Abstract
Introduction:
Chloride channel (CLC) proteins are crucial anion channels that play a vital role in plant adaptation to abiotic stresses. Potato (Solanum tuberosum L.) is a major global staple crop; however, the CLC gene family in potato (StCLC) remains poorly characterized, and its specific functions in salt tolerance are unclear. This study aimed to systematically identify and characterize the StCLC gene family and analyze its expression patterns under salt stress.
Methods:
Using bioinformatics methods based on the potato genome, transcriptome, and qRT-PCR data, we analyzed the protein structures, physicochemical properties, phylogenetic relationships, gene structures, conserved domains, chromosomal locations, collinearity, GO annotations, and promoter cis-acting elements of StCLC members.
Results:
Seven StCLC genes (StCLC1–7) were identified and unevenly distributed across four chromosomes. Based on protein structures and phylogenetic relationships with Arabidopsis thaliana CLCs, the seven StCLCs were classified into three clusters. Gene structure analysis revealed that StCLC genes contain 6–9 exons, and Motifs 6, 7, 8, and 9 were conserved across all seven StCLC proteins, suggesting their functional importance. Collinearity analysis indicated that the StCLC family has its own collinear genes and shares a close evolutionary relationship with the tomato (Solanum lycopersicum L) CLC family. GO annotation indicated that CLCs are primarily involved in chloride ion transport. Thirty-five cis-acting regulatory elements were identified in the promoter regions, predominantly associated with light response, abiotic stress, hormone regulation, and growth and development, implying potential roles in various physiological processes. RNA-seq data showed distinct expression patterns of StCLC genes across different tissues, indicating tissue-specific expression. Furthermore, qRT-PCR results demonstrated that under NaCl treatment, the expression levels of all seven StCLC genes, including StCLC3 and StCLC6, were significantly upregulated in roots, suggesting their active involvement in the response to salt stress.
Discussion:
These findings elucidate the structural, evolutionary, and functional diversity of the StCLC gene family and highlight its important role in salt stress response, thereby providing a theoretical foundation for the genetic improvement of salt tolerance in potato.
1 Introduction
Anion channels have been extensively studied in various tissues, cell types, and membranes of algae and higher plants, where they play central roles in cell signaling, osmotic regulation, plant nutrition, and metabolism (Barbier-Brygoo et al., 2000). In plant cells, these channels and transporters are essential for nutrient absorption, ion balance, and resistance to biotic or abiotic stress (Nguyen et al., 2016). Among them, chloride channel proteins (CLC) represent an important class of anion transporters distributed across organelles and plasma membranes, where they regulate key metabolic and physiological processes (Zifarelli and Pusch, 2010). The first CLC protein, CLC-0, was discovered in Torpedo marmorata in 1990 (Jentsch et al., 1990), and subsequent studies have revealed that CLC family genes are widely present in plants (Zifarelli and Pusch, 2010). Their physiological functions are broadly divided into two types: channel-type proteins that mediate passive Cl- transport, and exchanger-type proteins that couple Cl- transport with proton gradients (Picollo and Pusch, 2005). CLC members have been identified in Arabidopsis thaliana (Lv et al., 2009), Oryza sativa (Diédhiou and Golldack, 2006), and Nicotiana tabacum (Zhang et al., 2018), and in these and other crops, CLC genes have been shown to participate in tolerance to salinity, drought, and heavy metals.
Potato (Solanum tuberosum) is the fourth most important food crop worldwide, after maize, wheat, and rice, and contributes significantly to global food security through its high nutritional value, particularly as a source of carbohydrates, fiber, vitamins, and potassium (Zhang et al., 2017). It is cultivated in diverse agro-ecological zones, ranging from temperate regions of Europe and North America to the highlands of South America, Africa, and Asia, highlighting its adaptability and global significance. However, potato is highly sensitive susceptible to environmental stresses, and its productivity and quality are substantially reduced under unfavorable conditions (Suttle, 2008). Among these, salt stress is one of the most serious abiotic constraints, especially in irrigated regions affected by soil salinization, which currently accounts for nearly 20% of irrigated land worldwide (FAO, 2024; Liu et al., 2024). Salt stress exerts multiple adverse effects on plants, including osmotic stress, ionic imbalance, and oxidative damage, ultimately reducing growth and yield. Chloride ions accumulate excessively under saline conditions, and their mismanagement can cause toxicity. Thus, CLC proteins are critical regulators of Cl- homeostasis, enabling efflux, sequestration, and redistribution to maintain cellular function under stress.
Evidence from other crops highlights the central role of CLC genes in salinity tolerance. In soybean, GmCLC1 enhances salt resistance by reducing Cl- transport to shoots and sequestering it in roots (Wei et al., 2016). In cotton, members of the GhCLC gene family are differentially expressed under salt stress, with GhCLC5 and GhCLC16 implicated in Cl- and NO3- homeostasis (Liu et al., 2020). In pomegranate, PgCLC antiporters contribute to vacuolar Cl- sequestration in leaves and restriction of Cl- uptake in roots, helping sustain ion balance during salt stress (Wu and Li, 2019; Liu et al., 2020). Recent work in Arabidopsis further demonstrates that AtCLCf can be relocalized from the Golgi to the plasma membrane under salinity, facilitating Cl- efflux from roots as an adaptive mechanism (Liu et al., 2023; Rajappa et al., 2024). These studies collectively illustrate that plant CLC proteins, whether functioning in sequestration, efflux, or balancing Cl- and other anions, play indispensable roles in conferring salt tolerance.
Despite the global significance of the potato and the well-established importance of CLC genes in other plants, systematic studies of the CLC gene family in potato remain limited. Addressing this gap, we carried out a comprehensive bioinformatics analysis of the StCLC gene family. Our study identified CLC members in the potato genome and examined their physicochemical properties, transmembrane topology, and predicted subcellular localization. Phylogenetic analyses were conducted to assess evolutionary relationships with CLCs in other species, while motif composition, gene structures, and conserved domains were analyzed to infer functional diversity. Chromosomal distribution and synteny analysis revealed patterns of duplication and conservation. cis-acting regulatory elements were identified in promoter regions, and Gene Ontology enrichment provided insights into potential biological roles. Finally, transcriptomic and qRT-PCR analyses were used to characterize StCLC expression profiles across tissues and in response to salt stress. Together, these integrated approaches provide new theoretical insights into the CLC gene family in potato and lay a foundation for future efforts to enhance salt tolerance in this globally important crop.
2 Materials and methods
2.1 Identification and physicochemical analysis of CLC members in the potato genome
The potato genome annotation files, nucleotide sequences, and amino acid sequence files were downloaded from the Potato Genome Sequencing Consortium (PGSC) database (Hardigan et al., 2016) (http://spuddb.uga.edu/index.shtml). Protein sequences of members of the Arabidopsis CLC gene family (AtCLC) were downloaded from the Arabidopsis TAIR database (http://www.arabidopsis.org/). Using the protein characteristic domains of the CLC gene family (labeled as PF00654) identified in the Pfam database (http://pfam.sanger.ac.uk/) for the model plant Arabidopsis CLC proteins, Using the specific structural domain (PF00654) of the CLC gene family, the HMMER (https://www.ebi.ac.uk/Tools/hmmer/) online software was used to search the potato genome sequences containing the CLC structural domain feature (E-value ≤ 1e-5). After removing redundant sequences, the obtained sequences were submitted to SMART (http://smart.embl-heidelberg.de) and NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd) for screening, and sequences not containing the CLC structural domain were removed, ultimately identifying the members of the StCLC gene family. The online program ExPasy (Artimo et al., 2012) (https://web.expasy.org/protparam) was used to analyze the amino acid count, molecular weight, theoretical pI, and hydrophilicity of potato CLC family members. Transmembrane structure prediction was performed using the online program TMHMM 2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/). Subcellular localization prediction was performed using the online program Plant-mPLoc (Chou and Shen, 2010) (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
2.2 Protein tertiary structure and systematic tree analysis
To further investigate the functional and structural characteristics of StCLC proteins, the amino acid sequence of StCLC was input into SWISS-MODEL (Waterhouse et al., 2018) (https://swissmodel.expasy.org). Subsequently, the “Build Model” option was selected, and the resulting structure was exported as an image, thereby achieving the visualization of its three-dimensional structure. Protein sequences of AtCLC family members were downloaded from the Arabidopsis TAIR database, while sequences of tomato SlCLC family members were retrieved from the Ensembl Plants database (https://plants.ensembl.org/index.html). Use MEGA v11 software (website: https://www.megasoftware.net/) to perform multiple sequence alignment with the ClustalW algorithm under default parameters and find the most suitable model (Tamura et al., 2021). Construct a phylogenetic tree using the Maximum Likelihood (ML) method with the following parameter settings: 1000 replicates for the bootstrap method, select the Jones - Taylor - Thornton (JTT) model for Model/Method, choose Gamma Distributed (G) for RATES AND PATTERNS, and select Complete deletion for Gaps/Missing Data Treatment. Keep other parameters as default, and export the Newick file (Saitou, 1988). The phylogenetic tree was beautified and visualized using tvBOT (Xie et al., 2023) (https://www.chiplot.online/tvbot.html).
2.3 Protein motifs, conserved domain, and gene structures of StCLCs
The conserved motifs of the StCLC gene family were analyzed using the online protein motif analysis tool MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). The amino acid sequences of the StCLCs genes were input, with the parameters set as follows: the optimal matching length of conserved motifs was 6–100 amino acids (aa), the number of conserved motifs was set to 10, and all other parameters were kept at their default values (Nystrom and McKay, 2021). For subsequent visualization analysis, the mast.xml file was selected from the output results. The gene structure diagram was drawn using the online program GSDS (Hu et al., 2015) (https://gsds.cbi.pku.edu.cn/index.php). The conserved domains of StCLCs were analyzed using the Conserved Domain Database (Marchler-Bauer et al., 2015) (CDD, https://www.ncbi.nlm.nih.gov/cdd). Finally, TBtools v2.357 was used for visualization analysis.
2.4 Chromosome distribution and colinearity analysis
Based on the information obtained about the StCLC gene family members, chromosome localization information was retrieved from the Ensembl Plants database. The online program MG2C-2.1 (Chao et al., 2021) (http://mg2c.iask.in/mg2c_v2.1/) was used to generate chromosome localization maps. Synteny analysis was conducted on the CLC genes in potato (Solanum tuberosum), tomato (Solanum lycopersicum), Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and pea (Pisum sativum) to determine the syntenic relationships among them. Visualization was achieved using the MCScanX algorithm and the Dual Synteny Plot program of TBtools v2.357 with default parameters (Chen et al., 2023).
2.5 Promoter cis-acting element analysis and GO annotation analysis
To reveal the regulatory characteristics of the StCLC promoter, the promoter sequence upstream of the StCLC start codon (2000 kb) was downloaded from NCBI. Trans-acting element analysis was performed using PlantCARE (Lescot, 2002) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) The results of the cis-acting regulatory element analysis were visualized using TBtools v2.357.Perform GO (Gene Ontology) enrichment analysis of StCLC protein sequences using the online program DAVID (Sherman et al., 2022) (https://david.ncifcrf.gov/home.jsp).
2.6 In-silico expression of StCLCs in different tissues
Download potato transcriptome data from the Potato Genome Sequencing Consortium (PGSC) database. Retrieve the gene IDs of StCLC from the database and retrieve their FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) values in different tissues (Mortazavi et al., 2008) (Leaves, Roots, Flowers, Petals, Buds, Sepals, Stolons, and Petioles). To eliminate technical biases between samples, the FPKM values were normalized using the TMM (Trimmed Mean of M-values) method implemented in the edgeR software package. The FPKM values were log10-transformed. Differentially expressed genes (DEGs) were identified using DESeq2 with a false discovery rate (FDR) adjusted p-value < 0.05. Genes with an average FPKM > 0 across all tissues were considered expressed. The expression heatmap was generated using TBtools v2.357 software.
2.7 Plant materials and treatments
Potato ‘Desiree’ seedlings were grown hydroponically under conditions of 25°C/20°C (day/night), relative humidity of 65%-75%, and a photoperiod of 16h/8h (day/night), using Hoagland nutrient solution for irrigation (Gong et al., 2023). The seedlings were irrigated with Hoagland nutrient solution every 2 days. After 3 weeks, they were treated with 0 (control) and 200 mmol/L NaCl solutions. Leaves and roots were collected from the plants at 0, 3, 6, 12, 24, and 48 hours after salt treatment (three biological replicates were performed at each time point to ensure statistical validity), rapidly frozen in liquid nitrogen, and stored at -80°C until further analysis.
2.8 RNA extraction, cDNA synthesis, and qRT-PCR
Total RNA was extracted from potato leaves and roots using Trizol reagent (Sangon Biotech, China) according to the manufacturer’s protocol. After extraction, the first strand of cDNA was synthesized using PrimeScript™RT Master Mix (Perfect Real Time) (Takara, Japan) according to the manufacturer’s protocol. Specific primers were designed using Primer Premier 5 (Singh et al., 1998) and synthesized by Sangon Biotech (Shanghai, China). ef1α (LOC102577640) was used as the housekeeping gene. Amplification was performed on the BIOER QuantGene 9600 Fluorescent Quantitative PCR Instrument, using the TB Green® Premix Ex Taq™ II Reagent Kit (Tli RNaseH Plus) (Takara, Dalian, China). Each treatment group included 3 biological replicates (independent samples taken from different plants), with each biological sample having 3 technical replicates (parallel reactions using the same cDNA template). The qRT-PCR procedure is as follows: (і) predenaturation at 95°C for 2min, (ii) 40 cycles of amplification: denaturation at 95°C for 5s, annealing at 55°C for 30s, extension at 72°C for 1min, and (iii) extension at 72°C for 10min. The expression levels of genes were analyzed using the 2−ΔΔCT method. Significance was analyzed using SPSS, and relative gene expression levels were visualized using GraphPad Prism 9.
3 Results
3.1 Identification of CLC gene family members in potato
Seven CLC genes were identified from the potato genome, and they were named StCLC1 to StCLC7 based on their chromosomal locations. The physicochemical properties of StCLC proteins (Table 1) indicate that the amino acid count encoded by StCLC family members ranges from 752 to 784 AA, with molecular weights between 80.21 and 86.45 kDa. The theoretical isoelectric point (pI) ranges from 6.18 to 8.84, and the total average hydrophilicity coefficient ranges from 0.053 to 0.394, indicating that all seven StCLC proteins are hydrophobic. They have 8 to 12 transmembrane domains, and subcellular localization analysis shows that all seven StCLC proteins are expressed on the cell membrane. Additionally, StCLC5 and StCLC6 are also expressed on the mitochondrial membrane, which suggests that StCLC may be a transmembrane protein.
Table 1
| Gene name | Transcript ID | AA | MW(kDa) | pI | GRAVY | TMHs | Subcellular localization |
|---|---|---|---|---|---|---|---|
| StCLC1 | PGSC0003DMT400064087 | 775 | 84.32 | 8.55 | 0.363 | 8 | Cell membrane |
| StCLC2 | PGSC0003DMT400017962 | 784 | 86.45 | 8.76 | 0.312 | 8 | Cell membrane |
| StCLC3 | PGSC0003DMT400052072 | 774 | 86.10 | 6.8 | 0.341 | 10 | Cell membrane |
| StCLC4 | PGSC0003DMT400078919 | 767 | 84.38 | 8.84 | 0.389 | 12 | Cell membrane |
| StCLC5 | PGSC0003DMT400057279 | 752 | 80.21 | 6.18 | 0.053 | 8 | Cell membrane Mitochondrion |
| StCLC6 | PGSC0003DMT400029204 | 756 | 80.81 | 6.23 | 0.089 | 10 | Cell membrane Mitochondrion |
| StCLC7 | PGSC0003DMT400025239 | 776 | 84.73 | 8.5 | 0.394 | 10 | Cell membrane |
The gene and protein characteristics of the StCLC gene family in Solanum tuberosum.
AA, amino acid number; MW, molecular weight; pI, isoelectric point; GRAVY, grand average of hydro-pathicity; TMHs, number of predicted TMHs.
3.2 Tertiary structure of StCLC proteins and phylogenetic analysis of the gene family
The prediction of the three-dimensional structures of StCLC proteins revealed distinct structural characteristics among StCLC proteins (Figure 1). StCLC1, StCLC2, StCLC3, StCLC4, and StCLC7 exhibited relatively compact and well-organized 3D architectures, with prominent helical domains (displayed in blue) and some disordered or flexible regions (in orange). In contrast, StCLC5 and StCLC6 showed more dispersed structural patterns. These structural variations among StCLC proteins may imply differences in their spatial folding patterns, which could further relate to their functional diversities in biological processes. To clarify the evolutionary relationships within the StCLC gene family, members of the CLC gene families from potato, tomato, and Arabidopsis were selected in this study to construct a phylogenetic tree (Figure 2). Based on the classification criteria of Arabidopsis and tomato CLC genes, the StCLC family was divided into 3 clusters: Cluster CLCII contains 2 StCLC family members, namely StCLC2 and StCLC3; Cluster CLCIII includes 2 StCLC family members, specifically StCLC5 and StCLC6; and Cluster CLCIV comprises 3 StCLC family members, which are StCLC1, StCLC4, and StCLC7.
Figure 1
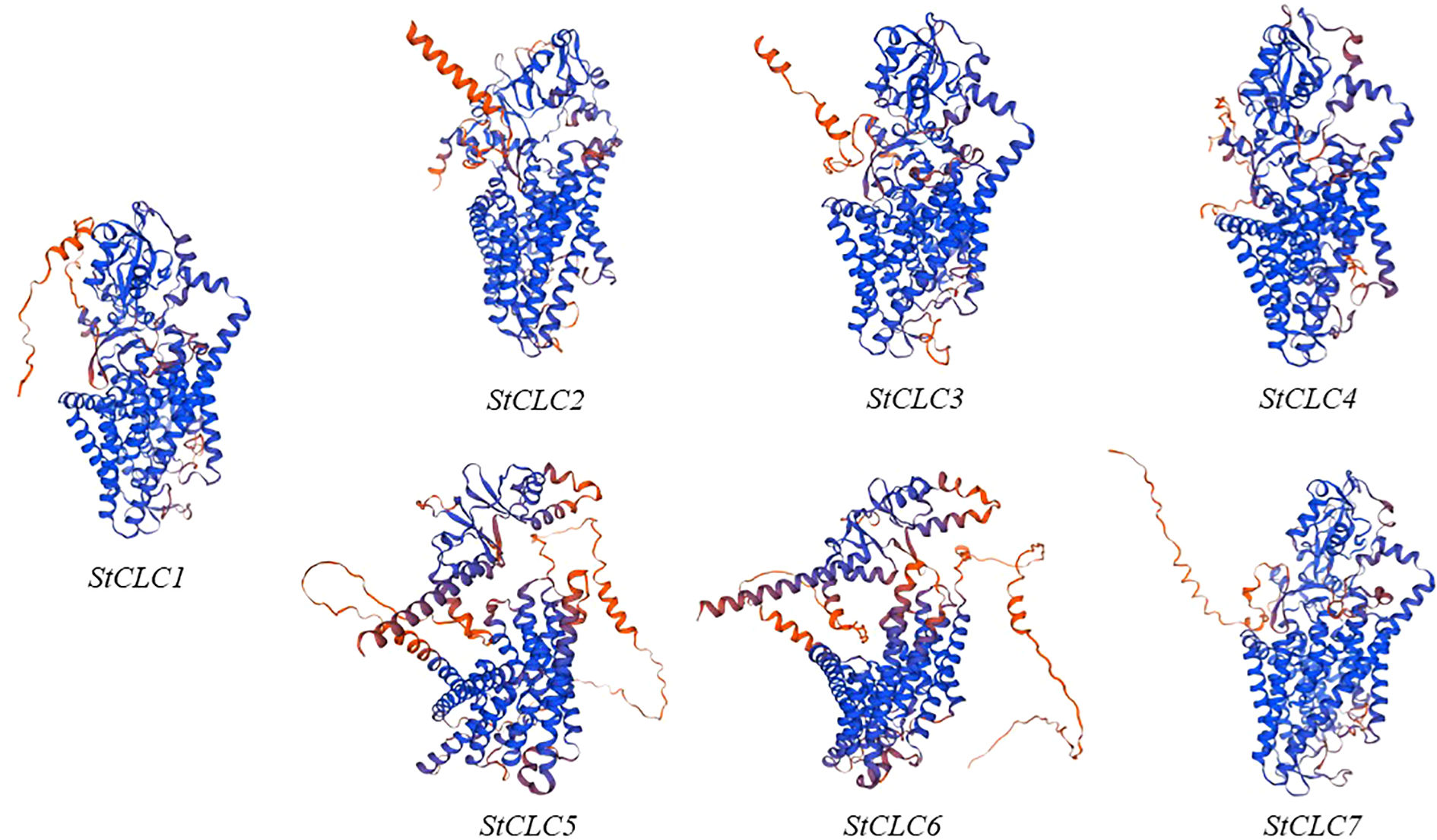
The three-dimensional structures of seven genes in the StCLCs family predicted using Swiss-Model.
Figure 2
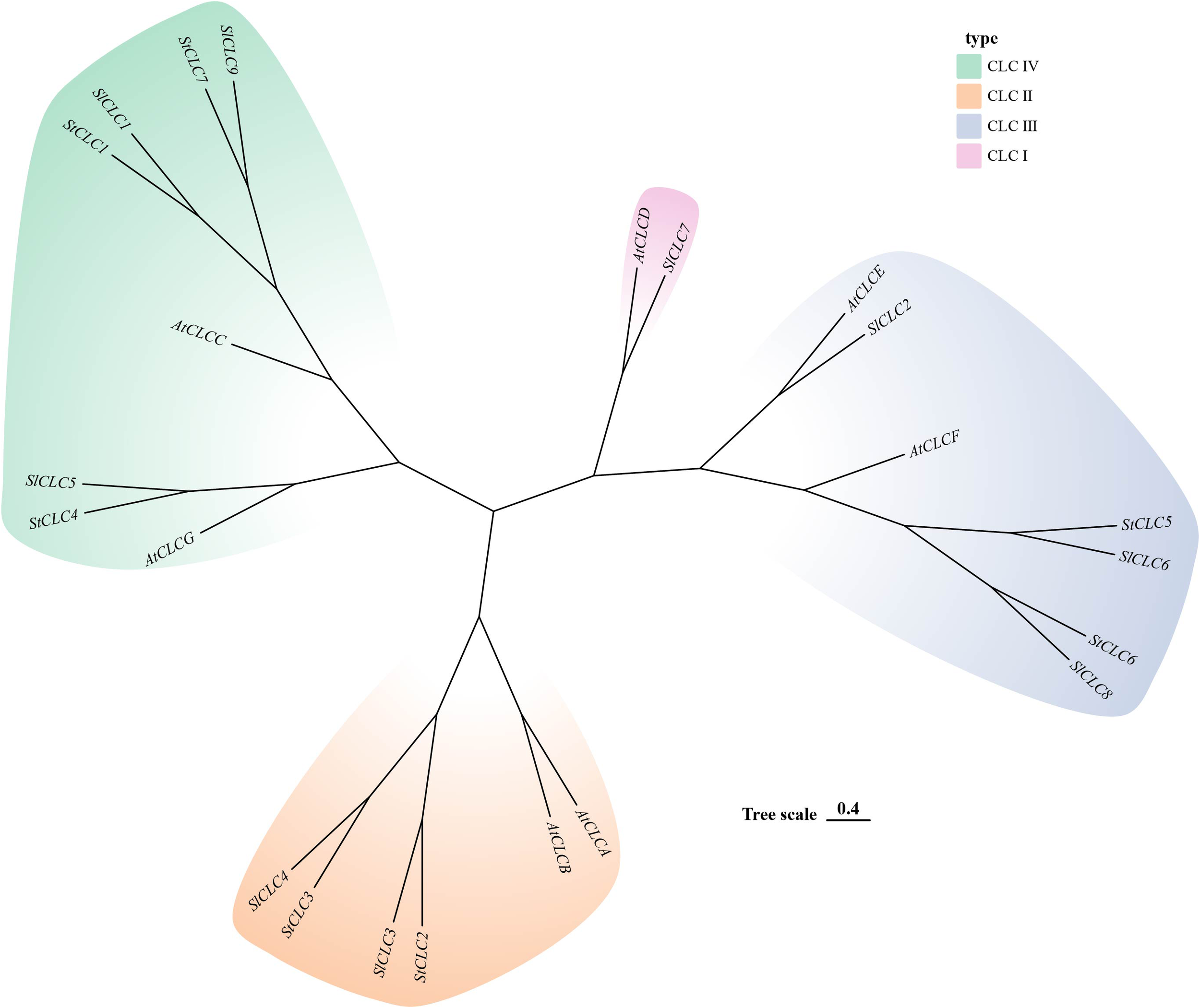
Phylogenetic analysis of potato (Solanum tuberosum, StCLC), Arabidopsis (Arabidopsis thaliana, AtCLC),and tomato (Solanum lycopersicum, SlCLC). Different colored regions represent four subfamilies: CLC IV (light green), CLC II (light orange), CLC III (light blue), and CLC I (pink). The “Tree scale 0.4” indicates the evolutionary distance.
3.3 Conserved motifs, conserved domains and gene structure analysis of the StCLCs
To further analyze the structure and function of StCLC members, we conducted conserved Motifs, conserved domain, and gene structure analyses on seven StCLC members. The results of the conserved Motif analysis (Figure 3a) showed that all seven StCLC members contained Motif6, Motif7, Motif8, and Motif9, which were in the middle part of the CLC domain and constituted most of the CLC domain. StCLC1, StCLC2, StCLC3, StCLC4, and StCLC7 all contain Motif1, 2, 3, 4, 5, 6, 7, 8, 9, and 10, which are respectively located at the N-terminal, C-terminal, or middle regions of the CLC domain. In addition to Motif6, Motif7, Motif8, and Motif9, StCLC5 also contains Motif3, and StCLC6 also contain Motif2. The results of the conserved domain analysis (Figure 3b) indicate that all StCLC proteins contain the CBS_pair_voltage-gated_CLC_euk_bac domain. Additionally, StCLC1, StCLC2, StCLC3, and StCLC7 all contain the CLC_6_like domain, StCLC4 contains the Voltage_gated_CLC superfamily domain, and StCLC5 and StCLC6 both contain the Voltage_gated_CLC domain. Gene structure analysis indicates (Figure 3c) that there is no significant difference in the number of exons and introns among StCLC genes. StCLC1, StCLC4, and StCLC7 contain 7 exons and 6 introns, while StCLC2 and StCLC3 contain 6 exons and 5 introns. StCLC5 and StCLC6 contain 9 exons and 8 introns.
Figure 3
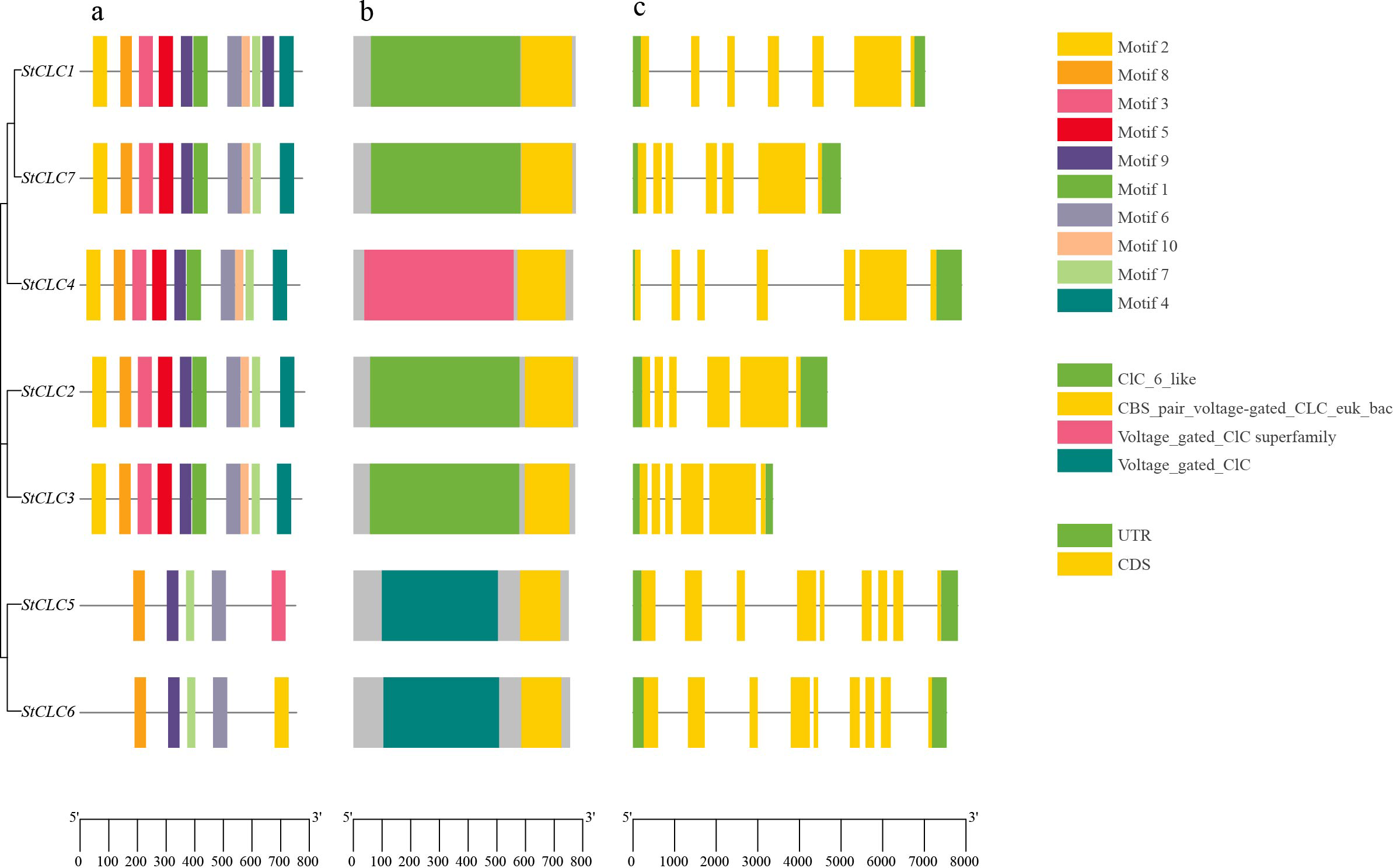
Conserved motifs, conserved domains, and gene structure analysis of the StCLCs gene family. (a) Schematic diagram of the amino acid motif of StCLCs. Different colors of the boxes represent different motifs. (b) Schematic representation of the conserved domains of StCLCs. Different colored boxes represent different conserved domains. (c) Illustration of the CDS/UTR of the corresponding StCLC genes. The yellow boxes indicate the CDS, while the green boxes indicate the UTR.
3.4 Chromosomal distribution and synteny analysis of the StCLCs
Members of the StCLC gene family are distributed across four chromosomes: Chr1, Chr2, Chr7, and Chr10 (Figure 4). There is one gene on Chr1, and two genes each on Chr2, Chr7, and Chr10. No genes are present on chromosomes Chr3, Chr4, Chr5, Chr6, Chr8, Chr9, Chr11, or Chr12.To further infer the evolutionary relationships among CLC genes in plants, we conducted intrachromosomal synteny analysis was performed on the CLC genes in potato (Solanaceae), tomato (Solanaceae), Arabidopsis thaliana (Brassicaceae), rice (Poaceae), and pea (Fabaceae). The results showed that there were 10 pairs of collinear CLC genes between potato and tomato, 6 pairs of collinear CLC genes between potato and Arabidopsis thaliana, 4 pairs of collinear CLC genes between potato and rice, and 2 pairs of collinear CLC genes between potato and pea (Figure 5). These findings indicate that the CLC genes of potato have a closer evolutionary relationship with those of tomato, suggesting that the two may share a common ancestor. Intraspecific synteny analysis of potato CLC genes revealed that the StCLC2 and StCLC3 genes exhibited synteny on Chr2; in contrast, the StCLC5 and StCLC6 genes showed synteny on Chr7 and Chr10, respectively (Figure 6). This result implies that the expansion in the number of members of the StCLC gene family may be attributed to gene duplication events.
Figure 4
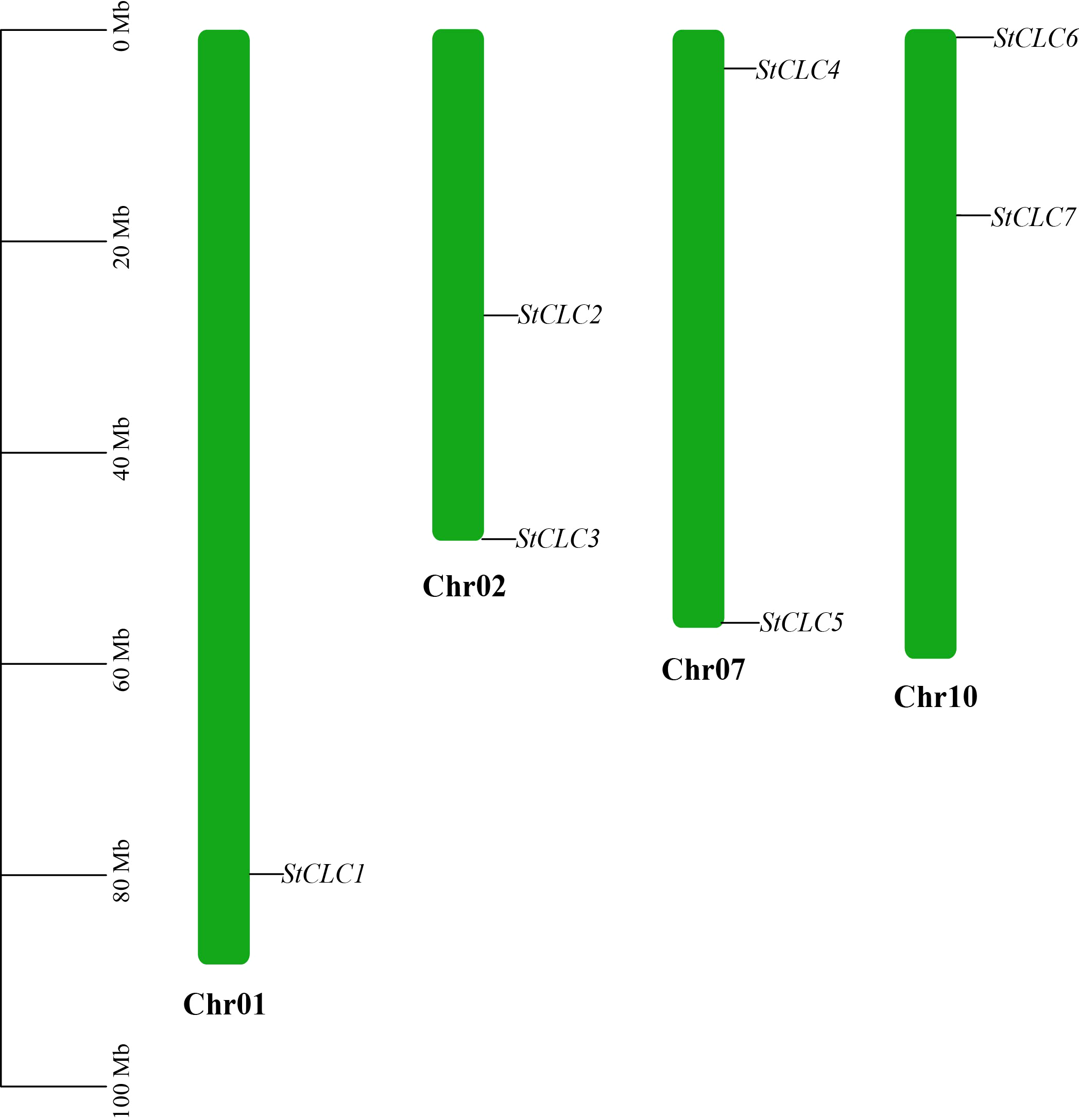
Chromosomal distribution of the StCLC gene family. The bars represented chromosomes, and chromosome numbers were indicated at the right of each bar with length marked at the left of whole figure.
Figure 5
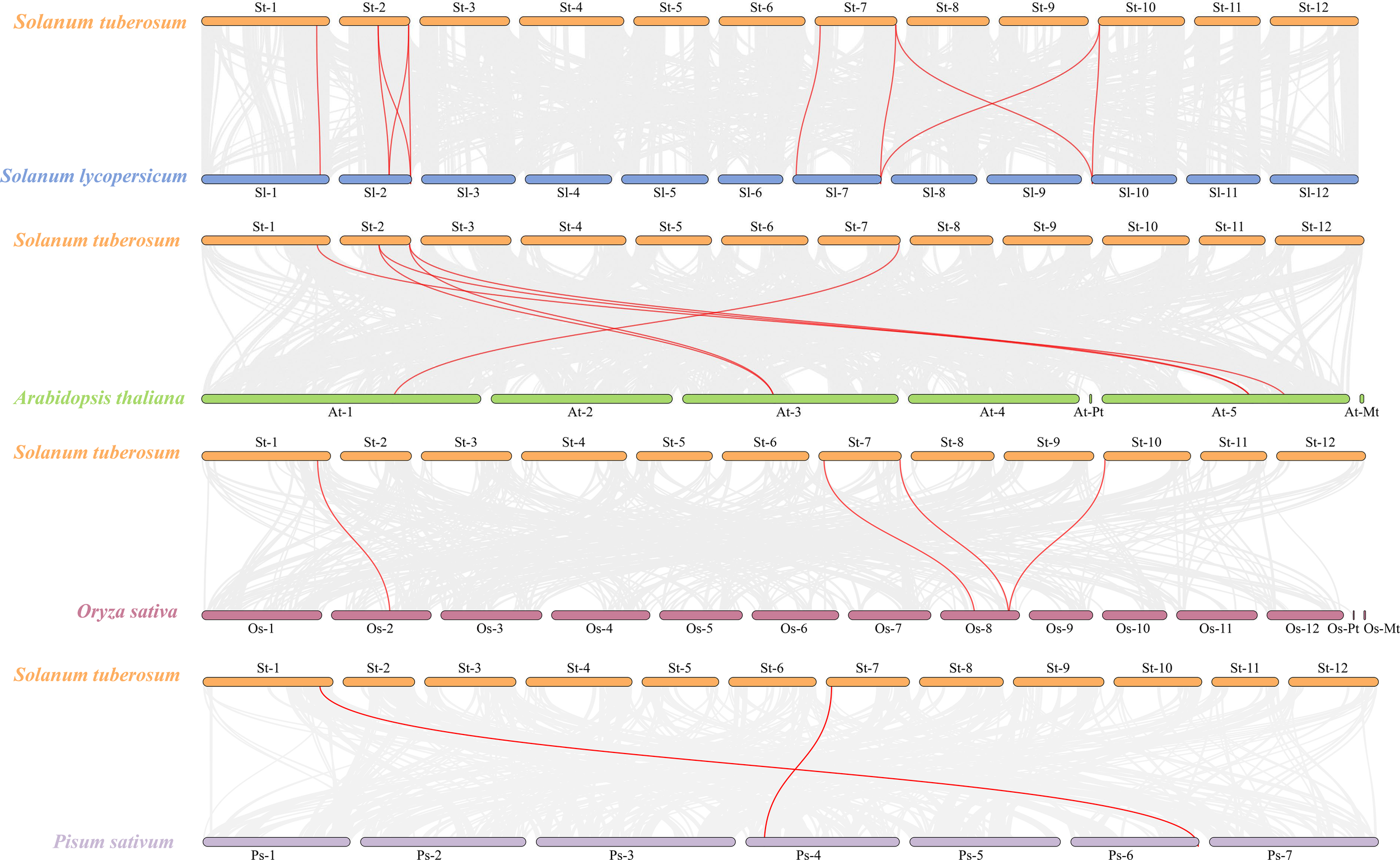
Synteny analysis of CLC genes between potato (Solanum tuberosum, St) and tomato (Solanum lycopersicum, Sl), Arabidopsis (Arabidopsis thaliana, At), rice (Oryza sativa, Os), and pea (Pisum sativum, Ps). The gray lines in the background represent collinear blocks in the genomes of potato, tomato, Arabidopsis, rice, and pea, while the red lines highlight the homologous pairs of CLC genes.
Figure 6
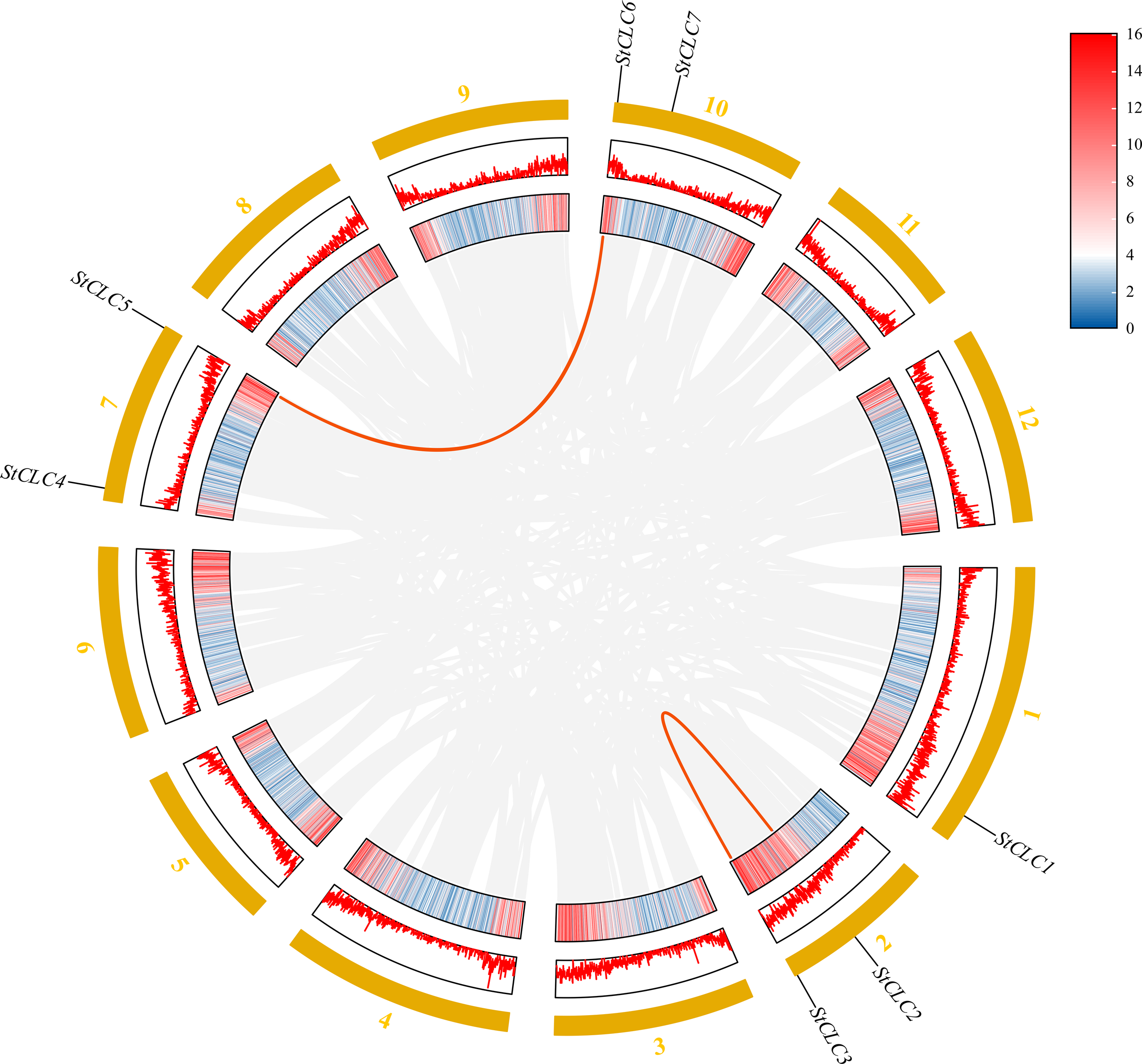
Homology analysis of StCLC genes in potatoes. Gray lines indicate all homologous blocks in the genome of potato, and the red lines indicate duplicated StCLC gene pairs.
3.5 Gene ontology enrichment analysis of StCLCs
To reveal the functional classification of StCLC genes, the GO enrichment pathways of StCLC were analyzed from three aspects: biological process (BP), cellular component (CC), and molecular function (MF) (Figure 7). In biological processes, most StCLC genes play a role in chloride transport and transmembrane transport. In terms of molecular function, it is associated with chloride channel complex and plant-type vacuole membrane. Among the cell components, all members are related to the activity of voltage-gated chloride channels. In addition, some members are related to the activity of chloride transmembrane transporters and the activity of nitrate proton symporters. The results indicate that the StCLC gene plays many important roles in potatoes, the most significant of which is its involvement in chloride ion transport.
Figure 7
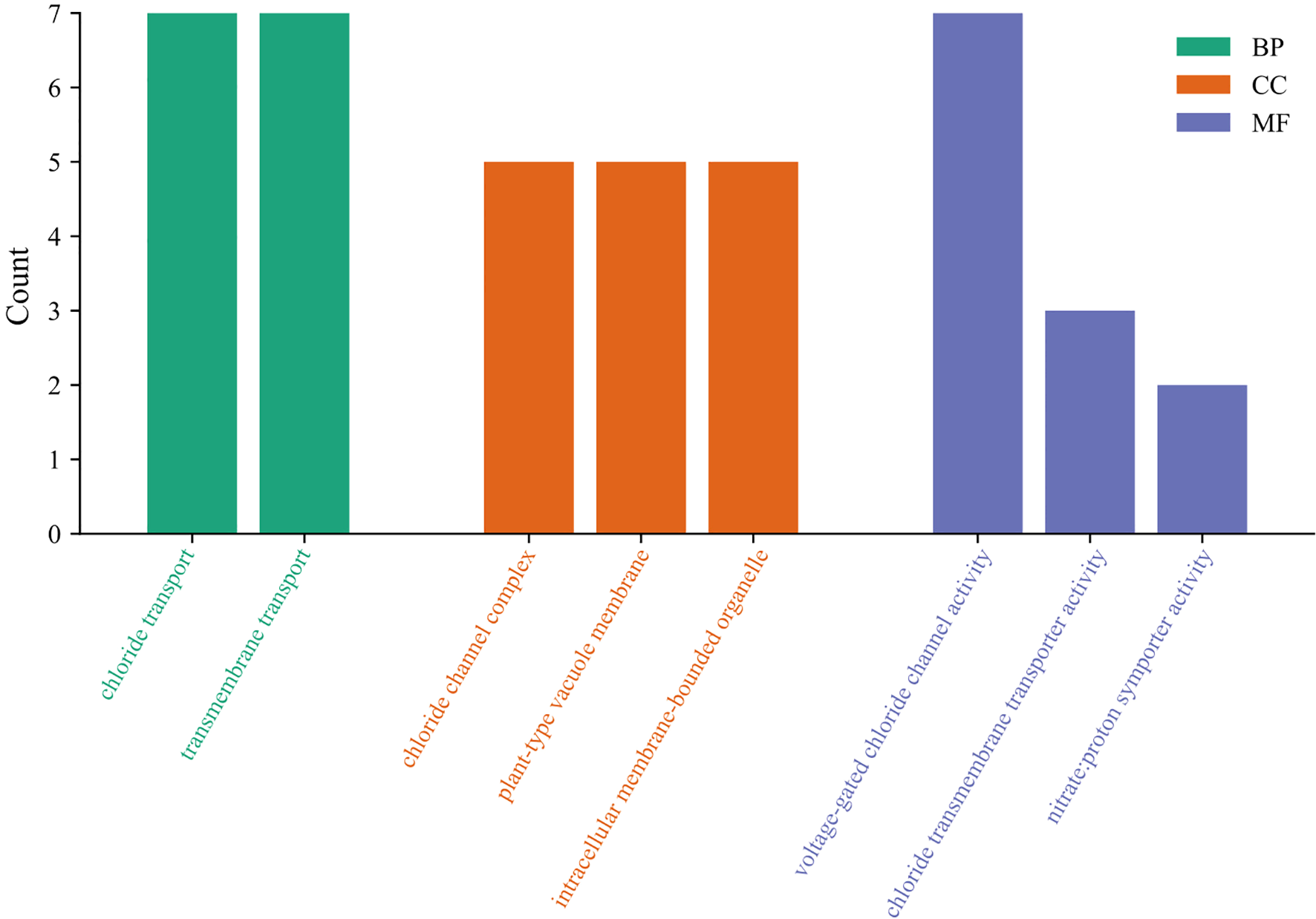
GO analysis of StCLC genes. The vertical axis represents the number of genes involved in the cor-responding classification, and the horizontal axis represents the functional categories of StCLC genes. Green rectangles represent biological processes (BP), orange rectangles represent cellular components (CC), and purple rectangles represent molecular functions (MF).
3.6 Analysis of promoter cis-acting elements in the StCLC gene family
To explore the cis-acting regulatory elements of StCLCs, the sequence upstream of the translation start site of StCLCs within 2000 bp was analyzed using the online tool PlantCARE. In this study, 35 distinct cis-acting regulatory elements were identified in the promoter region of StCLCs (Figure 8). All cis-acting regulatory elements can be broadly categorized into four major classes: light responsive elements, stress responsive elements, hormone responsive elements, and plant growth and development responsive elements. Among these, StCLCs primarily contain light responsive elements Box4, G-box, and abscisic acid response elements (ABRE). Additionally, they contain methyl jasmonate response elements (CGTCA-motif, TGACG-motif), light responsive elements (GA-motif, GATA-motif), corn solubilin protein metabolic regulation response elements (O2-site), antioxidant response elements (ARE), low temperature response elements (LTR), drought induced response elements (MBC), and defense and stress response elements (TC-rich repeats) et al., These results suggest that the expression of StCLCs genes may be regulated by light, environmental, and hormonal signals, among others, and that StCLCs may play an important role in potato growth and development under abiotic stress conditions.
Figure 8
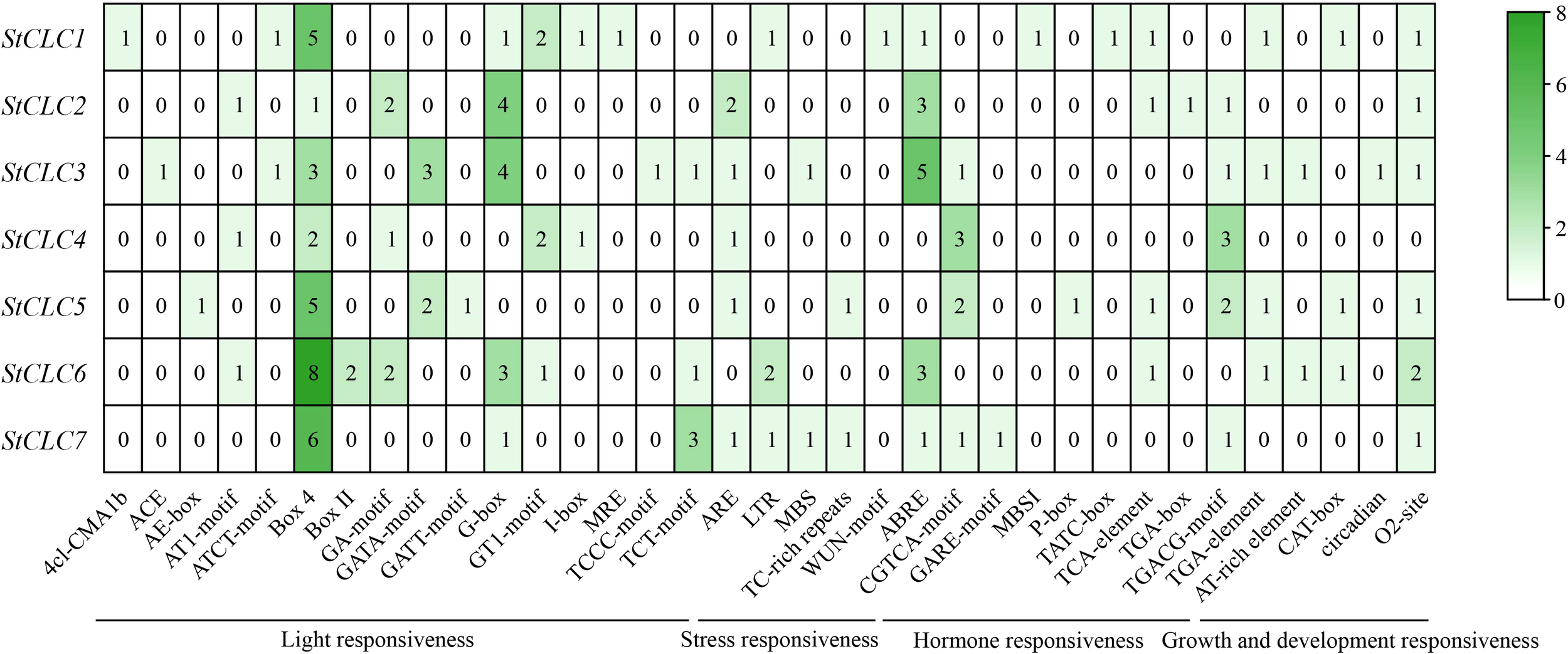
Analysis of cis-acting regulatory elements upstream of the StCLCs promoter within 2000 bp. The rows represent different StCLC genes (from StCLC1 to StCLC7), and the columns represent various cis - acting elements. These elements are categorized into functional groups such as light responsiveness, stress responsiveness, hormone responsiveness, and growth and development responsiveness. The color gradient (from white to dark green) and the numerical values in the cells indicate the frequency or abundance of each cis-acting element in the promoter region of the corresponding StCLC gene.
3.7 Analysis of StCLC gene expression in different tissues
To elucidate the potential function of the StCLC gene in potato growth and development, RNA-Seq data for the StCLC gene in different tissues (leaves, roots, flowers, petals, buds, sepals, stolons, and petioles) were downloaded from the potato transcriptome database. The analysis results showed that the StCLC gene is expressed in most tissues, with higher expression levels in sepals, petals, and roots (Figure 9). StCLC1 is highly expressed in flowers and petals, StCLC2 is highly expressed in petals, StCLC3, StCLC5, and StCLC6 are highly expressed in roots (with StCLC6 showing the highest expression in roots), StCLC4 is highly expressed in sepals, and StCLC7 is highly expressed in buds.
Figure 9
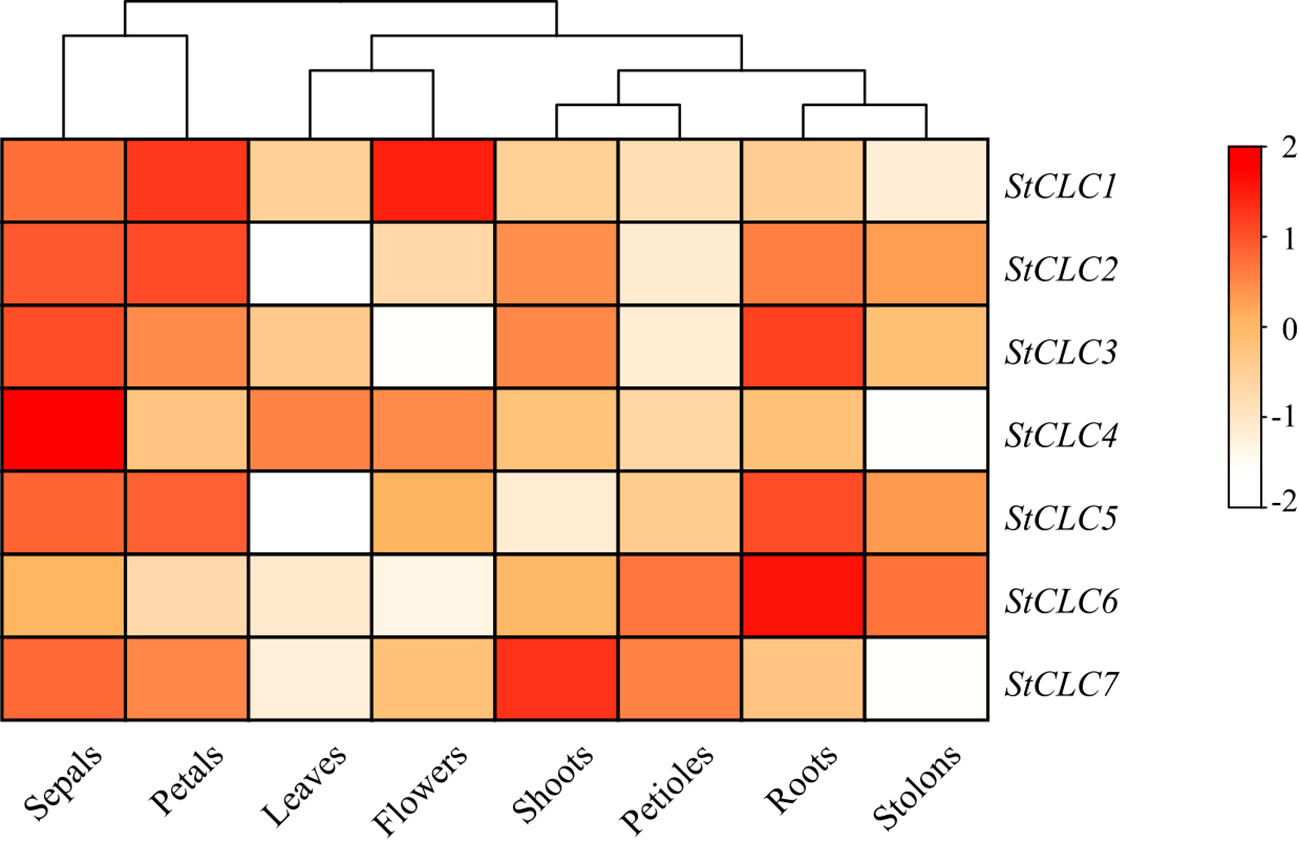
Expression analysis of potato StCLC in eight tissues. The heatmap presents the relative expression levels of genes through a color gradient (from white to dark red). The color scale on the right shows that the redder the color, the higher the gene expression level (the value range is from -2 to 2; the larger the positive value and the darker the red color, the higher the expression level; negative values or lighter colors indicate lower expression levels). The relative expression levels of StCLC genes were normalized using Log10.
3.8 Expression pattern of StCLC genes under salt stress
To analyze the response of StCLCs genes to salt stress treatment, qRT-PCR was used to analyze the gene expression of StCLCs in potato material ‘Desiree’ under NaCl treatment at 0, 3, 6, 12, 24, and 48 hours. The results showed (Figure 10) that in roots, the expression levels of StCLC1, StCLC2, StCLC3, StCLC4, and StCLC5 exhibited the same trend, increasing with increasing stress duration and reaching a peak at 48 h. StCLC6 and StCLC7 showed a significant increase in expression levels from 0 to 3 hours of salt stress treatment, a significant decrease from 3 to 6 hours, stable expression levels from 6 to 24 hours, a significant increase from 24 to 48 hours, and peak expression levels at 48 hours. In leaves, the expression levels of StCLC1, StCLC2, and StCLC5 showed the same trend, increasing initially and then decreasing with increasing salt stress duration. Among these, StCLC1 reached its peak at 12 hours, StCLC2 and StCLC5 reached their peaks at 3 hours, and StCLC3, StCLC6 showed an overall increase in expression levels with increasing salt stress duration, with StCLC3 reaching its peak at 12 hours and StCLC6 at 48 hours. StCLC4 and StCLC7 exhibited the same trend, with expression levels significantly increasing from 0 to 3 hours, decreasing from 3 to 12 hours, and then significantly increasing again from 12 to 48 hours, reaching their peak at 48 hours. These results indicate that StCLC genes are significantly induced by salt stress. Like other crop CLC gene family members, StCLCs may actively respond to salt stress and participate in the process of resisting salt stress.
Figure 10
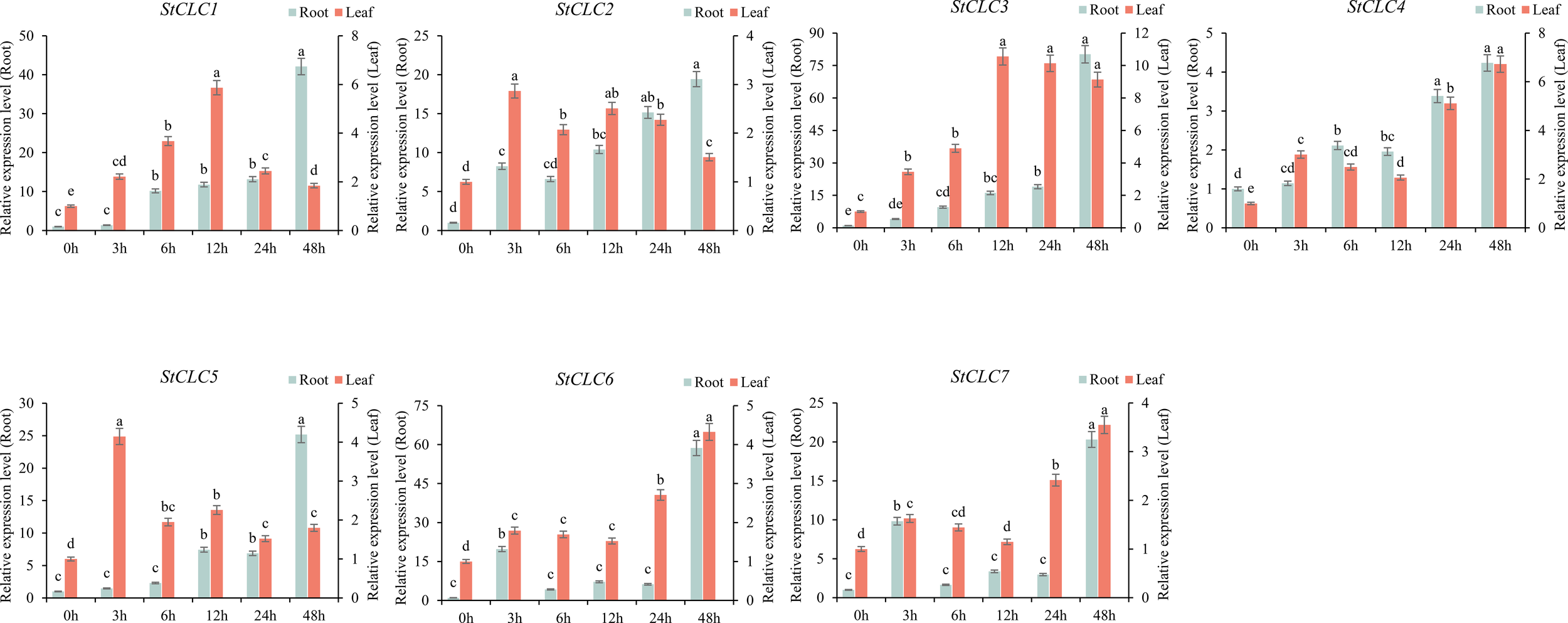
Relative expression levels of the StCLC gene in roots and leaves under 200 mmol/L NaCl treatment. Data are expressed as mean ± standard error of the mean (SEM). Bar charts with different letters within the same group indicate significant differences between different treatment times (p < 0.05).
4 Discussions
The identification of seven StCLC genes in the potato genome aligns with the number of CLC genes reported in Arabidopsis (7 AtCLC genes; Hechenberger et al., 1996) but is fewer than in tomato (9 SlCLC genes; Ma et al., 2025), rice (10 OsCLC genes; Diédhiou and Golldack, 2006), and tobacco (17 NtCLC genes; Zhang et al., 2018). This variation in gene family size likely reflects differences in genome size, ploidy levels, and gene duplication events across species. For instance, potato, a tetraploid species, has undergone extensive segmental and tandem duplications, which may have shaped the StCLC gene family size (Wang et al., 2018). The physicochemical properties of StCLC proteins (e.g., amino acid counts of 752–784, molecular weights of 80.21–86.45 kDa, and hydrophobic nature) are consistent with those of CLC proteins in other plants, indicating conserved structural roles as transmembrane anion transporters (Zifarelli and Pusch, 2010). The presence of 8–12 transmembrane domains and their localization primarily to the cell membrane, with StCLC5 and StCLC6 also localized to the mitochondrial membrane, suggests functional specialization. Mitochondrial localization may indicate roles in regulating anion fluxes in energy-related processes, as observed in Arabidopsis AtCLCd and AtCLCf (De Angeli et al., 2009).
Protein three - dimensional structure prediction shows that all StCLC proteins (StCLC1-StCLC7) in potato exhibit a similar overall folding pattern, which is consistent with the characteristics of the CLC protein family. As shown in the research on CLC protein structure in relevant literature (Dutzler et al., 2002), this conserved folding mode indicates that they retain the core functional domains responsible for anion transport. The presence of multiple α - helical structures (represented by the coiled blue parts in the figure1) is consistent with the typical transmembrane domain architecture of CLC proteins. In plants, like the CLC proteins in Arabidopsis (Yang et al., 2023), the transmembrane structure is the key to mediating the flow of anions across cell membranes, which is crucial for maintaining cellular ion homeostasis. In potato, this transmembrane structure of StCLC proteins is also very likely to play a similar role in the transmembrane transport of anions such as chloride ions.
During the long - term evolution of potato, gene loss events may have occurred, resulting in the absence of Cluster I genes. According to studies on plant gene family evolution, gene loss is usually related to the adaptation of species to specific environments. As a tuber-bearing crop, potato has unique physiological characteristics. Compared with Arabidopsis, it may have developed alternative mechanisms for nitrate storage and utilization. For example, Arabidopsis relies on AtCLCa in Cluster I to regulate nitrate accumulation in vacuoles (De Angeli et al., 2006). In potato, other genes or pathways in the CLC gene family or other gene families may undertake the function of nitrate regulation. This makes Cluster I genes no longer necessary for the survival and development of potato, so they are gradually lost during evolution (Helsen et al., 2020). Although the potato genome has been sequenced, there may still be problems of incomplete genome assembly and annotation. Current genome assembly algorithms may have difficulties in accurately assembling repetitive regions or regions with complex structures. If Cluster I genes in potato are located in these regions, they may not be accurately identified or annotated. Some studies have shown that genes with low expression levels or specific expression patterns are more likely to be missed during the annotation process (Li et al., 2022). Clustering analysis of StCLC genes with CLC genes from tomato (Solanaceae), Arabidopsis thaliana (Brassicaceae), rice (Poaceae), and pea (Fabaceae) showed that the homology between StCLC genes and tomato CLC genes (with 10 collinear gene pairs) was higher than that between StCLC genes and Arabidopsis thaliana CLC genes (6 collinear gene pairs), rice CLC genes (4 collinear gene pairs), and pea CLC genes (2 collinear gene pairs) (Figure 5). There exists a conserved evolutionary relationship of CLC genes within Solanaceae plants. Compared with CLC genes from non-Solanaceae families (Brassicaceae, Poaceae, Fabaceae), StCLC genes (from potato, a Solanaceae plant) show a closer clustering tendency with tomato CLC genes (also from Solanaceae). This clustering pattern is consistent with the taxonomic and phylogenetic hierarchy of plants, reflecting that CLC genes in plants of the same family have retained more ancestral sequence characteristics and evolutionary consistency during long-term evolution (Spooner et al., 2025; Ma et al., 2025) The syntenic relationship between potato StCLC genes and CLC genes from Arabidopsis thaliana (At, Brassicaceae), Oryza sativa (Os, Poaceae), and Pisum sativum (Ps, Fabaceae) is significantly sparser. Among these, the number of collinear gene pairs between StCLC genes and Arabidopsis thaliana CLC genes is greater than that between StCLC genes and rice or pea CLC genes, while the number of collinear gene pairs with rice and pea is even smaller (as indicated by the relatively sparse red connecting lines between St and At, Os, Ps in the figure). This phenomenon demonstrates that the taxonomic family difference among plants increases—especially when crossing from dicotyledonous plants (Arabidopsis thaliana) to monocotyledonous plants (rice) and across different dicotyledonous families (pea)—the synteny of CLC genes decreases significantly. It further verifies that for the clustering distance and synteny degree between StCLC genes and CLC genes from plants of different families, the more distant the phylogenetic relationship of the species, the weaker the gene synteny. Meanwhile, although the synteny between potato CLC genes and CLC genes from non-Solanaceae plants is relatively weak, there are still a certain number of collinear gene pairs (e.g., the red connecting lines between St and At, Os, and Ps in the figure). This also implies that the CLC gene family has an ancient origin, and these collinear genes may be derived from a common ancestral gene of early land plants, which provides a foundation for maintaining the conserved functions (such as chloride ion transport) of CLC genes across different plant groups (Wang et al., 2010). Intraspecific synteny analysis revealed tandem duplications (e.g., StCLC2/StCLC3 on Chr2 and StCLC5/StCLC6 on Chr7/Chr10; Figure 6), which likely contributed to the expansion of the StCLC gene family. Such duplications are common in plant genomes and often lead to functional redundancy or neofunctionalization (Panchy et al., 2016). For instance, StCLC2 and StCLC3, being syntenic, may share similar roles in chloride transport, as tandemly duplicated genes often retain conserved functions (Cannon et al., 2004; Zhou et al., 2024).
The conserved motif and domain analyses (Figures 3a, b) revealed that all StCLC proteins possess the CBS_pair_voltage-gated_CLC_euk_bac domain, a hallmark of CLC proteins involved in anion selectivity and gating (Estévez and Jentsch, 2002). The presence of additional domains, such as CLC_6_like in StCLC1, StCLC2, StCLC3, and StCLC7, and Voltage_gated_CLC in StCLC4, StCLC5, and StCLC6, suggests functional diversification within the family. For example, the Voltage_gated_CLC domain is associated with voltage-dependent anion transport, which may be critical for ion homeostasis under stress conditions (Zifarelli and Pusch, 2010). The gene structure analysis (Figure 3c) showed variability in exon-intron organization, with StCLC5 and StCLC6 having more exons (9) than others (6–7). This structural divergence may reflect adaptations in gene regulation or alternative splicing, as observed in rice OsCLC genes (Diédhiou and Golldack, 2006). The conserved motifs (Motif6, Motif7, Motif8, Motif9) across all StCLCs indicate a core functional region within the CLC domain, likely responsible for anion transport, while additional motifs in specific StCLCs (e.g., Motif3 in StCLC5, Motif2 in StCLC6) suggest potential functional specialization.
The distribution of StCLC genes across four chromosomes (Chr1, Chr2, Chr7, Chr10) with no genes on other chromosomes (Figure 4) suggests a non-random genomic organization, possibly driven by evolutionary constraints or selection pressures. The clustering of StCLC2/StCLC3 on Chr2 and StCLC5/StCLC6 on Chr7/Chr10 supports the hypothesis of tandem and segmental duplications as mechanisms for gene family expansion. Such duplications are critical for generating genetic diversity and enabling plants to adapt to environmental stresses (Hanada et al., 2008). The synteny analysis (Figures 5, 6) further corroborates that gene duplication events, particularly tandem duplications, have shaped the StCLC gene family, like patterns observed in tomato and Arabidopsis (Lv et al., 2009; Ma et al., 2025).
Gene ontology (GO) enrichment analysis (Figure 7) highlighted the primary roles of StCLC genes in chloride transport, transmembrane transport, and voltage-gated chloride channel activity. These functions are consistent with the roles of CLC genes in other plants, such as Arabidopsis AtCLCc, which regulates chloride homeostasis in roots under salt stress (Jossier et al., 2010). The association of StCLCs with nitrate proton symporter activity suggests a dual role in chloride and nitrate transport, as observed in rice OsCLC1, which facilitates nitrate accumulation under salt stress (Diédhiou and Golldack, 2006). This dual functionality could be critical for potato, an economically important crop, as nitrate uptake is essential for growth and tuber development (Marschner, 2012). The involvement of StCLCs in plant-type vacuole membranes further supports their role in vacuolar ion sequestration, a key mechanism for maintaining cellular ion balance under salt stress (Blumwald, 2000; Mansour, 2023).
The identification of 35 cis-acting regulatory elements in StCLC promoter regions (Figure 8) underscores their complex regulation under various environmental and hormonal signals. The prevalence of light-responsive elements (e.g., Box4, G-box) suggests that StCLC expression is modulated by light, which may influence diurnal ion transport cycles, as reported in Arabidopsis (Marmagne et al., 2007).Stress-responsive elements, such as abscisic acid response elements (ABRE), drought-induced elements (MBC), and low-temperature response elements (LTR), indicate that StCLCs are integral to abiotic stress responses. ABRE elements are linked to ABA-mediated stress signaling, which is critical for salt and drought tolerance (Nakashima and Yamaguchi-Shinozaki, 2013). The presence of MYC-binding elements (G-box) and methyl jasmonate response elements (CGTCA-motif, TGACG-motif) suggests regulation by jasmonic acid (JA), which is known to mediate defense and stress responses in plants (Wasternack and Hause, 2013). These findings align with studies in soybean, where GsCLC genes are regulated by ABA and JA under salt stress (Liu et al., 2021; Leung et al., 2023). The diversity of cis elements suggests that StCLCs are part of a complex regulatory network, enabling potatoes to adapt to multiple abiotic stresses.
The tissue-specific expression analysis (Figure 9) revealed high StCLC expression in sepals, petals, and roots, with StCLC3, StCLC5, and StCLC6 showing elevated expression in roots. This pattern suggests specialized roles in root ion homeostasis, which is critical for nutrient uptake and stress tolerance in potatoes. Roots are primary sites for ion transport, and CLC genes in other species, such as soybean GsCLC-e2, enhance salt tolerance by regulating Cl- accumulation (Liu et al., 2021). The high expression of StCLC1 in flowers and StCLC2 in petals may indicate roles in reproductive development, possibly related to ion balance during pollen tube growth or flower senescence, as reported in Arabidopsis (Song et al., 2009). The differential expression across tissues highlights the functional diversity of StCLCs and their involvement in both vegetative and reproductive development.
The qRT-PCR analysis of StCLC expression under salt stress (200 mmol/L NaCl; Figure 10) revealed dynamic and tissue-specific responses. In roots, the upregulation of StCLC1, StCLC2, StCLC3, StCLC4, and StCLC5 with increasing stress duration, peaking at 48 hours, suggests a robust response to salt stress, likely involving chloride sequestration to mitigate ionic toxicity (Munns and Tester, 2008). StCLC6 and StCLC7 exhibited biphasic expression patterns, with peaks at 3 and 48 hours, indicating possible roles in early and late stress responses. In leaves, the expression patterns were more variable, with StCLC1, StCLC2, and StCLC5 peaking early (3–12 hours) and declining, while StCLC3 and StCLC6 showed sustained increases. StCLC1, StCLC2, and StCLC5 show large differences in expression across different tissues under salt treatment at 48 h. This variability may reflect tissue-specific functional requirements, as leaves prioritize photosynthesis and gas exchange, whereas roots focus on ion uptake and exclusion (Shabala and Cuin, 2008). In numerous plant studies, tissue-specific gene expression is a universally observed phenomenon. Under salt stress, the expression levels of certain genes involved in ion transport and osmotic regulation—such as AtHKT1—are significantly higher in roots than in leaves. This is because roots, as organs that directly come into contact with soil salts, need to regulate the expression of these genes to maintain intracellular ion homeostasis and reduce the toxic effects of sodium ions (Wang et al., 2018). Although the expression differences of StCLC1, StCLC2, and StCLC5 in roots and leaves are different from those in other species, they all reflect the response of the CLC gene family to salt stress. The lower expression in leaves compared to roots aligns with findings in longan, where CLC genes show tissue-specific stress responses (Liu et al., 2020). The induction of StCLCs under salt stress is consistent with patterns in tobacco (NtCLC genes; Zhang et al., 2018) and soybean (GmCLC-g, GsCLC-g; Liu et al., 2021), where CLC genes are upregulated by salt stress and regulated by ABA and JA signaling pathways (Valenzuela et al., 2016).
The regulation of StCLC genes by ABA, JA, and potentially ethylene, as inferred from cis-elements and expression patterns, highlights their integration into broader stress signaling networks. ABA is a central regulator of salt stress responses, activating pathways such as the Salt Overly Sensitive (SOS) pathway, which modulates ion homeostasis (Zhu, 2002). In rice, OsCLC genes are regulated by ABA-responsive transcription factors like OsERF (Ponce et al., 2021), and similar mechanisms may regulate StCLCs. The presence of JA-responsive elements suggests that StCLCs are also modulated by JA-mediated defense pathways, which are critical for salt tolerance in maize (Ponce et al., 2021). Ethylene, another stress-related hormone, may indirectly influence StCLC expression by regulating ion homeostasis, as seen in maize (Ponce et al., 2021). These hormonal interactions suggest that StCLCs are part of a multifaceted stress response network, integrating signals from multiple pathways to enhance potato resilience.
The significant induction of StCLC genes under salt stress, particularly in roots, suggests their potential as targets for improving salt tolerance in potatoes. Chloride accumulation in roots can prevent toxic ion buildup in shoots, a strategy employed by salt-tolerant plants (Wu and Li, 2019). The dual role of StCLCs in chloride and nitrate transport, as inferred from GO analysis, could be leveraged to enhance nutrient uptake efficiency under stress conditions, a critical trait for potato yield stability (Rosales et al., 2020). Overexpression of CLC genes, such as AtCLCc in Arabidopsis, has been shown to enhance salt tolerance by improving chloride homeostasis (Wei et al., 2016). Similar strategies could be applied to StCLC3, StCLC5, and StCLC6, given their high expression in roots under salt stress. Additionally, the identification of stress-responsive cis-elements provides targets for genetic engineering to fine-tune StCLC expression under specific environmental conditions.
Plant CLC family proteins, acting as chloride channels or transporters, play a core role in maintaining chloride ion (Cl-) homeostasis under salt stress. Previous studies have confirmed that the expression level of the AtCLCf gene in Arabidopsis thaliana is significantly upregulated by 3-fold in roots after salt treatment; under sodium chloride (NaCl) stress, the AtCLCf mutant of the AtCLCf gene shows a sensitive phenotype, specifically characterized by shortened root meristematic and elongation zones and a significant reduction in root length, while complemented plants obtained by overexpressing the AtCLCf gene can restore the salt tolerance of wild-type Arabidopsis, with the core mechanism being that the AtCLCf protein promotes Cl- efflux from roots through Cl-/H+ antiport activity, reducing Cl- accumulation in the aboveground parts of plants to alleviate ionic toxicity (Rajappa et al., 2024). Evolutionary analysis of the StCLC family in potato reveals that members such as StCLC6 are orthologous to Arabidopsis AtCLCf; in addition, potato and tomato belong to the genus Solanum in the Solanaceae family, sharing high conservation in genome structure, gene family evolution, and stress response mechanisms, which provides a reliable basis for inferring the function of StCLC genes using research findings on SlCLC genes. Many studies have confirmed that tomato SlCLC genes are centrally involved in plant responses to salt stress by regulating Cl- homeostasis (Ma et al., 2025) and combined with the conserved characteristics of gene functions in Solanaceae plants, the key role of StCLC genes in potato salt stress adaptation can be confirmed. The cis-elements in the gene promoter region determine the ability of genes to respond to external signals: the promoters of tomato SlCLC genes generally contain salt stress-responsive cis-elements such as ABRE (abscisic acid-responsive element), MYB (MYB transcription factor binding site), and MYC (MYC transcription factor binding site), whose presence is directly related to their transcriptional activation characteristics under salt stress (Ma et al., 2025); similarly, the promoter regions of the potato StCLC gene family are also enriched with a large number of abiotic stress-responsive elements, including salt stress-related MYB binding sites and hormone-responsive elements, and the activation of these elements is the key regulatory basis for the upregulation of StCLC gene expression under salt stress. Combined with transcriptome data and real-time quantitative PCR (qRT-PCR) results, StCLC genes exhibit differential expression in different potato tissues, and their expression patterns change under salt stress: in roots, some StCLC genes may be rapidly upregulated in the early stage of salt stress, promoting Cl- efflux or transport to aboveground parts to reduce Cl- accumulation in root cells; in leaves, StCLC genes may be involved in transporting Cl- to vacuoles to maintain ion balance in mesophyll cells and normal photosynthesis. Integrating research results on the structural characteristics, salt stress response patterns, and regulatory mechanisms of tomato SlCLC genes and Arabidopsis AtCLC genes, as well as the conservation between potato and tomato in Solanaceae evolution, it can be fully inferred that StCLC genes are centrally involved in potato salt stress adaptation by maintaining Cl- homeostasis.
Future studies should focus on functional validation of StCLC genes using techniques such as CRISPR/Cas9-mediated knockout or overexpression to elucidate their specific roles in ion transport and stress tolerance. Multi-omics approaches, including transcriptomics, proteomics, and metabolomics, could provide a comprehensive understanding of StCLC interactions with other stress-responsive pathways. For example, integrating transcriptomic data with ionomic profiling could clarify the roles of StCLCs in chloride and nitrate transport under salt stress. Additionally, exploring the interaction of StCLCs with transcription factors, such as MYC or ERF family members, could reveal regulatory mechanisms underlying their stress responses. Comparative studies with other Solanaceae species, such as tomato and eggplant, could further elucidate the evolutionary and functional divergence of CLC genes. Finally, field-based studies under varying salinity levels are needed to translate these findings into practical applications for potato breeding, particularly for developing salt-tolerant cultivars suitable for saline soils.
5 Conclusions
This study utilized potato genome sequencing data and bioinformatics methods to identify seven members of the potato StCLC gene family, each with distinct physicochemical properties. Through phylogenetic analysis, the seven StCLC genes were grouped into three clusters, with differences in gene structure between clusters. Colinearity analysis indicated that potato CLC genes are more closely related to tomato CLC genes. RNA-seq analysis showed that StCLC3, StCLC5, and StCLC6 were highly expressed in roots. qRT-PCR results indicated that StCLC genes were significantly induced by salt stress. Like other crop CLC gene family members, StCLCs may actively respond to salt stress and participate in the process of resisting salt stress. Identification and characterization of the StCLC gene family provides a foundation for understanding their roles in potato growth, development, and salt stress responses. Their conserved structures, tissue-specific expression, and dynamic regulation under stress highlight their importance in ion homeostasis and abiotic stress tolerance. These insights pave the way for targeted genetic improvements to enhance potato resilience, supporting sustainable agriculture in challenging environments.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HG: Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing. HW: Formal Analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. JC: Resources, Writing – review & editing. LD: Writing – review & editing. ZF: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by National Natural Science Foundation of China (32560435), and the Key Program of Natural Science Foundation of Gansu Province in China (22JR5RA228).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1703856/full#supplementary-material
References
1
Artimo P. Jonnalagedda M. Arnold K. Baratin D. Csardi G. de Castro E. et al . (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res.40, W597–W603. doi: 10.1093/nar/gks400
2
Barbier-Brygoo H. Vinauger M. Colcombet J. Ephritikhine G. Frachisse J.-M. Maurel C. (2000). Anion channels in higher plants: functional characterization, molecular structure and physiological role. Biochim. Biophys. Acta (BBA)-Biomembranes1465, 199–218. doi: 10.1016/s0005-2736(00)00139-5
3
Blumwald E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol.12, 431–434. doi: 10.1016/S0955-0674(00)00112-5
4
Cannon S. B. Mitra A. Baumgarten A. Young N. D. May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol.4, 10. doi: 10.1186/1471-2229-4-10
5
Chao J. Li Z. Sun Y. Aluko O. O. Wu X. Wang Q. et al . (2021). MG2C: A user-friendly online tool for drawing genetic maps. Mol. horticulture1, 16. doi: 10.1186/s43897-021-00020-x
6
Chen C. Wu Y. Li J. Wang X. Zeng Z. Xu J. et al . (2023). TBtools-II: A “one for all, all for one. Bioinf. platform Biol. big-data Min. Mol. Plant16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
7
Chou K.-C. Shen H.-B. (2010). Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One5, e11335. doi: 10.1371/journal.pone.0011335
8
De Angeli A. Monachello D. Ephritikhine G. Frachisse J. M. Thomine S. Gambale F. et al . (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature442, 939–942. doi: 10.1038/nature05013
9
De Angeli A. Monachello D. Ephritikhine G. Frachisse J.-M. Thomine S. Gambale F. et al . (2009). CLC-mediated anion transport in plant cells. Philos. Trans. R. Soc. B: Biol. Sci.364, 195–201. doi: 10.1098/rstb.2008.0128
10
Diédhiou C. J. Golldack D. (2006). Salt-dependent regulation of chloride channel transcripts in rice. Plant Sci.170, 793–800. doi: 10.1016/j.plantsci.2005.11.014
11
Dutzler R. Campbell E. B. Cadene M. Chait B. T. MacKinnon R. (2002). X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature415, 287–294. doi: 10.1038/415287a
12
Estévez R. Jentsch T. (2002). CLC chloride channels: correlating structure with function. Curr. Opin. Struct. Biol.12, 531–539. doi: 10.1016/S0959-440X(02)00358-5
13
FAO (2024). FAO launches first major global assessment of salt-affected soils in 50 years (Rome, Italy: Food and Agriculture Organization of the United Nations).
14
Gong H.-L. Liu J.-B. Igiraneza C. Dusengemungu L. (2023). Sucrose transporter stSUT2 affects potato plants growth, flowering time, and tuber yield. Curr. Issues Mol. Biol.45, 2629–2643. doi: 10.3390/cimb45030172
15
Hanada K. Zou C. Lehti-Shiu M. D. Shinozaki K. Shiu S. H . (2008). Importance of Lineage-Specific Expansion of Plant Tandem Duplicates in the Adaptive Response to Environmental Stimuli. Plant Physiol.148, 993–1003. doi: 10.1104/pp.108.122457
16
Hardigan M. A. Crisovan E. Hamilton J. P. Kim J. Laimbeer P. Leisner C. P. et al . (2016). Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated solanum tuberosum. Plant Cell28, 388–405. doi: 10.1105/tpc.15.00538
17
Hechenberger M. Schwappach B. Fischer W. N. Frommer W. B. Jentsch T. J. Steinmeyer K. (1996). A family of putative chloride channels from arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. J. Biol. Chem.271, 33632–33638. doi: 10.1074/jbc.271.52.33632
18
Helsen J. Voordeckers K. Vanderwaeren L. Santermans T. Tsontaki M. Verstrepen K. J. et al . (2020). Gene loss predictably drives evolutionary adaptation. Mol. Biol. Evol.37, 2989–3002. doi: 10.1093/molbev/msaa172
19
Hu B. Jin J. Guo A.-Y. Zhang H. Luo J. Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics31, 1296–1297. doi: 10.1093/bioinformatics/btu817
20
Jentsch T. J. Steinmeyer K. Schwarz G. (1990). Primary structure of Torpedo marmorata chloride channel isolated by expression cloning in Xenopus oocytes. Nature348, 510–514. doi: 10.1038/348510a0
21
Jossier M. Kroniewicz L. Dalmas F. Le Thiec D. Ephritikhine G. Thomine S. et al . (2010). The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J.64, 563–576. doi: 10.1111/j.1365-313X.2010.04352.x
22
Lescot M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res.30, 325–327. doi: 10.1093/nar/30.1.325
23
Leung H.-S. Chan L.-Y. Law C.-H. Li M.-W. Lam H.-M. (2023). Twenty years of mining salt tolerance genes in soybean. Mol. Breed.43, 45. doi: 10.1007/s11032-023-01383-3
24
Li X. Zhang P. Wang H. Yu Y. (2022). Genes expressed at low levels raise false discovery rates in RNA samples contaminated with genomic DNA. BMC Genomics23, 554. doi: 10.1186/s12864-022-08785-1
25
Liu C. Jiang X. Yuan Z. (2024). Plant responses and adaptations to salt stress: A review. Horticulturae10, 1–19. doi: 10.3390/horticulturae10111221
26
Liu L. Li X. Wang C. Ni Y. Liu X. (2023). The role of chloride channels in plant responses to naCl. Int. J. Mol. Sci.25, 19. doi: 10.3390/ijms25010019
27
Liu X. Liu F. Zhang L. Cheng C. Wei P. Yu B. (2021). GsCLC-c2 from wild soybean confers chloride/salt tolerance to transgenic Arabidopsis and soybean composite plants by regulating anion homeostasis. Physiologia Plantarum172, 1867–1879. doi: 10.1111/ppl.13396
28
Liu X. Pi B. Pu J. Cheng C. Fang J. Yu B. (2020). Genome-wide analysis of chloride channel-encoding gene family members and identification of CLC genes that respond to Cl–/salt stress in upland cotton. Mol. Biol. Rep.47, 9361–9371. doi: 10.1007/s11033-020-06023-z
29
Liu C. Zhao Y. Zhao X. Dong J. Yuan Z. (2020). Genome-wide identification and expression analysis of the CLC gene family in pomegranate (Punica granatum) reveals its roles in salt resistance. BMC Plant Biol.20, 560. doi: 10.1186/s12870-020-02771-z
30
Lv Q. Tang R. Liu H. Gao X. Li Y. Zheng H. et al . (2009). Cloning and molecular analyses of the Arabidopsis thaliana chloride channel gene family. Plant Sci.176, 650–661. doi: 10.1016/j.plantsci.2009.02.006
31
Ma J. Li S. Zaman S. Anwar A. (2025). CLC gene family in Solanum lycopersicum: genome-wide identification, expression, and evolutionary analysis of tomato in response to salinity and Cd stress. Front. Plant Sci.16. doi: 10.3389/fpls.2025.1547723
32
Mansour M. M. F. (2023). Role of vacuolar membrane transport systems in plant salinity tolerance. J. Plant Growth Regul.42, 1364–1401. doi: 10.1007/s00344-022-10655-9
33
Marchler-Bauer A. Derbyshire M. K. Gonzales N. R. Lu S. Chitsaz F. Geer L. Y. et al . (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res.43, D222–D226. doi: 10.1093/nar/gku1221
34
Marmagne A. Vinauger-Douard M. Monachello D. de Longevialle A. F. Charon C. Allot M. et al . (2007). Two members of the Arabidopsis CLC (chloride channel) family, AtCLCe and AtCLCf, are associated with thylakoid and Golgi membranes, respectively. J. Exp. Bot.58, 3385–3393. doi: 10.1093/jxb/erm187
35
Marschner H . (2012). Marschner’s Mineral Nutrition of Higher Plants. Amsterdam: Elsevier.
36
Mortazavi A. Williams B. A. McCue K. Schaeffer L. Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods5, 621–628. doi: 10.1038/nmeth.1226
37
Munns R. Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol.59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
38
Nakashima K. Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep.32, 959–970. doi: 10.1007/s00299-013-1418-1
39
Nguyen C. T. Agorio A. Jossier M. Depré S. Thomine S. Filleur S. (2016). Characterization of the chloride channel-like, atCLCg, involved in chloride tolerance in arabidopsis thaliana. Plant Cell Physiol.57, 764–775. doi: 10.1093/pcp/pcv169
40
Nystrom S. L. McKay D. J. (2021). Memes: A motif analysis environment in R using tools from the MEME Suite. PloS Comput. Biol.17, e1008991. doi: 10.1371/journal.pcbi.1008991
41
Panchy N. Lehti-Shiu M. Shiu S.-H. (2016). Evolution of gene duplication in plants. Plant Physiol.171, 2294–2316. doi: 10.1104/pp.16.00523
42
Picollo A. Pusch M. (2005). Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature436, 420–423. doi: 10.1038/nature03720
43
Ponce K. S. Guo L. Leng Y. Meng L. Ye G. (2021). Molecular sciences advances in sensing, response and regulation mechanism of salt tolerance in rice. Int. J. Mol. Sci.22, 2254. doi: 10.3390/ijms22052254
44
Rajappa S. Krishnamurthy P. Huang H. Yu D. Friml J. Xu J. et al . (2024). The translocation of a chloride channel from the Golgi to the plasma membrane helps plants adapt to salt stress. Nat. Commun.15, 3978. doi: 10.1038/s41467-024-48234-z
45
Rosales M. A. Franco-Navarro J. D. Peinado-Torrubia P. Díaz-Rueda P. Álvarez R. Colmenero-Flores J. M. (2020). Chloride improves nitrate utilization and NUE in plants. Front. Plant Sci.11. doi: 10.3389/fpls.2020.00442
46
Saitou N. (1988). Property and efficiency of the maximum likelihood method for molecular phylogeny. J. Mol. Evol.27, 261–273. doi: 10.1007/BF02100082
47
Shabala S. Cuin T. A. (2008). Potassium transport and plant salt tolerance. Physiologia Plantarum133, 651–669. doi: 10.1111/j.1399-3054.2007.01008.x
48
Sherman B. T. Hao M. Qiu J. Jiao X. Baseler M. W. Lane H. C. et al . (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists, (2021 update). Nucleic Acids Res.50, W216–W221. doi: 10.1093/nar/gkac194
49
Singh V. K. Mangalam A. K. Dwivedi S. Naik S. (1998). Primer premier: program for design of degenerate primers from a protein sequence. BioTechniques24, 318–319. doi: 10.2144/98242pf02
50
Song L.-F. Zou J.-J. Zhang W.-Z. Wu W.-H. Wang Y. (2009). Ion transporters involved in pollen germination and pollen tube tip-growth. Plant Signaling Behav.4, 1193–1195. doi: 10.4161/psb.4.12.10063
51
Spooner D. M. McLean K. Ramsay G. Waugh R. Bryan G. J. (2005). A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci.102, 14694–14699. doi: 10.1073/pnas.0507400102
52
Suttle J. (2008). Symposium introduction: enhancing the nutritional value of potato tubers. Am. J. Potato Res.85, 266–266. doi: 10.1007/s12230-008-9033-3
53
Tamura K. Stecher G. Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.38, 3022–3027. doi: 10.1093/molbev/msab120
54
Valenzuela C. E. Acevedo-Acevedo O. Miranda G. S. Vergara-Barros P. Holuigue L. Figueroa C. R. et al . (2016). Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot.67, 4209–4220. doi: 10.1093/jxb/erw202
55
Wang L. Liu Y. Feng S. Wang Z. Zhang J. Zhang J. et al . (2018). AtHKT1 gene regulating K+ state in whole plant improves salt tolerance in transgenic tobacco plants. Sci. Rep.8, 16585. doi: 10.1038/s41598-018-34660-9
56
Wang P. Moore B. M. Panchy N. L. Meng F. Lehti-Shiu M. D. Shiu S.-H. (2018). Factors influencing gene family size variation among related species in a plant family, solanaceae. Genome Biol. Evol.10, 2596–2613. doi: 10.1093/gbe/evy193
57
Wang B. Yeun L. H. Xue J. Liu Y. Ané J. Qiu Y. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol.186, 514–525. doi: 10.1111/j.1469-8137.2009.03137.x
58
Wasternack C. Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot.111, 1021–1058. doi: 10.1093/aob/mct067
59
Waterhouse A. Bertoni M. Bienert S. Studer G. Tauriello G. Gumienny R. et al . (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res.46, W296–W303. doi: 10.1093/nar/gky427
60
Wei P. Wang L. Liu A. Yu B. Lam H.-M. (2016). GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci.7. doi: 10.3389/fpls.2016.01082
61
Wu H. Li Z. (2019). The importance of cl– exclusion and vacuolar cl– sequestration: revisiting the role of cl– transport in plant salt tolerance. Front. Plant Sci.10. doi: 10.3389/fpls.2019.01418
62
Xie J. Chen Y. Cai G. Cai R. Hu Z. Wang H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res.51, W587–W592. doi: 10.1093/nar/gkad359
63
Yang Z. Zhang X. Ye S. Zheng J. Huang X. Yu F. et al . (2023). Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids. Nat. Commun.14, 4879. doi: 10.1038/s41467-023-40624-z
64
Zhang H. Xu F. Wu Y. Hu H. Dai X . (2017). Progress of potato staple food research and industry development in China. Journal of Integrative Agriculture16, 2924–2932. doi: 10.1016/S2095-3119(17)61736-2
65
Zhang H. Jin J. Jin L. Li Z. Xu G. Wang R. et al . (2018). Identification and analysis of the chloride channel gene family members in tobacco (Nicotiana tabacum). Gene676, 56–64. doi: 10.1016/j.gene.2018.06.073
66
Zhou Z. Huang J. Wang Y. He S. Yang J. Wang Y. et al . (2024). Genome-wide identification and expression analysis of the DA1 gene family in sweet potato and its two diploid relatives. Int. J. Mol. Sci. 25, 3000. doi: 10.3390/ijms25053000
67
Zhu J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol.53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
68
Zifarelli G. Pusch M. (2010). CLC transport proteins in plants. FEBS Lett.584, 2122–2127. doi: 10.1016/j.febslet.2009.12.042
Summary
Keywords
potato, CLC, salt stress, expression analysis, qRT-PCR
Citation
Gong H, Wang H, Chen J, Dusengemungu L and Feng Z (2025) Genome-wide identification and expression pattern analysis of the chloride channel gene family in potato (Solanum tuberosum L.) under salt stress. Front. Plant Sci. 16:1703856. doi: 10.3389/fpls.2025.1703856
Received
12 September 2025
Accepted
06 October 2025
Published
22 October 2025
Volume
16 - 2025
Edited by
Mizanur Rahman, The University of Texas Rio Grande Valley, United States
Reviewed by
Kun Liu, Inner Mongolia Agricultural University, China
Amaal Maghraby, Cairo University, Egypt
Updates
Copyright
© 2025 Gong, Wang, Chen, Dusengemungu and Feng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiling Gong, gonghl@lut.edu.cn
†These authors share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.