- 1Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Department of Genetics, Hazara University, Mansehra, Pakistan
- 3Guangxi Collaborative Innovation Center of Sugarcane Industry, Guangxi University, Nanning, China
The L-ascorbate peroxidase 6 gene (APX6) is one of the most important genes for scavenging H2O2 and plays a vital role in plant resistance to environmental stresses. In this study, a novel ScAPX6 gene (GenBank Accession No. KT907352) was obtained from a sugarcane variety (ROC22). Bioinformatics analysis showed that ScAPX6 has a cDNA length of 1,086 bp and encoded 333 amino acid residues. Subcellular localization confirmed that ScAPX6 was located in the chloroplast. Enhanced growth of Escherichia coli BL21 cells that expressed ScAPX6 showed high tolerance under copper (Cu) stress. Real-time quantitative PCR analysis revealed that ScAPX6 was constitutively expressed wherein with the highest expression levels in sugarcane pith and leaf and the lowest in the root. ScAPX6 was down-regulated by salicylic acid (SA), hydrogen peroxide (H2O2), polyethylene glycol (PEG) and sodium chloride (NaCl) stimuli. Interestingly, it was significantly up-regulated under the stresses of abscisic acid (ABA) and methyl jasmonate (MeJA) wherein with the highest inducible expression levels at 6 h at 6.0- and 70.0-times higher, respectively than that of control. Overexpression of ScAPX6 in Nicotiana benthamiana leaves enhanced the resistance to the infection of tobacco pathogens Pseudomonas solanacearum and Fusarium solani var. coeruleum. These results implied that ScAPX6 might positively respond to ABA, MeJA, and Cu, but might negatively respond to the stresses of SA, H2O2, PEG, and NaCl. Keeping in view the current investigation, ScAPX6 could be associated with the hypersensitive response (HR) or immunity of sugarcane, which will provide a baseline for the function identification of sugarcane ScAPX6.
Introduction
In addition to Oryza sativa, Triticum aestivum, and Zea mays, sugarcane is the fourth largest staple food for the people of China. Sugarcane planting and production are of great significance in sugar supply (Li, 2000). However, the growth and development of sugarcane is severely affected by various abiotic and biotic stresses, such as drought, cold, salinity, heavy metals, high temperature, viruses, fungi, and so on (Li, 2000; Xu et al., 2008). As reported, environmental stimuli can induce active oxygen system which may cause injury to plant cells (Mittler et al., 2004, 2011). Peroxidases (EC number 1.11.1.x), including glutathione peroxidase (GPX), catalase (CAT), and ascorbate peroxidase (APX), are widespread in organisms and can remove the reactive oxygen (Shigeoka et al., 2002; Apel and Hirt, 2014). APX, belonging to type I heme peroxidase and copper oxidase family, is widely employed in plants and can rapidly scavenge hydrogen peroxide (H2O2) in the ascorbic acid (ASA) and glutathione (GSH) cycle (Shigeoka et al., 2002). The role of APX is highly specific to ascorbic acid, that is, to help electron donor to oxidation (Chen and Asada, 1989; Mittler, 2002; Foyer and Noctor, 2005).
According to the orientation characteristic, there are three mainly APX subfamilies in plants, such as cytoplasm APX (cAPX), thylakoid APX (tAPX), and APx-R (Apx-Related) (Mano et al., 1997; Shigeoka et al., 2002; Chew et al., 2003; Dunand et al., 2011). APXs gene have been reported in several plants such as Nelumbo nucifera (Chen et al., 2011), Hordeum vulgare (Shi et al., 2001), Solanum tuberosum (Kawakami et al., 2002), Z. mays (Breusegem et al., 1995), and Vitis pseudoreticulata (Lin et al., 2006). The expression of APX can be regulated by environmental stimuli, such as salt (Badawi et al., 2004), temperature (Kawakami et al., 2002), high light (Maruta et al., 2010), and heavy metal stresses (Pallavi and Dubey, 2007). Research has also shown that APX induced by adversity stress can regulate the content of H2O2 in the cell and redox signaling, and then affect plant tolerance to the osmotic stress (Andréia et al., 2012). This characteristic of APX enzyme activity may be treated as one of the physiological and biochemical indexes measuring crop resistance to biotic and abiotic stresses, for instance, water deficit (Nayyar and Gupta, 2006) and high temperature (Almeselmani et al., 2006). Kornyeyev et al. (2001) transferred the chloroplast APX gene into Gossypium spp., and it was found that APX activity in the transgenic cotton leaf was higher than that of the wild type. Overexpression of tAPX genes increased the resistance of Nicotiana tabacum and Arabidopsis thaliana to the oxidative stress induced by methyl violet essence (Yabuta et al., 2002; Murgia et al., 2004).
Until now, there are four nucleotide sequences of APX genes, which have been identified in sugarcane. Wang Z. Q. et al. (2015) indicated that the APX enzyme activity in sugarcane smut resistant variety Yacheng05-179 was significantly higher than the susceptible variety Liucheng03-182 after inoculated with Sporisorium scitamineum. As a result the expression level of one sugarcane ScAPX gene (GenBank Acc. No. KJ7565501) increased under the stresses of salicylic acid (SA), methyl jasmonate (MeJA), abscisic acid (ABA), H2O2, sodium chloride (NaCl), and polyethylene glycol (PEG). Wang S. et al. (2015) demonstrated that TAPX gene (GenBank Acc. No. JQ958327) played a part in sugarcane resistance to osmotic stress. Huang et al. (2013) found that sugarcane S-APX2 gene showed highly homologous with rice APX (GenBank Acc. No. XP_002463451.1) and mazie APX (GenBank Acc. No. DAA41857.1). Another sugarcane APX gene (GenBank Acc. No. KX235995) was found in Saccharum arundinaceum, but its function was unclear.
From all the above, cloning the APXs gene of different isozymes is necessary to better understand the APX gene family and know more about their expression levels under different stress conditions. In the present study, a putative APX6 unigene, named as ScAPX6, was cloned and identified based on our previous transcriptome data of sugarcane in response to sorghum mosaic virus (SrMV) infection (Bioproject number: PRJNA379719). The sequence characters of ScAPX6 was analyzed by bioinformatics analysis, and the expression patterns of ScAPX6 gene after exposure to various stresses, such as ABA, MeJA, SA, H2O2, PEG, NaCl, and copper (Cu), were detected by real-time quantitative polymerase chain reaction (qRT-PCR). Furthermore, its expression in Escherichia coli, subcellular localization and transient overexpression in Nicotiana benthamiana were also investigated. This study will be helpful to understand the gene function of ScAPX6 in sugarcane.
Materials and Methods
Plant Material and Treatments
For the analysis of the tissue-specific expression of ScAPX6, six healthy plants of 10 months old of ROC22 were used, and then +1 leaf, root, bud, skin and pith were collected. The samples were fixed in liquid nitrogen and stored at −80°C until the extraction of total RNA.
For the abiotic treatment, uniform tissue cultured plantlets of ROC22 at the five or six leaf stage were transferred to water for one week and then treated by the following six stress conditions with root dipping of 5 mmol·L−1 SA, 100 μmol·L−1 MeJA, 100 μmol·L−1 ABA, 10 μmol·L−1 H2O2, 25.0% PEG 8000, 250 mmol·L−1 NaCl, and 100 mmol·L−1 copper chloride (CuCl2), at 28°C with 16 h light and 8 h darkness (Su et al., 2014a). The whole plantlets under SA, MeJA, H2O2, PEG, and NaCl stresses were harvested at 0, 6, 12, and 24 h, respectively. Another set of plantlets under Cu stress was harvested at 0, 12, 24, and 48 h, respectively. Three plantlets per time point were gathered and immediately fixed in liquid nitrogen, and stored at −80°C until the extraction of total RNA.
Total RNA Extraction and the First-strand cDNA Synthesis
Total RNA of the treated samples was extracted by Trizol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The first-strand cDNA synthesis was performed using Prime-ScriptTM RT Reagent Kit (TaKaRa, Dalian, China) following manufacturer's instructions and tested by 1% agarose gel electrophoresis.
Sugarcane ScAPX6 Gene Isolation and Gateway Entry Vector Construction
The sequence of a putative APX6 unigene (ScAPX6) from our previous transcriptome data of sugarcane in response to SrMV infection was used to design the cloning primer APX6-1F/1R (Table 1). The first-strand cDNA of ROC22 was used as amplification template. The reverse transcription–polymerase chain reaction (RT-PCR) procedure was 94°C for 4 min; 94°C for 30 s, 55°C for 30 s, 72°C for 2 min, 35 cycles; and 72°C for 10 min. RT-PCR products were gel-purified and cloned into pMD19-T vector (TaKaRa, Dalian, China), and then transformed into E. coli DH5α competent cells and sequenced (Sangon, Shanghai, China).
The open reading frame (ORF) of ScAPX6 with Gateway entry adapters attB1 and attB2 was amplified from the plasmid of pMD19-T-ScAPX6 by the primers (APX6-3F/3R) (Table 1). The touchdown PCR procedure was 94°C for 4 min; 94°C for 30 s, 70°C for 30 s and then each loop drop 0.5°C, 72°C for 1 min and 30 s, 35 cycles; and 72°C for 10 min. The PCR amplification products were gel-purified and transformed into the Gateway@ donor vector of pDONR221 (Invitrogen, USA) following the manufacturer's instructions of Gateway® BP Clonase™ II Enzyme Mix (Invitrogen, USA). The mixture of BP reaction was transformed into DH5α competent cells and sequenced (Sangon, Shanghai, China). The positive plasmid pDONR221-ScAPX6 was achieved and then used for the constructions of prokaryotic expression vector and eukaryotic expression vector.
Bioinformatics Analysis
The ORF was translated and analyzed by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Conserved domain of ScAPX6 was predicted by the SMART program (http://smart.embl-heidelberg.de/) and NCBI Conserved Domains Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The ExPASy tool (http://us.expasy.org/tools) was used to predict the basic physical and chemical properties of ScAPX6. The cleavage sites of the signal peptides were predicted by SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/). Prediction of transmembrane helices in ScAPX6 protein was performed by TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Psort software was used to predict the subcellular localization of ScAPX6. GOR IV software (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html) was used to analyze the secondary structure of ScAPX6. The protein 3D model was predicted by SWISSMODEL software (http://swissmodel.expasy.org/). The homologous sequences of ScAPX6 were obtained using Blastp in NCBI. DNAMAN software was used for the multiple sequence alignment. The phylogenetic tree of ScAPX6 was constructed with amino acid sequences from other species by the neighbor-joining (NJ) method (1,000 bootstrap replicates) using the MEGA 6.06 (Saitou and Nei, 1987).
Subcellular Colocalization Assay
The ORF of ScAPX6 was amplified by the primer APX6–4F/4R, and then was inserted into the BsaI and Eco31I restriction sites of the pBWA(V)HS-ccdb-GLosgfp vector. Then the recombinant vector pBWA(V)HS-ScAPX6-GLosgfp and the chloroplast marker vector were co-transformed in rice protoplasts with PEG solution (40% W/V PEG 4000, 0.2 mol·L−1 mannitol and 0.1 mol·L−1 calcium chloride). The mixture was cultured in dark for 30 min, and then the protoplasts was gathered and cultured in dark for 16–24 h. The method of transformation of rice protoplasts was followed by Datta and Datta (1999). The subcellular localization of the fusion protein was observed by a confocal laser scanning microscope Leica TCS SP5 (Germany).
Expression of ScAPX6 in E. coli BL21 (DE3) Strain
The plasmid of pDONR221-ScAPX6 was digested with AvaI and then gel-purified for LR reaction with prokaryotic expressive vector of pEZYHb according to the manufacturer's instructions of LR Clonase™ II Enzyme Mix (Invitrogen, USA). The recombinant plasmid of pEZYHb-ScAPX6 was transformed into the competent cells E. coli BL21 (DE3) and then induced by 1.0 mmol·L−1 isopropyl β–D-thiogalactoside (IPTG) at 28°C for 0, 2, 4, and 8 h (Guo et al., 2008). LB medium with E. coli BL21 (blank) and BL21+pEZYHb (control) were induced by 1.0 mmol·L−1 IPTG for 0 and 8 h, respectively. The collected bacterial protein was analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Spot assay was conducted to study the responses of E. coli BL21 cells expressing the ScAPX6 gene under abiotic stress, such as NaCl, Cu and PEG. When OD600 of E. coli BL21 cells in LB medium (containing 80 μg·mL−1 ampicillin) reached to 0.6, IPTG with a concentration of 1.0 mmol·L−1 was added, and then the cells were grown at 37°C for 12 h. The cultures were diluted to OD600 = 0.6, and then diluted to two levels of 10−3 and 10−4 (Guo et al., 2012). Ten microliters from each level was spotted on LB plates containing NaCl (250, 500, and 750 mmol·L−1), CuCl2 (250, 500, and 750 μmol·L−1) and PEG (15, 30, and 45%), respectively (Su et al., 2013). All plates were cultured in 37°C overnight and photographed.
Gene Expression Patterns of ScAPX6
SYBR Green Master (ROX) (Roche, China) and a 7500 qRT-PCR system (Applied Biosystems, South San Francisco, CA, USA) were applied to analyze gene expression levels of ScAPX6 in different tissues and in response to various stresses. The sequence-specific primer of ScAPX6 (APX6-2F/2R) (Table 1) was designed by Premier 5.0 software. The primer combination of clathrin adaptor complex (CAC) and cullin (CUL) (Table 1) was regarded as the internal control (Guo et al., 2014). The 20 μL reaction system containing 10 μL SYBR Green Master Mix, 0.8 μL each of 10 μmol·L−1 upstream and downstream primers, 2 μL cDNA templates (20 × diluted cDNA) and 6.4 μL double distilled water. Each qRT-PCR was conducted in triplicate. The qRT-PCR procedure was 50°C for 2 min; 95°C for 10 min; 35 cycles of 95°C for 15 s, and 60°C for 1 min. The 2−ΔΔCt method (Livak and Schmittgen, 2001) was employed to analyze the qRT-PCR data.
Transient Overexpression of ScAPX6 in N. benthamiana
To study the role of ScAPX6 in response to pathogen infection and its hypersensitive reaction in plant, an overexpressed vector pEarleyGate 203-ScAPX6 was constructed by Gateway cloning technique according to the manufacturer's instructions of LR Clonase™ II Enzyme Mix (Invitrogen, USA). N. benthamiana leaves was inoculated with the vector of pEarleyGate 203-ScAPX6 by an Agrobacterium-mediated transient expression method conducted by Su et al. (2014b). Two important tobacco pathogens, Pseudomonas solanacearum and Fusarium solani var. coeruleum, were cultured in potato dextrose water (PDW) liquid medium at 28°C. When the two pathogens cells were cultured to an OD600 of 0.8, they were separately infected into the treated leaves that were agroinfiltrated with pEarleyGate 203-ScAPX6 for 24 h. Then the 3,3′-diaminobenzidine (DAB) staining, trypan blue staining and transcript analysis of the eight tobacco immunity-associated marker genes (Table 1), were conducted by the treated N. benthamiana leaves according to Su et al. (2016). RT-PCR was used to detect whether ScAPX6 has been overexpressed in N. benthamiana, with the RNA of treated leaves and ScAPX6 specific primer (APX6-3F/3R, Table 1), the NtEF1-α was treated as control. RT-PCR procedure was 94°C for 4 min; 94°C for 30 s, 72°C for 30 s, 72°C for 2 min, 35 cycles; and 72°C for 10 min. All treatment materials were cultured at 24°C (16 h light/8 h darkness) and then photographed at 1 and 7 day (d) separately. Each test was repeated three times.
DAB and trypan blue staining were used for histochemical analysis of Agrobacterium-infiltrated leaves. The leaves was soaked in DAB-HCl solution (1.0 mg·mL−1, pH 5.8), and then cultured in the dark for 12 h. The leaves were destained with 95% ethanol at 100°C for 10 min (Su et al., 2014a), and then was imaged for H2O2 detection with a stereoscopic microscope (Nikon, Tokyo, Japan) and a light microscope (Leica, Wetzlar, Germany). Three biological replicates were prepared. The leaves were also deal with trypan blue mixture, containing 10 mL lactic acid, 10 g phenol, 10 mL glycerol, 30 mL absolute ethanol, 10 mg trypan blue, and 10 mL distilled water, and then was boiled for 5 min. After staining, the leaves were soaked in a chloral hydrate solution (2.5 g·mL−1) for decoloring (Dang et al., 2013). The blue color of the leaves for the cell death was also imaged with a stereoscopic microscope (Nikon, Tokyo, Japan) and a light microscope (Leica, Wetzlar, Germany).
Results
Cloning and Bioinformatics Analysis of ScAPX6
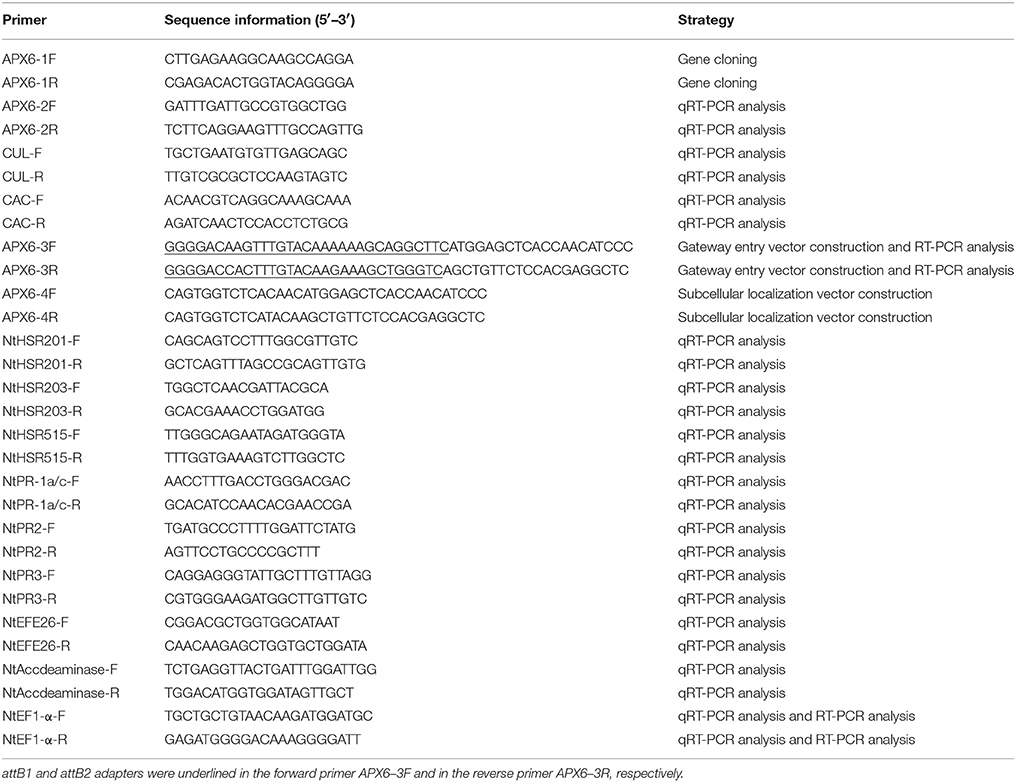
In the present study, a full-length cDNA of APX6 unigene, which was named as ScAPX6 (GenBank Acc. No. KT907352), was isolated from ROC22. The cDNA sequence length of ScAPX6 was 1,086 bp (Figure S1) with a complete ORF (1,002 bp, from position 27 to position 1,025), encoding 333 amino acid residues. ScAPX6 had a molecular mass of 36.21 kDa and an isoelectric point (pI) of 6.91. CDD search of NCBI showed that ScAPX6 belonged to a member of the plant-peroxidase-like superfamily (Figure 1). The instability index of ScAPX6 protein was 48.10, suggesting that ScAPX6 might be an unstable acid hydrophilic protein (Walker, 2005). Secondary structure prediction of ScAPX6 predicted that the percentages of alpha-helix, random coil, and extended strand were 44.74, 42.94, and 12.31%, respectively.
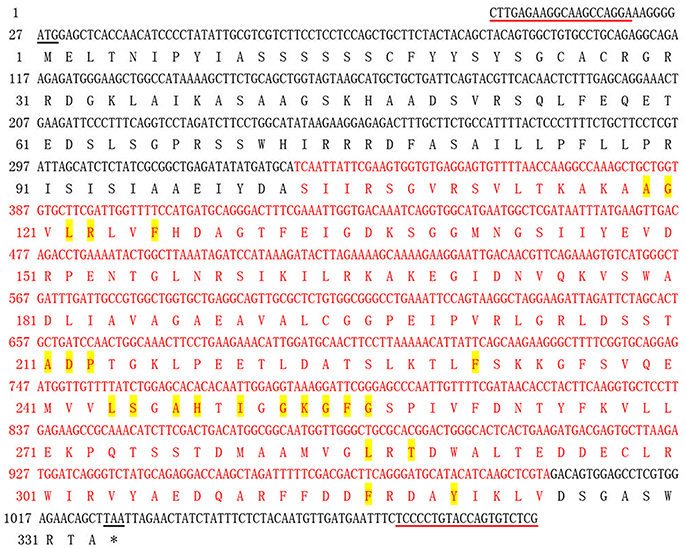
Figure 1. Nucleotide acid sequence and deduced amino acid sequence of sugarcane ScAPX6 gene obtained by RT-PCR. The start codon and termination codon were underlined in black. The primer used in RT-PCR was underlined in red line. The peroxidase like superfamily domain contains 222 amino acids (from 103 to 324) was highlighted in red. The amino acids highlighted in yellow represented the heme binding site. *, Stop codon.
Furthermore, SWISSMODEL program showed that the main spatial structures of ScAPX6 were alpha-helix and random coil (Figure 2). Comparing ScAPX6 with O. sativa Japonica Group APX6 (EAZ43377.1), S. italic APX6 (XP_004973913.1), and S. bicolor APX6 (XP_002445876.1), we found that the spatial structure of these four APX6 was basically in line with each other, suggesting that ScAPX6 owned high conservation of spatial structure with different plant species. Psort software predicted that ScAPX6 might be located in the chloroplast thylakoid membrane, plasma membrane, chloroplast stroma, and chloroplast thylakoid space with the probabilities of 71.9, 65.0, 56.1, and 56.1%, respectively.
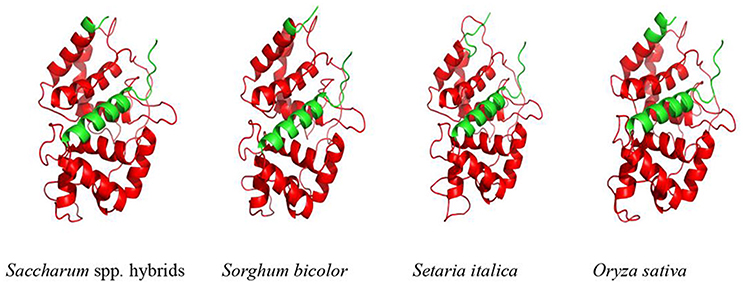
Figure 2. Predicted 3D structure of ScAPX6. The plant-peroxidase-like domain was in red. Saccharum spp. hybrids (AMQ80947.1), Sorghum bicolor (XP_002445876.1), Setaria italica (XP_004973913.1), Oryza sativa Japonica Group (EAZ43377.1).
According to the classification method by Teixeira et al. (2004), the phylogenetic tree was separated into three groups, including cytosolic isoforms, Apx-R isoforms and chloroplastic isforms (Figure 3). ScAPX6 was clustered into group chloroplastic isforms. Two APX proteins reported in Saccharum hybrid cultivar, which were ScAPX (AIG52216.1) and TAPX (AGD80596.1), were also clustered into the same clade as ScAPX6.
Figure 3. Phylogenetic analysis of deduced amino acid sequence from ScAPX6 and other ascorbate peroxidases proteins. The GenBank accession number of proteins were according to Teixeira et al. (2004) and downloaded from NCBI. The neighbor-joining method with 1,000 bootstrap replications was used.
Subcellular Localization
The recombinant vector pBWA(V)HS-ScAPX6-GLosgfp was constructed to understand the subcellular location of ScAPX6. The results showed that ScAPX6 and the chloroplast marker were located in the same place, so it was confirmed that ScAPX6 was located in the chloroplast, which is in accordance with the results of prediction (Figure 4).
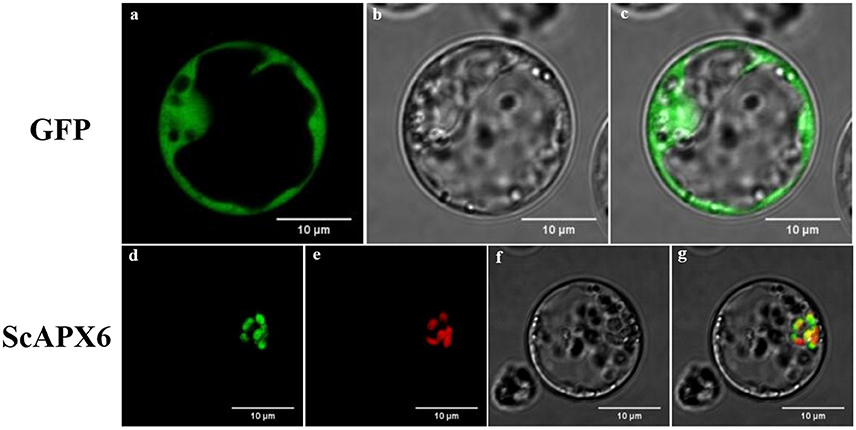
Figure 4. Subcellular localization analysis of ScAPX6 in rice protoplasts. (a,d) green fluorescence; (b,f) visible light; (e) red fluorescence from chloroplast marker; (c,g) merged light.
Expression of ScAPX6 in E. coli BL21 (DE3) Strain
The ScAPX6 gene was combined with the expression vector pEZYHb and then was transformed into E. coli BL21 cell. The SDS-PAGE analysis (Figure 5) showed that ScAPX6 was expressed as a recombinant protein in the BL21 cells. In Figure 5, after induced by 1.0 mmol·L−1 IPTG at 28°C for 2, 4, and 8 h, an obvious accumulation protein (including the 6× His-tag) at approximate 55 kDa was observed.
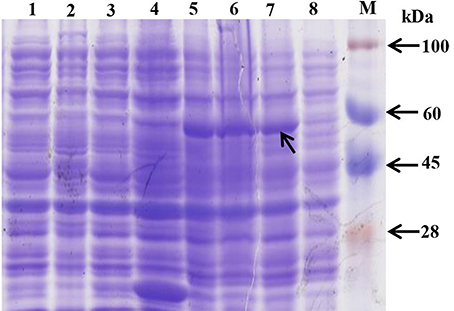
Figure 5. Prokaryotic expression of pEZY-Hb-ScAPX6 fusion protein in Escherichia coli BL21 (DE3). M, protein marker; 1, BL21 cell induction for 8 h; 2, BL21 cell without induction; 3, pEZY-Hb without induction; 4, pEZY-Hb induction for 8 h; 5–8, pEZY-Hb-ScAPX6 induction for 8, 4, 2, and 0 h, respectively.
Overexpression of ScAPX6 in E. coli Enhances Cell Growth under Cu Stress
It has been reported that the APX activity was up-regulated by abiotic stress, such as heavy metal, high salinity, drought, high temperature, and wounding (Shi et al., 2001). In this study, spot assay was performed to study the function of ScAPX6 in response to abiotic stress in vivo. The control (BL21+pEZY-Hb) and the gene-expressed cells (BL21+pEZY-Hb-ScAPX6) grew in LB plates containing NaCl, CuCl2, and PEG were performed (Figure 6). It had been recorded that the recombinant ScAPX6 cells showed a more rapid growth than the control in LB plates with PEG and CuCl2 supplement, but not with NaCl, suggesting that the overexpression of ScAPX6 in E. coli could enhance its tolerance to PEG and Cu stress.
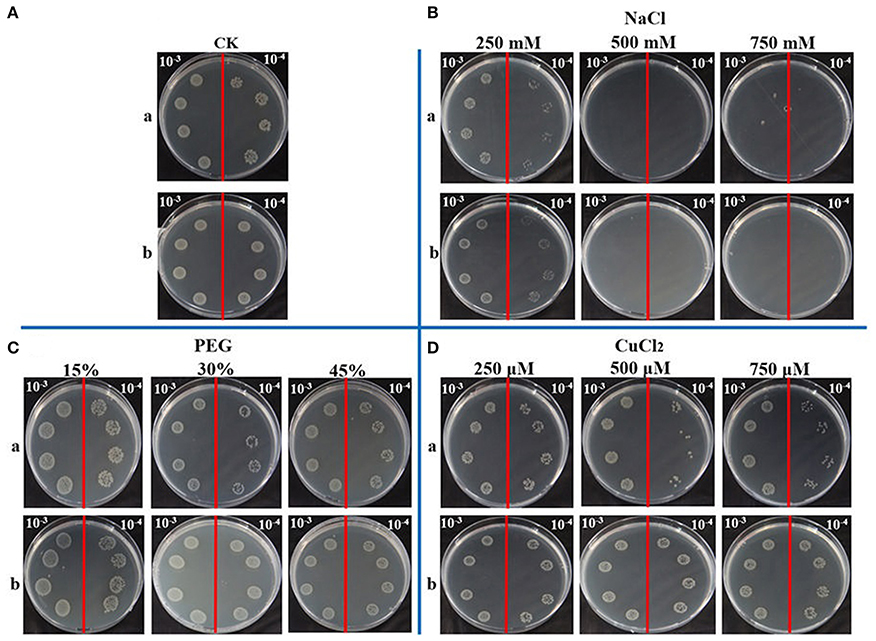
Figure 6. Spot assays of BL21+pEZY-Hb-ScAPX6 (b) and BL21+pEZY-Hb (control) (a) on LB plates with NaCl, PEG and CuCl2. Isopropyl β-D-thiogalactoside (IPTG) was added to the cultures of BL21+pEZY-Hb-ScAPX6 and BL21+pEZY-Hb to induce the expression of recombinant protein. The cultures were adjusted to OD600 = 0.6. Ten microliters from 10−3 (left side of the red line on the plate) to 10−4 (right side of the red line on the plate) dilutions were spotted onto LB plates without any supplement (CK) (A) or with NaCl (250, 500, and 750 mmol·L−1) (B), PEG (15, 30, and 45%) (C) and CuCl2 (250, 500, and 750 μmol·L−1) (D), respectively. NaCl, sodium chloride; PEG, polyethylene glycol; CuCl2, copper chloride.
Tissue-Specific Expression of ScAPX6
qRT-PCR analysis showed that ScAPX6 was constitutively expressed in all five kinds of sugarcane tissues, including root, bud, skin, leaf, and pith, but with different expression levels (Figure 7). ScAPX6 showed the highest expression levels in the pith and leaf, and then on the skin, while the transcript in root was at the lowest level.
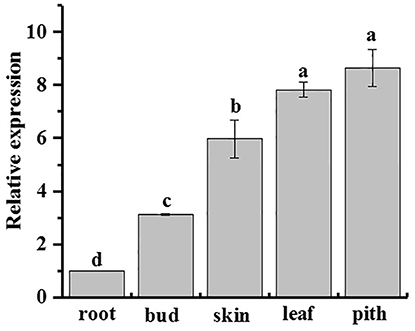
Figure 7. Tissue-specific expression analysis of ScAPX6 in sugarcane. The error bars represented the standard error of each treating group (n = 3). Data were normalized to the CAC and CUL expression level. All data points were means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the Duncan's new multiple range test (p < 0.05).
Gene Expression Patterns of ScAPX6 in Response to Abiotic Stress
qRT-PCR analysis revealed that the ScAPX6 gene exhibited different expression characteristics in response to ABA, MeJA, SA, H2O2, PEG, NaCl, and Cu stimuli (Figures 8A,B). As shown in Figure 8A, the transcripts of ScAPX6 were remarkably up-regulated under the stresses of ABA and MeJA, and with the highest inducible expression levels at 6 h, which were 6.0- and 70.0-times higher than that of control, respectively. However, ScAPX6 was down-regulated during the SA treatment and rapidly decreased at 6 h. Under the stress of PEG, the expression of ScAPX6 showed no change at 6 h, and then decreased at 12 h. ScAPX6 was down-regulated after the treatment of H2O2 and NaCl, but was up-regulated by the CuCl2. These results demonstrated that ScAPX6 might positively respond to ABA, MeJA, and Cu stresses but negatively respond to SA, H2O2, PEG, and NaCl stresses.
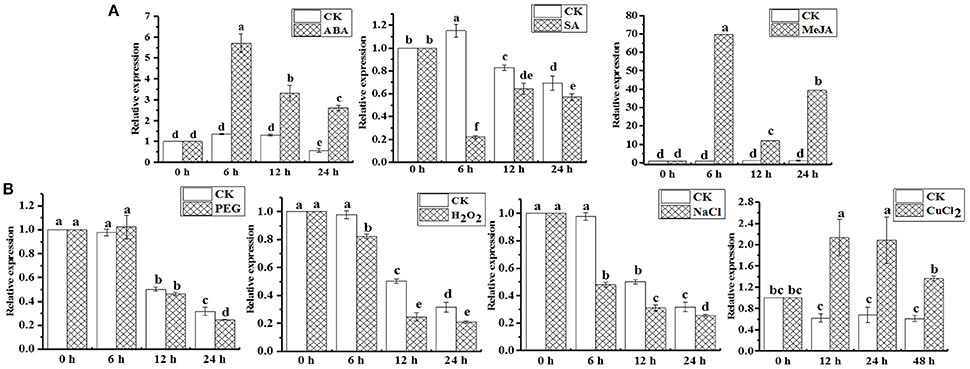
Figure 8. Gene expression patterns of ScAPX6 in sugarcane under various plant hormones (A) and abiotic stresses (B). Data were normalized to the CAC and CUL expression level. All data points were means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the Duncan's new multiple range test (p < 0.05). ABA, abscisic acid; SA, salicylic acid; MeJA, methyl jasmonate; PEG, polyethylene glycol; H2O2, hydrogen peroxide; NaCl, sodium chloride; CuCl2, copper chloride.
Transient Overexpression of ScAPX6 Induces a Defense Response in N. benthamiana
After transient overexpression of ScAPX6 in N. benthamiana leaves for 1 day (d), the transcripts of ScAPX6 were detected by qRT-PCR (Figure 9A). As shown in Figures 9A,B darker DAB staining color and more intense trypan blue staining cells were observed in ScAPX6 leaves than that in the control (35S::00) after infiltration for 2 and 6 d, respectively. The eight immunity-associated marker genes in N. benthamiana were induced by transient overexpression of ScAPX6 (Figure 9C). The hypersensitive response (HR) marker genes, NtHSR201 and NtHSR203, showed no change in transcript, while NtHSR515 was up-regulated. The expression level of SA-responsive gene NtPR2 remained unchanged, while NtPR-1a/c and NtPR3 and two ethylene synthesis dependent genes, NtEFE26 and NtAccdeaminase, were all down-regulated. Compared with the control leaves, 35S::ScAPX6 exhibited darker color reflecting high levels of H2O2 accumulation and intense hypersensitivity response.
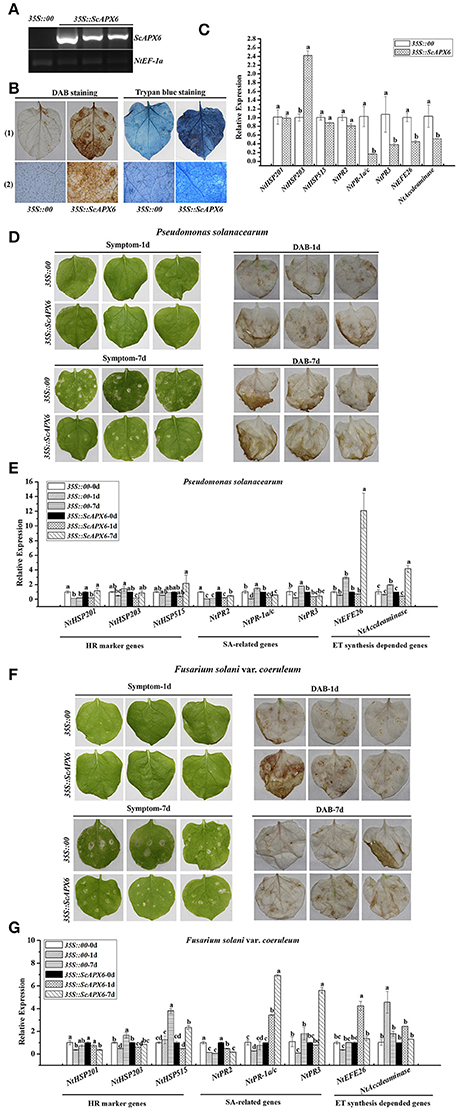
Figure 9. Transient overexpression of ScAPX6 in Nicotiana benthamiana leaves. (A) RT-PCR analysis of ScAPX6 in the N. benthamiana leaves after 1 d infiltration by Agrobacterium strain GV3101 carrying pEarleyGate 203-ScAPX6 and the empty vector (35S::00). (B) DAB (3,3′-diaminobenzidinesolution) staining and trypan blue staining of N. benthamiana leaves at 48 h and 6 d after Agrobacterium strain infiltration, respectively, (1) represented a stereoscopic microscope and (2) represented a light microscope. (C) The transcripts of eight immunity-associated marker genes in the N. benthamiana leaves at 24 h after infiltration. (D,F) Disease symptoms and DAB staining results of N. benthamiana leaves by P. solanacearum and F. solani var. coeruleum infection after infiltration with 35S::00 (control) or 35S::ScAPX6-containing Agrobacterium strain. Disease symptoms of infected leaves were observed at 1 and 7 d post-inoculation. (E,G) The transcripts of immunity-associated marker genes in the N. benthamiana leaves after inoculation with P. solanacearum or F. solani var. coeruleum for 1 and 7 d. NtEF1-α was used for normalization of the transcript levels. All data points were expressed as the mean ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the Duncan's new multiple range test (p < 0.05). NtHSR201, NtHSR203, and NtHSR515, hypersensitive response marker genes; NtPR2, NtPR-1a/c, and NtPR3, a salicylic acid pathway-related gene; NtEFE26 and NtAccdeaminase, the ethylene synthesis-dependent genes. Control, the Agrobacterium strain carrying 35S::00.
To further investigate the response of ScAPX6 to pathogen, two tobacco pathogens, P. solanacearum and F. solani var. coeruleum, were separately injected into N. benthamiana containing 35S::ScAPX6 or the control. After inoculation with P. solanacearum, no disease symptom was found between 35S::ScAPX6 and the control leaves for 1 d, while 35S::ScAPX6 exhibited darker color than the control at 1 d by DAB staining. With elongated treatment time, although the DAB staining showed no difference between 35S::ScAPX6 and the control leaves, the leaves in the control showed slight yellow phenomenon and necrotic spot, while the 35S::ScAPX6 only exhibited the faint wilting symptom after inoculation at 7 d (Figure 9D). After challenging with P. solanacearum for 1 and 7 d (Figure 9E), the expression levels of NtHSR201, NtHSR203, and NtPR2, were unchanged or down-regulated in the control and 35S::ScAPX6 leaves. The transcripts of NtPR-1a/c and NtPR3 were significantly down-regulated at 1 d and increased at 7 d in the control leaves, but were down-regulated or remained unchanged in the 35S::ScAPX6 leaves. The expression levels of NtHSR515 and NtEFE26 were unchanged at 1 and 7 d and NtAccdeaminase was up-regulated at 7 d in the control leaves, while these three genes in the 35S::ScAPX6 leaves were all unchanged at 1 d and reached the peak values at 7 d after inoculation.
Likewise, for DAB staining, the 35S::ScAPX6 leaves showed darker color than the control after inoculation with F. solani var. coeruleum for 1 d and exhibited no difference at 7 d. No apparent disease symptom differences between 35S::ScAPX6 and the control were found at 1 d after inoculation (Figure 9F). Some symptoms, such as wilting, decay phenomenon and necrotic spot, were observed in the control leaves at 7 d, but not in the 35S::ScAPX6 leaves. After challenging with F. solani var. coeruleum for 1 and 7 d (Figure 9G), the expression levels of NtHSR201 and NtPR2 were unchanged or down-regulated in the control and 35S::ScAPX6 leaves. The transcripts of NtAccdeaminase, NtHSR203, and NtHSR515, were significantly up-regulated at 1 or 7 d in the control leaves, while stayed stable or showed a small rise in the 35S::ScAPX6 leaves. The transcripts of NtPR-1a/c, NtPR3, and NtEFE26, were down-regulated or remained unchanged in the control leaves, but were all significantly up-regulated in the 35S::ScAPX6 leaves.
Discussion
Plant APXs are a multi-gene family (Shigeoka et al., 2002). Many plants APXs genes have been cloned and identified, including eight in O. sativa (Teixeira et al., 2005, 2006), four in Vigna unguiculata (D'Arcylameta et al., 2006) and Spinacia oleracea, respectively (Ishikawa et al., 1995, 1996, 1998), six in Eucalyptus grandis (Teixeira et al., 2005), seven in Lycopersicon esculentum (Najami et al., 2008), and nine in A. thaliana (Panchuk et al., 2002; Mittler et al., 2004; Narendra et al., 2006). In the present study, based on a putative APX6 unigene sequence from our previous transcriptome data, a sugarcane ScAPX6 gene (GenBank Acc. No. KT907352) was cloned (Figure S1), which was different from the other already reported sugarcane APX genes in NCBI (ScAPX: GenBank Acc. No. KJ7565501; TAPX: GenBank Acc. No. JQ958327; APX: GenBank Acc. No. KX235995), and shared only 19.79% identity at the amino acid sequence level. Najami et al. (2008) found that in Solanum lycopersicum three cytosolic SlAPX genes, SlAPX1, SlAPX2, and SlAPX3 showed a high sequence identity (>90%). Teixeira et al. (2004) found that APX contained two isoforms, chloroplastic and nonchloroplastic isoforms. Furthermore, APx-R was found as a new heme-containing protein functionally associated with ascorbate peroxidase (Lazzarotto et al., 2011). In this study, ScAPX6 was clustered in chloroplastic isoforms (Figure 3), which was consistent with the study conducted by Teixeira et al. (2004). Subcellular localization of ScAPX6 in rice protoplast showed that ScAPX6::GFP was targeted at chloroplast (Figure 4), which was consistent with the result of bioinformatics predicted localization. Similar to other plant species, such as the APXs from Cucumis melo (Cheng et al., 2009) and A. andraeanum (Liu et al., 2013), ScAPX6 also contained a plant peroxidase like superfamily and the heme binding site and shared 94.29 and 82.93% similarities with the APX homologs from S. italic APX6 (XP_004973913.1) and S. bicolor APX6 (XP_002445876.1), suggesting that ScAPX6 belongs to a member of APX family.
APX plays an important role in response to biotic and abiotic stresses (Andréia et al., 2012). Cheng et al. (2009) observed that in C. melo, the gene expression level of CmAPX varied in different tissues, and with the highest expression in leaves and roots. Chen et al. (2011) found that NuAPX showed higher expression levels in leaf stalks than in root, due to the reasons that the tissues of the leaf stalks and leaf were rich in chloroplast and mitochondria, which was the leading source of reactive oxygen species (ROS) through the electron-transport chain of photosynthesis. In this study, ScAPX6 was constitutively expressed in sugarcane tissues and with the highest expression in pith but the lowest in root (Figure 7).
For abiotic stress, Agarwal et al. (2005) have found that the enzyme activities of APX, SOD, and CAT in wheat seedlings could be increased by 500 mmol·L−1 ABA treatment. It was shown previously that in H. vulgare, the transcript level of HvAPX1 was remarkably up-regulated by the treatments of ABA and NaCl (Shi et al., 2001). In Brassica oleracea var. italica, Jiang et al. (2012) have found that the expression of BoAPX2 increased after H2O2, SA, and NaCl treatments. Previous research on sugarcane showed that the transcripts of ScAPX increased under the treatment of ABA, MeJA, SA, H2O2, PEG, and NaCl (Wang Z. Q. et al., 2015) In this study, the expression of ScAPX6 was also up-regulated by both ABA and MeJA, but down-regulated by SA and H2O2 treatments (Figure 8A). As reported, the expression of TAPX gene in sugarcane was significantly induced by NaCl and PEG stresses (Wang S. et al., 2015). Previous investigations have identified the transcripts of OsAPX7 and OsAPX8, which were separately down regulated by 300 mmol·L−1 NaCl in rice roots and leaves (Hong and Kao, 2007; Yamane et al., 2010). Similarly, in this study, down regulation of the transcripts of ScAPX6 were noted with NaCl and PEG treatment (Figure 8B), which was consistent with the results of the spot assay that the recombinant protein of ScAPX6 expressed in E. coli BL21 did not show better growth than the control under both two treatments (Figure 6). Previous reports have provided evidence that over-expressed plant stress tolerance genes in E. coli cells could enhance their growth under abiotic stress (Gupta et al., 2010; Guo et al., 2013). For example, Su et al. (2014b) have tested a chitinase gene ScChi in E. coli, which showed better growth under NaCl, Cu, CdCl2, and ZnSO4 treatments. Duan et al. (2006) have indicated that the transgenic O. sativa with HvAPX1 gene was more tolerant to cadmium stress when compared with the wild type. In this study, the transcript of ScAPX6 was also up-regulated by the treatment of Cu (Figure 8B), which was in line with the results that the recombinant protein of ScAPX6 expressed in E. coli BL21 resulted in a better growth under Cu stress (Figure 6). Therefore, it was predicted that ScAPX6 could be helpful for the tolerance of sugarcane to Cu. These findings suggested ScAPX6 might be a positive response to ABA, MeJA, and Cu stresses, while showed the negative response to SA, H2O2, PEG, and NaCl stresses. However, what should be pointed out here is that, we only use treatment with some hormones, such as SA, ABA, and MeJA with one concentration, and we cannot directly link effect of our treatment with specific hormone pathway because the specific concentration of the hormones was not determined. These points need to be considered in future.
Previous studies have revealed that the overexpression of tApx gene in tobacco enhanced tolerance to chilling, methylvioiogen, and high-intensity light (Yabuta et al., 2002). Transgenic potato with simultaneous overexpression of APX, choline oxidase (codA), and SOD, increased tolerance of SSAC plants and lower levels of H2O2 under methylvioiogen, drought and salt-mediated oxidative stresses (Ahmad et al., 2010). Investigations have revealed that cell death could induce R gene expression, ion fluxes, stimulation of ROS, and defense-related hormones, which can efficiently restrict pathogen growth and development (Li et al., 2010; Melech-Bonfil and Sessa, 2010; Du et al., 2012). Thordal-Christensen et al. (1997) proved that DAB-uptake method can serve H2O2 detection at a subcellular level. Although the DAB reactions reflect increases in local H2O2, this method has been adapted to many other plant species for in situ detection of H2O2. Thordal-Christensen et al. (1997) speculated that this could be related either to the fact that the optimal pH (5.5–6.0) for DAB precipitation coincides with the expected pH level in the leaf, or to the strong polymerization observed in the plant tissue. Lai et al. (2013) found that 35S::BrERF11 transgenic tobacco plants showed significantly increased HR and H2O2 accumulation compared with wild-type plants according to trypan blue and DAB staining. In the present study, a darker DAB staining color was found after overexpression of ScAPX6 in N. benthamiana leaves and after inoculation with P. solanacearum and F. solani var. coeruleum at 1 d compared with that in the leaves of control (Figures 9B,D,F), which was indicative of the accumulation of H2O2 and resulted in intense hypersensitivity response, but with elongated treatment time, the DAB staining color became lighter, which was consistent with the fact that APX can rapidly scavenge H2O2 in the plant by the ASA-GSH cycle (Shigeoka et al., 2002). However, since the DAB buffer is far from real physiological conditions, whether itself results in the accumulation of H2O2 remains an open question. Together, the antimicrobial action against the tobacco pathogens after overexpression of ScAPX6 in N. benthamiana, suggesting that ScAPX6 may enhance the resistance to P. solanacearum and F. solani var. coeruleum.
Conclusions
In this study, a novel ascorbate peroxidase gene, ScAPX6 (GenBank Acc. No. KT907352), was isolated and characterized. The cDNA of ScAPX6 gene was 1,086 bp long with a complete 1,002 bp ORF, encoding 333 amino acids. Subcellular localization revealed that ScAPX6 was targeted in chloroplast. After inducing by IPTG, the accumulation protein of pEZYHb-ScAPX6 at 55 KDa led to a better growth of E. coli BL21 under Cu stress. ScAPX6 was constitutively expressed in sugarcane tissues. Besides, ScAPX6 showed positive response to ABA, MeJA, and Cu stresses, but negative response to the stresses of SA, H2O2, PEG, and NaCl. The overexpression of ScAPX6 in N. benthamiana leaves showed positive response against the attack of P. solanacearum and F. solani var. coeruleum. These results suggested that ScAPX6 plays an important role in the HR or immunity of sugarcane.
Author Contributions
FL, YS, and YQ: Conceived, designed, and initiated the project; NH, HL, and SG: Prepared materials; FL, NH, LW, TS, WA, and JG: Performed experiments and contributed to data analysis and validation; FL and YS: Drafted the manuscript; LX, KM, YS, and YQ: Revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Natural Science Foundation of Fujian province, China (2015J06006), the National Natural Science Foundation of China (31501363), the Research Funds for Distinguished Young Scientists in Fujian Agriculture and Forestry University (xjq201630), the Research Funds for Distinguished Young Scientists in Fujian Provincial Department of Education, the earmarked fund for China Agricultural Research System (CARS-17) and the Program for New Century Excellent Talents in Fujian Province University (JA14095).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02262/full#supplementary-material
Figure S1. Amplification of ScAPX6 gene in sugarcane. M: DNA marker 2,000 bp; 1: RT-PCR product.
References
Agarwal, S., Sairam, R. K., Srivastava, G. C., Tyagi, A., and Meena, R. C. (2005). Role of ABA, salicylic acid, calcium and hydrogen peroxide on antioxidant enzymes induction in wheat seedlings. Plant Sci. 169, 559–570. doi: 10.1016/j.plantsci.2005.05.004
Ahmad, R., Kim, Y. H., Kim, M. D., Kwon, S. Y., Cho, K., Lee, H. S., et al. (2010). Simultaneous expression of choline oxidase, superoxide dismutase and ascorbate peroxidase in potato plant chloroplasts provides synergistically enhanced protection against various abiotic stresses. Physiol. Plant. 138:520. doi: 10.1111/j.1399-3054.2010.01348.x
Almeselmani, M., Deshmukh, P. S., Sairam, R. K., Kushwaha, S. R., and Singh, T. P. (2006). Protective role of antioxidant enzymes under high temperature stress. Plant Sci. 171, 382–388. doi: 10.1016/j.plantsci.2006.04.009
Andréia, C., Gisele, P., Silvia Barcellos, R., Carolina Werner, R., Fernanda, L., and Márcia, M. P. (2012). Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 35, 1011–1019.
Apel, K., and Hirt, H. (2014). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 728–749. doi: 10.1146/annurev.arplant.55.031903.141701
Badawi, G. H., Kawano, N., Yamauchi, Y., Shimada, E., Sasaki, R., Kubo, A., et al. (2004). Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant. 121, 231–238. doi: 10.1111/j.0031-9317.2004.00308.x
Breusegem, F. V., Villarroel, R., Montagu, M. V., and Inzé, D. (1995). Ascorbate peroxidase cDNA from maize. Plant Physiol. 107, 649–650. doi: 10.1104/pp.107.2.649
Chen, D., Zheng, X. F., Li, G. L., Pan, C., Zhou, M. Q., and Hu, Z. L. (2011). Cloning and expression of one chloroplastic ascorbate peroxidase gene from Nelumbo nucifera. Biochem. Genet. 49, 656–664. doi: 10.1007/s10528-011-9440-x
Chen, G. X., and Asada, K. (1989). Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol. 30, 987–998.
Cheng, H., He, Q. W., Huo, Y. M., Hou, L. X., and Lv, J. F. (2009). Molecular cloning, characterization and expression analysis of CmAPX. Mol. Biol. Rep. 36, 1531–1537. doi: 10.1007/s11033-008-9345-x
Chew, O., Whelan, J., and Millar, A. H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877. doi: 10.1074/jbc.M307525200
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
D'Arcylameta, A., Ferrariiliou, R., Contouransel, D., Phamthi, A. T., and Zuilyfodil, Y. (2006). Isolation and characterization of four ascorbate peroxidase cDNAs responsive to water deficit in cowpea leaves. Ann. Bot. 97, 133–140. doi: 10.1093/aob/mcj010
Datta, K., and Datta, S. K. (1999). Transformation of rice via PEG-mediated DNA uptake into protoplasts. Methods Mol. Biol. 111, 335–347. doi: 10.1385/1-59259-583-9:335
Du, S. C., Hwang, I. S., and Hwang, B. K. (2012). Requirement of the cytosolic interaction between pathogenesis-related protein10 and leucine-rich repeat protein1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. doi: 10.1105/tpc.112.095869
Duan, S. R., Ling, Y., and Xi, S. A. (2006). Overexpressing peroxisomal ascorbate peroxidase gene in rice enhanced tolerance to cadmium stress. Acta Pedol. Sinica 43, 111–116.
Dunand, C., Mathé, C., Lazzarotto, F., Margis, R., and Margispinheiro, M. (2011). Ascorbate peroxidase-related (APx-R) is not a duplicable gene. Plant Signal. Behav. 6, 1908–1913. doi: 10.4161/psb.6.12.18098
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Guo, F., Chiang, M. Y., Wang, Y. T., and Zhang, Y. Z. (2008). An in vitro recombination method to convert restriction- and ligation-independent expression vectors. Biotechnol. J. 3, 370–377. doi: 10.1002/biot.200700170
Guo, J. L., Ling, H., Wu, Q. B., Xu, L. P., and Que, Y. X. (2014). The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 4, 7042–7051. doi: 10.1038/srep07042
Guo, J. L., Xu, L. P., Fang, J. P., Su, Y. C., Fu, H. Y., Que, Y. X., et al. (2012). A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 31, 1801–1812. doi: 10.1007/s00299-012-1293-1
Guo, J. L., Xu, L. P., Su, Y. C., Wang, H. B., Gao, S. W., Xu, J. S., et al. (2013). ScMT2-1-3, a metallothionein gene of sugarcane, plays an important role in the regulation of heavy metal tolerance/accumulation. Biomed. Res. Int. 2013, 86–89. doi: 10.1155/2013/904769
Gupta, K., Agarwal, P. K., Reddy, M. K., and Jha, B. (2010). SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli. Plant Cell Rep. 29, 1131–1137. doi: 10.1007/s00299-010-0896-7
Hong, C. Y., and Kao, C. H. (2007). Expression of ascorbate peroxidase 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 58, 3273–3283. doi: 10.1093/jxb/erm174
Huang, F., Zhang, Y. H., and Li, X. M. (2013). In silico cloning and characterization of ascorbate peroxidase gene from Saccharum using bioinformatics tools. J. Shenyang Norm. Univ. 31, 295–300
Ishikawa, T., Sakai, K., Takeda, T., and Shigeoka, S. (1995). Cloning and expression of cDNA encoding a new type of ascorbate peroxidase from spinach. FEBS Lett. 367, 28–32. doi: 10.1016/0014-5793(95)00539-L
Ishikawa, T., Sakai, K., Yoshimura, K., Takeda, T., and Shigeoka, S. (1996). cDNAs encoding spinach stromal and thylakoid-bound ascorbate peroxidase, differing in the presence or absence of their 3′-coding regions. FEBS Lett. 384, 289–293. doi: 10.1016/0014-5793(96)00332-8
Ishikawa, T., Yoshimura, K., Sakai, K., Tamoi, M., Takeda, T., and Shigeoka, S. (1998). Molecular characterization and physiological role of a glyoxysome-bound ascorbate peroxidase from spinach. Plant Cell Physiol. 39, 23–34. doi: 10.1093/oxfordjournals.pcp.a029285
Jiang, M., Zhang, Z. X., and Yuan, L. J. (2012). Cloning and expression analysis of an ascorbate peroxidase gene BoAPX2 from Brassica oleracea var. italica. Acta Phytopathol. Sin. 42, 374–380.
Kawakami, S., Matsumoto, Y., Matsunaga, A. S., and Mizuno, M. (2002). Molecular cloning of ascorbate peroxidase in potato tubers and its response during storage at low temperature. Plant Sci. 163, 829–836. doi: 10.1016/S0168-9452(02)00232-7
Kornyeyev, D., Logan, B. A., Payton, P., Allen, R. D., and Holaday, A. S. (2001). Enhanced photochemical light utilization and decreased chilling-induced photoinhibition of photosystem II in cotton overexpressing genes encoding chloroplast-targeted antioxidant enzymes. Physiol. Plant. 113, 323–331. doi: 10.1034/j.1399-3054.2001.1130304.x
Lai, Y., Dang, F. F., Lin, J., Yu, L., Shi, Y. L., Xiao, Y. H., et al. (2013). Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol. Biochem. 62, 70–78. doi: 10.1016/j.plaphy.2012.10.010
Lazzarotto, F., Teixeira, F. K., Rosa, S. B., Dunand, C., Fernandes, C. L., Fontenele, A. D. V., et al. (2011). Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 191, 234–250. doi: 10.1111/j.1469-8137.2011.03659.x
Li, Q. W. (2000). Modern Technology for Sugarcane Improvement. Guangzhou: South China Science and Technology Press.
Li, Y. Z., Tessaro, M. J., Li, X., and Zhang, Y. L. (2010). Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 153, 1425–1434. doi: 10.1104/pp.110.156240
Lin, L., Wang, X. P., and Wang, Y. J. (2006). cDNA clone, fusion expression and purification of the novel gene related to ascorbate peroxidase from Chinese wild Vitis pseudoreticulata in E. coli. Mol. Biol. Rep. 33, 197–206. doi: 10.1007/s11033-006-0008-5
Liu, H. C., Tian, D. Q., Liu, J. X., Ma, G. Y., Zou, Q. C., and Zhu, Z. J. (2013). Cloning and functional analysis of a novel ascorbate peroxidase (APX) gene from Anthurium andraeanum. J. Zhejiang Univ. 14, 1110–1120. doi: 10.1631/jzus.B1300105
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mano, S., Yamaguchi, K., Hayashi, M., and Nishimura, M. (1997). Stromal and thylakoid-bound ascorbate peroxidases are produced by alternative splicing in pumpkin. FEBS Lett. 413, 21–26. doi: 10.1016/S0014-5793(97)00862-4
Maruta, T., Tanouchi, A., Tamoi, M., Yabuta, Y., Yoshimura, K., Ishikawa, T., et al. (2010). Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol. 51, 190–200. doi: 10.1093/pcp/pcp177
Melech-Bonfil, S., and Sessa, G. (2010). Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 64, 379–391. doi: 10.1111/j.1365-313X.2010.04333.x
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mittler, R., Sandy, V., Martin, G., and Frank, V. B. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mittler, R., Sandy, V., Nobuhiro, S., Gad, M., Tognetti, V. B., Klaas, V., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Murgia, I., Tarantino, D., Vannini, C., Bracale, M., Carravieri, S., and Soave, C. (2004). Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J. 38, 940–953. doi: 10.1111/j.1365-313X.2004.02092.x
Najami, N., Janda, T., Barriah, W., Kayam, G., Tal, M., Guy, M., et al. (2008). Ascorbate gene family in tomato: its identification and characterization. Mol. Genet. Genomics 279, 171–182. doi: 10.1007/s00438-007-0305-2
Narendra, S., Venkataramani, S., Shen, G., Wang, J., Pasapula, V., Lin, Y., et al. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57, 3033–3042. doi: 10.1093/jxb/erl060
Nayyar, H., and Gupta, D. (2006). Differential sensitivity of C3 and C4 plants to water deficit stress: association with oxidative stress and antioxidants. Environ. Exp. Bot. 58, 106–113. doi: 10.1016/j.envexpbot.2005.06.021
Pallavi, S., and Dubey, R. S. (2007). Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 26, 2027–2038. doi: 10.1007/s00299-007-0416-6
Panchuk, I. I., Volkov, R. A., and Friedrich, S. F. (2002). Heat stress and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Braz. J. Microbiol. 129, 838–853. doi: 10.1104/pp.001362
Saitou, N. N. M., and Nei, M. C. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Shi, W. M., Muramoto, Y., Ueda, A., and Takabe, T. (2001). Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene 273, 23–27. doi: 10.1016/S0378-1119(01)00566-2
Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., and Yabuta, Y. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. doi: 10.1093/jexbot/53.372.1305
Su, Y. C., Guo, J. L., Ling, H., Chen, S. S., Wang, S. S., Xu, L. P., et al. (2014a). Isolation of a novel peroxisomal catalase gene from sugarcane, which is responsive to biotic and abiotic stresses. PLoS ONE 9:e84426. doi: 10.1371/journal.pone.0084426
Su, Y. C., Wang, Z. Q., Liu, F., Li, Z., Peng, Q., Guo, J. L., et al. (2016). Isolation and characterization of ScGluD2, a new sugarcane beta-1,3-glucanase D family gene induced by Sporisorium scitamineum, ABA, H2O2, NaCl, and CdCl2 stresses. Front. Plant Sci. 7:1348. doi: 10.3389/fpls.2016.01348
Su, Y. C., Xu, L. P., Fu, Z. W., Yang, Y. T., Guo, J. L., Wang, S. S., et al. (2014b). ScChi, encoding an acidic class III chitinase of sugarcane, confers positive responses to biotic and abiotic stresses in sugarcane. Int. J. Mol. Sci. 15, 2738–2760. doi: 10.3390/ijms15022738
Su, Y. C., Xu, L. P., Xue, B. T., Wu, Q. B., Guo, J. L., Wu, L. G., et al. (2013). Molecular cloning and characterization of two pathogenesis-related β-1,3-glucanase genes ScGluA1 and ScGluD1 from sugarcane infected by Sporisorium scitamineum. Plant Cell Rep. 32, 1503–1519. doi: 10.1007/s00299-013-1463-9
Teixeira, F. K., Menezes-Benavente, L., Galvão, V. C., and Margis-Pinheiro, M. (2005). Multigene families encode the major enzymes of antioxidant metabolism in Eucalyptus grandis L. Genet. Mol. Biol. 28, 529–538. doi: 10.1590/S1415-47572005000400007
Teixeira, F. K., Menezes-Benavente, L., Galvão, V. C., Margis, R., and Margis-Pinheiro, M. (2006). Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 224, 300–314. doi: 10.1007/s00425-005-0214-8
Teixeira, F. K., Menezes-Benavente, L., Margis, R., and Margis-Pinheiro, M. (2004). Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J. Mol. Evol. 59, 761–770. doi: 10.1007/s00239-004-2666-z
Thordal-Christensen, H., Zhang, Z. G., Wei, Y. D., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Wang, S., Zhang, K. K., Huang, X., Fan, Y. J., Yang, L. T., and Li, Y. R. (2015). Cloning and functional analysis of thylakoidal ascorbate peroxidase (TAPX) gene in sugarcane. Sugar Tech 17, 131–135. doi: 10.1007/s12355-014-0354-x
Wang, Z. Q., Chen, Y., Yang, Y. T., Su, Y. C., Chen, S. S., Wu, Q. B., et al. (2015). Cloning and expression analysis of ascorbate peroxidase gene (ScAPX) in sugarcane (Saccharum officinarum). J. Agric. Biotechnol. 23, 170–180.
Xu, D. L., Park, J. W., Mirkov, T. E., and Zhou, G. H. (2008). Viruses causing mosaic disease in sugarcane and their genetic diversity in southern China. Arch. Virol. 153, 1031–1039. doi: 10.1007/s00705-008-0072-3
Yabuta, Y., Motoki, T. K., Takeda, T., Ishikawa, T., and Shigeoka, S. (2002). Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J. 32, 915–925. doi: 10.1046/j.1365-313X.2002.01476.x
Keywords: sugarcane, L-ascorbate peroxidase 6 gene, subcellular localization, biotic and abiotic stresses, transient overexpression
Citation: Liu F, Huang N, Wang L, Ling H, Sun T, Ahmad W, Muhammad K, Guo J, Xu L, Gao S, Que Y and Su Y (2018) A Novel L-ascorbate Peroxidase 6 Gene, ScAPX6, Plays an Important Role in the Regulation of Response to Biotic and Abiotic Stresses in Sugarcane. Front. Plant Sci. 8:2262. doi: 10.3389/fpls.2017.02262
Received: 25 July 2017; Accepted: 27 December 2017;
Published: 17 January 2018.
Edited by:
Sagadevan G. Mundree, Queensland University of Technology, AustraliaReviewed by:
Jian Li Yang, Zhejiang University, ChinaTaras P. Pasternak, Albert Ludwigs University of Freiburg, Germany
Copyright © 2018 Liu, Huang, Wang, Ling, Sun, Ahmad, Muhammad, Guo, Xu, Gao, Que and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youxiong Que, cXVleW91eGlvbmdAMTI2LmNvbQ==
Yachun Su, c3ljMjAwOW1haWxAMTYzLmNvbQ==
†These authors have contributed equally to this work.
 Feng Liu1†
Feng Liu1† Waqar Ahmad
Waqar Ahmad Liping Xu
Liping Xu Youxiong Que
Youxiong Que