- Laboratory of Genetics, University of Wisconsin – Madison, Madison, WI, United States
Introduction: A classical forward genetic screen for Arabidopsis mutants with altered morphology identified a pleiotropic mutant, orbiculata (orb1), that has phenotypes including rounded leaves, chlorosis, and reduced growth. orb1 mapped to one of the Arabidopsis genes that encodes glutamate synthase, fd-gogat1 (ferredoxin-dependent glutamine oxoglutarate aminotransferase or Fd-GOGAT).
Methods: To discover why this glutamate synthase impacts development, we conducted a forward genetic screen for suppressors of orb1. In the primary mutagenized generation, we identified a dominant mutant, which we call Lettuce, that rescues orb1 chlorosis but causes new pleiotropic defects that closely resemble the classical Arabidopsis cabbage and dwarf mutants that are defective in brassinosteroid or gibberellin signaling. Here, we take a chemical genetic approach to phenocopy Lettuce and investigate how gibberellins and brassinosteroids impact the development and physiology of fd-gogat1.
Results: We found that inhibiting brassinosteroid synthesis significantly increases chlorophyll content in fd-gogat1, which is chlorotic due to defects in the photorespiratory pathway.
Discussion: This discovery highlights how crosstalk among phytohormones (brassinosteroids) and core metabolic processes (amino acid biosynthesis and photorespiration) converge to regulate plant development and physiology.
1 Introduction
Plants are photoautotrophs that can synthesize all twenty proteinogenic amino acids from inorganic precursors, primarily CO2 assimilated by RuBisCO, NH4+ assimilated by glutamine synthetase, and SO42- assimilated by O-acetylserine thiollyase. The enzymes responsible for amino acid metabolism in plants have been elucidated over several decades using a combination of biochemical and genetic approaches. Amino acid synthesis in plants is deeply intertwined with other metabolic pathways, however, which complicates genetic analysis due to pleiotropic effects of disrupting the genes that encode enzymes involved in amino acid metabolism. Moreover, many enzymes are encoded by several paralogues in plant genomes that may be semi-redundant or may play specialized, subfunctionalized roles in metabolism (Maeda, 2019).
Illustrating this complexity, in the model plant Arabidopsis thaliana, there are three bona fide glutamate synthases each encoded by their own genes: FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1 (Fd-GOGAT1), Fd-GOGAT2, and NICOTINAMIDE ADENINE DINUCLEOTIDE-DEPENDENT GLUTAMATE SYNTHASE 1 (NADH-GOGAT1). There are an additional three genes that encode semi-redundant GLUTAMATE DEHYDROGENASE (GDH) enzymes, which are biochemically capable of synthesizing glutamate from α-ketoglutarate (αKG) and ammonium in vitro. In vivo, however, GDHs are understood to primarily catalyze the reverse reaction, deaminating glutamate to αKG to support the tricarboxylic acid cycle, and GDHs are therefore not typically involved in glutamate synthesis (Fontaine et al., 2012). GOGATs work intimately with glutamine synthetase (GS) to assimilate nitrogen in the GS-GOGAT cycle: GOGAT makes glutamate, which GS condenses with ammonia to yield glutamine (Figure 1). Fd-GOGAT2 and NADH-GOGAT1 are highly expressed in roots, where they drive nitrogen assimilation from the soil in the GS/GOGAT cycle (Lancien et al., 2002). Fd-GOGAT1 is instead highly expressed in leaves, where it plays a critical role in reassimilating carbon and nitrogen that are lost during photorespiration (Coschigano et al., 1998).
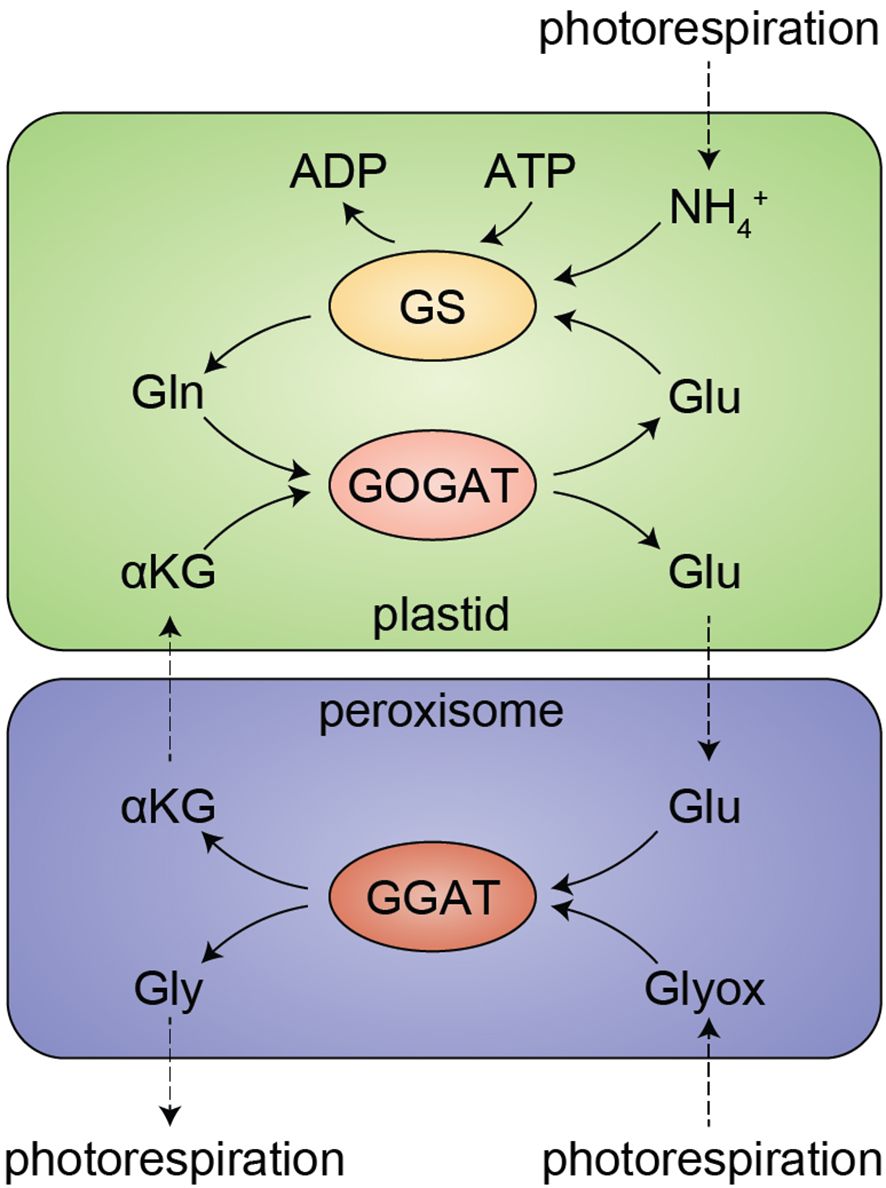
Figure 1. The GS/GOGAT cycle and photorespiration in leaves. Ammonium (NH4+), a byproduct of photorespiration, is salvaged by glutamine synthase (GS) in an ATP-dependent reaction to synthesize glutamine (Gln) from glutamate (Glu). Gln is then condensed with α-ketoglutarate (αKG, 2-oxoglutarate, or 2-OG) by glutamine oxoglutarate aminotransferase (GOGAT), yielding two Glu. In fd-gogat1 mutants lacking this recycling pathway, Glu levels in leaves are depleted within minutes during the day (Somerville and Ogren, 1980), leading to toxic overaccumulation of NH4+. Moreover, as part of the salvage pathway that rescues carbon during photorespiration, Glu is directly consumed in the peroxisome, where glutamate:glyoxylate aminotransferase (GGAT) converts Glu and glyoxylate (Glyox) to αKG and glycine (Gly). Thus, the GS/GOGAT cycle is required to rescue both nitrogen and carbon under photorespiratory conditions.
Photorespiration occurs in normal air conditions when RuBisCO oxygenates ribulose bisphosphate, yielding the waste product 2-phosphoglycolate (2-PG), instead of carboxylating ribulose bisphosphate, yielding the sugar precursor 3-phosphoglycerate (3-PG). The carbons “wasted” to 2-PG are recovered through photorespiration, a complicated metabolic process that requires glutamate, alanine, various other metabolites, and over a dozen enzymes that are spread across the cell in the cytosol, plastids, perxoisomes, and mitochondria (Bauwe et al., 2010; Peterhansel et al., 2010; Eisenhut et al., 2019; Timm and Hagemann, 2020). Photorespiration yields 3-PG and also releases NH3, which is then re-assimilated by the GS-GOGAT cycle. fd-gogat1 mutants accumulate toxic levels of NH3 in leaves and are unable to sustain photosynthesis, resulting in smaller, yellow plants when grown in standard conditions.
Arabidopsis first came to prominence among modern molecular biologists through the forward genetic screens that established the mechanisms of photorespiratory metabolism (Somerville and Ogren, 1980, p. 198; Somerville, 2001), but Arabidopsis already had a long history of investigation by developmental geneticists who researched how genes can influence leaf shape (Micol, 2009). Although leaf shape mutant phenotypes are often caused by disruptions in regulatory genes that encode, e.g., transcription factors that drive patterning or phytohormone signaling pathways (Moon and Hake, 2011), a surprising number of classical leaf shape mutants mapped to genes involved in primary metabolism or ribosome biogenesis (Martinez et al., 2025). For example, orbiculata 1 (orb1), a mutant with dramatically rounder leaves than wild-type siblings, is caused by loss of fd-gogat1 (Muñoz-Nortes et al., 2017).
Here, we set out to investigate how disrupting FD-GOGAT1 causes the orb1 leaf shape phenotype by conducting a forward genetic screen for orb1 suppressors. Unexpectedly, we discovered a strong, dominant, and ultimately lethal suppressor of the fd-gogat1 chlorotic phenotype in the mutagen-treated M1 population. Based on phenotypic comparison of this suppressor to well-studied Arabidopsis mutants, we explored how two major phytohormones, gibberellins (gibberellic acids, GAs) and brassinosteroids (BRs), impact the development and physiology of fd-gogat1 mutants. We show that inhibiting BR biosynthesis partially restores chlorophyll levels in fd-gogat1 plants, demonstrating how amino acid metabolism in plants intersects with other metabolic networks (such as photorespiration) and phytohormone signaling to determine physiological and developmental outcomes.
2 Materials and methods
2.1 Plant materials and growth conditions
Unless otherwise stated, plants were grown under standard conditions with 16 h day/8 h night at ~120 µE/m2s, 23°C, and 50% humidity. The fd-gogat1 line, SALK_011035C (previously called orb1-4 (Muñoz-Nortes et al., 2017)), and the Col-0 (wild-type) line were obtained from the Arabidopsis Biological Resource Center.
2.2 Forward genetic fd-gogat1 suppressor screen
Mutagenesis of fd-gogat1 seeds was carried out as previously described (Gillmor and Lukowitz, 2020). 100 mg fd-gogat1 seeds (~5,000 seeds) were weighed and then washed with 0.01% Tween 20 (VWR 97063-872) in Milli-Q H2O for 15 minutes. Tween 20 solution was removed and seeds were washed with Milli-Q H2O four times until no more bubbles formed. 40 mL of Milli-Q H2O were added to the seeds, which were then placed at 4°C on a rocker for gentle agitation overnight. The next day, Milli-Q H2O was removed from seeds. 1.2% Ethyl methanesulfonate (EMS) (Sigma-Aldrich M0880-5G) solution was prepared and poured into pre-treated seeds. Seeds were next placed on a rocker at room temperature to incubate for three hours under gentle agitation, ensuring full coverage of seeds by EMS solution. Seeds were washed 3 times in the fume hood with Milli-Q H2O and an additional 10 washes with tap water were performed outside of the hood over the span of 1 hour. Seeds were then suspended in 40 mL of a dilute agar slurry (0.4% and 0.1% agar solutions mixed in a 50/50 ratio) and left at room temperature overnight. 1 mL of seed slurry was drizzled on top of wet potted soil to get about 100 seeds planted per pot. Seeds were grown in a growth chamber under long day 16-hour light/8-hour dark conditions with a light intensity of 120 µE/m2s, at 23°C and 50% humidity. Plants were observed and screened over time for phenotypes to indicate EMS was successful (Figures 2).

Figure 2. Lettuce suppresses fd-gogat1. (A) Lettuce was discovered in the EMS-mutagenized M1 generation during a screen for genetic suppressors of fd-gogat1. Lettuce is shown here alongside its fd-gogat1 siblings in the M1 generation. (B) Lettuce continued to grow and eventually flowered weeks after fd-gogat1 siblings had completed their life cycle. Lettuce flowers were stunted, did not produce any pollen, and could not be fertilized by Col-0 or fd-gogat1 pollen. (C) Comparison of Col-0, fd-gogat1, and the double mutant Lettuce;fd-gogat1 rosettes.
2.3 Phytohormone and chemical inhibitor treatments
Seeds from Arabidopsis thaliana mutant fd-gogat1 (obtained from the Arabidopsis Biological Resource Center, line SALK_011035C) and Col-0 were sterilized using 30% bleach and 0.1% tween for 15 minutes and washed with sterile Milli-Q H2O 5 times to remove any excess bleach solution. Seeds were stratified at 4°C in 1 mL sterile Milli-Q H2O for 48 hours. Seeds were plated on ½ MS (RPI M10200-50.0) with 1% sucrose (RPI S24065-5000.0) and 0.8% agar (Fisher BioReagents BP9744-5). In addition to mock-treated “control” plates, seeds were sown on plates with gibberellic acid 3 (GA) (Dot Scientific DSG32020-5) or paclobutrazol (PBZ) (TCI America P2299) at 10 µM and 30 µM concentrations, propiconazole (PCZ) (Cayman Chemical 18853) at 1 µM and 5 µM concentrations, brassinazole (BRZ) (TCI America B2829) at 1 µM, 5 µM, or 10 µM concentrations, or brassinolide (BL) (Cayman Chemical 21594) at 0.5 µM or 1 µM concentrations. Plates were sealed with micropore tape and placed in growth chambers under a long day 16-hour light/8-hour dark cycle with a light intensity of 150-180 µmol/m2s for 14 or 20 days.
2.4 Chlorophyll quantifications
Seedlings were pooled, weighed (~100 mg of tissue per pool), and placed into tubes with 3 steel beads, 8 plants per pool for mock-treated and GA plates or 16 plants per pool for BL, BRZ, and PCZ plates, and flash frozen in liquid nitrogen. Plant tissue was ground using a homogenizer at 1,500 rpm for 1 minute. Chlorophyll was extracted by washing ground tissue with 100% acetone, vortexing for 30 seconds, centrifuging at 10,000 x g for 1 min, and then removing and saving the supernatant. This process was repeated 5 times to extract all of the chlorophyll from each sample. 50 µL extract was diluted in 800 µL ice cold 80% acetone solution and centrifuged for 5 min at 14,000 x g. UV-vis spectrometry was conducted on each sample using wavelengths of 647 nm, 664 nm, and 750 nm.
2.5 Growth measurements and data analysis
Plates were photographed beside a ruler for scaling under laboratory light on bench top. Rosette radius was then measured in ImageJ. Leaf number was counted at the indicated time, excluding cotyledons (which developed prior to treatments). Total chlorophyll concentrations and chlorophyll a/b ratios were calculated using the standard formula (Porra et al., 1989). Data were analyzed with R software using Student’s t-test for chlorophyll comparisons or ANOVA with Fisher’s exact test for rosette radius. Statistically-significant groupings indicated in Figures 3 and 4 were determined using Tukey’s HSD test with confidence level 0.95. Plant figures were made by removing the background and replacing with a black background. The brightness and contrast of the plants were uniformly adjusted across all images, with no other modifications. Scales were set based on a ruler in each picture.
![Plant growth and chlorophyll content comparison in Col-0 and fd-gogat1 genotypes. Panel A shows rosette images under mock, 10 µM, and 30 µM [GA] conditions; Panel D shows rosettes under [PBZ]. Panels B and E display box plots of rosette radius, while C and F present chlorophyll content. Col-0 (green) and fd-gogat1 (yellow) are distinguished, with significant differences marked by letters.](https://www.frontiersin.org/files/Articles/1680431/fpls-16-1680431-HTML/image_m/fpls-16-1680431-g003.jpg)
Figure 3. Inhibiting GA synthesis only moderately rescues fd-gogat1 chlorosis. (A) Representative images of Col-0 and fd-gogat1 seedlings grown on plates with indicated GA concentrations. (B) GA somewhat reduced Col-0 rosette diameter and had minimal effect on fd-gogat1 rosette diameter. (C) GA significantly reduced chlorophyll levels in both Col-0 or fd-gogat1. (D) Representative images of Col-0 and fd-gogat1 seedlings grown on plates with indicated PBZ concentrations. (E) PBZ significantly reduced rosette diameter in both genotypes, with a more pronounced effect on Col-0. (F) 30 µM PBZ decreased chlorophyll in Col-0 and significantly increased chlorophyll levels in fd-gogat1, although the effect on fd-gogat1 was very slight. Letters indicate significance groups as determined by Tukey’s HSD test, p < 0.05.
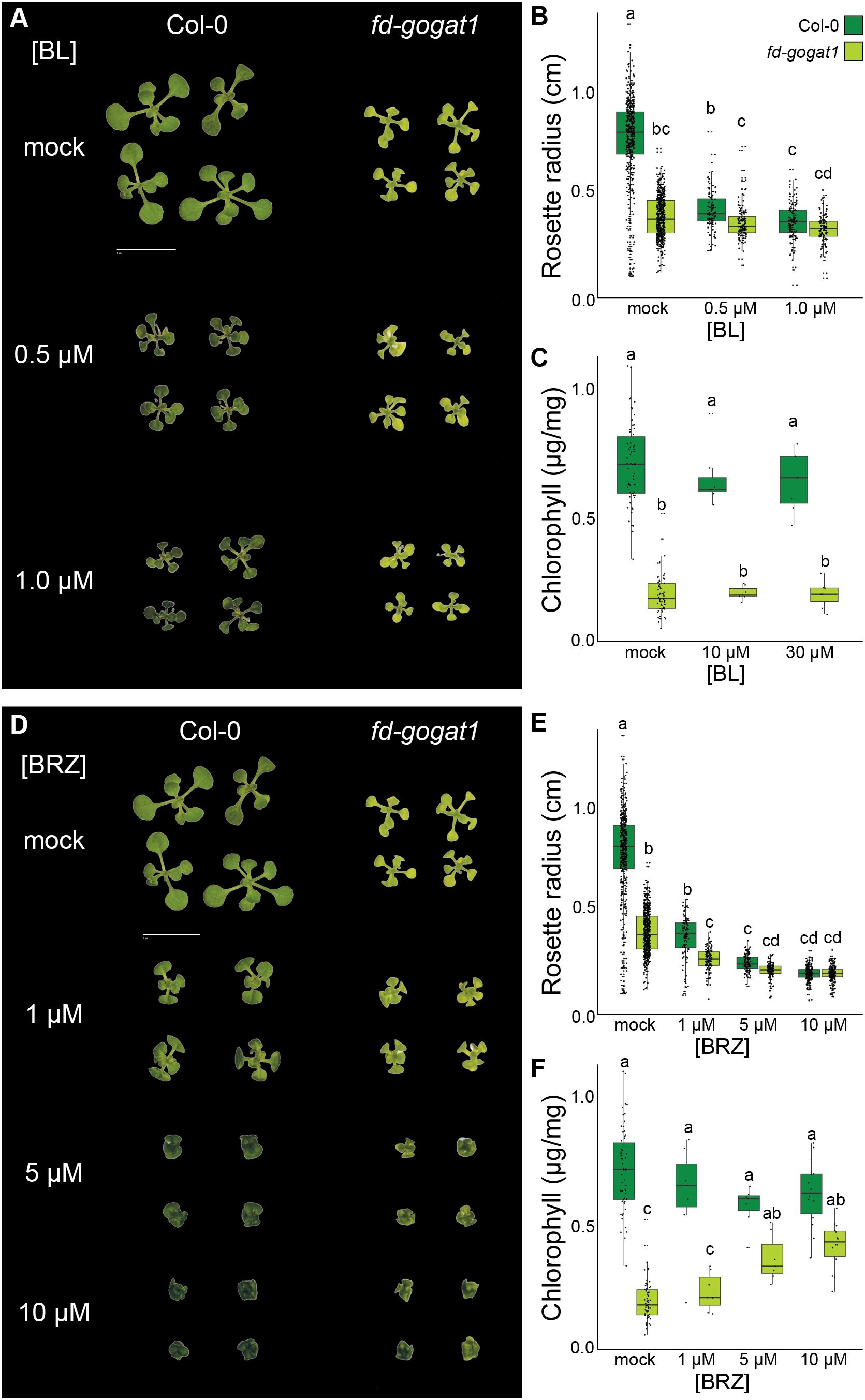
Figure 4. Inhibiting BR synthesis phenocopies Lettuce and partially rescues fd-gogat1 chlorosis. (A) Representative images of Col-0 and fd-gogat1 seedlings grown on plates with indicated BL concentrations. (B) BL significantly reduced Col-0 rosette diameter but had no substantial effect on fd-gogat1. (C) BL had not substantial effect on chlorophyll levels in Col-0 or fd-gogat1. (D) Representative images of Col-0 and fd-gogat1 seedlings grown on plates with indicated BRZ concentrations. fd-gogat1 mutants treated with 5 µM to 10 µM BRZ closely resembled the Lettuce suppressor mutant. (E) BRZ significantly reduced rosette diameter in both genotypes, with a more pronounced effect on Col-0. (F) BRZ slightly decreased chlorophyll in Col-0 but significantly increased chlorophyll levels in fd-gogat1. Letters indicate significance groups as determined by Tukey’s HSD test, p < 0.05.
3 Results
3.1 Lettuce suppresses fd-gogat1 chlorosis
To identify genetic mechanisms that drive the pleiotropic orbiculata syndrome in fd-gogat1 mutants, we conducted a forward genetic suppressor screen. fd-gogat1 seeds were mutagenized with ethyl methanesulfonate (EMS), which causes G/C to A/T transitions (Greene et al., 2003). The point mutations introduced by EMS can have diverse effects on gene function, including (but not limited to) missense mutations, nonsense mutations, or mutation of regulatory features (like splice sites) that broadly disrupt gene function. To validate that the EMS mutagenesis was successful, we screened the M1 generation for mosaic leaf color phenotypes; to ensure an efficient Arabidopsis genetic screen, we expect to observe as many as ~1% of plants with mutant sectors that have yellow or white leaves (Maple and Møller, 2007). During this analysis, we made a surprising discovery: a single plant with dramatically altered phenotypes from its siblings, which we named Lettuce (Figure 2A).
Lettuce was small, compact, and bright green, with curling leaves, slowed shoot development, multiple inflorescence stems, and delayed flowering compared to its fd-gogat1 siblings (Figure 2B). None of the bright green leaves of Lettuce yellowed or senesced after flowering, unlike Col-0 and fd-gogat1 plants. We confirmed that Lettuce was homozygous for the SALK_011035 T-DNA insertion in fd-gogat1 and was therefore a true suppressor and not a contaminant. Lettuce was sterile: flowers produced no pollen and could not be fertilized with pollen from other plants, which prevented us from propagating the genotype for deeper phenotypic analysis and genetic mapping. Since Lettuce was discovered in the M1 generation, it is almost certainly caused by a dominant mutation, which are less frequently encountered in genetic screens but can be powerful tools for discovery, especially when a dominant allele is also epistatic to redundant paralogues.
Since we could not map Lettuce to a causal mutation, we instead probed the literature for similar mutant phenotypes that could illuminate how Lettuce suppresses the chlorotic phenotype in fd-gogat1. Although there are many possible candidates, we noted that Lettuce is remarkably similar to mutants defective in GA and BR signaling. For example, Arabidopsis gid1a;gid1b;gid1c mutants lacking all three paralogues of GIBBERELLIN INSENSITIVE DWARF 1 (GID1), which encode the GA receptors (Murase et al., 2008), are extremely small, slow-growing, dark green, and infertile (Griffiths et al., 2006). Comparable phenotypes are also observed in the Arabidopsis ga20ox1;ga20ox2;ga20ox3 (Plackett et al., 2012) mutants that lack most of the GIBBERELLIN 20-OXIDASES that are required for synthesis of bioactive GA. Lettuce is also strikingly reminiscent of the cabbage (cbb) mutants (Kauschmann et al., 1996), which were shown to encode the BR receptor (cbb2 is an allele of BRASSINOSTEROID INSENSITIVE 1; other alleles are called bri1, bin1, and dwarf2) (Clouse et al., 1996; Li and Chory, 1997; Choe et al., 1998; Wang et al., 2001; Nam and Li, 2002; Hothorn et al., 2011; She et al., 2011) and enzymes involved in BR synthesis (cbb1, also called dwarf1 and diminuto, encodes a sterol C-24 reductase; cbb3, also called dwarf3 and constitutive photomorphogenic dwarf, encodes a cytochrome P450 enzyme, CYP90A) (Szekeres et al., 1996; Klahre et al., 1998; Choe et al., 1999a; Ohnishi et al., 2012). Lettuce similarly resembled other mutants defective in BR signaling, including the semidominant mutant bin2 (brassinosteroid insensitive 2, also called dwarf12) that constitutively represses BR responses (Li et al., 2001; Choe et al., 2002), and other mutants defective in BR synthesis, such as dwarf4 (which encodes CYP90B1) (Choe et al., 1998; Fujita et al., 2006), dwarf5 (Choe et al., 2000), det2 (de-etiolated 2, also called dwarf6) (Chory et al., 1991; Noguchi et al., 1999), and dwarf7 (also called sterol1 and boule1) (Gachotte et al., 1996; Choe et al., 1999b; Catterou et al., 2001). Inspired by the close similarities between these mutants and Lettuce, we speculated that disrupting GA and/or BR synthesis might be sufficient to phenocopy Lettuce and increase chlorophyll levels in fd-gogat1 mutants.
3.2 Inhibiting GA biosynthesis slightly rescues chlorophyll levels in fd-gogat1
Building on our observation that the Lettuce suppressor of fd-gogat1 resembles mutants defective in GA signaling and biosynthesis, we tested how treating plants with GA or paclobutrazol (PBZ), a selective inhibitor of GA biosynthesis (Hedden and Graebe, 1985), impacts fd-gogat1 growth, development, and physiology. We grew fd-gogat1 and Col-0 plants on 0.8% agar plates with ½ × Murashige & Skoog (MS) nutrients and 1% sucrose, plus GA, PBZ, or mock treatment. 20 days after germination, seedlings were photographed to measure rosette diameter and seedlings of each genotype from different plates were pooled for chlorophyll extraction and quantification (Figure 3). GA and PBZ were both supplied at either 10 µM or 30 µM, which are common effective concentrations for these hormones; in trial experiments, we tested lower concentrations and observed no substantial effects.
As expected, fd-gogat1 mutants were significantly smaller (p < 10-15, n ≥ 77) with significantly less chlorophyll (p < 0.01, n ≥ 4) than Col-0 plants (Figure 3). When treated with 10 µM GA, however, the rosette diameter was indistinguishable between the two genotypes (p = 0.50, n ≥ 78), because 10 µM GA increased fd-gogat1 rosette diameter but had the opposite effect on Col-0 plants (Figure 3B). 30 µM GA slightly reduced the rosette diameter of both genotypes compared to treatment with 10 µM GA, but fd-gogat1 mutants were significantly larger with this treatment than wild-type plants (p < 0.01, n ≥ 77) (Figure 3B). The GA biosynthesis inhibitor, PBZ, decreased rosette diameter in both genotypes (p < 10-11, n ≥ 40), with a marginally stronger effect at 30 µM (Figure 3E).
Whereas GA had opposite effects on rosette diameter for the two genotypes, GA reduced chlorophyll levels in both fd-gogat1 and Col-0 (Figure 3C). In contrast, PBZ slightly but significantly increased chlorophyll levels in fd-gogat1 (p < 0.01, n ≥ 5 pools of seedlings) (Figure 3F), supporting the hypothesis that disrupting GA biosynthesis could partially suppress fd-gogat1 chlorosis. PBZ also increased chlorophyll levels in Col-0 plants when treated with the lower concentration of 10 µM PBZ (p = 0.04, n ≥ 4), but treatment with 30 µM PBZ drastically reduced Col-0 chlorophyll levels (p < 10-6, n ≥ 4) (Figure 3F).
Overall, these experiments validated that a chemical genetic approach can replicate mutant analysis, since PBZ-treated Col-0 plants closely resembled the phenotypes of previously-studied GA signaling mutants, such as gid1a;gid1b;gid1c and ga20ox1;ga20ox2;ga20ox3. PBZ-treated fd-gogat1 mutants, however, did not closely resemble the Lettuce suppressor, suggesting that a disruption to GA biosynthesis or signaling is unlikely to be the cause of Lettuce phenotypes. PBZ did increase chlorophyll in fd-gogat1 mutants, hinting at possible crosstalk between GA biosynthesis and photorespiration, but the effect was very minor compared to the bright green phenotype of Lettuce.
3.3 Inhibiting brassinosteroid biosynthesis phenocopies the lettuce suppressor of fd-gogat1
Next, based on the similarity of Lettuce phenotypes to the cabbage and dwarf BR biosynthesis mutants, we tested how treating plants with the biologically-active BR brassinolide (BL) or the highly selective BR biosynthesis inhibitor brassinazole (BRZ) (Asami et al., 2000) impacted fd-gogat1 phenotypes compared to Col-0 and mock-treated controls. Again, we grew fd-gogat1 and Col-0 plants on 0.8% agar plates with ½ × MS nutrients and 1% sucrose, plus BL, BRZ, or mock treatment. 13 days after germination, seedlings were photographed to measure rosette diameter and then ~15 seedlings from each plate were pooled for chlorophyll extraction and quantification (Figure 4). BL was supplied at 0.5 µM or 1.0 µM and BRZ was supplied at 1.0 µM, 5.0 µM, or 10 µM, which are commonly used dose ranges for these chemicals.
Under these experimental conditions, BL significantly reduced rosette diameter for both genotypes (p < 0.01, n ≥ 115), although the effect was more pronounced for Col-0 than for fd-gogat1 mutants (Figure 4B). BL had no significant effect on chlorophyll accumulation in either genotype, however (p > 0.20, n ≥ 7 pools of seedlings) (Figure 4C). Inhibiting BR biosynthesis with the selective inhibitor BRZ caused similar phenotypes in both Col-0 and fd-gogat1 that very closely resembled the Lettuce mutant (Figure 4D). BRZ significantly reduced rosette diameter in both genotypes (p < 10-10, n ≥ 110) (Figure 4E). The higher concentrations of BRZ (5 µM and 10 µM) also mildly reduced chlorophyll levels in Col-0 plants (p < 0.05, n ≥ 7 pools of seedlings) (Figure 4F). Oppositely, these concentrations of BRZ dramatically increased chlorophyll levels in fd-gogat1 (p < 0.01, n ≥ 7 pools of seedlings), an effect that was strikingly similar to the Lettuce mutant (Figure 4F).
To validate and confirm these findings, we conducted an additional experiment with propiconazole (PCZ), a broad cytochrome P450 inhibitor that is thought to primarily interfere with BR biosynthesis in plants (Hartwig et al., 2012). Col-0 and fd-gogat1 seedlings were grown for 20 days on 0.8% agar plates with ½ × MS nutrients and 1% sucrose, supplemented with either 1.0 µM PCZ, 5.0 µM PCZ, or mock controls. 5.0 µM PCZ was sufficient to significantly increase chlorophyll levels 1.8-fold in fd-gogat1 (p = 0.02, n ≥ 5 pools of seedlings), with the opposite effect on Col-0 plants, significantly decreasing chlorophyll levels 2.0-fold (p < 0.01, n ≥ 5 pools of seedlings). This supported the hypothesis that inhibiting BR biosynthesis partially rescues chlorophyll accumulation in the fd-gogat1 photorespiration mutant.
4 Discussion
Photorespiration is responsible for major metabolic inefficiencies in plants, cutting net photosynthetic efficiency by ~50% (Zhu et al., 2008) and reducing many crop yields by ~20-40% (Walker et al., 2016). To overcome these losses, some photosynthetic lineages evolved carbon-concentrating mechanisms that reduce photorespiration by isolating RuBisCO in high CO2/low O2 environments, such as C4 photosynthesis found in several plant lineages, including the major crops maize, sorghum, and sugarcane (Sage, 2004; Kellogg, 2013; Schlüter and Weber, 2020); CAM photosynthesis, also found in several plant lineages, including many agave, pineapple, and cacti (Bräutigam et al., 2017); and pyrenoids in algae (He et al., 2023). Restricting or bypassing photorespiration, inspired by these evolutionary examples, is a promising target for breeders and synthetic biologists seeking to establish the resilient, high-yielding crops we will need for a sustainable agricultural future (Walker et al., 2016; Springmann et al., 2018; Bailey-Serres et al., 2019; Eisenhut et al., 2019; South et al., 2019; Lutt and Brunkard, 2022; Meacham-Hensold et al., 2024; Hadjikakou et al., 2025).
As demonstrated with the fd-gogat1 mutant, however, photorespiration is deeply intertwined with manifold core metabolic and developmental pathways, including amino acid biosynthesis, phytohormone signaling, redox homeostasis, and even leaf patterning. A deeper understanding of the crosstalk among these interconnected processes will be needed to guide efforts to engineer photorespiration bypasses in crops. Here, we showed that inhibiting brassinosteroid synthesis can rescue the chlorotic fd-gogat1 phenotype at the cost of reducing plant size and fertility. This trade-off could be mitigated by modulating BR signaling in specific cell types or in response to specific cues, rather than broadly inhibiting BR biosynthesis. For instance, ubiquitously overexpressing the BR response transcription factor, BRASSINAZOLE RESISTANT 1 (BZR1), drastically reduces fertility and photosynthetic efficiency, but overexpressing BZR1 exclusively in bundle sheath cells increases chloroplast area without negative trade-offs (Cackett et al., 2025).
Forward genetic screens in Arabidopsis for mutants that are visibly unhealthy in low CO2 environments but healthy in high CO2 environments (Somerville and Ogren, 1980) were among the first demonstrations that Arabidopsis genetics could be leveraged to resolve fundamental questions about plant physiology and biochemistry (Somerville, 2001), paving the way for the burst in Arabidopsis research in the 1990s. Despite >40 years of extensive research on these mutants, however, we still do not have a unified mechanistic understanding of why mutants defective in photorespiration exhibit such diverse phenotypes, ranging from mild growth defects to complete lethality (Timm and Bauwe, 2013). These phenotypes are not only suppressed by growing plants in high CO2 environments or, as we have shown here for the chlorotic phenotype of fd-gogat1, by inhibiting brassinosteroid synthesis, but sometimes by other conditions, including fluctuating light environments (von Bismarck et al., 2023). Whereas the suppression of photorespiration by high CO2 environments is easily explained, the suppressive effects of other treatments is less obvious; inhibiting BR synthesis, for example, might be directly regulating expression of chlorophyll biosynthesis and photosynthesis-associated genes or indirectly impacting fd-gogat1 photorespiration phenotypes through more complex effects on, e.g., nitrogen uptake, recycling, and metabolism, which are known to be sensitive to BR (Wang et al., 2019; Xing et al., 2022; Yadav et al., 2023; Yang et al., 2024). A combination of genetic and physiological approaches to unravel the functional roles of photorespiration in metabolism (Timm et al., 2024) and the signaling pathways that contribute to photorespiratory mutant phenotypes will be needed to build strong predictive models of how changing environmental CO2/O2 levels will impact plant health and agricultural yields.
fd-gogat1 is not only defective in photorespiration, but also in amino acid metabolism due to its role in glutamate synthesis from glutamine and 2-oxoglutarate. Across all eukaryotes, amino acid metabolism is monitored by the TARGET OF RAPAMYCIN (TOR), a serine/threonine kinase that is activated when conditions are favorable (Valvezan and Manning, 2019; Brunkard, 2020). TOR activity coordinates metabolism with nutrient availability by, among other mechanisms, driving ribosome biogenesis and protein synthesis when amino acids, nucleotides, and ATP are abundant in cells (Xiong et al., 2013; Scarpin et al., 2020, 2022; Busche et al., 2021). In mammals, TOR is particularly sensitive to the levels of essential amino acids that heterotrophs rely on consuming in their diets, especially leucine, arginine, and methionine (Jewell and Guan, 2013; Lutt and Brunkard, 2022). Several molecular sensors and mediating signal transduction pathways upstream of TOR have been identified in mammals and yeast (Goul et al., 2023), but these sensors and mediators are not conserved to plants (Brunkard, 2020). A handful of studies have demonstrated that TOR does sense amino acids in plants (Cao et al., 2019; Liu et al., 2021), but the molecular mechanisms have not yet been defined. Here, we illustrated a major challenge for biologists seeking to understand how plants sense and respond to amino acids: mutating one of the genes responsible for glutamate synthesis causes broad, unintended side effects. Adding to this complexity, the plant TOR network has evolved to interact various plant-specific signaling networks, including phytohormones like BR (Zhang et al., 2016; Liao et al., 2023) and metabolite transport via plasmodesmata (Brunkard et al., 2020). Establishing growth conditions, genetic approaches, or other methods to disentangle amino acid synthesis from other metabolic pathways and signaling networks will be needed to eventually elucidate how TOR monitors amino acid levels in plants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
KK: Writing – review & editing, Writing – original draft. AF: Writing – review & editing, Writing – original draft. SM: Writing – review & editing. LC-W: Writing – review & editing. CD: Writing – review & editing. LC: Writing – review & editing. MC: Writing – review & editing. MM: Writing – review & editing. JP: Writing – review & editing. DI: Writing – review & editing. MB: Writing – review & editing. JB: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by NIH R01-GM145814 to JOB, by NIH T32-GM007133 that supported KAK, and by NSF #2050567 that supported LCW.
Acknowledgments
We thank the Laboratory of Genetics for supporting AF and JP with Genetics & Genomics Undergraduate Distinguished Research Fellowship Awards and the UW Madison Summer Undergraduate Research Fellowship for supporting AF and JP. We thank Allison Karnitz for research support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Asami, T., Min, Y. K., Nagata, N., Yamagishi, K., Takatsuto, S., Fujioka, S., et al. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor1. Plant Physiol. 123, 93–100. doi: 10.1104/pp.123.1.93
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D., and Schroeder, J. I. (2019). Genetic strategies for improving crop yields. Nature 575, 109–118. doi: 10.1038/s41586-019-1679-0
Bauwe, H., Hagemann, M., and Fernie, A. R. (2010). Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336. doi: 10.1016/j.tplants.2010.03.006
Bräutigam, A., Schlüter, U., Eisenhut, M., and Gowik, U. (2017). On the evolutionary origin of CAM photosynthesis. Plant Physiol. 174, 473–477. doi: 10.1104/pp.17.00195
Brunkard, J. O. (2020). Exaptive evolution of target of rapamycin signaling in multicellular eukaryotes. Dev. Cell 54, 142–155. doi: 10.1016/j.devcel.2020.06.022
Brunkard, J. O., Xu, M., Regina Scarpin, M., Chatterjee, S., Shemyakina, E. A., Goodman, H. M., et al. (2020). TOR dynamically regulates plant cell-cell transport. Proc. Natl. Acad. Sci. United States America 117, 5049–5058. doi: 10.1073/pnas.1919196117
Busche, M., Scarpin, M. R., Hnasko, R., and Brunkard, J. O. (2021). TOR coordinates nucleotide availability with ribosome biogenesis in plants. Plant Cell 33, 1615–1632. doi: 10.1093/plcell/koab043
Cackett, L., Luginbuehl, L. H., Hendron, R.-W., Plackett, A. R. G., Stanley, S., Hua, L., et al. (2025). Increased chloroplast area in the rice bundle sheath through cell-specific perturbation of brassinosteroid signaling. Plant Physiol. 197, kiaf108. doi: 10.1093/plphys/kiaf108
Cao, P., Kim, S. J., Xing, A., Schenck, C. A., Liu, L., Jiang, N., et al. (2019). Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis. eLife 8, e50747. doi: 10.7554/eLife.50747
Catterou, M., Dubois, F., Schaller, H., Aubanelle, L., Vilcot, B., Sangwan-Norreel, B. S., et al. (2001). Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. I. Molecular, cellular and physiological characterization of the Arabidopsis bul1 mutant, defective in the Δ7-sterol-C5-desaturation step leading to brassinosteroid biosynthesis. Planta 212, 659–672. doi: 10.1007/s004250000466
Choe, S., Dilkes, B. P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K. A. (1998). The DWF4 gene of arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. doi: 10.1105/tpc.10.2.231
Choe, S., Dilkes, B. P., Gregory, B. D., Ross, A. S., Yuan, H., Noguchi, T., et al. (1999a). The arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis1. Plant Physiol. 119, 897–908. doi: 10.1104/pp.119.3.897
Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C. P., Gregory, B. D., et al. (1999b). The arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11, 207–221. doi: 10.1105/tpc.11.2.207
Choe, S., Schmitz, R. J., Fujioka, S., Takatsuto, S., Lee, M.-O., Yoshida, S., et al. (2002). Arabidopsis brassinosteroid-insensitive dwarf12Mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol. 130, 1506–1515. doi: 10.1104/pp.010496
Choe, S., Tanaka, A., Noguchi, T., Fujioka, S., Takatsuto, S., Ross, A. S., et al. (2000). Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 21, 431–443. doi: 10.1046/j.1365-313x.2000.00693.x
Chory, J., Nagpal, P., and Peto, C. A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in arabidopsis. Plant Cell 3, 445–459. doi: 10.1105/tpc.3.5.445
Clouse, S. D., Langford, M., and McMorris, T. C. (1996). A brassinosteroid-insensitive mutant in arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. doi: 10.1104/pp.111.3.671
Coschigano, K. T., Melo-Oliveira, R., Lim, J., and Coruzzi, G. M. (1998). Arabidopsis gls mutants and distinct fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell 10, 741–752. doi: 10.1105/tpc.10.5.741
Eisenhut, M., Roell, M.-S., and Weber, A. P. M. (2019). Mechanistic understanding of photorespiration paves the way to a new green revolution. New Phytol. 223, 1762–1769. doi: 10.1111/nph.15872
Fontaine, J.-X., Tercé-Laforgue, T., Armengaud, P., Clément, G., Renou, J.-P., Pelletier, S., et al. (2012). Characterization of a NADH-dependent glutamate dehydrogenase mutant of arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24, 4044–4065. doi: 10.1105/tpc.112.103689
Fujita, S., Ohnishi, T., Watanabe, B., Yokota, T., Takatsuto, S., Fujioka, S., et al. (2006). Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. 45, 765–774. doi: 10.1111/j.1365-313X.2005.02639.x
Gachotte, D., Husselstein, T., Bard, M., Lacroute, F., and Benveniste, P. (1996). Isolation and characterization of an Arabidopsis thaliana cDNA encoding a Δ7-sterol-C-5-desaturase by functional complementation of a defective yeast mutant. Plant J. 9, 391–398. doi: 10.1046/j.1365-313X.1996.09030391.x
Gillmor, C. S. and Lukowitz, W. (2020). EMS mutagenesis of arabidopsis seeds. Methods Mol. Biol. 2122, 15–23. doi: 10.1007/978-1-0716-0342-0_2
Goul, C., Peruzzo, R., and Zoncu, R. (2023). The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease. Nat. Rev. Mol. Cell Biol. 24, 857–875. doi: 10.1038/s41580-023-00641-8
Greene, E. A., Codomo, C. A., Taylor, N. E., Henikoff, J. G., Till, B. J., Reynolds, S. H., et al. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in arabidopsis. Genetics 164, 731–740. doi: 10.1093/genetics/164.2.731
Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.-L., Powers, S. J., et al. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in arabidopsis. Plant Cell 18, 3399–3414. doi: 10.1105/tpc.106.047415
Hadjikakou, M., Bowles, N. I., Geyik, O., Conijn, S. J. G., Mogollón, J. M., Bodirsky, B. L., et al. (2025). Ambitious food system interventions required to mitigate the risk of exceeding Earth’s environmental limits. One Earth 8, 101351. doi: 10.1016/j.oneear.2025.101351
Hartwig, T., Corvalan, C., Best, N. B., Budka, J. S., Zhu, J.-Y., Choe, S., et al. (2012). Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for arabidopsis and maize. PLoS One 7, e36625. doi: 10.1371/journal.pone.0036625
He, S., Crans, V. L., and Jonikas, M. C. (2023). The pyrenoid: the eukaryotic CO2-concentrating organelle. Plant Cell 35, 3236–3259. doi: 10.1093/plcell/koad157
Hedden, P. and Graebe, J. E. (1985). Inhibition of gibberellin biosynthesis by paclobutrazol in cell-free homogenates ofCucurbita maxima endosperm andMalus pumila embryos. J. Plant Growth Regul. 4, 111–122. doi: 10.1007/BF02266949
Hothorn, M., Belkhadir, Y., Dreux, M., Dabi, T., Noel, J. P., Wilson, I. A., et al. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471. doi: 10.1038/nature10153
Jewell, J. L. and Guan, K. L. (2013). Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 38, 233–242. doi: 10.1016/j.tibs.2013.01.004
Kauschmann, A., Jessop, A., Koncz, C., Szekeres, M., Willmitzer, L., and Altmann, T. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9, 701–713. doi: 10.1046/j.1365-313X.1996.9050701.x
Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., et al. (1998). The arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690. doi: 10.1105/tpc.10.10.1677
Lancien, M., Martin, M., Hsieh, M.-H., Leustek, T., Goodman, H., and Coruzzi, G. M. (2002). Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 29, 347–358. doi: 10.1046/j.1365-313X.2002.01218.x
Li, J. and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, J., Nam, K. H., Vafeados, D., and Chory, J. (2001). BIN2, a new brassinosteroid-insensitive locus in arabidopsis. Plant Physiol. 127, 14–22. doi: 10.1104/pp.127.1.14
Liao, C.-Y., Pu, Y., Nolan, T. M., Montes, C., Guo, H., Walley, J. W., et al. (2023). Brassinosteroids modulate autophagy through phosphorylation of RAPTOR1B by the GSK3-like kinase BIN2 in Arabidopsis. Autophagy 19, 1293–1310. doi: 10.1080/15548627.2022.2124501
Liu, Y., Duan, X., Zhao, X., Ding, W., Wang, Y., and Xiong, Y. (2021). Diverse nitrogen signals activate convergent ROP2-TOR signaling in Arabidopsis. Dev. Cell 56, 1283–1295.e5. doi: 10.1016/j.devcel.2021.03.022
Lutt, N. and Brunkard, J. O. (2022). Amino acid signaling for TOR in eukaryotes: sensors, transducers, and a sustainable agricultural fuTORe. Biomolecules 12, 387–387. doi: 10.3390/biom12030387
Maeda, H. A. (2019). Evolutionary diversification of primary metabolism and its contribution to plant chemical diversity. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00881
Maple, J. and Møller, S. G. (2007). “Mutagenesis in arabidopsis,” in Circadian Rhythms: Methods and Protocols. Ed. Rosato, E. (Humana Press, Totowa, NJ), 197–206. doi: 10.1007/978-1-59745-257-1_14
Martinez, R. E., Klimpel, K. A., Busche, M., and Brunkard, J. O. (2025). Plant ribosomopathies: New insights and a critical re-evaluation of ribosomal protein gene mutants in plants. Curr. Opin. Plant Biol. 88, 102791. doi: 10.1016/j.pbi.2025.102791
Meacham-Hensold, K., Cavanagh, A. P., Sorensen, P., South, P. F., Fowler, J., Boyd, R., et al. (2024). Shortcutting photorespiration protects potato photosynthesis and tuber yield against heatwave stress. Global Change Biol. 30, e17595. doi: 10.1111/gcb.17595
Micol, J. L. (2009). Leaf development: time to turn over a new leaf? Curr. Opin. Plant Biol. 12, 9–16. doi: 10.1016/j.pbi.2008.11.001
Moon, J. and Hake, S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol. 14, 24–30. doi: 10.1016/j.pbi.2010.08.012
Muñoz-Nortes, T., Pérez-Pérez, J. M., Sarmiento-Mañús, R., Candela, H., and Micol, J. L. (2017). Deficient glutamate biosynthesis triggers a concerted upregulation of ribosomal protein genes in Arabidopsis. Sci. Rep. 7, 1–14. doi: 10.1038/s41598-017-06335-4
Murase, K., Hirano, Y., Sun, T., and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Nam, K. H. and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Noguchi, T., Fujioka, S., Takatsuto, S., Sakurai, A., Yoshida, S., Li, J., et al. (1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis1. Plant Physiol. 120, 833–840. doi: 10.1104/pp.120.3.833
Ohnishi, T., Godza, B., Watanabe, B., Fujioka, S., Hategan, L., Ide, K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of arabidopsis, catalyzes C-3 oxidation *. J. Biol. Chem. 287, 31551–31560. doi: 10.1074/jbc.M112.392720
Peterhansel, C., Horst, I., Niessen, M., Blume, C., Kebeish, R., Kürkcüoglu, S., et al. (2010). Photorespiration. Arabidopsis Book 8, e0130. doi: 10.1199/tab.0130
Plackett, A. R. G., Powers, S. J., Fernandez-Garcia, N., Urbanova, T., Takebayashi, Y., Seo, M., et al. (2012). Analysis of the developmental roles of the arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24, 941–960. doi: 10.1105/tpc.111.095109
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta (BBA) - Bioenergetics 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Sage, R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. doi: 10.1111/j.1469-8137.2004.00974.x
Scarpin, M. R., Leiboff, S., and Brunkard, J. O. (2020). Parallel global profiling of plant tor dynamics reveals a conserved role for larp1 in translation. eLife 9, 1–34. doi: 10.7554/eLife.58795
Scarpin, M. R., Simmons, C. H., and Brunkard, J. O. (2022). Translating across kingdoms: target of rapamycin promotes protein synthesis through conserved and divergent pathways in plants. J. Exp. Bot. 73, 7016–7025. doi: 10.1093/jxb/erac267
Schlüter, U. and Weber, A. P. M. (2020). Regulation and evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 71, 183–215. doi: 10.1146/annurev-arplant-042916-040915
She, J., Han, Z., Kim, T.-W., Wang, J., Cheng, W., Chang, J., et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476. doi: 10.1038/nature10178
Somerville, C. R. (2001). An early arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol. 125, 20–24. doi: 10.1104/pp.125.1.20
Somerville, C. R. and Ogren, W. L. (1980). Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature 286, 257–259. doi: 10.1038/286257a0
South, P. F., Cavanagh, A. P., Liu, H. W., and Ort, D. R. (2019). Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077. doi: 10.1126/science.aat9077
Springmann, M., Clark, M., Mason-D’Croz, D., Wiebe, K., Bodirsky, B. L., Lassaletta, L., et al. (2018). Options for keeping the food system within environmental limits. Nature 562, 519–525. doi: 10.1038/s41586-018-0594-0
Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Timm, S. and Bauwe, H. (2013). The variety of photorespiratory phenotypes – employing the current status for future research directions on photorespiration. Plant Biol. 15, 737–747. doi: 10.1111/j.1438-8677.2012.00691.x
Timm, S. and Hagemann, M. (2020). Photorespiration—how is it regulated and how does it regulate overall plant metabolism? J. Exp. Bot. 71, 3955–3965. doi: 10.1093/jxb/eraa183
Timm, S., Sun, H., and Huang, W. (2024). Photorespiration – emerging insights into photoprotection mechanisms. Trends Plant Sci. 29, 1052–1055. doi: 10.1016/j.tplants.2024.04.011
Valvezan, A. J. and Manning, B. D. (2019). Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat. Metab. 1, 321–333. doi: 10.1038/s42255-019-0038-7
von Bismarck, T., Wendering, P., Perez de Souza, L., Ruß, J., Strandberg, L., Heyneke, E., et al. (2023). Growth in fluctuating light buffers plants against photorespiratory perturbations. Nat. Commun. 14, 7052. doi: 10.1038/s41467-023-42648-x
Walker, B. J., VanLoocke, A., Bernacchi, C. J., and Ort, D. R. (2016). The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 67, 107–129. doi: 10.1146/annurev-arplant-043015-111709
Wang, Y., Cao, J.-J., Wang, K.-X., Xia, X.-J., Shi, K., Zhou, Y.-H., et al. (2019). BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 179, 671–685. doi: 10.1104/pp.18.01028
Wang, Z.-Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Xing, J., Wang, Y., Yao, Q., Zhang, Y., Zhang, M., and Li, Z. (2022). Brassinosteroids modulate nitrogen physiological response and promote nitrogen uptake in maize (Zea mays L.). Crop J. 10, 166–176. doi: 10.1016/j.cj.2021.04.004
Xiong, Y., McCormack, M., Li, L., Hall, Q., Xiang, C., and Sheen, J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. doi: 10.1038/nature12030
Yadav, R. K., Analin, B., Panda, M. K., Ranjan, A., and Singh, A. P. (2023). Brassinosteroids-regulated nitrogen metabolism fine-tunes growth physiology and low nitrogen response in tomato. Environ. Exp. Bot. 216, 105528. doi: 10.1016/j.envexpbot.2023.105528
Yang, W., Wan, G.-F., Zhou, J.-Q., Song, G.-C., Zhao, J., Huang, F.-L., et al. (2024). The effects of brassinosteroids on nitrogen utilization in rice. Agronomy 14, 604. doi: 10.3390/agronomy14030604
Zhang, Z., Zhu, J. Y., Roh, J., Marchive, C., Kim, S. K., Meyer, C., et al. (2016). TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in arabidopsis. Curr. Biol. 26, 1854–1860. doi: 10.1016/j.cub.2016.05.005
Keywords: glutamate synthase, photorespiration, gibberellin, brassinosteroid, plant hormones, plant metabolism, plant genetics
Citation: Klimpel KA, Findlay A, Menon S, Cytron-Walker L, Dedow CA, Camou L, Cadarso M, McGuire MF, Pietroske J, Idowu D, Busche M and Brunkard JO (2025) Genetic suppressor of fd-gogat1 reveals crosstalk among brassinosteroids, photorespiration, and amino acid metabolism. Front. Plant Sci. 16:1680431. doi: 10.3389/fpls.2025.1680431
Received: 06 August 2025; Accepted: 31 October 2025;
Published: 01 December 2025.
Edited by:
Fernando De La Torre, University of Malaga, SpainReviewed by:
Amar Pal Singh, National Institute of Plant Genome Research (NIPGR), IndiaMarko Kolaksazov, Institute of Forage Crops (Bulgaria), Bulgaria
Copyright © 2025 Klimpel, Findlay, Menon, Cytron-Walker, Dedow, Camou, Cadarso, McGuire, Pietroske, Idowu, Busche and Brunkard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacob O. Brunkard, YnJ1bmthcmRAd2lzYy5lZHU=
 Katherine A. Klimpel
Katherine A. Klimpel Michael Busche
Michael Busche Jacob O. Brunkard
Jacob O. Brunkard