Abstract
The micronutrient iron (Fe) is essential for photosynthesis, respiration, and many other processes, but it is only sparingly soluble in aqueous solution, making adequate acquisition by plants a serious challenge. Fe is a limiting factor for plant growth on approximately 30% of the world’s arable lands. Moreover, Fe deficiency in humans is a global health issue, affecting 1.62 billion people, or about 25% of the world’s population. It is imperative that we gain a better understanding of the mechanisms that plants use to regulate iron homeostasis, since these will be important targets for future biofortification and crop improvement strategies. Grasses and non-grasses have evolved independent mechanisms for primary iron uptake from the soil. The grasses, which include most of the world’s staple grains, have evolved a distinct ‘chelation’ mechanism to acquire iron from the soil. Strong iron chelators called phytosiderophores (PSs) are synthesized by grasses and secreted into the rhizosphere where they bind and solubilize Fe(III). The Fe(III)-PS complex is then taken up into root cells via transporters specific for the Fe(III)-PS complex. In this study, 31 novel, uncharacterized striped maize mutants available through the Maize Genetics Cooperation Stock Center (MGCSC) were analyzed to determine whether their mutant phenotypes are caused by decreased iron. Many of these proved to be either pale yellow or white striped mutants. Complementation tests were performed by crossing the MGCSC mutants to ys1 and ys3 reference mutants. This allowed assignment of 10 ys1 alleles and 4 ys3 alleles among the novel mutants. In addition, four ys∗ mutant lines were identified that are not allelic to either ys1 or ys3. Three of these were characterized as being non-allelic to each other and as having low iron in leaves. These represent new genes involved in iron acquisition by maize, and future cloning of these genes may reveal novel aspects of the grass iron acquisition mechanism.
Introduction
The global demand for crops with high concentrations of nutrients in edible tissues is increasing due to current trends in population growth, global climate change, and decreasing arable land resources (Eckardt et al., 2009). Iron (Fe) deficiency in humans is a global health issue, affecting 1.62 billion people, or about 25% of the world’s population, and it is imperative that we gain a better understanding of the mechanisms that plants use to regulate iron homeostasis, since these will be important targets for future biofortification strategies (McLean et al., 2009; Murgia et al., 2012). Quantitative trait loci (QTL) have been identified for maize grain iron accumulation (Zhang et al., 2017) and identification of additional components of the maize iron homeostatic apparatus may help to elucidate the genes underlying such QTL. Although we have learned a great deal through the study of model organisms such as Arabidopsis, it is important to note that the grasses, which include most of the world’s staple grains, use phytosiderophores (PSs) that are secreted into the rhizosphere where they bind and solubilize Fe(III) (Tagaki, 1976; Tagaki et al., 1984, 1988). PSs are not made or used by non-grass species.
Biofortification of crops has been restricted by our limited knowledge of the molecular mechanisms controlling iron uptake, translocation, accumulation, and deposition in the grain. Attempts to increase iron content have been promising, but these efforts have been focused on the relatively small set of known genes that are involved in iron homeostasis. The iron-storage protein, ferritin (Briat and Lobreaux, 1998; Briat et al., 1999), has been expressed in rice endosperm, to increase iron and zinc content (Goto et al., 1999; Drakakaki et al., 2005). The iron uptake machinery has been a target for biofortification by engineering key enzymes involved in PS synthesis (Higuchi et al., 1999; Takahashi et al., 1999, 2001; Suzuki et al., 2008). These efforts have been only partially successful, suggesting that identifying additional genes involved in mobilization and translocation within the plant could be helpful to develop additional strategies for the production of biofortified crops.
In plants, iron is essential for photosynthesis, respiration, and many other processes, but is only sparingly soluble in aqueous solution, making adequate acquisition by plants a serious challenge (Marschner, 1995). Furthermore, iron is highly reactive and if over-accumulated can cause cellular damage. As a response to these key properties of iron, plants have evolved highly regulated iron mechanisms to ensure efficient and tightly controlled acquisition from the soil. Most plants use a combination of rhizosphere acidification, iron reduction, and uptake via the ZIP (ZRT, IRT-like protein) family transporter, IRT1 (iron-regulated transporter). In this strategy, iron is first solubilized and then taken up from the soil, as reviewed in Walker and Connolly (2008), Jeong and Guerinot (2009), and Morrissey and Guerinot (2009). In contrast, the grasses, which include most of the world’s staple grains, have evolved a distinct ‘chelation’ mechanism to acquire iron from the soil. PSs are synthesized by grasses and secreted into the rhizosphere where they bind and solubilize Fe(III) (Tagaki, 1976; Tagaki et al., 1984, 1988). The Fe(III)-PS complex is then taken up into root cells via transporters specific for the Fe(III)-PS complex (Romheld and Marschner, 1986; von Wiren et al., 1994). This mechanism is also known as ‘Strategy II.’ The Fe(III)-PS uptake transporter Yellow Stripe1 (YS1) has been studied extensively (Curie et al., 2001; Yen et al., 2001; Roberts et al., 2004; Schaaf et al., 2004; Murata et al., 2006; Harada et al., 2007; Inoue et al., 2009; Lee et al., 2009), and is a proton-coupled symporter of Fe(III)-PS complexes (Schaaf et al., 2004).
Phytosiderophores are chemically quite distinct from bacterial and fungal siderophores (Miethke and Marahiel, 2007) and belong to a class of compounds called mugineic acids (Ma and Nomoto, 1996), with a well-worked out biosynthesis (Mori and Nishizawa, 1987; Kawai et al., 1988; Shojima et al., 1990; Ma et al., 1995; Takahashi et al., 1999; Kobayashi et al., 2001). In contrast to the details established for PS biosynthesis and Fe-PS uptake, the molecular details of PS secretion have not been as well-characterized. In several grass species, PSs are secreted according to a diurnal cycle, with release occurring several hours after sunrise (Zhang et al., 1991; Walter et al., 1995; Ma et al., 2003; Reichman and Parker, 2007; Ueno et al., 2007; Nagasaka et al., 2009; Bernards et al., 2014). Large numbers of vesicles have been observed in barley roots just prior to the daily release of PS, suggesting that PSs are secreted by exocytosis (Nishizawa and Mori, 1987; Sakaguchi et al., 1999; Negishi et al., 2002). Furthermore, microarray analysis of barley roots indicated that expression of genes associated with polar vesicle transport increases in the early morning (Negishi et al., 2002). The anion channel blockers anthracene-9-carboxylic acid and phenylglyoxal were shown to inhibit PS secretion by barley roots (Sakaguchi et al., 1999), potentially indicating that anion channels are involved in loading PS to secretory vesicles. Alternatively, anion channels in the plasma membrane (PM) could be responsible for PS transport across the PM. Major facilitator superfamily transporters with PS efflux activity were recently identified in rice and barley, and have been called transporter of mugineic acid (TOM1) (Nozoye et al., 2011).
A classically known mutation in maize called yellow stripe3 (ys3; Wright, 1961) renders plants unable to secrete PSs, even though PSs are synthesized in normal amounts (Basso et al., 1994; Lanfranchi et al., 2002). The Ys3 gene in maize is located between 85,618,053 and 114,789,459 on chromosome 3 based on two genetic markers (IDP3861 and IDP4688) on the IBM2 2008 Neighbors map. A partial gene with similarity to TOM1 (GRMZM2G063306 also called ZEAMMB73_058478) is located within this interval in the maize reference sequence version 3 but the sequence contained two sequence gaps in the region occupied by GRMZM2G063306. Based on sequence similarity and strong expression during iron deficiency, this gene was suggested as a candidate for the locus affected in ys3 mutants (Nozoye et al., 2013; Li et al., 2014), but genetic evidence for this assignment has not been presented.
In spite of this progress in understanding the process of PS synthesis, release, and uptake of Fe-PS complexes, there are many gaps in our understanding of what makes a particular grass species or cultivar ‘iron efficient.’ In Kentucky bluegrass, for example, the amount of PS release does not correlate well with resistance to iron deficiency (Buxton et al., 2012). Because of this complexity, we sought to understand the genes in Zea mays (maize) that contribute to iron efficiency, by examining the set of maize mutants available through the Maize Genetics Cooperation Stock Center (MGCSC) that have been described as ‘yellow striped’ or ‘green striped.’ Both these descriptions may refer to iron deficiency chlorosis that is typical in both ys1 and ys3 maize mutants, and is characterized by yellow interveinal regions and green veins. By performing allelism tests with ys1 and ys3 reference mutant plants, we have identified novel yellow striped mutants (that we designate as ys∗, pending gene identification and assignment of new nomenclature) that may shed light on additional components contributing to iron efficiency in maize. We further characterized the sequence of GRMZM2G063306 in the WT B73 genome and the ys3 reference mutants. We have identified four new alleles of ys3. Based on the evidence from our sequencing of GRMZM2G063306 in multiple independent ys3 mutants, we present strong genetic evidence that GRMZM2G063306 (ZmTOM1) corresponds to the Ys3 gene of maize.
Materials and Methods
Plant Material and Growth Conditions
Maize (Zea mays) plants of B73 and W22 inbred lines were used as WT reference in our experiments, as indicated in the text. Uncharacterized yellow striped mutants were obtained from the MGCSC1.
For all experiments involving genetic crosses, stocks were grown at the University of Massachusetts Crop and Animal Research and Education Center, South Deerfield, MA, United States, during the summer season between May and September. Mutant plants were supplemented with foliar iron (Fe-EDDHA) through growing season to alleviate chlorosis. For the purposes of initial phenotyping, plants were grown in the greenhouse in a 4:1 v/v mix of potting soil and Turface. All phenotyping was also repeated under field conditions. Supplemental light was supplied with high-pressure sodium lamps to give a 20 h light period each day. For quantitative polymerase-chain-reaction (PCR) analysis, plants were grown in a sand:Turface mix (9:1 v/v) irrigated with water until germination and then irrigated with modified Hoagland’s nutrient solutions, with 1 mM KH2PO4, 3.75 mM KOAc, 5 mM Ca(NO3)2, 1.25 mM KNO3, 2 mM MgSO4, 3.75 mM NH4OAc, 46 uM H3BO3, 9.1 uM MnCl2, 0.77 uM ZnSO4, 0.32 uM CuSO4, and 0.83 uM H2MoO4 (Yordem et al., 2011) containing 100 μM FeSO4-EDTA every 48 h. Plants were grown for 10 days after germination before the root tissue was collected.
PCR and Sequencing of ZmTOM1 in ys3 Mutant Lines
Genomic DNA was extracted from leaves of ys3 mutant plants (ys3:04HI-A632GN-144, ys3:67-2403, ys3:04HI-Oh43xA632GN-187, and ys3:07IL-B73GN-279) and the exons of ZmTOM1 (GRMZM2G063306/Zm00001d041111) were amplified using primers listed in Supplementary Table 1. Amplifications were performed using ExTaq polymerase (Takara, Madison, WI, United States), with cycling conditions of 95°C, 60 s followed by 35 cycles of 95°C, 15 s, 55°C, 30 s, and 72°C, 60 s, with a final extension step at 72°C for 5 min. PCR products were gel purified before sequencing.
Real-Time PCR Analysis (qRT-PCR)
The root tissue was flash frozen in liquid nitrogen after harvesting. The frozen root tissue was ground using a Tissuelyser (QIAGEN, Valencia, CA, United States) in 2 ml tubes containing 3.2 mm chrome steel beads (BioSpect Products, Bartlesville, OK, United States). Total RNA was extracted using QIAGENR®Neasy Plant Mini Kit (QIAGEN, Valencia, CA, United States), and on-column DNAse treatment step was included for all samples. cDNA was synthesized from 750 ng of total RNA using SuperScript IV VILO (Life Technologies, Carlsbad, CA, United States). For real-time PCR (RT-PCR) analysis, Quantprime primer design webtool (Arvidsson et al., 2008) was used to design ZmTOM1 primers. The primer efficiency of each set of primers (Supplementary Table 2) was evaluated empirically by serial dilution curve of cDNA. PowerUPTM SYBRTMGreen Master Mix (Life Technologies, Carlsbad, CA, United States) was used in quantitative RT-PCR experiments. A two-step PCR protocol was used with the following conditions: initial cycle of 50°C, 120 s, and 95°C, 120 s, and 40 cycles of 95°C, 15 s, and 60°C, 60 s. After two-step cycling was completed, melting curve was performed to ensure that single amplicon was obtained from each reaction. To determine transcript levels, the threshold cycle (Ct) values from target gene was normalized to ZmGAPDH reference gene for each sample and by the ΔΔCt method, we calculated fold change compared to B73 WT. Data represent three biological replicates.
Inverse PCR
Genomic DNA (∼1 μg) was digested with AciI and NlaIII (New England Biolabs) for 2.5 h at 37°C and reaction was stopped by incubating for 20 min at 65°C. The DNA was then diluted 25-fold, and ligation was performed using 20 units of Epicenter®T4 DNA ligase (Illumina, Inc., Madison, WI, United States) overnight at either 20°C for blunt ends or 4°C for sticky ends. The resulting ligation was purified using phenol/chloroform (1:1, v/v) and ethanol precipitation in the presence of 40 μg of glycogen. Then, 1/6 of the purified ligation was used as template for the 1st round of PCR, with primers oZmTOM1_4456 and oZmTOM1_4504 for AciI restriction digest, or primers oZmTOM1_3641 and oZmTOM1_5012 for NlaIII restriction digestions. The 2nd round of PCR was performed using 1 ul of a 1:100 dilution of the PCR product from the 1st round as template using nested primers oZmTOM1_4338 and oZmTOM1_4573for AciI digested DNA or oZmTOM1 4774 and oZmTom1_5154, for NlaIII digested DNA. Amplifications were performed using ExTaq polymerase (Takara, Madison, WI, United States), with cycling conditions of 95°C, 2 min, and 25 cycles of 95°C, 15 s, 57°C, 30 s, 72°C for 2 min, and a final elongation step for 10 min. Primer sequences for inverse PCR (iPCR) are listed in Supplementary Table 3.
Metal Measurement
Leaves of at least 10 individual plants were collected from 19-day-old plants grown in the greenhouse and samples were dried at 65°C for 72 h. In every experiment, all controls and mutants were grown simultaneously and using the same soil batch. Metal concentrations were determined by inductively couple plasma mass spectrometry (ICP-MS) at the Donald Danforth Plant Research Institute.
Results
Complementation Testing of Yellow Striped Mutants from MGCSC
We obtained 31 mutants classified as having a yellow striped phenotype from the MGCSC (Table 1). These were planted in the field and phenotypic analysis indicated that 21 of the lines showed the phenotype typical of iron deficiency chlorosis. In the other 10 lines, we either did not observed stripes at all or else observed a solid yellow or white striped phenotype (Table 1). To identify new genes involved in iron uptake in maize, and to identify new alleles for ys3, we performed complementation tests between the uncharacterized yellow striped mutants and the reference maize mutants ys1:ref and ys3:ref. Due to stunting or sterility of some mutant stocks, not all crosses were obtained. From these crosses, we identified 10 new alleles for ys1 and 4 new alleles for ys3. Moreover, we found four novel yellow stripe mutants, ys∗-PI262172, ys∗:N2398, ys∗:PI228180, and ys∗:04HI-A632XOh43GN-18, that are not allelic to ys1 or ys3, and thus may represent new maize genes involved in iron uptake or homeostasis.
Table 1
| MGCSC | Mutation name | Phenotype | Allelic to | Allelic to |
|---|---|---|---|---|
| number | ys3:ref | ys1:ref | ||
| 503A | ys1:ref | Chlorotic, often sterile | – | – |
| 503B | ||||
| 3812I | ys1-N2261 | Small, chlorotic | No | Yes |
| 6003B | ys1-8912 | Small, chlorotic | No | Yes |
| 6003G | ys1-PI267219 | Small plant, chlorotic | No | Yes |
| 6003I | ys1-5-8(5575) | Small plant, chlorotic | No | Yes |
| 3812C | ys1-N139B | Small, severely chlorotic | No | Yes |
| 6003L | ys1-03HI-B73GN-182 | Chlorotic | No | Yes |
| 6505A | ys1-03HI-B73xMo17GN-210 | Small plant, chlorotic | NT | Yes |
| 6505C | ys1-04HI-A632xOh43GN-137 | Yellow striped plants also had crinkled leaves | No | Yes |
| 3812B | ys1-N71B | Chlorotic, asynchronous flowering | No | Yes |
| 304A | ys3:ref | Chlorotic, sometimes sterile | – | – |
| 311F | ||||
| 311G | ||||
| 6505E | ys3-07IL-B73GN-279 | Chlorotic | Yes | No |
| 6003C | ys3-67-2403 | Small, chlorotic | Yes | No |
| 6412B | ys3-04HI-A632GN-144 | Small plant, chlorotic | Yes | No |
| 6505D | ys3-04HI-Oh43xA632GN-187 | Chlorotic | Yes | No |
| 3812O | ys∗-N2398 | Chlorotic, chlorosis easily reversed by spraying leaves with Fe | No | No |
| 6003D | ys∗-PI228180 | Chlorotic, chlorosis easily reversed by spraying leaves with Fe. Yellow stripes appear again at time of flowering | No | No |
| 6505B | ys∗-04HI-A632xOh43GN-18 | Small plant, chlorotic | No | No |
| 3812D | ys-N326A | Pale yellow | NT | NT |
| 3812G | ys-N634B | Pale yellow | NT | NT |
| 3812H | ys-N2000 | Pale yellow | NT | NT |
| 3812J | ys-N2268 | Oil yellow – not striped | No | No |
| 3812K | ys-N2300 | White striped | NT | NT |
| 3812M | ys-N2361 | No yellow or striped phenotype observed | NT | NT |
| 3812N | ys-N2379 | No yellow or striped phenotype observed | NT | NT |
| 6003K | ys-03HI-B73xMo17GN-1123 | Poor germination; stripes not observed | NT | NT |
| 6005L | yel-str-W23 | Pale yellow | NT | NT |
| 6003F | ys-PI262475 | Small plant, yellow, not striped | No | No |
| 6003A | ys1-1479 | Chlorotic | No | Yes |
| 6003E | ys∗-PI262172 | Stunted, chlorotic | No | No |
| 6003J | ys?-68-1354 | Small plant, chlorotic | No | NT |
| 6412C | ys?-07IL-B73GN-171 | Yellow striped plants died | NT | No |
| 3812L | ys?-N2303 | Small, chlorotic, poor fertility | No | NT |
Yellow or green striped mutants from the Maize Genetics Cooperation Stock Center (MGCSC) and results of complementation tests with ys3:ref and ys1:ref.
NT indicates that allelism was not tested.
Analysis of ZmTOM1 Coding Sequence in ys3 Alleles
Because a gene with similarity to TOM1 (GRMZM2G063306 also called ZEAMMB73_058478, here designated ZmTOM1) is located within the genomic interval containing Ys3, this gene has been suggested as a candidate for the locus affected in ys3 mutants (Nozoye et al., 2013; Li et al., 2014). To determine whether the suggested candidate gene, ZmTOM1, underlies the long-known ys3 mutant, we sequenced the exons of ZmTOM1 in all five alleles of ys3 (reference allele and the four novel alleles identified through complementation tests; Figure 1) to identify causative mutations. The MGCSC holds three stocks designated as ys3:ref mutants (304A, 311F, and 311G). In all three lines, ZmTOM1 sequences were identical, and contained a 4 bp insertion in exon 9. This insertion causes a frame shift followed by 11 novel amino acids before introducing a premature stop codon (Figure 1). We found a different 4 bp insertion in exon 11 of ZmTOM1 in the ys3:67-2403 allele (Figure 1). This 4 bp insertion causes a frame shift followed by 107 new amino acids before a stop codon occurs to terminate the protein prematurely. For the ys3:04HI-Oh43XA632-GN-187 allele, we could not amplify fragments containing exon 10 and 11, but partial sequences from both exons could be obtained. We hypothesized that an insertion could be present between these two exons causing failure to amplify that region. Using iPCR, we identified both left and right borders of an insertion containing the characteristic elements of a transposon. The inserted sequences were flanked by 8 bp direct repeats and contained 130 bp terminal inverted repeats (TIRs). We aligned the TIR sequences with the maize reference sequence and identified two regions in chromosome 7 annotated as Far1-related sequence 5, which corresponds to a mutator-like transposable element (MULE). MULE transposons generate 8–10 bp target sequence duplications and have TIRs of >100 bp. Thus, the insertion has all the elements expected for a MULE transposon inserted in the ys3:04HI-Oh43XA632-GN-187 allele (Figure 1). A one nucleotide change at the exon–intron border for exon 5 of ys3:04HI-A632GN-144 was observed. Likewise, a one nucleotide change near the 3′ end of exon 9 was observed in ys3:07IL-B73GN-279. We hypothesized that splicing could be affected these two alleles, and so investigated ZmTOM1 gene expression in the roots of these plants using Q-RT-PCR. Expression of ZmTOM1 was observed in both ys3:04HI-A632GN-144 and ys3:07IL-B73GN-279 (data not shown). Since the ZmTOM1 transcript was observed, we speculated that altered splicing due to the mutations might be leading to aberrant ZmTOM1 mRNA, so we sequenced the full-length cDNA from each mutant line to test this. We confirmed that the one nucleotide change at the ys3:04HI-A632GN-144 exon–intron junction altered the splice donor site. The mutation causes splicing to occur at a new donor site 3 nucleotides into the adjacent intron (Figure 2). As a result, one additional amino acid is inserted without affecting the reading frame. The amino acid is inserted in a strongly conserved region that could lead to a non-functional protein. In the ys3:07IL-B73GN-279 allele, the single nucleotide change occurred at the first nucleotide of intron 9, changing the splice donor site from GT to AT. In the mRNA produced by this allele, a new splice donor site is recognized in exon 9, 21 nucleotides upstream from the original donor site (Figure 2). The resulting amino acid sequence is thus missing seven residues in a strongly conserved region. Our results show clear genetic evidence that the Ys3 gene is ZmTOM1.
FIGURE 1

ZmTOM1 sequence analysis. (A) Overview of the ys3 locus and gene model of ZmTOM1 showing ys3 mutant alleles. Exons are indicated by black boxes and the positions of mutations are indicated by black triangles. (B) Genomic sequences and predicted translation of ys3:ref and ys3:67-2403 alleles. WT sequence is shown for comparison. For ys3:04HI-Oh43XA632-GN-187, genomic sequence of the site of insertion is shown, with the 8 bp direct repeat flanking the insertion underlined.
FIGURE 2

ys3 mutations causing altered splicing. Boxes indicate exon sequences. Nucleotide changes relative to B73 reference sequence are indicated in red. Red underlined sequences are the splice donor and acceptor sites used in WT and the two mutant ys3 alleles. Genomic DNA and cDNA sequences with their predicted translation are shown. (A)ys3:04HI-A632GN-144 and (B)ys3:07IL-B73GN-279 alleles. The inset rectangles show alignments of the corresponding amino acid sequences of transporter of mugineic acid (TOM1) from barley, rice, and maize showing conserved regions. ∗ indicates an amino acid identical in all three species and indicates an amino acid that is conserved in all three species.
Analysis of Novel Yellow Striped Maize Mutants
To evaluate whether the yellow striped phenotype in ys∗ mutants is due to low iron, we analyzed metal levels in leaves of three of the mutants. The ys∗-PI262172 mutant was not included in this analysis, because its stunted growth prevented our obtaining sufficient material for this experiment. Visual inspection of the leaves of 12-day-old WT and mutant plants indicates differences in the severity of the observed chlorosis, with ys∗:PI228180 having very mild chlorosis and ys∗:04HI-A632xOh43GN-18, ys1:ref, and ys3:ref having the most marked chlorosis (Figure 3). In all three ys∗ mutants tested, the levels of iron were significantly lower than WT (Figure 3) indicating that the plants are iron-deficient. Control ys1 and ys3 plants are also low in iron, as expected. In a segregating population of ys∗:04-04HI-A632xOh43GN-18 mutants, the iron concentration in yellow striped siblings was less than half (42%) that of WT siblings. The iron concentration in ys∗:04HI-A632xOh43GN-18 was significantly lower even than ys1 and ys3, indicating a very substantial alteration in iron homeostasis in these plants. For ys∗:PI228180 and ys∗:N2398, iron levels were higher than either ys1 or ys3, but were still significantly lower than the amount in WT control plants.
FIGURE 3
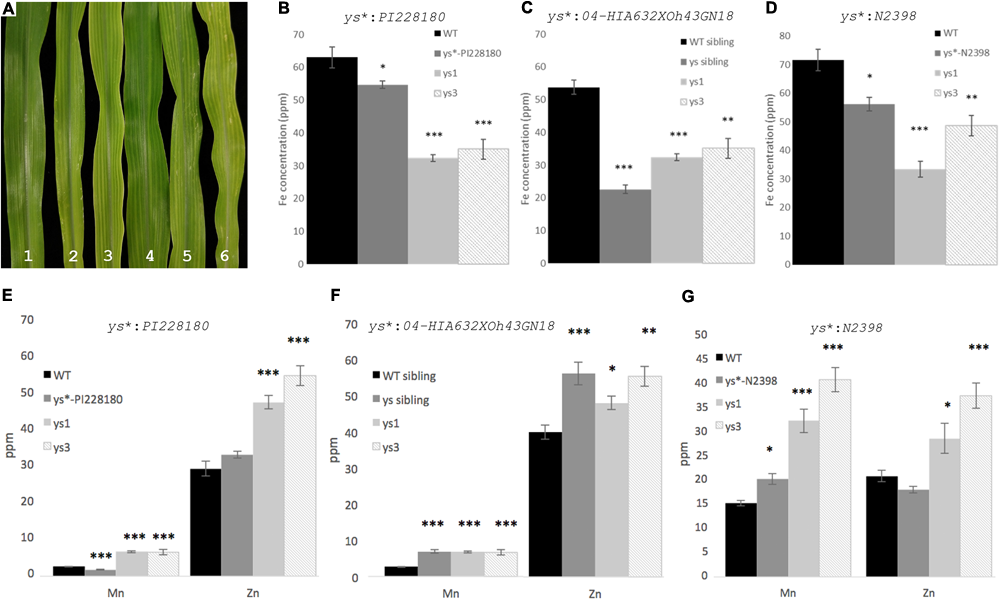
ys∗ mutant phenotypes and iron concentration. (A) Leaves of 12-day-old greenhouse grown plants. (1) WT; (2) ys∗:N2398; (3) ys∗:04HI-A632xOh43GN-18; (4) ys∗:PI228180; (5) ys1:ref; and (6) ys3:ref. Inductively couple plasma mass spectrometry (ICP-MS) measurement of the iron content of (B)ys∗:PI228180, (C)ys∗:04HI-A632xOh43GN-18, and (D)ys∗:N2398. ICP-MS measurements of the Mn and Zn content of (E)ys∗:PI228180, (F)ys∗:04HI-A632xOh43GN-18, and (G)ys∗:N2398. Error bars represent the standard error of the mean (n = 10). Significance was addressed using the Mann–Whitney test. Asterisk indicates significant difference from WT (∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001).
We also measured the Zn and Mn concentration in the leaves of the mutant plants (Figure 3). We note that altered iron homeostasis often causes alterations to multiple metals. For example, ys1 and ys3 mutants, which are clearly impaired in iron uptake, have higher Zn and Mn than WT control plants (Figure 3). It is possible that this occurs because PS secretion or uptake directly affects Mn and Zn uptake or translocation, but it is also possible that the mechanism is indirect. Like ys1 and ys3 mutants, ys∗:04HI-A632xOh43GN-18 and ys∗:N2398 plants have higher Mn and Zn than WT control plants. For ys∗:PI228180, the Zn concentration in leaves is not significantly different from WT control plants, and the Mn concentration is slightly but significantly lower than that of the WT controls, and much lower that the Mn concentration in the ys1 and ys3 mutants. We note that the soil batch used for growth of the ys∗:04HI-A632xOh43GN-18 and ys∗:N2398 plants and their controls was different from the batch used to grow ys∗:PI228180 and its controls.
Complementation Tests among ys∗ Mutants
We performed crosses among three of the four identified ys∗ mutants to determine how many loci are represented by these three mutants. The ys∗-PI262172 mutant was not included in this analysis, because its stunted growth prevented our obtaining the appropriate crosses. F1 seeds were grown in the greenhouse and the phenotypes were recorded. We found complementation among all crosses performed in ys∗ mutants, indicating that they do not represent alleles. These results show that we have identified three novel genes involved in iron homeostasis (Table 2).
Table 2
| MGCSC number | Mutation name | Allelic to ys∗:N2398 | Allelic to ys∗:04HI-A632xOh43GN-18 | Allelic to ys∗:PI228180 |
|---|---|---|---|---|
| 3812O | ys∗:N2398 | – | No | No |
| 6505B | ys∗:04HI-A632xOh43GN-18 | No | – | No |
| 6003D | ys∗:PI228180 | No | No | – |
Complementation test results among ys∗ mutants.
Discussion
The Rationale for Gene Discovery in Zea mays
Much of the molecular work on iron uptake and homeostasis in grasses has been performed using rice, both because of its properties as a model organism and also because of the fundamental importance of this species as a crop. Still, the Fe(III)-PS uptake transporter, YS1, was first identified in maize by making use of the excellent genetic resources available in this species (Curie et al., 2001). The YS1 gene has been used directly as a strategy for engineering biofortification with mixed results. In an early study using constitutive expression of barley YS1 in rice, plants showed superior growth in alkaline soil conditions but did not contain significantly more iron in grains (Gomez-Galera et al., 2012). Later, barley YS1 expressed in rice was shown to promote the preferential mobilization and loading of Fe in seeds while displacing Cd and Cu (Banakar et al., 2017). At present, two key uptake genes, Ys1 and Ys3 (TOM1), for the grass specific mechanism are understood, as are the genes involved in PS synthesis, but it is unclear whether additional grass specific components exist. If they do, they will need to be discovered directly in grass species such as maize.
Identification of the Ys3 Gene
Maize ys3 mutants lack the ability to secrete PS (Lanfranchi et al., 2002). ZmTOM1 has been proposed as candidate gene for Ys3 because of its function as PS effluxer (Nozoye et al., 2011) and its location within the same map interval as the genetically identified ys3 mutant allele. Previous reports analyzing the ys3 transcriptome during iron deficiency suggested reduced expression and alternative splicing of ZmTOM1 (Nozoye et al., 2013). However, this approach could not definitively assign ZmTOM1 as the Ys3 gene, since other genes (ZmMATE3/ZmPEZ1) had reduced expression in ys3 mutants, and mutations in ZmTOM1 were not identified. Here, we were able to show that multiple alleles of ys3 could be found among the yellow striped mutants held at the MGCSC, and that each of these carries a unique mutation that is expected to abolish the function of the ZmTOM1 protein.
Three Novel Yellow Striped Maize Mutants
In this study, we identified three novel yellow striped mutants whose phenotype is apparently caused by low iron content. These mutants represent three different loci involved in iron homeostasis. Genetic mapping to identify the underlying genes responsible for the yellow striped phenotype in these mutants will reveal unknown elements of the iron homeostasis machinery and may provide new options for biofortification. Initially, it appeared as though our screening of the MGCSC mutant collection had reached saturation since multiple alleles for both ys1 and ys3 were obtained. However, three novel loci contributing to iron content in leaves were identified, indicating that saturation mutagenesis has likely not been reached and additional genes causing an iron deficiency induced yellow striped phenotypes in maize could be uncovered. Genetic mapping of the three ys∗ mutants is underway to discover the genes responsible for these interesting metal homeostasis phenotypes. Future work will also include tests to indicate whether additional iron supply or direct iron supply to the leaves can alleviate the ys∗ phenotypes, and tests of the iron concentration in grains of the mutant plants to see whether the grain concentration of iron is altered in the mutants.
Statements
Author contributions
EW conceived the project, was responsible for the experimental design, and also performed some of the genetics crosses and phenotyping in the field. DC-R conducted screening and genetics crosses and performed all of the molecular work on the project.
Funding
This work was supported by grants to EW (USDA AFRI Grant Nos. 2009-02268 and NSF IOS-153980).
Acknowledgments
We would like to express our sincere gratitude to the lab members, past and present who helped in the cornfield, especially Rakesh K. Kumar and Harry Klein, to Mary Sachs, of the MGCSC, who answered numerous questions and provided us with details pertaining to the stocks used in these studies, and to Dan Jones and Chris Phillips for their expert assistance in the greenhouse.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00157/full#supplementary-material
Footnotes
References
1
ArvidssonS.KwasniewskiM.Riano-PachonD. M.Mueller-RoeberB. (2008). QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR.BMC Bioinformatics9:465. 10.1186/1471-2105-9-465
2
BanakarR.Alvarez FernandezA.AbadiaJ.CapellT.ChristouP. (2017). The expression of heterologous Fe (III) phytosiderophore transporter HvYS1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport.Plant Biotechnol. J.15423–432. 10.1111/pbi.12637
3
BassoB.BagnaresiP.BracaleM.SoaveC. (1994). The yellow-stripe-1 and -3 mutants of maize: nutritional and biochemical studies.Maydica3997–105.
4
BernardsM. L.BuxtonE. A.JolleyV. D.ShifflerA. K.StangerT. F.RandsJ. (2014). Maize hybrids differ in their 24-h patterns of phytosiderophore release.J. Plant Nutr.37575–594. 10.1080/01904167.2013.867984
5
BriatJ.-F.LobreauxS. (1998). “Iron storage and ferritin in plants,” inIron Transport and Storage in Microorganisms, Plants, and AnimalsedsSigelA.SigelH. (New York, NY: Marcel Dekker, Inc.) 563–584.
6
BriatJ. F.LobreauxS.GrignonN.VansuytG. (1999). Regulation of plant ferritin synthesis: how and why.Cell. Mol. Life Sci.56155–166. 10.1007/s000180050014
7
BuxtonE. A.HopkinsB. G.JolleyV. D.WebbB. L.ChristensenR. C. (2012). Iron efficiency in kentucky bluegrass not related to phytosiderophore release.J. Plant Nutr.35311–329. 10.1080/01904167.2012.636133
8
CurieC.PanavieneZ.LoulergueC.DellaportaS. L.BriatJ. F.WalkerE. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake.Nature409346–349. 10.1038/35053080
9
DrakakakiG.MarcelS.GlahnR. P.LundE. K.PariaghS.FischerR.et al (2005). Endosperm-specific co-expression of recombinant soybean ferritin and Aspergillus phytase in maize results in significant increases in the levels of bioavailable iron.Plant Mol. Biol.59869–880. 10.1007/s11103-005-1537-3
10
EckardtN. A.CominelliE.GalbiatiM.TonelliC. (2009). The future of science: food and water for life.Plant Cell21368–372. 10.1105/tpc.109.066209
11
Gomez-GaleraS.SudhakarD.PelachoA. M.CapellT.ChristouP. (2012). Constitutive expression of a barley Fe phytosiderophore transporter increases alkaline soil tolerance and results in iron partitioning between vegetative and storage tissues under stress.Plant Physiol. Biochem.5346–53. 10.1016/j.plaphy.2012.01.009
12
GotoF.YoshiharaT.ShigemotoN.TokiS.TakaiwaF. (1999). Iron fortification of rice seed by the soybean ferritin gene.Nat. Biotechnol.17282–286. 10.1038/7029
13
HaradaE.SugaseK.NambaK.IwashitaT.MurataY. (2007). Structural element responsible for the Fe(III)-phytosiderophore specific transport by HvYS1 transporter in barley.FEBS Lett.5814298–4302. 10.1016/j.febslet.2007.08.011
14
HiguchiK.SuzukiK.NakanishiH.YamaguchiH.NishizawaN. K.MoriS. (1999). Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores.Plant Physiol.119471–480. 10.1104/pp.119.2.471
15
InoueH.KobayashiT.NozoyeT.TakahashiM.KakeiY.SuzukiK.et al (2009). Rice OsYSL15 is an iron-regulated Iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings.J. Biol. Chem.2843470–3479. 10.1074/jbc.M806042200
16
JeongJ.GuerinotM. L. (2009). Homing in on iron homeostasis in plants.Trends Plant Sci.14280–285. 10.1016/j.tplants.2009.02.006
17
KawaiS.ItohK.TakagiS.IwashitaT.NomotoK. (1988). Studies on phytosiderophores - biosynthesis of mugineic acid and 2′-deoxymugineic acid in Hordeum vulgare l var Minorimugi.Tetrahedron Lett.291053–1056. 10.1016/0040-4039(88)85333-4
18
KobayashiT.NakanishiH.TakahashiM.KawasakiS.NishizawaN. K.MoriS. (2001). In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2′-deoxymugineic acid to mugineic acid in transgenic rice.Planta212864–871. 10.1007/s004250000453
19
LanfranchiS.BassoB.SoaveC. (2002). The yellow stripe 3 mutant of maize is defective in phytosiderophore secretion.Maydica47181–184.
20
LeeS.ChieckoJ. C.KimS. A.WalkerE. L.LeeY.GuerinotM. L.et al (2009). Disruption of OsYSL15 leads to iron inefficiency in rice plants.Plant Physiol.150786–800. 10.1104/pp.109.135418
21
LiY.WangN.ZhaoF.SongX.YinZ.HuangR.et al (2014). Changes in the transcriptomic profiles of maize roots in response to iron-deficiency stress.Plant Mol. Biol.85349–363. 10.1007/s11103-014-0189-6
22
MaJ. F.NomotoK. (1996). Effective regulation of iron acquisition in graminaceous plants. The role of mugineic acids as phytosiderophores.Physiol. Plant.97609–617. 10.1111/j.1399-3054.1996.tb00522.x
23
MaJ. F.ShinadaT.MatsudaC.NomotoK. (1995). Biosynthesis of phytosiderophores, mugineic acids, associated with methionine cycling.J. Biol. Chem.27016549–16554. 10.1074/jbc.270.28.16549
24
MaJ. F.UenoH.UenoD.RombolaA. D.IwashitaT. (2003). Characterization of phytosiderophore secretion under Fe deficiency stress in Festuca rubra.Plant Soil256131–137. 10.1023/A:1026285813248
25
MarschnerH. (1995). Mineral Nutrition of Higher Plants.San Diego, CA: Academic Press, Inc.
26
McLeanE.CogswellM.EgliI.WoidylaD.De BenoistB. (2009). Worldwide prevalence of anaemia, WHO vitamin and mineral nutrition information system, 1993-2005.Public Health Nutr.12444–454. 10.1017/S1368980008002401
27
MiethkeM.MarahielM. A. (2007). Siderophore-based iron acquisition and pathogen control.Microbiol. Mol. Biol. Rev.71413–451. 10.1128/MMBR.00012-07
28
MoriS.NishizawaN. (1987). Methionine as a dominant precursor of phytosiderophores in graminaceae plants.Plant Cell Physiol.281081–1092.
29
MorrisseyJ.GuerinotM. L. (2009). Iron uptake and transport in plants: the good, the bad, and the lonome.Chem. Rev.1094553–4567. 10.1021/cr900112r
30
MurataY.MaJ. F.YamajiN.UenoD.NomotoK.IwashitaT. (2006). A specific transporter for iron(III)-phytosiderophore in barley roots.Plant J.46563–572. 10.1111/j.1365-313X.2006.02714.x
31
MurgiaI.ArosioP.TarantinoD.SoaveC. (2012). Biofortification for combating ‘hidden hunger’ for iron.Trends Plant Sci.1747–55. 10.1016/j.tplants.2011.10.003
32
NagasakaS.TakahashiM.Nakanishi-ItaiR.BashirK.NakanishiH.MoriS.et al (2009). Time course analysis of gene expression over 24 hours in Fe-deficient barley roots.Plant Mol. Biol.69621–631. 10.1007/s11103-008-9443-0
33
NegishiT.NakanishiH.YazakiJ.KishimotoN.FujiiF.ShimboK.et al (2002). cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots.Plant J.3083–94. 10.1046/j.1365-313X.2002.01270.x
34
NishizawaN.MoriS. (1987). The particular vesicle appearing in barley root-cells and its relation to mugineic acid-secretion.J. Plant Nutr.101013–1020. 10.1080/01904168709363629
35
NozoyeT.NagasakaS.KobayashiT.TakahashiM.SatoY.UozumiN.et al (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants.J. Biol. Chem.2865446–5454. 10.1074/jbc.M110.180026
36
NozoyeT.NakanishiH.NishizawaN. K. (2013). Characterizing the crucial components of iron homeostasis in the maize mutants ys1 and ys3.PLoS One8:e62567. 10.1371/journal.pone.0062567
37
ReichmanS. M.ParkerD. R. (2007). Probing the effects of light and temperature on diurnal rhythms of phytosiderophore release in wheat.New Phytol.174101–108. 10.1111/j.1469-8137.2007.01990.x
38
RobertsL. A.PiersonA. J.PanavieneZ.WalkerE. L. (2004). Yellow stripe1. Expanded roles for the maize iron-phytosiderophore transporter.Plant Physiol.135112–120. 10.1104/pp.103.037572
39
RomheldV.MarschnerH. (1986). Evidence for a specific uptake system for iron phytosiderophores in roots of grasses.Plant Physiol.80175–180. 10.1104/pp.80.1.175
40
SakaguchiT.NishizawaN. K.NakanishiH.YoshimuraE.MoriS. (1999). The role of potassium in the secretion of mugineic acids family phytosiderophores from iron-deficient barley roots.Plant Soil215221–227. 10.1023/A:1004546112140
41
SchaafG.LudewigU.ErenogluB. E.MoriS.KitaharaT.WirénN. V. (2004). ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals.J. Biol. Chem.2799091–9096. 10.1074/jbc.M311799200
42
ShojimaS.NishizawaN. K.FushiyaS.NozoeS.IrifuneT.MoriS. (1990). Biosynthesis of phytosiderophores – in vitro biosynthesis of 2′-deoxymugineic acid from L-methionine and nicotianamine.Plant Physiol.931497–1503. 10.1104/pp.93.4.1497
43
SuzukiM.MorikawaK. C.NakanishiH.TakahashiM.SaigusaM.MoriS.et al (2008). Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil.Soil Sci. Plant Nutr.5477–85. 10.1111/j.1747-0765.2007.00205.x
44
TagakiS. (1976). Naturally occurring iron-chelating compounds in oat-and rice-root washings: I. Activity measurement and preliminary characterization.Soil Sci. Plant Nutr.22423–433. 10.1080/00380768.1976.10433004
45
TagakiS.KameiS.YuM. H. (1988). Efficiency of iron extraction from soil by mugineic acid family phytosiderophores.J. Plant Nutr.11643–651. 10.1080/01904168809363830
46
TagakiS.NomotoK.TakemotoT. (1984). Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants.J. Plant Nutr.7469–477. 10.1080/01904168409363213
47
TakahashiM.NakanishiH.KawasakiS.NishizawaN. K.MoriS. (2001). Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes.Nat. Biotechnol.19466–469. 10.1038/88143
48
TakahashiM.YamaguchiH.NakanishiH.ShioiriT.NishizawaN. K.MoriS. (1999). Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (strategy II) in graminaceous plants.Plant Physiol.121947–956. 10.1104/pp.121.3.947
49
UenoD.RombolaA. D.IwashitaT.NomotoK.MaJ. F. (2007). Identification of two novel phytosiderophores secreted by perennial grasses.New Phytol.174304–310. 10.1111/j.1469-8137.2007.02056.x
50
von WirenN.MoriS.MarschnerH.RomheldV. (1994). Iron inefficiency in maize mutant ys1 (Zea mays L. cv Yellow-Stripe) is caused by a defect in uptake of iron phytosiderophores.Plant Physiol.10671–77. 10.1104/pp.106.1.71
51
WalkerE. L.ConnollyE. L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants.Curr. Opin. Plant Biol.11530–535. 10.1016/j.pbi.2008.06.013
52
WalterA.PichA.ScholzG.MarschnerH.RomheldV. (1995). Effects of iron nutritional-status and time of day on concentrations of phytosiderophores and nicotianamine in different root and shoot zones of barley.J. Plant Nutr.181577–1593. 10.1080/01904169509365005
53
WrightJ. (1961). A new yellow stripe on chromosome 3.Maize Newslett.35:111.
54
YenM.-R.TsengY.-H.SaierM. H.Jr. (2001). Maize Yellow Stripe1, and iron-phytosiderophore uptake transporter, is a member of the oligopeptide transporter (OPT) family.Microbiology1472881–2883. 10.1099/00221287-147-11-2881
55
YordemB. K.ConteS. S.MaJ. F.YokoshoK.VasquesK. A.GopalsamyS. N.et al (2011). Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters.Ann. Bot.108821–833. 10.1093/aob/mcr200
56
ZhangF. S.RomheldV.MarschnerH. (1991). Diurnal rhythm of release of phytosiderophores and uptake rate of zinc in iron-deficient wheat.Soil Sci. Plant Nutr.37671–678. 10.1080/00380768.1991.10416935
57
ZhangH. D.LiuJ. X.JinT. T.HuangY. Q.ChenJ. T.ZhuL. Y.et al (2017). Identification of quantitative trait locus and prediction of candidate genes for grain mineral concentration in maize across multiple environments.Euphytica213:90. 10.1007/s10681-017-1875-7
Summary
Keywords
iron, phytosiderophores, yellow stripe, maize, mutants
Citation
Chan-Rodriguez D and Walker EL (2018) Analysis of Yellow Striped Mutants of Zea mays Reveals Novel Loci Contributing to Iron Deficiency Chlorosis. Front. Plant Sci. 9:157. doi: 10.3389/fpls.2018.00157
Received
01 December 2017
Accepted
29 January 2018
Published
20 February 2018
Volume
9 - 2018
Edited by
Felipe Klein Ricachenevsky, Universidade Federal de Santa Maria, Brazil
Reviewed by
Yoshiko Murata, Suntory Foundation for Life Sciences, Japan; Sebastien Thomine, Centre National de la Recherche Scientifique (CNRS), France
Updates
Copyright
© 2018 Chan-Rodriguez and Walker.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elsbeth L. Walker, ewalker@bio.umass.edu
This article was submitted to Plant Nutrition, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.