- 1Microbial Engineering, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
- 2Plant Stress Biology, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
- 3Stress Physiology and Molecular Biology Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
Response to stress represents a highly complex mechanism in plants involving a plethora of genes and gene families. It has been established that plants use some common set of genes and gene families for both biotic and abiotic stress responses leading to cross-talk phenomena. One such family, Meprin And TRAF Homology (MATH) domain containing protein (MDCP), has been known to be involved in biotic stress response. In this study, we present genome-wide identification of various members of MDCP family from both Arabidopsis and rice. A large number of members identified in Arabidopsis and rice indicate toward an expansion and diversification of MDCP family in both the species. Chromosomal localization of MDCP genes in Arabidopsis and rice reveals their presence in a few specific clusters on various chromosomes such as, chromosome III in Arabidopsis and chromosome X in rice. For the functional analysis of MDCP genes, we used information from publicly available data for plant growth and development as well as biotic stresses and found differential expression of various members of the family. Further, we narrowed down 11 potential candidate genes in rice which showed high expression in various tissues and development stages as well as biotic stress conditions. The expression analysis of these 11 genes in rice using qRT-PCR under drought and salinity stress identified OsM4 and OsMB11 to be highly expressed in both the stress conditions. Taken together, our data indicates that OsM4 and OsMB11 can be used as potential candidates for generating stress resilient crops.
Introduction
Abiotic stress is considered as one of the major factors affecting growth, biomass, and productivity in plants (Singh A. et al., 2015; Joshi et al., 2016a). Among several abiotic stresses, salinity and drought are the key factors for the downfall of yield in the agricultural sector due to reduced productivity in both irrigated and non-irrigated agricultural lands (Gupta et al., 2015). In plants, a high degree of similarity has been reported in salinity and drought stress responses with respect to their physiological, molecular and genetic effects (Joshi et al., 2014). Elevated levels of salt in the soil limits the water uptake because of low water potential, thereby initiating drought stress (Ahmed et al., 2015). It has been well established that osmotic stress in plants triggers turgor loss, membrane disorganization, protein denaturation and production of reactive oxygen species (Joshi et al., 2014). This situation further causes inhibition of photosynthesis, damage of cellular organelles and metabolic dysfunction resulting in growth retardation, reduced fertility, and premature senescence, thus causing severe yield losses (Joshi et al., 2016b). Plants use common pathways and components in response to these stresses (Pastori and Foyer, 2002). Hence, plants tolerant to salinity may also be tolerant to the drought stress or vice-versa (Farooq and Azam, 2001).
Over the years, a number of attempts have been made to improve stress tolerance in crop plants (Singh B. et al., 2015). One of the strategies adopted worldwide for this purpose is the identification of genes that can assist tolerant plants to survive under harsh conditions and using such genes to engineer similar trait in stress sensitive genotypes (Kumar et al., 2012). Though, remarkable progress has been made in developing transgenic plants that can tolerate various stresses (Joshi et al., 2016a), it has been well accepted that these tolerance mechanisms are synchronized by a complex signaling network and orchestrated stress-regulated gene expression (Bohnert et al., 2006; Sreenivasulu et al., 2007; Ramegowda et al., 2014). Thus, identification and characterization of overlapping signal transduction pathways between both salt and drought stresses is essential for getting a holistic view of the response.
One of the major food crops consumed by more than half of the world's population is rice (Oryza sativa L.; Bohra et al., 2015). Sensitivity toward abiotic stresses in rice varies with the growth stage, as young seedlings and reproductive stages are highly sensitive to salt and drought stress (Basu and Roychoudhury, 2014). The sensitivity toward these stresses in rice also varies considerably across genotypes. Comparative analysis of various genotypes in rice has been exploited as a successful strategy to discover novel genes and proteins which contribute toward abiotic stress tolerance (Gehan et al., 2015). Earlier, we had employed comparative transcriptomics approach between two contrasting rice genotypes to identify salinity tolerance related genes (Kumari et al., 2009). By employing subtractive hybridization using two contrasting rice genotypes, Pokkali (salt tolerant) and IR64 (salt sensitive), a total of 1194 ESTs (584 from IR64 and 610 from Pokkali) were identified. Analysis of these ESTs led to the identification of various novel genes playing a possible role in salt stress specific response. In fact, an EST identified from this study led to the characterization of CDCP genes in Arabidopsis and rice (Kushwaha et al., 2009; Singh et al., 2012). Another protein identified from the analysis of these ESTs is the MATH (Meprin And TRAF Homology) domain containing protein (MDCP) which has been analyzed in the present study.
Biotic stress is also reported to contribute to 50–80% yield loss in the absence of control measures (Foyer et al., 2016). Previously, available data on biotic stress along with the changing climatic conditions project toward an increase in reproductive potential and geographical expansion of different pathogen strains with higher chances of plants encountering biotic stresses in future (Kissoudis et al., 2014). MDCPs were earlier known for their role in the plant/microbe interaction. They are the early responsive membrane bound receptor kinases reported in Arabidopsis, which gets transiently up-regulated during the fungal interaction, and decrease thereafter when the interaction is established (Peškan-Berghöfer et al., 2004; Shahollari et al., 2005). The TRAF-C domain of TRAF protein and C-terminal region of meprin A and B constitutes the MATH domain of MDCPs (Sunnerhagen et al., 2002). Meprins are tissue-specific and membrane-associated oligomeric zinc endopeptidases that belong to the Astacin family of Metzincin superfamily. These are the largest extracellular proteases in the animal system which cleaves various peptides including growth factors, cytokines and extracellular matrix proteins (Broder and Becker-Pauly, 2013). Tumor necrosis factor-Receptor Associated Factors (TRAFs) belong to the adaptor protein family, and are characterized by a carboxy-terminal homology domain of about 180 amino acids, forming 7–8 antiparallel β-sheets defined as TRAF domain (TD) (Bradley and Pober, 2001; Zapata et al., 2007; Zhou et al., 2015). They are the key factors of the Toll-Like Receptor (TLR) family and Tumor Necrosis Factor (TNF) family, which regulate downstream signaling pathways and finally activate various transcription factors related to cell survival and stress responses (Huang B. et al., 2016). It also triggers the downstream components of signaling pathways, controls the sub-cellular localization of the receptor-ligand complexes, and modifies the response by controlling the degradation of proteins (Zapata et al., 2007). Recently, two redundant TRAF proteins were identified which play a role in the turnover of the nucleotide-binding domain and leucine-rich repeat-containing (NLR) immune receptors SNC1 and RPS2 (Huang S. et al., 2016).
Various other sets of protein domains such as peptidases, RING and zinc finger, filamin and RluA domains, BTB (Broad-complex, Tramtrack, and Bric a brac) domain, tripartite motif (TRIM) and astacin domains are known to be present in conjunction with the MATH domain (Zapata et al., 2007). The number of MDCPs in Arabidopsis and Brassica rapa have been found to be similar to C. elegans but their role in plants is still unknown (Oelmüller et al., 2005; Zhao et al., 2013). These MATH domain containing proteins have been hypothesized for having a role in the regulation of protein processing (Zapata et al., 2007). The MATH-BTB proteins have been in fact found to play a role in ABA signaling (Lechner et al., 2011). Further, MDCPs are reported to localize in various subcellular compartments such as endoplasmic reticulum, Golgi apparatus, cytosol, nucleus, and organellar membranes, especially peroxisomes.
In the present study, we have identified and characterized MDCP-encoding gene family members in Arabidopsis and rice. A detailed comparison has been made in terms of phylogeny and their genome organization. Expression profile for all the MDCP family members in various tissues, developmental stages as well as biotic and abiotic stress conditions has been studied using the publicly available database. Further, eleven biotic stress-responsive MDCP encoding genes have been analyzed for their expression under salt and drought stress by qRT-PCR. Based on the analysis presented here, we have highlighted the possible role of MDCP-encoding gene family members in both biotic and abiotic stress response in plants.
Materials and Methods
Identification of MDC Proteins
The MDC protein sequences were fetched and classified using Arabidopsis (TAIR release 10.0; Berardini et al., 2015) and Oryza sativa (TIGR release 7.0; Kawahara et al., 2013) whole genome sequences. Profiles unique to the MATH domain (accession no. PF00917) were obtained from Pfam database (Finn et al., 2014) and were used to screen the whole genome protein sequences of both Arabidopsis and rice, using the HMMER software (version 3.0) deploying default parameters (Eddy, 1998). The protein sequences obtained from the profile search were manually checked for the presence of additional domains along with the MATH domain. We have assigned names to these protein sequences following the domains observed in the individual protein sequences, where “At” denote Arabidopsis thaliana and “Os” denote Oryza sativa. This is followed by a number of times the MATH “M” or BTB “B” domains are present in the sequence.
Analysis of MDC Proteins
Further, the protein sequences identified were analyzed for their characteristics such as pI, molecular weight using TAIR (release 10.0; Berardini et al., 2015) and TIGR (release 7.0; Kawahara et al., 2013) for Arabidopsis and rice respectively. The subcellular localization of the MDC proteins of Arabidopsis was predicted based on SUBA database (http://suba3.plantenergy.uwa.edu.au/) while that of rice was predicted using subCELlular LOcalization predictor (CELLO v. 2.5: http://cello.life.nctu.edu.tw/) (Yu et al., 2006) and re-confirmed using WoLF PSORT, an advanced protein subcellular localization prediction tool (http://www.genscript.com/wolf-psort.html) (Horton et al., 2007; Nielsen, 2016).
Chromosomal Localization of MDCP Encoding Genes and Phylogenetic Analysis
In order to identify the localization of MDCP encoding genes on various chromosomes we used publicly available information resources, that is, TAIR for Arabidopsis and TIGR for rice. The chromosomal positions were plotted using Dia diagram editor (Dia 0.97.2). The rooted ML tree was build using PhyML 3.0 (Guindon et al., 2010) and the final tree was plotted using FigTree 1.4.2 (Rambaut, 2012). To build phylogenies, bootstrap analysis was conducted using 1000 replicates. The sequence analysis was performed using Seaview (version 4) multiple sequence alignment editor (Gouy et al., 2010).
In silico Gene Expression Analysis
Expression pattern for each gene model of MATH domain encoding genes were analyzed in different tissues (such as, callus, seedling, coleoptiles, root, inflorescence, panicle, spikelet, stamen, anther, pollen, stigma, ovary, caryopsis, embryo, endosperm, culm, node, internode, stele, pith, parenchyma, peduncle, leaf, blade, sheath, flag leaf, collar, rhizome, primary root, and root tip; Table S1), at various developmental stages (such as, germination, seedling, tillering, stem elongation, booting, heading, flowering, milk, and dough; Table S2), and under different abiotic stresses (such as, cold, drought, heat, and salinity; Table S3), and biotic stresses (Table S4) were obtained from Affymetrix GeneChip database using Response Viewer (https://www.genevestigator.com) (Hruz et al., 2008). For Arabidopsis, 22 K ATH1 genome array was chosen and pre-existing microarray data of Arabidopsis was considered for further analysis. In the case of rice, microarray datasets of OS_51 K: Rice Genome 51 K array were analyzed.
Further, the same dataset was used for analysis under various biotic stresses i.e., various nematodes and insect pests in rice. In Arabidopsis, various mutants were analyzed along with their response to various bacterial elicitors. The expression of MDC proteins in Arabidopsis was also analyzed in response to various bacterial and fungal infections.
Plant Material and Stress Treatments
Seeds of Oryza sativa ssp. indica, cv. IR64 were surface sterilized with bavistin solution (0.1%), rinsed with distilled water and germinated hydroponically in half strength Yoshida medium as described previously (Mustafiz et al., 2011). Seedlings were grown under 16 h/8 h photoperiod at 28 ± 2°C with 70% humidity in the growth chamber (Panasonic, Japan). Ten day old seedlings were subjected to various stress treatments for 6 h (Tripathi et al., 2012). For salinity stress, seedlings were supplemented with half strength Yoshida medium containing 200 mM NaCl and for drought stress, seedlings were air-dried between folds of tissue paper as described (Singh V. K. et al., 2015). Untreated seedlings grown in half strength Yoshida medium were taken as control. The shoot tissues were harvested and immediately frozen in liquid nitrogen and stored at −80°C for RNA isolation.
Quantitative Real-Time PCR Analysis
Total RNA was isolated from shoot tissues using TRIzol reagent (Thermo Fisher Scientific, USA) according to the manufacturer's protocol. RNA quality and integrity was determined using NanoDrop spectrophotometer and agarose gel electrophoresis. Total RNA was treated with 2 μg of DNase (Thermo Fisher Scientific, USA) and reverse transcribed with RevertAid® RNase H minus cDNA synthesis kit (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Using Primer Express Software v3.0 (Applied Biosystems, USA), the primers for qRT-PCR analysis were designed from the 3′-UTR region of the selected genes (Table S5). The specificity of amplification was further confirmed by Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The qRT-PCR assay was performed in 20 μl final reaction mixture according to the instructions for Power SYBR® Green PCR Master Mix (Applied Biosystems, USA) using 7500TM Real-Time PCR system and software (Applied Biosystems, USA). The reaction was performed using three biological and three technical replicates as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Elongation factor 1-α (eEf-1α) was used as reference gene for normalization (Tripathi et al., 2015). Dissociation curve analysis and gel electrophoresis was carried out to check the specificity of amplification. Relative change in fold expression was calculated using comparative CT value (Livak and Schmittgen, 2001) and two-tailed Student's t-test was used to analyze statistical significance at p < 0.05.
Results
Identification and Characterization of MDC Proteins
To identify the MDC proteins in Arabidopsis and rice, the profile of MATH domain (accession no. PF00917) was obtained from the Pfam database using HMM-based method (see Materials and Methods). The method used for the identification of MDC proteins remains same as used earlier for the identification and classification of various other gene families such as TCS (Pareek et al., 2006; Singh A. et al., 2015), CDCP (Kushwaha et al., 2009), glyoxalase I and II (Mustafiz et al., 2011), cyclophilins (Kumari et al., 2015), NCX (Singh A. K. et al., 2015), histone chaperones (Tripathi et al., 2015), and glyoxalase III (Ghosh et al., 2016).
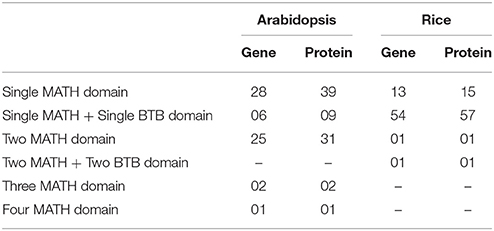
Genome-wide analysis search of MDC proteins revealed the presence of 62 MDC genes coding for 82 proteins in Arabidopsis. Similarly, in rice, 69 genes were found to be coding for 74 MDC proteins. Classification of these proteins was based on the presence of MATH domain either as a single domain or multiple domains or along with BTB domain (Accession No. PF00651; Figure S1). The POZ (POxvirus and Zinc finger) domain, renamed as BTB (Broad-Complex, Tramtrack, and Bric à brac) domain is evolutionarily conserved and plays a role in the regulation of gene expression through protein-protein interactions (Ahmad et al., 1998). The proteins having MATH domain have been named as “M” (for the single MATH domain), “2M” (for two MATH domains), “3M” (for three MATH domains), “4M” (for four MATH domains), “MB” (for single MATH and single BTB domain), and “2M2B” (for two MATH along with two BTB domains) followed by a number which represents the sequence order in which they were found in the search. Each name is preceded by the name of the species in which they were identified such as, “At” representing Arabidopsis and “Os” representing Oryza sativa. Further, the postscript alphabets were assigned like “a,” “b” etc for representing the alternative splice proteins in both the species.
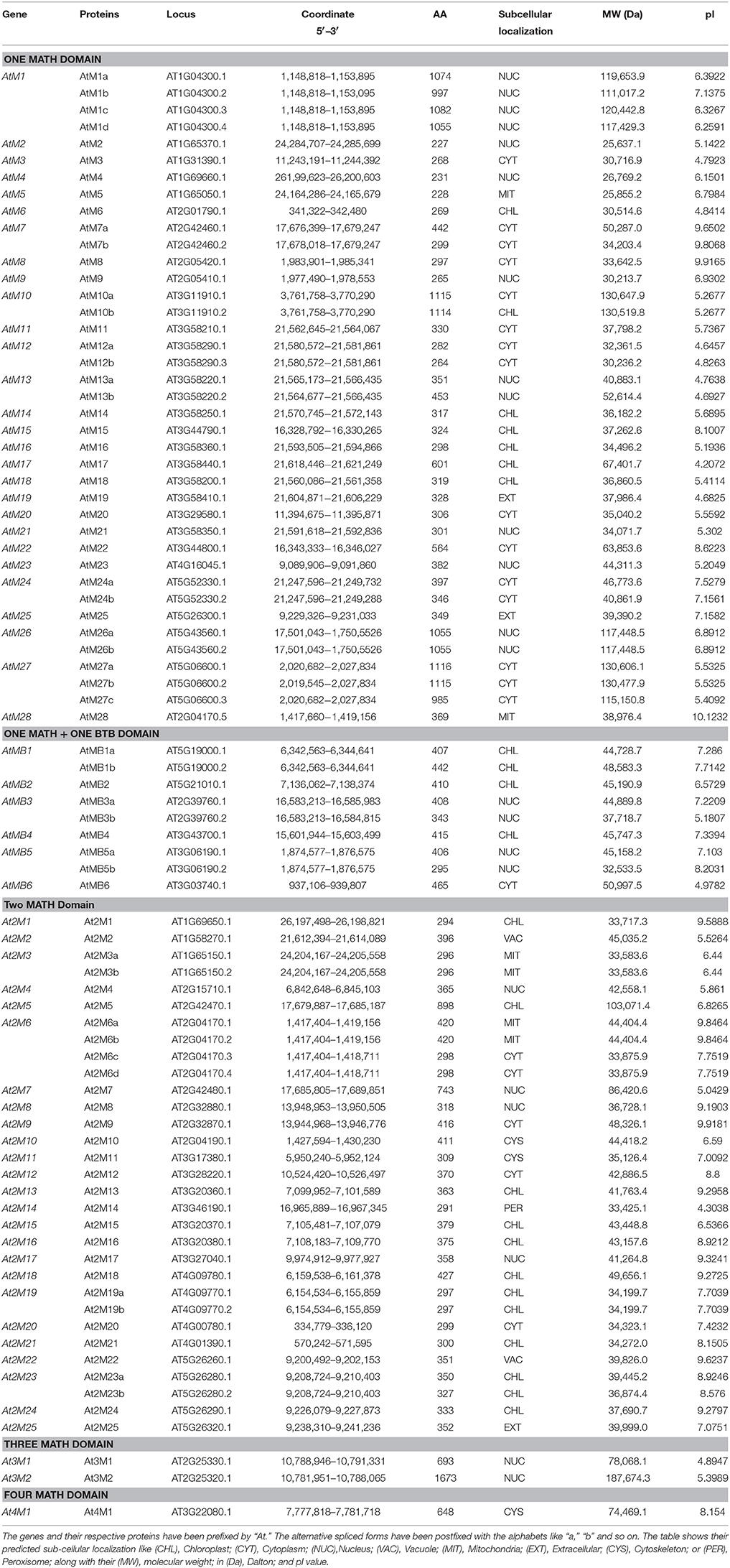
In Arabidopsis, 39 single domain proteins were encoded by 28 genes, while in rice, 13 such genes code for 15 proteins (Table 1). In the group of proteins having two MATH domains, 25 genes in Arabidopsis were found to code 31 proteins, while in rice, only a single such instance was observed. Only 2 proteins, encoded by 2 genes in Arabidopsis were found to possess three MATH domains and only 1 protein possessed 4 MATH domains. However, in rice, no protein was identified having 3 or 4 MATH domains.
The alternative splicing mechanism has been considered as the major source of diversity and complexity in various species (Brett et al., 2002; Ghosh et al., 2016). In Arabidopsis, 15 instances of alternative splicing have been observed generating 35 MDC proteins (Table 2) while in rice, 9 MDC proteins have been observed as a result of four alternative splicing events (Table 3).
Phylogenetic Analysis of MDC Proteins
To analyze the phylogenetic relationship between the MDC proteins in both Arabidopsis and rice, a rooted tree was prepared by aligning full-length protein sequence (Figure 1). This analysis gave a comprehensive picture of MDC protein classification. The single MATH domain containing proteins (AtM and OsM) were observed to be clustered in two groups (Clade 2 and 4) and the other major clade (Clade 1) was of proteins having one MATH domain along with one BTB domain (AtMB and OsMB). The third major clade was of the proteins having two MATH domains (At2M and Os2M). The three MATH domain containing proteins (At3M) formed a separate cluster with the group of proteins having two MATH domain containing proteins. Further, proteins with two MATH domains and two BTB domains of rice were found in the same clade as those of protein sequences with one MATH and one BTB domain. In addition, single-domain MDC proteins from Arabidopsis (AtM2, AtM4, AtM5, and At4M1) were found in the same clade as two MATH domain containing proteins from Arabidopsis. Similarly, proteins containing two MATH domains in Arabidopsis (At2M17, At2M7, and At2M5) were found to be present in the clade belonging to MDC proteins containing one domain.
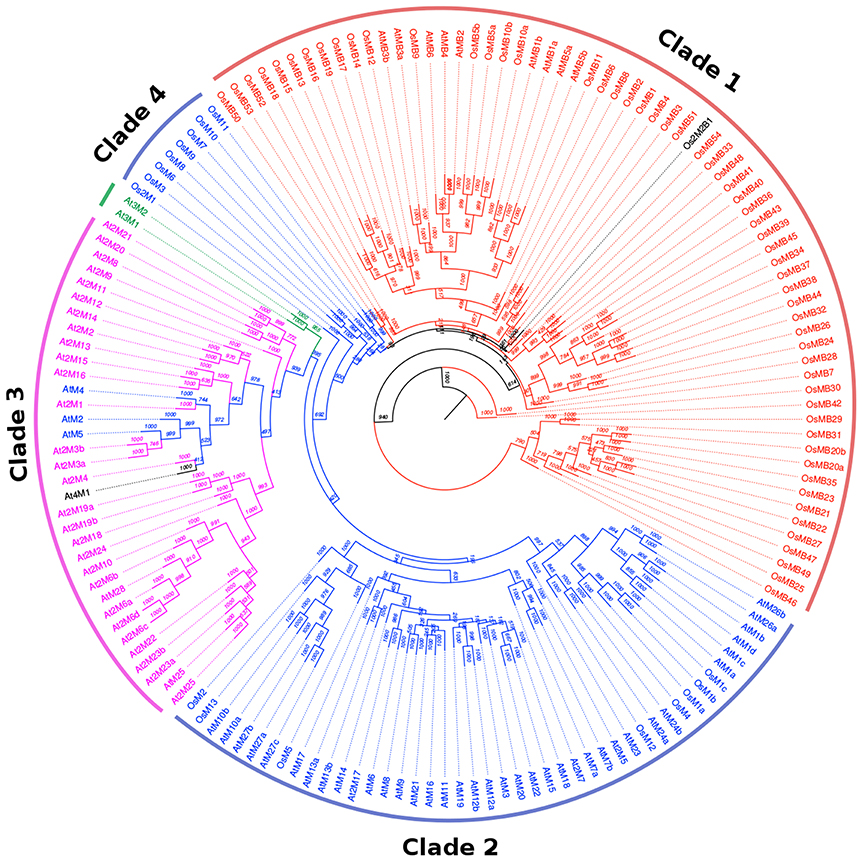
Figure 1. Rooted phylogenetic relationship tree of the MDC protein members of both Arabidopsis and rice. The rooted tree shows the presence of MDC proteins having single MATH domain (blue), MATH domain along with BTB domain (Red), two MATH domains (pink), three MATH domains (green), four MATH domains (black), and two MATH and two BTB domains (black). The bootstrap values are marked on the rooted tree.
Sequence Analysis of MDC Proteins
Amino acid sequence analysis of the MDC proteins revealed that single MATH domain containing protein OsM7 shared a very low level of identity with other single MATH domain proteins in rice (ranging from 19 to 28%) while it was found to be closer to the proteins with single MATH and single BTB domain (28–32% identity). Interestingly, all single MATH domain containing proteins from Arabidopsis showed significant identity (30–77%) with other members of their group, except for a few single MATH domain proteins from rice such as, OsM3, OsM6, OsM8, OsM9, OsM10, and OsM11, which showed only 15–22% identity (Figure S2). This was also evident from the phylogenetic tree where these protein sequences were found to lie in the separate clade from other single MATH domain containing protein sequences. The amino acid sequences of MDCPs containing single MATH domain along with single BTB domain (OsMB) were found to have 27–77% identity within their group. The two MATH domain containing proteins were found to be sharing 26–41% identity within their group. The single MATH domain containing protein AtM5 was found to possess 25–60% identity with the two MATH domain containing members. Similarly, AtM28, AtM2, and AtM4 shared 28–62% identity with the members having two MATH domains (Figure S3). The protein with two MATH domains along with two BTB domains in rice (Os2M2B1) was observed to be having 34–56% homology with the protein sequences having one MATH and one BTB domain. The two MATH domain containing proteins were observed to have 26–46% identity within their group (Figure S4). The four MATH domain containing protein in Arabidopsis, At4M1 was found to be sharing 28–51% identity with the members having two MATH domains. Analysis of alignment of all the MATH domain protein sequences suggests large-scale insertion in various protein sequences leading to low sequence identity between the sequences.
Chromosomal Localization of MDC Protein Encoding Genes
The analysis of the localization of MDC protein encoding genes on the chromosomes of Arabidopsis and rice reveals an interesting pattern. In Arabidopsis, the majority (28) of single MATH domain containing protein encoding genes were found to be localized uniformly on all the chromosomes (Figure 2A). Interestingly, maximum i.e., thirteen number of MDC proteins encoding genes were found to be present on chromosome III in Arabidopsis. Out of these, nine were forming a cluster. Further, five single domain MDC protein encoding genes namely, AtM1, AtM2, AtM3, AtM4, and AtM5 were located on chromosome I. The chromosome II and V were observed to contain four single domain MDC protein encoding genes. In Arabidopsis, 4 genes encoding MDC proteins were duplicated in the genome. The single domain MDC protein coding gene, AtM1, present on chromosome I was found to be duplicated on chromosome V as single domain MDC protein encoding gene AtM26. Another gene, AtM10 from chromosome III was found to be duplicated as AtM27 on chromosome V. Among the group of MDC proteins having BTB domain, AtMB4 present on chromosome III was found to be duplicated as AtMB2 on chromosome V. Another gene of the same group AtMB1 from chromosome V was found to be duplicated with AtMB5 on chromosome III (Figure 2A).
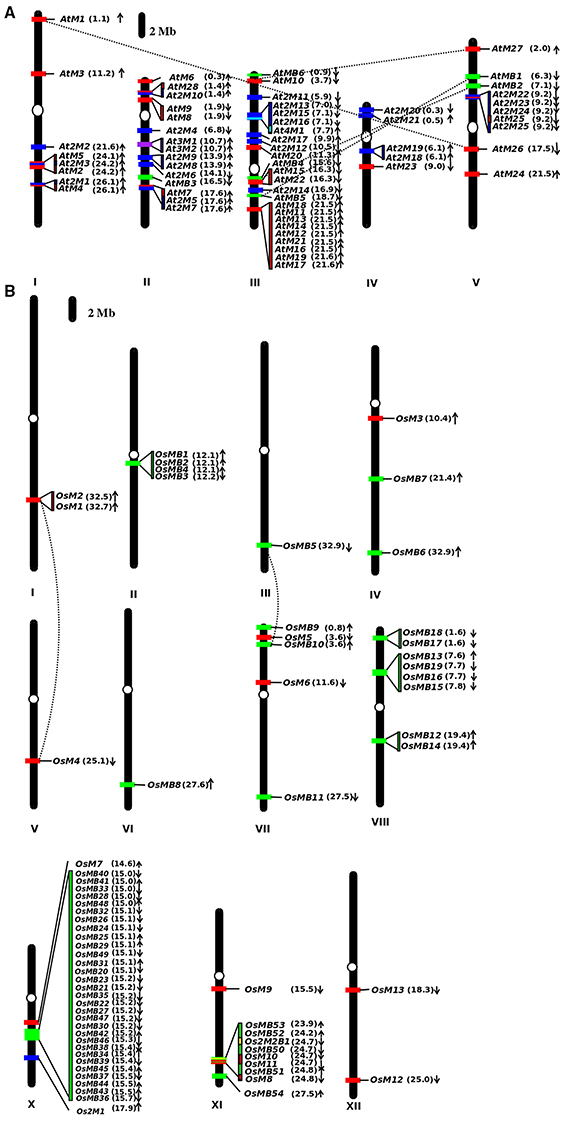
Figure 2. Graphical scaled representation of the location of MDC protein encoding genes on the chromosomes of (A) Arabidopsis and (B) rice. The centromeres are marked by ovals on the chromosomes. The position of MDC protein encoding genes has been marked in Mb in the parenthesis along with the direction of the ORF. The figure shows the MDC proteins having single MATH domain (red), MATH domain along with BTB domain (green), two MATH domains (blue), three MATH domains (purple), four MATH domains (cyan), and two MATH and two BTB domains (yellow).
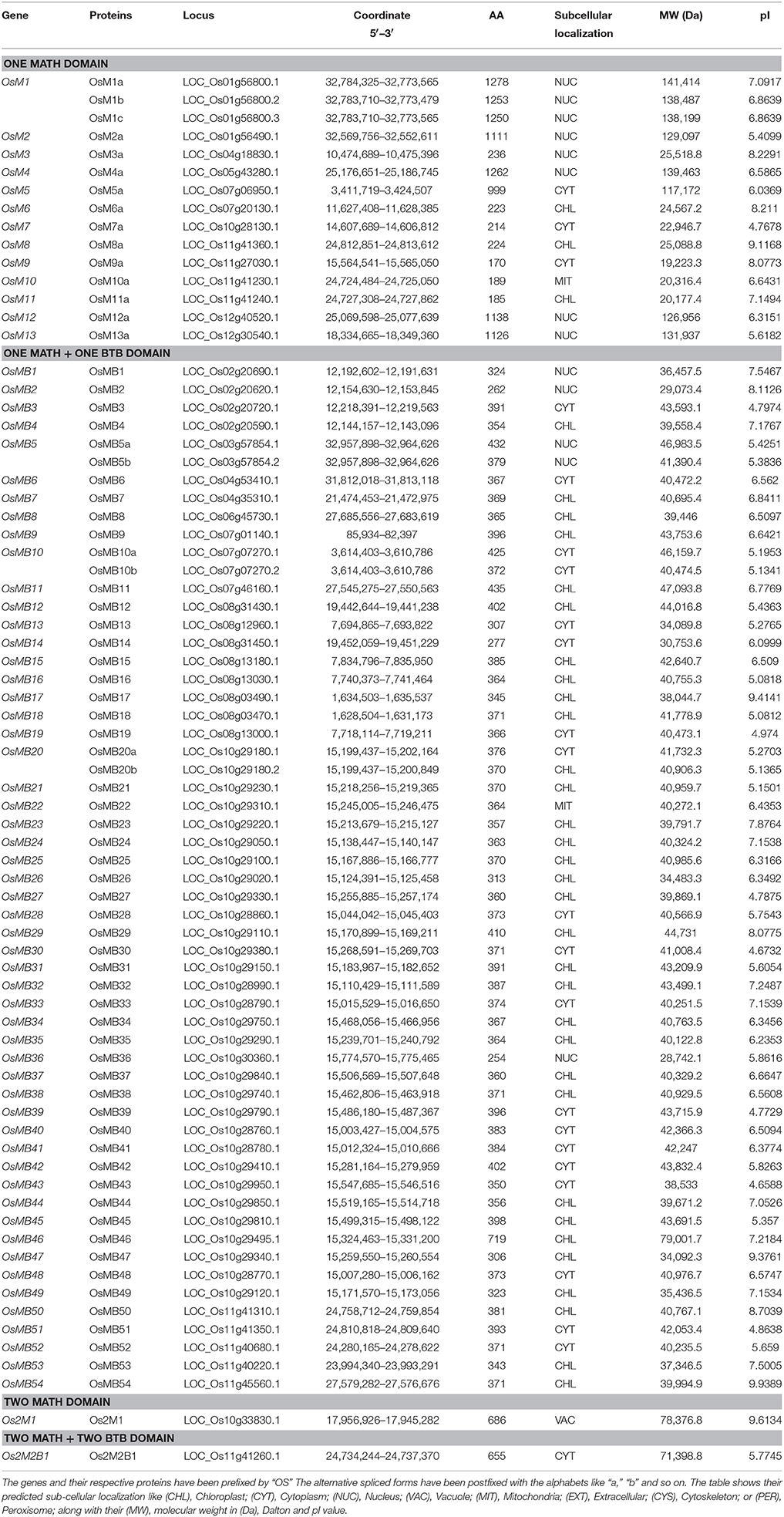
In rice, genes coding for one domain MDC were found to be scattered on various chromosomes (Figure 2B). It was found that out of 13 single domain containing genes, chromosome XI contained 4 genes (i.e., OsM8, OsM9, OsM10, and OsM11), chromosome I, VII, and XII contained 2 genes each, while chromosome IV, V, and IX contained only one single MATH domain coding gene. However, chromosome II, III, VI, and VIII did not contain any single MATH domain protein encoding gene. Surprisingly, in rice chromosome IX does not contain any MDC protein coding gene. Analysis of segmental duplications in MDC proteins revealed only two events of gene duplication in rice. The first instance where single-domain MDC protein encoding gene OsM1 present on chromosome I was found to be duplicated as OsM4 present on chromosome V. The other duplicated gene was MDC protein with a BTB domain, OsMB5 present on chromosome III was found duplicated on chromosome VII as OsMB10.
In rice, genes coding for proteins having single MATH domain along with single BTB domain were found in large numbers (54) unlike Arabidopsis (6). In Arabidopsis, genes that belong to this group were found on chromosome II, III, and V. Interestingly, the maximum number (3) of genes are present on the chromosome III namely, AtMB4, AtMB5, and AtMB6 followed by two genes present on chromosome V namely, AtMB1 and AtMB2 (Figure 2A). In rice, striking observation was noticed with respect to these genes where most of the genes of the group (30) are present on the chromosome X in a cluster within the same region. Further, eight genes of the group were found on chromosome VIII followed by five on chromosome XI, three on chromosome VII and two on chromosome IV. Chromosome III and VI contains only single gene each belonging to this group only. The chromosome II was found to have genes (four in number) from the group in a small cluster (Figure 2B).
The genes encoding proteins having two MATH domains in Arabidopsis (25) are found to be distributed between all chromosomes while in rice, only one gene from this group is located on chromosome X. Further, their distribution on the chromosome in Arabidopsis also presents an interesting pattern. A large number of such genes (total seven in number) were found to be present on chromosome II and III and further four genes were present on chromosome IV and V while chromosome I was observed to have three genes encoding for proteins having two MATH domains.
With only single instance of a protein having two MATH domains along with two BTB domains (2M2B) in rice, the gene was found to be present on chromosome XI while none of the protein of this group was present in Arabidopsis. However, in Arabidopsis, two proteins having three MATH domains were found and the genes encoding both these proteins were located together on chromosome II. Further, only one protein that too in Arabidopsis, having four MATH domains was observed. The gene encoding this protein was found to be localized on chromosome III.
Sub-Cellular Localization of MDC Proteins
Analysis of the sub-cellular localization of MDC proteins in Arabidopsis and rice presented an interesting pattern (Figure 3). Twenty-two MDC proteins in Arabidopsis were predicted to be localized in the nucleus, 21 in the cytoplasm and 20 in the chloroplast (Table 2). In contrast, majority of the rice MDC proteins were predicted to be present in either the chloroplast (35) or in the cytoplasm (23) (Table 3). Further analysis in rice revealed that mostly single MDC proteins were predicted to be localized in the nucleus. However, proteins containing MATH domain along with the BTB domain were predicted to be localized in the cytoplasm and the chloroplast. In Arabidopsis, six MDC proteins were predicted to be localized in the mitochondria in comparison to two in rice. The MDC proteins in Arabidopsis were also predicted to be localized in other sub-cellular locations such as, cytoskeleton, peroxisome, and extracellular matrix. These were mainly one and two MATH domain containing proteins. Similarly, two MATH domain protein of rice (Os2M1) was specifically predicted to be localized in the vacuole.
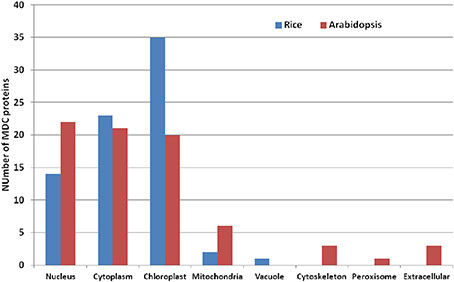
Figure 3. Bar diagram showing predicted sub-cellular localization of the members of MDC proteins in Arabidopsis and rice.
Expression Analysis of MDC Protein Encoding Genes
In Various Tissues
The expression analysis of MDCP encoding genes in Arabidopsis using 22 K ATH1 genome array dataset showed that most of the MDCPs encoding genes showed transcript at low levels in various tissues (Figure 4A, Table S1). The genes coding for BTB domain containing MDCPs showed low or no expression in Arabidopsis except AtMB1, AtMB3, and AtMB5 which showed increased expression in the inflorescence. Even in rice such genes showed similar levels of expression, except for OsMB5, OsMB9, OsMB10, and OsMB11 which were up-regulated in various tissues (Figure 4B, Table S1). Expression analysis in calli showed increased levels of AtMB1, AtMB3, and AtMB5 from Arabidopsis and OsMB9, OsMB10, and OsMB11 from rice. In Arabidopsis, single-domain MDCP encoding genes showed low expression in various tissues, except for AtM1 and AtM2 which were found to be up-regulated in the inflorescence. Another single-domain MDCP encoding gene AtM10 was found to be up-regulated in callus but also maintained a minimum level of expression across various tissues. In rice, six of the single-domain MDCP encoding genes viz. OsM1, OsM2, OsM4, OsM5, OsM12, and OsM13 were found to be highly up-regulated in various tissues. The two MDCP encoding genes At2M2 and At2M15 in Arabidopsis showed high expression in roots. Further, At2M23 showed variability in expression in roots but remained at low levels in other tissues. This analysis indicated that in rice at least 10 MDCP encoding genes were highly expressed throughout all tissues suggesting their possible role in the combinatorial transcriptional regulation of a broad set of genes in various tissues.
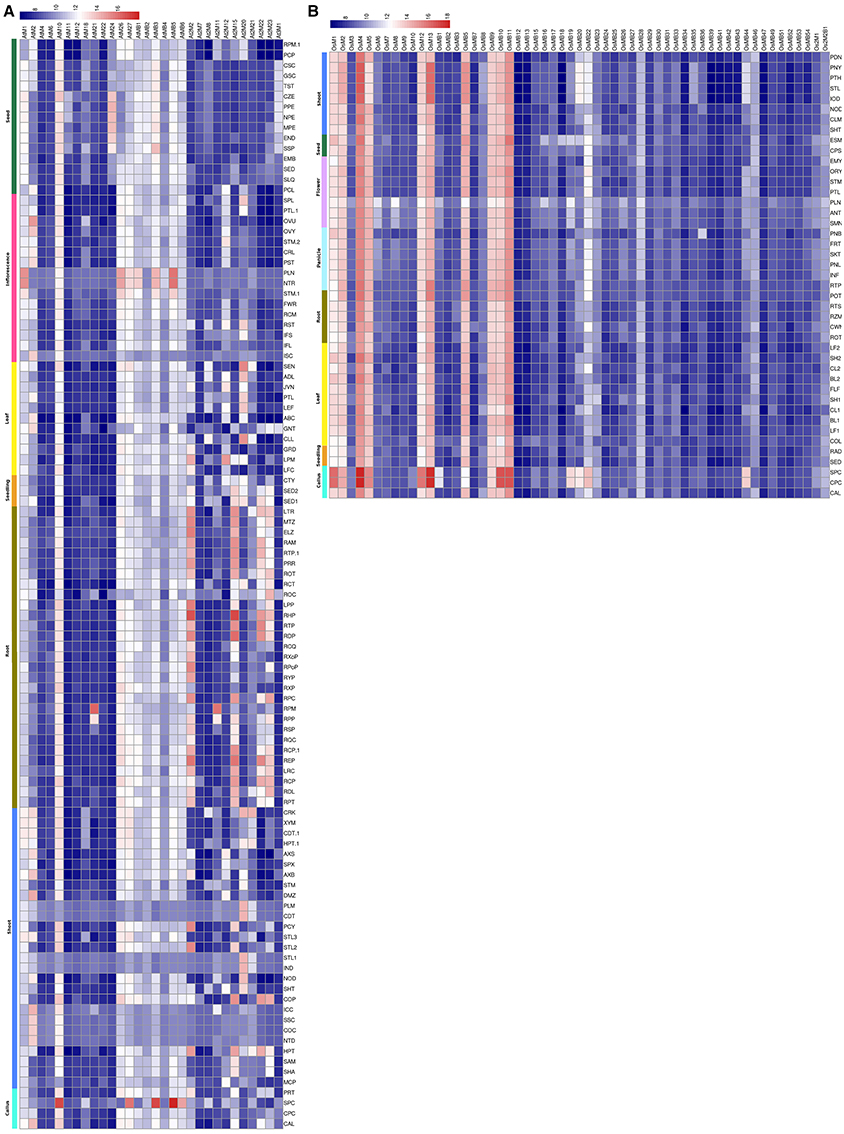
Figure 4. Heatmap representation of the expression of MDC protein encoding genes in various tissues in Arabidopsis (A) and rice (B). The expression values were obtained from Affymetrix array databases using Genevestigator Response Viewer (https://www.genevestigator.com). For Arabidopsis, 22 K ATH1 genome array was chosen along with pre-existing microarray and in case of rice, microarray results of OS_51 K: Rice Genome 51 K pre-existing microarrays were chosen. The details of the libraries used in the current analysis are presented in Table S1.
At Various Developmental Stages
To check the transcript levels of MDCPs encoding genes at various developmental stages of Arabidopsis and rice, publicly available microarray data was analyzed. In Arabidopsis, single-domain MDCPs encoding genes AtM1, AtM10, AtM26, and AtM27 were found to be up-regulated during all the developmental stages while AtM18 showed higher expression only during the senescence stage (Figure 5A, Table S2). Further, AtM2 showed variable expression during various developmental stages except for senescence and germinating seed stage where its levels remained low. In rice, the single-domain OsM4 showed significantly high expression at different developmental stages. The OsM1, OsM2, OsM5, OsM12, and OsM13 also showed high expression throughout all the developmental stages (Figure 5B, Table S2). The genes encoding MDCPs with BTB domain showed comparatively higher expression during all the developmental stages in Arabidopsis. While in rice, only four genes viz. OsMB5, OsMB9, OsMB10, and OsMB11 showed high expression during all the developmental stages. Rest of the other similar genes showed relatively lower expression in all the developing tissues in rice except for OsMB22 gene which showed variable expression. The two domain MDCPs coding genes in Arabidopsis showed differential expression in various tissues. The At2M2 gene showed comparatively high expression in the young rosette and seedling stage of the plant while maintaining variable expression in other tissues. Similarly, At2M15 gene showed higher expression during seed germination and seedling stage, while maintaining lower levels in most of the other developing tissues. The At2M20 showed minimal to high expression in all the developing tissues except for senescence and germinating seeds. The genes encoding two MATH domain MDC proteins in rice (Os2M1 and Os2M2B1) were observed to be expressed at lower levels in all the developmental tissues.
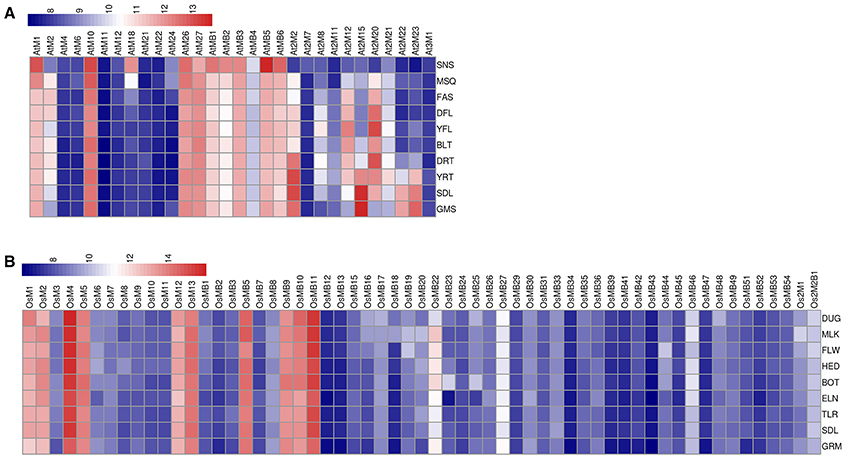
Figure 5. Heatmap representation of the expression of MDC protein encoding genes at various developmental stages in (A) Arabidopsis and (B) rice. The expression values were obtained from Affymetrix array databases using Genevestigator Response Viewer (https://www.genevestigator.com). For Arabidopsis, 22 K ATH1 genome array was chosen along with pre-existing microarray and in case of rice, microarray results of OS_51 K: Rice Genome 51 K pre-existing microarrays were chosen. The conditions considered for analysis in Arabidopsis are: (SNS), senescence; (MSQ), mature siliques;(FAS), flowers and siliques; (DFL), developed flower; (YFL), young flower;(BLT), bolting;(DRT), developed rosette; (YRT), young rosette; (SDL), seedling; (GMS), germinated seeds. The conditions considered for analysis in rice are: (DUG), dough stage; (MLK), milk stage; (FLW), flowering stage; (HED), heading stage; (BOT), booting stage; (ELN), stem elongation stage; (TLR), tillering stage;(SDL), seedling; (GRM), germination. The details of the libraries used in the current analysis are presented in Table S2.
In Response to Various Abiotic Stress Conditions
In Arabidopsis, most of the MDCP-coding genes maintain minimal expression under various abiotic stress conditions, while in rice the expression of MDC protein coding genes gets down-regulated (Figure S5A,B, Table S3). Interestingly, gene encoding two domain MDC protein At2M2, was found to be up-regulated in both root and shoot tissues during the late phase of both salinity and osmotic stress. Another gene At2M12 showed high expression under drought stress condition in both early and late phase in shoots. Similarly, At2M23 showed higher expression in shoots during the late phase of wounding. On the other hand in rice, gene encoding MATH-BTB domain containing proteins i.e., OsMB10 and OsMB11 showed high expression under salinity as well as drought stress. However, slight up-regulation was observed for OsMB12 and OsMB5 under salinity and drought stress and for OsMB19, OsMB20, OsMB22, and OsMB46 under heat stress. Interestingly, all the MATH domain encoding genes showed down-regulation under cold stress.
In Response to Various Biotic Stress Conditions
Under the biotic stresses, all the genes encoding single MDC proteins and also genes coding for proteins containing MATH with BTB domain showed very low expression in Arabidopsis (Figure 6A, Table S4). However, only genes coding for two MATH domain containing proteins showed differential expression under biotic stresses. On the other hand, MDCP encoding genes in rice showed an interesting pattern of expression. Single domain MDC protein encoding genes such as OsM1, OsM2, OsM4, OsM5, OsM12, and OsM13 showed significant up-regulation in response to various biotic stress conditions studied here (Figure 6B, Table S4). The genes coding for MDC proteins having BTB domain such as OsMB9, OsMB10, and OsMB11 also showed high up-regulation under various biotic stress conditions. All the other MDC genes showed little response toward the biotic stresses.
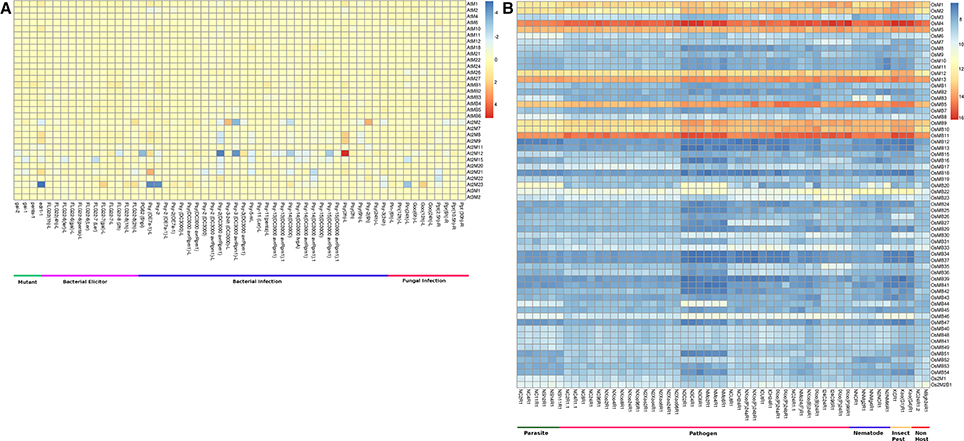
Figure 6. Heatmap representation of the expression of MDC protein encoding genes in response to various biotic stresses in (A) Arabidopsis and (B) rice. The expression values were obtained from Affymetrix array databases using Genevestigator Response Viewer (https://www.genevestigator.com). For Arabidopsis, 22 K ATH1 genome array was chosen along with pre-existing microarray and in case of rice, microarray results of OS_51 K: Rice Genome 51 K pre-existing microarrays were chosen. The details of the libraries used in the current analysis are presented in Table S4.
qRT-PCR Based Expression Analysis of MDCP Coding Genes under Abiotic Stresses
Expression analysis of large gene family members through the publicly available database and validation of selected gene expression pattern using qRT-PCR, is a useful approach, which provides primary information about the newly identified gene function (Singh et al., 2013). However, in few incidences, data retrieved through different resources may vary. Thus, to confirm the expression profile of MDCP encoding genes, we chose 11 representative OsMDCP encoding genes which were reported to be highly up-regulated in different tissues (Figure 4B), at different developmental stages (Figure 5B) as well as under different biotic stresses (Figure 6B). The level of expression of these selected 11 genes was further checked under abiotic stress conditions such as salinity (200 mM NaCl) and drought (air dry) to study their cross-inducibility. Our qRT-PCR results under these stresses corroborated with the expression pattern obtained by publicly available microarray data (Figure S5B). For instance, OsM4, OsM5, and OsM12 expression was up-regulated after 6 h of salt and drought stress, while OsM1 and OsM2 were up-regulated under drought stress only (Figure 7A). Similarly, an up-regulation in OsMB5, OsMB6 and OsMB11 levels and down-regulation in OsMB9 levels was observed under both salinity and drought stress (Figure 7B). The levels of OsM13 and OsMB10 could not detected in the qRT-PCR analysis. Our qRT-PCR results for OsM2, OsM12, OsMB5, OsMB9, and OsMB11 under salinity stress and OsM12, OsMB5, and OsMB9 under drought effectively validate the expression profile obtained from the publicly available database, thereby providing more authentic expression picture of MDCP family members. However, the transcript profile of OsM1, OsM4, OsM5, OsMB6, OsM13, and OsMB13 under salinity stress, and OsM1, OsM2, OsM4, OsM5, OsM13, OsMB6, and OsMB10 under drought stress did not corroborate well with their respective microarray data. These differences in expression levels in the publicly available microarray and qRT-PCR may be either due to genotypic differences between the samples or due to differences in the plant developmental stages.
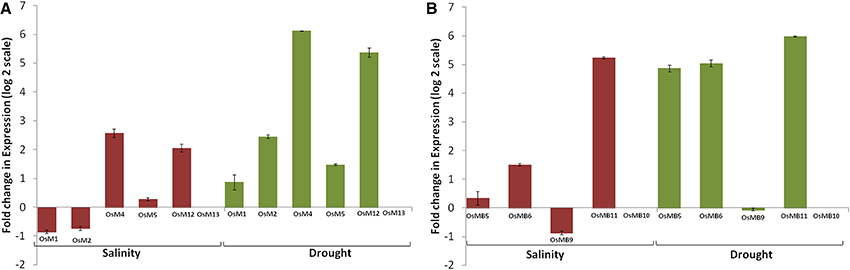
Figure 7. qRT-PCR confirms altered expression of selected biotic stress responsive genes under abiotic stress conditions. Bar diagram depicting fold change (log2 scale) in expression of selected single MATH domain containing genes (A) and single MATH along with single BTB domain containing genes (B) under salinity and drought stress conditions based on qRT-PCR analysis. For this analysis, 10 day old seedlings of IR64 variety (a moderately sensitive cultivar) of rice were subjected to stress treatments for 6 h followed by RNA isolation, first strand cDNA synthesis and real-time PCR. Error bars show standard deviation.
In addition, when we compare our qRT-PCR data with the biotic stress data from the publicly available database we found that most of the salt stress-responsive MDCP encoding genes namely, OsM4, OsM5, OsM12, OsMB5, OsMB6, and OsMB11 showed a positively correlated response to biotic stress. Similarly, most of the drought stress-responsive MDCP encoding genes namely, OsM1, OsM2, OsM4, OsM5, OsM12, OsMB5, OsMB6, and OsMB11 showed a positively correlated response to biotic stress. This indicates toward a significant role of these genes in both abiotic and biotic stress response. However, certain genes showed an inverse correlation between biotic and abiotic stress response. These genes are OsM1, OsM2, OsM13, OsMB9, and OsMB10 under salinity stress while OsM13, OsMB9, and OsMB10 under drought stress. Importantly, most of the genes i.e., OsM4, OsM5, OsM12, OsMB5, OsMB6, and OsMB11 showed positive correlation under all biotic and abiotic stress conditions, while OsM13, OsMB9 and OsMB10 showed an inverse correlation among biotic and abiotic stress response.
Discussion
Using subtractive hybridization approach in two contrasting cultivars of rice, Pokkali (salt tolerant) and IR64 (salt sensitive) 1194 high-throughput ESTs (584 from IR64 and 610 from Pokkali) were obtained in our previous study (Kumari et al., 2009). These ESTs were believed to be playing a significant role in salt stress tolerance in rice at the seedling stage. The MDC proteins were identified through this study as potential candidates that may play a role in both abiotic and biotic stress response. Earlier, the MDC proteins have been reported and analyzed for their role in plant-microbe interaction (Oelmüller et al., 2005). The analysis suggested that the MATH domain containing protein located at the plasma membrane in roots of Arabidopsis perceives the first signal for the presence of basidiomycete Piriformospora indica (Shahollari et al., 2005). In the present analysis, we have identified and classified the MATH domain containing proteins in Arabidopsis and rice and further, analyzed their potential role in the abiotic stress response. We have identified a total of 156 MDC proteins, with 62 genes encoding 82 MDC proteins in Arabidopsis and 69 genes encoding 74 MDC proteins in rice in comparison to an earlier report by Oelmüller et al. (2005), which identified 59 genes in Arabidopsis. Another previous study has reported the presence of 6 MATH-BTB genes in Arabidopsis and 69 MATH-BTB genes in rice while analyzing BTB superfamily in grasses (Juranić and Dresselhaus, 2014). Similar analysis between Brassica, rice and Arabidopsis showed 90 genes encoding MATH-domain proteins from B. rapa, 63 genes in Arabidopsis and 36 genes in rice (Zhao et al., 2013). Further, BTB superfamily has been characterized in various dicots species and comprises protein members from MATH-BTB family (Gingerich et al., 2007). Analysis of domains present in the MDCPs in both Arabidopsis and rice showed the presence of BTB domain along with the MATH domain. The BTB domain (POZ domain) has been earlier known for its protein-protein interaction modules with its ability to self-associate and also to interact with other non-BTB proteins (Stogios et al., 2005). As reported earlier, the BTB domain was also found at the carboxy-terminal in the MDC proteins in both Arabidopsis and rice. MDCP family members were earlier shown to mediate the interaction of BTB/POZ-MATH (BPM) proteins with ethylene response factor/Apetala2 transcription factor family members (Weber and Hellmann, 2009).
In this study, we show that MDC proteins along with BTB domain are found in large number in rice than in Arabidopsis. This large number of members in rice can be attributed to major expansion and diversification events in monocots including rice, which have probably occurred after the split of monocot and dicot (Gingerich et al., 2007). The low sequence conservation within the group signifies the evolution of monocots as a component of an innate immunity system owing to sophisticated mechanisms developed by the pathogens (Gingerich et al., 2005, 2007). Phylogenetic relationship tree of the MDC proteins in Arabidopsis and rice showed a distinct evolution of these proteins in plants. This shows that BTB domains in the MDC proteins might have been evolving distinctly to the MATH domain contributing to the overall distinctness to the MDC proteins having BTB domain. Previously, a phylogenetic analysis in mosses, eudicots, and grasses has shown that the expansion in MATH-BTB gene family occurred largely due to local gene duplications (Juranić and Dresselhaus, 2014). The localization of the MDC protein encoding genes in both Arabidopsis and rice shows that the MDC genes lie in a cluster on various chromosomes. Interestingly in rice, the maximum number (30) of genes coding for MDC proteins having BTB domains were found to be clustered on the chromosome X. However, one of the earlier studies showed MATH domain proteins as part of the syntenic region on chromosome VIII (Juranić and Dresselhaus, 2014). However, these proteins possessed only the BTB domain in their sequence and lacked MATH domain. In contrast, a large number of genes (24) encoding MDC protein were found clustered on the chromosome III in Arabidopsis which is known for the presence of clustered gene families (Salanoubat et al., 2000). Thus, the clade-specific expansion in MATH-BTB gene family occurred largely due to tandem or segmental duplications (Juranić and Dresselhaus, 2014).
Plants frequently encounter various biotic and abiotic stresses throughout their life cycle (Singh V. K. et al., 2015). The transcriptome analysis of the molecular response in plants toward multiple stresses (abiotic and biotic) has identified several overlapping genes which are identified and proposed to be responsible for generalized stress response or found to be the points of cross-talk between signaling pathways (Atkinson and Urwin, 2012; Kissoudis et al., 2014; Foyer et al., 2016). MDCPs of BTB superfamily, function as substrate-specific adaptors of CULLIN (CUL3)-based ubiquitin E3 ligase to target protein for ubiquitination (Weber et al., 2005). Ubiquitin significantly affects physiology, development and homeostasis of all eukaryotes including embryogenesis, cell cycle, hormonal balance, photomorphogenesis, circadian rhythms, flower development, self-incompatibility, ecological adaptation, disease resistance as well as cell death (Gingerich et al., 2007; Zapata et al., 2007; Qi et al., 2009; Zhao et al., 2013). Moreover, types of recognition motifs in BTB protein are mostly conserved between Arabidopsis and rice indicating that similar substrates exist in both the species (Gingerich et al., 2007; Juranić and Dresselhaus, 2014). Therefore, to gain preliminary insight into the potential function of plant MDCP genes during stress response and development, we have explored publicly available microarray data for Arabidopsis and rice. Expression analysis of MDCP gene family members using rice microarray data revealed that all the 11 highly expressed genes under biotic stress also showed high transcript levels in all the tissues as well as at all the development stages in rice. These findings highlight the role of MDCP genes in overall plant growth and development.
In order to analyze the correlated response under biotic and abiotic stress, MDC protein encoding genes which are highly up-regulated in all biotic stresses were analyzed for salt and drought stress response. Interestingly, these selected MDC genes showed positively correlated response for abiotic and biotic stress which further signifies the coordinated response of various gene families pertaining to various types of stress (abiotic or biotic). Similarly, BTB/POZ protein ETO1 (ethylene overproducer 1) was found to interact with ethylene biosynthesis protein ACS5 and negatively affects ethylene biosynthesis (Wang et al., 2004). In contrast, MATH-BTB proteins were also shown to directly interact with a class I homeodomain leucine zipper (HD-ZIP) transcription factor ATHB6, which negatively regulates ABA responses (Lechner et al., 2011). ABA regulates different phases of plant development including seed dormancy, germination, and reproduction and also acts as a key factor in biotic and abiotic stress responses in plants, particularly salinity and drought (Ton et al., 2009; Raghavendra et al., 2010). It was also reported earlier that MDC proteins located on the plasma membrane primarily respond to fungal infection in Arabidopsis roots and are also involved in nodule formation in Medicago (Oelmüller et al., 2005). Similarly, Cosson et al. (2010) found that one of the restricted TEV movement (RTM) genes i.e., RTM3 which restricts the long-distance movement of various potyviruses in Arabidopsis, encodes an unknown protein containing MATH domain in its amino-terminal region. In maize, MATH-BTB genes were shown to be expressed in zygote and control spindle length during meiosis as well as nuclei identity during first pollen mitosis (Juranič et al., 2012). An analysis suggested that some genes in the plants are universally stress responsive which leads to the evolution of effective strategies toward understanding the stress behavior in plants (Narsai et al., 2013). Earlier, disease resistant pathway similar to the Arabidopsis NPR1 (AtNPR1), which also showed negative effects on viral infections, showed negative regulation of this gene in plants under salt and drought stress response (Quilis et al., 2008). These observations indicate toward possibly diverse roles of MDCP genes throughout the plant development and stress response in rice.
Conclusions
The strategy of comparative genomics and transcriptomics had led to the discovery of many novel genes and gene families playing a role in various stress responses. One of the members identified in such strategic analysis toward salt stress led to the identification of MATH-domain family which has been earlier known for their role in the plant/microbe interaction. Apart from characterizing the family in both Arabidopsis and rice, we have attempted to establish their role in overall plant growth and development as well as abiotic and biotic stresses using the high-throughput expression data available in the public domain. Further, we narrowed down 11 potential candidate genes in rice which showed higher expression in all the developmental stages, tissues, as well as biotic stresses in rice. These genes were further validated through qRT-PCR with drought and salinity stress in rice. Combining the publicly available data and our study, we identified OsM4 and OsMB11 as the potential candidate genes ubiquitously expressed in all the tissues, developmental stages, biotic as well as abiotic stresses. This needs to be comprehensively analyzed further for functional validation of their specific roles in plant development and stress response in increasing environmental resilience in crops.
Author Contributions
SLS-P, AP conceived the idea and designed the experiments. RJ did the real time PCR work and its analysis. HK performed the MPSS and microarray database analysis. RJ, HK wrote the manuscript. SLS-P, AP edited the manuscript. All the authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
HK acknowledges Department of Science and Technology, Government of India for the grants received as DST-INSPIRE award. RJ acknowledges the Start-Up research grant (Young Scientist) from the Science and Engineering Research Board, Government of India. SLS-P acknowledges the support of research funds from the Department of Biotechnology, Government of India, and internal grants of International Center for Genetic Engineering and Biotechnology.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00923
Figure S1. Representative (unscaled) domain architecture of the MDC proteins in Arabidopsis and rice. All the MDC proteins in rice and Arabidopsis were found to consist of MATH domain (PF00917) while few MDC proteins in both Arabidopsis and rice were found to contain BTB domains (PF00651) also.
Figure S2. Multiple sequence alignment of full length sequences having single MATH domain in Arabidopsis and rice. The sequence analysis was performed using Seaview (version 4) multiple sequence alignment editor (Gouy et al., 2010).
Figure S3. Multiple sequence alignment of full length sequences having two MATH domains in Arabidopsis and rice. The sequence analysis was performed using Seaview (version 4) multiple sequence alignment editor (Gouy et al., 2010).
Figure S4. Multiple sequence alignment of full length sequences having MATH and BTB domains in Arabidopsis and rice. The sequence analysis was performed using Seaview (version 4) multiple sequence alignment editor (Gouy et al., 2010).
Figure S5. Heatmap representation of the expression of MDC protein encoding genes in response to various abiotic stresses (A) such as cold, drought, genotoxic, heat, osmotic, oxidative, salinity and wound in Arabidopsis and (B) salinity, heat, drought and cold in rice. The expression values were obtained from Affymetrix array databases using Genevestigator Response Viewer (https://www.genevestigator.com). For Arabidopsis, 22 K ATH1 genome array was chosen along with pre-existing microarray and in case of rice, microarray results of OS_51 K: Rice Genome 51 K pre-existing microarrays were chosen. The details of the libraries used in the current are presented in Table S3.
Table S1. List of libraries of different tissues with their abbreviations used in the expression analysis of MDC protein encoding genes in (a) rice and (b) Arabidopsis.
Table S2. List of libraries of different developmental stages with their abbreviations used in the expression analysis of MDC protein encoding genes in (a) rice and (b) Arabidopsis.
Table S3. List of libraries of various abiotic stresses with their abbreviations used in the expression analysis of MDC protein encoding genes in (a) rice and (b) Arabidopsis.
Table S4. List of libraries of various biotic stresses with their abbreviations used in the expression analysis of MDC protein encoding genes in (a) rice and (b) Arabidopsis.
Table S5. List of primers used for qRT-PCR analysis in the present study.
References
Ahmad, K. F., Engel, C. K., and Privé, G. G. (1998). Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. U.S.A. 95, 12123–12128. doi: 10.1073/pnas.95.21.12123
Ahmed, I. M., Nadira, U. A., Bibi, N., Cao, F., He, X., Zhang, G., et al. (2015). Secondary metabolism and antioxidants are involved in the tolerance to drought and salinity, separately and combined, in Tibetan wild barley. Environ. Expt. Bot. 111, 1–12. doi: 10.1016/j.envexpbot.2014.10.003
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Basu, S., and Roychoudhury, A. (2014). Expression profiling of abiotic stress-inducible genes in response to multiple stresses in rice (Oryza sativa L.) varieties with contrasting level of stress tolerance. BioMed. Res. Int. 2014:706890. doi: 10.1155/2014/706890
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Bohnert, H. J., Gong, Q., Li, P., and Ma, S. (2006). Unravelling abiotic stress tolerance mechanisms-getting genomic going. Curr. Opin. Plant Biol. 9, 180–188. doi: 10.1016/j.pbi.2006.01.003
Bohra, A., Sahrawat, K. L., Kumar, S., Joshi, R., Parihar, A. K., Singh, U., et al. (2015). Genetics and genomics based interventions for nutritional enhancement of grain-legume crops: status and outlook. J. Appl. Genet. 56, 151–161. doi: 10.1007/s13353-014-0268-z
Bradley, J. R., and Pober, J. S. (2001). Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20, 6482–6491. doi: 10.1038/sj.onc.1204788
Brett, D., Pospisil, H., Valcárcel, J., Reich, J., and Bork, P. (2002). Alternative splicing and genome complexity. Nat. Genet. 30, 29–30. doi: 10.1038/ng803
Broder, C., and Becker-Pauly, C. (2013). The metalloproteases meprin α and meprin β: unique enzymes in inflammation, neurodegeneration, cancer and fibrosis. Biochem. J. 450, 253–264. doi: 10.1042/BJ20121751
Cosson, P., Sofer, L., Schurdi-Levraud, V., and Revers, F. (2010). A member of a new plant gene family encoding a meprin and TRAF homology (MATH) domain-containing protein is involved in restriction of long distance movement of plant viruses. Plant Signal Behav. 5, 1321–1323. doi: 10.4161/psb.5.10.13244
Eddy, S. R. (1998). Profile hidden markov models. Bioinformatics 14, 755–763. doi: 10.1093/bioinformatics/14.9.755
Farooq, S., and Azam, F. (2001). Co-existence of salt and drought tolerance in Triticeae. Hereditas 135, 205–210. doi: 10.1111/j.1601-5223.2001.00205.x
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42(Database issue), D222–D230. doi: 10.1093/nar/gkt1223
Foyer, C. H., Rasool, B., Davey, J. W., and Hancock, R. D. (2016). Cross-tolerance to biotic and abiotic stresses in plants: a focus on resistance to aphid infestation. J. Exp. Bot. 67, 2025–2037. doi: 10.1093/jxb/erw079
Gehan, M. A., Greenham, K., Mockler, T. C., and McClung, C. R. (2015). Transcriptional networks-crops, clocks, and abiotic stress. Curr. Opin. Plant Biol. 24, 39–46. doi: 10.1016/j.pbi.2015.01.004
Ghosh, A., Kushwaha, H. R., Hasan, M. R., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2016). Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 6:18358. doi: 10.1038/srep18358
Gingerich, D. J., Gagne, J. M., Salter, D. W., Hellmann, H., Estelle, M., Ma, L., et al. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280, 18810–18821. doi: 10.1074/jbc.M413247200
Gingerich, D. J., Hanada, K., Shiu, S. H., and Vierstra, R. D. (2007). Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19, 2329–2348. doi: 10.1105/tpc.107.051300
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gupta, B., Tripathi, A. K., Joshi, R., Pareek, A., and Singla-Pareek, S. L. (2015). “Designing climate-smart future crops employing signal transduction components,” in Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives, Vol. 2., ed G. K. Pandey (New York, NY: Springer), 393–414.
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucl. Acids Res. 35(Web Server issue), W585–W587. doi: 10.1093/nar/gkm259
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. doi: 10.1155/2008/420747
Huang, B., Zhang, L., Du, Y., Li, L., Tang, X., and Zhang, G. (2016). Molecular characterization and functional analysis of tumor necrosis factor receptor-associated factor 2 in the Pacific oyster. Fish Shellfish Immunol. 48, 12–19. doi: 10.1016/j.fsi.2015.11.027
Huang, S., Chen, X., Zhong, X., Li, M., Ao, K., Huang, J., et al. (2016). Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe 19, 204–215. doi: 10.1016/j.chom.2016.01.005
Joshi, R., Karan, R., Singla-Pareek, S. L., and Pareek, A. (2016a). Ectopic expression of Pokkali phosphoglycerate kinase-2 (OsPGK2-P) improves yield in tobacco plants under salinity stress. Plant Cell Rep. 35, 27–41. doi: 10.1007/s00299-015-1864-z
Joshi, R., Ramanarao, V. M., Lee, S., Kato, N., and Baisakh, N. (2014). Ectopic expression of ADP Ribosylation Factor1 (SaARF1) from smooth cordgrass (Spartina alterniflora) confers drought and salt tolerance in transgenic rice and Arabidopsis. Plant Cell Tiss. Org. Cult. 117, 17–30. doi: 10.1007/s11240-013-0416-x
Joshi, R., Singh, B., Bohra, A., and Chinnusamy, V. (2016b). “Salt stress signalling pathways: specificity and crosstalk,” in Managing Salinity Tolerance in Plants: Molecular and Genomic Perspectives, eds S. H. Wani and M. A. Hossain (Boca Raton, FL: CRC Press), 51–78.
Juranić, M., and Dresselhaus, T. (2014). Phylogenetic analysis of the expansion of the MATH-BTB gene family in the grasses. Plant Signal. Behav. 9:e28242. doi: 10.4161/psb.28242
Juranič, M., Srilunchang, K. O., Krohn, N. G., Leljak-Levanic, D., Sprunck, S., and Dresselhaus, T. (2012). Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24, 4974–4991. doi: 10.1105/tpc.112.107169
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Kissoudis, C., van de Wiel, C., Visser, R. G., and van der Linden, G. (2014). Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 5:207. doi: 10.3389/fpls.2014.00207
Kumar, G., Kushwaha, H. R., Punjabi-Sabharwal, V., Kumari, S., Joshi, R., Karan, R., et al. (2012). Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-P confers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol. 12:107. doi: 10.1186/1471-2229-12-107
Kumari, S., Joshi, R., Singh, K., Roy, S., Tripathi, A. K., Singh, P., et al. (2015). Expression of a cyclophilin OsCyp2-P isolated from a salt tolerant landrace of rice in tobacco alleviates stress via ion homeostasis and limiting ROS accumulation. Funct. Integr. Genom. 15, 395–412. doi: 10.1007/s10142-014-0429-5
Kumari, S., Panjabi Nee Sabharwal, V., Kushwaha, H. R., Sopory, S. K., Singla-Pareek, S. L., and Pareek, A. (2009). Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct. Integr. Genom. 9, 109–123. doi: 10.1007/s10142-008-0088-5
Kushwaha, H. R., Singh, A. K., Sopory, S. K., Singla-Pareek, S. L., and Pareek, A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 10:200. doi: 10.1186/1471-2164-10-200
Lechner, E., Leonhardt, N., Eisler, H., Parmentier, Y., Alioua, M., Jacquet, H., et al. (2011). MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell. 21, 1116–1128. doi: 10.1016/j.devcel.2011.10.018
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mustafiz, A., Singh, A. K., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 11, 293–305. doi: 10.1007/s10142-010-0203-2
Narsai, R., Wang, C., Chen, J., Wu, J., Shou, H., and Whelan, J. (2013). Antagonistic, overlapping and distinct responses to biotic stress in rice (Oryza sativa) and interactions with abiotic stress. BMC Genom. 14:93. doi: 10.1186/1471-2164-14-93
Nielsen, H. (2016). “Predicting subcellular localization of proteins by bioinformatic algorithms”, in the series Current Topics in Microbiology and Immunology, eds F. Bagnoli, and R. Rappuoli (New York, NY: Springer), 1–30. doi: 10.1007/82_2015_5006
Oelmüller, R., Peskan-Berghöfer, T., Shahollari, B., Trebicka, A., Sherameti, I., and Varma, A. (2005). MATH-domain proteins represent a novel protein family in Arabidopsis thaliana, and at least one member is modified in roots during the course of a plant-microbe interaction. Physiol. Plant. 124, 152–166. doi: 10.1111/j.1399-3054.2005.00505.x
Pareek, A., Singh, A., Kumar, M., Kushwaha, H. R., Lynn, A. M., and Singla-Pareek, S. L. (2006). Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 142, 380–397. doi: 10.1104/pp.106.086371
Pastori, G. M., and Foyer, C. H. (2002). Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 129, 460–468. doi: 10.1104/pp.011021
Peškan-Berghöfer, T., Markert, C., Varma, A., and Oelmüller, R. (2004). Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmatic reticulum and at the plasma membrane. Physiol. Plant. 122, 465–477. doi: 10.1111/j.1399-3054.2004.00424.x
Qi, P., Yan, H., PeiFen, D., QingXuan, X., Ying, R., and ChunLin, L. (2009). Construction of SSH library with different stages of seeds development in Brassica napus L. Acta Agron. Sin. 35, 1576–1583. doi: 10.3724/SP.J.1006.2009.01576
Quilis, J., Peñas, G., Messeguer, J., Brugidou, C., and San Segundo, B. (2008). The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant Microbe Interact. 21, 1215–1231. doi: 10.1094/MPMI-21-9-1215
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Rambaut, A. (2012). FigTree_v1.4.0 2012. Available online at: http://tree.bio.ed.ac.uk/software/figtree
Ramegowda, V., Basu, S., Krishnan, A., and Pereira, A. (2014). Rice GROWTH UNDER DROUGHT KINASE is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 166, 1634–1645. doi: 10.1104/pp.114.248203
Salanoubat, M., Lemcke, K., Rieger, M., Ansorge, W., Unseld, M., Fartmann, B., et al. (2000). European Union Chromosome 3 Arabidopsis sequencing consortium; institute for genomic research; Kazusa DNA Research Institute. Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408, 820–822. doi: 10.1038/35048706
Shahollari, B., Varma, A., and Oelmüller, R. (2005). Expression of a receptor kinase in Arabidopsis roots is stimulated by the basidiomycete Piriformospora indica and the protein accumulates in Triton X-100 insoluble plasma membrane microdomains. J. Plant Physiol. 162, 945–958. doi: 10.1016/j.jplph.2004.08.012
Singh, A. K., Kumar, R., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2012). Overexpression of rice CBS domain containing protein improves salinity, oxidative and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 52, 205–216. doi: 10.1007/s12033-011-9487-2
Singh, A. K., Kumar, R., Tripathi, A. K., Gupta, B. K., Pareek, A., and Singla-Pareek, S. L. (2015). Genome-wide investigation and expression analysis of Sodium/Calcium exchanger gene family in rice and Arabidopsis. Rice. 8, 54. doi: 10.1186/s12284-015-0054-5
Singh, A. K., Sharma, V., Pal, A. K., Acharya, V., and Ahuja, P. S. (2013). Genome-Wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20, 403–423. doi: 10.1093/dnares/dst019
Singh, A., Kushwaha, H. R., Soni, P., Gupta, H., Singla-Pareek, S. L., and Pareek, A. (2015). Tissue specific and abiotic stress regulated transcription of histidine kinases in plants is also influenced by diurnal rhythm. Front. Plant Sci. 6:711. doi: 10.3389/fpls.2015.00711
Singh, B., Bohra, A., Mishra, S., Joshi, R., and Pandey, S. (2015). Embracing new-generation ‘omics’ tools to improve drought tolerance in cereal and food-legume crops. Biol. Plant. 59, 413–428. doi: 10.1007/s10535-015-0515-0
Singh, V. K., Jain, M., and Garg, R. (2015). Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front Plant Sci. 5:789. doi: 10.3389/fpls.2014.00789
Sreenivasulu, N., Sopory, S. K., and Kavi Kishor, P. B. (2007). Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388, 1–13. doi: 10.1016/j.gene.2006.10.009
Stogios, P. J., Down, G. S., Jauhal, J. J., Nandra, S. K., and Privé, G. G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6:R82. doi: 10.1186/gb-2005-6-10-r82
Sunnerhagen, M., Pursglove, S., and Fladvad, M. (2002). The new MATH: homology suggests shared binding surfaces in meprin tetramers and TRAF trimers. FEBS Lett. 530, 1–3. doi: 10.1016/S0014-5793(02)03330-6
Ton, J., Flors, V., and Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi: 10.1016/j.tplants.2009.03.006
Tripathi, A. K., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2012). Narrowing down the targets for yield improvement in rice under normal and abiotic stress conditions via expression profiling of yield-related genes. Rice 5:37. doi: 10.1186/1939-8433-5-37
Tripathi, A. K., Singh, K., Pareek, A., and Singla-Pareek, S. L. (2015). Histone chaperones in Arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol. 15:42. doi: 10.1186/s12870-015-0414-8
Wang, K. L., Yoshida, H., Lurin, C., and Ecker, J. R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 428, 945–950. doi: 10.1038/nature02516
Weber, H., Bernhardt, A., Dieterle, M., Hano, P., Mutlu, A., Estelle, M., et al. (2005). Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137, 83–93. doi: 10.1104/pp.104.052654
Weber, H., and Hellmann, H. (2009). Arabidopsis thaliana BTB/ POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 276, 6624–6635. doi: 10.1111/j.1742-4658.2009.07373.x
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Zapata, J. M., Martínez-García, V., and Lefebvre, S. (2007). Phylogeny of the TRAF/MATH domain. Adv. Exp. Med. Biol. 597, 1–24. doi: 10.1007/978-0-387-70630-6_1
Zhao, L., Huang, Y., Hu, Y., He, X., Shen, W., Liu, C., et al. (2013). Phylogenetic Analysis of Brassica rapa MATH-Domain Proteins. Curr. Genom. 14, 214–223. doi: 10.2174/1389202911314030007
Keywords: abiotic stress, biotic stress, MATH domain, BTB domain, rice, Arabidopsis
Citation: Kushwaha HR, Joshi R, Pareek A and Singla-Pareek SL (2016) MATH-Domain Family Shows Response toward Abiotic Stress in Arabidopsis and Rice. Front. Plant Sci. 7:923. doi: 10.3389/fpls.2016.00923
Received: 08 April 2016; Accepted: 10 June 2016;
Published: 28 June 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Anil Kumar Singh, Indian Council of Agricultural Research (ICAR)-Indian Institute of Agricultural Biotechnology, IndiaJohn Paul Délano-Frier, Centro de Investigación y de Estudios Avanzados del I.P.N., Mexico
Copyright © 2016 Kushwaha, Joshi, Pareek and Singla-Pareek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sneh L. Singla-Pareek, c25laEBpY2dlYi5yZXMuaW4=; c25laHBhcmVla0BnbWFpbC5jb20=
†These authors have contributed equally to this work.
 Hemant R. Kushwaha
Hemant R. Kushwaha Rohit Joshi
Rohit Joshi Ashwani Pareek
Ashwani Pareek Sneh L. Singla-Pareek
Sneh L. Singla-Pareek