Abstract
Plasma membrane intrinsic proteins (PIPs) are involved in plant growth and stress adaptation through their dynamic gating mechanism. Nevertheless, pinpointing the specific roles of individual isoforms remains challenging because of their functional pleiotropy and integrated responses to diverse cues. In this study, we characterized a salt-responsive aquaporin gene, designated SmPIP1;3, isolated from a salt-resistant Salix matsudana variety. Bioinformatics analysis confirmed that it encodes a protein that possesses canonical PIP features with six transmembrane domains, five interhelical loops, and seven serine phosphorylation sites involved in phosphorylation-mediated regulation. The SmPIP1;3 gene was introduced in tobacco plants, and its heterologous expression conferred significant morphological improvements, including taller plant height, larger leaves, longer roots, and increased biomass. Under salt, drought, cold, and heat stresses, transgenic plants showed substantially alleviated membrane damage, as evidenced by weakened Evans blue staining. Consistently, their malondialdehyde contents were 1.48-, 1.47-, 1.57-, and 1.62-fold lower, while relative electrolyte leakage values were 1.56-, 1.35-, 1.53-, and 1.61-fold lower than those of wild-type plants, respectively. SmPIP1;3 orchestrates multi-stress tolerance by sustaining physiological homeostasis and limiting membrane damage. Its performance positions it as a valuable genetic asset for molecular breeding programs.
Introduction
Water uptake and transmembrane water transport are fundamental physiological processes underlying plant growth and development and also serve as a crucial regulatory pathway for plant adaptation to abiotic stress (Gill et al., 2021). Water movement across cellular membranes is a well-established, osmotic-driven process. Aquaporins (AQPs), specialized water-channel proteins located in biological membranes, selectively facilitate water flux while largely preventing proton leakage and, in some cases, allowing the passage of small solutes such as glycerol or CO2 (Eisenbarth and Weig, 2005; Sohail et al., 2022). Based on the phylogenetic relationship and subcellular localization, AQPs in higher plants can be classified into five subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), and X-intrinsic proteins (XIPs) (Kaldenhoff and Fischer, 2006). Among these, PIPs are among the most extensively studied groups, exhibiting high sequence conservation, and can be systematically classified into two phylogenetic clades, designated as PIP1s and PIP2s (Törnroth-Horsefield et al., 2006).
Abiotic stress factors, such as drought, salinity, cold, and heavy metals, can disrupt a plant’s water homeostasis, leading to severe dehydration and death, thereby limiting plant growth, development, and productivity (Suslov et al., 2024). Recent studies have highlighted the functional versatility of AQPs, particularly PIPs, which exhibit bidirectional water transport driven by transmembrane osmotic gradients, with dynamic gating mechanisms being regulated by phosphorylation and osmotic stimuli (Horie et al., 2011). The expression patterns of PIPs vary substantially across various plant organs (e.g., roots and leaves) and along different durations of stress (Horie et al., 2011). For example, a study focused on Arabidopsis found that most PIP genes were downregulated in the leaves under drought, except for AtPIP1;4 and AtPIP2;5, which were upregulated (Alexandersson et al., 2005). The overexpression of AtPIP1;4 or AtPIP2;5 in Arabidopsis also improved resistance to cold stress (Rahman et al., 2020). The GoPIP1 gene identified from Galega orientalis was significantly induced by 200 mmol/L NaCl or 20% polyethylene glycol (PEG) 6000 treatment. The overexpression of this gene in Arabidopsis increased vulnerability to drought stress but not to salinity (Li et al., 2015). Transgenic tobacco expressing BnPIP1 from Brassica napus exhibited enhanced drought tolerance, improved growth, and higher seed germination rates, whereas antisense plants demonstrated developmental defects and heightened drought susceptibility (Yu et al., 2005). The overexpression of OsPIP1;1, a salt-inducible PIP gene from rice, conferred improved resistance to salinity in Arabidopsis (Guo et al., 2006). Moreover, transgenic bananas overexpressing MusaPIP1;2 demonstrated enhanced tolerance to cold, salinity, and drought compared to wild-type (WT) plants (Sreedharan et al., 2013). While substantial experimental evidence underscores the roles of PIP aquaporins in stress responses, elucidating the precise contributions of individual genes or isoforms is still difficult owing to their functional pleiotropy and integrated regulation under different environmental stimuli (Afzal et al., 2016). Consequently, systematic characterization of PIP family members is imperative to decipher the molecular basis of plant stress resilience.
Salix matsudana is an important afforestation and industrial timber tree species. A previous transcriptome analysis revealed that the expression of SmPIP1;3 was induced by salt stress in the salt-resistance 9901 variety of S. matsudana (Wang et al., 2024). In this study, we aimed to characterize the SmPIP1;3 gene to elucidate its evolutionary attributes and to assess its functional roles in plant growth and environmental stress resistance through genetic transformation. Our findings enrich the existing understanding of the stress resistance process in willow trees and offer a valuable gene resource for future molecular breeding strategies.
Materials and methods
Plant materials and treatment
Cuttings of Salix mandshurica 9901 were cultivated in pots (20 cm × 20 cm) filled with sandy soil in a growth chamber (2,000 lux light intensity, a 16-hour light/8-hour dark cycle, 70% relative humidity, 25°С). Plants were irrigated with water once every 4 days and with Hoagland solution (Hoagland and Arnon, 1950) once a week. After 1 month, they were irrigated with 300 mL of 200 mmol/L NaCl solution per pot. Leaves and roots were harvested at 0 and 24 h after salt application for RNA extraction.
Expression assessment of SmPIP1;3 in willow
The total RNAs were extracted from the leaves and root samples using the polysaccharide polyphenol kit (Tiangen, Beijing). Then, RNA was transcribed into cDNA using the Hifair®III 1st Strand cDNA Synthesis SuperMix for polymerase chain reaction (PCR) kit (Yeasen Biotech, Shanghai, China). Based on the genome sequence data of S. mandshurica, the specific primers were designed for SmPIP1;3 gene fragment (SmPIP1;3F, 5′-TGGCCTTGGTGCTGAGATAA-3′; SmPIP1;3R, 5′-CTCCTTGCTGGGTTAATGCC-3′). The qRT-PCR assay was conducted for expression test with the following procedure: 95°C for 5 min, 40 cycles of 95°C for 10 s, and 58°C for 30 s. Ubiquitin (UBQ) was used as the reference gene (UBQF, 5′-AGAAGGAGTCAGCAACGATG-3′; UBQR, 5′-CATTAGGTTCTGAACAGCAGG-3′).
Gene cloning of SmPIP1;3
Based on the genome data of S. mandshurica, the specific primers were designed at both sides of SmPIP1;3 gene fragment with additional restriction enzyme digestion sites (SmPIP1;3HindIIIF, 5′-cccaagcttATGGAAGGCAAAGAAGAGGA-3′, the lowercase letters mean HindIII digestion bases; SmPIP1;3XbaIR, 5′-tgctctagaTTAAGCTCTGCTCTTGAAAG-3′; the lowercase letters mean XbaI digestion bases). The full-length gene fragment was amplified from the cDNA sample of 9901 willow at 4-day salt treatment and ligated to the pGEM-T easy vector (Promega, Madison, WI, USA) following the manufacturer’s instructions. The recombinant plasmid was transformed into the bacterial strain TOP10, and the positive strain was used for plasmid sequencing.
Sequence characterization of SmPIP1;3
The PIP1 sequences in Populus trichocarpa v3.1 genome were obtained (https://phytozome-next.jgi.doe.gov/info/Ptrichocarpa_v3_1), and homologs of PIP1;3 from seven plant species (P. trichocarpa, S. matsudana, Hevea brasiliensis, Solanum lycopersicum, Vitis vinifera, Oryza sativa, and Zea mays) were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/). Multiple sequence alignment was conducted using the DNAMAN v7.0 software. The phylogenetic tree was performed using MEGA7 with the bootstrap value set at 1,000 repetitions. The online software ProtParam (https://web.expasy.org/protparam/) was used to analyze the physical and chemical properties of SmPIP1;3 proteins. Transmembrane helices were predicted using TMHMM Server v 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
SmPIP1;3 gene transformation to tobacco plants
The recombinant plasmids and gene expression vector pEZR(K)-LC were digested with restriction enzymes HindIII and XbaI and ligated to form the recombinant expression plasmids. The resulting constructs were introduced into Agrobacterium tumefaciens strain GV3101 using the electroporation method. The positive strain was identified and cultured in Yeast Extract Broth (YEB) medium containing 50 mg/L kanamycin and 10 mg/L rifampicin at 28 °C overnight and then transferred to antibiotic-free YEB liquid medium until OD600 0.6–0.8. The healthy tobacco (Nicotiana tabacum L.) leaves were cut and immersed in the bacterial solution for 10 min, transferred to the co-cultivation medium [Murashige and Skoog (MS) medium supplemented with 2 mg/L 6-benzylaminopurine (6-BA), 0.2 mg/L 1-naphthaleneacetic acid (NAA), 3% sucrose, and 0.55% agar] in darkness for 3 days, and transferred to the screening medium (MS with 2 mg/L 6-BA, 0.2 mg/L NAA, 3% sucrose, 0.55% agar, 100 mg/L kanamycin, and 200 mg/L timentin) in light for 30 days. The regenerated buds were transferred to the rooting medium (MS with 3% sucrose, 0.55% agar, 50 mg/L kanamycin, and 200 mg/L timentin) for 20 days to obtain the transgenic seedlings.
Genomic DNA was extracted from the leaves of regenerated plants and screened by PCR tests using the specific primer pair SmPIP1;3HindIIIF/SmPIP1;3XbaIR. Further, the total RNAs were extracted from the DNA-positive lines, reverse-transcribed into cDNA, and assayed for SmPIP1;3 gene expression by RT-PCR. The lines with the overexpression of SmPIP1;3 were recognized as transgenic and used for subsequent assays.
Physiological index tests of tobacco plants
The transgenic and WT tobacco plants were propagated by stem cutting. Seedlings were transplanted into pots (10 cm × 10 cm) filled with vermiculite and maintained in the growth chamber (2,000 lux light intensity, a 16-hour light/8-hour dark cycle, 70% relative humidity, 25°С) for 1 month. The plants were irrigated with water every 3 days and supplied with Hoagland solution once a week. Salt treatment was conducted with 150 mL NaCl solution (200 mmol/L) per pot. Leaves were harvested after 0- and 4-day treatments for physiological index tests. Three replicates for each line/treatment were conducted. Statistical analyses were performed using the SPSS 23.0 software. One-way ANOVA followed by Duncan’s multiple-range test was applied, * and ** indicate significant differences at p < 0.05 and p < 0.01, respectively.
Relative electrolyte leakage (REL): 0.1 g fresh leaves were cut into small pieces (0.5 cm × 0.5 cm), soaked in 30 mL deionized water, and shaken at 25 °C at 180 rpm overnight. The electrical conductivity was measured as R1 using the electrical conductivity meter (DDS-11C). The solutions with the samples were then autoclaved at 121°C for 20 min, cooled naturally to room temperature, and then re-measured as R2. Relative conductivity was calculated as R1/R2.
Malondialdehyde content (MDA): 0.1 g of fresh leaves were homogenized in 1 mL of 10% trichloroacetic acid (TCA). The homogenate was centrifuged at 12,000 rpm for 5 min. The supernatant was collected, mixed with 1 mL of 0.6% thiobarbituric acid (TBA), and boiled for 15 min. After centrifugation at 12,000 rpm for 5 min, the supernatant was collected for absorbance measurement at 450, 532, and 600 nm. MDA content was calculated as follows: [6.45 × (OD532 − OD600) − 0.56 × OD450] × total extract volume/fresh weight of sample.
Results
Expression pattern of SmPIP1;3 in S. matsudana
The expression pattern of SmPIP1;3 in the salt-resistant S. matsudana 9901 variety was examined using gene-specific primers SmPIP1;3-F/SmPIP1;3-R (Figure 1). SmPIP1;3 transcripts were expressed at comparable levels in the leaves and roots under control conditions. When subjected to salt treatment, the SmPIP1;3 transcript abundance significantly increased in both leaf and root tissues by more than 1.30- and 1.33-fold, respectively. This indicates that SmPIP1;3 may be involved in resisting salt stress in S. matsudana.
Figure 1
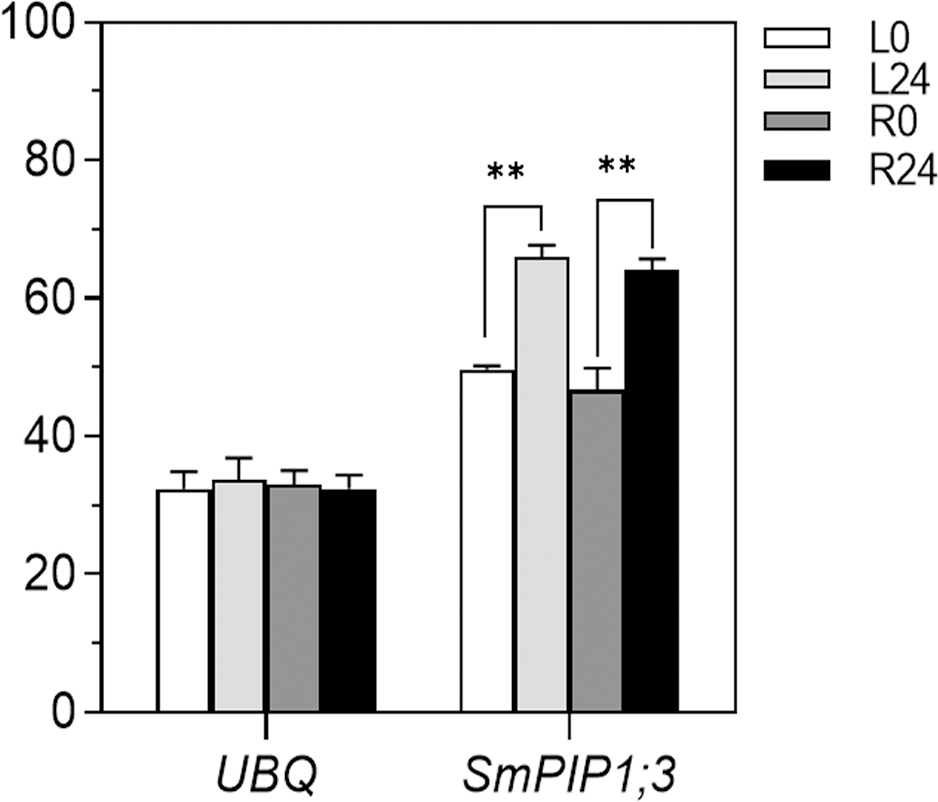
Expression pattern of SmPIP1;3 in 9901 willow under salt treatment. UBQ, ubiquitin gene used as an internal reference for normalization; L0 and L24, leaf samples at 0 and 24 h, respectively, of salt treatment; R0 and R24, root samples at 0 and 24 h, respectively, of salt treatment. ** indicates a statistically significant difference (p < 0.01) between 0 and 24 h of salt treatment.
Characterization of the properties of SmPIP1;3
The full-length fragment of SmPIP1;3 was amplified and cloned from the 9901 cDNA sample with the SmPIP1;3HindIIIF/SmPIP1;3XbaIR primers. The sequencing results revealed a length of 867 bp, encoding 288 amino acids (Figure 2A). Amino acid composition analysis showed that Gly, Ala, and Ile were abundantly present in SmPIP1;3 in proportions of 12.2%, 11.8%, and 8.7%, respectively. The two amino acids that are critical for PIP function, Ser and His, were present in proportions of 4.5% and 2.8%, respectively. SmPIP1;3 had a predicted molecular weight of 30.73 kDa, an isoelectric point of 8.99, an aliphatic index of 96.25, a grand average of hydropathicity of 0.374, and an instability index of 25.07. These parameters indicate that SmPIP1;3 is a basic, moderately hydrophobic, thermostable, and overall stable protein.
Figure 2
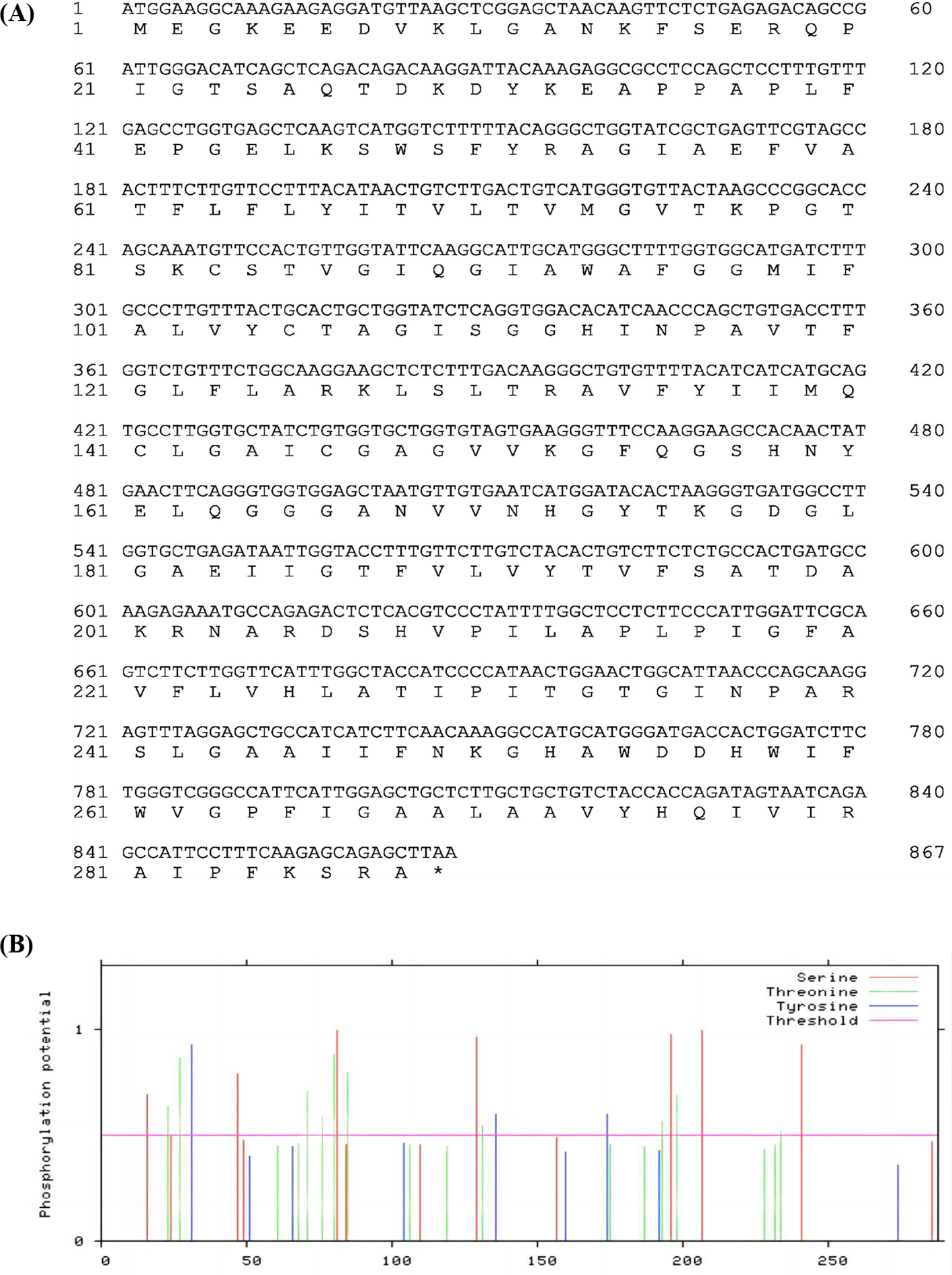
SmPIP1;3 nucleotide and amino acid sequence (A) and phosphorylation amino acids (B). The x-axis represents the amino acid order.
Phosphorylation is a gating mechanism that regulates the plant plasma membrane AQP activity. In SmPIP1;3, 19 putative phosphorylation sites were predicted (Figure 2B), including seven Ser (S) residues (S16, S49, S81, S129, S196, S207, and S241), three Tyr (Y) residues (Y31, Y136, and Y147), and nine Thr (T) residues (T23, T27, T71, T76, T85, T131, T193, T198, and T234), spreading in the protein sequence randomly.
Further sequence alignment revealed a high level of conservation among the PIPs. SmPIP1;3 showed the highest sequence identity (98.61%) with PtPIP1;3 (Figure 3A). Several amino acids were revealed to be specific to the Salicaceae family, such as T82, P84, and G85, or the dicot family, such as D28, A100, E165, F252, D259, and I281. PIP1;3 thus exhibited evolutionary characteristics, and it could be used for species classification.
Figure 3
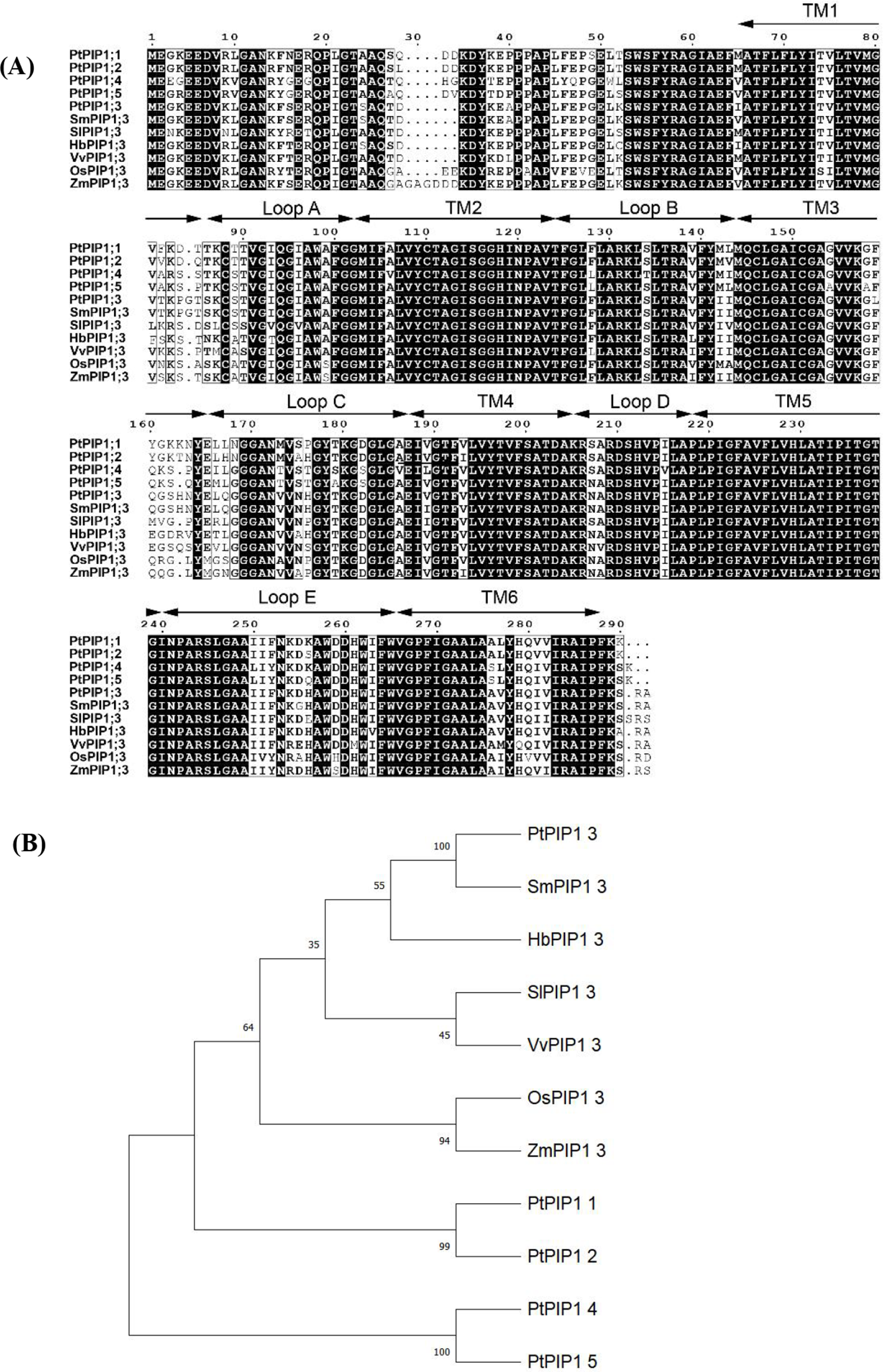
Sequence alignment of plasma membrane intrinsic proteins (PIPs) from seven species (A) and phylogenetic tree (B). Note: Pt, Populus trichocarpa; Sm, Salix matsudana; Sl, Solanum lycopersicum; Hb, Hevea brasiliensis; Vv, Vitis vinifera; Os, Oryza sativa; Zm, Zea mays.
Based on protein sequences from seven species, a phylogenetic tree was constructed (Figure 3B). SmPIP1;3 was found to cluster closely with PtPIP1;3, and they are both members of the Salicaceae family but are far from the other family species. The PIP1;3 proteins from the dicot species were grouped further from those of monocot species, such as maize and rice. All PIP1;3 proteins were grouped and separated from other PIP1 subfamily members, such as PIP1;1, PIP1;2, PIP1;4, and PIP1;5, indicating that each PIP1 subfamily has its own evolutionary identity.
The secondary protein structure prediction showed that SmPIP1;3 mainly included random coils in the proportion of 48.61%, while α-helix, extended chain, and β-bend were in the ratios of 28.82%, 19.79%, and 2.78%, respectively (Figure 4A). Further protein domain analysis revealed SmPIP1;3 as a typical PIP1 aquaporin, containing six transmembrane domains (TM1–TM6) and five interhelical loops (Loop A–Loop E) (Figure 4B). Each α-helix was associated with a membrane-intrinsic connecting loop and a membrane-extrinsic connecting loop on both sides to form an hourglass and a rotating symmetric structure. The transmembrane α-helices were not parallelly arranged but staggered at an angle between 25° and 40°, forming a structure connecting the cells inside and outside.
Figure 4
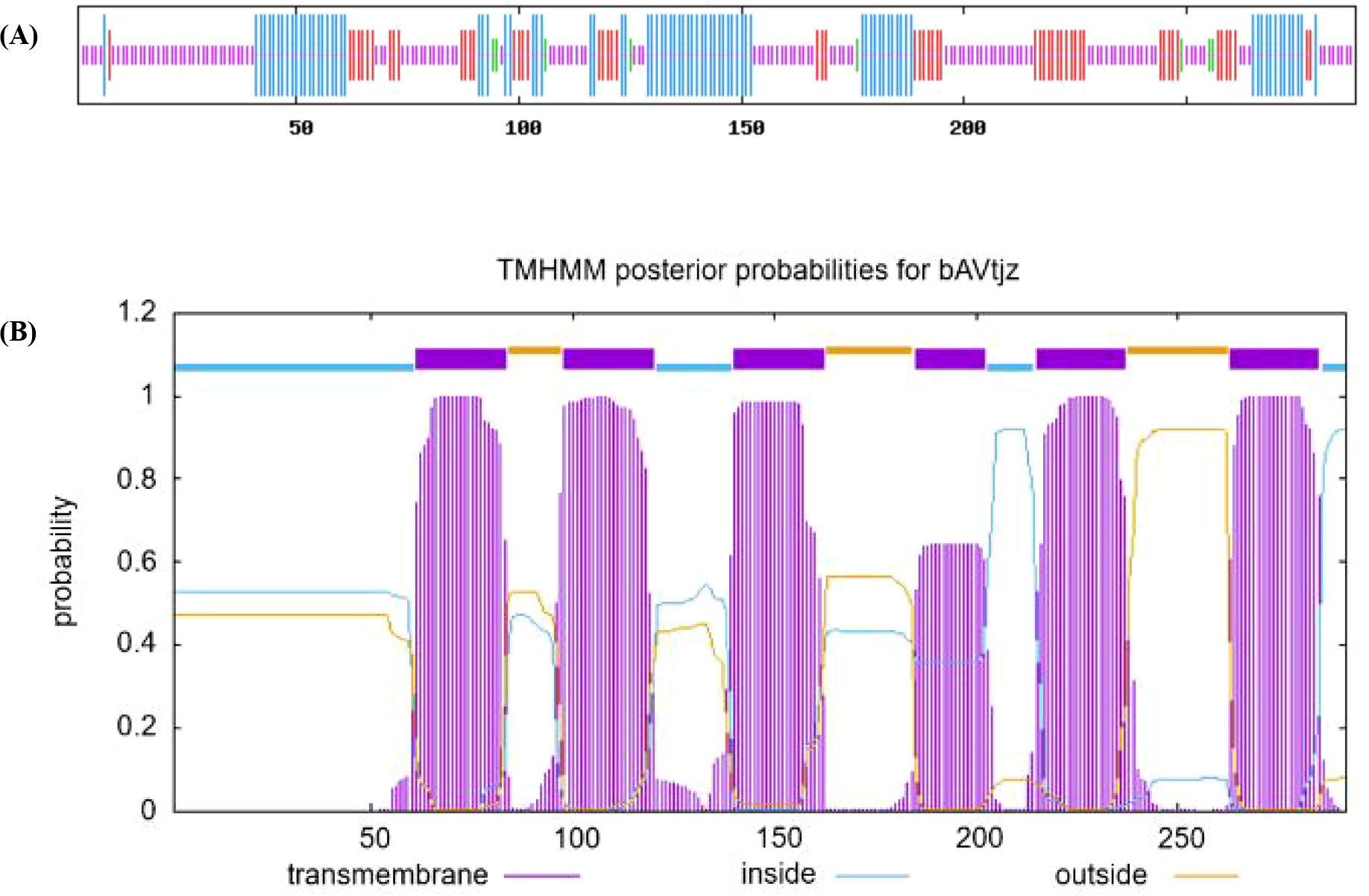
SmPIP1;3 protein secondary structure (A) and transmembrane structural domain (B).
Effect of SmPIP1;3 on plant growth
SmPIP1;3 was successively introduced in tobacco plants, and three independent SmPIP1;3-overexpressed tobacco lines (L1, L2, and L3) were obtained and further propagated (Figure 5C). One-month-old seedlings were used for the growth index evaluation (Figures 5A, B). Under normal conditions, the transgenic lines had a height between 3.0 and 3.8 cm, which was significantly higher than that of WT (2.6 cm). Meanwhile, the transgenic plants’ leaf length ranged from 5.0 to 5.6 cm and leaf width from 3.2 to 4.0 cm—an average increase of 1.15- and 1.16-fold compared to WT, respectively. The root length of the transgenic plants ranged from 11.5 to 12.3 cm—an increase of 1.44- to 1.54-fold over WT. The number of roots, however, was similar in both transgenic plants and WT. In addition, the average fresh weight and dry weight of the transgenic plants were 1.43 and 0.05 g, respectively—1.70- and 2.30-fold increase over WT, respectively (Figure 5D). Collectively, these results demonstrate that SmPIP1;3 overexpression significantly improved plant growth.
Figure 5
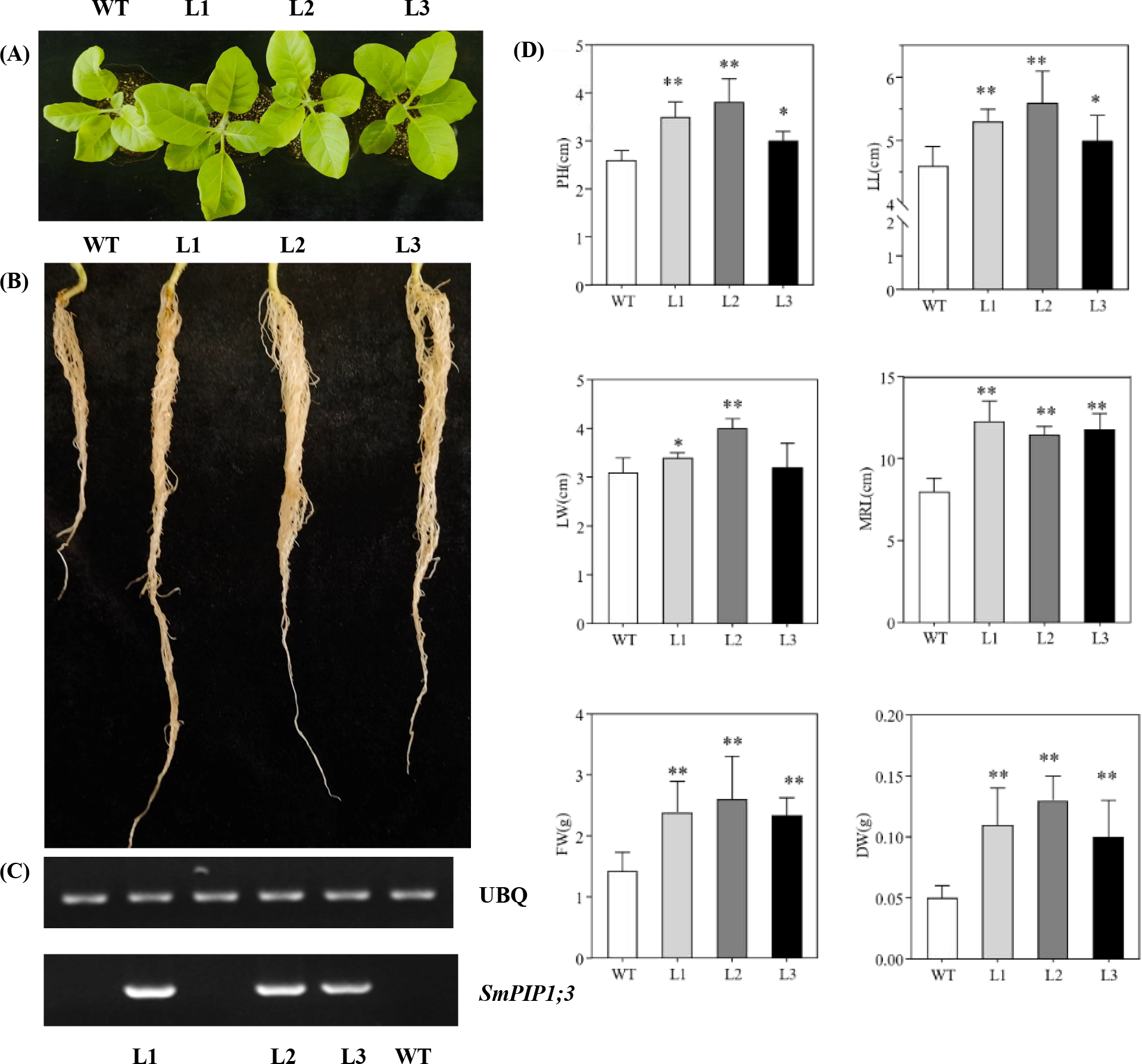
Growth performance of SmPIP1;3-transgenic lines (L1, L2, and L3) and wild-type plants (WT). (A) One-month-old tobacco leaves. (B) One-month-old tobacco roots. (C) RT-PCR detection of SmPIP1;3 in transgenic lines. (D) Growth indexes. PH, plant height; LL, leaf length; LW, leaf width; MRL, maximum root length; FW, fresh weight; DW, dry weight. The data were obtained in three replicates. * and ** mean the significant difference between the transgenic lines and WT at p < 0.05 and p < 0.01, respectively.
SmPIP1;3 enhanced plant resistance to abiotic stresses
Under normal growth conditions, both transgenic and WT tobacco seedlings performed well and showed no obvious morphological abnormalities. When exposed to stress, the WT tobacco plants manifested severe symptoms, including wilted leaves, chlorosis, and turgor loss, while the transgenic plants fared better (Figure 6A). Moreover, the Evans blue staining assays showed no background signal under control conditions; however, after being exposed to stress conditions, a few interspersed blue spots appeared (Figure 6B). Compared with WT, the transgenic lines had less stained dye, especially under salt treatment, indicating reduced cell damage.
Figure 6
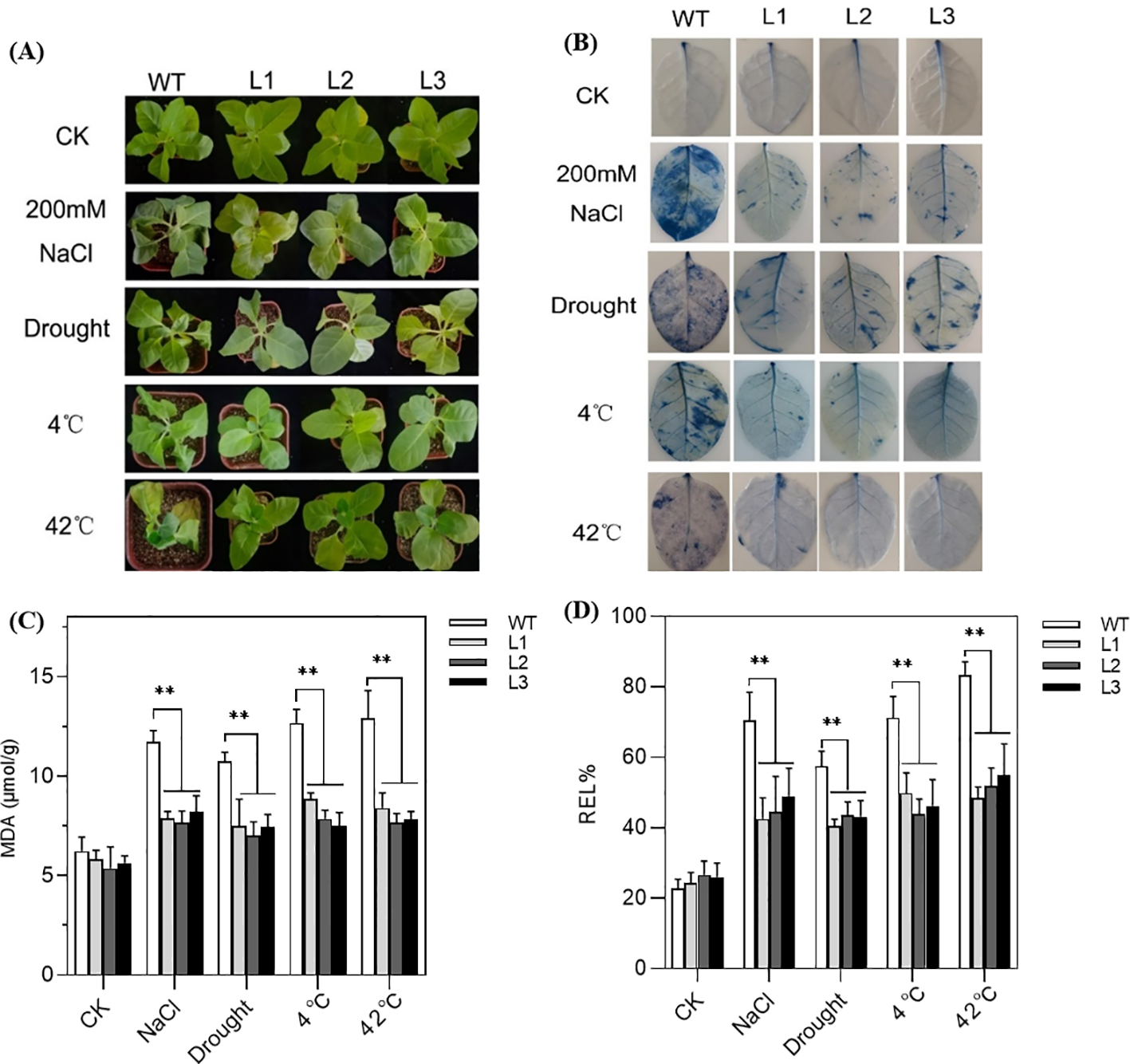
Performance of SmPIP1;3-overexpressing tobacco lines (L1–L3) and wild-type (WT) plants grown under control conditions or exposed to abiotic stresses: 200 mmol/L NaCl for 6 days, drought for 6 days, 4°C for 12 h, or 42°C for 4 days. (A) Morphological performance. (B) Evans blue dyeing. (C) Malondialdehyde content (MDA). (D) Relative electrolyte leakage (REL). ** mean the significant difference between the transgenic tobacco lines and WT at p < 0.01.
Physiological measurements further supported these observations. While no significant differences were detected between transgenic and WT plants under normal conditions, clear differences emerged under stress. Of them, MDA content in the transgenic plants was reduced by 1.48-, 1.47-, 1.57-, and 1.62-fold than WT under salt, drought, cold, and heat treatment, respectively (Figure 6C). Similarly, REL values of the transgenic plants showed decreases of 1.56-, 1.35-, 1.53-, and 1.61-fold compared with WT under the same stress (Figure 6D). Evidently, the overexpression of SmPIP1;3 effectively improves plants’ physiological state and enhances their tolerance to various stresses.
Discussion
PIPs are widely involved in environmental stress resistance
As pivotal regulators of transmembrane water flux, PIPs play crucial roles in plant adaptation to environmental challenges. The members of this aquaporin family not only significantly enhance bidirectional membrane water permeability but also facilitate the transport of glycerol, urea, metalloids, CO2, and H2O2, maintaining water homeostasis, thereby establishing a fundamental adaptive mechanism for abiotic stress tolerance (Gautam and Pandey, 2021; Kumar et al., 2018). Abiotic stresses such as salinity, drought, and cold exposure differentially modulate PIP gene expression profiles. In Arabidopsis thaliana, most PIPs exhibited downregulated expression, except for AtPIP1;3, AtPIP1;4, AtPIP2;1, and AtPIP2;5, whose expression increased after 4 h or 2 days of drought treatment (0.25 mol/L mannitol) (Jang et al., 2004). In Z. mays, salt stress induced a general downregulation of ZmPIP1 and ZmPIP2, while ZmPIP1;1, ZmPIP1;5, and ZmPIP2;4 exhibited transient upregulation patterns (Zhu et al., 2005). In S. lycopersicum, coordinated upregulation of SlPIP1;1, SlPIP1;3, SlPIP1;7, SlPIP2;10, and SlPIP2;12 was observed during salt stress and the recovery phase, implying the critical role of PIPs in regulating water transport (Jia et al., 2020). In O. sativa, the expression level of OsPIP1–3 was upregulated in response to 150 mmol/L salt treatment (Abdelkader et al., 2012). Similarly, this study observed significant induction of SmPIP1;3 transcripts in both leaf and root tissues of the salt-tolerant willow species following 200 mmol/L salt stress. These stress-induced modulation patterns of PIP isoforms indicate their adaptive regulation mechanisms for preserving water balance during abiotic stress.
Comprehensive functional studies employing transgenic approaches, including both overexpression and loss-of-function mutant analyses, have provided compelling evidence for the pivotal role of PIPs in mediating plant responses to abiotic stress (Kumar et al., 2018). For example, SpPIP1 is a drought-induced gene in Stipa purpurea, and its ectopic expression in Arabidopsis bestowed plants with superior drought tolerance as evidenced by longer roots, improved photosynthetic efficiency, increased survival rates, elevated leaf Relative Water Content (RWC), and reduced electrolyte leakage (Chen et al., 2018). Similarly, VfPIP1-overexpressing transgenic Arabidopsis plants exhibited a faster growth rate and enhanced drought resistance by reducing transpiration rates through stomatal closure (Cui et al., 2008). The overexpression of EuPIP1;2 conferred significant drought and salinity tolerance in transgenic tobacco, manifesting as superior germination, root development, and survival rates, which were mechanistically linked to reduced oxidative membrane damage and improved osmotic adjustment. In this study, the overexpression of SmPIP1;3 in transgenic tobacco lines enhanced growth under normal conditions, including taller plant height, larger leaves, longer roots, and increased biomass when compared with the wild type. Notably, these lines demonstrated robust multi-stress tolerance when subjected to salt, drought, cold, and heat challenges, as quantified by substantially reduced membrane damage indicators compared to wild-type controls.
These findings highlight PIP aquaporins’ functions as master regulators of abiotic stress responses, acting through mechanisms such as maintenance of membrane integrity, facilitation of osmotic adjustment, and promotion of stress-resilient growth. Further investigations are required to elucidate the regulatory networks and signaling pathways by which PIPs mediate these adaptive functions.
SmPIP1;3 displays pleiotropism against various stresses
Plant abiotic stress tolerance relies on complex molecular networks in which different genes contribute through distinct mechanisms. For example, expansin genes have been demonstrated to relax the cell wall components and release stress. The overexpression of the S. matsudana expansin gene SmEXPA23 in tobacco decreased REL and MDA contents by 9.87% and 19.26%, respectively, compared with WT plants under salt treatment (Yang et al., 2023). Similarly, DNA damage repair tolerance (DRT) genes can promote the repair of stress-induced DNA damage, thus maintaining cell viability. The overexpression of ZsDRT102 from Zelkova schneideriana significantly improved the plants’ chilling resistance, with MDA and REL levels decreasing by 10.3%–26.2% and 24.5%–35.3%, respectively, in transgenic plants relative to WT (Wang et al., 2025). PIPs constitute an efficient gene resource that can facilitate stress resistance by improving the water channel of the cell membrane (Maurel et al., 2008). In this study, the overexpression of SmPIP1;3 in tobacco alleviated stress-induced injury by salt, drought, cold, or heat stress: the REL values declined by 1.56-, 1.35-, 1.53-, and 1.61-fold, and the MDA content fell by 1.48-, 1.47-, 1.57-, and 1.62-fold compared with WT, together with reduced Evans blue staining, indicating diminished cell damage.
Although pleiotropic genes are common, their stress spectra and directions differ markedly. PttEXPA8 from Populus tomentosa was introduced into tobacco plants, and it improved the plants’ resistance to drought, heat, and cold; however, it had no significant contribution to salt and Cd stress resistance (Zhang et al., 2019). Cotton GhWRKY25 elevated salt tolerance but reduced drought resistance (Liu et al., 2016). Moreover, the overexpression of AtbZIP60 in tobacco enhanced its salt tolerance (Tang et al., 2012), but the overexpression of wheat TabZIP6 and tea CsbZIP6 in Arabidopsis lowered the plants’ tolerance to cold (Wang et al., 2017), thus suggesting that the homologous genes play different roles in different stress conditions. SmPIP1;3 had a broad-spectrum resistance to all four tested stresses, making it a more versatile candidate for molecular breeding. The synergistic potential of these multi-mechanism genes warrants further exploration.
PIPs’ function depends on their sequences and structures
Numerous studies have indicated that AQP activity is modulated by phosphorylation (Chaumont et al., 2005), which may affect AQP’s trafficking through the secretory pathway to the plasma membrane or the opening and closing of pores (Törnroth-Horsefield et al., 2006). In particular, the phosphorylation at Ser residues within the N- or C-terminal regions of PIP was more closely examined and recognized as crucial to the protein function. For example, phosphorylation at S115, S188, and S274 of SoPIP2;1 promoted the expansion of water-conducting pores in spinach (Johansson et al., 1996). In wheat, TaPIP2;10 phosphorylated at S280 facilitated CO2 transport into the cells, thereby resulting in enhanced photosynthesis and increasing yield, whereas the same protein phosphorylated at S121 was related to H2O2 transport into the cytoplasm (Lu et al., 2022). Several mutant assays have also confirmed the role of serine phosphorylation. The substitution of S115E in SoPIP2;1 led to a half-turn extension of transmembrane helix 1, thereby disrupting the divalent cation binding site in the gating mechanism (Nyblom et al., 2009). By contrast, the S188E mutant increased the water transport activity and supported the contribution of S188 phosphorylation to open channel formation. In our study, 19 phosphorylation sites in SmPIP1;3—seven Ser, three Tyr, and nine Thr—were determined. A sequence alignment among SmPIP1;3 and the PIPs from seven species revealed that most of them were conserved. A previous study found that the homologous mutations of S129 and S207 in ZmPIP2;1, S126A, and S203A did not affect the targeting of the plasma membrane but decreased its activity by 30% to 50% (Wilder et al., 2008). Further exploration of the changes in amino acid phosphorylation in SmPIP1;3 is necessary to better understand how the water channel activity responds to various growth conditions.
The loop feature was also recognized as an important factor for the functioning of the PIP channel. One study revealed that loop C residues contribute to the selectivity of solute transport (Wilder et al., 2008). We compared the residues of the PIP1 proteins and found that loop C contained a large number of Gly residues (6–8), indicating that these PIP1s are much more flexible members (Casino et al., 2014). The sequence alignment of PIP1;3 among the seven species also demonstrated more polymorphism in loop C than the others. These performances may support the diversity of PIP1;3 against various environmental stresses, like the transported solutes. More research attention on loop C and the other domains is essential for future gene modification and molecular breeding programs.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XX: Funding acquisition, Investigation, Methodology, Writing – original draft. PY: Investigation, Methodology, Writing – original draft. LW: Investigation, Writing – original draft. XL: Investigation, Writing – original draft. JX: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 32401296).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abdelkader A. F. El-Khawas S. El-Din El-Sherif N. A. S. Hassanein R. A. Emam M. A. Hassan R. E. S. (2012). Expression of aquaporin gene (OsPIP1-3) in salt-stressed rice (Oryza sativa L.) plants pre-treated with the neurotransmitter (dopamine). Plant Omics5, 532–541.
2
Afzal Z. Howton T. C. Sun Y. Mukhtar M. S. (2016). The roles of aquaporins in plant stress responses. J. Dev. Biol.4, 9. doi: 10.3390/jdb4010009
3
Alexandersson E. Fraysse L. Sjövall-Larsen S. Gustavsson S. Fellert M. Karlsson M. et al . (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol.59, 469–484. doi: 10.1007/s11103-005-0352-1
4
Casino P. Miguel-Romero L. Marina A. (2014). Visualizing autophosphorylation in histidine kinases. Nat. Commun.5, 3258. doi: 10.1038/ncomms4258
5
Chaumont F. Moshelion M. Daniels M. J. (2005). Regulation of plant aquaporin activity. Biol. Cell97, 749–764. doi: 10.1042/BC20040133
6
Chen Q. Yang S. Kong X. Wang C. Xiang N. Yang Y. et al . (2018). Molecular cloning of a plasma membrane aquaporin in Stipa purpurea, and exploration of its role in drought stress tolerance. Gene665, 41–48. doi: 10.1016/j.gene.2018.04.056
7
Cui X. H. Hao F. S. Chen H. Chen J. Wang X. C. (2008). Expression of the Vicia faba VfPIP1 gene in Arabidopsis thaliana plants improves their drought resistance. J. Plant Res.121, 207–214. doi: 10.1007/s10265-007-0130-z
8
Eisenbarth D. A. Weig A. R. (2005). Dynamics of aquaporins and water relations during hypocotyl elongation in Ricinus communis L. seedlings. J. Exp. Bot.56(417), 1831–1842. doi: 10.1093/jxb/eri173
9
Gautam A. Pandey A. K. (2021). Aquaporins responses under challenging environmental conditions and abiotic stress tolerance in plants. Botanical Rev.87, 467–495. doi: 10.1007/s12229-021-09249-z
10
Gill R. A. Ahmar S. Ali B. Saleem M. H. Khan M. U. Zhou W. et al . (2021). The role of membrane transporters in plant growth and development, and abiotic stress tolerance. Int. J. Mol. Sci.22, 12792. doi: 10.3390/ijms222312792
11
Guo L. Wang Z. Y. Lin H. Cui W. E. Chen J. Liu M. et al . (2006). Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res.16, 277–286. doi: 10.1038/sj.cr.7310035
12
Hoagland D. R. Arnon D. I. (1950). The water-culture method for growing plants without soil. (Berkeley: California Agricultural Experiment Station, University of California).
13
Horie T. Kaneko T. Sugimoto G. Sasano S. Panda S. K. Shibasaka M. et al . (2011). Mechanisms of water transport mediated by PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell Physiol.52, 663–675. doi: 10.1093/pcp/pcr027
14
Jang J. Y. Kim D. G. Kim Y. O. Kim J. S. Kang H. (2004). An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol. Biol.54, 713–725. doi: 10.1023/B:PLAN.0000040900.61345.a6
15
Jia J. Liang Y. Hou T. Hu Y. Zhu Y. Huo H. et al . (2020). The expression response of plasma membrane aquaporins to salt stress in tomato plants. Environ. Exp. Bot.178, 104190. doi: 10.1016/j.envexpbot.2020.104190
16
Johansson I. Larsson C. Ek B. Kjellbom P. (1996). The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell8, 1181–1191.
17
Kaldenhoff R. Fischer M. (2006). Functional aquaporin diversity in plants. Biochim. Biophys. Acta (BBA) - Biomembranes1758, 1134–1141. doi: 10.1016/j.bbamem.2006.03.012
18
Kumar K. Mosa K. A. Meselhy A. G. Dhankher O. P. (2018). Molecular insights into the plasma membrane intrinsic proteins roles for abiotic stress and metalloids tolerance and transport in plants. Indian J. Plant Physiol.23, 721–730. doi: 10.1007/s40502-018-0425-1
19
Li J. Ban L. Wen H. Wang Z. Dzyubenko N. Chapurin V. et al . (2015). An aquaporin protein is associated with drought stress tolerance. Biochem. Biophys. Res. Commun.459, 208–213. doi: 10.1016/j.bbrc.2015.02.052
20
Liu X. Song Y. Xing F. Wang N. Wen F. Zhu C. (2016). Ghwrky25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma253, 1265–1281. doi: 10.1007/s00709-015-0885-3
21
Lu K. Chen X. Yao X. An Y. Wang X. Qin L. et al . (2022). Phosphorylation of a wheat aquaporin at two sites enhances both plant growth and defense. Mol. Plant15, 1772–1789. doi: 10.1016/j.molp.2022.10.003
22
Maurel C. Verdoucq L. Luu D. T. Santoni V. (2008). Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol.59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
23
Nyblom M. Frick A. Wang Y. Ekvall M. Hallgren K. Hedfalk K. et al . (2009). Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating. J. Mol. Biol.387, 653–668. doi: 10.1016/j.jmb.2009.01.065
24
Rahman A. Kawamura Y. Maeshima M. Uemura M. (2020). Plasma membrane aquaporin members PIPs act in concert to regulate cold acclimation and freezing tolerance responses in Arabidopsis thaliana. Plant Cell Physiol.61, 787–802. doi: 10.1093/pcp/pcaa005
25
Sohail H. Noor I. Nawaz M. A. Ma M. R. Shireen F. Huang Y. et al . (2022). Genome-wide identification of plasma-membrane intrinsic proteins in pumpkin and functional characterization of CmoPIP1–4 under salinity stress. Environ. Exp. Bot.202, 104995. doi: 10.1016/j.envexpbot.2022.104995
26
Sreedharan S. Shekhawat U. K. S. Ganapathi T. R. (2013). Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1;2 display high tolerance levels to different abiotic stresses. Plant Biotechnol. J.11, 942–952. doi: 10.1111/pbi.12086
27
Suslov M. Daminova A. Egorov J. (2024). Real-time dynamics of water transport in the roots of intact maize plants in response to water stress: The role of aquaporins and the contribution of different water transport pathways. Cells13, 154. doi: 10.3390/cells13020154
28
Tang W. Page M. Fei Y. Liu L. Xu F. Cai X. et al . (2012). Overexpression of AtbZIP60deltaC gene alleviates salt-induced oxidative damage in transgenic cell cultures. Plant Mol. Biol. Rep.30, 1183–1195. doi: 10.1007/s11105-012-0437-3
29
Törnroth-Horsefield S. Wang Y. Hedfalk K. Johanson U. Karlsson M. Tajkhorshid E. et al . (2006). Structural mechanism of plant aquaporin gating. Nature439, 688–694. doi: 10.1038/nature04316
30
Wang L. Cao H. Qian W. Yao L. Hao X. Li N. et al . (2017). Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic Arabidopsis. Ann. Bot.119, 1195–1209. doi: 10.1093/aob/mcx011
31
Wang X. Gong L. Zhang J. Wang L. Wu D. Xu J. (2024). The genome-wide profiling of alternative splicing in willow under salt stress. Forests15, 30. doi: 10.3390/f15010030
32
Wang L. Liu X. Gong L. Hu J. Lu X. Xu J. (2025). Overexpression of ZsDRT102 from Zelkova schneideriana increases the chilling resistance of plants. Plant Cell Tissue Organ Culture160, 9. doi: 10.1007/s11240-024-02947-7
33
Wilder V. V. Miecielica U. Degand H. Derua R. Waelkens E. Chaumont F. (2008). Maize plasma membrane aquaporins belonging to the PIP1 and PIP2 subgroups are in vivo phosphorylated. Plant Cell Physiol.49, 1364–1377. doi: 10.1093/pcp/pcn112
34
Yang R. Yang L. Wang X. Wang Y. Zhang J. Xu J. (2023). Over-expression of the Salix matSudana expansin gene SmEXPA23 enhances plant salt tolerance. Plant Cell Tissue Organ Culture152, 309–316. doi: 10.1007/s11240-022-02407-0
35
Yu Q. Hu Y. Li J. Wu Q. Lin Z. (2005). Sense and antisense expression of plasma membrane aquaporin BnPIP1 from Brassica napus in tobacco and its effects on plant drought resistance. Plant Sci.169, 647–656. doi: 10.1016/j.plantsci.2005.04.013
36
Zhang H. Liu H. Yang R. Xu X. Liu X. Xu J. (2019). Over-expression of PttEXPA8 gene showed various resistances to diverse stresses. Int. J. Biol. Macromolecules130, 50–57. doi: 10.1016/j.ijbiomac.2019.02.115
37
Zhu C. Schraut D. Hartung W. Schaffner A. R. (2005). Differential responses of maize MIP genes to salt stress and ABA. J. Exp. Bot.56, 2971–2981. doi: 10.1093/jxb/eri294
Summary
Keywords
plasma membrane intrinsic proteins (PIPs), Salix matsudana , SmPIP1;3, overexpression, abiotic stress tolerance
Citation
Xu X, Yin P, Wang L, Lu X and Xu J (2025) Overexpression of the Salix matsudana aquaporin gene SmPIP1;3 enhances plant resistance to abiotic stresses. Front. Plant Sci. 16:1685356. doi: 10.3389/fpls.2025.1685356
Received
13 August 2025
Accepted
28 September 2025
Published
14 October 2025
Volume
16 - 2025
Edited by
Kai-Hua Jia, Shandong Academy of Agricultural Sciences, China
Reviewed by
Hongwei Cai, China Agricultural University, China
Guanghuai Jiang, Chinese Academy of Sciences (CAS), China
Updates
Copyright
© 2025 Xu, Yin, Wang, Lu and Xu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jichen Xu, jcxu282@sina.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.