operates downstream of the REF1-PORK1 module. Wounding signal significantly induces WIND1 expression in WT tomato plants but not in prp and pork1 mutants. The wind1 tomato mutant exhibits severely compromised callus formation and shoot regeneration capacities, whereas WIND1 overexpression plants show enhanced regenerative capabilities. Moreover, the regenerative deficiency in wind1 mutants cannot be ameliorated by REF1 peptides. Importantly, wound-induced WIND1 binds to the PRP promoter via the wound-responsive cis-element (VWRE) motif, promoting PRP transcription and amplifying REF1 signaling during regeneration. Notably, the utilization of the REF1 peptide has been shown to enhance both the regeneration and transformation efficiencies in soybean, wheat, and maize, indicating that REF1 peptide may substantially improve the recalcitrant crop regeneration and transformation processes. In conclusion, the REF1-PORK1-WIND1 module constitutes a regulatory loop that finely tunes the plant regeneration potential (Figure 1C; Yang et al., 2024).
4 RALF33-FER module regulates TPR4-ERF115 dynamics in root regeneration
RALF peptides, belonging to the cysteine-rich peptide family, are typically recognized by the Catharanthus roseus Receptor-Like Kinase 1-Like proteins (CrRLK1Ls), particularly FERONIA (FER) (Cheung, 2024; Pratyusha and Sarada, 2025). RALF peptides play a pivotal role in integrating developmental and environmental cues, thereby orchestrating optimal cellular and physiological responses (Cheung, 2024; Pratyusha and Sarada, 2025). Recent research highlights the significant function of the RALF33 peptide in root regeneration (Figure 1D; Shen et al., 2025).
RALF33 peptide shows rapid and substantial accumulation near the cut sites in the root within an hour post-resection, indicating its role as an early signaling molecule in response to root tip injury (Shen et al., 2025). Compared to WT plants, ralf33 mutant exhibits reduced regeneration rates when cuts are made at the transition zone. Moreover, exogenous application of synthetic RALF33 peptide enhances regeneration rates. These findings collectively demonstrate that RALF33 acts as a positive regulator of root regeneration capacity. The fer mutant shows an increased regeneration capacity compared to WT plants. Furthermore, synthetic RALF33 peptide does not further enhance the regeneration rate in the fer mutant. Collectively, wounding induces the accumulation of RALF33 peptide, which subsequently suppresses FER activity, thereby enhancing root regeneration capacity.
Subsequent investigations have elucidated that TOPLESS-RELATED4 (TPR4) and the transcription factor (TF) ERF115 act downstream of the RALF33-FER signaling module in root regeneration processes. FER interacts with TPR4, and this interaction inhibits the localization of TPR4 in nucleus. The mis-localized TPRE4 is unable to suppress the activity of ETHYLENE RESPONSE FACTOR 115 (ERF115), a critical regenerative TF (Zhou et al., 2019; Matosevich et al., 2020). The release of ERF115 then activates its downstream targets, thereby impairing root regeneration. (Figure 1D; Shen et al., 2025).
In addition, CLE46 is reported to have an inhibitory effect on DNRR originating from leaf explants (Ito et al., 2025). Spatial expression analysis indicates that the activity of the CLE46 promoter-GUS fusion is confined to the shoot apex in young Arabidopsis seedlings. Upon the excision of petioles in leaves, CLE46 activity is induced in response to wound, exerting a negative regulatory effect on root regeneration. This is evidenced by the increased root regeneration rate observed at the leaf cut end in the cle46 mutant. In hypocotyl-excised seedlings, CLE46 activity remains uninduced, while cle46 mutant still displays a significantly higher root regeneration rate compared to WT seedlings. Collectively, these findings suggest that CLE46, expressed in the shoot apex, suppresses root regeneration in the lower regions of the plant tissue (Ito et al., 2025). However, the molecule mechanism of CLE46-mediated DNRR regulation requires further investigations.
5 PSK peptide regulates somatic embryogenesis
Somatic embryogenesis (SE) is widely employed for the transformation and regeneration of diverse plant species (Martínez and Corredoira, 2024), this process is regulated phytosulfokine (PSK) peptide (Hao et al., 2023; Luo et al., 2024). Application of synthetic PSK peptide markedly stimulates the transitions of proembryogenic masses (PEMs) and enhances SE development in Cunninghamia lanceolata in a genotype-independent manner (Hao et al., 2023). Transcriptomic analyses reveal that PSK treatment results in a disruption of reactive oxygen species (ROS), specifically by reducing H2O2 levels through modulation of PEROXIDASEs (PRXs) expression. This reduction in ROS promotes the expression of SE-related genes, including WUS, WUSCHEL HOMEOBOX 2 (WOX2), BABY BOOM (BBM), AINTEGU MENTA (ANT) in the early phases of SE induction (Hao et al., 2023). Consistent with these findings, the overexpression of ClPSK, a homolog of PSK genes in Cunninghamia lanceolata (Wu et al., 2019), also facilitates enhanced SE induction and decreases ROS levels (Hao et al., 2023). PSK peptide can also trigger SE formation in Pinus massoniana by lowering H2O2 concentrations (Luo et al., 2024). Collectively, these results underscore the pivotal and conserved role of PSK peptide in the promotion of SE may via a genotype-independent manner (Figure 1E).
6 Future perspectives
Genetic transformation in plants is essential for advancing crop enhancement and commercial cultivation. Nevertheless, the issue of limited plant regeneration capacity continues to be a critical impediment in the precise generation of genetically modified plants (Mei et al., 2024; Wang et al., 2025; Youngstrom et al., 2025). Although numerous genes, such as BBM, GROWTH-REGULATING FACTORs (GRFs), and WUS, have been identified in promoting the regeneration of transgenic or gene-edited plants (Youngstrom et al., 2025). Nonetheless, these conventional approaches are frequently genotype-dependent and constrained by plants’ inherent recalcitrance to low regeneration capacity. The discovery of small signaling peptides in plant regeneration holds a potential to overcome this limitation. However, several questions need to be addressed to elucidate the underlying molecular mechanisms, thereby facilitating their widespread application in agriculture.
(1) How does wound signaling orchestrate the biosynthesis and post-translational modifications to yield functional peptides? Tissue injury triggers the synthesis of CLE1-7, CLE9/10, CLE46, REF1, RALF33 and PSK peptides, thus initiating regenerative processes (Figure 1), though the precise activation mechanisms remain elusive. It has been documented that proteolytic processing, proline hydroxylation, and arabinosylation are essential for the formation of an active CLE peptide (Stührwohldt et al., 2020a, 2020b). Key enzymes involved in CLE peptide processing and post-modification include SUBTILASEs (SBTs), PROLYL-4 HYDROXYLASE (P4H), O-arabinosyltransferase (HPAT), and arabinosyl transferases such as REDUCED RESIDUAL ARABINOSE 3 (RRA3) and XYLOGLUCANASE113 (XEG113) (Olsson et al., 2019; Stührwohldt and Schaller, 2019). RALF peptides, characterized by multiple cysteine residues, necessitate proper disulfide bond formation for their activity and receptor binding (Frederick et al., 2019; Moussu et al., 2020). The plant disulfide isomerase OaPDI, isolated from Oldenlandia affinis, facilitates the formation of disulfide bonds, thereby generating bioactive peptides (Gruber et al., 2007). Furthermore, metacaspases (MCs) and SUMOylation have been implicated in the proteolytic processing of PROPEPs, the precursors of Pep peptides (Hander et al., 2019; Shen et al., 2019; Zhang et al., 2025), leading to the production of functional Pep peptides. It is imperative to elucidate how wounding activates these enzymatic pathways to generate functional small signaling peptides that finely regulate the regenerative processes.
(2) What is the interplay between small signaling peptides and phytohormonal signaling pathways in plant regeneration? It is well-established that phytohormones are crucial in the plant regeneration process (García-Gómez and Ten Tusscher, 2024; Asghar et al., 2025; Xu and Yang, 2025). Additionally, small signaling peptides are responsive to phytohormones (Ji et al., 2025; Xiao et al., 2025). Specifically, jasmonate (JA) and auxin modulate root regeneration via ERF115 (Zhou et al., 2019), indicating that RALF33 may potentially integrate JA and auxin signaling to regulate root regeneration. These data further suggest a possible crosstalk between small signaling peptides and phytohormones in plant regeneration. However, the exact mechanisms remain to be clarified. Furthermore, what are the dynamics of these small signaling peptides, as they could induce either antagonistic or synergistic effects in plant regeneration processes? A comprehensive analysis of these intricate interactions in future research will aid in their effective application in agricultural practices.
(3) How do small signaling peptides modulate epigenetic mechanisms during plant regeneration? At the cellular level, the regeneration process entails dynamic alterations in gene expression that redirect cell fate transitions. Notable epigenetic modifications implicated in plant regeneration include DNA methylation, trimethylation at lysine 27 of histone H3 (H3K27me3), trimethylation at lysine 4 of histone H3 (H3K4me3), and acetylation of histones H3 and H4 (H3/H4 acetylation) (Chen et al., 2023; Li et al., 2024). During this process, the expression of WUS is subject to complex epigenetic regulation involving both DNA methylation and histone modifications. The WUS promoter is methylated in both CG and CHG contexts (Li et al., 2011; Shemer et al., 2015), and its activation is facilitated by the removal of the repressive histone mark H3K27me3 (Zhang et al., 2017). The H3K9 methyltransferase KRYPTONITE (KYP), the H3K4 demethylase JUMONJI 14 (JMJ14), and the histone acetyltransferase HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1 (HAC1) have all been shown to regulate WUS expression (Li et al., 2011). Additionally, the rapid induction of WIND1 following wounding is orchestrated by elevated levels of H3K9/14 acetylation and H3K4 trimethylation (Rymen et al., 2019). Histone acetyltransferases such as ACETYLTRANSFERASES1 (HAG1), HAG3, and General Control Non-depressible 5 (GCN5) also play roles in the transcriptional regulation of WIND1 (Rymen et al., 2019). POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) facilitates the repression of H3K27me3 to enhance WIND1 transcription (Ikeuchi et al., 2015). Nevertheless, the precise mechanisms by which small signaling peptides influence these epigenetic regulations to reprogram the transcriptional landscape of key regulators in plant regeneration warrant further investigation. The deployment of advanced epigenetic methodologies such as CRISPR-based activation/interference/epigenetic systems (Goell and Hilton, 2021; Cai et al., 2023; Joshi and Wang, 2024), will facilitate the construction of unprecedented transcriptional networks that are mediated by small signaling peptides in plant regeneration processes.
(4) How to boost plant regeneration capacity by engineered small signaling peptides? Genetic modifications of peptide precursors have proven to ensure optimal plant growth and development under fluctuating environmental conditions. Nonetheless, the extraction of endogenous small signaling peptides remains technically challenging due to their exceedingly low concentrations in planta. The exogenous application of chemically synthesized small signaling peptides offers a straightforward and efficient approach to modulate plant plasticity. Previous studies have shown that PSK peptide analogues incorporating diastereomers or N-methylation can enhance regeneration efficiency (van de Sande et al., 2024). Moreover, antagonistic CLE peptides have been developed by introducing an amino acid substitution at the sixth conserved glycine residue within the CLE motif (Song et al., 2013; Czyzewicz et al., 2015). Additionally, such amino acid substitutions in the CLE motif result in the generation of non-natural CLE-like peptides with bifunctional properties (Hirakawa et al., 2017). Consequently, there is considerable interest in creating more effective peptide variants, such as agonists, antagonists, chemically modified peptides, or non-natural peptide-like molecules, to enhance plant regeneration potential through targeted chemical modifications or substitutions. High-performance liquid chromatography (HPLC) or synthetic biology (Wu et al., 2021; Ji et al., 2025) could be utilized to produce these bioactive modified small signaling peptides. Additionally, the efficient delivery of small signaling peptides into plant systems presents a significant challenge. To address this issue, the development of nanomaterials (Santana et al., 2020), hydrogels (Zhang et al., 2023), and viral vectors (Abrahamian et al., 2020) may provide effective strategy for encapsulating and transporting small signaling peptides into plant tissues. The integration of these innovative techniques would enhance the application of small signaling peptides in the precise molecular breeding of crops and horticultural plants by improving regeneration and transformation efficiency.
Author contributions
MW: Conceptualization, Funding acquisition, Writing – review & editing. RL: Writing – original draft. ZZ: Writing – original draft. LL: Writing – original draft. YC: Writing – original draft. HH: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work is supported by the National Natural Science Foundation of China (32260058, 32460081) and Jiangxi Provincial Natural Science Foundation (20223BCJ25037, 20232BAB215008) and Jiangxi Agricultural University (9232308314).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahamian, P., Hammond, R. W., and Hammond, J. (2020). Plant Virus derived vectors: Applications in agricultural and medical biotechnology. Annu. Rev. Virol. 7, 513–535. doi: 10.1146/annurev-virology-010720-054958
Asghar, S., Hayat, F., Zhao, Z., Zheng, Z., Ghori, N., Lu, Z., et al. (2025). De novo root regeneration from leaf explant: a mechanistic review of key factors behind cell fate transition. Planta 261, 33. doi: 10.1007/s00425-025-04616-1
Cai, R., Lv, R., Shi, X., Yang, G., and Jin, J. (2023). CRISPR/dCas9 Tools: Epigenetic mechanism and application in gene transcriptional regulation. Int. J. Mol. Sci. 24, 14865. doi: 10.3390/ijms241914865
Chen, C., Hu, Y., Ikeuchi, M., Jiao, Y., Prasad, K., Su, Y. H., et al. (2024). Plant regeneration in the new era: from molecular mechanisms to biotechnology applications. Sci. China Life Sci. 67, 1338–1367. doi: 10.1007/s11427-024-2581-2
Chen, Y., Hung, F. Y., and Sugimoto, K. (2023). Epigenomic reprogramming in plant regeneration: Locate before you modify. Curr. Opin. Plant Biol. 75, 102415. doi: 10.1016/j.pbi.2023.102415
Cheung, A. Y. (2024). FERONIA: A receptor kinase at the core of a global signaling network. Annu. Rev. Plant Biol. 75, 345–375. doi: 10.1146/annurev-arplant-102820-103424
Czyzewicz, N., Wildhagen, M., Cattaneo, P., Stahl, Y., Pinto, K. G., Aalen, R. B., et al. (2015). Antagonistic peptide technology for functional dissection of CLE peptides revisited. J. Exp. Bot. 66, 367–374. doi: 10.1093/jxb/erv284
Doll, Y. and Ikeuchi, M. (2025). All roads lead to dome: Multicellular dynamics during de novo meristem establishment in shoot regeneration. Curr. Opin. Plant Biol. 85, 102733. doi: 10.1016/j.pbi.2025.102733
Eshed Williams, L. (2021). Genetics of shoot meristem and shoot regeneration. Annu. Rev. Genet. 55, 661–681. doi: 10.1146/annurev-genet-071719-020439
Frederick, R. O., Haruta, M., Tonelli, M., Lee, W., Cornilescu, G., Cornilescu, C. C., et al. (2019). Function and solution structure of the Arabidopsis thaliana RALF8 peptide. Protein Sci. 28, 1115–1126. doi: 10.1002/pro.3628
García-Gómez, M. L. and Ten Tusscher, K. (2024). Multi-scale mechanisms driving root regeneration: From regeneration competence to tissue repatterning. Plant J. 119, 1183–1196. doi: 10.1111/tpj.16860
Glazunova, A., Hu, Z., Ji, X., Chen, K., Zhang, L., Duan, B., et al. (2025). The CLE9/CLE10 signaling peptides control de novo shoot regeneration in Arabidopsis. Plant Physiol. Biochem. 229, 110399. doi: 10.1016/j.plaphy.2025.110399
Goell, J. H. and Hilton, I. B. (2021). CRISPR/Cas-based epigenome editing: Advances, applications, and clinical utility. Trends Biotechnol. 39, 678–691. doi: 10.1016/j.tibtech.2020.10.012
Gruber, C. W., Cemazar, M., Clark, R. J., Horibe, T., Renda, R. F., Anderson, M. A., et al. (2007). A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 282, 20435–20446. doi: 10.1074/jbc.M700018200
Hander, T., Fernández-Fernández, Á. D., Kumpf, R. P., Willems, P., Schatowitz, H., Rombaut, D., et al. (2019). Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363, eaar7486.
Hao, Z., Wu, H., Zheng, R., Li, R., Zhu, Z., Chen, Y., et al. (2023). The plant peptide hormone phytosulfokine promotes somatic embryogenesis by maintaining redox homeostasis in Cunninghamia lanceolata. Plant J. 113, 716–733. doi: 10.1111/tpj.16077
Hirakawa, Y., Shinohara, H., Welke, K., Irle, S., Matsubayashi, Y., Torii, K. U., et al. (2017). Cryptic bioactivity capacitated by synthetic hybrid plant peptides. Nat. Commun. 8, 14318.
Ikeuchi, M., Iwase, A., Rymen, B., Harashima, H., Shibata, M., Ohnuma, M., et al. (2015). PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1, 15089.
Ince, Y.Ç and Sugimoto, K. (2023). Illuminating the path to shoot meristem regeneration: Molecular insights into reprogramming cells into stem cells. Curr. Opin. Plant Biol. 76, 102452. doi: 10.1016/j.pbi.2023.102452
Islam, M. K., Mummadi, S. T., Liu, S., and Wei, H. (2023). Regulation of regeneration in Arabidopsis thaliana. aBIOTECH 4, 332–351.
Ito, T., Otsuki, K., Fukuda, H., and Endo, S. (2025). Shoot Apex-derived CLE46 peptide signal spatiotemporally restricts root regeneration in Arabidopsis. Plant Cell Physiol. 66, 1184–1191. doi: 10.1093/pcp/pcaf065
Iwase, A., Harashima, H., Ikeuchi, M., Rymen, B., Ohnuma, M., Komaki, S., et al. (2017). WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in arabidopsis. Plant Cell 29, 54–69. doi: 10.1105/tpc.16.00623
Ji, C., Li, H., Zhang, Z., Peng, S., Liu, J., Zhou, Y., et al. (2025). The power of small signaling peptides in crop and horticultural plants. Crop J. 13, 656–667. doi: 10.1016/j.cj.2024.12.020
Joshi, K. and Wang, D. O. (2024). epidecodeR: a functional exploration tool for epigenetic and epitranscriptomic regulation. Brief Bioinform. 25, bbad521. doi: 10.1093/bib/bbad521
Kang, J., Wang, X., Ishida, T., Grienenberger, E., Zheng, Q., Wang, J., et al. (2022). A group of CLE peptides regulates de novo shoot regeneration in Arabidopsis thaliana. New Phytol. 235, 2300–2312. doi: 10.1111/nph.18291
Kim, J. W. and Seo, P. J. (2025). The early hormone signaling network underlying wound-induced de novo root regeneration. J. Exp. Bot. 76, 1996–2004. doi: 10.1093/jxb/erae422
Li, W., Liu, H., Cheng, Z. J., Su, Y. H., Han, H. N., Zhang, Y., et al. (2011). DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PloS Genet. 7, e1002243. doi: 10.1371/journal.pgen.1002243
Li, J., Zhang, Q., Wang, Z., and Liu, Q. (2024). The roles of epigenetic regulators in plant regeneration: Exploring patterns amidst complex conditions. Plant Physiol. 194, 2022–2038. doi: 10.1093/plphys/kiae042
Luo, Q., Hu, S., Deng, Z., Gu, Z., Liu, Q., Zhou, G., et al. (2024). Plant peptide hormone phytosulfokine promotes embryo development of mass in Pinus massoniana. Plant Cell Tiss. Organ. Cult. 158, 58. doi: 10.1007/s11240-024-02857-8
Martínez, M. and Corredoira, E. (2024). Recent advances in plant somatic embryogenesis: Where we stand and where to go? Int. J. Mol. Sci. 25, 8912. doi: 10.3390/ijms25168912
Matosevich, R., Cohen, I., Gil-Yarom, N., Modrego, A., Friedlander-Shani, L., Verna, C., et al. (2020). Local auxin biosynthesis is required for root regeneration after wounding. Nat. Plants 6, 1020–1030.
Mei, G., Chen, A., Wang, Y., Li, S., Wu, M., Hu, Y., et al. (2024). A simple and efficient in planta transformation method based on the active regeneration capacity of plants. Plant Commun. 5, 4. doi: 10.1016/j.xplc.2024.100822
Moussu, S., Broyart, C., Santos-Fernandez, G., Augustin, S., Wehrle, S., Grossniklaus, U., et al. (2020). Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc. Natl. Acad. Sci. U.S.A. 117, 7494–7503. doi: 10.1073/pnas.2000100117
Olsson, V., Joos, L., Zhu, S., Gevaert, K., Butenko, M. A., and De Smet, I. (2019). Look closely, the beautiful may be small: Precursor-derived peptides in plants. Annu. Rev. Plant Biol. 70, 153–186. doi: 10.1146/annurev-arplant-042817-040413
Pratyusha, D. S. and Sarada, D. V. L. (2025). Rapid Alkalinization Factor - A cryptide regulating developmental and stress responses. Plant Sci. 359, 112600. doi: 10.1016/j.plantsci.2025.112600
Rymen, B., Kawamura, A., Lambolez, A., Inagaki, S., Takebayashi, A., Iwase, A., et al. (2019). Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun. Biol. 2, 404. doi: 10.1038/s42003-019-0646-5
Santana, I., Wu, H., Hu, P., and Giraldo, J. P. (2020). Targeted delivery of nanomaterials with chemical cargoes in plants enabled by a biorecognition motif. Nat. Commun. 11, 2045. doi: 10.1038/s41467-020-15731-w
Selby, R. and Jones, D. S. (2023). Complex peptide hormone signaling in plant stem cells. Curr. Opin. Plant Biol. 75, 102442. doi: 10.1016/j.pbi.2023.102442
Shemer, O., Landau, U., Candela, H., Zemach, A., and Eshed Williams, L. (2015). Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci. 238, 251–261. doi: 10.1016/j.plantsci.2015.06.015
Shen, W., Liu, J., and Li, J. F. (2019). Type-II metacaspases mediate the processing of plant elicitor peptides in arabidopsis. Mol. Plant 12, 1524–1533. doi: 10.1016/j.molp.2019.08.003
Shen, Y., Xie, Q., Wang, T., Wang, X., Xu, F., Yan, Z., et al. (2025). RALF33-FERONIA signaling orchestrates post-wounding root-tip regeneration via TPR4-ERF115 dynamics. Plant Cell 37, koaf098. doi: 10.1093/plcell/koaf098
Shpak, E. D. and Uzair, M. (2025). WUSCHEL: The essential regulator of the Arabidopsis shoot Apical Meristem. Curr. Opin. Plant Biol. 85, 102739. doi: 10.1016/j.pbi.2025.102739
Song, X. F., Guo, P., Ren, S. C., Xu, T. T., and Liu, C. M. (2013). Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiol. 161, 1076–1085. doi: 10.1104/pp.112.211029
Stührwohldt, N., Ehinger, A., Thellmann, K., and Schaller, A. (2020a). Processing and formation of bioactive CLE40 peptide are controlled by posttranslational Proline hydroxylation. Plant Physiol. 184, 1573–1584. doi: 10.1104/pp.20.00528
Stührwohldt, N. and Schaller, A. (2019). Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol. 21, 49–63. doi: 10.1111/plb.12881
Stührwohldt, N., Scholl, S., Lang, L., Katzenberger, J., Schumacher, K., and Schaller, A. (2020b). The biogenesis of CLEL peptides involves several processing events in consecutive compartments of the secretory pathway. Elife 9, e55580. doi: 10.7554/eLife.55580
van de Sande, J., Streefkerk, D., Immink, R., Fiers, M., and Albada, B. (2024). Phytosulfokine peptide library: Chemical synthesis and biological evaluation on protoplast regeneration. New J. Chem. 48, 8055–8063. doi: 10.1039/D3NJ05996K
Wang, P., Si, H., Li, C., Xu, Z., Guo, H., Jin, S., et al. (2025). Plant genetic transformation: achievements, current status and future prospects. Plant Biotechnol. J. 23, 2034–2058. doi: 10.1111/pbi.70028
Wang, G., Zhang, Y., Li, C., Wang, X., and Fletcher, J. C. (2022). Signaling peptides direct the art of rebirth. Trends Plant Sci. 27, 516–519. doi: 10.1016/j.tplants.2022.03.009
Wu, Z., Li, Y., Zhang, L., Ding, Z., and Shi, G. (2021). Microbial production of small peptide: pathway engineering and synthetic biology. Microb. Biotechnol. 14, 2257–2278. doi: 10.1111/1751-7915.13743
Wu, H., Zheng, R., Hao, Z., Meng, Y., Weng, Y., Zhou, X., et al. (2019). Cunninghamia lanceolata PSK Peptide Hormone Genes Promote Primary Root Growth and Adventitious Root Formation. Plants 8, 520.
Xiao, F., Zhou, H., and Lin, H. (2025). Decoding small peptides: Regulators of plant growth and stress resilience. J. Integr. Plant Biol. 67, 596–631. doi: 10.1111/jipb.13873
Xie, H., Zhao, W., Li, W., Zhang, Y., Hajný, J., and Han, H. (2022). Small signaling peptides mediate plant adaptions to abiotic environmental stress. Planta 255, 72. doi: 10.1007/s00425-022-03859-6
Xu, D. and Yang, L. (2025). Regeneration and defense: unveiling the molecular interplay in plants. New Phytol. 246, 2484–2494. doi: 10.1111/nph.70171
Yang, W., Zhai, H., Wu, F., Deng, L., Chao, Y., Meng, X., et al. (2024). Peptide REF1 is a local wound signal promoting plant regeneration. Cell 187, 3024–3038.e14. doi: 10.1016/j.cell.2024.04.040
Youngstrom, C., Wang, K., and Lee, K. (2025). Unlocking regeneration potential: harnessing morphogenic regulators and small peptides for enhanced plant engineering. Plant J. 121, e17193. doi: 10.1111/tpj.17193
Zhai, N. and Xu, L. (2021). Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants 7, 1453–1460. doi: 10.1038/s41477-021-01015-8
Zhang, T. Q., Lian, H., Zhou, C. M., Xu, L., Jiao, Y., and Wang, J. W. (2017). A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29, 1073–1087. doi: 10.1105/tpc.16.00863
Zhang, Q., Liu, Y., Yang, G., Kong, H., Guo, L., and Wei, G. (2023). Recent advances in protein hydrogels: From design, structural and functional regulations to healthcare applications. Chem. Eng. J. 451, 138494. doi: 10.1016/j.cej.2022.138494
Zhang, C., Wu, Y., Liu, J., Song, B., Yu, Z., Li, J. F., et al. (2025). SUMOylation controls peptide processing to generate damage-associated molecular patterns in Arabidopsis. Dev. Cell 60, 696–705.e4. doi: 10.1016/j.devcel.2024.11.010
Keywords: CLE peptide, REF1 peptide, RLAF33, PSK, PORK1, BAM, plant regeneration
Citation: Wang M, Lu R, Zhang Z, Li L, Chen Y and Han H (2025) Unlocking plant regeneration capacity: small signaling peptides are on aboard. Front. Plant Sci. 16:1679487. doi: 10.3389/fpls.2025.1679487
Received: 04 August 2025; Accepted: 24 September 2025;
Published: 13 October 2025.
Edited by:
Vishal Tripathi, Graphic Era University, IndiaReviewed by:
Noor Alam Chowdhary, Banaras Hindu University, IndiaCopyright © 2025 Wang, Lu, Zhang, Li, Chen and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meixia Wang, bWVpeGlhd2FuZ0BqeGF1LmVkdS5jbg==
 Meixia Wang
Meixia Wang Rong Lu2
Rong Lu2 Huibin Han
Huibin Han