- 1College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, China
- 2College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 3Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang Agricultural University, Shenyang, China
Target leaf spot (TLS), which is caused by Corynespora cassiicola (C. cassiicola), is one of the most important diseases in cucumber (Cucumis sativus L.). Our previous research identified several C. cassiicola-responsive miRNAs in cucumber by high-throughput sequencing, including two known miRNAs and two novel miRNAs. The target genes of these miRNAs were related to secondary metabolism. In this study, we verified the interaction between these miRNAs and target genes by histochemical staining and fluorescence quantitative assays of GUS. We transiently expressed the candidate miRNAs and target genes in cucumber cotyledons to investigate the resistance to C. cassiicola. Transient expression of miR164d, miR396b, Novel-miR1, and Novel-miR7 in cucumber resulted in decreased resistance to C. cassiicola, while transient expression of NAC (inhibited by miR164d), APE (inhibited by miR396b), 4CL (inhibited by Novel-miR1), and PAL (inhibited by Novel-miR7) led to enhanced resistance to C. cassiicola. In addition, overexpression of 4CL and PAL downregulated lignin synthesis, and overexpression of Novel-miR1 and Novel-miR7 also downregulated lignin synthesis, indicating that the regulation of 4CL and PAL by Novel-miR1 and Novel-miR7 could affect lignin content. The tobacco rattle virus (TRV) induced short tandem target mimic (STTM)-miRNA silencing vector was successfully constructed, and target miRNAs were successfully silenced. The identification of disease resistance and lignin content showed that silencing candidate miRNAs could improve cucumber resistance to C. cassiicola.
Introduction
Cucumber (Cucumis sativus L.) target leaf spot (TLS) is caused by Corynespora cassiicola (C. cassiicola), which is an obligate oomycete pathogen (Teramoto et al., 2011; Li et al., 2012). TLS affects a wide range of cucumbers worldwide, including those in China (Li et al., 2012; Yang et al., 2012), the United States (Ishii et al., 2007), Japan (Miyamoto et al., 2010), and South Korea (Kwon et al., 2003). TLS is a foliar disease that can occur throughout the growing period of cucumber, but it is more serious in the middle and late stages of growth than in other stages (Wen et al., 2015). There are abundant variations in the size and structure of the C. cassiicola conidia, as well as the color of the colonies. These variations are reflected not only in different colonies of the same strain but also among strains of different geographical origins and different hosts (Nghia et al., 2008; Qi et al., 2011). Therefore, it is important to study the molecular mechanisms of cucumber resistance to C. cassiicola and find new resistance gene resources.
Plant microRNAs (miRNAs) are a class of small non-coding single stranded RNAs encoded by endogenous genes that are mainly involved in gene expression and regulation at the post-transcriptional level (Waterhouse and Hellens, 2015). In plants, miRNAs can function by interacting with target genes, mainly through degradation and inhibition of target mRNAs (Slack et al., 2000; Khraiwesh et al., 2012). Plants are often subjected to various environmental stresses during their growth. These stresses can result in accumulation of various substances and induce plant-related gene expression and metabolic pathways to strengthen plant resistance. Recently, many genes encoding stress-related proteins have been discovered, and miRNAs have been shown to have important regulatory roles in the expression of these genes (Sanan-Mishra et al., 2009; Baxter et al., 2014; Liu et al., 2015; Zhang and Wang, 2015). Many plant miRNAs can be induced after infection by pathogens, and these miRNAs can participate in plant disease resistance by interacting with their targets (Thiebaut et al., 2015). miR393 was the first miRNA reported to participate in the interaction between plants and pathogens (Navarro et al., 2006). In Arabidopsis thaliana, miR824, miR843, miR852, miR166, miR156, and miR159 could respond to Pseudomonas syringae (Zhang et al., 2011). Overexpression of miR482b in tomato reduced resistance to Phytophthora infestans by inhibiting expression of NBS-LRR, while silencing miR482b increased resistance (Jiang et al., 2018). In cucumber, several studies have identified many miRNAs that are related to growth and stress responses (Martínez et al., 2011; Mao et al., 2012; Li et al., 2014; Jin and Wu, 2015; Burkhardt and Day, 2016), but there are currently no reports of miRNAs related to resistance to C. cassiicola.
After infection by pathogens, plants will produce secondary metabolites to resist pathogen invasion (Pateraki and Kanellis, 2010; Pusztahelyi et al., 2015). In plants, there are two main types of secondary metabolites that affect disease resistance. One is the inherent substances of plants, such as lignin, callose, and keratin. These substances can reinforce cell walls and prevent pathogens from damaging plant tissues (Zhao and Dixon, 2011). The other group includes alkaloids, terpenes, and phenols that are induced by pathogens. These substances have a direct bactericidal effect (Agrawal and Weber, 2015). Lignin is an important component of the cell wall, with a complex structure and induced properties. Increasing the lignin content can enhance the ability of plant cells to resist penetration and dissolution and inhibit the spread of pathogenic bacteria in plants (Zhao and Dixon, 2011; Li et al., 2015). Phenylalanine ammonia-lyase (PAL) and 4-coumarate: CoA ligase (4CL) are the two key genes involved in phenylpropanoid synthesis. They can promote the synthesis of lignin to enhance plant disease resistance (Kim and Hwang, 2014; Li et al., 2015).
In our previous study, high-throughput sequencing was performed to investigate the differentially expressed miRNAs in cucumber inoculated with C. cassiicola, including two known miRNAs (miR164d and miR396b) and two novel miRNAs (Novel-miR1 and Novel-miR7) (Wang et al., 2018). Based on the analyses of target genes function, we believe that these miRNAs play important roles in the interaction between cucumber and C. cassiicola. For further elucidation of cucumber miRNAs, it is necessary to effectively and accurately demonstrate the interaction of miRNAs and target genes. In this study, candidate miRNAs and target genes were transiently expressed in tobacco, and the interaction between miRNAs and target genes was determined by analysis of GUS histochemical staining and fluorescence quantification. Meanwhile, the candidate miRNAs and target genes were transiently expressed in the cucumber cotyledons to investigate the resistance of the transgenic cucumber to C. cassiicola. We analyzed the mechanism of cucumber resistance to C. cassiicola based on the regulation of miRNAs, which provided a theoretical reference for further stable genetic transformation and breeding research of plants.
Due to the short size of miRNAs and the redundancy of family functions, traditional gene silencing methods are not suitable for miRNA research. Target mimics (TMs) can block the inhibition of target genes by miRNAs, thereby inhibiting the regulatory function of miRNAs (Franco-Zorrilla et al., 2007; Meng et al., 2012). Tang et al. (2012) discovered a new miRNA silencing regulatory mechanism, short tandem target mimic (STTM), which has high silencing efficiency and can be widely used in the functional studies of miRNAs. The STTM is composed of two TMs and a 48 nt linkage sequence. There are three nucleotides forming a bulge between the 10th and 11th nucleotides of the TM on both sides of the STTM, and thus, the binding sites can capture miRNAs without being cleaved by them. Compared to TM, STTM has a better inhibitory effect on miRNAs (Yan et al., 2012). Virus-induced gene silencing (VIGS) can induce plant endogenous gene silencing and alter the plant phenotype to explore gene function (Sha et al., 2014). Tobacco rattle virus (TRV) is an RNA virus that can infect a variety of plants. TRV-based vectors have mild infection symptoms and high gene silencing efficiency (MacFarlane et al., 1999). TRV-based vectors have been widely used in VIGS to inhibit gene expression in a variety of plants (Bachan and Dinesh-Kumar, 2012; Huang et al., 2012; Zhou et al., 2012). However, virus-based miRNA silencing (VBMS) has not been reported in cucumber. In this study, a TRV-based VBMS system that can effectively inhibit the activity of endogenous miRNAs in cucumber for a certain period of time was developed and may provide a strategy for further analysis of the resistance mechanism of cucumber to C. cassiicola.
Materials and Methods
Plant Materials
Nicotiana benthamiana (N. benthamiana) were planted in a greenhouse at 25°C under 16:8 light/dark cycles for 20 days for agroinfiltration. The cucumber variety used in the experiments was Xintaimici, which was planted in a greenhouse at 28°C under 16:8 light/dark cycles for 10 days for agroinfiltration.
DNA and RNA Extraction and cDNA Synthesis
The DNA of the cucumber leaves was extracted using a Plant Genomic DNA Kit (Tiangen, Beijing, China). Total RNA was extracted by using a RNAprep Pure Plant Kit (Tiangen, Beijing, China) and synthesized into cDNA using a QuantScript RT Kit (Tiangen, Beijing, China). First-strand cDNA synthesis corresponding to miRNAs was performed using a miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China).
Construction of Transient Expression Vectors of Candidate miRNAs and Their Targets
Based on the precursor sequences of miR164d, miR396b, Novel-miR1, and Novel-miR7 (Supplementary Table S1), primers were designed according to the In-Fusion principles. Similarly, based on the NAC, APE, PAL, and 4CL open reading frame sequences (Supplementary Table S1), primers were also designed by the In-Fusion principles. The primers were synthesized by GENEWIZ (Suzhou, China) and are listed in Supplementary Table S2. The target genes were annotated via BLAST against the cucumber genome database1. The mRNA raw data were deposited in the NCBI Sequence Read Archive (SRA) with the accession number SRP117262; the small RNA raw data were deposited in the NCBI Sequence Read Archive (SRA) with the accession number SRP117230.
pRI-101 AN (TaKaRa, Dalian, China) is a binary vector for plant transformation that can efficiently express exogenous genes. The plant expression vectors pRI-101 AN and pRI-101 AN-GUS were constructed to verify the interaction between miRNAs and targets. For cloning cDNA into the vector, an InFusion HD cloning kit (Clontech, CA, United States) was used. All constructed vectors were confirmed by sequencing before transformation into Agrobacterium tumefaciens (A. tumefaciens) strain EHA105.
Co-transformation of Candidate miRNAs and Targets in N. benthamiana
To verify the interaction between candidate miRNAs and target genes, we transiently expressed constructed vectors in N. benthamiana leaves by A. tumefaciens transformation. The agrobacteria carrying constructs were diluted to OD600 = 0.5 with suspension buffer (10 mM MES, 10 mM MgCl2 and 200 μM acetosyringone). pRI-101 AN-miRNA and pRI-101 AN-GUS-target genes were mixed in equal volumes to test the cleavage function of tae-miR408. Normal tobacco (Control) and tobacco injected with pRI-101 AN-GUS were used as controls. After 48 h of infiltration, GUS staining and fluorescence quantitative analysis were performed as described by Bradford (1976) and Jefferson et al. (1987). All experiments were performed with three biological repeats.
Transient Expression in Cucumber Cotyledons and Disease Resistance Assays
Transient expression in cucumber cotyledons was performed as described by Shang et al. (2014). The agrobacteria carrying constructs were diluted to OD600 = 0.4 with suspension buffer (10 mM MES, 10 mM MgCl2 and 200 μM acetosyringone) for cotyledon infiltration. We overexpressed miRNA and target genes to verify disease resistance. Leaves were collected 48 h after agroinfiltration to analyze the expression of miRNAs and target genes by qRT-PCR.
For disease resistance assays, inoculation of C. cassiicola was performed by pipetting multiple 10 μL droplet spore suspensions (2 × 105 sporangia/mL) onto cucumber cotyledons. The inoculated samples were kept at 100% relative humidity for 5 days, and disease was assessed by measuring lesion size and quantifying fungal biomass by qRT-PCR quantification of C. cassiicola Actin. The primers used were CoActin-F and CoActin-R (Supplementary Table S2). All experiments were performed with three biological repeats. Normal cucumber (Control) and cucumber injected with pRI-101 AN were used as controls. All experiments were performed with three biological repeats.
Construction of the TRV-Induced Silencing Vector
The sequences of STTM-miR164d, STTM-miR396b, STTM-Novel-miR1, and STTM-Novel-miR7 used in this study were synthesized by GENEWIZ (Suzhou, China). The STTM primers were designed according to the In-Fusion principles and are listed in Supplementary Table S2. The pTRV vector can be efficiently expressed in plants. For cloning STTM into the vector, an InFusion HD cloning kit (Clontech, CA, United States) was used. All constructed vectors were confirmed by sequencing before transformation into A. tumefaciens strain EHA105.
Virus-Based MicroRNA Silencing (VBMS)
Cucumber cotyledons grown for approximately 10 days were used for VBMS, and the suspension method of Agrobacterium was the same as described previously. The mixed suspension containing pTRV1 and pTRV2-STTM was injected into the cucumber cotyledons by a sterile syringe. Cotyledons at 7 days post-injection were used for C. cassiicola inoculation and disease resistance assays.
Quantitative Real-Time RT-PCR Assay
qRT-PCR analyses of genes were conducted using a SuperReal PreMix Plus Kit (SYBR Green) (Tiangen, Beijing, China), and the cucumber Actin gene was used as the internal control. qRT-PCR analyses of miRNAs were performed using the miRcute miRNA qRT-PCR Detection Kit (SYBR Green) (Tiangen, Beijing, China), and the U6 snRNA was used as a reference to normalize the data. All qRT-PCR assays were performed on a LightCycler 480 system (Roche, CA, United States), and the primers used are listed in Supplementary Table S2. The relative expression was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001), and the standard deviation was calculated with three biological repeats.
Determination of Lignin in Cucumber Cotyledons
The determination of lignin content was performed as described by Morrison (1972) with slight modifications. The lignin content is defined as the absorbance at 280 nm per gram of fresh weight. The 1 g cucumber cotyledon sample was homogenized with 95% ethanol and centrifuged at 5000 rpm for 5 min, and the sediments were washed three times with 95% ethanol. After the samples were washed twice with a mixture of ethanol and n-hexane (1:2, v/v), sediments were collected by centrifugation. The dried samples were dissolved in 2 mL of bromoacetyl and glacial acetic acid (1:3, v/v) solution. After a 30 min water bath at 70°C for 30 min, 0.9 mL 2 mM NaOH was added to stop the reaction. Then, 2 mL glacial acetic acid and 0.1 mL 7.5 M hydroxylamine hydrochloride were added to the sample, and the sample was diluted to 5 mL with glacial acetic acid. After centrifugation at 5000 rpm for 5 min, the supernatants were collected to determine the absorbance at 280 nm.
Statistical Analysis
All data are the mean (±SD) of three biological replicates. Statistical analysis was carried out by one-way analysis of variance (ANOVA) using the IBM SPSS Statistics 22 software, and the significant differences were determined by Duncan’s multiple range test (P < 0.05) and indicated in alphabetical notation.
Results
Validation of the Interaction Between Candidate miRNAs and Target Genes
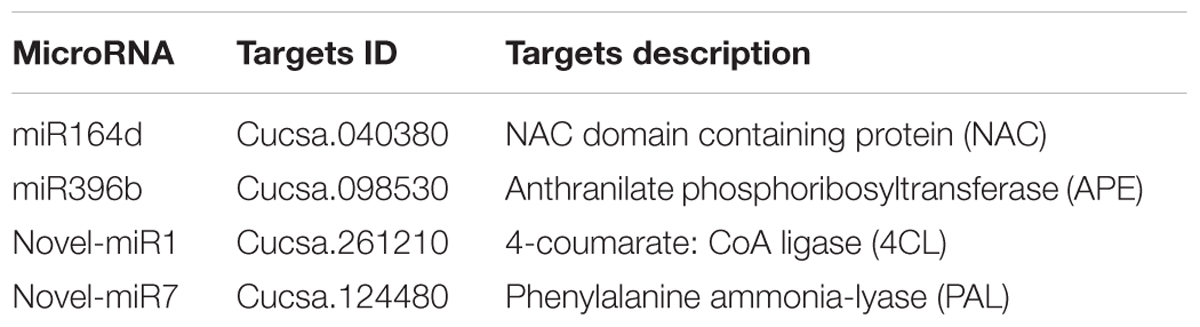
Our previous research found that secondary metabolism plays an important role in cucumber in response to C. cassiicola infection (Wang et al., 2018). Based on previous study, two known miRNAs and two novel miRNAs with their targets were selected, as they may be involved in enhanced C. cassiicola resistance in cucumber (Table 1). In plants, miRNAs regulate their target gene mainly by recognizing a specific site on the target gene mRNA and binding to it to form a silencing complex, thereby inhibiting translation of the target gene mRNA. Candidate miRNAs and target gene binding sites are shown in Figure 1.
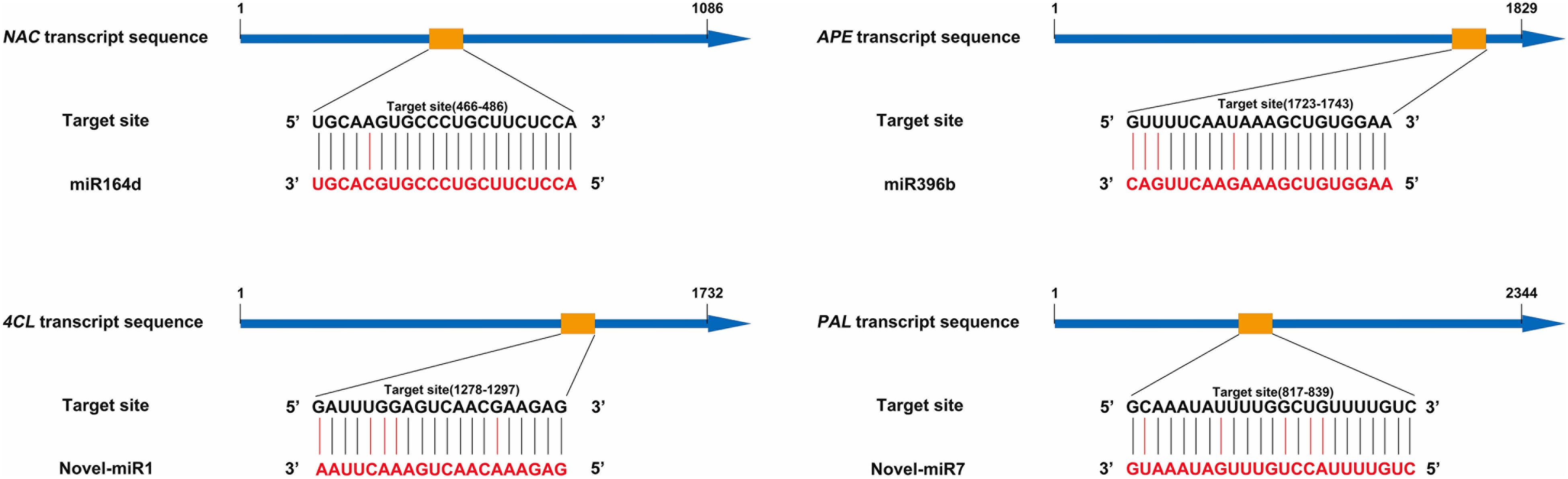
To verify the interaction, we co-transformed candidate miRNAs and target genes into tobacco leaves. Using the vector pRI-101 AN-GUS containing the GUS gene, we tested the inhibitory effect of miRNAs on target genes with GUS histochemical staining and fluorescence quantification. Normal tobacco (Control) and tobacco inoculated with pRI-101 AN-GUS were used as controls. The phenotypes of GUS histochemical staining are shown in Figure 2A. The GUS phenotypes were not observed in tobacco leaves inoculated with the recombination vector pRI-101 AN-miRNA (miR164d, miR396b, Novel-miR1, and Novel-miR7) and normal tobacco (Control). The GUS phenotypes were observed in tobacco leaves inoculated with pRI-101 AN-GUS, while leaves inoculated with pRI-101 AN-GUS-target gene (NAC, APE, 4CL, and PAL), in which the target gene was fused upstream of the GUS gene, showed similar phenotypes. However, GUS phenotypes were markedly reduced in leaves co-transformed with pRI-101 AN-miRNA (miR164d, miR396b, Novel-miR1, and Novel-miR7) and pRI-101 AN-GUS-target gene (NAC, APE, 4CL, and PAL).
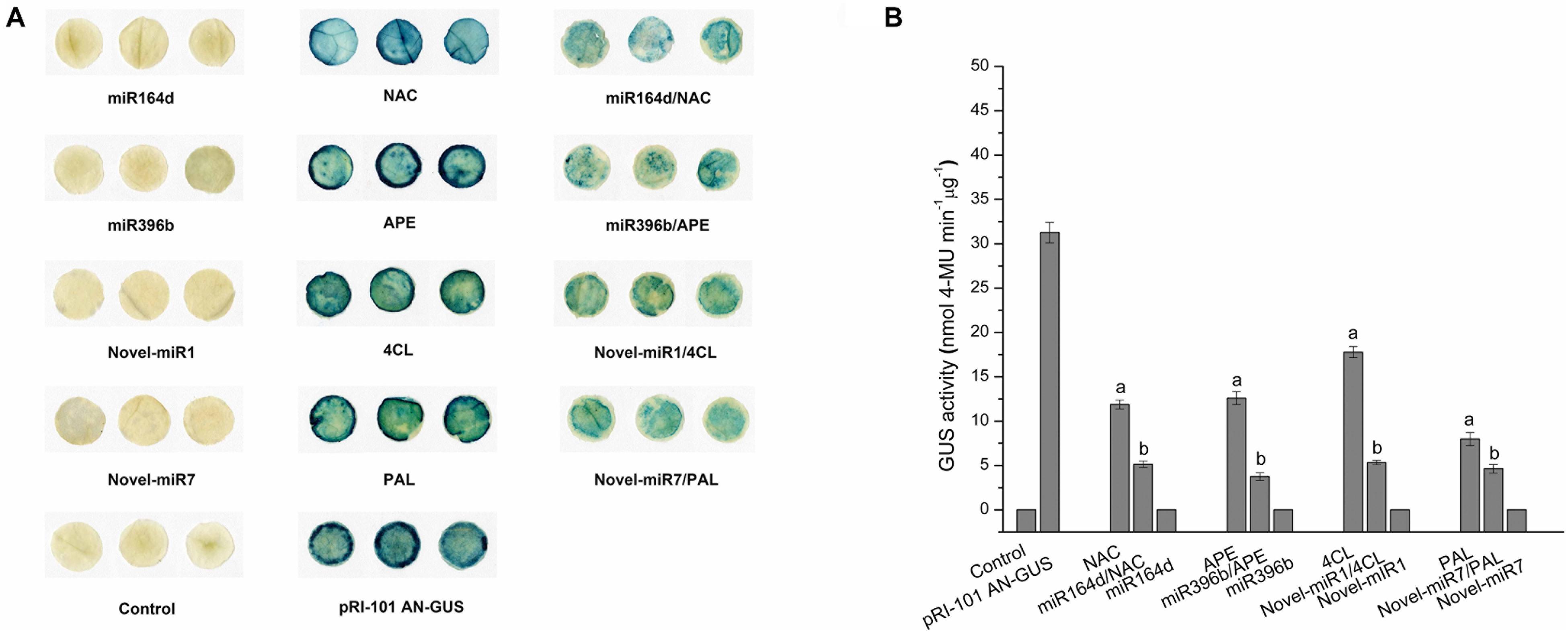
Figure 2. GUS assay of transiently transformed tobacco leaves. (A) GUS accumulation by histochemical staining. (B) GUS fluorescence quantitative assay. Significance was determined by Duncan’s multiple range test (P < 0.05).
The GUS protein activity in leaves inoculated with different recombinant vectors was measured by fluorescence quantitative assays (Figure 2B). GUS fluorescence was not detected in normal tobacco and leaves inoculated with pRI-101 AN-miRNA (miR164d, miR396b, Novel-miR1, and Novel-miR7). There were increased GUS fluorescence values in leaves inoculated with pRI-101 AN-GUS and pRI-101 AN-GUS-target genes (NAC, APE, 4CL, and PAL). The GUS fluorescence values of tobacco leaves co-transformed by pRI-101 AN-miRNA (miR164d, miR396b, Novel-miR1, and Novel-miR7) and pRI-101 AN-GUS-target gene (NAC, APE, 4CL, and PAL) were determined. The quantitative GUS fluorescence analysis supported the results of GUS histochemical staining. These experiments indicated the existence of a negative regulatory relationship between candidate miRNAs and target genes.
Transient Expression Levels of Candidate miRNAs and Target Genes
In this experiment, miR164d, miR396b, Novel-miR1, Novel-miR7, Novel-miR1/Novel-miR7 (1:1, v/v), NAC, APE, 4CL, PAL, and 4CL/PAL (1:1, v/v) were transiently expressed in cucumber cotyledons via Agrobacterium infiltration. Normal tobacco (Control) and tobacco injected with pRI-101 AN were used as controls. Expression levels of candidate miRNAs and target genes in transgenic cucumbers were detected by qRT-PCR. The results are shown in Figure 3. The gene expression levels were similar in the normal tobacco and pRI-101 AN experimental groups. The gene expression level of the overexpression group was significantly higher than that of the controls, which proved that the transient transformation was successful and could be used for further experiments.
Figure 3. Transient expression levels of miRNAs and target genes in cucumber cotyledons. Significance was determined by Duncan’s multiple range test (P < 0.05).
Function of Candidate miRNAs and Target Genes in the Interaction Between Cucumber and C. cassiicola
We performed analyses of transient expression in cucumber cotyledons to explore the functions of candidate miRNAs and target genes in disease resistance to C. cassiicola. Normal cucumber cotyledons (Control) and cucumber cotyledons injected with pRI-101 AN were used as controls. Two days after agroinfiltration for transient expression, the agroinfiltrated cotyledons were collected for disease assays by dropping spore suspensions of C. cassiicola onto cucumber cotyledons. The disease phenotype at 5 days after inoculation showed that the C. cassiicola-induced lesions in the experimental groups of miR164d, miR396b, Novel-miR7, and Novel-miR1/Novel-miR7 were significantly larger than those in the controls (Figures 4A,B), indicating that transient expression of these miRNAs in cucumber cotyledons reduced the resistance to C. cassiicola. However, the C. cassiicola-induced lesions on NAC-, APE-, 4CL-, and 4CL/PAL-infiltrated leaves were markedly smaller (Figures 4A,B) than those of the controls, showing that transient expression of these genes in cucumber cotyledons improved the resistance to C. cassiicola. Expression of the C. cassiicola Actin gene was used as a standard of fungal growth. The growth of C. cassiicola in the miR164d-, miR396b-, Novel-miR1-, and Novel-miR7-infiltrated leaves was significantly higher than that of the control, while the growth in the NAC-, APE-, 4CL-, and 4CL/PAL-infiltrated leaves was markedly lower than that of the control (Figure 4C). In addition, the disease lesions of the Novel-miR1 group did not increase significantly, but the growth of C. cassiicola increased significantly (Figures 4A–C). Transient expression assays showed that overexpression of candidate miRNAs could reduce the resistance to C. cassiicola, and overexpression of target genes corresponding to candidate miRNAs could improve the resistance to C. cassiicola. These results were consistent with those of our previous analysis.
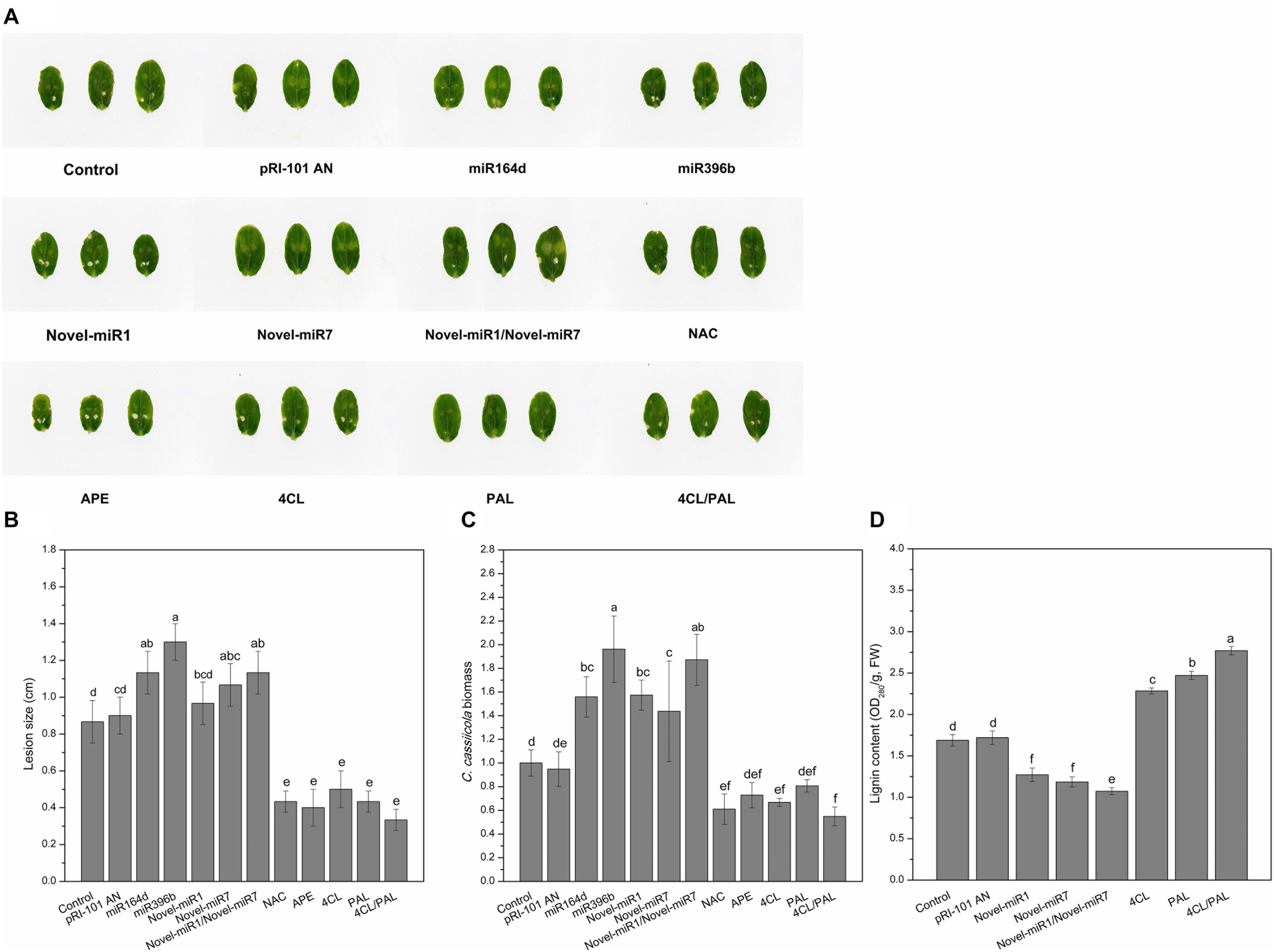
Figure 4. Disease phenotype in cucumber cotyledons transiently expressing miRNAs and target genes after inoculation with C. cassiicola. (A) Disease symptoms of cucumber cotyledons. (B) Disease lesion size of cucumber cotyledons. (C) Biomass of C. cassiicola of cucumber cotyledons. (D) Effect of transient expression of miRNAs and targets on lignin content in cucumber cotyledons. Significance was determined by Duncan’s multiple range test (P < 0.05).
The previous analysis shows that Novel-miR1 and Novel-miR7 can inhibit 4CL and PAL, respectively. The 4CL and PAL genes are two key genes in the lignin synthesis pathway, and the lignin content is positively correlated with disease resistance. In this experiment, Novel-miR1, Novel-miR7, Novel-miR1/Novel-miR7 (1:1, v/v), 4CL, PAL, and 4CL/PAL (1:1, v/v) were transiently expressed in cucumber cotyledons through agroinfiltration. Normal tobacco (Control) and tobacco injected with pRI-101 AN were used as controls. Two days after agroinfiltration for transient expression, the agroinfiltrated cotyledons were prepared for determination of lignin content (Figure 4D). The lignin content was determined to further analyze the effect of overexpression of candidate miRNAs and target genes on lignin accumulation in cucumber cotyledons. As shown in Figure 4D, the lignin content was significantly increased in 4CL-, PAL-, and 4CL/PAL-infiltrated samples compared to that of the control, and the lignin content in Novel-miR1-, Novel-miR7-, and Novel-miR1/Novel-miR7-infiltrated leaves was obviously decreased compared to that of the control. The lignin content of 4CL/PAL-infiltrated leaves was the highest, while the lignin content of Novel-miR1/Novel-miR7-infiltrated leaves was the lowest. The results showed that overexpression of 4CL and PAL could increase the lignin content in cucumber leaves, and overexpression of Novel-miR1 and Novel-miR7 could reduce lignin in cucumber leaves.
TRV-Induced VBMS
In this study, STTM-miR164d, STTM-miR396b, STTM-Novel-miR1, and STTM-Novel-miR7 recombinant vectors were constructed based on TRV. The STTM structures are shown in Figure 5A. The sequences of STTM-miR164d, STTM-miR396b, STTM-Novel-miR1, and STTM-Novel-miR7 used in this study are listed in Supplementary Table S1. The sides of the structure are TMs, and the restriction sites at both ends are EcoRI and SacI. STTM and pTRV2 were used to construct recombinant vectors by In-Fusion technology. Recombinant viruses TRV: 00 (pTRV1+pTRV2), TRV: STTM-miR164d, TRV: STTM-miR396b, TRV: STTM-Novel-miR1, and TRV: STTM-Novel-miR7 were infiltrated into cucumber cotyledons. After inoculation for 7 days, chlorosis and a few virus spots appeared in the cotyledons of cucumber inoculated with TRV recombinant virus, but no obvious phenotype was observed on the cotyledons of Control and A. tumefaciens EHA105 (Figure 5B), indicating that TRV had successfully replicated and proliferated in the cotyledons of cucumber.
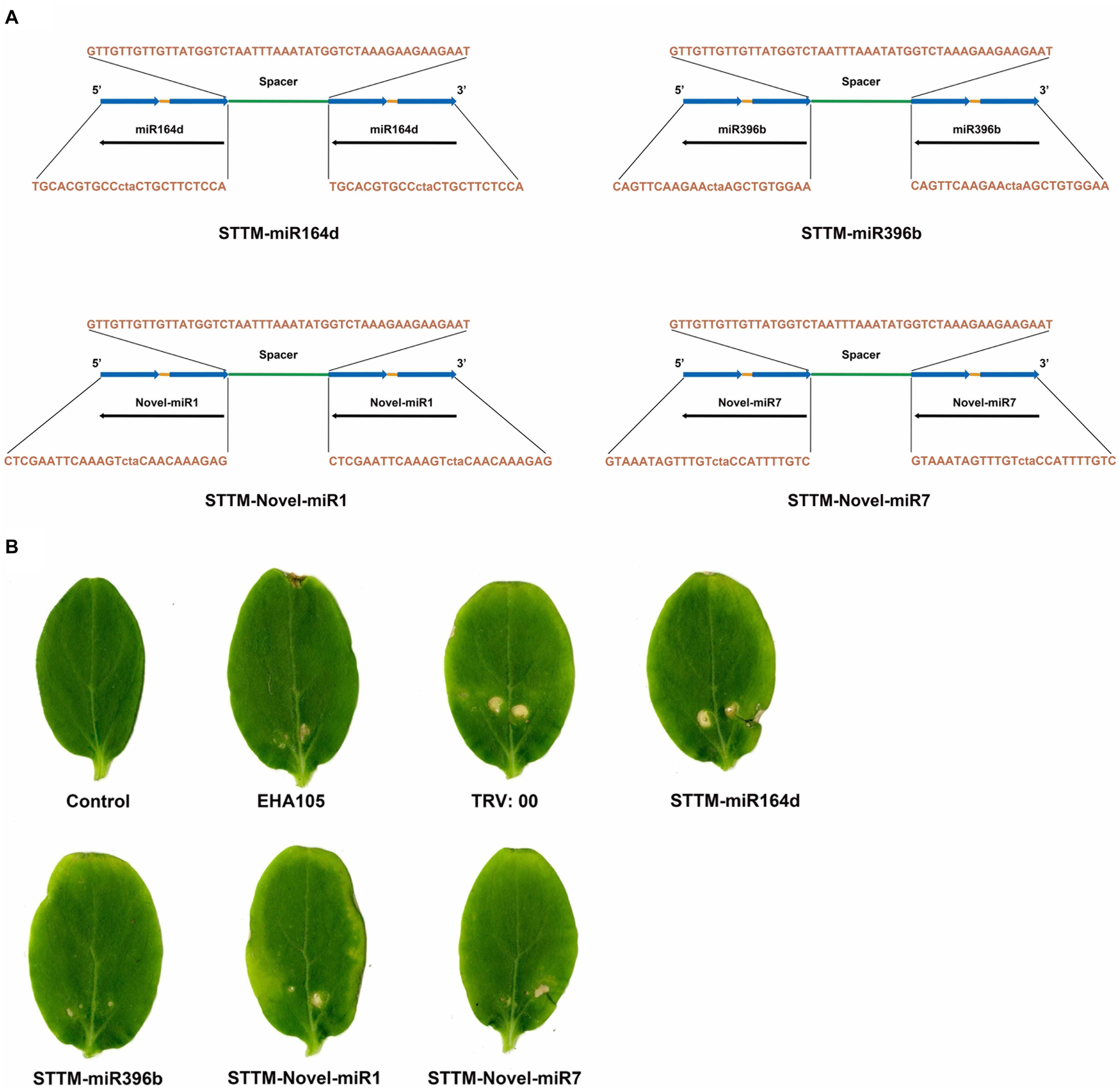
Figure 5. TRV induced STTM-miRNA silencing. (A) Diagram of STTM. (B) Symptoms in cucumber cotyledons following TRV-induced genes silencing.
To explore the silencing efficacy of candidate miRNAs, we detected the expression levels of candidate miRNAs and corresponding target genes in cucumber cotyledons injected with TRV: STTM by qRT-PCR. As shown in Figure 6, the expression levels of candidate miRNAs in the cotyledons infiltrated with TRV: STTM-miR164d, TRV: STTM-miR396b, TRV: STTM-Novel-miR1, and TRV: STTM-Novel-miR7 were significantly decreased, and the expression levels of target genes corresponding to these miRNAs were upregulated. These results indicated that candidate miRNAs in cucumber cotyledons were successfully inhibited.
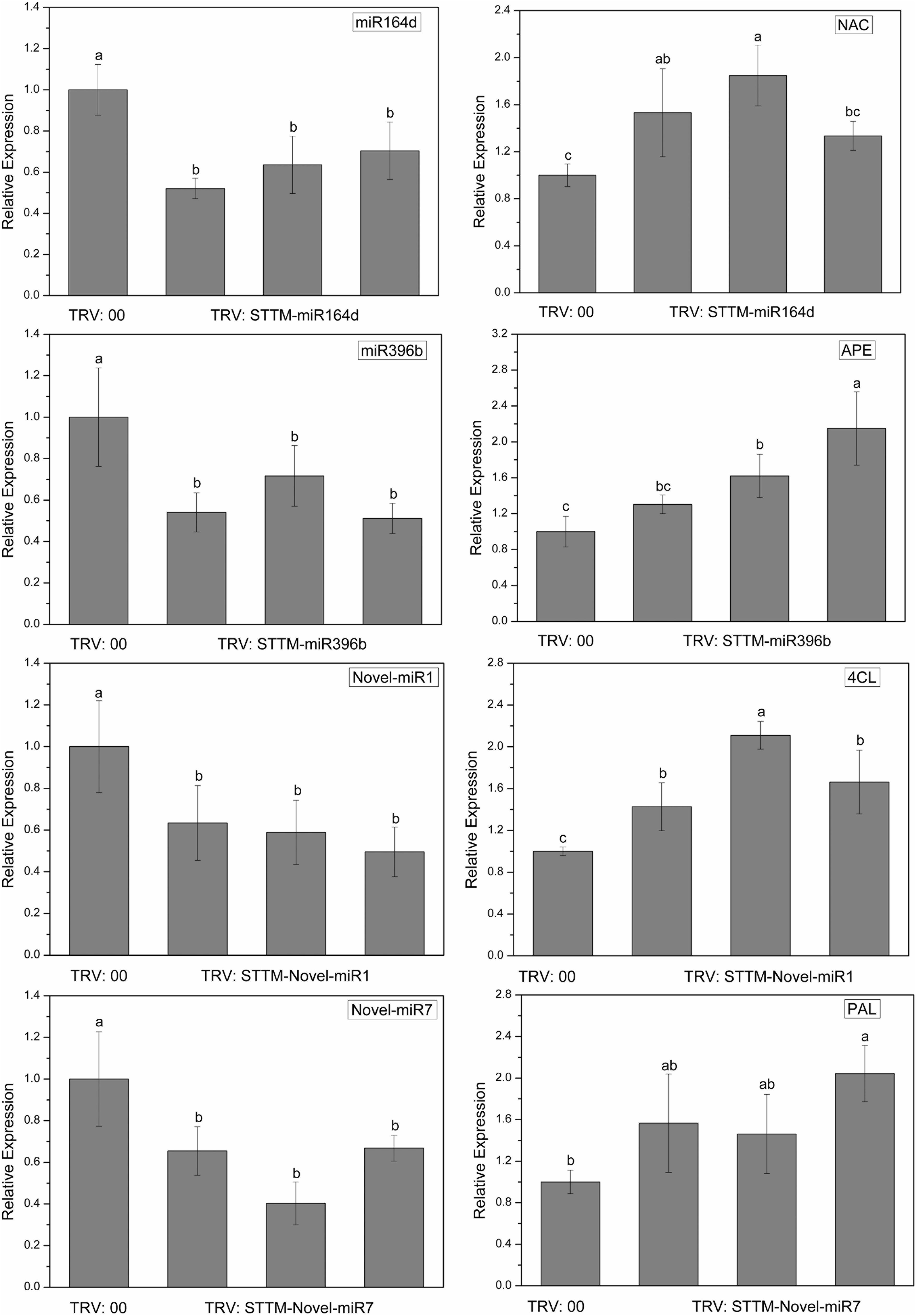
Figure 6. Expression levels of miRNAs and targets in cucumber cotyledons. Significance was determined by Duncan’s multiple range test (P < 0.05).
Response of Cucumber Cotyledons to C. cassiicola After Candidate miRNAs Silencing
To identify the role of STTM-miRNA in the interaction between cucumber and C. cassiicola, we inoculated the TRV: 00 (pTRV1+pTRV2)-, TRV: STTM-miR164d-, TRV: STTM-miR396b-, TRV: STTM-Novel-miR1-, and TRV: STTM-Novel-miR7-infiltrated cucumber cotyledons with C. cassiicola to observe phenotypic changes. After 5 days of inoculation of C. cassiicola, there was no significant difference between the disease lesions of TRV: 00 (pTRV1+pTRV2)-infiltrated cotyledons and control cotyledons. However, the C. cassiicola-induced lesions of miR164d-, miR396b-, Novel-miR1-, and Novel-miR7-silenced cucumber cotyledons were markedly smaller (Figures 7A,B) than those of the control. The biomass of C. cassiicola in the infected cotyledons of cucumber was detected by qRT-PCR (Figure 7C), and the expression of the C. cassiicola Actin gene in candidate miRNA-silenced cucumber cotyledons decreased significantly. Thus, silencing of miR164d, miR396b, Novel-miR1 and Novel-miR7 increased cucumber resistance to C. cassiicola. The lignin content was determined to further analyze the effect of silencing candidate miRNAs on lignin accumulation in cucumber cotyledons (Figure 7D). The lignin content was significantly upregulated in all silenced plants, especially in Novel-miR1- and Novel-miR7-silenced plants. Because 4CL (inhibited by Novel-miR1) and PAL (inhibited by Novel-miR7) are upstream and downstream genes in the phenylpropane pathway, which is related to lignin synthesis, this finding is also consistent with our previous experimental results.
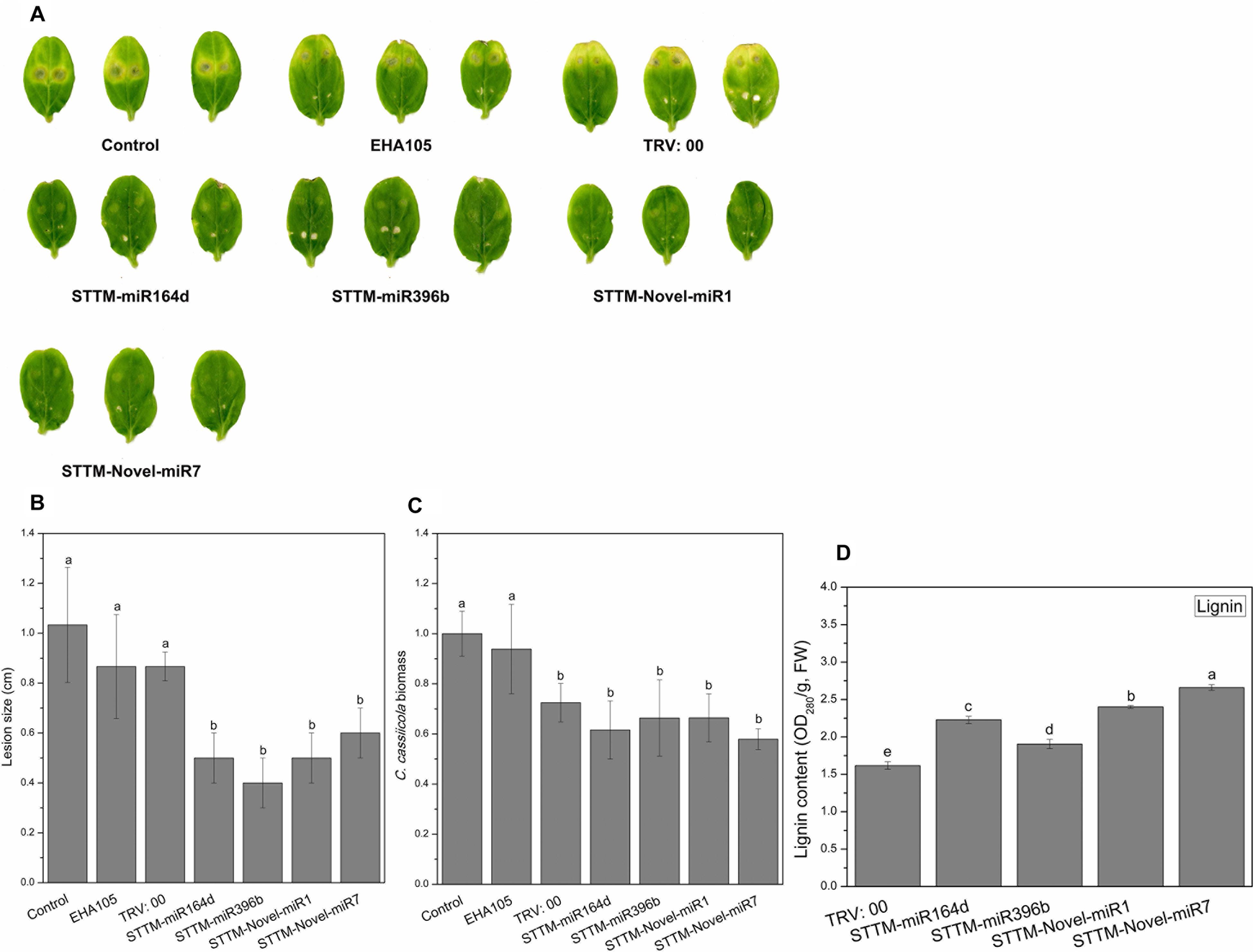
Figure 7. Disease phenotype in STTM transgenic cucumber cotyledons after inoculation with C. cassiicola. (A) Disease symptoms of cucumber cotyledons. (B) Disease lesion size of cucumber cotyledons. (C) Biomass of C. cassiicola of cucumber cotyledons. (D) Effect of candidate miRNAs silencing on lignin content in cucumber cotyledons. Significance was determined by Duncan’s multiple range test (P < 0.05).
Discussion
As miRNAs have no coding function, they can only act by inhibiting or degrading the corresponding targets. Genes related to abiotic and biotic stresses have been proven to be targets of miRNAs and can be used to determine the function of miRNAs (Jagadeeswaran et al., 2009; Katiyar-Agarwal and Jin, 2010). In plants, the interaction between miRNAs and target genes was verified by 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE) (Llave et al., 2002). The RACE method is cumbersome and does not visualize the interaction between the miRNA and target gene. Agrobacterium-mediated transient expression of tobacco is highly efficient and has a long expression time, which is suitable for the study of gene interactions in plants (Prabu and Prasad, 2012; Yin et al., 2017). This method is mainly used to verify the interaction between miRNAs and target genes by detecting the expression of the GUS gene in tobacco after transient expression of miRNAs and target genes (Feng et al., 2013; Han et al., 2016).
NAC is a plant-specific transcription factor that is involved in plant development and stress responses in plants. Previous studies have shown that many members of the NAC gene family exhibit differential expression characteristics after pathogen infection in plants, which indicates that NAC genes have specific biological functions in response to disease resistance in plants (Jensen and Skriver, 2014; Qin et al., 2014). In a recent study, researchers analyzed cucumber NAC genes related to the response to abiotic stresses (Zhang et al., 2017). However, there are few studies on the role of NAC genes in the interaction between cucumber and pathogens. BLAST analysis of existing data showed that the NAC gene we identified was csNAC30. Some studies have shown that NAC affects the synthesis of secondary metabolites, but the detailed mechanism is not clear (Zhao et al., 2010; Xu et al., 2015).
Tryptophan not only promotes endogenous jasmonic acid (JA) biosynthesis and triggers plant signal transduction pathways but also participates in the terpenoid indole alkaloid (TIA) pathway, which in turn regulates the plant response to stress (Sun et al., 2016). Anthranilate phosphoribosyltransferase (APE) is an important enzyme in the tryptophan synthesis pathway, which is indirectly involved in the synthesis of plant secondary metabolites (Dharmawardhana et al., 2013).
Phenylalanine ammonia-lyase (PAL) and 4-coumarate: CoA ligase (4CL) are two key genes in the phenylpropane synthesis pathway and are closely related to plant resistance to external stresses. First, PAL catalyzes phenylalanine to cinnamic acid; then, cinnamic acid participates in the synthesis of ubiquinone and is catalyzed by 4CL to form cinnamoyl-CoA; finally, cinnamoyl-CoA further synthesizes lignin and flavonoids (Kim and Hwang, 2014; Li et al., 2015).
After analyzing the function of the target genes, we selected miR164d, miR396b, Novel-miR1, and Novel-miR7 and their target genes NAC, APE, 4CL, and PAL for interaction verification. Histochemical staining and quantitative detection of GUS genes showed that these target genes could be suppressed by the corresponding miRNA, but the inhibition efficiency was not 100%. There are two possible causes for this result. First, there is a balance between the transcription of target genes and nucleotide cleavage, and this balance is affected by the expression level of miRNAs (Nikovics et al., 2006). Second, it may be that only a portion of the target mRNA was cleaved and degraded by the miRNA, and the remaining target mRNAs were detached from the cleavage system and normally transcribed (Adam et al., 2010).
To explore the functions of miR164d, miR396b, Novel-miR1, and Novel-miR7 and their target genes NAC, APE, 4CL, and PAL in cucumber response to C. cassiicola, we transiently expressed candidate miRNAs and target genes in cucumber cotyledons. After vaccination with C. cassiicola, phenotypic changes were observed. Because 4CL and PAL are upstream and downstream genes in the phenylpropanoid metabolic pathway, we coexpressed 4CL and PAL to establish the experimental group. Similarly, Novel-miR1 and Novel-miR7 were also coexpressed. Figure 4 shows that the disease resistance of the target gene overexpression groups was significantly higher than that of the control groups, indicating that these genes play an important role in cucumber resistance to C. cassiicola. The disease resistance of the candidate miRNAs overexpression group was lower than that of the control group, probably because these miRNAs inhibited the expression of their target genes. These results were also confirmed by measurement of the biomass of C. cassiicola in cucumber cotyledons after infection. The data showed that the coexpressed 4CL and PAL groups had the highest disease resistance, and the disease resistance of the corresponding Novel-miR1 and Novel-miR7 coexpression groups was the lowest. Because the phenylpropanoid metabolic pathway affects the synthesis of lignin, and 4CL and PAL are important genes in this pathway, we performed lignin content determination after transient expression of candidate miRNAs and genes in cucumber cotyledons for 2 days. Figure 4D shows that the lignin content of 4CL/PAL-infiltrated leaves was the highest; the lignin content of Novel-miR1-, Novel-miR7-, and Novel-miR1/Novel-miR7-infiltrated leaves was lower than that of the normal tobacco, and the lignin content reduction was more severe in the Novel-miR1/Novel-miR7-infiltrated leaves. STTM is an effective tool for studying miRNA function in plants and animals. Tang et al. (2012) developed STTM technology based TM. Overexpression of STTM has been used to identify the functions of miRNAs in a variety of plants, such as Arabidopsis and wheat (Jia et al., 2015; Jiao et al., 2015), but it has not been reported in cucumber. To further explore the function of candidate miRNAs, we developed a TRV-induced VBMS to silence cucumber endogenous miRNAs. The results showed that silencing of miR164d, miR396b, Novel-miR1 and Novel-miR7 increased the resistance of cucumber cotyledons to C. cassiicola, indicating that these miRNAs played a negative regulatory role in cucumber resistance to C. cassiicola. Notably, lignin content is highest in Novel-miR1- and Novel-miR7-silenced cucumber cotyledons. These findings also indicate that the interaction between Novel-miR1 and Novel-miR7 and their target genes affects the synthesis of lignin, which in turn affects the resistance of cucumber to C. cassiicola (Figure 8).
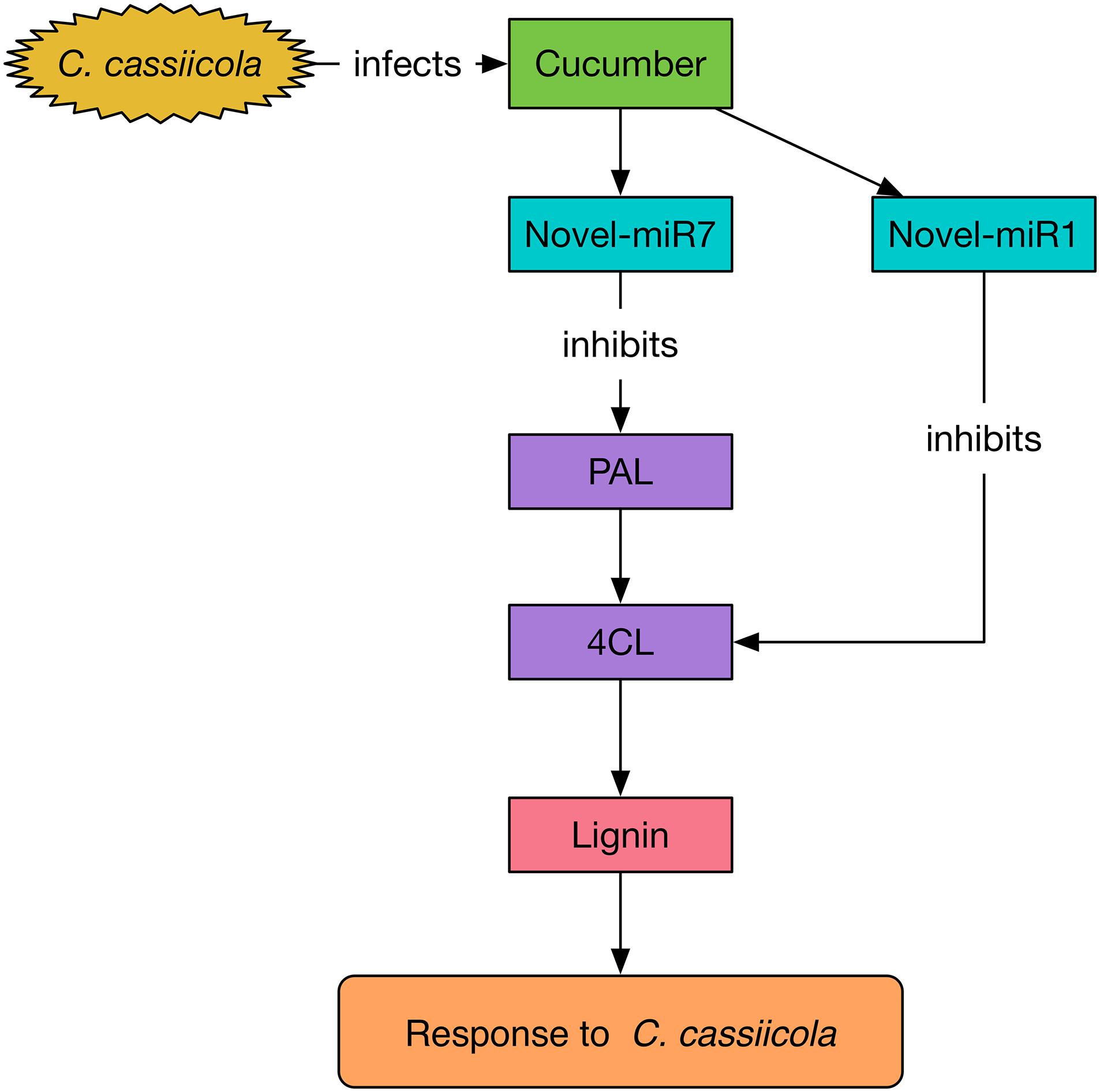
Figure 8. Mechanism of Novel-miR1 and Novel-miR7 regulation network involved in the response to C. cassiicola.
Conclusion
In our study, the expression vectors were constructed by In-Fusion technology, and the negative relationships of miR164d, miR396b, Novel-miR1, and Novel-miR7 and their targets were verified by the Agrobacterium-mediated tobacco transient expression system. Meanwhile, we found that overexpression of NAC, APE, 4CL, and PAL could improve the resistance to C. cassiicola, and overexpression of miR164d, miR396b, Novel-miR1, and Novel-miR7 could reduce the disease resistance in cucumber. We confirmed that silencing candidate miRNAs could improve the disease resistance of cucumber, and the lignin content in Novel-miR1- and Novel-miR7-silenced cucumber cotyledons was significantly increased. These candidate miRNAs and targets are closely related to cucumber lignin synthesis, and the data combined with previous analyses demonstrate the important role of secondary metabolism, especially the lignin metabolism pathway, in the process of cucumber resistance to C. cassiicola.
Data Availability
The datasets generated for this study can be found in NCBI Sequence Read Archive (SRA), SRP 117262 and SRP117230.
Author Contributions
XW and HF conceived and designed the research. XW performed the experiments and wrote the manuscript. All authors analyzed the data and read and approved the final manuscript. HF revised the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (31772314), Key Project of Natural Science Foundation of Liaoning Province (2017012023-301), and Science and Technology NOVA Program of Shenyang (RC170439).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00668/full#supplementary-material
Footnotes
References
Adam, H., Marguerettaz, M., Qadri, R., Adroher, B., Richard, F., Collin, M., et al. (2010). Divergent expression patterns of miR164 and CUP-SHAPED COTYLEDON genes in palms and other monocots: implication for the evolution of meristem function in angiosperms. Mol. Biol. Evol. 28, 1439–1454. doi: 10.1093/molbev/msq328
Agrawal, A. A., and Weber, M. G. (2015). On the study of plant defence and herbivory using comparative approaches: how important are secondary plant compounds. Ecol. Lett. 18, 985–991. doi: 10.1111/ele.12482
Bachan, S., and Dinesh-Kumar, S. P. (2012). Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods Mol. Biol. 89, 83–92. doi: 10.1007/978-1-61779-882-5_6
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Burkhardt, A., and Day, B. (2016). Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome 9, 1–19. doi: 10.3835/plantgenome2015.08.0069
Dharmawardhana, P., Ren, L., Amarasinghe, V., Monaco, M., Thomason, J., Ravenscroft, D., et al. (2013). A genome scale metabolic network for rice and accompanying analysis of tryptophan, auxin and serotonin biosynthesis regulation under biotic stress. Rice 6:15. doi: 10.1186/1939-8433-6-15
Feng, H., Zhang, Q., Wang, Q., Wang, X., Liu, J., Li, M., et al. (2013). Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol. Biol. 83, 433–443. doi: 10.1007/s11103-013-0101-9
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Han, L., Weng, K., Ma, H., Xiang, G., Li, Z., Wang, Y., et al. (2016). Identification and characterization of erysiphe necator-responsive microRNAs in Chinese wild vitis pseudoreticulata by high-throughput sequencing. Front. Plant Sci. 7:621. doi: 10.3389/fpls.2016.00621
Huang, C. J., Qian, Y. J., Li, Z. H., and Zhou, X. P. (2012). Virus-induced gene silencing and its application in plant functional genomics. Sci. China Life Sci. 55, 99–108. doi: 10.1007/s11427-012-4280-4
Ishii, H., Yano, K., Date, H., Furuta, A., Sagehashi, Y., Yamaguchi, T., et al. (2007). Molecular characterization and diagnosis of QoI resistance in cucumber and eggplant fungal pathogens. Phytopathology 97, 1458–1466. doi: 10.1094/PHYTO-97-11-1458
Jagadeeswaran, G., Saini, A., and Sunkar, R. (2009). Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229, 1009–1014. doi: 10.1007/s00425-009-0889-3
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1089/dna.1987.6.583
Jensen, M. K., and Skriver, K. (2014). NAC transcription factor gene regulatory and protein-protein interaction networks in plant stress responses and senescence. IUBMB Life 66, 156–166. doi: 10.1002/iub.1256
Jia, X. Y., Ding, N., Fan, W. X., Yan, J., Gu, Y. Y., Tang, X. Q., et al. (2015). Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 233, 11–21. doi: 10.1016/j.plantsci.2014.12.020
Jiang, N., Meng, J., Cui, J., Sun, G., and Luan, Y. (2018). Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic. Res. 5:9. doi: 10.1038/s41438-018-0017-2
Jiao, J., Wang, Y., Selvaraj, J. N., Xing, F., and Liu, Y. (2015). Barley stripe mosaic virus (BSMV) induced microRNA silencing in common wheat (Triticum aestivum L.). PLoS One 10:e0126621. doi: 10.1371/journal.pone.0126621
Jin, W. B., and Wu, F. L. (2015). Identification and characterization of cucumber microRNAs in response to Pseudoperonospora cubensis infection. Gene 569, 225–232. doi: 10.1016/j.gene.2015.05.064
Katiyar-Agarwal, S., and Jin, H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246. doi: 10.1146/annurev-phyto-073009-114457
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. BBA Gene Regul. Mech. 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kim, D. S., and Hwang, B. K. (2014). An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 65, 2295–2306. doi: 10.1093/jxb/eru109
Kwon, M. K., Kang, B. R., Cho, B. H., and Kim, Y. C. (2003). Occurrence of target leaf spot disease caused by Corynespora cassicolaon cucumber in Korea. Plant Pathol. 52:424. doi: 10.1046/j.1365-3059.2003.00846.x
Li, B., Gao, W., Shi, Y. X., and Xie, X. W. (2012). Progress in researches on Corynespora leaf spot. Acta Phytophylacica Sin. 39, 171–176.
Li, C. H., Li, Y., Bai, L., Zhang, T., He, C., Yan, Y., et al. (2014). Grafting-responsive miRNAs in cucumber and pumpkin seedlings identified by high-throughput sequencing at whole genome level. Physiol. Plant. 151, 406–422. doi: 10.1111/ppl.12122
Li, Y., Kim, J. I., Pysh, L., and Chapple, C. (2015). Four isoforms of Arabidopsis thaliana 4-coumarate: CoA ligase (4CL) have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. doi: 10.1104/pp.15.00838
Liu, Q., Wang, H., Hu, H., and Zhang, H. (2015). Genome-wide identification and evolutionary analysis of positively selected miRNA genes in domesticated rice. Mol. Genet. Genom. 290, 593–602. doi: 10.1007/s00438-014-0943-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
MacFarlane, S. A., Vassilakos, N., and Brown, D. J. (1999). Similarities in the genome organization of tobacco rattle virus and pea early-browning virus isolates that are transmitted by the same vector nematode. J. Gen. Virol. 80, 273–276. doi: 10.1099/0022-1317-80-1-273
Mao, W. H., Li, Z. Y., Xia, X. J., Li, Y. D., and Yu, J. Q. (2012). A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS One 7:e33040. doi: 10.1371/journal.pone.0033040
Martínez, G., Forment, J., Llave, C., Pallás, V., and Gómez, G. (2011). High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS One 6:e19523. doi: 10.1371/journal.pone.0019523
Meng, Y., Shao, C., Wang, H., and Jin, Y. (2012). Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genom. 13:197. doi: 10.1186/1471-2164-13-197
Miyamoto, T., Ishii, H., Stammler, G., Koch, A., Ogawara, T., Tomita, Y., et al. (2010). Distribution and molecular characterization of Corynespora cassiicola isolates resistant to boscalid. Plant Pathol. 59, 873–881. doi: 10.1111/j.1365-3059.2010.02321.x
Morrison, I. M. (1972). A semi-micro method for the determination of lignin and its use in predicting the digestibility of forage crops. J. Sci. Food Agr. 23, 455–463. doi: 10.1002/jsfa.2740230405
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Nghia, N. A., Kadir, J., Sunderasan, E., Puad, A. M., Malik, A., and Napis, S. (2008). Morphological and inter simple sequence repeat (ISSR) markers analyses of Corynespora cassiicola isolates from rubber plantations in Malaysia. Mycopathologia 166, 189–201. doi: 10.1007/s11046-008-9138-8
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Pateraki, I., and Kanellis, A. K. (2010). Stress and developmental responses of terpenoid biosynthetic genes in Cistus creticus subsp. creticus. Plant Cell Rep. 29, 629–641. doi: 10.1007/s00299-010-0849-1
Prabu, G., and Prasad, D. T. (2012). Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormone. Plant Cell Rep. 31, 661–669. doi: 10.1007/s00299-011-1183-y
Pusztahelyi, T., Holb, I. J., and Pócsi, I. (2015). Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6:573. doi: 10.3389/fpls.2015.00573
Qi, Y. X., Zhang, X., Pu, J. J., Liu, X. M., Lu, Y., Zhang, H., et al. (2011). Morphological and molecular analysis of genetic variability within isolates of Corynespora cassiicola from different hosts. Eur. J. Plant Pathol. 130, 83–95. doi: 10.1007/s10658-010-9734-6
Qin, X., Zheng, X., Huang, X., Lii, Y., Shao, C., Xu, Y., et al. (2014). A novel transcription factor JcNAC1 response to stress in new model woody plant Jatropha curcas. Planta 239, 511–520. doi: 10.1007/s00425-013-1993-y
Sanan-Mishra, N., Kumar, V., Sopory, S. K., and Mukherjee, S. K. (2009). Cloning and validation of novel miRNA from basmati rice indicates cross talk between abiotic and biotic stresses. Mol. Genet. Genom. 282, 463. doi: 10.1007/s00438-009-0478-y
Sha, A., Zhao, J., Yin, K., Tang, Y., Wang, Y., Wei, X., et al. (2014). Virus-based microRNA silencing in plants. Plant Physiol. 164, 36–47. doi: 10.1104/pp.113.23110
Shang, Y., Ma, Y., Zhou, Y., Zhang, H., Duan, L., Chen, H., et al. (2014). Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346, 1084–1088. doi: 10.1126/science.1259215
Slack, F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R., and Ruvkun, G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669. doi: 10.1016/S1097-2765(00)80245-2
Sun, J. Y., Manmathan, H., Sun, C., and Peebles, C. A. M. (2016). Examining the transcriptional response of overexpressing anthranilate synthase in the hairy roots of an important medicinal plant Catharanthus roseus by RNA-seq. BMC Plant Biol. 16:108. doi: 10.1186/s12870-016-0794-4
Tang, G. L., Yan, J., Gu, Y. Y., Qiao, M. M., Fan, R. W., Mao, Y. P., et al. (2012). Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58, 118–125. doi: 10.1016/j.ymeth.2012.10.006
Teramoto, A., Martins, M. C., Ferreira, L. C., and Cunha, M. G. (2011). Reaction of hybrids, inhibition in vitro and target spot control in cucumber. Hortic. Bras. 29, 342–348. doi: 10.1590/S0102-05362011000300014
Thiebaut, F., Grativol, C., Hemerly, A. S., and Ferreira, P. C. G. (2015). MicroRNA networks in plant-microorganism interactions. Trop. Plant Biol. 8, 40–50. doi: 10.1007/s12042-015-9149-9
Wang, X., Zhang, D., Cui, N., Yu, Y., Yu, G., and Fan, H. (2018). Transcriptome and miRNA analyses of the response to Corynespora cassiicola in cucumber. Sci. Rep. 8:7798. doi: 10.1038/s41598-018-26080-6
Waterhouse, P. M., and Hellens, R. P. (2015). Coding in non-coding RNAs. Nature 520, 41–42. doi: 10.1038/nrg3074
Wen, C., Mao, A., Dong, C., Liu, H., Yu, S., Guo, Y. D., et al. (2015). Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber, Cucumis sativus L. Theor. Appl. Genet. 128, 2495–2506. doi: 10.1007/s00122-015-2604-z
Xu, Q., Wang, W., Zeng, J., Zhang, J., Grierson, D., Li, X., et al. (2015). A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 102, 25–31. doi: 10.1016/j.postharvbio.2015.02.002
Yan, J., Gu, Y., Jia, X., Kang, W., Pan, S., Tang, X., et al. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427. doi: 10.1105/tpc.111.094144
Yang, L. M., Koo, D. H., Li, Y. H., Zhang, X. J., Luan, F. S., Havey, M. J., et al. (2012). Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 71, 895–906. doi: 10.1111/j.1365-313X.2012.05017.x
Yin, J. L., Wong, W. S., Jang, I. C., and Chua, N. H. (2017). Co-expression of peppermint geranyl diphosphate synthase small subunit enhances monoterpene production in transgenic tobacco plants. New Phytol. 213, 1133–1144. doi: 10.1111/nph.14280
Zhang, B., and Wang, Q. (2015). MicroRNA-based biotechnology for plant improvement. J. Cell Physiol. 230, 1–15. doi: 10.1002/jcp.24685
Zhang, X., Zhao, H., Gao, S., Wang, W. C., Katiyar-Agarwal, S., Huang, H. D., et al. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393∗-mediated silencing of a Golgi-localized snare gene, MEMB12. Mol. Cell 42, 356–366. doi: 10.1016/j.molcel.2011.04.010
Zhang, X. M., Yu, H. J., Sun, C., Deng, J., Zhang, X., Liu, P., et al. (2017). Genome-wide characterization and expression profiling of the NAC genes under abiotic stresses in Cucumis sativus. Plant Physiol. Biochem. 113, 98–109. doi: 10.1016/j.plaphy.2017.01.023
Zhao, Q., and Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhao, Q., Gallego-Giraldo, L., Wang, H., Zeng, Y., Ding, S. Y., Chen, F., et al. (2010). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63, 100–114. doi: 10.1111/j.1365-313X.2010.04223.x
Keywords: cucumber, Corynespora cassiicola, microRNA, transient transformation, short tandem target mimic, lignin
Citation: Wang X, Yu G, Zhao J, Cui N, Yu Y and Fan H (2019) Functional Identification of Corynespora cassiicola-Responsive miRNAs and Their Targets in Cucumber. Front. Plant Sci. 10:668. doi: 10.3389/fpls.2019.00668
Received: 09 March 2019; Accepted: 02 May 2019;
Published: 31 May 2019.
Edited by:
Yiqun Weng, University of Wisconsin–Madison, United StatesCopyright © 2019 Wang, Yu, Zhao, Cui, Yu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Fan, aHlmYW43NEAxNjMuY29t
 Xiangyu Wang
Xiangyu Wang Guangchao Yu1,2
Guangchao Yu1,2 Na Cui
Na Cui Haiyan Fan
Haiyan Fan