- State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai’an, China
Phytochrome-interacting factors (PIFs) play important roles in photomorphogenesis, the shade avoidance response, and other aspects of plant growth and development. PIF family proteins have been well-studied in Arabidopsis thaliana, but little is known about their physiological functions and molecular mechanisms in maize (Zea mays). In this study, we investigated the physiological functions of ZmPIF4 and ZmPIF5, two highly conserved members of the PIF gene family. RT-qPCR and western blot analyses revealed that ZmPIF4 and ZmPIF5 expression and ZmPIF4 and ZmPIF5 levels peak at night and remain low during the day. Overexpression of ZmPIF4 and ZmPIF5 in Arabidopsis partially rescued the reduced hypocotyl elongation and defective response to gravity in pif1 pif3 pif4 pif5 quadruple mutants (pifq). In addition, under high red: far-red light conditions, Arabidopsis lines overexpressing ZmPIF4 exhibited a constitutive shade avoidance response, including early flowering, slender leaves and inflorescences, plant lodging and precocious leaf senescence. Furthermore, ZmPIF4 physically interacted with the Arabidopsis DELLA protein REPRESSOR OF GA1-3 (RGA), indicating a potential interaction between ZmPIF4 and gibberellin signaling pathway on plant growth. Taken together, our results revealed that ZmPIF4 and ZmPIF5 are functionally conserved proteins that may play conserved roles in the response to phytochrome signaling in plants.
Highlights:
In this study, the functions of ZmPIF4 and ZmPIF5 were characterized by expression in Arabidopsis, revealing conserved roles of PIF family proteins in photomorphogenesis and the shade avoidance response in land plants.
Introduction
Shade avoidance is mainly triggered by the reduced ratio of R:FR, which is sensed by the phytochrome family of photoreceptors (Franklin et al., 2003; Casal, 2013). The phytochrome family of Arabidopsis thaliana includes five members (phyA–phyE), and phyB is the primarily photoreceptor involved in the shade avoidance response (Franklin et al., 2003; Li et al., 2011). Under high R:FR light conditions, active phyB translocates into the nucleus and interacts with multiple downstream signaling proteins to mediate light-regulated changes in plant growth and development (Quail, 1991; Kami et al., 2010; Hornitschek et al., 2012). Under low R:FR conditions, phyB is largely inactivated and located in the cytosol (Kircher et al., 1999). Disruption of phyB in Arabidopsis, or both phyB1 and phyB2 in maize, caused constitutive shade avoidance response, even under high R:FR conditions (Robson et al., 1993; Sheehan et al., 2007).
Phytochrome-interacting factors (PIFs) are basic helix-loop-helix (bHLH) transcription factors that are involved in seed germination, photomorphogenesis, shade responses, flowering time, and leaf senescence (Leivar and Quail, 2011; Casal, 2013; Sakuraba et al., 2014). As critical factors act at downstream of phyB, they positively regulate the shade avoidance response (Josse et al., 2008; Leivar and Quail, 2011). Comparison of the protein sequences of PIF family members has shown that they have evolutionarily conserved bHLH domains located at the C-terminal and these domains function in DNA binding and dimer formation. In addition to the bHLH domain, PIFs also have active phytochrome A binding domains (APA) and/or active phytochrome B binding domains (APB) at the N-terminal. Biochemical analyses have revealed that PIFs physically interact with phyA or phyB through their APA or APB domains, respectively (Choi et al., 1999; Castillon et al., 2007; Shen et al., 2008). Under high R:FR light conditions, the interaction between PIFs and phyB leads to PIF phosphorylation, ubiquitination, and then degradation via the 26S proteasome (Lorrain et al., 2008). Under low R:FR light conditions, PIFs promotes cell elongation by increasing the transcription of growth-promoting genes (Paik et al., 2017).
Multiple PIFs family proteins (PIF1, PIF3, PIF4, PIF5, PIF7) have been well-characterized in Arabidopsis thaliana. Disruption of PIF1, PIF3, PIF4, and PIF5 (the pifq quadruple mutant) causes constitutive photomorphogenesis under dark conditions and reduces the sensitively to shade signals (Leivar et al., 2008). Overexpression of AtPIF4 and AtPIF5 leads to a constitutive shade avoidance response, with plants displaying long hypocotyls and petioles, even under high R:FR conditions (Lorrain et al., 2008). PIF4, PIF5, and PIF7 directly regulate the expression of these genes that promote cell elongation and mediate the response to shade signals (Hornitschek et al., 2012; Li et al., 2012; Sakuraba et al., 2014). Genome-wide analyses of PIF targets have revealed that PIFs directly target 100s of genes involved in auxin homeostasis (TAA1 and YUC), signaling responses (GH3, IAA, and ARF), cell wall modification and elongation (EXB, XTH) (Zhang et al., 2013; Pfeiffer et al., 2014).
In addition to the auxin signaling pathway, PIFs are thought to be involved in a variety of hormone-response pathways, including gibberellin (GA), brassinosteroid (BR), jasmonic acid (JA), ethylene, and nitric oxide (Lau and Deng, 2010; Mazzella et al., 2014; Paik et al., 2017). For instance, GA can induce the expression of PIFs by promoting the degradation of DELLAs via the 26S proteasome system. In the DELLA mutant, exogenous GA prolongs hypocotyl elongation in the dark (Dill et al., 2001; Tyler et al., 2004; Li et al., 2016).
Although PIF family proteins have been identified as playing important roles in many aspects of growth in Arabidopsis, little is known about their physiological roles in other plants. There are about 300 putative members of the bHLH family existed in maize, of which about 200 putative members have the complete bHLH domain (Kumar et al., 2016). Previous studies have shown that ZmPIF3.1 (GRMZM2G115960) and ZmPIF3.2 (GRMZM2G387528) physically interact with the Pfr form of ZmphyB1, and ZmPIF3.1 can also interact with the Pfr of phyB of Arabidopsis (Kumar et al., 2016). Over-expression of ZmPIF3 (ZmPIF3.2) in rice (Oryza sativa) can enhance tolerance to drought and salt stress (Gao et al., 2015). These results show that ZmPIFs play important roles in phytochrome signal transduction and plant growth. However, the physiological and biochemical functions of other ZmPIFs are still unclear. In this study, we conducted a genome-wide analysis to identify 15 putative PIF family proteins in maize. We also cloned the genes encoding two ZmPIF family members from the maize inbred line B73 and transformed them into the Arabidopsis wild-type Col-0 and the Arabidopsis quadruple pifq mutant, to determine their roles in the shade avoidance response in plants.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana pifq quadruple mutant (pif1, pif3, pif4, pif5) used in this study was described by Shin et al. (2009), and the wild-type control plants used in this study were Arabidopsis ecotype Columbia-0 (Col-0). Arabidopsis seeds were surface-sterilized with 20% bleach for 20 min and then washed four times with sterile distilled-deionized H2O. After vernalization for 2 days at 4°C, seeds were germinated on GM plates (4.74 g/L Murashige and Skoog [MS] salts, 10 g/L 1% sucrose, 0.5 g/L MES, 8 g/L agar, pH 5.8). Generally, the Arabidopsis plants were grown under long-day conditions (16 h light/8 h dark, light intensity 100 μmol m-2 s-1, 22°C).
Seedlings of maize inbred line B73 and Nicotiana benthamiana were grown in growth chambers under a 12-h light/12-h dark cycle at 210 μmol m-2 s-1 of light at 28°C. Two weeks after planting, seedlings of maize inbred line B73 were transferred to constant light conditions and then harvested at different Zeitgeber times to measure the diurnal expression of ZmPIF4 and ZmPIF5. Three weeks after planting, seedlings of the maize inbred line B73 were harvested and separated into roots, coleoptiles, stems, and leaves for detection of the tissue expression patterns of ZmPIFs.
Total RNA Extraction and RT-qPCR Assays
Plant total RNA was extracted with an Ultrapure RNA Kit (CWBIO, China) following the manufacturer’s instructions. The first-strand cDNA was synthesized using EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGEN Biotech, China). The cDNA was then diluted to 60 ng/μL and 2 μL was used for qPCR. Quantitative PCR was performed using UltraSYBR Mixture (CWBIO, China) in an ABI QuantStudio 6 Flex Real-Time PCR System (ABI). The RT-qPCR procedure was described in Ma et al. (2016). The expression level of UBIQUITIN1 and 18S rRNA were used as internal controls for RT-qPCR in Arabidopsis and maize, respectively. Plant material for RT-qPCR was collected from three biological replicates, and three technical replicates were performed for each experiment.
Starch Staining and Gravitropism in Arabidopsis
Seedlings of Col-0, pifq mutants, and transgenic lines of ZmPIF4 and ZmPIF5 were grown on medium in the dark for 4 days, and then the direction of gravity was changed by 90° and the bending angle of the hypocotyl elongation zone was measured every hour using ImageJ program. The contents of amyloplasts in the endodermis of the hypocotyl elongation zone and the columella cells in the root cap of these seedlings were detected by I2–KI staining following the method described by Ma et al. (2017).
Plasmid Construction and Generation of Transgenic Plants of Arabidopsis
To generate transgenic ZmPIF4-OE and ZmPIF5-OE lines in the pifq or wild-type Col-0 background, the coding regions of ZmPIF4 and ZmPIF5 were PCR-amplified from cDNA of the maize inbred line B73 with the primer pairs ZmPIF4-F and ZmPIF4-R, and ZmPIF5-F and ZmPIF5-R (see Supplementary Table S2 for more information on all of the primers used in this study). The fragments of ZmPIF4 and ZmPIF5 were then inserted into a BamHI and SpeI digested pPZP211-35Spro::3FLAG empty binary vector (Ma et al., 2016) to produce 35Spro::ZmPIF4-3FLAG and 35Spro:ZmPIF5-3FLAG. Finally, these two plasmids were transformed into the pifq mutant plants and the Col-0 plants, to generate ZmPIF4-OE/pifq, ZmPIF5-OE/pifq, ZmPIF4-OE, and 35Spro:ZmPIF5-OE lines. More than 20 independent lines of each transformation were selected with kanamycin and verified by RT-qPCR and western blot assays. The procedure for the western blot was described previously (Ma et al., 2017). All immunoblots were repeated at least twice and a representative experiment is shown (Supplementary Figure S5).
Subcellular Localization of ZmPIF4 and ZmPIF5
Plasmids 35Spro::ZmPIF4-GFP and 35Spro:ZmPIF5-GFP were generated by PCR-amplifying the coding region of ZmPIF4 and ZmPIF5 from 35Spro::ZmPIF4-3FLAG or 35Spro:ZmPIF5-3FLAG using primer pairs GFP-ZmPIF4-F and GFP-ZmPIF4-R, and GFP-ZmPIF5-F and GFP-ZmPIF5-R. The fragments containing the coding region of ZmPIF4 and ZmPIF5 were then inserted into pPZP211-35Spro::GFP digested with BamHI and XbaI. The various plasmids were then transformed into the Agrobacterium strain GV3101 and infiltrated into leaves of N. benthamiana together with the 35Spro::H2B-Cherry (Howe et al., 2012). Three days after infiltration, the N. benthamiana plants were transferred into dark conditions for 12 h, the lower epidermis of the infiltrated leaf was peeled, and the fluorescence signals were observed with a two-photon laser confocal microscope (Zeiss).
For subcellular localization analysis of ZmPIF4 and ZmPIF5 in protoplasts, 30-day-old Arabidopsis wild-type plants (Col-0) grown under short day (8-h light/16-h dark) conditions were used to isolate the protoplasts. Plasmids 35Spro::ZmPIF4-GFP and 35Spro:ZmPIF5-GFP were transformed into the protoplasts using PEG method (Yoo et al., 2007). Incubated for 18 h in darkness to allow the constructs to be expressed, and then the fluorescent signals were observed with a laser confocal microscope (Zeiss).
Yeast Two-Hybrid Assay
The plasmids pGBKT7-ZmPIF4 and pGBKT7-ZmPIF5 were generated by PCR-amplifying the coding region of ZmPIF4 and ZmPIF5 from 35Spro::ZmPIF4-3FLAG and 35Spro::ZmPIF5-3FLAG using the primer pairs ADBD-ZmPIF4-F and ADBD- ZmPIF4-R, and ADBD-ZmPIF5-F and ADBD-ZmPIF5-R. The fragments containing the coding region of ZmPIF4 and ZmPIF5 were inserted into pGBKT7 (Clontech) digested with BamHI and NdeI to produce pGBKT7-ZmPIF4 and pGBKT7-ZmPIF5.
The plasmid pGADT7-RGA was generated by PCR-amplifying the coding region of RGA from Arabidopsis Col-0 with the primer pair ADBD-RGA-F and ADBD-RGA-R. Then, the fragment containing the RGA coding region was inserted into pGADT7 (Clontech) digested with BamHI and SacI to produce pGADT7-RGA. The yeast two-hybrid procedure was performed following the manufacturer’s instructions (Clontech).
Luciferase Complementation Imaging Assays
The plasmids nLUC-ZmPIF4, cLUC-ZmPIF4, nLUC-ZmPIF5, and cLUC-ZmPIF5 were generated by PCR-amplifying the coding regions of ZmPIF4 and ZmPIF5 from 35Spro::ZmPIF4-3FLAG and 35Spro::ZmPIF5-3FLAG using the primer pairs LCI- ZmPIF4-F and LCI-ZmPIF4-R, and LCI-ZmPIF5-F and LCI-ZmPIF5-R. The fragments containing the coding regions of ZmPIF4 and ZmPIF5 were inserted into KpnI and BamHI digested pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors (Chen et al., 2008) to produce nLUC-ZmPIF4, cLUC-ZmPIF4, nLUC-ZmPIF5, and cLUC-ZmPIF5. The coding region of RGA was PCR-amplified from pGADT7-RGA with the primer pair LCI-RGA-F and LCI-RGA-R, and inserted into KpnI and SalI digested pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors to produce nLUC-RGA and cLUC-RGA. Each of the nLUC- or cLUC- fused plasmids was transformed into Agrobacterium strain GV3101 and then infiltrated into leaves of N. benthamiana. Three days after infiltration the infiltrated plants were transferred into darkness for 12 h and the luciferase activity was measured using an in vivo imaging system (XENOGEN) with potassium luciferin as the substrate.
Bimolecular Fluorescence Complementation (BiFC) Assay
To generate plasmids ZmPIF4-YFPN and RGA-YFPC, ZmPIF4 and RGA were amplified with the primer pairs YFP-ZmPIF4-F and YFP-ZmPIF4-R, and YFP-RGA-F and YFP-RGA-R. Full-length cDNAs of ZmPIF4 and ZmPIF5 were subcloned into the TOPO vectors and then recombined into pSITE-nEYFP and pSITE-cEYFP (Martin et al., 2009), respectively. The plasmids ZmPIF4-YFPN and RGA-YFPC were then transformed into the Agrobacterium strain GV3101 and infiltrated into leaves of N. benthamiana. Three days after infiltration, the fluorescent signals were observed with a laser confocal microscope (Zeiss).
Results
Identification and Classification of PIF Family Proteins in Maize
To identify the putative PIFs family proteins in maize, we performed BLASTP analysis using the PIF protein sequences of Arabidopsis as query sequences. This identified 15 putative ZmPIF family proteins in maize (Supplementary Table S1). A phylogenetic tree with the protein sequences of PIF family proteins from Arabidopsis and maize showed that the putative ZmPIFs were closely related to AtPIFs (Figure 1A). Analysis of the conservation of the protein sequence of each putative PIF showed that seven proteins (ZmbHLH16, ZmbHLH27, ZmbHLH36, ZmbHLH76, ZmbHLH115, ZmbHLH165, and ZmbHLH198) have a highly-conserved APB domain, and three (ZmbHLH76, ZmbHLH165, and ZmbHLH198) have a highly-conserved APA domain (Figure 1B). Both APA and APB domains were also identified in the ZmPIF3.1 and ZmPIF3.2, two previously reported PIF family proteins in maize encoded by ZmbHLH76 and ZmbHLH165, respectively (Figure 1B, left lower panel). Multiple alignments with the full-length protein sequences of AtPIF4, AtPIF5, ZmbHLH16, ZmbHLH27, and the putative PIF4 and PIF5 proteins in other land plants showed that ZmbHLH16 (GRMZM5G865967) and ZmbHLH27 (GRMZM2G165042) are very similar to AtPIF4 and AtPIF5 (Figure 1C and Supplementary Figure S1). ZmbHLH16 and ZmbHLH27 were therefore renamed ZmPIF4 and ZmPIF5, respectively.
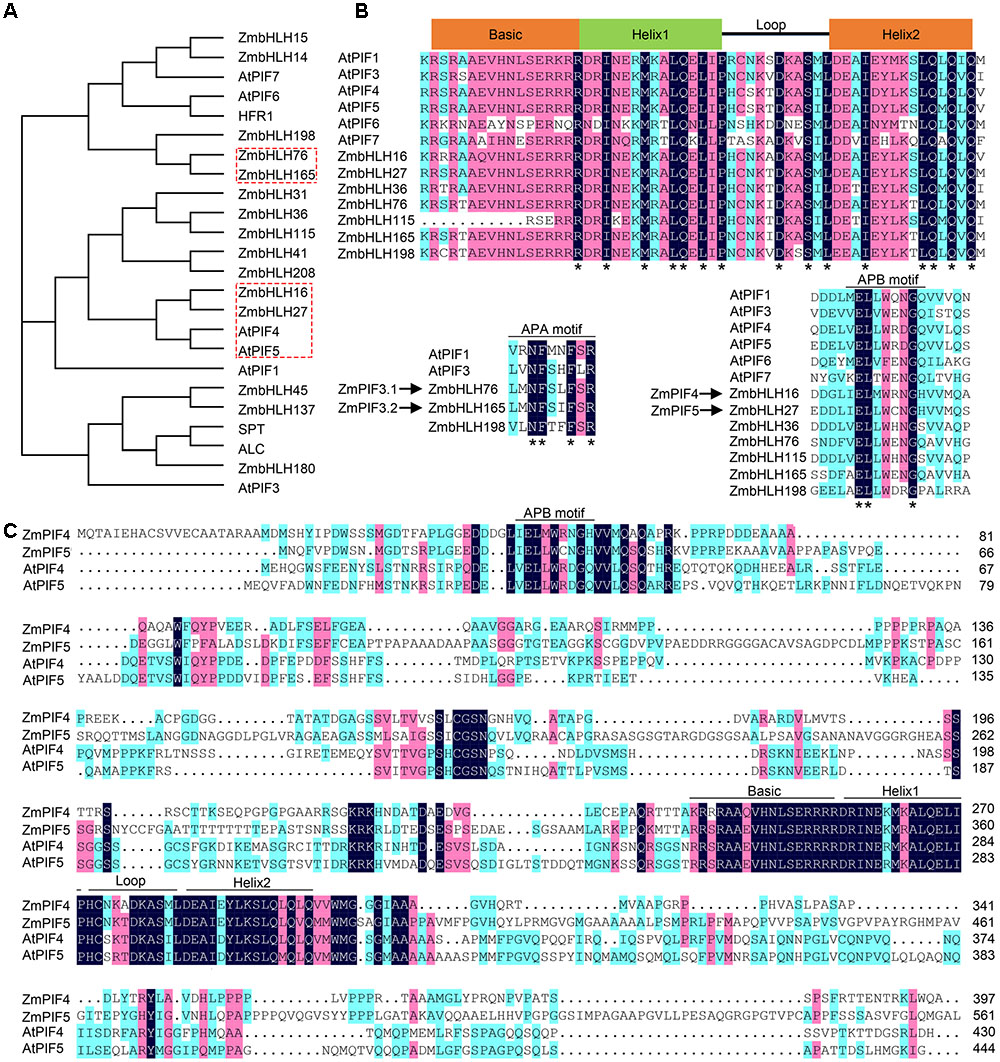
FIGURE 1. Sequence comparison of PIF family proteins in maize and Arabidopsis. (A) Phylogenetic tree of PIFs, PIF-like and bHLH family proteins in Zea mays (Zm) and Arabidopsis thaliana (At). The neighbor-joining method was used to construct the phylogenetic tree. The boxes with the red dashed line indicate the clade containing ZmPIF3, ZmPIF4, and ZmPIF5. (B) Multiple sequence alignment of bHLH (top panel), APA (left, lower panel), and APB (right, lower panel) domains of PIF family proteins. Asterisks, conserved amino acids. (C) Multiple sequence alignment of the protein sequences of AtPIF4, AtPIF5, ZmPIF4 (ZmbHLH16), and ZmPIF5 (ZmbHLH27). The conserved bHLH and APB domains are underlined.
Analysis of ZmPIF4 and ZmPIF5 Expression and Protein Subcellular Localization
RT-qPCR analysis showed that both ZmPIF4 and ZmPIF5 are expressed in the roots, stems, coleoptiles, and leaves at the six-leaf stage (Figure 2A). Under 12 h light/12 h dark diurnal conditions, RT-qPCR analysis also demonstrated that ZmPIF4 and ZmPIF5 have similar temporal expression patterns, with their transcript levels increasing overnight, peaking at dawn, and decreasing during the day (Figure 2B). In addition, to verify whether the transcript level of ZmPIF4 and ZmPIF5 is regulated by R or FR light, three-leaf stage seedlings of maize inbred line B73 grown under white light conditions were transferred to FR light for 1 h and then transferred to R light for various times. RT-qPCR analyses revealed that the ZmPIF4 and ZmPIF5 transcript levels were rapidly induced by FR light, but repressed by R light (Supplementary Figure S2A).
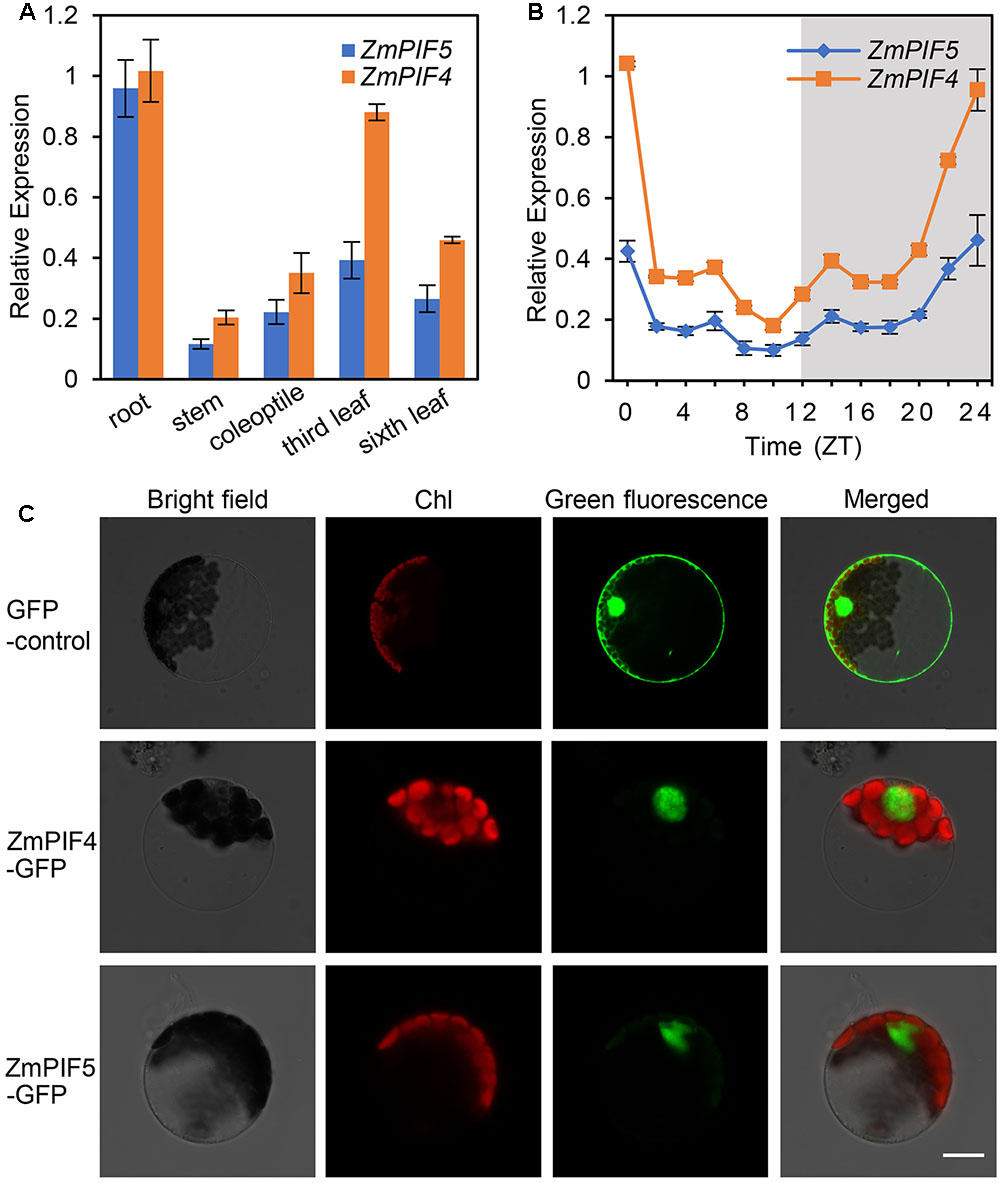
FIGURE 2. Gene expression and protein subcellular localization of ZmPIF4 and ZmPIF5. RT-qPCR analysis indicated that ZmPIF4 and ZmPIF5 are expressed in multiple tissues (A), and expressed at high levels at dawn and low levels at dusk (B). Six-leaf stage seedling plants of maize inbred line B73 were used to harvest different tissues (A), or whole plants were harvested at different times (B), and then used to perform RT-qPCR analysis. The 18S ribosomal RNA was used as an internal control for RT-qPCR analysis. Data are means and SD of three independent biological replicates. (C) Transient expression analysis in Arabidopsis protoplasts indicated that ZmPIF4-GFP and ZmPIF5-GFP localize in the nucleus. The expression of H2B-mCherry was used to show the position of the nucleus. Scale bar: 20 μm.
Further, we investigated the protein subcellular localization of ZmPIF4 and ZmPIF5 in Arabidopsis protoplasts and N. benthamiana epidermal cells. As shown in Figure 2C and Supplementary Figure S2B, the florescent signals of ZmPIF4-GFP and ZmPIF5-GFP fusion proteins were only observed in the nucleus. This suggests that both ZmPIF4 and ZmPIF5 localize in the nucleus, consistent with a potential function as transcription factors.
Overexpression of ZmPIF4 and ZmPIF5 Partially Rescued the Phenotype of Arabidopsis pifq Mutants
In Arabidopsis, the pifq quadruple mutant displays a constitutive photomorphogenic phenotype under continuous dark conditions, including short hypocotyls, open cotyledons, and the loss of negative gravitropism (Shin et al., 2009). To examine whether ZmPIF4 and ZmPIF5 could have PIF function, we tested whether they could complement the pifq phenotype by generating ZmPIF4-OE/pifq and ZmPIF5-OE/pifq plants. RT-qPCR analysis showed that the transcripts of ZmPIF4 and ZmPIF5 were present at high levels in the transgenic ZmPIF4-OE/pifq and ZmPIF5-OE/pifq plants (Figure 3E). The hypocotyl length of pifq was significantly shorter than that of Col-0, consistent with previous reports (Leivar et al., 2008). When ZmPIF4 and ZmPIF5 were over-expressed in the pifq background, their hypocotyls were significantly elongated relative to pifq (Figures 3A–D). The cotyledons were completely closed in ZmPIF4-OE4/pifq and ZmPIF4-OE5/pifq, and only some of the plants in the ZmPIF5 transgenic line had open cotyledons. This may be related to the level of protein expression. The partial complementation supports the hypothesis that both ZmPIF4 and ZmPIF5 have physiological functions similar to those of AtPIF4 and AtPIF5.
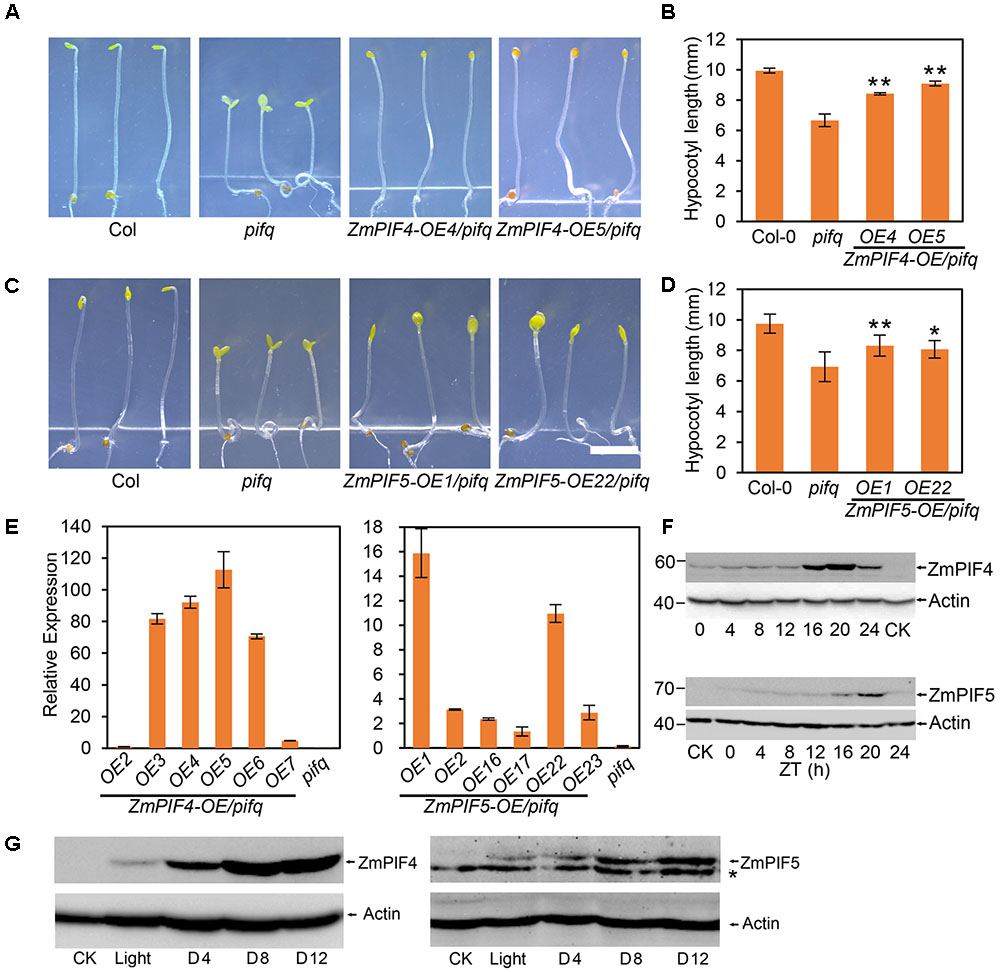
FIGURE 3. Overexpression of ZmPIF4 and ZmPIF5 partially rescued the reduced hypocotyl elongation of the pifq mutant in Arabidopsis. Overexpression of ZmPIF4 (A,B) and ZmPIF5 (C,D) partially rescued the reduced hypocotyl elongation of pifq mutants under continuous dark conditions in Arabidopsis. (A,C) The phenotype of 4-day-old dark-grown transgenic lines of ZmPIF4-OE (OE4 and OE5) and ZmPIF5-OE (OE1 and OE22) in the pifq mutant background. Scale bar: 3 mm. (B,D) The quantification of hypocotyl length of the seedling plants showed in (A,C), respectively. Data represent the mean and SD of at least 30 seedlings. Statistical significance analyses were performed between the transgenic plants and pifq mutant plants. ∗P < 0.05; ∗∗P < 0.01. (E) RT-qPCR analyses revealed that ZmPIF4 and ZmPIF5 were highly expressed in Arabidopsis plants overexpressing ZmPIF4 (left panel) and ZmPIF5 (right panel) in the pifq mutant background. Four-day-old seedling plants were used to perform RT-qPCR analysis. UBQ1 was used as the internal control for RT-qPCR analysis. Data are means and SD of three independent biological replicates. Western blot analyses of transgenic Arabidopsis plants revealed that the ZmPIF4 and ZmPIF5 proteins accumulated to high levels at midnight (F, ZT20), and were induced by darkness (G). Seven-day-old seedlings of ZmPIF4-OE4/pifq and ZmPIF5-OE1/pifq grown under LD conditions (16-h light/8-h dark) were harvested at different times (F), or transferred from light conditions (at ZT4) to darkness for 4, 8, 12 h (G), and then used to perform western blot analysis. ACTIN was used as the internal control for western blots. CK, control plant; ∗, non-specific bands.
Next, we investigated whether the ZmPIF4 and ZmPIF5 proteins are light labile using the Arabidopsis transgenic lines expressing ZmPIF4 and ZmPIF5. As shown in Figure 3F, ZmPIF4 and ZmPIF5 proteins accumulate to high levels at night, peak at ZT20, and are present at low levels during the day. Furthermore, we checked whether the levels of ZmPIF4 and ZmPIF5 proteins are more stable under dark conditions. The ZmPIF4 and ZmPIF5 transgenic seedlings were transferred from light (at ZT4) to darkness for various times and then used to perform western blots. As shown in Figure 3G, the accumulation of ZmPIF4 and ZmPIF5 proteins increased in the darkness. All these results indicate that ZmPIF4 and ZmPIF5 might be light labile and subject to degradation during the day, similar to AtPIF4 and AtPIF5.
Overexpression of ZmPIF4 Partially Rescued the Negative Gravitropism Response of pifq Hypocotyls
In the Arabidopsis pifq mutant, the negative gravitropism of hypocotyls is completely disrupted in dark conditions (Kim et al., 2011). Overexpression of ZmPIF4 and ZmPIF5 partially restored the negative gravitropism of hypocotyls in darkness, especially in ZmPIF4-OE/pifq (Figure 4A). This was further supported by growing seedlings of Col-0, pifq mutants, and transgenic lines of ZmPIF4 and ZmPIF5 on medium for 4 days in the dark, then changing the direction of gravity by 90° and measuring the bending angle of the hypocotyl elongation zone every hour (Figures 4B,C). The ZmPIF4 transgenic lines were able to respond quickly to gravity, but the pifq mutant and the ZmPIF5-OE transgenic lines responded more slowly (Figures 4B,C).
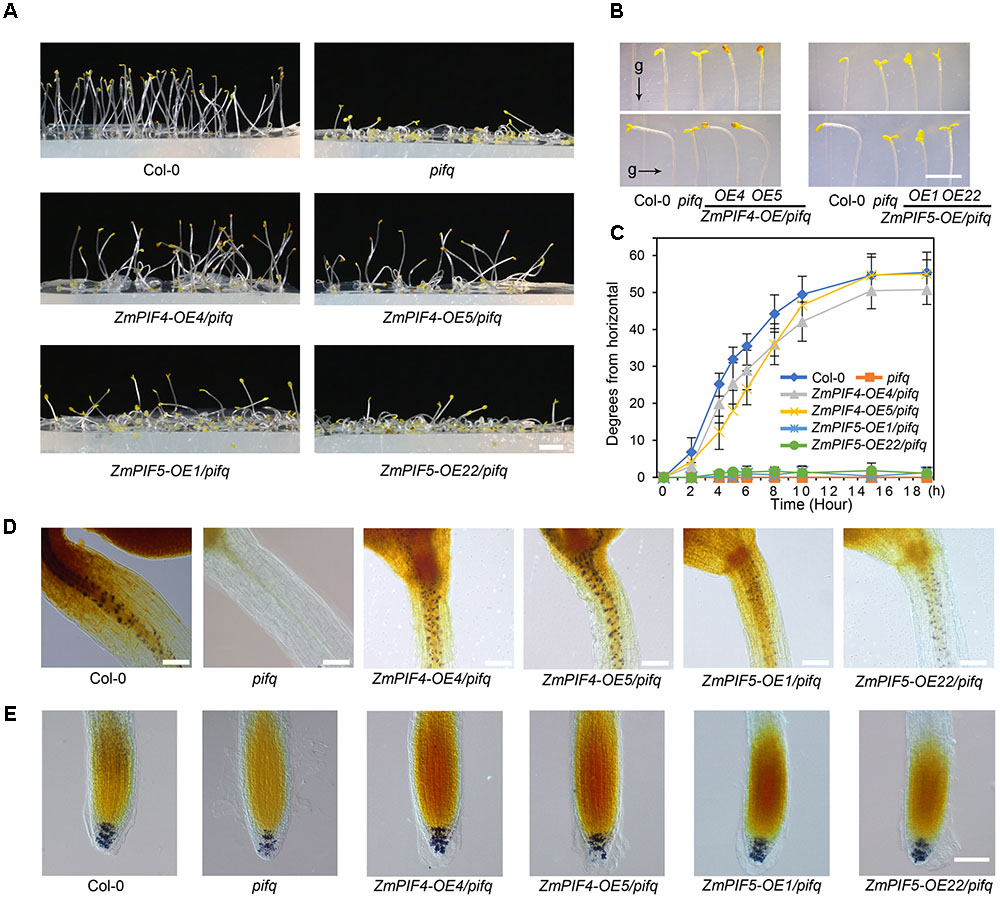
FIGURE 4. Overexpression of ZmPIF4 but not ZmPIF5 partially rescued the negative gravitropism response of the hypocotyl in Arabidopsis pifq mutants. (A) ZmPIF4 but not ZmPIF5 restored the negative gravitropism of pifq mutant. Seedlings of wild-type control (Col-0), pifq mutant, and transgenic lines of ZmPIF4 and ZmPIF5 in the pifq background were grown in continuous darkness for 4 days. Scale bar: 3 mm. (B,C) ZmPIF4, but not ZmPIF5, affects the negative gravitropism of hypocotyls. Four-day-old seedlings vertically grown in continuous darkness conditions were turned 90° for 10 h (B), or for various times (C). g, the direction of gravity. Data represent the mean and SD of 30 seedling plants. Scale bar: 3 mm. I2–KI staining of amyloplasts in the hypocotyl (D) and root (E). The seedlings were grown in white light for 24 h and then transferred to darkness for 72 h. Scale bar: 100 μm.
In darkness, accumulation of PIF proteins suppresses the conversion of starch-filled endodermal amyloplasts to plastids and thus plays an important role in the plant’s response to gravity (Kim et al., 2011). To test whether ZmPIF4 or ZmPIF5 could complement the amyloplast defect in pifq mutants, we detected amyloplasts by I2–KI staining. Amyloplasts were detected in both the hypocotyl and the root cap in Col-0 plants, but only in the root caps of the pifq mutants, consistent with previous studies (Kim et al., 2011). In the seedling plants of ZmPIF4-OE4/pifq and ZmPIF4-OE5/pifq, amyloplasts were detected in both the endodermis of the hypocotyl elongation zone and in the columella cells of the root cap. Indeed, the transgenic lines of ZmPIF4 stained more strongly, compared to the ZmPIF5 transgenic lines (Figures 4D,E). This indicates that both ZmPIF4 and ZmPIF5 might be involved in the plant response to gravity, with ZmPIF4 playing a primary role.
ZmPIF4 and ZmPIF5 Can Affect Seedling Development in Arabidopsis
To test whether ZmPIFs could regulate skotomorphogenesis and the shade avoidance response, we next overexpressed FLAG-tagged versions of the ZmPIFs (ZmPIF4-3FLAG and ZmPIF5-3FLAG) under the control of the constitutive 35S promoter in the Arabidopsis wild-type control Col-0 plants. Three independent transgenic ZmPIF4 over-expression lines (OE8, OE10, and OE11), and ZmPIF5 over-expression lines (OE2, OE3, and OE12) were selected and used to perform further analysis (Supplementary Figure S3A). The hypocotyls of all the transgenic lines were significantly longer than those of Col-0 in continuous dark conditions, which indicates that both ZmPIF4 and ZmPIF5 can participate in skotomorphogenesis in Arabidopsis (Figures 5A–D).
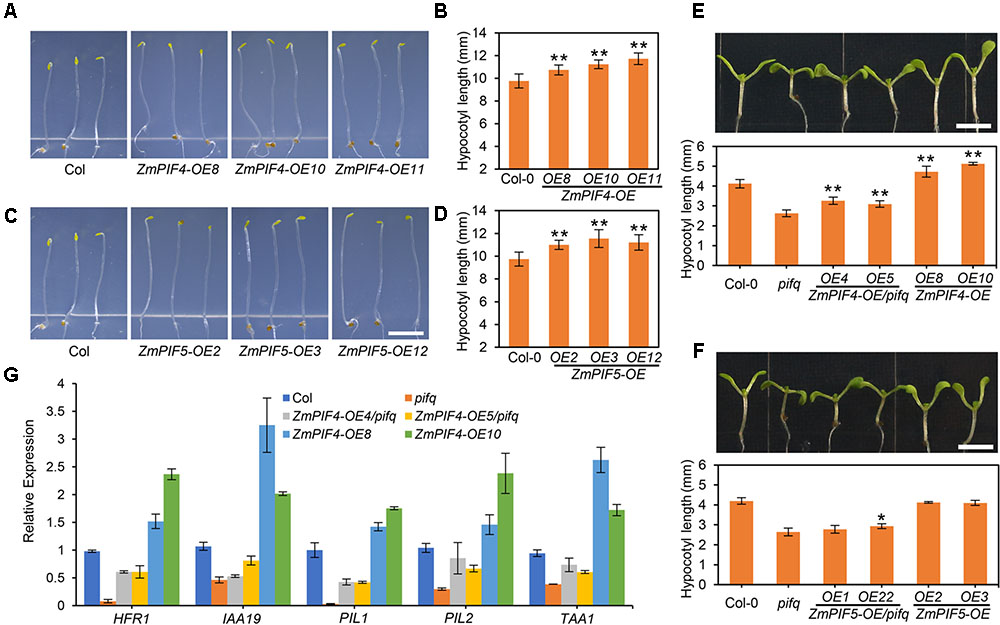
FIGURE 5. Overexpression of ZmPIF4 and ZmPIF5 in wild-type Arabidopsis Col-0 plants. (A–D) Overexpression of ZmPIF4 and ZmPIF5 promoted hypocotyl elongation in continuous darkness, compared with wild-type Col-0 control plants. (A,C) The phenotype of 4-day-old dark grown transgenic lines of ZmPIF4-OE and ZmPIF5-OE. Scale bar: 4 mm. (B,D) Quantification of hypocotyl length of the seedling plants shown in (A,C). Data represent the mean and SD of at least 30 seedlings. ∗P < 0.05; ∗∗P < 0.01. (E,F) The phenotype (upper) and quantification (lower panel) of 7-day-old seedling of ZmPIF4 and ZmPIF5 transgenic lines under LD with low R/FR light (R/FR = 0.35) conditions. Scale bar: 3 mm. Data represent the mean and SD. n = 30; ∗P < 0.05; ∗∗P < 0.01. (G) RT-qPCR analysis indicated that overexpression of ZmPIF4 enhanced the expression level of shade response related genes HFR1, IAA19, PIL1, PIL2, and TAA1. Seven-day-old seedlings grown under LD with low R/FR light conditions were used to perform RT-qPCR analysis. UBQ1 was used as the internal control. Data are means and SD of three independent biological replicates.
Further, we checked the hypocotyl elongation phenotype of all the ZmPIF4 and ZmPIF5 transgenic lines in the pifq mutant and Col-0 backgrounds under long-day conditions with high (Supplementary Figure S3B) or low R:FR (Figures 5E,F), respectively. Under long-day conditions with high R:FR (white light, R:FR = 5), the ZmPIF4 and ZmPIF5 transgenic lines displayed shorten hypocotyls, without significant differences between ZmPIF4 and ZmPIF5 (Supplementary Figure S3B). By contrast, under long-day conditions with low R:FR (F:FR = 0.35), the transgenic lines of ZmPIF4, but not ZmPIF5, in both pifq mutant and Col-0 backgrounds displayed longer hypocotyls, compared with the control plants (Figures 5E,F). This suggests that ZmPIF4 is involved in shade avoidance responses. Furthermore, RT-qPCR analyses revealed that the transcript levels of PIL1, PIL2, HFR1, IAA19, and TAA1 in 7-day-old seedlings grown under low R:FR conditions were significantly increased in ZmPIF4-OE4/pifq and ZmPIF4-OE5/pifq, and ZmPIF4-OE8 and ZmPIF4-OE10, compared with pifq mutant or Col-0 wild-type control plants, respectively (Figure 5F). These observations suggested that ZmPIF4 regulates cell elongation of the hypocotyl under low R:FR conditions, probably by promoting the expression of genes related to the shade avoidance response.
Overexpression of ZmPIF4 Produced a Constitutive Shade Avoidance Response in Arabidopsis
To further verify the physiological function of ZmPIF4 in the shade avoidance response, we examined the phenotype of adult plants of the ZmPIF4 transgenic line grown under long-day with high R:FR conditions. As shown in Figures 6A,B, three independent transgenic overexpression lines of ZmPIF4 showed earlier flowering times and had fewer rosette leaves at flowering, compared with wild-type control plant Col-0. In addition, the phenotypes of continuous shade avoidance, including elongated petioles, reduced leaf area, accelerated leaf senescence, slender inflorescences, and plant lodging were observed in the ZmPIF4 over-expression lines (Figures 6C–F and Supplementary Figure S4), but not the ZmPIF5 overexpression lines (data not shown). The chlorophyll and carotenoid contents were lower in the ZmPIF4 overexpression lines (Figure 6E), and RT-qPCR analysis revealed that the transcript levels of the chlorophyll biosynthesis genes GUN4, HEMA1, and CHLH were significantly decreased in adult plants of the ZmPIF4 overexpression lines (OE8 and OE10), compared with wild-type control plant Col-0 (Figure 6G). All these results suggested that overexpression of ZmPIF4 resulted in a constitutive shade avoidance response, even under high R:FR conditions.
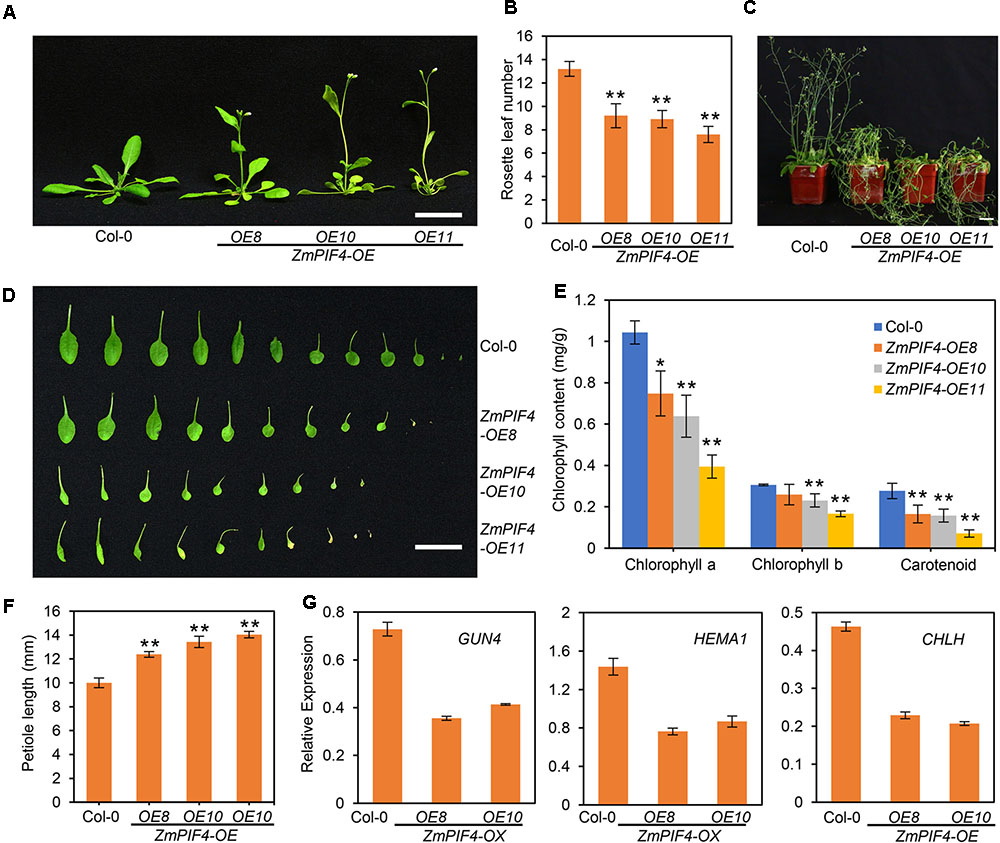
FIGURE 6. Overexpression of ZmPIF4 in Arabidopsis produces a constitutive shade avoidance response. (A,B) Arabidopsis overexpressing ZmPIF4 flowered early, compared with wild-type Col-0 plants. (A) 25-day-old plants grown under LD conditions (white light, 100 μmol m-2 s-1, R/FR = 5) were used to take a photograph. (B) The numbers of rosette leaves at bolting. Scale bar: 3 cm. ∗P < 0.05; ∗P < 0.01; n = 20. (C) Overexpression of ZmPIF4 resulted in plant lodging, compared with wild-type Col-0. 45-day-old plants grown under LD conditions are shown. Scale bar: 3 cm. Overexpression of ZmPIF4 altered leaf shape (D), promoted petiole elongation (F), and reduced the contents of chlorophyll and carotenoids (E), compared with wild-type Col-0 plants. 25-day-old mature plants grown under LD conditions wee used to perform the analyses in (D–F). Scale bar: 3 cm. ∗P < 0.05; ∗∗P < 0.01. (G) RT-qPCR analysis revealed that Arabidopsis plants overexpressing ZmPIF4 had reduced transcript levels of chlorophyll biosynthesis-related genes (GUN4, HEMA1, and CHLH). Seven-day-old seedlings grown under LD conditions were used to perform RT-qPCR analysis. UBQ1 was used as the internal control. Data are means and SD of three replicates.
ZmPIF4-Regulated Hypocotyl Elongation Involves the GA Signaling Pathway
To examine whether ZmPIF4 can interact with the GA signaling pathway to coordinately regulate hypocotyl elongation and the shade avoidance response, we first tested whether it can interact with the DELLA protein REPRESSOR OF GA1-3 (RGA), a negative regulator of GA signaling that interacts with Arabidopsis PIFs. Yeast two-hybrid assays indicated that ZmPIF4 can interact with RGA directly in vitro (Figure 7A). Further, luciferase complementation imaging (LCI) assays and bimolecular fluorescence complementation (BiFC) assays showed that ZmPIF4 can interact with RGA in planta (Figures 7B,C). In addition, the hypocotyl elongation of pifq mutant is less responsive to GA treatment, compared with wild-type control plant Col-0 under darkness conditions. Overexpression of ZmPIF4 in the pifq mutant background completely rescued the lack of response to GA treatment (Figures 7D,E), which suggests that ZmPIF4 might affect the GA signaling pathway, possibly by interacting with RGA to influence hypocotyl elongation.
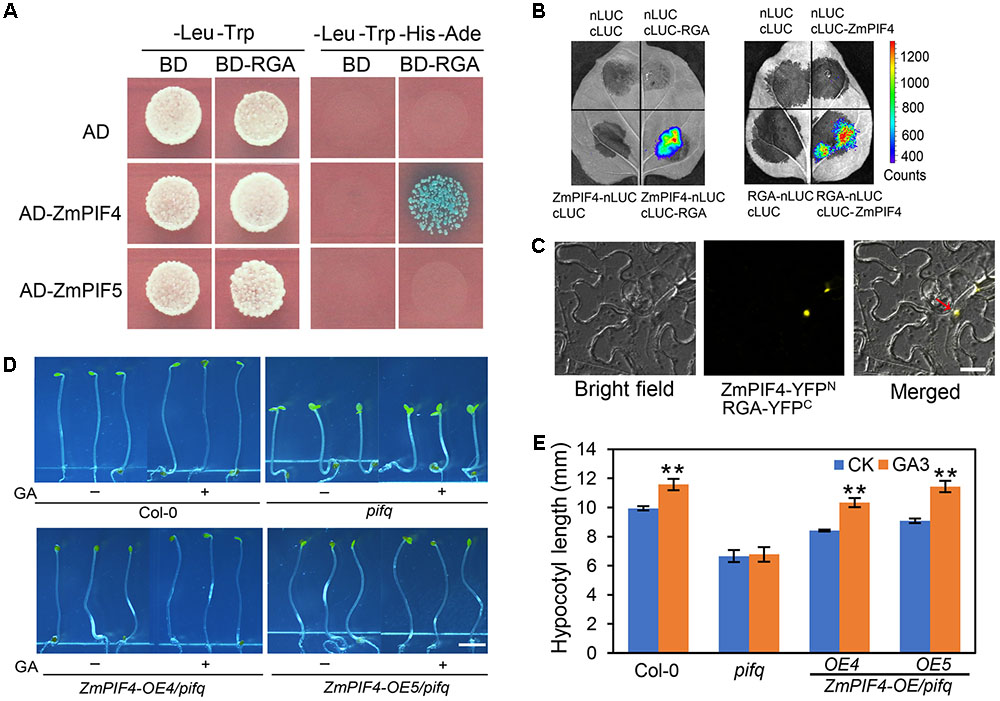
FIGURE 7. The effect of ZmPIF4 on hypocotyl elongation involves the GA signaling pathway. (A) Yeast two-hybrid analysis showed the protein–protein interaction between ZmPIF4 and Arabidopsis RGA. (B) Luciferase complementation imaging (LCI) assays showed ZmPIF4 interacts with RGA in vivo. ZmPIF4 and RGA were fused with the N- or C-terminal of LUC (indicated by nLUC or cLUC) and then infiltrated into Nicotiana benthamiana leaves. nLUC, cLUC, empty vector. (C) Bimolecular fluorescence complementation (BiFC) assay showed ZmPIF4 interacts with RGA in vivo. ZmPIF4 and RGA were fused with the N- or C-terminal of YFP (indicated by YFPN or YFPC) and then infiltrated into N. benthamiana leaves. The arrow indicates the YFP signal in the nucleus. Scale bar: 25 μm. (D) Phenotypes of 4-day-old seedlings grown under darkness with or without 10 μM GA3. Scale bar: 3 mm. (E) Quantification of hypocotyl length of the plants shown in (D). Data represent the mean and SD of at least 30 seedlings. ∗P < 0.05, ∗∗P < 0.01.
Discussion
Shade tolerance is a beneficial trait in crops such as maize, as trends in modern agriculture continue to increase planting density. Members of the PIF family of proteins play a critical role in plant responses to shading, and are highly conserved in land plants (Lee and Choi, 2017). The physiological functions and underlying molecular mechanisms of PIFs have been studied extensively in Arabidopsis, but little is known in maize and other plant species. In this study, we conducted a genome-wide analysis of maize and identified 15 putative PIF family proteins. Alignment of the protein sequences of AtPIFs and ZmPIFs showed that the seven putative ZmPIF proteins we identified have a highly conserved bHLH domain and an APB motif, which is essential for interacting with phytochromes in plants. Three of these (ZmbHLH76, ZmbHLH165, ZmbHLH198) also have an APA motif. The strong conservation of these motifs is consistent with their importance roles for PIF function in Arabidopsis and maize. These seven putative PIFs might interact with phyA or phyB and be involved in light signal transduction directly. The other eight members may function indirectly in light signal transduction by interacting with the seven members that containing APA or APB domains. Indeed, this kind of interaction has been identified in previous studies in Arabidopsis. For example, HFR1, an atypical bHLH type transcriptional regulator, directly interacts with PIF4 or PIF5 and forms non-DNA-binding bHLH heterodimers, thus mediating plant responses to shade (Hornitschek et al., 2009).
Previous studies have revealed that ZmPIF3.1 and ZmPIF3.2 can interact with ZmphyB, and affect responses to stress in rice (Gao et al., 2015; Kumar et al., 2016). Besides ZmPIF3.1 and ZmPIF3.2, the physiological roles of other ZmPIF members have remained largely unknown. Here, our results revealed that the transcript levels of ZmPIF4 and ZmPIF5 peaked at dawn and were low at dusk (Figure 2B). The ZmPIF4 and ZmPIF5 protein levels are light labile, accumulating at night and decreasing in the day, similar to the pattern of AtPIF4 and AtPIF5 in Arabidopsis (Figures 3F,G). By expressing ZmPIF4 and ZmPIF5 in pifq, the quadruple mutant of PIF1, PIF3, PIF4, and PIF5 in Arabidopsis, we showed that overexpression of these two genes partly rescued the phenotype of pifq in the dark (Figures 3A–D). Ectopic expression of ZmPIF4 also rescued the disrupted negative gravitropism of hypocotyls in pifq mutants, which might occur through inhibition of the conversion of endodermal amyloplasts to etioplasts. In addition, overexpression ZmPIF4 in Arabidopsis caused a moderate constitutive shade response, including early flowering, reduced chlorophyll, and premature leaf senescence, similar to the phenotype of plants overexpressing AtPIF4 and AtPIF5 (Figures 5, 6). Further, RT-qPCR indicated that ZmPIF4 or ZmPIF5 can regulate the transcript levels of selected downstream target genes of AtPIFs. In addition, ZmPIF4 can interact with RGA (a main member of the DELLA family) of Arabidopsis, and overexpression of ZmPIF4 partially rescued the defective response to GA in pifq, which suggested that ZmPIF4 may regulate plant growth by interacting with components of the GA pathway (Figure 7). All this genetic evidence revealed that ZmPIF4 and ZmPIF5 act similarly to AtPIF4 and AtPIF5, and participate in photomorphogenesis and the shade avoidance response in Arabidopsis.
Although the protein–protein interaction between ZmPhyB1 (or ZmPhyB2) and ZmPIF4 (or ZmPIF5) still needs to be tested, genetic evidence revealed that these two-putative maize PIF proteins, ZmPIF4 and ZmPIF5 play important roles in shade avoidance response. We also noticed that ZmPIF4 had a stronger effect on the Arabidopsis response to shade and gravity, compared with ZmPIF5. This may be due to the lower protein level of ZmPIF5 in the Arabidopsis transgenic lines, or other mechanisms that influence the protein or transcript level of ZmPIF5 in Arabidopsis. To further exam the physiological roles of ZmPIF4 and ZmPIF5, genetic materials including mutant plants or stable transgenic lines of ZmPIF4 and ZmPIF5 in maize are require. Our study revealed that ZmPIF4 and ZmPIF5 can affect photomorphogenesis and the shade avoidance response in Arabidopsis, similar to the physiological functions to AtPIF4 and AtPIF5, which indicate that PIF4 and PIF5 might play evolutionarily conserved roles in maize and Arabidopsis.
Author Contributions
QS, HZ, XS, YJ, RL, and GL designed the research. QS and XS performed the most of experiments and analyzed data. QS and GL wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the State Key Basic Research and Development Program of China (2014CB147301 and 2016YFD0101003), Funds of Shandong “Double Tops” Program (to GL), and State Key Laboratory of Crop Biology (DXKT201706).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02273/full#supplementary-material
FIGURE S1 | Phylogenetic tree of PIF4 and PIF5 in land plants. Phylogenetic tree analysis of PIF4 and PIF5 in land plants was conducted with SMART and output was generated with MEGA.
FIGURE S2 | Gene expression and protein subcellular localization analysis of ZmPIF4 and ZmPIF5. (A) RT-qPCR analysis indicated that ZmPIF4 and ZmPIF5 are highly induced by far-red light and repressed by red light. Three-leaf stage seedlings of maize inbred line B73 grown under white light were transferred to far-red and then transferred to red light for various times. The seedlings were harvested at different times and used to perform RT-qPCR analysis. Actin was used as internal control for RT-qPCR. Data are means and SD of three independent biological replicates. (B) Plasmids containing ZmPIF4-GFP and ZmPIF5-GFP were infiltrated into N. benthamiana and microscopy revealed that the fusion proteins localized in the nucleus. H2B- mCherry was used as internal control to indicate the position of the nucleus. Scale bar: 25 μm.
FIGURE S3 | Phenotype of the ZmPIF4 and ZmPIF5 transgenic Arabidopsis seedlings under high R/FR conditions. (A) Western blot analyses revealed that ZmPIF4 and ZmPIF5 were highly expressed in the transgenic plants of ZmPIF4 (left) and ZmPIF5 (right), respectively. Four-day-old seedlings were used for western blot analysis. The bands of RbcL stained with Ponceau S were used as the internal control. (B) The phenotype (upper) and quantification of hypocotyl length (lower) of the transgenic lines of ZmPIF4-OE and ZmPIF5-OE under long-day with low R:FR conditions (white light, R:FR = 5). Scale bar: 1.5 mm. Data represent the mean and SD. n = 30; ∗P < 0.05; ∗∗P < 0.01.
FIGURE S4 | Overexpression of ZmPIF4 enhanced the shade avoidance response in Arabidopsis. (A,B) Overexpression of ZmPIF4 (OE8, OE10, OE11) led to plant lodging and leaf senescence, compared with wild-type Col-0. Scale bar: 3 cm. Overexpression lines of ZmPIF4 (OE11) led to the inflorescence soft and lodging (C), and altered the length and thickness of the mature silique (D), compared with wild-type Col-0. Scale bar: 1 cm in (C) and 5 mm in (D). 45-day-old mature plants grown under LD conditions are shown in (A–D).
FIGURE S5 | Scanned original images of immunoblots and gels.
TABLE S1 | Annotation information for the putative PIF family proteins in maize.
TABLE S2 | Primers used in this study.
References
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521. doi: 10.1016/j.tplants.2007.10.001
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Choi, G., Yi, H., Lee, J., and Kwon, Y.-K. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. doi: 10.1038/44176
Dill, A., Jung, H. S., and Sun, T. P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. U.S.A. 98, 14162–14167. doi: 10.1073/pnas.251534098
Franklin, K. A., Praekelt, U., Stoddart, W. M., Billingham, O. E., Halliday, K. J., and Whitelam, G. C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131, 1340–1346. doi: 10.1104/pp.102.015487
Gao, Y., Jiang, W., Dai, Y., Xiao, N., Zhang, C., Li, H., et al. (2015). A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Mol. Biol. 87, 413–428. doi: 10.1007/s11103-015-0288-z
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., López-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Hornitschek, P., Lorrain, S., Zoete, V., Michielin, O., and Fankhauser, C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. doi: 10.1038/emboj.2009.306
Howe, E. S., Clemente, T. E., and Bass, H. W. (2012). Maize histone H2B-mCherry: a new fluorescent chromatin marker for somatic and meiotic chromosome research. DNA Cell Biol. 31, 925–938. doi: 10.1089/dna.2011.1514
Josse, E. M., Foreman, J., and Halliday, K. J. (2008). Paths through the phytochrome network. Plant Cell Environ. 31, 667–678. doi: 10.1111/j.1365-3040.2008.01794.x
Kami, C., Lorrain, S., Hornitschek, P., and Fankhauser, C. (2010). Chapter two-light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66. doi: 10.1016/S0070-2153(10)91002-8
Kim, K., Shin, J., Lee, S. H., Kweon, H. S., Maloof, J. N., and Choi, G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 108, 1729–1734. doi: 10.1073/pnas.1011066108
Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., et al. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. doi: 10.2307/3870974
Kumar, I., Swaminathan, K., Hudson, K., and Hudson, M. E. (2016). Evolutionary divergence of phytochrome protein function in Zea mays PIF3 signaling. J. Exp. Bot. 67, 4231–4240. doi: 10.1093/jxb/erw217
Lau, O. S., and Deng, X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577. doi: 10.1016/j.pbi.2010.07.001
Lee, N., and Choi, G. (2017). Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 35, 54–60. doi: 10.1016/j.pbi.2016.11.004
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. doi: 10.1016/j.cub.2008.10.058
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, J., Li, G., Wang, H., and Wang Deng, X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9:e0148. doi: 10.1199/tab.0148
Li, K., Yu, R., Fan, L. M., Wei, N., Chen, H., and Deng, X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7:11868. doi: 10.1038/ncomms11868
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Ma, L., Tian, T., Lin, R., Deng, X. W., Wang, H., and Li, G. (2016). Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 9, 541–557. doi: 10.1016/j.molp.2015.12.013
Ma, L., Xue, N., Fu, X., Zhang, H., and Li, G. (2017). Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 213, 1682–1696. doi: 10.1111/nph.14300
Martin, K., Kopperud, K., Chakrabarty, R., Banerjee, R., Brooks, R., and Goodin, M. M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162. doi: 10.1111/j.1365-313X.2009.03850.x
Mazzella, M. A., Casal, J. J., Muschietti, J. P., and Fox, A. R. (2014). Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 5:52. doi: 10.3389/fpls.2014.00052
Paik, I., Kathare, P. K., Kim, J. I., and Huq, E. (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035–1046. doi: 10.1016/j.molp.2017.07.002
Pfeiffer, A., Shi, H., Tepperman, J. M., Zhang, Y., and Quail, P. H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7, 1598–1618. doi: 10.1093/mp/ssu087
Quail, P. H. (1991). Phytochrome: a light-activated molecular switch that regulates plant gene expression. Annu. Rev. Genet. 25, 389–409. doi: 10.1146/annurev.ge.25.120191.002133
Robson, P., Whitelam, G. C., and Smith, H. (1993). Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in phytochrome B. Plant Physiol. 102, 1179–1184. doi: 10.1104/pp.102.4.1179
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Sheehan, M. J., Kennedy, L. M., Costich, D. E., and Brutnell, T. P. (2007). Subfunctionalization of PhyB1 and PhyB2 in the control of seedling and mature plant traits in maize. Plant J. 49, 338–353. doi: 10.1111/j.1365-313X.2006.02962.x
Shen, H., Zhu, L., Castillon, A., Majee, M., Downie, B., and Huq, E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586–1602. doi: 10.1105/tpc.108.060020
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Tyler, L., Thomas, S. G., Hu, J., Dill, A., Alonso, J. M., Ecker, J. R., et al. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019. doi: 10.1104/pp.104.039578
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, Y., Mayba, O., Pfeiffer, A., Shi, H., Tepperman, J. M., Speed, T. P., et al. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLOS Genet. 9:e1003244. doi: 10.1371/journal.pgen.1003244
Keywords: maize, photomorphogenesis, phytochrome-interacting factors (PIFs), shade avoidance response, ZmPIF4, ZmPIF5
Citation: Shi Q, Zhang H, Song X, Jiang Y, Liang R and Li G (2018) Functional Characterization of the Maize Phytochrome-Interacting Factors PIF4 and PIF5. Front. Plant Sci. 8:2273. doi: 10.3389/fpls.2017.02273
Received: 07 October 2017; Accepted: 29 December 2017;
Published: 18 January 2018.
Edited by:
Daoxin Xie, Tsinghua University, ChinaReviewed by:
Shangwei Zhong, Peking University, ChinaHsu-Liang Hsieh, National Taiwan University, Taiwan
Enamul Huq, University of Texas at Austin, United States
Copyright © 2018 Shi, Zhang, Song, Jiang, Liang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Li, Z2FuZ2xpQHNkYXUuZWR1LmNu
†These authors have contributed equally to this work.
 Qingbiao Shi
Qingbiao Shi Haisen Zhang†
Haisen Zhang† Gang Li
Gang Li