- 1Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding/Zhongshan Biological Breeding Laboratory/Key Laboratory of Plant Functional Genomics of the Ministry of Education, Agricultural College of Yangzhou University, Yangzhou, China
- 2Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops/Jiangsu Key Laboratory of Crop Genetics and Physiology, Yangzhou University, Yangzhou, China
Granule-bound starch synthase I (GBSSI) is encoded by the Waxy (Wx) gene, which is responsible for amylose biosynthesis and is an important factor determining rice cooking and eating quality in rice grains. Although some studies have edited the promoter of Wx to fine-tuning gene expression level and amylose content, few studies have focused on the distal allelic variations of Wx. In this study, we identified and confirmed one distal cis-regulatory element (CRE) related to the Wx gene. Furthermore, the cre mutants were generated in the Nipponbare background carrying the Wxb or Wxa allele. The amylose content of Wxb-cre mutant decreased by 21.43%–31.51% compared with that of wild-type rice with the Wxb allele. However, no significant difference in amylose content was observed between wild-type plants and Wxa-cre mutants which carried the Wxa allele. The altered mature mRNA transcript and protein level of Wx gene caused by CRE showed a high consistency with their amylose content. Meanwhile, lower gelatinization temperature and final viscosity were found in the Wxb-cre mutants, whereas the grain shape and chalkiness and major agronomic traits were not affected. Possibly, the cre mutant causes the transcriptional reprogramming of starch metabolism. Overall, we identified a distal CRE to finely regulate the Wx gene expression and amylose content, which provided a potentially useful allele for quality improvement in rice breeding.
1 Introduction
Gene expression is regulated at multiple levels in eukaryotes. Transcriptional regulation is a complex biological process that directly determines gene expression patterns and levels, influencing the phenotypes, stress tolerance/resistance, and productivity of plants (Cui et al., 2023). A key mechanism of gene transcriptional regulation is the recognition of cis-regulatory elements (CREs) by transcription factors, which controls transcription, and CREs are short DNA sequence motifs within the proximal genome region of a gene (Biłas et al., 2016). Variations in CREs play crucial roles in fine-tuning gene expression (Cui et al., 2023). Targeting the coding regions and CREs of genes through the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system has been well established, enabling the modulation of target gene expression and the improvement of plant traits (Saeed et al., 2022).
As one of the most important food crops in the world, rice provides energy sources for over half of the global population (Mohapatra and Sahu, 2022). The improvement of grain quality, especially cooking, taste, and appearance quality, is an important goal in rice breeding (Lau et al., 2015). Rice grain contains over 80% starch, including amylose and amylopectin. The amylose content (AC) is considered the key component controlling the eating and cooking quality of the rice endosperm, and the Waxy (Wx) gene has been identified to control AC (Li et al., 2016; Sano, 1984). The Wx gene encodes granule-bound starch synthase I (GBSSI), and at least nine natural Wx alleles have been identified in rice, causing differences in AC and rice quality (Teng et al., 2012; Zhang et al., 2021, 2019; Zhou et al., 2021). Due to specialized rice consumption, the demand for AC in rice is becoming increasingly diversified, which requires the regulation of AC or the generation of new Wx alleles.
Gene editing CRE is an improved method for fine-tuning Wx gene expression and AC compared with searching for natural or mutated alleles (Ding et al., 2021). There have been several successful reports of fine-tuning AC in the Wxb background by editing CREs in the promoter region of the Wx gene (Zhao et al., 2024). A previous study demonstrated that editing the promoter regulates Wx expression, changing AC in transgenic lines (Zeng et al., 2020; Huang et al., 2020). However, these studies have mainly focused on the promoter region within 2 kb upstream and the first intron region of the Wx gene but have rarely identified distal CREs (>2 kb from their nearest genes).
In this study, we combined the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) data of developing seeds and bioinformatics analysis to identify CREs that regulate the expression of the key gene in starch biosynthesis, Wx, and obtained CRE mutants using CRISPR/Cas gene editing technology. Additionally, we analyzed the grain morphology, physicochemical quality traits, and agronomic traits of Wx-cre mutants and Nipponbare (NIP) near-isogenic lines with Wxa and Wxb alleles.
2 Method
2.1 Plant materials, transgene constructs, and rice transformation and growth conditions
In this study, wild-type Oryza sativa L. ssp. japonica Nipponbare (NIP) was used for transformation, which carried the Wxb allele and thereby was named as NIP-Wxb. The near-isogenic line NIP-Wxa was derived from the indica cultivars GuiChao2 (GC2) through traditional rice hybridization and generated after at least six rounds of backcrossing with the recurrent parent NIP (Zhang et al., 2019). To detect the genetic backgrounds, NIP-Wxa was genotyped by whole-genome sequencing, as previously reported by our research group (Zhang et al., 2019). The gRNA target site was selected manually, according to the sequence information of CRE (Figure 1). To check for specificity, BLAST analyses of the gRNA target site were performed against the rice genome. The sgRNA expression cassette, including the CRE-targeting sequence, was constructed from SK-gRNA by PCR amplification with primers containing the gRNA sequence (Supplementary Table S1) and then subcloned into the CRISPR/Cas 9 vector by standard Golden Gate assembly (Engler et al., 2008). Rice calli from mature embryos were used as explants for Agrobacterium-mediated transformation, according to a previously published procedure (Liu et al., 1998).
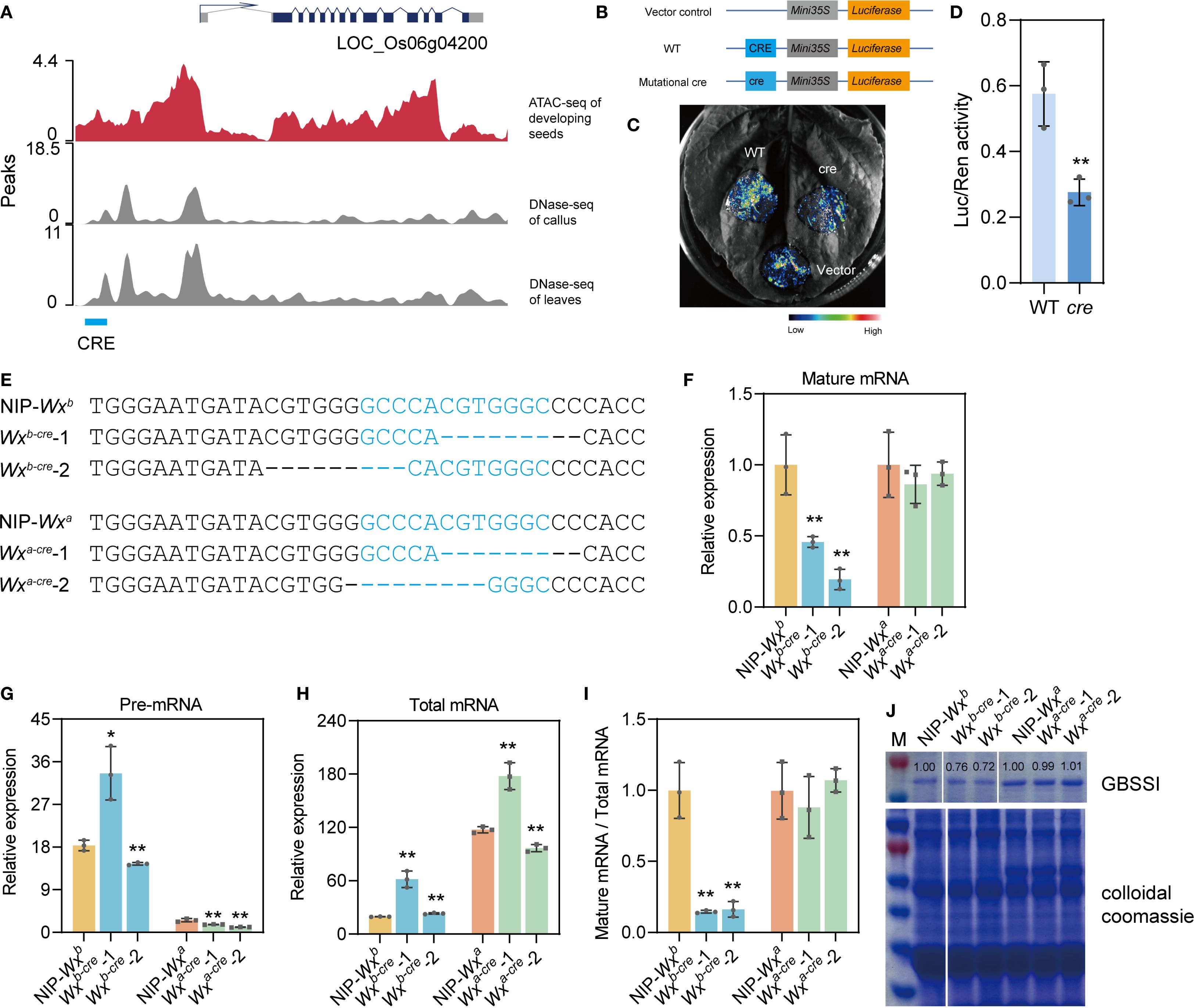
Figure 1. Mining and identification of cis-regulatory elements (CREs) related to the Wx gene. (A) Schematic diagram of predicted CREs of the Wx gene via ATAC-seq in developing seeds at 10 days after flowering (DAF), DNase-seq of leaves and callus (https://plantdhs.org/), and genome sequence analysis (http://rice.plantbiology.msu.edu/pub/data/). (B–D) Transient expression validation of screened CREs in the dual-luciferase reporter system. In detail, the tested sequence of WT CRE is 5′-GATACGTGGGGCCCACGTGGGCCCCACCATTT-3′, therein the underlined sequence is the predicted CRE; the tested sequence of mutational cre, 5′-GATACGTGGGCCCACCATT-3′, corresponds to a deletion of the predicted CRE. (E) Nucleotide variations of different homozygous mutants. The target CRE sequence is highlighted in blue font. “-” indicates a deletion. (F–H) Expression levels of mature Wx mRNA (F), precursor Wx mRNA (G), and total Wx mRNA (H) in developing seeds at 10 DAF of the wild-type and cre mutants. (I) Proportion of mature Wx mRNA of the total Wx mRNA. After normalization to Actin gene, the mean value of the control group was set to 1. (J) GBSSI protein level analyzed using SDS-PAGE. Rice total protein was used as the internal control. White lines represent the images from discontinuous lanes in a film. All data are presented as the mean ± SD. * and ** indicate significant differences from wild-type plants (*P < 0.05 and **P < 0.01) using two-tailed Student’s t-tests.
Total genomic DNA was extracted from transformed rice leaves, as described previously (Yang et al., 2021). The targeted sequences were obtained using PCR with specific primers (Supplementary Table S1), and Sanger sequencing was performed to identify mutations in the target region of CRE. Three or more independent mutants were screened for further analysis, and all selected transgenic lines were homozygous.
All rice materials were planted in the paddy field of Yangzhou University (Yangzhou, Jiangsu Province, China) from April to October in 2021–2024 under safety supervision for genetically modified materials and the same climate and management conditions.
2.2 Transient assay verification in Nicotiana benthamiana
To examine the effects of predicted CREs, the wild-type CRE fragments with flanking sequences from the upstream region of the Wx gene (known as WT) and artificially mutated cre sequence (excluding CRE sequences, also known as cre) were each cloned upstream of the luciferase (LUC) coding region in an expression cassette driven by the Mini35S promoter in the pGreenII0800-LUC vector by using a dual-luciferase reporter system with two reporter genes, LUC and Renilla luciferase (REN) (Xing et al., 2020). The generated plasmids were transformed into Agrobacterium tumefaciens strain EHA105 for transient expression, as reported previously (Xiong et al., 2022). Briefly, A. tumefaciens strains with the constructed vector were resuspended in infection buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 200 μM acetosyringone) to an optical density at 600 nm (0.4) and infiltrated into 5-week-old N. benthamiana leaves. After incubation for 48 h, the LUC signal was visualized with a CCD system of Tanon 5200 Multi Imager (Tanon, Shanghai, China), and the infected leaves were harvested and used for measuring LUC/REN activities.
2.3 RT-PCR, qRT-PCR, and SDS-PAGE
Total RNA was extracted from young panicles of wild-type and mutated plants by using the Plant Total RNA Kit (TaKaRa, Beijing, China). The cDNAs were synthesized using Perfect Real Time PrimeScript RT reagent (TaKaRa). Real-time PCR was performed using TB Green® Premix Ex Taq™ II (TaKaRa) and a LightCycler 480 system (Roche). The rice Actin gene was used as the internal reference to measure the relative expression levels of rice Wx and other 25 genes involved in starch synthesis. Gene-specific primers used for RT-PCR and qRT-PCR are listed in Supplementary Table S1.
For GBSSI activity and SDS-PAGE measurement, total protein was isolated from rice mature seeds, as described previously (Yang et al., 2021). The homogenate of total protein was then shaken for 30 min at room temperature and centrifuged at 10,000 ×g for 10 min. Then, the pellet was washed three times with the same buffer followed by twice with acetone and dried under vacuum. The dried pellet (50 mg) was then used for the extraction of granule-bound GBSSI according to Liu et al. (2014) and Zhang et al. (2019). SDS-PAGE was performed using standard procedures. Gels were stained with Coomassie Brilliant Blue R250 to examine the protein bands.
2.4 Agronomic trait analyses
The main agronomic traits, such as plant height, main panicle length, grains per panicle, effective tiller number, and seed setting rate, were investigated at maturity. The above samples have five biological replicates. At least 200 fully filled grains from panicles of a rice plant were used to measure 1,000-grain weight. The 1,000-grain weight data for each line were derived from the average of three panicles.
2.5 Grain physicochemical properties analyses
Harvested rice grains were dried in an oven at 37 °C for 1 week before analysis. Rice preparation and subsequent general quality measurements, such as apparent amylose content and rapid viscosity analyzer profile, were performed according to a previous study (Zhang et al., 2021). Differential scanning calorimetry (DSC) was performed using a DSC 200 F3 thermal analyzer (Netzsch Instruments NA LLC, Burlington, MA, USA) to analyze the gelatinization temperature of rice flour and isolated starches according to previous report (Zhang et al., 2013). The total starch content of the milled rice flour was analyzed using a total starch assay kit (K-TSTA, Megazyme, Ireland) according to the manufacturer’s assay procedure.
2.6 Statistical analysis
All data are presented as mean ± standard deviation (SD). Comparison of multiple transgenic and wild-type plants was performed using two-tailed Student’s t-tests. * and ** indicate statistical significance between transgenic and wild-type plants at P < 0.05 and P < 0.01, respectively.
3 Results and discussion
3.1 Identification of the distal CRE regulating Wx gene expression and the development of mutant varieties
In this study, we identified one distal CRE (occurring >2 kb upstream from the Wx gene) related to Wx by combining ATAC-seq data and bioinformatics analysis of developing seeds at 10 days after flowering (DAF) in Oryza sativa L. spp. japonica Nipponbare and comparative analysis of DNase I high-sensitivity sites obtained from published DNase-seq data of rice leaves and callus (https://plantdhs.org/) (Figure 1A). Therefore, this study focused on the distal CRE (5′-GCCCACGTGGGC-3′), which had not been reported in previous studies about Wx non-coding region editing (Huang et al., 2020; Zeng et al., 2020; Zhang et al., 2022). Preliminary assessment using the dual-fluorescence transient expression system showed that the CRE sequence enhanced the luciferase (LUC) activity in tobacco leaves compared with the plasmid control (Figures 1B–D).
In addition, we conducted sequence analysis using PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and found that this CRE contains four motifs, i.e., G-box (CACGTG), ABRE2 (CCACGTGG), ABRE (ACGTG), and an unnamed motif (CGTGG). G-box is a ubiquitous element, which was identified in many various plant genes (Menkens et al., 1995). Multiple copies of ABREs (with a core ACGT) generally occur in the upstream of ABA/abiotic stress inducible genes (Hossain et al., 2010). These motifs are generally bound by the bZIP or bHLH transcription factors, playing a central regulatory role in plant growth and development, stress response, and signal transduction (Kong et al., 2018). Two predicted transcription factors (an MYB family transcription factor and an HD-ZIP III transcription factor) that can bind to this CRE sequence are being studied, which might help us to understand the regulation network of the Wx gene in rice.
To investigate the effect of Wx-CRE on AC in rice grain, we obtained Wx-CRE mutants using CRISPR/Cas9 technology. Transgenic plants were generated from two near-isogenic lines, NIP-Wxb and NIP-Wxa, via Agrobacterium-mediated transformation. Two homozygous cre mutant plants of NIP-Wxb or NIP-Wxa were selected for further analysis (Figure 1E). Sequencing analysis revealed that 7- and 3-bp deletions occurred in the CRE sequence of the Wxb-cre-1 and Wxb-cre-2 lines, respectively. Wxa-cre-1 had a 7-bp deletion, and Wxa-cre-2 had an 8-bp deletion in the CRE sequence (Figure 1E). The obtained Wx allelic variations have not been reported in previous studies on Wx non-coding region editing.
We then evaluated GBSSI mRNA and protein levels in developing seeds at 10 DAF and mature seeds of mutants and the wild type (WT). In the Wxa background, the expression levels of GBSSI mRNA and protein were almost unchanged compared with WT (Figures 1F, J). Wxb-cre mutants expressed lower mRNA and protein levels than WT (Figures 1F, J).
In the developing endosperm, the Wx gene had two mRNA transcripts, namely, a 3.3-kb precursor mRNA (pre-mRNA) and a 2.3-kb mature mRNA (mature-mRNA, which is translated into the Wx protein). The endosperm AC and Wx protein content were significantly correlated with the mature mRNA content and the ability to excise intron I from the leader sequence of the Wx transcript (Wang et al., 1995). Thus, the relative amounts of Wx gene pre-mRNA, mature mRNA, and total mRNA (including the 3.3-kb precursor mRNA and 2.3-kb mature mRNA) in developing seeds of different mutant and WT plants at 10 DAF were detected using quantitative real-time PCR with specific primers (Supplementary Figure S1, Supplementary Table S1). As expected, the relative expression level of mature mRNA was reduced in the Wxb-cre mutants compared with WT plants that carried the Wxb allele, but similar results were not observed in Wxa-cre mutants (Figure 1F). The relative expression levels of total mRNA and pre-mRNA of the Wx gene exhibited inconsistent changes between the mutant and the WT no matter in the Wxa or Wxb background (Figures 1G, H). The proportion of mature Wx mRNA to total Wx mRNA significantly decreased in the mutants from the Wxb background after normalization, but no change was observed in the Wxa-cre mutant plants (Figure 1I). However, the total Wx mRNA showed a higher level in Wxb-cre mutants than that of WT (Figure 1H). The likely reason is that CRE editing may alters the chromatin conformation, in turn affecting the efficiency of transcription machinery (Rippe and Papantonis, 2025). These results suggest that this CRE regulates the transcription and protein levels of the Wx gene by influencing transcriptional splicing, thereby fine-tuning AC in rice via the Wxb allele.
3.2 Physicochemical property analysis of mutant grains
AC was analyzed to confirm the effect of CRE on different backgrounds (Wxa or Wxb). The apparent AC in the rice flour of the Wxb-cre mutant decreased by 21.43%–31.51% compared with WT with the Wxb allele (Figure 2A), but there was no significant difference in the total starch content between the mutants and WT (Figure 2B). There were no significant differences in the AC and total starch content between Wxa-cre mutants and WT plants, which carried the Wxa allele (Figures 2A, B).
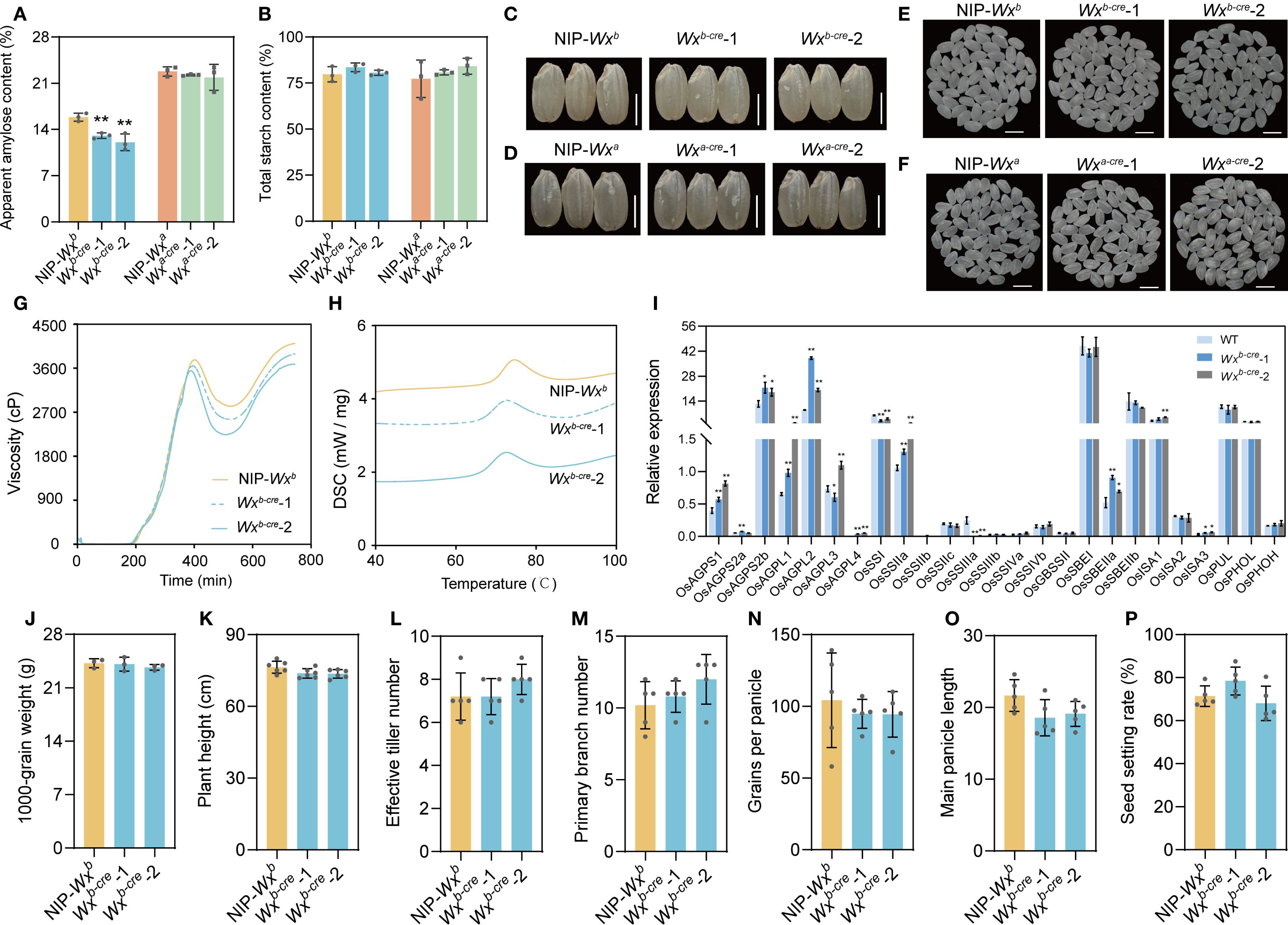
Figure 2. Effects of Wxb and Wxa allele backgrounds on cre mutants. (A, B) Contents of amylose (A) and total starch (B) in rice flour of mutants and WT. (C) Rice grain morphology of WT and Wxb-cre mutants with a Wxb allele. Bar, 3 mm. (D) Rice grain morphology of WT and Wxa-cre mutants with a Wxa allele. Bar, 3 mm. (E) Polished rice appearance of WT and Wxb-cre mutants with a Wxb allele. Bar, 5 mm. (F) Polished rice appearance of WT and Wxa-cre mutants with a Wxa allele. Bar, 5 mm. (G) Rapid viscosity profiles of rice flour. (H) Thermal properties of rice flour. (I) Expression levels of genes involved in starch biosynthesis in Wxb-cre mutants. The reference gene was Actin. Each gene name is indicated by a simplified representation. (J–P) Comparative analysis of main grain agronomic traits between Wxb-cre mutants and WT in rice with a Wxb allele. All data are presented as the mean ± SD. * and ** indicate significant differences from wild-type plants (*P < 0.05 and **P < 0.01) using two-tailed Student’s t-tests.
AC is closely linked to grain appearance quality and physicochemical properties in rice (Tian et al., 2009). Therefore, we evaluated the grain appearance quality and physicochemical properties of mutant lines and their corresponding WTs. The mutants exhibited a normal seed morphology, chalkiness rate, and chalkiness degree compared with WT (Figures 2C–F, Supplementary Figure S2). In the rice mutant with the Wxb background, the rapid viscosity analysis indicated that the decrease in AC caused the slightly lower final viscosity, suggesting an increased gelatinous consistency (Figure 2G). Compared with grains of WT plants, those of mutant plants had a lower gelatinization temperature in Wxb-cre lines (Figure 2H, Supplementary Table S2). These results indicate that the reduction in AC results in a softer texture in mutant rice compared with WT-Wxb.
3.3 Main agronomic traits were not affected in mutants
We assessed whether agronomic traits were affected in the cre mutant plants. The cre mutant lines and WT plants grew normally during the growing season. We investigated the main agronomic traits of the mutant lines and WT after maturation, and there were no statistically significant differences in plant height, main panicle length, or effective tiller number between the cre mutant and WT plants (Figures 2J–P). In addition, the rice grain yield traits, namely, grain number per panicle, seed setting rate grain size, and 1,000-grain weight, were similar in the mutants and the corresponding WT plants (Figures 2J–P). These findings indicate that editing the CRE(s) of Wx can fine-tune AC without affecting other agronomic traits.
3.4 CRE mutant causes the transcriptional reprogramming of starch metabolism
To determine whether changes to the Wx expression level also affected starch metabolism, we compared the expression profiles of 25 genes involved in starch biosynthesis, representing five classes of enzymes (AGPase, SS, SBE, DBE, and Pho), in the seeds at 10 DAF between Wxb-cre mutants and WT.
A significant increase in the expression of AGPase genes was observed in Wxb-cre mutants compared with WT. OsAGPS2b and OsAGPL2 were strongly expressed in the seeds and were significantly upregulated in the Wxb-cre lines compared with WT (Figure 2I). This might be because ADP-glucose accumulation within cells induces AGPase activity (Pérez et al., 2019). OsSSI, OsSSIIIa, and OsSSIIIa were significantly downregulated in developing seeds of Wxb-cre-1 and Wxb-cre-1 lines, but OsSSIIa and OsSSIIb were upregulated in these mutants (Figure 2I). These results suggest that transcriptional reprogramming of genes encoding starch synthases resulted in the regulation of the Wx gene. For starch branched enzyme genes, only the expression of SBEI showed a slight increase in the mutants (Figure 2I). In the Wxb-cre mutants, OsISA1 and OsISA3 were significantly upregulated, but no significant changes were observed in OsPUL, OsPHOL, or OsPHOH (Figure 2I). The changes in genes such as SSs, SBEs, DBEs, and Phos suggested that the amylopectin properties have been fine tuned. The above results indicated that starch biosynthesis metabolism is a complex regulatory network that may be regulated by common transcription factors or transcription factor complexes (Huang et al., 2021).
In summary, we present a novel and effective strategy for decreasing AC in rice inbred lines carrying a Wxb allele. This study shows that CREs have different effects depending on the Wx allele background. The results of this study provide invaluable insights into the regulation of Wx gene expression and a strategy for improving the cooking and tasting quality using varieties with different Wx alleles.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
QY: Funding acquisition, Supervision, Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. FD: Data curation, Formal analysis, Writing – review & editing. WZ: Data curation, Formal analysis, Writing – review & editing. DZ: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32270586) and the Program of Jiangsu Province Government (BE2023331), Jiangsu Science Association Young Science and Technology Talent Support Project (JSTJ-2023-037) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Jiangsu Qinglan Project and High Talent Supporting Program of Yangzhou University to Q.Y.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1694305/full#supplementary-material
Abbreviations
CRE, cis-regulatory element; AC, amylose content; GBSSI, granule-bound starch synthase I; Wx, Waxy; kb, kilobases; ATAC-seq, Assay for Transposase-Accessible Chromatin with high-throughput sequencing; DHSs, DNase I high-sensitivity sites; LUC, luciferase; DAF, days after flowering; WT, wild type; AGPase, ADP-glucose pyrophosphorylase; SS, starch synthases; SBE, starch branching enzymes; DBE, starch-debranching enzymes; PHO, starch phosphorylase; NIP, Nipponbare; DNase-seq, DNAse I hypersensitive sites sequencing; mRNA, messenger RNA.
References
Biłas, R., Szafran, K., Hnatuszko-Konka, K., and Kononowicz, A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tiss Organ Cult. 127, 269–287. doi: 10.1007/s11240-016-1057-7
Cui, Y., Cao, Q., Li, Y., He, M., and Liu, X. (2023). Advances in cis-element- and natural variation-mediated transcriptional regulation and applications in gene editing of major crops. J. Exp. Bot. 74, 5441–5457. doi: 10.1093/jxb/erad248
Ding, Y., Zhu, J., Zhao, D., Liu, Q., Yang, Q., and Zhang, T. (2021). Targeting cis-regulatory elements for rice grain quality improvement. Front. Plant Sci. 12, 705834. doi: 10.3389/fpls.2021.705834
Engler, C., Kandzia, R., and Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PloS One 3, 7. doi: 10.1371/journal.pone.0003647
Hossain, M. A., Cho, J. I., Han, M., Ahn, C. H., Jeon, J. S., An, G., et al. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167, 1512–1520. doi: 10.1016/j.jplph.2010.05.008
Huang, L., Li, Q., Zhang, C., Chu, R., Gu, Z., Tan, H., et al. (2020). Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 18, 2164–2166. doi: 10.1111/pbi.13391
Huang, L., Tan, H., Zhang, C., Li, Q., and Liu, Q. (2021). Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2, 100237. doi: 10.1016/j.xplc.2021.100237
Kong, W., Ding, L., Jia Cheng, J., and Wang, B. (2018). Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Rice 11, 52. doi: 10.1186/s12284-018-0243-0
Lau, W. C., Rafii, M. Y., Ismail, M. R., Puteh, A., Latif, M. A., and Ramli, A. (2015). Review of functional markers for improving cooking, eating, and the nutritional qualities of rice. Front. Plant Sci. 6, 832. doi: 10.3389/fpls.2015.00832
Li, H., Prakash, S., Nicholson, T. M., Fitzgerald, M. A., and Gilbert, R. G. (2016). The importance of amylose and amylopectin fine structure for textural properties of cooked rice grains. Food Chem. 196, 702–711. doi: 10.1016/j.foodchem.2015.09.112
Liu, D. R., Wang, W., and Cai, X. L. (2014). Modulation of amylose content by structure-based modification of OsGBSS1 activity in rice (Oryza sativa L.). Plant Biotechnol. J. 12, 1297–1307. doi: 10.1111/pbi.12228
Liu, Q. Q., Zhang, J. L., Wang, Z. Y., Hong, M. M., and Gu, M. H. (1998). A highly efficient transformation system mediated by agrobacterium tumefaciens in rice (Oryza sativa L.). Acta Photophysiol. Sin. 24, 259–271.
Menkens, A. E., Schindler, U., and Cashmore, A. R. (1995). The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. doi: 10.1016/S0968-0004(00)89118-5
Mohapatra, P. K. and Sahu, B. B. (2022). “Importance of rice as human food,” in Panicle architecture of rice and its relationship with grain filling (Springer, Cham). doi: 10.1007/978-3-030-67897-5_1
Pérez, L., Soto, E., Farré, G., Juanos, J., Villorbina, G., Bassie, L., et al. (2019). CRISPR/Cas9 mutations in the rice Waxy/GBSSI gene induce allele-specific and zygosity-dependent feedback effects on endosperm starch biosynthesis. Plant Cell Rep. 38, 417–433. doi: 10.1007/s00299-019-02388-z
Rippe, K. and Papantonis, A. (2025). RNA polymerase II transcription compartments-from factories to condensates. Nat. Rev. Genet. in press. doi: 10.1038/s41576-025-00859-6
Saeed, S., Usman, B., Shim, S. H., Khan, S. U., Nizamuddin, S., Saeed, S., et al. (2022). CRISPR/Cas-mediated editing of cis-regulatory elements for crop improvement. Plant Sci. 324, 111435. doi: 10.1016/j.plantsci.2022.111435
Sano, Y. (1984). Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68, 4670–4473. doi: 10.1007/BF00254822
Teng, B., Zeng, R., Wang, Y., Liu, Z., Zhang, Z., Zhu, H., et al. (2012). Detection of allelic variation at the Wx locus with singlesegment substitution lines in rice (Oryza sativa L.). Mol. Breed. 30, 583–595. doi: 10.1007/s11032-011-9647-x
Tian, Z., Qian, Q., Liu, Q., Yan, M., Liu, X., Yan, C., et al. (2009). Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. P. Natl. Acad. Sci. U.S.A. 106, 21760–21765. doi: 10.1073/pnas.0912396106
Wang, Z. Y., Zheng, G. Q., Shen, G. Z., Gao, J. P., Snustad, D. P., Li, M. G., et al. (1995). The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 7, 613–622. doi: 10.1046/j.1365-313X.1995.7040613.x
Xing, S., Chen, K., Zhu, H., Zhang, R., Zhang, H., Li, B., et al. (2020). Fine-tuning sugar content in strawberry. Genome Biol. 21, 230. doi: 10.1186/s13059-020-02146-5
Xiong, M., Yu, J., Wang, J., Gao, Q., Huang, L., Chen, C., et al. (2022). Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 189, 402–418. doi: 10.1093/plphys/kiac043
Yang, Q. Q., Yu, W. H., Wu, H. Y., Zhang, C. Q., Sun, S. S. M., and Liu, Q. Q. (2021). Lysine biofortification in rice by modulating feedback inhibition of aspartate kinase and dihydrodipicolinate synthase. Plant Biotechnol. J. 19, 490–501. doi: 10.1111/pbi.13478
Zeng, D. C., Liu, T. L., Ma, X. L., Wang, B., Zheng, Z. Y., Zhang, Y. L., et al. (2020). Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18, 2385–2387. doi: 10.1111/pbi.13427
Zhang, C., Yang, Y., Chen, S., Liu, X., Zhu, J., Zhou, L., et al. (2021). A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 63, 889–901. doi: 10.1111/jipb.13010
Zhang, C., Zhu, J., Chen, S., Fan, X., Li, Q., Lu, Y., et al. (2019). Wx(lv), the ancestral allele of rice Waxy gene. Mol. Plant 12, 1157–1166. doi: 10.1016/j.molp.2019.05.011
Zhang, C., Zhu, L., Shao, K., Gu, M., and Liu, Q. (2013). Toward underlying reasons for rice starches having low viscosity and high amylose: Physiochemical and structural characteristics. J. Sci. Food Agric. 93, 1543–1551. doi: 10.1002/jsfa.5987
Zhang, Q., Zhang, S., Yu, X., Wei, X., Huang, X., and Zhou, X. (2022). Fine-tuning grain amylose contents by genome editing of Waxy cis-regulatory region in rice. Mol. Breed. 42, 72. doi: 10.1007/s11032-022-01342-4
Zhao, P., Liu, Y., Deng, Z., Liu, L., Yu, T., Ge, G., et al. (2024). Creating of novel Wx allelic variations significantly altering Wx expression and rice eating and cooking quality. J. Plant Physiol. 303, 154384. doi: 10.1016/j.jplph.2024.154384
Keywords: rice, Wx, amylose content, cis-regulatory element, grain quality
Citation: Yang Q, Dong F, Zhu W and Zhao D (2025) A distal regulatory element regulates Wx gene expression and the amylose content in rice. Front. Plant Sci. 16:1694305. doi: 10.3389/fpls.2025.1694305
Received: 28 August 2025; Accepted: 23 September 2025;
Published: 08 October 2025.
Edited by:
Yue Feng, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Kunyong Huang, China National Rice Research Institute, ChinaKai Lu, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2025 Yang, Dong, Zhu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingqing Yang, cXF5YW5nQHl6dS5lZHUuY24=; Dongsheng Zhao, ZHN6aGFvQHl6dS5lZHUuY24=
 Qingqing Yang
Qingqing Yang Fuhua Dong1
Fuhua Dong1 Dongsheng Zhao
Dongsheng Zhao