- 1Department of Plant Science, Plant Genomics and Breeding Institute, and Vegetable Breeding Research Center, College of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 2Center for Eco-friendly New Materials, Korea Research Institute of Chemical Technology, Daejeon, South Korea
- 3Department of Plant Science, College of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
Powdery mildew, caused by Leveillula taurica, is a major fungal disease affecting greenhouse-grown pepper (Capsicum annuum). Powdery mildew resistance has a complex mode of inheritance. In the present study, we investigated a novel powdery mildew resistance locus, PMR1, using two mapping populations: 102 ‘VK515' F2:3 families (derived from a cross between resistant parental line ‘VK515R' and susceptible parental line ‘VK515S') and 80 ‘PM Singang' F2 plants (derived from the F1 ‘PM Singang' commercial hybrid). Genetic analysis of the F2:3 ‘VK515' and F2 ‘PM Singang' populations revealed a single dominant locus for inheritance of the powdery mildew resistance trait. Genetic mapping showed that the PMR1 locus is located on syntenic regions of pepper chromosome 4 in a 4-Mb region between markers CZ2_11628 and HRM4.1.6 in ‘VK515R'. Six molecular markers including one SCAR marker and five SNP markers were localized to a region 0 cM from the PMR1 locus. Two putative nucleotide-binding site leucine-rich repeat (NBS-LRR)-type disease resistance genes were identified in this PMR1 region. Genotyping-by-sequencing (GBS) and genetic mapping analysis revealed suppressed recombination in the PMR1 region, perhaps due to alien introgression. In addition, a comparison of species-specific InDel markers as well as GBS-derived SNP markers indicated that C. baccatum represents a possible source of such alien introgression of powdery mildew resistance into ‘VK515R'. The molecular markers developed in this study will be especially helpful for marker-assisted selection in pepper breeding programs for powdery mildew resistance.
Introduction
Leveillula taurica, an obligate fungal plant pathogen belonging to the ascomycetes, causes powdery mildew in various vegetable crops, resulting in significant quality and yield losses. In the past few decades, the incidence of L. taurica powdery mildew has been increasing in both greenhouse- and open field-grown peppers worldwide (Damicone, 2009; Sudha and Lakshmanan, 2009; Cerkauskas et al., 2011). Premature defoliation caused by powdery mildew severely reduces crop yields and makes fruits unfavorable for marketing. Unlike most fungi that cause powdery mildew, which are epiphytic, L. taurica is an endophytic fungus, which hinders the efficacy of chemical control (Elad et al., 2007). Therefore, developing powdery mildew disease resistance in pepper is one of the main objectives of genetic and breeding programs.
Introgressing disease resistance from wild species into popular cultivars greatly increases crop yields and quality. Resistance sources for various pepper diseases have been reported in several wild species, and resistance has been successfully introgressed into commercial pepper cultivars, including resistance to tobamoviruses from Capsicum chacoense and C. chinense (Boukema, 1980; Berzal-Herranz et al., 1995; de la Cruz et al., 1997), resistance to tomato spotted wilt virus (TSWV) from C. chinense and C. baccatum (Boiteux et al., 1993; Hoang et al., 2013; Soler et al., 2015), resistance to anthracnose fruit rot from C. chinense (Voorrips et al., 2004), resistance to Phytophthora capsici from C. annuum cv. CM334 (Mallard et al., 2013; Liu et al., 2014) and resistance to bacterial leaf spot disease from C. annuum and C. chacoense (Cook and Guevara, 1984; Kim and Hartmann, 1985; Hibberd et al., 1987; Vallejos et al., 2010).
Pepper genotypes showing varied resistance levels against powdery mildew have been identified in C. annuum, C. frutescens, C. baccatum, and C. chinense (Ullassa et al., 1981; Deshpande et al., 1985; Pochard et al., 1986; Anand et al., 1987; De Souza and Café-Filho, 2003). Pepper genotypes ‘H-V-12' and ‘4638' (C. annuum), ‘IHR 703' (C. frutescens), and CNPH 36, 38, 50, 52, 279, and 288 (C. baccatum) are resistant to L. taurica (Anand et al., 1987; De Souza and Café-Filho, 2003). According to De Souza and Café-Filho (2003), most C. annuum species are moderately to highly susceptible to powdery mildew, whereas C. baccatum, C. chinense, and C. frutescens species are often resistant, suggesting that among Capsicum species, resistance to powdery mildew is primarily found in taxa other than C. annuum.
Powdery mildew resistance in pepper is reported to be a dominant and polygenic trait (Anand et al., 1987; Murthy and Deshpande, 1997; Blat et al., 2005). Genetic analyses have also indicated that relatively few genetic factors with significant additive and epistatic effects confer resistance to powdery mildew in different pepper genetic backgrounds (Daubèze et al., 1995; Murthy and Deshpande, 1997; Blat et al., 2005, 2006). At least three pairs of incompletely dominant genes are thought to confer resistance to powdery mildew in the C. frutescens line ‘IHR 703' (Anand et al., 1987). The most important and durable source of powdery mildew resistance reported is in the small-fruited pungent C. annuum accession ‘H3' from Ethiopia (Daubèze et al., 1995; Lefebvre et al., 2003). In addition, the L. taurica-resistant Israeli pepper line ‘H-V-12' was derived from a cross between the resistant cultivar ‘H3' and the susceptible cultivar ‘Vania' (Shifriss et al., 1992); at least three genes appear to control resistance to L. taurica in ‘H3' (Shifriss et al., 1992; Daubèze et al., 1995). Lefebvre et al. (2003) identified seven genomic regions, including additive quantitative trait loci (QTLs) and epistatic loci, that contribute to the resistance of the cultivar ‘H3' using a double-haploid population derived from a cross between ‘H3' and “Vania.”
In the present study, we identified and mapped a novel powdery mildew resistance locus, PMR1, to pepper chromosome 4 using two segregating populations: 102 ‘VK515' F2:3 families derived from a cross between resistant parental line ‘VK515R' and susceptible parental line ‘VK515S' and 80 ‘PM Singang' F2 lines derived from the F1 ‘PM Singang' commercial hybrid. Based on genetic analysis, the PMR1 locus was localized to 1.0 and 5.1 cM genetic intervals on lower arm of chromosome 4 in ‘VK515R' and ‘PM Singang', respectively. In addition, we performed InDel marker sequence comparisons and GBS analyses to infer the origin of PMR1. Identification of molecular markers linked to the PMR1 locus in pepper in the present study should facilitate marker-assisted selection (MAS) in pepper breeding programs aimed at introgressing the powdery mildew disease resistance trait.
Materials and Methods
Plant Materials
C. annuum ‘VK515R' and ‘VK515S' are powdery mildew-resistant and susceptible parental lines, respectively, kindly provided by In-Tae Kim (Samsung Seeds Co., Ltd., Korea). C. annuum ‘PM Singang' is a powdery mildew resistant commercial F1 hybrid cultivar (Nongwoo Bio Co., Ltd., Korea). C. annuum ‘Bukang' is a susceptible commercial cultivar (Hungnong Seed Co., Korea). ‘VK515' families and the ‘PM Singang' F2 population were used to map the PMR1 locus. ‘VK515' F1 plants were derived from a cross between ‘VK515R' and ‘VK515S' lines. The ‘VK515' and ‘PM Singang' F1 hybrids were self-pollinated to produce F2 seeds in a greenhouse in 2012 and 2014, respectively. ‘VK515' F2 plants were grown in separate pots without selection and harvested individually to produce F2:3 families in 2015. 80 ‘PM Singang' F2 individuals and 102 ‘VK515' F2:3 families with 20 individuals per family were used for subsequent disease evaluation and mapping.
Inoculum Preparation and Disease Infection
For inoculum preparation, white powdery mildew spores collected from naturally-infected plants were used to inoculate the abaxial sides of leaves via the dropping method (Kim et al., 2009). Powdery mildew-infected ‘Bukang' and ‘VK515S' plants were maintained in greenhouses (Seoul National University, Suwon, Korea) and used as a source of L. taurica. At the time of sowing of ‘VK515' F2:3 families in plastic trays (50 cell trays), infectious ‘Bukang' and ‘VK515S' plants were introduced at regular intervals. In the case of the ‘PM Singang' F2 population, plants were raised in 20 cm diameter plastic pots and evaluated for resistance to powdery mildew under natural infection conditions (Korea Research Institute of Chemical Technology, Daejeon, Korea).
Disease Evaluation
‘VK515' F2:3 families and ‘PM Singang' F2 plants were evaluated for powdery mildew disease resistance 60 days after sowing. ‘VK515R' and ‘PM Singang' served as the resistant controls, whereas ‘VK515S' and ‘Bukang' served as the susceptible controls. In ‘VK515' F2:3 families, the presence or absence of white fungal hyphae on infected leaves at 60 days after sowing was used as a measure of disease infection. For ‘PM Singang' F2 plants, intensity of white fungal hyphae observed on infected leaves was visually scored using following disease scale: 1 = no sign of disease; 2 = minute necrotic lesions with no detectable sporulation; 3 = few large sporulating lesions; 4 = numerous large sporulating lesions.
Genomic DNA Extraction
Total genomic DNA (gDNA) was extracted from young leaf tissue using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). The quality and quantity of the gDNA were analyzed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Chromosomal Localization
Four hundred twelve previously reported SNP markers (Kang et al., 2014) were used to detect polymorphism between ‘VK515R' and ‘VK515S'. A total of 96 SNP markers with uniform distribution in 12 pepper chromosomes and showing clear polymorphism between ‘VK515R' and ‘VK515S' were selected for the localization study (Table S1). A total of 92 plants were randomly selected from the ‘VK515' 102 F2 population and used for the SNP genotyping assay. Genotyping was performed with the Fluidigm® EP1™ system according to the manufacturer's protocol (Fluidigm, San Francisco, CA, USA). Briefly, specific target amplification (STA) was performed in a 5 μL reaction containing 60 ng of the gDNA according to the manufacturer's recommendations (Wang et al., 2009). PCR cycling conditions were as follows: 95°C for 15 min, followed by 14 cycles of amplification consisting of 95°C for 15 s and 60°C for 2 min. SNPtype assays were carried out using STA products according to the manufacturer's protocol (Fluidigm, San Francisco, CA, USA). PCR thermal cycling conditions were as follows: 95°C for 15 s, 64°C for 45 s and 72°C for 15 s with a touchdown of −1°C per cycle from 64 to 61°C, followed by 34 cycles of 95°C for 15 s, 60°C for 45 s and 72°C for 15 s. Genotyping data were automatically generated from the end-point image of the genotyping chip using the Fluidigm SNP Genotyping Analysis tool.
Marker Development
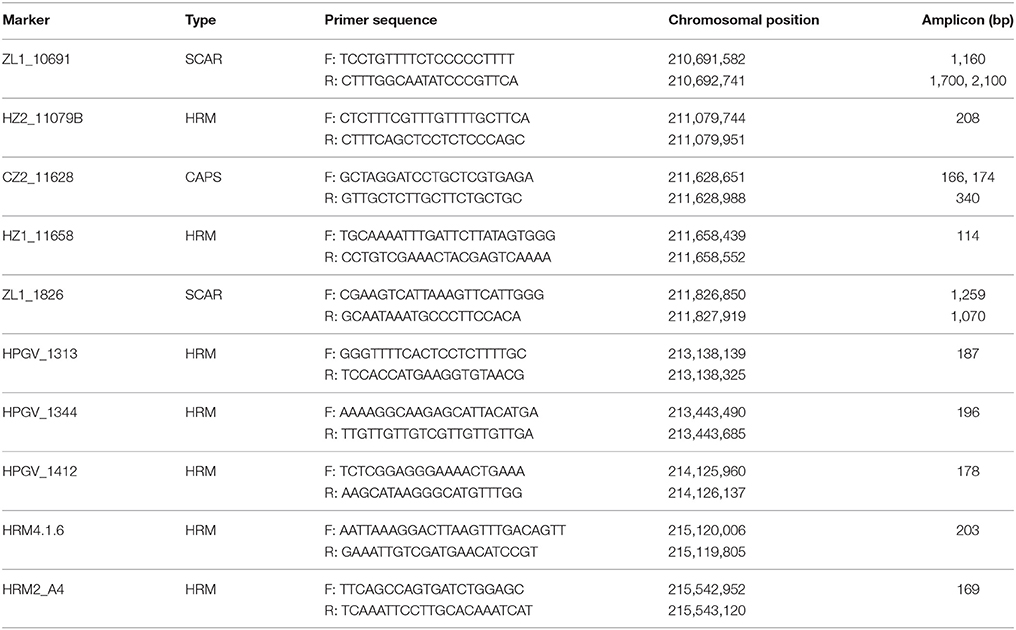
Based on our initial chromosomal localization study, SNP marker KS16052G01 was found to be linked to the powdery mildew resistance trait. To develop additional markers, PCR primers (Tables S2, S3) were designed in a 3.0 cM region around marker KS16052G01 using genomic information for Capsicum (Kim et al., 2014; Qin et al., 2014). To detect polymorphism between the parents, direct sequencing of gDNA PCR products was carried out. PCR analyses were performed according to Liu et al. (2016). The PCR products were gel eluted and sequenced at the National Instrumentation Center for Environmental Management (NICEM), Seoul National University, Seoul, Korea. Polymorphic PCR bands were used to develop SCAR markers, and SNPs were used to develop cleaved amplified polymorphic sequence (CAPS) or high-resolution melt (HRM) markers. CAPS markers were designed using CAPS Designer (https://solgenomics.net/tools/caps_designer/caps_input.pl), and HRM markers were designed according to Park et al. (2009). A total of ten markers including seven HRM, two SCAR and one CAPS markers were used for mapping the PMR1 locus (Table 1).
Genotyping and Molecular Mapping
A total of 102 ‘VK515' F2 individuals, 102 ‘VK515' F2:3 families, and 80 ‘PM Singang' F2 individuals were used to map the PMR1 locus. For genotyping with SCAR markers, PCR was performed in 25 μL reactions, including 2.5 μL 10 × buffer, 2 μL 10 mM dNTPs, 1 μL 10 μM each of forward and reverse primer, 15.2 μL distilled H2O, 0.3 μL Taq DNA polymerase, and 3 μL of 20 ng/μL gDNA. The PCR program was 95°C for 5 min, 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 60 s, followed by a final extension of 72°C for 10 min. For genotyping with CAPS marker (CZ2_11628), PCR was performed as mentioned above, but with an extension time of 72°C for 30 s. Amplified PCR products were digested with the restriction enzyme TaqαI according to the manufacturer's protocol (New England Biolabs, Beverly, MA). Digested PCR products were separated by gel electrophoresis and observed under a UV transilluminator (Bio-Rad, USA).
HRM analysis was carried out using a LightScanner® (Idaho Technology Inc., USA). For HRM analysis, PCR was performed in 20 μL of reaction mixture, including 2 μL 10 × buffer, 0.25 mM dNTPs, 5 pmol primer mix, 1 unit Taq polymerase, 1.25 μM Syto9, and 50 ng gDNA. The PCR program was 95°C for 5 min, 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s, followed by a final extension of 72°C for 10 min. HRM was run using 0.1°C increments between 70 and 90°C, with each increment held for 1 s.
Linkage analysis was performed using CarthaGene Software (Schiex and Gaspin, 1997). To construct a linkage group, the LOD threshold was set at 5.0, and the maximum distance was set at 30 cM. Members of linkage groups were determined based on the physical positions of the markers in ‘L_Zunla-1'. Genetic distances between markers were determined in cM using the Kosambi mapping function. The resulting genetic linkage map was drawn using MapChart 2.3 software (Voorrips, 2002).
Sequence Comparison of PMR1 Locus-Specific InDel Markers
To compare the PMR1 locus-specific InDel markers, the sequences of two PMR1 locus-specific InDel markers (ZL1_1826 and Chr4.1.6) from C. annuum ‘VK515' parents were PCR amplified using ZL1_1826 and Chr4.1.6 primer sets (Tables S2, S3) as described above and sequenced at NICEM, Seoul National University, Seoul, Korea. The PMR1 locus-specific InDel markers sequences from C. annuum ‘L_Zunla-1' (http://peppersequence.genomics.cn), C. chinense ‘PI159236' (version 1.2), and C. baccatum ‘PBC81' (version 1.2) (http://peppergenome.snu.ac.kr/download.php) were retrieved from corresponding genome databases (Kim et al., 2017). InDel marker sequences were aligned using the MAFFT multiple sequence alignment program (http://www.ebi.ac.uk/Tools/msa/mafft/). The sequence alignments were illustrated using the Jalview tool (Waterhouse et al., 2009).
Prediction of PMR1 Candidate Genes
Gene predictions were performed with the FGENESH tool (Solovyev et al., 2006). The predicted protein sequences from the PMR1 region were BLASTP searched against the NCBI database for disease resistance-related genes. Protein domain analysis of the candidate resistance (R) genes from the PMR1 locus was carried out at te Pfam database (http://pfam.xfam.org/).
Genotyping-by-Sequencing (GBS)
GBS was performed using 12 ‘VK515' F2 individuals, including 11 homozygous resistant genotypes and one heterozygous genotype, along with two replications of parents ‘VK515R' and ‘VK515S'. To construct comprehensive and representative libraries for Illumina sequencing, 400 ng of gDNA was used. Libraries for GBS were constructed as described by Truong et al. (2012). Briefly, gDNA from the parental lines (‘VK515R' and ‘VK515S') and 12 F2 lines was digested with PstI and MseI. MseI adapters and the PstI adapter with different barcodes for each sample were ligated to the digested gDNA fragments. After amplification, the quality and quantity of the libraries were evaluated using a Bioanalyzer DNA 1000 Chip (Agilent Technologies, Santa Clara, CA, USA). The same amount of adapter-ligated fragments from each sample was pooled for sequencing.
Sequencing was performed at Macrogen (Seoul, Korea) using a HiSeq 2000 (Illumina, San Diego, CA, USA). Raw reads were demultiplexed in accordance with individual barcodes, and the adapter and barcode sequences were removed using commercially available CLC genomics workbench software version 8.0 (CLC Bio, Aarhus, Denmark). Trimmed reads were mapped to genomic sequences of C. annuum ‘L_Zunla-1' version 2.0 (Qin et al., 2014; http://peppersequence.genomics.cn) using Burrows-Wheeler Aligner (BWA) version 0.7.12 (Li, 2013). Picard Tools version 1.119 and SAMtools version 1.1 were used for read grouping and sorting (Li et al., 2009). For genome-wide SNP calling, Genome Analysis Toolkit (GATK) UnifiedGenotyper version 3.3 was used. High-quality SNPs with QUAL value larger than 30 and minimum depth 3 were selected for further analysis.
The sequence regions corresponding to the PMR1 locus from C. annuum ‘L_Zunla-1', C. chinense, and C. baccatum chromosome sequences were aligned with GBS-SNP data from the ‘VK515' genome. Phylogenetic analysis was carried out using DARwin 6.0.9 (Perrier and Jacquemoud-Collet, 2006).
Results
Inheritance Analysis of Powdery Mildew Resistance
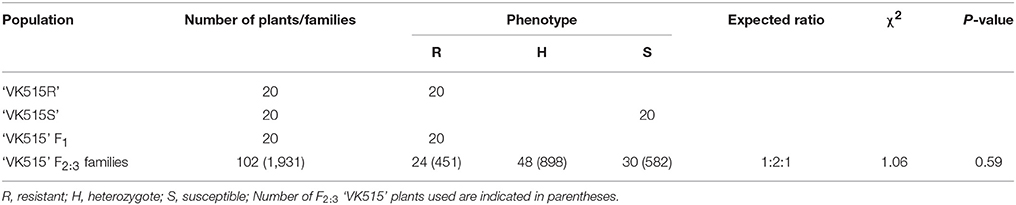
We evaluated ‘VK515' F2:3 families along with resistant (‘VK515R' and ‘PM Singang') and susceptible (‘VK515S' and ‘Bukang') pepper lines infected with L. taurica for powdery mildew disease resistance 60 days after infection. Powdery mildew infection was evident by the appearance of mycelial growth on the leaf surface of susceptible controls. Powdery mildew-resistant ‘PM Singang' and ‘VK515R' failed to show disease symptoms, as no white fungal hyphae were observed on the abaxial surfaces of leaves. Similar results were obtained for ‘VK515' F1 plants, whereas susceptible ‘Bukang' and ‘VK515S' plants showed white fungal hyphae on the abaxial sides of leaves (Figure 1). Among the 102 ‘VK515' F2:3 families, 24 families (total of 451 plants) were homozygous resistant, 48 families (total of 898 plants) were segregating for this trait, and 30 families (total of 582 plants) were homozygous susceptible, which showed a good fit to a 1:2:1 ratio (χ2 = 1.06; P = 0.59) (Table 2). In the case of the ‘PM Singang' F2 population, plants displaying a disease score value of one or two were considered as resistant, whereas plants scored as three or four were considered as susceptible (Figure S1). Out of 80 ‘PM Singang' F2 lines, 59 plants were resistant and 21 plants were susceptible, which fit to a 3:1 ratio (χ2 = 0.02; P = 0.80) (Table S4). Overall, these results suggest that resistance to powdery mildew is controlled by a single dominant locus, PMR1, in these pepper populations.
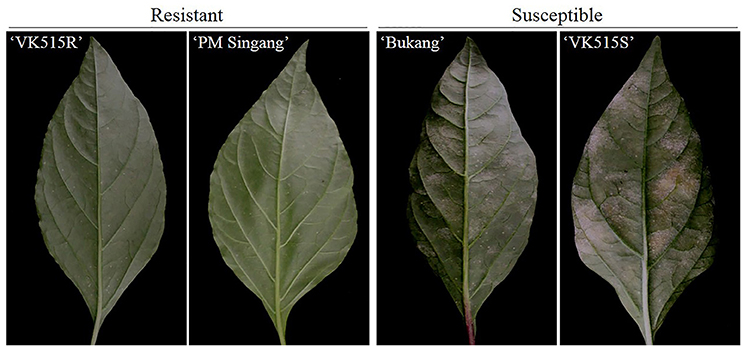
Figure 1. Phenotypic analysis of powdery mildew resistance in pepper. Comparison of the phenotypes of resistant (‘VK515R') and susceptible (‘VK515S') parental lines and resistant (‘PM Singang') and susceptible (‘Bukang') commercial pepper cultivars infected with L. taurica.
Marker Development and Genetic Mapping of the PMR1 Locus
Initial genotyping and linkage analysis of ‘VK515' F2 population with 96 SNP markers (Kang et al., 2014) identified seven SNP markers (KS16052G01, CAPS_CONTIG.12621, CAPS_CONTIG.6527, CAPS_CONTIG.7592, CAPS_CONTIG.8288, CAPS_CONTIG.4984, and CAPS_CONTIG.9996) linked to the PMR1 locus on linkage group 4 (Figure 2A). The closest markers, KS16052G01 and CAPS_CONTIG.12621 were 0 and 24.8 cM away from the PMR1 locus, respectively (Figure 2B). The physical interval between KS16052G01 and CAPS_CONTING.6527 markers was approximately 176.6 Mb on chromosome 4 of ‘L_Zunla-1' (Figure 2C). Therefore, more markers were required to saturate the PMR1 locus.
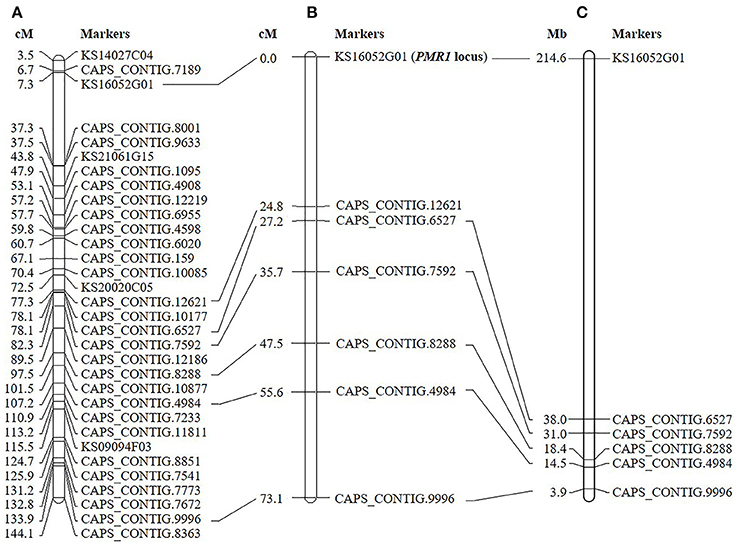
Figure 2. Linkage map of the pepper PMR1 locus. (A) Genetic map of C. annuum reported by Kang et al. (2014). (B) Seven markers linked to the PMR1 locus based on the ‘VK515' F2:3 mapping population are shown. (C) Physical locations of SNP markers on ‘L_Zunla-1' chromosome 4.
We developed additional markers linked to the PMR1 locus using genomic information for Capsicum. To find SNPs by direct sequencing, we designed 12 and 26 primer sets around the PMR1 region (176.6 Mb) on chromosome 4, respectively based on the ‘CM334' (Table S2) and ‘L_Zunla-1' (Table S3) genome sequence (Kim et al., 2014; Qin et al., 2014). Seven SNPs were identified through direct sequencing; one SNP with Chr4.1.6, two SNPs with each of the A4, ZL1_11079B, and ZL1_11658 primer sets. These SNPs were converted into four HRM markers, HRM4.1.6, HRM2_A4, HZ2_11079B, and HZ1_11658, respectively. Of these markers, HRM2_A4 and HRM4.1.6 showed polymorphism and co-segregated with the resistance phenotype in ‘VK515' population (Figure S2, Figure 3A). Additionally, we developed two SCAR markers, ZL1_1826 and ZL1_10691, and one CAPS marker, CZ2_11628 (Table 1, Figure S2) based on ‘L_Zunla-1' genome sequence information.
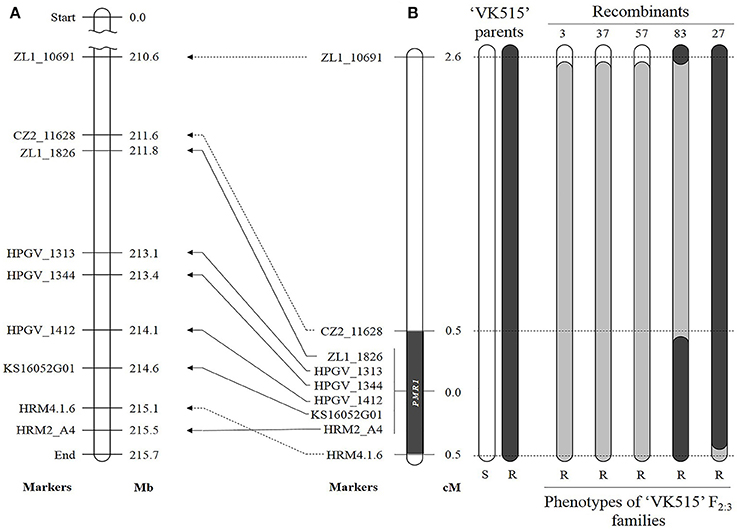
Figure 3. Comparative genetic linkage and physical maps of the powdery mildew resistance gene PMR1. (A) Physical locations of linked markers on ‘L_Zunla-1' chromosome 4. (B) Genetic map of ‘VK515' F2:3 families. Recombinant heterozygous resistant plants 3, 37, and 57; homozygous resistant plants 27 and 83. Eight SNP markers linked to the PMR1 locus are indicated next to pepper chromosome 4. Numbers on the right indicate genetic distances (cM). Black and white rectangles indicate the homozygous intervals of ‘VK515R' and ‘VK515S' chromosome 4, and gray rectangles indicate heterozygous intervals. The PMR1 locus was delimited to a 4 Mb-region between CZ2_11628 and HRM4.1.6 on ‘L_Zunla-1' chromosome 4.
A total of six polymorphic markers (ZL1_10691, CZ2_11628, ZL1_1826, HRM2_A4, HRM4.1.6, and KS16052G01) were mapped in the 102 ‘VK515' F2:3 families (Table 1, Figure 3B). Based on genotyping analysis of the 102 ‘VK515' F2:3 families (1931 plants), three recombinants with ZL1_10691 and one recombinant for each of the CZ2_11628 and HRM4.1.6 markers were identified (Figure 3B). Based on genetic analysis, the PMR1 locus was delimited to a 1.0 cM-region between markers CZ2_11628 and HRM4.1.6 on chromosome 4 (Figure 3B). We found that this 1 cM-region corresponded to a DNA fragment of an approximately 4 Mb-region in the lower arm of ‘L_Zunla-1' chromosome 4. The physical location of HRM4.1.6 showed no collinearity with the corresponding position in ‘L_Zunla-1' (Figure 3), perhaps due to mis-assembly of the contig sequence. Among the six markers used, three markers, ZL1_1826, KS16052G01, and HRM2_A4, co-segregated with the powdery mildew resistance phenotype and were found to be at a genetic distance of 0 cM from the PMR1 locus (Figure 3B).
For ‘PM Singang', seven molecular markers, which were found to be polymorphic (ZL1_10691, HZ2_11079B, CZ2_11628, HZ1_11658, ZL1_1826, HRM2_A4, and HRM4.1.6) were used to map the PMR1 locus (Table 1, Figure S3). The PMR1 locus was mapped in 5.1 cM interval flanked by HZ2_11079B and HRM2_A4. Marker order was found to be identical to that of ‘VK515', with the exception of markers HZ2_11079B and HZ1_11658, which were not polymorphic in the ‘VK515' population. The total genetic lengths covered by the flanking markers in the ‘VK515' and ‘PM Singang' population were 3.1 and 8.9 cM, respectively (Figure S3).
Investigating the Origin of the PMR1 Locus
To investigate the origin of the PMR1 locus, we performed sequence comparisons of the InDel markers (ZL1_1826 and Chr4.1.6) among Capsicum species. The ZL1_1826 and Chr4.1.6 marker sequences from ‘VK515R' and ‘VK515S' were aligned with corresponding sequences from C. annuum ‘L_Zunla-1', C. chinense, and C. baccatum (Figure 4, Figure S4). Sequence alignment of ZL1_1826 revealed an InDel of 236 bp that was specific to C. annuum ‘L_Zunla-1' and ‘VK515S'; a 34 bp InDel was found only in the ‘VK515R' and C. baccatum genomes. A 20 bp InDel was found in all species except C. chinense (Figure 4A, Figure S4), whereas 6 and 7 bp InDels were specific to C. chinense, C. baccatum, and ‘VK515R'. The Chr4.1.6 marker showed a 141 bp InDel specific to C. annuum ‘VK515R', C. chinense, and C. baccatum, whereas C. annuum ‘VK515S' and ‘L_Zunla-1' showed a 33 bp InDel. The presence of ZL1_1826 and Chr4.1.6 marker sequences with species-specific InDels indicated that the PMR1 locus from ‘VK515R' was more closely related to those of C. baccatum and C. chinense than those of ‘VK515S' and C. annuum, suggesting that the PMR1 region in ‘VK515R' may have resulted from an alien introgression of chromosomal segments from either C. chinense or C. baccatum (Figure 4A, Figure S4).
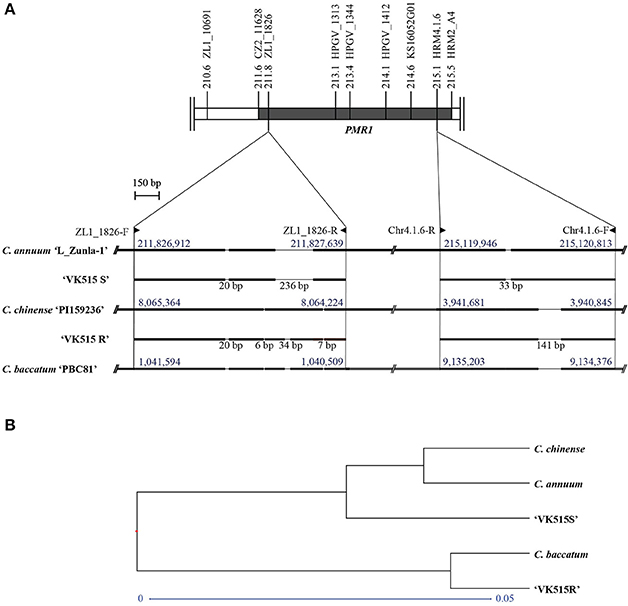
Figure 4. Sequence analysis of the PMR1 locus. (A) Schematic representation of ‘L_Zunla-1', C. chinense, and C. baccatum PMR1-specific InDels and corresponding sequences of ‘VK515' parental lines. InDel lengths in bp are shown below the solid lines. Numbers (blue font) above blocks indicate location on chromosome 4. Primer positions of the PMR1 linked markers are indicated with triangles. (B) Phylogenetic analysis of the PMR1 locus based on GBS data.
To further confirm the origin of the PMR1 locus, as well as the suppressed recombination in the PMR1 locus, we aligned the GBS data with genomic sequences from C. annuum ‘L_Zunla-1', C. chinense, and C. baccatum. Based on GBS data analysis, 22 SNPs (Table S5) were detected in the PMR1 locus, nine of which were converted into three HRM markers, HPGV_1313 (SNP4, SNP5, and SNP6), HPGV_1344 (SNP7), and HPGV_1412 (SNP8, SNP9, SNP10, SNP11, and SNP12) for genotyping. However, no recombinants were found with these markers, further confirming the suppressed recombination in the PMR1 region. Furthermore, phylogenetic analysis of the GBS data (Figure 4B) from the PMR1 locus revealed clustering of ‘VK515S', C. annuum, and C. chinense, whereas ‘VK515R' and C. baccatum shared a common node, suggesting that the PMR1 locus in ‘VK515R' is closely related to that of C. baccatum. These results suggest that the PMR1 locus might have been introgressed from C. baccatum.
Discussion
In the present study, we investigated the genetic factor responsible for powdery mildew resistance in two independent sources of powdery mildew resistance, C. annuum ‘VK515R' and ‘PM Singang'. Genetic analysis confirmed that a single dominant locus, PMR1, which mapped to chromosome 4 of the pepper genome, is responsible for the powdery mildew resistance in these cultivars. Synteny analysis between the pepper PMR1 locus and the tomato genome showed that much of the PMR1-region is syntenic with tomato chromosome 3 (Figure S5). The PMR1 locus was mapped to a syntenic region in two independent pepper populations, ‘VK515' and ‘PM Singang'. The genetic distance between the flanking markers CZ2_11628 and HRM4.1.6 in ‘VK515' F2:3 families and HZ2_11079B and HRM2_A4 in the ‘PM Singang' population were 1.0 and 5.1 cM with total map distances of 3.1 and 8.9 cM, respectively. The difference in genetic distance between the ‘VK515' and ‘PM Singang' populations is likely due to the difference in the size and nature of the genetic populations (having 102 F2:3 families vs. 80 F2 individuals).
Previously, Lefebvre et al. (2003) mapped powdery mildew resistance QTLs in a double-haploid population derived from a cross between ‘H3' (resistant) and ‘Vania' (susceptible). Five additive QTLs (located on chromosomal regions P5, P6, P9, P10, and P12) and two epistatic interactions together explaining more than 50% of the genotypic variance were detected (Lefebvre et al., 2003). Eggink et al. (2016) reported a QTL on LG1/8 explaining about 57% phenotypic variance from an unknown pepper population in their patent application. However, none of these correspond to the dominant PMR1 locus identified in this study. Recently, Gabor et al. (2017) reported a powdery mildew-resistance QTL located on chromosome 4 (in an interval of about < 40 cM between markers NE0235653 and NE0240958) in pepper line PBC167. Thus, previous genetic analyses indicate a complex mode of inheritance for powdery mildew resistance in pepper. Genetic analyses have also indicated dominant inheritance of powdery mildew resistance in certain pepper backgrounds (Anand et al., 1987). The dominant resistance sources in ‘VK515' and ‘PM Singang' are different from other known powdery mildew resistance factors reported. Overall, previous findings and the results presented here indicate that the nature of genetic resistance to powdery mildew in pepper genotypes is highly variable, and it is likely that different genes are involved in conferring the powdery mildew resistance in different genetic backgrounds (Daubèze et al., 1995; Murthy and Deshpande, 1997; Lefebvre et al., 2003; Blat et al., 2005, 2006).
Several studies have been carried out to identify and analyze the powdery mildew disease resistance trait in other crops. In particular, powdery mildew resistance conferred by the loss-of-function alleles of specific MLO (Mildew Locus O) genes due to recessive mutations, such as in Arabidopsis (AtMLO2), barley (Mlo), and tomato (SlMlo1) and their mutants have been utilized in breeding programs to provide effective mlo-mediated powdery mildew resistance (Büschges et al., 1997; Bai et al., 2008; Pavan et al., 2011; Zheng et al., 2013). MLO genes encode proteins with seven transmembrane domains and have a recessive inheritance pattern (Pessina et al., 2014). Unlike MLO genes, PMR1 showed dominant inheritance. Several dominant R genes, such as RPW8 from Arabidopsis (Xiao et al., 2001), Pm3b from wheat (Yahiaoui et al., 2004; Srichumpa et al., 2005), and Mla6, Mla1, and Mla13 from barley (Halterman et al., 2001, 2003; Zhou et al., 2001), belong to the NB-ARC domain-containing R gene family. In the present study, among the 622 predicted genes from the 4-Mb region of the PMR1 locus, two genes (FGENESH: 408 and FGENESH: 556) were found to encode R proteins sharing sequence similarity with NBS-LRR domain-containing R proteins (Table S6). These genes represent potential candidates for pepper powdery mildew resistance genes. However, the possible role of leucine-rich repeat receptor kinases in plant powdery mildew resistance cannot be ruled out, as these proteins play crucial roles in a wide variety of plant developmental and defense-related processes, including both host-specific and non-host-specific defense responses (Torii, 2004), and several of the predicted genes from the pepper PMR1 region were found to share sequence similarity with leucine-rich repeat receptor kinase genes.
The flanking markers CZ2_11628 and HRM4.1.6 delimited the PMR1 locus in ‘VK515R' to an interval of 1.0 cM corresponding to a DNA fragment of an approximately 4-Mb region in the lower arm of ‘L_Zunla-1' chromosome 4. Suppressed recombination and segregation distortion are two common problems in populations segregating for alien introgressions, which can affect genetic mapping and linkage analyses (Chetelat et al., 2000; Neu et al., 2002; Pertuzé et al., 2003; Canady et al., 2006; Nagy et al., 2010). Consistent with this notion, the present analysis, which is based on genotyping as well as GBS analysis, indicated suppressed recombination between markers CZ2_11628 and HRM4.1.6, each spanning a genetic distance of 0.5 cM from the PMR1 locus, which might have been introgressed from a wild Capsicum species. In accordance with this possible introgression, phylogenetic analysis of the GBS data from the PMR1 locus showed that ‘VK515R' shared a common node with C. baccatum. Similar results were obtained with phylogenetic analysis of the PMR1 locus-specific InDel marker sequences (data not shown). These results further substantiate the idea that the PMR1 locus might have been introgressed from C. baccatum. Nevertheless, a possible role of chromosomal rearrangements in the suppressed recombination at the PMR1 region cannot be ruled out (Neu et al., 2002).
Screening for powdery mildew resistance in numerous pepper genotypes (De Souza and Café-Filho, 2003) has suggested that resistance to powdery mildew most likely originated from species other than those of the C. annuum taxon. Furthermore, the dominant pattern of inheritance of powdery mildew resistance in ‘VK515R', which is similar to that of C. baccatum (Blat et al., 2005), supports the notion that powdery mildew resistance in ‘VK515R' might have introgressed from C. baccatum, possibly using C. chinense as a bridge species, since C. annuum and C. baccatum are not cross-compatible (Manzur et al., 2015; Martins et al., 2015). Indeed, phylogenetic analysis based on GBS data and InDel markers demonstrated the close relatedness of the PMR1 region from C. baccatum and C. annuum ‘VK515R', further supporting the introgression of the PMR1 locus from C. baccatum.
In summary, phenotypic and genetic analysis of powdery mildew resistance in pepper cultivars, VK515R and PM Singang identified a single dominant locus for powdery mildew resistance. We mapped the PMR1 locus to 1.0 and 5.1 cM intervals on chromosome 4 of ‘VK515R' and ‘PM Singang'. Six and three molecular markers co-segregating with the powdery mildew resistance were identified from ‘VK515R' and ‘PM Singang', respectively. The molecular markers that we developed in this study will be especially useful for MAS and pyramiding of powdery mildew-resistance genes into elite cultivars, as they are tightly linked to the PMR1 locus, and efficient SNP marker detection platforms are currently available. Further fine mapping and candidate gene analyses are needed to uncover the relationship between the predicted receptor kinase/NBS-LRR genes and PMR1.
Author Contributions
Conceived and designed the experiments: B-CK and JJ. Participated in phenotyping: JJ and GC. Participated in marker development and genetic mapping: JJ. Performed GBS analysis: JJ, KH, H-YL. Drafted the manuscript: JJ, JV. Revised the manuscript: JV, B-CK, HL, DC.
Funding
This research was supported by the Golden Seed Project (213002-04-3-CG900), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA), and Korea Forest Service (KFS), Republic of Korea, and a grant (710001-07) from the Vegetable Breeding Research Center through the Agriculture, Food and Rural Affairs Research Center Support Program, Ministry of Agriculture, Food and Rural Affairs. A patent has been submitted for the PMR locus described in this investigation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.02090/full#supplementary-material
Figure S1. Scoring of powdery mildew resistance in F2 ‘PM Singang' population. Disease score values 1, 2, 3, and 4 indicate no sign of disease development, minute necrotic lesions with no detectable sporulation, few large sporulating lesions, and numerous large sporulating lesions, respectively.
Figure S2. Analysis of molecular markers. (A) PCR analysis of SCAR marker ZL1_1826 and CAPS marker CZ2_11628. (B) Normalized HRM curves of SNP-based markers HRM4.1.6 and HRM2_A4. RR: genotype of resistant line, rr: genotype of susceptible line, Rr: genotype of heterozygous F1 plants.
Figure S3. Comparative map of the PMR1 locus in two pepper populations, ‘VK515' and ‘PM Singang'.
Figure S4. Sequence comparisons of InDel markers flanking PMR1.
Figure S5. Syntenic relationship between pepper PMR1 locus and tomato chromosome 4.
Table S1. 96 SNP markers used in the Fluidigm genotyping assay.
Table S2. ‘CM334' genome-based primer sequences used for polymorphism detection.
Table S3. ‘L_Zunla-1' genome-based primer sequences used for polymorphism detection.
Table S4. Segregation analysis of powdery mildew resistance in ‘PM Singang' population.
Table S5. Genotyping-by-sequencing of the PMR1 locus.
Table S6. Predicted genes from the PMR1 region.
References
Anand, N., Deshpande, A. A., and Sridhar, T. S. (1987). Resistance to powdery mildew in an accession of Capsicum frutescens and its inheritance pattern. Capsicum Eggplant Newsl. 6, 77–78.
Bai, Y., Pavan, S., Zheng, Z., Zappel, N. F., Reinstädler, A., Lotti, C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol. Plant Microbe Interact. 21, 30–39. doi: 10.1094/MPMI-21-1-0030
Berzal-Herranz, A., de la Cruz, A., Tenllado, F., Díaz-Ruíz, J. R., López, L., Sanz, A. I., et al. (1995). The Capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology 209, 498–505. doi: 10.1006/viro.1995.1282
Blat, S. F., da Costa, C. P., Vencovsky, R., and Sala, F. C. (2005). Inheritance of reaction to Leveillula taurica (Lev.) Arn. in Capsicum baccatum. Crop Breed. Appl. Biotechnol. 5, 467–472. doi: 10.1590/S0103-90162005000100008
Blat, S. F., da Costa, C. P., Vencovsky, R., and Sala, F. C. (2006). Hot pepper (Capsicum chinense, Jacq.) inheritance of reaction to powdery mildew. Sci. Agric. 63, 471–474. doi: 10.1590/S0103-90162006000500008
Boiteux, L. S., Cupertino, F. P., and Reifschneider, F. J. B. (1993). Capsicum chinense PI159236: a source of resistance to Phytophthora capsici and tomato spotted wilt virus. Capsicum Eggplant Newsl. 12, 76.
Boukema, I. W. (1980). Allelism of genes controlling resistance to TMV in Capsicum L. Euphytica 29, 433–439. doi: 10.1007/BF00025143
Büschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. doi: 10.1016/S0092-8674(00)81912-1
Canady, M. A., Ji, Y., and Chetelat, R. T. (2006). Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174, 1775–1788. doi: 10.1534/genetics.106.065144
Cerkauskas, R. F., Ferguson, G., and Banik, M. (2011). Powdery mildew (Leveillula taurica) on greenhouse and field peppers in Ontario: host range, cultivar response and disease management strategies. Can. J. Plant Pathol. 33, 485–498. doi: 10.1080/07060661.2011.619828
Chetelat, R. T., Meglic, V., and Cisneros, P. (2000). A genetic map of tomato based on BC1 Lycopersicon esculentum × Solanum lycopersicoides reveals overall synteny but suppressed recombination between these homeologous genomes. Genetics 154, 857–867.
Cook, A. A., and Guevara, Y. G. (1984). Hypersensitivity in Capsicum chacoense to race 1 of the bacterial spot pathogen of pepper. Plant Dis. 68, 329–330. doi: 10.1094/PD-69-329
Damicone, J. (2009). Fungicide resistance management. Fact Sheets EPP-7663. Oklahoma State University Division of Agricultural Sciences and Natural Resources website. Available online at: http://pods.dasnr.okstate.edu/docushare/dsweb/Get/Rendition-3508/F-7663web.pdf
Daubèze, A. M., Hennart, J. W., and Palloix, A. (1995). Resistance to Leveillula taurica in pepper (Capsicum annuum) is oligogenically controlled and stable in Mediterranean regions. Plant Breed. 114, 327–332. doi: 10.1111/j.1439-0523.1995.tb01243.x
de la Cruz, A., López, L., Tenllado, F., Díaz-Ruíz, J. R., Sanz, A. I., Vaquero, C., et al. (1997). The coat protein is required for the elicitation of the Capsicum L2 gene-mediated resistance against the tobamoviruses. Mol. Plant Microbe Interact. 10, 107–113
Deshpande, A. A., Anand, N., Pathak, C. S., and Sridhar, T. S. (1985). New sources of powdery mildew resistance in Capsicum species. Capsicum Eggplant Newsl. 4, 75–76.
De Souza, V. L., and Café-Filho, A. C. (2003). Resistance to Leveillula taurica in the genus Capsicum. Plant Pathol. 52, 613–619. doi: 10.1046/j.1365-3059.2003.00920.x
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15
Eggink, P. M., D'hoop, B. B., Brouwer, M., and Deniau, A. X. (2016). Resistance against Leveillula taurica in Pepper. U.S. Patent No 9,351,451. Washington, DC: U.S. Patent and Trademark Office.
Elad, Y., Messika, Y., Brand, M., David, D. R., and Sztejnberg, A. (2007). Effect of microclimate on Leveillula taurica powdery mildew of sweet pepper. Phytopathology 97, 813–824. doi: 10.1094/PHYTO-97-7-0813
Gabor, B. K., Just, B. J., Huang, C., Jones, C. M., Vreugdenhil, D., Kniskern, J. M., et al. (2017). Methods and Compositions for Producing Capsicum Plants with Powdery Mildew Resistance. U.S. Patent No 9,689,045. Washington, DC: U.S. Patent and Trademark Office
Halterman, D. A., Wei, F., and Wise, R. P. (2003). Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131, 558–567 doi: 10.1104/pp.014407
Halterman, D., Zhou, F., Wei, F., Wise, R. P., and Schulze-Lefert, P. (2001). The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. doi: 10.1046/j.1365-313x.2001.00982.x
Hibberd, A. M., Bassett, M. J., and Stall, R. E. (1987). Allelism tests of three dominant genes for hypersensitive resistance to bacterial spot of pepper. Phytopathology 77, 1304–1307. doi: 10.1094/Phyto-77-1304
Hoang, N. H., Yang, H. B., and Kang, B. C. (2013). Identification and inheritance of a new source of resistance against Tomato spotted wilt virus (TSWV) in Capsicum. Sci. Hortic. 161, 8–14. doi: 10.1016/j.scienta.2013.06.033
Kang, J. H., Yang, H. B., Jeong, H. S., Choe, P., Kwon, J. K., and Kang, B. C. (2014). Single nucleotide polymorphism marker discovery from transcriptome sequencing for marker-assisted backcrossing in Capsicum. Korean J. Hortic. Sci. Technol. 32, 535–543. doi: 10.7235/hort.2014.14109
Kim, B. S., and Hartmann, R. W. (1985). Inheritance of a gene (Bs3) conferring hypersensitive resistance to Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum). Plant Dis. 69, 233–235.
Kim, D. H., Park, J. H., Lee, J. S., Han, K. S., Han, Y. K., and Hwang, J. H. (2009). Effect of temperature, relative humidity on germination and development of powdery mildew (Leveillula taurica) on pepper and its inoculation method. Res. Plant Dis. 15, 187–192. doi: 10.5423/RPD.2009.15.3.187
Kim, S., Park, J., Yeom, S. I., Kim, Y. M., Seo, E., Kim, K. T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18:210. doi: 10.1186/s13059-017-1341-9
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Lefebvre, V., Daubèze, A. M., Rouppe van der Voort, J., Peleman, J., Bardin, M., and Palloix, A. (2003). QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor. Appl. Genet. 107, 661–666. doi: 10.1007/s00122-003-1307-z
Li, H. (2013). Aligning Sequence Reads, Clone Sequences and Assembly Contigs with Bwa-Mem. arXiv:1303.3997v2 [q-bio.GN] Available online at: https://arxiv.org/abs/1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, L., Venkatesh, J., Jo, Y. D., Koeda, S., Hosokawa, M., Kang, J. H., et al. (2016). Fine mapping and identification of candidate genes for the sy-2 locus in a temperature-sensitive chili pepper (Capsicum chinense). Theor. Appl. Genet. 129, 1541–1556. doi: 10.1007/s00122-016-2723-1
Liu, W. Y., Kang, J. H., Jeong, H. S., Choi, H. J., Yang, H. B., Kim, K. T., et al. (2014). Combined use of bulked segregant analysis and microarrays reveals SNP markers pinpointing a major QTL for resistance to Phytophthora capsici in pepper. Theor. Appl. Genet. 127, 2503–2513. doi: 10.1007/s00122-014-2394-8
Mallard, S., Cantet, M., Massire, A., Bachellez, A., Ewert, S., and Lefebvre, V. (2013). A key QTL cluster is conserved among accessions and exhibits broad-spectrum resistance to Phytophthora capsici: a valuable locus for pepper breeding. Mol. Breed. 32, 349–364. doi: 10.1007/s11032-013-9875-3
Manzur, J. P., Fita, A., Prohens, J., and Rodríguez-Burruezo, A. (2015). Successful wide hybridization and introgression breeding in a diverse set of common peppers (Capsicum annuum) using different cultivated Ají (C. baccatum) accessions as donor parents. PLoS ONE 10:e0144142. doi: 10.1371/journal.pone.0144142
Martins, K. C., Nair, T., Pereira, S., Alessandro, S., Souza, M., and Rodrigues, R. (2015). Crossability and evaluation of incompatibility barriers in crosses between Capsicum species. Crop Breed. Appl. Biotechnol. 15, 139–145. doi: 10.1590/1984-70332015v15n3a25
Murthy, H. M. K., and Deshpande, A. A. (1997). Studies on genetics of powdery mildew [Leveillula taurica (Lev.) Arn.] resistance in chilli (Capsicum annuum L.). Vegetable Sci. 24, 127–131.
Nagy, E. D., Chu, Y., Guo, Y., Khanal, S., Tang, S., Li, Y., et al. (2010). Recombination is suppressed in an alien introgression in peanut harboring Rma, a dominant root-knot nematode resistance gene. Mol. Breed. 26, 357–370. doi: 10.1007/s11032-010-9430-4
Neu, C., Stein, N., and Keller, B. (2002). Genetic mapping of the Lr20Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45, 737–744. doi: 10.1139/g02-040
Park, S. W., An, S. J., Yang, H. B., Kwon, J. K., and Kang, B. C. (2009). Optimization of high resolution melting analysis and discovery of single nucleotide polymorphism in Capsicum. Hortic. Environ. Biotech. 50, 31–39.
Pavan, S., Schiavulli, A., Appiano, M., Marcotrigiano, A. R., Cillo, F., Visser, R. G., et al. (2011). Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 123, 1425–1431. doi: 10.1007/s00122-011-1677-6
Perrier, X., and Jacquemoud-Collet, J. P. (2006). DARwin Software, Version 5.0.158. Available online at: http://darwin.cirad.fr/darwin
Pertuzé, R. A., Ji, Y., and Chetelat, R. T. (2003). Transmission and recombination of homeologous Solanum sitiens chromosomes in tomato. Theor. Appl. Genet. 107, 1391–1401. doi: 10.1007/s00122-003-1384-z
Pessina, S., Pavan, S., Catalano, D., Gallotta, A., Visser, R. G., Bai, Y., et al. (2014). Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics 15:618. doi: 10.1186/1471-2164-15-618
Pochard, E., Palloix, A., and Daubèze, A. M. (1986). “The use of androgenetic autodiploid lines for the analysis of complex resistance systems in the pepper,” in VIth Eucarpia Meeting on Genetics and Breeding on Capsicum and Eggplant (Saragossa), 105–109.
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.4172/2168-9881.S1.013
Schiex, T., and Gaspin, C. (1997). Carthagene: constructing and joining maximum likelihood genetic maps. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 258–267.
Shifriss, C., Pilovsky, M., and Zack, J. M. (1992). “Resistance to Leveillula taurica mildew (Oidiopsis taurica) in Capsicum annuum L.,” in VIIIth Eucarpia Meeting on Genetics and Breeding on Capsicum and Eggplant (Rome), 172–177.
Soler, S., Debreczeni, D. E., Vidal, E., Aramburu, J., López, C., Galipienso, L., et al. (2015). A new Capsicum baccatum accession shows tolerance to wild-type and resistance-breaking isolates of Tomato spotted wilt virus. Ann. Appl. Biol. 167, 343–353. doi: 10.1111/aab.12229
Solovyev, V., Kosarev, P., Seledsov, I., and Vorobyev, D. (2006). Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7(Suppl. 1), S10.1–12. doi: 10.1186/gb-2006-7-s1-s10
Srichumpa, P., Brunner, S., Keller, B., and Yahiaoui, N. (2005). Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139, 885–895. doi: 10.1104/pp.105.062406
Sudha, A., and Lakshmanan, P. (2009). Integrated disease management of powdery mildew (Leveillula taurica (Lev.) Arn.) of chilli (Capsicum annuum L.). Arch. Phytopathol. Plant Protect. 42, 299–317. doi: 10.1080/03235400601037198
Torii, K. U. (2004). Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234, 1–46. doi: 10.1016/S0074-7696(04)34001-5
Truong, H. T., Ramos, A. M., Yalcin, F., de Ruiter, M., van der Poel, H. J., et al. (2012). Sequence-based genotyping for marker discovery and co-dominant scoring in germplasm and populations. PLoS ONE 7:e37565. doi: 10.1371/journal.pone.0037565
Ullassa, B. A., Rawal, R. D., Sohi, H. S., and Sing, D. P. (1981). Reaction of sweet pepper genotypes to anthracnose leaf spot and powdery mildew. Plant Dis. 65, 600–601. doi: 10.1094/PD-65-600
Vallejos, C. E., Jones, V., Stall, R. E., Jones, J. B., Minsavage, G. V., Schultz, D. C., et al. (2010). Characterization of two recessive genes controlling resistance to all races of bacterial spot in peppers. Theor. Appl. Genet. 121, 37–46. doi: 10.1007/s00122-010-1289-6
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Voorrips, R. E., Finkers, R., Sanjaya, L., and Groenwold, R. (2004). QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annuum and C. Chinense. Theor. Appl. Genet. 109, 1275–1282. doi: 10.1007/s00122-004-1738-1
Wang, J., Lin, M., Crenshaw, A., Hutchinson, A., Hicks, B., Yeager, M., et al. (2009). High-throughput single nucleotide polymorphism genotyping using nanofluidic Dynamic Arrays. BMC Genomics 10:561. doi: 10.1186/1471-2164-10-561
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2- a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M., et al. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. doi: 10.1126/science.291.5501.118
Yahiaoui, N., Srichumpa, P., Dudler, R., and Keller, B. (2004). Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37, 528–538. doi: 10.1046/j.1365-313X.2003.01977.x
Zheng, Z., Nonomura, T., Appiano, M., Pavan, S., Matsuda, Y., Toyoda, H., et al. (2013). Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS ONE 8:e70723. doi: 10.1371/journal.pone.0070723
Keywords: Capsicum annuum, Leveillula taurica, marker-assisted selection, molecular markers, PMR1, powdery mildew resistance
Citation: Jo J, Venkatesh J, Han K, Lee H-Y, Choi GJ, Lee HJ, Choi D and Kang B-C (2017) Molecular Mapping of PMR1, a Novel Locus Conferring Resistance to Powdery Mildew in Pepper (Capsicum annuum). Front. Plant Sci. 8:2090. doi: 10.3389/fpls.2017.02090
Received: 28 July 2017; Accepted: 23 November 2017;
Published: 08 December 2017.
Edited by:
Laurent Gentzbittel, National Polytechnic Institute of Toulouse, FranceReviewed by:
Seonghee Lee, University of Florida, United StatesUmesh K. Reddy, West Virginia State University, United States
Copyright © 2017 Jo, Venkatesh, Han, Lee, Choi, Lee, Choi and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byoung-Cheorl Kang, Yms1NEBzbnUuYWMua3I=
 Jinkwan Jo
Jinkwan Jo Jelli Venkatesh
Jelli Venkatesh Koeun Han1
Koeun Han1 Hea-Young Lee
Hea-Young Lee Gyung Ja Choi
Gyung Ja Choi Doil Choi
Doil Choi Byoung-Cheorl Kang
Byoung-Cheorl Kang