- State Key Laboratory of Tropical Crop Breeding, Coconut Research Institute, Chinese Academy of Tropical Agricultural Sciences, Sanya/Wenchang, Hainan, China
Introduction: Climate change has exacerbated cold stress, which severely impairs plant development. Oil palm (Elaeis guineensis), a tropical crop highly sensitive to low temperatures, exhibits stunted growth and yield reductions under such conditions.
Methods: To investigate its cold stress response, oil palm seedlings were subjected to cold treatments, and their physiological and genetic adaptations were analyzed using fresh leaf samples. Key parameters, including antioxidant enzyme activity, reactive oxygen species (ROS) levels, photosynthetic pigment ratios, photosynthetic efficiency, and gene expression, were evaluated across exposure durations. Sequencing of the samples was performed using Illumina NovaSeq X Plus platform. Raw reads were processed using fastp (v0.18.0) to remove adapter-containing reads, exclude reads with >10% unidentified nucleotides (N), and eliminate reads where >50% of bases had Phred scores ≤20. The genome reference version is GCF_000442705.2 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000442705.2/).
Results and discussion: Under cold stress, seedlings displayed a significant increase in superoxide dismutase (SOD, 546.08 U/g min FW) and peroxidase (POD, 153.27 U/g min FW) activities within 4 h compared with the control. Prolonged exposure (8 h) further elevated soluble sugar content (406.27 μg/g FW), malondialdehyde (MDA, 80.22 nmol/g), relative electrical conductivity (109.71%), and the carotenoid-to-chlorophyll ratio, indicating oxidative damage and membrane instability. RNA-seq analysis identified 144, 392, and 6,585 differentially expressed genes (DEGs) after 1, 4, and 8 h of cold exposure, respectively. KEGG pathway enrichment highlighted predominant associations with plant–pathogen interaction, plant hormone signal transduction, and the mitogen-activated protein kinase (MAPK) signaling pathway. Functional analysis revealed DEGs involved in four major hormone signaling pathways (auxin (AUX/IAA), jasmonic acid (JA), abscisic acid (ABA), and brassinosteroid (BR)), which also interact with the MAPK cascade to collectively regulate oil palm cold stress adaptation and growth adjustments. This study provides comprehensive insights into the genetic and molecular mechanisms underlying cold tolerance in oil palm, offering a basis for breeding cold-resistant cultivars.
1 Introduction
Elaeis guineensis Jacq., commonly referred to as oil palm, is a long-lived woody monocot in the Arecaceae family. Recognized as the most productive oil crop worldwide, it plays a major role in the global vegetable oil industry. Palm oil, extracted from its fruit, is the most widely consumed edible oil, contributing nearly 35% to total global vegetable oil production, which surpassed 70 million metric tons in 2023 (Abubakar and Ishak, 2024; VanderWilde et al., 2023). The oil palm industry forms a high-value agricultural supply chain encompassing cultivation, oil extraction, and downstream processing for applications in food, cosmetics, biofuels, and other sectors (Al-Madani et al., 2023; Yılmaz and Ağagündüz, 2022; Chaparro et al., 2023). Malaysia and Indonesia dominate global production, jointly contributing over 85% of palm oil output (Danylo et al., 2021). Beyond economic significance, oil palm plantations support livelihoods for millions across Southeast Asia, Africa, and Latin America (Lai et al., 2022). Currently, the domestic cultivation of oil palm in China has not yet developed into a large-scale industry, with a total planting area of approximately 667 hectares scattered across regions such as Hainan, Yunnan, Guangdong, Guangxi, and Fujian. Expanding oil palm cultivation could significantly increase China’s self-sufficiency rate in edible oils, thereby enhancing national food security. It would also improve the efficiency of land resource utilization, boost farmers’ incomes, and promote regional economic development. Moreover, the growth of the oil palm industry could drive progress in related sectors such as agricultural technology, seed breeding, oil processing, and bio-new energy applications (John Martin et al., 2022). In China, oil palm cultivation is limited to tropical regions, and commercial expansion has been constrained by climatic limitations (particularly cold sensitivity) and suboptimal yields, necessitating heavy reliance on imports (Sarkar et al., 2020; Zhou et al., 2022).
Cold stress significantly hinders crop growth, development, and productivity and induces complex physiological responses. It suppresses growth processes, impairing seed germination, seedling establishment, flowering, and fruit set, ultimately leading to reduced germination rates, stunted seedling growth, and diminished seed set (Shahan, 2020). These adverse effects are especially detrimental to crops in tropical and subtropical regions, often causing substantial yield losses and declines in quality (Matsukura et al., 2024). At the cellular level, cold stress promotes the excessive accumulation of ROS, including superoxide (O2–), hydrogen peroxide (H2O2), and hydroxyl radicals (•OH), which induce membrane lipid peroxidation and damage to proteins and DNA (Xiang et al., 2025). To counteract oxidative stress, plants activate antioxidant defenses involving key enzymes such as Superoxide Dismutase (SOD), peroxidase (POD), and catalase (CAT) (Amini et al., 2021). Notably, at low concentrations, ROS act as signaling molecules that activate pathways such as MAPK and calcium (Ca2+) signaling cascades to regulate cold-responsive gene expression (Zhang et al., 2023).
Cold temperatures also disrupt the balance between plant growth and stress tolerance by activating a complex hormonal signaling network. Key hormones such as ABA, BR, and auxin (IAA) play pivotal roles in regulating these adaptive responses (Waadt et al., 2022). This hormonal coordination facilitates defense mechanisms through interconnected ROS–hormone networks, including the ABA–ROS–CBF signaling axis. Deciphering cold signal perception and multi-hormone crosstalk is essential for uncovering novel targets to engineer stress-resilient crops (Li et al., 2022; Shi and Chan, 2014).
Transcriptome sequencing has emerged as a pivotal tool in plant science, providing comprehensive insights into gene expression dynamics across developmental stages, environmental stresses, and treatments (Liu et al., 2022). In oil palm, this technology has advanced genetic improvement by enabling the discovery of genes associated with fatty acid biosynthesis, cold stress responses, and complex metabolic pathways (Lei et al., 2014; Xiao et al., 2014; Li et al., 2020, 2022; Saand et al., 2022; Apriyanto and Ajambang, 2022; Lee et al., 2024);. Despite progress in high-throughput sequencing, the molecular mechanisms underlying cold adaptation remain incompletely characterized in oil palm, and the identification and functional analysis of cold-tolerance markers and pathways are still limited. This gap impedes molecular breeding for high-yielding, cold-resilient varieties. Here, we employ high-throughput transcriptome sequencing to dissect gene regulatory networks in oil palm under cold stress and identify key molecular components governing stress adaptation, with the goal of establishing a molecular framework for genetic enhancement of this crop.
2 Materials and methods
2.1 Experimental material and cold exposure
Six-month-old African oil palm (Elaeis guineensis var. pisifera) seedlings were obtained from Wenchang, Hainan Province, China (19.63° N, 110.94° E) and acclimatized under nursery conditions. Oil palm seeds were initially germinated in a growth chamber under controlled conditions: a constant temperature of 29°C, 85% relative humidity, and a 12-hour photoperiod. Following germination, the sprouts were moved to a pre-nursery stage and placed in small polyethylene pots filled with a sterile peat–perlite mixture (3:1 v/v). At the two-leaf stage, the seedlings were transplanted into larger bags (35 cm × 50 cm) in the main nursery. The nursery environment was maintained at an average temperature of 28–32°C under natural sunlight. Seedlings received daily watering and were fertilized every two weeks using a balanced N-P-K fertilizer (mass ratio: 15-15-15). The bags were arranged in a triangular pattern with 90 cm spacing between them to ensure sufficient air circulation and light exposure. Twenty-four uniformly healthy seedlings were randomly divided into two experimental groups (n = 12 per group): (1) a control group maintained at 26°C with a 16 h light/8 h dark photoperiod, and (2) a treatment group exposed to 8°C for 1, 4, or 8 h under identical light (250 µmol/m2 s) and relative humidity (72-75%) conditions. The plant growth chamber (Baowen YL1, Shanghai, China) is equipped with built-in fluorescent lamps, whose light intensity is monitored in real time using a quantum sensor and automatically adjusted by the control system through regulating the lamp output. The chamber also incorporates humidification and dehumidification systems, which dynamically modulate relative humidity based on real-time feedback from high-precision humidity sensors, maintaining it within the range of 72–75%. Cold stress was induced in oil palm seedlings by exposing them to an artificial climate chamber pre-set to 8°C. All treatments were done at the same time of day to control the circadian effects. Each group had three biological replicates (one seedlings per replicate). Immediately following treatment, spear leaves were collected from both control and cold-treated seedlings, flash-frozen in liquid nitrogen, and stored at –80°C for RNA isolation and transcriptomic analysis.
2.2 Physiological and biochemical analyses
To evaluate cold stress responses, leaf samples were collected from the treated and control plants at 0, 1, 4, and 8 h post-exposure. Antioxidant enzyme activities, including SOD and POD, were determined using commercial assay kits (Micro SOD/POD Activity Assay Kits, Sangon Biotech, Shanghai, China). Photosynthetic parameters, including chlorophyll a/b ratio, carotenoid/chlorophyll ratio, net photosynthetic rate (Pn), intercellular CO2 concentration (Ci), and stomatal conductance (Gs), were measured according to established protocols (Wang et al., 2015; Porra, 2002). Stress-related metabolites were quantified as follows: soluble sugars (Teixeira et al., 2012), MDA (Micro MDA Assay Kit, Sangon Biotech), and relative electrical conductivity (REC) (He et al., 2018). To estimate leaf cell membrane damage, REC was measured using an electric conductivity meter EC215 (Hanna Instruments Romania Srl, Nusfalau, Romania) (Niu et al., 2016). REC (%) was calculated the following equation, REC (%) = (S1/S2) × 100, where S1 and S2 refer to electric conductivity of live leaf sections and boiled leaf sections, respectively.
2.3 RNA extraction and transcriptome analysis
Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA quality was evaluated by RNase-free agarose gel electrophoresis and quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Poly(A)+ mRNA was enriched using mRNA capture beads and fragmented at elevated temperature. First-strand cDNA was synthesized using reverse transcriptase, followed by second-strand synthesis with end repair and A-tailing. Adapters were ligated to the cDNA fragments, which were then size-selected using Hieff NGS DNA Selection Beads. The final library was prepared by PCR amplification and sequenced on the Illumina NovaSeq X Plus platform. Raw reads were processed using fastp (v0.18.0) to remove adapter-containing reads, exclude reads with >10% unidentified nucleotides (N), and eliminate reads where >50% of bases had Phred scores ≤20. HISAT2 was used before StringTie assembly. To validate the consistency and reproducibility among biological replicates, we conducted a principal component analysis (PCA) of all samples.
Transcript quantification used a reference-based approach, with mapped reads from each sample assembled via StringTie (v1.3.1). Expression levels were quantified as FPKM (Fragments Per Kilobase of transcript per Million mapped reads) using RSEM, enabling standardized comparison of transcript abundance across samples. The FPKM calculation is: FPKM = (106 × C)/(N × L/103). where C is the number of fragments aligned to gene i, N is the total number of fragments mapped to all reference genes, and L is the length of gene i (bp). Differential expression analysis used DESeq2. Transcripts meeting false discovery rate (FDR) < 0.05 and |log2 fold change| ≥ 1 (linear fold change ≥ 2) were classified as DEGs. Gene Ontology (GO) enrichment DEGs were mapped to GO terms (http://www.geneontology.org/), and enrichment was evaluated using the hypergeometric test with FDR correction (FDR ≤ 0.05).
2.4 KEGG pathway enrichment
KEGG enrichment used the same hypergeometric framework as GO, with FDR correction (FDR ≤ 0.05). Protein–protein interaction (PPI) analysis was conducted using STRING v12.0 (https://string-db.org/) with an interaction confidence cutoff ≥ 0.700.
2.5 Expression profiles of DEGs by qRT-PCR
Eight randomly selected DEGs associated with cold stress (primers in Supplementary Table 1) were validated by qRT-PCR. Total RNA was extracted using TRIzol (Invitrogen), and first-strand cDNA was synthesized from 1 µg of RNA using MightyScript First Strand cDNA Synthesis Master Mix (Sangon Biotech) with oligo(dT)18 primers. qRT-PCR was performed in 10 µL reactions using 2 × SYBR Green qPCR ProMix (low ROX) on a Mastercycler ep realplex4 system under the following conditions: 95°C for 5 s, 55°C for 15 s, and 68°C for 20 s, followed by a melt curve from 60°C to 95°C over 20 min. All reactions were run in triplicate at both biological and technical levels. The expression levels of genes were normalized to those of ELF, which was previously found to be a stable reference gene under abiotic stress (Xia et al., 2014). Gene expression changes were calculated using the 2–ΔΔCt method (Livak et al., 2001), with final Ct values representing the means of nine measurements. Statistical significance (p < 0.05) under cold treatments was assessed by one-way ANOVA with LSD post hoc tests (SPSS).
3 Results
3.1 Cold stress alters physiological and biochemical parameters in oil palm
Plants were subjected to cold treatment for 0, 1, 4, and 8 h. Key physiological and biochemical parameters were analyzed, including antioxidant enzyme activities, ROS-related damage (MDA), soluble sugars, pigment content (chlorophyll and carotenoids), and gas exchange traits. SOD and POD activities increased during cold exposure, peaking at 4 h and decreasing by 8 h (Figures 1A, B). MDA content, soluble sugars, and REC increased progressively (Figures 1C–E). Conversely, chlorophyll content, Ci, Gs, and Pn declined (Figures 1F–I). Carotenoid/chlorophyll ratios were higher at 4 h relative to the control (Figure 1J). The phenotype of oil palm seedlings exhibited minimal change compared to the 0-hour baseline following one hour of cold stress. After four hours, however, brown spots emerged on the leaves, suggesting the onset of chilling injury. Within eight hours, brown spots had spread across nearly the entire leaf surface, indicating severe cold damage (Figure 1P).
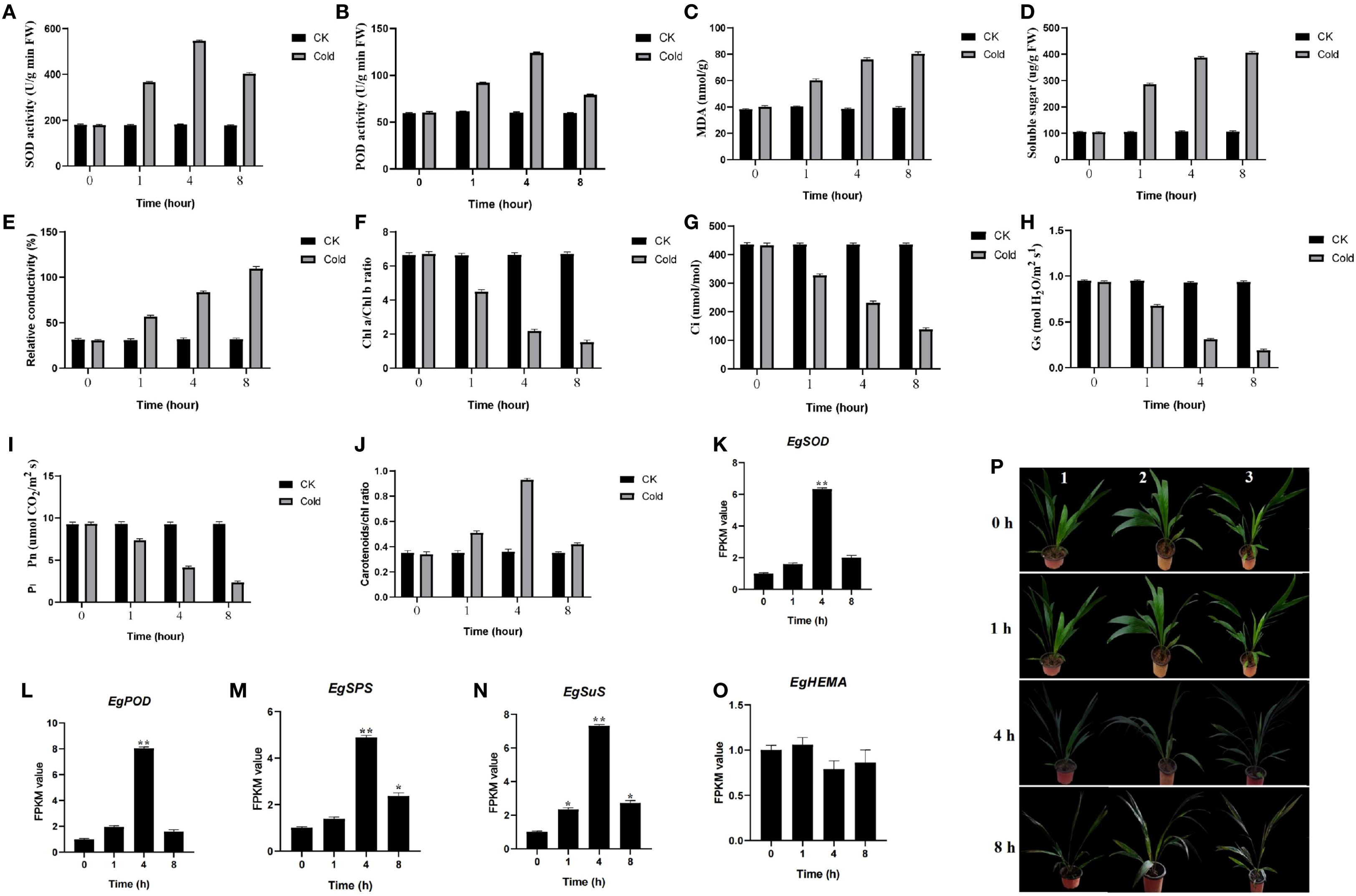
Figure 1. Effects of cold stress on the growth phenotype and physiological indices of oil palm seedlings. (A–J) Assessment of physiological and biochemical parameters in oil palm subjected to control and cold stress conditions at 0, 1, 4, and 8 hours of exposure. Data are presented as mean ± SD (n = 3). (K–N) FPKM value of SOD, POD, SPS, SuS, and HEMA genes under cold stress conditions. Asterisks represent significant difference at p ≤ 0.05(*) and p ≤ 0.01(**). (P) Growth phenotypes of seedlings from different treatment groups after the stress period. 1–3 means three biological replicates.
3.2 Cold stress induces differential transcriptional reprogramming in oil palm
Libraries generated 42.03–46.14 million high-quality paired-end reads (2 × 150 bp) per sample (~6.26 GB per replicate) with an average Q30 of 94.54% and a clean-reads percentage of 88.49% (Supplementary Table 2). The length of the DNA insertion fragment is 200–300 bp. The genome reference version is GCF_000442705.2 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000442705.2/). Employing thresholds of FDR < 0.05 and |fold change| ≥ 2, we identified DEGs across three pairwise comparisons (CK 1 h vs Cold 1 h, CK 4 h vs Cold 4 h, and CK 8 h vs Cold 8 h). The number of DEGs increased progressively with the duration of cold exposure, reaching a maximum of 6,585 DEGs in the CK 8 h vs Cold 8 h comparison (Figures 2A, B), reflecting a time-dependent expansion of transcriptional reprogramming. Venn analysis uncover both time-point-specific and shared DEGs, among which 68 were consistently regulated across all cold treatments, suggesting their role as potential core cold-response regulators (Figure 2C). Principal component analysis (PCA) showed clear separation among treatment groups, with PC1 and PC2 accounting for 90.3% and 4.0% of the total variance, respectively (Figure 2D).
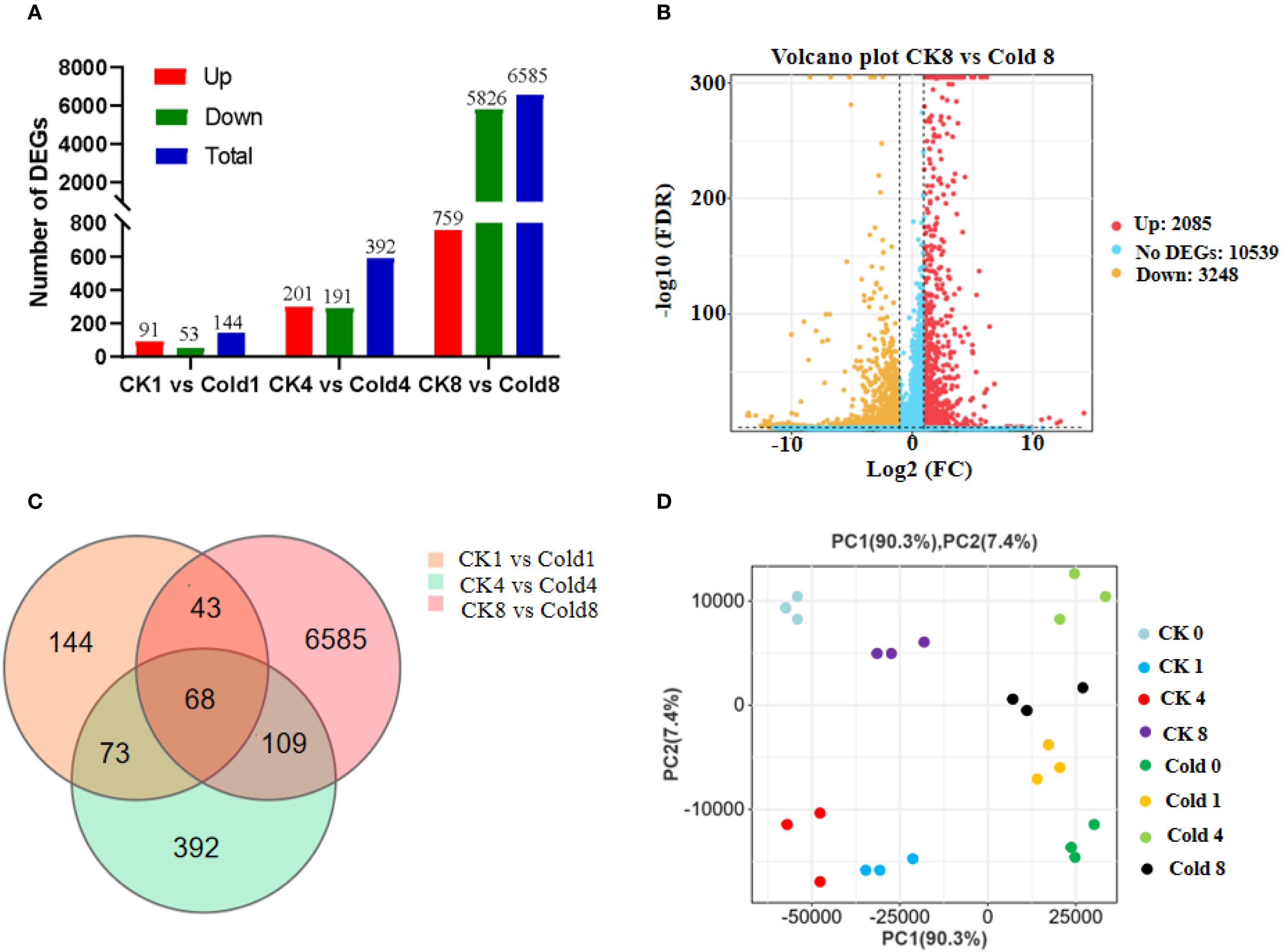
Figure 2. Transcriptional reprogramming in oil palm during cold stress. (A) Temporal dynamics of significantly upregulated (red) and downregulated (blue) differentially expressed genes (DEGs; FDR < 0.05, |fold change| ≥ 2). (B) Volcano plot of DEGs (8 h cold vs. control; red: upregulated, yellow: downregulated). (C) Venn diagram of DEG overlaps across time points; 68 genes consistently responded at all durations. (D) Principal Component Analysis (PCA) showing transcriptome separation by treatment time (PC1: 90.3%, PC2: 4.0% of variance).
3.3 Functional annotation of cold-responsive DEGs
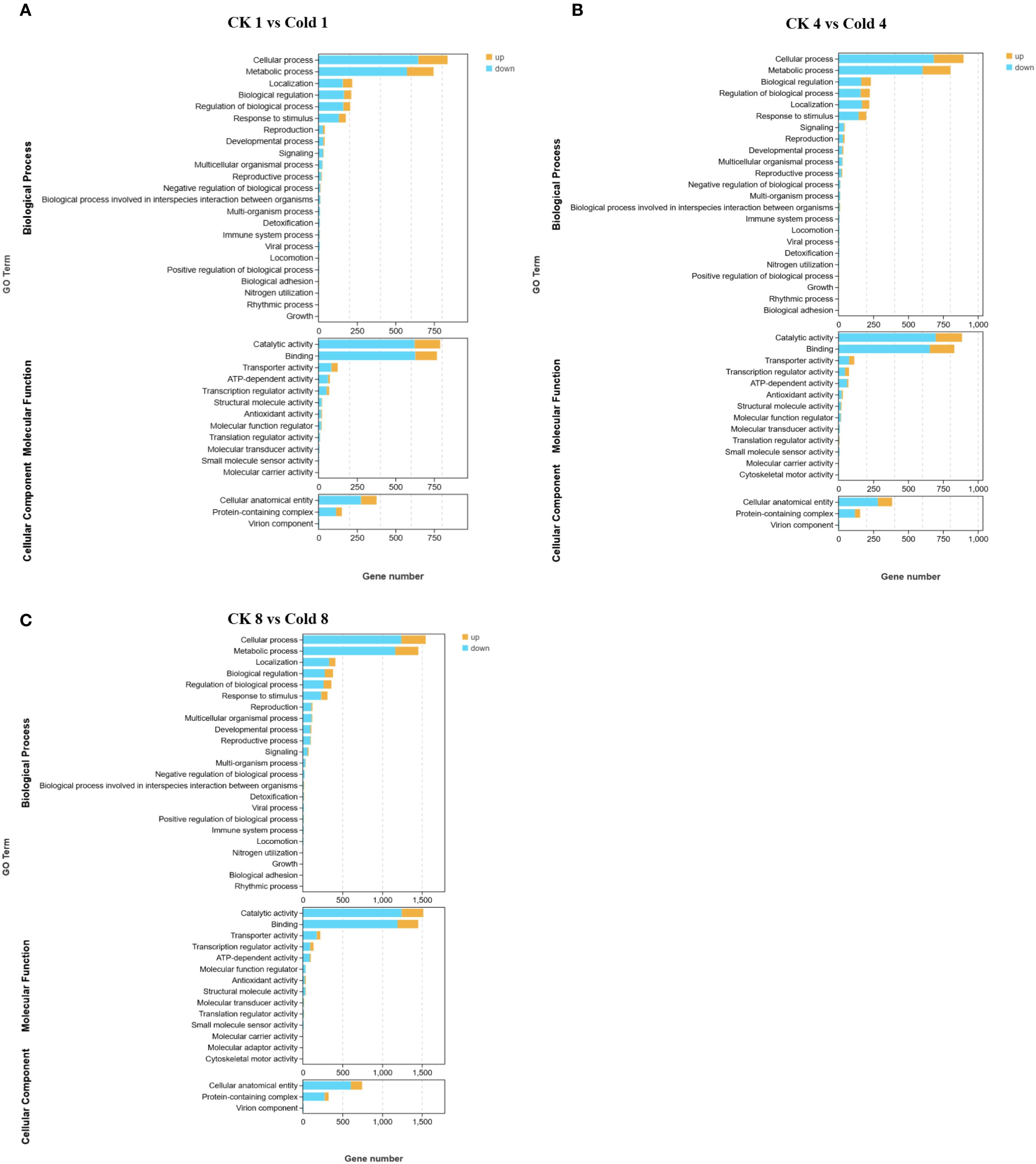
GO enrichment identified 35 significantly enriched terms (FDR < 0.05) at 8 h: 20 biological process (BP), 12 molecular function (MF), and 3 cellular component (CC) terms. Stress-response-related categories dominated BP, while oxidoreductase activity was prominent in MF. MF terms included binding, catalytic activity, transporter activity, transcription regulator activity, ATP-dependent activity, and antioxidant activity. BP terms included cellular processes, metabolic processes, biological regulation, response to stimulus, and localization. CC terms included cellular anatomical structures and protein-containing complexes (Figures 3A–C) (Supplementary Table 3). These results suggest coordinated adjustments at biological, cellular, and molecular levels (Figure 4). The comparison between CK 8 h and cold 8 h revealed significant biological characteristics. DEGs were primarily enriched in metabolic processes, cellular processes, and response to stimulus (Figure 4A), suggesting that cold stress may activate energy metabolism and secondary metabolite synthesis pathways as adaptive mechanisms. These DEGs likely participate in cell cycle regulation and cold stress responses, including oxidative damage mitigation and hormone-related signaling, highlighting cold stress’s disruptive effects on cellular equilibrium. In the MF category, most DEGs were strongly associated with catalytic activity and binding (Figure 4B), indicating potential alterations in enzymatic functions and molecular interactions during cold exposure. For the CC category, enriched terms were primarily related to cellular anatomical structures and protein complexes (Figure 4C), suggesting cold-induced modifications in cellular architecture and protein organization. Functional prediction and GO enrichment analysis underscored the critical involvement of diverse biological processes, molecular functions and cellular components in oil palm’s cold stress response. Notably, genes associated with metabolic pathways, enzymatic activities, and cellular structures emerged as key players in cold tolerance, likely contributing to cellular stability maintenance and enhanced stress resistance under cold conditions.
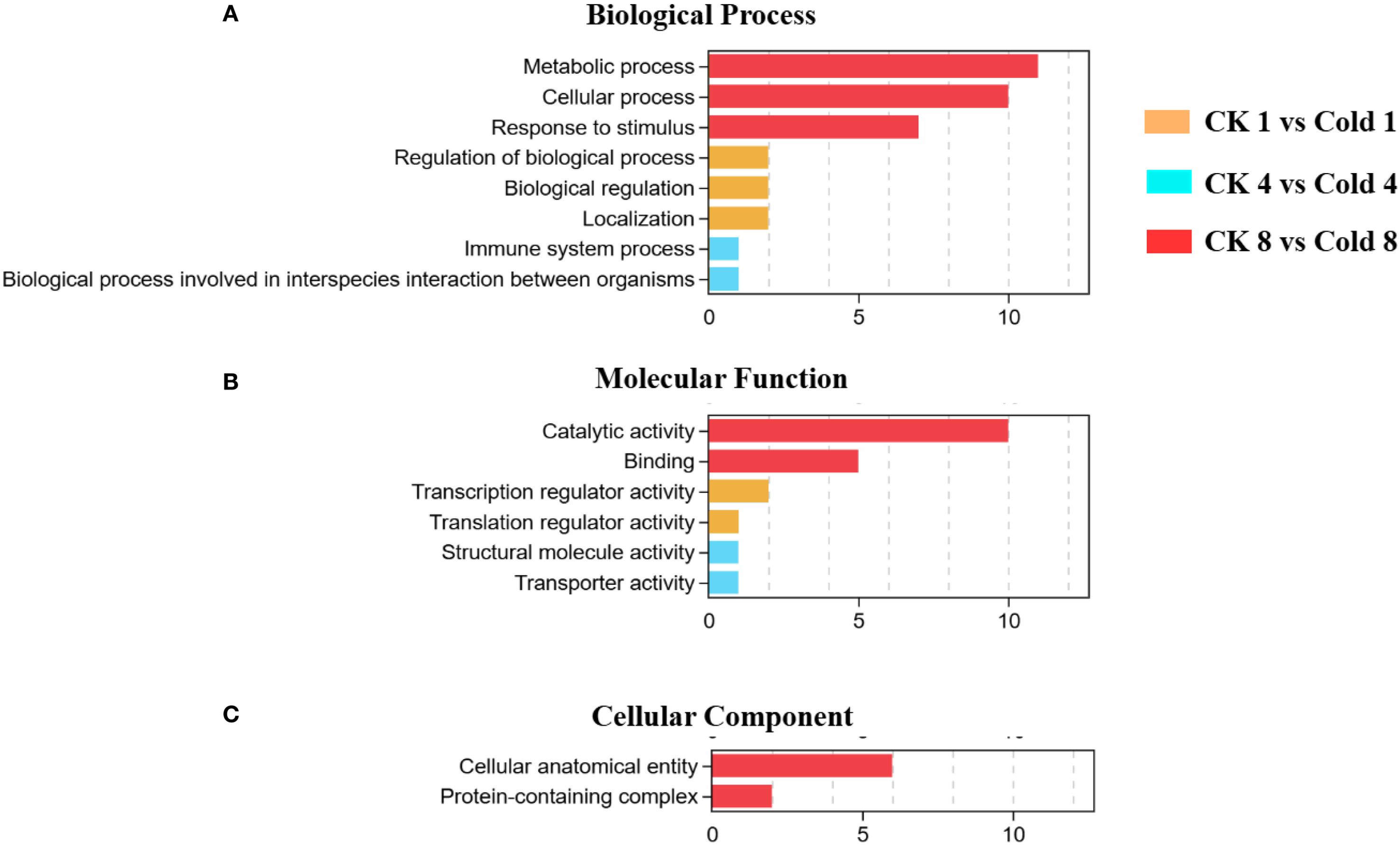
Figure 4. GO enrichment analysis of oil palm under cold stress. (A) Biological Process (BP), (B) Molecular Function (MF), and (C) Cellular Component (CC).
3.4 KEGG pathway enrichment and functional annotation
Significantly enriched KEGG pathways (Q < 0.05) were classified into organismal systems, cellular processes, environmental information processing, genetic information processing, and metabolism (Figure 5), comprising 19 subcategories. Both up- and downregulated DEGs accumulated with prolonged cold exposure, with the highest numbers at 8 h (Supplementary Table 4).
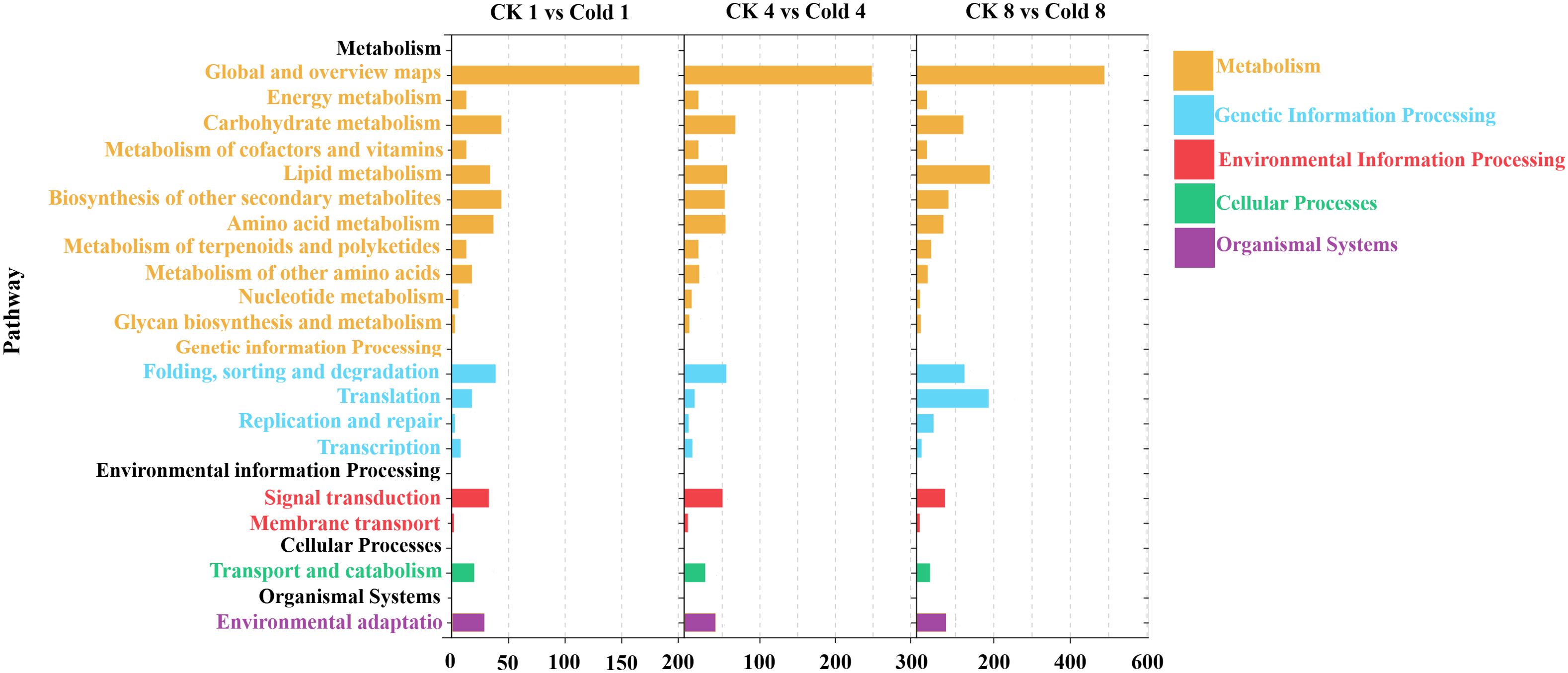
Figure 5. Comparative KEGG pathway enrichment analysis of DEGs in oil palm under cold stress was performed at 1 h (CK 1 h vs Cold 1 h), 4h (CK 4 h vs Cold 4 h), and 8 h (CK 8 h vs Cold 8 h).
Metabolic pathways dominated the transcriptome changes, including carbohydrate metabolism, secondary metabolite biosynthesis, and amino acid metabolism. Genetic information processing showed predominant upregulation. Environmental information processing, especially plant hormone signaling, the MAPK pathway, the phosphatidylinositol signaling system, and ABC transporters, was consistently downregulated. Cellular processes displayed initial downregulation (1 h) followed by increased upregulation (4–8 h), whereas organismal systems (environmental adaptation) were consistently downregulated (Figure 5). Across all times, the top 20 enriched pathways included MAPK signaling, plant–pathogen interaction, and phenylpropanoid biosynthesis (Figures 6A–C); secondary metabolite biosynthesis showed minimal expression changes at 8 h.
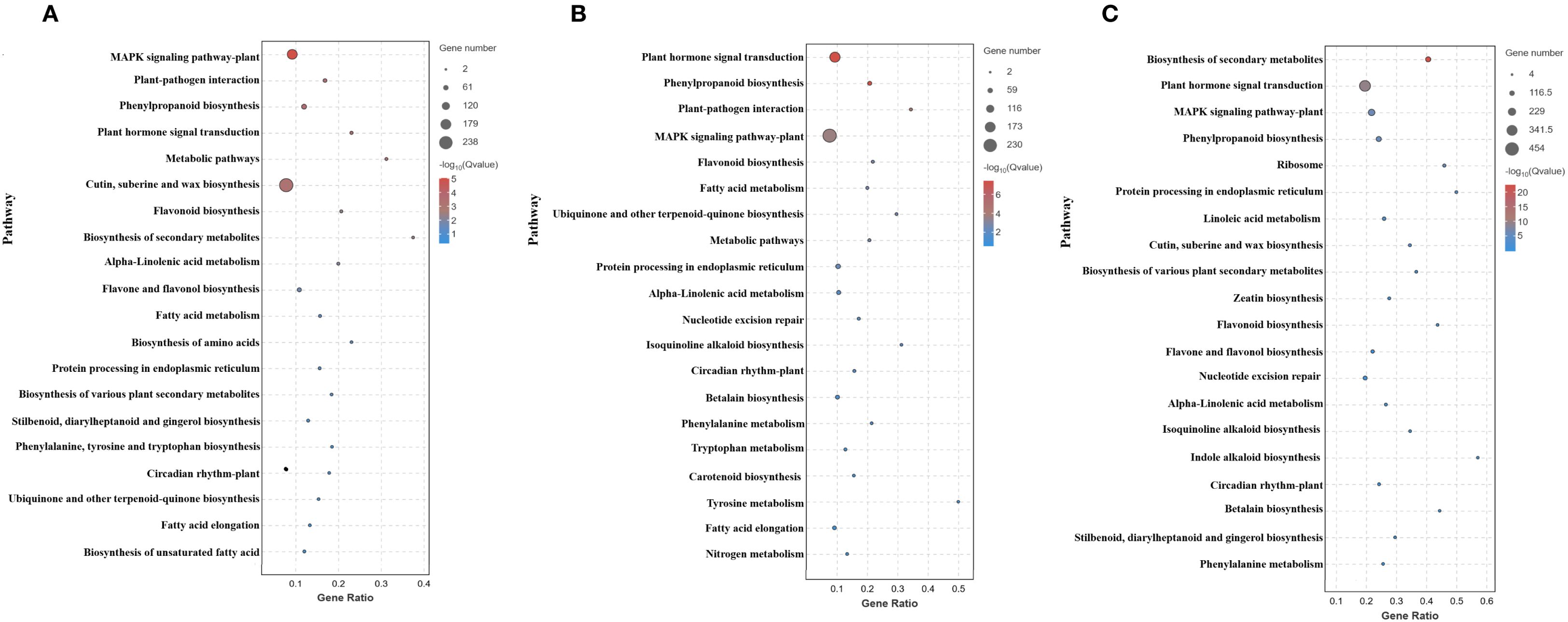
Figure 6. Top 20 enriched KEGG pathways: (A) 1 h, (B) 4 h, (C) 8 h cold stress. Each bubble represents a pathway, with its size indicating the number of enriched genes, while the color of the bubble reflects the significance level of the corresponding pathway.
3.5 Cold-induced DEGs in plant hormone signaling
Cold stress significantly altered expression of 45 genes in hormone signaling pathways (ABA, JA, auxin, BR, and SA) (Supplementary Table 5). The ABA pathway showed the strongest response (11 upregulated, 7 downregulated); auxin followed (8 up, 5 down); JA displayed moderate changes (3 up, 5 down); BR showed balanced regulation (2 up, 2 down); SA was minimally affected (1 up, 1 down). Overall, ABA, auxin, and JA pathways contained the most downregulated genes, with ABA also showing the greatest upregulation (Figure 7). Expression peaked at 4 h for representative proteins, whereas the largest number of DEGs occurred at 8 h, suggesting early regulation may involve post-translational mechanisms and later responses rely more on transcriptional modulation.
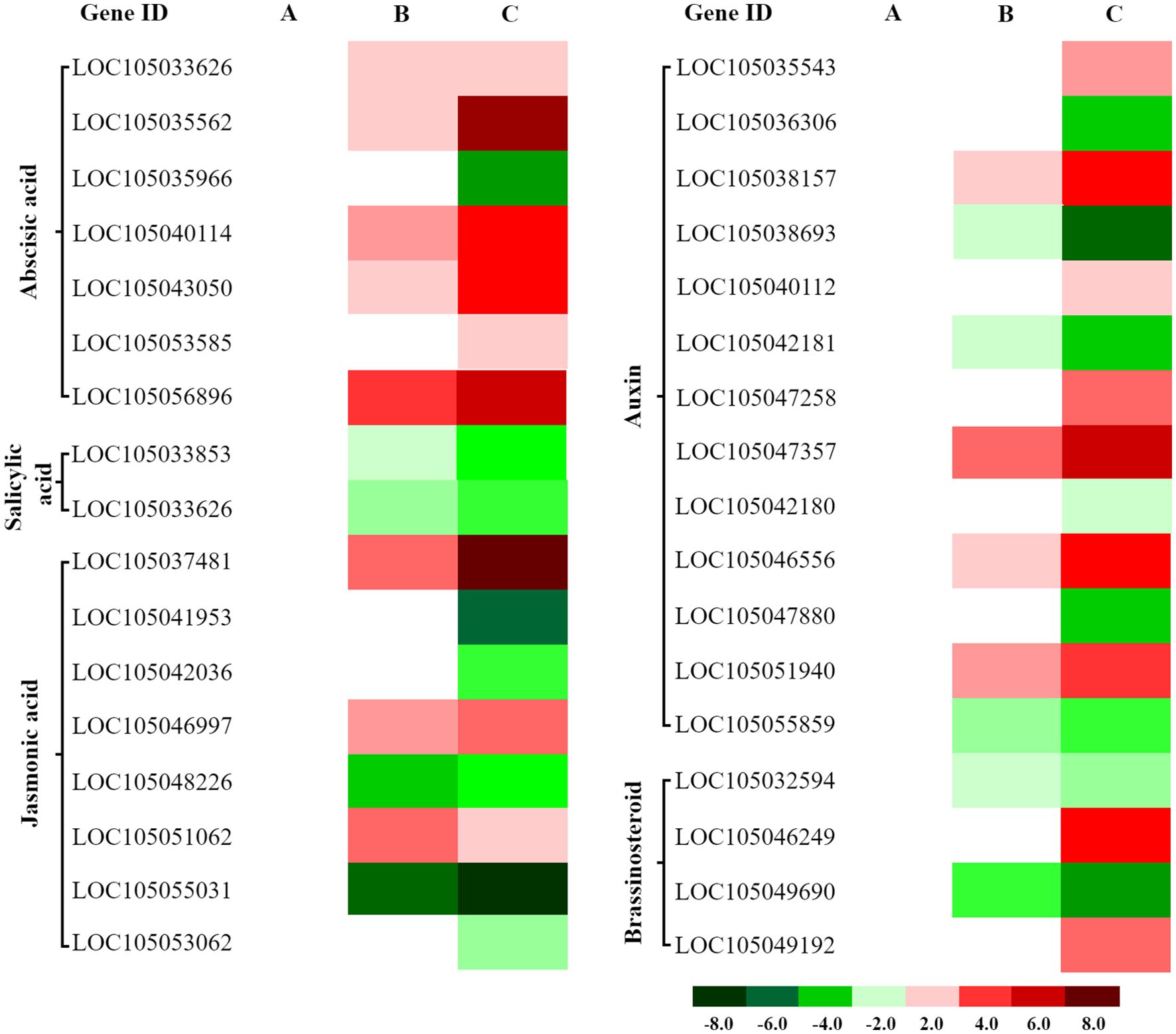
Figure 7. Differential expression of hormone signaling-related genes in oil palm under cold stress. Heatmaps display expression patterns of differentially expressed genes (DEGs) associated with abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), auxin, and brassinosteroid (BR) signaling pathways at three time points: (A) 1 hour (CK 1 h vs. Cold 1 h), (B) 4 hours (CK 4 h vs. Cold 4 h), and (C) 8 hours (CK 8 h vs. Cold 8 h) of cold exposure. Red: Upregulated genes (log2FC > 1, FDR < 0.05). Green: Downregulated genes (log2FC < −1, FDR < 0.05).
Cold stress initially induces a large-scale transcriptional suppression in oil palm seedlings, suggesting a conservative “niche adaptation” strategy that reduces overall metabolic activity to conserve energy and promote survival, rather than broadly activating defense pathways. This study reveals that low-temperature stress triggers a complex and coordinated reprogramming of hormone signaling. The central adaptive mechanism involves an “energy-saving defense” mode, rather than a global upregulation of gene expression. During later stress stages (8 h), transcriptomic reprogramming is characterized by a widespread downregulation of multiple signaling pathways, including auxin and jasmonic acid, likely to minimize metabolic expenditure and extend survival time. Conversely, the ABA pathway is specifically and strongly activated under low temperatures, functioning as a core regulatory hub for cold defense and mediating physiological responses such as stomatal closure. This marked contrast between the suppression of growth-related pathways (e.g., auxin, JA) and the activation of defense-related ABA signaling underscores a strategic ecological trade-off, sacrificing growth to ensure survival. The cold response also displays a temporal hierarchy, with early-stage (4 h) mechanisms relying on rapid post-translational protein modifications, and later-stage (8 h) adaptations driven by systematic transcriptional changes. Collectively, these results demonstrate that cold stress predominantly suppresses transcriptomic expression in oil palm seedlings, rather than enhancing individual gene expression.
3.6 Cold-regulated genes in the MAPK pathway
Cold stress can induce the MAPK signaling cascade, which is involved in regulating cell division, differentiation, programmed cell death, and stress responses. We identified 15 cold-responsive DEGs associated with hormone signaling, stress defense, and pathogen resistance, including nine linked to ABA signaling (7 up, 2 down), four to plant–pathogen interaction (2 up, 2 down), and two to phytohormone signaling (both down) (Figure 8; Supplementary Table 6). These findings suggest that cold stress induces MAPK to coordinate downstream adaptive responses, These responses may include the enhancement of ABA-dependent stress resistance, the recruitment of pathogen defense mechanisms, and the suppression of certain hormone signaling pathways, potentially contributing to more efficient energy allocation.
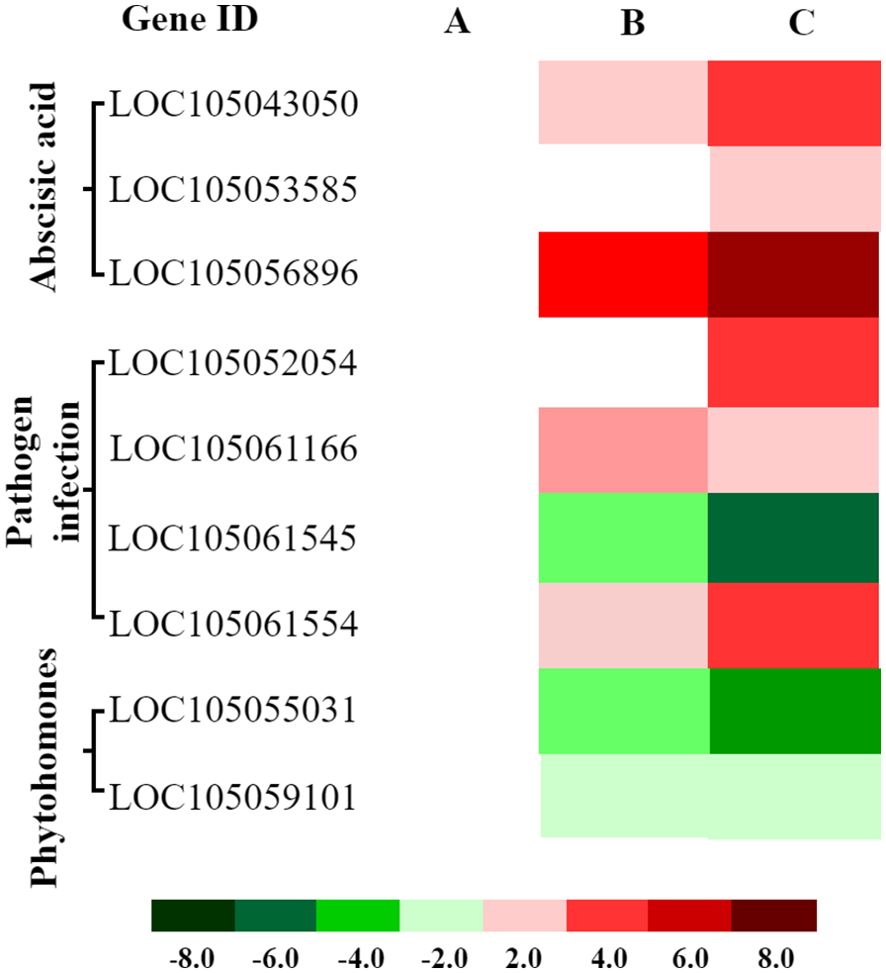
Figure 8. Expression patterns of cold-responsive DEGs in hormone signaling and stomatal development pathways mediated by MAPK cascade. Heatmaps display expression patterns of differentially expressed genes (DEGs) associated with ABA signaling, pathogen defense, and phytohormone signaling pathways at three time points: (A) 1 hour (CK 1 h vs. Cold 1 h), (B) 4 hours (CK 4 h vs. Cold 4 h), and (C) 8 hours (CK 8 h vs. Cold 8 h) of cold exposure. Red: Upregulated genes (log2FC > 1, FDR < 0.05). Green: Downregulated genes (log2FC < −1, FDR < 0.05).
3.7 Protein–protein interactions under cold stress
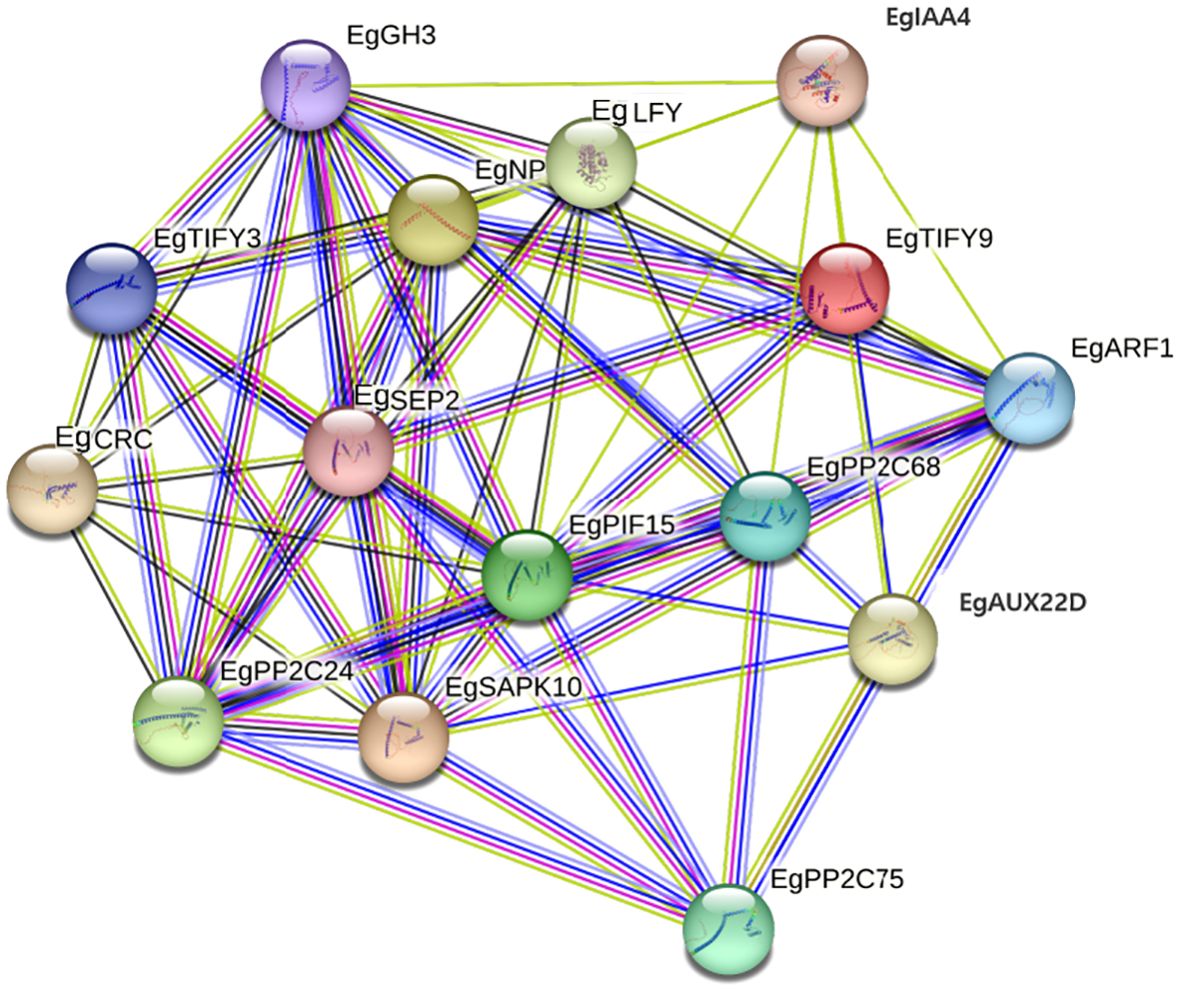
PPI analysis identified 15 core interacting proteins (EgIAA4, EgGH3, EgARF1, EgTIFY9, EgLFY, EgNP, EgTIFY3, EgCRC, EgSEP2, EgPP2C24/68/75, EgARF1, EgAUX22D, and EgSAPK10) constituting a potential cold-responsive regulatory module (Figure 9). The prevalence of PP2C family members highlights their roles in ABA signaling as negative regulators. The presence of auxin-associated proteins (EgIAA4, EgGH3, EgARF1) implies crosstalk between auxin and stress-responsive pathways (e.g., EgSAPK10). Collectively, these proteins may form an integrated network that coordinates hormonal and stress responses, with EgARF1 and PP2Cs as potential key nodes.
3.8 qRT-PCR validation
The eight differentially expressed genes (DEGs) selected for qRT-PCR validation were chosen based on specific and representative criteria, not randomly, to ensure a rigorous assessment of RNA-Seq reliability. These genes were intentionally selected to span a broad spectrum of fold-change values, in order to evaluate the accuracy of the transcriptome data across varying expression levels. As shown in Figure 10, the qRT-PCR results demonstrated high consistency with the RNA-Seq expression trends, strongly supporting the validity of our sequencing data. We are therefore confident that this strategically chosen gene set effectively represents the overall DEG profile and provides robust validation for the RNA-Seq findings.
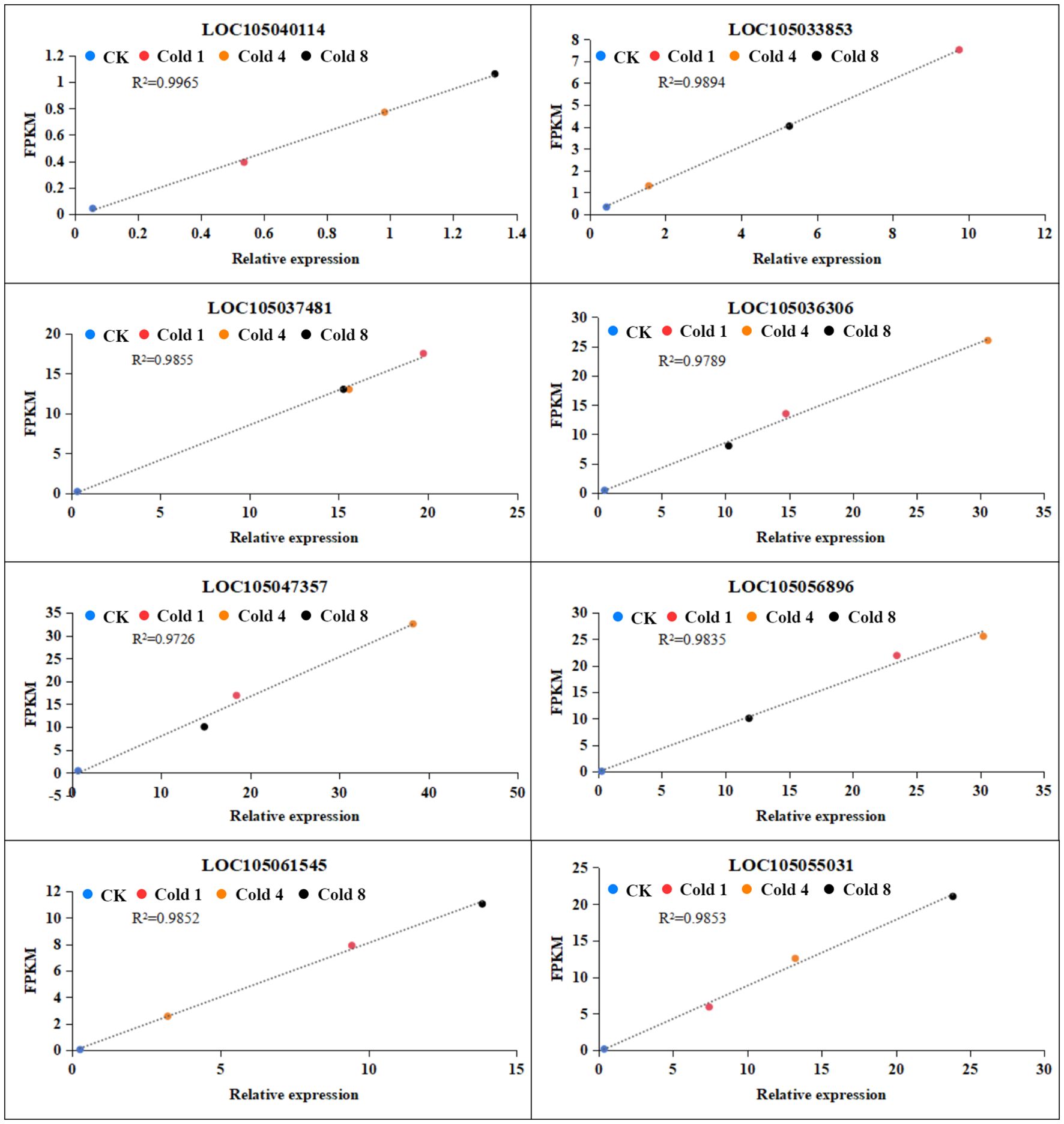
Figure 10. Expression analysis of selected genes under cold stress. Blue dots represent CK, red dots represent Cold 1, orange dots represent Cold 4, and black dots represent Cold 8, data represent mean values ± SD.
3.9 Hormone signaling and cold adaptation
The regulatory network governing plant responses to cold stress involves a tightly coordinated interplay between gene expression control, hormonal signaling, and environmental perception. These interconnected mechanisms collectively enable plants to adapt to cold conditions. Phytohormones serve as key mediators, playing a central role in orchestrating physiological and molecular responses to cold stress. In Arabidopsis, cold exposure disrupts auxin distribution by impairing the intracellular trafficking of auxin efflux transporters (Shibasaki et al., 2009). Similarly, in tea plants, three R2R3-MYB transcription factors (CsMYB45, CsMYB46, and CsMYB105) respond to cold stress by activating the JA signaling pathway (Han et al., 2022). Additionally, the plasma membrane-localized protein AtCor413pm1 in Arabidopsis functions as an ABA-responsive regulator, facilitating cold adaptation through ABA signal transduction (Hu et al., 2021). Further integrating immune and cold responses, the key transcription factor ICE1 in Arabidopsis interacts with the SA signaling pathway to modulate plant immunity during cold stress (Li et al., 2024). In wheat, the SA methyltransferase TaSAMT1 was recently identified, revealing a novel regulatory mechanism where the brassinosteroid (BR) signaling master regulator TaBZR1 epigenetically modulates SA metabolism (Chu et al., 2024). These findings illustrate the complexity of plant cold stress responses, which integrate hormonal crosstalk, transcriptional regulation, and epigenetic modifications to enhance cold tolerance. Such insights provide a valuable foundation for improving crop cold resistance through genetic engineering or targeted breeding strategies. Specifically, we observed a simultaneous and dramatic induction of ICE1 (LOC105051080) (12.33-fold), MYB (LOC105052782) (11.08-fold), and CBF (LOC105054150) (3.76-fold) homologs under cold stress condition for 8 h, highlighting the potential conservation and activation of this key pathway in a tropical woody oil crop. Our finding showed that the MAPK homolog (LOC140856367) was highly upregulated (8.24-fold) presents an intriguing new angle for oil palm cold tolerance (Figure 11). We hypothesize that this induced MAPK may serve as a critical upstream regulator that regulate the ICE1-MYB-CBF cascade in oil palm, potentially through post-translational modification. This represents a novel and species-specific regulatory layer atop the conserved transcriptional circuit. Therefore, our work not only confirms the activation of universal cold-responsive players in oil palm but also delineates a potential species-specific regulatory hierarchy, with MAPK acting as a putative master switch. This discovery provides valuable candidate genes (particularly MAPK and ICE1) for genetic engineering strategies aimed at enhancing cold tolerance in this critically important, yet cold-sensitive, tropical crop.
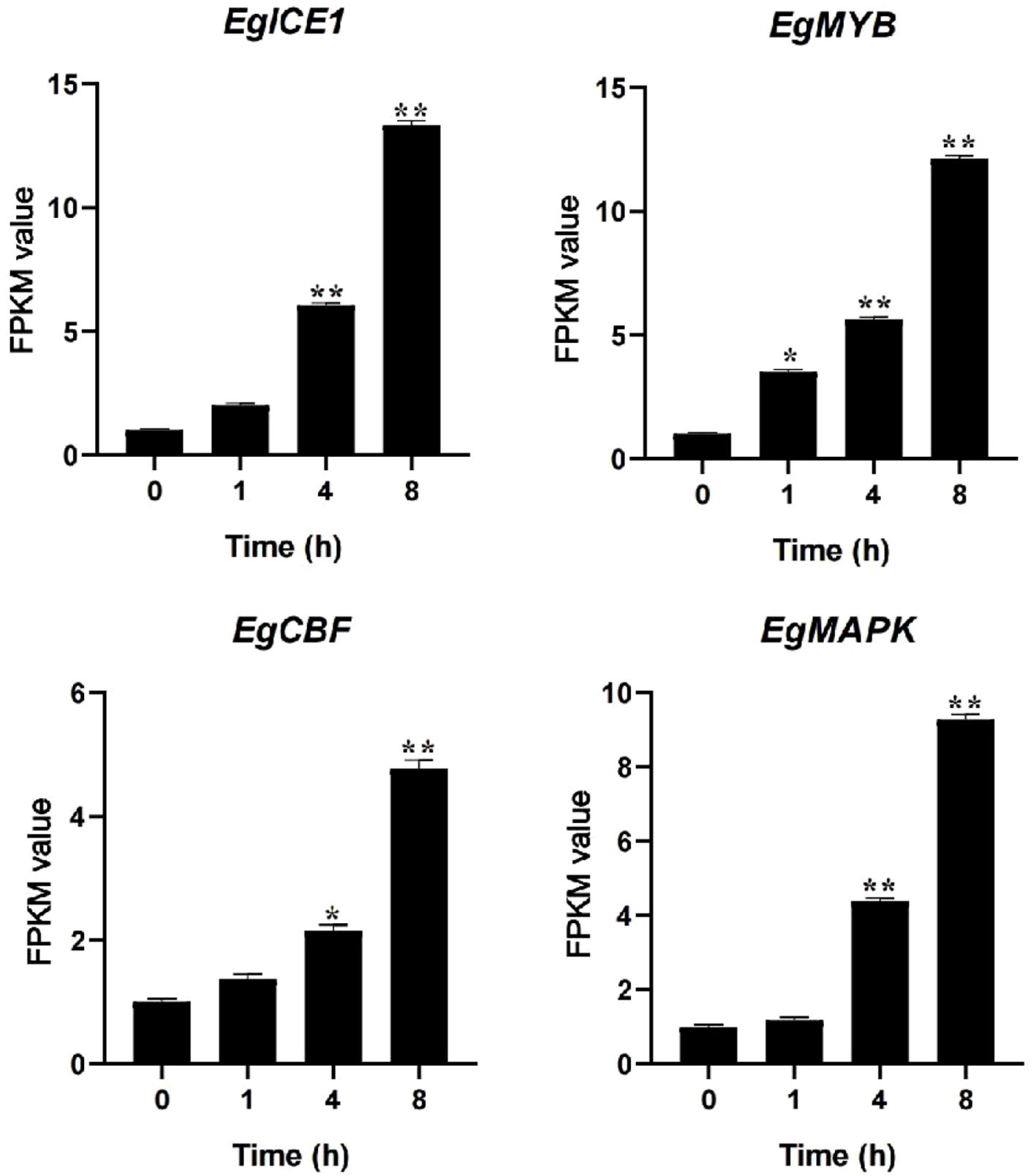
Figure 11. FPKM value of ICE1, MYB, CBF and MAPK genes under cold stress conditions. Asterisks represent significant difference at p ≤ 0.05(*) and p ≤ 0.01(**).
3.9.1 Auxin
Core components (TIR1/AFB–Aux/IAA–ARF) integrate auxin with ABA and ROS under cold (Rahman, 2013; Yu et al., 2017; Sharma et al., 2021; Du et al., 2022). We identified AUX1 (LOC1050536306), TIR1 (LOC105038693), IAA (LOC105042181, LOC105042180), ARF (LOC105047880, LOC105035543, LOC105038157), GH3 (LOC105055859, LOC105040112), and SAUR (LOC105047357, LOC105046556, LOC105051940) (Figure 12). Downregulation of AUX1/TIR1 suggests reduced transport/perception; SAUR genes were upregulated, whereas AUX1, TIR1, IAA, ARF, and GH3 were downregulated, indicating auxin pathway suppression to conserve energy.
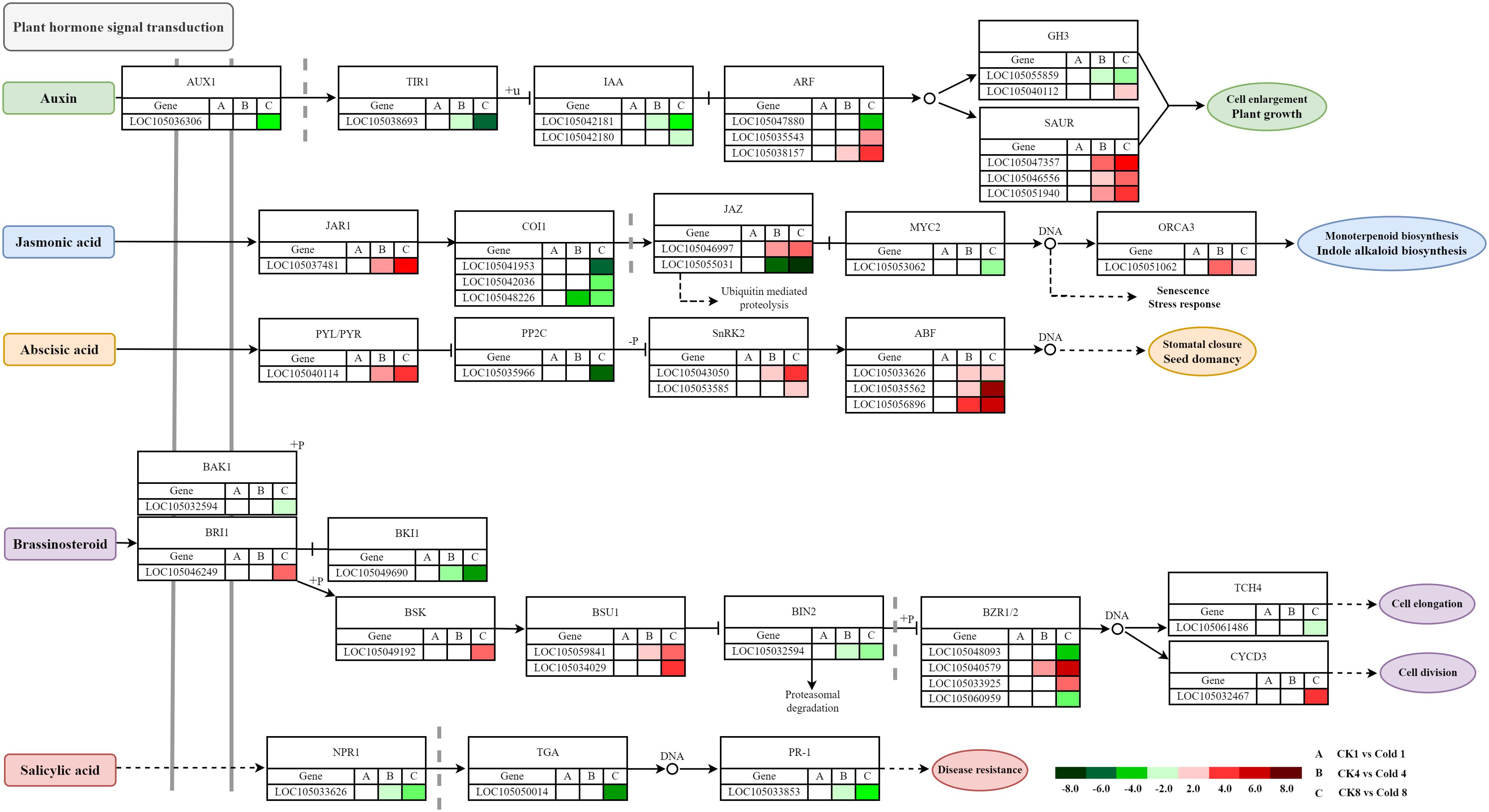
Figure 12. Heatmap visualization of gene expression dynamics in plant hormone signal transduction. Red and green hues denote up- and down-regulated DEGs, respectively, based on FPKM values.
3.9.2 Jasmonic acid
JA signaling (JAR1–COI1–JAZ–MYC2) coordinates stress responses (Carvalhais et al., 2017; Hu et al., 2017; Huang et al., 2024). Under cold, JAR1 (LOC105037481) was upregulated, COI1 (LOC105041953, LOC105042036, and LOC105048226) downregulated, and JAZ genes were differentially expressed (Figure 12), suggesting possible COI1-independent activation. Downregulation of MYC2 may release repression on stress-responsive genes (e.g., ORCA3), promoting adaptation.
3.9.3 Abscisic acid
PYR/PYL/RCAR receptors bind ABA to inhibit PP2Cs, releasing SnRK2s, which activate ABF transcription factors (Chen et al., 2020; Kou et al., 2024). Under cold, upregulation of PYL/PYR (LOC105040114) and downregulation of PP2C (LOC105035966) activate SnRK2 (LOC105043050, LOC105053585) and amplify ABA signaling, increasing ABF (LOC105033636, LOC105035562, and LOC105056896) activity and ABA-responsive gene expression (Figure 12), aligning with the classical model (Li et al., 2011). These results identify that oil palm responds to cold stress via a conserved ABA-dependent mechanism. The findings provide molecular evidence for ABA signaling in tropical crops like oil palm and highlight potential targets for cold-tolerance breeding.
3.9.4 Brassinosteroids
BRI1–BAK1 receptor activation initiates signaling through BSK–BSU1–BIN2–BZR1/2, with TCH4 and CYCD3 as effectors (Hafeez et al., 2021; Han et al., 2023; Kim and Russinova, 2020). Increased expression of BRI1 (LOC105046249), BSK (LOC105049192), BSU1 (LOC105059841, LOC105034029), and CYCD3 (LOC105032467), and decreased BAK1 (LOC105032594), BKI1 (LOC105049690), BIN2 (LOC105054741), and TCH4 (LOC105061486) support BR pathway activation with nuanced regulation. BZR1/2-related genes showed mixed regulation, suggesting paralog-specific responses (Figure 12).
3.9.5 Salicylic acid
NPR1–TGA–PR1 is central to SA signaling (Chen et al., 2019; Ding et al., 2018). Reduced expression of NPR1 (LOC105033626), TGA (LOC105050014), and PR1 (LOC105033853) suggests that SA-dependent SAR may not be triggered under cold (Figure 12).
3.10 MAPK pathway–mediated cold tolerance
Key cold-responsive genes in the MAPK cascade participate in pathogen defense and ABA/ethylene/JA responses (Iqbal et al., 2022). In pathogen signaling, FLS2 (LOC105049067) was downregulated; BAK1 genes (LOC105058366, LOC105056517, LOC105041665) were upregulated; MEKK1 (LOC105052054) and MKK1/2 (LOC105061166) were upregulated, while MKK4/5 (LOC105061545), MPK4 (LOC105059999), and MPK3/6 (LOC105052113) were downregulated, indicating activation of the MEKK1–MKK1/2 branch and suppression of the MEKK2–MKK4/5–MPK3/6/4 branch (Figure 13). Ethylene biosynthesis (ACS6; LOC105049327) was upregulated. VIP1 (LOC105048846) was upregulated, whereas WRKY22/29 (LOC105050724, LOC105046637) and PR1 (LOC105053324) were downregulated; FRK1 (LOC105051058) and PAD3 (LOC105037726, LOC105037358) were upregulated, suggesting prioritization of JA/ET-mediated rapid defenses over SA-dependent long-term resistance. In ABA signaling, PYL/PYR (e.g., LOC105040114) was upregulated, PP2C (LOC105035966) downregulated, SnRK2 (LOC105043050, LOC105053585) upregulated, and MAPK17/18 (LOC105047258) upregulated. Selective downregulation of MKK3 (LOC105056896) and MHK10 (LOC105059744) suggests pathway-specific modulation. In ethylene signaling, upregulation of ETR/ERS (LOC105040872), RAN1 (LOC105037481), EIN3/EIL (LOC105049190), ERF1 (LOC105051940), PDF1.2 (LOC105058257), and ChiB (LOC105053062), along with downregulation of RTE1 (LOC105055031) and EBF1/2 (LOC105049227), indicates activation of canonical and MAPK-linked branches, with post-transcriptional regulation (XRN4; LOC105050710). In JA signaling, increased MKK3, MPK6, MYC2, and VSP2 suggest MAPK-mediated JA activation and JA–ethylene co-regulation via ERF1 and PDF1.2. MYC2 and MPK6 emerge as targets for improving cold resilience, consistent with SlMYC2–SlCBF1/2 regulation in tomato (Li et al., 2025).
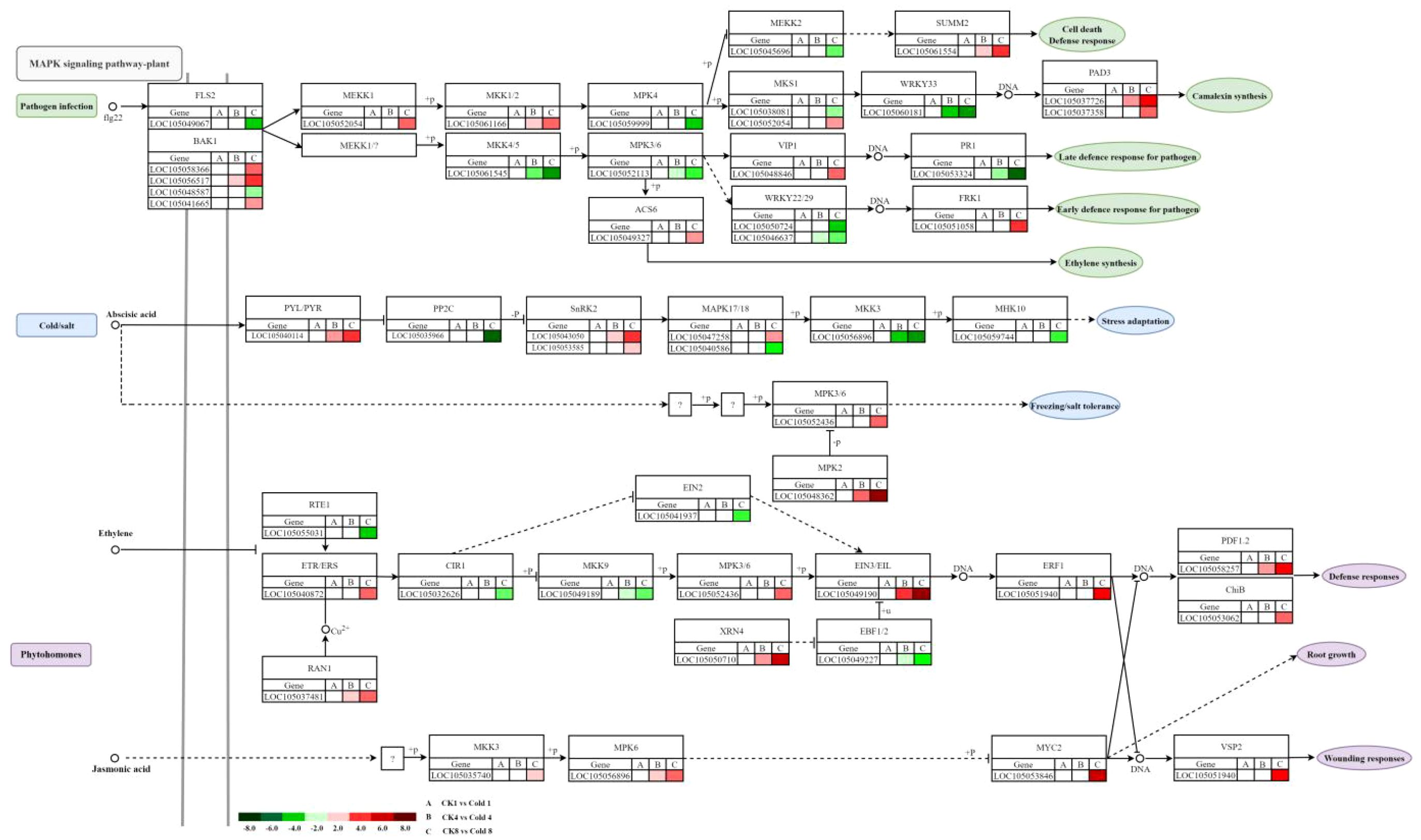
Figure 13. Heatmap visualization of different genes in MAPK signaling pathway. Red and green hues denote up- and downregulated DEGs, respectively, based on FPKM values.
4 Discussion
4.1 Integration of physiological and transcriptomic responses reveals a coordinated cold defense strategy
Cold exposure induces ROS accumulation, causing membrane destabilization and energy dysregulation (Xing et al., 2022). Plants activate antioxidant systems (SOD, POD) to restore redox homeostasis and mitigate damage (Figures 1A, B) (Liu et al., 2023; Foyer and Noctor, 2005). Transcriptome results indicated that the expression levels of the oil palm SOD gene (LOC105059069) and POD gene (LOC105040036) were up-regulated after 4 hours of cold stress (Figures 1K, L). The FPKM values were 5.32-fold and 7.03-fold higher than those in the control group, respectively. The observed increase in antioxidant enzyme activities is underpinned by the transcriptional activation of their corresponding genes, demonstrating a multi-level strategy to combat oxidative burst. Soluble sugars (e.g., sucrose, glucose, fructose) accumulate as compatible osmolytes to stabilize osmotic potential (Figure 1D). The expression levels of key sugar-synthesis-related genes, Sucrose Phosphate Synthase (SPS, LOC105036668) and Sucrose Synthase (SuS, LOC105050124), were significantly up-regulated after 4 hours of cold stress, with FPKM values 3.88-fold and 6.27-fold higher than those in the control group, respectively (Figures 1M, N). The massive accumulation of soluble sugars is not merely a passive consequence but an active process, facilitated by the transcriptional reprogramming of both biosynthetic and transport machinery. Cold increases membrane permeability, accelerates unsaturated fatty acid degradation, promotes lipid peroxidation, elevates MDA, and increases REC (Figures 1C, E). Low temperatures suppress key enzymes in chlorophyll biosynthesis, impair chloroplast development, and reduce chlorophyll a/b (Figures 1F–J). Transcriptome analysis revealed that the expression of Glutamyl-tRNA reductase (HEMA, LOC105054529) was down-regulated, with an FPKM value 0.21-fold that of the control group (Figure 1O). The cold-induced chlorophyll degradation results from a concerted transcriptional suppression of anabolic pathways and induction of catabolic pathways, effectively dismantling the photosynthetic apparatus to conserve resources and mitigate photo-oxidative damage. CO2 assimilation declines as electron transport is disrupted and lower stomatal conductance limits gas exchange (Li et al., 2020), while respiratory ATP production is hindered (Kang et al., 2023), collectively inhibiting growth. The cold-induced chlorophyll degradation results from a concerted transcriptional suppression of anabolic pathways and induction of catabolic pathways, effectively dismantling the photosynthetic apparatus to conserve resources and mitigate photo-oxidative damage.
4.2 Transcriptomic signatures of cold stress
The number of differentially expressed genes (DEGs) increased over the course of treatment, peaking at 8 hours post-cold exposure with 6,585 DEGs identified in the CK 8 h vs Cold 8 h comparison (Figures 2A, B). Notably, at this time point, the number of down-regulated genes exceeded that of up-regulated genes. This result suggested that this pattern as a central adaptive mechanism employed by plants under low-temperature stress, attributable largely to energy reallocation and extensive transcriptional reprogramming: i) In response to stress, plants suppress a range of energy-demanding basal biological processes, including photosynthesis, cell division, and ribosome biogenesis, to reallocate finite resources toward stress adaptation and survival mechanisms (Liu et al., 2022; Zhang et al., 2020). Consequently, genes associated with these pathways are broadly down-regulated. Our results are consistent with this phenomenon, showing pronounced enrichment and suppression of genes related to photosynthetic and ribosomal pathways. ii) In order to response cold stress, a select set of functionally important genes is up-regulated to enact protective responses, such as the production of osmoregulators (e.g., proline, soluble sugars), antioxidant enzymes (e.g., SOD, POD) (Zhou et al., 2022, 2025). This characteristic transcriptomic profile, marked by widespread down-regulation alongside targeted up-regulation, is well-documented in studies of plant abiotic stress. In conclusion, the prevalence of down-regulated genes at 8h should not be interpreted as a diminished stress response; rather, it reflects a strategic and efficient shift from a growth-oriented state to a defense-oriented state at the transcriptional level. These insights provide valuable temporal resolution for understanding molecular mechanisms underlying cold tolerance in oil palm.
RNA-seq profiling across 0–8 h cold exposure revealed DEGs in abiotic stress adaptation, developmental plasticity, and pathogen defense crosstalk, aligning with findings in other species (Li et al., 2024; Zhang et al., 2023; Peng et al., 2023). Enriched BP terms included cellular activity, metabolism, biological regulation, regulation of biological processes, and localization (Figure 3), indicating reorganization of energy distribution, activation of regulatory networks (e.g., transcription factors), and maintenance of cellular structures (Zhu, 2016; He et al., 2024; Kim et al., 2024). At 8 h, BP terms “metabolic process”, “cellular process”, and “response to stimulus” were prominent (Figure 4A), consistent with metabolic reprogramming, secondary metabolite synthesis, and homeostatic regulation (Wu et al., 2023; Hotamisligil and Davis, 2016). MF terms (catalytic activity, binding) reflected roles for enzymes and binding proteins (e.g., calmodulin, transcription factors) in stress adaptation (Jan et al., 2023), while CC terms implicated structural remodeling (Hu et al., 2024). KEGG analysis highlighted activation of plant–pathogen interactions, MAPK signaling, and hormone signaling (Figure 6). Under cold, MAPK components (e.g., MPK3/6) regulate transcription factors (ICE1, MYB/CBF) to modulate cold-responsive genes (Wen et al., 2023). ABA biosynthesis/signaling (PYR/PYL–PP2C–SnRK2) induces stomatal closure and stress gene expression (Li et al., 2023); ethylene via EIN3/EIL affects cell elongation and antifreeze proteins (Su et al., 2024); JA and SA bridge cold and immune signaling (Wang et al., 2020; Wu et al., 2019); auxin shifts reflect growth–defense trade-offs (Liu et al., 2024).
This study reveals, for the first time in oil palm, a differential regulatory phenomenon within the MAPK cascade, marked by pronounced activation of the MEKK1–MKK1/2 branch and concurrent suppression of the MEKK2–MKK4/5–MPK3/6/4 branch (Zhou et al., 2023). This branch-specific pattern may underlie a key cold adaptation strategy in oil palm as a tropical crop (Ye et al., 2017). Transcriptomic analyses further demonstrate that oil palm downregulates salicylic acid (SA) pathway markers (WRKY22/29, PR1) while upregulating jasmonic acid/ethylene (JA/ET)-mediated defense markers (FRK1, PAD3, PDF1.2), indicating a distinct preference in defense signaling compared to model plants (Bi et al., 2021; Kovács et al., 2025). This reprogramming may reflect a specialized mechanism for coping with low-temperature stress. The study further delineates key nodes of crosstalk between MAPK and hormone signaling pathways, including: i) The central role of the MPK6–MYC2 module in activating JA signaling. ii) The specific involvement of MAPK17/18 in ABA signal transduction. iii) A post-transcriptional regulatory mechanism mediated by the EIN3–EBF1 node within ethylene signaling. Together, these findings construct a unique MAPK–hormone regulatory network in oil palm. It is confirmed that MYC2 and MPK6 occupy central positions in this network, exhibiting both evolutionary conservation and species-specific features when compared to the SlMYC2–SlCBF1/2 regulatory axis in tomato. These results provide concrete targets for molecular breeding aimed at enhancing cold resilience in oil palm. These insights not only clarify the architecture of cold stress signaling in oil palm but also offer novel theoretical foundations and genetic resources for improving cold adaptation in tropical crops.
4.3 Prospects for molecular breeding
The findings elucidate the precise molecular mechanisms underlying oil palm’s response to cold stress, providing a clear roadmap for molecular breeding and biotechnological intervention. In this context, the widespread downregulation of genes in growth-promoting pathways (e.g., auxin, brassinosteroid) is not a passive impairment but an active, energy-saving strategy. By shutting down non-essential metabolic processes, the plant reallocates precious resources to activate and sustain critical defense responses, primarily orchestrated by ABA. This reprogramming ensures survival at the expense of temporary growth arrest. Our data, showing coordinated downregulation across multiple pathways, provide a comprehensive transcriptomic portrait of this conserved ecological trade-off phenomenon. The genes we identified as downregulated in auxin, JA, and BR pathways represent prime candidates for knockdown or CRISPR/Cas-mediated gene silencing in crops (Maharajan et al., 2022). Creating alleles with reduced expression of these genes could genetically predispose plants to a “primed” state, constitutively allocating more resources and thus enhancing hardness (Ma et al., 2024). The development of molecular markers associated with key genes significantly related to cold resistance, such as the superior allelic variations of LOC105059069 (SOD) and LOC105040036 (POD), can be used to screen breeding materials with enhanced ROS-scavenging capacity (Wang et al., 2025). Constructing expression vectors containing SOD or POD genes driven by stress-inducible promoters and transforming them into oil palm enables targeted and efficient ROS clearance, avoiding potential growth penalties associated with constitutive overexpression. Overexpressing core positive regulators in the JA signaling pathway, or using gene editing technologies (e.g., CRISPR/Cas9) to knock out negative regulators in the ABA pathway, can remodel the hormone network to favor cold resistance activation. Enhancing the expression or activity of positive regulatory branches in the MAPK cascade amplifies the cold warning signal, prompting earlier and stronger activation of downstream defense mechanisms. These strategies break the limitations of the natural gene pool, creating novel or rare superior alleles that enable leapfrog improvements in cold tolerance. Given that cold tolerance is a complex trait controlled by multiple genes, the effect of modifying a single gene may be limited. These findings suggest a strategy for synergistic multi-gene manipulation.
Compared to advances in cold tolerance research on tropical crops like cassava and banana, oil palm research can benefit from establishing a well-defined evaluation framework based on physiological indicators, antioxidant profiles, and osmolyte accumulation to enable precise assessment of cold tolerance across diverse germplasms (Santanoo et al., 2022; Wei et al., 2022). Both cassava and banana boast large germplasm collections, with core sets of cold-tolerant accessions already identified. Likewise, a systematic screening of oil palm genetic resources should be carried out to build a curated cold-tolerant core collection, serving as a critical source of parental material for conventional hybrid breeding. Moreover, a comparative biology strategy should be actively employed (Xu et al., 2022; Li et al., 2022). Oil palm research can efficiently target and adapt key pathways, substantially reducing redundant exploratory work and accelerating progress. Together, these approaches will deliver a robust scientific foundation for developing new cold-resilient oil palm varieties and strengthening agricultural sustainability.
5 Conclusions
Cold stress triggers significant physiological and molecular responses in oil palm, including enhanced antioxidant activity and the accumulation of soluble sugars and MDA. Transcriptomic analysis revealed time-dependent regulation of DEGs, with notable enrichment in hormone signaling (IAA, JA, ABA, BR) and the MAPK cascade. Together, these pathways orchestrate cold adaptation through extensive transcriptional reprogramming, predominantly characterized by the downregulation of energy-costly processes, coupled with the upregulation of specific protective mechanisms. These findings, and especially the identification of key genes within these regulated pathways, offer a foundation for targeted breeding and genetic engineering strategies aimed at enhancing oil palm resilience.
Data availability statement
The data presented in this study are publicly available. The data can be found here in the China National Gene Bank Database (https://db.cngb.org/cnsa) under accession numbers CNP0006920 and CNP0007547.
Author contributions
QW: Data curation, Resources, Writing – review & editing, Formal analysis. XY: Writing – review & editing, Formal analysis, Data curation. QH: Validation, Writing – review & editing. RL: Formal analysis, Data curation, Writing – review & editing. XZ: Formal analysis, Writing – review & editing. QL: Data curation, Writing – review & editing. ZL: Formal analysis, Writing – review & editing. DF: Writing – review & editing, Formal analysis. HC: Writing – review & editing. XNL: Data curation, Writing – review & editing. XOL: Writing – review & editing. LZ: Writing – original draft, Conceptualization, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was financially supported by the Project of State Key Laboratory of Tropical Crop Breeding (No. NKLTCB202402, NKLTCB-HZ06) and the National Natural Science Foundation of China (No. 32460412).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1687366/full#supplementary-material
References
Abubakar, A. and Ishak, M. Y. (2024). Exploring the intersection of digitalization and sustainability in oil palm production: challenges, opportunities, and future research agenda. Environ. Sci. pollut. Res. Int. 31, 50036–50055. doi: 10.1007/s11356-024-34535-9
Al-Madani, M. H. M., Fernando, Y., Tseng, M. L., and Abideen, A. Z. (2023). Uncovering four domains of energy management in palm oil production: a sustainable bioenergy production trend. Environ. Sci. pollut. Res. Int. 30, 38616–38633. doi: 10.1007/s11356-022-24973-8
Amini, S., Maali-Amiri, R., Kazemi-Shahandashti, S. S., López-Gómez, M., Sadeghzadeh, B., Sobhani-Najafabadi, A., et al. (2021). Effect of cold stress on polyamine metabolism and antioxidant responses in chickpea. J. Plant Physiol. 258-259, 153387. doi: 10.1016/j.jplph.2021.153387
Apriyanto, A. and Ajambang, W. (2022). Transcriptomic dataset for early inflorescence stages of oil palm in response to defoliation stress. Data Brief. 41, 107914. doi: 10.1016/j.dib.2022.107914
Bi, M., Li, X., Yan, X., Liu, D., Gao, G., Zhu, P., et al. (2021). Chrysanthemum WRKY15–1 promotes resistance to Puccinia horiana Henn. via the salicylic acid signaling pathway. Hortic. Res. 8, 6. doi: 10.1038/s41438-020-00436-4
Carvalhais, L. C., Schenk, P. M., and Dennis, P. G. (2017). Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 37, 42–47. doi: 10.1016/j.mib.2017.03.009
Chaparro, L. M., Neiram, L. F., Molina, D., Rivera-Barrera, D., Castañeda, M., López-Giraldo, L. J., et al. (2023). Biowaxes from palm oil as promising candidates for cosmetic matrices and pharmaceuticals for human use. Materials (Basel). 16, 4402. doi: 10.3390/ma16124402
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., and Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Chen, J., Mohan, R., Zhang, Y., Li, M., Chen, H., Palmer, I. A., et al. (2019). NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 181, 289–304. doi: 10.1104/pp.19.00124
Chu, W., Chang, S., Lin, J., Zhang, C., Li, J., Liu, X., et al. (2024). Methyltransferase TaSAMT1 mediates wheat freezing tolerance by integrating brassinosteroid and salicylic acid signaling. Plant Cell. 36, 2607–2628. doi: 10.1093/plcell/koae100
Danylo, O., Pirker, J., Lemoine, G., Ceccherini, G., See, L., McCallum, I., et al. (2021). A map of the extent and year of detection of oil palm plantations in Indonesia, Malaysia and Thailand. Sci. Data. 8, 96. doi: 10.1038/s41597-021-00867-1
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 173, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Du, W., Lu, Y., Li, Q., Luo, S., Shen, S., Li, N., et al. (2022). TIR1/AFB proteins: Active players in abiotic and biotic stress signaling. Front. Plant Sci. 13, 1083409. doi: 10.3389/fpls.2022.1083409
Foyer, C. H. and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 17, 1866–1875. doi: 10.1105/tpc.105.033589
Hafeez, M. B., Zahra, N., Zahra, K., Raza, A., Batool, A., Shaukat, K., et al. (2021). Brassinosteroids: molecular and physiological responses in plant growth and abiotic stresses. Plant Stress. 2, 100029. doi: 10.1016/j.stress.2021.100029
Han, C., Wang, L., Lyu, J., Shi, W., Yao, L., Fan, M., et al. (2023). Brassinosteroid signaling and molecular crosstalk with nutrients in plants. J. Genet. Genomics 50, 541–553. doi: 10.1016/j.jgg.2023.03.004
Han, Z., Zhang, C., Zhang, H., Duan, Y., Zou, Z., Zhou, L., et al. (2022). CsMYB transcription factors participate in jasmonic acid signal transduction in response to cold stress in tea plant (Camellia sinensis). Plants (Basel). 11, 2869. doi: 10.3390/plants11212869
He, A. L., Niu, S. Q., Zhao, Q., Li, Y. S., Gou, J. Y., and Gao, H. J. (2018). Induced salt tolerance of Perennial ryegrass by a novel bacterium strain from the rhizosphere of a desert shrub Haloxylon ammodendron. Int. J. Mol. Sci. 19, 469–472. doi: 10.3390/ijms19020469
He, Y., Zhang, Y., Li, J., Ren, Z., Zhang, W., Zuo, X., et al. (2024). Transcriptome dynamics in Artemisia annua provides new insights into cold adaptation and de-adaptation. Front. Plant Sci. 15, 1412416. doi: 10.3389/fpls.2024.1412416
Hotamisligil, G. S. and Davis, R. J. (2016). Cell signaling and stress responses. Cold Spring Harb. Perspect. Biol. 8, a006072. doi: 10.1101/cshperspect.a006072
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Hu, X., Liu, J., Liu, E., Qiao, K., Gong, S., Wang, J., et al. (2021). Arabidopsis cold-regulated plasma membrane protein Cor413pm1 is a regulator of ABA response. Biochem. Biophys. Res. Commun. 561, 88–92. doi: 10.1016/j.bbrc.2021.05.032
Hu, D., Yao, Y., Lv, Y., You, J., Zhang, Y., Lv, Q., et al. (2024). The OsSRO1c-OsDREB2B complex undergoes protein phase transition to enhance cold tolerance in rice. Mol. Plant 17, 1520–1538. doi: 10.1016/j.molp.2024.08.006
Huang, D., Li, J., Chen, J., Yao, S., Li, L., Huang, R., et al. (2024). Genome-wide identification and characterization of the JAZ gene family in Gynostemma pentaphyllum reveals the COI1/JAZ/MYC2 complex potential involved in the regulation of the MeJA-induced gypenoside biosynthesis. Plant Physiol. Biochem. 214, 108952. doi: 10.1016/j.plaphy.2024.108952
Iqbal, S., Wang, X., Mubeen, I., Kamran, M., Kanwal, I., Díaz, G. A., et al. (2022). Zin El-Abedin TK. Phytohormones trigger drought tolerance in crop plants: outlook and future perspectives. Front. Plant Sci. 12, 799318. doi: 10.3389/fpls.2021.799318
Jan, N., Wani, U. M., Wani, M., Qazi, H. A., and John, R. (2023). Comparative physiological, antioxidant and proteomic investigation reveal robust response to cold stress in Digitalis purpurea L. Mol. Biol. Rep. 50, 7319–7331. doi: 10.1007/s11033-023-08635-7
John Martin, J. J., Yarra, R., Wei, L., and Cao, H. (2022). Oil palm breeding in the modern era: challenges and opportunities. Plants (Basel). 11, 1395. doi: 10.3390/plants11111395
Kang, Z., Zhang, Y., Cai, X., Zhang, Z., Xu, Z., Meng, X., et al. (2023). Crosstalk between 5-aminolevulinic acid and abscisic acid adjusted leaf iron accumulation and chlorophyll synthesis to enhance the cold tolerance in solanum lycopersicum seedlings. Int. J. Mol. Sci. 24, 10781. doi: 10.3390/ijms241310781
Kim, J. S., Kidokoro, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2024). Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 195, 170–189. doi: 10.1093/plphys/kiae105
Kim, E. J. and Russinova, E. (2020). Brassinosteroid signalling. Curr. Biol. 30, R294–R298. doi: 10.1016/j.cub.2020.02.011
Kou, H., Zhang, X., Jia, J., Xin, M., Wang, J., Mao, L., et al. (2024). Research progress in the regulation of the ABA signaling pathway by E3 ubiquitin ligases in plants. Int. J. Mol. Sci. 25, 7120. doi: 10.3390/ijms25137120
Kovács, S., Nagy, Á., Major, G., Flors, V., Rácz, A., Mauch-Mani, B., et al. (2025). The role of PRLIP2 in the defence and growth of Arabidopsis Insertional mutagenesis of PRLIP2 gene alters hormone balance and defence responses in Arabidopsis. J. Plant Physiol. 314, 154609. doi: 10.1016/j.jplph.2025.154609
Lai, J. Y., Mardiyaningsih, D. I., Rahmadian, F., and Hamzah, N. (2022). What evidence exists on the impact of sustainability initiatives on smallholder engagement in sustainable palm oil practices in Southeast Asia: a systematic map protocol. Environ. Evid. 11, 28. doi: 10.1186/s13750-022-00283-x
Lee, F. C., Yeap, W. C., Kee, S. Y., Kulaveerasingam, H., and Ross Appleton, D. (2024). Core transcriptome network modulates temperature (heat and cold) and osmotic (drought, salinity, and waterlogging) stress responses in oil palm. Front. Plant Sci. 15, 1497017. doi: 10.3389/fpls.2024.1497017
Lei, X., Xiao, Y., Xia, W., Mason, A. S., Yang, Y., Ma, Z., et al. (2014). RNA-seq analysis of oil palm under cold stress reveals a different C-repeat binding factor (CBF) mediated gene expression pattern in Elaeis guineensis compared to other species. PloS One 9, e114482. doi: 10.1371/journal.pone.0114482
Li, S., He, L., Yang, Y., Zhang, Y., Han, X., Hu, Y., et al. (2024). INDUCER OF CBF EXPRESSION 1 promotes cold-enhanced immunity by directly activating salicylic acid signaling. Plant Cell. 36, 2587–2606. doi: 10.1093/plcell/koae096
Li, C., Jia, H., Chai, Y., and Shen, Y. (2011). Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening. Plant Signal Behav. 6, 1950–1953. doi: 10.4161/psb.6.12.18024
Li, W., Wen, Y., Quan, J., Gao, M., Shang, C., Liu, X., et al. (2025). Regulation of jasmonic acid signalling in tomato cold stress response: Insights into the MYB15-LOXD and MYB15-MYC2-LOXD regulatory modules. Plant Biotechnol. J., 70201. doi: 10.1111/pbi.70201
Li, J., Yang, Y., Sun, X., Liu, R., Xia, W., Shi, P., et al. (2022). Development of intron polymorphism markers and their association with fatty acid component variation in oil palm. Front. Plant Sci. 13, 885418. doi: 10.3389/fpls.2022.885418
Li, S. Y., Zhang, Q., Jin, Y. H., Zou, J. X., Zheng, Y. S., and Li, D. D. (2020). A MADS-box gene, EgMADS21, negatively regulates EgDGAT2 expression and decreases polyunsaturated fatty acid accumulation in oil palm (Elaeis guineensis Jacq.). Plant Cell Rep. 39, 1505–1516. doi: 10.1007/s00299-020-02579-z
Li, C., Zhang, H., Qi, Y., Zhao, Y., Duan, C., Wang, Y., et al. (2023). Genome-wide identification of PYL/PYR-PP2C (A)-SnRK2 genes in Eutrema and their co-expression analysis in response to ABA and abiotic stresses. Int. J. Biol. Macromol. 253, 126701. doi: 10.1016/j.ijbiomac.2023.126701
Liu, D., He, Y., Wang, Y., Chen, W., Yang, J., Zhang, Y., et al. (2024). Tetrad stage transient cold stress skews auxin-mediated energy metabolism balance in Chinese cabbage pollen. Plant Physiol. 195, 1312–1332. doi: 10.1093/plphys/kiae123
Liu, J., Li, J., Wang, Z., and Yang, H. (2022). Transcriptome sequencing and bioinformatics analysis of ovarian tissues from pomacea canaliculata in guangdong and hunan. Mediators Inflamm. 2022, 3917036. doi: 10.1155/2022/3917036
Liu, Y., Xiong, Y., Zhao, J., Bai, S., Li, D., Chen, L., et al. (2023). Molecular mechanism of cold tolerance of centipedegrass based on the transcriptome. Int. J. Mol. Sci. 24, 1265. doi: 10.3390/ijms24021265
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25(4):402-8. doi: 10.3390/ijms24021265
Ma, H., Fu, M., Xu, Z., Chu, Z., Tian, J., Wang, Y., et al. (2024). Allele-specific expression of AP2-like ABA repressor 1 regulates iron uptake by modulating rhizosphere pH in apple. Plant Physiol. 196, 2121–2136. doi: 10.1093/plphys/kiae452
Maharajan, T., Krishna, T. P. A., Rakkammal, K., Ceasar, S. A., and Ramesh, M. (2022). Application of CRISPR/Cas system in cereal improvement for biotic and abiotic stress tolerance. Planta. 256, 106. doi: 10.1007/s00425-022-04023-w
Matsukura, K., Mizutani, N., Tanaka, S., and Tanaka, Y. (2024). Evaluation of overwintering risk of tropical and subtropical insect pests in temperate regions. Sci Rep. 14(1):31333.
Niu, S. Q., Li, H. R., Paré, P. W., Aziz, M., Wang, S. M., Shi, H. Z., et al. (2016). Induced growth promotion and higher salt tolerance in the halophyte grass Puccinellia tenuiflora by beneficial rhizobacteria. Plant Soil. 407, 217–230. doi: 10.1007/s11104-015-2767-z
Peng, T., Guo, C., Yang, J., Wan, X., Wang, W., Zhang, J., et al. (2023). Transcriptome analysis revealed molecular basis of cold response in Prunus mume. Mol. Breed. 43, 34. doi: 10.1007/s11032-023-01376-2
Porra, R. J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynthesis Res. 73, 149–156. doi: 10.1023/A:1020470224740
Rahman, A. (2013). Auxin: a regulator of cold stress response. Physiol. Plant 147, 28–35. doi: 10.1111/j.1399-3054.2012.01617.x
Saand, M. A., Li, J., Wu, Y., Zhou, L., Cao, H., and Yang, Y. (2022). Integrative omics analysis of three oil palm varieties reveals (Tanzania × Ekona) TE as a cold-resistant variety in response to low-temperature stress. Int. J. Mol. Sci. 23, 14926. doi: 10.3390/ijms232314926
Santanoo, S., Vongcharoen, K., Banterng, P., Vorasoot, N., Jogloy, S., Roytrakul, S., et al. (2022). Physiological and proteomic responses of cassava to short-term extreme cool and hot temperature. Plants (Basel). 11, 2307. doi: 10.3390/plants11172307
Sarkar, M. S. K., Begum, R. A., and Pereira, J. J. (2020). Impacts of climate change on oil palm production in Malaysia. Environ. Sci. pollut. Res. Int. 27, 9760–9770. doi: 10.1007/s11356-020-07601-1
Shahan, R. (2020). The cold never bothered me anyway: DELLA-interacting GROWTH REGULATING FACTORS mediate plant growth in cold stress. Plant Cell. 32, 797–798. doi: 10.1105/tpc.20.00079
Sharma, E., Borah, P., Kaur, A., Bhatnagar, A., Mohapatra, T., Kapoor, S., et al. (2021). A comprehensive transcriptome analysis of contrasting rice cultivars highlights the role of auxin and ABA responsive genes in heat stress response. Genomics. 113, 1247–1261. doi: 10.1016/j.ygeno.2021.03.007
Shi, H. and Chan, Z. L. (2014). AtHAP5A modulates freezing stress resistance in Arabidopsis independent of the CBF pathway. Plant Signal Behav. 9, e29109. doi: 10.4161/psb.29109
Shibasaki, K., Uemura, M., Tsurumi, S., and Rahman, A. (2009). Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell. 21, 3823–3838. doi: 10.1105/tpc.109.069906
Su, X., Wang, J., Sun, S., Peng, W., Li, M., Mao, P., et al. (2024). Genome-wide identification of the EIN3/EIL transcription factor family and their responses under abiotic stresses in Medicago sativa. BMC Plant Biol. 24, 898. doi: 10.1186/s12870-024-05588-2
Teixeira, R. S., da Silva, A. S., Ferreira-Leitao, V. S., and da Silva Bon, E. P. (2012). Amino acids interference on the quantification of reducing sugars by the 3,5-dinitrosalicylic acid assay mislead carbohydrase activity measurements. Carbohydr Res. 363, 33–37. doi: 10.1016/j.carres.2012.09.024
VanderWilde, C. P., Newell, J. P., Gounaridis, D., and Goldstein, B. P. (2023). Deforestation, certification, and transnational palm oil supply chains: Linking Guatemala to global consumer markets. J. Environ. Manage. 344, 118505. doi: 10.1016/j.jenvman.2023.118505
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., and Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi: 10.1038/s41580-022-00479-6
Wang, X., Li, Y., Chen, Z., Li, L., Li, Q., Geng, Z., et al. (2025). MbWRKY50 confers cold and drought tolerance through upregulating antioxidant capacity associated with ROS scavenging. J. Plant Physiol. 310, 154526. doi: 10.1016/j.jplph.2025.154526
Wang, B., Ma, M., and Lu, H. (2015). Photosynthesis sucrose metabolism, and starch accumulation in two NILs of winter wheat. Photosynth. Res. 126, 363–373. doi: 10.1007/s11120-015-0126-9
Wang, J., Song, L., Gong, X., Xu, J., and Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21, 1446. doi: 10.3390/ijms21041446
Wei, J., Liu, D., Liu, Y., and Wei, S. (2022). Physiological Analysis and Transcriptome Sequencing Reveal the Effects of Salt Stress on Banana (Musa acuminata cv. BD) Leaf. Front. Plant Sci. 13, 822838. doi: 10.3389/fpls.2022.822838
Wen, Z., Li, M., Meng, J., Miao, R., Liu, X., Fan, D., et al. (2023). Genome-wide identification of the MAPK and MAPKK gene families in response to cold stress in prunus mume. Int. J. Mol. Sci. 24, 8829. doi: 10.3390/ijms24108829
Wu, G., Baumeister, R., and Heimbucher, T. (2023). Molecular mechanisms of lipid-based metabolic adaptation strategies in response to cold. Cells. 12, 1353. doi: 10.3390/cells12101353
Wu, Z., Han, S., Zhou, H., Tuang, Z. K., Wang, Y., Jin, Y., et al. (2019). Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway. Plant Cell Environ. 42, 2645–2663. doi: 10.1111/pce.13579
Xia, W., Mason, A. S., Xiao, Y., Liu, Z., and Yang, Y. (2014). Analysis of multiple transcriptomes of the African oil palm (Elaeis guineensis) to identify reference genes for RT-PCR. J. Biotechnol. 184, 63–73. doi: 10.1016/j.jbiotec.2014.05.008
Xiang, Z., Zhang, L., Zhang, M., Yao, Y., Qian, Q., Wei, Z., et al. (2025). OsNCED5 confers cold stress tolerance through regulating ROS homeostasis in rice. Plant Physiol. Biochem. 220, 109455. doi: 10.1016/j.plaphy.2024.109455
Xiao, Y., Zhou, L., Xia, W., Mason, A. S., Yang, Y., Ma, Z., et al. (2014). Exploiting transcriptome data for the development and characterization of gene-based SSR markers related to cold tolerance in oil palm (Elaeis guineensis). BMC Plant Biol. 14, 384. doi: 10.1186/s12870-014-0384-2
Xing, C., Li, J., Yuan, H., and Yang, J. (2022). Physiological and transcription level responses of microalgae Auxenochlorella protothecoides to cold and heat induced oxidative stress. Environ. Res. 211, 113023. doi: 10.1016/j.envres.2022.113023
Xu, Y., Hu, W., Song, S., Ye, X., Ding, Z., Liu, J., et al. (2022). MaDREB1F confers cold and drought stress resistance through common regulation of hormone synthesis and protectant metabolite contents in banana. Hortic. Res. 10, uhac275. doi: 10.1093/hr/uhac275
Ye, J., Yang, H., Shi, H., Wei, Y., Tie, W., Ding, Z., et al. (2017). The MAPKKK gene family in cassava: Genome-wide identification and expression analysis against drought stress. Sci. Rep. 7, 14939. doi: 10.1038/s41598-017-13988-8
Yılmaz, B. and Ağagündüz, D. (2022). Fractionated palm oils: emerging roles in the food industry and possible cardiovascular effects. Crit. Rev. Food Sci. Nutr. 62, 1990–1998. doi: 10.1080/10408398.2020.1869694
Yu, C., Dong, W., Zhan, Y., Huang, Z. A., Li, Z., Kim, I. S., et al. (2017). Genome-wide identification and expression analysis of ClLAX, ClPIN and ClABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 18, 33. doi: 10.1186/s12863-017-0500-z
Zhang, Q., Li, Y., Cao, K., Xu, H., and Zhou, X. (2023). Transcriptome and proteome depth analysis indicate ABA, MAPK cascade and Ca2+ signaling co-regulate cold tolerance in Rhododendron chrysanthum Pall. Front. Plant Sci. 14, 1146663. doi: 10.3389/fpls.2023.1146663
Zhang, H., Zhao, Y., and Zhu, J. K. (2020). Thriving under stress: how plants balance growth and the stress response. Dev. Cell. 55, 529–543. doi: 10.1016/j.devcel.2020.10.012
Zhou, L., Li, R., Yang, X., Peng, Y., Wang, Y., Xu, Q., et al. (2025). Interaction of R2R3-MYB transcription factor EgMYB111 with ABA receptors enhances cold tolerance in oil palm. Int. J. Biol. Macromol. 305, 141223. doi: 10.1016/j.ijbiomac.2025.141223
Zhou, F., Singh, S., Zhang, J., Fang, Q., Li, C., Wang, J., et al. (2023). The MEKK1-MKK1/2-MPK4 cascade phosphorylates and stabilizes STOP1 to confer aluminum resistance in Arabidopsis. Mol. Plant 16, 337–353. doi: 10.1016/j.molp.2022.11.010
Zhou, L., Yarra, R., Yang, Y., Liu, Y., Yang, M., and Cao, H. (2022). The oil palm R2R3-MYB subfamily genes EgMYB111 and EgMYB157 improve multiple abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Rep. 41, 377–393. doi: 10.1007/s00299-021-02814-1
Keywords: oil palm, cold stress, transcriptome, differentially expressed genes (DEGs), signaling pathway
Citation: Wu Q, Yang X, Huang Q, Li R, Zeng X, Li Q, Li Z, Fu D, Cao H, Li X, Liu X and Zhou L (2025) Comparative transcriptomics reveals key regulatory networks underlying cold stress adaptation in oil palm (Elaeis guineensis). Front. Plant Sci. 16:1687366. doi: 10.3389/fpls.2025.1687366
Received: 17 August 2025; Accepted: 31 October 2025;
Published: 26 November 2025.
Edited by:
Ahmad A. Omar, University of Florida, United StatesReviewed by:
Mumtaz Ali Saand, Zhejiang University, ChinaChunxia Wu, Shandong Normal University, China
Zafar Iqbal, Independent Researcher, Bahawalpur, Pakistan
Hesham Oraby, Zagazig University, Egypt
Copyright © 2025 Wu, Yang, Huang, Li, Zeng, Li, Li, Fu, Cao, Li, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixia Zhou, Z2x6el8yMDA5QDE2My5jb20=
†These authors have contributed equally to this work
 Qiufei Wu†
Qiufei Wu† Lixia Zhou
Lixia Zhou