- College of Life Sciences, Qufu Normal University, Qufu, China
Flowering time is a critical determinant of crop yield and adaptability, regulated by the integration of environmental cues, phytohormones, and genetic networks. Abiotic stresses such as drought, waterlogging, salinity, and high temperature, together with biotic stresses including pathogens, viruses, and herbivores, profoundly reshape flowering phenology through microRNAs, transcription factors, epigenetic modifications, and hormonal crosstalk. Phytohormones, especially ABA and GA, act as regulatory hubs coordinating stress adaptation and floral transition, though their effects vary across species and conditions. This review synthesizes recent advances in stress-mediated flowering regulation and emphasizes the challenges of balancing stress tolerance with yield stability. We propose that integrating multi-omics data, regulatory network modeling, and artificial intelligence will accelerate the breeding of stress-resilient cultivars with stable productivity.
1 Introduction
Recent abrupt environmental shifts have forced plants to undergo significant evolutionary adaptations to ensure survival. To mitigate the impacts of both internal and external environmental fluctuations on growth and reproduction, plants have developed sophisticated mechanisms to sense and respond to external cues such as light, temperature, and stress. Among these adaptive strategies, the regulation of flowering time plays a central role in coping with environmental challenges (Gu et al., 2022) and serves as a critical determinant of agricultural productivity (Ying et al., 2023).
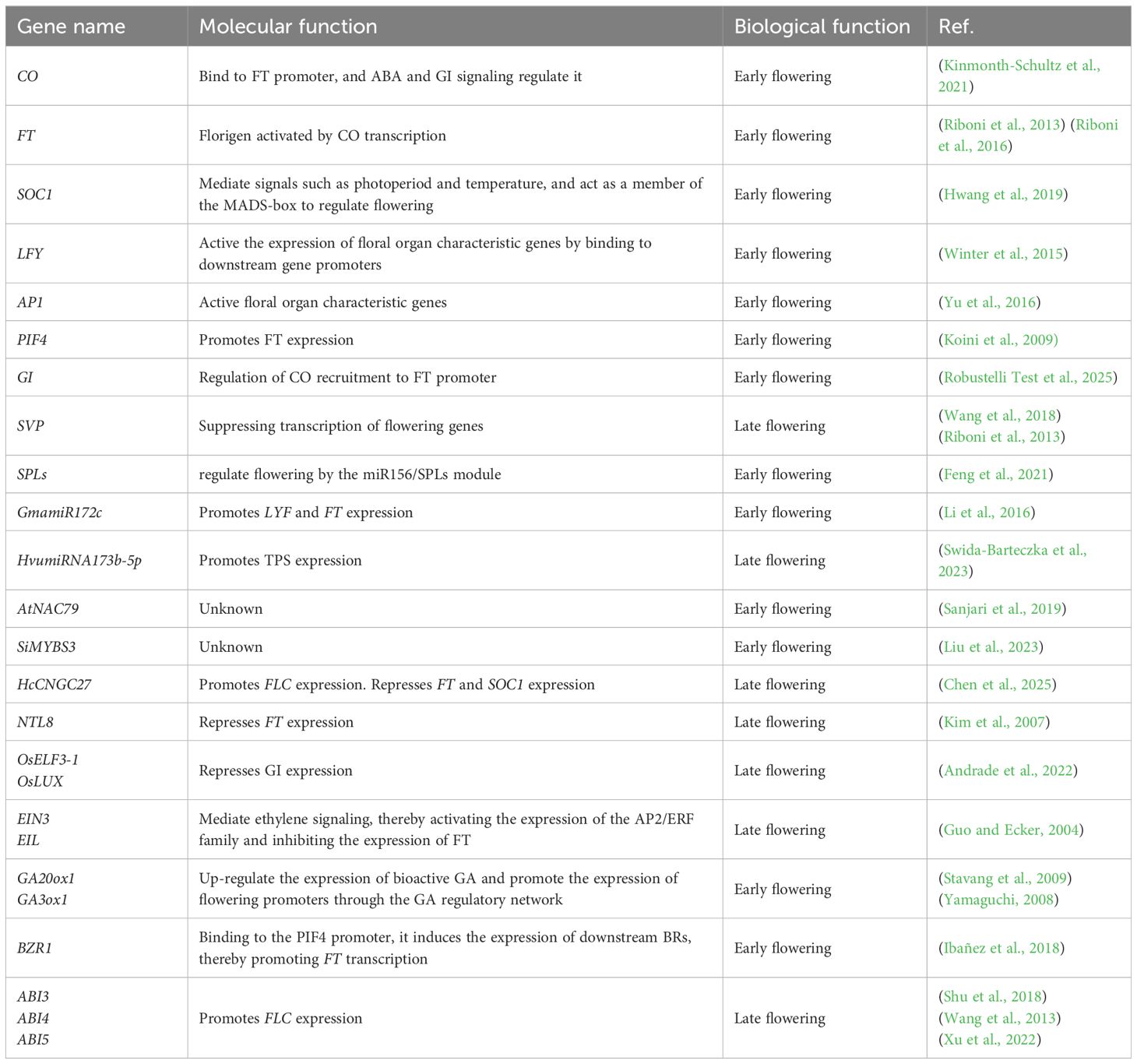
Flowering is controlled by the integration of exogenous signals—including temperature, photoperiod and endogenous cues such as developmental stage, phytohormone levels, and nutrient status (Zhao et al., 2023). These signals converge on key floral integrators, including FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1). The integrated signals are subsequently transmitted to the shoot apical meristem, where they activate floral identity genes such as LEAFY (LFY) and APETALA1 (AP1), thereby precisely triggering the floral transition (Winter et al., 2015; Yu et al., 2016) (Table 1). This review systematically examines the molecular mechanisms through which plants and crops integrate light, temperature, phytohormones, and environmental stresses to fine-tune flowering. By doing so, we establish a comprehensive framework for understanding adaptive flowering networks under climatic perturbations and provide a conceptual basis for targeted flowering regulation and crop improvement strategies.
2 Abiotic stress-mediated regulation of flowering
Crop yield is highly sensitive to environmental conditions. The strategic adjustment of flowering phenology under stress represents a core adaptive mechanism for balancing reproductive fitness across generations while enhancing crop resilience. Within this regulatory framework, diverse biotic and abiotic stressors coordinately modulate flowering timing and stress tolerance through integrative signaling networks that converge on key developmental regulators (Figure 1).
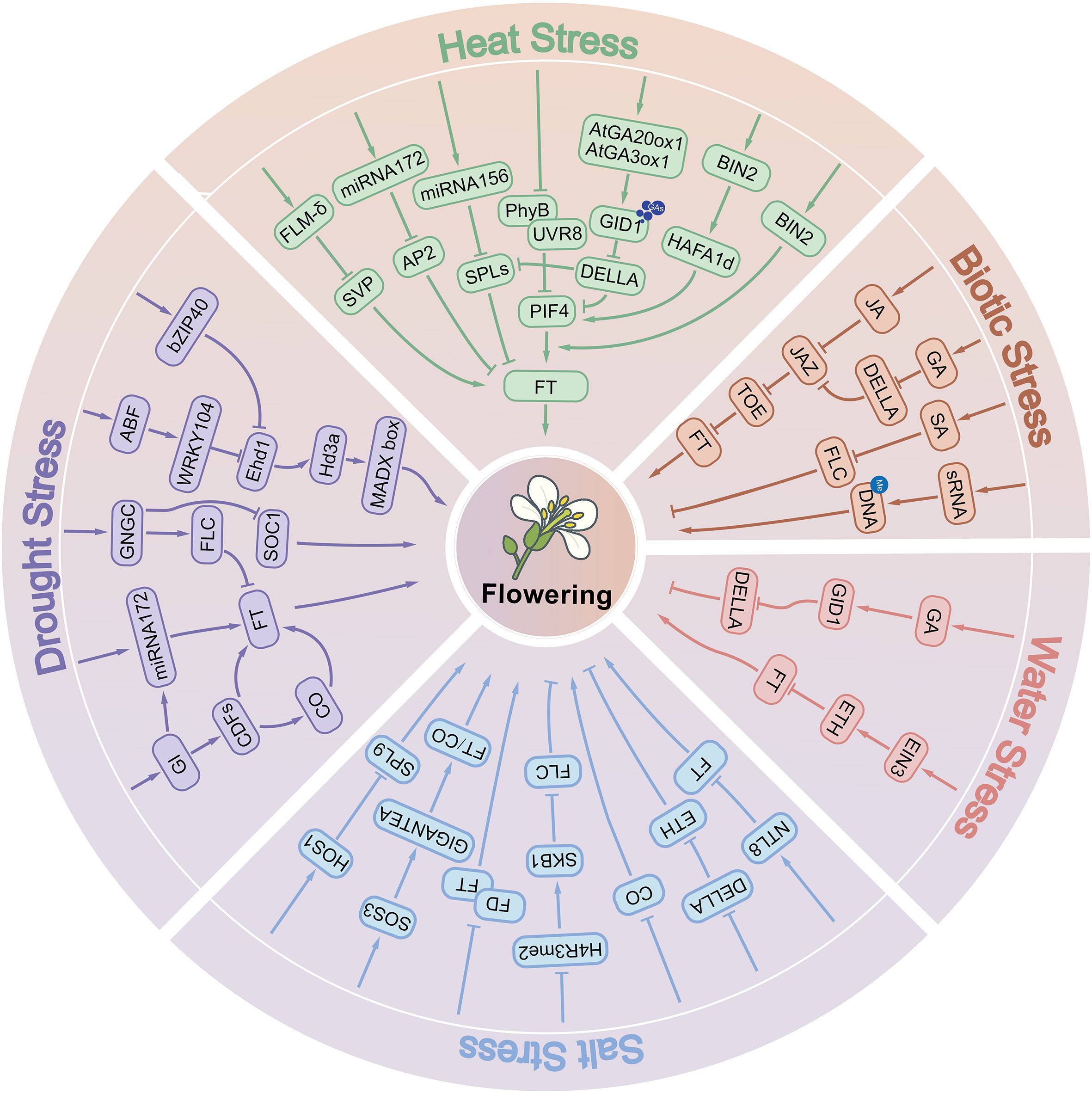
Figure 1. Molecular mechanisms underlying abiotic and biotic stress-mediated regulation of flowering time in plants.
2.1 Drought stress and flowering regulation
MicroRNAs (miRNAs) play crucial roles in mediating crop responses to drought stress. For example, in soybean, drought stress and abscisic acid (ABA) signaling synergistically induce gma-miR172c, which enhances ABA sensitivity by transcriptionally activating ABI3 and ABI5 (Li et al., 2016). This regulatory cascade promotes the accumulation of the flowering integrator FT, thereby accelerating floral transition under water-deficit conditions (Figure 1). A similar mechanism has been reported in barley, where drought-induced downregulation of miRNA173b-5p elevates trehalose accumulation. The metabolic intermediate trehalose-6-phosphate (T6P) subsequently promotes flowering via the trehalose biosynthetic pathway (Swida-Barteczka et al., 2023).
In parallel, transcription factors (TFs) play central roles in coordinating drought-mediated flowering. The NAC family, one of the largest TF families in crops, functions as a key regulator at the intersection of flowering time control and stress tolerance (Sanjari et al., 2019). For instance, overexpression of NAC79 in Arabidopsis confers both drought tolerance and an early-flowering phenotype (Guo et al., 2017). Similarly, MYB TFs are widely involved in abiotic stress responses (Ai et al., 2023), with MYBS3 overexpression shown to enhance drought tolerance while simultaneously promoting early flowering (Liu et al., 2023). By contrast, CNGC family genes exhibit an opposing effect: they enhance drought resistance through reactive oxygen species (ROS) scavenging and stress-gene activation, but delay floral transition by repressing FT and SOC1 (Chen et al., 2025) (Figure 1). This highlights the complex regulatory trade-offs between stress adaptation and reproductive timing under drought conditions.
2.2 Waterlogging stress and flowering regulation
Crop responses to waterlogging stress represent a complex physiological process, with root hypoxia acting as the primary stress factor. Ethylene, a key gaseous phytohormone, functions not only as a central stress signal but also as a regulator of flowering, influencing floral timing through multilayered signaling networks (Guo and Ecker, 2004). At the molecular level, ethylene signaling is mediated by EIN3/EIL transcription factors, which activate AP2/ERF family proteins such as ERF1, while simultaneously repressing the flowering integrator FT, ultimately leading to delayed flowering (Figure 1). This ethylene-dependent regulation is tightly coupled with gibberellin (GA) metabolism and DELLA protein stability (Figure 1). Reduced GA levels inhibit DELLA degradation, resulting in DELLA accumulation and suppression of floral initiation (Achard et al., 2007). Furthermore, under combined waterlogging and shading stress in maize (Zea mays), ethylene overaccumulation interferes with flowering through a dual mechanism: repression of FT expression and reduced activity of the CONSTANS (CO) protein, both of which markedly delay the floral transition (Zhou et al., 2021). A comprehensive understanding of how waterlogging stress interacts with flowering pathways is essential for breeding cultivars with enhanced tolerance to both drought and waterlogging, thereby minimizing yield penalties under adverse environmental conditions.
2.3 Salt stress and flowering regulation
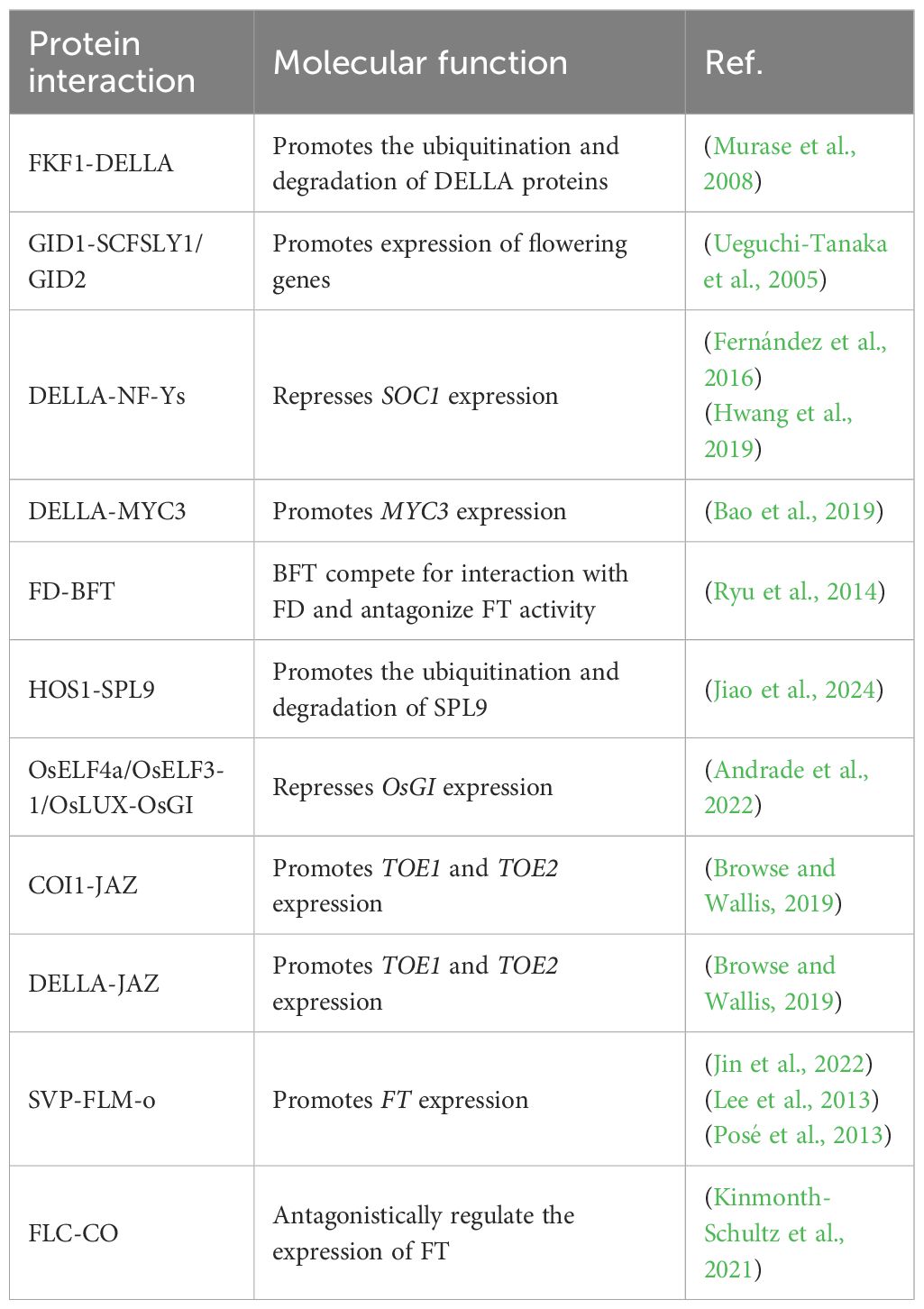
Salt stress represents a major abiotic constraint compromising crop growth, yield, and quality. As a leading contributor to global crop losses, salinity profoundly regulates flowering phenology through identified molecular pathways, providing foundational insights into the mechanistic basis of floral transition under salt stress. With respect to specific regulatory mechanisms, multiple molecular pathways and related proteins play key roles in salt stress-induced flowering delay in Arabidopsis. The plant-specific transcription factor NTL8 represses FT expression under high salinity (Kim et al., 2007), while salt stress concurrently suppresses CO transcription (Li et al., 2007) (Figure 1). Additionally, molecules related to floral initiation are closely involved in salt stress-regulated flowering. The floral initiator Shk1 kinase-binding protein 1 (SKB1) and symmetric dimethylation of histone H4 arginine 3 (H4R3sme2) play important roles in regulating flowering time under salt stress (Figure 1). Under salt stress, H4R3sme2 levels decrease, leading to the dissociation of SKB1 from chromatin, which in turn induces the expression of FLOWERING LOCUS C (FLC), ultimately delaying flowering (Zhang et al., 2011). The bZIP transcription factor FD, a positive regulator of flowering, interacts with the flowering repressors BROTHER OF FT and TFL1 (BFT). This interaction interferes with the normal interaction between FT and FD, resulting in delayed flowering of Arabidopsis under salt stress (Ryu et al., 2014) (Figure 1). The osmotic response protein HOS1 ubiquitinates the transcription factor SPL9 under salt stress, promoting its degradation and thereby delaying flowering in Arabidopsis (Jiao et al., 2024) (Figure 1). In rice, components of the Evening Complex (EC), including OsELF4a, OsELF3-1, and OsLUX, play important roles in response to salt stress (Table 1). Under salt stress, these proteins bind to the promoter of OsGI (GIGANTEA), repress its expression, and delay flowering (Andrade et al., 2022). Most studies on flowering regulation under salt stress focus mainly on the model plant Arabidopsis, whereas studies on important crop species, such as rice, remain limited and are in the early stages. Therefore, further research on the mechanisms underlying the regulation of flowering under salt stress in economically important crops, such as rice, soybean, and maize, is highly important for the breeding of stress-resistant and early-maturing varieties.
2.4 High temperature-mediated regulation of flowering
Global warming is profoundly altering the flowering times of plant species worldwide. Failure to complete the floral transition at the optimal developmental stage compromises reproductive success and often results in yield reduction in crops. High temperature generally promotes flowering, primarily through the activation of flowering inducers-such as PIF4, miR172, and SPLs (Feng et al., 2021; Koini et al., 2009) and through the transcriptional suppression of flowering repressors, including miR156, SVP, and DELLA proteins. These regulatory inputs converge on FT, the central integrator of flowering signals, which mediates the temperature-responsive floral transition (Figure 1, Table 2). At the molecular level, the transcriptional regulation of FT involves FLOWERING LOCUS M (FLM) and SHORT VEGETATIVE PHASE (SVP). FLM promotes FT expression, whereas SVP acts as a repressor. Notably, FLM undergoes temperature-dependent alternative splicing, generating two isoforms: FLM-β and FLM-δ. Under high temperatures, FLM-δ becomes predominant and forms a SVP–FLM-δ complex, which interferes with the ability of SVP to repress FT, thereby accelerating flowering (Jin et al., 2022; Lee et al., 2013; Posé et al., 2013).
Phytohormones also play critical roles in high temperature–induced flowering. Elevated temperatures upregulate the expression of GA biosynthesis genes AtGA20ox1 and AtGA3ox1, resulting in increased accumulation of the bioactive gibberellin GA4 (Stavang et al., 2009; Yamaguchi, 2008). Binding of GA4 to its receptor GID1 facilitates recruitment of the SCF^SLY1/GID2 complex, which targets DELLA proteins—negative regulators of GA signaling—for ubiquitin-dependent degradation. DELLA removal releases their repression on flowering activators such as miR172, SPLs, and PIF4, thus promoting flowering under high temperature (Galvão et al., 2012; Sun, 2011; Van De Velde et al., 2017; Yu et al., 2012) (Figure 1). In addition, brassinosteroids (BRs) contribute to heat-responsive flowering regulation. High temperature induces nuclear accumulation of the BR signaling transcription factor BZR1, which directly binds to the PIF4 promoter to enhance its expression (Ibañez et al., 2018). This activation further promotes FT transcription, collectively accelerating flowering under elevated temperatures.
3 Biotic stress and flowering regulation
Biotic stresses, including pathogens and pests, can profoundly influence flowering by altering the expression of key flowering genes (e.g., FLC, FT, GI), disrupting floral organogenesis (e.g., stigma, filament, anther), and impairing pollen viability (Lyons et al., 2015; Fan et al., 2022; Li et al., 2025) (Table 1).
Fungal pathogens exhibit strong tissue specificity in their infection strategies. Root-colonizing fungi such as Piriformospora indica and Pochonia chlamydosporia systemically accelerate flowering by manipulating phytohormonal pathways and directly regulating flowering gene expression (Cheng et al., 2004; Kim et al., 2017; Pan et al., 2017). In contrast, floral-infecting fungi such as Ustilaginoidea virens and Claviceps purpurea employ highly localized strategies, interfering with gametophyte development and seed formation through effector proteins and physical replacement of reproductive structures (Sun et al., 2020).
Viruses display distinct infection and transmission strategies. Unlike most pathogens, viruses are often excluded from meristematic tissues. However, species such as PNRSV and ToBRFV exploit pollen as transmission vectors, impairing pollen viability and tube growth (Amari et al., 2009; Avni et al., 2022). Owing to their systemic nature, viruses can also modulate flowering time by perturbing phytohormone signaling networks. In addition, biotic stress imposed by viral infection can induce early flowering. For example, the Foxtail Mosaic Virus–based VIF system (FoMViF) promotes early flowering in monocots and cereals, largely through upregulation of FT, although the role of DNA methylation in this process remains unclear. Moreover, small RNAs (sRNAs) have been shown to mediate methylation of both host and viral DNA, ultimately promoting precocious flowering in infected hosts (Zhang et al., 2015).
Insect herbivory further adds to the complexity of biotic stress–flowering interactions. In Arabidopsis, herbivore attack activates the jasmonic acid (JA) signaling pathway, which plays a dual role in defense and developmental regulation. JA accumulation promotes COI1-dependent degradation of JASMONATE-ZIM DOMAIN (JAZ) repressors, thereby releasing repression on TOE transcription factors. As a result, TOE1 and TOE2 more strongly suppress FT, leading to delayed flowering (Browse and Wallis, 2019; Li et al., 2004). In parallel, DELLA proteins physically interact with JAZ, alleviating JAZ-mediated inhibition of TOE activity and reinforcing FT repression, thus establishing a genetic link between the GA and JA pathways in the regulation of flowering (Figure 1, Table 2). Collectively, biotic stresses regulate flowering through diverse mechanisms—including transcriptional reprogramming, hormonal crosstalk, epigenetic modification, and reproductive organ disruption. These findings underscore the intricate balance plants must maintain between defense and reproduction when challenged by pathogens and herbivores.
4 Phytohormone-mediated regulation of flowering
4.1 Regulation of flowering by abscisic acid
Abscisic acid (ABA), a central stress-responsive phytohormone, restricts crop growth and development under adverse conditions while simultaneously modulating flowering time through multiple pathways across species. However, the role of ABA in floral transition remains debated, as it exerts both promotive and inhibitory effects depending on the environmental context. This functional divergence arises primarily from its differential regulatory actions under distinct photoperiods and across crop species.
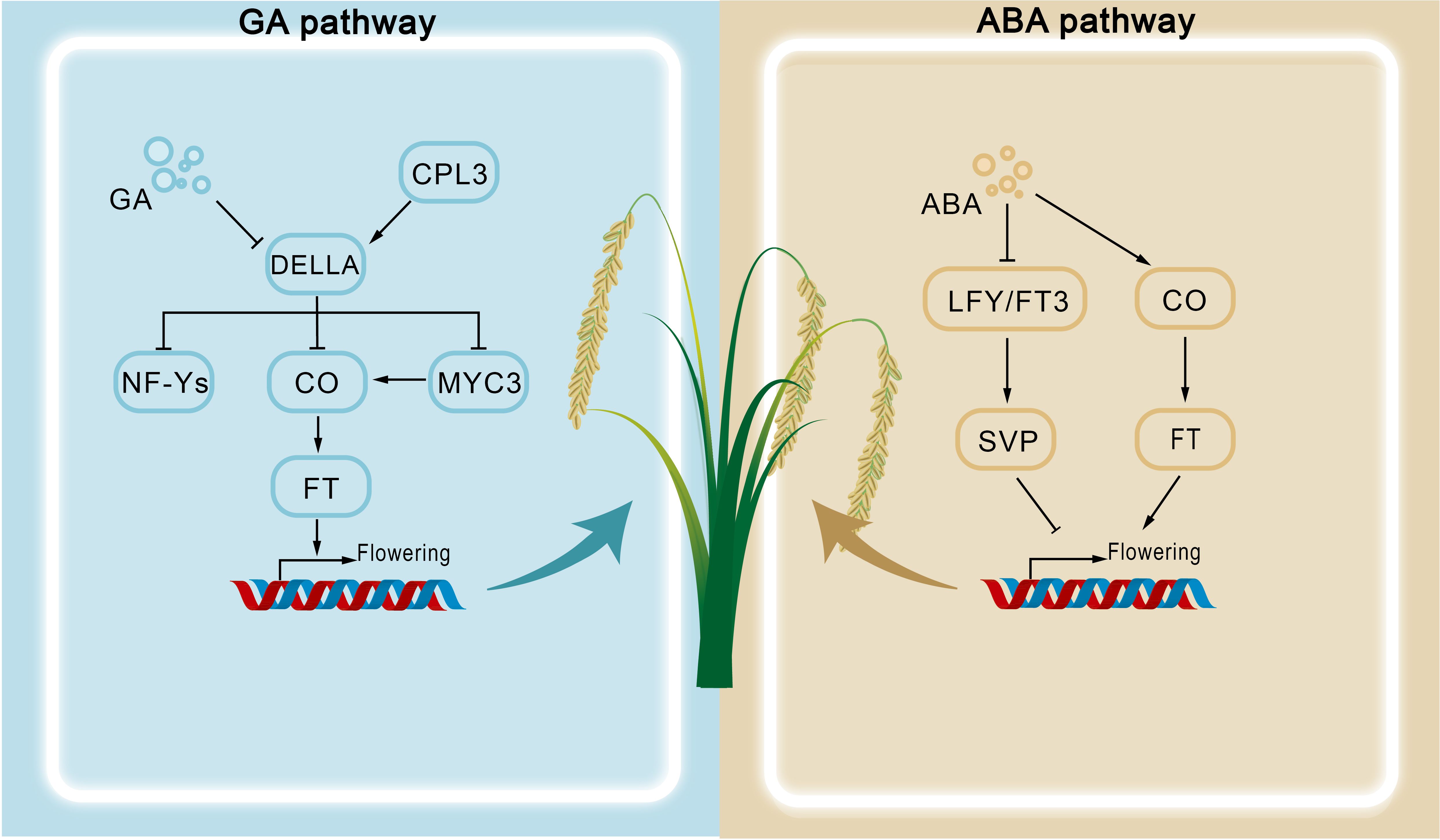
In Arabidopsis, ABA promotes early flowering under drought stress by interacting with components of the photoperiod pathway, such as GIGANTEA (GI) and CONSTANS (CO), thereby ensuring reproductive success. Specifically, ABA signaling enhances CO binding to the CORE cis-element of the FLOWERING LOCUS T (FT) promoter, inducing FT expression and accelerating flowering (Kinmonth-Schultz et al., 2021) (Figure 2). This ABA-induced pathway may be further coordinated by GI, although the precise molecular mechanism remains to be clarified (Robustelli Test et al., 2025). Supporting this view, mutants of ABA biosynthesis genes ABA DEFICIENT 1 (ABA1) and ABA2 exhibit delayed flowering under long-day (LD) conditions, but not under short-day (SD) conditions, highlighting ABA's promotive role under LD photoperiods (Riboni et al., 2013). Mechanistically, this ABA-dependent acceleration occurs largely through upregulation of FT and TWIN SISTER OF FT (TSF) during drought stress (Riboni et al., 2013, Riboni et al., 2016). By contrast, under SD conditions, drought stress delays flowering. This inhibitory effect is mediated by ABA-dependent upregulation of flowering repressors such as SHORT VEGETATIVE PHASE (SVP) (Wang et al., 2018), which suppress downstream floral identity genes (Riboni et al., 2013) (Figure 2). Consistently, loss-of-function mutants of ABA signaling components ABI3, ABI4, and ABI5 display accelerated flowering, whereas their overexpression delays floral initiation (Table 1). These findings emphasize the complexity of ABA concentration–dependent regulation and pathway integration, which remain incompletely understood.
Importantly, ABA's role in flowering exhibits significant interspecies divergence. In Crocus sativus, for instance, exogenous ABA application inhibits flowering by repressing both floral induction and development, a striking contrast to the ABA-promoted drought escape mechanism in Arabidopsis (Singh et al., 2023). This suggests evolutionarily conserved yet species-specific regulatory frameworks. Accumulating evidence also highlights extensive hormonal crosstalk involving ABA. For example, ABI4 activates the gibberellin catabolic gene GA2ox7, reducing bioactive GA levels and thereby indirectly modulating flowering via GA homeostasis (Shu et al., 2016). Moreover, strigolactones (SLs) and ABA share a common biosynthetic precursor and demonstrate reciprocal regulation during abiotic stress responses. Under favorable growth conditions, ABA–SL equilibrium maintains developmental homeostasis, ensuring normal crop growth (Chi et al., 2021). In contrast, drought stress suppresses SLs while elevating ABA, activating defense pathways and accelerating flowering (Bai et al., 2024).
4.2 Regulation of flowering time by gibberellins
Gibberellins (GAs) are a major class of phytohormones that profoundly influence crop growth and development, particularly stem elongation and the floral transition. GA generally promotes flowering by targeting DELLA proteins—transcriptional repressors that constrain floral initiation—for degradation (Sun, 2011). In many crop species, GA functions as a key flowering promoter, especially in those requiring vernalization or LD photoperiods to induce floral development.
DELLA proteins serve as central repressors within GA signaling, with their activity tightly regulated at the cellular level. Under stress conditions, enhanced expression of GA biosynthetic enzymes GA20ox and GA3ox increases the accumulation of bioactive GA (Ito et al., 2018). Bioactive GA binds to its receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1), inducing a conformational change that facilitates recruitment of the SCF^SLY1/GID2 E3 ubiquitin ligase complex. This interaction forms the GA–GID1–DELLA ternary complex, leading to ubiquitination and proteasomal degradation of DELLAs (Ueguchi-Tanaka et al., 2005; Murase et al., 2008). Depletion of DELLA proteins relieves transcriptional repression on flowering regulators, thereby accelerating flowering under LD conditions (Ito et al., 2018) (Table 2). Intriguingly, FKF1, a flowering regulator, promotes DELLA ubiquitination, while DELLAs suppress FKF1 transcription, establishing a negative feedback loop that fine-tunes floral initiation (Yan et al., 2020).
Beyond protein degradation, GA also regulates the transcription of core flowering genes such as FT and SOC1. The nuclear factor Y (NF-Y) transcription complex integrates photoperiodic and GA signals to activate SOC1 and FT. Under GA-deficient conditions, stabilized DELLAs sequester NF-Y subunits, repressing SOC1 expression. GA-mediated DELLA degradation liberates NF-Y, enabling it to complex with CONSTANS (CO) and activate SOC1 and FT, thus promoting flowering (Fernández et al., 2016; Hwang et al., 2019). In addition, DELLAs interact with the bHLH transcription factor MYC3, stabilizing it and allowing MYC3 to compete with CO for binding to the FT promoter. This antagonism impairs CO-mediated activation of FT, thereby delaying the floral transition (Bao et al., 2019) (Figure 2, Table 2).
The promotive effects of GA on flowering hold significant agronomic potential. For instance, exogenous GA application accelerates development and flowering in barley (Hordeum vulgare L.), whereas trinexapac-ethyl, a GA biosynthesis inhibitor, markedly delays flowering in a dose-dependent manner, independent of application timing (Kupke et al., 2021). Such vegetative phase extension through GA suppression enables strategic alignment of flowering with favorable reproductive windows, thereby maximizing yield potential. Conversely, GA promotion can be leveraged for precocious maturity where early flowering is desirable. These findings highlight GA modulation as a versatile agronomic strategy for optimizing crop productivity and adaptability.
5 Discussion
Floral transition at the appropriate developmental stage is critical for plant survival and plays a decisive role in improving crop yield. Yet, diverse biotic and abiotic stresses increasingly disrupt flowering schedules in many crops, thereby compromising productivity. This review synthesizes the molecular mechanisms underlying flowering regulation under the combined influences of photothermal cues, phytohormones, and environmental stresses, aiming to provide an integrative framework for understanding floral transition under adverse conditions and to inform strategies for crop improvement.
Despite recent advances, significant challenges remain in breeding stress-resilient cultivars. Flowering responses to stress are complex quantitative traits, governed by multiple genes (Atlin et al., 2017), and further complicated by the intricate cross-talk between stress signaling pathways and gene regulatory networks. This complexity hampers genetic dissection and hinders the precise identification of causal loci. Moreover, breeders face the persistent challenge of balancing stress tolerance with yield stability, not only within a single generation but also across successive breeding cycles. The simultaneous enhancement of stress resistance, flowering time optimization, and yield potential remains an elusive goal. Another pressing issue is the limited adaptability of many stress-resilient cultivars. Such genotypes often exhibit narrow ecological adaptation, resulting in significant performance variation across different geographical regions and production environments. This restricts their large-scale deployment and practical utility.
Looking forward, future efforts should emphasize the systematic integration of multi-omics approaches—including genomics, transcriptomics, proteomics, and metabolomics—to unravel the functional and mechanistic basis of gene clusters that govern stress resilience and flowering time. Building comprehensive gene regulatory network models will provide new insights into the dynamic interplay between environmental cues and developmental programs. Furthermore, leveraging big data analytics and artificial intelligence (AI) offers substantial promise for enhancing the precision and efficiency of breeding pipelines. By coupling multi-omics datasets with advanced computational tools, breeders can accelerate the development of cultivars that combine multi-stress resilience with superior agronomic performance, ultimately contributing to global food security under changing climates.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
WD: Writing – original draft. YZ: Writing – original draft. YX: Writing – original draft. KC: Writing – original draft. NQ: Writing – review & editing. YS: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant nos. 32371759, 32070304).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achard, P., Baghour, M., Chapple, A., Hedden, P., van der Straeten, D., Genschik, P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U.S.A. 104, 6484–6489. doi: 10.1073/pnas.0610717104
Ai, P., Xue, J., Shi, Z., Liu, Y., Li, Z., Li, T., et al. (2023). Genome-wide characterization and expression analysis of MYB transcription factors in Chrysanthemum nankingense. BMC Plant Biol. 23, 140. doi: 10.1186/s12870-023-04137-7
Amari, K., Burgos, L., Pallás, V., and Sánchez-Pina, M. A. (2009). Vertical transmission of Prunus necrotic ringspot virus: hitch-hiking from gametes to seedling. J. Gen. Virol. 90(Pt 7), 1767–1774. doi: 10.1099/vir.0.009647-0
Andrade, L., Lu, Y., Cordeiro, A., Costa, J. M. F., Wigge, P. A., Saibo, N. J. M., et al. (2022). The evening complex integrates photoperiod signals to control flowering in rice. Proc. Natl. Acad. Sci. U.S.A. 119, e2122582119. doi: 10.1073/pnas.2122582119
Atlin, G. N., Cairns, J. E., and Das, B. (2017). Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Glob Food Sec 12, 31–37. doi: 10.1016/j.gfs.2017.01.008
Avni, B., Gelbart, D., Sufrin-Ringwald, T., Zemach, H., Belausov, E., Kamenetsky-Goldstein, R., et al. (2022). ToBRFV infects the reproductive tissues of tomato plants but is not transmitted to the progenies by pollination. Cells. 11, 2864. doi: 10.3390/cells11182864
Bai, J., Lei, X., Liu, J., Huang, Y., Bi, L., Wang, Y., et al. (2024). The strigolactone receptor DWARF14 regulates flowering time in Arabidopsis. Plant Cell 36, 4752–4767. doi: 10.1093/plcell/koae248
Bao, S., Hua, C., Huang, G., Cheng, P., Gong, X., Shen, L., et al. (2019). Molecular basis of natural variation in photoperiodic flowering responses. Dev. Cell 50, 90–101.e103. doi: 10.1016/j.devcel.2019.05.018
Browse, J. and Wallis, J. G. (2019). Arabidopsis flowers unlocked the mechanism of jasmonate signaling. Plants (Basel) 8 (8), E285. doi: 10.3390/plants8080285
Chen, C., Xiao, H., Yue, J., Wang, X., Wang, C., Wei, R., et al. (2025). Kenaf cyclic nucleotide-gated channel gene HcCNGC27 confers plant drought stress tolerance and involved in flowering regulation. Mol. Genet. Genomics 300, 65. doi: 10.1007/s00438-025-02272-4
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 131, 1055–1064. doi: 10.1242/dev.00992
Chi, C., Xu, X., Wang, M., Zhang, H., Fang, P., Zhou, J., et al. (2021). Strigolactones positively regulate abscisic acid-dependent heat and cold tolerance in tomato. Hortic. Res. 8, 237. doi: 10.1038/s41438-021-00668-y
Fan, S., Liu, H., Liu, J., Hua, W., and Li, J. (2022). BnGF14-2c positively regulates flowering via the vernalization pathway in semi-winter rapeseed. Plants (Basel). 11, 2312. doi: 10.3390/plants11172312
Feng, G., Han, J., Yang, Z., Liu, Q., Shuai, Y., Xu, X., et al. (2021). Genome-wide identification, phylogenetic analysis, and expression analysis of the SPL gene family in orchardgrass (Dactylis glomerata L.). Genomics 113, 2413–2425. doi: 10.1016/j.ygeno.2021.05.032
Fernández, V., Takahashi, Y., Le Gourrierec, J., and Coupland, G. (2016). Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 86, 426–440. doi: 10.1111/tpj.13183
Galvão, V. C., Horrer, D., Küttner, F., and Schmid, M. (2012). Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139, 4072–4082. doi: 10.1242/dev.080879
Gu, H., Zhang, K., Chen, J., Gull, S., Chen, C., Hou, Y., et al. (2022). OsFTL4, an FT-like gene, regulates flowering time and drought tolerance in rice (Oryza sativa L.). Rice (N Y) 15, 47. doi: 10.1186/s12284-022-00593-1
Guo, H. and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Guo, Y., Pang, C., Jia, X., Ma, Q., Dou, L., Zhao, F., et al. (2017). An NAM domain gene, ghNAC79, improves resistance to drought stress in upland cotton. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01657
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., and Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505. doi: 10.1016/j.molp.2019.01.002
Ibañez, C., Delker, C., Martinez, C., Bürstenbinder, K., Janitza, P., Lippmann, R., et al. (2018). Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 28, 303–310.e303. doi: 10.1016/j.cub.2017.11.077
Ito, T., Okada, K., Fukazawa, J., and Takahashi, Y. (2018). DELLA-dependent and -independent gibberellin signaling. Plant Signal Behav. 13, e1445933. doi: 10.1080/15592324.2018.1445933
Jiao, Z., Shi, X., Xu, R., Zhang, M., Chong, L., and Zhu, Y. (2024). HOS1 ubiquitinates SPL9 for degradation to modulate salinity-delayed flowering. J. Integr. Plant Biol. 66, 2600–2612. doi: 10.1111/jipb.13784
Jin, S., Kim, S. Y., Susila, H., Nasim, Z., Youn, G., and Ahn, J. H. (2022). FLOWERING LOCUS M isoforms differentially affect the subcellular localization and stability of SHORT VEGETATIVE PHASE to regulate temperature-responsive flowering in Arabidopsis. Mol. Plant 15, 1696–1709. doi: 10.1016/j.molp.2022.08.007
Kim, D., Abdelaziz, M. E., Ntui, V. O., Guo, X., and Al-Babili, S. (2017). Colonization by the endophyte Piriformospora indica leads to early flowering in Arabidopsis thaliana likely by triggering gibberellin biosynthesis. Biochem. Biophys. Res. Commun. 490, 1162–1167. doi: 10.1016/j.bbrc.2017.06.169
Kim, S. G., Kim, S. Y., and Park, C. M. (2007). A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226, 647–654. doi: 10.1007/s00425-007-0513-3
Kinmonth-Schultz, H., Lewandowska-Sabat, A., Imaizumi, T., Ward, J. K., Rognli, O. A., and Fjellheim, S. (2021). Flowering times of wild arabidopsis accessions from across Norway correlate with expression levels of FT, CO, and FLC genes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.747740
Koini, M. A., Alvey, L., Allen, T., Tilley, C. A., Harberd, N. P., Whitelam, G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413. doi: 10.1016/j.cub.2009.01.046
Kupke, B. M., Tucker, M. R., Able, J. A., and Porker, K. D. (2021). Manipulation of barley development and flowering time by exogenous application of plant growth regulators. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.694424
Lee, J. H., Ryu, H. S., Chung, K. S., Posé, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Li, K. X., Wang, Y. N., Han, C. Y., Zhang, W. S., Jia, H. Z., and Li, X. (2007). GA signaling and CO/FT regulatory module mediate salt-induced late flowering in Arabidopsis thaliana. Plant Growth Regul. 53, 195–206. doi: 10.1007/s10725-007-9218-7
Li, Z., Wang, S., Wang, Y., Zhang, H., Liu, L., Su, S., et al. (2025). The effects of biotic stress on the sexual reproduction process of flowering plants. PeerJ . 13, e19880. doi: 10.7717/peerj.19880
Li, W., Wang, T., Zhang, Y., and Li, Y. (2016). Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 67, 175–194. doi: 10.1093/jxb/erv450
Li, L., Zhao, Y., McCaig, B. C., Wingerd, B. A., Wang, J., Whalon, M. E., et al. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143. doi: 10.1105/tpc.017954
Liu, X., Zhang, S., Sun, M., Guo, Y., Zhao, S., Zhou, X., et al. (2023). SiMYBS3, encoding a setaria italica heterosis-related MYB transcription factor, confers drought tolerance in arabidopsis. Int. J. Mol. Sci. 24 (6), 5418. doi: 10.3390/ijms24065418
Lyons, R., Rusu, A., Stiller, J., Powell, J., Manners, J. M., and Kazan, K. (2015). Investigating the Association between Flowering Time and Defense in the Arabidopsis thaliana-Fusarium oxysporum Interaction. PloS One 10, e0127699. doi: 10.1371/journal.pone.0127699
Murase, K., Hirano, Y., Sun, T. P., and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Pan, R., Xu, L., Wei, Q., Wu, C., Tang, W., Oelmüller, R., et al. (2017). Piriformospora indica promotes early flowering in Arabidopsis through regulation of the photoperiod and gibberellin pathways. PloS One 12, e0189791. doi: 10.1371/journal.pone.0189791
Posé, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Riboni, M., Galbiati, M., Tonelli, C., and Conti, L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 162, 1706–1719. doi: 10.1104/pp.113.217729
Riboni, M., Robustelli Test, A., Galbiati, M., Tonelli, C., and Conti, L. (2016). ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 67, 6309–6322. doi: 10.1093/jxb/erw384
Robustelli Test, A., Perrella, G., Landoni, B., Colanero, S., Sutti, A., Korwin Krukowski, P., et al. (2025). Abscisic acid and GIGANTEA signalling converge to regulate the recruitment of CONSTANS to the FT promoter and activate floral transition. J. Exp. Bot. 76 (14), 4085–4100. doi: 10.1093/jxb/eraf199
Ryu, J. Y., Lee, H. J., Seo, P. J., Jung, J. H., Ahn, J. H., and Park, C. M. (2014). The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol. Plant 7, 377–387. doi: 10.1093/mp/sst114
Sanjari, S., Shirzadian-Khorramabad, R., Shobbar, Z. S., and Shahbazi, M. (2019). Systematic analysis of NAC transcription factors' gene family and identification of post-flowering drought stress responsive members in sorghum. Plant Cell Rep. 38, 361–376. doi: 10.1007/s00299-019-02371-8
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. doi: 10.1111/tpj.13109
Shu, K., Chen, F., Zhou, W., Luo, X., Dai, Y., Shuai, H., et al. (2018). ABI4 regulates the floral transition independently of ABI5 and ABI3. Mol. Biol. Rep. 45, 2727–2731. doi: 10.1007/s11033-018-4290-9
Singh, D., Sharma, S., Jose-Santhi, J., Kalia, D., and Singh, R. K. (2023). Hormones regulate the flowering process in saffron differently depending on the developmental stage. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1107172
Stavang, J. A., Gallego-Bartolomé, J., Gómez, M. D., Yoshida, S., Asami, T., Olsen, J. E., et al. (2009). Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60, 589–601. doi: 10.1111/j.1365-313X.2009.03983.x
Sun, T. P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, R338–R345. doi: 10.1016/j.cub.2011.02.036
Sun, W., Fan, J., Fang, A., Li, Y., Tariqjaveed, M., Li, D., et al. (2020). Ustilaginoidea virens: Insights into an Emerging Rice Pathogen. Annu. Rev. Phytopathol. 58, 363–385. doi: 10.1146/annurev-phyto-010820-012908
Swida-Barteczka, A., Pacak, A., Kruszka, K., Nuc, P., Karlowski, W. M., Jarmolowski, A., et al. (2023). MicroRNA172b-5p/trehalose-6-phosphate synthase module stimulates trehalose synthesis and microRNA172b-3p/AP2-like module accelerates flowering in barley upon drought stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1124785
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Van De Velde, K., Ruelens, P., Geuten, K., Rohde, A., and Van Der Straeten, D. (2017). Exploiting DELLA signaling in cereals. Trends Plant Sci. 22, 880–893. doi: 10.1016/j.tplants.2017.07.010
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., and Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64, 675–684. doi: 10.1093/jxb/ers361
Wang, Z., Wang, F., Hong, Y., Yao, J., Ren, Z., Shi, H., et al. (2018). The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol. Plant 11, 1184–1197. doi: 10.1016/j.molp.2018.06.009
Winter, C. M., Yamaguchi, N., Wu, M. F., and Wagner, D. (2015). Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol. Plant 155, 55–73. doi: 10.1111/ppl.12357
Xu, G., Tao, Z., and He, Y. (2022). Embryonic reactivation of FLOWERING LOCUS C by ABSCISIC ACID-INSENSITIVE 3 establishes the vernalization requirement in each Arabidopsis generation. Plant Cell 34, 2205–2221. doi: 10.1093/plcell/koac077
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yan, J., Li, X., Zeng, B., Zhong, M., Yang, J., Yang, P., et al. (2020). FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis. J. Integr. Plant Biol. 62, 1717–1740. doi: 10.1111/jipb.12971
Ying, S., Scheible, W. R., and Lundquist, P. K. (2023). A stress-inducible protein regulates drought tolerance and flowering time in Brachypodium and Arabidopsis. Plant Physiol. 191, 643–659. doi: 10.1093/plphys/kiac486
Yu, S., Galvão, V. C., Zhang, Y. C., Horrer, D., Zhang, T. Q., Hao, Y. H., et al. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24, 3320–3332. doi: 10.1105/tpc.112.101014
Yu, Y., Liu, Z., Wang, L., Kim, S. G., Seo, P. J., Qiao, M., et al. (2016). WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 85, 96–106. doi: 10.1111/tpj.13092
Zhang, C., Wu, Z., Li, Y., and Wu, J. (2015). Biogenesis, function, and applications of virus-derived small RNAs in plants. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01237
Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396–411. doi: 10.1105/tpc.110.081356
Zhao, H., Huang, X., Yang, Z., Li, F., and Ge, X. (2023). Synergistic optimization of crops by combining early maturation with other agronomic traits. Trends Plant Sci. 28, 1178–1191. doi: 10.1016/j.tplants.2023.04.011
Keywords: stress resistance, flowering time, phytohormone, environmental stress, regulation mechanism
Citation: Dong W, Zhang Y, Xing Y, Chang K, Qiu N and Song Y (2025) Optimizing fitness: plastic flowering time in variable environments. Front. Plant Sci. 16:1690015. doi: 10.3389/fpls.2025.1690015
Received: 21 August 2025; Accepted: 23 September 2025;
Published: 07 October 2025.
Edited by:
Guangqiang Zhang, Heze University, ChinaReviewed by:
Chen Meng, Chinese Academy of Agricultural Sciences, ChinaYuebin Jia, Shandong Normal University, China
Copyright © 2025 Dong, Zhang, Xing, Chang, Qiu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuguang Song, c3l1Z3VhbmdAcWZudS5lZHUuY24=; Nianwei Qiu, bmlhbndlaXFpdUAxNjMuY29t
†These authors have contributed equally to this work
 Wei Dong
Wei Dong Yalin Zhang†
Yalin Zhang† Nianwei Qiu
Nianwei Qiu Yuguang Song
Yuguang Song