- 1INRA, University of Nice Sophia Antipolis, CNRS, UMR 1355-7254, Institut Sophia Agrobiotech, Sophia Antipolis, France
- 2INRA, UR1052, Génétique et Amélioration des Fruits et Légumes, Montfavet, France
With the banning of most chemical nematicides, the control of root-knot nematodes (RKNs) in vegetable crops is now based essentially on the deployment of single, major resistance genes (R-genes). However, these genes are rare and their efficacy is threatened by the capacity of RKNs to adapt. In pepper, several dominant R-genes are effective against RKNs, and their efficacy and durability have been shown to be greater in a partially resistant genetic background. However, the genetic determinants of this partial resistance were unknown. Here, a quantitative trait loci (QTL) analysis was performed on the F2:3 population from the cross between Yolo Wonder, an accession considered partially resistant or resistant, depending on the RKN species, and Doux Long des Landes, a susceptible cultivar. A genetic linkage map was constructed from 130 F2 individuals, and the 130 F3 families were tested for resistance to the three main RKN species, Meloidogyne incognita, M. arenaria, and M. javanica. For the first time in the pepper-RKN pathosystem, four major QTLs were identified and mapped to two clusters. The cluster on chromosome P1 includes three tightly linked QTLs with specific effects against individual RKN species. The fourth QTL, providing specific resistance to M. javanica, mapped to pepper chromosome P9, which is known to carry multiple NBS–LRR repeats, together with major R-genes for resistance to nematodes and other pathogens. The newly discovered cluster on chromosome P1 has a broad spectrum of action with major additive effects on resistance. These data highlight the role of host QTLs involved in plant-RKN interactions and provide innovative potential for the breeding of new pepper cultivars or rootstocks combining quantitative resistance and major R-genes, to increase both the efficacy and durability of RKN control by resistance genes.
Introduction
Root-knot nematodes (RKNs), Meloidogyne spp., are major plant pathogens worldwide. These extremely polyphagous endoparasites can infest more than 5,500 plant species, including many field and greenhouse crops (Goodey et al., 1965). Since the banning of most chemical nematicides, due to environmental and public health issues, resistant cultivars have become an increasing important weapon against these pests. This method efficiently controls RKN populations, and is economically sustainable, innocuous to health and environment-friendly. RKN resistance is generally mediated by single, major resistance genes (R-genes), which can easily be introgressed into cultivars through back-crossing and phenotypic or marker-assisted selection (MAS). For this reason, major R-genes are widely used in the breeding of RKN-resistant cultivars and/or rootstocks. However, their efficacy is threatened by the capacity of RKNs to adapt. Indeed, R-genes apply a selective pressure on nematode populations, increasing the risk of virulent nematode populations emerging (Castagnone-Sereno, 2006), and this greatly limits their use. Several management strategies have been developed, to prevent the breakdown of resistance by pathogens. Most of these approaches are based on spatiotemporal management of the deployment of R-genes: (i) alternation of different R-genes in the crop rotation, (ii) use of mixtures of cultivars with different R-genes, or (iii) pyramiding, the introduction of several R-genes into the same cultivar (Kiyosawa, 1982; Mundt, 2002; Pink, 2002). The use of such strategies requires several R-genes to be available, with no emergence of cross-virulent pathogens. Recent experimental studies have shown that the pyramiding of R-genes is the best method for promoting effective, durable RKN resistance in pepper (Djian-Caporalino et al., 2014).
Several recent studies, on very different pathosystems, have shown that the genetic background of the plant greatly influences R-gene efficiency, potentially slowing the adaptation of pathogen populations to R-gene-carrying cultivars (Palloix et al., 2009; Brun et al., 2010; Fournet et al., 2013; Barbary et al., 2014). In some pathosystems, this greater durability has been shown to result from quantitative trait loci (QTLs), which slow the selection of variants virulent against the R-gene and decrease the size of the pathogen population (e.g., Quenouille et al., 2014). Plant genetic background is rarely considered in breeding programs for RKN resistance, despite its contribution to R-gene efficiency and durability. Indeed, breeding for resistance with QTLs is more complex and costly than the use of R-genes. In particular, the introgression of QTLs into elite cultivars must not impair other agronomically important crop traits, such as yield, quality criteria and adaptation, or other physiological characteristics.
In pepper (Capsicum annuum L.), several dominant R-genes, the Me genes and the N gene, have been characterized in detail (Hare, 1956; Hendy et al., 1985; Djian-Caporalino et al., 1999; Thies and Fery, 2000). These genes map to a genetic cluster on pepper chromosome P9 (Djian-Caporalino et al., 2007; Fazari et al., 2012). Three of these genes, Me3, Me1, and N, are routinely used in breeding programs. These genes are effective against a wide range of RKN species, including M. arenaria, M. incognita, and M. javanica, the most common species in temperate and tropical areas. They differ in their mode of action: Me3 and Me1 are stable at high temperature (Djian-Caporalino et al., 1999), whereas the efficacy of N is temperature-dependent (Thies and Fery, 1998). In addition, Me3 and Me1 differ in the spatiotemporal location of the resistance response triggered by a nematode attack. Me3 triggers an early hypersensitive response in the root epidermis at the nematode penetration site, whereas Me1 triggers a later response in the root vascular cylinder (Bleve-Zacheo et al., 1998; Pegard et al., 2005). It was long thought that there was a relationship between the mode of action of these R-genes and their durability, as the emergence of virulent populations has been reported only for N and Me3 (Castagnone-Sereno et al., 1996; Thies, 2011). However, the risk of Me1-virulent RKN populations emerging and of the development of multi-virulent populations might increase with the extensive deployment of these resistance genes in agriculture. The efficacy of these genes has been shown to be higher when they are introgressed into a partially resistant background and lower if they are introgressed into a highly susceptible background (Barbary et al., 2014). These same genetic backgrounds were also previously shown to affect the durability of field resistance (Djian-Caporalino et al., 2014). However, no quantitative resistance loci that could be combined with major genes to increase the efficacy and durability of genetic RKN control have yet been identified in the pepper germplasm.
We report here a QTL analysis dissecting the genetic backgrounds previously shown to modulate the efficacy and durability of resistance. An F2:3 progeny derived from a cross between a partially resistant (Yolo Wonder, YW) and a highly susceptible (Doux Long des Landes, DLL) pepper inbred line was tested for quantitative resistance to the three main RKN species (M. incognita, M. arenaria, and M. javanica). Four new major QTLs were mapped to two separate clusters. The first, containing one QTL, colocalized with the cluster of Me genes on pepper chromosome P9. The second included three QTLs against the three Meloidogyne species located on pepper chromosome P1. This new cluster could potentially be used for innovative breeding strategies to increase R-gene efficacy and durability for the control of RKNs.
Materials and Methods
Plant Material
A population of 130 F2:3 families derived from a cross between YW and DLL was used. These pepper cultivars were selected from the INRA pepper germplasm collection at Avignon, France (CRB-Leg, INRA-GAFL), on the basis of their different levels of resistance to nematode species. DLL is highly susceptible to the three main RKN species: M. arenaria, M. javanica, and M. incognita. YW is partially resistant (i.e., low-level symptoms) to M. incognita (Figure 1), totally resistant to M. javanica and has variable levels of resistance to M. arenaria (i.e., totally or partially resistant), depending on the RKN isolate considered (Djian-Caporalino et al., 1999). A single F1 hybrid plant was self-pollinated to generate 130 F2 plants, which were used to construct the genetic map. The 130 F2 plants were self-pollinated to generate 130 F3 families, which were used to assess disease resistance.
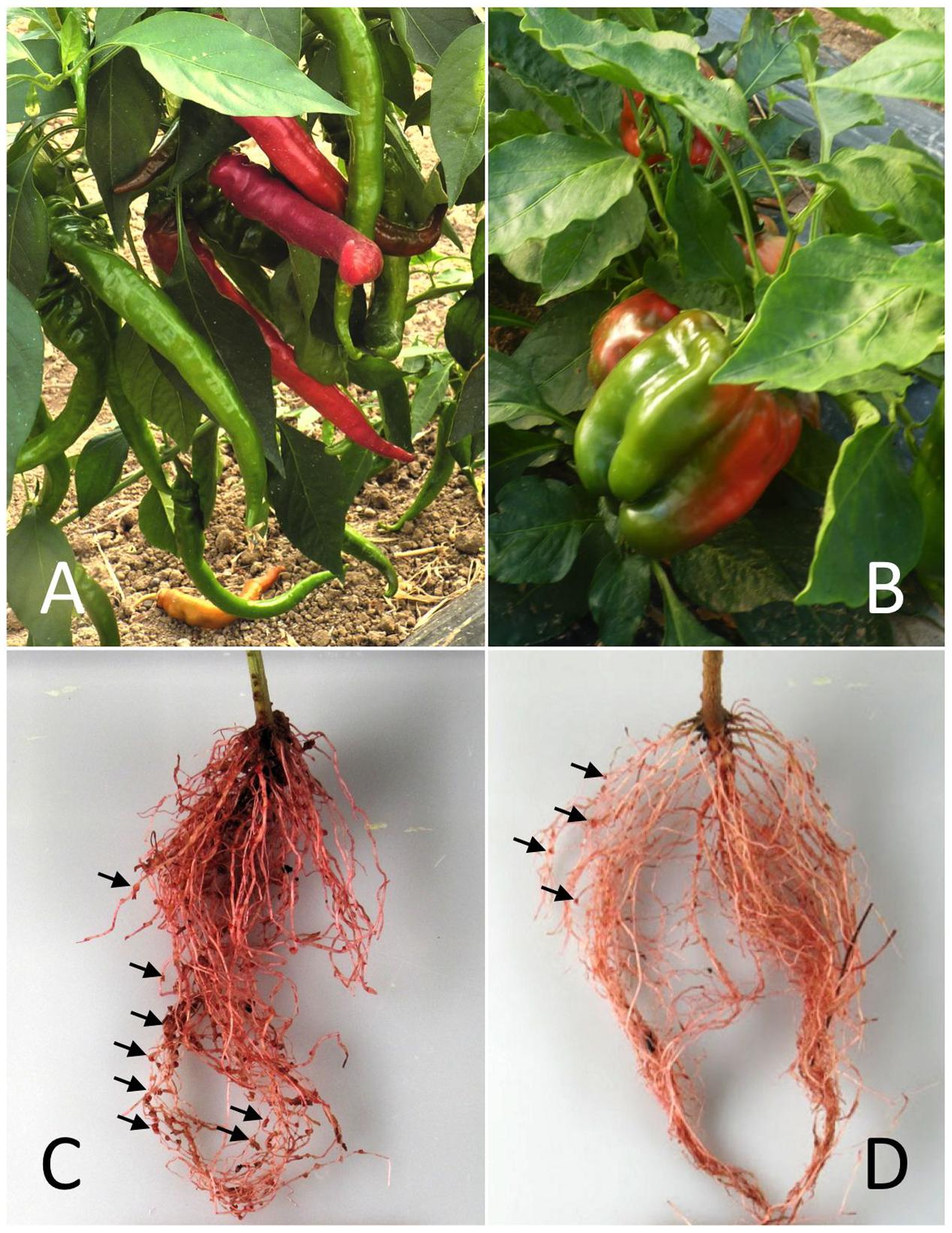
FIGURE 1. The pepper/Meloidogyne incognita pathosystem used in this study. The susceptible and resistant pepper cultivars Doux Long des Landes (DLL) (A) and Yolo Wonder (YW) (B) and their respective root systems (C,D) 6 weeks after inoculation with the nematodes. Egg masses (EMs; arrows) were stained with cold eosin (red).
Nematode Isolates
Three RKN species were used for resistance tests in controlled conditions. The first, M. incognita (Morelos isolate), causes disease on DLL, which is susceptible, whereas YW is partially resistant. The other two species used were M. arenaria (Marmande isolate) and M. javanica (Avignon isolate). DLL is susceptible and YW is highly resistant to these two species (Djian-Caporalino et al., 1999). These nematodes were obtained from the INRA Meloidogyne collection maintained at Institut Sophia Agrobiotech in Sophia Antipolis, France. These three Meloidogyne species have a mitotic parthenogenetic mode of reproduction. We therefore considered all the second-stage juveniles (J2s) hatching from a single egg mass to constitute a clonal line. Before multiplication, these isolates were specifically identified on the basis of isoesterase electrophoresis (Dalmasso and Berge, 1978) or sequence characterized amplified region (SCAR) PCR (Zijlstra et al., 2000).
DNA Extraction, Genotyping of Molecular Markers, and Linkage Map
Genomic DNA was isolated from the young leaves of both parents, the F1 and the individuals of the mapping population, as described by Fulton et al. (1995). After RNAse treatment, the concentration and purity of DNA were assessed with a NanoDrop 2000 spectrophotometer (Thermo Scientific), and the final DNA concentration was adjusted to 20 ng/μL for PCR.
The F2 mapping population was genotyped with 58 markers previously used in other populations: one B94 SCAR marker (Djian-Caporalino et al., 2007), 13 simple sequence repeat (SSR) markers previously mapped in pepper (Alimi et al., 2013), 44 single-nucleotide polymorphism (SNP) markers from Nicolaï et al. (2012). In addition, 272 new SNP markers were provided by Syngenta Seeds. We used these markers to construct a genetic linkage map with Mapmaker software version 3.0b (Lander et al., 1987), using a LOD score threshold of 3.0 and a maximum recombination fraction of 0.3. Distances between markers were calculated with the Kosambi mapping function. For each linkage group (LG), marker order was checked with the ‘ripple’ command and markers were retained only if the LOD score value was greater than 2.0. The LGs were assigned to pepper chromosomes on the basis of the positions of SSR and SNP markers common to the genetic maps for pepper published by Alimi et al. (2013) and Quenouille et al. (2014).
Experimental Procedures for Evaluating Nematode Resistance
Resistance was assessed on the F3 progenies. For each RKN isolate, 16 F3 seeds per F2:3 family were sown individually in 9 cm plastic pots containing steam-sterilized sandy soil covered with a 1 cm layer of loam. F3 plants were split evenly (i.e., eight plants per F3) between two growth chambers maintained at 24°C (±2°C) with a 12-h light12-h dark cycle and a relative humidity of 60–70%. Parental lines, the F1 progeny and two resistant controls (HD149 and HD330) were included in the experimental design. The 130 F2:3 families and controls were randomly arranged within each growth chamber. Six to seven-week-old plants (4–6 true leaves) were each inoculated with 500 freshly hatched J2s suspended in water, for experiments with M. arenaria and M. javanica, and with 1,000 J2s suspended in water for experiments with M. incognita. This difference in inoculum was based on the behavior of YW with respect to the species used (resistant or partially resistant). It has been shown that a higher inoculum density is required to reveal the differences between the partially resistant parent (YW) and the highly susceptible parent (DLL). J2s were obtained in a mist chamber, from previously inoculated susceptible tomato roots (cultivar Saint Pierre). Six to seven weeks after inoculation (i.e., a period long enough for completion of the nematode life cycle), plants were harvested, carefully washed individually with tap water and frozen at –20°C until scoring. Before analysis, the roots were thawed and stained by incubation for 10 min in a cold aqueous solution of eosin (0.1 g/l water), for the specific staining of egg masses (EMs). The roots were rinsed and examined under a magnifying glass. The number of EMs was counted for each plant and the median number of EMs per plant for each F3 family (and for the control genotype) was determined, for estimation of the phenotypic value of each F2.
Statistical Analyses
R software1 was used for descriptive statistics. Analyses of variance (ANOVA) were carried out for each phenotypic trait (i.e., for resistance to each RKN species), to estimate genotypic/environmental effects. For each phenotypic trait, narrow-sense heritability (h2) was estimated with the formula h2 = 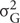 /(
/(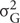 +
+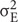 /n), where
/n), where 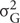 corresponds to the genotypic variance and
corresponds to the genotypic variance and 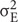 to the environment variance (including block, interaction, and error effects) and n is the number of replicates per F2:3 family (two growth rooms). Additive and dominance effects were calculated as described by Stuber et al. (1987). The normality of phenotype distributions was assessed with Shapiro–Wilks tests (α = 0.05).
to the environment variance (including block, interaction, and error effects) and n is the number of replicates per F2:3 family (two growth rooms). Additive and dominance effects were calculated as described by Stuber et al. (1987). The normality of phenotype distributions was assessed with Shapiro–Wilks tests (α = 0.05).
Quantitative trait loci analyses were performed with the R/qtl package of R software (Broman et al., 2003). QTLs were detected by regression analysis, SIM, CIM, and non-parametric interval mapping (model = “np” in the R/qtl package) for the non-Gaussian phenotype distributions, although the residues were normal, making it possible to carry out a regression analysis to estimate additive and R2 values. All the methods yielded similar results (the same QTLs at the same positions; data not shown), although the QTL peaks were slightly less sharp with the non-parametric procedure. A permutation test was performed with 1,000 replicates to determine the genome-wide LOD threshold empirically at the 5% probability level, for each phenotypic trait individually. The LOD threshold was estimated at 3.6 for the three traits. For each QTL, the confidence interval (CI) was defined as a 2-LOD drop-off around the maximum LOD score. R2 coefficients were calculated with the ‘fitqtl’ function of R/qtl.
Results
Linkage Map
Four of the 330 markers tested on the F2 mapping population remained unlinked. The genetic linkage map was therefore constructed with 326 markers: 13 SSRs, 312 SNPs, and 1 SCAR. Markers displaying significant deviation from the expected Mendelian ratio of 1:2:1 were retained (indicated by asterisks in Supplementary Data 1). The map comprised 12 LGs, corresponding to the 12 pepper chromosomes (P1–P12) and covering an overall length of 1436 cM (Supplementary Data 1).
Segregation of Resistance to the Three RKN Species
The frequency distributions of resistance to M. incognita, M. arenaria, and M. javanica in the F2:3 families are shown in Figure 2. For M. incognita, the effects of both genotype and block were significant (p-value = 0.000653 and p-value < 2.00e-16, respectively) as revealed by ANOVA. The regression of F3 values from block 1 over block 2 was significant (RPearson = 0.32, p = 0.00027) with higher values in block 1. Individual EM data were therefore adjusted by multiplying the data from block 2 by the regression coefficient (value: 1.4). This linear adjustment removed the block effect and the data were pooled for further analyses. The phenotypes of the control pepper lines were consistent with the expected results (Table 1): YW presented a lower infestation rate than DLL (mean of 250 and 330 EMs/plant, respectively). The F1 progeny was skewed toward DLL (319 EMs/plant), with a d/a ratio of –0.72, indicating additive to partly dominant inheritance in favor of susceptibility. A Shapiro–Wilk test showed that the values for the F2:3 families were normally distributed (W = 0.99, p-value = 0.2029; Figure 2A). The h2 value was 0.48.
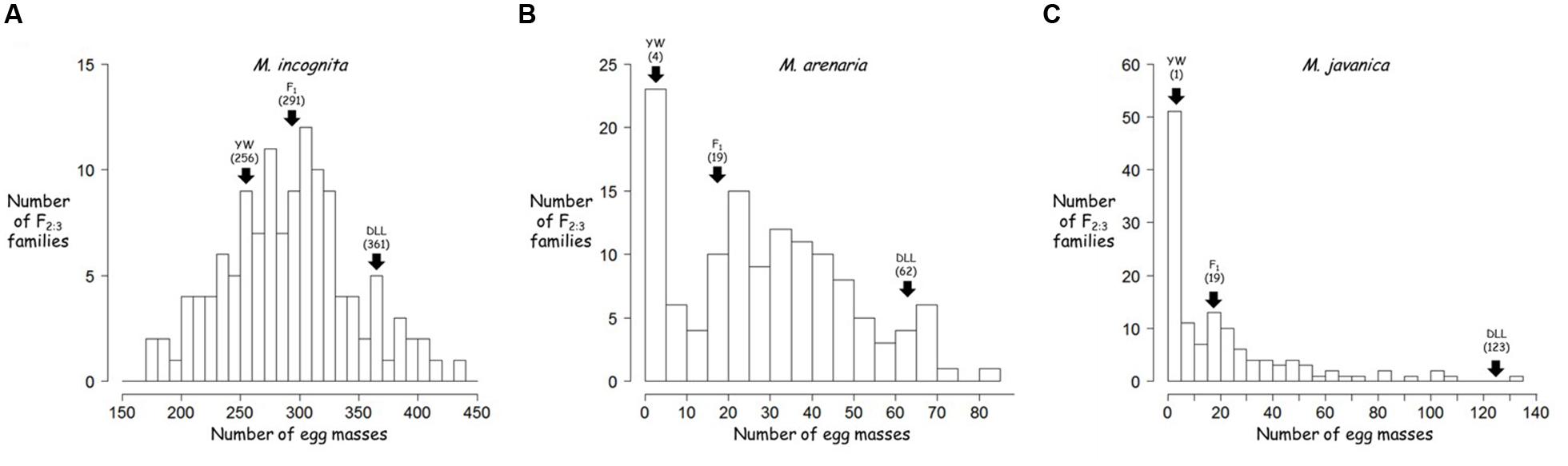
FIGURE 2. Frequency distribution of the resistance to different root-knot nematode (RKN) species of the pepper F2:3 families: (A) M. incognita; (B) M. arenaria; and (C) M. javanica. Arrows indicate the position of each parent and the F1 in the phenotypic distribution, with their values indicated in brackets. YW, Yolo Wonder; DLL, Doux Long des Landes; F1, (YW × DLL).
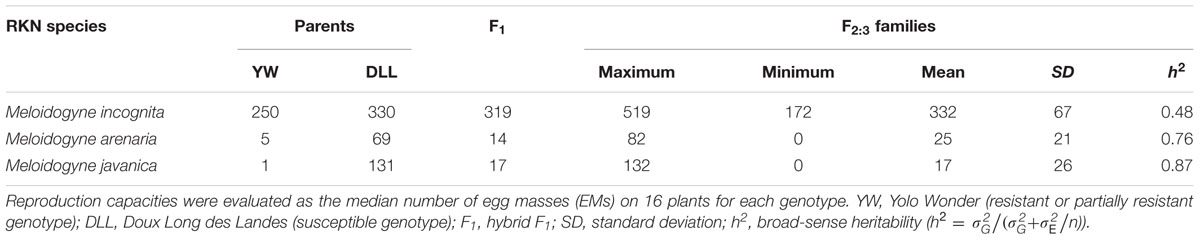
TABLE 1. Summary of root-knot nematode (RKN) reproduction capacity in the parental lines, F1 and F2:3 progeny (130 F2:3 families) from the pepper cross (YW × DLL), for different RKN species.
For the experiment with M. arenaria, ANOVA revealed a significant effect of genotype (p-value = 3.26e-15), but no significant block effect (p-value = 0.0805). YW and DLL were resistant and susceptible (mean values of 5 and 69 EMs/plant, respectively), as expected (Table 1). The F1 phenotype was skewed toward YW (14 EMs/plant), with a d/a ratio of 0.72, indicating that resistance was additive to partly dominant in favor of resistance. The values for the F2:3 families were not normally distributed, as confirmed by a Shapiro–Wilk test (W = 0.92, p-value = 8.63e-07; Figure 2B). The distribution was skewed toward resistance. Neither logarithmic nor square-root transformation resulted in normality (data not shown). Resistance to M. arenaria was highly heritable, as h2 was 0.76.
For the experiment with M. javanica, ANOVA showed a significant effect of genotype (p-value < 2.00e-16) but no significant block effect (p > 0.183). YW was highly resistant and DLL was susceptible, as expected (means of 1 and 131 EMs/plant, respectively; Table 1). The F1 displayed an intermediate phenotype, with skewing toward YW (mean of 17 EMs/plant), with a d/a ratio equal to 0.75, indicating that resistance was mostly dominant, but slightly additive. The Shapiro–Wilk test indicated that the data for the F2:3 families were not normally distributed (W = 0.66, p-value = 2.80e-15; Figure 2C). Neither logarithmic nor square-root transformation yielded normality. Heritability was high for resistance to M. javanica (h2 = 0.87).
Mapping QTLs for Resistance to the Three Meloidogyne Isolates
Simple interval mapping (SIM), composite interval mapping (CIM) and non-parametric (“np” or Kruskal–Wallis) analysis were performed to identify QTLs for resistance. However, as CIM and non-parametric analyses did not improve QTL detection, only the results for SIM are shown. Only one QTL for resistance to M. incognita was detected on pepper chromosome P1, with a LODmax at 179.2 cM and an R2 value of 40.9, corresponding to 85% of the heritability (h2 = 0.48; Figure 3; Table 2). This QTL was named Minc-P1. The d/a ratio of –0.38 at the closest marker (SP1790) indicates a mostly additive effect of this QTL, with a partial dominance effect for susceptibility.
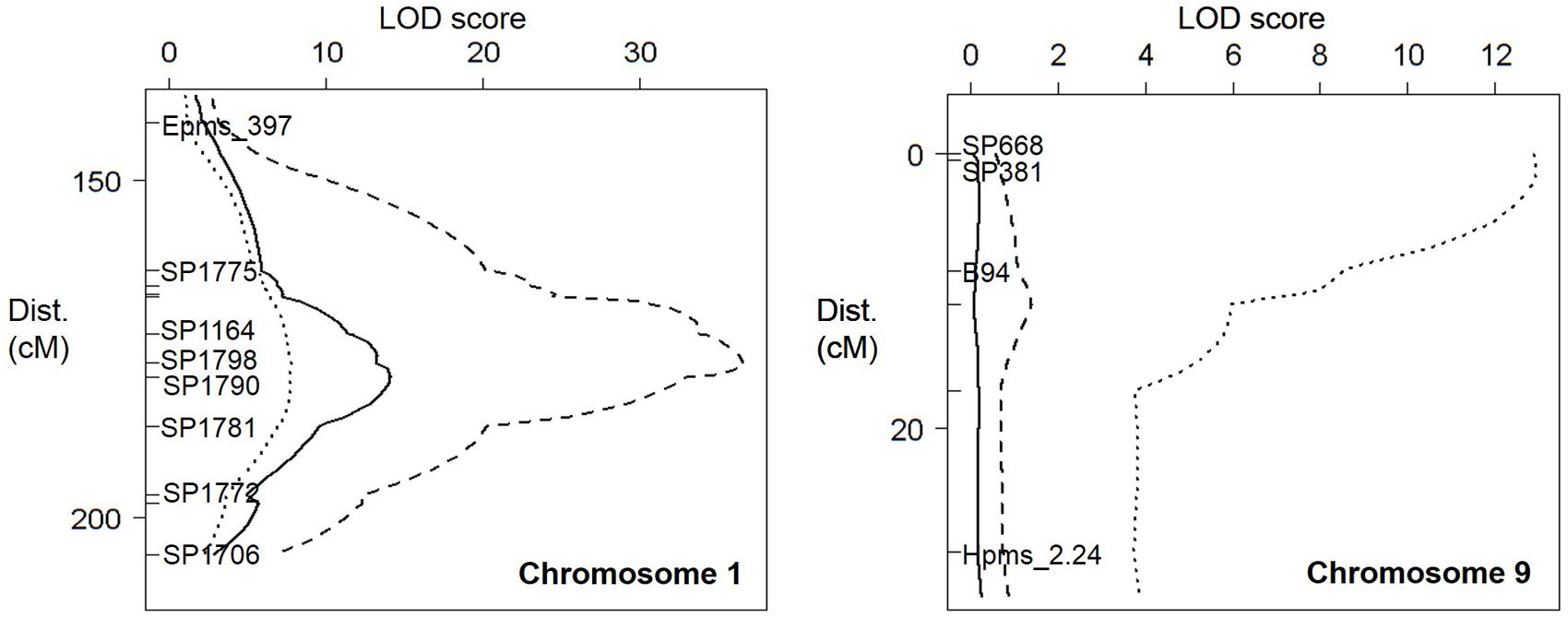
FIGURE 3. Quantitative trait loci (QTLs) against M. incognita (solid line), M. arenaria (dashed line), and M. javanica (dotted line) on pepper chromosome 1 (Left) and chromosome 9 (Right). On the left of each linkage group (LG), distances in centimorgans, flanking markers, and the marker closest to the resistance factors are indicated.
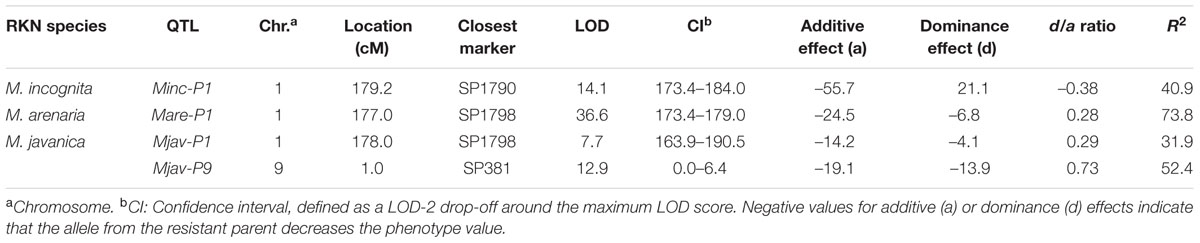
TABLE 2. Quantitative trait loci (QTLs) for resistance to the different RKN species in the pepper F2:3 progeny.
Similarly, only one QTL for resistance to M. arenaria was detected and mapped to chromosome P1 (Mare-P1) with a LODmax at 177.0 cM, close to Minc-P1. This QTL, with an R2 value of 73.8 (97% of the h2 value) and a d/a ratio of 0.28, acts as a major additive QTL, with a weak dominance effect in favor of resistance.
Two QTLs for resistance to M. javanica were detected. The first, Mjav-P1, was located on chromosome P1 with a LODmax at 178.0 cM, an R2 of 31.9 and a d/a ratio of 0.29, indicating a mostly additive effect. The second QTL, Mjav-P9, was detected on the distal part of chromosome P9 (closest marker SP381) with an R2 of 52.4 and a d/a of 0.73, indicating a mostly dominant effect in favor of resistance. Together, Mjav-P1 and Mjav-P9 explained 61.2% of the phenotypic variance, corresponding to 71% of the h2 value.
All the resistance-conferring alleles at these four QTLs originated from the partially resistant parent YW.
Looking for Recombinant Individuals Within the P1 QTL Cluster
F2 individuals with recombinant genotypes for the QTL cluster on chromosome P1 were surveyed, focusing on their genotypes for the markers and the phenotypes of their F3 progenies (homogeneous resistant, homogeneous susceptible, or segregating). Four F2 individuals were found to be recombinant for the alleles at the markers within the P1 QTL cluster (recombination between SP1790 and SP1798). Two F2:3 progenies for these individuals clearly corresponded to phenotypic recombinants in terms of their resistance to M. arenaria and M. incognita, as attested by the phenotypes of the F3 progenies (Table 3). Resistance to M. javanica was less informative, probably due to the major effect of the second locus Mjav-P9.
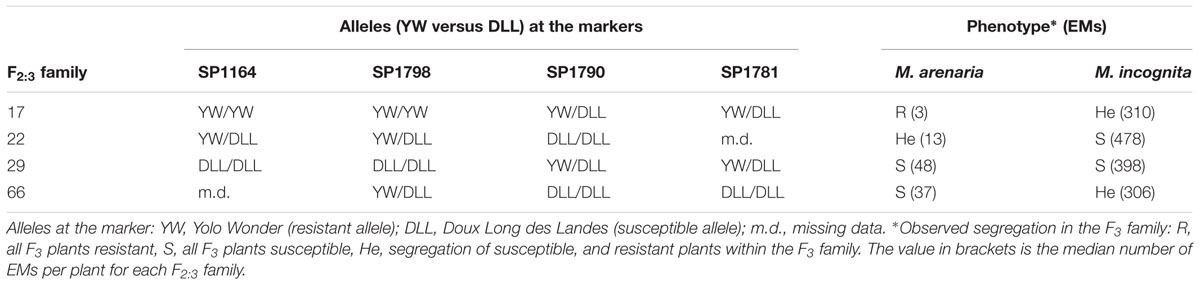
TABLE 3. Recombinant genotypes within the cluster of QTLs on chromosome P1 containing Minc-P1 and Mare-P1.
Discussion
The pepper genetic map constructed in this study with 130 F2 plants from the cross between YW and DLL comprised 12 LGs, consistent with the known number of chromosomes in pepper, and it covered a total length of 1436 cM, consistent with previous maps for pepper (Lefebvre et al., 2002; Paran et al., 2004; Wu et al., 2009). However, as only a few of the previously used markers proved to be polymorphic between the parental lines YW and DLL, new SNPs had to be developed to complete the map. The sequences supporting the SNPs targeting the QTLs are provided in Supplementary Data 2. This new mapping population was developed because it was shown in a previous study that R-genes are more effective and durable against RKN attacks when introgressed into the YW genetic background than when introgressed into the DLL genetic background (Barbary et al., 2014; Djian-Caporalino et al., 2014). This difference is thought to result from the partial resistance alleles carried by YW, which seem to protect the R-genes. This new map was constructed to identify these resistance factors and associated molecular markers, which should constitute valuable resources for further MAS.
The 130 F2:3 [YW × DLL] families were tested against the three main RKN species, M. incognita, M. arenaria, and M. javanica. The QTL analyses identified four new major QTLs affecting reproductive capacity, located in two separate clusters. No minor-effect QTL was detected, and the phenotypic variance explained by the major QTLs for each resistance trait closely fitted the h2 values (71–97%), indicating that almost all the genetic variance was explained by these major QTLs. Three of these QTLs were grouped on pepper chromosome P1, with overlapping CIs, in a 30 cM region. The YW alleles at these QTLs each confer resistance to a single species of RKN: M. incognita, M. arenaria, or M. javanica. These are the first QTLs conferring resistance to RKNs to have been detected in pepper, but QTLs conferring resistance to Meloidogyne spp., have already been mapped in soybean (Li et al., 2001), cotton (Shen et al., 2006), and cowpea (Muchero et al., 2009). This is also the first report of nematode resistance factors mapping to a genomic location other than chromosome P9 in pepper (i.e., on chromosome P1). All the RKN R-genes previously identified in pepper mapped to a cluster on P9 (Djian-Caporalino et al., 2007; Fazari et al., 2012). It is unclear whether Minc-P1, Mare-P1, and Mjav-P1 on P1 are all part of a single gene with a broad spectrum of action, or whether they belong to separate genes forming a new cluster, as observed on P9 for Me3, Me1, and N, which have different spectra of action against RKN species (Hendy et al., 1985; Thies and Fery, 2000). However, our results provide two lines of evidence in support of these QTLs belonging to different genes within a cluster. Firstly, for the Mare-P1 and Mjav-P1 QTLs, the resistant YW allele displayed partial dominance, whereas partial dominance of the susceptible allele from DLL was observed for Minc-P1, suggesting different modes of action. Secondly, F2 individuals displaying genetic recombination between the markers at the peak values of the Minc-P1 and Mare-P1 loci were detected and the phenotypes of the corresponding F3 families confirmed recombination in the F2 plant, with a homozygous resistant or susceptible genotype at one QTL and a heterozygous genotype at the other QTL (Table 3). Despite their very tight linkage, these QTLs thus probably belong to different genes conferring different specificities against RKN populations. These findings provide further evidence that broad-spectrum quantitative resistance can result from the combination of race-specific resistance factors.
An additional major QTL, Mjav-P9, with a major dominant effect for resistance against M. javanica, was mapped to pepper chromosome P9, close to the B94 marker. The detection of a new resistance factor at this site was not unexpected, because the P9 genomic region also carries the Me and N genes (Fazari et al., 2012), together with the R-gene Bs2, which confers resistance to the bacterial pathogen Xanthomonas campestris pv. Vesicatoria (Mazourek et al., 2009), and QTLs for resistance to potyvirus PVY (Wang et al., 2008) and to the oomycete Phytophthora capsici (Thabuis et al., 2004). The genomic sequence of this P9 chromosomal region also has a high density of NBS–LRR genes from the Bs2 subclass (82 genes), highlighting the “explosive expansion of the pepper genome” relative to those of other Solanaceae species (Kim et al., 2014). This expansion, which probably involved tandem duplications, resulted in diversification and a clustering of the R-genes on chromosome P9, and the Mjav-P9 QTL, which has a mostly dominant effect, probably belongs to this cluster.
Broad-spectrum R-genes are often preferentially used in breeding programs, but previous studies have shown that the use of R-genes in an inappropriate genetic background may decrease their efficacy, in turn affecting their durability. The strategy of combining an R-gene with a partially resistant genetic background (i.e., a background carrying relevant QTLs) to increase its durability has been evaluated and validated in other pathosystems (Palloix et al., 2009; Brun et al., 2010; Fournet et al., 2013). Quenouille et al. (2012) suggested that this effect was due mostly to the additional resistance conferred by QTLs from the genetic background, decreasing the size of the pathogen population and, thus, the risk of emergence and of the further selection of virulent variants. For interactions between pepper and RKNs, YW proved to be a better genetic background than DLL for strengthening the efficacy of Me1 or Me3 (Barbary et al., 2014) and reducing the frequency of Me3 resistance breakdown (Djian-Caporalino et al., 2014). However, the genetic determinants of this partial resistance had never been characterized. The new QTLs identified in this study (i.e., Minc-P1, Mare-P1, Mjav-P1, and Mjav-P9) are, thus, good candidates for pyramiding with Me1, Me3, or N, providing new opportunities for combining major and partial resistance factors together in pepper cultivars.
In terms of plant breeding, the location of the newly identified resistance factors on pepper chromosome P1 should facilitate their introgression by MAS, alongside current R-genes. Indeed, all the resistance genes effective against RKNs mapped to date are closely linked on pepper chromosome P9 (Fazari et al., 2012). However, they are in repulsion phases, with the different genes carried by different pepper accessions. Minc-P1, Mare-P1, and Mjav-P1 are independent of this cluster and all are carried by the same accession (YW). This should make it easier to generate homozygous plant genotypes harboring resistance factors from both the P9 and P1 clusters. Breeders could also make use of Minc-P1, Mare-P1, and Mjav-P1, which do not act as fully dominant R-genes and may act through different resistance mechanisms. He et al. (2014) reported two QTLs affecting two different processes in RKN attack on cotton plants: root galling and egg production. These QTLs provided strong resistance to RKNs when combined in the same genotype. We are currently investigating this aspect in our pathosystem, by performing histological time-course studies to explore the spatiotemporal induction of plant responses conferred by these new resistance factors in pepper. In particular, the kinetics of J2 invasion in the roots, and the timing and location of cell necrosis (if any) will be investigated. On the basis of our preliminary observations, we hypothesize that the newly identified QTLs will induce defense reactions different from the classical HR triggered by the major R-genes Me1 and Me3. We therefore anticipate that the successful combination of these qualitative and quantitative resistance factors into elite cultivars will provide new opportunities for enhanced and durable RKN resistance in pepper.
Author Contributions
AB, PC-S, CD-C, AP, and BC conceived the project, contribute to technical work, to data treatment and article writing, AF and NM organized and performed technical work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Anne-Marie Sage-Palloix and Ghislaine Nemouchi for plant breeding and for providing F3 families seeds. We also acknowledge Delphine Angella, Chami Kim, Cristina Martin Jimenez, and Pierre Bautheac for technical assistance. This work was financially supported by the French Ministry of Higher Education and Research (MESR) and the seed companies Gautier Semences, Rijk Zwaan France, Sakata Vegetables Europe, Syngenta Seeds, Takii France, and Vilmorin & Cie in the frame of a CIFRE contract.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00632
Footnotes
References
Alimi, N. A., Bink, M., Dieleman, J. A., Magan, J. J., Wubs, A. M., Palloix, A., et al. (2013). Multi-trait and multi-environment QTL analyses of yield and a set of physiological traits in pepper. Theor. Appl. Genet. 126, 2597–2625. doi: 10.1007/s00122-013-2160-3
Barbary, A., Palloix, A., Fazari, A., Marteu, N., Castagnone-Sereno, P., and Djian-Caporalino, C. (2014). The plant genetic background affects the efficiency of the pepper major nematode resistance genes Me1 and Me3. Theor. Appl. Genet. 127, 499–507. doi: 10.1007/s00122-013-2235-1
Bleve-Zacheo, T., Bongiovanni, M., Melillo, M. T., and Castagnone-Sereno, P. (1998). The pepper resistance genes Me1 and Me3 induce differential penetration rates and temporal sequences of root cell ultrastructural changes upon nematode infection. Plant Sci. 133, 79–90. doi: 10.1016/S0168-9452(98)00021-1
Broman, K., Wu, W. H., Sen, S., and Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. doi: 10.1093/bioinformatics/btg112
Brun, H., Chevre, A. M., Fitt, B. D., Powers, S., Besnard, A. L., Ermel, M., et al. (2010). Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 185, 285–299. doi: 10.1111/j.1469-8137.2009.03049.x
Castagnone-Sereno, P. (2006). Genetic variability and adaptive evolution in parthenogenetic root-knot nematodes. Heredity (Edinb.) 96, 282–289. doi: 10.1038/sj.hdy.6800794
Castagnone-Sereno, P., Bongiovanni, M., Palloix, A., and Dalmasso, A. (1996). Selection for Meloidogyne incognita virulence against resistance genes from tomato and pepper and specificity of the virulence/resistance determinants. Eur. J. Plant Pathol. 102, 585–590. doi: 10.1007/BF01877026
Dalmasso, A., and Berge, J. B. (1978). Molecular polymorphism and phylogenetic relationship in some Meloidogyne spp.: application to the taxonomy of Meloidogyne. J. Nematol. 10, 323–332.
Djian-Caporalino, C., Fazari, A., Arguel, M. J., Vernie, T., Van de Casteele, C., Faure, I., et al. (2007). Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor. Appl. Genet. 114, 473–486. doi: 10.1007/s00122-006-0447-3
Djian-Caporalino, C., Palloix, A., Fazari, A., Marteu, N., Barbary, A., Abad, P., et al. (2014). Pyramiding, alternating or mixing: comparative performances of deployment strategies of nematode resistance genes to promote plant resistance efficiency and durability. BMC Plant Biol. 14:53. doi: 10.1186/1471-2229-14-53
Djian-Caporalino, C., Pijarowski, L., Januel, A., Lefebvre, V., Daubèze, A. M., Palloix, A., et al. (1999). Spectrum of resistance to root-knot nematodes and inheritance of heat-stable resistance in pepper (Capsicum annuum L.). Theor. Appl. Genet. 99, 496–502. doi: 10.1007/s001220051262
Fazari, A., Palloix, A., Wang, L. H., Hua, M. Y., Sage-Palloix, A. M., Zhang, B. X., et al. (2012). The root-knot nematode resistance N-gene co-localizes in the Me-genes cluster on the pepper (Capsicum annuum L.) P9 chromosome. Plant Breed. 131, 665–673. doi: 10.1111/j.1439-0523.2012.01994.x
Fournet, S., Kerlan, M. C., Renault, L., Dantec, J. P., Rouaux, C., and Montarry, J. (2013). Selection of nematodes by resistant plants has implications for local adaptation and cross-virulence. Plant Pathol. 62, 184–193. doi: 10.1111/j.1365-3059.2012.02617.x
Fulton, T. M., Chunwongse, J., and Tanksley, S. D. (1995). Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13, 207–209. doi: 10.1007/BF02670897
Goodey, T., Goodey, J. B., Franklin, M. T., and Hooper, D. J. (1965). The Nematode Parasites of Plants Catalogued Under Their Hosts, 3rd Edn. Farnham Royal: Commonwealth Agricultural Bureaux, 214.
Hare, W. W. (1956). Resistance in pepper to Meloidogyne incognita acrita. Phytopathology 46, 98–104.
He, Y., Kumar, P., Shen, X., Davis, R. F., Van Becelaere, G., May, O. L., et al. (2014). Re-evaluation of the inheritance for root-knot nematode resistance in the upland cotton germplasm line M-120 RNR revealed two epistatic QTLs conferring resistance. Theor. Appl. Genet. 127, 1343–1351. doi: 10.1007/s00122-014-2302-2
Hendy, H., Pochard, E., and Dalmasso, A. (1985). Transmission héréditaire de la résistance aux Meloidogyne portée par deux lignées de Capsicum annuum: études de descendances d’homozygotes issues d’androgénèse. Agronomie 5, 93–100. doi: 10.1051/agro:19850201
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., and Lee, H. A. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kiyosawa, S. (1982). Genetics and epidemiological modeling of breakdown of plant disease resistance. Annu. Rev. Phytopathol. 20, 93–117. doi: 10.1146/annurev.py.20.090182.000521
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Lefebvre, V., Pflieger, S., Thabuis, A., Caranta, C., Blattes, A., Chauvet, J. C., et al. (2002). Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome 45, 839–854. doi: 10.1139/g02-053
Li, Z., Jakkula, L., Hussey, R. S., Tamulonis, J. P., and Boerma, H. R. (2001). SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. Theor. Appl. Genet. 103, 1167–1173. doi: 10.1007/s001220100672
Mazourek, M., Cirulli, E. T., Collier, S. M., Landry, L. G., Kang, B. C., Quirin, E. A., et al. (2009). The fractionated orthology of Bs2 and Rx/Gpa2 supports shared synteny of disease resistance in the Solanaceae. Genetics 182, 1351–1364. doi: 10.1534/genetics.109.101022
Muchero, W., Matthews, W. C., Diop, N. N., Bhat, P. R., Wanamaker, S., Ehlers, J. D., et al. (2009). QTL mapping of root-knot nematode resistance in cowpea (Vigna unguicula) using EST derived SNP markers. J. Nematol. 41, 361–362.
Mundt, C. C. (2002). Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Phytopathol. 40, 381–410. doi: 10.1146/annurev.phyto.40.011402.113723
Nicolaï, M., Pisani, C., Bouchet, J. P., Vuylsteke, M., and Palloix, A. (2012). Discovery of a large set of SNP and SSR genetic markers by high-throughput sequencing of pepper (Capsicum annuum). Genet. Mol. Res. 11, 2295–2300. doi: 10.4238/2012.August.13.3
Palloix, A., Ayme, V., and Moury, B. (2009). Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 183, 190–199. doi: 10.1111/j.1469-8137.2009.02827.x
Paran, I., Rouppe Van Der Voort, J., Lefebvre, V., Jahn, M., Landry, L., Van Schriek, M., et al. (2004). An integrated genetic linkage map of pepper (Capsicum spp.). Mol. Breed. 13, 251–261.
Pegard, A., Brizzard, G., Fazari, A., Soucaze, O., Abad, P., and Djian-Caporalino, C. (2005). Histological characterization of resistance to different root-knot nematode species related to phenolics accumulation in Capsicum annuum. Phytopathology 95, 158–165. doi: 10.1094/PHYTO-95-0158
Pink, D. (2002). Strategies using genes for non-durable disease resistance. Euphytica 124, 227–236. doi: 10.1023/A:1015638718242
Quenouille, J., Montarry, J., Palloix, A., and Moury, B. (2012). Farther, slower, stronger: how the plant genetic background protects a major resistance gene from breakdown. Mol. Plant Pathol. 14, 109–118. doi: 10.1111/j.1364-3703.2012.00834.x
Quenouille, J., Paulhiac, E., Moury, B., and Palloix, A. (2014). Quantitative trait loci from the host genetic background modulate the durability of a resistance gene: a rational basis for sustainable resistance breeding in plants. Heredity (Edinb.) 112, 579–587. doi: 10.1038/hdy.2013.138
Shen, X. L., Van Becelaere, G., Kumar, P., Davis, R. F., May, O. L., and Chee, P. (2006). QTL mapping for resistance to root-knot nematodes in the M-120 RNR upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theor. Appl. Genet. 113, 1539–1549. doi: 10.1007/s00122-006-0401-4
Stuber, C. W., Edwards, M. D., and Wendel, J. F. (1987). Molecular marker-facilitated investigations of quantitative trait loci in maize. II. Factors influencing yield and its component traits. Crop Sci. 27, 639–648. doi: 10.2135/cropsci1987.0011183X002700040006x
Thabuis, A., Lefebvre, V., Bernard, G., Daubèze, A. M., Phaly, T., Pochard, E., et al. (2004). Phenotypic and molecular evaluation of a recurrent selection program for a polygenic resistance to Phytophthora capsici in pepper. Theor. Appl. Genet. 109, 342–351. doi: 10.1007/s00122-004-1633-9
Thies, J. A. (2011). Virulence of Meloidogyne incognita to expression of N gene in pepper. J. Nematol. 43, 90–94.
Thies, J. A., and Fery, R. L. (1998). Modified expression of the N gene for southern root-knot nematode resistance in pepper at high soil temperatures. J. Am. Soc. Hortic Sci. 123, 1012–1015.
Thies, J. A., and Fery, R. L. (2000). Characterization of resistance conferred by the N gene to Meloidogyne arenaria Races 1 and 2, M. hapla, and M. javanica in two sets of isogenic lines of Capsicum annuum L. J. Am. Soc. Hortic. Sci. 125, 71–75.
Wang, L. H., Zhang, B., Caranta, C., Mao, S., and Palloix, A. (2008). Molecular markers assisted selection for three QTLs resistant to PVY in pepper (Capsicum annuum L.). Acta Hortic. Sin. 35, 53–58.
Wu, F., Eannetta, N. T., Xu, Y., Durrett, R., Mazourek, M., Jahn, M. M., et al. (2009). A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor. Appl. Genet. 118, 1279–1293. doi: 10.1007/s00122-009-0980-y
Keywords: Capsicum annuum, Meloidogyne spp., quantitative resistance, major resistance, resistance durability
Citation: Barbary A, Djian-Caporalino C, Marteu N, Fazari A, Caromel B, Castagnone-Sereno P and Palloix A (2016) Plant Genetic Background Increasing the Efficiency and Durability of Major Resistance Genes to Root-knot Nematodes Can Be Resolved into a Few Resistance QTLs. Front. Plant Sci. 7:632. doi: 10.3389/fpls.2016.00632
Received: 04 December 2015; Accepted: 25 April 2016;
Published: 10 May 2016.
Edited by:
Rex Brennan, James Hutton Institute, ScotlandCopyright © 2016 Barbary, Djian-Caporalino, Marteu, Fazari, Caromel, Castagnone-Sereno and Palloix. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippe Castagnone-Sereno, cGhpbGlwcGUuY2FzdGFnbm9uZUBzb3BoaWEuaW5yYS5mcg==
 Arnaud Barbary1
Arnaud Barbary1 Bernard Caromel
Bernard Caromel Philippe Castagnone-Sereno
Philippe Castagnone-Sereno Alain Palloix
Alain Palloix