- 1Hunan Provincial Key Laboratory for Biology and Control of Plant Diseases and Insect Pests, College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2Hunan Provincial Key Laboratory for Germplasm Innovation and Utilization of Crop, Changsha, China
- 3Agricultural College, Hunan Agricultural University, Changsha, China
The hypersensitive response (HR) is a mechanism by which plants prevent the spread of pathogen. Despite extensive study, the molecular mechanisms underlying HR remain poorly understood. Lesion mimic mutants (LMMs), such as LIL1 that was identified in an ethylmethane sulfonate mutagenized population of Indica rice (Oryza sativa L. ssp. Indica) 93-11, can be used to study the HR. Under natural field conditions, the leaves of LIL1 mutant plants exhibited light-induced, small, rust-red lesions that first appeared at the leaf tips and subsequently expanded throughout the entire leaf blade to the leaf sheath. Histochemical staining indicated that LIL1 lesions displayed an abnormal accumulation of reactive oxygen species (ROS) and resulted from programmed cell death (PCD). The LIL1 mutants also displayed increased expression of defense-related genes and enhanced resistance to rice blast fungus (Magnaporthe grisea). Genetic analysis showed that mutation of LIL1 created a semi-dominant allele. Using 1,758 individuals in the F2 population, LIL1 was mapped in a 222.3 kb region on the long arm of chromosome 7. That contains 12 predicted open reading frames (ORFs). Sequence analysis of these 12 candidate genes revealed a G to A base substitution in the fourth exon of LOC_Os07g30510, a putative cysteine-rich receptor-like kinase (CRK), which led to an amino acid change (Val 429 to Ile) in the LIL1 protein. Comparison of the transcript accumulation of the 12 candidate genes between LIL1 and 93-11 revealed that LOC_Os07g30510 was up-regulated significantly in LIL1. Overexpression of the LOC_Os07g30510 gene from LIL1 induced a LIL1-like lesion phenotype in Nipponbare. Thus, LIL1 is a novel LMM in rice that will facilitate the further study of the molecular mechanisms of HR and the rice blast resistance.
Introduction
Plants combat pathogens with a sophisticated immune system consisting of two branches. The first uses transmembrane pattern recognition receptors (PRRs) to recognize conserved pathogen-associated molecular patterns (PAMPs) and activate PAMP-triggered immunity (PTI). The second uses nucleotide binding site-leucine rich repeat (NBS-LRR) proteins to recognize effectors produced by the pathogens and activate effector triggered immunity (ETI). PTI and ETI elicit similar defense responses with ETI more frequently accompanied by a hypersensitive response (HR) (Jones and Dangl, 2006). A defining feature of strong, local defenses associated with the HR is programmed cell death (PCD) of host plant cells. Other events correlated with the HR include ion fluxes, active oxygen bursts, defense-related gene expression, accumulation of antimicrobial compounds and cell wall fortifications (Yin et al., 2000). The HR also activates a whole plant, long-lasting, broad-spectrum resistance to subsequent pathogens infection, referred to as systemic acquired immunity (SAR) (Hunt et al., 1996).
The HR is considered one of the most important reactions of plant resistance to pathogens (Tomiyama, 1963). However, the molecular mechanism regulating the HR remain mysterious. An efficient way to study the molecular mechanisms of HR and disease resistance is to use lesion mimic mutants (LMMs), which exhibit spontaneous, disease-like lesions without pathogen attack. These lesions, which can resemble either disease symptoms or pathogen-inducible HR, frequently are associated with enhanced plant disease resistance to a wide range of pathogens (Lorrain et al., 2003). Numerous LMMs have been identified in many plants, including Arabidopsis (Dietrich et al., 1994; Bowling et al., 1997; Mou et al., 2000), maize (Walbot, 1991; Johal et al., 1995), wheat (Yao et al., 2009; Du et al., 2014; Wang et al., 2016), barley (Wolter et al., 1993; Rostoks et al., 2006) and rice (Takahashi et al., 1999; Yin et al., 2000; Mizobuchi et al., 2002; Campbell and Ponald, 2005; Mori et al., 2007; Wu et al., 2008; Qiao et al., 2010; Chen et al., 2012; Fekih et al., 2015; Li et al., 2016). The notion that complex and diverse pathways underlie the phenotypes of LMMs is supported by the mutated genes falling into many different functional groups, including membrane-associated proteins (Büschges et al., 1997; Lorrain et al., 2004; Noutoshi et al., 2006), ion channel proteins (Balague et al., 2003; Rostoks et al., 2006), an U-Box/Armadillo repeat protein (Zeng et al., 2004), a splicing factor 3b subunit 3 (Chen et al., 2012), a zinc-finger protein (Dietrich et al., 1997), a heat stress transcription factor (Yamanouchi et al., 2002), a clathrin-associated adaptor protein (Qiao et al., 2010) and proteins involved in the biosynthesis and metabolic pathways of phenolic compounds (Gray et al., 1997), such as porphyrin (Hu et al., 1998; Ishikawa et al., 2001) and fatty acids or lipids (Kachroo et al., 2001; Brodersen et al., 2002). Although the phenotypes of some LMMs result from disorder in pathways unrelated to defense responses (Hu et al., 1998; Yamanouchi et al., 2002), extensive studies of other LMMs and their genes has shed light on complex and diverse pathways regulating both the initiation and containment of HR-associated cell death in plants (Lorrain et al., 2003).
In this study, we cloned the gene for which mutation and over-expression in the light-induced lesion mimic mutant 1 (LIL1) caused necrotic spots around 3-leaf stage and significantly reduced plant height. Diaminobenzidine (DAB) and trypan blue staining analysis revealed that ROS accumulation and death of plant cells, respectively, accompanied the lesions in the LIL1 mutant. Meanwhile, we also found that LIL1 plants had elevated expression of defense-related genes and increased resistance to Magnaporthe grisea. Genetic analysis of LIL1 indicated that the lesion phenotype is controlled by a semi-dominant gene located in a 222.3 kb interval of chromosome 7 that contains 12 predicted open reading frames (ORFs). Sequencing of these ORFs identified a missense mutation from G to A in the fourth exon of a predicted CRK, LOC_Os07g30510, that converts Val 429 to Ile within the serine/threonine protein kinase catalytic (STKc) domain. The expression levels of the 12 ORFs within the cloning interval were also compared between LIL1 and 93-11 and LOC_Os07g30510 was up-regulated significantly in LIL1. Notably, over-expression of the LOC_Os07g30510 gene caused Nipponbare to exhibit a lesion mimic phenotype similar to LIL1. Our identification of a novel LMM in a CRK gene, LIL1, in rice will facilitate further study of the molecular mechanisms of HR and rice blast resistance.
Materials and Methods
Plant Materials and Growth Conditions
The LIL1 mutant was derived from an M1 population of the indica rice cultivar 93-11 after treatment with 0.1% EMS. The leaf lesion phenotype was genetically stable for more than four generations under greenhouse and field conditions. Therefore, the F1 hybrid and F2 or BC1 progenies derived from crossing LIL1 with TeQing or Nipponbare were used for genetic analysis and molecular mapping. All the plants were grown at the Hunan Agricultural University in Changsha, or at Lingshui, Hainan, China.
To determine the occurrence of the lesion phenotype under aseptic conditions, the seeds of wild-type (WT) 93-11 and LIL1 were surface sterilized for 30 min in 20% bleach, rinsed three times in sterile water, and germinated in autoclaved MS medium in a 1000 mL glass cylinder at 25°C and with 14 h light/10 h dark.
Trypan Blue and DAB Staining
Trypan blue staining is a histochemical method to detect cell death or irreversible membrane damage. Leaves of LIL1 were harvested for trypan blue staining when visible lesions were apparent. Trypan blue staining was performed as previously described (Bowling et al., 1997).
3,3′-diaminobenzidine (DAB) staining was used to detect H2O2 accumulation in the leaves as previously described (Wang et al., 2007). Leaves of LIL1 were collected for the DAB staining prior to the appearance of macroscopic lesions.
Evaluation of Rice Blast Resistance
The LIL1 mutants were evaluated for blast resistance following manual inoculation in a growth chamber. The seedlings of LIL1 and 93-11 were grown in plastic trays filled with fertile soil in a greenhouse at 27–30°C. A 5 μL droplet of a spore suspension of M. grisea (∼2.0 × 105 conidia/mL) was placed onto the leaf blades of 4 week old seedlings followed by incubation in a growth chamber for 24 h in the dark at 26–27°C. Following inoculation, the leaf blades were misted with water after every 6-7 h for 4–5 days to maintain moisture and facilitate infection. The disease symptoms were observed after 7 days after inoculation.
RT-qPCR and Semi-quantitative RT-PCR Analysis
Total RNA was extracted with Trizol reagent (Invitrogen Biotechnology Co., Shanghai, China) according to the protocol. After DNaseI treatment, 1 μg of RNA was added to a 20 μL reaction system to synthesize first-strand cDNA using the Reverse Transcription System (Invitrogen Biotechnology Co., Shanghai, China) according to the instructions. Relative quantitative reverse transcription polymerase chain reaction (RT-qPCR) to assess the transcript abundances of defense-related genes were performed in 25 μL reactions using 1.0 μL of 1:10 diluted cDNA as template with 3 technical replicates within each biological replicate. Semi-Quantitative RT-PCR on candidate genes within the LIL1 mapping interval and on Nipponbare lines over-expressing LOC_Os07g30510 from LIL1 were performed in 25 μL reactions using 1.0 μL cDNA as template. In each case, the rice actin gene was used as the internal control. All primers for RT-PCR are list in Supplementary Table S1.
Genetic Analysis of LIL1
In November 2012, F1 hybrids, BC1 or F2 progenies derived from the reciprocal crosses between 93-11 and LIL1, TeQing and LIL1, Nipponbare and LIL1 were planted in Lingshui, Hainan Province. Segregation ratios of the leaf lesions were examined in the F2 and BC1 progenies at the booting stage.
DNA Extraction and SSR Analysis
Total rice genomic DNA from both parents and each individual of the F2 was extracted from mature leaves as described by McCouch et al. (1988). Microsatellite (simple sequence repeats, SSR) primer sequences were adopted from Temnykh et al. (2000) and McCouch et al. (2002) and synthesized by Invitrogen Biotechnology Co., Shanghai, China. The basic SSR procedure was as follows: 10× reaction buffer 2.0 μL, 2.0 mM of each dNTP, 2.0 U of Taq polymerase, 50 ng of template DNA, 30 ng of each primer, and distilled water in a 20 μL reaction that was overlaid with a drop of mineral oil. The amplification reactions were performed as follows: 95°C for 5 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s; and 72°C for 10 min. All PCR reactions were performed in an MJ PTC-100 thermocycler (Waltham, MA, United States). The amplification products were observed in a 8% polyacrylamide gels or 5% agarose gels stained with silver or ethidium bromide, respectively.
Development of New Markers
Simple sequence repeats sequences in BACs near the target gene were identified using SSRHunter (Li and Wan, 2005). CAPS markers were obtained according to the rice DNA polymorphic database between Nipponbare (O. sativa ssp. japonica) and 93-11 (O. sativa ssp. indica) (Shen et al., 2004). Primer Premier 5.0 was used to design all of the primers. Those markers that exhibited polymorphisms between TeQing and LIL1 were applied in the F2 mapping population.
Data Analysis and Linkage Mapping
The molecular data were analyzed by MAPMAKER/EXP 3.0b (Lander et al., 1987). The band types identical with that of TeQing, LIL1, and F1 were recorded as 1, 2, and 3, respectively. MAPDRAW2.1 (Liu and Meng, 2003) was used to a construct the linkage map.
Gene Annotation and Sequence Analysis
The candidate region was annotated by the Rice Genome Annotation Project1 in japonica rice Nipponbare. The candidate genes were sequenced by the Shanghai Invitrogen Biotechnology Co., China. PCR reactions were performed using KOD-Plus-Neo Taq polymerase (TOYOBO life science, Japan) and amplified fragments were separated on a 1.0% agarose gels. ContigExpress was used to assemble sequences and DNAMAN 5.2.2 was used to align the sequences.2
Mutant Validation
To confirm the role of LOC_Os07g30510 in the mutant phenotype of LIL1, we cloned the coding sequence of LOC_Os07g30510 from LIL1 by PCR amplification with gene-specific primers (5′-CGGAATTCATGGCCATTTTGACCGTTCTG-3′ and 5′-CTGTCTAGATTACCTGCCGTTCACAATTGTT-3′). Underlining indicates EcoRI and XbaI sites in the forward and reverse primers, respectively. LOC_Os07g30510 from LIL1 was cloned into the EcoR1 and Xba1 sites downstream from the rice Actin1 promoter in the binary vector pCAMBIA2300 and transformed into Nipponbare by Agrobacterium tumefaciens-mediated transformation. Primers for identifying the transgenic plants were 5′-GAATCCCTCAGCATTGTTC-3′ and 5′-TCACGAGGGTGACGAAGT-3′, with annealing sites on the rice Actin1 promoter and LOC_Os07g30510, respectively.
Result
Phenotypes of LIL1 Mutant
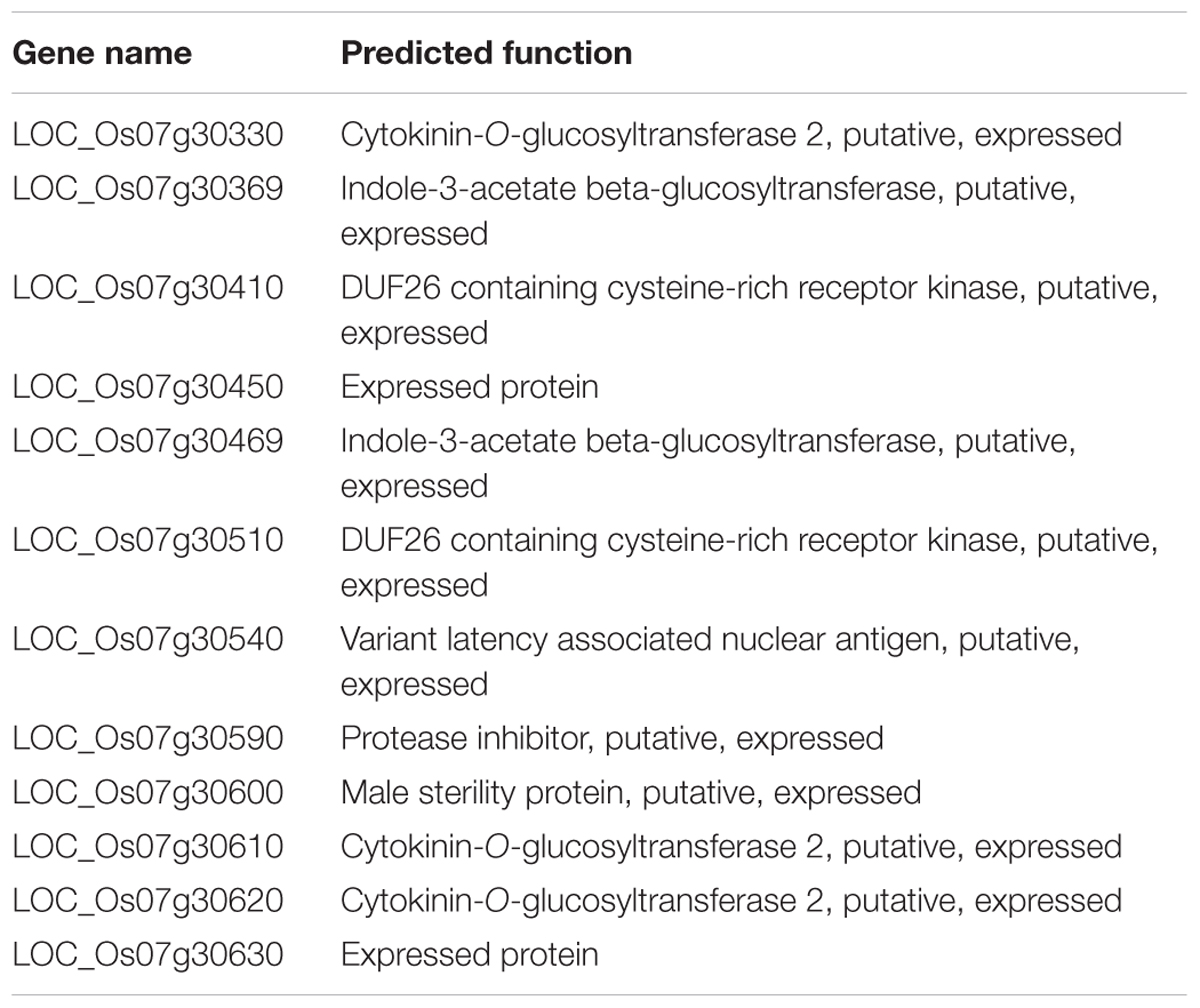
Compared with the WT, the plant height of the LIL1 mutant was reduced significantly (Figures 1A,B). Lesions of the LIL1 mutant first appeared on leaf tips at approximately the 3-leaf stage and then expanded to the whole leaf surface and eventually the leaf sheath. The lesions were small and rust red in color. The older leaves with many lesions turned yellow and showed early senescence. Ultimately, the whole LIL1 plants dried up earlier than the WT plants (Figure 1A). The lesion formation was controlled by a semi-dominant gene (Figure 1C), which decreased remarkably when light was intercepted by covering the leaf with aluminum foil (Figure 1D). To determine whether lesion formation in LIL1 required an exogenous biotic trigger, we grew LIL1 under sterile conditions. The initiation of lesion in these mutants was the same as plants grown in greenhouse conditions (data not shown). The result suggests that the lesion formation in LIL1 mutant is not caused by other biotic agents. To determine if H2O2 accumulation is associated with the onset of lesions, we conducted DAB staining on leaf tips from 2 weeks old plants that did not yet show any visible lesions. Brownish spots appeared on the pre-symptomatic leaves of LIL1 indicating that there is a burst of active oxygen species that precedes the onset of visible lesions (Figure 1E). To determine if the lesions on LIL1 are associated with plant cell death, we conducted trypan blue staining on lesioned leaves from 3.5 weeks old plants. The appearance of deep blue staining at the lesion sites in LIL1 mutants indicates the occurrence of extensive cell death (Figure 1F).
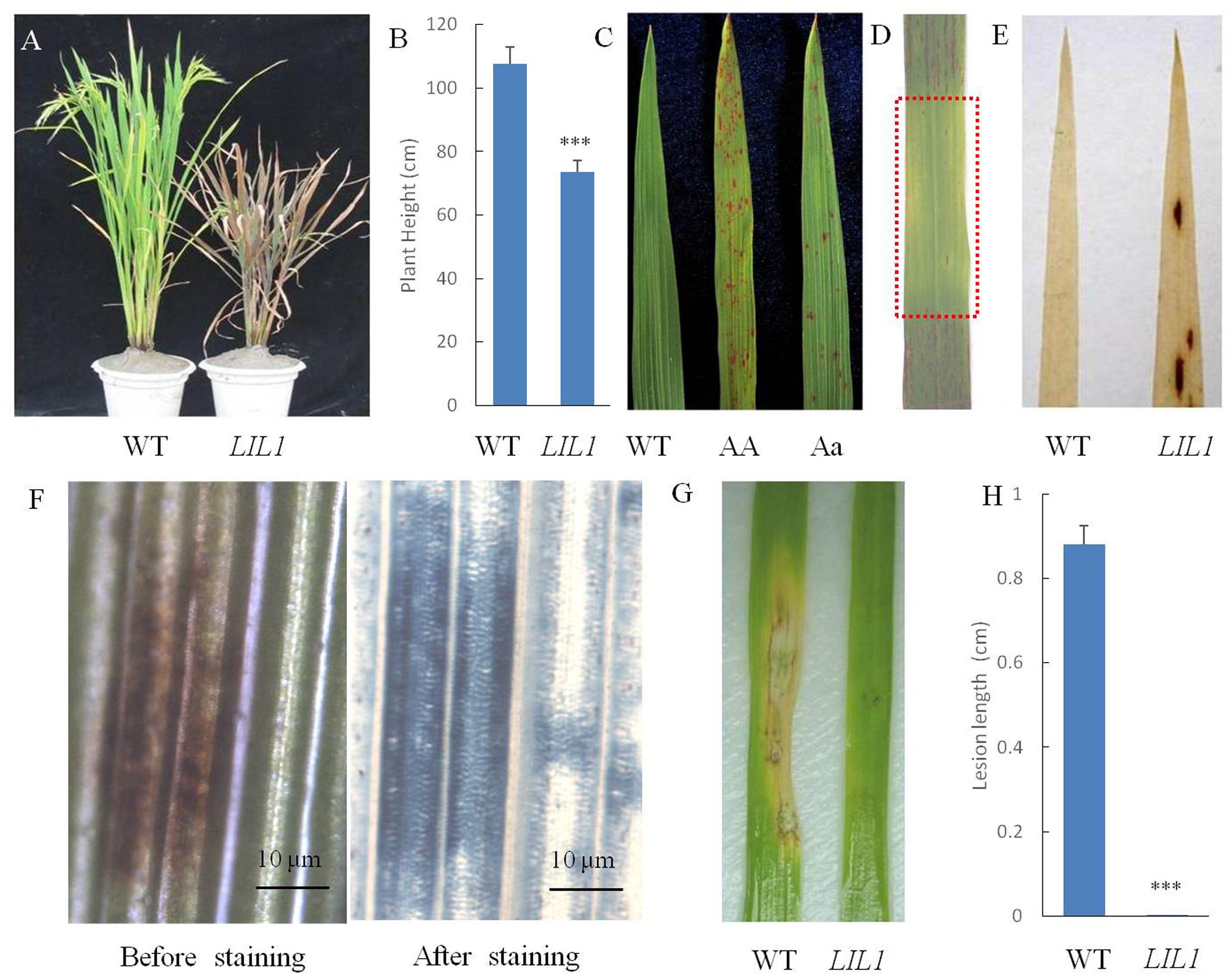
FIGURE 1. Phenotype characterization of the LIL1 mutant. (A) Photographs of representative plants of the WT (right) and LIL1 mutant (left) at filling stage; (B) Plant height of WT and LIL1. Values are means ± standard errors of 10 replications. Significance was determined at ∗∗∗P < 0.0001 with a Student’s t-test; (C) LIL1 is partially dominant: 93-11 (WT), LIL1 Homozygous (AA), LIL1 Heterozygous (Aa); (D) Determination of whether the LIL1 phenotype was light dependent. The area covered with aluminum is foil boxed with a red rectangle; (E) DAB staining results; (F) Trypan blue staining results. (G) Disease phenotypes of WT and LIL1 after inoculation with M. grisea isolate ZB25; (H) Lesion length of WT and LIL1 after inoculation with ZB25. Values are means ± standard errors of 10 replications. Significance was determined at ∗∗∗P < 0.0001 with a student’s t-test.
Enhanced Resistance of LIL1 Mutant to Rice Blast Pathogens
Many LMMs exhibit enhanced resistance to pathogens. To determine whether the mutation of LIL1 led to enhanced resistance to pathogens, we inoculated 4-week-old LIL1 and 93-11 plants with M. grisea isolates RB2, RB17, RB13 and ZB25. While 93-11 and LIL1 did not show a difference in resistance to M. grisea isolates RB2, RB17 or RB13 (data not shown), there was a significant difference in resistance to M. grisea isolate ZB25. The rice blast symptoms caused by ZB25 on leaves of LIL1 were almost invisible in stark contrast to the strong symptoms apparent on 93-11 (Figures 1G,H). Thus, LIL1 plants display significantly increased resistance against M. grisea.
In plants with enhanced disease resistance, defense genes frequently are expressed constitutively at elevated levels. To determine whether this is the case in LIL1, we analyzed the expression of pathogenesis-related (PR) genes and peroxidase genes in the flag leaf of LIL1 plants at the filling stage using quantitative RT-PCR. Consistent with the enhanced disease resistance of LIL1, the transcripts of the PR genes, PR1 and PR10, and the peroxidase genes, POC-1 and POX22.3, were significantly increased in LIL1 compared to 93-11 (Figure 2).
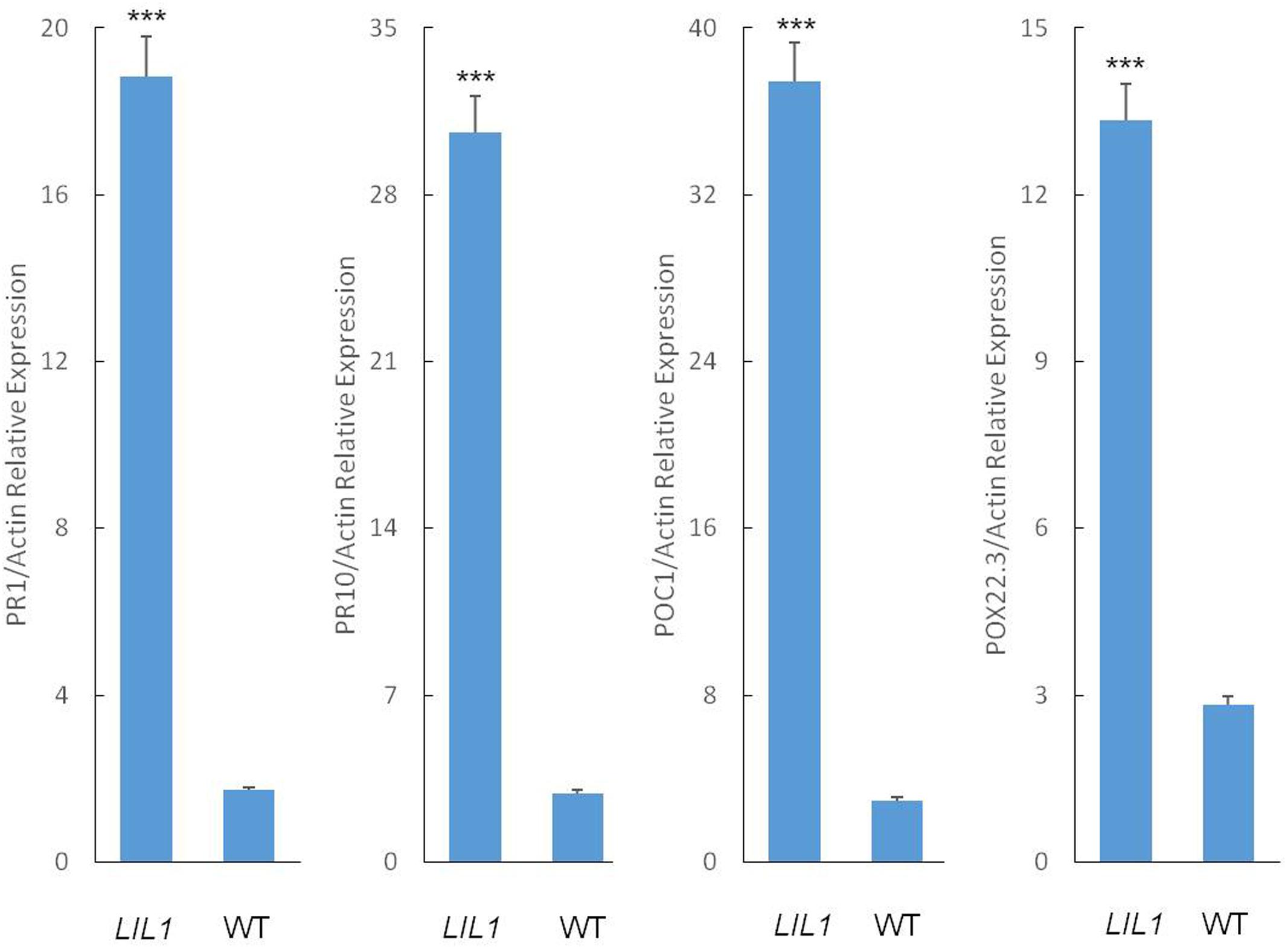
FIGURE 2. Defense-related gene expression in the LIL1 mutant. Transcript abundance was determined via RT-qPCR. The error bars represent the standard deviation between three biological replicates. Mean fold-changes in the transcript abundance were calculated using the ΔΔCt method between biological replicates ± standard deviation. Significance was determined at ∗∗∗P < 0.0001 with a student’s t-test.
Genetic Analysis
The LIL1 mutant was inter-crossed with two indica rice cultivars (TeQing, 93-11) and a japonica cultivar (Nipponbare). The F1 hybrid, BC1 and F2 populations were obtained from the reciprocal crosses between 93-11 and LIL1, TeQing and LIL1, Nipponbare and LIL1. All of the F1 hybrids exhibited lesions on the leaves but, consistent with the semi-dominant nature of the mutation, lesioning in the F1 hybrids was significantly less severe than in LIL1 homozygotes (data not shown). The segregation ratios in the F2 and BC1 were fitted to 3:1 and 1:1 (Table 1). These results indicated that the lesion mimic phenotype was controlled by a semi-dominant gene and not affected by the cytoplasm.
A mapping population consisting of 67 dominant individuals and 45 SSR markers distributed on different chromosomes were used to analysis the linkage relation between markers and the LIL1 locus. From the TeQing ×LIL1 cross, the “TeQing” homozygote, heterozygote, and “LIL1” homozygote segregation ratios of markers on chromosome 7 (RM1243, RM5481, and OSR22) were 4:37:26, 1:38:28, and 2:36:29, respectively, indicating that the LIL1 locus is located on the rice chromosome 7.
Based on these results, we used additional markers on chromosome 7 to more finely map the LIL1 locus. First, using 198 individuals from the F2 population and seven pairs of polymorphic SSR markers detected from 89 pairs of SSR markers published on a website3, the LIL1 gene was mapped between RM5793 and RM3186 (Figure 3A). And then, two SSR markers and two CAPS markers developed by the laboratory (Supplementary Table S2) and more 1,758 individuals from the F2 population were used for mapping. Finally, the LIL1 gene was mapped in a 222.3 kb region between CAPS H and RM7-3 on the long arm of chromosome 7 (Figure 3B).
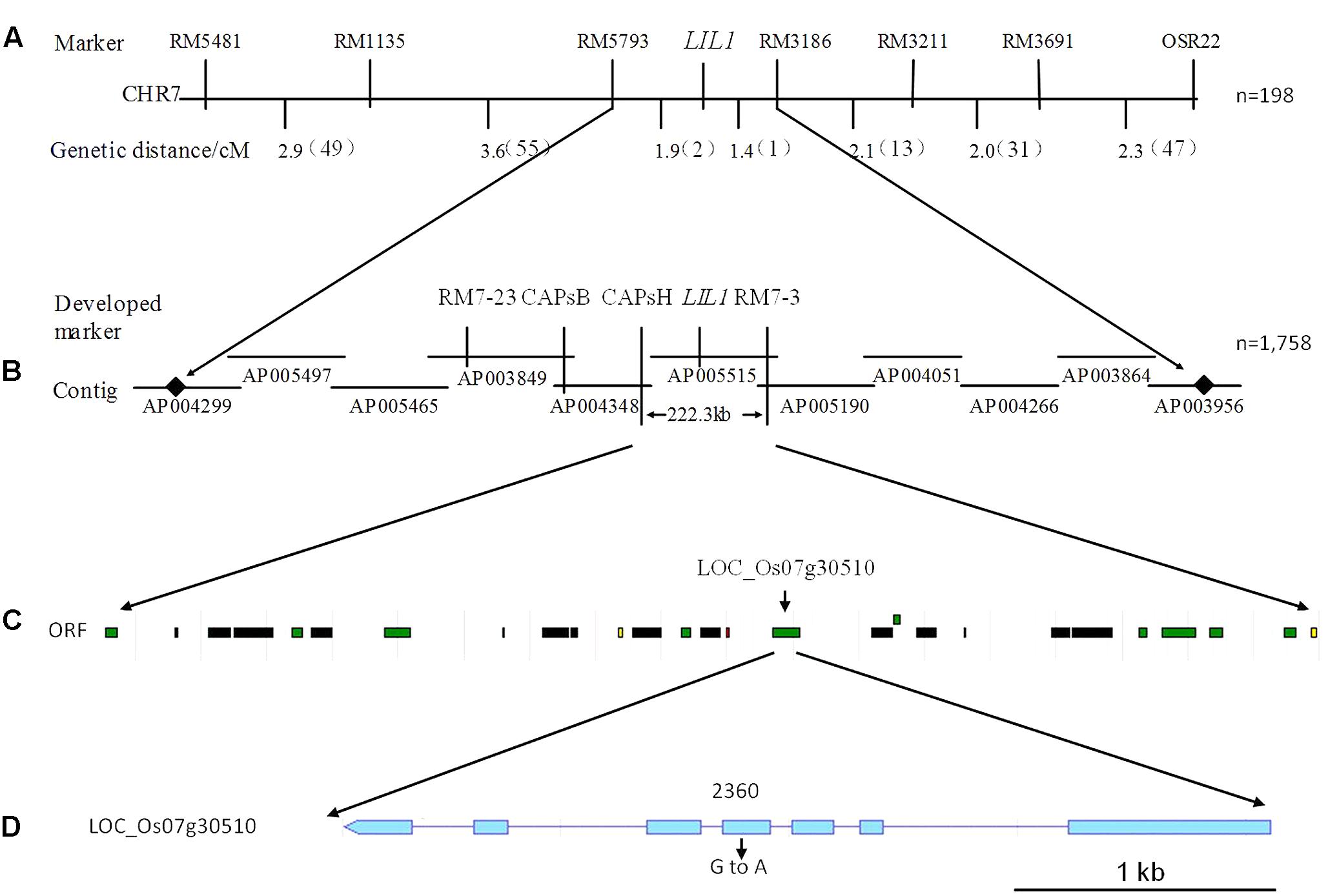
FIGURE 3. Fine mapping of the LIL1 gene. (A) Primary mapping of the interval between the RM5793 and RM3186 markers on chromosome 7 from LIL1; (B) Fine mapping of the interval between CAPs H and RM7-3 markers from LIL1; (C) Putative ORFs predicted in the interval between CAPs H and RM7-3. Green boxes indicate predicted proteins, black boxes indicate transposon or retrotransposon proteins, yellow boxes indicate expressed proteins and red boxes indicate hypothetical proteins; (D) Structure of the candidate gene for LIL1. Boxes indicate exons and lines indicate introns.
Candidate Gene for LIL1
The Rice Genome Annotation Project4 predicts 12 ORFs within the LIL1 mapping interval (Table 2 and Figure 3C). Sequencing these ORFs revealed that a base G was substituted with A at 2360-nt into the genomic sequence for LOC_Os07g30510, whereas no base changes occurred in the other genes. LOC_Os07g30510 is predicted to encode a 687 amino acid CRK protein consisting of a signal-peptide domain, two domains of unknown function 26 (DUF26), a transmembrane domain, and a STKc domain, the latter of which contains the mutated amino acid (Val 429/Ile) in LIL1 (Figure 4). To test whether the mutation is induced by ethylmethane sulfonate, 10 individuals of LIL1, 93-11 and 10 other varieties, including five cultivated rice and five wild rice, were sequenced at this site. All LIL1 individuals have an A and all others have a G at nt 2630 of LOC_Os07g30510 (Supplementary Figure S1). The expression levels of 12 ORFs of LIL1 and 93-11 were compared and significant differences were not observed for genes other than LOC_Os07g30510 (data not shown). However, the expression level of LOC_Os07g30510 was up-regulated significantly in LIL1 (Supplementary Figure S2). These results focused our attention on LOC_Os07g30510 as the candidate gene for LIL1.

FIGURE 4. Deduced amino acid sequence of LIL1 protein kinase. Protein domains, indicated by highlighted colors, are defined below the protein sequence.
Mutant Validation
We sought to confirm that LOC_Os07g30510 is the LIL1 mutant. Given that overexpression of LOC_Os07g30510 in the mutant may account for the phenotype, we surmised that a traditional complementation experiment, in which the WT gene is added to the mutant, would unlikely be informative. Instead, we asked if overexpression of the mutant allele of LOC_Os07g30510 from LIL1 was sufficient to cause the lesion mimic phenotype in an otherwise WT background. The full-length cDNA sequence of LOC_Os07g30510 from LIL1 was inserted into vector pCAMBIA2300 under control of the rice Actin1 promoter and was transformed into Nipponbare by Agrobacterium-mediated transformation. Of 32 regenerated T0 plants, 21 were positive transformants and each of those 21 exhibited the mutant phenotype (e.g., three transgenic lines shown in Figure 5). The transgenic Nipponbare lines exhibited the mutant phenotype later than LIL1, with lesions first observed at the booting stage compared to the three-leaf stage in LIL1. Semi-quantitative RT-PCR indicated that the tested transgenic lines over-expressed LOC_Os07g30510 relative to Nipponbare, where expression was not detected (Figure 5). Expression of LOC_Os07g30510 in the transgenic lines was slightly less than in the LIL1 mutant, possibly accounting for the delayed onset of lesions in the trangenics. Therefore, we confirmed that over-expression of the mutant form of LOC_Os07g30510 from LIL1 is sufficient to cause the lesion mimic phenotype of LIL1.
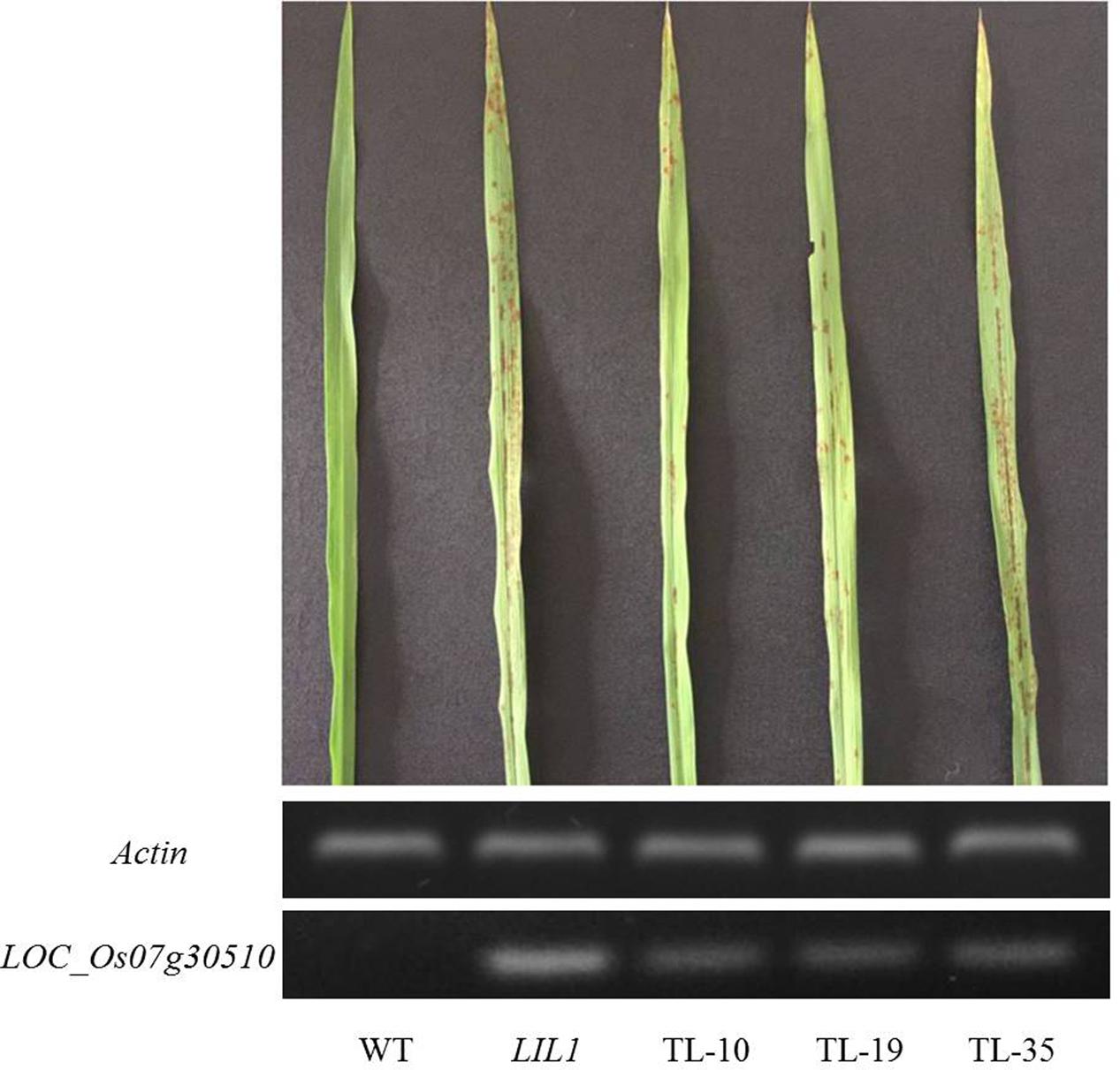
FIGURE 5. Induction of a lesion mimic phenotype in Nipponbare by over-expression of the LIL1 mutant-type of LOC_Os07g30510. TL-10, TL-19, and TL-35 are T0 plants of three independent transgenic lines in which LOC_Os07g30510 from LIL1 is expressed under control of the Actin1 promoter. Phenotypes are representative of an additional 18 lines.
Discussion
Plants have developed complicated signaling pathways and defense mechanisms to protect themselves against invasion of pathogens. The HR is an output of many of these pathways and a component of effective defense against biotrophic and hemi-biotrophic pathogens. However, the molecular mechanisms regulating the HR remain largely unknown (Hunt et al., 1996). LMMs, which include a broad group of phenotypes displaying spontaneous cell death in the absence of biotic challenge, are interesting genetic materials for dissecting the pathways of HR and disease resistance. Lesion appearance in these mutants differs in timing, induction conditions, extent of lesion spreading, color and size (Yin et al., 2000). So far, dozens of genes that control the lesion mimic phenotype have been cloned, and the functions of these genes fall into various groups (Dietrich et al., 1997; Yamanouchi et al., 2002; Balague et al., 2003; Zeng et al., 2004; Qiao et al., 2010; Chen et al., 2012; Fekih et al., 2015; Li et al., 2016; Wang et al., 2017). These results indicate that lesion mimic phenotypes are regulated by different biological processes, thus hinting at the complexity of molecular mechanisms and signaling networks involved in HR and disease resistance. (Lorrain et al., 2003).
Spontaneous lesion formation in the LIL1 mutant correlates with the expression of disease resistance response genes. The induction of PR genes is strongly correlated with the initiation of systemic acquired immunity in both dicot and monocot LMMs (Dietrich et al., 1994; Morris et al., 1998; Takahashi et al., 1999). The PR1 gene is induced in the rice blast resistance reaction (Schweizer et al., 1998). The PR10 gene, which encodes an intracellular protein, is induced by the chemical probenazole (3-allyloxy-1,2-benzisothiazole-1,1-dioxide) that also induces host resistance against rice blast (Midoh and Iwata, 1996). Indeed, the PR1 and PR10 were highly expressed in the LIL1 mutant relative to WT plants (Figure 2). Consistent with the enhanced expression of PR1 and PR10, the LIL1 mutant did display enhanced resistance to one of four tested isolates of M. grisea. The lack of phenotypic difference between LIL1 and 93-11 with the other three isolates may result from the strong disease resistance of 93-11 against those isolates. The generation of active oxygen species is an important step during plant-pathogen interactions (Baker and Orlandi, 1995; Low and Merida, 1995). Peroxidases (EC1.11.1.7, H2O2 oxidoreductase) catalyze the oxidation of various inorganic and organic substrates at the expense of H2O2, which is both a signal molecule and executor of many plant environmental and developmental responses (Gaspar et al., 1982). Chittoor et al. (1997) reported the induction of rice peroxidases POX22.3 and POC1 is correlated with the resistant interactions between rice and the bacterial blight strain Xanthomonas oryzae pv. oryzae. The POX 22.3 and POC1 genes were highly activated in LIL1 relative to WT plants (Figure 2), although the mutant and WT plants did not differ in their resistance to X. oryzae in our tests (data not show).
So far, at least 37 of the 49 rice LMMs have been reported to enhance resistance to at least one pathogen (Huang et al., 2011). However, the details of the underlying mechanism of HR-like lesion mimic formation and the resulting specificity of disease resistance remains poorly understood. In the present study, we cloned a rice LMM, LIL1, which exhibited spontaneous cell death without pathogen attack. Genetic analysis and molecular mapping indicated that LIL1 was controlled by a semi-dominant gene located on the long arm of chromosome 7. To our knowledge, this is the first rice LMM semi-dominant gene mapped on chromosome 7. Therefore, the LIL1 mutant is a novel LMM of rice. Moreover, most of the LMMs are controlled by recessive genes (Yin et al., 2000; Yamanouchi et al., 2002; Qiao et al., 2010; Wang et al., 2015), only a few mutations are regulated by dominant genes (Mori et al., 2007; Wang et al., 2016). In the homozygous LIL1 mutant, the lesions were much more abundant than those in the heterozygous, indicating that the lesion mimic phenotype of LIL1 was controlled by a semi-dominant gene. The LIL1 mutant was mapped to a 222.3 kb region containing 12 putative ORFs on the long arm of chromosome 7. Sequencing these 12 ORFs revealed one base substitution mutation (G/A) (Figure 3D) leading to an amino acid change (Val 429/Ile) in the ORF of LOC_Os07g30510. The mutation was observed in 10 more LIL1 individuals and not observed in 10 individuals of 93-11. Moreover, an additional five cultivated and five wild rice varieties showed the sequence matching to 93-11, indicating that this site is evolutionarily conserved. The expression level of LOC_Os07g30510 was up-regulated significantly in LIL1; however, expression of the other 11 ORFs in our mapping interval showed no obvious difference between LIL1 and 93-11. LOC_Os07g30510 encodes a predicted cysteine-rich repeat kinase (CRK), which is a member of a sub-family characterized by one or more extracellular DUF26 domains containing a C-X8-C-X2-C motif. The DUF26 domain is known as stress-antifungal domain (PF01657) (Sawano et al., 2007). Over-expressing the mutant-type gene of LOC_Os07g30510 is sufficient to cause the lesion mimic phenotype in transgenic lines. Therefore, the up-regulated expression and/or the point mutation in the kinase domain of LOC_Os07g30510 is likely responsible for LIL1 phenotype. Both overexpression of CRKs and the point mutation in the kinase domain could result in the HR-like cell death and increased plant resistance observed. Overexpression of AtCRK5, AtCRK13 and AtCRK20 from Arabidopsis leads to cell death and increased resistance to the bacterial pathogen Pseudomonas syringae (Chen et al., 2003, 2004; Acharya et al., 2007; Ederli et al., 2011). Further, a missense mutation on an Arabidopsis RLK gene (snc4-1D) at the kinase domain increases its expression and results in enhanced pathogen resistance (Bi et al., 2010). The snc4-1D is a gain-of-function semidominant mutant gene, similar to LIL1. It is currently unclear whether the phenotype of LIL1 is due to overexpression of LOC_Os07g30510 or the point mutation in the kinase domain. The point mutation could be the binding region for a microRNA or non-coding RNA, which results in the accumulation of LIL1 transcript. Based on this hypothesis, the region was used to search the microRNA database “miRbase” and “psTarget” and the non-coding RNA database “NONCODE” and “lncRNAdb”; however, no match was found. In WT plants, microRNA or non-coding RNA may bind and degrade the LIL1 transcript, this may explain why the LIL1 transcript could not be detected in WT plants, and potentially why there was no match in the four RNA databases. Further work needs to be performed to confirm this hypothesis. Functions of LIL1 in HR regulation and disease resistance also need to be clarified in future work.
Conclusion
Here, we identified and cloned a rice LMM LIL1 from an ethylmethane sulfonate mutagenized population of Indica rice (Oryza sativa L. ssp. Indica) 93-11. Characterization and molecular mapping revealed that LIL1 is a novel rice LMM with enhanced resistance to rice blast fungus (M. grisea). Genetic analysis illustrated that the mutation was controlled by a semi-dominant gene. Using 1,758 individuals in the F2 progenies, the mutant gene was located in a 222.3 kb region on the long arm of chromosome 7. The candidate genes on the interval were sequenced, and it was found that a G for A substitution in the fourth exon of LOC_Os07g30510 led to an amino acid change (Val 429 to Ile) in the serine/threonine kinase catalytic domain of the encoded protein. LOC_Os07g30510 was determined also to be over-expressed in LIL1 mutant plants. Rice lines transgenically over-expressing the mutant-type gene of LOC_Os07g30510 exhibit a lesion mimic phenotype similar to the LIL1 plants. Therefore, over-expression of LOC_Os07g30510 and/or the Val429Ile substitution is responsible for the LIL1 phenotype. Our results provide the basis for additional function analysis of this gene to advance broad understanding of mechanisms underlying HR and disease resistance in rice.
Author Contributions
QZ and XX conceived and designed the research. QZ collected samples, generated experimental data, performed the entire data analysis, and drafted earlier versions of the manuscript. ZZ and BG partially revised the manuscript. ZZ and TL was involved in the sample collection. All authors read, reviewed, and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31201502).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Thanks to professor Chuanqing Sun of the China Agricultural University provide LIL1 mutant and professor David Mackey of Ohio State University to revise the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02122/full#supplementary-material
FIGURE S1 | Comparison of the mutation sites at 10 additional WT and LIL1 individuals as well as 10 other rice varieties. 93-11 is WT, LIL1 is mutant, Guichao 2, Minghui 63, Weiyou 402, Shanyou 46, Zhonghua 11 are cultivated rice and Ledong, Gaozhou, Dongxiang, Chaling, Yuanjiang are wild varieties of rice.
FIGURE S2 | LOC_Os07g30510 expression comparison in WT and LIL1. WT is 93-11, AA is LIL1 Homozygous, Aa is LIL1 Heterozygous.
TABLE 1 | Details of the genes and primer sequences used.
TABLE 2 | Polymorphic markers developed on chromosome 7 of rice.
Footnotes
- ^ http://rice.plantbiology.msu.edu/
- ^ http://www.lynnon.com
- ^ www.gramene.org
- ^ http://rice.plantbiology.msu.edu
References
Acharya, B. R., Raina, S., Maqbool, S. B., Jagadeeswaran, G., Mosher, S. L., Appel, H. M., et al. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50, 488–499. doi: 10.1111/j.1365-313X.2007.03064.x
Baker, C. J., and Orlandi, E. W. (1995). Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 33, 299–322. doi: 10.1146/annurev.py.33.090195.001503
Balague, C., Lin, B., Alcon, C., Flottes, G., Malmstrom, S., Kohler, C., et al. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15, 365–379. doi: 10.1105/tpc.006999
Bi, D., Cheng, Y. T., Li, X., and Zhang, Y. (2010). Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol. 153, 1771–1779. doi: 10.1104/pp.110.158501
Bowling, S. A., Clarke, J. D., Liu, Y., Klessig, D. F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. doi: 10.1105/tpc.9.9.1573
Brodersen, P., Petersen, M., Pike, H. M., Olszak, B., Skov, S., Odum, N., et al. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Gene. Dev. 16, 490–502. doi: 10.1101/gad.218202
Büschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. doi: 10.1016/S0092-8674(00)81912-1
Campbell, M. A., and Ponald, P. C. (2005). Characterization of four rice mutants with alterations in the defence response pathway. Mol. Plant Pathol. 6, 11–21. doi: 10.1111/1468-0025.00206-i1
Chen, K., Du, L., and Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53, 61–74. doi: 10.1023/B:PLAN.0000009265.72567.58
Chen, K., Fan, B., Du, L., and Chen, Z. (2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56, 271–283. doi: 10.1007/s11103-004-3381-2
Chen, X., Hao, L., Pan, J., et al. (2012). Spl5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol. Breed. 30, 939–949. doi: 10.1007/s11032-011-9677-4
Chittoor, J. M., Leach, J. E., and White, F. F. (1997). Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 10, 861–871. doi: 10.1094/MPMI.1997.10.7.861
Dietrich, R. A., Delaney, T. P., Uknes, S. J., Ward, E. R., Ryals, J. A., Dangl, J. L., et al. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. doi: 10.1016/0092-8674(94)90218-6
Dietrich, R. A., Richberg, M. H., Schmidt, R., Dean, C., and Dangl, J. L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. doi: 10.1016/S0092-8674(00)81911-X
Du, L. F., Li, M. F., Liu, L. X., Wang, C. J., Liu, Y., Xu, X. T., et al. (2014). Physiological characteristics and genetic analysis on a spotted-leaf wheat derived from chemical mutation. Acta Agron. Sin. 40, 1020–1026. doi: 10.3724/SP.J.1006.2014.01020
Ederli, L., Madeo, L., Calderini, O., Gehring, C., Moretti, C., Buonaurio, R., et al. (2011). The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. J. Plant. Physiol. 168, 1784–1794. doi: 10.1016/j.jplph.2011.05.018
Fekih, R., Tamiru, M., Kanzaki, H., Abe, A., Yoshida, K., Kanzaki, E., et al. (2015). The rice (Oryza sativa L.) lesion mimic resembling, which encodes an AAA-type ATPase, is implicated in defense response. Mol. Genet. Genomics 290, 611–622. doi: 10.1007/s00438-014-0944-z
Gaspar, T., Penel, C., Thorpe, T., and Greppin, H. (1982). Peroxidases: A Survey of Their Biochemical and Physiological Roles in Higher Plants. Geneva: University of Geneva Press.
Gray, J., Close, P. S., Briggs, S. P., and Johal, G. S. (1997). A novel suppressor of cell death in plants encoded by the Llsl gene of maize. Cell 89, 25–31. doi: 10.1016/S0092-8674(00)80179-8
Hu, G., Yamada, K., Briggs, S. P., and Johal, G. S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. doi: 10.1105/tpc.10.7.1095
Huang, Q. N., Shi, Y. F., Yang, Y., Feng, B. H., Wei, Y. L., Chen, J., et al. (2011). Characterization and genetic analysis of a light- and temperature-sensitive spotted-leaf mutant in rice. J. Integr. Plant Biol. 53, 671–681. doi: 10.1111/j.1744-7909.2011.01056.x
Hunt, M. D., Neuenschwander, U. H., Delaney, T. P., Weymann, K. B., Friedrich, L. B., Lawton, K. A., et al. (1996). Recent advances in systemic acquired resistance research–a review. Gene 179, 89–95. doi: 10.1016/S0378-1119(96)00429-5
Ishikawa, A., Okamoto, H., Iwasaki, Y., and Asahi, T. (2001). A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 27, 89–99. doi: 10.1046/j.1365-313x.2001.01058.x
Johal, G., Hulbert, S., and Briggs, S. (1995). Disease lesion mimics of maize: a model for cell death in plants. Bioessays 17, 685–692. doi: 10.1002/bies.950170805
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kachroo, P., Shanklin, J., Shah, J., Whittle, E. J., and Klessig, D. F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl Acad. Sci. U.S.A. 98, 9448–9453. doi: 10.1073/pnas.151258398
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Li, L., Shi, X., Zheng, F., Li, C., Wu, D., Bai, G., et al. (2016). A novel nitrogen-dependent gene associates with the lesion mimic trait in wheat. Theor. Appl. Genet. 129, 2075–2084. doi: 10.1007/s00122-016-2758-3
Li, Q., and Wan, J. M. (2005). SSRHunter: development of a local searching software for SSR sites. Hereditas 27, 808–810.
Liu, R. H., and Meng, J. L. (2003). MapDraw: a Microsoft Excel Macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan 25, 317–321.
Lorrain, S., Lin, B., Auriac, M. C., Kroj, T., Saindrenan, P., Nicole, M., et al. (2004). Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16, 2217–2232. doi: 10.1105/tpc.104.022038
Lorrain, S., Vailleau, F., Balague, C., and Roby, D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271.
Low, P. S., and Merida, J. R. (1995). The oxidative burst in plant defense: function and signal transduction. Physiol. Plant. 96, 533–542. doi: 10.1111/j.1399-3054.1996.tb00469.x
McCouch, S. R., Kouchert, G., Yu, Z. H., Wang, Z. Y., Khush, G. S., Coffman, W. R., et al. (1988). Molecular mapping of rice chromosome. Theor. Appl. Genet. 76, 815–829. doi: 10.1007/BF00273666
McCouch, S. R., Teytelman, L., Xu, Y. B., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Midoh, N., and Iwata, M. (1996). Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol. 37, 9–18. doi: 10.1093/oxfordjournals.pcp.a028918
Mizobuchi, R., Hirabayashi, H., Kaji, R., Nishizawa, Y., Yoshimura, A., Satoh, H., et al. (2002). Isolation and characterization of rice lesion mimic mutants with enhanced resistance to rice blast and bacterial blight. Plant Sci. 163, 345–353. doi: 10.1016/S0168-9452(02)00134-6
Mori, M., Tomita, C., Sugimoto, K., Hasegawa, M., Hayashi, N., Dubouzet, J. G., et al. (2007). Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol. Biol. 63, 847–860. doi: 10.1007/s11103-006-9130-y
Morris, S. W., Vernooij, B., Titatarn, S., Starret, M., Thomas, S., Wiltse, C. C., et al. (1998). Induced resistance responses in maize. Mol. Plant Microbe Interact. 11, 643–658. doi: 10.1094/MPMI.1998.11.7.643
Mou, Z., He, Y., Dai, Y., Liu, X., and Li, J. (2000). Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–417. doi: 10.1105/tpc.12.3.405
Noutoshi, Y., Kuromori, T., Wada, T., Hirayama, T., Kamiya, A., Imura, Y., et al. (2006). Loss of NECROTIC SPOTTED LESIONS 1 associates with cell death and defense responses in Arabidopsis thaliana. Plant Mol. Biol. 62, 29–42. doi: 10.1007/s11103-006-9001-6
Qiao, Y., Jiang, W., Lee, J., Park, B., Choi, M., Piao, R., et al. (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit l1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185, 258–274. doi: 10.1111/j.1469-8137.2009.03047.x
Rostoks, N., Schmierer, D., Mudie, S., Drader, T., Brueggeman, R., Caldwell, D. G., et al. (2006). Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Mol. Genet. Genomics 275, 159–168. doi: 10.1007/s00438-005-0073-9
Sawano, Y., Miyakawa, T., Yamazaki, H., Tanokura, M., and Hatano, K. (2007). Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 388, 273–280. doi: 10.1515/BC.2007.030
Schweizer, P., Buchala, A., Dudler, R., and Metraux, J. P. (1998). Induced systemic resistance in wounded rice plants. Plant J. 14, 475–481. doi: 10.1046/j.1365-313X.1998.00141.x
Shen, Y. J., Jiang, H., Jin, J. P., Zhang, Z. B., Xi, B., He, Y. Y., et al. (2004). Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 135, 1198–1205. doi: 10.1104/pp.103.038463
Takahashi, A., Kawasaki, T., Henmi, K., Shi, I. K., Kodama, O., Satoh, H., et al. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17, 535–545. doi: 10.1046/j.1365-313X.1999.00405.x
Temnykh, S., Park, W. D., Ayres, N. S., Cartinhour, N., Hauck, L., Lipovich, Y. G., et al. (2000). Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100, 697–712. doi: 10.1007/s001220051342
Tomiyama, K. (1963). Physiology and biochemistry of disease resistance of plants. Annu. Rev. Phytopathol. 1, 295–324. doi: 10.1146/annurev.py.01.090163.001455
Walbot, V. (1991). Maize mutants for the 21st century. Plant Cell 3, 851–856. doi: 10.1534/genetics.111.130039
Wang, C. F., Huang, L. L., Buchenauer, H., Han, Q. M., Zhang, H. C., et al. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2- and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. Tritici. Physiol. Mol. Plant. Pathol. 71, 230–239. doi: 10.1016/j.pmpp.2008.02.006
Wang, F., Wu, W., Wang, D., Yang, W., Sun, J., Liu, D., et al. (2016). Characterization and genetic analysis of a novel light-dependent lesion mimic mutant, lm3, showing adult-plant resistance to powdery mildew in common wheat. PLOS ONE 11:e0155358. doi: 10.1371/journal.pone.0155358
Wang, J., Ye, B., Yin, J., Yuan, C., Zhou, X., Li, W., et al. (2015). Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol. Biochem. 97, 44–51. doi: 10.1016/j.plaphy.2015.09.001
Wang, S., Lei, C., Wang, J., Ma, J., Tang, S., Wang, C., et al. (2017). SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 68, 899–913. doi: 10.1093/jxb/erx001
Wolter, M., Hollricher, K., Salamini, F., and Schulzee-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128.
Wu, C., Bordeos, A., Madamba, M. R., Baraoidan, M., Ramos, M., Wang, G. L., et al. (2008). Rice lesion mimic mutants with enhanced resistance to diseases. Mol. Genet. Genomics 279, 605–619. doi: 10.1007/s00438-008-0337-2
Yamanouchi, U., Yano, M., Lin, H., Ashikari, M., and Yamada, K. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7530–7535. doi: 10.1073/pnas.112209199
Yao, Q., Zhou, R. H., Fu, T. H., Wu, W. R., Zhu, Z. D., Li, A. L., et al. (2009). Characterization and mapping of complementary lesion-mimic genes lm1 and lm2 in common wheat. Theor. Appl. Genet. 119, 1005–1012. doi: 10.1007/s00122-009-1104-4
Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G. S., et al. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 13, 869–876. doi: 10.1094/MPMI.2000.13.8.869
Keywords: lesion mimic mutant (LMM), hypersensitive response (HR), map-based cloning, rice (Oryza sativa L.), rice blast resistance
Citation: Zhou Q, Zhang Z, Liu T, Gao B and Xiong X (2017) Identification and Map-Based Cloning of the Light-Induced Lesion Mimic Mutant 1 (LIL1) Gene in Rice. Front. Plant Sci. 8:2122. doi: 10.3389/fpls.2017.02122
Received: 29 December 2016; Accepted: 29 November 2017;
Published: 19 December 2017.
Edited by:
Xiaowu Wang, Biotechnology Research Institute (CAAS), ChinaReviewed by:
Turgay Unver, iBG-Izmir, International Biomedicine and Genome Institute, TurkeyCai Lin Lei, Chinese Academy of Agricultural Sciences, China
Copyright © 2017 Zhou, Zhang, Liu, Gao and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingyao Xiong, eGlvbmd4aW5neWFvQDE2My5jb20=
 Qian Zhou
Qian Zhou Zhifei Zhang3
Zhifei Zhang3 Tiantian Liu
Tiantian Liu