- 1Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3State Key Laboratory of Cotton Biology, Institute of Plant Stress Biology, Henan University, Kaifeng, China
- 4Department of Biology, East Carolina University, Greenville, NC, United States
The spread of photosynthesis is one of the most important but constantly debated topics in eukaryotic evolution. Various hypotheses have been proposed to explain the plastid distribution in extant eukaryotes. Notably, the chromalveolate hypothesis suggested that multiple eukaryotic lineages were derived from a photosynthetic ancestor that had a red algal endosymbiont. As such, genes of plastid/algal origin in aplastidic chromalveolates, such as oomycetes, were considered to be important supporting evidence. Although the chromalveolate hypothesis has been seriously challenged, some of its supporting evidence has not been carefully investigated. In this study, we re-evaluate the “algal” genes from oomycetes with a larger sampling and careful phylogenetic analyses. Our data provide no conclusive support for a common photosynthetic ancestry of stramenopiles, but show that the initial estimate of “algal” genes in oomycetes was drastically inflated due to limited genome data available then for certain eukaryotic lineages. These findings also suggest that the evolutionary histories of these “algal” genes might be attributed to complex scenarios such as differential gene loss, serial endosymbioses, or horizontal gene transfer.
Introduction
How photosynthesis evolved in eukaryotes has been a subject of tremendous scientific interest. Oxygenic photosynthesis was first invented by cyanobacteria (Gould et al., 2008). During the early evolution of eukaryotes, a cyanobacterial cell was engulfed by a heterotrophic eukaryote (Margulis, 1970; Martin and Kowallik, 1999; McFadden, 2001; Palmer, 2003), spawning the origin of primary plastids and Plantae (also called Archaeplastida, including green plants, red algae, and glaucophytes) (Cavalier-Smith, 1981; Delwiche and Palmer, 1997; Gould et al., 2008). This process was accompanied by massive cyanobacterial gene loss and transfer to the host nucleus. Subsequently, the photosynthetic capacity was spread to multiple other eukaryotic lineages through higher-order endosymbioses (secondary, tertiary, or quaternary) (Delwiche, 1999), that is, these eukaryotes acquired plastids by engulfing another photosynthetic eukaryote instead of a cyanobacterial cell. Although it is clear that the spread of photosynthetic capacity in eukaryotic lineages represents a history of reticulate evolution involving multiple endosymbiotic events, the exact number and the nature of historical endosymbioses remain controversial.
Among eukaryotic lineages involved in higher-order endosymbioses, it was generally accepted that plastids of euglenids and chlorarachniophytes are derived from green algal endosymbionts (Gibbs, 1978; Ludwig and Gibbs, 1989; Van de Peer et al., 1996), whereas plastids of cryptophytes, alveolates, stramenopiles and haptophytes (CASH lineages) are from red algal endosymbionts (Cavalier-Smith, 1995; Cavalier-Smith et al., 1996; Palmer and Delwiche, 1998; Delwiche, 1999). For a long period of time, plastid gains through endosymbiotic events were considered to be extremely rare and plastid losses, on the other hand, were thought to be relatively common. Such a belief also formed the foundation of the Cabozoa hypothesis and the chromalveolate hypothesis (Cavalier-Smith, 1999; Cavalier-Smith and Chao, 2003). The Cabozoa hypothesis argued that plastids of euglenids and chlorarachniophytes could be traced back to a common secondary endosymbiotic event involving a green alga (Cavalier-Smith, 1999; Cavalier-Smith and Chao, 2003). Similarly, the chromalveolate hypothesis proposed that plastids in CASH lineages were vertically derived from a common ancestor that engulfed a red algal endosymbiont and, as such, aplastidic organisms in these lineages were interpreted as resulting from secondary plastid losses (Cavalier-Smith, 1999). However, multiple lines of evidence (Baldauf et al., 2000; Archibald et al., 2003; Leander, 2004; Gilson et al., 2006), including the complete chloroplast genome of chlorarachniophyte Bigelowiella natans (Rogers et al., 2007), rejected the Cabozoa hypothesis. Thus far, it is commonly believed that the plastids of euglenids and chlorarachniophytes were acquired from two independent green algal endosymbiotic events (Gould et al., 2008; Keeling, 2013). The chromalveolate hypothesis has long been under debate and now in jeopardy in face of recent data (Bodył, 2005; Burki et al., 2008, 2012b, 2016; Kim and Graham, 2008; Yoon et al., 2008; Bodył et al., 2009). Several other hypotheses have been proposed, each of which is supported by different lines of evidence (Bodył and Moszczyński, 2006; Sanchez-Puerta and Delwiche, 2008; Okamoto et al., 2009; Stiller et al., 2014). Therefore, how plastids evolved in red plastid lineages remains unsettled.
Stramenopiles (also known as heterokonts), as a major eukaryotic clade, include a wide variety of organisms (Cavalier-Smith, 1986; Patterson, 1989). This lineage contains not only many important algae, such as diatoms that are a major producer of oxygen and consumer of carbon dioxide in marine ecosystems, but also a significant fraction of aplastidic or heterotrophic organisms, including pathogens like Phytophthora infestans, the causative agent of potato late blight that triggered the Great Irish Famine in the 1840s. Whether these diverse organisms originated from a common photosynthetic ancestor is crucial for understanding the evolution of stramenopiles as well as eukaryotes in general. This in turn led to many studies on the existence of potential historical plastids in heterotrophic stramenopiles (Keeling, 2013).
Oomycetes are fungus-like eukaryotic microorganisms that often have a saprophytic or pathogenic lifestyle. Oomycetes were once placed within fungi in earlier classification systems, but are now widely considered as part of stramenopiles (Baldauf et al., 2000; Yoon et al., 2002). Although there are different views about the phylogenetic relationships within stramenopiles (Brown and Sorhannus, 2009; Riisberg et al., 2009; Yang et al., 2012; Cavalier-Smith and Scoble, 2013; Ševčíková et al., 2015), the most recent phylogenomic analyses suggest that oomycetes form a clade closely related to ochrophytes, a monophyletic group of photosynthetic stramenopiles (Derelle et al., 2016). Unlike ochrophytes, oomycetes do not contain plastids (Tyler et al., 2006; Derelle et al., 2016), not even vestigial ones like those in apicomplexan parasites (called apicoplast) (Maréchal and Cesbron-Delauw, 2001). If all stramenopiles are derived from a single photosynthetic ancestor, plastids would have been lost in oomycetes.
In 2006, draft genome sequences of two oomycete species, Phytophthora sojae and P. ramorum, were published (Tyler et al., 2006). In this study, 855 genes of putatively algal origin (“algal” genes hereafter) were identified based on their unusually high similarities to sequences from algae and/or cyanobacteria, 30 of which were considered the most likely cases after detailed phylogenetic analyses. These “algal” genes were interpreted as the relic from a red algal endosymbiont (plastid) and subsequent endosymbiotic gene transfer (EGT) or endosymbiotic gene replacement (EGR). As key evidence for historical plastids in oomycetes, these “algal” genes were further used to support the hypothesis that all stramenopiles were derived from a photosynthetic ancestor. Such evidence, however, has been called into question by a more recent statistical genomic analysis that found no unusual contribution from a red algal endosymbiont to Phytophthora genomes (Stiller et al., 2009).
Like in many earlier studies on gene transfer, insufficient taxonomic sampling was a potential caveat for the identification of “algal” genes in oomycetes. This is evidenced by the fact that, although the identified “algal” genes were interpreted as derived from a red algal endosymbiont, their sequences, on the other hand, were often found to be closely related to green algal homologs, presumably due to the lack of sufficient sequence data from red algae. As more genome sequence data from various major eukaryotic lineages become available in recent years, we now revisit the “algal” genes identified in oomycete genomes. Our goal is to provide a better understanding of the nature of these genes and the potential interactions of oomycetes/stramenopiles with other organisms, particularly primary photosynthetic eukaryotes.
Materials and Methods
Data Sources
In the original Phytophthora genome analyses, 855 genes were considered to be of algal or cyanobacterial origin, and 30 most likely candidates were subject to further detailed analyses (Tyler et al., 2006). We downloaded the protein sequences of these 30 genes of Phytophthora ramorum from http://www.jgi.doe.gov/Pramorum, and used them as queries to search the National Center for Biotechnology Information (NCBI) non-redundant (nr) protein sequences database (E-value cutoff 1e-7). Additional searches were also performed against over 650 transcriptomes in the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) (Keeling et al., 2014), the fungal genome database at the Joint Genome Institute1, and our internal customized database (Supplementary Table 1). Particularly, a total of six red algal genomes of five genera were used to search for P. ramorum gene homologs, including Cyanidioschyzon merolae, Porphyridium purpureum, Chondrus crispus, Galdieria sulphuraria, G. phlegrea, and Pyropia yezoensis. Complete genome sequence data of multiple photosynthetic stramenopiles (including Aureococcus anophagefferens, Ectocarpus siliculosus, Fragilariopsis cylindrus, Nannochloropsis gaditana, Phaeodactylum tricornutum, Saccharina japonica, and Thalassiosira pseudonana) were also searched in our analyses.
Re-analyses of BLAST Results
In the original Phytophthora genome paper (Tyler et al., 2006), the search of “algal” genes was based on significant matches to sequences from Plantae and cyanobacteria (that is, these sequence matches had the highest bit scores and the lowest E-values outside the stramenopiles). The Phytophthora “algal” genes identified from the BLAST search were shared by other stramenopiles (or chromalveolates), and they had stronger BLAST matches to homologous genes of red algae and/or cyanobacteria than to sequences from archaea, opisthokonts or non-cyanobacterial bacteria. Additionally, a complementary approach based on Smith–Waterman alignment was also used to identify candidates with significantly higher similarities to red algal or green plant homologs than to those from opisthokonts or amoebozoans. Because Cyanidioschyzon merolae, which also happens to have a streamlined genome, was the only red alga whose complete nuclear genome sequence was then available, the matches to green plant homologs were included and interpreted as resulting from the lack of sufficient red algal genome data (Tyler et al., 2006).
In the current study, BLAST search was performed against NCBI nr, MMETSP and our internal customized databases for each of the 30 most likely “algal” genes, followed by re-analyses of its phylogenetic distribution and gene structure. Following the criteria used by the Phytophthora genome paper (Tyler et al., 2006), we compared the best BLAST matches (represented by the highest bit scores) between homologs from red algae, cyanobacteria, photosynthetic stramenopiles and those from archaea, opisthokonts, amoebozoans, and non-cyanobacterial bacteria. Because more red algal genomes and transcriptomic data were included in our analyses, the matches to green plant homologs were no longer included and used as proxy for red algal homologs.
Phylogenetic Analyses
For each of the 30 “algal” genes identified in Phytophthora genome sequencing project, we performed further phylogenetic analyses. In order to attain a broad and balanced sampling, we selected protein sequences from representative groups of three domains of life (eukaryotes, bacteria, and archaea). The same sampling strategy was also used to ensure sufficient coverage of representative taxa within each major eukaryotic group. This was done using a Perl script followed by manual inspection and additional sequence sampling if needed. Particular attention was paid to groups under-sampled in the previous analyses, such as chromalveolates and other protists. Multiple alignments of sampled sequences were performed using MUSCLE (Edgar, 2004), followed by careful manual inspection of alignment quality, gene structure, shared insertions/deletions (indels), and conserved amino acid residues. Gaps, ambiguously aligned sites, and sequences whose real identity could not be confirmed were removed from alignments. Phylogenetic analyses were performed with maximum likelihood method using PhyML 3.1 (Guindon et al., 2010) and distance method using neighbor of PHYLIP-3.695 (Felsenstein, 2013). The optimal model of protein substitution and rate heterogeneity were chosen based on the result of ModelGenerator (Keane et al., 2006). Bootstrap analyses were performed using 1,000 replicates.
Results
The Identity of “Algal” Genes in Oomycetes
If the previously identified “algal” genes in Phytophthora are indeed derived from a red algal endosymbiont acquired by the ancestor of stramenopiles, their homologs might also be found in photosynthetic stramenopiles. Given their presumably red algal nature, these stramenopile sequences theoretically should have a closer relationship to homologs from red algae (or red algae and other photosynthetic eukaryotes plus cyanobacteria) than to those from other organisms (e.g., opisthokonts, amoebozoans, non-cyanobacterial bacteria, and archaea). Our BLAST results with a larger taxonomic sampling showed that, for all of the 30 most likely “algal” genes previously identified in Phytophthora, only 10 of them (about 33%) were more similar to sequences of red algae, photosynthetic eukaryotes and/or cyanobacteria (Table 1); these 10 genes were also the viable candidate genes of red algal origin.
Of the 30 “algal” genes in Phytophthora, nine (30%) showed stronger BLAST matches (represented by higher bit scores) to homologs from opisthokonts, amoebozoans, non-cyanobacterial bacteria or archaea than to those from photosynthetic stramenopiles (Table 1). Another gene encoding methylthioadenosine phosphorylase (P. ramorum Gene ID 86425) had no detectable homologs in sequenced photosynthetic stramenopiles. Although the possibility of differential gene losses cannot be ruled out, genes with such a distribution pattern may also suggest an independent origin in oomycetes, such as horizontal gene transfer (HGT) from prokaryotes or other eukaryotes to oomycetes. Moreover, 16 of these 30 genes (about 53%) had more significant matches to homologs from opisthokonts, amoebozoans, non-cyanobacterial bacteria or archaea than to those from red algae and cyanobacteria (Table 1). Particularly, four genes had the strongest BLAST matches in non-cyanobacterial bacteria, and two in opisthokonts or amoebozoans. This observation based on simple pairwise comparisons suggests that many of these “algal” genes in oomycetes have no stronger similarity to photosynthetic stramenopile, red algal or cyanobacterial sequences. If sequence similarity is largely correlated with sequence relatedness, as commonly believed, the nature of these “algal” genes might be seriously questioned.
We further performed phylogenetic analyses on each of these 30 “algal” genes to evaluate its origin. If an oomycete gene is of red algal origin, the gene and its stramenopile (or chromalveolate) homologs are expected to form a clade sister to red algal and/or cyanobacterial sequences. This, however, was not the pattern uncovered in our study. Tree topologies for 21 (70%) genes were poorly supported overall (or the position of oomycete sequences couldn’t be confidently determined), thus providing no sufficient evidence for any evolutionary scenarios (Supplementary Materials). These poorly supported tree topologies might be caused by multiple issues, for example, insufficient phylogenetic signal or heterogeneity in evolutionary rates. Nevertheless, such topologies, combined with the information of phylogenetic distribution from BLAST search, should not be interpreted as evidence for a red algal origin of involved Phytophthora genes. The remaining genes had relatively well-resolved phylogenies and will be detailed in the following sections.
Algal or Cyanobacterial Genes in Oomycetes
In our analyses, several of these “algal” genes indeed showed a close affinity with algal or cyanobacterial sequences. In addition, for 12 “algal” genes previously identified in Phytophthora, the protein products of their plant and/or algal homologs are localized in plastids (Tyler et al., 2006), as predicted by TargetP (Emanuelsson et al., 2000) (Table 1). It is well known that proteins of organelles-derived genes are often re-imported into the original organelles (mitochondria or plastids) to participate in related biochemical activities (Bogorad, 1975; Ellis, 1981; Weeden, 1981; Timmis et al., 2004). This information has been frequently used as supplemental evidence for genes of organellar origin. However, such affinity with algal/cyanobacterial sequences or functionality in other plastids might not necessarily support the suggestion of a historical red algal endosymbiont in the ancestral stramenopile.
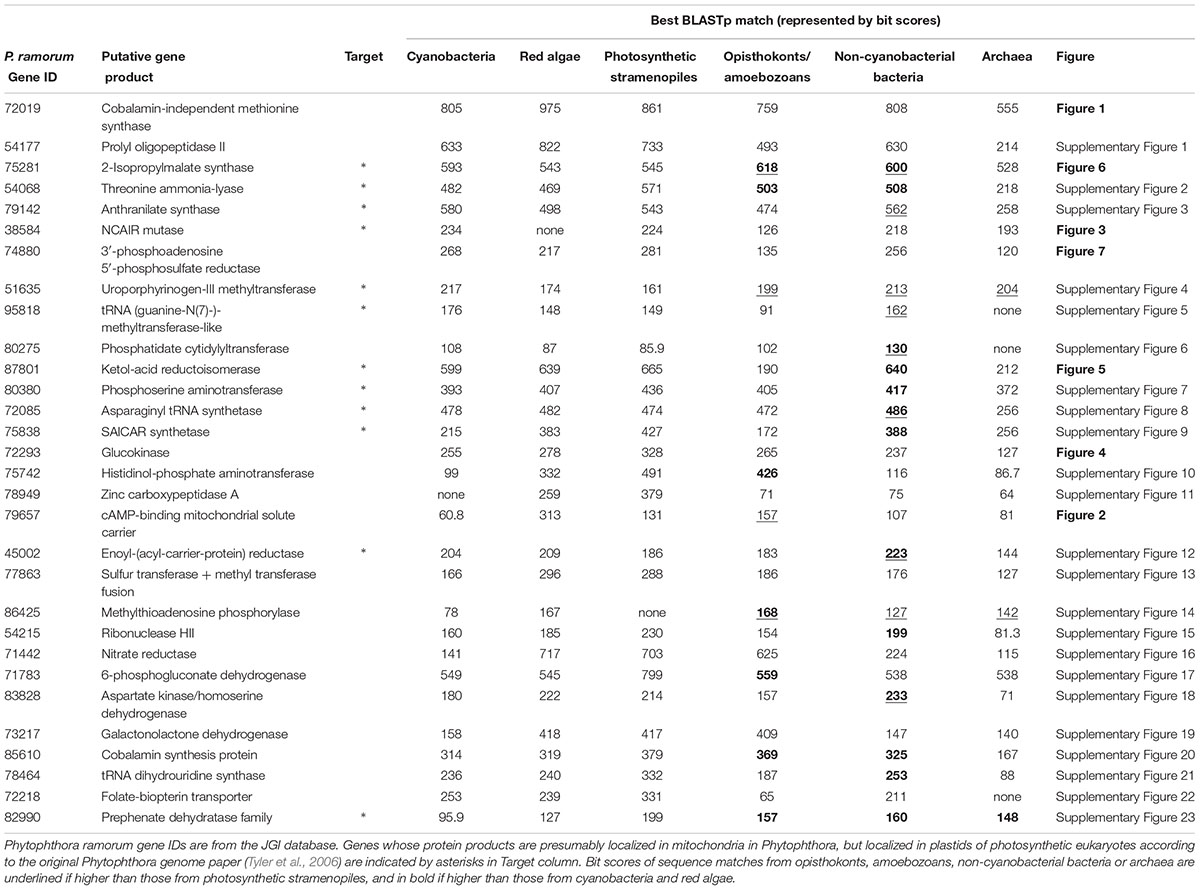
The most likely “algal” gene uncovered in our current study encodes cobalamin-independent methionine synthase (MetE). Our phylogenetic analyses indicated that MetE sequences from oomycetes, photosynthetic stramenopiles, chlorarachniophytes, chromerids and cryptophytes formed a large group with homologs of red algae, green algae, and cyanobacteria (Figure 1). Within this group, oomycete MetE sequences formed a strongly supported clade with red algal instead of other stramenopile homologs. Although the overall molecular phylogeny of MetE is consistent with an algal origin of oomycetes and, to a certain extent, an algal/cyanobacterial origin of all stramenopiles, the strength of this evidence is somewhat compromised by the fact that oomycete and other stramenopile sequences didn’t form a monophyletic group (see Discussion). Two other similar cases are related to the genes encoding prolyl oligopeptidase II (Supplementary Figure 1) and cAMP-binding mitochondrial solute carrier (Figure 2). For both genes, their molecular phylogenies showed that oomycete and red algal sequences were closely related. Particularly in the latter case, a NLPC_P60 and two CAP_ED domains are uniquely shared by oomycetes and red algae, but are absent from other stramenopiles (Figure 2). A parsimonious explanation for these findings would be that oomycetes obtained this gene from red algae directly or vice versa.
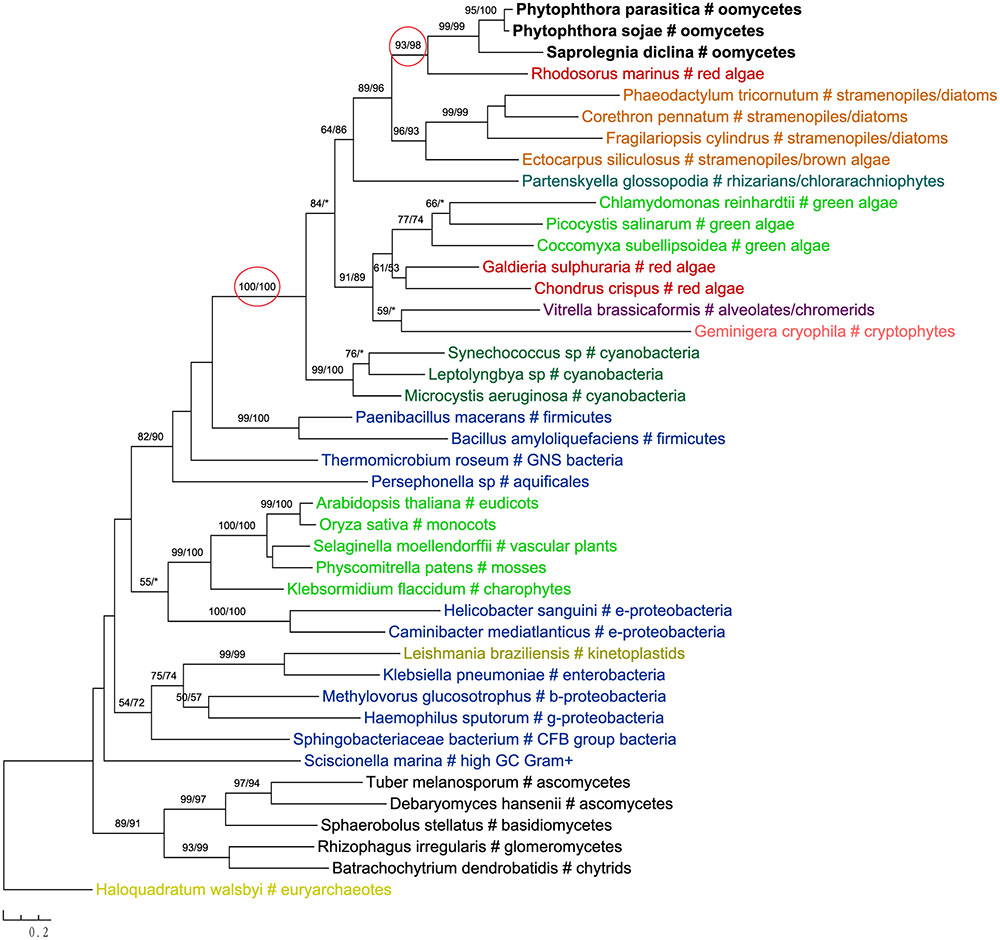
FIGURE 1. Molecular phylogeny of cobalamin-independent methionine synthase (MetE). Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks.
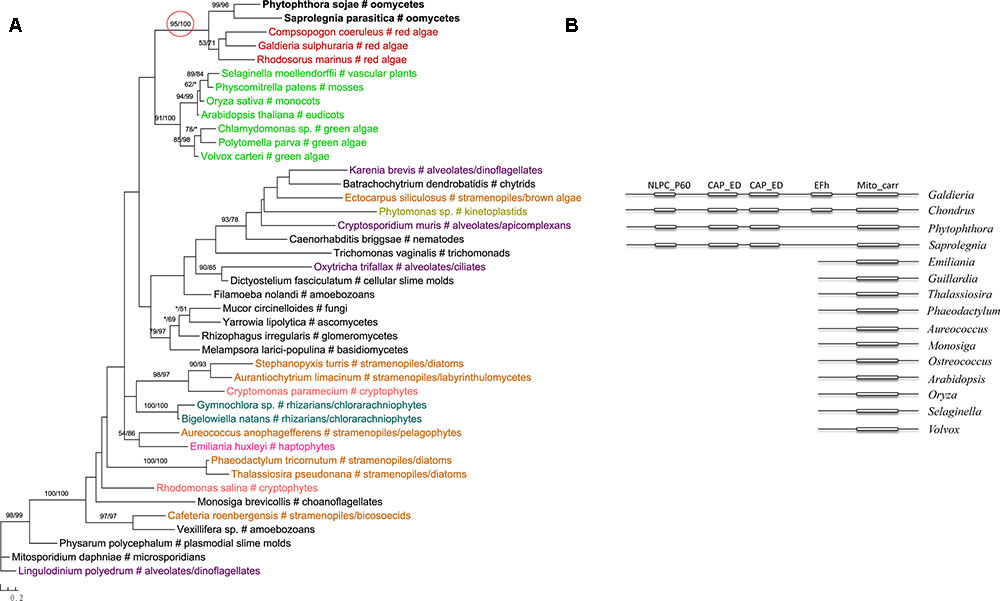
FIGURE 2. (A) Molecular phylogeny of cAMP-binding mitochondrial solute carrier. Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks. (B) Schematic gene structure and domain composition of the cAMP-binding mitochondrial solute carrier gene in different lineages. Boxes show individual domains.
Two genes in our analyses were found to be specifically related to green plant sequences, which is in disagreement with the suggestion of a red algal plastid in the ancestral stramenopile. The gene encoding NCAIR mutase does not have detectable homologs in red algae. Phylogenetic analyses of NCAIR mutase supported a monophyletic group including sequences from oomycetes, photosynthetic stramenopiles, dinoflagellates, green algae and cyanobacteria (Figure 3). Because of the lack of detectable NCAIR mutase homologs in red algae, a red algal origin of this gene in all stramenopiles cannot be concluded. On the other hand, an independent green algal endosymbiont in stramenopiles might potentially explain such a distribution pattern (Moustafa et al., 2009; Dorrell and Smith, 2011). The other green plants-related gene in oomycetes encodes a probable folate-biopterin transporter (Supplementary Figure 22). Our analyses showed that sequences from oomycetes, diatom Thalassionema frauenfeldii and land plants formed a strongly supported clade, whereas other photosynthetic sequences, including red algae and cyanobacteria, formed another large group with only modest support.
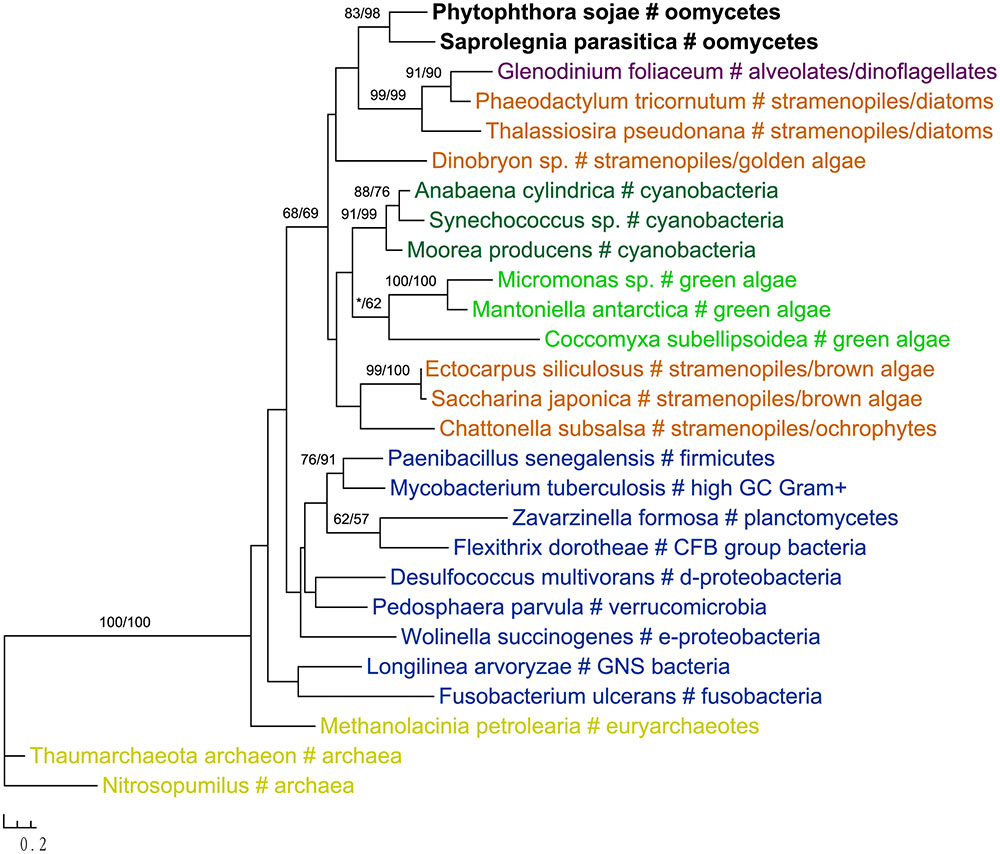
FIGURE 3. Molecular phylogeny of NCAIR mutase. Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks.
In addition to primary algae and cyanobacteria, several groups of eukaryotes that have secondary plastids through higher-order endosymbioses might also be potential donors for genes in oomycetes. For instance, phylogenetic analyses of glucokinase indicated that sequences from oomycetes, haptophytes and ciliates formed a well-supported clade, which in turn grouped with homologs from photosynthetic stramenopiles, red algae, green algae, choanoflagellates and dinoflagellates (Figure 4). A similar case was also observed for the gene encoding ketol-acid reductoisomerase (Figure 5). As it is known that ciliates contain sequences of algal origin (Reyes-Prieto et al., 2008), this topology might suggest HGT from haptophytes-related groups to oomycetes, and again provides no support for a common photosynthetic origin between oomycetes and other stramenopiles.
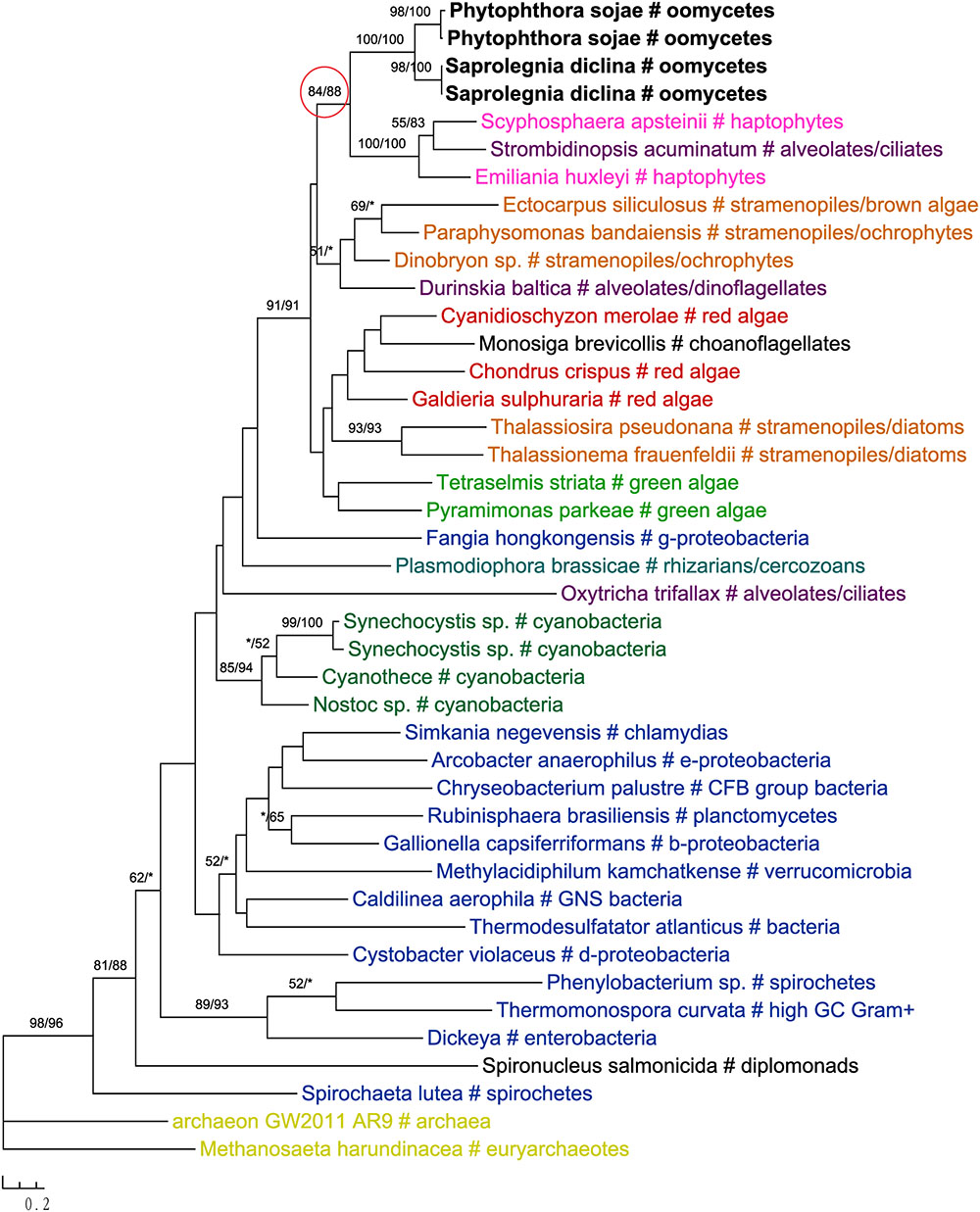
FIGURE 4. Molecular phylogeny of glucokinase. Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks.
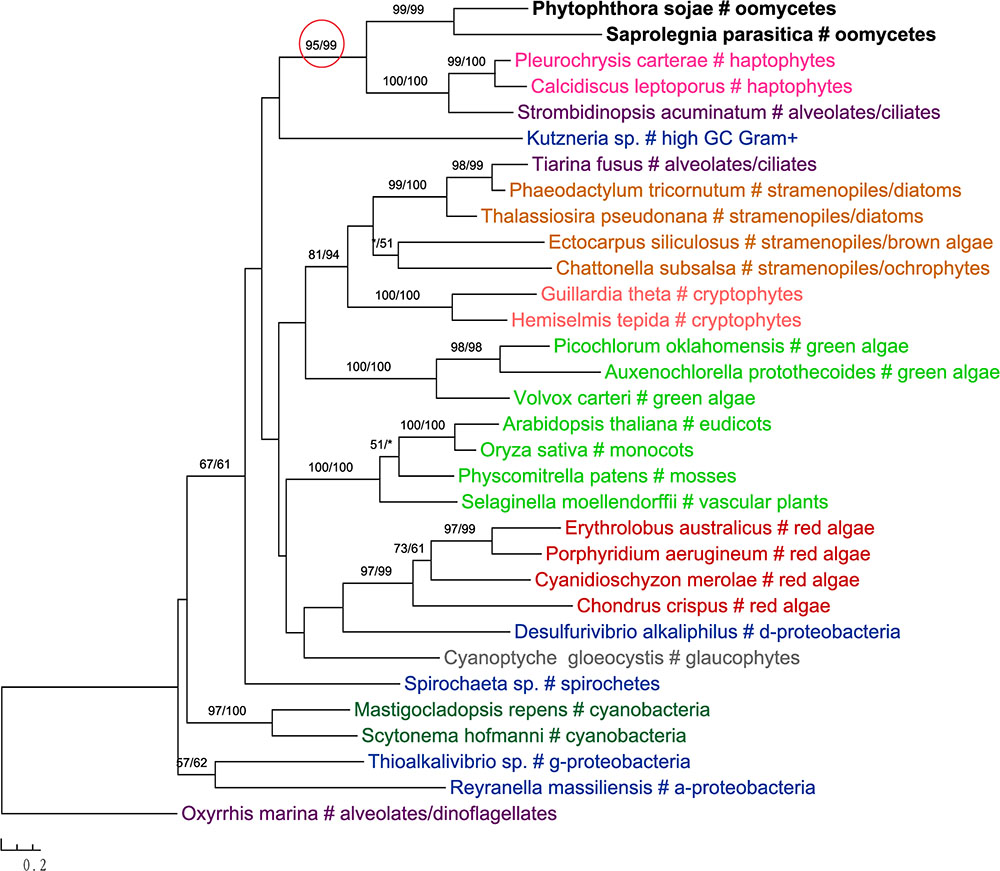
FIGURE 5. Molecular phylogeny of ketol-acid reductoisomerase. Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks.
Other Potential Evolutionary Scenarios
As indicated above, a large fraction of “algal” genes in oomycetes/stramenopiles showed stronger matches in our BLAST search to homologs from opisthokonts, amoebozoans or non-cyanobacterial bacteria rather than those from red algae and cyanobacteria. For several of these genes, this relationship was also confirmed by subsequent phylogenetic analyses.
One of these genes encodes 2-isopropylmalate synthase in leucine biosynthesis and was previously detailed in the Phytophthora genome paper (Tyler et al., 2006). According to the authors, this gene was subject to at least two transfer events in eukaryotes: sequences of primary photosynthetic eukaryotes and stramenopiles (including oomycetes) were derived from cyanobacteria, whereas sequences of fungi were from α-proteobacteria. Specifically, diatom sequences were found to group with green plant rather than red algal homologs, which was interpreted as a separate ancestry or artifacts due to incomplete sampling (Tyler et al., 2006). Our current analyses support the previous conclusion that this gene in stramenopiles might have different origins, but also suggest a potentially more complicated evolutionary scenario. While sequences from brown algae and cryptophytes indeed grouped with red algal homologs, those from diatoms and Aureococcus with green plant sequences instead (Figure 6). The relationships between brown algae, cryptophytes and red algae uncovered here is in line with the suggestion of serial endosymbioses by Stiller et al. (2014), where a red alga was first adopted by a cryptophyte that was in turn engulfed by ochrophytes. The sequence affiliation between diatoms, Aureococcus and green algae might point to separate origins of this gene in other photosynthetic stramenopiles [e.g., from a potential green algal endosymbiont (Moustafa et al., 2009; Dorrell and Smith, 2011) or an independent HGT event]. Nevertheless, unlike previously reported in the Phytophthora genome paper (Tyler et al., 2006), oomycete sequences grouped with labyrinthulomycetes, another group of heterotrophic stramenopiles, and other eukaryotes, rather than being affiliated with diatoms, primary photosynthetic eukaryotes and cyanobacteria (Figure 6).
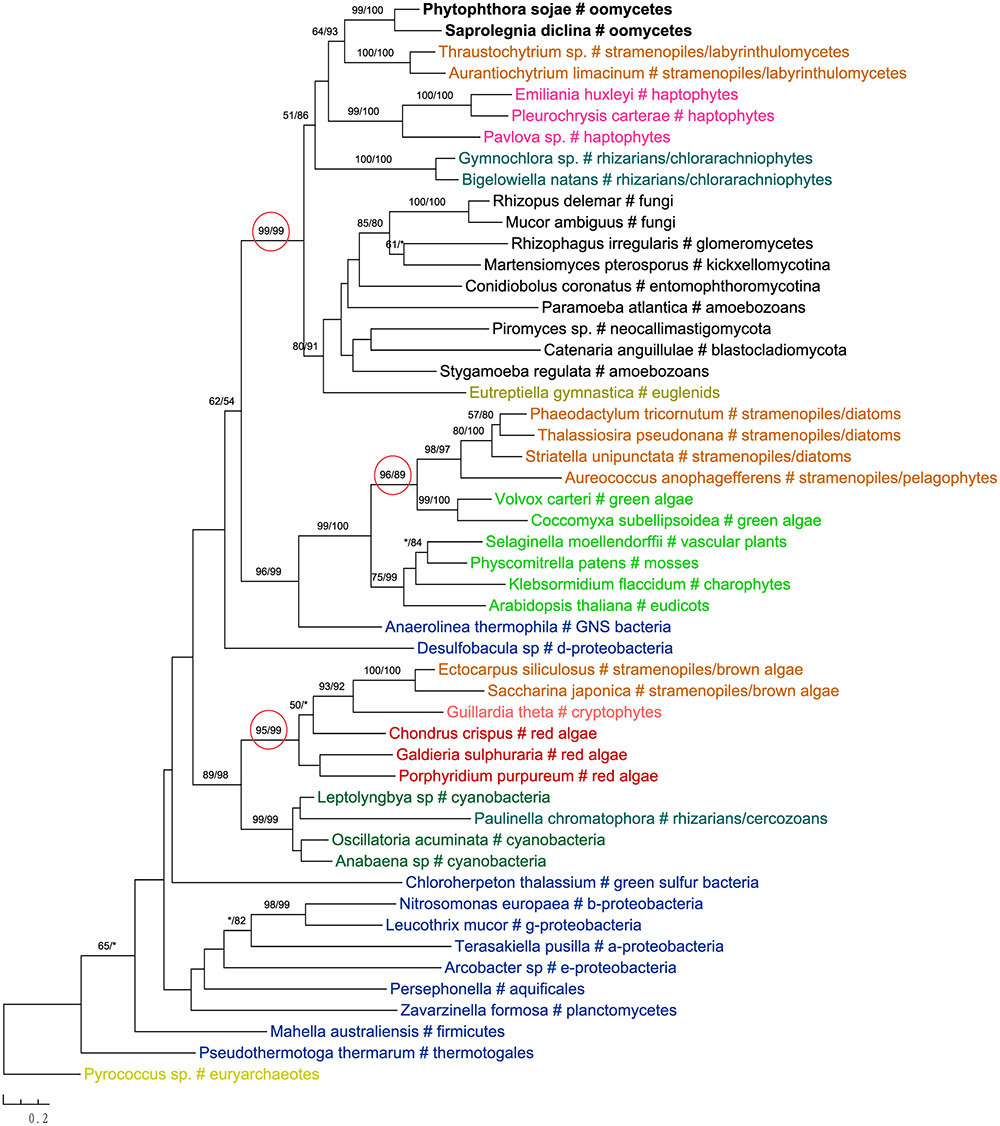
FIGURE 6. Molecular phylogeny of 2-isopropylmalate synthase. Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks.
The gene encoding 3′-phosphoadenosine 5′-phosphosulfate reductase (PAPR), an enzyme in the sulfate assimilation pathway, is another example highlighting the potential pitfalls of insufficient sampling. PAPR and adenosine 5′-phosphosulfate reductase (APR) are homologous proteins and have a complex evolutionary history in eukaryotes (Kopriva et al., 2002; Kopriva and Koprivova, 2004; Patron et al., 2008). The APR gene was previously thought to exist in land plants, algae, and phototrophic bacteria. PAPR, on the other hand, was initially identified mainly in fungi and bacteria (Kopriva et al., 2002). Several more recent studies reported PAPR sparely in phototrophic eukaryotes, suggesting potential HGT events (Kopriva and Koprivova, 2004; Kopriva et al., 2007; Patron et al., 2008). Particularly, the study of Patron et al. (2008) indicated a potential bacterial origin of PAPR in P. sojae. With a much larger taxonomic sampling, our analyses showed that sequences from some stramenopiles (including oomycetes), bacteria (both cyanobacteria and non-cyanobacteria) and Paulinella chromatophora formed a major PAPR clade (Figure 7). The cyanobacterial origin of PAPR in P. chromatophora is somewhat expected, as this species contains an independently evolved plastid organelle (cyanobacterial endosymbiont) (Marin et al., 2005). As the overall topology of this clade is poorly supported, whether PAPR in stramenopiles was derived from a red algal endosymbiont or a separate HGT event could not be answered by our study.
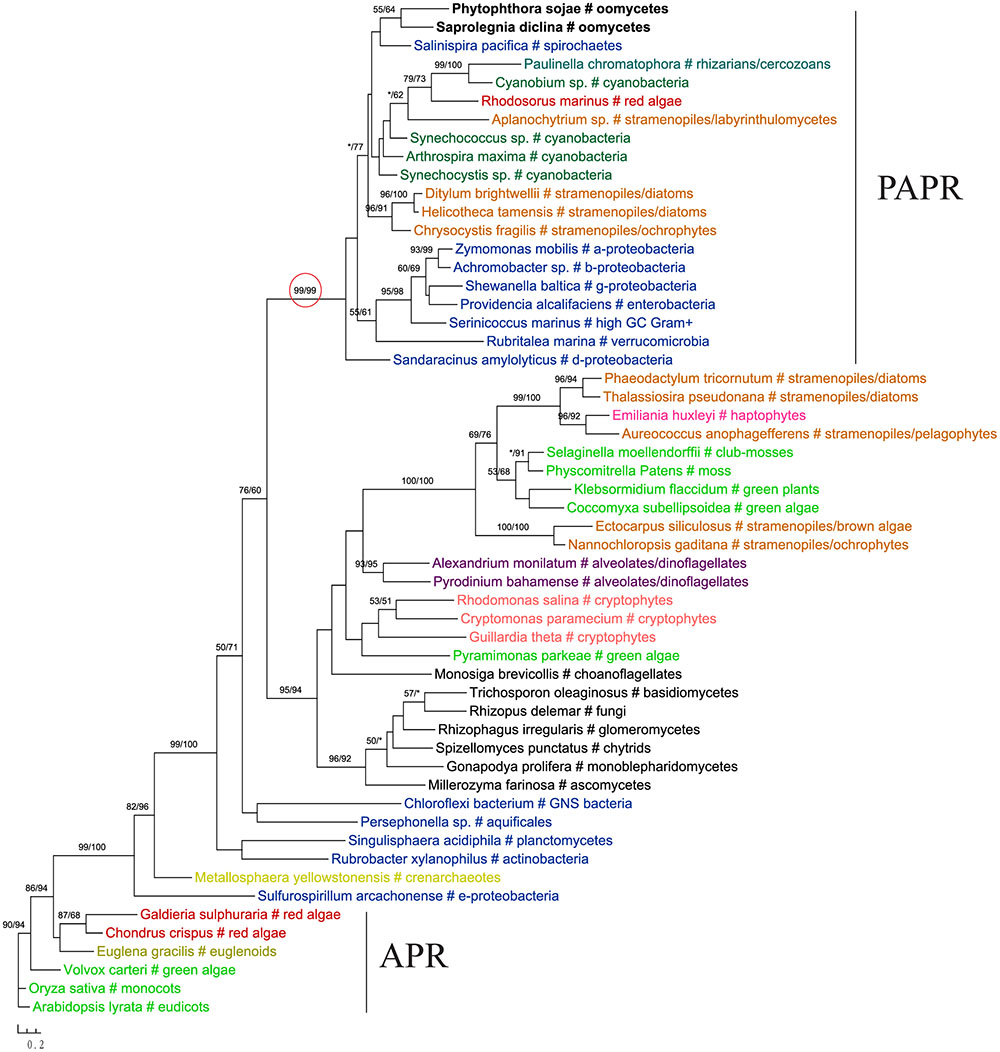
FIGURE 7. Molecular phylogeny of 3′-phosphoadenosine 5′-phosphosulfate reductase (PAPR) and adenosine 5′-phosphosulfate reductase (APR). Numbers above branches show bootstrap values in percentage for maximum likelihood and distance analyses, respectively. Values below 50% are indicated by asterisks.
Discussion
As evidence of historical plastids in oomycetes, the “algal” genes identified in Phytophthora genomes were used to support a common photosynthetic ancestry of stramenopiles, and the chromalveolate hypothesis in general. In the Phytophthora genome paper (Tyler et al., 2006), the identification of “algal” genes was heavily based on significant matches to sequences from red algae or cyanobacteria. Because the identification of foreign genes in eukaryote can be affected by taxonomic samplings and methods of analyses, studies on algal genes in different eukaryotes sometimes led to different interpretations after re-analyses. For example, 263 red algal genes and 250 green plant genes were reported in Chromera velia (Woehle et al., 2011), but only 23 and nine of them, respectively, were confirmed after re-evaluation (Burki et al., 2012a). When more stringent criteria were applied, the number of putative green algal genes in diatoms decreased from 1,700 (Moustafa et al., 2009) to only 144 (Dorrell and Smith, 2011). While an algal endosymbiont in the common ancestor of stramenopiles or any other lineages could certainly be a significant source of foreign genes, other issues, notably phylogenetic artifacts, insufficient sampling, differential gene losses and independent HGT events, could all lead to the same or similar atypical gene distributions or relationships.
With a much larger sampling and careful phylogenetic analyses, we revisited the 30 most likely “algal” genes identified in the Phytophthora genomes (Tyler et al., 2006). Our results show that the identification of these “algal” genes, to a great extent, was affected by limited genome data then available for certain eukaryotic lineages. Almost none of these 30 genes confidently supports the hypothesis of a red algal endosymbiont in the common ancestor of stramenopiles. Although the molecular phylogeny of MetE is indeed consistent with the suggestion of a photosynthetic ancestry of stramenopiles, its topology does not strictly support a historical red plastid in this lineage. As such, our current study is largely consistent with the statistical genome analyses of Stiller et al. (2009), which found no evidence for a red algal endosymbiont in the ancestral stramenopile. However, we should also caution here that, because the parasitic nature of oomycetes, the possibility of plastid loss during oomycete evolution cannot be entirely excluded based on our data. Furthermore, given the fact that many of the sampled sequences in our analyses were from transcriptomic data, it is unclear whether and how the data quality, for example potential sequencing contamination, might have affected our results. Additional investigations are needed to resolve this significant, nevertheless controversial, issue of eukaryotic evolution.
On the other hand, our results also indicate that the abnormal phylogenetic signal of these “algal” genes might be caused by a complex evolutionary history of oomycetes or stramenopiles. Although the origins of these 30 genes in oomycetes or stramenopiles are not always clear, several of them were found to be related to miscellaneous algae. To a certain extent, such sequence relatedness to various lineages might be attributed to other potential historical endosymbioses or independent HGT events involving oomycetes or stramenopiles. For instance, in lieu of the chromalveolate hypothesis, serial endosymbioses between different photosynthetic lineages have been proposed to explain the evolution of red algal plastids (Sanchez-Puerta and Delwiche, 2008; Stiller et al., 2014). A potential green algal endosymbiont was also suggested in stramenopiles (Moustafa et al., 2009; Dorrell and Smith, 2011). Furthermore, horizontally acquired genes have been reported in different eukaryotic lineages (Richardson and Palmer, 2007; Keeling and Palmer, 2008; Andersson, 2009; Dunning Hotopp, 2011; Huang and Yue, 2012; Soucy et al., 2015; Qiu et al., 2016), even though some of the earlier reports might turn out to be false positives as suggested by our current study. Especially for microbial eukaryotes, the importance and frequency of HGT in their evolution is increasingly being appreciated (Keeling and Palmer, 2008; Andersson, 2009), and there is evidence that microbial eukaryotes might have frequently acquired genes from various organisms, instead of a specific source of endosymbiotic relationship (Huang et al., 2004; Loftus et al., 2005; Carlton et al., 2007; Bowler et al., 2008; Sun et al., 2010; Yue et al., 2013). Oomycetes originated in marine environments and gradually spread to freshwater and terrestrial environments (Beakes and Sekimoto, 2009; Beakes et al., 2012). Bacteria, miscellaneous algae or other organisms in a common habitat could be potential sources of foreign genes in oomycetes. Additionally, feeding activities of their ancestors in aquatic environments or the parasitic feature of modern species [many oomycetes are parasites; for instance, the early diverging species Eurychasma dicksonii is an obligate parasite of marine brown algae (Küpper and Müller, 1999; Gachon et al., 2009; Strittmatter et al., 2009)] might have also facilitated genes acquisition in oomycetes. Indeed, several studies have already reported gene acquisition events in oomycetes and other stramenopiles, including fungi to oomycetes (Richards et al., 2006, 2011), bacteria to diatoms (Bowler et al., 2008), and different prokaryotic or eukaryotic sources to Blastocystis (Tsaousis et al., 2012; Eme et al., 2017). In this regard, our finding of multiple foreign genes in oomycetes might reflect the interactions among red/green algae, oomycetes/stramenopiles, and other microbes, as well as their ensuing genetic integration.
Author Contributions
JH conceived the study and wrote the manuscript. QW performed the analyses and wrote the manuscript. HS contributed to the analyses.
Funding
This work is supported by the Major Program of National Natural Science Foundation of China (grant no. 31590823) to HS, CAS “Light of West China” Program to JH, and a joint Ph.D. training program of University of Chinese Academy of Sciences (UCAS[2015]37) to QW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01540/full#supplementary-material
Footnotes
References
Andersson, J. O. (2009). Gene transfer and diversification of microbial eukaryotes. Annu. Rev. Microbiol. 63, 177–193. doi: 10.1146/annurev.micro.091208.073203
Archibald, J. M., Longet, D., Pawlowski, J., and Keeling, P. J. (2003). A novel polyubiquitin structure in Cercozoa and Foraminifera: evidence for a new eukaryotic supergroup. Mol. Biol. Evol. 20, 62–66. doi: 10.1093/molbev/msg006
Baldauf, S. L., Roger, A. J., Wenk-Siefert, I., and Doolittle, W. F. (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290, 972–977. doi: 10.1126/science.290.5493.972
Beakes, G. W., Glockling, S. L., and Sekimoto, S. (2012). The evolutionary phylogeny of the oomycete “fungi”. Protoplasma 249, 3–19. doi: 10.1007/s00709-011-0269-2
Beakes, G. W., and Sekimoto, S. (2009). “The evolutionary phylogeny of Oomycetes—insights gained from studies of holocarpic parasites of algae and invertebrates,” in Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools, eds K. Lamour and S. Kamoun (New York, NY: Wiley), 1–24.
Bodył, A. (2005). Do plastid-related characters support the chromalveolate hypothesis? J. Phycol. 41, 712–719. doi: 10.1111/j.1529-8817.2005.00091.x
Bodył, A., and Moszczyński, K. (2006). Did the peridinin plastid evolve through tertiary endosymbiosis? A hypothesis. Eur. J. Phycol. 41, 435–448. doi: 10.1080/09670260600961080
Bodył, A., Stiller, J. W., and Mackiewicz, P. (2009). Chromalveolate plastids: direct descent or multiple endosymbioses? Trends Ecol. Evol. 24, 119–121. doi: 10.1016/j.tree.2008.11.003
Bogorad, L. (1975). Evolution of organelles and eukaryotic genomes. Science 188, 891–898. doi: 10.1126/science.1138359
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Brown, J. W., and Sorhannus, U. (2009). A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): substantive underestimation of putative fossil ages. PLoS ONE 5:e12759. doi: 10.1371/journal.pone.0012759
Burki, F., Flegontov, P., Oborník, M., Cihlář, J., Pain, A., Lukeš, J., et al. (2012a). Re-evaluating the green versus red signal in eukaryotes with secondary plastid of red algal origin. Genome Biol. Evol. 4:evs049. doi: 10.1093/gbe/evs049
Burki, F., Kaplan, M., Tikhonenkov, D. V., Zlatogursky, V., Minh, B. Q., Radaykina, L. V., et al. (2016). Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. Biol. Sci. 283, 20152802. doi: 10.1098/rspb.2015.2802
Burki, F., Okamoto, N., Pombert, J.-F., and Keeling, P. J. (2012b). The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc. Biol. Sci 279, 2246–2254. doi: 10.1098/rspb.2011.2301
Burki, F., Shalchian-Tabrizi, K., and Pawlowski, J. (2008). Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol. Lett. 4, 366–369. doi: 10.1098/rsbl.2008.0224
Carlton, J. M., Hirt, R. P., Silva, J. C., Delcher, A. L., Schatz, M., Zhao, Q., et al. (2007). Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212. doi: 10.1126/science.1132894
Cavalier-Smith, T. (1986). The kingdom Chromista: origin and systematics. Prog. Phycol. Res. 4, 309–347.
Cavalier-Smith, T. (1995). “Membrane heredity, symbiogenesis, and the multiple origins of algae,” in Biodiversity and Evolution, eds R. Arai, M. Kato, and Y. Doi (Tokyo: The National Science Museum Foundation), 75–114.
Cavalier-Smith, T. (1999). Principles of protein and lipid targeting in secondary Symbiogenesis: Euglenoid, Dinoflagellate, and Sporozoan plastid origins and the eukaryote family tree1, 2. J. Eukaryot. Microbiol. 46, 347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x
Cavalier-Smith, T., and Chao, E. E.-Y. (2003). Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J. Mol. Evol. 56, 540–563. doi: 10.1007/s00239-002-2424-z
Cavalier-Smith, T., Couch, J., Thorsteinsen, K., Gilson, P., Deane, J., Hill, D., et al. (1996). Cryptomonad nuclear and nucleomorph 18S rRNA phylogeny. Eur. J. Phycol. 31, 315–328. doi: 10.1080/09670269600651541
Cavalier-Smith, T., and Scoble, J. M. (2013). Phylogeny of Heterokonta: incisomonas marina, a uniciliate gliding opalozoan related to Solenicola (Nanomonadea), and evidence that Actinophryida evolved from raphidophytes. Eur. J. Protistol. 49, 328–353. doi: 10.1016/j.ejop.2012.09.002
Delwiche, C. F. (1999). Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 154, S164–S177.
Delwiche, C. F., and Palmer, J. D. (1997). The Origin of Plastids and their Spread via Secondary Symbiosis. Vienna: Springer.
Derelle, R., López-García, P., Timpano, H., and Moreira, D. (2016). A phylogenomic framework to study the diversity and evolution of stramenopiles (= heterokonts). Mol. Biol. Evol. 33, 2890–2898. doi: 10.1093/molbev/msw168
Dorrell, R. G., and Smith, A. G. (2011). Do red and green make brown?: perspectives on plastid acquisitions within chromalveolates. Eukaryot. Cell 10, 856–868. doi: 10.1128/EC.00326-10
Dunning Hotopp, J. C. (2011). Horizontal gene transfer between bacteria and animals. Trends Genet. 27, 157–163. doi: 10.1016/j.tig.2011.01.005
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ellis, R. J. (1981). Chloroplast proteins: synthesis, transport, and assembly. Annu. Rev. Plant Physiol. 32, 111–137. doi: 10.1146/annurev.pp.32.060181.000551
Emanuelsson, O., Nielsen, H., Brunak, S., and Von, H. G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Eme, L., Gentekaki, E., Curtis, B., Archibald, J. M., and Roger, A. J. (2017). Lateral gene transfer in the adaptation of the anaerobic parasite Blastocystis to the gut. Curr. Biol. 27, 807–820. doi: 10.1016/j.cub.2017.02.003
Gachon, C. M., Strittmatter, M., Müller, D. G., Kleinteich, J., and Küpper, F. C. (2009). Detection of differential host susceptibility to the marine oomycete pathogen Eurychasma dicksonii by real-time PCR: not all algae are equal. Appl. Environ. Microbiol. 75, 322–328. doi: 10.1128/AEM.01885-08
Gibbs, S. P. (1978). The chloroplasts of Euglena may have evolved from symbiotic green algae. Can. J. Bot. 56, 2883–2889. doi: 10.1139/b78-345
Gilson, P. R., Su, V., Slamovits, C. H., Reith, M. E., Keeling, P. J., and McFadden, G. I. (2006). Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc. Natl. Acad. Sci. U.S.A. 103, 9566–9571. doi: 10.1073/pnas.0600707103
Gould, S. B., Waller, R. F., and Mcfadden, G. I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517. doi: 10.1146/annurev.arplant.59.032607.092915
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Huang, J., Mullapudi, N., Lancto, C. A., Scott, M., Abrahamsen, M. S., and Kissinger, J. C. (2004). Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 5, R88.
Huang, J. L., and Yue, J. P. (2012). Horizontal gene transfer in the evolution of photosynthetic eukaryotes. J. Syst. Evol. 51, 13–29. doi: 10.1111/j.1759-6831.2012.00237.x
Keane, T. M., Creevey, C. J., Pentony, M. M., Naughton, T. J., and Mclnerney, J. O. (2006). Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6:29. doi: 10.1186/1471-2148-6-29
Keeling, P. J. (2013). The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 64, 583–607. doi: 10.1146/annurev-arplant-050312-120144
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
Keeling, P. J., and Palmer, J. D. (2008). Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618. doi: 10.1038/nrg2386
Kim, E., and Graham, L. E. (2008). EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata. PLoS ONE 3:e2621. doi: 10.1371/journal.pone.0002621
Kopriva, S., Büchert, T., Fritz, G., Suter, M., Benda, R., Schünemann, V., et al. (2002). The presence of an iron-sulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadenosine 5′-phosphosulfate for sulfate assimilation. J. Biol. Chem. 277, 21786–21791. doi: 10.1074/jbc.M202152200
Kopriva, S., Fritzemeier, K., Wiedemann, G., and Reski, R. (2007). The putative moss 3′-phosphoadenosine-5′-phosphosulfate reductase is a novel form of adenosine-5′-phosphosulfate reductase without an iron-sulfur cluster. J. Biol. Chem. 282, 22930–22938. doi: 10.1074/jbc.M702522200
Kopriva, S., and Koprivova, A. (2004). Plant adenosine 5′-phosphosulphate reductase: the past, the present, and the future. J. Exp. Bot. 55, 1775–1783. doi: 10.1093/jxb/erh185
Küpper, F. C., and Müller, D. G. (1999). Massive occurrence of the heterokont and fungal parasites Anisolpidium, Eurychasma and Chytridium in Pylaiella littoralis (Ectocarpales, Phaeophyceae). Nova Hedw. 69, 381–389.
Leander, B. S. (2004). Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 12, 251–258.
Loftus, B., Anderson, I., Davies, R., Alsmark, U. C. M., Samuelson, J., Amedeo, P., et al. (2005). The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868. doi: 10.1038/nature03291
Ludwig, M., and Gibbs, S. P. (1989). Evidence that the nucleomorphs of Chlorarachnion reptans (Chlorarachniophyceae) are vestigial nuclei: morphology, division and DNA-DAPI fluorescence. J. Phycol. 25, 385–394. doi: 10.1111/j.1529-8817.1989.tb00135.x
Maréchal, E., and Cesbron-Delauw, M.-F. (2001). The apicoplast: a new member of the plastid family. Trends Plant Sci. 6, 200–205. doi: 10.1016/S1360-1385(01)01921-5
Margulis, L. (1970). Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. New Haven, CT: Yale University Press.
Marin, B., Nowack, E. C., and Melkonian, M. (2005). A plastid in the making: evidence for a second primary endosymbiosis. Protist 156, 425–432. doi: 10.1016/j.protis.2005.09.001
Martin, W., and Kowallik, K. (1999). Annotated English translation of Mereschkowsky’s 1905 paper ‘Über Natur und Ursprung der Chromatophoren im Pflanzenreiche’. Eur. J. Phycol. 34, 287–295. doi: 10.1080/09670269910001736342
McFadden, G. I. (2001). Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 37, 951–959. doi: 10.1046/j.1529-8817.2001.01126.x
Moustafa, A., Beszteri, B., Maier, U. G., Bowler, C., Valentin, K., and Bhattacharya, D. (2009). Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726. doi: 10.1126/science.1172983
Okamoto, N., Chantangsi, C., Horák, A., Leander, B. S., and Keeling, P. J. (2009). Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS ONE 4:e7080. doi: 10.1371/journal.pone.0007080
Palmer, J. D. (2003). The symbiotic birth and spread of plastids: how many times and whodunit? J. Phycol. 39, 4–12. doi: 10.1046/j.1529-8817.2003.02185.x
Palmer, J. D., and Delwiche, C. F. (1998). “The origin and evolution of plastids and their genomes,” in Molecular Systematics of Plants II, eds D. E. Soltis, P. S. Soltis, and J. J. Doyle (Boston, MA: Springer), 375–409.
Patron, N. J., Durnford, D. G., and Kopriva, S. (2008). Sulfate assimilation in eukaryotes: fusions, relocations and lateral transfers. BMC Evol. Biol. 8:39. doi: 10.1186/1471-2148-8-39
Patterson, D. (1989). “Stramenopiles: chromophytes from a protistan perspective,” in The Chromophyte Algae: Problems and Perspectives, eds J. C. Green, B. S. C. Leadbeater, and W. I. Diver (Oxford: Clarendon Press), 357–379.
Qiu, H., Cai, G., Luo, J., Bhattacharya, D., and Zhang, N. (2016). Extensive horizontal gene transfers between plant pathogenic fungi. BMC Biol. 14:41. doi: 10.1186/s12915-016-0264-3
Reyes-Prieto, A., Moustafa, A., and Bhattacharya, D. (2008). Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr. Biol. 18, 956–962. doi: 10.1016/j.cub.2008.05.042
Richards, T. A., Dacks, J. B., Jenkinson, J. M., Thornton, C. R., and Talbot, N. J. (2006). Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16, 1857–1864. doi: 10.1016/j.cub.2006.07.052
Richards, T. A., Soanes, D. M., Jones, M. D. M., Olga, V., Guy, L., Konrad, P., et al. (2011). Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. U.S.A. 108, 15258–15263. doi: 10.1073/pnas.1105100108
Richardson, A. O., and Palmer, J. D. (2007). Horizontal gene transfer in plants. J. Exp. Bot. 58, 1–9. doi: 10.1093/jxb/erl148
Riisberg, I., Orr, R. J. S., Kluge, R., Shalchian-Tabrizi, K., Bowers, H. A., Patil, V., et al. (2009). Seven gene phylogeny of heterokonts. Protist 160, 191–204. doi: 10.1016/j.protis.2008.11.004
Rogers, M. B., Gilson, P. R., Su, V., McFadden, G. I., and Keeling, P. J. (2007). The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol. 24, 54–62. doi: 10.1093/molbev/msl129
Sanchez-Puerta, M., and Delwiche, C. F. (2008). A hypothesis for plastid evolution in chromalveolates. J. Phycol. 44, 1097–1107. doi: 10.1111/j.1529-8817.2008.00559.x
Ševčíková, T., Horák, A., Klimeš, V., Zbránková, V., Demirhilton, E., Sudek, S., et al. (2015). Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 5:10134. doi: 10.1038/srep10134
Soucy, S. M., Huang, J., and Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Stiller, J. W., Huang, J., Ding, Q., Tian, J., and Goodwillie, C. (2009). Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? BMC Genomics 10:484. doi: 10.1186/1471-2164-10-484
Stiller, J. W., Schreiber, J., Yue, J., Guo, H., Ding, Q., and Huang, J. (2014). The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 5, 5764. doi: 10.1038/ncomms6764
Strittmatter, M., Gachon, C. M., and Küpper, F. C. (2009). Ecology of Lower Oomycetes. Oomycete Genetics and Genomics: Diversity, Interactions and Research Tools. Hoboken, NJ: John Wiley and Sons Inc, 25–46. doi: 10.1002/9780470475898.ch2
Sun, G., Yang, Z., Ishwar, A., and Huang, J. (2010). Algal genes in the closest relatives of animals. Mol. Biol. Evol. 27, 2879–2889. doi: 10.1093/molbev/msq175
Timmis, J. N., Ayliffe, M. A., Huang, C. Y., and Martin, W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Tsaousis, A. D., Ollagnier, D. C. S., Gentekaki, E., Long, S., Gaston, D., Stechmann, A., et al. (2012). Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc. Natl. Acad. Sci. U.S.A. 109, 10426–10431. doi: 10.1073/pnas.1116067109
Tyler, B. M., Sucheta, T., Xuemin, Z., Paramvir, D., Jiang, R. H. Y., Andrea, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
Van de Peer, Y., Rensing, S., Maier, U., and De Wachter, R. (1996). Substitution rate calibration of small subunit ribosomal RNA identifies chlorarachniophyte endosymbionts as remnants of green algae. Proc. Natl. Acad. Sci. U.S.A. 93, 7732–7736. doi: 10.1073/pnas.93.15.7732
Weeden, N. F. (1981). Genetic and biochemical implications of the endosymbiotic origin of the chloroplast. J. Mol. Evol. 17, 133–139. doi: 10.1007/BF01733906
Woehle, C., Dagan, T., Martin, W. F., and Gould, S. B. (2011). Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol. Evol. 3, 1220–1230. doi: 10.1093/gbe/evr100
Yang, E. C., Boo, G. H., Kim, H. J., Cho, S. M., Boo, S. M., Andersen, R. A., et al. (2012). Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist 163, 217–231. doi: 10.1016/j.protis.2011.08.001
Yoon, H. S., Grant, J., Tekle, Y. I., Wu, M., Chaon, B. C., Cole, J. C., et al. (2008). Broadly sampled multigene trees of eukaryotes. BMC Evol. Biol. 8:14. doi: 10.1186/1471-2148-8-14
Yoon, H. S., Hackett, J. D., Pinto, G., and Bhattacharya, D. (2002). From the cover: the single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. U.S.A. 99, 15507–15512. doi: 10.1073/pnas.242379899
Keywords: plastid evolution, stramenopiles, endosymbiosis, horizontal gene transfer, eukaryotic evolution
Citation: Wang Q, Sun H and Huang J (2017) Re-analyses of “Algal” Genes Suggest a Complex Evolutionary History of Oomycetes. Front. Plant Sci. 8:1540. doi: 10.3389/fpls.2017.01540
Received: 25 May 2017; Accepted: 22 August 2017;
Published: 06 September 2017.
Edited by:
Tatiana Matveeva, Saint Petersburg State University, RussiaReviewed by:
David John Studholme, University of Exeter, United KingdomHuan Qiu, Rutgers University, United States
Copyright © 2017 Wang, Sun and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Sun, c3VuaGFuZ0BtYWlsLmtpYi5hYy5jbg== Jinling Huang, aHVhbmdqQGVjdS5lZHU=
 Qia Wang
Qia Wang Hang Sun
Hang Sun Jinling Huang
Jinling Huang