- 1Department of Nematology, University of California, Riverside, Riverside, CA, United States
- 2Key Laboratory of Mollisols Agroecology, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin, China
- 3Plant Stress and Germplasm Development Research, PA, CSRL, USDA-ARS, Lubbock, TX, United States
Transgressive segregation in cotton (Gossypium spp.) provides an important approach to enhance resistance to the major pest root-knot nematode (RKN) Meloidogyne incognita. Our previous studies reported transgressive RKN resistance in an intraspecific Gossypium hirsutum resistant NemX × susceptible SJ-2 recombinant inbred line (RIL) population and early generations of interspecific cross Gossypium barbadense (susceptible Pima S-7) × G. hirsutum (NemX). However, the underlying functional mechanisms for this phenomenon are not known. In this study, the region of RKN resistance gene rkn1 on chromosome (Chr) 11 and its homoeologous Chr 21 was fine mapped with G. raimondii D5 genome reference sequence. Transgressive resistance was found in the later generation of a new RIL population F2:7 (Pima S-7 × NemX) and one interspecific F2 (susceptible Pima S-7 × susceptible SJ-2). QTL analysis revealed similar contributions to root-galling and egg-production resistance phenotypes associated with SSR marker CIR316 linked to resistance gene rkn1 in NemX on Chr 11 in all seven populations analyzed. In testcross NemX × F1 (Pima S-7 × SJ-2) marker allele CIR069-271 from Pima S-7 linked to CIR316 contributed 63% of resistance to galling phenotype in the presence of rkn1. Similarly, in RIL population F2:8 (NemX × SJ-2), SJ-2 markers closely linked to CIR316 contributed up to 82% of resistance to root-galling. These results were confirmed in BC1F1 SJ-2 × F1 (NemX × SJ-2), F2 (NemX × SJ-2), and F2 (Pima S-7 × SJ-2) populations in which up to 44, 36, and 15% contribution in resistance to galling was found, respectively. Transgressive segregation for resistance was universal in all intra- and inter-specific populations, although stronger transgressive resistance occurred in later than in early generations in the intraspecific cross compared with the interspecific cross. Transgressive effects on progeny from susceptible parents are possibly provided in the rkn1 resistance region of chromosome 11 by tandemly arrayed allele (TAA) or gene (TAG) interactions contributing to transgressive resistance. Complex TAA and TAG recombination and interactions in the rkn1 resistance region provide three genes and a model to study disease and transgressive resistance in polyploid plants, and novel genotypes for plant breeding.
Introduction
Host-plant resistance is a highly effective strategy to manage root-knot nematode (RKN, Meloidogyne spp.) damage in crops. The RKN Meloidogyne incognita is one of the most important pests of cotton (Goodell and Montez, 1994), and effective resistance is available for improving cotton cultivar performance (Starr et al., 2010). In addition, transgressive segregation is one of the approaches to enhance resistance, in which segregating hybrids exhibit extreme or novel phenotypes compared to the phenotypes of parental lines (Rieseberg et al., 1999, 2003). Improved resistance traits generated by transgressive segregation in progenies derived from interspecific and intraspecific crosses have been reported (Cherif and Harrabi, 1993; Zhang et al., 2001; Imtiaz et al., 2003a,b; Navabi et al., 2004; Bell and Travis, 2005; Zhao et al., 2005; Staal et al., 2006; Aghnoum and Niks, 2011). Transgressive segregation provides one of the major selection sources for enhanced resistance to RKN in cotton (Gossypium hirsutum L.) (Shepherd, 1974; Wang et al., 2006a; Wang C. et al., 2008, 2012; Ulloa et al., 2016). Other cotton-pathogen systems also displayed transgressive resistance, such as Fusarium wilt (Wang and Roberts, 2006a; Ulloa et al., 2011, 2013, 2016), Verticillium wilt (Bolek et al., 2005; Wang H. M. et al., 2008), and bacterial blight (Bayles et al., 2005).
Three major germplasm sources of RKN resistance have been utilized in Upland cotton G. hirsutum, NemX (Ogallo et al., 1999; Wang et al., 2006a; Roberts and Ulloa, 2010), Clevewilt 6 and derived lines Stoneville LA887 and Paymaster H1560 (Robinson et al., 2001), and Auburn 623 and its derivatives (Shepherd, 1974; McPherson et al., 2004). Highly resistant Auburn 623 RNR was derived from two moderately resistant parental lines, Clevewilt 6-1 and Mexico Wild Jack Jones. The RKN resistance sources Auburn 623 RNR, Auburn 634 RNR, and their derived N-lines (Hyer and Jorgenson, 1984) were reported to be transgressive segregants (Shepherd, 1974; Hyer et al., 1979). Wang C. et al. (2008) reported that a segregating factor (RKN2) from susceptible parent Gossypium barbadense Pima S-7 could not function alone but interacted with a major recessive gene rkn1 in G. hirsutum NemX (Wang et al., 2006b) to produce a higher resistance phenotype than resistant parental line NemX in progeny of the interspecific cross between Pima S-7 and NemX. Interestingly, the two genes RKN2 and rkn1 were mapped to the same region on Chr 11 (Wang C. et al., 2008). Transgressive resistance was also observed in some intraspecific F2:7 (NemX × SJ-2) RI (recombinant inbred) homozygous resistant lines, indicating susceptible parent SJ-2 contributed to higher resistance in progeny than the rkn1 resistance contributed by NemX alone (Wang et al., 2006a). Genetic mapping and quantitative trait loci (QTL) analysis suggested a major telomeric segment on Chr 11 harbors RKN resistance genes from these different resistance sources (Bezawada et al., 2003; Shen et al., 2006, 2010; Wang et al., 2006b; Wang C. et al., 2008; Gutiérrez et al., 2010; Roberts and Ulloa, 2010; Ulloa et al., 2010). A microsatellite marker (SSR) CIR 316 tightly linked to resistance gene rkn1 and other RKN resistance genes on Chr 11 was identified in different segregating populations (Shen et al., 2006, 2010; Wang and Roberts, 2006b; Wang et al., 2006b; Ynturi et al., 2006; Gutiérrez et al., 2010; Roberts and Ulloa, 2010; Ulloa et al., 2010). A resistance gene contributing to suppression of nematode egg production originally derived from Wild Mexico Jack Jones also was identified on Chr 14 (Gutiérrez et al., 2010; He et al., 2014; Kumar et al., 2016), which in combination with a resistance gene on Chr 11 derived from Clevewilt produced transgressive resistance in Auburn 623.
In a parallel study using an interspecific RIL population from a cross between two susceptible parents (G. hirsutism TM-1 × G. barbadense Pima 3-79), Wang C. et al. (2012) identified four major QTLs (on Chr 3, 4, 11, and 17) and two major QTLs (Chr 14 and 23) which contributed 8–12% transgressive resistance to nematode root-galling (galling index, GI) and nematode reproduction (eggs per gram root, EGR), respectively. In addition, 19 and 15 minor QTLs were identified in the TM-1 × Pima 3-79 population with each QTL accounting for 4–7% of phenotypic variance in GI and EGR, respectively (Wang C. et al., 2012). Although each of these QTLs contributed minor effects on phenotype, combinations of two to four major and/or minor QTLs were shown to dramatically reduce root-galling and nematode egg production by >50%, suggesting epistatic effects among these QTLs (Wang C. et al., 2012).
While these studies establish that transgressive segregation is common in cotton for disease resistance, the underlying functional mechanisms for this phenomenon are not known. For example, the location of the transgressive factor in SJ-2 in RIL population F2:8 (NemX × SJ-2) is not known, nor whether it would play a positive role on resistance in other recombinant populations. Other knowledge gaps include the nature of inheritance of the transgressive factor RKN2 in G. barbadense Pima S-7 and how it behaves in later generations of the RIL F2:7 (Pima S-7 × NemX) population; and how genes interact in the region of rkn1 in different genetic backgrounds. Previous studies demonstrated that a QTL mapping approach could be informative for studying inheritance and gene action to determine further the mechanism of transgressive segregation (Ulloa et al., 2010; Wang C. et al., 2012). Therefore, in this study, we report the development of a new RIL population (Pima S-7 × NemX) and its use in fine-mapping the rkn1 region on Chr 11 and characterizing transgressive resistance with new molecular markers developed from the G. raimondii D5 genome reference sequence (Paterson et al., 2012). Through QTL analysis the transgressive segregation for RKN resistance was characterized further in six other genetic populations derived from three parental lines, G. barbadense Pima S-7, G. hirsutum NemX and SJ-2, and a simple genetic model for transgressive segregation in the Chr 11 region is proposed.
Materials and Methods
Plant Materials and Crosses
Three highly inbred homogeneous parental lines, susceptible G. hirsutum cv. SJ-2 (USDA-ARS), resistant G. hirsutum cv. NemX (Bayer Corp., formerly by CPCSD; Oakley, 1995), and susceptible G. barbadense Pima S-7 (USDA-ARS) were utilized. Three F1 populations were developed from crosses between these genotypes. The F2 population was developed by self-pollinating the F1 hybrid between two parents. The RIL population was derived by single-seed descent from the F2 generation to develop highly homozygous generations. Interspecific crosses included 106 F2 (Pima S-7 × SJ-2) plants for nematode phenotyping of which 100 were used for mapping and QTL analysis, 108 RI Pima S-7 × NemX lines (90 F2:7, 8 F2:6, and 10 F2:5) for nematode phenotyping and QTL mapping, plus 165 F2 (Pima S-7 × NemX) plants and 51 plants of testcross NemX × F1 (Pima S-7 × SJ-2; Wang C. et al., 2008) for QTL mapping. Intraspecific crosses included 96 F2 (NemX × SJ-2) plants, 69 F2:8 (NemX × SJ-2) RIL families, 97 BC1F1 (NemX × F1) plants, and 48 BC1F1 (SJ-2 × F1) plants (Wang et al., 2006b).
Nematode Resistance Screening
Nematode resistance phenotyping followed the method of Wang et al. (2006a,b). Nematode resistance in cotton populations was screened under controlled greenhouse conditions at 28–35°C during the day and 24°C at night. Five replicate plants of each line in the RIL population and of parental lines were arranged in a complete randomized design. One seed per plastic pot (10-cm-diam. × 17-cm-deep) was sown in blow sand and grown for 3 weeks. Then each plant was inoculated with ~50,000 eggs of M. incognita race 3 (isolate Project 77) and fertilized with slow-release fertilizer (Scotts-Sierra Horticultural Products Co) (Wang et al., 2006a). Egg extraction was conducted in NaOCl (Hussey and Barker, 1973). Infection response on cotton plants was evaluated at 60 days post-inoculation using the root-galling scale modified from Bridge and Page (1980), with 0 (no symptom) −10 (severe symptom). The number of nematode eggs per gram fresh root was also utilized for nematode infection response on cotton. Resistance threshold in each test was determined by the mean values of galling index and eggs per gram root of parents plus standard deviation (SD) (Wang et al., 2006a).
Data Analysis
Data analysis followed the methods of Wang C. et al. (2008). One-way ANOVA was used to analyze data. The treatment means were compared with Fisher's Protected LSD test. The transformed log10 (x + 1) data for nematode egg production were used for analysis. Chi-square (goodness of fit) test was used to predict Mendelian inheritance ratios.
Marker Analysis
Since chromosomes 11 and 14 are associated with root-galling index and/or nematode egg production (Shen et al., 2006; Wang et al., 2006b; Gutiérrez et al., 2010; He et al., 2014), markers on Chr11 and its homoeologous chromosome 21, and markers on Chr 14 were screened for nematode resistance linkage in the RIL population Pima S-7 × NemX. In addition, polymorphic markers linked to nematode resistance or Fusarium wilt resistance in the RIL population Pima 3-79 × TM1 (Ulloa et al., 2011, 2013; Wang C. et al., 2012; Wang et al., 2015; www.cottonmarker.org) were also used for screening RIL population Pima S-7 × NemX. A total of 186 SSR markers containing 366 polymorphic alleles across the whole genome were obtained for QTL analysis of nematode resistance.
DNA was extracted from fresh or frozen (−80°C) young cotton leaves using the DNeasy® Plant Mini kit (Qiagen, Valencia, CA, USA; Wang et al., 2006b). Primers were synthesized by IDT (IDT, Coralville, IA, USA). The forward primers were synthesized with an M13 forward sequence on the 5′-end (Wang et al., 2006b). The IRD labeled M13 primer (700 or 800 channel: CACGACGTTGTAAAACGAC) was made by LI-COR (LI-COR, Lincoln, NE, USA). The methods of PCR amplification of cotton molecular markers and of electrophoresis and detection were described by Ulloa et al. (2016). The SCAR marker GHACC1 and AFLP markers M9E2, M5E1, and M4E5 linked to rkn1 were from Wang and Roberts (2006b).
Fine-Mapping Region rkn1 on Chr 11 and Chr 21 with D5 Genome
In order to fine-map the resistance region of rkn1, 68 primers (Supplemental file 1: Table S1) were designed from about 2.2 M bp of sequence which extended from 58.2 to 60.4 M bp on chromosome 7 (corresponds to Chr 21 in tetraploid cotton) of the diploid D5 genome (G. raimondii v.1.1; Paterson et al., 2012; https://phytozome.jgi.doe.gov) containing markers MUCS088, CIR316 and CIR069 which are associated with RKN resistance (Wang et al., 2006b; Wang C. et al., 2008). The primers were named as UCR+ number, such as UCR1 or UCR2. The 2.2 M bp sequence contained markers CIR316 and CIR069 which were linked to rkn1 or other resistance genes from different resistance sources on Chr 11 (Shen et al., 2006; Wang et al., 2006b). The parental lines Pima S-7, NemX and SJ-2 were screened for polymorphism with the 71 primers. The identified polymorphic markers were used to screen segregating populations F2 (NemX × SJ-2), F2:8 (NemX × SJ-2), RIL Pima S-7 × NemX, and test-cross NemX × F1 (Pima S-7 × SJ-2).
Genetic Linkage and QTL Analysis
The linkage groups for chromosomes were developed by using the JoinMapR version 4.0 program (Van Ooijen, 2006). The Kosambi map function was used to examine Logarithm of odds (LOD) scores of 3–15 for each population. To determine linkage between any two markers, a maximum distance of 40 centiMorgans (cM) and a LOD threshold score >4.0 were used. QTL analyses were conducted on galling index (GI) and egg production per gram root (LogEGR) using MapQTL 5.0 (Van Ooijen, 2004). Non-parametric mapping [Kruskal-Wallis analysis (K*)] test equivalent to the one-way analysis of variance was used for single-marker analysis and interval mapping for analysis of pairs of linked markers. Significant QTLs were set with a more stringent P < 0.005 for the K*-test (Wang C. et al., 2012).
Results
Phenotyping F2:7 (Pima S-7 × NemX)
Fifty-six of 108 RI lines had lower (P < 0.05) GI than parent NemX (2.7 ± 0.27, SD) accounting for 51.9 % transgressive resistance (Figure 1A) and 39 lines had lower (P < 0.05) LogEGR (1.35 ± 0.81, SD) accounting for 36.1% transgressive resistance (Figure 1B). Seventy-one lines with GI < 3 accounted for 66% resistance in the RIL population. Ten of 108 lines had higher LogEGR than parent Pima S-7 (3.33 ± 0.16, SD) accounting for 9.3% transgressive susceptibility (Figure 1A). A high correlation (R2 = 0.70) was found between GI and LogEGR. These results confirmed that genes from both parents play a role in determining the resistance phenotype in these transgressive segregants from the F2:7 population.
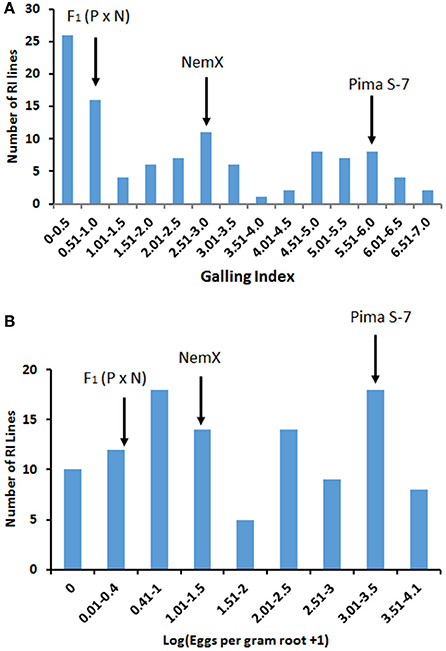
Figure 1. Distribution of Meloidogyne incognita root-galling (galling index, GI) in the Pima S-7 (P) × NemX (N) F2:7 RIL population (A), and M. incognita egg production [Log(EGR+1)] in the Pima S-7 × NemX F2:7 RIL population (B). The F2:7 population was evaluated at 60 days after inoculation. Galling index: 0–10 scale; 0 no galling, and 10 severe galling. Arrows point to the mean phenotypic reaction of the parents and F1 (P × N).
Fine Mapping Region rkn1 in RIL Populations F2:7 (Pima S-7 × NemX) and F2:8 (NemX × SJ-2) with D5 Genome Sequence
Twenty of 68 SSR primers designed from D5 sequence produced 29 polymorphic markers between Pima S-7 and NemX and six polymorphic markers between NemX and SJ-2 [the primer sequences, their positions on Gossypium raimondii (v.2.1) (https://phytozome.jgi.doe.gov), G. hirsutum (pre-released v.1.1) (https://phytozome.jgi.doe.gov), and G. arboreum (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and primer sequence function with JGI-Blast tool are provided in Supplemental file 1: Table S1]. Five polymorphic markers (UCR46, UCR49, UCR61, UCR102, and UCR108) were mapped to the rkn1 region on Chr 11 over 11.3 cM and 15 mapped to the homoeologous region over 12.4 cM on Chr 21 in the F2:7 (Pima S-7 × NemX) (Figure 2A), with both regions containing a homozygous allele pair of SSR marker CIR316 associated with RKN resistance. Twenty-two of 32 markers on Chr 11 showed homozygous allele content on Chr 21 (Figure 2A). Five (UCR49, UCR56, UCR61, UCR90, and UCR91) of six polymorphic markers mapped to the region of CIR316 closely linked to rkn1 expanded 2.3 cM in the F2:8 (NemX × SJ-2; Figure 2B). Of these, UCR90 and UCR91 produced 3 polymorphic markers between NemX and SJ-2, but not between Pima S-7 and NemX (Figure 2B).
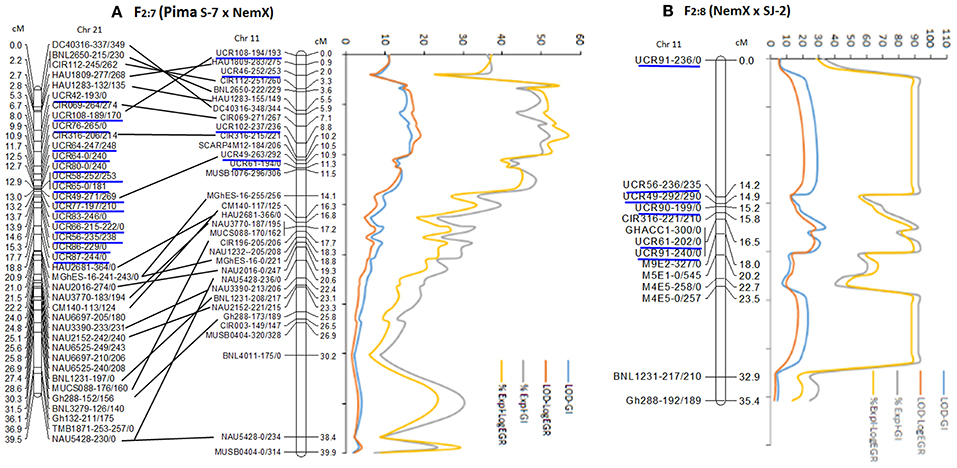
Figure 2. Fine mapping of rkn1 region on chromosome (Chr) 11 and its homoeologous Chr 21 in RIL population F2:7 (Pima S-7 × NemX) (A) and rkn1 region on Chr 11 in F2:8 (NemX × SJ-2) (B) using D5 genome sequence [41]. New markers are underlined in blue. Numbers after the marker name indicate the allele sizes after PCR amplification from the two parent lines, Pima S-7/NemX in the Pima S-7 × NemX RIL population and NemX/SJ-2 in the F2:8 (NemX × SJ-2) population. For example, for marker UCR108-194/193 in F2:7 (Pima S-7 × NemX), 194 bp amplified from Pima S-7 and 193 bp from NemX. All allelic sizes include the M13 primer tail.
QTL Mapping in F2:7 (Pima S-7 × NemX) and F2 (Pima S-7 × NemX) Populations
F2:7 (Pima S-7 × NemX)
A total of 395 (366 plus 29 listed above) polymorphic markers were used to genotype the F2:7 (Pima S-7 × NemX) for QTL mapping. Both Kruskal-Wallis analysis and interval mapping revealed that the significant (P < 0.0001) QTL(s) spanned 38 cM (0–38 cM) on Chr 11, associated with both GI and LogEGR phenotypes (Figure 2A, Supplemental file 2: Table S2A). The contribution to resistance to both galling and EGR was >40% in a 8.1 cM region spanning from 3.3 cM (CIR112-251/260) to 11.5 cM (MUSB1076-296/306) containing marker CIR316 linked to rkn1 (Figure 2A). The region between the two flanked SSR markers CIR069-271/269 and UCR102-237/236 contributed 52% phenotypic variance of resistance to galling and 57% to egg production (Figure 2A), indicating the rkn1 region contributed to both GI and LogEGR. Interval mapping demonstrated strong additive effects for resistance to both galling (1.62 for UCR102-237/236) and egg production (1.08 for CIR069-271/269; Supplemental file 2: Table S2A), suggesting an epistatic effect on phenotype. Resistance contribution of the locus CIR316-215/221 to both GI and LogEGR is shown in Table 1. Two other significant (P < 0.005) QTLs from Pima S-7 alleles, on Chr 3 (BNL3792-540/0) to GI, Chr 3 (MUSS396-0/300 to LogEGR, and Chr 1 (NAU4045-185/195) to both GI and LogEGR, accounted for ~7–10% phenotypic variance (Supplemental file 2: Table S2A). The homoeologous region on Chr 21 had no resistance contribution to GI or LogEGR even though the order of the marker alleles was similar to those on Chr11, and also no contribution to resistance was found on Chr 14.
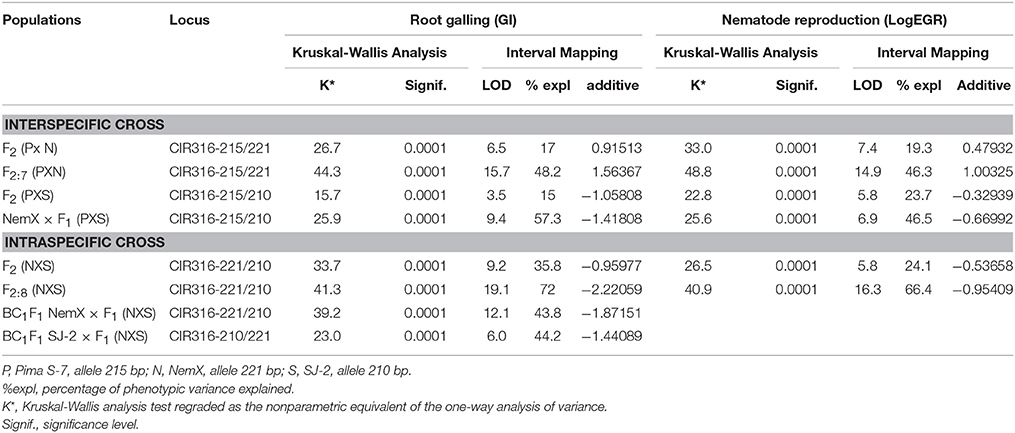
Table 1. Resistance contribution of CIR316 locus in seven interspecific and intraspecific populations.
F2 (Pima S-7 × NemX)
In order to confirm the rkn1 region on Chr11, markers CIR316, CIR069, and NAU2016 were used to screen 165 F2 (Pima S-7 × NemX) plants which were phenotyped in our previous study (Wang C. et al., 2008). QTL mapping confirmed that the three markers on chr11 were associated with resistance. CIR069 contributed 18.5% phenotypic variance to GI and 23% to LogEGR with additive effects (GI: 0.91454; LogEGR: 0.47609) based on interval mapping.
QTL Mapping in Testcross NemX × F1 (Pima S-7 × SJ-2)
In order to confirm the location of RKN2 in the same region of rkn1 [17], 32 SSR primers on Chr 11 were screened with the testcross population NemX × F1 (Pima S-7 × SJ-2) and produced 53 polymorphic markers between Pima S-7 and SJ-2. Of these, 32 were mapped on Chr 11 and 16 on Chr 21 (Figure 3). Interestingly, 18 markers including CIR316 and CIR069 on Chr 11-1 were clustered together in a 3.9-cM interval but over a 16.1 cM interval on the homoeologous region of Chr 21 (Figure 3). Marker MUCS088 closely linked to RKN2 was mapped to the same position as CIR316. Fourteen other markers without clustering were mapped to a region further from rkn1 spanning 42.9 cM (Figure 3, Chr 11-2).
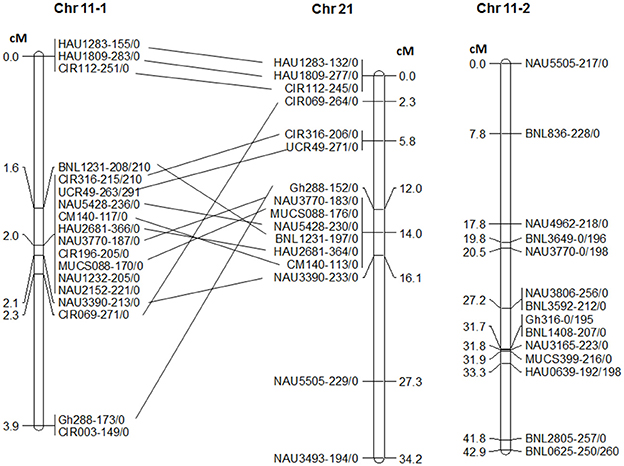
Figure 3. Linkage groups of chromosome (Chr) 11 and homoeologous Chr 21 in testcross population NemX × F1 (Pima S-7 × SJ-2). Chr 11-1 represents the rkn1 region at the telomeric end of Chr 11 and Chr 11-2 represents the lower part of this chromosome close to the centromere. Numbers after the marker name indicate the allele sizes after PCR amplification from the two lines Pima S-7/SJ-2. All allelic sizes include the M13 primer tail.
QTL analysis confirmed that the mapped rkn1 region (3.9 cM) was significantly (P < 0.0001) involved in both GI and LogEGR phenotypes in the testcross population (Supplemental file 2: Table S2B). Interval mapping analysis revealed that the CIR069 region on Chr 11 contributed up to 62.5% phenotypic variance in GI and 50.3% in LogEGR. Again, there was no contribution to resistance phenotype from Chr 21 and Chr 14.
Among the 18 markers clustered around rkn1, only CIR316 and BNL1231 had polymorphism among three parental lines (Pima S-7, NemX, and SJ-2). The application pattern of CIR316 clearly showed that all lines carried the heterozygous allele (221 bp) linked to rkn1 from NemX (Figure 4). The Pima S-7 allele band (215 bp) was present in the resistant lines and the SJ-2 allele band (210 bp) was present in the susceptible lines except for seven out of 51 lines for which recombination had occurred between CIR316 and the resistance gene based on galling index (Figure 4). An additional Pima S-7 allele band (206 bp) was mapped on Chr 21 (Figure 4).
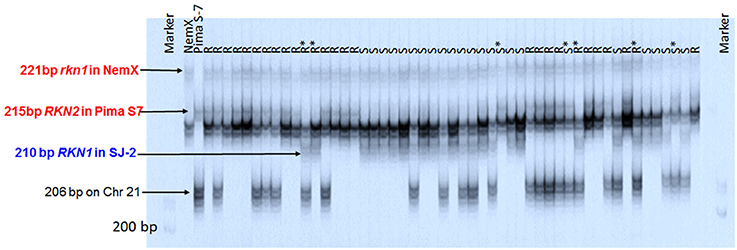
Figure 4. Image of amplification products with SSR marker CIR316 in the segregating population NemX × F1 (Pima S-7 × SJ-2) on polyacrylamide gel with model 4000 LI-COR IR2 automated sequencer. R, resistant; S, susceptible, all based on phenotype. R* and S*, recombinant line with resistant phenotype (R) and susceptible marker profile or susceptible phenotype (S) and resistant marker profile, respectively. Marker: 50–350 bp sizing standard. Arrows point to the marker allele positions. All allelic sizes include the M13 primer tail.
Phenotyping and QTL Mapping an F2 (Pima S-7 × SJ-2) Population Derived from Susceptible Parents
Since the interactions in the rkn1 region are complex, we examined whether transgressive segregation occurred in the F2 (Pima S-7 × SJ-2) population derived from two susceptible parents and determined whether transgressive factor RKN2 functions in the F2 (Pima S-7 × SJ-2) without the rkn1 allele. The correlation (R2) between galling index and egg production per gram root was 0.5235 (Figure 5). The phenotypic test revealed that 52 out of 106 (49%) F2 (Pima S-7 × SJ-2) plants showed less GI than Pima S-7 (5.85 ± 0.22, SD), SJ-2 (7 ± 0.61, SD), and their F1 (Pima S-7 × SJ-2; 6.9 ± 0.42, SD). Twenty out of 106 F2 plants had low GI < 3 (range 1–3) accounting for 19% resistance. The results indicated that both susceptible parents contributed to transgressive resistance. QTL mapping indicated that the CIR 316-215 bp allele from Pima S-7 on Chr 11 contributed 15% phenotypic variance to GI and 24% to LogEGR, respectively (Table 1, Supplemental file 2: Table S2C).
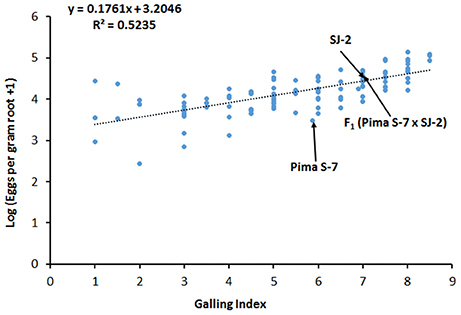
Figure 5. The relationship between galling index and egg production in the F2 (Pima S-7 × SJ-2) population 60 days after inoculation. Log10(x+1) transformed data were used for analysis of eggs per gram of root. Galling index: 0–10 scale; 0 no galling, and 10 severe galling. Mean data points for the Pima S-7 and SJ-2 parents and their F1 are indicated.
QTL Mapping of Intraspecific Cross NemX × SJ-2
F2:8 (NemX × SJ-2)
A set of 62 genome-wide markers polymorphic between NemX and SJ-2, including one SCAR marker (GHACC1) and four AFLP markers [37] associated with the rkn1 locus in NemX were screened for QTL analysis. QTL mapping showed that markers UCR91, UCR61, and GHACC1 tightly linked to the CIR316-221/210 marker contributed 82.1% phenotypic variance for GI and 81.7% for LogEGR around the region of rkn1 (Figure 2B, Table 1, Supplemental file 2: Table S2D), confirming that this same rkn1 region played a similar role in resistance to both GI and LogEGR. No resistance contribution was found from Chr 21 and Chr 14.
F2 (NemX × SJ-2)
QTL mapping using 40 polymorphic markers indicated that the CIR316 region on Chr 11 had a major effect on both galling and EGR (Supplemental file 2: Table S2E), which confirmed a previous report (Wang et al., 2006b). The CIR 316-221/210 marker contributed 36% phenotypic variance in galling and 24% in LogEGR (Table 1), and the AFLP-derived SCAR marker GHACC1 [37] which was 1.6 cM from CIR316 showed 36% phenotypic variance in galling and 33% in LogEGR (Supplemental file 2: Table S2E), emphasizing that the same rkn1 region contributed to both GI and EGR. No resistance contribution was found from Chr 14 and Chr 21.
BC1F1 NemX × F1 (NemX × SJ-2) and BC1F1 SJ-2 × F1 (NemX × SJ-2)
The CIR316-221/210 marker in BC1F1 population NemX × F1 (NemX × SJ-2) closely linked to rkn1 contributed 43.8% to phenotypic variance in galling (Table 1). The CIR316-215/210 marker in the BC1F1 population SJ-2 × F1 (NemX × SJ-2) in which lower galling index was found in progenies than both susceptible parental lines contributed 44.2% phenotypic variance in GI (Table 1), also indicating the rkn1 region from both NemX and SJ-2 plays an important role for nematode transgressive resistance in the intraspecific cross.
The increase in resistance by SJ-2 in progenies from both the Pima S-7 crosses with SJ-2 and the NemX crosses with SJ-2 is revealed by the CIR316 210 bp allele from SJ-2 and confirms that the transgressive factor in SJ-2 (designated as RKN3) is linked to rkn1 from NemX and to RKN2 from Pima S-7.
Discussion
Analysis of RKN resistance phenotypes in multiple segregating cotton populations in this study established the significant and common occurrence of transgressive nematode resistance among progenies of both interspecific and intraspecific populations. Transgressive segregation occurred in both interspecific crosses between G. barbadense Pima S-7 and G. hirsutum NemX, and between Pima S-7 and G. hirsutum SJ-2, and also in the G. hirsutum intraspecific cross between NemX and SJ-2. The results also establish that in the case of the crosses with resistant NemX which carries the recessive R gene rkn1 (Wang et al., 2006b), the susceptible parent in each case, whether intraspecific (SJ-2) or interspecific (Pima S-7) contributes at least one transgressive factor that enhances resistance. More unexpectedly, the interspecific cross between two susceptible parents [Pima S-7 (RKN2) (Wang C. et al., 2008) and SJ-2 (designated as RKN3)] also results in combinations of transgressive factors which produce some progenies with resistance phenotypes. Beside the R gene rkn1, these resistance-enhancer factors may be the results of epistasis or complex recombinations and interactions of tandemly arrayed alleles (TAA) or genes (TAG). Copy number of multiple genes in a 31-kb segment at the Rhg1 locus for resistance to cyst nematode in soybean has been shown to determine the level of expressed resistance, with copy number ranging from 1 (susceptible), to 3 (partially resistant) to 10 (highly resistant) in different genotypes (Cook et al., 2012). Whether or not a comparable arrangement of multiple genes with varying copy number determining the level of resistance expression occurs in the cotton rkn1 region will require identification of the gene sequences determining resistance phenotype.
The refined mapping by QTL analysis of the genome region harboring rkn1 on Chr 11 and its homoeologous region on Chr 21 provided further insight into the transgressive resistance control by this region. Of the original 40 cM mapped region of rkn1 which contributed resistance to both root-galling and egg production phenotypes in the F2:7 (Pima S-7 × NemX) population, 8 cM of the region containing marker CIR316 accounted for more than 40% of the phenotypic variance. This result indicated that genes cluster together in the region but cannot be separated only by phenotype. Strong positive additive effects on resistance phenotypes were found in both the F2 and F2:7 (Pima S-7 × NemX) RIL populations, suggesting that epistasis plays a role in the RKN transgressive resistance in this interspecific cross. Although the interval bracketed by markers HAU1809 and Gh288 in the rkn1 region was 25–27 cM in length on Chr 11 and Chr 21, in the testcross population we found at least three-fold shorter interval on Chr 11 than on Chr 21, indicating complex recombination between the rkn1 region and its homoeologous Chr 21. This shorter genetic distance on Chr 11 in the testcross population, which contributed 50–60% phenotypic variance, suggested that the complex recombination in the rkn1 region produced transgressive resistance and that the level of resistance depends on the specific parent combination (Ulloa et al., 2010). The non-separated markers might result from homologous/repetitive sequence or repetitive transposable elements in the rkn1 region (Wang et al., 2015). In addition, more QTLs around the rkn1 region in RIL population NemX × SJ-2 (Figure 2B) might exist but were not detected because of fewer polymorphic markers available between NemX and SJ-2, suggesting more complexity in this region.
The homoeologous region of chromosome 21 consistently failed to show any detectable resistance contribution even though the order of marker alleles amplified from the same primer pair appeared similar in various segregating populations developed from crosses between the three parental lines. Although the sequence is highly conserved between Chr 11 and Chr 21 (Paterson et al., 2012; Wang K. et al., 2012; Li et al., 2014, 2015; Zhang et al., 2015), only Chr 11 contributed to RKN resistance, emphasizing that the unique structure and gene combination on Chr 11 is the primary basis for RKN resistance control and that minor differences between Chr 11 and Chr 21 result in phenotypic change, as reported earlier with only one nucleotide difference (SCAR marker GHACC1) between NemX and SJ-2 causing phenotypic change for RKN resistance (Wang and Roberts, 2006b). An additional result of importance from this study was that no contribution of resistance to root-galling or egg production was found on Chr 14, supporting that the resistance in NemX is based on rkn1, and thus different from the resistance in other cotton lines such as Wild Mexico Jack Jones, Auburn 623 and its derived M-lines which all carry a resistance gene on Chr 14 effecting egg production but not galling response (Gutiérrez et al., 2010; He et al., 2014; Kumar et al., 2016).
Analysis of the CIR316-215/210 marker alleles in the F2 (Pima S-7 × SJ-2) population, which accounted for 15–24% of phenotypic variance, confirmed that the 215 bp allele in Pima S-7 enhanced resistance to both root-galling and egg production. We identified F2 progenies with GI phenotypes outside the range of the susceptible parents Pima S-7 and SJ-2, including ones with high resistance (GI < 3) and others with about 50% higher GI than the susceptible parents. Thus, both transgressive resistant and transgressive susceptible progenies were present in the F2, providing evidence that both parents contributed to the transgressive resistance. In this F2 population, which lacks the rkn1 locus resistance, the transgressive factor in Pima S-7 (RKN2) might be dominant over the transgressive factor(s) in SJ-2 (RKN3) and allele interaction might occur in the region of CIR316, as found in the testcross. The behavior of RKN2 from Pima S-7 showed dominant action in our previous study (Wang C. et al., 2008), where plants heterozygous or homozygous for RKN2 were effective in enhancing resistance phenotype in the presence of rkn1. Comparing different generations of Pima S-7 × NemX progenies, about 25% transgressive resistant lines were found in the F2 population and in F2:3 families (Wang C. et al., 2008) and 52% in the F2:7 RIL population in this study. It is likely that this is also the situation in progenies with RKN2 coupled with RKN3 in Pima S-7 × SJ-2 crosses, but to result in transgressive resistant phenotypes, the SJ-2 factor (RKN3) must be in the homozygous condition. This would explain transgressive progenies occurring in the F2 but not in the F1 which is susceptible to RKN (Wang et al., 2006b).
Further aspects of the transgressive behavior contributed by the susceptible SJ-2 factor RKN3 were revealed in the NemX × SJ-2 RIL population, in which SJ-2 markers closely linked to CIR316 contributed up to 82% of resistance to root-galling. This result indicated that the RKN3 factor from SJ-2 also interacts with rkn1-based resistance in NemX in the rkn1 resistance region. In our previous study of this population, about 62% of the RILs carrying the homozygous rkn1 resistance were significantly more resistant than the level of resistance in the rkn1 donor parent NemX and up to 30% of total RI lines showed transgressive resistance (Wang et al., 2006a,b). It is likely that this highly resistant set of RILs, which we confirmed to be homozygous for the recessive rkn1 from NemX, were also homozygous for the RKN3 transgressive resistance factor contributed by SJ-2. This was also confirmed in the BC1F1 population SJ-2 × F1 (NemX × SJ-2) in which the marker CIR316-210/221 contributed up to 44% of phenotypic variance to GI resistance. In the BC1F1, transgressive progenies showed less galling than both susceptible parent lines [SJ-2 and F1 (NemX × SJ-2)]. The stronger transgressive resistance which occurred in later than in early generations in the intraspecific NemX × SJ-2 cross compared with interspecific crosses between Pima S-7 and NemX or SJ-2 may reflect the increased levels of homozygosity of the interacting factors. Further, the presence of either homozygous or heterozygous rkn1 improves resistance, and the increasing contribution of resistance is influenced by different recombination backgrounds.
Based on GI and LogEGR phenotypic data in the rkn1 region in different populations and QTL analysis, we constructed a simple genetic model to better conceptualize the transgressive resistance among the three parental lines and their progenies (Table 2). The inferred genetic model is based on the linkage of the pairs of loci (rkn1 with RKN2, rkn1 with RKN3, and RKN2 with RKN3) as indicated by mapping the respective alleles of the closely linked CIR316 marker.
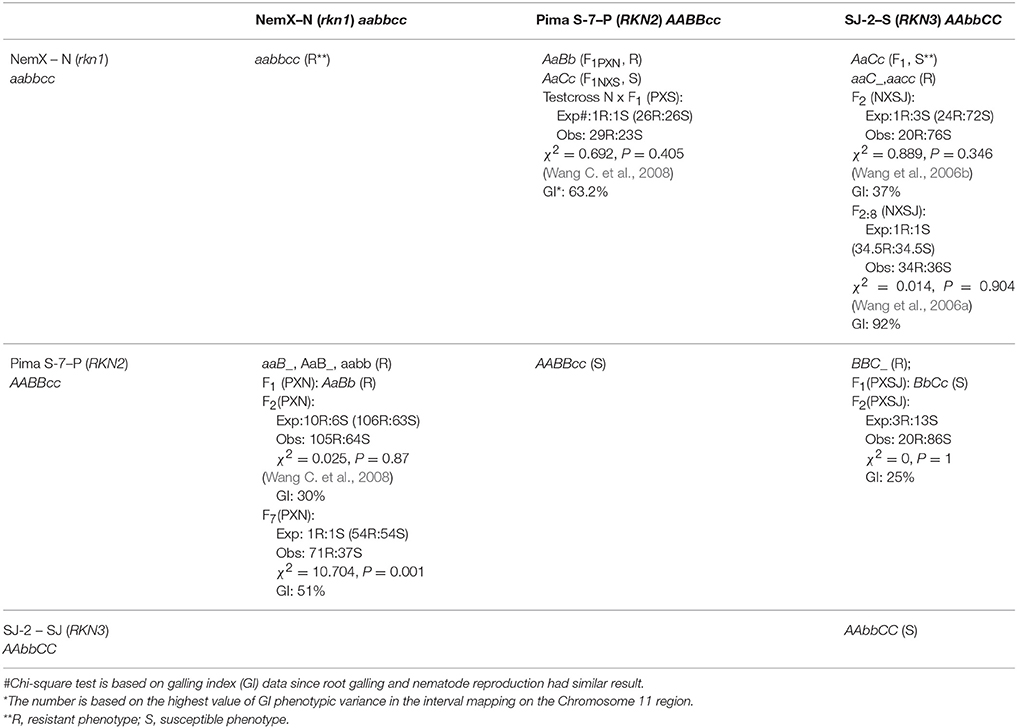
Table 2. The three gene model in five segregating populations based on phenotypic data and QTL analysis.
Allele interactions and/or linked gene interactions in the rkn1 region clearly play a major role in controlling transgressive resistance. We labeled the NemX resistance gene rkn1 as aa, Pima S-7 transgressive factor RKN2 as BB, and the SJ-2 transgressive factor RKN3 as CC. Analysis of the F2 population (Pima S-7 × NemX) published previously (Wang C. et al., 2008), showed that the segregation for root-galling response followed a two-gene model for resistance (R, genotypes AaB-, aaB-, and aabb) and susceptibility (S, genotypes AAB- and A-bb). However, 66% resistant lines (71R:37S) based on GI were observed in the new F2:7 (Pima S-7 × NemX) RIL population which did not match the expected 1:1 ratio if only rkn1 was involved. Since the transgressive resistance depended on the recombination background as discussed above, the greater proportion of resistant lines in this RIL population indicated that multiple genes or alleles are present in the rkn1 region, which was also supported by significant effect QTL(s) spanning 38 cM (0–38 cM) in the rkn1 region associated with both GI and LogEGR phenotypes. Extending the model, the exact fit to a 3R:13S ratio between resistant genotypes (BBC-) and susceptible genotypes (BBcc, BbC-, bbC-, and bbcc) in the F2 (Pima S-7 × SJ-2) population was predicted because both parents (BBcc and bbcc) and the F1 (BbCc) had susceptible phenotypes. The RKN3 gene in SJ-2 (AACC), as with RKN2 in Pima S-7, was expected not to express resistance on its own but produce transgressive resistance in the presence of homozygous rkn1 because the F1 (NemX × SJ-2) (AaCc) was susceptible. This result might explain why in our earlier study (Wang et al., 2006a), 21 (aaC-) out of 34 resistant lines had higher resistance than the resistant parent NemX (aacc) in the NemX × SJ-2 RIL population for which a 3:1 ratio (25.5HR: 8.5R) for GI (χ2 = 3.176, P = 0.075) was found between the highly resistant (HR) genotype (aaC-) and resistant NemX type (aacc).
Considering the resistance contribution of the rkn1 region in all seven segregating populations, the three resistance genes might represent multiple genes in tandem linkage, TAG or multiple alleles, TAA of unknown arrangement in one gene, possibly similar to the arrangement of resistance locus Rhg1 in soybean (Cook et al., 2012). Although we did not have a population which allowed analysis of the three genes together, based on the interaction of the three possible gene pairs which we have analyzed here, the three possible combination pairs of the different loci or genes produce novel resistance (aaB-, A-B-, aaC- and BBCC) or susceptibility (AAbb, AAcc, bbcc) phenotypes in the progenies. The interspecific crosses (aa + BB, BB + CC) produced transgressive resistance (aaB-, A-B-, aaC- and BBCC) in earlier progeny generations than intraspecific crosses (aa + CC). In the presence of rkn1 (a-B- or aaCC), stronger transgressive resistance occurred than without the rkn1 gene (AABB and BBCC).
Sequence tandem repeats result from unequal crossing over during genetic recombination, and these tandem repeats (TR), TAAs, or gene cluster-TAGs are abundant across all domains of life. Little is known about their distribution and contribution to proteins. However, it is known that TR enriched leucine-rich repeats (LRRs) are commonly found in R genes (Schaper and Anisimova, 2015). Evidence of complex recombination between the rkn1 region and its homoeologous chromosome 21 is presented in this study. The behavior of RKN2 and RKN3 showed the presence of TAAs or TAGs enhancing resistance phenotype in the presence of rkn1, resulting in transgressive resistant phenotypes. In addition, resistance to other soil-borne diseases mapped to Chr 11 also indicated the unique structure and resistance gene clustering on Chr 11 (Wang et al., 2015), such as found in resistance to reniform nematode (Dighe et al., 2009), Fusarium wilt (Ulloa et al., 2011, 2013), Verticillium wilt (Bolek et al., 2005), and black root rot (Niu et al., 2008). More research is needed to determine if TRs (e.g., SSRs), TAAs, or TAGs play a major role in transgressive resistance and facilitate resistance to emerging pathogen diseases.
Recently published genome-wide SNP or SSR linkage maps for fiber, yield, or other trait analysis (Yu et al., 2011, 2012; Zhao et al., 2012; Hulse-Kemp et al., 2015; Li et al., 2016; Iqbal and Rahman, 2017; Khan et al., 2017) and the available marker or genome sequence information might shed more light on the mechanism of transgressive resistance. For example, segregation distortion was commonly observed in cotton and greater genetic distance between parents could result in higher distortion (Yu et al., 2011; Zhao et al., 2012; Khan et al., 2017). The distortion could result from translocations, chromosome rearrangements, and other genomic structure variations (Khan et al., 2017). Therefore, deeper sequencing of Chr 11 would increase understanding of resistance gene clusters on Chr 11.
From a practical plant breeding standpoint, the SSR marker CIR316 not only has proven to be a powerful marker to study transgressive resistance in cotton, but also this marker plus the newly developed markers in the rkn1 region reported here are important for marker-based breeding selection to develop nematode resistant cotton varieties. Moreover, through selection of highly resistant progenies resulting from transgressive segregation, valuable novel sources of resistance are available which can be tracked for breeding advancement by the reported marker profiles in the rkn1 region. Although not proven, it is likely that the novel transgressive resistant progenies will be more durable than single source resistance when utilized in cotton production systems, because nematode populations frequently exposed to the resistance would have to overcome the combined effects of multiple allele or multiple gene action.
Conclusions
In this study, QTL mapping of seven intraspecific and interspecific segregating populations generated from Pima S-7, NemX, and SJ-2 crosses revealed the allele profile of marker CIR316 in all tested populations corresponding with phenotypic profile, demonstrating that a transgressive factor (designated as RKN3) from susceptible SJ-2 exists in the region of rkn1. Allele or gene interactions between NemX (rkn1), SJ-2 (RKN3), and Pima S-7 (RKN2) contributed to transgressive resistance. Stronger transgressive resistance occurred in later than in early generations in the intraspecific cross NemX × SJ-2 but not in the interspecific cross. All positive contributions to resistance phenotype came from the rkn1 region on Chr 11, while no evidence was found for any resistance contribution from the homoeologous region of Chr 21. The rkn1 region had similar resistance contribution to both root-galling and egg production in each population. The complex TAA and/or TAG recombination and interactions in the rkn1 resistance region in the NemX background provide a model to study transgressive resistance in plants.
Author Contributions
CW, PR, and MU conceived and designed the study. CW and TD performed the laboratory work. CW, PR, and MU analyzed the data and wrote the manuscript. All the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Teresa Mullens for technical help. Partial support for this work was provided by grants from Cotton Incorporated to PR and MU and from University of California Discovery Grant Program to PR. This research was also partially supported by USDA-ARS (30968-21000-019-00). Partial support also was provided by the National Science Foundation of China (31471749) and the One Hundred Talent Grant Program, Chinese Academy of Sciences to CW. Mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The U.S. Department of Agriculture is an equal opportunity provider and employer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01979/full#supplementary-material
Abbreviations
Chr, Chromosome; EGR, Eggs per gram root; GI, Galling Index; IM, Interval Mapping; K* test, Kruskal-Wallis Analysis; RIL, Recombinant Inbred Line; RKN, Root-knot nematodes; QTL, Quantitative Trait Locus.
References
Aghnoum, R., and Niks, R. E. (2011). Transgressive segregation for very low and high levels of basal resistance to powdery mildew in barley. J. Plant Physiol. 168, 45–50. doi: 10.1016/j.jplph.2010.09.005
Bayles, M. B., Verhalen, L. M., McCall, L. L., Johnson, W. M., and Barnes, B. R. (2005). Recovery of recurrent parent traits when backcrossing in cotton. Crop Sci. 45, 2087–2095. doi: 10.2135/cropsci2004.0631
Bell, M. A., and Travis, M. P. (2005). Hybridization, transgressive segregation, genetic covariation, and adaptive radiation. Trends Ecol. Evol. 20, 358–361. doi: 10.1016/j.tree.2005.04.021
Bezawada, C., Saha, S., Jenkins, J. N., Creech, R. G., and McCarty, J. C. (2003). SSR marker(s) associated with root-knot nematode resistance gene(s) in cotton. J. Cotton Sci. 7, 179–184.
Bolek, Y., El-Zik, K. M., Pepper, A. E., Bell, A. A., Magill, C. M., Thaxton, P. M., et al. (2005). Mapping of Verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. doi: 10.1016/j.plantsci.2005.02.008
Bridge, J., and Page, S. L. J. (1980). Estimation of root-knot nematode infestation levels on roots using a rating chart. Trop. Pest Manage. 26, 296–298. doi: 10.1080/09670878009414416
Cherif, M., and Harrabi, M. (1993). Transgressive segregation for resistance to Pyrenophora teres in barley. Plant Pathol. 42, 617–621. doi: 10.1111/j.1365-3059.1993.tb01542.x
Cook, D. E., Lee, T. G., Guo, X., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
Dighe, N., Robinson, A. F., Bell, A., Menz, M., Cantrell, R., and Stelly, D. (2009). Linkage mapping of resistance to reniform nematode in cotton (Gossypium hirsutum L.) following introgression from G. longicalyx (Hutch & Lee). Crop Sci. 49, 1151–1164. doi: 10.2135/cropsci2008.03.0129
Goodell, P. B., and Montez, G. H. (1994). “Acala cotton tolerance to southern root-knot nematode, Meloidogyne incognita,” in Proc. of Beltwide Cotton Prod. Res. Conf. Natl. Cotton Council of Am. (Memphis, TN). 265–267.
Gutiérrez, O. A., Jenkins, J. N., Wubben, M. J., Hayes, R. W., and Callahan, F. E. (2010). SSR markers closely associated with genes for resistance to root-knot nematode on chromosomes 11 and 14 of Upland cotton. Theor. Appl. Genet. 121, 1323–1337. doi: 10.1007/s00122-010-1391-9
He, Y., Kumar, P., Shen, X., Davis, R. F., Van Becelaere, G., May, O. L., et al. (2014). Re-evaluation of the inheritance for root-knot nematode resistance in the Upland cotton germplasm line M-120 RNR revealed two epistatic QTLs conferring resistance. Theor. Appl. Genet. 127, 1343–1351. doi: 10.1007/s00122-014-2302-2
Hulse-Kemp, A. M., Lemm, J., Plieske, J., Ashrafi, H., Buyyarapu, R., Fang, D. D., et al. (2015). Development of a 63K SNP array for cotton and high-density mapping of intra-and inter-specific populations of Gossypium spp. Genes Genomes Genet. 5, 1187–1209. doi: 10.1534/g3.115.018416
Hussey, R. S., and Barker, K. R. (1973). A comparison of methods of collecting inocula of Meloidogyne spp. including a new technique. Plant Dis. Rep. 57, 1025–1028.
Hyer, A. H., and Jorgenson, E. C. (1984). “Root-knot nematode resistance in cotton breeding, techniques and results,” in Proc. of Beltwide Cotton Prod. Res. Conf. Natl. Cotton Council of Am. (Memphis, TN). 377–379.
Hyer, A. H., Jorgenson, E. C., Garber, R. H., and Smith, S. (1979). Resistance to root-knot nematode in control of root-knot nematode Fusarium wilt disease complex in cotton Gossypium hirsutum. Crop Sci. 19, 898–901. doi: 10.2135/cropsci1979.0011183X001900060036x
Imtiaz, M., Cromey, M. G., Hampton, J. G., and Ahmad, M. (2003a). Inheritance of seedling resistance to stripe rust (Puccinia striiformis f. sp. tritici) in ‘Otane’ and ‘Tiritea’ wheat (Triticum aestivum). N. Zeal. J. Crop Hortic. Sci. 31, 15–22. doi: 10.1080/01140671.2003.9514231
Imtiaz, M., Cromey, M. G., Hampton, J. G., and Ahmad, M. (2003b). Inheritance of durable adult plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in ‘Otane’ wheat (Triticum aestivum). N. Zeal. J. Crop Hortic. Sci. 31, 23–31. doi: 10.1080/01140671.2003.9514232
Iqbal, M. A., and Rahman, M. (2017). Identification of marker-trait associations for lint traits in cotton. Front. Plant Sci. 8:86. doi: 10.3389/fpls.2017.00086
Khan, M. K. R., Chen, H., Zhou, Z., Ilyas, M. K., Wang, X., Cai, X., et al. (2017). Genome wide SSR high density genetic map construction from an interspecific cross of Gossypium hirsutum x Gossypium tomentosum. Front. Plant Sci. 7:436. doi: 10.3389/fpls.2016.00436
Kumar, P., He, Y., Singh, R., Davis, R. F., Guo, H., Paterson, A. H., et al. (2016). Fine mapping and identification of candidate genes for a QTL affecting Meloidogyne incognita reproduction in Upland cotton. BMC Genomics 201:567. doi: 10.1186/s12864-016-2954-1
Li, C., Dong, Y., Zhao, T., Li, L., Li, C., Yu, E., et al. (2016). Genome-wide SNP linkage mapping and QTL analysis for fiber quality and yield traits in the Upland cotton recombinant inbred lines population. Front. Plant Sci. 7:1356. doi: 10.3389/fpls.2016.01356
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33:524. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
McPherson, M. G., Jenkins, J. N., Watson, C. E., and McCarty, J. C. Jr. (2004). Inheritance of root-knot nematode resistance in M-315 RNR and M78-RNR cotton. J. Cotton Sci. 8, 154–161.
Navabi, A., Singh, R. P., Tewari, J. P., and Briggs, K. G. (2004). Inheritance of high levels of adult-plant resistance to stripe rust in five spring wheat genotypes. Crop Sci. 44, 1156–1162. doi: 10.2135/cropsci2004.1156
Niu, C., Lister, H. E., Nguyen, B., Wheeler, T. A., and Wright, R. J. (2008). Resistance to Thielaviopsis basicola in the cultivated A genome cotton. Theor. Appl. Genet. 117, 1313–1323. doi: 10.1007/s00122-008-0865-5
Oakley, S. R. (1995). “CPCSD Acala C-225, a new nematode-resistant Acala variety for California's San Joaquin Valley,” in Proc. of Beltwide Cotton Prod. Res. Conf. Natl. Cotton Council of Am. (Memphis, TN), 39.
Ogallo, J. L., Goodell, P. B., Eckert, J. W., and Roberts, P. A. (1999). Management of root-knot nematodes with resistant cotton cv. NemX. Crop Sci. 39, 418–421. doi: 10.2135/cropsci1999.0011183X0039000200020x
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers. Nature 492, 423–427. doi: 10.1038/nature11798
Rieseberg, L. H., Archer, M. A., and Wayne, R. K. (1999). Transgressive segregation, adaption and speciation. Heredity 83, 363–372. doi: 10.1038/sj.hdy.6886170
Rieseberg, L. H., Widmer, A., Arntz, A. M., and Burke, J. M. (2003). The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 1141–1147. doi: 10.1098/rstb.2003.1283
Roberts, P. A., and Ulloa, M. (2010). Introgression of root-knot nematode resistance into tetraploid cotton. Crop Sci. 50, 940–951. doi: 10.2135/cropsci2009.05.0281
Robinson, A. F., Bowman, D. T., Cook, C. G., Jenkins, J. N., Jones, J. E., May, L. O., et al. (2001). “Nematode resistance,” in Compendium of Cotton Diseases, eds T. L. Kirkpatrick and C. S. Rothrock (St. Paul, MN: APS Press), 68–79.
Schaper, E., and Anisimova, M. (2015). The evolution and function of protein tandem repeats in plants. New Phytol. 206, 397–410. doi: 10.1111/nph.13184
Shen, X., He, Y., Lubbers, E. L., Davis, R. F., Nichols, R. L., and Chee, P. W. (2010). Fine mapping QMi-C11 a major QTL controlling root-knot nematodes resistance in Upland cotton. Theor. Appl. Genet. 121, 1623–1631. doi: 10.1007/s00122-010-1415-5
Shen, X., Van Becelaere, G., Kumar, P., Davis, R. F., May, L. O., and Chee, P. (2006). QTL mapping for resistance to root-knot nematodes in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theor. Appl. Genet. 113, 1539–1549. doi: 10.1007/s00122-006-0401-4
Shepherd, R. L. (1974). Transgressive segregation for root-knot nematode resistance in cotton. Crop Sci. 14, 872–875. doi: 10.2135/cropsci1974.0011183X001400060029x
Staal, J., Kaliff, M., Bohman, S., and Dixelius, C. (2006). Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 46, 218–230. doi: 10.1111/j.1365-313X.2006.02688.x
Starr, J. L., Moresco, E., Smith, C. W., Nichols, R. W., Roberts, P. A., and Chee, P. (2010). Inheritance of resistance to Meloidoygne incognita in primitive cotton accessions from Mexico. J. Nematol. 42, 352–358.
Ulloa, M., Hutmacher, R. B., Roberts, P. A., Wright, S. D., Nichols, R. L., and Davis, R. M. (2013). Inheritance and QTL mapping of Fusarium wilt race 4 resistance in cotton. Theor. Appl. Genet. 126, 1405–1418. doi: 10.1007/s00122-013-2061-5
Ulloa, M., Wang, C., Hutmacher, R. B., Wright, S. D., Davis, R. M., Saski, C. A., et al. (2011). Mapping Fusarium wilt race 1 resistance genes in cotton by inheritance, QTL and sequencing composition. Mol. Genet. Genomics 286, 21–36. doi: 10.1007/s00438-011-0616-1
Ulloa, M., Wang, C., and Roberts, P. A. (2010). Gene action analysis by inheritance and quantitative trait loci mapping of resistance to root-knot nematodes in cotton. Plant Breed. 129, 541–550. doi: 10.1111/j.1439-0523.2009.01717.x
Ulloa, M., Wang, C., Saha, S., Hutmacher, R. B., Stelly, D. M., Jenkins, J. N., et al. (2016). Analysis of root-knot nematode and Fusarium wilt disease resistance in cotton (Gossypium spp.) using chromosome substitution lines from two alien species. Genetica 144, 167–179. doi: 10.1007/s10709-016-9887-0
Van Ooijen, J. W. (2004). MapQTL® 5, Software for the Calculation of Quantitative Trait Loci in Experimental Populations. Wageningen: Kyazma BV.
Van Ooijen, J. W. (2006). JoinMap® 4.0 Software for the Calculations of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma, BV.
Wang, C., Matthews, W. C., and Roberts, P. A. (2006a). Phenotypic expression of rkn1-mediated Meloidogyne incognita resistance in Gossypium hirsutum populations. J. Nematol. 38, 250–257.
Wang, C., and Roberts, P. A. (2006a). A Fusarium wilt resistance gene in Gossypium barbadense and its effect on root-knot nematode-wilt disease complex. Phytopathology 96, 727–734. doi: 10.1094/PHYTO-96-0727
Wang, C., and Roberts, P. A. (2006b). Development of AFLP and derived CAPS markers for root-knot nematode resistance in cotton. Euphytica 152, 185–196. doi: 10.1007/s10681-006-9197-1
Wang, C., Ulloa, M., Mullens, T. R., Yu, J. Z., and Roberts, P. A. (2012). QTL analysis for transgressive resistance to root-knot nematode in interspecific cotton (Gossypium spp.) progeny derived from susceptible parents. PLoS ONE 7:e34874. doi: 10.1371/journal.pone.0034874
Wang, C., Ulloa, M., and Roberts, P. A. (2006b). Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 112, 770–777. doi: 10.1007/s00122-005-0183-0
Wang, C., Ulloa, M., and Roberts, P. A. (2008). A transgressive segregation factor (RKN2) in Gossypium barbadense for nematode resistance clusters with gene rkn1 in G. hirsutum. Mol. Gen. Genomics 279, 41–52. doi: 10.1007/s00438-007-0292-3
Wang, C., Ulloa, M., Shi, X., Yuan, X., Saski, C., Yu, J. Z., et al. (2015). Sequence composition of BAC clones and SSR markers mapped to Upland cotton chromosomes 11 and 21 targeting resistance to soil-borne pathogens. Front. Plant Sci. 6:791. doi: 10.3389/fpls.2015.00791
Wang, H. M., Lin, Z. X., Zhang, X. L., Chen, W., Guo, X. P., Nie, Y. C., et al. (2008). Mapping and quantitative trait loci analysis of Verticillium wilt resistance genes in cotton. J. Integr. Plant Biol. 50, 174–182. doi: 10.1111/j.1744-7909.2007.00612.x
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1104. doi: 10.1038/ng.2371
Ynturi, P., Jenkins, J. N., McCarty, J. C. Jr., Gutiérrez, O. A., and Saha, S. (2006). Association of root-knot nematode resistance genes with simple sequence repeat markers on two chromosomes in cotton. Crop Sci. 46, 2670–2674. doi: 10.2135/cropsci2006.05.0319
Yu, J. Z., Kohel, R. J., Fang, D. D., Cho, J., Van Deynze, A., Ulloa, M., et al. (2012). A high-density simple sequence repeat and single nucleotide polymorphism genetic map of the tetraploid cotton genome. Genes Genomes Genet. 2, 43–58. doi: 10.1534/g3.111.001552
Yu, Y., Yuan, D. J., Liang, S. G., Li, X. M., Wang, X. Q., Lin, Z. X., et al. (2011). Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 12:15. doi: 10.1186/1471-2164-12-15
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33:531. doi: 10.1038/nbt.3207
Zhang, Z. J., Yang, G. H., Li, G. H., Jin, S. L., and Yang, X. B. (2001). Transgressive segregation, heritability, and number of genes controlling durable resistance to stripe rust in one Chinese and two Italian wheat cultivars. Phytopathology 91, 680–686. doi: 10.1094/PHYTO.2001.91.7.680
Zhao, G., Ablett, G. R., Anderson, T. R., Rajcan, I., and Schaafsma, A. W. (2005). Inheritance and genetic mapping of resistance to Rhizoctonia root and hypocotyl rot in soybean. Crop Sci. 45, 1441–1447. doi: 10.2135/cropsci2004.0560
Keywords: root-knot nematodes (RKN), Meloidogyne incognita, Gossypium spp., upland, Pima, allele interactions
Citation: Wang C, Ulloa M, Duong TT and Roberts PA (2017) QTL Analysis of Transgressive Nematode Resistance in Tetraploid Cotton Reveals Complex Interactions in Chromosome 11 Regions. Front. Plant Sci. 8:1979. doi: 10.3389/fpls.2017.01979
Received: 18 August 2017; Accepted: 02 November 2017;
Published: 20 November 2017.
Edited by:
Nicolas Rispail, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Mehboob-ur-Rahman, NIBGE, PakistanTuanjie Zhao, Nanjing Agricultural University, China
Copyright © 2017 Wang, Ulloa, Duong and Roberts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip A. Roberts, cGhpbGlwLnJvYmVydHNAdWNyLmVkdQ==
 Congli Wang
Congli Wang Mauricio Ulloa
Mauricio Ulloa Tra T. Duong1
Tra T. Duong1 Philip A. Roberts
Philip A. Roberts