- 1Departamento de Bioquímica, Biología Celular y Molecular de Plantas, Estación Experimental del Zaidín, Consejo Superior de Investigaciones Científicas, Granada, Spain
- 2Departamento de Bioquímica y Biología Molecular, Facultad de Ciencias Experimentales, Universidad de Jaén, Jaén, Spain
Nitric oxide (NO), peroxynitrite (ONOO−), and S-nitrosoglutathione (GSNO) are components of a family of molecules which have important signaling functions in higher plants under physiological and stress conditions because directly or indirectly can mediate post-translational modifications including binding to metal centers, S-nitrosylation of thiol groups and nitration of tyrosine (Lamattina et al., 2003; Besson-Bard et al., 2008; Baudouin, 2011).
During the last 10 years or so, different sets of data indicate surprising new findings in relation to the enzymatic composition and functions of plant peroxisomes. One of these discoveries was the presence of an L-arginine-dependent NO synthase activity which initially shows that these organelles house a complete NO metabolism that participates in the physiology of whole plants under normal and adverse environmental conditions (Barroso et al., 1999; Corpas et al., 2004).
Peroxisomal Nitric Oxide Participates in a Whole Array of Physiological Processes
Larger than a mitochondrion and smaller that a chloroplast, plant peroxisomes observed under an electron microscope are characterized as having a very simple structure made up of a single membrane that includes a granular matrix and sometimes a crystal structure (Tenberge and Eising, 1995; Usuda et al., 1999). However, from a metabolic perspective, peroxisomes possess an important and complex enzymatic composition characterized by plasticity which is adaptable to the plant organ, development stage, and/or environmental conditions (Fulda et al., 2004; Ma et al., 2006; León, 2008; Babujee et al., 2010; Hu et al., 2012). However, with the unexpected identification of peroxisomal proteins, new functions for these organelles have been proposed (Nowak et al., 2004; Reumann et al., 2007; Sørhagen et al., 2013). Given peroxisomal L-arginine-nitric oxide synthase (NOS) activity has the same cofactor requirements as animal NOS responsible for the endogenous generation of NO (Figures 1A–C) (Corpas et al., 2009 and references therein), it has been suggested that these organelles are a source of NO which can regulate peroxisomal metabolism and also be a source of long-distance signal molecules that participate in cross-talk between the different subcellular compartments (del Río, 2011). In addition, data from different areas of plant research suggest that peroxisomal NO participates in an array of physiological functions and in different organs such as leaf senescence, pollen tube growth (Prado et al., 2004), auxin-induced root organogenesis (Schlicht et al., 2013) as well as being involved in the mechanism of response to abiotic stress conditions such as salinity and cadmium stress (Corpas et al., 2009; Corpas and Barroso, 2014).
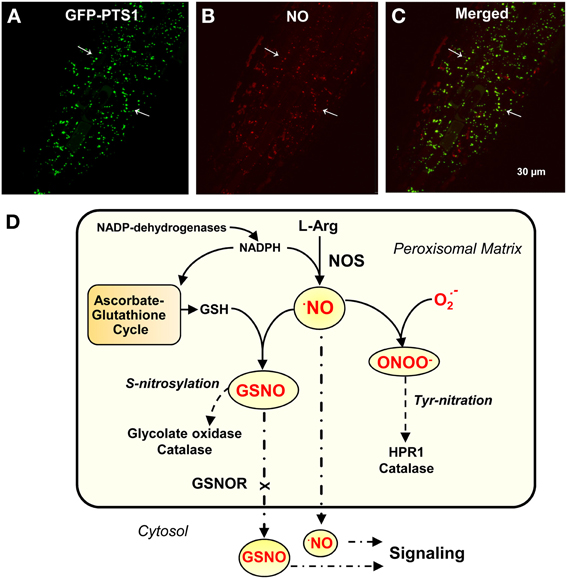
Figure 1. Peroxisomal localization by confocal laser scanning microscope (CLSM) of molecules involved in NO metabolism. (A) CLSM in vivo detection of peroxisomes (green color) in primary roots of Arabidopsis seedlings expressing GFP-PTS1 (green fluorescent protein fused with peroxisomal targeting signal 1). (B) CLSM in vivo detection of NO (red color) with DAR-AM AM in the field showed in panel (A). (C) merged images of panels (A,B). Arrows indicate representative punctate spots corresponding to NO and peroxisome localization. (D) Model proposed for the metabolism and signaling function of nitric oxide (NO) and S-nitrosoglutathione (GSNO) in plant peroxisomes. Reproduced, with permission, from Plant Physiol. 151:2083–2094 (Copyright American Society of Plant Biologists) for panels (A–C).
Peroxisomal Nitric Oxide Metabolism
Recently, cellular and biochemical approaches have shown the presence of new components involved in the peroxisomal metabolism of NO such as ONOO−, S-nitrosoglutathione (GSNO), GSNO reductase and protein nitration (Heijnen et al., 2006; Barroso et al., 2013; Corpas and Barroso, 2014) which contribute to a more complete picture of the peroxisomal metabolism. It has also been demonstrated that Arabidopsis plants under cadmium stress, peroxisomal peroxynitrite, NO, and superoxide anion (O·−2) are overproduced, suggesting that peroxisomes participate in the nitro-oxidative stress response to this heavy mental (Corpas and Barroso, 2014). Given that NO and related molecules can mediate post-translational modifications, mainly nitration and S-nitrosylation, several proteomic studies of different plant species have also shown that some peroxisomal proteins are potential targets of post-translation modifications mediated by NO-derived molecules. Thus, a proteomic analysis of isolated pea leaf peroxisomes has shown that the NADH-dependent hydroxypyruvate reductase (HPR1) involved in the photorespiration pathway is negatively modulated by tyrosine nitration, specifically Tyr198, which affects the binding of the coenzyme (Corpas et al., 2013). This suggests a clear connection between NO metabolism and photorespiration. In vitro assays using NO donors also revealed that several peroxisomal enzymes, including catalase, malate dehydrogenase, glycolate oxidase, and hydroxypyruvate reductase, are also potential candidates of S-nitrosylation (Ortega-Galisteo et al., 2012).
Concluding Remarks and Future Research
Accumulated data obtained during the last 10 years or so confirm that plant peroxisomes possess a whole metabolic mechanism related to NO metabolism which complements our knowledge of the reactive oxygen species (ROS) metabolism. Therefore, our understanding of the biological chemistry of NO in peroxisomes presented in Figure 1D will now provide a framework to comprehend how these NO-derived molecules participate in the plant development process (Prado et al., 2004; Schlicht et al., 2013) and to understand the mechanism of response to environmental stress (Corpas and Barroso, 2014). Further research is required to elucidate the nature of finely-tuned endogenous regulation of peroxisomal components.
Acknowledgments
Work in our laboratories is supported by ERDF-cofinanced grants from the Ministry of Science and Innovation (BIO2009-12003-C02-01, BIO2009-12003-C02-02, and BIO2012-33904) and Junta de Andalucía (groups BIO192 and BIO 286).
References
Babujee, L., Wurtz, V., Ma, C., Lueder, F., Soni, P., van Dorsselaer, A., et al. (2010). The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin K1) biosynthesis in plant peroxisomes. J. Exp. Bot. 61, 1441–1453. doi: 10.1093/jxb/erq014
Barroso, J. B., Corpas, F. J., Carreras, A., Sandalio, L. M., Valderrama, R., Palma, J. M., et al. (1999). Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 274, 36729–36733. doi: 10.1074/jbc.274.51.36729
Barroso, J. B., Valderrama, R., and Corpas, F. J. (2013). Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol. Plant. 35, 2635–2640. doi: 10.1007/s11738-013-1291-0
Baudouin, E. (2011). The language of nitric oxide signalling. Plant Biol. (Stuttg.) 13, 233–242. doi: 10.1111/j.1438-8677.2010.00403.x
Besson-Bard, A., Pugin, A., and Wendehenne, D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. doi: 10.1146/annurev.arplant.59.032607.092830
Corpas, F. J., Barroso, J. B., Carreras, A., Quirós, M., León, A. M., Romero-Puertas, M. C., et al. (2004). Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 136, 2722–2733. doi: 10.1104/pp.104.042812
Corpas, F. J., and Barroso, J. B. (2014). Peroxynitrite (ONOO−) is endogenously produced in Arabidopsis peroxisomes and is overproduced under cadmium stress. Ann. Bot. 113, 87–96. doi: 10.1093/aob/mct260
Corpas, F. J., Hayashi, M., Mano, S., Nishimura, M., and Barroso, J. B. (2009). Peroxisomes are required for n vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol. 151, 2083–2094. doi: 10.1104/pp.109.146100
Corpas, F. J., Leterrier, M., Begara-Morales, J. C., Valderrama, R., Chaki, M., López-Jaramillo, J., et al. (2013). Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim. Biophys. Acta 1830, 4981–4989. doi: 10.1016/j.bbagen.2013.07.002
del Río, L. A. (2011). Peroxisomes as a cellular source of reactive nitrogen species signal molecules. Arch. Biochem. Biophys. 506, 1–11. doi: 10.1016/j.abb.2010.10.022
Fulda, M., Schnurr, J., Abbadi, A., Heinz, E., and Browse, J. (2004). Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16, 394–405. doi: 10.1105/tpc.019646
Heijnen, H. F., van Donselaar, E., Slot, J. W., Fries, D. M., Blachard-Fillion, B., Hodara, R., et al. (2006). Subcellular localization of tyrosine-nitrated proteins is dictated by reactive oxygen species generating enzymes and by proximity to nitric oxide synthase. Free Radic. Biol. Med. 40, 1903–1913. doi: 10.1016/j.freeradbiomed.2005.09.006
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24, 2279–2303. doi: 10.1105/tpc.112.096586
Lamattina, L., García-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
León, J. (2008). Peroxisome proliferation in Arabidopsis: the challenging identification of ligand perception and downstream signaling is closer. Plant Signal. Behav. 3, 671–673. doi: 10.4161/psb.3.9.5780
Ma, C., Haslbeck, M., Babujee, L., Jahn, O., and Reumann, S. (2006). Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 141, 47–60. doi: 10.1104/pp.105.073841
Nowak, K., Luniak, N., Witt, C., Wüstefeld, Y., Wachter, A., Mendel, R. R., et al. (2004). Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol. 45, 1889–1894. doi: 10.1093/pcp/pch212
Ortega-Galisteo, A. P., Rodríguez-Serrano, M., Pazmiño, D. M., Gupta, D. K., Sandalio, L. M., and Romero-Puertas, M. C. (2012). S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J. Exp. Bot. 63, 2089–2103. doi: 10.1093/jxb/err414
Prado, A. M., Porterfield, D. M., and Feijo, J. A. (2004). Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131, 2707–2714. doi: 10.1242/dev.01153
Reumann, S., Babujee, L., Ma, C., Wienkoop, S., Siemsen, T., Antonicelli, G. E., et al. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193. doi: 10.1105/tpc.107.050989
Schlicht, M., Ludwig-Müller, J., Burbach, C., Volkmann, D., and Baluska, F. (2013). Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytol. 200, 473–482. doi: 10.1111/nph.12377
Sørhagen, K., Laxa, M., Peterhänsel, C., and Reumann, S. (2013). The emerging role of photorespiration and non-photorespiratory peroxisomal metabolism in pathogen defence. Plant Biol. (Stuttg.) 15, 723–736. doi: 10.1111/j.1438-8677.2012.00723.x
Tenberge, K. B., and Eising, R. (1995). Immunogold labelling indicates high catalase concentrations in amorphous and crystalline inclusions of sunflower (Helianthus annuus L.) peroxisomes. Histochem. J. 27, 184–195. doi: 10.1007/BF02389885
Keywords: nitric oxide, nitrosative stress, nitrosoglutathione, oxidative stress, peroxynitrite, peroxisomes, reactive nitrogen species
Citation: Corpas FJ and Barroso JB (2014) Functional implications of peroxisomal nitric oxide (NO) in plants. Front. Plant Sci. 5:97. doi: 10.3389/fpls.2014.00097
Received: 13 January 2014; Accepted: 27 February 2014;
Published online: 17 March 2014.
Edited by:
Dan Szymanski, Purdue University, USAReviewed by:
Jianping Hu, Michigan State University, USACopyright © 2014 Corpas and Barroso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:amF2aWVyLmNvcnBhc0BlZXouY3NpYy5lcw==
 Francisco J. Corpas
Francisco J. Corpas Juan B. Barroso
Juan B. Barroso