- 1Jiangxi Key Laboratory for Sustainable Utilization of Chinese Materia Medica Resources, Institute of Traditional Chinese Medicine Health Industry, China Academy of Chinese Medical Sciences, Nanchang, China
- 2Jiangxi Institute of Traditional Chinese Medicine Health Industry, Nanchang, China
- 3Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
Artemisia stolonifera, identified as an original source of mugwort leaf during the fourth national medicinal resource investigation in China, remains considerably understudied compared to the well-characterized Artemisia argyi, despite its distinctive mild aroma and potential therapeutic value. The lack of systematic comparative analyses on their active compounds and underlying biosynthetic mechanisms has limited the application and development of A. stolonifera. To address this gap, we conducted integrated metabolomic and transcriptomic analyses of leaves from both species. Using GC-MS for targeted profiling of volatile organic compounds and UPLC-ESI (-Q TRA)-MS/MS for flavonoids and lignin pathway intermediates, we identified 1,728 differentially accumulated metabolites (DAMs). Transcriptome sequencing generated 37.61 Gb of clean data, revealing 18,000 differentially expressed genes (DEGs). Kyoto Encyclopedia of Genes and Genomes analysis demonstrated significant divergence in terpenoid and flavonoid biosynthesis pathways between the two species. A. stolonifera exhibited accumulation sesquiterpenoids, consistent with the concerted upregulation of mevalonate pathway genes (e.g., AACT1-4, HMGR1-3) and sesquiterpene synthases (BAS1, LUP2, CAMS1, XF1). Conversely, A. argyi exhibited enrichment of monoterpenoids and flavonoids, associated with elevated expression of methylerythritol phosphate pathway genes (DXS2, DXR1-5), monoterpenoid biosynthesis genes (SDR2-4, TPS14), and flavonoid biosynthesis genes (e.g., CHS1-2, CHI, F3H1-3). This study elucidates the divergence of genetic and metabolic basis governing bioactive compound biosynthesis between these species, revealing that the characteristically mild aroma of A. stolonifera results from its lower volatile oil content and reduced levels of intense monoterpenoids. These insights provide a critical foundation for evaluating the medicinal value and supporting the safe utilization of A. stolonifera.
1 Introduction
Moxibustion, a cornerstone therapeutic modality in traditional Chinese medicine (TCM) (Tian et al., 2020), utilizes moxa wool derived primarily from the dried leaves of Artemisia argyi (designated “mugwort leaf” in the Chinese Pharmacopoeia) as its combustion. During the fourth national medicinal resource investigation in China, our research team identified Artemisia stolonifera as an original plant source for mugwort leaf due to its superior combustion properties. Multidisciplinary analyses, including textual examination of classical pharmacopeias, geographical provenance assessment, and field investigations, confirmed that A. stolonifera corresponds to the historical Jiu-Niu-Cao documented in classical pharmacopeia records (Ben Cao Tu Jing, 1061 CE) (Luo et al., 2020b). However, clear botanical descriptions distinguishing A. stolonifera from A. argyi remain relatively scarce, impeding accurate species identification and quality control. Importantly, while both A. argyi and A. stolonifera have been historically used for moxibustion and medicine, only A. argyi is currently recorded in the Chinese Pharmacopoeia for the medicinal use of Artemisiae Argyi Folium and has the largest cultivation scale. This is largely due to insufficient comparative data on their chemical composition profiles and associated bioactivities.
Chemical investigations have demonstrated that the bioactive constituents in Artemisia species predominantly consist of terpenoids, flavonoids, polysaccharides, coumarins, sesquiterpene lactones, and organic acids (Abad et al., 2012; Anwar et al., 2016; Han et al., 2017; Zhang et al., 2018; Jiang et al., 2019). These secondary metabolites exhibit a wide range of pharmacological properties, including antioxidant, antitumor, anti-inflammatory, anticoagulant, anti-osteoporotic, and immunomodulatory activities (Kim et al., 2015b, 2015a; Ge et al., 2016; Yun et al., 2016; Lv et al., 2018). Consequently, the medicinal efficacy of these plants correlates directly with their metabolite profiles. However, systematic comparative investigations of the specialized secondary metabolites, particularly volatile terpenoids and non-volatile bioactive components such as flavonoids, between A. stolonifera and A. argyi remain notably limited. Although A. stolonifera has documented therapeutic uses, its application remains restricted compared to A. argyi, primarily due to the absence of comprehensive comparative metabolomic studies. Current research on A. stolonifera has predominantly focused on characterizing its volatile components using techniques such as HS-SPME-GC-MS, revealing significant differences when compared to A. argyi (Li et al., 2022, 2023). Notably, A. stolonifera is distinguished by a mild and fresh aroma, a characteristic attributed to its lower volatile oil content. However, despite the recognized importance of terpenoids and flavonoids in the therapeutic efficacy of A. stolonifera, comprehensive metabolomic profiling targeting these key constituents is notably lacking. This deficiency was the primary reason terpenoids and flavonoids were utilized as comparative indices in this investigation. Previous studies on A. argyi have often prioritized the volatile secondary metabolite profiling of its various tissues; although fewer in number, studies examining the biosynthetic pathways have established a foundational understanding, particularly concerning the main flavonoid biosynthetic pathway, which is known to critically influence the quality of A. argyi. In contrast, the biosynthetic pathways and regulatory networks governing the production of these critical metabolites in A. stolonifera, especially in comparison to A. argyi, remain poorly elucidated. This lack of understanding regarding the metabolic machinery of A. stolonifera hinders a comprehensive assessment of its full potential for medicinal or moxibustion applications.
Headspace solid-phase microextraction coupled with gas chromatography-mass spectrometry (HS-SPME-GC-MS) provides exceptional sensitivity in characterizing volatile terpenoids in A. stolonifera. However, its inability to detect non-volatile metabolites necessitates the use of complementary analytical platforms. To address the limitations of GC-based methods for non-volatile metabolites, this study employs ultra-performance liquid chromatography coupled with mass spectrometry (UPLC-MS). Specifically, UPLC-ESI-MS facilitates comprehensive quantification of polar bioactive compounds, while UPLC-ESI-QTRAP-MS enhances the precision of targeted biomarker quantification through multiple reaction monitoring (MRM). This methodological integration, validated by Luo et al. (2020a), who identified chlorogenic acids and flavonoids as dominant non-volatiles, underscores the necessity of employing UPLC techniques to construct comprehensive flavonoid metabolomes for comparative structural characterization. This integrated metabolomic platform is vital for developing a detailed metabolome, particularly for flavonoids and other essential non-volatile phytochemicals, thereby enabling high-resolution structural characterization crucial for establishing bioactivity-compound relationships and quality control protocols for A. stolonifera. When integrated with transcriptomics, a multi-omics strategy offers systems-level insights into the regulatory mechanisms that govern metabolite accumulation and gene expression. Such multi-omics integration can effectively reveal the chemical basis of pharmacological properties and species-specific metabolic patterns (Chen et al., 2021; Du et al., 2021; Zhang et al., 2022).
In summary, this study employs integrated transcriptomic and metabolomic analyses of functional leaves to systematically investigate the secondary metabolism of A. stolonifera in comparison to A. argyi. We hypothesized that the distinct mild aroma and metabolic profile of A. stolonifera, compared to A. argyi, are primarily governed by species-specific differences in the expression of key genes involved in the terpenoid and flavonoid biosynthesis pathways, leading to significantly different abundances of volatile compounds. To validate this hypothesis, our study comprehensively characterizes and compares the metabolite profiles of volatile terpenoids and non-volatile key metabolites between the two species. We aim to delineate species-specific transcriptional regulation patterns, with particular focus on genes involved the terpenoid backbone biosynthesis pathways and the flavonoid biosynthetic pathway. Furthermore, we will construct integrated gene-metabolite correlation networks to elucidate the molecular mechanisms responsible for the observed metabolic divergence between A. stolonifera and A. argyi. These findings will significantly advance our understanding of the genetic and metabolic basis governing the biosynthesis of bioactive compounds in economically and medicinally significant Artemisia species, providing fundamental insights for evaluating the potential and safe utilization of A. stolonifera.
2 Materials and methods
2.1 Plant materials
Fresh leaves of A. stolonifera (AS) and A. argyi (AA) were collected for this study, identified by Academician Luqi Huang from the China Academy of Chinese Medical Sciences. Both species were initially cultivated in February 2020 at the Standardized Planting Base of Jiu-Niu-Cao in Sunjia Village, Zhangshu City, Jiangxi Province. After 20 days of growth, the plants were transplanted to Shennongling Herb Science and Technology Park (28.94°N, 115.81°E) in Nanchang City, Jiangxi Province, for field acclimatization and domestication. The plants were grown under standardized cultivation conditions for over 45 days prior to sampling in October 2024. The samples were immediately placed in 15 mL centrifuge tubes, flash-frozen in liquid nitrogen, and stored at -80°C until analysis. Three biological replicates per species were processed.
2.2 RNA-sequencing based on transcriptomic detection
Total RNA was extracted from 100 mg of leaf tissue using the RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China), following the manufacturer’s instructions. The integrity of the RNA was verified through 1.5% agarose gel electrophoresis and assessed using a NanoDrop 2000 (OD260/280 ≥ 1.8). Libraries were constructed from 3 μg of total RNA per sample and sequenced on the Illumina NovaSeq 6000.
Six RNA libraries (AS-1, AS-2, AS-3, AA-1, AA-2, and AA-3) were constructed and analyzed. The HISAT2 v2.0.5 software (Mortazavi et al., 2008) was utilized to map reads to the reference genome of A. argyi. The assembly of new transcripts was performed using StringTie v1.3.0 (Pertea et al., 2015). Gene expression was quantified as fragments per kilobase of transcript per million mapped reads (FPKM) (Bray et al., 2016) using featureCounts v1.5.0-p3 (Liao et al., 2014; Love et al., 2014). Differentially expressed genes (DEGs) were identified using DESeq2 v1.20.0 (Anders and Huber, 2010) (|log2FC| ≥ 1 and a false discovery rate (FDR) < 0.05). Functional annotation was conducted using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Pfam databases. Enrichment analysis was performed with clusterProfiler v3.8.1 (p < 0.05). Heatmaps were generated by TBtools-II (Chen et al., 2023).
2.3 Volatile components detection using HS-SPME-GC-MS metabolomic method
Volatile organic compounds (VOCs) were extracted from 500 mg of leaf powder using 20 mL of head-space l in a SPME process with a 120 μm DVB/CAR/PDMS fiber (Supelco). During the SPME analysis, each vial was maintained at 60°C for 5 min, after which the fiber was exposed to the headspace at the same temperature for 15 minutes. GC-MS (Agilent Technologies Inc., CA, USA) analysis was conducted using an Agilent 8890/7000D system equipped with a DB-5MS column (30 m × 0.25 mm × 0.25 μm). Helium (purity > 99.999%) was used as the carrier gas flowing at a constant rate of 1.2 mL·min-1. The injector temperature was set to 250°C. The temperature program was as follows: the column thermostat was held at 40°C for 3.5 min, then increased to 100°C at a rate of 10°C·min-1, further increased to 180°C at 7°C·min-1, escalated to 280°C at 25°C min-1, and held at this final temperature for 5 min. Mass spectra were recorded in electron impact (EI) ionization mode at 70 eV. The temperatures of the MS electron impact ion source and quadrupole were set to 230 and 150°C, respectively. Mass data were collected in ion monitoring (SIM) mode for the identification and quantification of analytes, with a solvent delay time of 5 min.
The raw data were obtained following the automatic and manual integration of the peak area using MassHunter Quantitative Analysis vB.07.00 (Yuan et al., 2022). Metabolites were identified by comparing mass spectra with the NIST20 library (similarity > 80%) and linear retention indices (RI) from the data system libraries.
2.4 Non-volatile components using UPLC-ESI (-Q TRA)-MS/MS metabolomic method
Fresh leaves of A. stolonifera and A. argyi were lyophilized using a vacuum freeze-dryer. The dried leaves were ground into a powder (30 Hz, 1.5 min) using a grinder. For UPLC-ESI-MS/MS analysis, 50 mg of the powder was extracted with 1.2 mL of 70% aqueous methanol. Following centrifugation at 12,000 rpm for 3 min, the supernatant was aspirated with a syringe and filtered through a 0.22 μm organic-phase membrane. For UPLC-ESI-Q TRAP-MS/MS analysis, 100 mg of the powder was extracted with 0.6 mL of 70% aqueous methanol at 4 °C for 12 h. After centrifugation (10, 000 rpm for 10 min), the supernatant was aspirated and filtered.
Chromatographic separation was achieved using Agilent SB-C18 columns (100 × 2.1 mm, 1.8 μm) on two platforms: (1) UPLC system (UPLC, ExionLC™ AD) coupled with a tandem mass spectrometry system (Sciex QTRAP 6500+), and (2) UPLC system (UPLC, Shim-pack UFLC SHIMADZU CBM30A system) with a tandem mass spectrometry system (Applied Biosystems 4500 Q TRAP). Both systems employed identical mobile phases: solvent A (pure water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid), with a flow rate of 0.35 mL·min-1 and a column temperature maintained at 40 °C. The analytical conditions were as follows: gradient program starting at 5% B at 0 min, increasing to 95% B at 9 min, maintaining 95% B for 1 min, then adjusting to 5.0% B within 1.1 min and maintaining it for 2.9 min.
For QQQ-MS/MS analysis (injection volume 2 μL), the ESI source conditions were as follows: ion source temperature at 500°C; ion spray voltage ranging from 5,500 to 4,500 V; ion source gas I, gas II, and curtain gas set at 50, 60, and 25.0 psi, respectively, with medium collision gas (N2) setting. For QTRAP-MS/MS analysis (4 μL injection), the ESI source conditions were as follows: ion source temperature at 550°C; ion spray voltage ranging from 5500 to 4500 V; ion source gas I, gas II, and curtain gas set at 50, 60, and 30.0 psi, respectively, with a collision gas setting of 5 psi. Both platforms acquired data in MRM mode. The qualitative and quantitative analyses of metabolites integrated both public databases and a self-built database of metabolite information.
The GC-/UPLC-/LC-MS data were imported into Microsoft Excel 2019 for data organization, processing, and statistical analysis. Multivariate analyses including principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA), were performed using R packages. Differentially accumulated metabolites (DAMs) were identified based on the thresholds of |log2(FC)| ≥ 1, FDR < 0.05, and variable importance in projection (VIP) ≥ 1.
2.5 Combined correlation analysis of DAMs and DEGs
Hierarchical clustering and KEGG pathway enrichment analyses were conducted using clusterProfiler (v4.0.5). All DEGs and DAMs that were associated with KEGG pathways were utilized. The Pearson correlation coefficients between DAMs and DEGs were calculated using the Psych package in R software. Significant pairs (r > 0.9, p < 0.05) were subsequently converted to mutual ranks using scripts available on GitHub (jwisecav/coexp-pipe).
2.6 Quantitative real-time polymerase chain reaction
Total RNA was extracted from 100 mg of frozen leaf tissue using the RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China) according to the manufacturer’s specifications, with integrity and concentration verified using a NanoDrop 2000 (Thermo Fisher Scientific). First-strand cDNA was synthesized from 1 μg of RNA with the PrimeScript™ RT Kit with gDNA Eraser (Perfect Real Time) (Takara Bio Inc., Shiga, Japan) under optimized conditions: 42°C for 2 min, 37°C for 15 min, and 85°C for 5 s. Amplification was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) and included initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s, concluding with melt curve analysis from 65°C to 95°C at a rate of 0.5°C·s-1, using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara Bio Inc., Shiga, Japan). The qPCR reactions (25 μL) contained 12.5 μL of TB Green Master Mix, 20 μM gene-specific primers (Supplementary Table S1, synthesized by Sangon Biotech, Shanghai, China), and 2 μL of diluted cDNA template. The constitutively expressed AaActin served as the internal control for Ct value normalization. Relative quantification was determined using the 2-ΔΔCT method with efficiency correction, employing three biological and technical replicates.
3 Results
3.1 Botanical description of two similar species between A. stolonifera and A. argyi
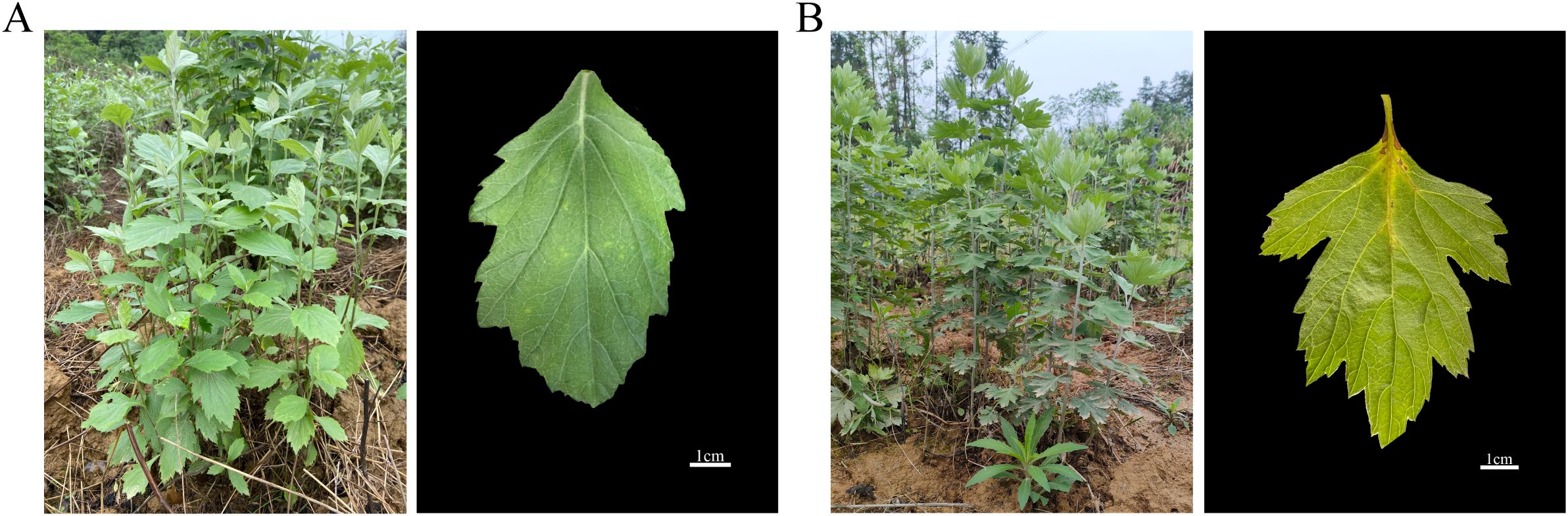
A. stolonifera and A. argyi are perennial herbs belonging to the genus Artemisia, exhibiting distinct morphological characteristics. A. stolonifera typically reaches a height of 50–120 cm, characterized by sparse gray arachnoid pubescence or a glabrescent appearance. The middle stem leaves are nearly sessile and can be described as obovate-elliptic, ovate-elliptic, or ovate, often exhibiting 2- or 3-cleft or -partite structures with coarse teeth, and are gland-dotted on the adaxial surface (Figure 1A). In contrast, A. argyi is taller, ranging from 80–250 cm, and features numerous lateral roots with short apical branching, also covered in gray arachnoid pubescence. Its middle stem leaves are ovate, triangular-ovate, or subrhombic, with the adaxial surfaces are incanous pubescent and white gland-dotted. These leaves are generally 1- or 2-pinnatipartite or -cleft (Figure 1B). Furthermore, we observed that A. stolonifera emits a mild and fresh aroma, which is distinctly different from the intense odor of A. argyi. To systematically investigate these phytochemical differences, we analyzed both the volatile and non-volatile components of each species. The volatile profiling aimed to elucidate the specific compositional basis for the characteristically mild aroma of A. stolonifera, while flavonoid quantification assessed its potential for non-volatile bioactive constituents. This combined approach provides a comprehensive chemical foundation for understanding the traditional and potential applications of both species, highlighting how the unique sensory characteristics of A. stolonifera arise from its distinctive volatile metabolome.
3.2 Differential expression of RNA sequencing between A. stolonifera and A. argyi
To elucidate the molecular mechanisms underlying the differences in secondary metabolism between A. stolonifera and A. argyi, we conducted comparative transcriptome analysis. A total of 38.25 Gb of raw bases were obtained (Supplementary Table S2). After quality filtering, 37.61 Gb of clean data were acquired, with individual sample data ranging from 5.87 to 6.75 Gb. High sequencing quality was confirmed, with Q30 scores exceeding 96.3% and an average GC content of 43.36% (Supplementary Table S2). De novo assembly of the clean reads yielded 76,717 expressed transcripts (unigenes), including 13,874 novel genes, with over 78.83% of clean reads mapping to the reference genome. GO analysis assigned 38,655 unigenes (50.39%) to functional categories, with the most abundant subcategories being “metabolic process” (Biological Process), “cellular anatomical entity” (Cellular Component), and “binding” (Molecular Function) (Supplementary Figure S1). KEGG pathway annotation assigned 36,468 unigenes (47.54%) to five categories, including 22 subcategories; those with the highest gene representation included “global and overview maps”, “carbohydrate metabolism”, and “translation” (Supplementary Figure S2). PCA confirmed clear separation between the transcriptomes of AS and AA (Supplementary Figure S3), which confirms the high quality of sampling, sequencing, and gene expression quantification in this study.
The comparative analysis between the AS and AA groups identified 9,554 upregulated and 8,446 downregulated DEGs in AS relative to AA (Figures 2A, B). This substantial set of DEGs likely underlies the observed differences in secondary metabolism between the two species. Among these DEGs, 4,656 were annotated with GO terms, with the most enriched subcategories in BP, CC, and MF being “arginyl-tRNA aminoacylation (GO:0006420)”, “eukaryotic translation initiation factor 3 complex (GO:0005852)”, and “translation initiation factor binding (GO:0031369)”, respectively (Figure 2C). To specifically investigate the molecular basis of secondary metabolic differences, we conducted KEGG enrichment analysis on the DEGs. This analysis revealed significant enrichment (p < 0.05) of DEGs in pathways directly related to secondary metabolism. Notably, the “phenylpropanoid biosynthesis” pathway was significantly enriched in both upregulated (80) and downregulated (66) gene sets (Figures 2D, E). Among the 18 significant pathways enriched for upregulated DEGs in AS, “biosynthesis of various plant secondary metabolites” (62), “linoleic acid metabolism” (23), “flavone/flavonol biosynthesis” (9), and “sesquiterpenoid/triterpenoid biosynthesis” (13) were prominent (Figure 2D). For downregulated DEGs in AS (12 significant pathways), the most enriched pathway was “glycolysis/gluconeogenesis” (71), while “monoterpenoid biosynthesis” (18) and “terpenoid backbone biosynthesis” (29) were also enriched (Figure 2E). Analysis of the top 20 most significantly enriched KEGG pathways further underscored distinct regulatory mechanisms within phenylpropanoid metabolism (“phenylpropanoid biosynthesis”) and terpenoid metabolism (“monoterpenoid biosynthesis” and “sesquiterpenoid/triterpenoid biosynthesis”) (Figure S4). Collectively, these transcriptomic analyses identified extensive DEGs and revealed significant alterations in key secondary metabolic pathways, particularly in phenylpropanoid and terpenoid biosynthesis, providing crucial insights into the molecular mechanisms responsible for the divergent secondary metabolite profiles between A. stolonifera and A. argyi.
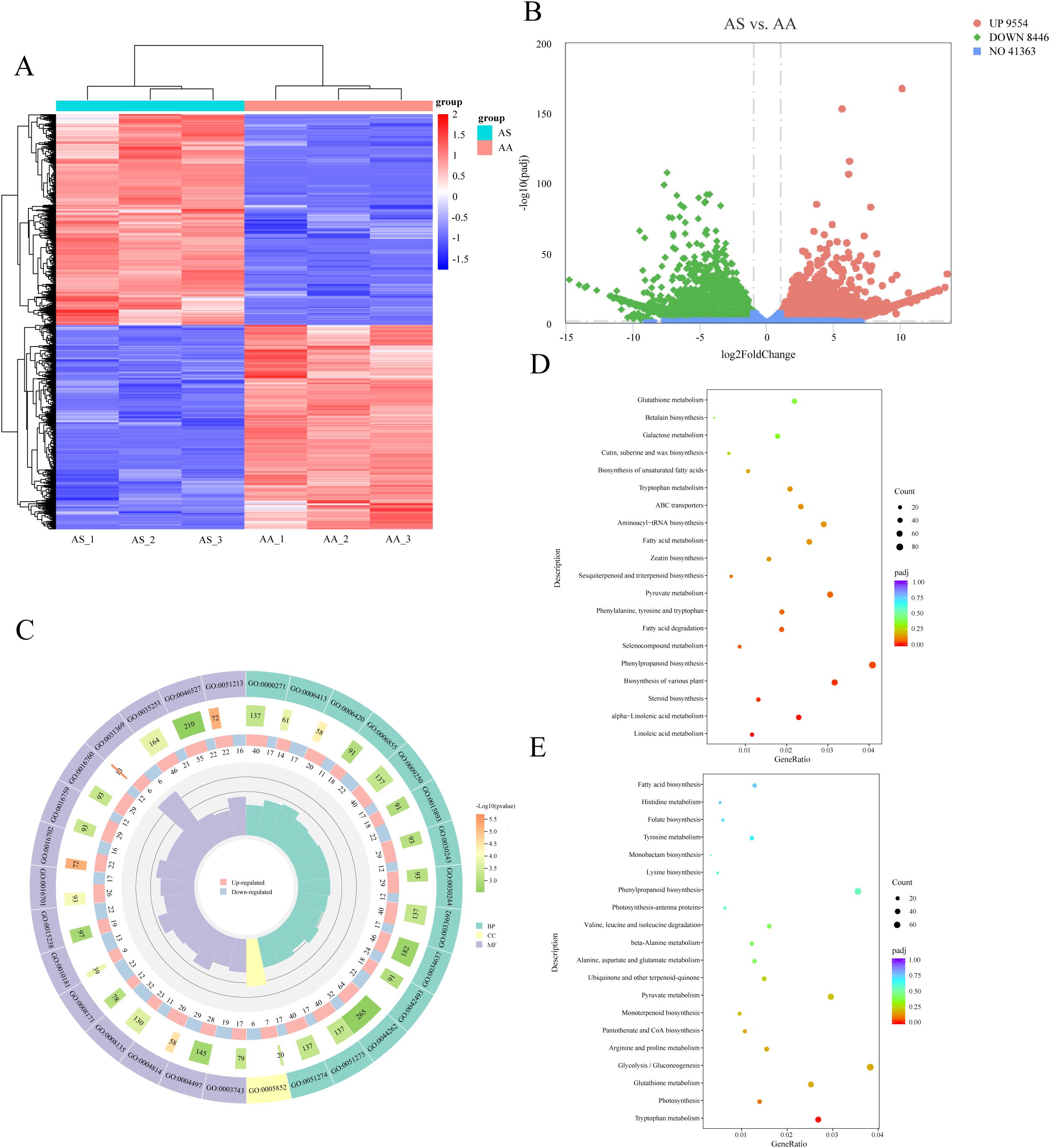
Figure 2. Functional classification of DEGs. (A) Hierarchical clustering heatmap of DEGs. (B) Volcano plot of DEGs. (C) Comparison of the distribution of DEGs at the GO level. KEGG enrichment analysis of upregulated (D) and downregulated (E) DEGs in AS.
3.3 Volatile terpenoids reveals metabolite changes between A. stolonifera and A. argyi
To comprehensively profile the metabolomic differences between AS and AA, we conducted a metabolic profiling analysis of VOCs using HS-SPME-GC-MS. An initial assessment of the total ion chromatograms (TICs) revealed strong analytical signals, high peak capacity, and good retention time reproducibility across all samples, indicating robust instrument performance (Supplementary Figure S5). A total of 1,577 metabolites were detected across all samples and categorized into 15 distinct groups. Terpenoids represented the most abundant class, accounting for 22.32%, followed by esters at 18.14%. Multivariate analysis utilizing PCA and OPLS-DA effectively segregated the samples into two distinct clusters corresponding to AS and AA, underscoring the significant metabolic divergence between A. stolonifera and A. argyi (Figures 3A-C).
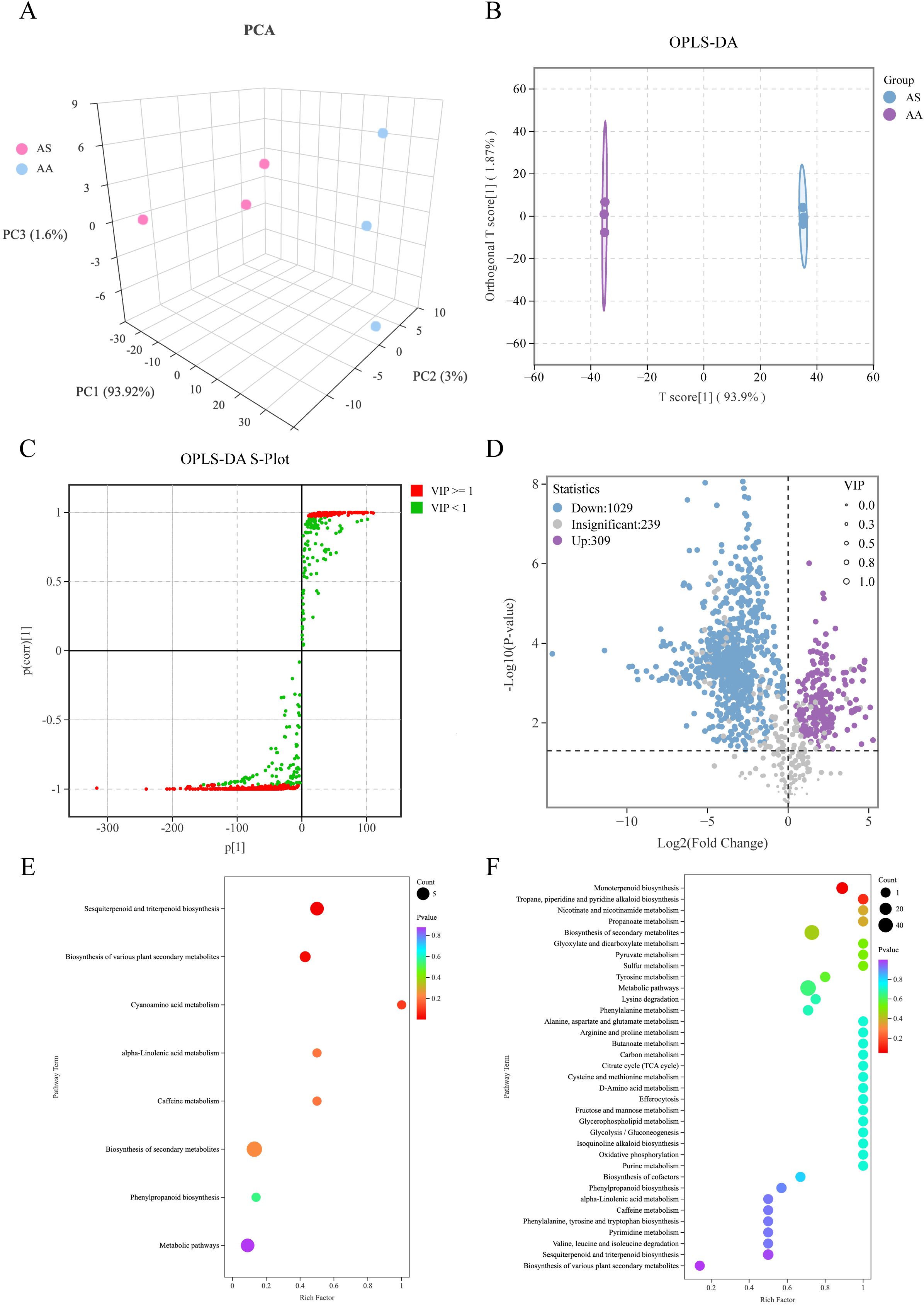
Figure 3. Multivariate statistical analysis of the DAMs based on HS-SPME-GC-MS. (A) Three-dimensional score plots of PCA, (B) OPLS-DA, (C) S-plot of OPLS-DA, (D) Volcano plot of DAMs, (E) Enrichment analysis of KEGG using upregulated DAMs, (F) Enrichment analysis of KEGG using downregulated DAMs in AS.
Employing both single- and multidimensional analytical approaches, we identified a total of 1,338 DAMs, of which 1,029 were significantly downregulated and 309 were significantly upregulated in AS compared to AA (Figure 3D). The top 30 DAMs exhibiting the highest absolute value of Log2FC were predominantly terpenoids (Supplementary Table S3). KEGG pathway enrichment analysis of the DAMs (Figures 3E, F) revealed significant alterations in specific metabolic pathways. Terpenoids constituted the dominant class among both the upregulated and downregulated DAMs, suggesting their key roles in differentiating the species. The upregulated DAMs in AS were extremely significantly enriched (p < 0.01) in “sesquiterpenoid/triterpenoid biosynthesis” (6) and significantly enriched (p < 0.05) in “biosynthesis of various plant secondary metabolites” (8) (Figure 3E). Conversely, the downregulated DAMs in AS were significantly enriched (p < 0.05) in “monoterpenoid biosynthesis” (17) (Figure 3F). Notably, this pattern of terpenoid metabolite enrichment aligns remarkably well with the transcriptomic data, which showed significant enrichment of DEGs in the corresponding terpenoid backbone and branch pathways (Figures 2D, E). Specifically, the upregulation of sesquiterpenoid/triterpenoid DAMs corresponds to the enrichment of upregulated DEGs in pathways such as “sesquiterpenoid/triterpenoid biosynthesis”, while the downregulation of monoterpenoid DAMs correlates with the observed downregulation of DEGs in “monoterpenoid biosynthesis” pathways. These findings strongly support the hypothesis that the differential accumulation of terpenoids in their biosynthetic pathways is a major contributor to the metabolic distinction between A. stolonifera and A. argyi.
3.4 Non-volatile flavonoids and lignin-related metabolites reveal metabolite changes between A. stolonifera and A. argyi
To investigate secondary metabolic divergence indicated by transcriptomic data, we performed targeted UPLC-ESI-MS analyses focusing on flavonoids and lignin biosynthesis intermediates. The assessment of base peak chromatograms confirmed the robustness of the instrument’s performance (Supplementary Figure S6). A total of 560 flavonoid metabolites were identified (417 neg and 143 pos), with flavones (179) and flavonols (130) constituting the predominant subclasses. PCA and OPLS-DA clearly separated AS and AA (Figures 4A–C), confirming substantial inter-species metabolic differentiation, as observed in the VOCs metabolomics and transcriptomics. We identified 390 DAMs in the flavonoid dataset, of which 273 were downregulated and 117 upregulated in AS (Figure 4D). Among the top 30 DAMs ranked by absolute value of Log2FC, flavones and flavonols showed the most pronounced alterations, with the majority being downregulated in AS (Supplementary Table S4). KEGG enrichment highlighted “flavone/flavonol biosynthesis” (21) and “flavonoid biosynthesis” (18) as the most significantly enriched pathways (Figure 4E), corroborating transcriptomic findings (Figure 2). This analysis illustrates that the predominantly downregulated accumulation of flavones and flavonols in AS significantly contributes to the metabolic differentiation between A. stolonifera and A. argyi.
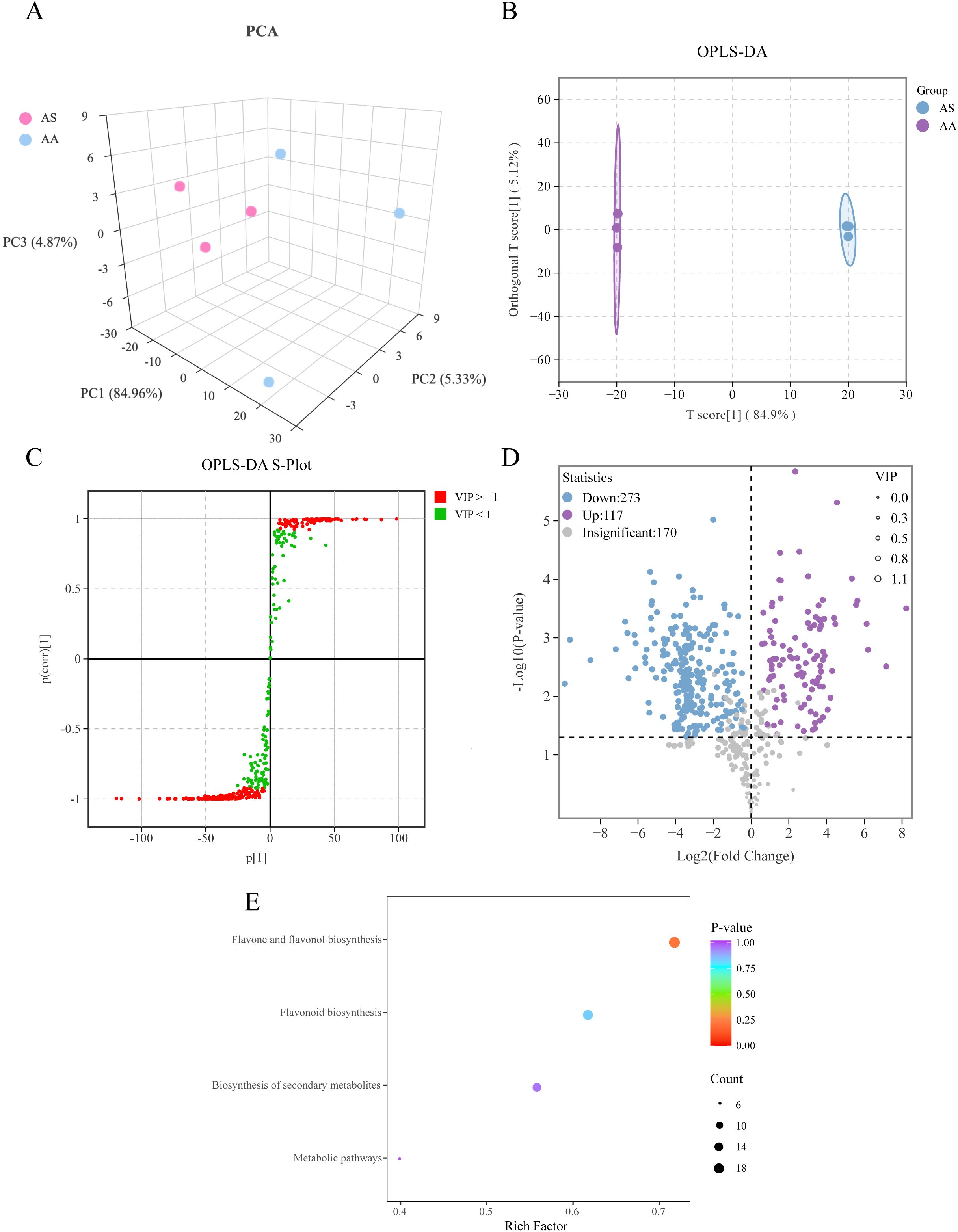
Figure 4. Multivariate statistical analysis of the DSMs based on UPLC-ESI-MS. (A) Three-dimensional score plots of PCA, (B) OPLS-DA, (C) S-plot of OPLS-DA, (D) Volcano plot of DAMs. (E) Enrichment analysis of KEGG of DAMs.
Complementary analysis of the lignin biosynthesis pathway identified 14 key intermediate metabolites, among which 5 DAMs were detected. Sinapic acid and sinapinaldehyde, precursors of S-lignin, were significantly upregulated in AS compared to AA, whereas p-coumaric acid, coniferyl alcohol, and p-coumaryl alcohol, precursors of G- and H-lignin, were significantly downregulated (Figure 5). These results suggest species-specific lignin subunit composition and illustrate coordinated regulation of phenylpropanoid-derived metabolism. Collectively, our UPLC-ESI-MS data delineate distinct flavonoid and lignin biosynthesis profiles between the two species, providing metabolite-level evidence.
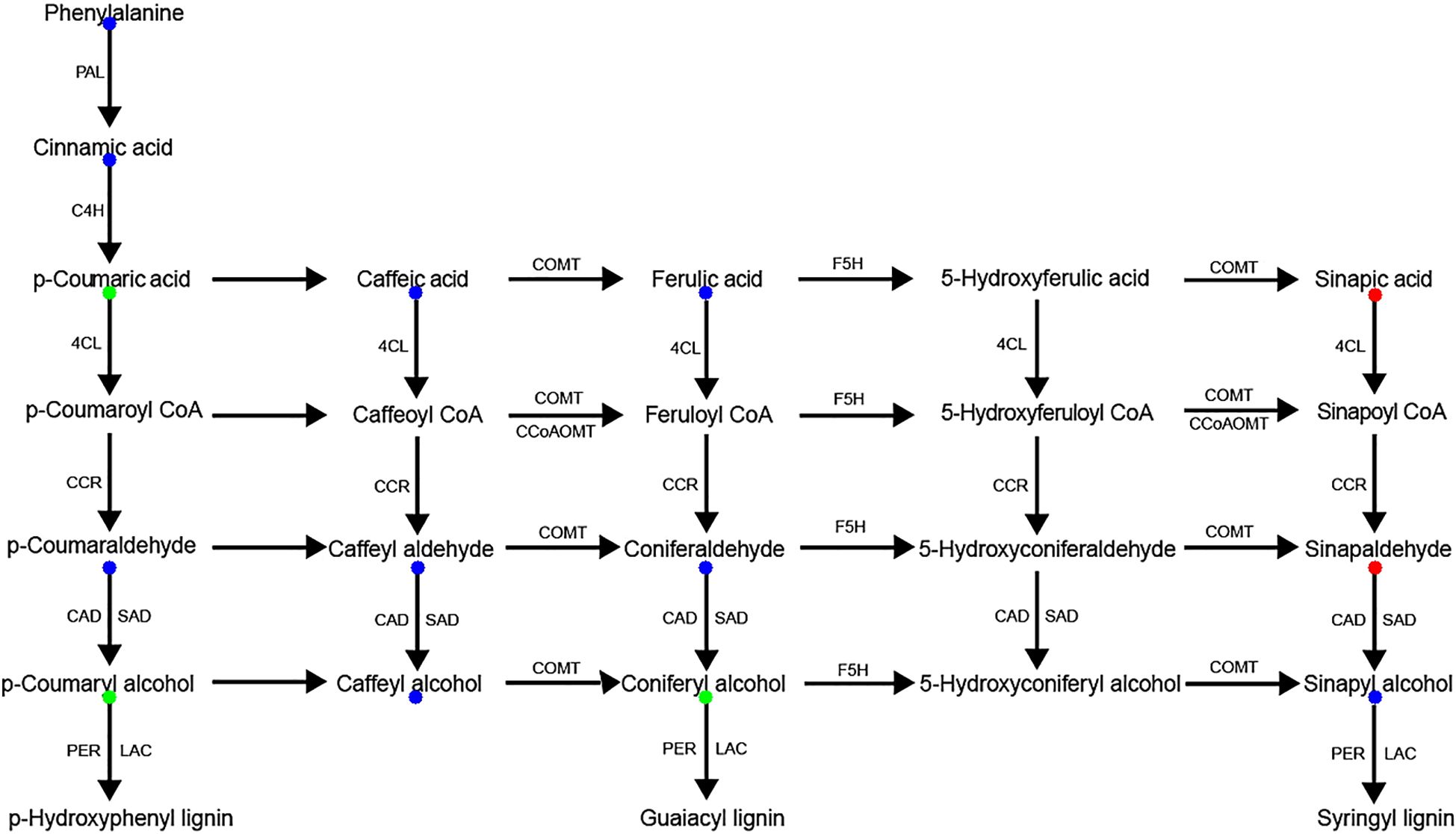
Figure 5. Lignin biosynthetic pathway in AS and AA. Intermediate metabolites detected with no significant difference between AS and AA are marked in blue. Red/green represents up/down-accumulated metabolites in the AS.
3.5 Correlation analysis between the transcriptome and metabolome data
To elucidate the regulation of secondary metabolism, integrated KEGG enrichment analysis revealed 33 pathways co-enriched in DEGs and DAMs. Figure 6A highlighted the top 20 pathways with the largest number of DAMs. Among these pathways, flavone/flavonol biosynthesis, flavonoid biosynthesis, monoterpenoid biosynthesis, and sesquiterpenoid/triterpenoid biosynthesis exhibited the highest numbers of DAMs. Notably, sesquiterpenoid/triterpenoid biosynthesis, monoterpenoid biosynthesis, terpenoid backbone biosynthesis, and flavone/flavonol biosynthesis were significantly enriched in the transcriptomic data, indicating their central roles in the differences observed between A. stolonifera and A. argyi. Specifically, 50 DEGs and 24 DAMs were identified as being associated with the terpenoid-related biosynthesis pathway, while 55 DEGs and 39 DAMs were linked to the flavonoid-related biosynthesis pathway.
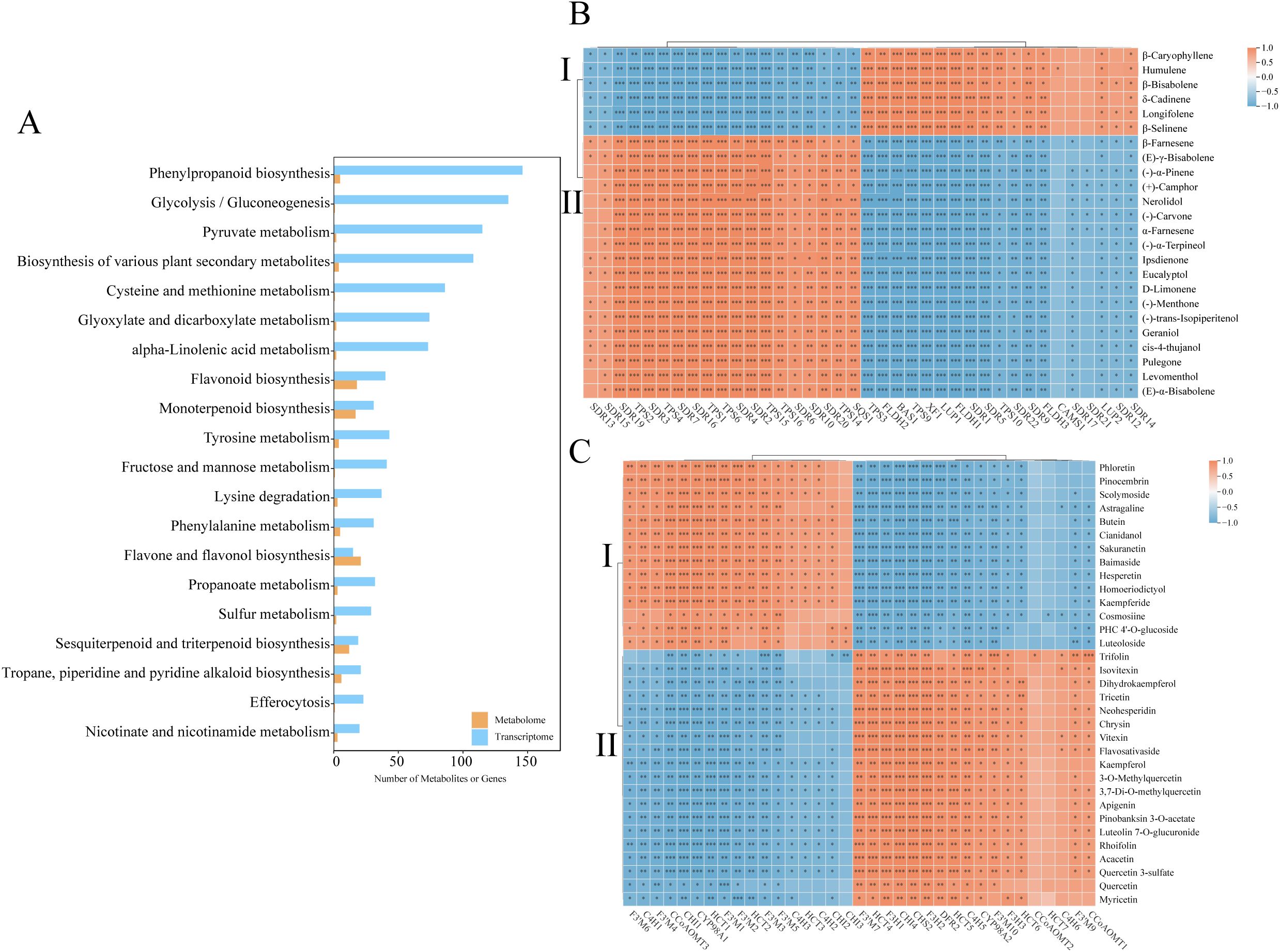
Figure 6. Co-enrichment analysis and correlation analysis of DAMs and DEGs shared in AS and AA. (A) Co-enrichment analysis of DAMs and DEGs, (B) Correlation analysis of the terpenoid-related DEGs and DSMs, (C) Correlation analysis of the flavonoid-related DEGs and DSMs. *pvalue<0.05;**pvalue<0.01; ***pvalue<0.001.
Crucially, Pearson correlations were computed using DEGs and DAMs, as visualized in Figures 6B, C. This analysis focused on terpenoid-related and flavonoid-related biosynthesis pathways, with a significance threshold set at p < 0.01 and |r| > 0.9. Among the findings, 24 terpenoids demonstrated significant correlations with 30 DEGs, while 33 flavonoids were significantly correlated with 27 DEGs. The co-expression analysis revealed that terpenoid metabolites could be distinctly categorized into two groups: Category I, which includes 6 sesquiterpenoids, and Category II, which comprises of 5 sesquiterpenoids and 13 monoterpenoids. Notably, 21 DEGs were positively correlated with Category I DAMs and negatively correlated with Category II DAMs (Figure 6B). Similarly, flavonoid-related metabolites were classified into two categories: Category I, consisting of 4 flavanones, 3 flavones, 3 flavonols, 3 chalcones and 1 flavanol, and Category II, which includes 9 flavones, 7 flavonols, 2 flavanonols and 1 flavanone. A total of 16 DEGs were positively correlated with Category I DAMs (Figure 6C). Within each category, metabolites exhibited positive correlations, while those from different categories typically displayed negative correlations. Collectively, we hypothesize that species-specific divergence is mechanistically linked to the differential regulation of terpenoid-related and flavonoid-related biosynthesis.
3.6 Identification of unigenes related to terpenoid-related, and flavonoid-related biosynthesis pathways
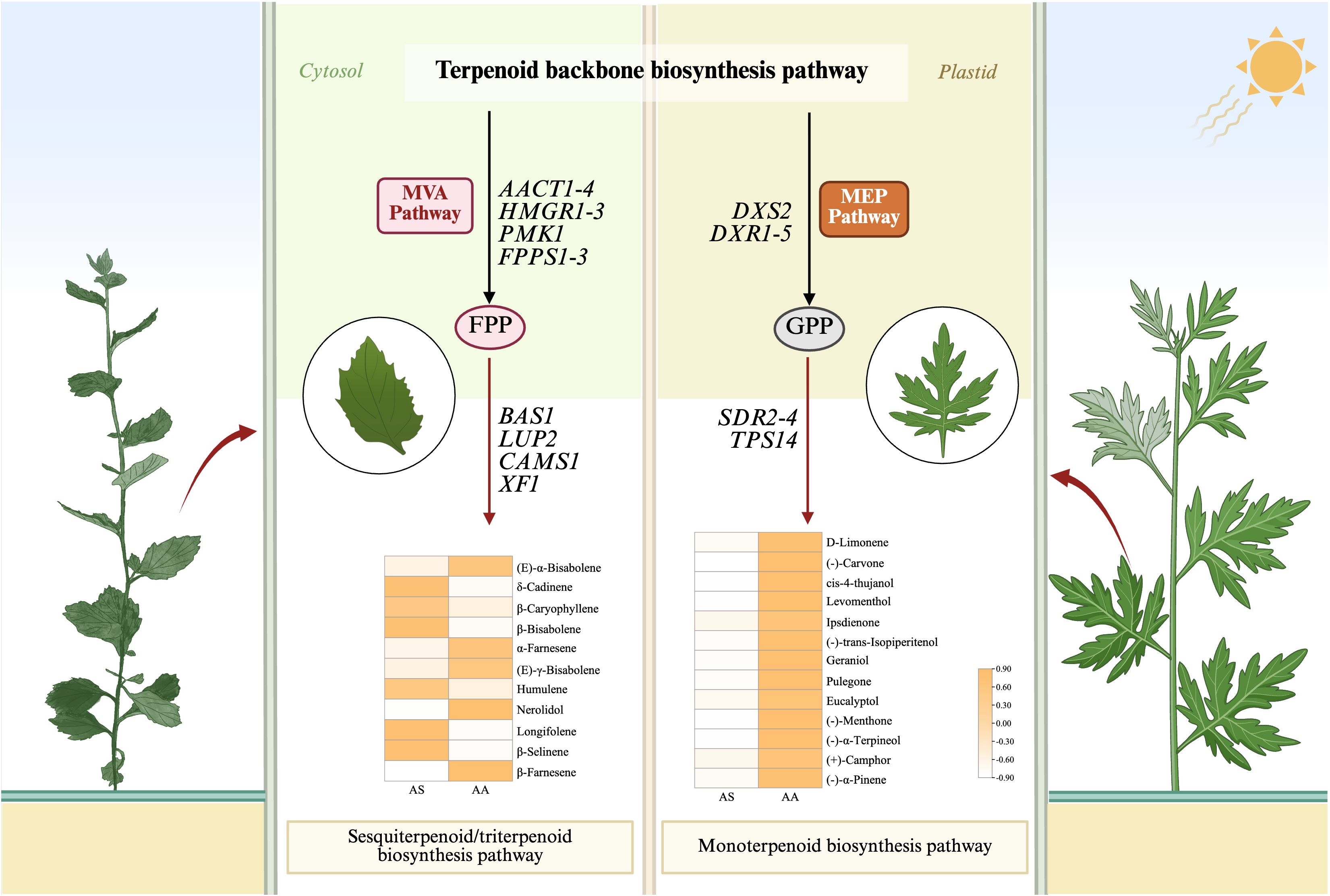
Our results, obtained through the integration of transcriptomics and metabolomics, indicate significant differences in the expression of genes related to terpenoid and flavonoid biosynthesis between the two species. The mevalonate (MVA) and methylerythritol phosphate (MEP) pathways are central to terpenoid biosynthesis, generating isoprenoid precursors including isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). To investigate the potential functions of genes involved in terpenoid backbone biosynthesis in leaves, which serve as the primary storage organ for essential oils, we identified candidate genes encoding enzymes within the MVA and MEP pathways from RNA-seq data. Transcriptomic analysis revealed 48 DEGs associated with terpenoid backbone biosynthesis, of which 19 unigenes exhibited upregulation in AS compared to AA. Within the MVA pathway, acetyl-CoA C-acetyltransferase (AACT), hydroxy methylglutaryl CoA reductase (HMGR), and phosphomevalonate kinase (PMK) showed species-specific regulation. Notably, the expression of AACT1–4 was markedly elevated in AS, corresponding to a 2.3- to 5.0-fold increase relative to AA. In contrast, genes associated with the MEP pathway, such as 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and reductoisomerase (DXR), displayed preferential expression in AA.
Downstream terpenoid synthesis revealed nuanced regulatory patterns. Farnesyl diphosphate synthase (FPPS) isoforms (FPPS1-3) exhibited statistically significant, albeit low-abundance, upregulation in AS, indicating a subtle enhancement of sesquiterpenoid precursor production. Additionally, four additional genes (FLDH1–3 and GPPS1) demonstrated AS-upregulated expression linked to specialized terpenoid diversification. Sesquiterpenoid/triterpenoid biosynthesis pathways contained 19 DEGs, predominantly upregulated in AS. In contrast, monoterpenoid and diterpenoid biosynthesis involved 31 and 12 DEGs respectively, with 18 and 8 unigenes upregulated in AA. Metabolomic validation confirmed species-specific accumulation. Notably, all 13 DAMs in monoterpenoid biosynthesis significantly increased in AA, with (-)-carvone, (-)-menthone, and cis-4-thujanol exhibiting 79.07-, 28.62-, and 24.57-fold higher contents respectively (Figure 7). Furthermore, 11 DAMs associated with sesquiterpenoid and triterpenoid biosynthesis were identified, predominantly upregulated in AS.
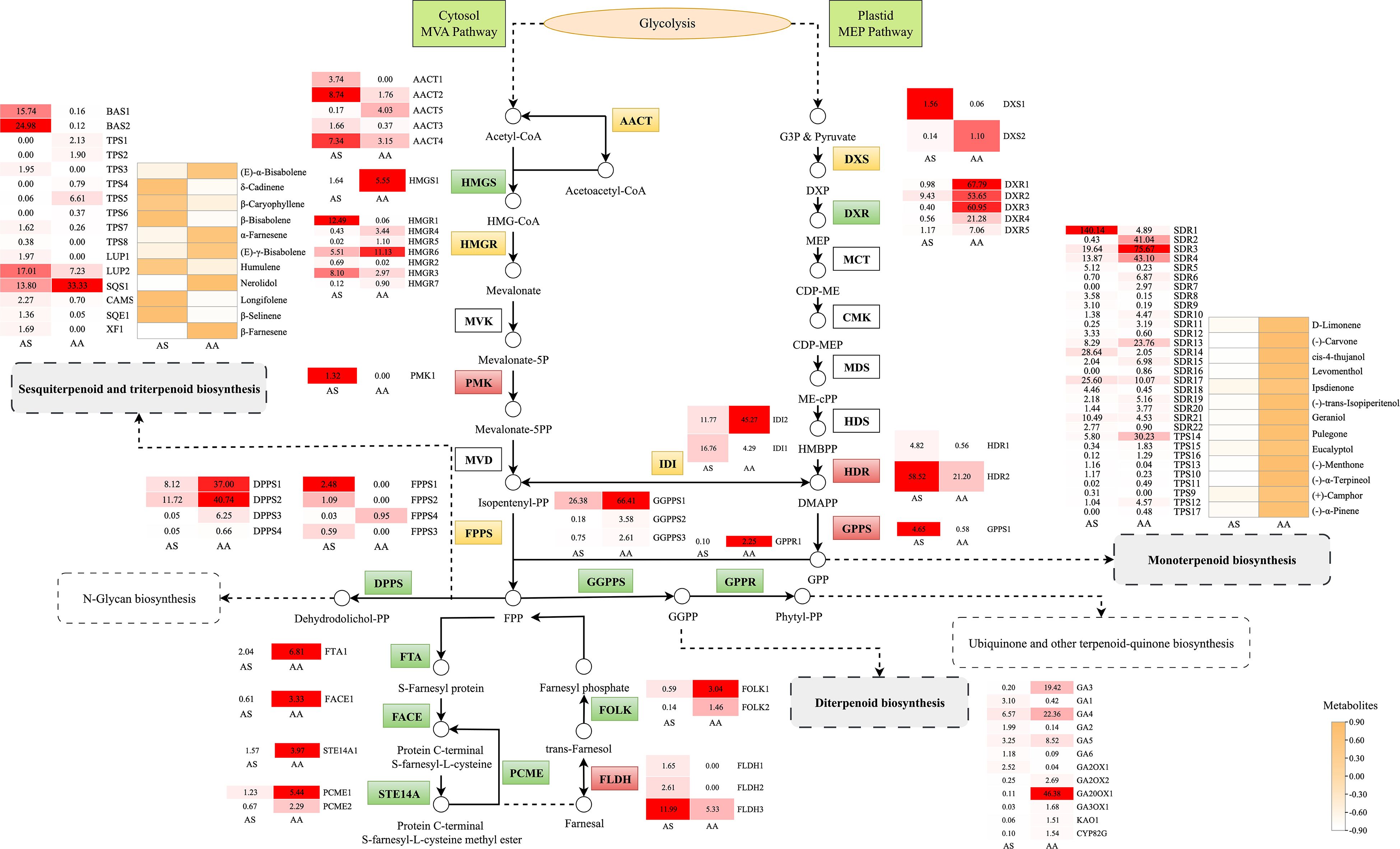
Figure 7. Metabolic pathway map of the terpenoid-related biosynthesis pathways. Small squares and circles indicate the expressed levels of unigenes/metabolites, and red/green/yellow graphics indicates up/down/both up- and downregulated genes or metabolites in the AS group.
Through combined transcriptomic and metabolomic profiling, along with KEGG pathway mapping, we reconstructed the species-divergent regulation of flavonoid biosynthesis in AS and AA. The phenylpropanoid pathway served as the central biosynthetic node, supplying precursors for both flavonoid and lignin production. In this study, we identified 145, 40, and 15 DEGs involved in phenylpropanoid, flavonoid, and flavone/flavonol biosynthesis, respectively, of which 80, 19, and 9 DEGs were significantly upregulated (p < 0.05) in AS. Correspondingly, 5, 18, and 15 metabolites were associated with phenylpropanoid, flavonoid, and flavone/flavonol biosynthesis (Figure 8). We identified distinct expression patterns for 39 peroxidases (PERs), 26 acetylserotonin O-methyltransferases (COMTs), 16 cinnamyl-alcohol dehydrogenases (CADs), 10 4-coumarate-CoA ligases (4CLs), 7 spermidine hydroxycinnamoyl transferases (HCTs), 7 lysophospholipases (LysoPLs), 6 cinnamate-4-hydroxylases (C4Hs), 6 UDP-glucosyl transferases 72E (UGT72Es), 5 aldehyde dehydrogenases (ALDH2Cs), 5 ferulate-5-hydroxylases (F5Hs), 3 caffeoyl-CoA O-methyltransferases (CCoAOMTs), 2 cytochrome P450 family 98 subfamily A8s (CYP98As), and 2 cinnamoyl-CoA reductases (CCRs) between the two species. This metabolic profile aligns with the observed transcriptomic differences, particularly the downregulation of key precursors for G-lignin (coniferyl alcohol) in AS, in conjunction with the upregulation of intermediates associated with S-lignin synthesis (sinapic acid, sinapinaldehyde). These monolignols are subsequently polymerized to form the complex lignin polymer.
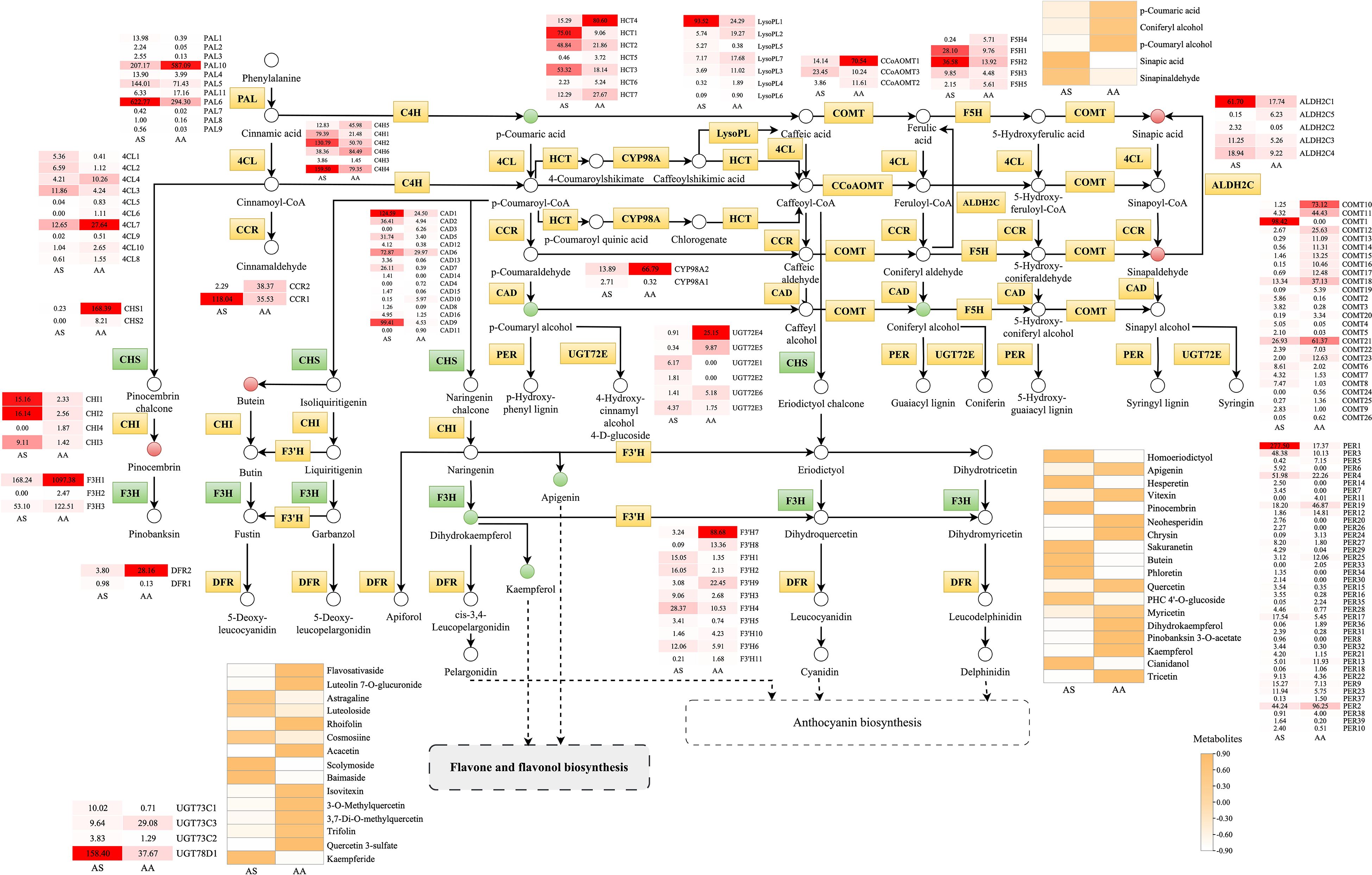
Figure 8. Metabolic pathway map of the flavonoid-related biosynthesis pathways. Small squares and circles indicate the expressed levels of unigenes/metabolites, and red/green/yellow graphics indicates up/down/both up- and downregulated genes or metabolites in the AS group.
The early steps of the flavonoid pathway exhibited complementary regulation among species. Chalcone synthase (CHS) genes showed significantly higher expression in AA, facilitating the accumulation of naringenin chalcone. In contrast, chalcone isomerase (CHI) isoforms displayed species-specific induction, with CHI2 expression increasing 6.3-fold, thereby enhancing the conversion to naringenin. Downstream hydroxylation modifications indicated pathway specialization. Flavanone 3-hydroxylase (F3H) expression increased by an average of 4.4-fold in AA, catalyzing the conversion of flavanones into flavanonols. The expression of flavonoids 3’-monooxygenase (F3’H) isoforms demonstrated divergent regulation: F3’H7–11 expression rose between 2.9 and 148.4-fold in AA, while F3’H1–6 expression by 2.0 to 11.1-fold in AS, facilitating the conversion of isoflavones into flavonols. Glycosylation processes played a crucial role in determining the final metabolite speciation. A total of 3 UDP-glycosyltransferases (UGT) 73Cs and 1 UGT78D were identified, with their expression levels varying between the leaves of the two Artemisia species. Metabolomic validation corroborated this divergence. Among 15 DAMs in flavone/flavonol biosynthesis, 9 compounds preferentially accumulated in AS, including kaempferide, while 6 metabolites enriched in AA, exemplified by naringenin. Therefore, the differential expression of UGT73Cs and UGT78D may be a critical factor directly influencing the type of flavone/flavonol synthesized.
3.7 qRT-PCR validation of DEG expression
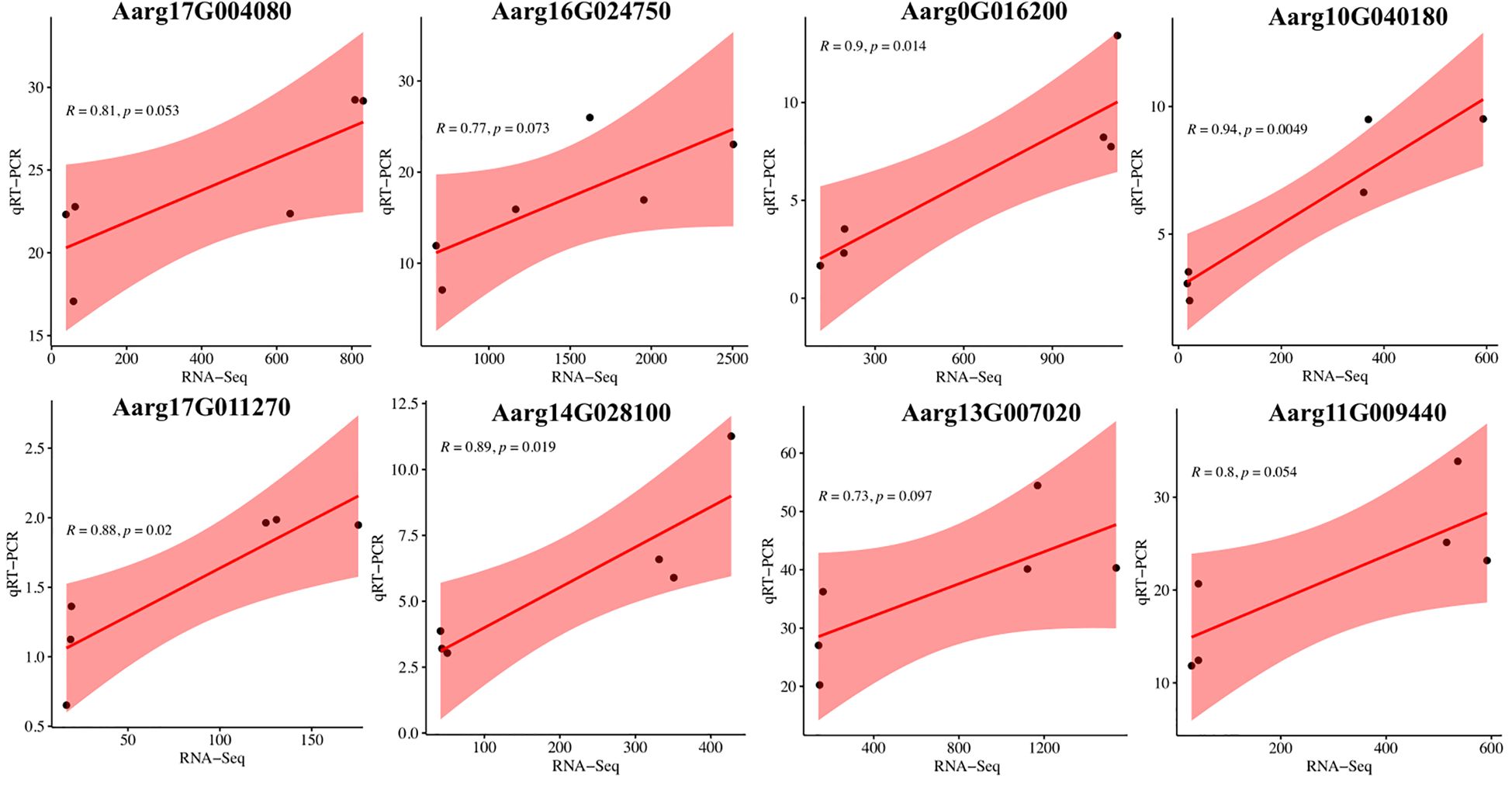
To validate the reliability and robustness of our transcriptome dataset, we conducted an independent verification by quantifying the expression patterns of eight DEGs using qRT-PCR. The expression profiles of these DEGs were consistent with those obtained from RNA sequencing analysis (Figure 9). This robust correlation establishes a reliable resource for elucidating species-specific divergence in specialized metabolite pathways between A. stolonifera and A. argyi, particularly in the terpenoid and flavonoid metabolic networks.
4 Discussion
Currently, A. argyi is officially documented in the Chinese Pharmacopoeia and is extensively cultivated across China for medicinal, food, and moxibustion applications, owing to its well-characterized efficacy and intensely aromatic properties. A. stolonifera exhibits a mild and delicate aroma, despite documented historical use, remains markedly underexplored and underutilized in modern applications, primarily due to insufficient comparative studies of its phytochemical aspects. Our integrated metabolomic platform characterized 2,153 metabolites (1,579 via GC-MS and 574 via UPLC- ESI (-QTRAP) -MS) with clear definitions. Among these, 1,657 metabolites were identified as DAMs, showing pronounced divergence between A. stolonifera and A. argyi (Supplementary Figures S2, S5). This represents an expansion in metabolite coverage compared to prior comparative studies. Li et al. (2023) employed HS-SPME-GC-MS to profile essential oils in A. stolonifera (59 volatile compounds) and A. argyi (64 volatile compounds). Luo et al. (2020a) utilized UPLC-Q-TOF-MS to analyze the chemical components of leaf and floss extracts from A. stolonifera and A. argyi sourced from different locations, identifying 18 compounds. Crucially, our non-targeted UPLC-ESI-MS and targeted UPLC-ESI-QTRAP-MS approaches, complementing the HS-SPME-GC-MS dataset, enabled unprecedented resolution of polar bioactive compounds, establishing a foundational framework for elucidating species-specific metabolic networks. Our results delineated distinct chemotypes: A. stolonifera accumulates sesquiterpenoids, while A. argyi enriches in monoterpenoids and flavonoids. The compositional differences identified through chemical analysis elucidate the contrasting olfactory profiles of the two species. A. stolonifera exhibits a much milder and more delicate aroma, a sensory characteristic closely linked to its significantly lower abundance of volatile constituents and reduced levels of strong-smelling monoterpenoids such as eucalyptol and camphor. Conversely, A. stolonifera contains higher proportions of lighter, less aggressive sesquiterpenoids, which contribute to its subtle and pleasant aroma. These characteristics suggest that A. stolonifera may be a suitable material for applications where milder odors are preferred, such as in sensitive-skin moxibustion or light-aroma therapeutics. In stark contrast, A. argyi produces a distinctively strong and penetrating aroma, which primarily arises from its high concentrations of monoterpenes. Species-specific metabolic have also been reported in other medicinal genera. In Coptis species, interspecific variation in alkaloid profiles directly influences pharmacological potency (Qi et al., 2022), supporting our hypothesis that the metabolic differences observed between Artemisia species have significant functional implications.
Field cultivation practices confirm that both A. argyi and A. stolonifera undergo stem lignification during development, a process supported by the positive correlation between lignin content and stem strength (Nan et al., 2022). Metabolomic profiling revealed higher accumulation of lignin pathway intermediates (p-coumaryl alcohols and coniferyl alcohols) in AA, indicating enhanced lignin deposition. Following the synthesis of monolignols (p-coumaryl alcohols, coniferyl alcohols, and sinapyl alcohols) in the cytoplasm, these compounds are transported to the apoplast where they are polymerized with lignin units (S, H and G units) by peroxidase (POD) and laccase (LAC) (Bonawitz and Chapple, 2010; Miao and Liu, 2010; Shah et al., 2017). Transcriptomic analysis further identified 145 DEGs involved in lignin biosynthesis, most exhibiting higher expression in AA, consistent with the metabolite data. In contrast, the lower accumulation of intermediate lignin metabolites in AS suggests that more phenylpropanoid precursors might be channeled to the biosynthesis of volatile terpenoids or aromatic esters, providing a metabolic explanation for its mild and fresh aroma profile. This divergence in phenylpropanoid partitioning provides a biochemical basis for the distinct aroma profiles of the two species.
Terpenoids are a class of important chemicals produced by plants, classified by carbon skeleton size into monoterpenes (C₁₀), sesquiterpenes (C15), diterpenes (C₂₀), triterpenes (C₃₀), tetraterpenes (C₄₀), and polyterpenes. Among these, sesquiterpenoids/triterpenoids are primarily biosynthesized via the cytosolic MVA pathway (Dubey et al., 2003). Focusing on terpenoid biosynthesis, structural genes within the MVA pathway exhibited significantly elevated expression (e.g., AACT1-4, HMGR1-3) in A. stolonifera. HMGR, the rate-limiting enzyme that converts HMG-CoA to mevalonate (Rodríguez-Concepción and Boronat, 2002; Rohmer, 2007), positively regulates terpenoid synthesis (Gondet et al., 1992; Zhang et al., 2019), which partially explains the accumulation of sesquiterpenoids observed in A. stolonifera. Conversely, genes of the MEP pathway displayed significantly higher expression (e.g., DXS2, DXR1-5) in A. argyi (Figure 7). DXS and DXR are key rate-limiting enzymes (Lange et al., 1998; Takahashi et al., 1998) that enhance the precursor supply for monoterpene biosynthesis (Lichtenthaler et al., 1997). The divergent terpenoid biosynthesis in A. stolonifera and A. argyi likely stems from evolutionary specialization. Evidence suggests that organellar genomes, in addition to nuclear genes, may influence metabolic differentiation. Mitochondrial genomic studies in Coptis species have demonstrated substantial organelle-level variation associated with interspecific metabolic divergence (Zhong et al., 2023), suggesting that cyto-nuclear interactions could similarly contribute to metabolic disparity in Artemisia. Transcriptome-metabolome correlation analysis identified key genes (TPS, SQS, BAS, SDR, LUP, CAMS, SQE, and XF) in terpenoid-related biosynthesis pathways regulating the differential synthesis of terpenoids in A. stolonifera and A. argyi (Figure 6B). Specifically, the enhanced expression of downstream terpenoid backbone biosynthesis genes (e.g., BAS1, LUP2, CAMS1, and XF1) in A. stolonifera showed a strong positive correlation with its significantly higher accumulation of specific sesquiterpenoids, including δ-cadinene, β-bisabolene, β-caryophyllene, humulene, longifolene, and β-selinene (Figures 6B, 7), indicating an enhanced flux toward sesquiterpenoid precursors. The elevated expression of key downstream genes (e.g., SDR2-4, TPS14) is closely associated with the preferential accumulation of monoterpenoids, such as eucalyptol and (+)-camphor. The complexity of this metabolic network, with characterized by numerous branches, underscores the necessity for further investigation into the cooperative expression and regulatory mechanisms of each gene. The terpenoids metabolic divergence between A. stolonifera and A. argyi may extend beyond transcriptional regulation to include post-translational modifications. PRMT5-mediated methylation significantly reshapes protein function and cell metastasis (Fan et al., 2025), suggesting that analogous mechanisms may similarly influence metabolic specialization in Artemisia species.
The differences identified between A. stolonifera and A. argyi emphasize the need for a deeper understanding of the properties of individual species used in various applications. This perspective echoes recent advances in medicinal plant resource evaluation, including innovative approaches to utilizing non-traditional plant parts as demonstrated in Coptis research (Ding et al., 2025). Previous studies have validated that monoterpenes exhibit potent antimicrobial and antioxidant activities, positioning them as ideal natural food preservatives that ensure the microbiological and oxidative stability of food products (Rehman et al., 2021; Kumar et al., 2022). Our metabolomic quantification confirmed that monoterpenes are primarily enriched in A. argyi (Figure 10), establishing this species as a premium source for natural food preservatives. Additionally, the higher levels of eucalyptol and camphor further highlight its suitability as a promising additive (Yazgan, 2020; Šojić et al., 2021). A. stolonifera is characterized by a high abundance of sesquiterpenoids, especially β-caryophyllene. Mechanistic studies demonstrate β-caryophyllene possesses a greater ability to retain in the stratum corneum compared to eucalyptol (Figure 10), thereby facilitating its disruptive effect on stratum corneum lipids (Tang et al., 2023). According to Zhang et al. (2024), seven flavonoids (5-desmethylnobiletin, rhoifolin, baicalin, biorobin, nepetin, eriodictyol, and isorhoifolin) and nine terpenoids (bornyl acetate, endo-borneol, L-fenchone, 2-pinen-7-one, caryophyllene, camphor, levomenthol, (+)-β-cedrene, and γ-terpinene) were identified as important biomarkers for tracking the aging time of A. argyi. The transformation of these compounds during post-harvest aging is critical for enhancing both the efficacy and safety of A. argyi, corroborating the traditional practice of prolonged storage. In contrast, the inherently mild properties of A. stolonifera suggest that it may not require extended aging to achieve optimal applicability. This fundamental difference not only indicates distinct post-harvest processing needs for the two species but also positions A. stolonifera as a promising resource with advantages for rapid utilization.
Flavonoids, which are low molecular weight secondary metabolites with significant pharmacological value, are predominantly biosynthesized via the phenylpropanoid pathway (Nigam et al., 2019). An analysis of flavonoid biosynthesis in A. argyi further demonstrated a significantly elevated expression of core structural genes (e.g., CHS1-2, CHI, F3H1-3) (Figure 8), correlating with the notably higher accumulation of specific flavonoids, including chrysin, neohesperidin, and isovitexin. These findings align with previous studies on the flavonoid profiles of Artemisia species (Liu et al., 2015; Song et al., 2017), confirming species-specific accumulation. Mechanistically, CHS catalyzes the stepwise condensation of 4-coumaroyl-CoA and malonyl-CoA to naringenin chalcone, thereby diverting phenylpropanoid flux toward flavonoids (Winkel-Shirley, 2001; Grotewold, 2006). The CHS genes constitute a polygenic family characterized by strong conservation (Park et al., 2020). Subsequent enzymatic conversions drive downstream synthesis: CHI catalyzes the isomerization of chalcone to naringenin (Dixon and Paiva, 1995), while the reaction catalyzed by F3H converts naringenin to dihydrokaempferol, which is subsequently converted to kaempferol by FLS (Winkel-Shirley, 2001). Thus, the coordinated upregulation of CHS, CHI, and F3H was positively associated with flavonoid synthesis in A. argyi. Collectively, these transcriptional changes underlie the significantly higher flavonoid accumulation in A. argyi relative to A. stolonifera. These biochemical advantages translate into substantial agricultural applications. The flavonoids from Artemisia have been shown to improve the growth performance and meat quality of broilers (Yang et al., 2021; Shi et al., 2022). Notably, 0.4 mg/mL of A. argyi flavonoids effectively prolongs the storage period of fresh-grade breast meat (Yang et al., 2023). The abundant flavonoid metabolites underlying these applications provide a biochemical basis for future innovations in deep processing beyond traditional moxibustion.
5 Conclusion
This study established a comprehensive multi-omics integration of metabolomic and transcriptomic profiles contrasting the historically significant A. stolonifera with the pharmacopeial standard A. argyi, revealing species-specific divergences. Comparative analysis identified 1,728 DAMs and 18,000 DEGs, demonstrating that species-specific divergence drives transcriptional and metabolic network reorganization in A. stolonifera and A. argyi. Transcriptomic profiling revealed significant regulatory divergence in terpenoid and flavonoid biosynthesis between the two species. In A. stolonifera, the coordinated upregulation of MVA pathway genes and sesquiterpene synthases strongly correlated with sesquiterpenoid accumulation. Conversely, A. argyi exhibited preferential induction of MEP pathway genes, monoterpenoid biosynthesis genes, and flavonoid biosynthesis genes, which are associated with monoterpenoid and flavonoid enrichment. These pronounced metabolic and transcriptional differences underscore the necessity for evaluating of A. stolonifera’s distinct phytochemical profile, particularly concerning its equivalence to A. argyi for medicinal applications reliant on monoterpenoids or specific flavonoids. Furthermore, while we elucidated novel molecular regulatory networks governing the biosynthesis of terpenoids and flavonoids, the potential regulatory characteristics still required systematic characterization. Our research team is currently investigating the precise functions of specific genes in A. stolonifera to decipher their mechanistic contributions to terpenoid biosynthesis. Our findings established a foundational framework for future research, including the functional validation of candidate genes involved in A. stolonifera’s terpenoid and flavonoid biosynthesis, which will inform strategies for its precision breeding and sustainable utilization in TCM.
Data availability statement
The datasets presented in the study are publicly available. This data can be found here: NCBI SRA repository under the project number PRJNA1363732.
Author contributions
YC: Writing – original draft, Methodology, Data curation, Validation, Software, Formal Analysis, Visualization. YR: Writing – review & editing. YW: Project administration, Conceptualization, Supervision, Writing – review & editing, Funding acquisition, Visualization. HL: Supervision, Project administration, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the Major Scientific and Technological Research and Development Projects in Jiangxi Province (20223AAF01007), Fundamental Research Funds for the Central Public Welfare Research Institutes (ZZ17-ND-12, ZZ17-XRZ-123, ZZ18-YQ-059), and Natural Science Foundation Project of Jiangxi Province (20242BAB20453).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1713657/full#supplementary-material
References
Abad, M. J., Bedoya, L. M., Apaza, L., and Bermejo, P. (2012). The Artemisia L. Genus: A review of bioactive essential oils. Molecules 17, 2542–2566. doi: 10.3390/molecules17032542
Anders, S. and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Anwar, F., Ahmad, N., Alkharfy, K. M., and Gilani, A. H. (2016). “Mugwort (Artemisia vulgaris) oils,” in Essential oils in food preservation, flavor and safety. Ed. Preedy, V. R. (Academic Press, San Diego), 573–579. doi: 10.1016/B978-0-12-416641-7.00065-1
Bonawitz, N. D. and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Chen, Y., Qi, L., Zhong, F., Li, Y., Ke, W., and Ma, Y. (2021). Integrated metabolomics and ligand fishing approaches to screen the hypoglycemic ingredients from four Coptis medicines. J. Pharm. BioMed. Anal. 192, 113655. doi: 10.1016/j.jpba.2020.113655
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Ding, Y. G., Feng, L. Q., Zhang, Z. N., Li, N., Huang, Y. M., Tang, Z. X., et al. (2025). Combining H2O auto-ML and traditional machine learning for the resource evaluation on non-medicinal parts of plants: taking Coptis deltoidea as an example. Talanta 298, 128874. doi: 10.1016/j.talanta.2025.128874
Dixon, R. A. and Paiva, N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. doi: 10.1105/tpc.7.7.1085
Du, Z., Lin, W., Yu, B., Zhu, J., and Li, J. (2021). Integrated metabolomic and transcriptomic analysis of the flavonoid accumulation in the leaves of Cyclocarya paliurus at different altitudes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.794137
Dubey, V. S., Bhalla, R., and Luthra, R. (2003). An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 28, 637–646. doi: 10.1007/BF02703339
Fan, S., Li, W. J., Qi, Y., Li, Z., He, Y. H., Luo, X., et al. (2025). PRMT5 encourages cell migration and metastasis of tongue squamous cell carcinoma through methylating ΔNp63α. Cell Death Differ. doi: 10.1038/s41418-025-01575-8
Ge, Y. B., Wang, Z. G., Xiong, Y., Huang, X. J., Mei, Z. N., and Hong, Z. G. (2016). Anti-inflammatory and blood stasis activities of essential oil extracted from Artemisia argyi leaf in animals. J. Nat. Med. 70, 531–538. doi: 10.1007/s11418-016-0972-6
Gondet, L., Weber, T., Maillot-Vernier, P., Benveniste, P., and Bach, T. J. (1992). Regulatory role of microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase in a tobacco mutant that overproduces sterols. Biochem. Biophys. Res. Commun. 186, 888–893. doi: 10.1016/0006-291x(92)90829-a
Grotewold, E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57, 761–780. doi: 10.1146/annurev.arplant.57.032905.105248
Han, B., Xin, Z., Ma, S., Liu, W., Zhang, B., Ran, L., et al. (2017). Comprehensive characterization and identification of antioxidants in Folium Artemisiae Argyi using high-resolution tandem mass spectrometry. J. Chromatogr B Analyt Technol. BioMed. Life Sci. 1063, 84–92. doi: 10.1016/j.jchromb.2017.08.021
Jiang, Z., Guo, X., Zhang, K., Sekaran, G., Cao, B., Zhao, Q., et al. (2019). The essential oils and eucalyptol from Artemisia vulgaris L. prevent acetaminophen-induced liver injury by activating Nrf2-Keap1 and enhancing APAP clearance through non-toxic metabolic pathway. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.00782
Kim, J. Y., Lee, M. S., Baek, J. M., Park, J., Youn, B. S., and Oh, J. (2015b). Massive elimination of multinucleated osteoclasts by eupatilin is due to dual inhibition of transcription and cytoskeletal rearrangement. Bone Rep. 3, 83–94. doi: 10.1016/j.bonr.2015.10.003
Kim, J. K., Shin, E.-C., Lim, H.-J., Choi, S. J., Kim, C. R., Suh, S. H., et al. (2015a). Characterization of nutritional composition, antioxidative capacity, and sensory attributes of seomae mugwort, a native korean variety of artemisia argyi H. Lév. & Vaniot. J. Anal. Methods Chem. 2015, 916346. doi: 10.1155/2015/916346
Kumar, A., Singh, P. P., and Prakash, B. (2022). Assessing the efficacy of chitosan nanomatrix incorporated with Cymbopogon citratus (DC.) Stapf essential oil against the food-borne molds and aflatoxin B1 production in food system. Pestic Biochem. Physiol. 180, 105001. doi: 10.1016/j.pestbp.2021.105001
Lange, B. M., Wildung, M. R., McCaskill, D., and Croteau, R. (1998). A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 2100–2104. doi: 10.1073/pnas.95.5.2100
Li, S. G., Wu, T., Lei, M., Chen, K. L., Liu, Y. M., Huang, X. Z., et al. (2022). Comparison of volatile components extracted from Artemisia stolonifera leaves by headspace-solid phase microextraction and steam distillation methods. J. Chin. Med. Mater. 45, 2399–2402. doi: 10.13863/j.issn1001-4454.2022.10.021
Li, S. G., Li, H., Huang, L. Q., Lei, M., Ma, L., Wu, T., et al. (2023). Analysis of volatile components in the leaves of Artemisia argyi and Artemisia stolonifera by HS-SPME-GC-MS coupling technique. J. Chin. Med. Mater. 46, 395–400. doi: 10.13863/j.issn1001-4454.2023.02.022
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient genera purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lichtenthaler, H. K., Schwender, J., Disch, A., and Rohmer, M. (1997). Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 400, 271–274. doi: 10.1016/s0014-5793(96)01404-4
Liu, X., Dong, Y., Yao, N., Zhang, Y., Wang, N., Cui, X., et al. (2015). De Novo sequencing and analysis of the safflower transcriptome to discover putative genes associated with safflor yellow in Carthamus tinctorius L. Int. J. Mol. Sci. 16, 25657–25677. doi: 10.3390/ijms161025657
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, D. D., Peng, H. S., Zhan, Z. L., Kang, L. P., Zhang, Y., Huang, X. Z., et al. (2020b). Textual research on traditional Chinese medicine Jiuniucao. China J. Chin. Mater Med. 45, 4081–4088. doi: 10.19540/j.cnki.cjcmm.20200626.102
Luo, D. D., Peng, H. S., Zhang, Y., Huang, X. Z., Zhan, Z. L., Liu, D. H., et al. (2020a). Comparison of chemical components between Artemisia stolonifera and Artemisia argyi using UPLC-Q-TOF-MS. China J. Chin. Mater Med. 45, 4057–4064. doi: 10.19540/j.cnki.cjcmm.20200602.203
Lv, J. L., Li, Z. Z., and Zhang, L. B. (2018). Two new flavonoids from Artemisia argyi with their anticoagulation activities. Nat. Prod Res. 32, 632–639. doi: 10.1080/14786419.2017.1332603
Miao, Y. C. and Liu, C. J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U.S.A. 107, 22728–22733. doi: 10.1073/pnas.1007747108
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nan, M., Li, J., Zhao, G. Q., Chai, J. K., and Liu, Y. M. (2022). Relationship between lodging resistance of oats and the basal internode stem characteristics and lignin synthesis. Acta Prataculturae Sinica 31, 172–180. doi: 10.11686/cyxb2021416
Nigam, M., Atanassova, M., Mishra, A. P., Pezzani, R., Devkota, H. P., Plygun, S., et al. (2019). Bioactive compounds and health benefits of Artemisia Species. Natural Product Commun. 14, 1934578X19850354. doi: 10.1177/1934578X19850354
Park, H. L., Yoo, Y., Bhoo, S. H., Lee, T. H., Lee, S. W., and Cho, M. H. (2020). Two chalcone synthase isozymes participate redundantly in UV-induced sakuranetin synthesis in rice. Int. J. Mol. Sci. 21, 3777. doi: 10.3390/ijms21113777
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Qi, L., Zhong, F., Liu, N., Wang, J., Nie, K., Tan, Y., et al. (2022). Characterization of the anti-AChE potential and alkaloids in Rhizoma Coptidis from different Coptis species combined with spectrum-effect relationship and molecular docking. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1020309
Rehman, A., Qunyi, T., Sharif, H. R., Korma, S. A., Karim, A., Manzoor, M. F., et al. (2021). Biopolymer based nanoemulsion delivery system: An effective approach to boost the antioxidant potential of essential oil in food products. Carbohydr. Polymer Technol. Appl. 2, 100082. doi: 10.1016/j.carpta.2021.100082
Rodríguez-Concepción, M. and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089. doi: 10.1104/pp.007138
Rohmer, M. (2007). Diversity in isoprene unit biosynthesis: The methylerythritol phosphate pathway in bacteria and plastids. Pure Appl. Chem. 79, 739–751. doi: 10.1351/pac200779040739
Shah, A. N., Tanveer, M., Rehman, A. U., Anjum, S. A., Iqbal, J., and Ahmad, R. (2017). Lodging stress in cereal-effects and management: An overview. Environ. Sci. pollut. Res. Int. 24, 5222–5237. doi: 10.1007/s11356-016-8237-1
Shi, L., Jin, X., Xu, Y., Xing, Y., Yan, S., Guo, Y., et al. (2022). Effects of total flavonoids of Artemisia ordosica on growth performance, oxidative stress, and antioxidant status of lipopolysaccharide-challenged broilers. Antioxid (Basel) 11, 1985. doi: 10.3390/antiox11101985
Šojić, B., Tomović, V., Savanović, J., Kocić-Tanackov, S., Pavlić, B., Jokanović, M., et al. (2021). Sage (Salvia officinalis L.) essential oil as a potential replacement for Sodium Nitrite in dry fermented sausages. Processes 9, 424. doi: 10.3390/pr9030424
Song, L., Ma, Q., Zou, Z., Sun, K., Yao, Y., Tao, J., et al. (2017). Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in Camellia sinensis cultivar “Huangjinya. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00803
Takahashi, S., Kuzuyama, T., Watanabe, H., and Seto, H. (1998). A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 9879–9884. doi: 10.1073/pnas.95.17.9879
Tang, Q., Xu, F., Wei, X., Gu, J., Qiao, P., Zhu, X., et al. (2023). Investigation of β-caryophyllene as terpene penetration enhancer: Role of stratum corneum retention. Eur. J. Pharm. Sci. 183, 106401. doi: 10.1016/j.ejps.2023.106401
Tian, R., Zhang, X., Li, S. Y., Aixinjueluo, Q. Y., Lam, W. C., and Bian, Z. X. (2020). Reporting quality of systematic reviews with moxibustion. Chin. Med. 15, 104. doi: 10.1186/s13020-020-00385-z
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Yang, Y., Huang, Y., and Zheng, W. (2023). Study on antioxidant effect of flavonoids from Artemisia argyi and its application in preservation of fresh grade breast. Food Machine 36, 122–127,142. doi: 10.13652/j.issn.1003-5788.2020.11.024
Yang, S., Zhang, J., Jiang, Y., Xu, Y. Q., Jin, X., Yan, S. M., et al. (2021). Effects of Artemisia argyi flavonoids on growth performance and immune function in broilers challenged with lipopolysaccharide. Anim. Biosci. 34, 1169–1180. doi: 10.5713/ab.20.0656
Yazgan, H. (2020). Investigation of antimicrobial properties of sage essential oil and its nanoemulsion as antimicrobial agent. LWT 130, 109669. doi: 10.1016/j.lwt.2020.109669
Yuan, H., Cao, G., Hou, X., Huang, M., Du, P., Tan, T., et al. (2022). Development of a widely targeted volatilomics method for profiling volatilomes in plants. Mol. Plant 15, 189–202. doi: 10.1016/j.molp.2021.09.003
Yun, C., Jung, Y., Chun, W., Yang, B., Ryu, J., Lim, C., et al. (2016). Anti-Inflammatory effects of Artemisia leaf extract in mice with contact dermatitis in vitro and in vivo. Mediators Inflammation 2016, 8027537. doi: 10.1155/2016/8027537
Zhang, P., Shi, B., Li, T., Xu, Y., Jin, X., Guo, X., et al. (2018). Immunomodulatory effect of Artemisia argyi polysaccharide on peripheral blood leucocyte of broiler chickens. J. Anim. Physiol. Anim. Nutr. (Berl) 102, 939–946. doi: 10.1111/jpn.12895
Zhang, L., Wang, X., Wang, W., Ning, E., Chen, L., Li, Z., et al. (2024). Metabolomic analysis reveals the changing trend and differential markers of volatile and nonvolatile components of Artemisiae argyi with different aging years. Phytochem. Anal. 35, 1286–1293. doi: 10.1002/pca.3359
Zhang, G., Wu, Y., Haq Muhammad, Z. U., Yang, Y., Yu, J., Zhang, J., et al. (2019). cDNA cloning, prokaryotic expression and functional analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) in Pogostemon cablin. Protein Expr Purif 163, 105454. doi: 10.1016/j.pep.2019.105454
Zhang, Y., Yang, L., Yang, J., Hu, H., Wei, G., Cui, J., et al. (2022). Transcriptome and metabolome analyses reveal differences in terpenoid and flavonoid biosynthesis in Cryptomeria fortunei needles across different seasons. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.862746
Keywords: Artemisia stolonifera, Artemisia argyi, metabolome, transcriptome, terpenoid biosynthesis, flavonoid biosynthesis
Citation: Cao Y, Ren Y, Wang Y and Li H (2025) Transcriptome and metabolome analyses revealed the main profiles contributing the mild aroma characteristics of Artemisia stolonifera. Front. Plant Sci. 16:1713657. doi: 10.3389/fpls.2025.1713657
Received: 26 September 2025; Accepted: 04 November 2025; Revised: 02 November 2025;
Published: 26 November 2025.
Edited by:
Bowei Chen, Jishou University, ChinaReviewed by:
Chaoqun Xu, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaFurong Zhong, Chengdu University of Technology, China
Copyright © 2025 Cao, Ren, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye Wang, eXdhbmdAaXRjbWhpLmFjLmNu; Hui Li, SExpMjAyMjAxQDEyNi5jb20=
 Ye Cao
Ye Cao Yan Ren
Yan Ren Ye Wang
Ye Wang Hui Li
Hui Li