- 1Key Laboratory of Oasis Eco-Agriculture, College of Agriculture, Shihezi University, Shihezi, China
- 2Agriculture and Food, CSIRO, Canberra, ACT, Australia
Verticillium wilt caused by Verticillium dahliae is a destructive cotton disease causing severe yield and quality losses worldwide. WRKY transcription factors play important roles in plant defense against pathogen infection. However, little has been reported on the functions of WRKYs in cotton's resistance to V. dahliae. Here, we identified 5, 5, and 10 WRKY70 genes in Gossypium arboreum, Gossypium raimondii, and Gossypium hirsutum, respectively, and investigated the expression profiles of all GhWRKY70 genes in various cotton tissues and in response to hormone treatment or V. dahliae infection. Reverse transcription-quantitative PCR analysis showed that GhWRKY70D13 was expressed higher in roots and stems than in other tissues, and up-regulated after V. dahliae inoculation. Knock-down of GhWRKY70D13 improved resistance to V. dahliae in both resistant and susceptible cotton cultivars. Comparative analysis of transcriptomes generated from wild-type and stable RNAi (RNA interference) plant with down-regulated GhWRKY70D13 showed that genes involved in ethylene (ET) and jasmonic acid (JA) biosynthesis and signaling were significantly upregulated in the GhWRKY70D13 RNAi plants. Consistently, the contents of 1-aminocyclopropane-1-carboxylic (ACC), JA, and JA-isoleucine levels were significantly higher in the GhWRKY70D13 RNAi plants than in wild-type. Following V. dahliae infection, the levels of ACC and JA decreased in the GhWRKY70D13 RNAi plants but still significantly higher (for ACC) than that in wild-type or at the same level (for JA) as in non-infected wild-type plants. Collectively, our results suggested that GhWRKY70D13 negatively regulates cotton's resistance to V. dahliae mainly through its effect on ET and JA biosynthesis and signaling pathways.
Introduction
Cotton (Gossypium hirsutum) produces important natural fibers and is one of the most important industrial crops globally. Cotton production is constantly threated by various biotic and abiotic stresses, such as cold, drought, insect pests, and pathogens. The soil-borne fungus Verticillium dahliae is one of the most destructive pathogens in cotton production areas worldwide (Cai et al., 2009; Wang et al., 2016; Gong et al., 2017). Verticillium wilt caused by V. dahliae is difficult to control since V. dahliae has a wide host range and its microsclerotia can survive for long periods in the soil even in the absence of suitable hosts (Fradin and Thomma, 2006; Shaban et al., 2018). Moreover, there is no effective fungicide that can prevent cotton from V. dahliae infection (Mol et al., 1995; Wang et al., 2016).
The most economical and efficient method to control V. dahliae is to improve host resistance. However, no germplasms of G. hirsutum cultivars have shown immune to Verticillium wilt (Yang et al., 2015). Thus, it is essential to identify Verticillium wilt resistance genes in cotton germplasm and incorporate them into elite cotton cultivars. In general, the V. dahliae strains are classified as defoliating or nondefoliating which have different pathogenic characteristics (Chen et al., 2018; Zhang et al., 2019). Although numerous V. dahliae-resistance genes have been characterized in cotton, including GbWRKY1 (Li et al., 2014a), GhCYP94C1 (Sun et al., 2014), GhERF6 (Yang et al., 2015), GhHDTF1 (Gao et al., 2016), GhERF1-like (Guo et al., 2016), GhbHLH171 (He et al., 2018), and GhJAZ2 (He et al., 2018), only a defoliating strain of V. dahliae was used in most of these studies. There is still limited knowledge of the genetic and molecular mechanisms on the interaction between cotton and V. dahliae.
Plant hormones, such as jasmonic acid (JA) and ethylene (ET), are crucial for plant defenses against pathogen infection. JA signaling plays a role in the resistance against necrotrophic pathogens (Glazebrook, 2005). JA has been found to induce degradation of Jasmonate ZIM domain (JAZ) proteins and to de-repress the MYC2 transcription factors to activate JA responsive gene expression (Chini et al., 2007; Chung and Howe, 2009; Zhang et al., 2015a). JA-related genes have a positive role in the defense of plant against V. dahliae in Arabidopsis thaliana and cotton (Gao et al., 2013; Li et al., 2014a; He et al., 2018). ET often synergizes with JA and increases plant tolerance to necrotrophic pathogens (Zhu et al., 2011). ET is a small gaseous hormone involved in plant innate immunity against pathogens, and various studies have revealed that ET plays a role in resistance to V. dahliae in tobacco and cotton (Yang et al., 2015; Guo et al., 2016; Liu et al., 2017).
Many transcription factors have important regulatory functions in plant defenses against pathogens (Nuruzzaman et al., 2013; Jiang et al., 2017). As stress-responsive transcription factors, plant WRKY proteins play important roles in plant immunity (Rushton et al., 2010; Jiang et al., 2017). The WRKY family belongs to one of the largest families of transcriptional regulators in plants and is characterized by a highly conserved WRKY domain (about 60 amino acids) at the N-terminus and an atypical zinc finger structure at the C-terminus (Rushton et al., 2010). Based on the number of WRKY domains and the structure of their zinc fingers, 74 WRKY transcription factors were identified and divided into three groups in Arabidopsis (Eulgem et al., 2000). The group III WRKY transcription factors are involved in plant resistance to pathogens (Li et al., 2004; Li et al., 2006; Murray et al., 2007; Higashi et al., 2008; Kim et al., 2008; Hu et al., 2012). In Arabidopsis, wrky53, wrky38, and wrky62 mutant plants displayed severer susceptibility to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) (Murray et al., 2007; Kim et al., 2008; Hu et al., 2012), and overexpression of AtWRKY41 enhances disease resistance to Pst DC3000 (Higashi et al., 2008). Similarly, overexpression of AtWRKY70 improves resistance to necrotrophic pathogens by activating the salicylic acid (SA) signaling pathway and suppressing the JA signaling pathway (Li et al., 2004; Li et al., 2006).
Recently, genome-wide identifications of the WRKY family genes have been carried out in Gossypium raimondii, Gossypium arboreum, and G. hirsutum (Dou et al., 2014; Ding et al., 2015), but only few studies have reported WRKY-regulated responses to V. dahliae infection in cotton (Li et al., 2014a; Xiong et al., 2019). Therefore, research to understand the biological functions and mechanisms of WRKY genes in resistance against V. dahliae in cotton remains an important goal. Our previous study showed that GhWRKY70A05a acted as a negative regulator in cotton's resistance against V. dahliae by repressing JA signaling pathway and promoting SA signaling pathway (Xiong et al., 2019). In order to get broader insight into the roles of GhWRKY70 genes in defense against V. dahliae, we investigated the expression of 10 GhWRKY70 genes in G. hirsutum in response to V. dahliae infection and to treatment with the plant hormones SA, methyl jasmonate (MeJA), and ethephon (ETH). We found that GhWRKY70D13 was highly expressed in both roots and stems, and was induced in roots of both V. dahliae-susceptible and resistant cotton cultivars upon V. dahliae infection. Using the VIGS and RNA interference (RNAi) approaches, we demonstrated that silencing the expression of GhWRKY70D13 enhanced cotton's resistance to V. dahliae, which was likely achieved through upregulating the JA and/or ET biosynthesis and signaling pathways, a potential mechanism different from that of GhWRKY70A05a.
Materials and Methods
Plant Materials
V. dahliae-resistant G. hirsutum cv. Zhongzhimian 2 and V. dahliae-susceptible G. hirsutum cv. Xinluzao 7 were used for gene expression analysis under V. dahliae, SA, MeJA, or ETH treatment. Zhongzhimian 2, Xinluzao 7 and another V. dahliae-susceptible G. hirsutum cv. Xincai 7 were used in virus-induced gene silencing (VIGS) experiments and disease assays. All plants were grown in a greenhouse with a 16-h-light/8-h-dark cycle at 24/23°C (light/dark), watered with Hoagland's nutrient solution weekly.
Identification of the GhWRKY70 Gene Family in Cotton
The protein sequences of Arabidopsis group III WRKYs were obtained from the The Arabidopsis Information Resource (TAIR) website (https://www.arabidopsis.org/). The AtWRKY70 protein sequence was used as a query to search the genome sequences of G. raimondii, G. arboreum, and G. hirsutum, which were retrieved from Cottongen (https://www.cottongen.org/), with an E-value less than 1 x 10−30. To identify WRKY70 genes in the three cotton genomes, the hits were further filtered using the Pfam database (http://pfam.xfam.org) and the National Center for Biotechnology Information (NCBI) Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) based on the presence of the WRKY domain and the C2HC zinc finger.
Gene Cloning, Multiple-Sequence Alignment, and Phylogenetic Analysis
The coding sequences of GhWRKY70A06 and GhWRKY70D13 were amplified from roots of Zhongzhimian 2 using gene specific primers (Supplementary Table S4). The amplified products were cloned into the pMDT-19 vector, and confirmed by sequencing (Sangon Biotech Co., Ltd., Shanghai, China). GhWRKY70A05a has been previously cloned (Xiong et al., 2019). The ClustalX software (ver. 1.83) was employed for alignment of the WRKY70 amino acid sequences from the three cotton species and the group III WRKY70 genes from Arabidopsis. The phylogenetic tree was constructed using the neighbor-joining method implemented in the Molecular Evolutionary Genetics Analysis (MEGA) software 7.1 with a bootstrap value of 1,000.
Analysis of Cis-Acting Elements in the Promoters of GhWRKY70 Genes
The 2-kb genomic sequences upstream the transcriptional start sites of GhWRKY70 genes were downloaded from Cottongen (https://www.cottongen.org/) and used in the analysis of cis-acting elements using the PlantCARE database.
Application of Plant Hormones
Two-leaf stage cotton seedlings were sprayed with 1 mmol L−1 SA, 100 μmol L−1 MeJA, or 5 mg L−1 ETH. Cotton seedlings sprayed with distilled water were used as controls. Roots were collected from the treated seedlings at 0, 0.5, 1, 3, 6, 9, 12, and 24 h after treatment, frozen immediately in liquid nitrogen and then stored at −80°C for future use. All treatments were done with three independent biological replicates.
Inoculations With Verticillium dahliae
A highly aggressive defoliating strain, V991, was kindly provided by Professor Longfu Zhu (Huazhong Agricultural University) and used in a disease assay. V. dahliae inoculation was performed according to a previously described method (Xiong et al., 2019). Two-leaf stage cotton plants were infected using 10 ml V. dahliae spore suspension (106 spores per ml) by irrigating injured roots. A similar routine was adopted for treating the control plants with distilled water. Cotton roots from five individual seedlings were harvested for RNA extraction at 0, 1, 3, 6, 12, 24, 48, and 72 h post-inoculation (hpi). Samples of three biological replicates were prepared. Disease index at 14 and 21 day post-inoculation (dpi) was calculated based on 30 VIGS or GhWRKY70D13 RNAi transgenic cotton plants. The fungal recovery assay and calculations of relative fungal biomass were performed as previously described (Ellendorff et al., 2009).
Ribonucleic Acid Isolation and Expression Pattern Analysis
Total RNA was extracted from cotton tissues with an RN09-EASYspin RNA Plant Mini Kit (Aidlab Biotechnologies Co. Ltd., Beijing, China) according to the manufacturer's instructions. All RNA samples were treated with DNase I (Takara Biotechnology Co., Ltd., Dalian, China) to remove genomic DNA. RNA quantity was determined using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, Wilmington, USA). First-strand complementary DNA (cDNA) was synthesized using the PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). The gene-specific primers used in reverse transcription-quantitative PCR (RT-qPCR) were designed using qPrimerDB (https://biodb.swu.edu.cn/qprimerdb/). The lengths of the amplified fragment were designed to be between 80 and 250 bp. qPCR reactions were performed on a Light Cycler 480II (Roche, Rotkreuz, Switzerland) sequence detection system using SYBR Green (Roche) including a pre-incubation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 15 s. The cotton UBQ7 (DQ116441) gene was used as an internal control. Each experiment included three biological replicates. The primers used in RT-qPCR are listed in Supplementary Table S4. The expression data of the 10 GhWRKY70 genes in other tissues were based on previously published transcriptomic data from TM-1 (Zhang et al., 2015b).
Virus-Induced Gene Silencing Assays
The vectors of tobacco rattle virus (TRV) were used for VIGS to investigate the function of GhWRKY70 in response to V. dahliae infection (Liu et al., 2002). The TRV: GhCHLI (encoding magnesium chelatase subunit I) was used as a positive control for the VIGS efficiency (Gu et al., 2014), because leaves of the TRV: GhCHLI plants would show a bleaching phenotype. Approximately 400 bp gene-specific fragment from the coding sequence of GhWRKY70D04, GhWRKY70D13, or GhCHLI was amplified from G. hirsutum cv. Zhongzhimian 2 and inserted into the TRV2 vector, respectively. The primers used in construction of the VIGS vectors are listed in Supplementary Table S4. TRV: GhWRKY70D04, TRV: GhWRKY70D13, TRV: GhCHLI or TRV1 construct was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. We then followed the same procedures as previously reported to carry out VIGS in cotton (Gao et al., 2013). RNA was extracted from the TRV: GhWRKY70D13 cotton leaves to measure the expression level of GhWRKY70.
Ribonucleic Acid Interference Construct and Cotton Transformation
The construct used in generating GhWRKY70D13 RNAi transgenics was generated by insert an amplified GhWRKY70D13 fragment (324-bp) into the PANDA35 HK vector through a recombination reaction. The RNAi vector was transformed into G. hirsutum cv. Xincai 7 via A. tumefaciens (LBA4404) as previously described (Jin et al., 2006). The regenerated plants were examined by antibiotic kanamycin and RT-qPCR to verify the presence and expression of the transgene. T3 homozygous lines were identified and used in all experiments.
Transcriptome Analysis
RNA-seq was used to investigate the effect of knocking-down of GhWRKY70D13 expression on disease responses. Root samples were collected from wild-type (WT) and the two GhWRKY70D13-RNAi lines (Ci1 and Ci2) at 0, 24, and 72 hpi following V. dahliae inoculation. The samples from Ci1 and Ci2 were mixed to form a single sample (designed Ci) based on time points and biological replicates. Finally, 18 samples (2 genotypes x 3 time points x 3 biological replicates) were used in transcriptome sequencing. Total RNA was extracted with a Tiangen RNA extraction kit (Tiangen, Beijing, China) according to the manufacturer's instructions. RNA purity and integrity were checked before library construction, using NanoPhotometer® spectrophotometer (Implen, CA, USA) and the Bioanalyzer 2100 System (Agilent Technologies, CA, USA), respectively. Approximately 3 μg RNA per sample was used for library construction. RNA sequencing was performed by Novogene (Novogene, Tianjin, China) with an Illumina HiSeq (Illumina, CA, USA) system. The RNA-seq raw data were checked for quality. After removing the low quality reads, the remaining clean reads were aligned to the G. hirsutum genome using TopHat v2.0.12 (Wang et al., 2019). The raw data of transcriptome sequencing were deposited in NCBI under PRJNA578842.
Identification and Functional Annotation of Differentially Expressed Genes
Gene expression levels were determined by FPKM (fragments per kilobase of transcript sequence per millions of base pairs sequenced) (Trapnell et al., 2010). Using DESeq, the transcripts with an adjusted P-value < 0.05 were considered to be differentially expressed genes (DEGs) (Anders and Huber, 2010). The DEGs were subjected to KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis to identify statistically enriched pathways (P-value < 0.05) (Kanehisa et al., 2008). Venn diagrams and heat-maps were generated using the tools available on the Novemagic server (https://magic.novogene.com).
Quantification of the Contents of 1-Aminocyclopropane-1-Carboxylic, Jasmonic Acid, Jasmonoyl Isoleucine, and Salicylic Acid
WT, Ci1 (GhWRKY70D13-RNAi line 1), and Ci2 (GhWRKY70D13-RNAi line 2) seedlings at the two-leaf stage were inoculated with V. dahliae through the root irrigation method. Fresh root samples of WT, Ci1, and Ci2 from three biological replicates were harvested at 0 and 72 hpi. Quantification of the content of each phytohormone was performed by Jiangxi Likon Science & Technology Co., Ltd (Nanchang, China). Extraction and measurement of endogenous ACC were performed as previously described (Datta et al., 2015).
The levels of JA, JA-isoleucine (JA-Ile), and SA were determined using a previously described method with slight modification (Sun et al., 2014). Approximately 0.2 g fresh samples were ground to a fine powder in liquid nitrogen and mixed with 2 ml pre-cooled extraction buffer (methanol:water, 80:20, v:v). After shaking at 4°C for 16 h in the dark, the samples were centrifuged at 12,000 rpm for 10 min at the same temperature with 0.3 ng added SA (Sigma) as an internal standard. The supernatant was dried under N2 at room temperature, then dissolved in 0.4 ml methanol and filtered with a 0.22 mm filter membrane. The JA and JA-Ile levels were quantified using the high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) system (AB SCIEX Triple Quad 5500) with JA (Sigma) and JA-Ile (Sigma) as the external standards.
Results
Identification of WRKY70 Transcription Factors in Cotton
Using AtWRKY70 as a query, we identified 5, 5, and 10 WRKY70 transcription factors in G. raimondii (Wang et al., 2012), G. arboreum (Li et al., 2014b), and G. hirsutum (Zhang et al., 2015b; Wang et al., 2019), respectively, indicating no gene loss occurred after polyploidization. They were named based on their chromosomal location (Supplementary Tables S1 and S2). Apart from GhWRKY70A05a, GhWRKY70A06, and GhWRKY70D13, other GhWRKY70 genes were annotated consistently in the two TM-1 genomes (G. hirsutum) (Zhang et al., 2015b; Wang et al., 2019) (Supplementary Table S2). To reconcile the annotation difference of GhWRKY70A05a, GhWRKY70A06, and GhWRKY70D13, we cloned their coding sequences (Supplementary Table S3), and confirmed one of the two annotations was correct for each gene (Supplementary Table S2).
Conserved Motifs, Phylogenetic and Cis-Acting Element Analysis of the GhWRKY70 Genes
Amino acid sequence alignment was performed using all the GhWRKY70 genes identified in G. raimondii, G. arboreum, and G. hirsutum together with AtWRKY70. Conserved domain analysis confirmed that all cotton WRKY70s identified belong to the group III with a well-conserved WRKY domain at the N-terminal region and a C2HC zinc finger at the C-terminus (Supplementary Figure S1A). The phylogenetic relationships among cotton WRKY70s and Arabidopsis group III WRKY proteins were further analyzed through construction of a phylogenetic tree using the maximum likelihood method. It was revealed that each pair of homeologs from the At and Dt subgenomes of G. hirsutum was unambiguously grouped with their orthologs in the diploids G. raimondii and G. arboreum (Supplementary Figure S1B), suggesting the conservation of each gene during the history of cotton evolution. GhWRKY70A02, GhWRKY70A05a, GhWRKY70A05b, GhWRKY70A06, and GhWRKY70A13 of At homologs were corresponding to GhWRKY70D02, GhWRKY70D05a, GhWRKY70D04, GhWRKY70D06, and GhWRKY70D13 of Dt homologs, respectively.
To identify the putative cis-acting elements in the promoters of GhWRKY70 genes, a 2-kb sequence upstream of the start codon of each GhWRKY70 was scanned using the online tool PlantCARE. The results showed that each GhWRKY70 contained at least one hormone responsive cis-element in its promoter (Supplementary Figure S2). Nine of the 10 GhWRKY70 genes contained SA (TCA-element) and MeJA responsive elements (TGACG-element) in their promoters. Eight contained the ET-responsive element (ERE). Five contained the gibberellin responsive element (GARE-motif) and abscisic acid responsive element (ABRE). The auxin-responsive element (TGA-element) was identified in three GhWRKY70 genes. GhWRKY70D05a contained cis-elements responsive to all hormones in its promoter. These data suggested that most GhWRKY70 genes might be regulated by one or more hormones, and could play important roles in hormonal responses.
Expression Patterns of GhWRKY70 Genes in Different Tissues
To investigate the expression patterns of the GhWRKY70 genes in different tissues, we used published RNA-seq data [G. hirsutum cv. TM-1; (Zhang et al., 2015b)] (Figure 1A), and did qPCR to quantify their expression levels in roots, stems, and leaves of Zhongzhimian 2 (V. dahliae resistant) and Xinluzao 7 (V. dahliae susceptible) (Figure 1B). The two observations were the tissue-specificity of individual genes and the similar expression profile of the two homeologs of most pairs of GhWRKY70 from the At and Dt subgenomes. Generally, the GhWRKY70 genes, except GhWRKY70D02 and GhWRKY70D05a, had a expression level higher in vegetative tissues than in reproductive tissues. Both GhWRKY70A06 and GhWRKY70D06 had higher expression levels in leaves than in other tissues (Figure 1A). GhWRKY70A05b, GhWRKY70D04, and GhWRKY70A13 had a higher expression level in roots than in other tissues. GhWRKY70D13 and GhWRKY70A02 exhibited higher expression levels in both roots and stems, while GhWRKY70A05a exhibited a higher expression level in stems and ovules (Figure 1A). Similar expression patterns of the two homologs suggest their possible redundant functions. The qPCR results were consistent with the RNA-seq results and some GhWRKY70 genes showed different expression profiles in the two G. hirsutum cultivars with different V. dahliae resistance (Figure 1B).
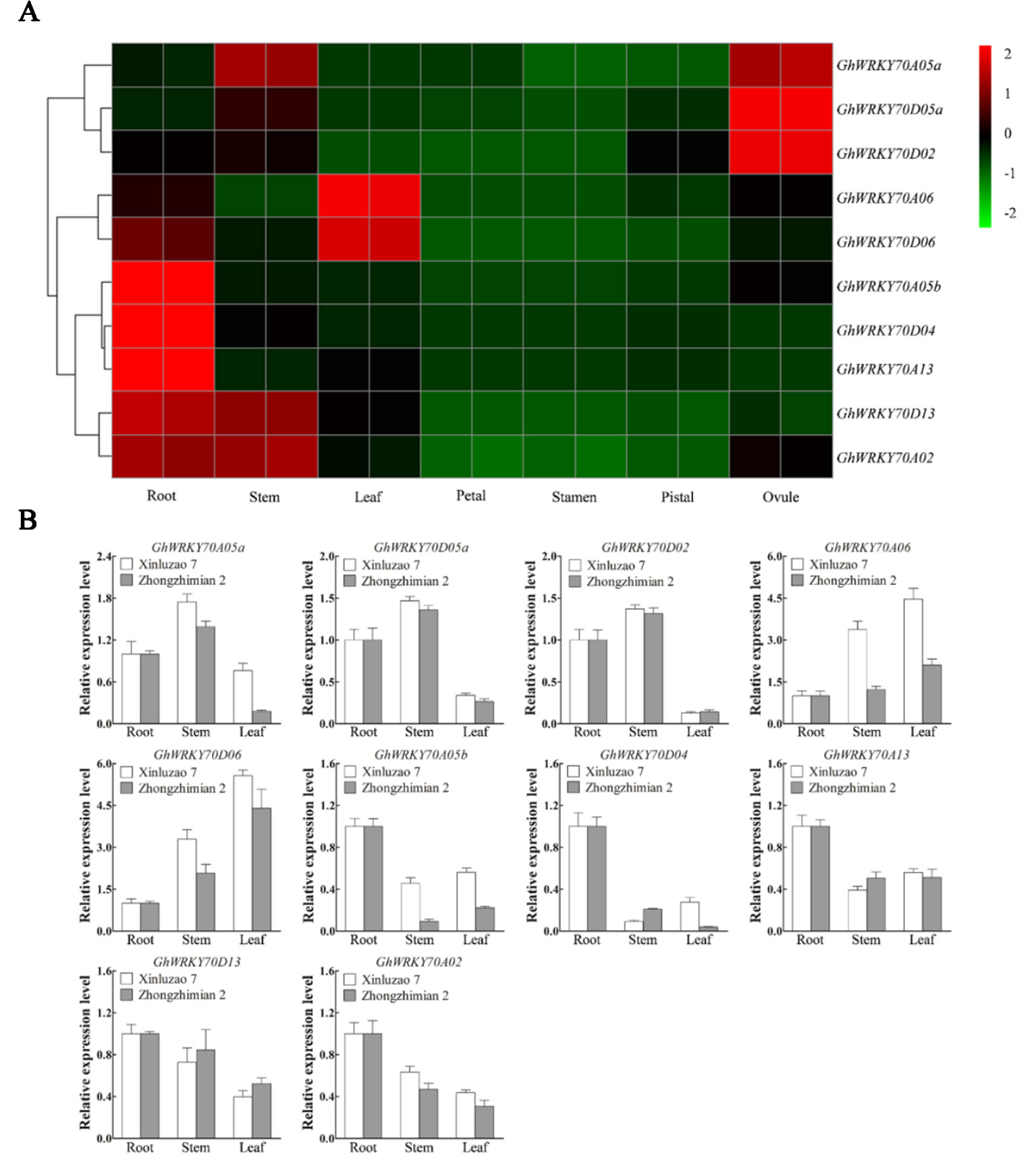
Figure 1 Expression patterns of GhWRKY70 genes in different cotton tissues. (A) Heat map showing the expression level of the 10 GhWRKY70 genes in various tissues (root, stem, leaf, petal, stamen, pistil, and ovule) of TM-1 based on RNA-seq data. (B) Expression profiling of GhWRKY70 genes in various tissues based on reverse transcription-quantitative PCR. Two-leaf stage cotton seedlings were used in the analyses. For both cultivars Xinluzao 7 and Zhongzhimian 2, root was used as the reference sample and its expression level was set to 1.0. Cotton GhUBQ7 was used as the internal control to normalize all data. Each experiment was performed using three independent biological replicates.
Expression Analysis of the GhWRKY70 Genes in Response to Hormone Treatments and Verticillium dahliae Inoculation
Analysis of hormone responsive elements in the promoters of GhWRKY70 genes suggests their expression might be induced by SA, MeJA, and/or ET. We thus investigated the effects of SA, MeJA, and ET on the changes of the transcript level of the 10 GhWRKY70 genes in Zhongzhimian 2 (resistant to V. dahliae) and Xinluzao 7 (susceptible to V. dahliae) using RT-qPCR. Upon SA treatment, the 10 GhWRKY70 genes were clustered into two groups. One group showed upregulation in Zhongzhimian 2 but not in Xinluzao 7, while another group showed upregulation in both cultivars (Figure 2A). GhWRKY70A02, GhWRKY70A05a, and GhWRKY70D05a were significantly (P < 0.01) downregulated by 3.0- to 87.4-fold, 1.4- to 5.9-fold, and 2.2- to 8.1-fold in the V. dahliae susceptible cultivar and upregulated by 1.5- to 2.7-fold, 1.6- to 2.1-fold, and 1.4- to 2.2-fold in the V. dahliae resistant cultivar, respectively (Figure 2A). In response to SA treatment, the expression level of GhWRKY70D13 increased gradually from 0.5 to 12 h, while significant (P < 0.01) upregulation of GhWRKY70A13 was observed at 24 h by 17.5-fold in the V. dahliae resistant cultivar (Figure 2A).
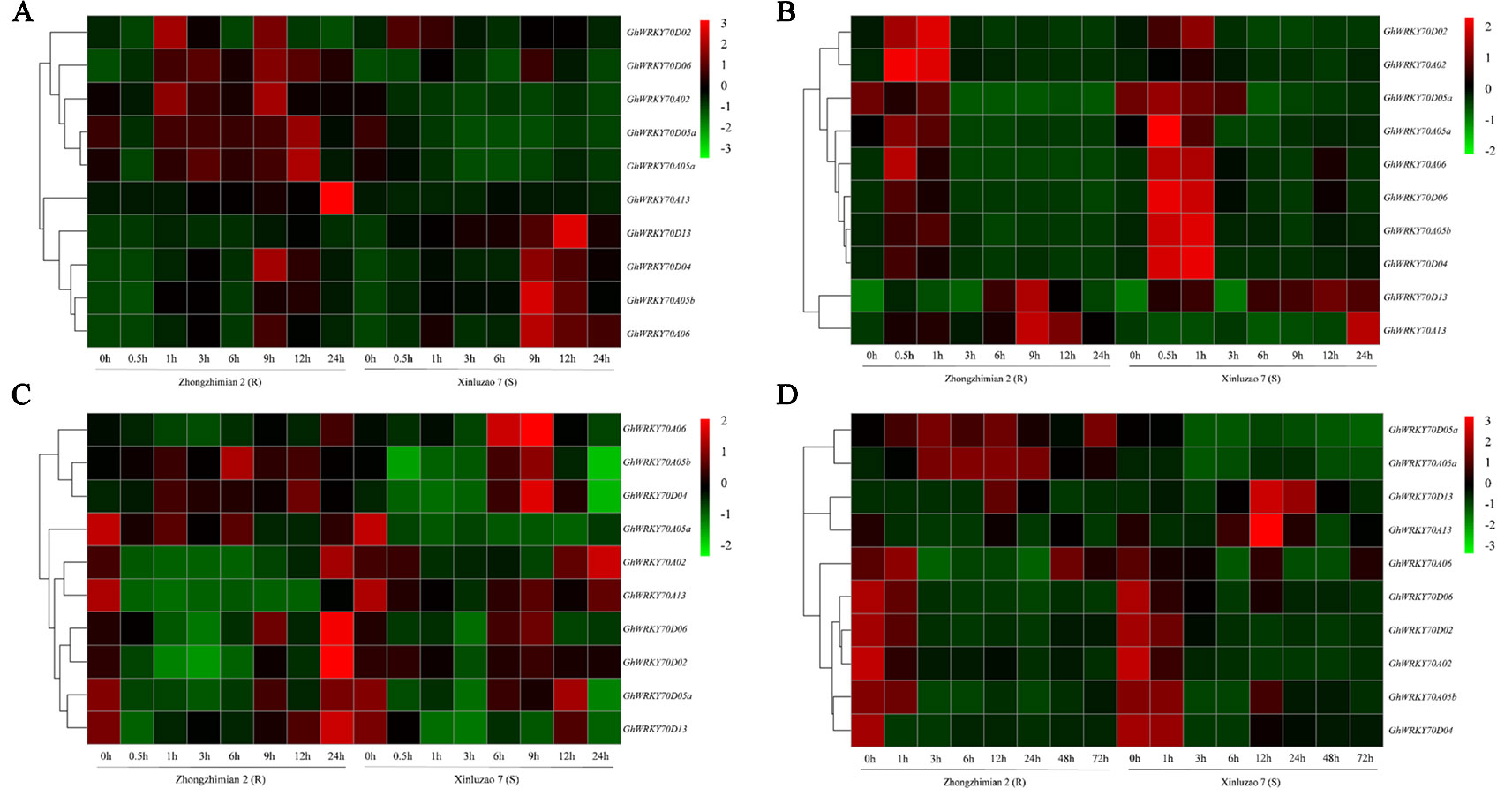
Figure 2 Expression patterns of the 10 GhWRKY70 genes from cotton under different hormone treatments and after Verticillium dahliae inoculation. (A–C) Expression changes of the 10 GhWRKY70 genes in roots of Zhongzhimian 2 (V. dahliae-resistant) and Xinluzao 7 (V. dahliae-susceptible) after treatment with (A) SA, (B) MeJA, and (C) ETH. Samples collected at 0, 0.5, 1, 3, 6, 9, 12, and 24 h after hormone treatment were used in real-time (RT)-qPCR. (D) Expression changes of the 10 GhWRKY70 genes in roots of Zhongzhimian 2 (V. dahliae-resistant) and Xinluzao 7 (V. dahliae-susceptible) after inoculation with V. dahliae strain Vd991. Samples collected at 0, 1, 3, 6, 12, 24, 48, and 72 h after inoculation were used in reverse transcription-quantitative PCR. Two-leaf stage cotton seedlings were used in the treatment of hormones or V. dahliae. Each experiment was performed using three independent biological replicates.
In response to MeJA treatment, the 10 GhWRKY70 genes generally also showed two patterns, one showing significant (P < 0.01) upregulation at 0.5 h and/or 1 h, and then downregulation afterwords in both cultivars, another (including GhWRKY70A13 and GhWRKY70D13) showing upregulation at 6–24 h, particularly GhWRKY70D13 increasing 7.6- to 38.3-fold and 23.2- to 29.1-fold at 6-24 h following MeJA treatment in Zhongzhimian 2 and Xinluzao 7, respectively (Figure 2B).
In response to ETH treatment, six GhWRKY70 genes in Zhongzhimian 2 and Xinluzao 7 were significantly (P < 0.01) suppressed at 0.5-24 h by 0.32- to 0.95-fold (Figure 2C). ETH treatment seemed to induce the expression of GhWRKY70A05a and GhWRKY70A06. In Xinluzao 7, GhWRKY70A05b and GhWRKY70D04 were significantly (P < 0.01) upregulated by 1.7- and 2.4-fold at 9 h. GhWRKY70D13 was downregulated by 1.5- to 2.3-fold from 0.5 to 9 h but restored its expression at 12 h in Zhongzhimian 2 (Figure 2C).
In response to V. dahliae infection, the expression patterns of the 10 GhWRKY70 genes could be divided into three groups (Figure 2D). Group I contains GhWRKY70A05a and GhWRKY70D05a. Both genes exhibited significant upregulation and downregulation in the V. dahliae-resistant and -susceptible cultivar, respectively. Group II includes GhWRKY70D13 and GhWRKY70A13, that were induced by 11.1- and 3.1-fold at 12 hpi in V. dahliae-susceptible cultivar Xinluzao 7 following V. dahliae infection. However, in V. dahliae-resistant cultivar Zhongzhimian 2, upregulation was only observed in GhWRKY70D13 (6.6-fold) but not in GhWRKY70A13 at 12 hpi. The remaining six genes belongs to group III. They were generally downregulated in response to V. dahliae infection (Figure 2D). These results suggest that the 10 GhWRKY70 genes may have different functions in defense against V. dahliae infection in cotton. Due to its consistent upregulation in both V. dahliae resistant and susceptible cultivars upon V. dahliae infection, GhWRKY70D13, together with a representative (GhWRKY70D04) from group III, was chosen for further functional characterization.
Silencing of GhWRKY70D13 Improves Resistance to Verticillium dahliae in Cotton
VIGS was employed to knock down the expression of GhWRKY70D13 and GhWRKY70D04 in two V. dahliae susceptible cultivars, Xinluzao 7 (both genes) and Xincai 7 (only GhWRKY70D13). Observation of the expected yellowing leaf phenotype in TRV: GhCHLI plants (Supplementary Figure S3) and notable reduction of GhWRKY70D04 and GhWRKY70D13 in the corresponding VIGS plants (Supplementary Figures S4A, B) suggested success of the VIGS experiment. Notably, expression of the corresponding At subgenome homologs of GhWRKY70D04 and GhWRKY70D13 was not affected in the corresponding VIGS plants (Supplementary Figures S4C, D), suggesting specificity of VIGS. At 14 days after V. dahliae inoculation, the control plants (TRV:00) and GhWRKY70D04-silenced plants displayed severe leaf yellowing and wilting (Figure 3A and Supplementary Figure S5A). The control plants (TRV:00) also showed obvious vascular browning (Figure 3D). In contrast, the GhWRKY70D13-silenced plants were much healthier and did not show obvious vascular browning (Figures 3B, D and Supplementary Figure S5A). The GhWRKY70D13-silenced plants had significantly (P < 0.05) lower disease indices (10.0 ± 2.8 and 33.1 ± 10.4%, respectively) than the control plants (24.2 ± 8.3% and 55.8 ± 8.2%, respectively) at 14 and 21 dpi in V. dahliae susceptible cultivar Xincai 7 (Figure 3C). In another V. dahliae susceptible cultivar Xinluzao 7, the GhWRKY70D13-silenced plants (46.1 ± 7.8%) had a significantly (P < 0.05) lower disease index than the TRV:00 plants (62.4 ± 6.0%) (Supplementary Figure S5B). Compared to the control plants, the GhWRKY70D13-silenced plants accumulated less fungal biomass (down by 0.28- and 0.19-fold in V. dahliae susceptible cultivar Xinluzao 7 and Xincai 7, respectively) in their stems and had a lower fungal recovery from the stem sections collected from the inoculated plants (Figures 3E, F and Supplementary Figures S5C, D). These results were further confirmed in the V. dahliae resistant cultivar Zhongzhimian 2 (Supplementary Figure S6), suggesting that the mechanism conferring the basal V. dahliae resistance in Zhongzhimian 2 does not involve GhWRKY70D13.
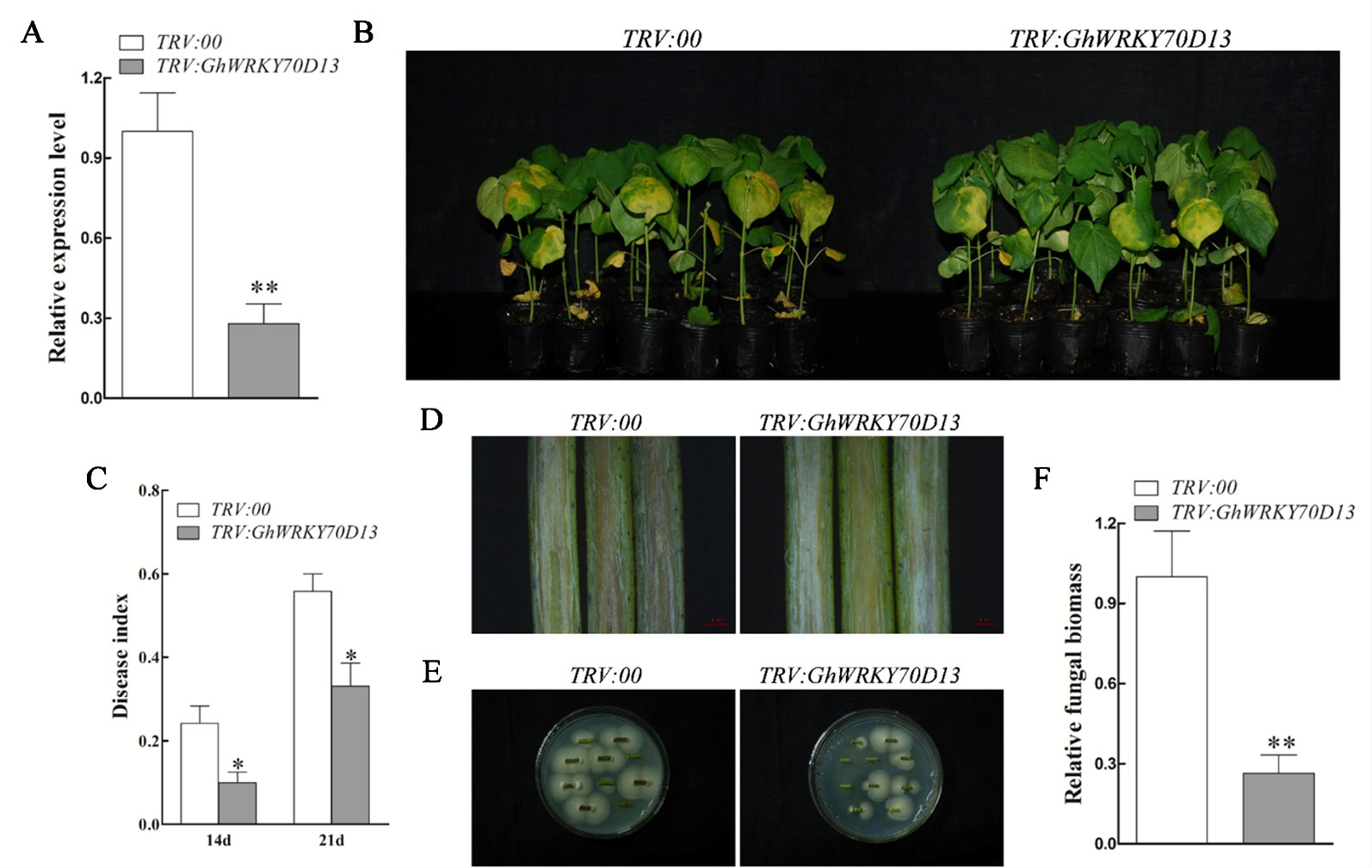
Figure 3 Silencing of GhWRKY70D13 enhanced resistance to Verticillium dahliae in V. dahliae-susceptible cotton cultivar Xincai 7. (A) The expression level of GhWRKY70D13 in the TRV:00 and TRV: GhWRKY70D13 plants. Total RNA was isolated from leaves at 10 days post-agroinfiltration. GhUBQ7 was used as the control. Each experiment was performed using three independent biological replicates. (B) Disease symptoms of the TRV:00 and TRV: GhWRKY70D13 plants after V. dahliae inoculation. Photographs were taken at 14 days after inoculation with 30 plants per treatment. (C) Disease index of the TRV:00 and TRV: GhWRKY70D13 plants at 14 days and 21 days after inoculation with V. dahliae. Each experiment was performed using three independent biological replicates. (D) Disease symptoms in stems of the TRV:00 and TRV: GhWRKY70D13 plants at 14 days after inoculation with V. dahliae. Vascular browning was observed in the TRV:00 plants but not in the TRV: GhWRKY70D13 plants. (E) Comparison of fungal growth in stem sections prepared from the TRV:00 and TRV: GhWRKY70D13 plants at 14 days after V. dahliae inoculation. PDA medium was used in the culture of stems. Photographs were taken after 7 days of culture at 25°C. (F) qPCR analysis of the relative fungal biomass in stems of the TRV:00 and TRV: GhWRKY70D13 plants at 14 days after inoculation with V. dahliae. Each experiment was performed using three independent biological replicates. Differences between groups were compared using the Student's t-test (*P < 0.05; **P < 0.01).
To further consolidate the results, we generated stable transgenic cotton lines (in the background of V. dahliae susceptible cultivar Xincai 7) with downregulated GhWRKY70D13 using RNAi and performed disease assay using two T3 lines (Ci1 and Ci2). As a result, we confirmed that downregulation of GhWRKY70D13 was able to enhance cotton's resistance to V. dahliae (Figure 4).

Figure 4 GhWRKY70D13 RNA interference (RNAi) cotton lines showed enhanced resistance to Verticillium dahliae in V. dahliae-susceptible cotton cultivar Xincai 7. (A) Comparison of the expression level of GhWRKY70D13 in WT, Ci1, and Ci2 plants. Total RNA was isolated from leaves of two-leaf stage seedlings. GhUBQ7 was used as the control. (B) Disease symptoms of the WT, Ci1, and Ci2 plants after V. dahliae inoculation. Photographs were taken at 14 days after inoculation with 30 plants per treatment. (C) Disease index of the WT, Ci1, and Ci2 plants at 14 days and 21 days after inoculation with V. dahliae. (D) Comparison of vascular browning in stems of the WT, Ci1, and Ci2 plants at 14 days after inoculation with V. dahliae. (E) Comparison of fungal growth in stem sections prepared from the WT, Ci1, and Ci2 plants at 14 days after V. dahliae inoculation. Stem sections were plated on potato dextrose agar (PDA) medium. Photographs were taken after 7 days of culture at 25°C. (F) Quantitative PCR (qPCR) analysis of the relative fungal biomass in stems from the WT, Ci1, and Ci2 plants at 14 days after inoculation with V. dahliae. WT, wild-type; Ci1 and Ci2, two independent GhWRKY70D13-RNAi cotton lines. Each experiment was performed using three independent biological replicates. Differences between wild-type and transgenic plants were compared using the Student's t-test (*P < 0.05; **P < 0.01).
Transcriptome Analysis of Wild Type and GhWRKY70D13-Ribonucleic Acid Interference Transgenic Line Infected by Verticillium dahliae
To explore the effect of downregulation of GhWRKY70D13 on cotton's response to V. dahliae infection, we compared the transcriptome profiles of the GhWRKY70D13-RNAi line (Ci, a mixture of Ci1 and Ci2) with those of WT using RNAs isolated from roots of 0, 24, and 72 hpi seedlings. Approximately 40–58 million clean reads per sample were generated (Supplementary Table S5). After mapping the clean reads to the G. hirsutum reference genome (Wang et al., 2019), we calculated the expression level of each expressed gene based on FPKM and identified DEGs between Ci and WT at each time point.
In total, 4,201 non-redundantly upregulated DEGS and 3,584 non-redundantly downregulated DEGs were identified (Supplementary Tables S6 and S7). The DEGs identified at 0 hpi were results of down-regulation of GhWRKY70D13. The DEGs identified at 24 or 72 hpi but not at 0 hpi were potential candidates responding to V. dahliae infection. Based on this rationale, down-regulation of GhWRKY70D13 resulted in 1,055 and 885 up- and down-regulated DEGs, respectively (Figure 5A). For the two time points, 1,240 up-regulated and 1,570 down-regulated DEGs were identified at 24 hpi (Figure 5B), 2,245 up-regulated and 1,312 down-regulated DEGs were identified at 72 hpi (Figure 5C).
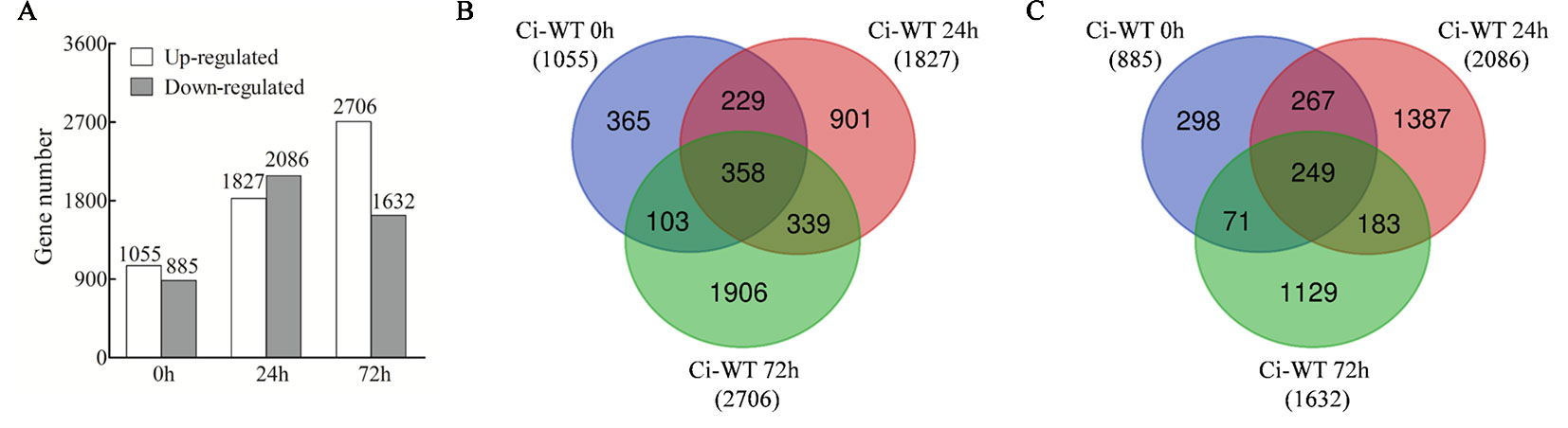
Figure 5 Comparative analysis of differentially expressed genes (DEGs) between the WT and Ci plants following V. dahliae inoculation at different time points. (A) Number of up-regulated or down-regulated DEGs between WT and Ci at 0, 24, and 72 h after V. dahliae inoculation. (B) Venn diagram showing up-regulated DEGs at different time points after V. dahliae inoculation in Ci compared with WT. (C) Venn diagram showing down-regulated DEGs at different time points after V. dahliae inoculation in Ci compared with WT. WT, wild-type; Ci, a mixed sample of GhWRKY70D13-RNAi line 1 (Ci1) and GhWRKY70D13-RNAi line 2 (Ci2).
Analysis of Enriched Pathways
To determine what pathways were affected in the roots of Ci and WT plants by V. dahliae infection; 3,146 upregulated genes (group I) and 2,699 downregulated genes (group II) due to V. dahliae infection were annotated using the KOBAS 2.0 software (Supplementary Tables S8 and S9). Of the top 15 KEGG enriched pathways from each group, 5 were found to be overlapped between the two groups (Supplementary Figure S7, Supplementary Tables S10 and S11). These pathways included limonene and pinene degradation, stilbenoid diarylheptanoid and gingerol biosynthesis, biosynthesis of secondary metabolites, alpha-linolenic acid metabolism, and plant hormone signaling transduction. Pathways unique to group II included carotenoid biosynthesis, carbon fixation in photosynthetic organisms, circadian rhythm–plant, cysteine and methionine metabolism, glycolysis/gluconeogenesis, ABC transporters, glucosinolate biosynthesis, brassinosteroid biosynthesis, flavonoid biosynthesis, and ribosome. Several pathways related to plant's response against V. dahliae were identified in group I, consistent with results previously reported (Zhang et al., 2013). Plant hormone signaling transduction pathways, including ET and JA, were the most enriched pathways in group I. Therefore, ET and JA signaling pathways might play critical roles in GhWRKY70D13-mediated regulation of V. dahliae resistance in cotton.
Silencing of GhWRKY70D13 Positively Regulates Cotton’s Resistance to Verticillium dahliae Through Activation of the Ethylene and Jasmonic Acid Signaling Pathways
To investigate the role of GhWRKY70D13-mediated regulation of V. dahliae resistance through ET and JA signaling, we quantified the expression levels of ET biosynthesis genes (GhACO1-1, GhACO1-2) and response genes (GhEIN3-1, GhEIN3-2, GhERF1-1, and GhERF1-2) in TRV: 00 and TRV: GhWRKY70D13 plants following V. dahliae infection using RT-qPCR. At 24 and 72 hpi, the expression levels of GhACO1, GhEIN3, and GhERF1 were higher in the TRV: GhWRKY70D13 plants than in the TRV: 00 plants (Supplementary Figure S8). Consistently, transcriptome data showed that numerous ET signaling pathway genes, including GhACO1, GhEIN3, and GhERF1, had an increased expression level in Ci than in WT (Figure 6A and Supplementary Table S12), and were further induced in both Ci1 and Ci2 at 24 hpi and/or 72 hpi (Figures 6B–G). Consequently, the production of endogenous ACC in Ci1 and Ci2 was significantly (P < 0.01) increased by 132.1 and 82.2%, respectively, compared to WT at 0 hpi (Figure 6H). Ci1 and Ci2 also showed 53.2 and 47.0% increase of the content of endogenous ACC at 72 hpi, respectively (Figure 6H). These results suggest that silencing of GhWRKY70D13 positively regulates the ET-mediated defense response.
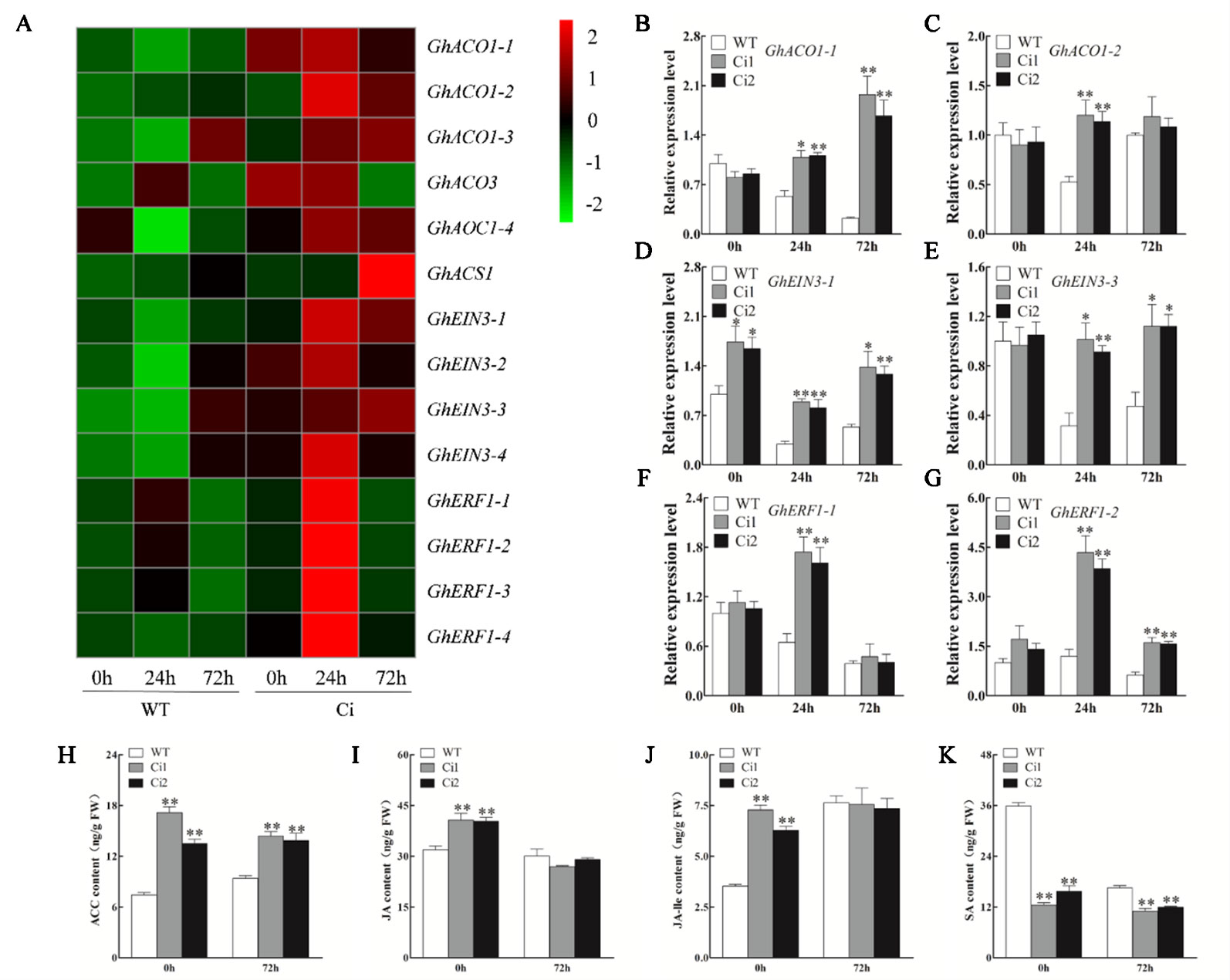
Figure 6 Reverse transcription-quantitative PCR analysis of the expression level of the ethylene (ET) biosynthesis and response genes, and the content of 1-aminocyclopropane-1-carboxylic (ACC), jasmonic acid (JA), JA-isoleucine (JA-Ile), and salicylic acid (SA) in the WT and GhWRKY70D13-RNA interference (RNAi) plants inoculated with Verticillium dahliae. (A) Comparison of the expression patterns of the ET biosynthesis and response genes in the WT and Ci plants infected by V. dahliae based on RNA-seq. (B–G) Relative expression levels of (B) GhACO1-1, (C) GhACO1-2, (D) GhEIN3-1, (E) GhEIN3-3, (F) GhERF1-1, (G) GhERF1-2 at 0, 24, and 72 h after inoculation with V. dahliae in the WT, Ci1, and Ci2 plants. (H–K) The content of (H) ACC, (I) JA, (J) JA-Ile, and (K) SA in the WT, Ci1 and Ci2 plants at 0 h and 72 h after inoculation with V. dahliae. WT, wild-type; Ci1 and Ci2, two independent GhWRKY70D13-RNAi cotton lines; Ci, a mixed sample of GhWRKY70D13-RNAi line 1 (Ci1) and GhWRKY70D13-RNAi line 2 (Ci2). Each experiment was performed using three independent biological replicates. Differences between wild-type and transgenic plants were compared using the Student's t-test (*P < 0.05; **P < 0.01).
Compared to the TRV: 00 control plants, the TRV: GhWRKY70D13 plants showed up-regulation of JA biosynthesis and response genes, such as GhAOS, GhJAZ1, and GhMYC2 in response to V. dahliae inoculation (Supplementary Figure S9). Both RNA-seq and qPCR results showed that JA signaling pathway genes were up-regulated in GhWRKY70D13-RNAi lines compared to WT following V. dahliae infection (Figure 7A, Supplementary Table S13, and Figures 7B–G). At 0 hpi, the GhWRKY70D13-silenced plants Ci1 and Ci2 produced 40.65 ± 3.55 and 40.36 ± 1.89 ng/g JA and 7.28 ± 0.42 and 6.28 ± 0.35 ng/g JA-Ile, respectively, which was significantly (P < 0.01) higher than the WT plants (31.88 ± 2.00 ng/g JA and 3.53 ± 0.16 ng/g JA-Ile) (Figures 6I, J). However, JA and JA-Ile contents were not significantly different between GhWRKY70D13-silenced and WT plants at 72 hpi (Figures 6I, J). Similarly, we analyzed the expression levels of SA signaling pathway genes and quantified the SA content in Ci1, Ci2, and WT before and after V. dahliae infection. The four potential genes (GhPAL1–GhPAL4) involved in SA biosynthesis seemed to have different expression patterns in response to down-regulation of GhWRKY70D13 and V. dahliae infection. Similar situation was observed for the SA response genes (Supplementary Figure S10 and Supplementary Table S14). Nevertheless, the content of SA was significantly (P < 0.01) lower in Ci1 and Ci2 than in WT at both 0 and 72 hpi (Figure 6K).
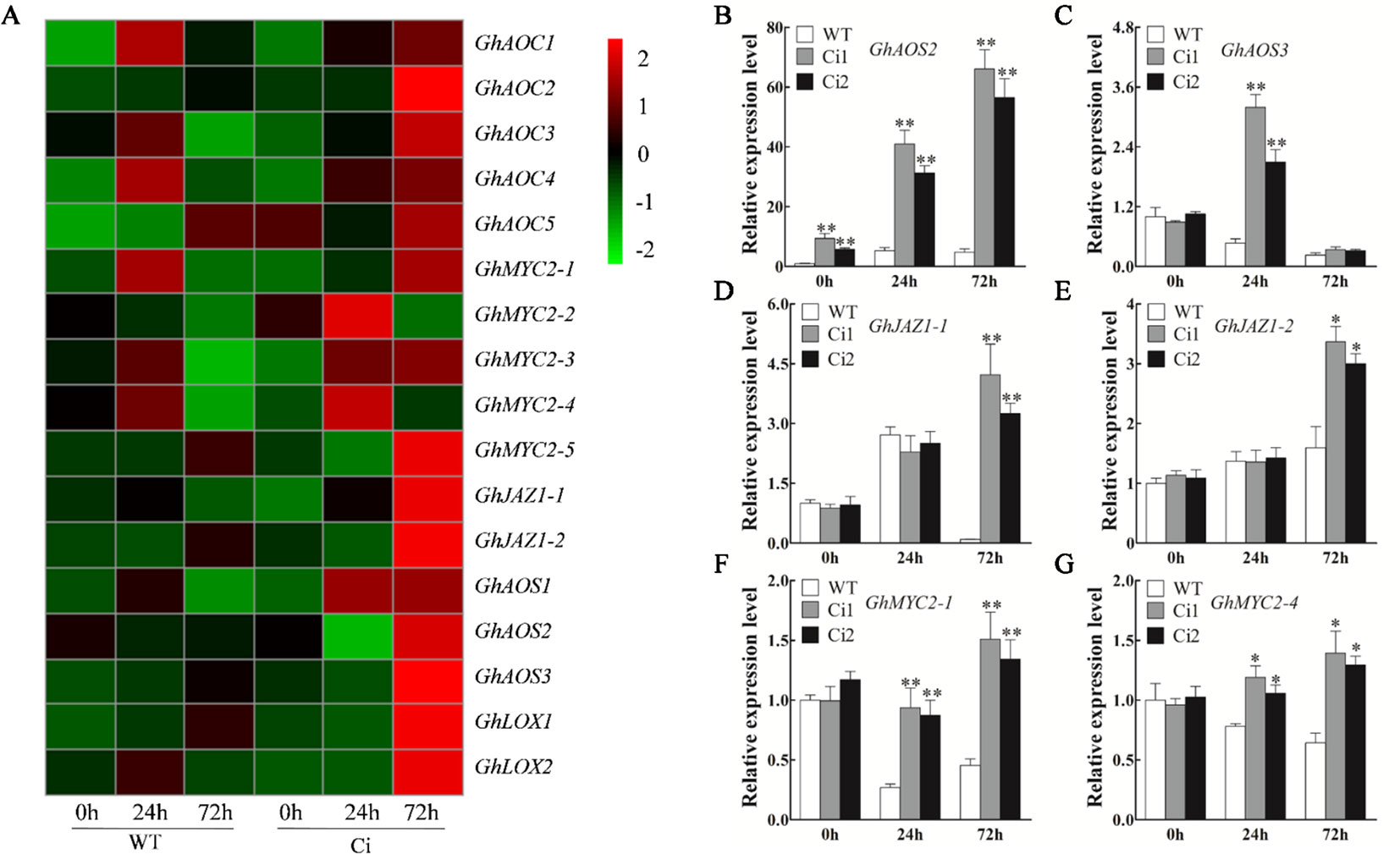
Figure 7 Reverse transcription-quantitative PCR analysis of the expression level of the jasmonic acid (JA) biosynthesis and response genes in the WT and GhWRKY70D13-RNAi plants inoculated with Verticillium dahliae. (A) Comparison of the expression patterns of the JA biosynthesis and response genes in the WT and Ci plants inoculated with V. dahliae based on RNA-seq. (B–G) RT-qPCR analysis of (B) GhAOS2, (C) GhAOS3, (D) GhJAZ1-1, (E) GhJAZ1-2, (F) GhMYC2-1, (G) GhMYC2-4 at 0, 24, and 72 h after inoculation with V. dahliae in the WT, Ci1, and Ci2 plants. WT, wild-type; Ci1 and Ci2, two independent GhWRKY70D13-RNAi cotton lines; Ci, a mixed sample of GhWRKY70D13-RNAi line 1 (Ci1) and GhWRKY70D13-RNAi line 2 (Ci2). Each experiment was performed using three independent biological replicates. Differences between wild-type and transgenic plants were compared using the Student's t-test (*P < 0.05; **P < 0.01).
Discussion
WRKY proteins play an important role in plant growth and development, as well as in regulating responses to pathogen infections. Numerous studies demonstrated that AtWRKY54 and AtWRKY70 co-operatively modulate plant growth, leaf senescence, drought response, and resistance to pathogens (Besseau et al., 2012; Chen et al., 2017; Li et al., 2017). We identified 10 GhWRKY70s in G. hirsutum using AtWRKY70 as a query and found that these GhWRKY70s are also closely related to AtWRKY54 (Supplementary Figure S1B), suggesting similar functions of GhWRKY70s as that of AtWRKY70 and AtWRKY54. Most GhWRKY70 genes had a much higher expression level in vegetative tissues than in reproductive tissues, indicating that those GhWRKY70 genes may act mainly in vegetative tissues. For example, similar to AtWRKY70 (Chen et al., 2017), GhWRKY70A06 and GhWRKY70D06 were highly expressed in leaves. It would be of interest to know whether they are involved in leaf senescence in cotton. GhWRKY70D02 and GhWRKY70D05a were significantly highly expressed in ovule, implying their important roles in ovule development. WRKY genes exhibited extensive cross-regulation with functional redundancy (Pandey and Somssich, 2009; Rushton et al., 2010). Similar expression patterns of the GhWRKY70 homologs, particularly the At and Dt homologs of each GhWRKY70, suggest their possible redundant functions. Analysis of gene expression pattern provides important clues of gene functions (Xue et al., 2008). Cis-regulatory elements found at the gene promoter regions directly influence gene expression and hence control plant development and responses to environmental stress (Wittkopp and Kalay, 2011; Puranik et al., 2012). Consistent with the presence of at least one hormone-responsive element in the promoters of the 10 GhWRKY70 genes (Supplementary Figure S2), most GhWRKY70 genes responded to SA, JA, and/or ETH treatment (Figures 2A–C).
Several studies have reported the involvement of WRKY70 in regulation of disease resistance. For example, Arabidopsis wrky70 mutant showed decreased response to biotroph Erysiphe cichoracearum infection (Li et al., 2004; Li et al., 2006). In wheat, TaWRKY70-silenced plants displayed reduced tolerance to Puccinia striiformis f. sp. tritici infection (Wang et al., 2017). Overexpression of PsnWRKY70 increased resistance to the leaf blight disease in transgenic Populus simonii × Populus nigra plants (Zhao et al., 2017). Our previous study found that GhWRKY70A05a (Gh_A05G2378) negatively regulates the defense response against V. dahliae (Xiong et al., 2019). Up-regulation of GhWRKY70A05a in V. dahliae resistant cotton cultivar and down-regulation of the gene in V. dahliae susceptible cotton cultivar suggest that GhWRKY70A05a is a positive regulator of a negative regulator of V. dahliae-resistance. Herein, two other GhWRKY70 genes, GhWRKY70D04 and GhWRKY70D13, with different expression patterns were silenced by VIGS to investigate their responses to V. dahliae infection. Silencing of GhWRKY70D13 but not GhWRKY70D04 resulted in improved tolerance to V. dahliae (Supplementary Figure S5). The role of GhWRKY70D13 in disease response was further confirmed in the stable transgenic RNAi cotton plants (Figure 4). These findings indicated that GhWRKY70D13 played a negative role in defense against V. dahliae in cotton. Consistent with this finding, we found that the expression levels of GhWRKY70D13 were much higher in the V. dahliae-susceptible cultivar Xinluzao 7 than in the V. dahliae-resistant cultivar Zhongzhimian 2 at 12 and 24 hpi (Figure 2D). V991 is an aggressive defoliating strain, it's necessary to use another nondefoliating strain to have a comprehensive understanding on the role of GhWRKY70D13 related to V. dahliae resistance. In Arabidopsis, AtWRKY70 regulates the susceptibility to necrotrophic Alternaria brassicicol (Li et al., 2004; Li et al., 2006). ET and JA signaling pathways are the major pathways involved in defense against necrotrophic pathogens (Glazebrook, 2005). AtWRKY70 was induced by SA and JA as well as by infection of several pathogens (Kalde et al., 2003; Li et al., 2004). In wheat, the expression of TaWRKY70 is increased upon exogenous application of SA and ET, but down-regulated by MeJA stimulation (Wang et al., 2017).
ACO facilitates ET biosynthesis in plant tissues (Nakatsuka et al., 1998; Ruduś et al., 2013). EIN3 and ERF1 are linked to the ET signaling pathway and have been implicated in regulating plant immune responses (Nie et al., 2012; Meng et al., 2013). Increasing evidence suggested that ET-associated genes regulate cotton resistance to V. dahliae. Cotton ACO1, ACO3, and ERF1 genes were activated upon V. dahliae infection (Xu et al., 2011). Overexpressing GhERF1-like in cotton enhanced resistance to V. dahliae (Guo et al., 2016). Knockdown of GhERF6 decreased cotton's tolerance to V. dahliae infection (Yang et al., 2015). We found that the expression levels of GhACO1, GhEIN3, and GhERF1 were increased in the roots of Ci1, Ci2, and TRV: GhWRKY70D13 plants (Figures 6A–G and Supplementary Figure S8). GhWRKY70D13-RNAi plant had a significantly increased level of ACC content compared to WT at both before and after V. dahliae inoculation (Figure 6H). These results demonstrated that GhWRKY70D13 negatively regulated the ET signaling pathway that positively regulates defense against V. dahliae infection in cotton.
Numerous studies have shown that ET and JA synergistically regulate plant tolerance to necrotrophic pathogens (Wang et al., 2015; Li et al., 2018). Our results showed that some JA biosynthesis genes and response genes were significantly up-regulated in TRV: GhWRKY70D13 plants after V. dahliae inoculation compared with TRV: 00 (Supplementary Figure S9). Consistently, the expression levels of GhAOS, GhJAZ1, and GhMYC2 were also up-regulated in the GhWRKY70D13-RNAi plant (Figure 7). Thus, silencing of GhWRKY70D13 might positively regulate the JA signaling pathway to increase cotton's resistance to V. dahliae. Our findings consist with those reported previously (Gao et al., 2013; Zhang et al., 2013). Suppression of GbWRKY1 and GhCYP94C1 increased the expression of JA-associated genes and improved resistance to V. dahliae (Li et al., 2014a; Sun et al., 2014). In contrast, GbSSI2-silenced cotton plants displayed decreased resistance to V. dahliae due to suppressed JA accumulation and signaling (Gao et al., 2013). Recently, Yang et al. reported that overexpression of JAZ inhibited JA signaling (Yang et al., 2012). Similar results were found in cotton, in which GhJAZ2 acted as a repressor of JA signaling and negatively regulated the resistance of cotton to V. dahliae (He et al., 2018). We found that the contents of JA and JA-Ile were significantly higher in the GhWRKY70D13-silenced plant (Ci1 and Ci2) than in WT but such difference vanished at 72 hpi (Figures 6I, J), which might be related to the expression patterns of the two GhJAZ1 genes that had a similar level in Ci1, Ci2, and WT uninfected by V. dahliae but were significantly up-regulated in Ci1 and Ci2 at 72 hpi (Figures 7D, E).
Studies in Arabidopsis showed that WRKY70 is a negative regulator of SA biosynthesis, but a positive regulator of SA-responsiveness (Wang et al., 2006). Knock-down the expression level of GhWRKY70D13 seemed to have different effect on SA biosynthesis and response genes in cotton (Supplementary Figure S10). Compared with WT, Ci1 and Ci2 had a significantly decreased SA content at both before and after V. dahliae infection, although the SA content in WT was dramatically reduced due to V. dahliae infection (Figure 6K). It is of interest to know whether this is a result of the antagonistic relationship between the JA and SA signaling pathways (Bari and Jones, 2009; Thaler et al., 2012). On the other hand, we showed previously that suppression of GhWRKY70A05a expression improved resistance to V. dahliae by down-regulating SA signaling pathways and up-regulating JA signaling pathways (Xiong et al., 2019). This is different to the function of GhWRKY70D13 which seems to negatively regulate ET and JA signaling pathways.
In summary, we showed that down-regulation of GhWRKY70D13 in cotton significantly enhanced accumulation of ACC, JA, and JA-Ile, and impaired SA biosynthesis, consequently enhanced cotton's resistance to V. dahliae. GhWRKY70D13 negatively regulates cotton's response to V. dahliae infection by downregulating the ET and JA signaling pathways (Figure 8). The role of cross-talk between the JA and ET signaling pathways in cotton's resistance to V. dahliae requires however further investigations.
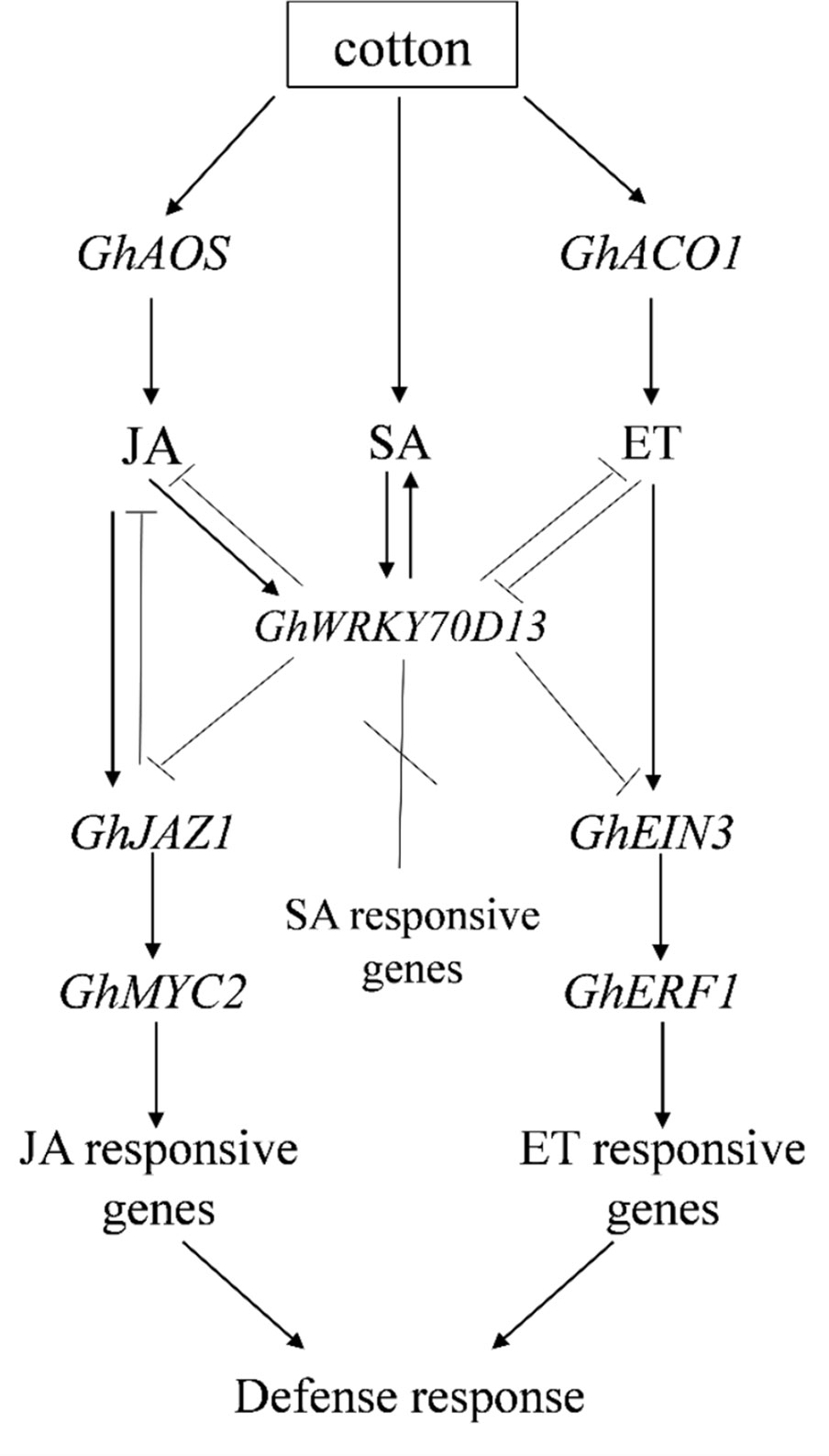
Figure 8 A working model describing the regulatory role of GhWRKY70D13 on salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) signaling in cotton upon infection of Verticillium dahliae.
Data Availability Statement
All sequencing data generated in this study can be found in the NCBI using accession number PRJNA578842.
Author Contributions
JS and FX conceived and designed the experiments. X-PX conducted most of the experiments, analyzed the data and wrote the manuscript. S-CS and X-YZ provided technical assistance to X-PX. X-YZ, Y-JL, and FL contributed reagent and materials. Q-HZ and JS revised the manuscript. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2016YFD0100200, 2016YFD0101900) and the Genetically Modified Organisms Breeding Major Project of China (Grant No. 2016ZX08005-005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00069/full#supplementary-material
References
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Besseau, S., Li, J., Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679. doi: 10.1093/jxb/err450
Cai, Y., He, X., Mo, J., Sun, Q., Yang, J., Liu, J. (2009). Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review. Afr. J. Biotechnol. 8, 7363–7372. doi: 10.5897/AJB2009.000-9571
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, J., Liu, C., Gui, Y., Si, K., Zhang, D., Wang, J., et al. (2018). Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 217, 756–770. doi: 10.1111/nph.14861
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chung, H. S., Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 21, 131–145. doi: 10.1105/tpc.108.064097
Datta, R., Kumar, D., Sultana, A., Hazra, S., Bhattacharyya, D., Chattopadhyay, S. (2015). Glutathione regulates 1-Aminocyclopropane-1-Carboxylate synthase transcription via WRKY33 and 1-Aminocyclopropane-1-Carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 169, 2963–2981. doi: 10.1104/pp.15.01543
Ding, M., Chen, J., Jiang, Y., Lin, L., Cao, Y., Wang, M., et al. (2015). Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Genet. Genomics 290, 151–171. doi: 10.1007/s00438-014-0904-7
Dou, L., Zhang, X., Pang, C., Song, M., Wei, H., Fan, S., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 289, 1103–1121. doi: 10.1007/s00438-014-0872-y
Ellendorff, U., Fradin, E. F., De Jonge, R., Thomma, B. (2009). RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. doi: 10.1093/jxb/ern306
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fradin, E. F., Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Gao, W., Long, L., Zhu, L., Xu, L., Gao, W., Sun, L., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Gao, W., Long, L., Xu, L., Lindsey, K., Zhang, X., Zhu, L. (2016). Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J. Integr. Plant Biol. 58, 503–513. doi: 10.1111/jipb.12432
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gong, Q., Yang, Z., Wang, X., Butt, H. I., Chen, E., He, S., et al. (2017). Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 17, 59. doi: 10.1186/s12870-017-1007-5
Gu, Z., Huang, C., Li, F., Zhou, X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649. doi: 10.1111/pbi.12169
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6
He, X., Zhu, L., Wassan, G. M., Wang, Y., Miao, Y., Shaban, M., et al. (2018). GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 19, 896–908. doi: 10.1111/mpp.12575
Higashi, K., Ishiga, Y., Inagaki, Y., Toyoda, K., Shiraishi, T., Ichinose, Y. (2008). Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol. Genet. Genomics 279, 303–312. doi: 10.1007/s00438-007-0315-0
Hu, Y., Dong, Q., Yu, D. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Jin, S., Zhang, X., Nie, Y., Guo, X., Liang, S., Zhu, H. (2006). Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 50, 519–524. doi: 10.1007/s10535-006-0082-5
Kalde, M., Barth, M., Somssich, I. E., Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant-Microbe Interact. 16, 295–305. doi: 10.1094/MPMI.2003.16.4.295
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, 480–484. doi: 10.1093/nar/gkm882
Kim, K. C., Lai, Z., Fan, B., Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 20, 2357–2371. doi: 10.1105/tpc.107.055566
Li, J., Brader, G., Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 16, 319–331. doi: 10.1105/tpc.016980
Li, J., Brader, G., Kariola, T., Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, C., He, X., Luo, X., Xu, L., Liu, L., Min, L., et al. (2014a). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014b). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genetics 46, 567–572. doi: 10.1038/ng.2987
Li, J., Zhong, R., Palva, E. T. (2017). WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS One 12, e0183731. doi: 10.1371/journal.pone.0183731
Li, J., Zhang, K., Meng, Y., Hu, J., Ding, M., Bian, J., et al. (2018). Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA59 transcription factor. Plant J. 95, 444–457. doi: 10.1111/tpj.13960
Liu, Y., Schiff, M., Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Liu, J., Wang, Y., Zhao, G., Zhao, J., Du, H., He, ,. X., et al. (2017). A novel Gossypium barbadense ERF transcription factor, GbERFb, regulation host response and resistance to Verticillium dahliae in tobacco. Physiol. Mol. Biol. Plants. 23, 125–134. doi: 10.1007/s12298-016-0402-y
Meng, X., Xu, J., He, Y., Yang, K. Y., Mordorski, B., Liu, Y., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 25, 1126–1142. doi: 10.1105/tpc.112.109074
Mol, L., Scholte, K., Vos, J. (1995). Effect of crop rotation and removal of crop debris on the soil population of two isolates of Verticillium dahliae. Plant Pathol. 44, 1070–1074. doi: 10.1111/j.1365-3059.1995.tb02666.x
Murray, S. L., Ingle, R. A., Petersen, L. N., Denby, K. J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant-Microbe Interact. 20, 1431–1438. doi: 10.1094/MPMI-20-11-1431
Nakatsuka, A., Murachi, S., Okunishi, H., Shiomi, S., Nakano, R., Kubo, Y., et al. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118, 1295–1305. doi: 10.1104/pp.118.4.1295
Nie, H., Zhao, C., Wu, G., Wu, Y., Chen, Y., Tang, D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158, 1847–1859. doi: 10.1104/pp.111.192310
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248. doi: 10.3389/fmicb.2013.00248
Pandey, S. P., Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Puranik, S., Sahu, P. P., Srivastava, P. S., Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Ruduś, I., Sasiak, M., Kępczyński, J. (2013). Regulation of ethylene biosynthesis at the level of 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene. Acta Physiol. Plant 35, 295–307. doi: 10.1007/s11738-012-1096-6
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Shaban, M., Miao, Y., Ullah, A., Khan, A. Q., Menghwar, H., Khan, A. H., et al. (2018). Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 125, 193–204. doi: 10.1016/j.plaphy.2018.02.011
Sun, L., Zhu, L., Xu, L., Yuan, D., Min, L., Zhang, X. (2014). Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5, 5372. doi: 10.1038/ncomms6372
Thaler, J. S., Humphrey, P. T., Whiteman, N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. doi: 10.1016/j.tplants.2012.02.010
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Wang, D., Amornsiripanitch, N., Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, e123. doi: 10.1371/journal.ppat.0020123
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, C., Ding, Y., Yao, J., Zhang, Y., Sun, Y., Colee, J., et al. (2015). Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 83, 1019–1033. doi: 10.1111/tpj.12946
Wang, Y., Liang, C., Wu, S., Zhang, X., Tang, J., Jian, G., et al. (2016). Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of Gastrodia antifungal proteins. Mol. Plant 9, 1436–1439. doi: 10.1016/j.molp.2016.06.013
Wang, J., Tao, F., An, F., Zou, Y., Tian, W., Chen, X., et al. (2017). Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 18, 649–661. doi: 10.1111/mpp.12425
Wang, M., Tu, L., Yuan, D., Zhu, ,. D., Shen, C., Li, J., et al. (2019). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi: 10.1038/s41588-018-0282-x
Wittkopp, P. J., Kalay, G. (2011). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Xiong, X., Sun, S., Lim, Y., Zhang, X., Sun, J., Xue, F. (2019). The cotton WRKY transcription factor GhWRKY70 negatively regulates the defense response against Verticillium dahliae. Crop J. 7, 393–402. doi: 10.1016/j.cj.2018.10.005
Xu, L., Zhu, L., Tu, L., Guo, X., Long, L., Sun, L., et al. (2011). Differential gene expression in cotton defence response to Verticillium dahliae by SSH. J. Phytopathol. 159, 606–615. doi: 10.1111/j.1439-0434.2011.01813.x
Xue, T., Wang, D., Zhang, S., Ehlting, J., Ni, F., Jakab, S., et al. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9, 550. doi: 10.1186/1471-2164-9-550
Yang, D., Yao, J., Mei, C., Tong, X., Zeng, L., Li, Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Nati. Acad. Sci. U. S. A. 109, E1192–E1200. doi: 10.1073/pnas.1201616109
Yang, C., Liang, S., Wang, H., Han, L., Wang, F., Cheng, H., et al. (2015). Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant 8, 399–411. doi: 10.1016/j.molp.2014.11.023
Zhang, Y., Wang, X., Ding, Z., Ma, Q., Zhang, G., Zhang, S., et al. (2013). Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics 14, 637. doi: 10.1186/1471-2164-14-637
Zhang, F., Yao, J., Ke, J., Zhang, L., Lam, V. Q., Xin, X., et al. (2015a). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. doi: 10.1038/nature14661
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015b). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, D., Wang, J., Wang, D., Kong, Z., Zhou, L., Zhang, G., et al. (2019). Population genomics demystifies the defoliation phenotype in the plant pathogen Verticillium dahliae. New Phytol. 222, 1012–1029. doi: 10.1111/nph.15672
Zhao, H., Jiang, J., Li, K., Liu, G. (2017). Populus simonii x Populus nigra WRKY70 is involved in salt stress and leaf blight disease responses. Tree Physiol. 37, 827–844. doi: 10.1093/treephys/tpx020
Keywords: Gossypium hirsutum, Verticillium dahliae, GhWRKY70D13, ribonucleic acid interference, jasmonic acid, ethylene
Citation: Xiong X-P, Sun S-C, Zhang X-Y, Li Y-J, Liu F, Zhu Q-H, Xue F and Sun J (2020) GhWRKY70D13 Regulates Resistance to Verticillium dahliae in Cotton Through the Ethylene and Jasmonic Acid Signaling Pathways. Front. Plant Sci. 11:69. doi: 10.3389/fpls.2020.00069
Received: 18 October 2019; Accepted: 17 January 2020;
Published: 25 February 2020.
Edited by:
Daguang Cai, University of Kiel, GermanyReviewed by:
Shan Lu, Nanjing University, ChinaPhilipp Franken, Friedrich Schiller University Jena, Germany
Steven J. Klosterman, United States Department of Agriculture (USDA), United States
Copyright © 2020 Xiong, Sun, Zhang, Li, Liu, Zhu, Xue and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Xue, eHVlZmVpQHNoenUuZWR1LmNu; Jie Sun, c3VuamllQHNoenUuZWR1LmNu
 Xian-Peng Xiong1
Xian-Peng Xiong1 Qian-Hao Zhu
Qian-Hao Zhu Jie Sun
Jie Sun