- 1Graduate School of Agriculture, Osaka Metropolitan University, Sakai, Osaka, Japan
- 2Research Institute of Environment, Agriculture and Fisheries, Habikino, Osaka, Japan
- 3Graduate School of Life and Environmental Sciences, Osaka Prefecture University, Sakai, Osaka, Japan
- 4School of Life and Environmental Sciences, Osaka Prefecture University, Sakai, Osaka, Japan
- 5United Graduate School of Agricultural Science, Tokyo University of Agriculture and Technology, Tokyo, Japan
- 6School of Agriculture, Meiji University, Kawasaki, Kanagawa, Japan
- 7Education and Research Field, School of Agriculture, Osaka Metropolitan University, Sakai, Osaka, Japan
Allopolyploidization plays an important role in the evolution of eukaryotes. Allopolyploids formed through interspecific hybridization and polyploidization undergo genetic and epigenetic changes in the early generations, known as ‘genome shock’. However, reproductive isolation often prevents interspecific hybridization. The mechanism by which a reproductively isolated species breaks reproductive barriers and crosses with other species is largely unknown, despite its importance in speciation and evolution. Here, we report the ultrahigh-frequency appearance of viable hybrids that overcame hybrid lethality, a type of reproductive isolation, in crosses between Nicotiana tabacum and N. amplexicaulis. Lethal hybrids exhibited Type II hybrid lethality characterized by browning of hypocotyl and roots at 28 °C, temperature sensitivity, and involvement of the Q chromosome from N. tabacum genome, indicating that N. amplexicaulis possesses the causal allele for hybrid lethality at the Hybrid Lethality A1 (HLA1) locus. Random amplified polymorphic DNA, amplified fragment length polymorphism, and methylation-sensitive amplified polymorphism analyses have detected genetic and epigenetic changes in viable and lethal hybrids, suggesting the occurrence of genome shock during interspecific hybridization. We found that many viable hybrids exhibited HLA1 locus deletion, indicating that it was the primary cause of overcoming hybrid lethality in these crosses. These findings demonstrate that genome shock-induced genetic changes promote the breakdown of reproductive barriers through the deletion of causal genes, providing insights into the mechanisms by which reproductively isolated species can overcome barriers and lead to the formation of new species.
1 Introduction
Allopolyploidization plays an important role in the evolution of eukaryotes (Rieseberg and Willis, 2007; Van de Peer et al., 2009). Allopolyploids formed through interspecific hybridization and polyploidization undergo genetic and epigenetic changes in early generations, as indicated by analyses of synthetic allopolyploids (Qiu et al., 2020). These changes, known as ‘genome shock’ (McClintock, 1984), have been widely studied in plant species such as rice (Wu et al., 2015), wheat (Shaked et al., 2001), rapeseed (Li et al., 2019), tobacco (Mhiri et al., 2019), and Arabidopsis (Madlung et al., 2002). However, reproductive isolation often prevents interspecific hybridization. If reproductive barriers are broken, new species can be established. Reproductive barriers are occasionally overcome; i.e., polyploidization, which balances homologous chromosome segregation and gene expression, has been reported as one mechanism for overcoming hybrid sterility, a type of reproductive barrier (Meeus et al., 2020; Mino et al., 2022; Ren et al., 2022). However, the mechanism by which reproductively isolated species break reproductive barriers and produce hybrid offspring remains largely unknown.
Reproductive isolation plays crucial roles in speciation and evolution. These barriers are divided into prezygotic and postzygotic barriers. Prezygotic isolation involves pollen–pistil incompatibility, conspecific pollen precedence, gametic incompatibility, and pistil-length mismatch (Rieseberg and Willis, 2007; Lee et al., 2008; Rieseberg and Blackman, 2010; Huang et al., 2023). Post-zygotic barriers include hybrid seed abortion, hybrid lethality, and hybrid sterility in the F1 generation, and hybrid breakdown in the F2 or later generations (Li J. et al., 2020; Dziasek et al., 2021; Xu et al., 2023). Hybrid lethality, which has been reported in many plant species, such as rice (Chen et al., 2014), wheat (Si et al., 2021), cotton (Deng et al., 2019), pepper (Shiragaki et al., 2020b), and Arabidopsis thaliana (Bomblies et al., 2007), is generally caused by the interaction between two dominant complementary genes from the parental plants (Bomblies and Weigel, 2007).
In the genus Nicotiana, many interspecific cross combinations result in hybrid lethality in seedlings. Hybrid lethality in this genus is classified into five types based on the early visible symptoms of hybrid seedlings: Type I, browning of the shoot apex and root tips; Type II, browning of the hypocotyl and roots; Type III, yellowing of true leaves; Type IV, formation of multiple shoots; and Type V, fading of shoot color (Yamada et al., 1999; Tezuka and Marubashi, 2012). Type II hybrid lethality occurs in most cross-combinations between the cultivated species N. tabacum and the wild species of the Nicotiana section Suaveolentes. Type II hybrid lethality is temperature-sensitive, observed at 28 °C but suppressed at elevated temperatures between 34 °C and 36 °C (Yamada et al., 1999; Tezuka et al., 2010), and is caused by the epistatic interaction between the dominant allele Hla1–1 at the HLA1 locus of Suaveolentes species and the dominant allele Hla2–1 at the HLA2 locus on the Q chromosome of N. tabacum (Iizuka et al., 2012; Ma et al., 2020; Nakata et al., 2021; Mino et al., 2022; He et al., 2023; Tezuka et al., 2024).
We previously reported that hybrid seedlings and plants that overcome Type II hybrid lethality and exhibit normal growth rarely appear spontaneously in the following cross combinations: N. africana × N. tabacum ‘Samsun NN’ (1.45% of hybrids), N. tabacum ‘Red Russian’ × N. debneyi (0.33%), N. megalosiphon × ‘Red Russian’ (0.60%), and N. suaveolens × ‘Red Russian’ (1.32%) (Tezuka and Marubashi, 2006b; Tezuka et al., 2010). Tezuka et al. (2012) reported that partial deletion of the Q chromosome, where the HLA2 locus is located, is responsible for the occurrence of viable hybrids in the N. tabacum × N. africana cross. Other authors have also reported that similar deletions in the terminal region of the Q chromosome are thought to result in viable hybrids in the N. tabacum ‘TN 90LC’ × N. africana (0.12%) and N. suaveolens × N. tabacum ‘Hicks-2’ (0.09%) crosses (Hancock et al., 2015; Nakata et al., 2021). One reason for these deletions is the reciprocal translocation between homoeologous chromosomes during pollen formation in the allotetraploid N. tabacum (Nakata et al., 2021). Despite these findings, the mechanism of overcoming hybrid lethality in interspecific crosses through allopolyploidization remains unknown.
In this study, we report that crosses between N. amplexicaulis (section Suaveolentes) and N. tabacum yield significantly more viable hybrids that overcome hybrid lethality than other lethal crosses. We characterized hybrid lethality based on phenotypic symptoms, temperature sensitivity, and the N. tabacum chromosome responsible for the hybrid lethality. To clarify the mechanism of the ultrahigh-frequency appearance of viable hybrids, we investigated the genetic changes in hybrids using random amplified polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP) analyses and the epigenetic changes in hybrids using methylation-sensitive amplified polymorphism (MSAP) analysis. In addition, we analyzed the genetic changes or alterations in the expression of the two causal genes for hybrid lethality. Based on these results, we discuss the effects of breaking reproductive isolation on natural plant evolution.
2 Materials and methods
2.1 Plant materials
Two cultivars of N. tabacum, ‘Red Russian’ and ‘Samsun NN,’ and three accessions of N. amplexicaulis, JT, PI 271989, and PI 555682, were used in this study. We also used F1 monosomic progeny from the cross between N. tabacum Q chromosome monosomic line Haplo-Q and disomic ‘Samsun NN’ (Tezuka et al., 2004). All plants were cultivated in a greenhouse under natural day lengths and used for crossing experiments.
2.2 Interspecific crosses and investigation of lethal symptoms in hybrid seedlings
In conventional crossing, the flowers of plants used as the maternal parent are emasculated one day before anthesis and pollinated with pollen from paternal parent plants. The obtained seeds were treated with a 0.5% gibberellic acid (GA3) solution for 30 minutes, soaked in a 5% sodium hypochlorite solution for 15 minutes, and rinsed three times with sterile water. The sterilized seeds were sown in Petri dishes containing 1/2 MS medium (Murashige and Skoog, 1962) supplemented with 1% sucrose and solidified with 0.2% Gelrite (pH 5.8) and then cultured at 28 °C under continuous illumination (approximately 150 μmol m−2 s−1).
Test-tube pollination and ovule culture were conducted as previously described (Tezuka and Marubashi, 2004). Hybrid seedlings obtained from the cross between ‘Samsun NN’ and N. amplexicaulis JT by the test-tube pollination and ovule culture were transferred to flat-bottomed test tubes (35 mm diameter, 120 mm length) containing 20 mL of 1/2 MS medium supplemented with 1% sucrose and solidified with 0.2% Gelrite (pH 5.8) immediately after germination and cultured at 28 °C under continuous illumination.
Hybrid seedlings obtained from conventional crossing and test-tube pollination with ovule culture were observed for lethal symptoms at 28 °C. To investigate temperature sensitivity of hybrid lethality, 20 hybrid seedlings from the cross N. amplexicaulis JT and ‘Red Russian’ were transferred into flat-bottomed test tubes (25 mm diameter, 100 mm) containing 10 mL of 1/2 MS medium supplemented with 1% sucrose and solidified with 0.2% Gelrite (pH 5.8) immediately after germination and were cultured at 34 °C. Hybrid seedlings cultured at 34 °C for 30 d after germination (DAG) were transferred to 28 °C under continuous illumination. Hybrid seedlings that survived for more than 30 d after transfer were potted and cultivated in a greenhouse under natural day lengths.
2.3 Investigation of N. tabacum Q-chromosome involvement in hybrid lethality
To investigate the involvement of the N. tabacum Q chromosome in hybrid lethality, hybrid seedlings from a cross between the F1 monosomic progeny of Haplo-Q × ‘Samsun NN’ and N. amplexicaulis JT were obtained by test-tube pollination with ovule culture. The hybrid seedlings were transferred to flat-bottomed test tubes (30 mm diameter, 120 mm length) containing 25 mL of 1/2 MS medium supplemented with 1% sucrose and solidified with 0.2% Gelrite (pH 5.8) immediately after germination and cultured at 34 °C under continuous illumination. Seedlings were subcultured in fresh medium every 3 weeks. After analysis using Q-chromosome-specific DNA markers, seedlings were transferred to 28 °C under continuous illumination.
2.4 PCR analyses
Total DNA was extracted from young leaves of each plant using a cetyltrimethylammonium bromide (CTAB)-based method (Murray and Thompson, 1980). The Q-chromosome-specific DNA markers QCS2, QCS3, and QCS4 were detected as previously described (Tezuka et al., 2004; Tezuka and Marubashi, 2006a). To investigate the HLA2 locus, the SSR markers PT30137, PT30342, and PT30365 (Bindler et al., 2011, 2007), were used. HLA2-specific sequence tagged site (STS) marker HLA2-STS was developed based on the nucleotide sequence of the Hla2–1 allele (Ma et al., 2020) (Supplementary Table 1). To investigate the HLA1 locus, three CAPS markers linked to HLA1, Nb14-CAPS, NbRGH1-CAPS, and Nb49-CAPS (Tezuka et al., 2021, 2024) were used. However, Nb14-CAPS and Nb49-CAPS were used as STS markers (Nb14-STS and Nb49-STS) because polymorphisms were detected between the species used in this study without restriction enzyme treatment. Because NbRGH1-CAPS treated with restriction enzyme BsrI showed no polymorphism between ‘Samsun NN’ and N. amplexicaulis, PCR products of NbRGH1-CAPS from both were completely sequenced. Subsequent digestion with restriction enzyme EcoT22I revealed a polymorphism (NbRGH1-EcoT22I). Five new STS markers, Nb138-STS, Nb88-STS, Nb89-STS, Nb93-STS, and Nb135-STS, were developed based on the scaffold sequence Nbe.v1.1.chr15 in N. benthamiana genome Nbe.v1.1. These markers were detected by PCR, using the same method as previously described (Tezuka et al., 2021).
2.5 RAPD, AFLP, and MSAP analysis
RAPD analysis was performed for viable hybrid seedlings using 20 random 10-mer oligonucleotide primers (Kit A; Operon Technologies, Inc., Alameda, CA, USA), as previously described (Tezuka et al., 2007). Only clear bands were observed.
For AFLP and MSAP analysis, hybrid seedlings obtained from the cross between ‘Samsun NN’ and N. amplexicaulis JT were transferred into flat-bottomed test tubes (35 mm diameter, 120 mm) containing 20 mL of 1/2 MS medium supplemented with 1% sucrose and solidified with 0.2% Gelrite (pH 5.8) immediately after germination at 28 °C and were cultured at 34 °C to suppress hybrid lethality. After extracting DNA for AFLP and MSAP analyses using the CTAB-based method (Murray and Thompson, 1980), the seedlings were transferred to 28 °C under continuous illumination and classified into viable and lethal hybrids based on their lethal phenotypes.
AFLP analysis was conducted as previously described by Vos et al. (1995), with a few modifications. Total DNA was double-digested with EcoRI and MseI and ligated to EcoRI and MseI adapters. The pre-amplification reaction was performed using EcoRI+A (E01) and MseI+C (M02) primers. The PCR products were then diluted tenfold with sterile water and used as templates for the second amplification reaction. Primers with three selective nucleotides, 7 EcoRI+ANN (N indicates A, T, G, or C; E37 to E43) and 16 MseI+CNN (M47 to M62) primers, were used for the second amplification reaction with 112 combinations (Supplementary Table 2). In both amplification reactions, KAPA Taq EXtra DNA polymerase (Kapa Biosystems, Wilmington, MA, USA) was used, and PCR amplification was performed using a PC-818A Program Temp Control System (Astec, Fukuoka, Japan). The second PCR product was separated by electrophoresis on an 8% polyacrylamide gel and visualized by staining with ethidium bromide. Only bands smaller than 800 bp were identified. Each band was named according to the primer combination, and the subsequent number indicated the estimated DNA fragment size.
MSAP analysis was conducted as previously described by Fulneček and Kovařík (2014), with a few modifications, using the same DNA used for AFLP analysis. Total DNA was double-digested with EcoRI and HpaII or MspI and ligated to EcoRI adapter and HpaII or MspI adapters. The isoschizomer pairs, HpaII and MspI, are methylation-sensitive restriction enzymes that cleave the CCGG sequence. HpaII can only cleave nonmethylated CCGG sequences or those hemi-methylated at the external cytosine. MspI can cleave non-methylated CCGG sequences and those hemi- or fully methylated at the internal cytosines. However, it cannot cleave CCGG sequences that are hemi-methylated or fully methylated at the external cytosine (Reyna-López et al., 1997). The preamplification reaction was performed using EcoRI+A (E01) and HpaII+T or MspI+T (HM01) primers. The PCR products were then diluted tenfold with sterile water and used as templates for the second amplification reaction. Primers with three selective nucleotides, 16 EcoRI+ANN (N indicates A, T, G, or C; E37–E46), and 4 HpaII+TAN or MspI+TAN (HM47–HM50) primers were used for the second amplification reaction with 64 combinations (Supplementary Table 2). Both amplification reactions and band detection were carried out as described for the AFLP analysis.
2.6 RT-PCR analysis of Hla2–1 expression
For RT-PCR analysis, hybrid seedlings obtained from the cross between ‘Samsun NN’ and N. amplexicaulis JT were cultured at 28 °C in vitro. At 4 DAG, the phenotype of each hybrid was evaluated and classified as either viable or lethal. Subsequently, before the lethal hybrids died, viable and lethal hybrids were transferred to flat-bottomed test tubes (35 mm diameter, 120 mm length) containing 20 mL of 1/2 MS medium supplemented with 1% sucrose, solidified with 0.2% Gelrite (pH 5.8), and cultured at 34 °C under continuous illumination to increase the growth of the hybrids. At 14 DAG, the seedlings were transferred to 28 °C under continuous illumination to induce hybrid lethality. One day after transfer, total RNA was extracted from the whole plant using TRIzol reagent (Invitrogen Inc., Carlsbad, USA) and then treated with RNase-free DNase (Promega Co., Madison, WI, USA) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 1 μg total RNA using oligo (dT)18 primers and ReverTra Ace (Toyobo Co., Ltd., Osaka, Japan).
To analyze Hla2–1 expression, primers for the HLA2-specific STS marker Nt6549g30-1 (Nakata et al., 2021) were used. Primers for Actin were designed by Shiragaki et al. (2020a). PCR reaction mixtures consisted of 1 × KAPA Taq EXtra Buffer (Kapa Biosystems, Wilmington, MA, USA), 0.2 mM each dNTP, 0.2 µM each primer, 0.25 U KAPA Taq EXtra DNA polymerase (Kapa Biosystems, Wilmington, MA, USA), and 1 µL of diluted cDNA (first-strand cDNA: nuclease-free water = 1:4) in a total volume of 10 µL. PCR amplification was performed using a PC-818A Program Temp Control System (Astec, Fukuoka, Japan) programmed for 3 min at 94 °C for initial denaturation, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 60 s, with a final 5-min extension at 72 °C. The PCR products were separated by electrophoresis on 3.0% agarose gels in TBE buffer and visualized by staining with ethidium bromide.
3 Results
3.1 Hybrid lethality observed in hybrid seedlings between N. amplexicaulis and N. tabacum
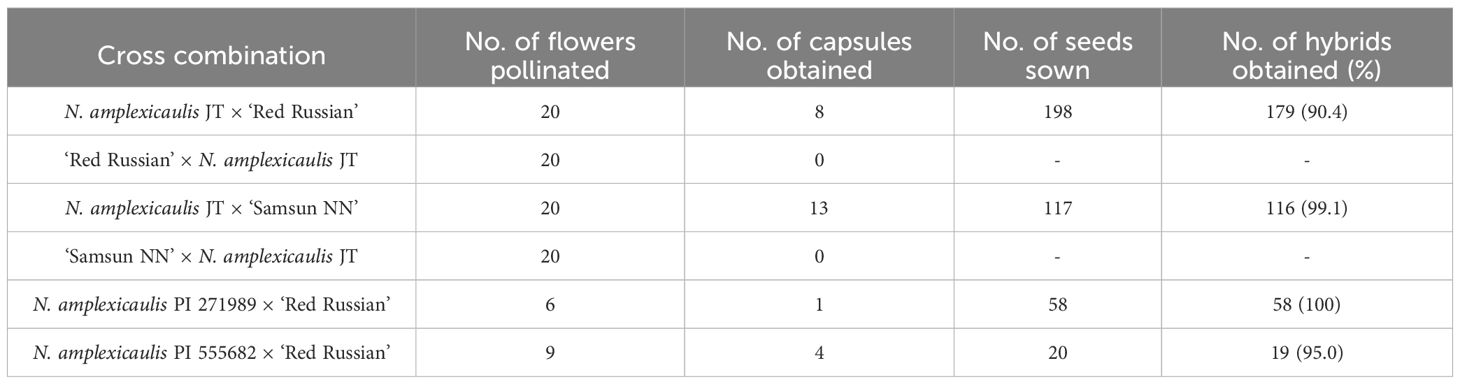
Three accessions of N. amplexicaulis (JT, PI 271989, and PI 555682) were crossed with N. tabacum. First, N. amplexicaulis JT was reciprocally crossed with two cultivars of N. tabacum, ‘Red Russian’ and ‘Samsun NN’. Seeds were only obtained in the crosses using N. tabacum as the male parent, and no capsules were produced from crosses using N. tabacum as the female parent, suggesting that fertilization was unsuccessful (Table 1). After sowing the seeds in vitro at 28 °C, 179 and 116 hybrid seedlings were obtained from crosses using ‘Red Russian’ and ‘Samsun NN’ as male parents, respectively. Of these, 159 and all 116 seedlings from each cross were subsequently cultured at 28 °C. Regardless of N. tabacum cultivar, more than half of the hybrid seedlings showed normal growth, whereas the remaining hybrid seedlings exhibited Type II hybrid lethality, characterized by early symptoms of hypocotyl and root browning (Table 2; Figure 1A). Based on these results, N. amplexicaulis PI 271989 and PI 555682 were crossed with ‘Red Russian’ as the male parent. PI 271989 yielded 17 viable hybrid seedlings (29.3%) and 41 lethal hybrid seedlings (70.7%) at 28 °C. Similarly, PI 555682 yielded five viable hybrid seedlings (26.3%) and 14 lethal hybrid seedlings (73.7%) at 28 °C (Table 1). The lethality of the hybrid seedlings from the two crosses was Type II. Viable hybrid seedlings from all four crosses were potted and cultivated in a greenhouse. All hybrids matured and flowered without any lethality symptoms. The morphological characteristics of the hybrid plants were uniform for each cross combination. The leaf shape, flower shape, and flower color of the hybrid plants were intermediate in appearance between those of the parents in all cross combinations (Figures 1B–E). Since the hybrid lethality observed in the four crosses appeared to be due to the same mechanism, subsequent studies focused on crosses between N. amplexicaulis JT and N. tabacum cultivars (‘Red Russian’ and/or ‘Samsun NN’).
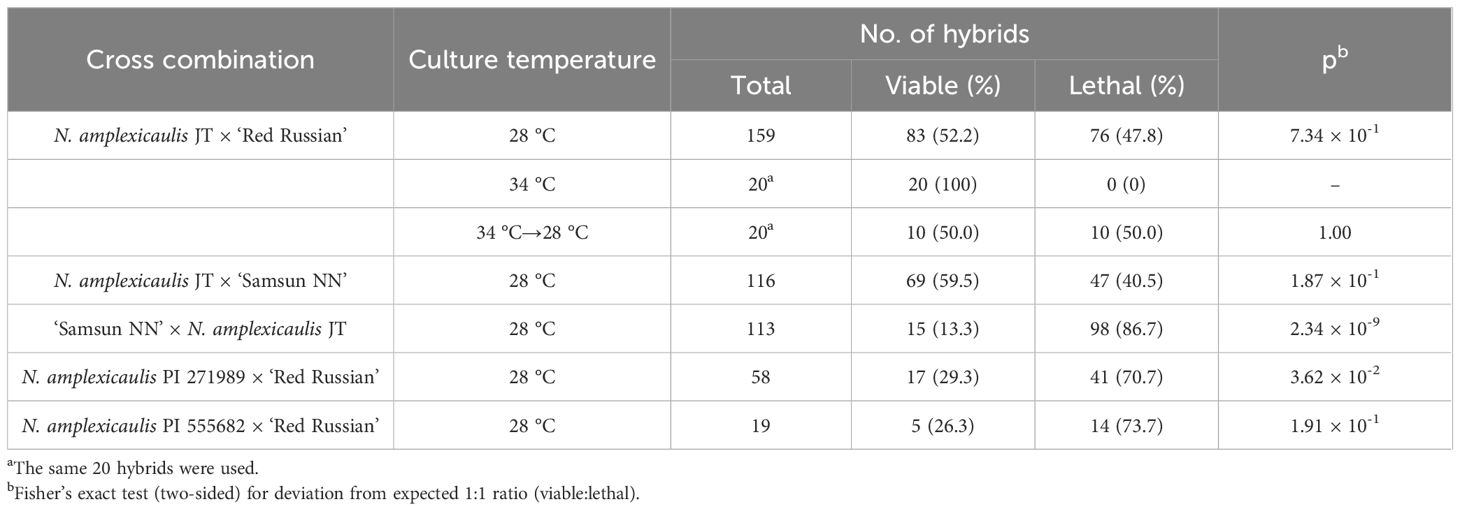
Table 2. Viability of hybrid seedlings between three N. amplexicaulis accessions and N. tabacum at 28 °C or 34 °C.

Figure 1. Hybrid seedlings and plants from the cross N. amplexicaulis JT × N. tabacum. (A) Characteristic early symptoms of hybrid lethality in the hybrid seedlings from the cross N. amplexicaulis × ‘Red Russian’. Arrowheads indicate browning of hypocotyl and roots, which is characteristic of type II hybrid lethality. (B) Shape of a viable hybrid plant from the cross N. amplexicaulis × ‘Samsun NN’ that has grown to maturity and flowered. (C, D) Flowers of N. amplexicaulis, a viable hybrid plant, and ‘Samsun NN’ (left to right). (E) Leaves of N. amplexicaulis, a viable hybrid plant, and ‘Samsun NN’ (left to right). Scale bars = 0.5 cm (A), 30 cm (B), and 2 cm (C-E).
To obtain hybrid seedlings of N. tabacum with N. amplexicaulis JT as the male parent, we performed test-tube pollination and ovule culture, as combined techniques are useful for bypassing the prezygotic barriers (Tezuka et al., 2010). Fifty-five placentas of N. tabacum ‘Samsun NN’ were pollinated with N. amplexicaulis JT pollen in vitro, resulting in 790 enlarged ovules. These ovules were cultured with fresh media at 28 °C, and 113 hybrid seedlings were obtained. Of these, 98 seedlings (86.7%) showed type II hybrid lethality and 15 seedlings (13.3%) showed normal growth (Table 2).
3.2 Effect of an elevated temperature on hybrid lethality
Hybrid lethality in crosses between species of section Suaveolentes and N. tabacum is suppressed at elevated temperatures (32 °C –36 °C) (Mino et al., 2002; Yamada and Marubashi, 2003; Tezuka et al., 2007, 2010). We investigated whether hybrid lethality in the cross between N. amplexicaulis JT and N. tabacum ‘Red Russian’ could also be suppressed under elevated temperature conditions. When the remaining 20 of 179 hybrid seedlings obtained by in vitro sowing were transferred to 34 °C, all of them exhibited normal growth without lethality until at least 30 DAG (Table 2). These hybrid seedlings were transferred to 28 °C at 30 DAG to assess whether lethality could be suppressed only during culture at elevated temperatures. Ten hybrid seedlings (50%) exhibited hybrid lethality and died, whereas the remaining 10 (50%) continued to grow.
3.3 Involvement of the N. tabacum Q chromosome in hybrid lethality
To investigate whether the Q chromosome of N. tabacum is responsible for hybrid lethality, monosomic analysis was conducted using monosomic plants lacking one copy of the Q chromosome, which can be easily identified from F1 progeny from the cross between N. tabacum Q chromosome monosomic line Haplo-Q and ‘Samsun NN’ using DNA markers (Tezuka et al., 2004). In the analysis, although test-tube pollination and ovule culture were required, monosomic plants were crossed with N. amplexicaulis as male parents because the probability of transmission of the monosomic chromosome through pollen was remarkably low (Olmo, 1935). Sixteen placentas of Q chromosome monosomic plants were pollinated with N. amplexicaulis JT pollen, resulting in 327 enlarged ovules. After ovule culture at 28 °C, 13 hybrid seedlings were obtained and transferred to 34 °C to suppress hybrid lethality. Hybrid seedlings were assessed for the presence or absence of the Q chromosome using two Q-chromosome-specific (STS) markers: QCS2 and QCS3 (Tezuka et al., 2004; Tezuka and Marubashi, 2006b). Both markers were detected in six hybrid seedlings, but neither marker was detected in seven hybrid seedlings. When these hybrid seedlings were transferred from 34 °C to 28 °C, all seedlings possessing the Q chromosome showed hybrid lethality and died, whereas all seedlings lacking the Q chromosome survived without lethal symptoms (Table 3). These results suggest that hybrid lethality was caused by the epistatic interaction between the Hla1–1 allele at the HLA1 locus in N. amplexicaulis and the Hla2–1 allele at the HLA2 locus on the Q chromosome in N. tabacum, similar to the Type II hybrid lethality observed in crosses between other Suaveolentes species and N. tabacum.
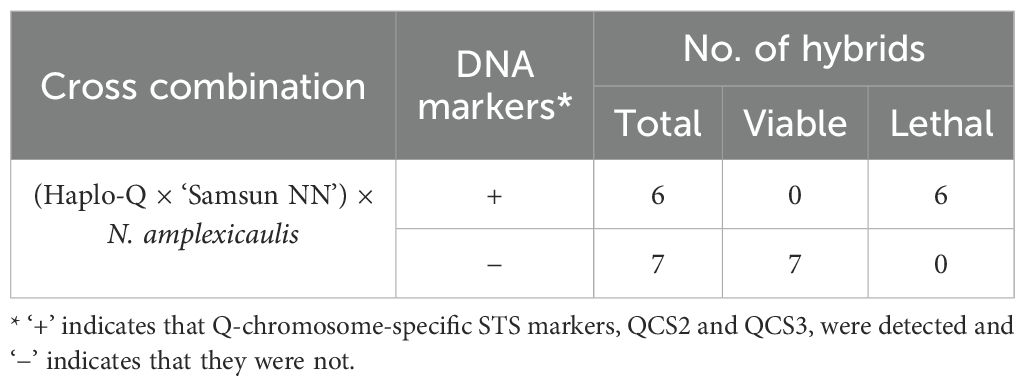
Table 3. Relationship between the Q chromosome and hybrid lethality observed in the cross between N. tabacum and N. amplexicaulis JT.
3.4 Involvement of N. amplexicaulis heterozygosity in viable hybrid formation
The occurrence rates of viable hybrids resulting from crosses between N. amplexicaulis accessions and N. tabacum were high (Table 1). Among the parental species used in this study, although N. tabacum cultivars were highly inbred, the homozygosity and heterozygosity of N. amplexicaulis remain unclear. Considering the results that the ratio of viable hybrids to lethal hybrids was close to 1:1 (Fisher’s exact test, two-sided, p > 0.05) in crosses between N. amplexicaulis JT and N. tabacum cultivars (Table 2), the heterozygosity of the HLA1 locus might be responsible for the occurrence of viable hybrids. To test this hypothesis, we newly prepared four plants of N. amplexicaulis JT as the first generation (S0) and their self-progenies (S1 generation; ten plants each from the four S0 plants) and crossed them with N. tabacum ‘Red Russian’ as the male parent. If the frequent occurrence of viable hybrids was due to the heterozygosity of HLA1, a quarter of the S1 plants (homozygous for the recessive hla1–2 allele) would produce only viable hybrids, half of the S1 plants (heterozygous) would produce viable and lethal hybrids in a 1:1 ratio, and the remaining quarter of the S1 plants (homozygous for Hla1–1 allele) would produce only lethal hybrids. After crosses with N. tabacum, four S0 plants produced viable hybrids at percentages of 8.9%–63.2%. All S1 plants produced viable hybrids with occurrence rates ranging from 3.2% to 49.2%; however, the rates varied among plants within each lineage, ruling out the possibility that the HLA1 locus was heterozygous (Supplementary Table 3).
3.5 Genetic changes in hybrid seedlings detected by RAPD and AFLP analyses
To investigate DNA mutations in viable hybrids, RAPD analysis was performed on five viable hybrids from the cross between S0 plants and N. tabacum ‘Red Russian’ (A0R-V1–5) and five viable hybrids from the cross between S0 plants and N. tabacum ‘Samsun NN’ (A0S-V1–5). Using 20 OPA primers, we detected 19 N. amplexicaulis-specific bands, 33 N. tabacum (‘Red Russian’ and ‘Samsun NN’) specific bands, and 24 common bands between the parents. No polymorphism was detected between ‘Red Russian’ and ‘Samsun NN’. In the two viable hybrids, A0S-V3 and A0S-V5, two N. amplexicaulis-specific bands detected using primers OPA-05 or OPA-19 were absent, suggesting that genetic changes occurred in the hybrid seedlings (Figures 2A, B; Table 2).
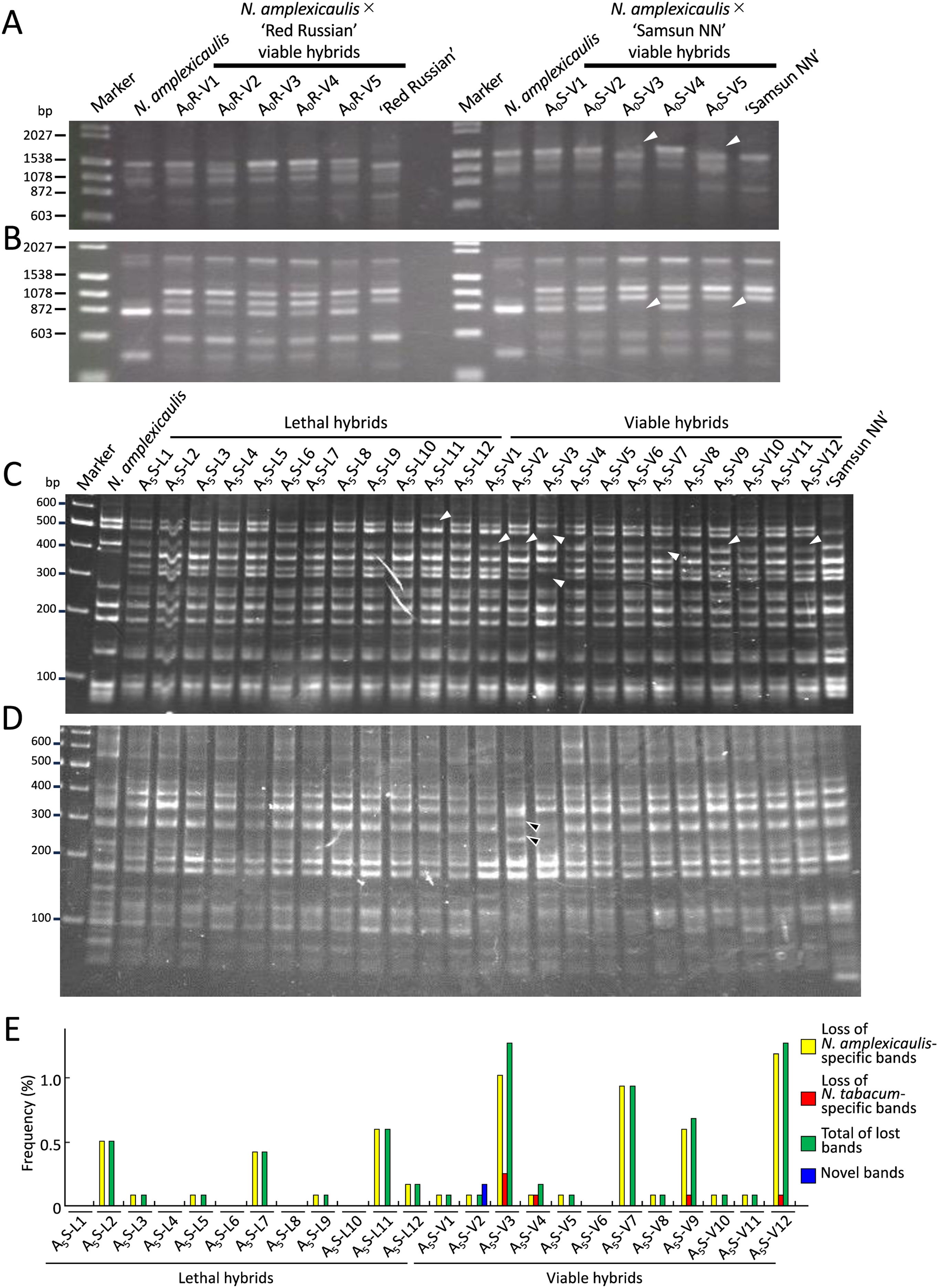
Figure 2. Genetic changes in hybrid plants identified by RAPD and AFLP analysis. (A, B) Loss of bands identified in hybrid plants by RAPD analysis using primers OPA-05 (A) and OPA-09 (B). White arrowheads indicate the disappeared bands. Marker lane, DNA size markers (λ/Hind III and φX174/Hae III). (C) Lost bands identified in hybrid plants by AFLP analysis using E40-M55 primer pair. White arrowheads indicate the disappeared bands. Marker lane, DNA size marker (GeneRuler DNA ladder mix). (D) Nobel bands identified in hybrid plants by AFLP analysis using E39-M49 primer pair. Black arrowheads indicate novel bands. Marker lane, DNA size marker (GeneRuler DNA ladder mix). (E) Frequency of lost or novel bands in each hybrid identified by AFLP analysis.
To investigate genetic changes more comprehensively, we conducted AFLP analysis using 112 primer sets on 12 viable hybrids (A5S-V1–12) and 12 lethal hybrids (A5S-L1–12) obtained from the cross between an N. amplexicaulis JT S5 plant and ‘Samsun NN’. AFLP analysis detected 224 N. amplexicaulis-specific bands, 323 N. tabacum-specific bands, and 588 common bands between the parents. Similar to RAPD analysis, no polymorphism was detected between ‘Red Russian’ and ‘Samsun NN’. Of the bands detected, 39 N. amplexicaulis-specific bands and four N. tabacum-specific bands were absent in 11 viable hybrids and seven lethal hybrids (Figures 2C, E; Supplementary Table 4). The number of absent bands varied depending on hybrids. Excluding six hybrids with no missing bands, the frequencies of lost bands in each hybrid ranged from 0.09% (one band) to 1.32% (15 bands) (Figure 2E). Among the lost bands, some were absent in only one hybrid, whereas others were absent in up to four hybrids (Supplementary Table 4). No bands were consistently missing across all the viable or lethal hybrids. In addition, two novel bands that were not present in the parents were observed in the viable hybrid A5S-V2 (Figures 2D, E).
3.6 Epigenetic changes in hybrid seedlings detected by MSAP analysis
To investigate whether methylation changes were involved in the occurrence of viable hybrids, we conducted MSAP analysis using EcoRI in combination with the isoschizomer pair HpaII and MspI, which allows for genome-wide determination of DNA methylation status by comparing band patterns generated using a pair of isoschizomer restriction enzymes with different sensitivities to methylation (Reyna-López et al., 1997). After MSAP analysis using 64 primer sets on 11 viable hybrids (A5S-V1–11) and 11 lethal hybrids (A5S-L1–11), which were also used for AFLP analysis, a total of 324 MSAP loci were detected, with 40 loci indicating de novo methylation and four loci indicating demethylation in hybrids compared to their parents (Figure 3A). The frequencies of de novo methylation and demethylation in the hybrids ranged from 5.25% to 8.64% and from 0% to 0.62% of the total MSAP loci, respectively (Figure 3B). Among the 44 loci showing methylation changes, 16 were consistently detected in all the hybrids. For the remaining 28 loci, the number of hybrids with methylation changes varied depending on the locus. The methylation changes most frequently observed in viable hybrids were detected in seven of the 11 viable hybrids, but this change was also detected in five of the 11 lethal hybrids. No methylation change was consistently detected across all viable or lethal hybrids.
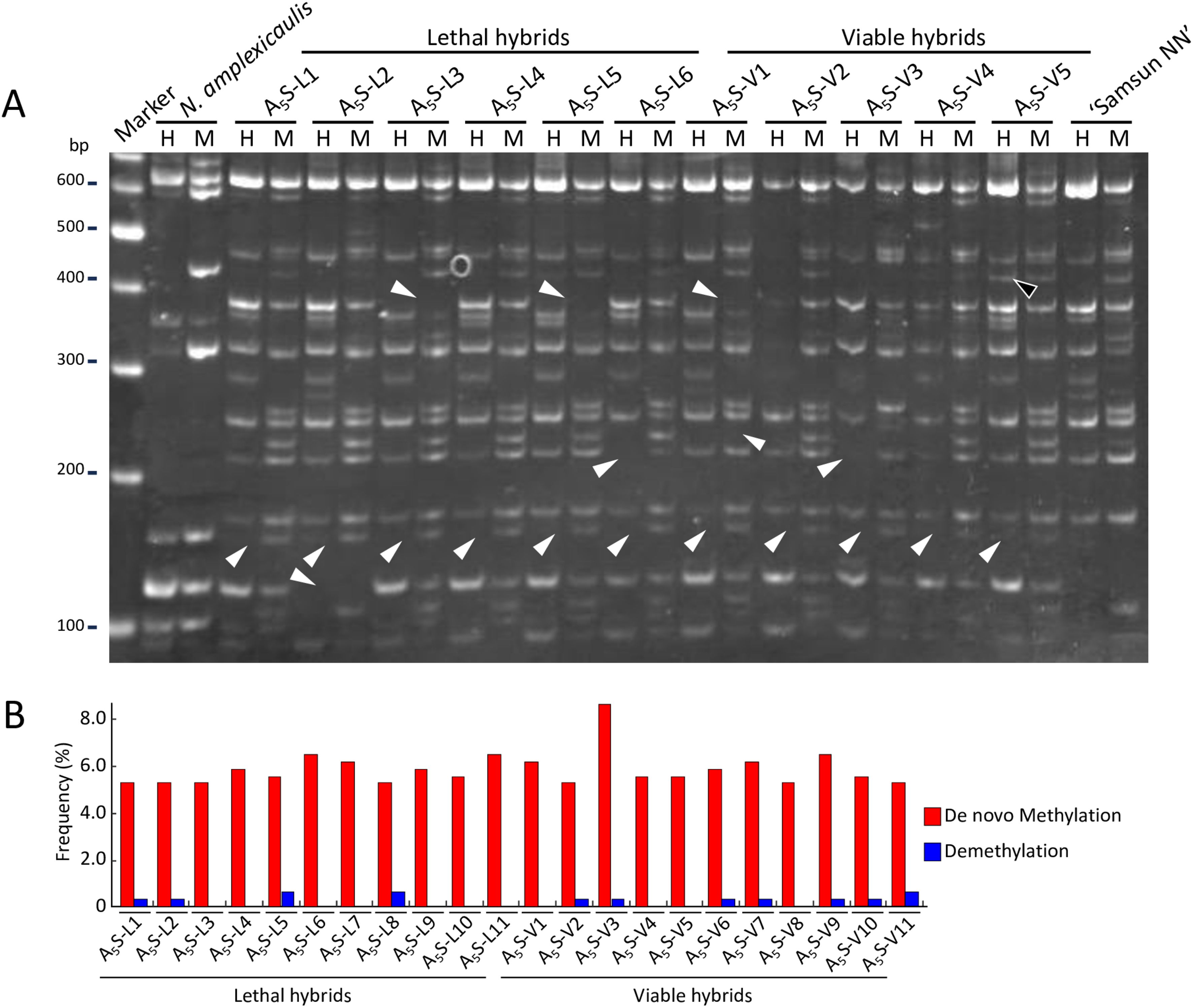
Figure 3. Epigenetic changes in hybrid plants identified by MSAP analysis. (A) An example of bands indicating variation of DNA methylation with the E35-HM49 primer pair. Lane H, PCR products obtained by digestion with EcoRI–HpaII; lane M, PCR products obtained by digestion with EcoRI–MspI; Marker lane, DNA size marker (GeneRuler DNA ladder mix). White arrowheads indicate MSAP bands showing de novo methylation, and black arrowheads indicate MSAP bands showing demethylation. (B) Frequency of methylation changes in each hybrid identified by MSAP analysis.
3.7 Investigation of loci causing hybrid lethality
Possible mutations or losses of Hla1–1 and Hla2–1 alleles were investigated in 10 viable (A8S-V1–10) and 10 lethal hybrids (A8S-L1–10) obtained from a cross between an S8 plant and N. tabacum ‘Samsun NN’. The N. tabacum HLA2 locus is closely linked to the SSR marker PT30342 on the Q chromosome (N. tabacum linkage group 11) (Ma, 2017; Bindler et al., 2007; Tezuka et al., 2012). We investigated the presence of three SSR markers linked to HLA2, including PT30342 (Bindler et al., 2007, 2011), and two unmapped Q chromosome-specific DNA markers, QCS3 and QCS4, (Tezuka et al., 2004; Tezuka and Marubashi, 2006a), in the hybrids. Additionally, an STS marker, HLA2-STS, which was newly developed based on the sequence of HLA2, was also investigated (Supplementary Table 1). All markers were detected in both the viable and lethal hybrids (Figure 4A).
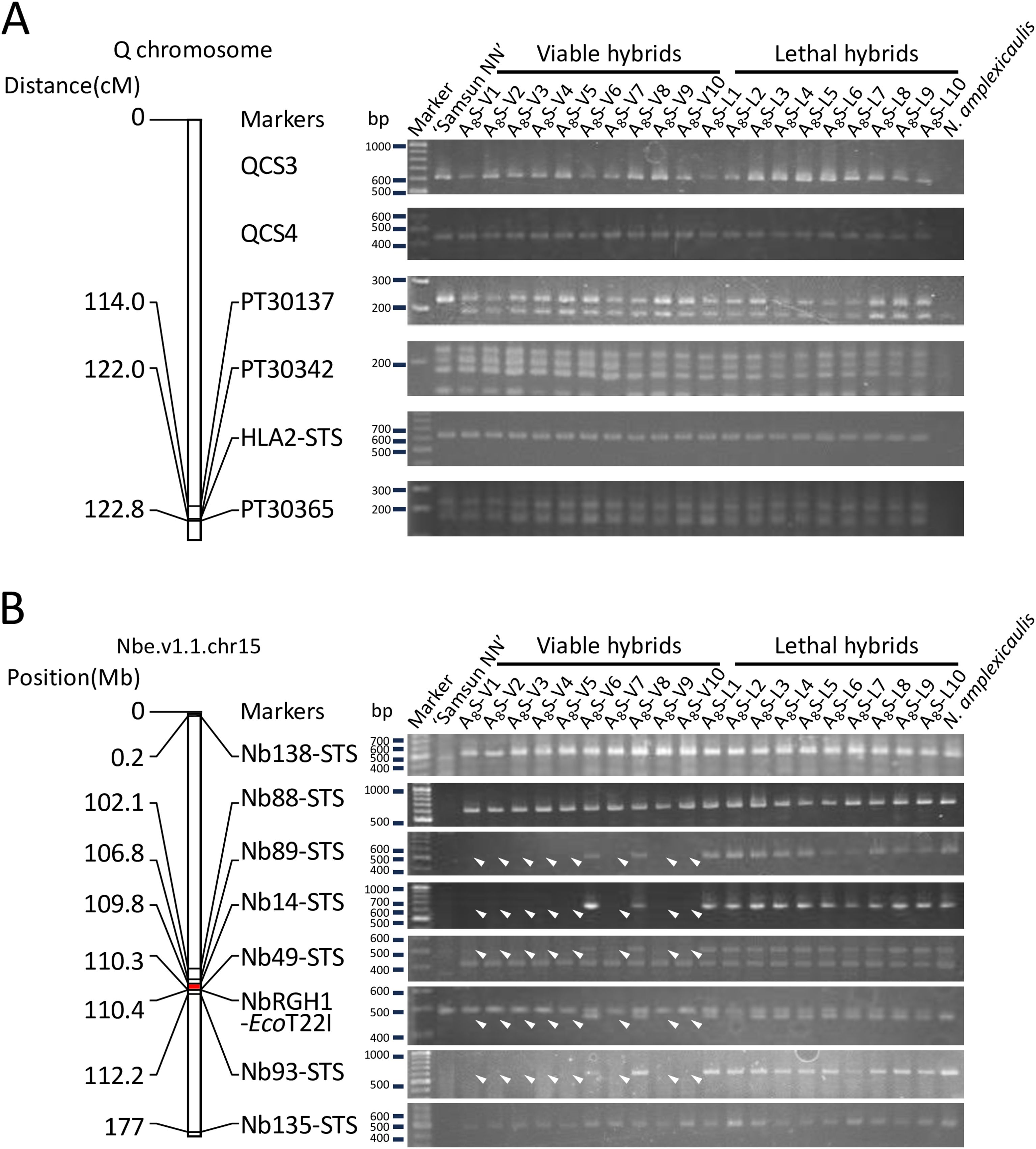
Figure 4. PCR analysis of the two hybrid lethality loci HLA2 (A) and HLA1 (B). (A) Linkage map showing marker positions on the Q chromosome involving HLA2 and the band patterns detected with the markers. (B) Physical map showing marker positions in Nbe.v1.1.chr15 scaffold, which involves HLA1, from the Nbe.v1.1 genome of N. benthamiana, and the band patterns detected with the markers. Position (Mb) indicates the approximate position of the markers in Nbe.v1.1.chr15 scaffold. The Red region indicates the region containing the HLA1 locus. Arrowheads indicate the disappeared bands. Marker lane, size marker (GeneRuler DNA ladder mix).
Another hybrid lethality gene HLA1 was mapped between two cleaved amplified polymorphic sequence (CAPS) markers, NbRGH1-CAPS and Nb14-CAPS, located on Nbe.v1.1.chr15 in N. benthamiana genome Nbe.v1.1 (Kurotani et al., 2023; Tezuka et al., 2021). In addition to these markers, we used the STS marker Nb49 located between NbRGH1-CAPS and Nb14-CAPS (Tezuka et al., 2024), and five other STS markers, Nb138-STS, Nb88-STS, Nb89-STS, Nb93-STS, and Nb135-STS, which were newly developed based on the scaffold sequence (Supplementary Table 1). Five markers from Nb89-STS to Nb93-STS in the physical map were detected in all lethal hybrids but not in eight of the 10 viable hybrids (Figure 4B). In contrast, markers located outside this region (Nb138-STS and Nb88-STS on the same side, and Nb135-STS on the other side) were detected in all hybrids.
Additionally, to investigate whether suppression of Hla2–1 expression contributed to the suppression of hybrid lethality, we conducted RT-PCR analysis. Because the 10 viable hybrids (A8S-V1–10) and 10 lethal hybrids (A8S-L1–10) used in the above marker analysis had died due to natural aging and hybrid lethality, respectively, 21 viable (A8S-V11–31) and three lethal (A8S-L11–13) hybrids were newly obtained from the cross between the S8 plant and ‘Samsun NN’, and used for the gene expression analysis. Hla2–1 was expressed in all the hybrids (Supplementary Figure 1).
4 Discussion
4.1 Characterization of hybrid lethality in crosses between N. amplexicaulis and N. tabacum
Hybrid lethality in reciprocal crosses between N. amplexicaulis and N. tabacum was characterized by lethal symptoms, such as browning of the hypocotyls and roots. In addition, hybrid lethality in the cross N. amplexicaulis × N. tabacum was temperature sensitive, and the Q chromosome in N. tabacum genome was involved in hybrid lethality. These characteristics are consistent with Type II hybrid lethality caused by epistatic interactions between the Hla1–1 allele from Suaveolentes species and the Hla2–1 allele from N. tabacum (Iizuka et al., 2012; Ma et al., 2020; Tezuka et al., 2024). Considering that species in the monophyletic section Suaveolentes are closely related (Chase et al., 2003; Clarkson et al., 2004; D’Andrea et al., 2023), the present results indicate that N. amplexicaulis also carries the Hla1–1 allele. This is also supported by a report by Ma et al. (2020), where all hybrids survived after crossing N. amplexicaulis with N. tabacum mutant for the HLA2 gene.
Crosses between N. amplexicaulis and N. tabacum produced a large number of normal hybrids compared to crosses using other Suaveolentes species. Previous studies have reported the occurrence of viable hybrids at extremely low frequencies in crosses between Suaveolentes and N. tabacum (Tezuka et al., 2010; Tezuka and Marubashi, 2006b; Hancock et al., 2015; He et al., 2019; Nakata et al., 2021) (Supplementary Table 5). In contrast, the present study demonstrated an ultrahigh frequency of viable hybrids, ranging from 3.2% to 63.2% (Table 2 and Supplementary Table 3). To date, such a high frequency of viable hybrids has not been reported in other plant species where hybrid lethality has been observed.
4.2 Causes of ultrahigh frequency appearance of viable hybrids
It is important to understand the mechanism of the ultrahigh frequency of viable hybrid occurrence from lethal crosses not only for plant breeding, which needs to overcome hybrid lethality, but also for understanding the evolution of species. Viable hybrids appeared at a high frequency in reciprocal crosses between N. amplexicaulis and N. tabacum, suggesting that the cause of viable hybrid occurrence was within the nuclear genome and not in the cytoplasm. Crossing experiments using the S0 and S1 plants of N. amplexicaulis suggested that the HLA1 gene was not heterozygous and that some other factor must have contributed to the survival of the hybrids.
In crosses using the S1 plants, the occurrence rate of viable hybrids varied among S1 plants derived from the same parents; 14.1%–29.8% in the pedigree of No. 1 (8.9%), 16.1%–49.2% in the pedigree of No. 2 (63.2%), 6.2%–34.4% in the pedigree of No. 3 (56.8%), and 3.2%–40.3% in the pedigree of No. 4 (51.2%) (Supplementary Table 3). If N. amplexicaulis is genetically fixed, the frequency is expected to remain constant between the parents and their progeny. Therefore, although the HLA1 gene was homozygous, some heterozygosity at other loci in N. amplexicaulis might have been involved in the fluctuations in the percentages. Alternatively, the frequency of viable hybrids may be influenced by environmental factors, such as temperature during fertilization or seed formation.
Genetic and epigenetic changes have been reported to occur in early generations of new allopolyploids (Shaked et al., 2001; Madlung et al., 2002; Wu et al., 2015; Li et al., 2019; Mhiri et al., 2019; Qiu et al., 2020). Such changes are also observed in interspecific hybrids. Wu et al. (2015) reported that genetic changes ranging from 0.7% to 6.9% and DNA methylation changes ranging from 0.2% to 5.8% were detected in interspecific F1 hybrids of Oryza using AFLP and MSAP analyses, respectively. In Nicotiana, RAPD analysis did not show genetic changes in F1 hybrids from several interspecific crosses between Suaveolentes species and N. tabacum (Tezuka and Marubashi, 2006b; Tezuka et al., 2010; Iizuka et al., 2012; Nakata et al., 2021). However, in the present study, RAPD and AFLP analyses indicated that genomic alterations occurred in the hybrids of N. amplexicaulis and N. tabacum (Figure 2, Table 2). In addition, MSAP revealed methylation changes in the hybrids (Figure 3). Because genetic and epigenetic changes common to the most viable or lethal hybrids were not detected, it was unclear whether these changes were related to the occurrence of viable hybrids. Nevertheless, the results demonstrated that large-scale genetic and epigenetic changes, commonly referred to as “genome shock” (McClintock, 1984), occurred in interspecific hybrids between N. amplexicaulis and N. tabacum, suggesting their involvements in ultrahigh frequency appearance of viable hybrids.
We investigated the potential involvement of mutations in HLA1 and HLA2, which are causal genes of hybrid lethality. No mutations were detected at the HLA2 locus or in the surrounding region in any of the (Figure 4). Expression of the Hla2–1 allele was also observed in all the hybrids (Supplementary Figure 1). In contrast, several DNA markers linked to HLA1 were not detected in eight of the ten viable hybrids, suggesting that this region could be frequently deleted (Figure 4). Therefore, one of the mechanisms underlying the ultrahigh-frequency appearance of viable hybrids is the partial deletion of the chromosome where the HLA1 locus resides.
Crossing results using the S1 plants provide insight into the timing of the partial deletion of the chromosome including the HLA1 locus. If the deletion of HLA1 occurred during gametogenesis in N. amplexicaulis, some S1 plants would produce either only viable hybrids (homozygous for the deletion of HLA1) or viable and lethal hybrids in a 1:1 ratio (heterozygous for the deletion of HLA1). Among the 40 S1 plants tested, eight plants produced viable and lethal hybrids in a ratio close to 1:1 (Supplementary Table 3, p > 0.05), indicating that the deletion frequency would be 10% (8 out of 80 gametes) if deletion occurred during gametogenesis. However, the overall frequency of viable hybrids from all S1 plants was 23.31% (597 out of 2561 F1 hybrids). This discrepancy suggests that the deletion of HLA1 occurs after fertilization rather than during gametogenesis in N. amplexicaulis.
Regarding the mechanism of deletion of HLA1, if the deletion were produced by reciprocal translocations between homoeologous chromosomes in N. amplexicaulis as reported by Nakata et al. (2021), either of the terminal markers on the chromosome carrying HLA1 would be lost. However, both terminal markers on Nbe.v1.1.chr15 were detected in all viable hybrids (Figure 4). Therefore, these hybrids appear to possess a mechanism for eliminating HLA1 that does not involve reciprocal translocation. Uniparental chromosome elimination following interspecific fertilization has been reported in several plant hybrids (Komeda et al., 2007; Ishii et al., 2010; Sanei et al., 2011). In the cross Hordeum vulgare × H. bulbosum, inactivation of H. bulbosum centromeres triggers uniparental chromosome elimination in the hybrid embryo (Sanei et al., 2011). However, to our knowledge, the mechanisms of large interstitial chromosomal deletions during hybrid formation have not been reported. Our findings suggest that hybrids of N. amplexicaulis and N. tabacum may possess a novel mechanism for inducing interstitial chromosome deletion through genome shock-induced changes.
In two of the ten viable hybrids examined, deletions at neither causal loci, HLA1 nor HLA2, were detected (Figure 4). Genetic and epigenetic changes, as revealed by the RAPD, AFLP, and MSAP analyses (Figures 2 and 3), may be involved in the survival of these hybrids. Alternatively, the suppression of causal gene expression may be considered a contributing factor to their occurrence. Based on Hla2–1 expression analysis, it is unlikely that suppression of Hla2–1 expression contributed to the occurrence of the two viable hybrids because Hla2–1 was expressed in all 21 viable hybrids investigated (Supplementary Figure 1). As HLA1 gene has not yet been identified, the expression level of Hla1–1 could not be assessed in the present study.
It is still unclear whether the epigenetic changes detected by MSAP analysis were linked to differences in gene expression or phenotypic variation among the hybrids. A future transcriptomic analysis, such as RNA-Seq of parental species, viable hybrids and lethal hybrids, could provide comprehensive insights into the molecular basis of hybrid lethality and its suppression. This analysis would help identify pathways downstream of the HLA1–HLA2 interaction and reveal how the loss of HLA1 alters gene regulation and restores hybrid viability in Nicotiana.
4.3 Evolutionary implications of ultrahigh frequency appearance of viable hybrids
Species are generally reproductively isolated from each other. However, the ability of species to overcome reproductive barriers and produce hybrid progenies is evolutionarily intriguing. We showed a phenomenon that enables many hybrids to overcome hybrid lethality and survive in lethal crosses. The section Suaveolentes is thought to have originated from the hybridization of a common ancestor of sections Alatae and Sylvestres with a common ancestor of sections Noctiflorae and Petunioides (D’Andrea et al., 2023). Considering that most Suaveolentes species, including N. amplexicaulis, carry the Hla1–1 allele, N. amplexicaulis likely independently acquired the ability to produce a large number of viable hybrids in lethal crosses with N. tabacum within the section Suaveolentes.
Suaveolentes species, which are mainly distributed in Australasia, and N. tabacum, which is believed to have originated in South America, are geographically isolated. Although there seems to be no opportunity for these species to intercross, the present study provides a model where sympatric species can overcome hybrid lethality through the ultrahigh-frequency appearance of viable hybrids. When crossed with N. tabacum, Suaveolentes species other than N. amplexicaulis rarely produce viable hybrids, while N. amplexicaulis frequently produce viable hybrids. Viable hybrids obtained from crosses between Suaveolentes species and N. tabacum are generally sterile, and fertility can be restored via amphiploidization through chromosome doubling (Lloyd, 1975; Tezuka and Marubashi, 2006b; Tezuka et al., 2010). In the present study, the viable hybrids obtained from the cross between N. amplexicaulis and N. tabacum were sterile. Given that speciation requires overcoming both hybrid lethality and hybrid sterility, the ultrahigh-frequency appearance of viable hybrids observed in N. amplexicaulis crosses provides a higher chance of overcoming hybrid sterility than crosses that rarely produce viable hybrids.
In plants, many new species are thought to have arisen through interspecific hybridization and whole-genome duplication, with genome shock serving as a driving force that rapidly generates diverse genetic combinations. For instance, field experiments using synthetic interspecific hybrids of the genus Helianthus have suggested that the genetic diversity generated by interspecific hybridization accelerates the pace of adaptive evolution (Mitchell et al., 2019). In Mimulus, artificially synthesized triploid hybrids overcome hybrid sterility through whole-genome duplication to produce fertile allohexaploids. While these synthetic lines displayed survival rates that were intermediate between those of their parental species, naturally formed triploids (M. × robertsii), and allohexaploids (M. peregrinus) often exhibited even higher survival rates, suggesting that further adaptive evolution under natural conditions can enhance the fitness of emerging polyploid lineages (Meeus et al., 2020). Thus, once a hybrid is established and the factors responsible for reproductive isolation are eliminated, such individuals can acquire novel adaptive capabilities in new environments that are not present in the parental species, leading to a new species.
Future studies could investigate transcriptomic regulation in the viable hybrids obtained from the cross between N. amplexicaulis and N. tabacum by analyzing expression level dominance (ELD) and homeolog expression bias (HEB), which describe how gene expression patterns are rebalanced between subgenomes following hybridization and polyploidization (Grover et al., 2012; Wu et al., 2018; Li M. et al., 2020). These analyses could reveal how the loss of HLA1 influences the global regulatory balance between parental subgenomes and whether specific ELD or HEB patterns in stress-related or developmental pathways correlate with the rescue of hybrid viability. This approach would provide broader insights into how interspecific hybridization and structural genomic changes contribute to polyploid genome evolution and phenotypic stabilization.
5 Conclusion
In conclusion, although some hybrid seedlings from the cross between N. amplexicaulis and N. tabacum showed hybrid lethality, others overcame this lethality at ultrahigh frequencies. Our findings indicate that this was primarily due to the elimination of the causal locus HLA1 for hybrid lethality. Moreover, interspecific hybridization between these species triggers genome shock, which facilitates the breakdown of reproductive barriers, including hybrid lethality. To date, there have been few reports on the mechanisms by which a once reproductively isolated species overcomes these barriers and produced hybrid progeny. The process by which strong hybrid incompatibility is overcome and novel genomic combinations become stabilized not only has significant implications for plant breeding but also serves as a critical model for understanding evolutionary processes in nature.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SN: Formal analysis, Writing – original draft, Writing – review & editing, Visualization, Conceptualization, Investigation. KKa: Writing – review & editing, Investigation, Formal analysis. HI: Investigation, Writing – review & editing, Formal analysis. KM: Writing – review & editing, Investigation, Formal analysis. TI: Writing – review & editing, Investigation, Formal analysis. KKo: Writing – review & editing, Investigation, Formal analysis. KN: Formal analysis, Writing – review & editing. TY: Supervision, Writing – review & editing. WM: Supervision, Writing – review & editing. MY: Writing – review & editing, Supervision. TM: Writing – review & editing, Supervision. SY: Supervision, Writing – review & editing. TT: Funding acquisition, Writing – review & editing, Writing – original draft, Project administration, Methodology, Supervision, Conceptualization, Investigation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was partly supported by JSPS KAKENHI Grant Numbers JP20880024, JP25870627, JP17K15224, JP20K05988, and JP25K09063 from the Japan Society for the Promotion of Science to TT.
Acknowledgments
We thank the Leaf Tobacco Research Center, Japan Tobacco Inc., Oyama, Japan, for providing seeds of N. tabacum ‘Red Russian’ and ‘Samsun NN’, and N. amplexicaulis accession JT. Seeds of N. amplexicaulis accessions PI 271989 and PI 555682 were kindly supplied by the United States Nicotiana Germplasm Collection. We also thank Dr. Tomoaki Kubo, a former director of the Iwata Tobacco Experiment Station of Japan Tobacco Inc., for the gift of Haplo-Q.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1690873/full#supplementary-material
Supplementary Figure 1 | Gene expression of Hla2–1 in the hybrids from the cross between N. amplexicaulis JT and ‘Samsun NN’ analyzed by RT-PCR.
Supplementary Table 1 | DNA markers developed in the present study.
Supplementary Table 2 | Primers used for AFLP and MSAP analysis.
Supplementary Table 3 | Viability of hybrid seedlings between N. amplexicaulis JT S0/S1 plants and N. tabacum.
Supplementary Table 4 | The number of hybrids lacking specific bands as detected by AFLP analysis.
References
Bindler, G., Plieske, J., Bakaher, N., Gunduz, I., Ivanov, N., Rutger Van der, H., et al. (2011). A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theor. Appl. Genet. 123, 219–230. doi: 10.1007/s00122-011-1578-8
Bindler, G., Rutger van der, H., Gunduz, I., Plieske, J., Ganal, M., Rossi, L., et al. (2007). A microsatellite marker based linkage map of tobacco. Theor. Appl. Genet. 114, 341–349. doi: 10.1007/s00122-006-0437-5
Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J. L., et al. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PloS Biol. 5, e236. doi: 10.1371/journal.pbio.0050236
Bomblies, K. and Weigel, D. (2007). Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8, 382–393. doi: 10.1038/nrg2082
Chase, M. W., Knapp, S., Cox, A. V., Clarkson, J. J., Butsko, Y., Joseph, J., et al. (2003). Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann. Bot. 92, 107–127. doi: 10.1093/aob/mcg087
Chen, C., Chen, H., Lin, Y.-S., Shen, J. B., Shan, J. X., Qi, P., et al. (2014). A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5, 3357. doi: 10.1038/ncomms4357
Clarkson, J. J., Knapp, S., Garcia, V. F., Olmstead, R. G., Leitch, A. R., and Chase, M. W. (2004). Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol. Phylogenet. Evol. 33, 75–90. doi: 10.1016/j.ympev.2004.05.002
D’Andrea, L., Sierro, N., Ouadi, S., Hasing, T., Rinaldi, E., Ivanov, N. V., et al. (2023). Polyploid Nicotiana section Suaveolentes originated by hybridization of two ancestral Nicotiana clades. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.999887
Deng, J., Fang, L., Zhu, X., Zhou, B., and Zhang, T. (2019). A CC-NBS-LRR gene induces hybrid lethality in cotton. J. Exp. Bot. 70, 5145–5156. doi: 10.1093/jxb/erz312
Dziasek, K., Simon, L., Lafon-Placette, C., Laenen, B., Wärdig, C., Santos-González, J., et al. (2021). Hybrid seed incompatibility in Capsella is connected to chromatin condensation defects in the endosperm. PloS Genet. 17, e1009370. doi: 10.1371/journal.pgen.1009370
Fulneček, J. and Kovařík, A. (2014). How to interpret Methylation Sensitive Amplified Polymorphism (MSAP) profiles? BMC Genet. 15, 2. doi: 10.1186/1471-2156-15-2
Grover, C. E., Gallagher, J. P., Szadkowski, E. P., Yoo, M. J., Flagel, L. E., and Wendel, J. F. (2012). Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196, 966–971. doi: 10.1111/j.1469-8137.2012.04365.x
Hancock, W. G., Kuraparthy, V., Kernodle, S. P., and Lewis, R. S. (2015). Identification of maternal haploids of Nicotiana tabacum aided by transgenic expression of green fluorescent protein: evidence for chromosome elimination in the N. tabacum × N. africana interspecific cross. Mol. Breed. 35, 179. doi: 10.1007/s11032-015-0372-8
He, H., Iizuka, T., Maekawa, M., Sadahisa, K., Morikawa, T., Yanase, M., et al. (2019). Nicotiana suaveolens accessions with different ploidy levels exhibit different reproductive isolation mechanisms in interspecific crosses with Nicotiana tabacum. J. Plant Res. 132, 461–471. doi: 10.1007/s10265-019-01114-w
He, H., Shiragaki, K., and Tezuka, T. (2023). Understanding and overcoming hybrid lethality in seed and seedling stages as barriers to hybridization and gene flow. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1219417
Huang, J., Yang, L., Yang, L., Wu, X., Cui, X., Zhang, L., et al. (2023). Stigma receptors control intraspecies and interspecies barriers in Brassicaceae. Nature 614, 303–308. doi: 10.1038/s41586-022-05640-x
Iizuka, T., Kuboyama, T., Marubashi, W., Oda, M., and Tezuka, T. (2012). Nicotiana debneyi has a single dominant gene causing hybrid lethality in crosses with N. tabacum. Euphytica 186, 321–328. doi: 10.1007/s10681-011-0570-3
Ishii, T., Ueda, T., Tanaka, H., and Tsujimoto, H. (2010). Chromosome elimination by wide hybridization between Triticeae or oat plant and pearl millet: pearl millet chromosome dynamics in hybrid embryo cells. Chrom. Res. 18, 821–831. doi: 10.1007/s10577-010-9158-3
Komeda, N., Chaudhary, H. K., Suzuki, G., and Mukai, Y. (2007). Cytological evidence for chromosome elimination in wheat × Imperata cylindrica hybrids. Genes. Genet. Syst. 82, 241–248. doi: 10.1266/ggs.82.241
Kurotani, K., Hirakawa, H., Shirasawa, K., Tanizawa, Y., Nakamura, Y., Isobe, S., et al. (2023). Genome sequence and analysis of Nicotiana benthamiana, the model plant for interactions between organisms. Plant Cell Physiol. 64, 248–257. doi: 10.1093/pcp/pcac168
Lee, C. B., Page, L. E., McClure, B. A., and Holtsford, T. P. (2008). Post-pollination hybridization barriers in Nicotiana section Alatae. Plant Reprod. 21, 183–195. doi: 10.1007/s00497-008-0077-9
Li, Q., Rana, K., Xiong, Z., Ge, X., Li, Z., Song, H., et al. (2019). Genetic and epigenetic alterations in hybrid and derived hexaploids between Brassica napus and B. oleracea revealed by SSR and MSAP analysis. Acta Physiol. Plant 41, 61. doi: 10.1007/s11738-019-2850-9
Li, M., Wang, R., Wu, X., and Wang, J. (2020). Homoeolog expression bias and expression level dominance (ELD) in four tissues of natural allotetraploid Brassica napus. BMC Genomics 21, 330. doi: 10.1186/s12864-020-6747-1
Li, J., Zhou, J., Zhang, Y., Yang, Y., Pu, Q., and Tao, D. (2020). New insights into the nature of interspecific hybrid sterility in rice. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.555572
Lloyd, R. (1975). Tissue culture as a means of circumventing lethality in an interspecific Nicotiana hybrid. Tob. Sci. 19, 4–6.
Ma, J. (2017). The fine mapping of two black shank resistance loci and identification of a hybrid lethality gene in tobacco. Ph.D thesis (Raleigh: North Carolina State University).
Ma, J., Hancock, W. G., Nifong, J. M., Kernodle, S. P., and Lewis, R. S. (2020). Identification and editing of a hybrid lethality gene expands the range of interspecific hybridization potential in Nicotiana. Theor. Appl. Genet. 133, 2915–2925. doi: 10.1007/s00122-020-03641-w
Madlung, A., Masuelli, R. W., Watson, B., Reynolds, S. H., Davison, J., and Comai, L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129, 733–746. doi: 10.1104/pp.003095
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. doi: 10.1126/science.15739260
Meeus, S., Šemberová, K., De Storme, N., Geelen, D., and Vallejo-Marín, M. (2020). Effect of whole-genome duplication on the evolutionary rescue of sterile hybrid monkeyflowers. Plant Commun. 1, 100093. doi: 10.1016/j.xplc.2020.100093
Mhiri, C., Parisod, C., Daniel, J., Petit, M., Lim, K. Y., Dorlhac de Borne, F., et al. (2019). Parental transposable element loads influence their dynamics in young Nicotiana hybrids and allotetraploids. New Phytol. 221, 1619–1633. doi: 10.1111/nph.15484
Mino, M., Maekawa, K., Ogawa, K., Yamagishi, H., and Inoue, M. (2002). Cell death processes during expression of hybrid lethality interspecific F1 hybrid between Nicotiana gossei Domin and Nicotiana tabacum. Plant Physiol. 130, 1776–1787. doi: 10.1104/pp.006023
Mino, M., Tezuka, T., and Shomura, S. (2022). The hybrid lethality of interspecific F1 hybrids of Nicotiana: a clue to understanding hybrid inviability–a major obstacle to wide hybridization and introgression breeding of plants. Mol. Breed. 42, 10. doi: 10.1007/s11032-022-01279-8
Mitchell, N., Owens, G. L., Hovick, S. M., Rieseberg, L. H., and Whitney, K. D. (2019). Hybridization speeds adaptive evolution in an eight-year field experiment. Sci. Rep. 9, 6746. doi: 10.1038/s41598-019-43119-4
Murashige, T. and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Murray, M. G. and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nakata, K., Nagashima, H., Inaba, N., Yamashita, H., Shinozaki, Y., Kanekatsu, M., et al. (2021). Analysis of the possible cytogenetic mechanism for overcoming hybrid lethality in an interspecific cross between Nicotiana suaveolens and Nicotiana tabacum. Sci. Rep. 11, 7812. doi: 10.1038/s41598-021-87242-7
Olmo, H. P. (1935). Genetical studies of monosomic types of Nicotiana tabacum. Genetics 20, 286–300. doi: 10.1093/genetics/20.3.286
Qiu, T., Liu, Z., and Liu, B. (2020). The effects of hybridization and genome doubling in plant evolution via allopolyploidy. Mol. Biol. Rep. 47, 5549–5558. doi: 10.1007/s11033-020-05597-y
Ren, L., Gao, X., Cui, J., Zhang, C., Dai, H., Luo, M., et al. (2022). Symmetric subgenomes and balanced homoeolog expression stabilize the establishment of allopolyploidy in cyprinid fish. BMC Biol. 20, 200. doi: 10.1186/s12915-022-01401-4
Reyna-López, G. E., Simpson, J., and Ruiz-Herrera, J. (1997). Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genom. 253, 703–710. doi: 10.1186/s12915-022-01401-4
Rieseberg, L. H. and Blackman, B. K. (2010). Speciation genes in plants. Ann. Bot. 106, 439–455. doi: 10.1093/aob/mcq126
Rieseberg, L. H. and Willis, J. H. (2007). Plant speciation. Science 317, 910–914. doi: 10.1126/science.1137729
Sanei, M., Pickering, R., Kumke, K., Nasuda, S., and Houben, A. (2011). Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. 108, E498–E505. doi: 10.1073/pnas.1103190108
Shaked, H., Kashkush, K., Ozkan, H., Feldman, M., and Levy, A. A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13, 1749–1759. doi: 10.1105/tpc.010083
Shiragaki, K., Nakamura, R., Nomura, S., He, H., Yamada, T., Marubashi, W., et al. (2020a). Phenylalanine ammonia-lyase and phenolic compounds are related to hybrid lethality in the cross Nicotiana suaveolens × N. tabacum. Plant Biotechnol. 37, 327–333. doi: 10.5511/plantbiotechnology.20.0606a
Shiragaki, K., Yokoi, S., and Tezuka, T. (2020b). A hypersensitive response-like reaction is involved in hybrid weakness in F1 plants of the cross Capsicum annuum × Capsicum chinense. Breed. Sci. 70, 430–437. doi: 10.1270/jsbbs.19137
Si, Y., Zheng, S., Niu, J., Tian, S., Gu, M., Lu, Q., et al. (2021). Ne2, a typical CC–NBS–LRR-type gene, is responsible for hybrid necrosis in wheat. New Phytol. 232, 279–289. doi: 10.1111/nph.17575
Tezuka, T., Kitamura, N., Imagawa, S., Hasegawa, A., Shiragaki, K., He, H., et al. (2021). Genetic mapping of the HLA1 locus causing hybrid lethality in Nicotiana interspecific hybrids. Plants 10, 2062. doi: 10.3390/plants10102062
Tezuka, T., Kuboyama, T., Matsuda, T., and Marubashi, W. (2007). Possible involvement of genes on the Q chromosome of Nicotiana tabacum in expression of hybrid lethality and programmed cell death during interspecific hybridization to Nicotiana debneyi. Planta 226, 753–764. doi: 10.1007/s00425-007-0522-2
Tezuka, T., Kuboyama, T., Matsuda, T., and Marubashi, W. (2010). Seven of eight species in Nicotiana section Suaveolentes have common factors leading to hybrid lethality in crosses with Nicotiana tabacum. Ann. Bot. 106, 267–276. doi: 10.1093/aob/mcq114
Tezuka, T. and Marubashi, W. (2004). Apoptotic cell death observed during the expression of hybrid lethality in interspecific hybrids between Nicotiana tabacum and N. suaveolens. Breed. Sci. 54, 59–66. doi: 10.1270/jsbbs.54.59
Tezuka, T. and Marubashi, W. (2006a). Hybrid lethality in interspecific hybrids between Nicotiana tabacum and N. suaveolens: evidence that the Q chromosome causes hybrid lethality based on Q-chromosome-specific DNA markers. Theor. Appl. Genet. 112, 1172–1178. doi: 10.1007/s00122-006-0219-0
Tezuka, T. and Marubashi, W. (2006b). Genomic factors lead to programmed cell death during hybrid lethality in interspecific hybrids between Nicotiana tabacum and N. debneyi. SABRAO. J. Breed. Genet. 32, 69–81.
Tezuka, T. and Marubashi, W. (2012). Genes in S and T subgenomes are responsible for hybrid lethality in interspecific hybrids between Nicotiana tabacum and Nicotiana occidentalis. PloS One 7, e36204. doi: 10.1371/journal.pone.0036204
Tezuka, T., Matsuo, C., Iizuka, T., Oda, M., and Marubashi, W. (2012). Identification of Nicotiana tabacum linkage group corresponding to the Q chromosome gene(s) involved in hybrid lethality. PloS One 7, e37822. doi: 10.1371/journal.pone.0037822
Tezuka, T., Nagai, S., Matsuo, C., Okamori, T., Iizuka, T., and Marubashi, W. (2024). Genetic cause of hybrid lethality observed in reciprocal interspecific crosses between Nicotiana simulans and N. tabacum. Int. J. Mol. Sci. 25, 1226. doi: 10.3390/ijms25021226
Tezuka, T., Onosato, K., Hijishita, S., and Marubashi, W. (2004). Development of Q-chromosome-specific DNA markers in tobacco and their use for identification of a tobacco monosomic line. Plant Cell Physiol. 45, 1863–1869. doi: 10.1093/pcp/pch204
Van de Peer, Y., Maere, S., and Meyer, A. (2009). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732. doi: 10.1038/nrg2600
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Wu, J., Lin, L., Xu, M., Chen, P., Liu, D., Sun, Q., et al. (2018). Homoeolog expression bias and expression level dominance in resynthesized allopolyploid Brassica napus. BMC Genomics 19, 586. doi: 10.1186/s12864-018-4966-5
Wu, Y., Sun, Y., Shen, K., Sun, S., Wang, J., Jiang, T., et al. (2015). Immediate genetic and epigenetic changes in F1 hybrids parented by species with divergent genomes in the rice genus (Oryza). PloS One 10, e0132911. doi: 10.1371/journal.pone.0132911
Xu, P., Xu, J., Guo, Q., Xu, Z., Ji, W., Yu, H., et al. (2023). A recessive LRR-RLK gene causes hybrid breakdown in cotton. Theor. Appl. Genet. 136, 189. doi: 10.1007/s00122-023-04427-6
Yamada, T. and Marubashi, W. (2003). Overproduced ethylene causes programmed cell death leading to temperature-sensitive lethality in hybrid seedlings from the cross Nicotiana suaveolens × N. tabacum. Planta 217, 690–698. doi: 10.1007/s00425-003-1035-2
Keywords: interspecific hybridization, hybrid lethality, reproductive isolation, genome shock, evolution, Nicotiana
Citation: Nagai S, Kawaguchi K, Itakura H, Matsumoto K, Iizuka T, Kobayashi K, Nakata K, Yamada T, Marubashi W, Yanase M, Morikawa T, Yokoi S and Tezuka T (2025) Genome-shock deletion of a hybrid lethality gene breaks a reproductive barrier and facilitates speciation in Nicotiana. Front. Plant Sci. 16:1690873. doi: 10.3389/fpls.2025.1690873
Received: 22 August 2025; Accepted: 03 November 2025;
Published: 19 November 2025.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Yingzheng Li, Sichuan Academy of Agricultural Sciences, ChinaApoloniusz Berbeć, Institute of Soil Science and Plant Cultivation, Poland
Copyright © 2025 Nagai, Kawaguchi, Itakura, Matsumoto, Iizuka, Kobayashi, Nakata, Yamada, Marubashi, Yanase, Morikawa, Yokoi and Tezuka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takahiro Tezuka, dGV6dWthQG9tdS5hYy5qcA==
†Present address: Kenji Kawaguchi, Kyushu Okinawa Agricultural Research Center, National Agricultural and Food Research Organization (NARO), Chikugo, Fukuoka, Japan
 Shota Nagai
Shota Nagai Kenji Kawaguchi3†
Kenji Kawaguchi3† Tetsuya Yamada
Tetsuya Yamada Shuji Yokoi
Shuji Yokoi Takahiro Tezuka
Takahiro Tezuka