Abstract
Rapeseed is one of the most important oilseed crops in the world. Improving the production of rapeseed is beneficial to relieve the shortage of edible vegetable oil. As the organ of support and transport, the main stem of rapeseed controls the plant architecture, transports the water and nutrients, and determines the number of inflorescence. Increasing the number of main stems would be helpful for the yield improvement in Brassica napus (B. napus). This attractive multi-main stem (MMS) trait was observed in the KN DH population. We investigated not only the frequency of MMS traits but also dissected the genetic basis with QTL mapping analysis and Gene-Fishing technique. A total of 43 QTLs were identified for MMS based on high-density linkage map, which explained 2.95–14.9% of the phenotypic variation, among which two environmental stable QTLs (cqMMS.A3-2 and cqMMS.C3-5) were identified in winter and semi-winter environments. Epistatic interaction analysis indicated cqMMS.C3-5 was an important loci for MMS. According to the functional annotation, 159 candidate genes within QTL confidence intervals, corresponding to 148 Arabidopsis thaliana (A. thaliana) homologous genes, were identified, which regulated lateral bud development and tiller of stem, such as shoot meristemless (STM), WUSCHEL-regulated-related genes, cytokinin response factors (CRF5), cytokinin oxidase (CKX4), gibberellin-regulated (RDK1), auxin-regulated gene (ARL, IAR4), and auxin-mediated signaling gene (STV1). Based on Gene-Fishing analysis between the natural plants and the double-main stem (DMS) plant, 31 differentially expressed genes (DEGs) were also obtained, which were related to differentiation and formation of lateral buds, biotic stimulus, defense response, drought and salt-stress responses, as well as cold-response functional genes. In addition, by combining the candidate genes in QTL regions with the DEGs that were obtained by Gene-Fishing technique, six common candidate genes (RPT2A, HLR, CRK, LRR-RLK, AGL79, and TCTP) were identified, which might probably be related to the formation of MMS phenotype. The present results not only would give a new insight into the genetic basis underlying the regulation of MMS but also would provide clues for plant architecture breeding in rapeseed.
Introduction
Plant architecture is a significant agronomic trait that indirectly influence the seed yield by affecting the formation of photosynthetic product and storage of grain-filling substance. Notably, the first “Green Revolution” was one of the great successes, which had improved plant architecture, especially in significant reduction plant height and an increase of the overall seed yield and harvest index (Khush, 2001). Seed yield per unit area is closely related to many plant architecture elements, including yield component, plant height, tiller number, intersection angle of tiller, and leaf shape. Especially in rice and wheat, the increase of the effective tiller number could improve their seed yield per plant and thus increase the overall yield. Some studies that are focused on the mechanism of plant architecture formation have been reported in rice, Arabidopsis thaliana (A. thaliana) and tomato (Leibfried et al., 2005; Jiang et al., 2013; Zhou et al., 2016). Therefore, improving the plant architecture of crops could effectively increase the crop yield.
Rapeseed (Brassica napus, AACC genome, 2n = 38) originated from a spontaneous hybridization between Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18) (Nagaharu, 1935). Rapeseed is cultivated throughout the world for the production of vegetable oil, animal feed, and biodiesel. At present, the proportion of rapeseed oil in the total oil production of crop is as high as 57.2% in China (Xie et al., 2005; Yin et al., 2010); in the meanwhile, 60% of edible oil have to be imported from abroad (Wang and Yin, 2014). Seed yield of rapeseed is directly determined by yield-component traits, including seed number per silique, seed weight, and the number of effective siliques per plant (Qzer et al., 1999; Quarrie et al., 2006). Several indirectly related traits, including plant height, the number of the first effective branch, and the silique number of main inflorescence, were also important contributions. The genetic basis of the seed yield components and seed yield–related traits have been dissected by quantitative trait loci (QTL) mapping (Chen et al., 2007; Ding et al., 2012; Zhao et al., 2016) and genome-wide association studies (GWAS) (Liu et al., 2016; Lu et al., 2016).
In recent years, the new agronomic traits, multi-main stem trait (MMS, including double-main stems, three main stems, and over three main stems), have been discovered in rapeseed. MMS is an important seed yield–related trait, which was differentiated from shoot meristem. The plants with MMS have many advantages, such as multiple main stems (or tiller number), higher growth potential, fewer branches, and more seed number, which play an important role for increasing seed yield in rice and wheat (Lu et al., 2015; Shao et al., 2019; Hu et al., 2017). Favorable main stem number can also increase planting density, which contributes to a higher yield per unit area. In the process of differentiation, the morphological structure, physiological status, and endogenous hormonal content of the shoot apical meristem (SAM) undergo substantial change (Gordon et al., 2009). The SAM must maintain a balance between stem cell niches updating and peripheral organ initiation in order to fulfill this function. The genes that cause MMS might have a relationship with WUSCHEL (WUS) and CLAVATA (CLV) (Brand et al., 2002; Müller et al., 2006). In normal condition, the gene WUS is always expressed in a small group of cells in the organization center of the SAM, underneath the three outer cell layers (Weigel and Jürgens, 2002; Williams and Fletcher, 2005). Cells in center zone proliferate to form two kinds of cells (Haecker and Laux, 2001): one is progeny of stem cell holding multipotency, which maintain the structure of center zone, and the other is a daughter cell, who will develop into perimeter zone and then differentiate into various organizations. Only when the two progresses develop at the same pace, SAM will develop into normal structure; otherwise, the SAM will abnormally develop (Wang and Li, 2008). During development process of SAM, WUS functions in maintaining the stem cells undifferentiated to keep natural structure and function of SAM (Schoof et al., 2000). Loss of function of WUS would lead to a large amount of stem cells developed into perimeter zone, and then giving rise to the leaf primordiums and secondary SAMs that will come into being (Wang and Li, 2008). Another important gene in SAM is CLV that can promote stem cells to develop into perimeter zone. A WUS-CLV feedback loop in plant exists, which is of great importance to maintain normal structure of SAM. CLV3 encodes a kind of secreted protein to activate CLV1/CLV2 complex when the number of stem cells in SAM increase. When WUS is inhibited, the number of stem cells will decrease which in turn restrains the expression of CLV3 (Fletcher and Meyerowitz, 2000; Dodsworth, 2009). Based on the previous reports, the maintenance homeostasis of SAM mainly includes WUS-CLV feedback loop (Fletche, 2018), phytohormone (Brand et al., 2000), redox regulation (Schippers et al., 2016), and network consisting of various genes.
QTL mapping has been proved to be an important approach for dissecting complex traits (Paran and Zamir, 2003). Many agricultural traits, for example, seed yield (Zhao et al., 2016), seed yield–related traits (Shi et al., 2009; Zhao et al., 2016), oil content (Wang et al., 2013), and fatty acid composition (Jiang et al., 2014; Bao et al., 2018), have been dissected by QTL mapping. In addition, some important genes that regulated agricultural traits of rapeseed were identified by fine mapping and map-based cloning, such as recessive genic male serility BNAMS1 (Yi et al., 2006), seed weight TSWA5a and TSWA5c (Fan et al., 2010), and seed number per silique BnaC9.SMG7b (Li et al., 2015), etc. Gene-Fishing technology, based on PCR technology, could identify the differentially expressed genes quickly (Kim et al., 2004). For example, Choi et al. (2008) found 31 differentially expressed fragments in the grapevines inoculated with Rhizobium and salicylic acid by using Gene-Fishing (Choi et al., 2008). Junaedi et al. (2007) explored the effects of high allelopathic rice varieties on barnyard grass and discovered nine differentially expressed fragments (Junaedi et al., 2007). Park et al. (2006) screened the related genes that regulated seed epidermal needling of carrot, and 11 differentially expressed fragments were identified (Park et al., 2006). In addition, some other genes, including HIP1, CSH1, PPTSPO1, and SLFN-2, were also identified by Gene-Fishing technique (Cho et al., 2006; Bradley et al., 2007; Frank et al., 2007; Sohn et al., 2007).
In the present study, the MMS characteristics were investigated in a doubled-haploid population named as KN DH population. We counted the number of multi-main stem in each DH line in Yangling and Wuhan for two successive years. Based on QTL mapping and Gene-Fishing technique, the purposes of the present study focused on two aspects: (1) identify QTLs for MMS and obtain candidate genes within QTL regions (2) based on differentially expressed genes and candidate gene within QTL regions and identify several common candidate genes that regulated the MMS.
Materials and Methods
Plant Materials and Field Experimental Design in Two Different Ecological Regions
The KN doubled-haploid (DH) population were derived from bi-parental segregating populations by microspore culture (Figure 1) (Wang et al., 2013). The KN population and its parents were sown in the middle of September in the experimental field of Hybrid Rape Research Center of Shaanxi Province, Yangling of Shaanxi province (YL) in northwest China (34°16’N, 108°5’E), and in the early October in the experimental field of Huazhong Agriculture University, Wuhan of Hubei province (WH) in central China (30°47’N, 114°35’E), for two consecutive years from the years 2014 to 2015, respectively. Yangling belonged to the winter ecological region, and Wuhan belonged to the semi-winter ecological region. The KN population and their two parents were planted in Yangling and Wuhan with two replications, respectively. Meanwhile, each particular combination of experimental year × location was defined as an independent experiment. Each independent experiment consisted of 348 DH lines. Each line was grown in a two-row plot with 40 cm between rows and 20 cm between individuals, and row length of 250 cm in all independent experiments. The field experiments followed a randomized complete block design and the normal agricultural practice.
Figure 1

The phenotype of the multi-main stem and their parents in KN population. The parents of KN population include the male parent KenC-8 and the female parent N53-2. The double-main stem and three main stems are located in the KN population.
Phenotypic Statistics and Analysis of MMS
The total number of each line corresponding to the number of plant with MMS in each line was investigated, and the ratio of the MMS was calculated in Yangling of Shaanxi province and Wuhan of Hubei province for two consecutive years. The ratio of the MMS was calculated as the ratio of the number of the MMS to the total number of each line. Not all of the DH lines had MMS in the KN population. In addition, the variance analysis and broad-sense heritability (h2) was calculated by using SAS 8 (SAS Institute Inc, 2000). The formula was h2 = б2g/(б2g + б2ge/n+ б2e/nr) × 100%, б2g is the variance among DH lines, б2ge is the interaction variance of the genotype with environment, б2e is the error variance, n is the number of environments, and r is the number of replications. Meanwhile, a DH lines with double-main stem (DMS) in KN DH population were selected for germination and morphological observation. The early morphology was observed in the five growth stages of seedling stage (15, 25, 30, 35, and 50 days). The seedlings for early morphological observation of the DMS materials and their natural plant type were grown in the culture chamber with 16 h light per day at 22°C ± 4°C.
QTL Mapping and Identification of Candidate Genes
The ratio of MMS in the KN population was collected as phenotype and used for QTL mapping. A high-density genetic linkage map of the KN population with 3,207 markers, including single-nucleotide polymorphism (SNP), SSR (simple sequence repeat), STS (sequence-tagged site), SRAP (sequence-related amplified polymorphism), and IFLP (intron fragment length polymorphism) markers, was constructed. The total length reached 3072.7 cM, and the average distance was only 0.96 cM between adjacent markers (Chao et al., 2017). Combining the ratio data of MMS with the high-density linkage map of KN population, QTL detection for MMS was performed using a composite interval mapping (CIM) with Windows QTL Cartographer 2.5 software (Wang et al., 2007). The scan walking speed was 1 cM; the window size was 10 cM with five background cofactors. According to the 1,000-permutation test corresponding to P = 0.05, the LOD value 2.0 was used as threshold for detection of significant QTLs. All detected QTLs were denoted as significant identified QTLs (Burns et al., 2003). The method for QTL nomenclature was as described by Wang et al. (2013). QTL integration was by meta-analysis using BioMercator 4.1 (Arcade et al., 2004). One QTL that was detected in at least two environments was taken to be a stable QTL, otherwise a specific QTL.
The alignment of the genetic map to the physical map and the identification of candidate genes were the same as those described by Chao et al. (2017). According to the collinearity of the high-density genetic map and B. napus “Darmor-bzh” reference genome (Chalhoub et al., 2014), genome regions corresponding to the QTL confidence intervals (CIs) were identified by using closely linked SNP within QTL CIs. The candidate genes within QTL CIs were regarded as candidate genes (Shi et al., 2015). The orthologs of A. thaliana involving SAM were gathered from previous reported (Wang and Li, 2008). The orthologous of candidate genes and their annotation were obtained by BLASTn based on A. thaliana database (http://www.arabidopsis.org/).
Additive × Additive Epistatic Interactions for MMS
To estimate the epistasis interaction of MMS, we used inclusive ICIM method with IciMapping software to analyze epistatic loci pairs (Li et al., 2008; Meng et al., 2015). To identify significant epistasis interaction loci, a walking speed was set to 1 cM, and significant LOD threshold was set at the same as default parameter of 5.0. The epistasis interaction loci were defined by abbreviation “Ep” of epistasis and trait name and chromosome number as well as location on chromosome (e.g., EpMMS.A8-115).
Acquisition of Differential Expression Genes Based on Gene-Fishing™ Technology
Differential expression genes (DEGs) were identified by using Gene-Fishing™ DEG Premix Kits (Seegene, Seoul, South Korea), with an annealing control primer (ACP)–based PCR method (Kim et al., 2004). The total RNA of the DMSs was extracted and then reversed transcription for cDNA by using ReverTra Ace-a-(code no. FSK-100, TOYOBO). According to Gene-Fishing™ PCR liquid system (20µl total volume system included cDNA 5µl, arbitrary ACP [5 µM] 2µl, dT-ACP2 [10 µM] 1µl, distilled water 2µl, 2×SeeAmp™ACP™Master Mix 10µl). The amplified PCR products were separated by 2% agarose gels electrophoresis with ethidium bromide, and differential expression fragments were discriminated. Differential expression fragments were isolated by using AxyPrep DNA Gel Extraction Kit (TIANGEN) and connected to a pMD 18-T Simple Vector (TaKaRa) and were sequenced. Sequencing data were confirmed with the GenBank database through the BlastX program of NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) and/or the A. thaliana and B. napus database. Then, the sequence differences of genes were compared in natural plant and the DMS plant.
Quantitative Real-Time PCR (qPCR) Analysis of Six Potential Candidate Genes Regulating MMS
In order to investigate the potential candidate genes regulating MMS, several common candidate genes were selected for quantitative real-time PCR analysis. The shoot meristem of the natural plant and DMS plant in 5, 10, 15, and 20 days after germination was collected and used for relative expression analysis. The total RNA extraction and reversed transcription were the same as above method. Gene-specific primers were designed by using Primer 5.0, and the primers of the six target genes and reference gene actin were listed in Table S1. The RT-PCR reaction system was performed with three technical replicates by using TOYOBO SYBR® R Green Realtime PCR Master Mix (code no. QPK-201) Kit. The amplification program was as follow: 95°C for 10 min, then 40 cycles with 95°C for 15s, 60°C for 15 s, and 72°C for 30 s, the last 60 to 95°C to do the melting curve.
Results
Phenotype Investigation of KN DH Population and Performance of Multiple Main Stem
We investigated the plant with MMS for each line in the KN DH population for two consecutive years from the years 2014 to 2015 in Wuhan and Yangling. According to investigations in the years 2014 and 2015, 45 and 76 DH lines showed MMS phenotype in Yangling; however, only 25 and 29 DH lines showed MMS phenotype in Wuhan (Figure 2, Table 1). Only three lines (QT22, QT229, and QT236) of the 348 lines were found with MMS in all four environments. According to the number of plant with MMS and the total number of each line, the ratio of the MMS occurrence was calculated, the highest ratio of MMS was 100% in Yangling in the years 2014 and 2015, and in Wuhan in the year 2015, the lowest ratio was 20.1% in the year 2015 in Yangling (Table 1). In addition, 7.18–21.84% of plants in KN population had the phenotypes of MMS (from two to three main stems), with more leaves in comparison to their parents (Figure 1, Table 1). Furthermore, broad-sense heritability (h2) was also calculated for MMS (Table S2). MMS had an h2 value 62.26% in Yangling, and an h2 value of 70.23% in Wuhan; these results indicated that the MMS had greater sensitivity and plasticity and was easily influenced by environmental condition.
Figure 2
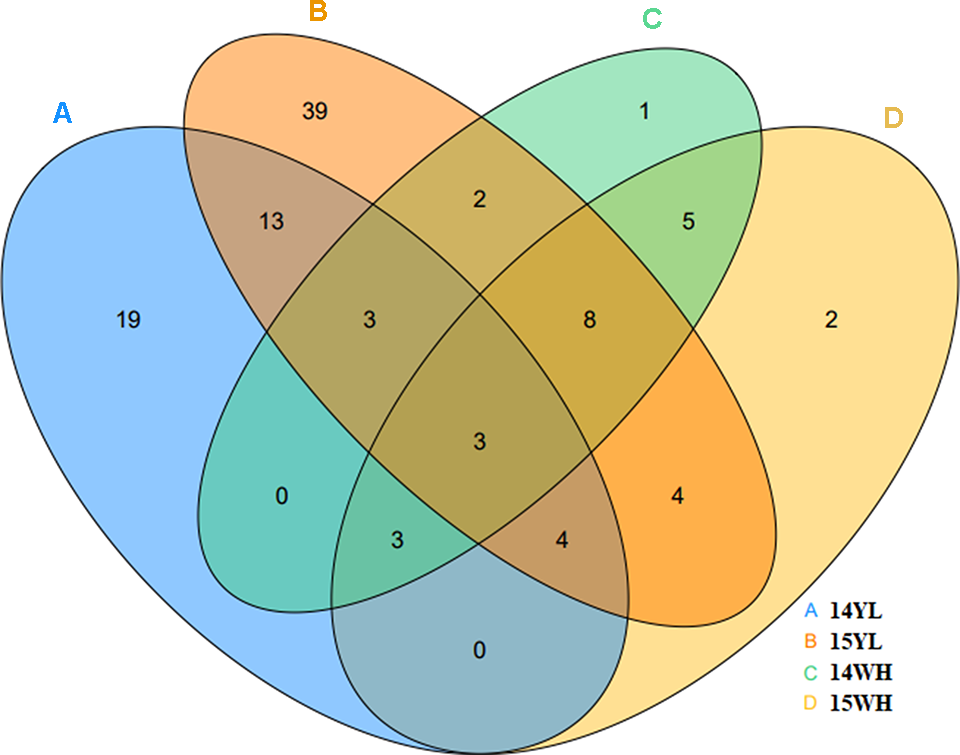
The number of multi-main stem in in different plant environments. (A) represents the number of multi-main stem in 2014 in Yangling of Shaanxi province. (B) represents the number of multi-main stem in 2015 in Yangling of Shaanxi province. (C) represents the number of multi-main stem in 2014 in Wuhan of Hubei province. (D) represented the number of multi-main stem in 2015 in Wuhan of Hubei province.
Table 1
| Year/location | 14 Yangling | 15 Yangling | 14 Wuhan | 15 Wuhan |
|---|---|---|---|---|
| Mean | 47.87% | 41.88% | 42.41% | 47.26% |
| Min | 21.70% | 20.10% | 20.50% | 22.20% |
| Max | 100.00% | 100.00% | 94.30% | 100.00% |
| MMS lines | 45 | 76 | 25 | 29 |
| Total lines | 348 | 348 | 348 | 348 |
| MMS ratio | 12.93% | 21.84% | 7.18% | 8.33% |
| h2 | 62.26% | 70.23% | ||
The multi-main stem phenotypic ratio of KN DH population in Yangling and Wuhan.
MMS, multiple main stem; 14 Yangling and 15 Yangling represent the years 2014 and 2015 in Yangling of Shaanxi province, respectively; 14 Wuhan and 15 Wuhan represent the years 2014 and 2015 in Wuhan of Hubei province; h2, broad-sense heritability.
Moreover, the plant with DMS and the natural normal plants from QT22 were used for germination and morphological development observation in 15, 25, 30, 35, and 50 days of different seedling stages (Figure 3). It was revealed that from germination to 25 days, no obvious differences were observed between the DMS plant and the normal plant, and DMS was first observed in 30 days. The stem base of the DMS plant hypertrophied gradually and had formed two main terminal buds. In 35 days, the DMS plant had formed two stems and could be obviously observed. In 50 days, the two main stems of DMS plants developed simultaneously and formed a plant type of DMS. These results indicated that the genes regulating the formation of the DMS function was in early developmental stages. The DMS plants had also grown more leaves and fewer branches compared to the natural plant (Figure 1, Figure 3E2).
Figure 3

Formation and development of the double-main stem in four seedling stages. (A1, B1, C1, D1, and E1) on the left represent the natural plant, and (A1, B1, C1, D1, and E1) on the right represent the double-main stem plant in 15, 25, 30, 35, and 50 days of seedling stages. (A2, B2, C2, D2, and E2) on the left show enlarge image of the natural plant, and (A2, B2, C2, D2, and E2) on the right show magnification of (A1, B1, C1, D1, and E1).
QTL Mapping for MMS in the KN Population and Candidate Genes Identification
The MMS ratio in Yangling and Wuhan of the years 2014 and 2015 was used for QTL identification. Totally, 43 identified QTLs for MMS were detected in the KN DH population, which were located on 14 chromosomes, except for A02, A09, C02, C04, and C08 (Figure 4, Table 2, Figure S2-1, Figure S2-2). It was revealed that q14WH7-2, q14WH15-1, and q15WH13-2 could explain the phenotypic variation more than 10%, which reached 11.38, 11.42, and 14.9%, respectively. Otherwise, QTL q14WH10-1 and q14WH10-2 were with the smallest phenotypic variation of 2.95%. In addition, the additive effects of QTLs were from −0.14 to 0.09, and the average CI was 4.77 cM (Table 2). QTLs for MMS with overlapping CIs were integrated into consensus QTLs by using BioMercator 4.2 software. Forty-three identified QTLs were integrated into 41 consensus QTLs, of which 39 consensus QTLs that were only expressed in Yangling or Wuhan, were considered as environmental specific QTLs (Table 2, Figure 4, Figure 5). The remaining two consensus QTLs (cqMMS.A3-2 and cqMMS.C3-5) were repeatedly detected in two successive years in Yangling and Wuhan (Table 2, Figure 5), respectively, which were considered to be environmental stable QTLs. These results indicated that the majority of consensus QTLs for MMS were expressed in a specific environment, and MMS was greatly affected by environmental conditions. Regretfully, no major QTL for MMS was identified in all environments.
Figure 4
Distribution of QTLs for multi-main stem on the KN high-density map. The 41 consensus QTLs for MMS are distributed on 14 linkage groups with the exception of A02, A09, C02, C04, and C08. : Red solid dot, QTL peak position. Blue bar length, QTL confidence interval.
Table 2
| Consensus QTL | Chr | Peak (cM) | CIs (cM) | Range (cM) | Identified QTL | LOD | PV (%) | A | E |
|---|---|---|---|---|---|---|---|---|---|
| cqMMS.A1-1 | A01 | 36.21 | 33.20–38.00 | 4.80 | q15YL1-1 | 2.88 | 4.99 | 0.06 | YL |
| cqMMS.A1-2 | A01 | 62.91 | 62.20–63.50 | 1.30 | q15YL1-2 | 2.38 | 9.18 | 0.08 | YL |
| cqMMS.A1-3 | A01 | 74.01 | 69.50–78.40 | 8.90 | q14YL1 | 2.12 | 6.31 | 0.07 | YL |
| cqMMS.A3-1 | A03 | 128.61 | 128.00–132.30 | 2.20 | q15YL3-1 | 3.66 | 6.05 | −0.04 | YL |
| cqMMS.A3-2 | A03 | 141.91 | 137.70–143.10 | 5.40 | q15YL3-2 | 2.93 | 4.88 | −0.04 | YL |
| q15WH3 | 2.82 | 5.02 | −0.04 | WH | |||||
| cqMMS.A4-1 | A04 | 34.61 | 33.10–35.50 | 2.40 | q14YL4-1 | 3.72 | 5.97 | 0.06 | YL |
| cqMMS.A4-2 | A04 | 42.41 | 39.20–46.40 | 7.20 | q14YL4-2 | 3.34 | 5.48 | 0.05 | YL |
| cqMMS.A5-1 | A05 | 44.71 | 41.00–45.20 | 4.20 | q14YL5-1 | 3.02 | 8.89 | 0.09 | YL |
| cqMMS.A5-2 | A05 | 53.31 | 51.50–54.60 | 3.10 | q14YL5-2 | 2.57 | 7.82 | 0.09 | YL |
| cqMMS.A6-1 | A06 | 46.21 | 45.50–46.70 | 1.20 | q14YL6-1 | 2.92 | 4.75 | 0.04 | YL |
| cqMMS.A6-2 | A06 | 49.81 | 48.90–50.0 | 1.70 | q15YL6 | 2.52 | 4.13 | 0.04 | YL |
| cqMMS.A6-3 | A06 | 52.31 | 51.20–53.00 | 1.80 | q14YL6-2 | 3.24 | 5.20 | 0.05 | YL |
| cqMMS.A7-1 | A07 | 19.61 | 17.60–20.60 | 3.00 | q14WH7-1 | 2.85 | 7.99 | −0.07 | WH |
| cqMMS.A7-2 | A07 | 25.81 | 24.80–26.30 | 1.50 | q14WH7-2 | 4.13 | 11.38 | −0.09 | WH |
| cqMMS.A7-3 | A07 | 106.91 | 104.80–108.30 | 3.50 | q15WH7 | 2.45 | 5.90 | −0.06 | WH |
| cqMMS.A8-1 | A08 | 40.51 | 38.60–43.40 | 4.80 | q14YL8-1 | 2.73 | 4.41 | −0.04 | YL |
| cqMMS.A8-2 | A08 | 49.01 | 43.40–54.00 | 10.60 | q14YL8-2 | 3.42 | 5.49 | −0.05 | YL |
| cqMMS.A8-3 | A08 | 59.01 | 54.00–62.20 | 8.20 | q14YL8-3 | 3.05 | 6.88 | −0.05 | YL |
| cqMMS.A10-1 | A10 | 9.31 | 7.50–9.70 | 2.20 | q14WH10-1 | 2.86 | 2.95 | 0.03 | WH |
| cqMMS.A10-2 | A10 | 12.51 | 12.2–12.90 | 0.70 | q14WH10-2 | 2.88 | 2.95 | 0.03 | WH |
| cqMMS.C1 | C01 | 85.71 | 82.70–90.10 | 7.40 | q14YL11 | 2.53 | 7.39 | −0.08 | YL |
| cqMMS.C3-1 | C03 | 4.21 | 4.00–5.90 | 1.90 | q14YL13-1 | 3.12 | 9.18 | −0.09 | YL |
| cqMMS.C3-2 | C03 | 14.71 | 14.30–16.20 | 1.90 | q14YL13-2 | 3.03 | 8.92 | −0.09 | YL |
| cqMMS.C3-3 | C03 | 47.91 | 44.30–49.70 | 5.40 | q15WH13-1 | 2.38 | 7.32 | 0.09 | WH |
| cqMMS.C3-4 | C03 | 62.51 | 61.00–64.10 | 3.10 | q15YL13 | 2.82 | 4.68 | −0.04 | YL |
| cqMMS.C3-5 | C03 | 65.31 | 64.20–66.30 | 2.10 | q14YL13-3 | 2.20 | 6.35 | −0.07 | YL |
| q15WH13-2 | 5.17 | 14.90 | −0.14 | WH | |||||
| cqMMS.C3-6 | C03 | 69.61 | 69.20–69.80 | 0.60 | q15WH13-3 | 2.53 | 7.66 | −0.10 | WH |
| cqMMS.C5-1 | C05 | 5.71 | 0.00–13.20 | 13.20 | q14WH15-1 | 3.86 | 11.42 | 0.09 | WH |
| cqMMS.C5-2 | C05 | 115.01 | 114.10–115.40 | 1.30 | q15WH15-1 | 3.28 | 8.86 | 0.07 | WH |
| cqMMS.C5-3 | C05 | 122.21 | 119.70–123.80 | 4.10 | q15WH15-2 | 2.22 | 6.11 | 0.06 | WH |
| cqMMS.C5-4 | C05 | 136.01 | 124.00–143.30 | 19.30 | q15WH15-3 | 3.03 | 7.31 | 0.04 | WH |
| cqMMS.C5-5 | C05 | 143.91 | 143.30–154.30 | 11.00 | q15WH15-4 | 2.76 | 4.93 | 0.03 | WH |
| cqMMS.C5-6 | C05 | 164.11 | 162.60–165.60 | 3.00 | q14WH15-2 | 2.52 | 6.85 | 0.07 | WH |
| cqMMS.C6-1 | C06 | 53.11 | 51.60–54.40 | 2.80 | q14WH16-1 | 2.29 | 6.27 | −0.07 | WH |
| cqMMS.C6-2 | C06 | 60.31 | 60.10–60.80 | 0.70 | q14WH16-2 | 2.15 | 5.85 | −0.07 | WH |
| cqMMS.C7-1 | C07 | 34.91 | 34.40–37.60 | 3.20 | q15WH17-1 | 3.08 | 5.56 | −0.03 | WH |
| cqMMS.C7-2 | C07 | 54.41 | 52.40–54.70 | 2.30 | q15WH17-2 | 3.11 | 5.60 | −0.03 | WH |
| cqMMS.C7-3 | C07 | 87.71 | 87.50–94.40 | 6.90 | q14WH17 | 4.07 | 11.17 | −0.09 | WH |
| cqMMS.C9-1 | C09 | 70.81 | 65.80–72.80 | 7.00 | q15WH19-1 | 3.30 | 9.19 | −0.08 | WH |
| cqMMS.C9-2 | C09 | 76.61 | 72.80–84.00 | 11.20 | q15WH19-2 | 3.97 | 10.91 | −0.08 | WH |
| cqMMS.C9-3 | C09 | 90.51 | 84.00–92.50 | 8.50 | q15WH19-3 | 3.42 | 10.29 | −0.08 | WH |
Information of QTLs for multi-main stem in the KN population.
Chr, chromosome; CIs, confidence interval; LOD, log odds score; PV, phenotypic variation; A, additive effect; E, environment; YL, Shaanxi Yali; WH, Hubei Wuhan.
Figure 5
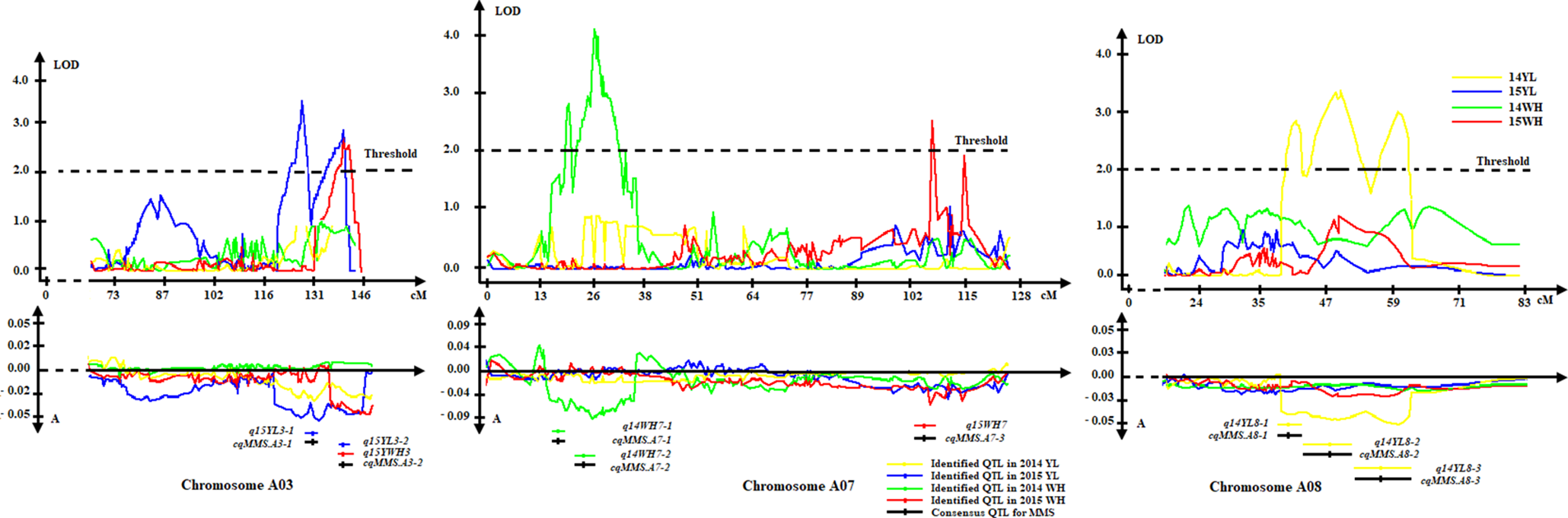
Environmental stable QTL and environmental specific QTL on A03, A07, and A08 chromosomes. In the left, orange QTL cqMMSa3-2 represents environmental stable QTL in Wuhan and Yangling. Middle green and red QTLs represent specific QTL in Wuhan. Right yellow QTLs represent specific QTL in Yangling.
According to the co-linearity relationship between the KN high-density genetic map and the B. napus reference genome (Chalhoub et al., 2014), 159 genes corresponding to 148 A. thaliana homologous genes within CIs of QTLs were identified; these genes were mainly involved in lateral bud development and tiller of stem (Table 3, Table S3). Both BnaC09g13580D (STM) and BnaC09g34650D (WOX2) were located in QTL region of cqMMS.C9-1, in which BnaC09g13580D participated in stem cell population maintenance (Müller et al., 2006) and cytokinin biosynthetic process (Yanai et al., 2005), and BnaC09g34650D belonged to homeobox genes family and mainly regulated the formation and maintenance of SAM (Clark et al., 1996; Laux et al., 1996). BnaA01g28990D (CUC1) was located in QTL region of cqMMS.A1-3, and it maintains boundaries between the SAM and lateral organ primordia (Aida et al., 1999), which was transcriptionally activated by gene STM (Spinelli et al., 2011). BnaA01g28990D and BnaC09g13580D (STM) are critical genes in lateral organ differentiation (Aida et al., 1999), of which STM promotes cell division by suppressing organ differentiation, and downregulation of STM is beneficial to the initiation of lateral organs (Heisler et al., 2005). BnaA03g50360D (BRS1), BnaA03g54090D (LBD39), and BnaA03g54110D (NPY5) were located in stable QTL region of cqMMS.A3-2 (Figure 5). BRS1 (BRI1 suppressor 1) is a serine carboxypeptidase that is involved in brassinosteroid signaling and was recognized to suppress the phenotypes of the brassinosteroid receptor weak mutant bri1-5 (Deng et al., 2017). NPY5 (naked pins in yuc mutants 5) was highly concentrated in cotyledons and was involved in auxin-mediated organogenesis (Cheng et al., 2008). LBD39 (LOB domain-containing protein 39) was a transcription factor in LOB family and could regulate lateral organ boundaries. BnaA07g02390D (WUS) within QTL region of cqMMS.A7-1 encodes a homeodomain protein and is expressed at the center of plant meristem, which induces cell proliferation of meristem and maintain the state of meristem (Kieffer et al., 2006; Sablowski, 2007). BnaC05g48100D (WOX11) was located in QTL region of qMMS.C5-2, and it was a transcription factor, which combines with WOX6 to control the shoot gravitropism and rice tiller angle by regulating asymmetric distribution of auxin (Zhang et al., 2018). BnaA10g02750D (YUC3) was located in QTL region of qMMS.A10-1, which is an important gene involved in auxin biosynthesis (Zhang et al., 2018; Zwack et al., 2016). Moreover, several related genes for cytokinins and auxins were found within QTL regions. BnaC03g29310D (CRF1) and BnaC03g25680D (CRF5) were located in cqMMS-C3-5 and cqMMS-C3-4 regions, respectively, and BnaC09g21990D (CRF6) were located in qMMS.C9-1 region, which were considered to be cytokinin response factors (Zwack et al., 2016; Striberny et al., 2017). BnaC09g33450D (CKX3) and BnaC09g36710D (CKX7) located in qMMS.C9-1 region, and BnaA03g49660D (CKX4) located in qMMS.A3-1 were considered to be involved in cytokinin catabolic process (Ding et al., 2015; Köllmer et al., 2014; Choi et al., 2014). In addition, BnaA06g14040D (MP) and BnaC05g11390D (EPR1) located in QTL region of qMMS.A6-2 and qMMS.C5-1, respectively, which were considered to be transcription factors and could regulate asymmetric distribution of auxin in upstream of auxin synthesis (Zhang et al., 2018; Krogan et al., 2016).
Table 3
| QTL name | Gene in B. napus | Homologous gene in A. thaliana | Gene name |
|---|---|---|---|
| cqMMS.A1-1 | BnaA01g08980D | AT1G20200 | EMB2719 |
| BnaA01g09110D | AT4G18370 | DEG5 | |
| BnaA01g09530D | AT4G18710 | BIN2 | |
| BnaA01g09700D | AT4G18890 | BEH3 | |
| BnaA01g24810D | AT4G23180 | CRK10 | |
| cqMMS.A1-2 | BnaA01g02910D | AT4G34220 | LRR-RLK |
| BnaA01g25310D | AT3G21180 | ACA9 | |
| BnaA01g24690D | AT3G22440 | FLA8 | |
| cqMMS.A1-3 | BnaA01g27090D | AT1G48620 | HON5 |
| BnaA01g28000D | AT1G51800 | ||
| BnaA01g30930D | AT3G55200 | CPSF | |
| BnaA01g28990D | AT3G15170 | CUC1 | |
| BnaA01g28930D | AT3G15500 | NAC3 | |
| BnaA01g28500D | AT3G15790 | MBD11 | |
| BnaA01g28410D | AT3G15880 | WSIP2 | |
| BnaA01g26700D | AT3G18550 | BRC1 | |
| BnaA01g07860D | AT4G29040 | RPT2A | |
| cqMMS.A3-1 | BnaA03g31060D | AT3G08770 | ERD10 |
| BnaA03g51000D | AT3G57230 | AGL16 | |
| BnaA03g58880D | AT4G29040 | RPT2A | |
| BnaA03g38750D | AT2G14900 | RDK1 | |
| BnaA03g51450D | AT4G31400 | CTF7 | |
| BnaA03g51900D | AT4G32040 | KNAT5 | |
| BnaA03g52120D | AT4G32551 | LUC | |
| BnaA03g52290D | AT4G32980 | ATH1 | |
| BnaA03g52520D | AT4G33430 | BAK1 | |
| BnaA03g50910D | AT4G34160 | CYCD3 | |
| BnaA03g53400D | AT4G35550 | WOX13 | |
| BnaA03g53680D | AT4G37180 | UIF1 | |
| BnaA03g49660D | AT4G29740 | CKX4 | |
| cqMMS.A3-2 | BnaA03g50360D | AT4G30610 | BRS1 |
| BnaA03g54090D | AT4G37540 | LBD39 | |
| BnaA03g54110D | AT4G37590 | NPY5 | |
| cqMMS.A4-2 | BnaA04g18690D | AT1G05230 | HDG2 |
| BnaA04g12130D | AT2G21330 | FBA1 | |
| BnaA04g18030D | AT2G31085 | CLE6 | |
| BnaA04g18380D | AT2G31310 | LBD14 | |
| BnaA04g27450D | AT3G62040 | HAD | |
| cqMMS.A5-1 | BnaA05g23180D | AT3G16640 | MYB73 |
| BnaA05g07510D | AT2G36890 | RAX2 | |
| BnaA05g07510D | AT2G36890 | RAX2 | |
| BnaA05g06540D | AT2G38120 | AUX1 | |
| BnaA05g06890D | AT2G37630 | AS1 | |
| BnaA05g07740D | AT3G53020 | STV1 | |
| cqMMS.A5-2 | BnaA05g04890D | AT2G45470 | FLA8 |
| BnaA05g09820D | AT2G33835 | FES1 | |
| BnaA05g09770D | AT2G33880 | HB-3 | |
| BnaA05g09360D | AT2G34440 | AGL29 | |
| BnaA05g09150D | AT2G34650 | PID | |
| BnaA05g09120D | AT2G34710 | PHB | |
| BnaA05g09060D | AT2G34780 | MEE22 | |
| BnaA05g09010D | AT2G34870 | MEE26 | |
| BnaA05g08960D | AT2G34925 | CLE42 | |
| cqMMS.A6-1 | BnaA06g24200D | AT4G01710 | CRK |
| BnaA06g22000D | AT1G78700 | BEH4 | |
| BnaA06g22420D | AT5G62940 | HCA2 | |
| BnaA06g06160D | AT1G10370 | ERD9 | |
| cqMMS.A6-2 | BnaA06g30630D | AT3G60900 | FLA10 |
| BnaA06g11350D | AT1G16880 | ||
| BnaA06g14040D | AT1G19850 | MP | |
| cqMMS.A6-3 | BnaA06g13550D | AT1G67720 | |
| cqMMS.A7-1 | BnaA07g02390D | AT2G17950 | WUS |
| cqMMS.A7-2 | BnaA07g11450D | AT1G20450 | ERD10 |
| BnaA07g11930D | AT5G67300 | MYBR1 | |
| BnaA07g27770D | AT1G69220 | SIK1 | |
| BnaA07g28930D | AT1G70830 | MLP28 | |
| BnaA07g26650D | AT1G67720 | ||
| cqMMS.A8-1 | BnaA08g14270D | AT4G26850 | VTC2 |
| BnaA08g15890D | AT4G37930 | SHMI | |
| BnaA08g15900D | AT4G37940 | AGL21 | |
| BnaA08g16780D | AT4G39070 | BZS1 | |
| BnaA08g16820D | AT4G38970 | FBA2 | |
| BnaA08g16720D | AT4G39110 | ||
| BnaA08g16520D | AT4G39400 | BRI1 | |
| BnaA08g16380D | AT4G39650 | GGT2 | |
| BnaA08g16660D | AT5G10720 | HK5 | |
| BnaA08g14480D | AT5G48820 | ICK6 | |
| cqMMS.A10-1 | BnaA10g02750D | AT1G04610 | YUC3 |
| BnaA10g12480D | AT5G59480 | HAD | |
| BnaA10g23990D | AT5G61410 | RPE | |
| BnaA10g29350D | AT5G57950 | HLR | |
| cqMMS.C1 | BnaC01g21630D | AT3G51030 | TRX1 |
| BnaC01g21860D | AT4G16280 | FCA | |
| BnaC01g22100D | AT4G16110 | RR2 | |
| BnaC01g22110D | AT1G27320 | HK3 | |
| BnaC01g24570D | AT3G46510 | PUB13 | |
| BnaC01g26510D | AT3G50070 | CYCD3.3 | |
| BnaC01g26840D | AT1G54990 | AXR4 | |
| BnaC01g27460D | AT5G13010 | EMB3011 | |
| BnaC01g32740D | AT3G20190 | PRK4 | |
| BnaC01g33180D | AT5G60440 | AGL62 | |
| BnaC01g35230D | AT3G16640 | TCTP | |
| cqMMS.C3-3 | BnaC03g13380D | AT5G56600 | PRF3 |
| BnaC03g13120D | AT5G57090 | EIR1 | |
| cqMMS.C3-4 | BnaC03g18890D | AT1G78380 | GSTU19 |
| cqMMS.C3-5 | BnaC03g25680D | AT2G46310 | CRF5 |
| BnaC03g29310D | AT4G11140 | CRF1 | |
| BnaC03g26730D | AT2G44080 | ARL | |
| cqMMS.C3-6 | BnaC03g20740D | AT2G38120 | AUX1 |
| BnaC03g45610D | AT2G14900 | ||
| cqMMS.C5-1 | BnaC05g07890D | AT1G10370 | ERD9 |
| BnaC05g11390D | AT1G18330 | EPR1 | |
| BnaC05g01780D | AT1G03170 | DUF3049 | |
| cqMMS.C5-2 | BnaC05g46270D | AT3G05530 | RPT2A |
| BnaC05g02370D | AT1G04250 | AXR3 | |
| cqMMS.C5-3 | BnaC05g46030D | AT3G05800 | AtBS1 |
| cqMMS.C5-4 | BnaC05g45960D | AT5G48670 | AGL80 |
| BnaC05g45360D | AT2G31530 | EMB2289 | |
| BnaC05g23630D | AT1G30610 | EMB2279 | |
| BnaC05g48100D | AT3G03660 | WOX11 | |
| cqMMS.C5-5 | BnaC05g47460D | AT3G04100 | AGL57 |
| cqMMS.C5-6 | BnaC05g46680D | AT3G05120 | GID1A |
| BnaC05g48320D | AT3G02310 | SEP2 | |
| cqMMS.C6-1 | BnaC06g23160D | AT1G74260 | PUR4 |
| BnaC06g35530D | AT1G74660 | MIF1 | |
| BnaC06g21810D | AT1G76520 | PILS3 | |
| BnaC06g20740D | AT1G77690 | LAX3 | |
| BnaC06g19100D | AT1G80350 | ERH3 | |
| BnaC06g19420D | AT1G80680 | SAR3 | |
| BnaC06g19460D | AT1G80730 | ZFP1 | |
| BnaC06g27630D | AT2G26670 | TED4 | |
| BnaC06g20020D | AT3G16830 | TPR2 | |
| BnaC06g30400D | AT1G69220 | SIK1 | |
| BnaC06g34100D | AT1G73190 | TIP3 | |
| BnaC06g28810D | AT1G67720 | ||
| BnaC06g16910D | AT3G58780 | SHP1 | |
| BnaC06g24760D | AT4G26000 | PEP | |
| cqMMS.C6-2 | BnaC06g31170D | AT1G24180 | IAR4 |
| BnaC06g29550D | AT1G68800 | TCP12 | |
| BnaC06g30690D | AT1G69500 | CYP704B1 | |
| BnaC06g31090D | AT1G69970 | CLE26 | |
| BnaC06g31830D | AT1G70700 | TIFY7 | |
| cqMMS.C7-1 | BnaC07g15380D | AT1G20450 | ERD10 |
| BnaC07g16030D | AT5G67300 | MYBR1 | |
| BnaC07g23630D | AT3G26520 | TIP2 | |
| BnaC07g25970D | AT3G30260 | AGL79 | |
| BnaC07g30600D | AT5G23720 | PHS1 | |
| BnaC07g26970D | AT5G24860 | FPF1 | |
| BnaC07g26450D | AT5G48380 | BIR1 | |
| cqMMS.C7-3 | BnaC03g77490D | AT4G30620 | RPT2A |
| cqMMS.C9-1 | BnaC09g32510D | AT1G33390 | FAS4 |
| BnaC09g31940D | AT1G34355 | PS1 | |
| BnaC09g13580D | AT1G62360 | STM | |
| BnaC09g21990D | AT3G61630 | CRF6 | |
| BnaC09g33500D | AT5G57950 | HLR | |
| BnaC09g34990D | AT3G51550 | FER | |
| BnaC09g32160D | AT5G16560 | KAN | |
| BnaC09g35030D | AT5G19140 | AILP1 | |
| BnaC09g36930D | AT5G20930 | TSL | |
| BnaC09g36710D | AT5G21482 | CKX7 | |
| BnaC09g36350D | AT5G22650 | HD2B | |
| BnaC09g36060D | AT5G23000 | MYB37 | |
| BnaC09g30190D | AT5G53760 | MLO11 | |
| BnaC09g31160D | AT5G54770 | THI1 | |
| BnaC09g32460D | AT5G55910 | D6PK | |
| BnaC09g32470D | AT5G55920 | OLI12 | |
| BnaC09g33450D | AT5G56970 | CKX3 | |
| BnaC09g32610D | AT5G57710 | SMAX1 | |
| BnaC09g34650D | AT5G59340 | WOX2 | |
| BnaC09g34750D | AT5G59370 | ACT4 |
Candidate genes in Brassica napus and homologous genes in Arabidopsis thaliana within QTL regions for MMS.
Additive × Additive Epistatic Interactions for MMS in Different Chromosomes
To estimate the epistasis interaction for MMS, epistatic loci pairs were detected by using IciMapping software. Totally, 26 statistically significant epistatic loci pairs were detected in three plant regions (Figure 6, Table S4), which were located on chromosome A01/A08, A01/C03, A02/C03, A02/A09, A03/A04, A04/A08, etc. which accounted for 1.76–17.70% of the phenotypic variance. Five epistatic interaction pairs, including EpMMS.A3-75/EpMMS.A3-25, EpMMS.A3-120/EpMMS.C2-105, EpMMS.A4-5/EpMMS.A8-115, EpMMS.A9-15/EpMMS.A9-160, and EpMMS.A9-160/EpMMS.C3-65, had positive epistatic effect, indicating that the effects of parents for MMS were larger than their the recombinant effects of epistatic interaction pairs. The remaining epistatic interaction pairs had negative epistatic effects, indicating that the recombinant effects of these epistatic interaction pairs for MMS were larger than their parental effects. Furthermore, we noted that seven epistatic interaction loci were located in the above-mentioned five QTL intervals. EpMMS.A7-105 was located in QTL region of cqMMS.A7-3, indicating that cqMMS.A7-3 also interacted with EpMMS.A7-125. In addition, three epistatic interaction pairs (EpMMS.A2-60/EpMMS.C3-65, EpMMS.C3-65/EpMMS.C3-75, and EpMMS.A9-160/EpMMS.C3-65) were closely related to cqMMS.C3-5, which indicated cqMMS.C3-5 was an important loci for MMS.
Figure 6
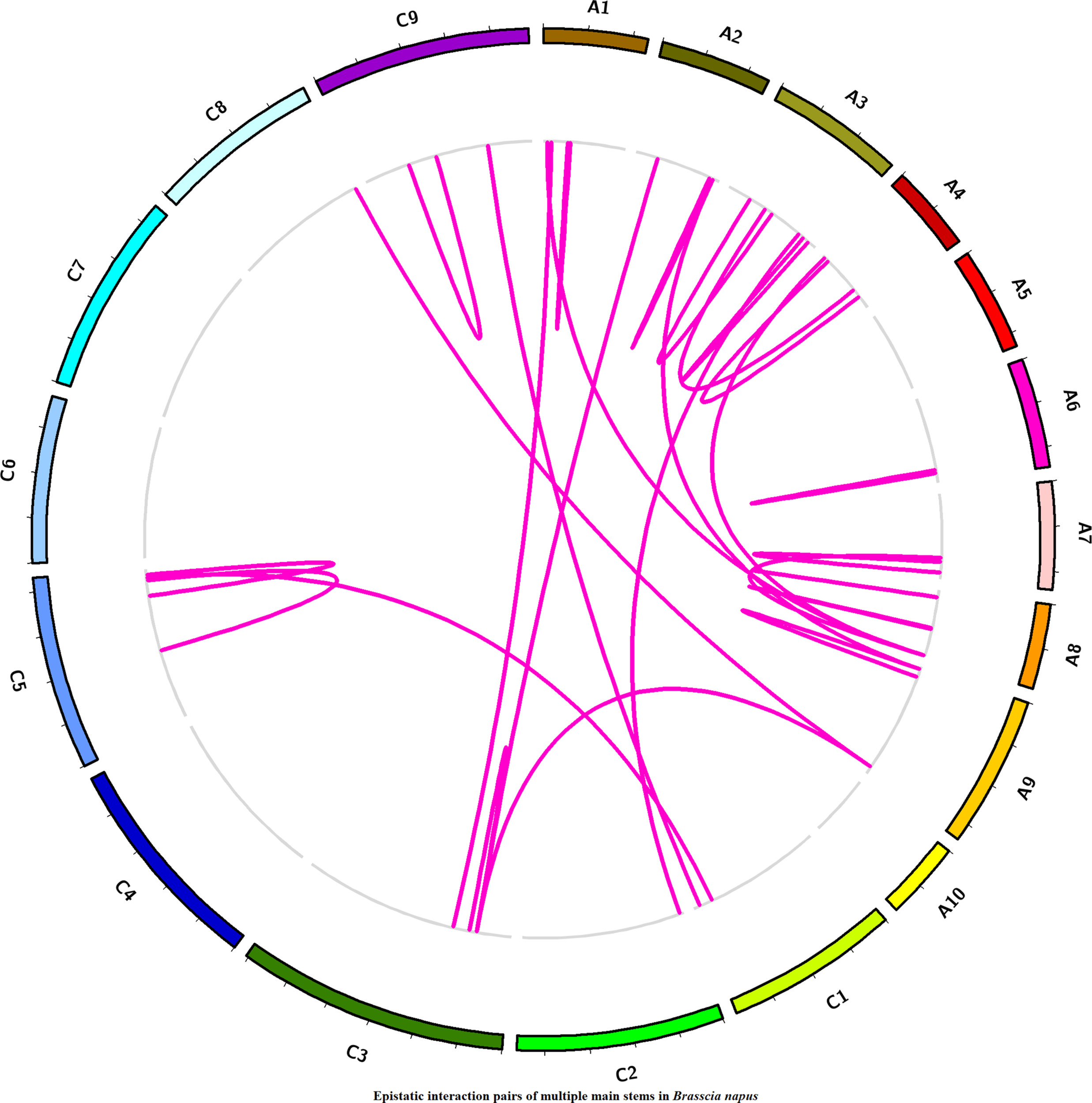
Distribution of epistatic interaction for MMS on different chromosomes. Violet lines represent epistatic interaction pairs of MMS; the epistatic interaction pairs were shown by links in the inside circle. The outside circle represents 19 chromosomes of Brassica napus.
Differentially Expressed Genes Between the Natural Plant and the DMS Plant by Gene-Fishing™ Technique
Total RNA of main stem apex of the natural plant and DMS in seedling of 20 days were extracted by using Invitrogen TRIzol Kit. The dT-ACP1 primers from Gene-Fishing technique were used as primers for reverse transcription. In order to identify differentially expressed genes (DEGs) in DMS and the natural plant of B. napus, the PCR amplification of cDNA using 36 random amplified primers was conducted. Totally, 46 differential expression bands were found, including 28 down-regulated and 18 up-regulated in the DMS compared to the natural plant (Figure 7, and a full scan of the entire original gels attached as Figures S1-1–Figure S1-6; Table S5). According to the sequence alignment results, 31 genes were obtained (Table 4). Based on functional annotation, seven underlying genes (DEG1, DEG2, DEG4, DEG5, DEG34, DEG36, and DEG46) were annotated to regulate the apical meristem, which might be related to the formation of DMS phenotypes (Table 4). In addition, some genes that are related to biotic stimulus, defense response, drought, and salt-stress responses, as well as cold-response functional genes were also identified (Table 4).
Figure 7
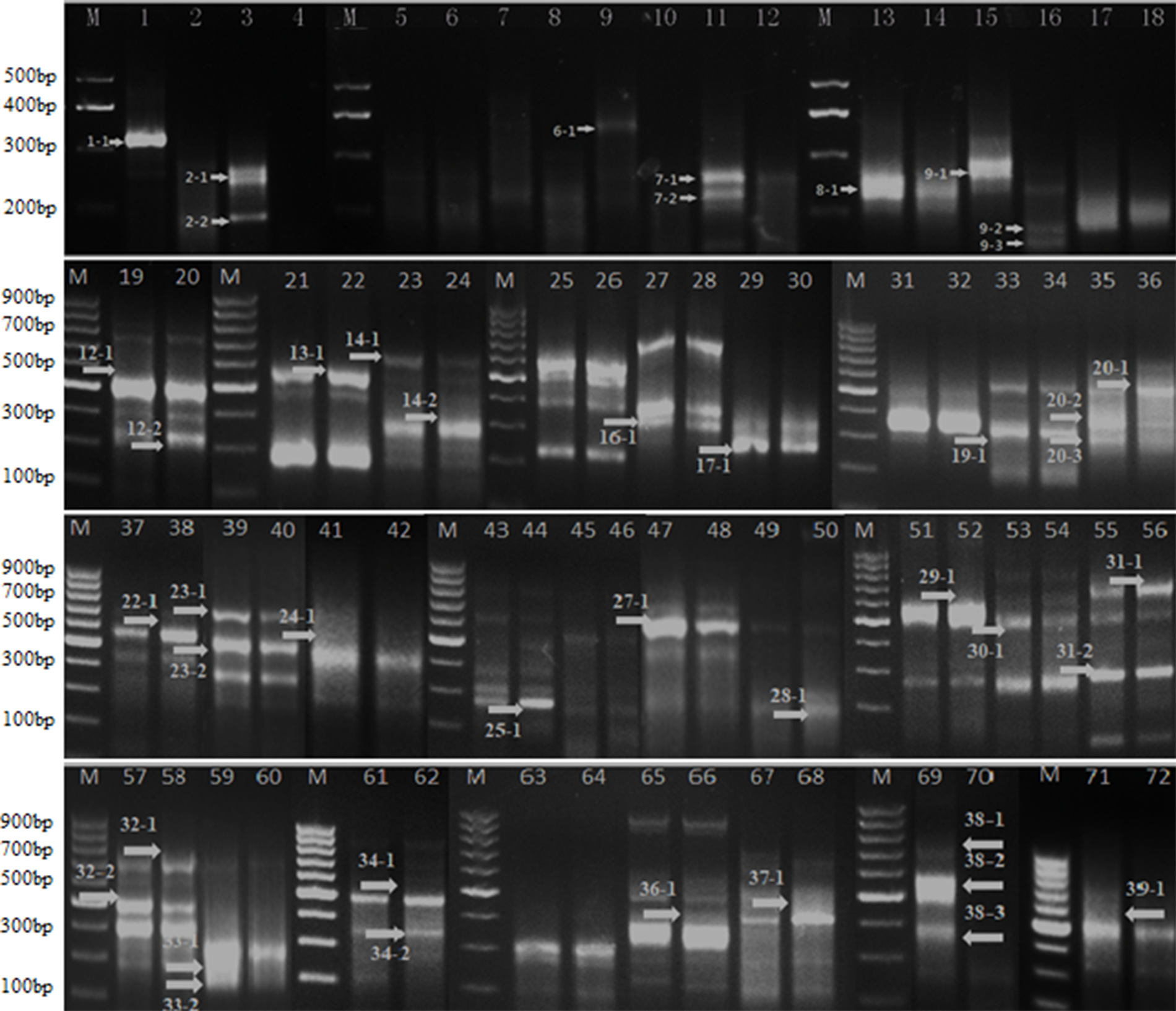
Differential expressed genes screen using ACPs primers based on Gene-Fishing. The numbers and arrows on image represent different differential expression genes. For better presentation, only cropped images of blots are shown here; a full scan of the entire original gels is submitted in the Supplementary Material (Figures S1-1–S1-6).
Table 4
| DEG number | Score | E-value | Max ident | Gene ID in B. napus | Chromosome | A. thaliana |
|---|---|---|---|---|---|---|
| DEG1 | 372 | 3.00E−99 | 99.00% | BnaA01g07860D | A01 | AT4G29040 |
| BnaA08g13550D | A08 | |||||
| BnaC01g09460D | C01 | |||||
| BnaC07g41720D | C07 | |||||
| BnaC08g13300D | C08 | |||||
| DEG2 | 811 | 0.00E+00 | 97.00% | BnaA10g29350D | A10 | AT5G57950 |
| BnaC09g33500D | C09 | |||||
| DEG3 | 457 | 1.00E−124 | 99.00% | BnaA01g02490D | A01 | AT4G34670 |
| BnaA03g28830D | A03 | |||||
| BnaA03g52970D | A03 | |||||
| BnaA08g11040D | A08 | |||||
| BnaC01g03750D | C01 | |||||
| BnaC01g39890D | C01 | |||||
| BnaC03g33960D | C03 | |||||
| BnaC03g65890D | C03 | |||||
| BnaC05g47000D | C05 | |||||
| BnaC07g45170D | C07 | |||||
| DEG4 | 555 | 4.00E−154 | 100.00% | BnaA06g24200D | A06 | AT4G01710 |
| DEG5 | 468 | 5.00E−128 | 100.00% | BnaA01g02910D | A01 | AT4G34220 |
| BnaC01g04180D | C01 | |||||
| DEG6 | 737 | 0.00E+00 | 99.00% | BnaA07g11450D | A07 | AT1G20450 |
| BnaC05g15780D | C05 | |||||
| BnaC07g15380D | C07 | |||||
| DEG8 | 697 | 0.00E+00 | 99.00% | BnaA07g11930D | A07 | AT5G67300 |
| BnaC02g16640D | C02 | |||||
| BnaC07g16030D | C07 | |||||
| DEG9 | 503 | 2.00E−138 | 96.00% | BnaA01g00540D | A01 | AT4G37410 |
| BnaA08g15660D | A08 | |||||
| BnaC01g01530D | C01 | |||||
| BnaC03g61420D | C03 | |||||
| DEG10 | 797 | 0.00E+00 | 99.00% | BnaA03g21640D | A03 | AT2G47130 |
| BnaA09g42190D | A09 | |||||
| BnaC03g72480D | C03 | |||||
| BnaC08g34620D | C08 | |||||
| DEG11 | 1177 | 0.00E+00 | 100.00% | BnaA03g00940D | A08 | AT5G03560 |
| BnaC03g01290D | C03 | |||||
| DEG12 | 736 | 0.00E+00 | 99.00% | BnaA02g15190D | A02 | AT1G70830 |
| BnaA07g28930D | A07 | |||||
| BnaA08g20650D | A08 | |||||
| BnaC02g20340D | C02 | |||||
| BnaC06g31980D | C06 | |||||
| BnaC07g13200D | C07 | |||||
| DEG14 | 503 | 2.00E−138 | 94.00% | BnaC01g07930D | C01 | AT4G30450 |
| DEG15 | 460 | 8.00E−26 | 96.00% | BnaA01g30930D | A01 | AT3G55200 |
| BnaA09g05750D | A09 | |||||
| DEG17 | 1050 | 0.00E+00 | 98.00% | BnaA06g05510D | A06 | AT1G09690 |
| BnaA08g26210D | A08 | |||||
| BnaA09g48320D | A09 | |||||
| BnaC05g07130D | C05 | |||||
| BnaC08g13710D | C08 | |||||
| DEG20 | 778 | 0.00E+00 | 99.00% | BnaA02g24900D | A02 | AT5G47030 |
| BnaA06g35640D | A06 | |||||
| BnaC04g00050D | C04 | |||||
| BnaC02g33050D | C02 | |||||
| BnaC07g19840D | C07 | |||||
| BnaC08g25530D | C08 | |||||
| BnaC09g19920D | C09 | |||||
| DEG21 | 342 | 2.00E−90 | 99.00% | BnaA06g06120D | A06 | AT1G10330 |
| BnaA06g06150D | A06 | |||||
| BnaC05g07870D | C05 | |||||
| DEG22 | 806 | 0.00E+00 | 99.00% | BnaA03g17080D | A03 | AT2G37220 |
| BnaA04g04760D | A04 | |||||
| BnaA05g07240D | A05 | |||||
| BnaC02g22410D | C02 | |||||
| BnaC04g08050D | C04 | |||||
| BnaC06g04460D | C06 | |||||
| BnaC09g41150D | C09 | |||||
| DEG23 | 630 | 8.00E−77 | 99.00% | BnaA01g26080D | A01 | AT2G14900 |
| BnaA03g38750D | A03 | |||||
| BnaC03g40940D | C03 | |||||
| BnaC03g45610D | C03 | |||||
| DEG25 | 497 | 6.00E−37 | 99.00% | BnaA04g17660D | A04 | AT2G30570 |
| BnaA05g11890D | A05 | |||||
| BnaC04g13890D | C04 | |||||
| BnaC04g41210D | C04 | |||||
| BnaC05g04700D | C05 | |||||
| BnaC09g18450D | C09 | |||||
| DEG26 | 942 | 0.00E+00 | 99.00% | BnaA07g29960D | A07 | AT1G72230 |
| BnaC02g21230D | C02 | |||||
| BnaC06g33270D | C06 | |||||
| DEG27 | 782 | 0.00E+00 | 98.00% | BnaA02g28130D | A02 | AT3G26520 |
| BnaA06g32840D | A06 | |||||
| BnaC02g36210D | C02 | |||||
| BnaC07g23630D | C07 | |||||
| DEG29 | 346 | 3.00E−91 | 87.00% | BnaA03g40170D | A03 | |
| BnaA10g23990D | A10 | |||||
| BnaC09g48610D | C09 | |||||
| DEG34 | 922 | 0.00E+00 | 98.00% | BnaA06g40690D | A06 | AT4G39270 |
| BnaC07g47290D | C07 | |||||
| DEG36 | 401 | 4.00E−108 | 100.00% | BnaA06g30700D | A06 | AT3G30260 |
| BnaA09g02740D | A09 | |||||
| BnaC02g38130D | C02 | |||||
| BnaC07g25980D | C07 | |||||
| BnaC09g02190D | C09 | |||||
| DEG37 | 523 | 1.00E−144 | 100.00% | BnaA04g26210D | A04 | AT2G45470 |
| BnaA05g04890D | A05 | |||||
| BnaC04g04230D | C04 | |||||
| BnaC04g50210D | C04 | |||||
| DEG39 | 1230 | 0.00E+00 | 100.00% | BnaA06g37230D | A06 | AT4G38970 |
| BnaA08g16820D | A08 | |||||
| BnaC01g00070D | C01 | |||||
| BnaC03g60280D | C03 | |||||
| BnaC07g47470D | C07 | |||||
| DEG41 | 571 | 4.00E−159 | 100.00% | BnaA03g31050D | A03 | AT3G08770 |
| BnaA05g29430D | A05 | |||||
| BnaC03g36390D | C03 | |||||
| BnaC05g43760D | C05 | |||||
| DEG42 | 446 | 2.00E−121 | 100.00% | BnaA02g17680D | A02 | AT4G30620 |
| BnaA08g12990D | A08 | |||||
| BnaC02g24220D | C02 | |||||
| BnaC03g77490D | C03 | |||||
| DEG43 | 431 | 6.00E−117 | 98.00% | BnaA03g50940D | A03 | AT4G34180 |
| BnaC07g44780D | C07 | |||||
| DEG44 | 562 | 3.00E−156 | 100.00% | BnaA02g14620D | A02 | AT1G69220 |
| BnaA07g27770D | A07 | |||||
| BnaC06g30400D | C06 | |||||
| DEG46 | 200 | 7.00E−48 | 99.00% | BnaA01g27710D | A01 | AT3G16640 |
| BnaA03g34250D | A03 | |||||
| BnaA05g23180D | A05 | |||||
| BnaC01g35230D | C01 | |||||
| BnaC03g39720D | C03 | |||||
| BnaC05g36580D | C05 |
Homology comparison of all differential expression genes and corresponding to Brassica napus genes and Arabidopsis thaliana genes.
DEG, different expression genes.
Subsequently, genes that regulate apical meristem were conducted for further analysis. DEG1 is homologous to the Arabidopsis gene AT4G29040 (RPT2A), and it had five copies distributed on A01 (BnaA01g07860D), A08 (BnaA08g13550D), C01 (BnaC01g09460D), C07 (BnaC07g41720D), and C08 (BnaC08g13300D). DEG2 had two copies found on A10 (BnaA10g29350D) and C09 (BnaC09g33500D) and are homologous to the Arabidopsis gene AT5G57950 (halted root, HLR). DEG4 (BnaA06g24200D) is homologous to the Arabidopsis gene AT4G01710 (crooked, CRK) and encodes ARP2/3 complex 16-kDa subunit. DEG5 had one copy on A01 (BnaA01g02910D), which is homologous to the Arabidopsis gene AT4G34220, and it was considered to be a leucine-rich repeat receptor-like protein kinase gene (LRR-RLK). DEG34 had two copies located on A06 (BnaA06g40690D) and C07 (BnaC07g47290D), and they were homologous to the Arabidopsis gene AT4G39270, which has the same function with DEG5 and was also considered to be leucine-rich repeat receptor-like protein kinase family gene (LRR-RLK). DEG36 is homologous to the Arabidopsis gene AT3G30260 (AGAMOUS-like MADS-box protein 79, AGL79), which has five copies found on A06 (BnaA06g30700D), A09 (BnaA09g02740D), C02 (BnaC02g38130D), C07 (BnaC07g25980D), and C09 (BnaC09g02190D). DEG46 (BnaC03g39720D) is homologous to the Arabidopsis gene AT3G16640 (TCTP), which is mainly expressed in meristematic regions of the shoot and root.
Relative Expression Level of Candidate Genes Based on QTL Mapping and Gene-Fishing
Based on the common candidate genes that were obtained from QTL mapping and Gene-Fishing technique, six commonly identified genes were annotated to be related to the formation of MMS, including RPT2A, HLR, CRK, LRR-RLK, AGL79, and TCTP, and these genes were further used to perform the relative expression analysis by quantitative real-time PCR between the normal plant and the DMS plant in four stages of seedling (5, 10, 15, and 20 days of seedling age) (Figure 8). It was revealed that the expression level of RPT2A was extremely and significantly higher in DMS plant than that of in natural plant in 15 days of seedling age, and in 10 and 20 days of seedling age, the expression level of RPT2A in DMS plant was extremely and significantly lower than that of in natural plant. For HLR, the expression level in DMS plant was much higher than that in natural plant in all four states of seedling age. Especially in 5, 10, and 15 days, the expression levels in DMS plant were significant or extremely and significantly higher than that in natural plant. In 10 to 20 days of seedling age, expression level were higher in natural plant than that in DMS plant, which were also consistent with Gene-Fishing results (Table 5). In addition, whether in natural plant or the DMS plant, expression of HLR also showed downward trend in all four states. For CRK, in 5 days of seedling age, expression level in DMS plant was extremely and significantly higher than that in natural plant; however, in 10 days of seedling age, the expression of CRK was severely inhibited in DMS plant. From 15 to 20 days, expressions of CRK were slowly rising again. The decline in gene expression of CRK might lead to a decrease in the activity of some regulated genes, which disrupted the balance of SAM. For LRR-RLK, the expression level in DMS plant was extremely and significantly lower than in natural plant of 10 and 20 days of seedling age; however, its expression level in DMS plant was extremely and significantly higher than in natural plant in 5 and 15 days of seedling age. In addition, the expression level of LRR-RLK in natural plant was the highest at fourth stage, which was compatible with result of Gene-Fishing (Table 5). For AGL79, the expression level in DMS plant was extremely and significantly lower than in natural plant in 5 days of seedling age. With the development of seedlings, the expression level in DMS plant was slightly higher (not significant) than in natural plant of 10 to 20 days of seedling age. For TCTP, the expression level in DMS was extremely and significantly lower than in natural plant of 5 to 15 days of seedling age. In 20 days of seedling age, the expression level of TCTP showed upward trend in DMS plant. Relative expression level of candidate genes showed that these six genes had different expression profile between the natural plant and the DMS plant in different seedling age stages.
Figure 8
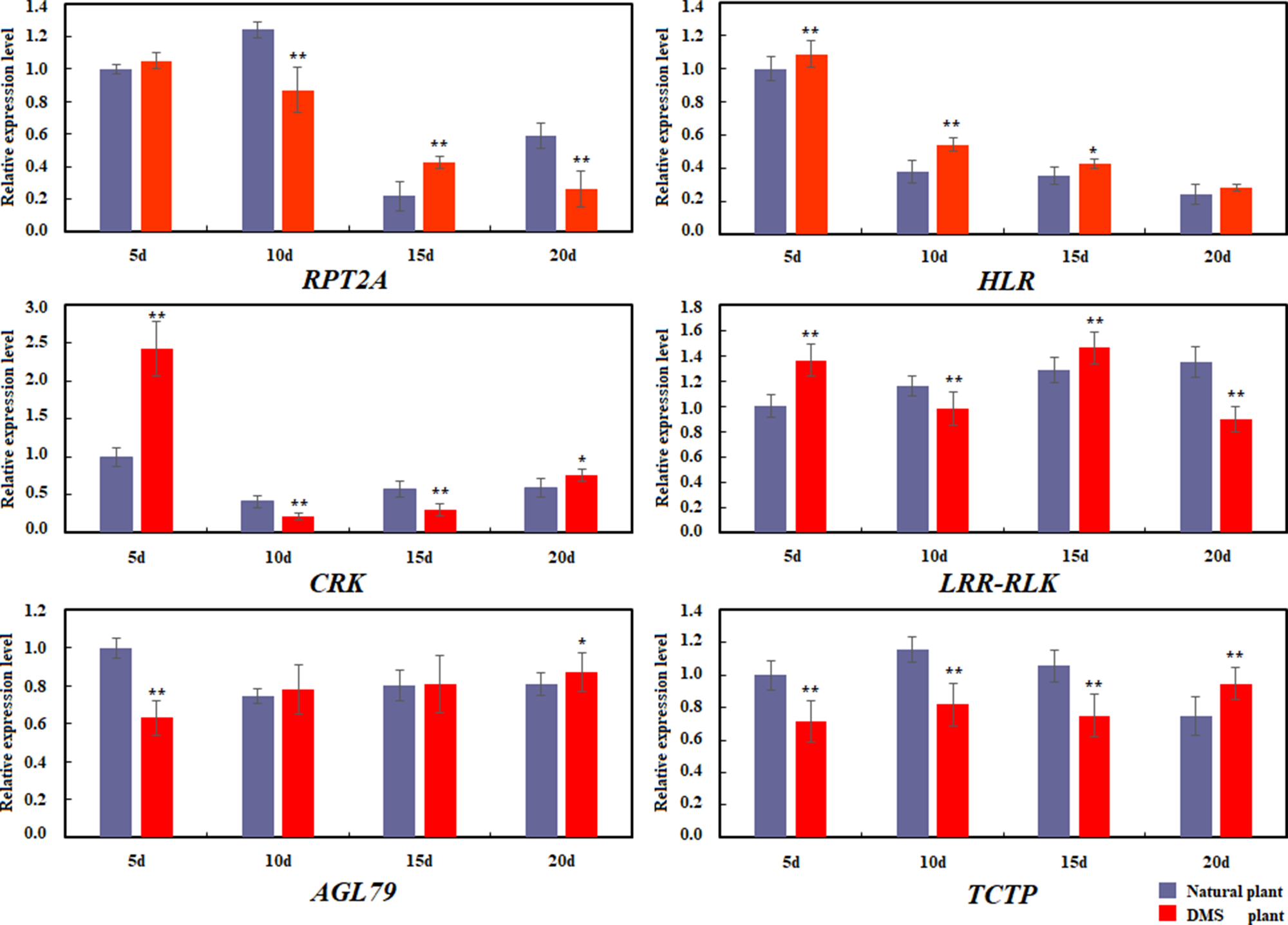
Differential expression of six common genes in natural plant and double-main stem plant. qRT-PCR validation of cloning results of six predictive genes. Left, natural plant (natural). Right, the double-main stem plant (DMS plant). **Significant at p < 0.01. *Significant at p < 0.05. No significance p > 0.05.
Table 5
| DEG number | Description | Up expression |
|---|---|---|
| DEG1 | RPA2A, Brassica napus 19S proteasome regulatory subunit 4 homolog A-like. | Natural |
| DEG2 | HLR, Brassica napus 26S proteasome non-ATPase regulatory subunit 9-like. | DMS |
| DEG4 | CRK, Brassica napus actin-related protein 2/3 complex subunit 5A-like. | DMS |
| DEG5 | LRR-RLK, Brassica napus receptor protein kinase-like protein At4g34220. | Natural |
| DEG36 | AGL79, Brassica napus uncharacterized LOC106436879. | DMS |
| DEG46 | TCTP, Brassica napus translationally controlled tumor protein | DMS |
Six differential expression gene description and differential expressed in natural plant and the double-main stem plant based on Gene-Fishing.
DEG, different expression genes; natural, natural plant; DMS, double-main plant.
Discussion
Rapeseed is an important oilseed crops worldwide (Cai et al., 2014). Although oil content and seed yield of rapeseed varieties have been effectively improved by rapeseed breeders (Zhao et al., 2011), the optimal plant architectural cultivated varieties are urgently needed for mechanized harvesting and high-density planting in the current agricultural management (Fu, 2008). Rapeseed plant architecture is characterized by plant height, branch number and angles, and inflorescence morphology (Li et al., 2018a), which indirectly influence rapeseed yield by affecting the major yield-component trait (Qiu et al., 2006; Zhou et al., 2014). Crop breeding has primarily focused on plant architecture, including plant height, branch or tiller number and angle, leaf shape, and size (Wang et al., 2018). MMS differentiated from shoot meristem, which is also one of the important factors for improving plant architecture and is conducive to increasing plant density and enhancing seed yield in rapeseed. In KN DH population, several MMS plants were found, and their main characteristics were composed of double- or three main stems from the base of a plant and had less lateral branches (Figure 1). Because of nutrient diversion, the MMS plant had slight lower plant height and stronger stem; thus, the MMS materials had stronger lodging resistance. In addition, under a small sowing amount of seeds, the multiple main stem materials not only can effectively increase plant density and the total number of siliques per unit, which finally might increase seed yield, but also, it is suitable for mechanized harvesting and to improve production efficiency. In total, the acquisition of the MMS material provided completely new ideas to improve the plant architecture and breed new varieties in crops.
According to the statistics of the number for MMS materials in different plant environments in several years, MMS was a complex quantitative trait and was significantly influenced by the environment. QTL mapping is considered to be an effective approach for dissecting quantitative traits (Paran and Zamir, 2003). In the present study, 43 QTLs for MMS that were located on 14 chromosomes were identified, explaining 2.95–14.9% of the phenotypic variation (Table 2). Two environment-stable QTLs, including qMMS.A3-2 and qMMS.C3-5, were expressed in both winter and semi-winter environments. More importantly, according to epistatic interaction analysis, QTL cqMMS.C3-5 had three epistatic interactions with other loci, which indicated that this stable QTL was an important loci for MMS. Several environment-stable QTLs for other agricultural traits in rapeseed have been reported, such as stable QTLs for flowering time (Li et al., 2018b; Shi et al., 2009) and thousand seed weight (Zhao et al., 2016). These environment-stable QTLs are suitable for developing new varieties of rapeseed with broad adaptability (Li et al., 2018b). Some genes within QTLs related to cell division and differentiation of shoot meristem were identified mainly included STM, WUS-related homeobox genes, cytokinin response factors, and auxin-related genes. Previous studies showed that cytokinin (CK) produced by root can promote cell division as well as maintenance of meristem, and the upward transport of CK in the root promotes axillary bud germination and the formation of lateral branches (Chatfield et al., 2000; Leyser, 2003; Ongaro and Leyser, 2008).
Gene-fishing technology has been widely used to identify DEGs in crops (Sanghoon et al., 2009; Liao et al., 2015). Some important differently expressed genes have been identified, such as salt-stress-induced up-regulated genes in barley leaves (Sanghoon et al., 2009) and pistillody genes in wheat (Liao et al., 2015). In the present study, 31 differentially expressed genes between DMS plants and the natural plants of B. napus were identified, in which six DEGs involved in the formation and regulation of DMS phenotype (Table 4), including RPT2A, HLR, CRK, LRR-RLK, AGL79 (AGAMOUS-LIKE 79), and TCTP (translationally controlled tumor protein). Among them, RPT2A is a 26S proteasome subunit and was located in QTL regions of cqMMS.A1-3, cqMMS.A3-1, cqMMS.C5-2, and cqMMS.C7-3 and was reported to be involved in maintenance of the post-embryonic meristems (Ueda et al., 2004). 26S proteasome is an important factor in signal transduction–mediated pathway; missing or mutation of 26S proteasome would lead to a decrease in strigolactones (SL) content, thereby promoting the excessive growth of axillary buds and other organs, such as branches and tillers (Wang and Bouwmeester, 2018). Therefore, changes of 26S proteasome may affect the growth of SAM and result in MMS phenotype. Abnormal expression of RPT2A in DMS plant might change the state of meristem and SL content in 5 to 20 days of seedling age, thereby forming MMS phenotype. HLR is located in QTL regions of cqMMS.A10-1 and cqMMS.C9-1, which encodes 26S proteasome regulatory subunit RPT2A, which is essential to maintain the size of SAM and might affect the growth of SAM by affecting the expression of WUS (Ueda et al., 2004). However, the expression level of HLR in DMS plant was much higher than that in natural plant in four states of seedling age, and overexpression of HLR might disturb the normal state of meristem and initiate bud differentiation. Meanwhile, HLR is also required to regulate the cell division pattern in SAM. In plants, the actin cytoskeleton is essential for various processes such as stomatal closure, cell proliferation, and cell morphogenesis (Akin and Zipursky, 2014). CRK encodes ARP2/3 complex 16-kDa subunit. In moss Physcomitrella, when the constituent part arpc4 of the CRK is lost, shoot growth of the apical cell slows down obviously. In the current study, CRK was located in QTL region of cqMMS.A6-1, and the decline in gene expression of CRK might lead to a decrease in the activity of some regulate genes in seedling. Studies in A. thaliana, when the CRK is absent, the actin of trichomes, hypocotyls cells, and epidermal pavement cells gather into bundles, downstream cell activity was interfered (Deeks and Hussey, 2003; Quatrano et al., 2006), which disrupted the balance of SAM. LRR-RLK represented a large family of protein kinases; it was located in QTL region of cqMMS.A1-2 (Gou et al., 2010). Expression of LRR-RLK had upward trend in DMS plant, which indicated that LRR-RLK might participate in the occurrence of axillary buds (Yaginuma et al., 2011). In addition, LRR-RLK contains ERECTA (ER) and CLV1. ER-family receptor kinases can regulate stem cell homeostasis to regulate organ shape and inflorescence architecture via cushioning its CK response in the SAM (Shchennikova et al., 2017). CLV1 interacts with the WUS gene to maintain the balance of differentiation of shoot and floral meristem cells in SAM (Gou et al., 2010). When CK stimulates the expression of WUS, the expression of CLV3 should have been inhibited to result in a SAM expansion; however, the morphology of SAM showed no difference with the normal plant. These phenomena could owe to the cushioning mechanism of ER family, which prevents the expression of CLV3 from increasing remarkably, thereby maintaining the normal development of stem meristem (Torii et al., 1996; Uchida et al., 2013). AGL79 is a transcription factor; relatively lower AGL79 overexpression would promote more lateral branches and rosette leaves (Gao et al., 2018). In addition, miR156/SPL10 modulated lateral root development and branching and leaf morphology in A. thaliana by silencing AGL79 (Gao et al., 2018). TCTP is a central regulator of cell proliferation and differentiation in animals and in plants, which is an important mitotic regulator (Amson et al., 2013; Roberto et al., 2015).
Furthermore, some genes also have similar functions in other crops. HLR gene is a homolog of the 26S proteasome subunit, which was strongly expressed both in the SAM and RAM of rice seedlings (Yanagawa et al., 2002). LRR-RLKs play an important role in regulating the SAM and microspores (Becraft, 2002), brassinosteroid perception, and floral abscission (Carles and Fletcher, 2003; Li, 2003). Rameneni et al. performed the whole-genome sequencing (WGS) of B. rapa and identified the LRR-RLK of B. rapa, which is involved in the plant morphological characters and plant stress resistance (Rameneni et al., 2015). ERECTA is one of LRR-RLK family, which coordinates Arabidopsis organ growth and flower development by promoting cell proliferation (Torii et al., 1996; Shpak et al., 2004). VEGETATIVE1 (VEG1) is an AGL79-like MADs-box gene; the secondary inflorescences in nutation were replaced by vegetative branches in pea (Berbel et al., 2012). In addition, AGL79 is involved in regulating leaf shape, shoot branching, and flowering time in A. thaliana (Gao et al., 2018). In addition, when maize is under stress response, TCTP expression showed a significant upward trend (Chen et al., 2014). Taken together, a series of balance will be broken when these genes mutated and may lead to disturbance in the growth of SAM and form abnormal organ morphology, such as clumping leaves and multiple axillary buds (Gou et al., 2010; Uchida et al., 2013).
Conclusion
The increase of yield and oil content is the ultimate goal in rapeseed breeding. Due to the lack of mechanized cultivated variety and high productive cost as well as low income, farmers prefer to idle the farmland rather than plant rapeseed, which seriously hampers the development of rapeseed in China. Importantly, the acquisition of MMS materials provides the possibility for breeding mechanized varieties, high-density planting varieties, and the yield improvement in rapeseed. In the present study, six common candidate genes within QTL regions and differentially expressed genes, including RPT2A, HLR, CRK, LRR-RLK, AGL79, and TCTP were identified by using QTL mapping and Gene-Fishing technique. Meanwhile, according to their functional annotation, these candidate genes might be related to the formation of MMS phenotype, in which genes might disrupt cell division and the differentiation of SAM and induce the formation of multiple buds, thereby promoting the formation and development of the MMS plant. MMS is an important agriculture trait, which is differentiated from shoot apical meristem. Relative expression level of candidate genes showed that these genes had different expression level between the natural plants and the DMS plants in different seedling age stages. Overall, the study would not only give a new insight into the genetic basis underlying the control of the MMS in rapeseed but also provide clues for plant architecture breeding in rapeseed.
Funding
This work was supported financially by the national key research project of China (2016YFD0101300, 2016YFD0101200), the Science and Technology Plan Project of Yangling Demonstration Zone of China (2017NY-20), Innovative Talents Promotion Plan of Shaanxi province- Key Technological Innovation Team Plan (2017KCT-23).
Statements
Data availability statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Ethics statement
The authors declare that the experiments comply with the current laws of the country in which they were performed.
Author contributions
WZ wrote the first draft of the manuscript. WZ and HC conducted all the field experiments, performed most of the experiments and data analysis for the overall study; HC, LnZ, NT, YZ, BL, KZ, ZG and HW helped and participated in the field trials and data collection for multiple-main stem. KC, LbZ and DH performed the phenotypic data processing and QTL detection. ML designed and conceived the overall study and revised the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01152/full#supplementary-material
Figure S1Differential expressed genes screen using ACPs amplified primer 1-9. Odd number lane represented natural plant and even number lane represented double-main stem plant.
Figure S2Differential expressed genes screen using ACPs amplified primer 10-15. Odd number lane represented natural plant and even number lane represented double-main stem plant.
Figure S3Differential expressed genes screen using ACPs amplified primer 16-20. Odd number lane represented natural plant and even number lane represented double-main stem plant.
Figure S4Differential expressed genes screen using ACPs amplified primer 21-25. Odd number lane represented natural plant and even number lane represented double-main stem plant.
Figure S5Differential expressed genes screen using ACPs amplified primer 26-31. Odd number lane represented natural plant and even number lane represented double-main stem plant.
Figure S6Differential expressed genes screen using ACPs amplified primer 32-36. Odd number lane represented natural plant and even number lane represented double-main stem plant.
Figure S7Distribution of QTLs for multi-main stem on A genome in B. napus
Figure S8Distribution of QTLs for multi-main stem on C genome in B. napus
Abbreviations
B. napus, Brassica napus; A. thaliana, Arabidopsis thaliana; QTL, quantitative trait locus; SNPs, single nucleotide polymorphisms; DL, Dali in Shannxi Province, China; WH, Wuhan in Hubei Province, China; MMS, multiple main stem; DMS, double-main stem; DEGs, different expression genes; DH, doubled haploid; qPCR, quantitative real-time PCR.
References
1
AidaM.IshidaT.TasakaM. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development126, 1563–1570. doi: 10.1007/s004290050234
2
AkinO.ZipurskyS. L. (2014). The shape of things to come. Cell156, 13–14. doi: 10.1016/j.cell.2013.12.037
3
AmsonR.PeceS.MarineJ. C.Di FioreP. P.TelermanA. (2013). TPT1/TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol.23, 37–46. doi: 10.1016/j.tcb.2012.10.002
4
ArcadeA.LabourdetteA.FalqueM.ManginB.ChardonF.CharcossetA.et al. (2004). BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics20, 2324–2326. doi: 10.1093/bioinformatics/bth230
5
BaoB. H.ChaoH. B.WangH.ZhaoW. G.ZhangL. N.RaboanatahiryN.et al. (2018). Stable, environmental specific and novel QTL identification as well as genetic dissection of fatty acid metabolism in Brassica napus. Front. Plant Sci.9, 1018. doi: 10.3389/fpls.2018.01018
6
BecraftP. W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol.18, 163–192. doi: 10.1146/annurev.cellbio.18.012502.083431
7
BerbelA.FerrándizC.HechtV.DalmaisM.LundO. S.SussmilchF. C.et al. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun.4, 797. doi: 10.1038/ncomms1801
8
BradleyS. V.SmithM. R.HyunT. S.LucasP. C.LiL.AntonukD.et al. (2007). Aberrant Huntingtin interacting protein 1 in lymphoid malignancies. Cancer Res.67, 8923–8931. doi: 10.1158/0008-5472.CAN-07-2153
9
BrandU.FletcherJ.HobeM.MeyerowitzE.SimonR. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science289, 617–619. doi: 10.1126/science.289.5479.617
10
BrandU.GrnewaldM.HobeM.SimonR. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol.129, 565–575. doi: 10.1104/pp.001867
11
BurnsM. J.BarnesS. R.BowmanJ. G.ClarkeM. H. E.WernerC. P.KearseyM. J. (2003). QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity90, 39–48. doi: 10.1038/sj.hdy.6800176
12
CaiD.XiaoY.YangW.YeW.WangB.YounasM.et al. (2014). Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor. Appl. Genet.127, 85–96. doi: 10.1007/s00122-013-2203-9
13
CarlesC. C.FletcherJ. C. (2003). Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci.8, 394–401. doi: 10.1016/S1360-1385(03)00164-X
14
ChaoH. B.WangH.WangX. D.GuoL. X.GuJ. W.ZhaoW. G.et al. (2017). Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep.7, 46295. doi: 10.1038/srep46295
15
ChalhoubB.DenoeudF.LiuS.ParkinI. A.TangH.WangX.et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science345, 950–953. doi: 10.1126/science.1253435
16
ChatfieldS. P.StirnbergP.FordeB. G.LeyserO. (2000). The hormonal regulation of axillary bud growth in Arabidopsis. Plant J.24, 159–169. doi: 10.1046/j.1365-313x.2000.00862.x
17
ChenW.ZhangY.LiuX.ChenB.TuJ.FuT.et al. (2007). Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor. Appl. Genet.115, 849–858. doi: 10.1007/s00122-007-0613-2
18
ChenY.ChenX.WangH. J.BaoY. Q.ZhangW. (2014). Examination of the leaf proteome during flooding stress and the induction of programmed cell death in maize. Proteome Sci.12, 33. doi: 10.1186/1477-5956-12-33
19
ChengY.QinG.DaiX.ZhaoY. (2008). NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci.105, 21017–21022. doi: 10.1073/pnas.0809761106
20
ChoS.ChaD. H.ChangJ. B.LeeC. N.KwakI. P.LeeS. H. (2006). Decreased chorionic somatomammotropin 1(CSH1) and nectin-3 gene expression of the placenta in the patient of pre-eclampsia. Fertil. Steril.86, 514–515. doi: 10.1016/j.fertnstert.2006.07.1429
21
ChoiY. I.NohE. W.KimH. J.ParkW. J. (2014). Differential regulation of cytokinin oxidase genes and cytokinin-induced auxin biosynthesis by cellular cytokinin level in transgenic poplars. Plant Cell Rep.33, 1737–1744. doi: 10.1007/s00299-014-1652-1
22
ChoiY.YunH.ParkK. (2008). Screening genes expressed by Rhizobium vitis inoculation and salicylic acid treatment in grapevines using GeneFishing. J. Jpn. Soc. Hortic. Sci.77, 137–142. doi: 10.2503/jjshs1.77.137
23
ClarkS. E.JacobsenS. E.LevinJ. Z.MeyerowitzE. M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development122, 1567–1575.
24
DeeksM. J.HusseyP. J. (2003). Arp2/3 and ‘The Shape of things to come. Curr. Opin. Plant Biol.6, 561–567. doi: 10.1016/j.pbi.2003.09.013
25
DengQ.WangX.ZhangD.WangX.FengC.XuS. (2017). BRS1 function in facilitating lateral root emergence in Arabidopsis. Int. J. Mol. Sci.18, E1549. doi: 10.3390/ijms18071549
26
DingG.ZhaoZ.LiaoY.HuY.ShiL.LongY.et al. (2012). Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Ann. Bot.-London109, 747–759. doi: 10.1093/aob/mcr323
27
DingL.YanS.JiangL.ZhaoW.NingK.ZhaoJ.et al. (2015). Hanaba taranu (han) bridges meristem and organ primordia boundaries through pinhead, jagged, blade-on-petiole2 and cytokinin oxidase 3 during flower development in arabidopsis. Plos Genet.11, e1005479. doi: 10.1371/journal.pgen.1005479
28
DodsworthS. (2009). A diverse and intricate signaling network regulates stem cell fate in the shoot apical meristem. Dev. Biol.336, 1–9. doi: 10.1016/j.ydbio.2009.09.031
29
FanC.CaiG.QinJ.LiQ.YangM.WuJ.et al. (2010). Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor. Appl. Genet.121, 1289–1301. doi: 10.1007/s00122-010-1388-4
30
FletcherJ.MeyerowitzE. (2000). Cell signaling within the shoot meristem. Curr. Opin. Plant Biol.3, 23–30. doi: 10.1016/S1369-5266(99)00033-3
31
FletcheJ. C. (2018). The CLV-WUS stem cell signaling pathway: a roadmap to crop yield optimization. Plants (Basel)7, 87. doi: 10.3390/plants7040087
32
FrankW.Kim-MiriamB.QudeimatE.WoriedhM.AlawadyA.RatnadewiD.et al. (2007). A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. Plant J.51, 1004–1018. doi: 10.1111/j.1365-313X.2007.03198.x
33
FuT. D. (2008). On research and application of heterosis in rapeseed. Agric. Sci. Technol. Equip.5, 10–11. doi: 10.3969/j.issn.1673-887X.2008.05.005
34
GaoR.WangY.GruberM. Y.HannoufaA. (2018). miR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front. Plant Sci.8, 2226. doi: 10.3389/fpls.2017.02226
35
GordonS.ChickarmaneV.OhnoC.MeyerowitzE. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci.106, 16529–16534. doi: 10.1073/pnas.0908122106
36
GouX.HeK.YangH.YuanT.LinH.ClouseS. D.et al. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics11, 1–15. doi: 10.1186/1471-2164-11-19
37
HaeckerA.LauxT. (2001). Cell-cell signaling in the shoot meristem. Curr. Opin. Plant Biol.4, 441–446. doi: 10.1016/S1369-5266(00)00198-9
38
HeislerM. G.OhnoC.DasP.SieberP.ReddyG. V.LongJ. A.et al. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol15, 1899–1911. doi: 10.1016/j.cub.2005.09.052
39
HuY.RenT.LiZ.TangY.RenZ.YanB. (2017). Molecular mapping and genetic analysis of a QTL controlling spike formation rate and tiller number in wheat. Gene634, 15–21. doi: 10.1016/j.gene.2017.08.039
40
JiangC.ShiJ.LiR.LongY.WangH.LiD.et al. (2014). Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor. Appl. Genet.127, 957–968. doi: 10.1007/s00122-014-2271-5
41
JiangL.LiuX.XiongG.LiuH.ChenF.WangL.et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature504, 401–405. doi: 10.1038/nature12870
42
JunaediA.JungW.ChungI.KimK. (2007). Differentially expressed gene of potentially allelopathic rice in response against Barnyardgrass. Crop Sci. Biotech. J.10, 231–236.
43
KhushG. (2001). Green revolution: the way forward. Nat. Rev. Genet.2, 815-822. doi: 10.1038/35093585
44
KiefferM.SternY.CookH.ClericiE.MaulbetschC.LauxT.et al. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for the irroles in meristem maintenance. Plant Cell18, 560–573. doi: 10.1105/tpc.105.039107
45
KimY.KwakC.GuY.HwangI.ChunJ. (2004). Annealing control primer system for identification of differentially expressed genes on agarose gels. Bio. Technol.36, 424–426. doi: 10.2144/04363ST02
46
KroganN. T.MarcosD.WeinerA. I.BerlethT. (2016). The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol.212, 42–50. doi: 10.1111/nph.14107
47
KöllmerI.NovákO.StrnadM.ThomasS.TomáW. (2014). Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. Cell Mol. Biol.78, 359–371. doi: 10.1111/tpj.12477
48
LauxT.MayerK. F.BergerJ.JurgensG. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development122, 87–96.
49
LeibfriedA.ToJ.BuschW.StehlingS.KehleA.DemarM.et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature438, 1172–1175. doi: 10.1038/nature04270
50
LeyserO. (2003). Regulation of shoot branching by auxin. Trends Plant Sci.8, 541–545. doi: 10.1016/j.tplants.2003.09.008
51
LiH.RibautJ.LiZ.WangJ. (2008). Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor. Appl. Genet.116, 243–260. doi: 10.1007/s00122-007-0663-5
52
LiH.LiJ.SongJ.ZhaoB.GuoC. H.WangB.et al. (2018a). An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol.11, 15632. doi: 10.1111/nph.15632
53
LiB.ZhaoW.LiD.ChaoH.ZhaoX.TaN.et al. (2018b). Genetic dissection of the mechanism of flowering time based on an environmentally stable and specific QTL in Brassica napus. Plant Sci.277, 296–310. doi: 10.1016/j.plantsci.2018.10.005
54
LiJ. (2003). Brassinosteroids signal through two receptor-like kinases. Curr. Opin. Plant Biol.6, 494–499. doi: 10.1016/S1369-5266(03)00088-8
55
LiS.ChenL.ZhangL.LiX.LiuY.WuZ.et al. (2015). Bna C9.SMG7b functions as a positive regulator of number of seeds per silique in Brassica napus by regulating the formation of functional female gametophytes. Plant Physiol.169, 2744–2760. doi: 10.1104/pp.15.01040
56
LiaoM. L.PengZ. S.YangZ. J.WeiS. H.MartinekP. (2015). Identification of differentially expressed genes in a pistillody common wheat mutant using an annealing control primer system. Genet. Mol. Res.14, 3995–4004. doi: 10.4238/2015.April.27.14
57
LiuS.FanC.LiJ.CaiG.YangQ.WuJ.et al. (2016). A genome wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet.129, 1203–1215. doi: 10.1007/s00122-016-2697-z
58
LuK.XiaoZ.JianH.PengL.QuC.FuM.et al. (2016). A combination of genome-wide association and transcriptome analysis reveals candidate genes controlling harvest index-related traits in Brassica napus. Sci. Rep.6, 36452. doi: 10.1038/srep36452
59
LuZ.ShaoG.XiongJ.JiaoY.WangJ.LiuG.et al. (2015). MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genomics42, 71–78. doi: 10.1016/j.jgg.2014.12.005
60
MengL.LiH.ZhangL.WangJ. (2015). QTL IciMapping Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J.3, 269–283. doi: 10.1016/j.cj.2015.01.001
61
MüllerR.BorghiL.KwiatkowskaD.LaufsP.SimonR. (2006). Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell18, 1188–1198. doi: 10.1105/tpc.105.040444
62
NagaharuU. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot.7, 389–452.
63
ParanI.ZamirD. (2003). Quantitative traits in plants: beyond the QTL. Trends Genet.19, 303–306. doi: 10.1016/S0168-9525(03)00117-3
64
ParkJ. S.KimI. S.ChoM. S.ParkS.ParkS. G. (2006). Identification of differentially expressed genes involved in spine formation on seeds of Daucus carota L. (carrot), using annealing control primer (ACP) system. J. Plant Biol.49, 133–140. doi: 10.1007/BF03031009
65
OngaroV.LeyserO. (2008). Hormonal control of shoot branching. J. Exp. Bot.59, 67–74. doi: 10.1093/jxb/erm134
66
QiuD.MorganC.ShiJ.LongY.LiuJ.LiR.et al. (2006). A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet.114, 67–80. doi: 10.1007/s00122-006-0411-2
67
QuarrieS.PekicQ. S.RadosevicR.KaminskaA.BarnesJ. D.LeveringtonM.et al. (2006). Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J. Exp. Bot.57, 2627–2637. doi: 10.1093/jxb/erl026
68
QuatranoR. S.PerroudP. F.HarriesP.BezanillaM.PanA.KluehR.et al. (2006). The role of ARP2/3 and Scar/WAVE complexes in polar growth of the apical cell of the moss Physcomitrella. Dev. Biol.295, 342–344. doi: 10.1016/j.ydbio.2006.04.059
69
QzerH.OralE.DogruU. (1999). Relationships between yield and yield components on currently improved spring rapeseed cultivars. Tr. J. Agric. For.23, 603–607.
70
RameneniJ. J.LeeY.DhandapaniV.YuX.ChoiS. R.OhM.-H.et al. (2015). Genomic and post-translational modification analysis of leucine-rich-repeat receptor-like kinases in Brassica rapa. Plos One10, e0142255. doi: 10.1371/journal.pone.0142255
71
RobertoT. M.BeatrizX. C.JoséL. C. P.JesúsH. M.JorgeL. R. S.SantiagoV. G. G.et al. (2015). AtTCTP2, an Arabidopsis thaliana homolog of translationally controlled tumor protein, enhances in vitro plant regeneration. Front. Plant Sci.6, 468. doi: 10.3389/fpls.2015.00468
72
SablowskiR. (2007). Flowering and determinacy in Arabidopsis. J. Exp. Bot.58, 899–907. doi: 10.1093/jxb/erm002
73
SanghoonL.KiwonL.KiyongK.GijunC.SeihyungY.JiH. C.et al. (2009). Identification of salt-stress induced differentially expressed genes in barley leaves using the annealing-control-primer-based gene fishing technique. Afr. J. Biotechnol.8, 1326–1331. doi: 10.5897/AJB2009.000-9212
74
SAS Institute Inc. (2000). SAS/STAT User’s Guide, Version 8. Cary, NC: SAS Institute Inc.
75
SchippersJ. H.FoyerC. H.Van DongenJ. T. (2016). Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol.29, 121–128. doi: 10.1016/j.pbi.2015.11.009
76
SchoofH.LenhardM.HaeckerA.MayerK.JürgensG.LauxT. (2000). The stem cell population of Arabidopsis shoot, meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
77
ShaoG.LuZ.XiongJ.WangB.JingY.MengX.et al. (2019). Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant19, 1–13. doi: 10.1016/j.molp.2019.04.008
78
ShchennikovaA. V.KochievaE. Z.BeletskyA. V.FilyushiM. A.ShulgaO. A.RavinN. V.et al. (2017). Identification and expression analysis of receptor-like kinase gene ERECTA in mycoheterotrophic plant Monotropa hypopitys. Mol. Biol.51, 681–686. doi: 10.1134/S002689331705017X
79
ShiJ.LiR.QiuD.JiangC.LongY.MorganC.et al. (2009). Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics182, 851–861. doi: 10.1534/genetics.109.101642
80
ShiJ.ZhanJ.YangY.YeJ.HuangS.LiR.et al. (2015). Linkage and regional association analysis reveal two new tightly linked major-QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci. Rep.5, 14481. doi: 10.1038/srep14481
81
ShpakE. D.BerthiaumeC. T.HillE. J.ToriiK. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development131, 1491–1501. doi: 10.1242/dev.01028
82
SohnW. J.KimD.LeeK. W.KimM. S.KwonS.LeeY.et al. (2007). Novel transcriptional regulation of the schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol. Immunol.44, 3273–3282. doi: 10.1016/j.molimm.2007.03.001
83
SpinelliS. V.MartinA. P.ViolaI. L.GonzalezD. H.PalatnikJ. F. (2011). A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol.156, 1894–1904. doi: 10.1104/pp.111.177709
84
StribernyB.MeltonA. E.SchwackeR.KrauseK.FischerK.GoertzenL. R.et al. (2017). Cytokinin response factor 5 has transcriptional activity governed by its C-terminal domain. Plant Signaling Behav.12, e1276684. doi: 10.1080/15592324.2016.1276684
85
ToriiK. U.MitsukawaN.OosumiT.MatsuuraY.YokoyamaR.WhittierR. F.et al. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell8, 735–746. doi: 10.1105/tpc.8.4.735
86
UchidaN.ShimadaM.TasakaM. (2013). ERECTA-family receptor kinases regulate stem cell homeostasis via buffering its cytokinin responsiveness in the shoot apical meristem. Plant Cell Physiol.54, 343–351. doi: 10.1093/pcp/pcs109
87
UedaM.MatsuiK.IshiguroS.SanoR.WadaT.PaponovL.et al. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development131, 2101–2111. doi: 10.1242/dev.01096
88
WangB.SmithM. S.LiJ. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol.69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
89
WangH.YinY. (2014). Analysis and strategy for oil crop industry in China. Chin. J. Oil Crop Sci.36, 414–421. doi: 10.7505/j.issn.1007-9084.2014.03.020
90
WangS.BastenC.ZengZ. (2007). Windows QTL Cartographer 2.5. Raleigh, NC: North Carolina State University.
91
WangY.LiJ. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol.59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
92
WangX.WangH.LongY.LiD.YinY.TianJ.et al. (2013). Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS One8, e80569. doi: 10.1371/journal.pone.0080569
93
WangY.BouwmeesterH. J. (2018). Structural diversity in the strigolactones. J. Exp. Bot.69, 2219–2230. doi: 10.1093/jxb/ery091
94
WeigelD.JürgensG. (2002). Stem cells that make stems. Nature415, 751–754. doi: 10.1038/415751a
95
WilliamsL.FletcherJ. (2005). Stem cell regulation in the Arabidopsis shoot apical meristem. Curr. Opin. Plant Biol.8, 582–586. doi: 10.1016/j.pbi.2005.09.010
96
XieL.LiP.ZhangW.YangM. (2005). A review on the development potential of bioenergy rapeseed. Chin. J. Bioprocess Eng.3, 28–31. doi: 10.3969/j.issn.1672-3678.2005.01.007
97
YaginumaH.HirakawaY.KondoY.Ohashi-ItoK.FukudaH. (2011). A novel function of TDIF-related peptides: promotion of axillary bud formation. Plant Cell Physiol.52, 1354–1364. doi: 10.1093/pcp/pcr081
98
YanaiO.ShaniE.DolezalK.TarkowskiP.SablowskiR.SandbergG.et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol.15, 1566–1571. doi: 10.1016/j.cub.2005.07.060
99
YanagawaY.KimuraS.TakaseT.SakaguchiK.UmedaM.KomamineA.et al. (2002). Spatial distribution of the 26S proteasome in meristematic tissues and primordia of rice (Oryza sativa L.). Planta214, 703–707. doi: 10.1007/s00425-001-0676-2
100
YiB.ChenY.LeiS.TuJ.FuT. (2006). Fine mapping of the recessive genie male sterile gene (Bnmsl) in Brassica napus. Theor. Appl. Genet.113, 643–650. doi: 10.1016/S1369-5266(99)00033-3
101
YinY.ChenZ.YuJ.WangH.FengZ. (2010). Analysis of potential for rapeseed production in China. J. Agric. Sci. Technol.12, 16–21. doi: 10.3969/j.issn.1008-0864.2010.03.03
102
ZhangN.YuH.YuH.CaiY.HuangL.XuC.et al. (2018). A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell30, 1461–1475. doi: 10.1105/tpc.18.00063
103
ZhaoW.WangH.LiD.TianJ.ZhaoY.LiB.et al. (2011). Fingerprinting construction of Brassica napus germplasm with super-high oil content and main rapeseed cultivars. J. Plant Genet. Resour.12, 904–909. doi: 10.13430/j.cnki.jpgr.2011.06.012
104
ZhaoW.WangX.WangH.TianJ. H.LiB. J.ChenL.et al. (2016). Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front. Plant Sci.7, 17. doi: 10.3389/fpls.2016.00017
105
ZhouF.LinQ.ZhuL.RenY.ZhouK.ShabekN.et al. (2016). Corrigendum: D14-SCF (D3)-dependent degradation of D53 regulates strigolactone signalling. Nature532, 406–410. doi: 10.1038/nature16537
106
ZhouQ.FuD.MasonA.ZengY.ZhaoC.HuangY. (2014). In silico integration of quantitative trait loci for seed yield and yield-related traits in Brassica napus. Mol. Breed.33, 881–894. doi: 10.1007/s11032-013-0002-2
107
ZwackP. J.De ClercqI.HowtonT. C.HallmarkH. T.HurnyA.KeshishianE. A.et al. (2016). Cytokinin response factor 6 represses cytokinin-associated genes during oxidative stress. Plant Physiol.172, 1249. doi: 10.1104/pp.16.00415
Summary
Keywords
Brassica napus, multi-main stem trait, QTL mapping, Gene-Fishing, candidate genes
Citation
Zhao W, Chao H, Zhang L, Ta N, Zhao Y, Li B, Zhang K, Guan Z, Hou D, Chen K, Li H, Zhang L, Wang H and Li M (2019) Integration of QTL Mapping and Gene Fishing Techniques to Dissect the Multi-Main Stem Trait in Rapeseed (Brassica napus L.). Front. Plant Sci. 10:1152. doi: 10.3389/fpls.2019.01152
Received
21 April 2019
Accepted
23 August 2019
Published
20 September 2019
Volume
10 - 2019
Edited by
Jianjun Chen, University of Florida, United States
Reviewed by
Zhongyun Piao, Shenyang Agricultural University, China; Rodrigo Gazaffi, Federal University of São Carlos, Brazil
Updates
Copyright
© 2019 Zhao, Chao, Zhang, Ta, Zhao, Li, Zhang, Guan, Hou, Chen, Li, Zhang, Wang and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maoteng Li, limaoteng426@hust.edu.cn
This article was submitted to Plant Breeding, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.