- 1State Key Laboratory of Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
- 2Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, College of Life Science, Zhejiang Sci-Tech University, Hangzhou, China
- 3Provincial Key Laboratory of Agrobiology, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 4State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Lysin-motif (LysM) receptor kinases (LYKs) play essential roles in recognition of chitin and activation of defense responses against pathogenic fungi in the model plants Arabidopsis and rice. The function of LYKs in non-model plants, however, remains elusive. In the present work, we found that the transcription of two LYK-encoding genes from cotton, Gh-LYK1 and Gh-LYK2, was induced after Verticillium dahliae infection. Virus-induced gene silencing (VIGS) of Gh-LYK1 and Gh-LYK2 in cotton plants compromises resistance to V. dahliae. As putative pattern recognition receptors (PRRs), both Gh-LYK1 and Gh-LYK2 are membrane-localized, and all three LysM domains of Gh-LYK1 and Gh-LYK2 are required for their chitin-binding ability. However, since Gh-LYK2, but not Gh-LYK1, is a pseudo-kinase and, on the other hand, the ectodomain (ED) of Gh-LYK2 can induce reactive oxygen species (ROS) burst in planta, Gh-LYK2 and Gh-LYK1 may contribute differently to cotton defense. Taken together, our results establish that both Gh-LYK1 and Gh-LYK12 are required for defense against V. dahliae in cotton, possibly through different mechanisms.
Introduction
Plants trigger immune responses upon recognition of conserved microbe-derived components, known as microbe-associated molecular patterns (MAMPs) or pathogen-associated molecular patterns (PAMPs), via pattern recognition receptors (PRRs) (Nürnberger and Brunner, 2002; Bittel and Robatzek, 2007; Boller and Felix, 2009; Macho and Zipfel, 2014). MAMPs are invariant structures originating from microbial cell components that are not host constituents. Several MAMPs, including bacterial lipopolysaccharide (LPS), peptidoglycan (PGN), and fungal chitin, have been shown to possess immunogenic activities in animals and plants (Aslam et al., 2009; Newman et al., 2013), suggesting evolutionary similarity of pattern recognition systems between these two kingdoms.
The lysine-motif (LysM) was first identified in secretory bacterial hydrolases and is known to be involved in bacterial cell wall biogenesis, modification, and degradation (Beliveau et al., 1991). Encompassing 42–48 amino acids, the LysM is a ubiquitous domain found in all living organisms except for Archaea (Bateman and Bycroft, 2000; Zhang et al., 2007, 2009). LysM-containing receptors have been shown to be involved in the recognition of glycans containing N-acetylglucosamine (Buist et al., 2008). In model plants such as Arabidopsis thaliana (Arabidopsis) and Oryza sativa (rice), LysM receptor kinases (LYKs) are, together with leucine-rich repeat receptor-like kinase, the best studied PRRs, and are crucial for innate immunity against fungi and bacteria (Miya et al., 2007; Wan et al., 2008, 2012; Fradin et al., 2009; Shimizu et al., 2010; Willmann et al., 2011; Shinya et al., 2012; Cao et al., 2014; Hayafune et al., 2014; Kouzai et al., 2014; Paparella et al., 2014). The ectodomain (ED) of plant LYKs usually contains a signal peptide (SP) and LysMs, while their intracellular domain (ID) is known to contain an active or inactive kinase domain (Gust et al., 2012). Previous studies have shown that LYKs are essential for plant recognition of chitin or Nod factors, leading to the activation of plant innate immunity or beneficial symbioses (Liang et al., 2013, 2014). In rice, the chitin oligosaccharide elicitor-binding protein CEBiP was firstly shown to be required for the activation of the chitin PRR signaling pathway (Kaku et al., 2006; Kaku and Shibuya, 2016). Subsequently, OsCERK1 was proven to interact with CEBiP to regulate chitin-triggered defense responses (Shinya et al., 2010). In Arabidopsis, LYKs, including CERK1, LYK4, and LYK5, act as chitin PRRs to regulate the chitin signaling pathway (Miya et al., 2007; Wan et al., 2012; Cao et al., 2014), while LYK3 is involved in the negative regulation of immunity (Liang et al., 2013; Paparella et al., 2014). Two recent studies demonstrated that dimerization of CERK1 or CEBiP is required to activate immune signaling (Liu et al., 2012; Hayafune et al., 2014). Notably, the ED of CERK1, which is shed from the receptor, is involved in cell death regulation (Petutschnig et al., 2014). Although the functions of LYKs in model plants are gradually being revealed, the roles of LYKs in cotton defense against pathogens remain unknown.
Cotton (Gossypium spp.) is an important crop used in fiber, oil, and biofuel products. Four important cotton genera are cultivated around the world, including two allotetraploids (Gossypium hirsutum and Gossypium barbadense) and two diploids (Gossypium herbaceum and Gossypium arboreum). The allotetraploid cottons, especially G. hirsutum, are globally cultivated due to their good fiber quality and high yield. Verticillium wilt, primarily caused by the soil-born fungus Verticillium dahliae (Fradin and Thomma, 2006), is one of the most devastating plant diseases worldwide and is a major threat to cotton yield and fiber quality. V. dahliae from infected cotton can be categorized into two subgroups: defoliating and non-defoliating isolates (Daayf et al., 1995). The defoliating isolates cause severe leaf wilting, shedding, and cell death in the infected plants, whereas the non-defoliating isolates induce leaf-wilting symptoms with only minor leaf defoliation during the disease progression (Daayf et al., 1995; Zhang et al., 2012, 2013). V. dahliae isolates BP2 and V991 are non-defoliating and defoliating isolates, respectively, found in China (Zhang et al., 2012).
Our development of a cotton leaf crumple virus (CLCrV)-based virus-induced gene silencing (VIGS) vector has been proven to be an efficient gene-silencing tool that can be used for reverse genetics and functional analyses of candidate genes in cotton (Zhang et al., 2012; Gu et al., 2014; Liu et al., 2014, 2015; Chen et al., 2015; Fu et al., 2015; Zhang H. et al., 2016; Zhang W. et al., 2016). In this study, we employed CLCrV-based VIGS methods to study the function of two members of the cotton LYK family, Gh-LYK1 and Gh-LYK2, in defense. Our results show that both genes are involved in chitin signaling perception and play important roles in resistance to V. dahliae. Furthermore, our findings indicate that Gh-LYK2 is involved in the regulation of cell death.
Materials and Methods
Plant Materials, V. dahliae Isolates, and Culture Conditions
Cotton (G. hirsutum) cultivars 3503 and ZN905 were cultivated at temperatures ranging from 20 to 25°C, under a 16 h:8 h, light: dark photoperiod at 80% relative humidity. The defoliating isolate V991 and the non-defoliating isolate BP2 of V. dahliae were maintained on potato dextrose agar (PDA) at 25°C for 7 days, followed by inoculation into Czapek's medium in 1 l of distilled water (Zhang et al., 2012). The conidial suspensions of V. dahliae were adjusted to 1 × 106 conidia ml−1 with distilled water and used for the inoculation of plants.
Gene Cloning
Full-length ORFs of Gh-LYK1, Gh-LYK2, and Gh-LYK5 were amplified from the G. hirsutum cultivar 3503 cDNA with three primer pairs: LYK1 full F and LYK1 full R; LYK2 full F and LYK2 full R; and LYK5 full F and LYK5 full R, respectively (Table S1). The PCR products were purified via gel electrophoresis and cloned into the pZeroback/blunt plasmid (Tiangen) to produce pLYK1, pLYK2, and pLYK5.
VIGS Vector Construction, Agroinfiltration, and V. dahliae Inoculation
To generate the VIGS vector for silencing of Gh-LYK1, a 325-bp fragment of Gh-LYK1 was amplified from pLYK1 via PCR with the primer pair LYK1 F SpeI and LYK1 R PacI (Table S1). The resulting PCR product was inserted into pCLCrVA to produce pCLCrV-LYK1. Another 333-bp fragment was amplified from pLYK2 with the primer pair LYK2 F SpeI and LYK2 R AvrII (Table S1), and the fragment was inserted into the SpeI-AvrII sites of pCLCrVA to produce pCLCrV-LYK2.
To generate a double-silencing vector for Gh-LYK1 and Gh-LYK2, two primers (LYK2 silencing F PacI and LYK2 silencing R AscI) (Table S1) were used for PCR amplification from the pCLCrV-LYK2 template. The PCR products were purified, digested with PacI and AscI and inserted into pCLCrV-LYK1 to produce pCLCrV-LYK1+2. All of the plasmids were sequenced; the primers used are listed in Table S1.
Transformed Agrobacterium cultures were grown to an approximate OD600 of 1.0 and incubated in induction buffer [10 mM MgCl2, 100 mM MES (pH 5.7), 100 μM acetosyringone] for 3 h at room temperature. Combinations of the induced cultures were infiltrated into the abaxial side of leaves of four-leaf-stage Nicotiana benthamiana plants or the abaxial side of cotyledons of 2-week-old cotton seedlings.
After agro-infiltration, the cotton plants were maintained in an incubator at 25°C with 80% relative humidity. At 15 dpi, all VIGS-treated plantlets were evaluated for CLCrV replication via PCR with the specific primers CLCrV-F and CLCrV-R. More than 15 positive plants per target were then inoculated with V. dahliae through soil drenching with 30 ml of a conidial suspension (1 × 106 conidia ml−1) in each pot (250 ml). Foliar damage was evaluated by rating the symptoms on the cotyledons and leaves of inoculated plants according to the following disease grades: 0 = healthy plant; 1 = yellowing or necrosis of 1–2 cotyledons; 2 = yellowing or necrosis of 1 true leaf; 3 = more than two wilted or necrotic leaves; and 4 = all leaves wilted or dead plant. The disease index was calculated according to the following formula: disease index = [∑disease grades × number of infected)/(total plants × 4)] × 100 (Zhang et al., 2012). The VIGS experiments and the V. dahliae infection assays were repeated three times with more than 15 plants analyzed per replicate.
PAMP Treatment
The roots of 2-week-old cotton seedlings were immersed in PAMP solutions containing 0.1% Tween-20 and 10 μg ml−1 PGN (Sigma), 10 μg ml−1 chitin (Sigma), 10 μg ml−1 LPS (Sigma), or 1 μM flg22 (Phytotech, USA), and were then subjected to vacuum while shaking at 90 rpm for 30 min.
Quantitative RT-PCR
The primers for quantitative RT-PCR were designed using the Primer5 software. The specificity of the primers was evaluated by subjecting the primer sequences to BLAST searches against the NCBI database. PCR amplification was performed in a qTOWER 2.0/2.2 real-time thermal cycler (Analytikjena, Germany) with a 25 μl final volume containing 2.5 μl of cDNA, 0.5 μl of each primer (10 μM), 9 μl of sterile water, and 12.5 μl (2 ×) of SYBR Premix ExTaqTM II Kit (TaKaRa). The conditions for amplification were as follows: 5 min denaturation at 95°C followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 10 s. The expression levels of selected genes were normalized to that of UBQ14, and relative gene expression was calculated using the 2−ΔΔCT method. Data are presented as the mean ± standard error (SE) of three independent experiments in which three independent plants were included per experiment. The primer sequences are listed in Table S1.
In Planta V. dahliae Biomass Quantification
Quantification of V. dahliae biomass was performed as described previously (Fradin et al., 2011). Essentially, four V. dahliae inoculated plants per genotype were harvested and pooled. The samples were ground into fine powder in liquid nitrogen, and DNA was extracted from 100 mg of powder. V. dahliae biomass was determined by real-time PCR. To assess V. dahliae biomass, the fungus specific ITS1-F primer and V. dahliae-specific reverse primer ST-Ve1-R (Fradin et al., 2011) were used. The primer pair AtRuBisCo-F3 and AtRuBisCo-R3, targeting the large subunit of the Rubisco gene (Fradin et al., 2011) in Arabidopsis, and the primer pair UBQ14F and UBQ14R, targeting the UBQ14 gene in cotton (Table S1) were used for sample calibration, respectively.
Subcellular Localization Analysis
To evaluate the subcellular localization of the Gh-LYKs, the full-length cDNAs of Gh-LYK1/2 were generated via PCR. The gene encoding At-PIP2A (AT3G53420), which is used as a plasma membrane (PM) marker (Cutler et al., 2000), was amplified from an Arabidopsis cDNA and cloned into the pCHF3-dsRED vector. The plasmids were individually introduced into Agrobacterium tumefaciens strain EHA105. To enhance ectopic expression, agrobacteria harboring TBSV P19 were co-infiltrated into tobacco leaves. The co-localization of GFP-labeled Gh-LYK derivative fusion proteins and dsRED-labeled At-PIP2A fusion proteins was analyzed with a ZEISS confocal microscope (LSM 780, Germany) at 72 h post-infiltration.
Protein Expression and Purification
To enhance the solubility of proteins expressed in Escherichia coli (E. coli), a pGTf2 plasmid (TAKARA) encoding chaperons was introduced into E. coli strain DE3 to produce strain DE3-Tf2. All of the prokaryotic expression assays described below were performed in this strain.
The EDs of Gh-LYK1 and Gh-LYK12 (minus signal peptide) were cloned into pET30a to form 6 × His tag fusion proteins. The fusion proteins were purified with Ni-NTA resin (Novagen) and used in the chitin-binding assay. To test the kinase activities of Gh-LYK1 and Gh-LYK2, the internal regions of Gh-LYK1 (residues 253–620) and Gh-LYK2 (residues 286–672) from cultivar 3503 were cloned into pMBP28 to form N-terminal maltose-binding protein (MBP) tag fusion proteins, designated Gh-LYK1-ID and Gh-LYK2-ID, respectively. The fusion proteins were then purified over amylose resin (New England Biolabs) and used in the kinase assays.
Chitin-Binding Assays
The in vitro chitin binding assay was performed as described previously (Petutschnig et al., 2010) with minor modifications. Each purified protein was incubated with the chitin beads (New England Biolabs) at 4°C on a rocking platform for 3h. The mixtures were then centrifuged for 5 min at 13,000 g. The pellet fractions were rinsed five times with CBD binding buffer (500 mM NaCl, 20 mM Tris-HCl, 1 mM EDTA 0.1% Tween-20, pH8.0 25°C). Both the pellet and supernatant fractions collected after the last rinse were boiled in SDS loading buffer for 10 min, and centrifuged for 10 min at 13,000 g. Twenty milliliters of supernatant fractions of the solutions were used to perform 12% SDS-PAGE and immunoblot analysis with anti-His antibody (Tiangen, Beijing, China).
Additionally, the EDs of Gh-LYK1, Gh-LYK2 and derivatives were transiently expressed in N. benthamiana leaves. At 72 h post-agroinfiltration, the infiltrated leaves were harvested. Homogenization buffer [50 mM HEPES-KOH (pH 7.4), 150 mM NaCl, 5 mM NaF, 1 mM Na3VO4, 0.5% Triton X-100, 1 mM PMSF, 1 × PI cocktail (Roche)] was added at 2 ml per g of plant material. The sample was then centrifuged with 5,000 × g at 4°C for 15 min, and the supernatant mixture was incubated with chitin beads at 4°C for 3 h. The incubated beads were rinsed five times with the homogenization buffer. The finally rinsed beads were subsequently boiled in SDS loading buffer for 10 min and centrifuged for 10 min at 13,000 g. The supernatant fractions of the solutions were used to perform SDS-PAGE and immunoblot analyses with an anti-Flag antibody (Sigma).
Kinase Assays
For the kinase assays, the purified Gh-LYK1-ID or Gh-LYK2-ID protein was individually incubated with γ-32P ATP at 30°C for 30 min, as previously described (Liu et al., 2011). The phosphorylated substrate was visualized with a Typhoon 9200 imager (GE Healthcare) after separation through SDS-PAGE.
DUAL Membrane Yeast Two-Hybrid System
To detect interactions between Gh-LYK1 and Gh-LYK2 in yeast, the full-length ORFs of Gh-LYK1 and Gh-LYK2 were amplified from pLYK1 and pLYK2 and cloned into pBT3-C and pPR3-C, respectively. DUAL membrane yeast two-hybrid assays were then performed according to the manufacturer's protocol (Dualsystems Biotech, Switzerland).
Homology Model Building
Homology modeling was performed to generate the 3D structure of Gh-LYK1. The crystal structure of the ED of a receptor-like kinase from Arabidopsis, which shares 51% sequence identity with Gh-LYK1, in complex with a chitin pentamer (PDB ID: 4EBZ) was used as template. Gh-LYK1 homology models were generated using Modeller 9.16 (Martí-Renom et al., 2000; Webb and Sali, 2016).
Reactive Oxygen Species (ROS) and Cell-Death Detection in Planta
The accumulation of H2O2 was detected using 3,3′-Diaminobenzidine as previously described, with minor modifications (Qian et al., 2015). Briefly, leaves were excised at the base of the petiole and supplied with a 1 mg/ml solution (pH 3–3.8) of DAB (Sigma-Aldrich) for 8 h at 25°C. Leaves were then immersed in boiling 96% ethanol for 5–10 min, cooled and preserved at room temperature in 70% ethanol, and photographed.
The trypan blue staining assay was performed as described previously, with minor modifications (Heese et al., 2007). Briefly, leaves were submerged in trypan blue staining solution (6 vol of ethanol, 1 vol of distilled water, 1 vol of lactic acid, 1 vol of glycerol, 1 vol of phenol, and 0.067% w/v trypan blue) for 45 min without boiling. After staining, leaves immersed in ethanol were shaken overnight at room temperature until the tissue had become completely colorless.
Bimolecular Fluorescence Complementation (BiFC) Assay
To detect the dimerization of Gh-LYK1/2 in tobacco leaves, the full length ORFs of Gh-LYK1 and Gh-LYK2 were cloned into p2YN and p2YC, respectively. The resulting plasmids were individually introduced into A. tumefaciens strain EHA105 by electroporation using a Gene Pulser Apparatus (Bio-Rad, Hercules, CA) as described (Huang et al., 2009). The bimolecular fluorescence complementation (BiFC) assay was then performed as described previously (Yang et al., 2007). To enhance the ectopic expression, Agrobacterium harboring the gene silencing suppressor P19 was co-infiltrated into tobacco leaves. Emission of YFP fluorescence was detected and the cells were imaged under a confocal microscope (LSM 780, Germany) at 48 h post infiltration.
Results
Gh-LYK1 and Gh-LYK2 Are Up-Regulated during V. dahliae Infection and Chitin Treatment
There are five LYK gene family members encoded in the Arabidopsis genome (At-LYK1–At-LYK5) (Zhang et al., 2007, 2009). To identify the homologs of Arabidopsis LYKs in cotton (G. hirsutum), we used the full-length open reading frames (ORFs) of At-LYK1 to At-LYK5 as queries in BLAST searches against the Gossypium unigene database (https://phytozome.jgi.doe.gov) and retrieved the candidates with highest BLAST scores. We identified two LYK1 homologs (D_cotton_018813, D_cotton_000932), one LYK2 homolog (D_cotton_035752), one LYK3 homolog (D_cotton_014206), two LYK4 homologs (D_cotton_017682, D_cotton_017683), and one LYK5 homolog (D_cotton_019720) encoded in the Gossypium genome. We monitored the expression of these seven putative Gh-LYK genes in the roots of the V. dahliae resistant cultivar 3503 during infections with defoliating and non-defoliating V. dahliae isolates. The expression of D_cotton_018813, D_cotton_014206, D_cotton_017682, and D_cotton_017683 in roots was too low to be detected (data not shown), while the expression of D_cotton_000932, D_cotton_035752, and D_cotton_019720 was up-regulated during V. dahliae infection (Figures 1A,B and Figure S1). Thus, these three putative genes (D_cotton_000932, D_cotton_035752, and D_cotton_019720) were designated as Gh-LYK1, Gh-LYK2, and Gh-LYK5, respectively, for further study. As the Gh-LYK5-silenced cotton plants exhibited a similar V. dahliae biomass compared with control plants after challenge with V. dahliae (Figure S2), we focused on the functional characterization of Gh-LYK1 and Gh-LYK2. The expression of Gh-LYK1 and Gh-LYK2 increased significantly at 2 days post inoculation with V. dahliae. At 12 days post inoculation, the relative mRNA levels of Gh-LYK1 and Gh-LYK2 increased by at least 2 and 9-fold, respectively, while there were no significant changes in the levels of Gh-LYK1 or Gh-LYK2 transcription in the non-inoculated control (Figures 1A,B). To define whether the expression of Gh-LYK1 and Gh-LYK2 was responsive to MAMPs, we analyzed the transcription of Gh-LYK1 and Gh-LYK2 in roots treated with Flg22, LPS, PGN, and chitin. As expected, the expression of Gh-LYK1 or Gh-LYK2 was up-regulated by both PGN and chitin treatments (Figure 1C and Figure S3A). Interestingly, LPS also induced the expression of Gh-LYK2 (Figure 1C). On the contrary, we did not see any significant changes in the transcription of Gh-LYKs, WRKY53, and MPK3 in cotton roots after a 30-min treatment with Flg22 (Figure S3A), or even a 24-h treatment (Figure S3B). The expression changes of Gh-LYK1 and Gh-LYK2 suggested that both of these LYKs might be involved in chitin-triggered immunity during the defense against V. dahliae in cotton.
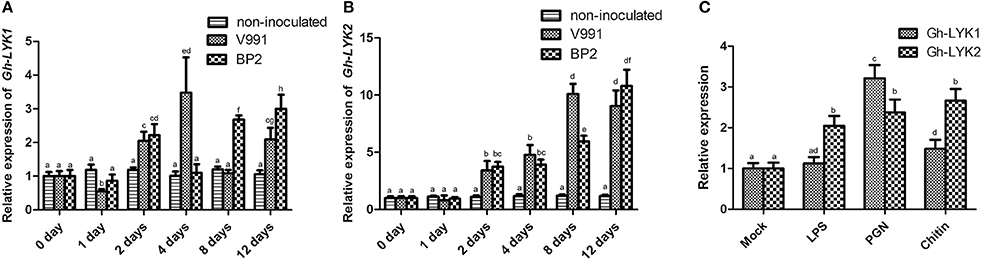
Figure 1. Expression of Gh-LYK1 and Gh-LYK2 is induced by challenge with V. dahliae. (A,B) Relative expression of Gh-LYK1 (A) and Gh-LYK2 (B) in roots determined through quantitative reverse transcription PCR (qRT-PCR) at 0, 1, 2, 4, 8, 12 days post V. dahliae inoculation. Values with the same lower case letter above the error bar were not significantly different according to Duncan's multiple range tests (P < 0.05). (C) Relative expression of Gh-LYK1 and Gh-LYK2 in roots determined through qRT-PCR in cotton treated with various PAMPs. Values with the same lower case letter above the error bar were not significantly different according to Duncan's multiple range tests (P < 0.05).
Next, we cloned the full-length cDNAs of Gh-LYK1 and GhLYK2 from G. hirsutum cultivar 3503. The ORF of Gh-LYK1 and Gh-LYK2 are 1,860 and 2,016 bp, respectively (GenBank accession numbers KU598832 and KU598833). Both of these proteins share a similar domain structure with an SP, extracellular LysM domains, a transmembrane (TM) domain, and an intracellular Ser/Thr kinase domain. Phylogenetic analysis and amino acid (aa) sequence alignment revealed that Gh-LYK1 shares 61.4% identity with At-LYK1, while Gh-LYK2 shares 41% identity with At-LYK2 (Figure S4).
Silencing of Gh-LYK1 and Gh-LYK2 Compromises Resistance to V. dahliae in Cotton
To determine the roles of Gh-LYK1 and Gh-LYK2 in defense, we used the CLCrV-based VIGS system to silence the transcription of Gh-LYK1 or Gh-LYK2 in cotton and tested the defense responses of the silenced plants upon V. dahliae infection. We generated the vectors pCLCrV-LYK1, pCLCrV-LYK2, and pCLCrV-LYK1+2, which can silence Gh-LYK1, Gh-LYK2, or both, in the VIGS system, respectively. Agrobacteria containing CLCrV, CLCrV-LYK1, CLCrV-LYK2, or CLCrV-LYK1+2 were infiltrated into the cotyledons of cotton cultivar 3503. At 15 days post-infiltration (dpi), silenced and non-silenced plants were challenged with V. dahliae isolates V991 or BP2. At 30 days post V. dahliae inoculation, we scored the disease index in the VIGS silenced plants using mock- and empty vector-infiltrated plants as controls. In general, the CLCrV-LYK1+2-silenced cotton plants were more susceptible to the infection than the rest (Figures 2A,B). Notably, defoliating strain V991 is more virulent than BP2 in most cotton cultivars, but not in cultivar 3503. After challenge with V991, the average disease index of the CLCrV-LYK1-, CLCrV-LYK2-, and CLCrV-LYK1+2-infiltrated plants were 27, 31, and 46, respectively, whereas the index of the mock- and CLCrV-infiltrated plants were 14 and 15, respectively. For the inoculation of non-defoliating isolate BP2, the average disease index of the CLCrV-LYK1-, CLCrv-LYK2-, and CLCrV-LYK1+2-infiltrated plants were 31, 34, and 42, respectively, whereas the index in both the mock- and CLCrV-infiltrated plants was 17 (Figure 2C). To evaluate the pathogen propagation in cotton plants, the biomass of V. dahliae was measured by qPCR, and the results were correlated with the disease index (Figure 2D). Silencing of the specific genes was confirmed via qRT-PCR analysis using RNA isolated from the roots (Figure 2E). Additionally, we monitored the expression of two defense-related genes, WRKY53 and MPK3 (Murray et al., 2007; Meng et al., 2013), in MAMP-treated Gh-LYK1/2/1+2-silenced or mock cotton plants. The inducible transcription of both WRKY53 and MPK3 by chitin or PGN was significantly decreased in the silenced cotton plants as compared to the controls (Figures 2F,G). These results indicate that Gh-LYK1 and Gh-LYK2 are important for basal resistance in cotton, and that they act additively.
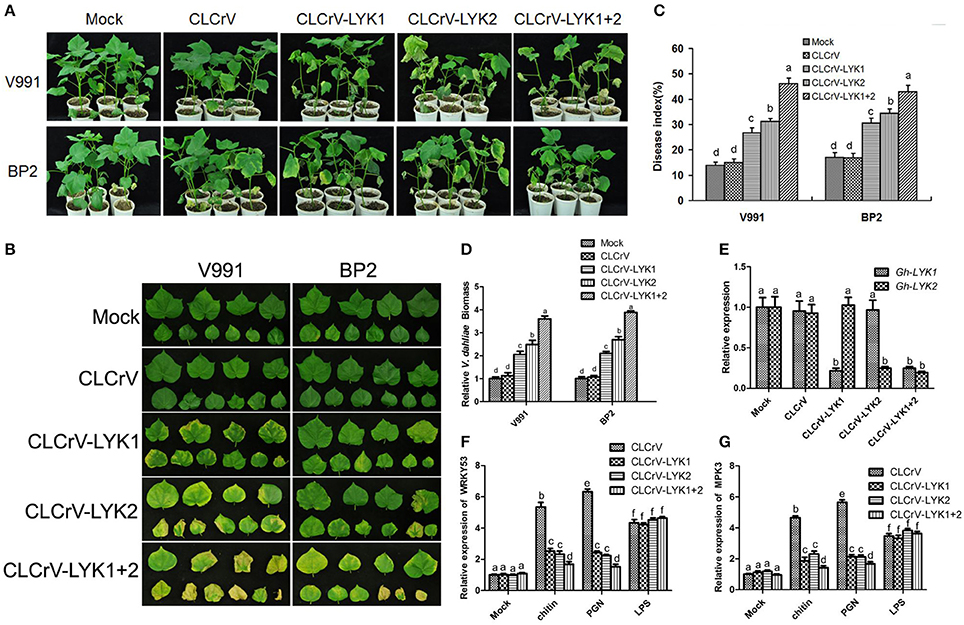
Figure 2. Silencing of Gh-LYK1 and Gh-LYK2 enhances the susceptibility of cultivar 3,503 to V. dahliae. (A,B) Typical symptoms of Gh-LYK1-silenced or Gh-LYK2-silenced whole plants (A) and leaves (B) in which Gh-LYK1 and Gh-LYK2 were simultaneously silenced, infected with two isolates of V. dahliae at 30 days post V. dahliae inoculation. Mock- and empty vector-infiltrated plants were used as controls. (C) Disease index of the silenced plants and non-silenced plants challenged with V. dahliae at 30 days post V. dahliae inoculation. The experiment was repeated three times. The error bars represent SE of the biological replicates. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05). (D) The V. dahliae biomass in the silenced and non-silenced plants challenged with V. dahliae was estimated at 30 days post V. dahliae inoculation. The error bars represent SE of the biological replicates. Values with the same lower case letter above the error bar were not significantly different according to Duncan's multiple range tests (P < 0.05). (E) Relative expression of Gh-LYK1 and Gh-LYK2 in the roots of the silenced and non-silenced plants measured through qRT-PCR. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05). (F,G) Relative expression of WRKY53 (F) and MPK3 (G) in the silenced and non-silenced plants measured through qRT-PCR. Roots from empty vector-infiltrated or VIGS-silenced seedlings were incubated with PAMPs or sterile water (mock) for 30 min and the induction of WRKY53 and MPK3 was determined by qPCR. Values with the same letter were not significantly different according to Duncan's multiple range tests (P < 0.05).
Subcellular Localization of Gh-LYK1 and Gh-LYK2
To investigate the subcellular localization of Gh-LYK1/2, we generated constructs to express the full-length sequences of Gh-LYK1 or Gh-LYK2 fused to the green fluorescent protein (GFP) at their C-terminus and driven by the CaMV 35S promoter, designated as Gh-LYK1-GFP and Gh-LYK2-GFP, and then transiently expressed in N. benthamiana leaves. Under the confocal microscope, the green florescence signal of Gh-LYK1-GFP or Gh-LYK2-GFP was localized to the PM (Figure 3A). To confirm these localization, an AtPIP2A-dsRed fusion construct, which has been used as a PM marker (Pumplin and Harrison, 2009; Pumplin et al., 2012; Sun et al., 2013), was co-expressed with Gh-LYK-GFP. Gh-LYK1/2-GFP co-localized with AtPIPA2-dsRed (Figure 3A), while GFP alone did not (Figure 3B). To further confirm their PM localization, we observed Gh-LYK1-GFP and Gh-LYK2-GFP in plasmolysed N. benthamiana leaves. After plasmolysis with 30% glycerol, Gh-LYK1-GFP and Gh-LYK2-GFP were visualized at the PM and in Hechtian strands (Figure S5).
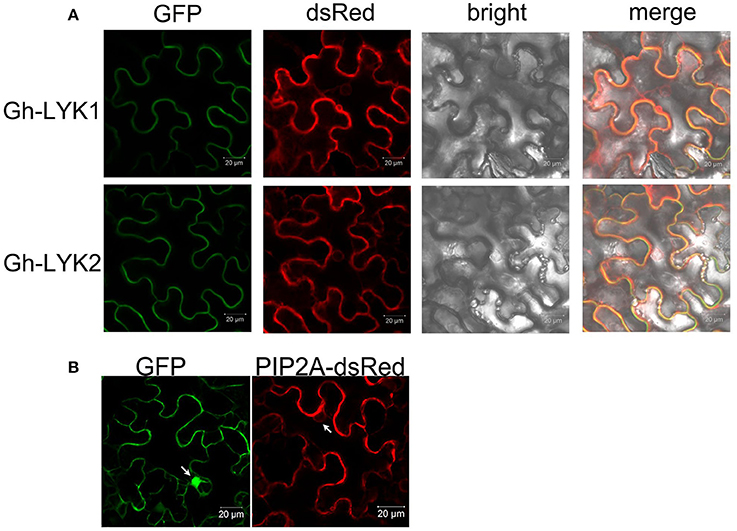
Figure 3. The subcellular localization of Gh-LYK1 and Gh-LYK2 (A) Confocal microscopy images of tobacco leaf cells infiltrated with Agrobacterium harboring the Gh-LYK1-GFP and Gh-LYK2-GFP fusion constructs. Fluorescent signals from AtPIP2A-dsRed fusion proteins were used as the membrane marker. (B) Fluorescent images of GFP alone and AtPIP2A-dsRed fusion proteins under a confocal microscope. White arrows indicate the nuclei.
All Three LysMs of Gh-LYK1 and Gh-LYK2 Are Required for Their Chitin-Binding Ability
Because the ED of Gh-LYK1 shares 51% sequence similarity with that of the chitin receptor kinase of Arabidopsis (AtCERK1-ED), we performed homology modeling of Gh-LYK1-ED based on a crystal structure of AtCERK1-ED in complex with chitin residues (PDB ID: 3KTI). As shown in Figure 4A, all of the three LysM domains of Gh-LYK1 pack tightly against each other, resulting in a globular structure that is similar to that of AtCERK1-ED. Previous publications have shown that all three LysMs are required for the chitin-binding ability of LYKs (Petutschnig et al., 2010; Cao et al., 2014). Hence, deletion of any LysM domain may disturb the structural integrity, resulting in loss of chitin-binding ability. Next, we determined the chitin-binding activities of Gh-LYK1 and Gh-LYK2 using in vitro chitin pull-down assays. The results showed that both purified recombinant EDs can directly bind to chitin beads (Figure 4B). To further confirm this result, we transiently expressed the full-length and LysM single-deletion Gh-LYKs-ED mutants fused to a C-terminal Flag-HA dual tag in N. benthamiana leaves and checked chitin-binding activity as previously described (Petutschnig et al., 2010). In accordance with the results of the in vitro assays, the full length Gh-LYK1-ED and Gh-LYK2-ED could bind chitin beads, while deletion of any of the LysM domains abolished their chitin-binding ability (Figures 4C,D).
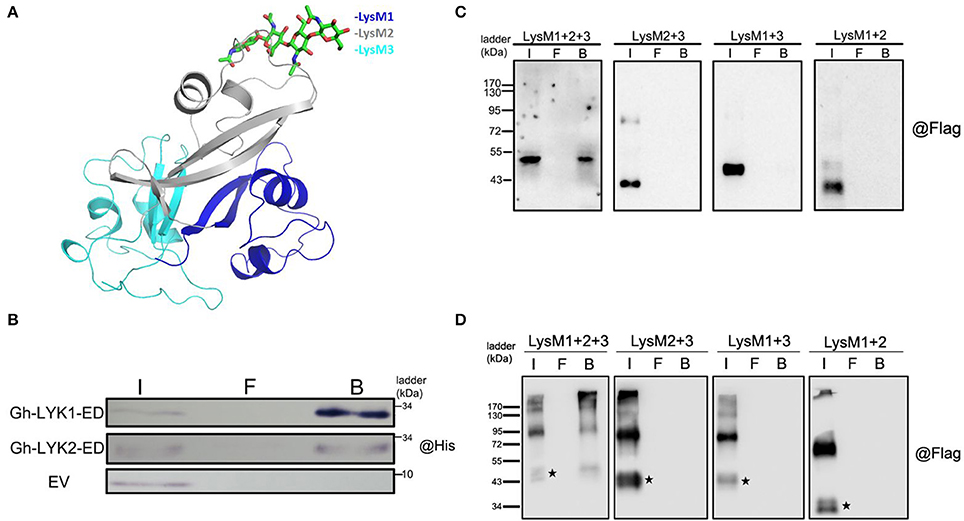
Figure 4. Modeled structure of Gh-LYK1 with chitin and chitin-binding assay. (A) Computer modeling of tight packing of the three LysM domains in Gh-LYK1 with chitin. Chitin is shown in green sticks. (B) Chitin beads binding assay of purified Gh-LYK1-ED and Gh-LYK2-ED. I: input, F: flow through, B: beads. (C) Chitin beads binding assay of Gh-LYK1-ED and single LysM domain deletion mutants expressed in N. benthamiana leaves. I: input, F: flow through, B: beads. (D) Chitin beads binding assay of Gh-LYK2-ED and single LysM domain deletion mutants expressed in N. benthamiana leaves. I: input, F: flow through, B: beads. The black pentagram indicates the monomer.
Gh-LYK2 Forms Chitin-Independent Dimers in Vivo
Previous studies have shown that homodimerization of AtCERK1 and Os-CEBiP is important for chitin perception (Liu et al., 2012; Hayafune et al., 2014). Unexpectedly, we found that a portion of Gh-LYK2-ED formed dimers or oligomers in N. benthamiana without induction by chitin (Figure 4D). To determine whether disulfide bonds contribute to the formation of the dimer, total protein extracts were treated with extra 100 mM Dithiothreitol (DTT) and then analyzed via immunoblotting. The results show that the dimers were not disrupted by DTT (Figure 5A), which suggests that this dimer does not depend on disulfide bond linkages.
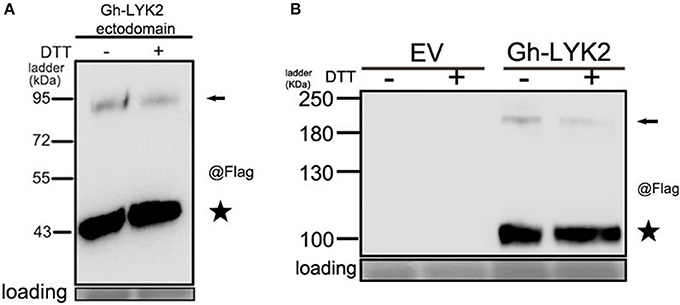
Figure 5. Gh-LYK2 forms dimer in N. benthamiana leaves. (A) The dimers of Gh-LYK2-ED were not disrupted by DTT. (B) The dimers of Gh-LYK2 were not disrupted by DTT. Black arrows indicate full-length dimer; black pentacles indicate the monomer.
We next tested whether the full-length Gh-LYK2 could also form dimers in the absence of chitin. The full-length sequence of Gh-LYK2 was transiently expressed in N. benthamiana fused to a C-terminal Flag-HA dual tag. Similar results to those with Gh-LYK2-ED were obtained (Figure 5B). To test the dimerization of Gh-LYK2 in vivo, we performed a BiFC assay. The yellow fluorescent protein (YFP) signal, absent in controls, indicated formation of a Gh-LYK2 homo-dimer (Figure S6A). Taken together, these results indicate that the homodimerization of Gh-LYK2 is chitin-independent and does not involve disulfide linkages.
Gh-LYK2, but Not Gh-LYK1, Contains a Pseudo-Kinase Domain
Previous reports have shown that Lotus LjNFR5, Medicago NFP, and Arabidopsis LYK4/5 all contain a dead kinase domain (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Wan et al., 2012). We compared the sequence of the kinase domains of the Gh-LYKs to that of typical kinases and found that several key residues were altered or absent, especially in subdomains I, III, and VII (Figure S7) in Gh-LYK2. These differences suggest that Gh-LYK2 is an atypical kinase. On the contrary, the Gh-LYK1 kinase domain is very similar to that of At-LYK1 in its subdomain alignment as well as in the key catalytic amino acid residues (Figure S7). To test whether Gh-LYK1-ID and Gh-LYK2-ID are active kinases, we expressed and purified Gh-LYK1-ID and Gh-LYK2-ID from E. coli and performed in vitro kinase assays. The results showed that, unlike Gh-LYK1-ID, Gh-LYK2-ID was not capable of catalyzing auto-phosphorylation (Figure 6). We next tested whether Gh-LYK2 can interact with Gh-LYK1: no interaction could be detected either in BiFC or dual membrane yeast two-hybrid assays (Figures S6A,B, S8).
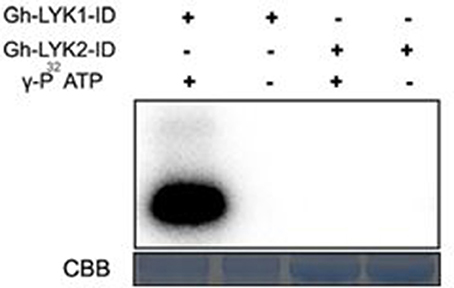
Figure 6. In vitro kinase activities of Gh-LYK1-ID and Gh-LYK2-ID. The auto-phosphorylation was measured by incorporation ofγ-[32P]-ATP. Up panel shows autoradiograph, and bottom panel shows gel stained with CBB. CBB, coomassie brilliant blue.
Overexpression of Gh-LYK2-ED Induces a Burst of Reactive Oxygen Species in N. benthamiana
A recent study reported that the ED and TM of AtCERK1 in an A. thaliana mutant (cerk1-4) could deregulate cell death independently of chitin signaling (Petutschnig et al., 2014). And the reactive oxygen species (ROS) is a signal in plant cell death (Van Breusegem and Dat, 2006). We next asked whether the cotton LYKs could induce a burst of ROS. When the Gh-LYKs and their truncated mutants were transiently expressed in N. benthamiana, we could not detect accumulation of ROS as determined by DAB staining in Gh-LYK2- (Figure S9A), Gh-LYK2-ID- (Figure 7A), Gh-LYK1-ED- (Figure S9B), and empty vector (EV)-infiltrated leaves, while this accumulation was detectable in Gh-LYK2-ED-infiltrated leaves (Figure 7A). As mentioned above, all three LysMs are important for Gh-LYK2 chitin-binding ability. To figure out whether the process of ROS burst induced by Gh-LYK2-ED is dependent on its chitin-binding activity, three single domain-deleted mutants of Gh-LYK2 were transiently expressed in tobacco leaves. The DAB staining results show that all mutants could induce ROS accumulation (Figure 7B), while the transfected leaves without DAB treatment showed no significant staining (Figure S9C). We also tested whether the ROS accumulation was associated with programed cell death (PCD). In the infiltrated areas, water soaking symptoms appeared 3 days post-infiltration when Gh-LYK2-ED or LysM deletion mutants were expressed (Figure 7C). Additionally, uneven blue spots in the infiltrated areas could be visualized upon trypan blue staining (Figure 7D). These results suggest that ectopic expression of Gh-LYK2-ED results in ROS generation and chitin-independent induction of cell-death.
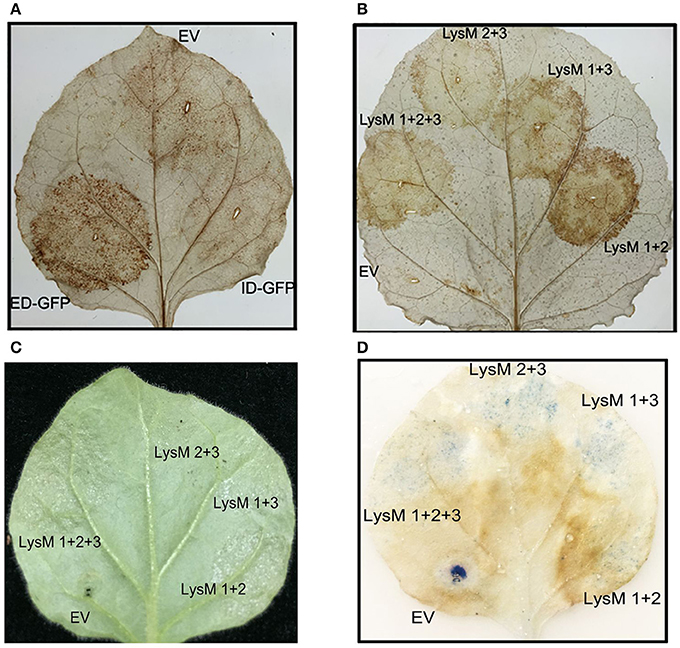
Figure 7. Transient expression of Gh-LYK2-ED can induce the accumulation of ROS and cell death of in N. benthamiana. (A) DAB staining of N. benthamiana leaf expression with Gh-LYK2-ED-GFP, Gh-LYK2-ID-GFP, and EV. EV, empty vector. (B) DAB staining of N. benthamiana leaf expression with Gh-LYK2-ED, single LysM domain deletion mutants of Gh-LYK2-ED and EV. EV: empty vector. (C) Phenotype of N. benthamiana leaf expression with Gh-LYK2-ED, single LysM domain deletion mutants of Gh-LYK2-ED and EV. (D) Trypan blue staining of the same leaf presented in (C).
Discussion
In recent years, the function of LysM receptors in plant innate immunity has been investigated in model plants such as Arabidopsis and rice (Buist et al., 2008; Gust et al., 2012; Tanaka et al., 2013; Shinya et al., 2015). In Arabidopsis, LysM-containing proteins, including CERK1, LYK4, and LYK5, act as essential chitin PRRs that regulate the chitin signaling pathway (Miya et al., 2007; Wan et al., 2012; Cao et al., 2014). AtLYK3 is reported to be responsible for the negative regulation of immunity (Liang et al., 2013; Paparella et al., 2014). However, the function of At-LYK2 in defense remains unclear, as a single atlyk2 mutant or a triple atlyk2/atlyk3/atlyk5 mutant display no significant changes in the expression of defense-related marker genes in response to chitin treatment (Wan et al., 2008, 2012). In this study, we found that both Gh-LYK1 and Gh-LYK2 are important for Verticillium wilt resistance in cotton, because silencing of these two genes compromised resistance to both defoliating and non-defoliating V. dahliae isolates. We also simultaneously knocked down the expression of Gh-LYK1 and Gh-LYK2 with a CLCrV VIGS vector. The double-silenced plants developed much more severe wilting symptoms compared to the single-silenced or control plants, suggesting that two members of the LYK family, Gh-LYK1 and Gh-LYK2, act additively to promote Verticillium wilt resistance in cotton. Recent studies have demonstrated that LysM proteins play roles in sensing chitin of fungal origin or PGN of bacterial origin to mediate plant basal immunity (Willmann et al., 2011; Ao et al., 2014; Kouzai et al., 2014; Mesnage et al., 2014). Based on our observations, the expression of Gh-LYK1 and Gh-LYK2 increased during either chitin or PGN treatment, and two plant downstream defense genes, namely WRKY53 and MPK3, were down-regulated in Gh-LYK1 or Gh-LYK2 knockdown plants after PAMP treatment. It is tempting to speculate that, like CERK1, Gh-LYK1, and Gh-LYK2 could play roles in both resistance to fungal and bacterial infection.
Although the molecular mechanism of chitin recognition by the LysM receptors in rice and Arabidopsis has been gradually revealed in recent years (reviewed in Böhm et al., 2014; Zipfel, 2014; Shinya et al., 2015), the function of LYK proteins in non-model plants remains obscure. Our chitin-binding assays showed that the ED of Gh-LYK1 and Gh-LYK2 could directly bind chitin, and that all three LysMs are required for their chitin-binding activity. The lack of kinase activity of Gh-LYK2 implies that it may interact directly with some other factors, rather than Gh-LYK1, to form a receptor complex involved in chitin perception.
The homodimerization of LysM-containing proteins has been described in plant PRRs such as AtCERK1, At-LYK5, and Os-CEBiP, as well as in the fungal effector Ecp6 (Liu et al., 2012; Sánchez-Vallet et al., 2013; Cao et al., 2014; Hayafune et al., 2014). These findings suggest that formation of homo-complexes in LysM-containing proteins is important for chitin-induced signaling. Based on our observations, both the ED of Gh-LYK2 and the full length protein can form homodimers in the absence of the chitin (Figures 5A,B and Figure S6A). And the homodimer formation does not depend on disulfide bonds, because extra treatment with DTT would not eliminate the homodimer bands, as shown in the immunoblot assay (Figures 5A,B). These data suggest that the homodimer form of Gh-LYK2 could exist in the cell in the abscence of chitin.
Recent work in Arabidopsis proved that the ED and TM of cerk1-4 were sufficient to deregulate cell death (Petutschnig et al., 2014). We found that ectopic expression of the ED of Gh-LYK2, but not Gh-LYK2-ID or Gh-LYK1-ED (Figure S9B), can induce ROS burst and PCD (Figure 7A). More interestingly, ectopic expression of LysM deletion mutants was sufficient to induce ROS accumulation in N. benthamiana (Figure 7B). This suggests that the LysM-related cell death regulation might be chitin-independent, because any single LysM deletion mutation led to a loss of the chitin-binding ability of Gh-LYK2 (Figure 4D). During the arms race between plants and pathogens, fungal and bacterial pathogens have evolved some effector molecules to interact with host receptors and subvert host immunity (Rosebrock et al., 2007; Shan et al., 2008; Xiang et al., 2008; Dou and Zhou, 2012; Macho and Zipfel, 2015). The best-characterized LYK in Arabidopsis, CERK1, has been reported to be targeted by AvrPtoB for degradation (Gimenez-Ibanez et al., 2009). In our study, the ED or the partial ED (LysM deletion mutants), rather than its full-length form, of Gh-LYK2 could induce ROS burst in plants without pathogen challenge (Figures 7A,B), and the expression of Gh-LYK2 was up-regulated during V. dahliae infection (Figure 1B). One possible scenario is that, during pathogen infection, Gh-LYK2, as an important receptor, is targeted by some specific effector to be degraded. Concomitantly, the integrity of Gh-LYK2 could be monitored in the plant so that the breakdown products, i.e., ED, might act as signals to activate the downstream PCD and restrict pathogen spread. In order to speculate about which part of Gh-LYK2-ED could induce PCD, we searched the SMART database (Letunic et al., 2015) and found a specific domain, belonging to the actin-crosslinking proteins superfamily, located between the SP and LysM1. Functional annotation indicates that members of this superfamily are involved in the ARP2/3-dependent actin polymerization pathway, which is important for forming autophagosomes (Goley and Welch, 2006; Höhfeld, 2016). In this pathway, filamin dimers and subsequent ubiquitination mediate the membrane isolation and motility of autophagosomes. This may be a possible explanation as to why Gh-LYK2-ED and derivatives show dimeric and smear bands in western blot upon transient expression in planta (Figure 4D). Using a specific antibody to differentially detect the ecto- and intracellular domains of Gh-LYK2 under physiological conditions may help to unravel the role of GH-LYK2 ED in cell death control.
In conclusion, we show that two LysM receptor-like kinases (Gh-LYK1 and Gh-LYK2) contribute to resistance against Verticillium wilt in cotton. Additionally, Gh-LYK2 might play a role in the regulation of PCD, which needs to be investigated further.
Author Contributions
ZG, TL, BZ, and XZ: planned and designed the research; ZG and TL: performed the experiments; ZG, TL, BD, FL, QW, SQ, FY, TC, YY, JW, GW, BZ, and XZ: analyzed the data; ZG, TL, BD, BZ, and XZ: wrote the manuscript.
Funding
This research was supported by grants from the National Science and Technology Major Project for Transgenic Breeding (2014ZX08009-003-001 and 2014ZX0800501B), the Natural Science Foundation of China (31371930, 31672096, 31600125), the Outstanding Youth Fund of Jiangsu Province (BK20160016), the Natural Science Foundation of Jiangsu Province (BK20140743), and the Science Foundation of Zhejiang Sci-Tech University (14042089-Y). The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02133/full#supplementary-material
Figure S1. Transcriptional analysis of Gh-LYK5 in cotton plant after V. dahliae infection. Relative expression of Gh-LYK5 in roots determined through quantitative reverse transcription PCR (qRT-PCR) at 0, 1, 2, 4, 8, 12 days post V. dahliae inoculation. Values with the same lower case letter above the error bar were not significantly different according to Duncan's multiple range tests (P < 0.05).
Figure S2. Relative fugal biomass in Gh-LYK5-silenced cotton plants after V. dahliae challenge. The V. dahliae biomass in the silenced and non-silenced plants challenged with V. dahliae was estimated at 30 days post V. dahliae inoculation. The error bars represent SE of the biological replicates. Values with the same lower case letter above the error bar were not significantly different according to Duncan's multiple range tests (P < 0.05).
Figure S3. Transcriptional analysis of Gh-LYKs, WRKY53, and MPK3 in cotton c.v. 3,503 root after PAMP treatment. (A) The relative expressions of Gh-LYKs, WRKY53 and MPK3 after 30 min treatment of flg22 or chitin. (B) The relative expressions of Gh-LYKs, WRKY53, and MPK3 after 24 h flg22-treated cotton root.
Figure 4. Phylogenetic analysis of Gh-LYKs with homologies to A. thaliana.
Figure S5. The subcellular localization of Gh-LYK1 and Gh-LYK2 after plasmolysis treatment. Confocal microscopy images of tobacco leaf cells infiltrated with Agrobacterium harboring the Gh-LYK1-GFP (A) and Gh-LYK2-GFP (B) fusion constructs before (control panel) or after plasmolysis treatment.
Figure S6. BiFC assays of Gh-LYK1 and Gh-LYK2 in N. benthamiana leaves. (A) YFP fluorescence (yellow) was observed as a consequence of self-interaction of Gh-LYK2 tagged with 2YN and 2YC, but not in the self-interaction of Gh-LYK1 tagged with 2YN and 2YC or interaction between Gh-LYK1and Gh-LYK2 tagged with 2YN and 2YC. (B) The immunoblotting of Gh-LYK1 fused proteins were detected with anti-HA epitope antibody in BiFC assays.
Figure S7. Alignment of Gh-LYK1 and Gh-LYK2 kinase domain. Alignment was performed using ClustalW with default parameters. The red arrows indicated the amino-acid residues missed or changed in the sub-kinase domains.
Figure S8. The split-ubiquitin yeast two hybrid assay of Gh-LYK1 and Gh-LYK 2.
Figure S9. Transient expression of Gh-LYK2 (A) or Gh-LYK1-ED (B) could not induce the accumulation of ROS in N. benthamiana leaf and the EV/GH-LYK2-ED and derivates infiltrated leaf showed no significant differences without DAB staining (C).
Table S1. Primers used in this study.
References
Ao, Y., Li, Z., Feng, D., Xiong, F., Liu, J., Li, J., et al. (2014). OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084. doi: 10.1111/tpj.12710
Arrighi, J. F., Barre, A., Amor, B. B., Bersoult, A., Soriano, L. C., Mirabella, R., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. doi: 10.1104/pp.106.084657
Aslam, S. N., Erbs, G., Morrissey, K. L., Newman, M. A., Chinchilla, D., Boller, T., et al. (2009). Microbe-associated molecular pattern (MAMP) signatures, synergy, size and charge: influences on perception or mobility and host defence responses. Mol. Plant Pathol. 10, 375–387. doi: 10.1111/j.1364-3703.2009.00537.x
Bateman, A., and Bycroft, M. (2000). The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299, 1113–1119. doi: 10.1006/jmbi.2000.3778
Beliveau, C., Potvin, C., Trudel, J., Asselin, A., and Bellemare, G. (1991). Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J. Bacteriol. 173, 5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991
Bittel, P., and Robatzek, S. (2007). Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10, 335–341. doi: 10.1016/j.pbi.2007.04.021
Böhm, H., Albert, I., Fan, L., Reinhard, A., and Nürnberger, T. (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant. Biol. 20, 47–54. doi: 10.1016/j.pbi.2014.04.007
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Buist, G., Steen, A., Kok, J., and Kuipers, O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido) glycans. Mol. Microbiol. 68, 838–847. doi: 10.1111/j.1365-2958.2008.06211.x
Cao, Y., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3:e03766. doi: 10.7554/eLife.03766
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., and Zhang, B. (2015). A Cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 56, 917–929. doi: 10.1093/pcp/pcv019
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP? cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723. doi: 10.1073/pnas.97.7.3718
Daayf, F., Nicole, M., and Geiger, J. (1995). Differentiation of Verticillium dahliae populations on the basis of vegetative compatibility and pathogenicity on cotton. Eur. J. Plant Pathol. 101, 69–79. doi: 10.1007/BF01876095
Dou, D., and Zhou, J.-M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Fradin, E. F., Abd-El-Haliem, A., Masini, L., Van Den Berg, G. C., Joosten, M. H., and Thomma, B. P. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Fradin, E. F., and Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Fradin, E. F., Zhang, Z., Ayala, J. C. J., Castroverde, C. D., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Fu, W., Shen, Y., Hao, J., Wu, J., Ke, L., Wu, C., et al. (2015). Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 5:11790. doi: 10.1038/srep11790
Gimenez-Ibanez, S., Hann, D. R., Ntoukakis, V., Petutschnig, E., Lipka, V., and Rathjen, J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. doi: 10.1016/j.cub.2009.01.054
Goley, E. D., and Welch, M. D. (2006). The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726. doi: 10.1038/nrm2026
Gu, Z., Huang, C., Li, F., and Zhou, X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649. doi: 10.1111/pbi.12169
Gust, A. A., Willmann, R., Desaki, Y., Grabherr, H. M., and Nürnberger, T. (2012). Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502. doi: 10.1016/j.tplants.2012.04.003
Hayafune, M., Berisio, R., Marchetti, R., Silipo, A., Kayama, M., Desaki, Y., et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U.S.A. 111, E404–E413. doi: 10.1073/pnas.1312099111
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. doi: 10.1073/pnas.0705306104
Höhfeld, J. (2016). Autophagy: press and push for destruction. Curr. Biol. 26, R703–R705. doi: 10.1016/j.cub.2016.06.017
Huang, C., Xie, Y., and Zhou, X. (2009). Efficient virus-induced gene silencing in plants using a modified geminivirus DNA1 component. Plant Biotechnol. J. 7, 254–265. doi: 10.1111/j.1467-7652.2008.00395.x
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Kaku, H., and Shibuya, N. (2016). Molecular mechanisms of chitin recognition and immune signaling by LysM-receptors. Physiol. Mol. Plant Pathol. 95, 60–65. doi: 10.1016/j.pmpp.2016.02.003
Kouzai, Y., Mochizuki, S., Nakajima, K., Desaki, Y., Hayafune, M., Miyazaki, H., et al. (2014). Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant Microbe Interact. 27, 975–982. doi: 10.1094/MPMI-03-14-0068-R
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Liang, Y., Cao, Y., Tanaka, K., Thibivilliers, S., Wan, J., Choi, J., et al. (2013). Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341, 1384–1387. doi: 10.1126/science.1242736
Liang, Y., Tóth, K., Cao, Y., Tanaka, K., Espinoza, C., and Stacey, G. (2014). Lipochitooligosaccharide recognition: an ancient story. New Phytol. 204, 289–296. doi: 10.1111/nph.12898
Liu, G., Xiao, G., Liu, N., Liu, D., Chen, P., Qin, Y., et al. (2015). Targeted lipidomics studies reveal that linolenic acid promotes cotton fiber elongation by activating phosphatidylinositol and phosphatidylinositol monophosphate biosynthesis. Mol. Plant 8, 911–921. doi: 10.1016/j.molp.2015.02.010
Liu, J., Elmore, J. M., Lin, Z.-J. D., and Coaker, G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9, 137–146. doi: 10.1016/j.chom.2011.01.010
Liu, T., Guo, S., Lian, Z., Chen, F., Yang, Y., Chen, T., et al. (2014). A P4-ATPase Gene GbPATP of cotton confers chilling tolerance in plants. Plant Cell Physiol. 56, 549–557. doi: 10.1093/pcp/pcu200
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Macho, A. P., and Zipfel, C. (2015). Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 23, 14–22. doi: 10.1016/j.mib.2014.10.009
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Martí-Renom, M. A., Stuart, A. C., Fiser, A., Sánchez, R., Melo, F., and Šali, A. (2000). Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325. doi: 10.1146/annurev.biophys.29.1.291
Meng, X., Xu, J., He, Y., Yang, K.-Y., Mordorski, B., Liu, Y., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Mesnage, S., Dellarole, M., Baxter, N. J., Rouget, J., Dimitrov, J. D., Wang, N., et al. (2014). Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 5:4269. doi: 10.1038/ncomms5269
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Murray, S. L., Ingle, R. A., Petersen, L. N., and Denby, K. J. (2007). Basal resistance against Pseudomonas syringae in arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 20, 1431–1438. doi: 10.1094/MPMI-20-11-1431
Newman, M.-A., Sundelin, T., Nielsen, J. T., and Erbs, G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4:139. doi: 10.3389/fpls.2013.00139
Nürnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. doi: 10.1016/S1369-5266(02)00265-0
Paparella, C., Savatin, D. V., Marti, L., De Lorenzo, G., and Ferrari, S. (2014). The Arabidopsis thaliana LYSM-CONTAINING RECEPTOR-LIKE KINASE 3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 165, 262–276. doi: 10.1104/pp.113.233759
Petutschnig, E. K., Jones, A. M. E., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Petutschnig, E. K., Stolze, M., Lipka, U., Kopischke, M., Horlacher, J., Valerius, O., et al. (2014). A novel Arabidopsis CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) mutant with enhanced pathogen-induced cell death and altered receptor processing. New Phytol. 204, 955–967. doi: 10.1111/nph.12920
Pumplin, N., and Harrison, M. J. (2009). Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 151, 809–819. doi: 10.1104/pp.109.141879
Pumplin, N., Zhang, X., Noar, R. D., and Harrison, M. J. (2012). Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc. Natl. Acad. Sci. U.S.A. 109, E665–E672. doi: 10.1073/pnas.1110215109
Qian, Y., Hou, H., Shen, Q., Cai, X., Sunter, G., and Zhou, X. (2015). RepA protein encoded by Oat dwarf virus elicits a temperature-sensitive hypersensitive response–type cell death that involves jasmonic acid–dependent signaling. Mol. Plant Microbe Interact. 29, 5–21. doi: 10.1094/MPMI-07-15-0149-R
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
Rosebrock, T. R., Zeng, L., Brady, J. J., Abramovitch, R. B., Xiao, F., and Martin, G. B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374. doi: 10.1038/nature05966
Sánchez-Vallet, A., Saleem-Batcha, R., Kombrink, A., Hansen, G., Valkenburg, D. -J., Thomma, B. P., and Mesters, J. R. (2013). Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife 2:e00790. doi: 10.7554/eLife.00790
Shan, L., He, P., Li, J., Heese, A., Peck, S. C., Nürnberger, T., et al. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27. doi: 10.1016/j.chom.2008.05.017
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Shinya, T., Motoyama, N., Ikeda, A., Wada, M., Kamiya, K., Hayafune, M., et al. (2012). Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706. doi: 10.1093/pcp/pcs113
Shinya, T., Nakagawa, T., Kaku, H., and Shibuya, N. (2015). Chitin-mediated plant–fungal interactions: catching, hiding and handshaking. Curr. Opin. Plant Biol. 26, 64–71. doi: 10.1016/j.pbi.2015.05.032
Shinya, T., Osada, T., Desaki, Y., Hatamoto, M., Yamanaka, Y., Hirano, H., et al. (2010). Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol. 51, 262–270. doi: 10.1093/pcp/pcp185
Sun, Z., Yang, D., Xie, L., Sun, L., Zhang, S., Zhu, Q., et al. (2013). Rice black-streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology 447, 131–139. doi: 10.1016/j.virol.2013.09.001
Tanaka, K., Nguyen, C. T., Liang, Y., Cao, Y., and Stacey, G. (2013). Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal. Behav. 8:e22598. doi: 10.4161/psb.22598
Van Breusegem, F., and Dat, J. F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141, 384–390. doi: 10.1104/pp.106.078295
Wan, J., Tanaka, K., Zhang, X. C., Son, G. H., Brechenmacher, L., Nguyen, T. H. N., et al. (2012). LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160, 396–406. doi: 10.1104/pp.112.201699
Wan, J., Zhang, X.-C., Neece, D., Ramonell, K. M., Clough, S., Kim, S.-Y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. doi: 10.1105/tpc.107.056754
Webb, B., and Sali, A. (2016). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37. doi: 10.1002/cpbi.3
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M., Kolb, D., Tsuda, K., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108, 19824–19829. doi: 10.1073/pnas.1112862108
Xiang, T., Zong, N., Zou, Y., Wu, Y., Zhang, J., Xing, W., et al. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. doi: 10.1016/j.cub.2007.12.020
Yang, X., Baliji, S., Buchmann, R. C., Wang, H., Lindbo, J. A., Sunter, G., et al. (2007). Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 81, 11972–11981. doi: 10.1128/JVI.00617-07
Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 7:e51091. doi: 10.1371/journal.pone.0051091
Zhang, H., Zhang, W., Jian, G., Qi, F., and Si, N. (2016). The genes involved in the protective effects of phytohormones in response to Verticillium dahliae infection in Gossypium hirsutum. J. Plant Biol. 59, 194–202. doi: 10.1007/s12374-016-0568-4
Zhang, W., Zhang, H., Qi, F., and Jian, G. (2016). Generation of transcriptome profiling and gene functional analysis in Gossypium hirsutum upon Verticillium dahliae infection. Biochem. Biophys. Res. Commun. 473, 879–885. doi: 10.1016/j.bbrc.2016.03.143
Zhang, X., Cannon, S., and Stacey, G. (2009). Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 9:183. doi: 10.1186/1471-2148-9-183
Zhang, X., Wu, X., Findley, S., Wan, J., Libault, M., Nguyen, H. T., et al. (2007). Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 144, 623–636. doi: 10.1104/pp.107.097097
Zhang, Z., Esse, H. P., Damme, M., Fradin, E. F., Liu, C. M., and Thomma, B. P. (2013). Ve1-mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol. Plant Pathol. 14, 719–727. doi: 10.1111/mpp.12042
Keywords: cotton, Verticillium dahliae, resistance, lysin-motif receptor kinases, VIGS
Citation: Gu Z, Liu T, Ding B, Li F, Wang Q, Qian S, Ye F, Chen T, Yang Y, Wang J, Wang G, Zhang B and Zhou X (2017) Two Lysin-Motif Receptor Kinases, Gh-LYK1 and Gh-LYK2, Contribute to Resistance against Verticillium wilt in Upland Cotton. Front. Plant Sci. 8:2133. doi: 10.3389/fpls.2017.02133
Received: 14 January 2017; Accepted: 01 December 2017;
Published: 13 December 2017.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Yuxia Hou, China Agricultural University, ChinaTomomi Nakagawa, National Institute for Basic Biology, Japan
Simone Ferrari, Sapienza Università di Roma, Italy
Copyright © 2017 Gu, Liu, Ding, Li, Wang, Qian, Ye, Chen, Yang, Wang, Wang, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baolong Zhang, emhibDIyNDhAaG90bWFpbC5jb20=
Xueping Zhou, enpob3VAemp1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Zhouhang Gu1,2†
Zhouhang Gu1,2† Tingli Liu
Tingli Liu Bo Ding
Bo Ding Yuwen Yang
Yuwen Yang Baolong Zhang
Baolong Zhang Xueping Zhou
Xueping Zhou