- 1Department of Computer Science, City University of Hong Kong, Hong Kong, China
- 2Pepper Institute, Zunyi Academy of Agricultural Sciences, Zunyi, China
- 3Guizhou Provincial College-based Key Lab for Tumor Prevention and Treatment with Distinctive Medicines, Zunyi Medical University, Zunyi, China
- 4BGI-Shenzhen, Shenzhen, China
- 5College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 6College of Horticulture, South China Agricultural University, Guangzhou, China
The microRNA (miRNA) can regulate the transcripts that are involved in eukaryotic cell proliferation, differentiation, and metabolism. Especially for plants, our understanding of miRNA targets, is still limited. Early attempts of prediction on sequence alignments have been plagued by enormous false positives. It is helpful to improve target prediction specificity by incorporating the other data sources such as the dependency between miRNA and transcript expression or even cleaved transcripts by miRNA regulations, which are referred to as trans-omics data. In this paper, we developed MiRTrans (Prediction of MiRNA targets by Trans-omics data) to explore miRNA targets by incorporating miRNA sequencing, transcriptome sequencing, and degradome sequencing. MiRTrans consisted of three major steps. First, the target transcripts of miRNAs were predicted by scrutinizing their sequence characteristics and collected as an initial potential targets pool. Second, false positive targets were eliminated if the expression of miRNA and its targets were weakly correlated by lasso regression. Third, degradome sequencing was utilized to capture the miRNA targets by examining the cleaved transcripts that regulated by miRNAs. Finally, the predicted targets from the second and third step were combined by Fisher's combination test. MiRTrans was applied to identify the miRNA targets for Capsicum spp. (i.e., pepper). It can generate more functional miRNA targets than sequence-based predictions by evaluating functional enrichment. MiRTrans identified 58 miRNA-transcript pairs with high confidence from 18 miRNA families conserved in eudicots. Most of these targets were transcription factors; this lent support to the role of miRNA as key regulator in pepper. To our best knowledge, this work is the first attempt to investigate the miRNA targets of pepper, as well as their regulatory networks. Surprisingly, only a small proportion of miRNA-transcript pairs were shared between degradome sequencing and expression dependency predictions, suggesting that miRNA targets predicted by a single technology alone may be prone to report false negatives.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs (~22-nt) that arise from short stem-loop precursors through the double-stranded ribonuclease (Bernstein et al., 2001). They are found widely exist in plants, animals, and bacteria. By directly binding to the transcripts, their regulations result in transcript cleavage or translational protein repression. The miRNAs serve as key regulators in cell proliferation, differentiation, metabolism, and apoptosis (Calin and Croce, 2006; Ameres and Zamore, 2013).
Although novel miRNAs remains to be discovered, exploring their targets is more crucial to understand the mechanisms of miRNA. Comparing with transcripts, miRNAs are of shorter lengths and they are supposed to bind their targets anywhere in the sequence. Many studies have been proposed to identify miRNA targets by sequence-based prediction tools (Zhang, 2005; Zhang et al., 2009; Dai and Zhao, 2011; Milev et al., 2011; Iossifov et al., 2014).
Sequence complementarity forms the basis of many miRNA target discovery tools. In plants, sequence complementarity is strongly observed, especially for the positions from the 2nd to the 13th of mature miRNA sequences (5′ end) with their targets (Dai and Zhao, 2011; Dai et al., 2011). Other characteristics of miRNA base-pairing were also exploited, e.g., target accessibility and the presence of multiple miRNA binding sites on the same transcript (Axtell et al., 2006; Brodersen and Voinnet, 2009). The free energy of sequence hybridization was also considered as an attribute of miRNA target prediction. The lower free energy required for binding, the more likely the binding site exists (Yue et al., 2009). RNAhybrid (Muckstein et al., 2006) was developed to utilize the free energy of hybridization, which successfully identified several novel targets of bantam in Drosophila, such as the hid, Nerfin-1, and Dll (Muckstein et al., 2006; Kertesz et al., 2007).
MiRNA targets predicted solely on sequence characteristics are glutted by false positives (Yue et al., 2009) since the sequences of miRNA-target pairs may be complementary by random chance. This may result in unproductive experimental validation. Alternative methods have been devised to eliminate false positives by investigating the expression correlations between miRNAs and their putative targets. The putative targets are removed if their expressions are weakly correlated with the miRNAs. These correlations are commonly computed by linear correlation (Xiao et al., 2009) or mutual information (Hsu et al., 2011). However, these two methods are unable to distinguish those false targets which co-express with the true ones. This issue can be alleviated by the predictive models such as multivariate linear regression (Jayaswal et al., 2009; Beck et al., 2011), regularized least square, and Bayesian network (Mootha et al., 2003; Carmona-Saez et al., 2007; Ragan et al., 2011). We applied transcriptome sequencing to quantify transcript expression, which was proved to be better than microarray (Iancu et al., 2012). On the other hand, degradome sequencing can help in identifying cleaved transcripts directly by sequencing the 5' ends of uncapped RNAs, which is considered as the miRNA regulation productions. Through this technology, Addo-Quaye et al. detected 100 potential miRNA targets in Arabidopsis (Addo-Quaye et al., 2008), which has been identified previously (Jones-Rhoades et al., 2006). Degradome sequencing also led to the identification of 160 targets of 53 miRNA families in rice (Li et al., 2010), including the CCS1, a novel and conserved target of miR398.
Most of the targets of a given miRNA are expected to be involved in the similar functions, especially for plant miRNAs. For instance, the targets of a human miRNA i.e., let-7b, are enriched in the genes (including PRDM2, DUSO9, OSMR, NDST2) from the same Gene Ontology annotation (Huang et al., 2007). This assumption allows one to refine miRNA targets by leveraging gene co-expression rank, as demonstrated in CoMeTa (Gennarino et al., 2012). Furthermore, for plants such as Arabidopsis, miRNA demonstrate a tendency to regulate genes from the same protein family. For example, miR156 and miR157 bind to the genes in Squamosa-promoter Binding Protein(SBP)-like proteins; the targets of miR170 and miR171 are enriched in GRAS domain proteins (SCARECROW-like) (Rhoades et al., 2002; Jones-Rhoades et al., 2006; Chen, 2009; Borges et al., 2011; Song et al., 2011).
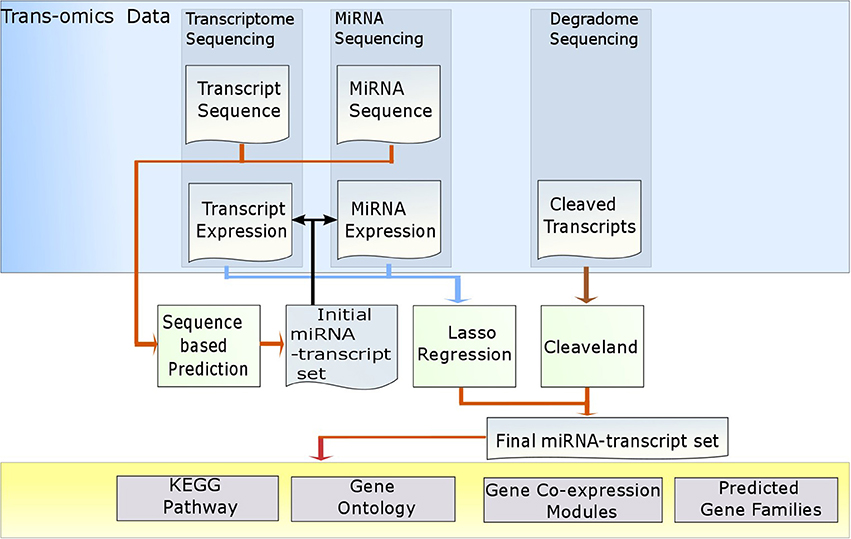
In this study, we developed MiRTrans, a program to predict MiRNA targets by Trans-omics data. The trans-omics data includes miRNA sequencing, transcriptome sequencing and degradome sequencing (Figure 1). MiRTrans consists of three steps. First, the potential miRNA targets were collected by combining the non-redundant targets predicted by sequence-based tools psRNATarget (Dai and Zhao, 2011) and Tapir (Bonnet et al., 2010). Second, the targets were eliminated if the expressions of miRNAs and their targets are weakly correlated by lasso regression. The significance of regression coefficients was evaluated by Wald test and they were usually zero for irrelevant targets through L1-norm constrain. Third, degradome sequencing was utilized to capture the miRNA targets by examining the cleaved transcripts regulated by miRNAs. The second and third steps were parallel and were based on the independent data sources. Finally, the predicted targets from these two steps were combined by Fisher's combination test. The p-values were further rectified by Bonferroni correction to deal with multiple testing issue. We reported MiRTrans predictions on Capsicum spp. the only plant with trans-omics data which we have full access to.
Our results showed MiRTrans produced more functional miRNA targets comparing with sequence-based predictions, where the targets from the same miRNA were prone to be enriched in Gene Ontology, KEGG pathway, gene co-expression modules, or predicted gene families. MiRTrans identified 58 miRNA-target pairs with high confidence for 18 miRNAs families conserved in eudicots. Most of these targets were transcription factors; this lent support to the role of miRNA as a key regulator in pepper. Our results also showed that only a small proportion of miRNA targets were shared between the predictions of miRNA-transcript expression dependency and degradome sequencing, which indicated that the miRNA targets may be lost if they were predicted by single data source (Section Results).
Methods
The MiRTrans Algorithm
The workflow of MiRTrans is described in Figure 1. To predict miRNA target more efficiently, MiRTrans incorporated the information from a variety of data sources including miRNA sequencing, transcriptome sequencing, and degradome sequencing. MiRTrans started from examining the characteristics of miRNA-transcript sequences by integrating the predictions from psRNATarget (Dai and Zhao, 2011) and Tapir (Bonnet et al., 2010). Next, lasso regression was applied to evaluate the expression dependency between miRNA-transcript pairs from sequence-based prediction. Weakly correlated pairs were considered to be less reliable and removed from initial pool (Section Methods). Parallelely, miRNA targets were also predicted by the cleaved transcripts which were thought as the productions of miRNA regulation (Section Methods). The two sets of miRNA targets were integrated and the p-values were calculated by Fisher's combination test followed by Bonferroni correction. Figure 2 illustrates an example on how MiRTrans works.
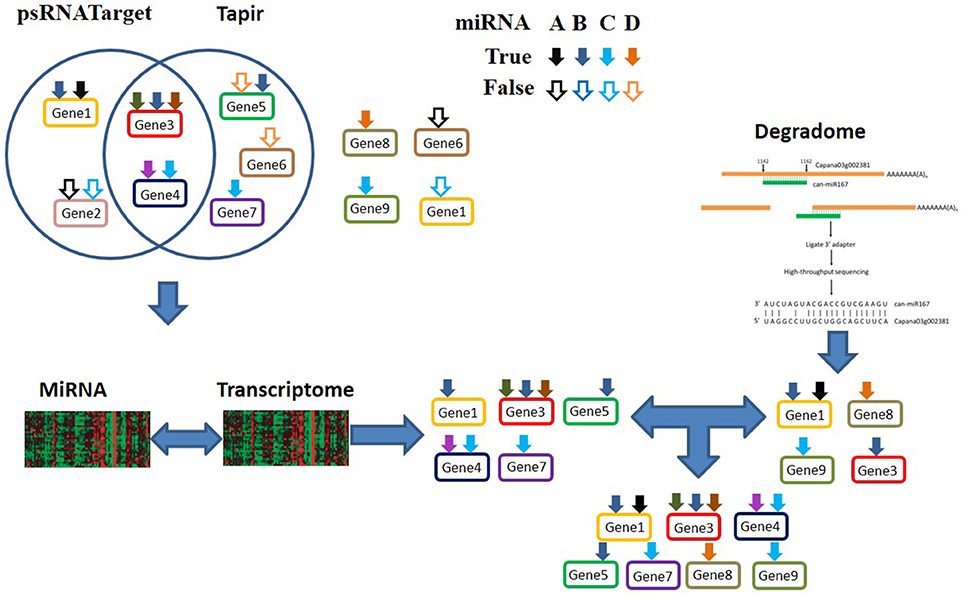
Figure 2. An example for the miRNA targets prediction by MiRTrans. Seven genes (Gene1 to Gene7) are assumed to be putative target genes of four miRNAs: A–D (demonstrated as black, dark blue, light blue and orange arrows), according to the non-redundant combination of psRNATarget and Tapir predictions. False positive miRNA-transcript pairs: A to Gene2, C to Gene2, D to Gene5, D to Gene6 are included the in sequence-based predictions. D to Gene8 and C to Gene9 are incorrectly missing in sequence-based predictions. By identifying the dependency between the expression of miRNAs and transcripts, MiRTrans refines the predictions by removing the false miRNA-transcript pairs from sequence-based predictions. Degradome sequencing data are incorporated to recoup those targets falsely removed by the previous steps (A to Gene1, B to Gene3, D to Gene8, C to Gene9).
Data Preparation
MiRNA Sequencing
The 17 samples for miRNA sequencing are derived from different development stages and tissues of Capsicum spp. The paired-end reads of five samples were available on Sequence Read Archive (SRP019257), and the other 12 sequenced samples were prepared for sequencing by Illumina HiSeq 2000.
Putative pre-miRNAs were inferred by aligning trimmed reads to the plant pre-miRNAs sequences in inmiRBase (Kozomara and Griffiths-Jones, 2014) by SOAP2 (Li et al., 2009). We removed the pre-miRNAs that failed in predicting miRNA secondary structure or positional overlaps and orientation of mature miRNA sequences within the respective stem-loop structure. The miRNA families were also limited if they were inconsistent with the confident miRNA annotation guidelines or current literature (Rhoades et al., 2002; Meyers et al., 2008; Hwang et al., 2013). We annotated 176 miRNAs with high confidence and their corresponding pre-miRNAs from 64 families (Table S1). The transcripts per million (TPM) was calculated for each miRNA followed by quantile normalization (Table S2).
Transcriptome Sequencing
The matched 17 samples were prepared for transcriptome sequencing. Raw reads of 14 samples were available on Sequence Read Archive (SRP019256) and the other three samples were newly sequenced. The sequences of pepper transcripts were downloaded from The Pepper Genome Database (http://peppersequence.genomics.cn/page/species/download.jsp). The transcripts were predicted by de novo gene prediction, homology searching and transcriptome inference (Qin et al., 2014). These reads were mapped to pepper reference genome by TopHat (Trapnell et al., 2009) allowing at most five mismatches; the RPKM was calculated for each transcript followed by quantile normalization (Table S3).
Degradome Sequencing
For degradome sequencing, we mixed equal amounts of five different tissues (flower, fruit, leaf, root, and stem) to capture the 5' ends of uncapped RNAs by using Illumina HiSeq 2000. CleaveLand classified miRNA hitting positions into four categories (Addo-Quaye et al., 2009). Category 0 and Category 1 were accorded on maximum depth and maximum value. If both maximum depth and maximum value were unique, these positions were labeled as Category 0. If multiple maximum depths and maximum values existed, these positions were labeled as Category 1. The positions with hit number larger than the median value and smaller than the maximum are labeled as Category 2. Other positions with more than one read coverage were labeled as Category 3. The positions covered by only one read were marked as Category 4. Targetfinder (Allen et al., 2005) aligned miRNAs to the cleaved transcripts of Category 0 or 1, followed by removing the alignments of distance more than 4.5 (Table S4).
MiRNA Target Predictions
Sequence-Based Predictions
For each miRNA, the putative miRNA targets were generated by combining two sequence-based prediction tools designed for plants, psRNATarget and Tapir. Because of a high degree of complementarity was required for miRNA binding in plants, we expected these two programs to arrive at similar targets. However, that turned out not to be the case (Figure 3). To avoid missing any putative targets, we merged the non-redundant predictions from the two programs.
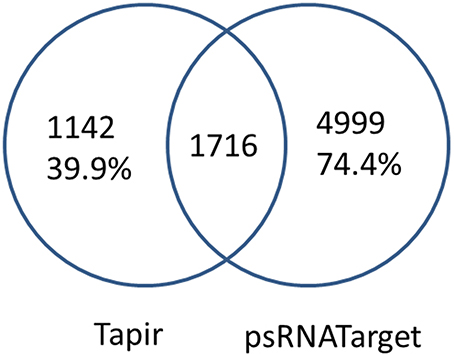
Figure 3. MiRNA targets prediction by psRNATarget and Tapir. Only 60.0% (Tapir) and 25.6% (psRNATarget) of the miRNA-transcript pairs are shared between Tapir and psRNATarget.
Lasso Regression to Determine Expression Dependency
Previous studies have shown sequence-based predictions are full of false positives. MiRTrans were eliminated these false positives by examining of the expression dependency between miRNAs and their targets. We assumed the expression of a particular transcript can be predicted by all the miRNAs that regulated it (Le et al., 2013). Lasso regression (Lu et al., 2011) was applied to alleviate over-fitting and remove irrelevant miRNAs. In lasso regression, the association between miRNAs and transcript was defined as “direct connection” by controlling the other miRNAs predicted to regulate the same transcript in sequence-based predictions.
For a transcript i(i = 1…L), its expression yi was modeled by a simple linear regression model, including those potential miRNA inferred to regulate transcript i in sequence-based predictions.
To address the over-fitting issue caused by incorporating too many predictors, lasso regression was applied by introducing L1-norm penalty to make the regression coefficient to be zero if the regulation did not exist.
where N is the number of sequenced samples (17 in this study). lasso regression was adopted to restrict by introducing the shrinkage parameter λ to control the sparsity of the regression model, which was determined by the minimum mean squared error from cross validation. There were no appropriate approximation approaches to evaluate the standard error of non-zero regression coefficients () from lasso regression. Therefore, we performed 10,000 bootstraps to estimate by fitting lasso regression with the same number of randomly selected miRNAs. The p-values to evaluate the expression dependency between miRNAs and the transcript were calculated by Wald test:
We assumed βnull = 0 for irrelevant miRNA and transcripts.
MiRNA Target Prediction by Degradome Sequencing
The potential transcript cleavage positions were determined by CleaveLand in conjunction with Targetfinder with default parameters. We chose the targets with alignment distance smaller than 4.5 and calculated the p-value as the likelihood of observing a degradome “peak” at the tenth nucleotide of the binding site. We adopted Fisher's combination test to incorporate the p-values from lasso regression and degradome sequencing followed by Bonferroni correction.
Validation Approaches
Because of lacking gold standard to evaluate the performance of MiRTrans, it was validated by calculating the functional enrichment of predicted targets. We assumed the real targets of a miRNA should be engaged in similar biological processes, and hence should be enriched in the particular functional modules. We collected the functional modules from four functional sources: Gene Ontology, KEGG pathway, gene co-expression modules, and predicted gene families. The Gene Ontology annotations for genes were determined by their InterPro (Mitchell et al., 2015) entries. All transcripts were aligned against KEGG (Kanehisa et al., 2014) (Release 58) proteins to determine the pathways they were involved in. We applied OrthoMCL (Li et al., 2003) to define the gene family as a group of genes that were descended from the identical gene in the most recent common ancestor of the considered species.
WGCNA (Weighted Gene Co-expression Network Analysis) (Langfelder and Horvath, 2008) was applied to identify the co-expression network of 16,357 expressed transcripts from 17 development stages (transcripts with RPKM>1 for all stages). The correlation between two transcripts xi and xj was calculated by the absolute value of the Pearson coefficient (|corr(xi, xj)|). WGCNA constructed the similarity matrix for all transcript pairs, which is further transformed to a weighted adjacency matrix by introducing power function , β = 12 was chosen to guarantee that the modules follow a scale-free topology. The total node connectivity of transcript i was defined as . WGCNA proposed a “topology overlap” (TO) to define the correlation between two transcripts by averaging the adjacency information over all their network “neighbors.” The TO between transcripts i and j was calculated as , where and it represented the degree of transcripts connection between i and j. The topological overlap matrix was composed by TO values and clustered by “dynamic tree cut” algorithm (Langfelder et al., 2008). The transcripts were clustered to co-expression modules according their topology overlap with the co-members in the same module.
Results
Sequence-Based Prediction
For sequence-based prediction, MiRTrans combined the non-redundant predictions from psRNATarget and Tapir (Figure 3), and assumed all the true miRNA-target pairs were included in the combined results. psRNATarget reported 6,715 pairs, 162 of which are found to have more than one potential binding sites in the same transcript. 76.5% of the targets were predicted as the cleavage effect of miRNA binding. Tapir created 2,858 miRNA-target pairs, of which merely 1,716 (60.0%) are shared with psRNATarget. Finally, 7,857 miRNA-target pairs are collected by combining the predictions of psRNATarget and Tapir.
The Expression Dependency between miRNAs and Transcripts
MiRTrans eliminated the miRNA-transcript pairs with insignificant p-values (Section Methods, p-value > 6.36e–6 = 0.05/7,857 from Bonferroni correction), which were calculated from their regression coefficients by Wald test. After removing the irrelevant targets removal, MiRTrans kept 363 pairs (4.62% in sequence-based predictions) by considering their expression dependency.
Degradome Sequencing Prediction
CleaveLand (Section Methods) was used to analyze the paired-end reads from degradome sequencing, which generated 586 miRNA-transcript pairs. The remaining 405 (69.1%) pairs, with p-values ≤ 0.05 and alignment distance <4.5, were selected for further analysis.
Comparing MiRTrans with Sequence-Based Predictions
MiRTrans produced 774 miRNA-transcript pairs by integrating the targets predicted by their expression dependencies and cleaved transcripts. To compare with sequence-based predictions, for each miRNA we generated 10,000 random networks, each including a miRNA ℜ and k targets ofℜ chosen from sequence-based predictions, where k was the number of targets of ℜ by MiRTrans. The functional enrichment from a variety of resources (KEGG pathway, Gene Ontology, Gene co-expression, and Predicted gene families) were applied to evaluate the performance of target prediction. We chose the functional modules with ≥ five genes from the four resources to avoid misleading significance. This resulted in 2,762 functional modules, including 275 KEGG pathways, 689 Gene Ontology terms, 112 gene co-expression modules and 1,686 predicted gene families. For the targets of each miRNA, the enrichment scores of the targets from MiRTrans were compared to the average value of 10,000 random networks. The significant functional modules (with p-value < , red circle in Figure 4) were illustrated to compare the predicted targets between MiRTrans and sequence-based predictions.
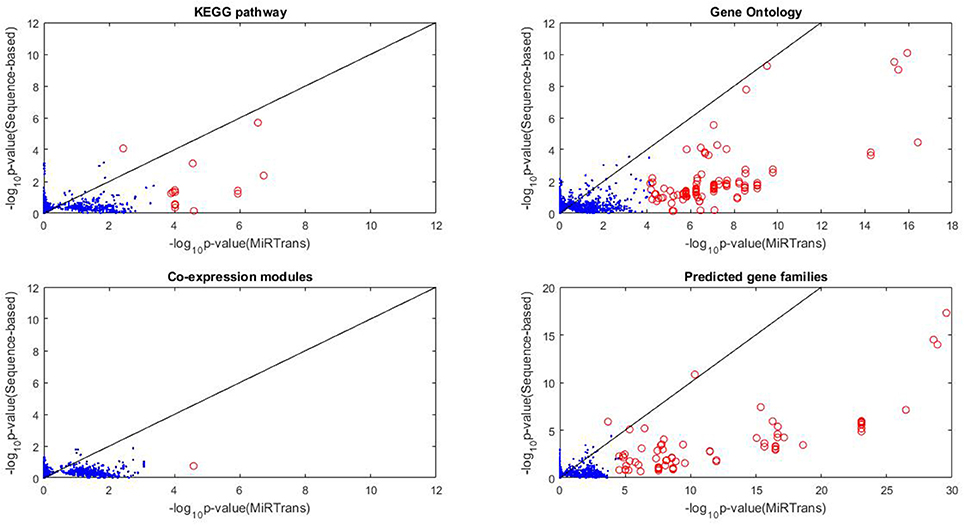
Figure 4. Functional enrichment comparison between MiRTrans and sequence-based predictions. MiRTrans achieved more functional miRNA targets than the sequence-based predictions.
Comparing with sequence-based predictions, MiRTrans produced more functional miRNA targets (p-value = 8.2252e-33 for predicted gene families, p-value = 7.8417e–32 for gene co-expression modules, and p-value = 3.7142e–15 for KEGG pathway). Furthermore, no significant difference was observed for Gene Ontology (p-value = 0.8173), because more sequence-based random networks were with marginal p-values. Nevertheless, more modules with significant p-values were prone to be included in MiRTrans predictions (Figure 4).
The predicted targets were also evaluated by the capability to infer the miRNA functions. We performed two experiments to compare the capability for predicting miRNA functions between MiRTrans and sequence-based predictions. In Figures 5, 6, we investigated the absolute number and the cumulative frequency of miRNAs, whose targets were significantly enriched in functional modules by an increment of p-value thresholds. Compared to sequence-based predications, more miRNAs were annotated by the functions of their targets involved in. The targets of a miRNA were more likely to be from the same gene family, not only because of their similar sequence characteristics, but also they participated in the same synthesis or growth procedure (Song et al., 2011). Using a p-value threshold of 1e–4, MiRTrans predicted the functions of 22% of total miRNAs, whereas sequence-based predictions achieved a rate of mere 3%. We cannot readily identify miRNA functions by co-expression modules or signaling pathways because they were usually regulated by multiple miRNAs.
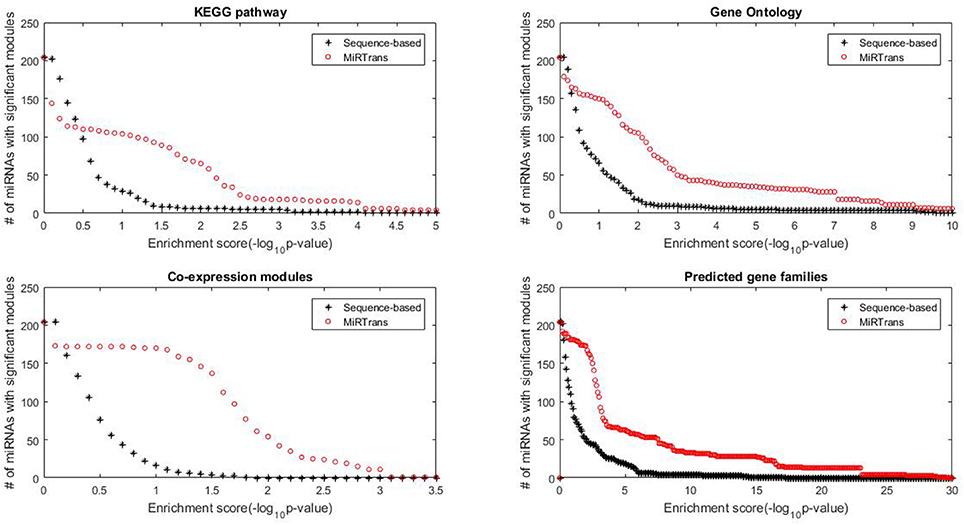
Figure 5. The absolute number of miRNAs, whose targets are enriched in at least one significant function module for MiRTrans and sequence-based predictions.
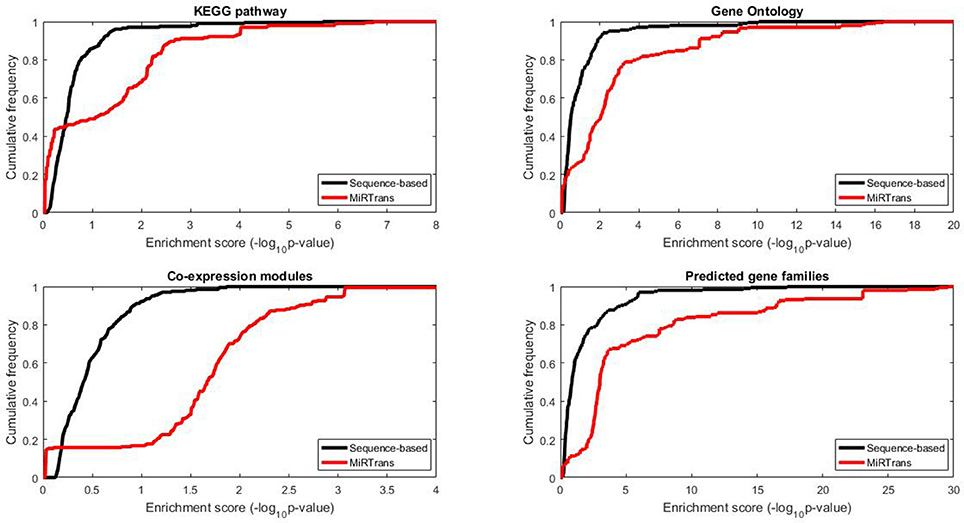
Figure 6. The cumulative frequency of miRNAs, whose targets are enriched in at least one significant function module for MiRTrans and sequence-based predictions.
It is interesting to distinguish that how many contributions of the targets predicted by expression dependency and degradome sequencing. We calculated the functional enrichment for the targets predicted by expression dependency and degradome sequencing, respectively. In Table 1, the p-values were calculated by Wilcoxon-Mann-Whitney test against sequence-based predictions. The targets predicted by expression dependency and degradome sequencing demonstrated consistent trend of functional enrichment even they only shared two miRNA-target pairs. This phenomenon suggested single data source was not enough to capture all the miRNA targets.
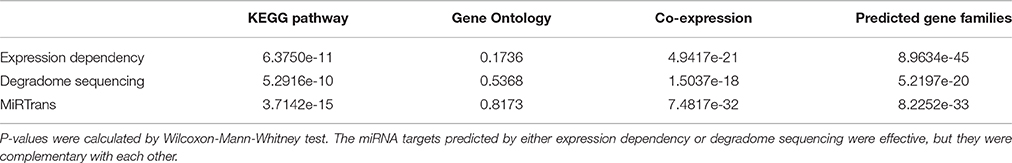
Table 1. Comparison of functional enrichment p-values of miRNA targets predicted by expression dependency and degradome sequencing.
Identification and Classification of the Targets for Pepper miRNAs
In our previous study, we reported 176 annotated miRNAs for Capsicum spp (Table S5) (Qin et al., 2014). In this study, after removing the transcripts shorter than 150 bp, MiRTrans revealed 186 targets of 18 conserved miRNA families and 68 targets of 15 pepper-specific miRNA families (Tables S6, S7). Additionally, there were 103 genes targeted by 26 pepper novel miRNA families (Tables S6, S7). Most of the targets of a conserved miRNA have similar sequence characteristics and belonging to the same gene family; this has been observed in the previous findings (Jones-Rhoades et al., 2006; Chen, 2009). We observed six conserved miRNAs, namely can-miR156, can-miR162, can-miR164, can-miR169, can-miR171, and can-miR172 with at least 10 targets, while other pepper-specific miRNAs appeared to have fewer targets except can-miR482, can-miR2873, and can-miR6149 (Tables S6, S7). We identified 18 out of 59 miRNA families binding to transcription factors; most of them were auxin response factors, TCP family transcription factors and NAC transcription factors (Table S7). These findings indicated the roles of these miRNA families in post-transcriptional regulation and transcriptional networks. Besides transcription factors, other identified targets were involved in macromolecule metabolic process, regulation of metabolic process, and nucleic acid binding etc. (Table S8).
The Comparison of miRNA Targets between Pepper and Other Plants
Several miRNA-transcript pairs predicted by MiRTrans were conserved across many plants, such as Arabidopsis, rice, soybean, and maize. Many predicted miRNA targets encoded regulatory proteins (Tables S7, S9), suggesting that miRNA served as a kind of master regulator in plant (Jones-Rhoades et al., 2006). MiRTrans predicted 58 conserved targets in pepper for 18 highly conserved miRNA families in eudicots (Arabidopsis, rice, maize, soybean, and pepper) with high confidence (Tables S9, S10). There were 46 targets of them that encoded transcription factors, which may play an important role in gene regulatory networks (Jones-Rhoades et al., 2006).
In addition, we retrieved putative orthologs of pepper miRNA targets based on the information from the EnsemblCompara gene trees (Vilella et al., 2009) on peppersequence.genomics.cn and gramene.org (Liang et al., 2008). We revealed 33 orthologs in Arabidopsis, 22 orthologs in soybean, 25 orthologs in rice, and 19 orthologs in maize (Table S7). Most of these orthologs were transcription factors and were known to regulate plant development (Table S9) (Jones-Rhoades et al., 2006). The miR156 family targeted SBP proteins and played a critical role in regulating phase change and floral induction (Kasschau et al., 2003; Chen et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Wu and Poethig, 2006; Addo-Quaye et al., 2008; Wu et al., 2009). Previous studies reported that miR172 bound to a set of AP2 transcription factors (Addo-Quaye et al., 2008; Li et al., 2010; Song et al., 2011; Lee et al., 2014; Liu et al., 2014; Wang et al., 2014) (Table S9), but presented a variety of functions in different plants: Controlled genes related to flowering time and floral organ in Arabidopsis (Aukerman and Sakai, 2003; Kasschau et al., 2003; Jung et al., 2007; Mathieu et al., 2009; Wang et al., 2009; Wu et al., 2009); regulated inflorescence development in maize (Chuck et al., 2007a,b); involved in the regulation of vegetative and reproductive branching in rice (Zhu et al., 2009; Lee et al., 2014; Wang et al., 2015). The shared miRNA targets among Arabidopsis, maize, rice, and pepper suggested the existence of a similar mechanism of phase change and flowering time control. Similarly, miR159 regulated MYB genes and had various roles in flower development of Arabidopsis (Palatnik et al., 2003; Achard et al., 2004; Millar and Gubler, 2005; Alonso-Peral et al., 2010) and rice (Li et al., 2010), while miR319 controlled floral organ size and shape in Arabidopsis (Palatnik et al., 2003; Nag et al., 2009). Our predictions were consistent with previous studies of miR164, which regulated NAC genes that function in organ boundary formation (Laufs et al., 2004; Guo et al., 2005; Peaucelle et al., 2007; Sieber et al., 2007) (Table S9). In those non-transcription factor targets (Table S10), DCL1 and AGO1, which were predicted as miR162 and miR168 targets, played a key role in tuning plant biogenesis and function (Xie et al., 2003; Vaucheret et al., 2004; Vazquez et al., 2004).
Discussion
We presented MiRTrans, a trans-omics based program, to infer miRNA targets from three data sources: miRNA sequencing, transcriptome sequencing, and degradome sequencing. To our best knowledge, this is the first investigation to analyze and compare the contributions of different available technologies and trans-omics data sources for predicting miRNA targets. MiRTrans extracted and utilized three types of information: First, sequence characteristics, such as miRNA-target sequence complementarity, the sequence hybridization energy, and target site multiplicity; second, the expression dependency between miRNA and transcript; third, the cleaved transcripts were detected by sequencing the 5' ends of uncapped RNAs. These cleaved transcripts were assumed to be the productions of miRNA regulation. We further evaluated the performance of MiRTrans by comparing the targets functional enrichment with sequence-based predictions. These results supported that MiRTrans can generate more functional relevant miRNA targets than sequence-based predictions. The source code of MiRTrans is publicly available on https://github.com/zhanglu295/MiRTrans.
Degradome sequencing, designed to capture cleaved transcripts, has been widely applied in plants to predict miRNA targets. Successful stories were reported in Arabidopsis (Addo-Quaye et al., 2008), rice (Li et al., 2010), soybean (Song et al., 2011), and apple (Xia et al., 2012), it has yet to be proved whether all the targets were captured. There are three limitations of degradome sequencing in miRNA target prediction. First, RISC-mediated transcript cleavage cannot explain all the miRNA regulation in plant. The miRNAs regulation is via two molecular mechanisms: transcriptional degradation and translational repression (Jones-Rhoades et al., 2006; Gandikota et al., 2007). Degradome sequencing can only capture those cleaved transcripts rather than those repressed proteins. For example, miR172 regulates flowering time and floral organ by repressing the protein of APETALA2 (AP2), rather than degrade its transcripts (Aukerman and Sakai, 2003; Chen, 2004; Schwab et al., 2005). Second, the sample for degradome sequencing were collected from mixed tissues in this study, the transcripts with low expression may be overwhelmed in the highly expressed transcripts; third, CleaveLand is the most famous program to analyze degradome sequencing data, but it applies Targetfinder to determine which miRNAs bind to the cleaved transcripts. Targetfinder is a sequence-based miRNA target prediction program for plant, and cannot avoid sequence supplementarity by random chance.
The expression dependency between miRNA and transcript is an indirect evidence in target prediction and is easily influenced by gene co-expression and miRNA temporospatial specificity. The transcripts may be incorrectly predicted as miRNA targets, if they are observed highly co-expressing with the real targets. miRNA regulation is unlikely to occur anytime and anywhere, which results in the unstable trend of expression correlations. Degradome sequencing alleviates these issues by directly sequencing the cleaved transcripts from mixed samples. Comparing the targets predicted by degradome sequencing and expression dependency, we found most of them were unique and had the same trend in functional enrichment, suggesting these two data sources were orthogonal. MiRTrans reduced the number of miRNA-target pairs from 7,857 to 774 (9.85%) and a significant reduction (from 148 to 20) of average targets for each miRNA from sequence-based predictions. The targets predicted by MiRTrans are more likely to be involved in the same predefined functional modules than those from sequence-based predictions.
The availability of next generation sequencing provides us an unprecedented opportunity to predict miRNA targets from trans-omics data. In this paper, we introduced MiRTrans, a trans-omics based program, that leveraged miRNA sequencing, transcriptome sequencing and degradome sequencing to predict miRNA targets. MiRTrans generated an atlas of the miRNA targets for pepper. Some of these targets have orthologs in other plants such as Arabidopsis, rice, maize, and soybean as validated by RLM-5' RACE, degradome, and/or miRNA-resistant/Agroinfiltration experiments. We discovered that data from different sources are orthogonal to each other, which suggested that previously reported miRNA targets that rely on a single data source may be incomplete.
Author Contributions
SL and KH supervised the work and together with LZ, CQ, and JM, developed MiRTrans and experiments. LZ, JM implemented the MiRTrans method, XC, ZW, XL, JC, and XT did the experiments on pepper. CQ and KH provided degradome sequencing data and other trans-omics data. LZ, CQ, and SL wrote the manuscript. All authors have read and approved the final manuscript.
Funding
The work described in this paper was supported by a grant of GRF Project from RGC General Research Fund [9041901 (CityU 118413)], the Zunyi City Natural Science Foundation of China (No. 201201), the Guizhou Province and Zunyi City Science and Technology Cooperation Project of China (No. 201307, 201542), the Zunyi County Technology Cooperation Project (SSX201407), the National Natural Science Foundation of China (31372076), and Key Lab Construction Project of the Educational Department of Guizhou Province (Guizhou Education Cooperation KY [2014] 212).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00495/full#supplementary-material
References
Achard, P., Herr, A., Baulcombe, D. C., and Harberd, N. P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. doi: 10.1242/dev.01206
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P., and Axtell, M. J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. doi: 10.1016/j.cub.2008.04.042
Addo-Quaye, C., Miller, W., and Axtell, M. J. (2009). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131. doi: 10.1093/bioinformatics/btn604
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Alonso-Peral, M. M., Li, J., Li, Y., Allen, R. S., Schnippenkoetter, W., Ohms, S., et al. (2010). The miR159 regulated GAMYB-like genes inhibit growth and promote Programmed Cell Death in Arabidopsis. Plant Physiol. 154, 757–771. doi: 10.1104/pp.110.160630
Ameres, S. L., and Zamore, P. D. (2013). Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488. doi: 10.1038/nrm3611
Aukerman, M. J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
Axtell, M. J., Jan, C., Rajagopalan, R., and Bartel, D. P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. doi: 10.1016/j.cell.2006.09.032
Beck, D., Ayers, S., Wen, J., Brandl, M. B., Pham, T. D., Webb, P., et al. (2011). Integrative analysis of next generation sequencing for small non-coding RNAs and transcriptional regulation in Myelodysplastic Syndromes. BMC Med. Genomics 4:19. doi: 10.1186/1755-8794-4-19
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Bonnet, E., He, Y., Billiau, K., and Van De Peer, Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26, 1566–1568. doi: 10.1093/bioinformatics/btq233
Borges, F., Pereira, P. A., Slotkin, R. K., Martienssen, R. A., and Becker, J. D. (2011). MicroRNA activity in the Arabidopsis male germline. J. Exp. Bot. 62, 1611–1620. doi: 10.1093/jxb/erq452
Brodersen, P., and Voinnet, O. (2009). Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10, 141–148. doi: 10.1038/nrm2619
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866. doi: 10.1038/nrc1997
Carmona-Saez, P., Chagoyen, M., Tirado, F., Carazo, J. M., and Pascual-Montano, A. (2007). GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 8:R3. doi: 10.1186/gb-2007-8-1-r3
Chen, J., Li, W. X., Xie, D., Peng, J. R., and Ding, S. W. (2004). Viral virulence protein suppresses RNA silencing–mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16, 1302–1313. doi: 10.1105/tpc.018986
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chen, X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. doi: 10.1146/annurev.cellbio.042308.113417
Chuck, G., Cigan, A. M., Saeteurn, K., and Hake, S. (2007a). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39, 544–549. doi: 10.1038/ng2001
Chuck, G., Meeley, R., Irish, E., Sakai, H., and Hake, S. (2007b). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517–1521. doi: 10.1038/ng.2007.20
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Dai, X., Zhuang, Z., and Zhao, P. X. (2011). Computational analysis of miRNA targets in plants: current status and challenges. Brief. Bioinform. 12, 115–121. doi: 10.1093/bib/bbq065
Gandikota, M., Birkenbihl, R. P., Hohmann, S., Cardon, G. H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49, 683–693. doi: 10.1111/j.1365-313X.2006.02983.x
Gennarino, V. A., D'angelo, G., Dharmalingam, G., Fernandez, S., Russolillo, G., Sanges, R., et al. (2012). Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 22, 1163–1172. doi: 10.1101/gr.130435.111
Guo, H.-S., Xie, Q., Fei, J.-F., and Chua, N.-H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Hsu, S. D., Lin, F. M., Wu, W. Y., Liang, C., Huang, W. C., Chan, W. L., et al. (2011). miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39, D163–D169. doi: 10.1093/nar/gkq1107
Huang, J. C., Babak, T., Corson, T. W., Chua, G., Khan, S., Gallie, B. L., et al. (2007). Using expression profiling data to identify human microRNA targets. Nat. Methods 4, 1045–1049. doi: 10.1038/nmeth1130
Hwang, D.-G., Park, J. H., Lim, J. Y., Kim, D., Choi, Y., Kim, S., et al. (2013). The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: a foundation for understanding microRNA functional roles in hot pepper. PLoS ONE 8:e64238. doi: 10.1371/journal.pone.0064238
Iancu, O. D., Kawane, S., Bottomly, D., Searles, R., Hitzemann, R., and Mcweeney, S. (2012). Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics 28, 1592–1597. doi: 10.1093/bioinformatics/bts245
Iossifov, I., O'roak, B. J., Sanders, S. J., Ronemus, M., Krumm, N., Levy, D., et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. doi: 10.1038/nature13908
Jayaswal, V., Lutherborrow, M., Ma, D. D., and Hwa Yang, Y. (2009). Identification of microRNAs with regulatory potential using a matched microRNA-mRNA time-course data. Nucleic Acids Res. 37, e60. doi: 10.1093/nar/gkp153
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Jung, J.-H., Seo, Y.-H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N.-H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A., et al. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. doi: 10.1016/S1534-5807(03)00025-X
Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., and Segal, E. (2007). The role of site accessibility in microRNA target recognition. Nat. Genet. 39, 1278–1284. doi: 10.1038/ng2135
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9:559. doi: 10.1186/1471-2105-9-559
Langfelder, P., Zhang, B., and Horvath, S. (2008). Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720. doi: 10.1093/bioinformatics/btm563
Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. doi: 10.1242/dev.01320
Le, T. D., Liu, L., Tsykin, A., Goodall, G. J., Liu, B., Sun, B. Y., et al. (2013). Inferring microRNA-mRNA causal regulatory relationships from expression data. Bioinformatics 29, 765–771. doi: 10.1093/bioinformatics/btt048
Lee, Y.-S., Lee, D.-Y., Cho, L.-H., and An, G. (2014). Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7, 1–13. doi: 10.1186/s12284-014-0031-4
Li, L., Stoeckert, C. J. Jr., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Li, R., Yu, C., Li, Y., Lam, T.-W., Yiu, S.-M., Kristiansen, K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Li, Y. F., Zheng, Y., Addo−Quaye, C., Zhang, L., Saini, A., Jagadeeswaran, G., et al. (2010). Transcriptome−wide identification of microRNA targets in rice. Plant J. 62, 742–759. doi: 10.1111/j.1365-313X.2010.04187.x
Liang, C., Jaiswal, P., Hebbard, C., Avraham, S., Buckler, E. S., Casstevens, T., et al. (2008). Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 36, D947–D953. doi: 10.1093/nar/gkm968
Liu, H., Qin, C., Chen, Z., Zuo, T., Yang, X., Zhou, H., et al. (2014). Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics 15:25. doi: 10.1186/1471-2164-15-25
Lu, Y., Zhou, Y., Qu, W., Deng, M., and Zhang, C. (2011). A Lasso regression model for the construction of microRNA-target regulatory networks. Bioinformatics 27, 2406–2413. doi: 10.1093/bioinformatics/btr410
Mathieu, J., Yant, L. J., Murdter, F., Kuttner, F., and Schmid, M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7:e1000148. doi: 10.1371/journal.pbio.1000148
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., et al. (2008). Criteria for annotation of plant MicroRNAs. Plant Cell 20, 3186–3190. doi: 10.1105/tpc.108.064311
Milev, I., Yahubyan, G., Minkov, I., and Baev, V. (2011). miRTour: plant miRNA and target prediction tool. Bioinformation 6, 248–249. doi: 10.6026/97320630006248
Millar, A. A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Mitchell, A., Chang, H. Y., Daugherty, L., Fraser, M., Hunter, S., Lopez, R., et al. (2015). The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221. doi: 10.1093/nar/gku1243
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. doi: 10.1038/ng1180
Muckstein, U., Tafer, H., Hackermuller, J., Bernhart, S. H., Stadler, P. F., and Hofacker, I. L. (2006). Thermodynamics of RNA-RNA binding. Bioinformatics 22, 1177–1182. doi: 10.1093/bioinformatics/btl024
Nag, A., King, S., and Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Peaucelle, A., Morin, H., Traas, J., and Laufs, P. (2007). Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134, 1045–1050. doi: 10.1242/dev.02774
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Ragan, C., Zuker, M., and Ragan, M. A. (2011). Quantitative prediction of miRNA-mRNA interaction based on equilibrium concentrations. PLoS Comput. Biol. 7:e1001090. doi: 10.1371/journal.pcbi.1001090
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. doi: 10.1016/j.devcel.2005.01.018
Sieber, P., Wellmer, F., Gheyselinck, J., Riechmann, J. L., and Meyerowitz, E. M. (2007). Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060. doi: 10.1242/dev.02817
Song, Q.-X., Liu, Y.-F., Hu, X.-Y., Zhang, W.-K., Ma, B., Chen, S.-Y., et al. (2011). Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 11:5. doi: 10.1186/1471-2229-11-5
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Vaucheret, H., Vazquez, F., Crété, P., and Bartel, D. P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. doi: 10.1101/gad.1201404
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Vilella, A. J., Severin, J., Ureta-Vidal, A., Heng, L., Durbin, R., and Birney, E. (2009). EnsemblCompara genetrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19, 327–335. doi: 10.1101/gr.073585.107
Wang, J.-W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, L., Sun, S., Jin, J., Fu, D., Yang, X., Weng, X., et al. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 15504–15509. doi: 10.1073/pnas.1521949112
Wang, Y., Wang, L., Zou, Y., Chen, L., Cai, Z., Zhang, S., et al. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26, 4782–4801. doi: 10.1105/tpc.114.131607
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Wu, G., Park, M. Y., Conway, S. R., Wang, J.-W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Xia, R., Zhu, H., An, Y. Q., Beers, E. P., and Liu, Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13:R47. doi: 10.1186/gb-2012-13-6-r47
Xiao, F., Zuo, Z., Cai, G., Kang, S., Gao, X., and Li, T. (2009). miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37, D105–D110. doi: 10.1093/nar/gkn851
Xie, Z., Kasschau, K. D., and Carrington, J. C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789. doi: 10.1016/S0960-9822(03)00281-1
Yue, D., Liu, H., and Huang, Y. (2009). Survey of computational algorithms for microRNA target prediction. Curr. Genomics 10, 478–492. doi: 10.2174/138920209789208219
Zhang, L., Chia, J.-M., Kumari, S., Stein, J. C., Liu, Z., Narechania, A., et al. (2009). A genome-wide characterization of microRNA genes in maize. PLoS Genet. 5:e1000716. doi: 10.1371/journal.pgen.1000716
Zhang, Y. (2005). miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 33, W701–W704. doi: 10.1093/nar/gki383
Keywords: pepper (Capsicum spp.), miRNA targets, miRNA sequencing, transcriptome sequencing, degradome sequencing, lasso regression
Citation: Zhang L, Qin C, Mei J, Chen X, Wu Z, Luo X, Cheng J, Tang X, Hu K and Li SC (2017) Identification of MicroRNA Targets of Capsicum spp. Using MiRTrans—a Trans-Omics Approach. Front. Plant Sci. 8:495. doi: 10.3389/fpls.2017.00495
Received: 27 July 2016; Accepted: 21 March 2017;
Published: 10 April 2017.
Edited by:
Chun Liang, Miami University, USAReviewed by:
Praveen Kumar Raj Kumar, Chan Soon-Shiong Institute of Molecular Medicine at Windber, USAHarsh Dweep, Heidelberg University, Germany
Copyright © 2017 Zhang, Qin, Mei, Chen, Wu, Luo, Cheng, Tang, Hu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kailin Hu, aHVrYWlsaW5Ac2NhdS5lZHUuY24=
Shuai Cheng Li, c2h1YWljbGlAY2l0eXUuZWR1Lmhr
†These authors have contributed equally to this work.
 Lu Zhang
Lu Zhang Cheng Qin
Cheng Qin Junpu Mei
Junpu Mei Xiaocui Chen2
Xiaocui Chen2 Zhiming Wu
Zhiming Wu Xirong Luo
Xirong Luo Jiaowen Cheng
Jiaowen Cheng Kailin Hu
Kailin Hu Shuai C. Li
Shuai C. Li