- 1Xinjiang Key Laboratory for Ecological Adaptation and Evolution of Extreme Environment Organisms, College of Life Sciences, Xinjiang Agricultural University, Urumqi, China
- 2College of Agronomy, Xinjiang Agricultural University, Urumqi, China
OVATE family proteins (OFPs), a class of plant-specific transcription factors, have been increasingly demonstrated to play pivotal roles in multiple aspects of plant growth and development. However, their functional characterization in soybean (Glycine max) remains largely unexplored. In this study, we conducted a genome-wide identification of OFP genes in soybean, followed by comprehensive analyses, including phylogenetic reconstruction, gene structure characterization, conserved motif and sequence alignment assessments, chromosomal localization, collinearity evaluation, promoter cis-acting element profiling, transcriptome-based expression pattern investigation, and quantitative polymerase chain reaction validation. Key findings revealed that the soybean genome harbors 42 GmOFP genes, all of which contain the conserved OVATE domain and are distributed unevenly across 19 chromosomes. Most members of this gene family exhibit single-exon architectures, with conserved motif analysis demonstrating that Motif 1 and Motif 2 collectively constitute the OVATE domain. Collinearity analysis indicated that a majority of GmOFPs underwent duplication events during evolution. Promoter analysis revealed abundant cis-regulatory elements associated with abiotic stress responses, hormonal regulation, light responsiveness, and growth-related processes. Expression profiling revealed that GmOFP genes exhibit tissue-specific expression patterns across various soybean organs, with several GmOFP genes showing differential responsiveness to salt stress. These findings provide crucial insights into the molecular characteristics and potential biological functions of GmOFPs, establishing a theoretical foundation for further investigations into their regulatory mechanisms in soybean growth, development, and stress adaptation.
1 Introduction
The OVATE family proteins (OFPs) represent a class of plant-specific transcription factors characterized by a conserved OVATE domain at their C-terminus, comprising approximately 70 amino acids. These proteins, which were initially identified in tomato (Solanum lycopersicum), are hydrophilic and contain nuclear localization signals (Liu et al., 2002). Studies have demonstrated that OFP family members play regulatory roles in diverse plant developmental processes, including organ morphogenesis, hormone signal transduction, abiotic stress responses, and secondary cell wall formation, highlighting their critical involvement in key physiological functions (Wang et al., 2022; Xia et al., 2023; Ding et al., 2020; Borovsky et al., 2022; Liu et al., 2018).
In plant organ morphogenesis, OFPs play pivotal regulatory roles. As a critical factor governing organ development, the OVATE gene in tomato, when harboring a premature termination codon mutation, not only transforms spherical fruits to a pear-shaped phenotype but also significantly reduces the floral organ size and diminishes leaf growth scales (Liu et al., 2002). In Arabidopsis, AtOFP1 functions as a cellular morphology regulator by suppressing the transcriptional activity of AtGA20ox1, a key gene in gibberellin (GA)biosynthesis, thereby negatively regulating cell elongation (Wang et al., 2007). The upregulation of the rice (Oryza sativa) OsOFP2 gene induces dwarfism, disrupts leaf development, causes grain morphological deformities, and reorganizes vascular bundle architecture in stems through its influence on the cellular polarity growth regulatory network (Schmitz et al., 2015). Similarly, knockdown of OsOFP6 leads to plant dwarfing, altered grain shape, and shortened lateral roots (Ma et al., 2017). In Arabidopsis, KNAT7 collaborates with AtOFP1 and AtOFP4 via a protein interaction network to increase the transcriptional repression of target genes synergistically, a molecular mechanism critically involved in regulating secondary cell wall biosynthesis (Li et al., 2011). Additionally, OVATE family proteins coordinate with TONNEAU1 to modulate fruit morphology by influencing cell division patterns (Wu et al., 2018). Although these studies span diverse species and tissues, they collectively support a unifying conclusion: OFPs primarily regulate plant growth and development through transcriptional repression mechanisms.
OFPs play crucial roles in multiple plant hormone signaling pathways and auxin regulation. In rice, OsOFP6 has been shown to potentially suppress the expression of auxin signaling-related genes, thereby modulating this pathway. Notably, RNA interference-mediated knockdown of this gene resulted in a significantly stronger regulatory effect on downstream target genes than did the phenotypic changes induced by its overexpression. This observation further supports the biological function of OsOFP6 as a key repressor in the auxin signaling network, suggesting that this protein may finely regulate auxin homeostasis in plants through negative feedback mechanisms (Ma et al., 2017). During lettuce (Lactuca sativa) development, the LsOFP6 gene exhibits marked spatiotemporal expression specificity, with its expression peaking at the critical transition stage from vegetative to reproductive growth. Recent studies revealed that the LsKN1-LsOFP6 molecular module regulatory network achieves precise control of bolting time by bidirectionally modulating both the biosynthetic pathway and signaling response system of GA (Qi et al., 2025). Research on the brassinosteroid (BR) signaling pathway has revealed more complex regulatory networks involving OFPs. Rice OFPs have been demonstrated to participate in BR signal transduction and influence grain size. Specifically, FBX206 interacts with OsOFP8 (a positive regulator) and OsOFP19 (a negative regulator) in the BR signaling pathway, forming a coordinated regulatory pathway that ultimately determines grain size and yield (Sun et al., 2024). Additionally, studies have shown that OsOFP19, OSH1, and DLT can form a functional complex critical for plant growth and development. This complex regulates BR signaling and determines cell division patterns, with DLT notably suppressing the individual activities of both OsOFP19 and OSH1 and their synergistic regulatory effects on gene expression (Yang et al., 2018). These findings collectively demonstrate that OFPs execute intricate and precise regulatory functions across diverse developmental stages and tissues by participating in multiple phytohormone signaling pathways. They likely act as molecular hubs to integrate different hormone signals, thereby coordinating plant growth and developmental processes.
OFP family members play crucial regulatory roles in plant responses to abiotic stresses. Studies have demonstrated that these proteins participate in plant adaptation to drought, salinity, and other adverse conditions through diverse molecular mechanisms. In rice, drought resistance is positively correlated with OsOFP6 expression levels. The OFP6-overexpressing lines presented a relatively low rate of water loss and reduced H2O2 accumulation, whereas the OsOFP6-knockdown plants presented the opposite phenotypes, suggesting that OsOFP6 may increase drought avoidance and tolerance by regulating oxidative stress responses (Ma et al., 2017). In woody plants, Populus trichocarpa PtOFP1 was identified as a transcriptional repressor. Heterologous overexpression of PtOFP1 in Arabidopsis enhanced tolerance to PEG-mediated drought stress at the seedling stage and increased survival rates at the mature stage, indicating that PtOFP1 may improve drought resistance through conserved regulatory mechanisms (Wang et al., 2021). Moreover, OFPs can interact with other transcription factors to regulate stress responses in a coordinated manner. For example, PpOFP1 interacts with PpZFHD1 to increase salt tolerance synergistically in transgenic tomato plants (Tan et al., 2021). In Arabidopsis, AtOFP8 modulates cuticular wax biosynthesis by regulating the expression of wax-related genes, potentially influencing leaf surface wax deposition to reduce transpirational water loss and improve drought resistance (Tang et al., 2018). Taken together, OFPs play multifaceted roles in plant responses to abiotic stress, including the regulation of oxidative stress, the modulation of water metabolism, the enhancement of salt tolerance, and the modification of epidermal structures.
Soybean, a globally vital legume crop, has substantial nutritional and economic value. However, its productivity is severely reduced due to the detrimental impacts of salt stress. OFP proteins play significant roles in mediating plant responses to salt stress. However, the function of the OFP gene in the response of soybean to salt stress has not yet been elucidated. Therefore, we conducted a genome-wide analysis of the soybean OFP gene family and systematically investigated its expression patterns under salt stress. The research included family member identification, chromosomal localization, phylogenetic tree construction, gene structure analysis, collinearity relationship assessment, and promoter cis-acting element characterization. Furthermore, we analyzed the tissue-specific expression profiles of the soybean OFP genes using genomic databases and publicly available RNA-seq data, with a particular focus on their expression dynamics under salt stress. The expression patterns of the soybean OFP genes were further validated through experimental approaches. In summary, our findings provide valuable insights and theoretical foundations for elucidating the functional roles of soybean OFP genes and their potential involvement in salt stress responses.
2 Materials and methods
2.1 Plant materials
The plant material used in this study was the soybean cultivar Williams 82. The seedlings were cultivated under controlled environmental conditions, with a temperature regimen of 26°C and a 14-hour light/10-hour dark photoperiod. At the cotyledon (VC) growth stage (approximately 14 days post-germination, which is characterized by full expansion of two unifoliate leaves), the plants were subjected to salt stress treatment with 250 mM NaCl. Root tissues were sampled at 0, 6, 12, and 24 hours post-treatment initiation, with three biological replicates collected at each time point. Harvested samples were immediately flash-frozen in liquid nitrogen and stored at −80°C for subsequent analyses.
2.2 Screening and identification of the OFP gene family in soybean
The soybean genome (Wm82.gnm4) sequence and corresponding annotation files were retrieved from the Phytozome database (G. max Wm82.a4.v1: Phytozome; accessed 15 February 2025). The hidden Markov model (HMM) profile of the OVATE domain (PF04844) was downloaded through the Pfam plugin in the InterPro database (https://www.ebi.ac.uk/interpro/entry/pfam/; accessed 15 February 2025) (Bateman et al., 2002). Genome-wide screening for OVATE domain-containing genes was performed using the HMM search plugin in TBtools software (https://github.com/CJ-Chen/TBtools/releases; accessed 15 February 2025) against the soybean proteome (Chen et al., 2023; Zhang et al., 2021). Candidate genes were subsequently validated through domain architecture analysis using the NCBI Conserved Domains Database (CDD; https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART database (https://smart.embl.de/; both accessed 17 February 2025) (Letunic et al., 2002). Proteins lacking conserved OVATE-associated domains were manually excluded. Comprehensive analysis of protein physicochemical properties of the identified GmOFP family members, including amino acid number, molecular weight (MW), isoelectric point (pI), instability index, aliphatic index, and grand average of hydropathicity (GRAVY), was conducted using TBtools (Fang et al., 2021).
2.3 Phylogenetic analysis of the OFP gene family in soybean
The protein sequences of the OFP gene family in Arabidopsis (retrieved from https://www.arabidopsis.org/, accessed on February 20, 2025) and Oryza sativa (obtained from https://rapdb.dna.affrc.go.jp/, accessed on February 20, 2025) were acquired using the methodology described in Section 2.2. These sequences were subsequently aligned through multisequence alignment performed with ClustalW software. An integrated dataset comprising OFP family members from Arabidopsis, rice, and soybean was then compiled for phylogenetic analysis. A neighbor-joining (NJ) phylogenetic tree was constructed using MEGA-7.0 software with 1000 bootstrap replicates to assess node reliability (Kumar et al., 2018; Belamkar et al., 2014). Final tree visualization and annotation were conducted through the iTOL online platform (https://itol.embl.de/, accessed on February 23, 2025) to enhance topological clarity and graphical presentation (Jiang et al., 2024).
2.4 Comprehensive analysis of gene structure, conserved motifs, and sequence alignment in the OFP gene family of soybean
The gene structures of the soybean OFP gene family members were analyzed and visualized using TBtools software on the basis of their DNA sequences, protein sequences, and soybean genome annotation information (Zhang et al., 2016). Conserved motifs in the soybean OFP gene family were predicted through the MEME suite (https://meme-suite.org/meme/, accessed on February 27, 2025) and subsequently visualized using TBtools software (Bailey et al., 2009). Multiple sequence alignment of the soybean OFP gene family was performed using Jalview software (https://www.jalview.org/, accessed on March 2, 2025) to investigate sequence conservation patterns (Waterhouse et al., 2009).
2.5 Chromosomal localization and collinearity analysis of the GmOFP gene
The chromosomal location of the GmOFP gene was visualized using the Gene Location Visualize plugin within the GTF/GFF annotation module of TBtools software (Chen et al., 2023). Furthermore, intra- and interspecies syntenic relationships of the soybean OFP gene family were analyzed using the MCScanX plugin in TBtools, with Arabidopsis and Oryza sativa selected as comparative species (Wang et al., 2012).
2.6 Comprehensive analysis of cis-acting elements in the promoter regions of the GmOFP gene family in soybean
The upstream 2000 bp promoter sequences of the soybean OFP family genes were extracted using TBtools software. The extracted promoter sequence files were subsequently submitted to the PlantCARE web server (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed on 7 March 2025) for systematic prediction of cis-acting regulatory elements. Lastly, visualization and bioinformatics analysis of the identified cis-regulatory elements were performed using TBtools software (Wang et al., 2024; Chen et al., 2020).
2.7 Transcriptome database-based expression profiling analysis of the GmOFP gene in soybean
The RNA-seq data for root tissues under salt stress in the soybean cultivar Williams 82 were retrieved from the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; accessed on March 7, 2025). Concurrently, tissue-specific expression patterns of GmOFP genes were acquired from the SoyBase genomic resource (https://www.soybase.org/; accessed on March 9, 2025) (Libault et al., 2010; Razzaq et al., 2023). Visualization of GmOFP gene expression profiles was performed through heat map generation using the HeatMap Illustrator module within the TBtools bioinformatics platform (Luo et al., 2022).
2.8 RNA extraction and quantitative polymerase chain reaction validation
Total RNA was isolated from soybean root tissues subjected to salt stress at 0, 6, 12, and 24 hours using a Plant Total RNA Extraction Kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China), with three biological replicates collected per time point. Genomic DNA removal and first-strand cDNA synthesis were performed according to the manufacturer’s protocol using the EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech (Beijing) Co., Ltd., Beijing, China). qPCR analysis was conducted on a LightCycler 96 system with PerfectStart® Green qPCR SuperMix (TransGen Biotech (Beijing) Co., Ltd., Beijing, China) in a 20 μL reaction volume. The soybean GmCYP2 gene served as an internal reference for normalizing the expression levels of selected GmOFP genes under salt stress. Relative gene expression levels were calculated using the 2−ΔΔCt method, with data processing performed in Excel 2019 (Cheng et al., 2024). The sequences of primers used for the qPCR analysis are detailed in Supplementary Table S1.
2.9 Statistical analysis
Statistical analyses, including Student’s t test and one-way ANOVA, were con-ducted using GraphPad Prism 10.1 (GraphPad Software), with column graphs subsequently generated to illustrate the relative expression levels.
3 Results
3.1 Screening and identification of the OFP gene family in soybean
In this study, we systematically identified genes encoding the OVATE domain in the soybean genome through initial screening followed by validation using the NCBI-CDD and the SMART database. Genes not encoding the OVATE domain were manually excluded, and redundant sequences were removed through rigorous filtering. This process yielded 42 non-redundant OFP gene family members in the soybean genome. These genes were designated GmOFP1 through GmOFP42 according to their chromosomal localization; comprehensive gene information is presented in Supplementary Table S2. Physicochemical characterization revealed that all the encoded proteins except GmOFP2 exhibited hydrophilic properties. The amino acid lengths of these proteins ranged from 152 to 414 residues. Further analyses of critical biochemical parameters, including the MW, pI, instability index, and aliphatic index, were conducted, and the complete data are summarized in Supplementary Table S2. Notably, the observed variations in these parameters suggest functional diversity within the OFP family, which may be associated with distinct biological roles in soybean development.
3.2 Phylogenetic analysis of the OFP gene family in soybean
To investigate the phylogenetic relationships of the OFP gene family in soybean, we retrieved OFP gene sequences from Arabidopsis (19 members) and Oryza sativa (31 members) and constructed an evolutionary tree using the NJ method (Figure 1). Phylogenetic analysis revealed that these OFP proteins clustered into eight distinct subfamilies, designated Groups A to H. Among the 42 soybean OFP genes, the members were distributed across seven subfamilies: Group B contained eight members, Group C one, Group D three, Group E twelve, Group F four, Group G four, and Group H ten. Notably, soybean OFP genes co-clustered with their Arabidopsis and rice orthologs in varying proportions within specific subfamilies, suggesting conserved evolutionary trajectories and functional diversification across these species. This shared phylogenetic topology implies potential homology and parallel evolutionary processes among OFP genes in soybean, Arabidopsis, and rice.
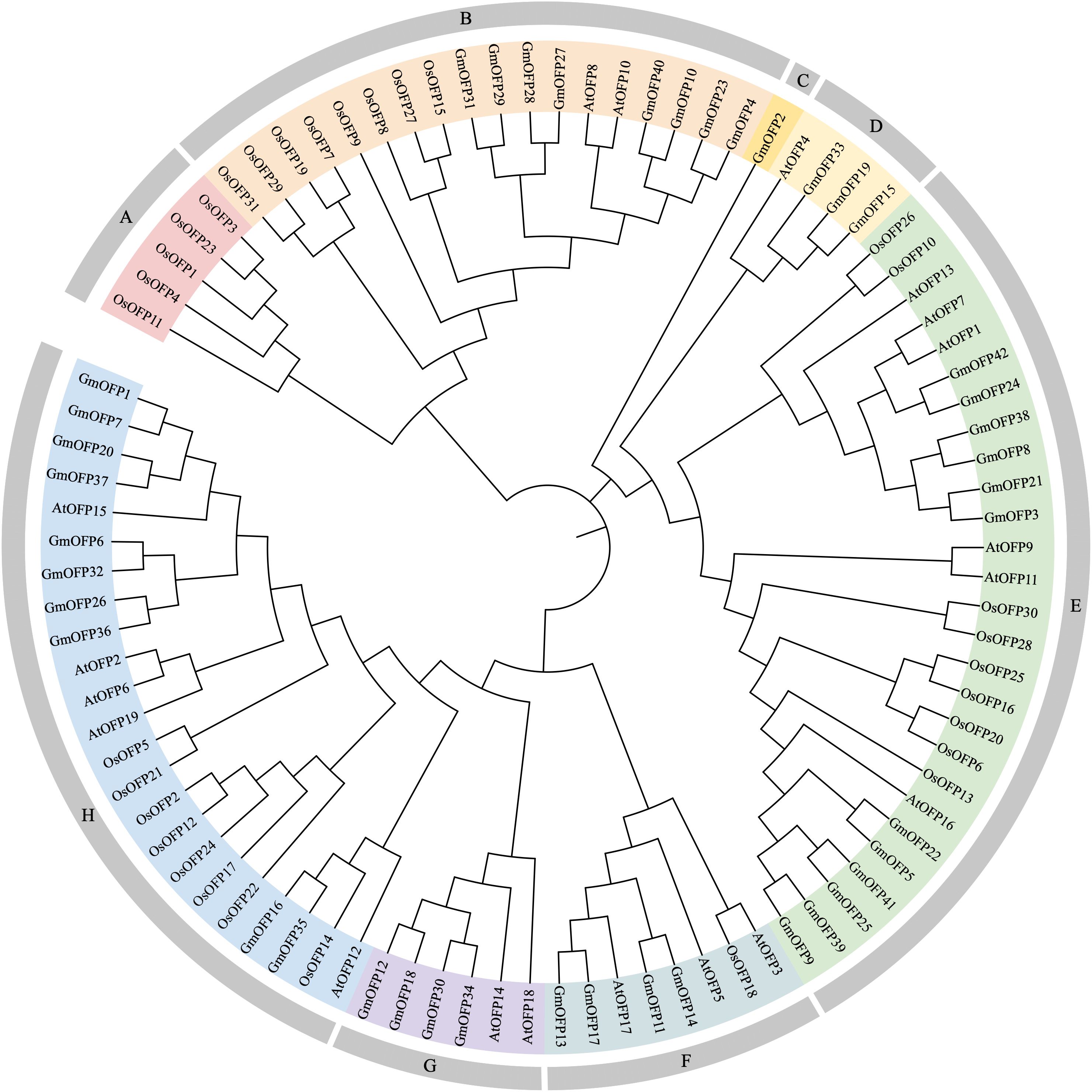
Figure 1. Phylogenetic trees of OFP gene family members in soybean, Arabidopsis, and rice. Phylogenetic analysis of OFP gene family members in soybean (42), Arabidopsis (19), and rice (31) was conducted using MEGA 7.0 software. Different uppercase letters represent different subfamilies.
3.3 Analysis of the gene structure, conserved motifs and sequence alignment of the soybean OFP gene family
To elucidate the fundamental architecture of soybean OFP gene family members, we conducted a comprehensive series of analyses. Initially, phylogenetic clustering on the basis of gene structure (Figure 2A) revealed that members with similar structural organization were grouped into distinct clades. Conserved motif analysis (Figure 2B, Supplementary Figure S1) revealed six characteristic motifs distributed across all 42 OFP family members. Notably, Motif 1 and Motif 2 exhibited universal conservation and were present in all the members with consistent spatial proximity. Gene structure characterization (Figure 2C) revealed that all the family members contained the diagnostic OVATE domain (including the A_thal_3678 domain) in their C-terminal regions. Through comparative analysis of motif distribution patterns and frequency, we hypothesize that Motif 1 and Motif 2 collectively constitute the core components of the OVATE domain. Intriguingly, structural examination revealed that a majority of these genes possess single-exon architectures, a feature potentially associated with evolutionary conservation and enhanced regulatory efficiency (Figure 2D). Multiple sequence alignment further confirmed the universal presence of OVATE protein domains across all family members, reinforcing their structural conservation within this gene family (Supplementary Figure S2).
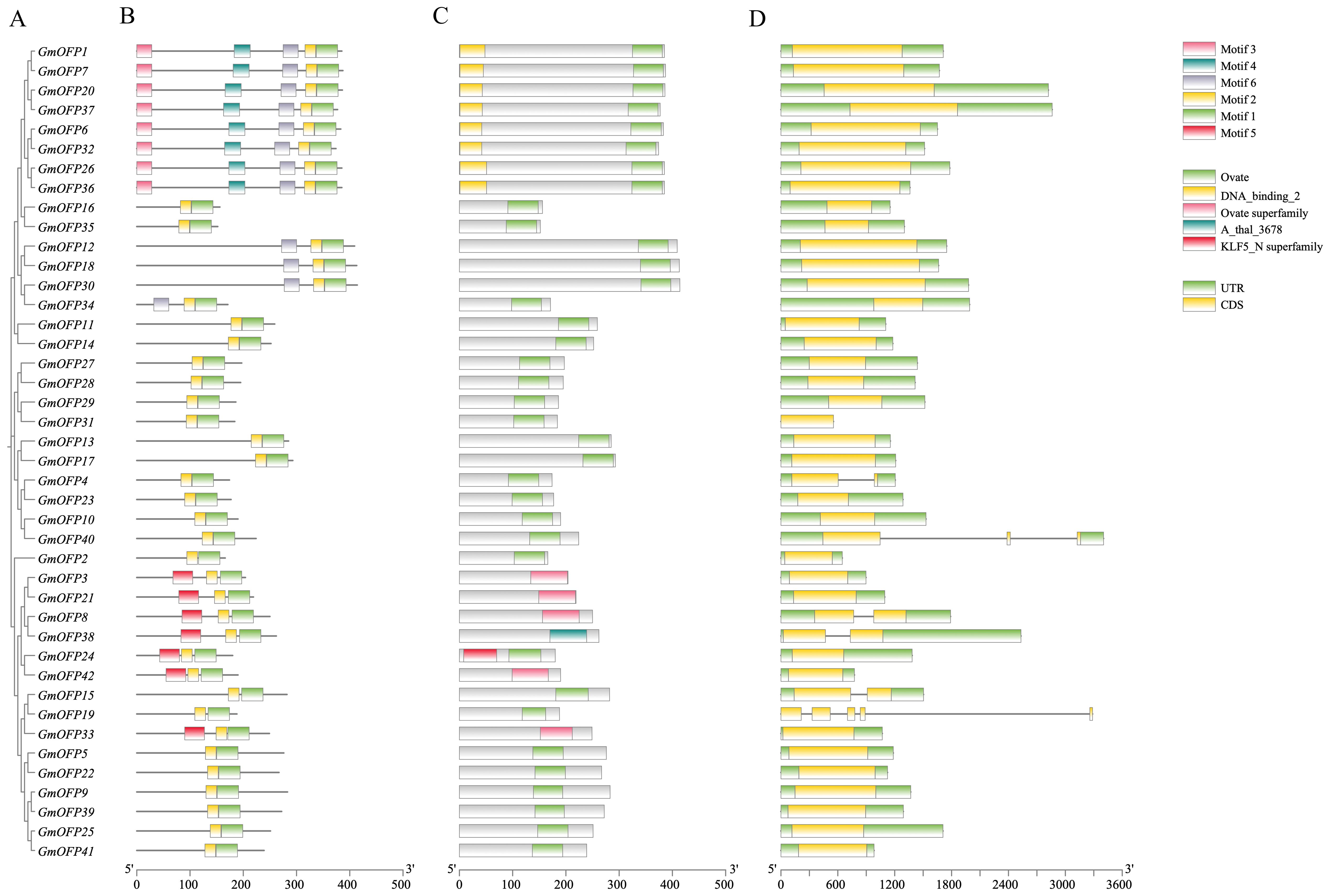
Figure 2. Conserved motif and gene structure analysis of the soybean OFP gene family. (A) Phylogenetic tree of the soybean OFP gene family. (B) Schematic diagram of the structural distribution of the conserved motifs of the soybean OFP gene, in which different colors represent different conserved motifs. (C) Distribution of the OVATE conserved domain of the soybean OFP gene. (D) Structure of the soybean OFP gene, where yellow represents exons and green represents the 5’UTR and 3’UTR.
3.4 Chromosomal localization and collinearity analysis of the GmOFP gene
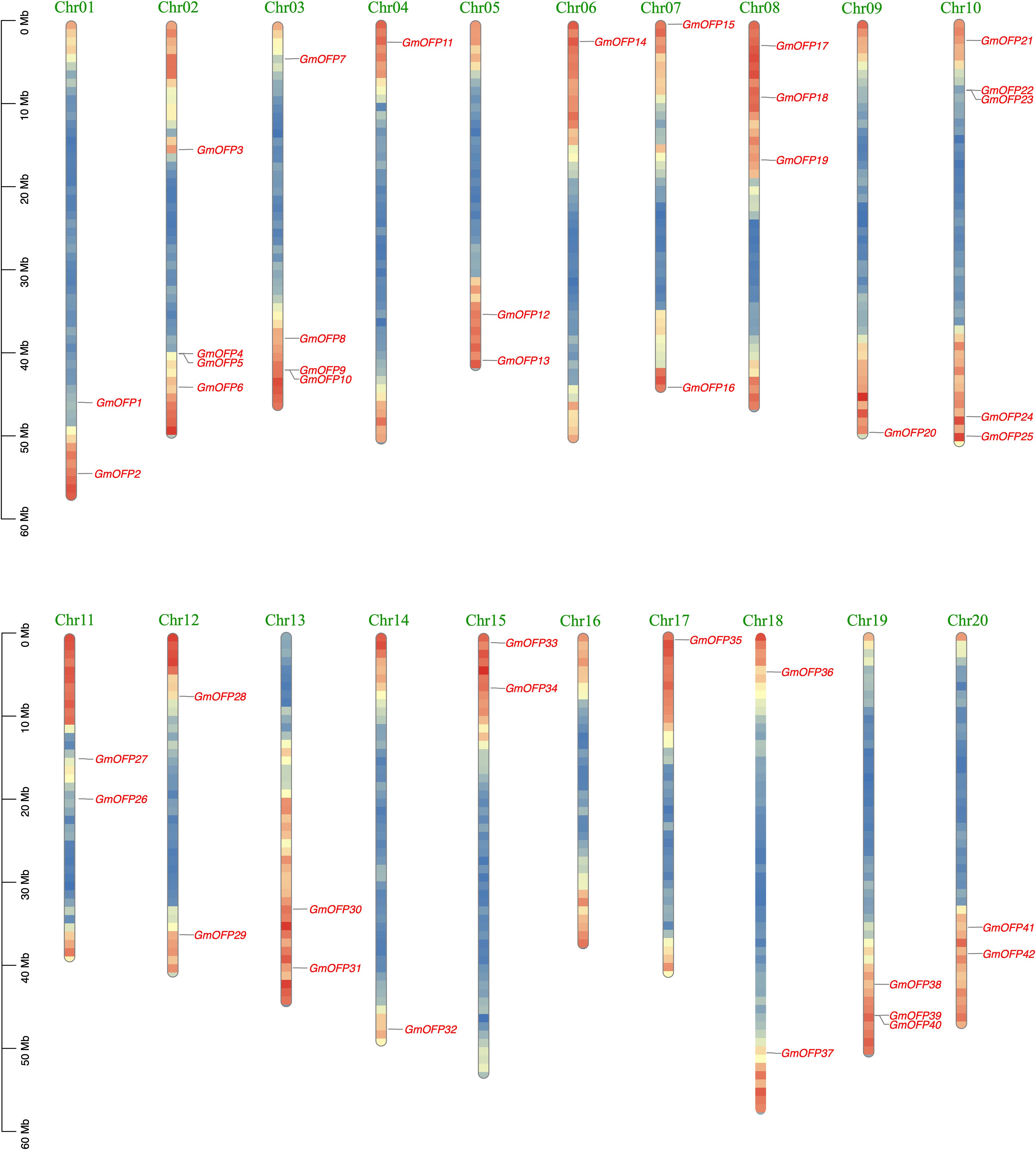
Through the chromosomal localization of the OFP gene in soybean, we found that the 42 members of the soybean OFP gene family were unevenly distributed on 19 chromosomes (Figure 3), and most of the genes were located on both sides of the head and tail of the chromosomes (head or tail). Among them, the GmOFP gene is not present on chromosome 16. One GmOFP gene is located on chromosomes 4, 6, 9, 14 and 17. There are two GmOFP genes on chromosomes 1, 5, 7, 11, 12, 13, 15, 18 and 20. There are three GmOFP genes on chromosomes 8 and 19. There are four GmOFP genes on chromosomes 2 and 3. There are five GmOFP genes on chromosome 10.
To investigate the evolutionary dynamics and gene duplication relationships among members of the soybean OFP gene family, we conducted intraspecies collinearity analysis of OFP family members in Glycine max (Figure 4). The results revealed numerous homologous gene pairs among the 42 identified OFP family members, demonstrating the conserved evolutionary characteristics of this gene family and indicating multiple gene duplication events during its evolutionary history. Furthermore, cross-species collinearity analyses between soybean and divergent species (Oryza sativa and Arabidopsis) were performed (Figure 5). These interspecific comparisons revealed conserved syntenic relationships and evolutionary conservation of duplication events across different lineages, suggesting that these collinear gene pairs may have originated from ancestral genomic regions prior to species divergence. The persistence of syntenic relationships across evolutionary timescales provides evidence for the functional conservation and ancient origins of OFP gene family expansion mechanisms in plants.
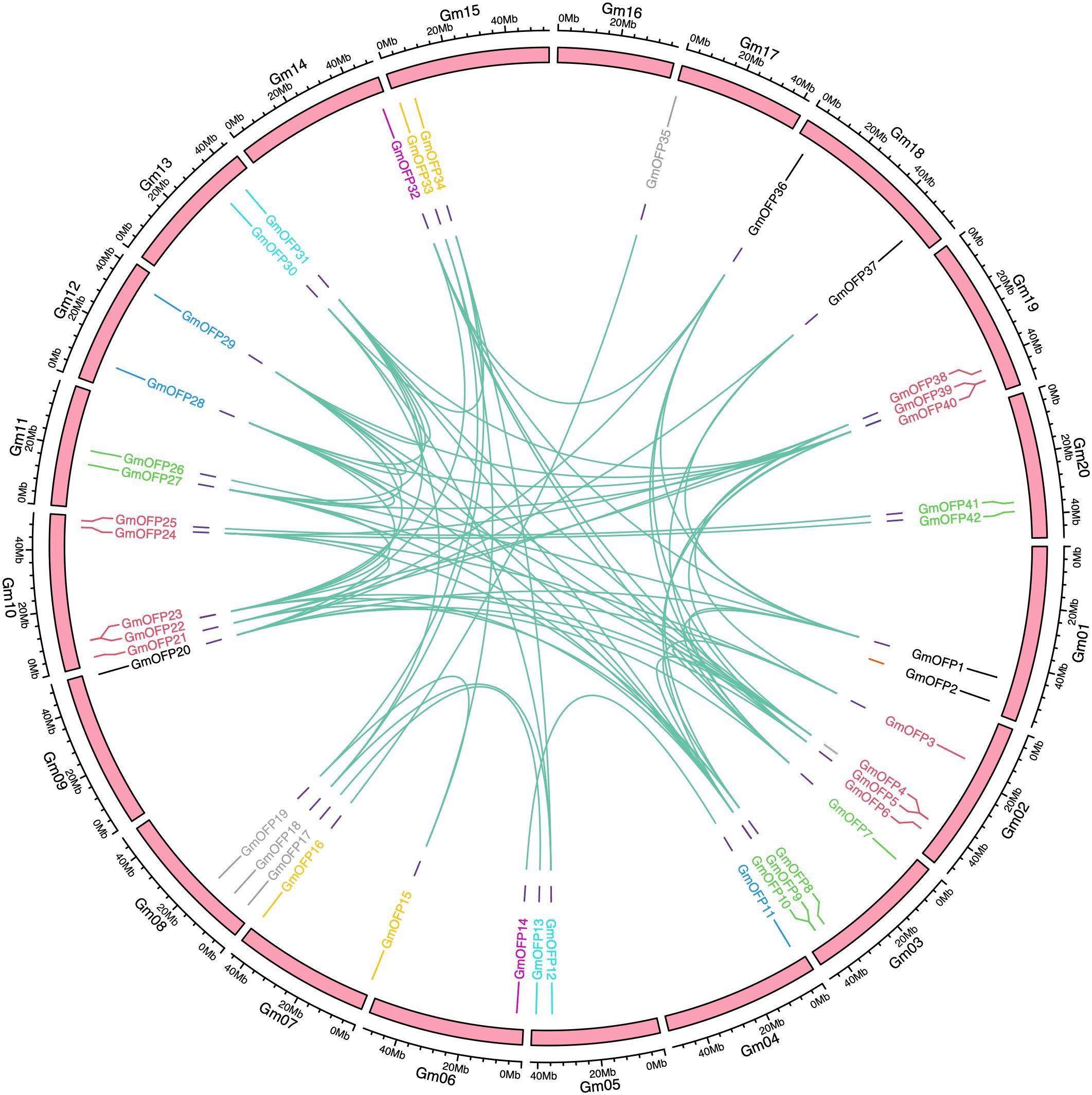
Figure 4. Intraspecies collinearity analysis of the soybean OFP gene family. The pink squares represent chromosomes, and the gene pairs with fragment repetitions are connected by green lines.
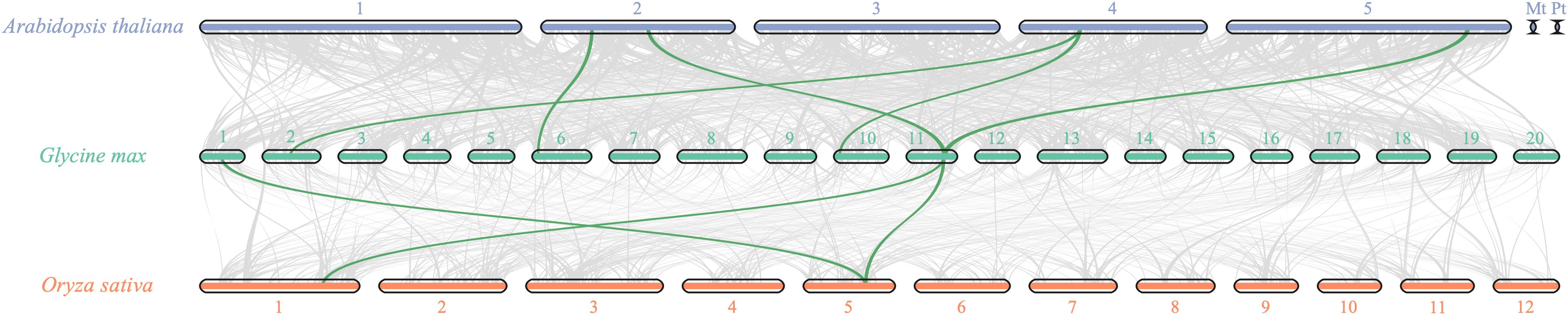
Figure 5. Collinearity analysis of OFP gene family members in soybean, Arabidopsis, and rice. The bar squares represent chromosomes (purple bar squares represent Arabidopsis chromosomes, green bar squares represent soybean chromosomes, and orange bar squares represent rice chromosomes), and gene pairs with fragment duplications are connected by green lines.
3.5 Analysis of cis-acting elements in the promoter of the GmOFP gene family
To investigate the regulation of soybean OFP gene expression, we extracted the 2000-bp promoter sequences upstream of its members and performed cis-acting element prediction using the PlantCARE database. Bioinformatics analysis revealed 25 distinct cis-regulatory elements associated with abiotic stress responses, hormonal regulation, light responsiveness, and plant growth/development (Figure 6). The coordinated presence of these diverse regulatory motifs reveals the molecular basis for functional diversification within the OFP gene family. Furthermore, the differential cis-regulatory element combinations among family members suggest potential mechanisms for precise transcriptional regulation through collaborative interactions with other gene family products during specific developmental stages or under particular environmental conditions. These findings collectively demonstrate that the soybean OFP gene family plays crucial regulatory roles in plant growth and developmental processes.
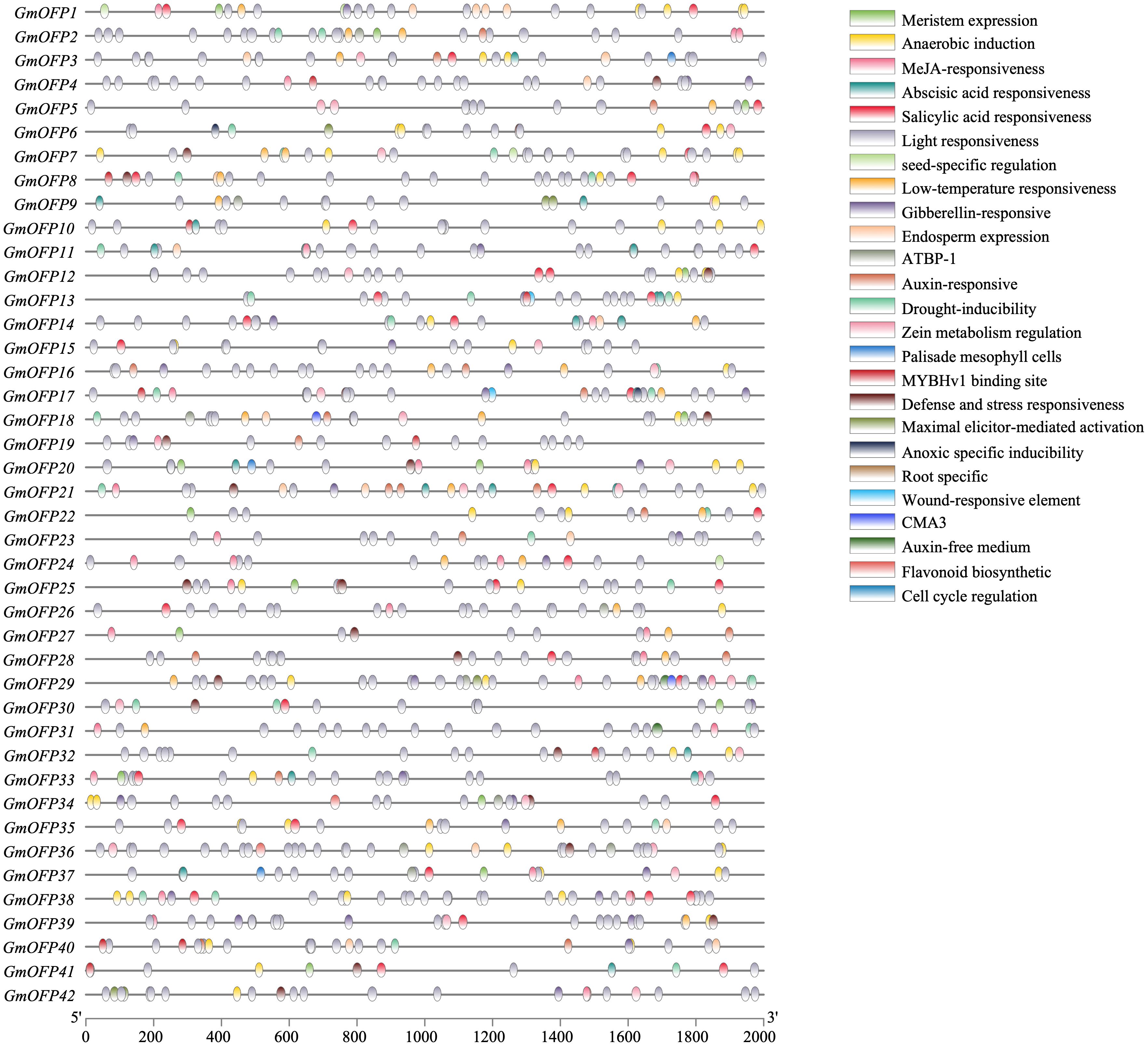
Figure 6. Transcriptional regulatory elements of the promoter region of soybean OFP genes. There are 25 types of transcriptional regulatory elements in the soybean OFP gene family. Different colors represent different types of regulatory elements.
3.6 Transcriptome database-based expression profiling analysis of the GmOFP gene in soybean
Through integrative analysis of RNA-seq data and the soybean genomic database, we systematically investigated the expression patterns of the Glycine max OFP gene family across diverse tissues and under rhizobial inoculation and salt stress conditions. Comparative expression profiling of GmOFP members between the rhizobium-inoculated (IN_RH) and uninoculated (UN_RH) treatments at 12, 24, and 48 hours post-inoculation revealed distinct responsiveness patterns. Multiple GmOFP genes, including GmOFP4, GmOFP23, GmOFP31, and GmOFP35, exhibited transcriptional responsiveness to rhizobial symbiosis, whereas a subset (e.g., GmOFP2, GmOFP8, GmOFP18, and GmOFP34) maintained constitutive expression unaffected by inoculation (Figure 7). Tissue-specific expression profiling demonstrated spatial regulation of GmOFP members: increased transcript accumulation was observed for GmOFP8, GmOFP18, GmOFP27, and GmOFP20 in apical meristem tissues, whereas GmOFP14, GmOFP26, GmOFP32, and GmOFP33 showed preferential expression in root tissues. Differential expression levels were further detected across reproductive and vegetative organs, with distinct GmOFP members displaying modulated transcriptional activity in flowers, green pods, leaves, root nodules, and root tips. These findings collectively suggest both functional diversification and context-dependent regulation occur within the GmOFP gene family during developmental processes and biotic interactions.

Figure 7. Expression patterns of the soybean OFP gene in inoculated rhizobia and different tissues. Cluster analysis was conducted on the expression patterns of 42 GmOFP genes in soybeans before and after rhizobium inoculation and in different tissues. Blue indicates low expression levels, and red indicates high expression levels.
In the analysis of expression patterns under salt stress, we observed that a majority of the soybean GmOFP genes responded to salt stress (Figure 8). Compared with those under the control conditions, the transcript levels of genes such as GmOFP9, GmOFP12, GmOFP25, and GmOFP33 were downregulated following salt stress treatment. Conversely, the transcript levels of genes such as GmOFP3, GmOFP7, GmOFP16, and GmOFP41 were markedly upregulated under the same stress conditions. Notably, the expression profiles of genes such as GmOFP11, GmOFP14, GmOFP19, and GmOFP23 remained unaltered or showed no detectable expression in response to salt stress exposure. These expression patterns indicate that these genes may exhibit functional diversity in regulating developmental processes in soybean under salt stress conditions.
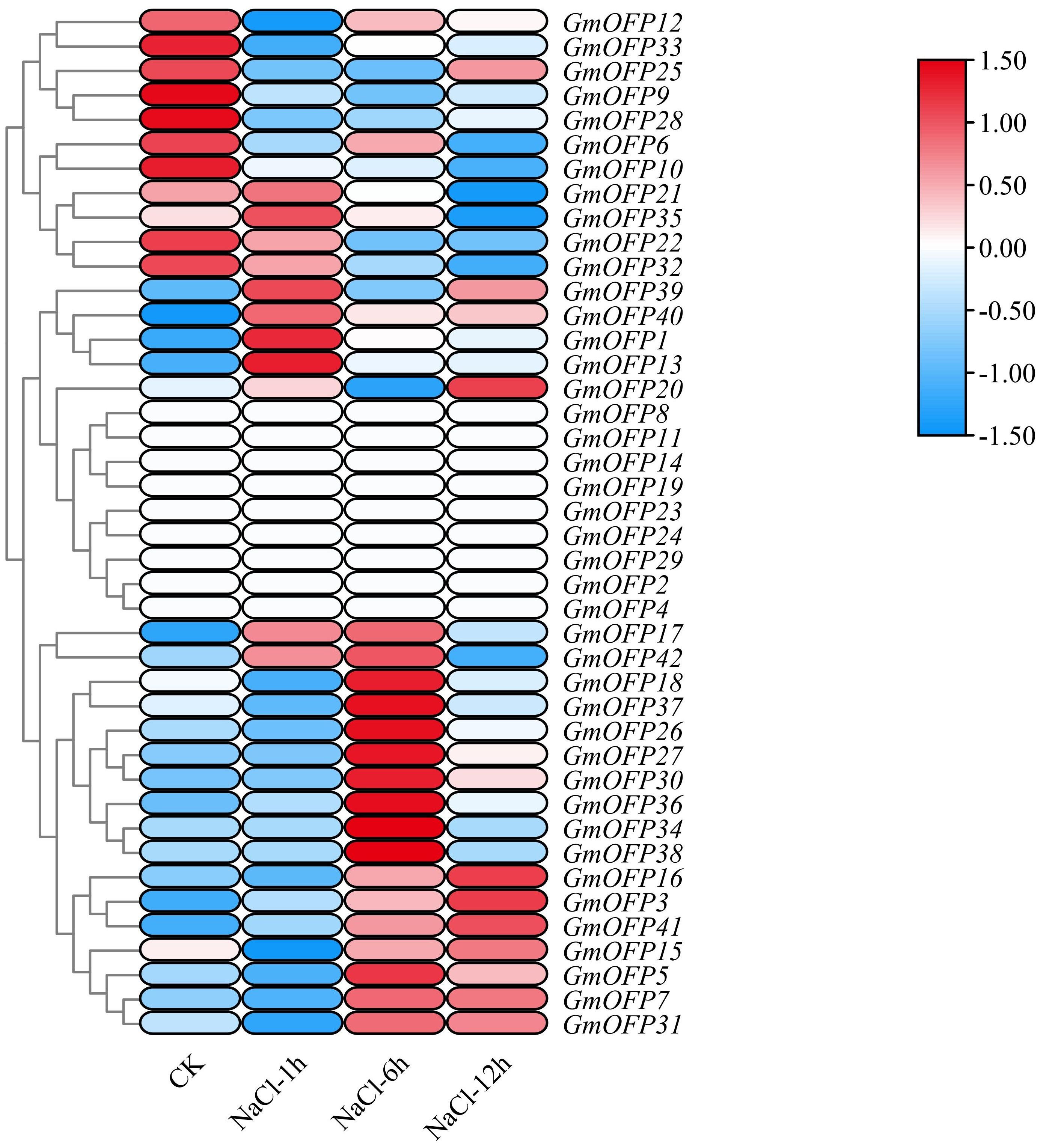
Figure 8. Expression pattern of the GmOFP gene in soybeans under salt stress. A cluster analysis was conducted on the expression patterns of 42 GmOFP genes in the transcriptome of soybean plants under salt stress. The blue color indicates low expression levels, whereas the red color indicates high expression levels.
3.7 qPCR detection of the GmOFP gene in soybean under salt stress
To investigate the expression patterns of soybean OFP gene family members under salt stress, we randomly selected 12 genes from 42 GmOFPs to validate the data in the transcriptome. we performed qPCR validation on a subset of salt-responsive genes identified from soybean salt stress transcriptomic data. Our analysis revealed differential responsiveness to salt stress across all 12 examined GmOFP genes (Figure 9). Comparative analysis with untreated controls (0-hour treatment) revealed temporally specific expression dynamics: At 6 hours post-treatment, GmOFP7 and GmOFP18 presented rapid salt stress induction with significant upregulation, whereas GmOFP9, GmOFP10, GmOFP16, GmOFP21, and GmOFP22 presented marked downregulation. No significant expression changes were detected in GmOFP3, GmOFP32, GmOFP34, GmOFP36, or GmOFP41 at this timepoint. After 12 hours of salt exposure, GmOFP3, GmOFP7, GmOFP18, GmOFP34, GmOFP36, and GmOFP41 were significantly upregulated, whereas GmOFP9, GmOFP10, GmOFP21, and GmOFP22 were markedly downregulated. GmOFP16 and GmOFP32 maintained baseline expression levels during this phase. After 24 hours of treatment, eight genes (GmOFP3, GmOFP7, GmOFP16, GmOFP18, GmOFP32, GmOFP34, GmOFP36, and GmOFP41) were significantly upregulated, whereas four members (GmOFP9, GmOFP10, GmOFP21, and GmOFP22) remained downregulated. Notably, the qPCR results showed substantial concordance with the transcriptomic data for 11 genes, with GmOFP32 being the sole exception. This validation confirms the reliability of transcriptome-based preliminary screening for identifying salt-responsive GmOFP genes and provides a valuable reference for subsequent mechanistic investigations into soybean salt stress adaptation.
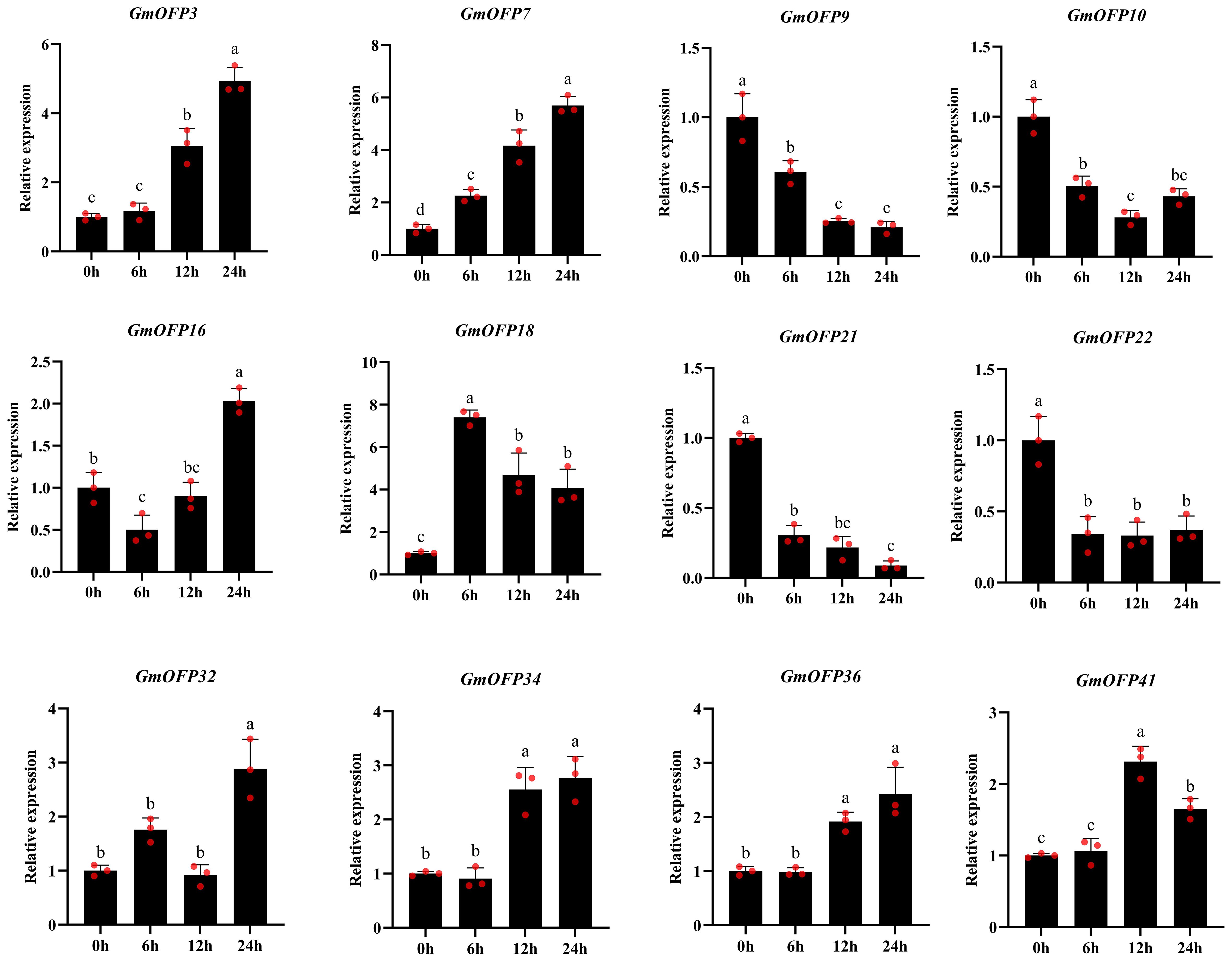
Figure 9. Relative expression levels of 12 GmOFP genes in soybean. The sample size was n = 3. The data are presented as the means ± SDs. Different lowercase letters indicate significant differences within the same treatment at different times (P < 0.05).
4 Discussion
The OFP family, as a plant-specific transcription factor family, have been extensively studied and characterized in various species, demonstrating their crucial roles in plant growth and development (Wang et al., 2010; Zhang et al., 2016; Liu et al., 2022; Wang et al., 2020b). The OFP gene family of soybean, a globally important economic crop and vital source of dietary protein, remains underexplored. Previous studies identified 19 AtOFPs in Arabidopsis (Liu et al., 2014), 31 OsOFPs in rice (Yu et al., 2015), 31 SlOFPs in tomato (Huang et al., 2013), and, remarkably, 100 TaOFPs in hexaploid wheat (Triticum aestivum L.), revealing significant family expansion in polyploid species (Wang et al., 2020a). Given that the complex paleopolyploid background of soybean is shaped by two whole-genome duplication events, our study identified 42 GmOFP members in the soybean genome. This number exceeds that of the diploid model plant Arabidopsis but remains lower than the 56 members reported in allotetraploid upland cotton (Gossypium hirsutum) (Zhang et al., 2022). Such quantitative differences likely reflect species-specific evolutionary trajectories involving gene duplication events and functional diversification during genome evolution.
During the structural analysis of the soybean GmOFP genes, we observed that most family members contained only a single exon (Figure 2). This characteristic single-exon architecture devoid of introns is highly conserved with OFP family members in Arabidopsis, rice, and other species (Liu et al., 2014; Yu et al., 2015), suggesting potential evolutionary maintenance of structural simplicity through functional selection. Single-exon genes demonstrate accelerated transcriptional activation in response to abiotic stresses, a phenomenon likely attributable to the absence of splicing regulation steps that could otherwise delay transcriptional initiation (Hu et al., 2023; Jorquera et al., 2018). Chromosomal distribution analysis revealed the preferential localization of GmOFPs in subtelomeric regions (Figure 3), which was potentially associated with the unique epigenetic regulatory milieu of the chromosomal termini. These telomere-proximal domains typically maintain open chromatin configurations, which may facilitate transcription factor accessibility and rapid transcriptional responses to environmental signals (Philimonenko et al., 2010).
OFPs play diverse roles across various growth and developmental processes. Molecular mechanistic studies in peach (Prunus persica) revealed that PpOFP4 and PpOFP5 exhibit self-assembly properties to form homodimers. Notably, PpOFP5 specifically binds with either PpOFP7 or PpOFP8 to form heterodimeric complexes (Li et al., 2019b). This multilayered protein interaction network potentially operates through dual regulatory pathways: it may modulate either the transcriptional activation potency of the proteins themselves or their DNA-binding efficiency, thereby collectively orchestrating the expression regulation of downstream target genes. Parallel investigations on the OFP family in apple (Malus pumila Mill.) indicated that MdOFP13, MdOFP16, MdOFP20, and MdOFP2 can assemble into heterodimers with other MdOFP members, while MdOFP16 additionally possesses a self-association capacity to form homodimers (Li et al., 2019a). In our study, based on tissue-specific expression pattern analysis and phylogenetic analysis, we speculate that analogous dimers may also occur in soybean. This possibility is suggested by the co-high expression of specific GmOFP members within distinct tissues, such as GmOFP8, GmOFP18, GmOFP27, and GmOFP28 in the shoot apical meristem and GmOFP14 and GmOFP33 in the roots (Figure 7). However, whether GmOFP proteins possess similar interaction networks remains to be experimentally validated and requires further investigation.
With respect to the regulation of plant fruit morphology, researchers have reported that CmFSI8/CmOFP13, a member of the OFP family in melon (Cucumis melo), has a potent inhibitory effect on organ elongation growth when overexpressed in the model plant Arabidopsis (Ma et al., 2022). This gene has ovary-specific expression patterns during reproductive development in melon, with genomic variations in its promoter region critically influencing transcriptional activity and function determination. In this study, our expression profiling revealed that a substantial number of GmOFP genes are expressed in soybean flowers (Figure 7). However, whether these genes play similar roles in floral organ morphogenesis remains unknown.
In previous studies, researchers focused primarily on the roles of OFPs in plant organogenesis and fruit morphological development. However, accumulating evidence has recently revealed their critical involvement in abiotic stress responses. In rice, multiple OsOFPs display responsive expression patterns under salt and drought stress conditions, suggesting their potential functional significance in mitigating these abiotic stresses (Ahmad et al., 2023). Furthermore, in rice, OsOFP6 not only plays essential roles in diverse growth stages and developmental processes but also acts as a positive regulator of drought and cold stress adaptation (Ma et al., 2017). Phylogenetic analysis revealed that specific GmOFPs in soybean (e.g., GmOFP5, GmOFP9, and GmOFP22) clustered within the same clade as OsOFP6 (Figure 1), indicating a close phylogenetic relationship. This high degree of homology suggests that these GmOFPs may share functional similarity with OsOFP6, although further experimental validation is needed. In this study, we also investigated the roles of soybean GmOFPs in the response to abiotic stress. Through systematic mining and screening of soybean transcriptomic data under salt stress combined with qPCR validation, we identified a substantial number of GmOFPs that presented significant alterations in relative expression levels under saline conditions. These findings imply that these GmOFPs may have crucial biological functions in mediating salt stress adaptation during soybean growth and development.
5 Conclusions
In this study, we conducted a comprehensive genome-wide identification and bioinformatics analysis of the OFP gene family in soybean. A total of 42 GmOFPs were systematically identified and found to be unevenly distributed across 19 soybean chromosomes. Notably, most members of this family are characterized as single-exon genes. Through integrated gene expression profiling and experimental validation, we revealed that GmOFPs exhibit varying expression levels across different soybean tissues. Furthermore, several GmOFPs demonstrated differential responsiveness to salt stress conditions, suggesting their potential functional involvement in regulating soybean growth and developmental processes under salinity challenges. These findings establish a crucial theoretical foundation for subsequent investigations into the molecular mechanisms underlying OFP-mediated stress adaptation in soybean.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
FZ: Methodology, Writing – review & editing, Investigation, Writing – original draft, Data curation, Formal Analysis. YW: Writing – review & editing, Data curation, Formal Analysis, Methodology. LZ: Resources, Writing – review & editing, Methodology. YY: Data curation, Funding acquisition, Formal Analysis, Writing – review & editing, Supervision, Investigation, Visualization. ZN: Funding acquisition, Formal Analysis, Writing – review & editing, Writing – original draft, Supervision.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the Tianshan Talent Training Program (2023TSYCCX0012) and the National Natural Science Foundation of China (32360454).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1682513/full#supplementary-material
Supplementary Table 1 | Primer sequences used for qPCR.
Supplementary Table 2 | Information on soybean OFP family members.
Supplementary Figure 1 | Conserved Motif-Logo graphs.
Supplementary Figure 2 | Multiple sequence alignment.
References
Ahmad, S., Ahmad, N., and Bayar, J. (2023). Genome-wide identification, characterization, and expression analysis of the Ovate family protein in Oryza sativa under biotic and abiotic stresses. Plant Stress 10, 100228. doi: 10.1016/j.stress.2023.100228
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bateman, A., Birney, E., Coin, L., Durbin, R., Finn, R. D., Hollich, V., et al. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. doi: 10.1093/nar/30.1.276
Belamkar, V., Weeks, N. T., Bharti, A. K., Farmer, A. D., Graham, M. A., and Cannon, S. B. (2014). Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics 15, 1–25. doi: 10.1186/1471-2164-15-950
Borovsky, Y., Raz, A., Doron-Faigenboim, A., Zemach, H., Karavani, E., and Paran, I. (2022). Pepper fruit elongation is controlled by Cap-sicum annuum ovate family protein 20. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.815589
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bio-informatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cheng, C., Zhang, F., Li, L., and Ni, Z. (2024). Identification and analysis of the plasma membrane H+-ATPase gene family in cotton and its roles in response to salt stress. Plants 13, 3510. doi: 10.3390/plants13243510
Ding, B., Hu, C., Feng, X., Cui, T., Liu, Y., and Li, L. (2020). Systematic analysis of the OFP genes in six Rosaceae genomes and their roles in stress response in Chinese pear (Pyrus bretschneideri). Physiol. Mol. Biol. Plants 26, 2085–2094. doi: 10.1007/s12298-020-00866-3
Fang, Y., Cao, D., Yang, H., Guo, W., Ouyang, W., Chen, H., et al. (2021). Genome-wide identification and characterization of soybean GmLOR gene family and expression analysis in response to abiotic stresses. Int. J. Mol. Sci. 22, 12515. doi: 10.3390/ijms222212515
Hu, H., Dong, B., Fan, X., Wang, M., Wang, T., and Liu, Q. (2023). Mutational bias and natural selection driving the synonymous codon usage of single-exon genes in rice (Oryza sativa L.). Rice (N Y) 16, 11. doi: 10.1186/s12284-023-00627-2
Huang, Z., Houten, J., Gonzalez, G., Xiao, H., and Van der Knaap, E. (2013). Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genomics 288, 111–129. doi: 10.1007/s00438-013-0733-0
Jiang, X., Cao, J., Cao, L., Wang, L., and Che, J. (2024). Genome-wide re-identification of the CrRLK1L family in soybean and functional characterization of GmCrRLK1L2 in salt stress response. Environ. Exp. Bot. 226, 105903. doi: 10.1016/j.envexpbot.2024.105903
Jorquera, R., González, C., Clausen, P., Petersen, B., and Holmes, D. S. (2018). Improved ontology for eukaryotic single-exon coding sequences in biological databases. Database (Oxford) 2018, 1–6. doi: 10.1093/database/bay089
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., Goodstadt, L., Dickens, N. J., Doerks, T., Schultz, J., Mott, R., et al. (2002). Recent im-provements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242–244. doi: 10.1093/nar/30.1.242
Li, H., Dong, Q., Zhao, Q., and Ran, K. (2019a). Genome-wide identification, expression profiling, and protein-protein interaction properties of ovate family proteins in apple. Tree Genet. Genomes 15, 45. doi: 10.1007/s11295-019-1354-5
Li, H., Dong, Q., Zhu, X., Zhao, Q., and Ran, K. (2019b). Genome-wide identification, expression, and interaction analysis for ovate family proteins in peach. Mol. Biol. Rep. 46, 3755–3764. doi: 10.1007/s11033-019-04817-4
Li, E., Wang, S., Liu, Y., Chen, J. G., and Douglas, C. J. (2011). OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates sec-ondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341. doi: 10.1111/j.1365-313X.2011.04595.x
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D., et al. (2010). An integrated tran-scriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. doi: 10.1111/j.1365-313X.2010.04222.x
Liu, D., Sun, W., Yuan, Y., Zhang, N., Hayward, A., Liu, Y., et al. (2014). Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 113, 1219–1233. doi: 10.1093/aob/mcu061
Liu, J., Van Eck, J., Cong, B., and Tanksley, S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 99, 13302–13306. doi: 10.1073/pnas.162485999
Liu, J., Wu, Y., Cui, X., Zhang, X., Xie, M., Liu, L., et al. (2022). Genome-wide characterization of ovate family protein gene family associated with number of seeds per silique in Brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.962592
Liu, J., Zhang, J., Wang, J., Zhang, J., Miao, H., Jia, C., et al. (2018). MuMADS1 and MaOFP1 regulate fruit quality in a tomato ovate mutant. Plant Biotechnol. J. 16, 989–1001. doi: 10.1111/pbi.12843
Luo, C., Yan, J., He, C., Liu, W., Xie, D., and Jiang, B. (2022). Genome-wide identification of the SAUR gene family in wax gourd (Benincasa hispida) and functional characterization of BhSAUR60 during fruit development. Int. J. Mol. Sci. 23, 14021. doi: 10.3390/ijms232214021
Ma, J., Li, C., Zong, M., Qiu, Y., Liu, Y., Huang, Y., et al. (2022). CmFSI8/CmOFP13 encoding an OVATE family protein controls fruit shape in melon. J. Exp. Bot. 73, 1370–1384. doi: 10.1093/jxb/erab510
Ma, Y., Yang, C., He, Y., Tian, Z., and Li, J. (2017). Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J. Exp. Bot. 68, 4885–4898. doi: 10.1093/jxb/erx309
Philimonenko, A. A., Janacek, J., Snyers, L., Almeder, M., Berger, W., Schmidt, W., et al. (2010). Chromosomal dynamics of cell cycle regulator gene p21 during transcriptional activation. J. Struct. Biol. 173, 382–390. doi: 10.1016/j.jsb.2010.10.010
Qi, Y., Shao, W., Chen, H., Ahmed, T., Zhao, X., Wang, Y., et al. (2025). LsKN1 and LsOFP6 synergistically regulate the bolting time by modulating the gibberellin pathway in lettuce. New Phytol. 246, 1049–1065. doi: 10.1111/nph.20307
Razzaq, M. K., Rani, R., Xing, G., Xu, Y., Raza, G., Aleem, M., et al. (2023). Genome-wide identification and analysis of the Hsp40/J-protein family reveals its role in soybean (Glycine max) growth and development. Genes 14, 1254. doi: 10.3390/genes14061254
Schmitz, A. J., Begcy, K., Sarath, G., and Walia, H. (2015). Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 241, 177–188. doi: 10.1016/j.plantsci.2015.10.011
Sun, X., Xie, Y., Xu, K., and Li, J. (2024). Regulatory networks of the F-box protein FBX206 and OVATE family proteins modulate brassi-nosteroid biosynthesis to regulate grain size and yield in rice. J. Exp. Bot. 75, 789–801. doi: 10.1093/jxb/erad397
Tan, Q., Jiang, S., Wang, N., Liu, X., Zhang, X., Wen, B., et al. (2021). OVATE family protein PpOFP1 physically interacts with PpZFHD1 and confers salt tolerance to tomato and yeast. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.759955
Tang, Y., Zhang, W., Yin, Y. L., Feng, P., Li, H. L., and Chang, Y. (2018). Expression of ovate family protein 8 affects epicuticular waxes ac-cumulation in Arabidopsis thaliana. Bot. Stud. 59, 1–9. doi: 10.1186/s40529-018-0228-8
Wang, D., Cao, Z., Wang, W., Zhu, W., Hao, X., Fang, Z., et al. (2020a). Genome-wide characterization of OFP family genes in wheat (Triticum aestivum L.) reveals that TaOPF29a-A promotes drought tolerance. BioMed. Res. Int. 1), 9708324. doi: 10.1155/2020/9708324
Wang, S., Chang, Y., Guo, J., and Chen, J. G. (2007). Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872. doi: 10.1111/j.1365-313X.2007.03096
Wang, Y. K., Chang, W. C., Liu, P. F., Hsiao, M. K., Lin, C. T., Lin, S. M., et al. (2010). Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Mol. Biol. 74, 453–466. doi: 10.1007/s11103-010-9685-5
Wang, H., Chen, J. G., and Chang, Y. (2021). Identification, expression, and interaction analysis of ovate family proteins in Populus trichocarpa reveals a role of PtOFP1 regulating drought stress response. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.650109
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Wang, Y., Wang, Q., Hao, W., Sun, H., and Zhang, L. (2020b). Characterization of the OFP gene family and its putative involvement of tu-berous root shape in radish. Int. J. Mol. Sci. 21, 1293. doi: 10.3390/ijms21041293
Wang, H., Zhang, D., Zhou, X., Zhou, G., Zong, W., Chen, L., et al. (2022). Transcription factor AtOFP1 involved in ABA-mediated seed germination and root growth through modulation of ROS homeostasis in Arabidopsis. Int. J. Mol. Sci. 23, 7427. doi: 10.3390/ijms23137427
Wang, Y., Zong, Z., Chen, J., Sun, X., Wang, J., Yu, Y., et al. (2024). Genome-wide identification of the GbUBC gene family in sea-island cotton (Gossypium barbadense) and the active regulation of drought resistance in cotton by GbUBC23. Int. J. Mol. Sci. 25, 12948. doi: 10.3390/ijms252312948
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wu, S., Zhang, B., Keyhaninejad, N., Rodríguez, G. R., Kim, H. J., Chakrabarti, M., et al. (2018). A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 9, 4734. doi: 10.1038/s41467-018-07216-8
Xia, K., Pan, X., Chen, H., Xu, X., and Zhang, M. (2023). Rice miR168a-5p regulates seed length, nitrogen allocation and salt tolerance by targeting OsOFP3, OsNPF2.4 and OsAGO1a, respectively. J. Plant Physiol. 280, 153905. doi: 10.1016/j.jplph.2022.153905
Yang, C., Ma, Y., He, Y., Tian, Z., and Li, J. (2018). OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 93, 489–501. doi: 10.1111/tpj.13793
Yu, H., Jiang, W., Liu, Q., Zhang, H., Piao, M., Chen, Z., et al. (2015). Expression pattern and subcellular localization of the ovate protein family in rice. PloS One 10, e0118966. doi: 10.1371/journal.pone.0118966
Zhang, X., Chen, B., Wang, L., Ali, S., Guo, Y., Liu, J., et al. (2021). Genome-wide identification and characterization of caffeic acid o-methyltransferase gene family in soybean. Plants 10, 2816. doi: 10.3390/plants10122816
Zhang, L., Tehseen Azhar, M., Che, J., and Shang, H. (2022). Genome-wide identification, expression and evolution analysis of OVATE family proteins in cotton (Gossypium spp.). Gene 834, 146653. doi: 10.1016/j.gene.2022.146653
Keywords: soybean, OVATE family protein, salt stress, growth, development
Citation: Zhan F, Wang Y, Zhang L, Yu Y and Ni Z (2025) Genome-wide identification of the OVATE gene family of proteins in soybean and expression profiling under salt stress. Front. Plant Sci. 16:1682513. doi: 10.3389/fpls.2025.1682513
Received: 09 August 2025; Accepted: 25 August 2025;
Published: 08 September 2025.
Edited by:
Lichao Zhang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Chengzhen Liang, Chinese Academy of Agricultural Sciences, ChinaJunji Su, Gansu Agricultural University, China
Copyright © 2025 Zhan, Wang, Zhang, Yu and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Ni, bml6aGl5b25nQDEyNi5jb20=; Yuehua Yu, eXV5dWVodWExMjEzQHNpbmEuY29t
 Fan Zhan1
Fan Zhan1 Yi Wang
Yi Wang Zhiyong Ni
Zhiyong Ni